- 1Department of Thyroid Surgery, The First Affiliated Hospital of China Medical University, Shenyang, Liaoning, China
- 2Department of Obstetrics and Gynecology, Shengjing Hospital of China Medical University, Shenyang, China
Natural killer cells (NK cells) are important immune cells within the tumor microenvironment (TME) and are considered the first line of defense in tumor immunity. Although many studies have focused on the role of NK cells in tumor therapy, the heterogeneity of NK cells complicates the investigation of the complex mechanisms within the tumor microenvironment. Single-cell sequencing technology, with its high-resolution capability, reveals the gene expression profiles of individual NK cells, highlighting their heterogeneity and providing more accurate information for NK cell therapy. This article begins by addressing the mechanisms underlying the formation of NK cell heterogeneity, emphasizing the significance of differentiation, development, and tissue residency in establishing this heterogeneity. It also summarizes the advances in the study of NK cell heterogeneity under physiological conditions and in tumor environments using single-cell sequencing technology. Finally, it analyzes the dynamic changes of NK cells within the tumor microenvironment under various therapeutic approaches to explore drug effects and resistance mechanisms, as well as to optimize therapeutic options. Investigating the mechanisms of tumor progression and drug intervention at the single-cell level will provide new perspectives for personalized treatment strategies centered around NK cells.
1 Introduction
Single-cell sequencing technology enables the tracking of cellular development and differentiation, and by analyzing single-cell data from various developmental stages, it elucidates cellular differentiation pathways and fate decisions. A study that integrated single-cell RNA sequencing, antigen receptor sequencing, and spatial transcriptomics reconstructed the developmental trajectory of the human immune system and mapped its distribution across multiple organs in the body (1). Beyond the enumeration of clusters, single-cell transcriptomics has significantly enhanced our capability to characterize cell states from an individual cell perspective (2). This technology is instrumental in uncovering cellular heterogeneity and has emerged as a powerful tool for investigating cellular diversity within complex biological systems. The basic sequencing process is illustrated in the following figure (Figure 1). Through single-cell sequencing, researchers can identify the heterogeneity within cell populations and discern the variations in cell states and functions. For instance, Han X et al. employed single-cell mRNA sequencing to determine the cellular composition of major human organs and established the Human Cell Landscape (HCL) schema (3). With the advent of microfluidic technology and integrated sequencing strategies, the application of single-cell sequencing has significantly expanded, particularly in the field of oncology, where it holds substantial promise. Historically, researchers have often equated the uniqueness of individual cells with the characteristics of the population. However, recent advancements in oncology research have highlighted the substantial heterogeneity among tumor cells from different sources and even within individual tumors, underscoring the importance of single-cell analysis (4). Innovative single-cell genomics techniques and spatially multiplexed in situ methods now offer an unparalleled level of resolution to comprehend the intricate interactions within tumor ecosystems. A growing body of research is contributing to the development of tumor atlases, which are anticipated to profoundly influence our understanding of cancer biology and potentially enhance the detection, prevention, and treatment of cancer, leading to precision medicine for cancer patients and those at risk (5). Innovative single-cell genomics techniques and spatially multiplexed in situ methods now offer an unparalleled level of resolution to comprehend the intricate interactions within tumor ecosystems. Since single-cell sequencing technology was selected as the Method of the Year by Nature magazine in 2013 (6), it has been increasingly utilized in both tumorology basic science and clinical research, successively characterizing the TME in glioblastoma, melanoma, breast tumors, colorectal cancer, squamous cell carcinoma of the skin, and lung cancer (7–12).
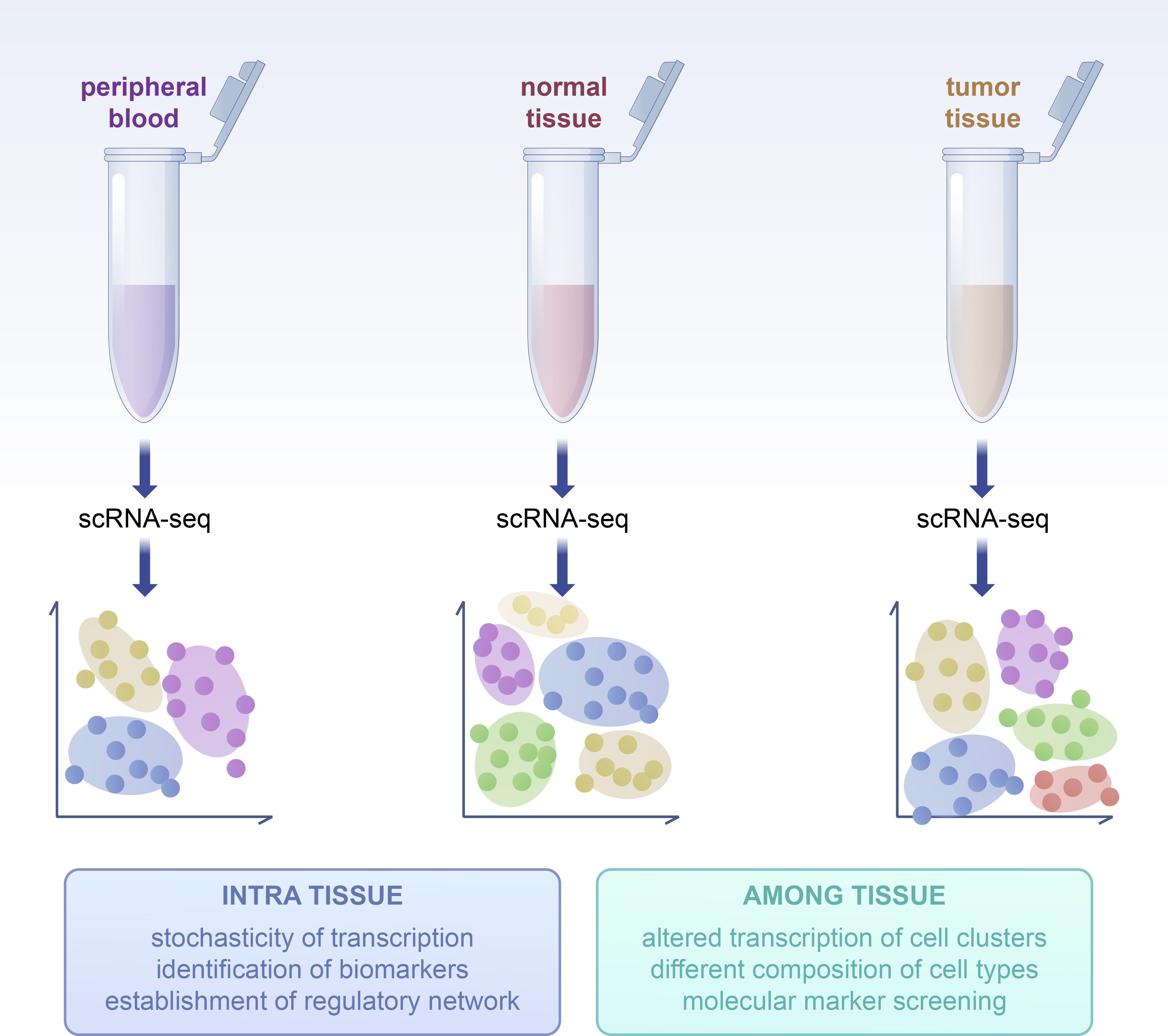
Figure 1. Basic process of scRNA-seq analysis. Including the following steps: 1. Dissociation of single cell 2. RNA hybridization 3. cDNA amplified by PCR 4. Library building 5. Sequencing 6. Bioinformatics analysis.
Natural killer (NK) cells were first identified in the 1970s through cytotoxicity assays conducted on cancer patients (13, 14). These cells demonstrated the capacity to lyse a diverse array of tumor cells. NK cells play a critical role in anti-tumor immunity and function as the primary line of defense in the immune response against tumors. (Figure 2) The killing mechanisms of NK cells are primarily categorized into two pathways. The first involves the secretion of cytoplasmic granule toxins, including perforin and granzymes, which activate cell death mechanisms. The second pathway leads to classical caspase-dependent apoptosis by the binding of death receptors, such as Fas/CD95, on target cells to their corresponding ligands, such as FasL, on NK cells, resulting in target cell killing (15, 16). NK cells are the first recognized subset of innate lymphoid cells (ILCs) and play a crucial role in the anti-tumor response of innate immunity (17), characterized by their autonomous ability to kill target cells and the high heterogeneity they exhibit within the tumor microenvironment (18).Recently, single-cell sequencing technology has proven instrumental in characterizing the heterogeneity and developmental processes of tumor-infiltrating T cells across various types of cancer (19). However, the understanding of NK cells remains incomplete. This review analyzes the sources of NK cell heterogeneity and provides a comprehensive overview of the development process of NK cells and the mechanisms of NK cell tissue residency to gain a deeper understanding of the core of tumor heterogeneity. It also explores advancements in the study of NK cell heterogeneity using single-cell sequencing technology in both physiological and tumor environments, particularly in solid tumors. Single-cell sequencing technology facilitates detailed subpopulation classification and functional studies of NK cells, constructs maps of the TME, and enhances research on intercellular interactions within the TME. Additionally, this review examines drugs and their interaction mechanisms, investigates the evolution of NK cells, and aims to establish efficacy evaluation models to study drug effects and resistance mechanisms.
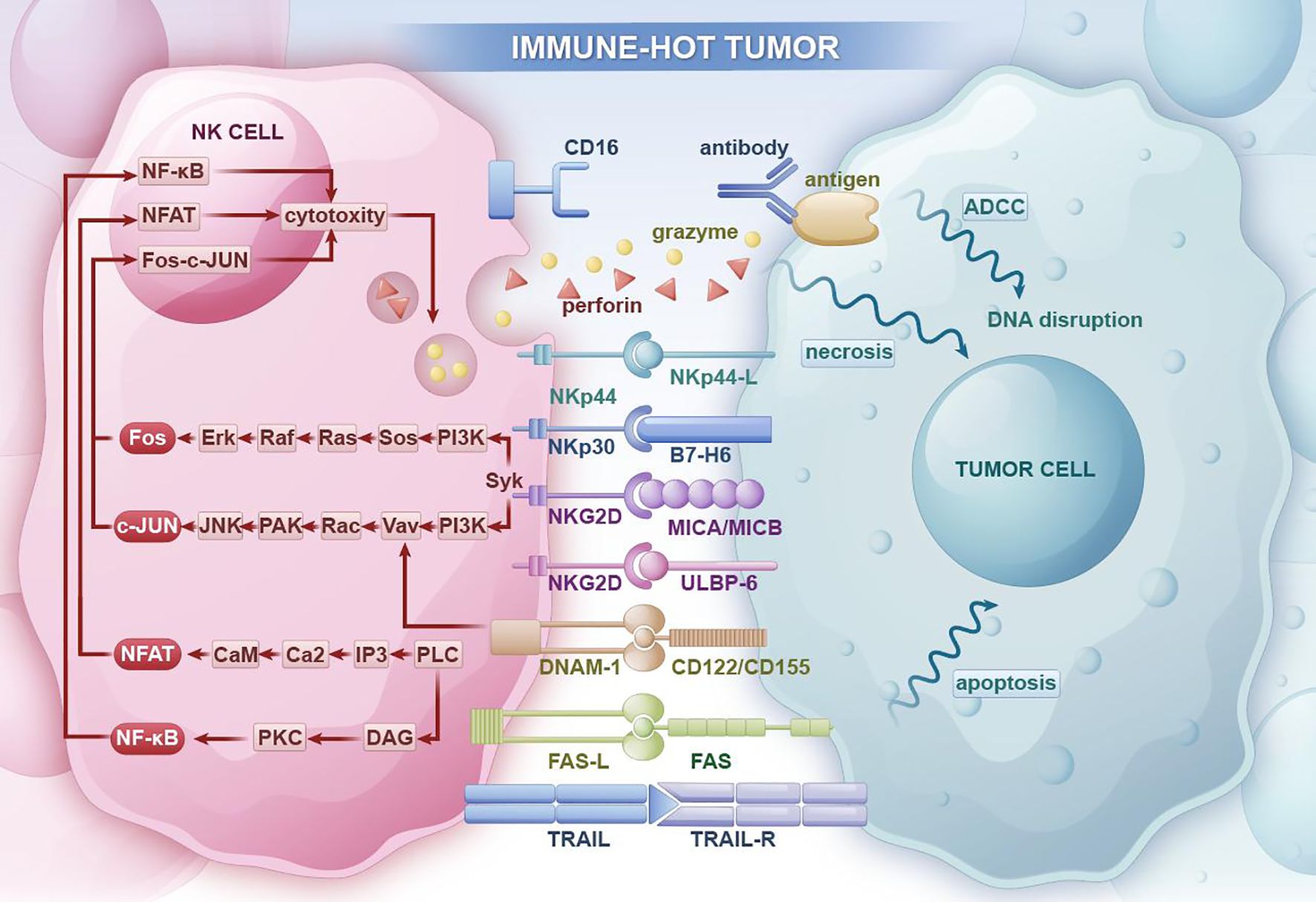
Figure 2. In immune-hot tumor regions, NK cells can promote tumor cell necrosis through antibody-dependent cellular cytotoxicity (ADCC) and the release of perforin and granzymes. They can also inhibit tumor occurrence and progression through apoptosis receptor ligands that induce tumor cell apoptosis pathways.
2 Differentiation and tissue residency endow NK cells with heterogeneity
2.1 The differentiation and development as prerequisite of heterogeneity
NK cells are known to derive from CD34+ hematopoietic stem cells (20) and progress through several developmental stages, including precursor, mature, and effector phases, ultimately forming the common NK cell populations observed in peripheral blood. This developmental trajectory is characterized by cellular differentiation into distinct NK cell types, influenced by the microenvironment and the signals they receive. The differentiation and maturation processes of NK cells contribute significantly to the heterogeneity of NK cell populations. Typically, NK cells are categorized into CD56dimCD16+ and CD56brightCD16- subsets based on the expression levels of CD56 and CD16. These subsets differ significantly in cytolytic activity, cytokine production, and proliferative capacity, and they fulfill unique roles in the immune response (21). CD56dim NK cells, which constitute approximately 90% of peripheral blood NK cells, express high levels of Fcγ receptor III (FcγRIII, CD16) and killer cell immunoglobulin-like receptors (KIR) and contain abundant perforin and granzymes, conferring high cytotoxicity but low cytokine production. In contrast, CD56bright NK cells have reduced quantities of perforin and granzymes, lower cytotoxicity, and a greater propensity for cytokine secretion, including IFN-γ, GM-CSF, and TNF-α (22, 23). This review initially approaches the formation of NK cell heterogeneity by examining the process of NK cell development, providing insights into the origins of NK cell heterogeneity.
2.1.1 The traditional view of NK cell development process
The linear model has traditionally been the prevailing perspective on NK cell development, and recent insights into ILC subsets have contributed to a more nuanced understanding of this developmental trajectory. (Figure 3) According to the linear model, hematopoietic stem cells (HSCs) differentiate into common lymphoid progenitors (CLPs). CLPs have the potential to commit to various ILC progenitor lineages, including B progenitor cells (pre-B), T progenitor cells (pre-T), NK progenitor cells (NKP), and others (24–26). CLPs subsequently differentiate through an intermediate stage of pre-NK progenitors (pre-NKPs) into NKPs (27, 28). The acquisition of the common β-chain of CD122 and IL-15 by NKPs represents a critical milestone in NK cell differentiation. The expression of CD122 signifies the irreversible commitment of NK cells from CLPs, while IL-15, produced by hematopoietic cells, monocytes, and dendritic cells, is thought to facilitate NK cell development in the bone marrow (29). Following this, NKP cells differentiate into immature NK (iNK) cells, which are characterized by increased expression of IL-1R1 and the emergence of CD314 (NKG2D), CD335 (NKp46), CD337 (NKp30), and CD161 (the human homolog of NK1.1 in mouse). The subsequent transitional stage is marked by the appearance of CD56bright NK cells, which display high levels of CD56 and peak expression of NKG2D, NKp46, NKp30, and CD161. NK cells at this stage can be subdivided into phase 4a NK cells, which express more IL-22, and phase 4b NK cells, which express higher levels of transcription factors T-BET and EOMES and are capable of producing interferon-γ and thus cytotoxicity (30). CD56bright NK cells eventually transition into CD56dim NK cells, distinguished by increased CD16 expression, decreased CD56 expression, and the expression of various CD158 (KIR) subtypes (24, 26). CD56dimCD16+ NK cells can be further classified into subpopulations based on CD57 expression, including CD56dimCD16+ NK cells and CD56dimCD16+CD57+ NK cells, with the CD56dimCD16+CD57+ subset being considered the terminally differentiated subset, generated by prolonged stimulation from antigens or inflammatory signals (26, 31, 32). As the human NK cell developmental system has been refined, the developmental process of NK cells in mouse models has also been better characterized, aligning with this linear model. A recent study proposed a four-stage developmental model for the further progression of NKP cells according to the expression of NKp46. The NKP population sequentially matures into the CD122+NK1.1−CD49b+NKp46− iNK-a population, the CD122+NK1.1+CD49b−/+NKp46− iNK-b population, and finally, the CD122+NK1.1+CD49b+NKp46+ mNK population. These four NK cell populations display distinct phenotypic differences in cell surface markers, transcription factors, and effector molecules (33).
