- 1Gansu University of Traditional Chinese Medicine, Lanzhou, China
- 2Affiliated Hospital of Gansu University of Traditional Chinese Medicine, Lanzhou, China
Gout is an arthritis characterized by the deposition of urate crystals, often accompanied by robust inflammatory responses. The gut microbiome profoundly influences gout pathogenesis, progression, and management by affecting uric acid metabolism, immune responses, and intestinal barrier function. Studies reveal that gut microorganisms exert a dual role in gout development. Gout patients typically exhibit increased harmful bacterial abundance and reduced beneficial species. Harmful bacteria and associated metabolites can influence systemic uric acid levels by regulating excretion and synthesis, thereby promoting gout manifestations. Conversely, beneficial bacteria interact with the host immune system to inhibit inflammation and modulate the inflammatory state of joints. Furthermore, the gut microbiome can significantly impact gout treatment efficacy through its influence on drug metabolism and absorption. Research highlighting the gut-joint-inflammation axis offers novel therapeutic strategies for gout, suggesting that future approaches may involve microbiome modulation to enhance clinical outcomes.
1 Introduction
Gouty arthritis (GA), a metabolic disorder arising from dysregulated purine metabolism culminating in elevated serum uric acid (UA) concentrations, constitutes a prevalent inflammatory arthritis form, characterized by tissue and organ-damaging modifications (1). Global GA prevalence is estimated at approximately 2-4%, with higher incidence in men over 40 years of age, frequently co-occurring with comorbidities, such as obesity, coronary artery disease, hypertension, and diabetes mellitus (1). GA originates from aberrant purine metabolism or diminished UA excretion, leading to monosodium urate (MSU) crystal deposition intra- and peri-articularly. Clinical manifestations encompass inflammatory symptomatology, including erythema, edema, calor, and dolor in joint soft tissues (2). Initial onset site is typically the first metatarsophalangeal joint, but it can extend to larger joints and instigate systemic acute inflammatory sequelae (3).
The human microbiome represents a critical ecosystem for sustaining health and mediating disease development (4–8). Gastrointestinal microorganisms constitute approximately 70% of the total microbial population within this ecosystem (9). The human gut microbiome is predominantly composed of Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria phyla (10), participating in diverse physiological processes, such as nutrient metabolism, xenobiotic detoxification, immune modulation, and intestinal barrier integrity maintenance (11). Investigations indicate that the gut microbiota influences various autoimmune disease pathogenesis by modulating the host immune system (12). Upon gut microbiota dysbiosis, detrimental bacterial taxa proliferate excessively, and elaborated metabolites, such as polyphenols, vitamins, and tryptophan, may trigger GA onset (13). Furthermore, gut microorganism and metabolite alterations not only impact GA therapeutic efficacy but may also affect pharmacological agent toxic side effects. In-depth exploration of gut microorganism and GA interplay is of paramount significance for elucidating disease mechanisms, enhancing diagnostic precision, and optimizing therapeutic strategies.
2 The association of gut microbiome with GA
The gastrointestinal tract, the most microbial-rich human habitat, encompasses a 250–400 square meter surface area and harbors approximately 1014 microorganisms (14). Given that roughly 30% of UA is excreted via the gastrointestinal tract, the gut microbiome-GA relationship is receiving escalating attention. Gut microbiota dysbiosis is implicated in various diseases (15), encompassing hyperuricemia and GA. Gut microbes participate in purine and UA metabolism via diverse pathways: Escherichia coli and Proteus spp. secrete xanthine dehydrogenase, fostering purine conversion to UA (16, 17); Lactobacillus spp. inhibit intestinal purine absorption, preventing serum UA level elevations; Pseudomonas spp. synthesize uricase, participating in UA catabolism (18); a multitude of gut microorganisms secrete UA transporters, influencing UA excretion (19).
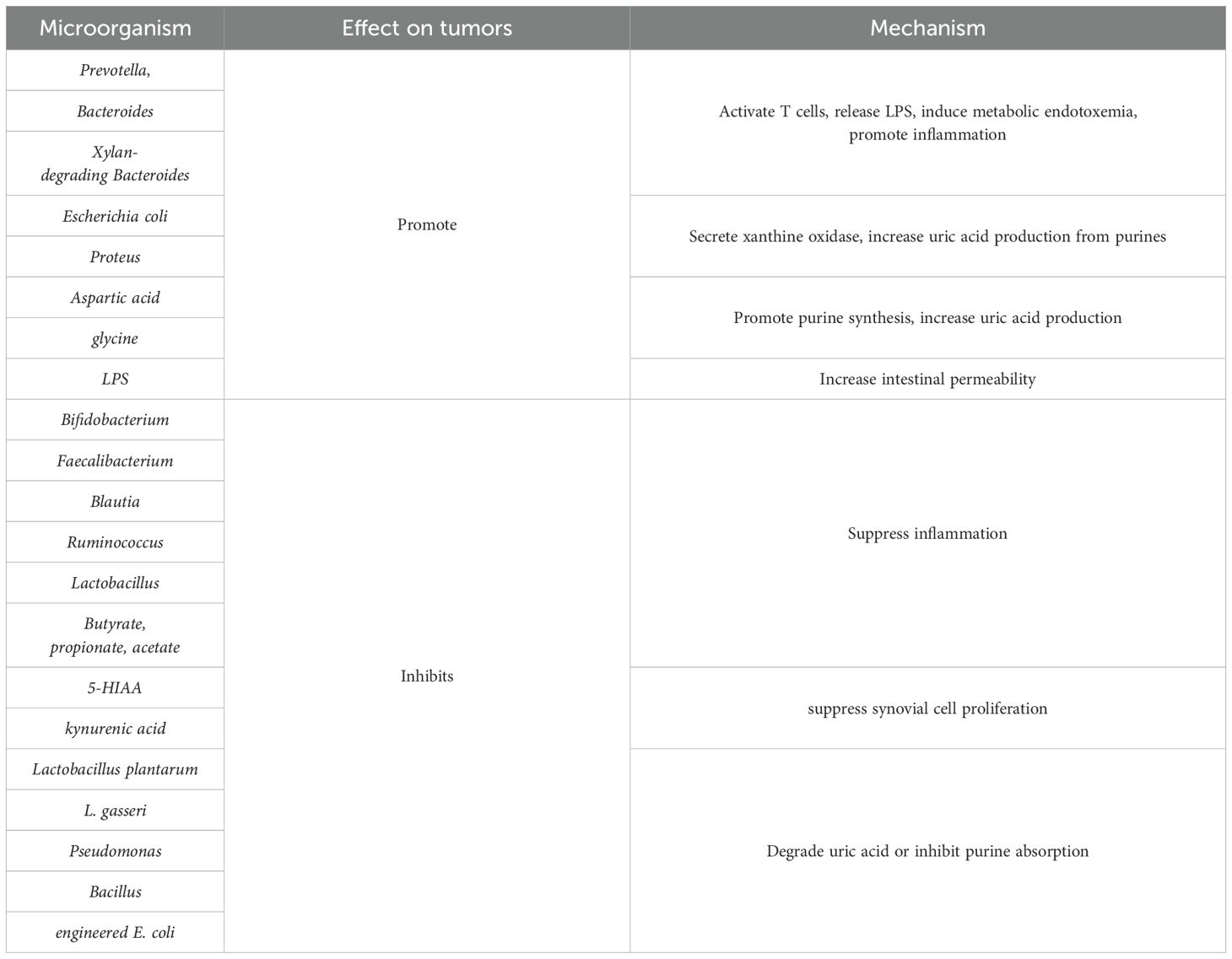
The gut microbiome in gout patients exhibits characteristic alterations: augmented Prevotella, Clostridium, Bacteroides, Megamonas, and Xylanibacter abundance, and diminished Enterobacteriaceae, butyrate-producing bacteria, Coprococcus, and Bifidobacteria abundance (20). Functional genomic analysis reveals increased fructose, mannose metabolism, and lipid A biosynthesis gene abundance in gout patients, and reduced UA degradation and short-chain fatty acids (SCFAs) production gene abundance (20). Diminished Enterobacteriaceae species abundance correlates with reduced amino acid metabolism and environmental sensing, collectively contributing to elevated serum UA and C-reactive protein concentrations (21). Liu et al., utilizing stable isotope tracing technology in a human intestinal bacterial library, identified 46 UA-degrading bacterial taxa, belonging to Actinobacteria, Firmicutes, Clostridia, and Proteobacteria phyla (22). These strains metabolize UA into xanthine or SCFAs (22). Transcriptomic analysis unveiled a highly conserved gene cluster (ygeX, ygeY, ygeW, ygfK, and ssnA) encoding pivotal UA degradation enzymes (23). Uricase-deficient murine model studies have confirmed that gut microbiota depletion exacerbates hyperuricemia, whereas UA-degrading bacteria colonization attenuates UA levels. Recent research indicates Phascolarctobacterium and Bacteroides enrichment in gout patients, forming a distinctive core microbiome encompassing three Bacteroides genera (24). Researchers have subsequently developed a 17 gout-associated bacteria diagnostic model based on these characteristics, providing novel biomarkers for gout early diagnosis and prognostic assessment (25). Table 1 shows the correlation between intestinal microorganisms and metabolites and GA.
