- 1The Laboratory of Medical Mycology, Jining No.1 People’s Hospital, Jining, Shandong, China
- 2Department of Microbiology & Immunology, Georgetown University Medical Center, Washington, DC, United States
- 3Department of pathology, Jining No.1 People’s Hospital, Jining, Shandong, China
- 4Department of Dermatology, Jining No.1 People’s Hospital, Jining, Shandong, China
Pentraxin-3 (PTX3) is a soluble pattern recognition molecule (PRM) characterized by a C-terminal pentraxin structural domain and a unique N-terminal structural domain. As a key component of the innate immune system, PTX3 can be rapidly released into the extracellular space during microbial invasion and inflammatory responses. It plays a crucial role in regulating complement activation, enhancing the ability of myeloid cells to recognize pathogens, and exerting various immune effects. PTX3 is integral to the regulation of innate immunity, inflammation, and tumor dynamics through its dual function as both a pro-inflammatory and anti-inflammatory mediator depending on the context. This role is closely linked to its diverse molecular and cellular targets. Additionally, PTX3 has been implicated in the pathogenesis of various lung diseases through its involvement in numerous physiological and pathological processes. In this paper, we summarize the complex immunological functions of PTX3 and review the multifaceted roles it plays in the development of infectious lung diseases. Our objective is to highlight the potential for clinical targeting of PTX3 as a biomarker in infectious diseases and to propose it as a viable alternative in future therapeutic strategies.
1 Introduction
Lung inflammation caused by various infectious pathogens-including bacteria, fungi, viruses, and atypical organisms-is a common condition affecting the alveoli, distal airways, and interstitium (1). The clinical outcomes of such infections vary widely, depending on the virulence factors of the pathogens and the host’s immune status. These outcomes can range from manageable conditions in otherwise healthy individuals to severe cases with high mortality rates, particularly among critically ill patients or high-risk groups (2). The lungs possess several defense mechanisms to protect the host from pathogens, with innate immunity playing a pivotal role. A key component of this defense involves soluble pattern recognition molecules (PRMs), which are crucial for recognizing and responding to microbial threats (3). However, overactivation of these molecules can lead to tissue damage, exacerbating lung inflammation.
Pentraxin 3 (PTX3), a key component of humoral immunity, is rapidly produced and released by macrophages, dendritic cells (DC), fibroblasts, and activated endothelial cells at sites of infection or inflammation (4, 5). The production PTX3 is regulated by various inflammatory signal molecules, such as IL-1β and TNF-α, as well as Toll-like receptor (TLR) agonists, and microbial components like lipopolysaccharides (LPS), lipoarabinomannan (LAM), and outer membrane proteins (6). These microbial components, which stimulate PTX3 production, are derived from diverse bacteria like Pseudomonas aeruginosa (P. aeruginosa), Shigella flexneri (S. flexneri), and urinary tract pathogenic Escherichia coli (UPEC); fungi like Aspergillus fumigatus (A. fumigatus); and viruses like the influenza virus (7).
While PTX3 enhances the body’s defense to clear these pathogens, it also plays an indispensable role in regulating inflammation, tissue remodeling, and anti-tumor responses (8–10). In the context of pulmonary infectious diseases, PTX3 not only promotes the clearance of pathogens by phagocytes but also activates mucosal immune responses against infectious agents. PTX3 thus serves multiple essential functions in respiratory diseases, facilitating both host defense and the regulation of inflammatory processes. In this paper, we summarized the structure, immune role of PTX3, as well as its mechanism in lung infection caused by three main pathogens: bacteria, fungi and viruses. Additionally, we discuss the potential of PTX3 modulators in manage lung infection.
2 PTX3 and its immune function
2.1 The structure of PTX3
The pentraxin superfamily is characterized by structural domains that are evolutionarily conserved. Based on the length of thei N-terminal regions, pentraxins are categorized into long-chain pentraxins (> 45 kDa) and short-chain pentraxins (25 kDa) (11). Short-chain proteins, including C-reactive protein (CRP) and serum amyloid P component (SAP), are predominately produced by hepatocytes and act as important mediators in regulation of the human immune system (12).
Long-chain pentraxins include Pentraxin 3 (PTX3), Pentraxin 4 (PTX4), neuronal pentraxin 1 (NP1), and neuronal pentraxin 2 (NP2). PTX3 is encoded by three exons, which correspond to a targeting signal peptide, a C-terminal structural domain homologous to classical short pentraxin, and a unique, extended long N-terminal structural pentraxin domain (13, 14). The C-terminal structural domain contains a single N-glycosylation site, crucial for regulating the biological function of the protein in inflammation and innate immunity (15, 16). The N-terminal structural domain contains a convoluted helix and an intrinsically disordered region (17). In addition, the proximal promoter region of the PTX3 gene contains several potential transcription factor binding sites, including specificity protein 1 (Sp1), transcription factor Pu. 1, activator protein-1 (AP-1), nuclear factor of IL-6 (NF-IL-6), and NF-κB (18). The specificity and complexity of PTX3 structure are closely linked to its versatility as a molecule with broad molecular and cellular targets.
2.2 Genetic variant of PTX3 and their association with Lung pathogens
The PTX3 gene is located in the q25 region of human chromosome 3 and comprises three exons separated by two introns. The first two exons encode the signal peptide and N-terminal domain, and the third exon encodes the C-terminal pentraxin domain, as shown in Figure 1 (19). Multiple single nucleotide polymorphisms (SNPs) in PTX3 have been identified (20), and these are strongly associated with susceptibility to, and the progression of, infectious and neoplastic diseases (21, 22). Among them, the PTX3 gene variant rs2120243, located at a transcription factor binding site, influences transcriptional regulation, thereby affecting circulating plasma PTX3 levels (21, 23).
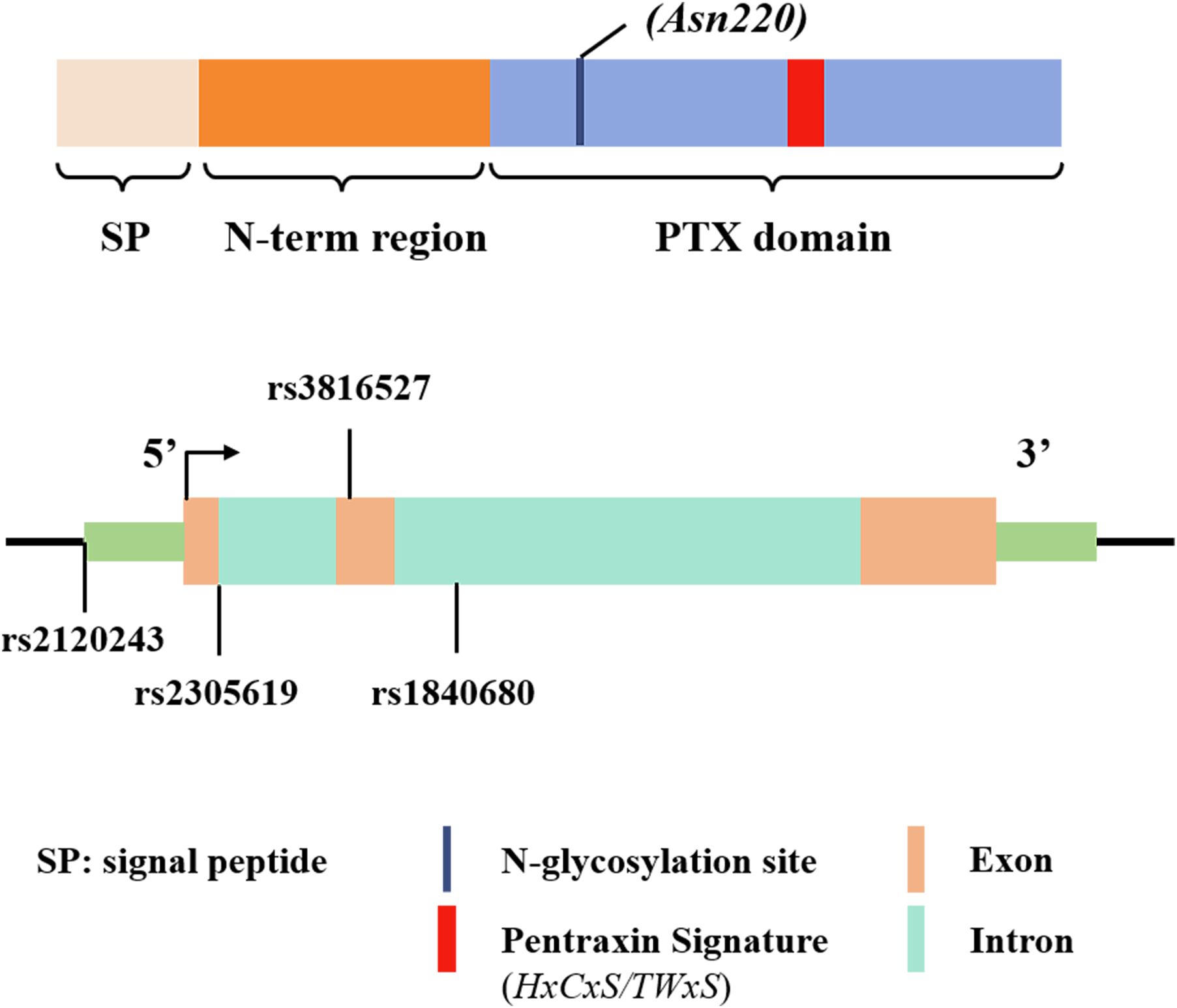
Figure 1. Schematic representation of the gene and protein structure of PTX3, including the approximate locations of the single nucleotide polymorphisms (SNPs) found in various infections. Exons are depicted as solid rectangles, while introns are hollow rectangles.
