- 1Department of Pediatrics, University of Minnesota Medical School, Minneapolis, MN, United States
- 2Center for Immunology, University of Minnesota, Minneapolis, MN, United States
- 3Microbiology, Immunology, and Cancer Biology, University of Minnesota, Minneapolis, MN, United States
- 4College of Veterinary Medicine, University of Minnesota, Saint Paul, MN, United States
- 5Division of Immunobiology, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, United States
- 6Department of Obstetrics and Gynecology, University of Toyama, Takaoka, Japan
- 7Departrment of Pediatrics, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, United States
- 8Center for Inflammation and Tolerance, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, United States
- 9Masonic Cancer Center, University of Minnesota, Minneapolis, MN, United States
Diverse T cell types accumulate at the human maternal fetal interface (MFI) during pregnancy to orchestrate immune tolerance for foreign fetal/placental tissues and immunity to pathogens. Yet, the dynamics of T cell influx and function at the MFI remain poorly defined. Conventional specific pathogen free (SPF) murine models fail to replicate the number and diversity of T cells in the human MFI, hindering mechanistic study of MFI T cells. Here we present an innovative use of a natural microbial exposure (NME) mouse model that enhances T cell influx and diversity in the MFI. We defined changes in the MFI of NME mice, relative to SPF mice and human tissues using transcriptomic and proteomic approaches. Physiological maternal microbial burden reproduced key features of human MFI immunology by i) significantly increasing the numbers and diversity of CD4 and CD8 effector and memory T cells at the MFI; ii) skewing the CD8 T cell composition towards tissue resident memory phenotypes with increased signatures of activation and dysfunction similar to human decidual T cells; and iii) expanding unconventional γδ T cells and Killer Lectin-like Receptors (KLR) expressing T cell types at the MFI, representative of an enhanced ability to interact with placental trophoblasts or infected cells. Thus, maternal microbial exposure induces vast changes to T cell numbers, diversity and functions at the MFI that models human MFI T cells with great fidelity. The NME model allows for improved translational investigation of the mechanisms of T cell tolerance, immunity, and inflammation in pregnancy.
Introduction
Balancing the requirements for host defense and tolerance to fetal antigens is critical for successful pregnancy outcomes (1, 2). A loss of placental immunity or tolerance can drive excessive placental inflammation, which is strongly associated with spontaneous preterm birth, fetal growth restriction (FGR), stillbirth, or congenital infection (3–5). T cells found at the maternal-fetal interface (MFI) play a central role in executing both antimicrobial and immune regulatory functions. Regulatory T (TREG) cells accumulate in decidual tissues of pregnant mice and humans; TREG cells have been shown to restrain immune responses to fetal allo-antigens in mice (6–8) and humans (9, 10). In addition, TREG frequencies and functions were shown to be diminished during placental inflammation and complications of pregnancy (11–14). By contrast, cytotoxic CD8 T cells are effective killers of cells expressing foreign antigens, including microbial or fetal allo-antigens (2, 15, 16). Paradoxically, despite the inherent risk of cytotoxic T cell responses, memory CD8 T cells accumulate at the MFI over the course of gestation in humans, becoming the most abundant immune cell at the MFI late in the third trimester (17–23). In addition, both decidual CD8 T cells with fetal and viral specificity expressing markers of T cell activation and dysfunction were found (22). In contrast to humans, CD8 T cells are rare in uterine tissues of conventional specific pathogen free (SPF) mice during healthy pregnancy, possibly due to epigenetic silencing of key T cell attracting chemokines in these mice (24). The paucity of CD8 T cells at the MFI in SPF mice limits their utility for immunologic study, underscoring the critical need for improved preclinical models of human reproductive immunology. Yet, upon depletion of TREGs, CD8 T cells can home to uterine tissues of pregnant dams, increasing resorptions, whereas adoptive transfer of pregnancy-induced TREGs prevents resorptions (6, 25). Similarly, viral infection during murine gestation allows recruitment of CD8 T cells to uterine and placental tissues, yet without effective clearance of placental infection (26).
Due to the importance of pregnancy for species survival, it is expected that a multitude of mechanisms contribute to reducing the risk of maternal T cell rejection of fetal and placental allo-antigens, including select expression of MHC by fetal trophoblasts to limit NK cell and T cell cytotoxicity (1, 18), select expression of chemokines to limit infiltration of pathogenic T cells (24), activation and induction of TREGs with specificity for fetal antigens (27–29), and peripheral deletion of fetal-reactive T cells in mice (30), but not in humans (15, 16). While these mechanisms of tolerance likely inhibit the effective clearance of infections from uterine tissues in pregnant mice and humans (26, 31), not all mechanisms of immunity are suppressed, as most pregnant people can mount effective immune responses to vaccines and pathogens that protect against congenital infections (32–34).
The constraints of human research and the lack of affordable animal models with T cell diversity reflecting human MFI tissues have hindered advancement in our understanding of T cell immunology at the MFI (1, 3, 18, 24, 35). We and others have demonstrated that the practice of raising mice in conventional SPF conditions stunts immune development relative to humans and wild mice, particularly the formation of a robust memory T cell compartment in tissues (36–42). Raising mice in diverse microbial environments does not negatively impact the health and survival of offspring provided the mother is acclimated to the environment; in fact, microbially exposed offspring demonstrate improved immune development and host defense relative to SPF pups (36).
Therefore, we sought to determine whether the paucity of T cells in SPF mouse MFI results from the lack of microbial exposure. To this end, we employed a natural microbial exposure (NME) model whereby laboratory mice are co-housed with pet store mice to passively share their diverse microbial community prior to breeding. We found, maternal microbial experience expanded T cell populations and increased T cell diversity at the MFI relative to SPF dams in a manner that closely phenocopied our observations in humans, displaying resident memory and mixed activation/dysfunction gene expression signatures. The experiments described here amend our understanding of the immune landscape at the MFI, underscoring the influence of maternal microbial experience. In a significant step forward for the field we have established an improved translational model of human pregnancy that permits mechanistic study of the immunology of the MFI.
Materials and methods
Mice
Male and female C57BL/6J (B6) were purchased from Jackson Laboratories (Bar Harbor, ME). Female pet store mice were purchased from local pet stores in the Minneapolis-Saint Paul, Minnesota metro area. To naturalize female B6 mice we cohoused them with pet store mice for >4 wks. During proestrus, naturalized B6 female mice are transferred to a cage housing a male B6 mouse for timed overnight mating. Successful mating is confirmed by the presence of a copulatory plug. The female B6 mouse is then returned to the pet store cage where she will remain for the duration of the experiment. Serological testing has revealed pet-store mice often carry some combination of rotavirus (epizootic diarrhea of infant mice), mouse hepatitis virus, murine norovirus, mouse parvovirus NS1, type 1/2, minute virus of mice, Theiler’s murine encephalomyelitis virus (TMEV), Sendai virus, lymphocytic choriomeningitis, mouse adenovirus types 1 and 2, mouse CMV, polyomavirus, pneumonia virus of mice, Mycoplasma pulmonis, Clostridium piliforme, pinworms, fur mites, and Encephalitozoon cuniculi. Pet-store mice were also tested for ectromelia virus (mousepox), reovirus, and cilia-associated respiratory Bacillus, but have not tested positive for these microbes. Our specific pathogen free (SPF) colony is routinely tested to ensure the absence of the following pathogens: mouse parvovirus, minute virus of mice, mouse hepatitis virus, mouse rotavirus-A (epizootic diarrhea of infant mice), Theiler’s murine encephalomyelitis virus (TMEV), Sendai virus, pneumonia virus of mice, reovirus, Ectromelia (Mousepox), mouse adenovirus types 1 and 2, polyomavirus, lymphocytic choriomeningitis virus, mouse CMV, Mycoplasma pulmonis, Clostridium piliforme (Tyzzer’s disease), cilia-associated respiratory Bacillus, fur mites (Myobia musculi, Radfordia affinis, Radfordia ensifera, Myocoptes musculinus), pinworms (Aspiculuris tetraptera, Syphacia obvelata, Syphacia muris), and Encephalitozoon cuniculi. Mice were sacrificed at 14.5 dg for experiments. All mice were housed in Association for Assessment and Accreditation of Laboratory Animal Care-approved animal facilities at the University of Minnesota (BSL-1 for SPF mice and BSL-3 for cohoused mice). All animal use was performed per a University of Minnesota Institutional Animal Care and Use Committee approved protocol (2404-42036A).
Human placental tissue collection and processing
Placental tissues and maternal blood samples from full-term pregnancies (gestational age >39 weeks) were collected from healthy women with uncomplicated pregnancies that delivered by elective aesarean sections under IRB approved protocols. To further ensure quality, the tissues were inspected for signs of placental inflammation (such as discoloration, significant infarctions, or bad odor), and only healthy tissue samples were selected for further use. Maternal blood samples were collected directly before delivery. Detailed procedures to isolate decidual, villous, and peripheral blood CD8+ T cells were recently described (21, 43). Briefly, decidua basalis tissue was dissected from the maternal side of the placenta and villi were removed from the decidua basalis. Villous tissue was collected separately to purify placental immune populations as described (43, 44). Collected decidual and villous tissues were washed extensively and then minced and digested with 0.1% collagenase type IV and 0.01% Dnase I (Sigma-Aldrich) for 75 minutes at 37°C in a gently shaking water bath. Following digestion, the cells were washed and filtered through 100-, 70-, and 40-μm cell strainers (BD, Labware). Lymphocytes were suspended in 20 mL of 1.023 g/mL Percoll (GE Healthcare) and subjected to a gradient centrifugation, layered with 10 mL of 1.080 g/mL and 15 mL of 1.053 g/mL Percoll. After 30 minutes of centrifugation at 800g, lymphocytes were harvested from the interface of the 1.080 and 1.053 g/mL layers and subsequently washed. Peripheral CD8+ T cells were isolated using RosetteSep (StemCell Technologies) and a Ficoll gradient (GE Healthcare) during a 20-minute centrifugation at 800g. Finally, the blood and lymphocyte samples were washed and stained for flow cytometry using a Cytek Aurora spectral flow cytometer.
IV labeling and tissue harvest
Mice were sedated with isoflurane prior to retroorbital intravenous (IV) injection of 5 μg of anti-CD45.2-APC (Proteintech) antibody to label leukocytes in contact with the circulation. Mice were then sacrificed after 3 min for tissue harvest. Concepti were removed and placenta and decidua were dissected. Tissue was mechanically separated and filtered through a 70 μm filter to form a single cell suspension. Cells are then stained with anti-CD45 conjugated to a different fluorophore (i.e. BV605), which binds surface CD45 on leukocytes not already occupied by the IV injected anti-CD45-APC. This method permits the delineation of leukocytes in circulation from those within tissue.
Flow cytometry
Mouse cells were washed with FACS buffer and stained for flow cytometry using antibodies referenced in Table 1. CD1d tetramer was prepared by incubating PBS-57-loaded CD1d biotinylated monomer (NIH Tetramer Core Facility) with Streptavidin PE-Cyanine7 conjugate (eBioscience, catalog number 25-4317-82). After extracellular staining, cells were washed, fixed, and permeabilized (BD Biosciences). Permeabilized cells were intracellularly stained using the antibodies referenced in Table 1. Cells were washed and flow cytometric analysis was performed on the Cytek Aurora (Cytek Biosciences). Data were analyzed with FlowJo Software version 10 (BD Biosciences). Surface staining of human cells was performed by incubating cells for 30 minutes in RPMI medium supplemented with penicillin/streptomycin and 10% newborn calf serum. For intracellular staining, cells were fixed and permeabilized using the CytoFix/CytoPerm kit (BD). Data acquisition was performed on a Cytek Aurora, and data were analyzed using FlowJo software (BD Biosciences).
