- 1Collenge of Veterinary Medicine, Jeonbuk National University, Iksan, Republic of Korea
- 2Vaccine Lab, WOOGENE B&G Co., LTD., Seoul, Republic of Korea
- 3Biosafety Research Institute, Jeonbuk National University, Iksan, Republic of Korea
- 4Animal Clinical Evaluation Center, Optipharm Inc, Cheongju, Republic of Korea
- 5Animal Disease Diagnostic Division, Animal and Plant Quarantine Agency, Gimcheon, Republic of Korea
- 6College of Veterinary Medicine, Chungbuk National University, Cheongju, Republic of Korea
Introduction: Porcine reproductive and respiratory syndrome virus (PRRSV) remains one of the most economically devastating pathogens in swine, primarily due to its extensive genetic diversity and lineage-dependent pathogenicity. Despite widespread vaccination, distinct PRRSV-2 lineages continue to circulate in Korea. This study aimed to elucidate the immunopathological features of two Korean-specific Lineage B (LKB) strains, GGYC45 and PJ10, compared with a vaccine-like L5 strain, M8.
Methods: Thirty, 4-week-old piglets were divided into M8-, GGYC45-, PJ10-infected groups, and control. After acclimatization, pigs were intramuscularly inoculated with PRRSV-2 strains. Pigs were monitored, and blood and nasal swabs were collected. At 12- and 28 days post-infection (dpi), pigs were euthanized for histopathological analysis and tissue collection. Histopathological evaluations were conducted on lung and brain tissues. Bronchoalveolar lavage (BAL) cells and lung tissues were analyzed for immune responses, including flow cytometry (FACS), cytokine expression, viral load, and expression of immune checkpoint molecules.
Results: Both LKB strains (GGYC45 and PJ10) observed moderate to severe clinical symptoms. Notably, PJ10-infected pigs exhibited high mortality accompanied by significantly (p < 0.05) low average daily weight gain (ADWG), high temperatures, and high levels of viremia and viral loads in various tissues. Immunopathological analysis showed severe respiratory and neurological lesions in PJ10-infected pigs. PJ10 destroyed over 90% of residential alveolar macrophages and increased infiltrated monocyte-derived cells and T lymphocytes in the lungs up to 12 dpi. Pigs infected with the GGYC45 strain exhibited a relatively lower virulence profile than those infected with the PJ10 strain; however, GGYC45 induced moderate pathogenicity in pigs. Regardless of the lineages or genotypes, pigs infected with PRRSV-2 increased immune checkpoint molecule expression, such as PD1, PDL1, CTLA4, IDO1, and LAG3 in BAL cells, resulting in insufficient T cell activation.
Conclusion: These results highlight the differential virulence and immunomodulatory profiles of genetically distinct PRRSV-2 strains circulating in Korea. The heightened immune checkpoint expression, particularly in PJ10-infected pigs, underscores a potential mechanism of PRRSV-induced immune suppression and viral persistence. This study provides critical insights into PRRSV pathogenesis and host-virus interactions and supports the need for lineage-adapted control strategies that account for both the genetic heterogeneity of PRRSV and immune evasion mechanisms.
1 Introduction
The porcine reproductive and respiratory syndrome (PRRS) virus has emerged as a formidable and lethal threat to the global swine industry since its initial outbreaks in the United States and Europe (1). The economic repercussions of this disease in the U.S. swine industry alone have been estimated to incur an annual loss of approximately $664 million (2). The causative agent, PRRSV, is a positive-sense single-stranded RNA virus (~15 kb) classified as Betaarterivirus by the International Committee on Taxonomy of Viruses (ICTV, 2018), belonging to the family Arteriviridae of the order Nidovirales (3–5). The PRRSV genome comprises at least 10 open reading frames (ORFs), encompassing seven structural genes (ORF1a/b, ORF2a/b, ORF3, ORF4, ORF5/5a, ORF6, and ORF7) and additional nonstructural genes (6). PRRSV is categorized into two divergent genotypes, Betaarterivirus suid-1 (formerly PRRSV-1, EU-type, prototype strain Lelystad virus) and Betaarterivirus suid-2 (formerly PRRSV-2, NA-type, prototype strain VR2332 virus), exhibiting approximately 60% similarity at the nucleotide level (7, 8). According to the ORF5 lineage classification system, PRRSV-2 is subdivided into nine lineages, lineage 1 (L1) to lineage 9 (L9) (9–13). Although the origins of these nation-specific lineages remain largely unknown, Korean lineages have been observed to form a cohesive cluster in whole-genome-based phylograms, exhibiting an identical deletion pattern in the hypervariable region of nonstructural protein 2 (nsp2), suggesting regional independent evolution (14).
Generally, PRRSV-2 strains of different lineages induce varying levels of pathogenicity and susceptibility in infected pigs (15–17). Notably, the past decade has seen an increase in sudden and severe outbreaks caused by highly pathogenic PRRSV-2 strains, such as the emergence of Highly Pathogenic PRRSV (HP-PRRSV) in China and NADC30-like or NADC34-like PRRSV strains (16, 18–22). Since 2014, the emergence of LKB (Korean Lineage B) in Korea is suspected to result from potential recombination between LKC (Korean Lineage C) and MLV vaccine strains, which have been extensively used nationwide for decades (14). Previous studies have conducted pathogenicity experiments on field isolates from Korea, revealing that strains belonging to the LKB (Korean Lineage B) exhibit relatively higher pathogenicity (23).
The immunopathological characteristics of PRRSV are strain-dependent. Upon PRRSV exposure, the virus replicates in alveolar macrophages (AMs) and swiftly disseminates throughout the host. Viremia peaks at approximately 7–10 days post-infection (dpi) and typically subsides by 28 dpi, contingent upon the strains, pig age, and host immune status (24). During high-pathogenic PRRSV infection, the lung environment of pigs undergoes significant alterations and collapse, leading to various local and peripheral immune responses (25–29). Among effector T lymphocytes, cytotoxic T lymphocytes (CTLs), alongside natural killer (NK) cells, play a crucial role in eliminating virus-infected cells. Despite advancements in our understanding of host immunity, the features and role of the immune response, both local and systemic, to genetically unique Korean PRRSV strains remain elusive.
Immune checkpoint molecules serve as negative regulatory receptors expressed on immune cells. Under normal physiological conditions, the host maintains self-tolerance, modulating the breadth of effector immune responses in peripheral tissues (30, 31). However, persistent immune checkpoint signaling leads to T-cell dysfunction, inducing an ‘exhausted state’ in effector immune cells (32, 33). Notable immune checkpoint molecules include programmed cell death 1 (PD1) and its ligand programmed cell death ligand (PDL-1), which are upregulated at various stages of viral infection and contribute to T-cell proliferation (34–36). Other immune checkpoint molecules have also been studied in the context of immunosuppressive viral diseases (37). Recent findings indicate upregulated expression of immune checkpoint molecules during highly virulent PRRSV strain infections, leading to the induction of non-effector T cells (38). However, the role of immune checkpoint molecules in genetically distinct PRRSV strains remains poorly understood.
This study aims to investigate the immunopathological characteristics of genetically unique Korean PRRSV-2 strains, including strain-specific clinical signs and pathology, gross lesions, strain-specific T-cell subset proliferation and differentiation, and expression of immune checkpoint molecules. Thereby, this research contributes crucial insights into highly virulent PRRSV-2 strains prevalent in Korea.
2 Materials and methods
2.1 Cell culture and virus propagation
The three Korean PRRSV-2 strains, previously categorized into distinct lineages, were obtained from field samples submitted to the JBNU Veterinary Diagnostic Center (JBNU-VDC). The PRRSV-2 (NA-type) strains utilized in this study were JB15-M8-GN (M8, GenBank: AF066183.4), GGYC45 (GenBank: MN073134.1), and JB15-PJ10-GN (PJ10, GenBank: MK057532.1). Strains belonging to Korean Lineage B (LKB), a subgroup of the Korean Lineage that occurs specifically in South Korea, have been consistently reported to account for over 10% of cases annually (39). Phylogenetic tree analyses based on the ORF5 region and whole-genome sequencing (WGS) revealed differing clustering patterns (12, 14). While ORF5-based analysis suggested that strains belonging to LKB grouped within the same clade, WGS analysis demonstrated their separation into distinct clades (Supplementary Figures 1A, B). The tested Korean PRRSV-2 strains represent distinct pathogenic phenotypes: M8 is a vaccine-like strain classified under Lineage 5 with low pathogenicity, GGYC45 belongs to Lineage Korean B (LKB) and exhibits moderate virulence, and PJ10, also classified as LKB, has been associated with severe clinical signs, mortality and is considered highly pathogenic based on previous challenge studies (23).
These viruses were propagated in primary porcine alveolar macrophages (PAMs) using RPMI-1640 medium (Gibco® RPMI-1640, Life Technologies, Carlsbad, USA) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Capricorn Scientific), 2 mM L-glutamine, and an antibiotic-antimycotic cocktail (Anti-Anti, Life Technologies) containing 100 IU/ml penicillin, 100 µg/ml streptomycin, and 0.25 µg/ml Fungizone® [amphotericin B] at 37°C in a humidified 5% CO2 environment. This culture medium is referred to as RPMI growth medium. Viral titration was conducted on PAM cells, and the titers were determined at 5 to 6 days post-inoculation based on cytopathic effects (CPE), expressed as TCID50/mL (40). The virus-containing cell lysate was clarified by centrifugation at 4000 rpm for 30 min at 4°C. Following virus propagation, the cell lysate containing the virus was clarified via centrifugation at 4000 rpm for 30 minutes at 4°C. The clarified samples were filtered through a 0.22μm-pore size filter and collected in a sterile container.
2.2 Animal study
Thirty 4-week-old piglets obtained from a PRRSV- and PCV2-seronegative farm were randomly assigned to four groups. An overview of the animal experiment is depicted in Figure 1. Following a 3-day acclimatization, the infected groups were administered 2 mL of PRRSV-2 strain diluted in sterile PBS. Eight pigs in each group were intramuscularly inoculated with M8, GGYC45, and PJ10 at a concentration of 1 x 103 TCID50/mL, respectively. Control pigs (n=6) remained uninfected. Daily monitoring of all pigs was conducted three days before inoculation and continued until the end of the experiment. The pigs were fed twice daily, and monitoring, including temperature measurement, was conducted during feeding times. Blood collection and nasal swab samplings were conducted at 0, 3, 7, 12, 21, and 28 dpi. Body weights of all pigs were recorded, while the average daily weight gain (ADWG) of euthanized pigs was assessed at 12 and 28 dpi. To elucidate the pathogenicity of Korean PRRSV-2, half of the animals in each group were humanely euthanized at 12 dpi, and the remaining half at 28 dpi for necropsy. Euthanasia was performed via electrocution following an intramuscular injection of 2 mL of azaperone (40 mg/mL, StressGuard®, Dong Bang Inc., Republic of Korea). The pigs were allowed to reach adequate sedation approximately 10–15 minutes post-injection. Once adequate sedation was confirmed, electrocution was performed using 60 Hz alternating current applied across the brain and heart for 5–10 seconds to ensure immediate loss of consciousness. Exsanguination was performed immediately following electrocution to complete the euthanasia process.
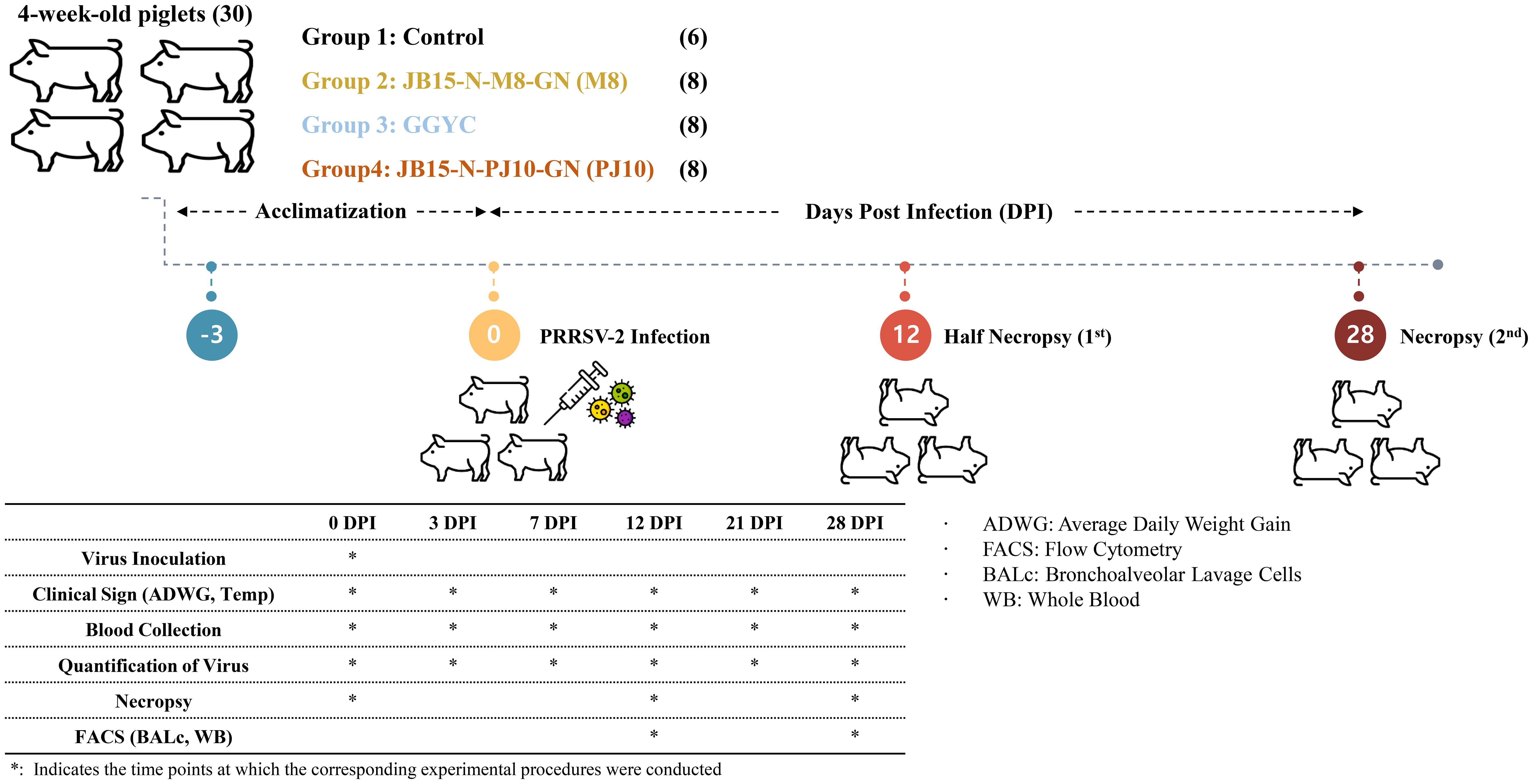
Figure 1. Experimental design overview. Thirty 4-week-old piglets from a PRRSV- and PCV2-seronegative farm were randomly divided into four groups. After a 3-day acclimatization, infected groups received 2 mL of PRRSV-2 strain via intramuscular inoculation. Daily monitoring, including temperature checks and clinical observation, was conducted. Blood samples were collected at various dpi intervals, and body weights were recorded. Euthanasia was performed at 12 and 28 dpi. Tissue samples were collected for histopathological examination, and bronchoalveolar lavage cells were isolated for further analysis.
