- 1Laboratory Medicine, First Affiliated Hospital of Gannan Medical University, Ganzhou, China
- 2Laboratory Medicine, People’s Hospital of Ganzhou Economic Development Zone, Ganzhou, China
- 3Department of Gastroenterology, First Affiliated Hospital of Gannan Medical University, Ganzhou, China
- 4The First School of Clinical Medicine, Gannan Medical University, Ganzhou, China
- 5Department of Cardiothoracic Surgery, First Affiliated Hospital of Gannan Medical University, Ganzhou, China
Exosomes are nanoscale, double-membraned vesicles released by a variety of living cells. A wide variety of cargoes, including proteins, DNA and RNA, are transported by exosomes to target cells, thereby transmitting biological signals. In addition to being an essential component of the exosomal cargo, exosomal proteins are a reflection of the physiological state of the originating cell, and play an essential part in intercellular communication in numerous diseases, including cancer. The present review provides a summary of the novel uses of exosomal proteins in cancer diagnosis and prognosis, and highlights the distinct qualities that exosomal proteins possess, when compared with typical serological measurements.
1 Introduction
Cancer is a leading cause of death worldwide. At present, tissue biopsy is considered the gold standard method for the clinical diagnosis of cancer. However, tissue collection may be complex, due to the close proximity of some tumors to major arteries in the body. In addition, obtaining tumor tissues from a specific location may not accurately represent the overall health of the patient (1). By contrast, liquid biopsies, which include biopsies of circulating tumor cells (CTCs), free DNA (cfDNA) and exosomes, provide further information regarding the status of the tumor, and do not require invasive procedures (2). This information may be obtained through frequent non-invasive sampling, which aids in the diagnosis of cancer and monitoring the pathophysiologic condition of the patient (3, 4).
Exosomes are biologically active extracellular vesicles that range from 50–200 nanometers in size, and are produced by live cells (5). Exosomes are located in the majority of bodily fluids, including urine, blood, saliva, ascites, breast milk, amniotic fluid and cerebrospinal fluid (6). Exosomes are responsible for transporting a variety of substances (7, 8), including proteins, nucleic acids, lipids, enzymes and metabolites, and these are regulated by the physiology of the original cell (9). Several mechanisms, including direct fusion with cell membranes, endocytosis routes and ligand-receptor interactions, are involved in the process of intercellular communication (10–12). Notably, exosomes may play a role in intercellular communication.
As tumor cells are primarily responsible for the production of tumor-derived exosomes that contain chemicals which reflect the properties of the parental tumor cells (13), they may exhibit potential as diagnostic markers for tumors (14). Importantly, liquid biopsy of exosomes may be more useful than CTC or circulating tumor DNA (ctDNA), as there is a large volume of exosomes in bodily fluids (up to 1011 exosomes per ml in blood), making them easier to detect. In addition, ~10% of exosomes detected in the bodily fluids of individuals who have advanced malignancies originate from tumor cells (15). At present, research is focused on exosomal RNAs, including microRNAs, circular RNAs (circRNA) and long non-coding RNAs (16–18). Exosomal proteins are either encased inside an inner lumen or embedded on the surface of the exosome. This allows for the categorization of exosome subtypes based on surface biomarkers, without causing any disruption to the structure of the exosomes. Compared with alternate exosomal cargoes, exosomal proteins exhibit numerous advantages, including i) a long half-life and stability inside the exosomes in which they are found (19), and ii) a relatively straightforward isolation procedure, due to identification of exosomal surface proteins in smaller sample sizes (20–22). Through the use of proteomics, protein coverage and sensitivity have been significantly enhanced, which has led to an increase in the breadth of exosomal proteomics data surrounding cancer causation, function and disease association (23). Further investigations are required to determine the specific process of carcinogenesis and the advancement of cancer. The present study aimed to provide a summary of the diagnostic and prognostic functions that exosomal proteins play in a variety of malignancies, and demonstrate the potential uses of exosomal proteins in clinical cancer treatment.
2 Biogenesis of exosomes
Extracellular vesicles (EVs) are a collective name that refers to nanoscale vesicles that are actively released by cells (24). Exosomes are formed through the fusion of multi-vesicular vesicles with the cell membrane, with a diameter of 50–200 nm, and microvesicles are formed through the direct outgrowth of the cell membrane, with a diameter of 200-2,000 nm. Moreover, apoptotic vesicles are formed through the shrinkage and fragmentation of apoptotic cells, with a diameter of 500-2,000 nm (25) (Figure 1A).
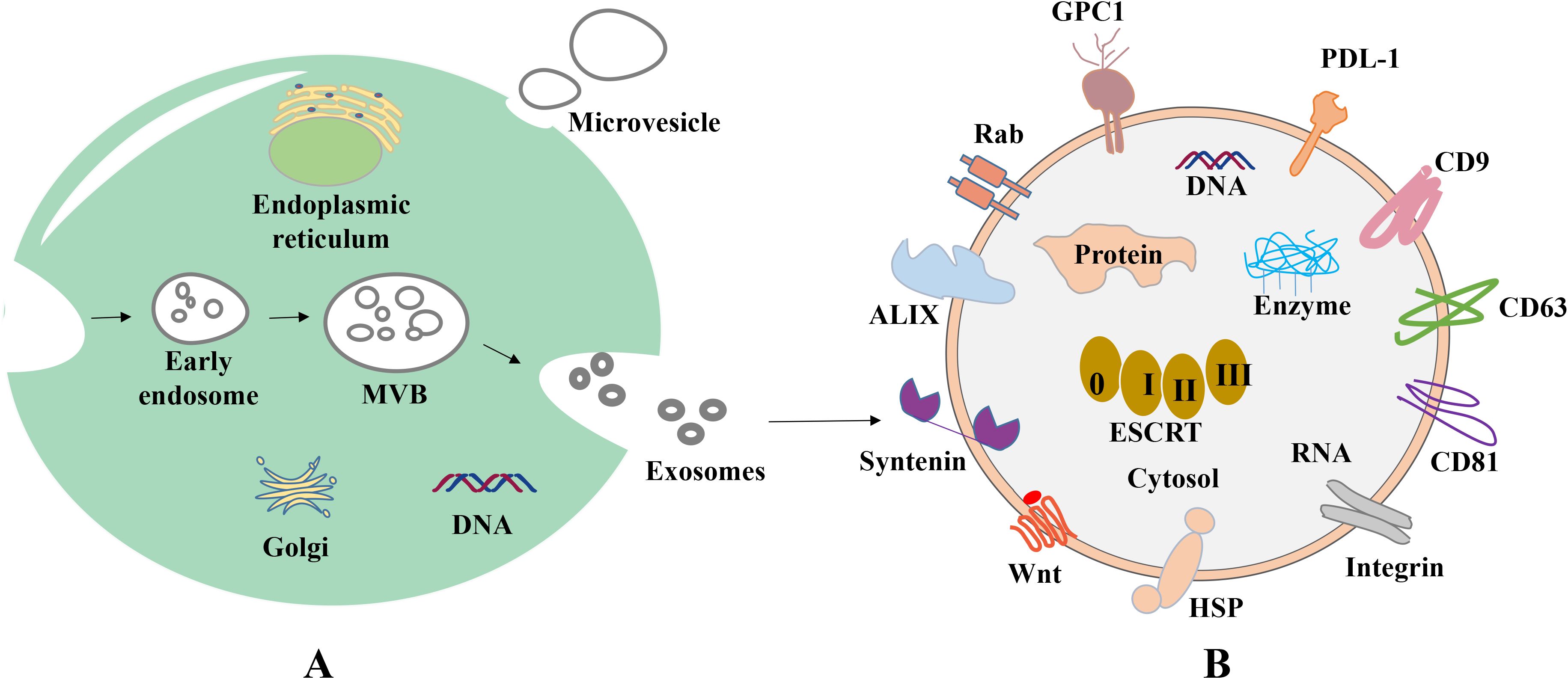
Figure 1. Biogenesis and composition of exosomes. (A) Origin of exosomes. First, plasma membrane invagination of donor cells forms early endosomes, which further mature into late endosomes. During the maturation, the membrane of early endosomes invaginates inwardly to form ILVs. Endosomes with ILVs are often referred to as MVBs. When MVBs fuse with the plasma membrane, ILVs are released into the extracellular space and termed as exosomes. (B) Common proteins of exosomes. Tetraspanin proteins (CD9, CD63, CD81), PD-L1, Integrins; Wnt protein, ALIX, Syntenin, HSPs, GPC1, Rabs, Flotillin, etc.
