- Immunoregulation Unit, Department of Immunology, Institut Pasteur, Université Paris Cité, Paris, France
Mucosal-associated invariant T (MAIT) cells are innate-like T cells that express a semi-invariant T cell receptor (TCR). These cells predominantly reside in tissues, such as the liver, lung, skin and the gastrointestinal tract. MAIT cells can be activated via their TCR that recognizes riboflavin metabolites presented by the MHC class I-related protein 1 (MR1). These cells can also be activated in a TCR-independent manner by cytokines, in particular IL-12 and IL-18, but also by type I interferons, IL-7, IL-15 and IL-23, underlining their innate-like characteristics. MAIT cells have important functions in antibacterial and viral immunity but also in tissue repair and homeostasis. Recent studies highlighted the plasticity of MAIT cells in response to cytokines, suggesting an important role of the cytokine milieu in modulating MAIT cell functions. Here, we discuss how cytokines control MAIT cell functions in various contexts.
1 Introduction
Mucosal-associated invariant T (MAIT) cells are a subset of innate-like T cells characterized by their semi-invariant T cell receptor (TCR) composed of TRAV1–2 paired with TRAJ33, TRAJ12 or TRAJ20 in human and Trav1 paired with Traj33 in mouse; with a CDR3 region of constant length (1, 2). They are restricted by the MHC class I-related protein 1 (MR1) presenting metabolites derived from the riboflavin (vitamin B2) synthesis pathway, the most potent ligand being 5-OP-RU (5-(2-oxopropylideneamino)-6-D-ribitylaminouracil) (3, 4). In mammals, there is a striking conservation of MR1 and the TRAV1–2 and TRAJ33 genes (5), indicating strong selective pressure and suggesting that MAIT cells have important non-redundant functions.
MAIT cells are found in tissues at various frequencies. In humans, they represent in average 3% of CD3+ peripheral blood cells, are enriched in barrier tissues such as the digestive tract and the lungs, and are particularly abundant in the liver (6).
The MAIT cell phenotype has been extensively described. In humans, they are mainly CD8+ (~80%) and double negative (CD4-CD8-, ~15%) with very few CD4+ cells (7, 8), while murine MAIT cells are mostly double negative (9). Based on co-receptor expression patterns, some groups identified distinct MAIT cell subsets (10, 11), while others only detected minor differences and suggest that they rather belong to a continuum (12). MAIT cells have a CD45RA-CD45RO+CD95highCD62Llow effector memory phenotype. They express a specific pattern of chemokine receptors, with high levels of CCR6, CXCR6 and CCR5, intermediate levels of CCR9 and heterogenous expression of CXCR4, which endow their tissue tropism. MAIT cells also express some natural killer (NK) cell markers at heterogeneous levels including NKp30, NKp80, CD56, NKG2A and NKG2D (7, 13). The MAIT cell phenotype is also characterized by the expression of a broad range of cytokine receptors including interleukin 7 receptor (IL-7R), IL-18R, IL-12R, IL-15R and IL-23R (7, 13–17). MAIT cells are also defined by the expression of a specific set of transcription factors. They express the Promyelocytic Leukemia Zinc Finger protein (PLZF, encoded by ZBTB16), a critical transcription factor for the acquisition of innate-like functions (7, 18, 19). Human MAIT cells express high levels of retinoic-acid related orphan receptor gamma (RORγt) and intermediate levels of T-bet (encoded by TBX21), conferring them mixed type 1/17 phenotype and functions (7, 13, 20). In the mouse, MAIT cells are differentiated into MAIT1 and MAIT17 subsets, which express either T-bet or RORγt, respectively (21, 22).
MAIT cell activation can be achieved in a TCR-dependent manner, via the presentation of riboflavin metabolites on MR1 by antigen presenting cells. However, TCR signaling alone is not sufficient to induce full MAIT cell activation and requires additional signals that can be provided through co-stimulatory molecules such as CD28, or innate cytokines (23). Additionally, MAIT cells can be activated in a TCR-independent manner by cytokines, such as the combination of IL-12 and IL-18 (24). MAIT cell activation and localization are also modulated by other factors such as chemokines or prostaglandins (25, 26). In response to these signals, MAIT cells can perform a broad range of functions, including antimicrobial and antiviral defense, or tissue repair (27–29). Here, we review how cytokines modulate these various MAIT cell functions.
2 Role of cytokines in MAIT cell development
MAIT cell development is a three-stage process that has recently been reviewed elsewhere (30). Here, we will focus on the importance of cytokine signals at different stages of this process.
In the thymus, murine MAIT cells are selected by double positive thymocytes expressing MR1 and are initially CD24+CD44- (stage 1). They differentiate into an immature stage 2 with CD24 downregulation and expression of CD62L. Stage 3 MAIT cells are CD24-CD44+, acquire PLZF expression and differentiate into MAIT1, expressing T-bet, and MAIT17, expressing RORγt, subsets (22, 30, 31). Similarly, human MAIT cell development stages are defined by the expression patterns of CD27 and CD161. Stage 1 MAIT cells are CD27-CD161-, differentiation into stage 2 is defined by the acquisition of CD27 expression, and stage 3 cells express CD161. Unlike in mice, human MAIT cells do not commit to type 1 or 17 lineages, but rather exhibit a homogeneous mixed 1/17 phenotype (13, 32–34).
In mouse, IL-18 is important for stage 2 to stage 3 transition as IL-18-deficient mice display reduced frequencies of stage 3 MAIT cells in the thymus and in peripheral tissues (31). Single cell transcriptomes and TCR repertoires analyses revealed that commitment to the MAIT1 or MAIT17 subsets is independent of TCR characteristics, suggesting the involvement of other signals which could likely be provided by cytokines (35). Supporting this model, a study identified IL-15 and IL-2 signaling through CD122 (IL-2Rβ) to be critical specifically for MAIT1 cell development and/or maintenance, while the co-stimulatory molecule ICOS was necessary for the MAIT17 subset development (36).
After thymus egress, murine MAIT cell subsets populate different tissues: MAIT1 preferentially colonize the spleen and liver; MAIT17 are mostly found at barrier sites including the lung, gut and skin (9, 37). The cues regulating this differential tissue colonization are not completely understood, yet there is some evidence that cytokine signals are involved. Indeed, IL-23R-deficient mice exhibit an impaired MAIT cell compartment in the skin (38). IL-23-deficient mice have reduced MAIT cell frequencies in the ileum and colon, and their type 1/17 tissue-specific phenotype is altered (33). Yet, MAIT cell numbers are normal in the lungs of Il23a-/- animals (39). Of note, MAIT cell numbers are also normal in the lungs of Ifng-/-, Il18-/-, Il6-/- and Il12a-/- mice under homeostatic conditions (39). Together, these studies point to a critical role for IL-23 signaling in the establishment of a MAIT cell population in the skin and in the gut but not in the lung, highlighting the importance of tissue-specific cues, including cytokines, in controlling MAIT cell tissue localization.
In human, inborn errors of immunity (IEI) - a heterogeneous group of diseases in which a germline variant causes defects in the immune system - provide invaluable insights into critical components of MAIT cell biology [reviewed in (40)]. A complete lack of MAIT cells has been observed in individuals with MR1 (41) or RORγt (42) deficiencies, highlighting that these proteins are essential for the development and/or maintenance of a MAIT cell population. In most IEI cases impacting the MAIT cell compartment, a reduced frequency of circulating MAIT cells is reported. These include variants in several cytokine receptors namely IL12RB1, IL12RB2, IL21R, IL23R and IL6ST (43–46). This suggests a critical role for these cytokines in the development and/or maintenance of human MAIT cells, yet the underlying mechanisms remain to be deciphered.
3 Tissue repair functions
In the past few years, tissue repair functions have been attributed to MAIT cells [reviewed in (47)]. Transcriptomic analyses of human and mouse MAIT cells have largely attributed the expression of this tissue repair program to TCR triggering (48–50). More recently in a mouse model of skin excision, du Halgouet et al. showed that MAIT cells exhibited a tissue repair program, were recruited to skin lesions and accelerated wound closure independently of TCR signaling. IL-18 was identified as an important inducer of amphiregulin production by MAIT cells, that was critical for their tissue repair functions (51). Thus, tissue repair functions of MAIT cells can be triggered by different modes of activation that could depend on the environment. More studies are required to fully understand the extent of MAIT cell tissue repair program, and its function in different contexts such as chronic inflammation versus acute lesion.
