- 1The Feinstein Institutes for Medical Research, Northwell Health, Manhasset, NY, United States
- 2Department of Molecular Medicine, Donald and Barbara Zucker School of Medicine at Hofstra/Northwell, Hempstead, NY, United States
- 3Division of Surgical Research, Brown University Health-Rhode Island Hospital, Providence, RI, United States
- 4Department of Surgery, the Warren Alpert School of Medicine at Brown University, Providence, RI, United States
- 5Department of Surgery, University of Pittsburgh, Pittsburgh, PA, United States
- 6Department of Pathobiological Science, School of Veterinary Medicine, Louisiana State University, Baton Rouge, LA, United States
- 7Department of Surgery, UT Southwestern Medical Center, Dallas, TX, United States
Introduction
Sepsis is a life-threatening organ dysfunction caused by a dysregulated host response to infection (1), as manifested by early activation of both pro- and anti-inflammatory responses (2), along with major alterations in non-immunologic pathways such as cardiovascular, neuronal, autonomic, hormonal, bioenergetic, metabolic, and coagulation (3). It accounts for almost 20% of total deaths worldwide (4), and annually costs more than $60 billion in the U.S. alone. The onset of the disease and the intricate interplay of various immune cells, inflammatory mediators, signaling pathways, and organ systems makes studying sepsis in humans ethically and logistically challenging. This necessitates the use of animal models to systematically dissect its intricate pathophysiology and evaluate potential therapies in a controlled setting, which has contributed to developing and implementing clinical therapies for other inflammatory diseases, notably rheumatoid arthritis.
The indispensable role of animal models in sepsis research
Animal models allow researchers to manipulate key variables such as infection type and severity, intervention timing, and the genetic background (e.g., gene knockout or knock in strategy) of experimental animals (5–8). This level of control enables researchers to isolate the effects of specific interventions and identify potential therapeutic targets, such as tumor necrosis factor (TNF) (9), high mobility group box 1 (HMGB1) (10), cold-inducible RNA-binding protein (CIRP) (11), sequestosome-1 (SQSTM1) (12), and procathepsin L (pCTS-L) (13). Moreover, these models allow for tracking the temporal progression of sepsis from initial insult to subsequent organ dysfunctions (14) and eventual outcomes (7, 8), offering invaluable insight into the complex interplay of multiple pathophysiological processes (15, 16), including hyperinflammation (17), immunocoagulation (18, 19), pyroptosis-mediated immune cell death (20), and immunosuppression (21, 22). Given the influence of comorbidities and other factors on disease progression and treatment response in human sepsis, it is essential to incorporate comorbidities (e.g., diabetes, hypertension, or coronary artery disease) and pre-existing injuries (e.g., smoke inhalation occurring in burn patients) into animal modeling, thereby improving the translatability of experimental findings into future clinical therapies (6–8, 23).
The challenges and complexities of translation
Despite advancements in understanding sepsis pathophysiology, translating preclinical findings into effective human therapies remains challenging, as exemplified by the failure of anti-TNF antibodies in clinical trials (3, 24). However, attributing this translational gap solely to the limitations of animal models is an oversimplification (6), because the inherent complexity and heterogeneity of human sepsis, coupled with challenges in clinical trial design, also contribute to this difficulty.
Animal models typically use a single, standardized insult in genetically homogeneous animals. However, this genetic and environmental homogeneity of laboratory animals contrasts sharply with the genetic and environmental diversity of human populations, as well as the variety of infections in clinical sepsis (8, 25). The inherent heterogeneity in septic patients is further compounded by other factors such as age, sex, underlying health conditions (comorbidities), environmental exposure/history, and time to treatment initiation (2). Because patient variability often creates a broad spectrum of pathophysiological endotypes, it is important to develop animal models to recapitulate some human sepsis endotypes. While comprehensive immune profiling (cytokine/chemokine levels, immune cell function, gene expression) can potentially characterize “endotypes” in animal models, their accuracy in reflecting human sepsis endotypes (such as hyper- or hypo-inflammatory states) remains unclear (6–8), presenting challenges for translational research. Thus, the failure of identifying and recruiting homogenous patient subgroups in previous clinical trials might have diluted treatment effects due to potential outcome variations (8, 23, 26).
