- 1UNSW School of Clinical Medicine, University of New South Wales, Sydney, NSW, Australia
- 2School of Medical Sciences, University of New South Wales, Sydney, NSW, Australia
- 3Blood Stem Cell and Cancer Research Group, St. Vincent’s Centre for Applied Medical Research, Sydney, NSW, Australia
- 4Department of Neurology, St. Vincent’s Hospital, Sydney, NSW, Australia
- 5Department of Gastroenterology and Hepatology, St. Vincent’s Hospital, Sydney, NSW, Australia
Background: MS is a chronic inflammatory and degenerative disease of the central nervous system (CNS) resulting in neurological deficits associated with physical and/or cognitive disability. The gut microbiome can interact with the CNS and immune system through various molecular pathways and has been previously implicated in MS. Autologous Haematopoietic Stem Cell Transplant (AHSCT) in MS arrests inflammatory disease and has evidence of long-term therapeutic benefit. To date, no study has explored the effect of AHSCT on the gut microbiome in people with MS.
Method: The microbiome of people with MS (pwMS) undergoing AHSCT was compared with pwMS on Natalizumab (NTZ). Longitudinal microbiome analysis was also conducted within the AHSCT cohort at two timepoints. Amplicon sequencing of the 16S ribosomal RNA V3–4 region (Illumina MiSeq) was used to evaluate alpha and beta diversity, oral-stool microbiota distances, and relative taxa abundances on both oral and stool microbiota.
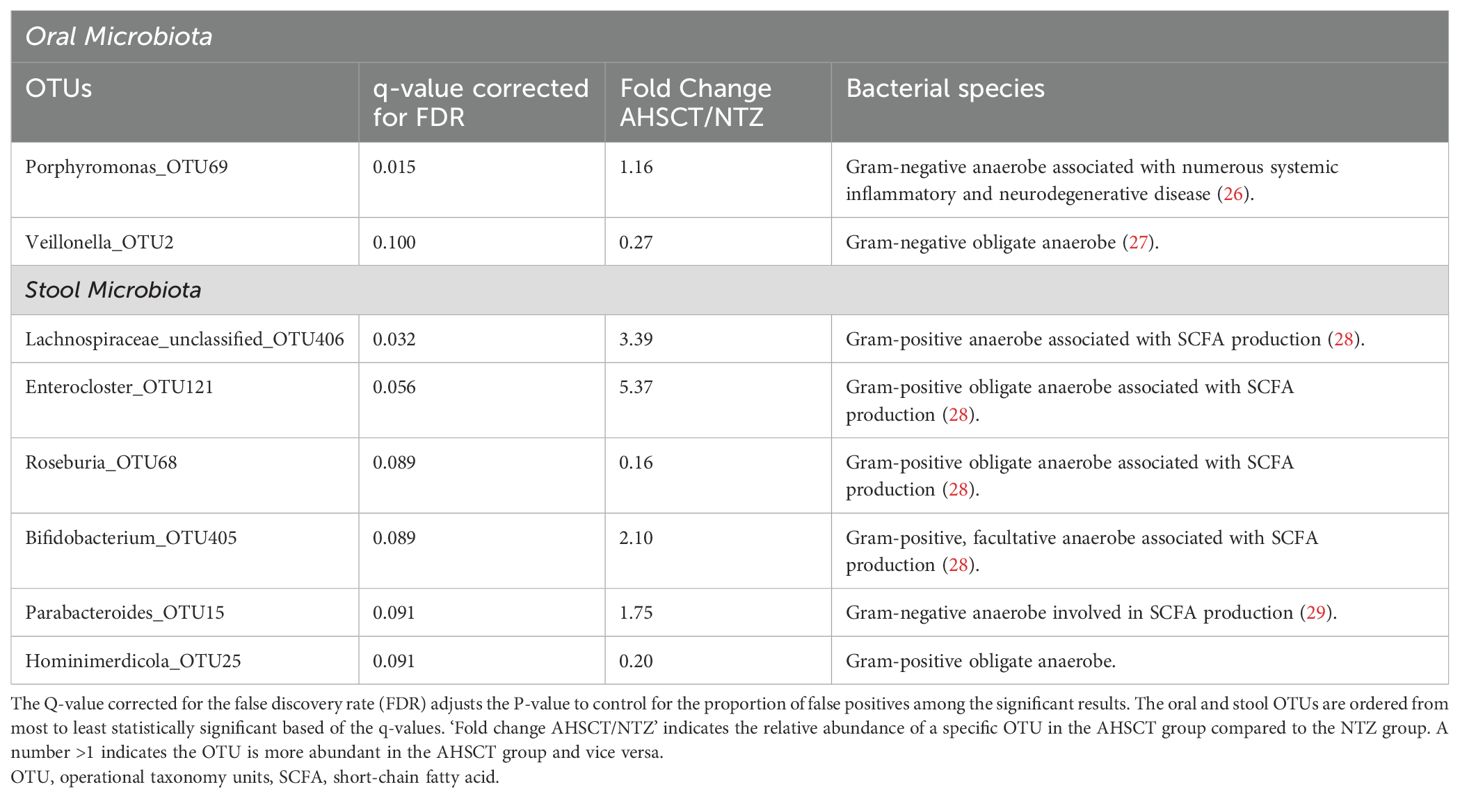
Results: The pre-transplant, baseline samples from the AHSCT cohort (n=8) was compared to the Natalizumab group (n=22). The AHSCT cohort had lower oral species richness compared to the NTZ cohort (p=0.026). There was a significant difference in oral beta diversity between the two cohorts (p=0.043). The oral taxa analysis of AHSCT subjects showed increased relative abundances of Porphyromonas and decreased Veillonella.
Conclusion: This pilot study identified specific microbiome changes, particularly in the oral alpha diversity and abundance of specific bacteria which may reflect treatment status or disease activity in MS.
1 Introduction
Multiple sclerosis (MS) is a chronic, immune-mediated disease of the central nervous system (CNS) which affects an estimated 2.8 million people worldwide (1). With the average age of diagnosis ranging from 30–40 years old (2), MS causes significant physical and cognitive disability (3).
Genes within the human leukocyte antigen (HLA) complex remain the strongest genetic risk factor for MS (4). However, other epidemiologic risk factors include exposure to Epstein Barr virus, vitamin D levels, geographical location such as latitude, obesity in adolescence and smoking (5). Intestinal microbiota has also emerged as a potential factor influencing MS pathogenesis (6). Activation of an abnormal adaptive immune response is central to disease initiation (7). Self-reactive T and B lymphocytes infiltrate the CNS, crossing the blood-brain barrier (BBB) and triggering inflammatory cascades that result in glial and neuronal injury (8).
Natalizumab (NTZ), is a monoclonal antibody which inhibits T-cell migration into the CNS via α4-integrin inhibition (9). However, its action is non-specific, also disrupting T-cell circulation within the gut (10). This makes NTZ an intriguing drug for studying the gut-immune axis and its potential alternate pathways of action in MS. Autologous hematopoietic stem cell transplant (AHSCT) has demonstrated the potential for immune reconstitution, particularly in patients with more severe, DMT-refractory MS (11). AHSCT involves high-dose chemotherapy to eradicate autoreactive immune cells, followed by the infusion of autologous stem cells to rebuild a novel immune system (7).
The gut microbiome is increasingly recognized as a key player in modulating systemic immunity and CNS inflammation. Microbial metabolites, such as short-chain fatty acids (SCFAs) and neurotransmitters, influence peripheral immune function, while cytokines and chemokines derived from gut bacteria can disrupt CNS homeostasis (12). Conversely, CNS-derived biochemical changes can alter gut microbial composition through the hypothalamic-pituitary-adrenal (HPA) axis (13). Specific gut bacteria, such as segmented filamentous bacteria (SFB), are known to drive T-helper 17 cell differentiation, which is implicated in autoimmune diseases, including MS (14). Dysbiosis, an imbalance in microbial composition or metabolites, can exacerbate neuroinflammation by disrupting intestinal and systemic immune homeostasis (15). Although numerous studies have identified differences in stool microbial taxa between people with MS (pwMS) and healthy controls (HC) (16–21), these findings are largely correlative. It remains unclear whether dysbiosis contributes to MS pathogenesis or is secondary to the disease and its treatments. There are also limited studies characterising the oral microbiome in pwMS. Furthermore, while research on gut microbiota’s role in immune reconstitution following hematopoietic stem cell transplantation (HSCT) in haematological malignancies has revealed intriguing associations, similar studies in pwMS have yet to be conducted.
This present study aims to firstly characterise the differences in the oral and stool microbiome of people with MS in remission on treatment (NTZ) compared with those pre-AHSCT, refractory to available DMT. Secondly, for pwMS undergoing AHSCT, analysis of longitudinal changes in oral and stool microbiota could help identify microbial shifts associated with disease activity or treatment effects, and explore their potential relevance to immune reconstitution.
2 Method
2.1 Study outline
An observational cohort study was conducted by comparing the oral and stool microbiome of pre-AHSCT pwMS with a control group of pwMS undergoing treatment with NTZ (St. Vincent’s Hospital, Sydney HREC 2021/ETH11173). This allowed for a comparison of the microbiome in active DMT-refractory MS and stable, treated MS. Within the AHSCT cohort, a longitudinal analysis of the oral and stool microbiome was then conducted throughout treatment. All patients provided written consent prior to sample collection.