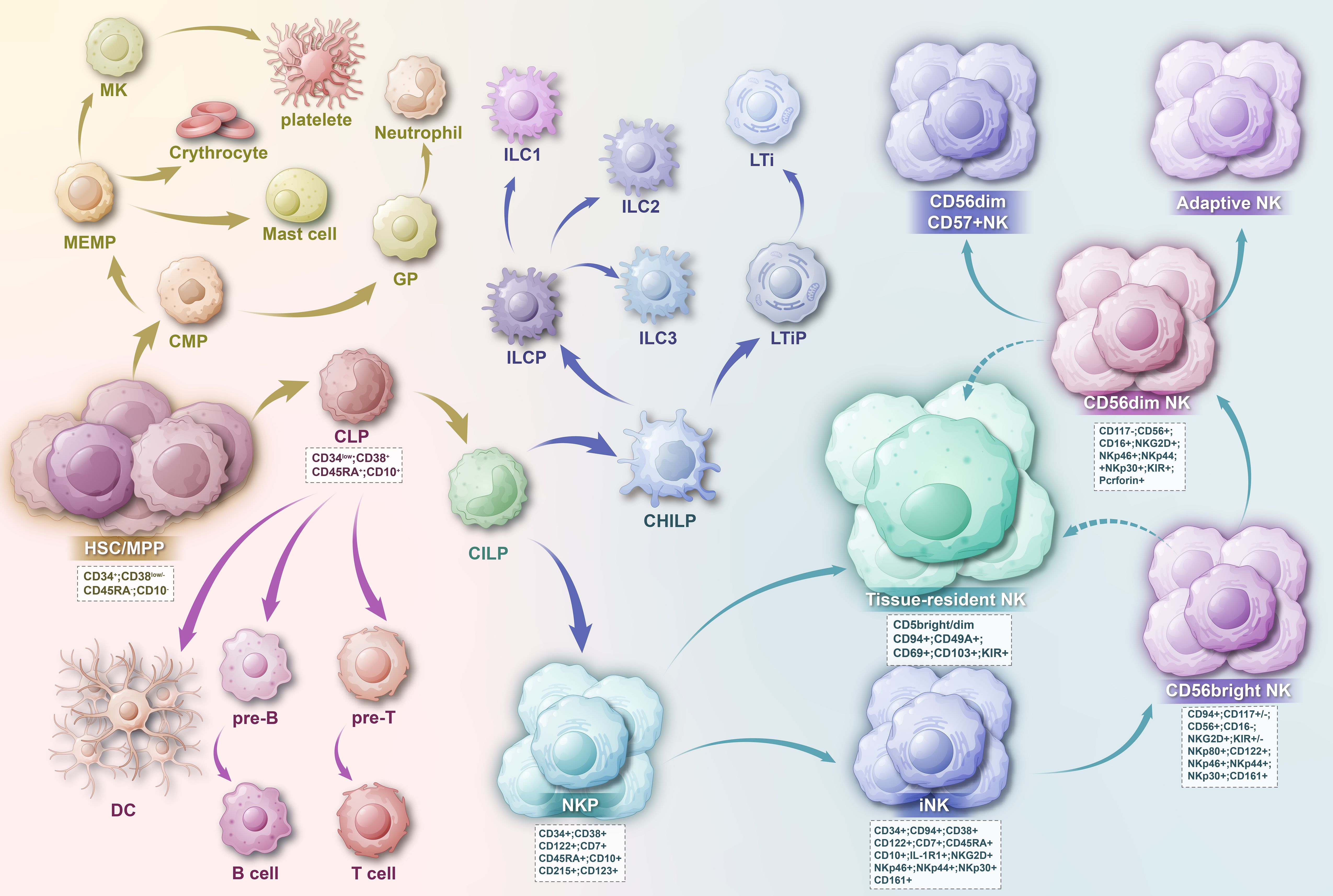
Figure 3. HSC cells develop into CMP and CLP, and the precursor of NK cells is the CHLP cell developed from CLP cell. CHLP cells then differentiate into CHILP cells, subsequently leading to the transition into ILC cells and NK cells. As NK cells approach maturity, they can migrate to tissues to form tissue-resident NK cells. The developmental outcome of NK cells is CD56dim CD57+ NK cells.
Notably, ILC cells share characteristic markers with NK cells, and the homology between them has been significantly enhanced through exploration, which has been pivotal in elucidating the developmental trajectory of NK cells and constructing the developmental lineage. It is recognized that the precursors of NK cells may diverge from those of ILC1s, ILC2s, and ILC3s. According to the development and functions of ILCs, including the production of cytokines and the expression profile of transcription factors, ILCs can be divided into five major subsets, including NK cells, ILC1s, ILC2s, ILC3s, and LTis (lymphoid tissue inducers) (34, 35). Common innate lymphoid progenitors (CILPs) are considered the intermediate stage between CLPs and the development of ILCs. CILPs can differentiate into NKPs or common helper innate lymphoid progenitors (CHILPs), which subsequently produce lymphoid tissue inducer progenitors (LTiPs) and innate lymphoid cell precursors (ILCPs). LTiPs differentiate into LTis, while ILCPs further differentiate into ILC1s, ILC2s, or ILC3s (34, 36), and NKP cells develop into mature NK cells along the well-known linear model. (Figure 3) However, this model cannot explain all experimental analytical conclusions, and many studies have supplemented this known developmental model, with many unknown directions yet to be explored. A study using global transcriptomics, flow cytometry analysis, and genetically engineered mouse models found that NK cells in the tumor microenvironment can depend on TGF-β to convert into intermediate ILC1 groups and ILC1 groups (37). Furthermore, emerging evidence suggests that previously identified ILCP cells retain the capacity to generate NK cells (38–41), indicating that the differentiation and commitment process within the ILC lineage is more intricate than previously thought. Additionally, Chen L et al. found that the CD34−CD117+ ILCP population can express CD56, produce NK cells and ILC3s, but not ILC2s. In vitro molecular and functional analysis and lineage differentiation analysis have proven that the final step of ILC2 development is independent and non-overlapping with NK cells and ILC3s (40). It is worth noting that NK cells and ILC1 together form group 1 ILCs, whose development and function rely on the T-box transcription factor T-bet and are characterized by the production of interferon-γ (IFN-γ) (34). An accurate understanding of the developmental relationships between NK cells and other cells within the ILC lineage, as well as their interactions, is essential for constructing a comprehensive NK cell developmental lineage.
Additionally, many questions remain unresolved. For example, whether there are still undefined subgroups in the development process of NK cells, from which stage NK cells separate from T/B cells and become independently developed individual cells, and the developmental process between ILCs and NK cells has not yet been precisely defined.
2.1.2 Further dissection of NK cell development process under single-cell sequencing
Deciphering the cellular state transitions behind immune adaptation is fundamental to advancing biology. Through the application of single-cell sequencing, researchers are able to track the fate of individual cells, revealing the genetic changes as cells transition from a single state to new cell states and types, which helps to understand the development process of multicellular organisms. Although single-cell sequencing has not been extensively applied to the study of NK cell development, its research achievements are still encouraging, as it is expected to construct a complete developmental system from a single-cell perspective and to deeply understand the ontogeny of NK cells. Meanwhile, scRNA technology plays a key role in exploring ILC heterogeneity (35, 42), and it is expected to provide clues for the origin and understanding of NK cells and ILCs.
scRNA-seq technology can further elucidate the reliability of the linear model, explore the continuity of the developmental process, discover new subpopulations, and define the developmental process based on gene expression patterns. Yang et al. (43) used scRNA-seq technology to study the heterogeneity of NK cells in the peripheral blood and bone marrow of healthy individuals, and transcriptomic and pseudotime analysis confirmed the progression from CD56bright NK to CD56dim NK. A study on liver cancer also identified the continuous process of transformation from CD56bright NK cells to CD56dim NK cells through single-cell analysis (44). Furthermore, Yang et al. (43) identified a transitional NK cell population that bridges CD56bright NK cells and CD56dim NK cells, characterized by intermediate expression of marker genes associated with either CD56bright or CD56dimCD57+ NK cells. More recently, Melsen JE et al. (45) discovered the CD56dimGZMK+ subpopulation, which is considered an intermediate stage between CD56bright and CD56dim NK cells. This subpopulation exhibits shared characteristics with both CD56bright and CD56dimGZMK- NK cells and displays higher expression of SELL (CD62L) and lower expression of GZMB, PRF1, FCGR3A (CD16), and FGFBP2 in comparison to CD56dimGZMK- cells.
Furthermore, single-cell sequencing has provided insights into the point of divergence of NK cells from the lymphoid lineage. A study on the single-cell transcriptomic atlas of human blood cells revealed through pseudotime analysis that memory B cells and NK cells are distributed on two different branches, which better reflects the distinct trajectories of differentiation between B cells and NK cell populations. They also found that NK cells and T cells lack continuous transcriptional transitions from progenitors to differentiated cells and inferred that these cells may deviate from T cell development during the macro-DN(Q) stage of gene expression acquisition (46). Another study, utilizing single-cell RNA sequencing and adaptive immune receptor (AIR) sequencing (scVDJ-seq), has employed TRBJ frequency to create a V(D)J feature space, as all T, ILC, and NK cells express TRBJ. The inferred trajectories suggest that ILC/NK cells diverge from T cell development at the DN (early) and DN (Q) stages, while also indicating that T cells and ILC/NK cells may share a common initial developmental stage before diverging prior to the production of productive TRB, TRG, and TRD (47). This provides better evidence for the separation of NK cells from the lymphoid lineage. Meanwhile, scRNA-seq research reveals a more continuous transcriptional differentiation landscape, although lacking classical defined precursor stages, it contributes to a more systematic understanding of NK cell development (48–50).
Notably, the relationship between the ILC and NK cell developmental lineages and the heterogeneity between them has been better authenticated under single-cell sequencing, and these studies are expected to provide clues for the origin and understanding of NK cells and ILCs. A study identified common ILC1/NK cell progenitors and referred to them as aceNKPs. Single-cell RNA sequencing of the descendants of aceNKPs in the lung confirmed that aceNKPs are not only progenitors of NK cells but also of ILC1 cells. It was found that the descendants of aceNKPs could not be separated according to the traditional dichotomy between ILC1 and NK cells, and no clear demarcation of the transcriptional profiles between the ILC1 and NK cell lineages was observed, indicating that these closely related group 1 innate lymphocytes are indistinguishable. Furthermore, a single-cell study on tissue-resident NK cells (trNK) in the skin found that trNK cells retained the expression of the transcription factor ectodermal protein EOMES and did not acquire Hobit expression, indicating that trNK cells do not transition into ILC1, and ILC1 do not convert into trNK cells (51), challenging the notion that NK cells can readily convert into ILC1. Moreover, the heterogeneity between NK cells and ILCs has been a hot topic in recent years, and a comprehensive understanding of this heterogeneity is crucial for studying the origin of both and for the identification and selection of cells in single-cell sequencing. As note, ILC cells share typical markers with NK cells, and in humans, NK cells (c-kit-) are typically distinguished from ILC3 (c-kit+) (52). In mice, the distinction between NK cells and ILC1s is often based on the expression of EOMES, with NK cells (CD49a- CD49b+) and ILC1s (CD49a+ CD49b-) being differentiated by the differential expression of integrins CD49a and CD49b (53). Lopes et al. (35) used single-cell RNA sequencing and CITE-seq to explore the heterogeneity between inter-tissue NK and ILC1 and defined syndecan-4 as a marker to distinguish mouse ILC1 from NK cells. They also found that the expression of EOMES, GZMA, IRF8, JAK1, NKG7, PLEK, PRF1, and ZEB2 defines NK cells, while IL7R, LTB, and RGS1 distinguish ILC1s from NK cells in both mice and humans. McFarland et al. (42) employed scRNA sequencing to clarify the gene signatures of mouse ILC1-NK cells obtained from tissue, tumors, and circulation, identifying unique phenotypic markers, transcription factors, and metabolic characteristics that distinguish tissue NK cells from circulating NK cells and ILC1. While these studies provide important clues for the investigation of NK cell development, it is acknowledged that the process of NK cells and ILCs developing from progenitors has not been thoroughly investigated. Therefore, it is imperative to further deepen our understanding of the origin of NK cells, as this knowledge will inform ongoing research into NK cell heterogeneity.
2.2 Tissue residency as a core factor in NK cell heterogeneity
Most of our knowledge of NK cells comes from studies on mouse spleen, bone marrow, and human peripheral blood mononuclear cells (PBMC). These cells are commonly referred to as conventional NK cells (cNK). In recent years, a distinct subset of NK cells, known as tissue-resident NK cells, has been identified and has garnered significant interest. Human NK cells typically comprise 5-15% of peripheral blood lymphocytes and are abundant in bone marrow (BM), liver, uterus, spleen, and lungs, with a smaller population also found in secondary lymphoid tissues (SLT) and mucosal-associated lymphoid tissue (MALT) (54–56). In 2016, Lughart G et al. identified a CD69+CXCR6+ NK cell subset in lymphoid tissues, which is considered an independent NK cell population with distinct phenotypes and functions (57). Unlike cNK, trNK are exclusively found in peripheral tissues. While there is phenotypic similarity between peripheral blood NK cells and trNK cells, unique transcriptional profiles indicate that the cellular environment is a key factor in NK cell heterogeneity within the same developmental stage (58). By tracing the origin of trNK cells and their unique developmental process, we can further investigate the diversity of NK cell expression and subpopulation ratios influenced by the tissue, as well as characterize the heterogeneity of tissue-resident NK cells under physiological conditions. It is important to note that previous studies have primarily focused on the heterogeneity of tumor NK cells, often overlooking the diversity of tissue cells under physiological conditions—a distinction that should not be disregarded. NK cells in the TME originate from various sources, including cNK and trNK. A comprehensive understanding of NK cell heterogeneity under physiological conditions is crucial for analyzing the differences between NK cells in the TME and non-diseased tissues, providing a significant reference for personalized immunotherapy strategies for tumors.
2.2.1 The unique generation process of tissue-resident NK cells contributes to the heterogeneity of NK cells
Previous studies have indicated that during differentiation, both immature and mature NK cells migrate from bone marrow parenchyma to sinusoids and subsequently enter the bloodstream (59, 60). The circulation of blood between lymphoid and non-lymphoid organs facilitates their entry into secondary lymphoid tissues and various other tissues at different stages of development, differentiation, and activation (54, 61, 62). Some specialized subsets are capable of returning to specific niches within the BM to fulfill functions such as control and surveillance of malignant cells (59). However, more detailed research has revealed the presence of trNK cell populations in certain tissues, which are believed to establish residency early in life without the need for microbial stimulation during development and are capable of self-maintenance (63). Moreover, due to the functional and residential similarities between ILC1 and NK cells, ILC1 found in organs has traditionally been considered a subtype of trNK cells (64). Despite sharing overlapping transcriptomic regions and dependence on IL-15, the developmental pathways of these two cell types are quite distinct (36). NK cells derive strictly from NKP cells, which are committed to producing NK cells and do not generate other ILC subsets. It is evident that the trNK cell population exhibits distinct developmental requirements from cNK cells and ILC1 and is considered unique in ontogeny (65).
Accurate knowledge of the origin of tissue-resident NK cells is crucial for understanding the roles of NK cells in various tissues and their physiological functions, and it forms the basis for further insights into the functions of NK cells under pathological conditions such as inflammation and tumors. Studies have suggested that trNK cells may originate from tissue-resident progenitor cells rather than following the bone marrow-bloodstream pathway. It has been observed that NK progenitor and immature cells mature in secondary lymphoid tissues such as spleen, lymph nodes, and tonsils (24, 25, 66). These progenitor and immature NK cells can also be inoculated into the lymph nodes and intestine, where they reside and act as reservoirs for the maintenance of NK cell populations (55). Furthermore, a study utilizing 12-color flow cytometry identified pre-NKPs, which represents an intermediate developmental stage between the upstream CLP cells and the downstream NKP cells and was previously considered the initial stage of commitment to the NK lineage. Unlike other transplant recipients, intrathymic injection of these progenitors did not produce NK cells, suggesting that thymic NK cells have a distinct developmental origin (28). Some studies have also provided evidence that DN1 thymic progenitor cells can differentiate into thymic NK cells, which are distinct from cNK cells (67). These findings indicate the presence of NK cell progenitors in the human thymus that can differentiate and develop independently to form tissue-resident NK cells within the thymus. Additionally, in mouse lymph nodes, certain NK cell subsets are thought to originate from NKP cells in the thymus, while others are thymus-independent NKP cells originated from lymph nodes, indicating the presence of NK progenitors in lymph nodes (68). The advent of single-cell sequencing technology, with its high-resolution capabilities, has further characterized the developmental process, providing compelling evidence for the existence of trNK progenitor cells. Recent studies combining single-cell transcriptomics, flow cytometry, and TCRD rearrangement data have demonstrated that non-T cell lineages, including NK cells, can develop in the human thymus (69), confirming the presence of NK cell progenitors in tissues capable of developing into trNK cells. In 2018, Eric Vivier et al. identified four subgroups in the human spleen at single-cell resolution, among which hNK_Sp3 and hNK_Sp4 cells did not resemble any blood NK subgroups and appeared to be spleen-specific (70). Three years later, they used the same technology for single-cell sequencing of the bone marrow and found that spleen NK 0 (hNK_Sp3) cells could also differentiate into spleen CD56dim NK cells and CD56bright NK cells. This contradicts the notion that NK cells develop exclusively in the bone marrow, suggesting that some precursors of NK cells are enriched in extramedullary tissues (71). Moreover, researchers have identified CD117+ ILC clusters lacking mature ILC markers in the colon and lung through single-cell sequencing, which, unlike circulating ILCs, exhibit a transcriptome consistent with tissue residency and may represent resident precursors of mature ILCs in these tissues (35). These studies confirm that the development of NK cells not only originates from the bone marrow but also involves some extramedullary tissues that harbor progenitors of the NK cell lineage, which differentiate into trNK cells with a distinct developmental process from cNK cells and present unique growth requirements.
Notably, in specific situations such as infection and tumors, some studies have also observed that cNK cells can be recruited to tissues to fulfill various roles. Recent investigations have revealed that cNK cells are redistributed and persistently located at previously virus- and bacteria-infected skin sites through a mechanism that promotes tissue retention, and they differentiate into TCF1hiCD69hi NK cells that are transcriptionally similar to CD56brightTCF1hi NK cells in human tissue (51, 65). Utilizing single-cell sequencing technology, Dean I et al. (72) discovered in mouse solid tumors that cNK cells recruited from the circulation can transform into CD49a+ NK cells that adapt to the tumor environment, establishing a tumor-resident state and expressing markers of tissue residency such as CD49a and CD69. These cells also exhibit effector functions, including the killing of cancer cells and the promotion of the recruitment and activation of dendritic cells (DCs). Another study, employing scRNA-seq, found that circulating NK cells are recruited to the salivary gland in a CX3CR1-dependent manner, where they develop into NKRM (tissue-resident memory-like NK) cells. These long-lived tissue-resident cells prevent autoimmunity by eliminating CD4+ T cells in a TRAIL-dependent manner, providing sentinel protection for the host and playing a crucial role in maintaining immune homeostasis, limiting immune responses, and preventing autoimmune damage (73).
By elucidating the origin and developmental pathways of trNK cells, we can ascertain that the tissue-resident nature of these cells may arise from pre-existing tissue-resident NK cell precursors within the tissue, thereby becoming a core element of NK cell heterogeneity. Concurrently, our research offers researchers valuable insights. For instance, in recent years, the application of NK cell therapy has expanded significantly. Beyond the challenge of poor targeting affecting NK cell efficacy, insufficient infiltration of NK cells into target tissues remains a substantial obstacle that must be addressed. Our research opens up new directions for researchers, as NK cell precursors can be used as research objects to regulate the tissue-resident capacity and infiltration level.
