- Department of Acupuncture, Longhua Hospital, Shanghai University of Traditional Chinese Medicine, Shanghai, China
Alzheimer’s disease (AD) is a common neurodegenerative disease, which is characterized by β-amyloid (Aβ) deposition, Tau hyperphosphorylation, synaptic dysfunction and chronic neuroinflammation. Despite significant advances in research in recent years, effective therapeutic options remain limited. The development of single-cell RNA sequencing (scRNA-seq) has made it possible to analyze cellular heterogeneity in AD brain tissues at high resolution, breaking through the limitation of signal averaging in traditional large-scale tissue analysis. This technology has led to the discovery of novel disease-associated cell subsets, such as pro-inflammatory microglia and reactive astrocytes, and the identification of key molecular markers linked to disease progression. Integrating scRNA-seq with AI-driven analytics and multi-omics platforms further enhances our ability to decode the intricate immune-inflammatory networks underlying AD. This strategy is expected to achieve accurate classification and early diagnosis of AD subtypes, and promote the development of individualized treatment strategies based on individual molecular and immune characteristics.
1 Introduction
Alzheimer’s disease (AD) is one of the leading causes of dementia and represents a major public health challenge in the 21st century (1, 2). As a progressive neurodegenerative disease, AD usually manifests as mild memory impairment in the early stage (2), and gradually leads to severe impairment of executive function and cognitive ability as the disease progresses. The pathogenesis of AD involves A variety of complex pathological processes, including β-amyloid (Aβ) deposition, neurofibrillary tangles (NFTs) formation caused by abnormal phosphorylation of tau protein, synaptic dysfunction, blood-brain barrier (BBB) destruction, and chronic neuroinflammation. Abnormal aggregation of Aβ and tau proteins is not only A core pathological feature of AD, but also activates the immune response and induces the formation of inflammasomes, thereby driving the continuous development of neuroinflammation. Mounting evidence implicates immune dysregulation—particularly involving microglia, astrocytes, and infiltrating immune cells—as a central driver of disease progression (3). Yet, the inflammatory response in AD is highly heterogeneous, making it challenging to identify universal therapeutic targets (4, 5). Recent advances in single-cell RNA sequencing (scRNA-seq) (6–8) have enabled high-resolution profiling of cellular diversity and gene expression in AD brains. Unlike bulk RNA-seq, these technologies can resolve rare cell populations and distinguish cellular states, offering new insights into immune–inflammatory interactions (9–11) and lay the groundwork for mechanistic studies and therapeutic innovation (12–14).
2 Advances in single-cell sequencing in AD research
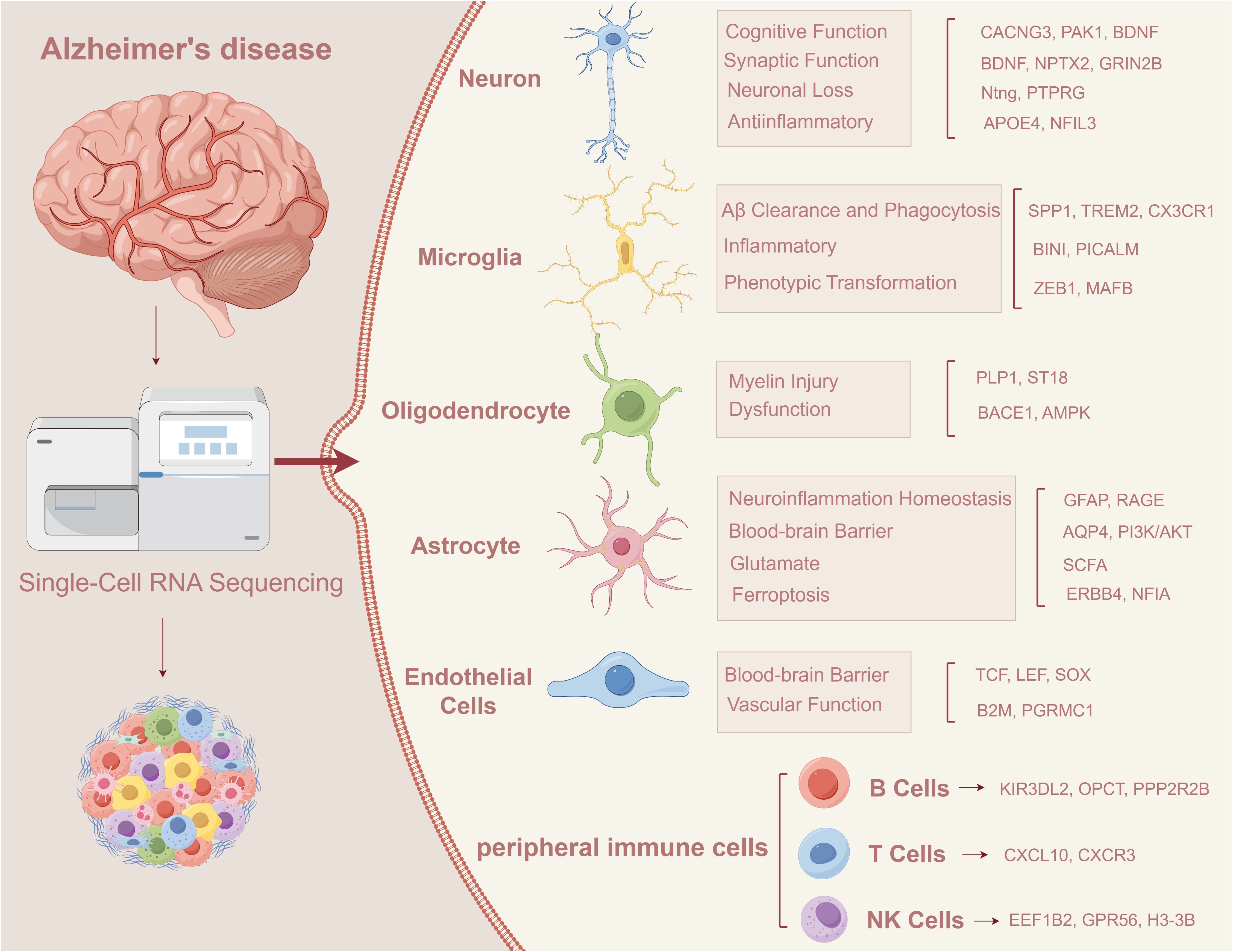
In recent years, the development of scRNA-seq (15–17) and spatial transcriptomics has greatly promoted our understanding of cellular diversity and immune dynamics in AD brain tissue (18, 19). These techniques have revealed specific transcriptional states of microglia, and region-specific subsets of astrocytes and endothelial cells associated with inflammatory signaling and neurovascular dysfunction. Moreover, multi-omics approaches integrating transcriptomics, epigenomics, proteomics, and metabolomics at the single-cell level are uncovering patient-specific molecular signatures that were previously obscured in bulk analyses (20–25). In addition, it identified cell type-specific markers associated with AD, providing new strategies for early diagnosis and targeted therapy (Figure 1).
