- Department of Critical Care Medicine, The Affiliated People’s Hospital of Jiangsu University, Zhenjiang, Jiangsu, China
In recent years, the study of the interaction between gut microbiota and distant organs such as the heart, lungs, brain, and liver has become a hot topic in the field of gut microbiology. With a deeper understanding of its immune regulation and mechanisms of action, these findings have increasingly highlighted their guiding value in clinical practice. The gut is not only the largest digestive organ in the human body but also the habitat for most microorganisms. Imbalances in gut microbial communities have been associated with various lung diseases, such as allergic asthma and cystic fibrosis. Furthermore, gut microbial communities have significant impacts on metabolic function and immune responses. Their metabolites not only regulate gastrointestinal immune systems but may also affect distant organs such as the lungs and brain. As one of the most common types of respiratory system diseases worldwide, pulmonary infections have high morbidity and mortality rates. Pulmonary infections caused by immune dysfunction can lead to gastrointestinal problems like diarrhea, further resulting in imbalances within complex interactions that are associated with abnormal manifestations under disequilibrium conditions. Meanwhile, clinical interventions can significantly modulate the composition of gut microbiota, and alteration in gut microbiota may subsequently indicate susceptibility to pulmonary infections and even contribute to the prevention or regulation of their progression. This review delves into the interaction between gut microbiota and pulmonary infections, elucidating the latest advancements in gut-lung axis research and providing a fresh perspective for the treatment and prevention of pneumonia.
1 Introduction
Pneumonia is defined as a lung parenchymal infection that typically presents as localized inflammation in the lungs. However, in severe cases, persistent inflammatory response can lead to sepsis, organ failure, shock, and even death (1). Furthermore, there still exists a high incidence and mortality rate of pneumonia globally. According to statistics, approximately 102,000 people die from community-acquired pneumonia in the United States each year, with mortality rates being correlated with hospitalization time (2). The pathogens responsible for pulmonary infections are highly diverse, encompassing bacteria, viruses, fungi, and parasites. Among these, bacterial pneumonia is the most prevalent type, primarily caused by Streptococcus pneumoniae, Haemophilus influenzae, Staphylococcus aureus, Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa. The predominant pathogen of community-acquired pneumonia (CAP) is Streptococcus pneumoniae, whose capsular polysaccharide facilitates immune evasion through mechanisms such as complement inhibition and blocking Toll-like receptor ligand recognition. Haemophilus influenzae enhances its colonization and infectivity in the respiratory tract via virulence factors like pili and capsules. As a common opportunistic pathogen, Staphylococcus aureus produces toxins and enzymes that induce tissue damage and inflammatory responses (3–5). In hospital-acquired pneumonia (HAP) and ventilator-associated pneumonia (VAP), Escherichia coli, Klebsiella pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa are key pathogens (6), characterized by high levels of drug resistance and adaptability, which pose significant challenges to clinical management. Viral pneumonia represents another critical form of pulmonary infection, mainly caused by influenza viruses, respiratory syncytial virus, adenoviruses, and coronaviruses. The annual seasonal influenza epidemic may be influenced by environmental factors affecting the stability of the influenza virus (7). Respiratory syncytial virus is the predominant pathogen responsible for acute lower respiratory tract infections in infants, young children, and the elderly, often leading to bronchiolitis and pneumonia. Adenovirus binds to host cell surface receptors via fiber proteins, exhibiting specific tissue tropism and causing respiratory infections. Furthermore, adenovirus evades the host immune response by inhibiting interferon activity, apoptosis, and the expression of major histocompatibility complex class I molecules (8). Coronaviruses are enveloped RNA viruses with lipid layers on their outer surfaces; their spike proteins bind to host cell surface receptors, enabling cellular invasion and subsequent infection (9). Notably, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has triggered the global public health crisis of the COVID-19 pandemic (10). Among these, the COVID-19 pandemic caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) has triggered a significant global public health crisis (11). Moreover, fungal pneumonia and pneumonia caused by opportunistic pathogens are also prevalent among patients with compromised immune function, such as pulmonary infections induced by Cryptococcus, Aspergillus, and Pneumocystis jirovecii (12).
In fact, pulmonary infections including bacterial infections, fungal infections, and viral infections are all associated with dysbiosis of the lung microbiota. This differs from the perception of a sterile state in the lungs because in reality, the pulmonary microbial community is constantly undergoing dynamic migration and clearance (13). It’s worth noting that human microbiota is not limited to just the lungs; it also exists in various parts of the body such as the gastrointestinal tract (GI), skin, oral cavity,respiratory tract,and vagina (14). The majority of microorganisms reside in the gastrointestinal (GI) tract, where their total gene count surpasses that of human genes by several hundred times. A large and complex microbial ecosystem, known as the gut microbiota, exists within the human intestinal tract. This community is primarily composed of bacteria, archaea, fungi, and viruses, with bacteria playing a dominant role. Among the human intestinal bacteria, the key phyla include Firmicutes (Bacillota), Bacteroidetes (Bacteroidota), Proteobacteria (Pseudomonadota), Actinobacteria, and Fusobacteria (15). In a healthy human intestinal tract, Firmicutes (Bacillota) and Bacteroidetes (Bacteroidota) typically dominate, exhibiting genetic diversity in cytochrome P450 monooxygenase and ferredoxin genes. These microbial communities are not only essential for maintaining intestinal microecological balance and host health but may also exert diverse influences on human health (16). Therefore, the large number and abundance of gut microbiota may play an important role in influencing lung microbiota and subsequently affecting prognosis when patients have respiratory-related diseases. Certainly, existing evidence highlights the critical role of gut microbiota and its metabolites in sustaining human health. Abnormal alterations in the gut microbiota, referred to as dysbiosis, encompass changes in both composition and function. Dysbiosis is generally characterized by a reduction in beneficial bacteria, an expansion of potentially pathogenic bacteria, and a decline in microbial diversity. A variety of factors can induce this imbalance, including antibiotic usage, modifications in dietary patterns, infections, and immune system fluctuations. Dysbiosis has been closely associated with the onset, progression, and exacerbation of numerous diseases (17–19). However, when these steady states are disrupted, it not only affects GI tract but also impacts distal organs such as lungs, brain, and liver (20, 21). It is this close connection that brings together gut microbial communities and cutaneous immune systems into what best exemplifies the concept known as “gut-lung axis”. In this review article, we will focus on recent advancements in the field of “gut-lung axis”.
2 The causal relationship between gut microbiota and lung infections
There is a complex interaction between the gut microbiota and lung diseases. On one hand, lung diseases and respiratory infections can cause changes in the diversity and abundance of gut microbiota, leading to an ecological imbalance in the gut microbial community. Recent studies have shown that infection with novel coronavirus increases the number of pathogenic bacteria in patients’ intestines while reducing beneficial bacteria. In addition, coronavirus infection also reduces transcriptional activity in patients’ feces and is associated with longitudinal changes in fecal microbiota composition (22). Furthermore, analysis of infant fecal samples using 16S rRNA gene sequencing has revealed that respiratory syncytial virus infection affects gut microbial composition. Severe respiratory syncytial virus-infected patients (such as S247 cases) show increased relative abundance of bacterial taxa such as Clostridium difficile, Moraxellaceae bacteria, Lactobacillus, and Actinobacteria compared to healthy infants; meanwhile, there is a decrease in the quantity of Moraxellaceae bacteria in severe respiratory syncytial virus children (23). These alterations in microbial composition and function may disrupt intestinal immune balance and increase susceptibility to inflammatory-related issues within the intestine. On the other hand, disrupted gut microbiota can also impact the progression and prognosis of pulmonary problems; for example, there is an association between low-abundance specific bacterial groups at ages 4-7 years old and high risk for asthma development. Importantly, it should be noted that intestinal microecology can enhance immune responses within respiratory mucosa by regulating production components within small bodies which subsequently improves host resistance against viral infections or secondary bacterial infections (24–26). Additionally, intake of inhibitors can lead to a decrease in specific species within gut microbiota thereby affecting the occurrence of pulmonary diseases and allergic inflammation (27). All these findings confirm that the gastrointestinal tract and lungs are complex organ systems that together maintain homeostasis within the body.
2.1 Pneumonia has the potential to induce dysbiosis in the intestinal microbiota
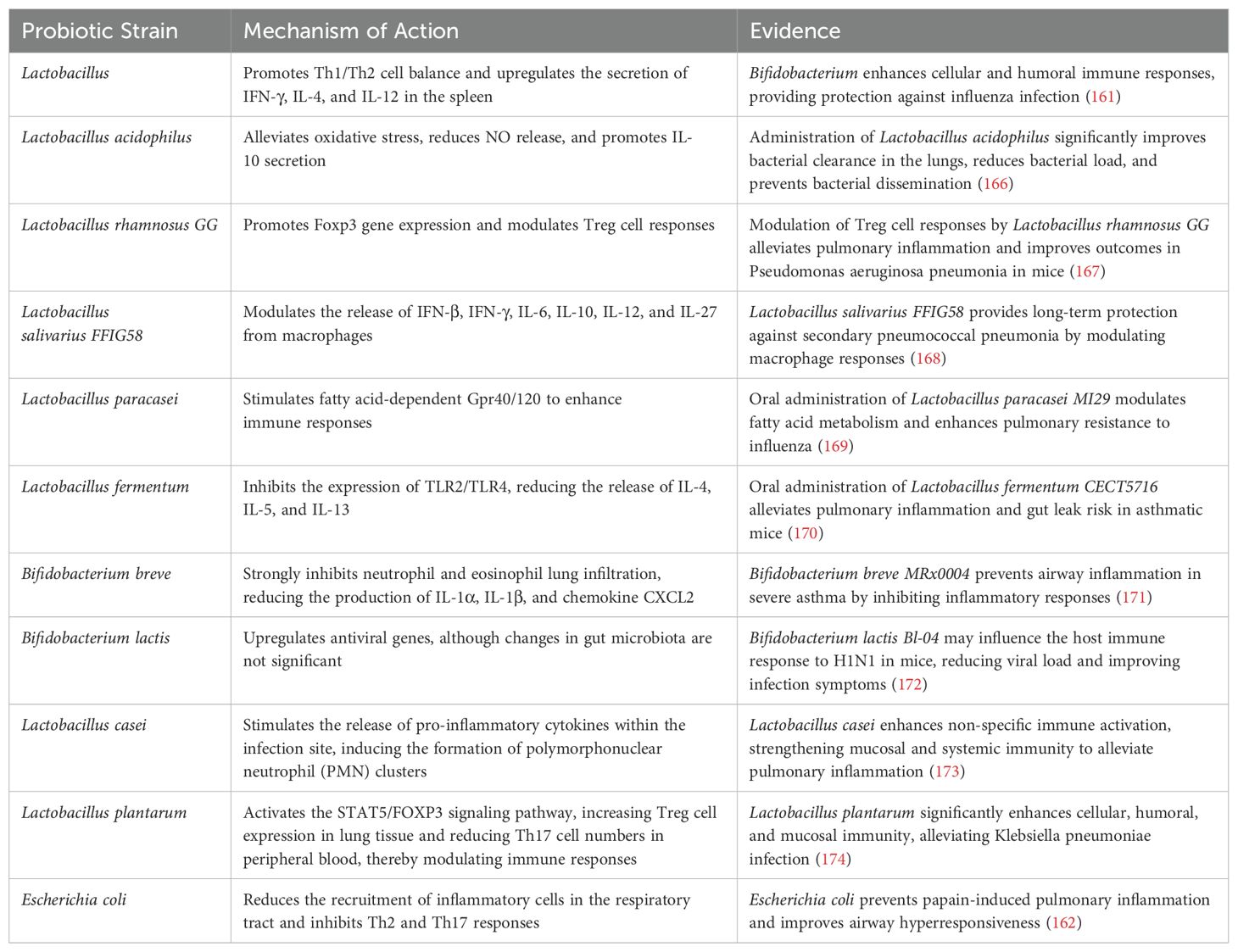
A growing body of evidence suggests the involvement of the gastrointestinal system in the pathogenesis, progression, and prognosis of pneumonia patients undergoing treatment. Gastrointestinal symptoms, including diarrhea, are observed in 2%-20% of COVID-19 infected individuals (28). Moreover, COVID-19 patients with gastrointestinal symptoms (particularly diarrhea) exhibit reduced tryptophan synthesis (29). To investigate specific alterations in gut microbiota composition, a study employing deep sequencing of fecal samples from children with community-acquired pneumonia (CAP) revealed a decline in Proteobacteria (Pseudomonadota) and Bacteroidetes (Bacteroidota) as well as Escherichia/Shigella, Prevotella, Clostridium sensu stricto groups and Enterobacteriaceae among children aged 0-3 years old. Conversely, Actinobacteria and Firmicutes (Bacillota) along with Bifidobacterium and Streptococcus showed varying degrees of increase. In children aged 4-5 years old, there was a significant decrease in Firmicutes (Bacillota) while an increase was observed for Actinobacteria and Streptomyces. The changes at the genus level also exhibited significant differences (30). Pseudomonas aeruginosa infection can elicit a robust inflammatory response, thereby altering the diversity and composition of the gut microbiota. Specifically, the abundances of Firmicutes (Bacillota), Campylobacterota, Actinobacteriota, and Cyanobacteria decreased, whereas the abundances of Proteobacteria (Pseudomonadota) and Verrucomicrobiota increased (31). Additionally, Klebsiella pneumoniae infection may modulate the host’s immune regulatory function by altering the metabolites of the gut microbiota, such as short-chain fatty acids (SCFAs) (32). Additionally, respiratory syncytial virus and influenza virus infections induced dysbiosis in mouse gut microbiota (33). In an acute lung injury model using mice, bacterial migration from lungs to intestines via blood circulation resulted in increased microbial load within the intestines leading to disruption of gut microbiota diversity (34). These findings highlight potential impacts of pulmonary infections on gut microbiota composition while potentially triggering gastrointestinal-related symptoms through immune regulatory pathways mediated by the ‘gut-lung’ axis. (Figure 1) However, whether gut microbiota dysbiosis is merely a “consequence” of pneumonia remains debated. For instance, a longitudinal study in stroke patients revealed that gut microbiota dysbiosis (e.g., reduced Roseburia levels) at admission independently predicted the risk of subsequent stroke-associated pneumonia (SAP). Moreover, the microbial dysbiosis index (MDI) was significantly correlated with both the severity and mortality of pneumonia (35).
