- Department of Orthopedics, The Fourth Hospital of Hebei Medical University, Shijiazhuang, Hebei, China
Human immunodeficiency virus (HIV) infection remains a major global public health challenge. Although highly active antiretroviral therapy (HAART or ART) can effectively control viral replication, it fails to eradicate latent viral reservoirs and poses limitations such as lifelong medication and cumulative drug toxicity. This study focuses on the pivotal role of C-C chemokine receptor 5 (CCR5) gene editing in HIV immunotherapy, particularly highlighting the natural resistance to R5-tropic HIV strains observed in the “Berlin” and “London” patients carrying the homozygous CCR5-Δ32 mutation. We further explore the synergistic potential of multiplex gene editing strategies—including CCR5, CXCR4, and HIV LTR loci—and the combinatorial mechanisms between gene editing technologies and immunotherapy. A personalized treatment framework is proposed to address the clinical heterogeneity among people living with HIV. In addition, we assess the balance between long-term safety and global accessibility of gene-editing approaches such as CRISPR/Cas9, emphasizing strategies to enhance therapeutic efficacy while reducing cost and off-target effects. Our findings suggest that the integration of CCR5-targeted gene editing with immune-based interventions holds great promise for overcoming current therapeutic limitations and achieving functional HIV cure. However, key challenges—such as immune rejection, viral tropism switching, and economic feasibility—must be resolved. This integrative approach provides a robust theoretical and technical foundation for the next generation of HIV treatment paradigms.
1 Introduction
Human immunodeficiency virus (HIV) has exerted a profound impact on global public health, claiming millions of lives (1). Highly active antiretroviral therapy (HAART or ART) has significantly altered the natural course of HIV infection, prolonging survival and improving quality of life for those affected (1, 2). However, ART is not curative: it cannot eliminate latent viral reservoirs (3–5), necessitates lifelong adherence, and is associated with cumulative drug toxicity and the emergence of resistant viral strains (2, 6).
Traditional immune-based strategies—such as the use of broadly neutralizing antibodies (bNAbs) to target circulating virus (7–9) or immunostimulatory agents to enhance host immune responses (10, 11)—have shown promise but remain limited in their ability to eliminate latent HIV. Similarly, allogeneic hematopoietic stem cell transplantation (allo-HSCT) is constrained by high procedural risk and donor scarcity (6, 12).
The discovery of C-C chemokine receptor 5 (CCR5) as a major HIV co-receptor represents a breakthrough in overcoming these limitations. HIV entry into CD4+ T cells and other host cells requires not only CD4 receptor binding but also co-receptors such as CCR5 or CXCR4 (3, 13). Case studies of the “Berlin” and “London” patients—who achieved viral remission following transplantation from CCR5-Δ32 homozygous donors—have provided compelling evidence that genetic disruption of CCR5 can confer natural resistance to R5-tropic HIV strains (6, 12). These findings have catalyzed the rapid development of gene editing technologies targeting CCR5, including CRISPR/Cas9, for potential curative therapy (9, 13–15).