3 Mechanisms of gut microbiota and metabolite promotion of GA development
The gut microbiota participates in GA pathogenesis via multiple regulatory mechanisms (26, 27). The abundance of specific bacteria, including Prevotella, Bacteroides fragilis, and Xylan-degrading Bacteroides, is significantly increased in patients with gout. This dysbiosis, marked by an expansion of pathogenic microbes, contributes to the development of GA through its impact on gut barrier integrity, metabolic regulation, and immune system function. Figure 1 schematically illustrates gut microbiota and metabolite promotion of gout development. In innate immunity, gut microbiota dysbiosis leads to aberrant pattern recognition receptor activation, pro-inflammatory mediator expression upregulation, and anti-inflammatory mediator level downregulation, disrupting local immune homeostasis (28). At the adaptive immunity level, dysbiotic microbiota mediates autoimmune responses by modulating antigen-presenting cell function, T cell subset differentiation, and B cell activation. Microbiota dysbiosis-elicited inflammatory responses impair tight junctions between intestinal epithelial cells, augmenting intestinal permeability. Increased permeability facilitates microorganism and metabolite entry, and antigenic components, into the circulatory system, triggering systemic immune responses. These mechanisms establish a positive feedback loop: immune dysfunction exacerbates barrier functional impairment, and barrier functional damage, in turn, potentiates immunostimulatory molecule release, further amplifying immune abnormalities and leading to sustained GA progression.
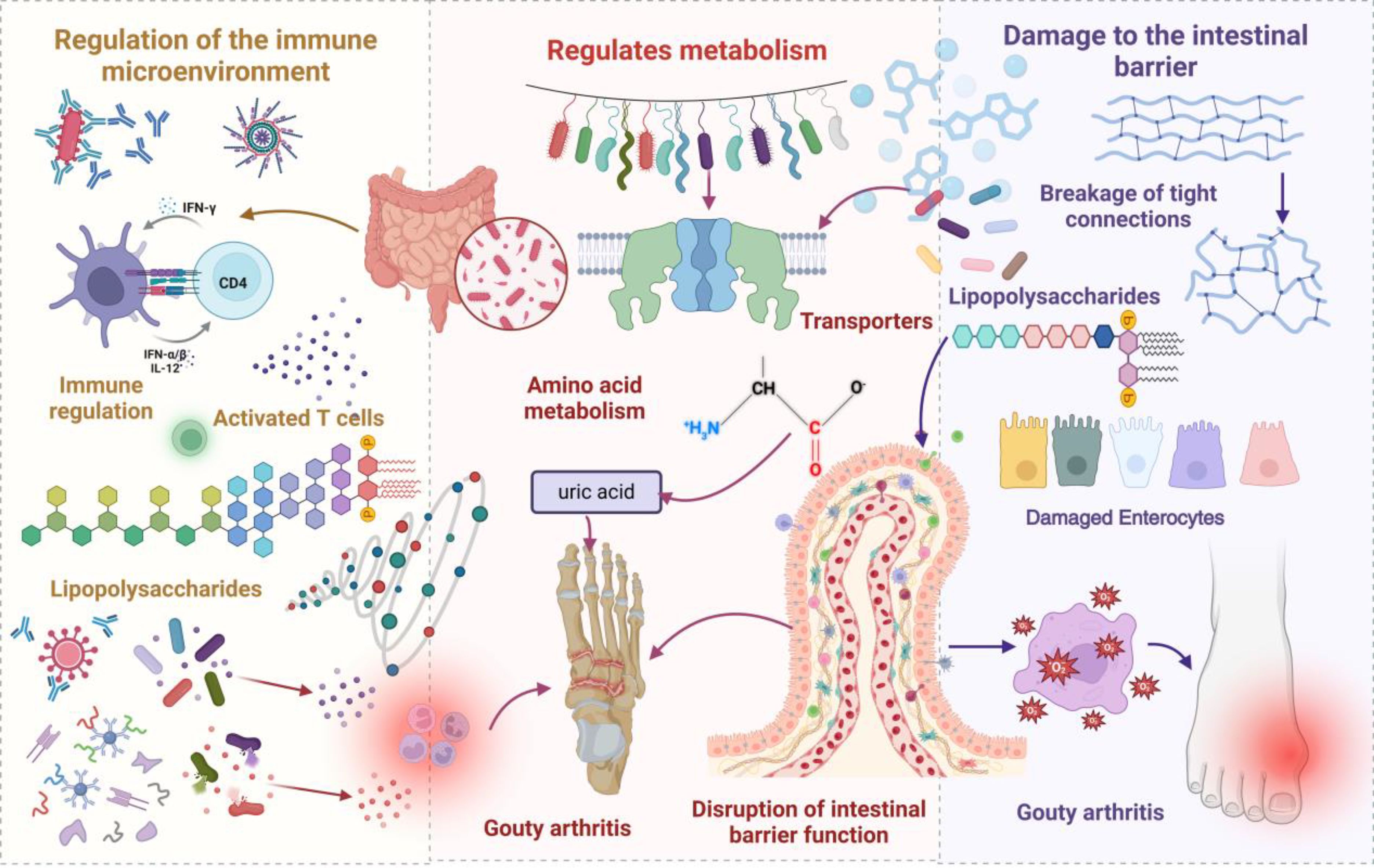
Figure 1. A schematic representation illustrating how gut microbiota and its metabolites contribute to the initiation and progression of gout. In the context of dysbiosis, an increase in pathogenic bacteria activates T cells, leading to the secretion of lipopolysaccharides (LPS), which triggers metabolic endotoxemia and induces systemic inflammation, thereby exacerbating gout flare-ups. Moreover, dysbiosis alters the expression of urate transporters, impeding uric acid excretion and elevating serum uric acid levels. Compromised barrier integrity facilitates the translocation of bacteria and their metabolites into the bloodstream, further amplifying inflammation and promoting the deposition of urate crystals, which increases the risk of GA.
3.1 Immunomicroenvironmental regulation
The gut microbiota orchestrates GA progression via the immune network. At the innate immunity level, gut-associated lymphoid tissue immune cells engage in signaling crosstalk with the microbiota, establishing an immune defense barrier (29). Gut microbiota dysbiosis precipitates aberrant innate immune cell activation, pro-inflammatory mediator upregulation, such as Interleukin-6, tumor necrosis factor-alpha, Interleukin-1β, Interleukin-12, and Interleukin-23, and anti-inflammatory mediator level downregulation, such as transforming growth factor-beta and Interleukin-10 (27, 30). In adaptive immune modulation, microbial antigens are recognized by dendritic cells and macrophages and presented to CD4+ T cells, inducing their differentiation (31, 32). Gut microbiota imbalance can augment intestinal permeability, leading to LPS translocation into the circulatory system, precipitating metabolic endotoxemia and inflammation (33). Elevated xanthine oxidase (XOD) activity is associated with increased serum LPS concentrations and chronic inflammation (34). Uox-KO mice exhibit significantly elevated inflammatory cytokine, LPS, and XOD activity levels (35). Compared with normouricemic mice, hyperuricemic murine models demonstrate diminished Bifidobacterium and Lactobacillus abundance in fecal samples, accompanied by elevated UA, XOD activity, and LPS levels (36), thereby accelerating GA pathogenesis and progression.
3.2 Metabolic Modulation
The gut microbiota and its metabolites play a crucial role in gout pathogenesis and progression by modulating UA metabolism. UA transporters in intestinal epithelial cells transport UA from the bloodstream to the intestinal lumen (37). The microbiota influences UA metabolism by modulating UA transporter expression, such as ABCG2 and SLC2A9 (38, 39). Purine, a precursor of uric acid, can accumulate during its metabolic processes, leading to excessive uric acid production. Escherichia coli and Proteus species are capable of secreting xanthine dehydrogenase, which catalyzes the conversion of purines to uric acid, thereby directly enhancing the host’s capacity for uric acid synthesis. This process further aggravates hyperuricemia and the development of GA. Amino acid metabolism plays a significant role in gout development. Amino acids, such as aspartic acid and glycine, participate in purine biosynthesis, augmenting UA production. Aberrant amino acid metabolism can diminish UA excretion. Furthermore, amino acid metabolites promote UA crystal deposition and exacerbate gout symptomatology by modulating immune and inflammatory responses. Gut microbiota dysbiosis may also accelerate gout onset by altering amino acid metabolism (38, 40). The gut microbiota promotes gout pathogenesis by influencing amino acid metabolism. The high-gout cluster microbiota manifests increased D/L-alanine and branched-chain amino acid metabolism, modulating UA biosynthesis (41). The abnormal accumulation of these metabolites may regulate uric acid production and levels through modulation of the purine synthesis pathway or by indirectly affecting uric acid excretion. Furthermore, amino acid metabolites may contribute to the deposition of urate crystals by modulating immune responses and inflammatory pathways, thereby aggravating the clinical manifestations of gout.
3.3 Intestinal barrier impairment
The intestinal barrier constitutes a multi-layered defense system, composed of the gut microbiota, mucus layer, epithelial cell monolayer, and lamina propria immune cells (42). Within the epithelial barrier, intercellular tight junction proteins play a pivotal role in maintaining barrier integrity by regulating transepithelial permeability and cellular mechanical junctions (43, 44). Increased intestinal permeability, resulting from diminished epithelial tight junction protein occludin and claudin-1 expression, exhibits a positive correlation with serum UA levels (45) (46) (47). Intestinal barrier impairment precipitates gut microbiota dysbiosis. Metabolites elaborated from gut microbiota dysbiosis, such as hydrogen sulfide, reactive oxygen species, and reactive nitrogen species, exert direct damage to intestinal epithelial cell structure and function (48), augmenting intestinal permeability, subsequently leading to bacterial translocation and increasing inflammation and gout incidence. LPS constitutes the principal Gram-negative bacteria cell wall component. Gut micro-ecological imbalance can markedly inhibit Gram-negative bacteria physiological activity, leading to increased LPS elaboration. Excessive LPS can induce pro-inflammatory cytokine production and augment intestinal barrier permeability, precipitating metabolic endotoxemia (49). In gout pathogenesis, gut microbiota dysbiosis promotes LPS translocation into the bloodstream by impairing intestinal barrier function, leading to metabolic endotoxemia. This process not only escalates the systemic inflammatory burden but can also exacerbate UA accumulation and gout onset by affecting renal function (50).