Histology and immunohistochemistry
Concepti, including the placenta and decidua, were placed in OCT compound and frozen at -80°C. Tissue blocks were then sectioned with a cryostat and prepared for slides. Serial sections were stained with hematoxylin and eosin (Vector Labs) or with AF488 anti-CD144/VE-cadherin (Thermo Fisher, reference # 53-1441-832), AF647 anti-CD324/E-cadherin (Biolegend, catalog # 147308), and Vectashield with DAPI (Vector Labs) and imaged for immunofluorescence. Slides were imaged with a Leica DM6000 Thunder Epifluorescent Microscope equipped with a K8 CCD camera or Leica SP8 inverted confocal/epifluorescence scope equipped with a Leica DFC7000 T Camera. Images were analyzed with Leica X software.
Single cell sequencing
MFI (placenta and decidua combined) from CD45.2 IV-labeled SPF and NME mice were isolated at 14.5 dg. Cells were stained with the antibodies referenced in Table 1. Live CD45+ leukocytes were analyzed by fluorescence-activated cell sorting (FACS) and sorted into IV+ and IV- populations. Cell captures and 5’ gene expression libraries were prepared using the 10X Chromium X instrument (10X Genomics) with the assistance of the University of Minnesota Genomics Core (UMGC). Samples underwent quality control sequencing on the MiSeq QC (Illumina) followed by full sequencing with the NovaSeq X Plus (Illumina). Samples were analyzed in R using Seurat V5 using standard methods and cell clusters were annotated with the help of scType cell-type identification package (45, 46).
Quantification and statistical analysis
Data were collected across multiple experiments performed over 2 years. GraphPad Prism 9 was used to perform statistical analysis. A two-tailed unpaired, nonparametric Mann-Whitney U test was used to compare two groups at a single time point. Where three groups were compared Kruskal-Wallis test was used with Dunn’s multiple comparisons test.
Results
Maternal microbial exposure is compatible with healthy pregnancy
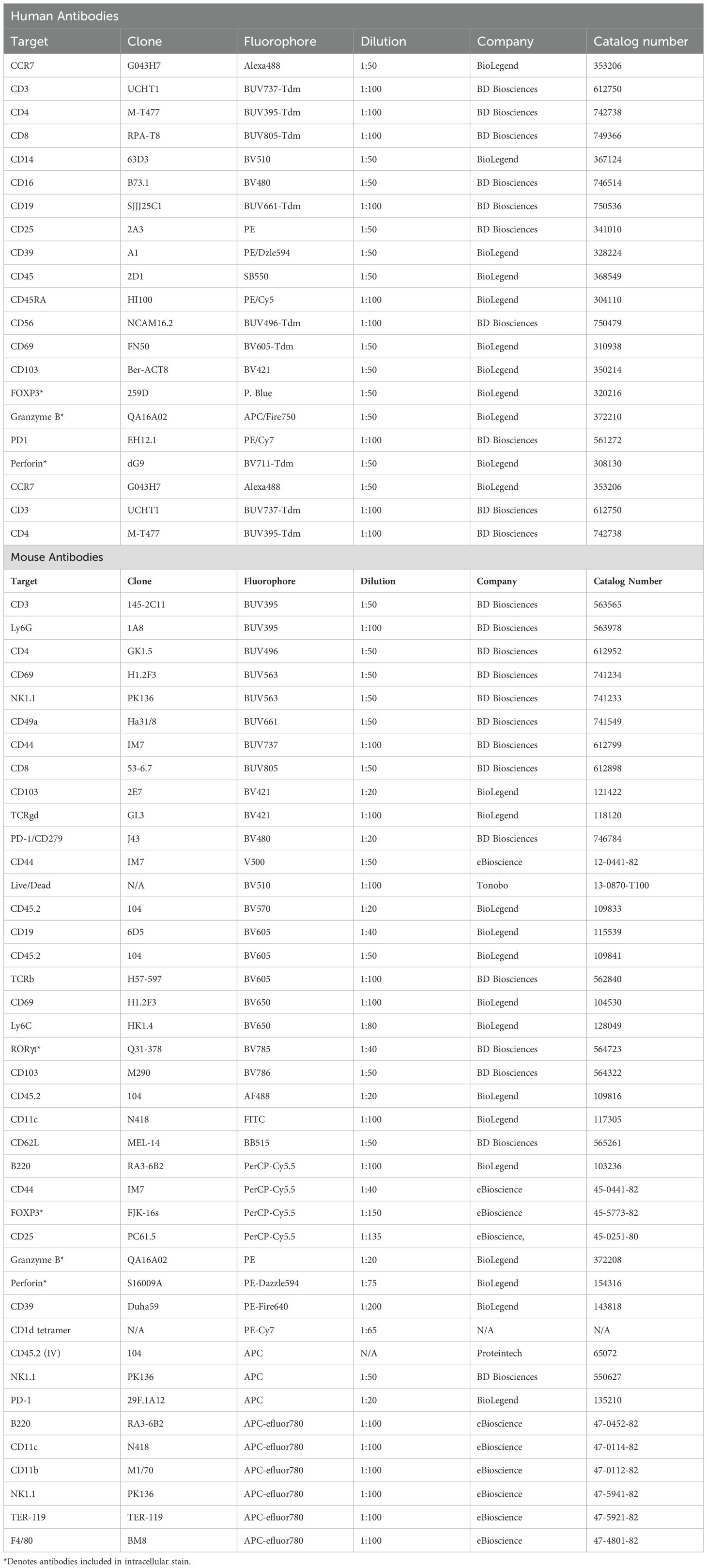
To investigate how maternal exposure to diverse microbial communities influence the immune populations at the MFI, we cohoused female C57BL/6 mice with female pet store mice for a minimum of 4 wks to passively share their diverse microbial communities. After this acclimation period, female C57BL/6 mice were transferred during proestrus to a separate cage with a male C57BL/6 mouse for timed overnight mating. The following morning, female mice were checked for a copulatory plug and returned to their cohousing cage where they remained for the duration of the experiment (Figure 1A) (36). This ensures the maternal immune system and other maternal factors influenced by microbial experience are normalized prior to conception and throughout gestation. NME did not impact the average size of the litter or the number of resorptions per litter (Figures 1B, C), confirming previous observations (36). In addition, there were no gross histologic differences between NME and SPF placenta and decidua (Figure 1D; Supplementary Figure S1). Interestingly, NME-exposed mice had a slight but significantly increased length of gestation compared to conventionally housed SPF mice (Figure 1E). Thus, cohousing female C57BL/6 mice with female pet store mice is an innovative pregnancy model compatible with healthy murine pregnancy and pup survival.
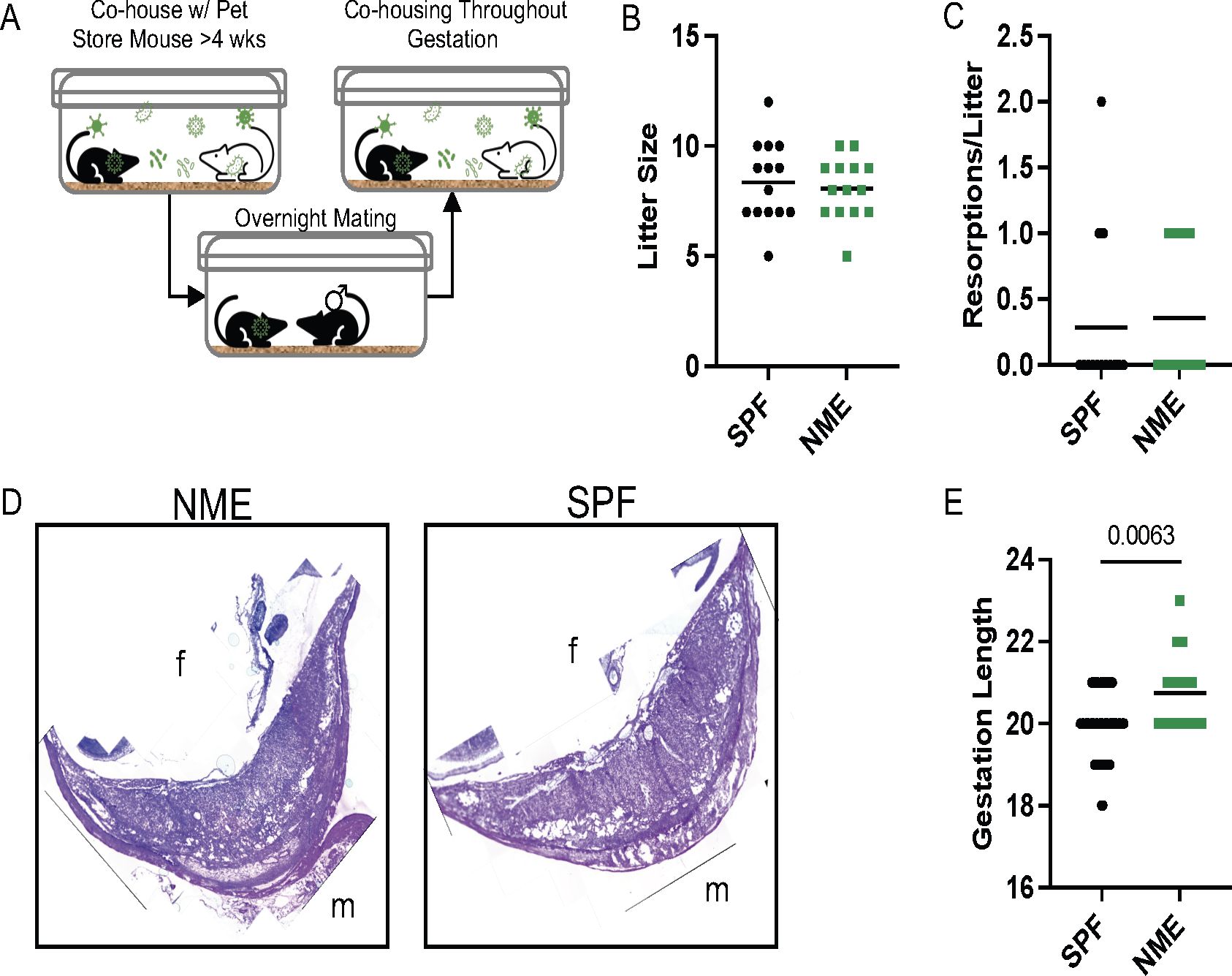
Figure 1. NME model of reproductive immunology. (A) Female C57BL/6 mice are cohoused with pet store mice for >4 weeks, referred to as natural microbial exposure (NME). Female NME mice are placed into a separate cage with a male C57BL/6 mouse during proestrus for overnight breeding. The following morning the NME female mouse is returned to her co-housing cage, where she will remain for the duration of the experiment. Pregnant NME and SPF female mice were euthanized at 14.5 dg. We observed (B) litter size and (C) concepti resorptions (n = 14 litters per group). (D) The maternal fetal interface (MFI) was flash frozen in OCT and sectioned for hematoxylin and eosin (H&E) staining. Images are labelled with maternal (m) and fetal (fetal) orientation. (E) Average length of gestation under SPF and NME conditions (n = 16 litters for NME and 51 litters for SPF). Mann-Whitney U test was used to determine significance.
