- 1Department of Fish Nutrition, West Bengal University of Animal and Fishery Sciences, Kolkata, India
- 2Department of Livestock Farm Complex, West Bengal University of Animal and Fishery Sciences, Kolkata, India
- 3Indian Council of Agriculture Research (ICAR)-Central Institute of Brakishwater Aquaculture, Kakdwip Research Centre, Kakdwip, India
- 4Department of Animal Nutrition, West Bengal University of Animal and Fishery Sciences, Kolkata, India
- 5West Bengal State Council of Science and Technology, Department of Science and Technology and Biotechnology, Kolkata, India
In the era of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the application of zinc has increased worldwide. It has the potential to increase the body’s antioxidant status and provide better immune makeup. Currently, zinc is indiscriminately used as a therapeutic agent, and this creates an unwanted antagonism with the other interacting micronutrients that are present in the gut and causes secondary deficiencies of other critical micronutrients, which often leads to various complications. In this study, our aim was to synthesize a nano-sized zinc followed by its dietary fortification. We found that the supplemental nano-zinc has an antiviral effect using milkfish (Chanos chanos) as a model organism. The IRF3 gene was chosen as a molecular marker for antiviral assessment, which has different integral zinc binding sites. For the first time, we have characterized the IRF3 gene in C. chanos and discovered certain important domains as zinc binding sites, as well as other important domains related to antiviral activity, such as the serine protease NS3 activity and the interferon regulatory factor (IRF) tryptophan pentad repeat domain. The expression profile of IRF3 was significantly improved among fish supplemented with dietary nano-zinc, and the best effect was observed in the group provided feed fortified with 40 ppm nano-zinc. The results of this study revealed that nano-zinc can directly be incorporated into IRF3, which increases its bioavailability and improves its antiviral activity through biochemical pathways, as described in the STRING and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses.
Introduction
The recent advances in nanobiotechnology have reshaped scientific innovations with newer molecular insights to understand the basis of different biological processes. The higher surface-to-charge ratio offers nanoscale materials some unique properties to carry out desired functions. Research is in vogue to design different varieties of nanostructures for processing the production of vaccines, drugs, feed additives, nutrient delivery, disease treatment, and water purification, among others (1).
Minerals, as micronutrients, play a crucial role in the life processes of all categories of animals, including fish, and there is a need for their regular intake in order to combat physiological deficiencies. Merely fulfilling the requirements of a mineral might not assure its adequacy at the cellular level. The bioavailability of minerals appears to be the most challenging issue (2) in the science of nutrition. The bioavailability of a mineral is mostly regulated by its source and the presence of other interacting micronutrients in the gut. Recently, different varieties of nano-minerals have been synthesized, which appear to be highly bioavailable and hence have the least interaction with other nutrients available in the gut. Supplemental nano-minerals may have the potential to enhance growth and immunity (3). Synthesized nanoparticles appear to have more efficacy than their bulk counterparts in combating pathogens. The antimicrobial activity of several metal oxide nanoparticles has been proven against a wide range of bacteria and fungi (4).
Zinc is the second most abundant trace element in the body. However, it cannot be stored in the body, which emphasizes the need for regular dietary intake to avoid any subclinical deficiencies (5, 6). Zinc plays a vital role in prostaglandin metabolism and provides structural rigidity to nucleoproteins (7). It serves as an essential part of approximately 20 metalloenzymes, including alkaline phosphatase, alcohol dehydrogenase, and carbonic anhydrase, among others. In addition, zinc plays a critical role in growth performance, fertility, immune function, wound healing, oxidative stress maintenance, etc. (5, 8, 9). Zinc oxide and nano-zinc oxide have the same chemical structures, which suggests similar zinc and oxygen ratios. However, nano-sized atoms have a wider energy level confinement and, due to size variations, become less interactive with other bulk-sized molecules available in the gut (10). The higher bioavailability of nanostructures ensures adequacy at the tissue level; hence, nano-zinc appears to be more potent for immunomodulation in fish. It can increase the immunoglobulin M level and upregulate the expression of interleukin 1 beta (IL-1β (11). This higher immunomodulation may be associated with the increased thymocytes and peripheral T cells, enhancing the production of interferon and interleukin 2. Supplemental dietary zinc can improve the immune biomarkers that lead to a better immune response (12). Zinc oxide nanostructures appear potent immunomodulators even at lower doses compared with their bulk counterparts and organic zinc sources (11). Nano-zinc oxide can also promote the immune response by increasing the extra thymulin activity, which helps in the enhancement of mature T lymphocytes and the activation of B lymphocytes by T helper cells (12, 13). It has often been associated with increased lymphocyte count and phagocytic activity (11).
Interferon regulatory factors (IRFs) are transcription factors of the interferon (IFN) inducible signaling pathway and are crucial for host immunity against antimicrobial infection by both viruses and bacteria (14). IRFs are transcriptional regulators in the antiviral signaling pathway of the cell. When microbes attack the cell, IRFs bind to the IFN regulatory element (IRF-E) within the promoter region and regularize the transcriptional levels of the target genes. Thus, they play a pivotal role in the response to antiviral infections. There are different IRFs that play critical roles in innate immunity. IRF1, IRF3, IRF5, IRF7, IRF8, and IRF9 are among the positive regulators of IFN and other immune-related genes (15). IRF3 was found as a regulator of virus-infected cells 20 years ago (16). It promotes IFN-α/β gene transcription in virus-infected cells (17) and plays a very important role in controlling the IFN response during viral infection in mammals (18). IRF3 can also mediate apoptosis-independent p53 and type l IFN (19). There are nine members of IRF3 present in mammals (20), 10 members present in birds (21), and 11 members found in fish (18, 22). Studies on the innate immunity of fish are of recent interest considering the repeated outbreaks of pathogens in recent times. Several studies have reported that fish, similar to mammals, possess a defense mechanism in their innate immune system (14). Fish utilize the IFN-induced system against viral infections (14, 23). The induction of fish IFN through viral infection promotes antiviral activity and increases the interferon-stimulated gene (ISG) expression that reduces viral replication (14, 24). Several representative IRFs have been identified in fish, among which Irf3 appears to play the key role in regulating the IFN response (14). Viral infection causes severe mortality in aquaculture, and it appears to cause great burden for farmers.
Attempts are in vogue to develop zinc-based feed additives with the aim of improving the body defense of fish. Being more bioavailable than and least antagonistic to other micronutrients, nano-zinc is presently gaining more attention. Fish are continuously being exposed to a plethora of stresses. The outbreak of various viral diseases is quite common in aquaculture and is a major issue for farmers. A number of sporadic experiments on dietary zinc for immunomodulation have been carried out in freshwater fish. However, there are scant reports available on brackish water fish. Milkfish (Chanos chanos) is rather delicious with an excellent nutrient profile and is thus quite popular in Southeast Asia and Taiwan (25). It is an important euryhaline finfish that can be cultured in freshwater, brackish water, or seawater (25, 26). In India, it has a high domestic market with good prices.
The present experiment aimed to compare the effect of formulated diets fortified with different sources of zinc (organic and nano-zinc) to evaluate its role in IRF3 gene expression, which is essential for the antiviral defense of milkfish.
Materials and methods
The present study aimed to determine the antiviral activity of nano-zinc in a milkfish (C. chanos) model. We synthesized the zinc nano-structures by employing an environment-benign colloidal chemistry route and characterized what has already been reported (27). They are suitable for mineral supplementation. IRF3 was employed as the molecular marker for assessment. We characterized the IRF3 gene in C. chanos for the first time and assessed the differential messenger RNA (mRNA) expression level in the liver with respect to organic zinc and 10, 20, and 40 ppm nano-zinc.
Experimental animals and dietary allocation
Apparently disease-free, healthy, motile C. chanos fries (0.35 ± 0.05 g) were procured from the pond of the Central Institute of Breakwater Aquaculture (CIBA), Kakdwip Research Centre, and were acclimatized for 15 days in 600-L fiber-reinforced plastic (FRP) tanks. The fish were fed twice daily (morning and evening) at 10% of their body weight. The waste was siphoned out and 80% water was exchanged at 3-day intervals to ensure the wellbeing of the fish with the least stress (28). No mortality was recorded during the present acclimatization period. The fish were observed for mortality daily, and dead fish were immediately removed. The water quality parameters (pH, dissolved oxygen, ammonia–nitrogen, etc.) were monitored regularly.
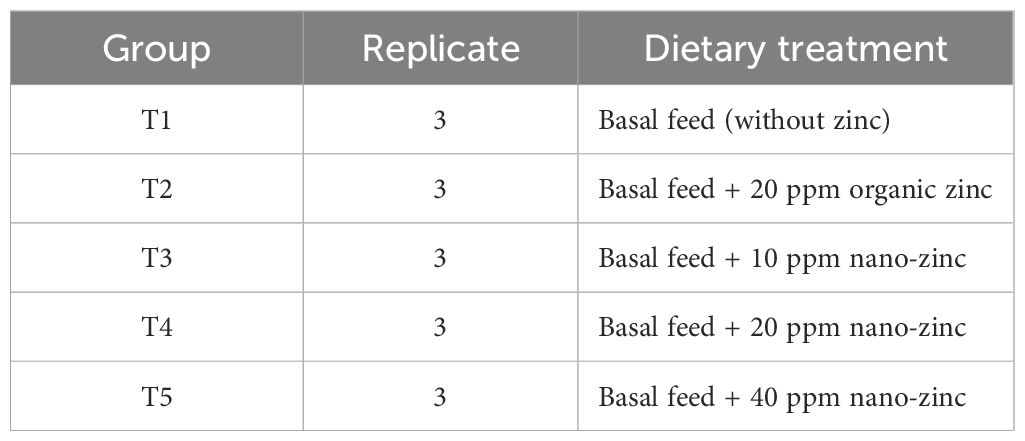
After the initial acclimatization, a total of 450 fish were randomly allocated into five treatment groups (Table 1), each with three replicates and each replicate containing 30 fish. The fish in T1 were fed the basal diet without any zinc supplementation; the fish in T2 were fed the basal diet supplemented with 20 ppm zinc proteinate; and the fish in the other three treatment groups (T3, T4, and T5) were supplemented with 10, 20, and 40 ppm nano-zinc oxide, respectively, as depicted in Table 1. The basal feeder remained isonitrogenous and isocaloric throughout the feeding trial across the treatment groups.
