- 1Institute of Respiratory Health, Frontiers Science Center for Disease-related Molecular Network, West China Hospital, Sichuan University, Chengdu, China
- 2West China School of Basic Medical Sciences & Forensic Medicine, Sichuan University, Chengdu, Sichuan, China
- 3College of Life Sciences, Sichuan University, Chengdu, Sichuan, China
- 4School of Clinical Medicine, North Sichuan Medical College, Nanchong, Sichuan, China
Tuberculosis (TB), caused by Mycobacterium tuberculosis (Mtb), remains a leading global health threat, exacerbated by drug resistance and inadequate vaccine efficacy. The PE/PPE protein family, unique to mycobacteria, constitutes ~10% of the Mtb genome and plays critical roles in bacterial physiology, immune evasion, and host-pathogen interactions. This review synthesizes advances in understanding the evolutionary expansion, structural diversity, and functional versatility of PE/PPE proteins, emphasizing their co-evolution with type VII secretion systems (T7SS). We highlight their roles in nutrient acquisition, immune modulation, and pathogenesis, alongside their potential as diagnostic and vaccine targets. Clinical progress in PE/PPE-based vaccines, such as M72/AS01E and ID93/GLA-SE, underscores their promise in combating TB, while challenges in epitope variability and functional redundancy demand innovative strategies. By integrating evolutionary, structural, and immunological insights, this review provides a roadmap for leveraging PE/PPE biology to develop next-generation TB interventions.
1 Introduction
Tuberculosis (TB), caused by Mycobacterium tuberculosis (Mtb), remains one of the world’s most persistent infectious diseases, with a disproportionate burden in low-resource settings (1). The World Health Organization (WHO) estimates that, in 2023, there were 10.8 million new TB cases and 1.25 million deaths globally, positioning TB as one of the top ten causes of death worldwide (2). The emergence of multidrug-resistant tuberculosis (MDR-TB) and TB associated with HIV further complicates efforts to control the disease, exacerbating the global public health crisis. Among the various strategies to combat TB, vaccination remains the most cost-effective approach (3). However, despite the use of the Bacille Calmette-Guérin (BCG) vaccine since 1921, no new, effective vaccines have been developed. While BCG provides substantial protection against childhood TB, its efficacy in adults and adolescents remains limited (4). Moreover, the effectiveness of BCG varies significantly across regions, influenced by factors such as climate, economics, and healthcare infrastructure (5). In response, the WHO has launched initiatives aimed at developing the next generation of TB vaccines.
The PE/PPE proteins of mycobacteria—defined by the highly conserved N-terminal Pro-Glu (PE) or Pro-Pro-Glu (PPE) motifs—represents a unique and highly diverse group of proteins (6). In Mtb, the 169 genes encoding PE and PPE proteins constitute approximately 7–10% of the genome’s coding capacity (7). Although similar genes exist in fast-growing mycobacteria, their amplification and functional specialization are particularly pronounced in slow-growing species (8). These proteins are predominantly secreted to the bacterial surface or extracellular space via the Type VII secretion system (T7SS) (9). PE/PPE proteins play essential roles in both bacterial physiology and host-pathogen interactions. In terms of bacterial physiology, they are involved in nutrient uptake, metabolism, enzymatic activity, and drug resistance (10, 11). In host-pathogen interactions, they modulate processes such as phagosome maturation, antigen presentation, autophagy, cell death regulation, and both innate and adaptive immune responses, interacting with Toll-like receptors (TLRs) (11). Importantly, PE/PPE proteins are rich in T-cell and B-cell epitopes, eliciting strong immune responses and thus representing promising candidates for diagnostic tools and subunit vaccine development (12).
Despite the biological significance of the PE/PPE family, considerable challenges remain in their study. The redundancy and sequence repetition inherent in these proteins complicate functional annotation (6). Their high content of low-complexity sequences further complicates structural analyses, limiting our understanding of their three-dimensional conformation and functional domains. Different PE/PPE proteins—or even distinct domains within the same protein—may have divergent roles in modulating host immunity, and the flexible interactions between these proteins contribute to their functional diversity, adding complexity to their study (11, 13). Although recombinant protein expression systems, such as Mycobacterium smegmatis (M. smegmatis), are commonly used for functional analysis, these models may fail to replicate the native localization and functionality of PE/PPE proteins in Mtb. Additionally, the limitations of animal models—ranging from mice to non-human primates—pose challenges in assessing vaccine efficacy, as these models often reflect only certain immunological features of TB (14, 15). Nevertheless, vaccines targeting PPE proteins, such as M72/AS01E and ID93/GLA-SE, have shown promise in clinical trials, providing valuable insights for the future development of PE/PPE-based vaccines.
Recent advances in evolutionary biology, functional genomics, and structural biology have significantly enhanced our understanding of the PE/PPE family. This review aims to synthesize these developments, focusing on the characteristics, classification, evolution, structure, subcellular localization, secretion mechanisms, and physiological and pathological functions of PE/PPE proteins, as well as their potential as vaccine antigens. By elucidating the diversity and biological relevance of PE/PPE proteins, we aim to lay a theoretical foundation for the next generation of anti-tuberculosis vaccines.
2 General characteristics of the PE/PPE protein family
The PE/PPE protein family is a distinctive and multi-domain protein family exclusive to mycobacteria (7). In the Mtb H37Rv reference strain, the genome encodes 100 members of the PE family and 69 members of the PPE family (6). The number of PE/PPE genes can vary across different Mtb clinical isolates, reflecting both genetic polymorphism and strain-specific expression patterns. The genomic distribution of pe/ppe genes in Mtb is notably non-random. At least 28 distinct pe/ppe operons have been identified, such as pe25-ppe41 and pe35-ppe68, with the majority of classical pe genes (excluding pe_pgrs members) located upstream of their corresponding ppe genes within these operons. By contrast, pe_pgrs genes are dispersed throughout the genome and generally do not form operonic structures with ppe genes, suggesting functional and regulatory divergence within the pe/ppe family (16, 17). Furthermore, these genes are often clustered near the ESX (ESAT-6 secretion system) gene clusters, particularly those associated with the ESX-1 and ESX-5 transcriptional units, suggesting an evolutionary and functional link with these secretion systems (8).
A defining feature of the PE/PPE family is the high GC content of their gene sequences, which results in relatively low transcriptional and translational efficiency. This may serve as a mechanism for tightly regulated gene expression in response to various environmental stresses encountered during infection (6). These stressors include conditions such as hypoxia, nutrient deprivation, heat shock, and acidic pH, all of which influence the tissue-specific expression of these genes during the infection process (18, 19).
The PE/PPE protein family is characterized by significant gene redundancy and polymorphism, particularly in their C-terminal regions. Many pe/ppe genes contain variable numbers of tandem repeat sequences (VNTRs), which are subject to mutations through mechanisms like homologous recombination, gene conversion, and repeat expansion. These genetic mechanisms provide the PE/PPE proteins with substantial adaptive potential (20). Among these, the PE_PGRS (Polymorphic GC-Rich Sequence) proteins exhibit the highest degree of polymorphism. A comparative analysis of 27 pe_pgrs genes across 94 clinical isolates revealed considerable genetic variation, particularly in comparison to other genomic regions. Nucleotide diversity, along with insertion/deletion events and distinct dN/dS ratios, indicates diverse selective pressures acting on these genes (21). Furthermore, a recent study has shown that pe/ppe transcripts are enriched in RNA G-quadruplex (rG4) structures, suggesting an additional layer of post-transcriptional regulation. These rG4 elements may enable Mtb to fine-tune PE/PPE protein expression in response to infection stage and environmental stress, thereby modulating the intensity of host–pathogen interactions (22).
From an immunological perspective, the PE/PPE proteins exhibit varying degrees of sequence conservation and variability, resulting in marked differences in their immunogenic potential. The N-terminal domains of these proteins are typically more conserved and enriched in T-cell epitopes, whereas the C-terminal regions, especially within the PE_PGRS and PPE_MPTR (Major Polymorphic Tandem Repeats) subfamilies, display higher genetic variability (7, 21). Additionally, antibodies against multiple PE/PPE proteins have been detected in the sera of TB patients, suggesting that these proteins are recognized by the immune system and can elicit humoral responses during natural infection (23, 24). Initially, these proteins were thought to primarily contribute to antigenic variation as a mechanism to evade host immune surveillance (25). However, emerging evidence indicates that not all pe/ppe genes are under positive selective pressure; many instead exhibit characteristics of neutral evolution or purifying selection, challenging the traditional view of antigenic variation as the primary driver of their evolution (20, 21).
3 Classification of the PE/PPE protein family
The PE/PPE protein family can be classified through several approaches, including analysis of C-terminal diversity, phylogenetic relationships, and functional roles.
A defining characteristic of the PE protein family is the considerable variability observed in the C-terminal region, which serves as a key basis for classification (6, 8). The PE family is divided into two subfamilies: PE and PE_PGRS (Polymorphic GC-Rich Sequence). PE proteins possess a conserved N-terminal structure with a relatively short, less variable C-terminal, whereas PE_PGRS proteins feature extended C-terminals rich in repeat sequences, such as glycine-rich polymers (Gly-Gly-Ala or Gly-Gly-Asn), contributing to their high variability (6, 8). Phylogenetic analysis further subdivides the PE family into five distinct subfamilies. Subfamilies I and II, representing the oldest members, include PE34 and PE35 (subfamily I) and PE5 and PE15 (subfamily II). Subfamily III includes PE22, PE25, and PE36, while subfamily IV contains PE proteins secreted via the ESX-5 system, which are pivotal in Mtb-host cell interactions. Subfamily V is primarily composed of PE_PGRS proteins, with some members containing unique C-terminal enzymes such as lipases (8, 11).
The classification of the PPE family is also determined by the diversity of the C-terminal region, with PPE proteins typically possessing extended C-terminals that vary significantly in structure and function. Based on these variations, the PPE family is categorized into four subfamilies: PPE proteins lacking distinctive C-terminal sequences, PPE proteins with the PxxPxxW motif (PPE_PPW), PPE proteins containing the GxxSVPxxW motif (PPE_SVP), and PPE proteins with major polymorphic tandem repeats (PPE_MPTR) (8). Phylogenetic analysis further divides the PPE family into five subfamilies. Subfamily I include members such as PPE68. Subfamily II is composed of PPE_PPW members, while subfamily III includes PPE36, PPE41, PPE57, PPE58, PPE59, and PPE69. Subfamilies III and IV are particularly involved in immune evasion. Subfamily V consists of PPE_MPTR members, which are typically large, with some secreted PPE proteins exceeding 3,000 amino acids in length (8, 26).
4 Evolution of the PE/PPE family proteins
4.1 Evolutionary differences of PE/PPE family proteins across species
The evolutionary trajectory of the PE/PPE protein family is indicative of the genome plasticity that has enabled Mycobacterium species to adapt to diverse ecological niches and develop pathogenicity. Comparative genomic analyses reveal distinct patterns of expansion within these gene families in mycobacterial species with differing lifestyles.
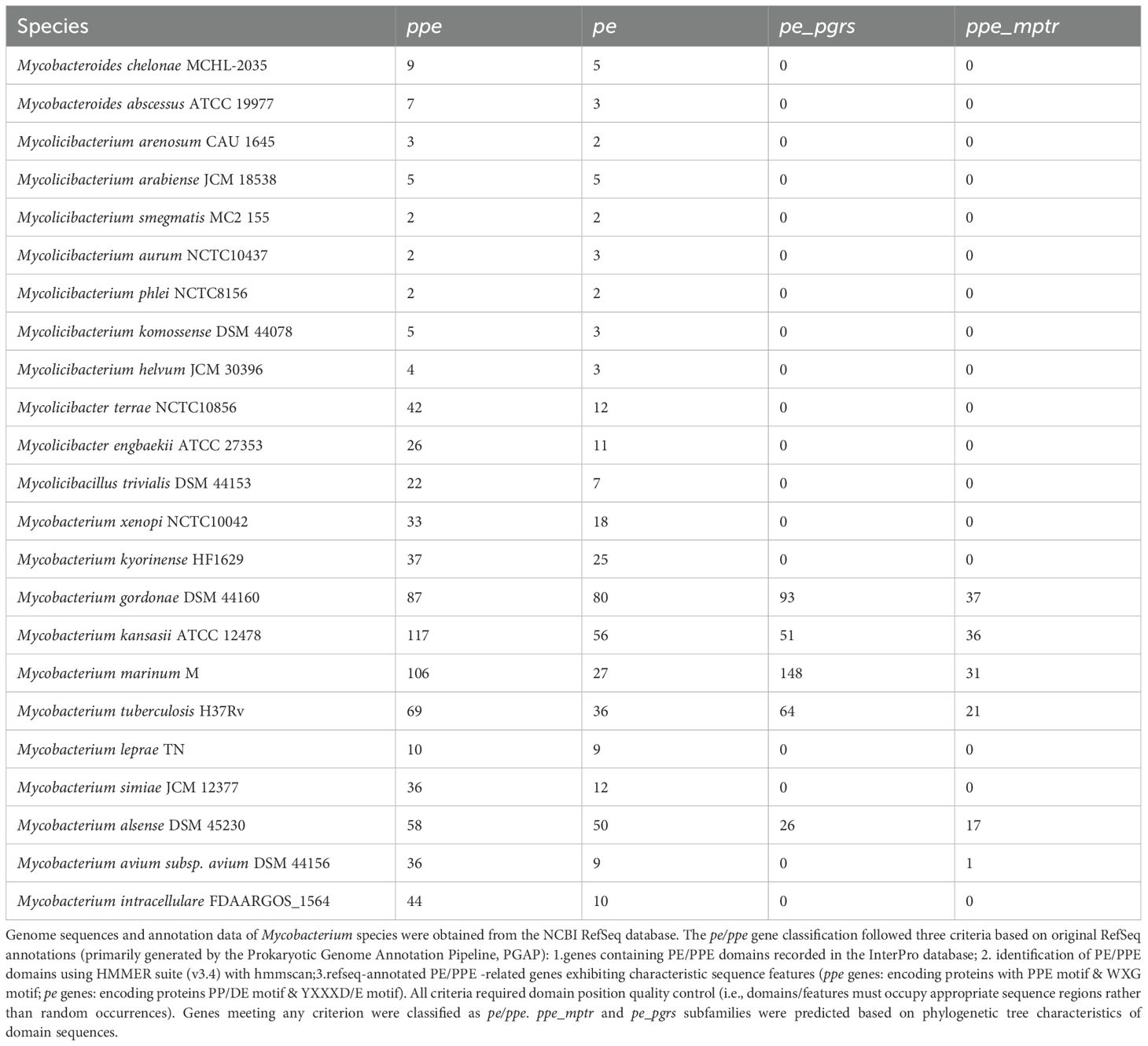
In free-living, fast-growing, non-pathogenic mycobacteria such as M. smegmatis, only two pairs of pe/ppe genes are present, which are primarily involved in basic metabolic functions (27). In contrast, pathogenic, slow-growing mycobacteria exhibit substantial expansion of these gene families (Table 1). For instance, the genome of Mycobacterium marinum contains 281 pe/ppe genes, encoding 27 PE, 148 PE_PGRS, and 106 PPE proteins (28). Notably, Mycobacterium leprae (M. leprae) represents an exceptional case: although it is a slow-growing pathogen, it has undergone significant genomic reduction. With a genome size of approximately 3.2 Mb and containing 1,129 pseudogenes, M. leprae harbors only 1,440 protein-coding genes, in stark contrast to other closely related mycobacterial species, which possess more than 4,000 protein-coding genes (29). In terms of pe/ppe genes, M. leprae retains just 9 complete pe genes and 10 complete ppe genes (30). This genomic reduction reflects the ecological niche contraction that accompanied the transition of M. leprae from a free-living ancestor to an obligate intracellular parasite.
Notably, the evolutionary expansion of the pe/ppe gene family is accompanied by a striking enrichment in RNA G-quadruplex (rG4) structures within their transcripts (22). Although pe/ppe genes comprise less than 8% of total transcript length, they account for over 50% of all rG4 motifs identified in Mtb transcripts. Comparative analyses further show that rG4 density exceeds putative quadruplex sequence (PQS) density in the genomes of slow-growing pathogenic mycobacteria (rG4/PQS ratio >1), but not in non-pathogenic species (ratio <1) (22). These findings suggest that during the evolution of pathogenic slow-growers, not only did the pe/ppe family expand, but their transcripts also acquired increased rG4 content—potentially enabling fine-tuned post-transcriptional regulation in response to the complex intracellular environment of the host.
