- 1Neuroscience Institute Cavalieri Ottolenghi (NICO), Department of Neuroscience Rita Levi-Montalcini, University of Turin, Turin, Italy
- 2EPIGET LAB, Department of Clinical Sciences and Community Health, University of Milan, Milan, Italy
- 3Neuroscience Institute Cavalieri Ottolenghi (NICO), Turin, Italy
- 4Koelliker Hospital, Turin, Italy
Epidemiological studies have highlighted the existence of population groups exhibiting a higher sensitivity to the impact of environmental factors, such as exposure to air pollution. In these regards, people with Multiple Sclerosis (MS) or predisposed to develop MS - an autoimmune disorder of the Central Nervous System (CNS) - appear as a more vulnerable cohort to the effects of particulate matter (PM) exposure. Here, we aimed at disclosing the biological substrate of such higher vulnerability, and specifically at understanding whether individuals primed to develop autoimmunity (as it occurs in MS and in the experimental autoimmune encephalomyelitis - EAE - animal model of MS) respond differently to PM compared to healthy subjects. To this purpose, we characterized plasmatic extracellular vesicles (EVs) and their microRNA (miRNA) cargo in healthy and presymptomatic EAE mice early after exposure to PM10, compared to unexposed healthy and EAE mice. Results showed that the response of EAE mice to PM10 did not differ in terms of EV number or source, compared to that of healthy mice. Yet, remarkable differences existed in the identity of deregulated EV-associated miRNAs, which, in EAE mice, were predicted to target several MS-relevant biological processes and nervous system-, immune- and inflammation-related pathways, possibly contributing to disease worsening.
Introduction
Despite increased public awareness and global improvements in air quality, air pollution remains a significant public health challenge for low-, middle-, and high-income countries. Particulate matter (PM) - a mix of solid particles and liquid droplets - is one of the main air pollutants, arising from natural sources (e.g. pollen, sea spray, and dust from ground erosion) and anthropic activities (e.g. combustion of fossil fuels, roadways and mining operations, agricultural activities; 1). Exposure to PM has been associated with an increased risk of cardiovascular and respiratory diseases, different types of cancer (2), and, more recently, pathologies affecting the central nervous system (CNS; 3–5).
PM includes coarse (PM10), fine (PM2.5), and ultrafine (PM0.1) particles - having an aerodynamic diameter smaller than 10, 2.5, and 0.1 μm, respectively - that typically comprise inorganic compounds, aromatic hydrocarbons, metals, and microbial components (6). When inhaled, PM penetrates the lungs and settles deep within, triggering a local inflammatory and defensive response that ultimately impacts all major organs and systems, including the CNS. (3, 4).
Although smaller particles - such as PM0.1 - could directly enter the blood circulation and overcome the blood-brain-barrier (BBB; 3), the disruption of the homeostasis of distant organs most likely relies on mechanisms other than a direct effect of PM particles. Alongside the secretion of proinflammatory cytokines, a well-established mechanism by which the effects of exposure to PM can extend beyond the respiratory compartment and trigger a systemic response, is the release of extracellular vesicles (EVs; 7–10). EVs are nano-sized (0.03–1 μm) plasma membrane fragments actively released by cells, which can be distributed to all systems, including the CNS (11) - via the blood and other biological fluids. EVs play a key role in the intercellular and inter-organ transfer of biological information. After reaching target cells through the recognition of surface-expressed ligands, EVs deliver their content, which can include proteins, lipids, metabolites, organelles, and genetic information. As regards this latter aspect, the most well studied molecules in EV cargo are microRNAs (miRNAs; 12), endogenous non-coding small RNAs, which play an important role in the post-transcriptional regulation of gene expression via either mRNA cleavage or translation inhibition. The power of miRNAs as regulatory mechanisms of cellular processes lies in their capability to tune the expression of multiple transcripts, exerting a broad control over functionally related target mRNAs. Moreover, different miRNAs can act together to cooperatively target mRNAs that encode proteins within the same functional network (13). Thus, deregulation of even a restricted set of miRNAs can significantly impact gene expression, and - as a consequence - EVs are able to profoundly influence the molecular state and function of target cells (14). Consistently, shuttling of miRNAs packaged in plasmatic EVs has emerged as one of the most powerful mechanisms linking PM exposure with the disruption of homeostasis in extrapulmonary compartments (7, 15, 16).
Epidemiological studies have strongly indicated that environmental factors play a role in shaping the geographical distribution and the timing of hospital admissions for a number of diseases. In individuals with a predisposing background, environmental factors have been proposed to “set the disease threshold” (17). Specifically, as regards PM, while most individuals can maintain homeostasis even upon chronic or high dosage exposure, certain vulnerable groups exhibit heightened sensitivity to PM impact (18). This increased susceptibility is attributed to genetic or acquired factors that make the biological responses to PM exposure differ from those elicited in “coping” individuals (18).
Lifestyle and environmental factors (e.g. smoking, limited sun exposure, low vitamin D, obesity, infections and exposure to environmental pollutants) have been associated with increased risk of Multiple Sclerosis (MS; 19–21). In particular, airborne PM peaks have been associated with higher rates of hospital admissions for MS onset or relapses (22–30), suggesting that people with MS (or predisposed to develop MS) are a PM-vulnerable cohort. MS is a chronic disease with autoimmune components, characterized by immune cell infiltration in the CNS, neuroinflammation, diffuse myelin loss, and, over time, neurodegeneration. These processes result in functional impairments and, in many cases, cognitive and psychiatric symptoms (31–33). Of note, PM exposure has been reported to trigger MS-relevant events (e.g. inflammation, endothelial and BBB alterations, CNS white matter damage, and changes in brain innate immune cell activity) even in healthy individuals (34, 35). This response seems to be more pronounced in MS patients (29). Yet, the biological substrate of such vulnerability to PM remains uninvestigated so far.
Here, we aimed at understanding whether individuals primed to develop autoimmunity against CNS myelin (as it occurs in MS and in the MOG35-55-induced experimental autoimmune encephalomyelitis - EAE - animal model of MS) respond differently to PM compared to healthy subjects. Specifically, we characterized EVs (i.e. number, size and cellular source) and EV-associated miRNA profile in the plasma of healthy (Ctrl) and presymptomatic EAE mice acutely exposed to PM10, compared to unexposed Ctrl and EAE mice. These analyses showed that the response of mice primed to develop EAE to PM exposure differed from that of healthy Ctrl mice and did not simply correspond to the amplification of differences already existing between EAE and healthy mice. Although the miRNA cargo of plasma EVs released following PM exposure had a signature relevant for MS in both healthy and EAE mice, deregulated miRNAs were distinct and the response of EAE mice appeared more “pathogenic” than that of healthy mice. In silico analyses were applied to identify target gene transcripts and biological processes possibly underlying PM effects, unveiling a significant enrichment in nervous system-, immune- and inflammation- related pathways. Overall, our data support the idea of a synergy between PM exposure and immune system priming toward autoimmunity, and suggest that shuttling of EV-associated miRNAs in response to PM exposure may participate in different aspects of MS pathogenesis and exacerbation.
Materials and methods
Animals and experimental design
Mice were housed in the vivarium under standard conditions (12-hr light/12-hr dark cycle at 21°C) with food and water ad libitum. The project was designed according to the guidelines of the NIH, the European Communities Council (2010/63/EU) and the Italian Law for Care and Use of Experimental Animals (DL26/2014). It was also approved by the Italian Ministry of Health (authorization 510/2020-PR to EB) and the Bioethical Committee of the University of Turin. The study was conducted according to the ARRIVE guidelines.
Chronic EAE induction
To induce chronic EAE, 8 week-old female C57BL/6J mice (Charles River, Calco, Italy) were immunized by 2 subcutaneous injections of 200 μg myelin oligodendrocyte glycoprotein 35–55 peptide (MOG35-55; Espikem, Florence, Italy) in incomplete Freund’s adjuvant (IFA; Sigma-Aldrich, Milan, Italy) containing 8 mg/ml Mycobacterium tuberculosis (strain H37Ra; Difco Laboratories Inc., Franklin Lakes, NJ, USA), followed by 2 intravenous injections of 500 ng of Pertussis toxin (Duotech, Milan, Italy) on the immunization day and 48 h later (36).
PM10 administration and blood collection
We exposed healthy (Ctrl) and EAE mice to a commercially available urban outdoor PM10 (NIST Standard Reference Material SRM1648a; Sigma-Aldrich), which is a chemically standardized compound (https://tsapps.nist.gov/srmext/certificates/1648a.pdf), widely used in toxicological studies (37–39). Stock suspensions of NIST SRM1648 PM were prepared in sterile ultrapure water and aliquots were thoroughly mixed under sonication for 1 hour prior to each experiment. Mice were randomly divided into control (saline, i.e. NaCl 0.9% solution) and treatment (PM) groups. The treatment group was treated by intratracheal instillations (Intubation stand, Kent Scientific Corporation) of a PM10 suspension (10 μg in 50 μl of saline), and the control group was treated with saline (50 μl), as in our previous study (40). We opted for the intratracheal administration of PM to assure low variability of absorption and exclude a direct nose-to-CNS transit of PM via the olfactory mucosa.
PM10 dose was calculated considering the daily respiratory volume of mice (0.04 m3) and the daily peaks of PM10 concentration in East Europe polluted areas (>250 μg/m3; 41). Therefore, in order to use a PM10 dose relevant for human exposure, the dose used in this study was set as 10 μg (0.04 m3/day × 250 μg/m3). PM10 was administered 4 days after EAE immunization, i.e. during the presymptomatic phase of the EAE disease course, where debilitating symptoms are yet to be displayed by the animals (i.e. clinical score=0, according to the conventional EAE clinical score assessment (36)). Peripheral blood of each mouse was collected in EDTA Vacutainer tubes (Becton Dickinson, New Jersey, USA), 6 hours after PM10/saline exposure, and processed within 3 hours from sampling. Blood was centrifuged at 1300xg for 15 min to separate plasma.
EVs isolation and characterization
For EV studies, we followed the MISEV2018 guidelines (42; Supplementary Table S1, Supplementary Figure S1A, B). EVs have been isolated from plasma pools from six mice of each condition: healthy mice which received saline (Ctrl); healthy mice which received PM10 (Ctrl PM); EAE mice which received saline (EAE); EAE mice which received PM10 (EAE PM). The resulting pooled samples were centrifuged three times at increasing speeds (1000xg, 2000xg, 3000xg) for 15 min at 4°C. After every centrifugation, the supernatant was decanted into a new tube and centrifuged again to remove cell debris and aggregates. Shortly after centrifugation, plasma samples were transferred into ultracentrifuge tubes (Polycarbonate Centrifuge Bottles, Beckman Coulter) and filled up to 10.5 ml with NaCl 0.9% solution. Ultracentrifugation was performed at 110,000xg for 75 min at 4°C to allow sedimentation of EVs. After ultracentrifugation, the supernatant was discarded and tubes were allowed to air dry for 2 min.
To confirm that this isolation protocol resulted in preparations enriched in EVs, western blotting (WB) was used to assess the expression of EV markers (i.e. the tetraspanins CD63/CD9) and absence of contamination from other cellular components (i.e. calnexin, a marker for endoplasmic reticulum; Supplementary Figure S1A). Briefly, plasma EVs lysates were obtained in RIPA buffer (150 mM NaCl, 1% Triton X-100, 0.5% Sodium deoxycholate, 0.1% sodium dodecyl sulfate, 50 mM Tris Base, 5mM EDTA, 1mM EGTA) containing protease and phosphatase inhibitors (100mM DTT, 1M NaF, 200 mM sodium orthovanadate, 100 mM PMSF), boiled in SDS buffer and separated using a 10% SDS-PAGE gel. Proteins were transferred to PVDF membranes, blocked in 5% non-fat milk in TBS (20 mM Tris-HCl, pH 7.5, 150 mM NaCl) + 0.1% Tween-20 (TBS-T) for 1h and then incubated with primary antibodies (anti-rabbit CD63, 1:500, Immunological Sciences, Italy; anti-mouse CD9, 1:250, BD Biosciences; anti-rabbit calnexin 1:250, Cell signaling, USA) overnight at 4°C. After washes with TBS-T, membranes were incubated with appropriate secondary antibodies (anti-rabbit or anti-mouse, 1:3000, Sigma, Italy) for 1h at room temperature. Protein extract from a mouse cerebral cortex was used as a positive control. The chemiluminescent signal was visualized using Westar Antares ECL Blotting Substrates (Cyanagen; Italy), acquired with Bio-Rad ChemiDocTM Imagers (Bio-Rad; Italy).