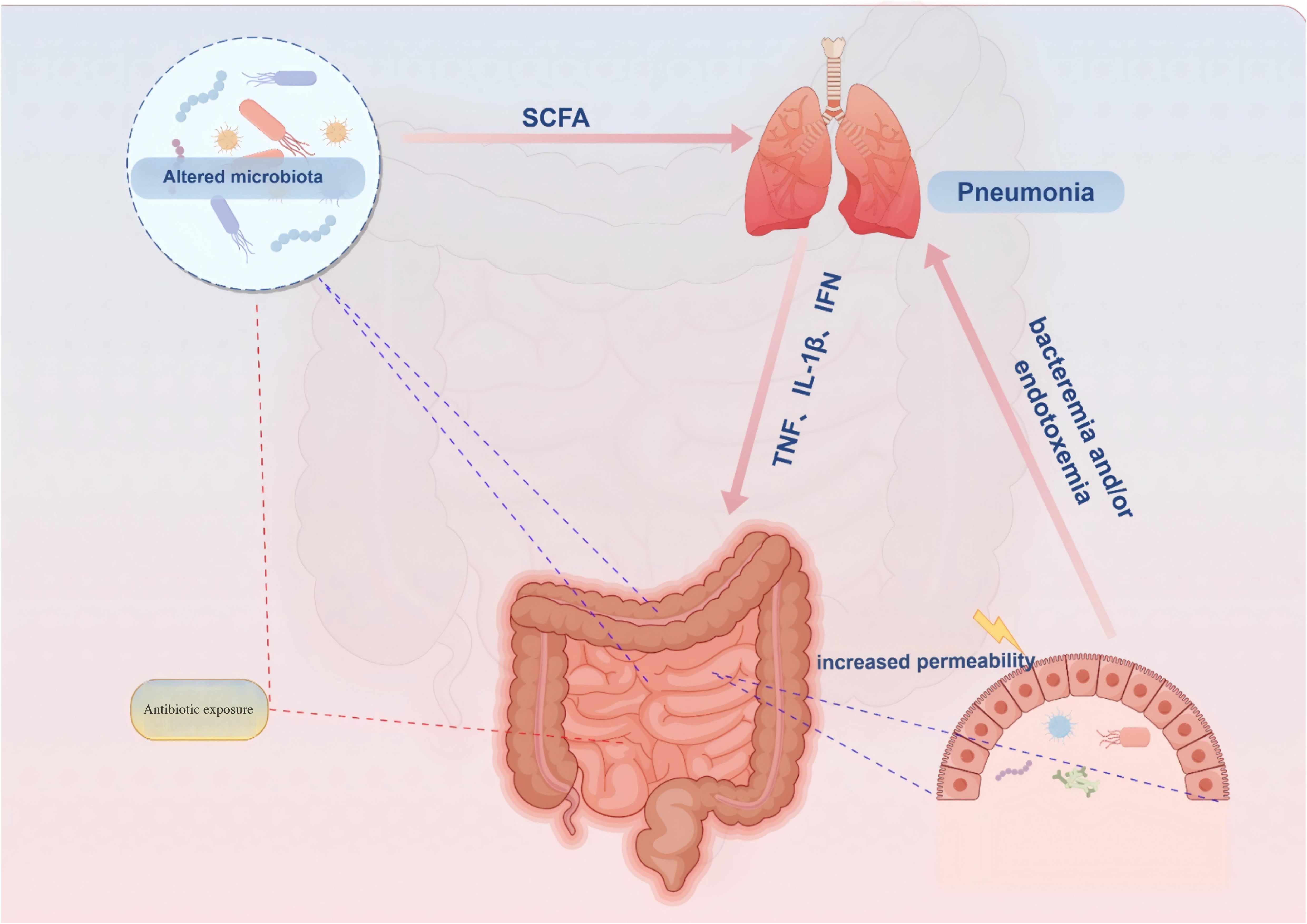
Figure 1. Proposed connections between the gut and lungs in pneumonia. Pneumonia is characterized by a pulmonary parenchymal infection, typically presenting as localized inflammation in the lungs and impaired epithelial barrier function. (1) Damaged and activated lung cells further stimulate the innate immune response through elevated proinflammatory mediators such as TNF-α, IFN-γ, and IL-1β. (2) These proinflammatory mediators can migrate to the gastrointestinal tract, partly via systemic circulation, exacerbating intestinal impairments including increased inflammatory immune infiltration and epithelial barrier damage. Additionally, antibiotic exposure and inflammatory immune cell infiltration in the gut can alter microbiota composition, leading to reduced abundance of health-promoting commensal bacteria that attenuate bacterial synthesis of immune-modulating compounds like SCFAs. (3) Impaired gut function not only increases production and entry of proinflammatory mediators into systemic circulation but also weakens protection against pathogens, thereby further worsening pneumonia symptoms by decreasing intestinal mucosa permeability while enhancing bacterial synthesis of immune-modulating compounds like SCFAs. SCFA = short-chain fatty acid.
2.2 Dysbiosis of the gut microbiota may impact pulmonary inflammation
Dysbiosis of the gut microbiota can alter the lung microbiota, exacerbate lung inflammation, and even serve as a negative prognostic factor for pulmonary infections. SARS-CoV-2 has been found to replicate in the gastrointestinal tract and potentially spread to other sites, such as the lungs, through its impact on inflammatory regulation and ACE2 expression in the gastrointestinal tract (36).Recent research has confirmed and clarified that tryptophan can enhance the expression of ACE2 and B0AT1, and activate the mTOR signaling pathway to repair intestinal injury (37).Research has indicated an association between specific gut microbial groups, such as Enterobacteriaceae, and community-acquired pneumonia (CAP) (30). Furthermore, an imbalance in the intestinal flora may influence brain-gut axis signaling by producing autoinducer-2 (AI-2), potentially contributing to the induction or exacerbation of post-stroke pneumonia (38). Conversely, certain gut microbial groups and their metabolites may play a protective role for the host. Reversing the restoration of the gut microbiota has been proven to mediate the de-colonization of Pseudomonas aeruginosa from the gastrointestinal tract, thus preventing its infection and spread (39). Short-chain fatty acids (SCFAs), commonly present in the gut, act as crucial immunoregulatory metabolites and have demonstrated protective effects in individuals with airway inflammation (40). Furthermore, depletion or loss of gut microbiota is believed to impact host immune response. A study by Schuijt et al. revealed that mice with depleted gut microbiota exhibited increased bacterial dissemination, inflammation, organ damage, and mortality compared to control mice. However, fecal microbiota transplantation (FMT) reversed diversity of gut microbiota and enhanced host defense against pneumonia (26). Those evidence implies that gut microbiota disorders might serve as an “antecedent” for certain types of pneumonia. It should be emphasized that the “causal effect” of gut microbiota on pulmonary infections requires validation through interventional studies. For example, fecal microbiota transplantation (FMT) in animal models has been shown to reverse antibiotic-induced microbiota dysbiosis, restore alveolar macrophage phagocytic function, and decrease pneumonia mortality rates (41). Additionally, research on pulmonary arterial hypertension patients indicates that gut microbiota metabolites (e.g., trimethylamine N-oxide [TMAO]) can indirectly exacerbate lung inflammation by promoting pulmonary vascular remodeling (42). These findings collectively support the notion that gut microbiota dysbiosis may act as a driving force behind lung diseases.
However, the causal relationship between gut microbiota dysbiosis and pulmonary infections could potentially be bidirectional: in immunosuppressed or chronically ill patients, microbiota dysregulation may enhance susceptibility to pneumonia by impairing mucosal immunity; conversely, acute pulmonary infections may secondarily disrupt intestinal homeostasis via inflammatory factor release, increased mucus secretion, or antibiotic use. Furthermore, future investigations should integrate multi-omics technologies and intervention experiments to comprehensively analyze this intricate interaction.
3 The distinct immunomodulatory characteristics of the gut microbiota in pneumonia
Increasing evidence supports the concept of a “common mucosal immune response,” highlighting the potential influence of gut microbial community and its metabolites on mucosal immunity, potentially extending to distant sites such as lung immune responses (43–45). Innate lymphoid cells (ILCs) are a population of antigen receptor-negative immune cells that reside on mucosal surfaces, playing a pivotal role in maintaining tissue homeostasis and eliciting immune responses. Despite their lower abundance in circulation, they exhibit high enrichment within mucosal barriers such as the intestines and airways. Within these mucosal tissues, ILCs play crucial roles in preserving internal equilibrium and participating in pathological alterations (46). In healthy individuals, ILCs actively participate in tissue repair processes. Over the past few years, research into the role of ILC2 cells in pneumonia and their interactions with the intestinal microbiota has gradually expanded. ILC2 cells regulate the inflammatory response in the lungs by secreting cytokines such as IL-5 and IL-13, which play pivotal roles in diseases like asthma and allergies (47). Studies have demonstrated that the activation and function of ILC2 cells are significantly modulated by the intestinal microbiota and its metabolites. It is noteworthy that activated intestinal ILC2 cells can employ lymphocyte function-associated antigen to egress from the intestine and migrate towards the lungs, thereby initiating airway inflammation (48). Furthermore, Huang et al.’s investigation has unveiled that type 2 innate lymphoid cells (ILC2s) can respond to inflammatory signals and subsequently migrate from the intestines to other organs including the lungs (49).These findings have offered a novel perspective on elucidating the role of ILC2s in pneumonia and their association with the intestinal microbiota. Likewise, Th17 and Treg cells are predominantly found on the mucosal surface, particularly in the lamina propria of the intestine. While Th17 cells induce autoimmune reactions and inflammation, Treg cells suppress these responses to maintain immune system homeostasis (50). However, pneumonia can disrupt the gut bacterial community, leading to an imbalance between Th17 and regulatory T (Treg) cells (51). Moreover, these commensal bacteria can enhance host resistance to bacterial pneumonia by upregulating IL-17A expression and promoting signaling pathways for granulocyte-macrophage colony-stimulating factor (GM-CSF) in lung epithelial cells (52).
Interestingly, the gut microbiota plays a crucial role in regulating the spatiotemporal dynamic balance of the mucosal immune system and exhibits distinct immune regulatory characteristics in bacterial, viral, and fungal pneumonia. The type of pathogenic microorganism directly influences the immune response pattern mediated by the gut microbiota.
3.1 The immunoregulatory characteristics of bacterial pneumonia: microbial flora metabolism and immune collaborative defense mechanisms
The immune regulation in bacterial pneumonia (e.g., infections caused by Streptococcus pneumoniae and Pseudomonas aeruginosa) is closely linked to the interaction between gut microbiota and host metabolism. Short-chain fatty acids (SCFAs), such as butyric acid, produced by the gut microbiota can circulate through the bloodstream to the lungs, where they enhance the bactericidal activity of alveolar macrophages and suppress excessive release of pro-inflammatory cytokines. For example, during Pseudomonas aeruginosa infection, butyrate modulates the maturation and effector functions of neutrophils, thereby influencing their recruitment and bactericidal capacity in the lungs and exerting complex regulation of the immune response (53). Furthermore, gut microbiota can upregulate the expression of Toll-like receptor 4 (TLR4) and activate the host’s defense mechanisms against Streptococcus pneumoniae infection (54). Additionally, gut microbiota enhances resistance to Streptococcus pneumoniae pneumonia and Klebsiella pneumoniae pneumonia via nucleotide-binding oligomerization domain protein 2 (NOD2) and granulocyte-macrophage colony-stimulating factor (GM-CSF) signaling pathways (52). Recent studies have demonstrated that abnormal proliferation of specific gut bacteria, such as Eggerthella lenta, impairs neutrophil function. These bacteria produce taurochenodeoxycholic acid (TUDCA), which inhibits the AMPK signaling pathway crucial for neutrophil energy metabolism, thereby reducing their phagocytic capacity against Pseudomonas aeruginosa. However, metformin, an AMPK activator, can reverse this defect, presenting a novel therapeutic approach for drug-resistant pneumonia (55). SFB can suppress the expression of pro-inflammatory genes in alveolar macrophages (AMs), thereby attenuating the inflammatory response and alleviating lung inflammation and injury. Moreover, SFB contributes to maintaining the homeostasis of AMs and prevents their depletion during viral infections, thus ensuring sustained protection of lung health and resistance to viral invasion (56). These findings suggest that the gut microbiota is involved in the immune regulation of bacterial pneumonia via multiple mechanisms, offering novel insights for the prevention and treatment of this condition.
3.2 The immunoregulatory characteristics of viral pneumonia: focus on the interferon pathway and immune cell migration
The immune regulation of viral pneumonia is intricately linked to the modulation of the interferon system by the gut microbiota. Deaminotyrosine (DAT), primarily produced by specific gut bacteria such as Clostridium orbiscindens, enhances type I interferon (IFN-I) signaling pathways, thereby promoting the expression of antiviral proteins like protein kinase R and MX1, which inhibit viral replication and dissemination. Animal studies have demonstrated that Lactobacillus pentosus CCFM1227 augments the IFN-I response, leading to reduced lung inflammation and enhanced resistance to influenza virus infection (57, 58). In COVID-19 infections, dysbiosis of the gut microbiota decreases short-chain fatty acid (SCFA) levels, thereby weakening antiviral immunity in the lungs. Research has shown that severe COVID-19 patients exhibit a significant reduction in the abundance of butyrate-producing bacteria, including Faecalibacterium, Roseburia, and Anaerostipes. Supplementing with butyric acid can mitigate the “cytokine storm” induced by the virus (59, 60). Furthermore, the gut microbiota influences viral clearance and tissue repair by regulating the migration of Th17 cells and group 2 innate lymphoid cells (ILC2s) to the lungs. For instance, IL-17 plays a critical role in granulocyte colony-stimulating factor (G-CSF) expression and neutrophil homeostasis, while IL-5 secreted by ILC2s contributes to maintaining antiviral immune homeostasis (61, 62).
3.3 The immunoregulatory characteristics of fungal pneumonia: Th17 polarization and metabolite-mediated immune equilibrium
The intestinal flora plays a critical role in immune regulation during fungal pneumonia by modulating systemic immune responses and preserving the integrity of the intestinal barrier. Relevant studies demonstrate that the gut microbiota can regulate the polarization of CD4+ T cells during pulmonary fungal infection, thereby influencing lung immune defense mechanisms. For instance, vancomycin-treated drinking water significantly decreases the number of Th17 cells in the lungs during acute fungal infection, suggesting that the symbiotic effects of Gram-positive bacteria can modulate the expression of local antibacterial factors in the intestine and type 17 cytokines in the lungs. Additionally, the gut microbiota regulates the accumulation of Th17 cells in the lungs by affecting the expression of RegIIIγ (an IL-22-induced antibacterial protein specific to Gram-positive bacteria) and IL-22, thus impacting the immune response during fungal infection (63). Further research indicates that colonization with segmented filamentous bacteria (SFB) can induce the expansion of peripheral SFB-specific Th17 cells (64), which may influence the number of Th17 cells in the lungs. Moreover, metabolites of the gut microbiota also play a pivotal role. For example, short-chain fatty acids (SCFAs) enhance the production of bactericidal reactive oxygen species (ROS) by macrophages through histone deacetylase (HDAC) inhibition (65). Indole-3-carboxaldehyde, a metabolite produced by Lactobacillus reuteri during tryptophan catabolism, is involved in regulating host physiological processes and exhibits anti-inflammatory properties in various inflammatory diseases. The latest research indicates that I3A not only sustains lung epithelial barrier function through AhR receptor activation and IL-22 secretion stimulation but also inhibits HDAC5/6, resulting in the inactivation of the NF-κB/NLRP3 signaling pathway and the subsequent reduction of inflammatory factor expression (66). Additionally, studies have demonstrated that 5-hydroxytryptamine (5-HT), secreted by mast cells, plays a critical role in immune homeostasis during pathogen clearance and infection. This is achieved by promoting the host’s protective indoleamine-2,3-dioxygenase 1/kynurenine pathway while restricting the microflora-activated indole/aryl hydrocarbon receptor pathway. 5-HT exerts these effects via regulation of microbiota and signaling pathways in both lungs and intestines. Notably, supplementation with 5-HT has been shown to restore dysregulated tryptophan metabolism in mouse disease models, potentially offering a novel therapeutic strategy for pulmonary fungal infections (67).
3.4 Other pneumonias: unique pathogenic mechanisms of mycoplasma, chlamydia, and mixed infections
For atypical pathogens, such as Mycoplasma pneumoniae, or in cases of mixed infections, the regulatory mechanisms of the intestinal flora exhibit significant diversity. For instance, acetate and butyrate demonstrate potent anti-inflammatory effects on Mycoplasma pneumoniae-induced THP-1 cells. These short-chain fatty acids effectively suppress the activation of the NLRP3 inflammasome and Caspase-1, inhibit the nuclear translocation of NF-κB p65 protein, and consequently reduce the production and release of inflammatory factors. Moreover, acetate and butyrate decrease the levels of reactive oxygen species (ROS), enhance the secretion of anti-inflammatory cytokine IL-10 and immunoregulatory factor IFN-γ, thereby further mitigating the inflammatory response (68). Dysbiosis of the intestinal flora can also influence immune responses and the progression of Chlamydia pneumoniae infections. For example, using 16S rDNA sequencing technology, it has been demonstrated that children with Chlamydia pneumoniae exhibit impaired intestinal flora diversity and a marked reduction in Bifidobacterium abundance. Supplementation with Bifidobacterium can partially restore fecal microbial diversity, strengthen the intestinal barrier function, and exert a therapeutic effect on mycoplasma pneumonia (69, 70).