scRNA-seq reveals key transcriptional changes in various cell types in Alzheimer’s disease, including neurons, microglia, astrocytes, oligodendrocytes, vascular endothelial cells and peripheral immune cells, which may jointly drive the occurrence and progression of AD (26). For example, neurons from AD patients show synaptic dysfunction, microglia shift to a proinflammatory state, an imbalance of A1/A2 subtypes of astrocytes (27), myelin damage to oligodendrocytes, and increased BBB permeability. Accompanied by abnormal peripheral immune cell infiltration, these changes may accelerate the process of neurodegeneration (28). Future studies should leverage AI-powered cell lineage tracing (29), spatial multi-omics, and functional genomics to identify critical regulatory nodes and develop personalized, stage-specific interventions for AD.
3 Neuroinflammation in AD: cellular and molecular landscape
3.1 scRNA-seq reveals inflammation-related cell state transitions
Neuroinflammation plays a crucial role in the progression of AD. With the progression of AD, the inflammatory response is usually aggravated, which is not only derived from the activation of immune cells, but also closely related to neuronal damage, neurodegeneration and loss of synaptic function. Neuroinflammation is triggered by interactions between microglia, astrocytes, endothelial cells, and peripheral immune cells. Neuroinflammation can further exacerbate cellular damage, thereby contributing to disease progression.
In recent years, studies based on AD mouse models (such as 5xFAD, APP/PS1) and human brain tissues have found that a variety of cell types associated with neuroinflammation have characteristic transcriptional patterns (30). Among them, disease-associated microglia (DAM) were first identified by Keren-Shaul et al. in the 5xFAD model, showing Trem2 dependence and up-regulating Apoe, Lpl, Cst7, Itgax and other genes (31). Building on this understanding of microglial function, studies PD-1 deficiency results in increased Aβ deposition, decreased microglia uptake, in APP/PS1 mice, suggesting that dysregulation of the PD-1/PD-L1 axis can exacerbate neuroinflammation and Aβ plaque formation (32). In addition, SLC11A1 was identified as an inflammatory gene associated with AD, and iron overload can induce its expression, suggesting that it may play an important role in iron metabolic-related inflammation (33). T cell infiltration is another key factor in AD neuroinflammation. scRNA-seq studies showed that CD8+ T cells activated microglia after entering AD brain tissue, triggering interferon-γ pathway and neuronal damage. CXCL10/CXCR3 axis plays an important regulatory role in this process (34). In conclusion, scRNA-seq can help to clarify the localization of inflammatory cells in brain tissue and its relationship with pathological changes.
3.2 Inflammatory signaling pathways and cytokine networks
Neuroinflammation is one of the central features in the pathogenesis of AD, which together with Aβ deposition and abnormal Tau protein drive neuronal dysfunction and neurodegeneration. Neuroinflammation, especially mediated by activated glia, neutrophils, and macrophages, also plays an important role in the pathogenesis of AD.
The occurrence of neuroinflammation is regulated by multiple signaling pathways. TREM2 signaling is critical for microglial metabolic reprogramming, plaque encapsulation, and inflammation regulation. In the CNS, TREM2 is expressed by microglia, and its expression is linked to the immunomodulatory function of APOE (35). NLRP3 inflammasome played a crucial role in AD-related pathology. NLRP3 induced interleukin-18 (IL-18) or interleukin-18 (1L-1B) to further enhance the progression of AD (36). NF-xB was a recognized inflammatory transcription factor that promotes neurodegeneration, was activated in a variety of cell types, and induced transcription of TNF-α, IL-6 and other inflammatory factors (37). The JAK/STAT signaling pathway was one of the key factors promoting neuroinflammation in AD and was affected by the excessive activation of microglia and astrocytes; it suppresses neuroinflammatory responses (38, 39). These inflammatory pathways together constructed a complex network of cytokines, which affect the function of neurons and vascular system.To better understand the role of neuroinflammation in AD, we next focus on cell-type-specific transcriptional changes revealed by scRNA-seq.
4 Cell-type-specific investigation of novel biomarkers in AD
4.1 Neuronal cells
In terms of cognitive recovery and memory improvement, scRNA-seq has identified key neuronal subtypes in AD and revealed some pathways that may contribute to memory recovery. Hansruedi Mathys et al. showed that cognitive recovery in AD patients was strongly associated with specific subtypes of inhibitory and excitatory neurons. In particular, the expression levels of genes such as CACNG3, PAK1, NPTX2, RPH3A, SVOP, and BDNF in excitatory neurons were positively correlated with overall cognitive function (1). In addition, the BDNF-regulated synapse-associated gene VGF, as well as FGF1 and FGF22 from the fibroblast growth factor (FGF) signaling pathway, were also associated with cognitive function in excitatory neurons (40).
For the regulation of synaptic function, scRNA-seq was used to investigate the role of synaptic and calcium homeostasis related genes in tau-induced neurotoxicity, which was a key factor leading to neuronal loss in AD. Analysis found that a variety of regulators were involved in synaptic function (such as Mctp, Prosap, DIP-t), neuronal excitability (such as Dpp10, GluRIA, Eaat2), and intracellular calcium regulation (such as Trpm, Calx, Cam, RyR) (1, 41).
Regarding the regulation of neuroinflammation, Antara Rao et al. identified two subtypes of proinflammatory microglia with high MHC-II gene expression and highlighted the synergistic role of neuronal APOE, especially APOE4, in AD pathogenesis through interactions with microglia (42). Transcriptome analysis in 5xFAD and trem2^R47H^ mutant models revealed transcriptional changes in microglia and astrocytes that shape microglial and astrocyte inflammatory responses, as well as neuronal activity and BDNF signaling pathway (43). In addition, the up-regulation of NF-κB signaling in inhibitory neurons and the expression of the transcription factor NFIL3 suggest that inflammatory and immune regulatory mechanisms may play a role in AD progression by modulating neuronal function (44). Multi-omics integration is gradually revealing the multi-dimensional regulatory mechanisms of different cell types in AD, and single-cell proteomics and metabolomics can provide functional evidence (45–48).