In 1983, exosomes were initially isolated from sheep reticulocytes. Through the investigation of the transferrin receptor during the maturation of reticulocytes, Johnston et al. discovered that the mechanism of the transferrin receptor is lost when exosomes are created during the maturation of erythrocytes (26). There are a variety of cell types that are capable of actively producing exosomes, including human umbilical vein endothelial cells, reticulocytes and immune cells, such as lymphocytes, macrophages, dendritic cells, natural killer cells, stem cells, endothelial cells and neuronal cells (27–30). Exosomes may be produced under both healthy and pathological conditions, and exosomal development includes initiation, endocytosis, the creation of multivesicular bodies and release. The creation of exosomes begins with the endocytosis of vesicles, leading to the development of early sorting endosomes via the invagination of the cell membrane. Subsequently, these endosomes evolve into late sorting endosomes (31), and these bud inward to form multivesicular bodies (MVBs). MVBs ultimately fuse with the plasma membrane and discharge their contents into the extracellular environment, and these are referred to as exosomes (Figure 1A). Moreover, released exosomes may be directed to other cells using a variety of cell surface proteins, including tetraspanning proteins (32–34).
Endosomal sorting complexes (ESCRT) are required for translocation-dependent processes. Significantly, ESCRT and non-ESCRT mechanisms (35) play a key role in the production of exosomes. ESCRT-0, ESCRT-I, ESCRT-II and ESCRT-III are the four functional subcomplexes involved in the ESCRT mechanism, and this is comprised of ~30 proteins. Particularly, the aforementioned subcomplexes are required for exosomal biogenesis. Lipids and associated proteins, such as the transmembrane tetraprotein CD63, play key roles in the ESCRT pathway, and these do not require any other proteins. Results of previous studies demonstrated that specific structures, such as lipid rafts and proteins with a four-transmembrane structural domain, may play a crucial role in the creation of certain exosomes (36, 37). Notably, exosomes transfer information to receptor cells via three primary routes (Figure 2); namely, i) Receptor-ligand interactions; ii) direct fusion with the cell membrane; and iii) endocytosis via phagocytosis.
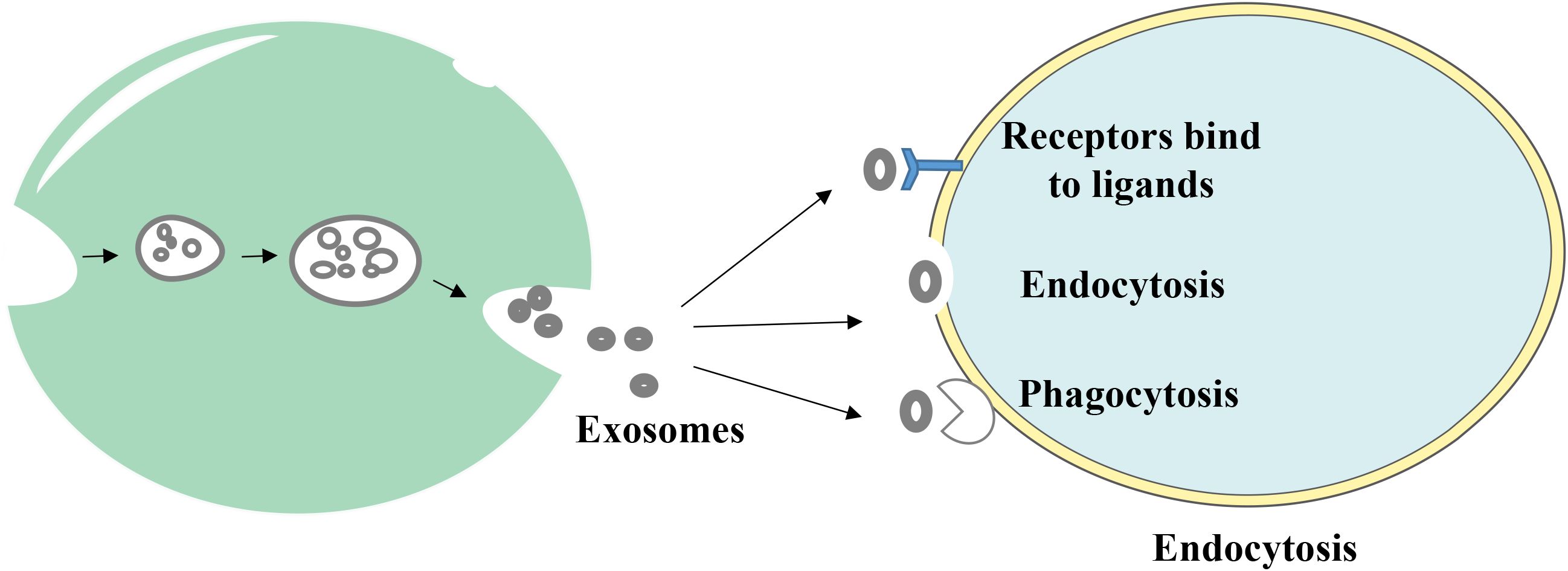
Figure 2. The way exosomes bind to receptor cells. Exosomes transmit information to receptor cells through three main pathways: (1) receptor-ligand interactions, (2) direct fusion with the cell membrane, and (3) endocytosis via phagocytosis.
3 Proteins of exosomes
At present, research is focused on exosomal proteins due to their unique biological functions and the key roles that they play in regulating the tumor microenvironment (TME). Some exosomal proteins are embedded on the surface of the membrane, and some are completely encased in the membrane (Figure 1B). Importantly, certain proteins, such as CD63, TSG101 and Alix, have been recognized as biomarkers for exosomes, while others, such as Calnexin, may function as negative markers for exosome identification (38). Specific proteins on exosomes, such as epidermal growth factor receptor (EGFR), Ephrin type-A receptor 2 (EphA2) and Epithelial cell adhesion molecule (EpCAM), are increasingly used to distinguish between tumor-derived exosomes and non-tumor-derived exosomes (39). Metastatic ovarian cancer cells release numerous exosomes carrying E-cadherin, which is an inducer of angiogenesis (40). Expression levels of exosomal lipopolysaccharide-binding protein and E-cadherin were used to identify non-small cell lung cancer (NSCLC) and ovarian cancer cells with metastatic phenotypes (41). Results of a previous study revealed that in patients with head and neck squamous cell carcinoma, exosomal Programmed cell death ligand 1 (PD-L1) expression levels were associated with disease progression, UICC stage and lymph node invasion (42). Moreover, the detection of PD-L1 positive exosomes in blood samples from patients with pancreatic ductal adenocarcinoma was associated with poorer survival (43). As a key immune checkpoint molecule, the expression levels of PD-L1 directly reflect the ability of tumor cells to evade immune surveillance. It has been found that revealed that PD-L1 inhibits T cell function and promotes immune escape through binding to PD-1 on the surface of T cells (44). In addition, epigenetic studies revealed that the demethylation status of the PD-L1 promoter is positively correlated with its expression levels (45). Collectively, these studies suggest that exosomal surface proteins may exhibit potential in the diagnosis and prognosis prediction of cancer; thus, exhibiting potential in monitoring treatment response. In addition, exosomal surface proteins may contribute to further understanding the mechanisms underlying exosome biogenesis (46–51), targeting (52, 53) and interaction (54–57). The main types of exosomal surface proteins and their functions are displayed in Table 1.
4 Function of exosomes in cancer
Research has focused on the role of exosomes in tumors due to the key role they play in intercellular communication. Exosomes that originate from tumor cells or stromal cells are associated with all phases of cancer development. These stages include tumor growth and cell proliferation, the prevention of cell death, angiogenesis, immune evasion, invasion and metastasis. It has been shown that exosomes may play a key role in the intricate biological interactions that take place between tumor cells and the TME (58). Interestingly, there are numerous different physiologically active chemicals that are carried by exosomes. These compounds are critical signals for reprogramming the TME for the induction of carcinogenesis (59). For example, delta-like 4 protein (DLL4) plays a key role in the promotion of cancer-associated TME alterations (60) and the expansion of vascular branching. The presence of DLL4 in individuals with colorectal cancer (CRC) may be associated with aggressiveness of the tumor and negative clinical outcomes (61). Moreover, tumor cell-derived exosomes may function as catalysts for epithelial-to-mesenchymal transition (EMT) and tumor metastasis. This is accomplished by stimulating quiescent cancer cells to actively metastasize via the use of a wide range of signaling molecules, such as Notch1 and HIF1α (62, 63). It has been shown that tumor-derived exosomes may assist tumor cells in evading monitoring of the immune system, developing resistance to chemotherapy and further promoting the growth of the tumor. Results of previous studies revealed that tumor-derived exosomes decrease the cytotoxicity of natural killer cells and cytotoxic T-lymphocytes. Particularly, these cells are essential components of the immune response, which plays a key role in preventing the growth of tumors (64, 65). Previous research has shown that tumor-derived exosomes may promote the polarization of immature macrophages into M2 macrophages and exhibit anti-inflammatory activity, both of which are beneficial to the continued spread of the tumor. Interestingly, angiogenesis is a fundamental physiological process during carcinogenesis that involves numerous steps. A wide range of angiogenic factors, including vascular endothelial growth factor, interleukin, transforming growth factor-β and fibroblast growth factor, are located inside exosomes (66). These factors play a crucial role in promoting the proliferation and migration of endothelial cells, and are essential in the generation of tumor angiogenesis (67, 68).