Previous studies have also shown that two intronic variants of the PTX3 SNPs —rs2305619 and rs1840680 —affect PTX3 function (21). The A allele of these variants is linked to elevated plasma PTX3 levels (23) and increased susceptibility to clinical conditions such as infections caused by P. aeruginosa and A. fumigatus (22, 24, 25). In addition, rs3816527is a non-synonymous mutation located on exon 2. This variant affects the function of PTX3 by interfering the N-terminal-mediated PTX3 binding to its ligand (26). These studies emphasize the significance of PTX3 genetic variants in modulating gene expression and protein levels. The detection of PTX3 gene polymorphisms thus may help predict the risk of infection by a variety of pathogens and provide insights on disease susceptibility.
2.3 PTX3 in innate and adaptive immune response to infections
PTX3 functions as a soluble pattern recognition receptor and “antibody precursor”, recognizing exogenous microorganisms and modulating the inflammatory responses. - PTX3 binds wide range of ligands, including complement components, growth factors, adhesion molecules (P-selectins), and extracellular matrix glycoproteins, thereby coordinating a interaction between innate immunity and inflammation (27). The complement system, comprising over 30 synergistic soluble proteins, membrane-bound proteins, and complement receptors such as C1q, ficolin, and factor H, is a highly conserved component of the innate immune system. PTX3 exhibits specific binding to these complement components, playing a crucial role in the inflammatory response.
PTX3 closely interacts with three pathways of complement system, generating distinct context-dependent results (Figure 2) (28). In the classical pathway (CP), PTX3 binds to complement component C1q (29) via its globular structural domain. This binding promotes the C3 and C4 deposition, facilitating complement activation on pathogens. Notably, this interaction does not require protein aggregation or the presence of calcium ions (27). However, pre-incubation of PTX3 with C1q in the liquid phase inhibits complement activation by blocking the C1q interaction with immunoglobulins. In the Lectin Pathway (LP), PTX3 directly interacts with ficolin-1, ficolin-2, and mannose-binding lectin (MBL). The Ficolin-1 bound PTX3 promotes the deposition of C4b (30) while the ficolin-2-mediated complement deposition has been observed during A. fumigatus infection (31). MBL binds PTX3 and SAP through its collagen-like structural domain, forming an MBL-PTX3 complex that not only recruits C1q, but also enhances C3 and C4 deposition, as well as the phagocytosis of C. albicans by polymorphonuclear leukocytes. This MBL- PTX3 complex interacts with both the Lectin and Classical Pathways, amplifying complement activation (32). Furthermore, PTX3 is a unique ligand for Factor H (FH), which recruits FH by recognizing two FH binding sites, SCR7 and SCR19-20. This interaction prevents complement over-activation by downregulating the Alternative Pathway (AP) of complement (29).
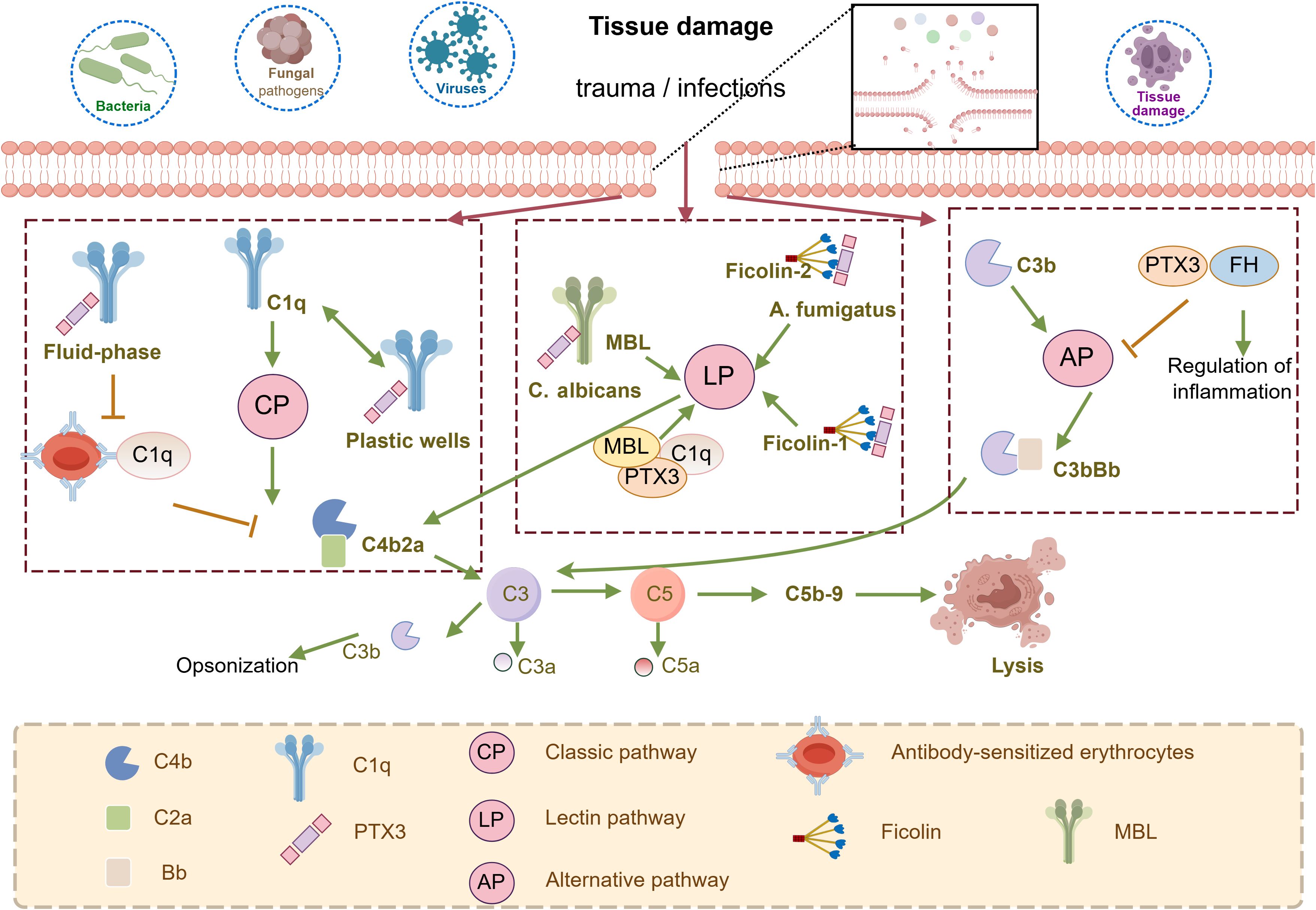
Figure 2. Schematic representation of the involvement of PTX3 in the regulation of complement activity. PTX3 can activate the classical complement pathway (CP) by binding to the C1q globular domain under specific conditions. PTX3 is also involved in the regulation of the lectin pathway (LP) through interactions with MBL and Ficolins. In addition, PTX3 recognizes regulators of complement activation, in particular the most important regulator of the alternative pathway (AP), FH, is involved in the regulation of AP. they trigger the hydrolysis of C3 via C3 convertases (C4b2a for CP and LP, C3bBb for AP) to form C3a, C3b, and C5a, followed by the formation of C5b-9, the membrane attack complex that leads to cell lysis. This figure is drawn by Figdraw.
In addition to its involvement in complement activation, PTX3 is involved in the recruitment of inflammatory cells. PTX3 has been reported to have significant pro-inflammatory effects, since silencing PTX3 inhibits LPS-induced expression of IL-6, IL-8, IL-1β, and Mucin 5AC (MUCSAC) through suppression of the PI3K/AKT pathway (33). In microglioma cells BV-2, PTX3 knockdown markedly reduces the expression of β-amyloid-induced cytokines TNF, IL-1β, IL-6, iNOS, and CD86 and COX-2 (34).
Interestingly, PTX3 also exerts anti-inflammatory effects under certain conditions. In a model of LPS-induced acute lung injury, PTX3 deficiency significantly exacerbates the lung injury, accompanied by increased neutrophil infiltration and elevated plasma levels of TNF-α and monocyte chemotactic protein-1 (MCP-1) (35). Similarly, exogenous PTX3 stimulation in human PBMCs increased the production of the anti-inflammatory cytokine IL-10, without affecting production of pro-inflammatory cytokines IL-1β, IL-6, and TNF-α (36). Beyond its direct induction of inflammatory cytokines, PTX3 also modulates inflammation-related signaling pathways. In A. fumigatus infection, PTX3 binds to myeloid differentiation protein 2 (MD-2) in vitro and exerts its protective antifungal activity in vivo by activating the anti-inflammatory TLR4/TRIF signaling pathway (37).