Nanoparticle tracking analysis (NTA) and flow cytometry were used to characterize EV number/size (Supplementary Figure S1B) and cellular source, respectively (as in 43). To determine EV cellular origins, immunophenotyping was achieved with the MACSQuant Analyser flow cytometer (Miltenyi Biotec, Bergisch Gladbach, DE) following the manufacturer’s protocol. The Fluoresbrite Carboxylate Size Range Kit I (0.2, 0.5, 0.75, and 1 µm) was used to set the calibration gate on the MACSQuant Analyser system. To evaluate the integrity and to highlight EV subsets, 60-µL sample aliquots were stained with 0.02 µM 5(6)-carboxyfluorescein diacetate N-succinimidyl ester (CFSE) at 37°C for 20 min in the dark. CFSE is a vital non-fluorescent dye that enters EVs, where intracellular esterase enzymes remove the acetate group and convert the molecule into the fluorescent ester form. To characterize and count EVs, the following panel of antibodies was used: monoclonal APC-anti-CD14 (Clone REA934; dilution 1:10) to identify EVs from macrophages and/or monocytes; APC-antiCD41 (Clone: REA1194; dilution 1:10) for EVs from platelets; APC-anti-CD25 (clone: REA568; dilution 1:10) for EVs from regulatory T (Treg) lymphocytes. All antibodies were purchased from Miltenyi Biotec. Quantitative multiparameter analysis of flow cytometry data was conducted using FlowJo software (Tree Star, Inc., Ashland, OR, USA) and antibody gating strategies were performed as previously described (43).
EV-miRNAs isolation
miRNA isolation from EV pellets in tubes was performed using the miRNeasy Mini Kit (Qiagen, Venlo, NL) according to the manufacturer’s protocol, with the addition of the “Rneasy Mini Elute Clean-up” kit (Qiagen). To assess the quality of miRNA purification, samples were analyzed by 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) using Agilent RNA 6000 Pico Kit (Supplementary Figure S2) and stored at -80°C.
miRNA profiling
Reverse Transcriptase reaction was performed using Megaplex™ RT Primers Pool A and B Rodent (Life Technologies, Foster City, CA) and TaqMan® MicroRNA Reverse Transcriptase Kit (Life Technologies). Briefly, two distinct reactions (A and B) were performed, to cover the reverse transcription of 758 target miRNAs, including 0.75 µl of Megaplex RT Pool, 0.15 µl of dNTPs (100 mM), 0.75 µl 10X RT Buffer, 0.90 µl of MgCl2 (25 mM), 0.1 µl of RNases Inhibitor (20 U/µl) and 1.5 µl of MultiScribe™ Reverse Transcriptase (50 U/µl). The reverse transcription thermal protocol consisted of 40 cycles at 16°C for 2 min, 42°C for 1 min, and 50°C for 1 s, plus one cycle at 85°C for 5 min and final stage at 4°C. The cDNA samples were pre-amplified using “Low Sample Input Protocol (LSI) for profiling human miRNA using OpenArray® Platform” (Application Note 2011- Life Technologies) with a specific Megaplex™ Preamp Primer Mix for Pool A and Pool B (Rodent). The pre-amplified samples were diluted 1:20 with nuclease-free water and were analyzed by QuantStudio™ 12K Flex Real-Time PCR System with OpenArray® Platform (Applied Biosystems) according to manufacturer’s instructions.
miRNA expression data analysis
The Gene Expression Suite Software was used to analyze the expression of 754 unique miRNAs. Four small RNA species (RNU48, RNU44, U6, and ath-miR159a), included in the array as internal technical controls to assess assay performance and RNA integrity, were also present but not used for normalization purposes. Only miRNAs with Relative Threshold Cycle (Crt) value< 27 or AmpScore> 1.24 were considered amplified. The expression data were normalized using the “Global Normalization Factor” which calculates the average Crt of all detected miRNAs in a given sample. This method is recommended for EV-derived RNA, where validated endogenous reference genes are lacking due to the heterogeneity of vesicle content. Relative miRNA expression (also referred as Fold Change - FC) was determined using the relative quantification RQ= 2(-Δcrt) formula, with -ΔCrt = measured Crt -medium normalization Crt (44).
Statistical analyses
Statistical analyses were carried out with GraphPad Prism 9 (GraphPad software, Inc, RRID: SCR_002798). As for EV characterization (i.e. EV concentration, mean EV size and percentage of marker-positive EVs; Figure 1), the Shapiro-Wilk test was first applied to test for a normal distribution of the data. Then, as data were normally distributed, a Two-ways ANOVA test followed by Bonferroni’s post-hoc analysis was used for multiple group comparisons. P<0.05 was considered as statistically significant. Statistical differences were indicated with * P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. The list of the applied tests and number of animals in each case are included in Supplementary Table S2. Differential expression analysis of 754 unique miRNAs between groups was assessed using unpaired t-tests followed by the Benjamini-Hochberg FDR correction for multiple testing. A threshold of 0.20 was applied to the FDR p-value significance level to identify the set of top miRNAs.
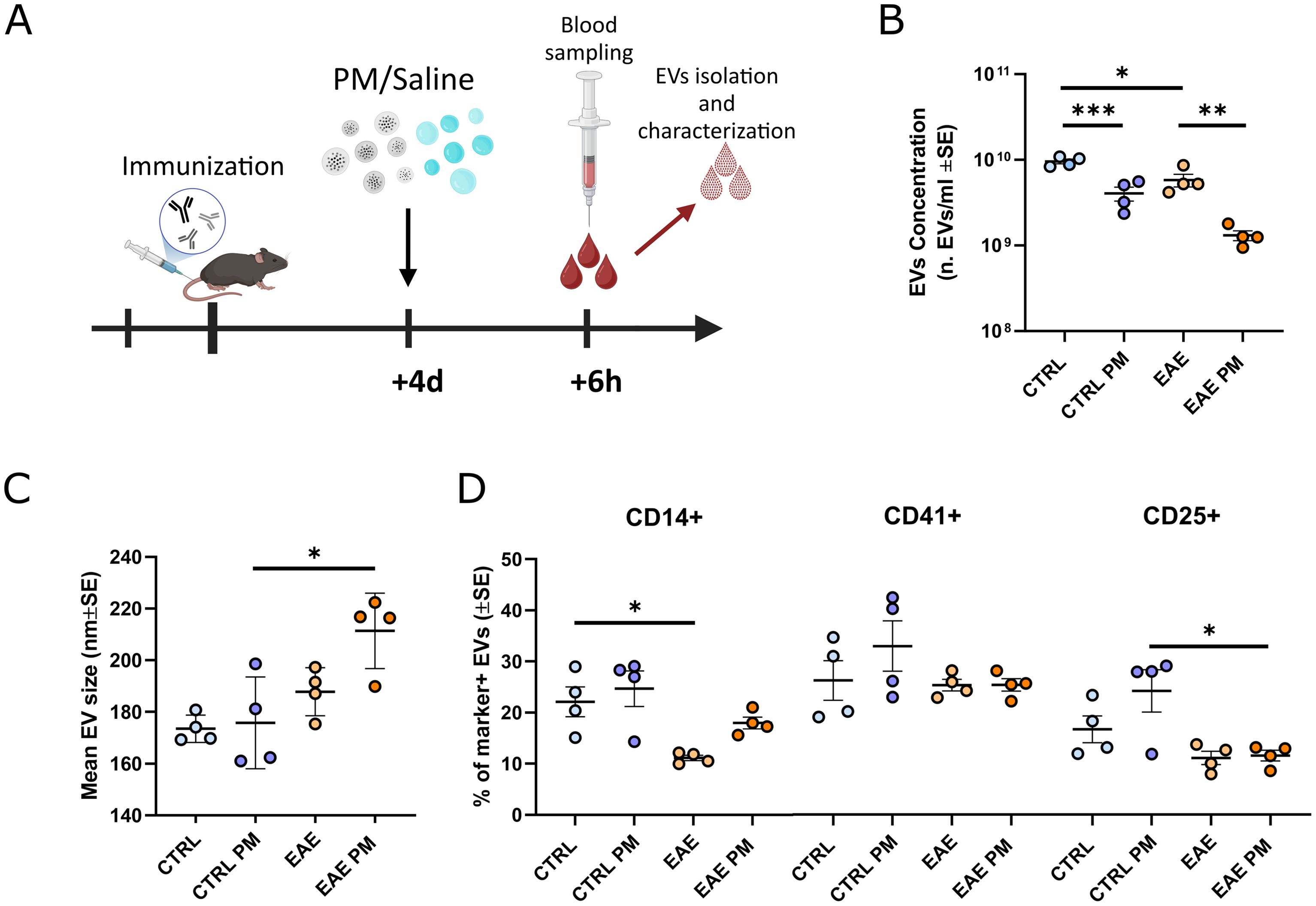
Figure 1. Characterization of mouse plasmatic EVs isolated after an acute PM10 exposure. (A) Schematic representation of the experimental design (graphics created with BioRender.com). (B, C) Mean EVs concentration (B) and size (C) in mouse plasma of CTRL and EAE mice exposed either to saline or PM10. (D) Percentage (%) of EVs positive for markers of monocytes/macrophages (CD14+), platelets (CD41+) and T-regs (CD25+). Each dot represents an individual mouse. Differences were assessed by Two-way ANOVA (see Supplementary Table S2 for P and F values of each comparison). *p<0.05; **p<0.01; ***p<0.001.
Network analysis of deregulated miRNAs and KEGG pathway enrichment analysis
The interactions between deregulated miRNAs and their target gene transcripts in each experimental condition were visualized using the web-based platform miRNet 2.0 (https://www.mirnet.ca/miRNet/home.xhtml45). Significantly upregulated or downregulated miRNAs from each of the four groups, identified by their miRBase IDs, were uploaded into the “miRNA” module of miRNet 2.0. The target list was specified as “Genes (miRTarBase v9.0)” to link the input miRNAs to their predicted target gene transcripts, generating interaction tables and network visualizations. In the resulting networks, miRNAs may appear as specific strands (e.g., miR-9-5p from the input miR-9) or as closely related variants (e.g., miR-451a from miR-451). For the sake of clarity, only the most highly connected miRNAs in each network were labeled in the visualizations. KEGG pathway enrichment analysis was performed on the target gene lists from each network using the “Function Explorer” toolbox within miRNet, which employed a hypergeometric test to identify enriched (p-value < 0.05) pathways by comparing the input genes against the full set of mouse genes annotated in the KEGG database. The resulting p-values were adjusted for multiple testing using the Benjamini-Hochberg procedure to control the false discovery rate (FDR).
Gene-to-disease association analysis
The list of target genes identified by the miRNA network analysis was used as input for a gene-to-disease association analysis using the DisGeNET Curated dataset. Genes associated with a disease class were displayed using the DisGeNET application in Cytoscape (46), operated through a DisGeNET specific R script. Edge thickness in the network graphical representation is proportional to the DisGeNET score for association robustness (SGDA, score of Gene to Disease Association), which takes into consideration the number and type of sources and number of publications that support the association, following the subsequent formula:
SGDA=C+M+I+L+T
C = score for number of curated sources supporting the GDA (range from 0 to 0.7)
M = score for number of model sources supporting the GDA (range from 0 to 0.1)
I = score for number of inferred sources supporting the GDA (range from 0 to 0.05)
L = score for number of publications supporting the GDA (range from 0 to 0.4)
T = score for number of clinical trials supporting the GDA (range from 0 to 0.1). Further information can be found at https://disgenet.com/About#metrics.
Results
Acute exposure to PM10 similarly affects circulating EV release in healthy and presymptomatic EAE mice
To assess whether individuals primed toward the development of autoimmunity against CNS myelin (i.e. immunized to develop EAE) react differently to PM10 exposure, we characterized plasmatic EVs (number/size, source and miRNAs cargo) in healthy (Ctrl) vs. presymptomatic (i.e. 4 days after immunization) EAE mice exposed to PM10, compared to Ctrl and presymptomatic EAE mice which received saline (Figure 1A). Although not resulting in an overt motor impairment, nor in demyelination, in the presymptomatic stage of EAE the biological processes leading to the pathology have already started, as exemplified by cytokine and chemokine alterations in the nervous tissue, cerebrospinal fluid (CSF), and plasma (47), changes in the expression levels of molecules involved in neuronal-microglial communication (48), as well as by mouse cognitive impairment (49), thus recapitulating the early stages of MS.
EAE mice basally showed a slightly lower concentration of plasmatic EVs compared to Ctrl. PM10 exposure induced a further significant decrease in plasmatic EV release in both Ctrl and EAE mice, compared to unexposed individuals (Figure 1B). Although reduced, about 1.2 x109 EVs/ml were still found in the plasma of PM10-exposed EAE mice (Figure 1B), showing a moderate increase in size, compared to those of unexposed EAE and PM10-exposed and unexposed Ctrl mice (Figure 1C). To characterize EV cellular source, we exploited a panel of markers specific for cell types reported to be at the origin of plasma EVs upon PM exposure in humans, i.e. monocytes/macrophages and platelets (8, 15), and important players in MS pathophysiology, i.e. T regulatory cells (Tregs; 50). While in Ctrl mice 20-25% of EVs were produced by monocytes/macrophages and by Tregs, only 10% of EVs have these cellular origins in EAE mice, as assessed by immunopositivity for CD14 and CD25 surface antigens respectively (Figure 1D). Although the fraction of monocyte/macrophage-derived EVs appeared slightly higher in PM10-exposed EAE mice compared to unexposed EAE, overall PM10 exposure did not alter the percentage of marker-positive EVs in both Ctrl and EAE mice (Figure 1D).
Taken together, these analyses showed that – in terms of plasmatic EV number and source - mice primed to develop autoimmunity do not respond differently to PM10 exposure, compared to healthy Ctrl mice.