4.2 Co-evolution of PE/PPE family proteins with the ESX system
The evolution of PE/PPE proteins exhibits a co-evolutionary trajectory with the expansion of esx gene clusters. Phylogenetic analyses suggest that the ancestral ESX-4 system initially lacked pe/ppe genes. Subsequent gene duplication events gave rise to five distinct ESX loci in Mtb (8). The earliest incorporation of pe35 (Rv3872) and its partner ppe68 (Rv3873) into ESX-1—Mtb’s second type VII secretion system—marked the origin of these protein families (31). Subsequent duplications led to the emergence of ESX-3, ESX-2, and ESX-5 (8). Comparative genomics between the fast-growing M. smegmatis and the slow-growing Mtb supports this evolutionary trajectory. M. smegmatis retains pe/ppe pairs associated with ESX-1 and ESX-3 but lacks pe_pgrs, ppe_svp and ppe_mptr subfamilies entirely (8). In contrast, ESX-5, the most recently evolved type VII secretion system, is exclusive to slow-growing mycobacteria and coincides with a significant expansion of PE_PGRS, PPE_SVP and PPE_MPTR subfamilies. This suggests that ESX-5 acquisition was a pivotal event in the evolution of mycobacterial pathogenicity (32).
The diversification of the pe/ppe gene family is driven by gene duplication, point mutations, and recombination. Notably, the PE_PGRS and PPE_MPTR subfamilies, characterized by repetitive sequences, undergo frequent recombination, generating extensive protein diversity. Karboul et al. demonstrated that homologous recombination rates within pe/ppe loci in clinical mycobacterial isolates significantly exceed those of other genomic regions, suggesting these loci as hotspots for genomic plasticity (33).
The co-evolution of PE/PPE proteins with the ESX system is likely shaped by host-pathogen interactions. Slow-growing pathogenic mycobacteria encounter complex immune pressures necessitating prolonged infection, favoring an expanded PE/PPE repertoire that enhances antigenic variation and immune modulation (27). Conversely, free-living, fast-growing mycobacteria retain a streamlined system, sufficient for their ecological niche.
5 Structure of the PE/PPE family proteins
The structural features of PE/PPE proteins are central to their functional roles in mycobacteria. Structural studies employing X-ray crystallography, nuclear magnetic resonance, and bioinformatics have provided insights into their molecular organization. While the conserved N-terminal regions of PE/PPE proteins, particularly those involved in heterodimer formation, have been well characterized, the highly variable C-terminal domains remain structurally elusive.
5.1 Basic structure of PE/PPE family proteins
PE proteins are defined by a conserved N-terminal domain (~100 amino acids) that adopts an antiparallel α-helix-turn-α-helix conformation. This region harbors the characteristic PE motif (Pro-Glu) and a YXXXD/E secretion signal within the first α-helix. The C-terminal domain exhibits significant variability, ranging from short extensions to >1400 amino acids. Members of the PE_PGRS subfamily contain glycine-rich repeats (e.g., Gly-Gly-Ala, Gly-Gly-Asn), which may confer structural flexibility (25). Recent computational structure predictions of PE_PGRS proteins using AlphaFold revealed the PGRS domain as tightly packed β-sandwiches, prompting the authors to propose a ‘sailing’ model wherein this domain acts as a mechanistic framework to diffuse along the mycomembrane, expose structural motifs mediating host interactions, and deliver functional C-terminal protein modules (34).
PPE proteins share a conserved N-terminal domain (~180 amino acids) composed of five α-helices arranged in a helical bundle. The PPE motif is localized in the first α-helix, while the WxG motif resides between the second and third helices. A hydrophobic hh motif, positioned between the fourth and fifth α-helices, is crucial for interactions with secretion chaperones (35). The PPE C-terminal domain is highly variable, exceeding 3000 amino acids in some cases. Members of the PPE_MPTR subfamily are distinguished by a conserved Asn-X-Gly-X-Gly-Asn-X-Gly (NXGXGXN) repeat motif (6).
5.2 PE-PPE heterodimer structure
Although definitive experimental evidence remains limited, PE and PPE proteins are thought to be secreted as heterodimers via the mycobacterial ESX system. Structural elucidation of the PE25/PPE41 heterodimer revealed that the α-helices of PE25 interact with those of PPE41 via hydrophobic and intermolecular forces, forming a stable four-helix bundle (36). This organization juxtaposes the PE YXXXD/E motif with the PPE WxG motif, likely generating a composite secretion signal for type VII secretion system (T7SS) recognition (37). Several PE-PPE pairs, including PE35-PPE68, PE18-PPE26, and PE5-PPE4, have been experimentally validated (16, 31, 38). While most PE-PPE pairs are encoded by adjacent genes, non-adjacent interactions, such as PE19-PPE51, indicate a degree of pairing flexibility (10). Moreover, PE-PE interactions, exemplified by PE9-PE10 surface-localized dimers, further diversify the functional landscape of this protein family (39).
Strikingly, PE-PPE heterodimers share structural homology with ESX substrates, including the EsxA/EsxB (ESAT-6/CFP-10) complex and EspB, folding into multi-helical bundles with conserved secretion-associated motifs. This structural conservation supports their role as canonical T7SS substrates and suggests shared functional mechanisms (40, 41).
5.3 PE-PPE-EspG heterotrimer structure
The secretion-associated protein EspG plays a pivotal role in PE-PPE protein export. Structural analysis of the PE25-PPE41-EspG5 complex revealed that EspG5 binds to the PPE hh motif, preventing aggregation and stabilizing the heterodimer during secretion (42). Subsequent studies on PE8-PPE15-EspG5 suggested a conserved interaction interface across PPE proteins (43). Comparative structural analyses indicate that while PE-PPE-EspG complexes share a conserved binding mode, subtle interaction differences exist. For example, in the ESX-3 system, EspG3 binds PPE4 at a distinct angle relative to EspG5 interactions with PPE15 and PPE41. Additionally, PPE4 exhibits an extended hh motif loop, highlighting potential ESX system-specific substrate adaptations (31).
6 Subcellular localization and secretion of PE/PPE proteins
The spatial organization and transport of PE-PPE proteins are integral to their functional specialization in mycobacteria. These proteins exhibit distinct subcellular distribution patterns, dictated by specialized secretion systems and molecular chaperones.
6.1 Subcellular distribution of PE/PPE proteins
High-throughput proteomic analyses have identified over 35 PE/PPE proteins in the membrane and/or cell wall of Mtb (7). For instance, PE_PGRS33 localizes to the cell wall of M. smegmatis and Mtb, guided by its N-terminal PE domain (44–46). Similarly, LipY and PE_PGRS30 utilize N-terminal sequences for secretion and membrane association (47, 48). Notably, PE19-PPE51, PE20-PPE31, and PE15-PPE20 form outer membrane-associated channels, suggesting roles in nutrient exchange or virulence (10, 49–51). Certain PE/PPE proteins are also secreted into the extracellular environment, as evidenced by proteomic detection of at least seven PE/PPE proteins in culture filtrates (7). Recently, Lepe et al. further expanded our understanding of the Mtb surface PE/PPE proteome through the development of protease shaving techniques coupled with quantitative mass spectrometry analysis (52). This study identified 167 proteins with significantly elevated abundance under protease treatment conditions, including multiple PE/PPE family members such as PE12, PE23, PPE10, PPE18, PPE20, PPE32, PPE33, PPE38, PPE40, PPE51, and PPE60. Notably, the researchers validated the surface localization of PPE18, PPE38, and PE23 through flow cytometry, with PPE18, a component of the M72 vaccine candidate, demonstrating particularly remarkable surface enrichment (52).
Intriguingly, subcellular localization may be dynamically regulated by environmental cues. For example, PPE37 undergoes proteolytic cleavage under iron-limiting conditions, leading to differential localization and functional specialization of its N- and C-terminal fragments (13). Such adaptive relocalization may contribute to mycobacterial persistence and host adaptation.
6.2 Mechanisms of PE/PPE translocation across the membrane
6.2.1 Inner membrane transport
The translocation of PE/PPE proteins across the inner membrane is orchestrated by the ESX secretion system, a multi-component apparatus that ensures substrate specificity and transport fidelity. PE-PPE heterodimers first associate with EspG, a dedicated chaperone that stabilizes the complex and prevents aggregation via interactions with the PPE hh motif (53, 54).
The ESX membrane translocon comprises five core components—EccB, EccC, EccD, EccE, and MycP—along with cytosolic factors EccA and EspG, forming a ~2 MDa secretion complex (55–57). Cryo-electron microscopy studies of ESX-3 and ESX-5 have provided structural insights into this machinery (56, 58). EccC, the key ATPase, drives substrate translocation via ATP hydrolysis, with its third nucleotide-binding domain (NBD3) interacting with the YXXXD/E secretion signal, while linker 2 mediates PE-PPE specificity (59, 60). Mutational analyses highlight the functional importance of EccC, as disruptions in its NBD1 domain abrogate PE_PGRS secretion (61). EccD, the central transmembrane conduit, features 11 transmembrane helices, while EccB, EccE, and EccC contribute to structural integrity and motor function. The protease MycP stabilizes the complex via a single transmembrane domain (56). Notably, while some PE_PGRS proteins (e.g., PE5, PE15, PE_PGRS12, PE_PGRS29) possess putative Sec-signal sequences, their dependence on the Sec pathway remains unconfirmed (62).
6.2.2 Outer membrane translocation
The mechanisms facilitating PE/PPE protein translocation across the outer membrane remain incompletely understood. Some evidence suggests that PE/PPE proteins contribute to outer membrane channel formation, potentially mediating their own export. In M. marinum, PPE68 and its functional homolog MMAR_2894 are essential for ESX-1 substrate secretion, suggesting a role in periplasmic or outer membrane transport (63, 64). However, PPE68 does not appear to form a stable membrane channel but is instead stored intracellularly and subsequently processed by PecABC and other proteases (64).
Recent work expressing the Mycobacterium xenopi esx-5 operon in M. smegmatis demonstrated that PPE proteins within the esx-5 gene cluster are indispensable for functional ESX-5 secretion, implicating these proteins in outer membrane translocation (32). In Mtb, PPE38 is critical for ESX-5-dependent PE_PGRS and PPE_MPTR secretion; loss-of-function mutations result in secretion defects and altered virulence (65). Strikingly, clinical isolates of the hypervirulent Beijing lineage exhibit ppe38 deletions, with reintroduction of PPE38 attenuating virulence, further underscoring its functional significance (65, 66). While the precise mechanism remains unresolved, PPE38 may function as a secretion facilitator, either directly forming an outer membrane channel or interacting with auxiliary factors. The ESX-5 system may employ additional, yet unidentified, components to mediate PE/PPE export, warranting further investigation.
7 Functions of the PE/PPE family proteins
7.1 Biological functions in bacterial physiology
7.1.1 Nutrient acquisition
The highly hydrophobic cell wall of Mycobacterium tuberculosis lacks classical porins for transmembrane transport (11). Recent findings suggest that PE/PPE family proteins function as outer membrane channels mediating nutrient uptake, despite the absence of direct structural evidence. PPE51 and PE19 facilitate the uptake of glucose, glycerol, maltose, trehalose, and certain low-molecular-weight drugs (10, 49, 50). The PE20-PPE31 and PE15-PPE20 complexes mediate magnesium and calcium uptake, respectively (10, 51). The PE5-PPE4 complex, a substrate of the ESX-3 secretion system, is crucial for mycobactin-bound iron uptake; its deletion renders Mtb incapable of growth under low-iron conditions (67). This system is transcriptionally regulated by the metal-ion-responsive transcription factors Zur and MntR (68, 69). However, no evidence currently supports the involvement of PE5-PPE4 in Zn or Mn uptake.
While Mitra et al. identified PPE36, along with PPE62, as essential for heme utilization (70, 71), Tullius et al. reported no impairment in heme uptake following ppe36 deletion (72). Instead, their findings implicated PPE37 in heme acquisition (72). This discrepancy may arise from strain differences, as Mitra et al. used H37Rv, whereas Tullius et al. employed the Erdman strain. Given the variability of ppe37 among clinical isolates (72, 73), PPE37 is unlikely essential for Mtb survival. Additionally, PPE37 and its paralogs contain a positively charged C-terminal segment that may serve as a nuclear localization signal (NLS), suggesting potential novel functions (13). PPE64 also exhibits heme-binding ability and channel-forming activity, likely contributing to heme-iron utilization (74). The cell wall-localized PE-PGRS3, with an arginine-rich C-terminal, interacts with negatively charged phospholipids on alveolar epithelial cells, facilitating phosphate acquisition under nutrient-limiting conditions (75).
7.1.2 Metabolism and enzyme activity
During infection, Mtb enters dormancy within alveolar macrophages, reactivating upon immune suppression. Dormancy maintenance and reactivation depend on the storage and metabolism of fatty acids and cholesterol within nutrient-limited phagolysosomes (76). PPE15 (mper1), upregulated during dormancy, is crucial for triglyceride accumulation and homeostasis (77). Deletion of mper1 prevents lipid droplet formation, thereby impairing dormancy establishment both in vitro and in human granuloma models (78). PE_PGRS63 (LipY), a member of the hormone-sensitive lipase family, preferentially hydrolyzes short to intermediate p-nitrophenyl esters and is highly induced under starvation and hypoxia. It serves as the primary lipase for stored triglyceride utilization, playing a critical role in dormancy exit (79, 80).
The surface-localized esterase PE11 (LipX) influences cell wall remodeling and virulence; its deletion alters cell wall composition and reduces intracellular survival within macrophages (81). In M. smegmatis, recombinant PE11 modulates fatty acid profiles in cell wall polar lipids, increasing hydrophobicity and enhancing resistance to stressors such as SDS, lysozyme, acidity, and anti-tuberculosis drugs (24). Other PE proteins, including PE16, PE1, and PE2, exhibit serine esterase activity in vitro, though their role in virulence remains unclear (82, 83). The C-terminal domain of PPE63 also possesses esterase activity, potentially modulating cell wall properties by altering lipid composition (84).
7.2 Functions in regulating host immune responses
7.2.1 Interaction with TLRs to modulate downstream signaling
During early infection, host pattern recognition receptors, particularly Toll-like receptors (TLRs), recognize mycobacterial ligands, initiating immune responses. Several PE/PPE proteins interact with TLRs, notably TLR2 and TLR4, to influence immune signaling cascades (Figure 1).
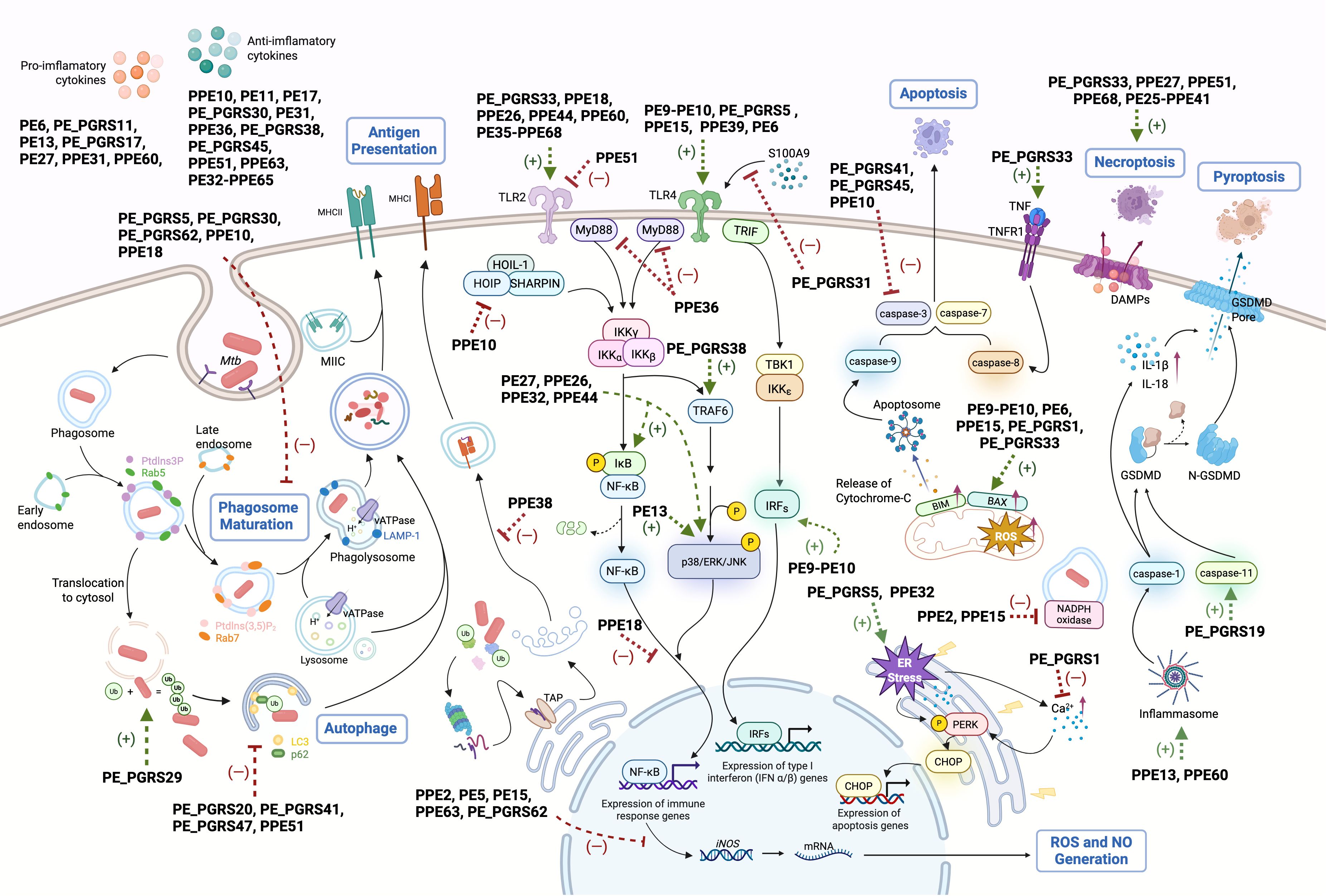
Figure 1. Schematic representation of PE/PPE-mediated immunomodulatory mechanisms in host immune cells. PE/PPE family proteins mediate diverse immunomodulatory functions across multiple phases of Mtb-host interactions. Key mechanisms include: (1) Toll-like receptor (TLR) engagement to modulate downstream signaling cascades; (2) Disruption of phagosome maturation; (3) Interference with antigen presentation; (4) Neutralization of macrophage-derived reactive oxygen species (ROS)/nitric oxide (NO); (5) Regulation of autophagy through ubiquitin (Ub)-dependent pathways; (6) Modulation of programmed cell death; (7) Cytokine network manipulation; (8) Shaping of adaptive immunity. Green arrows: PE/PPE-induced processes, red blunt lines: PE/PPE-inhibited pathways.
