- 1Pancreatic Center, Department of Gastroenterology, The Affiliated Hospital of Yangzhou University, Yangzhou University, Yangzhou, China
- 2Department of Gastroenterology, Huai’an Hospital Affiliated to Yangzhou University (The Fifth People’s Hospital of Huai’an), Yangzhou University, Huai’an, China
- 3Department of Gastroenterology, Clinical Medical College, Yangzhou University, Yangzhou, China
- 4Clinical Research Division, Fred Hutchinson Cancer Research Center, Seattle, WA, United States
- 5Faculty of Pharmaceutical Sciences, Toho University, Funabashi, Chiba, Japan
- 6Department of Gastroenterology, Kunshan Hospital of Traditional Chinese Medicine, Suzhou Key Laboratory of Integrated Traditional Chinese and Western Medicine of Digestive Diseases, Kunshan Affiliated Hospital of Yangzhou University, Kunshan, China
Introduction: Macrophage activation is closely associated with Acute pancreatitis (AP). We screened and found that Growth factor receptor bound protein 2 (Grb2) is highly expressed in macrophages during AP. However, the relationship between Grb2 and AP is still poorly understood. In this study, we explored the role of Grb2 in AP.
Methods: We screened for gene affecting macrophage activation in AP by combining transcriptomics with Single-cell RNA-sequence analysis. Next, the expression of Grb2 in M1/M2 macrophage activation was detected by Single-cell RNA-sequence analysis and western blot. Furthermore, the effect of Grb2 on M1/M2 macrophage activation was detected by flow cytometry. The severity of AP was assessed by histological analysis, serum amylase, serum lipase and serum inflammatory factors in vivo. NOD-like receptor thermal protein domain associated protein 3 (Nlrp3) and Nuclear factor kappa-B (NF-kB) signaling pathways were also evaluated.
Results: Grb2 is mainly expressed in macrophages of pancreas in AP and up-regulated in M1 macrophage activation. Inhibiting Grb2 could alleviate AP by preventing M1 macrophage activation through down-regulating Nlrp3 and NF-κB.
Discussion: Inhibition of Grb2 can effectively prevent M1 macrophage activation and alleviate AP. Grb2 may potentially be an effective target of macrophage activation for the treatment of AP.
Introduction
AP is a prevalent gastrointestinal disease marked by acinar cell death and the infiltration of inflammatory cells (1, 2). The incidence of AP is increasing globally. Mounting evidence suggest that the infiltration of inflammatory immune cells impacts the severity of AP. The inflammatory cells infiltrated in acute pancreatitis are predominantly neutrophils and macrophages (3–6). Neutrophil forming Neutrophil Extracellular Traps (NETs) can lead to pancreatic tissue damage (7, 8). The activated macrophages in AP are mainly classified into two types. The macrophages infiltrating in the early stage of AP are generally pro-inflammatory M1 macrophage, while anti-inflammatory M2 macrophages typically play a role in the repair period of pancreatic tissue (9, 10). Hence, targeting macrophage activation could be a strategy for the treatment of AP.
Grb2 is a non-enzymatic adaptor protein consisting of a single Src homology 2 (SH2) domain and two Src homology 3 (SH3) domains (11, 12). The classic Grb2 signaling pathway is initiated by receptor tyrosine kinases (RTKs) and plays an important role in cell signal transduction. Studies have indicated that Grb2 plays a crucial role in cell proliferation (13–15),cell death (16–19) and cell differentiation (20, 21). In addition, Grb2 is closely related to immune regulation or inflammatory diseases. Treg cell-specific Grb2 deletion abrogates the augmentation of allergic airway inflammation by Il4raR576 (22). In the inflammatory lesions of the infected liver, Grb2 was positive in both hepatocytes and infiltrating leukocytes (23). The expression of GRB2 was increased in psoriatic lesions compared with normal skin adjacent to psoriasis and healthy controls. Inhibition of GRB2 inhibits the secretion of inflammatory mediators in keratinocytes (24).In this research, Grb2 was primarily expressed in macrophages of pancreas in AP. Inhibition of Grb2 can prevent the transformation of macrophage into M1 macrophage and alleviated pancreatic tissue injury caused by caerulein-induced AP. Thus, Grb2 may become an potential target of macrophage activation in AP.
Experimental methods
Antibodies and reagents
All reagents and antibodies involved in this study are shown in Supplementary Tables S1, S2.
Animals
C57BL/6 mice were purchased from GemPharmatech Co., Ltd. (Nanjing, China). All mice were raised in a suitable environment (20-25°C, 12h light/12h dark) without specific pathogen. They were provided with sufficient water and food. All studies were approved by Yangzhou University (No.202307018).
Construction of the AP model and Grb2 inhibitors administration
Caerulein (Cae) model, acute pancreatitis model was established by intraperitoneal (i.p.) injection of Caerulein (100 mg/kg, 1h interval, 10 times). At 0 hour, GRB2 inhibitor prexigebersen (20 mg/kg, 40 mg/kg, 80mg/kg) was administered via intravenous (i.v.) injection. 12 hours later, mice were anesthetized and euthanized. Serum samples were collected for serological analysis and ELISA assays. Pancreatic tissues were harvested and stored at -80°C for subsequent experiments.
Pancreatic duct ligation (PDL)-induced AP model: All mice were anesthetized and placed on a horizontal surgical table. A longitudinal incision of 1–2 centimeters was made on the abdomen to expose the abdominal cavity. The duodenum was turned over to expose the pancreatic duct. At a position 1 centimeter above the duodenal papilla, the tissue around the pancreatic duct was bluntly dissected, and the pancreatic duct was ligated with a ligation thread. The silk thread completely blocked it to simulate cholestasis and biliary pancreatitis caused by cholelithiasis. Then, the abdominal cavity was sutured in layers. At 1 hour, GRB2 inhibitor prexigebersen (40 mg/kg) was administered via intravenous (i.v.) injection. 24 hours later, mice were anesthetized and euthanized.
Serum samples and pancreatic tissues were harvested or subsequent experiments.
ELISA assays
Following the manufacturer’s protocol, the plates were coated with antibody, sealed, and incubated overnight at 4°C. The following day, the liquid was aspirated, and the plates were washed three times with phosphate-buffered saline containing 0.05% Tween 20 (PBST). Blocking was performed using ELISPOT blocking buffer for 1 h at room temperature (RT). After removing the blocking solution and washing with PBST, serum samples and standards were added to the plates. The plates were sealed and incubated at RT for 2 h. Subsequently, the liquid was discarded, and the plates were washed three times with PBST. Detection Antibody (diluted according to the datasheet) was added, followed by sealing and incubation at RT for 1 h. After additional PBST washes, Streptavidin-HRP conjugate (1:250 dilution) was introduced, and the plates were sealed and incubated in the dark at RT for 30 min. Following further PBST washing, TMB Substrate Solution was added, and the plates were incubated in the dark at RT for 15 min. The reaction was terminated by adding Stop Solution. Absorbance was measured at 450 nm using a microplate reader.
Extraction and induction of bone marrow-derived macrophages (BMDMs)
The leg bones of the mice were isolated and soaked in 75% alcohol for 1 min. Subsequently, the bone marrow cavity was washed with 1640 medium. The cells were collected after centrifugation at 500g for 5 minutes. Macrophages were cultured in 1640 medium containing 20ng/ml macrophage-stimulating factor (M-CSF), 10% fetal bovine serum (FBS), 1% penicillin and 1% streptomycin for 7 days in 37°C, 5% CO2 incubator. The medium was changed every other day and mature macrophages were obtained after 7 days.
Flow cytometry
Mature BMDMs were stimulated with LPS (100ng/ml) and INFγ (10ng/ml) for 24h.The BMDMs were incubated with flow cytometry antibody F4/80 (1:200 dilution) and CD86 (1:200 dilution), washed with PBS and resuspended with PBS. M1 activation of BMDM was detected by flow cytometry. Mature BMDMs were stimulated with IL-4 (50ng/ml) for 24h, incubated with flow cytometry antibody F4/80 (1:200 dilution) and CD206 (1:200 dilution) at 4°C for 30min, washed with PBS and resuspended with PBS. M2 activation of BMDM was detected by flow cytometry.
Western blot
RIPA lysis buffer and protease inhibitor (1:10) were mixed to lyse BMDMs. The BMDMs were sonicated, centrifuged at 12000 rpm at 4°C for 30 min. The protein supernatant was collected, added protein loading buffer, and boiled at 100°C for 10 min. The protein samples were subjected to SDS-PAGE, transferred to PVDF membrane. The membranes were blocked with 5% skim milk for 2 h, washed with TBST (3 times, 5 min each time), incubated with primary antibody (1:1000 dilution) overnight at 4°C. The next day, the membranes were washed with TBST (3 times, 15min each time), and incubated with second antibody(1:5000 dilution) at room temperature for 2 h. TBST washing (3 times, 10min each time). The protein bands were detected by ECL Luminometer and ImageJ was used to analyze the gray value.