Selective deregulation of extracellular vesicle-packaged miRNAs in EAE mice after short-term exposure to PM10
To assess whether the response to PM10 of presymptomatic EAE mice differed in terms of EV miRNA cargo, miRNAs have been extracted and quantified from plasmatic EVs. This analysis provided a list of deregulated miRNAs for each of the following comparisons: PM-exposed Ctrl (i.e. Ctrl PM) vs. unexposed Ctrl (i.e. Ctrl); unexposed EAE (i.e. EAE) vs. unexposed Ctrl mice; PM-exposed EAE (i.e. EAE PM) vs. unexposed EAE; PM-exposed EAE vs. PM-exposed Ctrl mice (Figure 2). Deregulated miRNAs are listed in Supplementary Table S3. In Ctrl mice, PM10 exposure induced changes in the expression of 59 miRNAs, among which the most downregulated ones were miR-2134, miR-712-5p and miR-29a-5p, while the most upregulated were miR-215-5p, miR-199a-3p and miR-146b-5p (7 miRNA species with FDR<0.05; Figures 2A, E). Of note, miR-2134 was also one of the most downregulated miRNAs among the 45 differentially expressed miRNAs in EAE vs. Ctrl mice, together with miR-1839-3p, miR-200b-3p and miR-451a. Top upregulated miRNAs in EAE vs. Ctrl mice instead included miR-223-3p, miR-1195 and miR-685 (6 miRNA species with FDR<0.05; Figures 2C, E).
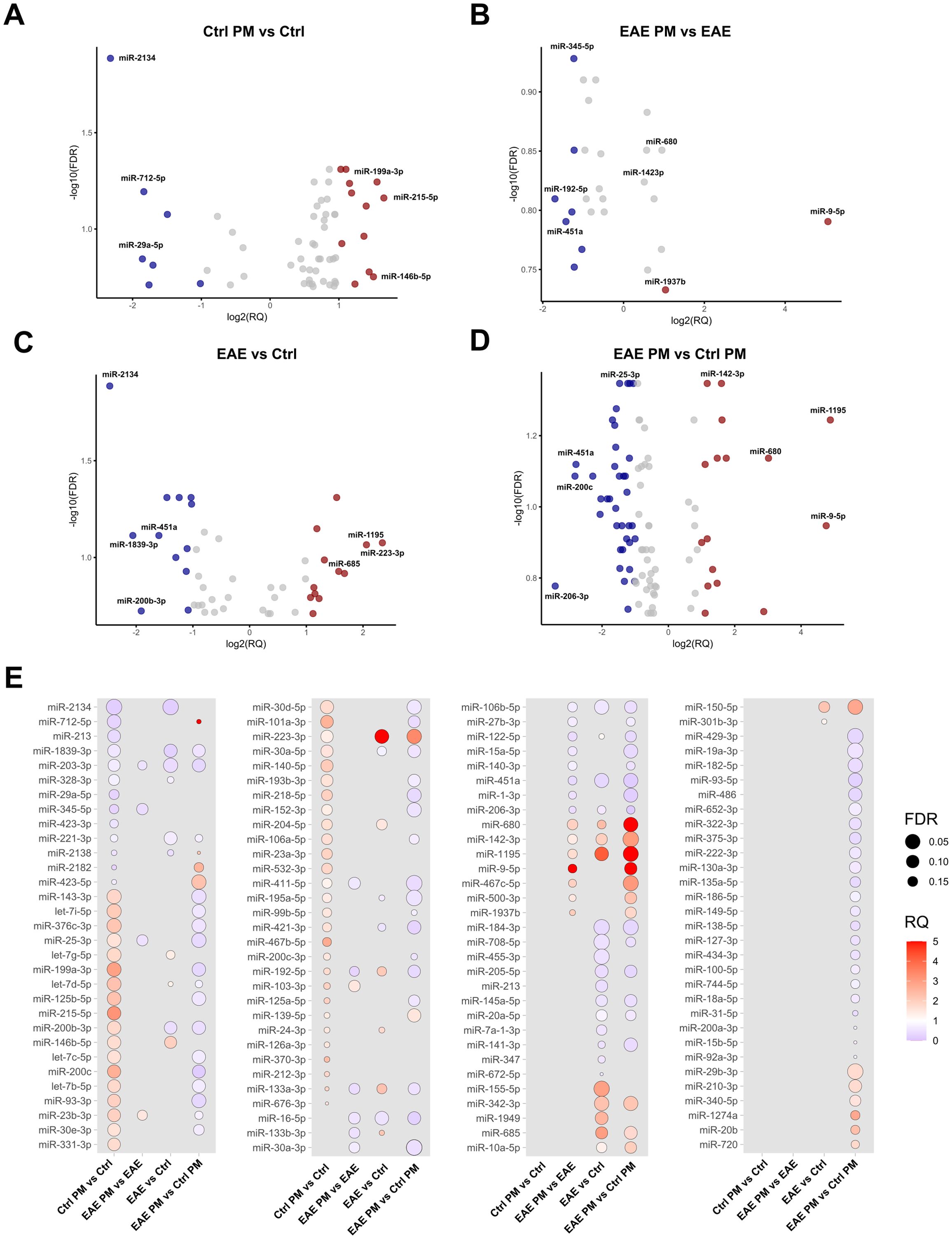
Figure 2. Deregulated EV-packaged miRNAs. Volcano Plots showing upregulated and downregulated miRNAs (p<0.05; FDR<0.2) in (A) Ctrl PM10 vs. Ctrl, (B) EAE PM10 vs. EAE, (C) EAE vs. Ctrl and (D) EAE PM10 vs. Ctrl PM10 mice. Top deregulated miRNAs are highlighted in red (upregulated, RQ >1) and blue (downregulated, RQ<0.5). (E) Dot plots showing differentially expressed miRNAs across comparisons, with dot size indicating FDR and color representing RQ. See Supplementary Table S3 for the entire list of deregulated miRNAs.
When comparing EAE mice exposed to PM10 to unexposed EAE mice, we observed 26 differentially expressed miRNAs (Figures 2B, E). In particular, miR-192-5p, miR-451a and miR-122-5p were the most downregulated, whereas miR-9-5p and miR-680 were the most up-regulated. Finally, 94 miRNAs were differentially expressed between EAE and healthy (Ctrl) mice exposed to PM10, with miR-200c, miR-451a and miR-25-3p among the most downregulated, and miR-9-5p, miR-1195 and miR142-3p among the most upregulated miRNAs (7 miRNA species with FDR<0.05; Figures 3D, E).
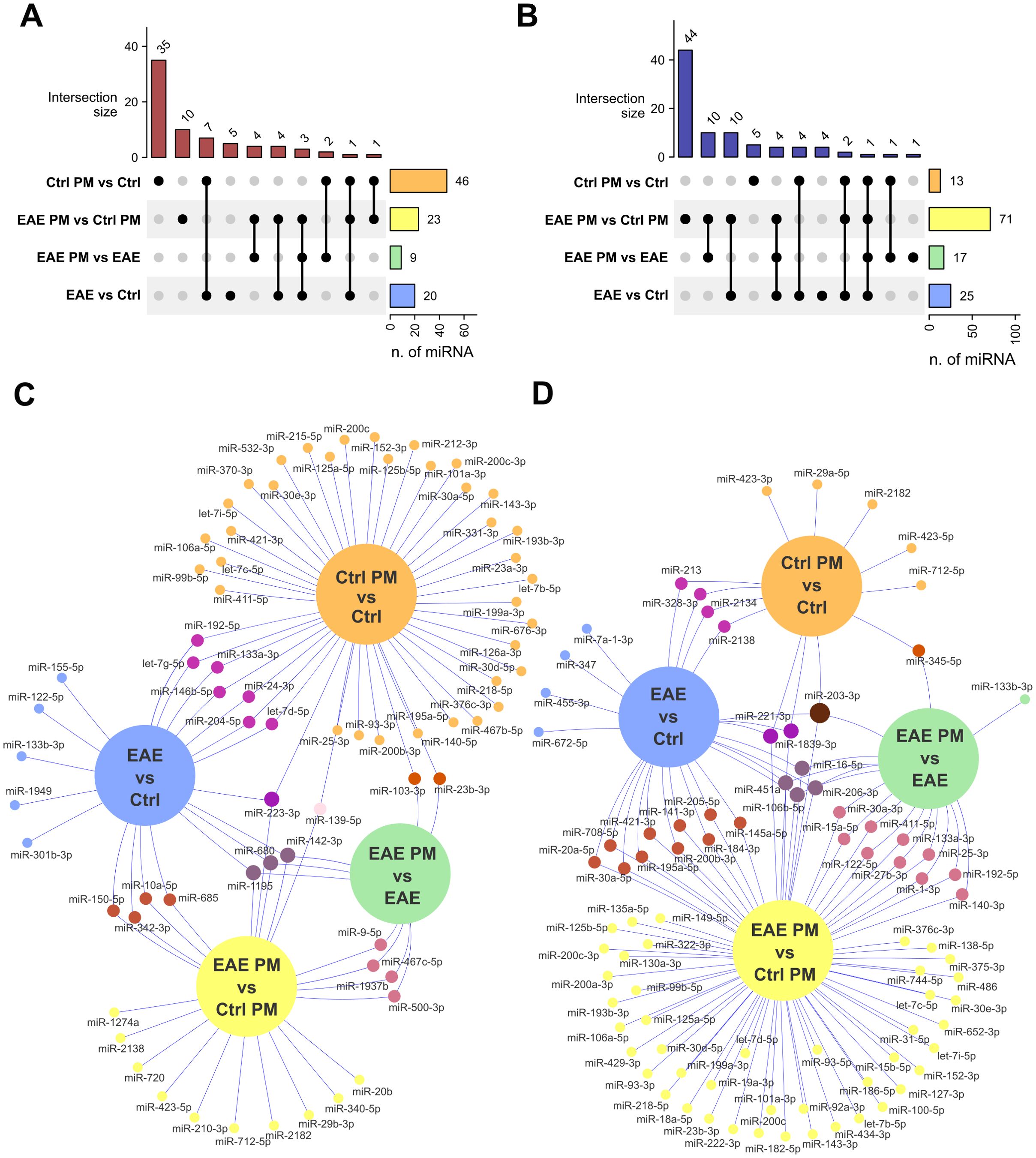
Figure 3. Overlap of deregulated EV-associated miRNAs among groups. (A, B) Upset plots showing the number of upregulated (A) and downregulated (B) miRNAs that are selectively or commonly expressed among comparisons. (C, D) Venn networks showing the identity of miRNAs upregulated (C) or downregulated (D) in all comparisons of interest. See Supplementary Table S4 for the entire list of deregulated miRNAs that are selectively or commonly expressed among comparisons.
Notably, a certain degree of overlap existed between the responses of healthy mice to immunization and to PM10, especially in terms of downregulated miRNAs (58.84% of the miRNAs downregulated in Ctrl PM vs. Ctrl mice overlapped with those downregulated in EAE vs. Ctrl mice; Figure 3; Supplementary Table S4), suggesting a similar biological effect. In contrast, the responses to PM10 of EAE and healthy mice were largely divergent, with only 22% of upregulated miRNAs and 12% of miRNAs downregulated in EAE PM vs. EAE overlapping with those downregulated in Ctrl PM vs. Ctrl mice (Figure 3; Supplementary Table S4).
Overall, these analyses show that the response to PM10 – in terms of deregulated EV-associated miRNAs – is qualitatively different in mice primed to develop autoimmunity compared to healthy Ctrl mice.
miRNA target prediction and network analysis
To further explore the potential functional impact of miRNA deregulations triggered by PM10 in healthy and EAE mice, we identified the target gene transcripts of miRNAs that were differentially expressed in the 4 comparisons (Supplementary Table S5). Notably, several targets were shared among multiple deregulated miRNAs, suggesting potential regulatory hubs. In line with this idea, network-based analyses performed on upregulated (Figure 4) and downregulated (Figure 5) miRNAs for each experimental comparison highlighted a significant number of interactions between the deregulated miRNAs (yellow squares) and their predicted target gene transcripts (red dots for upregulated miRNAs in Figure 4 and blue dots for downregulated miRNAs in Figure 5). Overall, the networks of upregulated miRNAs (Figure 4) were remarkably more complex than those of the downregulated miRNAs (Figure 5), which revealed relatively few target gene connections (especially for Ctrl PM vs Ctrl and EAE vs Ctrl, with fewer than 50 interactions; Figures 5A, C). In contrast, the networks of downregulated miRNAs in EAE PM vs both EAE (Figure 5B) and Ctrl PM (Figure 5D) exhibited more intricate interactions, with a few miRNAs having over 300 connections each.