4 Gut microbiota and metabolite mechanisms inhibiting GA development
Gut microbiota dysbiosis exerts a dual regulatory role in GA pathological progression. Beneficial bacterial communities play a protective role by modulating immune responses and maintaining intestinal barrier integrity. Investigations have revealed significantly reduced Faecalibacterium and Bifidobacterium catenulatum abundance in gout patients (21). Metagenomic analysis has corroborated decreased Enterobacteriaceae bacteria and butyrate-producing bacteria numbers within gout patient intestines (38). In a hyperuricemic nephropathy rat model, beneficial bacterial taxa, such as SCFA-producing Blautia and Ruminococcus genera, were markedly diminished. Figure 2 schematically illustrates gut microbiota and metabolite inhibition of gout development.
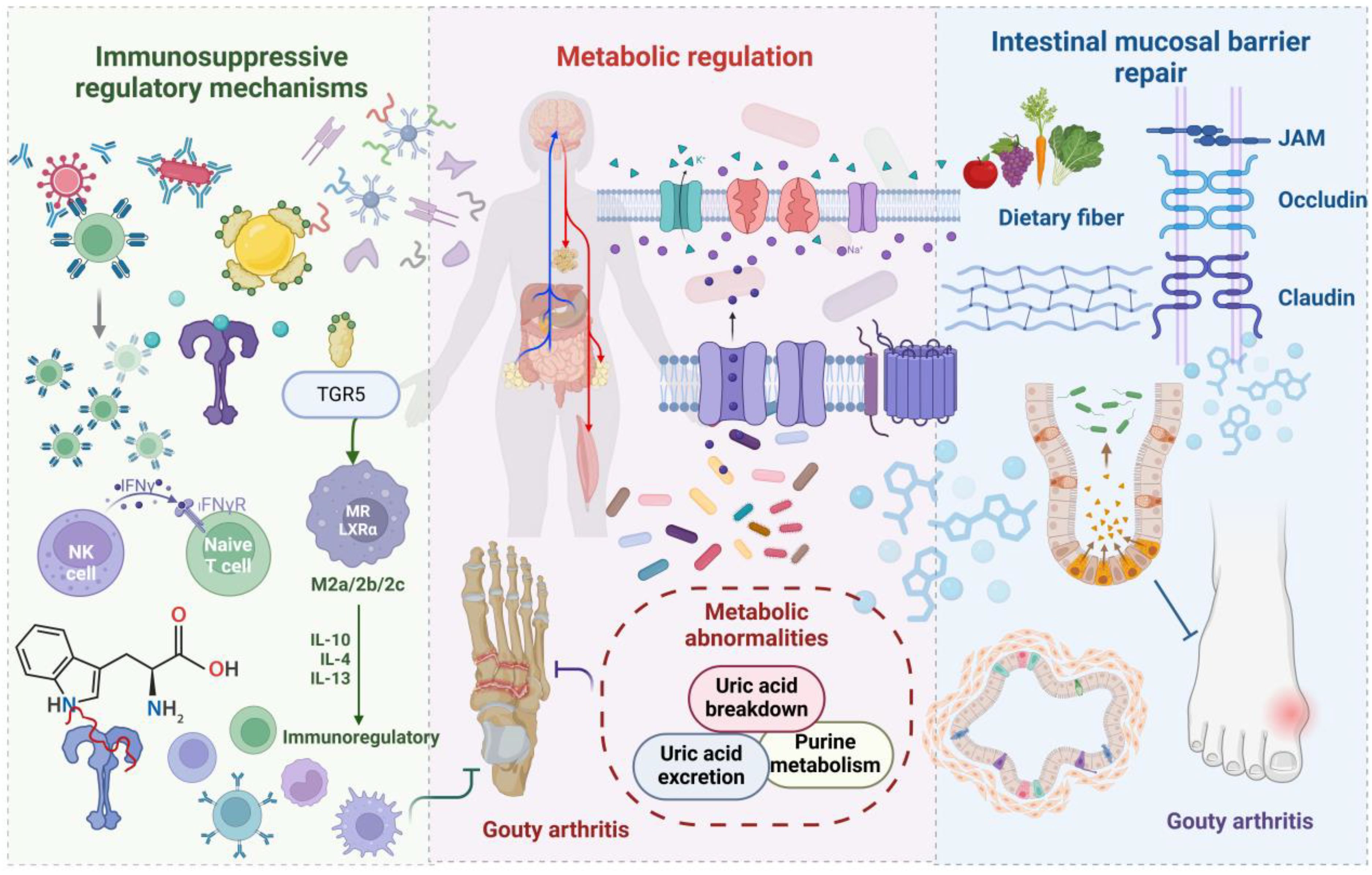
Figure 2. A schematic representation illustrating how gut microbiota and its metabolites suppress the onset and progression of gout. Beneficial bacteria and their metabolites mitigate inflammation through immune modulation mediated by TGR5, while uricase reduces systemic uric acid accumulation. Nutritional factors, such as dietary fiber, enhance microbiota composition, preserve gut barrier integrity, and decrease the permeability to harmful substances. These mechanisms work in concert to inhibit the development and recurrence of GA.
4.1 Immunosuppressive regulatory mechanisms
Gut microbiota and their metabolites inhibit GA pathogenesis and progression by modulating immune cell function and inflammatory cytokine expression. The NLR family pyrin domain Containing 3 (NLRP3) inflammasome, functioning as a metabolic stress sensor, participates in gout development (51–53). Lactobacillus spp. inhibit NLRP3 activation by restoring mitochondrial membrane potential, and Bifidobacterium longum and Bacteroides fragilis suppress NLRP3 activation via inflammatory signaling pathway inhibition (54–56). Lactobacillus casei elicits anti-inflammatory responses by interleukin-10 expression upregulation and Interleukin-6 and tumor necrosis factor-α level downregulation. Lactobacillus rhamnosus mediates immunosuppression by modulating Interleukin-1 expression. Phascolarctobacterium praecalvibacter reduces pro-inflammatory mediator concentrations, Interleukin-17, Interleukin-1β, and tumor necrosis factor-α, while concurrently promoting symbiotic microbiota proliferation, such as Akkermansia and Bifidobacteria (57, 58). Microbial metabolites play a crucial role in immunomodulation. Butyrate, a SCFA, promotes follicular regulatory T cell differentiation (59). Parabacteroides distasonis-produced bile acid metabolites induce macrophage M2 polarization and inhibit Th17 cell differentiation via TGR5 receptors (60). The tryptophan metabolic network plays a salient role in GA immunomodulation: 5-hydroxyindoleacetic acid induces regulatory T cell differentiation via aromatic hydrocarbon receptors (61); kynurequinolic acid inhibits synovial cell proliferation (62); 3-hydroxyanthranilic acid suppresses inflammatory responses by blocking the NF-κB signaling pathway (63). Blautia- and Roseburia-elaborated butyrate attenuates GA flares by inhibiting MSU crystal-induced Interleukin-1β, Interleukin-6, and Interleukin-8 production.
4.2 Metabolic regulation
Gut microorganisms regulate UA metabolism via multiple enzymatic systems, and certain beneficial bacterial taxa exhibit UA oxidase and xanthine dehydrogenase inhibitory activities. Lactobacillus and Pseudomonas spp. promote UA catabolism and excretion by producing SCFAs (64). The Lactobacillus genus catabolizes inosine and guanosine to inhibit UA biosynthesis (65–67). Lactobacillus OL-5, Lactobacillus plantarum Mut-7, and Lactobacillus plantarum Dad-13 exhibit elevated uricase activity and can degrade UA via uricase, thereby reducing in vivo UA accumulation (37). Lactobacillus gallinarum reduces intestinal purine concentrations (68). Lactobacillus gasseri diminishes purine absorption (18). Propionate and butyrate promote UA excretion by supplying adenosine triphosphate (69, 70). Bacillus, Proteus mirabilis, Escherichia coli, and various other microorganisms can degrade UA via uricase, thereby reducing in vivo UA accumulation (71) (72), consequently inhibiting GA development.
4.3 Intestinal mucosal barrier restoration
Aberrant intestinal barrier function is intimately associated with autoimmune disorders. Nutritional factors participate in barrier repair by modulating microbial composition and metabolic pathways. Dietary fiber maintains barrier integrity by reducing serum Zonulin and calprotectin levels (73) (74). Vitamin D modulates epithelial cell tight junctions and apoptosis (75). Vitamin E promotes butyrate-producing bacteria proliferation (76). Glutamine and tryptophan deficiency result in barrier function impairment (77, 78). Plant polyphenols confer barrier function protection by enhancing transepithelial electrical resistance and ZO-1 and claudin-1 expression upregulation (79). Butyrate repairs intestinal epithelial cells and stabilizes the epithelial mucosal barrier (72). Acetate provides energy substrates for intestinal epithelial cells and promotes UA transport (72). Bifidobacteria ameliorate mucosal barrier function by inhibiting detrimental strain proliferation (80), and SCFAs mediate mucosal barrier repair (81), thereby inhibiting GA progression.