4 Fundamental studies on the effects of cytokines on MAIT cell functions
4.1 Combination of IL-12 and IL-18
The first studies identifying MAIT cell TCR-independent activation revealed that the combination of IL-12 and IL-18, that was previously shown to mediate NK cell activation (52), could potently activate MAIT cells and induce production of interferon γ (IFN-γ) (24, 53, 54). Further studies aimed at characterizing this TCR-independent response in vitro. TCR- and cytokine-mediated activation of MAIT cells have different kinetics and induce distinct responses. Cytokine-mediated activation peaks after 20–24 hours while TCR signaling is faster with the activation peak reached after 6 hours (24, 49). IL-12/IL-18 stimulation leads mainly to the production of IFN-γ, while TCR stimulation induces a more polyfunctional profile. The induced cytotoxic profiles are also different with increased production of granzymes A, K and M by cytokine-activated MAIT cells (49). Additionally, although activation of MAIT cells results in a core activated transcriptomic signature, the two modes of activation lead to distinct transcriptomic profiles. Finally, it is noteworthy that there is a synergy between the two modes of activation, that results in enhanced effector functions and a specific transcriptomic profile (49, 50).
Further studies have aimed at deciphering specific mechanisms underlying IL-12 and IL-18-mediated MAIT cell activation. Specifically, it has been demonstrated that IL-12 and IL-18 synergize together and with TCR signaling for optimal IL-17 production (55). IL-15 and tumor necrosis factor (TNF)-like protein 1A (TL1A) also enhanced effector functions of human blood and gut MAIT cells stimulated in vitro with suboptimal concentrations of IL-12 and IL-18 (50). A cocktail of inflammatory cytokines containing IL-12, IL-18 and IL-15 promoted sustained CTLA-4 expression on MAIT cells (56).
4.2 Individual cytokines
Besides IL-12R and IL-18R, MAIT cells express a broad range of cytokine receptors that prompted the analysis of the effects of other cytokines on their activation. The effects of these cytokines on MAIT cell responses are summarized in Figure 1. Specifically, MAIT cells express very high levels of IL-7R (14). IL-7 stimulation induced proliferation of peripheral CD161highIL-18Rα+CD8+ T cells and liver-derived MAIT cells (57, 58). Several studies identified IL-7 as an important cytokine promoting IL-17A production by MAIT cells. Indeed, priming of human peripheral and liver MAIT cells with IL-7 before TCR stimulation resulted in enhanced MAIT cell activation and secretion of IFN-γ and IL-17A, while priming with the classical type 17 inducing cytokines IL-1β and IL-23 mostly induced IFN-γ production (14). Similarly, priming of peripheral MAIT cells from axial spondyloarthritis patients with IL-7 but not with IL-23 increased activation and IL-17A production (59). A study on MAIT cells from the salivary glands of primary Sjogren’s Syndrome patients revealed that IL-17A production was induced in vitro by two different pathways. IL-7 induced IL17A concomitantly with STAT3, HIF1A and a decrease of RORC, while IL-23 increased IL17A expression together with the master type 17 transcription factor RORC (60). Additionally, IL-7 stimulation with a low dose of fixed E. coli induced production of IL-17A, IFN-γ and cytotoxic mediators by MAIT cells (20).
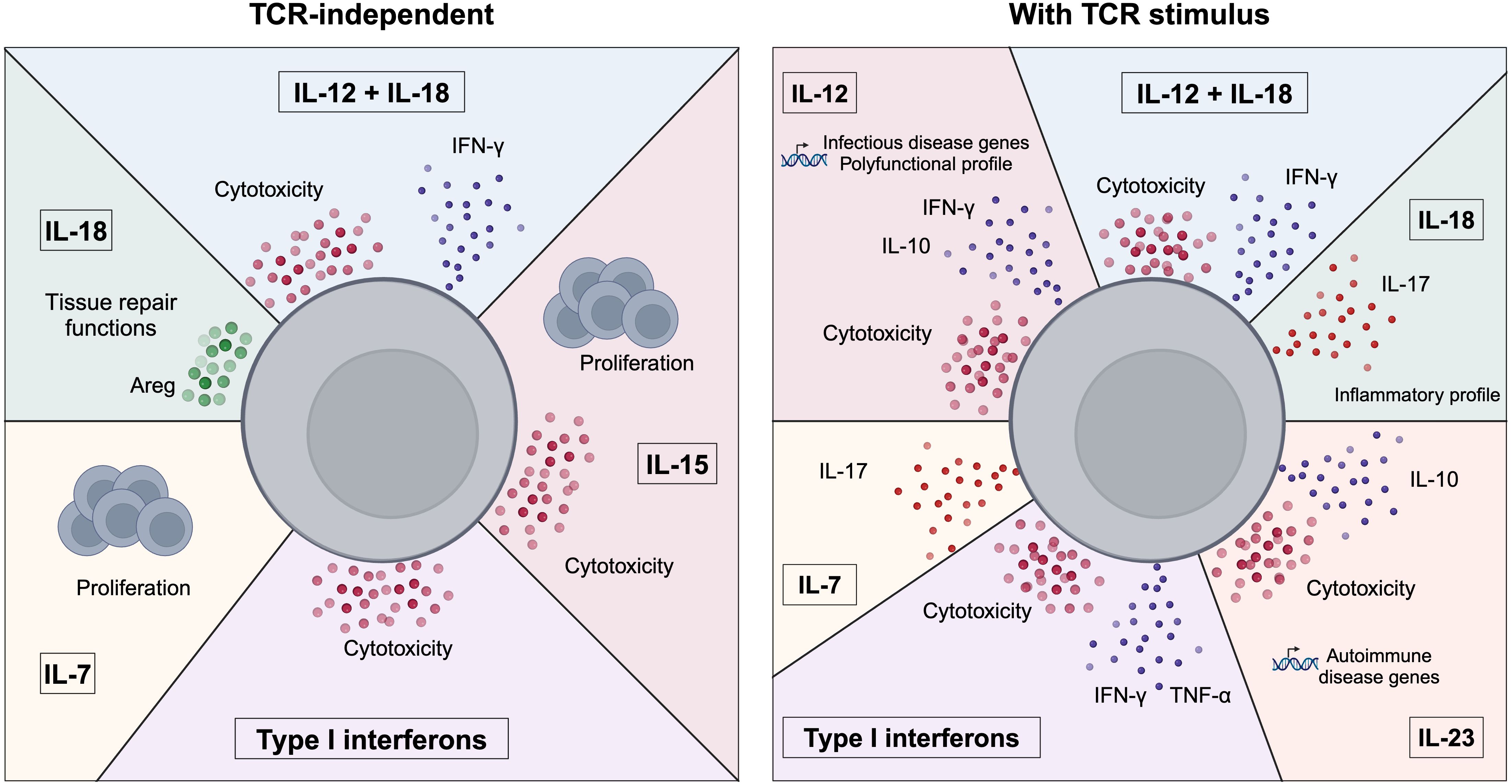
Figure 1. Control of MAIT cell functions by cytokines. Summary of the effects of cytokines on MAIT cell functions. MAIT cells can be activated in the absence of a TCR stimulus (left panel) by cytokines such as IL-7, IL-18, IL-15, type I interferons or the combination of IL-12+IL-18. MAIT cells exhibit various responses to cytokines as they can secrete pro-inflammatory cytokines (e.g. IFN-γ), cytotoxic mediators or adopt a tissue repair program. The cytokine milieu can also influence MAIT cell functions in the presence of a TCR stimulus (right panel). MAIT cells can adopt distinct functional and transcriptional profiles in response to different cytokines, highlighting their plasticity.