Two pigs infected with PJ10 died during the experiment due to severe clinical signs, high fever, and reduced body weight from the PJ10 group. The experiment for the PJ10 group was terminated at 12 dpi as the pigs were deemed to be in a moribund state. Post-euthanasia, respiratory organs, including lungs, trachea, and bronchi, were aseptically excised and rinsed with 75 mL of sterile PBS. Bronchoalveolar lavage cells (BAL cells) were isolated from the collected lung lavage fluid by centrifugation at 1000 ×g at room temperature for 10 minutes. BAL cells were stored in tubes containing cell banker (CELLBANKER® Cell Freezing Media, Amsbio, UK) and immediately frozen at -80°C for subsequent RNA extraction and immune analysis. Lung and brain tissues were preserved in 10% neutral-buffered formalin for histopathological examination.
The animal experiment protocol was approved by the Jeonbuk National University Institutional Animal Care and Use Committee (JBNU 2021-095) and performed according to the guidelines and regulations detailed by the committee.
2.3 Quantification of viral load
Serum viremia levels were assessed at designated days, and viral loads in the tissues (Nasal swab, lung, and brain) were quantified in euthanized pigs at 12 and 28 dpi. Viral RNA extraction from serum and tissues was conducted using the MagMAX viral RNA isolation kit (Ambion; Applied Biosystems, Life Technologies, Inc.). The viral loads were quantified through real-time reverse transcription-polymerase chain reaction (RT–qPCR) utilizing the Prime-Q PCV2, PRRSV Detection Kit (Genet Bio, Republic of Korea).
2.4 Anti-PRRSV specific antibody detection
Serum samples from all groups were assessed for anti-PRRSV-specific antibody (IgG) using a commercially available ELISA kit (PRRS Ab ELISA 4.0; Bionote Inc., South Korea) following the manufacturer’s instructions. Serum samples exhibiting an S/P ratio (the ratio of the net optical density of the test samples to the net optical density of the positive controls) ≥ 0.4 were classified as positive for PRRSV-specific antibody, as previously described (26, 28).
2.5 Cytokine immunoassay
Cytokine levels in the serum from all groups were evaluated utilizing a commercially available porcine-specific ProcartaPlex™ Multiplex Immunoassay (ThermoFisher Scientific, Vienna-1030, Austria) following the manufacturer’s instructions. The concentration of each cytokine was determined by analyzing the samples on the Luminex® 200TM system (Luminex Corporation, Austin, TX, USA). The provided standards in the kit were utilized. The machine underwent verification and calibration using a Luminex® 100/200TM verification kit and a Luminex® 100/200™ calibration kit following the manufacturer’s guidelines (26).
2.6 Histopathological evaluations
The lungs and brains of necropsied pigs in all experimental groups performed a pathologically assessment at 12 and 28 dpi. Macroscopic and microscopic lung lesions were evaluated following established protocols (23, 26, 41). A macroscopic lung lesion was scored for the percentage of lungs affected by pneumonia in the right lung lobes using a predefined evaluation system. Microscopic lung lesions from three distinct lung lobes were graded on a scale from 0 to 3, reflecting the absence of lesions or mild interstitial pneumonia, moderate-multifocal interstitial pneumonia, moderate diffuse interstitial pneumonia, and severe interstitial pneumonia, respectively. Microscopic brain lesions were similarly scored on a scale from 0 to 3, indicating the absence of lesions or minimal-multifocal perivascular inflammatory cell infiltration, mild-multifocal perivascular inflammatory cell infiltration, moderate-multifocal perivascular inflammatory cell infiltration, and severe multifocal perivascular inflammatory cell infiltration.
Immunohistochemistry (IHC) was employed to detect PRRSV-specific antigens in infected brain tissue. For IHC staining, tissue embedded on a glass slide was treated with an endogenous peroxidase inhibitory reagent (3% H2O2 in phosphate-buffered saline (PBS), pH 7.2) at room temperature for 10 minutes, followed by incubation with 0.05% protease type XIV (Sigma, USA) at 37°C for 20 minutes for antigen retrieval. Subsequently, the primary antibody (monoclonal mouse anti-PRRSV, SDOW17) antibody, RTI, LLC, USA) was applied at a dilution ratio of 1:1000 and incubated at 37°C for 1 hour. Next, the secondary antibody (EnVision™/HRP, Rabbit/Mouse (EVN) reagent, DAKO, Denmark) was added and incubated at 37°C for 40 minutes. The IHC analysis was then conducted using 3,3-diaminobenzidine tetrahydrochloride (DAB) staining (DAKO, Denmark).
2.7 Flow cytometry
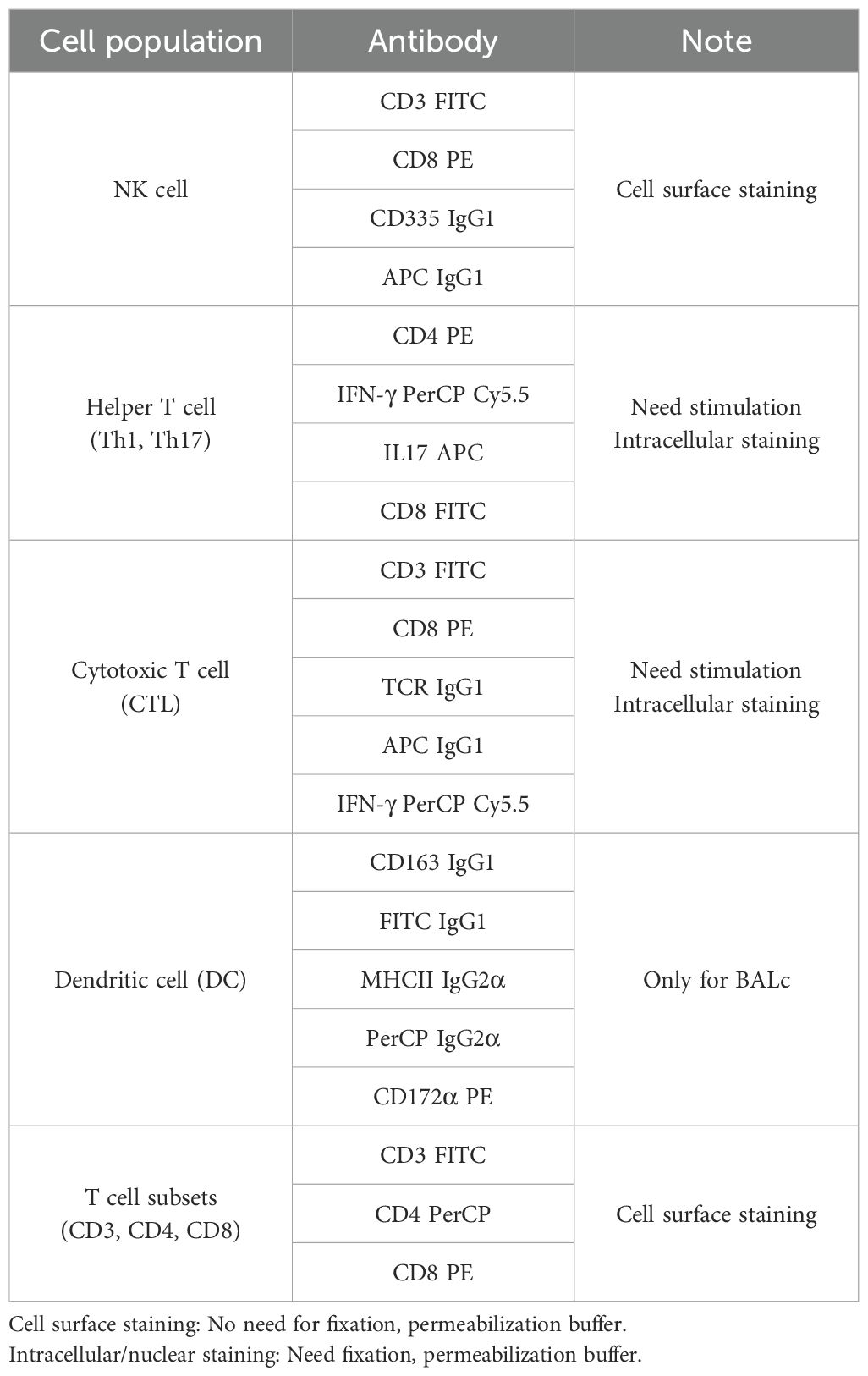
Flow cytometry was utilized to analyze cell surface, intracellular, and intranuclear staining for each cell type. Single-cell suspensions underwent cell surface staining by incubating with specific antibodies listed in Table 1 on ice for 30 minutes, followed by three washes with FACS buffer (3% FBS in PBS). NK cells, DCs, and macrophages (MǾs) were analyzed post-cell surface staining, while other T cell subsets required intranuclear and intracellular staining. BAL cells were preferentially subjected to cell surface staining for the CD163 surface marker, followed by the identification of DCs and MǾs in the BAL cell population using staining and gating strategies (42, 43).
Five functionally and phenotypically defined subpopulations were distinguished from the MHCII+ cell population via CD163 and CD172α surface markers, including alveolar macrophages (AMs, CD172α+/CD163high), monocyte-derived macrophages (moMǾs, CD172α+/CD163int), monocyte-derived dendritic cells (moDCs, CD172α+/CD163low), conventional dendritic cells 1 (cDC1s, CD172α+/CD163-), and conventional dendritic cells 2 (cDC2s, CD172α-/CD163-) (26, 27, 43).
Two subsets of NK cells were analyzed based on NKp46 marker expression, classified as NKp46+ and NKp46- NK cells (44, 45). CD3- lymphocytes were preferentially sorted and further analyzed for CD8α and NKp46 expression. Through this process, two populations were found in FACS analysis. NK cells were classified as NKp46+ (CD8+/NKp46+/CD3-) and NKp46− (CD8-/NKp46+/CD3-) NK cells and both of these cells were CD8α+. In addition, the T-cell subsets subjected to intracellular staining were stained according to previous studies (26). CD3- lymphocytes were sorted and further analyzed for CD8α and NKp46 expression, resulting in the identification of NK cells as NKp46+ (CD8+/NKp46+/CD3-) and NKp46− (CD8-/NKp46+/CD3-) NK cells, both expressing CD8α+. Intracellular staining was performed for T cell subsets such as cytotoxic T cells (CTLs).
Stained cells were suspended in FACS buffer and analyzed using an Accuri C6 flow cytometer (BD Accuri™ C6 Plus, BD Biosciences, MD, USA). Data analysis was performed on 100,000 events using FLOWJO version 10.6.1 (LLC, Ashland, OR 97520), excluding unstained doublets and irrelevant populations. Auto compensation was set based on color control and isotype control stains. FACS data are presented as percentages of all cell subsets.