Maternal microbial experience alters leukocyte populations at the MFI
To determine how maternal microbial exposure alters immune cells at the MFI, we performed single cell RNA sequencing (scRNAseq) on CD45.2+ cells purified from the placental tissues of SPF and NME dams at 14.5 days of gestation (dg). The tissue included both decidual and villous tissues to capture all MFI immune cell types. In addition, IV labeling with anti-CD45.2-APC was performed to purify IV+ circulating and IV– resident leukocytes by FACS sort before making cDNA libraries (see Materials and Methods). Interestingly, ~80% of decidual immune cells (~90% of T cells) were IV–, indicating these consist of predominant tissue resident immune populations. In contrast, and likely due to the vascular nature of the placenta, a high level of IV+ staining (~80% of immune cells and ~90% of T cells) was found in the placental tissue (Supplementary Figures S2A–D). NME conditions did not appear to alter the proportions of IV labeled cells (Supplementary Figures S2B, D). Thus, IV labeling may serve as a proxy to separate decidual and placental immune cells in the scRNA-seq data.
Uniform manifold approximation and projection (UMAP) graphs and the ScType cell classification package in R were used to help identify and annotate 13 immune cell clusters (Figure 2A) (46). Immune cell cluster identities were confirmed by expression of several key lineage markers (Figure 2B; Supplementary Figure S3A). SPF and NME mice tissues contained immune cells spanning all clusters (Figures 2C, D) but had marked differences. Using IV labeling to enrich for placental and decidual immune cells, we found that NME i) increased frequencies of several memory CD8 and CD4 T cell types in both placental and decidual tissues; ii) increased frequencies of neutrophils/granulocytes in the placenta and mast cells/granulocytes in the decidua; iii) decreased frequencies of macrophages in the decidua; and iv) decreased frequencies of dendritic cells in the placenta (Figures 2E, F). The transcriptomic signature of NME MFI macrophages showed an increase in expression of pro-inflammatory genes associated with M1 polarization (i.e., Socs3, FosB, Jun, Hspa1a, and Il1b), as well as an increase in expression of the anti-inflammatory/M2-associated Dusp1 gene relative to SPF dams (Supplementary Figure S4A). Interestingly, while the frequencies of NK and B cells in the MFI were largely the same in SPF and NME mice, NK and B cells from the MFI of NME dams were also marked by an increase in the expression of Jun, suggesting enhanced activation in NME mice (Supplementary Figures S4B, C). Thus, maternal microbial experience induces significant changes in decidual, and placental leukocyte composition and activation states compared to SPF housed pregnant mice.
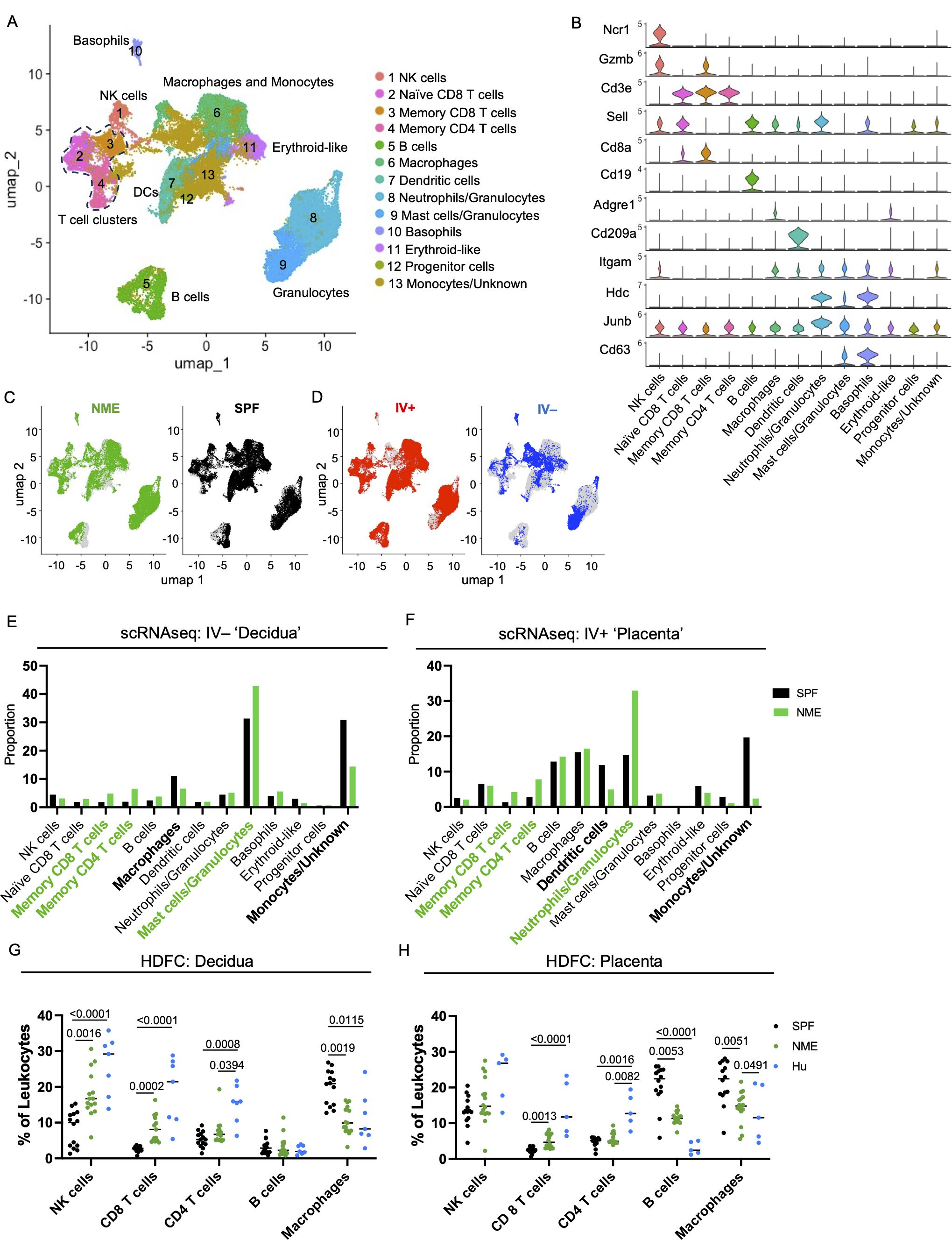
Figure 2. NME alters immune cell composition at the MFI. IV-labeled CD45+ immune cells were sorted from the MFI of NME and SPF dams at 14.5 dg and used for single cell transcriptomic analysis (scRNAseq) and high dimensional flow cytometry (HDFC) (n = 3 litters/group). (A) We performed dimensional reduction of the scRNAseq dataset using uniform manifold approximation and projection (UMAP) in Seurat and the scType cell identification R package to identify 13 immune cell clusters. T cell clusters are outlined with dotted line. (B) Key markers were used to confirm and refine cluster annotation. The location of (C) NME (green) and SPF (black) and (D) IV-labeled immune cells (red IV+ and blue IV negative) on the UMAP plot revealed cluster biases. Immune cell composition of (E) decidua and (F) placenta in NME (green) and SPF (black) mice determined by scRNAseq. Cell populations enriched in the NME and SPF samples are in bolded green and black, respectively. HDFC of immune cell composition in the (G) decidua and (H) placenta of NME mouse (green), SPF mouse (black), and human (blue) samples (n = 14–15 MFI from 4–5 separate litters, n = 8 for human samples). Mann-Whitney U test was used to determine significance.
Leukocyte composition in MFI of NME mice reflect human MFI leukocytes
Next, we performed high dimensional flow cytometry (HDFC) on mechanically separated placental and decidual tissues harvested from SPF and NME dams at 14.5 dg to enumerate immune cells, confirm the scRNAseq data, and compare immune populations with human decidual and placental tissues (Figures 2G, H, Supplementary Figures S3A–C). In agreement with the scRNAseq data, HDFC demonstrated decidual and placental T cells were expanded in NME dams relative to SPF dams (Figures 2G, H). Specifically, CD8 T cells were virtually absent from MFI tissues in SPF dams and present in MFI tissues of NME dams in a range that more closely resembles the CD8 T cell frequencies found in human decidual and placental tissues (Figure 2G, H). In addition, a marked increase in the frequency of decidual NK cells was observed in NME dams relative to SPF dams, with a range that resembled the decidual NK cell frequency in human tissues with improved fidelity. Confirming the scRNAseq data, a decrease in the frequency of decidual and placental macrophages in NME dams relative to SPF dams was observed (Figures 2G, H). A comparison with human macrophage and B cell frequencies again shows that the range of frequencies observed in NME dams resemble human MFI macrophage frequencies with greater fidelity than those observed in SPF dams. CD8 T cells were also found to be more numerous and macrophages less numerous in the decidua and placenta, while B cells were less numerous in the placenta of NME dams relative to SPF dams, demonstrating NME induces both a change in frequency and absolute number for these populations (Supplementary Figures 3B, C). Altogether, these data demonstrate maternal microbial exposure alters the immune cell composition of the murine MFI and phenocopy the human MFI immune cell populations with great fidelity.
Maternal microbial experience influences T cell composition at the MFI
CD4 and CD8 memory T cell populations are dominant immune populations in human MFI tissues at term pregnancy (17–23). Consequently, the significant expansion of T cells under NME conditions compared to SPF conditions has high translational significance. To provide further resolution of how NME changes T cell diversity at the MFI, scRNAseq resolved T cell clusters were selected for further analysis. UMAPs of all T cells from SPF and NME MFI tissues identified 13 separate T cell clusters (Figure 3A). T cell cluster identities were confirmed by expression of key markers (Figures 3A, B; Supplementary Figure 6). Stark differences in the distribution of T cells across NME and SPF dams as well as decidual and placental tissues were observed. Overall, a strong bias towards activated T cells (clusters 4, 5, 8, 9, and 13) in the NME tissues was found (Figures 3C, D). Of the IV– decidual T cells in NME samples, a greater proportion of i) CD8 TEM and TRM cells (clusters 4 and 5); ii) γδ T cells (cluster 13); iii) CD4 TCM cells (cluster 8) were found; combined with iv) lower frequencies of Klra6 (Ly49F)+ Ikzf2 (Helios)+ T cells (cluster 3) and cytotoxic CD4 cells (cluster 12) were observed (Figure 3E). NME IV+ placental T cells demonstrated an even greater skewing toward memory T cell populations, with increased proportions of CD4 and CD8 memory T cells (clusters 4, 5, 8, and 9) and γδ T cells (cluster 13) (Figure 3F). While proportions of naïve T cells (clusters 1, 2, 6, and 7) and Klra6 (Ly49F)+ Ikzf2 (Helios)+ T cells (cluster 3) were decreased in NME mice relative to SPF mice (Figure 3F). These observations were confirmed by HDFC where we also observed increased proportions of memory T cell subsets in both the decidua and placenta and corresponding decreases in naïve T cell populations in the placenta (Figures 3G, H). Direct comparison of SPF, NME, and human T cell diversity by HDFC showed NME conditions replicated human conditions more accurately than SPF conditions. Specifically, the increased proportion of CD8 TEM/TEFF and TRM subsets in the decidua and placenta and the decrease of CD4 and CD8 naïve T cells in both tissues of NME mice match human T cell diversity in more detail (Figures 3G, H). CD4 and CD8 TEM/TEFF cells were also more numerous in both decidua and placenta, and CD4 and CD8 TCM cells were more numerous in the placenta of NME dams than SPF dams (Supplementary Figures 6B, C). Thus, maternal microbial exposure induces profound changes in the composition of T cells at the MFI, with stark increases in tissue memory CD8, CD4, and γδ T cell types. These data establish NME mice as an improved translational model for human immunology at the MFI.