Another important cytokine in MAIT cell biology is IL-15. Similarly to IL-7, stimulation with IL-15 alone induced proliferation of peripheral and liver-derived MAIT cells (57, 58). In liver-derived MAIT cells, IL-15 induced the highest cytotoxic killing capabilities compared to IL-2, IL-7 and IL-12 which induced modest killing. This mechanism is TCR-independent and mediated by NKG2D and granzyme B (58). IL-15 stimulation of peripheral blood mononuclear cells (PBMC) also induced cytotoxicity in MAIT cells, accompanied by IFN-γ production, through a MR1-independent mechanism and mediated by IL-18, mainly secreted by monocytes in this context. However, adding IL-18 alone to PBMC cultures was not sufficient to recapitulate MAIT cell activation induced by IL-15 and trigger IFN-γ production, highlighting the importance of the integration of multiple signals to control MAIT cell effector functions (61). In a co-culture model of MAIT cells with mesenchymal stem cells, MAIT cell activation was enhanced by enhancing autophagy in these cells via IL-15 and independently of MR1 and cell-cell contact (62).
Type I interferons, mainly IFN-α and IFN-β, are important activators of MAIT cells. Lamichhane et al. revealed that type I interferons alone were able to activate cytotoxic responses of MAIT cells, and combined with TCR stimulation additionally enhanced effector cytokine secretion (63). Type I interferons are also important in mediating the effects of toll like receptor (TLR) 7 and 8 signaling in PBMC cultures. In particular, IFN-α enhanced MAIT cell response to anti-CD3 stimulation only when added first, indicating a priming role for type I interferons rather than a co-activator role per se (64).
5 Cytokines drive functional MAIT cell plasticity
Although human (as well as sheep, cattle, opossum and bat) MAIT cells are a rather homogeneous population (33, 65), they exhibit a broad spectrum of functions, suggesting that their responses can be adapted to their environment. Bulk transcriptional analyses revealed that TCR and cytokine stimulations elicited distinct transcriptional programs in MAIT cells (49, 50). This was further validated using single cell transcriptomics (12). Specifically, the transcriptional responses to TCR or IL-12/IL-18 stimulation followed distinct activation trajectories. Analysis of changes in transcription factor activity identified specific regulons for each of the two modes of activation, for example STAT1 and IKZF1 were specific for cytokine-mediated activation. Stimulation with both TCR and cytokines induced an IL-17-expressing population, that was not detected with either of the single stimulation, highlighting the synergy of these signals. Except for the expression of IL-17, this population was not transcriptionally, or clonally different from the non-expressing ones, suggesting that they do not represent a distinct subset of cells but result from functional plasticity (12). We and others also profiled MAIT cell responses to various cytokines in the presence of a TCR stimulus and identified different functional polarizations (17, 66). IL-23 enhanced IL-10 production, cytotoxicity and expression of autoimmune-related genes, mediated by the AP-1 family member Basic Leucine Zipper ATF-Like Transcription Factor (BATF). IL-18 polarized MAIT cells to an inflammatory profile, and drove IL-17 production (66) while IL-12 induced a diverse profile, including immunoregulatory mediators such as IL-10 and infectious disease related genes (17, 66). IL-12 induced IL-10 secretion in a c-Maf-dependent manner (66). These studies underline the functional and transcriptional adaptability of MAIT cells to the cytokine environment, and their capability to adopt not only inflammatory profiles but also to mediate anti-inflammatory responses.
A report in the mouse also highlighted functional plasticity of MAIT cells. In this model, MAIT17 cells can convert into functional MAIT1 cells that protected mice against bacterial infections (67). Altogether, the cytokine environment is important in driving MAIT cell plastic responses.
6 Cytokines modulate antibacterial activity of MAIT cells
The most appreciated role of MAIT cell is their antibacterial activity, mediated by the recognition of bacteria through MR1 presenting 5-OP-RU derived from bacteria (4, 27). However, the antibacterial responses do not entirely rely on antigen-dependent activation of MAIT cells and involve additional signals that can be provided by cytokines.
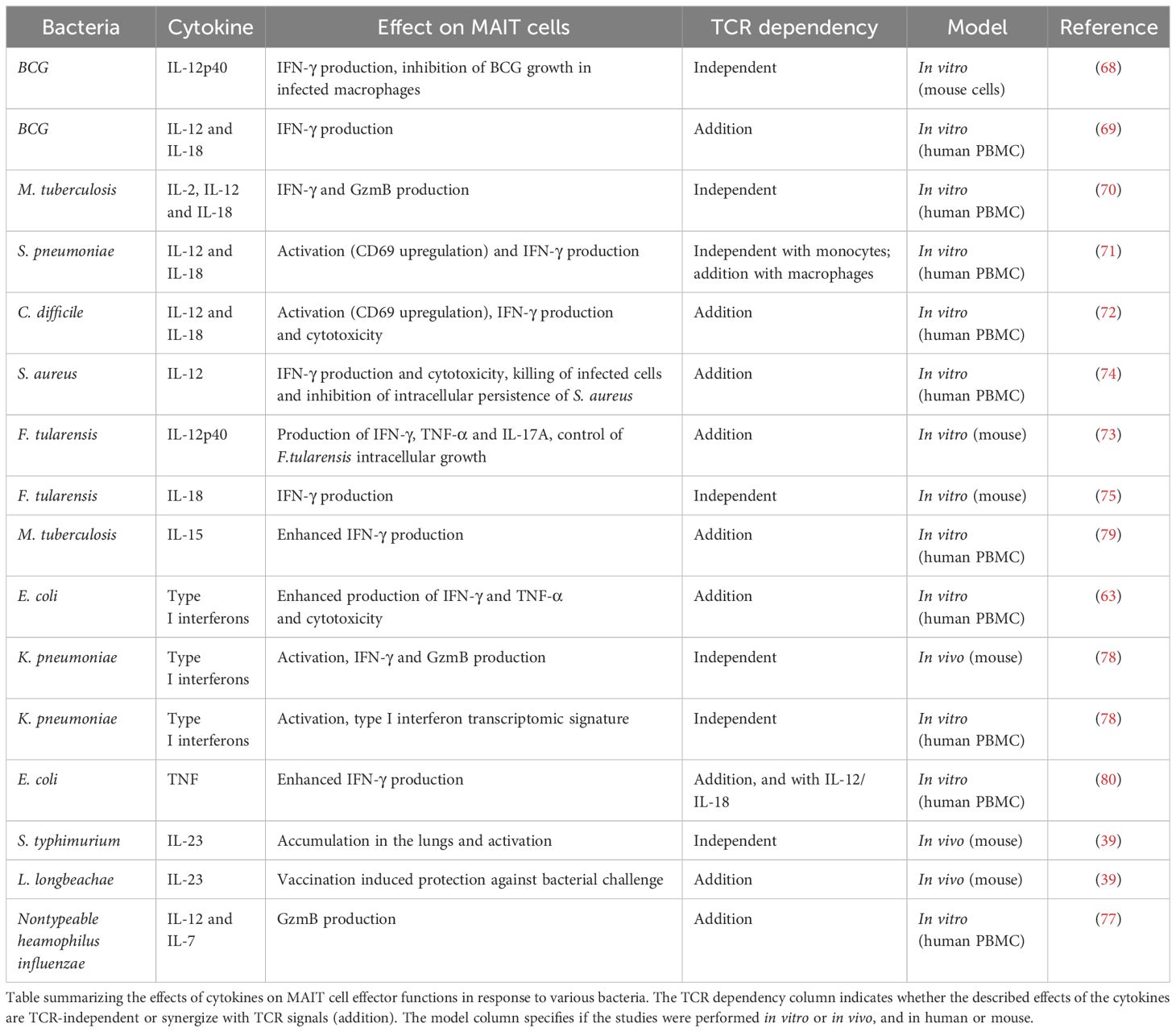
Several studies identified IL-12 and IL-18 as key cytokines mediating MAIT cell antibacterial activity by promoting IFN-γ production and cytotoxicity. These interleukins are important for in vitro MAIT cell responses to various bacteria including Bacillus Calmette-Guérin (BCG, 59, 60), M. tuberculosis (70), S. pneumoniae (71), C. difficile (72), F. tularensis (73) or S. aureus (74). It is noteworthy that the need for additional TCR signal depends on the context since in response to BCG one study on mouse cells described a MR1-independent mechanism (68), while another one using human PBMC revealed a synergy of TCR and cytokine signaling (69). Interestingly, the relative importance of TCR or cytokine signaling for MAIT cell activation was dependent on the antigen presenting cell (71). Further highlighting the importance of the environment in controlling MAIT cell functions, a study investigating the role of IL-18 in Francisella infections demonstrated that this cytokine was critical for in vitro production of IFN-γ by MAIT cells, but was dispensable in vivo (75). IL-12 and IL-23 were crucial in promoting MAIT cell responses in vivo upon F. tularensis infection as mice lacking either one of these cytokines exhibited impaired MAIT1 cell numbers in the lung. It is noteworthy that Il12p40-/- mice, lacking both cytokines, had a more severe defect than Il12p35-/- or Il23p19-/- deficient in IL-12 or IL-23 respectively; pointing to nonredundant or synergistic functions of these interleukins in MAIT1 cell responses (76). Administration of IL-12 during L. longbeachae (MAIT17-polarizing) infection resulted in dose-dependent increased MAIT1 responses, highlighting the modulation of MAIT cell responses to bacterial infection by the cytokine milieu (76). IL-12 was also shown to synergize with IL-7 secreted by macrophages and MR1 signaling upon Nontypeable Haemophilius influenzae infection, to induce granzyme B production by MAIT cells (77).