Cell siRNA transfection
For one well of a 24-well plate: Diluted 2 μL Lipofectamine 3000 (Lipo3000) in 50 μL serum-free RPMI 1640 medium, mixed gently, and incubated at room temperature for 5 minutes. Diluted 4 μL siRNA in 50 μL serum-free RPMI 1640 medium, then combined this mixture with the prepared Lipo3000/1640 mixture. Mixed thoroughly and incubated at room temperature for 20 minutes to allow complex formation. Added the resulting transfection complex mixture to the cell culture plate. After 24 hours, replaced the medium with fresh RPMI 1640 medium supplemented with 10% FBS. Performed subsequent experiments 48 hours post-transfection.
Histological analysis
Pancreatic tissue samples were fixed in 4% paraformaldehyde, paraffin-embedded, and sectioned into 5-μm-thick slices. After deparaffinization and dehydration, the sections were stained with hematoxylin and eosin (H&E). Histopathological examination was performed using a light microscope. The severity of acute pancreatitis was evaluated according to previously established methodologies (25).
Immunofluorescence
The slices were treated with dewaxing, dehydration, membrane rupture, antigen repair, blocking. The slices were incubated with primary antibody (1:100 dilution) overnight at 4°C. On the second day, the slices were washed with PBS (3 times, 5 minutes each), incubated with secondary antibody (1:1000 dilution) at room temperature for 2 hours and washed again with PBS (3 times, 5 minutes each). Added DAPI and anti-quenching agent, then sealing. The results were detected by laser scanning confocal microscope (Nikon, Ti2-E-A1 HD25).
Isolation of Peripheral blood mononuclear cells (PBMCs)
The anticoagulated blood was layered over Ficoll separation medium, and density gradient centrifugation was performed. The PBMC layer was collected. The cells were washed with PBS, centrifuged to remove the supernatant, and lysed with lysis buffer to extract proteins.
Microarray analysis
The microarray data of AP GSE65146, GSE109227 were downloaded from Gene Expression Omnibus (GEO) database (26). Both data had been log2 transformed. Limma package (27) was employed for identifying differentially expressed genes (DEGs). All genes (|log2FoldChange|>0) were analyzed by Gene Set Enrichment Analysis (GSEA) via clusterProfiler package (28, 29).
RNA-seq analysis
The RNA-seq data of macrophage GSE138134 (9) and RNA-seq data of M1 macrophage activation GSE247106 (30) were downloaded from GEO database. The DEGs (log2FoldChange>0.5, padj<0.01) were analyzed by DESeq2 package (31) or edgeR package (32–34). The pheatmap package was employed to display DEGs.
Single-cell RNA-sequence analysis
The scRNA-seq data of AP GSE188819 (35) and scRNA-seq data of macrophage activation GSE117176 (36),GSE158094 (37) were downloaded from GEO database. Downstream analysis was performed using Seurat package (V5) (38).
GSE188819: Following quality control (QC), cells with fewer than 200 expressed genes and >10% mitochondria-related genes were excluded. Harmony R package (39) was used to eliminate batch effects between different samples. As a result, a SeuratObject with 31080 cells and 22508 genes was subsequently normalized and scaled for the subsequent analysis, including RunPCA and RunTSNE. The cells were then clustered using the FindNeighbors and FindClusters functions with a resolution of 0.4. The generated clusters were visualized using T-distributed stochastic neighbor embedding (tSNE) plot.
GSE117176: Following quality control (QC), cells with fewer than 200 expressed genes and >10% mitochondria-related genes were excluded. Merge different samples. As a result, a SeuratObject with 17661 cells and 15217 genes was subsequently normalized and scaled for the subsequent analysis, including RunPCA and RunTSNE. The cells were then clustered using the FindNeighbors and FindClusters functions with a resolution of 0.1. The generated clusters were visualized using T-distributed stochastic neighbor embedding (tSNE) plot. FindMarkers was used to analyze DEGs (log2FoldChange>0.5, padj<0.01).
GSE158094: Three samples (24h_M0, 24h_M1 and 24h_M2) were selected for analysis. Following quality control (QC), cells with fewer than 200 expressed genes and >10% mitochondria-related genes were excluded. Merge different samples. As a result, a SeuratObject with 8492 cells and 12523 genes was subsequently normalized and scaled for the subsequent analysis, including RunPCA and RunTSNE. The cells were then clustered using the FindNeighbors and FindClusters functions with a resolution of 0.1. The generated clusters were visualized using T-distributed stochastic neighbor embedding (tSNE) plot. FindMarkers was used to analyze DEGs (log2FoldChange>0.5, padj<0.01).
Spearman Correlation Analysis was performed to assess the relationship between the two genes. KEGG (Kyoto Encyclopedia of Genes and Genomes) enrichment analysis were performed to analyze Genes positively associated with Grb2.
Short time-series expression miner analysis