This paper seeks to address four key questions:
1. Synergistic Multi-target Editing: How can simultaneous editing of CCR5, CXCR4, and HIV LTR collectively establish a comprehensive viral blockade to counteract tropism switching and latent reactivation?
2. Gene-Immune Synergy: How can gene editing augment the anti-HIV capacity and persistence of immune cells? Conversely, how can immunotherapy complement gene editing to eradicate latent reservoirs more effectively?
3. Personalized Approaches for Clinical Heterogeneity: Given the high variability in viral subtypes, host immunity, and genetic background among HIV-infected individuals, how can we design broadly applicable yet individually adaptable treatment regimens?
4. Balancing Safety and Accessibility: How can we ensure long-term safety while enhancing global accessibility through technological optimization and innovative payment models?
Figure 1 outlines the integrated framework for HIV treatment, which systematically combines multi-target gene editing with synergistic immunotherapy, not only intervening at key stages of HIV infection but also addressing implementation challenges in clinical applications.
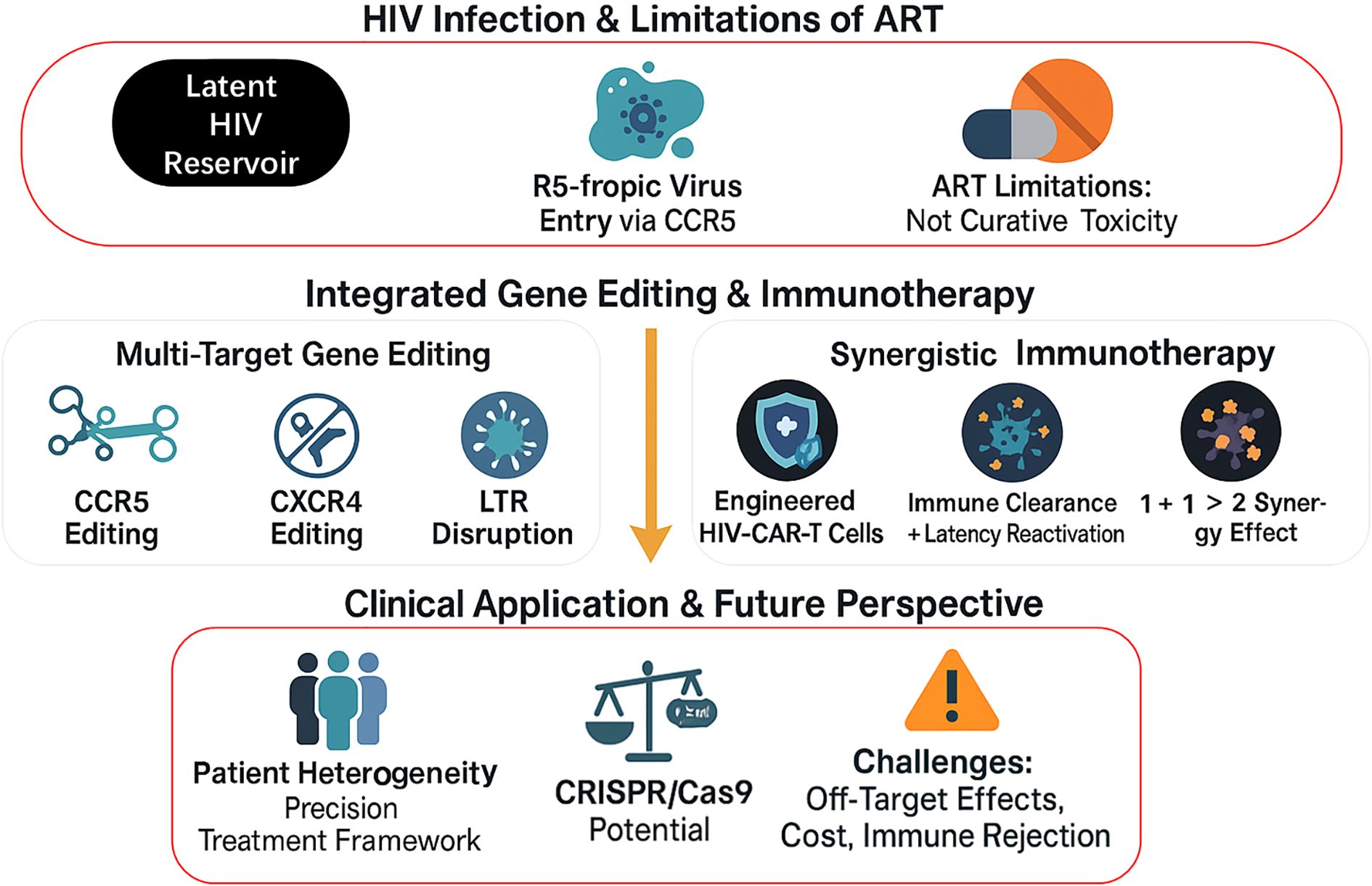
Figure 1. Integrated Strategy of Gene Editing and Immunotherapy for HIV Treatment. This figure presents an integrated framework for HIV treatment, structured in three levels from top to bottom: First, it outlines the characteristics of HIV infection and limitations of current antiretroviral therapy (ART); the middle section showcases two core therapeutic approaches—multi-target gene editing (CCR5, CXCR4, and LTR) and synergistic immunotherapy (HIV-CAR-T cells, etc.); the bottom displays clinical application considerations, including patient heterogeneity, CRISPR/Cas9 technology potential, and implementation challenges.
2 Current advances in integrating gene editing with immunotherapy for HIV
For R5-tropic HIV-1 strains—which dominate during the early and chronic phases of infection—C-C chemokine receptor 5 (CCR5) is an essential co-receptor for viral entry into CD4+ T cells and macrophages (3, 13). Its expression directly determines the susceptibility of these target cells to HIV. Individuals with naturally occurring CCR5 deletions, such as the homozygous CCR5-Δ32 mutation, exhibit high resistance to HIV-1 infection, providing a theoretical rationale for CCR5-targeted gene editing as a therapeutic strategy (6, 12).
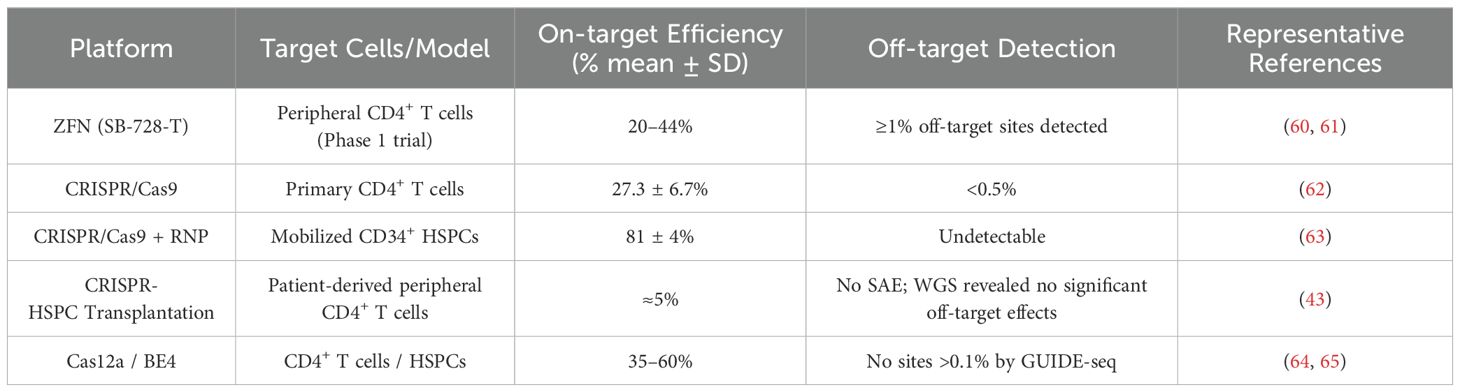
In recent years, molecular tools including zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas system have enabled precise targeting and editing of the CCR5 gene (14, 16, 17). Each technology offers unique features and therapeutic potential in the context of HIV treatment (see Table 1). Notably, CCR5 editing using CRISPR/Cas9 has progressed to early-phase clinical trials, including NCT03164135, which assessed CRISPR/Cas9-mediated CCR5 editing in hematopoietic stem cells for patients with both HIV and acute lymphoblastic leukemia—demonstrating feasibility and safety (18).