5 Potential use of exosomal proteins in the diagnosis of cancer
Given their disease-specific alterations, exosomal proteins have emerged as potential biomarkers for cancer. Exosomes that are produced from tumors carry a large amount of data regarding the biology of cancer cells (69). Levels of exosomal proteins in patients with cancer are considerably higher than those observed in healthy individuals. As a result, the identification of proteins in exosomes is sensitive and advantageous for the early diagnosis of cancer. Thus, the identification of exosomal proteins may exhibit potential in the diagnosis of cancer and in predicting patient prognosis (70). The tumor-derived exosomal protein analysis process is displayed in Figure 3, and Table 2 summarizes the exosomal proteins that may exhibit potential as diagnostic markers in various tumors.
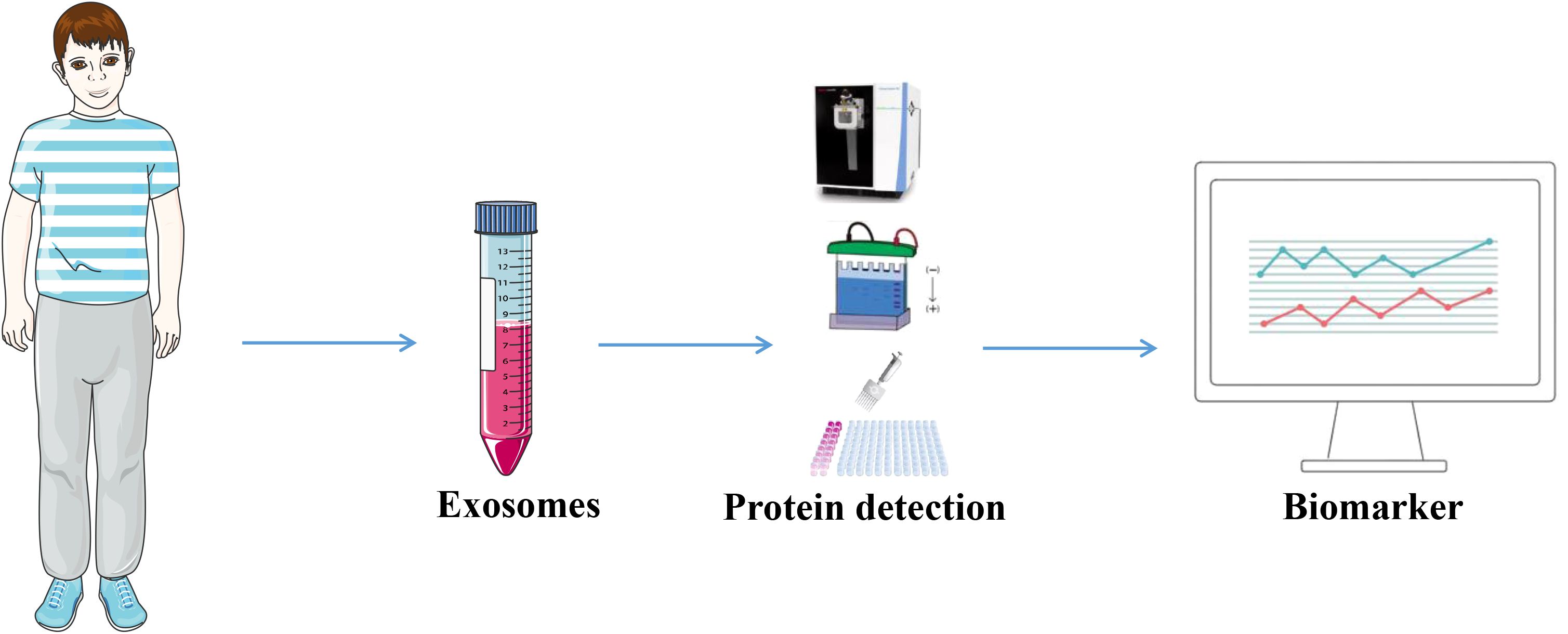
Figure 3. Tumor-derived exosomal proteins were used as diagnostic markers. Exosomes were extracted from body fluids from tumor patients and analyzed by Western Blotting, ELISA, proteomics and other exosomal proteins for diagnosis.
5.1 Digestive cancers, including colorectal cancer, stomach, liver and pancreatic cancer
Exosomes include a variety of proteins that may exhibit potential as diagnostic markers for CRC. These proteins include carcinoembryonic antigen (CEA), epidermal growth factor receptor, mitogen-activated protein kinase and keratin (71). Previous studies have found that these proteins may be used for the diagnosis of CRC. In the previous study, Glypican-1+ (GPC1+) exosomes were successfully separated from the tissues and plasma of CRC. Compared with controls, the expression of GPC1+ exosomes were markedly higher in the tumor tissues and plasma of patients with CRC prior to surgical treatment (72). CD147-positive exosomes have been shown to be effective as a diagnostic indicator for colorectal cancer, with an AUC of 0.827 (73). In addition, exosomes produced from CRC cells have been subjected to proteomic analysis, which revealed the unique expression of a number of metastatic factors. Importantly, these factors include hepatocyte growth factor receptor, S100A8, S100A9 and tenascin C (74, 75). A histological examination of phosphorylated proteins in exosomes obtained from metastatic CRC cells revealed that the levels of phosphorylated proteins in these exosomes were higher than in those located in exosomes derived from non-metastatic CRC cells (76, 77). Further investigations are required to determine the potential use of exosomal proteins in the detection and treatment of CRC.
According to the World Cancer Report, gastric cancer (GC) is the fourth most common cancer worldwide, and the second highest cause of cancer-associated death (78). Yoon et al. revealed that the blood GKN1 levels of healthy controls were markedly higher than those of patients with GC. Especially, the sensitivity and specificity of GC were 91.2 and 96.0%, respectively (79). In addition, results of a previous study revealed that serum GKN1 levels may aid in distinguishing individuals with CRC from those with CRC, liver, lung, breast, pancreatic, ovarian or prostate cancers that exhibited area under the curve (AUC) values of >0.94 (80). Interestingly, exosomes produced from GC cells possess the ability to promote activation of the NF-ÙB pathway in macrophages, leading to the advancement of cancer (81). It has been found that the involvement of exosomes carrying tetraspanin 8 may be associated with the proliferation and invasion of cells in GC, and that tetraspanin 8 is an independent factor in determining the prognosis of patients with GC (82). Serum-derived exosomes of HER2 as a promising biomarker for advanced gastric cancer had an area under the ROC curve of 0.746, a sensitivity of 66.7% and a specificity of 74.2% (83). Collectively, these results imply that exosomal proteins may exhibit potential as diagnostic and prognostic indicators for GC.
When liver cancer is in its early stages, the symptoms that patients experience are often non-specific, and late diagnosis leads to limited treatment options. Protein levels in serum exosomes obtained from patients with hepatocellular carcinoma (HCC) and a healthy cohort were analyzed by Arbelaiz et al, who demonstrated that the expression levels of G3BP and PIGR were significantly elevated in patients with HCC. Moreover, exosomal proteins were more effective than AFP in predicting HCC (84). Fu et al. revealed that the amount of SMAD3-positive exosomes generated from HCC cells was positively correlated with the staging and pathologic grading of HCC; however, this was negatively correlated with the disease-free survival of patients with HCC following surgery (85). In addition, 14-3–3 protein expression levels were associated with a larger tumor size, poorer tumor differentiation and more advanced TNM staging. Wang et al. revealed that the amount of exosomal 14-3–3 proteins were elevated in HCC cell sources. Notably, 14-3–3 protein expression levels were also associated with increased tumor size (86).
In addition, exosomal proteins may exhibit potential in the pathological detection of pancreatic cancer (PC). Buscail et al. revealed that exosomes produced from PC cells that were positive for GPC1 exhibited a high level of accuracy, with an AUC of 1.0, a sensitivity of 100% and a specificity of 80% (87). Combined detection of the exosomal membrane proteins EGFR and HER2 improves the diagnostic sensitivity of pancreatic cancer (85%) and is particularly effective in CA19-9-negative patients (88). Individuals with metastatic pancreatic cancer exhibited markedly increased levels of macrophage migration inhibitory factors (MIFs), compared with those who did not experience progression of PC. These findings suggested that exosomal MIFs may exhibit potential as predictive markers for liver metastasis (89). Thus, further investigations are required to determine the specific function of exosomal proteins in PC.
5.2 Urinary cancers, including prostate, bladder and ovarian cancer
Plasma prostate-specific antigen, also referred to as prostate specific antigen (PSA), is a biomarker that is frequently used for the purpose of detecting and monitoring prostate cancer. Nilsson et al. revealed that urine exosomes derived from individuals with prostate cancer exhibited high expression levels of β-catenin, prostate cancer gene-3 (PCA-3) and several other markers associated with prostate cancer. These findings highlight the potential for urine exosomes in identifying and monitoring cancer (90). Moreover, Øverbye et al. conducted a study on urine exosomal proteins in a group consisting of 16 individuals diagnosed with prostate cancer and 15 healthy controls. Results of this previous study revealed that 246 proteins were differently expressed in both groups of patients. Out of 17 of these proteins, all 17 exhibited a sensitivity of >60% and a specificity of 100%, with TM256 exhibiting the highest level of sensitivity at 94% (91). Proteomes of urine exosomes obtained from patients with prostate cancer and healthy participants were compared, and the results revealed that TM256 and ADIRF exhibited the greatest diagnostic value (91). Collectively, these results revealed that urine exosomal proteins may exhibit potential as a source of enrichment for prostate cancer indicators.
