- 1Ignaz Semmelweis Institute, Interuniversity Institute for Infection Research, Medical University of Vienna, Vienna, Austria
- 2G5 Unit Structural Biology of Infectious Diseases, Institut Pasteur, Université Paris Cité, Paris, France
- 3Department of Microbiology, Icahn School of Medicine at Mount Sinai, New York, NY, United States
- 4Center for Vaccine Research and Pandemic Preparedness (C-VaRPP), Icahn School of Medicine at Mount Sinai, New York, NY, United States
- 5Department of Internal Medicine, Division of Infectious Diseases, Medical University of Graz, Graz, Austria
- 6Department of Pathology, Molecular and Cell-Based Medicine, Icahn School of Medicine at Mount Sinai, New York, NY, United States
Puumala orthohantavirus (PUUV) is an emerging zoonotic virus that was first discovered in the Puumala region of Finland in the early 1980s and is the primary etiological agent of nephropathia epidemica (NE), a milder form of a life-threatening disease known as hemorrhagic fever with renal syndrome (HFRS). PUUV and other members of the Old World hantaviruses (OWHVs) predominantly circulate in rodents or insectivores across Eurasia, accounting for several thousand of reported HFRS cases every year (with many more unreported/misdiagnosed cases suspected). The rodent reservoir of PUUV is the common bank vole (Myodes (M.) glareolus), and transmission of the virus to humans occurs via inhalation of contagious aerosols and through contact with contaminated droppings or urine. Although PUUV is the subject of extensive research, due to its potential to cause severe disease outcomes in humans and its considerable economic and social impact, neither licensed vaccines nor specific antiviral treatments are available against PUUV. However, many important advancements have been made in terms of PUUV research over the last years. This included the elucidation of its glycoproteins, the discovery of broadly neutralizing hantavirus antibodies as therapeutic candidates and expanded research on the mRNA vaccine technology which will likely enable the development of strong PUUV vaccine candidates in the near future. Currently, there is still a lack of suitable animal models for the preclinical evaluation of experimental vaccines and antivirals, which hampers vaccine and antiviral development. Current attempts to decrease hantavirus-associated human infections rely primarily on prevention and countermeasures for rodent control, including reduced contact to droppings, saliva and urine, and disinfection of areas that are contaminated with rodent excreta. Here, we review these recent advances and other aspects including PUUV prevalence, virus biology, diagnosis and clinical features, and current animal models for vaccine and treatment development.
1 Introduction
Hantaviruses are a diverse family of single-stranded, trisegmented RNA viruses within the order Bunyavirales. Currently, Bunyavirales encompass eight genera (Agnathovirus, Loan virus, Actinovirus, Percilovirus, Mobatvirus, Thottimvirus, Reptillovirus and Orthohantavirus) (1, 2), which are known to infect different rodents and insectivores, with each strain being specific for a certain host species (3). The genus orthohantavirus includes species which vary in their geographic distribution and are capable of causing asymptomatic or mild to severe/lethal disease outcomes in humans. Generally, hantaviruses are classified into New World hantaviruses (NWHVs) and Old World hantaviruses (OWHVs) according to the geographic location of their respective rodent reservoir and the type of clinical manifestation upon infection of humans (4). NWHVs, such as Sin Nombre orthohantavirus (SNV) or Andes orthohantavirus (ANDV), mainly affect the human lung, causing a disease called hantavirus cardiopulmonary syndrome (HCPS) (5), and circulate in North and South America. OWHVs, such as Dobrava-Belgrade orthohantavirus (DOBV), Hantaan orthohantavirus (HTNV) and Puumala orthohantavirus (PUUV), mainly affect human kidneys, causing a disease called hemorrhagic fever with renal syndrome (HFRS) (6), and predominantly circulate in Eurasia. Furthermore, PUUV is associated with a third clinical phenotype, termed as nephropathia epidemica (NE) (7, 8), which is a less severe and milder form of HFRS. The case fatality rate (CFR) of PUUV infections ranges between 0.1-0.4% (9, 10), which is relatively low compared to the estimated high CFR of infections with NWHVs (30-60%) (11). Although most patients fully recover from an acute PUUV infection after weeks or months (12), several long-term sequelae (e.g., glomerular hyperfiltration, hypertension, stroke) (13) are observed. In addition, studies have indicated an increased risk to develop lymphatic/hematopoietic malignancies within the first years after recovering from a PUUV infection (14, 15).
Unlike other members of the order Bunyavirales, hantaviruses are not transmitted via obligate intermediate vectors, such as ticks, mosquitoes, flies or arthropods. However, they are directly or indirectly transmitted via hosts during close interactions, via inhalation of infectious aerosols or via contact with droppings or urine of infected animals (16–18). It has been shown that hantaviruses are more infectious via parental injection than aerosol transmission, thus, bite wounds and scratches caused by rodents present a risk for transmission that needs to be taken into account (19). Furthermore, observations of experimental ANDV and PUUV infections in Syrian hamsters indicate a potential transmission via the intragastric route (20, 21), thus, consumption of hantavirus contaminated food might be another conceivable way of transmission. The risk of direct human-to-human transmission of hantaviruses at this point is relatively low and almost neglectable, as humans are mostly dead-end hosts for the virus (2). So far, virus transmission from infected to naïve individuals has only been reported during ANDV-caused HCPS cases in Argentina (22, 23). In addition, one suspicious case of PUUV transmission via blood products in Finland has been reported recently (24) and mother-to-child transmission of ANDV through breast milk in Chile (25).
However, several questions regarding transmission to and pathogenesis in humans, but also the mechanisms of replication in both, humans and rodent hosts, including entry and tissue and organ tropism, still remain to be answered. Given the broad nature of this topic, it is beyond the scope of this review to describe all aspects of PUUV in-depth. Rather we aim to provide fundamental information about PUUV prevalence, its natural rodent reservoir, its viral biology, its diagnosis and clinical outcome, and treatment development, with an emphasis on current animal models and vaccine research.
2 PUUV prevalence and epidemiology
The initial discovery of hantaviruses dates back to in the 1950s during the Korean war (1951–1953) (7), where more than 3,000 military staff members suffered from a severe hemorrhagic fever disease of unknown origin. The etiologic agent of this hemorrhagic fever disease was isolated more than 25 years later, in 1978, from a striped field mouse (Apodemus agrarius) near the Hantan River in South Korea and was named HTNV (26). The second outbreak of severe hantavirus infections occurred in 1993 around the Four Corners region (New Mexico, Arizona, Utah, Colorado) in the United States of America. Individuals suffered from a hemorrhagic fever disease with pulmonary involvement, initially named Four Corners disease, later renamed as HCPS (27). In the same year, SNV was identified as the causative agent of the HCPS outbreak in the Four Corners region (28). In subsequent years, many other hantavirus species were identified globally in rodents or insectivores, e.g., ANDV in Argentina (1995) (29), or DOBV in Slovenia (1992) (30).
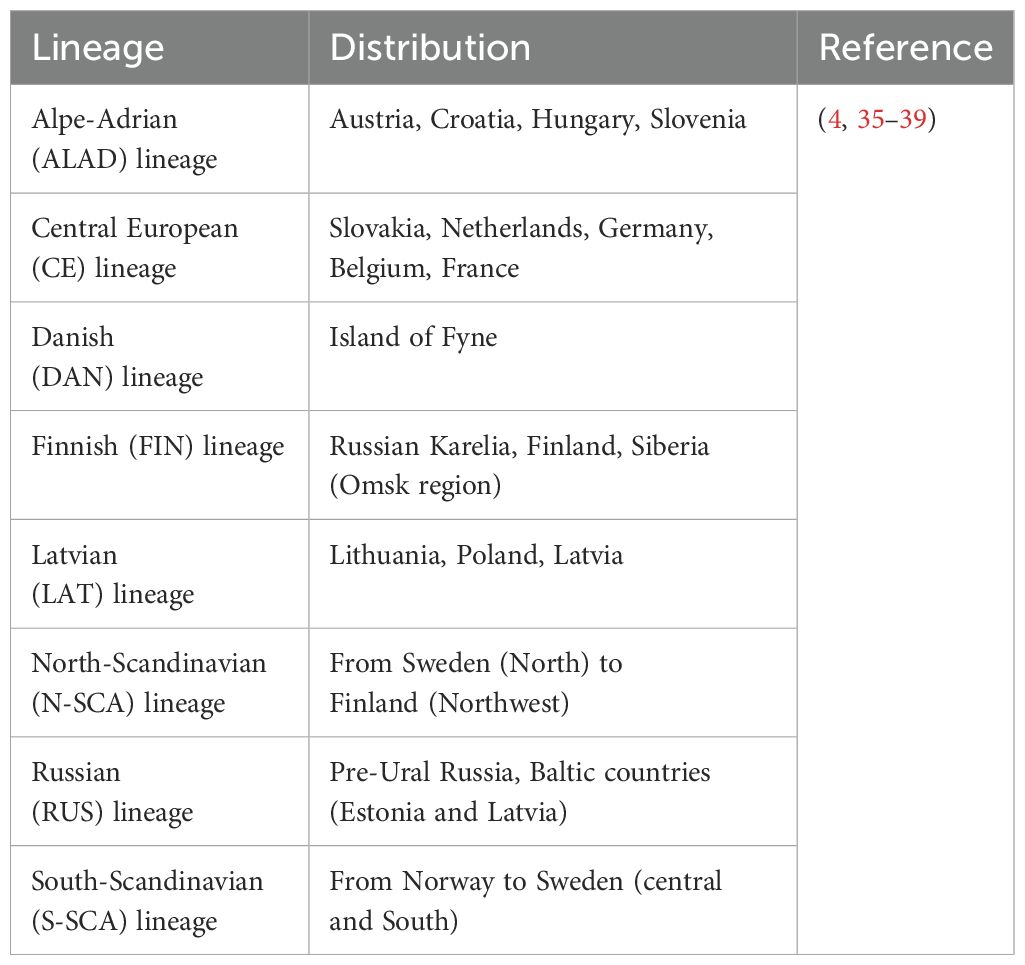
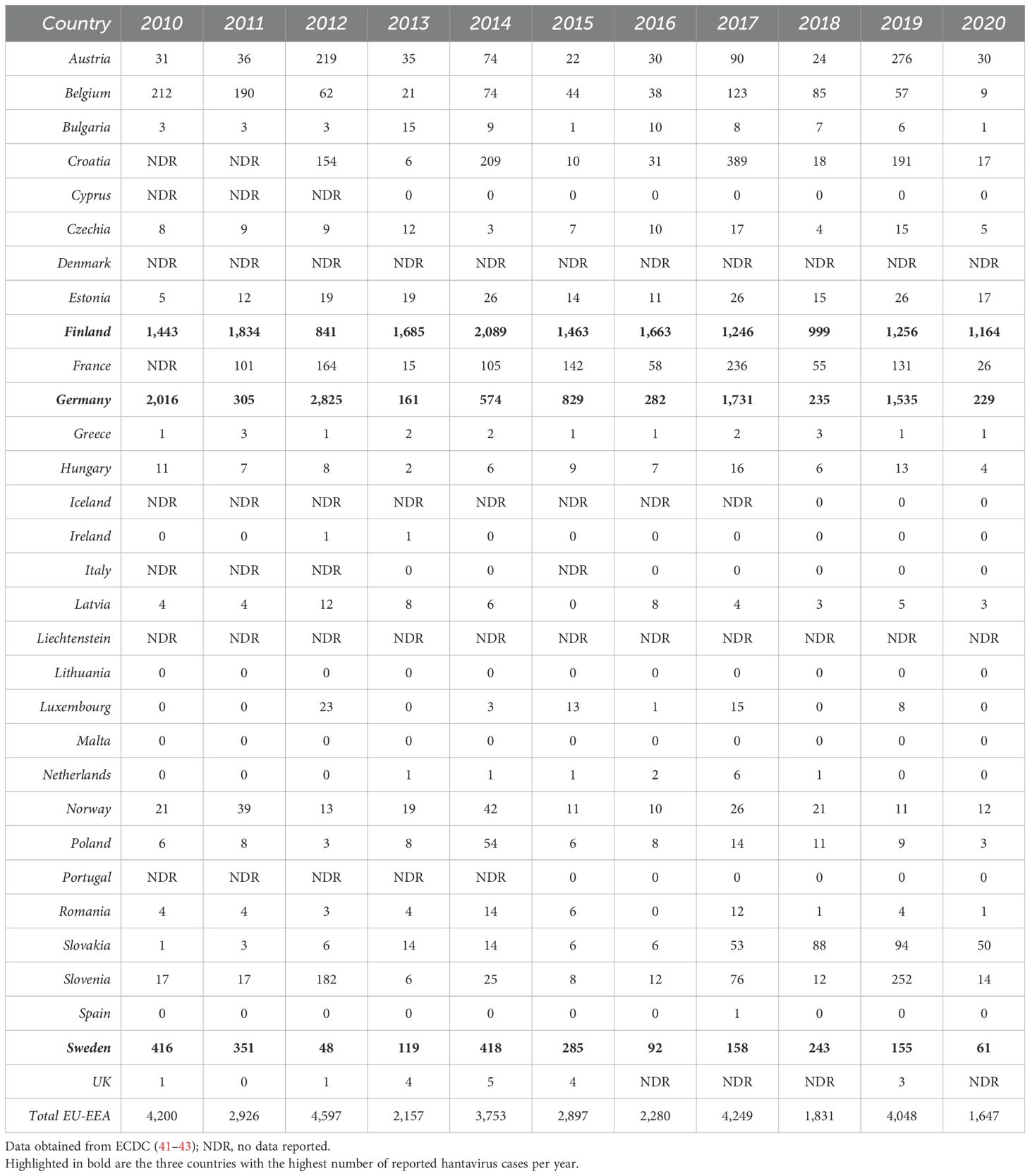
PUUV was first described (31) in the early 1980s, when the virus was detected in bank voles (Myodes glareolus) in the Puumala region of Finland. It is the causative agent of the vast majority of hantavirus infections in Europe within the last years (>98% of reported cases). Besides, other hantavirus species, such as TULV, DOBV, HTNV and Saaremaa orthohantavirus (SAAV), which persistently infect other rodents (e.g., Microtus voles, Apodemus mice), account for human HFRS infections in Europe (32–34). Eight PUUV lineages (4, 35–37) (Table 1) have been detected widely across Europe (with the exception of Southern Mediterranean coastal areas, British Isles, and the very Northern regions (8, 40)). However, only three European countries, Finland, Germany and Sweden, accounted for more than 85% of the annually reported cases (41–43) (Tables 2, 3) in Europe within the last years. From 2010-2020, between 1,647 (in 2020) and 4,597 (in 2013) cases of hantavirus infections were reported in Europe (mean: 3,100). Interestingly, the reported case numbers follow a cyclical pattern, with a significant increase every two to three years. Notably, Finland has by far the highest infection rate per 100,000 population, ranging between 18.1 (in 2018) and 38.3 (in 2014) every year. Germany and Sweden, reporting the second and third most annual cases, respectively, have an infection rate per 100,000 population ranging between 0.2 (in 2013) and 3.5 (in 2012) and between 0.5 (in 2013) and 4.5 (in 2010), respectively (Table 3). However, epidemiological data are incomplete as many European countries do not report cases of hantavirus infection. In addition, low numbers of reported cases in regions with high hantavirus seroprevalence in the rodent population clearly demonstrate an underdiagnosis of hantavirus infections in Europe (44, 45). The incidence of PUUV infections in Europe varies considerably across time, from year to year, seasonally, and across countries, but also within each country (2, 32, 46) (Tables 2, 3), and is strongly associated with the presence of its respective rodent host. Outbreaks of HFRS during spring and summer seasons are associated with close human contact with infected rodents during crop planting or harvesting, but also with increased travels of urban dwellers and camping tourists during the summer holiday season (42). In Northern Europe, hantavirus cases are frequently associated with close contact with infected rodents in the countryside (e.g., forest worker, soldiers) (27, 47).
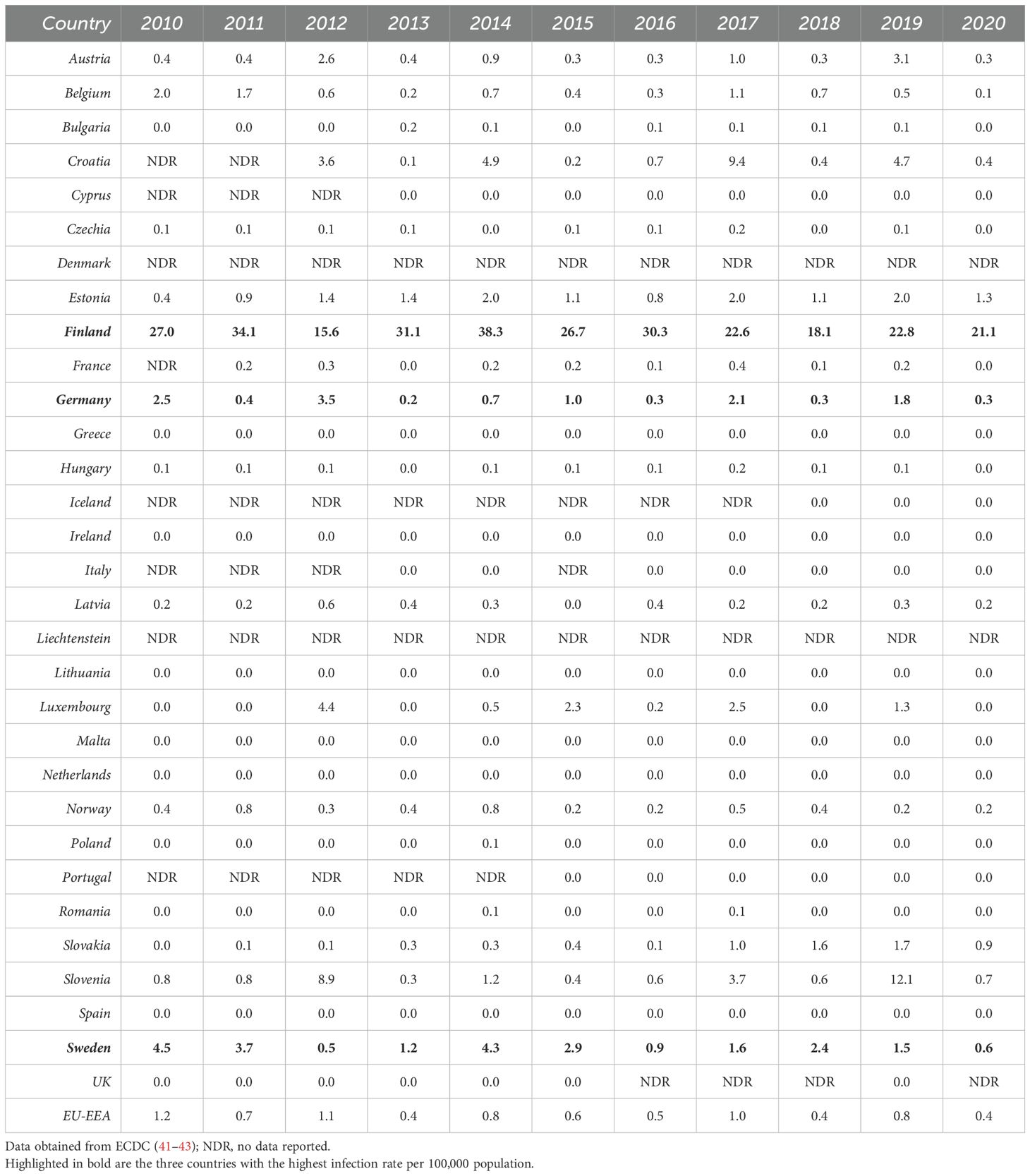
Table 3. Distribution of hantavirus infection rates per 100,000 population by country and year, EU/EEA, 2010-2020.
3 Natural reservoir
Hantaviruses have been detected in different families of rodents (e.g., Muridae and Cricetidae) (26), bats (e.g., Vespertilionidae, Rhinolophidae, and Nycteridae) (48) and insectivores (e.g., Talpidae, Soricidae) (49). They seem to be very strictly associated with one or very few closely related reservoir species and follow the distribution of the respective reservoir (8, 50). The main, and in Central Europe exclusive, reservoir for PUUV are common bank voles (M. glareolus), which are small rodents that are found in temperate and boreal forests (taiga) (51), but also in urban gardens, parks and hedges (46). Interestingly, genetically closely related PUUV species from Asia (Japan, China) have been found in vole species other than M. glareolus, but did not show any pathogenicity in humans (51, 52) so far.
There is in fact a strong relationship between the bank vole population density, PUUV prevalence, and the number of PUUV infections in humans in a specific area (53, 54). The population dynamics of bank voles change intra-and inter-annually within Europe and depend on climate changes (55) and variations in the landscape attributes (56), but also on other extrinsic factors, such as social behavior, the presence of predators (e.g., weasels) (57) or the availability of food (18). In temperate Europe rodent population increases mainly due to mast years, which occur when a substantial number of nuts from beech (Fagus sylvatica) or oak trees (Quercus petraea and Q. robur) pile up on the ground (58), providing sufficient nutrition for the rodents. Due to this substantial higher supply of food, the survival rate of the voles increases with an earlier breeding throughout the winter, causing a fluctuation of the bank vole population that can be 10-fold higher in these years compared to normal years (58, 59). In addition, studies have shown a correlation between an increase of bank vole abundance and an increase of beech fructification (60) or bilberry production (61) the year prior. Infections of bank voles and other rodents with hantaviruses apparently causes a prolonged or persistent infection (8, 62), which can last several months (46, 63) and is characterized by a subclinical or asymptomatic course (63). Apparent symptoms have not been detected in PUUV infected rodents; however, host survival and maturation (64) might be impaired. Infected rodents shed the virus through feces, urine and saliva (65, 66), causing subsequent infections of hosts via bites and scratches or contact with contagious excreta (67). Vertical transmission of the virus is less unlikely, as maternal antibodies protect the offspring (27). Studies reported a transient viraemia in infected bank voles (66, 68, 69), and infectious virus (66), PUUV antigen (62, 66, 68) or viral RNA (68, 69) could be detected several weeks or months after infection in various organs.
4 Virology
4.1 Virus structure and genome organization
Generally, hantaviruses display a spherical to pleomorphic shape (70), with a diameter of the virions broadly ranging between 80-160 nm (71, 72). Virions are relatively stable and survive for a few days at room temperature and up to several weeks at 4°C and -20°C (8, 65). The negative-sense and tri-segmented RNA genome is found within a lipid bilayer-based, enveloped virion, comprising of a large segment (L), a medium segment (M) and a small segment (S) (Figure 1A) (73), which display different sizes among the hantaviruses. The PUUV L segment is ~ 6,550 nucleotides (nt) in size and encodes for a ~ 2,156 amino acid (aa) long L protein. The L protein is an RNA-dependent RNA polymerase (RdRp), which mediates transcription and replication of the viral RNA genome. The PUUV M segment is ~ 3,682 nt in size and encodes for a ~ 1,148 aa long precursor glycoprotein (GPC). GPC is co-translationally processed into two envelope proteins, Gn and Gc, which are important for binding to the respective host cell receptor and the subsequent entry of the virus (74). The PUUV S segment is ~ 1,830 nt in size and encodes for a ~ 433 aa long nucleoprotein (N), that encapsidates the viral RNA genome (75). PUUV, TULV and other hantavirus species infecting members of the family Cricetidae (e.g., lemmings, New World mice/rats) (73, 76) additionally encode for a non-structural (NSs) protein (PUUV NSs: ~90 aa in size), which is located on the S segment in an overlapping open reading frame (ORF) (Figure 1B) (73, 77), and expressed via leaky scanning. Leaky scanning is a wide spread process among viruses to express polycistronic RNA, in which scanning ribosomes skip the first start codon and initiate protein synthesis at downstream located start codons (78). NSs is thought to be a non-essential protein, however, if expressed, it plays a role in the viral pathogenesis and immune evasion of infected hosts (79).
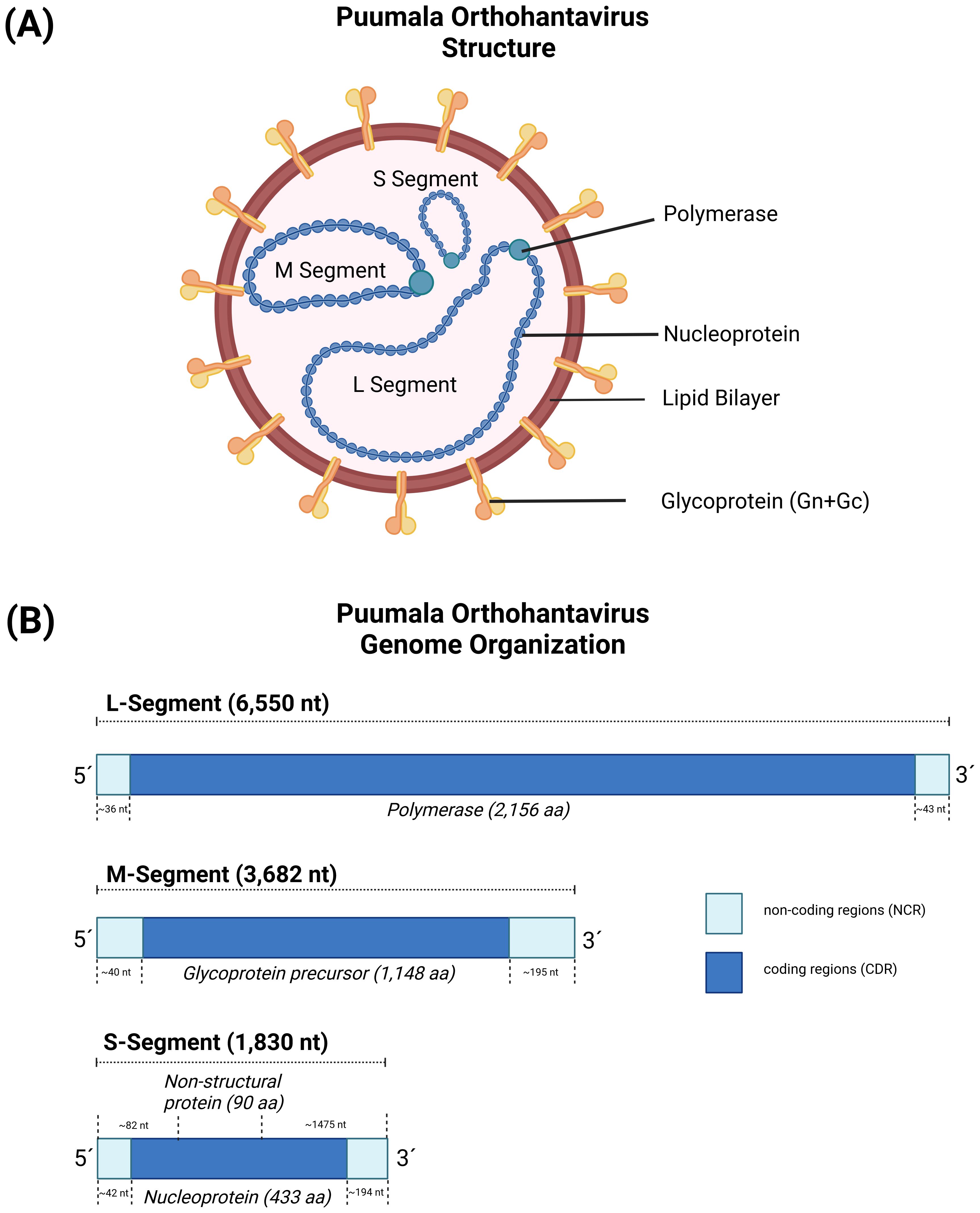
Figure 1. The virus particle and genome structure of Puumala orthohantavirus (PUUV). (A) The genome structure of orthohantaviruses, based on PUUV strain Sotkamo, accession numbers MN832782.1, MN832783.1, and MN832784.1 for the L, M, and S segment, respectively. (B) The S segment encodes for the nucleoprotein (433 aa) and the non-structural protein (90 aa), the M segment encodes for a glycoprotein precursor (1,148 aa), and L segment encodes for an RNA-dependent RNA polymerase (2,156 aa). Created with BioRender.com.