2.2.2 Tissue influences NK cell differentiation leading to heterogeneity
As previously discussed, growing evidence supports the notion that the tissue microenvironment influences the expression profile of NK cells. We can discern the role of the tissue environment in the development of trNK cells by comparing the environmental differences between trNK cells and cNK cells, as well as by examining the variations in trNK cells across different tissues.
Tissue-resident trNK cells and conventional cNK cells may originate from distinct precursor cells and exhibit significant disparities in their differentiation, development, and transcriptional profiles. Unique transcriptional signatures indicate that the cellular environment is a key factor in generating NK cell heterogeneity within the same developmental stage (58, 74, 75). A consistent feature among tissue-resident NK cells across various tissues is their expression of tissue-resident markers and a less mature phenotype compared to cNK cells, with their development being less reliant on the transcription factor NFIL3 (76). In both trNK cells and cNK cells, the distinct cytokine milieus influence their activation profiles. IL-21R is highly expressed in trNK cells, while IL-18R is predominant in cNK cells, and their respective ligands, IL-21 and IL-18, primarily facilitate the activation of trNK and cNK (77). Notably, cNK cells are absent in NFIL3-deficient mice, whereas trNK cells persist in the liver, skin, and uterus of these mice, suggesting differential requirements for transcription factors during development (78). The different tissue environments also result in distinct cytokine expression profiles between trNK cells and cNK cells. For instance, liver trNK cells exhibit higher expression of TNF-α and GM-CSF compared to cNK cells (78). A recent study on liver cancer demonstrated that the local immunosuppressive microenvironment during the chronic liver injury phase affects peripheral NK cells but does not impact tissue-resident NK cell subsets (79). This suggests that different NK cell types exhibit varying tolerances to the immune microenvironment.
Additionally, significant variations exist among NK cells across different tissues, which are evident in the differential distribution of NK cell subsets, as well as in the disparities in phenotype, function, and transcription factor requirements of NK cells in various tissues. These differences reflect the impact of the local microenvironment on the acquisition of tissue-specific traits by trNK cells (76). Studies have demonstrated that the abundance of NK cells varies across different tissues. The prevalence of CD56dimCD16+ NK cells expressing CD57, a marker of terminal differentiation, differs between locations, with higher frequencies observed in tissues rich in CD56dimCD16+ NK cells such as blood, spleen, bone marrow and lung, and lower frequencies in locations like lymph nodes and intestines (55). In humans, CD56brightCD16− NK cells exhibit tissue-resident traits in multiple locations, while CD56dimCD16+ NK cells display characteristics of circulating cells in blood, spleen and bone marrow but can adopt a resident phenotype in lymph nodes and mucosal tissues. This distribution suggests that NK cells play a role in controlling viral infection and tumor progression in tissues accessible to circulation, while intestinal and secondary lymphoid sites may be less conducive to NK cell-mediated immune surveillance (55). Moreover, the expression profile of NK cells differs significantly across tissues. A recent study on mouse utilizing scRNA-seq and flow cytometry identified gene expression patterns in NK1.1+NKp46+ cells across various tissues, revealing substantial differences in the proportion of marker-positive cells in blood, spleen, inguinal lymph nodes, CD49α− liver, intestinal epithelial lymphocytes, intestinal lamina propria lymphocytes, liver, salivary gland, and uterus (42). In blood and other tissue sites, there is a bias towards the expression of GZMB in CD56dim cells. Compared to CD56dim NK cells in lymph nodes and intestines, the frequency of GZMB+ in CD56dim NK cells in the lungs is significantly higher (55). Similarly, the transcriptional characteristics, phenotype, and function of ILCs are inherently influenced by their tissue microenvironment (80). Furthermore, the expression of transcription factors in trNK cells varies between species. For instance, trNK cells in livers of human and pigs express high levels of EOMES and low levels of TBox transcription factor 21 (TBX21), while trNK cells in mouse livers express high levels of TBX21 and low levels of EOMES (81). Tissue-resident NK cells exhibit significant differences in gene expression compared to conventional NK cells, and there is also substantial heterogeneity in receptor expression. (Table 1) These findings highlight the role of the tissue microenvironment in shaping the distinct expressions and heterogeneity of NK cells.
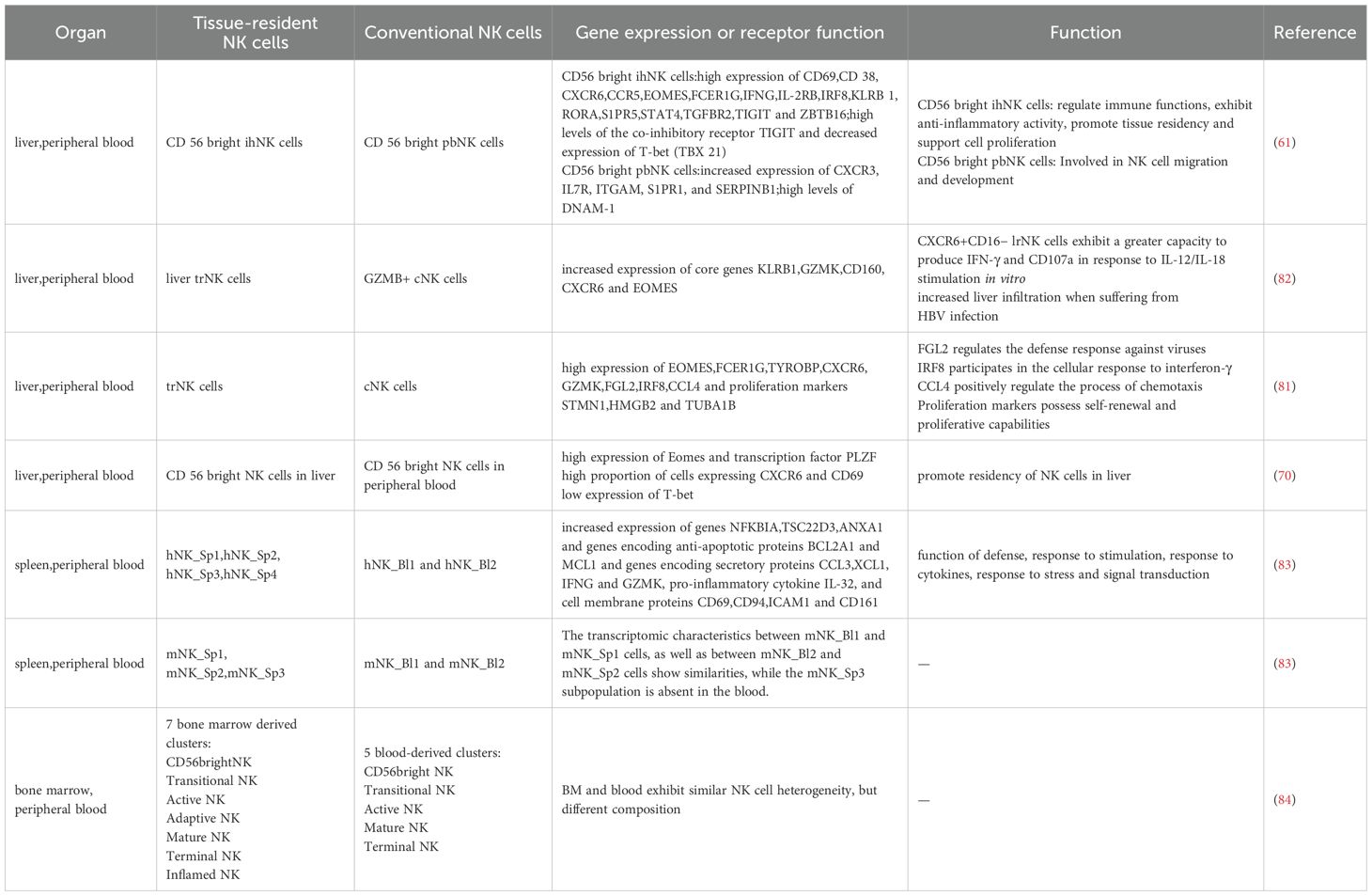
Table 1. The differential gene expression and receptor heterogeneity between tissue-resident NK cells and conventional NK cells under single-cell sequencing technology.
3 Advances in the study of NK cell heterogeneity through single-cell sequencing
3.1 Advances in the study of NK cell heterogeneity in physiological conditions through scRNA-seq technology
NK cells’ capacity to recognize and eliminate malignant or infected cells through germline-encoded and nonclonal receptors offers clinical promise. However, this straightforward paradigm is complicated by the considerable heterogeneity observed within NK cell populations. Extensive research has been dedicated to understanding the heterogeneity of trNK cells (54, 61). Single-cell sequencing technology, particularly single-cell transcriptomics, offers an unbiased approach to elucidating cellular heterogeneity and cell states (82), thereby providing the technical framework necessary for identifying cell subpopulations and for gaining a deeper understanding of the heterogeneity of trNK cells both between and within different tissues. In this review, we summarize the recent studies on NK cell heterogeneity facilitated by single-cell sequencing, which contributes to a more profound understanding of the impact of tissue microenvironments on NK cell expression and the adaptive changes of NK cells. This knowledge enhances our grasp of the role of NK cell distribution and expression in physiological functions and establishes a foundation for investigating NK cell heterogeneity in the tumor environment.
In 2018, Adeline Crinier et al. conducted scRNA sequencing on spleens and peripheral blood of humans and mice, respectively, and compared the results. They identified hNK_Sp3 and hNK_Sp4 in human spleen and mNK_Sp3 in mouse spleen, which were not present in peripheral blood. These cells exhibited high expression of anti-apoptotic proteins and pro-inflammatory cytokines, indicating that tissue-resident NK cells possess enhanced defense responses, sensitivity to stimuli, cytokine responsiveness, stress tolerance, and signal transduction capabilities, all of which are associated with high NK functional activity (70). Another study revealed that compared to peripheral blood, bone marrow NK cell-derived clusters exhibited two additional derived clusters, termed “adaptive NK cells” and “inflammatory NK cells”, suggesting a functional diversity and inter-group heterogeneity (43). In 2020, Zhao J et al. (83) demonstrated that liver-resident NK cells exhibited increased expression of core genes KLRB1, GZMK, CD160, GZMK, and CXCR6 compared to GZMB+ cNK cells and displayed a greater capacity to produce CD107a and IFN-γ in response to IL-12/IL-18 stimulation in vitro, as well as enhanced infiltration in HBV-infected livers. In 2023, a single-cell sequencing study on pig liver revealed that trNK cells, compared to peripheral blood, showed increased expression of EOMES, FCER1G, TYROBP, GZMK, and CXCR6, as well as high expression levels of fibrinogen-like 2 (FGL2), interferon regulatory factor 8 (IRF8), and CCL4, which are involved in positive regulation of the defense response to viruses, the response to IFN-γ, and chemotaxis. Additionally, they also highly expressed proliferation markers HMGB2, STMN1, and TUBA1B, indicating that these cells possess proliferation and self-renewal capabilities (81).
At the same time, researchers have further classified NK cells based on their expression patterns, further exploring the heterogeneity of different NK cell subpopulations within tissues. Smith SL et al. (84) utilized scRNA-seq to identify seven blood NK subgroups (cluster0.1.2.3.4.6.8), each displaying distinct expression patterns of cytokines and receptors. These subgroups were categorized based on gene expression as CD56dim NK cells, terminally differentiated CD57+ NK cells, CD56bright NK cells, CD56-CD16+CD7+ NK cells, CIML NK cells, and cells undergoing senescence or death. The researchers also divided bone marrow NK cells into seven bone marrow-derived clusters based on their expression patterns and further investigated their respective functions. Among these, “inflammatory NK cells” respond to interferon stimulation and promote positive regulation of the inflammatory response; “activated NK cells” respond rapidly to stimulation; “adaptive NK cells” can enhance the positive regulation of gene sets involved in endoplasmic reticulum-to-Golgi transport vesicle formation and IFN-γ production, while “mature NK cells” exhibit potent cytotoxic functions 43. Moreover, the proportion and function of NK cell subpopulations can vary under different physiological conditions. Studies have shown that the proportion of NK/ILC1 cells in the uterus of pregnant mice is higher than that in non-pregnant mice, and the expression of GZMB, PRF1 and SPP1 is increased in the trNK#3 and Mki67+ trNK#3 subgroups, which is associated with cell lysis, endoplasmic reticulum stress response, and regulation of tissue remodeling (85). Notably, new subpopulations also emerge following HIV infection (86).
The study of NK cell heterogeneity under physiological conditions has delineated the differences between trNK cells and cNK cells, and it has detailed the diversity of NK cell subpopulations within various tissues. (Table 2) This research also suggests that when examining NK cell heterogeneity in the tumor microenvironment, it is crucial to distinguish between the differences between tumors and peripheral blood, as well as between tumors and unaffected normal tissues, thereby avoiding a simplistic view of the tumor microenvironment and fostering a more comprehensive, multi-dimensional understanding of these differences. In recent years, single-cell sequencing has primarily focused on the liver, spleen, uterus, and bone marrow, while organs such as the lung, thymus, lymph nodes, tonsils, kidney, and adipose tissue have been less explored. Previous studies have analyzed the heterogeneity of NK cells in these organs (57, 75, 93), and single-cell sequencing technology is expected to provide a more comprehensive and deeper understanding in the future.
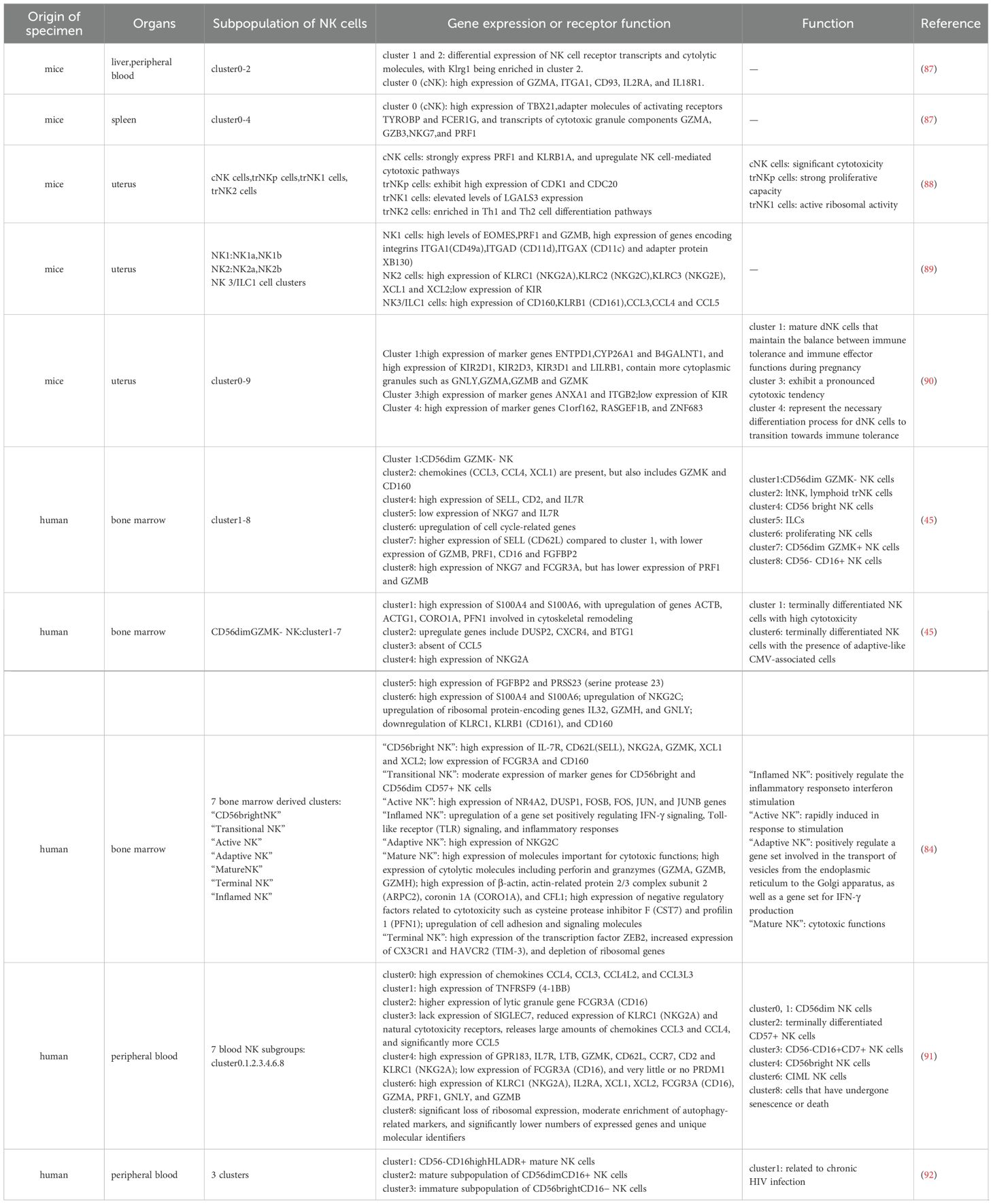
Table 2. The classification of NK cell subpopulations and their corresponding functions in various tissues under non-tumor conditions using single-cell sequencing technology.
3.2 Advances in the study of NK cell heterogeneity in the tumor microenvironment through scRNA-seq technology
Traditional methods are insufficient for accurately assessing the functional state of individual NK cells within tumors. Therefore, higher resolution techniques are necessary to delineate the heterogeneity of NK cell subsets, their spatial distribution, and functional status within TME. scRNA-seq possesses the capability to identify specific cell populations and subpopulations, making it a potent tool for analyzing the heterogeneity present in the TME (42). (Figure 4).
Figure 4. NK cells in tumor tissues exhibit expression differences not only compared to peripheral blood but also show significant differences compared to adjacent normal tissues. This disparity underscores the necessity to focus on the distinctions between tumor tissues and adjacent normal tissues. The diversity of subpopulations within tumors and the heterogeneity of expression among different tissues contribute to the overall heterogeneity of NK cells in the TME.