PE_PGRS33 promotes Mtb entry into macrophages via a TLR2-dependent mechanism, activating the PI3K-mediated adhesion pathway (85). Inactivation of PE_PGRS33 in BCG Pasteur results in impaired bacterial growth in liquid medium and macrophages (86). Similarly, PPE26 and PPE44 activate TLR2-mediated p38 MAPK and NF-κB signaling, inducing IL-6 and IL-12p40 production in a dose-dependent manner (87, 88). PPE60 promotes dendritic cell maturation and Th1/Th17 responses via TLR2 activation (89). In contrast, PPE51 acts as a TLR2 antagonist, inhibiting autophagy, cytokine secretion, and antigen presentation (90). Mtb also exploits TLR2 signaling to suppress host immunity; PPE18 triggers TLR2-mediated p38 MAPK activation, enhancing IL-10 secretion and attenuating bacterial virulence (91, 92). The PE35-PPE68 complex similarly induces IL-10 and MCP-1 while suppressing IL-12p40 via TLR2-mediated MAPK activation (93). PPE36 inhibits NF-κB activation through Smurf1-mediated MyD88 degradation, although direct PPE36-TLR interactions remain unconfirmed (94).
Several PE/PPE proteins modulate TLR4 signaling. The PE9-PE10 complex activates the TRIF pathway via TLR4, enhancing IFN-β secretion (39). PE_PGRS31 inhibits TLR4-MyD88-NF-κB signaling by blocking S100A9 binding, promoting Mtb survival (95). Conversely, PE6 and PPE15 interact with TLR4 to upregulate NF-κB and induce TNF-α and IL-1β secretion (96, 97). PPE39 enhances DC maturation and Th1 polarization via TLR4 (98).
7.2.2 Interference with phagosome maturation
Host macrophages are the preferred niche of Mtb. Once macrophages internalize pathogens, the phagocytic vesicles need to fuse with lysosomes and mature to create an acidic environment that kills pathogens (99). Mtb can successfully avoid phagosome maturation by manipulating multiple strategies, with some PE/PPE involved. PE_PGRS30 inhibits phagosome-lysosome fusion by reducing LAMP-1 expression, and its deletion lowers bacterial burden and attenuates lung pathology in mice (47, 100). PE_PGRS5 and PE_PGRS62 promote survival under acidic stress by suppressing Rab7 and cathepsin D (101, 102). Homologs of these proteins in M. marinum are enriched in granulomas, suggesting conserved roles in persistence (103). While M. avium MAV_2928 blocks phagosome-lysosome fusion, functional evidence for Mtb’s PPE25 remains elusive (104, 105).
7.2.3 Interfering with antigen presentation
Mtb with PE_PGRS47 mutation exhibits enhanced MHC II-restricted antigen presentation during infection in mice. Intriguingly, of the tested antigens, PE_PGRS47 was found to inhibit the presentation of TB9.8 and Ag85B P25, but not ESAT-6, indicating that PE_PGRS47 was ineffective during the early stages of infection when ESAT-6 was expressed, or because of that ESAT-6 can directly break down the phagosome membrane and access other cellular compartments (106). Studies involving the M. marinum PPE38 knockout strain and PPE38 recombinant M. smegmatis indicated that PPE38 decreases the expression of MHC class I proteins, and consequently, the count of effector/memory CD8 T-cells in mice (107–110). Given that PPE38 is among the most highly expressed proteins in Mtb after 90 days of infection in guinea pigs, it may play a significant role in infection persistency (80).
7.2.4 Counteracting macrophage ROS and NO killing
PE-PPE proteins neutralize ROS/NO defenses. PPE2 translocates to host nuclei via an NLS, repressing iNOS transcription and sequestering p67phox to inhibit NADPH oxidase assembly (111, 112). MtbΔPPE2 exhibits reduced virulence in mice (112). PE5, PE15, PPE63 and PE_PGRS62 similarly suppress iNOS/ROS in M. smegmatis and M. marinum (102, 113). PE13 enhance bacterial survival under oxidative stress, correlating with stress-induced expression (114).
7.2.5 Regulating cell death
7.2.5.1 Apoptosis
While macrophage apoptosis restricts Mtb growth and promotes antigen cross-presentation, the pathogen exploits apoptosis for dissemination via apoptotic bodies (115). PE/PPE proteins regulate apoptosis through ER stress, mitochondrial pathways, and TNF signaling. PE_PGRS5 induces caspase-8-dependent apoptosis via ER stress, whereas PE_PGRS62 suppresses it (116, 117). PE9-PE10 and PE_PGRS33 activate mitochondrial pathways (e.g., Bax upregulation, caspase-3) or TNFRI signaling (39, 118, 119). Conversely, PE_PGRS1 inhibits apoptosis by blocking PERK-mediated stress (120).
7.2.5.2 Necrosis
Necrosis facilitates bacterial dissemination and is exploited by Mycobacterium tuberculosis (Mtb) via PE-PPE proteins. Mtb Δ ppe51 strain induces necrosis via elevated ROS, exceeding levels in wild-type or complemented strains (90) PE_PGRS33 amplifies necrosis in M. smegmatis-infected macrophages by elevating TNF, reflected in increased LDH and nucleosome release (121). The PE25-PPE41 complex drives dose-dependent necrosis independently of TNF-α, NO, or NF-κB signaling (122). PPE68 and PPE27 promote necrosis, as Mtb Δppe68 reduces LDH release, while Msmeg-PPE27 elevates it (123, 124) However, LDH and Annexin V/PI staining alone cannot exclude concurrent apoptosis or pyroptosis.
7.2.5.3 Pyroptosis
PE/PPE proteins drive pyroptosis to amplify inflammation. PPE13 activates NLRP3 inflammasome-mediated pyroptosis, while PE_PGRS19 enhances caspase-11-dependent GSDMD cleavage (125, 126). PPE60 upregulates NLRP3 and GSDMD, suggesting a pro-pyroptotic role (127).
7.2.5.4 Autophagy
PE/PPE proteins modulate autophagy to balance bacterial persistence and host defense. PPE51 inhibits autophagy; its deletion enhances autophagic flux and reduces Mtb burden (90). PE_PGRS47 and PE_PGRS20 suppress autophagy by disrupting Rab1A-ULK1 interactions, impairing autophagosome formation (106, 128). PE6 inactivates mTORC1-ULK1 signaling, while PE_PGRS41 blocks LC3-II conversion (96, 129). Paradoxically, the surface-localized PE_PGRS29 promotes host autophagy by recruiting ubiquitin and engaging ubiquitin-binding receptors such as p62 and NBR1, potentially modulating intracellular bacterial burden to facilitate persistent infection (130).
7.2.6 Modulation of cytokine production
7.2.6.1 Cytokine dynamics in infection
Early Mtb infection triggers IL-1β production by macrophages and dendritic cells (DCs), essential for granuloma formation through macrophage migration to lymph nodes (131). While critical for initiating adaptive immunity, excessive IL-1β exacerbates tissue damage during chronic phases (132). TNF-α maintains granuloma integrity by promoting lymphocyte-macrophage clustering and enhancing NADPH oxidase-mediated ROS production (109, 133). IL-6 exhibits dual roles: early deficiency increases susceptibility, yet sustained expression impairs Th1 polarization and IFNγ production (110, 134, 135).
Th1 responses dominate chronic-phase protection, though Th17 contributions emerge through IL-17-mediated neutrophil recruitment and CXCL13-driven lymphoid organization (136, 137). While IL-12p40 from APCs drives Th1 polarization, IL-10 suppresses this process by blocking IL-12p40 production (138). Late-stage IL-10R1 blockade enhances bacterial control in mice, suggesting temporal regulation of IL-10’s immunosuppressive effects (139–141). Paradoxically, IL-23/Th17 axis amplifies IL-12p40 secretion but risks pathological inflammation through neutrophil infiltration (142, 143).
7.2.6.2 Proinflammatory cytokine induction
Mtb virulence factors orchestrate proinflammatory responses through distinct signaling mechanisms. PE13 enhances IL-6/IL-1β production in macrophages via p38/ERK/NF-κB activation while suppressing SOCS3 expression (114). PPE60 promotes Th1/Th17 polarization through DC-derived IL-12p70 and IL-23p19 (89, 127), whereas PE6 modulates TLR4 signaling to elevate TNF-α/IL-6/IL-12 levels (96, 144). Structural PE_PGRS proteins (Rv0978c, Rv0754) induce DC maturation and proinflammatory cytokine secretion, correlating with CD4+ T-cell activation (145). PE27 specifically activates MAPK/NF-κB pathways to drive TNF-α/IL-6/IL-1β production (146). These coordinated mechanisms suggest evolutionary optimization of Mtb’s capacity to manipulate host inflammatory cascades.
7.2.6.3 Anti-inflammatory modulation
Mtb counterbalances inflammation through sophisticated immunosuppressive strategies. PPE51 deletion elevates IL-6/IL-1β/ROS and impairs bacterial survival, revealing its anti-inflammatory function (90). PE_PGRS38 destabilizes TRAF6 via HAUSP-mediated interference with K48-polyUb deubiquitination, suppressing TNF-α/IL-6/IL-1β to enhance intracellular persistence (147). PPE10 inhibits NF-κB by downregulating LUBAC component HOIP (148), while PE_PGRS45/PE31/PPE36 shift cytokine profiles toward IL-10 dominance (149–151). PE11 exacerbates tissue damage through TNF-α/Th2 cytokine induction (24), contrasting with PPE65/PE32-PPE65’s dose-dependent IL-10 promotion and IL-6 suppression (152). This multi-layered regulation enables Mtb to establish chronic infection by modulating both pro- and anti-inflammatory axes.
7.3 Roles in drug resistance
Emerging evidence suggests that PE/PPE family proteins may contribute to drug resistance in Mtb. Early whole-genome analyses of drug-resistant clinical isolates identified mutations in pe/ppe genes in the absence of canonical resistance-conferring mutations. For example, certain kanamycin-resistant strains lacking alterations in rrs, rpsL or eis were found to harbor mutations in ppe60. Similarly, mutations in pe_pgrs9 or ppe54/55 were observed in pyrazinamide-resistant strains without known resistance mutations (153). Subsequent analysis of 161 drug-resistant Mtb isolates revealed that several pe/ppe genes—including pe_pgrs4, pe_pgrs9, ppe13, ppe20, and ppe9—exhibited significantly elevated mutation frequencies in resistant compared to susceptible strains (154). Enrichment of mutations was also reported in pe_pgrs genes (pe_pgrs3, 6, 9, 10, 19, 33, and 49) among 37 extensively drug-resistant (XDR) strains from Pakistan (155). In parallel, variants in ppe18, ppe19, ppe46, and ppe47 were found to be associated with the spread of isoniazid resistance (156).
Gene interaction studies further implicated PE/PPE proteins in resistance phenotypes. Resistance-associated gene pairs frequently included a known drug target and a pe/ppe gene, such as katG–ppe54 and rpoB–ppe54 (isoniazid and rifampicin resistance, respectively), and embA–ppe68 and embB–ppe54 (ethambutol resistance) (157). A large-scale analysis of 1,170 clinical isolates found that 36% of homoplastic SNPs—variants recurrent across independent lineages—resided in pe/ppe genes, and identified a novel mutation in pe_pgrs7 linked to streptomycin resistance (158).
While the mechanistic basis of these associations remains unclear, these findings suggest that pe/ppe mutations may modulate drug susceptibility. Functional studies support this hypothesis: heterologous expression of PE/PPE proteins, such as PPE63 (84), PE11 (24), and PE_PGRS41 (129), in M. smegmatis altered cell wall lipid profiles and surface hydrophobicity, impacting drug permeability. However, whether such changes directly contribute to resistance in Mtb warrants further investigation.
7.4 Considerations in studying the functions of PE/PPE proteins
Although numerous studies have reported diverse roles for PE/PPE proteins in mycobacterial physiology and host interactions, several limitations warrant caution in interpreting these findings. Many investigations rely on overexpression or heterologous expression of PE/PPE proteins in M. smegmatis, a species that lacks the ESX-5 secretion system required for the proper export of most native PE/PPE proteins (159). These strategies can produce nonspecific effects due to protein aggregation or cellular stress, potentially leading to artefactual phenotypes and misleading suggestions. Subcellular localization remains unresolved for many PE/PPE proteins, further complicating functional interpretation (160). Engineered M. smegmatis with functional ESX-5 may help bridge this gap (32).
In addition, many investigations remain descriptive, offering limited mechanistic insight and often failing to reconcile conflicting findings. Several PE/PPE proteins are critical for maintaining mycobacterial cell wall integrity, particularly those with intrinsic enzymatic activity—including lipases and glyco- or proteohydrolases—raising the possibility that phenotypes observed in gene deletion mutants reflect indirect alterations in the bacterial surface, rather than direct modulation of host immunity. Ex vivo models—including the treatment of host cells with purified proteins or the ectopic expression of bacterial genes in host systems—often ignore the native expression levels, spatial distribution, and secretion dynamics of PE/PPE proteins during infection. These limitations can obscure the physiological relevance of observed effects and overlook the essential role of bacterial secretion machinery in delivering these proteins.
Functional redundancy across PE/PPE family members also complicates interpretation of single-gene knockout studies, although such models remain essential for delineating context-specific functions within the broader protein network. Recent evidence emphasizes the biological relevance of co-regulated PE–PPE operons—such as PE35–PPE68 and PE25–PPE41—whose co-expression enhances solubility and amplifies immune responses relative to individual proteins (122, 161). This partnership flexibility may enable dynamic host adaptation through stage-specific interactions (11).
Future work should prioritize systems-level approaches to uncover coordinated functions, temporal regulation, and functional cooperativity among PE/PPE proteins. Accurate modeling of native expression and secretion will be critical to advancing our understanding of their multifaceted roles in Mycobacterium tuberculosis pathogenesis.
This review synthesizes current knowledge of PE/PPE functions based on published literature and adopts a functional classification scheme for clarity. However, this framework is necessarily artificial and may not fully capture the biological diversity or contextual complexity of these proteins.