2.2 Cohorts
NTZ Control Cohort: Patients, aged 18-65, with MS as determined by a neurologist according to the 2017 revised McDonald’s criteria (22) were suitable for inclusion in the control cohort. Recruited patients had been receiving NTZ for 3 or more months, 300mg of intravenous NTZ every 4–6 weeks. Pre- and post-AHSCT samples were collected from these patients.
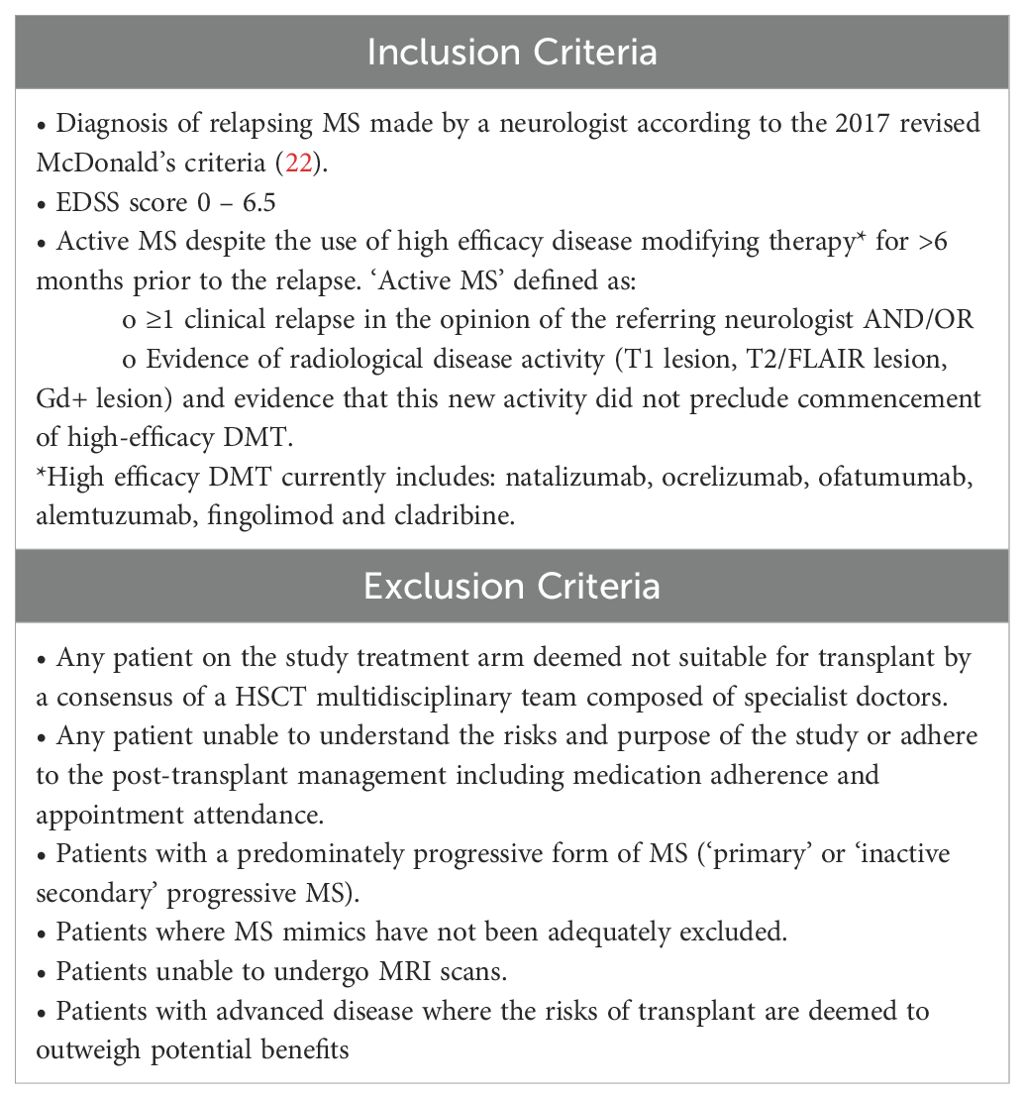
AHSCT Cohort: All patients undergoing AHSCT as part of the AHSCT in Multiple Sclerosis (AIMS) study were required to contribute stool and saliva samples as part of the interventional clinical trial. Participants were off immunotherapy at clinically appropriate wash-out periods prior to the collection of baseline, pre-ASHCT samples (Appendix 1). This allowed for a snapshot of the gut microbiome in ‘active’, DMT-refractory MS. See Table 1. for the inclusion and exclusion criteria of this cohort.
2.3 Patient recruitment
Patients were recruited between 1 Jan 2022 and 1 July 2024. Each patient was assigned a unique study ID, with relevant clinical and demographic data recorded in the institutional REDCap database.
2.4 Sample collection and storage
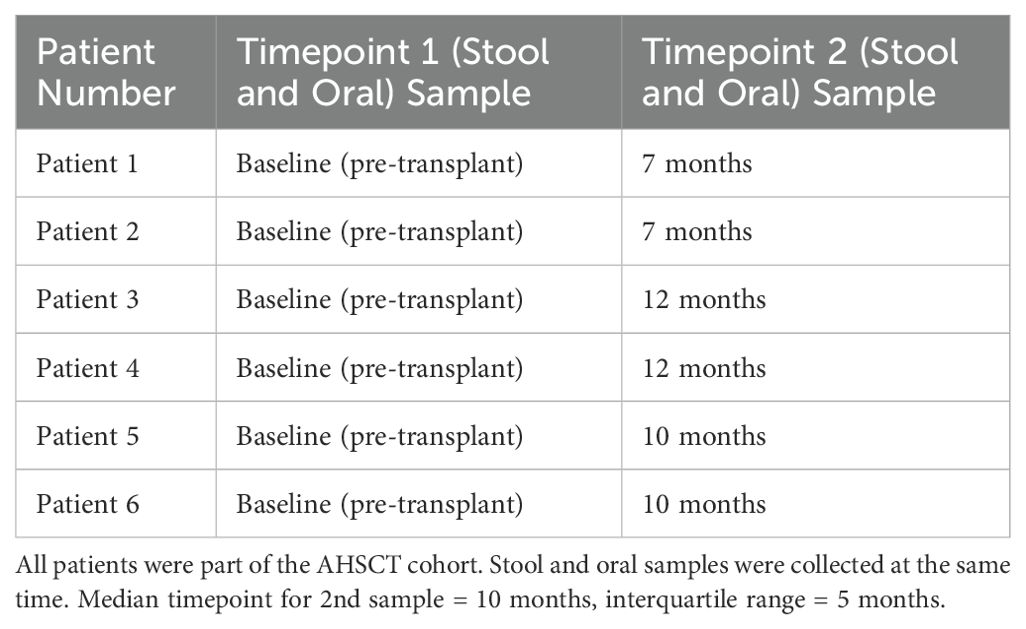
INVITEK Molecular kits were used to collect faecal samples and DNAGenotek kits were used to collect oral samples. For AHSCT patients, sample collection was scheduled before AHSCT treatment (baseline samples), and 6 months post-AHSCT. AHSCT participants were provided with self-collection kits at the 6-month post-AHSCT appointment. However, as samples were self-collected at home and returned via post, actual collection times ranged from 7 to 12 months. Samples were aliquoted and stored at -80°C, which is regarded as the gold standard (23).
2.5 Nucleic acid extraction
DNA extraction was conducted at the Microbiome Research Centre, Kogarah, NSW. Frozen stool samples were processed using the INVITEK™ PSP DNA extraction kit. Frozen oral samples were processed using the Qiagen™ DSP pathogen DNA extraction kit.
2.6 Sequencing and data analysis
The 16S rRNA V4 regions were amplified by PCR and sequenced on the MiSeq platform (Illumina) using the 2×300 bp paired-end protocol. Sequencing was performed at the UNSW Ramaciotti Centre for Genomics. Raw sequencing data was analysed using Mothur v1.48.1 and vSearch v2.22.1 on the UNSW Katana server (24). The Ribosomal Database Project (RDP) v19 was used as the reference database for taxonomic classification. Operational taxonomic units (OTUs) were used to group clusters of uncultivable microorganisms with sequence similarity as an analytical unit. The numbers represented the degree of importance; the lower the number the more prevalent the genus (i.e. OTU05 is more prevalent than OTU26). Similarity was detected by the 16S taxonomic marker gene and was used to classify microbial individuals at different taxonomic levels.
Primer-E v6 was used to calculate alpha diversity measures including Margalef’s species richness, Pielou’s evenness and Shannon’s diversity index. For both stool and oral samples, the Shaprio-Wilk test was used to test for normality of data. Alpha diversity of normally distributed data was analysed with unpaired t-tests while data for which didn’t fit a normal distribution, Mann-Whitney U tests were used. Beta diversity was calculated with Bray-Curtis dissimilarities from OUT relative abundances (%) that were transformed by square root. Principal-coordinate analysis (PCoA) and permutational multivariate analysis of variance (PERMANOVA) was then calculated. The comparison between the oral and stool microbiota was performed with a PCoA and PERMANOVA on a Bray-Curtis resemblance matrix, and the distance relationship between the oral and stool microbiota was calculated with a Mann-Whitney U test. GraphPad Prism v9.4 was used for statistical analysis.