4.2 Microglial cells
In terms of Aβ clearance and phagocytosis, SPP1 was found to be an important factor in regulating microglia-mediated synaptic phagocytosis. In a mouse model of AD, knockout of SPP1 effectively prevented synaptic loss, highlighting its critical role in neurodegeneration (49). Similarly, impaired TREM2 function impaired the ability of microglia to clear Aβ plaques. In contrast, overexpression of TREM2 was able to alleviate pathological changes, showing its potential in AD treatment (50). TREM2 was not only involved in Aβ clearance, but also inhibits excessive neuroinflammation by regulating the phenotype switching of microglia. In addition, disruption of CX3CR1 signaling also reduced Aβ deposition, further underscoring the importance of the microglial receptor pathway in regulating amyloid pathology (31).
Neuroinflammation plays a key role in regulating disease progression. Single-cell analysis revealed several inflammatory response regulators, such as BINI and RELB, which were highly expressed in a variety of microglial states and were involved in the immune activation process (51). In addition, genes such as APOE, BINI and PICALM are considered as risk genes for AD, suggesting their broader regulatory roles in AD pathogenesis. The use of therapeutic antibody AL002c can effectively reduce neurofilament damage and amyloid load, while alleviating inflammatory response, providing a promising intervention (26).
During the progression of AD, the dynamic changes of microglia phenotype reflected their adaptability to pathological signals (52). scRNA-seq analysis has shown that microglia gradually change from a homeostatic state to a DAM, which was dependent on TREM2 signaling pathway, and TREM2 mutations increase the risk of AD (31). One of the DAM subgroups with neuroprotective potential was characterized by up-regulation of Trem2, Tyrobp, Lpl, and Cst7 genes, and down-regulation of P2ry12 and Cx3cr1. In addition, transcription factors such as ZEB1 and MAFB have been found to regulate AD-specific microglial and neuronal transcription patterns, suggesting that cross-type regulatory networks may exist between cells (53).
4.3 Oligodendrocytes
In the myelin damage study, Joel W. Blanchard et al. found that promoting cholesterol transport enhanced ApoE4-mediated myelination and improved cognitive function. Studies have shown that APOE4 disrupts cholesterol homeostasis in oligodendrocytes, thereby impairing the ability to generate myelin (54). In addition, activation of Erk1/2 signaling promotes oligodendrocyte (DAO) repair of damaged axonal myelin, ameliorated Aβ-related pathology and cognitive decline in A male APP^NL-G-F^ mouse model (55). Junjie Sun et al. further emphasized that specific marker genes in myelinogenic oligodendrocytes (MOL), such as Mbp, Mobp, Olig2, Mag and Mog, played key roles in myelination, axonal support and signaling (56). Notably, scRNA-seq analysis also identified novel oligodendrocyte markers PLP1 and ST18 (57).
In terms of dysfunction, BACE1 deficiency was found to upregulate the expression of ADAM10, Ano4, ApoE, Il33, and Sort1, which were closely related to Aβ production and clearance. Therefore, targeted inhibition of BACE1 in oligodendrocytes was proposed as a potential strategy to alleviate AD-related Aβ pathology (58). Shiyun Yang et al. demonstrated that overexpression of AK5 in oligodendrocytes could activate AMPK signaling pathway, thereby inhibiting neuroinflammation and apoptosis and promoting oxidative phosphorylation and overall energy metabolism (59). In contrast, elevated PIP4K2A levels may contribute to cellular dysfunction in AD (60). In addition, oligodendrocyte regulatory modules related to APOE and CLU were also identified, providing a new perspective for further understanding the role of these cells in the pathogenesis of AD (61).
4.4 Astrocytes
In the regulation of neuroinflammation, astrocytes, especially GfAP-positive subsets (such as AST1 and AST6), play an important role in neuroinflammation, normal aging and a variety of neurological diseases such as AD. These cells act as reactive astrocytes and respond to neuronal injury (62). For example, inhibition of STAT3 signaling in astrocytes can reduce amyloid plaque deposition, improve memory function, and inhibit the activation of proinflammatory factors, thereby slowing the progression of AD (63). Similarly, C5aR1 antagonists were able to suppress glial inflammatory responses, modulate cellular signaling, and prevent cognitive decline (64). In addition, β-amyloid induced the NF-κB pathway by activating RAGE, thereby promoting the inflammatory state, which provides another target for therapy. In AD, reactive astrocytes can release inflammatory factors such as IL-6 and TNF-α and participate in neuronal injury response. Targeting the STAT3 pathway has been shown to slow down neuroinflammation in AD and improve cognitive function (65). In addition, astrocytes also maintained glutamate homeostasis through interactions with neurons, which was essential for reducing neuroinflammation.
Astrocytes also play a key role in maintaining the integrity of the BBB. Studies have found that AQP4 in astrocytes promotes BBB permeability by regulating water transport and assists in the removal of brain waste, thus providing a potential target for AD treatment (66). In addition, studies have revealed that insulin signaling is closely related to BBB function in AD. The interaction between RAGE and the PI3K/AKT pathway is upregulated with age in wild-type mice and may lead to insulin resistance, thereby increasing the risk of AD (67).
Regarding glutamate homeostasis, Yan Sun and his team found that short-chain fatty acids (SCFAs) promote glutamine transport between astrocytes and neurons and alleviate AD symptoms. SCFAs could enhance the communication between astrocytes and neurons, improve glutamate-glutamine circulation, mainly acted on astrocytes to combat nerve oxidative damage, reduce Aβ deposition and Tau protein hyperphosphorylation, and reduced cognitive impairment by remodeling intestinal flora homeostasis (68).