The feeding trial continued for 120 days and was followed by a digestibility trial extending for 5 days. At the end of the feeding trial, three fish from each replicate were collected in compliance with the guidelines for experimentation on fishes set by the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA), Government of India. At the end of the feeding trial, three fish from each replicate were sacrificed by a registered veterinarian (PNC). The fish were firstly taken out from the tank and were euthanized using 0.1% of 100 ml clove oil at a rate of per liter of sterile water. The liver was stored in a sterile vial by keeping submerged in RNAlater solution (Invitrogen by Thermo Fisher Scientific, Waltham, MA, USA).
Characterization of the IRF3 gene in C. chanos
Total RNA was isolated from the liver using the TRIzol method and was subsequently converted to complementary DNA (cDNA) as per the standard protocol developed in our laboratory (29–32).
Materials
Taq DNA polymerase, 10× buffer, and dNTP were purchased from Invitrogen (Carlsbad, CA, USA). The SYBR Green qPCR Master Mix (2×) was obtained from Thermo Fisher Scientific Inc. (Allentown, PA, USA). l-Glutamine (Glutamax 100×) was purchased from Invitrogen Corp., (Carlsbad, CA, USA), while penicillin-G and streptomycin were from Amresco (Solon, OH, USA). Filters (Millex GV., 0.22 µm) were purchased from Millipore Pvt. Ltd., (Billerica, MA, USA). All other reagents were of analytical and molecular biology grade.
Synthesis, confirmation of cDNA, and PCR amplification of the gene
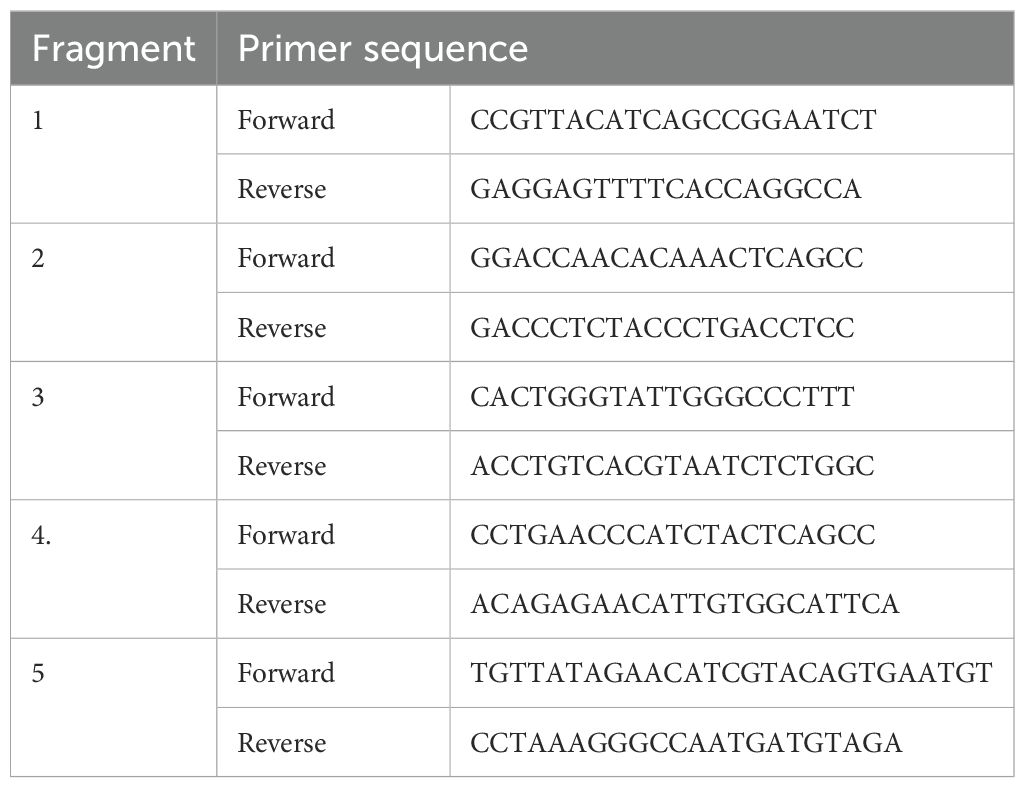
The 20-μl reaction mixture contained 5 μg of total RNA, 0.5 μg of oligo dT primer (16–18 mer), 40 U of ribonuclease inhibitor, 10 M of dNTP mix, 10 mM of DTT, and 5 U of MuMLV reverse transcriptase in reverse transcriptase buffer. The reaction mixture was gently mixed and incubated at 37°C for 1 h. The reaction was stopped by heating the mixture at 70°C for 10 min and chilling on ice. The integrity of the cDNA was assessed by PCR. The primers used are listed in Tables 2, 3. The 25-μl reaction mixture contained 80–100 ng cDNA, 3.0 μl 10× PCR assay buffer, 0.5 μl of 10 mM dNTP, 1 U Taq DNA polymerase, 60 ng of each primer (as in Table 2), and 2 mM MgCl2. The PCR reactions were carried out in a thermocycler (PTC-200; MJ Research, Hercules, CA, USA), with cycling conditions as follows: initial denaturation at 94°C for 3 min, denaturation at 94°C for 30 s, annealing temperature of 60°C for 35 s, and extension at 72°C for 3 min carried out for 35 cycles, followed by a final extension at 72°C for 10 min.
Study of the predicted gene using bioinformatics tools
The predicted peptide sequences of the genes of milkfish were derived using Edit sequence (Lasergene software; DNASTAR, Madison, WI, USA). Prediction of the signal peptide of the genes was conducted using the software SignalP3.0 Server—prediction results (Technical University of Denmark). Analysis of the O-linked glycosylation sites was carried out using the NetOGlyc 4 server (https://www.expassy.org/), whereas the N-linked glycosylation sites were detected using NetNGlyc 1.0 software (https://www.expassy.org/). The regions for the alpha-helix and beta-sheet were predicted using NetSurfP-Protein Surface Accessibility and Secondary Structure Predictions, Technical University of Denmark (33). Domain linker prediction was performed according to the software developed (34). The lipopolysaccharide (LPS) binding sites (35), as well as the LPS signaling sites (36), were predicted based on homology studies with other species of polypeptide.
Three-dimensional structure prediction and model quality assessment
The templates with the highest sequence ID entity to our target template were identified using PSI-BLAST (http://blast.ncbi.nlm.nih.gov/Blast). Homology modeling was used to build a three-dimensional (3D) structure based on homologous template structures using the PHYRE2 server (37). The 3D structures were visualized with PyMOL (http://www.pymol.org/), which is an open-source molecular visualization tool. Subsequently, the mutant model was generated using PyMoL tools. The Swiss PDB Viewer was employed to control energy minimization. Structural evaluation, along with a stereochemical quality assessment of the predicted model, was carried out using SAVES (Structural Analysis and Verification Server), which is an integrated server (http://nihserver.mbi.ucla.edu/SAVES/). The ProSA (Protein Structure Analysis) web server (https://prosa.services.came.sbg.ac.at/prosa) was used for the refinement and validation of the protein structure (38). ProSA was used to evaluate the structural quality of the model with potential errors, and the program shows a plot of its residue energies and Z-scores, which determine the overall quality of the model. The solvent accessibility surface area of the genes was generated using the NetSurfP server (https://www.cbs.dtu.dk/services/NetSurfP/) (33). This calculates the relative surface accessibility, the Z-fit score, the probabilities for both the alpha-helix and the beta-strand, and the coil score, among others. TM-align software was used for the alignment of the 3D structure of the IR proteins for different species and for the estimation of the root mean square deviation (RMSD) to assess structural differentiation (39). I-Mutant analysis was conducted for the mutations detected to assess the thermodynamic stability. PROVEAN analysis was conducted to assess the deleterious nature of the mutant amino acid. The PDB structure for the 3D structural prediction of the gene for milkfish was carried out using PHYRE software (38). Protein–protein interactions were examined using STRING analysis (40).
Differential mRNA expression profiling of the IRF3 gene in C. chanos
Real-time PCR
Total RNA was estimated from the liver of milkfish from the six treatment groups using the TRIzol method. First-strand cDNA was synthesized using reverse transcriptase polymerase chain reaction (RT-PCR) in an automated temperature-maintained thermocycler machine. M-MLVRT (200 U/µl) was used as the reverse transcriptase enzyme. The primer was obtained from a published journal (25). The primers used are listed in Table 3. Equal amounts of RNA (quantified with a Qubit fluorometer; Invitrogen, Carlsbad, CA, USA), whenever applicable, were used for cDNA preparation (SuperScript III cDNA Synthesis Kit; Invitrogen, Carlsbad, CA, USA). All qRT-PCR reactions were conducted on an ABI 7500 Fast system. Each reaction consisted of a 2-µl cDNA template, 5 µl of 2× SYBR Green PCR Master Mix, 0.25 µl each of forward and reverse primers (10 pmol/µl), and nuclease-free water for a final volume of 10 µl. Each sample was run in triplicate. Analysis of real-time PCR (qRT-PCR) was performed using the delta–delta Ct (ΔΔCt) method (29, 30, 32, 41).