2.8 Quantitative real-time polymerase chain reaction of BAL cells
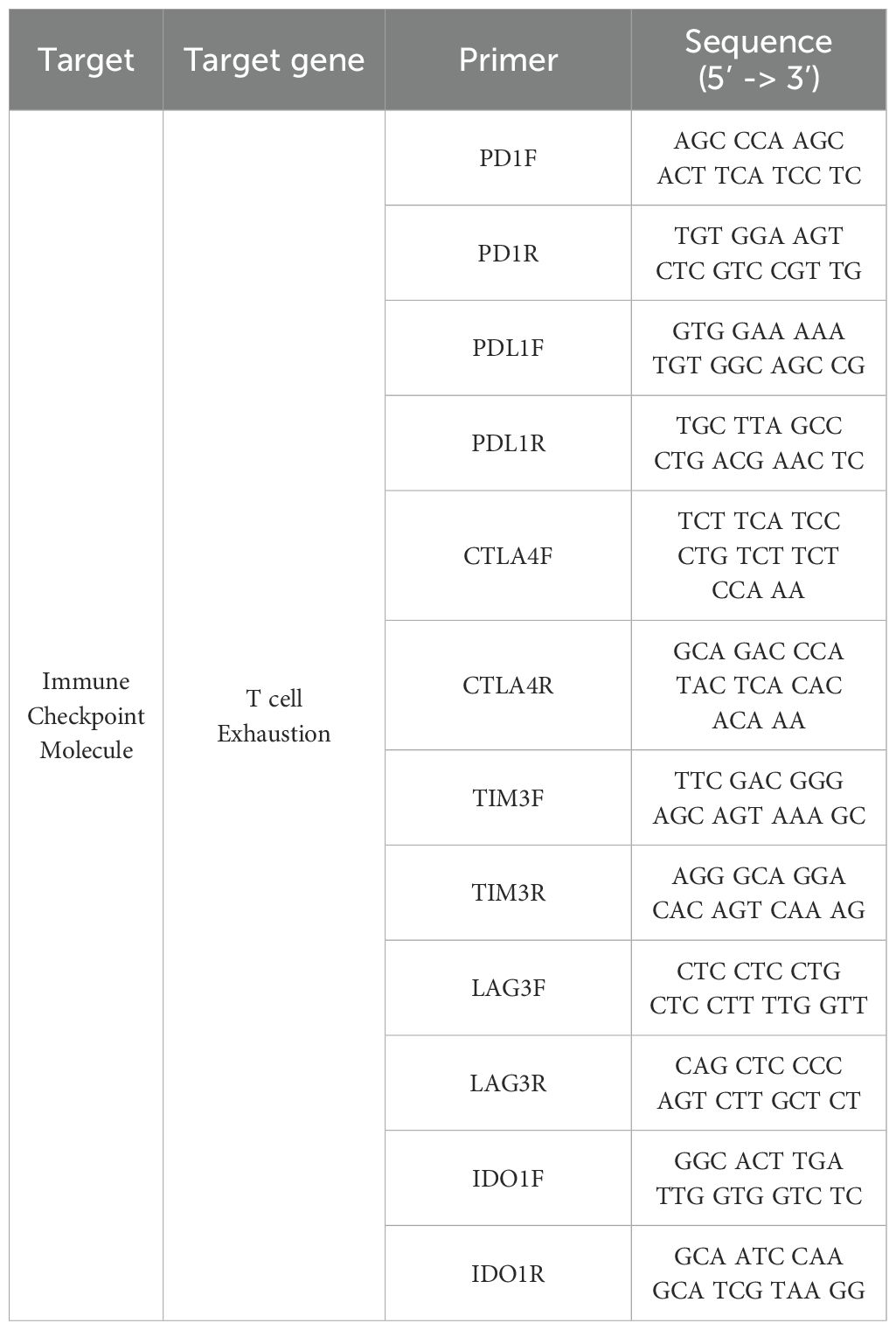
RNA was extracted using the GeneAll® RiboEx™ kit (GeneAll, Republic of Korea) following the manufacturer’s instructions. The concentration and purity of the isolated RNA were determined spectrophotometrically using a Nanodrop (Biospec Nano, Shimadzu, Japan), with samples meeting the 260/280 ratio criteria. Subsequently, cDNA was synthesized utilizing the WizScript™ cDNA Synthesis Kit - High Capacity (Wizbio, Republic of Korea) following the manufacturer’s protocol. Quantitative reverse-transcription PCR (RT-qPCR) was conducted using the extracted samples, employing primers detailed in Table 2.
2.9 Data analysis
Graphical representations of data were generated using GraphPad Prism 9.00 (GraphPad, USA), and statistical analyses were conducted using SPSS Advanced Statistics 17.0 software (SPSS, USA). Two-way ANOVA with Tukey’s multiple comparison tests was employed to assess the significance of variability within animal experimental groups for clinical data, including viremia, viral load, temperature, and anti-PRRSV antibodies. For comparisons of average daily weight gain (ADWG), phenotypes of various cell subsets, and mRNA expression, a nonparametric one-way ANOVA (Kruskal–Wallis test) was utilized. Statistically significant differences are denoted by asterisks and different letters over the bars.
3 Results
3.1 Clinical and pathological features of Korean PRRSV-2 infected pigs
Genetically distinct Korean PRRSV-2 induced various clinical and pathological symptoms. Especially, the PJ10, a Korean Lineage B (LKB) PRRSV-2 strain, induced severe clinical symptoms, including severe weight loss, respiratory distress, and high fever, resulting in significant morbidity and mortality.
3.1.1 Mortality
Notably, two pigs in the PJ10-infected group, belonging to Korean Lineage B (LKB), died at 10 and 12 dpi. The other pigs also displayed severe weight loss, respiratory signs, high fever, and severe nervous signs such as ataxia (Figure 2A). To comply with animal ethics, the remaining pigs in the PJ10 infected group were humanely euthanized at 12 dpi due to the severity of clinical symptoms. In contrast to PJ10-infected pigs, the M8- and GGYC45-infected pigs did not observe death until the end of the experiment.
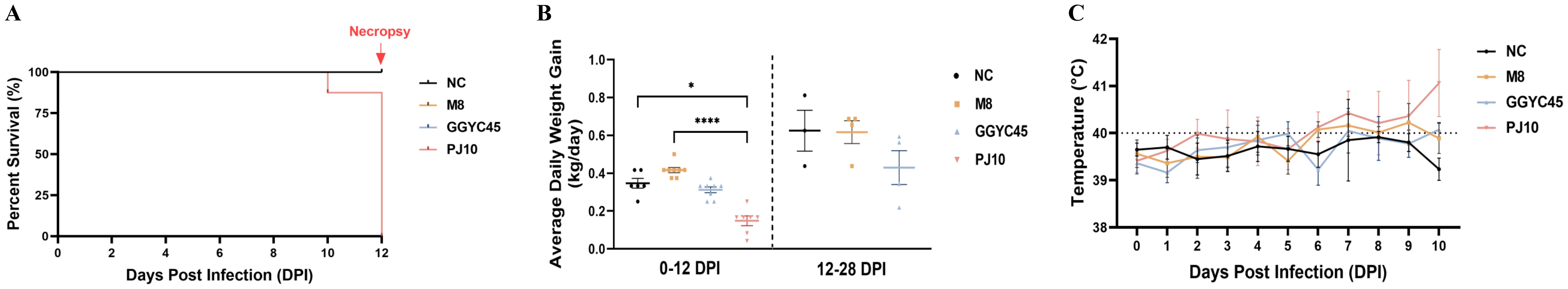
Figure 2. Clinical and pathological features of Korean PRRSV-2 infected pigs. (A) Survival curve of pigs infected with the PRRSV-2 strain PJ10, a Korean Lineage B (LKB) strain. Infected pigs exhibited severe clinical signs, including significant weight loss, respiratory distress, and persistent high fever, which ultimately led to sudden death. (B) Average daily weight gain (ADWG) comparison among pig groups challenged with genetically distinct PRRSV-2 strains M8, GGYC45, and PJ10 at 12 and 28 days post-infection (dpi). (C) Body temperature profiles of pigs infected with different PRRSV-2 strains, highlighting elevated temperatures exceeding 40°C observed in the PJ10-infected group at 6-10 dpi. The bars represent the mean, and the error bars represent the standard error of the mean (SEM). Bars with asterisks (*) indicate significantly different values (*indicates p < 0.05, ****indicates p<0.0005).
3.1.2 Weight gain
The average daily weight gain (ADWG) of the PJ10-infected group was significantly lower than that of the control and other infected groups at 12 dpi (p < 0.05 and p < 0.0005, respectively). Specifically, while the ADWG per control pig was 0.347 kg, the ADWG in the infected groups was recorded at 0.417, 0.313, and 0.149 kg for M8, GGYC45, and PJ10 at 12 dpi, respectively. At 28 dpi, the ADWG in the M8-, GGYC45-infected pigs increased by approximately 0.11 kg and 0.07 kg, respectively; however, both remained significantly lower than the control pigs (Figure 2B).
3.1.3 Body temperature
Pigs infected with PJ10 exhibited elevated body temperatures compared to other infected groups. The M8- and GGYC45-infected pigs showed relatively high fever (around 40°C), however, the PJ-10-infected pigs observed exceeding 40°C observed at 6–10 dpi (Figure 2C).
3.1.4 Viremia and virus shedding
The PJ10-infected group demonstrated the highest viremia and nasal swab shedding between 3 and 12 dpi. Conversely, the control group maintained an uninfected status throughout the experiment. The mean peak of viral titer in serum (viremia) was recorded as 103.12, 103.59, and 105.37 TCID50/mL in the M8, GGYC45, and PJ10 infected groups at 3 dpi, respectively. While the PJ10-infected group showed a similarly high level of viremia and died until 12 dpi, the viremia of the other two groups was observed to increase until 7 dpi and then decreased thereafter. In the nasal virus shedding results, the PJ10 showed the highest virus level (101.22 TCID50/mL) at 3 dpi, while M8 showed the highest virus titre at 7 dpi, and GGYC45 showed the highest virus titre at 12 dpi. (Figures 3A, B).
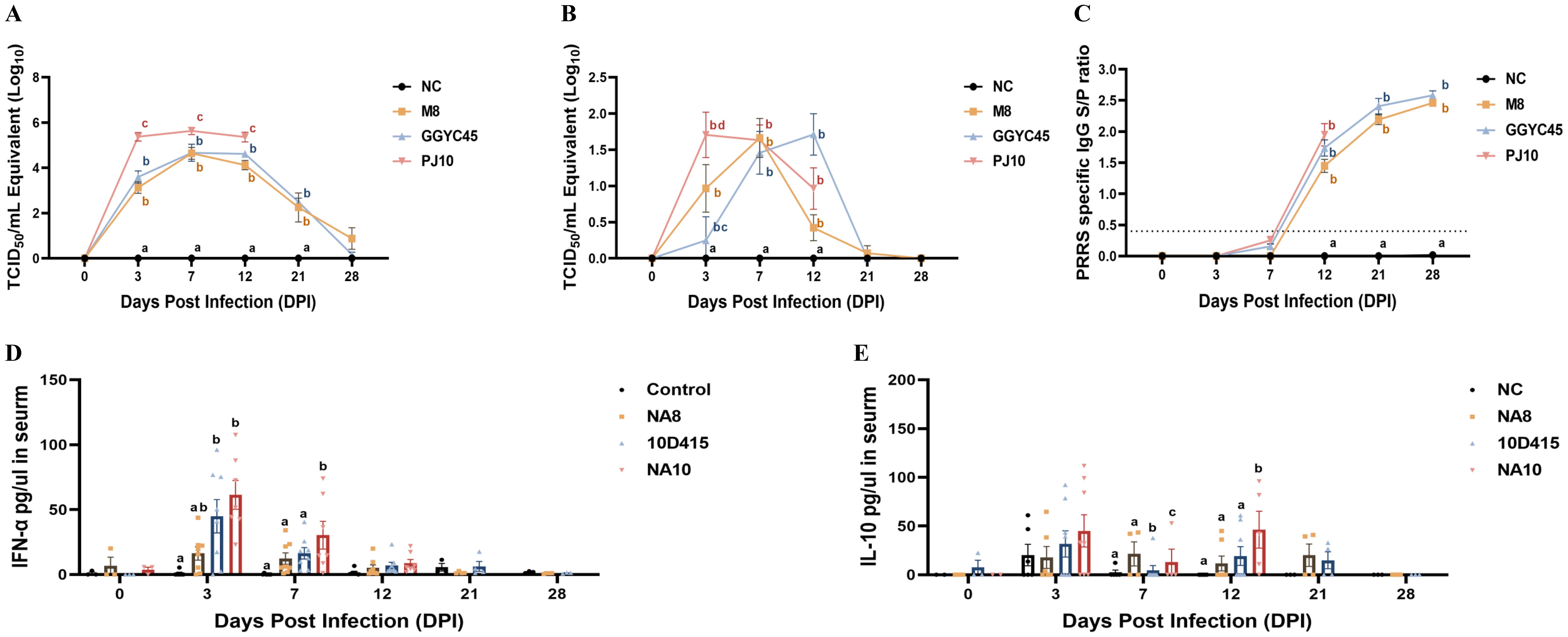
Figure 3. Pathogenicity of Korean PRRSV-2: viremia, nasal shedding, and antibody, cytokine response. (A) Mean peak virus titers in serum (viremia) measured as TCID 50/mL at 3 dpi were recorded as 103.12, 103.59, and 105.37 in the M8, GGYC45, and PJ10 infected groups, respectively. (B) Mean peak virus titers in nasal virus shedding measured as TCID 50/mL were detected at 3, 7, and 12 dpi with values of 100.69, 100.22, and 101.22 in the M8, GGYC45, and PJ10 infected groups at 3dpi, respectively. (C) PRRSV-specific antibody (IgG) levels measured by ELISA at 12 dpi with mean peaks of the S/P ratio were 1.448, 1.736, and 1.948 in the M8, GGYC45, and PJ10 infected groups, respectively. (D) Serum cytokine protein levels comparison between uninfected and infected groups showing significant (p < 0.0005) induction of interferon-a (IFN-a), a proinflammatory cytokine, in PJ10 infected pigs at 3 and 7 dpi. (E) Significant (p < 0.0005) induction of interleukin-10 (IL-10), an anti-inflammatory cytokine, at 7 and 14 dpi in infected groups compared to uninfected controls. The error bars represent standard deviations.
3.1.5 PRRSV-specific antibody
PRRSV-specific antibody (IgG) levels were measured by commercialized ELISA, with the control group showing no detectable PRRSV-specific IgG. There were no significant differences among the challenged group, however, the PJ10-infected pigs showed robust antibody responses compared to other strains. The M8, GGYC45, and PJ10 exhibited mean peaks of the S/P ratio of 1.448, 1.736, and 1.948 at 12 dpi, respectively, with antibody levels increasing up to 28 dpi (Figure 3C).
3.1.6 Cytokine
Serum cytokine protein levels were compared after genetically distinct PRRSV-2 infections. Interferon-α (IFN-α), a proinflammatory cytokine, was significantly induced (p < 0.005) in LKB-infected groups (GGYC45 and PJ10) at 3 and 7 dpi. Especially, the PJ10-infected groups showed high levels of IFN-α at the early time of infection (Figure 3D). Interestingly, the level of interleukin-10 (IL-10), an anti-inflammatory cytokine, was also slightly or significantly elevated in the LKB-infected (GGYC45 and PJ10) groups until 12 dpi (Figure 3E).