Astrocyte heterogeneity is a key factor in neurodegeneration, where reactive astrocytes can exhibit either a neurotoxic (type A1) or a neuroprotective (type A2) phenotype. Type A1 has neurotoxic characteristics and is induced by activated microglia through TNF-α, IL-1α and C1q. It is manifested as up-regulation of C3, Serping1 and other genes, which can promote neuronal death. However, type A2 upregulates neurotrophins such as S100A10 and PTX3, which are involved in the repair process (69). Specific markers such as GFAP, vimentin, and nestin have emerged as novel therapeutic targets to promote astrocyte-mediated neuroprotection (63). In addition, the pathological down-regulation of ERBB4 and transcription factor NFIA in reactive astrocytes has been found to affect cell-cell interactions, neuronal development and synaptic regulation, thereby aggravating the condition of AD (70). Other studies have shown that NGFR can promote the differentiation of astrocytes into neurogenesis, stimulate cell proliferation and nerve regeneration (71). The continuous expression of NGFR could reduce Aβ plaque and Tau protein phosphorylation, and alleviate the pathological changes of AD. It has also been found that its downstream co-regulators such as PFKP can enhance cell proliferation and neurogenesis (71).
4.5 Endothelial cells
In terms of BBB regulation, integrated multi-omics analysis revealed the core regulatory role of TCF/LEF, SOX and ETS transcription factor families in the maintenance of BBB function, providing potential therapeutic targets for nervous system diseases such as AD (72). TCF/LEF, SOX (such as SOX17), and ETS (such as ERG, FLI1) transcription factors maintained the stability of BBB by regulating tight junctions, endothelial differentiation, and anti-inflammatory pathways. In addition, single-cell transcriptomic and immunohistochemical studies showed that IQGAP2, a key molecule that maintains the immune immunity of BBB, was significantly down-regulated in AD. IQGAP2 null mice exhibited a pronounced inflammatory phenotype of brain endothelial cells, as shown- by upregulation of adhesion receptors and antigen presentation related molecules, suggesting impaired BBB function and increased immune cell penetration (73). After the BBB was impaired, cerebrovascular endothelial cells mediated the infiltration and activation of peripheral immune cells through adhesion molecules and chemokines, and acted as antigen-presenting cells to exacerbate neuroinflammation.
In terms of the regulation of vascular function, Shun-Fat Lau et al. observed that endothelial cells in the prefrontal cortex of AD patients showed angiogenic and immune response characteristics. The up-regulation of angiogenic factors and their receptors (such as EGR, FLT1, VWE) and genes related to antigen presentation (such as B2M, HLA-E) suggests that they play an important role in regulating angiogenesis and immune responses in AD pathology (74). In addition, single-cell RNA sequencing results showed that overexpression of PGRMC1 significantly enhanced the proliferation, migration and angiogenesis of endothelial cells, indicating its potential role in regulating the function of cerebrovascular endothelial cells in AD (75).
Studies also found that EIF1 and HSPA1B were key genes closely related to the progression of AD, which were involved in the regulation of peripheral immune and inflammatory responses (76). EIF1 was found to be closely related to cognitive function. Guo et al. integrated single-cell data from ischemic stroke, hemorrhagic stroke, and AD models, and identified Lef1, Elk3, and Fosl1 as potential upstream transcription factors involved in metabolic regulation, suggesting their potential value for therapeutic intervention (77). In addition, the expression of CR1, which was necessary for the clearance of immune complexes, is significantly down-regulated in cerebrovascular endothelial cells of AD patients, suggesting that dysfunction of immune surveillance exacerbated the imbalance of immune homeostasis in the neurovascular unit (78).
4.6 Peripheral immune cells
In AD, B cells exhibit significant transcriptomic alterations. The analysis of differentially expressed genes showed that the expression of KIR3DL2, OPCT and PPP2R2B was up-regulated, while FRAT2, WWC3 and SPG20 were down-regulated, and these genes were closely related to the neurodegenerative process (19). scRNA-seq further identified a novel B-cell phenotype with high expression of CD45, enhanced phagocytosis and chemotaxis, and the release of multiple chemokines to recruit peripheral immune cells through the CCL signaling pathway. This phenotypic change may be driven by up-regulation of myeloid-associated transcription factors such as the CEBP family and down-regulation of lymphoid transcription factors such as Pax5 (79).
On the T cell side, the CXCL10-CXCR3 axis played a key role in mediating T cell infiltration and neuronal injury. In particular, the infiltration of CD8+ T cells promoted the activation of microglia and further aggravates neuroinflammation and neurodegeneration (34). In addition, cis-regulatory elements co-accessible with the CXCR3 promoter in peripheral CD8+ T cells identified, suggesting an epigenetic mechanism associated with AD susceptibility (80). The infiltration of T cells, especially CD8+ T cells, activated microglia and further worsens neuroinflammation and neurodegeneration.
Natural killer (NK) cells also play an important role in the pathogenesis of AD. One study identified 17 marker genes associated with AD (such as EEF1B2, GPR56, H3-3B, ZEB2), which may affect immune cell infiltration (81). Through cell communication analysis, researcher identified NK cell subset modules related to AD, and the characteristic genes included RPLP2, RPSA and RPL18A. One specific subgroup was characterized by upregulation of CX3CR1, TBX21, MYOM2, DUSP1, and ZFP36L2, and was negatively correlated with cognitive function in AD patients (82). Moreover, NK cells interacted with other immune cells, such as dendritic cells and macrop-hages, to shape the immune landscape. Activated NK cells induced dendritic cell maturation or apoptosis, thereby indirectly regulating T cell priming (83). In addition, peripheral NK cells infiltrated the brain tissue and regulate the transcription of immune response genes by activating STAT3 signaling pathway, further amplifying the neuroinflammatory response (84).
5 Conclusion
Using scRNA-seq, researchers have uncovered transcriptional changes in neurons, glial, and immune cells in AD, elucidating mechanisms of neuroinflammation, synaptic damage, and neurodegeneration. These insights lay a foundation for precision diagnosis and targeted therapy. Integrating AI with single-cell multi-omics and spatial transcriptomics may enable early biomarker discovery and AD subtype identification, facilitating personalized treatment.