The tumor-associated calcium signaling 2 (TACSTD2) protein, which exhibits potential in the detection of bladder cancer (92), is one of the 29 urine exosomal proteins that have emerged as novel prospective biomarkers. Lee et al. carried out proteome characterization of urine-derived exosomes from ten healthy controls and ten patients with bladder cancer. Results of this previous study revealed that 56 proteins were highly expressed in the urinary exosomes of patients with bladder cancer (93). In addition, the expression levels of CD36 and CD44 in exosomes were identified via immunoblotting and flow cytometry, and results of this previous study revealed substantial differences in the expression of CD36 and CD44 between healthy individuals and patients with bladder cancer (94).
It is estimated that >70% of ovarian cancer diagnoses are made at an advanced stage (95), leading to high levels of mortality in female patients. Research focused on epithelial cell adhesion molecules and CD24 in exosomes produced from ovarian cancer has led to a novel option for the early diagnosis of ovarian cancer (96). The study shows that serum-derived exosome Claudin 4 steadily increased with the advancement of cancer in patients with ovarian cancer (97). In addition, expression levels of the exosome surface marker, HSP70, were higher in exosomes formed from OC cells, compared with those obtained from healthy controls. Moreover, study find that the serum of individuals with OC exhibited a significant quantity of exosomes that expressed HER2. Thus, further investigations are required to determine and distinguish exosomal populations, and investigate the biological activities of exosomal proteins in biological organisms (98).
5.3 Thoracic cancers, including lung and breast cancer
Exosomal proteins may exhibit potential in the diagnosis of non-small cell lung cancer (NSCLC). Significantly, 2% of exosomes were located in chronic inflammatory lung tissue (99). Wu et al. revealed that 80% of exosomes recovered from NSCLC biopsies were positive for EGFR. Interestingly, EGFR, K-Ras, claudin1, claudin3 and RAB family proteins were among the potential diagnostic indicators discovered by Park et al (100), when exosomes were recovered from pleural effusions of patients with NSCLC. Results of proteomic mass spectrometry (101) revealed that human leucine-rich alpha-2-glycoprotein 1 (LRG1) was concentrated in urine exosomes, and was expressed at higher levels in patients with NSCLC, compared with healthy individuals. Sandfeld et al. used 49 antibodies to identify EV proteins obtained from 431 patients with lung cancer and 150 healthy individuals (102). Moreover, a diagnostic model containing 30 exosomal proteins was developed in a further study, with a sensitivity and specificity of ~75% (103). The model was constructed using an exosome array to harvest exosomes from the blood of patients with NSCLC. Yang et al. identified plasma-derived exosomal immunoglobulins IGHV4–4 and IGLV1–40 as new non-small cell lung cancer biomarkers (104). A recent study has identified plasma exosomal versican as a potential diagnostic marker for non-small cell lung cancer (105). Thus, exosomes and associated components may exhibit potential in the early identification of lung cancer.
At present, breast cancer is considered the most prevalent form of cancer among females. In patients with metastatic breast cancer, the 5-year survival rate is ~20%, despite >50% of patients with breast cancer developing metastases following systemic intervention. Melo et al. demonstrated that 75% of patients with breast cancer exhibited greater levels of exosomal GPC1 expression, compared with healthy controls (106). The diagnostic value of fibronectin and developmental endothelial locus-1 in exosomes produced from breast cancer cells was reported by Moon et al, with an AUC of 0.961, a sensitivity of 94.70%, and a specificity of 86.36% (107, 108). Moreover, the unique expression pattern of exosomal survivin-2B in serum may exhibit potential as an indicator for breast cancer in its early stages (109).
5.4 Other cancers
In a wide range of malignancies, exosomal proteins have been proposed as potential diagnostic and prognostic indicators. The study noted that numerous clinical samples may include a high number of exosomal proteins that are specific to melanoma (110), such as caveolin-1. Serum exosomes were analyzed in patients with glioblastoma, and results of a previous study revealed that EGFR, EGFRvIII and CD63 were expressed at high levels (111). Fraser et al. discovered that leucine-rich repeat kinase 2 (LRRK2), which is abundant in urine exosomes, may exhibit potential as a biomarker for Parkinson’s disease (112). In addition, the existence of spongioblastoma-specific EGFR variant type III (EGFRV III) was discovered through the detection of serum exosomes from 25 individuals with spongioblastoma (113). Reale recently showed that proteomic characterization of human plasma extracellular vesicles provides important implications for the development of multiple myeloma diagnostics (114). A recent study identified 199 common proteins in exomes secreted from Synovial sarcoma cells, with the monocarboxylate transporter 1 (MCT1) as a novel surface marker, highly expressed in SS patient-derived exosomes compared with healthy individuals (115). Wang et al. found that DIO3OS could be a potential biomarker for thyroid-like cancer (116). With further research, exosomal proteins are increasingly used in various cancer diagnostics.
6 Potential therapeutic value of exosomal proteins in cancer
Exosomal proteins not only exhibit potential as a molecular marker for the diagnosis of cancer, but also demonstrate therapeutic potential due to their unique biological characteristics. At present, research is focused on use of engineered exosomes for the targeted distribution of anti-cancer therapy. Results of a previous study revealed that modified exosomes that include adriamycin may be more effective in targeting tumors and inhibiting their growth (117). An additional investigation was conducted in which the parental cells of exosomes were modified to produce Lamp2b linked to αv integrin-specific iRGD peptide. Importantly, this peptide demonstrated sufficient tumor-targeting characteristics in prostate, breast and cervical cancer models (118). In addition, The study noted that a subtype of MVBs, known as inhibitory protein structural domain 1-mediated microvesicles, aids in the transportation of NOTCH receptors to target cells and the stimulation of downstream gene expression (119). Votteler et al. discovered an innovative method for hybridizing exosomes using envelope protein nanocages, also known as EPNs. Through the process of membrane attachment and self-assembly, extracellular polymeric nanoparticles (EPNs) transfer their payload into the cytoplasm of recipient cells (120). This is accomplished through binding a variety of designed proteins.
In addition, the immunological activity of exosomes may allow them to be used as drug transporters in cancer immunotherapy or as vaccines (121). As exosomes are responsible for transporting a large number of tumor antigens, they may play a role in antigen presentation, exhibiting potential as cancer vaccines (122). Moreover, Hsp70 plays a key role in activating dendritic cells and monocytes, which in turn stimulates immunological responses that are mediated by tumor-derived exosomes (123). Hsp70 also plays a key role in promoting the release of granzyme B from natural killer cells, ultimately leading to the induction of apoptosis in tumor cells (124). Interestingly, HSP70 may function as an exosome surface antigen, eliciting anti-tumor antibody responses. Thus, exosomal proteins exhibit potential in the treatment of a wide range of malignancies. Clinical trials of exosomes in cancer therapy have focused on the following areas, with specific applications including as drug delivery vehicles, immunomodulators, diagnostic markers, and combination therapy tools. Most of the trials are currently in the early stages (phase I/II), but the multifunctional properties of exosomes make them have great potential in personalized cancer therapy (Table 3). With advances in bioengineering technology, more precise clinical applications may be realized in the future.
7 Discussion
As a novel form of liquid biopsy markers, exosomal proteins may demonstrate potential in the diagnosis of cancer. In recent years, tumor-derived exosomal proteins have demonstrated unique diagnostic value, and key advantages of these proteins include the following factors: i) Disease-specific enrichment, where the protein composition of exosomes highly reflects the physiological and pathological status of their parent cells, particularly tumor cell-derived exosomes (TEXs). These exosomes carry a large number of tumor-related proteins, such as EGFR, HER2, PD-L1, MET and GPC1, which may be used as cancer-specific markers; ii) a high level of stability, making them suitable for long-term storage and the detection of clinical samples. The lipid bilayer membrane structure of exosomes protects the internal proteins from proteases in the blood. Notably, this is more stable than free proteins or ctDNA due to the strong anti-degradation ability; iii) the potential clinical application of minimally invasive/non-invasive testing. Especially, small volumes of bodily fluid, such as 1 ml of blood or urine, is required for routine tests, mitigating limitations associated with tissue biopsy and sampling; and iv) multi-dimensional diagnostic information integration. Multiple protein marker combinations may be analyzed at the same time to improve diagnostic accuracy.