8 Vaccine potential of the PE/PPE family proteins
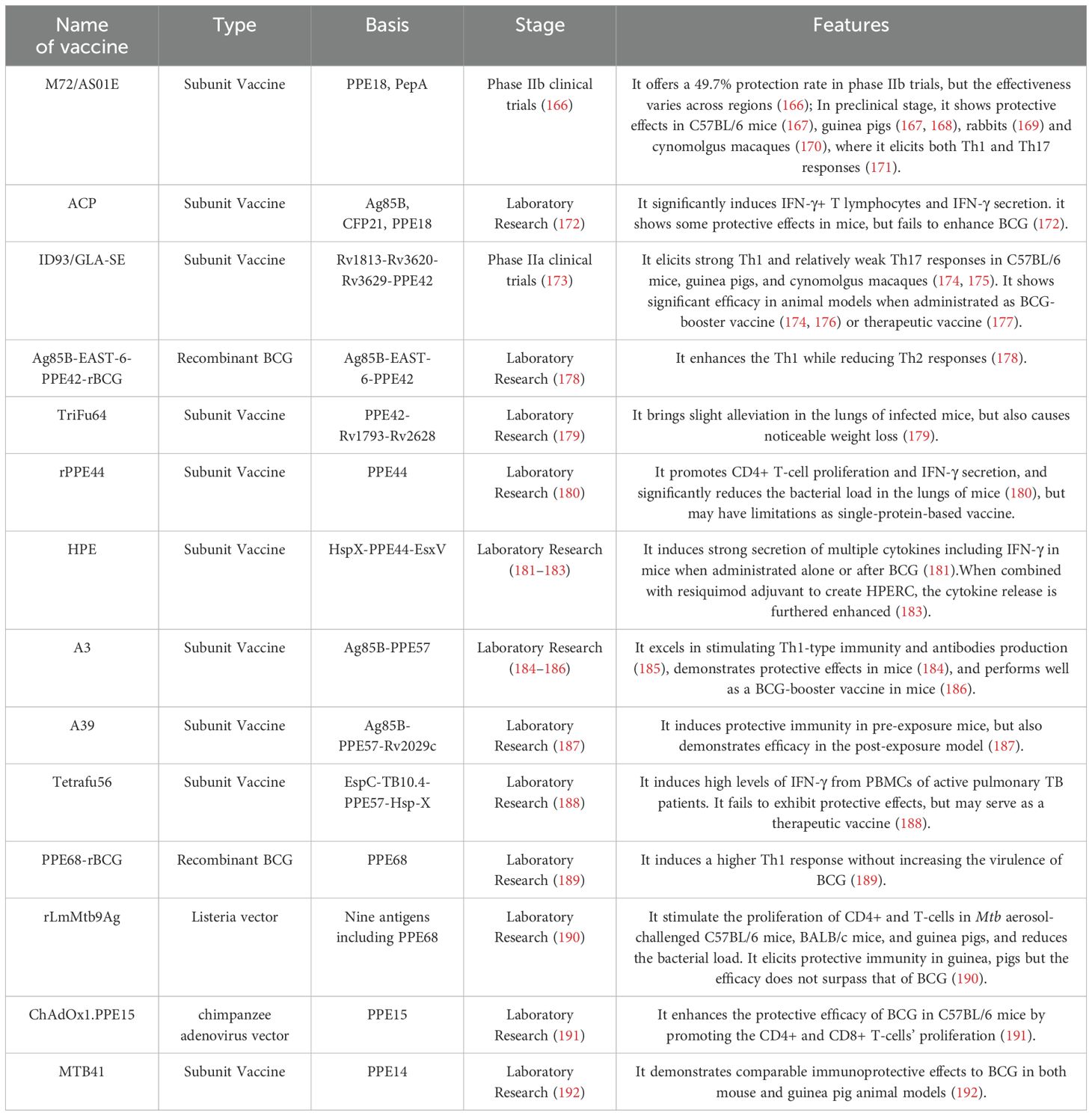
The failure of some individuals to control Mtb infection highlights the need for improved vaccines (3). PE/PPE-based subunit vaccines combine immunodominant T-cell epitopes from selected PE/PPE proteins with other antigens, delivered via adjuvants or vectors (162). These proteins are compelling vaccine targets due to their dense T-cell epitopes and capacity to induce cross-reactive immunity. PPE proteins, in particular, contain numerous confirmed and predicted MHC-binding epitopes that drive robust T-cell responses in humans and animal models, serving as key T-cell targets during Mtb infection (11, 159, 163). Furthermore, epitope redundancy among PE/PPE proteins enables broad CD4+ T-cell cross-reactivity, potentially sustaining immune responses despite shifting PE/PPE expression during infection (164, 165). Current progress in PE/PPE-based vaccine development—spanning protective and therapeutic candidates—is summarized below (Table 2).
8.1 PPE18
The M72/AS01E subunit vaccine (GlaxoSmithKline), containing a fusion protein (M72, derived from PPE18 and PepA) and AS01E adjuvant, represents a leading post-BCG tuberculosis vaccine candidate. A phase IIb trial involving 3,573 HIV-negative adults in South Africa, Zambia, and Kenya demonstrated 49.7% efficacy over three years in preventing latent-to-active TB progression, though efficacy varied regionally (166, 193). Currently in phase III trials, M72 faces challenges due to PPE18 variability across clinical strains, particularly in putative T-cell epitope regions (194, 195). Computational analyses, however, reveal conserved T-cell epitopes under positive selection, contrasting with hypervariable B-cell epitopes—a disparity that may compromise clinical recognition by M72-induced immunity (73, 196, 197).
Despite incomplete understanding of TB-protective immunity and inconsistent vaccine evaluation frameworks (198), M72 preclinical data offer critical insights. The vaccine conferred protection in murine, guinea pig, rabbit, and cynomolgus macaque models (167–170). In guinea pigs, M72 alone matched BCG’s one-year efficacy, while BCG coadministration extended survival beyond two years post-challenge (168). Macaque studies linked protection to elevated Th1 cytokines (IFN-γ, TNF-α, IL-2), suppressed Th2 signals (IL-4, IL-5), and elevated IFN-γ/IL-6 ratios (170). Phase I trials showed M72/AS01E drives durable M72-specific CD4+ T-cells co-expressing IFN-γ, IL-2, TNF-α, and/or IL-17 (171). Its efficacy may stem from balanced Th1/Th17 induction—critical for chronic infection control (199).
An alternative peptide vaccine (ACP: Ag85B12-26, CFP2112-26, PPE18149-163 epitopes) boosted IFN-γ+ T-cells and antibodies in mice but provided limited lung CFU reduction and failed to synergize with BCG (172), underscoring the need for biomarkers beyond IFN-γ.
8.2 PPE42
PPE42 (Rv2608) is a promising vaccine target. The ID93/GLA-SE subunit vaccine combines four Mtb antigens (PPE42, Rv3619, Rv3620, Rv1813) with the TLR4 agonist GLA-SE. Preclinically, it drives Th1 responses and modest IL-17 production in mice, guinea pigs, and macaques, while enhancing BCG’s long-term efficacy. In BCG-primed guinea pigs, ID93/GLA-SE prevented lung lesions for 432 days post-challenge, matching M72/AS02A (168, 174). ID93/GLA-SE-boosted/BCG-primed mice exhibited sustained Th1 CD4+ T-cell and IgG responses, reducing lung/spleen bacterial loads and inflammation after Mtb challenge (176).
Phase I trials in the U.S. and South Africa confirmed ID93/GLA-SE safety and robust Th1/IgG induction in BCG-naive and -vaccinated adults (200, 201). PPE42 dominated T-cell responses, while Rv1813 elicited stronger IgG (201). A Phase 2a trial highlighted its potential to improve TB treatment outcomes (173). As a chemotherapy adjunct, ID93/GLA-SE increased survival in mice and reduced bacterial loads/lesions in macaques (177).
The recombinant 85B-ESAT-6-PPE42-rBCG vaccine elevated Th1 cytokines and T-cell proliferation in mice but lacks in vivo protection data (178). TriFu64, a tri-antigen fusion (EsxN, PPE42, Rv2628), modestly lowered murine lung bacterial loads but induced weight loss, potentially linked to elevated TNFα/IL-17 ratios (179).
8.3 PPE44
PPE44 (Rv2770c) drives macrophage secretion of proinflammatory cytokines (IL-12 p40, IL-6) and Th1 responses, linked to its high expression across Mtb infection stages and utility as a cross-protective epitope source (88, 180). The gene is conserved clinically, with elevated expression in Beijing genotype strains versus H37Rv (202).
A recombinant PPE44 (rPPE44) subunit vaccine with DDA adjuvant matches BCG’s protection in mice, reducing lung bacterial loads via CD4+ T-cell proliferation and IFN-γ production (180). To overcome single-antigen limitations, multi-fusion strategies emerged. The HspX-PPE44-EsxV (HPE) trivalent vaccine, delivered as DNA, doubles IFN-γ levels versus BCG alone and elevates IL-12, IL-4, and TGF-β; BCG prime-boost amplifies cytokine responses (181). Liposomal HPE with DDA/TDB adjuvants enhances Th1 polarization and BCG immunological memory (182).
Further innovation yielded HPERC: HPE co-encapsulated with the TLR7/8 agonist resiquimod in size- and charge-optimized particles. HPERC improves APC uptake efficiency and Th1 activation in mice, marked by elevated IFN-γ. As a BCG booster, it augments IFN-γ, IL-17, and IgG2a—suggesting combined Th1/Th17 immunity (183).
8.4 PPE57
PPE57 displays strong immunogenicity but is encoded in the RD11 region, absent in BCG strains (203). The A3 vaccine—a fusion of Ag85B and PPE57—exemplifies multistage vaccine design. Delivered via plasmid DNA, protein, or lentiviral vectors, A3 induces Th1-polarized immunity in mice, activating CD4+/CD8+ T-cells, IFN-γ/TNF-α production, and antigen-specific antibodies (184, 185). In BCG prime-boost regimens, A3 reduces lung/spleen bacterial loads and lesion severity (186). Its successor, A39 (adding latency antigen Rv2029c), broadens protection to acute and latent TB, inhibiting bacterial reactivation and inflammation in pre- and post-exposure murine models (187).
The tetravalent Tetrafu56 vaccine (EspC, TB10.4, PPE57, HspX) targets replicative and dormant Mtb phases (188). While lacking prophylactic efficacy, it triggers robust IFN-γ in PBMCs from active TB patients—unlike healthy controls—suggesting therapeutic potential for Mtb-exposed individuals (162, 188). Notably, PPE57 exhibits structural variability across clinical strains, warranting further evaluation of population-level efficacy (73).
8.5 PPE68
The P9 peptide (aa 21–145) within PPE68’s RD1 region elicits IFN-γ production in PBMCs from TB patients and BCG-vaccinated individuals (73, 204). PPE68-expressing BCG (PPE68-rBCG) enhances Th1 responses without elevating virulence (189). The multivalent rLmMtb9Ag vaccine, which express nine immunoprotective Mtb antigens including PPE68 and is delivered via Listeria monocytogenes, activates CD4+/CD8+ T-cell proliferation and reduces lung/spleen bacterial loads in murine and guinea pig models. However, its protection against the Erdman strain remains inferior to BCG (190).
8.6 PPE15
PPE15, notable for conserved epitopes and roles in Mtb dormancy entry, is a vaccine candidate (73, 78, 97). Intranasal ChAdOx1.PPE15 boosts BCG efficacy in mice by amplifying CD4+/CD8+ T-cell responses and accelerating lung bacterial clearance (191).
8.7 PPE14
The rMTB41/AS02A subunit vaccine (PPE14-based) matches BCG’s protection in mice and guinea pigs. Immunized mice show Th1-polarized CD4+ and CD8+ T-cell responses with reduced lung CFUs; 60% of vaccinated guinea pigs survive 50 weeks post-challenge, contrasting with complete mortality in controls by 20 weeks (192).
8.8 Other PE/PPEs
Several additional PE/PPE proteins show immunomodulatory potential. The PPE39 homolog MTBK_24820 from Mtb Beijing/K drives Th1/IL-17 immunity in mice, matching BCG’s short-term protection but showing superior durability at 10 weeks post-infection (205). PE4 triggers IL-2/TNF/IL-6 production in mice, outperforming BCG in reducing lung CFUs and pathology by 45 days post-challenge (206). PPE26 promotes Th1 responses (elevated IFN-γ/IL-2) but lacks conservation and fails to protect mice despite inducing effector memory T-cells (207).
PE13, regulated by Rv0485 (which also controls PPE18), is conserved across lineages and elicits stronger T-cell responses in Mtb controllers versus progressors, suggesting protective potential (12, 208). PPE27, co-localized with PPE26 in the esx-5 locus, enhances M.smegmatis survival in murine tissues (124). PPE36-flagellin fusion induces Th1 immunity and splenocyte proliferation in mice (209). PE27 stimulates IFN-γ and memory T-cells in infected mice, though antigen specificity remains unconfirmed (146).
8.9 Challenges in targeting PE/PPE proteins for vaccination
Notably, the use of PE/PPE proteins as vaccine antigens also presents conceptual and practical challenges. As known immunomodulators, PE/PPE proteins can influence both innate and adaptive immune responses, raising the possibility that vaccines based on these proteins might dysregulate immunity rather than induce protective responses. Compounding this concern, pe/ppe genes in clinical Mtb isolates exhibit high genetic variability, potentially leading to antigenic divergence that may compromise vaccine efficacy across genetically diverse strain populations (65). Moreover, expression of these proteins is temporally and spatially dynamic, varying with infection stage and host environment—potentially limiting consistent antigen presentation during infection.
Future studies should address whether individual PE/PPE proteins in clinical isolates undergo antigenic variation, are differentially expressed, or are lost altogether. In parallel, detailed characterization of expression kinetics, antigen abundance, and cross-reactive immune responses is essential to evaluate their suitability as vaccine candidates.
8.10 Broader considerations in TB vaccine antigen selection
Clinical trial failures underscore the need to refine antigen selection criteria. Historically, immunodominant T-cell antigens—identified using PBMCs from active TB patients—guided target prioritization. This approach enabled Dillon et al. to isolate PPE18, leading to the M72/AS01E vaccine’s phase IIb success (166, 210). However, conserved immunodominant epitopes may benefit Mtb by diverting immune responses toward non-protective “decoys” (21, 211). Supporting this, weakly immunogenic antigens can confer stronger protection than dominant ones when adjuvanted (212), and most TB patient T-cell responses lack disease-modulating effects (12). Thus, immunodominance alone is an insufficient selection metric.
Further challenges arise from TB’s pulmonary tropism. Lung-resident memory T-cells (Trm), critical for local pathogen control, are underrepresented in peripheral blood analyses (213). Preclinical models must therefore incorporate lung-specific immunity assessments. Consensus favors tiered efficacy evaluation across mice, guinea pigs/rabbits, and nonhuman primates to improve translatability (198). Additionally, while prophylactic vaccines dominate research, therapeutic candidates—particularly for multidrug-resistant TB—demand exploration to shorten treatments and reduce relapse (214).
9 Diagnostic potential of PE/PPE family proteins
PE/PPE proteins show promise as serological or cellular biomarkers for tuberculosis. Several members stimulate Mtb-specific antibodies or cytokine profiles in PBMCs, distinguishing latent and active infections. PPE2 (Rv0256c), overexpressed during macrophage infection, outperforms PPD and ESAT-6 in detecting active TB across clinical subtypes (215). PPE17 demonstrates superior sensitivity for smear-negative TB and better distinguishes active TB from BCG-vaccinated individuals than PPE2 (216, 217). PPE42’s C-terminal MPTR sequence improves sensitivity in sputum/culture-negative cases, particularly when fused with other antigens (218). PE11, a virulence factor, elicits stage-specific antibodies; its co-transcription with PPE17 suggests complex formation, though diagnostic utility remains unexplored (219, 220).
PE35 and PPE68 trigger IFN-γ/IL-2 responses that differentiate active (IFN-γ-dominant) and latent (IL-2/dual-positive) TB via effector vs. memory T-cell profiles (11, 221). PPE41, particularly when complexed with PE25, boosts diagnostic sensitivity to 75% via enhanced B-cell responses (161). PE32-PPE65 induces Th2 responses that may compromise protection but could serve as diagnostic markers (152). PPE36-specific IgA (absent IgG) may offer TB-specific serological signatures (222). Recombinant PPE57 elicits IgG levels surpassing ESAT-6 and matching CFP-10, distinguishing TB patients from BCG-vaccinated controls (223).
10 Concluding remarks
The Mtb PE/PPE protein family, occupying ~10% of its genome, evolved from ancestral PE35-PPE68 pairs into diverse subfamilies through co-evolution with ESX secretion systems (7). These proteins form heterodimers or EspG-bound trimers for transport to the cell surface or secretion, with ESX-5 specifically exporting virulence-linked PE_PGRS and PPE_MPTR members (11, 61, 224). PE/PPE proteins mediate diverse roles in bacterial physiology and host-pathogen interactions, yet their evolutionary drivers—particularly the selective pressures driving their expansion and epitope conservation—remain enigmatic.
Structural and functional insights remain limited. While solved PE/PPE-EspG complexes reveal organizational principles (37, 225), most structures and proposed porin-like roles lack direct evidence. Similarly, their hypothesized role as ESX substrate channels awaits validation. Technical hurdles—including heterologous expression artifacts and functional redundancy—necessitate innovative approaches to study collective PE/PPE actions during infection.
Despite challenges, PE/PPE proteins offer untapped potential for TB interventions. Their conserved, cross-reactive T-cell epitopes underpin vaccine candidates like M72/AS01E, while clinical trial failures underscore the need to refine antigen selection beyond immunodominance. Therapeutic vaccines, critical for curbing drug-resistant TB, warrant urgent exploration.
Future research must bridge evolutionary, structural, and immunological gaps to unravel PE/PPE contributions to Mtb’s success and leverage these insights for next-generation diagnostics and vaccines.