Figure 3. NME alters T cell composition at the MFI. (A) T cells were sub-gated from the scRNAseq dataset and UMAP plots were made and annotated as before. (B) Key markers were used to confirm and refine cluster annotation. (C) NME (green) and SPF (black) and (D) IV-labeled cells (red IV+ and blue IV negative) were identified on the UMAP plot. scRNAseq data revealed NME conditions skewed the T cell population toward activated subsets and away from naïve subsets in both the (E) decidua (IV–) and (F) placenta (IV+). T cell composition of (E) decidua and (F) placenta in NME (green) and SPF (black) mice determined by scRNAseq. T cell populations enriched in the NME and SPF samples are in bolded green and black, respectively. HDFC of T cell populations in the (G) decidua and (H) placenta of NME mouse (green), SPF mouse (black), and human (blue) samples. Sample size is 14–15 MFI from 4–5 separate litters. Sample size is 8 for human samples. Mann-Whitney U test was used to determine significance.
Maternal microbial experience increases CD8 T cell activation and diversity at the MFI
To further define functional changes in NME-expanded CD8 T cell clusters, differential gene expression analysis between total NME and SPF MFI CD8 T cells was performed using Seurat. Gene Set Enrichment Analysis (GSEA) found several hallmark pathways including “TNFα-signaling via NFKβ“, “Complement”, “IFNα Response”, and “IL-2 STAT5 Signaling” significantly enriched in CD8 T cells of NME MFI, a finding in line with the expectation of increased basal immune activation in NME mice (Figure 4A). Hallmark pathways in NME CD8 T cells further indicated i) a marked increase in apoptosis, glycolysis, cholesterol, adipogenesis, and hypoxia pathways, suggesting a skewing of CD8 T cell viability and metabolic pathways; and ii) an increase in late estrogen response genes, suggesting that pregnancy hormones have a stronger influence on CD8 T cell activation and differentiation in NME compared to SPF dams (Figure 4A). To further define how the activation state of the NME expanded MFI CD8 T cells compare to human MFI T cells, we applied GSEA using curated datasets of ‘Activation’, ‘Dysfunction’, and ‘Activation/Dysfunction’ as we described previously (21, 47). Similar to what we demonstrated in humans, MFI CD8 T cells from NME dams overexpressed a mixed signature of ‘Activation/Dysfunction’ as compared to SPF mice, further confirming the greater similarity of MFI immunology between NME mice and humans (Figure 4B) (21, 47).
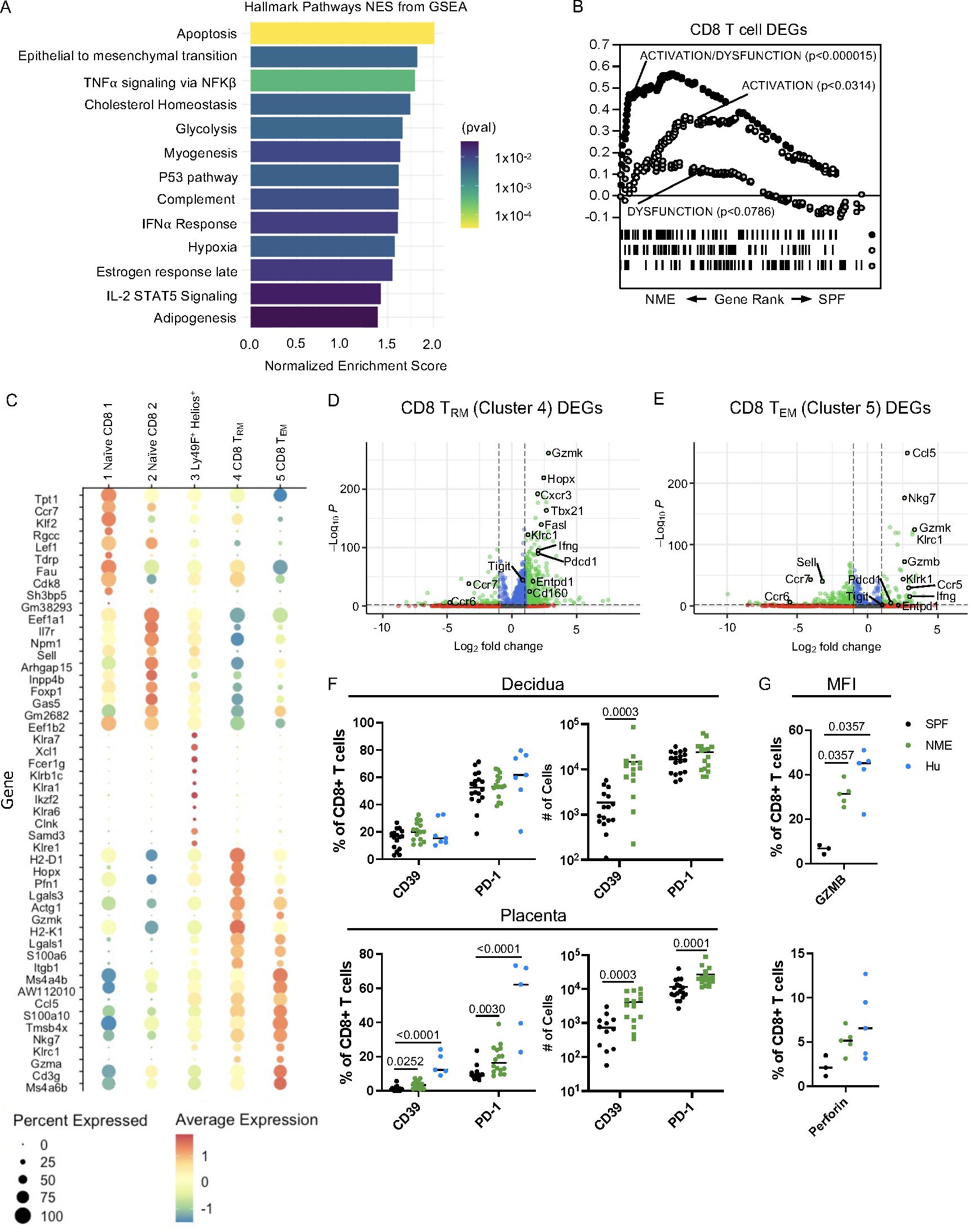
Figure 4. NME MFI CD8 T cells phenocopy those of humans. (A) GSEA was used to identified Hallmark Pathways associated with NME CD8 T cells. (B) Gene sets for activation/dysfunction, activation, and dysfunction were compared to DEGs from NME CD8 T cells using gene rank. (C) The top ten DEGs defining each CD8 T cell cluster were determined using Seurat. Volcano plots showing DEGs from (D) CD8 TRM (cluster 4) and (E) CD8 TEM (cluster 5). (F) Proportion and number of CD39 and PD-1 expressing CD8 T cells in the decidua (top) and placenta (bottom) determined flow cytometrically of NME mouse (green), SPF mouse (black), and human (blue) samples ((n = 15–18 MFI from 4–5 separate litters, 7 for human samples). (G) Granzyme B (left) and Perforin (right) protein expression in T cells harvested from the MFI (decidua and placenta combined) of NME mouse (green), SPF mouse (black), and human (blue) samples (n = 3 for SPF, 5 for NME, and 5 for human). Mann-Whitney U test was used to determine significance.
Differentially expressed genes (DEGs) from CD8 TRm and Tem cells (clusters 4 and 5), which were enriched in NME dams relative to SPF dams, revealed increased expression of multiple markers associated with cytotoxicity including Nkg7, Gzmk, Gzma, Gzmb, and Ifng as well as markers associated with regulation (Pdcd1, Cd160, and Entpd1) (Figures 4C–E). In agreement with the transcriptomic data, a greater proportion and number of CD8 T cells expressing PD-1 or CD39 protein in the placenta of NME dams relative to SPF dams was detected by HDFC (Figure 4F). Likewise, intracellular expression of granzyme B and perforin protein by CD8 T cells at the MFI (combined decidua and placenta) was higher in NME dams relative to SPF dams (Figure 4G). Both an increase in markers of CD8 dysfunction (PD-1 and CD39) and in markers of activation (GZMB and PFN) align the CD8 T cell phenotype of NME dams more closely with human MFI CD8 T cells (21, 22).
Maternal microbial experience increases CD8 T cell residency at the MFI
Besides the strong increase in CD8 activation and dysfunction, our prior studies of human MFI CD8 T cells demonstrated a predisposition towards the tissue residency phenotype (18, 19, 22). Using GSEA and published gene sets of ‘Resident’ and ‘Circulating’ T cells curated by Milner et al., we expectedly found that the NME CD8 T cell DEGs strongly aligned with the ‘Resident’ gene set (Figure 5A) (48). We measured the expression of commonly used markers of residency (CD69 and CD103) in decidual and placental CD44+ CD62L– effector CD8 T cells and observed an increased in the number of these cells in NME tissues relative to SPF tissues (Figure 5B), that more closely aligned with human CD8 T cells (Figure 5C). In mice, we found CD69 and CD49a expression were more associated with IV– cells than IV+ CD8 T cells in the decidua and placenta in agreement with these being markers of residency (Figure 5D). However, in the placenta, a greater proportion of IV+ cells were CD103+ than IV– cells, while IV– cells were more likely to be CD103+ than IV+ cells in the decidua (Figure 5D). CD103 expression was very rare in CD44+ effector/memory CD8 T cells in the peripheral blood, demonstrating placental IV+ T cells are unique from circulating T cells and may contain ‘resident’ T cells.