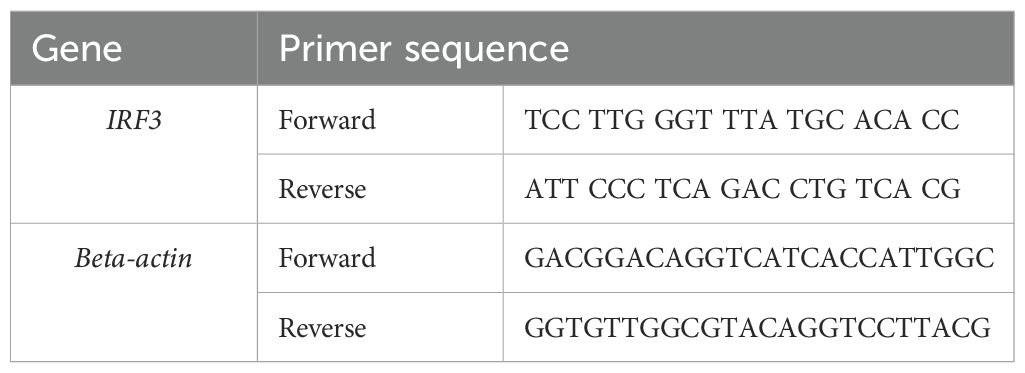
Table 3. PCR primers for differential mRNA expression profiling of the IRF3 gene and the housekeeping gene in Chanos chanos (25).
The entire reactions were performed in triplicate (as per the MIQE guidelines), and the experiment was repeated twice, in a 20-µl reaction volume, using FastStart Essential DNA Green Master (HiMedia, Mumbai, India) on the ABI 7500 system.
Statistical analysis
Descriptive statistics with the mean and standard error were estimated with the SYSTAT package for the expression levels analyzed using real-time PCR and accordingly presented in a graph. The expression level with real-time PCR was estimated using 2−ΔΔCt.
Results
Characterization of the IRF3 gene in C. chanos
Characterization of the IRF3 gene: in silico studies and identification of the important domains
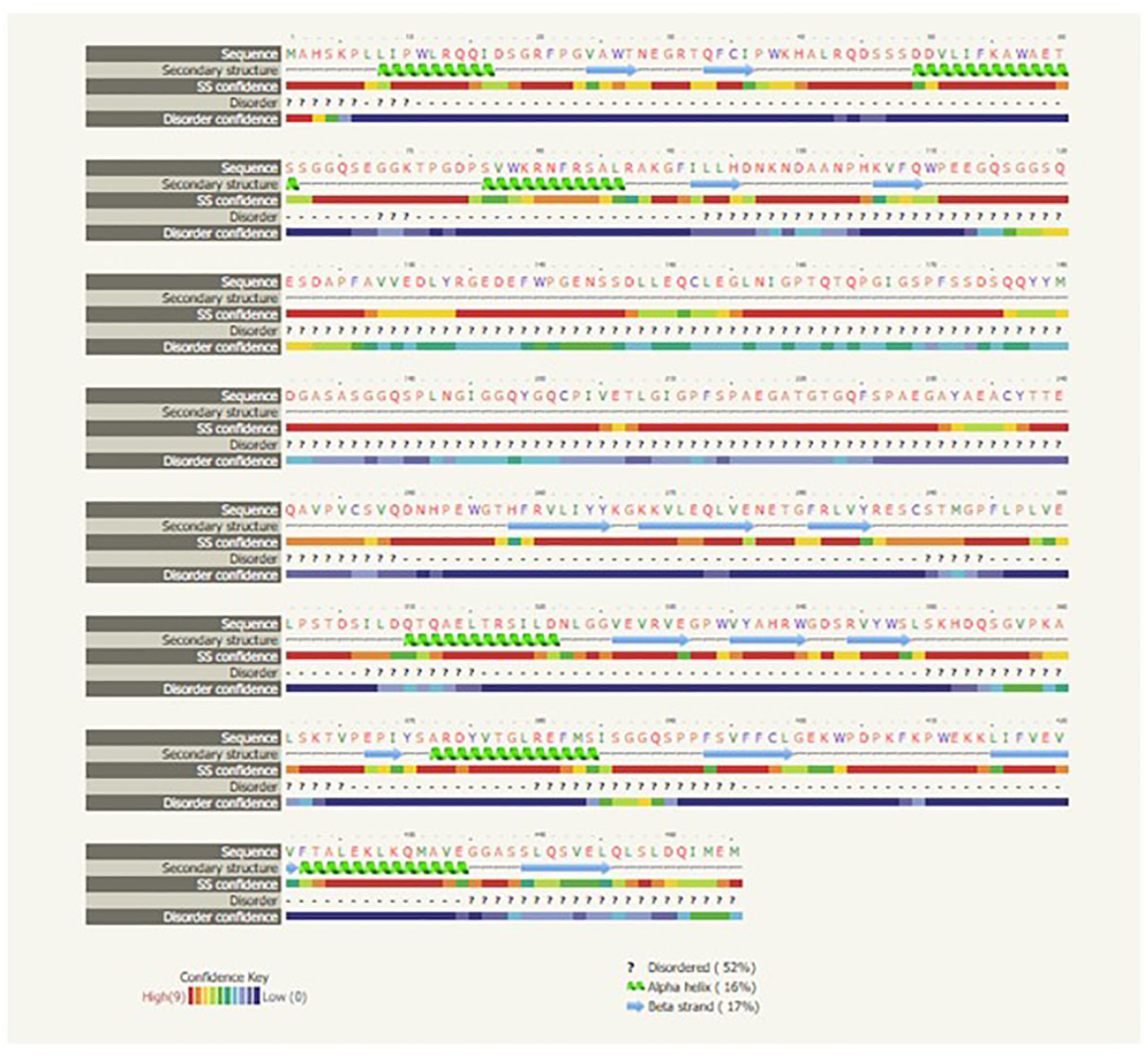
The IRF3 gene has been employed in current studies as an important molecular marker against viral infections. The IRF3 gene has been characterized and the predicted 3D structure for the peptide sequence visualized (Figure 1), with a predicted interferon regulatory factor (IRF) tryptophan pentad repeat domain at amino acid positions 5–102 (Figure 2, orange surface). The serine protease NS3 domain is also depicted in Figure 3. The zinc binding sites were detected at amino acid position 97 (Figure 1, green sphere), position 99 (Figure 1, red sphere), position 145 (Figure 1, blue sphere), and position 149 (Figure 1, yellow sphere). The secondary structure of the IRF3 gene of C. chanos is depicted in Figure 4.
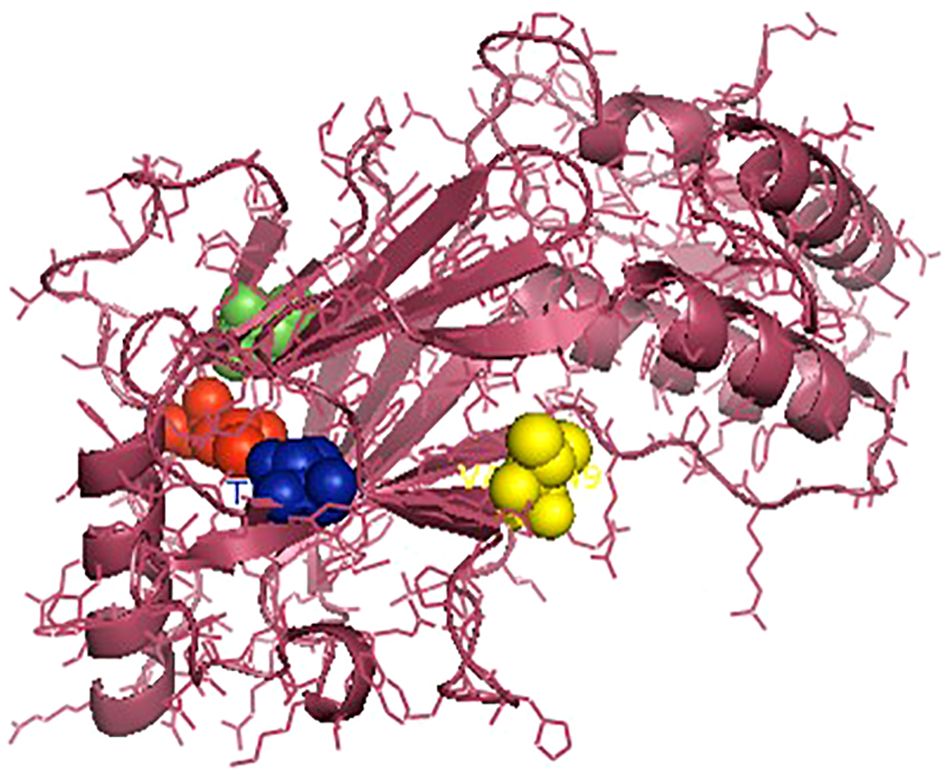
Figure 1. Three-dimensional structure of IRF3 in Chanos chanos with the domains for the zinc binding sites. Zinc binding sites: amino acid (aa) 97 (green sphere); aa 99 (red sphere); aa 145 (blue sphere); and aa 149 (yellow sphere).