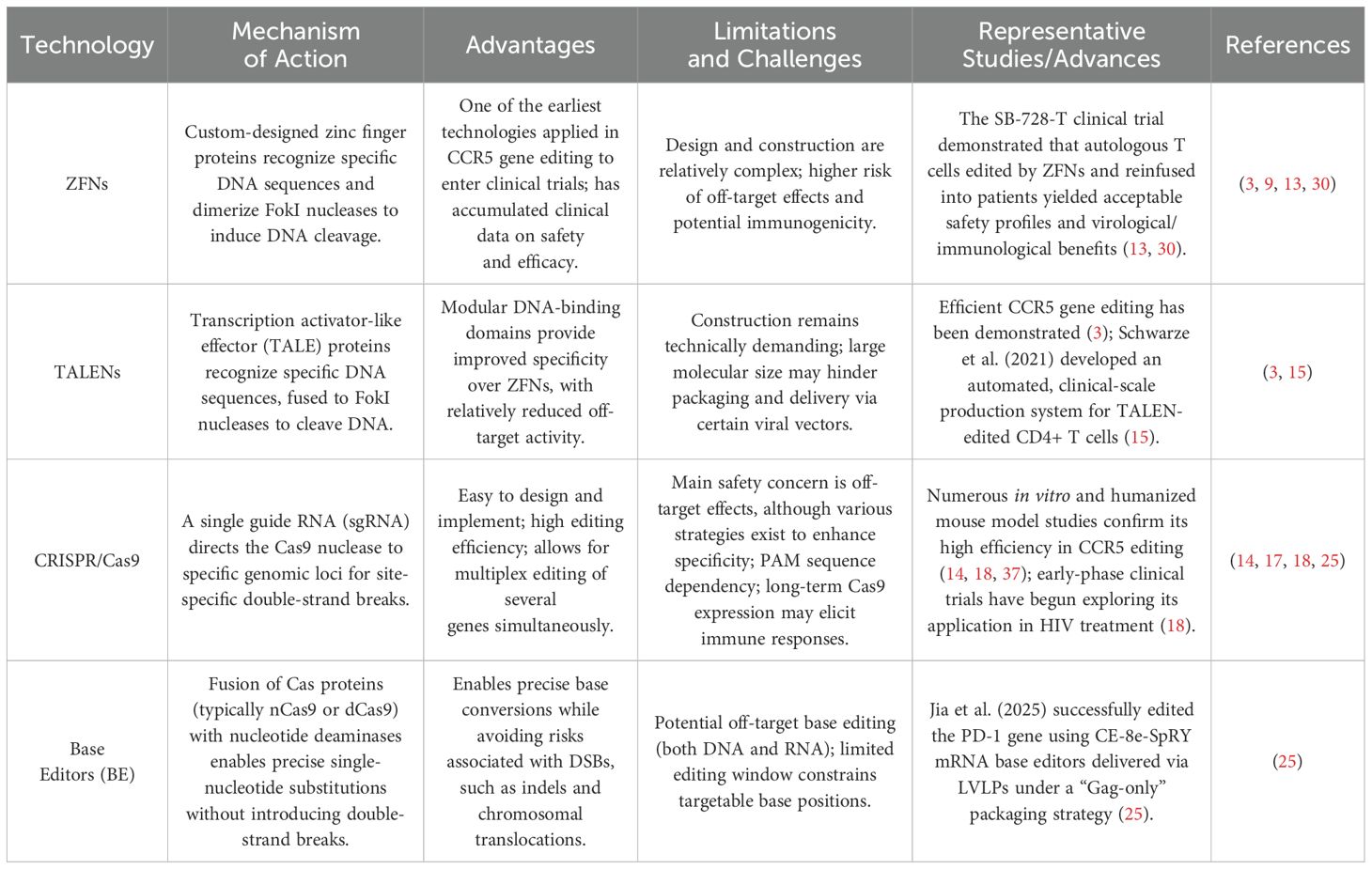
Table 1. Comparative characteristics of major gene editing technologies for CCR5-targeted HIV therapy.
Chronic viral infections such as HIV, HBV, and HCV share the common hallmark of progressive T cell exhaustion, which is closely linked to sustained expression of immune checkpoint molecules like programmed cell death protein 1 (PD-1), PD-L1, and cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) (19). Evidence suggests that PD-1/PD-L1 blockade may restore the function of HIV-specific CD8+ T cells, improving their ability to clear infected cells and potentially reactivating latent reservoirs (20, 21). Additionally, anti-PD-1 chimeric antigen receptor (CAR) T cells have shown efficacy in targeting SIV-infected CD4+ T cells in germinal centers of non-human primate models (19), raising interest in the use of immune checkpoint inhibitors in HIV treatment.
Although CCR5 gene editing and immunotherapies—such as checkpoint inhibitors and CAR-T cells—have each demonstrated distinct promise, studies combining these approaches in a synergistic and systematic manner remain limited. Current research often involves engineering CAR-T cells (21–24)to co-express anti-HIV shRNAs or edited CCR5 genes (21, 22, 24), primarily to enhance the therapeutic durability of the immune cells. Similarly, allogeneic HIV-specific CAR-T cells engineered to secrete PD-1-blocking scFv have shown increased cytotoxicity against HIV Env+ cells (13), but such efforts still focus on incorporating immune-modulatory elements into CAR-T cell functionality rather than exploring dynamic synergy between gene editing and immunotherapy.
Most existing studies remain centered on either optimizing gene editing efficiency and safety (7, 15, 25) or developing novel immune-based therapies for HIV (4, 10, 11, 19, 22, 24, 26–29), with few addressing their integrated or synergistic potential.
3 Cutting-Edge Developments
Due to the high genetic variability of HIV, single-target CCR5 editing strategies alone are insufficient. Thus, recent innovations in the field are moving toward two main directions: (1) constructing a comprehensive viral defense through multi-target gene editing, and (2) enhancing viral clearance through synergistic integration of gene editing and immunotherapy.
3.1 Multi-target gene editing to construct a comprehensive viral barrier
Following effective CCR5 disruption, HIV may switch coreceptor usage to CXCR4 (X4-tropic strains), enabling continued infection (30). Furthermore, once HIV integrates into the host genome, the virus can be reactivated via the long terminal repeat (LTR) region, which contains strong promoter and enhancer elements. This allows viral reactivation even in cells lacking CCR5 or CXCR4 expression (31, 32). Activation of LTR promotes Gag expression, enhances viral particle assembly (33), and facilitates reverse transcription (34), followed by integrase-mediated insertion of viral DNA into host chromosomes (35), ultimately driving viral replication (36).
Thus, multi-target gene editing strategies—targeting both host and viral genes—are critical for combating tropism switching, latent reactivation, and escape mutations. ZFNs and TALENs can be paired for multi-locus editing, as demonstrated by Schwarze et al. (2021), who achieved efficient CCR5 editing using TALENs (15). The CRISPR/Cas9 system offers more versatility by co-delivering Cas9 with multiple single-guide RNAs (sgRNAs) targeting CCR5, CXCR4, HIV LTR, and viral structural genes (e.g., Gag, Pol) (14, 18).
Simultaneous knockout of CCR5 and CXCR4 prevents infection by both R5- and X4-tropic viruses. Editing the HIV LTR suppresses transcriptional activation, while targeting Gag disrupts particle assembly. Collectively, such multi-site CRISPR/Cas9 interventions show superior efficacy in inhibiting viral replication and transmission (17).
The CRISPR/Cas12 system—specifically Cas12a (formerly Cpf1)—has unique features such as recognition of TTTN PAM sites and sticky-end cleavage. A crRNA array can generate multiple mature crRNAs for multiplex editing. This makes Cas12a well-suited for simultaneous targeting of diverse loci. Additionally, base editors (BEs) and prime editors (PEs) offer precise nucleotide modifications or small insertions/deletions without inducing double-strand breaks, minimizing the risk of chromosomal translocations and large deletions.
