- 1State Key Laboratory of Swine and Poultry Breeding Industry Guangdong Key Laboratory of Animal Breeding and Nutrition Institute of Animal Science, Guangdong Academy of Agricultural Sciences, Guangzhou, China
- 2College of Natural and Computational Science, Aksum University, Aksum, Tigray, Ethiopia
Newcastle disease (ND) is one of the most serious viral diseases affecting chickens and is caused by Newcastle disease virus (NDV), an avian paramyxovirus serotype-1. The virus contains five structural proteins and two nonstructural proteins that interact with the host proteins involved in viral infection and host antiviral responses. Currently, several NDV strains have been molecularly characterized; however, a comprehensive overview of NDV in chickens that addresses recent findings is lacking. This review summarizes the current report of the molecular structure of NDV, including candidate genes and genomic regions, virulence and route of infection, mechanisms of resistance, host immune response, disease resistance mechanisms and effects of NDV on chicken immune performance. Therefore, this review can be used by researchers seeking a comprehensive understanding that can be applied in future breeding programs aimed at enhancing disease resistance.
1 Introduction
Newcastle disease (ND) is a devastating and highly contagious disease that causes significant economic losses in the poultry industry by generating digestive, neurological, and respiratory symptoms (1). ND is caused by avian orthoavulavirus 1 (AOAV-1), commonly referred to as Newcastle disease virus (NDV) (2). The virus belongs to the order Mononegavirales, family Paramyxoviridae, and genus Orthoavulavirus (3). The genome is composed of a negative-sense, single-stranded RNA of approximately 15.2 kb coding for six structural proteins: nucleocapsid protein (NP), phosphoprotein (P), matrix protein (M), fusion protein (F), hemagglutinin-neuraminidase (HN), and large polymerase protein (L), arranged in the order of 3’-NP-P-M-F-HN-L-5’ (4). Variations in the hemagglutinin-neuraminidase (HN) and fusion (F) proteins lead to differences in the strains, affecting the virus’s ability to infect and cause disease (2). NDV strains vary in virulence and are categorized into three pathotypes based on their pathogenicity in chickens: lentogenic (low or avirulent), mesogenic (moderate virulence), and velogenic (high virulence) (5). Among the various strains of NDV, high levels (velogenic) strains typically cause severe neurological and respiratory symptoms, such as tremors, incoordination, paralysis, and sudden death (5). The virus can infect multiple tissues, including respiratory, digestive, and nervous systems, leading to destructive effects on poultry farms. The high morbidity and mortality rates of velogenic NDV strains are often associated with significant financial losses due to the mass culling of infected flocks, decreased production, and the cost of implementing control measures to prevent further spread (6). This genetic diversity of NDV strains complicates disease management and mitigation, emphasizing the need for a deeper understanding of the virus (7). The high genetic variability of NDV, virulent strains that may evade vaccine protection are more likely to emerge, potentially leading to larger epidemics with significant economic losses. Therefore, monitoring circulating NDV genotypes is crucial to reducing the impact of ND. Recently, various diagnostic strategies have been introduced, with continuous surveillance and strain characterization being key to maintaining the effectiveness of diagnostic tools and vaccines as NDV evolves (8).
In recent years, several approaches such as genome-wide association studies (GWAS), gene editing, deep sequencing, and transcriptomics have been developed to understand the viral evolution, host-pathogen interactions, vaccine development, and resistance mechanisms in chickens (6). These advancements allow for the identification of genetic markers linked to disease resistance across different chicken breeds by analyzing immune responses to NDV infection, which involves both innate and adaptive immunity (5). The involvement of genes and signaling pathways in NDV resistance highlights the potential for genetic selection based on these markers, which could significantly enhance the immune response in chickens (9).
Considering the expanding global livestock and poultry industry and the increasing volume of international trade, effective ND prevention strategies are particularly important. These strategies include strict biosecurity measures, rigorous vaccination programs, active surveillance, and proper disposal of infected carcasses (10). As the poultry industry evolves, the implications of ND extend beyond immediate health concerns to broader economic impacts. Frequent outbreaks also threaten animal health and disrupt trade relationships and markets. Therefore, understanding the persistence of NDV strains highlights the necessity for continuous molecular characterization of the virus, provision of updated information, and ongoing research into more effective vaccines and innovative control measures (11). Globalization facilitates the rapid movement of goods and animals, increasing the risk of disease spread and necessitating effective surveillance and response strategies to mitigate potential outbreaks of new diseases (11). Addressing these challenges is important for protecting public health and ensuring the sustainability of the poultry industry in an interconnected world. This approach not only addresses existing health issues but also supports long-term strategies for managing ND outbreaks globally. This review will concentrate on the current research findings on NDV, including insights into its molecular characteristics, genomic structure, pathogenicity, and mechanisms of infection and disease resistance. Additionally, we will highlight host genomics and innovative tools for NDV detection, including viral diagnostic methods and the impact of NDV.
2 Molecular characterization of NDV
Molecular characterization of NDV refers to the assessment of the virus’s genetic material, protein constituents, and molecular mechanisms that facilitate its replication, pathogenesis, and interaction with host cells. Understanding these molecular characteristics is critical for the development of vaccines, effective disease control strategies, precise diagnostic tools, and therapeutic approaches (12). More interestingly, molecular characterization encompasses several important aspects, including pathogenesis, mechanisms of infection, structures, host genomic interactions, and diagnostic approaches. The following sections provide a detailed overview of these aspects.
2.1 NDV classification
Based on phylogenetic analyses, NDV strains are classified into two primary classes: Class I, which generally includes avirulent strains primarily found in wild birds, and Class II, which encompasses both virulent and avirulent strains that affect various avian species, including domestic chickens (13). Within Class II, over 21 genotypes (I–XXI) have been identified, with notable variations in virulence that necessitate a classification system based on the severity of the diseases they cause (13). Similarly, Jia et al. (14) reported that class I NDVs were the most dominant population, with the identification of sub-genotype 1.1.2-accumulating mutations that potentially increase virulence in central China. A comprehensive study of 1,065 farms in 18 provinces revealed that the prevalence of NDV in chickens gradually declined from 1.49% in 2019 to 0.44% in 2022 at the bird level in China (1). A previous report from the Moscow region of Russia showed approximately 15 outbreaks of NDV in August 2022, with an additional three cases documented in 2023 (15). Qiu et al. (16) evaluated the biological properties of genotype VII, particularly its pathogenicity and viral replication patterns, following different inoculation routes, including intranasal, intraocular, and cloacal infections. Their findings revealed that intravenous injection induced higher morbidity and mortality, whereas intranasal, intraocular, and cloacal routes led to slower disease progression and less severe clinical symptoms than other routes of administration. Similarly, Sarika et al. (17) highlighted an important genetic relation between velogenic (highly virulent) strains of NDV and the XIII.2.2 sub-genotype circulating in India, suggesting that the XIII.2.2 sub-genotype may be associated with high pathogenicity.
To date, about half of the NDV class II genotypes, including I, IV, V, VI, VII, XI, XIII, XIV, XVII, XVIII, and XXI, with genotype VII being the most prevalent, have been reported in Africa (18). A study by Amoia et al. (19) revealed the high genetic diversity of virulent NDV strains circulating in almost all African countries, with genotypes II, IV, V, and VII being the most prevalent in East Africa, including Sudan, Somalia, Seychelles, Kenya, Uganda, South Sudan, Djibouti, Burundi, Ethiopia, Tanzania, Eritrea, Mauritius, Rwanda, and Comoros. Genotype VII has emerged as a particularly virulent variant and can be further subdivided into three subgenotypes: VII.1.1, VII.1.2, and VII.2 (5). Sub-genotypes VII.1.1 and VII.2 circulate predominantly in southern Africa, including Angola, Malawi, Eswatini, Lesotho, Zimbabwe, Zambia, Namibia, Botswana, Mozambique, and South Africa, and are predominant in northern Africa, particularly in Tunisia, Morocco, Western Sahara, Algeria, Egypt, and Libya (7, 20). Furthermore, these sub-genotypes (VII.1.1 and VII.2) were involved in the fourth NDV panzootic during the 1990s in Europe, Asia, and the Middle East, whereas VII.2 was first identified in Indonesia (21). Genotype XIII has also emerged, affecting regions such as southern, western, and central Asia and Africa (21). Similarly, genotype XVII is the most geographically dispersed genotype in Central Africa (Equatorial Guinea, Chad, São Tomé and Príncipe, Central African Republic, Democratic Republic of Congo, Cameroon, Gabon, and Congo-Brazzaville) and West Africa (Nigeria, Niger, Burkina Faso, Benin, and Mali) (19). Molecular analyses of the fusion (F) protein-encoding gene have revealed the presence of velogenic genotypes, including genotype VII, in Egypt and Saudi Arabia (22, 23).
Phylogenetic and network analyses have provided insights into the evolutionary dynamics and transmission patterns of NDV in chicken populations (24). A study using phylogenetic tree analysis revealed that NDV belongs to genotype VII, ranging from 99.7% to 98.5% with isolates from Bangladesh, Iran, and India (25). Furthermore, a recent investigation in Tanzania reported a 25.23% positivity rate for NDV in backyard chickens, emphasizing the ongoing issue in live bird markets (21). Bahoussi et al. (26) identified potential recombination events among NDV strains circulating in different provinces of China, involving strains from various genotypes and host species (chickens, ducks, and swine). Furthermore, phylogenetic analysis has identified two distinct major groups: Group I (GI), which includes a single genotype (Ib), and Group II (GII), which consists of eight genotypes (I, II, III, VI, VII, VIII, IX, and XII) (26. Notably, the Ib genotype is predominant in China, accounting for 34% of cases, with a strong concentration in the southern and eastern regions. Following Ib, genotypes VII and VI are also prevalent, representing 24% and 22% of cases, respectively (26). Sultan et al. (27) assessed a genotype VII-matched vaccine in commercial layers and found that the combination of a recombinant, genotype-matched inactivated vaccine and a live attenuated vaccine effectively reduced virus shedding and enhanced egg production in layers challenged with a velogenic genotype VII virus under field conditions. Overall, in this review, we have assessed and summarized the various types of NDV strains, their prevalence, and regional distribution (Table 1).
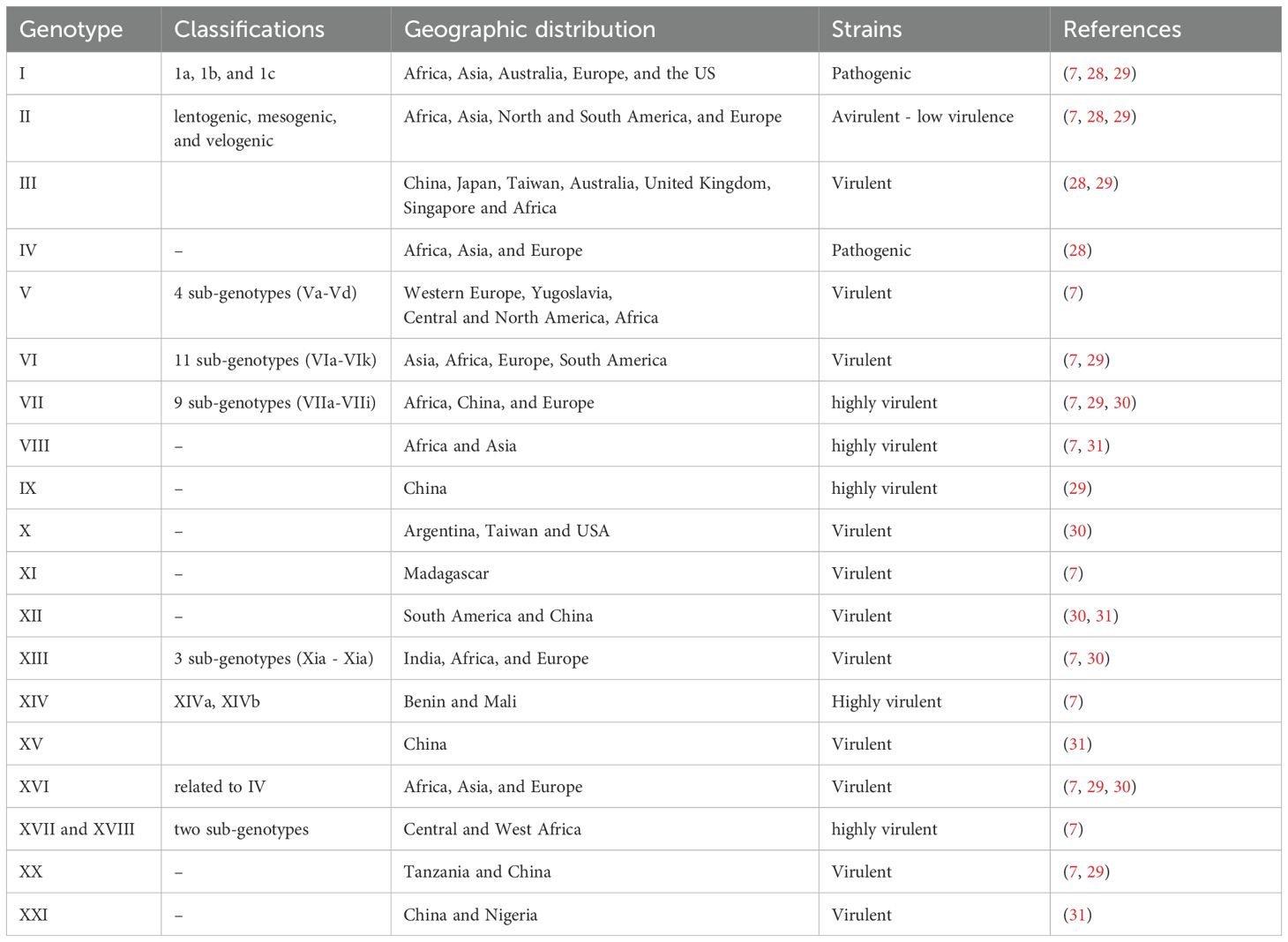
Table 1. The Newcastle disease virus genotype, genetic diversity and geographic distribution across different regions.