The 5´and 3´ non-coding regions of the three segments have different lengths (Figure 1B), ranging between 40-50 nt (5´ of all three segments) to 300-700 nt (3´ of S and M segment). Interestingly, the very terminal part of the sequences (consensus sequence AUCAUCAUCUG) (80) is conserved within the hantaviruses and can form panhandle-like structures (81, 82), which is a hallmark of the respective genus and shared with other genera in the Bunyavirales. Sequence analysis of the L, M and S segments showed a high degree of genetic diversity between the different hantavirus species, which is most likely caused by the accumulation of point mutations in combination with deletions and insertions mainly in the non-coding areas of the viral RNA segments (81). In addition, there is evidence for genetic shift, which occurs through the reassortment and recombination of genome RNA segments (83, 84).
4.2 The structure of the glycoprotein shell
Hantaviruses have a glycoprotein shell that orchestrates all the steps required for viral entry and is the primary target for neutralizing antibodies (85–88). This shell is composed of the two membrane glycoproteins Gn and Gc, which are encoded on the M segment as a polyprotein precursor and processed to form a tetrameric (Gn/Gc)4 spike (89). Gn acts as a folding chaperone for Gc and regulates fusion timing, Gc is the protein responsible for mediating fusion. Gn is about 650 amino acids long and is composed of two globular regions (GnH and GnB), two transmembrane (TM) regions and an intraviral domain. The second TM ends in a conserved motif that is cleaved to produce the Gc amino (N)-terminus (90). Gc is a class-II fusion protein of about 450 amino acids length, featuring an elongated ectodomain, a transmembrane region, and a short intraviral tail.
The structures of the GnH/Gc, GnH, GnB and Gc have been extensively studied by x-ray crystallography (87, 91–95) and the structure of the spike and its organization on the viral particle using cryo-electron microscopy (72, 96–98). The ectodomain of Gn is formed by three domains (A, B, C) and a membrane proximal region (MPRN). Domains A and B form the GnH region, which interact with Gc to stabilize its prefusion conformation and prevent premature association of Gc with cell membranes. Domain C and the MPRN constitute the GnB region, which functions as the tetramerization domain of the spike. Gc is a class-II fusion protein formed by a central β-sandwich (domain I) flanked by domains II and III. Domain III connects to the transmembrane region via the stem, a flexible region that is about 30 amino acids long, and the Gc membrane proximal region (MPRC). Like other class-II fusion proteins, the prefusion complex GnH/Gc dissociates at low pH, and Gc undergoes conformational changes that extend domain II toward the endosomal membrane to insert a hydrophobic region (termed the target membrane insertion surface (TMIS) in the hantavirus), reassemble into homotrimers, and refold domain III and the stem to approach the viral and endosomal membranes and induce fusion.
Despite these similarities, there are important structural and mechanistic differences between Gc and other class II fusion proteins. Notably, the TMIS of hantaviruses is composed of three flexible loops rather than a single rigid one. These loops coordinate to adopt two different conformations through an allosteric mechanism that is regulated by pH and the presence of GnH (92, 93). At neutral pH, GnH/Gc forms a stable complex in which the side chains of three key hydrophobic residues (W766, Y745, and F900) are buried. In this conformation, the tip of domain II exposes a polar surface that cannot interact with membranes. At acidic pH, GnH dissociates from Gc, triggering a reorganization of the tip of domain II which exposes the key residues, allowing Gc to insert into the endosomal membrane. The precise mechanism controlling the reorganization of domain II remains unclear, but some evidence suggest that an unusual acidic hydrogen bond forms at low pH between the side chains of the conserved E757 and D759 residues and plays a critical role. The formation of this bond is required to structure the TMIS in the post-fusion conformation and the presence of GnH induces a major reorganization of the loop containing these residues, which prevents the formation of the acidic hydrogen bond.
Another notable difference with other class-II fusion machineries is how Gn and Gc are organized on the viral surface. Cryo-electron microscopy (72, 96, 99) and biochemical studies (100, 101) revealed that the glycoprotein shell does not exhibit an icosahedral symmetry. Instead, it is composed of tetrameric (Gn/Gc)4 spikes, with four molecules of Gn at the center and four of Gc at the periphery, which interact laterally to form a grid-like pattern. This unique organization generates a topological problem because square-shaped spikes are incompatible with the formation of a closed, curved surface. Consequently, hantavirus particles display a distinctive pattern characterized by areas of ordered lattices coexisting with regions containing lattice-free spikes (Figure 2). Interestingly, a cryo-ET study (99) using the neutralizing antibody P4G2, which targets the interspike Gc/Gc interface and can only bind to isolated spikes, showed that this antibody induces the accumulation of isolated spikes on the viral surface. This finding reveals that the distribution between isolated and lattice-associated spikes is dynamic and can be influenced by the immune response. Along the same lines, ADI-42898, a cross-neutralizing antibody isolated from a patient infected with PUUV, binds to a quaternary epitope at the tip of Gc (85, 91). Structural modeling of ADI-42898 IgG molecules shows that they cannot bivalently bind to isolated spikes but cross-link neighboring tetramers within the virion lattice, likely promoting the accumulation of lattice-associated spikes. It remains to be seen if antibodies targeting the tip of domain II (like ADI-42898) interfere with the activity of those targeting the inter-spike regions (like P4G2).
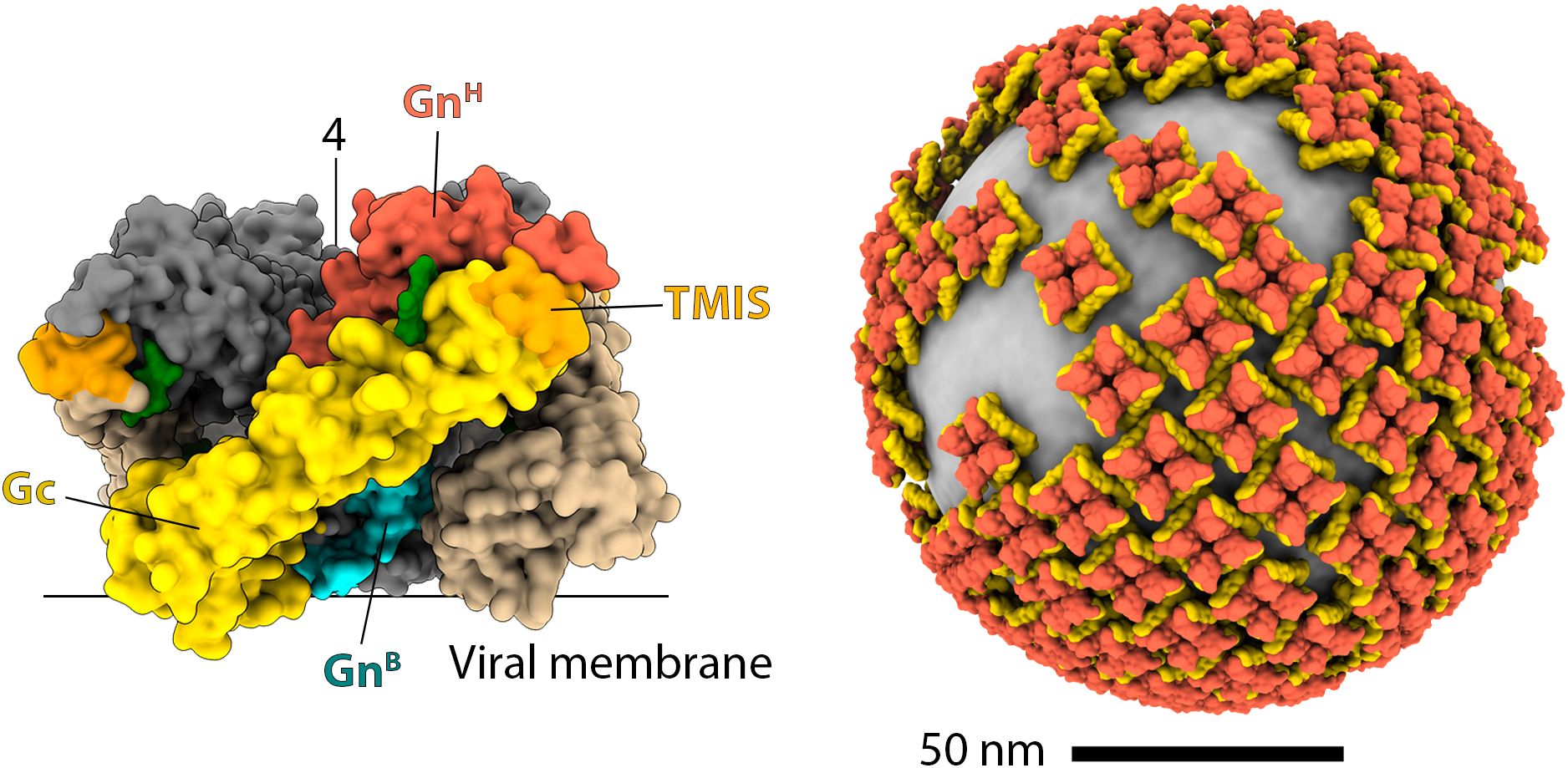
Figure 2. The organization of hantavirus spikes and the glycoprotein shell. The left panel shows a surface representation of the hantavirus spike in a side view. In the front protomer, GnH, GnB, and Gc are colored red, cyan, and yellow, respectively, as indicated. The TMIS is colored orange, and the N-glycans are shown in green. For clarity, the other protomers are colored differently: Gn in gray, and Gc in brown. The approximate positions of the viral membrane and the symmetry axis are indicated with lines. The right panel is a reconstruction of the hantavirus glycoprotein shell, with Gn and Gc colored red and yellow, respectively.
Combining the cryo-ET map and x-ray crystallography models of GnH/Gc and GnB tetramer produced a quasi-atomic model for the (Gn/Gc)4 spike (Figure 2) (92). In this model, GnB –the most conserved region of the polyprotein – is at the core of the spike, inaccessible for the immune system, and mediating most of the intra-spike interactions. GnH, which is much more variable, is exposed at the membrane distal surface, where it makes extensive contacts with Gc. A distinctive feature that emerges from the model is that all N-linked glycans play a structural role, either stabilizing the interaction between GnH/Gc or filling the internal cavities of the spike. Consistently, the removal of any of these glycans has been shown to impair the intracellular trafficking of the spike (102). Biochemical analysis revealed that the N-linked glycans remained of the high-mannose type in secreted particles (102). This observation suggests that spikes assemble early in the endoplasmic reticulum (ER), prior to transport to the Golgi apparatus, where glycan chains would otherwise undergo modification. Interestingly, some hantaviruses, such as DOBV, HTNV, and Thottapalayam virus (TPMV), possess additional N-glycosylation motifs that may help them to evade the immune system. However, the acquisition of new N-glycosylation sites appears to be a rare event in hantaviruses.
The structural studies conducted in recent years have provided important insights into how to design better immunogens and optimize the neutralizing activity of antibodies. Antibodies targeting Gn have potent neutralizing activity, but are serotype-specific, while antibodies targeting Gc are broadly neutralizing, but have weaker activity and tend to leave unneutralized fractions (85). The only cross-clade neutralizing antibody reported to date is ADI-42898, but it has reduced activity against ANDV. Structural studies (91) have shown that ADI-42898 recognizes a quaternary epitope that is only present in the prefusion conformation of the Gn/Gc heterodimer. Mechanistic studies have shown that ADI-42898 blocks viral membrane fusion by stapling together the Gn and Gc subunits, preventing them from dissociating at the acidic pH of the endosomes. These studies have also shown that ADI-42898 rapidly dissociates from the ANDV heterodimer at acidic pH, which limits its activity against this virus. In vitro affinity maturation experiments have identified mutations of this antibody that correct this defect and neutralize ANDV more effectively. Overall, these results suggest that stabilized heterodimers in the prefusion formation are better immunogens than Gn or Gc alone, leading to the development of various approaches to stabilize them, including the insertion of a linker between Gn and Gc, the design of disulfide bonds crosslinking Gn and Gc, or the introduction of mutations in Gc that interfere with the adoption of the post-fusion form (92).
4.3 Viral entry
In vitro studies indicated that integrins (β1-3) are potential candidate receptors, and complement factors (e.g., gC1qR/P32), decay acceleration factors (e.g., DAF/CD55) or protocadherin-1 are critical (co)-factors for viral attachment (103, 104) of NWHVs and OWHVs. Despite ongoing research in this field, the role of the suggested candidate receptors in pathogenesis and host range restriction is poorly understood [reviewed in (103)].
The primary targets for hantavirus replication in humans are macrophages, dendritic cells, (micro)vascular endothelial cells (69, 105), and pulmonary cells (106). Interestingly, in vitro studies have shown that the hantavirus tropism for cells belonging to the mononuclear phagocyte system is not exclusively limited to human, as also dendritic cells of the rodent reservoir can be productively infected (107). Apparently, the viral replication does not directly kill or damage the cells and the vascular endothelium; however the endothelial barrier integrity is impaired due to excessive and uncontrolled innate and adaptive immune responses (108–110).
4.4 Replication cycle
The two envelope glycoproteins Gn/Gc are the only viral proteins that are exposed on the virus surface and are essential for attachment to and entry into host cells (Figure 3). After attachment to its respective cellular receptor, the virus is internalized into the host cell. Interestingly, OWHVs, such as PUUV, enter the target cells via clathrin-dependent receptor-mediated endocytosis (111, 112), whereas NWHVs use clathrin-independent mechanisms (104, 113), such as macropinocytosis (114) or cholesterol-mediated micropinocytosis (103). Upon entry, viral particles are transported from early endosomes to late endosomal compartments. During endosomal maturation, the intra-luminal pH changes, from mildly acidic (early endosome) to strong acidic (endolysosome). This acidification process is required by the virus to detach from the bound integrin receptor and to undergo fusion of the viral with the endosomal membrane, which is mediated by a conformational change within the Gc glycoprotein (92). This fusion process consequently leads to an uncoating (112) of the virion. The hantavirus genome in form of ribonucleoproteins (RNPs) is released into the cytoplasm (115) and transcribed into S, M and L mRNAs, which are subsequently translated into proteins that are essential to hijack the host cell machinery. S and L mRNAs are translated via episomal ribosomes, whereas M-specific mRNA is translated into a glycoprotein precursor (GPC) at the rough endoplasmic reticulum (27). GPC is co-translational cleaved into Gn and Gc, most likely by host cell derived signal peptidases that are located in the lumen of the ER (116). It is thought that the cleavage site is located downstream of the conserved WAASA amino acid motif (90, 117). Shortly after the initial transcription of viral mRNA, synthesis of complementary RNA (cRNA) occurs, which serve as a template to synthesize viral RNA (vRNA) (118, 119). Both processes, transcription and replication, are mediated by the RdRp. The N-terminal part of the RdRp harbors an endonuclease activity, which allows for the cleavage and utilization of capped primers from host cell mRNAs to synthesize the viral mRNA (cap-snatching) (119). Once replication and amplification of viral genome is completed, vRNA is subsequently encapsulated by the nucleoprotein (120) and assembly of viral particles either occurs at the Golgi complex (OWHVs) (121) or at the plasma membrane (NWHVs) (122). It is assumed that the newly assembled virions bud into the Golgi complex, are transported to the cell membrane and released via exocytosis (OWHVs) (123). When assembly occurs at the plasma membrane, it is thought that viral vesicles and cell membrane fuse, and virions are released (NWHVs) (123).
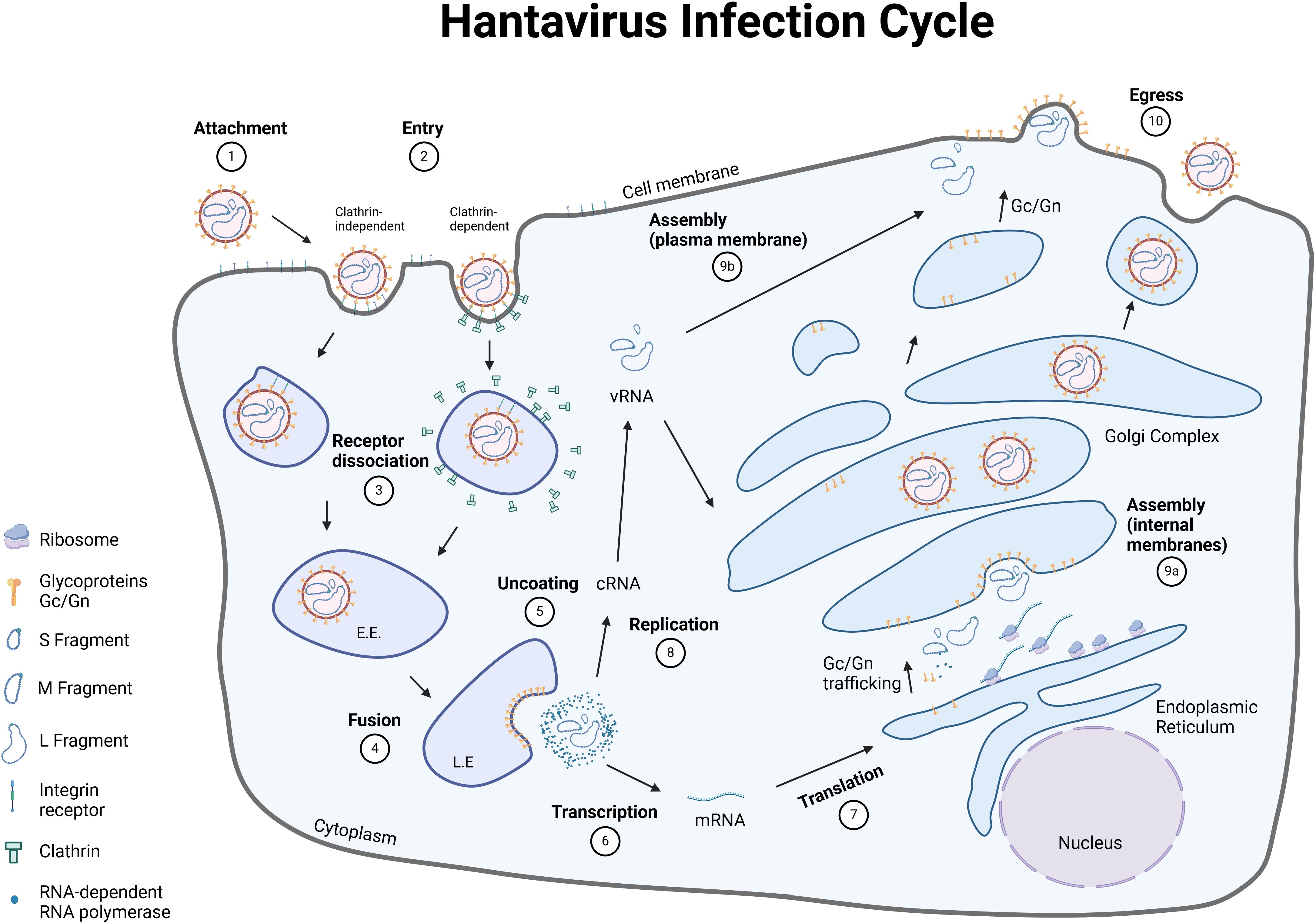
Figure 3. Hantavirus life cycle. The hantavirus life cycle consists of ten major steps, that are necessary to release new viral particles. [1] Hantaviruses bind to their respective receptor on the surface of the host cell with the envelope glycoproteins Gn/Gc. [2] Entry of the viral particles occur either via clathrin-dependent (OWHVs, e.g., PUUV) or clathrin-independent endocytosis (NWHVs). [3] The viral glycoproteins dissociate from the cellular receptors and traffic through the endocytic pathway. [4] Low pH of the endosomes and other cellular factors trigger a membrane-fusion process between viral and cellular membranes. [5] Viruses are uncoated and viral genome and proteins are released into the cytoplasm. [6] Viral RNA (vRNA) is transcribed by the RNA-dependent RNA polymerase (RdRp) and [7] mRNA is subsequently translated into different viral proteins, which are necessary to hijack the host cell machinery. [8] vRNA is synthesized and [9] new viral particles are assembled at the [9a] Golgi-complex (OWHVs, e.g., PUUV) or at the [9b] cell membrane (NWHVs). [10] Viral particles are released by fusion of the Golgi-complex (OWHVs, e.g., PUUV) or viral vesicle (NWHVs) with host cell membrane. E.E., early endosome; L.E., late endosome. Created with BioRender.com.
4.5 Evasion of the human innate immune system
As a response to viral infection, the host innate immune system is activated to provide a first line of defense to eliminate the virus and clear the infection [for review see (124, 125)]. Recognition of pathogen-associated molecular patterns (PAMPs), which can be viral components or by-products (e.g., double-stranded RNA during replication) occurs via pattern recognition receptors (PRRs), such as retinoic acid-inducible gene I-like RNA helicases (RLHs; e.g., melanoma differentiation-associated gene 5 helicase (MDA-5) or retinoic acid-inducible gene I helicase (RIG-I)) and Toll-like receptors (TLRs). TLRs recognize pathogens in endosomal or extracellular compartments, whereas RLHs recognize viral double-stranded RNA in the cytoplasm of infected cells (126). Upon recognition of and binding to PAMPs, the receptors mediate a signal cascade resulting in the activation of TANK-binding kinase 1 (TBK1) and IkappaB kinase (IKK) to produce type I interferons (IFNs). The released IFNs bind to their respective type I interferon receptors, resulting in the activation of the Janus kinases/signal transducer and activator of transcription proteins (JAK/STAT) pathway, which causes the expression of IFN-stimulated genes (126) (Figure 4). Hantaviruses have evolved several strategies to evade the host´s defense mechanism (in particular the type I interferon pathway) in order to efficiently replicate and spread in the infected host. Interestingly, different hantavirus species interfere with different modulators and regulatory factors of the type I interferon pathway, which differ also within OWHV and NWHV, and are independent of their virulence in humans (summarized in Figure 4). In vitro studies demonstrated that PUUV Gn/Gc antagonizes the IFN pathway that is stimulated via activated RIG-I (128) and interfere with the activation of IFN-stimulated response elements (ISRE) (128). In addition, PUUV NSs inhibits the activation of MDA5 (128), TBK1 (128) and interferes with the activation of interferon regulatory factor 3 (IRF3) responsive promoters and IFN-β promoters (129). TULV N inhibits the activation of RIG-I (128), and so does ANDV N (137), which additionally blocks the activation of protein kinase R (PKR) (133), TBK1 (137) and the phosphorylation of STAT1/2 (134). Furthermore, HTNV N interferes with the interaction of TRIM 25 (127) and RIG-I (127), thereby inhibiting the downstream activation of RIG-I. However, studies demonstrating the capability of PUUV N to downregulate the type I interferon pathway are lacking. Contrarily, Gallo and colleagues could confirm enhanced IFN-β promoter activity driven by PUUV N (128).