Jia et al. (2025) used lentiviral-like particles (LVLPs) to deliver CE-8e-SpRY mRNA, an adenine base editor, effectively targeting the PD-1 gene—highlighting both the precision and delivery feasibility of such systems (25). Given the differences in editing efficiency and off-target risks among platforms, combinatorial multi-platform strategies are encouraged, as they can cover over 90% of HIV strains and significantly enhance therapeutic efficacy (see Table 2).
3.2 Synergistic integration of gene editing and immunotherapy
The concept of gene-immune synergy leverages the long-term protection conferred by gene editing and the potent viral clearance enabled by immunotherapy, aiming for a “1 + 1 > 2” therapeutic outcome. This can be achieved through two key pathways:
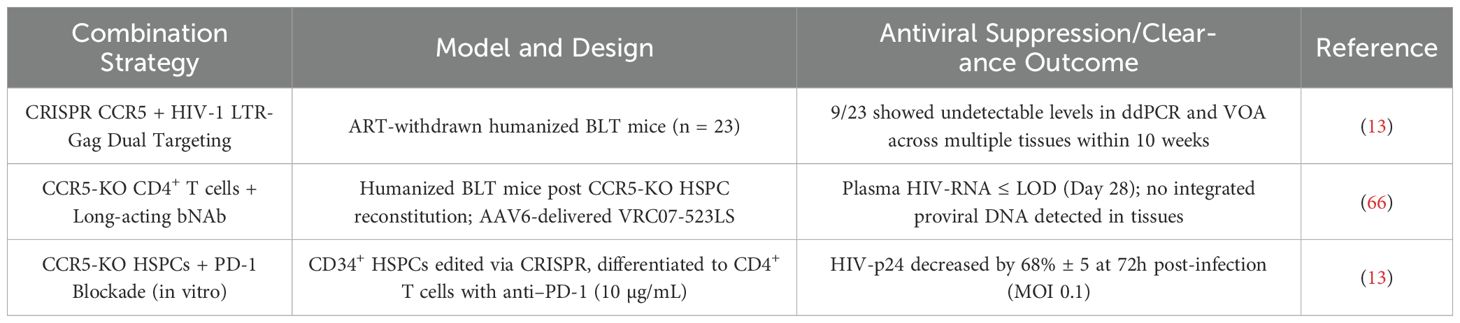
First, immune effector cells such as T cells and NK cells can be genetically modified using CRISPR/Cas9 to enhance their resistance to HIV infection. For example, when engineering HIV-targeted CAR-T cells, concurrent CCR5 knockout can protect them from HIV-mediated depletion post-infusion, extending their in vivo persistence and enhancing antiviral durability (21, 24). Studies have shown that such modifications significantly reduce viral load and may directly contribute to reservoir clearance (see Table 3). Current research is increasingly focused on generating CAR-T cells with dual functions: intrinsic HIV resistance and active antiviral cytotoxicity (37).
Second, the combination of gene editing and immune checkpoint modulation can further improve viral eradication. Allogeneic HIV-specific CAR-T cells engineered to secrete PD-1-blocking scFv exhibited heightened cytotoxicity against HIV Env-expressing cells (13), demonstrating a clear case of synergy. Additionally, stem cell-derived CAR-T cells have shown effective migration and infiltration into viral reservoirs located in germinal centers, the central nervous system, and gut-associated lymphoid tissue in macaque models (22), offering strong experimental support for this strategy.
4 Challenges and limitations
As a retrovirus, HIV presents formidable challenges to therapeutic strategies due to its high mutation rate and ability to establish latent reservoirs. Certain HIV strains—particularly X4-tropic variants—can utilize CXCR4 as a coreceptor to enter host cells. This tropism shift from R5 to X4 is especially common in late-stage disease or after CCR5-targeted interventions (3). Furthermore, regulatory elements within the HIV long terminal repeat (LTR) region can independently drive viral transcription and replication, posing a risk of reactivation even in the absence of coreceptor expression.
Latent HIV reservoirs are anatomically dispersed throughout various tissues and cell types, including lymph nodes, spleen, gut-associated lymphoid tissue (GALT), and the central nervous system (CNS) (1, 3–5). These reservoirs predominantly reside in resting memory CD4+ T cells but also include macrophages and dendritic cells, which exhibit low metabolic activity, long half-lives, and resistance to conventional antiretroviral therapy (ART) (4, 12). The complexity and heterogeneity of HIV persistence across tissue compartments represent a major barrier to viral eradication (4, 5).
Moreover, HIV-infected individuals exhibit significant heterogeneity in terms of viral load, subtype, genotype, resistance history, CD4+ T cell counts, immune activation, and host genetic background (27). Such variability complicates treatment response, as a uniform therapeutic approach may not achieve comparable efficacy across different patients. In cases of severe immunosuppression, gene-edited cells may lack adequate immune surveillance support, diminishing their antiviral potential. This underscores the urgent need for diverse, personalized therapeutic strategies (1).
Although gene editing technologies—such as CRISPR/Cas9—have made substantial progress in improving target specificity, off-target effects remain a critical safety concern (14, 16, 17). The long-term in vivo persistence, genetic stability, and potential late-onset adverse effects of edited cells must be rigorously evaluated through large-scale, longitudinal clinical trials. These safety issues highlight the necessity of long-term surveillance frameworks (38).