For taxonomic classification, per taxon analysis was performed using ZicoSeq (25). Counts were log-transformed and only taxa that contributed to at least 1% to the oral and stool microbiota were included. An unpaired t-test was applied and p-values were corrected for false discovery rates (FDR) using the Benjamini- Hochberg method (BH) to form a q-value.
3 Results
3.1 Subject characteristics
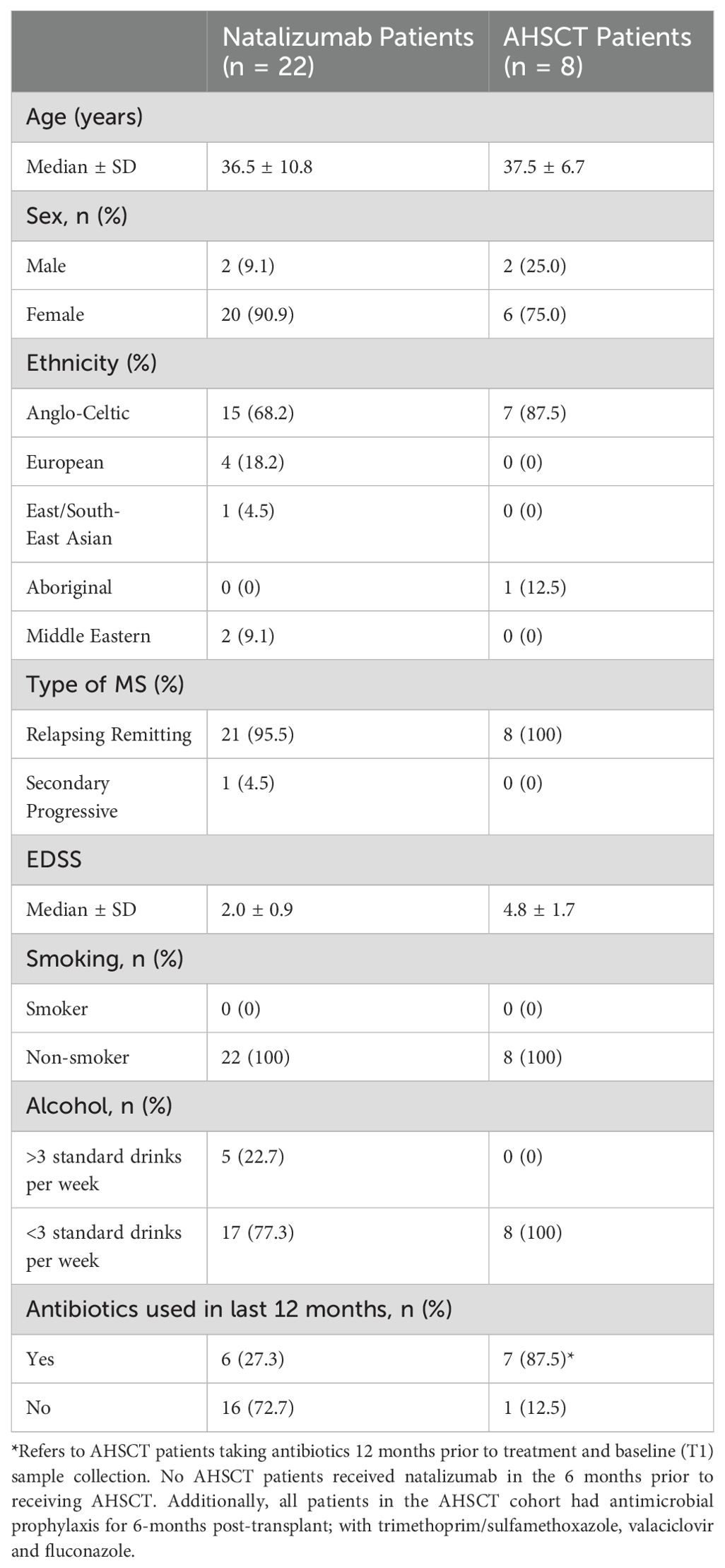
A total of 30 participants with MS were enrolled into the study: 22 in the NTZ cohort and 8 in the AHSCT cohort. The two cohorts were well-matched in terms of sex and age. The AHSCT cohort had a higher Expanded Disability Status Scale (EDSS) median score and higher 12-month antibiotic exposure. No patients in the AHSCT cohort received NTZ within 6-months of treatment. Demographic information about the cohorts is summarised in Table 2. Baseline oral and stool samples were compared from all patients in the AHSCT and the NTZ control cohorts. Of the 8 AHSCT patients who provided baseline oral and stool samples (T1 samples), 6 provided longitudinal samples at a second timepoint (T2 samples) with a median time of 10-months (interquartile range: 5-months) (see Table 3).
3.2 Comparison of baseline oral and stool microbiota
To analyse the overall interindividual difference between stool and oral samples (beta diversity), PCoA and a non-parametric multivariate statistical permutation test (PERMANOVA) was performed. There was a highly significant difference between oral and stool microbiota (p=0.0001, PERMANOVA) (see Figure 1).
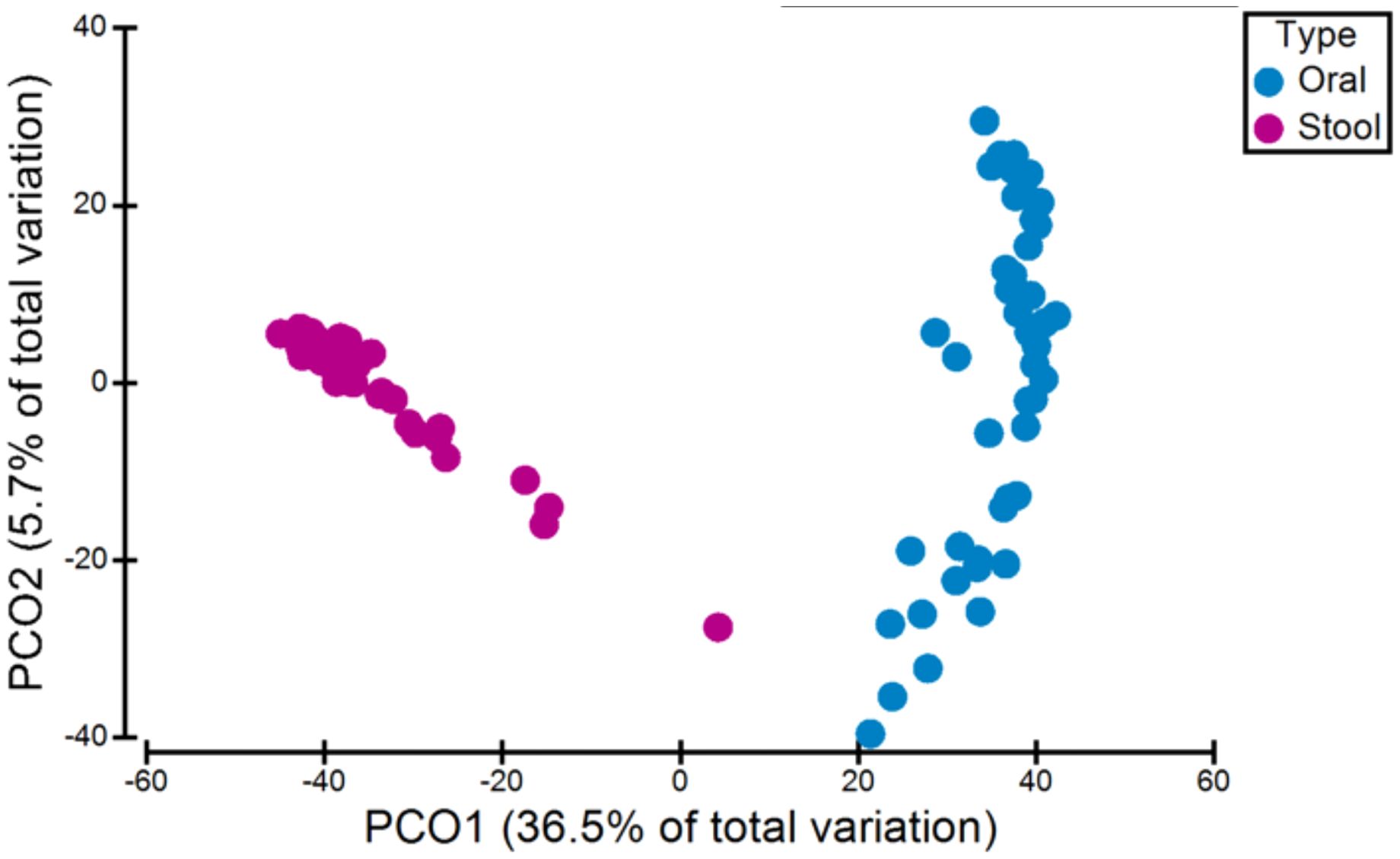
Figure 1. Variation in microbiota (beta-diversity) between all oral and stool samples. There was a significant difference between oral and stool microbiota (p=0.0001, PERMANOVA).