This dual immune effects of PTX3 suggests its critical function in regulating antimicrobial activity and controlling immunopathogenesis.
3 Role of PTX-3 in bacterial infections of the lungs
PTX3 plays a protective role in bacterial lung infections through multiple mechanisms, including enhancing pathogen recognition, promoting phagocytosis, and modulating inflammatory responses. Additionally, PTX3 serves as a valuable biomarker for assessing disease severity and prognosis. Below we highlight a few typical bacterial infection examples.
3.1 Pseudomonas aeruginosa
Pseudomonas aeruginosa is a common lung colonizer that can cause acute and chronic respiratory infections, particularly in patients with underlying conditions such as cystic fibrosis (CF). Several studies have demonstrated that P. aeruginosa infection significantly upregulates PTX3 expression (25, 38–40). Notably, serum PTX3 levels are significantly higher in CF patients compared to healthy individuals (38). Furthermore, an animal study has confirmed that PTX3-deficient mice exhibit increased susceptible to infection with laboratory strains of P. aeruginosa.
Mechanistically, PTX3 protects against P. aeruginosa infections by modulating inflammatory responses and enhancing pathogen clearance. Moalli F et al.demonstrated that PTX3 reduces lung infections in mice by decreasing levels of pro-inflammatory cytokines (e.g.,IL-1β) and chemokines (e.g., CXCL1, CXCL2, and CCL2), as well as by reducing airway neutrophil recruitment in a mouse model infected with the clinical strain RP73, isolated from CF patients (41). PTX3 enhances regulatory phagocytosis in a complement- and Fc gamma receptor (FCGR)-dependent manner. Genetic variations in PTX3 also play a role in P. aeruginosa colonization. Chiarini M et al. (25)found significant differences in the frequency of PTX3 haplotypes among 172 Caucasian CF patients, suggesting that genetic variants in PTX3 influence the lung colonization of P. aeruginosa in CF patients.
Heat shock proteins (HSP)70 chaperone proteins, conserved across prokaryotic and eukaryotic cells, induced under various stresses and infections, have been implicated in PTX3 regulation (42). DnaK,a homologue of HSP70, was shown to stimulate PTX3 expression through the NF-κB and extracellular signal-regulated kinase (ERK) signaling pathways. Notably, ERK activation is negatively regulated by NF-κB, indicating crosstalk between these pathways. The same study suggests that P. aeruginosa DnaK acts as a pathogen-associated molecular pattern that enhances host defense responses by promoting PTX3 production (43).
3.2 Klebsiella pneumoniae
Nosocomial Klebsiella pneumoniae (K. pneumoniae) is a leading cause of community-acquired pneumonia, particularly in individuals with impaired lung defenses and functions (44, 45). Similar to other bacterial infections, PTX3 levels have been found to correlate with the progression of K. pneumoniae infection. Using a PTX3 transgenic mouse model, Soares et al. (46) demonstrated that PTX3 exerts dual effects during lung infection. In mice receiving low-dose bacterial inoculation, PTX3 overexpression led to increased TNF-α expression and neutrophil infiltration, which significantly reduced bacterial loads and enhanced resistance to respiratory infections. Conversely, in mice challenging with high-dose inoculations, PTX3 overexpression was associated with increased lethality. This highlights that the effects of PTX3 are dose-dependent and influenced by the severity of the inflammatory response, suggesting a fine balance between protective and detrimental outcomes in K. pneumoniae infections.
Using PTX3-deficient mice, PTX3’s role in innate immunity against K. pneumoniae was investigated in a recent study. Consistent with previous findings, local and systemic PTX3 expression was induced following lung infection, accompanied by increased TNF-α and IL-1β levels. However, unlike in P. aeruginosa infections, PTX3 did not directly interact with K. pneumoniae or promote conditioned phagocytosis. This discrepancy may be contributed to the pathogen’s escape mechanisms, such as polysaccharide capsule of K. pneumoniae (47).
Analysis of the susceptibility of wild-type, PTX3-deficient, complement C3-deficient and Ptx3-/-/C3-/- mice to infection revealed that PTX3 exerts its protective effects against K. pneumoniae in a complement-independent manner. Histopathological analysis of the infected lungs showed increased fibrin deposition and greater consumption of circulating fibrinogen in the lungs. These findings suggest that PTX3 controls K. pneumoniae infection primarily by modulating the inflammatory response and tissue damage (48).
3.3 Streptococcus pneumoniae
Streptococcus pneumoniae(S. pneumoniae)is a gram-positive diplococcus and a major pathogen responsible for approximately 2 million deaths annually worldwide (49). Toxigenic strains of S. pneumoniae secrete pneumolysin, a toxin that upregulates PTX3 expression in mouse epithelial cells and lungs via the JNK1/2 MAPK pathway. This interaction enhances the inflammatory response and induces the expression of inflammatory cytokines in the lungs (50).
PTX3 expression is significantly elevated in patients with bacterial infections, including chronic obstructive pulmonary disease and community-acquired pneumonia (51, 52). Notably, PTX3 levels in pleural fluid are valuable for identifying parapneumonic effusion (53). Furthermore, studies measuring PTX3 levels in the plasma of patients with bacterial infections and aseptic meningoencephalitis revealed significantly higher PTX3 levels in those with bacterial infection (54). These findings suggest that PTX3 has a potential as a biomarker for S. pneumoniae infections, providing valuable support for clinical diagnosis. However, further studies are needed to clarify the molecular mechanisms underlying PTX3-mediated inflammatory regulation during S. pneumoniae infections.
3.4 M. tuberculosis
A study investigating 70 biomarkers associated with infection and inflammation in tuberculosis patients identified PTX3 as one of seven elevated proteins linked to disease severity, smear grading, and extensive imaging lesions (55). Similarly, a separate study involving 220 tuberculosis patients demonstrated significantly higher plasma PTX3 levels in tuberculosis patients compared to healthy controls (56). Notably, PTX3 levels decreased significantly in 186 patients who were cured by the end of treatment, whereas levels rose again in all 34 patients who experienced treatment failure. Although PTX3 is not specific for diagnosing tuberculosis, its detection can help monitor disease activity and treatment efficacy.
PTX3 gene polymorphisms have also been associated with increased susceptibility to M. tuberculosis infections (57). In a study of 213 tuberculosis patients in West Africa, two SNPs—rs1840680 and rs2305619 (Figure 1) —in the PTX3 gene were found to be less frequent in the TB group compared to healthy controls, indicating that these genotypes may confer protection against tuberculosis infection (58). Additionally, exposure to the Bacillus Calmette-Guérin (BCG) vaccine was associated with increased PTX3 levels in human lung disease specimens (59), suggesting that PTX3 might play a role not only as a marker of disease state but also as a potential protective mechanism, though this mechanism remains to be fully understood.
Collectively, these studies advance our understanding of immune response due to bacterial infections and highlights PTX3 as a potential target for therapeutic strategies to control bacterial infections.
4 Role of PTX3 in fungal infections of the lungs
PTX3 plays a crucial role in immune defense, particularly innate immunity, against fungal infections in the lungs by enhancing macrophages phagocytosis, promoting complement deposition, and forming protein complexes to facilitate fungal clearance (Figure 3).