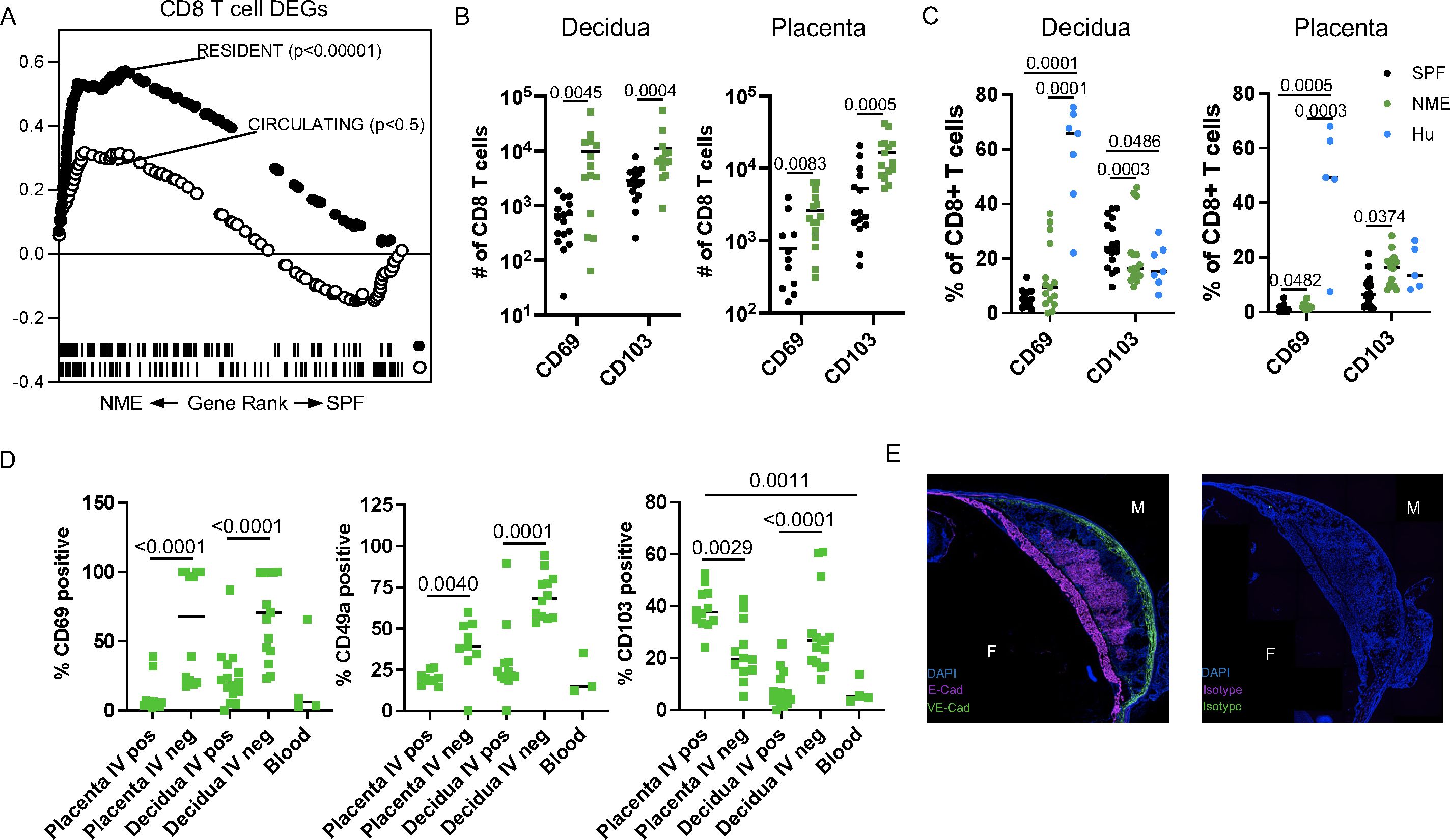
Figure 5. NME induces the expansion of cells expressing a residency phenotype. (A) GSEA using gene sets for resident and circulating cells was performed on NME differentially expressed genes (DEGs) from MFI CD8 T cells. (B, C) Number (B) and proportion (C) of CD69 and CD103 expressing CD8 T cells from SPF (black), NME (green) and human (blue) decidua (left) and placentas (right). Sample size is 14–15 MFI from 4–5 separate litters. (D) Flow cytometric measurement of common markers of T cell residency (CD69, CD49a, and CD103) in CD45.2 IV+ and IV- NME CD8 T cells from decidua and placenta compared to maternal blood (n = 9–12 from 4 separate litters). (E) Immunohistochemistry staining of CD103 ligand, E-cadherin, VE-cadherin, or isotype expression with DAPI from NME frozen MFI sections.
Immunofluorescence staining confirmed the expression of the CD103 ligand, E-cadherin, in the chorionic plate and placental labyrinth of both NME and SPF mice, similar to humans, suggesting CD103 expression may help recruit and/or retain T cells at the MFI (Figure 5E) (49). Altogether, these data demonstrate that NME expands T cell subsets with a resident memory-like phenotype in both placental and decidual tissues. These data also demonstrate that placental T cells, despite IV labeling, are unique from peripheral blood and express receptors for placental tissue ligands that may promote recruitment and residency.
Maternal microbial experience increases CD4 T cell diversity at the MFI
CD4 T cell populations have been extensively studied in murine and human pregnancy with a predominant focus on TREG function (6, 9, 29). In contrast to CD8 T cells, which are virtually absent in the MFI of SPF dams, CD4 TREG have been found and studied in detail in SPF dams (7, 50, 51). Surprisingly, the scRNAseq data analysis suggests that the frequency of CD4 TREG may be decreased in the placenta of NME dams compared to SPF dams (Figure 3F). In contrast, HDFC showed an increase in the proportion of TREGs in both the decidua and placenta of NME dams relative to SPF dams and human samples (Figure 6A). Enumeration of the Treg numbers in SPF and NME dams confirms the increase in TREG in NME dams (Figure 6A). To define differences in memory CD4 T cells between SPF and NME dams, we selected memory CD4 T cell clusters 8 and 9 for DEG analysis. Both memory CD4 T cell types that were enriched by NME expressed high levels of transcripts associated with TH17 cells (Rorc, Tmem176a, Tmem176b, Il17a, Il17re, Ccr4, Ccr6, and Il22) (Figure 6B). CD4 TCM (Cluster 8) was further defined by the expression of Sell, Klf2 and Tle5, while the decreased expression of these genes in cluster 9 is consistent with an effector memory phenotype (Figure 6B). HDFC analysis confirmed NME placenta and decidua contained greater numbers of TEM and TCM CD4 memory and TH17 cells (Figures 3G, H and 6C). Thus, natural microbial exposure expands TREG numbers and activates CD4+ memory T cell populations at the MFI.
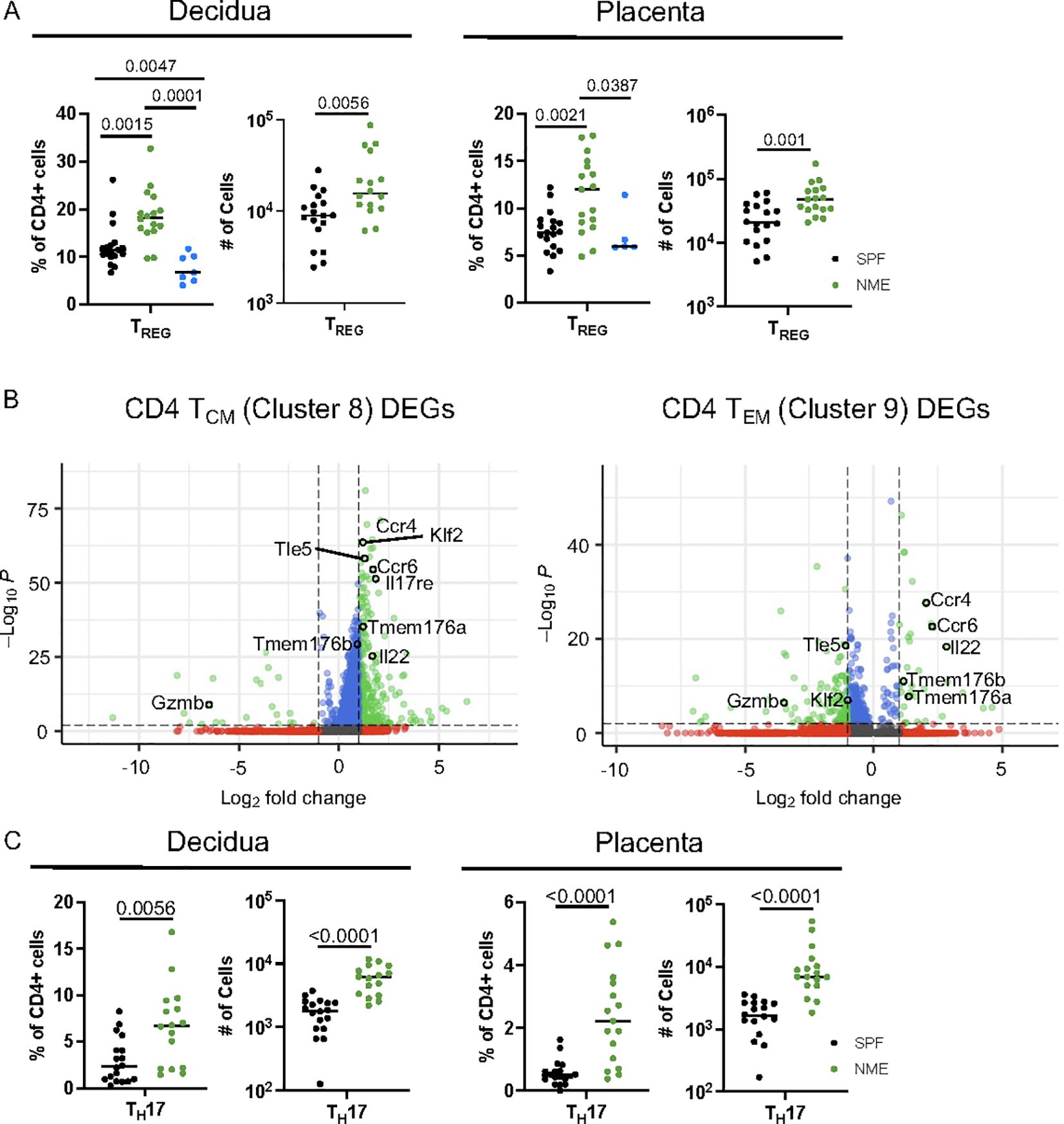
Figure 6. NME expands activated CD4 T cell at the MFI. (A) Proportion and number of TREGs from SPF (black), NME (green) and human (blue) decidua (left) and placentas (right) determined flow cytometrically. (n = 16-18, three separate litters). (B) DEGs from scRNAseq data for CD4 TCM (cluster 8) and CD4 TEM (cluster 9). (C) Proportion and number of TH17 cells from SPF (black) and NME (green) in decidua (left) and placenta (right) determined flow cytometrically (n = 16-18, three separate litters).
Maternal Microbial exposure increase expression of killer lectin-like receptors on T cells
Notably, the NME-expanded CD8 T cell populations at the MFI expressed high levels of multiple genes commonly associated with natural killer cells. Previously, we observed higher levels of Natural Killer Receptors (NKRs) including Killer Immunoglobulin-like Receptors (KIRs), CD94-NKG2A (inhibitory), and CD94-NKG2C (activating) receptors on human decidual T cells than peripheral blood T cells (52, 53). In mice, Ly49 receptors or killer lectin-like receptors (KLRs), while structurally dissimilar, have evolved similarly to human NKRs and also recognize MHC class I molecules to promote or inhibit NK and CD8 T cell cytotoxicity. The scRNAseq data presented here identified multiple activating and inhibiting KLRs upregulated in MFI CD8 (clusters 3, 4, 5), and CD4 T cells (cluster 12) (Figure 7). Of these receptors, NKG2A (Klrc1) in mice recognizes the non-classical non-polymorphic MHC class I molecule Qa-1 providing them with a receptor to recognize placental trophoblasts; whereas NKG2D (Klrk1) binds RAE1 (Rea-1), H60 (H60), and MULT-1 (Ulbp1) in mice, NKG2D binds MICA/B in humans, which are upregulated on stressed or infected target cells and provide strong pro-cytolytic signaling. Thus, maternal microbial exposure activates T cells at the MFI and upregulates their potential for TCR-independent recognition of trophoblasts as well as infected or stressed target cells. Altogether, these data demonstrate that NME conditions expand T cell diversity that phenocopies human MFI T cells and provide maternal-fetal tolerance and anti-microbial responses (2, 19–22, 54, 55).