5 Translational gut microbiome applications in GA diagnosis and therapeutics
Gut microbiota composition and diversity alterations participate in GA pathogenesis and progression via immune system modulation (82). Gut microbiome-based intervention strategies have demonstrated preliminary therapeutic efficacy in GA clinical investigations. Animal studies have substantiated that probiotic supplementation and intestinal barrier stability maintenance, among other measures, possess potential therapeutic value for GA (83), suggesting that precision microbiome composition modulation may facilitate novel diagnostic biomarker and personalized therapeutic regimen development.
5.1 Diagnostic biomarkers
Predicated on the gut microbiome-gout development correlation, gout-specific microbiota may serve as potential diagnostic biomarkers. Lin et al., based on bacterial genera exhibiting significant disparities between healthy individuals and gout patients, constructed a classification model with an area under the receiver operating characteristic curve reaching 0.973 (84). Another cohort study established a 17 gout-associated bacteria diagnostic model, achieving 88.9% accuracy (20). Chu et al., via metagenomic analysis, identified three genes significantly enriched in the gout cohort, with development and validation cohort area under the receiver operating characteristic curve of 0.91 and 0.80, respectively (21). Gout-characteristic gut microbiota imbalance may serve as a non-invasive diagnostic tool for gout and asymptomatic hyperuricemia, providing novel prevention and intervention targets. The significance of gut microbiota characteristics extends beyond current diagnostic applications, with dynamic changes also being explored as potential prognostic indicators for disease progression and treatment response. Monitoring shifts in the gut microbiota structure and function in gout patients undergoing urate-lowering therapy or probiotic interventions can provide valuable insights into treatment efficacy, while also offering the potential to predict disease progression and relapse risk. To more deeply investigate the complex interplay between gut microbiota and the immune-pathological processes of gout, advanced technologies are increasingly being applied to identify relevant biomarkers (85). Spatial transcriptomics and spatial proteomics offer the ability to perform high-resolution analyses of local microenvironments—such as synovial and renal tissues—while preserving spatial context, allowing for precise mapping of microbial components, immune cell populations, inflammatory mediators, and urate crystal distribution, along with their interactions. By correlating this localized microenvironmental data with global gut microbiota profiles obtained via high-throughput sequencing, we can uncover how distal gut microbial signals impact local joint inflammation or renal damage, thereby identifying spatial biomarkers more tightly associated with gout onset, progression, and prognosis (86). These integrative approaches offer novel insights into how gut microbiota modulate host immunity and regulate local microenvironments in the context of gout, paving the way for the discovery of mechanistically targeted biomarkers with potential clinical diagnostic, prognostic, and predictive applications.
5.2 Gut Microbiota and Metabolite-Based GA Therapies
Non-steroidal anti-inflammatory drugs, glucocorticoids, and colchicine constitute first-line pharmacological agents for acute gout management (87). XOD inhibitors and uricosuric agents serve as first- and second-line choices, respectively, for UA-lowering therapy (88). Gut microbiota dysbiosis can impact therapeutic outcomes (89): GA patient gut microbiota composition undergoes post-treatment alterations, promoting SCFA and acetate production (90).
5.2.1 Post-traditional pharmacotherapy gut microecology alterations
Non-steroidal anti-inflammatory drugs can disrupt gut microbiota equilibrium, fostering Gram-negative bacteria proliferation and inhibiting Gram-positive bacteria growth. Microbiota dysbiosis activates inflammatory responses via the TLR4 pathway and augments intestinal permeability (91, 92). Colchicine blocks microtubule protein polymerization, precluding inflammasome activation (93). However, colchicine demonstrably impacts gastrointestinal architecture, altering gut microbiota diversity and composition, resulting in pro-inflammatory mediator downregulation and intestinal barrier impairment (94). Allopurinol therapy can augment Bifidobacteria abundance and reduce anaerobic bacteria numbers (89). Bilophila genera, as the sole reducing genus, can induce systemic inflammation (95). Benzbromarone reduces UA concentrations by blocking URAT-1 (96), while concurrently modifying gut microbiota composition, increasing Bifidobacteria and reducing butyrate-producing bacteria (89). Febuxostat inhibits XOD activity and can partially restore gut microbiota diversity in gout patients (66). Functional analysis reveals enhanced purine metabolism potential of gut microorganisms in post-treatment patients (84), and micro-inflammation suppression (97).
5.2.2 Microbiome-mediated therapeutic efficacy modulation mechanisms
Novel therapeutic modalities, such as natural products, probiotics, and fecal microbiota transplantation (FMT), regulate gout pathogenesis and progression via multiple mechanisms (98–100). Figure 3 schematically illustrates microbiome-mediated therapeutic approaches. These modalities inhibit purine metabolism and inflammasome activation, regulate transporter protein expression, and maintain intestinal barrier integrity. Concurrently, they can augment SCFA-producing bacteria abundance, thereby inhibiting XOD activity and achieving UA-lowering effects (101). These interventions provide novel paradigms and potential targets for gout therapy.
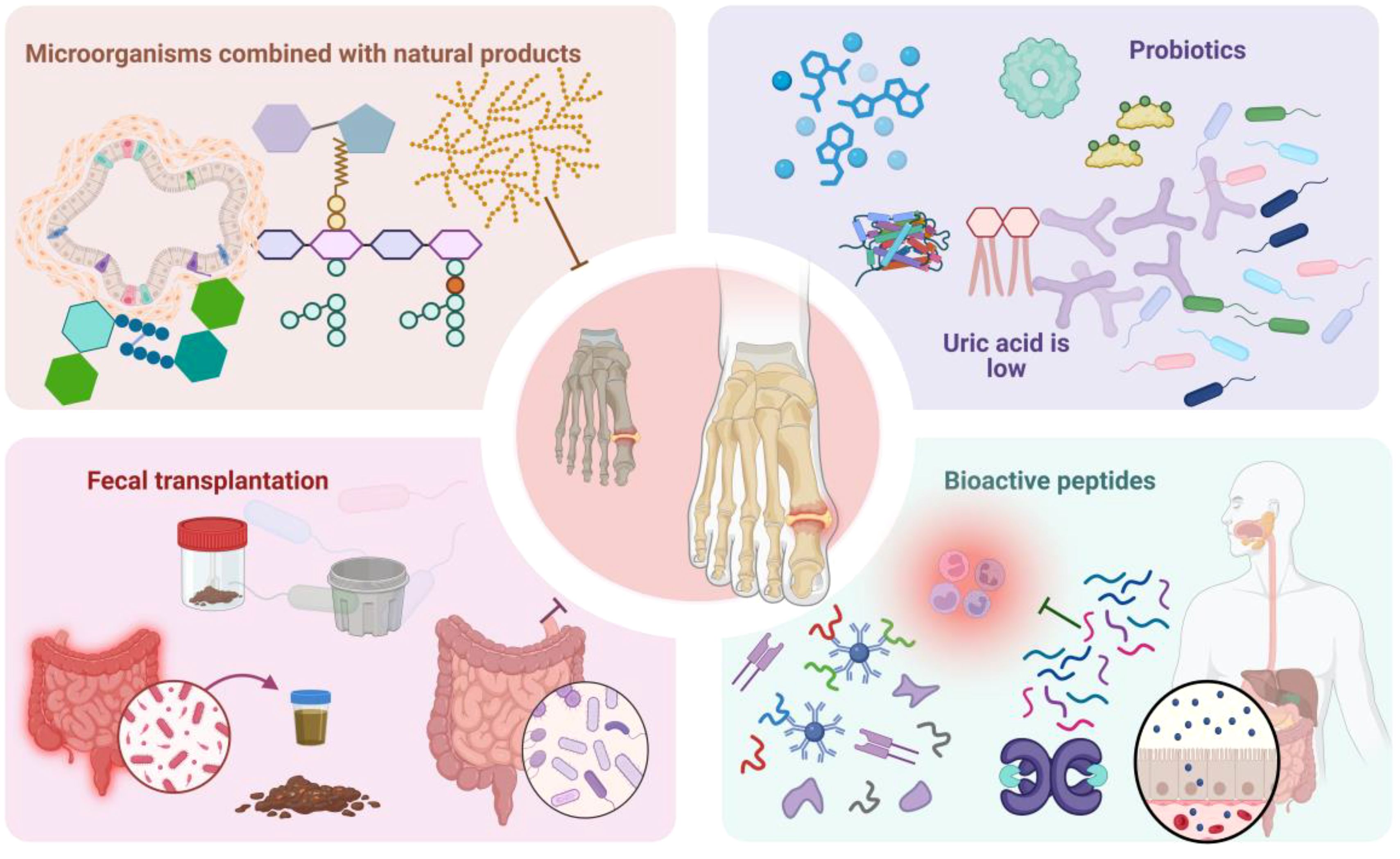
Figure 3. Schematically illustrates microbiome-mediated therapeutic approaches. Microbiome-mediated therapies encompass probiotic modulation, microbial combination with natural products, FMT, bioactive peptides, and other modalities. Among these, probiotics regulate gut microbiota structure and function to ameliorate gout symptomatology. Microbial combination with natural products, FMT, bioactive peptides, and other therapeutic interventions can effectively reduce serum UA levels, attenuate inflammatory responses, and inhibit GA pathogenesis and progression, providing novel paradigms for GA treatment.