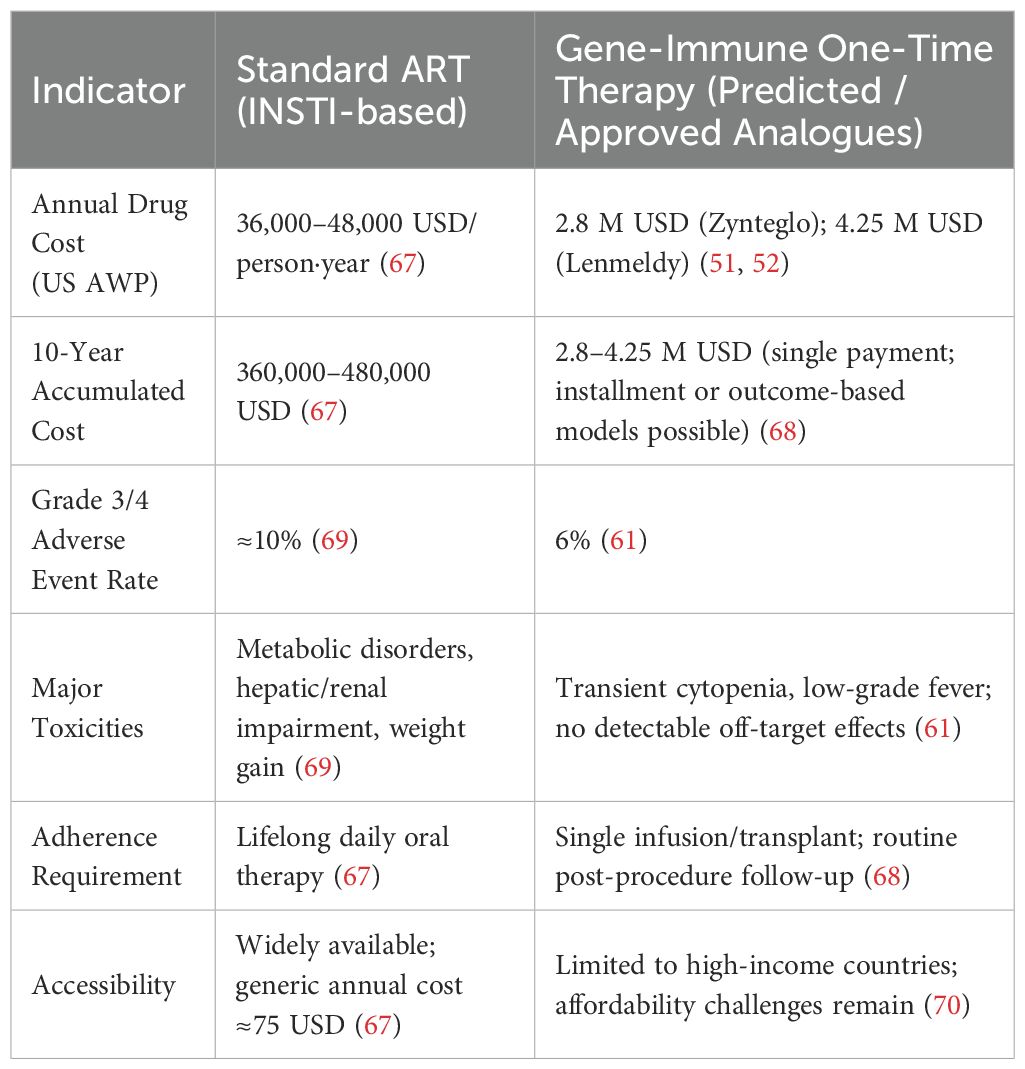
Additionally, the economic burden of gene editing is considerable. One-time gene therapies currently cost 5–10 times more than conventional treatments (see Table 4) (30). This high upfront cost and limited scalability constitute significant barriers to widespread implementation, especially in low-resource settings.
5 Discussion
Over the next five years, research will likely focus on strengthening the scientific basis for gene–immune synergistic therapies and refining preliminary treatment paradigms. Current humanized mouse models (2, 26, 37, 39, 40) and non-human primate models (7, 8, 22, 41) provide critical platforms for evaluating novel strategies. Safer and more efficient delivery systems—such as Gag-only lentiviral-like particles (LVLPs) (25) and optimized adeno-associated viral vectors (AAVs) (18)—should be explored in early-phase clinical trials using stepwise dose escalation and real-time safety monitoring to validate CCR5-targeted gene–immune combinatorial strategies, adhering to a “safety-first” principle (42).
Lessons learned from the first-in-human CRISPR-HSPC transplantation trial, which reported no serious adverse events during 19-month follow-up (43), can inform early efficacy assessments and data collection for future trials. Moreover, genetic heterogeneity among HIV patients must be considered: CXCR4-tropic viruses are associated with poorer ART response and accelerated CD4+ decline (44), while protective alleles such as HLA-B57 (45) and homozygous CCR5Δ32 (46) correlate with better viral control. Thus, stratified trials that enroll patients based on viral tropism, baseline viral load, and immune status are essential.
Promising preclinical findings—such as duoCAR-T cells eliminating over 90% of HIV-infected cells in humanized mice (47), PD-1 blockade reducing viral reservoirs and enhancing CD8+ T cell function in macaques (48), and dual-CRISPR strategies (CCR5 + LTR-Gag) achieving complete viral clearance in 39% of BLT-mice (49)—suggest that deeper investigation of combinatorial approaches (e.g., CCR5 editing + multi-specific CAR-T or CCR5 editing + PD-1 blockade) is warranted.
Patient preferences regarding the risk–benefit tradeoffs of gene therapy can be quantitatively integrated into clinical pathway design (50), supporting the development of truly individualized treatment strategies (41). To facilitate broad translation, future protocols must be optimized not only for biological efficacy but also for clinical feasibility and acceptability.
In the next decade, the high cost of gene therapy will necessitate parallel efforts to build scalable, cost-efficient manufacturing platforms. Approved therapies such as β-thalassemia gene treatments currently range from $2.8 to $4.25 million per patient (51, 52), often exceeding the annual health expenditure of many low- and middle-income countries. Current payment models rely on large, one-time upfront payments (53), underscoring the need for cost reduction.
Strategies such as serum-free suspension cultures, high-density perfusion, continuous chromatography, and optimized transfection workflows can reduce lentiviral vector production costs by up to 50% (54–56). Payment innovations—such as outcomes-based agreements and installment plans—could further improve affordability (57).
Importantly, technical breakthroughs must be accompanied by robust ethical and regulatory oversight, lifelong follow-up systems (58), and multi-stakeholder coordination to ensure sustained monitoring (3). Continued innovation in low-cost vectors, miniaturized CRISPR platforms (59), and automated manufacturing—coupled with forward-thinking reimbursement models—will be essential for overcoming current barriers and enabling widespread implementation of gene–immune strategies in HIV treatment.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author/s.