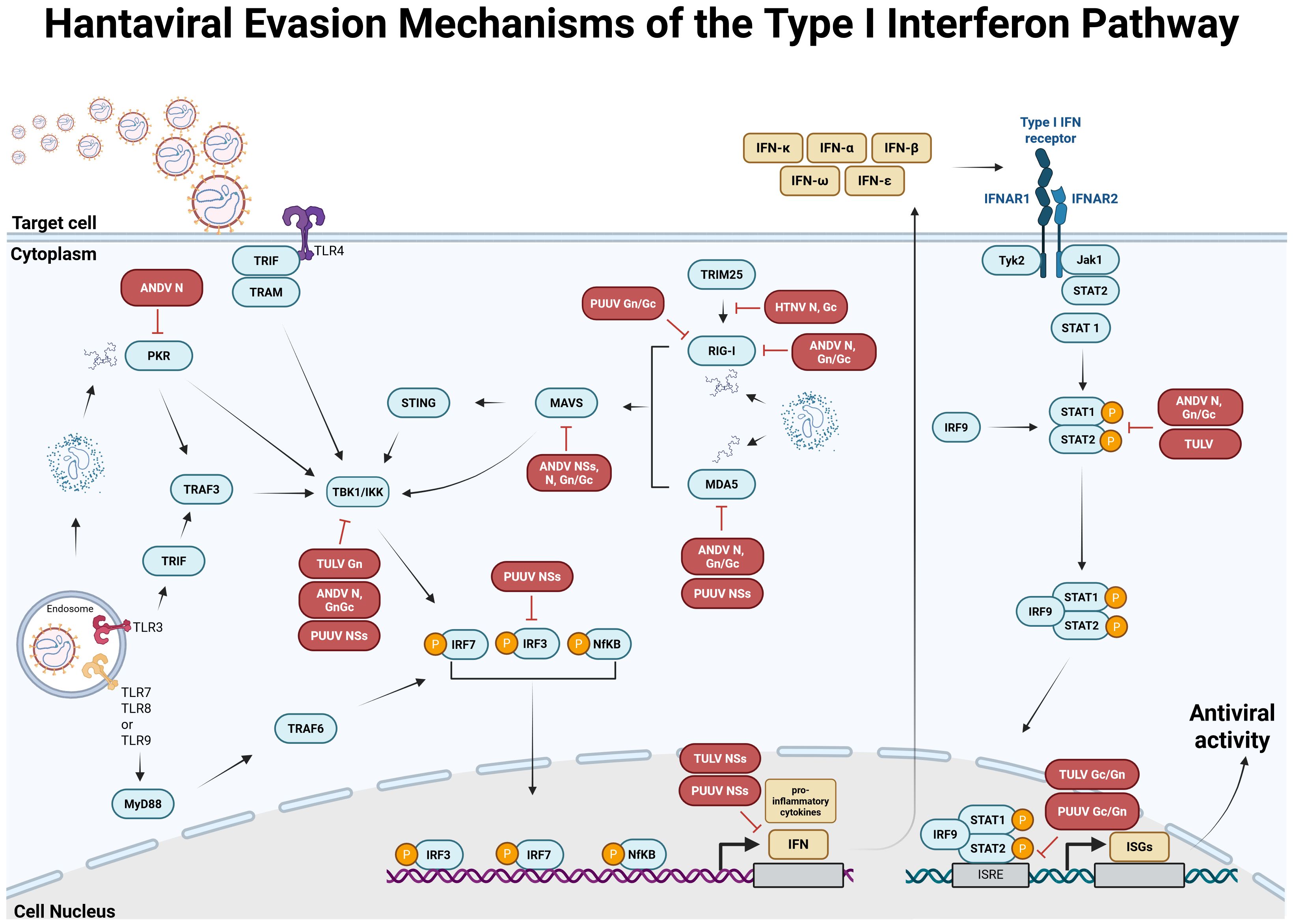
Figure 4. Antiviral type I interferon (IFN) response pathway and known evasion mechanisms of orthohantaviruses. Based on data from Hantaan orthohantavirus (HTNV) (127), Puumala orthohantavirus (PUUV) (128–131), Tula orthohantavirus (TULV) (128, 129, 131, 132) and Andes orthohantavirus (ANDV) (133–136). In infected cells, viral components or by-products, called pathogen-associated molecular patterns (PAMPs) are recognized by pathogen recognition receptors (PRRs), such as melanoma differentiation-associated gene 5 helicase (MDA-5), retinoic acid-inducible gene I helicase (RIG-I) or Toll-like receptors (TLRs). Receptor-ligand binding activates the type I IFN pathway resulting in the activation of TANK-binding kinase 1 (TBK1) and IkappaB kinase (IKK), which causes the phosphorylation and activation of IFN regulatory factors (IRF) 3/IRF7 and/or NfκB, leading to the expression of different type I IFNs. IFNs are released and bind to type I IFN receptors, thereby activating Janus kinase 1 (Jak1) and tyrosine kinase 2 (Tyk2). Once activated, the two proteins activate signal transducers and activators of transcription 1 and 2 (STAT1 and STAT2), which become phosphorylated and form a complex with IRF9, subsequently inducing the expression of different IFN-stimulated genes (ISGs). Hantaviruses evolved different immune evasion mechanisms to avoid detection by PRRs or interfere with downstream factors of the type I IFN pathway. These antagonisms are associated with hantavirus N (127, 133, 135–137), Gc/Gn (127, 132, 136) or NSs (128, 129, 131). MAVS, mitochondrial antiviral-signaling protein; STING, stimulator of interferon genes; TRIF, TIR-domain-containing adaptor inducing IFN-β; TRAM, TRIF-related adaptor molecule; TRAF, tumor necrosis factor receptor associated factor; TRIM, tripartite motif-containing; IFNAR, interferon α/β receptor. Created with BioRender.com.
5 Clinical presentation and pathogenesis
The clinical presentation of PUUV infection varies from subclinical, mild, and moderate to even severe courses (138–140). The proportion of different severities is difficult to assess, as the reported numbers of PUUV infections are quite low compared to infections estimated from sero-surveillance studies (139, 141). Thus, most of the mild cases are likely missed and consequently, the clinical characteristics have been mainly derived from hospitalized patients (139). Approximately 8% of all PUUV infected patients diagnosed in a tertiary care center have been admitted to the intensive care unit (ICU) for oxygen supply (intubation and mechanical ventilation was necessary in 66%) and renal replacement therapy (applied in 66%) (142). In general, HFRS caused by DOBV is more severe with mortality rates from 5% to 15%, whereas SEOV causes moderate and PUUV and SAAV cause mild forms of disease with mortality rates of <1%. Whereas the overall mortality of PUUV infection is reported to be low, the 30-day death rate of PUUV infected patients treated at ICUs was 14% (142). The definitive reasons for the individual differences in the clinical course and outcome of PUUV infection remain unclear, but have been considered rather to be determined by factors in the human host (severe courses were associated with certain HLA alleles and genetic variation in cytokines) than by variations in PUUV virulence (143, 144). Severe PUUV infections are mainly described in male patients, but this might be explained by the exerted activities considered as risk factors for acquisition of PUUV that are more common in men (142).
The incubation period of PUUV infection is usually 2-6 weeks (Figure 5), followed by unspecific symptoms and, in severe cases, organ dysfunction (139, 146). The main clinical findings of PUUV infection include fever, myalgia, headache, backache, abdominal pain, vomiting, diarrhea, cough and blurred vision (summarized in Figure 6) (139, 146). Vascular leakage can cause edema in many organ tissues and hypotension (139, 147). In severe cases, renal failure, marked hypotension or circulatory failure, petechiae and hemorrhages might occur (139). In case of acute kidney injury, the course of HFRS is divided into five stages (febrile, hypotensive, oliguric, diuretic, and convalescent (Figure 5) (138, 139). Whereas these phases are usually present in DOBV or HTNV infection, the five phases are not easily distinguishable in NE caused by PUUV (138). The urinary excretion of interleukin-6 (IL-6) correlates with the amount of proteinuria in NE (148). It has been hypothesized, that urinary IL-6 levels might reflect the production of this proinflammatory cytokine in the kidneys (147). Ultrastructural changes decrease the barrier functions of the kidney resulting in proteinuria in NE (145). Although HCPS, the disease caused by NWHV, and HFRS are separated entities, they share some common clinical characteristics. Both are characterized by the strong systemic inflammation and affection of vascular endothelial cells, leading to organ dysfunction. HCPS is characterized by respiratory symptoms, hypoxia and pulmonary infiltration in radiological examination, but HFRS can also affect the respiratory system in approximately one third to half of the patients (139, 149). In case of pulmonary involvement in PUUV infection, patients show cough, tachypnea, and dyspnea. To address the pathophysiological role of bradykinin in severe capillary leakage, the bradykinin receptor antagonist icatibant was used as treatment in some cases of severely ill NE patients (150). In ICU patients, invasive aspergillosis might complicate the course of critically ill PUUV infected patients (142). Hemophagocytic lymphohistocytosis associated with PUUV infections has also been reported and was treated with anti-inflammatory and immunosuppressive medication in one case (151, 152). A detailed description of further clinical features, findings in clinical laboratory tests and treatment are provided in the Supplementary Material.
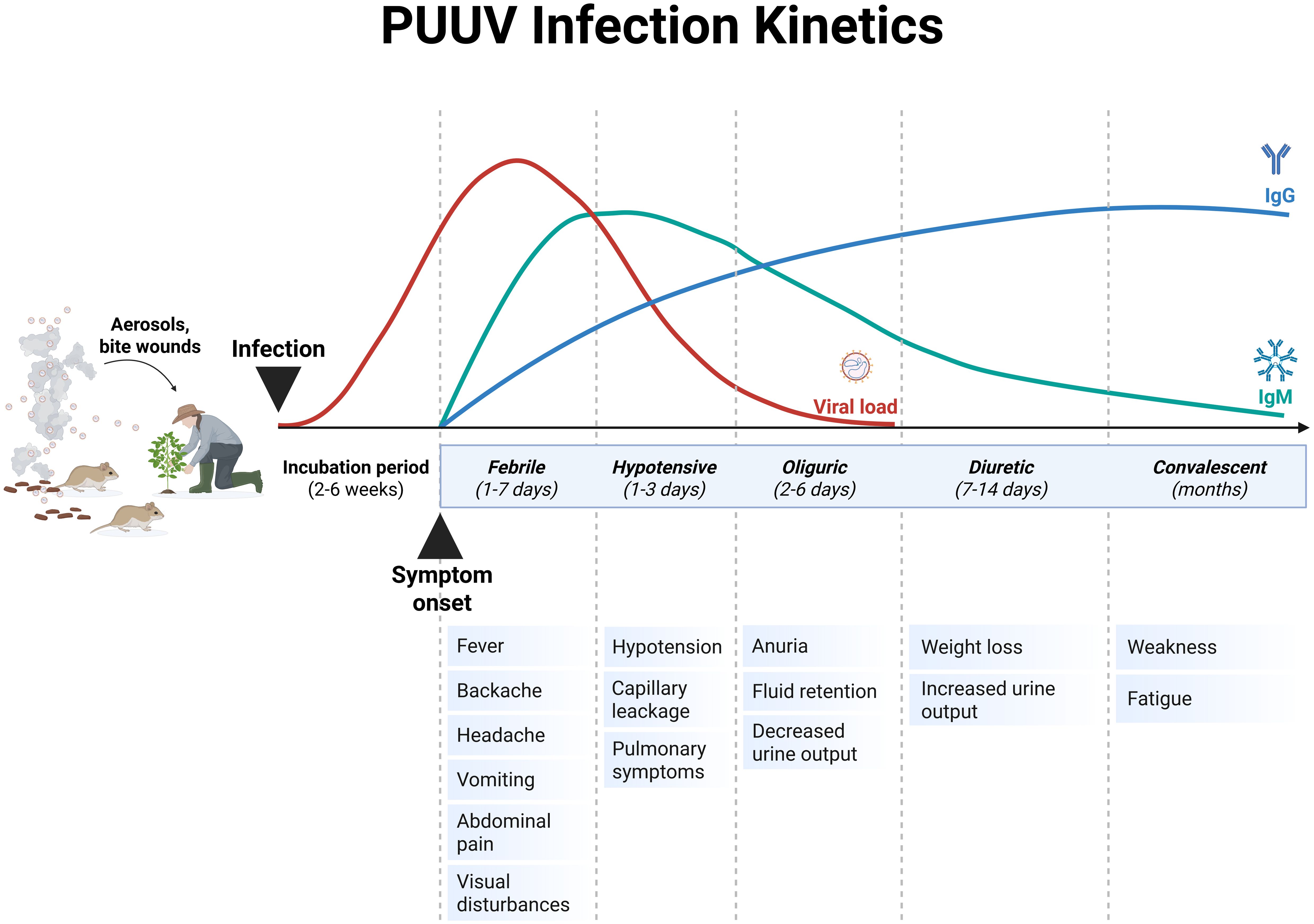
Figure 5. Schematic representation of the Puumala virus (PUUV) infection kinetics in humans. Typically, the severe clinical course of nephropathia epidemica (NE) that is caused by PUUV can be divided into five stages, which are not easily distinguishable: febrile, hypotensive, oliguric, diuretic and convalescent. The incubation period of PUUV infections ranges between 2-6 weeks, and is associated with an increase in viral load. The onset of the first symptoms is accompanied with an increase in antibody titers. Adapted from Avšič-Županc T et al. (138) and Mustonen et al. (145). Created with BioRender.com.
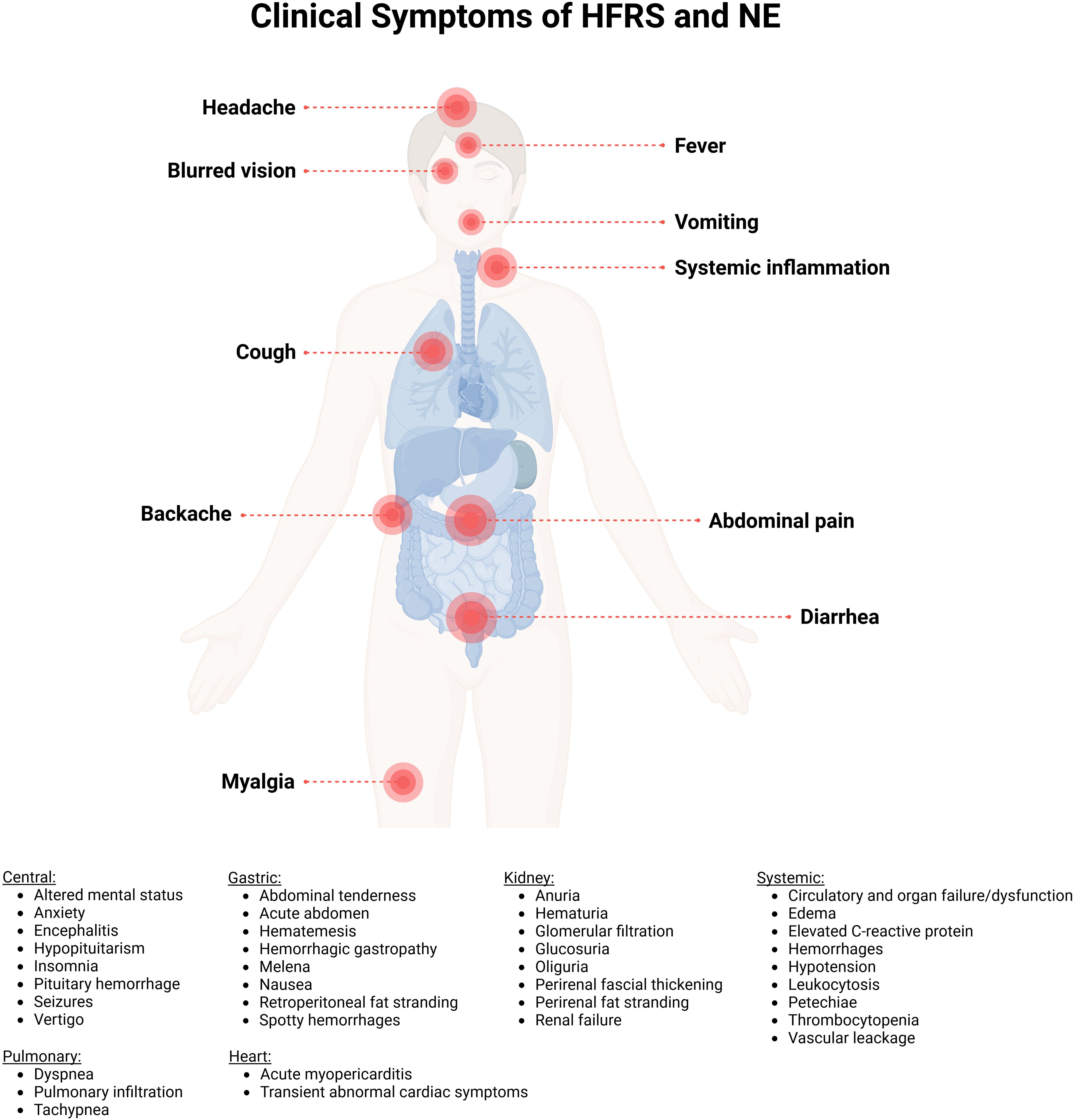
Figure 6. Clinical representation of hemorrhagic fever with renal syndrome (HFRS) and nephropathia epidemica (NE) caused by Puumala orthohantavirus. The main clinical symptoms are myalgia, backache, abdominal pain, vomiting, diarrhea, cough, headache, fever, systemic inflammation and blurred vision. Created with BioRender.com.
6 Detection, diagnostics, and treatment
6.1 Detection and diagnostics
Several methods for PUUV detection and diagnosis have been developed. They are either based on the direct detection of PUUV genome via nucleic acid testing or on detection of antibody responses to the virus. Serological responses to virus proteins can typically already be detected at symptom onset. One of the most widely used ways to confirm PUUV infections in clinical laboratories is to measure IgM to N using enzyme-linked immunosorbent assays (ELISAs) or other immune-assays like immune-blotting or immunofluorescence assays (IFAs) and even neutralization assays (153–156).
However, nucleic acid-based detection methods have also been widely used (140, 157–160), especially in research settings and those assays may potentially detect the presence of virus genome before the onset of an antibody response (161). The challenge in diagnosing PUUV cases might often be based on insufficient awareness of physicians in areas with low or unknown PUUV prevalence, resulting in missing suspicion of PUUV infection and lack of testing. This may lead to underreporting. However, in case of availability of specific treatment options in the future (e.g., monoclonal antibodies (mAbs) or antivirals), early diagnosis may be essential to enable early treatment of patients.
6.2 Antivirals and immunotherapy
Currently, there are no specific targeted treatments for hantavirus infection and current strategies predominantly focus on the management of clinical symptoms. Treatment is usually symptomatic and in severe cases include oxygen supply, non-invasive or invasive mechanical ventilation, renal replacement therapy and extracorporeal membrane oxygenation (162). In individual cases, treatments with icantibant or glucocorticoids, immunoglobulins and ruxolitinib in PUUV associated hemophagocytic lymphohistocytosis have been reported (162). Several pre-clinical studies have been conducted investigating the use of both antiviral drugs and mAbs as post-exposure therapeutics.
Ribavirin (1-β-D-ribofuranosyl-1, 2, 4-triazole-3-carboxamide) is a synthetic guanosine nucleoside analog which has displayed potent broad antiviral activity against a range of RNA viruses, including hantaviruses. This antiviral activity is thought to be exerted through multiple mechanisms of action. Ribavirin has been shown to exert antiviral effects via the interruption of viral capping (163, 164) and polymerase activity (165, 166). Ribavirin also abrogates inosine monophosphate dehydrogenase (IMPDH), which results in the depletion of intracellular guanosine triphosphate (GTP) (167, 168), thereby reducing viral replication. Conversely, the antiviral effects of ribavirin against arenaviruses are independent of GTP depletion (169), and may instead be dependent on the reduction of inflammatory responses via the protection of infected cells from death (170). The inhibition of cellular GTP by ribavirin has also been shown to restrict viral infection via the induction of spermine-spermidine acetyltransferase (SSAT1) (171). SSAT1 decreases intracellular polyamine levels, which have been shown to be vital for the replication of Zika virus (ZIKV) and Chikungunya virus (CHIKV) (172). Ribavirin also acts as a mutagen, promoting the accruing of mutations in viral genomes which result in the production of defective particles and “error catastrophe” (172–175). This has been demonstrated in vitro to be one of the mechanisms through which ribavirin restricts HTNV infection (176), as opposed to GTP depletion (177). The protective effects of ribavirin against HTNV were first shown in vivo in the 1980s using a suckling mouse challenge model, wherein daily 50 mg/kg ribavirin treatment promoted survival (178). Following these promising early results, a placebo-controlled, double-blinded clinical trial was conducted using 242 HFRS patients in China which demonstrated that ribavirin therapy administered within 7 days of symptom onset reduced mortality seven-fold, prevented the induction of the oliguric phase of disease, and reduced hemorrhagic manifestations (179). These findings were recapitulated in a smaller study using US Department of Defense personnel stationed in Korea, wherein ribavirin treatment was found to restrict the renal complications of HFRS (180). The protective effects of ribavirin for treating the hantavirus cardiopulmonary syndrome caused by NWHVs are less well established. Ribavirin was found to be a potent inhibitor of ANDV infection in vitro and in vivo in the Syrian golden hamster challenge model of infection (181, 182), and in the deer mouse model of SNV infection (183). Based on these findings, an open-label clinical trial of ribavirin was conducted in the US from 1993-1994, however the results were inconclusive and no differences in mortality were observed (184). A follow-up randomized, double-blinded, placebo-controlled clinical trial was conducted in 2004 where ribavirin treatment did not show improvements in 28-day survival compared to the control group (185). Unfortunately, this study recruited only 36 patients which did not allow for robust comparisons between groups (185). Additionally, intravenous ribavirin treatment was found to be ineffective at lowering viral loads in a randomized, open-label study conducted on HFRS caused by PUUV infection (186). While the predominant side-effect of ribavirin in the treatment of HTNV was limited to a reversible hemolytic anemia (178), its usage in the treatment of PUUV infection also resulted in increased incidence of hyperbilirubinemia, sinus bradycardia, and rash (186). A phase II clinical trial (NCT00868946) was set to be carried out in Germany to investigate the efficacy of ribavirin in the treatment of HFRS, however this was withdrawn due to poor patient enrollment. Several additional antiviral drugs have been tested in combination with ribavirin to improve efficacy and limit the emergence of drug-resistant viral variants. Lactoferrin, an iron-binding glycoprotein naturally secreted in milk and saliva, has been shown to exhibit broad antiviral effects. Bovine lactoferrin has been demonstrated to inhibit SEOV cell entry in vitro, with complete abrogation of viral replication when used in combination with ribavirin (187). Lactoferrin has also been shown to inhibit SEOV infection in vivo in the suckling mouse challenge model, wherein pre-treatment with 160 mg/kg 48- and 24-hours prior to infection promoted a survival rate of 94% (188). While these results are encouraging, it is unclear how efficacious post-exposure treatment with lactoferrin would be, especially considering that the timing of therapeutic intervention is critical in the treatment of HFRS and HCPS.
Favipiravir (T-705), like ribavirin, is a synthetic nucleotide analogue which inhibits viral RNA-dependent RNA polymerases (189), abrogates viral RNA transcription (189), and promotes lethal mutagenesis (190). Favipiravir has been demonstrated to lower viral loads and promote survival in the ANDV Syrian golden hamster challenge model and to limit viral replication of hamster-adapted SNV (191). In vitro studies have shown that favipiravir potently inhibits HTNV and works synergistically in combination with ribavirin (192), and that it is also effective against DOBV and Maporal virus (MPRLV) (193), a NWHV which is closely related to ANDV. The synergistic effects of combination therapies with ribavirin and favipiravir have been demonstrated in vivo and in vitro for other hemorrhagic fever viruses such as filoviruses, arenaviruses (194, 195), and bunyaviruses (196, 197). While these preclinical results are promising, clinical trials are required to determine if favipiravir, either alone or in combination with ribavirin, is effective in the treatments of HFRS and HCPS including PUUV infections. An additional nucleoside analogue, 1-β-d-ribofuranosyl-3-ethynyl-[1,2,4]triazole (ETAR), has also been demonstrated to possess antiviral activity against hantaviruses. Like ribavirin, ETAR reduces intracellular GTP pools and a single study has shown that it effectively inhibits HTNV and ANDV in vitro and conferred improved survival in a suckling mouse HTNV challenge model (198).
Hantaviruses, as members of the Bunyavirales family, share some aspects of their biology with other negative sense, segmented viruses, such as the Orthomyxoviridae. As such, some antiviral drugs which have been characterized for the treatment of influenza virus may exhibit activity against hantaviruses. Like the Orthomyxoviridae, hantaviruses rely on cap-snatching for viral transcription, with the RdRp acting as a cap-dependent endonuclease (CEN) (119). CENs represent attractive targets for antiviral drug design, and a wealth of compounds have been identified for the treatment of influenza virus infection. In one study, the authors screened a library of CEN inhibitory compounds and identified several drugs which were potently antiviral against a number of bunyaviruses in vitro and in vivo (199). Of these drugs, two candidates displayed antiviral effects against TPMV, an OWHV which is apathogenic in humans, though to a lesser degree than other bunyaviruses tested (199). The authors speculate that this is due to differences in the cap snatching machinery employed by these viruses, which necessitates further study for effective hantavirus CEN inhibitors. In a separate study, one such CEN inhibitor, baloxavir acid (BXA), was found to inhibit HTNV in vitro and had comparable activity to favipiravir (200).
An alternative strategy for the treatment of hantavirus infection is the targeting of host proteins which are required by the virus for replication and pathogenesis. Using a small interfering RNA (siRNA) screen approach, one study identified several pro viral host proteins which promote the replication of influenza A virus (IAV). By selecting known, approved, drugs that target these proteins, the authors identified the urea-based kinase inhibitors (UBKIs) regorafenib and sorafenib as potent antiviral agents against IAV (201). These compounds also exhibited robust activity against HTNV in vitro, possibly via the interruption of the early stages of viral replication. In another study, authors identified a compound, 8G1, as possessing anti-HTNV activity by screening a library of kinase-inhibitors. Like regorafenib and sorafenib, 8G1 was found to inhibit the early stages of viral infection in vitro and effectively reduced intracellular N protein levels when administered 2-12 hours after infection (202). Using similar techniques, the same authors have further identified N6, a coumarin derivative, which inhibited HTNV replication in vitro, reduced organ viral titers in vivo, and moderately improved weight loss and survival in a suckling mice challenge model (203).
Another strategy through which host proteins can be targeted to alleviate the symptoms of HCPS is via the targeting of vascular endothelial growth factor (VEGF). It has been shown that pathogenic hantaviruses modulate the expression of VEGF as a strategy to enhance lung endothelial vascular permeability (204). Once activated, the VEGF receptor (VEGFR2) promotes the internalization and subsequent degradation of VE-cadherin, an endothelial cell junction protein which is responsible for maintaining vascular barrier function (205, 206), via a signaling pathway mediated by Src family kinases (SFKs) (207). To combat this loss of barrier function, one study utilized a panel of FDA-approved VEGFR2 and SFK inhibitors which identified several drugs that inhibited the cell permeability induced by ANDV infection in vitro (208). An additional study utilizing the ANDV Syrian golden hamster challenge model showed that vandetanib, a VEGFR2 antagonist, delayed the onset of severe disease, increased survival, and decreased the accumulation of fluid in the lungs (209). While these results are encouraging, there was only a moderate decrease in lung, heart, and blood virus titers three days post infection which then increased to comparable levels with control treated animals, and treatment with high doses resulted in severe side-effects (209).