In addition, potential differences in immune responses between animals and humans may pose another significant challenge (27). Although genomic comparisons between mouse models and human sepsis have revealed significant similarities (28, 29), they also highlight some noticeable differences (30), underscoring the complexity and difficulty of extrapolating experimental findings across species (Table 1). Therefore, developing more diverse and sophisticated animal models that incorporate polymicrobial infections, comorbidities, and genetic variability is crucial to improving translatability (7, 8). For example, a refined murine sepsis model that adhered the Minimum Quality Threshold in Pre-Clinical Sepsis Studies (MQTiPSS) guidelines (5, 31) by incorporating daily chronic stress closely recapitulated the genomic and phenotypic responses observed in human surgical sepsis (32). Similarly, humanized mice that express human genes or possess a humanized immune system may similarly offer a more promising approach to enhance translatability (7, 8, 33). Conversely, stratifying patients based on specific biomarkers (34, 35) indicative of the unique pathobiology one is hoping to modify, along with relevant clinical parameters beyond overall mortality, should similarly enhance the precision and power of future clinical trials (3, 35–37).
The rationale for targeting TNF in sepsis stemmed from its early and prominent role in initiating the inflammatory response (9, 38). However, sepsis is a highly heterogeneous syndrome characterized by variable timing for the release and pathogenic actions of various cytokines (17). While TNF may be critical in the initial hyperinflammatory phase, its importance diminishes over time. Therefore, blocking it at a wrong time could even be detrimental to the host by impairing essential immunities needed for pathogen clearance (39–42). Accordingly, shifting the focus to some later-acting mediators such as HMGB1 (10) and pCTS-L (13), which have relatively wider therapeutic windows (43), may present a more promising avenue for future sepsis trials. Given the multifaceted nature of sepsis, combinatorial therapies targeting multiple mediators may be more effective than targeting a single cytokine. Such combination therapies, if tailored to the specific cytokine profile and disease stage of individual patients, may offer a more personalized and effective approach to sepsis treatment (8, 35, 44), unlocking the therapeutic potential of cytokine-targeting therapies for this devastating condition.
The dynamic nature of sepsis requires timely interventions and optimal dosing regimens (45, 46), which are also difficult to translate from animal models to human clinical trials. Simplified dosing regimens used in animal models (32) often struggle to capture the complex pharmacokinetics and pharmacodynamics observed in humans (3). Beyond the heterogeneity of sepsis patients, interspecies differences in these kinetic and dynamic parameters further complicate translation, necessitating personalized dosing algorithms based on individual patient characteristics such as age, sex, weight, comorbidities, and disease severity.
The unexpected value of animal sepsis research in human therapies
Although directly translating animal sepsis research to human sepsis therapies remains challenging, the knowledge gained from animal models has advanced treatments for other inflammatory diseases, such as rheumatoid arthritis (RA) (47), Crohn’s diseases (48), and ulcerative colitis (49). RA, a chronic autoimmune disease affecting 0.5–1% of the global population (50), shares unexpected commonalities with sepsis in its inflammatory pathways, particularly the involvement of TNF (51, 52) and other cytokines. The identification of TNF as an early mediator of sepsis, largely through animal models (9, 38), paved the way for the development of anti-TNF biologics like infliximab, etanercept, and adalimumab (53, 54) as cornerstone therapies for RA (53, 55), Crohn’s diseases (48), and ulcerative colitis (49). These success stories have exemplified the broader impact of sepsis research using animal models, extending beyond sepsis itself to benefit patients with other inflammatory conditions. It highlights the value of fundamental research in revealing unexpected connections between seemingly disparate fields and potentially driving significant therapeutic advances for many inflammation disorders.