Although exosomal proteins exhibit potential in the diagnosis of cancer, limitations may remain, leading to limited use in clinical practice. Limitations may include the following factors: i) Complexities in the standardization of isolation and purification techniques. At present, exosome isolation requires numerous techniques, such as ultracentrifugation, size-exclusion chromatography, polymer precipitation and immunoaffinity capture. However, these methods exhibit notable differences in recovery rate, purity and integrity of exosomes. Contaminants, such as lipoproteins and protein aggregates in blood samples, often co-precipitate with exosomes, affecting downstream analysis. Although the International Society for Extracellular Vesicles (ISEV) has proposed the MISEV guideline, a globally recognized standardized process is yet to be developed, resulting in poor comparability of data; ii) analytical challenges associated with tumor heterogeneity. Importantly, exosomes in bodily fluids may be derived from tumor cells, immune cells and platelets. Tumor-derived exosomes may account for >1% of total exosomes, and therefore require enrichment with highly specific markers. In addition, the composition of exosomal proteins changes continuously with tumor progression and treatment response, and a dynamic monitoring system is required; and iii) complexities in large-scale application and the translation to clinical practice. The majority of studies include small-scale cohorts and lack validation through multi-center, prospective clinical trials. High-precision separation and detection technologies, such as nanoflow or single exosome analysis are costly, and there may be barriers associated with their use in clinical practice.
Exosomal proteins exhibit high levels of potential in the diagnosis of cancer; however, interdisciplinary collaboration, optimization of technical processes and the establishment of a globally uniform standardization system are required for their use in clinical practice. Future research should focus on developing high-purity and high-throughput separation technologies, such as microfluidic chips and aptamer capture. In addition, further investigations focused on the establishment of a multi-omics integrated analysis process are required, to integrate proteins, RNA and metabolites. Future research should also focus on the use of AI-assisted marker screening to improve the efficiency of data analysis. The aforementioned investigations may lead to evolution of the field of liquid biopsy, revolutionizing the early diagnosis and treatment of cancer.
As emerging tumor markers and therapeutic carriers, exosomal proteins have demonstrated revolutionary potential in the diagnosis and treatment of cancer. In the diagnosis of cancer, the unique molecular features and high stability of exosomes may provide a novel basis for early screening, precise typing and the dynamic monitoring of tumors. As a natural delivery system, exosomal proteins may exhibit potential in the treatment of cancer, leading to the development of targeted therapies and immunomodulation. However, future investigations are required to overcome key challenges, including isolation standardization, up-scaling of production and clinical validation. With the integration and development of interdisciplinary technologies, such as nanotechnology, multi-omics analysis and artificial intelligence, exosomal proteins may lead to considerable developments in the diagnosis and treatment of cancer, aiding the development of precise and personalized medicines. Notably, future investigations should focus on establishing a standardized technical system through multi-center clinical trials, and exploring the specific molecular mechanisms associated with exosomes.
Author contributions
HS: Data curation, Writing – original draft. ML: Data curation, Writing – original draft. WY: Data curation, Writing – original draft. DX: Investigation, Writing – original draft. ZP: Investigation, Writing – original draft. DR: Investigation, Writing – review & editing. DH: Funding acquisition, Investigation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Ganzhou Science and Technology Plan Project (2023LNS37081 and GZ2024YLJ137), Jiangxi Provincial Health Commission Technology Plan Project (202510463) and Jiangxi Province Administration of Traditional Chinese Medicine Science and Technology Plan Project (2024B0303).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Oshi M, Murthy V, Takahashi H, Huyser M, Okano M, Tokumaru Y, et al. Urine as a source of liquid biopsy for cancer. Cancers (Basel). (2021) 13. doi: 10.3390/cancers13112652
2. Watanabe K, Nakamura Y, and Low SK. Clinical implementation and current advancement of blood liquid biopsy in cancer. J Hum Genet. (2021) 66:909–26. doi: 10.1038/s10038-021-00939-5
3. Zhang W, Xia W, Lv Z, Ni C, Xin Y, and Yang L. Liquid biopsy for cancer: circulating tumor cells, circulating free DNA or exosomes? Cell Physiol Biochem. (2017) 41:755–68.
4. Wu L, Wang Y, Zhu L, Liu Y, Wang T, Liu D, et al. Aptamer-based liquid biopsy. ACS Appl Bio Mater. (2020) 3:2743–64. doi: 10.1021/acsabm.9b01194
5. Thery C, Amigorena S, Raposo G, and Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. (2006). doi: 10.1002/0471143030.2006.30.issue-1
6. Li QL, Bu N, Yu YC, Hua W, and Xin XY. Exvivo experiments of human ovarian cancer ascites-derived exosomes presented by dendritic cells derived from umbilical cord blood for immunotherapy treatment. Clin Med Oncol. (2008) 2:461–7. doi: 10.4137/CMO.S776
7. Taylor DD and Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. (2008) 110:13–21. doi: 10.1016/j.ygyno.2008.04.033
8. Keller S, Rupp C, Stoeck A, Runz S, Fogel M, Lugert S, et al. CD24 is a marker of exosomes secreted into urine and amniotic fluid. Kidney Int. (2007) 72:1095–102. doi: 10.1038/sj.ki.5002486
9. Kalluri R and LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. (2020) 367. doi: 10.1126/science.aau6977
10. Jan AT, Rahman S, Khan S, Tasduq SA, and Choi I. Biology, pathophysiological role, and clinical implications of exosomes: A critical appraisal. Cells. (2019) 8.
11. Meckes DG Jr. Exosomal communication goes viral. J Virol. (2015) 89:5200–3. doi: 10.1128/JVI.02470-14
12. Kucharzewska P and Belting M. Emerging roles of extracellular vesicles in the adaptive response of tumour cells to microenvironmental stress. J Extracell Vesicles. (2013) 2.
13. Barteneva NS, Maltsev N, and Vorobjev IA. Microvesicles and intercellular communication in the context of parasitism. Front Cell Infect Microbiol. (2013) 3:49. doi: 10.3389/fcimb.2013.00049
14. Aslan C, Maralbashi S, Salari F, Kahroba H, Sigaroodi F, Kazemi T, et al. Tumor-derived exosomes: Implication in angiogenesis and antiangiogenesis cancer therapy. J Cell Physiol. (2019) 234:16885–903. doi: 10.1002/jcp.v234.10
15. Vaidyanathan R, Soon RH, Zhang P, Jiang K, and Lim CT. Cancer diagnosis: from tumor to liquid biopsy and beyond. Lab Chip. (2018) 19:11–34. doi: 10.1039/C8LC00684A
16. Zhang Y, Han T, Feng D, Li J, Wu M, Peng X, et al. Screening of non-invasive miRNA biomarker candidates for metastasis of gastric cancer by small RNA sequencing of plasma exosomes. Carcinogenesis. (2020) 41:582–90. doi: 10.1093/carcin/bgz186
17. Chen M, Xu R, Ji H, Greening DW, Rai A, Izumikawa K, et al. Transcriptome and long noncoding RNA sequencing of three extracellular vesicle subtypes released from the human colon cancer LIM1863 cell line. Sci Rep. (2016) 6:38397. doi: 10.1038/srep38397
18. Del Re M, Marconcini R, Pasquini G, Rofi E, Vivaldi C, Bloise F, et al. PD-L1 mRNA expression in plasma-derived exosomes is associated with response to anti-PD-1 antibodies in melanoma and NSCLC. Br J Cancer. (2018) 118:820–4. doi: 10.1038/bjc.2018.9
19. Schey KL, Luther JM, and Rose KL. Proteomics characterization of exosome cargo. Methods. (2015) 87:75–82. doi: 10.1016/j.ymeth.2015.03.018
20. Su YY, Sun L, Guo ZR, Li JC, Bai TT, Cai XX, et al. Upregulated expression of serum exosomal miR-375 and miR-1307 enhance the diagnostic power of CA125 for ovarian cancer. J Ovarian Res. (2019) 12:6. doi: 10.1186/s13048-018-0477-x
21. Meng X, Muller V, Milde-Langosch K, Trillsch F, Pantel K, and Schwarzenbach H. Diagnostic and prognostic relevance of circulating exosomal miR-373, miR-200a, miR-200b and miR-200c in patients with epithelial ovarian cancer. Oncotarget. (2016) 7:16923–35. doi: 10.18632/oncotarget.v7i13
22. Tang Y, Zhao Y, Song X, Song X, Niu L, and Xie L. Tumor-derived exosomal miRNA-320d as a biomarker for metastatic colorectal cancer. J Clin Lab Anal. (2019) 33:e23004. doi: 10.1002/jcla.23004