3.2.1 Functional analysis of NK cell subsets and identification of new subgroups
scRNA-seq enables the detection of heterogeneity between cells, allowing for a more detailed classification of NK cells that is not achievable with bulk sequencing methods. (Table 3) Recent studies have utilized scRNA-seq to analyze the pan-cancer landscape of NK cells. Tang F et al. identified five subgroups of CD56brightCD16low NK cells and nine subgroups of CD56dimCD16high NK cells and compared them with adjacent non-tumor tissues. The study revealed reduced expression of the cytokines CCL3 and CCL4 in the c3-CCL3 NK cells of tumor tissues and decreased expression of XCL1 and XCL2 in the c5-CREM NK cells, indicating their functional transition within tumors (100). Xu L et al. characterized the heterogeneity of NK cells in the breast TME under single-cell analysis, categorizing NK-0 to NK-5 as “memory-like” NK cells, NK cells with reduced IFN-γ production, CD56dim bone marrow NK cells, tissue-resident NK cells, activated NK cells involved in direct anti-tumor responses, and NK cell activity inhibitors (101). Song G et al. sequenced NK cells in HBV/HCV-related hepatocellular carcinoma and non-tumor liver tissue, and divided them into six subgroups (NK_c1, 2, 3, 4, 5, 6). Based on differential gene expression, they defined an terminal NK subgroup (NK_c1), an exhausted NK subgroup (NK_c4), and two undefined NK subgroups (NK_c2 and NK_c6) and noted that NK_c5 was absent in the non-HBV population and played an additional antiviral role (44). Liu H et al. used single-cell sequencing to compare NK cells in liver cancer with NK cells in normal liver tissue and found that the L3-NK-HLA and L4-LrNK-FCGR3A subgroups were absent in tumor tissues, while L2 and L5 subgroups were significantly increased. They observed high expression of inhibitory receptor genes TIGIT, NKG2A, TIM3, and CD96 in L1-NK-CD56bright cell, TIM3 and CD96 in L2-NK-CD56dim cells, and KLRC1 and TIGIT in L5-LrNK-XCL1 cells. Furthermore, common tumor-related genes (RHOB, TALDO1, TKT, and HLA-DPA1) are overexpressed in L5-LrNK-XCL1 cells, exhibiting inhibitory anti-tumor activity (87). A study re-clustered all NK cells from 40 tumor samples and adjacent normal tissues in non-small cell lung cancer (NSCLC) patients into 12 subgroups (c0-c11), showing differential expression of cytotoxic markers and varying enrichment in lung squamous cell carcinoma(LUSC) and lung adenocarcinoma(LUAD) (88). Another study conducted cluster analysis on NK cells in triple-negative breast cancer (TNBC), HER2+ and luminal-like breast cancer and divided them into six groups. They defined two cytotoxic NK cell subgroups and two exhausted NK cell subgroups as well as two NKT cell groups. The study found that TNBC had a higher proportion of cytotoxic NK cells and an increased expression level of “cytotoxic” marker genes (GZMA, GZMB, and CST7) in NK_c03_Cytotoxic_GZMB while HER2+ and luminal-like breast cancer lost this feature, which provided new insights for NK cell immunotherapy (89). Additionally, de Andrade LF et al. sequenced NK cell population in melanoma metastases and defined seven NK cell subgroups. They then categorized them into proliferating NK cells, high cytotoxic NK cells and low cytotoxic NK cells based on their expression (90).
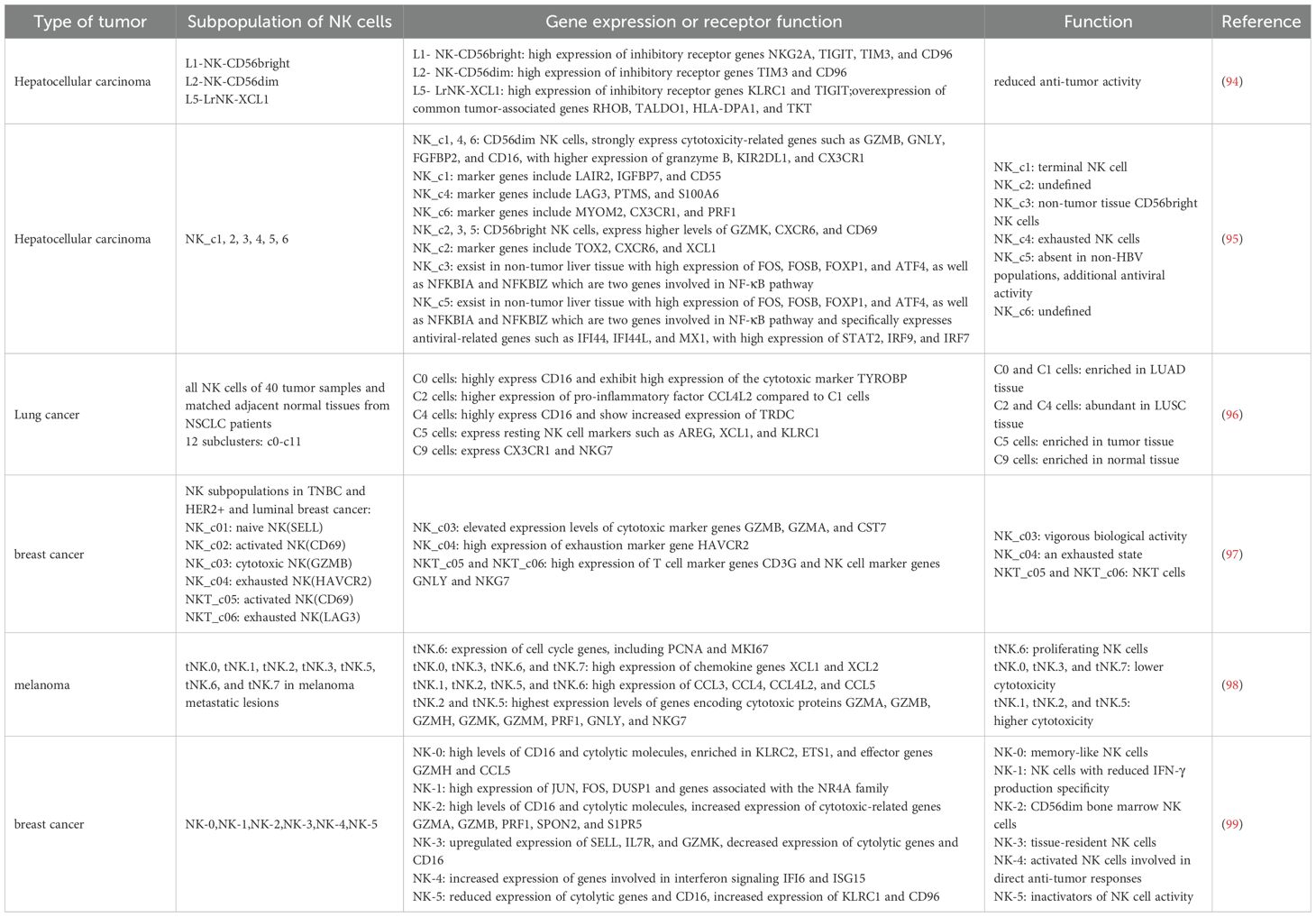
Table 3. The classification of NK cell subpopulations and their corresponding functions in different tumors under single-cell sequencing technology.
Additionally, single-cell sequencing technology can also identify new subgroups, thereby laying the foundation for a deeper understanding of tumor microenvironment changes. As previously mentioned, Tang F et al. (100) identified a c6-DNAJB1 subgroup (TaNK cells) specifically enriched in tumors. This subgroup exhibited significantly higher expression of transcripts such as KLF6 and EGR3, which are associated with the inhibition of cytotoxic function and inferred to represent a terminal state based on RNA velocity analysis. A high abundance of TaNK cells in tumors is associated with poor prognosis in cancer patients. Ding S et al. (89) observed a unique subgroup of SOCS3 highCD11b-CD27- immature NK cells that were found only in TNBC samples. These cells exhibited reduced cytotoxic granule characteristics and were responsible for activating cancer cells through Wnt signaling pathway in mice, thereby promoting tumor progression. Another study revealed that peripheral circulating NK cells differentiate along two distinct pathways within the tumor microenvironment, leading to different final states. One of these pathways results in a phenotype similar to intraepithelial ILC1s, which represents the NK cell phenotype with the highest anti-tumor activity (91). Furthermore, Lin Q et al. (92) discovered a CD8+ NK cell population specific to the TME of nasopharyngeal carcinoma, providing new insights into the complexity of the immune landscape in nasopharyngeal carcinoma.
3.2.2 Functional heterogeneity of NK cell populations within TME
Single-cell RNA sequencing has the capability to generate single-cell level maps of TME, thereby providing the most comprehensive composition and transcriptomic information of immune cell populations. By utilizing scRNA-seq, researchers can summarize and analyze the differences between tumor and normal tissue, the diversity of TME maps across different subtypes of the same tumor, and the variations within different regions of tumors and at various stages of tumor progression.
Xiao J et al. (102) utilized spatial and single-cell transcriptomics to uncover the tumor heterogeneity of colorectal cancer, revealing that the proportion of NK cells and B cells in tumors was reduced by approximately 0.3-0.4 times compared to normal tissue. A study on melanoma showed that cell clusters with shared key genes and similar transcriptional characteristics had significantly different proportions in tumors. Tumor-infiltrating NK cells exhibited significantly higher expression of FOS and JUN compared to blood NK cells, and their expression of the KLRC1 gene (encoding the inhibitory receptor NKG2A protein) also elevated (90). In lung cancer, the c5 cell cluster enriched in lung cancer tissues expressed resting NK cell markers XCL1, KLRC1 and AREG, while the c9 cell cluster expressing CX3CR1 and NKG7 was abundant in normal tissue (88). In lung adenocarcinoma lesions, NK cells were the most abundant immune cell lineage compared to non-affected lung tissue (nLung), with the CD16+ NK cell subset showing the most significant reduction. NK cells infiltrating tumor lesions expressed higher levels of CXCR3, which is necessary for NK cell infiltration, while lower levels of GZMB and CD57, and reduced IFN-γ production, indicated their decreased cytolytic activity (99). Furthermore, overall exhaustion of NK cells was observed in neuroblastoma and lung adenocarcinoma metastasis, indicating that the tumor immune response shifts towards immunosuppression (94, 95).
Additionally, related studies have also focused on the differences in TME maps between different subtypes of the same tumor category. Studies have demonstrated significant differences in the distribution and expression of NK cells in various types of lung cancer, including LUAD and LUSC. Specifically, c0 and c1 NK cell clusters were abundant in LUAD tissues, while c2 and c4 NK cell clusters were enriched in LUSC tissues (88). De Zuani M et al. also observed reduced cytotoxicity of NK cells in LUAD and LUSC, with significant differences in the co-expression of various immune checkpoint inhibitors (96). A study on human lung adenocarcinoma analyzed two distinct microenvironmental patterns formed by the transcriptomes of heterogeneous cancer cells. The inert N3MC microenvironment, compared to the immunologically activated CP2E microenvironment, contained normal-like myofibroblasts, non-inflammatory monocyte-derived macrophages, NK cells, bone marrow-derived dendritic cells, and conventional T cells, and was associated with a favorable prognosis (83). A study on mouse gliomas revealed increased infiltration of CD4+ T cells, CD8+ T cells and NK cells in low-grade gliomas (LGGs), while high-grade gliomas (HGGs) lacked this infiltration (98). Conversely, the abundance of neutrophils, T cells, and NK cells in samples from human high-grade gliomas showed an increasing trend compared to low-grade gliomas (103), suggesting that this trend may be related to the staging and malignancy of tumor progression and is independent of species differences. Ding S et al. (104) analyzed tumor samples from patients with TNBC and non-TNBC, finding that HER2+ and luminal-like breast cancer exhibited significantly lower levels of cytotoxic NK cells compared to TNBC, indicating a pro-tumor environment of NK cell exhaustion, and suggesting that these patients may benefit more from NK-based immunotherapy.
Furthermore, the enrichment and expression of NK cell subpopulations vary across different spatial areas within tumor tissue. A study examining the immune microenvironment of cervical squamous cell carcinoma (CSCC) revealed that different tumor areas exhibited varying degrees of cell infiltration. High-metabolic tumor areas displayed stronger signals for CD56+ NK cells and immature dendritic cells, while low-metabolic tumor areas showed stronger signals for eosinophils, immature B cells, and Treg cells (97). A single-cell study on nasopharyngeal carcinoma by Chen YP et al. (105) demonstrated that immune cells, particularly CD57+ NK cells, CD20+ B cells and IL3RA+ plasmacytoid DCs (pDCs), were more abundant in the tumor stroma compared to tumor epithelial nests.
What’s more, the level of NK cell infiltration and expression is dynamic and changes with tumor progression, recurrence, and metastasis. Studies have shown that chromosomal instability (CIN) in the precancerous stage can activate atypical NF-κB signaling, leading to anti-tumor immunity by NK cells. However, when CIN exceeds a certain threshold, the functional capacity of NK cells is compromised, and their effector capabilities are diminished compared to tumors with low CIN, thus creating a permissive microenvironment for tumor growth (104). Zhu J et al. characterized the cellular map of lung adenocarcinoma invasion trajectories through integrated analysis of single-cell RNA sequencing and spatial transcriptomics. The study revealed that cancer cells, NK cells, and mast cells were co-located in the cancer area in adenocarcinoma in situ (AIS), and NK cells and B cells increased during the progression from AIS to minimally invasive adenocarcinoma (MIA), enriching the study of cell ecology and spatial niches involved in the dynamic and sequential processes from AIS to MIA and subsequent invasive adenocarcinoma (IAC) (106). Tumor recurrence and metastasis are associated with shifts in the TME. In the study of chordomas, it was observed that NKT and NK cells had higher scores in recurrent chordomas compared to primary chordomas, and NK cells exhibited higher exhaustion scores in primary chordomas, providing a more comprehensive understanding of the microenvironment of primary and recurrent chordomas (107). Deng JY et al. analyzed the transcriptomic characteristics of metastatic NSCLC and found that the lung lesion group was enriched in NK cells and NK-mediated cytotoxicity and T-NK cell receptor signaling pathways, with NK cells, helper T cells, IFN-γ-related T cells, and proliferating T cells exhibiting higher cytotoxic activity than that of liver metastasis group (108). Similarly, the number and proportion of immune cells with tumor immune function, such as CD8+ T cells, NK cells, and monocytes, were significantly reduced in prostate cancer lymph node metastases compared to primary prostate cancer lesions (109). Gene mutation subtypes also influence cellular infiltration and transcriptional characteristics. BRAF mutant melanomas showed a significant reduction in the infiltration of CD8+ T cells and macrophages compared to BRAF wild-type melanomas, but an increase in the infiltration of NK cells, B cells, and NKT cells (110).
3.2.3 NK cell-related intercellular interaction
The dynamic and heterogeneous nature of the tumor microenvironment contributes to insufficient infiltration and diminished efficacy of NK cell immunotherapy, which is a significant factor in tumor progression. Understanding the interactions between NK cells and other cells in the TME provides new insights for optimizing NK cell immunotherapy. Single-cell sequencing technology presents complex interactions of NK cells in a higher dimension, providing crucial insights for the selection of NK cell immune checkpoints and the development of drug intervention strategies.
Firstly, interactions between NK cells and tumor cells can inhibit the cytotoxic function of NK cells, enabling tumor cells to evade immune surveillance and promoting tumor recurrence and metastasis. (Figure 5) NKG2A, an inhibitory receptor on NK cells, acts as an immune checkpoint receptor. Jiang H et al. (111) utilized single-cell sequencing technology to reveal that NK cells exhibited the highest expression of NKG2A, with over half of NK cells in TME expressing KLRC1. Malignant cells interact with NK cells through the HLA-E-NKG2A pathway, which presents new opportunities for the clinical treatment of gastric cancer. In the study of pancreatic ductal adenocarcinoma circulating tumor cells (CTCs), Liu X et al. also demonstrated that CTCs interact with NK cells via the immune checkpoint molecule HLA-E-NKG2A, protecting CTCs from NK-mediated immune surveillance and thereby promoting the metastasis of pancreatic ductal adenocarcinoma (112). CD39, a transmembrane protein encoded by the ENTPD1 gene, in synergy with CD73, can convert ATP and ADP into the immunosuppressive adenosine through a cascade reaction. Liu L et al. (113) found that the expression of CD39 and LAG3 of NK cells was significantly upregulated in most tumor samples, creating an immunosuppressive state in the TME. Cao G et al. (114) conducted cell interaction analysis to investigate the potential mechanisms underlying T/NK cell exhaustion and identified poliovirus receptor (PVR)-like protein signaling as the primary co-inhibitory interaction between NK cells (CD96 and TIGIT) and malignant cells (PVR/CD155 and NECTIN1). Recent single-cell RNA sequencing data suggests that high expression of platelet-related genes in circulating tumor cells CTC directly upregulates CD155 expression on tumor cells through platelet adhesion. This upregulation protects CTCs from NK cell killing through the interaction between CD155 and the immune receptor TIGIT, thereby promoting the metastatic dissemination of tumor cells (115). The functional state of NK cells in the TME is significantly suppressed, while unsuppressed NK cells can exert cytotoxic effects and resist tumors. A study using scRNA-seq to analyze various immune cells within tumors found that NK cells in tumor sites expressed cytotoxic molecules such as GZMA, PRF1, and chemokines XCL2, CCL5, CCL3, and CCL4, indicating the potential for an antitumor response. Additionally, NK cells also expressed several inhibitory and co-stimulatory molecules, including CD96, TNFRSF18, and KIR2DL4, representing targets for regulating their function (116).