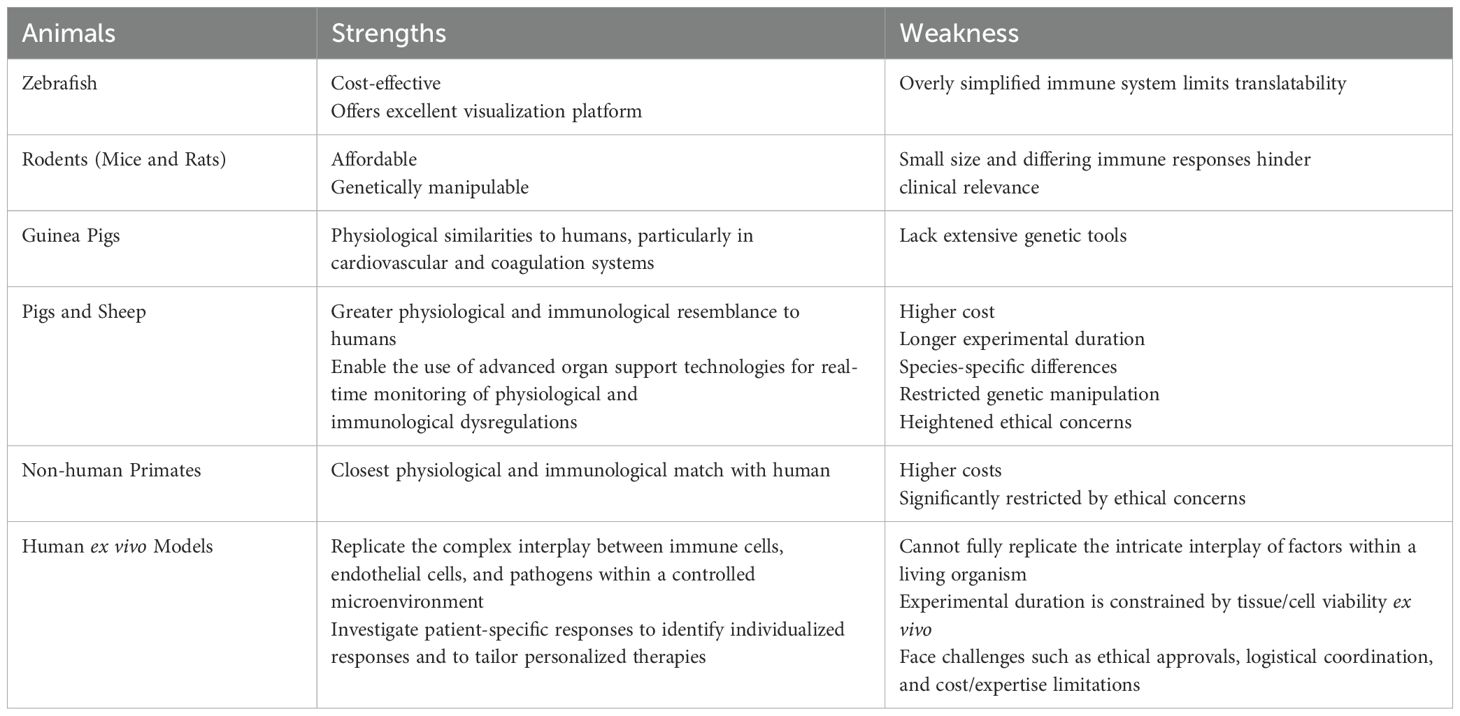
The path forward: refining models and experimental approaches
Animal models remain indispensable in sepsis research, providing a controlled setting to unravel complex pathophysiological mechanisms, identify therapeutic targets, and evaluate novel interventions. While acknowledging their limitations and actively refining these models is crucial, abandoning animal research would be a short-sighted setback hindering scientific progress (6). Therefore, the future of sepsis research still hinges on developing more sophisticated and clinically relevant animal models that incorporate age, sex, polymicrobial infections, comorbidities (e.g., diabetes, hypertension, or coronary artery disease), pre-existing injuries (e.g., smoke inhalation in burn patients) (56), and genetic diversity, thereby better reflecting the complexity and heterogeneity of human sepsis. Animal models of sepsis can be developed in a vast array of different animal species (Table 1), but each species possesses unique strengths and weaknesses that influence the translatability of research findings. Therefore, model selection depends on the specific research question and a balanced assessment of species-specific strengths and weaknesses. For instance, a multi-species approach, strategically incorporating larger animal models like pigs or non-human primates when necessary, might be needed to bridge this species gap.
Traditional animal sepsis studies rely on limited number of physiological and biochemical markers, and thus lack the granularity to capture the complex molecular landscape of clinical sepsis. It is thus critical to refine animal sepsis models (57) by integrating transcriptomic, epigenomic, metabolomic, and proteomic analyses across various body compartments at single-cell level (58). Comparing these multi-omics profiles between animal models and human sepsis patients will allow for rigorous model validation, ensuring an accurate reflection of dysregulated molecular pathways of human sepsis (32). This improved approach may also help identify key pathways and biomarkers conserved across species, guiding relevant therapeutic target selection to improve the predictive power of preclinical studies.