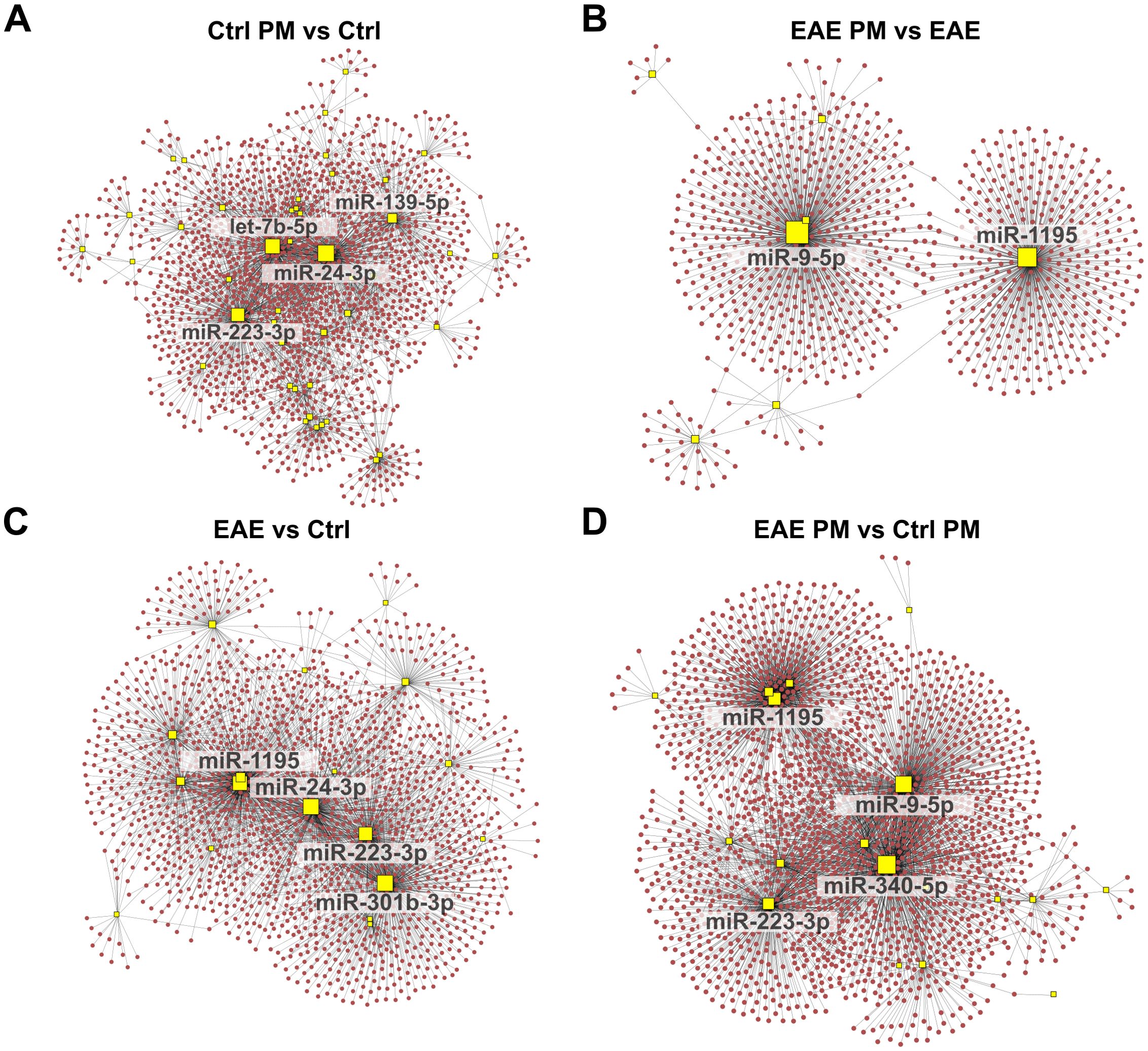
Figure 4. Network analysis of upregulated miRNAs and their predicted target gene transcripts. Network diagrams represent the interactions between upregulated miRNAs and their predicted target gene transcripts in the following conditions: (A) Ctrl PM10 vs Ctrl, (B) EAE PM10 vs EAE, (C) EAE vs Ctrl, and (D) EAE PM10 vs Ctrl PM10. Yellow squares indicate miRNAs, and red dots represent target gene transcripts. For clarity, only the names of miRNAs with more connections (threshold varies across networks) are labeled. See Supplementary Table S5 for the entire list of upregulated miRNAs and their predicted target gene transcripts.
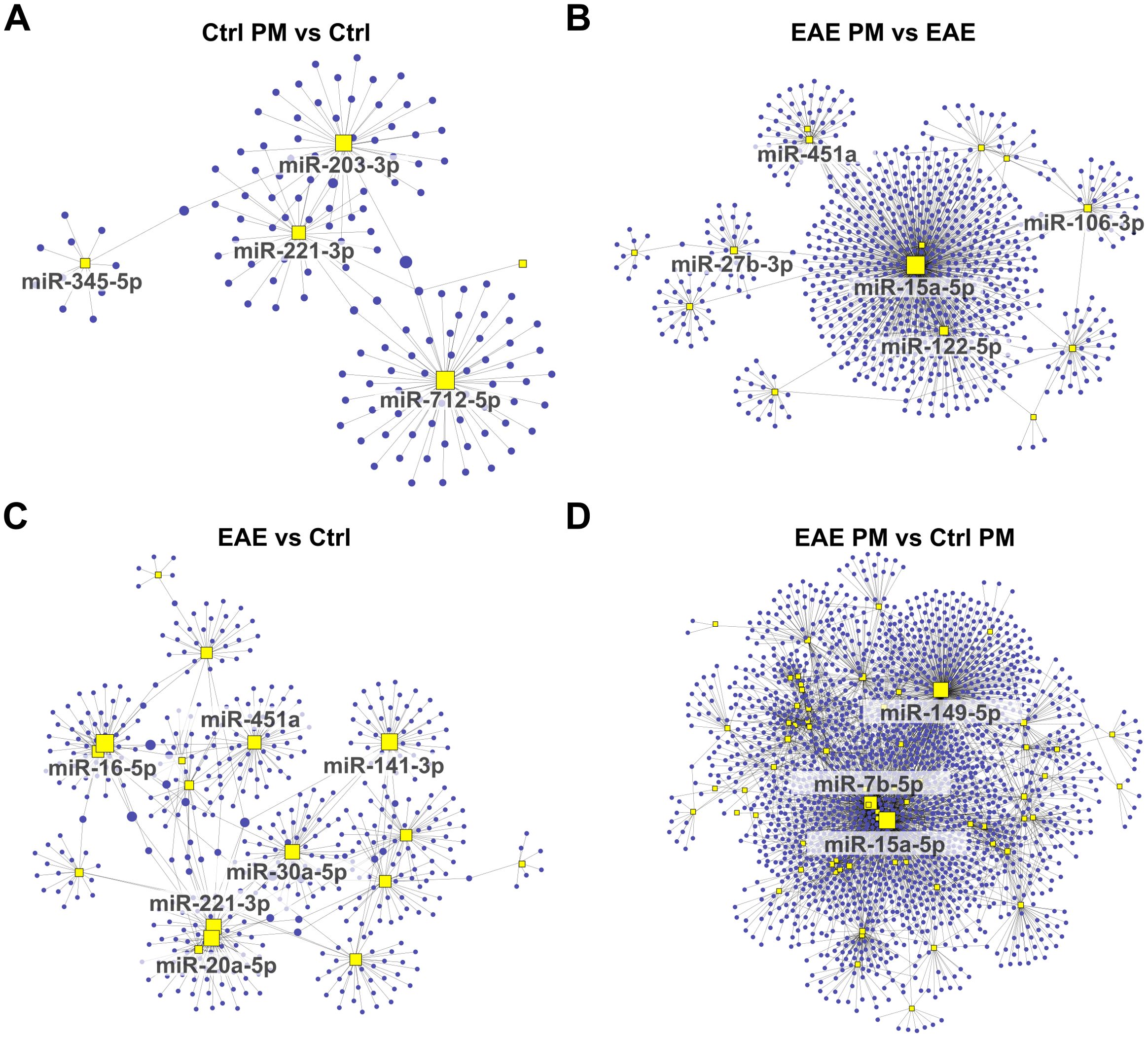
Figure 5. Network analysis of downregulated miRNAs and their predicted target gene transcripts. Network diagrams represent the interactions between downregulated miRNAs and their predicted target gene transcripts in the following conditions: (A) Ctrl PM10 vs Ctrl, (B) EAE PM10 vs EAE, (C) EAE vs Ctrl, and (D) EAE PM10 vs Ctrl PM10. Yellow squares indicate miRNAs, and blue dots represent target gene transcripts. For clarity, only the names of miRNAs with more connections (threshold varies across networks) are labeled. See Supplementary Table S5 for the entire list of downregulated miRNAs and their predicted target gene transcripts.
Examining the shared miRNAs within these networks allowed us to provide more insightful information about the actual biological consequences of miRNA deregulation, accounting for both the miRNA-target gene transcript interactions and their collective regulatory impact, rather than simply considering the shared miRNAs across conditions. Interestingly, among the upregulated miRNAs displaying the highest number of connections, a partial overlap appeared again between the response of healthy mice to immunization (EAE vs Ctrl) and to PM10 (Ctrl PM vs Ctrl; e.g. miR-24-3p, and miR-223-3p among the upregulated miRNAs and miR-221-3p among the downregulated miRNAs; Figures 4A, C, 5A, C), suggesting that these miRNAs may lead to similar cascades in response to both environmental and autoimmune triggers. Except for miR-1195, miR223-3p and miR-451a which were shared across EAE vs Ctrl and EAE PM vs Ctrl PM (Figures 4B–D), “hub” miRNAs deregulated in EAE mice responding to PM10 were exclusive. Among them, miR-9-5p (Figures 4B, D) and miR-15a-5p (Figures 5B, D) were the most connected upregulated and downregulated miRNAs, respectively.
To further investigate the biological pathways potentially influenced by the detected miRNA deregulation, KEGG pathway enrichment analysis was performed on the predicted target genes from the upregulated and downregulated miRNA networks across the different comparisons. Of relevance in the context of an autoimmune CNS disease like MS, this analysis revealed a significant enrichment of terms related to synapse function, nervous system, and immune response, for both up- and downregulated miRNAs (Figures 6A, B, respectively, Supplementary Table S6). Terms such as Glutamatergic synapse, GABAergic synapse, Dopaminergic synapse, Cholinergic synapse suggest that miRNA deregulation may play a role in modulating synaptic activity and plasticity, as further supported by the enrichment of Long-term potentiation and Long-term depression pathways. Moreover, several terms associated with addiction - such as Morphine addiction, Cocaine addiction, Amphetamine addiction, and Nicotine addiction - emerged across the comparisons, suggesting a possible influence of deregulated miRNAs on neurotransmitter systems. Besides nervous system-related pathways, immune- and inflammation-related pathways were also significantly enriched. These pathways were significantly enriched in the networks of upregulated miRNAs across the different comparisons (Figure 6A; Supplementary Table S5). In contrast, for downregulated miRNAs, nervous system- and immune-related pathways emerge almost exclusively in the comparisons involving EAE mice (i.e. EAE vs Ctrl, EAE PM vs Ctrl PM and EAE PM vs EAE comparisons). Although this might be due only to the very small size of the network of downregulated miRNAs in Ctrl PM vs Ctrl comparison (Figure 5A; Supplementary Table S5), this pointed again to the special character of the response of EAE mice to PM10. In line with this idea, while the pathways described so far were largely shared across the four comparisons, several terms associated with fatty acid (FA) metabolism (Biosynthesis of unsaturated fatty acids, Fatty acid metabolism, Fatty acid elongation) were instead specifically enriched for the upregulated miRNA networks of EAE mice responding to PM (i.e. PM vs Ctrl PM and EAE PM vs EAE comparisons; Figure 6A; Supplementary Table S6).
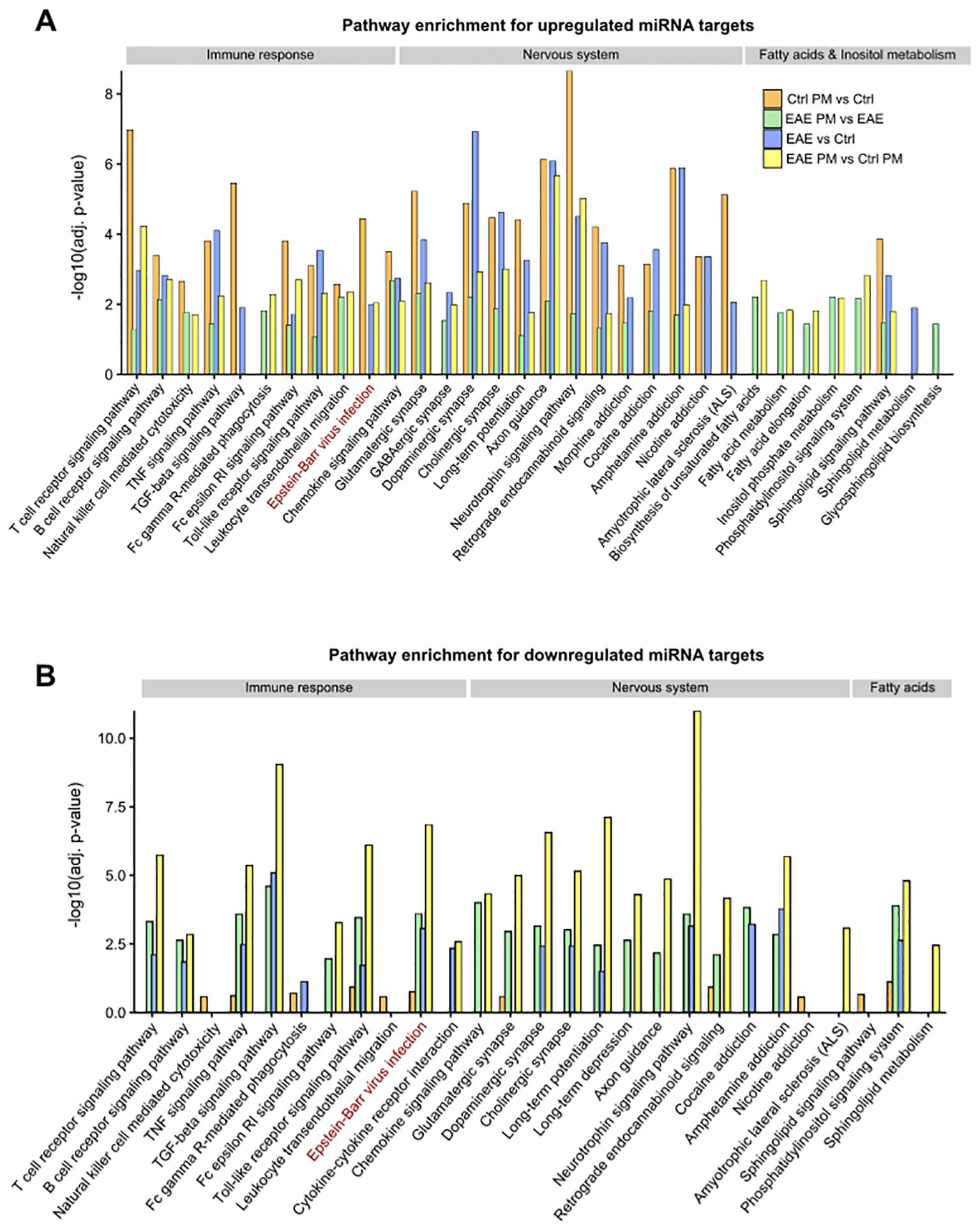
Figure 6. KEGG Pathway enrichment analysis of miRNA target networks. Bar plots showing selected significantly enriched pathways (adjusted p < 0.05) for the target genes of upregulated (A) and downregulated (B) miRNAs across the distinct comparisons. The y-axis indicates the –log10 of the adjusted p-values (adj. p-value). The displayed pathways represent those most relevant to nervous system function, immune response, and fatty acid/inositol phosphate metabolism. See Supplementary Table S6 for the entire list of enriched pathways.