Author contributions
SL: Conceptualization, Writing – original draft, Visualization, Formal analysis. YZ: Formal analysis, Conceptualization, Visualization, Writing – original draft. RW: Formal analysis, Writing – original draft, Visualization, Conceptualization. JP: Writing – original draft, Funding acquisition, Validation, Writing – review & editing, Project administration.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. Key Scientific Research Program of Shanghai Municipal Science and Technology Committee in China (No.22Y11922900, 18401970500); TCM Genre Program of Shanghai Health Bureau [No. ZY(2021-2023)-0209-10, ZY (2018-2020)-CCCX-1006]; Clinical Research Plan of SHDC (No. SHDC2020CR3091B).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Mathys H, Peng Z, Boix CA, Victor MB, Leary N, Babu S, et al. Single-cell atlas reveals correlates of high cognitive function, dementia, and resilience to Alzheimer’s disease pathology. Cell. (2023) 186:4365–85. doi: 10.1016/j.cell.2023.08.039
2. Scheltens P, Blennow K, Breteler MM, de Strooper B, Frisoni GB, Salloway S, et al. Alzheimer’s disease. Lancet. (2016) 388:505–17. doi: 10.1016/S0140-6736(15)01124-1
3. Ju Y and Tam KY. Pathological mechanisms and therapeutic strategies for Alzheimer’s disease. Neural Regener Res. (2022) 17:543–9. doi: 10.4103/1673-5374.320970
5. Liu S, Fan M, Zheng Q, Hao S, Yang L, Xia Q, et al. MicroRNAs in Alzheimer’s disease: Potential diagnostic markers and therapeutic targets. BioMed Pharmacother. (2022) 148:112681. doi: 10.1016/j.biopha.2022.112681
6. Shi J, Liu J, Tu X, Li B, Tong Z, Wang T, et al. Single-cell immune signature for detecting early-stage HCC and early assessing anti-PD-1 immunotherapy efficacy. J Immunother Cancer. (2022) 10:1. doi: 10.1136/jitc-2021-003133
7. Du H, Li S, Lu J, Tang L, Jiang X, He X, et al. Single-cell RNA-seq and bulk-seq identify RAB17 as a potential regulator of angiogenesis by human dermal microvascular endothelial cells in diabetic foot ulcers. Burns Trauma. (2023) 11:tkad020. doi: 10.1093/burnst/tkad020
8. Fehlings M, Kim L, Guan X, Yuen K, Tafazzol A, Sanjabi S, et al. Single-cell analysis reveals clonally expanded tumor-associated CD57(+) CD8 T cells are enriched in the periphery of patients with metastatic urothelial cancer responding to PD-L1 blockade. J Immunother Cancer. (2022) 10:e004759. doi: 10.1136/jitc-2022-004759
9. Lin Z, Wang F, Yin R, Li S, Bai Y, Zhang B, et al. Single-cell RNA sequencing and immune microenvironment analysis reveal PLOD2-driven Malignant transformation in cervical cancer. Front Immunol. (2024) 15:1522655. doi: 10.3389/fimmu.2024.1522655
10. Lin Z, Zou J, Sui X, Yao S, Lin L, Wang J, et al. Necroptosis-related lncRNA signature predicts prognosis and immune response for cervical squamous cell carcinoma and endocervical adenocarcinomas. Sci Rep. (2022) 12:16285. doi: 10.1038/s41598-022-20858-5
11. Lin Z, Sui X, Jiao W, Wang Y, and Zhao J. Exploring the mechanism and experimental verification of puerarin in the treatment of endometrial carcinoma based on network pharmacology and bioinformatics analysis. BMC Complement Med Ther. (2022) 22:150. doi: 10.1186/s12906-022-03623-z
12. Huang W, Kim BS, Zhang Y, Lin L, Chai G, and Zhao Z. Regulatory T cells subgroups in the tumor microenvironment cannot be overlooked: Their involvement in prognosis and treatment strategy in melanoma. Environ Toxicol. (2024) 39:4512–30. doi: 10.1002/tox.24247
13. Lin L, Zou J, Pei S, Huang W, Zhang Y, Zhao Z, et al. Germinal center B-cell subgroups in the tumor microenvironment cannot be overlooked: Their involvement in prognosis, immunotherapy response, and treatment resistance in head and neck squamous carcinoma. Heliyon. (2024) 10:e37726. doi: 10.1016/j.heliyon.2024.e37726
14. Zhao Z, Ding Y, Tran LJ, Chai G, and Lin L. Innovative breakthroughs facilitated by single-cell multi-omics: manipulating natural killer cell functionality correlates with a novel subcategory of melanoma cells. Front Immunol. (2023) 14:1196892. doi: 10.3389/fimmu.2023.1196892
15. Zhang Y, Zhao Z, Huang W, Kim BS, Lin L, Li X, et al. Pan-cancer single-cell analysis revealing the heterogeneity of cancer-associated fibroblasts in skin tumors. Curr Gene Ther. (2024). doi: 10.2174/0115665232331353240911080642
16. Hou M, Zhao Z, Li S, Zhang Z, Li X, Zhang Y, et al. Single-cell analysis unveils cell subtypes of acral melanoma cells at the early and late differentiation stages. J Cancer. (2025) 16:898–916. doi: 10.7150/jca.102045
17. Li H, Bian Y, Xiahou Z, Zhao Z, Zhao F, and Zhang Q. The cellular signaling crosstalk between memory B cells and tumor cells in nasopharyngeal carcinoma cannot be overlooked: Their involvement in tumor progression and treatment strategy is significant. J Cancer. (2025) 16:288–314. doi: 10.7150/jca.101420
18. Julian A, Dugast E, Ragot S, Krolak-Salmon P, Berrut G, Dantoine T, et al. There is no correlation between peripheral inflammation and cognitive status at diagnosis in Alzheimer’s disease. Aging Clin Exp Res. (2015) 27:589–94. doi: 10.1007/s40520-015-0332-5
19. Xiong LL, Xue LL, Du RL, Niu RZ, Chen L, Chen J, et al. Single-cell RNA sequencing reveals B cell-related molecular biomarkers for Alzheimer’s disease. Exp Mol Med. (2021) 53:1888–901. doi: 10.1038/s12276-021-00714-8
20. Wang M, Song WM, Ming C, Wang Q, Zhou X, Xu P, et al. Guidelines for bioinformatics of single-cell sequencing data analysis in Alzheimer’s disease: review, recommendation, implementation and application. Mol Neurodegener. (2022) 17:17. doi: 10.1186/s13024-022-00517-z