To fully replicate organ dysfunctions characteristic of human sepsis, it is paramount to integrate organ support technologies (e.g., mechanical ventilation, fluid resuscitation, and vasopressor support) into larger animal models by creating “ICU-like” experimental conditions that allow real-time monitoring and modulation of immunological and physiological parameters that mirror clinical sepsis management. Large animal models, like pigs and sheep (56, 59), offer a unique platform for sepsis research due to their larger size, as well as immunological and physiological similarities to humans (e.g., heart rate, blood pressure, and lung mechanics), enabling the use of advanced organ support technologies for real-time monitoring of immune and physiological dysregulations during sepsis (Table 1). However, these models are also limited by higher cost, longer experimental duration, species-specific differences, restricted genetic manipulation, and heightened ethical concerns (Table 1).
To overcome the translational limitations of animal models in sepsis research, several human ex vivo models have also been developed, including i) whole blood assays (60); ii) precision-cut tissue slices (lung and others) (61); iii) human blood-perfused organ models (62, 63); and iv) miniaturized “organ-on-a-chip” systems (64, 65). These models can replicate the complex interplay between immune cells, endothelial cells, and pathogens within a controlled microenvironment, offering a more human-relevant platform for studying sepsis (Table 1). Another key strength is their capacity to dissect patient-specific responses, using samples from diverse cohorts (varying ages, comorbidities, genetic backgrounds) to identify individualized responses and to potentially tailor personalized therapies. Despite these advantages, human ex vivo models suffer from many limitations (Table 1), such as the inability to fully replicate in vivo complexity, restricted experimental duration due to ex vivo tissue/cell viability, and challenges in obtaining/utilizing human samples, including ethical approvals, logistical coordination, and cost/expertise limitations.
Conclusions
Refining animal models of sepsis requires a multifaceted approach encompassing: 1) the development of more sophisticated and clinically relevant animal models that incorporate age, sex, polymicrobial infections, and comorbidities; 2) the integration of organ support technologies and multi-omics calibration; 3) the strategic use of multiple species; and 4) the selection of multiple more feasible therapeutic targets including HMGB1, CIRP, and pCTS-L. By implementing these refinements, future animal research can more accurately reflect the complexity of human sepsis, thereby enhancing the predictive validity of preclinical studies and accelerating the development of effective therapies for this devastating condition. In addition to refining animal models, improving clinical trial design through better patient stratification based on biomarkers and cytokine profiles may be equally important. The continued pursuit of knowledge through well-designed animal models, coupled with rigorous clinical research, holds the key to unlocking effective therapies for sepsis and other inflammatory diseases. The unexpected success of anti-TNF therapies in RA, born from sepsis research, serves as a powerful testament to the value of animal research.
Author contributions
HW: Writing – review & editing, Conceptualization, Writing – original draft. AA: Writing – review & editing. MA: Writing – review & editing. TB: Writing – review & editing. CD: Writing – review & editing. SJ: Writing – review & editing. DT: Writing – review & editing. PW: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. Research in Dr. Haichao Wang’s laboratory was partly supported by the National Institutes of Health (NIH) grants R01AT005076 and R35GM145331. Research in Dr. Alfred Ayala’s laboratory was partly supported by the National Institutes of Health (NIH) grant R35 GM118097.