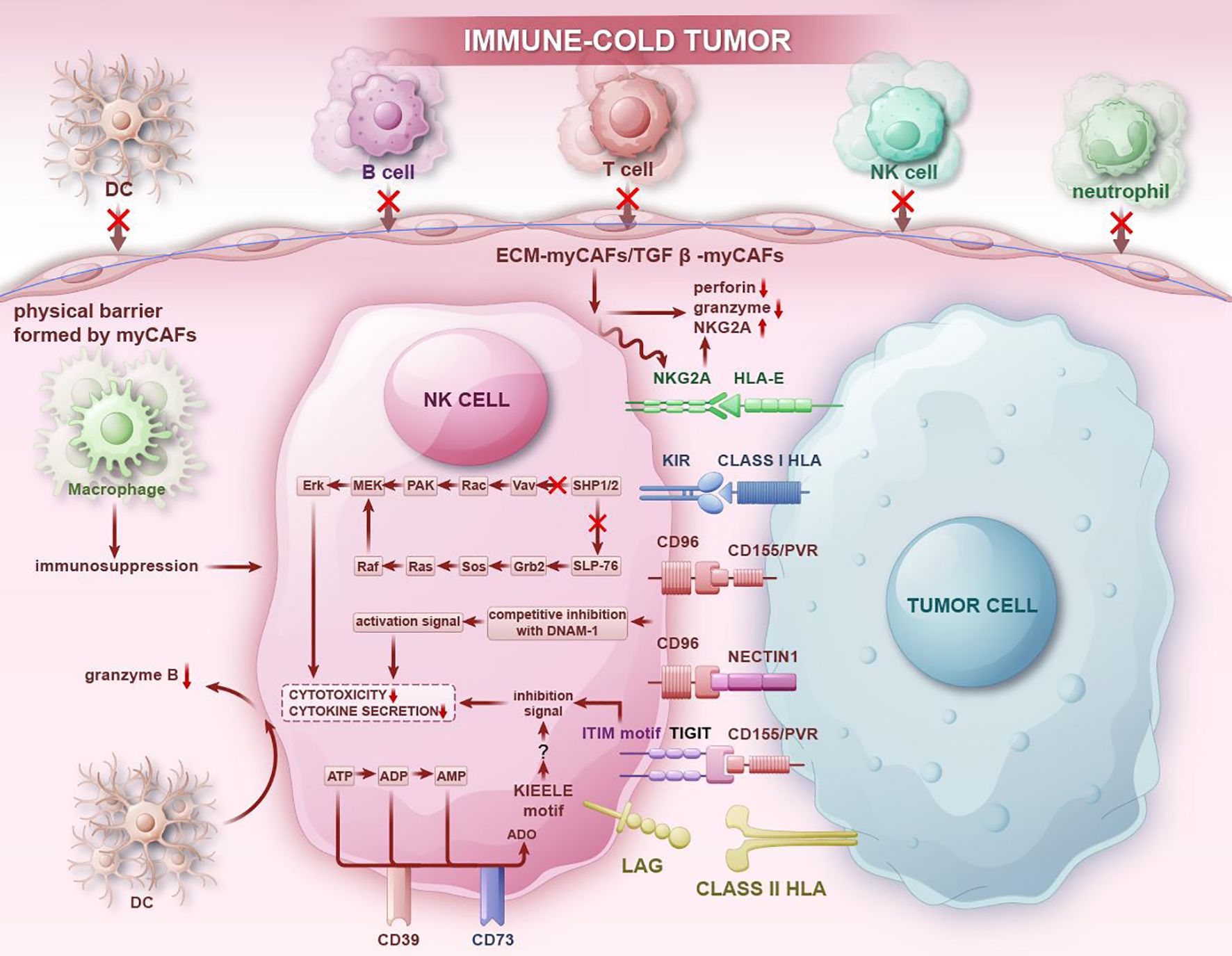
Figure 5. In immune-cold tumor regions, NK cells transmit immunosuppressive signals through immunosuppressive receptors and interact with immune cells in the immunosuppressive TME, thereby reducing the secretion of cytokines and decreasing cytotoxic activity, allowing tumors to evade NK cell attacks. At the same time, interactions between NK cells and other immune cells inhibit the chemotaxis of other immune cells, promoting tumor progression and the formation of an immunosuppressive microenvironment.
NK cells can also interact with immune cells, forming a complex immune network that together constructs an immunosuppressive tumor microenvironment. The study of NK cell-immune cell interactions provides foundational information for deepening the understanding of the immune network and immunotherapy. Research has found a strong negative correlation between the frequency of STAB1+ Mφ/AI Mφ and T/NK cells, further validating that Mφ acquire an immunosuppressive phenotype and inhibit NK cell infiltration in non-small cell lung cancer (96). Using single-cell transcriptomic analysis, Kim HJ et al. (117) found that CD169+ macrophages in human and mouse gliomas produce pro-inflammatory chemokines which can lead to the accumulation of NK cells, and NK cell-derived IFN-γ is crucial for the accumulation of CD169+ macrophages derived from blood monocytes in gliomas. In a single-cell sequencing study of primary tumors and adjacent non-tumor samples from different organs or tissues and metastases, Jiang H et al. (111) found that when macrophages and NK cells were co-cultured, treatment with an anti-NKG2A antibody inhibited HLA-KLRC1 (NKG2A) signaling and the expression of IFN-γ was significantly improved. Additionally, researchers showed through scRNA-seq of glioblastoma tumor-infiltrating immune cells that the loss of Dicer in macrophages reduced the proportion of cluster 3 of glioma-associated macrophage (GAM) and reprogrammed the remaining GAM to a pro-inflammatory activation state, interacting positively with tumor-infiltrating T cells and NK cells through TNF paracrine signaling, creating a pro-inflammatory immune microenvironment for tumor cell elimination (112). A study compared receptor-ligand pairs in LUAD and LUSC to evaluate the interaction strength between immune cell subsets in the tumor microenvironment and found that the SPP1 pathway showed greater interaction between SPP1-Mφ clusters and NK cells in LUSC than in LUAD, and further analysis revealed different pathways in LUSC, such as TIGIT and cell proliferation-related LCK, which can enhance the interaction between NK cells and SELENOP-Mφ together with SPP1-Mφ clusters (88). Similarly, the interactions between NK cells and other immune cells, including cancer-associated fibroblasts (CAF) and dendritic cells, have been thoroughly investigated. A study deconvoluted spatial transcriptomics at the single-cell level and performed functional analysis to generate a high-resolution integrated map of breast cancer. The study found that co-culture of FAP+ CAF clusters with NK cells reduced the levels of perforin and GZMB in NK cells and significantly increased the proportion of CD56high NK cells and their surface NKG2A levels, thereby reducing the cytotoxicity of NK cells (118). Additionally, Ou Z et al. used single-nucleus RNA sequencing (snRNA-seq) and spatial enhanced resolution omics sequencing to study the immune microenvironment of CSCC and found that the number of B cells, T cells, NK cells, neutrophils, DCs and Th1 cells was significantly reduced in myCAF+ tumors, proving that myCAF may act as a physical barrier to prevent the infiltration of pro-immune cells (97). What’s more, mature cDCs (LAMP3+ DCs) which are recently characterized showed the strongest interaction with CD56dim NK cells. Tumor-infiltrating LAMP3+ DCs showed lower IL-15 expression compared to those in non-tumor tissues, and NK cells physically close to LAMP3+ DCs expressed lower levels of GZMB, suggesting that LAMP3+ DCs have a compromised activation effect on CD56dim NK cells in TME (100).
4 Drug intervention and resistance mechanism research guide new treatment strategies
4.1 Pharmacological and non-pharmacological intervention mechanisms
Recent studies on drug mechanisms have shown that certain pharmacological and non-pharmacological interventions can inhibit tumor growth, reduce tumor invasion, and improve prognosis. This effect is closely associated with the activation of NK cells within the tumor microenvironment and the interactions between NK cells and other cells. Notably, combination therapy also demonstrates significant advantages.(Table 4).
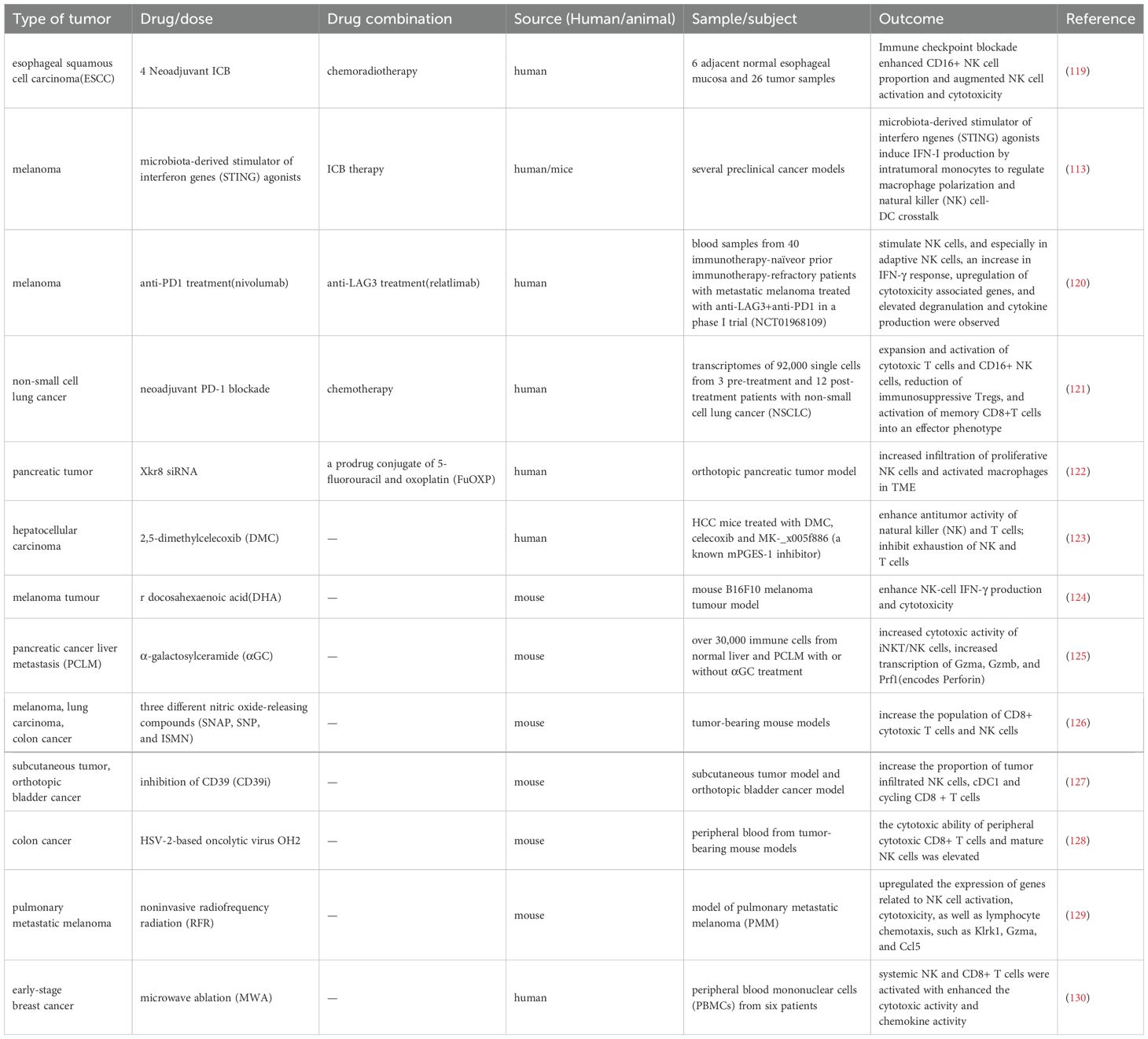
Table 4. The application of single therapy and combination therapies influences activation of NK cells within the TME, thereby affecting the therapeutic efficacy against tumors.
4.1.1 Immune checkpoint inhibitors
Immune checkpoint blockade (ICB) can disrupt the immune surveillance of cancer cells and has significantly transformed the landscape of cancer treatment (131). A research conducted single-cell RNA and T cell receptor sequencing on PBMCs from 60 patients with stage IV non-small cell lung cancer and divided NK cells into CD56high NK cells (c1), CD56low NK cells (c2), and CD56low NK_IFNG cells (c3). The results revealed that after immune checkpoint inhibitor (ICI) treatment, the proportion of the c1 subgroup increased, and in the durable clinical benefit (DCB) group after ICI treatment, the proportion of the c3 subgroup also increased (132). Another study on esophageal squamous cell carcinoma analyzed the dynamic changes in the tumor immune microenvironment and found that immune checkpoint blockade enhanced the activation and cytotoxicity of NK cells by increasing the frequency of CD56dimCD16+ NK cells (133). Shang J et al. (134)analyzed single-cell transcriptomic data from patients with non-small cell lung cancer treated with anti-PD-1and they observed six different NK cell clusters, among which immature NK cells were enriched in the responder group and expressed a series of marker genes associated with anti-PD-1 response and were related to antigen processing, Th1 activation, Th17 cell activation and other immune regulatory processes. Additionally, scRNA sequencing also demonstrated that microbial-derived interferon gene stimulators (STING) agonists can regulate macrophage polarization, induce IFN-I production from intratumoral monocytes, and facilitate crosstalk between NK cells and DCs. The microbiome triggers the IFN-I-NK cell-DC axis within the tumor, improving the efficacy of ICB (135).
The combination of ICB therapy with other treatment modalities has yielded promising results. Relatlimab combined with nivolumab (anti-LAG-3 and anti-PD-1) has been approved by the FDA as a first-line treatment for stage III/IV melanoma. Utilizing single-cell RNA and TCR sequencing in conjunction with other multi-omics analyses, researchers found that adaptive NK cells were enriched in responders after treatment and underwent significant transcriptomic changes, leading to an activated phenotype (136). Single-cell sequencing compared the transcriptomes of approximately 92,000 single cells from three pre-treatment and twelve post-treatment NSCLC patients who received neoadjuvant PD-1 blockade combined with chemotherapy and researchers concluded that the neoadjuvant PD-1 blockade combined with chemotherapy therapy can promote the expansion and activation of cytotoxic T cells and CD16+ NK cells (137). Additionally, a study on a new type of high-grade serous ovarian cancer (HGSC) compared samples treated with a bispecific anti-PD-1/PD-L1 antibody to a control group treated with monospecific anti-PD-1 or anti-PD-L1. The results demonstrated that the bispecific antibody induced superior cellular state changes in T cells and NK cells, uniquely prompting NK cells to transition from a quiescent phenotype to an active and cytotoxic phenotype. This transition enhanced the efficacy of immunotherapy for ovarian cancer (138). Another study identified 207 liver cancer NK cell marker genes based on single-cell sequencing, which are primarily involved in immune functions and found that the degree of immune cell infiltration and function in the low-risk group were higher than those in the high-risk group, indicating that the low-risk group is more suitable for ICI and anti-PD-1 treatment (139).
4.1.2 Other biochemical methods
Some single-cell sequencing studies have shown that NK cells in the TME after chemotherapy show increased infiltration and enhanced killing efficacy, indicating good anti-tumor effects. A study conducted whole exome sequencing, large-scale RNA sequencing, and single-cell transcriptomics on paired pretreatment and treatment samples from gastric cancer patients and explained the remodeling of the TME after platinum chemotherapy, characterized by the recruitment of NK cells, the reduction of tumor-associated macrophages, the repolarization of M1 macrophages, and the increase in effector T cell infiltration (140). The scramblase Xkr8 plays a key role in promoting tumor immunosuppression and the use of Xkr8 small interfering RNA (siRNA) targeting Xkr8 combined with the chemotherapeutic prodrug FuOXP in the TME of in situ pancreatic tumors caused an increase in the infiltration of proliferative NK cells and activated macrophages which could enhance the antitumor immune response and showing good antitumor effects (141). Additionally, a study based on high-dimensional cell counting deeply analyzed the effects and mechanisms of 2,5-dimethyl-celecoxib (DMC) on the infiltration of immune cells in liver cancer. The study found that after using DMC, NK cells and T cells in the TME showed strong anti-tumor activity and effectively inhibited the exhaustion of NK cells and T cells, thereby inhibiting the growth of liver cancer cells and improving the prognosis of mice (142).
The application of certain chemical agents has also been observed to affect NK cells through single-cell sequencing technology, providing insights into clinical efficacy and guiding treatment strategies. Wu S et al. concluded from single-cell sequencing in a mouse B16F10 melanoma model that docosahexaenoic acid (DHA) can enhance the effector function and oxidative phosphorylation of NK cells, effectively inhibiting the growth of B16F10 melanoma in the lungs (143). Yi Q et al. analyzed pancreatic cancer liver metastasis (PCLM) models with or without α-galactosylceramide (αGC) treatment, characterizing the overall changes in immune cells in the TME after αGC treatment. They found that the cytotoxic activity of invariant natural killer T cells (iNKT) and NK cells increased (119). Li CY et al. studied the immune mechanisms of the anti-tumor effects of low-dose nitric oxide donors in three ways, among which scRNA sequencing showed that low-dose nitric oxide donors increased the number of CD8+ T cells and NK cells while reducing central memory T cells (144). Through single-cell RNA sequencing, Liu L et al. found that sodium polyoxotungstate inhibited the increase of NK cells within tumors. They also performed basic analysis of NK cell phenotypes through flow cytometry, revealing that treatment with CD39i significantly increased the proportion of mature CD27+CD11b+ and CD27-CD11b+ NK cells, confirming that CD39 is a potential target for immunotherapy in bladder cancer (113).
Additionally, some treatment strategies using oncolytic viruses have led the way in biological treatment concepts. For example, in a mouse model treated with the oncolytic virus OH2, Wu Q et al. (120) found that OH2 could enhance the cytotoxic capabilities of peripheral CD8+ T cells and mature NK cells, effectively activate the systemic immune response and induce a sustained anti-tumor immune response.
4.1.3 Non-pharmacological intervention mechanisms
Non-pharmacological interventions, such as radiation and microwave ablation therapy, have also been better explored for their efficacy through single-cell RNA sequencing. Jiao JZ et al. investigated the impact of non-invasive radiofrequency radiation (RFR) exposure on the tumor immunosuppressive microenvironment. ScRNA-seq analysis of infiltrating immune cells in lung metastatic melanoma revealed that RFR exposure could increase NK cell subsets, enhance the effector functions of tumor-infiltrating CD8+ T cells and NK cells, induce anti-tumor remodeling of TME and thereby inhibit tumor progression (121). Another study conducted scRNA-seq on PBMCs before and after microwave ablation (MWA) for breast cancer. The study found that XCL2+ NK cells specifically enriched after MWA were associated with genes related to cytokine production pathways, lymphocyte differentiation pathways and T cell activation pathways and presented higher chemokine function. While in GZMB+ NK cells, IFITM1, MYOM2, JUN, IFITM3, CCL4, and RBM25 were upregulated, indicating higher cytotoxic activity (145).
4.2 NK cell exhaustion as an important factor in the mechanism of tumor resistance
Immunotherapy and emerging treatments have revolutionized treatment for cancer but most patients either do not respond to immunotherapy or show resistance, and the underlying mechanisms remain to be fully elucidated. Single-cell sequencing technology has provided a robust technical foundation for us to gain a deeper understanding of the resistance mechanisms within the tumor microenvironment under various therapeutic interventions.