2.2 Genomic structure
The genomic structure of NDV is characterized by a single-stranded, negative-sense RNA genome of approximately 15,186, 15,192, and 15,198 nucleotides, depending on the genotype (32). The negative-sense nature of the NDV genome means that it is complementary to positive-sense mRNA and must be transcribed into positive-sense mRNA by the viral RNA-dependent polymerase before it can be translated into viral proteins within the host cells (33). As illustrated in Figure 1, NDV encodes six structural and two nonstructural (V and W) proteins: nucleocapsid protein (NP), phosphoprotein (P), matrix protein (M), fusion protein (F), hemagglutinin-neuraminidase (HN), and large polymerase protein (L), arranged in the order of 3’-NP-P-M-F-HN-L-5’ (4). The fusion (F) and hemagglutinin-neuraminidase (HN) glycoproteins are primary determinants of virulence, facilitating viral entry and attachment to host cells (34). The matrix (M) protein regulates viral replication and inhibits host protein synthesis, while the polymerase-associated proteins (NP, P, and L) significantly impact virulence, especially when combined, with the L protein being a major contributor (34). The P protein, essential for viral RNA synthesis, interacts with NP to form the ribonucleoprotein complex and inhibits illicit mRNA encapsidation (35).
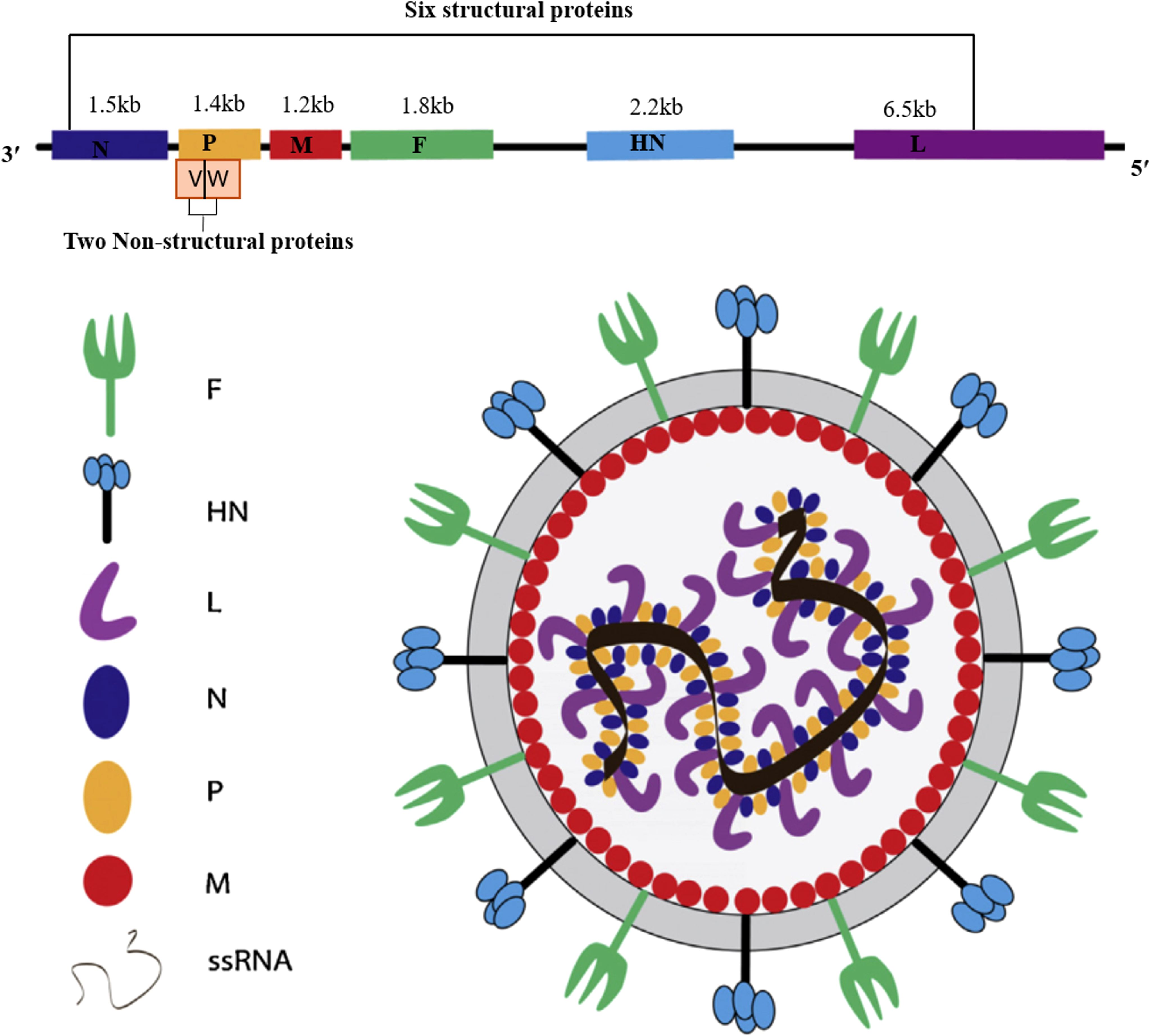
Figure 1. Schematic representation of the Newcastle disease virus structure, illustrating its six structural proteins: nucleoprotein (NP), phosphoprotein (P), matrix protein (M), fusion protein (F), hemagglutinin-neuraminidase (HN), and polymerase (L); its two non-structural proteins (V and W), along with the lengths of their corresponding nucleotide sequences.
More importantly, we also summarize the molecular structure, molecular weight, and protein length of key NDV proteins, as these factors play a crucial role in understanding viral pathogenicity and the potential for virotherapy applications (Table 2). The genome also features a leader (55 nucleotides) and trailer (114 nucleotides) sequence at the terminal ends (38), which play roles in viral transcription and pathogenesis. The intergenic sequences between these genes are essential for efficient transcription and have potential applications in chicken vaccine development (39, 40). NDV virulence is determined by multiple factors, including the amino acid sequence of the F protein cleavage site, tissue tropism, ability to evade the host immune response, and replication efficiency (5). The specific amino acid composition of F and the HN protein cleavage site play roles in the virulence and tissue tropism of NDV strains (5). The F protein must be cleaved by host proteases into F1 and F2 subunits for the virus to become infectious and virulent. The HN glycoprotein, which is responsible for virus attachment to host cell receptors, varies in length and is associated with pathogenicity (41). The protein contains 14 cysteine residues, 12 of which are conserved and form intramolecular disulfide linkages (37). Moreover, the M protein regulates viral replication and transcription, contributing to pathogenicity, whereas NP, P, and L proteins increase viral replication and virulence (42). In contrast, the F, M, and NP proteins are relatively conserved across NDV strains, whereas the length and molecular weight of the HN protein are more variable because of differences in the C-terminal region (39). The NP protein has been found to be highly conserved in the N-terminal region but highly variable in the C-terminal region (36). These proteins are essential for NDV virulence and pathogenesis, with the L gene identified as a key modulator of virulence (4). Furthermore, the two nonstructural proteins, W and V, are produced through RNA editing of the P gene and exhibit varying molecular weights and lengths depending on the specific strain of NDV (39). The V protein, commonly referred to as the antagonist protein, helps the virus evade the host innate immune response by facilitating escape from interferon (IFN) detection. The distribution of messenger RNA (mRNA) for these proteins is as follows: 68% for the P protein, 29% for the V protein, and 2% for the W protein (42). Although these nonstructural proteins have identical N-terminal domains (NTDs), their C-terminal domains (CTDs) differ from those of the P protein (11). The authors also noted that the V and W proteins contribute to NDV infection by antagonizing the host immune response.
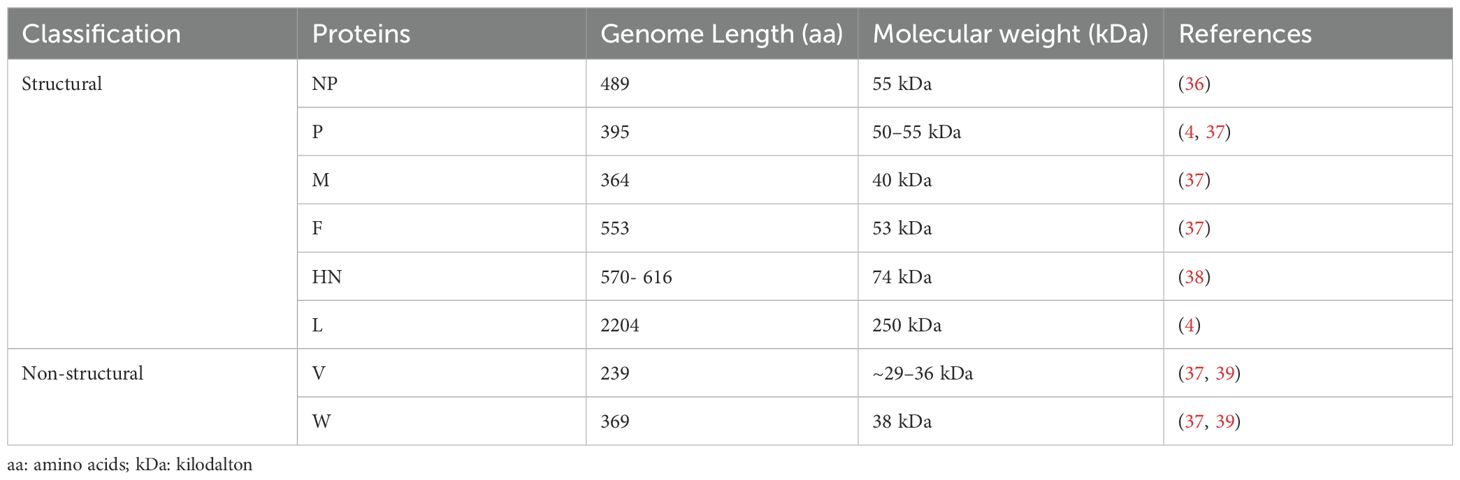
Table 2. Molecular characteristics, including structural and nonstructural proteins, exhibit a range of molecular weights and lengths.
2.3 Pathogenicity
The terms “pathogenicity” and “virulence” are often used interchangeably, but they have distinct meanings in microbial pathology. Pathogenicity is currently conceptualized as a qualitative assessment of a microorganism’s capacity to induce disease, encompassing variables such as infection, host entry mechanisms, proliferation, evasion of host defenses, and resultant tissue damage (43). In chickens, the pathogenicity of NDV strains varies greatly, ranging from mild respiratory or enteric disease to severe and often fatal infections (44). It depends on multiple factors, including the genetic type of the infecting virus, age, immune status, and host susceptibility (5). Botchway et al. (45) further confirmed that a virulent strain of NDV may be less lethal for a highly resistant host than for a susceptible host. Conversely, younger immunocompromised or genetically susceptible chickens are more vulnerable to severe diseases, even those caused by less virulent strains (46).
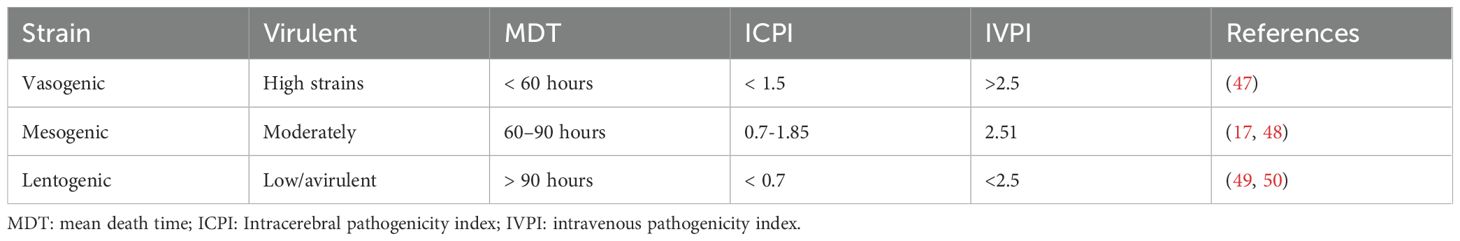
NDV is classified into different pathotypes, such as velogenic, mesogenic, and lentogenic, based on its pathogenicity (Table 3). Velogenic strains cause severe neurological and respiratory signs with high mortality rates and specific histopathological changes, whereas mesogenic strains cause respiratory signs with lower mortality rates (47). Lentogenic strains cause subclinical infections with mild respiratory or enteric diseases and are considered low-virulence strains (17). The vasogenic pathotype can be further classified into two subtypes: velogenic viscerotropic (VVNDV), which causes acute hemorrhagic lesions throughout the gastrointestinal tract, and velogenic neurotropic (VNNDV), which can lead to high mortality in chickens, with clinical signs ranging from respiratory distress and diarrhea to neurological disorders and sudden death (5).
Furthermore, the intracerebral pathogenicity index (ICPI), intravenous pathogenicity index (IVPI), and mean death time (MDT) are important measures for assessing NDV pathogenicity in chickens (6). Among these methods, ICPI is the most accurate and sensitive method for determining the virulence of NDV strains in day-old chicks, whereas IVPI is effective in six-week-old birds (47). Specifically, the ICPI values for the vaccine strains ranged from 0.0 to 0.37, indicating low virulence or non-pathogenic strains. Similarly, MDT has been used to assess the virulence of these viruses, with some strains causing death in chickens within a short period (47). Based on their MDT values, all live vaccine strains used, including the La Sota and B1 strains, were classified as lentogenic (low virulence), with the embryos remaining alive for more than 90 h (17). Studies on Brazilian commercial NDV vaccine strains in chicken clones revealed that 30 strains had an ICPI value of 0.11 and an MDT of 104 h, indicating their lentogenic (low virulence) nature (6). The author noted that the Ulster strain presented an ICPI of 0 and an MDT greater than 150 h, classifying it as lentogenic. Similarly, the Villegas-Glisson (VG/GA) strain presented an ICPI of 0.03 and an MDT of 140 h and was classified as lentogenic, indicating low virulence. The C2 strain had an ICPI of 0.04 and an MDT greater than 144 h, further confirming its lentogenic characteristics (6).