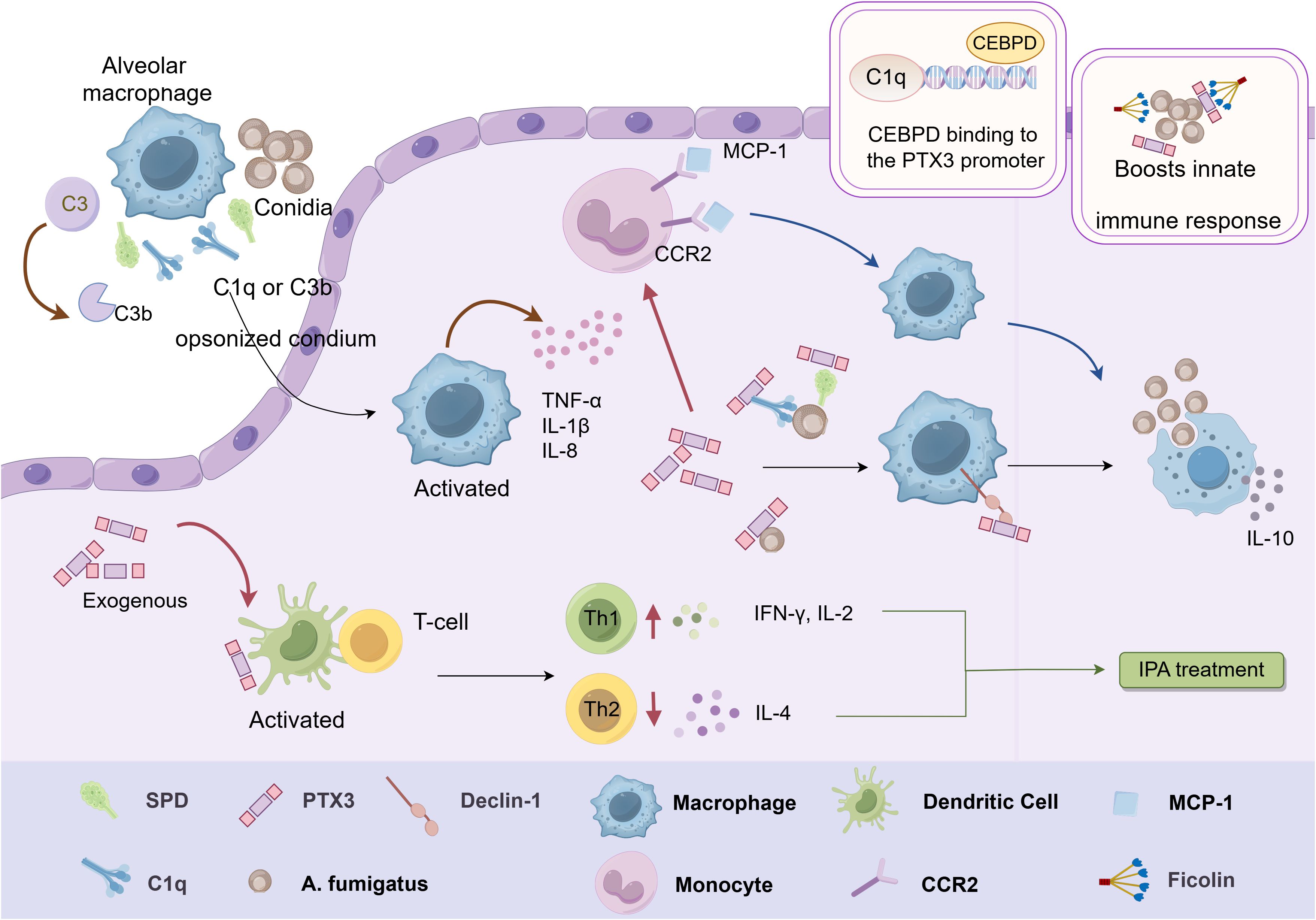
Figure 3. Schematic representation of the immunomodulatory function of PTX3 in Aspergillus. Inhaled conidia conditioned by SP-D, C1q or C3b are phagocytosed by alveolar macrophages, which stimulate the release of various cytokines from macrophages. PTX3 induces mononuclear phagocytes to release the chemokine MCP-1 in conjunction with CCR2, which potentiates the macrophage antifungal response. Exogenous PTX3 activates dendritic cells and induces an imbalance in the Th1/Th2 response, resulting in effective mitigation of IPA. PTX3 also interacts with ficolin on the surface of conidia and enhances the activation of LPs, which promotes innate immunity against A. fumigatus. In addition, CEBPD can activate PTX3 by binding to the PTX3 promoter region, thereby enhancing PTX3 expression and promoting phagocytosis by macrophages. This figure is drawn by Figdraw.
Invasive pulmonary aspergillosis (IPA) is a severe condition characterized by the dissemination of fungal hyphae in lung and surrounding tissues, with high morbidity and mortality rates (60–62). The host’s intrinsic immune system is critical for clearing inhaled Aspergillus fumigatus spores, primarily through immune cells such as macrophages, neutrophils, and dendritic cells (63). Studies have shown that PTX3-deficient mice exposed to A. fumigatus are highly susceptible to IPA, with all mice succumbing within three days. These PTX3-deficient mice displayed significantly elevated fungal loads and severe inflammatory injury in the lung. Notably, the administration of recombinant PTX3 could reverse these outcomes. Mechanistic studies revealed that PTX3 selectively recognizes and binds galactomannan (GM), a polysaccharide in A. fumigatus cell wall. This binding activates the complement system and promotes macrophage phagocytosis, thereby enhancing resistance to A. fumigatus infection (64).
Susceptibility to IPA is also associated with an imbalance in Th1/Th2 response. PTX3-deficient mice exhibited increased levels of Th2-type cytokine IL-4 and decreased levels of Th1-type cytokine IL-2 and IFN-γ. Administration of exogenous PTX3 in PTX3-deficient mice activated DCs and induced Th1 responses, effectively mitigating IPA (64). PTX3 further enhances phagocytosis by interacting with dendritic cell-associated C-type lectin-1 (Dectin-1), amplifying the immune response against A. fumigatus. Notably, blocking the Dectin-1 receptor impairs phagocytosis in PTX3 transgenic mice (65). In addition, PTX3 induces mononuclear phagocytes to release chemokine MCP-1/CCL-2, which recruits monocytes to lung tissues, enhancing antifungal responses (66).
PTX3 mitigates harmful inflammation by recognizing A. fumigatus directly or indirectly through interactions with other humoral PRMs. Among these PRMs, fibronectin (ficolin) glycoproteins play a role by binding oligosaccharides on pathogenic microorganisms and activating the complement lectin pathway (LP) (67). However, both ficolin-1 binding complex and LP activation with PTX3 were not observed during exposure to A. fumigatus conidia (68). Instead, PTX3 interacts with ficolin-2 on the conidial surface, enhancing LP activation and contributing to innate immunity against A. fumigatus (69). Although the interaction between PTX3 and ficolin-3 has been identified, its functional implications in antifungal immunity remain unclear (31). A recent study confirmed that PTX3 recognizes germinating conidia via galactosaminogalactan, a polysaccharide on the A. fumigatus cell wall. For dormant conidia, other humoral PRMs such as Surfactant Associated Protein D (SP-D) and complement proteins C1q and C3b facilitate PTX3 binding. Interestingly, PTX3 binding to A. fumigatus conidia significantly decreased pro-inflammatory cytokine/chemokine production and increased IL-10 release (70). This finding suggests a nuanced role of PTX3 in modulating inflammation by balancing immune defense with the prevention of excessive inflammatory damage.
Furthermore, PTX3 gene polymorphisms significantly influence PTX3 expression levels and the risk of IPA (71–75). Recent studies have shown that specific polymorphisms in PTX3 promoter may affect its binding with transcriptional enhancer-binding proteins (76), such as CCAAT/enhancer-binding proteins (CEBPs), influencing PTX3 transcription efficiency. It has been shown that CEBPD activates PTX3 by binding to its promoter region following A. fumigatus infection, thereby enhancing PTX3 expression and facilitating macrophage phagocytosis (77).
IPA mainly affects immunocompromised patients, including solid organ transplant recipients and those undergoing hematopoietic stem cell transplantation (78). In bone marrow transplantation patients, PTX3 promotes the accumulation of myeloid and lymphoid cells in the lung tissue, accelerating the immune function recovery. The efficacy of antifungal treatment for pulmonary aspergillosis is enhanced when PTX3 is administered alongside antifungal drugs (79). Additionally, PTX3 gene polymorphisms are associated with the incidence of IPA in hematopoietic stem cell transplant recipients, potentially due to defects in neutrophil antifungal activity.
Recent reports indicate a rising incidence of IPA in patients with chronic obstructive pulmonary disease (COPD), highlighting COPD as a significant risk factor for IPA (22, 80). Studies on PTX3 gene polymorphisms and susceptibility to Aspergillus in patients with COPD have revealed that the frequency of the rs1840680 AA genotype is significantly higher in those with comorbid IPA compared to COPD patients without IPA. Additionally, peripheral blood PTX3 levels are lower in COPD patients with the AA phenotype at the rs1840680 locus, suggesting an increased risk of IPA in this population (81). These findings support that potential of plasma PTX3 levels as a promising diagnostic indicator for IPA in COPD patients.
Although the results of the available studies confirmed the potential of PTX3 as a biomarker for early diagnosis of IPA in non-neutropenic patients and COPD combined with A. fumigatus infection,. studies on the correlation between PTX3 and the prognosis of IPA are still limited. The prognostic value of PTX3 in pulmonary aspergillosis needs to be further explored.
5 The role of PTX3 in viral infections
Studies also confirmed that PTX3 plays a vital role in the immune response to variable viruses, including SARS-CoV-2, H3N2 influenza, and human cytomegalovirus, through activating complement cascades and promoting pathogen recognition by myeloid cells.
5.1 The role of PTX3 in SARS-CoV-2 lung infection
SARS-CoV-2, the virus responsible for the COVID-19 pandemic, elicits a robust immune response of the innate immune system (82, 83). Several studies have shown PTX3 levels as a biomarker for predicting disease severity and mortality risk in COVID-19 patients (57, 84, 85). Immunohistochemical analysis of autopsy samples from COVID-19 lungs revealed high PTX3 expression in bone marrow monocytes and endothelial cells. Plasma PTX3 level was even identified as a significant independent predictor of 28-day mortality in hospitalized patients, demonstrating superior over traditional inflammatory markers (85).