Zhang Y et al. discovered that patients with MET amplification exhibit resistance to ICB in lung cancer. Through deep single-cell RNA sequencing, they found that MET amplified patients exhibited immunosuppressive characteristics, characterized by the increase in XIST+ and CD96+ exhausted NK cell subsets and reduction in CD8+ T cells and NK cell populations. The study also demonstrated that inhibition of MET can overcome the decreased efficacy of ICB caused by MET amplification (146). A study identified a new podoplanin-positive (PDPN+) CAFs subset enriched in trastuzumab-resistant tumor tissues through single-cell sequencing. This subset induced trastuzumab resistance in HER2+ breast cancer by inhibiting the NK cell-mediated antibody-dependent cellular cytotoxicity (ADCC) immune response (147). Li XY et al. conducted scRNA sequencing on tumors of patients with metastatic melanoma or head and neck squamous cell carcinoma and found that NKG7 was highly correlated with the cytotoxicity of CD8+ T cells and NK cells. Tumors expressing new antigens grew faster in mice lacking NKG7. Furthermore, the efficacy of single or combined ICB was significantly reduced in NKG7-deficient mice (122). As previously mentioned, scRNA sequencing revealed that conditional deletion of HIF-1α in mouse tumor-infiltrating NK cells resulted in reduced tumor growth, increased expression of activation markers and effector molecules, and enriched the NF-κB pathway in tumor-infiltrating NK cells. Furthermore, long-term culture with a HIF-1α inhibitor also increased the efficacy of human NK cells (123). Additionally, through single-cell transcriptomics and gene reporter mice, Bi J et al. found that the expression of TIPE2 in the human and mouse TME was associated with NK cell exhaustion, and TIPE2 deletion promoted the anti-tumor activity of human NK cells in mice, enhancing NK cell infiltration and effector function, thereby improving the survival rate of tumor patients (124). A study using high-dimensional flow cytometry, mass cytometry, and single-cell RNA sequencing combined with functional analysis created a comprehensive map of human NK cell migration phenotypes and explained the reasons for the systemic rejection of NK cells from the tumor microenvironment, providing a basis for designing the next generation of NK cell therapy for malignant tumors (125). Drug resistance largely depends on the sensitivity and efficacy of different tumor subgroups to treatment. Through the integration of scRNA-seq data and TCGA data in the study of head and neck squamous cell carcinoma (HNSCC), Yang F et al. (126) found that, compared to the HPV+ HNSCC cohort, the IL6/IL6R and CCL2/CCR2 signaling pathways had a more significant impact on NK cells evading immune attack in the HPV- HNSCC cohort. The combination of CCR2 chemokine receptor antagonists with IL-6 blockers had a significant anti-tumor effect in HPV- HNSCC, which was associated with larger number of activated intratumoral NK cells.
4.3 Selection of therapeutic target
Through single-cell sequencing technology, researchers can also achieve the identification of cell markers, thereby providing better targets and ideas for drug treatment. In a pan-cancer analysis utilizing single-cell sequencing technology, Tang F et al. (127) identified regulator of G protein signaling 1 (RGS1) as a marker expressed specifically in NK cells within tumor and adjacent non-tumor tissues, while being nearly undetectable in blood. RGS1 was widely expressed across analyzed patients and cancer types, exhibiting higher sensitivity and specificity than other tumor tissue resident markers. Another study in mice identified CCRL2 as a marker of general alveolar and pulmonary capillary endothelial cells through scRNA-seq and found that upregulating CCRL2 increased NK cell recruitment and reduced lung tumor growth, thus establishing CCRL2 as an NK cell lung homing molecule and providing new insights into NK cell-mediated lung tumor immune surveillance (128). Zhang Y et al. investigated the potential role of pyroptosis-related genes (PRGs) in preventing malignant melanoma through the study of melanoma pyroptosis, identifying CD57+ NK cells with downregulated cell pyroptosis expression, particularly GSDMB+, as a protective prognostic predictor in clinical settings (129). A separate study demonstrated that the endoplasmic reticulum-regulated secretory protein SCUBE2 was enriched in osteoblasts within early bone metastatic niches, promoting osteoblast differentiation. Osteoblast deposition of collagen inhibited NK cells by inhibiting LAIR1 signaling, thereby promoting tumor colonization (130). Slattery K et al. (148) conducted single-cell analysis, metabolic flux, and confocal analysis on NK cells from patients with metastatic breast cancer and the healthy control groups, identifying TGF-β as a factor involved in the metabolic dysfunction of circulating NK cells in patients with metastatic breast cancer. Blocking the GARP-TGF-β axis can restore the metabolism and function of NK cells, representing an important target for enhancing NK cell immunotherapy.
5 Concluding remarks
Despite the continuous emergence of new technologies and a deeper understanding of interactions within tumors, information regarding the sources of NK cell heterogeneity, the individual developmental processes of NK cells, and the role of tumor programming in regulating the functional diversity of NK cells remains insufficiently comprehensive. With ongoing advancements in technology and the continual exploration of interactions within tumors, many questions may be addressed in the coming years, potentially providing promising targets for the development of candidate therapies. The application of single-cell sequencing technology offers a promising avenue to tackle these challenges. Single-cell sequencing technology plays a significant role in identifying new NK cell subpopulations, analyzing differences among various subpopulations, and investigating intercellular interaction relationships in depth. Single-cell RNA sequencing studies on NK cell heterogeneity have identified correlation maps between NK cell subtypes in the TME and tumor progression, providing evidence for patient prognosis and therapeutic efficacy. These studies reveal promising cellular and molecular markers as new targets for cancer treatment. They offer immune cell marker genes for tumor prognosis, which can serve as a reference for immunotherapy decisions in cancer patients. Additionally, scRNA-seq provides valuable insights into the transcriptional heterogeneity of the TME, the overall intercellular interaction network, and new molecular mechanisms of action and resistance related to immune checkpoint inhibitors and targeted therapies. Single-cell sequencing enables dimensionality reduction and clustering of NK cells into distinct subpopulations, each exhibiting unique transcriptional profiles. This facilitates a paradigm shift in prognostic stratification from population-level analysis to single-cell precision. Through scRNA-seq, we identify NK-specific immune checkpoints (e.g., NKG2A, KIR), which inform monoclonal antibody targeting strategies (e.g. anti-NKG2A monalizumab) for precision patient selection. Spatial proximity analysis of interactions between NK cells and tumor cells coupled with expression quantitation of receptor-ligand pair directly correlates with immune evasion capacity and metastatic potential, providing actionable biomarkers for personalized immunotherapies (e.g. CAR-NK design, checkpoint inhibitor combination). Furthermore, single-cell sequencing can also compare the intervention effects of regulatory molecule dynamics and metabolic reprogramming interventions, evaluate the superiority of synergistic immunotherapy to guide intervention measures, overcome tumor drug resistance and enhance antitumor efficacy.
At the same time, we must also acknowledge the limitations of scRNA-seq technology. Firstly, there are technical issues. RNA instability compromises library construction quality, and some key but rare subpopulations, such as drug-resistant cells and migratory cells, may escaped detection by most methods, including scRNA-seq. Secondly, conventional scRNA-seq captures cellular states but fails to preserve spatial information. The continuous improvements in spatially resolved techniques and multimodal data integration are expected to significantly advance our understanding of intercellular interactions and spatial effects in the coming years. Moreover, there are also challenges in clinical translation. Issues such as insufficient drug target potency, the need for multi-target combination to enhance anti-tumor ability, and limitations in adoptive cell therapies including poor in vivo persistence and insufficient tumor infiltration, all collectively impede translational progress in the field. David’s research summarized the eleven major challenges faced by single-cell data science, highlighting various issues with current single-cell sequencing data, such as insufficient sequencing coverage and depth, high overall costs, and sequencing biases arising from model construction and metric selection (149). We believe that the research benefits provided by single-cell sequencing technology will accelerate the standardization of analytical methods, thereby promoting technological development, while the accuracy and stability of sequencing data will continue to be optimized to meet the needs of clinical accessibility. The combination of single-cell sequencing technology with spatial transcriptomics can generate high-resolution tissue maps, delineate developmental pathways within specific spatial contexts, and elucidate gene expression activity in particular tissue regions, thereby enriching the dimension of single-cell sequencing technology (150).In recent years, NK cell-based cellular therapies, such as chimeric antigen receptor (CAR)-NK cell therapy, have demonstrated certain advantages over CAR-T cell therapy and have been partially validated for their roles in the treatment of various hematological malignancies (151).Single-cell RNA sequencing has been employed to evaluate the efficacy of CAR-NK cells in acute myeloid leukemia (AML). Studies revealed upregulation of cell proliferation, protein folding, immune response, and key metabolic pathways in CAR-NK cells, leading to tumor-specific, CAR-dependent activation and functionality against AML target cells (152). Furthermore, scRNA-seq has provided critical theoretical insights for gene-editing strategies in CAR-NK cells to counteract tumor immunosuppression in AML (153). Additionally, this technology enables comparative analysis of how agonists or cytokines influence CAR-NK cell activity and therapeutic outcomes. For instance, STING agonists were found to enhance the migration and cytotoxic capacity of mesothelin-targeted CAR-NK cells (154) and IL-15 helps overcome the loss of NK cell metabolic fitness induced by tumor interactions (155). Notably, ongoing innovations in single-cell sequencing methodologies and precision have not only strengthened the technical foundation for CAR-NK cell therapy applications but also provided unique insights into the multifaceted mechanisms underlying CAR-NK cell cytotoxicity (156, 157). Although research on CAR-NK cells in solid tumors remains limited, and their application faces multiple challenges, such as the degree of targeted infiltration, understanding NK cell biology is essential for the development of improved therapies, whether through the use of potential CAR-NK cells or by directly targeting NK cells. Therefore, we summarize the progress in the study of NK cell heterogeneity using single-cell sequencing to gain deeper insights into the biological characteristics of NK cells within the tumor microenvironment. Additionally, as recent studies frequently compare NK cell expression levels in the tumor microenvironment with those in peripheral blood to analyze the differences, we examine NK cell expression from both physiological and tumor-centric pathological perspectives. We also acknowledge the importance of studying NK cell expression in non-tumor tissues under physiological conditions. We also dissect the origins and developmental processes of tissue-resident NK cells and gather evidence regarding the enrichment of NK cell precursors in extramedullary tissues, providing new ideas and directions for NK cell-targeted therapies. We believe that the integration of various omics technologies, along with the construction of the latest data from cellular dynamics research, molecular profiling, and spatial localization, can create a platform for the development of new and more effective anti-tumor NK cell-targeted therapies. Excitingly, certain preclinical results are impressive and are expected to start generating clinical impact.
Author contributions
MS: Conceptualization, Writing – review & editing, Writing – original draft, Methodology, Validation. YL: Writing – review & editing, Conceptualization, Methodology, Visualization. LS: Visualization, Validation, Funding acquisition, Writing – review & editing. MQ: Conceptualization, Writing – review & editing, Validation, Visualization. SS: Writing – review & editing, Validation, Methodology. HZ: Writing – review & editing, Methodology, Funding acquisition, Visualization, Conceptualization, Validation. WS: Visualization, Funding acquisition, Validation, Writing – review & editing, Methodology.
Funding
The author(s) declare that fifinancial support was received for the research and/or publication of this article. This study was fifinancially supported by: Science Foundation of Liaoning Province (2024-MS-052); Applied Basic Research Program of Liaoning Province (2022020225 JH2/1013); Basic Research Project of Liaoning Provincial Department of Education for Universities (LJ242410159046).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Author disclaimer
We hereby confirm that neither the manuscript nor any part of it has been published or is being considered for publication elsewhere. We acknowledge that all authors sufficiently participated in the study and take responsibility for its content.
Glossary
NK cells: Natural killer cells
TME: tumor microenvironment
scRNA-seq: single-cell RNA sequencing
HCL: Human Cell Landscape
ILCs: innate lymphoid cells
FcγRIII: Fcγ receptor III
KIR: killer cell immunoglobulin-like receptor
HSCs: hematopoietic stem cell
CLPs: common lymphoid progenitors
pre-NKPs: pre-NK progenitors
NKPs: NK progenitor cells
iNK: immature NK
LTis: lymphoid tissue inducers
CILPs: common innate lymphoid progenitors
CHILPs: common helper innate lymphoid progenitors
LTiPs: lymphoid tissue inducer progenitors
ILCPs: innate lymphoid cell precursors
IFN-γ: interferon-γ
AIR: adaptive immune receptor
trNK: tissue-resident NK cells
PBMC: peripheral blood mononuclear cells
cNK: conventional NK cells
BM: bone marrow
SLT: secondary lymphoid tissues
MALT: mucosal-associated lymphoid tissue
DC: dendritic cell
NKRM: tissue-resident memory-like NK
TBX21: TBox transcription factor 21
FGL2: fibrinogen-like 2;IRF8, interferon regulatory factor 8
NSCLC: non-small cell lung cancer
LUSC: lung squamous cell carcinoma
LUAD: lung adenocarcinoma
TNBC: triple-negative breast cancer
LGGs: low-grade gliomas
HGGs: high-grade gliomas
CSCC: cervical squamous cell carcinoma
pDC: plasmacytoid DCs
CIN: chromosomal instability
AIS: adenocarcinoma in situ
MIA: minimally invasive adenocarcinoma
IAC: invasive adenocarcinoma
CTCs: circulating tumor cells
PVR: poliovirus receptor
GAM: glioma-associated macrophage
CAF: cancer-associated fibroblasts
snRNA-seq: single-nucleus RNA sequencing
ICB: immune checkpoint blockade
ICI: immune checkpoint inhibitor
DCB: durable clinical benefit
HGSC: high-grade serous ovarian cancer
siRNA: small interfering RNA
DMC: 2,5-dimethyl-celecoxib
DHA: docosahexaenoic acid
PCLM: pancreatic cancer liver metastasis
αGC: α-galactosylceramide
iNKT: invariant natural killer T cells
RFR: radiofrequency radiation
MWA: microwave ablation
ADCC: antibody-dependent cellular cytotoxicity
HNSCC: head and neck squamous cell carcinoma
RGS1: regulator of G protein signaling 1
PRGs: pyroptosis-related genes
CAR: chimeric antigen receptor
AML: acute myeloid leukemia
References
1. Suo C, Dann E, Goh I, Jardine L, Kleshchevnikov V, Park J-E, et al. Mapping the developing human immune system across organs. Science. (2022) 376:eabo0510. doi: 10.1126/science.abo0510
2. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, et al. Comprehensive integration of single-cell data. Cell. (2019) 177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031
3. Han X, Zhou Z, Fei L, Sun H, Wang R, Chen Y, et al. Construction of a human cell landscape at single-cell level. Nature. 581(7808):303–9. doi: 10.1038/s41586-020-2157-4
4. Ren X, Kang B, and Zhang Z. Understanding tumor ecosystems by single-cell sequencing: promises and limitations. Genome Biol. (2018) 19:211. doi: 10.1186/s13059-018-1593-z
5. Rozenblatt-Rosen O, Regev A, Oberdoerffer P, Nawy T, Hupalowska A, Rood JE, et al. The Human Tumor Atlas Network: Charting Tumor Transitions across Space and Time at Single-Cell Resolution. Cell. (2020) 181:236–49. doi: 10.1016/j.cell.2020.03.053
7. Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. (2014) 344:1396–401. doi: 10.1126/science.1254257
8. Azizi E, Carr AJ, Plitas G, Cornish AE, Konopacki C, Prabhakaran S, et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell. (2018) 174:1293–1308.e36. doi: 10.1016/j.cell.2018.05.060
9. Bian S, Hou Y, Zhou X, Li X, Yong J, Wang Y, et al. Single-cell multiomics sequencing and analyses of human colorectal cancer. Science. (2018) 362:1060–3. doi: 10.1126/science.aao3791
10. Ji AL, Rubin AJ, Thrane K, Jiang S, Reynolds DL, Meyers RM, et al. Multimodal analysis of composition and spatial architecture in human squamous cell carcinoma. Cell. (2020) 182:1661–2. doi: 10.1016/j.cell.2020.08.043
11. Maynard A, McCoach CE, Rotow JK, Harris L, Haderk F, Kerr DL, et al. Therapy-induced evolution of human lung cancer revealed by single-cell RNA sequencing. Cell. (2020) 182:1232–1251.e22. doi: 10.1016/j.cell.2020.07.017
12. Tirosh I, Izar B, Prakadan SM, Wadsworth MH, Treacy D, Trombetta JJ, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. (2016) 352:189–96. doi: 10.1126/science.aad0501
13. Kiessling R, Petranyi G, Kärre K, Jondal M, Tracey D, and Wigzell H. Killer cells: A functional comparison between natural, immune T-cell and antibody-dependent in vitro systems. J Exp Med. (1976) 143:772–80. doi: 10.1084/jem.143.4.772
14. Kiessling R, Klein E, Pross H, and Wigzell H. Natural” Killer cells in the mouse. II. Cytotoxic cells with specificity for mouse moloney leukemia cells. Characteristics of the killer cell. Eur J Immunol. (1975) 5:117–21. doi: 10.1002/eji.1830050209
15. Krzewski K and Coligan JE. Human NK cell lytic granules and regulation of their exocytosis. Front Immunol. 3:335. doi: 10.3389/fimmu.2012.00335
16. Zamai L, Ahmad M, Bennett IM, Azzoni L, Alnemri ES, and PeRussia B. Natural killer (NK) cell–mediated cytotoxicity: differential use of TRAIL and fas ligand by immature and mature primary human NK cells. J Exp Med. (1998) 188:2375–80. doi: 10.1084/jem.188.12.2375
17. Huntington ND, Vosshenrich CAJ, and Santo JPD. Developmental pathways that generate natural-killer-cell diversity in mice and humans. Nat Rev Immunol. (2007) 7(9):703–14. doi: 10.1038/nri2154
18. Wu S-Y, Fu T, Jiang Y-Z, and Shao Z-M. Natural killer cells in cancer biology and therapy. Mol Cancer. (2020) 19:120. doi: 10.1186/s12943-020-01238-x
19. Zheng L, Qin S, Si W, Wang A, Xing B, Gao R, et al. Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science. (2021) 374:abe6474. doi: 10.1126/science.abe6474
20. Miller JS, Alley KA, and McGlave P. Differentiation of natural killer (NK) cells from human primitive marrow progenitors in a stroma-based long-term culture system: identification of a CD34+7+ NK progenitor. Blood. (1994) 83(9):2594–601.