3.3 AHSCT vs NTZ cohorts
The AHSCT cohort had significantly lower species richness of oral microbiota compared to the NTZ cohort (p=0.026) (Figure 2A). While there was no significant difference in evenness (p=0.0872) and diversity (p=0.0581) between the two cohorts, there was a trend to lower evenness and diversity in the AHSCT group. While generally lower in the AHSCT cohort, there was no significant difference in the alpha diversity of the stool microbiota richness (p=0.2676), evenness (p=0.0564) and diversity (p=0.0871) between the AHSCT and NTZ cohorts (Figure 2B). There was a significant difference in the oral beta diversity (p=0.043, PERMANOVA) (Figure 3A) and stool beta diversity (p=0.0086, PERMANOVA) (Figure 3B) between the AHSCT and NTZ groups.
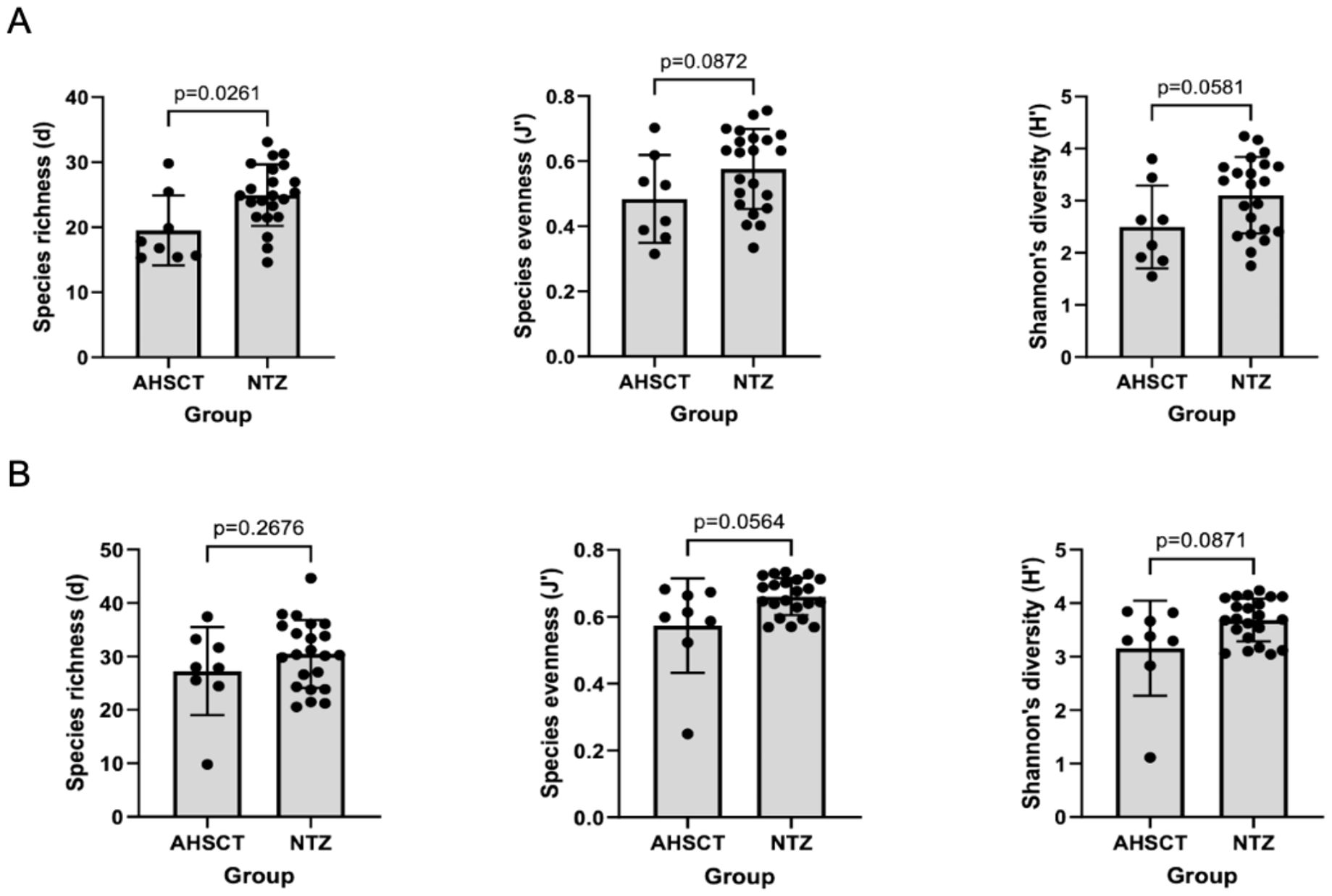
Figure 2. (A) Comparison of oral alpha diversity between AHSCT and NTZ cohort; (B) Comparison of stool alpha diversity between AHSCT and NTZ cohorts. Normality of datasets was determined using Shapiro-Wilk tests. Unpaired t-tests (parametric) or Mann-Whitney U tests (non-parametric) were then used to compare alpha diversity statistics, including Margalef’s richness, Pielou’s evenness and Shannon’s diversity. For the oral samples, there was a trend towards lower alpha diversity in the AHSCT cohorts. However, only species richness was statistically significant (p=0.0261).
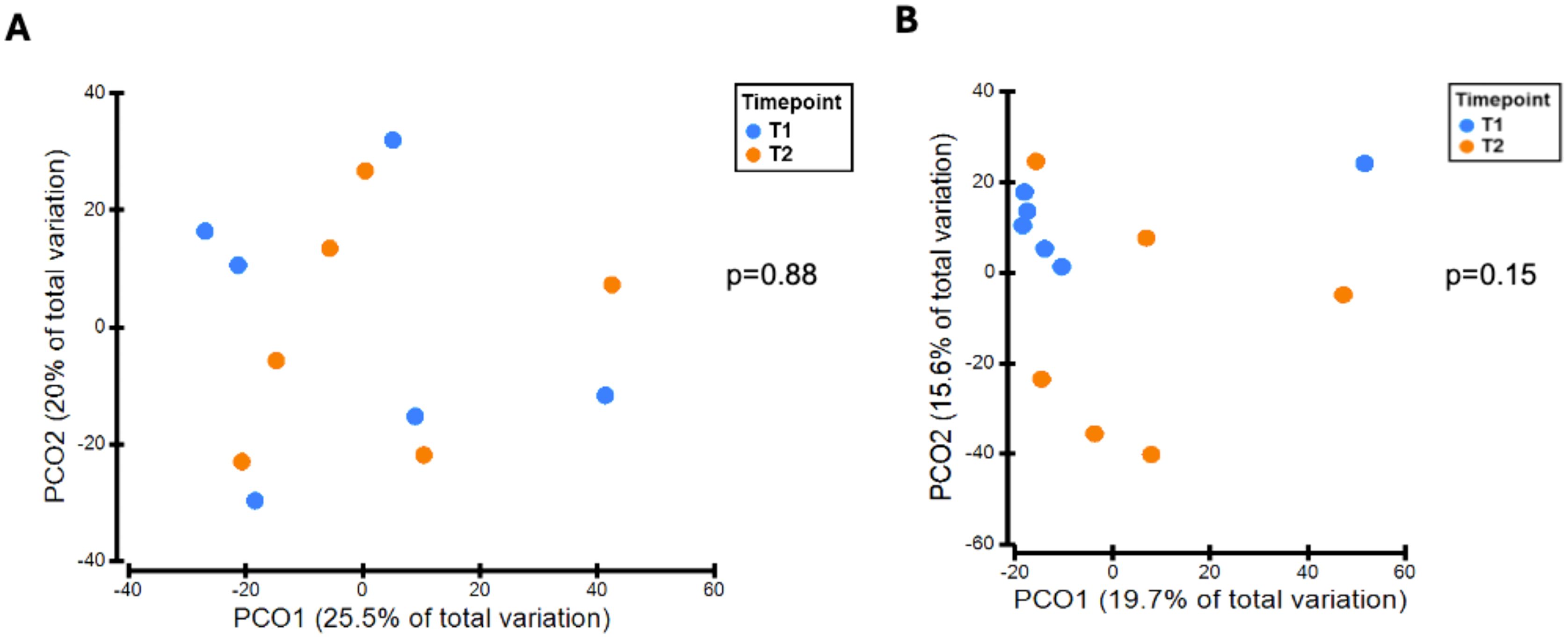
Figure 3. (A) Oral microbiota variation between AHSCT and NTZ cohorts, (B) Stool microbiota variation between AHSCT and NTZ cohorts. Beta diversity statistics were generated using PCoA and PERMANOVA. PCoA was performed on Bray Curtis resemblance matrix of square root transformed relative abundances. There were significant differences in beta-diversity for oral (p=0.043, PERMANOVA) and stool (p=0.0086, PERMANOVA) samples between AHSCT and NTZ cohorts.
3.4 Longitudinal AHSCT analysis
There was no significant difference in alpha diversity between T1 and T2 oral microbiota samples (Figure 4A). T1 and T2 stool microbiota samples also revealed no significant differences in alpha diversity (Figure 4B). Comparison of oral samples at T1 and T2 showed no differences in beta diversity (p=0.88, PERMANOVA) (Figure 5A). Similarly, comparison of stool samples at T1 and T2 showed no differences in beta diversity (p=0.15, PERMANOVA) (Figure 5B).