Furthermore, type I interferons have been shown to be important to enhance MAIT cell effector functions in response to E. coli in vitro (63). In K. pneumoniae infection in the mouse, MAIT cells were activated by type I interferon, produced IFN-γ and granzyme B and had a protective role. In human MAIT cells, transcriptomic analyses revealed that K. pneumoniae induced a type I interferon signature; and MAIT cell activation was dependent on type I interferon. Interestingly, K. pneumoniae-mediated activation of both murine and human MAIT cells was MR1-independent, even though this bacteria possesses the riboflavin pathway (78). Other studies also highlighted the role of IL-15 and TNF-α in enhancing MAIT cell effector responses to M. tuberculosis and E. coli challenges respectively (79, 80). Additionally, IL-23 was important for MAIT cell activation and accumulation in the lungs in mice infected with S. typhimurium, and vaccination using a combination of 5-OP-RU and IL-23 induced protection against L. longbeachae (39).
Altogether, various cytokines can modulate antibacterial activity of MAIT cells. The effects of cytokines on MAIT cells in response to bacteria are summarized in Table 1. Remarkably, there is a complex integration of antigen and cytokine signals that depend on the context and allow fine tuning of MAIT cell responses.
7 Cytokines drive MAIT cell responses to viral infections
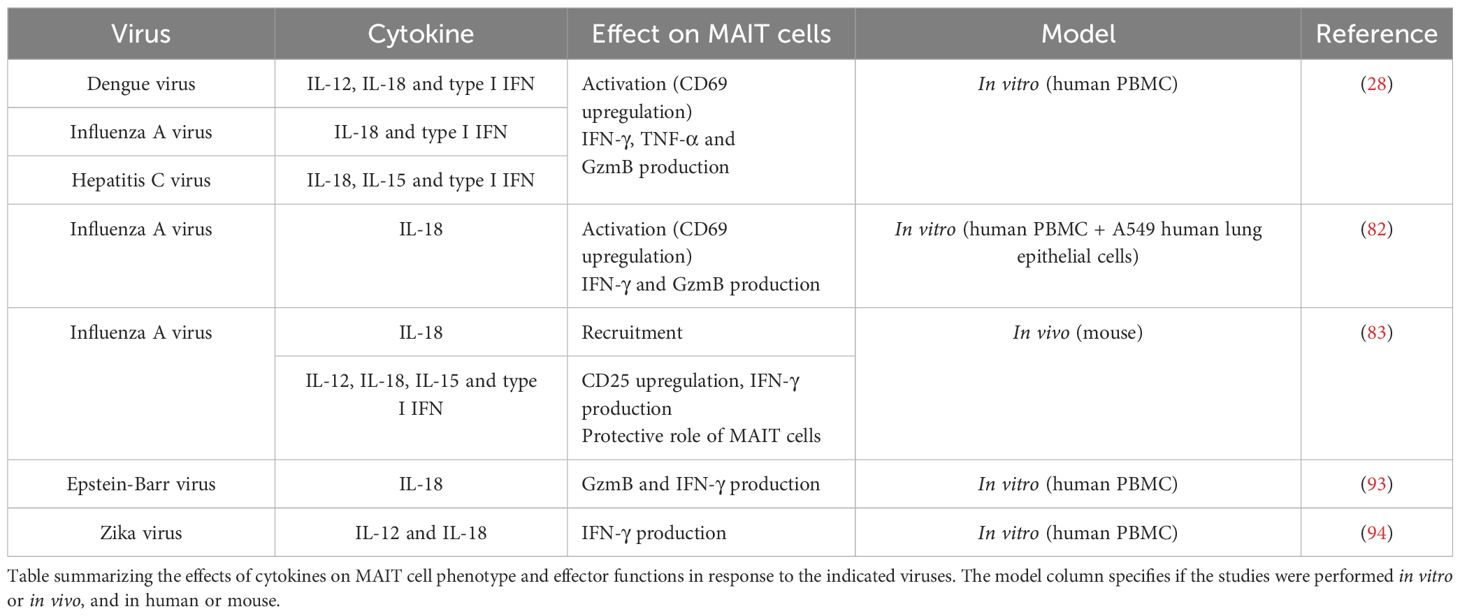
Albeit initial studies suggested that MAIT cells could not respond to viral infections but were rather antibacterial (27, 81), it is now clear that MAIT cells can be activated by viruses through cytokines, in a TCR-independent manner. Table 2 summarizes the effects of cytokines on MAIT cells in response to various viruses.
Van Wilgenburg et al. showed that human MAIT cells are activated in vitro by antigen presenting cells infected with various viruses including dengue virus, influenza or hepatitis C virus. Viral activation of MAIT cells was not dependent on TCR signaling, but mediated by cytokines (IL-18, IL-15 and type I interferons) with a central role for IL-18. Importantly, the activation required more than one cytokine, and the ones involved depended on the virus (28). Furthermore, co-culture of PBMC with lung epithelial cells infected with influenza A virus (IAV) elicited MAIT cell activation, which was mediated by IL-18 and required CD14+ monocytes (82). Upon IAV infection in vivo, MAIT cell recruitment was impaired by IL-18 deficiency, while their activation was affected by deficiency of IL-15, IL-18, IFNαR and most dramatically of IL-12; indicating the involvement of various cytokines in coordinating MAIT cell responses to viral infections. In this context, MAIT cells protected against lethal influenza infection, at least in part through production of IFN-γ (83). In a mouse model of bleomycin-induced sterile lung injury, MAIT cells were similarly recruited and activated by IFN-α and IL-18 and had protective functions (84).
Further studies have demonstrated the antiviral functions of MAIT cells against other viruses. Notably, MAIT cells were shown to decline in the circulation of patients with active Covid-19 disease, and exhibited a strongly activated phenotype (85–88). The MAIT cell phenotype correlated with disease severity and IL-18 plasma concentration, and the authors identified a monocyte/macrophage shift from IFN-α to IL-18 production. This suggested that MAIT cell functions were altered by the pro-inflammatory environment, in particular IL-18, and may contribute to disease severity (86). Another study identified defects in MAIT cell responses from Covid-19 patients and proposed a mechanism by which IFN-α triggered important IL-10 production by suppressive monocytes, which impaired MAIT cell responses (88). Altered responses to IL-12/IL-18 stimulation were observed in vitro with MAIT cells from Covid-19 patients, and were partially rescued by IL-7 stimulation (87).
In human immunodeficiency virus (HIV) infections, several studies identified reduced frequencies and impaired responses of MAIT cells in the peripheral blood of patients (20, 89–91). IL-7 was important in arming MAIT cells with cytotoxic capabilities that were defective in HIV patients (20), and IL-7 treatment restored MAIT cell frequency in HIV-infected patients (92). Variants in IL7RA were also associated with the frequency and functionality of MAIT cells in HIV patients (90). A study suggested a mechanism to explain impaired MAIT cell functionality in HIV-infected patients: sustained type I interferon signaling induced IL-10 production by monocytes, and reduced IL-12 that together reduced MAIT cell antibacterial responsiveness (89).
MAIT cells also exhibited decreased frequency and activated phenotype in the blood of patients with Epstein-Barr virus (EBV)-associated T/NK lymphoprolipherative disorder, in correlation with disease severity and IL-18 plasma concentration. In vitro infection of PBMC with EBV induced IL-18 secretion by monocytes, and blocking this cytokine reduced MAIT cell activation suggesting that IL-18 may be important in MAIT cell phenotype in this disease (93). Similarly, in dengue infected patients, MAIT cells had an activated phenotype, and their activation in vitro by Zika virus was blocked when antibodies blocking IL-12 and IL-18 were added to the culture (94).