3.2 Comprehensive analysis of respiratory pathology in PRRSV-2 infection: gross pathology, histopathology, and lung viral load
3.2.1 Macroscopic and microscopic pathology
The PJ10-infected group exhibited more severe gross lesions, characterized such as diffuse dark-red consolidation and firmness, than the M8- and GGYC45-infected groups. Microscopically, the PJ10-infected lungs displayed more extensive interstitial thickening, alveolar septal infiltration, and mononuclear cell accumulation, consistent with severe diffuse interstitial pneumonia, whereas the M8 and GGYC45 groups showed mild to moderate multifocal lesions. In contrast, no macroscopic (lung-gross) lesions were observed in the negative control groups (Figure 4A). The average lung lesion (Lung consolidation) score was significantly higher (p < 0.05) in the LKB-infected group than in the M8-infected group. Notably, among the infected groups, the PJ10-infected group demonstrated significantly higher levels than the other strains (Figure 4B). Histopathological evaluations revealed that the PJ10-infected group displayed mild to severe multifocal/diffuse interstitial pneumonia with alveolar wall thickening due to pneumocyte proliferation, along with inflammatory cell infiltration in the infected groups at 12 dpi (Figure 4C). The lung microscopic lesions in the M8- and GGYC45-infected groups showed a slight increase due to PRRSV infection. Specifically, the highest microscopic lung lesion was observed at 12 dpi in the PJ10-infected group, and the lung lesion score of pigs was also significantly (p < 0.05) higher. Subsequently, the inflammatory cell infiltration and inflammatory response induced by PRRSV infection gradually decreased until 28 dpi (Figure 4D).
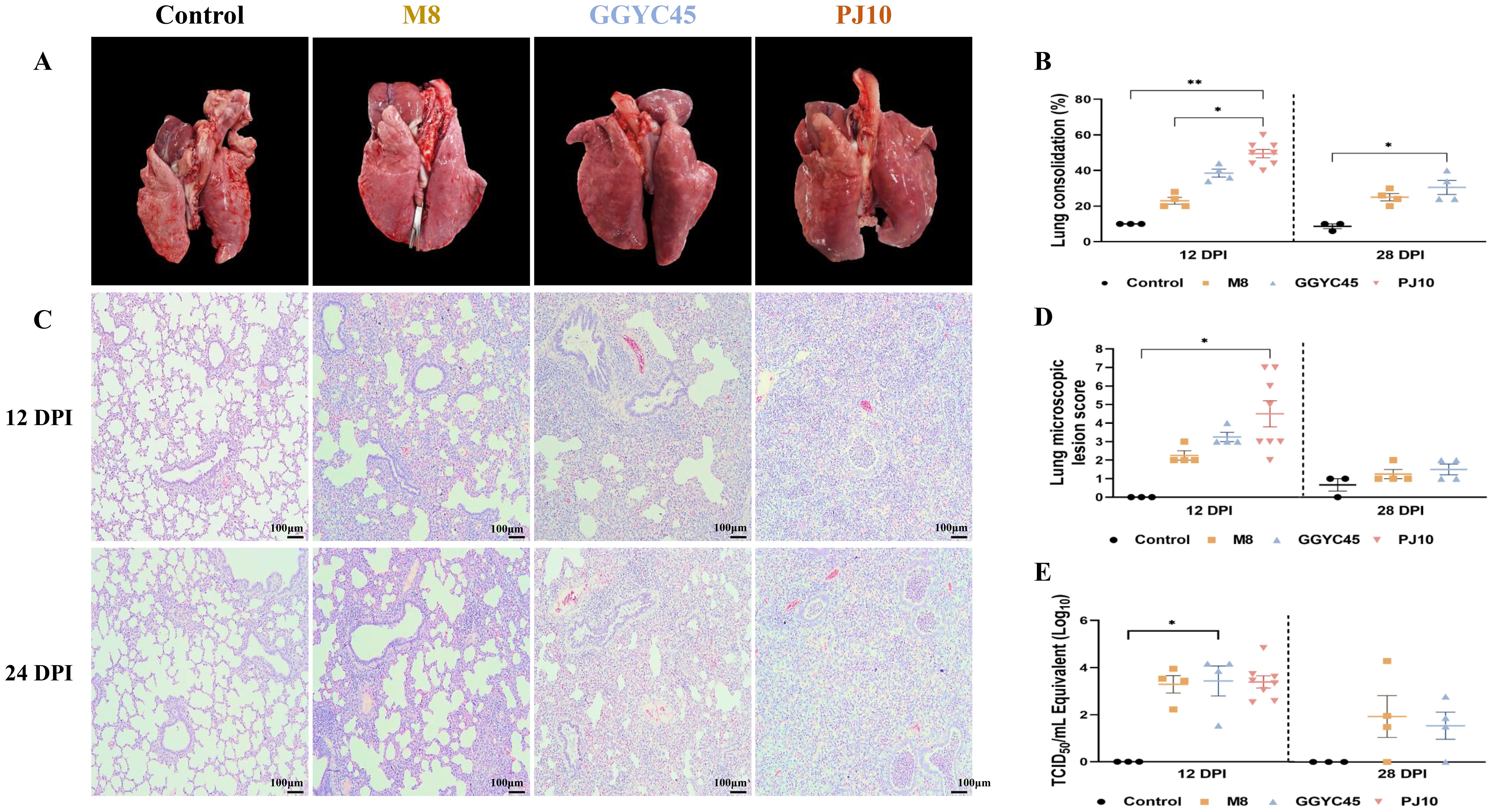
Figure 4. Comprehensive analysis of respiratory pathology in PRRSV-2 infection: gross pathology, histopathology, and lung viral load (A) Gross examination of lung tissues from control and PRRSV-infected pigs at 12 and 28 days post-infection (dpi). Pigs infected with GGYC45 and PJ10 strains exhibited severe lung consolidation compared to the M8-infected and control group. (B) Average lung lesion scores indicated significantly higher scores in infected groups compared to the control group, with the PJ10 infected group showing the highest scores among the infected groups. (C) Histopathological analysis observed interstitial pneumonia with alveolar wall thickening and inflammatory cell infiltration in PRRSV-infected groups at 12 dpi (Bar =100μm). (D) Microscopic lung lesion scores at 12 dpi demonstrate significantly higher levels in the PJ10 infected group than the other infected groups, with a subsequent decrease in inflammatory response by 28 dpi. (E) Viral titration in lung tissues showed slightly higher mean titers in the GGYC45 and PJ10 infected groups compared to the M8-infected group at 12 dpi The bars represent the mean and the error bars represent the standard error of the mean (SEM). Bars with asterisks (*) indicate significantly different values (*indicates p < 0.05, ****indicates p<0.0005).
3.2.2 Lung viral load
Viral load was quantified to reveal distinct characteristics of viral replication in the lungs. The groups infected with the PJ10 and GGYC45 strains demonstrated slightly elevated mean titer (103.43 and 103.38 TCID50/mL, respectively) compared to those observed in the M8-infected group at 12 dpi. “At 28 dpi, the lung viral loads of the M8- and GGYC45-infected groups were analyzed as 101.69 and 101 TCID50/mL, respectively; however, no significant differences were observed between infected groups (Figure 4E).
3.3 Comprehensive analysis of neurological pathology in PRRSV-2 infection: histopathology and brain viral load
3.3.1 Histopathological examination
To investigate PRRSV-induced brain lesions, brain tissues collected at 12 dpi and 28 dpi were analyzed. However, pathological findings associated with PRRSV infection were not observed in any groups except for the PJ10-infected group. Therefore, brain tissue analysis was performed on the PJ10-infected group at 12 dpi. Histopathological analysis using the H&E assay unveiled mild levels of perivascular inflammatory cell infiltration and infiltrated glial nodules in the Virchow-Robin space surrounding the brain in PJ10-infected pigs. Notably, distinctive inflammatory cell-infiltrated lesions attributed to PRRSV infection were exclusively observed in the PJ10-infected group, with positive reactions confirmed in the cytoplasm of cells located within some perivascular inflammatory cell infiltration sites (Figure 5A). Immunohistopathological evaluation (IHC) of the brain revealed no lesions associated with PRRSV infection in any group except for the PJ10-infected group. Remarkably, three PJ10-infected pigs exhibited a positive IHC reaction in the cytoplasm within the perivascular inflammatory cell-infiltrated lesion (Figure 5B). Microscopic examination revealed a slightly higher lesion score in the brains of the PJ10-infected group at 12 dpi (Figure 5C).
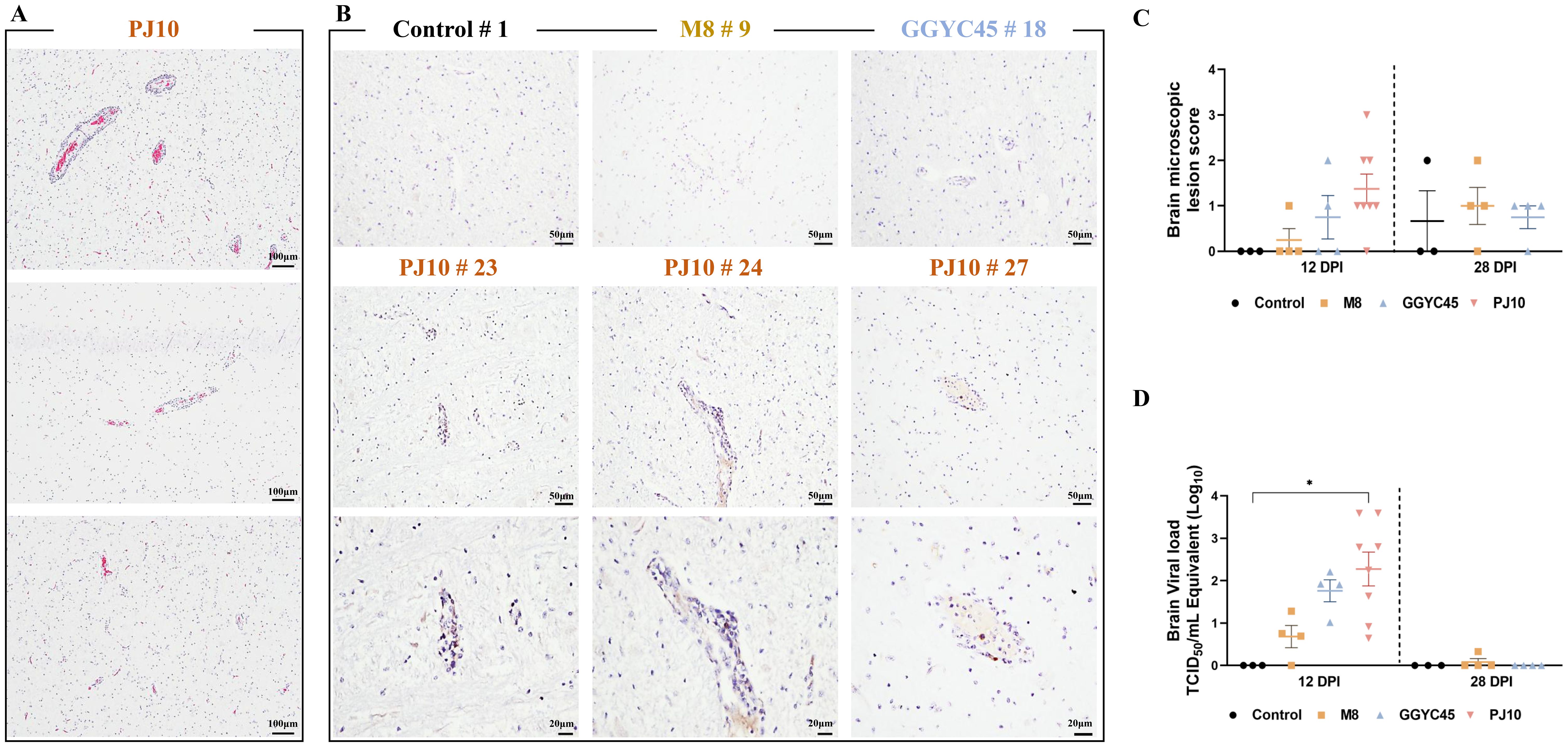
Figure 5. Comparison of brain viral load and histopathological findings in PRRSV-infected pigs H&E staining and IHC were performed to observe the histopathological features of brain tissue sections obtained from samples collected at 12 dpi after PJ10 infection (A) H&E assay reveals mild levels of perivascular inflammatory cell infiltration and infiltrated glial nodules in the Virchow-Robin space surrounding the brain in PJ10-infected pigs. Distinct inflammatory cell-infiltrated lesions were exclusively observed in the PJ10 infected group, with positive reactions confirmed in the cytoplasm of cells located within some perivascular inflammatory cell infiltration sites (Bar=100μm). (B) Three PJ10-infected pigs exhibit a PRRSV-specific IHC reaction in the cytoplasm located within the perivascular inflammatory cell-infiltrated lesion (Bar =50μm, 20μm). (C) Microscopic lesion scores indicate a slight increase in the infected group at 12 and 28 dpi. The bars represent the mean and the error bars represent the standard error of the mean (SEM). (D) Mean titers of the PJ10-infected group (102.27 TCID 50/mL) show a significant increase compared to other infected groups (100.68 and 101.76 TCID 50/mL, respectively). Bars with asterisks (*) indicate significantly different values (*indicates p < 0.05).
3.3.2 Brain viral load
The PJ10-infected group exhibited significantly (p < 0.05) higher mean titers (102.27 TCID50/mL) in the brain compared to the other infected groups (100.68 and 101.76 TCID50/mL, M8 and GGYC45, respectively) (Figure 5D).
3.4 Temporal dynamics of immune cell populations in PRRSV-2 infected pigs: comprehensive analysis of monocytes, T lymphocytes, and NK Cells in BAL cells
3.4.1 DC/macrophage networks
In bronchoalveolar lavage (BAL) cell flow cytometry, the control group exhibited a stable subset of the cell population. Monocyte populations constituted 64.9% in the control group; however, at 12 dpi, they were significantly reduced in the infected groups, with percentages of 21.6% in the M8-infected group, 17.0% in the GGYC-infected group, and 8.2% in the PJ10-infected group. By 28 dpi, the mixture of monocyte, neutrophils, and lymphocyte populations in the M8-infected group and GGYC45-infected group showed partial recovery, reaching approximately 21% and 16%, respectively, although lower than the control group (Figure 6A).