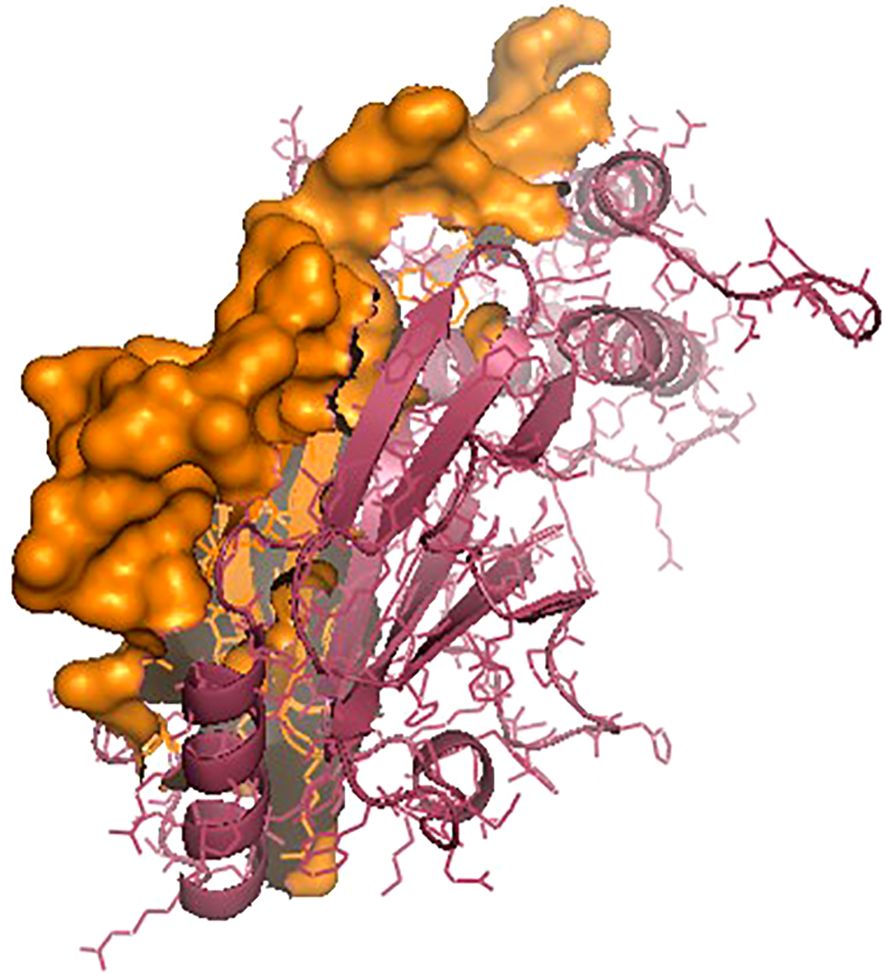
Figure 2. IRF3 protein with the interferon regulatory factor (IRF) tryptophan pentad repeat domain (amino acids 5–102) as the orange surface.
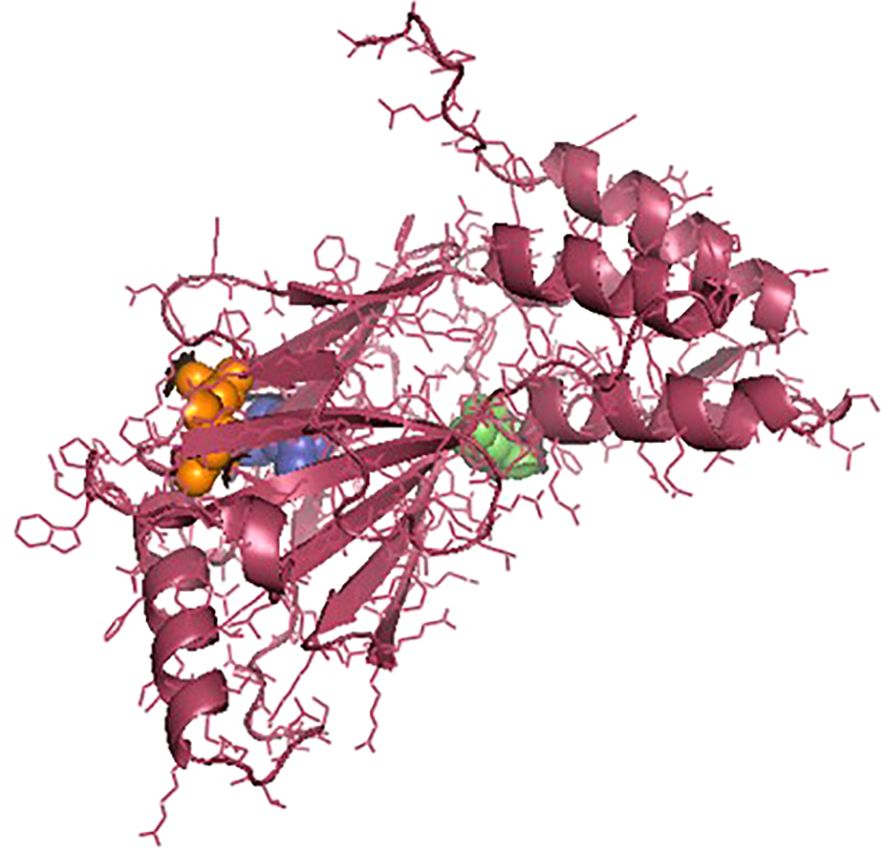
Figure 3. Three-dimensional structure of IRF3 in Chanos chanos with the domains for serine protease NS3 activity. Amino acid (aa) 57 (green sphere); aa 81 (blue sphere); and aa 139 (orange sphere).
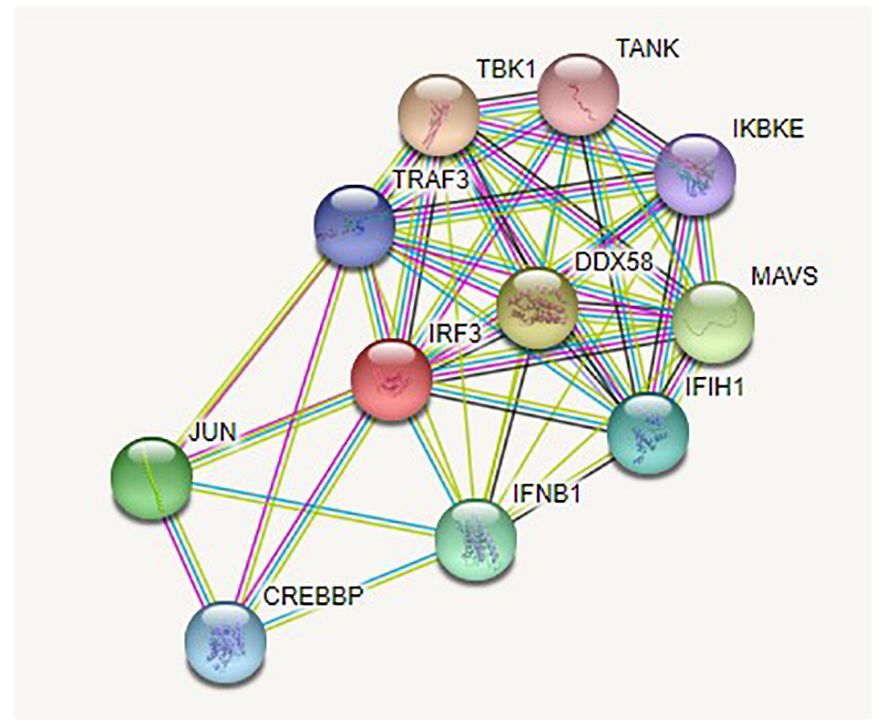
STRING analysis
The analysis revealed that IRF3 interacts with many other genes in a complex biological process. According to their interaction scores, these genes included DDX58, MAVS, IFNB1, IFIH1, TRAF3, TBK1, TANK, IKBKE, JUN, and CREBBP (Figures 5, 6).
KEGG analysis
The roles of IRF3 in NOD-like receptor signaling, the Toll-like receptor signaling pathway, and the cytosolic DNA sensing pathway are described in Figures 7–9, respectively. The analysis also provided proof of the antiviral effect of IRF3 through these aforementioned pathways.
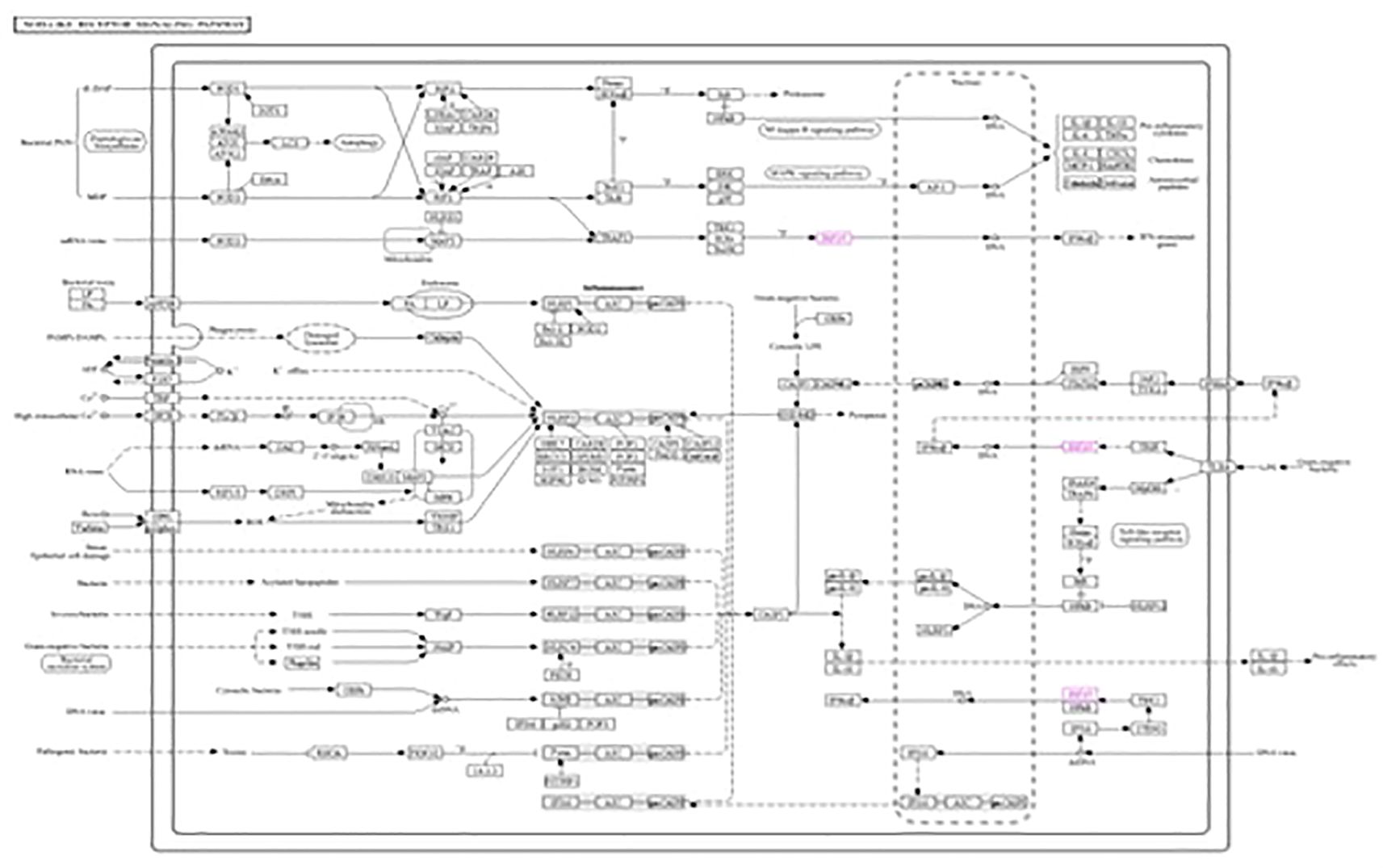
Figure 7. Kyoto Encyclopedia of Genes and Genomes (KEGG1) analysis of IRF3 showing the antiviral signaling pathway through the NOD-like receptor pathway.