21. Lin Z, Sui X, Jiao W, Chen C, Zhang X, and Zhao J. Mechanism investigation and experiment validation of capsaicin on uterine corpus endometrial carcinoma. Front Pharmacol. (2022) 13:953874. doi: 10.3389/fphar.2022.953874
22. Lin Z, Fan W, Yu X, Liu J, and Liu P. Research into the mechanism of intervention of SanQi in endometriosis based on network pharmacology and molecular docking technology. Med (Baltimore). (2022) 101:e30021. doi: 10.1097/MD.0000000000030021
23. Lin W, Wang Q, Chen Y, Wang N, Ni Q, Qi C, et al. Identification of a 6-RBP gene signature for a comprehensive analysis of glioma and ischemic stroke: Cognitive impairment and aging-related hypoxic stress. Front Aging Neurosci. (2022) 14:951197. doi: 10.3389/fnagi.2022.951197
24. Sun C, Ma S, Chen Y, Kim NH, Kailas S, Wang Y, et al. Diagnostic value, prognostic value, and immune infiltration of LOX family members in liver cancer: Bioinformatic analysis. Front Oncol. (2022) 12:843880. doi: 10.3389/fonc.2022.843880
25. Sun C, Chen Y, Kim NH, Lowe S, Ma S, Zhou Z, et al. Identification and verification of potential biomarkers in gastric cancer by integrated bioinformatic analysis. Front Genet. (2022) 13:911740. doi: 10.3389/fgene.2022.911740
26. Wang S, Mustafa M, Yuede CM, Salazar SV, Kong P, Long H, et al. Anti-human TREM2 induces microglia proliferation and reduces pathology in an Alzheimer’s disease model. J Exp Med. (2020) 217:e20200785. doi: 10.1084/jem.20200785
27. Xu X, Wang J, Chen T, Wang S, Wang F, He J, et al. Deciphering novel mitochondrial signatures: multi-omics analysis uncovers cross-disease markers and oligodendrocyte pathways in Alzheimer’s disease and glioblastoma. Front Aging Neurosci. (2025) 17:1536142. doi: 10.3389/fnagi.2025.1536142
28. Fujikawa R and Tsuda M. The functions and phenotypes of microglia in Alzheimer’s disease. Cells. (2023) 12:1207. doi: 10.3390/cells12081207
29. Dong J, Qian J, Yu K, Huang S, Cheng X, Chen F, et al. Rational design of organelle-targeted fluorescent probes: insights from artificial intelligence. Res (Wash D C). (2023) 6:75. doi: 10.34133/research.0075
30. Gardner RS, Kyle M, Hughes K, and Zhao LR. Single-cell RNA sequencing reveals immunomodulatory effects of stem cell factor and granulocyte colony-stimulating factor treatment in the brains of aged APP/PS1 mice. Biomolecules. (2024) 14:827. doi: 10.3390/biom14070827
31. Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell. (2017) 169:1276–90. doi: 10.1016/j.cell.2017.05.018
32. Kummer MP, Ising C, Kummer C, Sarlus H, Griep A, Vieira-Saecker A, et al. Microglial PD-1 stimulation by astrocytic PD-L1 suppresses neuroinflammation and Alzheimer’s disease pathology. EMBO J. (2021) 40:e108662. doi: 10.15252/embj.2021108662
33. Zhou H, Peng Y, Huo X, Li B, Liu H, Wang J, et al. Integrating bulk and single-cell transcriptomic data to identify ferroptosis-associated inflammatory gene in Alzheimer’s disease. J Inflammation Res. (2025) 18:2105–22. doi: 10.2147/JIR.S497418
34. Jorfi M, Park J, Hall CK, Lin CJ, Chen M, von Maydell D, et al. Infiltrating CD8(+) T cells exacerbate Alzheimer’s disease pathology in a 3D human neuroimmune axis model. Nat Neurosci. (2023) 26:1489–504. doi: 10.1038/s41593-023-01415-3
35. Shi Y and Holtzman DM. Interplay between innate immunity and Alzheimer disease: APOE and TREM2 in the spotlight. Nat Rev Immunol. (2018) 18:759–72. doi: 10.1038/s41577-018-0051-1
36. Barczuk J, Siwecka N, Lusa W, Rozpedek-Kaminska W, Kucharska E, and Majsterek I. Targeting NLRP3-mediated neuroinflammation in Alzheimer’s disease treatment. Int J Mol Sci. (2022) 23:8979. doi: 10.3390/ijms23168979
37. Sun E, Motolani A, Campos L, and Lu T. The pivotal role of NF-kB in the pathogenesis and therapeutics of Alzheimer’s disease. Int J Mol Sci. (2022) 23:8972. doi: 10.3390/ijms23168972
38. Rusek M, Smith J, El-Khatib K, Aikins K, Czuczwar SJ, and Pluta R. The role of the JAK/STAT signaling pathway in the pathogenesis of Alzheimer’s disease: new potential treatment target. Int J Mol Sci. (2023) 24:864. doi: 10.3390/ijms24010864
39. Chen Y, Peng F, Xing Z, Chen J, Peng C, and Li D. Beneficial effects of natural flavonoids on neuroinflammation. Front Immunol. (2022) 13:1006434. doi: 10.3389/fimmu.2022.1006434
40. Alder J, Thakker-Varia S, Bangasser DA, Kuroiwa M, Plummer MR, Shors TJ, et al. Brain-derived neurotrophic factor-induced gene expression reveals novel actions of VGF in hippocampal synaptic plasticity. J Neurosci. (2003) 23:10800–8. doi: 10.1523/JNEUROSCI.23-34-10800.2003
41. Praschberger R, Kuenen S, Schoovaerts N, Kaempf N, Singh J, Janssens J, et al. Neuronal identity defines alpha-synuclein and tau toxicity. Neuron. (2023) 111:1577–90. doi: 10.1016/j.neuron.2023.02.033
42. Rao A, Chen N, Kim MJ, Blumenfeld J, Yip O, Liang Z, et al. Microglia depletion reduces human neuronal APOE4-related pathologies in a chimeric Alzheimer’s disease model. Cell Stem Cell. (2025) 32:86–104. doi: 10.1016/j.stem.2024.10.005
43. Johnston KG, Berackey BT, Tran KM, Gelber A, Yu Z, MacGregor GR, et al. Single-cell spatial transcriptomics reveals distinct patterns of dysregulation in non-neuronal and neuronal cells induced by the Trem2(R47H) Alzheimer’s risk gene mutation. Mol Psychiatry. (2025) 30:461–77. doi: 10.1038/s41380-024-02651-0
44. Soelter TM, Howton TC, Clark AD, Oza VH, and Lasseigne BN. Altered glia-neuron communication in Alzheimer’s Disease affects WNT, p53, and NFkB Signaling determined by snRNA-seq. Cell Commun Signal. (2024) 22:317. doi: 10.1186/s12964-024-01686-8
45. Zhao Z, Dong Y, Zhao Z, Xiahou Z, and Sun C. Single-cell atlas of endothelial cells in atherosclerosis: identifying C1 CXCL12+ ECs as key proliferative drivers for immunological precision therapeutics in atherosclerosis. Front Immunol. (2025) 16.