Acknowledgments
Research in Dr. Haichao Wang’s laboratory was partly supported by the National Institutes of Health (NIH) grants R01AT005076 and R35GM145331.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA. (2016) 315:801–10. doi: 10.1001/jama.2016.0287
2. Dyson A, Singer M. Animal models of sepsis: why does preclinical efficacy fail to translate to the clinical setting? Crit Care Med. (2009) 37:S30–7. doi: 10.1097/CCM.0b013e3181922bd3
3. Marshall JC, Leligdowicz A. Gaps and opportunities in sepsis translational research. EBioMedicine. (2022) 86:104387. doi: 10.1016/j.ebiom.2022.104387
4. Rudd KE, Johnson SC, Agesa KM, Shackelford KA, Tsoi D, Kievlan DR, et al. Global, regional, and national sepsis incidence and mortality, 1990-2017: analysis for the global burden of disease study. Lancet. (2020) 395:200–11. doi: 10.1016/S0140-6736(19)32989-7
5. Osuchowski MF, Ayala A, Bahrami S, Bauer M, Boros M, Cavaillon JM, et al. Minimum quality threshold in pre-clinical sepsis studies (Mqtipss): an international expert consensus initiative for improvement of animal modeling in sepsis. Intensive Care Med Exp. (2018) 6:26. doi: 10.1186/s40635-018-0189-y
6. Osuchowski MF, Remick DG, Lederer JA, Lang CH, Aasen AO, Aibiki M, et al. Abandon the mouse research ship? Not just yet! Shock. (2014) 41:463–75. doi: 10.1097/shk.0000000000000153
7. Stortz JA, Raymond SL, Mira JC, Moldawer LL, Mohr AM, Efron PA. Murine models of sepsis and trauma: can we bridge the gap? Ilar J. (2017) 58:90–105. doi: 10.1093/ilar/ilx007
8. Cavaillon JM, Singer M, Skirecki T. Sepsis therapies: learning from 30 years of failure of translational research to propose new leads. EMBO Mol Med. (2020) 12:e10128. doi: 10.15252/emmm.201810128
9. Tracey KJ, Fong Y, Hesse DG, Manogue KR, Lee AT, Kuo GC, et al. Anti-cachectin/TNF monoclonal antibodies prevent septic shock during lethal bacteraemia. Nature. (1987) 330:662–4. doi: 10.1038/330662a0
10. Wang H, Bloom O, Zhang M, Vishnubhakat JM, Ombrellino M, Che J, et al. Hmg-1 as a late mediator of endotoxin lethality in mice. Science. (1999) 285:248–51. doi: 10.1126/science.285.5425.248
11. Qiang X, Yang WL, Wu R, Zhou M, Jacob A, Dong W, et al. Cold-inducible RNA-binding protein (CIRP) triggers inflammatory responses in hemorrhagic shock and sepsis. Nat Med. (2013) 19:1489–95. doi: 10.1038/nm.3368
12. Zhou B, Liu J, Zeng L, Zhu S, Wang H, Billiar TR, et al. Extracellular SQSTM1 mediates bacterial septic death in mice through insulin receptor signalling. Nat Microbiol. (2020) 5:1576–87. doi: 10.1038/s41564-020-00795-7
13. Zhu CS, Qiang X, Chen W, Li J, Lan X, Yang H, et al. Identification of procathepsin L (pCTS-L)-neutralizing monoclonal antibodies to treat potentially lethal sepsis. Sci Adv. (2023) 9:eadf4313. doi: 10.1126/sciadv.adf4313
14. Radermacher P, Billiar TR, Ghezzi P, Martin L, Thiemermann C. Editorial: translational insights into mechanisms and therapy of organ dysfunction in sepsis and trauma. Front Immunol. (2020) 11:1987. doi: 10.3389/fimmu.2020.01987
15. Matthay MA, Schmidt EP, Bastarache JA, Calfee CS, Frevert CW, Martin TR. The translational value of rodent models of sepsis. Am J Respir Crit Care Med. (2024) 209:488–90. doi: 10.1164/rccm.202308-1489VP
16. Cecconi M, Evans L, Levy M, Rhodes A. Sepsis and septic shock. Lancet. (2018) 392:75–87. doi: 10.1016/s0140-6736(18)30696-2
17. Ayala A, Chaudry IH. Immune dysfunction in murine polymicrobial sepsis: mediators, macrophages, lymphocytes and apoptosis. Shock. (1996) 6 Suppl 1:S27–38. doi: 10.1097/00024382-199606001-00007
18. Tang D, Wang H, Billiar TR, Kroemer G, Kang R. Emerging mechanisms of immunocoagulation in sepsis and septic shock. Trends Immunol. (2021) 21):10. doi: 10.1016/j.it.2021.04.001