Author contributions
QW: Funding acquisition, Writing – review & editing, Supervision, Resources, Formal analysis, Project administration, Conceptualization. ZZ: Writing – original draft, Data curation, Methodology, Investigation, Validation. LD: Methodology, Investigation, Writing – original draft, Data curation, Validation. XL: Writing – review & editing, Data curation, Formal analysis. TD: Writing – review & editing, Data curation, Formal analysis.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by grants from the Natural Science Foundation of Sichuan Province (Grant No. 2025ZNSFSC0675).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Schito M, Migliori GB, Fletcher HA, McNerney R, Centis R, D'Ambrosio L, et al. Perspectives on advances in tuberculosis diagnostics, drugs, and vaccines. Clin Infect Dis. (2015) 61Suppl 3:S102–18. doi: 10.1093/cid/civ609
3. Schrager LK, Vekemens J, Drager N, Lewinsohn DM, and Olesen OF. The status of tuberculosis vaccine development. Lancet Infect Dis. (2020) 20:e28–37. doi: 10.1016/S1473-3099(19)30625-5
4. Ahmed A, Rakshit S, Adiga V, Dias M, Dwarkanath P, D'Souza G, et al. A century of bcg: impact on tuberculosis control and beyond. Immunol Rev. (2021) 301:98–121. doi: 10.1111/imr.12968
5. Dockrell HM and Smith SG. What have we learnt about bcg vaccination in the last 20 years? Front Immunol. (2017) 8:1134. doi: 10.3389/fimmu.2017.01134
6. Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. Deciphering the biology of mycobacterium tuberculosis from the complete genome sequence. Nature. (1998) 393:537–44. doi: 10.1038/31159
7. Fishbein S, van Wyk N, Warren RM, and Sampson SL. Phylogeny to function: pe/ppe protein evolution and impact on mycobacterium tuberculosis pathogenicity. Mol Microbiol. (2015) 96:901–16. doi: 10.1111/mmi.12981
8. Gey van Pittius NC, Sampson SL, Lee H, Kim Y, van Helden PD, and Warren RM. Evolution and expansion of the mycobacterium tuberculosis pe and ppe multigene families and their association with the duplication of the esat-6 (Esx) gene cluster regions. BMC Evol Biol. (2006) 6:95. doi: 10.1186/1471-2148-6-95
9. Abdallah AM, Gey van Pittius NC, Champion PA, Cox J, Luirink J, Vandenbroucke-Grauls CM, et al. Type vii secretion–mycobacteria show the way. Nat Rev Microbiol. (2007) 5:883–91. doi: 10.1038/nrmicro1773
10. Wang Q, Boshoff HIM, Harrison JR, Ray PC, Green SR, Wyatt PG, et al. Pe/ppe proteins mediate nutrient transport across the outer membrane of mycobacterium tuberculosis. Science. (2020) 367:1147–51. doi: 10.1126/science.aav5912
11. D'Souza C, Kishore U, and Tsolaki AG. The pe-ppe family of mycobacterium tuberculosis: proteins in disguise. Immunobiology. (2023) 228:152321. doi: 10.1016/j.imbio.2022.152321
12. Musvosvi M, Huang H, Wang C, Xia Q, Rozot V, Krishnan A, et al. T cell receptor repertoires associated with control and disease progression following mycobacterium tuberculosis infection. Nat Med. (2023) 29:258–69. doi: 10.1038/s41591-022-02110-9
13. Ahmad J, Farhana A, Pancsa R, Arora SK, Srinivasan A, Tyagi AK, et al. Contrasting function of structured N-terminal and unstructured C-terminal segments of mycobacterium tuberculosis ppe37 protein. mBio. (2018) 9:e01712–17. doi: 10.1128/mBio.01712-17
14. Gong W, Liang Y, and Wu X. Animal models of tuberculosis vaccine research: an important component in the fight against tuberculosis. BioMed Res Int. (2020) 2020:4263079. doi: 10.1155/2020/4263079
15. Lai R, Ogunsola AF, Rakib T, and Behar SM. Key advances in vaccine development for tuberculosis-success and challenges. NPJ Vaccines. (2023) 8:158. doi: 10.1038/s41541-023-00750-7
16. Tundup S, Akhter Y, Thiagarajan D, and Hasnain SE. Clusters of pe and ppe genes of mycobacterium tuberculosis are organized in operons: evidence that pe rv2431c is co-transcribed with ppe rv2430c and their gene products interact with each other. FEBS Lett. (2006) 580:1285–93. doi: 10.1016/j.febslet.2006.01.042
17. Xie Y, Zhou Y, Liu S, and Zhang XL. Pe_Pgrs: vital proteins in promoting mycobacterial survival and modulating host immunity and metabolism. Cell Microbiol. (2021) 23:e13290. doi: 10.1111/cmi.13290
18. Ramakrishnan P, Aagesen AM, McKinney JD, and Tischler AD. Mycobacterium tuberculosis resists stress by regulating pe19 expression. Infect Immun. (2015) 84:735–46. doi: 10.1128/IAI.00942-15
19. Voskuil MI, Schnappinger D, Rutherford R, Liu Y, and Schoolnik GK. Regulation of the mycobacterium tuberculosis pe/ppe genes. Tuberculosis (Edinb). (2004) 84:256–62. doi: 10.1016/j.tube.2003.12.014
20. McEvoy CR, Cloete R, Muller B, Schurch AC, van Helden PD, Gagneux S, et al. Comparative analysis of mycobacterium tuberculosis pe and ppe genes reveals high sequence variation and an apparent absence of selective constraints. PloS One. (2012) 7:e30593. doi: 10.1371/journal.pone.0030593
21. Copin R, Coscolla M, Seiffert SN, Bothamley G, Sutherland J, Mbayo G, et al. Sequence diversity in the pe_Pgrs genes of mycobacterium tuberculosis is independent of human T cell recognition. mBio. (2014) 5:e00960–13. doi: 10.1128/mBio.00960-13
22. Kumar A, Kamuju V, and Vivekanandan P. Rna G-quadruplexes inhibit translation of the pe/ppe transcripts in mycobacterium tuberculosis. J Biol Chem. (2024) 300:105567. doi: 10.1016/j.jbc.2023.105567
23. Bhat KH, Das A, Srikantam A, and Mukhopadhyay S. Ppe2 protein of mycobacterium tuberculosis may inhibit nitric oxide in activated macrophages. Ann N Y Acad Sci. (2013) 1283:97–101. doi: 10.1111/nyas.12070
24. Singh P, Rao RN, Reddy JR, Prasad RB, Kotturu SK, Ghosh S, et al. Pe11, a pe/ppe family protein of mycobacterium tuberculosis is involved in cell wall remodeling and virulence. Sci Rep. (2016) 6:21624. doi: 10.1038/srep21624
25. Brennan MJ and Delogu G. The pe multigene family: A 'Molecular mantra' for mycobacteria. Trends Microbiol. (2002) 10:246–9. doi: 10.1016/s0966-842x(02)02335-1
26. Hermans PW, van Soolingen D, and van Embden JD. Characterization of a major polymorphic tandem repeat in mycobacterium tuberculosis and its potential use in the epidemiology of mycobacterium kansasii and mycobacterium gordonae. J Bacteriol. (1992) 174:4157–65. doi: 10.1128/jb.174.12.4157-4165.1992
27. McGuire AM, Weiner B, Park ST, Wapinski I, Raman S, Dolganov G, et al. Comparative analysis of mycobacterium and related actinomycetes yields insight into the evolution of mycobacterium tuberculosis pathogenesis. BMC Genomics. (2012) 13:120. doi: 10.1186/1471-2164-13-120
28. Stinear TP, Seemann T, Harrison PF, Jenkin GA, Davies JK, Johnson PD, et al. Insights from the complete genome sequence of mycobacterium marinum on the evolution of mycobacterium tuberculosis. Genome Res. (2008) 18:729–41. doi: 10.1101/gr.075069.107
29. Gomez-Valero L, Rocha EP, Latorre A, and Silva FJ. Reconstructing the ancestor of mycobacterium leprae: the dynamics of gene loss and genome reduction. Genome Res. (2007) 17:1178–85. doi: 10.1101/gr.6360207
30. Cole ST, Eiglmeier K, Parkhill J, James KD, Thomson NR, Wheeler PR, et al. Massive gene decay in the leprosy bacillus. Nature. (2001) 409:1007–11. doi: 10.1038/35059006
31. Williamson ZA, Chaton CT, Ciocca WA, Korotkova N, and Korotkov KV. Pe5-ppe4-espg(3) heterotrimer structure from mycobacterial esx-3 secretion system gives insight into cognate substrate recognition by esx systems. J Biol Chem. (2020) 295:12706–15. doi: 10.1074/jbc.RA120.012698
32. Bunduc CM, Ding Y, Kuijl C, Marlovits TC, Bitter W, and Houben ENG. Reconstitution of a minimal esx-5 type vii secretion system suggests a role for ppe proteins in the outer membrane transport of proteins. mSphere. (2023) 8:e0040223. doi: 10.1128/msphere.00402-23
33. Karboul A, Mazza A, Gey van Pittius NC, Ho JL, Brousseau R, and Mardassi H. Frequent homologous recombination events in mycobacterium tuberculosis pe/ppe multigene families: potential role in antigenic variability. J Bacteriol. (2008) 190:7838–46. doi: 10.1128/JB.00827-08
34. Berisio R and Delogu G. Pgrs domain structures: doomed to sail the mycomembrane. PloS Pathog. (2022) 18:e1010760. doi: 10.1371/journal.ppat.1010760
35. Poulsen C, Panjikar S, Holton SJ, Wilmanns M, and Song YH. Wxg100 protein superfamily consists of three subfamilies and exhibits an alpha-helical C-terminal conserved residue pattern. PloS One. (2014) 9:e89313. doi: 10.1371/journal.pone.0089313
36. Strong M, Sawaya MR, Wang S, Phillips M, Cascio D, and Eisenberg D. Toward the structural genomics of complexes: crystal structure of a pe/ppe protein complex from mycobacterium tuberculosis. Proc Natl Acad Sci U.S.A. (2006) 103:8060–5. doi: 10.1073/pnas.0602606103
37. Daleke MH, Ummels R, Bawono P, Heringa J, Vandenbroucke-Grauls CM, Luirink J, et al. General secretion signal for the mycobacterial type vii secretion pathway. Proc Natl Acad Sci U.S.A. (2012) 109:11342–7. doi: 10.1073/pnas.1119453109
38. Ehtram A, Shariq M, Quadir N, Jamal S, Pichipalli M, Zarin S, et al. Deciphering the functional roles of pe18 and ppe26 proteins in modulating mycobacterium tuberculosis pathogenesis and immune response. Front Immunol. (2025) 16:1517822. doi: 10.3389/fimmu.2025.1517822
39. Tiwari B, Ramakrishnan UM, and Raghunand TR. The mycobacterium tuberculosis protein pair pe9 (Rv1088)-pe10 (Rv1089) forms heterodimers and induces macrophage apoptosis through toll-like receptor 4. Cell Microbiol. (2015) 17:1653–69. doi: 10.1111/cmi.12462
40. Ates LS, Houben ENG, and Bitter W. Type vii secretion: A highly versatile secretion system. Microbiol Spectr. (2016) 4:eVMBF-0011-2015. doi: 10.1128/microbiolspec.VMBF-0011-2015
41. Solomonson M, Setiaputra D, Makepeace KAT, Lameignere E, Petrotchenko EV, Conrady DG, et al. Structure of espb from the esx-1 type vii secretion system and insights into its export mechanism. Structure. (2015) 23:571–83. doi: 10.1016/j.str.2015.01.002
42. Ekiert DC and Cox JS. Structure of a pe-ppe-espg complex from mycobacterium tuberculosis reveals molecular specificity of esx protein secretion. Proc Natl Acad Sci U.S.A. (2014) 111:14758–63. doi: 10.1073/pnas.1409345111
43. Chen X, Cheng HF, Zhou J, Chan CY, Lau KF, Tsui SK, et al. Structural basis of the pe-ppe protein interaction in mycobacterium tuberculosis. J Biol Chem. (2017) 292:16880–90. doi: 10.1074/jbc.M117.802645
44. Cascioferro A, Delogu G, Colone M, Sali M, Stringaro A, Arancia G, et al. Pe is a functional domain responsible for protein translocation and localization on mycobacterial cell wall. Mol Microbiol. (2007) 66:1536–47. doi: 10.1111/j.1365-2958.2007.06023.x
45. Cascioferro A, Daleke MH, Ventura M, Donà V, Delogu G, Palù G, et al. Functional dissection of the pe domain responsible for translocation of pe_Pgrs33 across the mycobacterial cell wall. PloS One. (2011) 6:e27713. doi: 10.1371/journal.pone.0027713
46. Zumbo A, Palucci I, Cascioferro A, Sali M, Ventura M, D'Alfonso P, et al. Functional dissection of protein domains involved in the immunomodulatory properties of pe_Pgrs33 of mycobacterium tuberculosis. Pathog Dis. (2013) 69:232–9. doi: 10.1111/2049-632x.12096
47. Iantomasi R, Sali M, Cascioferro A, Palucci I, Zumbo A, Soldini S, et al. Pe_Pgrs30 is required for the full virulence of mycobacterium tuberculosis. Cell Microbiol. (2011) 14:356–67. doi: 10.1111/j.1462-5822.2011.01721.x
48. Daleke MH, Cascioferro A, de Punder K, Ummels R, Abdallah AM, van der Wel N, et al. Conserved pro-glu (Pe) and pro-pro-glu (Ppe) protein domains target lipy lipases of pathogenic mycobacteria to the cell surface via the esx-5 pathway. J Biol Chem. (2011) 286:19024–34. doi: 10.1074/jbc.m110.204966
49. Babu Sait MR, Koliwer-Brandl H, Stewart JA, Swarts BM, Jacobsen M, Ioerger TR, et al. Ppe51 mediates uptake of trehalose across the mycomembrane of mycobacterium tuberculosis. Sci Rep. (2022) 12:2097. doi: 10.1038/s41598-022-06109-7
50. Korycka-Machała M, Pawełczyk J, Borówka P, Dziadek B, Brzostek A, Kawka M, et al. Ppe51 is involved in the uptake of disaccharides by mycobacterium tuberculosis. Cells. (2020) 9:603. doi: 10.3390/cells9030603
51. Boradia V, Frando A, and Grundner C. The mycobacterium tuberculosis pe15/ppe20 complex transports calcium across the outer membrane. PloS Biol. (2022) 20:e3001906. doi: 10.1371/journal.pbio.3001906
52. Lepe BA, Zheng CR, Leddy OK, Allsup BL, Solomon SL, and Bryson BD. Protease shaving of mycobacterium tuberculosis facilitates vaccine antigen discovery and delivery of novel cargoes to the mtb surface. Microbiol Spectr. (2025) 13:e0227724. doi: 10.1128/spectrum.02277-24
53. Ekiert DC and Cox JS. Structure of a pe–ppe–espg complex from mycobacterium tuberculosis reveals molecular specificity of esx protein secretion. Proc Natl Acad Sci. (2014) 111:14758–63. doi: 10.1073/pnas.1409345111
54. Korotkova N, Creekmore CC, and Korotkov KV. Structure of the mycobacterium tuberculosis type vii secretion system chaperone espg5 in complex with pe25-ppe41 dimer. Worldwide Protein Data Bank. (2014) 94:367–82. doi: 10.2210/pdb4kxr/pdb
55. Granados-Tristán AL, Hernández-Luna CE, González-Escalante LA, Camacho-Moll ME, Silva-Ramírez B, Bermúdez de León M, et al. Esx-3 secretion system in mycobacterium: an overview. Biochimie. (2024) 216:46–55. doi: 10.1016/j.biochi.2023.10.013
56. Bunduc CM, Fahrenkamp D, Wald J, Ummels R, Bitter W, Houben ENG, et al. Structure and dynamics of a mycobacterial type vii secretion system. Nature. (2021) 593:445–8. doi: 10.1038/s41586-021-03517-z
57. Poweleit N, Czudnochowski N, Nakagawa R, Trinidad DD, Murphy KC, Sassetti CM, et al. The structure of the endogenous esx-3 secretion system. Elife. (2019) 8:e52983. doi: 10.7554/eLife.52983
58. Famelis N, Rivera-Calzada A, Degliesposti G, Wingender M, Mietrach N, Skehel JM, et al. Architecture of the mycobacterial type vii secretion system. Nature. (2019) 576:321–5. doi: 10.1038/s41586-019-1633-1
59. Rosenberg Oren S, Dovala D, Li X, Connolly L, Bendebury A, Finer-Moore J, et al. Substrates control multimerization and activation of the multi-domain atpase motor of type vii secretion. Cell. (2015) 161:501–12. doi: 10.1016/j.cell.2015.03.040
60. Bunduc CM, Ummels R, Bitter W, and Houben ENG. Species-specific secretion of esx-5 type vii substrates is determined by the linker 2 of eccc(5). Mol Microbiol. (2020) 114:66–76. doi: 10.1111/mmi.14496