(1) Probiotics
Probiotics, such as Bifidobacteria and Lactobacillus genera, ameliorate gout symptomatology by modulating gut microbiota structure and function (102). Novel probiotics, including Faecalibacterium prausnitzii, Akkermansia muciniphila, and Clostridium spp., also demonstrate therapeutic potential (103). Lactobacillus fermentum JL-3 can rectify gut microbiota dysbiosis associated with hyperuricemia (99). Probiotic strain DM9218 diminishes serum UA concentrations and hepatic XOD activity (104). Uricase-elaborating bacteria-containing probiotics can improve hyperuricemia and confer renal function protection (105). Prebiotics, functioning as selective substrates for host microorganisms (106), can augment Lactobacillus and Bifidobacteria abundance and foster butyrate and propionate production (107), providing novel therapeutic avenues for gout. In a diet-induced hyperuricemic murine model, EH-JAP and EH-LEU improved hyperuricemia and GA by inhibiting UA biosynthesis and promoting UA excretion. Concurrently, these two therapeutic regimens improved gut microbiota functionality by increasing beneficial Lactobacillus and SCFA-producing bacteria abundance and reducing opportunistic pathogen numbers (37).
(2)Microbial combination with natural products
Microbial combination with natural products can effectively diminish serum UA levels and inhibit GA pathogenesis and progression. Luteolin attenuates serum UA concentrations in GA mice by modulating gut microbiota composition, reversing Bacteroidetes and Firmicutes phyla dysbiosis. Investigations suggest that luteolin also ameliorates renal function in GA-induced chronic kidney disease mice by modulating gut microbiota-mediated tryptophan metabolism (108). Metabolomic analysis indicates that nuciferine can inhibit hyperuricemia development by modulating impaired metabolic pathways and gut microbiota composition (109). Ulva pertusa polysaccharide, the most prevalent nuciferine family green algae extract, not only reduces serum UA levels but also significantly enhances gut microbiota diversity, particularly increasing Alistipes and Parasutterella genera abundance. Correlation analysis reveals Parasutterella content exhibiting a negative correlation with UA levels (110). Therefore, microbial combination with natural products offers novel gout therapy strategies.
(3) Fecal microbiota transplantation
FMT can reduce serum UA concentrations in gout patients and diminish acute gout flare frequency and duration (37). Simultaneously, FMT can reduce diamine oxidase and endotoxin levels and ameliorate impaired intestinal barrier function (100). Animal experiments further substantiate FMT potential value in hyperuricemia therapy. Furthermore, FMT effectively alleviates hyperuricemia in mice by selectively modulating AJOP-related metabolic pathways, suggesting AJOP protective effect partially contingent upon microbiota modulation (111). Goose essence-treated mice FMT also demonstrates significant anti-hyperuricemic effects, with mechanisms encompassing gut microbiota equilibrium restoration, intestinal epithelial barrier repair, and SCFA production promotion (112). Although FMT long-term human health effects necessitate further clinical validation, it offers a novel gout treatment option and possesses significant clinical application prospects.
(4)Bioactive peptides
Bioactive peptides constitute a key research domain in food, health products, and specialized medical foods. These polypeptides are not only elaborated from intestinal microbial proteases acting on dietary proteins but also significantly modulate gut microbiota structure, consequently impacting host health. Tuna oligopeptides ameliorate hyperuricemia and GA by reprogramming UA metabolic pathways, inhibiting NLRP3 inflammasome and TLR4/myeloid differentiation primary response 88/nuclear factor-kappa beta signaling pathway activation, and p65-nuclear factor-kappa beta phosphorylation. Concurrently, tuna oligopeptides repair the intestinal epithelial barrier, rectify gut microbiota dysbiosis, and promote SCFA production (113). Investigations reveal that hexapeptides GPAGPR and GPSGRP, as potential microbiota modulators, reduce serum UA levels by inhibiting UA biosynthesis and reabsorption, and attenuate renal inflammation by inhibiting NLRP3 inflammasome activation. These two peptides also diminish gut microbiota richness and diversity, altering phylum and genus level microbiota composition (114). Sea cucumber oligopeptides significantly alleviate hyperuricemia, with action mechanisms encompassing UA metabolism modulation, NLRP3 inflammasome and nuclear factor-kappa beta-related signaling pathway activation inhibition, and m6A methylation level restoration (111).
6 Conclusion
GA is a complex autoimmune disorder with multifactorial pathogenesis, wherein gut micro-ecological imbalance plays a pivotal role in disease progression. GA patients exhibit characteristic alterations, including detrimental bacteria increase and beneficial bacteria decrease. Gut microbiota and metabolites participate in GA regulation through multiple pathways: detrimental bacteria promote GA pathogenesis and progression by fostering inflammation and disrupting the intestinal barrier, whereas beneficial bacteria inhibit GA pathogenesis and progression via inflammation suppression and intestinal barrier repair. Gut microbiota and metabolites possess potential applicability in GA diagnosis and therapy. Microbiota composition analysis can predict GA susceptibility and has emerged as an efficacious methodology for morbidity prediction and control. Furthermore, gut microbiota and enzymatic products can directly or indirectly influence pharmaceutical agent bioavailability, clinical efficacy, and toxicity, while therapeutic drugs and active compounds can also modulate immune cell function by normalizing gut microbiota composition.
While therapeutic strategies targeting the gut microbiome have demonstrated potential for modulating the immune system, current research faces several limitations. The molecular mechanisms by which specific microbial communities or metabolites regulate host immune responses and drive the pathogenesis of GA remain elusive. Precise mechanisms linking gut microbiota dysbiosis and impaired intestinal barrier function to the induction of systemic inflammatory responses also warrant further investigation. Moreover, applying microbiota modulation strategies for GA therapy is challenging, primarily due to significant inter-individual variations in gut microbial composition, immune responses, and therapeutic efficacy, which can substantially impact intervention outcomes. However, leveraging continuous advancements in genomics, microbiology, and immunology, coupled with multidisciplinary integrated analyses, the development of personalized microbiome-based interventions holds promise for enhancing the precision and effectiveness of GA therapy. Future efforts must prioritize exploring the intricate microbiota-immune interactions to facilitate the development of truly personalized therapies. Rigorous systematic clinical studies and robust experimental data are essential to fully realize the potential of microbiota modulation as a novel therapeutic strategy for GA.
Author contributions
PQ: Methodology, Writing – original draft, Writing – review & editing. LL: Writing – review & editing. JZ: Writing – review & editing. LR: Writing – review & editing. XX: Conceptualization, Funding acquisition, Methodology, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was supported by the National Nature Fund Regional Project (82160911, 81860864); Central University Basic Scientific Research Business Expenses Project (31920210041); The special project of science and technology development under the guidance of the central government (YDZX20206200002356).
Acknowledgments
Acknowledgments The authors acknowledge the use of Biorender to create schematic representations in Figure 1, Figure 2 and Figure 3.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
GA, Gouty arthritis; SCFAs, short-chain fatty acids; FMT, fecal microbiota transplantation; XOD, Xanthine oxidase; LPS, Lipopolysaccharide; UA, Uric Acid; MSU, Monosodium Urate; NLRP3, NLR family pyrin domain Containing 3.
References
1. Dalbeth N, Gosling AL, Gaffo A, and Abhishek A. Gout. Lancet Lond Engl. (2021) 397:1843–55. doi: 10.1016/S0140-6736(21)00569-9
2. Punzi L, Scanu A, Galozzi P, Luisetto R, Spinella P, Scirè CA, et al. One year in review 2020: gout. Clin Exp Rheumatol. (2020) 38:807–21.
3. Allam-Ndoul B, Castonguay-Paradis S, and Veilleux A. Gut microbiota and intestinal trans-epithelial permeability. Int J Mol Sci. (2020) 21:6402. doi: 10.3390/ijms21176402
4. Almeida A, Mitchell AL, Boland M, Forster SC, Gloor GB, Tarkowska A, et al. A new genomic blueprint of the human gut microbiota. Nature. (2019) 568:499–504.
5. Morrison DJ and Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. (2016) 7:189–200. doi: 10.1080/19490976.2015.1134082
7. Gomaa EZ. Human gut microbiota/microbiome in health and diseases: a review. Antonie Van Leeuwenhoek. (2020) 113:2019–40.
8. Durack J and Lynch SV. The gut microbiome: Relationships with disease and opportunities for therapy. J Exp Med. (2019) 216:20–40. doi: 10.1084/jem.20180448
9. Sekirov I, Russell SL, Antunes LCM, and Finlay BB. Gut microbiota in health and disease. Physiol Rev. (2010) 90:859–904. doi: 10.1152/physrev.00045.2009
10. Song M, Chan AT, and Sun J. Influence of the gut microbiome, diet, and environment on risk of colorectal cancer. Gastroenterology. (2020) 158:322–40.
11. Sun J and Chang EB. Exploring gut microbes in human health and disease: Pushing the envelope. Genes Dis. (2014) 1:132–9. doi: 10.1016/j.gendis.2014.08.001
12. Attur M, Scher JU, Abramson SB, and Attur M. Role of intestinal dysbiosis and nutrition in rheumatoid arthritis. Cells. (2022) 11:2436.