23. Repetto O, Lovisa F, Elia C, Enderle D, Romanato F, Buffardi S, et al. Proteomic exploration of plasma exosomes and other small extracellular vesicles in pediatric Hodgkin lymphoma: A potential source of biomarkers for relapse occurrence. Diagn (Basel). (2021) 11. doi: 10.3390/diagnostics11060917
24. Rao D, Huang D, Sang C, Zhong T, Zhang Z, and Tang Z. Advances in mesenchymal stem cell-derived exosomes as drug delivery vehicles. Front Bioeng Biotechnol. (2021) 9:797359. doi: 10.3389/fbioe.2021.797359
25. Xiao Y, Zheng L, Zou X, Wang J, Zhong J, and Zhong T. Extracellular vesicles in type 2 diabetes mellitus: key roles in pathogenesis, complications, and therapy. J Extracell Vesicles. (2019) 8:1625677. doi: 10.1080/20013078.2019.1625677
26. Pan BT and Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. (1983) 33:967–78. doi: 10.1016/0092-8674(83)90040-5
27. Raposo G and Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. (2013) 200:373–83. doi: 10.1083/jcb.201211138
28. Cheng L, Zhang K, Wu S, Cui M, and Xu T. Focus on mesenchymal stem cell-derived exosomes: opportunities and challenges in cell-free therapy. Stem Cells Int. (2017), 6305295. doi: 10.1155/2017/6305295
29. Zhang Y, Bi J, Huang J, Tang Y, Du S, and Li P. Exosome: A review of its classification, isolation techniques, storage, diagnostic and targeted therapy applications. Int J Nanomed. (2020) 15:6917–34. doi: 10.2147/IJN.S264498
30. Ha D, Yang N, and Nadithe V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: current perspectives and future challenges. Acta Pharm Sin B. (2016) 6:287–96. doi: 10.1016/j.apsb.2016.02.001
31. Zhao R, Chen X, Song H, Bie Q, and Zhang B. Dual role of MSC-derived exosomes in tumor development. Stem Cells Int. (2020), 8844730. doi: 10.1155/2020/8844730
32. Willis GR, Kourembanas S, and Mitsialis SA. Toward exosome-based therapeutics: isolation, heterogeneity, and fit-for-purpose potency. Front Cardiovasc Med. (2017) 4:63. doi: 10.3389/fcvm.2017.00063
33. Zhang X, Yuan X, Shi H, Wu L, Qian H, and Xu W. Exosomes in cancer: small particle, big player. J Hematol Oncol. (2015) 8:83. doi: 10.1186/s13045-015-0181-x
34. Schorey JS, Cheng Y, Singh PP, and Smith VL. Exosomes and other extracellular vesicles in host-pathogen interactions. EMBO Rep. (2015) 16:24–43. doi: 10.15252/embr.201439363
35. Kowal J, Tkach M, and Thery C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol. (2014) 29:116–25. doi: 10.1016/j.ceb.2014.05.004
36. Christ L, Raiborg C, Wenzel EM, Campsteijn C, and Stenmark H. Cellular functions and molecular mechanisms of the ESCRT membrane-scission machinery. Trends Biochem Sci. (2017) 42:42–56. doi: 10.1016/j.tibs.2016.08.016
37. Tschuschke M, Kocherova I, Bryja A, Mozdziak P, Angelova Volponi A, Janowicz K, et al. Inclusion biogenesis, methods of isolation and clinical application of human cellular exosomes. J Clin Med. (2020) 9. doi: 10.3390/jcm9020436
38. Liang Y, Lehrich BM, Zheng S, and Lu M. Emerging methods in biomarker identification for extracellular vesicle-based liquid biopsy. J Extracell Vesicles. (2021) 10:e12090. doi: 10.1002/jev2.12090
39. Hoshino A, Kim HS, Bojmar L, Gyan KE, Cioffi M, Hernandez J, et al. Extracellular vesicle and particle biomarkers define multiple human cancers. Cell. (2020) 182:1044–1061 e18. doi: 10.1016/j.cell.2020.07.009
40. Tang MKS, Yue PYK, Ip PP, Huang RL, Lai HC, Cheung ANY, et al. Soluble E-cadherin promotes tumor angiogenesis and localizes to exosome surface. Nat Commun. (2018) 9:2270. doi: 10.1038/s41467-018-04695-7
41. Wang N, Song X, Liu L, Niu L, Wang X, Song X, et al. Circulating exosomes contain protein biomarkers of metastatic non-small-cell lung cancer. Cancer Sci. (2018) 109:1701–9. doi: 10.1111/cas.2018.109.issue-5
42. Theodoraki MN, Yerneni SS, Hoffmann TK, Gooding WE, and Whiteside TL. Clinical significance of PD-L1(+) exosomes in plasma of head and neck cancer patients. Clin Cancer Res. (2018) 24:896–905. doi: 10.1158/1078-0432.CCR-17-2664
43. Lux A, Kahlert C, Grutzmann R, and Pilarsky C. c-met and PD-L1 on circulating exosomes as diagnostic and prognostic markers for pancreatic cancer. Int J Mol Sci. (2019) 20. doi: 10.3390/ijms20133305
44. Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. (2012) 366:2443–54. doi: 10.1056/NEJMoa1200690
45. Yi M, Niu M, Xu L, Luo S, and Wu K. Regulation of PD-L1 expression in the tumor microenvironment. J Hematol Oncol. (2021) 14:10. doi: 10.1186/s13045-020-01027-5
46. Larios J, Mercier V, Roux A, and Gruenberg J. ALIX- and ESCRT-III-dependent sorting of tetraspanins to exosomes. J Cell Biol. (2020) 219. doi: 10.1083/jcb.201904113
47. Mazurov D, Barbashova L, and Filatov A. Tetraspanin protein CD9 interacts with metalloprotease CD10 and enhances its release via exosomes. FEBS J. (2013) 280:1200–13. doi: 10.1111/febs.2013.280.issue-5
48. Malla RR, Pandrangi S, Kumari S, Gavara MM, and Badana AK. Exosomal tetraspanins as regulators of cancer progression and metastasis and novel diagnostic markers. Asia Pac J Clin Oncol. (2018) 14:383–91. doi: 10.1111/ajco.2018.14.issue-6
49. Verweij FJ, Bebelman MP, Jimenez CR, Garcia-Vallejo JJ, Janssen H, Neefjes J, et al. Quantifying exosome secretion from single cells reveals a modulatory role for GPCR signaling. J Cell Biol. (2018) 217:1129–42. doi: 10.1083/jcb.201703206
50. Blanc L and Vidal M. New insights into the function of Rab GTPases in the context of exosomal secretion. Small GTPases. (2018) 9:95–106. doi: 10.1080/21541248.2016.1264352
51. Phuyal S, Hessvik NP, Skotland T, Sandvig K, and Llorente A. Regulation of exosome release by glycosphingolipids and flotillins. FEBS J. (2014) 281:2214–27. doi: 10.1111/febs.2014.281.issue-9
52. Mashouri L, Yousefi H, Aref AR, Ahadi AM, Molaei F, and Alahari SK. Exosomes: composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol Cancer. (2019) 18:75. doi: 10.1186/s12943-019-0991-5
53. Lynch S, Santos SG, Campbell EC, Nimmo AM, Botting C, Prescott A, et al. Novel MHC class I structures on exosomes. J Immunol. (2009) 183:1884–91. doi: 10.4049/jimmunol.0900798
54. Berenguer J, Lagerweij T, Zhao XW, Dusoswa S, van der Stoop P, Westerman B, et al. Glycosylated extracellular vesicles released by glioblastoma cells are decorated by CCL18 allowing for cellular uptake via chemokine receptor CCR8. J Extracell Vesicles. (2018) 7:1446660. doi: 10.1080/20013078.2018.1446660
55. Chauhan S, Danielson S, Clements V, Edwards N, Ostrand-Rosenberg S, and Fenselau C. Surface glycoproteins of exosomes shed by myeloid-derived suppressor cells contribute to function. J Proteome Res. (2017) 16:238–46. doi: 10.1021/acs.jproteome.6b00811
56. Tian T, Zhang HX, He CP, Fan S, Zhu YL, Qi C, et al. Surface functionalized exosomes as targeted drug delivery vehicles for cerebral ischemia therapy. Biomaterials. (2018) 150:137–49. doi: 10.1016/j.biomaterials.2017.10.012
57. Munich S, Sobo-Vujanovic A, Buchser WJ, Beer-Stolz D, and Vujanovic NL. Dendritic cell exosomes directly kill tumor cells and activate natural killer cells via TNF superfamily ligands. Oncoimmunology. (2012) 1:1074–83. doi: 10.4161/onci.20897
58. Figueroa J, Phillips LM, Shahar T, Hossain A, Gumin J, Kim H, et al. Exosomes from Glioma-Associated Mesenchymal Stem Cells Increase the Tumorigenicity of Glioma Stem-like Cells via Transfer of miR-1587. Cancer Res. (2017) 77:5808–19. doi: 10.1158/0008-5472.CAN-16-2524
59. Tai YL, Chen KC, Hsieh JT, and Shen TL. Exosomes in cancer development and clinical applications. Cancer Sci. (2018) 109:2364–74. doi: 10.1111/cas.2018.109.issue-8
60. Sheldon H, Heikamp E, Turley H, Dragovic R, Thomas P, Oon CE, et al. New mechanism for Notch signaling to endothelium at a distance by Delta-like 4 incorporation into exosomes. Blood. (2010) 116:2385–94. doi: 10.1182/blood-2009-08-239228
61. Zhang Z, Li X, Yan X, Qiu H, Li G, Guo X, et al. Delta-like ligand 4 level in colorectal cancer is associated with tumor aggressiveness and clinical outcome. Cancer biomark. (2022) 33:415–22. doi: 10.3233/CBM-200986
62. Aga M, Bentz GL, Raffa S, Torrisi MR, Kondo S, Wakisaka N, et al. Exosomal HIF1alpha supports invasive potential of nasopharyngeal carcinoma-associated LMP1-positive exosomes. Oncogene. (2014) 33:4613–22. doi: 10.1038/onc.2014.66
63. Yoshizaki T, Kondo S, Wakisaka N, Murono S, Endo K, Sugimoto H, et al. Pathogenic role of Epstein-Barr virus latent membrane protein-1 in the development of nasopharyngeal carcinoma. Cancer Lett. (2013) 337:1–7. doi: 10.1016/j.canlet.2013.05.018
64. Berchem G, Noman MZ, Bosseler M, Paggetti J, Baconnais S, Le Cam E, et al. Hypoxic tumor-derived microvesicles negatively regulate NK cell function by a mechanism involving TGF-beta and miR23a transfer. Oncoimmunology. (2016) 5:e1062968.