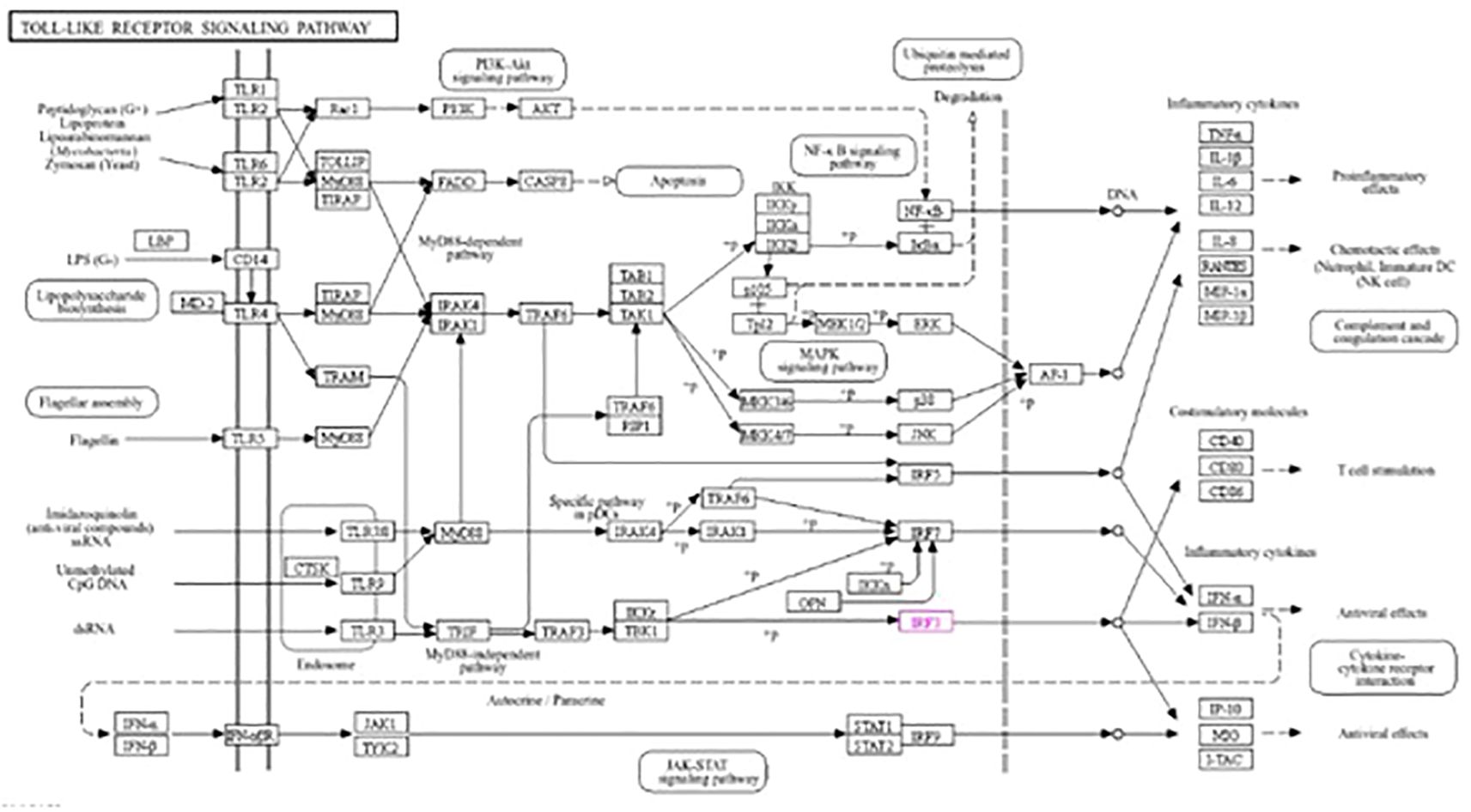
Figure 8. Kyoto Encyclopedia of Genes and Genomes (KEGG2) analysis of IRF3 showing the antiviral signaling pathway through the Toll-like receptor pathway.
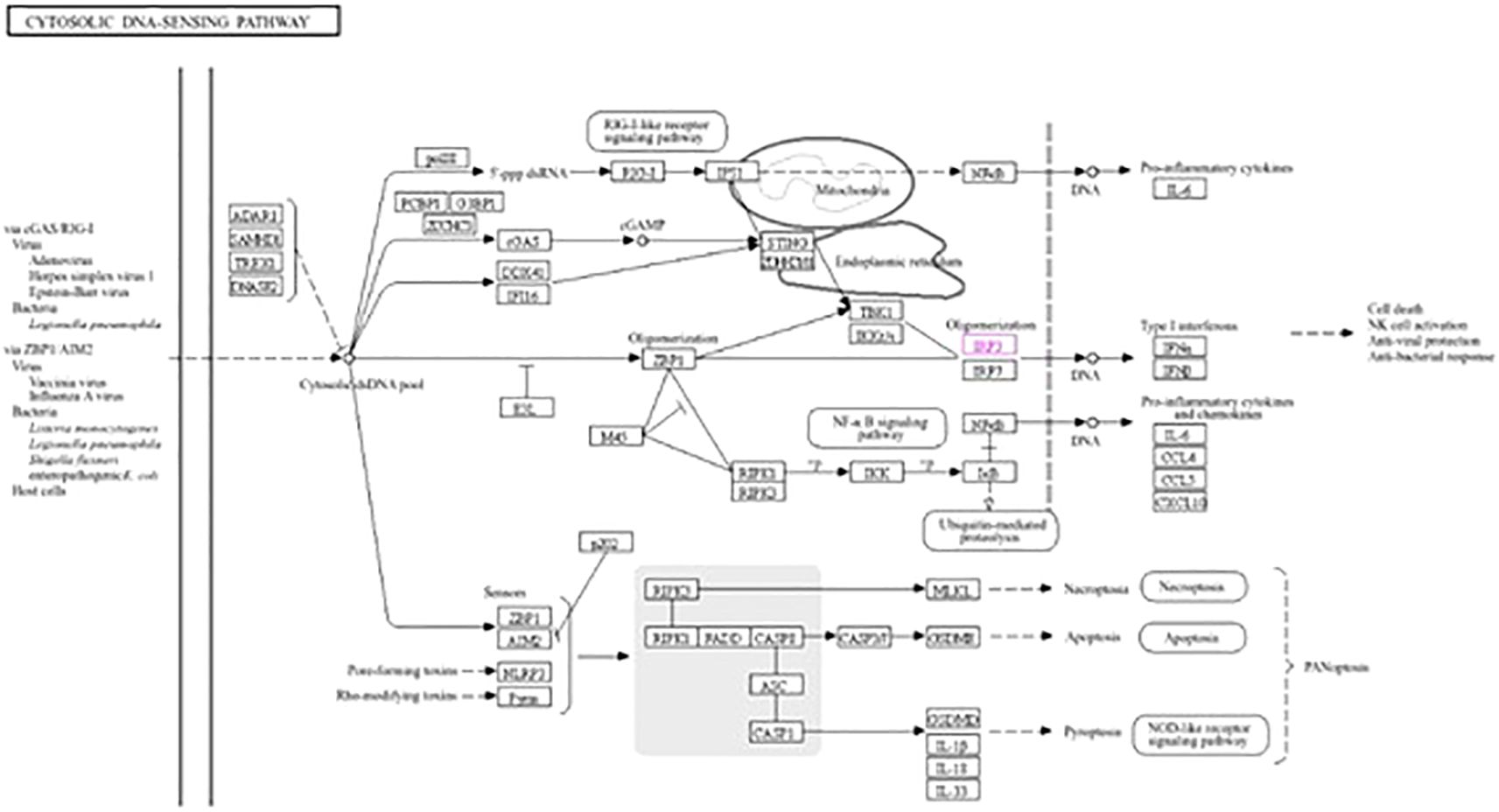
Figure 9. Kyoto Encyclopedia of Genes and Genomes (KEGG3) analysis of IRF3 showing the antiviral signaling pathway through the cytosolic DNA sensing pathway.