The detection of gene clustering profiles were conducted by using the STEM (40, 41) clustering algorithm to identify temporal gene expression profiles, with the maximum number of model profiles set to 50 and maximum unit change in model profiles between time points set to 3.
Statistical analysis
Statistical analysis was performed by GraphPad Prism 8 software (GraphPad, San Diego, CA, USA) and the results were presented as mean ± standard deviation (SD). The difference between two groups was analyzed by t-test, and the difference between more than two groups was analyzed by one-way ANOVA test. P < 0.05 was considered statistically significant (two-tailed).
Results
Grb2 is mainly expressed in pancreatic tissue macrophages in AP
According to the results of GSEA of pancreatic tissue microarray data of AP (GSE65146 and GSE109227), it was found that macrophage activation involved in immune response, regulation of macrophage activation, macrophage activation, positive regulation of macrophage activation were significantly up-regulated in AP (Figures 1A, B and Supplementary Table S3, S4). This indicates that macrophages in pancreas are activated when AP occurs.
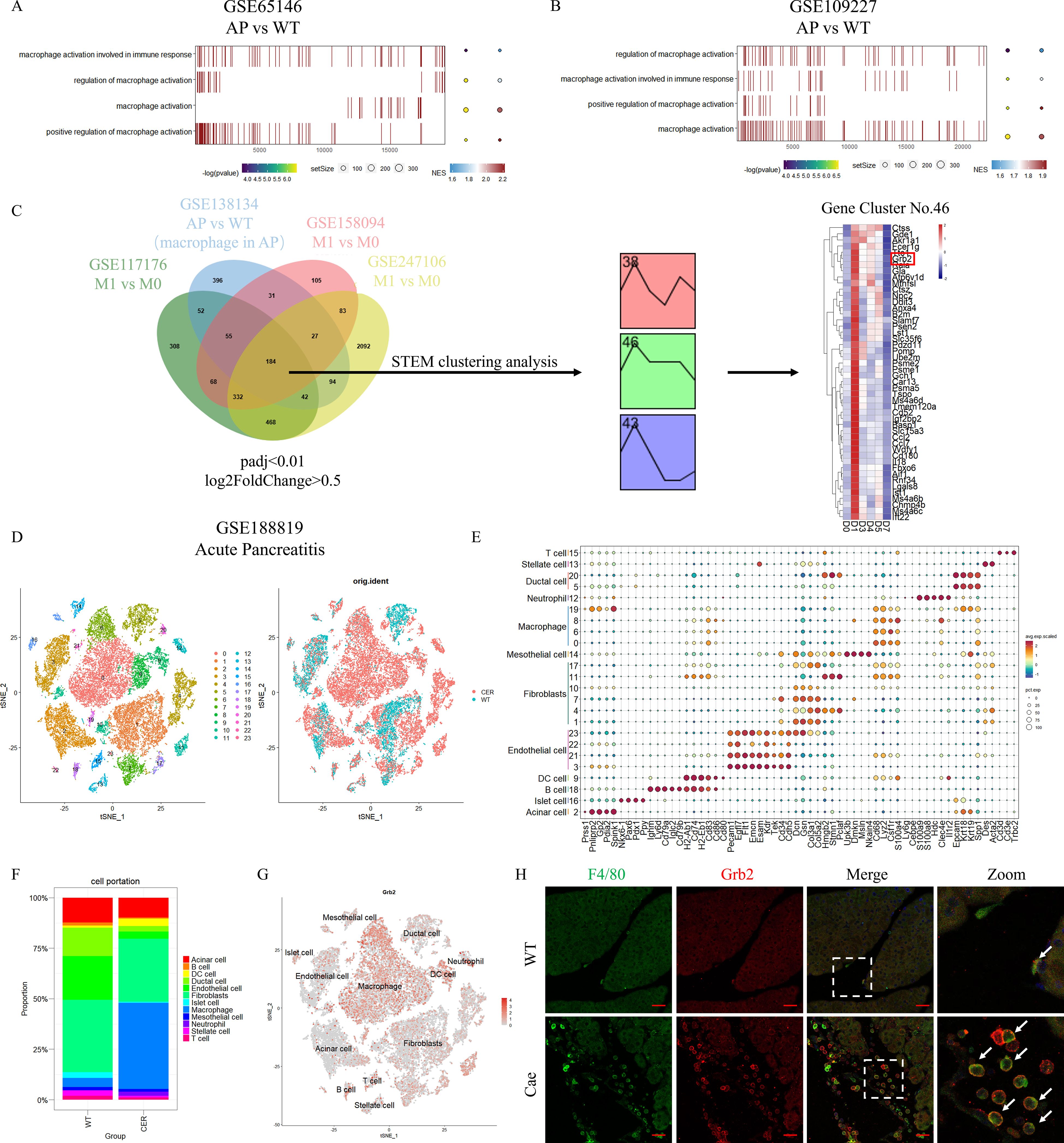
Figure 1. Grb2 is mainly expressed in macrophages of pancreas in AP. (A,B) GSEA pathway enrichment analysis of AP microarray data GSE65146 and GSE109227. (AP versus WT). (C) The flowchart of screening differentially expressed genes. (D) The t-distributed Stochastic Neighbor Embedding (tSNE) visualization of AP scRNA-seq GSE188819.(E) DotPlot showing the expression of marker genes for each annotated cell type: T cell (Cd3d, Cd3e,Trbc2), Stellate cell (Des, Acta2), Ductal cell (Epcam, Krt18, Krt19, Spp1), Neutrophil (Ly6g, Cebpe, S100a9, S100a8, Hdc, Clec4e, Il1r2), Macrophage (Cd68, Lyz2, Csf1r, S100a4), Mesothelial cell (Upk3b, Dmkn, Msln, Nkain4), Fibroblasts (Dcn, Gsn, Col3a1, Col5a2, Hmgb2, Stmn1, Pclaf), Endothelial cell (Pecam1, Egfl7, Flt1, Emcn, Esam, Kdr, Tek, Cd34, Cdh5), DC cell (H2-Ab1, Cd74, H2-Eb1, Cd83, Cd86, Cd80), B cell (Ighm, Ly6d, Cd79a, Iglc2, Cd79b), Islet cell (Nkx6-1, Pax6, Pdx1, Ppy), Acinar cell (Prss1, Pnliprp2, Gp2, Pdia2, Spink1). (F) The cell proportion of cells derived from different cell type. (G) The tSNE visualization of the expression of Grb2. (H) Representative Immunofluorescence images for Grb2 and F4/80 expression in macrophage of AP mice. The white arrow represents colocalization of Grb2 and F4/80. Scale Bar = 50μM.
We conducted a joint analysis of RNA-seq and scRNA-seq datasets (GSE117176, GSE138134, GSE158094 and GSE247106). As a result, we found a total of 184 common up-regulated DEGs. Next, a total of 3 clusters were screened by STEM clustering analysis (Supplementary Table S5). Among them, cluster 46 gene expression increased in the early stage and gradually decreased to return to the baseline level. We screened Grb2 for subsequent experiments through heatmap analysis (Figure 1C).
We obtained 24 cell clusters through cell clustering analysis on AP scRNA-seq data (GSE188819) (Figure 1D). Then we annotated different clusters based on cell-specific, highly expressed genes (Figure 1E and Supplementary Table S6). Next, we carried out cell proportion analysis. The number of macrophages increased significantly during AP (Figure 1F). FeaturePlot demonstrated that Grb2 was mainly expressed in pancreatic macrophages (Figure 1G). The results of immunofluorescence were consistent with those of scRNA-seq (Figure 1H). In addition, we detected PBMC in peripheral blood of AP patients and found that Grb2 was up-regulated in PBMC of AP patients compared with PBMC of healthy people (Supplementary Figures S1A, B). Therefore, we speculated that Grb2 may play a role in macrophage activation during AP.
Grb2 is up-regulated in M1 macrophage activation
To verify the role of Grb2 in macrophage, we initially analyzed the scRNA-seq data of macrophage activation (GSE117176) and obtained 5 cell clusters (Figure 2A). Subsequently, we performed cell annotation. M1 macrophages highly expressed Nos2, Cd86, Tnf and M2 macrophages highly expressed Chil3, Arg1, Retnla, Mgl2. M0 macrophages only expressed Ptprc, Adgre1, Lyz2, and did not express M1M2-specific genes. (Figure 2B). Nos2, Cd86 and Tnf were mainly expressed in M1 macrophage, while Chil3, Arg1, Retnla and Mgl2 were highly expressed in M1 macrophage (Figure 2B). The VlnPlot demonstrated that Grb2 was up-regulated in M1 macrophages and M2 macrophages (Figure 2C). Western blot results showed expression of Grb2 was up-regulated in M1 macrophages, but not in M2 macrophages. (Figures 2D, E). Thus, we hypothesized that Grb2 only exerts an effect on M1 macrophage activation.
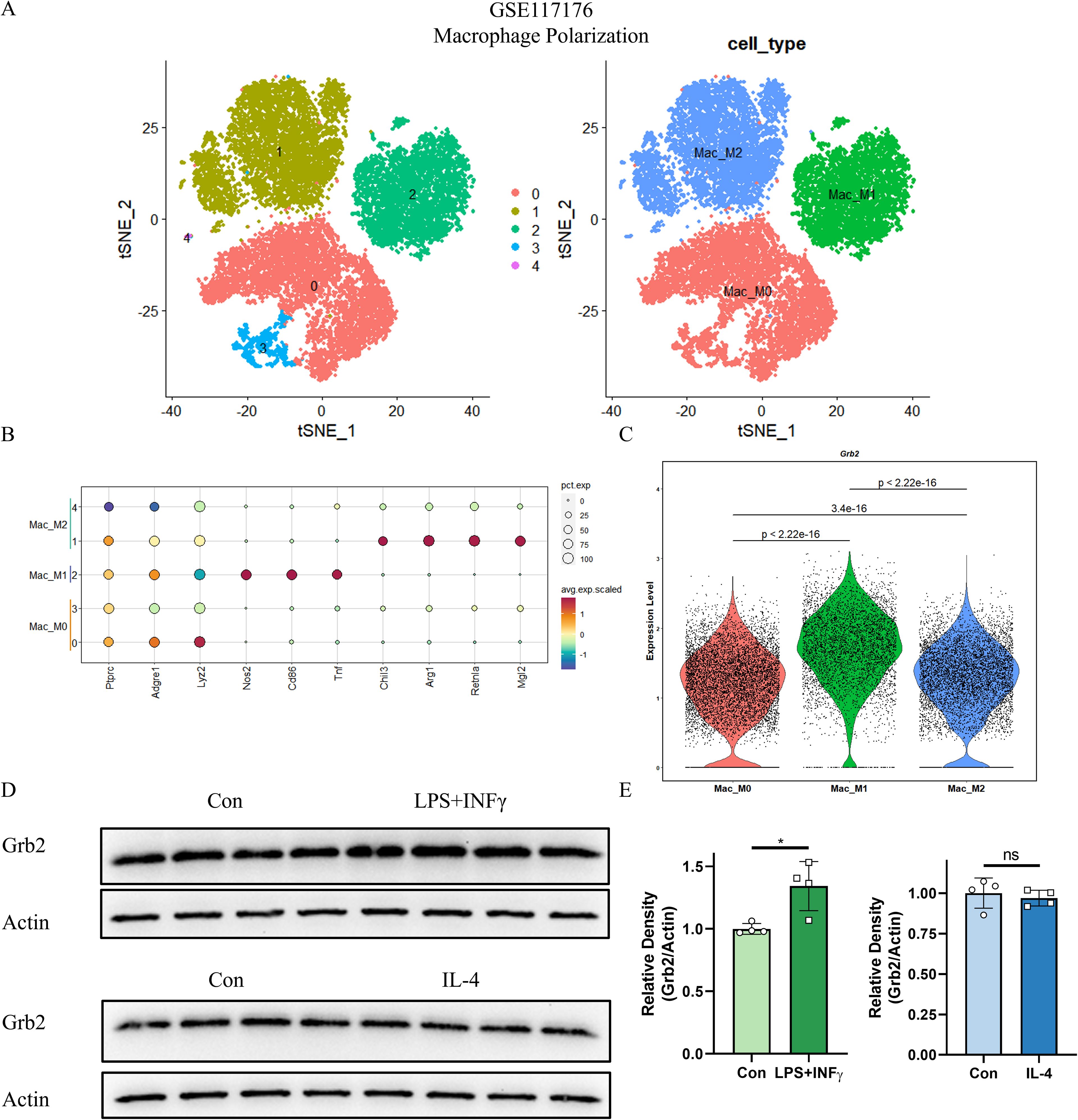
Figure 2. Grb2 is up-regulated in M1 macrophage activation. (A) The tSNE visualization of macrophage activation scRNA-seq GSE117176. (B) DotPlot of the three major cell types (Columns) and their marker genes (Rows). (C) VlnPlot showed the expression of Grb2. (D) Protein levels of Grb2 in macrophage activation were analyzed by western blotting. (E) Relative density of Grb2.Beta-actin was used as control for protein loading, N=4 each group. *P < 0.05. LPS, Lipopolysaccharide; INFγ, Interferon-γ; IL-4, Interleukin 4.