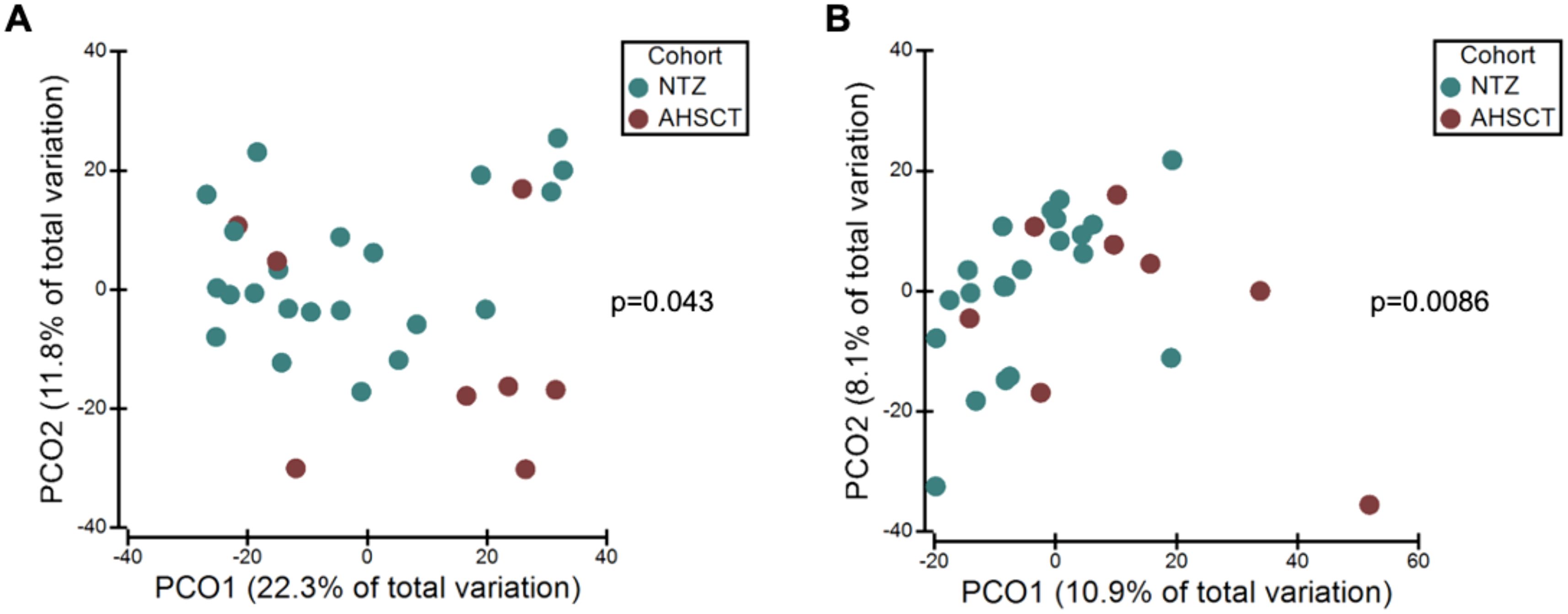
Figure 4. (A) Comparison of oral alpha diversity between T1 and T2 samples, (B) Comparison of stool alpha diversity between T1 and T2 samples. The differences in alpha diversity, including Margalef’s richness, Pielou’s evenness and Shannon’s diversity, between T1 and T2 samples was determined using unpaired t-tests and Mann Whitney U tests. Analysis of both oral and stool microbiota revealed no differences in alpha diversity between T1 and T2 samples.
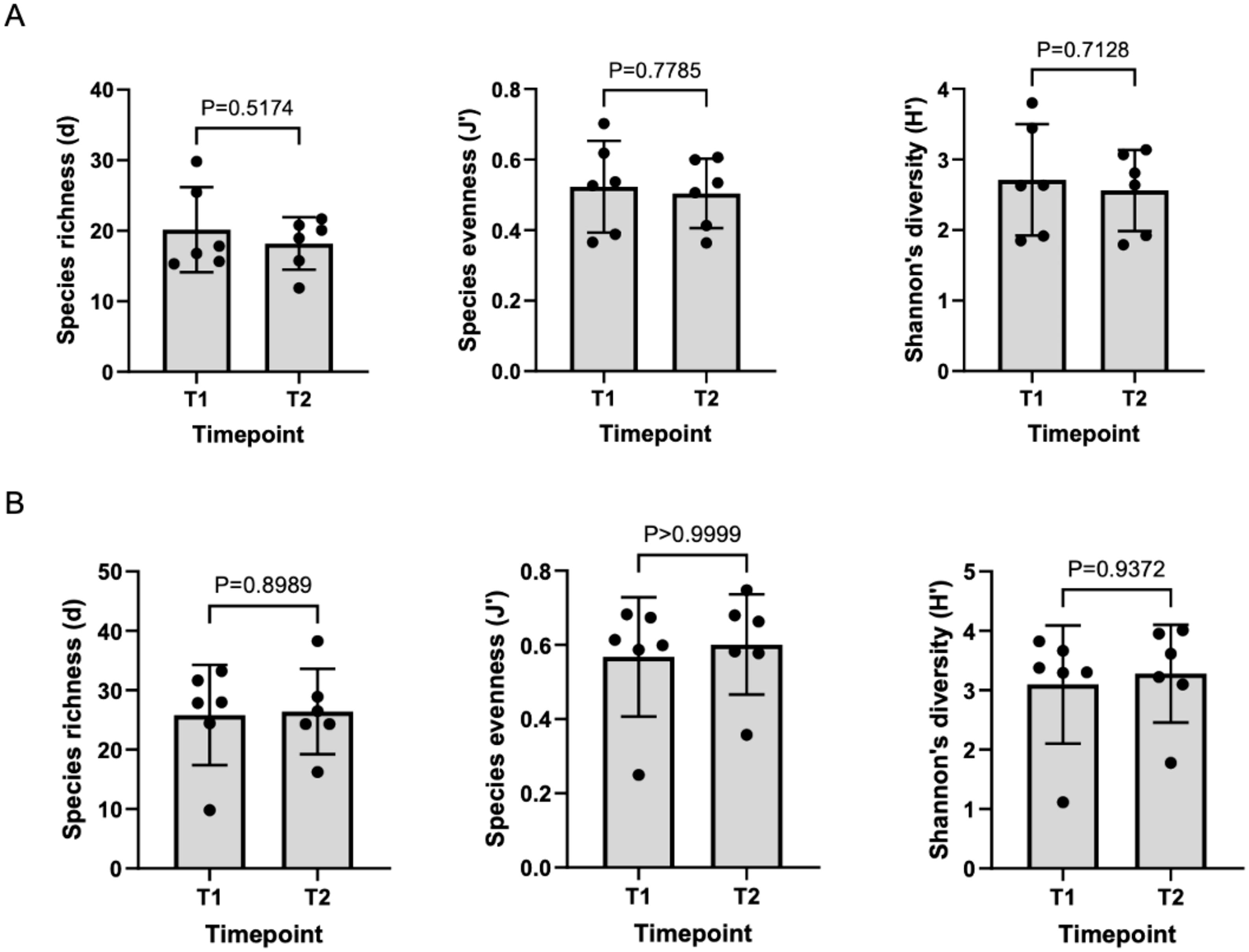
Figure 5. (A) Variation in oral microbiota between T1 and T2 samples, (B) Variation in stool microbiota between T1 and T2 samples. There was no significant difference in oral (p=0.88, PERMANOVA) or stool (p=0.15, PERMANOVA) beta-diversity between T1 and T2 samples.
3.5 Taxa analysis of NTZ and AHSCT cohorts at baseline
A per taxa analysis was performed to identify operational taxonomy units (OTUs) which contributed to differences in beta diversity between AHSCT and NTZ cohorts. The taxa were identified at the genus level, their lowest possible level of classification (Table 4). For stool samples, the AHSCT group had greater relative abundances of an unclassified genus of Lachnospiraceae, Enterocloster, Bifidobacterium and Parabacteroides and lower abundances of Roseburia and Hominimerdicola. For oral microbiota, the AHSCT group has greater relative abundances of Porphyromonas and decreased abundances of Veillonella.
4 Discussion
4.1 Comparison of AHSCT and NTZ cohorts
Despite emerging evidence linking oral bacteria to various systemic diseases (30), the role of oral microbiota in MS remains underexplored. While previous research predominantly compares individuals with MS to healthy controls (HC) (16–21), this study offers a novel approach by examining microbiome differences across MS patients with differing treatment exposures and disease activity levels. Specifically, we focused on patients undergoing AHSCT, who typically have active, treatment-refractory MS, and compared them to patients receiving NTZ, with controlled, non-active MS.
A key finding in this study was the lower species richness of oral microbiota in the ASHCT group compared to the NTZ group. The correlation of reduced alpha diversity in the more active AHSCT cohort raises the possibility that increased oral diversity may have a protective effect in MS. This supports the observations of a longitudinal study comparing the microbiome of pwMS undergoing ocrelizumab therapy. The study reported decreased oral alpha diversity in MS patients compared to HC, revealing that diversity patterns were most pronounced in patients with higher disease activity and disability (31). It is unclear whether these oral microbial changes are an effect of MS therapy or indirectly modulate MS disease activity and this requires further investigation.