19. Levi M, van der Poll T. Inflammation and coagulation. Crit Care Med. (2010) 38:S26–34. doi: 10.1097/CCM.0b013e3181c98d21
20. Wu C, Lu W, Zhang Y, Zhang G, Shi X, Hisada Y, et al. Inflammasome activation triggers blood clotting and host death through pyroptosis. Immunity. (2019) 50:1401–11.e4. doi: 10.1016/j.immuni.2019.04.003
21. Hotchkiss RS, Monneret G, Payen D. Sepsis-induced immunosuppression: from cellular dysfunctions to immunotherapy. Nat Rev Immunol. (2013) 13:862–74. doi: 10.1038/nri3552
22. Pei F, Yao RQ, Ren C, Bahrami S, Billiar TR, Chaudry IH, et al. Expert consensus on the monitoring and treatment of sepsis-induced immunosuppression. Mil Med Res. (2022) 9:74. doi: 10.1186/s40779-022-00430-y
23. Marshall JC, Deitch E, Moldawer LL, Opal S, Redl H, van der Poll T. Preclinical models of shock and sepsis: what can they tell us? Shock. (2005) 24 Suppl 1:1–6. doi: 10.1097/01.shk.0000191383.34066.4b
24. Abraham E, Wunderink R, Silverman H, Perl TM, Nasraway S, Levy H, et al. Efficacy and safety of monoclonal antibody to human tumor necrosis factor alpha in patients with sepsis syndrome. A randomized, controlled, double-blind, multicenter clinical trial. TNF-alpha MAb sepsis study group. JAMA. (1995) 273:934–41. doi: 10.1001/jama.1995.03520360048038
25. Angus DC, Wax RS. Epidemiology of sepsis: an update. Crit Care Med. (2001) 29:S109–S16. doi: 10.1097/00003246-200107001-00035
26. Seymour CW, Kennedy JN, Wang S, Chang CH, Elliott CF, Xu Z, et al. Derivation, validation, and potential treatment implications of novel clinical phenotypes for sepsis. Jama. (2019) 321:2003–17. doi: 10.1001/jama.2019.5791
27. Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J Immunol. (2004) 172:2731–8. doi: 10.4049/jimmunol.172.5.2731
28. Hackert NS, Radtke FA, Exner T, Lorenz HM, Müller-Tidow C, Nigrovic PA, et al. Human and mouse neutrophils share core transcriptional programs in both homeostatic and inflamed contexts. Nat Commun. (2023) 14:8133. doi: 10.1038/s41467-023-43573-9
29. Takao K, Miyakawa T. Genomic responses in mouse models greatly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. (2015) 112:1167–72. doi: 10.1073/pnas.1401965111
30. Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. (2013) 110:3507–12. doi: 10.1073/pnas.1222878110
31. Osuchowski MF, Ayala A, Bahrami S, Bauer M, Boros M, Cavaillon JM, et al. Minimum quality threshold in pre-clinical sepsis studies (MQTiPSS): an international expert consensus initiative for improvement of animal modeling in sepsis. Shock. (2018) 50:377–80. doi: 10.1097/shk.0000000000001212
32. Efron PA, Darden DB, Wang Z, Nacionales DC, Lopez MC, Hawkins RB, et al. Transcriptomic responses from improved murine sepsis models can better mimic human surgical sepsis. FASEB J. (2021) 35:e21156. doi: 10.1096/fj.202002150R
33. Shultz LD, Lyons BL, Burzenski LM, Gott B, Chen X, Chaleff S, et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J Immunol. (2005) 174:6477–89. doi: 10.4049/jimmunol.174.10.6477
34. Billiar TR. Biomarkers to distinguish sepsis from sterile inflammation. Ann Surg. (2020) 272:611. doi: 10.1097/sla.0000000000004376
35. Gordon AC, Alipanah-Lechner N, Bos LD, Dianti J, Diaz JV, Finfer S, et al. From ICU syndromes to ICU subphenotypes: consensus report and recommendations for developing precision medicine in the ICU. Am J Respir Crit Care Med. (2024) 210:155–66. doi: 10.1164/rccm.202311-2086SO
36. Marshall JC. Why have clinical trials in sepsis failed? Trends Mol Med. (2014) 20:195–203. doi: 10.1016/j.molmed.2014.01.007