Grb2 is associated with immune and inflammatory pathways
To verify the relationship between and macrophage activation. In GSE117176, correlation analysis revealed that Grb2 was positively correlated with M1 macrophage activation marker Nos2, CD86 and Tnf (Figures 3A–C), but not with M2 macrophage activation markers Chil3, Arg1, Retnla and Mgl2 (Supplementary Figures S2A-D). Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of genes positively related to Grb2 in the scRNA-seq data of M1 macrophage activation (GSE117176 and GSE158094) demonstrated that Grb2 was associated with immune and inflammatory pathways such as the NOD-like receptor signaling pathway and the TNF signaling pathway (Figures 3D, E). Additionally, in the scRNA-seq data of AP (GSE188819), Grb2 was also found to be related to immune and inflammatory pathways such as the Chemokine signaling pathway and the Cytokine-cytokine receptor interaction (Figure 3F).
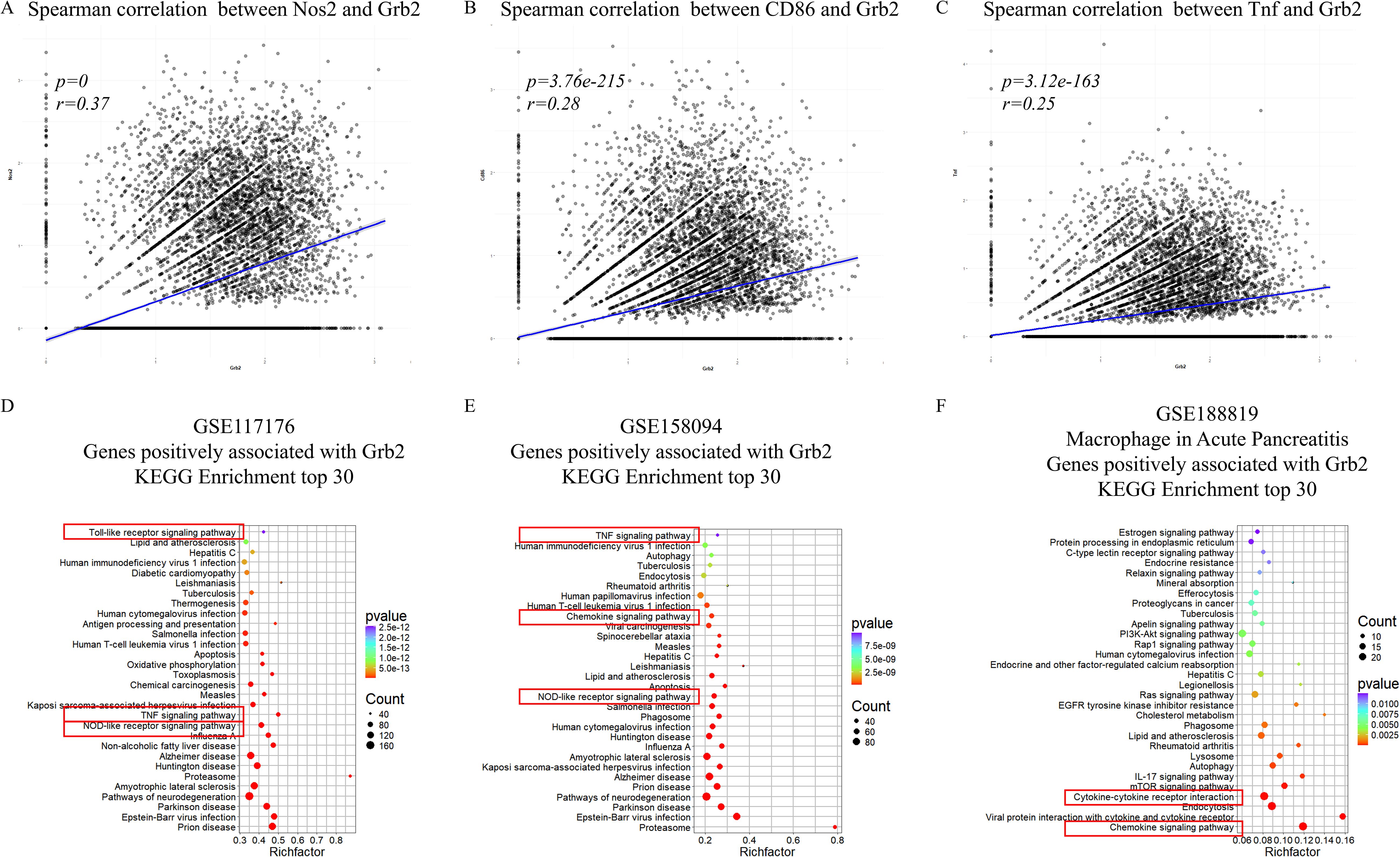
Figure 3. Grb2 is associated with immune and inflammatory pathway. (A) The Spearman correlation between Nos2 and Grb2. (B) The Spearman correlation between CD86 and Grb2. (C) The Spearman correlation between Tnf and Grb2. (D) KEGG analysis of genes positively associated with Grb2 of macrophage activation scRNA-seq GSE117176. (E) KEGG analysis of genes positively associated with Grb2 of macrophage activation scRNA-seq GSE158094. (F) KEGG analysis of genes positively associated with Grb2 of AP scRNA-seq GSE188819.
Inhibition of Grb2 inhibits M1 macrophage activation
To clarify the role of Grb2 in macrophage activation, BMDMs were extracted to induce M1/M2 macrophage activation (Figure 4A). SiRNA was employed to silence the expression of Grb2 in BMDMs, and western blot was utilized to verify the silencing efficiency (Figure 4B). The silencing of Grb2 significantly inhibited M1 macrophage activation but had no significant impact on M2 activation (Figures 4C–F). Subsequently, the Grb2 inhibitor prexigebersen was used, and it was found that the results of the Grb2 inhibitor prexigebersen were consistent with those of Grb2 silencing (Figures 4G–J). All these results confirmed that Grb2 affected M1 macrophage activation and had little effect on M2.