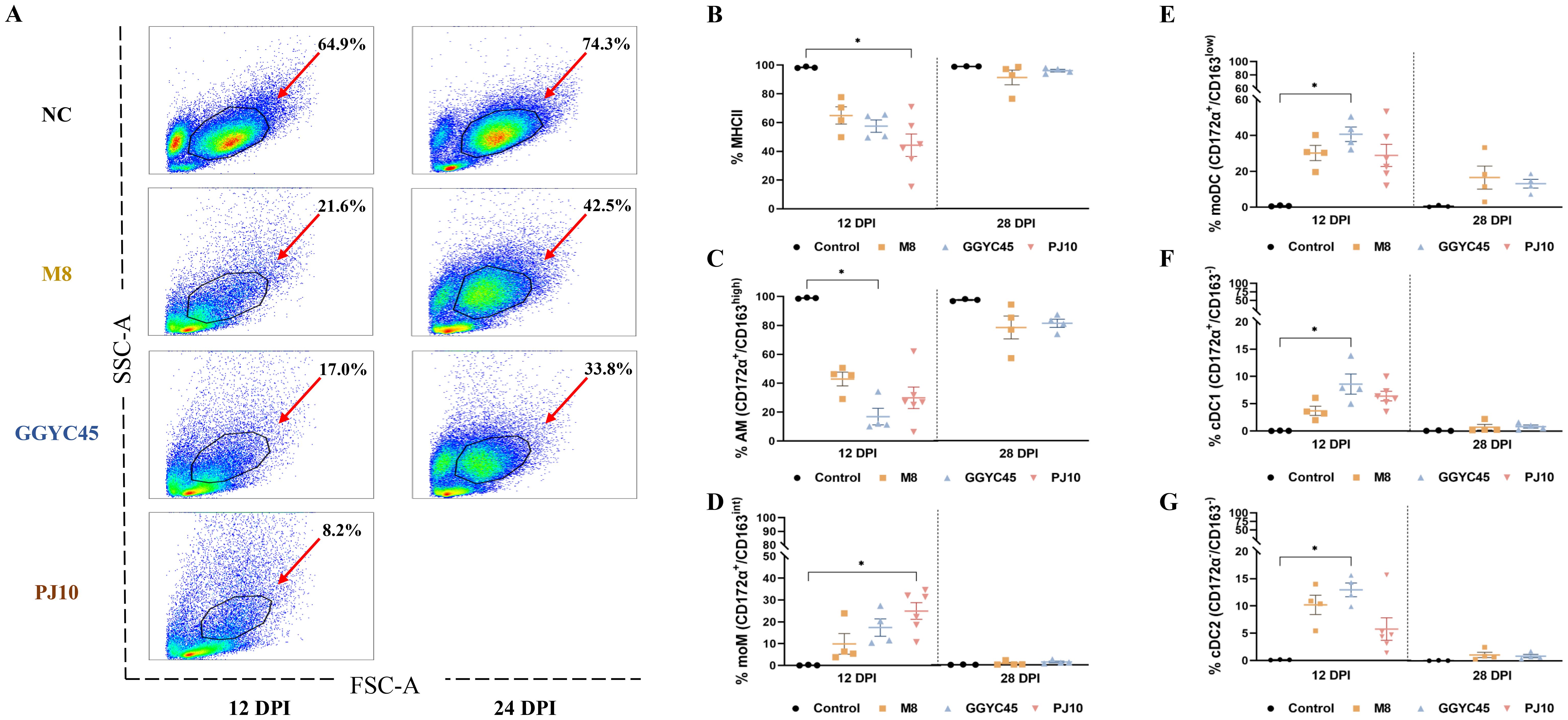
Figure 6. The dynamics of the lung immune cell population are altered after genetically unique PRRSV infection in BAL cells: Monocytes (A) Flow cytometric analysis of bronchoalveolar lavage (BAL) cells. (A) Distribution of cell populations in BAL fluid across experimental groups. (B) Changes in major histocompatibility complex class II positive (MHCII) cell populations at 12 days post-infection (dpi). (C) Percentage of alveolar macrophages (AMS) (CD172a/CD163high) among MHCII cells, indicating a significant decrease in infected groups at 12 dpi, particularly noticeable in GGYC45-infected groups (p<0.05). (D) Increase in monocyte-derived macrophages (moMs, CD172a/CD163int), notably higher in PJ10-infected group at 12 dpi (p < 0.05). (E-G) Significant increase in monocyte-derived dendritic cells (moDCs) and conventional dendritic cells (DC1/2) at 12 dpi, with the GGYC45-infected group exhibiting the highest rise compared to other infected groups (p<0.05). By 28 dpi a slight decrease in these cell populations is observed, approaching levels similar to the control group. The bars represent the mean and the error bars represent the standard error of the mean (SEM). Bars with asterisks (*) indicate significantly different values (*indicates p < 0.05, **indicates p<0.005).
3.4.2 Monocytes
Following an initial evaluation of the overall monocyte population, a comprehensive analysis was performed on the individual monocyte subpopulations. The population of major histocompatibility complex class II positive (MHCII+) cells decreased at 12 dpi in the infected groups. The average percentage of MHCII+ cells in the control group was 97.9%, while in the M8-infected group, GGYC45-infected group, and PJ10-infected group, it was 62.9%, 56.2%, and 51.2%, respectively. Although no significant differences were observed between the infected groups, it was noted that the PJ10-infected group showed a greater reduction compared to the other infected groups (Figure 6B). The percentages of alveolar macrophages (AMs; CD172α+/CD163high), constituting the major MHCII+ cells, were significantly decreased in all infected groups at 12 dpi. The average percentage of AM in the control group was 93.5%, while in the M8-infected group, GGYC45-infected group, and PJ10-infected group, it was 20.8%, 5.6%, and 10.1%, respectively. A significant difference (p < 0.05) was observed in the GGYC45-infected group. By 28 dpi, the decreased AM ratio had been restored to approximately 50% (Figure 6C).
3.4.3 Monocyte-derived cell populations
In contrast to MHCII+ cells and AMs, the proportion of the monocyte-derived macrophage (moMs; CD172α+/CD163int) infiltrating the lung was significantly increased in PRRSV-infected groups. The average percentage of moMs in the control group was 0.5%, while in the M8-infected group, GGYC45-infected group, and PJ10-infected group, it was 5.6%, 9.6%, and 13.1%, respectively. Notably, a significant (p < 0.05) influx of moMs was observed in the PJ10-infected group at 12 dpi (Figure 6D). Similar to the moM findings, monocyte-derived dendritic cells (moDCs; CD172α+/CD163low) and conventional DC1/2 (cDC1; CD172α+/CD163-, cDC2; CD172α-/CD163-) also significantly increased in the infected group at 12 dpi (p < 0.05). Notably, the GGYC45-infected group showed a significant (p < 0.05) influx of moDCs and cDC1 compared to the other infected groups. Although not statistically significant, a similar trend was observed in the PJ10-infected group, resembling the GGYC45-infected group. (Figures 6E–G).
3.4.4 T lymphocyte subsets
The frequencies of T lymphocyte subsets, including CD3+CD4+, CD3+CD8+, and CD3+CD4+CD8+ populations, in BAL cells were analyzed. Regardless of the PRRSV strains, all infected groups exhibited an increase in these subpopulations. Specifically, CD3+CD4+ T lymphocytes in the infected groups showed a slight increase at 12 dpi compared to levels in the control group (Figure 7A). Notably, CD3+CD8+ T lymphocytes showed a significant (p < 0.05) increase, especially in the M8-infected group, at 12 dpi, persisting slightly higher up to 28 dpi (Figure 7B). Additionally, CD3+CD4+CD8+ T lymphocytes exhibited a significant (p < 0.05) increase, particularly in the M8 and PJ10 infected groups; however, at low percentages compared to CD3+CD4+ and CD3+CD8+ populations (Figure 7C).
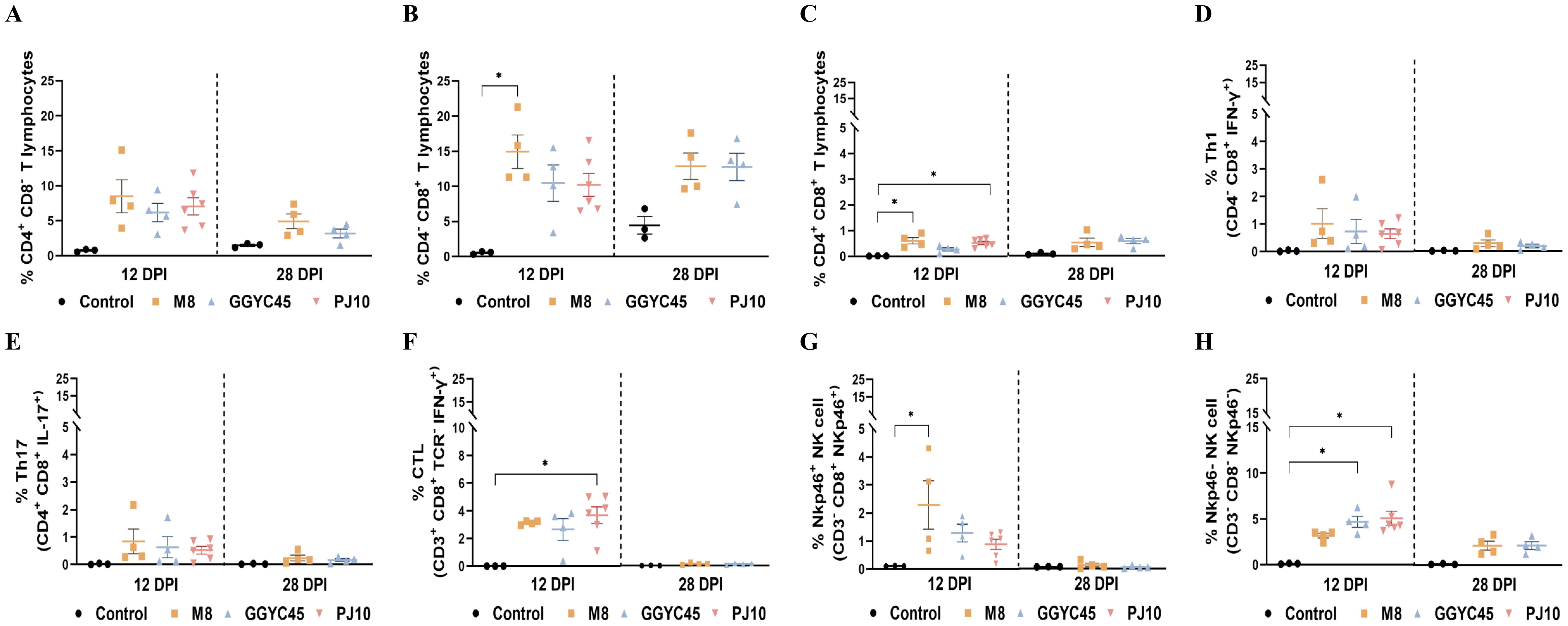
Figure 7. The dynamics of the lung immune cell population are altered after genetically unique PRRSV infection in BAL cells: T Cells, NK cells (A) Analysis of CD3+CD4+ T lymphocytes showing a slight increase at 12 days post-infection (dpi) compared to control levels. (B) Significant increase (p < 0.05) in CD3+*CD8+ T lymphocytes, especially in the M8-infected group, persisting up to 28 dpi. (C) Significant (p < 0.05) increase in CD3+CD4+CD8+ T lymphocytes, particularly in the M8 and PJ10 infected groups. (D) Th1 (IFN-y+/CD4/CD8+) and (E) Th17 (IL-17/CD4+/CD8+) responses slightly higher than controls at 12 dpi, with no significant difference among infected groups. (F) Significantly (p<0.05) higher frequency of cytotoxic T lymphocytes (CTL) (IFN-y+/CD3+/TCR-/CD8+) in BAL cells of PJ10 infected group at 12 dpi, returning to normal at 28 dpi. (G) Slightly higher percentages of NKp46+ (CD8+/Nkp46/CD3-) NK cells in infected group, notably significantly (p<0.05) higher in M8 infected group at 12 dpi. (H) Significant (p<0.05) increase in percentages of NKp46- (CD8-/Nkp46+/CD3-) NK cells in BAL cell population of infected pigs, especially in PJ10 infected group at 12 dpi. The bars represent the mean and the error bars represent the standard error of the mean (SEM). Bars with asterisks (*) indicate significantly different values (*indicates p < 0.05).
3.4.5 Effector T lymphocytes and natural killer cells
Overall, the percentages of various T-cell responses were significantly (p < 0.05) induced in BAL cells at 12 dpi. Th1 (IFN-γ+/CD4-/CD8+) and Th17 (IL-17+/CD4+/CD8+) responses in the infected groups were slightly higher than those in the control groups at 12 dpi, with no significant difference observed among the infected groups (Figures 7D, E). Intriguingly, the frequency of cytotoxic T lymphocytes (CTL; IFN-γ+/CD3+/TCR-/CD8+) was significantly (p < 0.05) higher in the PJ10-infected group at 12 dpi (Figure 7F). To observe the alteration of NK cells in pigs upon PRRSV infection, we analyzed the frequency and pattern of two different NK-cell subsets in the BAL cell population. Interestingly, the percentages of NKp46+ (CD8+/Nkp46+/CD3-) NK cells were slightly higher in the infected group, notably significantly (p < 0.05) higher in the M8 infected group compared to the control group at 12 dpi (Figure 7G). In contrast, the percentages of NKp46- (CD8-/Nkp46+/CD3-) NK cells in the BAL cell population of infected pigs, especially in the GGYC45 and PJ10 infected groups, were significantly (p < 0.05) higher at 12 dpi (Figure 7H).
3.5 Temporal profiling of T lymphocyte and natural killer cell subsets in PRRSV-2 infected pigs: insights from whole blood flow cytometric analysis
The characteristics of systemic immunity were evaluated by analyzing NK and T-cell populations in whole blood (WB) samples collected from uninfected and infected pigs at 0, 12, and 28 dpi. The Th1 and Th17 responses in infected pigs showed no significant differences compared to those in control pigs (Figures 8A, B). Among the strains within the LKB lineage, pigs infected with GGYC45 and PJ10 exhibited a significantly increased CTL response (p < 0.005) compared to the M8-infected group at 12 dpi, which was maintained until 28 dpi (Figure 8C). Consistent with the results from BAL cell flow cytometry, a trend in the responses of NK cell subsets (NKp46+ and NKp46- cells) was observed in WB from infected pigs. While the percentage of NKp46+ NK cell subsets significantly increased (p < 0.05) at 12 dpi in the GGYC45 and PJ10 infected groups, the overall magnitude of increase in NKp46- NK cells was higher compared to NKp46+ NK cells (Figures 8D, E).