Gene-to-disease association of miRNA targets
To link EV-packaged miRNA dysregulation with possible pathological alterations elicited by PM exposure, we used the list of predicted target gene transcripts to perform a “gene-to-disease association” analysis. Again of relevance in the context of MS, and consistent with the KEGG pathway analysis, many of the targets of the miRNAs selectively deregulated in PM-exposed EAE vs PM-exposed healthy mice (i.e. EAE PM vs Ctrl PM; see Supplementary Table S5) are involved in nervous systems diseases and immune system diseases, and particularly in autoimmune diseases including MS (Figures 7A–D; Supplementary Table S7). Consistently, when assessing the fraction of gene transcripts targeted by miRNAs selectively deregulated in PM-exposed EAE mice over the entire number of genes listed in each of the disease classes included in the DisGeNET database, “Nervous System Diseases” was the most represented class, followed by Mental Disorders, Hemic and Lymphatic Diseases, Behavior and Behavior Mechanisms and Immune System Diseases (Figures 7E, F).
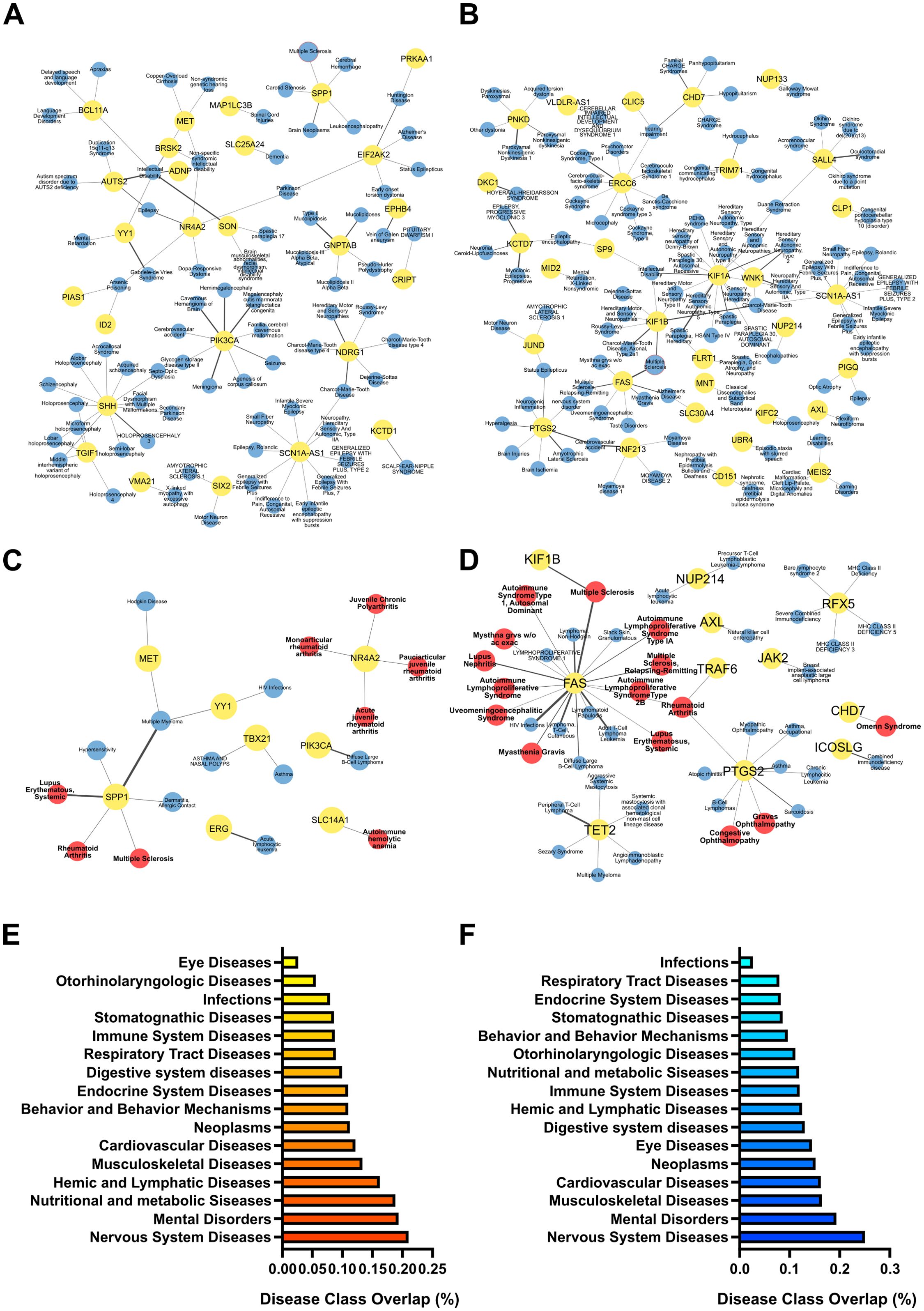
Figure 7. Pathological relevance of the selective response of EAE mice to PM10. Gene-to-disease association performed with DisGeNET performed for the target genes of miRNAs significantly upregulated (A, C) and downregulated (B, D) specifically in EAE PM vs Ctrl PM. (A-D) Networks of target genes (in yellow dots) associated with pathologies (in blue dots) in the DisGeNET disease class “Nervous Systems Diseases” (A, B) and “Immune System Diseases” (C, D). Multiple Sclerosis (MS) node is highlighted with a red border in (A, B). In the “Immune System Diseases” Network, Autoimmune diseases are represented by red dots. Thickness of links is proportional to the DisGeNET score, as a measure of quality/number of evidence supporting the gene-to-disease association. (E, F) Gene enrichment in disease classes found in the DisGeNET database. Bars represent the fraction of target genes over the entire number of genes listed in each disease class. Source data are provided in Supplementary Table S7.
Overall, this analysis showed that gene transcripts targeted by miRNAs selectively altered in EAE mice exposed to PM10 were associated with nervous system disorders and with pathologies closely related to MS, suggesting that the plasmatic EV cargo may mediate the transfer of PM-induced damage to the CNS and synergize with the immune setting of the individual, possibly contributing to immune dysregulation in the CNS via yet unknown mechanisms.
Discussion
With the aim of disclosing the biological bases of the higher vulnerability of immunologically-primed subjects to PM exposure, here we characterized plasmatic EVs and their miRNA cargo in healthy and presymptomatic EAE mice following a single exposure to high level PM10. We focused on PM10 because it has been consistently associated with MS-related outcomes in epidemiological studies (see above), and it includes finer fractions such as PM2.5. Results showed that the response of EAE mice to PM did not differ in terms of EV number and source, compared to that of healthy mice. Yet, remarkable differences existed in the identity of deregulated EV-associated miRNAs, which were predicted to target MS-relevant pathways and might contribute to disease worsening.
As regards EV abundance and features, PM10 exposure induced a significant decrease in plasmatic EV release in both healthy and EAE mice - accompanied by a moderate increase in size in PM10-exposed EAE mice - compared to unexposed individuals. These findings clash with what observed in humans - where short-term PM exposure has been associated with increases in plasmatic EVs (8, 15, 51), and may suggest a species-specific response, with possible change in EV release mechanisms in PM10-exposed EAE mice (42). Yet, EV numbers and size remained in the range of those reported in humans following PM exposure (8, 15) and EV size did not reach that of apoptotic bodies (42), thus not indicating higher toxicity of PM10 to murine cells.
As regards EV cargo, here we focused on EV-associated miRNAs as informative post-transcriptional markers, given their regulatory role and the interpretability of their dysregulation through known mRNA targets. In both healthy and EAE mice, the miRNA cargo of EVs released following PM exposure had a signature relevant for MS. However, deregulated miRNAs were distinct and the response of EAE mice appeared remarkably more “pathogenic” than that of healthy mice. Specifically, in Ctrl mice, exposure to PM10 led to a complex and “ambivalent” miRNA signature, including decreased “protective” miRNAs (e.g. miR-29a-5p; 52–55; miR-2134; 56), but also decreased proinflammatory miRNAs (e.g. miR-712-5p, 57) and increased anti-inflammatory miRNAs (e.g. miR-215-5p; 58, 59; miR146b-5p; 60, 61; miR-199a-3p; 62, 63). Of note, among miRNAs upregulated in healthy mice exposed to PM10, miR-146b-5p was formerly found upregulated in the blood and white matter tissue of MS patients and MS animal models (64), presenting a peak of expression in peripheral blood mononuclear cells (PBMCs) of treatment-naïve relapsing and remitting MS (RRMS) patients during the remission phase (65), in line with miR-146b-5p role as a molecular brake for T-cell B-cell crosstalk restraining autoimmunity (66). Similarly, miR-199a-3p expression in PBMCs was proposed to mediate the effects of disease-modifying therapies (DMTs) in RRMS patients (67), in line with its role in promoting immune tolerance (68).
Among these deregulated miRNAs, miR-215-5p and miR-199a-3p have been already reported as EV-packaged miRNAs in human serum (69) and mouse bronchoalveolar lavage fluid (70), respectively. Moreover, increased expression of miR-215-5p, miR-146b-5p and miR-199a-3p was detected in human and mouse lung epithelial cells upon exposure to PM2.5 and other pollutants (70–72).
All in all, these findings point to a prevalent compensatory/protective response elicited in healthy mice by short term exposure to PM10.
In contrast, in EAE mice, PM exposure resulted in a strong enrichment in miR-9-5p, whose level in the serum was correlated with clinical activity, brain atrophy and disability progression in MS patients (73–75). Such an association appears to rely on miR-9-5p role in microglia activation and neuroinflammation (76, 77), dendritic cell activation (78), Th17/Treg differentiation (79, 80), and oligodendroglia maturation (81). Notably, miR-9-5p was also one of the “hub” upregulated miRNAs highlighted by the network analysis in EAE mice exposed to PM10 (vs unexposed EAE mice and vs PM10 exposed healthy mice), corroborating the idea of its critical involvement in the response of EAE mice to PM10. In addition, EAE mice showed a strong decrease of anti-inflammatory miRNAs, such as miR-345-5p (82), miR-192-5p (83, 84) and miR-451a (85), compared to unexposed EAE mice. Of note, miR451a was formerly shown to play a wide array of immunoregulatory actions, by attenuating macrophage and dendritic cell responses (86), inhibiting T cell activation (87), suppressing NK cell activation and cytotoxicity (88) and regulating myeloid-derived suppressor cells differentiation in the context of autoimmune disease (85). In line with this, miR-451a was proposed to contribute to the remission phase of a number of autoimmune diseases, such as familial Mediterranean fever (89), rheumatoid arthritis (RA, 90) and systemic lupus erythematosus (SLE, 85, 91). Similar to miR-9-5p, miR-451a was also one of the “hub” downregulated miRNAs highlighted by the network analysis in EAE mice exposed to PM10 (vs unexposed EAE mice and vs PM10 exposed healthy mice), in line with a key role in EAE mouse response to PM10.