21. Moretta L, Ferlazzo G, Bottino C, Vitale M, Pende D, Mingari MC, et al. Effector and regulatory events during natural killer–dendritic cell interactions. Immunol Rev. (2006) 214:219–28. doi: 10.1111/j.1600-065X.2006.00450.x
22. Jacobs R, Hintzen G, Kemper A, Beul K, Kempf S, Behrens G, et al. CD56bright cells differ in their KIR repertoire and cytotoxic features from CD56dim NK cells. Eur J Immunol. (2001) 31:3121–6. doi: 10.1002/1521-4141(2001010)31:10<3121::AID-IMMU3121>3.0.CO;2-4
23. Poli A, Michel T, Thérésine M, Andrès E, Hentges F, and Zimmer J. CD56 bright natural killer (NK) cells: an important NK cell subset. Immunology. (2009) 126:458–65. doi: 10.1111/j.1365-2567.2008.03027.x
24. Yu J, Freud AG, and Caligiuri MA. Location and cellular stages of natural killer cell development. Trends Immunol. (2013) 34:573–82. doi: 10.1016/j.it.2013.07.005
25. Scoville SD, Freud AG, and Caligiuri MA. Modeling human natural killer cell development in the era of innate lymphoid cells. Front Immunol. (2017) 8:360. doi: 10.3389/fimmu.2017.00360
26. Cichocki F, Grzywacz B, and Miller JS. Human NK cell development: one road or many? Front Immunol. (2019) 10:2078. doi: 10.3389/fimmu.2019.02078
27. Rosmaraki EE, Douagi I, Roth C, Colucci F, Cumano A, and Di Santo JP. Identification of committed NK cell progenitors in adult murine bone marrow. Eur J Immunol. (2001) 31:1900–9. doi: 10.1002/1521-4141(200106)31:6<1900::AID-IMMU1900>3.0.CO;2-M
28. Fathman JW, Bhattacharya D, Inlay MA, Seita J, Karsunky H, and Weissman IL. Identification of the earliest natural killer cell–committed progenitor in murine bone marrow. Blood. (2011) 118:5439–47. doi: 10.1182/blood-2011-04-348912
29. Abe S, Asahi T, Hara T, Cui G, Shimba A, Tani-ichi S, et al. Hematopoietic cell-derived IL-15 supports NK cell development in scattered and clustered localization within the bone marrow. Cell Rep. (2023) 42:113127. doi: 10.1016/j.celrep.2023.113127
30. Freud AG, Keller KA, Scoville SD, Mundy-Bosse BL, Cheng S, Youssef Y, et al. NKp80 defines a critical step during human natural killer cell development. Cell Rep. (2016) 16:379–91. doi: 10.1016/j.celrep.2016.05.095
31. Björkström NK, Riese P, Heuts F, Andersson S, Fauriat C, Ivarsson MA, et al. Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK-cell differentiation uncoupled from NK-cell education. Blood. (2010) 116:3853–64. doi: 10.1182/blood-2010-04-281675
32. Lopez-Vergès S, Milush JM, Pandey S, York VA, Arakawa-Hoyt J, Pircher H, et al. CD57 defines a functionally distinct population of mature NK cells in the human CD56dimCD16+ NK-cell subset. Blood. (2010) 116:3865–74. doi: 10.1182/blood-2010-04-282301
33. Ma S, Caligiuri MA, and Yu JA. Four-stage model for murine natural killer cell development in vivo. J Hematol Oncol. (2022) 15:31. doi: 10.1186/s13045-022-01243-1
34. Vivier E, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate lymphoid cells: 10 years on. Cell. (2018) 174:1054–66. doi: 10.1016/j.cell.2018.07.017
35. Lopes N, Galluso J, Escalière B, Carpentier S, Kerdiles YM, and Vivier E. Tissue-specific transcriptional profiles and heterogeneity of natural killer cells and group 1 innate lymphoid cells. Cell Rep Med. (2022) 3:100812. doi: 10.1016/j.xcrm.2022.100812
36. Constantinides MG, McDonald BD, Verhoef PA, and Bendelac A. A committed precursor to innate lymphoid cells. Nature. (2014) 508:397–401. doi: 10.1038/nature13047
37. Gao Y, Souza-Fonseca-Guimaraes F, Bald T, Ng SS, Young A, Ngiow SF, et al. Tumor immunoevasion by the conversion of effector NK cells into type 1 innate lymphoid cells. Nat Immunol. (2017) 18:1004–15. doi: 10.1038/ni.3800
38. Walker JA, Clark PA, Crisp A, Barlow JL, Szeto A, Ferreira ACF, et al. Polychromic reporter mice reveal unappreciated innate lymphoid cell progenitor heterogeneity and elusive ILC3 progenitors in bone marrow. Immunity. (2019) 51:104–118.e7. doi: 10.1016/j.immuni.2019.05.002
39. Xu W, Cherrier DE, Chea S, Vosshenrich C, Serafini N, Petit M, et al. An id2RFP-reporter mouse redefines innate lymphoid cell precursor potentials. Immunity. (2019) 50:1054–1068.e3. doi: 10.1016/j.immuni.2019.02.022
40. Chen L, Youssef Y, Robinson C, Ernst GF, Carson MY, Young KA, et al. CD56 expression marks human group 2 innate lymphoid cell divergence from a shared NK cell and group 3 innate lymphoid cell developmental pathway. Immunity. (2018) 49:464–476.e4. doi: 10.1016/j.immuni.2018.08.010
41. Zhong C, Zheng M, Cui K, Martins AJ, Hu G, Li D, et al. Differential expression of the transcription factor GATA3 specifies lineage and functions of innate lymphoid cells. Immunity. (2020) 52:83–95.e4. doi: 10.1016/j.immuni.2019.12.001
42. McFarland AP, Yalin A, Wang S-Y, Cortez VS, Landsberger T, Sudan R, et al. Multi-tissue single-cell analysis deconstructs the complex programs of mouse natural killer and type 1 innate lymphoid cells in tissues and circulation. Immunity. (2021) 54:1320–1337.e4. doi: 10.1016/j.immuni.2021.03.024
43. Yang C, Siebert JR, Burns R, Gerbec ZJ, Bonacci B, Rymaszewski A, et al. Heterogeneity of human bone marrow and blood natural killer cells defined by single-cell transcriptome. Nat Commun. (2019) 10:3931. doi: 10.1038/s41467-019-11947-7
44. Song G, Shi Y, Zhang M, Goswami S, Afridi S, Meng L, et al. Global immune characterization of HBV/HCV-related hepatocellular carcinoma identifies macrophage and T-cell subsets associated with disease progression. Cell Discov. (2020) 6:90. doi: 10.1038/s41421-020-00214-5
45. Melsen JE, Van Ostaijen-ten Dam MM, Schoorl DJA, Schol PJ, Van Den Homberg DAL, Lankester AC, et al. Single-cell transcriptomics in bone marrow delineates CD56dimGranzymeK+ Subset as intermediate stage in NK cell differentiation. Front Immunol. (2022) 13:1044398. doi: 10.3389/fimmu.2022.1044398
46. Xie X, Liu M, Zhang Y, Wang B, Zhu C, Wang C, et al. Single-cell transcriptomic landscape of human blood cells. Natl Sci Rev. (2021) 8:nwaa180. doi: 10.1093/nsr/nwaa180
47. Suo C, Polanski K, Dann E, Lindeboom RGH, Vilarrasa-Blasi R, Vento-Tormo R, et al. Dandelion uses the single-cell adaptive immune receptor repertoire to explore lymphocyte developmental origins. Nat Biotechnol. (2024) 42:40–51. doi: 10.1038/s41587-023-01734-7
48. Rodda LB, Lu E, Bennett ML, Sokol CL, Wang X, Luther SA, et al. Single-cell RNA sequencing of lymph node stromal cells reveals niche-associated heterogeneity. Immunity. (2018) 48:1014–1028.e6. doi: 10.1016/j.immuni.2018.04.006
49. Hay SB, Ferchen K, Chetal K, Grimes HL, and Salomonis N. The human cell atlas bone marrow single-cell interactive web portal. Exp Hematol. (2018) 68:51–61. doi: 10.1016/j.exphem.2018.09.004
50. Pellin D, Loperfido M, Baricordi C, Wolock SL, Montepeloso A, Weinberg OK, et al. Comprehensive single cell transcriptional landscape of human hematopoietic progenitors. Nat Commun. (2019) 10:2395. doi: 10.1038/s41467-019-10291-0
51. Narni-Mancinelli E, Berruyer C, and Vivier E. On blood and tissue-resident natural killer cells. Immunity. (2024) 57:6–8. doi: 10.1016/j.immuni.2023.12.013
52. Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate lymphoid cells — a proposal for uniform nomenclature. Nat Rev Immunol. (2013) 13:145–9. doi: 10.1038/nri3365
53. Colonna M. Innate lymphoid cells: diversity, plasticity, and unique functions in immunity. Immunity. (2018) 48:1104–17. doi: 10.1016/j.immuni.2018.05.013
54. Björkström NK, Ljunggren H-G, and Michaëlsson J. Emerging insights into natural killer cells in human peripheral tissues. Nat Rev Immunol. (2016) 16:310–20. doi: 10.1038/nri.2016.34
55. Dogra P, Rancan C, Ma W, Toth M, Senda T, Carpenter DJ, et al. Tissue determinants of human NK cell development, function, and residence. Cell. (2020) 180:749–763.e13. doi: 10.1016/j.cell.2020.01.022
56. Hughes T, Becknell B, McClory S, Briercheck E, Freud AG, Zhang X, et al. Stage 3 immature human natural killer cells found in secondary lymphoid tissue constitutively and selectively express the TH17 cytokine interleukin-22. Blood. (2009) 113:4008–10. doi: 10.1182/blood-2008-12-192443
57. Lugthart G, Melsen JE, Vervat C, Van Ostaijen-ten Dam MM, Corver WE, Roelen DL, et al. Human lymphoid tissues harbor a distinct CD69+CXCR6+ NK cell population. J Immunol. (2016) 197:78–84. doi: 10.4049/jimmunol.1502603
58. Mazzurana L, Czarnewski P, Jonsson V, Wigge L, Ringnér M, Williams TC, et al. Tissue-specific transcriptional imprinting and heterogeneity in human innate lymphoid cells revealed by full-length single-cell RNA-sequencing. Cell Res. (2021) 31:554–68. doi: 10.1038/s41422-020-00445-x
59. Peng H and Tian Z. NK cell trafficking in health and autoimmunity:A comprehensive review. Clin Rev Allerg Immunol. (2014) 47:119–27. doi: 10.1007/s12016-013-8400-0
60. Yao X and Matosevic S. Chemokine networks modulating natural killer cell trafficking to solid tumors. Cytokine Growth Factor Rev. (2021) 59:36–45. doi: 10.1016/j.cytogfr.2020.12.003
61. Freud AG, Mundy-Bosse BL, Yu J, and Caligiuri MA. The broad spectrum of human natural killer cell diversity. Immunity. (2017) 47:820–33. doi: 10.1016/j.immuni.2017.10.008
62. Ran GH, Lin YQ, Tian L, Zhang T, Yan DM, Yu JH, et al. Natural killer cell homing and trafficking in tissues and tumors: from biology to application. Sig Transduct Target Ther. (2022) 7:205. doi: 10.1038/s41392-022-01058-z
63. Gray JI and Farber DL. Tissue-resident immune cells in humans. Annu Rev Immunol. (2022) 40:195–220. doi: 10.1146/annurev-immunol-093019-112809
64. the Immunological Genome Consortium, Robinette ML, Fuchs A, Cortez VS, Lee JS, Wang Y, et al. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat Immunol. (2015) 16:306–17. doi: 10.1038/ni.3094
65. Torcellan T, Friedrich C, Doucet-Ladevèze R, Ossner T, Solé VV, Riedmann S, et al. Circulating NK cells establish tissue residency upon acute infection of skin and mediate accelerated effector responses to secondary infection. Immunity. (2024) 57:124–140.e7. doi: 10.1016/j.immuni.2023.11.018
66. Bai L, Vienne M, Tang L, Kerdiles Y, Etiennot M, Escalière B, et al. Liver type 1 innate lymphoid cells develop locally via an interferon-γ–dependent loop. Science. (2021) 371:eaba4177. doi: 10.1126/science.aba4177
67. Vargas CL, Poursine-Laurent J, Yang L, and Yokoyama WM. Development of thymic NK cells from double negative 1 thymocyte precursors. Blood. (2011) 118:3570–8. doi: 10.1182/blood-2011-06-359679
68. Luther C, Warner K, and Takei F. Unique progenitors in mouse lymph node develop into CD127+ NK cells: thymus-dependent and thymus-independent pathways. Blood. (2011) 117:4012–21. doi: 10.1182/blood-2010-07-298901
69. Cordes M, Canté-Barrett K, Van Den Akker EB, Moretti FA, Kiełbasa SM, Vloemans SA, et al. Single-cell immune profiling reveals thymus-seeding populations, T cell commitment, and multilineage development in the human thymus. Sci Immunol. (2022) 7:eade0182. doi: 10.1126/sciimmunol.ade0182
70. Crinier A, Milpied P, Escalière B, Piperoglou C, Galluso J, Balsamo A, et al. High-dimensional single-cell analysis identifies organ-specific signatures and conserved NK cell subsets in humans and mice. Immunity. (2018) 49:971–986.e5. doi: 10.1016/j.immuni.2018.09.009
71. Crinier A, Dumas P-Y, Escalière B, Piperoglou C, Gil L, Villacreces A, et al. Single-cell profiling reveals the trajectories of natural killer cell differentiation in bone marrow and a stress signature induced by acute myeloid leukemia. Cell Mol Immunol. (2021) 18:1290–304. doi: 10.1038/s41423-020-00574-8
72. Dean I, Lee CYC, Tuong ZK, Li Z, Tibbitt CA, Willis C, et al. Rapid functional impairment of natural killer cells following tumor entry limits anti-tumor immunity. Nat Commun. (2024) 15:683. doi: 10.1038/s41467-024-44789-z
73. Schuster IS, Sng XYX, Lau CM, Powell DR, Weizman O-E, Fleming P, et al. Infection induces tissue-resident memory NK cells that safeguard tissue health. Immunity. (2023) 56:531–546.e6. doi: 10.1016/j.immuni.2023.01.016
74. Hegewisch-Solloa E, Seo S, Mundy-Bosse BL, Mishra A, Waldman EH, Maurrasse S, et al. Differential integrin adhesome expression defines human NK cell residency and developmental stage. J Immunol. (2021) 207:950–65. doi: 10.4049/jimmunol.2100162
75. Marquardt N, Kekäläinen E, Chen P, Lourda M, Wilson JN, Scharenberg M, et al. Unique transcriptional and protein-expression signature in human lung tissue-resident NK cells. Nat Commun. (2019) 10:3841. doi: 10.1038/s41467-019-11632-9
76. Peng H and Tian Z. Diversity of tissue-resident NK cells. Semin Immunol. (2017) 31:3–10. doi: 10.1016/j.smim.2017.07.006
77. Hu L, Han M, Deng Y, Gong J, Hou Z, Zeng Y, et al. Genetic distinction between functional tissue-resident and conventional natural killer cells. iScience. (2023) 26:107187. doi: 10.1016/j.isci.2023.107187
78. Sojka DK, Plougastel-Douglas B, Yang L, Pak-Wittel MA, Artyomov MN, Ivanova Y, et al. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. eLife. (2014) 3:e01659. doi: 10.7554/eLife.01659
79. You Z, Ling S, Zhao S, Han H, Bian Y, He Y, et al. Tissue damage from chronic liver injury inhibits peripheral NK cell abundance and proinflammatory function. J Leukocyte Biol. (2024) 115:1042–52. doi: 10.1093/jleuko/qiae027
80. Ricardo-Gonzalez RR, Van Dyken SJ, Schneider C, Lee J, Nussbaum JC, Liang H-E, et al. Tissue signals imprint ILC2 identity with anticipatory function. Nat Immunol. (2018) 19:1093–9. doi: 10.1038/s41590-018-0201-4
81. Rao L, Cai L, and Huang L. Single-cell dynamics of liver development in postnatal pigs. Sci Bull. (2023) 68:2583–97. doi: 10.1016/j.scib.2023.09.021
82. Papalexi E and Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol. (2018) 18:35–45. doi: 10.1038/nri.2017.76
83. Bischoff P, Trinks A, Obermayer B, Pett JP, Wiederspahn J, Uhlitz F, et al. Single-cell RNA sequencing reveals distinct tumor microenvironmental patterns in lung adenocarcinoma. Oncogene. (2021) 40:6748–58. doi: 10.1038/s41388-021-02054-3
84. Smith SL, Kennedy PR, Stacey KB, Worboys JD, Yarwood A, Seo S, et al. Diversity of peripheral blood human NK cells identified by single-cell RNA sequencing. Blood Adv. (2020) 4:1388–406. doi: 10.1182/bloodadvances.2019000699
85. Han M, Hu L, Wu D, Zhang Y, Li P, Zhao X, et al. IL-21R-STAT3 signalling initiates a differentiation program in uterine tissue-resident NK cells to support pregnancy. Nat Commun. (2023) 14:7109. doi: 10.1038/s41467-023-42990-0
86. Vallejo J, Saigusa R, Gulati R, Armstrong Suthahar SS, Suryawanshi V, Alimadadi A, et al. Combined protein and transcript single-cell RNA sequencing in human peripheral blood mononuclear cells. BMC Biol. (2022) 20:193. doi: 10.1186/s12915-022-01382-4
87. Liu H, Zhao R, Qin R, Sun H, Huang Q, Liu L, et al. Panoramic comparison between NK cells in healthy and cancerous liver through single-cell RNA sequencing. Cancer Biol Med. (2022) 19(9):1334–51. doi: 10.20892/j.issn.2095-3941.2022.0050
88. Wang C. The heterogeneous immune landscape between lung adenocarcinoma and squamous carcinoma revealed by single-cell RNA sequencing. Signal Transduct Target Ther. (2022) 7(1):289. doi: 10.1038/s41392-022-01130-8
89. Ding S, Qiao N, Zhu Q, Tong Y, Wang S, Chen X, et al. Single-cell atlas reveals a distinct immune profile fostered by T cell-B cell crosstalk in triple negative breast cancer. Cancer Commun (Lond). (2023) 43(6):661–84. doi: 10.1002/cac2.12429
90. De Andrade LF, Lu Y, Luoma A, Ito Y, Pan D, Pyrdol JW, et al. Discovery of specialized NK cell populations infiltrating human melanoma metastases. JCI Insight. (2019) 4:e133103. doi: 10.1172/jci.insight.133103
91. Moreno-Nieves UY, Tay JK, Saumyaa S, Horowitz NB, Shin JH, Mohammad IA, et al. Landscape of innate lymphoid cells in human head and neck cancer reveals divergent NK cell states in the tumor microenvironment. Proc Natl Acad Sci USA. (2021) 118:e2101169118. doi: 10.1073/pnas.2101169118
92. Lin Q, Zhou Y, Ma J, Han S, Huang Y, Wu F, et al. Single-cell analysis reveals the multiple patterns of immune escape in the nasopharyngeal carcinoma microenvironment. Clin Trans Med. (2023) 13:e1315. doi: 10.1002/ctm2.1315