A prospective cohort study also confirmed that plasma PTX3 effectively predicts the risk of respiratory failure and death within 30 days for COVID-19 patients, regardless of treatment with remdesivir or dexamethasone (86). Additionally, PTX3 has been identified as a low-cost/low-tech early biomarker for detecting secondary bacterial or fungal infections in COVID-19 patients (87), aiding in identification of individuals at high risk for community- or hospital-acquired infections and guiding an early antimicrobial therapy.
Mechanistically, PTX3 binds specifically to the nucleocapsid protein (NP) of SARS-CoV-2 through its N-terminal structural domain in a dose-dependent manner. However, it does not exhibit direct anti-SARS-CoV-2 activity. Whether PTX3 participates in nucleocapsid-mediated complement activation and cytokine production remains to be elucidated (81). SNP analysis revealed that individuals with the rs1840680 (1449A/G) AG genotype (Figure 1) have greater protection against severe COVID-19 compared to those with the AA genotype. Additionally, PTX3 level was positively correlated with the expression of inflammatory cytokines such as IL-6, IL-8, and IL-10, suggesting a role of PTX3 in modulating the complement system and inflammatory cytokines (88). Differences in PTX3 expression were observed between deteriorating and recovering COVID-19 patients, with associated changes in the JAK-STAT, NF-κB, and MAPK signaling pathways, all contributing to inflammatory responses (89). These findings demonstrate the critical role of PTX3 in the immunopathology of COVID-19, however, the specific mechanism of action remains unclear and further studies are needed.
5.2 The role of PTX3 in influenza lung infection
Respiratory influenza viruses are one of the most common human pathogens and are responsible for seasonal morbidity and mortality worldwide. PTX3, a soluble pattern-recognition receptor, exhibits potent antiviral activity against influenza virus both in vivo and ex vivo, mediating a range of antiviral responses. These responses include inhibition of hemagglutination, neutralization of the virus and inhibition of viral neuraminidase activity. Studies have demonstrated that PTX3 is rapidly upregulated following H3N2 influenza infection. It binds to viral hemagglutinin and prevents viral attachment to host cells, thereby inhibiting viral replication in the lung. Animal models further confirm PTX3’s importance since PTX3 knockout mice are more susceptible to the virus (90).
PTX3 also participates in neutralization of influenza A viruses (IAV). The anti-IAV activity is achieved by increasing salivary acids that can neutralize the hemagglutinin (HA) glycoprotein (91). Studies have revealed that the specificity of HA and viral neuraminidase for particular sialic acid linkages influences the susceptibility of H1N1, H3N2, and H7N9 strains to PTX3’s antiviral effects, suggesting a variable effectiveness of PTX3 to different IAV subtypes (92).
5.3 The role of PTX3 in human cytomegalovirus lung infection and other viruses
Human cytomegalovirus (HCMV), a member of the Herpesviridae family, is a ubiquitous opportunistic pathogen capable of entering a latent phase after the clearance of primary infection. It often reactivates when the body’s immunity is suppressed (93–95). PTX3 inhibits the activity of HCMV virus upon binding to hemagglutinin on viral surface and reduces the invasiveness and infectivity of the virus in vitro. This mechanism appears to be mediated by TLR9/MyD88-independent viral recognition that activates interferon regulatory factor 3 (IRF3) in DCs and promotes IL-12/IFN-dependent immune pathways. In CMV-infected mice, exogenous PTX3 treatment significant reduced lung viral titers and virus-induced inflammatory cell infiltration while mitigating lung parenchymal destruction (96).
Clinical studies have also highlighted the role of PTX3 gene polymorphisms in the context of HCMV infection. For instance, patients receiving allogeneic transplants, such as stem cell and kidney transplants (97, 98), especially exhibit an increased risk of CMV reactivation if their donors carry PTX3 haplotypes such as h2/h2 (rs2305619, rs3816527) (Figure 1). Genetic variation in donor PTX3 in patients undergoing hematopoietic stem cell transplantation thus has the prognostic value to predicate the risk of CMV reactivation (97).
Using PTX3 knockout mice infected with Mouse Hepatitis Virus-1 (MHV-1) has demonstrates that PTX3 is essential for modulating recruitment of neutrophils and macrophages to the lungs. In its absence, this recruitment significantly aggravates lung injury. The elevation of inflammatory factors such as IL-6, MCP-1 and Macrophage Inflammatory Protein 1Beta (MIP) were observed in knockout mice. In contrast, treatment with PTX3 effectively reversed these inflammatory symptoms, reducing cytokine levels and ameliorating airway injury (99). In addition, significant PTX3 expression has also been observed in infections caused by hepatitis B virus, hepatitis C virus and dengue virus infections (100–102).
In conclusion, PTX3 responds rapidly to viral invasion, exerting antiviral effects by regulating complement activation and enhancing pathogen recognition by myeloid cells. Following viral infection, PTX3 concentrations increase quickly and correlate closely with disease severity, suggesting it as a potential biomarker for the diagnosis and prognosis of viral infections in the lungs. In addition, PTX3 may serve a therapeutic role by binding to viruses to reduce intracellular viral load and activating relevant immune pathways.
6 Conclusion and perspective
PTX3 has recently garnered significant attention due to its multiple functions in innate immunity. As a soluble pattern recognition receptor and acute phase response protein, PTX3 serves as a crucial component of immune response against infectious pathogens, showing promise as a novel biomarker for diagnosing various lung diseases, assessing severity, and predicting prognosis. Unlike traditional markers like CRP, PTX3 occurs earlier at the site of inflammation and injury.
PTX3 recognizes and binds to various pathogens, including bacteria, fungi, and viruses, thereby promoting phagocytosis by macrophages and other immune cells. It enhances complement activation, regulates inflammatory responses, amplifies immune signaling, activates adaptive immunity, and aids in pathogen clearance. These characteristics position as a potential therapeutic target for various infectious and inflammatory diseases. However, the dual anti-inflammatory and pro-inflammatory roles of PTX3 present a complex dynamic require further investigation.
In conclusion, while PTX3 holds great promise as a biomarker and therapeutic target, more in-depth studies are required to fully unlock its clinical value and address its complex roles in infectious and inflammatory diseases. Key studies should focus on establishing its reliability, optimizing its clinical applications, and exploring strategies to harness its therapeutic potential.
Author contributions
LM: Methodology, Software, Validation, Writing – original draft. DL: Conceptualization, Data curation, Writing – original draft. YW: Formal Analysis, Investigation, Writing – review & editing. DS: Funding acquisition, Project administration, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by grants from the National Nature Science Foundation of China (NM 82272358), Key R&D Program of Jining (2023YXNS001, 2023YXNS055, 2023YXNS247 and 2024YXNS183).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Mizgerd JP. Lung infection–a public health priority. PLoS Med. (2006) 3:e76. doi: 10.1371/journal.pmed.0030076
2. Aliberti S, Dela CC, Amati F, Sotgiu G, Restrepo MI. Community-acquired pneumonia. Lancet. (2021) 398:906–19. doi: 10.1016/S0140-6736(21)00630-9
3. Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. (2001) 1:135–45. doi: 10.1038/35100529
4. Wang H, Atkins SJ, Fernando R, Wei RL, Smith TJ. Pentraxin-3 is a TSH-inducible protein in human fibrocytes and orbital fibroblasts. Endocrinology. (2015) 156:4336–44. doi: 10.1210/en.2015-1399
5. Daigo K, Inforzato A, Barajon I, Garlanda C, Bottazzi B, Meri S, et al. Pentraxins in the activation and regulation of innate immunity. Immunol Rev. (2016) 274:202–17. doi: 10.1111/imr.12476
6. Bottazzi B, Garlanda C, Cotena A, Moalli F, Jaillon S, Deban L, et al. The long pentraxin PTX3 as a prototypic humoral pattern recognition receptor: interplay with cellular innate immunity. Immunol Rev. (2009) 227:9–18. doi: 10.1111/j.1600-065X.2008.00719.x
7. Qin S, Chen X, Gao M, Zhou J, Li X. Prenatal exposure to lipopolysaccharide induces PTX3 expression and results in obesity in mouse offspring. Inflammation. (2017) 40:1847–61. doi: 10.1007/s10753-017-0626-1
8. Bottazzi B, Doni A, Garlanda C, Mantovani A. An integrated view of humoral innate immunity: pentraxins as a paradigm. Annu Rev Immunol. (2010) 28:157–83. doi: 10.1146/annurev-immunol-030409-101305
9. Ying TH, Lee CH, Chiou HL, Yang SF, Lin CL, Hung CH, et al. Knockdown of Pentraxin 3 suppresses tumorigenicity and metastasis of human cervical cancer cells. Sci Rep. (2016) 6:29385. doi: 10.1038/srep29385
10. Kim MJ, Lee HS, Sol IS, Kim MN, Hong JY, Lee KE, et al. Sputum pentraxin 3 as a candidate to assess airway inflammation and remodeling in childhood asthma. Med (Baltimore). (2016) 95:e5677. doi: 10.1097/MD.0000000000005677
11. van der Meer-van KC, Siezen R, Kramer E, Reinders M, Blokzijl H, van der Meer R, et al. Dietary modulation and structure prediction of rat mucosal pentraxin (Mptx) protein and loss of function in humans. Genes Nutr. (2007) 2:275–85. doi: 10.1007/s12263-007-0058-x
12. Richards DB, Cookson LM, Barton SV, Liefaard L, Lane T, Hutt DF, et al. Repeat doses of antibody to serum amyloid P component clear amyloid deposits in patients with systemic amyloidosis. Sci Transl Med. (2018) 10(422):eaan3128. doi: 10.1126/scitranslmed.aan3128
13. Garlanda C, Jaillon S, Doni A, Bottazzi B, Mantovani A. PTX3, a humoral pattern recognition molecule at the interface between microbe and matrix recognition. Curr Opin Immunol. (2016) 38:39–44. doi: 10.1016/j.coi.2015.11.002
14. Bottazzi B, Inforzato A, Messa M, Barbagallo M, Magrini E, Garlanda C, et al. The pentraxins PTX3 and SAP in innate immunity, regulation of inflammation and tissue remodeling. J Hepatol. (2016) 64:1416–27. doi: 10.1016/j.jhep.2016.02.029
15. Doni A, Musso T, Morone D, Bastone A, Zambelli V, Sironi M, et al. An acidic microenvironment sets the humoral pattern recognition molecule PTX3 in a tissue repair mode. J Exp Med. (2015) 212:905–25. doi: 10.1084/jem.20141268
16. Ronca R, Giacomini A, Di Salle E, Coltrini D, Pagano K, Ragona L, et al. Long-pentraxin 3 derivative as a small-molecule FGF trap for cancer therapy. Cancer Cell. (2015) 28:225–39. doi: 10.1016/j.ccell.2015.07.002
17. Inforzato A, Baldock C, Jowitt TA, Holmes DF, Lindstedt R, Marcellini M, et al. Correction: The angiogenic inhibitor long pentraxin PTX3 forms an asymmetric octamer with two binding sites for FGF2. J Biol Chem. (2020) 295:1768. doi: 10.1074/jbc.AAC120.012568
18. Altmeyer A, Klampfer L, Goodman AR, Vilcek J. Promoter structure and transcriptional activation of the murine TSG-14 gene encoding a tumor necrosis factor/interleukin-1-inducible pentraxin protein. J Biol Chem. (1995) 270:25584–90. doi: 10.1074/jbc.270.43.25584
19. Garlanda C, Bottazzi B, Bastone A, Mantovani A. Pentraxins at the crossroads between innate immunity, inflammation, matrix deposition, and female fertility. Annu Rev Immunol. (2005) 23:337–66. doi: 10.1146/annurev.immunol.23.021704.115756
20. Sun YH, Chou YH, Wang CH, Hsiao YH, Lee CY, Yang SF, et al. Impact of pentraxin 3 genetic variants on uterine cervical cancer clinicopathologic characteristics. Int J Med Sci. (2021) 18:2339–46. doi: 10.7150/ijms.57886
21. Yeh CM, Lin CW, Chuang CY, Liu YF, Chou CH, Yang SF, et al. Functional genetic variant of long pentraxin 3 gene is associated with clinical aspects of oral cancer in male patients. Front Oncol. (2019) 9:581. doi: 10.3389/fonc.2019.00581
22. He Q, Li H, Rui Y, Liu L, He B, Shi Y, et al. Pentraxin 3 gene polymorphisms and pulmonary aspergillosis in chronic obstructive pulmonary disease patients. Clin Infect Dis. (2018) 66:261–67. doi: 10.1093/cid/cix749
23. Diamond JM, Meyer NJ, Feng R, Rushefski M, Lederer DJ, Kawut SM, et al. Variation in PTX3 is associated with primary graft dysfunction after lung transplantation. Am J Respir Crit Care Med. (2012) 186:546–52. doi: 10.1164/rccm.201204-0692OC
24. Carmo RF, Aroucha D, Vasconcelos LR, Pereira LM, Moura P, Cavalcanti MS. Genetic variation in PTX3 and plasma levels associated with hepatocellular carcinoma in patients with HCV. J Viral Hepat. (2016) 23:116–22. doi: 10.1111/jvh.12472
25. Chiarini M, Sabelli C, Melotti P, Garlanda C, Savoldi G, Mazza C, et al. PTX3 genetic variations affect the risk of Pseudomonas aeruginosa airway colonization in cystic fibrosis patients. Genes Immun. (2010) 11:665–70. doi: 10.1038/gene.2010.41
26. Inforzato A, Rivieccio V, Morreale AP, Bastone A, Salustri A, Scarchilli L, et al. Structural characterization of PTX3 disulfide bond network and its multimeric status in cumulus matrix organization. J Biol Chem. (2008) 283:10147–61. doi: 10.1074/jbc.M708535200
27. Inforzato A, Peri G, Doni A, Garlanda C, Mantovani A, Bastone A, et al. Structure and function of the long pentraxin PTX3 glycosidic moiety: fine-tuning of the interaction with C1q and complement activation. Biochemistry. (2006) 45:11540–51. doi: 10.1021/bi0607453
28. Inforzato A, Doni A, Barajon I, Leone R, Garlanda C, Bottazzi B, et al. PTX3 as a paradigm for the interaction of pentraxins with the complement system. Semin Immunol. (2013) 25:79–85. doi: 10.1016/j.smim.2013.05.002
29. Nauta AJ, Bottazzi B, Mantovani A, Salvatori G, Kishore U, Schwaeble WJ, et al. Biochemical and functional characterization of the interaction between pentraxin 3 and C1q. Eur J Immunol. (2003) 33:465–73. doi: 10.1002/immu.200310022
30. Gout E, Moriscot C, Doni A, Dumestre-Pérard C, Lacroix M, Pérard J, et al. M-ficolin interacts with the long pentraxin PTX3: a novel case of cross-talk between soluble pattern-recognition molecules. J Immunol. (2011) 186:5815–22. doi: 10.4049/jimmunol.1100180
31. Ma YJ, Doni A, Hummelshøj T, Honoré C, Bastone A, Mantovani A, et al. Synergy between ficolin-2 and pentraxin 3 boosts innate immune recognition and complement deposition. J Biol Chem. (2009) 284:28263–75. doi: 10.1074/jbc.M109.009225
32. Ma YJ, Doni A, Skjoedt MO, Honoré C, Arendrup M, Mantovani A, et al. Heterocomplexes of mannose-binding lectin and the pentraxins PTX3 or serum amyloid P component trigger cross-activation of the complement system. J Biol Chem. (2011) 286:3405–17. doi: 10.1074/jbc.M110.190637
33. Li A, Zhao F, Yang T, Zhao Y, Liu H, Yang S, et al. PTX3/TWIST1 feedback loop modulates lipopolysaccharide-induced inflammation via PI3K/akt signaling pathway. J Interferon Cytokine Res. (2022) 42:161–69. doi: 10.1089/jir.2021.0183
34. Ham HJ, Lee YS, Yun J, Son DJ, Lee HP, Han SB, et al. K284-6111 alleviates memory impairment and neuroinflammation in Tg2576 mice by inhibition of Chitinase-3-like 1 regulating ERK-dependent PTX3 pathway. J Neuroinflammation. (2020) 17:350. doi: 10.1186/s12974-020-02022-w
35. Han B, Haitsma JJ, Zhang Y, Bai X, Rubacha M, Keshavjee S, et al. Long pentraxin PTX3 deficiency worsens LPS-induced acute lung injury. Intensive Care Med. (2011) 37:334–42. doi: 10.1007/s00134-010-2067-2