Author contributions
J-WW: Conceptualization, Data curation, Investigation, Writing – original draft. J-HL: Investigation, Writing – review & editing. J-JX: Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Alum EU, Uti DE, Ugwu OP, and Alum BN. Toward a cure - Advancing HIV/AIDs treatment modalities beyond antiretroviral therapy: A Review. Med (Baltimore). (2024) 103:e38768. doi: 10.1097/MD.0000000000038768
2. Abeynaike S and Paust S. Corrigendum: humanized mice for the evaluation of novel HIV-1 therapies. Front Immunol. (2021) 12:808068. doi: 10.3389/fimmu.2021.808068
3. Brown R, Deeks SG, and Eyal N. Maximising the global health impact of future HIV cure-related interventions through advance planning. J Virus Erad. (2018) 4:182–5. doi: 10.1016/S2055-6640(20)30266-1
4. Henderson LJ, Reoma LB, Kovacs JA, and Nath A. Advances toward curing HIV-1 infection in tissue reservoirs. J Virol. (2020) 94:e00375–00319. doi: 10.1128/JVI.00375-19
5. Li K and Zhang Q. Eliminating the HIV tissue reservoir: current strategies and challenges. Infect Dis (Lond). (2024) 56:165–82. doi: 10.1080/23744235.2023.2298450
6. Pathak MP, Pathak K, Saikia R, Gogoi U, Ahmad MZ, Patowary P, et al. Immunomodulatory effect of mushrooms and their bioactive compounds in cancer: A comprehensive review. BioMed Pharmacother. (2022) 149:112901. doi: 10.1016/j.biopha.2022.112901
7. Martinez-Navio JM, Fuchs SP, Mendes DE, Rakasz EG, Gao G, Lifson JD, et al. Long-term delivery of an anti-SIV monoclonal antibody with AAV. Front Immunol. (2020) 11:449. doi: 10.3389/fimmu.2020.00449
8. Martinez-Navio JM, Fuchs SP, Pantry SN, Lauer WA, Duggan NN, Keele BF, et al. Adeno-associated virus delivery of anti-HIV monoclonal antibodies can drive long-term virologic suppression. Immunity. (2019) 50:567–575.e5. doi: 10.1016/j.immuni.2019.02.005
9. Wise MC, Xu Z, Tello-Ruiz E, Beck C, Trautz A, Patel A, et al. In vivo delivery of synthetic DNA-encoded antibodies induces broad HIV-1-neutralizing activity. J Clin Invest. (2020) 130:827–37. doi: 10.1172/JCI132779
10. Guo Y, Luan L, Patil NK, and Sherwood ER. Immunobiology of the IL-15/IL-15Rα complex as an antitumor and antiviral agent. Cytokine Growth Factor Rev. (2017) 38:10–21. doi: 10.1016/j.cytogfr.2017.08.002
11. Jain A, Canepa GE, Liou ML, Fledderman EL, Chapoval AI, Xiao L, et al. Multiple treatment interruptions and protecting HIV-specific CD4 T cells enable durable CD8 T cell response and viral control. Front Med (Lausanne). (2024) 11:1342476. doi: 10.3389/fmed.2024.1342476
12. Xiao Q, He S, Wang C, Zhou Y, Zeng C, Liu J, et al. Deep thought on the HIV cured cases: where have we been and what lies ahead? Biomolecules. (2025) 15:378. doi: 10.3390/biom15030378
13. Pan H, Yang X, Wang J, Liang H, Jiang Z, Zhao L, et al. Allogeneic gene-edited HIV-specific CAR-T cells secreting PD-1 blocking scFv enhance specific cytotoxic activity against HIV Env(+) cells invivo. Virol Sin. (2023) 38:285–95. doi: 10.1016/j.virs.2023.01.003
14. Ernst M, Broeders M, Herrero-Hernandez P, Oussoren E, van der Ploeg AT, Pijnappel PWM, et al. Ready for repair? Gene editing enters the clinic for the treatment of human disease. Mol Ther Methods Clin Dev. (2020) 18:532–57. doi: 10.1016/j.omtm.2020.06.022
15. Schwarze LI, Sonntag T, Wild S, Schmitz S, Uhde A, Fehse B, et al. Automated production of CCR5-negative CD4(+)-T cells in a GMP-compatible, clinical scale for treatment of HIV-positive patients. Gene Ther. (2021) 28:572–87. doi: 10.1038/s41434-021-00259-5
16. Broeders M, Herrero-Hernandez P, Ernst M, van der Ploeg AT, Pijnappel PWM, et al. Sharpening the molecular scissors: advances in gene-editing technology. iScience. (2020) 23:100789. doi: 10.1016/j.isci.2019.100789
17. Davenport MP, Khoury DS, Cromer D, Lewin SR, Kelleher AD, and Kent SJ. Functional cure of HIV: the scale of the challenge. Nat Rev Immunol. (2019) 19:45–54. doi: 10.1038/s41577-018-0085-4
18. Zhan W, Muhuri M, Tai P, and Gao G. Vectored immunotherapeutics for infectious diseases: can rAAVs be the game changers for fighting transmissible pathogens? Front Immunol. (2021) 12:673699. doi: 10.3389/fimmu.2021.673699
19. Rogers GL and Cannon PM. Genome edited B cells: a new frontier in immune cell therapies. Mol Ther. (2021) 29:3192–204. doi: 10.1016/j.ymthe.2021.09.019
20. Cao S and Woodrow KA. Nanotechnology approaches to eradicating HIV reservoirs. Eur J Pharm Biopharm. (2019) 138:48–63. doi: 10.1016/j.ejpb.2018.06.002
21. Eichholz K, Fukazawa Y, Peterson CW, Mukherjee I, Balogun BM, Huaman-Vergara H, et al. Anti-PD-1 chimeric antigen receptor T cells efficiently target SIV-infected CD4+ T cells in germinal centers. J Clin Invest. (2024) 134:e169309. doi: 10.1172/JCI169309
22. Barber-Axthelm IM, Barber-Axthelm V, Sze KY, Galvin JA, Kumar PN, Bordon J, et al. Stem cell-derived CAR T cells traffic to HIV reservoirs in macaques. JCI Insight. (2021) 6:e141502. doi: 10.1172/jci.insight.141502
23. Kranz E, Kuhlmann CJ, Chan J, Kim PY, Chen ISY, Kamata M, et al. Efficient derivation of chimeric-antigen receptor-modified T(SCM) cells. Front Immunol. (2022) 13:877682. doi: 10.3389/fimmu.2022.877682
24. Mu W, Carrillo MA, and Kitchen SG. Engineering CAR T cells to target the HIV reservoir. Front Cell Infect Microbiol. (2020) 10:410. doi: 10.3389/fcimb.2020.00410
25. Jia J, Hao Y, Zhang L, Cao X, An L, Wang H, et al. Development and validation of optimized lentivirus-like particles for gene editing tool delivery with Gag-Only strategy. Eur J Med Res. (2025) 30:242. doi: 10.1186/s40001-025-02499-2