Differential mRNA expression profiling of the gene
The IRF3 gene was observed to be upregulated in group T5 (40 ppm nano-zinc-supplemented group) compared with that of the other treatment groups (Figure 10). The lowest expression was observed in the organic zinc-supplemented treatment group, i.e., group T2 (20 ppm zinc supplementation). However, the 10- and 20-ppm nano-zinc-supplemented groups showed better expression of IRF3 than the T2 group. When the concentration of nano-zinc increased to 40 ppm, the expression of the IRF3 gene was also highly upregulated. The expression of the IRF3 gene gradually increased in three nano-zinc supplement treatments: T3 (10 ppm nano-zinc supplementation), T4 (20 ppm nano-zinc supplementation), and T5 (40 ppm nano-zinc supplementation).
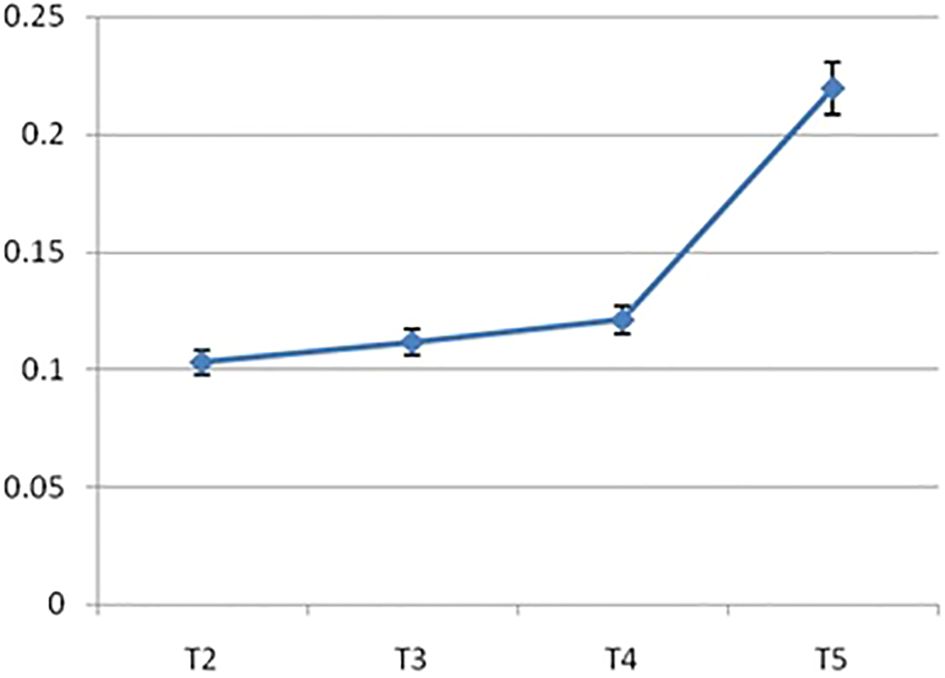
Figure 10. Differential mRNA Expression profiling of IRF3 gene of Chanos chanos upon feeding with zinc of different sources. (T2: Organic zinc, T3: Nano zinc 10ppm, T4: Nano zinc 20ppm, T5: Nano zinc 40 ppm).
Discussion
Zinc is an extremely critical micronutrient needed in an adequate amount at the tissue level in order to impart proper antioxidant status and optimum immunity to fight against pathogens. The antibacterial effects of nano-zinc have already been established (27, 42, 43). The bioavailability of zinc has been recorded to be quite high and with good antimicrobial potential (27). For dietary fortification, synthesized nano-ZnO was used at up to 40 ppm based on the recommendation of our earlier studies (27, 44).
To overcome the issues of bioavailability and to impart a higher antioxidant status, attempts are in vogue (27, 45, 46) to develop different varieties of nano-zinc for use in dietary fortification. A number of researchers have synthesized a zinc nanoparticle (206 nm) and observed its positive impact on the immune response and antioxidant status of fish (47). Different varieties of zinc nanoparticles (size range between 105.7 and 122.4 nm) have also been devised and evaluated for their efficacy in different fish models (48). One special variety of dietary nano-zinc oxide was synthesized with a particle size of 50–60 nm and fortified the diet of grass carp with the aim of imparting better body defense (49). The particle sizes of the synthesized nano-zinc ranged from 10 to 50 nm and from 20 to 120 nm, as confirmed by transmission electron microscopy (TEM) and dynamic light scattering (DLS) studies (50).
Zinc has already been reported to possess an antiviral effect against retroviruses, such as severe acute respiratory syndrome coronavirus (SARS-CoV) and SARS-CoV-2, among others (51). Several studies have proven that zinc has antiviral activity against hepatitis virus (52). Zinc oxide nanoparticles protected the Madin–Darby bovine kidney (MDBK) cell culture and experimental rabbits that were infected with herpesvirus (53). It also has the potential to induce varied gene expression related to immunity, including Toll-like receptors, tumor necrosis factor (TNF), and IL-1β, among others (54). However, research has been limited at a certain level, and there is no report elucidating the molecular mechanism.
We found several reports demonstrating the effect of nano-zinc on viruses and the mechanism of how they destroy a particular virus. However, our aim was to determine the molecular mechanism and the effect of synthesized nano-zinc on the expression of a gene that initiates signaling against viral infection through studying the detailed molecular structure using bioinformatics tools. This experiment attempted to assess the antiviral potential of nano-zinc, with IRF3 as a marker gene. IRF3 is considered to be one of the most important factors in antiviral signaling (15). In aquaculture, virus infection is one of the major threats (55), which often causes serious economic losses (56). When fishes are infected with a virus, severe mortality occurs (57), along with associated morbidity losses. It is a challenge to fight viral diseases in aquaculture. Therefore, there is an urgent need to search for a natural ingredient/additive to provide a considerable degree of protection against sudden viral infections.
This study showed that nano-zinc upregulates the expression of the IRF3 gene. IRF3 can fight against both RNA and DNA viruses (58, 59). Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis showed that IRF3 is involved in three antiviral signaling pathways: the cytosolic DNA sensing pathway, the NOD-like receptor signaling pathway, and the Toll-like receptor signaling pathway. When an RNA virus infects the cell, the NOD-like receptor signaling pathway is activated in the cytosol by the activation of IRF3 through MAVS and TRAF3 (60, 61). This activated IRF3 then stimulates the activation of IFNαβ and initiates antiviral activity. When the double-stranded DNA is recognized by TLR3 in the endosome in the Toll-like receptor pathway, IRF3 is stimulated through TRIF, TRAF3, and TBK1 by interacting with TLR3 (62). Subsequently, IFNα and IFNβ are activated by IRF3 and initiate an antiviral signaling pathway. In the case of the cytosolic DNA binding pathway, when the cell is infected with a virus, IRF3 is activated through ZBP1, TBK1, and IKK. Now activated, IRF3 then stimulates the production of IFNα and IFNβ and provides cell antiviral protection (58, 63). IRF3 initiates antiviral activity by activating two factors, i.e., IFNα and IFNβ (14, 15).
IRF3 has a tryptophan pentad domain that helps in the cellular response to viruses. This domain can recognize any change in the state or activity of a cell that is mediated by the stimulus of a virus (https://www.ebi.ac.uk/QuickGO/term/GO:0098586). There is another domain present in IRF3, i.e., the NS3 domain. It plays an important role in viral infection. NS3 hydrolyzes the four peptides bonded in the viral precursor polyprotein; thus, NS3 can destroy the viruses that infect the host cell (https://ftp.expasy.org/databases/prosite). The activated IRF3 functions against viral pathogens through the tryptophan pentad domain and the NS3 domain.
Organic zinc and nano-zinc differ in particle size. The smaller size of nano-zinc provides a high surface area, bioavailability, and more efficacy than organic zinc. Organic zinc is always in competition with other micronutrients and cannot be properly absorbed. Due to this, bulk-sized zinc cannot easily enter the IRF3 protein and cannot be placed properly through all the different zinc binding sites of the protein. This is why organic zinc fails to upregulate the expression of the IRF3 gene. On the other hand, nano-zinc is small and there is no chance to compete with other micronutrients. It can also easily enter the protein and properly bind with the different zinc binding sites of the protein. When nano-zinc binds with the IRF3 protein, it activates IRF3 against viral pathogens by activating the tryptophan pentad domain and the NS3 domain and stimulating its expression. When the concentration of nano-zinc increases, more zinc particles can accumulate with the protein and increase its expression.
From this experiment, it can be revealed that nano-zinc stimulates the signaling against viral pathogens by upregulating the IRF3 protein expression, and IRF3 stimulates IFNα and IFNβ, which then initiate antiviral signaling. Nano-zinc has the potential to fight against viral infection in the cell.
Conclusion
The IRF3 gene plays an extremely crucial role in the antiviral pathway. In the present study, the highest expression was recorded among fish that received feed fortified with 40 ppm nano-zinc. The corresponding STRING and KEGG pathway analyses revealed the possible mechanism of the higher IRF3 expression as induced by nano-zinc. Due to the higher bioavailability of nano-zinc, it can saturate the zinc binding sites present in IRF3 and can modulate its expression profiling in C. chanos. The present study explored the possible mechanism of action of nano-zinc in conferring antiviral immunity via the upregulation of the IRF3 gene, which could be used as a molecular marker to assess the antiviral protection in C. chanos. Nano-zinc oxide not only acts as an antibacterial agent but can also be used as a potential antiviral agent.
Future scope
In the future, this study could be propagated with challenging studies against pathogenic viruses by maintaining an appropriate biosecurity protocol.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
The research has been carried out as per the research norms and existing ethical guidelines for fish followed in ICAR-CIBA.