4 The role mechanism of intestinal microbiota and their metabolites in the regulation of pulmonary infection
4.1 Gut bacteria-derived Metabolites
Metabolites serve as the critical mediators of the interaction (crosstalk) between the gut microbiota and the human body. The gut microbiota generates a diverse array of metabolites, including volatile small molecules, lipids, proteins and peptides, sugars, secondary bile acids, terpenoids, biogenic amines, oligosaccharides, glycolipids, organic acids, and amino acids, among others (71). These metabolites are produced by the gut microbiota upon the absorption of specific substances and function as signaling molecules and reaction substrates during metabolic processes, playing a crucial role in the onset and progression of lung diseases, including lung infections. Furthermore, compared to microbial taxa, metabolites may be more suitable for research and intervention, as they not only reflect the functional attributes of microorganisms but also actively participate in the metabolic regulation between the host and microbiota (72). In subsequent sections, we will review several key gut microbial metabolites and investigate their potential promoting or inhibitory effects on lung infections.
4.1.1 Short-chain fatty acids
Short-chain fatty acids (SCFAs) are pivotal metabolites generated by the intestinal microbiota via the fermentation of carbohydrates, particularly dietary fiber. These include acetic acid, propionic acid, and butyric acid. SCFAs exert a significant influence on both host intestinal health and systemic metabolism. Research has demonstrated that specific microbial groups are responsible for producing distinct types of SCFAs. For instance, members of the Ruminococcaceae and Lachnospiraceae families within the Firmicutes (Bacillota) are primary producers of butyric acid, whereas certain species in the Bacteroidetes (Bacteroidota) are crucial for the synthesis of acetic acid and propionic acid (73). Moreover, SCFAs produced in the gut not only achieve systemic distribution but also serve as an energy source and act as signaling molecules (74). Recent study has revealed that SCFAs can interact with G protein-coupled receptors to sense metabolites, thereby activating signaling pathways and inducing anti-inflammatory effects within the host’s immune response (75). Specifically, SCFAs generated by the intestinal microbiota through the fermentation of dietary fiber primarily enter systemic circulation via the portal vein and subsequently distribute to various organs, including the lungs (76). Within the lungs, SCFAs are absorbed and utilized by pulmonary immune cells, modulating the local immune response (77). Additionally, a study employed high-performance liquid chromatography (HPLC) to analyze cecal and serum samples from mice fed different dietary combinations. The findings revealed that increased intake of soluble dietary fiber significantly elevated SCFA levels in both cecal and serum samples. However, SCFAs were not detected in lung tissue samples (78). In contrast, another investigation identified SCFAs in the bronchoalveolar lavage fluid (BALF) of lung cancer patients, albeit at relatively low concentrations (79).
The presence of short-chain fatty acids (SCFAs) in the gut can effectively lower the pH of the intestinal lumen, thereby exerting inhibitory effects on the proliferation of pathogenic microorganisms. For instance, experimental evidence has demonstrated that acetate produced by Bifidobacterium is capable of suppressing pathogen growth within mouse intestines (80). Moreover, elevated levels of butyrate have been shown to enhance mucus production, leading to reduced bacterial adhesion and improved integrity of epithelial cells (80, 81). Oral supplementation with SCFAs can facilitate their entry into the bloodstream, potentially promoting the generation of protective regulatory T cells (Tregs) (82). Further investigations conducted on mice have revealed a correlation between butyrate production and an increase in Treg numbers. Additionally, in vitro experiments have elucidated the pivotal role played by butyrate in regulating forkhead box protein 3 (FOXP3), a lineage specification factor. As an inhibitor of histone deacetylases (HDACs), butyrate facilitates acetylation at FOXP3 gene promoter regions on histone H3 (83).SCFAs also exert an influence on dendritic cell development and inflammatory cytokine production, thereby modulating the function of intestinal macrophages (84). Moreover, SCFAs play critical roles in energy metabolism, which are essential for supporting B cell activation, differentiation, and antibody synthesis (85). Specifically, experimental studies have demonstrated that vancomycin administration in mice exacerbates allergic airway disease symptoms by altering the composition and abundance of gut microbiota, leading to reduced SCFA production. Conversely, supplementation with SCFAs via a high-fiber diet can effectively mitigate this adverse effect induced by vancomycin and alleviate disease severity (86, 87). Therefore, oral supplementation with SCFA supplements may also exhibit potential therapeutic efficacy in treating pulmonary diseases.
4.1.2 Microbial tryptophan catabolites
As an essential aromatic amino acid, tryptophan primarily originates from dietary proteins. Its absorption predominantly occurs in the small intestine for protein synthesis. Concurrently, various bacteria in the colon can directly metabolize tryptophan into multiple derivatives, including indole, indole ethanol (IE), indole propionic acid (IPA), indole lactic acid (ILA), indole acetic acid (IAA), indole aldehyde (IAld), indole acrylic acid (IA), skatole, and tryptamine (88). Tryptophan and its derivatives are pivotal in the metabolism of the intestinal microbiota, modulating the host immune response and intestinal barrier function via diverse mechanisms. These metabolites also serve as critical signaling molecules in host-microbe cross-talk, preserving the integrity of the intestinal mucosal barrier and supporting the maintenance of intestinal and systemic homeostasis (89). For instance, IPA specifically binds to methionine adenosyltransferase 2A (MAT2A), enhancing S-adenosylmethionine synthesis and subsequently inducing DNA methylation of the deubiquitinating enzyme USP16. This process promotes ubiquitination and degradation of Toll-like receptor 4 (TLR4), thereby inhibiting the NF-κB signaling pathway and reducing IL-1β expression in M1 macrophages, ultimately alleviating lung inflammatory responses (90). Additionally, IPA functions as an allosteric inhibitor of TrpE by mimicking tryptophan, blocking the catalytic step of tryptophan synthesis mediated by TrpE, thus disrupting metabolic pathways in pathogens such as Mycobacterium tuberculosis and suppressing their proliferation (91). Animal studies demonstrate that influenza infection significantly alters the intestinal tryptophan metabolic pathway, characterized by a marked reduction in blood IPA levels; however, IPA supplementation effectively decreases viral load and mitigates both local (lung) and systemic inflammatory responses (92). IAA activates the aryl hydrocarbon receptor (AhR) to regulate pulmonary immune responses and reduce inflammation (93). ILA inhibits the polarization of mouse Th17 cells in vitro and stimulates the secretion of IL-22 by lamina propria lymphocytes through activation of the aryl hydrocarbon receptor (AhR), thereby promoting the proliferation of intestinal epithelial cells and repairing damaged intestinal mucosa (94). This regulatory mechanism not only aids in controlling infections but also effectively mitigates excessive inflammatory responses, thus protecting lung tissue. Recent studies indicate that in OVA-induced mouse asthma models, tryptophan metabolites such as KYN, ILA, I3C, and IAA can modulate the Th17/Treg balance via AHR activation, significantly reducing inflammation and improving survival rates. Specifically, KYN primarily restores the diversity of the gut microbiota, whereas ILA and I3C markedly enhance the proliferation of bacteria associated with tryptophan metabolism, such as Akkermansia and Ruminococcus_1. While KYN focuses on restoring overall microbial diversity, ILA and I3C specifically increase the relative abundance of these bacteria (95). Additionally, tryptophan metabolites regulate the interaction between intestinal commensal bacteria and the intestinal barrier by activating nuclear receptors, including the aryl hydrocarbon receptor (AhR) and the pregnane X receptor (PXR), further maintaining intestinal homeostasis and host health. Decreased levels of these metabolites may result in immune dysregulation. These metabolites can function independently or interact synergistically to jointly regulate intestinal homeostasis, offering novel therapeutic strategies for intestinal diseases.
4.1.3 Conjugated linoleic acid
Conjugated Linoleic Acid (CLA) is a type of polyunsaturated fatty acid that is metabolized by intestinal flora and primarily derived from dietary linoleic acid. Certain gut microbiota, such as Bifidobacterium and Lactobacillus, can convert linoleic acid into CLA, thereby modulating host metabolism and immune function (96, 97). CLA plays an immunoregulatory role in the gut-lung axis by regulating the activity of immune cells and inflammatory responses. For instance, CLA can effectively promote the expression of IL-35 in macrophages via the Gαq/11-mediated STAT1/4 signaling pathway (98). Notably, a meta-analysis of the effects of CLA on inflammatory factors in adults indicates that CLA reduces levels of pro-inflammatory cytokines (e.g., TNF-α and IL-6), while slightly increasing C-reactive protein (CRP) levels (99). This suggests that CLA may exhibit complex pleiotropic effects in inflammatory responses, demonstrating both anti-inflammatory properties and potential pro-inflammatory characteristics depending on the isomer type or dose. Therefore, further studies are required to elucidate its specific mechanisms and biological significance. This regulatory effect aids in maintaining immune homeostasis in the lungs and mitigating inflammatory responses. Additionally, CLA enhances the immune system’s anti-infective capacity by regulating macrophage activity and differentiation and inhibiting excessive reactive oxygen species (ROS) synthesis (100). The mechanism by which conjugated linoleic acid (CLA) regulates immune responses is both intricate and multifaceted. CLA can directly modulate the activity of immune cells while also indirectly influencing immune regulation through enhancement of intestinal barrier function and modulation of the gut microbiota composition. The Jiang research group successfully established a neonatal rat model of necrotizing enterocolitis (NEC) by simulating the intestinal microenvironment of preterm infants using a multi-factorial induction approach that combined hypoxia, hyperosmolar formula feeding, and lipopolysaccharide (LPS) stimulation. Experimental findings revealed that the CLA intervention group significantly suppressed the overexpression of pro-inflammatory cytokines (IL-6, TNF-α) by activating the peroxisome proliferator-activated receptor γ (PPARγ) signaling pathway. Simultaneously, it restored the expression levels of intestinal tight junction proteins (e.g., occludin and claudin-1), thereby effectively enhancing intestinal barrier function (101). This may reduce the translocation of pathogens and toxins across the intestinal barrier, potentially lowering the risk of lung infections. Furthermore, under inflammatory conditions, CLA may influence host metabolism and immune status by reshaping the gut microbiota composition (increasing the abundance of beneficial bacteria such as Peptostreptococcaceae and reducing harmful bacteria like Desulfovibrionaceae). Huang et al. stimulated TNF-α-induced human bronchial epithelial (BEAS-2B) cells with varying doses of cis-9, trans-11-conjugated linoleic acid (c9,t11-CLA) and demonstrated that c9,t11-CLA significantly inhibited the secretion of inflammatory cytokines (e.g., IL-6, IL-8, MCP-1, and CCL5) and reduced the expression level of intercellular adhesion molecule 1 (ICAM-1), thereby attenuating monocyte adhesion to inflamed bronchial epithelial cells (102). Subsequent investigations revealed that c9,t11-CLA effectively suppresses the expression of pro-inflammatory factors, chemokines, and ICAM-1 by inhibiting NF-κB transcriptional regulation and dampening MAPK signaling pathway activity, thus exhibiting potent anti-inflammatory properties. Moreover, in a prospective, randomized, double-blind, placebo-controlled trial, conjugated linoleic acid (CLA) as a dietary supplement has been shown to offer potential benefits for overweight patients with mild asthma. The study indicated that CLA not only improves airway hyperresponsiveness (AhR) but also reduces body mass index (BMI), highlighting its critical role in modulating immune responses and inflammatory processes (103). Additionally, CLA may support the improvement of chronic obstructive pulmonary disease (COPD) conditions by suppressing oxidative stress production and regulating serum matrix metalloproteinase-9 (MMP-9) levels. Despite the limited number of studies on the specific mechanisms of CLA in pulmonary infections, its status as an intestinal flora metabolite suggests that CLA may indirectly influence lung health by modulating gut microbiota composition and function. Therefore, future research should further explore the potential application value of CLA in pulmonary infections and inflammatory diseases, which could facilitate the development of novel CLA-based therapeutic strategies.
4.1.4 Secondary bile acids
Bile acids are synthesized from cholesterol in the liver via oxidative pathways. The primary bile acids include cholic acid (CA) and chenodeoxycholic acid (CDCA). These bile acids conjugate with taurine or glycine to form conjugated bile acids. The intestinal microbiota metabolically modifies bile acids through diverse pathways, specifically involving the following mechanisms (1): Deconjugation: Bile salt hydrolase (BSH), secreted by gut microbes such as Firmicutes (Bacillota), Bacteroidetes (Bacteroidota), Actinobacteria, Proteobacteria (Pseudomonadota), and archaea, hydrolyzes conjugated bile acids into free bile acids (2). Dehydroxylation: Certain gut bacteria, particularly Clostridium species, convert primary bile acids into secondary bile acids, such as deoxycholic acid (DCA) and lithocholic acid (LCA), via 7α/β-dehydroxylase activity (3). Oxidation and epimerization: Gut microbes further diversify bile acid structures through oxidation and epimerization reactions (4). Sulfation and esterification: Specific gut bacteria alter the solubility and toxicity of bile acids through sulfation and esterification, thereby influencing their excretion (104). As key metabolites of the gut microbiota, bile acids enhance the solubilization of dietary lipids in the small intestine by forming micelles, thus promoting lipid absorption and excretion. Moreover, bile acids function as signaling molecules that interact with nuclear receptors (e.g., FXR) and G protein-coupled receptors (e.g., TGR5). Through these interactions, they regulate bile acid synthesis and metabolism, modulate lipid and glucose homeostasis, and participate in immune response regulation (105). Compared with the healthy control group, the intestinal metabolite profile of patients with community-acquired pneumonia (CAP) showed significant changes. Among them, the level of arachidonic acid significantly increased, while secondary bile acids (such as deoxycholic acid, lithocholic acid, and ursodeoxycholic acid) significantly decreased. This phenomenon suggests that the alteration of bile acid metabolism may play an important role in regulating the pulmonary inflammatory response (106), providing a potential direction for anti-inflammatory treatment strategies targeting bile acids. In vitro studies have shown that bile acids can regulate the expression level of angiotensin-converting enzyme 2 (ACE2) by activating the farnesoid X receptor (FXR), thereby influencing the pulmonary inflammatory response (107). Additionally, in the acute lung injury model, significantly upregulating the expression of FXR can inhibit the phosphorylation of p38 MAPK and NF-κB p65, reduce the release of inflammatory factors, and thereby protect lung tissue (108). At the same time, in the lipopolysaccharide-induced acute lung injury (ALI) model in rats, ursodeoxycholic acid (UDCA) and chenodeoxycholic acid (CDCA) not only enhanced the activity of antioxidant enzymes (including superoxide dismutase [SOD], catalase [CAT], and glutathione [GSH]), effectively alleviated oxidative stress, but also inhibited the inflammatory response by downregulating the expression of the pro-inflammatory transcription factor NF-κB. Specifically, it manifested as a reduction in the production of pro-inflammatory cytokines TNF-α, IL-6, and IL-1β, which play a key role in the pathogenesis of ALI. Furthermore, UDCA and CDCA can also upregulate the expression of aquaporin AQP1 and AQP5, improve the fluid balance in the lungs, and alleviate pulmonary edema. The aforementioned findings demonstrate that UDCA and CDCA exert multi-faceted protective effects on ALI by modulating oxidative stress, inflammatory responses, and fluid balance (109). Furthermore, bile acids exhibit concentration-dependent regulation of respiratory tract antimicrobial peptides and innate immune responses. At low concentrations, bile acids can enhance the expression of antimicrobial peptides in the respiratory epithelium and strengthen innate immunity via activation of relevant signaling pathways (e.g., TGR5-ERK1/2). Conversely, at higher concentrations, this promoting effect is suppressed, indicating a dual regulatory role of bile acids on respiratory immune responses depending on their concentration (110). In summary, bile acids hold potential therapeutic significance for maintaining pulmonary immune homeostasis.