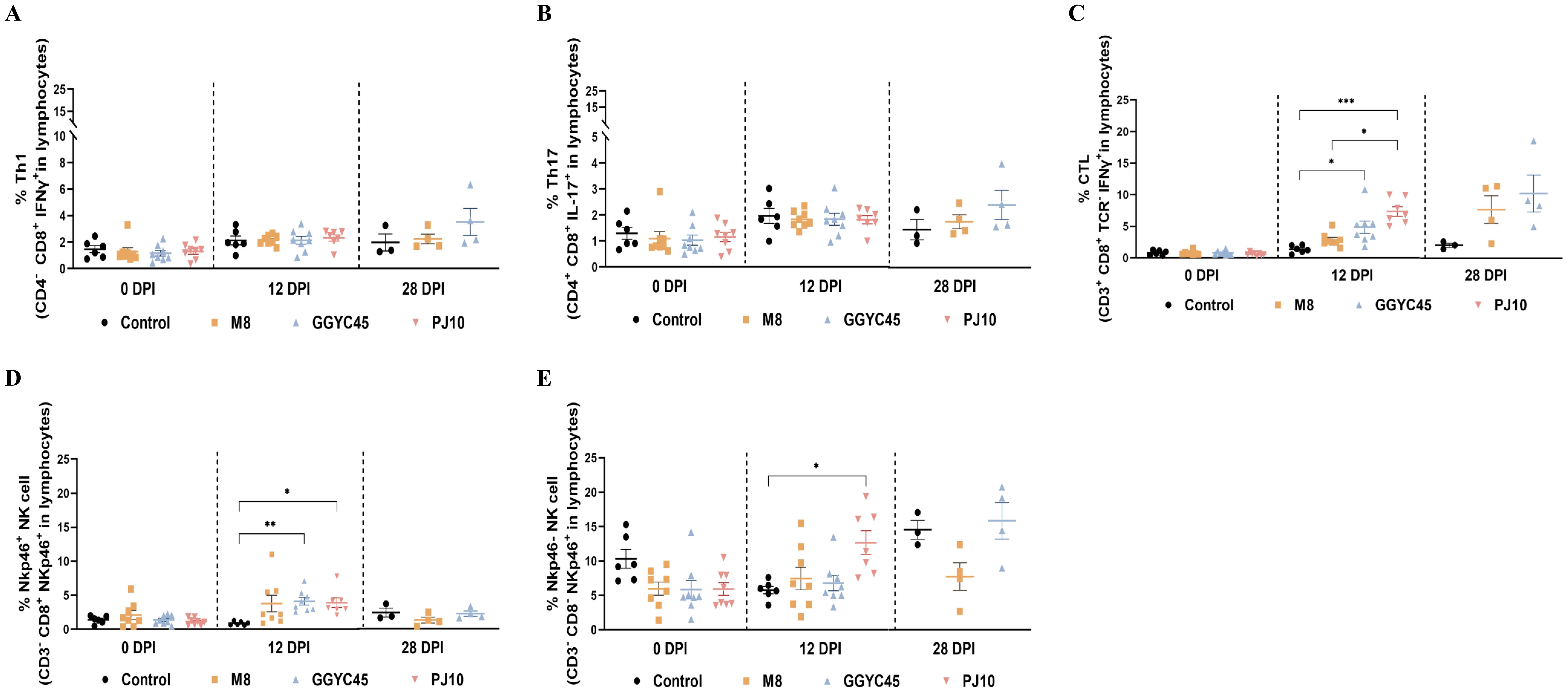
Figure 8. Temporal profiling of NK and T lymphocyte subsets in whole blood of PRRSV-2 infected pigs. (A) Thi and (B) Th17 responses in whole blood of PRRSV-2 infected pigs at 0, 12, and 28 days post-infection (dpi), with no significant differences observed compared to control pigs. (C) Cytotoxic T lymphocyte (CTL) response in pigs infected with different LKB lineage strains, including GGYC45, PJ10, and M8. A significantly increased CTL response (p < 0.005) was detected in GGYC45 and PJ10- infected pigs at 12 dpi, which persisted until 28 dpi, compared to the M8-infected group. (D, E) NK cell subset analysis in whole blood of infected pigs. A significant increase in the percentage of NKp46+ NK cells (p < 0.05) was observed in the GGYC45 and PJ10-infected groups at 12 dpi, whereas the NKp46- subset exhibited a higher overall magnitude of increase relative to the NKp46+ NK cells across all time points. Bars with asterisks (*) indicate significantly different values (*indicates p < 0.05, **indicates p<0.005, ***indicates p<0.0005).
3.6 Immune checkpoint molecules in PRRSV-2 infected pigs: temporal expression and dynamics
Overall, the expression of immune checkpoint molecules was observed to be upregulated regardless of the PRRSV strains. At 12 dpi, immune checkpoint molecules were significantly upregulated in the BAL cells of the GGYC45- and PJ10-infected groups within the LKB. Significant (p < 0.05) upregulation of PD1 was observed in BAL cells following PRRSV infection with levels exceeding 200-fold compared to controls at 12 dpi (Figure 9A). Similarly, expression of PDL1 increased at 12 dpi, with significant (p < 0.005) elevations observed in the GGYC45-infected group (Figure 9B). TIM3 expression also significantly (p < 0.05) increased in infected groups at 12 dpi (Figure 9C). A progressive upregulation of CTLA4 and IDO1 was observed. In the case of CTLA4, a fold change exceeding 250 was observed across all infected groups. IDO1 exhibited a similar trend to CTLA4; however, no statistical significance was observed (Figures 9D, E). Notably, LAG3 expression was significantly (p < 0.005) upregulated at 12 dpi in infected groups (Figure 9F).
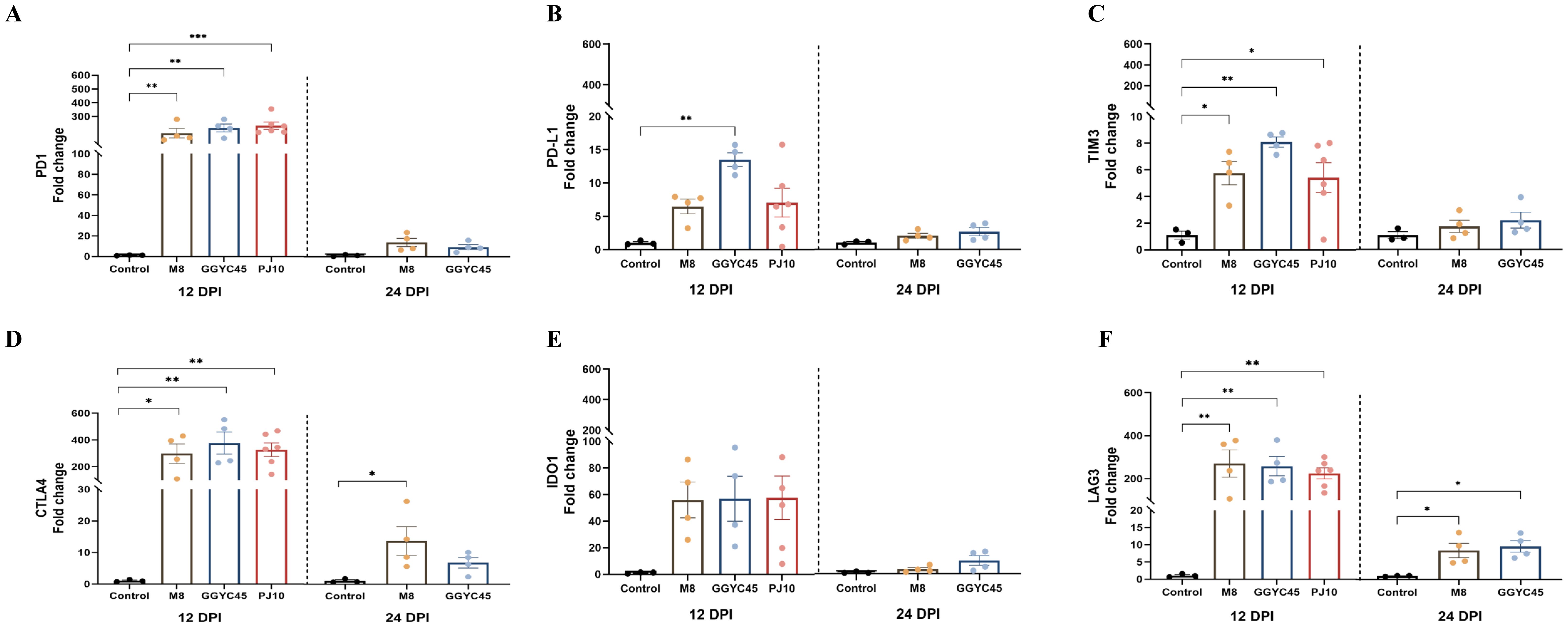
Figure 9. Temporal expression of immune checkpoint molecules in BAL cells of PRRSV-2 infected pigs. Expression levels of various immune checkpoint molecules in bronchoalveolar lavage (BAL) cells were analyzed in genetically diverse PRRSV-2-infected pigs. (A) PD1 expression levels in BAL cells from PRRSV-2 infected pigs, with a significant (p < 0.05) upregulation observed in PJ10 and GGYC45-infected pigs, both from lineage LKB, reaching over 200-fold compared to controls at 12 dpi. (B) PDL1 expression significantly increased (p < 0.05) in PJ10 and GGYC45-infected pigs at 12 dpi. (C) TIM3 expression in infected groups, showing a significant elevation (p < 0.05) at 12 dpi, with the GGYC45- infected group exhibiting the most pronounced increase. (D, E) Progressive upregulation of CTLA4 and IDO1, respectively, across infection time points, although no significant differences were observed between the infected groups. (F) LAG3 expression was markedly upregulated at 12 dpi, with the M8-infected group showing the highest levels compared to the other strains. Bars with asterisks (*) indicate significantly different values (*indicates p<0.05, **indicates p<0.005, ***indicates p<0.0005).
4 Discussions
Herein, we investigated the pathogenicity and immune characteristics associated with infection with genetically distinct Korean-specific PRRSV strains. Prior to 2003, Ingelvac PRRS MLV variants (Lineage 5) predominated in the epidemiology of PRRSV-2 in Korea. Despite the use of the MLV vaccine, recombination between wild-type PRRSV and MLV variants led to the emergence of novel PRRSV-2 Korean lineages (14). In the previous study, GGYC45 was considered the potential ancestor of the current circulating LKB strain as it possessed the LKB-like ORF5 gene with other genetic compartments originating from the recombination between LKC and MLV variants (39). LKB strains, especially PJ10-like strains became one of the major PRRSV-2 populations circulating in Korea since its initial identification in 2014 (12, 14). These recombination-derived genetic differences, particularly in the ORF5 gene and non-structural regions, are hypothesized to influence viral replication, immune evasion, and tissue tropism. Such genetic alterations may partially explain the distinct pathogenic profiles observed in this study, where the PJ10 strain exhibited higher virulence compared to GGYC45 and M8, despite sharing a common LKB lineage background (23).
These genomic traits appear to translate into distinct clinical outcomes, particularly in the PJ10-infected group, as reflected in differential disease severity and virological responses Based on clinical results, the PJ10 strain belonging to LKB demonstrates more severe pathogenicity than the other Korean-type PRRSV-2 strains, evidenced by severe clinical signs, including hyperthermia decreased weight gain, and higher viral replication. Notably, moderate to severe lung consolidation accompanied by a significant decrease in ADWG, characteristic of highly pathogenic PRRSV strains, was observed in the LKB-infected group (46, 47). Moreover, a significant negative correlation (p < 0.0001) between ADWG and viremia AUC at 12 dpi in the PRRSV-2 infected group was observed (Supplementary Figures 2A, B). In general, highly virulent PRRSV strains are characterized by high temperatures and viremia at early times of infection (48). LKB strains observed high viremia accompanied by high temperature, these strains induced negatively impact the ADWG in pigs. These findings align with previous studies analyzing the association among ADWG, viremia AUC, and host immune responses such as T cell response (26, 49, 50). Additionally, PRRSV and inflammation scores of the LKB-infected groups were markedly higher than the MLV variant strain (M8). This underscores the severity of clinical symptoms in pigs infected with LKB.
PRRSV-associated brain lesions have been rarely confirmed except for PRRSV infections with highly pathogenic strains such as Chinese HP-PRRSV and PRRSV 1-4–4 Lineage 1C variants (18, 51). Notably, with virus antigens confirmed in brain tissues with IHC, pigs infected with PRRSV 1-4–4 L1C variants exhibited neurological signs including ataxia, incoordination, posterior paresis, and convulsion without any bacterial co-infection (51). Additionally, it is significant that the highly pathogenic PRRSV antigens were able to penetrate the blood-brain barrier (BBB), thereby reaching the brain (51). Intriguingly, the PRRSV-positive antigens were detected in the Virchow-Robin (perivascular) space in PJ10-infected pigs. Virchow-Robin spaces are physiological structures in normal brain parenchyma that contain interstitial fluid filled with macrophages (52). In a previous study, PRRSV-positive antigens were identified in field cases (53). The affected farm exhibited a high morbidity rate (85%) and mortality rate (12%), with clinical signs indicative of nervous system involvement, including ataxia, posterior paresis, and convulsions. Immunohistochemical analysis revealed PRRS antigens in the brain tissue of weaning pigs, with prominent brown staining of perivascular and intramural mononuclear cells in small arteries and capillaries throughout the brain. Likewise, pigs infected with the PJ10 strain exhibited neurological symptoms, including immobility, resembling those observed in PRRSV infections with highly pathogenic strains. Immunohistopathological evaluations revealed the presence of PJ10 antigens, indicating potential dissemination to the brain. Highly pathogenic PRRSV strains appear to affect multiple tissues and organs beyond the respiratory system (54, 55). Consistent with findings from previous studies, the PJ10 strain exhibits characteristics similar to those of highly pathogenic PRRSV. Therefore, detailed studies on viral infection mechanisms and host immune responses are essential to understand its pathogenesis.