An alternative to antiviral drugs is the use of neutralizing antibodies for the treatment of hantavirus infection. Early studies demonstrated that the passive transfer of sera from rabbits (20), ducks (210), rhesus macaques (211), and geese (212) vaccinated using DNA vaccine technology was protective in the ANDV Syrian golden hamster challenge model. Polyclonal alpaca IgG has also been generated via the DNA vaccination of alpacas (213). Camelid-derived IgG has the advantage of its small size which, as it is composed of only heavy chains with no light chains, allows enhanced binding to epitopes usually inaccessible to human IgG (214).
More recent studies have utilized transchromosomic cattle, in which the bovine immunoglobulin G (IgG) locus has been replaced with the human locus, for the production of anti-hantavirus polyclonal sera (215, 216). In one study, transchromosomic cattle were immunized four times using DNA vaccines encoding the M segment of either PUUV or HTNV, plasma was then drawn from the cattle, and the anti-hantavirus human IgG was purified (215). This purified polyclonal IgG was found to be potently neutralizing against HTNV and PUUV in vivo, protective against HTNV infection in the Syrian golden hamster challenge model, and limited infection in a marmoset model of HTNV infection (215). Similar results were achieved when the vaccination protocol was modified to focus on ANDV and SNV, resulting in polyclonal IgG which could protect from HCPS (217). In a follow- up study, transchromosomal cattle were vaccinated five times with either ANDV and SNV or HTNV and PUUV DNA vaccines prior to plasma harvest and IgG purification. The resulting polyclonal IgG exhibit strong neutralizing activity against HTNV, PUUV, ANDV, SNV, SEOV, DOBV, and Choclovirus (CHOV) and protected against infection with HTNV, PUUV and SNV in vivo (216). The use of transchromosomal cattle is advantageous as it allows for the rapid production of potently neutralizing human IgG in large volumes that, as it is polyclonal, targets multiple epitopes on the hantavirus glycoproteins.
MAbs have also been investigated as a potential therapeutic avenue for the treatment of hantavirus infections. Early experiments with recombinantly produced murine antibodies identified using hybridoma technology showed that neutralizing mAbs targeting the HTNV glycoprotein could promote protection from infection in suckling mice (218, 219). These studies indicated that mAbs targeting either the Gn or Gc domain of the HTNV glycoprotein were sufficient to protect from infection, a finding which was later recapitulated using hybridoma-derived mAbs obtained from mice vaccinated against ANDV (220). More recent studies have focused on the production of recombinantly produced human monoclonal antibodies derived from survivors of hantavirus infection. By screening B cells from ANDV patients, one study identified two mAbs that exhibited strong neutralization against ANDV and protected hamsters from infection when used individually and in combination (221). Strikingly, these mAbs, when administered together, provided 50% protection even when given at a later stage of infection (8- and 10-days post-infection) (222). MAbs derived from HCPS survivors have also exhibited broad activity against a range of hantaviruses, with mAbs cloned from SNV survivors showing broader neutralization than those from ANDV survivors (88). Indeed, mAbs derived from PUUV patients have exhibited exceptional cross-neutralizing activity against both OWHVs and NWHVs and represent promising therapeutics when used alone or in combination with other mAbs (85). However, so far clinical development of these mAbs has not been initiated.
7 Animal models
Despite ongoing research on hantaviruses since their first emergence decades ago, suitable animal models that closely mimic human HFRS or HCPS disease outcomes are still lacking for the vast majority of hantaviruses. Indeed, there are few animal models available to study different aspects of viral pathogenicity, e.g., a lethal Syrian golden hamster model for ANDV infection (223) or lethal mouse models for HTNV infection (224, 225). Even though the clinical outcome in these models do not closely recapitulate human HFRS and HCPS, parameters of a persistent or acute infection, including high viral loads and the presence of viral genome in different organs (e.g., spleen, kidney, lungs, brain), as well as seroconversion can be observed [for review (226–228)]. Nevertheless, animal models that resemble human disease outcomes more faithfully are urgently needed to investigate the efficacy of vaccines and antivirals as well as the pathogenicity of hantavirus infection in humans.
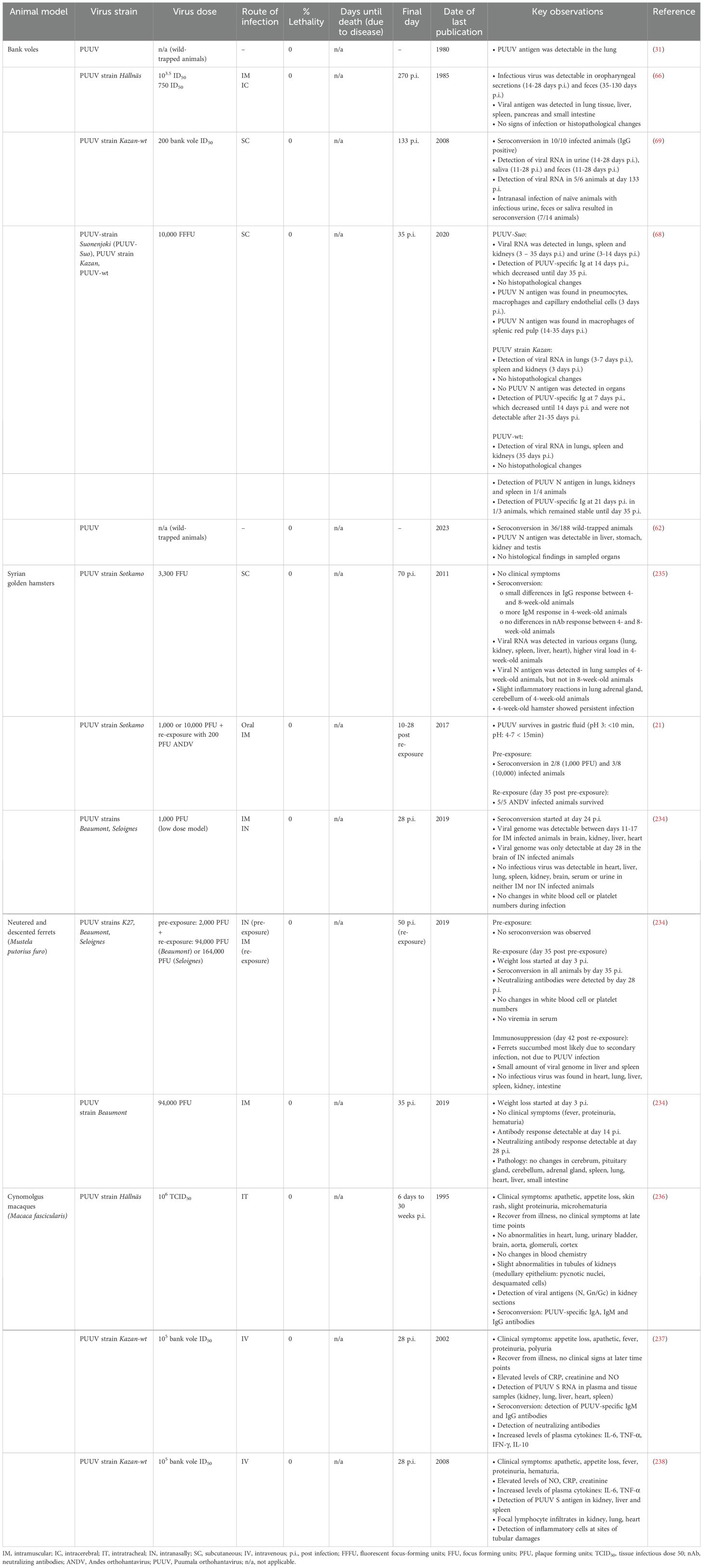
Preclinical studies on PUUV infections are typically performed in rodents and other small animal models. However, neither mice (66, 68, 69, 229–233) nor hamsters (21, 234, 235) develop clinical symptoms after PUUV infection, which is a limitation of these animal models. Nevertheless, they seroconvert after infection, and viral loads or viral antigens can be detected. Larger animals, such as non-human primates (236–238) (NHPs; e.g., cynomolgus macaques), develop clinical symptoms after PUUV infection, which partially resemble disease outcome in humans. However, due to the limited access to NHPs, their high costs and ethical reasons, they will most likely not become the standard model to study PUUV disease outcome in humans. Novel approaches besides immunocompetent small animals and non-human primates are currently being tested for both NWHVs and OWHVs. This includes animal models, in which hantavirus host receptors are artificially introduced into the animals, immunodeficient mice (239) or humanized/xenografted animal models (240–242). Those animals are more susceptible to hantavirus infection, develop clinical symptoms after hantavirus infection and partially resemble clinical signs observed in humans.
In this part, we aim to provide an overview on current animal models for hantaviruses with the focus on PUUV (summarized in Table 4).
7.1 Rodent models
Rodents are the natural reservoir of many of the hantaviruses, in which the viruses mainly cause a persistent infection. Investigations on disease outcome and the presence of viral RNA or infectious virus are either done by using wild-trapped animals or animals, which are infected in a controlled laboratory setting. Experimental infection of bank voles with PUUV resulted in seroconversion. Furthermore, viral RNA and infectious virus could be detected in serum, lung, spleen, kidneys, urine, feces and saliva; however, the animals did not succumb to the infection (66, 68, 69). No specific histological findings or signs of infection were observed in wild-trapped or experimentally infected bank voles, and PUUV N antigen was detectable in several organs, including kidney, lung, testis, liver and stomach, indicating a broad organ tropism (31, 62, 66, 68). Studies on hantavirus infections using rodent models demonstrated a strong correlation between the age of rodents and the disease outcome. Infection of 3-day-old suckling mice (Mus musculus) with HTNV caused 100% lethality, only 50% lethality in 1-week-old mice and no lethality in 2-week-old mice (229). Similar effects were observed upon infection of newborn rats with HTNV (230). Commonly used laboratory mouse strains (BALB/c, C57BL/6, SJL/J) have been susceptible to HTNV infection, however, mice had to be infected intraperitoneally (IP) with high doses, which do not mimic the natural route of infection in humans. Additionally, infected animals died due to acute encephalitis, which is not a typical symptom of HFRS (231). Due to the short time window in which rodents are susceptible to infection and the differences in the clinical outcome compared to human hantavirus infections, these models are not suitable to investigate the efficacy of antivirals and the protective capacity of vaccine candidates for translation to humans. Immunodeficient mice, such as Nlrc3-/- mice were more susceptible to IP infection with HTNV compared to C57BL/6 wild-type mice (232), indicated by higher weight loss and higher viral load in different organs (e.g., spleen, kidney). Humanized mice, such as hNSG/HLA-A2 mice (240) were highly susceptible to infection with HTNV. Mice showed weight loss, ruffled fur, decreased activity and inflammatory activities in the lung tissue. Furthermore, these animals showed reduced numbers of platelets in the blood, which has been also observed in hantavirus-infected humans (233).
7.2 Syrian golden hamster
Studies have shown that Syrian golden hamsters do not develop clinical symptoms upon intramuscular (IM) (234), subcutaneous (SC) (235), or intranasal (IN) (234) infection with PUUV. In addition, no changes in white blood cell numbers or platelet numbers, which are associated with leukocytosis and thrombocytopenia in human HFRS patients (243), respectively, were observed. However, animals seroconverted and viral genome could be detected in various organs, such as brain, kidney, liver and heart of IM (21) and SC (235) infected animals. In contrast, hamsters that were IN (234) infected showed only detectable amounts of viral genome in the brain, but not in other organs. Witkowski and colleagues (21) could demonstrate seroconversion in hamsters that were experimentally infected with PUUV via the gastric route, which subsequently provided protection against lethal ANDV infection. Sanada and colleagues (235) demonstrated an age-dependent effect on PUUV persistence. Four-week-old hamsters showed persistence up to 70 days post infection, higher viral load in various organs compared to 8-week-old hamster, and slightly increased inflammatory responses in lung, adrenal gland and cerebellum. Moreover, viral N antigen was detectable in 4-week-old hamster, but not in 8-week-old hamster.
7.3 Ferrets
Studies have shown, that ferrets do not develop clinical symptoms except for weight loss upon infection with PUUV (234). In addition, no pathological changes were observed in different organs, including lung, heart, liver, cerebrum or small intestine. However, seroconversion could be confirmed, including the detection of neutralizing antibodies (234).
7.4 Cynomolgus macaques
Cynomolgus macaques are more susceptible to PUUV infection compared to other animals, however, they do not succumb the infection (236–238). Intratracheal (IT) (236) or intravenous (IV) (237, 238) inoculation with PUUV led to the development of clinical symptoms (appetite loss, apathy, skin rash, proteinuria, microhematuria, fever, polyuria). However, the animals fully recovered after a few days. No abnormalities were observed in various organs, such as the liver, brain, heart, or urinary bladder, but slight abnormalities were seen in the tubules of the kidneys, which were limited to the medullary epithelium. Viral antigens could be detected in kidney sections and other tissues, such as lung, liver, heart or spleen (237, 238). Seroconversion was confirmed by detecting PUUV-specific IgG, IgA, IgM and neutralizing antibodies (237). Increased levels of plasma cytokines (IL-6, TNF-α, IFN-γ, IL-10) could be detected (237), which is typically found in human NE patients (244). Inflammatory cells were detected at sites of tubular damage, indicating that PUUV replication provokes immunopathology induced by activated T cells. Furthermore, the authors observed a correlation between high viral load and disease severity (238).
8 Vaccine approaches
Multiple vaccine candidates to prevent HFRS, mainly targeting HTNV or SEOV (245), have been developed using inactivated virus grown in cell culture or rodent brains, and were evaluated in preclinical and clinical trials in Asia. However, none of them were approved for human use in the US or Europe, mainly because of the used vaccine platforms employed and the targeted hantavirus species. Due to safety concerns, rodent brain-derived vaccines are no longer suitable for use in humans (246). In addition, there is only little cross-reactivity among certain hantavirus species (247), and as PUUV is the primary circulating hantavirus species in Europe, vaccines based on HTNV or SEOV would not be effective in Europe.
The very first candidate vaccine for prevention of HFRS, Hantavax, was already developed in 1988 by Lee and colleagues (248), by propagating the Hantaan virus ROK 84-105 strain on suckling mouse brains, followed by an inactivation step with 0.05% formalin. By demonstrating seroconversion with both ELISA and IFA as a surrogate for the efficacy of the vaccine, Hantavax was approved in 1990 in Korea for human use. However, the premise was to demonstrate in the subsequent years the protective efficacy of Hantavax in a controlled clinical trial compared to a placebo control group, and to demonstrate long-term maintenance of protection (249). The recommended vaccination schedule was a primary immunization with two doses one month apart, followed by a booster immunization one year later (0-1-13 schedule). However, by 2018 the Ministry of Food and Drug Safety of Korea changed this recommendation from three to four immunizations (250). Since its licensing more than 30 years ago, several million doses of Hantavax were administrated (251). However, its effectiveness, which is primarily determined by measuring humoral immune responses as a correlate of protection, is still debated (252, 253). Clinical trials with Hantavax had demonstrated a need for optimization for both, the recommended doses and immunization schedule, as the rate of seroconversion in the vaccinees was low, followed by a swift decline in titers of neutralizing antibodies (254–256). Song and colleagues (254) performed a phase III, multi-center clinical trial by immunizing healthy adults with Hantavax according to the recommended 0-1-13 immunization schedule. One month after the primary immunization with two doses, seroconversion was detected in 90% of the vaccinees via indirect IFA and in only 23% of the vaccinees via plaque-reduction serum neutralization assay (PRNT50). The rate of seroconversion declined to the pre-vaccination level after one year, however, the booster immunization led to an increase of the seroconversion rate by 87% (IFA) and 45.07% (PRNT50). Based on these observations, Song and colleagues (255) performed an additional multi-center phase III clinical trial immunizing healthy adults with Hantavax using a modified immunization schedule with three doses for primary vaccination followed by a booster immunization one year later (0–1–2–13 schedule). One month after the third primary vaccination, the seroconversion rate was 92.81% (IFA) and 80.97% (PRNT50) and declined to almost pre-vaccination level before the booster immunization. One month after the booster vaccination, seroconversion was detectable in 96% (IFA) and 67% (PRNT50) of the vaccinees. However, it decreased to around 40% a few months later.
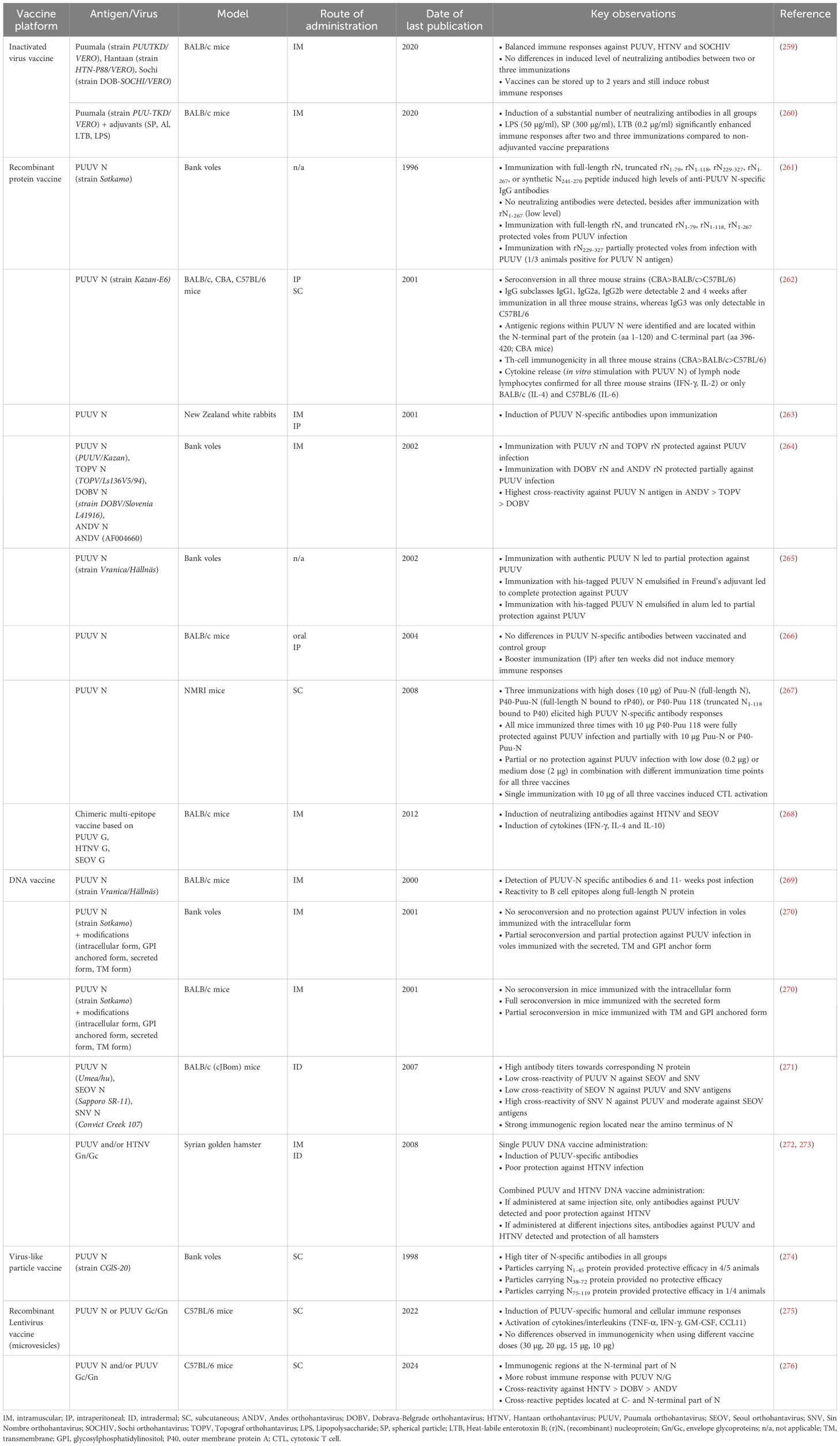
Over the last years, several new strategies, such as virus-like particles (VLP) vaccines, recombinant protein vaccines, subunit vaccines, recombinant viral-vector vaccines, and nucleic acid-based vaccines, were developed and served to generate vaccine candidates, mainly targeting ANDV, DOBV, HTNV or PUUV [for review (257, 258)]. In this part, we aim to provide detailed information about the current status of preclinical (summarized in Table 5) and clinical testing (summarized in Table 6) of vaccines targeting PUUV. The main targets for HFRS vaccine research are Gn/Gc and N. Gn/Gc was found to induce high levels of neutralizing antibodies (280), which are thought to be the main correlate of protection against hantavirus infection (281). N is thought to induce mainly cellular immune responses (265, 280), and although N-specific antibodies are induced upon immunization, they show poor neutralizing activity (264, 274). However, N has the advantage of inducing immunogenicity independent from post-translational modifications, which allows for an efficient production in cost-effective expression systems, such as Escherichia (E.) coli. Furthermore, the amino acid sequence of N among hantavirus serotypes is more conserved compared to Gn/Gc, thus, N might be a good target to generate cross-protective vaccines (282).
8.1 Inactivated whole virus vaccines
Dzagurova and colleagues (259) established a polyvalent vaccine, based on β-propiolacton inactivated cell culture preparation of the Hantaan HTN-P88/VERO strain, the Puumala PUU-TKD/VERO strain and the Sochi DOB-SOCHI/VERO strain (SOCHIV), and evaluated its immunogenicity in BALB/c mice. Mice were immunized intramuscularly two to three times two weeks apart with 0.5 ml (52 µg total protein/ml) of the vaccine, either undiluted or diluted (1:2, 1:8, 1:32), and the level of induced neutralizing antibodies were determined two weeks after the last immunization. In general, the polyvalent vaccine elicited neutralizing antibodies equally to SOCHIV, PUUV, and HTNV, providing a balanced immune response. There was no difference in the level of induced neutralizing antibodies and cytokines in mouse sera (IL-1β, IL-12, IFN-γ) between two and three immunizations.
Kurashova and colleagues (260) generated an inactivated PUUV vaccine, based on the propagation of the Puumala PUU-TKD/VERO strain on Vero cells and subsequent inactivation with β-propiolacton, and tested the beneficial effect of different adjuvants (subunit of an E. coli derived heat-labile enterotoxin (0.2 µg/ml, 7.5 µg/ml), aluminum hydroxide (1 mg/ml), spherical particles of coat protein from tobacco mosaic virus (100 µg/ml, 150 µg/ml, 300 µg/ml, and lipopolysaccharide (low endotoxic) from Shigella sonnei (50 µg/ml)) upon vaccination of BALB/c mice. Mice were immunized intramuscularly three times two weeks apart with the vaccines, either undiluted or diluted (1:2, 1:4, 1:8). All vaccines induced a substantial titer of neutralizing antibodies after two and three vaccinations. Interestingly, aluminum hydroxide (283), which is commonly used as adjuvant in inactivated vaccine preparations, did not lead to an increase of induced neutralizing antibodies compared to the non-adjuvanted vaccine group. Immunization with PUUV vaccines adjuvanted with either the lipopolysaccharide, the B subunit of heat-labile enterotoxin or spherical particles (300 µg/ml) significantly increased the humoral immune responses, also when administered in a diluted formulation.
Cho and colleagues (251) reported immunogenicity data from a small clinical study with 10 participants, who received three times four weeks apart a combined PUUV/HTNV vaccine, which was developed by propagation of the viruses on suckling hamster brains with subsequent formalin inactivation. The vaccine was well tolerated and induced high levels of neutralizing antibodies against HTNV and PUUV after the second and third immunization.
8.2 Recombinant protein vaccines
Maes and colleagues (267) linked the outer membrane protein A of Klebsiella pneumoniae (rP40) to a full-length (P40-Puu-N) or a truncated (P40-Puu 118) form of PUUV N and compared their immunogenic properties to an unmodified full-length PUUV N vaccine (Puu N). Outbred NMRI mice were immunized subcutaneously (SC) once, twice or three times with different doses (0.2 µg, 2 µg, 10 µg) of the three vaccines. NMRI mice were chosen, because they have been described previously as a suitable non-lethal rodent model, as the mice readily seroconvert and show detectable levels of neutralizing antibodies after infection (284). Overall, there was a dose and frequency of immunizations depended effect on the induction of antibody responses, with three immunizations with 10 µg of each vaccine eliciting the highest responses. In addition, full protection against PUUV was only seen with three immunizations of 10 µg P40-Puu 118. All three vaccines induced substantial numbers of cytotoxic T lymphocytes (CTLs) after a single immunization with 10 µg of each vaccine.
De Carvalho Nicacio and colleagues (262) evaluated the immunogenic properties of recombinant E. coli expressed PUUV N in three different mouse strains (CBA, BALB/c and C57BL/6), focusing on IgG subclasses and T-helper (Th) lymphocyte responses. Mice were immunized with 20 µg via the intraperitoneal (IP) route to determine antibody responses and with 50 µg via the SC route to determine Th lymphocyte responses. Overall, seroconversion was observed in all three mouse strains, with the highest titers for CBA > BALB/c > C57BL/6. All IgG subtypes were detectable 2 and 6 weeks after immunization in all three mouse strains, besides IgG3, which was only found in small amounts in C57BL/6 mice 2 weeks after immunization. Th lymphocyte responses could be confirmed in all three mouse strains, and immunogenic epitopes were identified across PUUV N, mainly located at the N-terminal part of the protein. In vitro stimulation of Th lymphocytes with PUUV N induced the release of different cytokines, including IFN-γ and IL-2 (all three mouse strains), as well as IL-4 (CBA) and IL-6 (C57BL/6).
Lindkvist and colleagues (261) used a full-length version (rN) and truncated versions of PUUV N (rN1-79, rN1-118, rN229-327, rN1-267) and aimed to investigate their immunogenic properties upon vaccination and challenge infection of bank voles. Voles were immunized with 50 µg of each vaccine three times three weeks apart and were infected with PUUV two weeks after the last immunization. Immunization with full-length rN, truncated rN1-79, rN1-118, rN229-327, rN1-267, or synthetic peptide N241-270 induced high levels of anti-PUUV N-specific IgG antibodies, however, these antibodies did not show neutralizing activity. Furthermore, immunization with full-length rN, and truncated rN1-79, rN1-118, rN1-267 fully protected voles against PUUV infection, whereas immunization with rN229-327 only partially protected voles against PUUV infection.