4.1.5 Polyamines
Polyamines are a class of metabolites produced by the intestinal microbiota, primarily comprising putrescine, spermidine, and spermine. These metabolites play crucial roles in maintaining intestinal and lung health. Polyamines are predominantly generated via the decomposition of dietary proteins and amino acids by the intestinal microbiota. For example, certain intestinal bacteria, such as Bifidobacterium, Clostridium, Enterococcus, Lactobacillus, and Pediococcus, can convert amino acids into polyamines through decarboxylation (111). Polyamines are essential for maintaining intestinal barrier function, regulating immune responses, and promoting cell proliferation (112). The composition and diversity of the intestinal flora significantly influence polyamine production. Studies have demonstrated that Bacteroides plays a pivotal role in the intestinal microbiota, and changes in its relative abundance are closely associated with polyamine biosynthesis levels. For instance, research conducted by Li’s team revealed that Bacteroides is the predominant genus involved in polyamine biosynthesis. In patients with inflammatory bowel disease (IBD), alterations in the relative abundance of Bacteroides are linked to variations in polyamine levels. Through single-bacteria stable isotope tracing analysis and computational simulations, the study further elucidated that Bacteroides synthesizes polyamines via specific metabolic pathways, underscoring the critical role of the intestinal microbiota in polyamine metabolism (113). Polyamines influence lung health through the gut-lung axis. Under pulmonary inflammatory conditions, the levels of different polyamine types may exhibit inconsistent changes. Research has shown that cadaverine levels increase in asthma patients, whereas spermidine levels decrease. Spermine and spermidine can modulate LPS-induced NF-κB activation, and oral administration of polyamines alleviates airway inflammation (114). Furthermore, polyamine metabolism plays a pivotal role in the growth, virulence, and stress response of Streptococcus pneumoniae, serving as a critical factor in its pathogenesis (115). Recent studies have demonstrated that spermidine effectively inhibits the phosphorylation of signal transducer and activator of transcription 1 (STAT1) by upregulating protein tyrosine phosphatase non-receptor 2 (PTPN2), thereby promoting macrophage polarization toward the anti-inflammatory M2 phenotype and suppressing excessive inflammatory responses. In a methicillin-resistant Staphylococcus aureus (MRSA) infection model, mice treated with spermidine exhibited significantly improved survival rates and enhanced bacterial clearance, providing robust experimental evidence for spermidine’s potential application in treating infectious diseases and laying the groundwork for its clinical use as an immunomodulator (116). Future research should focus on elucidating the precise mechanisms by which polyamines influence lung diseases. By unraveling these mechanisms, more effective therapeutic strategies can be developed, offering novel insights and approaches for the clinical management of lung diseases.
4.1.6 Trimethylamine-N-oxide
Trimethylamine (TMA) is produced through the metabolism of specific dietary compounds by the gut microbiota and subsequently absorbed into the bloodstream (117). It primarily arises from the catabolism of nutritional substrates such as phosphatidylcholine, choline, carnitine, betaine, dimethylglycine, and ergothioneine by the colonic microbiota (118). Once absorbed, TMA can be oxidized to trimethylamine N-oxide (TMAO) by hepatic flavin monooxygenases (FMO1 and FMO3). Additionally, a portion of TMA may undergo further degradation in the colon, yielding methylamine, dimethylamine (DMA), and ammonia. The production of TMA is modulated by the balance and diversity of the gut microbiota. The higher ratio of Firmicutes (Bacillota) to Bacteroidetes (Bacteroidota) is associated with a greater response to the dietary precursors of TMAO, suggesting that TMAO production may be influenced by individual variations in the intestinal microbiota. An imbalance in the intestinal flora induces vascular damage by activating macrophages, which promotes atherosclerosis progression, thereby increasing the risk of coronary heart disease (119). In the rat model of diabetic nephropathy, dietary supplementation with trimethylamine-N-oxide (TMAO) and choline was found to significantly enhance the activation of the NLRP3 inflammasome. This, in turn, promoted the increased secretion of IL-1β and IL-18, triggering renal inflammatory responses and exacerbating oxidative stress through the overproduction of reactive oxygen species (ROS) (120). Futhermore, TMAO exacerbates TNF-α-mediated renal inflammatory responses by promoting the release of cytokines, chemokines, inflammatory mediators, and growth factors from renal fibroblasts (121). Also, study has demonstrated that in the presence of TMAO, the addition of nobiletin can inhibit vascular oxidative stress and suppress the expression and phosphorylation of ERK/NF-κB. Consequently, this results in an upregulation of Bcl-2 mRNA expression, a downregulation of Bax mRNA expression, and a reduction in downstream inflammatory factors, ultimately leading to the inhibition of inflammation (122).
4.1.7 Polysaccharide A
Polysaccharide A (PSA) is a carbohydrate antigen secreted by the symbiotic bacterium Bacteroides fragilis. Prior studies have demonstrated that T cells exposed to PSA can synergize with tissue-resident FoxP3+ regulatory T cells (Tregs) to promote the secretion of the inhibitory cytokine IL-10 by Tregs, thereby effectively suppressing inflammatory cascade reactions and safeguarding lung health (123). Furthermore, a high-protein diet markedly elevates the level of the phenylalanine derivative PAGln produced by the gut microbiota, exacerbating colitis symptoms in mouse models. Notably, Proteobacteria (Pseudomonadota) harboring the phenylpyruvate decarboxylase (PPDC) gene are regarded as the primary contributors to PAGln production in patients with Crohn’s disease (CD) (124). Based on current research findings, both PSA and PAGln play potentially critical roles in the immune regulation of infectious diseases. Specifically, PSA can suppress inflammatory responses via its interaction with T cells, whereas the metabolic levels of PAGln may be influenced by alterations in the composition of the gut microbiota and exhibit pro-inflammatory effects in inflammatory bowel disease (IBD). Future investigations should delve deeper into the immune regulatory mechanisms of PSA and PAGln in specific infection models (e.g., pneumonia) and their functional interplay in the context of host-microbiota interactions. These research outcomes will offer crucial theoretical foundations and novel research avenues for the development of personalized medical strategies and the refinement of immunotherapies.
To sum up, the intestinal microbiota and its metabolites play a pivotal role in the immune regulation of various types of pneumonia. They modulate immune cell functions and inflammatory responses via multiple pathways, thereby influencing pathogen clearance and lung tissue repair. However, current research still has certain limitations. First, most studies concentrate on specific pathogens or single immune pathways, with limited exploration of mixed infections or multi-pathway synergistic effects. Second, clinical studies are relatively scarce; the majority of research relies on animal models or in vitro experiments, and the translatability of their findings to humans requires further validation. Future research directions should encompass the following aspects: conducting more clinical trials to assess the therapeutic efficacy and safety of interventions targeting the gut microbiota (e.g., probiotics, fecal microbiota transplantation) for various types of pneumonia; investigating the regulatory mechanisms of the gut microbiota in mixed infections and multi-pathogen scenarios to elucidate its immunomodulatory roles in complex infections; and considering factors such as individual variability and microbial diversity that may influence research outcomes, thereby enhancing the precision and reproducibility of studies.
4.2 Microbial translocation
The maintenance of intestinal barrier function relies on the equilibrium of gut microbiota, integrity of the mucosa, and normal functioning of the immune system. Once one or more of these protective mechanisms are compromised, bacteria or their products such as endotoxins (lipopolysaccharides, LPS) may breach the intestinal epithelium and disseminate to mesenteric lymph nodes and even distant organs like liver and lungs. This phenomenon is known as bacterial translocation (125). During bacterial translocation, bacteria initially adhere to intestinal epithelial cells, followed by rupture of these cells’ membranes allowing bacteria to infiltrate and reach the basement membrane. Subsequently, through drainage from the intestine, bacteria can migrate to mesenteric lymph nodes where they have potential for further dissemination to other organs and tissues. Studies have revealed an enrichment of gut bacteria in lung microbiota among patients with acute respiratory distress syndrome (ARDS), providing novel evidence for a pivotal role played by bacterial translocation in the gut-lung axis (126). Furthermore, bacterial translocation is closely associated with alterations in intestinal barrier function, bacterial overgrowth, and impairment of host defense mechanisms. (Figure 2).
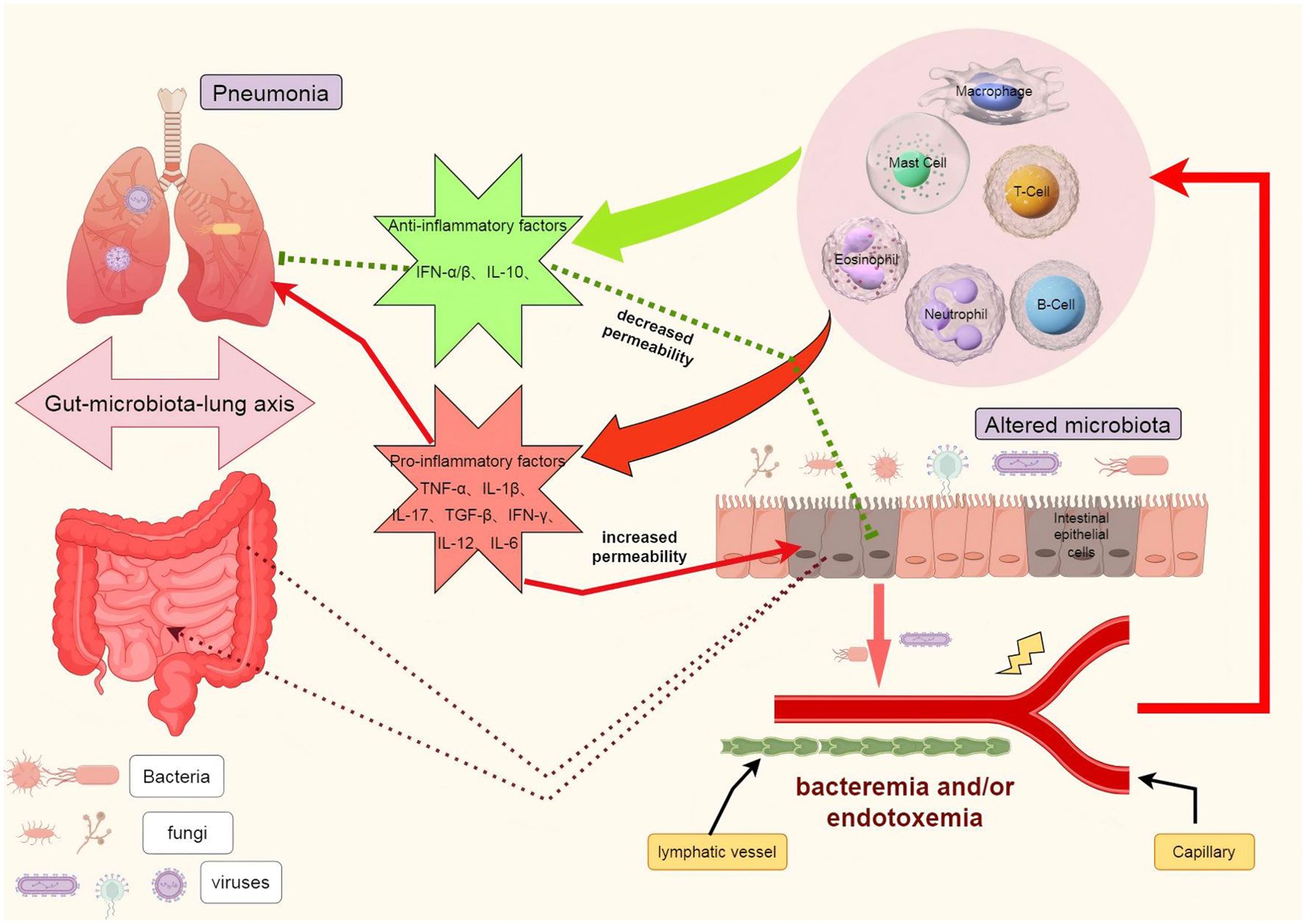
Figure 2. The role of bacterial translocation in the gut-lung axis. In the context of illness, the disruption of the intestinal barrier facilitates the translocation of pathogenic bacteria, leading to dysbiosis in the intestinal flora. These bacteria can be transported to the lungs via the lymphatic system and/or bloodstream, where they activate immune cells such as macrophages, neutrophils, T cells, and B cells. This activation results in the release of cytokines including IL-1, IL-6, IL-10, IL-12, TNFα, and IFN, which may either exacerbate or alleviate pulmonary infections and acute lung injury. Furthermore, these inflammatory factors can also reciprocally influence the intestinal barrier, potentially worsening or mitigating its damage. TNF-α tumor necrosis factor α, IL interleukin, IFN-γ interferon γ.
The healthy intestinal mucosal mechanical barrier consists of intact epithelial cells and the cell-to-cell junction complex, which includes tight junction proteins (TJ proteins). The function of the intestinal mucosal barrier is closely associated with the regulation of TJ protein levels. Indeed, the permeability of the intestinal mucosa is considered a crucial factor in innate defense, and disruption of barrier function can lead to excessive activation of immune cells (127). Inflammatory cytokines such as tumor necrosis factor-alpha (TNF-α), interferon-gamma (IFN-γ), interleukin-1 beta (IL-1β), and IL-12 disrupt the TJ barrier in the intestine during inflammation, thereby increasing TJ permeability (128). Moreover, IL-6 modulates claudin-2 protein expression through IL-6Rα-associated signaling transducers and gp130 signaling pathway, consequently enhancing TJ permeability in intestinal epithelium (129). Additionally, anti-inflammatory cytokine IL-10 plays a critical role in maintaining intestinal homeostasis by preserving intestinal barrier integrity; decreased levels of IL-10 and increased levels of IL-17 may exacerbate inflammation in a specific manner (130, 131).
In the gastrointestinal tract, a well-balanced symbiotic microbial community can confer health benefits to the host, while reduced microbial diversity may serve as an early indicator of certain pathological conditions. Both bacterial and viral-induced lung infections have the potential to disrupt the respiratory microbiota, leading to dysbiosis in the host gut microbiome along with compromised mucosal barrier and immune function, thereby promoting the development of lung infections. Deng et al.’s study unveiled that Ge-Gen-Qin-Lian decoction plays a protective role in safeguarding mice against influenza virus-induced pneumonia by partially improving the gut microbiota composition (such as increasing Akkermansia, Desulfovibrio, and Lactobacillus abundance while reducing Escherichia coli), inhibiting NOD/RIP2/NF-κB signaling pathway activation, and modulating expression of inflammatory cytokines in mesenteric lymph nodes (mLNs) and serum (132).
Gut-associated lymphoid tissue (GALT), being the largest immune organ in the human body, constitutes 25% of mucosal cell mass. In a thymus-deficient mouse model, spontaneous transfer of certain naturally occurring bacteria (e.g., Escherichia coli) to mesenteric lymph nodes (MLN), spleen, and liver has been observed; however, this phenomenon was not evident in heterozygous mice or nude mice receiving thymus transplantation (125). Subsequent investigations have demonstrated that T cell depletion leads to bacterial accumulation in rat MLN, while alcohol consumption and burns exacerbate bacterial alterations in rat MLN and increase the risk of bacterial translocation (133). Clearly, there exists a discernible correlation between host immune status and bacterial translocation.
Under healthy conditions, the host immune system efficiently prevents bacterial translocation by preserving intestinal barrier integrity, modulating microbiota balance, and regulating inflammatory responses. Gut-associated lymphoid tissue (GALT) plays a pivotal role in this process. It not only monitors pathogens within the intestine but also maintains intestinal homeostasis by generating immune cells and factors. For example, T cells and B cells in GALT can recognize and eliminate translocated bacteria while simultaneously producing anti-inflammatory cytokines such as IL-10 to sustain intestinal immune tolerance (134, 135).Moreover, innate immune cells recognize pathogen-associated molecular patterns via pattern recognition receptors (e.g., Toll-like receptors) and activate signaling pathways like NF-κB, thereby modulating the intensity and duration of the immune response (136). Conversely, under pathological conditions, the host immune system’s ability to regulate bacterial translocation becomes compromised. For instance, patients with chronic intestinal inflammatory diseases, such as inflammatory bowel disease (IBD), often exhibit impaired intestinal barrier function and increased mucosal permeability, which facilitates bacterial translocation (137).