To evaluate the cell population modulation within the lung in response to various PRRSV infections, collected BAL cells were analyzed using flow cytometry (FACS). Consistent with previous studies, the DC/Macrophage network in the lung environment undergoes alterations following PRRSV infection (26, 27). During the initial stages of PRRSV infection, a continuous decrease is observed in MHCII+ and AM cells, which are significant constituents of pulmonary alveolar macrophages (PAM) cells. In response to PRRSV infection, the host immune system initiates a defense by recruiting monocyte-derived cells to the lungs (25). The PJ10 infection resulted in significant alterations in the dendritic cell/macrophage network during the early stages of infection, including a decrease of over 50% in the monocyte population. In pigs infected with PRRSV-1 Lena, a severely disrupted monocyte network was observed from early infection stages, persisting without recovery (56). Similar findings in PJ10-infected pigs suggest that PJ10 may exhibit pathogenic characteristics comparable to those of highly pathogenic PRRSV strains, such as the Lena strain. Contrary to the marked reduction observed in AM, a noticeable increase in the influx of monocyte-derived cells was observed. Among them, a significant increase in the ratio of moM and moDC, associated with viral defense mechanisms, was observed in the PJ10-infected group. Unlike the aforementioned studies (26, 27), this study did not investigate alterations in the DC/macrophage network during the early stages of PRRSV infection (3 dpi and 7 dpi). Nevertheless, distinct patterns of monocyte collapse were observed, varying with viral strain and pathogenicity. Furthermore, the characteristics of monocyte-derived cells infiltrating the lungs differed depending on the pathogenicity of the virus. Despite the activation of a vigorous host immune response associated with monocytes following PJ10 strain infection, the PJ10-infected group exhibited severe clinical symptoms and mortality. Therefore, additional analysis was conducted on cellular immune responses, specifically focusing on T lymphocytes and NK cells.
The Cytotoxic T Lymphocyte (CTL) reaction is a host reaction to respond to virus infection (57), but interesting results were obtained in LKB-infected pigs. The PJ10 strain, which caused higher mortality in pigs, appears to have triggered both local (BAL cells) and systemic (PBMC) host immune responses to the virus; however, it is likely that these responses ultimately failed to mount an effective defense. In addition, there is a notable influx of CD3+CD4+ T cells into the lungs during the early stages of infection, less than 1% of these cells differentiate into effecter T lymphocytes (Th1, Th17). This low proportion of effector T cells suggests that an effective host immune response does not occur, which appears to be influenced by the expression of immune checkpoint molecules that inhibit T-cell activation (30, 58). In general, various immune checkpoint molecules were shown to be expressed in several cell types, and overexpression in T cells is known to cause T-cell dysfunction (59–63). Also, an analysis of immune checkpoint molecule expression in lymph nodes during PRRSV high- and low-pathogenic strain infection has already been performed (38). The characterization of immune checkpoint molecules in local organs (PAMs) has been analyzed (64). Due to PRRSV infection, the T-cell exhaustion pathway was exclusively activated. Consistent with previous findings (65), our results also demonstrated an increase in the expression of T cell exhaustion markers, including PD1, LAG3, TIM3, CTLA4, IDO1, and PDL1 which are known to downregulate T cell activation, significantly (p < 0.05) increased at 12 dpi. Recent studies, including single-cell transcriptomic analyses of bronchoalveolar lavage cells during PRRSV infection, have further highlighted that CTLA4 expression is significantly upregulated in alveolar macrophages, contributing to PRRSV-induced immunosuppressive environments (64, 66, 67). Integrative transcriptomic profiling also supports the critical involvement of CTLA4 in the impairment of T cell responses during PRRSV infection (65). These findings indicate that PRRSV infection involves complex mechanisms in modulating host immune responses and may represent a key factor in the failure of the host immune system to mount an effective defense against PRRSV.
Taken together, the PJ10 strain is considered a highly pathogenic variant of PRRSV-2, characterized by severe clinical symptoms, including neurological disorders, high mortality, and significant alterations in immune cell populations, particularly monocytes and T cell subsets. These observations suggest that the failure of the host immune system to effectively respond to highly pathogenic PRRSV infection is likely driven by complex immunological mechanisms. Despite the limitation of focusing on the later stages of infection, the potential role of immune checkpoint molecules in suppressing effective T-cell responses has been observed. Therefore, further research is needed to investigate the early phase of infection to fully elucidate the initial host-pathogen interactions. Moreover, the substantial upregulation of immune checkpoint molecules observed in this study highlights the need for further investigation to determine their precise role and potential as therapeutic targets for managing PRRSV-induced immunopathogenesis.
5 Conclusion
This study provides valuable insights into the pathogenic characteristics and immune response alterations of genetically distinct Korean PRRSV-2. Notably, the PJ10 strain which belongs to LKB exhibited severe clinical signs, including high viremia, severe lung consolidation, and significant reductions in ADWG, setting it apart from other PRRSV-2 strains. Alongside PJ10, our research also analyzed three genetically distinct PRRSV strains, which presented a challenging experimental framework and offered a comprehensive view of the varying pathogenic profiles. Despite belonging to the same LKB lineage, the patterns of DC/macrophage network alterations differed between the GGYC45 and PJ10 strains, with PJ10 showing more severe disruptions. This finding aligns with clinical signs, including ADWG, high temperature, and highest viral load, which were more severe in PJ10-infected pigs. Although the host immune response, such as an increase in effector T cells like cytotoxic T lymphocytes (CTLs) in BAL cells (BALc) and Whole blood (WB), was upregulated, it failed to prevent mortality associated with PJ10 infection. Furthermore, the upregulation of immune checkpoint molecules suggests a critical role in the suppression of effective T-cell responses, which is considered to be the reason for the inadequate defense against PRRSV. These findings highlight the importance of further investigation into the immunopathogenesis of PRRSV, particularly the mechanisms of infection and host immune evasion, better to understand the underlying reasons for inadequate immune responses.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal studies were approved by Jeonbuk National University Institutional Animal Care and Use Committee (JBNU 2021-095). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
G-SP: Methodology, Investigation, Writing – review & editing, Writing – original draft, Project administration. S-CKi: Writing – review & editing, Project administration, Writing – original draft, Investigation, Methodology. H-JK: Writing – review & editing, Investigation, Methodology, Writing – original draft, Project administration. C-GJ: Writing – review & editing, Investigation, Methodology. S-CKa: Investigation, Writing – review & editing, Methodology. G-ES: Investigation, Writing – review & editing. S-HK: Investigation, Writing – review & editing. H-YJ: Writing – review & editing, Investigation. K-KL: Funding acquisition, Conceptualization, Writing – review & editing. S-ML: Methodology, Writing – review & editing. W-IK: Resources, Supervision, Project administration, Writing – review & editing, Conceptualization, Funding acquisition.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by a grant (Z-1543069-2017-20-1) from the Animal and Plant Quarantine Agency (QIA), Technology Commercialization Support Program (RS-2024-00398685, RS-2025-02304147) of the Ministry of Agriculture, Food and Rural Affairs, Republic of Korea.
Acknowledgments
The authors gratefully thank the laboratory technicians at the Veterinary Diagnostic Center of Jeonbuk National University (VDC-JBNU) for their constant assistance throughout the study.
Conflict of interest
Authors G-SP was employed by the company WOOGENE B&G Co., LTD. Author S-CK was employed by the company Optipharm Inc.
The remaining authors declare that the research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1599468/full#supplementary-material
References
1. Rossow K. Porcine reproductive and respiratory syndrome. Vet Pathol. (1998) 35:1–20. doi: 10.1177/030098589803500101
2. Neumann EJ, Kliebenstein JB, Johnson CD, Mabry JW, Bush EJ, Seitzinger AH, et al. Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. J Am Vet Med Assoc. (2005) 227:385–92. doi: 10.2460/javma.2005.227.385
3. Cavanagh D. Nidovirales: A new order comprising coronaviridae and arteriviridae. Arch Virol. (1997) 142:629–33.
4. Collins JE, Benfield DA, Christianson WT, Harris L, Hennings JC, Shaw DP, et al. Isolation of swine infertility and respiratory syndrome virus (Isolate atcc vr-2332) in north america and experimental reproduction of the disease in gnotobiotic pigs. J Vet Diagn Invest. (1992) 4:117–26. doi: 10.1177/104063879200400201
5. Wensvoort G, Terpstra C, Pol J, Ter Laak E, Bloemraad M, De Kluyver E, et al. Mystery swine disease in the Netherlands: the isolation of lelystad virus. Vet Q. (1991) 13:121–30. doi: 10.1080/01652176.1991.9694296
6. Han M and Yoo D. Engineering the prrs virus genome: updates and perspectives. Vet Microbiol. (2014) 174:279–95. doi: 10.1016/j.vetmic.2014.10.007
7. Adams MJ, Lefkowitz EJ, King AM, Harrach B, Harrison RL, Knowles NJ, et al. Changes to taxonomy and the international code of virus classification and nomenclature ratified by the international committee on taxonomy of viruses (2017). Arch Virol. (2017) 162:2505–38. doi: 10.1007/s00705-017-3358-5
8. Nelsen CJ, Murtaugh MP, and Faaberg KS. Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. J Virol. (1999) 73:270–80. doi: 10.1128/JVI.73.1.270-280.1999
9. Kang H, Yu JE, Shin J-E, Kang A, Kim W-I, Lee C, et al. Geographic distribution and molecular analysis of porcine reproductive and respiratory syndrome viruses circulating in swine farms in the Republic of Korea between 2013 and 2016. BMC Vet Res. (2018) 14:1–11. doi: 10.1186/s12917-018-1480-6
10. Shi M, Lam TT-Y, Hon C-C, Murtaugh MP, Davies PR, Hui RK-H, et al. Phylogeny-based evolutionary, demographical, and geographical dissection of north american type 2 porcine reproductive and respiratory syndrome viruses. J Virol. (2010) 84:8700–11. doi: 10.1128/JVI.02551-09
11. Lee J-A, Lee N-H, Lee J-B, Park S-Y, Song C-S, Choi I-S, et al. Genetic diversity of the korean field strains of porcine reproductive and respiratory syndrome virus. Infection Genet Evol. (2016) 40:288–94. doi: 10.1016/j.meegid.2015.11.001
12. Kim S-C, Jeong C-G, Park G-S, Park J-Y, Jeoung H-Y, Shin G-E, et al. Temporal lineage dynamics of the orf5 gene of porcine reproductive and respiratory syndrome virus in korea in 2014–2019. Arch Virol. (2021) 166:2803–15. doi: 10.1007/s00705-021-05169-w
13. Pegu SR, Sonowal J, Das PJ, Rajkhowa S, Choudhury M, Deb R, et al. Molecular genetic characterization of porcine reproductive and respiratory syndrome virus outbreak in assam, India and neighbouring regions. Braz J Microbiol. (2024), 1–9. doi: 10.1007/s42770-024-01592-8
14. Kim S-C, Moon S-H, Jeong C-G, Park G-S, Park J-Y, Jeoung H-Y, et al. Whole-genome sequencing and genetic characteristics of representative porcine reproductive and respiratory syndrome virus (Prrsv) isolates in korea. Virol J. (2022) 19:1–13. doi: 10.1186/s12985-022-01790-6
15. Meng X. Heterogeneity of porcine reproductive and respiratory syndrome virus: implications for current vaccine efficacy and future vaccine development. Vet Microbiol. (2000) 74:309–29. doi: 10.1016/S0378-1135(00)00196-6
16. Li Y, Wang X, Bo K, Wang X, Tang B, Yang B, et al. Emergence of a highly pathogenic porcine reproductive and respiratory syndrome virus in the mid-eastern region of China. Vet J. (2007) 174:577–84. doi: 10.1016/j.tvjl.2007.07.032
17. Obregon-Gutierrez P, Cortey M, Martín-Valls GE, Clilverd H, Correa-Fiz F, Aragón V, et al. Nasal microbial diversity is associated with survival in piglets infected by a highly virulent prrsv-1 strain. Anim Microbiome. (2025) 7:9. doi: 10.1186/s42523-024-00371-y