8 MAIT cells in inflammatory diseases
It has been reported in many inflammatory diseases, such as inflammatory bowel disease (IBD, 85, 86), multiple sclerosis (97), type 1 diabetes (98) or primary Sjogren’s Syndrome (60, 99), that MAIT cell frequency is reduced in the periphery [reviewed in (100)]. Given the chemokine receptor pattern expressed by MAIT cells, it was suggested that these cells were recruited to the inflamed tissues. Supporting this hypothesis, MAIT cells are reduced in the blood and enriched in the inflamed mucosa of IBD patients (95, 96). The plasma levels of pro-inflammatory cytokines such as IL-18 have been correlated with MAIT cell frequencies and phenotype (97). These observations together with the known responsiveness of MAIT cells to cytokine stimuli suggest that they could have a role in these diseases. However, to our knowledge, no mechanism of MAIT cell activation by cytokines in the context of inflammatory diseases has been described to date. The precise functions of MAIT cells in inflammatory diseases and the underlying mechanisms remain to be deciphered.
9 Discussion
Cytokines are key modulators of MAIT cell functions, as they can drive MAIT cell activation in the absence of TCR signaling in an innate-like manner. Since they modulate or synergize with the effects of TCR signaling, cytokines are also important in MAIT cell responses to antigen recognition. Many reports have focused on IL-12 and IL-18 to identify the TCR-independent activation programs of MAIT cells, or in the context of bacterial and viral infections. Yet, MAIT cells express a broad range of cytokine receptors, rendering them responsive to many other signals that remain underexplored. For example, IEI suggest important roles for IL-21 in human MAIT cell biology (46). However, to our knowledge, the effects of IL-21 on MAIT cell have not been assessed.
Furthermore, many studies highlight the importance of the context and the integration of multiple signals to fully activate MAIT cells and fine tune their functions. Thus, it would be of interest to further explore how different combinations of cytokines, other than IL-12/IL-18, in the presence or absence of TCR stimulus, can regulate the functions of MAIT cells. Additionally, further work is required to decipher how cytokine signals are integrated with various cues such as chemokines, interactions with other cells or the microbiota. These are indeed important for MAIT cells to adopt their broad effector functions in tissue-specific contexts.
There is an increasing number of reports denoting functional plasticity of MAIT cells, driven by the cytokine milieu. This sensing of the microenvironment enables MAIT cells to finely adapt their functions, either pro- or anti-inflammatory, to their tissue localization and to the homeostatic or inflammatory contexts. More work is required to decipher the underlying mechanisms, including the transcriptional networks and epigenetic processes possibly involved.
Author contributions
LC: Visualization, Writing – original draft. EB: Writing – review & editing. LR: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. LC is supported by a PhD fellowship from the Université Paris Cité. Work in the authors’ laboratory is supported by institutional funds from Institut Pasteur, grants from the Fondation de la Recherche Médicale (Equipe FRM, EQU202303016264). The authors declare that this study received funding from Janssen Pharmaceuticals (Madeleine project to LR). The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article, or the decision to submit it for publication.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Tilloy F, Treiner E, Park S-H, Garcia C, Lemonnier F, de la Salle H, et al. An invariant T cell receptor α Chain defines a novel TAP-independent major histocompatibility complex class Ib–restricted α/β T cell subpopulation in mammals. J Exp Med. (1999) 189:1907–21. doi: 10.1084/jem.189.12.1907
2. Treiner E, Duban L, Bahram S, Radosavljevic M, Wanner V, Tilloy F, et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature. (2003) 422:164–9. doi: 10.1038/nature01433
3. Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. (2012) 491:717–23. doi: 10.1038/nature11605
4. Corbett AJ, Eckle SBG, Birkinshaw RW, Liu L, Patel O, Mahony J, et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. (2014) 509:361–5. doi: 10.1038/nature13160
5. Boudinot P, Mondot S, Jouneau L, Teyton L, Lefranc M-P, and Lantz O. Restricting nonclassical MHC genes coevolve with TRAV genes used by innate-like T cells in mammals. Proc Natl Acad Sci. (2016) 113:E2983–92. doi: 10.1073/pnas.1600674113
6. Provine NM and Klenerman P. MAIT cells in health and disease. Annu Rev Immunol. (2020) 38:203–28. doi: 10.1146/annurev-immunol-080719-015428
7. Gherardin NA, Souter MNT, Koay HF, Mangas KM, Seemann T, Stinear TP, et al. Human blood MAIT cell subsets defined using MR1 tetramers. Immunol Cell Biol. (2018) 96:507–25. doi: 10.1111/imcb.12021
8. Souter MNT, Awad W, Li S, Pediongco TJ, Meehan BS, Meehan LJ, et al. CD8 coreceptor engagement of MR1 enhances antigen responsiveness by human MAIT and other MR1-reactive T cells. J Exp Med. (2022) 219:e20210828. doi: 10.1084/jem.20210828
9. Rahimpour A, Koay HF, Enders A, Clanchy R, Eckle SBG, Meehan B, et al. Identification of phenotypically and functionally heterogeneous mouse mucosal-associated invariant T cells using MR1 tetramers. J Exp Med. (2015) 212:1095–108. doi: 10.1084/jem.20142110
10. Dias J, Boulouis C, Gorin J-B, van den Biggelaar RHGA, Lal KG, Gibbs A, et al. The CD4–CD8– MAIT cell subpopulation is a functionally distinct subset developmentally related to the main CD8+ MAIT cell pool. Proc Natl Acad Sci. (2018) 115:E11513–22. doi: 10.1073/pnas.1812273115
11. Vorkas CK, Krishna C, Li K, Aubé J, Fitzgerald DW, Mazutis L, et al. Single-cell transcriptional profiling reveals signatures of helper, effector, and regulatory MAIT cells during homeostasis and activation. J Immunol. (2022) 208:1042–56. doi: 10.4049/jimmunol.2100522
12. Garner LC, Amini A, FitzPatrick MEB, Lett MJ, Hess GF, Filipowicz Sinnreich M, et al. Single-cell analysis of human MAIT cell transcriptional, functional and clonal diversity. Nat Immunol (2023) 24:1565–78. doi: 10.1038/s41590-023-01575-1
13. Dusseaux M, Martin E, Serriari N, Péguillet I, Premel V, Louis D, et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161 hi IL-17-secreting T cells. Blood. (2011) 117:1250–9. doi: 10.1182/blood-2010-08-303339
14. Tang X-Z, Jo J, Tan AT, Sandalova E, Chia A, Tan KC, et al. IL-7 licenses activation of human liver intrasinusoidal mucosal-associated invariant T cells. J Immunol. (2013) 190:3142–52. doi: 10.4049/jimmunol.1203218
15. Billerbeck E, Kang Y-H, Walker L, Lockstone H, Grafmueller S, Fleming V, et al. Analysis of CD161 expression on human CD8+ T cells defines a distinct functional subset with tissue-homing properties. Proc Natl Acad Sci. (2010) 107:3006–11. doi: 10.1073/pnas.0914839107