61. Ates LS, Ummels R, Commandeur S, van der Weerd R, Sparrius M, Weerdenburg E, et al. Essential role of the esx-5 secretion system in outer membrane permeability of pathogenic mycobacteria. PloS Genet. (2015) 11:e1005190. doi: 10.1371/journal.pgen.1005190
62. de Souza GA, Leversen NA, Målen H, and Wiker HG. Bacterial proteins with cleaved or uncleaved signal peptides of the general secretory pathway. J Proteomics. (2011) 75:502–10. doi: 10.1016/j.jprot.2011.08.016
63. Cronin RM, Ferrell MJ, Cahir CW, Champion MM, and Champion PA. Proteo-genetic analysis reveals clear hierarchy of esx-1 secretion in mycobacterium marinum. Proc Natl Acad Sci. (2022) 119:e2123100119. doi: 10.1073/pnas.2123100119
64. Damen MPM, Meijers AS, Keizer EM, Piersma SR, Jiménez CR, Kuijl CP, et al. The esx-1 substrate ppe68 has a key function in esx-1-mediated secretion in mycobacterium marinum. mBio. (2022) 13:e02819–22. doi: 10.1128/mbio.02819-22
65. Ates LS, Dippenaar A, Ummels R, Piersma SR, van der Woude AD, van der Kuij K, et al. Mutations in ppe38 block pe_Pgrs secretion and increase virulence of mycobacterium tuberculosis. Nat Microbiol. (2018) 3:181–8. doi: 10.1038/s41564-017-0090-6
66. Bunduc CM, Bitter W, and Houben ENG. Structure and function of the mycobacterial type vii secretion systems. Annu Rev Microbiol. (2020) 74:315–35. doi: 10.1146/annurev-micro-012420-081657
67. Tufariello JM, Chapman JR, Kerantzas CA, Wong K-W, Vilchèze C, Jones CM, et al. Separable roles for mycobacterium tuberculosis esx-3 effectors in iron acquisition and virulence. Proc Natl Acad Sci. (2016) 113:E348–57. doi: 10.1073/pnas.1523321113
68. Maciag A, Dainese E, Rodriguez GM, Milano A, Provvedi R, Pasca MR, et al. Global analysis of the mycobacterium tuberculosis zur (Furb) regulon. J Bacteriology. (2007) 189:730–40. doi: 10.1128/jb.01190-06
69. Pandey R, Russo R, Ghanny S, Huang X, Helmann J, and Rodriguez GM. Mntr (Rv2788): A transcriptional regulator that controls manganese homeostasis in mycobacterium tuberculosis. Mol Microbiol. (2015) 98:1168–83. doi: 10.1111/mmi.13207
70. Mitra A, Speer A, Lin K, Ehrt S, and Niederweis M. Ppe surface proteins are required for heme utilization by mycobacterium tuberculosis. mBio. (2017) 8:e01720–16. doi: 10.1128/mbio.01720-16
71. Mitra A, Ko Y-H, Cingolani G, and Niederweis M. Heme and hemoglobin utilization by mycobacterium tuberculosis. Nat Commun. (2019) 10:4260. doi: 10.1038/s41467-019-12109-5
72. Tullius MV, Nava S, and Horwitz MA. Ppe37 is essential for mycobacterium tuberculosis heme-iron acquisition (Hia), and a defective ppe37 in mycobacterium bovis bcg prevents hia. Infection Immun. (2019) 87:e00540–18. doi: 10.1128/iai.00540-18
73. Gómez-González PJ, Grabowska AD, Tientcheu LD, Tsolaki AG, Hibberd ML, Campino S, et al. Functional genetic variation in pe/ppe genes contributes to diversity in mycobacterium tuberculosis lineages and potential interactions with the human host. Front Microbiol. (2023) 14:1244319. doi: 10.3389/fmicb.2023.1244319
74. Sankey N, Merrick H, Singh P, Rogers J, Reddi A, Hartson SD, et al. Role of the mycobacterium tuberculosis esx-4 secretion system in heme iron utilization and pore formation by ppe proteins. mSphere. (2023) 8:e00573–22. doi: 10.1128/msphere.00573-22
75. De Maio F, Salustri A, Battah B, Palucci I, Marchionni F, Bellesi S, et al. Pe_Pgrs3 ensures provision of the vital phospholipids cardiolipin and phosphatidylinositols by promoting the interaction between M. Tuberculosis Host Cells Virulence. (2021) 12:868–84. doi: 10.1080/21505594.2021.1897247
76. Lovewell RR, Sassetti CM, and VanderVen BC. Chewing the fat: lipid metabolism and homeostasis during M. Tuberculosis infection. Curr Opin Microbiol. (2016) 29:30–6. doi: 10.1016/j.mib.2015.10.002
77. Murphy DJ and Brown JR. Identification of gene targets against dormant phase mycobacterium tuberculosis infections. BMC Infect Dis. (2007) 7:84. doi: 10.1186/1471-2334-7-84
78. Daniel J, Kapoor N, Sirakova T, Sinha R, and Kolattukudy P. The perilipin-like ppe15 protein in mycobacterium tuberculosis is required for triacylglycerol accumulation under dormancy-inducing conditions. Mol Microbiol. (2016) 101:784–94. doi: 10.1111/mmi.13422
79. Deb C, Daniel J, Sirakova TD, Abomoelak B, Dubey VS, and Kolattukudy PE. A novel lipase belonging to the hormone-sensitive lipase family induced under starvation to utilize stored triacylglycerol in mycobacterium tuberculosis. J Biol Chem. (2006) 281:3866–75. doi: 10.1074/jbc.m505556200
80. Kruh NA, Troudt J, Izzo A, Prenni J, and Dobos KM. Portrait of a pathogen: the mycobacterium tuberculosis proteome in vivo. PloS One. (2010) 5:e13938. doi: 10.1371/journal.pone.0013938
81. Rastogi S, Singh AK, Pant G, Mitra K, Sashidhara KV, and Krishnan MY. Down-regulation of pe11, a cell wall associated esterase, enhances the biofilm growth of mycobacterium tuberculosis and reduces cell wall virulence lipid levels. Microbiology. (2017) 163:52–61. doi: 10.1099/mic.0.000417
82. Sultana R, Vemula MH, Banerjee S, and Guruprasad L. The pe16 (Rv1430) of mycobacterium tuberculosis is an esterase belonging to serine hydrolase superfamily of proteins. PloS One. (2013) 8:e55320. doi: 10.1371/journal.pone.0055320
83. Divya MB, Vemula M, Balakrishnan K, Banerjee S, and Guruprasad L. Mycobacterium tuberculosis pe1 and pe2 proteins carrying conserved A/B-serine hydrolase domain are esterases hydrolyzing short to medium chain P-nitrophenyl esters. Prog Biophysics Mol Biol. (2018) 140:90–102. doi: 10.1016/j.pbiomolbio.2018.04.012
84. Anand PK and Kaur J. Rv3539 (Ppe63) of mycobacterium tuberculosis promotes survival of mycobacterium smegmatis in human macrophages cell line via cell wall modulation of bacteria and altering host’s immune response. Curr Microbiol. (2023) 80:267. doi: 10.1007/s00284-023-03360-7
85. Palucci I, Camassa S, Cascioferro A, Sali M, Anoosheh S, Zumbo A, et al. Pe_Pgrs33 contributes to mycobacterium tuberculosis entry in macrophages through interaction with tlr2. PloS One. (2016) 11:e0150800. doi: 10.1371/journal.pone.0150800
86. Brennan MJ, Delogu G, Chen Y, Bardarov S, Kriakov J, Alavi M, et al. Evidence that mycobacterial pe_Pgrs proteins are cell surface constituents that influence interactions with other cells. Infection Immun. (2001) 69:7326–33. doi: 10.1128/iai.69.12.7326-7333.2001
87. Su H, Kong C, Zhu L, Huang Q, Luo L, Wang H, et al. Ppe26 induces tlr2-dependent activation of macrophages and drives th1-type T-cell immunity by triggering the cross-talk of multiple pathways involved in the host response. Oncotarget. (2015) 6:38517–37. doi: 10.18632/oncotarget.5956
88. Yu Z, Zhang C, Zhou M, Li Q, Li H, Duan W, et al. Mycobacterium tuberculosis ppe44 (Rv2770c) is involved in response to multiple stresses and promotes the macrophage expression of il-12 P40 and il-6 via the P38, erk, and nf-Kb signaling axis. Int Immunopharmacol. (2017) 50:319–29. doi: 10.1016/j.intimp.2017.06.028
89. Su H, Zhang Z, Liu Z, Peng B, Kong C, Wang H, et al. Mycobacterium tuberculosis ppe60 antigen drives th1/th17 responses via toll-like receptor 2–dependent maturation of dendritic cells. J Biol Chem. (2018) 293:10287–302. doi: 10.1074/jbc.ra118.001696
90. Strong EJ, Wang J, Ng TW, Porcelli SA, and Lee S. Mycobacterium tuberculosis ppe51 inhibits autophagy by suppressing toll-like receptor 2-dependent signaling. mBio. (2022) 13:e02974–21. doi: 10.1128/mbio.02974-21
91. Nair S, Ramaswamy PA, Ghosh S, Joshi DC, Pathak N, Siddiqui I, et al. The ppe18 of mycobacterium tuberculosis interacts with tlr2 and activates il-10 induction in macrophage. J Immunol. (2009) 183:6269–81. doi: 10.4049/jimmunol.0901367
92. Bhat KH, Ahmed A, Kumar S, Sharma P, and Mukhopadhyay S. Role of ppe18 protein in intracellular survival and pathogenicity of mycobacterium tuberculosis in mice. PloS One. (2012) 7:e52601. doi: 10.1371/journal.pone.0052601
93. Tiwari B, Soory A, and Raghunand TR. An immunomodulatory role for the mycobacterium tuberculosis region of difference 1 locus proteins pe35 (Rv3872) and ppe68 (Rv3873). FEBS J. (2014) 281:1556–70. doi: 10.1111/febs.12723
94. Peng Z, Yue Y, and Xiong S. Mycobacterial ppe36 modulates host inflammation by promoting E3 ligase smurf1-mediated myd88 degradation. Front Immunol. (2022) 13:690667. doi: 10.3389/fimmu.2022.690667
95. Liu S, Xie Y, Luo W, Dou Y, Xiong H, Xiao Z, et al. Pe_Pgrs31-S100a9 interaction promotes mycobacterial survival in macrophages through the regulation of nf-Kb-tnf-A Signaling and arachidonic acid metabolism. Front Microbiol. (2020) 11:845. doi: 10.3389/fmicb.2020.00845
96. Sharma N, Shariq M, Quadir N, Singh J, Sheikh JA, Hasnain SE, et al. Mycobacterium tuberculosis protein Pe6 (Rv0335c), a Novel Tlr4 Agonist, Evokes an Inflammatory Response and Modulates the Cell Death Pathways in Macrophages to Enhance Intracellular Survival. Front Immunol. (2021) 12:696491. doi: 10.3389/fimmu.2021.696491
97. Priyanka, Sharma S, and Varma-Basil M. Sharma M. C-terminal region of rv1039c (Ppe15) protein of mycobacterium tuberculosis targets host mitochondria to induce macrophage apoptosis. Apoptosis. (2024) 29:1757–79. doi: 10.1007/s10495-024-01965-2
98. Choi H-H, Kwon KW, Han SJ, Kang SM, Choi E, Kim A, et al. Ppe39 of the mycobacterium tuberculosis strain beijing/K induces th1-cell polarization through dendritic cell maturation. J Cell Sci. (2019) 132:jcs228700. doi: 10.1242/jcs.228700
99. Krishnan V, Nath S, Nair P, and Das B. Mycobacterium tuberculosis and its clever approaches to escape the deadly macrophage. World J Microbiol Biotechnol. (2023) 39:300. doi: 10.1007/s11274-023-03735-9
100. Chatrath S, Gupta VK, Dixit A, and Garg LC. Pe_Pgrs30 of mycobacterium tuberculosis mediates suppression of proinflammatory immune response in macrophages through its pgrs and pe domains. Microbes Infection. (2016) 18:536–42. doi: 10.1016/j.micinf.2016.04.004
101. Sharma T, Grover S, Arora N, P M, Ehtesham NZ, and Hasnain SE. Pgrs domain of rv0297 of mycobacterium tuberculosis is involved in modulation of macrophage functions to favor bacterial persistence. Front Cell Infection Microbiol. (2020) 10:451. doi: 10.3389/fcimb.2020.00451
102. Thi EP, Hong CJH, Sanghera G, and Reiner NE. Identification of the mycobacterium tuberculosis protein pe-pgrs62 as a novel effector that functions to block phagosome maturation and inhibit inos expression. Cell Microbiol. (2012) 15:795–808. doi: 10.1111/cmi.12073
103. Ramakrishnan L, Federspiel NA, and Falkow S. Granuloma-specific expression of mycobacterium virulence proteins from the glycine-rich pe-pgrs family. Science. (2000) 288:1436–9. doi: 10.1126/science.288.5470.1436
104. Li Y, Miltner E, Wu M, Petrofsky M, and Bermudez LE. A mycobacterium avium ppe gene is associated with the ability of the bacterium to grow in macrophages and virulence in mice. Cell Microbiol. (2004) 7:539–48. doi: 10.1111/j.1462-5822.2004.00484.x
105. Jha SS, Danelishvili L, Wagner D, Maser J, Li Y-J, Moric I, et al. Virulence-related mycobacterium avium subsp hominissuis mav_2928 gene is associated with vacuole remodeling in macrophages. BMC Microbiol. (2010) 10:100. doi: 10.1186/1471-2180-10-100
106. Saini NK, Baena A, Ng TW, Venkataswamy MM, Kennedy SC, Kunnath-Velayudhan S, et al. Suppression of autophagy and antigen presentation by mycobacterium tuberculosis pe_Pgrs47. Nat Microbiol. (2016) 1:16133. doi: 10.1038/nmicrobiol.2016.133
107. Meng L, Tong J, Wang H, Tao C, Wang Q, Niu C, et al. Ppe38 protein of mycobacterium tuberculosis inhibits macrophage mhc class I expression and dampens cd8+ T cell responses. Front Cell Infection Microbiol. (2017) 7:68. doi: 10.3389/fcimb.2017.00068
108. Wang H, Dong D, Tang S, Chen X, and Gao Q. Ppe38 of mycobacterium marinum triggers the cross-talk of multiple pathways involved in the host response, as revealed by subcellular quantitative proteomics. J Proteome Res. (2013) 12:2055–66. doi: 10.1021/pr301017e
109. Stutz MD, Allison CC, Ojaimi S, Preston SP, Doerflinger M, Arandjelovic P, et al. Macrophage and neutrophil death programs differentially confer resistance to tuberculosis. Immunity. (2021) 54:1758–71.e7. doi: 10.1016/j.immuni.2021.06.009
110. Rincón M, Anguita J, Nakamura T, Fikrig E, and Flavell RA. Interleukin (Il)-6 directs the differentiation of il-4–producing cd4+ T cells. J Exp Med. (1997) 185:461–70. doi: 10.1084/jem.185.3.461
111. Bhat KH, Srivastava S, Kotturu SK, Ghosh S, and Mukhopadhyay S. The ppe2 protein of mycobacterium tuberculosis translocates to host nucleus and inhibits nitric oxide production. Sci Rep. (2017) 7:39706. doi: 10.1038/srep39706
112. Srivastava S, Battu MB, Khan MZ, Nandicoori VK, and Mukhopadhyay S. Mycobacterium tuberculosis ppe2 protein interacts with P67phox and inhibits reactive oxygen species production. J Immunol. (2019) 203:1218–29. doi: 10.4049/jimmunol.1801143
113. Tiwari BM, Kannan N, Vemu L, and Raghunand TR. The mycobacterium tuberculosis pe proteins rv0285 and rv1386 modulate innate immunity and mediate bacillary survival in macrophages. PloS One. (2012) 7:e51686. doi: 10.1371/journal.pone.0051686
114. Li H, Li Q, Yu Z, Zhou M, and Xie J. Mycobacterium tuberculosis pe13 (Rv1195) manipulates the host cell fate via P38-erk-nf-Kb axis and apoptosis. Apoptosis. (2016) 21:795–808. doi: 10.1007/s10495-016-1249-y
115. Behar SM and Briken V. Apoptosis inhibition by intracellular bacteria and its consequence on host immunity. Curr Opin Immunol. (2019) 60:103–10. doi: 10.1016/j.coi.2019.05.007
116. Grover S, Sharma T, Singh Y, Kohli S, P M, Singh A, et al. The pgrs domain of mycobacterium tuberculosis pe_Pgrs protein rv0297 is involved in endoplasmic reticulum stress-mediated apoptosis through toll-like receptor 4. mBio. (2018) 9:e01017–18. doi: 10.1128/mbio.01017-18
117. Long Q, Xiang X, Yin Q, Li S, Yang W, Sun H, et al. Pe_Pgrs62 promotes the survival of mycobacterium smegmatis within macrophages via disrupting er stress-mediated apoptosis. J Cell Physiol. (2019) 234:19774–84. doi: 10.1002/jcp.28577