Kehm and colleagues (263) used transgenic tobacco and potato plants to express PUUV N protein and immunized New Zealand white rabbits with leaf tissue extracts intramuscularly (IM) and IP four times two weeks apart. When using the collected rabbit antisera in a Western blot, the authors were able to confirm immunogenicity against authentic PUUV N protein.
De Carvalho Nicacio and colleagues (264) immunized bank voles with recombinant nucleoprotein (rN) derived from PUUV, ANDV, DOBV or Topograf orthohantavirus (TOPV) and screened for cross-reactivity and protective efficacy upon challenge infection with PUUV. Bank voles were immunized three times three weeks apart with 50 µg of rN and challenge infected with wild-type PUUV (strain Kazan) two weeks after the last immunization. Bank voles do not succumb to PUUV infection and thus, protective efficacy was determined by analyzing lung tissue samples for the presence of viral RNA. Voles immunized with TOPV rN or PUUV rN showed complete protection, whereas with ANDV rN or DOBV rN partial protection was observed. The highest cross-reactivity against PUUV antigen was observed in sera of ANDV rN immunized bank voles, followed by those vaccinated with TOPV rN and DOBV rN.
Dargeviciute and colleagues (265) used the yeast expression system (Saccharomyces cerevisiae strain FH4C) to generate recombinant PUUV N, either in its authentic form or fused with a his-tag, and evaluated its immunogenicity in a bank vole challenge model. Voles were immunized with 50 µg of each vaccine three weeks apart and challenge infected with PUUV two weeks after the last immunization. Voles immunized with the authentic PUUV N antigen were only partially protected, whereas immunization with the his-tagged PUUV N antigen fully protected voles. In a subset of experiments, the his-tagged PUUV N antigen was emulsified in alum and bank voles were immunized and infected as described above. Six out of eight voles were completely protected, whereas the remaining two voles showed only partial protection.
In a follow up study, Khattak and colleagues (266) used the PUUV N protein, which was expressed in transgenic tobacco and potato plants, to evaluate the immunogenicity upon oral administration. BALB/c mice were fed with cheese balls containing the air-dried tobacco leaves or with pieces of the potato plants on days 1, 2, 17 and 31. In addition, mice received a booster administration (IP) ten weeks after the first immunization to test memory immune responses. Overall, no anti-PUUV specific antibody responses were induced upon oral administration of the recombinant proteins. Furthermore, the booster immunization did not induce memory immune responses.
Zhao and colleagues designed (268) a multi-epitope-based vaccine based on potential immunodominant B and T cell epitopes from HTNV, PUUV and SEOV Gn/Gc and immunized BALB/c mice once intramuscularly with 100 µg of the multi-epitope vaccine. Cytokine profile analysis of collected splenocytes revealed an increase in IFN-γ, IL-10, and IL-4 until days 31 (IFN-γ, IL-10) and 60 (IL-4), however, the levels of cytokines decreased swiftly until day 90. In addition, low level of neutralizing antibodies against HTNV and SEOV could be detected (neutralization against PUUV was not tested). Furthermore, they observed IgG binding antibody responses against the designed multi-epitope sequence, which increased until day 31 and remained stable until day 90.
8.3 DNA based vaccines
Koletzki and colleagues (269) immunized BALB/c mice with plasmid pcDNA3 encoding for the full-length N sequence via the IM route. Serum samples were collected six and eleven weeks after immunization and high titers of PUUV-N specific antibodies could be detected. Further analysis revealed that reactive B cell epitopes are distributed along the whole N protein.
Bucht and colleagues (270) developed modified DNA-based vaccines targeting PUUV N using an intracellular version, a secreted version (addition of an N-terminal secretion signal) and two membrane associated versions (carboxyl (C) -terminal addition of a glycosylphosphatidylinositol (GPI) anchor or a transmembrane (TM) signal) of the protein. BALB/c mice were immunized intramuscularly four times three weeks apart with 50 µg of each vaccine and seroconversion was confirmed in all mice which received the secreted form of PUUV N. In contrary, no seroconversion was observed in mice which received the intracellular form of PUUV N. Immunization with any of the two transmembrane associated forms resulted in partial seroconversion. Furthermore, bank voles were immunized intramuscularly four times three weeks apart with 50 µg of each vaccine and were challenge infected with PUUV (strain Kazan-wt). No seroconversion and no protection against PUUV were observed in any mice which were immunized with the intracellular form of PUUV N. Seroconversion and partial protection against PUUV were observed in voles immunized with the secreted, TM and GPI anchor form.
Lindkvist and colleagues (271) generated DNA-based vaccines targeting PUUV N, SEOV N or SNV N and designed full-length or truncated/deleted versions of the respective proteins. BALB/c (cJBom) mice were immunized five times two weeks apart with the DNA vaccines via the gene-gun method and they mainly focused on the analysis of cross-reactive antibody responses as outcome. As expected, they observed high antibody titers towards the corresponding full-length N protein. Interestingly, they observed high cross-reactivity of SNV N serum against PUUV N antigen and a moderate cross-reactivity against SEOV N antigen. When they screened all other possible combinations, only low level of cross-reactivity was observed. In addition, they immunized BALB/c (cJBom) mice with truncated/deleted versions of PUUV N, SEOV N, and SNV N and aimed to determine the location of B cell epitopes, which they identified to be located at the N-terminal part of the N protein.
Spik and colleagues (272) tested the immunogenicity of DNA-based vaccines encoding for the PUUV or HTNV Gn/Gc glycoprotein and evaluated their protective efficacy against HTNV challenge infection in Syrian golden hamsters. The vaccines (single or combined as mixture) were either administrated using the IM (via electroporation) or the intradermal (ID) route (via particle-mediated epidermal delivery (PMED)). Hamsters were immunized three times with 100 µg DNA each three weeks apart using the electroporation approach, with the combined PUUV/HTNV DNA vaccine being administrated either at the same injection site or at separate sites. Hamsters were immunized three times with 5-10 µg of DNA each three to four weeks apart using the PMED approach, with the combined PUUV/HTNV DNA vaccine being administrated 1) separate at adjacent injection sites, 2) by coating both plasmids onto the same gold beads or 3) by coating both plasmids onto different gold beads before mixing together. In addition, a subset of hamsters was infected with HTNV (273) via the IM route and as the animal do not develop clinical symptoms, the level of N-specific antibodies (which is not part of the vaccine) were determined as a surrogate of protection (272). When analyzing the sera four weeks after the last immunization, neutralizing antibodies against PUUV or HTNV were detected for both methods when individual vaccines were administrated. However, when analyzing the sera of hamsters that received both vaccines as a mixture, the elicited level of neutralizing antibodies against both HTNV or PUUV was strongly reliant on the administration mode. A substantial titer of neutralizing antibodies could be detected when immunizing the animals at different injection sites or coating the plasmids on different beads. The same effect could be observed when analyzing the sera of HTNV challenge infected hamsters. The PUUV DNA vaccine alone induced only poor humoral responses (13% for PMED and 38% for electroporation), which was inferior compared to the HTNV DNA vaccine alone (63% for PMED and 88% for electroporation). However, when used as a mixture and administrated at a separate injection site or coated on different gold beads, all hamsters showed protection.
Based on these promising observations, three open-label, single-center phase I studies were conducted, immunizing volunteers with either the PUUV or HTNV DNA vaccine or combined as a mixture using PMED (246), intramuscular electroporation (IM-EP) (277) or intramuscular delivery via the PharmaJet Stratis® needle-free injection system (279). In the first study (246), a total of 27 individuals were immunized three times via the PMED route with 8 µg of the PUUV DNA vaccine, the HTNV DNA vaccine or a mixture of both (half-dose of each), following a 0-1-2 immunization schedule. All vaccines were found to be safe and were well tolerated with no observed severe adverse events related to the study procedures or the vaccines. Serum samples were analyzed for neutralizing antibodies by PRNT50 and determined as seropositive if measurable titers were found within at least one of the collected serum samples. Overall, the rate of seroconversion was low, with 30% for the HTNV DNA vaccine group, 44% for PUUV DNA vaccine group, and 56% for the PUUV/HTNV DNA group. In the second study (277), a total of 27 individuals were immunized three times via the IM-EP route with 2 mg of the PUUV DNA vaccine, the HTNV DNA vaccine or combined as a mixture (half-dose each), following a 0-1-2 immunization schedule. Serum samples were analyzed for neutralizing antibodies by PRNT50. Seroconversion was observed in 5/9 and 7/9 vaccinees who received all vaccinations with the HTNV or PUUV vaccines, respectively, and in 7/9 vaccinees against PUUV who received the combined vaccine preparation. Interestingly, more individuals responded to the PUUV vaccine than to the HTNV vaccine in the combined vaccine group, however, the three individuals with the highest PRNT50 titer against PUUV had also high amounts of neutralizing antibodies directed against HTNV. In the third study (279), a total of 27 individuals were immunized four times via the PharmaJet Stratis® needle-free injection system with 2 mg of the PUUV DNA vaccine, HTNV DNA vaccine or combined as a mixture (half-dose each), following a 0-1-2-6 immunization schedule. Serum samples from 22 individuals were analyzed for neutralizing antibodies using PRNT50 and pseudovirion neutralization assay (PsVNA), where titers determined with PRNT50 were found to be lower compared to PsVNA titers. Seroconversion was observed in 7/7 and 6/6 individuals receiving the HTNV or PUUV DNA vaccines, respectively, and only in 4/9 individuals who received the combined PUUV/HTNV vaccine. As already observed in the previous study, individuals who received the combined PUUV/HTNV vaccine responded more to PUUV than to HTNV. Cross-reactivity against HTNV was observed in individuals who received the PUUV DNA vaccine and vice versa. In addition, little cross-reactivity against DOBV was observed in samples from all three vaccine groups.
As a follow up, a randomized, double-blinded, phase IIa study was conducted (278), focusing on an optimized, combined PUUV/HTNV DNA vaccine to determine an optimal dose and immunization schedule. A total of 120 subjects were divided into 4 cohorts and subsequently immunized three or four times (0-1-2-6 immunization schedule) with either 2 mg (1 mg per vaccine) or 1 mg (0.5 mg per vaccine) of the PUUV/HTNV DNA vaccine. Cohorts 1 and 3 received four administrations, whereas cohorts 2 and 4 received a phosphate buffered saline vehicle at day 28. Serum samples were screened for neutralizing antibodies by HTNV and PUUV PsVNAs or PRNT50. The PUUV/HTNV DNA vaccine induced strong anti-HTNV neutralizing antibodies in the presence of the PUUV vaccine. Overall, only 9% of the subjects did not respond to the PUUV/HTNV vaccine at any point during the study. The second vaccination at day 28 induced higher seropositivity rates in cohorts 1 and 3 compared to the cohorts which received only phosphate buffered saline, however, analysis of subsequent time points could not demonstrate significant differences between three and four vaccinations. Overall, subjects from cohort 3 (four immunizations with 0.5 mg vaccine each) showed the highest rate of seropositivity and highest median neutralizing titers against both PUUV and HTNV.
8.4 Virus-like particle vaccines
Ulrich and colleagues (274) generated three chimeric VLP vaccine candidates based on the core antigen of hepatitis B virus which presented PUUV N fragments (N1-45, N38-72, or N75-119) on their surface. Bank voles were immunized subcutaneously three times three weeks apart with 50 µg of each vaccine and challenge infected with PUUV (strain Kazan) two weeks after the last immunization. All vaccinated animals showed high titers of N-specific antibodies. Bank voles immunized with VLP carrying fragment N1-45 showed the highest protection, followed by N75-119. Immunization with VLP carrying fragment N38-72 did not induce protective immunity.
8.5 Recombinant lentivirus vaccines
Shkair and colleagues (275) developed PUUV candidate vaccines based on microvesicles (MVs) that carry PUUV N and/or PUUV Gn/Gc glycoproteins and evaluated their immunogenicity in C57BL/6 mice. Mice were immunized SC with MVs (15 µg/50 µL) containing PUUV N or PUUV Gn/Gc or both, PUUV N and PUUV Gn/Gc. They detected elevated levels of anti-orthohantavirus-specific IgG in sera of all vaccinated mice at days 14 and 28 post immunization. The highest level of seroconversion was found in the sera of mice vaccinated with MVs carrying both PUUV N and PUUV Gn/Gc. In addition, they confirmed cellular immune responses upon vaccination with all three vaccines by measuring IFN-γ secretion of activated cytotoxic T lymphocytes. Furthermore, they confirmed the induction of cytokines (TNF-α, IL-6, GM-CSF, and G-CSF), which are important to stimulate proliferation and/or differentiation of leukocytes and stimulating phagocytosis by other immune cells, such as macrophages.
In a follow up study, Shkair and colleagues (276) used the developed and characterized PUUV N and PUUV N/G vaccines to screen for specific immunogenic regions of the N protein by using synthesized N peptide fragments. C57BL/6 mice were immunized SC with the two vaccines and collected sera were screened for immunoreactivity towards PUUV N, HTNV N, DOBV N and ANDV N peptides. Vaccination with PUUV N/G induced the selection of more immunogenic PUUV N-specific epitopes compared to PUUV N alone (eleven vs. seven, respectively). In addition, sera of PUUV N vaccinated mice reacted with seven HTNV, two DOBV and two ANDV N peptide fragments, whereas PUUV N/G reacted with only four HTNV N peptide fragments, which were none of the seven HTNV N peptide fragments that reacted with the PUUV N vaccine alone. Overall, the identified reacting and cross-reacting N peptides were located at the N- and C-terminal part of the full-length hantavirus N protein, which are important for the replication process.
9 Reverse genetics
Reverse genetics (RG) systems are well-established and highly efficient molecular tools. RG systems allow for replication and transcription of either full-length viral RNA genomes or truncated analogues from a complementary DNA (cDNA), with the aim to produce a full-length infectious viral clone or minigenome systems. Contrary to classical genetics approaches, in which a certain phenotype is analyzed for its causative genotype, RG systems aim to manipulate a viral genotype and analyze the resulting changes in the phenotype (285). One major advantage of using minigenome systems is the possibility to handle and manipulate them under biosafety level (BSL) 1/2 conditions. Especially for working with highly pathogenic viruses, such as Orthohantaviridae and Filoviridae (e.g., Ebola-virus, Marburg-virus), which require BSL3 and 4 laboratories, respectively, RG offers a convenient way to analyze their pathology, life cycle or molecular biology potentially under lower biosafety requirements if attenuated versions can be generated [for review (286–288)]. In addition, animal studies involving hantaviruses and other highly pathogenic viruses require BSL3 and 4 animal facilities, which are expensive in maintenance and are associated with many restrictions and conditions. Suitable RG systems would potentially allow the efficacy testing of vaccines or therapeutics under lower biosafety requirements.
To obtain recombinant viruses using RG, the first step is the generation of cDNA plasmid intermediates that contain the viral genome or reporter proteins (e.g., to study the function of non-coding regions). The transcription of biologically active molecules is placed under the control of a DNA-dependent RNA polymerase (DdRP), such as RNA polymerase I/II (289) or T7 RNA polymerase (290). Upon transfection of cells with the cDNA plasmids and the co-expression of the respective DdRP, an unencapsidated genomic RNA is transcribed. However, a hallmark of negative-strand RNA viruses is the necessity of viral RNA to be encapsidated by the nucleoprotein to serve as a template for the viral polymerase (286). Therefore, the nucleoprotein has to be co-expressed and once encapsidated, other ribonucleoprotein components, that are provided by either helper virus co-infection or helper plasmid co-transfection, recognize the genomic RNA and initiate replication and transcription into mRNAs. Subsequently, all viral proteins required to start the viral replication cycle are translated, which ultimately leads to the generation of infectious viruses (286).
Reverse genetics systems have been established for several Bunyavirales (Peribunyaviridae, Nairoviridae or Arenaviridiae), based on either the bacteriophage T7 RNA polymerase system or RNA polymerase I/II system (291). However, attempts to establish RG systems for hantaviruses were only partially successful, but mostly failed (292). In the late 1990s, Welzel and colleagues (293) efficiently expressed PUUV and HTNV N in mammalian cells, a first step towards the establishment of an eukaryotic system for hantavirus reverse genetics. Flick and colleagues (294) successfully established the first HTNV minigenome, thereby demonstrating the expression of a functional recombinant hantavirus polymerase and the rescue of HTNV minigenomes without superinfection with infectious hantavirus, allowing for handling of the recombinant viral clones outside a BSL3 facility. However, the established system was not further used, suggesting suboptimal tractability (295). The first minigenome system for ANDV was described more than 10 years ago by Brown and colleagues (292), however, unsuccessful expression of the L-protein and lack of reproducibility were observed. Infectious virus has not been rescued so far.
Overall, successful and tractable RG systems for hantaviruses are urgently required for a better understanding of certain molecular pathways and to establish effective antivirals and vaccines. However, to develop effective RG systems for hantaviruses, several obstacles and hurdles have to be overcome as shown by the previous failed attempts (292, 294): (I) the type of promoter (weak or strong) used to express the viral genome, (II) the presence of a potential cryptic promoter in the 3´ non-coding regions, (III) hantavirus- and/or host-cell specific factors that might inhibit rescue of an infectious clone, (IV) the determination/consideration of a correct biological ratio for the expressed viral proteins, (V) the choice of suitable cell lines for transfection; e.g., Vero E6 cells are suitable for viral infection and propagation, but BHK-21 cells show a higher transfection efficiency, (VI) the potential need of cellular factors that only exist in the reservoir but are not present in available cell lines.
Intra-and inter-lineage reassortment events of naturally circulating strains are reported frequently [for review: (296)] and in vitro reassortment systems are a suitable alternative to reverse genetic systems for culturing hantavirus species and analyze e.g., growth characteristics or innate immune responses. Cell cultures are co-infected with two closely related hantavirus species which allows the two viruses to exchange genomic segments. Subsequently, the infected cell cultures are screened to identify novel reassortant viruses. Interestingly, current attempts were only successful in exchanging the M segment between two hantavirus species, indicating that certain species-specific determinants (297) inhibit heterologous reassortment. Furthermore, the high instability of reassortant viruses suggests a co-requirement of L and S segments from the same hantavirus species (298). The RdRp which is encoded by the L segment interacts with the encapsidated RNA in a sequence specific manner, and interaction with the nucleocapsid protein (encoded by the S segment) is required for viral replication and transcription (299). In the late 1990s, Rodriguez and colleagues (83) generated reassortant viruses by co-infecting SNV and Black Creek Canal virus (BCCV), thereby obtaining virus plaques that appeared diploid, containing S or M segments originating from parental SNV and BCCV. However, most of these diploid virus genotypes were unstable and only one reassortant virus, based on L and S segments from BCCV, and M segment from SNV appeared stable. McElroy and colleagues (298) recovered a reassortant virus (SAS-11) based on SNV L and S segments and ANDV M segment. Other combinations, e.g., ANDV L and S segments and SNV M segment turned out to be unstable and were lost during plaque isolation. SAS-11 showed similar plaque morphology and growth characteristics as ANDV, but failed to induce a lethal infection in Syrian hamsters. Handke and colleagues (297) generated a reassortant virus (PHPUV) by co-infecting PUUV and Prospect Hill virus (PHV), which contained the PUUV M segment and the PHV L and S segments. PHPUV showed growth characteristics and ability to stimulate innate immune responses in vitro similar to the parental PHV.
10 Discussion and future perspectives
Nephropathia epidemica is an important and significant disease in Europe that is caused by PUUV infections. While thousands of cases are reported every year, it is likely also underdiagnosed and underreported. In recent years, major advances to a better understanding of this viral infection have been made. These include the elucidation of the structure of the viral surface glycoproteins and the discovery of broadly protective hantavirus mAbs. However, much remains to be done. A reverse genetics system for hantaviruses in general has not been established and is urgently needed to better understand PUUV biology. A reverse genetics system could also be used to rationally design live attenuated vaccines. In addition, better animal models for PUUV that more closely reflect human disease and can be used for evaluation of therapeutics and prophylactics are also urgently needed. Furthermore, no human vaccines or therapeutics for PUUV are available. MAbs as therapeutic candidates have been developed but still need to enter clinical development. In addition, no PUUV vaccines are available for human use. While several vaccine candidates already exist, mRNA vaccine development also opens up exciting new avenues for the development of PUUV vaccines. However, while thousands of cases occur every year, it will potentially be difficult to perform late-stage clinical trials to establish efficacy of vaccines or treatments. We do believe that this is possible by focusing on risk groups and areas in Europe where clinicians are very experienced with diagnosing the disease, like parts of Sweden, Finland, Germany, Austria or Slovenia. While there is certainly a medical need, it is also unclear if there is a business case for commercial entities to develop vaccines or therapeutics for PUUV infections. It is likely that these interventions have to be developed through public-private partnerships and with the help of public funding bodies like the European Union.
Author contributions
AT: Writing – original draft, Writing – review & editing. PG-C: Writing – original draft, Writing – review & editing. JJC: Writing – original draft, Writing – review & editing. RK: Writing – original draft, Writing – review & editing. FK: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. Work in the Krammer laboratory at the Ignaz Semmelweis Institute at the Medical University of Vienna is supported by institutional funds. Work in the Krammer laboratory at the Icahn School of Medicine at Mount Sinai is also supported by institutional funds. Some Puumala studies in the Division of Infectious Diseases Graz were in part supported by the Styrian government, Austria (project no. ABT12-106729/2022-13).
Conflict of interest
FK declares the following conflicts of interest. The Icahn School of Medicine at Mount Sinai has filed patent applications regarding influenza virus vaccines on which FK is listed as inventor. The Icahn School of Medicine at Mount Sinai has filed patent applications relating to SARS-CoV-2 serological assays, NDV-based SARS-CoV-2 vaccines influenza virus vaccines and influenza virus therapeutics which list FK as co-inventor and FK has received royalty payments from some of these patents. Mount Sinai has spun out a company, Kantaro, to market serological tests for SARS-CoV-2 and another company, Castlevax, to develop SARS-CoV-2 vaccines. FK is co-founder and scientific advisory board member of Castlevax. FK has consulted for Merck, GSK, Sanofi, Curevac, Seqirus and Pfizer and is currently consulting for 3rd Rock Ventures, Gritstone and Avimex. The Krammer laboratory is also collaborating with Dynavax on influenza vaccine development and with VIR on influenza virus therapeutics.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1575112/full#supplementary-material
References
1. Bradfute SB, Calisher CH, Klempa B, Klingström J, Kuhn JH, Laenen L, et al. ICTV virus taxonomy profile: hantaviridae 2024. J Gen Virol. (2024) 105. doi: 10.1099/jgv.0.001975
2. Mustonen J, Vapalahti O, Henttonen H, Pasternack A, Vaheri A. Epidemiology of hantavirus infections in Europe. Nephrol Dialysis Transplant. (1998) 13:2729–31. doi: 10.1093/ndt/13.11.2729
3. Monroe MC, Morzunov SP, Johnson AM, Bowen MD, Artsob H, Yates T, et al. Genetic diversity and distribution of peromyscus-borne hantaviruses in north America. Emerging Infect Dis. (1999) 5:75–86. doi: 10.3201/eid0501.990109
4. Kabwe E, Davidyuk Y, Shamsutdinov A, Garanina E, Martynova E, Kitaeva K, et al. Orthohantaviruses, emerging zoonotic pathogens. Pathog (Basel Switzerland). (2020) 9. doi: 10.3390/pathogens9090775
5. Warner BM, Dowhanik S, Audet J, Grolla A, Dick D, Strong JE, et al. Hantavirus cardiopulmonary syndrome in Canada. Emerging Infect Dis. (2020) 26:3020–4. doi: 10.3201/eid2612.202808
6. Gajdusek DC. Virus hemorrhagic fevers. Special reference to hemorrhagic fever with renal syndrome (Epidemic hemorrhagic fever). J Pediatr. (1962) 60:841–57. doi: 10.1016/s0022-3476(62)80170-x
7. Lee HW, Lee PW, Johnson KM. Isolation of the etiologic agent of Korean hemorrhagic fever. J Infect Dis. (1978) 137:298–308. doi: 10.1093/infdis/137.3.298
8. Vapalahti O, Mustonen J, Lundkvist A, Henttonen H, Plyusnin A, Vaheri A. Hantavirus infections in Europe. Lancet Infect Dis. (2003) 3:653–61. doi: 10.1016/s1473-3099(03)00774-6
9. Hjertqvist M, Klein SL, Ahlm C, Klingström J. Mortality rate patterns for hemorrhagic fever with renal syndrome caused by Puumala virus. Emerging Infect Dis. (2010) 16:1584–6. doi: 10.3201/eid1610.100242
10. Szabó R. Antiviral therapy and prevention against hantavirus infections. Acta Virol. (2017) 61:3–12. doi: 10.4149/av_2017_01_3
11. Jacob AT, Ziegler BM, Farha SM, Vivian LR, Zilinski CA, Armstrong AR, et al. Sin Nombre virus and the emergence of other hantaviruses: A review of the biology, ecology, and disease of a zoonotic pathogen. Biology. (2023) 12. doi: 10.3390/biology12111413
12. Furberg M, Anticona C, Schumann B. Post-infectious fatigue following Puumala virus infection. Infect Dis (London England). (2019) 51:519–26. doi: 10.1080/23744235.2019.1605191
13. Mustonen J, Vaheri A, Pörsti I, Mäkelä S. Long-term consequences of Puumala hantavirus infection. Viruses. (2022) 14. doi: 10.3390/v14030598
14. Kääriäinen S, Ollgren J, Dub T, Laine O, Sinisalo M, Hepojoki J, et al. Risk of lymphoid Malignancies increased after Puumala virus infection in Finland, 2009-2019: A retrospective register-based cohort study. Int J Infect Dis. (2023) 131:1–6. doi: 10.1016/j.ijid.2023.03.026
15. Klingström J, Granath F, Ekbom A, Björkström NK, Ljunggren H-G. Increased risk for lymphoma following hemorrhagic fever with renal syndrome. Clin Infect Dis. (2014) 59:1130–2. doi: 10.1093/cid/ciu488
16. Forbes KM, Sironen T, Plyusnin A. Hantavirus maintenance and transmission in reservoir host populations. Curr Opin Virol. (2018) 28:1–6. doi: 10.1016/j.coviro.2017.09.003
17. Plyusnin A, Morzunov SP. Virus evolution and genetic diversity of hantaviruses and their rodent hosts. Curr Topics Microbiol Immunol. (2001) 256:47–75. doi: 10.1007/978-3-642-56753-7_4
18. Tagliapietra V, Rosà R, Rossi C, Rosso F, Hauffe HC, Tommasini M, et al. Emerging rodent-borne viral zoonoses in trento, Italy. EcoHealth. (2018) 15:695–704. doi: 10.1007/s10393-018-1335-4
19. Nuzum EO, Rossi CA, Stephenson EH, LeDuc JW. Aerosol transmission of hantaan and related viruses to laboratory rats. Am J Trop Med Hygiene. (1988) 38:636–40. doi: 10.4269/ajtmh.1988.38.636
20. Hooper JW, Ferro AM, Wahl-Jensen V. Immune serum produced by DNA vaccination protects hamsters against lethal respiratory challenge with Andes virus. J Virol. (2007) 82:1332–8. doi: 10.1128/jvi.01822-07
21. Witkowski PT, Perley CC, Brocato RL, Hooper JW, Jürgensen C, Schulzke J-D, et al. Gastrointestinal tract as entry route for hantavirus infection. Front Microbiol. (2017) 8:1721. doi: 10.3389/fmicb.2017.01721
22. Martínez VP, Di Paola N, Alonso DO, Pérez-Sautu U, Bellomo CM, Iglesias AA, et al. Super-spreaders” and person-to-person transmission of Andes virus in Argentina. New Engl J Med. (2020) 383:2230–41. doi: 10.1056/NEJMoa2009040
23. Martinez VP, Bellomo C, San Juan J, Pinna D, Forlenza R, Elder M, et al. Person-to-person transmission of Andes virus. Emerging Infect Dis. (2005) 11:1848–53. doi: 10.3201/eid1112.050501
24. Sinisalo M, Vapalahti O, Ekblom-Kullberg S, Laine O, Mäkelä S, Rintala H, et al. Headache and low platelets in a patient with acute leukemia. J Clin Virol. (2010) 48:159–61. doi: 10.1016/j.jcv.2010.02.015
25. Ferrés M, Martínez-Valdebenito C, Angulo J, Henríquez C, Vera-Otárola J, Vergara MJ, et al. Mother-to-child transmission of Andes virus through breast milk, Chile1. Emerging Infect Dis. (2020) 26:1885–8. doi: 10.3201/eid2608.200204
27. Jonsson CB, Figueiredo LTM, Vapalahti O. A global perspective on hantavirus ecology, epidemiology, and disease. Clin Microbiol Rev. (2010) 23:412–41. doi: 10.1128/cmr.00062-09
28. Nichol ST, Spiropoulou CF, Morzunov S, Rollin PE, Ksiazek TG, Feldmann H, et al. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Sci (New York NY). (1993) 262:914–7. doi: 10.1126/science.8235615
29. López N, Padula P, Rossi C, Lázaro ME, Franze-Fernández MT. Genetic identification of a new hantavirus causing severe pulmonary syndrome in Argentina. Virology. (1996) 220:223–6. doi: 10.1006/viro.1996.0305
30. Avsic-Zupanc T, Xiao SY, Stojanovic R, Gligic A, van der Groen G, LeDuc JW. Characterization of Dobrava virus: A hantavirus from Slovenia, yugoslavia. J Med Virol. (1992) 38:132–7. doi: 10.1002/jmv.1890380211
31. Brummer-Korvenkontio M, Vaheri A, Hovi T, Bonsdorff C, Vuorimies J, Manni T, et al. Nephropathia epidemica: detection of antigen in bank voles and serologic diagnosis of human infection. J Infect Dis. (1980) 141:131–4. doi: 10.1093/infdis/141.2.131
32. Vaheri A, Henttonen H, Voutilainen L, Mustonen J, Sironen T, Vapalahti O. Hantavirus infections in Europe and their impact on public health. Rev Med Virol. (2013) 23:35–49. doi: 10.1002/rmv.1722
33. European Centre for Disease Prevention and Control. Disease Information About Hantavirus: Factsheet. Available online at: https://www.ecdc.europa.eu/en/hantavirus-infection/facts (Accessed November 18, 2024).