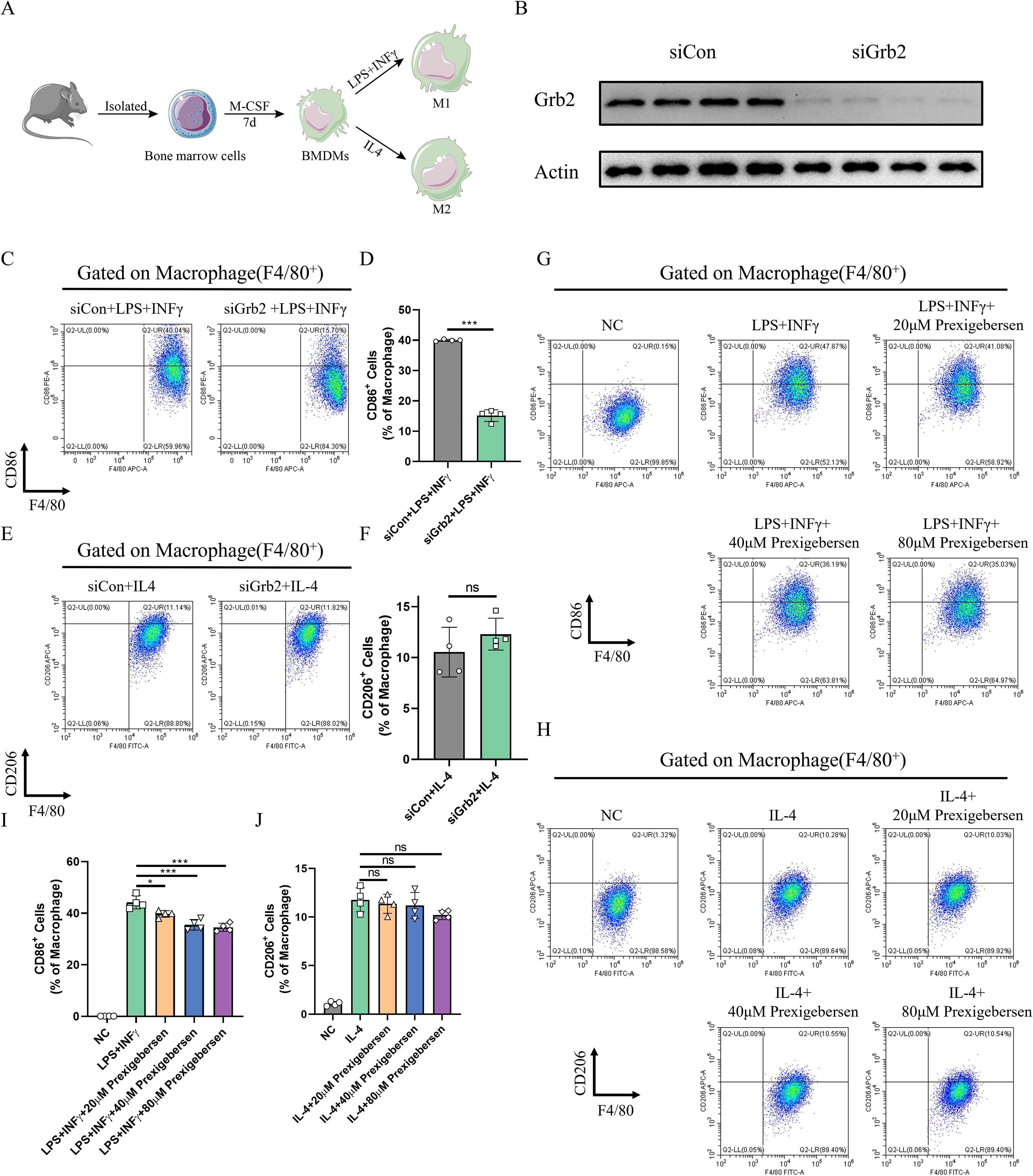
Figure 4. Inhibition of Grb2 inhibits M1 macrophage activation. (A) The flowchart of macrophage activation. (B) The silencing efficiency of Grb2 was analyzed by western blotting. (C, D) CD86 (M1 macrophage marker) and F4/80 (macrophage marker) expression levels in macrophages treated with Grb2 siRNA were detected by flow cytometry, N=4 each group. (E, F) CD206 (M2 macrophage marker) and F4/80 (macrophage marker) expression levels in macrophages treated with Grb2 siRNA were detected by flow cytometry, N=4 each group. (G) CD86 (M1 macrophage marker) and F4/80 (macrophage marker) expression levels in macrophages treated with Grb2 inhibitor prexigebersen were detected by flow cytometry, N=4 each group. (H) CD206 (M2 macrophage marker) and F4/80 (macrophage marker) expression levels in macrophages treated with Grb2 inhibitor prexigebersen were detected by flow cytometry, N=4 each group. (I, J) Quantification of M1/M2 macrophage activation detected by flow cytometry. *P < 0.05 and ***P < 0.001.
Grb2 inhibitor alleviates AP in vivo
To explore the role of Grb2 in vivo, we established a classic acute pancreatitis model using caerulein and treat with the Grb2 inhibitor prexigebersen at 0h. In the Cae group, the pancreas of mice showed edema, inflammatory cell infiltration and cell necrosis. In the medium-dose prexigebersen treatment group, the edema, inflammatory cell infiltration and cell necrosis of pancreas of mice were significantly improved (Figures 5A, B). Additionally, the levels of serum amylase and lipase levels in the medium-dose Prexigebersen group were lower than those in the Cae group (Figures 5C, D). We detected serum inflammatory factors Interleukin-6 (IL-6) and Monocyte Chemoattractant Protein-1 (MCP-1) in mice, reflecting inflammation in mice. IL-6 and MCP-1 were significantly increased in AP mice. Grb2 Inhibitor prexigebersen reduces serum IL-6 and MCP-1 levels (Figure 5D). We further verified in another PDL model, and the results showed that Grb2 inhibitor prexigebersen also alleviated the severity of PDL-induced acute pancreatitis (Supplementary Figures S3A-C). All these results indicate that the Grb2 inhibitor prexigebersen alleviates AP in vivo.
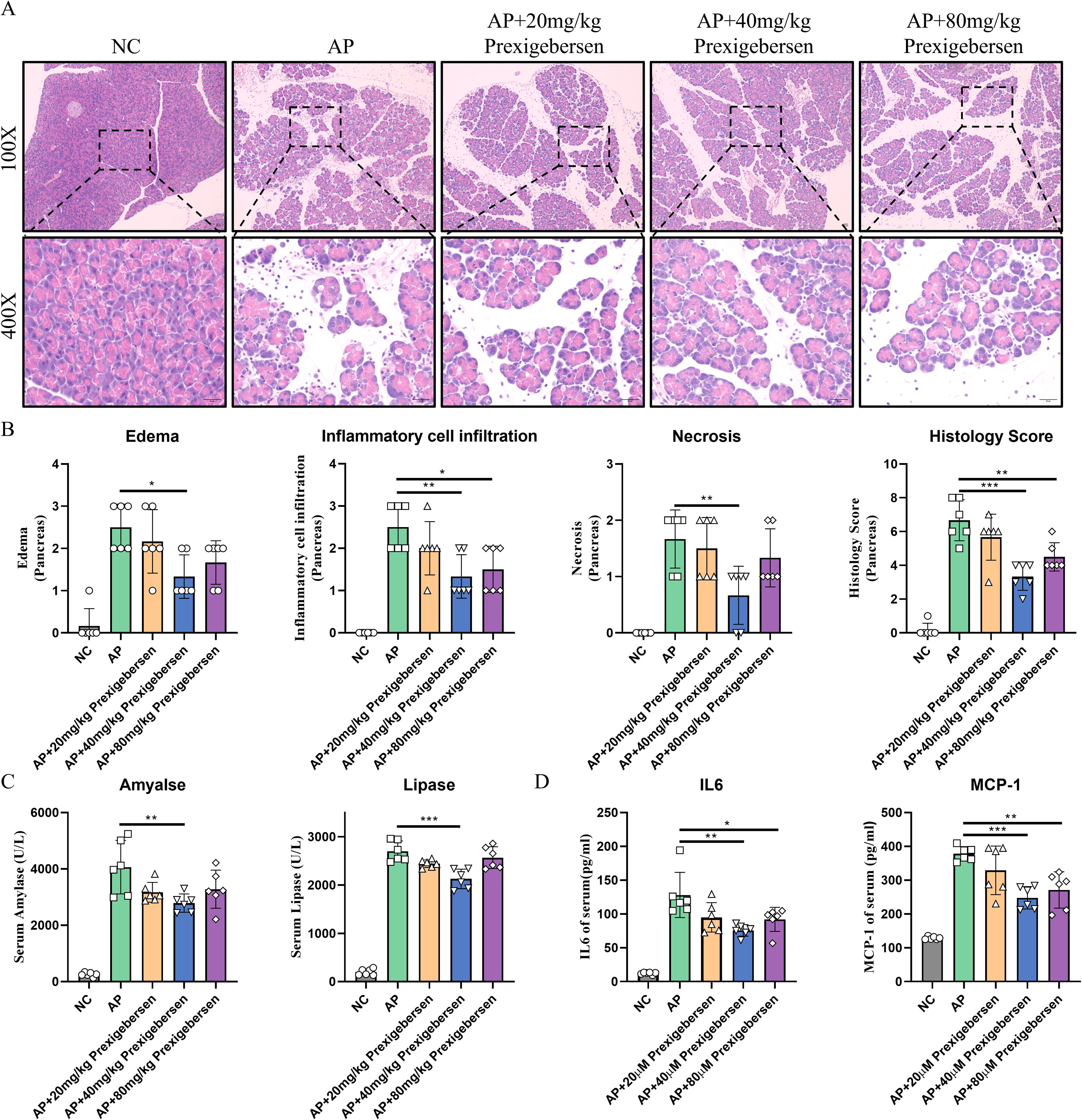
Figure 5. Grb2 inhibitor alleviates Cae-induced AP. (A) Representative HE staining of pancreatic tissues in magnifications 100x and 400x. Scale Bar = 50μM. (B) The pathological scores of pancreatic tissues. (C) Serum levels of amylase and lipase, N=6 each group. (D) The serum levels of IL-6, MCP-1 were detected by ELISA. N= 6 each group. *P < 0.05, **P < 0.01 and ***P < 0.001.