37. Biron BM, Ayala A, Lomas-Neira JL. Biomarkers for sepsis: what is and what might be? Biomark Insights. (2015) 10:7–17. doi: 10.4137/bmi.S29519
38. Beutler B, Milsark IW, Cerami AC. Passive immunization against cachectin/tumor necrosis factor protects mice from lethal effect of endotoxin. Science. (1985) 229:869–71. doi: 10.1126/science.3895437
39. Remick D, Manohar P, Bolgos G, Rodriguez J, Moldawer L, Wollenberg G. Blockade of tumor necrosis factor reduces lipopolysaccharide lethality, but not the lethality of cecal ligation and puncture. Shock. (1995) 4:89–95. doi: 10.1097/00024382-199508000-00002
40. Chen W, Havell EA, Harmsen AG. Importance of endogenous tumor necrosis factor alpha and gamma interferon in host resistance against pneumocystis carinii infection. Infect Immun. (1992) 60:1279–84. doi: 10.1128/iai.60.4.1279-1284.1992
41. Havell EA. Evidence that tumor necrosis factor has an important role in antibacterial resistance. J Immunol. (1989) 143:2894–9. doi: 10.4049/jimmunol.143.9.2894
42. Osuchowski MF, Welch K, Siddiqui J, Remick DG. Circulating cytokine/inhibitor profiles reshape the understanding of the sirs/cars continuum in sepsis and predict mortality. J Immunol. (2006) 177:1967–74. doi: 10.4049/jimmunol.177.3.1967
43. Li J, Zhu CS, He L, Qiang X, Chen W, Wang H. A two-decade journey in identifying high mobility group box 1 (HMGB1) and procathepsin L (pCTS-L) as potential therapeutic targets for sepsis. Expert Opin Ther Targets. (2023) 27:575–91. doi: 10.1080/14728222.2023.2239495
44. Angus DC, van der Poll T. Severe sepsis and septic shock. N Engl J Med. (2013) 369:840–51. doi: 10.1056/NEJMra1208623
45. Slim MA, van Amstel RBE, Müller MCA, Cremer OL, Vlaar APJ, van der Poll T, et al. Clinical subtype trajectories in sepsis patients admitted to the ICU: A secondary analysis of an observational study. Crit Care Explor. (2024) 6:e1176. doi: 10.1097/cce.0000000000001176
46. Seymour CW, Kerti SJ, Lewis AJ, Kennedy J, Brant E, Griepentrog JE, et al. Murine sepsis phenotypes and differential treatment effects in a randomized trial of prompt antibiotics and fluids. Crit Care. (2019) 23:384. doi: 10.1186/s13054-019-2655-7
47. Elliott MJ, Maini RN, Feldmann M, Kalden JR, Antoni C, Smolen JS, et al. Randomised double-blind comparison of chimeric monoclonal antibody to tumour necrosis factor alpha (Ca2) versus placebo in rheumatoid arthritis. Lancet. (1994) 344:1105–10. doi: 10.1016/s0140-6736(94)90628-9
48. Sands BE, Anderson FH, Bernstein CN, Chey WY, Feagan BG, Fedorak RN, et al. Infliximab maintenance therapy for fistulizing crohn’s disease. N Engl J Med. (2004) 350:876–85. doi: 10.1056/NEJMoa030815
49. Rutgeerts P, Sandborn WJ, Feagan BG, Reinisch W, Olson A, Johanns J, et al. Infliximab for induction and maintenance therapy for ulcerative colitis. N Engl J Med. (2005) 353:2462–76. doi: 10.1056/NEJMoa050516
50. Di Matteo A, Bathon JM, Emery P. Rheumatoid arthritis. Lancet. (2023) 402:2019–33. doi: 10.1016/s0140-6736(23)01525-8
51. Sewell KL, Trentham DE. Pathogenesis of rheumatoid arthritis. Lancet. (1993) 341:283–6. doi: 10.1016/0140-6736(93)92627-6
52. McInnes IB, Schett G. Pathogenetic insights from the treatment of rheumatoid arthritis. Lancet. (2017) 389:2328–37. doi: 10.1016/s0140-6736(17)31472-1
53. Burmester GR, Pope JE. Novel treatment strategies in rheumatoid arthritis. Lancet. (2017) 389:2338–48. doi: 10.1016/s0140-6736(17)31491-5
54. Feldmann M, Maini RN. Anti-TNF alpha therapy of rheumatoid arthritis: what have we learned? Annu Rev Immunol. (2001) 19:163–96. doi: 10.1146/annurev.immunol.19.1.163
55. Taylor PC, Feldmann M. Anti-TNF biologic agents: still the therapy of choice for rheumatoid arthritis. Nat Rev Rheumatol. (2009) 5:578–82. doi: 10.1038/nrrheum.2009.181