34. Mertens M, Hofmann J, Petraityte-Burneikiene R, Ziller M, Sasnauskas K, Friedrich R, et al. Seroprevalence Study in forestry workers of a non-endemic region in eastern Germany reveals infections by Tula and Dobrava-Belgrade hantaviruses. Med Microbiol Immunol. (2011) 200:263–8. doi: 10.1007/s00430-011-0203-4
35. Razzauti M, Plyusnina A, Sironen T, Henttonen H, Plyusnin A. Analysis of Puumala hantavirus in a bank vole population in northern Finland: evidence for co-circulation of two genetic lineages and frequent reassortment between strains. J Gen Virol. (2009) 90:1923–31. doi: 10.1099/vir.0.011304-0
36. Nemirov K, Leirs H, Lundkvist A, Olsson GE. Puumala hantavirus and myodes glareolus in northern Europe: no evidence of co-divergence between genetic lineages of virus and host. J Gen Virol. (2010) 91:1262–74. doi: 10.1099/vir.0.016618-0
37. Blinova E, Deviatkin A, Makenov M, Popova Y, Dzagurova T. Evolutionary formation and distribution of Puumala virus genome variants, Russia. Emerging Infect Dis. (2023) 29:1420–4. doi: 10.3201/eid2907.221731
38. Sironen T, Vaheri A, Plyusnin A. Molecular evolution of Puumala hantavirus. J Virol. (2001) 75:11803–10. doi: 10.1128/jvi.75.23.11803-11810.2001
39. Davidyuk YN, Kabwe E, Shakirova VG, Martynova EV, Ismagilova RK, Khaertynova IM, et al. Characterization of the Puumala orthohantavirus strains in the northwestern region of the republic of Tatarstan in relation to the clinical manifestations in hemorrhagic fever with renal syndrome patients. Front Pharmacol. (2019) 10:970. doi: 10.3389/fphar.2019.00970
40. European Centre for Disease Prevention and Control. Facts About Hantavirus. Available online at: https://www.ecdc.europa.eu/en/hantavirus-infection/facts(Accessed September 29, 2024).
41. European Centre for Disease Prevention and Control. Annual Epidemiological Report 2016- Hantavirus infection. Stockholm: ECDC (2018). Available at: http://ecdc.europa.eu/en/healthtopics/hantavirus/Pages/Annualepidemiologicalreport2016.aspx(Accessed September 25, 2024).
42. European Centre for Disease Prevention and Control. Hantavirus infection. In: ECDC. Annual epidemiological report for 2018. ECDC, Stockholm (2020). Available at: https://www.ecdc.europa.eu/en/publications-data/hantavirus-infection-annual-epidemiologicalreport-2018 (Accessed September 25, 2024).
43. European Centre for Disease Prevention and Control. Hantavirus infection. In: ECDC. Annual epidemiological report for 2020. ECDC, Stockholm (2023). Available at: https://www.ecdc.europa.eu/sites/default/files/documents/Hantavirus-AER-2020.pdf (Accessed September 25, 2024).
44. Heyman P, Ceianu CS, Christova I, Tordo N, Beersma M, João Alves M, et al. A five-year perspective on the situation of haemorrhagic fever with renal syndrome and status of the hantavirus reservoirs in Europe, 2005-2010. Euro Surveillance. (2011) 16. doi: 10.2807/ese.16.36.19961-en
45. Lupuşoru G, Andronesi AG, Lupuşoru M, Ailincăi I, Sfeatcu R, Văcăroiu I, et al. Hantavirus infections in the South−Eastern European countries: A study of two cases and literature review. Exp Ther Med. (2023) 26. doi: 10.3892/etm.2023.12129
46. Reil D, Rosenfeld UM, Imholt C, Schmidt S, Ulrich RG, Eccard JA, et al. Puumala hantavirus infections in bank vole populations: host and virus dynamics in central Europe. BMC Ecol. (2017) 17. doi: 10.1186/s12898-017-0118-z
47. Manigold T, Vial P. Human hantavirus infections: epidemiology, clinical features, pathogenesis and immunology. Swiss Med Weekly. (2014) 144:w13937. doi: 10.4414/smw.2014.13937
48. Arai S, Yanagihara R. Genetic diversity and geographic distribution of bat-borne hantaviruses. Curr Issues Mol Biol. (2020) 39:1–28. doi: 10.21775/cimb.039.001
49. Gu SH, Miñarro M, Feliu C, Hugot J-P, Forrester NL, Weaver SC, et al. Multiple lineages of hantaviruses harbored by the iberian mole (Talpa occidentalis) in Spain. Viruses. (2023) 15. doi: 10.3390/v15061313
50. Reusken C, Heyman P. Factors driving hantavirus emergence in Europe. Curr Opin Virol. (2013) 3:92–9. doi: 10.1016/j.coviro.2013.01.002
51. Clement J, Maes P, van Ypersele de Strihou C, van der Groen G, Barrios JM, Verstraeten WW, et al. Beechnuts and outbreaks of nephropathia epidemica (NE): of mast, mice and men. Nephrol Dialysis Transplant. (2010) 25:1740–6. doi: 10.1093/ndt/gfq122
52. Thuy DTN, Sasaki M, Orba Y, Thammahakin P, Maezono K, Kobayashi S, et al. Molecular evolution of Hokkaido virus, a genotype of orthohantavirus puumalaense, among Myodes rodents. Virology. (2024) 597:110168. doi: 10.1016/j.virol.2024.110168
53. Guterres A, Lemos E. Hantaviruses and a neglected environmental determinant. One Health. (2018) 5:27–33. doi: 10.1016/j.onehlt.2017.12.002
54. Swart A, Bekker DL, Maas M, Vries Ad, Pijnacker R, Reusken CBEM, et al. Modelling human Puumala hantavirus infection in relation to bank vole abundance and masting intensity in the Netherlands. Infect Ecol Epidemiol. (2017) 7. doi: 10.1080/20008686.2017.1287986
55. Amirpour Haredasht S, Barrios M, Farifteh J, Maes P, Clement J, Verstraeten WW, et al. Ecological niche modelling of bank voles in Western Europe. Int J Environ Res Public Health. (2013) 10:499–514. doi: 10.3390/ijerph10020499
56. Linard C, Lamarque P, Heyman P, Ducoffre G, Luyasu V, Tersago K, et al. Determinants of the geographic distribution of Puumala virus and Lyme borreliosis infections in Belgium. Int J Health Geographics. (2007) 6:15. doi: 10.1186/1476-072x-6-15
57. Wang YXG, Voutilainen L, Aminikhah M, Helle H, Huitu O, Laakkonen J, et al. The impact of wildlife and environmental factors on hantavirus infection in the host and its translation into human risk. Proc Biol Sci. (2023) 290:20222470. doi: 10.1098/rspb.2022.2470
58. Clement J, Vercauteren J, Verstraeten WW, Ducoffre G, Barrios JM, Vandamme A-M, et al. Relating increasing hantavirus incidences to the changing climate: the mast connection. Int J Health Geographics. (2009) 8:1. doi: 10.1186/1476-072x-8-1
59. Clement J, Maes P, van Ranst M. Hantaviruses in the old and New World. In: Emerging Viruses in Human Populations. Perspectives in Medical Virology, vol. 16. Elsevier (2006). p. 161–77.
60. Reil D, Imholt C, Eccard JA, Jacob J. Beech fructification and bank vole population dynamics - combined analyses of promoters of human Puumala virus infections in Germany. PloS One. (2015) 10. doi: 10.1371/journal.pone.0134124
61. Selås V. Evidence for different bottom-up mechanisms in wood mouse (Apodemus sylvaticus) and bank vole (Myodes glareolus) population fluctuations in Southern Norway. Mammal Res. (2020) 65:267–75. doi: 10.1007/s13364-020-00476-0
62. Schlohsarczyk EK, Drewes S, Koteja P, Röhrs S, Ulrich RG, Teifke JP, et al. Tropism of Puumala orthohantavirus and endoparasite coinfection in the bank vole reservoir. Viruses. (2023) 15. doi: 10.3390/v15030612
63. Bernshtein AD, Apekina NS, Mikhailova TV, Myasnikov YA, Khlyap LA, Korotkov YS, et al. Dynamics of Puumala hantavirus infection in naturally infected bank voles (Clethrinomys glareolus). Arch Virol. (1999) 144:2415–28. doi: 10.1007/s007050050654
64. Tersago K, Crespin L, Verhagen R, Leirs H. Impact of Puumala virus infection on maturation and survival in bank voles: A capture-mark-recapture analysis. J Wildlife Dis. (2012) 48:148–56. doi: 10.7589/0090-3558-48.1.148
65. Kallio ER, Klingström J, Gustafsson E, Manni T, Vaheri A, Henttonen H, et al. Prolonged survival of Puumala hantavirus outside the host: evidence for indirect transmission via the environment. J Gen Virol. (2006) 87:2127–34. doi: 10.1099/vir.0.81643-0
66. Yanagihara R, Amyx HL, Gajdusek DC. Experimental infection with Puumala virus, the etiologic agent of nephropathia epidemica, in bank voles (Clethrionomys glareolus). J Virol. (1985) 55:34–8. doi: 10.1128/jvi.55.1.34-38.1985
67. Schönrich G, Rang A, Lütteke N, Raftery MJ, Charbonnel N, Ulrich RG. Hantavirus-induced immunity in rodent reservoirs and humans. Immunol Rev. (2008) 225:163–89. doi: 10.1111/j.1600-065X.2008.00694.x
68. Strandin T, Smura T, Ahola P, Aaltonen K, Sironen T, Hepojoki J, et al. Orthohantavirus isolated in reservoir host cells displays minimal genetic changes and retains wild-type infection properties. Viruses. (2020) 12. doi: 10.3390/v12040457
69. Hardestam J, Karlsson M, Falk KI, Olsson G, Klingström J, Lundkvist A. Puumala hantavirus excretion kinetics in bank voles (Myodes glareolus). Emerging Infect Dis. (2008) 14:1209–15. doi: 10.3201/eid1408.080221
70. Parvate A, Williams EP, Taylor MK, Chu Y-K, Lanman J, Saphire EO, et al. Diverse morphology and structural features of old and New World hantaviruses. Viruses. (2019) 11. doi: 10.3390/v11090862
71. McCormick JB, Sasso DR, Palmer EL, Kiley MP. Morphological identification of the agent of Korean haemorrhagic fever (Hantaan virus)as a member of the bunyaviridae. Lancet (London England). (1982) 1:765–8. doi: 10.1016/s0140-6736(82)91812-8
72. Huiskonen JT, Hepojoki J, Laurinmäki P, Vaheri A, Lankinen H, Butcher SJ, et al. Electron cryotomography of Tula hantavirus suggests a unique assembly paradigm for enveloped viruses. J Virol. (2010) 84:4889–97. doi: 10.1128/jvi.00057-10
73. Plyusnin A. Genetics of hantaviruses: implications to taxonomy. Arch Virol. (2002) 147:665–82. doi: 10.1007/s007050200017
74. Fulhorst CF, Koster FT, Enría DA, Peters CJ. Hantavirus infections. In: Tropical Infectious Diseases: Principles, Pathogens and Practice. Elsevier (2011). p. 470–80.
75. Vapalahti O, Kallio-Kokko H, Salonen EM, Brummer-Korvenkontio M, Vaheri A. Cloning and sequencing of Puumala virus Sotkamo strain S and M RNA segments: evidence for strain variation in hantaviruses and expression of the nucleocapsid protein. J Gen Virol. (1992) 73:829–38. doi: 10.1099/0022-1317-73-4-829
76. Binder F, Gallo G, Bendl E, Eckerle I, Ermonval M, Luttermann C, et al. Inhibition of interferon I induction by non-structural protein NSs of Puumala virus and other vole-associated orthohantaviruses: phenotypic plasticity of the protein and potential functional domains. Arch Virol. (2021) 166:2999–3012. doi: 10.1007/s00705-021-05159-y
77. Hedil M, Kormelink R. Viral RNA silencing suppression: the enigma of bunyavirus NSs proteins. Viruses. (2016) 8. doi: 10.3390/v8070208
78. Ryabova LA, Pooggin MM, Hohn T. Translation reinitiation and leaky scanning in plant viruses. Virus Res. (2006) 119:52–62. doi: 10.1016/j.virusres.2005.10.017
79. Bridgen A, Weber F, Fazakerley JK, Elliott RM. Bunyamwera bunyavirus nonstructural protein NSs is a nonessential gene product that contributes to viral pathogenesis. Proc Natl Acad Sci United States America. (2001) 98:664–9. doi: 10.1073/pnas.98.2.664
80. Schmaljohn CS, Hasty SE, Dalrymple JM, LeDuc JW, Lee HW, Bonsdorff C, et al. Antigenic and genetic properties of viruses linked to hemorrhagic fever with renal syndrome. Sci (New York NY). (1985) 227:1041–4. doi: 10.1126/science.2858126
81. Plyusnin A, Vapalahti O, Vaheri A. Hantaviruses: genome structure, expression and evolution. J Gen Virol. (1996) 77:2677–87. doi: 10.1099/0022-1317-77-11-2677
82. Mir MA, Panganiban AT. The hantavirus nucleocapsid protein recognizes specific features of the viral RNA panhandle and is altered in conformation upon RNA binding. J Virol. (2005) 79:1824–35. doi: 10.1128/jvi.79.3.1824-1835.2005
83. Rodriguez LL, Owens JH, Peters CJ, Nichol ST. Genetic reassortment among viruses causing hantavirus pulmonary syndrome. Virology. (1998) 242:99–106. doi: 10.1006/viro.1997.8990
84. Sibold C, Meisel H, Krüger DH, Labuda M, Lysy J, Kozuch O, et al. Recombination in Tula hantavirus evolution: analysis of genetic lineages from Slovakia. J Virol. (1999) 73:667–75. doi: 10.1128/JVI.73.1.667-675.1999
85. Mittler E, Wec AZ, Tynell J, Guardado-Calvo P, Wigren-Byström J, Polanco LC, et al. Human antibody recognizing a quaternary epitope in the Puumala virus glycoprotein provides broad protection against orthohantaviruses. Sci Trans Med. (2022) 14:eabl5399. doi: 10.1126/scitranslmed.abl5399
86. Engdahl TB, Binshtein E, Brocato RL, Kuzmina NA, Principe LM, Kwilas SA, et al. Antigenic mapping and functional characterization of human New World hantavirus neutralizing antibodies. eLife. (2023) 12. doi: 10.7554/eLife.81743
87. Rissanen I, Krumm SA, Stass R, Whitaker A, Voss JE, Bruce EA, et al. Structural basis for a neutralizing antibody response elicited by a recombinant Hantaan virus Gn immunogen. mBio. (2021) 12. doi: 10.1128/mBio.02531-20
88. Engdahl TB, Kuzmina NA, Ronk AJ, Mire CE, Hyde MA, Kose N, et al. Broad and potently neutralizing monoclonal antibodies isolated from human survivors of New World hantavirus infection. Cell Rep. (2021) 35:109086. doi: 10.1016/j.celrep.2021.109086
89. Antic D, Wright KE, Kang CY. Maturation of Hantaan virus glycoproteins G1 and G2. Virology. (1992) 189:324–8. doi: 10.1016/0042-6822(92)90709-x
90. Löber C, Anheier B, Lindow S, Klenk HD, Feldmann H. The Hantaan virus glycoprotein precursor is cleaved at the conserved pentapeptide WAASA. Virology. (2001) 289:224–9. doi: 10.1006/viro.2001.1171
91. Mittler E, Serris A, Esterman ES, Florez C, Polanco LC, O’Brien CM, et al. Structural and mechanistic basis of neutralization by a pan-hantavirus protective antibody. Sci Trans Med. (2023) 15:eadg1855. doi: 10.1126/scitranslmed.adg1855
92. Serris A, Stass R, Bignon EA, Muena NA, Manuguerra J-C, Jangra RK, et al. The hantavirus surface glycoprotein lattice and its fusion control mechanism. Cell. (2020) 183:442–56.e16. doi: 10.1016/j.cell.2020.08.023
93. Guardado-Calvo P, Bignon EA, Stettner E, Jeffers SA, Pérez-Vargas J, Pehau-Arnaudet G, et al. Mechanistic insight into bunyavirus-induced membrane fusion from structure-function analyses of the hantavirus envelope glycoprotein Gc. PloS Pathog. (2016) 12. doi: 10.1371/journal.ppat.1005813
94. Rissanen I, Stass R, Zeltina A, Li S, Hepojoki J, Harlos K, et al. Structural transitions of the conserved and metastable hantaviral glycoprotein envelope. J Virol. (2017) 91. doi: 10.1128/jvi.00378-17
95. Willensky S, Bar-Rogovsky H, Bignon EA, Tischler ND, Modis Y, Dessau M. Crystal structure of glycoprotein C from a hantavirus in the post-fusion conformation. PloS Pathog. (2016) 12:e1005948. doi: 10.1371/journal.ppat.1005948
96. Li S, Rissanen I, Zeltina A, Hepojoki J, Raghwani J, Harlos K, et al. A molecular-level account of the antigenic hantaviral surface. Cell Rep. (2016) 15:959–67. doi: 10.1016/j.celrep.2016.03.082
97. Martin ML, Lindsey-Regnery H, Sasso DR, McCormick JB, Palmer E. Distinction between bunyaviridae genera by surface structure and comparison with Hantaan virus using negative stain electron microscopy. Arch Virol. (1985) 86:17–28. doi: 10.1007/bf01314110
98. Battisti AJ, Chu Y-K, Chipman PR, Kaufmann B, Jonsson CB, Rossmann MG. Structural studies of Hantaan virus. J Virol. (2010) 85:835–41. doi: 10.1128/jvi.01847-10
99. Rissanen I, Stass R, Krumm SA, Seow J, Hulswit RJ, Paesen GC, et al. Molecular rationale for antibody-mediated targeting of the hantavirus fusion glycoprotein. eLife. (2020) 9. doi: 10.7554/eLife.58242
100. Hepojoki J, Strandin T, Vaheri A, Lankinen H. Interactions and oligomerization of hantavirus glycoproteins. J Virol. (2010) 84:227–42. doi: 10.1128/jvi.00481-09
101. Bignon EA, Albornoz A, Guardado-Calvo P, Rey FA, Tischler ND. Molecular organization and dynamics of the fusion protein Gc at the hantavirus surface. eLife. (2019) 8. doi: 10.7554/eLife.46028
102. Shi X, Elliott RM. Analysis of N-linked glycosylation of Hantaan virus glycoproteins and the role of oligosaccharide side chains in protein folding and intracellular trafficking. J Virol. (2004) 78:5414–22. doi: 10.1128/jvi.78.10.5414-5422.2004
103. Mittler E, Dieterle ME, Kleinfelter LM, Slough MM, Chandran K, Jangra RK. Hantavirus entry: perspectives and recent advances. Adv Virus Res. (2019) 104:185–224. doi: 10.1016/bs.aivir.2019.07.002
104. Jangra RK, Herbert AS, Li R, Jae LT, Kleinfelter LM, Slough MM, et al. Protocadherin-1 is essential for cell entry by New World hantaviruses. Nature. (2018) 563:559–63. doi: 10.1038/s41586-018-0702-1
105. Yanagihara R, Silverman DJ. Experimental infection of human vascular endothelial cells by pathogenic and nonpathogenic hantaviruses. Arch Virol. (1990) 111:281–6. doi: 10.1007/bf01311063
106. Hägele S, Nusshag C, Müller A, Baumann A, Zeier M, Krautkrämer E. Cells of the human respiratory tract support the replication of pathogenic Old World orthohantavirus puumala. Virol J. (2021) 18:169. doi: 10.1186/s12985-021-01636-7
107. Au RY, Jedlicka AE, Li W, Pekosz A, Klein SL. Seoul virus suppresses NF-κb-mediated inflammatory responses of antigen presenting cells from Norway rats. Virology. (2010) 400:115–27. doi: 10.1016/j.virol.2010.01.027
108. Schönrich G, Krüger DH, Raftery MJ. Hantavirus-induced disruption of the endothelial barrier: neutrophils are on the payroll. Front Microbiol. (2015) 6:222. doi: 10.3389/fmicb.2015.00222
109. Klingstrom J, Smed-Sorensen A, Maleki KT, Sola-Riera C, Ahlm C, Bjorkstrom NK, et al. Innate and adaptive immune responses against human Puumala virus infection: immunopathogenesis and suggestions for novel treatment strategies for severe hantavirus-associated syndromes. J Intern Med. (2019) 285:510–23. doi: 10.1111/joim.12876
110. Terajima M, Vapalahti O, Van Epps HL, Vaheri A, Ennis FA. Immune responses to Puumala virus infection and the pathogenesis of nephropathia epidemica. Microbes Infect. (2004) 6:238–45. doi: 10.1016/j.micinf.2003.10.017
111. Bauherr S, Larsberg F, Petrich A, Sperber HS, Klose-Grzelka V, Luckner M, et al. Macropinocytosis and clathrin-dependent endocytosis play pivotal roles for the infectious entry of Puumala virus. J Virol. (2020) 94. doi: 10.1128/jvi.00184-20
112. Jin M, Park J, Lee S, Park B, Shin J, Song K-J, et al. Hantaan virus enters cells by clathrin-dependent receptor-mediated endocytosis. Virology. (2002) 294:60–9. doi: 10.1006/viro.2001.1303
113. Ramanathan HN, Chung D-H, Plane SJ, Sztul E, Chu Y-K, Guttieri MC, et al. Dynein-dependent transport of the Hantaan virus nucleocapsid protein to the endoplasmic reticulum-golgi intermediate compartment. J Virol. (2007) 81:8634–47. doi: 10.1128/jvi.00418-07
114. Torriani G, Mayor J, Zimmer G, Kunz S, Rothenberger S, Engler O. Macropinocytosis contributes to hantavirus entry into human airway epithelial cells. Virology. (2019) 531:57–68. doi: 10.1016/j.virol.2019.02.013
115. Cifuentes-Muñoz N, Salazar-Quiroz N, Tischler ND. Hantavirus Gn and Gc envelope glycoproteins: key structural units for virus cell entry and virus assembly. Viruses. (2014) 6:1801–22. doi: 10.3390/v6041801
116. Compans RW, Cooper M, Ito Y, Koprowski H, Melchers F, Oldstone M, et al eds. Hantaviruses. Berlin, Heidelberg: Springer Berlin Heidelberg (2001).