Grb2 inhibitor inhibits Nlrp3 and NF-kB in macrophages
To further investigate the downstream mechanism of Grb2, we discovered that Nlrp3 and NF-kB (Rela) were significantly up-regulated in the NOD-like receptor signaling pathway (Figure 6A). Recently, increasing evidence has suggested that Nlrp3 and NF-kB play a crucial role in M1 macrophage activation (42–45). We observed that Nlrp3 and NF-kB were upregulated during M1 macrophage activation in vitro. The Grb2 inhibitor prexigebersen reversed the upregulation of Nlrp3 and NF-kB (Figures 6B–D). Consequently, we performed immunofluorescence analysis of pancreas of AP mice. The results demonstrated that Nlrp3 and NF-kB were up-regulated in macrophages during AP, and Grb2 inhibitor prexigebersen inhibited the expression of Nlrp3 and NF-KB in macrophage (Figures 6E, F). The above results indicate that Grb2 may influence M1 macrophage activation by regulating Nlrp3 and NF-kB.
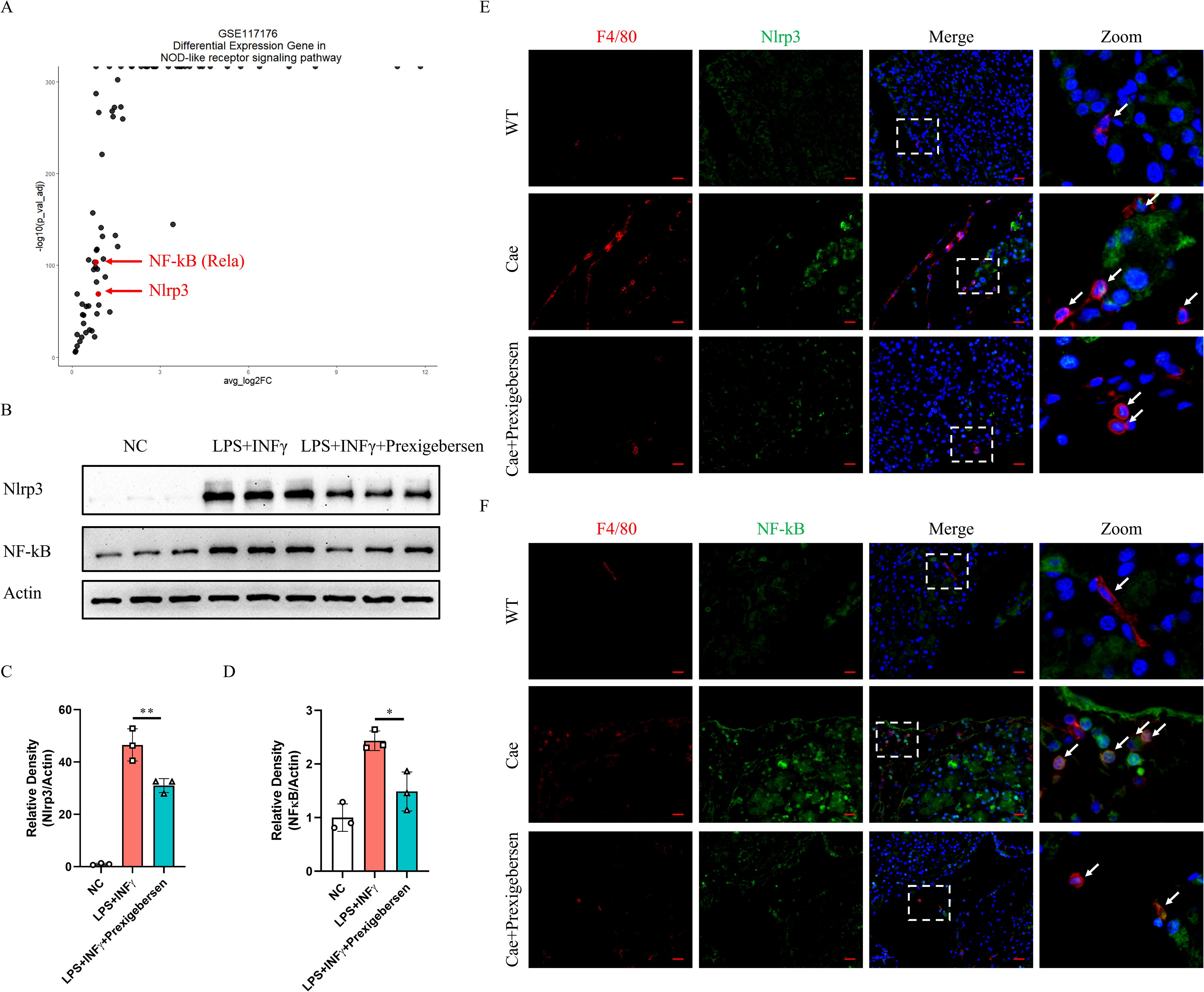
Figure 6. Nlrp3 and NF-κB were inhibited by the Grb2 inhibitor in macrophages of AP mice. (A) The volcano plot of Differential Expression Gene in NOD-like receptor signaling pathway of GSE117176. (B) Protein levels of Nlrp3 and NF-kB in M1 macrophage activation were analyzed by western blotting. (C, D) Relative density of Nlrp3 and NF-kB. Beta-actin was used as control for protein loading, N=3 each group. *P < 0.05, **P < 0.01. (E) Representative Immunofluorescence images for Nlrp3 and F4/80 expression in macrophage of AP mice. (F) Representative Immunofluorescence images for NF-kB and F4/80 expression in macrophage of AP mice. Scale Bar = 20μM.
Discussion
It is widely acknowledged that macrophages are one of essential cells during AP inflammation (9, 46–48). Through joint analysis of multiple RNA-seq and scRNA-seq databases, we discovered that Grb2 was highly expressed in macrophages during AP. Researchers have found that inhibition of Grb2 repolarized alveolar macrophages from M1 to M2 phenotype (49). Consequently, we hypothesize that Grb2 may play a role in AP by regulating macrophage activation.
We discovered up-regulation of Grb2 in M1 macrophage activation through single-cell RNA-sequence analysis, whereas there was no significant alteration in M2. The result was validated by western blot. Subsequently, we further investigated the function of Grb2. Inhibition of Grb2 hindered M1 macrophage activation, but had no impact on M2. M1 macrophage activation affects AP. Inhibition of M1 macrophage activation can effectively mitigate AP (42, 50, 51). To study the effect of Grb2 in the pathological process of AP, we employed the Grb2 inhibitor prexigebersen to treat caerulein-induced AP mice and found that prexigebersen alleviated the severity of AP and decreased the inflammatory response in AP mice.
A number of studies have demonstrated that Nlrp3 and NF-kB play a crucial role in M1 macrophage activation (52–55). We discovered that Nlrp3 and NF-kB were up-regulated in M1 macrophage activation. Both in vitro and in vivo experiments have proved that Nlrp3 and NF-kB were significantly up-regulated in M1 macrophage activation or macrophages of AP group, and Grb2 inhibitor prexigebersen inhibited the expression of Nlrp3 and NF-kB in macrophages. In summary, Inhibition of Grb2 can alleviate AP by down-regulating Nlrp3 and NF-kB of macrophage.
Conclusion
Overall, our study firstly demonstrates that inhibition of Grb2 can effectively prevent M1 macrophage activation and alleviate AP. Grb2 may potentially be an effective target of macrophage activation for the treatment of AP.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was approved by Laboratory Animal Ethics Committee of Yangzhou University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
QS: Validation, Visualization, Writing – original draft, Writing – review & editing. SW: Validation, Writing – original draft, Writing – review & editing. KW: Validation, Writing – original draft, Writing – review & editing. LW: Visualization, Writing – review & editing. WG: Supervision, Writing – review & editing. GL: Funding acquisition, Supervision, Writing – review & editing. WC: Funding acquisition, Supervision, Writing – review & editing. CY: Funding acquisition, Supervision, Writing – review & editing. BT: Supervision, Writing – review & editing. WL: Supervision, Writing – review & editing. YW: Conceptualization, Funding acquisition, Supervision, Writing – review & editing. WY: Conceptualization, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by the National Natural Science Foundation of China (No. 82270680, 82200721, 82300732); Jiangsu Funds for Distinguished Young Scholars (No. BK20240028); Policy guidance program (International science and technology cooperation) project of Yangzhou (No. YZ2023265); Yangzhou medical key disciplines incubation program (No. YZYXZDXK-11); Suzhou Key Laboratory of Digestive diseases of integrated Traditional Chinese and Western medicine (No. SZS2023001).
Acknowledgments
We are grateful to all the funds that have supported our research.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1575880/full#supplementary-material
Supplementary Table 1 | Reagent table.