2.4 Insights into the NDV infection mechanism
Recent studies have provided insights into the NDV infection mechanisms in chickens. It involves a complex interplay between molecular interactions and cellular responses (51). The initial step of NDV infection is the attachment of the virus to specific receptors on the surface of avian cells. NDVs primarily utilize sialic acid-containing glycoproteins, specifically hemagglutinin–neuraminidase (HN) and fusion (F) proteins, as receptors (42). When the HN protein binds to the receptor, structural changes lead to the breakage of the HN and F interactions. These proteins are abundantly present on the surfaces of respiratory and intestinal epithelial cells in birds (51). Moreover, the HN and F proteins play major roles in NDV virulence, as they are involved in the assembly and budding of the virus RNA genome (42). Following attachment, the viral envelope fuses with the host cell membrane, allowing the virus to enter the cell. Once inside the host cell, the RNA genome of NDV is released for replication and transcription. As depicted in Figure 2, NDV consists of three processes: viral infection and replication, release of TAAs, PAMPs, and DAMPs, and finally, an activated immune response. For example, viral RNA polymerase transcribes negative-sense single-stranded RNA (ssRNA) into positive-sense ssRNA, which serves as a template for translating viral proteins and replicating the genome. Newly synthesized viral proteins, such as HN and F, are processed and assembled, resulting in the budding and release of new virions that can infect other tumor cells (37, 38). During the replication cycle, the viral ssRNA genome is detected by pattern recognition receptors, such as retinoic acid-inducible gene I (RIG-I). This recognition triggers downstream signaling cascades that activate type I interferons and proinflammatory responses. As the infection progresses and tumor cells undergo lysis, they release tumor-associated antigens (TAAs), pathogen-associated molecular patterns (PAMPs), including viral proteins and RNA, and danger-associated molecular patterns (DAMPs), such as surface calreticulin, cellular DNA, and ATP (52). These molecules then activate antigen-presenting cells (APCs), such as dendritic cells, which prime CD4+, CD8+, and NK cells against tumor and viral antigens, thereby stimulating an antitumor immune response. Notably, the mechanism of NDV infection in chickens is characterized by the high virulence of the virus, which causes severe respiratory, neurological, and digestive symptoms, along with its ability to induce autophagy and inflammatory pathways that exacerbate the disease (5).
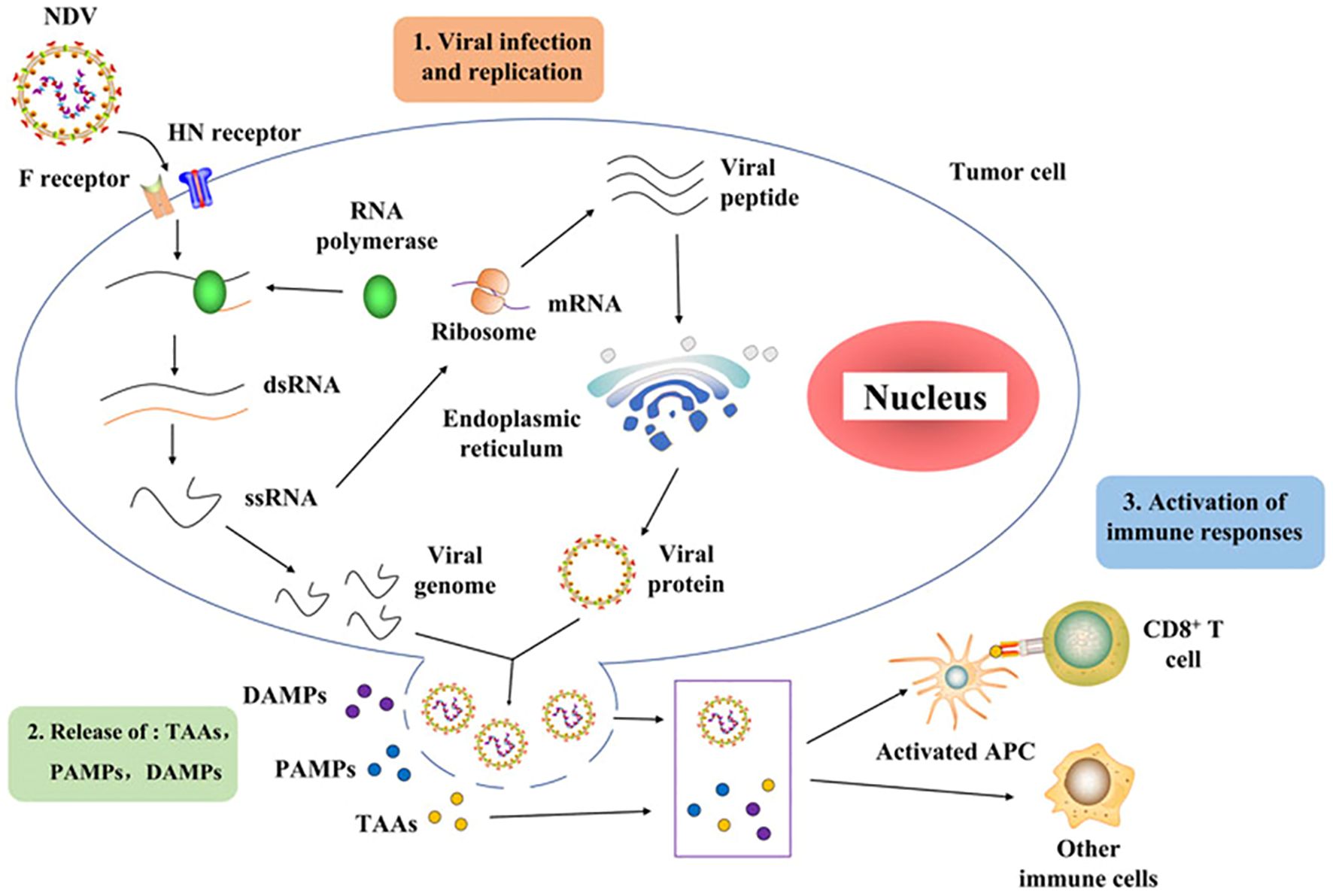
Figure 2. Schematic diagram of Newcastle disease virus infection cycle adapted from (52).
NDV primarily infects and replicates in the respiratory and digestive systems of chickens (51). Virulent strains can cause severe respiratory distress, nervous system disorders, and even sudden death. Chickens are the most susceptible domestic poultry species to NDV. NDV infection activates autophagy in chicken cells, promoting viral replication and the expression of inflammatory cytokines and chemokines, including interleukin 1 beta (IL-1β), interleukin 8 (IL-8), interleukin 18 (IL-18), C-C motif chemokine ligand 5 (CCL-5), and tumor necrosis factor alpha (TNF-α) (53). NDV-induced autophagy is positively correlated with the activation of the Caspase-1 inflammasome pathway (NLRP3) and p38/MAPK signaling pathway, resulting in increased inflammation. Finally, NDV infection can lead to mitochondrial damage and mitophagy in chicken cells, although this does not appear to significantly contribute to inflammatory responses (53).
2.5 NDV host genomics insights
Genomic studies have shed light on how host genetics influence immune responses to NDV infections in poultry. Host genomic insights in chickens refer to a comprehensive understanding and analysis of their genetic makeup (54), with a particular focus on the interactions between chicken DNA and various factors, including pathogens, immune responses, and other environmental or biological influences (55). In recent years, various advanced genomic approaches have been employed to identify genes associated with disease resistance, including deep RNA sequencing, whole-genome sequencing, signature of selection analysis, and genome-wide association studies (GWAS) (55). These approaches have enabled comprehensive analyses of differential gene expression, immune responses, and host-pathogen interactions in chickens. Transcriptomic studies have identified genes associated with immune regulation mechanisms and pathogen biology in chickens, including those involved in innate and adaptive immunity (56). Moreover, different NDV strains can induce varying levels of immune responses, with some strains leading to greater upregulation of avian β-defensins, cytokines, Toll-like receptors, and other immune-related genes (57). These studies provide insights into the potential genetic markers for resistance and the complex signaling pathways involved in the chicken immune response to NDV infection. Research utilizing whole-genome data from different chicken lines revealed approximately 60 variants of interest in interferon pathway genes that are known to affect susceptibility to viral pathogens (58), whereas GWAS revealed nine significant single-nucleotide polymorphisms associated with the heterophil/lymphocyte (H/L) ratio, located at 6.85 Mb on chromosome 19 (GGA19) in chickens. Furthermore, recent evidence has demonstrated that different chicken genetic lines present varying levels of resistance to NDV, which can be attributed to differences in the expression of host translation initiation factors (9, 59). For example, genes related to the integrated stress response (ISR) in chickens involve a complex interaction of eukaryotic translation initiation factor 2α (eIF2α) kinases in response to various stressors, regulating translation and cellular responses (9). The regulation of the eIF2α-mediated stress response is controlled by four key eIF2α kinases: protein kinase R (PKR), PKR-like ER kinase (PERK), general control nonderepressible 2 (GCN2), and heme-regulated inhibitor (HRI) (60). These kinases serve as central regulators of translation initiation, integrating various stress signals to reduce global protein synthesis as an adaptive response in chickens (9). The protein kinase R (PKR) pathway plays a vital role in the cellular response to viral infections and is activated upon the detection of viral components (60). Once activated, PKR phosphorylates eIF2α, leading to a decrease in global protein translation and effectively inhibiting the viral replication. In contrast, PKR, like PERK, is induced by endoplasmic reticulum (ER) stress and phosphorylates eIF2α, triggering the unfolded protein response (UPR) to alleviate ER stress (61). Similarly, the GCN2 kinase is activated by amino acid deprivation and phosphorylates eIF2α, reducing global translation while selectively upregulating stress response gene translation (9). HRI detects heme deficiency and phosphorylates eIF2α, resulting in decreased protein synthesis as an adaptive response to cellular stress (60). Through the coordinated action of these four eIF2α kinases, chickens mount an integrated stress response to a range of stimuli, including viral infection, ER stress, nutrient deprivation, and haem deficiency (60).
Modulating eIF2α phosphorylation and the expression of eIF2 subunit genes is a vital mechanism in chickens resistant to viral infection, as it shuts down host protein synthesis and induces antiviral defenses. As illustrated in Figure 3, the eIF2α signaling pathway and its function are closely associated with the global attenuation of protein synthesis, with eIF2α phosphorylation serving as a key regulatory mechanism. Studies have shown that the eIF family of genes encode three subunits, eIF2α, eIF2β, and eIF2γ, produced by the EIFS1, EIFS2, and EIFS3 genes, respectively, which are essential for protein synthesis and the innate immune response in chickens (60). Phosphorylation of eIF2α following NDV infection in chicken cells leads to increased translation of activating transcription factor 4 (ATF4) and growth arrest and DNA damage-inducible 34 (GADD34), linking eIF2α phosphorylation to the immune response (60). NDV can manipulate the PKR/eIF2α signaling cascade to favor viral replication by capturing cellular mRNA during periods of stress. NDV infection downregulates the expression of EIF2B5 and EIF2S3 in chickens, potentially facilitating the shut-off of protein synthesis during the infection (60).
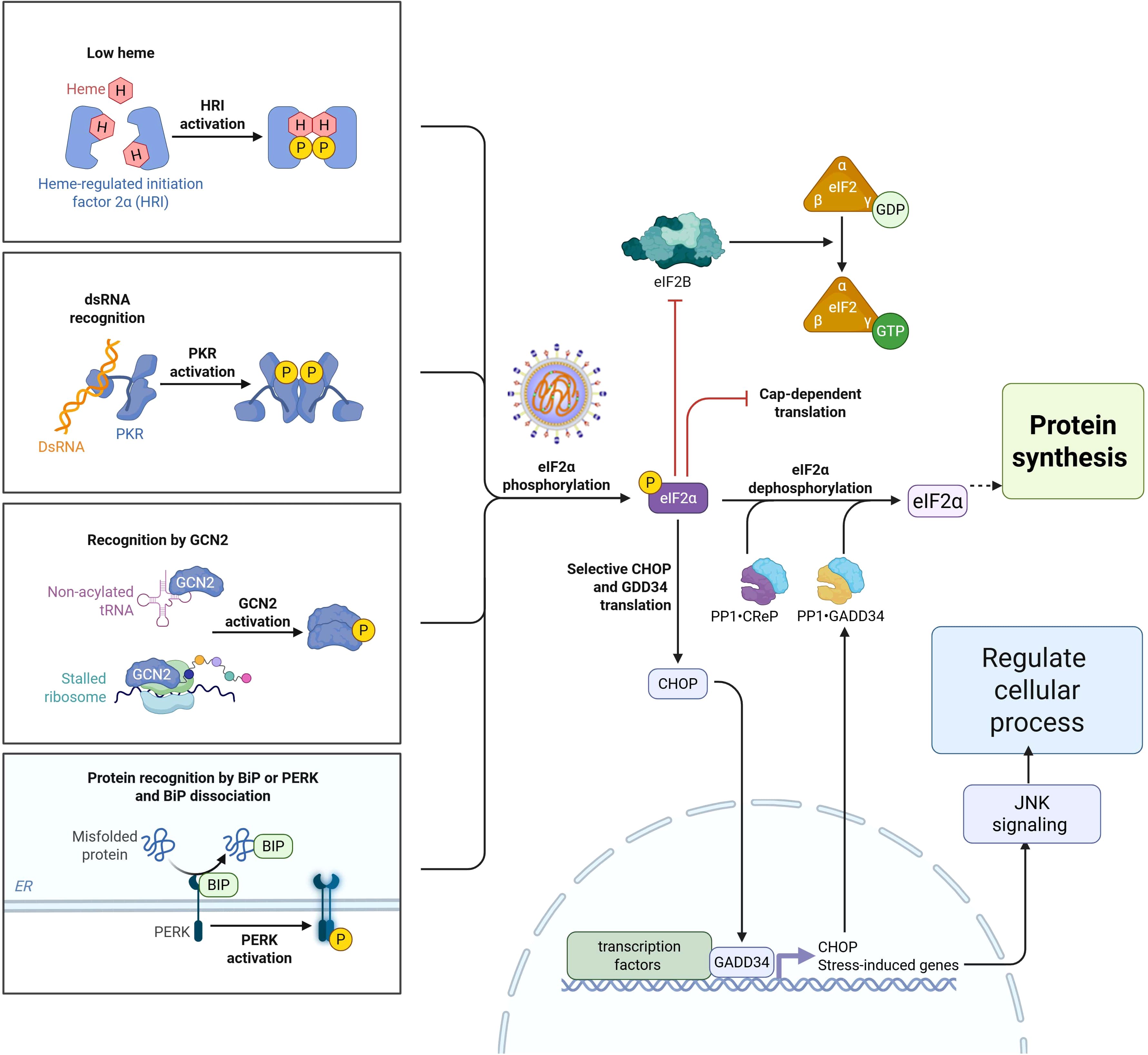
Figure 3. Schematic diagram of eIF2α phosphorylation signaling pathway family in response to Newcastle disease virus during protein synthesis.