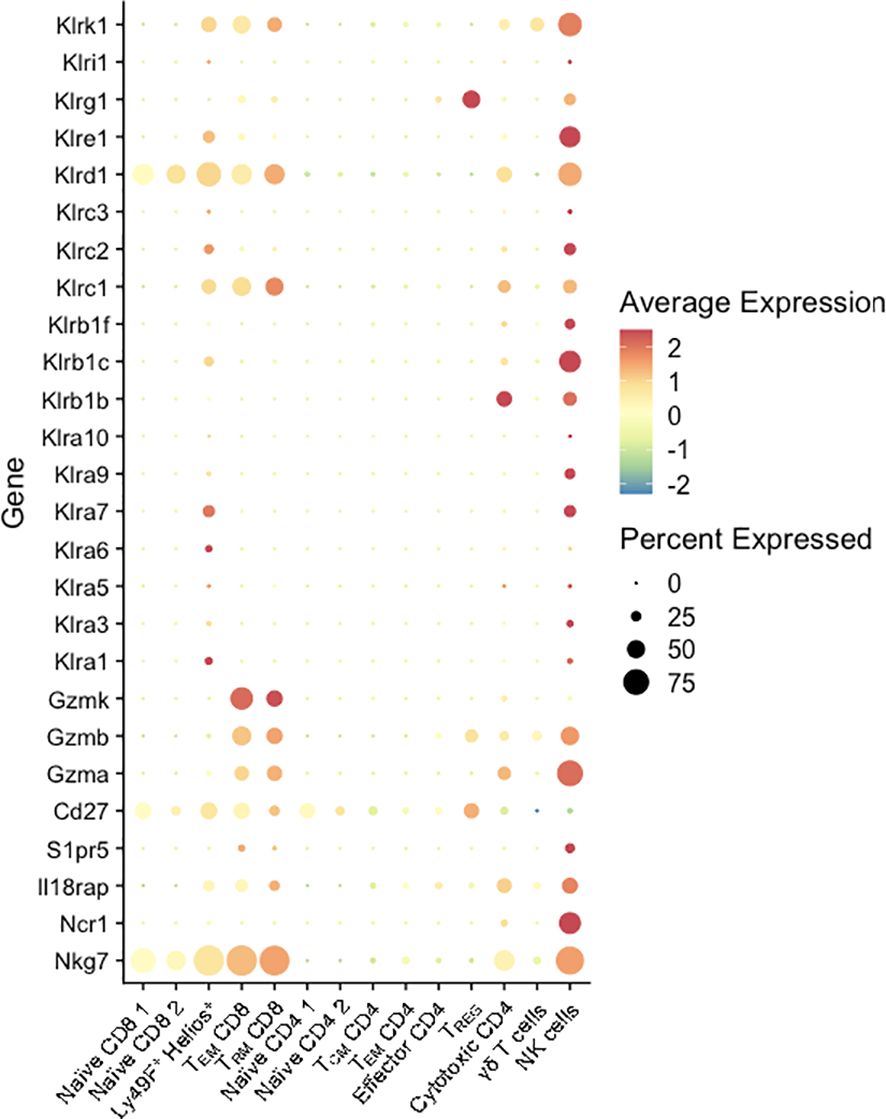
Figure 7. KLR and NK marker expression by T cells at the MFI. Percent expressed (size of the circle) and average expression (based on color spectrum) of select KLR genes and markers associated with NK cells across all T cell clusters from scRNAseq data determined using Seurat.
Discussion
Using an innovative model of natural microbial exposure during murine pregnancy, we demonstrate the profound effects of microbial exposure driving increased T cell activation, differentiation, and tissue residency at the maternal-fetal interface. One of the profound changes observed was the significant increase in CD4 and CD8 memory T cells, whereas TREG, NK, and B cell populations had fewer differences between SPF and NME dams. Most notably, NME conditions reduced frequencies of naïve T cells, significantly expanded CD8 T cell numbers and memory CD8 and CD4 T cell diversity with signatures of activation and dysfunction unique to the MFI that phenocopy signatures observed in humans. We and others have demonstrated NME enhances peripheral TEFF and TEM cells, which characteristically traffic through a broad range of non-lymphoid tissues (36, 37, 41). These observations are consistent with the hypothesis that TEM cells have greater ability to access decidual and placental tissues through CXCR3 expression. The relative paucity of these cell types in SPF mice likely limit T cell accumulation at the MFI. However, NME conditions may also enhance T cell recruitment and differentiation by altering the expression of integrins, cytokines, and chemokines at the MFI. Once in the MFI, T cells may acquire further unique phenotypes. Whether NME alters the MFI microenvironment to augment MFI T cell differentiation needs to be investigated further.
In this study, we used syngeneic matings to focus on the effects of natural microbial exposure on the MFI immune cell states. In most cases, NME generated immune cell states in the MFI that were much more, but not completely, aligned with human MFI immune cell diversity. The remaining gap between NME and human T cell metrics may be due to additional immune activation and regulation induced by paternal alloantigen exposure that is much reduced in syngeneic breeding. In addition, the NME and SPF dams were of similar age, healthy weight, and only primary pregnancies were used. These clinical variables likely influence T cell characteristics and should be investigated in further detail. Most notably, expanding these studies to include parity as a factor driving immune activation and memory diversity as has been observed in humans would be of high interest (56, 57).
We took a microbial agnostic approach to our experimental design, choosing to focus on the diversity of microbial exposure over the presence of individual microbes. The microbial communities of pet store mice are diverse and variable, containing bacteria, fungi, viruses, and parasites. NME dams were co-housed with a single pet store mouse throughout mating and gestation. Therefore, while the microbial exposure of an individual dam may be unique from that of others in the group, it represents a very high diversity of commensal and pathogenic microbial communities. Data from our group and others using microbial naturalization models demonstrate that the effects of diverse microbial exposure are notably consistent and generally have a similar deviation as SPF mice (36, 37, 39–41, 58). While it is entirely possible that individual microbes have specific effects on the immunology of the MFI, our data shows remarkable stability across litters co-housed with different pet store mice, and we have not identified any obvious immunological effects associated with a particular microbe.
The placenta and decidua are unique in that they are temporary tissues; as a result, markers of tissue residency are difficult to define. A large proportion of human decidual CD8 T cells are described as TRM based on expression of CD69, CD103, and CD49A, however, residency markers can be tissue dependent (19, 22). Here, we demonstrate that decidual T cells are consistent with this convention, as IV– decidual T cells have a resident transcriptomic profile and express higher levels of CD69, CD103, and CD49a in the decidua. We also describe placental T cells that, while in contact with the circulation, express features that indicate they are unique from circulating T cells and may be resident in the placenta. For example, we observed IV+ placental T cells expressing higher levels of CD103 than T cells in the blood. E-cadherin, a ligand for CD103, is strongly expressed in the placental labyrinth and chorionic plate, supporting a placental tissue-specific role for CD103 in T cell homing. KLF2 is a transcription factor that regulates migration of T cells by inducing the expression of S1PR1, which permits egress from lymphoid tissues via binding to S1P. Therefore, KLF2 is considered a marker of circulating T cells, i.e., naïve and central memory T cells. Unexpectedly, high Klf2 expression was associated with the TEM population (Sell low Cd44+ cluster 4) at the MFI. However, S1P is expressed in the placenta; therefore, KLF2 expression may be important for T cell migration to and retention in the MFI (59). Altogether, both HDFC and scRNAseq data demonstrate maternal microbial exposure increases T cells with a residency phenotype at the MFI.
To evade killing by maternal CD8 T cells, human fetal trophoblasts reduce the expression of HLA-A and HLA-B in favor of HLA-C and non-classical HLA-E and HLA-G, which are thought to induce tolerance in T cells via modulation of myeloid cells or direct engagement of inhibitory NK cell receptors or KIRs (10, 52). In mice, the non-classical MHC molecules Qa-1b and Qa-2 are expressed on trophoblasts. Qa-1b is a homologue of HLA-E and provides surveillance of antigen processing defects (60). Qa-2 is proposed as a functional homologue to human HLA-G and involved in immune tolerance (61). We have previously demonstrated KIR expression is increased in human MFI CD4 and CD8 T cells relative to peripheral T cells (52). Here, we discovered high levels of KLR (Ly49) expression (similar function to human KIRs) in subsets of murine CD4 and CD8 T cells at the MFI. The Klra6 (Ly49F)+ Ikzf2 (Helios)+ cluster 3 exhibited similarities to intraepithelial lymphocytes (IELs) and appeared to make up a smaller proportion of T cells under NME conditions, as did the cytotoxic CD4 T cells cluster (Klrb1+ Gzma+). Conversely, memory CD8 T cells in the MFI (clusters 4 and 5) were expanded by NME conditions and expressed multiple cytotoxic molecules and a mix of activating and inhibiting KLRs. Whether the expression of KLRs increases the potential for TCR-independent recognition of infected and stressed cells or modulates T cell activity to maintain tolerance or enhance cytotoxicity requires further investigation.
In addition to activated CD8 T cells, maternal microbial exposure also enhanced activated CD4 T cell subsets at the MFI. TREGs are critical for restraining active immune responses and maintaining tolerance at the MFI (6, 11, 62). Unsurprisingly, HDFC analysis showed TREGs increased in number and proportion at the MFI in response to maternal immune activation, presumably to regulate active immune cells and maintain tolerance. This was not evident in the scRNAseq data, likely owing to differences in sensitivity to detect bona fide TREGs between the two methods. TH17 cells were found to be preferentially expanded by NME conditions at the MFI in both scRNAseq and HDFC datasets. TH17 cells secrete proinflammatory cytokines (IL-17, IL-21, IL-22, and IL-26) to assist in the defense against extracellular bacteria and fungi (63). Overactive TH17 responses can lead to inflammation and tissue destruction and are linked to several pregnancy complications (e.g., preeclampsia, gestational diabetes mellitus, and preterm birth) (64). TH17 cells can also play a pathologic role in allograft rejection as the balance between TH17 and TREG cell responses is crucial in reducing the risk of rejection (65, 66). While the expansion of TREGs at the MFI may be sufficient to control cytotoxic and TH17 cells, we also observed a corresponding expansion of Il23r expressing γδ T cells, which have been described to identify a subset of γδ T cells that suppress TH1 and TH17 cells by absorbing the available IL-23 (67). Altogether, maternal microbial exposure enhanced anti-microbial and immune regulatory T cell populations providing insight into the cellular mechanisms that balance host defense and tolerance at the MFI in the context of physiological microbial exposure and a healthy pregnancy.
Our data demonstrate maternal microbial exposure dramatically enhanced T cell heterogeneity to replicate the T cell landscape of the human MFI more accurately. However, there still exist differences between human and NME mouse MFI T cells. While we observed that NME increases the number and proportion of T cells at the MFI, T cells only comprised ~6% of immune cells at 14.5 dg, well below the 20-60% at the human MFI at the beginning of the third trimester (3, 17, 68). This could reflect differences in the length of gestation between humans and mice, as memory T cells accumulate in the MFI over time. Differences in the anatomy of human and murine MFI and the average number of concepti per pregnancy are limitations of the model that are not feasibly altered but may also impact MFI T cell immunology.
Cumulatively, this study presents an improved model of human pregnancy, creating opportunities for detailed mechanistic studies of microenvironmental signals driving T cell function and recruitment. The extent to which maternal microbial exposure alters T cell influx and diversity to the MFI is vast, and careful and comprehensive investigation into how these cells contribute to tolerance, immunity, and inflammation in healthy and pathogenic pregnancies is warranted.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, GSE293451.
Ethics statement
The studies involving humans were approved by Institutional Review Board, Cincinnati Children’s Hospital Medical Center. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. The animal study was approved by Institutional Animal Care and Use Committee, University of Minnesota. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
AW: Methodology, Conceptualization, Investigation, Writing – review & editing, Writing – original draft, Data curation, Formal analysis. PS: Formal analysis, Data curation, Investigation, Writing – review & editing. SB: Investigation, Writing – review & editing, Data curation. AS: Data curation, Investigation, Writing – review & editing. AL: Formal analysis, Data curation, Investigation, Writing – review & editing. JB: Data curation, Formal analysis, Writing – review & editing, Investigation. ST: Writing – review & editing, Formal analysis, Data curation, Investigation. SM: Writing – review & editing, Data curation, Formal analysis, Investigation. TT: Investigation, Resources, Writing – original draft, Writing – review & editing, Funding acquisition, Visualization, Formal analysis, Conceptualization, Validation, Data curation, Project administration, Supervision. NS: Investigation, Conceptualization, Resources, Funding acquisition, Project administration, Writing – review & editing, Validation, Visualization, Data curation, Writing – original draft, Methodology, Formal analysis, Supervision.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by NIH grants R01HD113192 (to N.J.S.), R01HD116852 (to T.T.) and S10OD025045 (to the Research Flow Cytometry Facility, Cincinnati Children’s), a University of Minnesota Medical School Department of Pediatrics ‘R’ award (to N.J.S), A Masonic Cross-Departmental Grant (to N.J.S), the Burroughs Wellcome Fund Next Gen Pregnancy Award #NGP10115 (to T.T.), and the Cincinnati Children’s Research Foundation (CCRF) Trustee Award (to T.T.).