117. Spiropoulou CF. Hantavirus maturation. In: Compans RW, Cooper M, Ito Y, Koprowski H, Melchers F, Oldstone M, et al, editors. Hantaviruses. Current topics in microbiology and immunology, vol. 256 . Springer Berlin Heidelberg, Berlin, Heidelberg (2001). p. 33–46.
118. Vaheri A, Strandin T, Hepojoki J, Sironen T, Henttonen H, Mäkelä S, et al. Uncovering the mysteries of hantavirus infections. Nat Rev Microbiol. (2013) 11:539–50. doi: 10.1038/nrmicro3066
119. Olschewski S, Cusack S, Rosenthal M. The cap-snatching mechanism of bunyaviruses. Trends Microbiol. (2020) 28:293–303. doi: 10.1016/j.tim.2019.12.006
120. Hepojoki J, Strandin T, Lankinen H, Vaheri A. Hantavirus structure–molecular interactions behind the scene. J Gen Virol. (2012) 93:1631–44. doi: 10.1099/vir.0.042218-0
121. Koehler FC, Di Cristanziano V, Späth MR, Hoyer-Allo KJR, Wanken M, Müller R-U, et al. The kidney in hantavirus infection—Epidemiology, virology, pathophysiology, clinical presentation, diagnosis and management. Clin Kidney J. (2022) 15:1231–52. doi: 10.1093/ckj/sfac008
122. Ravkov EV, Compans RW. Hantavirus nucleocapsid protein is expressed as a membrane-associated protein in the perinuclear region. J Virol. (2001) 75:1808–15. doi: 10.1128/jvi.75.4.1808-1815.2001
123. Meier K, Thorkelsson SR, Quemin ERJ, Rosenthal M. Hantavirus replication cycle—an updated structural virology perspective. Viruses. (2021) 13. doi: 10.3390/v13081561
124. McNab F, Mayer-Barber K, Sher A, Wack A, O’Garra A. Type I interferons in infectious disease. Nat Rev Immunol. (2015) 15:87–103. doi: 10.1038/nri3787
125. Ji L, Li T, Chen H, Yang Y, Lu E, Liu J, et al. The crucial regulatory role of type I interferon in inflammatory diseases. Cell Biosci. (2023) 13:230. doi: 10.1186/s13578-023-01188-z
126. Thompson AJV, Locarnini SA. Toll-like receptors, RIG-I-like RNA helicases and the antiviral innate immune response. Immunol Cell Biol. (2007) 85:435–45. doi: 10.1038/sj.icb.7100100
127. Zhao Y, Che L, Pan M, Huang Y, Fang S, Wang M, et al. Hantaan virus inhibits type I interferon response by targeting RLR signaling pathways through TRIM25. Virology. (2024) 589:109942. doi: 10.1016/j.virol.2023.109942
128. Gallo G, Caignard G, Badonnel K, Chevreux G, Terrier S, Szemiel A, et al. Interactions of viral proteins from pathogenic and low or non-pathogenic orthohantaviruses with human type I interferon signaling. Viruses. (2021) 13. doi: 10.3390/v13010140
129. Jääskeläinen KM, Kaukinen P, Minskaya ES, Plyusnina A, Vapalahti O, Elliott RM, et al. Tula and Puumala hantavirus NSs ORFs are functional and the products inhibit activation of the interferon-beta promoter. J Med Virol. (2007) 79:1527–36. doi: 10.1002/jmv.20948
130. Bourquain D, Bodenstein C, Schürer S, Schaade L. Puumala and Tula virus differ in replication kinetics and innate immune stimulation in human endothelial cells and macrophages. Viruses. (2019) 11. doi: 10.3390/v11090855
131. He Y, Shen M, Wang X, Yin A, Liu B, Zhu J, et al. Suppression of interferon response and antiviral strategies of bunyaviruses. Trop Med Infect Dis. (2024) 9. doi: 10.3390/tropicalmed9090205
132. Matthys V, Gorbunova EE, Gavrilovskaya IN, Pepini T, Mackow ER. The C-terminal 42 residues of the Tula virus Gn protein regulate interferon induction. J Virol. (2011) 85:4752–60. doi: 10.1128/jvi.01945-10
133. Wang Z, Mir MA. Andes virus nucleocapsid protein interrupts protein kinase R dimerization to counteract host interference in viral protein synthesis. J Virol. (2015) 89:1628–39. doi: 10.1128/jvi.02347-14
134. Spiropoulou CF, Albariño CG, Ksiazek TG, Rollin PE. Andes and Prospect Hill hantaviruses differ in early induction of interferon although both can downregulate interferon signaling. J Virol. (2007) 81:2769–76. doi: 10.1128/jvi.02402-06
135. Simons MJ, Gorbunova EE, Mackow ER. Unique interferon pathway regulation by the Andes virus nucleocapsid protein is conferred by phosphorylation of serine 386. J Virol. (2019) 93. doi: 10.1128/jvi.00338-19
136. Levine JR, Prescott J, Brown KS, Best SM, Ebihara H, Feldmann H. Antagonism of type I interferon responses by New World hantaviruses. J Virol. (2010) 84:11790–801. doi: 10.1128/jvi.00916-10
137. Cimica V, Dalrymple NA, Roth E, Nasonov A, Mackow ER. An innate immunity-regulating virulence determinant is uniquely encoded by the Andes virus nucleocapsid protein. mBio. (2014) 5. doi: 10.1128/mbio.01088-13
138. Avšič-Županc T, Saksida A, Korva M. Hantavirus infections. Clin Microbiol Infect. (2019) 21S:e6–e16. doi: 10.1111/1469-0691.12291
139. Vial PA, Ferrés M, Vial C, Klingström J, Ahlm C, López R, et al. Hantavirus in humans: A review of clinical aspects and management. Lancet Infect Dis. (2023) 23:e371–e82. doi: 10.1016/s1473-3099(23)00128-7
140. Hoier S, Aberle SW, Langner C, Schnedl W, Högenauer C, Reisinger EC, et al. Puumala virus RNA in patient with multiorgan failure. Emerging Infect Dis. (2006) 12:356–7. doi: 10.3201/eid1202.050634
141. Makary P, Kanerva M, Ollgren J, Virtanen MJ, Vapalahti O, Lyytikäinen O. Disease burden of Puumala virus infections, 1995-2008. Epidemiol Infect. (2010) 138:1484–92. doi: 10.1017/s0950268810000087
142. Hatzl S, Scholz L, Posch F, Eller P, Reisinger AC, Zacharias M, et al. Invasive pulmonary aspergillosis in critically ill patients with hantavirus infection, Austria. Emerging Infect Dis. (2024) 30:1275–8. doi: 10.3201/eid3006.231720
143. Charbonnel N, Pagès M, Sironen T, Henttonen H, Vapalahti O, Mustonen J, et al. Immunogenetic factors affecting susceptibility of humans and rodents to hantaviruses and the clinical course of hantaviral disease in humans. Viruses. (2014) 6:2214–41. doi: 10.3390/v6052214
144. Vaheri A, Smura T, Vauhkonen H, Hepojoki J, Sironen T, Strandin T, et al. Puumala hantavirus infections show extensive variation in clinical outcome. Viruses. (2023) 15. doi: 10.3390/v15030805
145. Mustonen J, Mäkelä S, Outinen T, Laine O, Jylhävä J, Arstila PT, et al. The pathogenesis of nephropathia epidemica: new knowledge and unanswered questions. Antiviral Res. (2013) 100:589–604. doi: 10.1016/j.antiviral.2013.10.001
146. Mustonen J, Brummer-Korvenkontio M, Hedman K, Pasternack A, Pietilä K, Vaheri A. Nephropathia epidemica in Finland: A retrospective study of 126 cases. Scand J Infect Dis. (1994) 26:7–13. doi: 10.3109/00365549409008583
147. Vaheri A, Henttonen H, Mustonen J. Hantavirus research in Finland: highlights and perspectives. Viruses. (2021) 13. doi: 10.3390/v13081452
148. Mäkelä S, Mustonen J, Ala-Houhala I, Hurme M, Koivisto A-M, Vaheri A, et al. Urinary excretion of interleukin-6 correlates with proteinuria in acute Puumala hantavirus-induced nephritis. Am J Kidney Dis. (2004) 43:809–16. doi: 10.1053/j.ajkd.2003.12.044
149. Turčinov D, Puljiz I, Markotić A, Kuzman I, Begovac J. Clinical and laboratory findings in patients with oliguric and non-oliguric hantavirus haemorrhagic fever with renal syndrome: an analysis of 128 patients. Clin Microbiol Infect. (2013) 19:674–9. doi: 10.1111/j.1469-0691.2012.03994.x
150. Vaheri A, Strandin T, Jääskeläinen AJ, Vapalahti O, Jarva H, Lokki M-L, et al. Pathophysiology of a severe case of Puumala hantavirus infection successfully treated with bradykinin receptor antagonist icatibant. Antiviral Res. (2014) 111:23–5. doi: 10.1016/j.antiviral.2014.08.007
151. Smet M, Bogaert S, Schauwvlieghe A, Dendooven A, Depuydt P, Druwé P. Case report: hemorrhagic fever with renal syndrome presenting as hemophagocytic lymphohistiocytosis. Front Med. (2022) 9:1096900. doi: 10.3389/fmed.2022.1096900
152. Scholz L, Posch F, Schulz E, Gornicec M, Wölfler A, Reisinger AC, et al. Ruxolitinib, IV immunoglobulin, and high-dose glucocorticoids for critically ill adults with secondary hemophagocytic lymphohistiocytosis: A single-center observational pilot study. Crit Care Explor. (2024) 6. doi: 10.1097/cce.0000000000001046
153. Hoornweg TE, Zutt I, de Vries A, Maas M, Hoogerwerf MN, Avsic-Zupanc T, et al. Development of a comparative European orthohantavirus microneutralization assay with multi- species validation and evaluation in a human diagnostic cohort. Front Cell Infect Microbiol. (2020) 10:580478. doi: 10.3389/fcimb.2020.580478
154. Schubert J, Tollmann F, Weissbrich B. Evaluation of a pan-reactive hantavirus enzyme immunoassay and of a hantavirus immunoblot for the diagnosis of nephropathia epidemica. J Clin Virol. (2001) 21:63–74. doi: 10.1016/s1386-6532(00)00187-6
155. Hujakka H, Koistinen V, Eerikainen P, Kuronen I, Mononen I, Parviainen M, et al. New immunochromatographic rapid test for diagnosis of acute Puumala virus infection. J Clin Microbiol. (2001) 39:2146–50. doi: 10.1128/JCM.39.6.2146-2150.2001
156. Acham-Roschitz B, Aberle SW, Pirker N, Kaulfersch W, Boehm M, Roedl S, et al. Nephropathia epidemica (Puumala virus infection) in Austrian children. Pediatr Infect Dis J. (2010) 29:874–6. doi: 10.1097/INF.0b013e3181dfbbe5
157. Lagerqvist N, Hagstrom A, Lundahl M, Nilsson E, Juremalm M, Larsson I, et al. Molecular diagnosis of hemorrhagic fever with renal syndrome caused by Puumala virus. J Clin Microbiol. (2016) 54:1335–9. doi: 10.1128/JCM.00113-16
158. Milhano N, Korslund L, Evander M, Ahlm C, Vainio K, Dudman SG, et al. Circulation and diagnostics of Puumala virus in Norway: nephropatia epidemica incidence and rodent population dynamics. APMIS. (2017) 125:732–42. doi: 10.1111/apm.12712
159. Reynes JM, Schaeffer L, Papadopoulos P, Ait-Ahmed M, Siby-Diakite D, Ripaux-Lefevre M, et al. Molecular detection of orthohantavirus puumalaense in plasma and urine samples from hospitalized patients presenting with a serologically confirmed acute hantavirus infection in France. J Clin Microbiol. (2023) 61:e0037223. doi: 10.1128/jcm.00372-23
160. Geeraedts F, Wevers M, Bosma F, Boer M, Brinkman JN, Delsing C, et al. Use of a diagnostic Puumala virus real-time RT-PCR in an orthohantavirus endemic region in the Netherlands. Microbiol Spectr. (2024) 12:e0381323. doi: 10.1128/spectrum.03813-23
161. Niskanen S, Jaaskelainen A, Vapalahti O, Sironen T. Evaluation of real-time RT-PCR for diagnostic use in detection of Puumala virus. Viruses. (2019) 11. doi: 10.3390/v11070661
162. Hatzl S, Posch F, Linhofer M, Aberle S, Zollner-Schwetz I, Krammer F, et al. Poor prognosis for Puumala virus infections predicted by lymphopenia and dyspnea. Emerging Infect Dis. (2023) 29:1038–41. doi: 10.3201/eid2905.221625
163. Bougie I, Bisaillon M. The broad spectrum antiviral nucleoside ribavirin as a substrate for a viral RNA capping enzyme. J Biol Chem. (2004) 279:22124–30. doi: 10.1074/jbc.M400908200
164. Goswami BB, Borek E, Sharma OK, Fujitaki J, Smith RA. The broad spectrum antiviral agent ribavirin inhibits capping of mRNA. Biochem Biophys Res Commun. (1979) 89:830–6. doi: 10.1016/0006-291x(79)91853-9
165. Wray SK, Gilbert BE, Noall MW, Knight V. Mode of action of ribavirin: effect of nucleotide pool alterations on influenza virus ribonucleoprotein synthesis. Antiviral Res. (1985) 5:29–37. doi: 10.1016/0166-3542(85)90012-9
166. Fernandez-Larsson R, Patterson JL. Ribavirin is an inhibitor of human immunodeficiency virus reverse transcriptase. Mol Pharmacol. (1990) 38:766–70. doi: 10.1016/S0026-895X(25)09567-7
167. Leyssen P, Balzarini J, Clercq Ed, Neyts J. The predominant mechanism by which ribavirin exerts its antiviral activity in vitro against flaviviruses and paramyxoviruses is mediated by inhibition of IMP dehydrogenase. J Virol. (2005) 79:1943–7. doi: 10.1128/jvi.79.3.1943-1947.2005
168. Debing Y, Emerson SU, Wang Y, Pan Q, Balzarini J, Dallmeier K, et al. Ribavirin inhibits in vitro hepatitis E virus replication through depletion of cellular GTP pools and is moderately synergistic with alpha interferon. Antimicrobial Agents Chemother. (2014) 58:267–73. doi: 10.1128/aac.01795-13
169. Ölschläger S, Neyts J, Günther S. Depletion of GTP pool is not the predominant mechanism by which ribavirin exerts its antiviral effect on Lassa virus. Antiviral Res. (2011) 91:89–93. doi: 10.1016/j.antiviral.2011.05.006
170. Carrillo-Bustamante P, Nguyen THT, Oestereich L, Günther S, Guedj J, Graw F. Determining ribavirin’s mechanism of action against Lassa virus infection. Sci Rep. (2017) 7:11693. doi: 10.1038/s41598-017-10198-0
171. Tate PM, Mastrodomenico V, Mounce BC. Ribavirin induces polyamine depletion via nucleotide depletion to limit virus replication. Cell Rep. (2019) 28:2620–33.e4. doi: 10.1016/j.celrep.2019.07.099
172. Mounce BC, Poirier EZ, Passoni G, Simon-Loriere E, Cesaro T, Prot M, et al. Interferon-induced spermidine-spermine acetyltransferase and polyamine depletion restrict Zika and Chikungunya viruses. Cell Host Microbe. (2016) 20:167–77. doi: 10.1016/j.chom.2016.06.011
173. Crotty S, Cameron CE, Andino R. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc Natl Acad Sci United States America. (2001) 98:6895–900. doi: 10.1073/pnas.111085598
174. Hofmann WP, Polta A, Herrmann E, Mihm U, Kronenberger B, Sonntag T, et al. Mutagenic effect of ribavirin on hepatitis C nonstructural 5b quasispecies in vitro and during antiviral therapy. Gastroenterology. (2007) 132:921–30. doi: 10.1053/j.gastro.2006.12.005
175. Loeb LA, Essigmann JM, Kazazi F, Zhang J, Rose KD, Mullins JI. Lethal mutagenesis of HIV with mutagenic nucleoside analogs. Proc Natl Acad Sci USA. (1999) 96:1492–7. doi: 10.1073/pnas.96.4.1492
176. Severson WE, Schmaljohn CS, Javadian A, Jonsson CB. Ribavirin causes error catastrophe during Hantaan virus replication. J Virol. (2003) 77:481–8. doi: 10.1128/jvi.77.1.481-488.2003
177. Sun Y, Chung D-H, Chu Y-K, Jonsson CB, Parker WB. Activity of ribavirin against Hantaan virus correlates with production of ribavirin-5′-triphosphate, not with inhibition of IMP dehydrogenase. Antimicrobial Agents Chemother. (2006) 51:84–8. doi: 10.1128/aac.00790-06
178. Huggins JW, Kim GR, Brand OM, McKee KT. Ribavirin therapy for Hantaan virus infection in suckling mice. J Infect Dis. (1986) 153:489–97. doi: 10.1093/infdis/153.3.489
179. Huggins JW, Hsiang CM, Cosgriff TM, Guang MY, Smith JI, Wu ZO, et al. Prospective, double-blind, concurrent, placebo-controlled clinical trial of intravenous ribavirin therapy of hemorrhagic fever with renal syndrome. J Infect Dis. (1991) 164:1119–27. doi: 10.1093/infdis/164.6.1119
180. Rusnak JM, Byrne WR, Chung KN, Gibbs PH, Kim TT, Boudreau EF, et al. Experience with intravenous ribavirin in the treatment of hemorrhagic fever with renal syndrome in Korea. Antiviral Res. (2008) 81:68–76. doi: 10.1016/j.antiviral.2008.09.007
181. Safronetz D, Haddock E, Feldmann F, Ebihara H, Feldmann H. In vitro and in vivo activity of ribavirin against Andes virus infection. PloS One. (2011) 6. doi: 10.1371/journal.pone.0023560
182. Ogg M, Jonsson CB, Camp JV, Hooper JW. Ribavirin Protects Syrian Hamsters against Lethal Hantavirus Pulmonary Syndrome — after Intranasal Exposure to Andes virus. Viruses. (2013) 5:2704–20. doi: 10.3390/v5112704
183. Medina RA, Mirowsky-Garcia K, Hutt J, Hjelle B. Ribavirin, human convalescent plasma and anti-beta3 integrin antibody inhibit infection by Sin Nombre virus in the deer mouse model. J Gen Virol. (2007) 88:493–505. doi: 10.1099/vir.0.82459-0
184. Chapman LE, Mertz GJ, Peters CJ, Jolson HM, Khan AS, Ksiazek TG, et al. Intravenous ribavirin for hantavirus pulmonary syndrome: safety and tolerance during 1 year of open-label experience. Antiviral Ther. (1999) 4:211–9. doi: 10.1177/135965359900400404
185. Mertz GJ, Miedzinski L, Goade D, Pavia AT, Hjelle B, Hansbarger CO, et al. Placebo-controlled, double-blind trial of intravenous ribavirin for the treatment of hantavirus cardiopulmonary syndrome in North America. Clin Infect Dis. (2004) 39:1307–13. doi: 10.1086/425007
186. Malinin OV, Platonov AE. Insufficient efficacy and safety of intravenous ribavirin in treatment of haemorrhagic fever with renal syndrome caused by Puumala virus. Infect Dis (London England). (2017) 49:514–20. doi: 10.1080/23744235.2017.1293841
187. Murphy ME, Kariwa H, Mizutani T, Yoshimatsu K, Arikawa J, Takashima I. In vitro antiviral activity of lactoferrin and ribavirin upon hantavirus. Arch Virol. (2000) 145:1571–82. doi: 10.1007/s007050070077
188. Murphy ME, Kariwa H, Mizutani T, Tanabe H, Yoshimatsu K, Arikawa J, et al. Characterization of in vitro and in vivo antiviral activity of lactoferrin and ribavirin upon hantavirus. J Vet Med Sci. (2001) 63:637–45. doi: 10.1292/jvms.63.637
189. Jin Z, Smith LK, Rajwanshi VK, Kim B, Deval J. The ambiguous base-pairing and high substrate efficiency of T-705 (Favipiravir) ribofuranosyl 5′-triphosphate towards influenza a virus polymerase. PloS One. (2013) 8. doi: 10.1371/journal.pone.0068347
190. Baranovich T, Wong S-S, Armstrong J, Marjuki H, Webby RJ, Webster RG, et al. T-705 (Favipiravir) induces lethal mutagenesis in influenza a H1N1 viruses in vitro. J Virol. (2013) 87:3741–51. doi: 10.1128/jvi.02346-12
191. Safronetz D, Falzarano D, Scott DP, Furuta Y, Feldmann H, Gowen BB. Antiviral efficacy of favipiravir against two prominent etiological agents of hantavirus pulmonary syndrome. Antimicrobial Agents Chemother. (2013) 57:4673–80. doi: 10.1128/aac.00886-13
192. Mayor J, Engler O, Rothenberger S. Antiviral efficacy of ribavirin and favipiravir against Hantaan virus. Microorganisms. (2021) 9. doi: 10.3390/microorganisms9061306
193. Buys KK, Jung K-H, Smee DF, Furuta Y, Gowen BB. Maporal virus as a surrogate for pathogenic New World hantaviruses and its inhibition by favipiravir. Antiviral Chem Chemother. (2011) 21:193–200. doi: 10.3851/imp1729
194. Westover JB, Sefing EJ, Bailey KW, van Wettere AJ, Jung K-H, Dagley A, et al. Low-dose ribavirin potentiates the antiviral activity of favipiravir against hemorrhagic fever viruses. Antiviral Res. (2015) 126:62–8. doi: 10.1016/j.antiviral.2015.12.006
195. Oestereich L, Rieger T, Lüdtke A, Ruibal P, Wurr S, Pallasch E, et al. Efficacy of favipiravir alone and in combination with ribavirin in a lethal, immunocompetent mouse model of Lassa fever. J Infect Dis. (2015) 213:934–8. doi: 10.1093/infdis/jiv522
196. Oestereich L, Rieger T, Neumann M, Bernreuther C, Lehmann M, Krasemann S, et al. Evaluation of antiviral efficacy of ribavirin, arbidol, and T-705 (Favipiravir) in a mouse model for Crimean-Congo hemorrhagic fever. PloS Negl Trop Dis. (2014) 8. doi: 10.1371/journal.pntd.0002804
197. Tipih T, Meade-White K, Rao D, Bushmaker T, Lewis M, Shaia C, et al. Favipiravir and ribavirin protect immunocompetent mice from lethal CCHFV infection. Antiviral Res. (2023) 218:105703. doi: 10.1016/j.antiviral.2023.105703
198. Chung D-H, Kumarapperuma SC, Sun Y, Li Q, Chu Y-K, Arterburn JB, et al. Synthesis of 1-B-D-ribofuranosyl-3-ethynyl-1,2,4triazole and its in vitro and in vivo efficacy against hantavirus. Antiviral Res. (2008) 79:19–27. doi: 10.1016/j.antiviral.2008.02.003
199. Toba S, Sato A, Kawai M, Taoda Y, Unoh Y, Kusakabe S, et al. Identification of cap-dependent endonuclease inhibitors with broad-spectrum activity against bunyaviruses. Proc Natl Acad Sci United States America. (2022) 119. doi: 10.1073/pnas.2206104119
200. Ye C, Wang D, Liu H, Ma H, Dong Y, Yao M, et al. An improved enzyme-linked focus formation assay revealed baloxavir acid as a potential antiviral therapeutic against hantavirus infection. Front Pharmacol. (2019) 10:1203. doi: 10.3389/fphar.2019.01203
201. Lesch M, Luckner M, Meyer M, Weege F, Gravenstein I, Raftery M, et al. RNAi-based small molecule repositioning reveals clinically approved urea-based kinase inhibitors as broadly active antivirals. PloS Pathog. (2019) 15:e1007601. doi: 10.1371/journal.ppat.1007601
202. Li Z, Wang F, Ying Q, Kong D, Zhang X, Dong Y, et al. In vitro anti-hantavirus activity of protein kinase inhibitor 8G1 targeting AKT/mTOR/eIF4E signaling pathway. Front Microbiol. (2022) 13:880258. doi: 10.3389/fmicb.2022.880258
203. Li Z, Wang F, Liu Y, Zhai D, Zhang X, Ying Q, et al. Coumarin derivative N6 as a novel anti-hantavirus infection agent targeting AKT. Front Pharmacol. (2021) 12:745646. doi: 10.3389/fphar.2021.745646
204. Gavrilovskaya IN, Gorbunova EE, Mackow NA, Mackow ER. Hantaviruses direct endothelial cell permeability by sensitizing cells to the vascular permeability factor VEGF, while angiopoietin 1 and sphingosine 1-phosphate inhibit hantavirus-directed permeability. J Virol. (2008) 82:5797–806. doi: 10.1128/jvi.02397-07
205. Shrivastava-Ranjan P, Rollin PE, Spiropoulou CF. Andes virus disrupts the endothelial cell barrier by induction of vascular endothelial growth factor and downregulation of VE-cadherin. J Virol. (2010) 84:11227–34. doi: 10.1128/jvi.01405-10
206. Gorbunova E, Gavrilovskaya IN, Mackow ER. Pathogenic hantaviruses Andes virus and Hantaan virus induce adherens junction disassembly by directing vascular endothelial cadherin internalization in human endothelial cells. J Virol. (2010) 84:7405–11. doi: 10.1128/jvi.00576-10
207. Gavard J, Gutkind JS. VEGF controls endothelial-cell permeability by promoting the beta-arrestin-dependent endocytosis of VE-cadherin. Nat Cell Biol. (2006) 8:1223–34. doi: 10.1038/ncb1486
208. Gorbunova EE, Gavrilovskaya IN, Pepini T, Mackow ER. VEGFR2 and src kinase inhibitors suppress Andes virus-induced endothelial cell permeability. J Virol. (2010) 85:2296–303. doi: 10.1128/jvi.02319-10