Author contributions
AT: Data curation, Methodology, Conceptualization, Investigation, Writing – original draft. AP: Data curation, Methodology, Formal Analysis, Software, Supervision, Validation, Visualization, Writing – review & editing. AM: Investigation, Methodology, Writing – original draft. DD: Methodology, Project administration, Supervision, Validation, Writing – review & editing. AC: Investigation, Methodology, Writing – original draft. MD: Methodology, Writing – original draft. SR: Project administration, Writing – review & editing. PNC: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. Support from the Department of Biotechnology, Ministry of Science and Technology, Govt. of India (project number: BT/PR25311/NER/95/1127/2017), and the Department of Science and Technology and Biotechnology, Govt. of West Bengal (project number: ST/P/S&T/1G-18/2017), is gratefully acknowledged.
Acknowledgments
The authors remain indebted to Dr. Sougata Sarkar, Ramakrishna Mission Vivekananda Centenary College, Rahara, Khardaha 700118, for his assistance in the synthesis of nano-zinc.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Bhat IA. Nanotechnology in reproduction, breeding and conservation of fish biodiversity: Current status and future potential. Rev Aquacult. (2023) 15:557–67. doi: 10.1111/raq.12736
2. Chatterjee PN, Mahato A, and Pal A. Nano zinc: it’s influence on health and production. Book chapter. In: Recent biotechnological Advances for Precision Feeding of Livestock and Poultry. Published by Director of Research, West Bengal University of Animal and Fishery Sciences, Kolkata, India (2019). p. 128–37.
3. Sharma R, M.R. S, and Ahmad MA. Nanotechnology: A novel tool for aquaculture and fisheries development. A prospective mini-review. FAJ. (2013) 02. doi: 10.4172/2150-3508.1000016
4. Swain P, Nayak SK, Sasmal A, Behera T, Barik SK, Swain SK, et al. Antimicrobial activity of metal based nanoparticles against microbes associated with diseases in aquaculture. World J Microbiol Biotechnol. (2014) 30:2491–502. doi: 10.1007/s11274-014-1674-4
5. Mondal AH, Behera T, Swain P, Das R, Sahoo SN, Mishra SS, et al. Nano zinc vis-à-vis inorganic Zinc as feed additives: Effects on growth, activity of hepatic enzymes and non-specific immunity in rohu, Labeo rohita (Hamilton) fingerlings. Aquacult Nutr. (2020) 26:1211–22. doi: 10.1111/anu.13077
6. Zalewski PD, Truong-Tran AQ, Grosser D, Jayaram L, Murgia C, and Ruffin RE. Zinc metabolism in airway epithelium and airway inflammation: basic mechanisms and clinical targets. A review. Pharmacol Ther. (2005) 105:127–49. doi: 10.1016/j.pharmthera.2004.09.004
7. Chanda S, Paul BN, Ghosh K, and Giri SS. Dietary essentiality of trace minerals in aquaculture-A Review. Agric Rev. (2015) 36:100. doi: 10.5958/0976-0741.2015.00012.4
8. Feng J, Ma WQ, Niu HH, Wu XM, Wang Y, and Feng J. Effects of zinc glycine chelate on growth, hematological, and immunological characteristics in broilers. Biol Trace Elem Res. (2010) 133:203–11. doi: 10.1007/s12011-009-8431-9
9. Zhao C-Y, Tan S-X, Xiao X-Y, Qiu X-S, Pan J-Q, and Tang Z-X. Effects of dietary zinc oxide nanoparticles on growth performance and antioxidative status in broilers. Biol Trace Elem Res. (2014) 160:361–7. doi: 10.1007/s12011-014-0052-2
10. Mahato A. Synthesis of different varieties Nano-dimensional Zinc particles and Biopotency Assay in Animal Model. M.V.S.C. Thesis. Kolkata-37: West Bengal University of Animal and Fishery Sciences (2017).
11. Hafez A, Nassef E, Fahmy M, Elsabagh M, Bakr A, and Hegazi E. Impact of dietary nano-zinc oxide on immune response and antioxidant defense of broiler chickens. Environ Sci pollut Res. (2020) 27:19108–14. doi: 10.1007/s11356-019-04344-6
12. Kidd MT, Ferket PR, and Qureshi MA. Zinc metabolism with special reference to its role in immunity. World’s Poult Sci J. (1996) 52:309–24. doi: 10.1079/WPS19960022
13. Abedini M, Shariatmadari F, Torshizi MAK, and Ahmadi H. Effects of zinc oxide nanoparticles on performance, egg quality, tissue zinc content, bone parameters, and antioxidative status in laying hens. Biol Trace Elem Res. (2018) 184:259–67. doi: 10.1007/s12011-017-1180-2
14. Chen X, Shen Y, Wu M, and Zhao J. Irf3 from mandarin fish thymus initiates interferon transcription. Fish Physiol Biochem. (2019) 45:133–44. doi: 10.1007/s10695-018-0543-8
15. Xu X, Lai Q, Gu M, Liu D, Hou Q, Liu X, et al. Fish IRF3 up-regulates the transcriptional level of IRF1, IRF2, IRF3 and IRF7 in CIK cells. Fish Shellfish Immunol. (2015) 47:978–85. doi: 10.1016/j.fsi.2015.10.039
16. Au WC, Moore PA, Lowther W, Juang YT, and Pitha PM. Identification of a member of the interferon regulatory factor family that binds to the interferon-stimulated response element and activates expression of interferon-induced genes. Proc Natl Acad Sci USA. (1995) 92:11657–61. doi: 10.1073/pnas.92.25.11657
17. Sato S, Sugiyama M, Yamamoto M, Watanabe Y, Kawai T, Takeda K, et al. Toll/IL-1 receptor domain-containing adaptor inducing IFN-β (TRIF) associates with TNF receptor-associated factor 6 and TANK-binding kinase 1, and activates two distinct transcription factors, NF-κB and IFN-regulatory factor-3, in the toll-like receptor signaling. J Immunol. (2003) 171:4304–10. doi: 10.4049/jimmunol.171.8.4304
18. Holland JW, Bird S, Williamson B, Woudstra C, Mustafa A, Wang T, et al. Molecular characterization of IRF3 and IRF7 in rainbow trout, Oncorhynchus mykiss: Functional analysis and transcriptional modulation. Mol Immunol. (2008) 46:269–85. doi: 10.1016/j.molimm.2008.08.265
19. Weaver BK, Ando O, Kumar KP, and Reich NC. Apoptosis is promoted by the dsRNA-activated factor (DRAF1) during viral infection independent of the action of interferon or p53. FASEB J. (2001) 15:501–15. doi: 10.1096/fj.00-0222com
20. Barnes B, Lubyova B, and Pitha PM. Review: on the role of IRF in host defense. J Interferon Cytokine Res. (2002) 22:59–71. doi: 10.1089/107999002753452665
21. Huang B, Qi ZT, Xu Z, and Nie P. Global characterization of interferon regulatory factor (IRF) genes in vertebrates: Glimpse of the diversification in evolution. BMC Immunol. (2010) 11:22. doi: 10.1186/1471-2172-11-22
22. Gu Y-F, Wei Q, Tang S-J, Chen X-W, and Zhao J-L. Molecular characterization and functional analysis of IRF3 in tilapia (Oreochromis niloticus). Dev Comp Immunol. (2016) 55:130–7. doi: 10.1016/j.dci.2015.10.011
23. Briolat V, Jouneau L, Carvalho R, Palha N, Langevin C, Herbomel P, et al. Contrasted innate responses to two viruses in zebrafish: insights into the ancestral repertoire of vertebrate IFN-stimulated genes. J Immunol. (2014) 192:4328–41. doi: 10.4049/jimmunol.1302611
24. López-Muñoz A, Roca FJ, Meseguer J, and Mulero V. New insights into the evolution of IFNs: zebrafish group II IFNs induce a rapid and transient expression of IFN-dependent genes and display powerful antiviral activities1. J Immunol. (2009) 182:3440–9. doi: 10.4049/jimmunol.0802528
25. Chang C-H, Lin J-Y, Lo W-Y, and Lee T-H. Hypothermal stress induced differential expression profiles of the immune response gene, warm-temperature-acclimation associated 65-kDa protein (Wap65), in the liver of fresh water and seawater milkfish, Chanos chanos. Fish Shellfish Immunol. (2017) 70:174–84. doi: 10.1016/j.fsi.2017.09.012
26. Yang W-K, Chung C-H, Cheng HC, Tang C-H, and Lee T-H. Different expression patterns of renal Na+/K+-ATPase α-isoform-like proteins between tilapia and milkfish following salinity challenges. Comp Biochem Physiol Part B: Biochem Mol Biol. (2016) 202:23–30. doi: 10.1016/j.cbpb.2016.07.008
27. Mahato A, Chatterjee PN, Sarkar S, Sen AR, Pal A, Roy S, et al. Effects of chemically and green synthesized zinc oxide nanoparticles on shelf life and sensory quality of minced fish (Pangasius hypophthalmus). Foods. (2024) 13:2810. doi: 10.3390/foods13172810
28. Sîrbu E, Dima MF, Tenciu M, Cretu M, Coadă MT, Ţoţoiu A, et al. Effects of dietary supplementation with probiotics and prebiotics on growth, physiological condition, and resistance to pathogens challenge in nile tilapia (Oreochromis niloticus). Fishes. (2022) 7:273. doi: 10.3390/fishes7050273