16. Rosine N, Rowe H, Koturan S, Yahia-Cherbal H, Leloup C, Watad A, et al. Characterization of blood mucosal-associated invariant T cells in patients with axial spondyloarthritis and of resident mucosal-associated invariant T cells from the axial entheses of non-axial spondyloarthritis control patients. Arthritis Rheumatol. (2022) 74:1786–95. doi: 10.1002/art.42090
17. Camard L, Stephen T, Yahia-Cherbal H, Guillemot V, Mella S, Baillet V, et al. IL-23 tunes inflammatory functions of human mucosal-associated invariant T cells. iScience. (2025) 28:111898. doi: 10.1016/j.isci.2025.111898
18. Kreslavsky T, Savage AK, Hobbs R, Gounari F, Bronson R, Pereira P, et al. TCR-inducible PLZF transcription factor required for innate phenotype of a subset of gammadelta T cells with restricted TCR diversity. Proc Natl Acad Sci U.S.A. (2009) 106:12453–8. doi: 10.1073/pnas.0903895106
19. Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. (2008) 9:1055–64. doi: 10.1038/ni.1641
20. Leeansyah E, Svärd J, Dias J, Buggert M, Nyström J, Quigley MF, et al. Arming of MAIT cell cytolytic antimicrobial activity is induced by IL-7 and defective in HIV-1 infection. PloS Pathog. (2015) 11:e1005072. doi: 10.1371/journal.ppat.1005072
21. Legoux F, Gilet J, Procopio E, Echasserieau K, Bernardeau K, and Lantz O. Molecular mechanisms of lineage decisions in metabolite-specific T cells. Nat Immunol. (2019) 20:1244–55. doi: 10.1038/s41590-019-0465-3
22. Koay H-F, Su S, Amann-Zalcenstein D, Daley SR, Comerford I, Miosge L, et al. A divergent transcriptional landscape underpins the development and functional branching of MAIT cells. Sci Immunol. (2019) 4:eaay6039. doi: 10.1126/sciimmunol.aay6039
23. Turtle CJ, Delrow J, Joslyn RC, Swanson HM, Basom R, Tabellini L, et al. Innate signals overcome acquired TCR signaling pathway regulation and govern the fate of human CD161hi CD8α+ semi-invariant T cells. Blood. (2011) 118:2752–62. doi: 10.1182/blood-2011-02-334698
24. Ussher JE, Bilton M, Attwod E, Shadwell J, Richardson R, de Lara C, et al. CD161++ CD8+ T cells, including the MAIT cell subset, are specifically activated by IL-12+IL-18 in a TCR-independent manner. Eur J Immunol. (2014) 44:195–203. doi: 10.1002/eji.201343509
25. Mehta H, Tasin I, Hackstein CP, Willberg C, and Klenerman P. Prostaglandins differentially modulate mucosal-associated invariant T-cell activation and function according to stimulus. Immunol Cell Biol. (2023) 101:262–72. doi: 10.1111/imcb.12617
26. Wu Z, Chen X, Han F, and Leeansyah E. MAIT cell homing in intestinal homeostasis and inflammation. Sci Adv. (2025) 11:eadu4172. doi: 10.1126/sciadv.adu4172
27. Le Bourhis L, Martin E, Péguillet I, Guihot A, Froux N, Coré M, et al. Antimicrobial activity of mucosal-associated invariant T cells. Nat Immunol. (2010) 11:701–8. doi: 10.1038/ni.1890
28. Van Wilgenburg B, Scherwitzl I, Hutchinson EC, Leng T, Kurioka A, Kulicke C, et al. MAIT cells are activated during human viral infections. Nat Commun. (2016) 7:11653. doi: 10.1038/ncomms11653
29. Salou M and Lantz O. A TCR-dependent tissue repair potential of MAIT cells. Trends Immunol. (2019) 40:975–7. doi: 10.1016/j.it.2019.09.001
30. Salou M, Paiva RA, and Lantz O. Development and functions of MAIT cells. Annu Rev Immunol. (2025) 43:253–83. doi: 10.1146/annurev-immunol-082323-025943
31. Koay H-F, Gherardin NA, Enders A, Loh L, Mackay LK, Almeida CF, et al. A three-stage intrathymic development pathway for the mucosal-associated invariant T cell lineage. Nat Immunol. (2016) 17:1300–11. doi: 10.1038/ni.3565
32. Chandra S, Ascui G, Riffelmacher T, Chawla A, Ramírez-Suástegui C, Castelan VC, et al. Transcriptomes and metabolism define mouse and human MAIT cell populations. Sci Immunol. (2023) 8:eabn8531. doi: 10.1126/sciimmunol.abn8531
33. Bugaut H, El Morr Y, Mestdagh M, Darbois A, Paiva RA, Salou M, et al. A conserved transcriptional program for MAIT cells across mammalian evolution. J Exp Med. (2024) 221:e20231487. doi: 10.1084/jem.20231487
34. Loh L, Carcy S, Krovi HS, Domenico J, Spengler A, Lin Y, et al. Unraveling the phenotypic states of human innate-like T cells: Comparative insights with conventional T cells and mouse models. Cell Rep. (2024) 43:114705. doi: 10.1016/j.celrep.2024.114705
35. Karnaukhov VK, Le Gac A-L, Bilonda Mutala L, Darbois A, Perrin L, Legoux F, et al. Innate-like T cell subset commitment in the murine thymus is independent of TCR characteristics and occurs during proliferation. Proc Natl Acad Sci. (2024) 121:e2311348121. doi: 10.1073/pnas.2311348121
36. Tao H, Pan Y, Chu S, Li L, Xie J, Wang P, et al. Differential controls of MAIT cell effector polarization by mTORC1/mTORC2 via integrating cytokine and costimulatory signals. Nat Commun. (2021) 12:2029. doi: 10.1038/s41467-021-22162-8
37. Salou M, Legoux F, Gilet J, Darbois A, du Halgouet A, Alonso R, et al. A common transcriptomic program acquired in the thymus defines tissue residency of MAIT and NKT subsets. J Exp Med. (2019) 216:133–51. doi: 10.1084/jem.20181483
38. Constantinides MG, Link VM, Tamoutounour S, Wong AC, Perez-Chaparro PJ, Han SJ, et al. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science. (2019) 366:eaax6624. doi: 10.1126/science.aax6624
39. Wang H, Kjer-Nielsen L, Shi M, D’Souza C, Pediongco TJ, Cao H, et al. IL-23 costimulates antigen-specific MAIT cell activation and enables vaccination against bacterial infection. Sci Immunol. (2019) 4:1–15. doi: 10.1126/sciimmunol.aaw0402
40. Howson LJ and Bryant VL. Insights into mucosal associated invariant T cell biology from human inborn errors of immunity. Front Immunol. (2022) 13:1107609. doi: 10.3389/fimmu.2022.1107609
41. Howson LJ, Awad W, Borstel AV, Lim HJ, McWilliam HEG, Sandoval-Romero ML, et al. Absence of mucosal-associated invariant T cells in a person with a homozygous point mutation in MR1. Sci Immunol. (2020) 5:1–14. doi: 10.1126/SCIIMMUNOL.ABC9492
42. Okada S, Markle JG, Deenick EK, Mele F, Averbuch D, Lagos M, et al. Impairment of immunity to Candida and Mycobacterium in humans with bi-allelic RORC mutations. Science. (2015) 349:606–13. doi: 10.1126/science.aaa4282
43. Wilson RP, Ives ML, Rao G, Lau A, Payne K, Kobayashi M, et al. STAT3 is a critical cell-intrinsic regulator of human unconventional T cell numbers and function. J Exp Med. (2015) 212:855–64. doi: 10.1084/jem.20141992
44. Martínez-Barricarte R, Markle JG, Ma CS, Deenick EK, Ramírez-Alejo N, Mele F, et al. Human IFN-γ immunity to mycobacteria is governed by both IL-12 and IL-23. Sci Immunol. (2018) 3:eaau6759. doi: 10.1126/sciimmunol.aau6759
45. Béziat V, Tavernier SJ, Chen Y-H, Ma CS, Materna M, Laurence A, et al. Dominant-negative mutations in human IL6ST underlie hyper-IgE syndrome. J Exp Med. (2020) 217:e20191804. doi: 10.1084/jem.20191804