In healthy mice miRNAs deregulated after immunization and after PM10 exposure partly overlapped, suggesting a similar outcome in response to autoimmune and environmental triggers. In contrast, the response of EAE mice to PM10 was largely segregated from the response of healthy mice. Notably, while comparing the response of EAE and healthy mice to PM10 (i.e. EAE PM vs Ctrl PM), one of the top enriched miRNAs was miR-142-3p, which was shown to impact MS progression, severity, and therapeutic outcomes by orchestrating neuronal toxicity and excitotoxic synaptic alterations, (92–95), and possibly by regulating also Treg cell differentiation (96) and myelin repair (97). On the other hand, in addition to miR-451a, one of the top downregulated miRNAs in EAE PM vs Ctrl PM was the anti-inflammatory miR-25-3p (98), formerly found down-regulated in Treg cells of MS patients and proposed to alter Treg cell activity in MS by targeting TGF-β biological functions (99).
Overall, these findings indicate that the response to PM10 – in terms of deregulated plasma EV-associated miRNAs – is qualitatively different in mice primed to develop autoimmunity compared to healthy mice, and includes a repertoire of miRNAs that, if also deregulated in PM10-exposed MS patients, might contribute to key pathological aspects, including autoimmunity, CNS infiltration, neuroinflammation, neurodegeneration and myelin repair.
The special character of the response of EAE mice to PM10 and its relevance for MS were also highlighted by miRNA target prediction and network analysis, as well as by the analysis of the biological pathways potentially influenced by the detected miRNA deregulation. In general, the networks of upregulated miRNAs were remarkably complex, in line with the idea that upregulated miRNAs might have a broad biological impact, influencing a wide range of cellular processes. Pathways related to synapse function, neurotransmitter systems, nervous system, immune system and inflammation were significantly enriched for the networks of upregulated miRNAs in both healthy and EAE mice, whereas pathways related to immune system and inflammation emerged almost exclusively for the networks of downregulated miRNAs in EAE mice responding to PM10. Interestingly, among immune-related pathways, Epstein Barr virus (EBV) infection emerged as one of the most enriched pathways for downregulated miRNAs in PM-exposed EAE mice, suggesting that exposure to PM10 might result in the upregulation of target genes belonging to this category and might mimic the anti-EBV response in EAE mice. This finding is particularly intriguing as EBV infection is epidemiologically linked with the development of autoimmune diseases - including SLE, RA and MS - and multiple mechanisms associated with EBV infections (e.g. molecular mimicry, B cell reprogramming, crosstalk between EBV-infected B cells and T cells, etc.) are now considered causally implicated in autoimmunity and MS development (100, 101). Moreover, as an additional element of uniqueness of EAE mouse response to PM10, terms associated with FA metabolism (Biosynthesis of unsaturated fatty acids, Fatty acid metabolism, Fatty acid elongation) were specifically enriched for the upregulated miRNA networks of EAE PM vs Ctrl PM and EAE PM vs EAE comparisons. Of note, altered lipid profiles were reported in MS patients, with FA serological concentration reflecting disease activity and disability score, and a number of FA metabolism-related enzyme single nucleotide polymorphisms (SNPs) associated with MS incidence (102). Even before onset, MS patients display a unique FA serological profile and FA intake and metabolism are thought to contribute to MS susceptibility (102). Availability and profiles of FA species might alter the disease course by influencing immune cell function and specifically by modulating the polarization, differentiation, and cytokine production of T cells, which largely contribute to MS pathogenesis (103). Thus, a clear MS-relevant signature of the response of presymptomatic EAE mice to PM10 emerged, as also corroborated by “gene-to-disease association” analysis.
Limitations of the study and future directions
Overall, our findings support the view that, in immunologically primed individuals - such as people with MS - exposure to PM10 can trigger a unique response through the delivery of EV-associated miRNAs, which can contribute to a worse manifestation or an unfavorable evolution of the pathology. Yet, our study has limitations that must be disclosed to correctly interpret our findings and to identify persisting open issues. Here, we combined exposure to PM10 with EAE, which is the closest animal model that approximates human MS pathogenesis by reflecting its autoimmune component (104). Yet, as our results point to a likely participation of exposure to PM10 in immune dysregulation, it is relevant to acknowledge that some differences exist in the innate and adaptive immune system of mice and humans (105), potentially limiting the translatability of our findings and calling for future investigations in MS patients. Moreover, besides the relatively small size of individuals used in this kind of in vivo experiments, here we used only female mice, as in most studies focusing on EAE. This choice relies on the well-known sexual dimorphism existing in MS, with women showing a higher susceptibility to the disease (106). Yet, disease progression appears worse in men (106) and mechanisms leading to EAE partially differ in female and male mice (107). Inclusion of male mice in future studies will validate the generalizability of our findings or, alternatively, unveil sex-specific aspects in the response to PM10 of immunologically primed individuals. Moreover, as regards the analysis of EV-packaged miRNAs, this study did not include EV subtype-specific miRNA profiling due to limited EV yield, and sample pooling - while necessary due to sample constraints and ethical (i.e. 3Rs) considerations - limited the assessment of inter-individual variability. Finally, our study investigated an acute (i.e. occurring within 6 hours) biological response triggered by a single exposure to PM10. While this was instrumental to investigate the short-term effect of the peaks of airborne PM10 which were associated with higher rates of hospital admissions for MS and MS relapses (22, 29), results obtained in this experimental setting might not reflect the biological responses elicited upon long-term or chronic exposure to PM10. Similarly, in our study PM10 dose was calculated to mimic daily peaks of PM10 concentration in the most polluted European areas (>250 μg/m3; 41). Whether exposure to higher PM10 concentrations - as detected in other geographical regions (108) - or to other risk factors epidemiologically associated with MS risk may converge on the same biological mechanisms unveiled by our study deserves further investigation.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was approved by the Italian Ministry of Health and the Bioethical Committee of the University of Turin. The study was designed according to the guidelines of the European Communities Council (2010/63/EU) and the Italian Law for Care and Use of Experimental Animals (DL26/2014), and was conducted according to the ARRIVE guidelines. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
MB: Data curation, Formal Analysis, Investigation, Methodology, Visualization, Writing – review & editing. VC: Data curation, Formal Analysis, Investigation, Methodology, Visualization, Writing – review & editing. LD: Formal Analysis, Investigation, Methodology, Visualization, Writing – review & editing. FM: Investigation, Methodology, Writing – review & editing. RP: Investigation, Methodology, Writing – review & editing. AB: Supervision, Writing – review & editing. VB: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing. LF: Conceptualization, Project administration, Supervision, Writing – original draft, Writing – review & editing. EB: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. Our work was supported by FISM - Fondazione Italiana Sclerosi Multipla (Italy) (ID:2019/PR-Multi/003) to EB and VB, and by Cassa di Risparmio di Torino (CRT) Foundation grant (ID: 2021.0657) and Ministero dell’Istruzione, dell’Università e della Ricerca—MIUR (Italy) PRIN (Research Programs of National Interest) - PNRR 2022 (ID: P20225Z3J5) to EB. MB was supported by a PON R&I 2014–2020 PhD Fellowship “Dottorati di ricerca su tematiche green e dell’innovazione”, financed by Ministero dell’Istruzione, dell’Università e della Ricerca—MIUR (Italy) in the frame of FSE–REACT EU. This study was also supported by Ministero dell’Istruzione, dell’Università e della Ricerca—MIUR (Italy) project “Dipartimenti di Eccellenza 2018–2022” and “Dipartimenti di Eccellenza 2023–2027” to Department of Neuroscience “Rita Levi Montalcini” of the University of Turin and to Department of Clinical Sciences and Community Health of the University of Milan.
Acknowledgments
We sincerely thank Dr. Vita Cardinale and Prof. Maurizio Giustetto, Department of Neuroscience “Rita Levi Montalcini” of the University of Turin, for fundamental technical support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1596935/full#supplementary-material
Supplementary Figure 1 | Global characterization of isolated EVs. (A) Western Blotting analyses on plasma EV lysates from 3 representative mice. WB for CD63 and CD9 shows an enrichment in EV samples. WB for markers of other cellular compartments (i.e., calnexin) shows no positivity in EV samples and a positive band in the mouse cerebral cortex extract (positive control). (B) Size profile of isolated EVs. Distribution of EV concentration across the 30–700 nm size range, as measured by NTA, for each experimental group (CTRL, CTRL PM, EAE, EAE PM). Each curve represents the mean relative abundance of EVs over replicate measurements, and the colored halo is defined by the standard error at each size.
Supplementary Figure 2 | Agilent profiles of isolated miRNAs. Representative profiles of the miRNAs isolated from 2 mice, showing RNA integrity and suitability for subsequent analyses.
References
1. Sierra-Vargas MP and Teran LM. Air pollution: Impact and prevention. Respirology. (2012) 17:1031–8. doi: 10.1111/j.1440-1843.2012.02213.x
2. Stanaway JD, Afshin A, Gakidou E, Lim SS, Abate D, Abate KH, et al. Global, regional, and national comparative risk assessment of 84 behavioural, environmental and occupational, and metabolic risks or clusters of risks for 195 countries and territories 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. (2018) 392:1923–94. doi: 10.1016/s0140-6736(18)32225-6
3. Boda E, Rigamonti AE, and Bollati V. Understanding the effects of air pollution on neurogenesis and gliogenesis in the growing and adult brain. Curr Opin Pharmacol. (2020) 50:61–6. doi: 10.1016/j.coph.2019.12.003
4. Peeples L. News Feature: How air pollution threatens brain health. Proc Natl Acad Sci. (2020) 117(25):13856–60. doi: 10.1073/pnas.2008940117
5. Flood-Garibay JA, Angulo-Molina A, and Méndez-Rojas MÁ. Particulate matter and ultrafine particles in urban air pollution and their effect on the nervous system. Environ Sci Processes Impacts. (2023) 25:704–26. doi: 10.1039/d2em00276k
6. Becker S, Fenton MJ, and Soukup JM. Involvement of microbial components and toll-like receptors 2 and 4 in cytokine responses to air pollution particles. Am J Respir Cell Mol Biol. (2002) 27:611–8. doi: 10.1165/rcmb.4868
7. Pavanello S, Bonzini M, Angelici L, Motta V, Pergoli L, Hoxha M, et al. Extracellular vesicle-driven information mediates the long-term effects of particulate matter exposure on coagulation and inflammation pathways. Toxicol Lett. (2016) 259:143–50. doi: 10.1016/j.toxlet.2016.08.002
8. Pergoli L, Cantone L, Favero C, Angelici L, Iodice S, Pinatel E, et al. Extracellular vesicle-packaged miRNA release after short-term exposure to particulate matter is associated with increased coagulation. Particle Fibre Toxicol. (2017) 14(1):32. doi: 10.1186/s12989-017-0214-4