Furthermore, activation of the eIF2α-CHOP-BCL-2/JNK and inositol-requiring enzyme 1 alpha (IRE1α) or Jun N-terminal kinase (JNK) signaling cascades by NDV promotes apoptosis and cytokine secretion, supporting viral proliferation (55). More specifically, breed-specific responses were observed in Fayoumi chickens, which exhibited activation of eIF2 signaling and downregulation of collagen-related genes (60). Collectively, these findings highlight the importance of eIF2α in protein synthesis during the innate immune response to NDV in chickens. NDV infection in chickens activates various signaling pathways that influence viral replication and host immune response. The phosphatidylinositol 3-kinase (PI3K)/protein kinase B (Akt) signaling pathway is transiently activated early during infection, promoting cell survival and facilitating viral replication (55). The p38 MAPK pathway may regulate the expression of avian β-defensin 2 (AvBD2), an important innate immune response to NDV (57).
Investigating the molecular mechanisms underlying the host immune response to NDV is crucial for improving disease resistance in chickens. This review presents several candidate genes and signaling pathways associated with NDV resistance in chickens (Table 4). For example, genes related to the major histocompatibility complex (MHC-B) locus, such as LEI0070 and ADL0146, play vital roles in modulating immune responses (66). The LEI0070 marker has been associated with resistance to various pathogens and may influence antibody responses to NDV (66). The genes encoding type I interferons (IFN-α), DEAD box helicase 1 (DDX-1), interferon-γ (IFN-γ), and interleukin 6 (IL-6) are integral to the antiviral defense mechanisms in chickens infected with NDV (67). In this regard, type I interferons, particularly interferon-alpha (IFN-α), are essential for the initial defense mechanisms against NDV infections. Upon NDV infection, pattern recognition receptors (PRRs), notably RIG-I, identify viral RNA, thereby activating the RIG-I/MAVS signaling pathway, leading to IFN-α production. This pathway subsequently binds to interferon receptors on adjacent cells, activating the JAK-STAT pathway. This activation induces the expression of interferon-stimulated genes (ISGs), which produce antiviral proteins that impede viral replication and enhance the immune response, thereby establishing a robust antiviral state in host cells (68). Candidate genes, such as DDX-1, enhance the immune response to NDV; however, excessive activation can result in excessive inflammation, potentially causing tissue damage and contributing to immune pathology during severe infections. IFN-γ is crucial for robust cellular immunity and viral clearance; however, excessive production can cause tissue damage, particularly in the lungs and other organs due to heightened inflammation (5). IL-6 gene helps in the activation of adaptive immunity and infection resolution; however, excessive production can lead to detrimental effects, including a cytokine storm that causes severe inflammation, tissue damage, and potentially fatal outcomes (5).
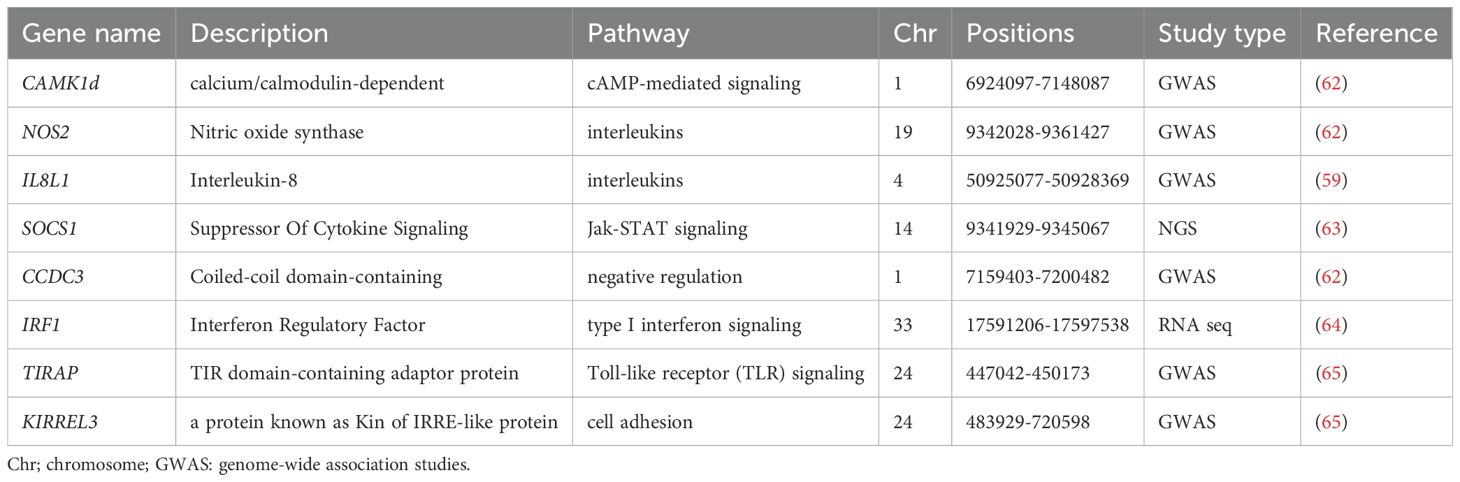
Table 4. Candidate genes and genomic regions associated with the Newcastle disease virus in chickens.
Other host genes, including CDC16, ZBED1, MX1, and GRAP2, are also located on chromosome 1 and are strongly associated with the chicken immune response (69). CDC16 interacts with viral proteins, influencing both viral replication and immune responses. ZBED1 enhances the transcription of genes that activate innate immune responses, including cytokines and interferons (69). Conversely, insufficient expression or viral antagonism by MX1 may lead to increased viral replication and aggravated infection (70). The GRAP2 gene modulates immune responses by regulating immune cell activation and promoting cytokine production (69). Although CAMK1D affects calcium signaling, immune responses, and cell survival, CCDC3 is crucial for efficient viral entry and spread because of its involvement in cellular trafficking and cytoskeletal dynamics (69). Furthermore, TIRAP, ETS1, and KIRREL3 are important in modulating the immune response to NDV infection, influencing both viral replication and the host’s ability to control the infection (70). The same author noted that infection with Newcastle disease virus (NDV) stimulates the recognition of viral components by host cells, particularly the recognition of RNA by Toll-like receptor 3 (TLR3), which subsequently activates the TIRAP-dependent signaling pathway. Furthermore, Zhang et al. (71) identified genes such as IFIT5 and SPRY1 that were strongly associated with NDV resistance.
2.6 Virus diagnostic methods
In the poultry industry, the demand for rapid and accurate diagnostic methods for NDV is vital because of its potential to cause extensive economic losses (72). Effective diagnostics not only facilitate timely intervention but also support broader biosecurity measures that are essential for maintaining global poultry health. As a result, recent research has emphasized the importance of proper sample collection from various organs, including tracheal and cloacal swabs of live birds, and from tissues such as the spleen, lungs, intestines, liver, kidneys, heart, thymus, and brain, which are commonly used for detecting NDV in birds (73) and are commonly used for detecting NDV in birds, as illustrated (Figure 4). These repetitive organs are the most used organs by various researchers to detect NDV infection in birds, utilizing both in vivo and in vitro methods for subsequent sample processing, library construction, and sequencing. Tracheal samples presented the highest detection rate (92.94%), followed by spleen (90.62%) and lung (86.95%) samples (73). Wang et al. (74) performed a transcriptome analysis of spleen and thymus tissue samples obtained from NDV-infected chickens and, revealing significant alterations in gene expression associated with various cellular and biological processes. Chellappa et al. (75) conducted in vivo studies to assess the immune response in chickens, comparing groups that received vaccination to control groups. Their findings revealed a significant reduction in antibody titers over time in vaccinated birds as compared to unvaccinated controls, suggests that the immune response may undergo modifications as time progresses, possibly reflecting a natural waning of immunity or changes in how the chicken immune systems respond to the vaccine. Another investigation utilizing single-cell transcriptome analysis of NDV in chickens, conducted both in vitro and in vivo, revealed that the Toll-like receptor signaling pathway was the primary pathway activated following viral infection (76).
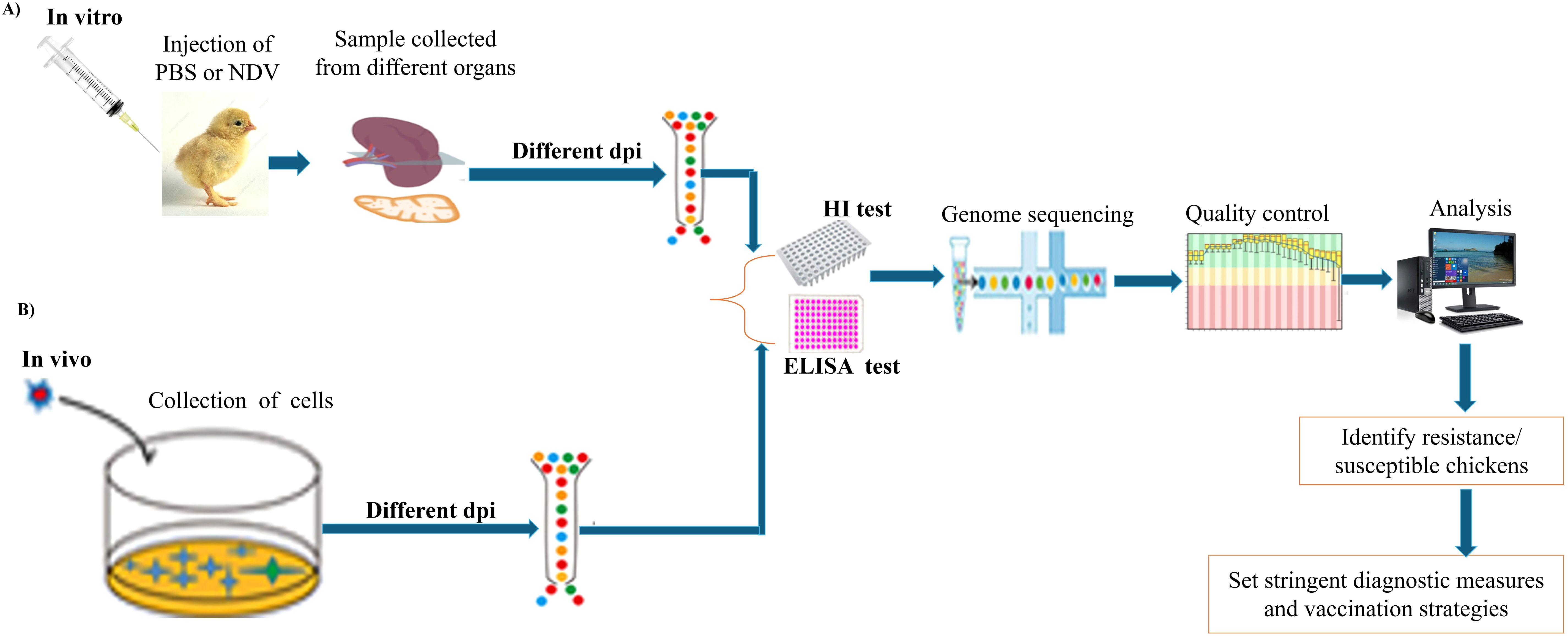
Figure 4. Schematic diagram of the experimental workflow for NDV diagnostic testing using different chicken organs. (A) In vitro procedure and key steps. (B) In vivo procedure and key steps.
The use of optimal tissue samples plays a crucial role in gaining insights into both pathogenesis and immune responses in chickens (11). The type of tissue selected is key to ensuring an accurate diagnosis of avian diseases. Additionally, the diagnostic process must carefully consider factors such as the selection of diagnostic tests, appropriate sample types, and choice of animals for testing, as multiple tests may be necessary for proper interpretation (77). Clinical examination, which remains essential in the diagnostic workflow, continues to guide both the selection of laboratory tests and the interpretation of the results (77).