46. Cagdas D, Mayr D, Baris S, Worley L, Langley DB, Metin A, et al. Genomic spectrum and phenotypic heterogeneity of human IL-21 receptor deficiency. J Clin Immunol. (2021) 41:1272–90. doi: 10.1007/s10875-021-01031-5
47. Gao M and Zhao X. Insights into the tissue repair features of MAIT cells. Front Immunol. (2024) 15:1432651. doi: 10.3389/fimmu.2024.1432651
48. Hinks TSC, Marchi E, Jabeen M, Olshansky M, Kurioka A, Pediongco TJ, et al. Activation and in vivo evolution of the MAIT cell transcriptome in mice and humans reveals tissue repair functionality. Cell Rep. (2019) 28:3249–3262.e5. doi: 10.1016/j.celrep.2019.07.039
49. Lamichhane R, Schneider M, de la Harpe SM, Harrop TWR, Hannaway RF, Dearden PK, et al. TCR- or cytokine-activated CD8+ Mucosal-associated invariant T cells are rapid polyfunctional effectors that can coordinate immune responses. Cell Rep. (2019) 28:3061–3076.e5. doi: 10.1016/j.celrep.2019.08.054
50. Leng T, Akther HD, Hackstein CP, Powell K, King T, Friedrich M, et al. TCR and inflammatory signals tune human MAIT cells to exert specific tissue repair and effector functions. Cell Rep. (2019) 28:3077–3091.e5. doi: 10.1016/j.celrep.2019.08.050
51. du Halgouet A, Darbois A, Alkobtawi M, Mestdagh M, Alphonse A, Premel V, et al. Role of MR1-driven signals and amphiregulin on the recruitment and repair function of MAIT cells during skin wound healing. Immunity. (2023) 56:78–92.e6. doi: 10.1016/j.immuni.2022.12.004
52. Gorski KS, Waller EL, Bjornton-Severson J, Hanten JA, Riter CL, Kieper WC, et al. Distinct indirect pathways govern human NK-cell activation by TLR-7 and TLR-8 agonists. Int Immunol. (2006) 18:1115–26. doi: 10.1093/intimm/dxl046
53. Fergusson JR, Smith KE, Fleming VM, Rajoriya N, Newell EW, Simmons R, et al. CD161 defines a transcriptional and functional phenotype across distinct human T cell lineages. Cell Rep. (2014) 9:1075–88. doi: 10.1016/j.celrep.2014.09.045
54. Provine NM, Binder B, FitzPatrick MEB, Schuch A, Garner LC, Williamson KD, et al. Unique and common features of innate-like human Vδ2+ γδT cells and mucosal-associated invariant T cells. Front Immunol. (2018) 9:756. doi: 10.3389/fimmu.2018.00756
55. Cole S, Murray J, Simpson C, Okoye R, Tyson K, Griffiths M, et al. Interleukin (IL)-12 and IL-18 synergize to promote MAIT cell IL-17A and IL-17F production independently of IL-23 signaling. Front Immunol. (2020) 11:585134. doi: 10.3389/fimmu.2020.585134
56. Berkson JD, Slichter CK, DeBerg HA, Delaney MA, Woodward-Davis AS, Maurice NJ, et al. Inflammatory cytokines induce sustained CTLA-4 cell surface expression on human MAIT cells. Immunohorizons. (2020) 4:14–22. doi: 10.4049/immunohorizons.1900061
57. Havenith SHC, Yong SL, Henson SM, Piet B, Idu MM, Koch SD, et al. Analysis of stem-cell-like properties of human CD161++IL-18Rα+ memory CD8+ T cells. Int Immunol. (2012) 24:625–36. doi: 10.1093/intimm/dxs069
58. Rha M-S, Han JW, Kim JH, Koh J-Y, Park HJ, Kim SI, et al. Human liver CD8+ MAIT cells exert TCR/MR1-independent innate-like cytotoxicity in response to IL-15. J Hepatol. (2020) 73:640–50. doi: 10.1016/j.jhep.2020.03.033
59. Gracey E, Qaiyum Z, Almaghlouth I, Lawson D, Karki S, Avvaru N, et al. IL-7 primes IL-17 in mucosal-associated invariant T (MAIT) cells, which contribute to the Th17-axis in ankylosing spondylitis. Ann Rheum Dis. (2016) 75:2124–32. doi: 10.1136/annrheumdis-2015-208902
60. Guggino G, Di Liberto D, Lo Pizzo M, Saieva L, Alessandro R, Dieli F, et al. IL-17 polarization of MAIT cells is derived from the activation of two different pathways. Eur J Immunol. (2017) 47:2002–3. doi: 10.1002/eji.201747140
61. Sattler A, Dang-Heine C, Reinke P, and Babel N. IL-15 dependent induction of IL-18 secretion as a feedback mechanism controlling human MAIT-cell effector functions. Eur J Immunol. (2015) 45:2286–98. doi: 10.1002/eji.201445313
62. Ye G, Wang P, Xie Z, Cao Q, Li J, Zheng G, et al. Autophagy-mediated activation of mucosal-associated invariant T cells driven by mesenchymal stem cell-derived IL-15. Stem Cell Rep. (2021) 16:926–39. doi: 10.1016/j.stemcr.2021.03.005
63. Lamichhane R, Galvin H, Hannaway RF, de la Harpe SM, Munro F, Tyndall JD, et al. Type I interferons are important co-stimulatory signals during T cell receptor mediated human MAIT cell activation. Eur J Immunol. (2020) 50:178–91. doi: 10.1002/eji.201948279
64. Pavlovic M, Gross C, Chili C, Secher T, and Treiner E. MAIT cells display a specific response to type 1 IFN underlying the adjuvant effect of TLR7/8 ligands. Front Immunol. (2020) 11:2097. doi: 10.3389/fimmu.2020.02097
65. Leeansyah E, Hey YY, Sia WR, Ng JHJ, Gulam MY, Boulouis C, et al. MR1-restricted T cells with MAIT-like characteristics are functionally conserved in the pteropid bat Pteropus alecto. iScience. (2020) 23:101876. doi: 10.1016/j.isci.2020.101876
66. Boulouis C, Mouchtaridi E, Müller TR, Mak JYW, Fairlie DP, Bergman P, et al. Human MAIT cell response profiles biased toward IL-17 or IL-10 are distinct effector states directed by the cytokine milieu. Proc Natl Acad Sci. (2025) 122:e2414230122. doi: 10.1073/pnas.2414230122
67. Wang H, Souter MNT, De Lima Moreira M, Li S, Zhou Y, Nelson AG, et al. MAIT cell plasticity enables functional adaptation that drives antibacterial immune protection. Sci Immunol. (2024) 9:eadp9841. doi: 10.1126/sciimmunol.adp9841
68. Chua W-J, Truscott SM, Eickhoff CS, Blazevic A, Hoft DF, and Hansen TH. Polyclonal mucosa-associated invariant T cells have unique innate functions in bacterial infection. Infect Immun. (2012) 80:3256–67. doi: 10.1128/iai.00279-12
69. Suliman S, Murphy M, Musvosvi M, Gela A, Meermeier EW, Geldenhuys H, et al. MR1-independent activation of human mucosal-associated invariant T cells by mycobacteria. J Immunol. (2019) 203:2917–27. doi: 10.4049/jimmunol.1900674
70. Jiang J, Chen X, An H, Yang B, Zhang F, and Cheng X. Enhanced immune response of MAIT cells in tuberculous pleural effusions depends on cytokine signaling. Sci Rep. (2016) 6:32320. doi: 10.1038/srep32320
71. Kurioka A, van Wilgenburg B, Javan RR, Hoyle R, van Tonder AJ, Harrold CL, et al. Diverse Streptococcus pneumoniae Strains Drive a Mucosal-Associated Invariant T-Cell Response Through Major Histocompatibility Complex class I–Related Molecule–Dependent and Cytokine-Driven Pathways. J Infect Dis. (2018) 217:988–99. doi: 10.1093/infdis/jix647
72. Bernal I, Hofmann JD, Bulitta B, Klawonn F, Michel A-M, Jahn D, et al. Clostridioides difficile activates human mucosal-associated invariant T cells. Front Microbiol. (2018) 9:2532. doi: 10.3389/fmicb.2018.02532
73. Meierovics A, Yankelevich W-JC, and Cowley SC. MAIT cells are critical for optimal mucosal immune responses during in vivo pulmonary bacterial infection. Proc Natl Acad Sci. (2013) 110:E3119–28. doi: 10.1073/pnas.1302799110
74. Cooper AJR, Clegg J, Cassidy FC, Hogan AE, and McLoughlin RM. Human MAIT cells respond to staphylococcus aureus with enhanced anti-bacterial activity. Microorganisms. (2022) 10:148. doi: 10.3390/microorganisms10010148