Acknowledgments
We would like to extend our gratitude to the University of Minnesota Flow Cytometry Core, The University of Minnesota Genomics Center (UMGC), the Minnesota Supercomputing Institute (MSI), Research Animal Resources (RAR), the Center for Immunology, the Dirty Mouse Colony Core Facility, and the Center for Immunology Imaging Center for their support, Sherry Thornton and the Research Flow Cytometry Core in the Division of Rheumatology at Cincinnati Children’s Hospital Medical Center for help with flow cytometry, and all nurses and physicians of participating hospitals for their efforts collecting human placental materials.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1616491/full#supplementary-material
Supplementary Figure 1 | Example H&E staining of NME and SPF MFI frozen sections. Images are labeled with maternal (m) and fetal (f) sides.
Supplementary Figure 2 | Frequency of IV labeled immune cells in the placenta and decidua. (A) Example CD45.2 IV+ labeling of leukocytes from decidua, placenta, and maternal blood. (B) Proportion of IV+ leukocytes in decidua, placenta, and maternal blood (n = 10–12 MFI from 4 separate litters). (C) Example CD45.2 IV+ labeling of T cells from decidua, placenta, and maternal blood. (D) Proportion of IV+ T cells in decidua, placenta, and maternal blood (n = 10–12 MFI from 4 separate litters).
Supplementary Figure 3 | Leukocyte composition at the MFI. (A) Top ten DEGs for each scRNAseq leukocyte cluster. (B, C) Cytometric enumeration of NK cells, CD8 T cells, CD4 T cells, B cells, and macrophages in the decidua (B) and placenta (C).
Supplementary Figure 4 | NME conditions associated with increased Jun expression. Volcano plot of DEGs from (A) Macrophage clusters, (B) B cell clusters, (C) NK cell clusters.
Supplementary Figure 5 | Example flow cytometry gating strategy. (A) gating strategy used to identify T cells populations. (B) gating strategy used for FACS sorting of CD45+ IV+ and IV- leukocytes in preparation for single cell sequencing.
Supplementary Figure 6 | T cell composition at the MFI. (A) Top ten DEGs for each scRNAseq T cell cluster. (B, C) Cytometric enumeration of T cell subsets in the decidua (B) and placenta (C).
References
1. Erlebacher A. Immunology of the maternal-fetal interface. Annu Rev Immunol. (2013) 31:387–411. doi: 10.1146/annurev-immunol-032712-100003
2. Tilburgs T and Strominger JL. CD8+ effector T cells at the fetal-maternal interface, balancing fetal tolerance and antiviral immunity. Am J Reprod Immunol. (2013) 69:395–407. doi: 10.1111/aji.12094
3. Yang F, Zheng Q, and Jin L. Dynamic function and composition changes of immune cells during normal and pathological pregnancy at the maternal-fetal interface. Front Immunol. (2019) 10:2317. doi: 10.3389/fimmu.2019.02317
4. Gomez-Lopez N, Stlouis D, Lehr MA, Sanchez-Rodriguez EN, and Arenas-Hernandez M. Immune cells in term and preterm labor. Cell Mol Immunol. (2014) 11:571–81. doi: 10.1038/cmi.2014.46
5. Parasar P, Guru N, and Nayak NR. Contribution of macrophages to fetomaternal immunological tolerance. Hum Immunol. (2021) 82:325–31. doi: 10.1016/j.humimm.2021.02.013
6. Aluvihare VR, Kallikourdis M, and Betz AG. Regulatory T cells mediate maternal tolerance to the fetus. Nat Immunol. (2004) 5:266–71. doi: 10.1038/ni1037
7. Severance AL, Kinder JM, Xin L, Burg AR, Shao TY, Pham G, et al. Maternal-fetal conflict averted by progesterone- induced FOXP3+ regulatory T cells. iScience. (2022) 25:104400. doi: 10.1016/j.isci.2022.104400
8. Moldenhauer LM, Diener KR, Thring DM, Brown MP, Hayball JD, and Robertson SA. Cross-presentation of male seminal fluid antigens elicits T cell activation to initiate the female immune response to pregnancy. J Immunol. (2009) 182:8080–93. doi: 10.4049/jimmunol.0804018
9. Tilburgs T, Roelen DL, van der Mast BJ, De Groot-Swings GM, Kleijburg C, Scherjon SA, et al. Evidence for a selective migration of fetus-specific CD4+CD25bright regulatory T cells from the peripheral blood to the decidua in human pregnancy. J Immunol. (2008) 180:5737–45. doi: 10.4049/jimmunol.180.8.5737
10. Tilburgs T, Scherjon SA, van der MAST BJ, Haasnoot GW, Versteeg-V D Voort-Maarschalk M, Roelen DL, et al. Fetal-maternal HLA-C mismatch is associated with decidual T cell activation and induction of functional T regulatory cells. J Reprod Immunol. (2009) 82:148–57. doi: 10.1016/j.jri.2009.05.003
11. Gomez-Lopez N, Arenas-Hernandez M, Romero R, Miller D, Garcia-Flores V, Leng Y, et al. Regulatory T cells play a role in a subset of idiopathic preterm labor/birth and adverse neonatal outcomes. Cell Rep. (2020) 32:107874. doi: 10.1016/j.celrep.2020.107874
12. Inada K, Shima T, Nakashima A, Aoki K, Ito M, and Saito S. Characterization of regulatory T cells in decidua of miscarriage cases with abnormal or normal fetal chromosomal content. J Reprod Immunol. (2013) 97:104–11. doi: 10.1016/j.jri.2012.12.001
13. Tsuda S, Shichino S, Tilburgs T, Shima T, Morita K, Yamaki-Ushijima A, et al. CD4. Front Immunol. (2024) 15:1401738. doi: 10.3389/fimmu.2024.1401738
14. Tsuda S, Zhang X, Hamana H, Shima T, Ushijima A, Tsuda K, et al. Clonally expanded decidual effector regulatory T cells increase in late gestation of normal pregnancy, but not in preeclampsia, in humans. Front Immunol. (2018) 9:1934. doi: 10.3389/fimmu.2018.01934
15. Verdijk RM, Kloosterman A, Pool J, Van De Keur M, Naipal AM, Van Halteren AG, et al. Pregnancy induces minor histocompatibility antigen-specific cytotoxic T cells: implications for stem cell transplantation and immunotherapy. Blood. (2004) 103:1961–4. doi: 10.1182/blood-2003-05-1625
16. Lissauer D, Piper K, Goodyear O, Kilby MD, and Moss PA. Fetal-specific CD8+ cytotoxic T cell responses develop during normal human pregnancy and exhibit broad functional capacity. J Immunol. (2012) 189:1072–80. doi: 10.4049/jimmunol.1200544
17. Dejong CS, Maurice NJ, Mccartney SA, and Prlic M. Human tissue-resident memory T cells in the maternal–fetal interface. Lost soldiers or special forces? Cells. (2020) 9:2699. doi: 10.3390/cells9122699
18. Nancy P and Erlebacher A. T cell behavior at the maternal-fetal interface. Int J Dev Biol. (2014) 58:189–98. doi: 10.1387/ijdb.140054ae
19. Liu L, Huang X, Xu C, Chen C, Zhao W, Li D, et al. Decidual CD8. J Transl Med. (2020) 18:221. doi: 10.1186/s12967-020-02371-3
20. Tilburgs T, Schonkeren D, Eikmans M, Nagtzaam NM, Datema G, Swings GM, et al. Human decidual tissue contains differentiated CD8+ effector-memory T cells with unique properties. J Immunol. (2010) 185:4470–7. doi: 10.4049/jimmunol.0903597
21. Van Der Zwan A, Bi K, Norwitz ER, Crespo Â.C, Claas FHJ, Strominger JL, et al. Mixed signature of activation and dysfunction allows human decidual CD8 + T cells to provide both tolerance and immunity. Proc Natl Acad Sci. (2018) 115:385–90. doi: 10.1073/pnas.1713957115
22. Mahajan S, Alexander A, Koenig Z, Saba N, Prasanphanich N, Hildeman DA, et al. Antigen-specific decidual CD8+ T cells include distinct effector memory and tissue-resident memory cells. JCI Insight. (2023) 8(17). doi: 10.1172/jci.insight.171806
23. Van Egmond A, van der Keur C, Swings GM, Scherjon SA, and Claas FH. The possible role of virus-specific CD8(+) memory T cells in decidual tissue. J Reprod Immunol. (2016) 113:1–8. doi: 10.1016/j.jri.2015.09.073
24. Nancy P, Tagliani E, Tay CS, Asp P, Levy DE, and Erlebacher A. Chemokine gene silencing in decidual stromal cells limits T cell access to the maternal-fetal interface. Science. (2012) 336:1317–21. doi: 10.1126/science.1220030
25. Zenclussen AC, Gerlof K, Zenclussen ML, Sollwedel A, Bertoja AZ, Ritter T, et al. Abnormal T-cell reactivity against paternal antigens in spontaneous abortion: adoptive transfer of pregnancy-induced CD4+CD25+ T regulatory cells prevents fetal rejection in a murine abortion model. Am J Pathol. (2005) 166:811–22. doi: 10.1016/S0002-9440(10)62302-4
26. Constantin CM, Masopust D, Gourley T, Grayson J, Strickland OL, Ahmed R, et al. Normal establishment of virus-specific memory CD8 T cell pool following primary infection during pregnancy. J Immunol. (2007) 179:4383–9. doi: 10.4049/jimmunol.179.7.4383
27. Svensson-Arvelund J, Mehta RB, Lindau R, Mirrasekhian E, Rodriguez-Martinez H, Berg G, et al. The human fetal placenta promotes tolerance against the semiallogeneic fetus by inducing regulatory T cells and homeostatic M2 macrophages. J Immunol. (2015) 194:1534–44. doi: 10.4049/jimmunol.1401536
28. Tilburgs T, Crespo Â, van der Zwan A, Rybalov B, Raj T, Stranger B, et al. Human HLA-G+ extravillous trophoblasts: Immune-activating cells that interact with decidual leukocytes. Proc Natl Acad Sci U.S.A. (2015) 112:7219–24. doi: 10.1073/pnas.1507977112
29. Salvany-Celades M, van der Zwan A, Benner M, Setrajcic-Dragos V, Bougleux Gomes HA, Iyer V, et al. Three types of functional regulatory T cells control T cell responses at the human maternal-fetal interface. Cell Rep. (2019) 27:2537–2547.e5. doi: 10.1016/j.celrep.2019.04.109
30. Jiang SP and Vacchio MS. Multiple mechanisms of peripheral T cell tolerance to the fetal “allograft. J Immunol. (1998) 160:3086–90. doi: 10.4049/jimmunol.160.7.3086
31. Crespo Â, Strominger JL, and Tilburgs T. Expression of KIR2DS1 by decidual natural killer cells increases their ability to control placental HCMV infection. Proc Natl Acad Sci U.S.A. (2016) 113:15072–7. doi: 10.1073/pnas.1617927114