46. Zhao Z, Zhao Z, Lin Z, Fan L, Xiahou Z, Dong Y, et al. Decoding multiple myeloma: single-cell insights into tumor heterogeneity, immune dynamics, and disease progression. Front Immunol. (2025) 16.
47. Feng X, Luo Z, Zhang W, Wan R, Chen Y, Li F, et al. Zn-DHM nanozymes enhance muscle regeneration through ROS scavenging and macrophage polarization in volumetric muscle loss revealed by single-cell profiling. Adv Funct Mater. (2025), 2506476. doi: 10.1002/adfm.202506476
48. He Y, Luo Z, Nie X, Du Y, Sun R, Sun J, et al. An injectable multi-functional composite bioactive hydrogel for bone regeneration via immunoregulatory and osteogenesis effects. Adv Compos Hybrid Mater. (2025) 8:128. doi: 10.1007/s42114-025-01213-4
49. De Schepper S, Ge JZ, Crowley G, Ferreira L, Garceau D, Toomey CE, et al. Perivascular cells induce microglial phagocytic states and synaptic engulfment via SPP1 in mouse models of Alzheimer’s disease. Nat Neurosci. (2023) 26:406–15. doi: 10.1038/s41593-023-01257-z
50. Tapia-Arancibia L, Aliaga E, Silhol M, and Arancibia S. New insights into brain BDNF function in normal aging and Alzheimer disease. Brain Res Rev. (2008) 59:201–20. doi: 10.1016/j.brainresrev.2008.07.007
51. Sun N, Victor MB, Park YP, Xiong X, Scannail AN, Leary N, et al. Human microglial state dynamics in Alzheimer’s disease progression. Cell. (2023) 186:4386–403. doi: 10.1016/j.cell.2023.08.037
52. Zhou Y, Song WM, Andhey PS, Swain A, Levy T, Miller KR, et al. Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nat Med. (2020) 26:131–42. doi: 10.1038/s41591-019-0695-9
53. Anderson AG, Rogers BB, Loupe JM, Rodriguez-Nunez I, Roberts SC, White LM, et al. Single nucleus multiomics identifies ZEB1 and MAFB as candidate regulators of Alzheimer’s disease-specific cis-regulatory elements. Cell Genom. (2023) 3:100263. doi: 10.1016/j.xgen.2023.100263
54. Blanchard JW, Akay LA, Davila-Velderrain J, von Maydell D, Mathys H, Davidson SM, et al. APOE4 impairs myelination via cholesterol dysregulation in oligodendrocytes. Nature. (2022) 611:769–79. doi: 10.1038/s41586-022-05439-w
55. Park H, Cho B, Kim H, Saito T, Saido TC, Won KJ, et al. Single-cell RNA-sequencing identifies disease-associated oligodendrocytes in male APP NL-G-F and 5XFAD mice. Nat Commun. (2023) 14:802. doi: 10.1038/s41467-023-36519-8
56. Sun J, Song Y, Chen Z, Qiu J, Zhu S, Wu L, et al. Heterogeneity and molecular markers for CNS glial cells revealed by single-cell transcriptomics. Cell Mol Neurobiol. (2022) 42:2629–42. doi: 10.1007/s10571-021-01159-3
57. Soreq L, Bird H, Mohamed W, and Hardy J. Single-cell RNA sequencing analysis of human Alzheimer’s disease brain samples reveals neuronal and glial specific cells differential expression. PloS One. (2023) 18:e0277630. doi: 10.1371/journal.pone.0277630
58. Ishii A, Pathoulas JA, MoustafaFathy OO, Ge Y, Yao AY, Pantalena T, et al. Contribution of amyloid deposition from oligodendrocytes in a mouse model of Alzheimer’s disease. Mol Neurodegener. (2024) 19:83. doi: 10.1186/s13024-024-00759-z
59. Yang S, Chen B, Zhang J, Zhou X, Jiang Y, Tong W, et al. Single-cell sequencing reveals that AK5 inhibits apoptosis in AD oligodendrocytes by regulating the AMPK signaling pathway. Mol Biol Rep. (2025) 52:213. doi: 10.1007/s11033-025-10311-x
60. Nguyen D, Jallow AW, Lin YF, and Lin YF. Exploring the potential role of oligodendrocyte-associated PIP4K2A in Alzheimer’s disease complicated with type 2 diabetes mellitus via multi-omic analysis. Int J Mol Sci. (2024) 25:6640. doi: 10.3390/ijms25126640
61. Morabito S, Miyoshi E, Michael N, Shahin S, Martini AC, Head E, et al. Single-nucleus chromatin accessibility and transcriptomic characterization of Alzheimer’s disease. Nat Genet. (2021) 53:1143–55. doi: 10.1038/s41588-021-00894-z
62. Su Y, Zhou Y, Bennett ML, Li S, Carceles-Cordon M, Lu L, et al. A single-cell transcriptome atlas of glial diversity in the human hippocampus across the postnatal lifespan. Cell Stem Cell. (2022) 29:1594–610. doi: 10.1016/j.stem.2022.09.010
63. Monterey MD, Wei H, Wu X, and Wu JQ. The many faces of astrocytes in Alzheimer’s disease. Front Neurol. (2021) 12:619626. doi: 10.3389/fneur.2021.619626
64. Schartz ND, Liang HY, Carvalho K, Chu SH, Mendoza-Arvilla A, Petrisko TJ, et al. C5aR1 antagonism suppresses inflammatory glial responses and alters cellular signaling in an Alzheimer’s disease mouse model. Nat Commun. (2024) 15:7028. doi: 10.1038/s41467-024-51163-6
65. Zhu H, Jian Z, Zhong Y, Ye Y, Zhang Y, Hu X, et al. Janus kinase inhibition ameliorates ischemic stroke injury and neuroinflammation through reducing NLRP3 inflammasome activation via JAK2/STAT3 pathway inhibition. Front Immunol. (2021) 12:714943. doi: 10.3389/fimmu.2021.714943
66. Steenberghen H, Beuckeleer S, Hellings N, Somers V, Breedam EV, Ponsaerts P, et al. Single-cell analysis of osmoregulation reveals heterogeneity of aquaporin 4 functionality in human astrocytes. Cytom A. (2024) 105:870–82. doi: 10.1002/cyto.a.24905