Our findings of increased oral Porphyromonas in the AHSCT group is also notable, given previous studies have reported an increased abundance of oral Porphyromonas in pwMS compared to HC (32, 33). Additionally, Porphyromonas gingivalis has been associated with several systemic diseases, including Alzheimer’s disease, various cancers, atherosclerosis, periodontitis, and systemic inflammation (34–36). Its pathogenic potential can be attributed to its lipopolysaccharides, particularly the lipid A structure, which can trigger proinflammatory responses by activating the NF-κB signalling pathways and promoting the secretion of inflammatory cytokines (37). In EAE, a murine model of demyelination, when immune-competent glial cells of the CNS are exposed to antigens of Porphyromonas gingivalis, immune responses and disability scores are exacerbated. However, the significance of this finding needs to be confirmed in human studies (38, 39).
We observed a significant depletion of oral Veillonella in ‘active’ MS (pre-AHSCT cohort) compared to treated MS (NTZ group). A recent study comparing pwMS to HC found increased abundance of oral Veillonella in MS populations. Several factors may explain these divergent findings. Veillonella is part of the core oral microbiome in healthy individuals and its abundance can be influenced by confounders such as periodontal disease, oral hygiene, and dietary habits (40, 41). Notably, a greater proportion of AHSCT patients had received antibiotics in the 12 months prior to sample collection, which may have contributed to the observed depletion, as systemic antibiotic use is known to disrupt oral microbial communities even when not directly targeting the oral cavity (42). It is also important to consider that our comparison involves two MS subgroups with differing disease activity and treatment exposure, rather than a comparison with healthy controls. As such, the observed differences in Veillonella abundance may reflect treatment effects, immunological state, or other systemic influences. Thus, further studies with larger MS cohorts with controlled oral confounders are required to are required to clarify Veillonella’s role and clinical relevance in the context of MS.
Our study revealed no differences in stool alpha diversity between AHSCT and NTZ groups. Similar results were observed in the iMSMS study (21) which found no differences in alpha diversity between treated and non-treated MS populations. Beta diversity between the AHSCT and NTZ cohorts, however, was significantly different, highlighting the possibility that varying MS disease states may be associated with stool microbial composition.
Specific taxa findings revealed increased relative abundances of an unclassified genus of Lachnospiraceae, Enterocloster, Bifidobacterium and Parabacteroides in the AHSCT group compared to the NTZ group. While the lack of HCs in this study makes it difficult to confirm whether these bacteria are significantly altered in MS populations compared to healthy individuals, evidence from numerous studies show they are likely to be implicated in MS (18, 20, 21, 43–45). Consistent with previous studies, we found lower abundances of Roseburia and Hominimerdicola in the AHSCT group. Roseburia has been noted to decrease in MS patients compared to healthy controls (45, 46), suggesting a protective role in MS. It is hypothesised that Roseburia may ameliorate MS through its production of the butyrate, an SCFA with anti-inflammatory properties (47). It should be acknowledged that prior work has conversely suggested that Lachnospiraceae, Bifidobacteirum and Parabacteroides have decreased abundance in untreated MS populations compared to HC (43–45). Specifically, Parabacteroides distasonis has been shown to induce regulatory T cells that produce anti-inflammatory IL-10 (48). Acetate, a SCFA highly produced by Bifidobacteirum, regulates intestinal inflammation by stimulating the GPR43 receptor which in turn, inhibits the secretion of pro-inflammatory IL-18 (49). These conflicting results highlight the complex relationship between specific bacterial taxa, MS and the immune system which can be modulated by other factors, such as disease stage and treatment status.
Interestingly, a consistent pattern emerging from both our study and previous research is the association of MS with SCFA-producing bacteria (46). Five out of the six taxa identified in our study responsible for the stool beta-diversity differences are involved in the production of SCFAs. SCFAs, particularly butyrate and acetate, can promote anti-inflammatory responses by supporting regulatory T cells but also influence pro-inflammatory Th1 and Th17 cell differentiation in certain contexts (50, 51). Increased acetate levels in MS patients correlated with higher disability levels measured by their EDSS (52). While our findings highlight a strong association between stool microbiota and their metabolites with MS, further investigation into bacterial metabolites like SCFAs in MS populations is required to elucidate the exact mechanisms involved.
A potential confounding factor is the effect of NTZ on both oral and gut microbiota. Currently, there is limited evidence on the impact of NTZ on oral microbial composition. However, research has demonstrated that NTZ treatment can reduce specific stool bacterial populations, including some that produce short-chain fatty acids (SCFAs), such as Bacteroides and Bifidobacterium species, which are key to gastrointestinal health (21, 53). While it is believed NTZ’s mechanism of action against MS involves the inhibition of T-cell trafficking to the CNS, it also impacts T-cell circulation within the GIT (54). This immunomodulatory action on the gut could influence microbial composition, possibly representing an alternative pathway by which this therapy acts against MS. NTZ-treated patients were chosen as the comparator group due to their clinical stability, consistent treatment duration, and lack of recent immunosuppressive induction, which make them a valuable reference for understanding microbiome profiles in MS under controlled treatment conditions. However, the differences in microbial composition between the AHSCT and NTZ cohorts may be due to treatment-specific effects, in addition to or instead of disease activity.
4.2 Longitudinal analysis
Analysis of T1 (pre-transplant) and T2 (post-transplant) samples for both oral and stool demonstrated no significant differences in alpha- or beta-diversity, revealing that the diversity of stool and oral microbial populations remained relatively stable post-AHSCT, despite the peri-transplant use of chemotherapy, steroids and antimicrobials. Our findings contrast numerous studies where a marked reduction in stool alpha diversity post-allogenic HSCT was observed (55, 56). In Khan et al., the largest and first multicentre study of stool microbiota composition in autologous HSCT, alpha diversity of post-transplant stool samples was also significantly decreased (57). A plausible explanation for the discrepancy in our results lies in the different post-transplant timeframes assessed across studies. While the aforementioned studies collected post-transplant stool samples within the at 0–100 days post-transplant, our study examined samples collected between 7–12 months post-transplant. This extended period may have allowed the stool microbial population to recover and return to pre-transplant diversity levels, and this return to ‘baseline’ is a fascinating observation in itself. This is supported by longitudinal data from Khan et al., where microbial diversity in stool samples reached its lowest point at 14–17 days post-transplant but subsequently increased (57).
The diversity of gut microbiome before and after transplant has also been linked to improved clinical outcomes. Notably, research has indicated that higher gut microbiota diversity pre-transplant correlates with improved overall survival and a lower incidence of acute graft-versus-host disease (GVHD) in allogeneic HSCT patients (58–60). Additionally, lower diversity during the peri-engraftment phase has been associated with worse outcomes in autologous HSCT patients (57). While these correlations have primarily been drawn from malignant-related HSCT cases, they suggest that the stability of the gut microbiome post-AHSCT in our cohort of MS patients may be linked to positive clinical outcomes, especially since all patients our AHSCT cohort have met the “no evidence of disease activity” (NEDA) criteria and have not progressed in terms of MS disability. However, a larger sample size and correlation to long-term follow up data is required to validate such relationships.
4.3 Limitations and future direction
This study was limited by the relatively small sample size of 30 patients. In particular, recruitment for the AHSCT cohort was restricted by the number of MS patients who met the inclusion criteria for the AiMS study at St Vincent’s Hospital, and ongoing challenges through 2022 with COVID-19. On average, only 5–10 patients undergo AHSCT for MS each year, which has limited the pace of recruitment. As this pilot study is ongoing, we anticipate that a more comprehensive analysis of longitudinal changes in the oral and gut microbiome will be possible as additional data are collected across multiple timepoints. These future analyses may enable correlation of microbial profiles with clinical outcomes, including MS severity and markers of immune reconstitution following AHSCT.
Although the NTZ and AHSCT cohorts were broadly matched for age, sex, and ethnicity, minimising the likelihood of major demographic confounding, statistical analyses to explore associations between individual demographic variables and microbiota composition were not conducted. This was due to the limited sample size and lack of statistical power for reliable subgroup analysis but can also be addressed as additional data is collected.
Due to small numbers, confounding factors in each cohort such as gender, diet or recent antibiotics couldn’t be accounted for and may influence GIT microbial composition. Oral health parameters such as periodontal status and oral hygiene practices were not assessed. Given the known influence of these factors on oral microbial composition, their omission may have impacted our findings (61–63). In future studies, adopting a household paired system whereby the patient with MS and controls live in the same household could address this issue as both individuals would have similar diets and lifestyle factors. Incorporating standardised oral health assessments may also better control for oral microbiome variability.
The variability in timing of post-AHSCT sample collection (between 7–12 months) is also acknowledged as a limitation. This arose due to the logistics of home sampling and postal return. While all samples were collected after the acute transplant phase, this range may have influenced microbiota composition. As more data are collected, we intend to stratify post-AHSCT samples into narrower timepoints (e.g. 6–8 months vs 9–12 months) to examine trends with greater granularity.
Additionally, Natalizumab’s effects on immune cell trafficking in the gastrointestinal tract may independently influence microbiota composition. This introduces a potential confounder when interpreting differences between cohorts and highlights the need for future studies incorporating additional control groups.