5 Key mediators in the immune regulation of the gut-lung axis: PAMPs and DAMPs
Pattern recognition receptors (PRRs) serve as key mediators in the immune regulation of the gut-lung axis by recognizing pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs) (138). PAMPs are molecular structures derived from pathogens, such as lipopolysaccharides or endotoxins from bacterial cell walls, which are recognized by intracellular pattern recognition receptors, including Toll-like receptors. Through the modulation of transcriptional initiation and signal cascade reactions, PRRs facilitate the elimination of pathogens (139). In contrast, the main difference of damage-associated molecular patterns is that they recognize endogenous molecular structures associated with damaged cells. Microbe-related molecular patterns can activate host pattern recognition receptors (such as NOD1 and NOD2), thereby activating downstream signaling pathways (140). Mutations in NOD proteins are associated with various inflammatory diseases affecting nuclear factor κB (NF-κB) activity, which is a key signaling pathway involved in apoptosis, inflammation, and immune response (141, 142).
TLRs are canonical transmembrane PRRs expressed on the surface of intestinal epithelial cells and alveolar macrophages, which specifically recognize pathogen-associated molecular components. For instance, upon recognizing LPS from Gram-negative bacteria, TLR4 activates NF-κB and MAPK signaling via the MyD88-dependent pathway, inducing the release of pro-inflammatory cytokines such as IL-6 and TNF-αto clear pathogens in the intestine or lungs (143, 144);Meanwhile, the TLR2/6 heterodimer recognizes bacterial toxins, promoting Th17 cell differentiation to enhance immune responses (145). It is worth noting that there exists a synergistic effect in the activation of Toll-like receptors (TLRs) in both the intestine and lungs. Specifically, TLR4 and TLR5 in the intestine recognize specific components of the gut microbiota (GM) and contribute to the immune defense mechanism against the influenza virus during this process (146). In contrast, intracellular NOD-like receptors (NLRs), such as NOD1/NOD2, primarily sense bacterial peptidoglycan fragments (e.g., iE-DAP and MDP). Their signaling activates the NF-κB and pro-inflammatory body pathways via RIP2 kinase, thereby regulating the production of antibacterial peptides in intestinal Paneth cells and the repair of the lung epithelial barrier (147, 148). Studies have demonstrated that mutations in the NOD2 gene are associated with susceptibility to Crohn’s disease and severe pneumonia, potentially due to lung immune imbalance caused by intestinal flora dysregulation (149, 150). Furthermore, recent research has uncovered that the gut microbiota regulates the metabolic reprogramming of alveolar macrophages through the NLRP6 inflammasome, enhancing neutrophil extracellular traps (NETs) and influencing bacterial clearance rates in the lungs (151, 152). This discovery offers a novel perspective on the immune interaction mediated by pattern recognition receptors (PRRs) within the gut-lung axis.
To sum up, PRRs function as a “two-way regulator” in the collaborative defense of intestinal and pulmonary immunity through tissue-specific expression and signal cross-talk. Nevertheless, their excessive activation may also result in immunopathological damage, indicating that targeting the PRR pathway necessitates careful consideration of both spatiotemporal specificity and the homeostasis of the host microenvironment.
6 The influence of clinical treatment intervention
6.1 Antibiotics
The physiological structure of the mucosal surfaces in the upper respiratory tract and intestines harbors distinct microbial communities. In a state of health, these microbial communities play a crucial role in preventing potential pathogen colonization and regulating immune responses to mitigate lung infections. However, the administration of antibiotics and intensive care procedures can disrupt the equilibrium of these microbial communities, compromising their protective functions and consequently elevating the susceptibility to lung infections.
Antibiotics may affect the nutritional metabolism of the gut by altering the abundance, diversity, and evenness of gut microbiota, reducing levels of short-chain fatty acids, regulating solute secretion and absorption, and decreasing secondary bile acid production. This can subsequently promote the occurrence of antibiotic-associated diarrhea (153). Research suggests that long-term exposure to antibiotics can damage the quantity and diversity of distal gut microbiota, even in the absence of clinically related gastrointestinal symptoms. The use of azithromycin significantly reduces gut microbial diversity in the short term, particularly leading to a significant decrease in Actinobacteria. Although there is some recovery over time, this dysbiosis may persist for up to three years (154). In contrast, disruption caused by macrolide antibiotics has a shorter duration but can still last up to two years. Similarly, studies on mouse models have revealed that after using neomycin sulfate, clearance of sensitive intestinal bacteria increases susceptibility to influenza virus infection in lungs (24, 155), and antibiotic use may lead to dysbiosis in gut microbiota communities which increases mortality risk due to bacterial infections (26). Conversely, Rifaximin boosts the expression of intestinal tight junction proteins and enhances intestinal barrier integrity, preventing the translocation of harmful substances and pathogens. It also inhibits gut dysbiosis caused by influenza virus, reduces conditionally pathogenic bacteria like E. coli, and lowers infection risks and disease severity (156).
6.2 Probiotics
Probiotics not only prevent intestinal dysbiosis caused by certain diseases and its impact on the intestines, but also exhibit anti-inflammatory effects in the lungs. Research has demonstrated that probiotics have a positive effect on relieving symptoms of influenza and COVID-19 (157). For instance, a study evaluating the anti-influenza activity of four strains of Lactobacillus plantarum and two strains of Bifidobacterium longum found that treatment with Lp 330, CK10, 920, and Lm 218 reduced the size of viral plaques for rK09 and rAH01 viruses (158). This provides strong evidence for the protective role of probiotics in human viral infections. In hospital intensive care units (ICUs), ventilator-associated pneumonia (VAP) is one of the most prevalent infections among mechanically ventilated patients and is associated with high mortality rates. Nevertheless, randomized controlled trials investigating probiotic treatment for VAP have reported inconsistent results regarding its efficacy (159, 160). Oral administration of probiotics can modulate respiratory immune responses through various signaling pathways. For instance, Bifidobacterium promotes Th1/Th2 cell balance and upregulates the secretion of IFN-γ, IL-4, and IL-12 from the spleen (161). Escherichia coli reduces recruitment of inflammatory cells in the respiratory tract and suppresses Th2 and Th17 responses (162). Lactobacillus plantarum decreases the number of innate immune cells (such as macrophages and neutrophils) as well as cytokine levels (such as IL-6 and TNF-α) in bronchoalveolar lavage fluid (BALF), while inducing immunosuppressive Treg responses in the lungs (163). The intervention effects of different probiotic strains on pulmonary infection via the microbiota-immune axis exhibit significant variation. The underlying mechanisms encompass immune regulation, barrier repair, and metabolic modulation (as detailed in Table 1). For critically ill patients, intestinal-targeted therapy necessitates a delicate balance between efficacy and safety. While probiotics may induce bacteremia in immunocompromised individuals (164), such occurrences are typically self-limiting in most cases. However, some probiotics harbor antibiotic resistance genes, such as ARGs (165), which underscores the importance of whole-genome screening to ensure strain safety. Consequently, further extensive studies are warranted to elucidate the potential mechanisms governing the selection of specific probiotic strains, optimal timing, and clinical trials must be carried out to validate their efficacy. Future research should prioritize the precise identification of probiotic strains suitable for treating specific pulmonary infections, the development of more efficient formulations or delivery methods, and the implementation of personalized medicine tailored to individual health statuses and risk factors.
6.3 Fecal microbiota transplantation
Fecal Microbiota Transplantation (FMT) is a therapeutic approach that involves transferring the microbial community from the feces of healthy donors into the patient’s intestinal tract via minimally invasive techniques. This process aims to restore the diversity of intestinal microbiota, suppress the proliferation of pathogenic bacteria within the intestinal environment, and establish a competitive exclusion effect against pathogenic bacteria within the local intestinal microbiota. Recent mouse experiments have validated the efficacy of fecal microbiota transplantation (FMT) in ameliorating colonic pathological changes and upregulated TLR expression induced by smoke exposure. Additionally, FMT has been shown to restore intestinal flora diversity and mitigate lung inflammation mediated by IL-17 in models of COPD and smoking-related lung injury. Moreover, fecal microbiota transplantation (FMT) alleviates the cigarette smoke (CS)-induced depletion of blood B cells and spleen CD8+ T cells, as well as the increased circulation of Ly6Clo monocytes. These effects contribute to the restoration of gut microbial balance and the mitigation of lung injury (175). Tang et al. utilized FMT and antibiotic treatment to demonstrate that FMT can enhance the structure of the gut microbiota, promote the production of beneficial metabolites, reduce inflammation, repair the barrier function of alveolar epithelial cells, alleviate lung pathological damage, and significantly improve the prognosis of mice with pneumonia-induced sepsis caused by Klebsiella pneumoniae, and FMT restores immune system homeostasis by introducing specific bacterial colonies that produce short-chain fatty acids (e.g., butyrate). Studies involving healthy volunteers have demonstrated that FMT can transiently attenuate the cytotoxicity of systemic immune cells, decrease the proportion of CD8+ T cells and natural killer cells in circulation, while concurrently increasing the proportion of helper T cells (CD4+). This ultimately leads to an elevated CD4+/CD8+ ratio (176). Furthermore, the combination of FMT and antibiotics demonstrates superior efficacy in reducing gut dysbiosis and decreasing the abundance of antibiotic resistance genes (177). Furthermore, FMT restores immune system homeostasis by introducing specific bacterial colonies that produce short-chain fatty acids (e.g., butyrate). Studies involving healthy volunteers have demonstrated that FMT can transiently attenuate the cytotoxicity of systemic immune cells, decrease the proportion of CD8+ T cells and natural killer cells in circulation, while concurrently increasing the proportion of helper T cells (CD4+). This ultimately leads to an elevated CD4+/CD8+ ratio [56]. Nevertheless, its clinical application requires further optimization regarding standardized preparation protocols (e.g., rigorous donor screening) and long-term safety concerns (e.g., potential risks of metabolic disorders).
7 Conclusion
As mucosal organs directly exposed to the external environment, the lungs and intestines may harbor similar microbial communities that can modulate local and systemic immune and inflammatory responses. Pulmonary diseases can disrupt intestinal microbial ecology, leading to clinical manifestations of gastrointestinal disorders such as diarrhea and abdominal pain. Moreover, gut-derived metabolites may exacerbate lung inflammation through systemic circulation. Conversely, dysbiosis of gut microbiota could enhance susceptibility to pulmonary diseases and potentially act as causative agents for respiratory infections. Furthermore, antibiotic-induced diarrhea highlights how clinical interventions like antibiotic usage can perturb the balance of gut microbiota, thereby impacting immune defense function and predisposing individuals to lung infections. In the future, alternative intervention methods such as probiotics and fecal microbiota transplantation (FMT) may hold unique potential in the treatment of pulmonary infections. Extensive research has unveiled intricate interactions and mechanisms between the gut and lung infections, encompassing bacterial translocation, short-chain fatty acids, and recognition receptors for pathogen-associated molecular patterns. A comprehensive comprehension of these interaction mechanisms between the gut and lungs is imperative since targeting gut treatments to combat pneumonia could significantly propel medical advancements and enhance global health standards. Therefore, it is crucial to further elucidate the pertinent mechanisms underlying the gut-lung axis. To explore novel avenues for pneumonia treatment, forthcoming experiments involving probiotics/prebiotics should incorporate metagenomic analysis, metabolite studies, assessment of bacterial abundance, as well as analysis of their impact on inflammatory markers from diverse perspectives that influence gut microbiota in experimental research on pneumonia treatment and prevention.
Author contributions
FY: Conceptualization, Data curation, Methodology, Writing – original draft. LL: Conceptualization, Data curation, Methodology, Writing – original draft. JW: Conceptualization, Data curation, Writing – original draft. HY: Conceptualization, Data curation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. Medical Education Collaborative Innovation Fund of Jiangsu University (No.JDY2022008) and Key Research and Development Program of Zhenjiang City - Social Development Project (SH2023005).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
GI: gastrointestinal
COVID-19: Corona Virus Disease 2019
SARS-CoV-2: Severe Acute Respiratory Syndrome Coronavirus-2
CAP: Community-acquired- pneumonia
SCFA: Short-chain fatty acids
ILC: Innate lymphoid cells
Th17: T helper cell 17
Treg: Regulatory T cells
FMT: Fecal microbiota transplantation
GM-CSF: Granulocyte macrophage-colony stimulating factor
NF-κβ: Nuclear Factor kappa-light-chain-enhancer of activated B cells
IFN: Interferon
SFB: segmented filamentous bacteria
H1N1: AH1N1 influenza
VAP: Ventilator associated pneumonia
IFN-γ: Interferon-γ
BALF: Bronchoalveolar lavage fluid
TNF-α: Tumor Necrosis Factor-α
CS: Cigarette Smoke
FOXP3: Transcription factor Forkhead box P3
HDAC: Histone deacetylase
ARDS: acute respiratory distress syndrome
IL-1β: Interleukin-1beta
TJ: tight junction
MLN: Mesenteric Lymph Node
GALT: Gut-Associated Lymphoreticular Tissue
AI-2: Autoinducer 2
MDI: Microbiota Dysbiosis Index
SAP: Stroke-associated pneumonia
TMAO: Trimethylamine N-oxide
GATA3: GATA Binding Protein 3
NOD: Nucleotide-binding oligomerization domain
TUDCA: Tauroursodeoxycholate
AMPK: Adenosine 5’-monophosphate (AMP)-activated protein kinase
AMs: Alveolar Macrophage;DAT, Desaminotyrosine
MX1: Myxovirus Resistance Protein 1
AhR: Aryl Hydrocarbon Receptor
5-HT: 5-hydroxytryptamine
NLRP3: Nucleotide- binding oligomerization domain, leucine- rich repeat and pyrin domain- containing 3
TLR: Toll-like receptors
ROS: Reactive Oxygen Species
CXCL2: C C motif ligand 2
PMN: Polymorphonuclear
ARGs: Antibiotics resistance genes
HPLC: High-performance liquid chromatography
PXR: Pregnane X Receptor
18-HEPE: 18-Hydroxyeicosapentaenoic Acid
GM: Gut Microbiota
iE-DAP: γ-D-Glu-mDAP
RIP2: Receptor-Interacting Protein 2
MDP: Muramyl Dipeptide
IE: indole ethanol
IPA: indole propionic acid
ILA: indole lactic acid
IAA: indole acetic acid
IAld: indole aldehyde
IA: indole acrylic acid
MAT2A: methionine adenosyltransferase 2A
USP16: ubiquitin specific peptidase 16
OVA: ovalbumin
KYN: kynurenine
I3C: Indole-3-carbinol
CLA: Conjugated Linoleic Acid
NEC: necrotizing enterocolitis
BEAS-2B: Human Bronchial Epithelioid Cells
MCP-1: Monocyte chemotactic protein-1
CCL5: chemokine (C-C motif) ligand 5
ICAM-1: Intercellular cell adhesion molecule-1
CA: cholic acid
CDCA: chenodeoxycholic acid
BSH: Bile salt hydrolase
DCA: deoxycholic acid
LCA: lithocholic acid
TGR5: Takeda G protein-coupled receptor 5
ACE2: angiotensin-converting enzyme 2
UDCA: ursodeoxycholic acid
SOD: superoxide dismutase
CAT: catalase
GSH: Glutathione
ALI: acute lung injury
AQP: Aquaporin
LPS: lipopolysaccharide
PPARγ: peroxisome proliferator-activated receptor γ
BMI: body mass index
PTPN2: protein tyrosine phosphatase non-receptor 2
MRSA: methicillin-resistant Staphylococcus aureus
TMAO: Trimethylamine-N-oxide
TMA: Trimethylamine
DMA: dimethylamine
FMO: Flavin MonoOxygenase
PSA: Polysaccharide A
PPDC: phenylpyruvate decarboxylase
CD: Crohn’s disease
IBD: inflammatory bowel disease
PAGln: Phenylacetylglutamine.