67. Zhou AL, Sharda N, Sarma VV, Ahlschwede KM, Curran GL, Tang X, et al. Age-dependent changes in the plasma and brain pharmacokinetics of amyloid-beta peptides and insulin. J Alzheimers Dis. (2022) 85:1031–44. doi: 10.3233/JAD-215128
68. Sun Y, Zhang H, Zhang X, Wang W, Chen Y, Cai Z, et al. Promotion of astrocyte-neuron glutamate-glutamine shuttle by SCFA contributes to the alleviation of Alzheimer’s disease. Redox Biol. (2023) 62:102690. doi: 10.1016/j.redox.2023.102690
69. Liddelow SA, Guttenplan KA, Clarke LE, Bennett FC, Bohlen CJ, Schirmer L, et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. (2017) 541:481–7. doi: 10.1038/nature21029
70. Dai DL, Li M, and Lee EB. Human Alzheimer’s disease reactive astrocytes exhibit a loss of homeostastic gene expression. Acta Neuropathol Commun. (2023) 11:127. doi: 10.1186/s40478-023-01624-8
71. Siddiqui T, Cosacak MI, Popova S, Bhattarai P, Yilmaz E, Lee AJ, et al. Nerve growth factor receptor (Ngfr) induces neurogenic plasticity by suppressing reactive astroglial Lcn2/Slc22a17 signaling in Alzheimer’s disease. NPJ Regener Med. (2023) 8:33. doi: 10.1038/s41536-023-00311-5
72. Yu M, Nie Y, Yang J, Yang S, Li R, Rao V, et al. Integrative multi-omic profiling of adult mouse brain endothelial cells and potential implications in Alzheimer’s disease. Cell Rep. (2023) 42:113392. doi: 10.1016/j.celrep.2023.113392
73. Katdare KA, Kjar A, O’Brown NM, Neal EH, Sorets AG, Shostak A, et al. IQGAP2 regulates blood-brain barrier immune dynamics. Iscience. (2025) 28:111994. doi: 10.1016/j.isci.2025.111994
74. Lau SF, Cao H, Fu A, and Ip NY. Single-nucleus transcriptome analysis reveals dysregulation of angiogenic endothelial cells and neuroprotective glia in Alzheimer’s disease. Proc Natl Acad Sci U S A. (2020) 117:25800–9. doi: 10.1073/pnas.2008762117
75. Xu X, Ruan X, Ju R, Wang Z, Yang Y, Cheng J, et al. Progesterone receptor membrane component-1 may promote survival of human brain microvascular endothelial cells in Alzheimer’s disease. Am J Alzheimers Dis Other Demen. (2022) 37:240468981. doi: 10.1177/15333175221109749
76. Gu Y, Chen N, Qi J, Zhu L, Chen X, Wang F, et al. The endothelial cell-related genes EIF1 and HSPA1B contribute to the pathogenesis of Alzheimer’s disease by modulating peripheral immunoinflammatory responses. Brain Sci. (2025) 15:205. doi: 10.3390/brainsci15020205
77. Guo G, Fan L, Yan Y, Xu Y, Deng Z, Tian M, et al. Shared metabolic shifts in endothelial cells in stroke and Alzheimer’s disease revealed by integrated analysis. Sci Data. (2023) 10:666. doi: 10.1038/s41597-023-02512-5
78. Crouch EE, Bhaduri A, Andrews MG, Cebrian-Silla A, Diafos LN, Birrueta JO, et al. Ensembles of endothelial and mural cells promote angiogenesis in prenatal human brain. Cell. (2022) 185:3753–69. doi: 10.1016/j.cell.2022.09.004
79. Wang R, Li H, Ling C, Zhang X, Lu J, Luan W, et al. A novel phenotype of B cells associated with enhanced phagocytic capability and chemotactic function after ischemic stroke. Neural Regener Res. (2023) 18:2413–23. doi: 10.4103/1673-5374.371365
80. Ramakrishnan A, Piehl N, Simonton B, Parikh M, Zhang Z, Teregulova V, et al. Epigenetic dysregulation in Alzheimer’s disease peripheral immunity. Neuron. (2024) 112:1235–48. doi: 10.1016/j.neuron.2024.01.013
81. Chen X, Hu F, Chi Q, and Rao C. The study of Alzheimer’s disease risk diagnosis based on natural killer cell marker genes in the multi-omics data. J Alzheimers Dis. (2024), 268770860. doi: 10.1177/13872877241295316
82. Qi C, Liu F, Zhang W, Han Y, Zhang N, Liu Q, et al. Alzheimer’s disease alters the transcriptomic profile of natural killer cells at single-cell resolution. Front Immunol. (2022) 13:1004885. doi: 10.3389/fimmu.2022.1004885
83. Sirkis DW, Warly SC, Johnson TP, Bonham LW, Oddi AP, Geier EG, et al. Expansion of highly interferon-responsive T cells in early-onset Alzheimer’s disease. Alzheimers Dement. (2024) 20:5062–70. doi: 10.1002/alz.13892
Keywords: single-cell RNA sequencing, Alzheimer’s disease (AD), heterogeneity, neuroinflammation, prognostic biomarkers
Citation: Lin S, Zhan Y, Wang R and Pei J (2025) Decoding neuroinflammation in Alzheimer’s disease: a multi-omics and AI-driven perspective for precision medicine. Front. Immunol. 16:1616899. doi: 10.3389/fimmu.2025.1616899
Received: 23 April 2025; Accepted: 02 June 2025;
Published: 18 June 2025.
Edited by:
Yongfu Shao, Ningbo University, ChinaReviewed by:
Yisheng Chen, Ningde Normal University, ChinaXiaojing Wang, Shanghai Jiao Tong University, China
Copyright © 2025 Lin, Zhan, Wang and Pei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian Pei, bG9uZ2h1YWFjdXBAYWxpeXVuLmNvbQ==
 Shiyu Lin
Shiyu Lin Yijun Zhan
Yijun Zhan Ruiqi Wang
Ruiqi Wang Jian Pei
Jian Pei