13. Johnson CH, Spilker ME, Goetz L, Peterson SN, and Siuzdak G. Metabolite and microbiome interplay in cancer immunotherapy. Cancer Res. (2016) 76:6146–52. doi: 10.1158/0008-5472.CAN-16-0309
14. yu FX, wei Q, feng CH, Gao P, Zhang Q, xue LR, et al. The interaction between dietary fructose and gut microbiota in hyperuricemia and gout. Front Nutr. (2022) 9:890730. doi: 10.3389/fnut.2022.890730
15. Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. Host-gut microbiota metabolic interactions. Science. (2012) 336:1262–7.
16. Sathisha KR, Khanum SA, Chandra JNNS, Ayisha F, Balaji S, Marathe GK, et al. Synthesis and xanthine oxidase inhibitory activity of 7-methyl-2-(phenoxymethyl)-5H-[1,3,4]thiadiazolo[3,2-a]pyrimidin-5-one derivatives. Bioorg Med Chem. (2011) 19:211–20. doi: 10.1016/j.bmc.2010.11.034
17. Crane JK. Role of host xanthine oxidase in infection due to enteropathogenic and Shiga-toxigenic Escherichia coli. Gut Microbes. (2013) 4:388–91.
18. Yamada N, Iwamoto C, Kano H, Yamaoka N, Fukuuchi T, Kaneko K, et al. Evaluation of purine utilization by Lactobacillus gasseri strains with potential to decrease the absorption of food-derived purines in the human intestine. Nucleosides Nucleotides Nucleic Acids. (2016) 35:670–6. doi: 10.1080/15257770.2015.1125000
19. Hosomi A, Nakanishi T, Fujita T, and Tamai I. Extra-renal elimination of uric acid via intestinal efflux transporter BCRP/ABCG2. PloS One. (2012) 7:e30456. doi: 10.1371/journal.pone.0030456
20. Guo Z, Zhang J, Wang Z, Ang KY, Huang S, Hou Q, et al. Intestinal microbiota distinguish gout patients from healthy humans. Sci Rep. (2016) 6:20602. doi: 10.1038/srep20602
21. Chu Y, Sun S, Huang Y, Gao Q, Xie X, Wang P, et al. Metagenomic analysis revealed the potential role of gut microbiome in gout. NPJ Biofilms Microbiomes. (2021) 7:66.
22. Liu Y, Jarman JB, Low YS, Augustijn HE, Huang S, Chen H, et al. A widely distributed gene cluster compensates for uricase loss in hominids. Cell. (2023) 186:3400–3413.e20.
23. Wang L and Ye J. Commentary: Gut microbiota reduce the risk of hyperuricemia and gout in the human body. Acta Pharm Sin B. (2024) 14:433–5. doi: 10.1016/j.apsb.2023.11.013
24. Méndez-Salazar EO, Vázquez-Mellado J, Casimiro-Soriguer CS, Dopazo J, Çubuk C, Zamudio-Cuevas Y, et al. Taxonomic variations in the gut microbiome of gout patients with and without tophi might have a functional impact on urate metabolism. Mol Med Camb Mass. (2021) 27:50.
25. Xiao N, Zhang X, Xi Y, Li Z, Wei Y, Shen J, et al. Study on the effects of intestinal flora on gouty arthritis. Front Cell Infect Microbiol. (2024) 14:1341953. doi: 10.3389/fcimb.2024.1341953
26. Fan Z, Ross RP, Stanton C, Hou B, Zhao J, Zhang H, et al. Lactobacillus casei CCFM1074 Alleviates Collagen-Induced Arthritis in Rats via Balancing Treg/Th17 and Modulating the Metabolites and Gut Microbiota. Front Immunol. (2021) 12:680073. doi: 10.3389/fimmu.2021.680073
27. Horta-Baas G, Romero-Figueroa MDS, Montiel-Jarquín AJ, Pizano-Zárate ML, García-Mena J, and Ramírez-Durán N. Intestinal dysbiosis and rheumatoid arthritis: A link between gut microbiota and the pathogenesis of rheumatoid arthritis. J Immunol Res. (2017) 2017:4835189. doi: 10.1155/2017/4835189
28. Jiang ZM, Zeng SL, Huang TQ, Lin Y, Wang FF, Gao XJ, et al. Sinomenine ameliorates rheumatoid arthritis by modulating tryptophan metabolism and activating aryl hydrocarbon receptor via gut microbiota regulation. Sci Bull. (2023) 68:1540–55. doi: 10.1016/j.scib.2023.06.027
29. Dong Y, Yao J, Deng Q, Li X, He Y, Ren X, et al. Relationship between gut microbiota and rheumatoid arthritis: A bibliometric analysis. Front Immunol. (2023) 14:1131933. doi: 10.3389/fimmu.2023.1131933
30. Zhang X, di CB, LD Z, and Li H. The gut microbiota: emerging evidence in autoimmune diseases. Trends Mol Med. (2020) 26:862–73. doi: 10.1016/j.molmed.2020.04.001
31. Flannigan KL and Denning TL. Segmented filamentous bacteria-induced immune responses: a balancing act between host protection and autoimmunity. Immunology. (2018) 154:537–46. doi: 10.1111/imm.2018.154.issue-4
32. Wang Y, Yin Y, Chen X, Zhao Y, Wu Y, Li Y, et al. Induction of intestinal th17 cells by flagellins from segmented filamentous bacteria. Front Immunol. (2019) 10:2750. doi: 10.3389/fimmu.2019.02750
33. Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. (2008) 57:1470–81. doi: 10.2337/db07-1403
34. Ramos MF de P, Monteiro de Barros A do CM, Razvickas CV, Borges FT, and Schor N. Xanthine oxidase inhibitors and sepsis. Int J Immunopathol Pharmacol. (2018) 32:2058738418772210. doi: 10.1177/2058738418772210
35. Guo Y, Yu Y, Li H, Ding X, Li X, Jing X, et al. Inulin supplementation ameliorates hyperuricemia and modulates gut microbiota in Uox-knockout mice. Eur J Nutr. (2021) 60:2217–30. doi: 10.1007/s00394-020-02414-x
36. Liu Y, Yu P, Sun X, and Di D. Metabolite target analysis of human urine combined with pattern recognition techniques for the study of symptomatic gout. Mol Biosyst. (2012) 8:2956–63. doi: 10.1039/c2mb25227a
37. Wang Z, Li Y, Liao W, Huang J, Liu Y, Li Z, et al. Gut microbiota remodeling: A promising therapeutic strategy to confront hyperuricemia and gout. Front Cell Infect Microbiol. (2022) 12:935723. doi: 10.3389/fcimb.2022.935723
38. Pan L, Han P, Ma S, Peng R, Wang C, Kong W, et al. Abnormal metabolism of gut microbiota reveals the possible molecular mechanism of nephropathy induced by hyperuricemia. Acta Pharm Sin B. (2020) 10:249–61. doi: 10.1016/j.apsb.2019.10.007
39. Maiuolo J, Oppedisano F, Gratteri S, Muscoli C, and Mollace V. Regulation of uric acid metabolism and excretion. Int J Cardiol. (2016) 213:8–14. doi: 10.1016/j.ijcard.2015.08.109
40. Mahbub MH, Yamaguchi N, Takahashi H, Hase R, Amano H, Kobayashi-Miura M, et al. Alteration in plasma free amino acid levels and its association with gout. Environ Health Prev Med. (2017) 22:7. doi: 10.1186/s12199-017-0609-8
41. Henson MA. Interrogation of the perturbed gut microbiota in gouty arthritis patients through in silico metabolic modeling. Eng Life Sci. (2021) 21:489–501. doi: 10.1002/elsc.202100003
42. Shi N, Li N, Duan X, and Niu H. Interaction between the gut microbiome and mucosal immune system. Mil Med Res. (2017) 4:14. doi: 10.1186/s40779-017-0122-9
43. Majka G, Więcek G, Śróttek M, Śpiewak K, Brindell M, Koziel J, et al. The impact of lactoferrin with different levels of metal saturation on the intestinal epithelial barrier function and mucosal inflammation. Biometals Int J Role Met Ions Biol Biochem Med. (2016) 29:1019–33. doi: 10.1007/s10534-016-9973-x
44. Assimakopoulos SF, Scopa CD, Charonis A, Spiliopoulou I, Georgiou C, Nikolopoulou V, et al. Experimental obstructive jaundice disrupts intestinal mucosal barrier by altering occludin expression: beneficial effect of bombesin and neurotensin. J Am Coll Surg. (2004) 198:748–57. doi: 10.1016/j.jamcollsurg.2003.12.017
45. Campisi L, Barbet G, Ding Y, Esplugues E, Flavell RA, and Blander JM. Apoptosis in response to microbial infection induces autoreactive TH17 cells. Nat Immunol. (2016) 17:1084–92. doi: 10.1038/ni.3512
46. Romero-Figueroa MDS, Ramírez-Durán N, Montiel-Jarquín AJ, and Horta-Baas G. Gut-joint axis: Gut dysbiosis can contribute to the onset of rheumatoid arthritis via multiple pathways. Front Cell Infect Microbiol. (2023) 13:1092118. doi: 10.3389/fcimb.2023.1092118
47. Van Itallie CM, Fanning AS, Bridges A, and Anderson JM. ZO-1 stabilizes the tight junction solute barrier through coupling to the perijunctional cytoskeleton. Mol Biol Cell. (2009) 20:3930–40. doi: 10.1091/mbc.e09-04-0320
48. Chen X, Ge HZ, Lei SS, Jiang ZT, Su J, He X, et al. Dendrobium officinalis six nostrum ameliorates urate under-excretion and protects renal dysfunction in lipid emulsion-induced hyperuricemic rats. BioMed Pharmacother Biomedecine Pharmacother. (2020) 132:110765.