Supplementary Table 2 | Antibody table
Supplementary Table 3 | The list of genes for pathways of GSE65146
Supplementary Table 4 | The list of genes for pathways of GSE109227
Supplementary Table 5 | The list of genes for cluster of STEM clustering analysis
Supplementary Table 6 | Markers for identifying cell types.
Supplementary Figure 1 | GRB2 is upregulated in PBMC of AP patients. (A) Protein levels of GRB2 in PBMC were analyzed by western blotting. (B) Relative density of Grb2.Beta-actin was used as control for protein loading, N=4 each group. *P < 0.05.
Supplementary Figure 2 | Grb2 is not associated with M2 marker. (A) The Spearman correlation between Chil3 and Grb2. (B) The Spearman correlation between Arg1 and Grb2. (C) The Spearman correlation between Retnla and Grb2. (D) The Spearman correlation between Mgl2 and Grb2.
Supplementary Figure 3 | Grb2 inhibitor alleviates PDL-induced AP. (A) Representative HE staining of pancreatic tissues in magnifications 100x and 400x. Scale Bar = 50μM. (B) Serum levels of amylase and lipase. (C) The pathological scores of pancreatic tissues. N=6 each group. N= 6 each group. *P < 0.05, **P < 0.01.
References
1. Trikudanathan G, Yazici C, Evans Phillips A, and Forsmark CE. Diagnosis and management of acute pancreatitis. Gastroenterology. (2024) 167:673–88. doi: 10.1053/j.gastro.2024.02.052
2. Zhou Y, Huang X, Jin Y, Qiu M, Ambe PC, Basharat Z, et al. The role of mitochondrial damage-associated molecular patterns in acute pancreatitis. BioMed Pharmacother. (2024) 175:116690. doi: 10.1016/j.biopha.2024.116690
3. Aney KJ, Jeong WJ, Vallejo AF, Burdziak C, Chen E, Wang A, et al. Novel approach for pancreas transcriptomics reveals the cellular landscape in homeostasis and acute pancreatitis. Gastroenterology. (2024) 166(6):1100–13. doi: 10.1053/j.gastro.2024.01.043
4. Fang Z, Li J, Cao F, and Li F. Integration of scRNA-seq and bulk RNA-seq reveals molecular characterization of the immune microenvironment in acute pancreatitis. Biomolecules. (2022) 13:78. doi: 10.3390/biom13010078
5. Zhu Q, Yuan C, Wang D, Tu B, Chen W, Dong X, et al. The TRIM28/miR133a/CD47 axis acts as a potential therapeutic target in pancreatic necrosis by impairing efferocytosis. Mol Ther. (2024) 32(9):3025–41. doi: 10.1016/j.ymthe.2024.06.005
6. Manohar M, Jones EK, Rubin SJS, Subrahmanyam PB, Swaminathan G, Mikhail D, et al. Novel circulating and tissue monocytes as well as macrophages in pancreatitis and recovery. Gastroenterology. (2021) 161(6):2014–2029.e14. doi: 10.1053/j.gastro.2021.08.033
7. Han F, Ding ZF, Shi XL, Zhu QT, Shen QH, Xu XM, et al. Irisin inhibits neutrophil extracellular traps formation and protects against acute pancreatitis in mice. Redox Biol. (2023) 64:102787. doi: 10.1016/j.redox.2023.102787
8. Li G, Liu L, Lu T, Sui Y, Zhang C, Wang Y, et al. Gut microbiota aggravates neutrophil extracellular traps-induced pancreatic injury in hypertriglyceridemic pancreatitis. Nat Commun. (2023) 14(1):6179. doi: 10.1038/s41467-023-41950-y
9. Wu J, Zhang L, Shi J, He R, Yang W, Habtezion A, et al. Macrophage phenotypic switch orchestrates the inflammation and repair/regeneration following acute pancreatitis injury. EBioMedicine. (2020) 58:102920. doi: 10.1016/j.ebiom.2020.102920
10. Yang J, Tang X, Li B, and Shi J. Sphingosine 1-phosphate receptor 2 mediated early stages of pancreatic and systemic inflammatory responses via NF-kappa B activation in acute pancreatitis. Cell Commun Signal. (2022) 20:157. doi: 10.1186/s12964-022-00971-8
11. Wang D, Liu G, Meng Y, Chen H, Ye Z, and Jing J. The configuration of GRB2 in protein interaction and signal transduction. Biomolecules. (2024) 14:259. doi: 10.3390/biom14030259
12. Dias RVR, Pedro RP, Sanches MN, Moreira GC, Leite VBP, Caruso IP, et al. Unveiling metastable ensembles of GRB2 and the relevance of interdomain communication during folding. J Chem Inf Model. (2023) 63(20):6344–53. doi: 10.1021/acs.jcim.3c00955
13. Zhou J, Tu D, Peng R, Tang Y, Deng Q, Su B, et al. RNF173 suppresses RAF/MEK/ERK signaling to regulate invasion and metastasis via GRB2 ubiquitination in Hepatocellular Carcinoma. Cell Commun Signal. (2023) 21(1):224. doi: 10.1186/s12964-023-01241-x
14. Lv J, Zhang S, Wu H, Lu J, Lu Y, Wang F, et al. Deubiquitinase PSMD14 enhances hepatocellular carcinoma growth and metastasis by stabilizing GRB2. Cancer Lett. (2020) 469:22–34. doi: 10.1016/j.canlet.2019.10.025
15. Wang LN, Zhang ZT, Wang L, Wei HX, and Zhang T. TGF-β1/SH2B3 axis regulates anoikis resistance and EMT of lung cancer cells by modulating JAK2/STAT3 and SHP2/Grb2 signaling pathways. Cell Death Dis. (2022) 13(5):472. doi: 10.1038/s41419-022-04890-x
16. Hou B, Huang H, Li Y, Liang J, Xi Z, Jiang X, et al. Grb2 interacts with necrosome components and is involved in rasfonin-induced necroptosis. Cell Death Discovery. (2022) 8(1):319. doi: 10.1038/s41420-022-01106-1
17. Montero-Vergara J, Plachetta K, Kinch L, Bernhardt S, Kashyap K, Levine B, et al. GRB2 is a BECN1 interacting protein that regulates autophagy. Cell Death Dis. (2024) 15(1):14. doi: 10.1038/s41419-023-06387-7
18. Zhao H and Mao H. ERRFI1 exacerbates hepatic ischemia reperfusion injury by promoting hepatocyte apoptosis and ferroptosis in a GRB2-dependent manner. Mol Med. (2024) 30:82. doi: 10.1186/s10020-024-00837-4
19. Chen Y, Wu J, Yan H, Cheng Y, Wang Y, Yang Y, et al. Lymecycline reverses acquired EGFR-TKI resistance in non-small-cell lung cancer by targeting GRB2. Pharmacol Res. (2020) 159:105007. doi: 10.1016/j.phrs.2020.105007
20. Choi S, Hatzihristidis T, Gaud G, Dutta A, Lee J, Arya A, et al. GRB2 promotes thymocyte positive selection by facilitating THEMIS-mediated inactivation of SHP1. J Exp Med. (2023) 220(7):e20221649. doi: 10.1084/jem.20221649
21. Zhao X, Cui P, Hu G, Wang C, Jiang L, Zhao J, et al. PIP5k1β controls bone homeostasis through modulating both osteoclast and osteoblast differentiation. J Mol Cell Biol. (2020) 12(11):55–70. doi: 10.1093/jmcb/mjz028
22. Benamar M, Harb H, Chen Q, Wang M, Chan TMF, Fong J, et al. A common IL-4 receptor variant promotes asthma severity via a T(reg) cell GRB2-IL-6-Notch4 circuit. Allergy. (2022) 77:3377–87. doi: 10.1111/all.15444
23. Diwan BA, Ramakrishna G, Anderson LM, and Ramljak D. Overexpression of Grb2 in inflammatory lesions and preneoplastic foci and tumors induced by N-nitrosodimethylamine in Helicobacter hepaticus-infected and -noninfected A/J mice. Toxicol Pathol. (2000) 28:548–54. doi: 10.1177/019262330002800407
24. Li J, Chang W, Li J, Zhao X, and Li X. IL-22-mediated microRNA-124-3p/GRB2 axis regulates hyperproliferation and inflammatory response of keratinocytes in psoriasis. Arch Dermatol Res. (2025) 317:227. doi: 10.1007/s00403-024-03668-9
25. Liu X, Zhu Q, Zhang M, Yin T, Xu R, Xiao W, et al. Isoliquiritigenin ameliorates acute pancreatitis in mice via inhibition of oxidative stress and modulation of the nrf2/HO-1 pathway. Oxid Med Cell Longev. (2018) 2018:7161592. doi: 10.1155/2018/7161592