References
1. Mandell LA. Community-acquired pneumonia: An overview. Postgrad. Med. (2015) 127:607–15. doi: 10.1080/00325481.2015.1074030
2. Ramirez JA, Wiemken TL, Peyrani P, Arnold FW, Kelley R, Mattingly WA, et al. Adults hospitalized with pneumonia in the United States: Incidence, epidemiology, and mortality. Clin Infect Dis. (2017) 65:1806–12. doi: 10.1093/cid/cix647
3. Paton JC, Trappetti C. Streptococcus pneumoniae capsular polysaccharide. Microbiol. Spectr. (2019) 7. doi: 10.1128/microbiolspec.GPP3-0019-2018
4. McChlery S, Ramage G, Bagg J. Respiratory tract infections and pneumonia. Periodontol. 2000. (2009) 49:151–65. doi: 10.1111/j.1600-0757.2008.00278.x
5. Pivard M, Moreau K, Vandenesch F. Staphylococcus aureus arsenal to conquer the lower respiratory tract. mSphere. (2021) 6:e00059-21. doi: 10.1128/mSphere.00059-21
6. Luyt CE, Hekimian G, Koulenti D, Chastre J. Microbial cause of ICU-acquired pneumonia: Hospital-acquired pneumonia versus ventilator-associated pneumonia. Curr Opin Crit Care. (2018) 24:332–8. doi: 10.1097/MCC.0000000000000526
7. Chatterjee A, Mavunda K, Krilov LR. Current state of respiratory syncytial virus disease and management. Infect Dis Ther. (2021) 10:5–16. doi: 10.1007/s40121-020-00387-2
8. Clementi N, Ghosh S, De Santis M, Castelli M, Criscuolo E, Zanoni I, et al. Viral respiratory pathogens and lung injury. Clin Microbiol Rev. (2021) 34:e00103-20. doi: 10.1128/CMR.00103-20
9. Dunn JJ, Miller MB. Emerging respiratory viruses other than influenza. Clin Lab Med. (2014) 34:409–30. doi: 10.1016/j.cll.2014.02.011
10. Jackson CB, Farzan M, Chen B, Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol. (2022) 23:3–20. doi: 10.1038/s41580-021-00418-x
11. Lamers MM, Haagmans BL. SARS-coV-2 pathogenesis. Nat Rev Microbiol. (2022) 20:270–84. doi: 10.1038/s41579-022-00713-0
12. Aleem MS, Sexton R, Akella J. Pneumonia in an immunocompromised patient. Treasure Island (FL: StatPearls (2025).
13. Natalini JG, Singh S, Segal LN. The dynamic lung microbiome in health and disease. Nat Rev Microbiol. (2023) 21:222–35. doi: 10.1038/s41579-022-00821-x
14. Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. (2012) 13:260–70. doi: 10.1038/nrg3182
15. Hillman ET, Lu H, Yao T, Nakatsu CH. Microbial ecology along the gastrointestinal tract. Microb Environ. (2017) 32:300–13. doi: 10.1264/jsme2.ME17017
16. Nkosi BVZ, Padayachee T, Gront D, Nelson DR, Syed K. Contrasting health effects of bacteroidetes and firmicutes lies in their genomes: Analysis of P450s, ferredoxins, and secondary metabolite clusters. Int J Mol Sci. (2022) 23:5057. doi: 10.3390/ijms23095057
17. Wei L, Singh R, Ro S, Ghoshal UC. Gut microbiota dysbiosis in functional gastrointestinal disorders: Underpinning the symptoms and pathophysiology. JGH Open. (2021) 5:976–87. doi: 10.1002/jgh3.12528
18. Frohlich EE, Farzi A, Mayerhofer R, Reichmann F, Jacan A, Wagner B, et al. Cognitive impairment by antibiotic-induced gut dysbiosis: Analysis of gut microbiota-brain communication. Brain Behav Immun. (2016) 56:140–55. doi: 10.1016/j.bbi.2016.02.020
19. Yan F, Zhang Q, Shi K, Zhang Y, Zhu B, Bi Y, et al. Gut microbiota dysbiosis with hepatitis B virus liver disease and association with immune response. Front Cell Infect Microbiol. (2023) 13:1152987. doi: 10.3389/fcimb.2023.1152987
20. Huang Z, Liu K, Ma W, Li D, Mo T, Liu Q. The gut microbiome in human health and disease-Where are we and where are we going? A bibliometric analysis. Front Microbiol. (2022) 13:1018594. doi: 10.3389/fmicb.2022.1018594
21. Bingula R, Filaire M, Radosevic-Robin N, Bey M, Berthon JY, Bernalier-Donadille A, et al. Desired turbulence? Gut-lung axis, immunity, and lung cancer. J Oncol. (2017) 2017:5035371. doi: 10.1155/2017/5035371
22. Zuo T, Liu Q, Zhang F, Lui GC, Tso EY, Yeoh YK, et al. Depicting SARS-CoV-2 faecal viral activity in association with gut microbiota composition in patients with COVID-19. Gut. (2021) 70:276–84. doi: 10.1136/gutjnl-2020-322294
23. Harding JN, Siefker D, Vu L, You D, DeVincenzo J, Pierre JF, et al. Altered gut microbiota in infants is associated with respiratory syncytial virus disease severity. BMC Microbiol. (2020) 20:140. doi: 10.1186/s12866-020-01816-5
24. Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A. (2011) 108:5354–9. doi: 10.1073/pnas.1019378108
25. Chen LW, Chen PH, Hsu CM. Commensal microflora contribute to host defense against Escherichia coli pneumonia through Toll-like receptors. Shock. (2011) 36:67–75. doi: 10.1097/SHK.0b013e3182184ee7
26. Schuijt TJ, Lankelma JM, Scicluna BP, de Sousa e Melo F, Roelofs JJ, de Boer JD, et al. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut. (2016) 65:575–83. doi: 10.1136/gutjnl-2015-309728
27. Metsala J, Lundqvist A, Virta LJ, Kaila M, Gissler M, Virtanen SM. Prenatal and post-natal exposure to antibiotics and risk of asthma in childhood. Clin Exp Allergy. (2015) 45:137–45. doi: 10.1111/cea.12356
28. Liang W, Feng Z, Rao S, Xiao C, Xue X, Lin Z, et al. Diarrhoea may be underestimated: a missing link in 2019 novel coronavirus. Gut. (2020) 69:1141–3. doi: 10.1136/gutjnl-2020-320832
29. Blackett JW, Sun Y, Purpura L, Margolis KG, Elkind MSV, O’Byrne S, et al. Decreased gut microbiome tryptophan metabolism and serotonergic signaling in patients with persistent mental health and gastrointestinal symptoms after COVID-19. Clin Transl Gastroenterol. (2022) 13:e00524. doi: 10.14309/ctg.0000000000000524
30. Ren X, Gamallat Y, Liu D, Zhu Y, Meyiah A, Yan C, et al. The distribution characteristics of intestinal microbiota in children with community-acquired pneumonia under five Years of age. Microb Pathog. (2020) 142:104062. doi: 10.1016/j.micpath.2020.104062
31. Wen L, Shi L, Kong X-L, Li K-Y, Li H, Jiang D-X, et al. Gut Microbiota Protected Against pseudomonas aeruginosa Pneumonia via Restoring Treg/Th17 Balance and Metabolism. Front Cell Inf Microbiol. (2022) 12. doi: 10.3389/fcimb.2022.856633
32. Wu T, Xu F, Su C, Li H, Lv N, Liu Y, et al. Alterations in the gut microbiome and cecal metabolome during klebsiella pneumoniae-induced pneumosepsis. Front Immunol. (2020) 11. doi: 10.3389/fimmu.2020.01331
33. Groves HT, Cuthbertson L, James P, Moffatt MF, Cox MJ, Tregoning JS. Respiratory disease following viral lung infection alters the murine gut microbiota. Front Immunol. (2018) 9:182. doi: 10.3389/fimmu.2018.00182
34. Sze MA, Tsuruta M, Yang SW, Oh Y, Man SF, Hogg JC, et al. Changes in the bacterial microbiota in gut, blood, and lungs following acute LPS instillation into mice lungs. PLoS One. (2014) 9:e111228. doi: 10.1371/journal.pone.0111228
35. Xia GH, Zhang MS, Wu QH, Wang HD, Zhou HW, He Y, et al. Dysbiosis of gut microbiota is an independent risk factor of stroke-associated pneumonia: A chinese pilot study. Front Cell Infect Microbiol. (2021) 11:715475. doi: 10.3389/fcimb.2021.715475
36. Viana SD, Nunes S, Reis F. ACE2 imbalance as a key player for the poor outcomes in COVID-19 patients with age-related comorbidities - Role of gut microbiota dysbiosis. Ageing Res Rev. (2020) 62:101123. doi: 10.1016/j.arr.2020.101123
37. Li J, Yan Y, Fu Y, Chen Z, Yang Y, Li Y, et al. ACE2 mediates tryptophan alleviation on diarrhea by repairing intestine barrier involved mTOR pathway. Cell Mol Biol Lett. (2024) 29:90. doi: 10.1186/s11658-024-00603-8
38. Zeng X, Yue H, Zhang L, Chen G, Zheng Q, Hu Q, et al. Gut microbiota-derived autoinducer-2 regulates lung inflammation through the gut-lung axis. Int Immunopharmacol. (2023) 124:110971. doi: 10.1016/j.intimp.2023.110971
39. Janapatla RP, Dudek A, Chen CL, Chuang CH, Chien KY, Feng Y, et al. Marine prebiotics mediate decolonization of Pseudomonas aeruginosa from gut by inhibiting secreted virulence factor interactions with mucins and enriching Bacteroides population. J BioMed Sci. (2023) 30:9. doi: 10.1186/s12929-023-00902-w
40. Cait A, Hughes MR, Antignano F, Cait J, Dimitriu PA, Maas KR, et al. Microbiome-driven allergic lung inflammation is ameliorated by short-chain fatty acids. Mucosal Immunol. (2018) 11:785–95. doi: 10.1038/mi.2017.75
41. Li M, Liang P, Li Z, Wang Y, Zhang G, Gao H, et al. Fecal microbiota transplantation and bacterial consortium transplantation have comparable effects on the re-establishment of mucosal barrier function in mice with intestinal dysbiosis. Front Microbiol. (2015) 6:692. doi: 10.3389/fmicb.2015.00692
42. Moutsoglou DM, Tatah J, Prisco SZ, Prins KW, Staley C, Lopez S, et al. Pulmonary arterial hypertension patients have a proinflammatory gut microbiome and altered circulating microbial metabolites. Am J Respir Crit Care Med. (2023) 207:740–56. doi: 10.1164/rccm.202203-0490OC
43. Ipci K, Altintoprak N, Muluk NB, Senturk M, Cingi C. The possible mechanisms of the human microbiome in allergic diseases. Eur Arch Otorhinolaryngol. (2017) 274:617–26. doi: 10.1007/s00405-016-4058-6
44. Date Y, Ebisawa M, Fukuda S, Shima H, Obata Y, Takahashi D, et al. NALT M cells are important for immune induction for the common mucosal immune system. Int Immunol. (2017) 29:471–8. doi: 10.1093/intimm/dxx064
45. Hauptmann M, Schaible UE. Linking microbiota and respiratory disease. FEBS Lett. (2016) 590:3721–38. doi: 10.1002/feb2.2016.590.issue-21
46. Carding SR, Egan PJ. Gammadelta T cells: Functional plasticity and heterogeneity. Nat Rev Immunol. (2002) 2:336–45. doi: 10.1038/nri797
47. Cui L, Wang Y. Regulatory factors of ILC2 are therapeutic targets for lung inflammation. iLABMED. (2024) 2:205–20. doi: 10.1002/ila2.v2.3
48. Hurrell BP, Howard E, Galle-Treger L, Helou DG, Shafiei-Jahani P, Painter JD, et al. Distinct roles of LFA-1 and ICAM-1 on ILC2s control lung infiltration, effector functions, and development of airway hyperreactivity. Front Immunol. (2020) 11:542818. doi: 10.3389/fimmu.2020.542818
49. Huang Y, Mao K, Chen X, Sun MA, Kawabe T, Li W, et al. S1P-dependent interorgan trafficking of group 2 innate lymphoid cells supports host defense. Science. (2018) 359:114–9. doi: 10.1126/science.aam5809
50. Littman DR, Rudensky AY. Th17 and regulatory T cells in mediating and restraining inflammation. Cell. (2010) 140:845–58. doi: 10.1016/j.cell.2010.02.021
51. Liu J, Huang Y, Liu N, Qiu H, Zhang X, Liu X, et al. The imbalance of pulmonary Th17/Treg cells in BALB/c suckling mice infected with respiratory syncytial virus-mediated intestinal immune damage and gut microbiota changes. Microbiol Spectr. (2024) 12:e0181224. Correction for Liu. doi: 10.1128/spectrum.01812-24
52. Brown RL, Sequeira RP, Clarke TB. The microbiota protects against respiratory infection via GM-CSF signaling. Nat Commun. (2017) 8:1512. doi: 10.1038/s41467-017-01803-x
53. Dang AT, Begka C, Pattaroni C, Caley LR, Floto RA, Peckham DG, et al. Butyrate regulates neutrophil homeostasis and impairs early antimicrobial activity in the lung. Mucosal. Immunol. (2023) 16:476–85. doi: 10.1016/j.mucimm.2023.05.005
54. Wang H, Lian P, Niu X, Zhao L, Mu X, Feng B, et al. TLR4 deficiency reduces pulmonary resistance to Streptococcus pneumoniae in gut microbiota-disrupted mice. PLoS One. (2018) 13:e0209183. doi: 10.1371/journal.pone.0209183
55. Wang LL, Shen X, Xie Y, Ge A, Lu H, Gu S, et al. A gut Eggerthella lenta-derived metabolite impairs neutrophil function to aggravate bacterial lung infection. Sci Transl Med. (2025) 17:eadq4409. doi: 10.1126/scitranslmed.adq4409
56. Ngo VL, Lieber CM, Kang H-j, Sakamoto K, Kuczma M, Plemper RK, et al. Intestinal microbiota programming of alveolar macrophages influences severity of respiratory viral infection. Cell Host Microbe. (2024) 32:335–48.e8. doi: 10.1016/j.chom.2024.01.002
57. Viox EG, Bosinger SE, Douek DC, Schreiber G, Paiardini M. Harnessing the power of IFN for therapeutic approaches to COVID-19. J Virol. (2024) 98:e0120423. doi: 10.1128/jvi.01204-23
58. Wang Q, Fang Z, Xiao Y, Wang H, Zhang P, Lu W, et al. Lactiplantibacillus pentoses CCFM1227 Produces Desaminotyrosine to Protect against Influenza Virus H1N1 Infection through the Type I Interferon in Mice. Nutrients. (2023) 15:3659. doi: 10.3390/nu15163659
59. Fan R, Liu S, Sun N, Yang Y, Deng X, Hu B, et al. Gut microbiota composition is associated with disease severity and host immune responses in COVID-19. Front Cell Infect Microbiol. (2023) 13:1274690. doi: 10.3389/fcimb.2023.1274690
60. Hagihara M, Yamashita M, Ariyoshi T, Eguchi S, Minemura A, Miura D, et al. Clostridium butyricum-induced omega-3 fatty acid 18-HEPE elicits anti-influenza virus pneumonia effects through interferon-lambda upregulation. Cell Rep. (2022) 41:111755. doi: 10.1016/j.celrep.2022.111755
61. Mei J, Liu Y, Dai N, Hoffmann C, Hudock KM, Zhang P, et al. Cxcr2 and Cxcl5 regulate the IL-17/G-CSF axis and neutrophil homeostasis in mice. J Clin Invest. (2012) 122:974–86. doi: 10.1172/JCI60588
62. Gogoi M, Clark PA, Ferreira ACF, Rodriguez Rodriguez N, Heycock M, Ko M, et al. ILC2-derived LIF licences progress from tissue to systemic immunity. Nature. (2024) 632:885–92. doi: 10.1038/s41586-024-07746-w
63. McAleer JP, Nguyen NL, Chen K, Kumar P, Ricks DM, Binnie M, et al. Pulmonary th17 antifungal immunity is regulated by the gut microbiome. J Immunol. (2016) 197:97–107. doi: 10.4049/jimmunol.1502566
64. Zegarra-Ruiz DF, Kim DV, Norwood K, Kim M, Wu WH, Saldana-Morales FB, et al. Thymic development of gut-microbiota-specific T cells. Nature. (2021) 594:413–7. doi: 10.1038/s41586-021-03531-1