Several detection methods have been employed for NDV, including virus isolation and molecular diagnostic techniques such as RT-PCR, serotyping, and sequencing, which have become promising alternatives (11). Real-time RT-PCR offers several advantages, including high sensitivity, specificity, rapid result turnaround, and applicability to a range of sample types for avian virus detection (78). This method surpasses the hemagglutination inhibition (HI) test in terms of sensitivity and allows for further sequencing to assess viral pathotyping (78). For the accurate detection and characterization of NDV isolates, a comprehensive diagnostic approach is recommended, which combines traditional methods, such as histopathology and immunohistochemistry, with molecular techniques (72). Early detection is essential for prompt treatment and implementation of effective control measures (11). Additionally, novel approaches, such as the coagglutination kit, present promising options for rapid, cost-effective diagnosis in field settings (79).
Serological assays, such as hemagglutination (HA) and HI tests or enzyme-linked immunosorbent assays (ELISA), are commonly used for the initial detection and characterization of NDV and its related antibodies (11). HI tests are particularly favored in diagnostic laboratories because of their simplicity, rapid results, and high sensitivity and specificity. This test quantifies the titer of specific antibodies in the serum, playing a critical role in NDV surveillance, vaccine development, and, in some cases, diagnosis. ELISA is another widely used method that can detect antibodies earlier than HI tests, often identifying antibodies as early as two days post-infection. ELISA results have been shown to correlate with protective immunity against NDV challenges in chickens (11). Furthermore, the competitive ELISA format has proven effective for detecting NDV antibodies in ducks, enhancing its utility across various avian species. These serological assays are effective in detecting NDV-specific antibodies in chicken sera, making them valuable tools for monitoring infection prevalence and assessing vaccine efficacy (11).
3 NDV disease resistance mechanisms
Disease resistance is a critical economic trait in farm animals, as it not only protects the animals but also ensures better returns on investment for farmers. Disease resistance refers to an animal’s ability to inhibit the growth, productivity, and spread of invading pathogens within its body (80). In chickens, it is defined as the ability of an individual to disrupt the pathogen life cycle, leading to decreased pathogen load and increased antibody titters. Disease resistance in chickens is mainly influenced by genetic factors, environmental stressors, and the dynamics of their immune response (80). For example, individual birds with genetic susceptibility to diseases may have a strong immune response. However, adverse environmental conditions can increase the risk of diseases. Genetic factors play crucial roles in shaping the immune response and determining how effectively a bird can combat pathogens. Therefore, understanding the mechanisms of resistance to virulent NDV pathogens in chickens is essential for developing effective breeding strategies. Gul et al. (80) characterized disease resistance in chickens as a multifaceted interaction among these three components, as detailed below.
3.1 Genetic factors
Research has indicated that genetic factors significantly contribute to the resistance of chickens to NDV. Genetic differences between chicken lines with varying levels of susceptibility are key to elucidating the mechanisms underlying the immune response and identifying potential genetic markers for selecting birds with increased resistance to NDV infection (81). A study estimating the breeding value of the antibody response to NDV (Ab-NDV) via pedigree-based and genomic prediction models estimated heritability for Ab-NDV, with a genetic correlation of 0.438 between the models (58). The author further estimated the heritability of the single-trait and multiple-trait models to be 0.478 and 0.487, respectively, whereas the accuracy of genomic prediction increased from 0.086 to 0.237. In chickens, the MHC-B region contains approximately 46 genes and is associated with genetic resistance or susceptibility to infectious diseases (82). In contrast, genetic variation in chicken populations plays a role in natural resistance to specific diseases, leading to increased disease resistance (83). For example, a comparative study between Fayoumi and Leghorn breed lines revealed that the Fayoumi line was more resistant to NDV than the Leghorn line, with fewer differentially expressed genes (DEGs), a lower viral load, greater viral clearance, and greater anti-NDV antibody levels (84).
3.2 Environmental factors
Environmental factors, such as temperature fluctuations, humidity levels, and housing conditions, weaken the immune response of chickens, increasing their susceptibility to infection and overall health (85). Hygiene and feeding practices influence the susceptibility of chickens to viral pathogens (86). For example, gut microbiota, which are influenced by environmental factors, play a role in the immune responses of chickens to infections (85). The composition of the gut microbiome is particularly affected by poor housing conditions, with cage-free environments promoting greater diversity (85, 87). For example, hens in cage systems have lower numbers of lymphocytes and higher heterophil-to-lymphocyte (H/L) ratios, indicating a compromised immune status. Changes in environmental conditions significantly influence the spread of pathogens and the incidence of disease in chicken populations (88). Various stressors, including temperature fluctuations, humidity, and housing conditions, can compromise the immune system of chickens, increasing their susceptibility to infections (87). Akinyemi and Adewole (86) highlighted the role of prior infection with avian paramyxoviruses in conferring protection against NDV, indicating the potential influence of environmental exposure on this resistance. A study on seventy-two 32-week-old DeKalb hens revealed that a warm perch system could effectively reduce cold stress during winter, helping hens maintain body temperature without excessive feed intake or weight loss, while also improving eggshell quality (89). Another recent study on 36-week-old Hy-Line Brown laying hens reported the physiological and stress responses of hens exposed to low temperatures, indicating that cold stress can lead to tissue damage and affect physiological function (90).
3.3 Immune response
The immune response to NDV in chickens is a complex polygenic trait, with multiple genes contributing to genetic variations in antibody response and disease resistance (91). A greater antibody response is associated with better resistance; however, an excessively strong immune response can sometimes lead to increased susceptibility and pathology (92). As shown in Figure 5, the immune response comprises both innate (first-line defense) and adaptive (second-line defense) components that work together to protect birds from infection and disease (93). The innate system provides an immediate, nonspecific response to contain infections, whereas the adaptive system offers a specific, long-lasting response that can remember and more effectively combat pathogens upon re-exposure (94). Like mammals, chickens initiate their adaptive immune response through the activation of antigen-presenting cells (APCs) (94). The immune system then triggers either a humoral response (such as antibody production) or a cell-mediated response (which activates antiviral and antitumor mechanisms) (53). Both responses, driven by T and B lymphocytes, are characterized by specificity and memory, essential for effective vaccination. In chickens, the immune system consists of various organs, tissues, cells, and molecules, all working together to generate appropriate immune responses and establish immunological memory (83). The humoral immunity is primarily mediated by B cells that differentiate into plasma cells that secrete antibodies specific to the encountered antigens. In contrast, cellular immunity is driven mainly by T cells and does not rely on antibodies; instead, it involves the direct action of T cells against infected cells (94). Chickens possess a strong immune response system that combats various threats, particularly viruses, by producing antibodies (IgA, IgM, and IgY) (5, 91). These antibodies, or immunoglobulins, have a Y-shaped structure with heavy and light chains. Importantly, IgY antibodies exhibit high specificity and effectiveness in neutralizing viral infections (95). IgA protects mucosal surfaces, whereas IgM enhances early immune responses by forming pentameric structures that facilitate opsonization and complement protein activation (96).
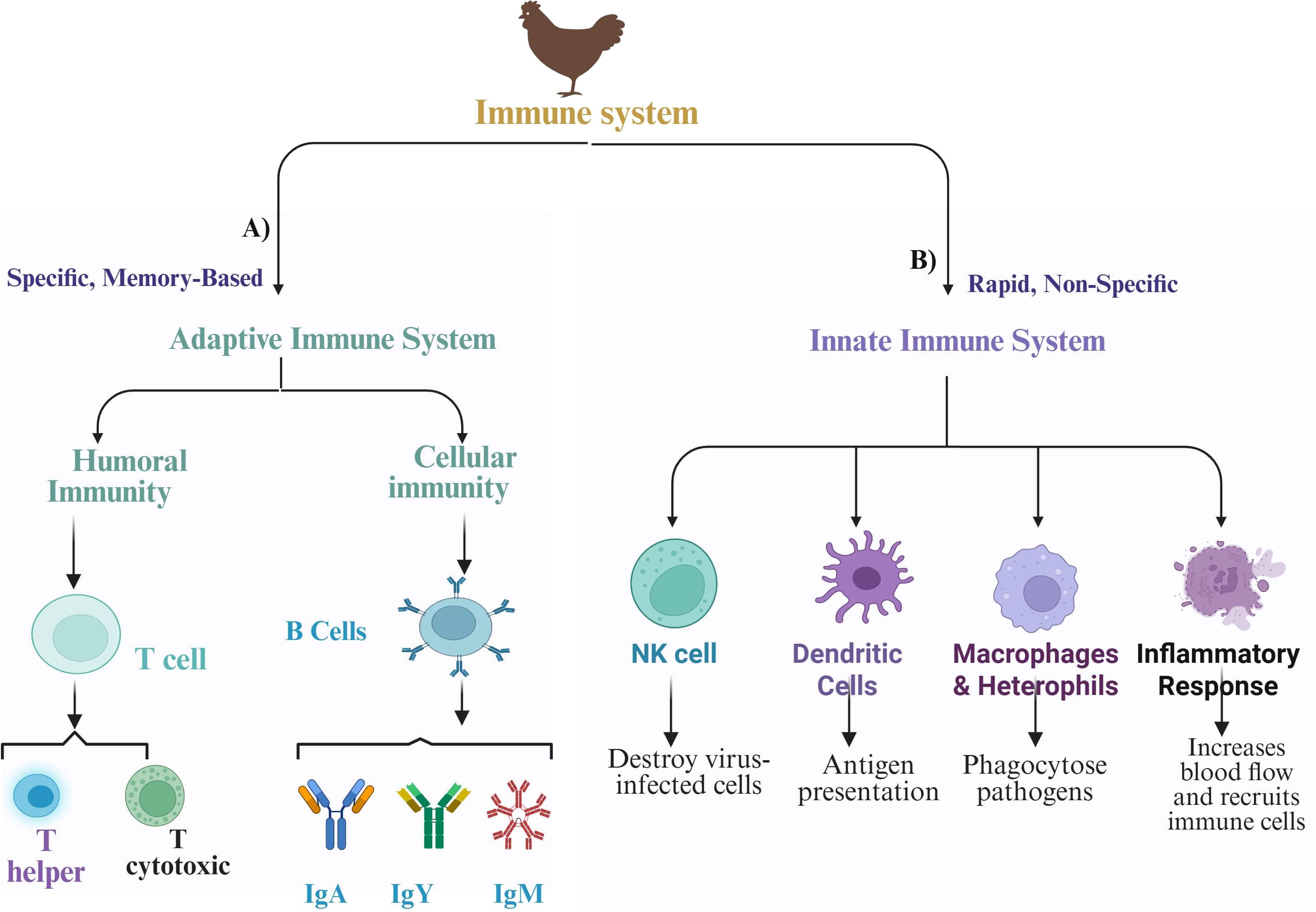
Figure 5. Schematic diagram of adaptive and innate immune responses against Newcastle disease virus (NDV) in chickens. (A) Adaptive immunity and its key components. (B) Innate immunity and its key components.
Currently, different researchers have evaluated NDV adaptive immune responses by measuring antibody titters at different ages and time points (Table 5) and have observed significant differences. For example, on day 0 (hatch), chicks primarily rely on maternally derived antibodies (MDA), and the live NDV LaSota strain vaccine does not substantially stimulate adaptive immunity at this stage (100). Within the first 1–2 weeks post-vaccination, the replicating live attenuated NDV triggers an innate immune response at the site of administration (93). Both humoral and cellular immune responses can be activated within 2–4 weeks following vaccination (100), with B cells producing NDV-specific antibodies and T cells differentiating into effector and memory T cells. By 4–6 weeks, strong antibody titers along with established populations of memory B and T cells provide long-term immune memory against the target antigen expressed by the NDV vector (101). At4 weeks of age, chicken MDA levels decrease below protective thresholds and then significantly increase at 7 and 14 days after vaccination with either the virulent ZG1999HDS strain or the lentogenic LaSota vaccine strain (98).
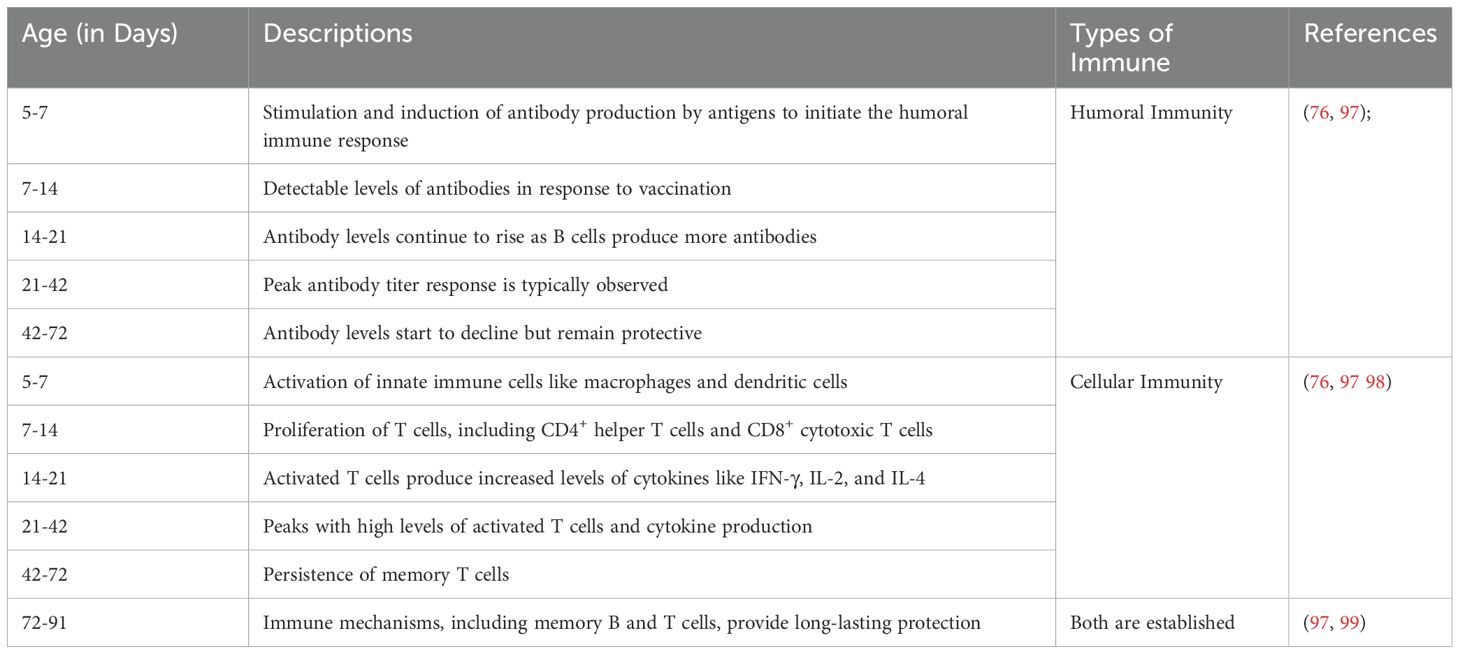
Table 5. Antibody titters of the adaptive immune response to Newcastle disease virus at different ages and time points.