93. O’Sullivan TE, Rapp M, Fan X, Weizman O-E, Bhardwaj P, Adams NM, et al. Adipose-resident group 1 innate lymphoid cells promote obesity-associated insulin resistance. Immunity. (2016) 45:428–41. doi: 10.1016/j.immuni.2016.06.016
94. Kim N, Kim HK, Lee K, Hong Y, Cho JH, Choi JW, et al. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat Commun. (2020) 11:2285. doi: 10.1038/s41467-020-16164-1
95. Wienke J, Visser LL, Kholosy WM, Keller KM, Barisa M, Poon E, et al. Integrative analysis of neuroblastoma by single-cell RNA sequencing identifies the NECTIN2-TIGIT axis as a target for immunotherapy. Cancer Cell. (2024) 42:283–300.e8. doi: 10.1016/j.ccell.2023.12.008
96. De Zuani M, Xue H, Park JS, Dentro SC, Seferbekova Z, Tessier J, et al. Single-cell and spatial transcriptomics analysis of non-small cell lung cancer. Nat Commun. (2024) 15:4388. doi: 10.1038/s41467-024-48700-8
97. Ou Z, Lin S, Qiu J, Ding W, Ren P, Chen D, et al. Single-nucleus RNA sequencing and spatial transcriptomics reveal the immunological microenvironment of cervical squamous cell carcinoma. Adv Sci. (2022) 9:2203040. doi: 10.1002/advs.202203040
98. Rajendran S, Hu Y, Canella A, Peterson C, Gross A, Cam M, et al. Single-cell RNA sequencing reveals immunosuppressive myeloid cell diversity during Malignant progression in a murine model of glioma. Cell Rep. (2023) 42:112197. doi: 10.1016/j.celrep.2023.112197
99. Lavin Y, Kobayashi S, Leader A, Amir ED, Elefant N, Bigenwald C, et al. Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell. (2017) 169:750–765.e17. doi: 10.1016/j.cell.2017.04.014
100. Tang F, Li J, Qi L, Liu D, Bo Y, Qin S, et al. A pan-cancer single-cell panorama of human natural killer cells. Cell. (2023) 186(19):4235–4251.e20. doi: 10.1016/j.cell.2023.07.034
101. Xu L, Saunders K, Huang SP, Knutsdotti H, Martinez-Algarin K, Terrazas I, et al. A comprehensive single-cell breast tumor atlas defines epithelial and immune heterogeneity and interactions predicting anti-PD-1 therapy response. Open Access. (2024) 5(5):101511. doi: 10.1016/j.xcrm.2024.101511
102. Xiao J, Yu X, Meng F, Zhang Y, Zhou W, Ren Y, et al. Integrating spatial and single-cell transcriptomics reveals tumor heterogeneity and intercellular networks in colorectal cancer. Cell Death Dis. (2024) 15:326. doi: 10.1038/s41419-024-06598-6
103. Zamler DB, Shingu T, Kahn LM, Huntoon K, Kassab C, Ott M, et al. Immune landscape of a genetically engineered murine model of glioma compared with human glioma. JCI Insight. (2022) 7:e148990. doi: 10.1172/jci.insight.148990
104. Kandala S, Ramos M, Voith von Voithenberg L, et al. Chronic chromosome instability induced by Plk1 results in immune suppression in breast cancer. Cell Rep. (2023) 42(12):113266. doi: 10.1016/j.celrep.2023.113266
105. Chen Y-P, Yin J-H, Li W-F, Li H-J, Chen D-P, Zhang C-J, et al. Single-cell transcriptomics reveals regulators underlying immune cell diversity and immune subtypes associated with prognosis in nasopharyngeal carcinoma. Cell Res. (2020) 30:1024–42. doi: 10.1038/s41422-020-0374-x
106. Zhu J, Fan Y, Xiong Y, et al. Delineating the dynamic evolution from preneoplasia to invasive lung adenocarcinoma by integrating single-cell RNA sequencing and spatial transcriptomics. Mol Med. (2022) 54(11):2060–76. doi: 10.1038/s12276-022-00896-9
107. Huo X, Ma S, Wang C, Song L, Yao B, Zhu S, et al. Unravelling the role of immune cells and FN1 in the recurrence and therapeutic process of skull base chordoma. Clin Trans Med. (2023) 13:e1429. doi: 10.1002/ctm2.1429
108. Deng J-Y, Gou Q, Yang L, Chen Z-H, Yang M-Y, Yang X-R, et al. Immune suppressive microenvironment in liver metastases contributes to organ-specific response of immunotherapy in advanced non-small cell lung cancer. J Immunother Cancer. (2023) 11:e007218. doi: 10.1136/jitc-2023-007218
109. Xin S, Liu X, Li Z, Sun X, Wang R, Zhang Z, et al. ScRNA-seq revealed an immunosuppression state and tumor microenvironment heterogeneity related to lymph node metastasis in prostate cancer. Exp Hematol Oncol. (2023) 12:49. doi: 10.1186/s40164-023-00407-0
110. Wang M, Zadeh S, Pizzolla A, Thia K, Gyorki DE, McArthur GA, et al. Characterization of the treatment-naive immune microenvironment in melanoma with BRAF mutation. J Immunother Cancer. (2022) 10:e004095. doi: 10.1136/jitc-2021-004095
111. Jiang H, Yu D, Yang P, Guo R, Kong M, Gao Y, et al. Revealing the transcriptional heterogeneity of organ-specific metastasis in human gastric cancer using single-cell RNA sequencing. Clin Trans Med. (2022) 12:e730. doi: 10.1002/ctm2.730
112. Liu YQ, Luo M, Shi Y, Guo Y, Zhang H, Yang K-D, et al. Dicer deficiency impairs proliferation but potentiates anti-tumoral effect of macrophages in glioblastoma. Oncogene. (2022) 41(30):3791–803. doi: 10.1038/s41388-022-02393-9
113. Liu L, Hou Y, Deng C, Tao Z, Chen Z, Hu J, et al. Single cell sequencing reveals that CD39 inhibition mediates changes to the tumor microenvironment. Nat Commun. (2022) 13:6740. doi: 10.1038/s41467-022-34495-z
114. Cao G, Yue J, Ruan Y, Han Y, Zhi Y, Lu J, et al. Single-cell dissection of cervical cancer reveals key subsets of the tumor immune microenvironment. EMBO J. (2023) 42:e110757. doi: 10.15252/embj.2022110757
115. Sun Y, Li T, Ding L, Wang J, Chen C, Liu T, et al. Platelet-mediated circulating tumor cell evasion from natural killer cell killing through immune checkpoint CD155-TIGIT. Hepatology. (2024) 81(3):791–807. doi: 10.1097/HEP.0000000000000934
116. Sathe A, Grimes SM, Lau BT, Chen J, Suarez C, Huang RJ, et al.Single cell genomic characterization reveals the cellular reprogramming of the gastric tumor microenvironment. Clin Cancer Res. (2020) 26(11):2640–53. doi: 10.1158/1078-0432.CCR-19-3231
117. Kim H-J. Blood monocyte-derived CD169+ Macrophages contribute to antitumor immunity against glioblastoma. Nat Commun. (2022) 13(1):6211. doi: 10.1038/s41467-022-34001-5
118. Croizer H. Deciphering the spatial landscape and plasticity of immunosuppressive fibroblasts in breast cancer. Nat Commun. (2024) 15(1):2806. doi: 10.1038/s41467-024-47068-z
119. Yi Q. scRNA-seq and imaging mass cytometry analyses unveil iNKT cells-mediated anti-tumor immunity in pancreatic cancer liver metastasis. Cancer Lett. (2023) 597:217077. doi: 10.1016/j.canlet.2023.216149
120. Wu Q, Hu X, Zhang X, Kong D, Yang Z, Li G, et al. Single-cell transcriptomics of peripheral blood reveals anti-tumor systemic immunity induced by oncolytic virotherapy. Theranostics. (2022) 12(17):7371–89. doi: 10.7150/thno.74075
121. Jiao J. Radiofrequency radiation reshapes tumor immune microenvironment into antitumor phenotype in pulmonary metastatic melanoma by inducing active transformation of tumor-infiltrating CD8+ T and NK cells. Acta Pharmacol Sin. (2024) 45(7):1492–505. doi: 10.1038/s41401-024-01260-5
122. Li X-Y, Corvino D, Nowlan B, Aguilera AR, Ng SS, Braun M, et al. NKG7 is required for optimal antitumor T-cell immunity. Cancer Immunol Res. (2022) 10(2):154–61. doi: 10.1158/2326-6066.CIR-20-0649
123. Ni J, Wang X, Stojanovic A, Zhang Q, Wincher M, Bu L, et al. Single-cell RNA sequencing of tumor-infiltrating NK cells reveals that inhibition of transcription factor HIF-1a unleashes NK cell activity. Immunity. (2020) 52(6):1075–1087.e8. doi: 10.1016/j.immuni.2020.05.001
124. Bi J, Huang C, Jin X, Zheng C, Huang Y, Zheng X, et al. TIPE2 deletion improves the therapeutic potential of adoptively transferred NK cells. J Immunother Cancer. (2023) 11(2):e006002. doi: 10.1136/jitc-2022-006002
125. Lachota M. Mapping the chemotactic landscape in NK cells reveals subset-specific synergistic migratory responses to dual chemokine receptor ligation. EBioMedicine. (2023) 96:104811. doi: 10.1016/j.ebiom.2023.104811
126. Yang F, Yuan C, Chen F, Qin ZS, Schmitt NC, Lesinski GB, et al. Combined IL6 and CCR2 blockade potentiates antitumor activity of NK cells in HPV-negative head and neck cancer. J Exp Clin Cancer Res. (2024) 43:76. doi: 10.1186/s13046-024-03002-1
127. Tang F, Li J, Qi L, Liu D, Bo Y, Qin S, et al. A pan-cancer single-cell panorama of human natural killer cells. Cell. (2023) 186:4235–4251.e20. doi: 10.1016/j.cell.2023.07.034
128. Sozio F, Schioppa T, Laffranchi M, Salvi V, Tamassia N, Bianchetto-Aguilera FM, et al. CCRL2 expression by specialized lung capillary endothelial cells controls NK-cell homing in lung cancer. Cancer Immunol Res. (2023) 11:1280–95. doi: 10.1158/2326-6066.CIR-22-0951
129. Zhang Y, Bai Y, Ma X-X, Song J-K, Luo Y, Fei X-Y, et al. Clinical-mediated discovery of pyroptosis in CD8+ T cell and NK cell reveals melanoma heterogeneity by single-cell and bulk sequence. Cell Death Dis. (2023) 14:553. doi: 10.1038/s41419-023-06068-5
130. Wu Q, Tian P, He D, Jia Z, He Y, Luo W, et al. SCUBE2 mediates bone metastasis of luminal breast cancer by modulating immune-suppressive osteoblastic niches. Cell Res. (2023) 33:464–78. doi: 10.1038/s41422-023-00810-6
131. Ribas A and Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science. (2018) 359:1350–5. doi: 10.1126/science.aar4060
132. Kim H, Park S, Han K-Y, Lee N, Kim H, Jung HA, et al. Clonal expansion of resident memory T cells in peripheral blood of patients with non-small cell lung cancer during immune checkpoint inhibitor treatment. J Immunother Cancer. (2023) 11:e005509. doi: 10.1136/jitc-2022-005509
133. Han D, Han Y, Guo W, Wei W, Yang S, Xiang J, et al. High-dimensional single-cell proteomics analysis of esophageal squamous cell carcinoma reveals dynamic alterations of the tumor immune microenvironment after neoadjuvant therapy. J Immunother Cancer. (2023) 11:e007847. doi: 10.1136/jitc-2023-007847
134. Shang J. Single-cell profiling reveals the heterogeneity of NK cells during anti-PD-1 therapy in non-small-cell lung cancer. Int Immunopharmacol. (2023) 124(Pt A):110743. doi: 10.1016/j.intimp.2023.110743
135. Lam KC. Microbiota triggers STING-type I IFN-dependent monocyte reprogramming of the tumor microenvironment. Cell. (2021) 184(21):5338–5356.e21. doi: 10.1136/jitc-2022-006002
136. Huuhtanen J, Kasanen H, Peltola K, Lönnberg T, Glumoff V, Brück O, et al. Single-cell characterization of anti-LAG3+anti-PD1 treatment in melanoma patients. J Clin Invest. (2023) 133(6):e164809. doi: 10.21417/JH2023JCI
137. Hu J. Tumor microenvironment remodeling after neoadjuvant immunotherapy in non-small cell lung cancer revealed by single-cell RNA sequencing. Genome Med. (2023) 15(1):14. doi: 10.1186/s13073-023-01164-9
138. Wan C, Keany MP, Dong H, Al-Alem LF, Pandya UM, Lazo S, et al. Enhanced efficacy of simultaneous PD-1 and PD-L1 immune checkpoint blockade in high-grade serous ovarian cancer. Cancer Res. (2021) 81:158–73. doi: 10.1158/0008-5472.CAN-20-1674
139. Yang D, Zhao F, Su Y, Zhou Y, Shen J, Yu B, et al. Integrated analysis of single-cell and bulk RNA-sequencing identifies a signature based on NK cell marker genes to predict prognosis and immunotherapy response in hepatocellular carcinoma. J Cancer Res Clin Oncol. (2023) 149:10609–21. doi: 10.1007/s00432-023-04965-y
140. Kim R, An M, Lee H, Mehta A, Heo YJ, Kim K-M, et al. Early tumor–immune microenvironmental remodeling and response to first-line fluoropyrimidine and platinum chemotherapy in advanced gastric cancer. Cancer Discov. (2022) 12:984–1001. doi: 10.1158/2159-8290.CD-21-0888
141. Chen Y, Chen C-Y, Huang H, Luo Z, Mu Y, Li S, et al. Knocking down of Xkr8 Enhances Chemotherapy Efficacy through Modulating Tumor Immune Microenvironment. J Controlled Release. (2024) 370:479–89. doi: 10.1016/j.jconrel.2024.04.041
142. Pan B, Chen Z, Zhang X, Wang Z, Yao Y, Wu X, et al. 2,5-dimethylcelecoxib alleviated NK and T-cell exhaustion in hepatocellular carcinoma via the gastrointestinal microbiota-AMPK-mTOR axis. J Immunother Cancer. (2023) 11:e006817. doi: 10.1136/jitc-2023-006817
143. Wu S, Peng H, Li S, Huang L, Wang X, Li Y, et al. The ω-3 polyunsaturated fatty acid docosahexaenoic acid enhances NK-cell antitumor effector functions. Cancer Immunol Res. (2024) 12:744–58. doi: 10.1158/2326-6066.CIR-23-0359
144. Li C-Y. Repurposing nitric oxide donating drugs in cancer therapy through immune modulation. J Exp Clin Cancer Res (2023) 42(1):22. doi: 10.1186/s13046-022-02590-0
145. Zhou W, Yu M, Mao X, Pan H, Tang X, Wang J, et al. Landscape of the peripheral immune response induced by local microwave ablation in patients with breast cancer. Adv Sci. (2022) 9(17):e2200033. doi: 10.1002/advs.202200033
146. Zhang Y, Yang Q, Zeng X, Wang M, Dong S, Yang B, et al. MET amplification attenuates lung tumor response to immunotherapy by inhibiting STING. Cancer Discov. (2021) 11(11):2726–37. doi: 10.1158/2159-8290.CD-20-1500
147. Du R, Zhang X, Lu X, Ma X, Guo X, Shi C, et al. PDPN positive CAFs contribute to HER2 positive breast cancer resistance to trastuzumab by inhibiting antibody-dependent NK cell-mediated cytotoxicity. Drug Resist Updates. (2023) 68:100947. doi: 10.1016/j.drup.2023.100947
148. Slattery K, Woods E, Zaiatz-Bittencourt V, Marks S, Chew S, Conroy M, et al. TGFβ Drives NK cell metabolic dysfunction in human metastatic breast cancer. J Immunother Cancer. (2021) 9:e002044. doi: 10.1136/jitc-2020-002044
149. Lähnemann D, Köster J, Szczurek E, McCarthy DJ, Hicks SC, Robinson MD, et al. Eleven grand challenges in single-cell data science. (2020) 21(1):31. doi: 10.1186/s13059-020-1926-6
150. Longo SK, Guo MG, Ji AL, and Khavari PA. Integrating single-cell and spatial transcriptomics to elucidate intercellular tissue dynamics. Nat Rev Genet. (2021) 22:627–44. doi: 10.1038/s41576-021-00370-8
151. Gong Y, Klein Wolterink RGJ, Wang J, Bos GMJ, and Germeraad WTV. Chimeric antigen receptor natural killer (CAR-NK) cell design and engineering for cancer therapy. J Hematol Oncol. (2021) 14:73. doi: 10.1186/s13045-021-01083-5
152. Dong H, Ham JD, Hu G, Xie G, Vergara J, Liang Y, et al. Memory-like NK cells armed with a neoepitope-specific CAR exhibit potent activity against NPM1 mutated acute myeloid leukemia. Proc Natl Acad Sci USA. (2022) 119:e2122379119. doi: 10.1073/pnas.2122379119
153. Bexte T, Albinger N, Al Ajami A, Wendel P, Buchinger L, Gessner A, et al. CRISPR/cas9 editing of NKG2A improves the efficacy of primary CD33-directed chimeric antigen receptor natural killer cells. Nat Commun. (2024) 15:8439. doi: 10.1038/s41467-024-52388-1
154. Knelson EH, Ivanova EV, Tarannum M, Campisi M, Lizotte PH, Booker MA, et al. Activation of tumor-cell STING primes NK-cell therapy. Cancer Immunol Res. (2022) 10:947–61. doi: 10.1158/2326-6066.CIR-22-0017
155. Li L, Mohanty V, Dou J, Huang Y, Banerjee PP, Miao Q, et al. Loss of metabolic fitness drives tumor resistance after CAR-NK cell therapy and can be overcome by cytokine engineering. Sci Adv. (2023) 9:eadd6997. doi: 10.1126/sciadv.add6997
156. Sullivan MR, Finocchiaro M, Yang Y, Thomas J, Ali A, Kaplan I, et al. An innovative single-cell approach for phenotyping and functional genotyping of CAR NK cells. J Immunother Cancer. (2024) 12:e008912. doi: 10.1136/jitc-2024-008912
Keywords: single-RNA sequencing, NK cell, heterogeneity, tumor microenvironment, drug intervention
Citation: Shen M, Liu Y, Shao L, Qu M, Song S, Sun W and Zhang H (2025) Unraveling NK cell heterogeneity through single-cell sequencing: insights from physiological and tumor contexts for clinical applications. Front. Immunol. 16:1612352. doi: 10.3389/fimmu.2025.1612352
Received: 15 April 2025; Accepted: 02 July 2025;
Published: 21 July 2025.
Edited by:
Elise Nassif, University of Texas MD Anderson Cancer Center, United StatesReviewed by:
Sara Passos, Century Therapeutics, United StatesSiyao Liu, Texas A and M University, United States
Copyright © 2025 Shen, Liu, Shao, Qu, Song, Sun and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hao Zhang, aGFvemhhbmdAY211LmVkdS5jbg==; Wei Sun, c3VuMTk4OTAyMDhAMTI2LmNvbQ==
 Mingxin Shen
Mingxin Shen Yutong Liu2
Yutong Liu2 Liang Shao
Liang Shao Wei Sun
Wei Sun Hao Zhang
Hao Zhang