26. Li H, Lahusen T, Xiao L, Mu N, Blazkova J, Chun TW, et al. Preclinical development and clinical-scale manufacturing of HIV gag-specific, lentivirusModified CD4 T cells for HIV functional cure. Mol Ther Methods Clin Dev. (2020) 17:1048–60. doi: 10.1016/j.omtm.2020.04.024
27. Lundstrom K. Self-replicating vehicles based on negative strand RNA viruses. Cancer Gene Ther. (2023) 30:771–84. doi: 10.1038/s41417-022-00436-7
28. Sandel DA, Rutishauser RL, and Peluso MJ. Post-intervention control in HIV immunotherapy trials. Curr Opin HIV AIDS. (2025) 20:70–9. doi: 10.1097/COH.0000000000000890
29. Zaongo SD, Wang Y, Ma P, Song FZ, Chen YK, et al. Selective elimination of host cells harboring replication-competent human immunodeficiency virus reservoirs: a promising therapeutic strategy for HIV cure. Chin Med J (Engl). (2021) 134:2776–87. doi: 10.1097/CM9.0000000000001797
30. Levine B, Leskowitz R, and Davis M. Personalized gene therapy locks out HIV, paving the way to control virus without antiretroviral drugs. Expert Opin Biol Ther. (2015) 15:831–43. doi: 10.1517/14712598.2015.1035644
31. Dahiya S, Liu Y, Williams JW, Pirrone V, Nonnemacher MR, Wigdahl B, et al. Role of downstream elements in transcriptional regulation of the HIV-1 promoter[. J Hum Virol Retrovirology. (2014) 1:28–33. doi: 10.15406/JHVRV.2014.01.00006
32. Limsirichai P, Gaj T, and Schaffer DV. CRISPR-mediated Activation of Latent HIV-1 Expression.Mol Ther. 2016 24:499–507.doi: 10.1038/mt.2015.213
33. Marie V and Gordon M. The HIV-1 gag protein displays extensive functional and structural roles in virus replication and infectivity. Int J Mol Sci. (2022) 23:7569–9. doi: 10.3390/ijms23147569
34. Andreola M, Parissi V, and Litvak S. DNA polymerases: reverse transcriptase integrase, and retrovirus replication. In: Lennarz WJ and Lane MD, eds. Encyclopedia of Biological Chemistry. 2nd ed. Vol 2. San Diego, CA: Academic Press (2013), 101–7. doi: 10.1016/B978-0-12-378630-2.00258-9
35. Craigie R, Hickman AB, and Engelman A. Integrase. In: Karn J, ed. HIV: A Practical Approach. Vol 2. Oxford: Oxford University Press (1995), 53–72. doi: 10.1093/oso/9780199634996.003.0004
36. Tekeste SS, Wilkinson TA, Weiner EM, Weiner JH, Hall AM, Weber RTA, et al. Interaction between reverse transcriptase and integrase is required for reverse transcription during HIV-1 replication. J Virol. (2015) 89:12058–69. doi: 10.1128/JVI.01471-15
37. Hale M, Mesojednik T, Romano Ibarra GS, Fisher KD, Mazar J, Peyser B, et al. Engineering HIV-resistant, anti-HIV chimeric antigen receptor T cells. Mol Ther. (2017) 25:570–9. doi: 10.1016/j.ymthe.2016.12.023
38. Marcucci KT, Jadlowsky JK, Hwang WT, Weber AD, Lancaster ER, Knight EM, et al. Retroviral and lentiviral safety analysis of gene-modified T cell products and infused HIV and oncology patients. Mol Ther. (2018) 26:269–79. doi: 10.1016/j.ymthe.2017.10.012
39. Abeynaike S and Paust S. Humanized mice for the evaluation of novel HIV-1 therapies. Front Immunol. (2021) 12:636775. doi: 10.3389/fimmu.2021.636775
40. Carrillo MA, Zhen A, and Kitchen SG. The use of the humanized mouse model in gene therapy and immunotherapy for HIV and cancer. Front Immunol. (2018) 9:746. doi: 10.3389/fimmu.2018.00746
41. Protiere C, Arnold M, Fiorentino M, Raho-Moussa F, Mokdad A, and Allard A. Differences in HIV cure clinical trial preferences of French people living with HIV and physicians in the ANRS-APSEC study: a discrete choice experiment. J Int AIDS Soc. (2020) 23:e25443. doi: 10.1002/jia2.25443
42. Tebas P, Stein D, Tang WW, Frank I, Levine BL, Humeau LM, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. New Engl J Of Med. (2014) 370:901–10. doi: 10.1056/NEJMoa1300662
43. Xu L, Wang J, Liu Y, Yu L, Rui S, Qian Q, et al. CRISPR-edited stem cells in a patient with HIV and acute lymphocytic leukemia. New Engl J Of Med. (2019) 381:1240–7. doi: 10.1056/NEJMoa1817426
44. Bader J, Däumer M, Schöni-Affolter F, Loretan C, Böni J, Klimkait T, et al. Therapeutic immune recovery and reduction of CXCR4-tropic HIV-1. Clin Infect Dis. (2016) 64:295–300. doi: 10.1093/cid/ciw737
45. Sugawara S, Reeves RK, and Jost S. Learning to be elite: lessons from HIV-1 controllers and animal models on trained innate immunity and virus suppression. Front Immunol. (2022) 13:858383. doi: 10.3389/fimmu.2022.858383
46. Ni J, Wang D, and Wang S. The CCR5-delta32 genetic polymorphism and HIV-1 infection susceptibility: a meta-analysis. Open Med (Wars). (2018) 13:467–74. doi: 10.1515/med-2018-0062
47. Anthony-Gonda K, Bardhi A, Ray A, Bardhan K, Ray R, Fauci JM, et al. Multispecific anti-HIV duoCAR-T cells display broad in vitro antiviral activity and potent in vivo elimination of HIV-infected cells in a humanized mouse model. Sci Transl Med. (2019) 11:eaav5685. doi: 10.1126/scitranslmed.aav5685
48. Mylvaganam GH, Chea LS, Tharp GK, Huntsberger GE, Girouard M-P, Francella N, et al. Combination anti-PD-1 and antiretroviral therapy provides therapeutic benefit against SIV. JCI Insight. (2018) 3:e122940. doi: 10.1172/jci.insight.122940
49. Dash PK, Chen C, Kaminski R, Sariyer IK, Bock PJ, Su HX, et al. CRISPR editing of CCR5 and HIV-1 facilitates viral elimination in antiretroviral drug-suppressed virus-infected humanized mice. Proc Natl Acad Sci U.S.A. (2023) 120:e2217887120. doi: 10.1073/pnas.2217887120
50. van Overbeeke E, Hauber B, Michelsen S, van Schaik I, Janssen M, Phillips PJ, et al. Patient preferences for gene therapy in haemophilia: Results from the PAVING threshold technique survey. Haemophilia. (2021) 27:957–66. doi: 10.1111/hae.14401
51. Barnes O. Rare genetic disease therapy becomes world’s most expensive drug at $4.25mn. London, United Kingdom: Financial Times (2024).