Moreover, studies have demonstrated that chicken immune development exhibits three distinct patterns: Down-Up, Up-Down, and Up-Up (102). These patterns are characterized by fluctuations in cytokine levels and immune cell function, with the immune system not fully maturing until a later stage. The down-up, down-up pattern is characterized by an initial decrease in cytokine levels and certain immune indicators, followed by a subsequent increase. Notably, the lowest levels of immune function were observed between days 6 and 13 (102). In the up-down pattern, the highest levels of various immune components were observed between days 30 and 34. These include nonspecific cellular immunity components, such as the ratio of peripheral blood mononuclear macrophages; specific cellular immunity components, such as the ratio of peripheral blood helper T cells and the proliferative activity of T and B cells; and mucosal immunity components, including ileal CD4, TGF-β1, and IgA mRNA levels (102). In the Up-Up pattern, various immune components increased from days 1 to 34. Nonspecific cellular immunity components, including serum nitric oxide, C3, and C4, were elevated, as were specific cellular immunity components, such as the spleen index, peripheral blood IL-2, IFN-γ/IL-4 ratio, cytotoxic T-cell ratio, and splenic NF-κB mRNA. Additionally, humoral immunity is enhanced, as evidenced by increased serum IgG levels, whereas the levels of mucosal immunity components, including ileal MHC-II and pIgR mRNAs and ileal mucosal IgA, also increase during this period (102).
Furthermore, the immune response to NDV is characterized by the expression of various cytokines. For example, infection with virulent strains of NDV results in elevated levels of interleukin-1β (IL-1β), which is a crucial mediator of inflammation (32). Research has demonstrated that the expression of IL-1β is regulated by the NLRP3/caspase-1 pathway during NDV infection, highlighting the importance of specific immune pathways in determining disease outcomes (32). The expression patterns of cytokines, such as type I interferons (IFN-α), interferon-γ (IFN-γ), and interleukin 6 (IL-6), in relation to the NDV strain and the timing of the immune response underline the importance of both genetic factors and the timing of immune activation (103). Highly virulent NDV strains are known to elicit rapid and robust expression of proinflammatory cytokines, particularly IL-1β and IL-6, in the lymphoid tissues of young chicks (104). The expression of IFN-γ is also upregulated, although to a lesser extent (104).
4 Impact of NDV on chicken immune performance
Newcastle disease virus infection can significantly impact chicken immune performance and health. Numerous studies have elucidated the mechanisms by which NDV influences immune responses and the efficacy of vaccination (105). NDV infection can negatively affect chicken performance by decreasing body weight gain and efficiency, particularly during the early growth stages (106, 107). The virus causes severe pancreatic damage, which reduces enzyme activity and expression, potentially contributing to impaired growth performance (106). One of the primary mechanisms by which NDV affects chicken performance is through its influence on the immune system. Infected chickens present elevated levels of corticosterone, a stress hormone that is associated with decreased growth rates, compromised antibody responses, and lower lymphocyte counts (106). This hormonal response may negatively impact the overall health of chickens, increasing their susceptibility to further infections and diseases, which in turn exacerbates production losses (106). Furthermore, NDV infection has been shown to disrupt the composition of the gut microbiota, which is essential for nutrient absorption and overall health. Such disruption of the gut microbiota can lead to impaired growth performance and increased susceptibility to systemic diseases (108). This relationship between NDV and the gut microbiota highlights the interaction between viral infections and the physiological state of the host.
Additionally, NDV has been shown to replicate effectively within chicken macrophages, which are key players in the immune system. This replication may result in the polarized activation of these immune cells, potentially altering the immune response and diminishing the chickens’ ability to fight off other pathogens (109). By manipulating immune responses, NDV not only ensures its own survival but also compromises host health, leading to reduced productivity and increased mortality rates in severe cases (71). The economic implications of NDV are profound, particularly in regions where poultry production is vital for income and nutrition. This disease can cause mass mortality and substantial declines in productivity, adversely affecting both meat and egg production (110). In many developing countries, the impact of NDV is particularly severe due to inadequate vaccination coverage and insufficient biosecurity measures, which worsen the spread of the virus and its economic consequences (110, 111). For example, in rural communities, outbreaks of NDV can devastate local poultry populations, resulting in food insecurity and financial distress for households that depend on chicken farming for their livelihoods (110).
Research indicates that vaccination against NDV enhances the immune response in chickens, leading to improved disease resistance. The selection for high antibody responsiveness to NDV vaccination has been shown to improve natural immunity, suggesting that genetic selection can increase disease tolerance in chickens. Vaccination-induced stress can increase serum stress hormones, affect growth performance, and trigger lymphocyte apoptosis, with optimal antibody titers achieved at lower doses (112). Lower doses of the NDV vaccine (2–6 doses) were found to be optimal, producing peak antibody titers without compromising growth performance (112), whereas higher doses led to increased lymphocyte apoptosis and reduced feed efficiency. Moreover, combining live and inactivated NDV vaccines provides superior protection compared with live vaccines alone (105).
Recombinant NDV vaccines have demonstrated effectiveness against NDV and other avian pathogens, enhancing overall chicken flock productivity (53). Various administration routes for NDV vaccines, such as oculonasal or nebulization, have been evaluated for their immunogenicity, indicating that these methods can effectively stimulate the immune system in day-old chicks. Innovative vaccine formulations, including those encapsulated in chitosan nanoparticles, have shown the capacity to induce effective immune responses while maintaining stability and efficacy (25). The immune response to NDV vaccination can be compromised by concurrent infections, such as chicken infectious anemia virus (CIAV), which causes immunosuppression and affects bird performance (105). These advancements in vaccine technology are essential for ensuring that chickens effectively withstand NDV challenges. However, the decline in these antibodies within several weeks can render chicks susceptible to infection if they are not adequately vaccinated (98). Consequently, the timing and effectiveness of vaccination programs are critical for managing NDV outbreaks and ensuring optimal performance in poultry production.
5 Conclusions
In this review, we have explored the molecular mechanisms of NDV, including its virulence determinants, infection routes, host resistance factors, and immune evasion strategies, as well as the key candidate genes and genomic regions involved in chickens. We also highlight the interaction between genetic factors, environmental influences, and immune responses that shape NDV resistance. While significant progress has been made in understanding the interaction of host immunity and cytokine regulation, NDV’s ability to evade immune defense remains a challenge. The virus significantly impacts the immune performance of chickens, often causing immunosuppression in severe cases and compromising overall poultry health and productivity. Understanding these interactions is essential for developing effective control strategies against this economically important pathogen. Consequently, a multidisciplinary approach combining genomics, immunology, and epidemiology will be key to will provide new insights into the replication and pathogenesis of NDV. Further research should focus on elucidating host-pathogen interactions at the molecular level, optimizing immune modulation strategies, and developing sustainable solutions tailored to diverse poultry production systems. This comprehensive overview underscores the advancements in understanding the molecular basis of NDV and the potential for developing immunological-based techniques for targeted disease control measures in the poultry industry.
Author contributions
HB: Methodology, Conceptualization, Writing – review & editing, Writing – original draft. WL: Writing – review & editing. AL: Writing – review & editing. JJ: Writing – review & editing. MC: Writing – review & editing. DS: Writing – review & editing. CL: Writing – review & editing, Supervision.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Special Fund for the Promotion of Breeding Industry in the 2024 Guangdong Provincial Rural Revitalization Strategy (2024-XPY-00-002), the Special Fund for Scientific Innovation Strategy-Construction of High-Level Academy of Agriculture Science (R2020PY-JG008), the Project of Collaborative Innovation Center of GDAAS (XT202217), the Earmarked fund for CARS (CARS-41). Modern Seed Industry Innovation Capacity Enhancement Program of Guangdong Academy of Agricultural Sciences.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
Akt, Protein Kinase B; CCL-5 C-C, Motif Chemokine Ligand 5; CD4, Cluster of differentiation 4; CD8, Cluster of differentiation 8; DDX-1, DEAD box helicase 1; eIF2α, eukaryotic translation initiation factor 2α; IgA, Immunoglobulin A; IgG, Immunoglobulin G,; IgM, Immunoglobulin M; IFNα, Type I Interferons; IFN-γ, Interferon gamma; IL-1β, Interleukin 1 beta; IL-6, Interleukin 6; IL-8, Interleukin 8; JNK, C-JUN N-terminal kinase; JNK, Jun N-terminal kinase; TNF-α, Tumor Necrosis Factor alpha; IRE1α, Inositol-requiring enzyme 1 alpha; PI3K, Phosphatidylinositol 3-Kinase; MHC-B, Histocompatibility Complex locus.