This study did not aim to compare MS cohorts with a healthy control population, as this has been extensively previously explored. However, the lack of an internal control is another limitation in our study. Consequently, it was questionable whether the differences between our two cohorts were due to the differing treatments or the varying severity of disease. However, this limitation was partially overcome by supporting evidence from studies which found that the specific bacterial taxa identified in our study were previously implicated in MS.
5 Conclusion
This pilot study identified specific microbiome changes, particularly in the oral alpha diversity and abundance of specific bacteria which may play roles in MS progression or pathogenesis. Furthermore, longitudinal analysis combined with clinical follow-up data alludes to a potential relationship between stool microbial diversity and positive AHSCT outcomes. Ongoing recruitment and further analysis will determine the validity of these findings and may provide a novel therapeutic target in MS treatment.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by St Vincent’s Hospital Human Research and Ethics Committee. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
JY: Data curation, Writing – original draft, Writing – review & editing. NK: Data curation, Formal Analysis, Methodology, Software, Writing – review & editing. JM: Conceptualization, Supervision, Writing – review & editing. MD: Conceptualization, Supervision, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Walton C, King R, Rechtman L, Kaye W, Leray E, Marrie RA, et al. Rising prevalence of multiple sclerosis worldwide: Insights from the Atlas of MS, third edition. Mult Scler. (2020) 26:1816–21. doi: 10.1177/1352458520970841
2. Habbestad A, Willumsen JS, Aarseth JH, Grytten N, Midgard R, Wergeland S, et al. Increasing age of multiple sclerosis onset from 1920 to 2022: a population-based study. J Neurol. (2024) 271:1610–7. doi: 10.1007/s00415-023-12047-9
3. Ford H. Clinical presentation and diagnosis of multiple sclerosis. Clin Med (Lond). (2020) 20:380–3. doi: 10.7861/clinmed.2020-0292
4. Moutsianas L, Jostins L, Beecham AH, Dilthey AT, Xifara DK, Ban M, et al. Class II HLA interactions modulate genetic risk for multiple sclerosis. Nat Genet. (2015) 47:1107–13. doi: 10.1038/ng.3395
5. Hollenbach JA and Oksenberg JR. The immunogenetics of multiple sclerosis: A comprehensive review. J Autoimmun. (2015) 64:13–25. doi: 10.1016/j.jaut.2015.06.010
6. Berer K and Krishnamoorthy G. Microbial view of central nervous system autoimmunity. FEBS Lett. (2014) 588:4207–13. doi: 10.1016/j.febslet.2014.04.007
7. Massey JC, Sutton IJ, Ma DDF, and Moore JJ. Regenerating immunotolerance in multiple sclerosis with autologous hematopoietic stem cell transplant. Front Immunol. (2018) 9:410. doi: 10.3389/fimmu.2018.00410
8. Voet S, Prinz M, and van Loo G. Microglia in central nervous system inflammation and multiple sclerosis pathology. Trends Mol Med. (2019) 25:112–23. doi: 10.1016/j.molmed.2018.11.005
9. Biswas S, Bryant RV, and Travis S. Interfering with leukocyte trafficking in Crohn’s disease. Best Pract Res Clin Gastroenterol. (2019) 38-39:101617. doi: 10.1016/j.bpg.2019.05.004
10. Nguyen Ky M, Duran A, Hasantari I, Bru A, Deloire M, Brochet B, et al. Natalizumab treatment induces proinflammatory CD4 T cells preferentially in the integrin β7+ Compartment. Neurol Neuroimmunol Neuroinflamm. (2023) 10. doi: 10.1212/NXI.0000000000200166
11. Cohen JA, Baldassari LE, Atkins HL, Bowen JD, Bredeson C, Carpenter PA, et al. Autologous hematopoietic cell transplantation for treatment-refractory relapsing multiple sclerosis: position statement from the american society for blood and marrow transplantation. Biol Blood Marrow Transpl. (2019) 25:845–54. doi: 10.1016/j.bbmt.2019.02.014
12. Chen Y, Xu J, and Chen Y. Regulation of neurotransmitters by the gut microbiota and effects on cognition in neurological disorders. Nutrients. (2021) 13. doi: 10.3390/nu13062099
13. Fülling C, Dinan TG, and Cryan JF. Gut microbe to brain signaling: what happens in vagus…. Neuron. (2019) 101:998–1002. doi: 10.1016/j.neuron.2019.02.008
14. Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. (2009) 139:485–98. doi: 10.1016/j.cell.2009.09.033
15. Correale J, Hohlfeld R, and Baranzini SE. The role of the gut microbiota in multiple sclerosis. Nat Rev Neurol. (2022) 18:544–58. doi: 10.1038/s41582-022-00697-8
16. Cantarel BL, Waubant E, Chehoud C, Kuczynski J, DeSantis TZ, Warrington J, et al. Gut microbiota in multiple sclerosis: possible influence of immunomodulators. J Investig Med. (2015) 63:729–34. doi: 10.1097/JIM.0000000000000192
17. Miyake S, Kim S, Suda W, Oshima K, Nakamura M, Matsuoka T, et al. Dysbiosis in the gut microbiota of patients with multiple sclerosis, with a striking depletion of species belonging to clostridia XIVa and IV clusters. PloS One. (2015) 10:e0137429. doi: 10.1371/journal.pone.0137429
18. Chen J, Chia N, Kalari KR, Yao JZ, Novotna M, Paz Soldan MM, et al. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci Rep. (2016) 6:28484. doi: 10.1038/srep28484
19. Jangi S, Gandhi R, Cox LM, Li N, von Glehn F, Yan R, et al. Alterations of the human gut microbiome in multiple sclerosis. Nat Commun. (2016) 7:12015. doi: 10.1038/ncomms12015
20. Berer K, Gerdes LA, Cekanaviciute E, Jia X, Xiao L, Xia Z, et al. Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc Natl Acad Sci U S A. (2017) 114:10719–24. doi: 10.1073/pnas.1711233114
21. Zhou X, Baumann R, Gao X, Mendoza M, Singh S, Katz Sand I, et al. Gut microbiome of multiple sclerosis patients and paired household healthy controls reveal associations with disease risk and course. Cell. (2022) 185:3467–86.e16. doi: 10.1016/j.cell.2022.08.021
22. Thompson AJ, Banwell BL, Barkhof F, Carroll WM, Coetzee T, Comi G, et al. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. (2018) 17:162–73. doi: 10.1016/S1474-4422(17)30470-2
23. Doki N, Suyama M, Sasajima S, Ota J, Igarashi A, Mimura I, et al. Clinical impact of pre-transplant gut microbial diversity on outcomes of allogeneic hematopoietic stem cell transplantation. Ann Hematol. (2017) 96:1517–23. doi: 10.1007/s00277-017-3069-8
24. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. (2009) 75:7537–41. doi: 10.1128/AEM.01541-09
25. Yang L and Chen J. A comprehensive evaluation of microbial differential abundance analysis methods: current status and potential solutions. Microbiome. (2022) 10:130. doi: 10.1186/s40168-022-01320-0
26. Olsen I. Porphyromonas gingivalis-induced neuroinflammation in Alzheimer’s disease. Front Neurosci. (2021) 15:691016. doi: 10.3389/fnins.2021.691016
27. Retnakumar RJ, Nath AN, Nair GB, and Chattopadhyay S. Chapter Three - Gastrointestinal microbiome in the context of Helicobacter pylori infection in stomach and gastroduodenal diseases. Prog Mol Biol Transl Sci. (2022) 192:53–95. doi: 10.1016/bs.pmbts.2022.07.001
28. Fusco W, Lorenzo MB, Cintoni M, Porcari S, Rinninella E, Kaitsas F, et al. Short-chain fatty-acid-producing bacteria: key components of the human gut microbiota. Nutrients. (2023) 15. doi: 10.3390/nu15092211
29. Cui Y, Zhang L, Wang X, Yi Y, Shan Y, Liu B, et al. Roles of intestinal Parabacteroides in human health and diseases. FEMS Microbiol Lett. (2022) 369. doi: 10.1093/femsle/fnac072
30. Peng X, Cheng L, You Y, Tang C, Ren B, Li Y, et al. Oral microbiota in human systematic diseases. Int J Oral Sci. (2022) 14:14. doi: 10.1038/s41368-022-00163-7
31. Troci A, Zimmermann O, Esser D, Krampitz P, May S, Franke A, et al. B-cell-depletion reverses dysbiosis of the microbiome in multiple sclerosis patients. Sci Rep. (2022) 12:3728. doi: 10.1038/s41598-022-07336-8
32. Boussamet L, Montassier E, Mathé C, Garcia A, Morille J, Shah S, et al. Investigating the metabolite signature of an altered oral microbiota as a discriminant factor for multiple sclerosis: a pilot study. Sci Rep. (2024) 14:7786. doi: 10.1038/s41598-024-57949-4