56. Maybauer MO, Maybauer DM, Fraser JF, Traber LD, Westphal M, Enkhbaatar P, et al. Recombinant human activated protein C improves pulmonary function in ovine acute lung injury resulting from smoke inhalation and sepsis. Crit Care Med. (2006) 34:2432–8. doi: 10.1097/01.Ccm.0000230384.61350.Fa
57. Seymour CW, Urbanek KL, Nakayama A, Kennedy JN, Powell R, Robinson RAS, et al. A prospective cohort protocol for the remnant investigation in sepsis study. Crit Care Explor. (2023) 5:e0974. doi: 10.1097/cce.0000000000000974
58. Yao RQ, Li ZX, Wang LX, Li YX, Zheng LY, Dong N, et al. Single-cell transcriptome profiling of the immune space-time landscape reveals dendritic cell regulatory program in polymicrobial sepsis. Theranostics. (2022) 12:4606–28. doi: 10.7150/thno.72760
59. Booke M, Hinder F, McGuire R, Traber LD, Traber DL. Nitric oxide synthase inhibition versus norepinephrine for the treatment of hyperdynamic sepsis in sheep. Crit Care Med. (1996) 24:835–44. doi: 10.1097/00003246-199605000-00018
60. Samuelsen AM, Halstead ES, Lehman EB, McKeone DJ, Bonavia AS. Predicting organ dysfunction in septic and critically ill patients: A prospective cohort study using rapid ex vivo immune profiling. Crit Care Explor. (2024) 6:e1106. doi: 10.1097/cce.0000000000001106
61. Yeh CT, Hsu CW, Chang ML, Sheen IS, Lin SM, Lin CJ, et al. A novel ex vivo assay of interferon-based suppression, to predict the outcome of antiviral therapy for hepatitis C. J Infect Dis. (2006) 193:1365–70. doi: 10.1086/503749
62. Frank JA, Briot R, Lee JW, Ishizaka A, Uchida T, Matthay MA. Physiological and biochemical markers of alveolar epithelial barrier dysfunction in perfused human lungs. Am J Physiol Lung Cell Mol Physiol. (2007) 293:L52–9. doi: 10.1152/ajplung.00256.2006
63. Lee JW, Fang X, Gupta N, Serikov V, Matthay MA. Allogeneic human mesenchymal stem cells for treatment of E. Coli endotoxin-induced acute lung injury in the ex vivo perfused human lung. Proc Natl Acad Sci U S A. (2009) 106:16357–62. doi: 10.1073/pnas.0907996106
64. Alonso-Roman R, Mosig AS, Figge MT, Papenfort K, Eggeling C, Schacher FH, et al. Organ-on-chip models for infectious disease research. Nat Microbiol. (2024) 9:891–904. doi: 10.1038/s41564-024-01645-6
65. Liu D, Langston JC, Prabhakarpandian B, Kiani MF, Kilpatrick LE. The critical role of neutrophil-endothelial cell interactions in sepsis: new synergistic approaches employing organ-on-chip, omics, immune cell phenotyping and in silico modeling to identify new therapeutics. Front Cell Infect Microbiol. (2023) 13:1274842. doi: 10.3389/fcimb.2023.1274842
Keywords: sepsis, animal model, human therapy, therapeutic targets, rheumatoid arthritis
Citation: Wang H, Ayala A, Aziz M, Billiar TR, Deutschman CS, Jeyaseelan S, Tang D and Wang P (2025) Value of animal sepsis research in navigating the translational labyrinth. Front. Immunol. 16:1593342. doi: 10.3389/fimmu.2025.1593342
Received: 13 March 2025; Accepted: 04 April 2025;
Published: 15 April 2025.
Edited by:
Guochang Hu, University of Illinois Chicago, United StatesReviewed by:
Jae Woo Lee, University of California, United StatesEric Schmidt, Massachusetts General Hospital, United States
Copyright © 2025 Wang, Ayala, Aziz, Billiar, Deutschman, Jeyaseelan, Tang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haichao Wang, aHdhbmdAbm9ydGh3ZWxsLmVkdQ==
 Haichao Wang
Haichao Wang Alfred Ayala
Alfred Ayala Monowar Aziz
Monowar Aziz Timothy R. Billiar
Timothy R. Billiar Clifford S. Deutschman
Clifford S. Deutschman Samithamby Jeyaseelan
Samithamby Jeyaseelan Daolin Tang
Daolin Tang Ping Wang
Ping Wang