49. Fuke N, Nagata N, Suganuma H, and Ota T. Regulation of gut microbiota and metabolic endotoxemia with dietary factors. Nutrients. (2019) 11:2277.
50. Xi Y, Yan J, Li M, Ying S, and Shi Z. Gut microbiota dysbiosis increases the risk of visceral gout in goslings through translocation of gut-derived lipopolysaccharide. Poult Sci. (2019) 98:5361–73. doi: 10.3382/ps/pez357
51. Martinon F, Pétrilli V, Mayor A, Tardivel A, and Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. (2006) 440:237–41.
52. So AK and Martinon F. Inflammation in gout: mechanisms and therapeutic targets. Nat Rev Rheumatol. (2017) 13:639–47. doi: 10.1038/nrrheum.2017.155
53. Jhang JJ, Lin JH, and Yen GC. Beneficial properties of phytochemicals on NLRP3 inflammasome-mediated gout and complication. J Agric Food Chem. (2018) 66:765–72. doi: 10.1021/acs.jafc.7b05113
54. Gu QY, Zhang J, and Feng YC. Role of NLRP3 inflammasome in Bifidobacterium longum-regulated visceral hypersensitivity of postinfectious irritable bowel syndrome. Artif Cells Nanomedicine Biotechnol. (2016) 44:1933–7. doi: 10.3109/21691401.2015.1111238
55. Saber S, Abd El-Fattah EE, Yahya G, Gobba NA, Maghmomeh AO, Khodir AE, et al. A novel combination therapy using rosuvastatin and lactobacillus combats dextran sodium sulfate-induced colitis in high-fat diet-fed rats by targeting the TXNIP/NLRP3 interaction and influencing gut microbiome composition. Pharm Basel Switz. (2021) 14:341. doi: 10.3390/ph14040341
56. Shao X, Sun S, Zhou Y, Wang H, Yu Y, Hu T, et al. Bacteroides fragilis restricts colitis-associated cancer via negative regulation of the NLRP3 axis. Cancer Lett. (2021) 523:170–81. doi: 10.1016/j.canlet.2021.10.002
57. Guo R, Li S, Zhang Y, Zhang Y, Wang G, Ullah H, et al. Dysbiotic oral and gut viromes in untreated and treated rheumatoid arthritis patients. Microbiol Spectr. (2022) 10:e0034822. doi: 10.1128/spectrum.00348-22
58. Krych Ł, Nielsen DS, Hansen AK, and Hansen CHF. Gut microbial markers are associated with diabetes onset, regulatory imbalance, and IFN-γ level in NOD mice. Gut Microbes. (2015) 6:101–9. doi: 10.1080/19490976.2015.1011876
59. Takahashi D, Hoshina N, Kabumoto Y, Maeda Y, Suzuki A, Tanabe H, et al. Microbiota-derived butyrate limits the autoimmune response by promoting the differentiation of follicular regulatory T cells. EBioMedicine. (2020) 58:102913.
60. Sun H, Guo Y, Wang H, Yin A, Hu J, Yuan T, et al. Gut commensal Parabacteroides distasonis alleviates inflammatory arthritis. Gut. (2023) 72:1664–77.
61. Agus A, Planchais J, and Sokol H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe. (2018) 23:716–24. doi: 10.1016/j.chom.2018.05.003
62. Yu D, Du J, Pu X, Zheng L, Chen S, Wang N, et al. The gut microbiome and metabolites are altered and interrelated in patients with rheumatoid arthritis. Front Cell Infect Microbiol. (2022) 11:763507. doi: 10.3389/fcimb.2021.763507
63. Lee K, Kwak JH, and Pyo S. Inhibition of LPS-induced inflammatory mediators by 3-hydroxyanthranilic acid in macrophages through suppression of PI3K/NF-κB signaling pathways. Food Funct. (2016) 7:3073–82. doi: 10.1039/C6FO00187D
64. Wrigley R, Phipps-Green AJ, Topless RK, Major TJ, Cadzow M, Riches P, et al. Pleiotropic effect of the ABCG2 gene in gout: involvement in serum urate levels and progression from hyperuricemia to gout. Arthritis Res Ther. (2020) 22:45. doi: 10.1186/s13075-020-2136-z
65. Álvarez-Lario B and Macarrón-Vicente J. Uric acid and evolution. Rheumatol Oxf Engl. (2010) 49:2010–5. doi: 10.1093/rheumatology/keq204
66. Mackenzie IS, Ford I, Nuki G, Hallas J, Hawkey CJ, Webster J, et al. Long-term cardiovascular safety of febuxostat compared with allopurinol in patients with gout (FAST): a multicentre, prospective, randomised, open-label, non-inferiority trial. Lancet Lond Engl. (2020) 396:1745–57. doi: 10.1016/S0140-6736(20)32234-0
67. Wu J, Wei Z, Cheng P, Qian C, Xu F, Yang Y, et al. Rhein modulates host purine metabolism in intestine through gut microbiota and ameliorates experimental colitis. Theranostics. (2020) 10:10665–79.
68. Li M, Yang D, Mei L, Yuan L, Xie A, and Yuan J. Screening and characterization of purine nucleoside degrading lactic acid bacteria isolated from Chinese sauerkraut and evaluation of the serum uric acid lowering effect in hyperuricemic rats. PloS One. (2014) 9:e105577. doi: 10.1371/journal.pone.0105577
69. Merriman TR. An update on the genetic architecture of hyperuricemia and gout. Arthritis Res Ther. (2015) 17:98. doi: 10.1186/s13075-015-0609-2
70. Xu X, Li C, Zhou P, and Jiang T. Uric acid transporters hiding in the intestine. Pharm Biol. (2016) 54:3151–5. doi: 10.1080/13880209.2016.1195847
71. Nakagawa S, Ishino S, and Teshiba S. Construction of catalase deficient Escherichia coli strains for the production of uricase. Biosci Biotechnol Biochem. (1996) 60:415–20. doi: 10.1271/bbb.60.415
72. Tong S, Zhang P, Cheng Q, Chen M, Chen X, Wang Z, et al. The role of gut microbiota in gout: Is gut microbiota a potential target for gout treatment. Front Cell Infect Microbiol. (2022) 12:1051682. doi: 10.3389/fcimb.2022.1051682
73. Sturgeon C and Fasano A. Zonulin, a regulator of epithelial and endothelial barrier functions, and its involvement in chronic inflammatory diseases. Tissue Barriers. (2016) 4:e1251384. doi: 10.1080/21688370.2016.1251384
74. Harre U, Georgess D, Bang H, Bozec A, Axmann R, Ossipova E, et al. Induction of osteoclastogenesis and bone loss by human autoantibodies against citrullinated vimentin. J Clin Invest. (2012) 122:1791–802.
75. Xiao M, Fu X, Ni Y, Chen J, Jian S, Wang L, et al. Protective effects of Paederia scandens extract on rheumatoid arthritis mouse model by modulating gut microbiota. J Ethnopharmacol. (2018) 226:97–104. doi: 10.1016/j.jep.2018.08.012
76. Guerreiro CS, Calado Â, Sousa J, and Fonseca JE. Diet, microbiota, and gut permeability-the unknown triad in rheumatoid arthritis. Front Med. (2018) 5:349. doi: 10.3389/fmed.2018.00349
77. Lima AAM, Brito LFB, Ribeiro HB, Martins MCV, Lustosa AP, Rocha EM, et al. Intestinal barrier function and weight gain in malnourished children taking glutamine supplemented enteral formula. J Pediatr Gastroenterol Nutr. (2005) 40:28–35. doi: 10.1002/j.1536-4801.2005.tb00922.x
78. De Santis S, Cavalcanti E, Mastronardi M, Jirillo E, and Chieppa M. Nutritional keys for intestinal barrier modulation. Front Immunol. (2015) 6:612. doi: 10.3389/fimmu.2015.00612
79. Finamore A, Massimi M, Conti Devirgiliis L, and Mengheri E. Zinc deficiency induces membrane barrier damage and increases neutrophil transmigration in Caco-2 cells. J Nutr. (2008) 138:1664–70. doi: 10.1093/jn/138.9.1664
80. Li B, Ding M, Liu X, Zhao J, Ross RP, Stanton C, et al. Bifidobacterium breve CCFM1078 Alleviates Collagen-Induced Arthritis in Rats via Modulating the Gut Microbiota and Repairing the Intestinal Barrier Damage. J Agric Food Chem. (2022) 70:14665–78. doi: 10.1021/acs.jafc.2c04602
81. Zhou L, Zhang M, Wang Y, Dorfman RG, Liu H, Yu T, et al. Faecalibacterium prausnitzii produces butyrate to maintain th17/treg balance and to ameliorate colorectal colitis by inhibiting histone deacetylase 1. Inflammation Bowel Dis. (2018) 24:1926–40. doi: 10.1093/ibd/izy182
82. Tajik N, Frech M, Schulz O, Schälter F, Lucas S, Azizov V, et al. Targeting zonulin and intestinal epithelial barrier function to prevent onset of arthritis. Nat Commun. (2020) 11:1995. doi: 10.1038/s41467-020-15831-7
83. Pan H, Guo R, Ju Y, Wang Q, Zhu J, Xie Y, et al. A single bacterium restores the microbiome dysbiosis to protect bones from destruction in a rat model of rheumatoid arthritis. Microbiome. (2019) 7:107. doi: 10.1186/s40168-019-0719-1
84. Lin S, Zhang T, Zhu L, Pang K, Lu S, Liao X, et al. Characteristic dysbiosis in gout and the impact of a uric acid-lowering treatment, febuxostat on the gut microbiota. J Genet Genomics Yi Chuan Xue Bao. (2021) 48:781–91.