118. Basu S, Pathak SK, Banerjee A, Pathak S, Bhattacharyya A, Yang Z, et al. Execution of macrophage apoptosis by pe_Pgrs33 of mycobacterium tuberculosis is mediated by toll-like receptor 2-dependent release of tumor necrosis factor-A. J Biol Chem. (2007) 282:1039–50. doi: 10.1074/jbc.m604379200
119. Cadieux N, Parra M, Cohen H, Maric D, Morris SL, and Brennan MJ. Induction of cell death after localization to the host cell mitochondria by the mycobacterium tuberculosis pe_Pgrs33 protein. Microbiology. (2011) 157:793–804. doi: 10.1099/mic.0.041996-0
120. Yu X, Huang Y, Li Y, Li T, Yan S, Ai X, et al. Mycobacterium tuberculosis pe_Pgrs1 promotes mycobacteria intracellular survival via reducing the concentration of intracellular free ca2+ and suppressing endoplasmic reticulum stress. Mol Immunol. (2023) 154:24–32. doi: 10.1016/j.molimm.2022.12.007
121. Dheenadhayalan V, Delogu G, and Brennan MJ. Expression of the pe_Pgrs 33 protein in mycobacterium smegmatis triggers necrosis in macrophages and enhanced mycobacterial survival. Microbes Infection. (2006) 8:262–72. doi: 10.1016/j.micinf.2005.06.021
122. Tundup S, Mohareer K, and Hasnain SE. Mycobacterium tuberculosis pe25/ppe41 protein complex induces necrosis in macrophages: role in virulence and disease reactivation? FEBS Open Bio. (2014) 4:822–8. doi: 10.1016/j.fob.2014.09.001
123. Danelishvili L, Everman J, and Bermudez LE. Mycobacterium tuberculosis ppe68 and rv2626c genes contribute to the host cell necrosis and bacterial escape from macrophages. Virulence. (2015) 7:23–32. doi: 10.1080/21505594.2015.1102832
124. Yang G, Luo T, Sun C, Yuan J, Peng X, Zhang C, et al. Ppe27 in mycobacterium smegmatis enhances mycobacterial survival and manipulates cytokine secretion in mouse macrophages. J Interferon &Cytokine Res. (2017) 37:421–31. doi: 10.1089/jir.2016.0126
125. Yang Y, Xu P, He P, Shi F, Tang Y, Guan C, et al. Mycobacterial ppe13 activates inflammasome by interacting with the natch and lrr domains of nlrp3. FASEB J. (2020) 34:12820–33. doi: 10.1096/fj.202000200rr
126. Qian J, Hu Y, Zhang X, Chi M, Xu S, Wang H, et al. Mycobacterium tuberculosis pe_Pgrs19 induces pyroptosis through a non-classical caspase-11/gsdmd pathway in macrophages. Microorganisms. (2022) 10:2473. doi: 10.3390/microorganisms10122473
127. Gong Z, Kuang Z, Li H, Li C, Ali MK, Huang F, et al. Regulation of host cell pyroptosis and cytokines production by mycobacterium tuberculosis effector ppe60 requires lubac mediated nf-Kb signaling. Cell Immunol. (2019) 335:41–50. doi: 10.1016/j.cellimm.2018.10.009
128. Strong EJ, Ng TW, Porcelli SA, and Lee S. Mycobacterium tuberculosis pe_Pgrs20 and pe_Pgrs47 proteins inhibit autophagy by interaction with rab1a. mSphere. (2021) 6:00549–21. doi: 10.1128/msphere.00549-21
129. Deng W, Long Q, Zeng J, Li P, Yang W, Chen X, et al. Mycobacterium tuberculosis pe_Pgrs41 enhances the intracellular survival of M. Smegmatis within macrophages via blocking innate immunity and inhibition of host defense. Sci Rep. (2017) 7:46716. doi: 10.1038/srep46716
130. Chai Q, Wang X, Qiang L, Zhang Y, Ge P, Lu Z, et al. A mycobacterium tuberculosis surface protein recruits ubiquitin to trigger host xenophagy. Nat Commun. (2019) 10:1973. doi: 10.1038/s41467-019-09955-8
131. Juffermans Nicole P, Florquin S, Camoglio L, Verbon A, Kolk Arend H, Speelman P, et al. Interleukin-1 signaling is essential for host defense during murine pulmonary tuberculosis. J Infect Dis. (2000) 182:902–8. doi: 10.1086/315771
132. Zhang G, Zhou B, Li S, Yue J, Yang H, Wen Y, et al. Allele-specific induction of il-1β Expression by C/ebpβ and pu.1 contributes to increased tuberculosis susceptibility. PloS Pathog. (2014) 10:e1004426. doi: 10.1371/journal.ppat.1004426
133. Pyle CJ and Tobin DM. People who lack the immune protein tnf can still fight infection. Nature. (2024) 633:293–4. doi: 10.1038/d41586-024-02657-2
134. Diehl S, Anguita J, Hoffmeyer A, Zapton T, Ihle JN, Fikrig E, et al. Inhibition of th1 differentiation by il-6 is mediated by socs1. Immunity. (2000) 13:805–15. doi: 10.1016/s1074-7613(00)00078-9
135. Li J, Cao C, Xiang Y, Hong Z, He D, Zhong H, et al. Tlt2 suppresses th1 response by promoting il-6 production in monocyte through jak/stat3 signal pathway in tuberculosis. Front Immunol. (2020) 11:2031. doi: 10.3389/fimmu.2020.02031
136. Rangel-Moreno J, Carragher DM, de la Luz Garcia-Hernandez M, Hwang JY, Kusser K, Hartson L, et al. The development of inducible bronchus-associated lymphoid tissue depends on il-17. Nat Immunol. (2011) 12:639–46. doi: 10.1038/ni.2053
137. Gopal R, Monin L, Slight S, Uche U, Blanchard E, A. Fallert Junecko B, et al. Unexpected role for il-17 in protective immunity against hypervirulent mycobacterium tuberculosis hn878 infection. PloS Pathog. (2014) 10:e1004099. doi: 10.1371/journal.ppat.1004099
138. Goriely S, Neurath MF, and Goldman M. How microorganisms tip the balance between interleukin-12 family members. Nat Rev Immunol. (2008) 8:81–6. doi: 10.1038/nri2225
139. Beamer GL, Flaherty DK, Assogba BD, Stromberg P, Gonzalez-Juarrero M, de Waal Malefyt R, et al. Interleukin-10 promotes mycobacterium tuberculosis disease progression in cba/J mice. J Immunol. (2008) 181:5545–50. doi: 10.4049/jimmunol.181.8.5545
140. Cyktor JC, Carruthers B, Kominsky RA, Beamer GL, Stromberg P, and Turner J. Il-10 inhibits mature fibrotic granuloma formation during mycobacterium tuberculosis infection. J Immunol. (2013) 190:2778–90. doi: 10.4049/jimmunol.1202722
141. Dwivedi V, Gautam S, Headley CA, Piergallini T, Torrelles JB, and Turner J. Il-10 receptor blockade delivered simultaneously with bacillus calmette–guérin vaccination sustains long-term protection against mycobacterium tuberculosis infection in mice. J Immunol. (2022) 208:1406–16. doi: 10.4049/jimmunol.2100900
142. Lin Y, Ritchea S, Logar A, Slight S, Messmer M, Rangel-Moreno J, et al. Interleukin-17 is required for T helper 1 cell immunity and host resistance to the intracellular pathogen francisella tularensis. Immunity. (2009) 31:799–810. doi: 10.1016/j.immuni.2009.08.025
143. Zelante T, De Luca A, Bonifazi P, Montagnoli C, Bozza S, Moretti S, et al. Il-23 and the th17 pathway promote inflammation and impair antifungal immune resistance. Eur J Immunol. (2007) 37:2695–706. doi: 10.1002/eji.200737409
144. Medha, Priyanka, Bhatt P, Sharma S, and Sharma M. Role of C-terminal domain of mycobacterium tuberculosis pe6 (Rv0335c) protein in host mitochondrial stress and macrophage apoptosis. Apoptosis. (2022) 28:136–65. doi: 10.1007/s10495-022-01778-1
145. Bansal K, Elluru SR, Narayana Y, Chaturvedi R, Patil SA, Kaveri SV, et al. Pe_Pgrs antigens of mycobacterium tuberculosis induce maturation and activation of human dendritic cells. J Immunol. (2010) 184:3495–504. doi: 10.4049/jimmunol.0903299
146. Kim WS, Kim J-S, Cha SB, Kim SJ, Kim H, Kwon KW, et al. Mycobacterium tuberculosis pe27 activates dendritic cells and contributes to th1-polarized memory immune responses during in vivo infection. Immunobiology. (2016) 221:440–53. doi: 10.1016/j.imbio.2015.11.006
147. Kim J-S, Kim HK, Cho E, Mun S-J, Jang S, Jang J, et al. Pe_Pgrs38 interaction with hausp downregulates antimycobacterial host defense via traf6. Front Immunol. (2022) 13:862628. doi: 10.3389/fimmu.2022.862628
148. Asaad M, Kaisar Ali M, Abo-kadoum MA, Lambert N, Gong Z, Wang H, et al. Mycobacterium tuberculosis ppe10 (Rv0442c) alters host cell apoptosis and cytokine profile via linear ubiquitin chain assembly complex hoip-nf-Kb signaling axis. Int Immunopharmacol. (2021) 94:107363. doi: 10.1016/j.intimp.2020.107363
149. Xu T, Wang C, Li M, Wei J, He Z, Qian Z, et al. Mycobacterium tuberculosis pe_Pgrs45 (Rv2615c) promotes recombinant mycobacteria intracellular survival via regulation of innate immunity, and inhibition of cell apoptosis. J Microbiol. (2024) 62:49–62. doi: 10.1007/s12275-023-00101-0
150. Ali MK, Zhen G, Nzungize L, Stojkoska A, Duan X, Li C, et al. Mycobacterium tuberculosis pe31 (Rv3477) attenuates host cell apoptosis and promotes recombinant M. Smegmatis intracellular survival via up-regulating gtpase guanylate binding protein-1. Front Cell Infection Microbiol. (2020) 10:40. doi: 10.3389/fcimb.2020.00040
151. Gong Z, Han S, Liang T, Zhang H, Sun Q, Pan H, et al. Mycobacterium tuberculosis effector ppe36 attenuates host cytokine storm damage via inhibiting macrophage M1 polarization. J Cell Physiol. (2021) 236:7405–20. doi: 10.1002/jcp.30411
152. Khubaib M, Sheikh JA, Pandey S, Srikanth B, Bhuwan M, Khan N, et al. Mycobacterium tuberculosis co-operonic pe32/ppe65 proteins alter host immune responses by hampering th1 response. Front Microbiol. (2016) 7:719. doi: 10.3389/fmicb.2016.00719
153. Farhat MR, Shapiro BJ, Kieser KJ, Sultana R, Jacobson KR, Victor TC, et al. Genomic analysis identifies targets of convergent positive selection in drug-resistant mycobacterium tuberculosis. Nat Genet. (2013) 45:1183–9. doi: 10.1038/ng.2747
154. Zhang H, Li D, Zhao L, Fleming J, Lin N, Wang T, et al. Genome sequencing of 161 mycobacterium tuberculosis isolates from China identifies genes and intergenic regions associated with drug resistance. Nat Genet. (2013) 45:1255–60. doi: 10.1038/ng.2735
155. Kanji A, Hasan Z, Ali A, McNerney R, Mallard K, Coll F, et al. Characterization of genomic variations in snps of pe_Pgrs genes reveals deletions and insertions in extensively drug resistant (Xdr) M. Tuberculosis strains from Pakistan. Int J Mycobacteriol. (2015) 4:73–9. doi: 10.1016/j.ijmyco.2014.11.049
156. Hang NTL, Hijikata M, Maeda S, Thuong PH, Ohashi J, Van Huan H, et al. Whole genome sequencing, analyses of drug resistance-conferring mutations, and correlation with transmission of mycobacterium tuberculosis carrying katg-S315t in hanoi, Vietnam. Sci Rep. (2019) 9:15354. doi: 10.1038/s41598-019-51812-7
157. Cui ZJ, Yang QY, Zhang HY, Zhu Q, and Zhang QY. Bioinformatics identification of drug resistance-associated gene pairs in mycobacterium tuberculosis. Int J Mol Sci. (2016) 17:1417. doi: 10.3390/ijms17091417
158. Tantivitayakul P, Ruangchai W, Juthayothin T, Smittipat N, Disratthakit A, Mahasirimongkol S, et al. Homoplastic single nucleotide polymorphisms contributed to phenotypic diversity in mycobacterium tuberculosis. Sci Rep. (2020) 10:8024. doi: 10.1038/s41598-020-64895-4
159. Ates LS. New insights into the mycobacterial pe and ppe proteins provide a framework for future research. Mol Microbiol. (2019) 113:4–21. doi: 10.1111/mmi.14409
160. Ates LS, Sayes F, Frigui W, Ummels R, Damen MPM, Bottai D, et al. Rd5-mediated lack of pe_Pgrs and ppe-mptr export in bcg vaccine strains results in strong reduction of antigenic repertoire but little impact on protection. PloS Pathog. (2018) 14:e1007139. doi: 10.1371/journal.ppat.1007139
161. Tundup S, Pathak N, Ramanadham M, Mukhopadhyay S, Murthy KJR, Ehtesham NZ, et al. The co-operonic pe25/ppe41 protein complex of mycobacterium tuberculosis elicits increased humoral and cell mediated immune response. PloS One. (2008) 3:e3586. doi: 10.1371/journal.pone.0003586
162. Guo F, Wei J, Song Y, Li B, Qian Z, Wang X, et al. Immunological effects of the pe/ppe family proteins of mycobacterium tuberculosis and related vaccines. Front Immunol. (2023) 14:1255920. doi: 10.3389/fimmu.2023.1255920
163. Ahmed A, Das A, and Mukhopadhyay S. Immunoregulatory functions and expression patterns of pe/ppe family members: roles in pathogenicity and impact on anti-tuberculosis vaccine and drug design. IUBMB Life. (2015) 67:414–27. doi: 10.1002/iub.1387
164. Sayes F, Sun L, Di Luca M, Simeone R, Degaiffier N, Fiette L, et al. Strong immunogenicity and cross-reactivity of mycobacterium tuberculosis esx-5 type vii secretion -encoded pe-ppe proteins predicts vaccine potential. Cell Host Microbe. (2012) 11:352–63. doi: 10.1016/j.chom.2012.03.003
165. Sayes F, Pawlik A, Frigui W, Gröschel MI, Crommelynck S, Fayolle C, et al. Cd4+ T cells recognizing pe/ppe antigens directly or via cross reactivity are protective against pulmonary mycobacterium tuberculosis infection. PloS Pathog. (2016) 12:e1005770. doi: 10.1371/journal.ppat.1005770
166. Tait DR, Hatherill M, van der Meeren O, Ginsberg AM, Van Brakel E, Salaun B, et al. Final analysis of a trial of M72/as01e vaccine to prevent tuberculosis. New Engl J Med. (2019) 381:2429–39. doi: 10.1056/nejmoa1909953
167. Skeiky YAW, Alderson MR, Ovendale PJ, Guderian JA, Brandt L, Dillon DC, et al. Differential immune responses and protective efficacy induced by components of a tuberculosis polyprotein vaccine, mtb72f, delivered as naked DNA or recombinant protein. J Immunol. (2004) 172:7618–28. doi: 10.4049/jimmunol.172.12.7618
168. Brandt L, Skeiky YAW, Alderson MR, Lobet Y, Dalemans W, Turner OC, et al. The protective effect of the mycobacterium bovis bcg vaccine is increased by coadministration with the mycobacterium tuberculosis 72-kilodalton fusion polyprotein mtb72f in M. Tuberculosis-infected Guinea pigs. Infection Immun. (2004) 72:6622–32. doi: 10.1128/iai.72.11.6622-6632.2004
169. Kumarasamy N, Poongulali S, Bollaerts A, Moris P, Beulah FE, Ayuk LN, et al. A randomized, controlled safety, and immunogenicity trial of the M72/as01 candidate tuberculosis vaccine in hiv-positive Indian adults. Medicine. (2016) 95:e2459. doi: 10.1097/md.0000000000002459
170. Reed SG, Coler RN, Dalemans W, Tan EV, DeLa Cruz EC, Basaraba RJ, et al. Defined tuberculosis vaccine, mtb72f/as02a, evidence of protection in cynomolgus monkeys. Proc Natl Acad Sci. (2009) 106:2301–6. doi: 10.1073/pnas.0712077106
171. Penn-Nicholson A, Geldenhuys H, Burny W, van der Most R, Day CL, Jongert E, et al. Safety and immunogenicity of candidate vaccine M72/as01e in adolescents in a tb endemic setting. Vaccine. (2015) 33:4025–34. doi: 10.1016/j.vaccine.2015.05.088
172. Gong W, Liang Y, Mi J, Xue Y, Wang J, Wang L, et al. A peptide-based vaccine acp derived from antigens of mycobacterium tuberculosis induced th1 response but failed to enhance the protective efficacy of bcg in mice. Indian J Tuberculosis. (2022) 69:482–95. doi: 10.1016/j.ijtb.2021.08.016
173. Day TA, Penn-Nicholson A, Luabeya AKK, Fiore-Gartland A, Du Plessis N, Loxton AG, et al. Safety and immunogenicity of the adjunct therapeutic vaccine id93 + Gla-se in adults who have completed treatment for tuberculosis: A randomised, double-blind, placebo-controlled, phase 2a trial. Lancet Respir Med. (2021) 9:373–86. doi: 10.1016/S2213-2600(20)30319-2