65. Taylor DD and Gercel-Taylor C. Exosomes/microvesicles: mediators of cancer-associated immunosuppressive microenvironments. Semin Immunopathol. (2011) 33:441–54. doi: 10.1007/s00281-010-0234-8
66. Ying X, Wu Q, Wu X, Zhu Q, Wang X, Jiang L, et al. Epithelial ovarian cancer-secreted exosomal miR-222-3p induces polarization of tumor-associated macrophages. Oncotarget. (2016) 7:43076–87. doi: 10.18632/oncotarget.v7i28
67. Adem B, Vieira PF, and Melo SA. Decoding the biology of exosomes in metastasis. Trends Cancer. (2020) 6:20–30. doi: 10.1016/j.trecan.2019.11.007
68. Abak A, Abhari A, and Rahimzadeh S. Exosomes in cancer: small vesicular transporters for cancer progression and metastasis, biomarkers in cancer therapeutics. PeerJ. (2018) 6:e4763. doi: 10.7717/peerj.4763
69. Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. (2012) 18:883–91. doi: 10.1038/nm.2753
70. He M and Zeng Y. Microfluidic exosome analysis toward liquid biopsy for cancer. J Lab Autom. (2016) 21:599–608. doi: 10.1177/2211068216651035
71. Xiao Y, Zhong J, Zhong B, Huang J, Jiang L, Jiang Y, et al. Exosomes as potential sources of biomarkers in colorectal cancer. Cancer Lett. (2020) 476:13–22. doi: 10.1016/j.canlet.2020.01.033
72. Li J, Chen Y, Guo X, Zhou L, Jia Z, Peng Z, et al. GPC1 exosome and its regulatory miRNAs are specific markers for the detection and target therapy of colorectal cancer. J Cell Mol Med. (2017) 21:838–47. doi: 10.1111/jcmm.2017.21.issue-5
73. Gu C, Shang A, Liu G, Zhu J, Zhang W, Jin L, et al. Identification of CD147-positive extracellular vesicles as novel non-invasive biomarkers for the diagnosis and prognosis of colorectal cancer. Clin Chim Acta. (2023) 548:117510. doi: 10.1016/j.cca.2023.117510
74. Suwakulsiri W, Rai A, Xu R, Chen M, Greening DW, and Simpson RJ. Proteomic profiling reveals key cancer progression modulators in shed microvesicles released from isogenic human primary and metastatic colorectal cancer cell lines. Biochim Biophys Acta Proteins Proteom. (2019) 1867:140171. doi: 10.1016/j.bbapap.2018.11.008
75. Novikova S, Shushkova N, Farafonova T, Tikhonova O, Kamyshinsky R, and Zgoda V. Proteomic approach for searching for universal, tissue-specific, and line-specific markers of extracellular vesicles in lung and colorectal adenocarcinoma cell lines. Int J Mol Sci. (2020) 21. doi: 10.3390/ijms21186601
76. Sun Z, Ji S, Wu J, Tian J, Quan W, Shang A, et al. Proteomics-based identification of candidate exosomal glycoprotein biomarkers and their value for diagnosing colorectal cancer. Front Oncol. (2021) 11:725211. doi: 10.3389/fonc.2021.725211
77. Li C, Sun YD, Yu GY, Cui JR, Lou Z, Zhang H, et al. Integrated omics of metastatic colorectal cancer. Cancer Cell. (2020) 38:734–747 e9. doi: 10.1016/j.ccell.2020.08.002
78. Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, and Lordick F. Gastric cancer. Lancet (London England). (2020) 396:635–48. doi: 10.1016/S0140-6736(20)31288-5
79. Yoon JH, Park YG, Nam SW, and Park WS. The diagnostic value of serum gastrokine 1 (GKN1) protein in gastric cancer. Cancer Med. (2019) 8:5507–14. doi: 10.1002/cam4.v8.12
80. Yoon JH, Ham IH, Kim O, Ashktorab H, Smoot DT, Nam SW, et al. Gastrokine 1 protein is a potential theragnostic target for gastric cancer. Gastric Cancer. (2018) 21:956–67. doi: 10.1007/s10120-018-0828-8
81. Wu L, Zhang X, Zhang B, Shi H, Yuan X, Sun Y, et al. Exosomes derived from gastric cancer cells activate NF-kappaB pathway in macrophages to promote cancer progression. Tumour Biol. (2016) 37:12169–80. doi: 10.1007/s13277-016-5071-5
82. Anami K, Oue N, Noguchi T, Sakamoto N, Sentani K, Hayashi T, et al. TSPAN8, identified by Escherichia coli ampicillin secretion trap, is associated with cell growth and invasion in gastric cancer. Gastric Cancer. (2016) 19:370–80. doi: 10.1007/s10120-015-0478-z
83. Li Q, Lv M, Lv L, Cao N, Zhao A, Chen J, et al. Identifying HER2 from serum-derived exosomes in advanced gastric cancer as a promising biomarker for assessing tissue HER2 status and predicting the efficacy of trastuzumab-based therapy. Cancer Med. (2023) 12:4110–24. doi: 10.1002/cam4.v12.4
84. Arbelaiz A, Azkargorta M, Krawczyk M, Santos-Laso A, Lapitz A, Perugorria MJ, et al. Serum extracellular vesicles contain protein biomarkers for primary sclerosing cholangitis and cholangiocarcinoma. Hepatology. (2017) 66:1125–43. doi: 10.1002/hep.29291
85. Fu Q, Zhang Q, Lou Y, Yang J, Nie G, Chen Q, et al. Primary tumor-derived exosomes facilitate metastasis by regulating adhesion of circulating tumor cells via SMAD3 in liver cancer. Oncogene. (2018) 37:6105–18. doi: 10.1038/s41388-018-0391-0
86. Wang X, Shen H, Zhangyuan G, Huang R, Zhang W, He Q, et al. 14-3-3zeta delivered by hepatocellular carcinoma-derived exosomes impaired anti-tumor function of tumor-infiltrating T lymphocytes. Cell Death Dis. (2018) 9:159. doi: 10.1038/s41419-017-0180-7
87. Buscail E, Alix-Panabieres C, Quincy P, Cauvin T, Chauvet A, Degrandi O, et al. High clinical value of liquid biopsy to detect circulating tumor cells and tumor exosomes in pancreatic ductal adenocarcinoma patients eligible for up-front surgery. Cancers (Basel). (2019) 11. doi: 10.3390/cancers11111656
88. Chen C, Zong S, Liu Y, Wang Z, Zhang Y, Chen B, et al. Profiling of exosomal biomarkers for accurate cancer identification: combining DNA-PAINT with machine- learning-based classification. Small. (2019) 15:e1901014. doi: 10.1002/smll.201901014
89. Casari I, Howard JA, Robless EE, and Falasca M. Exosomal integrins and their influence on pancreatic cancer progression and metastasis. Cancer Lett. (2021) 507:124–34. doi: 10.1016/j.canlet.2021.03.010
90. Nilsson J, Skog J, Nordstrand A, Baranov V, Mincheva-Nilsson L, Breakefield XO, et al. Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br J Cancer. (2009) 100:1603–7. doi: 10.1038/sj.bjc.6605058
91. Overbye A, Skotland T, Koehler CJ, Thiede B, Seierstad T, Berge V, et al. Identification of prostate cancer biomarkers in urinary exosomes. Oncotarget. (2015) 6:30357–76. doi: 10.18632/oncotarget.v6i30
92. Chen CL, Lai YF, Tang P, Chien KY, Yu JS, Tsai CH, et al. Comparative and targeted proteomic analyses of urinary microparticles from bladder cancer and hernia patients. J Proteome Res. (2012) 11:5611–29. doi: 10.1021/pr3008732
93. Lee J, McKinney KQ, Pavlopoulos AJ, Niu M, Kang JW, Oh JW, et al. Altered proteome of extracellular vesicles derived from bladder cancer patients urine. Mol Cells. (2018) 41:179–87.