36. Slusher AL, Mischo AB, Acevedo EO. Pentraxin 3 is an anti-inflammatory protein associated with lipid-induced interleukin 10 in vitro. Cytokine. (2016) 86:36–40. doi: 10.1016/j.cyto.2016.07.012
37. Bozza S, Campo S, Arseni B, Inforzato A, Ragnar L, Bottazzi B, et al. PTX3 binds MD-2 and promotes TRIF-dependent immune protection in aspergillosis. J Immunol. (2014) 193:2340–48. doi: 10.4049/jimmunol.1400814
38. Hamon Y, Jaillon S, Person C, Giniès JL, Garo E, Bottazzi B, et al. Proteolytic cleavage of the long pentraxin PTX3 in the airways of cystic fibrosis patients. Innate Immun. (2013) 19:611–22. doi: 10.1177/1753425913476741
39. Shin H, Jeon J, Lee JH, Jin S, Ha UH. Pseudomonas aeruginosa groEL stimulates production of PTX3 by activating the NF-κB pathway and simultaneously downregulating microRNA-9. Infect Immun. (2017) 85(3):e00935-16. doi: 10.1128/IAI.00935-16
40. Paroni M, Moalli F, Nebuloni M, Pasqualini F, Bonfield T, Nonis A, et al. Response of CFTR-deficient mice to long-term chronic Pseudomonas aeruginosa infection and PTX3 therapy. J Infect Dis. (2013) 208:130–38. doi: 10.1093/infdis/jis636
41. Moalli F, Paroni M, Véliz RT, Riva F, Polentarutti N, Bottazzi B, et al. The therapeutic potential of the humoral pattern recognition molecule PTX3 in chronic lung infection caused by Pseudomonas aeruginosa. J Immunol. (2011) 186:5425–34. doi: 10.4049/jimmunol.1002035
42. Bruemmer-Smith S, Stüber F, Schroeder S. Protective functions of intracellular heat-shock protein (HSP) 70-expression in patients with severe sepsis. Intensive Care Med. (2001) 27:1835–41. doi: 10.1007/s00134-001-1131-3
43. Jeon J, Lee Y, Yu H, Ha U. Pseudomonas aeruginosa DnaK Stimulates the Production of Pentraxin 3 via TLR4-Dependent NF-κB and ERK Signaling Pathways. Int J Mol Sci. (2021) 22(9):4652. doi: 10.3390/ijms22094652
44. Granton JT, Grossman RF. Community-acquired pneumonia in the elderly patient. Clinical features, epidemiology, and treatment. Clin Chest Med. (1993) 14:537–53. doi: 10.1016/S0272-5231(21)00914-X
45. Maloney SA, Jarvis WR. Epidemic nosocomial pneumonia in the intensive care unit. Clin Chest Med. (1995) 16:209–23. doi: 10.1016/S0272-5231(21)00988-6
46. Soares AC, Souza DG, Pinho V, Vieira AT, Nicoli JR, Cunha FQ, et al. Dual function of the long pentraxin PTX3 in resistance against pulmonary infection with Klebsiella pneumoniae in transgenic mice. Microbes Infect. (2006) 8:1321–29. doi: 10.1016/j.micinf.2005.12.017
47. Paczosa MK, Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. (2016) 80:629–61. doi: 10.1128/MMBR.00078-15
48. Asgari F, Supino D, Parente R, Polentarutti N, Stravalaci M, Porte R, et al. The long pentraxin PTX3 controls klebsiella pneumoniae severe infection. Front Immunol. (2021) 12:666198. doi: 10.3389/fimmu.2021.666198
49. Dockrell DH, Whyte M, Mitchell TJ. Pneumococcal pneumonia: mechanisms of infection and resolution. Chest. (2012) 142:482–91. doi: 10.1378/chest.12-0210
50. Koh SH, Shin SG, Andrade MJ, Go RH, Park S, Woo CH, et al. Long pentraxin PTX3 mediates acute inflammatory responses against pneumococcal infection. Biochem Biophys Res Commun. (2017) 493:671–76. doi: 10.1016/j.bbrc.2017.08.133
51. Shi GQ, Yang L, Shan LY, Yin LZ, Jiang W, Tian HT, et al. Investigation of the clinical significance of detecting PTX3 for community-acquired pneumonia. Eur Rev Med Pharmacol Sci. (2020) 24:8477–82. doi: 10.26355/eurrev_202008_22645
52. Mallia P, Footitt J, Sotero R, Jepson A, Contoli M, Trujillo-Torralbo MB, et al. Rhinovirus infection induces degradation of antimicrobial peptides and secondary bacterial infection in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. (2012) 186:1117–24. doi: 10.1164/rccm.201205-0806OC
53. Yeo CD, Kim JW, Cho MR, Kang JY, Kim SJ, Kim YK, et al. Pleural fluid pentraxin-3 for the differential diagnosis of pleural effusions. Tuberc Respir Dis (Seoul). (2013) 75:244–49. doi: 10.4046/trd.2013.75.6.244
54. Zatta M, Di Bella S, Bottazzi B, Rossi F, D’Agaro P, Segat L, et al. Determination of pentraxin 3 levels in cerebrospinal fluid during central nervous system infections. Eur J Clin Microbiol Infect Dis. (2020) 39:665–70. doi: 10.1007/s10096-019-03767-w
55. Sigal GB, Segal MR, Mathew A, Jarlsberg L, Wang M, Barbero S, et al. Biomarkers of tuberculosis severity and treatment effect: A directed screen of 70 host markers in a randomized clinical trial. EBioMedicine. (2017) 25:112–21. doi: 10.1016/j.ebiom.2017.10.018
56. Azzurri A, Sow OY, Amedei A, Bah B, Diallo S, Peri G, et al. IFN-gamma-inducible protein 10 and pentraxin 3 plasma levels are tools for monitoring inflammation and disease activity in Mycobacterium tuberculosis infection. Microbes Infect. (2005) 7:1–08. doi: 10.1016/j.micinf.2004.09.004
57. Brunetta E, Folci M, Bottazzi B, De Santis M, Gritti G, Protti A, et al. Macrophage expression and prognostic significance of the long pentraxin PTX3 in COVID-19. Nat Immunol. (2021) 22:19–24. doi: 10.1038/s41590-020-00832-x
58. Tang T, Dai Y, Zeng Q, Bu S, Huang B, Xiao Y, et al. Pentraxin-3 polymorphisms and pulmonary fungal disease in non-neutropenic patients. Ann Transl Med. (2020) 8:1142. doi: 10.21037/atm-20-5454
59. Aranday CE, Kaveh D, Nunez-Garcia J, Hogarth PJ, Vordermeier HM. Mycobacterium bovis-BCG vaccination induces specific pulmonary transcriptome biosignatures in mice. PLoS One. (2010) 5:e11319. doi: 10.1371/journal.pone.0011319
60. Lin SJ, Schranz J, Teutsch SM. Aspergillosis case-fatality rate: systematic review of the literature. Clin Infect Dis. (2001) 32:358–66. doi: 10.1086/318483
61. Danion F, Duval C, Séverac F, Bachellier P, Candolfi E, Castelain V, et al. Factors associated with coinfections in invasive aspergillosis: a retrospective cohort study. Clin Microbiol Infect. (2021) 27:1644–51. doi: 10.1016/j.cmi.2021.02.021
62. De Pauw B, Walsh TJ, Donnelly JP, Stevens DA, Edwards JE, Calandra T, et al. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin Infect Dis. (2008) 46:1813–21. doi: 10.1086/588660
63. Camargo JF, Husain S. Immune correlates of protection in human invasive aspergillosis. Clin Infect Dis. (2014) 59:569–77. doi: 10.1093/cid/ciu337
64. Garlanda C, Hirsch E, Bozza S, Salustri A, De Acetis M, Nota R, et al. Non-redundant role of the long pentraxin PTX3 in anti-fungal innate immune response. Nature. (2002) 420:182–86. doi: 10.1038/nature01195
65. Diniz SN, Nomizo R, Cisalpino PS, Teixeira MM, Brown GD, Mantovani A, et al. PTX3 function as an opsonin for the dectin-1-dependent internalization of zymosan by macrophages. J Leukoc Biol. (2004) 75:649–56. doi: 10.1189/jlb.0803371
66. Runza VL, Schwaeble W, Männel DN. Ficolins: novel pattern recognition molecules of the innate immune response. Immunobiology. (2008) 213:297–306. doi: 10.1016/j.imbio.2007.10.009
67. Munthe-Fog L, Hummelshøj T, Ma YJ, Hansen BE, Koch C, Madsen HO, et al. Characterization of a polymorphism in the coding sequence of FCN3 resulting in a Ficolin-3 (Hakata antigen) deficiency state. Mol Immunol. (2008) 45:2660–66. doi: 10.1016/j.molimm.2007.12.012
68. Rosbjerg A, Genster N, Pilely K, Skjoedt MO, Stahl GL, Garred P. Complementary Roles of the Classical and Lectin Complement Pathways in the Defense against Aspergillus fumigatus. Front Immunol. (2016) 7:473. doi: 10.3389/fimmu.2016.00473
69. Dellière S, Chauvin C, Wong S, Gressler M, Possetti V, Parente R, et al. Interplay between host humoral pattern recognition molecules controls undue immune responses against Aspergillus fumigatus. Nat Commun. (2024) 15:6966. doi: 10.1038/s41467-024-51047-9
70. Wójtowicz A, Bochud PY. Host genetics of invasive Aspergillus and Candida infections. Semin Immunopathol. (2015) 37:173–86. doi: 10.1007/s00281-014-0468-y