26. Norberg KJ, Nania S, Li X, Gao H, Szatmary P, Segersvärd R, et al. RCAN1 is a marker of oxidative stress, induced in acute pancreatitis. Pancreatology. (2018) 18(7):734–41. doi: 10.1016/j.pan.2018.08.005
27. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. (2015) 43(7):e47. doi: 10.1093/nar/gkv007
28. Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. . Innovation (Camb). (2021) 2(3):100141. doi: 10.1016/j.xinn.2021.100141
29. Yu G, Wang LG, Han Y, and He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. (2012) 16:284–7. doi: 10.1089/omi.2011.0118
30. Mishra AK, Rodriguez M, Torres AY, Smith M, Rodriguez A, Bond A, Morrissey MA, et al. Hyperactive Rac stimulates cannibalism of living target cells and enhances CAR-M-mediated cancer cell killing. Proc Natl Acad Sci U S A. (2023) 120(52):e2310221120. doi: 10.1073/pnas.2310221120
31. Love MI, Huber W, and Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. (2014) 15:550. doi: 10.1186/s13059-014-0550-8
32. Chen Y, Lun AT, and Smyth GK. From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Res. (2016) 5:1438. doi: 10.12688/f1000research.8987.2
33. McCarthy DJ, Chen Y, and Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. (2012) 40:4288–97. doi: 10.1093/nar/gks042
34. Robinson MD, McCarthy DJ, and Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. (2010) 26:139–40. doi: 10.1093/bioinformatics/btp616
35. Melendez E, Chondronasiou D, Mosteiro L, Martínez de Villarreal J, Fernández-Alfara M, Lynch CJ, et al. Natural killer cells act as an extrinsic barrier for in vivo reprogramming. Development. (2022) 149(8):dev200361. doi: 10.1242/dev.200361
36. Li C, Menoret A, Farragher C, Ouyang Z, Bonin C, Holvoet P, et al. Single cell transcriptomics based-MacSpectrum reveals novel macrophage activation signatures in diseases. JCI Insight. (2019) 5(10):e126453. doi: 10.1172/jci.insight.126453
37. Liu SX, Gustafson HH, Jackson DL, Pun SH, and Trapnell C. Trajectory analysis quantifies transcriptional plasticity during macrophage polarization. Sci Rep. (2020) 10:12273. doi: 10.1038/s41598-020-68766-w
38. Hao Y, Stuart T, Kowalski MH, Choudhary S, Hoffman P, Hartman A, et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat Biotechnol. (2024) 42(2):293–304. doi: 10.1038/s41587-023-01767-y
39. Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. (2019) 16(12):1289–96. doi: 10.1038/s41592-019-0619-0
40. Ernst J, Nau GJ, and Bar-Joseph Z. Clustering short time series gene expression data. Bioinformatics. (2005) 21 Suppl 1:i159–68. doi: 10.1093/bioinformatics/bti1022
41. Ernst J and Bar-Joseph Z. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinf. (2006) 7:191. doi: 10.1186/1471-2105-7-191
42. Han X, Ni J, Wu Z, Wu J, Li B, Ye X, et al. Myeloid-specific dopamine D(2) receptor signalling controls inflammation in acute pancreatitis via inhibiting M1 macrophage. Br J Pharmacol. (2020) 177(13):2991–3008. doi: 10.1111/bph.v177.13
43. He R, Li Y, Han C, Lin R, Qian W, and Hou X. L-Fucose ameliorates DSS-induced acute colitis via inhibiting macrophage M1 polarization and inhibiting NLRP3 inflammasome and NF-kB activation. Int Immunopharmacol. (2019) 73:379–88. doi: 10.1016/j.intimp.2019.05.013
44. Fang C, Zhong R, Lu S, Yu G, Liu Z, Yan C, et al. TREM2 promotes macrophage polarization from M1 to M2 and suppresses osteoarthritis through the NF-κB/CXCL3 axis. Int J Biol Sci. (2024) 20(6):1992–2007. doi: 10.7150/ijbs.91519
45. Zhang S, Hou B, Xu A, Wen Y, Zhu X, Cai W, et al. Ganlu formula ethyl acetate extract (GLEE) blocked the development of experimental arthritis by inhibiting NLRP3 activation and reducing M1 type macrophage polarization. J Ethnopharmacol. (2024) 332:118377. doi: 10.1016/j.jep.2024.118377
46. Zhang L, Shi J, Du D, Niu N, Liu S, Yang X, et al. Ketogenesis acts as an endogenous protective programme to restrain inflammatory macrophage activation during acute pancreatitis. EBioMedicine. (2022) 78:103959. doi: 10.1016/j.ebiom.2022.103959
47. Peng C, Tu G, Wang J, Wang Y, Wu P, Yu L, et al. MLKL signaling regulates macrophage polarization in acute pancreatitis through CXCL10. Cell Death Dis. (2023) 14(2):155. doi: 10.1038/s41419-023-05655-w
48. Wang LJ, Jin YL, Pei WL, Li JC, Zhang RL, Wang JJ, et al. Amuc_1100 pretreatment alleviates acute pancreatitis in a mouse model through regulating gut microbiota and inhibiting inflammatory infiltration. Acta Pharmacol Sin. (2024) 45(3):570–80. doi: 10.1038/s41401-023-01186-4
49. Wang Q, Hong L, Chen M, Shi J, Lin X, Huang L, et al. Targeting M2 Macrophages Alleviates Airway Inflammation and Remodeling in Asthmatic Mice via miR-378a-3p/GRB2 Pathway. Front Mol Biosci. (2021) 8:717969. doi: 10.3389/fmolb.2021.717969
50. Tang DS, Cao F, Yan CS, Cui JT, Guo XY, Cheng L, et al. Acinar cell-derived extracellular vesicle miRNA-183-5p aggravates acute pancreatitis by promoting M1 macrophage polarization through downregulation of foxO1. Front Immunol. (2022) 13:869207. doi: 10.3389/fimmu.2022.869207
51. Song H, Zhang J, Lou N, Jiang X, Cui Y, Liu J, et al. Emodin nanocapsules inhibit acute pancreatitis by regulating lipid metabolic reprogramming in macrophage polarization. Phytomedicine. (2024) 130:155763. doi: 10.1016/j.phymed.2024.155763
52. Sun J, Zeng Q, Wu Z, Huang L, Sun T, Ling C, et al. Berberine inhibits NLRP3 inflammasome activation and proinflammatory macrophage M1 polarization to accelerate peripheral nerve regeneration. Neurotherapeutics. (2024) 21(4):e00347. doi: 10.1016/j.neurot.2024.e00347
53. Zhao YX, Sun YY, Li LY, Li XF, Li HD, Chen X, et al. Rab11b promotes M1-like macrophage polarization by restraining autophagic degradation of NLRP3 in alcohol-associated liver disease. Acta Pharmacol Sin. (2024) 46(1):134–46. doi: 10.1038/s41401-024-01333-5
54. Yang S, Yin Y, Sun Y, Ai D, Xia X, Xu X, et al. AZGP1 aggravates macrophage M1 polarization and pyroptosis in periodontitis. J Dent Res. (2024) 103(6):631–41. doi: 10.1177/00220345241235616
Keywords: GRB2, acute pancreatitis, macrophage activation, scRNA-seq, inflammation
Citation: Shen Q, Wang S, Wu K, Wang L, Gong W, Lu G, Chen W, Yuan C, Tu B, Li W, Wang Y and Yang W (2025) Identification of Grb2 protein as a potential mediator of macrophage activation in acute pancreatitis based on bioinformatics and experimental verification. Front. Immunol. 16:1575880. doi: 10.3389/fimmu.2025.1575880
Received: 13 February 2025; Accepted: 05 May 2025;
Published: 26 May 2025.
Edited by:
Yu-Chan Chang, National Yang Ming Chiao Tung University, TaiwanReviewed by:
Imran Mohammad, Stanford University, United StatesJieliang Zuo, Tongji University, China
Copyright © 2025 Shen, Wang, Wu, Wang, Gong, Lu, Chen, Yuan, Tu, Li, Wang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaodong Wang, ZGF5LndhbmdAbGl2ZS5jbg==; Weixuan Yang, eXd4NzgwMkAxNjMuY29t
†These authors have contributed equally to this work
 Qinhao Shen1†
Qinhao Shen1† Shuai Wang
Shuai Wang Weijuan Gong
Weijuan Gong Guotao Lu
Guotao Lu Chenchen Yuan
Chenchen Yuan Bo Tu
Bo Tu Wei Li
Wei Li Yaodong Wang
Yaodong Wang Weixuan Yang
Weixuan Yang