85. Luo J and Solit DB. Leveraging real-world data to advance biomarker discovery and precision oncology. Cancer Cell. (2025) 43:606–10. doi: 10.1016/j.ccell.2025.03.012
86. Mi H, Sivagnanam S, Ho WJ, Zhang S, Bergman D, Deshpande A, et al. Computational methods and biomarker discovery strategies for spatial proteomics: a review in immuno-oncology. Brief Bioinform. (2024) 25:bbae421. doi: 10.1093/bib/bbae421
87. Kiltz U, Alten R, Fleck M, Krüger K, Manger B, Müller-Ladner U, et al. Full version of the S2e guidelines on gouty arthritis: Evidence-based guidelines of the German Society of Rheumatology (DGRh). Z Rheumatol. (2016) 75 Suppl 2:11–60.
88. Engel B, Just J, Bleckwenn M, and Weckbecker K. Treatment options for gout. Dtsch Arzteblatt Int. (2017) 114:215–22. doi: 10.3238/arztebl.2017.0215
89. Yu Y, Liu Q, Li H, Wen C, and He Z. Alterations of the gut microbiome associated with the treatment of hyperuricaemia in male rats. Front Microbiol. (2018) 9:2233. doi: 10.3389/fmicb.2018.02233
90. Park HK and Lee SJ. Treatment of gouty arthritis is associated with restoring the gut microbiota and promoting the production of short-chain fatty acids. Arthritis Res Ther. (2022) 24:51. doi: 10.1186/s13075-022-02742-9
91. Montalto M, Gallo A, Curigliano V, D’Onofrio F, Santoro L, Covino M, et al. Clinical trial: the effects of a probiotic mixture on non-steroidal anti-inflammatory drug enteropathy - a randomized, double-blind, cross-over, placebo-controlled study. Aliment Pharmacol Ther. (2010) 2):209–14. doi: 10.1111/j.1365-2036.2010.04324.x
92. Wang X, Tang Q, Hou H, Zhang W, Li M, Chen D, et al. Gut microbiota in NSAID enteropathy: new insights from inside. Front Cell Infect Microbiol. (2021) 11:679396. doi: 10.3389/fcimb.2021.679396
93. Parker A, Fonseca S, and Carding SR. Gut microbes and metabolites as modulators of blood-brain barrier integrity and brain health. Gut Microbes. (2020) 11:135–57. doi: 10.1080/19490976.2019.1638722
94. Shi Y, Li J, Yang P, Niu Z, Wei L, Chen L, et al. Colchicine increases intestinal permeability, suppresses inflammatory responses, and alters gut microbiota in mice. Toxicol Lett. (2020) 334:66–77. doi: 10.1016/j.toxlet.2020.09.018
95. Feng Z, Long W, Hao B, Ding D, Ma X, Zhao L, et al. A human stool-derived Bilophila wadsworthia strain caused systemic inflammation in specific-pathogen-free mice. Gut Pathog. (2017) 9:59. doi: 10.1186/s13099-017-0208-7
96. Azevedo VF, Kos IA, Vargas-Santos AB, da Rocha Castelar Pinheiro G, and Dos Santos Paiva E. Benzbromarone in the treatment of gout. Adv Rheumatol Lond Engl. (2019) 59:37. doi: 10.1186/s42358-019-0080-x
97. Tu Y, Fang QJ, Sun W, Liu BH, Liu YL, Wu W, et al. Total flavones of abelmoschus manihot remodels gut microbiota and inhibits microinflammation in chronic renal failure progression by targeting autophagy-mediated macrophage polarization. Front Pharmacol. (2020) 11:566611. doi: 10.3389/fphar.2020.566611
98. Zhao H, Lu Z, and Lu Y. The potential of probiotics in the amelioration of hyperuricemia. Food Funct. (2022) 13:2394–414. doi: 10.1039/D1FO03206B
99. Wu Y, Ye Z, Feng P, Li R, Chen X, Tian X, et al. Limosilactobacillus fermentum JL-3 isolated from “Jiangshui” ameliorates hyperuricemia by degrading uric acid. Gut Microbes. (2021) 13:1–18. doi: 10.1080/19490976.2021.1897211
100. Xie WR, Yang XY, Deng ZH, Zheng YM, Zhang R, Wu LH, et al. Effects of washed microbiota transplantation on serum uric acid levels, symptoms, and intestinal barrier function in patients with acute and recurrent gout: A pilot study. Dig Dis Basel Switz. (2022) 40:684–90. doi: 10.1159/000521273
101. Ni C, Li X, Wang L, Li X, Zhao J, Zhang H, et al. Lactic acid bacteria strains relieve hyperuricaemia by suppressing xanthine oxidase activity via a short-chain fatty acid-dependent mechanism. Food Funct. (2021) 12:7054–67. doi: 10.1039/D1FO00198A
102. Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. (2014) 11:506–14. doi: 10.1038/nrgastro.2014.66
103. Vallianou N, Stratigou T, Christodoulatos GS, Tsigalou C, and Dalamaga M. Probiotics, prebiotics, synbiotics, postbiotics, and obesity: current evidence, controversies, and perspectives. Curr Obes Rep. (2020) 9:179–92. doi: 10.1007/s13679-020-00379-w
104. Wang H, Mei L, Deng Y, Liu Y, Wei X, Liu M, et al. Lactobacillus brevis DM9218 ameliorates fructose-induced hyperuricemia through inosine degradation and manipulation of intestinal dysbiosis. Nutr Burbank Los Angel Cty Calif. (2019) 62:63–73.
105. García-Arroyo FE, Gonzaga G, Muñoz-Jiménez I, Blas-Marron MG, Silverio O, Tapia E, et al. Probiotic supplements prevented oxonic acid-induced hyperuricemia and renal damage. PloS One. (2018) 13:e0202901. doi: 10.1371/journal.pone.0202901
106. Swanson KS, Gibson GR, Hutkins R, Reimer RA, Reid G, Verbeke K, et al. The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of synbiotics. Nat Rev Gastroenterol Hepatol. (2020) 17:687–701. doi: 10.1038/s41575-020-0344-2
107. Holscher HD. Dietary fiber and prebiotics and the gastrointestinal microbiota. Gut Microbes. (2017) 8:172–84. doi: 10.1080/19490976.2017.1290756
108. Ren Q, Cheng L, Guo F, Tao S, Zhang C, Ma L, et al. Fisetin improves hyperuricemia-induced chronic kidney disease via regulating gut microbiota-mediated tryptophan metabolism and aryl hydrocarbon receptor activation. J Agric Food Chem. (2021) 69:10932–42. doi: 10.1021/acs.jafc.1c03449
109. Wang LM, Wang P, Teka T, Zhang YC, Yang WZ, Zhang Y, et al. 1H NMR and UHPLC/Q-orbitrap-MS-based metabolomics combined with 16S rRNA gut microbiota analysis revealed the potential regulation mechanism of nuciferine in hyperuricemia rats. J Agric Food Chem. (2020) 68:14059–70. doi: 10.1021/acs.jafc.0c04985
110. Li X, Gao X, Zhang H, Liu Y, Sarker MMR, Wu Y, et al. The anti-hyperuricemic effects of green alga Enteromorpha prolifera polysaccharide via regulation of the uric acid transporters in vivo. Food Chem Toxicol Int J Publ Br Ind Biol Res Assoc. (2021) 158:112630. doi: 10.1016/j.fct.2021.112630
111. Lu C, Tang S, Han J, Fan S, Huang Y, Zhang Z, et al. Apostichopus japonicus oligopeptide induced heterogeneity in the gastrointestinal tract microbiota and alleviated hyperuricemia in a microbiota-dependent manner. Mol Nutr Food Res. (2021) 65:e2100147. doi: 10.1002/mnfr.202100147
112. Han J, Wang Z, Lu C, Zhou J, Li Y, Ming T, et al. The gut microbiota mediates the protective effects of anserine supplementation on hyperuricaemia and associated renal inflammation. Food Funct. (2021) 12:9030–42. doi: 10.1039/D1FO01884A
113. Han J, Wang X, Tang S, Lu C, Wan H, Zhou J, et al. Protective effects of tuna meat oligopeptides (TMOP) supplementation on hyperuricemia and associated renal inflammation mediated by gut microbiota. FASEB J Off Publ Fed Am Soc Exp Biol. (2020) 34:5061–76. doi: 10.1096/fj.201902597RR
Keywords: gut microbiota, gut metabolites, gout, new therapeutic perspective, dual role
Citation: Qi P, Li L, Zhang J, Ren L and Xie X (2025) The dual regulatory effects of intestinal microorganisms and their metabolites in gouty arthritis pathogenesis: a balance between promotion and inhibition. Front. Immunol. 16:1591369. doi: 10.3389/fimmu.2025.1591369
Received: 11 March 2025; Accepted: 26 May 2025;
Published: 18 June 2025.
Edited by:
Teresa Zelante, University of Perugia, ItalyCopyright © 2025 Qi, Li, Zhang, Ren and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xingwen Xie, cTEzMzA5MzI5OTQ5QDE2My5jb20=
 Peng Qi
Peng Qi Longcan Li1
Longcan Li1 Jianrong Zhang
Jianrong Zhang Xingwen Xie
Xingwen Xie