174. Bertholet S, Ireton GC, Ordway DJ, Windish HP, Pine SO, Kahn M, et al. A defined tuberculosis vaccine candidate boosts bcg and protects against multidrug-resistant mycobacterium tuberculosis. Sci Trans Med. (2010) 2:53ra74–53ra74. doi: 10.1126/scitranslmed.3001094
175. Baldwin SL, Reese VA, Larsen SE, Beebe E, Guderian J, Orr MT, et al. Prophylactic efficacy against mycobacterium tuberculosis using id93 and lipid-based adjuvant formulations in the mouse model. PloS One. (2021) 16:e0247990. doi: 10.1371/journal.pone.0247990
176. Kwon KW, Lee A, Larsen SE, Baldwin SL, Coler RN, Reed SG, et al. Long-term protective efficacy with a bcg-prime id93/gla-se boost regimen against the hyper-virulent mycobacterium tuberculosis strain K in a mouse model. Sci Rep. (2019) 9:15560. doi: 10.1038/s41598-019-52146-0
177. Coler RN, Bertholet S, Pine SO, Orr MT, Reese V, Windish HP, et al. Therapeutic immunization against mycobacterium tuberculosis is an effective adjunct to antibiotic treatment. J Infect Dis. (2012) 207:1242–52. doi: 10.1093/infdis/jis425
178. Lu Y, Xu Y, Yang E, Wang C, Wang H, and Shen H. Novel recombinant bcg coexpressing ag85b, esat-6 and rv2608 elicits significantly enhanced cellular immune and antibody responses in C57bl/6 mice. Scandinavian J Immunol. (2012) 76:271–7. doi: 10.1111/j.1365-3083.2012.02726.x
179. Sulman S, Savidge BO, Alqaseer K, Das MK, Nezam Abadi N, Pearl JE, et al. Balance between protection and pathogenic response to aerosol challenge with mycobacterium tuberculosis (Mtb) in mice vaccinated with trifu64, a fusion consisting of three mtb antigens. Vaccines. (2021) 9:519. doi: 10.3390/vaccines9050519
180. Romano M, Rindi L, Korf H, Bonanni D, Adnet P-Y, Jurion F, et al. Immunogenicity and protective efficacy of tuberculosis subunit vaccines expressing ppe44 (Rv2770c). Vaccine. (2008) 26:6053–63. doi: 10.1016/j.vaccine.2008.09.025
181. Moradi B, Sankian M, Amini Y, Gholoobi A, and Meshkat Z. A new DNA vaccine expressing hspx-ppe44-esxv fusion antigens of mycobacterium tuberculosis induced strong immune responses. Iran J Basic Med Sci. (2020) 23:909–14. doi: 10.22038/ijbms.2020.38521.9171
182. Mansury D, Ghazvini K, Amel Jamehdar S, Badiee A, Tafaghodi M, Nikpoor AR, et al. Enhancement of the effect of bcg vaccine against tuberculosis using dda/tdb liposomes containing a fusion protein of hspx, ppe44, and esxv. Artif Cells Nanomedicine Biotechnol. (2019) 47:370–7. doi: 10.1080/21691401.2018.1557674
183. Hoseinpur R, Hasani A, Baradaran B, Abdolalizadeh J, Amini Y, Salehi R, et al. Chitosan nanoparticles containing fusion protein (Hspx–ppe44–esxv) and resiquimod adjuvant (Hperc) as a novel booster vaccine for mycobacterium tuberculosis. J Biomaterials Appl. (2022) 37:40–7. doi: 10.1177/08853282221079105
184. Xu Y, Yang E, Wang J, Li R, Li G, Liu G, et al. Prime–boost bacillus calmette–guérin vaccination with lentivirus-vectored and DNA-based vaccines expressing antigens ag85b and rv3425 improves protective efficacy against mycobacterium tuberculosis in mice. Immunology. (2014) 143:277–86. doi: 10.1111/imm.12308
185. Wang J, Qie Y, Zhang H, Zhu B, Xu Y, Liu W, et al. Ppe protein (Rv3425) from DNA segment rd11 of mycobacterium tuberculosis: A novel immunodominant antigen of mycobacterium tuberculosis induces humoral and cellular immune responses in mice. Microbiol Immunol. (2008) 52:224–30. doi: 10.1111/j.1348-0421.2008.00029.x
186. Yang E, Gu J, Wang F, Wang H, Shen H, and Chen ZW. Recombinant bcg prime and ppe protein boost provides potent protection against acute mycobacterium tuberculosis infection in mice. Microbial Pathogenesis. (2016) 93:1–7. doi: 10.1016/j.micpath.2016.01.006
187. Su H, Zhu S, Zhu L, Kong C, Huang Q, Zhang Z, et al. Mycobacterium tuberculosis latent antigen rv2029c from the multistage DNA vaccine A39 drives th1 responses via tlr-mediated macrophage activation. Front Microbiol. (2017) 8:2266. doi: 10.3389/fmicb.2017.02266
188. Arif S, Akhter M, Khaliq A, and Akhtar MW. Fusion peptide constructs from antigens of M. Tuberculosis producing high T-cell mediated immune response. PloS One. (2022) 17:e0271126. doi: 10.1371/journal.pone.0271126
189. Dong Z, Xu L, Yang J, He Y, She Q, Wang J, et al. Primary application of ppe68 of mycobacterium tuberculosis. Hum Immunol. (2014) 75:428–32. doi: 10.1016/j.humimm.2014.02.017
190. Jia Q, Masleša-Galić S, Nava S, and Horwitz MA. Listeria-vectored multi-antigenic tuberculosis vaccine protects C57bl/6 and balb/C mice and Guinea pigs against mycobacterium tuberculosis challenge. Commun Biol. (2022) 5:1388. doi: 10.1038/s42003-022-04345-1
191. Stylianou E, Harrington-Kandt R, Beglov J, Bull N, Pinpathomrat N, Swarbrick GM, et al. Identification and evaluation of novel protective antigens for the development of a candidate tuberculosis subunit vaccine. Infection Immun. (2018) 86:00014–18. doi: 10.1128/iai.00014-18
192. Skeiky YAW, Alderson MR, Ovendale PJ, Lobet Y, Dalemans W, Orme IM, et al. Protection of mice and Guinea pigs against tuberculosis induced by immunization with a single mycobacterium tuberculosis recombinant antigen, mtb41. Vaccine. (2005) 23:3937–45. doi: 10.1016/j.vaccine.2005.03.003
193. Van Der Meeren O, Hatherill M, Nduba V, Wilkinson RJ, Muyoyeta M, Van Brakel E, et al. Phase 2b controlled trial of M72/as01e vaccine to prevent tuberculosis. New Engl J Med. (2018) 379:1621–34. doi: 10.1056/nejmoa1803484
194. Hebert AM, Talarico S, Yang D, Durmaz R, Marrs CF, Zhang L, et al. DNA polymorphisms in the pepaand ppe18 genes among clinical strains of mycobacterium tuberculosis: implications for vaccine efficacy. Infection Immun. (2007) 75:5798–805. doi: 10.1128/iai.00335-07
195. Homolka S, Ubben T, and Niemann S. High sequence variability of the ppe18 gene of clinical mycobacterium tuberculosis complex strains potentially impacts effectivity of vaccine candidate M72/as01e. PloS One. (2016) 11:e0152200. doi: 10.1371/journal.pone.0152200
196. Hakim JMC and Yang Z. Predicted structural variability of mycobacterium tuberculosis ppe18 protein with immunological implications among clinical strains. Front Microbiol. (2021) 11:595312. doi: 10.3389/fmicb.2020.595312
197. Vargas R, Freschi L, Marin M, Epperson LE, Smith M, Oussenko I, et al. In-host population dynamics of mycobacterium tuberculosis complex during active disease. Elife. (2021) 10:e61805. doi: 10.7554/eLife.61805
198. Li J, Zhao A, Tang J, Wang G, Shi Y, Zhan L, et al. Tuberculosis vaccine development: from classic to clinical candidates. Eur J Clin Microbiol Infect Dis. (2020) 39:1405–25. doi: 10.1007/s10096-020-03843-6
199. Torrado E and Cooper AM. Il-17 and th17 cells in tuberculosis. Cytokine Growth Factor Rev. (2010) 21:455–62. doi: 10.1016/j.cytogfr.2010.10.004
200. Coler RN, Day TA, Ellis R, Piazza FM, Beckmann AM, Vergara J, et al. The tlr-4 agonist adjuvant, gla-se, improves magnitude and quality of immune responses elicited by the id93 tuberculosis vaccine: first-in-human trial. NPJ Vaccines. (2018) 3:34. doi: 10.1038/s41541-018-0057-5
201. Penn-Nicholson A, Tameris M, Smit E, Day TA, Musvosvi M, Jayashankar L, et al. Safety and immunogenicity of the novel tuberculosis vaccine id93 + Gla-se in bcg-vaccinated healthy adults in South Africa: A randomised, double-blind, placebo-controlled phase 1 trial. Lancet Respir Med. (2018) 6:287–98. doi: 10.1016/s2213-2600(18)30077-8
202. Rindi L, Peroni I, Lari N, Bonanni D, Tortoli E, and Garzelli C. Variation of the expression of mycobacterium tuberculosis ppe44 gene among clinical isolates. FEMS Immunol Med Microbiol. (2007) 51:381–7. doi: 10.1111/j.1574-695x.2007.00315.x
203. Wang J, Qie Y, Liu W, and Wang H. Protective efficacy of a recombinant bcg secreting antigen 85b/rv3425 fusion protein against mycobacterium tuberculosis infection in mice. Hum Vaccin Immunother. (2012) 8:1869–74. doi: 10.4161/hv.21817
204. Mustafa AS. Characterization of a cross-reactive, immunodominant and hla-promiscuous epitope of mycobacterium tuberculosis-specific major antigenic protein ppe68. PloS One. (2014) 9:e103679. doi: 10.1371/journal.pone.0103679
205. Kim A, Hur Y-G, Gu S, and Cho S-N. Protective vaccine efficacy of the complete form of ppe39 protein from mycobacterium tuberculosis beijing/K strain in mice. Clin Vaccine Immunol. (2017) 24:e00219–17. doi: 10.1128/cvi.00219-17
206. Singh SK, Tripathi DK, Singh PK, Sharma S, and Srivastava KK. Protective and survival efficacies of rv0160c protein in murine model of mycobacterium tuberculosis. Appl Microbiol Biotechnol. (2012) 97:5825–37. doi: 10.1007/s00253-012-4493-2
207. García-Bengoa M, Vergara EJ, Tran AC, Bossi L, Cooper AM, Pearl JE, et al. Immunogenicity of pe18, pe31, and ppe26 proteins from mycobacterium tuberculosis in humans and mice. Front Immunol. (2023) 14:1307429. doi: 10.3389/fimmu.2023.1307429
208. Goldstone RM, Goonesekera SD, Bloom BR, and Sampson SL. The transcriptional regulator rv0485 modulates the expression of a pe and ppe gene pair and is required for mycobacterium tuberculosis virulence. Infection Immun. (2009) 77:4654–67. doi: 10.1128/iai.01495-08
209. Le Moigne V, Robreau G, and Mahana W. Flagellin as a good carrier and potent adjuvant for th1 response: study of mice immune response to the P27 (Rv2108) mycobacterium tuberculosis antigen. Mol Immunol. (2008) 45:2499–507. doi: 10.1016/j.molimm.2008.01.005
210. Dillon DC, Alderson MR, Day CH, Lewinsohn DM, Coler R, Bement T, et al. Molecular characterization and human T-cell responses to a member of a novel mycobacterium tuberculosis mtb39 gene family. Infection Immun. (1999) 67:2941–50. doi: 10.1128/iai.67.6.2941-2950.1999
211. Comas I, Chakravartti J, Small PM, Galagan J, Niemann S, Kremer K, et al. Human T cell epitopes of mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet. (2010) 42:498–503. doi: 10.1038/ng.590
212. Orr MT, Ireton GC, Beebe EA, Huang P-WD, Reese VA, Argilla D, et al. Immune subdominant antigens as vaccine candidates against mycobacterium tuberculosis. J Immunol. (2014) 193:2911–8. doi: 10.4049/jimmunol.1401103
213. Ogongo P, Tezera LB, Ardain A, Nhamoyebonde S, Ramsuran D, Singh A, et al. Tissue-resident-like cd4+ T cells secreting il-17 control mycobacterium tuberculosis in the human lung. J Clin Invest. (2021) 131:e142014. doi: 10.1172/jci142014
214. Karanika S, Wang T, Yilma A, Castillo JR, Gordy JT, Bailey H, et al. Therapeutic DNA vaccine targeting mycobacterium tuberculosis persisters shortens curative tuberculosis treatment. bioRxiv. (2024). doi: 10.1101/2024.09.03.611055
215. Abraham PR, Latha GS, Valluri VL, and Mukhopadhyay S. Mycobacterium tuberculosis ppe protein rv0256c induces strong B cell response in tuberculosis patients. Infection Genet Evol. (2014) 22:244–9. doi: 10.1016/j.meegid.2013.06.023
216. Khan N, Alam K, Nair S, Valluri VL, Murthy KJR, and Mukhopadhyay S. Association of strong immune responses to ppe protein rv1168c with active tuberculosis. Clin Vaccine Immunol. (2008) 15:974–80. doi: 10.1128/cvi.00485-07
217. Abraham PR, Udgata A, Latha GS, and Mukhopadhyay S. The mycobacterium tuberculosis ppe protein rv1168c induces stronger B cell response than rv0256c in active tb patients. Infection Genet Evol. (2016) 40:339–45. doi: 10.1016/j.meegid.2015.09.005
218. Sulman S, Shahid S, Khaliq A, Ambreen A, Khan IH, Cooper AM, et al. Enhanced serodiagnostic potential of a fusion molecule consisting of rv1793, rv2628 and a truncated rv2608 of mycobacterium tuberculosis. PloS One. (2021) 16:e0258389. doi: 10.1371/journal.pone.0258389
219. Narayana Y, Joshi B, Katoch VM, Mishra KC, and Balaji KN. Differential B-cell responses are induced by mycobacterium tuberculosis pe antigens rv1169c, rv0978c, and rv1818c. Clin Vaccine Immunol. (2007) 14:1334–41. doi: 10.1128/cvi.00181-07
220. Donà V, Ventura M, Sali M, Cascioferro A, Provvedi R, Palù G, et al. The ppe domain of ppe17 is responsible for its surface localization and can be used to express heterologous proteins on the mycobacterial surface. PloS One. (2013) 8:e57517. doi: 10.1371/journal.pone.0057517
221. Sargentini V, Mariotti S, Carrara S, Gagliardi MC, Teloni R, Goletti D, et al. Cytometric detection of antigen-specific ifn-Γ/il-2 secreting cells in the diagnosis of tuberculosis. BMC Infect Dis. (2009) 9:99. doi: 10.1186/1471-2334-9-99
222. Le Moigne V, Le Moigne D, and Mahana W. Antibody response to mycobacterium tuberculosis P27-ppe36 antigen in sera of pulmonary tuberculosis patients. Tuberculosis. (2013) 93:189–91. doi: 10.1016/j.tube.2012.10.006
223. Zhang H, Wang J, Lei J, Zhang M, Yang Y, Chen Y, et al. Ppe protein (Rv3425) from DNA segment rd11 of mycobacterium tuberculosis: A potential B-cell antigen used for serological diagnosis to distinguish vaccinated controls from tuberculosis patients. Clin Microbiol Infection. (2007) 13:139–45. doi: 10.1111/j.1469-0691.2006.01561.x
224. Abdallah AM, Verboom T, Weerdenburg EM, Gey van Pittius NC, Mahasha PW, Jiménez C, et al. Ppe and pe_Pgrs proteins of mycobacterium marinum are transported via the type vii secretion system esx-5. Mol Microbiol. (2009) 73:329–40. doi: 10.1111/j.1365-2958.2009.06783.x
Keywords: Mycobacterium tuberculosis, PE/PPE, evolution, outer membrane, porin, vaccine
Citation: Zhang Z, Dong L, Li X, Deng T and Wang Q (2025) The PE/PPE family proteins of Mycobacterium tuberculosis: evolution, function, and prospects for tuberculosis control. Front. Immunol. 16:1606311. doi: 10.3389/fimmu.2025.1606311
Received: 05 April 2025; Accepted: 30 May 2025;
Published: 17 June 2025.
Edited by:
Selvakumar Subbian, The State University of New Jersey, United StatesReviewed by:
Perumal Vivekanandan, Indian Institute of Technology Delhi, IndiaSamantha Lynn Bell, The State University of New Jersey, United States
Copyright © 2025 Zhang, Dong, Li, Deng and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qinglan Wang, d2FuZ3FpbmdsYW5Ac2N1LmVkdS5jbg==
†These authors have contributed equally to this work
 Zhijing Zhang1,2†
Zhijing Zhang1,2† Le Dong
Le Dong Qinglan Wang
Qinglan Wang