References
1. Hu Z, He X, Deng J, Hu J, and Liu X. Current situation and future direction of Newcastle disease vaccines. Vet Res. (2022) 53:99. doi: 10.1186/s13567-022-01118-w
2. Zereen F, Rahman MA, Hossain MG, Alam J, Shimada M, Rahman MT, et al. First report of the emergence of novel sub-genotype XIII. 2.3 of Newcastle disease virus in chickens from selected regions of Bangladesh. Infection Genet Evol. (2025) 130:105742. doi: 10.1016/j.meegid.2025.105742
3. Reid SM, Skinner P, Sutton D, Ross CS, Drewek K, Weremczuk N, et al. Understanding the disease and economic impact of avirulent avian paramyxovirus type 1 (APMV-1) infection in Great Britain. Epidemiol. Infect. (2023) 151:1–7. doi: 10.1017/S0950268823001255
4. Cong J, Feng X, Kang H, Fu W, Wang L, Wang C, et al. Structure of the Newcastle Disease Virus L protein in complex with tetrameric phosphoprotein. Nat Commun. (2023) 14:1324. doi: 10.1038/s41467-023-37012-y
5. Zhang D, Ding Z, and Xu X. Pathologic mechanisms of the newcastle disease virus. Viruses. (2023) 15:864. doi: 10.3390/v15040864
6. Zhou J, Duan Y, Liu M, Liu J, Hu Z, and Duan Z. Recent advancements in the diverse roles of polymerase-associated proteins in the replication and pathogenesis of Newcastle disease virus. Veterinary Res. (2025) 56:8. doi: 10.1186/s13567-024-01429-0
7. Amoia CF, Hakizimana JN, Chengula AA, Munir M, Misinzo G, and Weger-Lucarelli J. Genomic diversity and geographic distribution of newcastle disease virus genotypes in africa: implications for diagnosis, vaccination, and regional collaboration. Viruses. (2024) 16:795. doi: 10.3390/v16050795
8. Yang H, Tian J, Zhao J, Zhao Y, and Zhang G. The application of newcastle disease virus (NDV): vaccine vectors and tumor therapy. Viruses. (2024) 16:886. doi: 10.3390/v16060886
9. Komar AA and Merrick WC. A retrospective on EIF2A—and not the alpha subunit of EIF2. Int J Mol Sci. (2020) 21:2054. doi: 10.3390/ijms21062054
10. Dai J, Qiu X, Cui X, Feng Y, Hou Y, Sun Y, et al. Newcastle disease virus infection remodels plasma phospholipid metabolism in chickens. Iscience. (2024) 27(17):108962. doi: 10.1016/j.isci.2024.108962
11. Mao Q, Ma S, Schrickel PL, Zhao P, Wang J, Zhang Y, et al. Review detection of Newcastle disease virus. Front.Vet.Sci. (2022) 9:936251. doi: 10.3389/fvets.2022.936251
12. Nooruzzaman M, Hossain I, Begum JA, Moula M, Khaled SA, Parvin R, et al. The first report of a virulent newcastle disease virus of genotype VII.2 causing outbreaks in chickens in Bangladesh. Viruses. (2022) 14:1–18. doi: 10.3390/v14122627
13. Li Y, Rehman ZU, Li M, Manzoor Z, Liu W, Qiu X, Sun Y, et al. Comparison of the protective antigen variabilities of prevalent Newcastle disease viruses in response to homologous/heterologous genotype vaccines. Poult.Sci. (2021) 100:101267. doi: 10.1016/j.psj.2021.101267
14. Jia L, Liang B, Wu K, Wang R, Liu H, Liu D, et al. Circulation, genomic characteristics, and evolutionary dynamics of class I Newcastle disease virus in China. Virulence. (2022) 13:414–27. doi: 10.1080/21505594.2022.2037342
15. Rtishchev A, Treshchalina A, Shustova E, Boravleva E, and Gambaryan A. An outbreak of newcastle disease virus in the moscow region in the summer of 2022. Vet Sci. (2023) 10:404. doi: 10.3390/vetsci10060404
16. Qiu X, Jia Y, Zhang Z, Fo X, and Wang W. Characterization of chicken-derived genotype VII newcastle disease virus isolates from Northwest China. J Poult. Sci. (2023) 60(2):2023010. doi: 10.2141/jpsa.2023010
17. Sarika N, John Kirubaharan J, Vidhya M, Shilpa P, and Rajasekaran R. Emergence of XIII.2.2 genotype of Avian Avulavirus-1 with unique FPCS site in India. Indian J Anim. Sci. (2022) 92:814–8. doi: 10.56093/ijans.v92i7.105529
18. Tsaxra JB, Abolnik C, Kelly TR, Chengula AA, Mushi JR, Msoffe PLM, et al. Molecular characterization of Newcastle disease virus obtained from Mawenzi live bird market in Morogoro, Tanzania in 2020–2021. Braz J Microbiol. (2023) 54:3265–73. doi: 10.1007/s42770-023-01159-z
19. Amoia CF, Hakizimana JN, Duggal NK, Chengula AA, Rohaim MA, Munir M, et al. Genetic diversity of newcastle disease virus involved in the 2021 outbreaks in backyard poultry farms in Tanzania. Vet Sci. (2023) 10:477. doi: 10.3390/vetsci10070477
20. Henriques AM, Neto A, Fagulha T, Almeida V, and Fevereiro M. Molecular characterization and phylogenetic analysis of Newcastle disease viruses isolated in southern Angola 2016-2018. Infect Genet Evol. (2023) 113:0–7. doi: 10.1016/j.meegid.2023.105481
21. Kariithi HM, Volkening JD, Chiwanga GH, Goraichuk IV, Olivier TL, Msoffe PLM, et al. Virulent Newcastle disease virus genotypes V.3, VII.2, and XIII.1.1 and their coinfections with infectious bronchitis viruses and other avian pathogens in backyard chickens in Tanzania. Front.Vet.Sci. (2023) 10:1272402. doi: 10.3389/fvets.2023.1272402
22. Al-Mubarak AIA, Al-Kubati AAG, Sheikh A, Abdelaziz AM, Hussen J, Kandeel M, et al. Detection of Avian Orthoavulavirus-1 genotypes VI.2.1 and VII.1.1 with neuro-viscerotropic tropism in some backyard pigeons (Columbidae) in Eastern Saudi Arabia. Front Vet Sci. (2024) 11:1352636. doi: 10.3389/fvets.2024.1352636
23. El-Morshidy Y, Abdo W, Elmahallawy EK, El-Dayem GAA, El-Sawak A, El-Habashi N, et al. Pathogenesis of velogenic genotype VII.1.1 newcastle disease virus isolated from chicken in Egypt via different inoculation routes: Molecular, histopathological, and immunohistochemical study. Animals. (2021) 11:3567. doi: 10.3390/ani11123567
24. Dewidar AAA, Kilany WH, El-Sawah AA, Shany SAS, Dahshan AHM, Hisham I, et al. Genotype VII.1.1-based newcastle disease virus vaccines afford better protection against field isolates in commercial broiler chickens. Animals. (2022) 12:1696. doi: 10.3390/ani12131696
25. Haque MA, Sadekuzzaman M, Enamul Haque M, Parvin K, Mostofa MK, Hayat S, et al. Characterization of the dominant strain (G-VII) of Newcastle disease viruses isolated from commercial chickens in Bangladesh during recent outbreaks. J Adv Vet Anim. Res. (2024) 11:408–17. doi: 10.5455/javar.2024.k790
26. Bahoussi AN, Shah PT, Zhao JQ, Wang PH, Guo YY, Wu C, et al. Multiple potential recombination events among Newcastle disease virus genomes in China between 1946 and 2020. Front.Vet. Sci. (2023) 10:1136855. doi: 10.3389/fvets.2023.1136855
27. Sultan HA, Talaat S, Elfeil WK, Selim K, Kutkat MA, Amer SA, et al. Protective efficacy of the Newcastle disease virus genotype VII–matched vaccine in commercial layers. Poult.Sci. (2020) 99:1275–86. doi: 10.1016/j.psj.2019.10.063
28. Goraichuk IV, Gerilovych A, Bolotin V, Solodiankin O, Dimitrov KM, Rula O, et al. Genetic diversity of Newcastle disease viruses circulating in wild and synanthropic birds in Ukraine between 2006 and 2015. Front Veterinary Sci. (2023) 10:1026296. doi: 10.3389/fvets.2023.1026296
29. Wang X, Lu X, Wang M, Zhou Q, Wang X, Yang W, Liu X, et al. RNA-seq profiling in chicken spleen and thymus infected with newcastle disease virus of varying virulence. Vet Sci. (2024) 11:569. doi: 10.3390/vetsci11110569
30. Moustapha A, Talaki E, Akourki A, and Ousseini M. Newcastle disease virus in poultry: current status and control prospects. World’s Veterinary J. (2023) 13(2):240–9. doi: 10.54203/scil.2023.wvj26
31. Wang Z, Geng Z, Zhou H, Chen P, Qian J, and Guo A. Genetic characterization, pathogenicity, and epidemiology analysis of three sub-genotype pigeon newcastle disease virus strains in China. Microorganisms. (2024) 12:738. doi: 10.3390/microorganisms12040738
32. Gao P, Chen L, Fan L, Ren J, Du H, Sun M, et al. Newcastle disease virus RNA-induced IL-1β expression via the NLRP3/caspase-1 inflammasome. Vet Res. (2020) 51:53. doi: 10.1186/s13567-020-00774-0
33. Larkina TA, Barkova OY, Peglivanyan GK, Mitrofanova OV, Dementieva NV, Stanishevskaya OI, et al. Evolutionary subdivision of domestic chickens: Implications for local breeds as assessed by phenotype and genotype in comparison to commercial and fancy breeds. Agric (Switzerland). (2021) 11:914. doi: 10.3390/agriculture11100914
34. Worku T and Teshome I. Review on the role of viral structural proteins on the pathogenicity of Newcastle disease virus in chickens. J Zool. (2020) 3:40–6. doi: 10.1128/JVI.77.16.8676-8685
35. Zhang G, Xue J, and Li X. The influence of Newcastle disease virus major proteins on virulence. Vet Sci Res. (2021) 3:10.30564. doi: 10.30564/vsr.v3i2.4098
36. Disease N, Ndv V, Naz Z, Ismat F, Saleem M, and Iqbal M. Single step detergent assisted extraction and development of ELISA for detection of anti-NDV. Pakistan J Zool. (2022) 54:2765–73. doi: 10.17582/journal.pjz/20210913180900
37. Ganar K, Das M, Sinha S, and Kumar S. Newcastle disease virus: Current status and our understanding. Trends Anaesth. Crit Care. (2021) 184:71–81. doi: 10.1016/j
38. Dementieva NV, Kudinov AA, Larkina TA, Mitrofanova OV, Dysin AP, Terletsky VP, et al. Genetic variability in local and imported germplasm chicken populations as revealed by analyzing runs of homozygosity. Animals. (2020) 10:1887. doi: 10.3390/ani10101887
39. Bashir Bello M, Yusoff K, Ideris A, Hair-Bejo M, Hassan Jibril A, Peeters BPH, et al. Exploring the prospects of engineered Newcastle disease virus in modern vaccinology. Viruses. (2020) 12:1–23. doi: doi:10.3390/v12040451.
40. Vakhrameev AB, Narushin VG, Larkina TA, Barkova OY, Peglivanyan GK, Dysin AP, et al. Disentangling clustering configuration intricacies for divergently selected chicken breeds. Sci Rep. (2023) 13:1–13. doi: 10.1038/s41598-023-28651-8
41. Rabiei M, Low WY, Ren Y, Cahyono MI, Doan PTK, Dharmayanti I, et al. Indicators of the molecular pathogenesis of virulent Newcastle disease virus in chickens revealed by transcriptomic profiling of spleen. Sci.Rep. (2021) 11:17570. doi: 10.1038/s41598-021-96929-w
42. Lu X, Wang X, Liu X, and Liu X. The multifaceted interactions between Newcastle disease virus proteins and host proteins: a systematic review. Virulence. (2024) 15(1). doi: 10.1080/21505594.2023.2299182
44. Afonso CL. Virulence during newcastle disease viruses cross species adaptation. Viruses. (2021) 13:110. doi: 10.3390/v13010110
45. Botchway PK, Amuzu-Aweh EN, Naazie A, Aning GK, Otsyina HR, Saelao P, et al. Host response to successive challenges with lentogenic and velogenic Newcastle disease virus in local chickens of Ghana. Poult.Sci. (2022) 101:102138. doi: 10.1016/j.psj.2022.102138
46. Li C, Wang L, and Zheng S. Editorial: Immunosuppressive disease in poultry. Front.Immuno. (2023) 14:1215513. doi: 10.3389/fimmu.2023.1215513
47. Hu J, Afayibo DJA, Zhang B, Zhu H, Yao L, Guo W, et al. Characteristics, pathogenic mechanism, zoonotic potential, drug resistance, and prevention of avian pathogenic Escherichia coli (APEC). Front Microbiol. (2022) 13:1049391. doi: 10.3389/fmicb.2022.1049391
48. Hejazi Z, Tabatabaeizadeh SE, Toroghi R, Farzin H, and Saffarian P. First detection and characterisation of sub-genotype XIII. 2.1 Newcastle disease virus isolated from backyard chickens in Iran. Veterinary Med Sci. (2022) 8:2521–31. doi: 10.1002/vms3.v8.6
49. Al-Shammari AM, Hamad MA, Al-Mudhafar MA, Raad K, and Ahmed A. Clinical, molecular and cytopathological characterization of a Newcastle disease virus from an outbreak in Baghdad, Iraq. Veterinary Med Sci. (2020) 6:477–84. doi: 10.1002/vms3.v6.3
50. Alazawy AK and Al Ajeeli KS. Isolation and molecular identification of wild Newcastle disease virus isolated from broiler farms of Diyala Province, Iraq. Veterinary World. (2020) 13:33–39. doi: 10.14202/vetworld.2020.33-39
51. Naz D, Rahman SU, Aslam MA, and Muhammad F. Newcastle disease virus in poultry with an interface as a human vector. Vet Vaccine. (2022) 1:100003. doi: 10.1016/j.vetvac.2022.100003
52. Huang F, Dai C, Zhang Y, Zhao Y, Wang Y, and Ru G. Development of molecular mechanisms and their application on oncolytic newcastle disease virus in cancer therapy. Front Mol Biosci. (2022) 9:889403. doi: 10.3389/fmolb.2022.889403
53. Cai J, Wang S, Du H, Fan L, Yuan W, Xu Q, et al. NDV-induced autophagy enhances inflammation through NLRP3/Caspase-1 inflammasomes and the p38/MAPK pathway. Vet Res. (2023) 54:43. doi: 10.1186/s13567-023-01174-w
54. Huang Q, Wen C, Gu S, Jie Y, Li G, Yan Y, et al. Synergy of gut microbiota and host genome in driving heterosis expression of chickens. J Genet Genomics. (2024) 51:1121–34. doi: 10.1016/j.jgg.2024.06.011
55. Zhang F, Chen H, Chang C, Zhou J, and Zhang H. Comparative genomic analysis across multiple species to identify candidate genes associated with important traits in chickens. Genes. (2025) 16:627. doi: 10.3390/genes16060627
56. Guan D, Bai Z, Zhu X, Zhong C, Hou Y, Zhu D, et al. Genetic regulation of gene expression across multiple tissues in chickens. Nat Genet. (2025) 57:1298–308. doi: 10.1038/s41588-025-02155-9
57. Lestari D, Khaerunnisa I, Murtini S, Ulupi N, Gunawan A, and Sumantri C. Variation and association of avian β-defensin 2 gene with the concentration of immunoglobulin Y and the titer of newcastle-disease antibody in IPB-D1 chicken. Trop Anim Sci J. (2022) 45:121–7. doi: 10.5398/tasj.2022.45.2.121