71. Rubino M, Kunderfranco P, Basso G, Greco CM, Pasqualini F, Serio S, et al. Epigenetic regulation of the extrinsic oncosuppressor PTX3 gene in inflammation and cancer. Oncoimmunology. (2017) 6:e1333215. doi: 10.1080/2162402X.2017.1333215
72. Cunha C, Aversa F, Lacerda JF, Busca A, Kurzai O, Grube M, et al. Genetic PTX3 deficiency and aspergillosis in stem-cell transplantation. N Engl J Med. (2014) 370:421–32. doi: 10.1056/NEJMoa1211161
73. Pana ZD, Farmaki E, Roilides E. Host genetics and opportunistic fungal infections. Clin Microbiol Infect. (2014) 20:1254–64. doi: 10.1111/1469-0691.12800
74. Wójtowicz A, Lecompte TD, Bibert S, Manuel O, Rüeger S, Berger C, et al. PTX3 polymorphisms and invasive mold infections after solid organ transplant. Clin Infect Dis. (2015) 61:619–22. doi: 10.1093/cid/civ386
75. Cunha C, Monteiro AA, Oliveira-Coelho A, Kühne J, Rodrigues F, Sasaki SD, et al. PTX3-based genetic testing for risk of aspergillosis after lung transplant. Clin Infect Dis. (2015) 61:1893–94. doi: 10.1093/cid/civ679
76. Liu L, Zhong J, Chen B, Wang W, Xi H, Su X. CCAAT/enhancer binding protein (C/EBP) delta promotes the expression of PTX3 and macrophage phagocytosis during A. fumigatus infection. J Leukoc Biol. (2022) 111:1225–34. doi: 10.1002/JLB.4MA1121-451RR
77. Maschmeyer G, Haas A, Cornely OA. Invasive aspergillosis: epidemiology, diagnosis and management in immunocompromised patients. Drugs. (2007) 67:1567–601. doi: 10.2165/00003495-200767110-00004
78. Gaziano R, Bozza S, Bellocchio S, Perruccio K, Montagnoli C, Pitzurra L, et al. Anti-Aspergillus fumigatus efficacy of pentraxin 3 alone and in combination with antifungals. Antimicrob Agents Chemother. (2004) 48:4414–21. doi: 10.1128/AAC.48.11.4414-4421.2004
79. Taccone FS, Van den Abeele AM, Bulpa P, Misset B, Meersseman W, Cardoso T, et al. Epidemiology of invasive aspergillosis in critically ill patients: clinical presentation, underlying conditions, and outcomes. Crit Care. (2015) 19:7. doi: 10.1186/s13054-014-0722-7
80. Izumikawa K, Takazono T, Kohno S. Chronic aspergillus infections of the respiratory tract: diagnosis, management and antifungal resistance. Curr Opin Infect Dis. (2010) 23:584–89. doi: 10.1097/QCO.0b013e32833ed694
81. Stravalaci M, Pagani I, Paraboschi EM, Pedotti M, Doni A, Scavello F, et al. Recognition and inhibition of SARS-CoV-2 by humoral innate immunity pattern recognition molecules. Nat Immunol. (2022) 23:275–86. doi: 10.1038/s41590-021-01114-w
82. Schultze JL, Aschenbrenner AC. COVID-19 and the human innate immune system. Cell. (2021) 184:1671–92. doi: 10.1016/j.cell.2021.02.029
83. van Meijgaarden KE, van Veen S, Tsonaka R, Ruibal P, Roukens A, Arbous SM, et al. Longitudinal soluble marker profiles reveal strong association between cytokine storms resulting from macrophage activation and disease severity in COVID-19 disease. Sci Rep. (2024) 14:12882. doi: 10.1038/s41598-024-63586-8
84. Phetsouphanh C, Jacka B, Ballouz S, Jackson K, Wilson DB, Manandhar B, et al. Improvement of immune dysregulation in individuals with long COVID at 24-months following SARS-CoV-2 infection. Nat Commun. (2024) 15:3315. doi: 10.1038/s41467-024-47720-8
85. Barbosa M, Da CAA, de Souza C, Do CR. Pentraxin-3 (PTX-3) as a potential biomarker for predicting death in hospitalized patients with COVID-19. Virol J. (2024) 21:233. doi: 10.1186/s12985-024-02501-z
86. Hansen CB, Sandholdt H, Møller M, Pérez-Alós L, Pedersen L, Bastrup IS, et al. Prediction of respiratory failure and mortality in COVID-19 patients using long pentraxin PTX3. J Innate Immun. (2022) 14:493–501. doi: 10.1159/000521612
87. Scavello F, Brunetta E, Mapelli SN, Nappi E, García MI, Sironi M, et al. The long Pentraxin PTX3 serves as an early predictive biomarker of co-infections in COVID-19. EBioMedicine. (2024) 105:105213. doi: 10.1016/j.ebiom.2024.105213
88. Feitosa TA, de Souza SM, Pereira VC, de Andrade CM, Pereira V, Da CAA, et al. Association of polymorphisms in long pentraxin 3 and its plasma levels with COVID-19 severity. Clin Exp Med. (2023) 23:1225–33. doi: 10.1007/s10238-022-00926-w
89. Han E, Youn S, Kwon KT, Kim SC, Jo HY, Jung I. Disease progression associated cytokines in COVID-19 patients with deteriorating and recovering health conditions. Sci Rep. (2024) 14:24712. doi: 10.1038/s41598-024-75924-x
90. Reading PC, Bozza S, Gilbertson B, Tate M, Moretti S, Job ER, et al. Antiviral activity of the long chain pentraxin PTX3 against influenza viruses. J Immunol. (2008) 180:3391–98. doi: 10.4049/jimmunol.180.5.3391
91. Pritchett TJ, Paulson JC. Basis for the potent inhibition of influenza virus infection by equine and Guinea pig alpha 2-macroglobulin. J Biol Chem. (1989) 264:9850–58. doi: 10.1016/S0021-9258(18)81736-7
92. Job ER, Bottazzi B, Short KR, Deng YM, Mantovani A, Brooks AG, et al. A single amino acid substitution in the hemagglutinin of H3N2 subtype influenza A viruses is associated with resistance to the long pentraxin PTX3 and enhanced virulence in mice. J Immunol. (2014) 192:271–81. doi: 10.4049/jimmunol.1301814
93. Hebart H, Einsele H. Clinical aspects of CMV infection after stem cell transplantation. Hum Immunol. (2004) 65:432–36. doi: 10.1016/j.humimm.2004.02.022
94. Peggs KS. Cytomegalovirus following stem cell transplantation: from pharmacologic to immunologic therapy. Expert Rev Anti Infect Ther. (2004) 2:559–73. doi: 10.1586/14787210.2.4.559
95. Gandhi MK, Khanna R. Human cytomegalovirus: clinical aspects, immune regulation, and emerging treatments. Lancet Infect Dis. (2004) 4:725–38. doi: 10.1016/S1473-3099(04)01202-2
96. Bozza S, Bistoni F, Gaziano R, Pitzurra L, Zelante T, Bonifazi P, et al. Pentraxin 3 protects from MCMV infection and reactivation through TLR sensing pathways leading to IRF3 activation. Blood. (2006) 108:3387–96. doi: 10.1182/blood-2006-03-009266
97. Campos CF, Leite L, Pereira P, Vaz CP, Branca R, Campilho F, et al. PTX3 polymorphisms influence cytomegalovirus reactivation after stem-cell transplantation. Front Immunol. (2019) 10:88. doi: 10.3389/fimmu.2019.00088
98. Parhizgari N, Zarei GM, Rezaei F, Maraashi SM, Khatami MR, Mokhtari-Azad T. Transcriptomic analysis of human cytomegalovirus to survey the indirect effects on renal transplant recipients. Transpl Immunol. (2023) 78:101746. doi: 10.1016/j.trim.2022.101746
99. Han B, Ma X, Zhang J, Zhang Y, Bai X, Hwang DM, et al. Protective effects of long pentraxin PTX3 on lung injury in a severe acute respiratory syndrome model in mice. Lab Invest. (2012) 92:1285–96. doi: 10.1038/labinvest.2012.92
100. Han Q, Deng H, Fan X, Wang X, Zhang X, Zhang K, et al. Increased serum pentraxin 3 levels are associated with poor prognosis of hepatitis B virus-related hepatocellular carcinoma. J Hepatocell Carcinoma. (2021) 8:1367–73. doi: 10.2147/JHC.S337936
101. Gorka-Dynysiewicz J, Pazgan-Simon M, Zuwala-Jagiello J. Pentraxin 3 detects clinically significant fibrosis in patients with chronic viral hepatitis C. BioMed Res Int. (2019) 2019:2639248. doi: 10.1155/2019/2639248
Keywords: pentraxin 3 (PTX3), lung infections, inflammation, complement, biomarkers
Citation: Ma L, Li D, Wen Y and Shi D (2025) Advances in understanding the role of pentraxin-3 in lung infections. Front. Immunol. 16:1575968. doi: 10.3389/fimmu.2025.1575968
Received: 13 February 2025; Accepted: 03 April 2025;
Published: 17 April 2025.
Edited by:
Francisco Jose Roig, Universidad San Jorge, SpainReviewed by:
Cristina Florescu, University of Medicine and Pharmacy of Craiova, RomaniaMehmet Emirhan Işık, Kosuyolu Yuksek Ihtisas Egitim ve Arastirma Hastanesi, Türkiye
Copyright © 2025 Ma, Li, Wen and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dongmei Shi, c2hpZG9uZ21laTI4QDE2My5jb20=; Yiyang Wen, d2VueXk1MzU3QGhvdG1haWwuY29t
 Li Ma1
Li Ma1 Dongmei Li
Dongmei Li Yiyang Wen
Yiyang Wen Dongmei Shi
Dongmei Shi