32. Giles ML, Krishnaswamy S, Macartney K, and Cheng A. The safety of inactivated influenza vaccines in pregnancy for birth outcomes: a systematic review. Hum Vaccin Immunother. (2019) 15:687–99. doi: 10.1080/21645515.2018.1540807
33. Tormen M, Taliento C, Salvioli S, Piccolotti I, Scutiero G, Cappadona R, et al. Effectiveness and safety of COVID-19 vaccine in pregnant women: A systematic review with meta-analysis. BJOG. (2023) 130:348–57. doi: 10.1111/1471-0528.17354
34. Fleming-Dutra KE, Jones JM, Roper LE, Prill MM, Ortega-Sanchez IR, Moulia DL, et al. Use of the pfizer respiratory syncytial virus vaccine during pregnancy for the prevention of respiratory syncytial virus-associated lower respiratory tract disease in infants: recommendations of the advisory committee on immunization practices - United States 2023. MMWR Morb Mortal Wkly Rep. (2023) 72:1115–22. doi: 10.15585/mmwr.mm7241e1
35. Li Y, Lopez GE, Vazquez J, Sun Y, Chavarria M, Lindner PN, et al. Decidual-placental immune landscape during syngeneic murine pregnancy. Front Immunol. (2018) 9:2087. doi: 10.3389/fimmu.2018.02087
36. Burger S, Stenger T, Pierson M, Sridhar A, Huggins MA, Kucaba TA, et al. Natural microbial exposure from the earliest natural time point enhances immune development by expanding immune cell progenitors and mature immune cells. J Immunol. (2023) 210:1740–51. doi: 10.4049/jimmunol.2300061
37. Beura LK, Hamilton SE, Bi K, Schenkel JM, Odumade OA, Casey KA, et al. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature. (2016) 532:512–6. doi: 10.1038/nature17655
38. Fiege JK, Block KE, Pierson MJ, Nanda H, Shepherd FK, Mickelson CK, et al. Mice with diverse microbial exposure histories as a model for preclinical vaccine testing. Cell Host Microbe. (2021) 29:1815–1827.e6. doi: 10.1016/j.chom.2021.10.001
39. Huggins MA, Sjaastad FV, Pierson M, Kucaba TA, Swanson W, Staley C, et al. Microbial Exposure Enhances Immunity to Pathogens Recognized by TLR2 but Increases Susceptibility to Cytokine Storm through TLR4 Sensitization. Cell Rep. (2019) 28:1729–1743.e5. doi: 10.1016/j.cerep.2019.07.028
40. Rosshart SP, Vassallo BG, Angeletti D, Hutchinson DS, Morgan AP, Takeda K, et al. Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell. (2017) 171:1015–1028.e13. doi: 10.1016/j.cell.2017.09.016
41. Rosshart SP, Herz J, Vassallo BG, Hunter A, Wall MK, Badger JH, et al. Laboratory mice born to wild mice have natural microbiota and model human immune responses. Science. (2019) 365(6452). doi: 10.1126/science.aaw4361
42. Reese TA, Bi K, Kambal A, Filali-Mouhim A, Beura LK, Bürger MC, et al. Sequential infection with common pathogens promotes human-like immune gene expression and altered vaccine response. Cell Host Microbe. (2016) 19:713–9. doi: 10.1016/j.chom.2016.04.003
43. Ikumi NM, Koenig Z, Mahajan S, Gray CM, and Tilburgs T. Purification of primary human placental leukocytes to study maternal-fetal interactions. STAR Protoc. (2023) 4:102277. doi: 10.1016/j.xpro.2023.102277
44. Ikumi NM, Pillay K, Tilburgs T, Malaba TR, Dzanibe S, Enninga EAL, et al. T-cell homeostatic imbalance in placentas from women with human immunodeficiency virus in the absence of vertical transmission. J Infect Dis. (2021) 224:S670–82. doi: 10.1093/infdis/jiab192
45. Hao Y, Stuart T, Kowalski MH, Choudhary S, Hoffman P, Hartman A, et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat Biotechnol. (2024) 42:293–304. doi: 10.1038/s41587-023-01767-y
46. Ianevski A, Giri AK, and Aittokallio T. Fully-automated and ultra-fast cell-type identification using specific marker combinations from single-cell transcriptomic data. Nat Commun. (2022) 13:1246. doi: 10.1038/s41467-022-28803-w
47. Singer M, Wang C, Cong L, Marjanovic ND, Kowalczyk MS, Zhang H, et al. A distinct gene module for dysfunction uncoupled from activation in tumor-infiltrating T cells. Cell. (2016) 166:1500–1511.e9. doi: 10:1016/j.cell.2017.11.006
48. Milner JJ, Toma C, Yu B, Zhang K, Omilusik K, Phan AT, et al. Runx3 programs CD8. Nature. (2017) 552:253–7. doi: 10.1038/nature24993
49. Kokkinos MI, Murthi P, Wafai R, Thompson EW, and Newgreen DF. Cadherins in the human placenta–epithelial-mesenchymal transition (EMT) and placental development. Placenta. (2010) 31:747–55. doi: 10.1016/j.placenta.2010.06.017
50. Moldenhauer LM, Hull ML, Foyle KL, Mccormack CD, and Robertson SA. Immune-metabolic interactions and T cell tolerance in pregnancy. J Immunol. (2022) 209:1426–36. doi: 10.4049/jimmunol.2200362
51. Lentz LS, Stutz AJ, Meyer N, Schubert K, Karkossa I, Von Bergen M, et al. Human chorionic gonadotropin promotes murine Treg cells and restricts pregnancy-harmful proinflammatory Th17 responses. Front Immunol. (2022) 13:989247. doi: 10.3389/fimmu.2022.989247
52. Tilburgs T, van der Mast BJ, Nagtzaam NM, Roelen DL, Scherjon SA, and Claas FH. Expression of NK cell receptors on decidual T cells in human pregnancy. J Reprod Immunol. (2009) 80:22–32. doi: 10.1016/j.jri.2009.02.004
53. Van Der Zwan A, Van Unen V, Beyrend G, Laban S, van der Keur C, Kapsenberg HJM, et al. Visualizing dynamic changes at the maternal-fetal interface throughout human pregnancy by mass cytometry. Front Immunol. (2020) 11:571300. doi: 10.3389/fimmu.2020.571300
54. Kinder JM, Turner LH, Stelzer IA, Miller-Handley H, Burg A, Shao TY, et al. CD8. Cell Rep. (2020) 31:107784. doi: 10.1016/j.celrep.2020.107784
55. Maurice NJ, Erickson JR, Dejong CS, Mair F, Taber AK, Frutoso M, et al. Converging cytokine and metabolite networks shape asymmetric T cell fate at the term human maternal-fetal interface. bioRxiv. (2024). doi: 10.1101/2024.06.10.598377
56. Kotzur R, Kahlon S, Isaacson B, Gamliel M, Charpak-Amikam Y, Lieberman J, et al. Pregnancy trained decidual NK cells protect pregnancies from harmful Fusobacterium nucleatum infection. PloS Pathog. (2024) 20:e1011923. doi: 10.1371/journal.ppat.1011923
57. Gamliel M, Goldman-Wohl D, Isaacson B, Gur C, Stein N, Yamin R, et al. Trained memory of human uterine NK cells enhances their function in subsequent pregnancies. Immunity. (2018) 48:951–962.e5. doi: 10:1016/j.immuni.2018.03.030
58. Liu Q, Pickett T, Hodge D, Rios C, Arnold M, Dong G, et al. Leveraging dirty mice that have microbial exposure to improve preclinical models of human immune status and disease. Nat Immunol. (2024) 25:947–50. doi: 10.1038/s41590-024-01842-9
59. Kono M, Allende ML, and Proia RL. Sphingosine-1-phosphate regulation of mammalian development. Biochim Biophys Acta. (2008) 1781:435–41. doi: 10.1016/j.bbalip.2008.07.001
60. Oliveira CC, Van Veelen PA, Querido B, De Ru A, Sluijter M, Laban S, et al. The nonpolymorphic MHC Qa-1b mediates CD8+ T cell surveillance of antigen-processing defects. J Exp Med. (2010) 207:207–21. doi: 10.1084/jem.20091429
61. Da Silva IL, Montero-Montero L, Ferreira E, and Quintanilla M. New insights into the role of qa-2 and HLA-G non-classical MHC-I complexes in Malignancy. Front Immunol. (2018) 9:2894. doi: 10.3389/fimmu.2018.02894
62. Krop J, Heidt S, Claas FHJ, and Eikmans M. Regulatory T cells in pregnancy: it is not all about foxP3. Front Immunol. (2020) 11:1182. doi: 10.3389/fimmu.2020.01182
63. Ouyang W, Kolls JK, and Zheng Y. The biological functions of T helper 17 cell effector cytokines in inflammation. Immunity. (2008) 28:454–67. doi: 10.1016/j.immuni.2008.03.004
64. Osborne LM, Brar A, and Klein SL. The role of Th17 cells in the pathophysiology of pregnancy and perinatal mood and anxiety disorders. Brain Behav Immun. (2019) 76:7–16. doi: 10.1016/j.bbi.2018.11.015
65. Abadja F, Sarraj B, and Ansari MJ. Significance of T helper 17 immunity in transplantation. Curr Opin Organ Transplant. (2012) 17:8–14. doi: 10.1097/MOT.0b013e32834ef4e4
66. Huang DL, He YR, Liu YJ, He HY, Gu ZY, Liu YM, et al. The immunomodulation role of Th17 and Treg in renal transplantation. Front Immunol. (2023) 14:1113560. doi: 10.3389/fimmu.2023.1113560
67. Liang D, Zuo A, Shao H, Born WK, O’brien RL, Kaplan HJ, et al. IL-23 receptor expression on γδ T cells correlates with their enhancing or suppressive effects on autoreactive T cells in experimental autoimmune uveitis. J Immunol. (2013) 191:1118–25. doi: 10.4049/jimmunol.1300626
Keywords: pregnancy, T cells, maternal fetal interface, dirty mice, microbial exposure, placenta, decidua
Citation: Whillock A, Smith P, Burger S, Sridhar A, Lindgren A, Berg J, Tsuda S, Mahajan S, Tilburgs T and Schuldt NJ (2025) Natural microbial exposure populates the maternal fetal interface with diverse T cells. Front. Immunol. 16:1616491. doi: 10.3389/fimmu.2025.1616491
Received: 22 April 2025; Accepted: 25 June 2025;
Published: 09 July 2025.
Edited by:
Roberta Bulla, University of Trieste, ItalyReviewed by:
Hamid Ahmadi, University of Pécs, HungaryMichael Eikmans, Leiden University Medical Center (LUMC), Netherlands
Copyright © 2025 Whillock, Smith, Burger, Sridhar, Lindgren, Berg, Tsuda, Mahajan, Tilburgs and Schuldt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nathaniel J. Schuldt, c2NodTEwNDhAdW1uLmVkdQ==; Tamara Tilburgs, dGFtYXJhLnRpbGJ1cmdzQGNjaG1jLm9yZw==
 Amy Whillock1
Amy Whillock1 Adhvaith Sridhar
Adhvaith Sridhar Alex Lindgren
Alex Lindgren James Berg
James Berg Tamara Tilburgs
Tamara Tilburgs Nathaniel J. Schuldt
Nathaniel J. Schuldt