18. Tian K, Yu X, Zhao T, Feng Y, Cao Z, Wang C, et al. Emergence of fatal prrsv variants: unparalleled outbreaks of atypical prrs in China and molecular dissection of the unique hallmark. PLoS One. (2007) 2:e526. doi: 10.1371/journal.pone.0000526
19. Karniychuk UU, Geldhof M, Vanhee M, Van Doorsselaere J, Saveleva TA, and Nauwynck HJ. Pathogenesis and antigenic characterization of a new east european subtype 3 porcine reproductive and respiratory syndrome virus isolate. BMC Vet Res. (2010) 6:1–10. doi: 10.1186/1746-6148-6-30
20. Rajkhowa T, Jagan Mohanarao G, Gogoi A, Hauhnar L, and Isaac L. Porcine reproductive and respiratory syndrome virus (Prrsv) from the first outbreak of India shows close relationship with the highly pathogenic variant of China. Vet Q. (2015) 35:186–93. doi: 10.1080/01652176.2015.1066043
21. Han D, Hu Y, Li L, Tian H, Chen Z, Wang L, et al. Highly pathogenic porcine reproductive and respiratory syndrome virus infection results in acute lung injury of the infected pigs. Vet Microbiol. (2014) 169:135–46. doi: 10.1016/j.vetmic.2013.12.022
22. Rajkhowa S, Sonowal J, Pegu SR, Sanger GS, Deb R, Das PJ, et al. Natural co-infection of pigs with african swine fever virus and porcine reproductive and respiratory syndrome virus in India. Braz J Microbiol. (2024) 55:1017–22. doi: 10.1007/s42770-023-01203-y
23. Jeong C-G, Kim S-C, Lee S, Kim H-J, Mattoo SUS, Nazki S, et al. Pathogenic characteristics of five different lineage of korean prrsv-2 isolates (Nadc30-like, vr2332-like, lka, lkb, and lkc). Transboundary Emerging Dis. (2024) 2024:1618472. doi: 10.1155/2024/1618472
24. Díaz I, Gimeno M, Darwich L, Navarro N, Kuzemtseva L, López S, et al. Characterization of homologous and heterologous adaptive immune responses in porcine reproductive and respiratory syndrome virus infection. Vet Res. (2012) 43:1–15. doi: 10.1186/1297-9716-43-30
25. Bordet E, Blanc F, Tiret M, Crisci E, Bouguyon E, Renson P, et al. Porcine reproductive and respiratory syndrome virus type 1.3 lena triggers conventional dendritic cells 1 activation and T helper 1 immune response without infecting dendritic cells. Front Immunol. (2018) 9:2299. doi: 10.3389/fimmu.2018.02299
26. Nazki S, Khatun A, Jeong C-G, Gu S, Lee S-I, Kim S-C, et al. Evaluation of local and systemic immune responses in pigs experimentally challenged with porcine reproductive and respiratory syndrome virus. Vet Res. (2020) 51:1–19. doi: 10.1186/s13567-020-00789-7
27. Rodríguez-Gómez IM, Sánchez-Carvajal JM, Pallarés FJ, Mateu E, Carrasco L, and Gómez-Laguna J. Virulent lena strain induced an earlier and stronger downregulation of cd163 in bronchoalveolar lavage cells. Vet Microbiol. (2019) 235:101–9. doi: 10.1016/j.vetmic.2019.06.011
28. Shabir N, Khatun A, Nazki S, Gu S, Lee S-M, Hur T-Y, et al. In vitro immune responses of porcine alveolar macrophages reflect host immune responses against porcine reproductive and respiratory syndrome viruses. BMC Vet Res. (2018) 14:1–13. doi: 10.1186/s12917-018-1675-x
29. Domingo-Carreño I, Serena MS, Martín-Valls GE, Clilverd H, Aguirre L, Cortey M, et al. The introduction of a highly virulent prrsv strain in pig farms is associated with a change in the pattern of influenza a virus infection in nurseries. Vet Res. (2024) 55:147. doi: 10.1186/s13567-024-01406-7
30. Cai H, Liu G, Zhong J, Zheng K, Xiao H, Li C, et al. Immune checkpoints in viral infections. Viruses. (2020) 12:1051. doi: 10.3390/v12091051
31. Wykes MN and Lewin SR. Immune checkpoint blockade in infectious diseases. Nat Rev Immunol. (2018) 18:91–104. doi: 10.1038/nri.2017.112
32. Yi JS, Cox MA, and Zajac AJ. T-cell exhaustion: characteristics, causes and conversion. Immunology. (2010) 129:474–81. doi: 10.1111/j.1365-2567.2010.03255.x
33. Kahan SM, Wherry EJ, and Zajac AJ. T cell exhaustion during persistent viral infections. Virology. (2015) 479:180–93. doi: 10.1016/j.virol.2014.12.033
34. Yue F, Zhu Y-P, Zhang Y-F, Sun G-P, Yang Y, Guo D-G, et al. Up-regulated expression of pd-1 and its ligands during acute classical swine fever virus infection in swine. Res Vet Sci. (2014) 97:251–6. doi: 10.1016/j.rvsc.2014.07.023
35. Yue F, Cheng A, Zhu Y, Li P, Zhang Y, Sun G, et al. Overexpression of programmed death ligands in naturally occurring postweaning multisystemic wasting syndrome. Viral Immunol. (2015) 28:101–6. doi: 10.1089/vim.2014.0097
36. Richmond O, Cecere T, Erdogan E, Meng X, Pineyro P, Subramaniam S, et al. The pd-L1/cd86 ratio is increased in dendritic cells co-infected with porcine circovirus type 2 and porcine reproductive and respiratory syndrome virus, and the pd-L1/pd-1 axis is associated with anergy, apoptosis, and the induction of regulatory T-cells in porcine lymphocytes. Vet Microbiol. (2015) 180:223–9. doi: 10.1016/j.vetmic.2015.09.014
37. Hulst M, Loeffen W, and Weesendorp E. Pathway analysis in blood cells of pigs infected with classical swine fever virus: comparison of pigs that develop a chronic form of infection or recover. Arch Virol. (2013) 158:325–39. doi: 10.1007/s00705-012-1491-8
38. Ruedas-Torres I, Rodríguez-Gómez IM, Sánchez-Carvajal JM, Guil-Luna S, Larenas-Muñoz F, Pallarés FJ, et al. Up-regulation of immune checkpoints in the thymus of prrsv-1-infected piglets in a virulence-dependent fashion. Front Immunol. (2021) 12:671743. doi: 10.3389/fimmu.2021.671743
39. Lee M-A, Jayaramaiah U, You S-H, Shin E-G, Song S-M, Ju L, et al. Molecular characterization of porcine reproductive and respiratory syndrome virus in korea from 2018 to 2022. Pathogens. (2023) 12:757. doi: 10.3390/pathogens12060757
40. Reed LJ and Muench H. A simple method of estimating fifty per cent endpoints12. Am J Epidemiol. (1938) 27:493–7. doi: 10.1093/oxfordjournals.aje.a118408
41. Jeong C-G, Khatun A, Nazki S, Kim S-C, Noh Y-H, Kang S-C, et al. Evaluation of the cross-protective efficacy of a chimeric prrsv vaccine against two genetically diverse prrsv2 field strains in a reproductive model. Vaccines. (2021) 9:1258. doi: 10.3390/vaccines9111258
42. Maisonnasse P, Bordet E, Bouguyon E, and Bertho N. Broncho alveolar dendritic cells and macrophages are highly similar to their interstitial counterparts. PLoS One. (2016) 11:e0167315. doi: 10.1371/journal.pone.0167315
43. Maisonnasse P, Bouguyon E, Piton G, Ezquerra A, Urien C, Deloizy C, et al. The respiratory dc/macrophage network at steady-state and upon influenza infection in the swine biomedical model. Mucosal Immunol. (2016) 9:835–49. doi: 10.1038/mi.2015.105
44. Mair KH, Essler SE, Patzl M, Storset AK, Saalmüller A, and Gerner W. Nkp46 expression discriminates porcine nk cells with different functional properties. Eur J Immunol. (2012) 42:1261–71. doi: 10.1002/eji.201141989
45. Mair KH, Müllebner A, Essler SE, Duvigneau JC, Storset AK, Saalmüller A, et al. Porcine cd8αdim/-nkp46high nk cells are in a highly activated state. Vet Res. (2013) 44:1–14. doi: 10.1186/1297-9716-44-13
46. Rossow K, Collins J, Goyal S, Nelson E, Christopher-Hennings J, and Benfield D. Pathogenesis of porcine reproductive and respiratory syndrome virus infection in gnotobiotic pigs. Vet Pathol. (1995) 32:361–73. doi: 10.1177/030098589503200404
47. Xie CZ, Ha Z, Zhang H, Zhang Y, Xie YB, Zhang H, et al. Pathogenicity of porcine reproductive and respiratory syndrome virus (Orf5 rflp 1-7-4 viruses) in China. Transboundary Emerg Dis. (2020) 67:2065–72. doi: 10.1111/tbed.13549
48. Ruedas-Torres I, Rodríguez-Gómez I, Sánchez-Carvajal JM, Larenas-Muñoz F, Pallarés F, Carrasco L, et al. The jigsaw of prrsv virulence. Vet Microbiol. (2021) 260:109168. doi: 10.1016/j.vetmic.2021.109168
49. Khatun A, Nazki S, Jeong C-G, Gu S, Lee S-I, Yang M-S, et al. Effect of polymorphisms in porcine guanylate-binding proteins on host resistance to prrsv infection in experimentally challenged pigs. Vet Res. (2020) 51:1–14. doi: 10.1186/s13567-020-00745-5
50. Boddicker NJ, Garrick DJ, Rowland R, Lunney JK, Reecy J, and Dekkers J. Validation and further characterization of a major quantitative trait locus associated with host response to experimental infection with porcine reproductive and respiratory syndrome virus. Anim Genet. (2014) 45:48–58. doi: 10.1111/age.2013.45.issue-1
51. Rawal G, Almeida MN, Gauger PC, Zimmerman JJ, Ye F, Rademacher CJ, et al. In vivo and in vitro characterization of the recently emergent prrsv 1-4–4 L1c variant (L1c. 5) in Comparison with Other Prrsv-2 Lineage 1 Isolates. Viruses. (2023) 15:2233. doi: 10.3390/v15112233
52. Reith W and Haußmann A. Importance of virchow-robin spaces. Der Radiol. (2018) 58:142–7. doi: 10.1007/s00117-017-0354-4
53. Thanawongnuwech R, Halbur PG, and Andrews JJ. Immunohistochemical detection of porcine reproductive and respiratory syndrome virus antigen in neurovascular lesions. J Vet Diagn Invest. (1997) 9:334–7. doi: 10.1177/104063879700900323
54. Cao J, Li B, Fang L, Chen H, and Xiao S. Pathogenesis of nonsuppurative encephalitis caused by highly pathogenic porcine reproductive and respiratory syndrome virus. J Vet Diagn Invest. (2012) 24:767–71. doi: 10.1177/1040638712445764
55. Suh J and Chae C. Genetic and pathogenic characteristics of an emerging highly virulent recombinant lineage korean clade C prrsv strain. Transboundary Emerging Dis. (2024) 2024:5785557. doi: 10.1155/2024/5785557
56. Sánchez-Carvajal J, Rodríguez-Gómez I, Ruedas-Torres I, Zaldívar-López S, Larenas-Muñoz F, Bautista-Moreno R, et al. Time series transcriptomic analysis of bronchoalveolar lavage cells from piglets infected with virulent or low-virulent porcine reproductive and respiratory syndrome virus 1. J Virol. (2022) 96:e01140–21. doi: 10.1128/JVI.01140-21
57. Newton AH, Cardani A, and Braciale TJ. The host immune response in respiratory virus infection: balancing virus clearance and immunopathology. Semin Immunopathol. (2016) 38:471–82. doi: 10.1007/s00281-016-0558-0
58. Kochin V and Nishikawa H. Meddling with meddlers: curbing regulatory T cells and augmenting antitumor immunity. Nagoya J Med Sci. (2019) 81:1. doi: 10.18999/nagjms.81.1.1
59. Collins AV, Brodie DW, Gilbert RJ, Iaboni A, Manso-Sancho R, Walse B, et al. The interaction properties of costimulatory molecules revisited. Immunity. (2002) 17:201–10. doi: 10.1016/S1074-7613(02)00362-X
60. Polesso F, Weinberg AD, and Moran AE. Late-stage tumor regression after pd-L1 blockade plus a concurrent ox40 agonistlarge tumors regress with anti–pd-L1 and anti-ox40 treatment. Cancer Immunol Res. (2019) 7:269–81. doi: 10.1158/2326-6066.CIR-18-0222
61. Linsley PS, Greene J, Tan P, Bradshaw J, Ledbetter JA, Anasetti C, et al. Coexpression and functional cooperation of ctla-4 and cd28 on activated T lymphocytes. J Exp Med. (1992) 176:1595–604. doi: 10.1084/jem.176.6.1595
62. Anderson AC, Joller N, and Kuchroo VK. Lag-3, tim-3, and tigit: co-inhibitory receptors with specialized functions in immune regulation. Immunity. (2016) 44:989–1004. doi: 10.1016/j.immuni.2016.05.001
63. Yeung AW, Terentis AC, King NJ, and Thomas SR. Role of indoleamine 2, 3-dioxygenase in health and disease. Clin Sci. (2015) 129:601–72. doi: 10.1042/CS20140392
64. Chaudhari J, Liew C-S, Riethoven J-JM, Sillman S, and Vu HL. Porcine reproductive and respiratory syndrome virus infection upregulates negative immune regulators and T-cell exhaustion markers. J Virol. (2021) 95:10. doi: 10.1128/JVI.01052-21
65. Ruedas-Torres I, Sánchez-Carvajal JM, Carrasco L, Pallarés FJ, Larenas-Muñoz F, Rodríguez-Gómez IM, et al. Prrsv-1 induced lung lesion is associated with an imbalance between costimulatory and coinhibitory immune checkpoints. Front Microbiol. (2023) 13:1007523. doi: 10.3389/fmicb.2022.1007523
66. Lim B, Kim S-C, Kim H-J, Kim J-H, Seo Y-J, Lim C, et al. Single-cell transcriptomics of bronchoalveolar lavage during prrsv infection with different virulence. Nat Commun. (2025) 16:1112. doi: 10.1038/s41467-024-54676-2
Keywords: porcine reproductive and respiratory syndrome virus, Korean Lineage, pathogenicity, immune characteristics, host immunity, immune checkpoint molecules
Citation: Park G-S, Kim S-C, Kim H-J, Jeong C-G, Kang S-C, Shin G-E, Kim S-H, Jeong H-Y, Lee K-K, Lee S-M and Kim W-I (2025) Immunopathological features of highly pathogenic Korean Lineage B PRRSV-2: insights into virulence indicators and host immune responses. Front. Immunol. 16:1599468. doi: 10.3389/fimmu.2025.1599468
Received: 25 March 2025; Accepted: 26 May 2025;
Published: 18 June 2025.
Edited by:
Lei Tan, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Pranab Jyoti Das, National Research Centre on Pig (ICAR), IndiaOuyang Peng, Sun Yat-sen University, China
Copyright © 2025 Park, Kim, Kim, Jeong, Kang, Shin, Kim, Jeong, Lee, Lee and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Won-Il Kim, a3dpMDYyMUBqYm51LmFjLmty
†These authors have contributed equally to this work and share first authorship
 Gyeong-Seo Park
Gyeong-Seo Park Seung-Chai Kim
Seung-Chai Kim Hwan-Ju Kim
Hwan-Ju Kim Chang-Gi Jeong1,3
Chang-Gi Jeong1,3 Sang-Myeong Lee
Sang-Myeong Lee Won-Il Kim
Won-Il Kim