65. Vinolo MA, Rodrigues HG, Hatanaka E, Sato FT, Sampaio SC, Curi R. Suppressive effect of short-chain fatty acids on production of proinflammatory mediators by neutrophils. J Nutr Biochem. (2011) 22:849–55. doi: 10.1016/j.jnutbio.2010.07.009
66. Wang P, Tao W, Li Q, Ma W, Jia W, Kang Y. Indole-3-Aldehyde alleviates lung inflammation in COPD through activating Aryl Hydrocarbon Receptor to inhibit HDACs/NF-kappaB/NLRP3 signaling pathways. J Mol Med (Berl). (2025) 103:157–74. doi: 10.1007/s00109-024-02506-9
67. Renga G, D’Onofrio F, Pariano M, Galarini R, Barola C, Stincardini C, et al. Bridging of host-microbiota tryptophan partitioning by the serotonin pathway in fungal pneumonia. Nat Commun. (2023) 14:5753. doi: 10.1038/s41467-023-41536-8
68. Wen X, Xiaoyue D, Longkun D, Yue X, Man Y, Min Z, et al. Three main short-chain fatty acids inhibit the activation of THP-1 cells by Mycoplasma pneumoniae. Biosci Biotechnol Biochem. (2021) 85:923–30. doi: 10.1093/bbb/zbaa110
69. Shi DW, Wang DM, Ning LH, Li J, Dong Y, Zhang ZK, et al. Using 16S rDNA Sequencing Technology to Preliminarily Analyze Intestinal Flora in Children with Mycoplasma pneumoniae Pneumonia. BioMed Environ Sci. (2022) 35:528–37. doi: 10.3967/bes2022.070
70. Ling Z, Liu X, Guo S, Cheng Y, Shao L, Guan D, et al. Role of probiotics in mycoplasma pneumoniae pneumonia in children: A short-term pilot project. Front Microbiol. (2018) 9:3261. doi: 10.3389/fmicb.2018.03261
71. Wong SH, Yu J. Gut microbiota in colorectal cancer: Mechanisms of action and clinical applications. Nat Rev GI Hepatol. (2019) 16:690–704. doi: 10.1038/s41575-019-0209-8
72. Krautkramer KA, Fan J, Bäckhed F. Gut microbial metabolites as multi-kingdom intermediates. Nat Rev Microbiol. (2020) 19:77–94. doi: 10.1038/s41579-020-0438-4
73. Vital M, Karch A, Pieper DH. Colonic butyrate-producing communities in humans: an overview using omics data. mSystems. (2017) 2:e00130-17. doi: 10.1128/mSystems.00130-17
74. Sun M, Wu W, Liu Z, Cong Y. Microbiota metabolite short chain fatty acids, GPCR, and inflammatory bowel diseases. J Gastroenterol. (2017) 52:1–8. doi: 10.1007/s00535-016-1242-9
75. Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. (2011) 469:543–7. doi: 10.1038/nature09646
76. Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. (1987) 28:1221–7. doi: 10.1136/gut.28.10.1221
77. Tian X, Hellman J, Horswill AR, Crosby HA, Francis KP, Prakash A. Elevated gut microbiome-derived propionate levels are associated with reduced sterile lung inflammation and bacterial immunity in mice. Front Microbiol. (2019) 10:159. doi: 10.3389/fmicb.2019.00159
78. Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. (2014) 20:159–66. doi: 10.1038/nm.3444
79. Zhang Y, Chen X, Wang Y, Li L, Ju Q, Zhang Y, et al. Alterations of lower respiratory tract microbiome and short-chain fatty acids in different segments in lung cancer: a multiomics analysis. Front Cell Infect Microbiol. (2023) 13:1261284. doi: 10.3389/fcimb.2023.1261284
80. Jung TH, Park JH, Jeon WM, Han KS. Butyrate modulates bacterial adherence on LS174T human colorectal cells by stimulating mucin secretion and MAPK signaling pathway. Nutr Res Pract. (2015) 9:343–9. doi: 10.4162/nrp.2015.9.4.343
81. Jiminez JA, Uwiera TC, Abbott DW, Uwiera RRE, Inglis GD. Butyrate Supplementation at High Concentrations Alters Enteric Bacterial Communities and Reduces Intestinal Inflammation in Mice Infected with Citrobacter rodentium. mSphere. (2017) 2:e00243-17. doi: 10.1128/mSphere.00243-17
82. Gill PA, Muir JG, Gibson PR, van Zelm MC. A randomized dietary intervention to increase colonic and peripheral blood SCFAs modulates the blood B- and T-cell compartments in healthy humans. Am J Clin Nutr. (2022) 116:1354–67. doi: 10.1093/ajcn/nqac246
83. Kespohl M, Vachharajani N, Luu M, Harb H, Pautz S, Wolff S, et al. The microbial metabolite butyrate induces expression of th1-associated factors in CD4(+) T cells. Front Immunol. (2017) 8:1036. doi: 10.3389/fimmu.2017.01036
84. Wu T, Li H, Su C, Xu F, Yang G, Sun K, et al. Microbiota-derived short-chain fatty acids promote LAMTOR2-mediated immune responses in macrophages. mSystems. (2020) 5:e00587-20. doi: 10.1128/mSystems.00587-20
85. Kim M, Qie Y, Park J, Kim CH. Gut microbial metabolites fuel host antibody responses. Cell Host Microbe. (2016) 20:202–14. doi: 10.1016/j.chom.2016.07.001
86. Chen Z, Xu Q, Liu Y, Wei Y, He S, Lin W, et al. Vancomycin-induced gut microbiota dysbiosis aggravates allergic rhinitis in mice by altered short-chain fatty acids. Front Microbiol. (2022) 13:1002084. doi: 10.3389/fmicb.2022.1002084
87. Ballout J, Akiba Y, Kaunitz JD, Schwiertz A, Mazzuoli-Weber G, Breves G, et al. Alteration of the microbiota with vancomycin and high-fibre diet affects short-chain fatty acid/free fatty acid receptor signalling in rat caecum. J Nutr Biochem. (2025), 109881. doi: 10.1016/j.jnutbio.2025.109881
88. Roager HM, Licht TR. Microbial tryptophan catabolites in health and disease. Nat Comms. (2018) 9:3294. doi: 10.1038/s41467-018-05470-4
89. Ranhotra HS. Discrete interplay of gut microbiota L-tryptophan metabolites in host biology and disease. Mol Cell Biochem. (2023) 479:2273–90. doi: 10.1007/s11010-023-04867-0
90. Han Z, Fu J, Gong A, Ren W. Bacterial indole-3-propionic acid inhibits macrophage IL-1β production through targeting methionine metabolism. Sci China Life Sci. (2025) 68(4):1118–31. doi: 10.1007/s11427-024-2789-1
91. Negatu DA, Yamada Y, Xi Y, Go ML, Zimmerman M, Ganapathy U, et al. Gut microbiota metabolite indole propionic acid targets tryptophan biosynthesis in mycobacterium tuberculosis. mBio. (2019) 10:e02781-18. doi: 10.1128/mBio.02781-18
92. Heumel S, de Rezende Rodovalho V, Urien C, Specque F, Brito Rodrigues P, Robil C, et al. Shotgun metagenomics and systemic targeted metabolomics highlight indole-3-propionic acid as a protective gut microbial metabolite against influenza infection. Gut Microb. (2024) 16:2325067. doi: 10.1080/19490976.2024.2325067
93. Li L, Xu Z, Ni H, Meng Y, Xu Y, Xu H, et al. Hydrogen-rich water alleviates asthma airway inflammation by modulating tryptophan metabolism and activating aryl hydrocarbon receptor via gut microbiota regulation. Free Radical Biol Med. (2024) 224:50–61. doi: 10.1016/j.freeradbiomed.2024.08.009
94. Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. (2013) 39:372–85. doi: 10.1016/j.immuni.2013.08.003
95. Wang H, He Y, Dang D, Zhao Y, Zhao J, Lu W. Gut microbiota-derived tryptophan metabolites alleviate allergic asthma inflammation in ovalbumin-induced mice. Foods. (2024) 13:1336. doi: 10.3390/foods13091336
96. Viladomiu M, Hontecillas R, Bassaganya-Riera J. Modulation of inflammation and immunity by dietary conjugated linoleic acid. Eur J Pharmacol. (2016) 785:87–95. doi: 10.1016/j.ejphar.2015.03.095
97. Wang K, Xin Z, Chen Z, Li H, Wang D, Yuan Y. Progress of conjugated linoleic acid on milk fat metabolism in ruminants and humans. Anim Open Access J MDPI. (2023) 13:3429. doi: 10.3390/ani13213429
98. Su X, Yang Y, Gao Y, Wang J, Hao Y, Zhang Y, et al. Gut microbiota CLA and IL-35 induction in macrophages through Gαq/11-mediated STAT1/4 pathway: an animal-based study. Gut Microb. (2024) 16:2437253. doi: 10.1080/19490976.2024.2437253
99. Rastgoo S, Shimi G, Shiraseb F, Karbasi A, Ashtary-Larky D, Yousefi M, et al. The effects of conjugated linoleic acid supplementation on inflammatory cytokines and adipokines in adults: A GRADE-assessed systematic review and dose–response meta-analysis. Front Immunol. (2023) 14. doi: 10.3389/fimmu.2023.1092077
100. Stachowska E, Baśkiewicz-Masiuk M, Dziedziejko V, Gutowska I, Baranowska-Bosiacka I, Marchlewicz M, et al. Conjugated linoleic acid increases intracellular ROS synthesis and oxygenation of arachidonic acid in macrophages. Nutr (Burbank Los Angeles County Calif). (2008) 24:187–99. doi: 10.1016/j.nut.2007.10.018
101. Jiang C, Zhang F, Zhang M, Yan X, Chen Y, Yu Q, et al. Conjugated linoleic acid ameliorates necrotizing enterocolitis by suppressing inflammatory responses and maintaining intestinal barrier integrity via the PPARγ/NFκB signaling pathway. J Funct Foods. (2024) 123:106581. doi: 10.1016/j.jff.2024.106581
102. Huang WC, Tu RS, Chen YL, Tsai YY, Lin CF, Liou CJ. Conjugated linoleic acids suppress inflammatory response and ICAM-1 expression through inhibition of NF-κB and MAPK signaling in human bronchial epithelial cells. Food Funct. (2016) 7:2025–33. doi: 10.1039/c5fo01037c
103. MacRedmond R, Singhera G, Attridge S, Bahzad M, Fava C, Lai Y, et al. Conjugated linoleic acid improves airway hyper-reactivity in overweight mild asthmatics. Clin Exp Allergy. (2010) 40:1071–8. doi: 10.1111/j.1365-2222.2010.03531.x
104. Larabi AB, Masson HLP, Bäumler AJ. Bile acids as modulators of gut microbiota composition and function. Gut Microb. (2023) 15:2172671. doi: 10.1080/19490976.2023.2172671
105. Fuchs CD, Trauner M. Role of bile acids and their receptors in gastrointestinal and hepatic pathophysiology. Nat Rev GI Hepatol. (2022) 19:432–50. doi: 10.1038/s41575-021-00566-7
106. Zhang F, Luan J, Suo L, Wang H, Zhao Y, Sun T, et al. Altered gut microbiota and metabolite profiles in community-acquired pneumonia: a metagenomic and metabolomic study. Microbiol Spectr. (2025) 13(4):e0263924. doi: 10.1128/spectrum.02639-24
107. Brevini T, Maes M, Webb GJ, John BV, Fuchs CD, Buescher G, et al. FXR inhibition may protect from SARS-CoV-2 infection by reducing ACE2. Nature. (2023) 615:134–42. doi: 10.1038/s41586-022-05594-0
108. He X, Shi J, Bu L, Zhou S, Wu K, Liang G, et al. Ursodeoxycholic acid alleviates fat embolism syndrome-induced acute lung injury by inhibiting the p38 MAPK/NF-κB signalling pathway through FXR. Biochem Pharmacol. (2024) 230:116574. doi: 10.1016/j.bcp.2024.116574
109. Milivojac T, Grabež M, Amidžić L, Prtina A, Krivokuća A, Malicevic U, et al. Ursodeoxycholic and chenodeoxycholic bile acids alleviate endotoxininduced acute lung injury in rats by modulating aquaporin expression and pathways associated with apoptosis and inflammation. Front Pharmacol. (2025) 16:1484292. doi: 10.3389/fphar.2025.1484292
110. Myszor IT, Lapka K, Hermannsson K, Rekha RS, Bergman P, Gudmundsson GH. Bile acid metabolites enhance expression of cathelicidin antimicrobial peptide in airway epithelium through activation of the TGR5-ERK1/2 pathway. Sci Rep. (2024) 14:6750. doi: 10.1038/s41598-024-57251-3
111. Pugin B, Barcik W, Westermann P, Heider A, Wawrzyniak M, Hellings P, et al. A wide diversity of bacteria from the human gut produces and degrades biogenic amines. Microb Ecol Health DZ. (2017) 28:1353881. doi: 10.1080/16512235.2017.1353881
112. Nakamura A, Kurihara S, Takahashi D, Ohashi W, Nakamura Y, Kimura S, et al. Symbiotic polyamine metabolism regulates epithelial proliferation and macrophage differentiation in the colon. Nat Commun. (2021) 12:2105. doi: 10.1038/s41467-021-22212-1
113. Li X, Xiao X, Wang S, Wu B, Zhou Y, Deng P. Uncovering de novo polyamine biosynthesis in the gut microbiome and its alteration in inflammatory bowel disease. Gut Microb. (2025) 17:2464225. doi: 10.1080/19490976.2025.2464225
114. Wawrzyniak M, Groeger D, Frei R, Ferstl R, Wawrzyniak P, Krawczyk K, et al. Spermidine and spermine exert protective effects within the lung. Pharmacol Res POV. (2021) 9:e00837. doi: 10.1002/prp2.v9.4
115. Nanduri B, Swiatlo E. The expansive effects of polyamines on the metabolism and virulence of Streptococcus pneumoniae. PNA (Nathan). (2021) 13:4. doi: 10.1186/s41479-021-00082-x
116. Li Q, Tian P, Guo M, Liu X, Su T, Tang M, et al. Spermidine associated with gut microbiota protects against MRSA bloodstream infection by promoting macrophage M2 polarization. ACS Infect DZ. (2024) 10:3751–64. doi: 10.1021/acsinfecdis.3c00669
117. Wang X, Li X, Dong Y. Vitamin D decreases plasma trimethylamine-N-oxide level in mice by regulating gut microbiota. BioMed Res Int. (2020) 2020:9896743. doi: 10.1155/2020/9896743
118. Gessner A, di Giuseppe R, Koch M, Fromm MF, Lieb W, Maas R. Trimethylamine-N-oxide (TMAO) determined by LC-MS/MS: Distribution and correlates in the population-based PopGen cohort. Clin Chem Lab Med. (2020) 58:733–40. doi: 10.1515/cclm-2019-1146
119. Hemmati M, Kashanipoor S, Mazaheri P, Alibabaei F, Babaeizad A, Asli S, et al. Importance of gut microbiota metabolites in the development of cardiovascular diseases (CVD). Life Sci. (2023) 329:121947. doi: 10.1016/j.lfs.2023.121947
120. Fang Q, Zheng B, Liu N, Liu J, Liu W, Huang X, et al. Trimethylamine N-oxide exacerbates renal inflammation and fibrosis in rats with diabetic kidney disease. Front Physiol. (2021) 12:682482. doi: 10.3389/fphys.2021.682482
121. Stefania K, Ashok KK, Geena PV, Katarina P, Isak D. TMAO enhances TNF-α mediated fibrosis and release of inflammatory mediators from renal fibroblasts. Sci Rep. (2024) 14:9070. doi: 10.1038/s41598-024-58084-w
122. Guignabert C, Humbert M. Targeting transforming growth factor-β receptors in pulmonary hypertension. Eur Respir J. (2021) 57:2002341. doi: 10.1183/13993003.02341-2020
123. Johnson JL, Jones MB, Cobb BA. Polysaccharide-experienced effector T cells induce IL-10 in FoxP3+ regulatory T cells to prevent pulmonary inflammation. Glycobiology. (2018) 28:50–8. doi: 10.1093/glycob/cwx093