52. Pagliarulo N. With $2.8M gene therapy, Bluebird sets new bar for US drug pricing. Washington, DC, USA: BioPharma Dive (2022).
53. Cornetta K, Bonamino M, Mahlangu J, Dotti G, High KA, Morgan RA, et al. Gene therapy access: Global challenges, opportunities, and views from Brazil, South Africa, and India. Mol Ther. (2022) 30:2122–9. doi: 10.1016/j.ymthe.2022.04.002
54. Bauler M, Roberts JK, Wu CC, Gilbert R, Manceur AP, Loignon M, et al. Production of lentiviral vectors using suspension cells grown in serum-free media. Mol Ther Methods Clin Dev. (2020) 17:58–68. doi: 10.1016/j.omtm.2019.11.011
55. Gouvarchin Ghaleh HE, Bolandian M, Dorostkar R, Etemadifar M, Pastorzadeh S, Khabiri A, et al. Concise review on optimized methods in production and transduction of lentiviral vectors in order to facilitate immunotherapy and gene therapy. BioMed Pharmacother. (2020) 128:110276. doi: 10.1016/j.biopha.2020.110276
56. Manceur AP, Loignon M, and Gilbert R. Cost-effective Biomanufacturing of Lentiviral Vectors. Erie, Pennsylvania, USA: Cell & Gene (2022).
57. Borchardt M and Cyr P. Pros and Cons of Various Reimbursement Models for Cell and Gene Therapies. Cranbury, New Jersey, USA: CGTLive (2024).
58. Organization WH. Human genome editing: recommendations. Geneva, Switzerland: World Health Organization (2021).
59. Wang L. Tiny CRISPR Tool Opens the Door to Faster, Simpler Plant Genome Editing. Berkeley, California, USA: Innovative Genomics Institute (2025).
60. DiGiusto DL, Cannon PM, Holmes MC, DeFemina RM, Kalos M, Ostertag AT, et al. Preclinical development and qualification of ZFN-mediated CCR5 disruption in human hematopoietic stem/progenitor cells. Mol Ther Methods Clin Dev. (2016) 3:16067. doi: 10.1038/mtm.2016.67
61. Tebas P, Jadlowsky JK, Shaw PA, Andrade VM, Mangan B, Tebas JR, et al. CCR5-edited CD4+ T cells augment HIV-specific immunity to enable post-rebound control of HIV replication. J Clin Invest. (2021) 131:e144486. doi: 10.1172/JCI144486
62. Xu L, Yang H, Gao Y, Chen B, Liu F, Liu J, et al. CRISPR/cas9-mediated CCR5 ablation in human hematopoietic stem/progenitor cells confers HIV-1 resistance in vivo. Mol Ther. (2017) 25:1782–9. doi: 10.1016/j.ymthe.2017.04.027
63. Claiborne DT, Detwiler Z, Docken SS, Johnston RM, Carlson JM, Permar SR, et al. High frequency CCR5 editing in human hematopoietic stem progenitor cells protects xenograft mice from HIV infection. Nat Commun. (2025) 16:446. doi: 10.1038/s41467-025-55873-3
64. Atkins A, Chung CH, Allen AG, Martinez-Murillo F, das Neves J, Patton D, et al. Off-target analysis in gene editing and applications for clinical translation of CRISPR/cas9 in HIV-1 therapy. Front Genome Ed. (2021) 3:673022. doi: 10.3389/fgeed.2021.673022
65. Lee B, Lozano RJ, and Dunbar CE. Understanding and overcoming adverse consequences of genome editing on hematopoietic stem and progenitor cells. Mol Ther. (2021) 29:3205–18. doi: 10.1016/j.ymthe.2021.09.001
66. Feist WN, Luna SE, Ben-Efraim K, Tiang A, Yang B, Jiang Y, et al. Multilayered HIV-1 resistance in HSPCs through CCR5 Knockout and B cell secretion of HIV-inhibiting antibodies. Nat Commun. (2025) 16:3103. doi: 10.1038/s41467-025-58371-8
67. McCann NC, Horn TH, Hyle EP, Lowry C, Walensky RP, Freedberg KA, et al. HIV antiretroviral therapy costs in the United States, 2012-2018. JAMA Intern Med. (2020) 180:601–3. doi: 10.1001/jamainternmed.2019.7108
68. Phares S, Trusheim M, Emond SK, Chien R, Alliance-QALY T, Pontin P, et al. Managing the Challenges of Paying for Gene Therapy: Strategies for Market Action and Policy Reform. Boston, MA, USA: Institute for Clinical and Economic Review (ICER)/NEWDIGS, Tufts Center for Biomedical System Design (2024).
69. Walmsley SL, Antela A, Clumeck N, Gathe J, Duiculescu D, Domingo P, et al. Dolutegravir plus abacavir–lamivudine for the treatment of HIV-1 infection. New Engl J Of Med. (2013) 369:1807–18. doi: 10.1056/NEJMoa1215541
Keywords: HIV, CCR5, gene editing, immunotherapy, synergistic strategy, viral reservoir, challenges, future directions
Citation: Wang J-W, Liu J-H and Xun J-J (2025) CCR5 gene editing and HIV immunotherapy: current understandings, challenges, and future directions. Front. Immunol. 16:1590690. doi: 10.3389/fimmu.2025.1590690
Received: 10 March 2025; Accepted: 02 June 2025;
Published: 18 June 2025.
Edited by:
Guido Poli, Vita-Salute San Raffaele University, ItalyReviewed by:
Daniela Cesana, San Raffaele Telethon Institute for Gene Therapy (SR-Tiget), ItalyCopyright © 2025 Wang, Liu and Xun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian-Jun Xun, NDY3MDA3MDRAaGVibXUuZWR1LmNu
 Jia-Wen Wang
Jia-Wen Wang Jian-Jun Xun
Jian-Jun Xun