75. Jesteadt E, Zhang I, Yu H, Meierovics A, Chua Yankelevich W-J, and Cowley S. Interleukin-18 is critical for mucosa-associated invariant T cell gamma interferon responses to francisella species in vitro but not in vivo. Infect Immun. (2018) 86:e00117–18. doi: 10.1128/IAI.00117-18
76. Wang H, Nelson AG, Wang B, Zhao Z, Lim XY, Shi M, et al. The balance of IL-12 and IL -23 determines the bias of MAIT1 versus MAIT17 responses during bacterial infection. Immunol Cell Biol. (2022) 100:547–61. doi: 10.1111/imcb.12556
77. Wallington JC, Williams AP, Staples KJ, and Wilkinson TMA. IL-12 and IL-7 synergize to control mucosal-associated invariant T-cell cytotoxic responses to bacterial infection. J Allergy Clin Immunol. (2018) 141:2182–2195.e6. doi: 10.1016/j.jaci.2017.08.009
78. López-Rodríguez JC, Hancock SJ, Li K, Crotta S, Barrington C, Suárez-Bonnet A, et al. Type I interferons drive MAIT cell functions against bacterial pneumonia. J Exp Med. (2023) 220:e20230037. doi: 10.1084/jem.20230037
79. Jiang J, Yang B, An H, Wang X, Liu Y, Cao Z, et al. Mucosal-associated invariant T cells from patients with tuberculosis exhibit impaired immune response. J Infect. (2016) 72:338–52. doi: 10.1016/j.jinf.2015.11.010
80. Bánki Z, Krabbendam L, Klaver D, Leng T, Kruis S, Mehta H, et al. Antibody opsonization enhances MAIT cell responsiveness to bacteria via a TNF-dependent mechanism. Immunol Cell Biol. (2019) 97:538–51. doi: 10.1111/imcb.12239
81. Gold MC, Cerri S, Smyk-Pearson S, Cansler ME, Vogt TM, Delepine J, et al. Human mucosal associated invariant T cells detect bacterially infected cells. PloS Biol. (2010) 8:e1000407. doi: 10.1371/journal.pbio.1000407
82. Loh L, Wang Z, Sant S, Koutsakos M, Jegaskanda S, Corbett AJ, et al. Human mucosal-associated invariant T cells contribute to antiviral influenza immunity via IL-18–dependent activation. Proc Natl Acad Sci. (2016) 113:10133–8. doi: 10.1073/pnas.1610750113
83. van Wilgenburg B, Loh L, Chen Z, Pediongco TJ, Wang H, Shi M, et al. MAIT cells contribute to protection against lethal influenza infection in vivo. Nat Commun. (2018) 9:4706. doi: 10.1038/s41467-018-07207-9
84. Zhang X, Li S, Lason W, Greco M, Klenerman P, and Hinks TSC. MAIT cells protect against sterile lung injury. Cell Rep. (2025) 44:115275. doi: 10.1016/j.celrep.2025.115275
85. Parrot T, Gorin J-B, Ponzetta A, Maleki KT, Kammann T, Emgård J, et al. MAIT cell activation and dynamics associated with COVID-19 disease severity. Sci Immunol. (2020) 5:eabe1670. doi: 10.1126/sciimmunol.abe1670
86. Flament H, Rouland M, Beaudoin L, Toubal A, Bertrand L, Lebourgeois S, et al. Outcome of SARS-CoV-2 infection is linked to MAIT cell activation and cytotoxicity. Nat Immunol. (2021) 22:322–35. doi: 10.1038/s41590-021-00870-z
87. Hubrack S, Al-Nesf MA, Agrebi N, Raynaud C, Khattab MA, Thomas M, et al. In vitro Interleukin-7 treatment partially rescues MAIT cell dysfunction caused by SARS-CoV-2 infection. Sci Rep. (2021) 11:14090. doi: 10.1038/s41598-021-93536-7
88. Yang Q, Wen Y, Qi F, Gao X, Chen W, Xu G, et al. Suppressive monocytes impair MAIT cells response via IL-10 in patients with severe COVID-19. J Immunol. (2021) 207:1848–56. doi: 10.4049/jimmunol.2100228
89. Tang X, Zhang S, Peng Q, Ling L, Shi H, Liu Y, et al. Sustained IFN-I stimulation impairs MAIT cell responses to bacteria by inducing IL-10 during chronic HIV-1 infection. Sci Adv. (2020) 6:eaaz0374. doi: 10.1126/sciadv.aaz0374
90. Han F, Gulam MY, Zheng Y, Zulhaimi NS, Sia WR, He D, et al. IL7RA single nucleotide polymorphisms are associated with the size and function of the MAIT cell population in treated HIV-1 infection. Front Immunol. (2022) 13:985385. doi: 10.3389/fimmu.2022.985385
91. Ryu A, Clagett BM, and Freeman ML. Inflammation and microbial translocation correlate with reduced MAIT cells in people with HIV. Pathog Immun. (2024) 10:19–46. doi: 10.20411/pai.v10i1.746
92. Sortino O, Richards E, Dias J, Leeansyah E, Sandberg JK, and Sereti I. IL-7 treatment supports CD8+ mucosa-associated invariant T-cell restoration in HIV-1-infected patients on antiretroviral therapy. AIDS. (2018) 32:825–8. doi: 10.1097/QAD.0000000000001760
93. Ishikawa Y, Yamada M, Wada N, Takahashi E, and Imadome K-I. Mucosal-associated invariant T cells are activated in an interleukin-18-dependent manner in Epstein-Barr virus-associated T/natural killer cell lymphoproliferative diseases. Clin Exp Immunol. (2022) 207:141–8. doi: 10.1093/cei/uxab004
94. Paquin-Proulx D, Avelino-Silva VI, Santos BAN, Barsotti NS, Siroma F, Ramos JF, et al. MAIT cells are activated in acute Dengue virus infection and after in vitro Zika virus infection. PloS Negl Trop Dis. (2018) 12:e0006154. doi: 10.1371/journal.pntd.0006154
95. Haga K, Chiba A, Shibuya T, Osada T, Ishikawa D, Kodani T, et al. MAIT cells are activated and accumulated in the inflamed mucosa of ulcerative colitis. J Gastroenterol Hepatol. (2016) 31:965–72. doi: 10.1111/jgh.13242
96. Serriari N-E, Eoche M, Lamotte L, Lion J, Fumery M, Marcelo P, et al. Innate mucosal-associated invariant T (MAIT) cells are activated in inflammatory bowel diseases. Clin Exp Immunol. (2014) 176:266–74. doi: 10.1111/cei.12277
97. Willing A, Leach OA, Ufer F, Attfield KE, Steinbach K, Kursawe N, et al. CD8+ MAIT cells infiltrate into the CNS and alterations in their blood frequencies correlate with IL-18 serum levels in multiple sclerosis. Eur J Immunol. (2014) 44:3119–28. doi: 10.1002/eji.201344160
98. Rouxel O, Da silva J, Beaudoin L, Nel I, Tard C, Cagninacci L, et al. Cytotoxic and regulatory roles of mucosal-associated invariant T cells in type 1 diabetes. Nat Immunol. (2017) 18:1321–31. doi: 10.1038/ni.3854
99. Hinrichs AC, Kruize AA, Leavis HL, and van Roon JAG. In patients with primary Sjögren’s syndrome innate-like MAIT cells display upregulated IL-7R, IFN-γ, and IL-21 expression and have increased proportions of CCR9 and CXCR5-expressing cells. Front Immunol. (2022) 13:1017157. doi: 10.3389/fimmu.2022.1017157
Keywords: MAIT cells, Cytokines, tissue repair, infectious diseases, inflammatory diseases, cell plasticity, MAIT cell activation, interleukins
Citation: Camard L, Bianchi E and Rogge L (2025) Control of MAIT cell functions by cytokines in health and disease. Front. Immunol. 16:1594712. doi: 10.3389/fimmu.2025.1594712
Received: 16 March 2025; Accepted: 13 May 2025;
Published: 30 May 2025.
Edited by:
Luc Van Kaer, Vanderbilt University Medical Center, United StatesReviewed by:
Mariolina Salio, Immunocore (United Kingdom), United KingdomSeokmann Hong, Sejong University, Republic of Korea
Edwin Leeansyah, Tsinghua University, China
Copyright © 2025 Camard, Bianchi and Rogge. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Laetitia Camard, bGFldGl0aWEuY2FtYXJkQHBhc3RldXIuZnI=; Lars Rogge, bGFycy5yb2dnZUBwYXN0ZXVyLmZy
 Laetitia Camard
Laetitia Camard Elisabetta Bianchi
Elisabetta Bianchi Lars Rogge
Lars Rogge