9. Bonzini M, Pergoli L, Cantone L, Hoxha M, Spinazzè A, Buono LD, et al. Short-term particulate matter exposure induces extracellular vesicle release in overweight subjects. Environ Res. (2017) 155:228–34. doi: 10.1016/j.envres.2017.02.014
10. Eckhardt CM, Baccarelli AA, and Wu H. Environmental exposures and extracellular vesicles: indicators of systemic effects and human disease. Curr Environ Health Rep. (2022) 9(3):465–76. doi: 10.1007/s40572-022-00357-5
11. Yuan D, Zhao Y, Banks WA, Bullock KM, Haney M, Batrakova E, et al. Macrophage exosomes as natural nanocarriers for protein delivery to inflamed brain. Biomaterials. (2017) 142:pp.1–12. doi: 10.1016/j.biomaterials.2017.07.011
12. Liu T, Zhang Q, Zhang J, Li C, Miao Y, Lei Q, et al. EVmiRNA: a database of miRNA profiling in extracellular vesicles. Nucleic Acids Res. (2018) 47:D89–93. doi: 10.1093/nar/gky985
13. Fabian MR, Sonenberg N, and Filipowicz W. Regulation of mRNA Translation and Stability by microRNAs. Annu Rev Biochem. (2010) 79:351–79. doi: 10.1146/annurev-biochem-060308-103103
14. Raposo G and Stoorvogel W. Extracellular vesicles: Exosomes, microvesicles, and friends. J Cell Biol. (2013) 200:pp.373–383. doi: 10.1083/jcb.201211138
15. Rota F, Ferrari L, Hoxha M, Favero C, Antonioli R, Pergoli L, et al. Blood-derived extracellular vesicles isolated from healthy donors exposed to air pollution modulate in vitro endothelial cells behavior. Sci Rep. (2020) 10:20138. doi: 10.1038/s41598-020-77097-9
16. Nicholson S, Baccarelli A, and Prada D. Role of brain extracellular vesicles in air pollution-related cognitive impairment and neurodegeneration. Environ Res. (2022) 204:112316. doi: 10.1016/j.envres.2021.112316
17. Ebers GC. Environmental factors and multiple sclerosis. Lancet Neurol. (2008) 7:268–77. doi: 10.1016/S1474-4422(08)70042-5
18. Hooper LG and Kaufman JD. Ambient air pollution and clinical implications for susceptible populations. Ann Am Thorac Soc. (2018) 15:pp.S64–S68. doi: 10.1513/AnnalsATS.201707-574MG
19. Alfredsson L and Olsson T. Lifestyle and environmental factors in multiple sclerosis. Cold Spring Harbor Perspect Med. (2018) 9:a028944. doi: 10.1101/cshperspect.a028944
20. Belbasis L, Bellou V, Evangelou E, Ioannidis JPA, and Tzoulaki I. Environmental risk factors and multiple sclerosis: an umbrella review of systematic reviews and meta-analyses. Lancet Neurol. (2015) 14:263–73. doi: 10.1016/s1474-4422(14)70267-4
21. Giovannoni G, Hawkes CH, Lechner-Scott J, Levy M, and Pohl D. Air pollution and multiple sclerosis risk. Multiple Sclerosis Related Disord. (2021) 48:102797. doi: 10.1016/j.msard.2021.102797
22. Angelici L, Piola M, Cavalleri T, Randi G, Cortini F, Bergamaschi R, et al. Effects of particulate matter exposure on multiple sclerosis hospital admission in Lombardy region, Italy. Environ Res. (2015) 145:68–73. doi: 10.1016/j.envres.2015.11.017
23. Jeanjean M, Bind M, Roux J, Ongagna J, De Sèze J, Bard D, et al. Ozone, NO2 and PM10 are associated with the occurrence of multiple sclerosis relapses. Evidence from seasonal multi-pollutant analyses. Environ Res. (2018) 163:43–52. doi: 10.1016/j.envres.2018.01.040
24. Roux J, Bard D, Le Pabic E, Segala C, Reis J, Ongagna J-C, et al. Air pollution by particulate matter PM10 may trigger multiple sclerosis relapses. Environ Res. (2017) 156:404–10. doi: 10.1016/j.envres.2017.03.049
25. Heydarpour P, Amini H, Khoshkish S, Seidkhani H, Sahraian MA, and Yunesian M. Potential impact of air pollution on multiple sclerosis in Tehran, Iran. Neuroepidemiology. (2014) 43:pp.233–238. doi: 10.1159/000368553
26. Oikonen M, Laaksonen M, Laippala P, Oksaranta O, Lilius E, Lindgren S, et al. Ambient air quality and occurrence of multiple sclerosis relapse. Neuroepidemiology. (2003) 22:95–9. doi: 10.1159/000067108
27. Gregory AC, Shendell DG, Okosun IS, and Gieseker KE. Multiple Sclerosis disease distribution and potential impact of environmental air pollutants in Georgia. Sci Total Environ. (2008) 396:pp.42–51. doi: 10.1016/j.scitotenv.2008.01.065
28. Noorimotlagh Z, Azizi M, Pan H-F, Mami S, and Mirzaee SA. Association between air pollution and Multiple Sclerosis: A systematic review. Environ Res. (2020) 196:110386. doi: 10.1016/j.envres.2020.110386
29. Bergamaschi R and Montomoli C. Air pollution is a risk factor for multiple sclerosis – Yes. Multiple Sclerosis J. (2021) 27:pp.2137–2138. doi: 10.1177/13524585211035953
30. Januel E, Dessimond B, Colette A, Annesi-Maesano I, and Stankoff B. Fine particulate matter related to multiple sclerosis relapse in young patients. Front Neurol. (2021) 12:651084. doi: 10.3389/fneur.2021.651084
31. Woo MS, Engler JB, and Friese MA. The neuropathobiology of multiple sclerosis. Nat Rev Neurosci. (2024) 25:493–513. doi: 10.1038/s41583-024-00823-z
32. Benedict RHB. Cognition in multiple sclerosis: Charcot was right. Lancet Neurol. (2020) 19:810. doi: 10.1016/S1474-4422(20)30306-9
33. Sparaco M, Lavorgna L, and Bonavita S. Psychiatric disorders in multiple sclerosis. J Neurol. (2019). doi: 10.1007/s00415-019-09426-6
34. Calderón-Garcidueñas L, Villarreal-Calderon R, Valencia-Salazar G, Henríquez-Roldán C, Gutiérrez-Castrellón P, Torres-Jardón R, et al. Systemic inflammation, endothelial dysfunction, and activation in clinically healthy children exposed to air pollutants. Inhalation Toxicol. (2008) 20:499–506. doi: 10.1080/08958370701864797
35. Babadjouni RM, Hodis DM, Radwanski R, Durazo R, Patel A, Liu Q, et al. Clinical effects of air pollution on the central nervous system; A review. J Clin Neurosci: Off J Neurosurg Soc Australasia. (2017) 43:pp.16–24. doi: 10.1016/j.jocn.2017.04.028
36. Montarolo F, Perga S, Martire S, and Bertolotto A. Nurr1 reduction influences the onset of chronic EAE in mice. Inflammation Res. (2015) 64:841–4. doi: 10.1007/s00011-015-0871-4
37. Akhtar US, McWhinney RD, Rastogi N, Abbatt JPD, Evans GJ, and Scott JA. Cytotoxic and proinflammatory effects of ambient and source-related particulate matter (PM) in relation to the production of reactive oxygen species (ROS) and cytokine adsorption by particles. Inhalation Toxicol. (2010) 22:37–47. doi: 10.3109/08958378.2010.518377
38. Kewcharoenwong C, Khongmee A, Nithichanon A, Palaga T, Prueksasit T, Mudway IS, et al. Vitamin D3 regulates PM-driven primary human neutrophil inflammatory responses. Sci Rep. (2023) 13(1):15850. doi: 10.1038/s41598-023-43252-1
39. Gałuszka-Bulaga A, Tkacz K, Węglarczyk K, and Maciej Siedlar and Baran J. Air pollution induces pyroptosis of human monocytes through activation of inflammasomes and Caspase-3-dependent pathways. J Inflammation. (2023) 20(1):26. doi: 10.1186/s12950-023-00353-y
40. Parolisi R, Montarolo F, Pini A, Rovelli S, Cattaneo A, Bertolotto A, et al. Exposure to fine particulate matter (PM2.5) hampers myelin repair in a mouse model of white matter demyelination. Neurochem Int. (2021) 145:104991. doi: 10.1016/j.neuint.2021.104991
41. Žibert J, Cedilnik J, and Pražnikar J. Particulate matter (PM10) patterns in Europe: An exploratory data analysis using non-negative matrix factorization. Atmospheric Environ. (2016) 132:217–28. doi: 10.1016/j.atmosenv.2016.03.005
42. Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. (2018) 7:1535750. doi: 10.1080/20013078.2018.1535750
43. Ferrari L, Iodice S, Cantone L, Solazzo G, Dioni L, Hoxha M, et al. Extracellular vesicles and their miRNA contents counterbalance the pro-inflammatory effect of air pollution during physiological pregnancy: A focus on Syncytin-1 positive vesicles. Environ Int. (2022) 169:107502–2. doi: 10.1016/j.envint.2022.107502
44. Livak KJ and Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods. (2001) 25:402–8. doi: 10.1006/meth.2001.1262
45. Chang L, Zhou G, Soufan O, and Xia J. miRNet 2.0: network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. (2020) 48:W244–51. doi: 10.1093/nar/gkaa467
46. Piñero J, Saüch J, Sanz F, and Furlong LI. The DisGeNET cytoscape app: Exploring and visualizing disease genomics data. Comput Struct Biotechnol J. (2021) 19:2960–7. doi: 10.1016/j.csbj.2021.05.015
47. Borjini N, Fernández M, Giardino L, and Calzà L. Cytokine and chemokine alterations in tissue, CSF, and plasma in early presymptomatic phase of experimental allergic encephalomyelitis (EAE), in a rat model of multiple sclerosis. J Neuroinflamm. (2016) 13(1):291. doi: 10.1186/s12974-016-0757-6
48. Valente T, Serratosa J, Perpiñá U, Saura J, and Solà C. Alterations in CD200-CD200R1 System during EAE Already Manifest at Presymptomatic Stages. Front Cell Neurosci. (2017) 11:129. doi: 10.3389/fncel.2017.00129
49. Rizzo FR, Guadalupi L, Sanna K, Vanni V, Fresegna D, De Vito F, et al. Exercise protects from hippocampal inflammation and neurodegeneration in experimental autoimmune encephalomyelitis. Brain Behav Immun. (2021) 98:13–27. doi: 10.1016/j.bbi.2021.08.212
50. Sumida TS, Cheru NT, and Hafler DA. The regulation and differentiation of regulatory T cells and their dysfunction in autoimmune diseases. Nat Rev Immunol. (2024) 24(7):503–17. doi: 10.1038/s41577-024-00994-x
51. Rodosthenous RS, Coull BA, Lu Q, Vokonas PS, Schwartz J, and Baccarelli AA. Ambient particulate matter and microRNAs in extracellular vesicles: a pilot study of older individuals. Part Fibre Toxicol. (2016) 13:13. doi: 10.1186/s12989-016-0121-0
52. Yang J, Zhao J, Chen Y, Wang R, Rong Z, Wang S, et al. miR-29a-5p rescues depressive-like behaviors in a CUMS-induced mouse model by facilitating microglia M2-polarization in the prefrontal cortex via TMEM33 suppression. J Affect Disord. (2024) 360:188–97. doi: 10.1016/j.jad.2024.05.156
53. Zhao F, Zhao H, Fan J, Wang R, Han Z, Tao Z, et al. MIR-29A knockout aggravates neurological damage by pre-polarizing M1 microglia in experimental RAT models of acute stroke. Front Genet. (2021) 12:642079. doi: 10.3389/fgene.2021.642079
54. Zhang A, Lu Y, Yuan L, Zhang P, Zou D, Wei F, et al. MIR-29A-5P alleviates traumatic brain injury- (TBI-) induced permeability disruption via regulating NLRP3 pathway. Dis Markers. (2021) 2021:1–11. doi: 10.1155/2021/9556513
55. Zhang L, Jiang S, Guo X, Xiao B, Li Q, Chen J, et al. MiR-146b-5p targets IFI35 to inhibit inflammatory response and apoptosis via JAK1/STAT1 signalling in lipopolysaccharide-induced glomerular cells. Autoimmunity. (2021) 54:430–8. doi: 10.1080/08916934.2020.1864730
56. Baltan S, Sandau US, Brunet S, Bastian C, Tripathi A, Nguyen H, et al. Identification of miRNAs that mediate protective functions of anti-Cancer drugs during white matter ischemic injury. ASN Neuro. (2021) 13:175909142110422–175909142110422. doi: 10.1177/17590914211042220
57. Qin Y, Zheng B, Yang G, Zhou J, Yang H, Nie Z, et al. Tanshinone IIA inhibits VSMC inflammation and proliferation in vivo and in vitro by downregulating miR-712-5p expression. Eur J Pharmacol. (2020) 880:173140. doi: 10.1016/j.ejphar.2020.173140
58. Zhu W, Peng F, Cui X, Li J, and Sun C. LncRNA SOX2OT facilitates LPS-induced inflammatory injury by regulating intercellular adhesion molecule 1 (ICAM1) via sponging miR-215-5p. Clin Immunol. (2022) 238:109006. doi: 10.1016/j.clim.2022.109006
59. Yao Y, Xu K, Sun Y, Tian T, Shen W, Sun F, et al. MiR-215-5p inhibits the inflammation injury in septic H9c2 by regulating ILF3 and LRRFIP1. Int Immunopharmacol. (2019) 78:106000. doi: 10.1016/j.intimp.2019.106000
60. Zhang M, Chen H, Yang Z, Zhang M, Wang X, Zhao K, et al. 17β-Estradiol attenuates LPS-Induced macrophage inflammation in vitro and Sepsis-Induced vascular inflammation in vivo by upregulating MIR-29A-5P expression. Mediators Inflammation. (2021) 2021:1–16. doi: 10.1155/2021/9921897
61. Wang C, Cheng H, Yan F, Zhang H, Zhang J, Li C, et al. MicroRNA-146b protects kidney injury during urinary tract infections by modulating macrophage polarization. mBio. (2023) 14(6):e0209423. doi: 10.1128/mbio.02094-23