58. Mountford J, Gheyas A, Vervelde L, and Smith J. Genetic variation in chicken interferon signalling pathway genes in research lines showing differential viral resistance. Anim Genet. (2022) 53:640–56. doi: 10.1111/age.13233
59. Wang J, Zhu B, Wen J, Li Q, and Zhao G. Genome-wide association study and pathway analysis for heterophil/lymphocyte (H/L) ratio in chicken. Genes. (2020) 11:1–11. doi: 10.3390/genes11091005
60. Vesco D, Paula A, Kaiser MG, Monson MS, Zhou H, and Lamont SJ. Genetic responses of inbred chicken lines illustrate importance of eIF2 family and immune-related genes in resistance to Newcastle disease virus. Sci Rep. (2020) 10:6155. doi: 10.1038/s41598-020-63074-9
61. Shi J, Li Z, Xu R, Zhang J, Zhou Q, Gao R, et al. The PERK/PKR-eIF2α pathway negatively regulates porcine hemagglutinating encephalomyelitis virus replication by attenuating global protein translation and facilitating stress granule formation. J Virol. (2022) 96:e01695–01621. doi: 10.1128/JVI.01695-21
62. Saelao P, Wang Y, Chanthavixay G, Gallardo RA, Wolc A, Dekkers JCM, et al. Genetics and genomic regions affecting response to newcastle disease virus infection under heat stress in layer chickens. Genes. (2019) 10:3390. doi: 10.3390/genes10010061
63. Truong AD, Hong Y, Tran HTT, Dang HV, Nguyen VK, Pham TT, et al. Characterization and functional analyses of novel chicken leukocyte immunoglobulin-like receptor subfamily B members 4 and 5. Poultry Science. (2019) 98:6989–7002. doi: 10.3382/ps/pez442
64. Feng H, Zhang YB, Gui JF, Lemon SM, and Yamane D. Interferon regulatory factor 1 (IRF1) and anti-pathogen innate immune responses. PLoS Pathogens. (2021) 17:1–22. doi: 10.1371/journal.ppat.1009220
65. Walugembe M, Mushi JR, Amuzu-Aweh EN, Chiwanga GH, Msoffe PL, Wang Y, et al. Genetic analyses of Tanzanian local chicken ecotypes challenged with newcastle disease virus. Genes. (2019). 10(7). doi: 10.3390/genes10070546
66. Fulton JE. Advances in methodologies for detecting MHC-B variability in chickens. Poult. Sci. (2020) 99:1267–74. doi: 10.1016/j.psj.2019.11.029
67. Lin Z, Wang J, Zhu W, Yu X, Wang Z, Ma J, et al. Chicken DDX1 acts as an RNA sensor to mediate IFN-β Signaling pathway activation in antiviral innate immunity. Front Immunol. (2021) 12:742074. doi: 10.3389/fimmu.2021.742074
68. Mckellar J, Rebendenne A, Wencker M, Moncorgé O, and Goujon C. Mammalian and avian host cell influenza A restriction factors. Viruses. (2021) 13:522. doi: 10.3390/v13030522
69. Habimana R, Ngeno K, Okeno TO, Hirwa CA, Keambou Tiambo C, and Yao NK. Genome-wide association study of growth performance and immune response to newcastle disease virus of indigenous chicken in Rwanda. Front Genet. (2021) 12:723980. doi: 10.3389/fgene.2021.723980
70. Wang L, Xue Z, Wang J, Jian Y, Lu H, Ma H, et al. Targeted knockout of Mx in the DF-1 chicken fibroblast cell line impairs immune response against Newcastle disease virus. Poultry Sci. (2023) 102:102855. doi: 10.1016/j.psj.2023.102855
71. Zhang J, Kaiser MG, Gallardo RA, Kelly TR, Dekkers JCM, Zhou H, et al. Transcriptome analysis reveals inhibitory effects of lentogenic newcastle disease virus on cell survival and immune function in spleen of commercial layer chicks. Genes. (2020) 11:1003. doi: 10.3390/genes11091003
72. Song HS, Kim HS, Kim JY, Kwon YK, and Kim HR. The development of novel reverse transcription loop-mediated isothermal amplification assays for the detection and differentiation of virulent newcastle disease virus. Int J Mol Sci. (2023) 24:13847. doi: 10.3390/ijms241813847
73. Deka P, Das S, Deka P, Nath MK, Hazarika R, Das G, et al. Molecular detection of newcastle disease virus from field outbreaks in assam, India. Indian J Vet Sci India. Indian J Vet Sci Biotechnol. (2022) 18:5–7. doi: 10.21887/ijvsbt.18.3.22
74. Wang S, Wei L, Wang J, and Zhang Z. Epidemiological study of Newcastle disease in chicken farms in China. Front. Vet. Sci. (2024) 11:1410878. doi: 10.3389/fvets.2024.1410878
75. Chellappa MM, Dey S, Pathak DC, Singh A, Ramamurthy N, Ramakrishnan S, et al. Newcastle disease virus vectored chicken infectious anaemia vaccine induces robust immune response in chickens. Viruses. (2021) 13:1985. doi: 10.3390/v13101985
76. Liu W, Xu Z, Qiu Y, Qiu X, Tan L, Song C, et al. Single-cell transcriptome atlas of newcastle disease virus in chickens both in vitro and in vivo. Microbiol Spectr. (2023) 11. doi: 10.1128/spectrum.05121-22
77. Burrough ER, Derscheid RJ, Mainenti M, Piñeyro P, and Baum DH. The diagnostic process for infectious disease diagnosis in animals. J Am Veterinary Med Assoc. (2025) 263:S6–S16. doi: 10.2460/javma.24.10.0657
78. Spackman E. Avian influenza virus detection and quantitation by real-time RT-PCR. In: Animal Influenza Virus: Methods and Protocols. Mol Biol. (2020). 2123:137–48. doi: 10.1007/978-1-0716-0346-8_11
79. Naf’An MK, Kurniasih K, Untari T, and Prakoso YA. Development of a coagglutination kit as a rapid test for diagnosing Newcastle disease in poultry. Vet World. (2020) 13:1719–24. doi: 10.14202/vetworld.2020.1719-1724
80. Gul H, Habib G, Khan IM, Rahman SU, Khan NM, Wang H, et al. Genetic resilience in chickens against bacterial, viral and protozoal pathogens and Safety Risk Assessment for Animal Products on Biohazards (Shanghai) of Ministry of Agriculture. Front.Vet.Sci. (2022) 9:1032983. doi: 10.3389/fvets.2022.1032983
81. Mo G, Wei P, Hu B, Nie Q, and Zhang X. Advances on genetic and genomic studies of ALV resistance. J Anim. Sci Biotechnol. (2022) 13:1–14. doi: 10.1186/s40104-022-00769-1
82. Idoko-Akoh A, Goldhill DH, Sheppard CM, Bialy D, Quantrill JL, Sukhova K, et al. Creating resistance to avian influenza infection through genome editing of the ANP32 gene family. Nat Commun. (2023) 14:6136. doi: 10.1038/s41467-023-41476-3
83. Kaufman J. Innate immune genes of the chicken MHC and related regions. Immunogenetics. (2022) 74:167–77. doi: 10.1007/s00251-021-01229-2
84. Abbas G, Yu J, and Li G. Novel and alternative therapeutic strategies for controlling avian viral infectious diseases: focus on infectious bronchitis and avian influenza. Front Vet Sci. (2022) 9:933274. doi: 10.3389/fvets.2022.933274
85. Liu ZL, Chen Y, Xue JJ, Huang XF, Chen ZP, Wang QG, et al. Effects of ambient temperature on the growth performance, fat deposition, and intestinal morphology of geese from 28 to 49 days of age. Poult.Sci. (2022) 101:101814. doi: 10.1016/j.psj.2022.101814
86. Akinyemi F and Adewole D. Environmental stress in chickens and the potential effectiveness of dietary vitamin supplementation. Front Anim. Sci. (2021) 2:775311. doi: 10.3389/fanim.2021.775311
87. Hofmann T, Schmucker SS, Bessei W, Grashorn M, and Stefanski V. Impact of housing environment on the immune system in chickens: A review. Animals. (2020) 10:1138. doi: 10.3390/ani10071138
88. Campbell AM, Johnson AM, Persia ME, and Jacobs L. Effects of housing system on anxiety, chronic stress, fear, and immune function in bovan brown laying hens. Animals. (2022) 12:1803. doi: 10.3390/ani12141803
89. Hu JY and Cheng HW. Warm perches: a novel approach for reducing cold stress effect on production, plasma hormones, and immunity in laying hens. Poult. Sci. (2021) 100(8):101294. doi: 10.1016/j.psj.2021.101294
90. Kim DH, Song JY, Park J, Kwon BY, and Lee KW. The effect of low temperature on laying performance and physiological stress responses in laying hens. Animals. (2023) 13:3824. doi: 10.3390/ani13243824
91. Wlaźlak S, Pietrzak E, Biesek J, and Dunislawska A. Modulation of the immune system of chickens a key factor in maintaining poultry production—a review. Poult.Sci. (2023) 102:102785. doi: 10.1016/j.psj.2023.102785
92. Zhou Y, Cao D, Liu J, Li F, Han H, Lei Q, et al. Chicken adaptive response to nutrient density: immune function change revealed by transcriptomic analysis of spleen. Front Immunol. (2023) 14:2023.1188940. doi: 10.3389/fimmu.2023.1188940
93. Soliman RM, Nishioka K, Murakoshi F, and Nakaya T. Use of live attenuated recombinant Newcastle disease virus carrying avian paramyxovirus 2 HN and F protein genes to enhance immune responses against species A rotavirus VP6 protein. Vet.Res. (2024) 55:16. doi: 10.1186/s13567-024-01271-4
94. Yang W, Liu X, and Wang X. The immune system of chicken and its response to H9N2 avian influenza virus. Vet Quart. (2023) 43:1–14. doi: 10.1080/01652176.2023.2228360
95. El-Kafrawy SA, Abbas AT, Oelkrug C, Tahoon M, Ezzat S, Zumla A, et al. IgY antibodies: The promising potential to overcome antibiotic resistance. Front Immunol. (2023) 14:1065353. doi: 10.3389/fimmu.2023.1065353
96. Matsumoto ML. Molecular mechanisms of multimeric assembly of igM and igA. Annu Rev Immunol. (2022) 40:221–47. doi: 10.1146/annurev-immunol-101320-123742
97. Mahmood S, Skinner P, Warren CJ, Mayers J, James J, Núñez A, et al. In vivo challenge studies on vaccinated chickens indicate a virus genotype mismatched vaccine still offers significant protection against NDV. Vaccine. (2024) 42:653–661. doi: 10.1016/j.vaccine.2023.12.037
98. Nedeljković G, Mazija H, Cvetić Ž, Jergović M, Bendelja K, and Gottstein Ž. Comparison of chicken immune responses to immunization with vaccine la sota or ZG1999HDS strain of newcastle disease virus. Life. (2022) 12:72. doi: 10.3390/life12010072
99. Oberländer B, Failing K, Jüngst CM, Neuhaus N, Lierz M, Palau-Ribes FM, et al. Evaluation of Newcastle Disease antibody titers in backyard poultry in Germany with a vaccination interval of twelve weeks. PLoS ONE. (2020) 15:1–14. doi: 10.1371/journal.pone.0238068
100. Sun W, Liu Y, Amanat F, González-Domínguez I, McCroskery S, Slamanig S, et al. A Newcastle disease virus expressing a stabilized spike protein of SARS-CoV-2 induces protective immune responses. Nat Commun. (2021) 12:1–14. doi: 10.1038/s41467-021-26499-y
101. Fulber JPC and Kamen AA. Development and scalable production of newcastle disease virus-vectored vaccines for human and veterinary use. Viruses. (2022) 14:975. doi: 10.3390/v14050975
102. Song B, Tang D, Yan S, Fan H, Li G, Shahid MS, et al. Effects of age on immune function in broiler chickens. J Anim. Sci Biotechnol. (2021) 12:42. doi: 10.1186/s40104-021-00559-1
103. Cheng S, Liu X, Mu J, Yan W, Wang M, Chai H, et al. Intense innate immune responses and severe metabolic disorders in chicken embryonic visceral tissues caused by infection with highly virulent newcastle disease virus compared to the avirulent virus: A bioinformatics analysis. Viruses. (2022) 14(5):911. doi: 10.3390/v14050911
104. Brown C, Zhang J, Pantin-Jackwood M, Dimitrov K, Ferreira HL, and Suarez D. In situ cytokine gene expression in early stage of virulent Newcastle disease in chickens. Vet.Pathol. (2022) 59:75–81. doi: 10.1177/03009858211045945
105. Abdel-Maksoud EM, Daha AAEF, Taha NM, Lebda MA, Sadek KM, Alshahrani MY, et al. Effects of ginger extract and/or propolis extract on immune system parameters of vaccinated broilers. Poult. Sci. (2023) 102:102903. doi: 10.1016/j.psj.2023.102903
106. Rehman ZU, Ren S, Butt SL, Manzoor Z, Iqbal J, Anwar MN, et al. Newcastle disease virus induced pathologies severely affect the exocrine and endocrine functions of the pancreas in chickens. Genes. (2021) 12:495. doi: 10.3390/genes12040495
107. Ju A, Duan A, Zhang Y, Qin Y, Xue L, Ma X, et al. Effects of orally administered recombinant lactobacillus casei expressing HN protein on early growth performance, intestinal health and protection against NDV challenge in chickens. International Journal of Biological Macromolecules (2020) 282:137006. doi: 10.21203/rs.3.rs-48868/v1
108. Tong L, Wang W, Ren S, Wang J, Wang J, Qu Y, et al. The 16S rRNA gene sequencing of gut microbiota in chickens infected with different virulent newcastle disease virus strains. Animals. (2022) 12:2558. doi: 10.3390/ani12192558
109. Ni J, Deng J, Chen Q, Liao T, Hu J, Chen Y, et al. Role of macrophages in the pathogenesis of genotype VII newcastle disease virus in chickens. Animals. (2023) 13:2239. doi: 10.3390/ani13132239
110. Walugembe M, Naazie A, Mushi JR, Akwoviah GA, Mollel E, Mang’enya JA, et al. Genetic analyses of response of local Ghanaian Tanzanian chicken ecotypes to a natural challenge with velogenic newcastle disease virus. Animals. (2022) 12:2755. doi: 10.3390/ani12202755
111. Apopo AA, Kariithi HM, Ateya LO, BiNepal YS, Sirya JH, Dulu TD, et al. A retrospective study of Newcastle disease in Kenya. Trop Anim. Health Prod. (2020) 52:699–710. doi: 10.1007/s11250-019-02059-x
Keywords: chicken, immune response, molecular structure, NDV, pathogenesis, genetic resistance
Citation: Berihulay H, Luo W, Lao A, Ji J, Cai M, Shu D and Luo C (2025) Exploring the genetic basis of Newcastle disease virus in chickens: a comprehensive review. Front. Immunol. 16:1614794. doi: 10.3389/fimmu.2025.1614794
Received: 19 April 2025; Accepted: 04 June 2025;
Published: 27 June 2025.
Edited by:
Lei Tan, Chinese Academy of Agricultural Sciences, ChinaCopyright © 2025 Berihulay, Luo, Lao, Ji, Cai, Shu and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chenglong Luo, bHVvY2hlbmdsb25nQGdkYWFzLmNu
†These authors have contributed equally to this work
 Haile Berihulay
Haile Berihulay Wei Luo
Wei Luo Ainong Lao1
Ainong Lao1 Jian Ji
Jian Ji