33. Zangeneh Z, Abdi-Ali A, Khamooshian K, Alvandi A, and Abiri R. Bacterial variation in the oral microbiota in multiple sclerosis patients. PloS One. (2021) 16:e0260384. doi: 10.1371/journal.pone.0260384
34. Rafiei M, Kiani F, Sayehmiri F, Sayehmiri K, Sheikhi A, and Zamanian Azodi M. Study of Porphyromonas gingivalis in periodontal diseases: A systematic review and meta-analysis. Med J Islam Repub Iran. (2017) 31:62. doi: 10.14196/mjiri.31.62
35. Gnanasekaran J, Binder Gallimidi A, Saba E, Pandi K, Eli Berchoer L, Hermano E, et al. Intracellular porphyromonas gingivalis promotes the tumorigenic behavior of pancreatic carcinoma cells. Cancers (Basel). (2020) 12. doi: 10.3390/cancers12082331
36. Dominy SS, Lynch C, Ermini F, Benedyk M, Marczyk A, Konradi A, et al. Porphyromonas gingivalis in Alzheimer’s disease brains: Evidence for disease causation and treatment with small-molecule inhibitors. Sci Adv. (2019) 5:eaau3333. doi: 10.1126/sciadv.aau3333
37. Nativel B, Couret D, Giraud P, Meilhac O, d’Hellencourt CL, Viranaïcken W, et al. Porphyromonas gingivalis lipopolysaccharides act exclusively through TLR4 with a resilience between mouse and human. Sci Rep. (2017) 7:15789. doi: 10.1038/s41598-017-16190-y
38. Shapira L, Ayalon S, and Brenner T. Effects of porphyromonas gingivalis on the central nervous system: activation of glial cells and exacerbation of experimental autoimmune encephalomyelitis. J Periodontol. (2002) 73:511–6. doi: 10.1902/jop.2002.73.5.511
39. Memedovski Z, Czerwonka E, Han J, Mayer J, Luce M, Klemm LC, et al. Classical and alternative activation of rat microglia treated with ultrapure porphyromonas gingivalis lipopolysaccharide in vitro. Toxins (Basel). (2020) 12. doi: 10.3390/toxins12050333
40. Perera M, Al-hebshi NN, Speicher DJ, Perera I, and Johnson NW. Emerging role of bacteria in oral carcinogenesis: a review with special reference to perio-pathogenic bacteria. J Oral Microbiol. (2016) 8:32762. doi: 10.3402/jom.v8.32762
41. Li X, Liu Y, Yang X, Li C, and Song Z. The oral microbiota: community composition, influencing factors, pathogenesis, and interventions. Front Microbiol. (2022) 13:895537. doi: 10.3389/fmicb.2022.895537
42. Cheng X, He F, Si M, Sun P, and Chen Q. Effects of antibiotic use on saliva antibody content and oral microbiota in sprague dawley rats. Front Cell Infect Microbiol. (2022) 12. doi: 10.3389/fcimb.2022.721691
43. Cekanaviciute E, Yoo BB, Runia TF, Debelius JW, Singh S, Nelson CA, et al. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc Natl Acad Sci. (2017) 114:10713–8. doi: 10.1073/pnas.1711235114
44. Cantoni C, Lin Q, Dorsett Y, Ghezzi L, Liu Z, Pan Y, et al. Alterations of host-gut microbiome interactions in multiple sclerosis. EBioMedicine. (2022) 76:103798. doi: 10.1016/j.ebiom.2021.103798
45. Ling Z, Cheng Y, Yan X, Shao L, Liu X, Zhou D, et al. Alterations of the fecal microbiota in chinese patients with multiple sclerosis. Front Immunol. (2020) 11:590783. doi: 10.3389/fimmu.2020.590783
46. Saresella M, Marventano I, Barone M, La Rosa F, Piancone F, Mendozzi L, et al. Alterations in circulating fatty acid are associated with gut microbiota dysbiosis and inflammation in multiple sclerosis. Front Immunol. (2020) 11:1390. doi: 10.3389/fimmu.2020.01390
47. Tamanai-Shacoori Z, Smida I, Bousarghin L, Loreal O, Meuric V, Fong SB, et al. Roseburia spp.: A marker of health? Future Microbiol. (2017) 12:157–70. doi: 10.2217/fmb-2016-0130
48. Ezeji JC, Sarikonda DK, Hopperton A, Erkkila HL, Cohen DE, Martinez SP, et al. Parabacteroides distasonis: intriguing aerotolerant gut anaerobe with emerging antimicrobial resistance and pathogenic and probiotic roles in human health. Gut Microbes. (2021) 13:1922241. doi: 10.1080/19490976.2021.1922241
49. Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. (2009) 461:1282–6. doi: 10.1038/nature08530
50. Li M, van Esch BCAM, Wagenaar GTM, Garssen J, Folkerts G, and Henricks PAJ. Pro- and anti-inflammatory effects of short chain fatty acids on immune and endothelial cells. Eur J Pharmacol. (2018) 831:52–9. doi: 10.1016/j.ejphar.2018.05.003
51. Ney LM, Wipplinger M, Grossmann M, Engert N, Wegner VD, and Mosig AS. Short chain fatty acids: key regulators of the local and systemic immune response in inflammatory diseases and infections. Open Biol. (2023) 13:230014. doi: 10.1098/rsob.230014
52. Pérez-Pérez S, Domínguez-Mozo MI, Alonso-Gómez A, Medina S, Villarrubia N, Fernández-Velasco JI, et al. Acetate correlates with disability and immune response in multiple sclerosis. PeerJ. (2020) 8:e10220. doi: 10.7717/peerj.10220
53. Kujawa D, Laczmanski L, Budrewicz S, Pokryszko-Dragan A, and Podbielska M. Targeting gut microbiota: new therapeutic opportunities in multiple sclerosis. Gut Microbes. (2023) 15:2274126. doi: 10.1080/19490976.2023.2274126
54. Lobb RR and Hemler ME. The pathophysiologic role of alpha 4 integrins in vivo. J Clin Invest. (1994) 94:1722–8. doi: 10.1172/JCI117519
55. Peled JU, Gomes ALC, Devlin SM, Littmann ER, Taur Y, Sung AD, et al. Microbiota as predictor of mortality in allogeneic hematopoietic-cell transplantation. N Engl J Med. (2020) 382:822–34. doi: 10.1056/NEJMoa1900623
56. Taur Y, Xavier JB, Lipuma L, Ubeda C, Goldberg J, Gobourne A, et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis. (2012) 55:905–14. doi: 10.1093/cid/cis580
57. Khan N, Lindner S, Gomes ALC, Devlin SM, Shah GL, Sung AD, et al. Fecal microbiota diversity disruption and clinical outcomes after auto-HCT: a multicenter observational study. Blood. (2021) 137:1527–37. doi: 10.1182/blood.2020006923
58. Masetti R, Leardini D, Muratore E, Fabbrini M, D’Amico F, Zama D, et al. Gut microbiota diversity before allogeneic hematopoietic stem cell transplantation as a predictor of mortality in children. Blood. (2023) 142:1387–98. doi: 10.1182/blood.2023020026
59. Vaitkute G, Panic G, Alber DG, Faizura-Yeop I, Cloutman-Green E, Swann J, et al. Linking gastrointestinal microbiota and metabolome dynamics to clinical outcomes in paediatric haematopoietic stem cell transplantation. Microbiome. (2022) 10:89. doi: 10.1186/s40168-022-01270-7
60. Liu C, Frank DN, Horch M, Chau S, Ir D, Horch EA, et al. Associations between acute gastrointestinal GvHD and the baseline gut microbiota of allogeneic hematopoietic stem cell transplant recipients and donors. Bone Marrow Transpl. (2017) 52:1643–50. doi: 10.1038/bmt.2017.200
61. Kim YS, Unno T, Kim BY, and Park MS. Sex differences in gut microbiota. World J Men’s Health. (2020) 38:48–60. doi: 10.5534/wjmh.190009
62. Patangia DV, Anthony Ryan C, Dempsey E, Paul Ross R, and Stanton C. Impact of antibiotics on the human microbiome and consequences for host health. MicrobiologyOpen. (2022) 11:e1260. doi: 10.1002/mbo3.1260
63. Singh RK, Chang HW, Yan D, Lee KM, Ucmak D, Wong K, et al. Influence of diet on the gut microbiome and implications for human health. J Transl Med. (2017) 15:73. doi: 10.1186/s12967-017-1175-y
Appendix 1
Washout periods off DMT prior to sample collection
● S1P receptor modulators (i.e. Fingolimod): 4 weeks from last dose
● Cladribine: 6 months from last dose
● Natalizumab: 6 weeks from last dose
● CD20 monoclonal antibodies (Ocrelizumab/Ofatumumab): 3 months from last dose
● Alemtuzumab: 6 months from last dose
Keywords: multiple sclerosis, gut microbiome, autologous haemopoietic stem cell transplant (AHSCT), dysbiosis, immune reconstitution (IR)
Citation: Yin J, Kaakoush NO, Massey J and Danta M (2025) Understanding the microbiome in autologous haemopoietic stem cell transplant for multiple sclerosis. Front. Immunol. 16:1590601. doi: 10.3389/fimmu.2025.1590601
Received: 10 March 2025; Accepted: 15 June 2025;
Published: 30 June 2025.
Edited by:
Ana Raquel Santiago, University of Coimbra, PortugalReviewed by:
Alice Mariottini, University of Florence, ItalyHajra Ashraf, University of Sassari, Italy
Copyright © 2025 Yin, Kaakoush, Massey and Danta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jennifer Massey, amVubmlmZXIubWFzc2V5QHN2aGEub3JnLmF1
 Jun Yin
Jun Yin Nadeem O. Kaakoush
Nadeem O. Kaakoush Jennifer Massey
Jennifer Massey Mark Danta
Mark Danta