62. Liu M, Cao Y, Hu Y, Zhang Z, Ji S, Shi L, et al. MIR-199A-3P restrains foaming and inflammation by regulating RUNX1 in macrophages. Mol Biotechnol. (2022) 64:1130–42. doi: 10.1007/s12033-022-00484-2
63. Gu X, Weng R, Hou J, and Liu S. Endothelial miR-199a-3p regulating cell adhesion molecules by targeting mTOR signaling during inflammation. Eur J Pharmacol. (2022) 925:174984. doi: 10.1016/j.ejphar.2022.174984
64. Ma X, Zhou J, Zhong Y, Jiang L, Mu P, Li Y, et al. Expression, regulation and function of MicroRNAs in multiple sclerosis. Int J Med Sci. (2014) 11:810–8. doi: 10.7150/ijms.8647
65. Baulina N, Kulakova O, Kiselev I, Osmak G, Popova E, Boyko A, et al. Immune-related miRNA expression patterns in peripheral blood mononuclear cells differ in multiple sclerosis relapse and remission. J Neuroimmunol. (2018) 317:67–76. doi: 10.1016/j.jneuroim.2018.01.005
66. Cho S, Lee H, Yu I, Choi YS, Huang H, Hashemifar SS, et al. Differential cell-intrinsic regulations of germinal center B and T cells by miR-146a and miR-146b. Nat Commun. (2018) 9(1):2757. doi: 10.1038/s41467-018-05196-3
67. Arisi I, Malimpensa L, Manzini V, Brandi R, Di Sturmeck TG, D’Amelio C, et al. Cladribine and ocrelizumab induce differential miRNA profiles in peripheral blood mononucleated cells from relapsing–remitting multiple sclerosis patients. Front Immunol. (2023) 14:1234869. doi: 10.3389/fimmu.2023.1234869
68. Xiong A, Wang J, Mao XL, Jiang Y, and Fan Y. MiR-199a-3p modulates the function of dendritic cells involved in transplantation tolerance by targeting CD86. HLA. (2019) 94:493–503. doi: 10.1111/tan.13677
69. Vázquez-Mera S, Martelo-Vidal L, Miguéns-Suárez P, Saavedra-Nieves P, Arias P, González-Fernández C, et al. Serum exosome inflamma-miRs are surrogate biomarkers for asthma phenotype and severity. Allergy. (2022) 78:pp.141–155. doi: 10.1111/all.15480
70. Carnino JM, Lee H, Smith LC, Sunil VR, Rancourt RC, Vayas K, et al. Microvesicle-Derived MIRNAs regulate proinflammatory macrophage activation in the lung following ozone exposure. Toxicol Sci. (2022) 187:162–74. doi: 10.1093/toxsci/kfac025
71. Cai Y, Li R, Zheng K, Wang B, Qin S, Li B, et al. Effect of c-fos gene silence on PM2.5-induced miRNA alteration in human bronchial epithelial cells. Environ Toxicol Pharmacol. (2021) 84:103607–7. doi: 10.1016/j.etap.2021.103607
72. Hou T, Chen Q, and Ma Y. Elevated expression of miR-146 involved in regulating mice pulmonary dysfunction after exposure to PM2.5. J Toxicol Sci. (2021) 46:437–43. doi: 10.2131/jts.46.437
73. Casanova I, Domínguez-Mozo MI, De Torres L, Aladro-Benito Y, García-Martínez Á, Gómez P, et al. MicroRNAs associated with disability progression and clinical activity in multiple sclerosis patients treated with glatiramer acetate. Biomedicines. (2023) 11:2760–0. doi: 10.3390/biomedicines11102760
74. Domínguez-Mozo MI, Casanova I, Monreal E, Costa-Frossard L, Sainz-de-la-Maza S, Sainz-Amo R, et al. Association of microRNA expression and serum neurofilament light chain levels with clinical and radiological findings in multiple sclerosis. Int J Mol Sci. (2024) 25:pp.10012–10012. doi: 10.3390/ijms251810012
75. Dominguez-Mozo MI, Casanova I, De Torres L, Aladro-Benito Y, Perez-Perez S, Garcia-Martínez A, et al. MicroRNA expression and its association with disability and brain atrophy in multiple sclerosis patients treated with glatiramer acetate. Front Immunol. (2022) 13:904683. doi: 10.3389/fimmu.2022.904683
76. Yao H, Ma R, Yang L, Hu G, Chen X, Duan M, et al. MiR-9 promotes microglial activation by targeting MCPIP1. Nat Commun. (2014) 5:4386. doi: 10.1038/ncomms5386
77. Yue P, Jing L, Zhao X, Zhu H, and Teng J. Down-regulation of taurine-up-regulated gene 1 attenuates inflammation by sponging miR-9-5p via targeting NF-κB1/p50 in multiple sclerosis. Life Sci. (2019) 233:116731. doi: 10.1016/j.lfs.2019.116731
78. Stumpfova Z, Hezova R, Meli AC, Slaby O, and Michalek J. MicroRNA Profiling of Activated and Tolerogenic Human Dendritic Cells. Mediators of Inflammation. (2014) 10:259689. doi: 10.1155/2014/259689
79. Majd M, Hosseini A, Ghaedi K, Kiani-Esfahani A, Tanhaei S, Shiralian-Esfahani H, et al. MiR-9-5p and miR-106a-5p dysregulated in CD4+ T-cells of multiple sclerosis patients and targeted essential factors of T helper17/regulatory T-cells differentiation. PubMed. (2018) 21:pp.277–283. doi: 10.22038/ijbms.2018.25382.6275
80. Shirani F, Baghi M, Rostamian Delavar M, Shoaraye Nejati A, Eshaghiyan A, Nasr-Esfahani MH, et al. Upregulation of miR-9 and miR-193b over human Th17 cell differentiation. Mol Genet Genom Med. (2020) 8(12):e1538. doi: 10.1002/mgg3.1538
81. Barca-Mayo O and Lu QR. Fine-tuning oligodendrocyte development by microRNAs. Front Neurosci. (2012) 6:13. doi: 10.3389/fnins.2012.00013
82. Liu J, Jiang Y, Han M, Jiang L, Liang D, Li S, et al. MicroRNA-345-5p acts as an anti-inflammatory regulator in experimental allergic rhinitis via the TLR4/NF-κB pathway. Int Immunopharmacol. (2020) 86:106522. doi: 10.1016/j.intimp.2020.106522
83. Tsukamoto H, Kouwaki T, and Oshiumi H. Aging-associated extracellular vesicles contain immune regulatory microRNAs alleviating hyperinflammatory state and immune dysfunction in the elderly. iScience. (2020) 23:101520. doi: 10.1016/j.isci.2020.101520
84. An L and Yin F. MiR-192-5p suppresses M1 macrophage polarization via epiregulin (EREG) downregulation in gouty arthritis. Tissue Cell. (2021) 73:101669. doi: 10.1016/j.tice.2021.101669
85. Shi G, Li D, Zhang D, Xu Y, Pan Y, Lu L, et al. IRF-8/miR-451a regulates M-MDSC differentiation via the AMPK/mTOR signal pathway during lupus development. Cell Death Discov. (2021) 7(1):179. doi: 10.1038/s41420-021-00568-z
86. Okamoto M, Fukushima Y, Kouwaki T, Daito T, Kohara M, Kida H, et al. MicroRNA-451a in extracellular, blood-resident vesicles attenuates macrophage and dendritic cell responses to influenza whole-virus vaccine. J Biol Chem. (2018) 293:18585–600. doi: 10.1074/jbc.ra118.003862
87. Zeng Z, Wang K, Li Y, Xia N, Nie S, Lv B, et al. Down-regulation of microRNA-451a facilitates the activation and proliferation of CD4+ T cells by targeting Myc in patients with dilated cardiomyopathy. J Biol Chem. (2017) 292:6004–13. doi: 10.1074/jbc.m116.765107
88. Li Y, Guan X, Lan T, Zhang Z, Zhang Y, Jiang S, et al. The miR-451a facilitates natural killer cell-associated immune deficiency after ischemic stroke. J Cereb Blood Flow Metab. (2025). doi: 10.1177/0271678x251321641
89. Terzi MY, Özcan O, Kimyon G, Okuyan HM, Arpacı A, and Doğan S. miR-451a and IL18 can differentiate familial Mediterranean fever patients in attack and remission periods: a prospective cross-sectional study. Clin Rheumatol. (2025) 44(4):1691–704. doi: 10.1007/s10067-025-07359-2
90. Mi L, Gao J, Li N, Liu Y, Zhang N, Gao Y, et al. Human umbilical cord mesenchymal stem cell-derived exosomes loaded miR-451a targets ATF2 to improve rheumatoid arthritis. Int Immunopharmacol. (2023) 127:111365. doi: 10.1016/j.intimp.2023.111365
91. Tan L, Zhao M, Wu H, Zhang Y, Tong X, Gao L, et al. Downregulated serum exosomal MIR-451A expression correlates with renal damage and its intercellular communication role in systemic lupus erythematosus. Front Immunol. (2021) 12:630112. doi: 10.3389/fimmu.2021.630112
92. De Vito F, Balletta S, Caioli S, Musella A, Guadalupi L, Vanni V, et al. MiR-142-3p is a critical modulator of TNF-mediated neuronal toxicity in multiple sclerosis. Curr Neuropharmacol. (2023) 21:2567–82. doi: 10.2174/1570159x21666230404103914
93. Mandolesi G, Vito F, Musella A, Gentile A, Bullitta S, Fresegna D, et al. miR-142-3p is a key regulator of IL-1β-dependent synaptopathy in neuroinflammation. J Neurosci. (2017) 37:546–61. doi: 10.1523/jneurosci.0851-16.2016
94. De Vito F, Musella A, Fresegna D, Rizzo FR, Gentile A, Stampanoni Bassi M, et al. MiR-142-3p regulates synaptopathy-driven disease progression in multiple sclerosis. Neuropathol Appl Neurobiol. (2021) 48(2):e12765. doi: 10.1111/nan.12765
95. Dolcetti E, Musella A, Balletta S, Gilio L, Bruno A, Bassi MS, et al. Interaction between miR-142-3p and BDNF val/met polymorphism regulates multiple sclerosis severity. Int J Mol Sci. (2024) 25:pp.5253–5253. doi: 10.3390/ijms25105253
96. Scherm MG, Serr I, Zahm AM, Schug J, Bellusci S, Manfredini R, et al. miRNA142-3p targets Tet2 and impairs Treg differentiation and stability in models of type 1 diabetes. Nat Commun. (2019) 10:5697. doi: 10.1038/s41467-019-13587-3
97. Hinman JD, Ngo KJ, Kim D, Chen C, Abraham CR, Ghanbari M, et al. miR-142-3p regulates cortical oligodendrocyte gene co-expression networks associated with tauopathy. Hum Mol Genet. (2021) 30:pp.103–118. doi: 10.1093/hmg/ddaa252
98. Luo X-Y, Ying J-H, and Wang Q-S. miR-25-3p ameliorates SAE by targeting the TLR4/NLRP3 axis. Metab Brain Dis. (2022) 37:1803–13. doi: 10.1007/s11011-022-01017-1
99. De Santis G, Ferracin M, Biondani A, Caniatti L, Tola MR, Castellazzi M, et al. Altered miRNA expression in T regulatory cells in course of multiple sclerosis. J Neuroimmunol. (2010) 226:165–71. doi: 10.1016/j.jneuroim.2010.06.009
100. Bjornevik K, Münz C, Cohen JI, and Ascherio A. Epstein–Barr virus as a leading cause of multiple sclerosis: mechanisms and implications. Nat Rev Neurol. (2023) 19(3):160–71. doi: 10.1038/s41582-023-00775-5
101. Gottlieb A, Pham HPT, Saltarrelli JG, and Lindsey JW. Expanded T lymphocytes in the cerebrospinal fluid of multiple sclerosis patients are specific for Epstein-Barr-virus-infected B cells. Proc Natl Acad Sci. (2024) 121(3):e2315857121. doi: 10.1073/pnas.2315857121
102. Yu H, Bai S, Hao Y, and Guan Y. Fatty acids role in multiple sclerosis as ‘metabokines’. J Neuroinflamm. (2022) 19:157. doi: 10.1186/s12974-022-02502-1
103. Pompura SL, Hafler DA, and Dominguez-Villar M. Fatty acid metabolism and T cells in multiple sclerosis. Front Immunol. (2022) 13:869197. doi: 10.3389/fimmu.2022.869197
104. Procaccini C, De Rosa V, Pucino V, Formisano L, and Matarese G. Animal models of multiple sclerosis. Eur J Pharmacol. (2015) 759:182–91. doi: 10.1016/j.ejphar.2015.03.042
105. Mestas J and Hughes CC. Of Mice and Not Men: Differences between Mouse and Human Immunology. The Journal of Immunology. (2014) 172(5):2731–38. doi: 10.4049/jimmunol.172.5.2731
106. Gilli F, DiSano KD, and Pachner AR. SEXX matters in multiple sclerosis. Front Neurol. (2020) 11:616. doi: 10.3389/fneur.2020.00616
107. Wiedrick J, Meza-Romero R, Gerstner G, Seifert H, Chaudhary P, Headrick A, et al. Sex differences in EAE reveal common and distinct cellular and molecular components. Cell Immunol. (2020) 359:104242. doi: 10.1016/j.cellimm.2020.104242
Keywords: air pollution, particulate matter, experimental autoimmune encephalomyelitis, multiple sclerosis, miRNA
Citation: Bonato M, Cerrato V, Dioni L, Montarolo F, Parolisi R, Bertolotto A, Bollati V, Ferrari L and Boda E (2025) Short-term exposure to particulate matter triggers a selective alteration of plasma extracellular vesicle-packaged miRNAs in a mouse model of multiple sclerosis. Front. Immunol. 16:1596935. doi: 10.3389/fimmu.2025.1596935
Received: 20 March 2025; Accepted: 16 June 2025;
Published: 03 July 2025.
Edited by:
Chris Wincup, King’s College Hospital NHS Foundation Trust, United KingdomReviewed by:
Miguel Angel Mendez-Rojas, University of the Americas Puebla, MexicoDennis Klose, Amsterdam University Medical Center, Netherlands
Sarah Ingrid Pinto Santos, University of São Paulo, Brazil
Copyright © 2025 Bonato, Cerrato, Dioni, Montarolo, Parolisi, Bertolotto, Bollati, Ferrari and Boda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luca Ferrari, bHVjYS5mZXJyYXJpQHVuaW1pLml0; Enrica Boda, ZW5yaWNhLmJvZGFAdW5pdG8uaXQ=
†These authors share first authorship
 Martino Bonato
Martino Bonato Valentina Cerrato
Valentina Cerrato Laura Dioni
Laura Dioni Francesca Montarolo
Francesca Montarolo Roberta Parolisi
Roberta Parolisi Antonio Bertolotto
Antonio Bertolotto Valentina Bollati
Valentina Bollati Luca Ferrari
Luca Ferrari Enrica Boda
Enrica Boda