29. Banerjee S, Pal A, Pal A, Mandal SC, Chatterjee PN, and Chatterjee JK. RIG-I Has a Role in Immunity Against Haemonchus contortus, a Gastrointestinal Parasite in Ovis aries: A Novel Report. Front Immunol. (2021) 11:534705. doi: 10.3389/fimmu.2020.534705
30. Pal S, Banerjee S, Pal A, Kalita DJ, Batabyal S, Debnath M, et al. Role of RIGI, MDA5 and interferon alpha of duck in Duck Plague infection – a novel report. bioRxiv (2022) 2022–01. doi: 10.1101/2022.01.26.477779
31. Pal A and Chatterjee PN. Molecular cloning and characterization of CD14 gene in goat. Small Ruminant Res. (2009) 82:84–7. doi: 10.1016/j.smallrumres.2008.11.016
32. Rawat K, Pal A, Banerjee S, Pal A, Mandal SC, and Batabyal S. Ovine CD14- an immune response gene has a role against gastrointestinal nematode haemonchus contortus—A novel report. Front Immunol. (2021) 12:664877. doi: 10.3389/fimmu.2021.664877
33. Petersen B, Petersen TN, Andersen P, Nielsen M, and Lundegaard C. A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct Biol. (2009) 9:51. doi: 10.1186/1472-6807-9-51
34. Ebina T, Toh H, and Kuroda Y. Loop-length-dependent SVM prediction of domain linkers for high-throughput structural proteomics. Biopolymers. (2009) 92:1–8. doi: 10.1002/bip.21105
35. Caroff M, Karibian D, Cavaillon J-M, and Haeffner-Cavaillon N. Structural and functional analyses of bacterial lipopolysaccharides. Microbes Infect. (2002) 4:915–26. doi: 10.1016/S1286-4579(02)01612-X
36. Muroi M, Ohnishi T, and Tanamoto K. Regions of the mouse CD14 molecule required for toll-like receptor 2- and 4-mediated activation of NF-κB. J Biol Chem. (2002) 277:42372–9. doi: 10.1074/jbc.M205966200
37. Kelley LA, Mezulis S, Yates CM, Wass MN, and Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. (2015) 10:845–58. doi: 10.1038/nprot.2015.053
38. Wiederstein M and Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. (2007) 35:W407–10. doi: 10.1093/nar/gkm290
39. Zhang Y and Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. (2005) 33:2302–9. doi: 10.1093/nar/gki524
40. Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. (2012) 41:D808–15. doi: 10.1093/nar/gks1094
41. Pal A, Pal A, Mallick AI, Biswas P, and Chatterjee PN. Molecular characterization of Bu-1 and TLR2 gene in Haringhata Black chicken. Genomics. (2020) 112:472–83. doi: 10.1016/j.ygeno.2019.03.010
42. El-Saadony MT, Alkhatib FM, Alzahrani SO, Shafi ME, El. Abdel-Hamid S, Taha TF, et al. Impact of mycogenic zinc nanoparticles on performance, behavior, immune response, and microbial load in Oreochromis niloticus. Saudi J Biol Sci. (2021) 28:4592–604. doi: 10.1016/j.sjbs.2021.04.066
43. Shaalan MI, El-Mahdy MM, Theiner S, El-Matbouli M, and Saleh M. In vitro assessment of the antimicrobial activity of silver and zinc oxide nanoparticles against fish pathogens. Acta Vet Scand. (2017) 59:49. doi: 10.1186/s13028-017-0317-9
44. Dey S. Exploiting the antioxidant efficacy of zinc based nano particles when supplemented in IMC (Cirrhinus mrigala) diet. M.F.S.C. Thesis. Kolkata-37: West Bengal University of Animal and Fishery Sciences (2023).
45. De I. Evaluation of zinc oxide nanoparticles to be used as a growth promoter in brackishwater fish Mystus gulio. M.F.S.C. Thesis. Kolkata-37: West Bengal University of Animal and Fishery Sciences (2023).
46. Pradhan A. Ameliorating efficacy of supplementing dietary nano-Zn fortified with Vitamin E on lead-induced stress in Rohu (Labeo rohita). M.F.S.C. Thesis. Kolkata-37: West Bengal University of Animal and Fishery Sciences (2024).
47. Kumar N, Krishnani KK, and Singh NP. Effect of dietary zinc-nanoparticles on growth performance, anti-oxidative and immunological status of fish reared under multiple stressors. Biol Trace Elem Res. (2018) 186:267–78. doi: 10.1007/s12011-018-1285-2
48. Swain P, Das R, Das A, Padhi SK, Das KC, and Mishra SS. Effects of dietary zinc oxide and selenium nanoparticles on growth performance, immune responses and enzyme activity in rohu, Labeo rohita (Hamilton): XXXX. Aquacult Nutr. (2019) 25:486–94. doi: 10.1111/anu.12874
49. Faiz H, Zuberi A, Nazir S, Rauf M, and Younus N. Zinc oxide, zinc sulfate and zinc oxide nanoparticles as source of dietary zinc: comparative effects on growth and hematological indices of juvenile grass carp (Ctenopharyngodon idella). IJAB. (2015) 17:568–74. doi: 10.17957/IJAB/17.3.14.446
50. Kaya H, Aydın F, Gürkan M, Yılmaz S, Ates M, Demir V, et al. A comparative toxicity study between small and large size zinc oxide nanoparticles in tilapia (Oreochromis niloticus): Organ pathologies, osmoregulatory responses and immunological parameters. Chemosphere. (2016) 144:571–82. doi: 10.1016/j.chemosphere.2015.09.024
51. Samad N, Sodunke TE, Abubakar AR, Jahan I, Sharma P, Islam S, et al. The implications of zinc therapy in combating the COVID-19 global pandemic. J Inflammation Res. (2021) 14:527–50. doi: 10.2147/JIR.S295377
52. Kumar S, Ansari S, Narayanan S, Ranjith-Kumar CT, and Surjit M. Antiviral activity of zinc against hepatitis viruses: current status and future prospects. Front Microbiol. (2023) 14:1218654. doi: 10.3389/fmicb.2023.1218654
53. Zeedan GSG, El-Razik KAA, Allam AM, Abdalhamed AM, and Zeina HAA. Evaluations of potential antiviral effects of green zinc oxide and silver nanoparticles against bovine herpesvirus-1. Adv Anim Vet Sci. (2020) 8:433–43. doi: 10.17582/journal.aavs/2020/8.4.433.443
54. Poon WL, Alenius H, Ndika J, Fortino V, Kolhinen V, Meščeriakovas A, et al. Nano-sized zinc oxide and silver, but not titanium dioxide, induce innate and adaptive immunity and antiviral response in differentiated THP-1 cells. Nanotoxicology. (2017) 11:936–51. doi: 10.1080/17435390.2017.1382600
55. Murray AG. Epidemiology of the spread of viral diseases under aquaculture. Curr Opin Virol. (2013) 3:74–8. doi: 10.1016/j.coviro.2012.11.002
56. Mugimba KK, Byarugaba DK, Mutoloki S, Evensen Ø, and Munang’andu HM. Challenges and solutions to viral diseases of finfish in marine aquaculture. Pathogens. (2021) 10:673. doi: 10.3390/pathogens10060673
57. Escobar LE, Escobar-Dodero J, and Phelps NBD. Infectious disease in fish: global risk of viral hemorrhagic septicemia virus. Rev Fish Biol Fish. (2018) 28:637–55. doi: 10.1007/s11160-018-9524-3
58. Abe T, Marutani Y, and Shoji I. Cytosolic DNA-sensing immune response and viral infection. Microbiol Immunol. (2019) 63:51–64. doi: 10.1111/1348-0421.12669
59. Chattopadhyay S, Yamashita M, Zhang Y, and Sen GC. The IRF-3/bax-mediated apoptotic pathway, activated by viral cytoplasmic RNA and DNA, inhibits virus replication. J Virol. (2011) 85:3708–16. doi: 10.1128/JVI.02133-10
60. Al Hamrashdi M and Brady G. Regulation of IRF3 activation in human antiviral signaling pathways. Biochem Pharmacol. (2022) 200:115026. doi: 10.1016/j.bcp.2022.115026
61. Almeida-da-Silva CLC, Savio LEB, Coutinho-Silva R, and Ojcius DM. The role of NOD-like receptors in innate immunity. Front Immunol. (2023) 14:1122586. doi: 10.3389/fimmu.2023.1122586
62. Zhu J, Smith K, Hsieh PN, Mburu YK, Chattopadhyay S, Sen GC, et al. High-throughput screening for TLR3–IFN regulatory factor 3 signaling pathway modulators identifies several antipsychotic drugs as TLR inhibitors. J Immunol. (2010) 184:5768–76. doi: 10.4049/jimmunol.0903559
Keywords: nano-zinc, antiviral immunity, IRF3 expression, Chanos chanos, immune gene
Citation: Tah A, Pal A, Chatterjee PN, De D, Mahato A, Chakraborty A, Debnath M and Roy S (2025) Dietary supplementation with nano-zinc modulates the expression of the antiviral immune gene IRF3: a novel report. Front. Immunol. 16:1583956. doi: 10.3389/fimmu.2025.1583956
Received: 28 February 2025; Accepted: 26 June 2025;
Published: 04 August 2025.
Edited by:
Maria Del Mar Ortega-Villaizan, Miguel Hernández University of Elche, SpainReviewed by:
Ganesh Yadagiri, The Ohio State University, United StatesLingmin Jiang, Sun Yat-sen University Cancer Center, China
Copyright © 2025 Tah, Pal, Chatterjee, De, Mahato, Chakraborty, Debnath and Roy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aruna Pal, YXJ1bmFjaGF0dGVyamVlQGdtYWlsLmNvbQ==; Paresh Nath Chatterjee, Y2hhdHRlcmplZW51dHJpdGlvbkBnbWFpbC5jb20=
 Amrita Tah
Amrita Tah Aruna Pal
Aruna Pal Paresh Nath Chatterjee
Paresh Nath Chatterjee Debasis De
Debasis De Achinta Mahato
Achinta Mahato Argha Chakraborty
Argha Chakraborty Manti Debnath
Manti Debnath Sovan Roy
Sovan Roy