94. Welton JL, Khanna S, Giles PJ, Brennan P, Brewis IA, Staffurth J, et al. Proteomics analysis of bladder cancer exosomes. Mol Cell Proteomics. (2010) 9:1324–38. doi: 10.1074/mcp.M000063-MCP201
95. Khalyfa A, Almendros I, Gileles-Hillel A, Akbarpour M, Trzepizur W, Mokhlesi B, et al. Circulating exosomes potentiate tumor Malignant properties in a mouse model of chronic sleep fragmentation. Oncotarget. (2016) 7:54676–90. doi: 10.18632/oncotarget.10578
96. Runz S, Keller S, Rupp C, Stoeck A, Issa Y, Koensgen D, et al. Malignant ascites-derived exosomes of ovarian carcinoma patients contain CD24 and EpCAM. Gynecol Oncol. (2007) 107:563–71. doi: 10.1016/j.ygyno.2007.08.064
97. Li J, Sherman-Baust CA, Tsai-Turton M, Bristow RE, Roden RB, and Morin PJ. Claudin-containing exosomes in the peripheral circulation of women with ovarian cancer. BMC Cancer. (2009) 9:244. doi: 10.1186/1471-2407-9-244
98. Chanteloup G, Cordonnier M, Isambert N, Bertaut A, Marcion G, Garrido C, et al. Membrane-bound exosomal HSP70 as a biomarker for detection and monitoring of Malignant solid tumours: a pilot study. Pilot Feasib Stud. (2020) 6:35. doi: 10.1186/s40814-020-00577-2
99. Wu S, Luo M, To KKW, Zhang J, Su C, Zhang H, et al. Intercellular transfer of exosomal wild type EGFR triggers osimertinib resistance in non-small cell lung cancer. Mol Cancer. (2021) 20:17. doi: 10.1186/s12943-021-01307-9
100. Park JO, Choi DY, Choi DS, Kim HJ, Kang JW, Jung JH, et al. Identification and characterization of proteins isolated from microvesicles derived from human lung cancer pleural effusions. Proteomics. (2013) 13:2125–34. doi: 10.1002/pmic.201200323
101. Li Y, Zhang Y, Qiu F, and Qiu Z. Proteomic identification of exosomal LRG1: a potential urinary biomarker for detecting NSCLC. Electrophoresis. (2011) 32:1976–83. doi: 10.1002/elps.201000598
102. Sandfeld-Paulsen B, Jakobsen KR, Baek R, Folkersen BH, Rasmussen TR, Meldgaard P, et al. Exosomal proteins as diagnostic biomarkers in lung cancer. J Thorac Oncol. (2016) 11:1701–10. doi: 10.1016/j.jtho.2016.05.034
103. Jakobsen KR, Paulsen BS, Baek R, Varming K, Sorensen BS, and Jorgensen MM. Exosomal proteins as potential diagnostic markers in advanced non-small cell lung carcinoma. J Extracell Vesicles. (2015) 4:26659. doi: 10.3402/jev.v4.26659
104. Yang P, Zhang Y, Zhang R, Wang Y, Zhu S, Peng X, et al. Plasma-derived exosomal immunoglobulins IGHV4–4 and IGLV1–40 as new non-small cell lung cancer biomarkers. Am J Cancer Res. (2023) 13:1923–37.
105. Chang W, Zhu J, Yang D, Shang A, Sun Z, Quan W, et al. Plasma versican and plasma exosomal versican as potential diagnostic markers for non-small cell lung cancer. Respir Res. (2023) 24:140. doi: 10.1186/s12931-023-02423-4
106. Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. (2015) 523:177–82. doi: 10.1038/nature14581
107. Moon PG, Lee JE, Cho YE, Lee SJ, Chae YS, Jung JH, et al. Fibronectin on circulating extracellular vesicles as a liquid biopsy to detect breast cancer. Oncotarget. (2016) 7:40189–99. doi: 10.18632/oncotarget.v7i26
108. Moon PG, Lee JE, Cho YE, Lee SJ, Jung JH, Chae YS, et al. Identification of developmental endothelial locus-1 on circulating extracellular vesicles as a novel biomarker for early breast cancer detection. Clin Cancer Res. (2016) 22:1757–66. doi: 10.1158/1078-0432.CCR-15-0654
109. Li K, Liu T, Chen J, Ni H, and Li W. Survivin in breast cancer-derived exosomes activates fibroblasts by up-regulating SOD1, whose feedback promotes cancer proliferation and metastasis. J Biol Chem. (2020) 295:13737–52. doi: 10.1074/jbc.RA120.013805
110. Logozzi M, De Milito A, Lugini L, Borghi M, Calabro L, Spada M, et al. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PloS One. (2009) 4:e5219. doi: 10.1371/journal.pone.0005219
111. Shao H, Chung J, Balaj L, Charest A, Bigner DD, Carter BS, et al. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat Med. (2012) 18:1835–40. doi: 10.1038/nm.2994
112. Fraser KB, Moehle MS, Alcalay RN, West AB, and Consortium LC. Urinary LRRK2 phosphorylation predicts parkinsonian phenotypes in G2019S LRRK2 carriers. Neurology. (2016) 86:994–9. doi: 10.1212/WNL.0000000000002436
113. Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. (2008) 10:1470–6. doi: 10.1038/ncb1800
114. Reale A, Khong T, Xu R, Chen M, Mithraprabhu S, Bingham N, et al. Human plasma extracellular vesicle isolation and proteomic characterization for the optimization of liquid biopsy in multiple myeloma. Methods Mol Biol. (2021) 2261:151–91.
115. Yokoo S, Fujiwara T, Yoshida A, Uotani K, Morita T, Kiyono M, et al. Liquid biopsy targeting monocarboxylate transporter 1 on the surface membrane of tumor-derived extracellular vesicles from synovial sarcoma. Cancers (Basel). (2021) 13. doi: 10.3390/cancers13081823
116. Wang Y, Wang J, Wang C, Chen Y, and Chen J. DIO3OS as a potential biomarker of papillary thyroid cancer. Pathol Res Pract. (2022) 229:153695. doi: 10.1016/j.prp.2021.153695
117. Schindler C, Collinson A, Matthews C, Pointon A, Jenkinson L, Minter RR, et al. Exosomal delivery of doxorubicin enables rapid cell entry and enhanced in vitro potency. PloS One. (2019) 14:e0214545. doi: 10.1371/journal.pone.0214545
118. Li Z, Zhou X, Gao X, Bai D, Dong Y, Sun W, et al. Fusion protein engineered exosomes for targeted degradation of specific RNAs in lysosomes: a proof-of-concept study. J Extracell Vesicles. (2020) 9:1816710. doi: 10.1080/20013078.2020.1816710
119. Wang Q and Lu Q. Plasma membrane-derived extracellular microvesicles mediate non-canonical intercellular NOTCH signaling. Nat Commun. (2017) 8:709. doi: 10.1038/s41467-017-00767-2
120. Wang Q, Yu J, Kadungure T, Beyene J, Zhang H, and Lu Q. ARMMs as a versatile platform for intracellular delivery of macromolecules. Nat Commun. (2018) 9:960. doi: 10.1038/s41467-018-03390-x
121. Xu Z, Zeng S, Gong Z, and Yan Y. Exosome-based immunotherapy: a promising approach for cancer treatment. Mol Cancer. (2020) 19:160. doi: 10.1186/s12943-020-01278-3
122. Pi YN, Xia BR, Jin MZ, Jin WL, and Lou G. Exosomes: Powerful weapon for cancer nano-immunoengineering. Biochem Pharmacol. (2021) 186:114487. doi: 10.1016/j.bcp.2021.114487
123. Albakova Z, Armeev GA, Kanevskiy LM, Kovalenko EI, and Sapozhnikov AM. HSP70 multi-functionality in cancer. Cells. (2020) 9. doi: 10.3390/cells9030587
Keywords: exosomes, exosomal proteins, biomarkers, cancer diagnosis, treatment
Citation: Shen H, Liu M, Yang W, Xiao D, Peng Z, Rao D and Huang D (2025) Exosomal proteins: new targets for early diagnosis and treatment of cancer. Front. Immunol. 16:1613494. doi: 10.3389/fimmu.2025.1613494
Received: 17 April 2025; Accepted: 09 June 2025;
Published: 19 June 2025.
Edited by:
Anil Kumar Kalvala, Texas Tech University Health Sciences Center, Abilene, United StatesReviewed by:
Ashok Silwal, Texas Tech University Health Sciences Center, United StatesBhaumik Patel, Texas Tech University Health Sciences Center, Abilene, United States
Copyright © 2025 Shen, Liu, Yang, Xiao, Peng, Rao and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dingyu Rao, MTgzMjkwMzc1MjFAMTYzLmNvbQ==; Defa Huang, YWEzNzE2MzQwQDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
 Haibin Shen
Haibin Shen Meijin Liu
Meijin Liu Wentai Yang
Wentai Yang Dewang Xiao4
Dewang Xiao4 Dingyu Rao
Dingyu Rao Defa Huang
Defa Huang