209. Bird BH, Shrivastava-Ranjan P, Dodd KA, Erickson BR, Spiropoulou CF. Effect of vandetanib on Andes virus survival in the hamster model of hantavirus pulmonary syndrome. Antiviral Res. (2016) 132:66–9. doi: 10.1016/j.antiviral.2016.05.014
210. Brocato R, Josleyn M, Ballantyne J, Vial P, Hooper JW. DNA vaccine-generated duck polyclonal antibodies as a postexposure prophylactic to prevent hantavirus pulmonary syndrome (Hps). PloS One. (2012) 7. doi: 10.1371/journal.pone.0035996
211. Custer DM, Thompson E, Schmaljohn CS, Ksiazek TG, Hooper JW. Active and passive vaccination against hantavirus pulmonary syndrome with Andes virus M genome segment-based DNA vaccine. J Virol. (2003) 77:9894–905. doi: 10.1128/jvi.77.18.9894-9905.2003
212. Haese N, Brocato RL, Henderson T, Nilles ML, Kwilas SA, Josleyn MD, et al. Antiviral biologic produced in DNA vaccine/goose platform protects hamsters against hantavirus pulmonary syndrome when administered post-exposure. PloS Negl Trop Dis. (2015) 9:e0003803. doi: 10.1371/journal.pntd.0003803
213. Sroga P, Sloan A, Warner BM, Tierney K, Lew J, Liu G, et al. Polyclonal alpaca antibodies protect against hantavirus pulmonary syndrome in a lethal Syrian hamster model. Sci Rep. (2021) 11:17440. doi: 10.1038/s41598-021-96884-6
214. Muyldermans S. Nanobodies: natural single-domain antibodies. Annu Rev Biochem. (2013) 82:775–97. doi: 10.1146/annurev-biochem-063011-092449
215. Perley CC, Brocato RL, Wu H, Bausch C, Karmali PP, Vega JB, et al. Anti-HFRS human IgG produced in transchromosomic bovines has potent hantavirus neutralizing activity and is protective in animal models. Front Microbiol. (2020) 11:832. doi: 10.3389/fmicb.2020.00832
216. Brocato RL, Wu H, Kwilas SA, Principe LM, Josleyn M, Shamblin J, et al. Preclinical evaluation of a fully human, quadrivalent-hantavirus polyclonal antibody derived from a non-human source. mBio. (2024) 15:e0160024. doi: 10.1128/mbio.01600-24
217. Hooper JW, Brocato RL, Kwilas SA, Hammerbeck CD, Josleyn MD, Royals M, et al. DNA vaccine-derived human IgG produced in transchromosomal bovines protect in lethal models of hantavirus pulmonary syndrome. Sci Trans Med. (2014) 6:264ra162. doi: 10.1126/scitranslmed.3010082
218. Arikawa J, Yao J-S, Yoshimatsu K, Takashima I, Hashimoto N. Protective role of antigenic sites on the envelope protein of Hantaan virus defined by monoclonal antibodies. Arch Virol. (1992) 126:271–81. doi: 10.1007/bf01309700
219. Liang M, Chu YK, Schmaljohn C. Bacterial expression of neutralizing mouse monoclonal antibody Fab fragments to Hantaan virus. Virology. (1996) 217:262–71. doi: 10.1006/viro.1996.0113
220. Duehr J, McMahon M, Williamson B, Amanat F, Durbin A, Hawman DW, et al. Neutralizing monoclonal antibodies against the Gn and the Gc of the Andes virus glycoprotein spike complex protect from virus challenge in a preclinical hamster model. mBio. (2020) 11. doi: 10.1128/mBio.00028-20
221. Garrido JL, Prescott J, Calvo M, Bravo F, Alvarez R, Salas A, et al. Two recombinant human monoclonal antibodies that protect against lethal Andes hantavirus infection in vivo. Sci Trans Med. (2018) 10. doi: 10.1126/scitranslmed.aat6420
222. Williamson BN, Prescott J, Garrido JL, Alvarez RA, Feldmann H, Barría MI. Therapeutic efficacy of human monoclonal antibodies against Andes virus infection in Syrian hamsters. Emerging Infect Dis. (2021) 27:2707–10. doi: 10.3201/eid2710.210735
223. Safronetz D, Ebihara H, Feldmann H, Hooper JW. The Syrian hamster model of hantavirus pulmonary syndrome. Antiviral Res. (2012) 95:282–92. doi: 10.1016/j.antiviral.2012.06.002
224. Seto T, Nagata N, Yoshikawa K, Ichii O, Sanada T, Saasa N, et al. Infection of Hantaan virus strain aa57 leading to pulmonary disease in laboratory mice. Virus Res. (2012) 163:284–90. doi: 10.1016/j.virusres.2011.10.016
225. Wei Z, Shimizu K, Sarii RS, Muthusinghe DS, Lokupathirage SMW, Nio-Kobayashi J, et al. Pathological studies on Hantaan virus-infected mice simulating severe hemorrhagic fever with renal syndrome. Viruses. (2022) 14. doi: 10.3390/v14102247
226. Golden JW, Hammerbeck CD, Mucker EM, Brocato RL. Animal models for the study of rodent-borne hemorrhagic fever viruses: arenaviruses and hantaviruses. BioMed Res Int. (2015) 2015. doi: 10.1155/2015/793257
227. Hartman AL, Myler PJ. Bunyavirales: scientific gaps and prototype pathogens for a large and diverse group of zoonotic viruses. J Infect Dis. (2023) 228:S376–S89. doi: 10.1093/infdis/jiac338
228. Smith DR, Holbrook MR, Gowen BB. Animal models of viral hemorrhagic fever. Antiviral Res. (2014) 112:59–79. doi: 10.1016/j.antiviral.2014.10.001
229. Kim GR, McKee KT. Pathogenesis of Hantaan virus infection in suckling mice: clinical, virologic, and serologic observations. Am J Trop Med Hygiene. (1985) 34:388–95. doi: 10.4269/ajtmh.1985.34.388
230. Zhang XK, Takashima I, Mori F, Hashimoto N. Comparison of virulence between two strains of rattus serotype hemorrhagic fever with renal syndrome (HFRS) virus in newborn rats. Microbiol Immunol. (1989) 33:195–205. doi: 10.1111/j.1348-0421.1989.tb01513.x
231. Wichmann D, Gröne H-J, Frese M, Pavlovic J, Anheier B, Haller O, et al. Hantaan virus infection causes an acute neurological disease that is fatal in adult laboratory mice. J Virol. (2002) 76:8890–9. doi: 10.1128/jvi.76.17.8890-8899.2002
232. Ma R, Zhang X, Shu J, Liu Z, Sun W, Hou S, et al. Nlrc3 knockout mice showed renal pathological changes after HTNV infection. Front Immunol. (2021) 12:692509. doi: 10.3389/fimmu.2021.692509
233. Lopez R, Vial C, Graf J, Calvo M, Ferres M, Mertz G, et al. Platelet count in patients with mild disease at admission is associated with progression to severe hantavirus cardiopulmonary syndrome. Viruses. (2019) 11. doi: 10.3390/v11080693
234. Perley CC, Brocato RL, Kwilas SA, Daye S, Moreau A, Nichols DK, et al. Three asymptomatic animal infection models of hemorrhagic fever with renal syndrome caused by hantaviruses. PloS One. (2019) 14:e0216700. doi: 10.1371/journal.pone.0216700
235. Sanada T, Kariwa H, Nagata N, Tanikawa Y, Seto T, Yoshimatsu K, et al. Puumala virus infection in Syrian hamsters (Mesocricetus auratus) resembling hantavirus infection in natural rodent hosts. Virus Res. (2011) 160:108–19. doi: 10.1016/j.virusres.2011.05.021
236. Groen J, Gerding M, Koeman JP, Roholl PJ, van Amerongen G, Jordans HG, et al. A macaque model for hantavirus infection. J Infect Dis. (1995) 172:38–44. doi: 10.1093/infdis/172.1.38
237. Klingström J, Plyusnin A, Vaheri A, Lundkvist Å. Wild-type Puumala hantavirus infection induces cytokines, C-reactive protein, creatinine, and nitric oxide in cynomolgus macaques. J Virol. (2002) 76:444–9. doi: 10.1128/jvi.76.1.444-449.2002
238. Sironen T, Klingström J, Vaheri A, Andersson LC, Lundkvist A, Plyusnin A. Pathology of Puumala hantavirus infection in macaques. PloS One. (2008) 3:e3035. doi: 10.1371/journal.pone.0003035
239. Dowall SD, Graham VA, Aram M, Findlay-Wilson S, Salguero FJ, Emery K, et al. Hantavirus infection in type I interferon receptor-deficient (A129) mice. J Gen Virol. (2020) 101:1047–55. doi: 10.1099/jgv.0.001470
240. Kobak L, Raftery MJ, Voigt S, Kühl AA, Kilic E, Kurth A, et al. Hantavirus-induced pathogenesis in mice with a humanized immune system. J Gen Virol. (2015) 96:1258–63. doi: 10.1099/vir.0.000087
241. Rissmann M, Noack D, Spliethof TM, Vaes VP, Stam R, van Run P, et al. A pan-orthohantavirus human lung xenograft mouse model and its utility for preclinical studies. PloS Pathog. (2025) 21:e1012875. doi: 10.1371/journal.ppat.1012875
242. Raftery MJ, Lalwani P, Lutteke N, Kobak L, Giese T, Ulrich RG, et al. Replication in the mononuclear phagocyte system (Mps) as a determinant of hantavirus pathogenicity. Front Cell Infect Microbiol. (2020) 10:281. doi: 10.3389/fcimb.2020.00281
243. Iheozor-Ejiofor R, Vapalahti K, Sironen T, Levanov L, Hepojoki J, Lundkvist Å, et al. Neutralizing antibody titers in hospitalized patients with acute puumala orthohantavirus infection do not associate with disease severity. Viruses (2022) 14(5). doi: 10.3390/v14050901
244. Linderholm M, Ahlm C, Settergren B, Waage A, Tärnvik A. Elevated plasma levels of tumor necrosis factor (TNF)-alpha, soluble TNF receptors, interleukin (IL)-6, and il-10 in patients with hemorrhagic fever with renal syndrome. J Infect Dis. (1996) 173:38–43. doi: 10.1093/infdis/173.1.38
245. Schmaljohn C. Vaccines for hantaviruses. Vaccine. (2009) 27 Suppl 4:D61–4. doi: 10.1016/j.vaccine.2009.07.096
246. Boudreau EF, Josleyn M, Ullman D, Fisher D, Dalrymple L, Sellers-Myers K, et al. A phase 1 clinical trial of Hantaan virus and Puumala virus M-segment DNA vaccines for hemorrhagic fever with renal syndrome. Vaccine. (2012) 30:1951–8. doi: 10.1016/j.vaccine.2012.01.024
247. Avižinienė A, Kučinskaitė-Kodzė I, Petraitytė-Burneikienė R, Žvirblienė A, Mertens ML, Schmidt S, et al. Characterization of a panel of cross-reactive hantavirus nucleocapsid protein-specific monoclonal antibodies. Viruses. (2023) 15. doi: 10.3390/v15020532
248. Lee HW, Chang Na. Development of vaccine against Hemorrhagic Fever with Renal Syndrome. (1988) 6:143–8. Butterworth & Co. (Publishers) Ltd.
249. Sohn YM, Rho HO, Park MS, Kim JS, Summers PL. Primary humoral immune responses to formalin inactivated hemorrhagic fever with renal syndrome vaccine (Hantavax): consideration of active immunization in South Korea. Yonsei Med J. (2001) 42:278–84. doi: 10.3349/ymj.2001.42.3.278
250. Bae J-M. Introduction of vaccinomics to develop personalized vaccines in light of changes in the usage of Hantaan virus vaccine (Hantavax®) in Korea. J Prev Med Public Health. (2019) 52:277–80. doi: 10.3961/jpmph.19.018
251. Cho H-W, Howard CR, Lee H-W. Review of an inactivated vaccine against hantaviruses. Intervirology. (2002) 45:328–33. doi: 10.1159/000067925
252. Park K, Kim CS, Moon K-T. Protective effectiveness of hantavirus vaccine. Emerging Infect Dis. (2004) 10:2218–20. doi: 10.3201/eid1012.040684
253. Yi Y, Park H, Jung J. Effectiveness of inactivated hantavirus vaccine on the disease severity of hemorrhagic fever with renal syndrome. Kidney Res Clin Pract. (2018) 37:366–72. doi: 10.23876/j.krcp.18.0044
254. Song JY, Woo HJ, Cheong HJ, Noh JY, Baek LJ, Kim WJ. Long-term immunogenicity and safety of inactivated Hantaan virus vaccine (Hantavax™) in healthy adults. Vaccine. (2016) 34:1289–95. doi: 10.1016/j.vaccine.2016.01.031
255. Song JY, Jeong HW, Yun JW, Lee J, Woo HJ, Bae J-Y, et al. Immunogenicity and safety of a modified three-dose priming and booster schedule for the Hantaan virus vaccine (Hantavax): A multi-center phase III clinical trial in healthy adults. Vaccine. (2020) 38:8016–23. doi: 10.1016/j.vaccine.2020.10.035
256. Cho HW, Howard CR. Antibody responses in humans to an inactivated hantavirus vaccine (Hantavax). Vaccine. (1999) 17:2569–75. doi: 10.1016/s0264-410x(99)00057-2
257. Brocato RL, Hooper JW. Progress on the prevention and treatment of hantavirus disease. Viruses. (2019) 11. doi: 10.3390/v11070610
258. Afzal S, Ali L, Batool A, Afzal M, Kanwal N, Hassan M, et al. Hantavirus: an overview and advancements in therapeutic approaches for infection. Front Microbiol. (2023) 14:1233433. doi: 10.3389/fmicb.2023.1233433
259. Dzagurova TK, Siniugina AA, Ishmukhametov AA, Egorova MS, Kurashova SS, Balovneva MV, et al. Pre-clinical studies of inactivated polyvalent HFRS vaccine. Front Cell Infect Microbiol. (2020) 10:545372. doi: 10.3389/fcimb.2020.545372
260. Kurashova SS, Ishmukhametov AA, Dzagurova TK, Egorova MS, Balovneva MV, Nikitin NA, et al. Various adjuvants effect on immunogenicity of Puumala virus vaccine. Front Cell Infect Microbiol. (2020) 10:545371. doi: 10.3389/fcimb.2020.545371
261. Lundkvist A, Kallio-Kokko H, Sjölander KB, Lankinen H, Niklasson B, Vaheri A, et al. Characterization of Puumala virus nucleocapsid protein: identification of B-cell epitopes and domains involved in protective immunity. Virology. (1996) 216:397–406. doi: 10.1006/viro.1996.0075
262. Carvalho Nicacio Cd, Sällberg M, Hultgren C, Lundkvist Å. T-helper and humoral responses to Puumala hantavirus nucleocapsid protein: identification of T-helper epitopes in a mouse model. J Gen Virol. (2001) 82:129–38. doi: 10.1099/0022-1317-82-1-129
263. Kehm R, Jakob NJ, Welzel TM, Tobiasch E, Viczian O, Jock S, et al. Expression of immunogenic Puumala virus nucleocapsid protein in transgenic tobacco and potato plants. Virus Genes. (2001) 22:73–83. doi: 10.1023/a:1008186403612
264. Carvalho Nicacio Cd, Della Gonzalez Valle M, Padula P, Björling E, Plyusnin A, Lundkvist Å. Cross-Protection against Challenge with Puumala virus after Immunization with Nucleocapsid Proteins from Different Hantaviruses. J Virol. (2002) 76:6669–77. doi: 10.1128/jvi.76.13.6669-6677.2002
265. Dargeviciute A, Brus Sjölander K, Sasnauskas K, Krüger DH, Meisel H, Ulrich R, et al. Yeast-expressed Puumala hantavirus nucleocapsid protein induces protection in a bank vole model. Vaccine. (2002) 20:3523–31. doi: 10.1016/s0264-410x(02)00341-9
266. Khattak S, Darai G, Rösen-Wolff A. Puumala virus nucleocapsid protein expressed in transgenic plants is not immunogenic after oral administration. Virus Genes. (2004) 29:109–16. doi: 10.1023/b:Viru.0000032794.24032.72
267. Maes P, Clement J, Cauwe B, Bonnet V, Keyaerts E, Robert A, et al. Truncated recombinant Puumala virus nucleocapsid proteins protect mice against challenge in vivo. Viral Immunol. (2008) 21:49–60. doi: 10.1089/vim.2007.0059
268. Zhao C, Sun Y, Zhao Y, Wang S, Yu T, Du F, et al. Immunogenicity of a multi-epitope DNA vaccine against hantavirus. Hum Vaccines Immunotherapeutics. (2012) 8:208–15. doi: 10.4161/hv.18389
269. Koletzki D, Schirmbeck R, Lundkvist A, Meisel H, Krüger DH, Ulrich R. DNA vaccination of mice with a plasmid encoding Puumala hantavirus nucleocapsid protein mimics the B-cell response induced by virus infection. J Biotechnol. (2001) 84:73–8. doi: 10.1016/s0168-1656(00)00329-1
270. Bucht G, Sjölander KB, Eriksson S, Lindgren L, Lundkvist A, Elgh F. Modifying the cellular transport of DNA-based vaccines alters the immune response to hantavirus nucleocapsid protein. Vaccine. (2001) 19:3820–9. doi: 10.1016/s0264-410x(01)00151-7
271. Lindkvist M, Lahti K, Lilliehöök B, Holmström A, Ahlm C, Bucht G. Cross-reactive immune responses in mice after genetic vaccination with cDNA encoding hantavirus nucleocapsid proteins. Vaccine. (2007) 25:1690–9. doi: 10.1016/j.vaccine.2006.09.082
272. Spik KW, Badger C, Mathiessen I, Tjelle T, Hooper JW, Schmaljohn C. Mixing of M segment DNA vaccines to Hantaan virus and Puumala virus reduces their immunogenicity in hamsters. Vaccine. (2008) 26:5177–81. doi: 10.1016/j.vaccine.2008.03.097
273. Hooper JW, Custer DM, Thompson E, Schmaljohn CS. DNA vaccination with the Hantaan virus M gene protects hamsters against three of four HFRS hantaviruses and elicits a high-titer neutralizing antibody response in rhesus monkeys. J Virol. (2001) 75:8469–77. doi: 10.1128/jvi.75.18.8469-8477.2001
274. Ulrich R, Lundkvist A, Meisel H, Koletzki D, Sjölander KB, Gelderblom HR, et al. Chimaeric HBV core particles carrying a defined segment of Puumala hantavirus nucleocapsid protein evoke protective immunity in an animal model. Vaccine. (1998) 16:272–80. doi: 10.1016/s0264-410x(97)00172-2
275. Shkair L, Garanina EE, Martynova EV, Kolesnikova AI, Arkhipova SS, Titova AA, et al. Immunogenic properties of MVs containing structural hantaviral proteins: an original study. Pharmaceutics. (2022) 14. doi: 10.3390/pharmaceutics14010093
276. Shkair L, Sharma D, Hamza S, Garanina E, Shakirova V, Khaertynova I, et al. Cross-reactivity of hantavirus antibodies after immunization with PUUV antigens. Biotechnol Appl Biochem. (2024) 71:1139–53. doi: 10.1002/bab.2604
277. Hooper JW, Moon JE, Paolino KM, Newcomer R, McLain DE, Josleyn M, et al. A phase 1 clinical trial of Hantaan virus and Puumala virus M-segment DNA vaccines for haemorrhagic fever with renal syndrome delivered by intramuscular electroporation. Clin Microbiol Infect. (2014) 20 Suppl 5:110–7. doi: 10.1111/1469-0691.12553
278. Hooper J, Paolino KM, Mills K, Kwilas S, Josleyn M, Cohen M, et al. A phase 2a randomized, double-blind, dose-optimizing study to evaluate the immunogenicity and safety of a bivalent DNA vaccine for hemorrhagic fever with renal syndrome delivered by intramuscular electroporation. Vaccines. (2020) 8. doi: 10.3390/vaccines8030377
279. Hooper JW, Kwilas SA, Josleyn M, Norris S, Hutter JN, Hamer M, et al. Phase 1 clinical trial of Hantaan and Puumala virus DNA vaccines delivered by needle-free injection. NPJ Vaccines. (2024) 9. doi: 10.1038/s41541-024-00998-7
280. Xu X, Ruo SL, McCormick JB, Fisher-Hoch SP. Immunity to hantavirus challenge in meriones unguiculatus induced by vaccinia-vectored viral proteins. Am J Trop Med Hygiene. (1992) 47:397–404. doi: 10.4269/ajtmh.1992.47.397
281. Engdahl TB, Crowe JE. Humoral immunity to hantavirus infection. mSphere. (2020) 5. doi: 10.1128/mSphere.00482-20
282. Elgh F, Linderholm M, Wadell G, Tärnvik A, Juto P. Development of humoral cross-reactivity to the nucleocapsid protein of heterologous hantaviruses in nephropathia epidemica. FEMS Immunol Med Microbiol. (1998) 22:309–15. doi: 10.1111/j.1574-695X.1998.tb01220.x
283. He P, Zou Y, Hu Z. Advances in aluminum hydroxide-based adjuvant research and its mechanism. Hum Vaccines Immunotherapeutics. (2015) 11:477–88. doi: 10.1080/21645515.2014.1004026
284. Maes P, Keyaerts E, Bonnet V, Clement J, Avsic-Zupanc T, Robert A, et al. Truncated recombinant Dobrava hantavirus nucleocapsid proteins induce strong, long-lasting immune responses in mice. Intervirology. (2006) 49:253–60. doi: 10.1159/000093454
285. Hoenen T, Groseth A, Kok-Mercado Fd, Kuhn JH, Wahl-Jensen V. Minigenomes, transcription and replication competent virus-like particles and beyond: reverse genetics systems for filoviruses and other negative stranded hemorrhagic fever viruses. Antiviral Res. (2011) 91:195–208. doi: 10.1016/j.antiviral.2011.06.003
286. Conzelmann K-K, Meyers G. Genetic engineering of animal RNA viruses. Trends Microbiol. (1999) 4:386–93. doi: 10.1016/0966-842x(96)10062-7
287. Stobart CC, Moore ML. RNA virus reverse genetics and vaccine design. Viruses. (2014) 6:2531–50. doi: 10.3390/v6072531
288. Walpita P, Flick R. Reverse genetics of negative-stranded RNA viruses: A global perspective. FEMS Microbiol Lett. (2005) 244:9–18. doi: 10.1016/j.femsle.2005.01.046
289. Zobel A, Neumann G, Hobom G. RNA polymerase I catalysed transcription of insert viral cDNA. Nucleic Acids Res. (1993) 21:3607–14. doi: 10.1093/nar/21.16.3607
290. Fuerst TR, Niles EG, Studier FW, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. (1986) 83:8122–6. doi: 10.1073/pnas.83.21.8122
291. Tercero B, Makino S. Reverse genetics approaches for the development of bunyavirus vaccines. Curr Opin Virol. (2020) 44:16–25. doi: 10.1016/j.coviro.2020.05.004
292. Brown KS, Ebihara H, Feldmann H. Development of a minigenome system for Andes virus, a New World hantavirus. Arch Virol. (2012) 157:2227–33. doi: 10.1007/s00705-012-1401-0
293. Welzel TM, Kehm R, Tidona CA, Muranyi W, Darai G. Stable expression of nucleocapsid proteins of Puumala and Hantaan virus in mammalian cells. Virus Genes. (1998) 17:185–98. doi: 10.1023/a:1008076926061
294. Flick K, Hooper JW, Schmaljohn CS, Pettersson RF, Feldmann H, Flick R. Rescue of Hantaan virus minigenomes. Virology. (2003) 306:219–24. doi: 10.1016/s0042-6822(02)00070-3
295. Kell AM. Innate immunity to orthohantaviruses: could divergent immune interactions explain host-specific disease outcomes? J Mol Biol. (2021) 434:167230. doi: 10.1016/j.jmb.2021.167230
296. Klempa B. Reassortment events in the evolution of hantaviruses. Virus Genes. (2018) 54:638–46. doi: 10.1007/s11262-018-1590-z
297. Handke W, Oelschlegel R, Franke R, Wiedemann L, Krüger DH, Rang A. Generation and characterization of genetic reassortants between Puumala and Prospect Hill hantavirus in vitro. J Gen Virol. (2010) 91:2351–9. doi: 10.1099/vir.0.021139-0
298. McElroy AK, Smith JM, Hooper JW, Schmaljohn CS. Andes virus M genome segment is not sufficient to confer the virulence associated with Andes virus in Syrian hamsters. Virology. (2004) 326:130–9. doi: 10.1016/j.virol.2004.05.018
Keywords: Puumala orthohantavirus, animal models, vaccine research, antiviral treatment, glycoprotein, nephropathia epidemica
Citation: Tscherne A, Guardado-Calvo P, Clark JJ, Krause R and Krammer F (2025) Puumala orthohantavirus: prevalence, biology, disease, animal models and recent advances in therapeutics development and structural biology. Front. Immunol. 16:1575112. doi: 10.3389/fimmu.2025.1575112
Received: 11 February 2025; Accepted: 21 March 2025;
Published: 08 May 2025.
Edited by:
Michael H. Lehmann, Ludwig-Maximilians-Universität München, GermanyReviewed by:
Tomas Strandin, University of Helsinki, FinlandMartin Raftery, Charité–Universitätsmedizin Berlin, Germany
Zhixin Liu, Hubei University of Medicine, China
Copyright © 2025 Tscherne, Guardado-Calvo, Clark, Krause and Krammer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Florian Krammer, Zmxvcmlhbi5rcmFtbWVyQG1lZHVuaXdpZW4uYWMuYXQ=; Zmxvcmlhbi5rcmFtbWVyQG1zc20uZWR1
 Alina Tscherne
Alina Tscherne Pablo Guardado-Calvo
Pablo Guardado-Calvo Jordan J. Clark
Jordan J. Clark Robert Krause
Robert Krause Florian Krammer
Florian Krammer