124. Feng R, Tian Z, Mao R, Ma R, Luo W, Zhao M, et al. Gut microbiome-generated phenylacetylglutamine from dietary protein is associated with crohn’s disease and exacerbates colitis in mouse model possibly via platelet activation. J Crohn’s Colitis. (2023) 17:1833–46. doi: 10.1093/ecco-jcc/jjad098
125. Krack A, Sharma R, Figulla HR, Anker SD. The importance of the gastrointestinal system in the pathogenesis of heart failure. Eur Heart J. (2005) 26:2368–74. doi: 10.1093/eurheartj/ehi389
126. Dickson RP, Singer BH, Newstead MW, Falkowski NR, Erb-Downward JR, Standiford TJ, et al. Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat Microbiol. (2016) 1:16113. doi: 10.1038/nmicrobiol.2016.113
127. Motta JP, Magne L, Descamps D, Rolland C, Squarzoni-Dale C, Rousset P, et al. Modifying the protease, antiprotease pattern by elafin overexpression protects mice from colitis. Gastroenterology. (2011) 140:1272–82. doi: 10.1053/j.gastro.2010.12.050
128. Al-Sadi R, Boivin M, Ma T. Mechanism of cytokine modulation of epithelial tight junction barrier. Front Biosci (Landmark Ed). (2009) 14:2765–78. doi: 10.2741/3413
129. Seyed Toutounchi N, Hogenkamp A, Varasteh S, Van’t Land B, Garssen J, Kraneveld AD, et al. Fusarium mycotoxins disrupt the barrier and induce IL-6 release in a human placental epithelium cell line. Toxins (Basel). (2019) 11:665. doi: 10.3390/toxins11110665
130. Burrello C, Giuffre MR, Macandog AD, Diaz-Basabe A, Cribiu FM, Lopez G, et al. Fecal microbiota transplantation controls murine chronic intestinal inflammation by modulating immune cell functions and gut microbiota composition. Cells. (2019) 8:517. doi: 10.3390/cells8060517
131. Shi Y, Liu Z, Cui X, Zhao Q, Liu T. Intestinal vitamin D receptor knockout protects from oxazolone-induced colitis. Cell Death Dis. (2020) 11:461. doi: 10.1038/s41419-020-2653-3
132. Deng L, Shi Y, Liu P, Wu S, Lv Y, Xu H, et al. GeGen QinLian decoction alleviate influenza virus infectious pneumonia through intestinal flora. BioMed Pharmacother. (2021) 141:111896. doi: 10.1016/j.biopha.2021.111896
133. Choudhry MA, Fazal N, Goto M, Gamelli RL, Sayeed MM. Gut-associated lymphoid T cell suppression enhances bacterial translocation in alcohol and burn injury. Am J Physiol Gastrointest Liver Physiol. (2002) 282:G937–47. doi: 10.1152/ajpgi.00235.2001
134. Gonnella PA, Waldner HP, Weiner HL. B cell-deficient (mu MT) mice have alterations in the cytokine microenvironment of the gut-associated lymphoid tissue (GALT) and a defect in the low dose mechanism of oral tolerance. J Immunol. (2001) 166:4456–64. doi: 10.4049/jimmunol.166.7.4456
135. Gu Y, Bartolome-Casado R, Xu C, Bertocchi A, Janney A, Heuberger C, et al. Immune microniches shape intestinal T(reg) function. Nature. (2024) 628:854–62. doi: 10.1038/s41586-024-07251-0
136. Wullaert A, Bonnet MC, Pasparakis M. NF-kappaB in the regulation of epithelial homeostasis and inflammation. Cell Res. (2011) 21:146–58. doi: 10.1038/cr.2010.175
137. Yu S, Sun Y, Shao X, Zhou Y, Yu Y, Kuai X, et al. Leaky gut in IBD: Intestinal barrier-gut microbiota interaction. J Microbiol Biotechnol. (2022) 32:825–34. doi: 10.4014/jmb.2203.03022
138. Nofi CP, Wang P, Aziz M. Chromatin-associated molecular patterns (CAMPs) in sepsis. Cell Death Dis. (2022) 13:700. doi: 10.1038/s41419-022-05155-3
139. Esmaealzadeh N, Ram M, Abdolghaffari A, Marques AM, Bahramsoltani R. Toll-like receptors in inflammatory bowel disease: A review of the role of phytochemicals. Phytomedicine. (2024) 123:155178. doi: 10.1016/j.phymed.2023.155178
140. Natividad JM, Petit V, Huang X, de Palma G, Jury J, Sanz Y, et al. Commensal and probiotic bacteria influence intestinal barrier function and susceptibility to colitis in Nod1-/-; Nod2-/- mice. Inflamm Bowel Dis. (2012) 18:1434–46. doi: 10.1002/ibd.22848
141. Sharma A, Maurya CK, Arha D, Rai AK, Singh S, Varshney S, et al. Nod1-mediated lipolysis promotes diacylglycerol accumulation and successive inflammation via PKCdelta-IRAK axis in adipocytes. Biochim Biophys Acta Mol Basis Dis. (2019) 1865:136–46. doi: 10.1016/j.bbadis.2018.10.036
142. Duggan BM, Foley KP, Henriksbo BD, Cavallari JF, Tamrakar AK, Schertzer JD. Tyrosine kinase inhibitors of Ripk2 attenuate bacterial cell wall-mediated lipolysis, inflammation and dysglycemia. Sci Rep. (2017) 7:1578. doi: 10.1038/s41598-017-01822-0
143. Wang S, Zhang K, Song X, Huang Q, Lin S, Deng S, et al. TLR4 overexpression aggravates bacterial lipopolysaccharide-induced apoptosis via excessive autophagy and NF-kappaB/MAPK signaling in transgenic mammal models. Cells. (2023) 12:1769. doi: 10.3390/cells12131769
144. Tang J, Xu L, Zeng Y, Gong F. Effect of gut microbiota on LPS-induced acute lung injury by regulating the TLR4/NF-kB signaling pathway. Int Immunopharmacol. (2021) 91:107272. doi: 10.1016/j.intimp.2020.107272
145. Simpson M, Frisbee A, Kumar P, Schwan C, Aktories K, Petri WA. Clostridioides difficile binary toxin is recognized by the toll-like receptor 2/6 heterodimer to induce a nuclear factor-kappaB response. J Infect Dis. (2022) 225:1296–300. doi: 10.1093/infdis/jiaa620
146. Ou G, Xu H, Wu J, Wang S, Chen Y, Deng L, et al. The gut-lung axis in influenza A: the role of gut microbiota in immune balance. Front Immunol. (2023) 14:1147724. doi: 10.3389/fimmu.2023.1147724
147. Trindade BC, Chen GY. NOD1 and NOD2 in inflammatory and infectious diseases. Immunol Rev. (2020) 297:139–61. doi: 10.1111/imr.12902
148. Tan G, Li RH, Li C, Wu F, Zhao XM, Ma JY, et al. Down-regulation of human enteric antimicrobial peptides by NOD2 during differentiation of the paneth cell lineage. Sci Rep. (2015) 5:8383. doi: 10.1038/srep08383149
149. Horowitz JE, Warner N, Staples J, Crowley E, Gosalia N, Murchie R, et al. Mutation spectrum of NOD2 reveals recessive inheritance as a main driver of Early Onset Crohn’s Disease. Sci Rep. (2021) 11:5595. doi: 10.1038/s41598-021-84938-8
150. Kapoor A, Forman M, Arav-Boger R. Activation of nucleotide oligomerization domain 2 (NOD2) by human cytomegalovirus initiates innate immune responses and restricts virus replication. PLoS One. (2014) 9:e92704. doi: 10.1371/journal.pone.0092704
151. Wang P, Zhu S, Yang L, Cui S, Pan W, Jackson R, et al. Nlrp6 regulates intestinal antiviral innate immunity. Science. (2015) 350:826–30. doi: 10.1126/science.aab3145
152. Cai S, Paudel S, Jin L, Ghimire L, Taylor CM, Wakamatsu N, et al. NLRP6 modulates neutrophil homeostasis in bacterial pneumonia-derived sepsis. Mucosal Immunol. (2021) 14:574–84. doi: 10.1038/s41385-020-00357-4
153. Mekonnen SA, Merenstein D, Fraser CM, Marco ML. Molecular mechanisms of probiotic prevention of antibiotic-associated diarrhea. Curr Opin Biotechnol. (2020) 61:226–34. doi: 10.1016/j.copbio.2020.01.005
154. Wei S, Mortensen MS, Stokholm J, Brejnrod AD, Thorsen J, Rasmussen MA, et al. Short- and long-term impacts of azithromycin treatment on the gut microbiota in children: A double-blind, randomized, placebo-controlled trial. EBioMedicine. (2018) 38:265–72. doi: 10.1016/j.ebiom.2018.11.035
155. Looft T, Allen HK. Collateral effects of antibiotics on mammalian gut microbiomes. Gut Microb. (2012) 3:463–7. doi: 10.4161/gmic.21288
156. Chen Y, Jiang Z, Lei Z, Ping J, Su J. Effect of rifaximin on gut-lung axis in mice infected with influenza A virus. Comp Immunol Microbiol Infect DZ. (2021) 75:101611. doi: 10.1016/j.cimid.2021.101611
157. Shahbazi R, Yasavoli-Sharahi H, Alsadi N, Ismail N, Matar C. Probiotics in treatment of viral respiratory infections and neuroinflammatory disorders. Molecules. (2020) 25:4891. doi: 10.3390/molecules25214891
158. Bae JY, Kim JI, Park S, Yoo K, Kim IH, Joo W, et al. Effects of Lactobacillus plantarum and Leuconostoc mesenteroides Probiotics on Human Seasonal and Avian Influenza Viruses. J Microbiol Biotechnol. (2018) 28:893–901. doi: 10.4014/jmb.1804.04001
159. Mahmoodpoor A, Hamishehkar H, Asghari R, Abri R, Shadvar K, Sanaie S. Effect of a probiotic preparation on ventilator-associated pneumonia in critically ill patients admitted to the intensive care unit: A prospective double-blind randomized controlled trial. Nutr Clin Pract. (2019) 34:156–62. doi: 10.1002/ncp.2019.34.issue-1
160. Johnstone J, Meade M, Lauzier F, Marshall J, Duan E, Dionne J, et al. Effect of probiotics on incident ventilator-associated pneumonia in critically ill patients: A randomized clinical trial. JAMA. (2021) 326:1024–33. doi: 10.1001/jama.2021.13355
161. Mahooti M, Abdolalipour E, Salehzadeh A, Mohebbi SR, Gorji A, Ghaemi A. Immunomodulatory and prophylactic effects of Bifidobacterium bifidum probiotic strain on influenza infection in mice. World J Microbiol Biotechnol. (2019) 35:91. doi: 10.1007/s11274-019-2667-0
162. Secher T, Maillet I, Mackowiak C, Le Berichel J, Philippeau A, Panek C, et al. The probiotic strain Escherichia coli Nissle 1917 prevents papain-induced respiratory barrier injury and severe allergic inflammation in mice. Sci Rep. (2018) 8:11245. doi: 10.1038/s41598-018-29689-9
163. Vareille-Delarbre M, Miquel S, Garcin S, Bertran T, Balestrino D, Evrard B, et al. Immunomodulatory Effects of Lactobacillus plantarum on Inflammatory Response Induced by Klebsiella pneumoniae. Infect Immun. (2019) 87:e00570-19. doi: 10.1128/IAI.00570-19
164. Salminen MK, Rautelin H, Tynkkynen S, Poussa T, Saxelin M, Valtonen V, et al. Lactobacillus bacteremia, clinical significance, and patient outcome, with special focus on probiotic L. rhamnosus GG. Clin Infect Dis. (2004) 38:62–9. doi: 10.1086/cid.2004.38.issue-1
165. Montassier E, Valdes-Mas R, Batard E, Zmora N, Dori-Bachash M, Suez J, et al. Probiotics impact the antibiotic resistance gene reservoir along the human GI tract in a person-specific and antibiotic-dependent manner. Nat Microbiol. (2021) 6:1043–54. doi: 10.1038/s41564-021-00920-0
166. Shoaib A, Xin L, Xin Y. Oral administration of Lactobacillus acidophilus alleviates exacerbations in Pseudomonas aeruginosa and Staphylococcus aureus pulmonary infections. Pak J Pharm Sci. (2019) 32:1621–30.
167. Khailova L, Baird CH, Rush AA, McNamee EN, Wischmeyer PE. Lactobacillus rhamnosus GG Improves Outcome in Experimental Pseudomonas aeruginosa Pneumonia: Potential Role of Regulatory T Cells. Shock. (2013) 40:496–503. doi: 10.1097/SHK.0000000000000066
168. Elean M, Raya Tonetti F, Fukuyama K, Arellano-Arriagada L, Namai F, Suda Y, et al. Immunobiotic Ligilactobacillus salivarius FFIG58 Confers Long-Term Protection against Streptococcus pneumoniae. Int J Mol Sci. (2023) 24:15773. doi: 10.3390/ijms242115773
169. Kim S, Lee S, Kim TY, Lee SH, Seo SU, Kweon MN. Newly isolated Lactobacillus paracasei strain modulates lung immunity and improves the capacity to cope with influenza virus infection. Microbiome. (2023) 11:260. doi: 10.1186/s40168-023-01687-8
170. Wang W, Li Y, Han G, Li A, Kong X. Lactobacillus fermentum CECT5716 alleviates the inflammatory response in asthma by regulating TLR2/TLR4 expression. Front Nutr. (2022) 9:931427. doi: 10.3389/fnut.2022.931427
171. Raftis EJ, Delday MI, Cowie P, McCluskey SM, Singh MD, Ettorre A, et al. Bifidobacterium breve MRx0004 protects against airway inflammation in a severe asthma model by suppressing both neutrophil and eosinophil lung infiltration. Sci Rep. (2018) 8:12024. doi: 10.1038/s41598-018-30448-z
172. Zabel B, Makela SM, Nedveck D, Hibberd AA, Yeung N, Latvala S, et al. The Effect of Bifidobacterium animalis subsp. lactis Bl-04 on Influenza A Virus Infection in Mice. Microbe. (2023) 11:2582. doi: 10.3390/microorganisms11102582
173. Racedo S, Villena J, Medina M, Aguero G, Rodriguez V, Alvarez S. Lactobacillus casei administration reduces lung injuries in a Streptococcus pneumoniae infection in mice. Microb Infect. (2006) 8:2359–66. doi: 10.1016/j.micinf.2006.04.022
174. Zeng Y, Li T, Chen X, Fang X, Fang C, Liang X, et al. Oral administration of Lactobacillus plantarum expressing aCD11c modulates cellular immunity alleviating inflammatory injury due to Klebsiella pneumoniae infection. BMC Vet Res. (2024) 20:399. doi: 10.1186/s12917-024-04248-9
175. Budden KF, Shukla SD, Bowerman KL, Vaughan A, Gellatly SL, Wood DLA, et al. Faecal microbial transfer and complex carbohydrates mediate protection against COPD. Gut. (2024) 73:751–69. doi: 10.1136/gutjnl-2023-330521
176. Goloshchapov OV, Olekhnovich EI, Sidorenko SV, Moiseev IS, Kucher MA, Fedorov DE, et al. Long-term impact of fecal transplantation in healthy volunteers. BMC Microbiol. (2019) 19:312. doi: 10.1186/s12866-019-1689-y
Keywords: pulmonary infection, gut microbiota, gut-lung axis, immunity, probiotics
Citation: Ye F, Li L, Wang J and Yang H (2025) Advances in gut-lung axis research: clinical perspectives on pneumonia prevention and treatment. Front. Immunol. 16:1576141. doi: 10.3389/fimmu.2025.1576141
Received: 13 February 2025; Accepted: 03 April 2025;
Published: 22 April 2025.
Edited by:
Teresa Zelante, University of Perugia, ItalyReviewed by:
Hideki Yoshimatsu, Osaka Dental University, JapanFelipe Melo-Gonzalez, Andres Bello University, Chile
Copyright © 2025 Ye, Li, Wang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongfeng Yang, ZmVuZzEwMjIyMEAxNjMuY29t
 Fang Ye
Fang Ye Linnan Li
Linnan Li