- 1Department of Laboratory Medicine, Tianjin Academy of Traditional Chinese Medicine Affiliated Hospital, Tianjin, China
- 2Department of Gastroenterology, No. 983 Hospital of People's Liberation Army Joint Logistics Support Force, Tianjin, China
Objective: This study explores the prognostic value of the CLEC4E gene in systemic lupus erythematosus (SLE) through bioinformatics analysis and evaluates its role in disease diagnosis and progression.
Methods: Gene expression datasets related to SLE (GSE17755, GSE50772, and GSE61635) were obtained from the GEO (Gene Expression Omnibus) database. Intersection analysis was performed using the Jvenn tool with a screening threshold of |log2FC| > 1 and P< 0.05 to identify differentially expressed genes (DEGs). The resulting DEGs were then cross-referenced with immune-related genes in the GeneCards database (relevance score > 8) to further prioritize candidates with immunological relevance. Peripheral blood from 360 SLE patients and 360 healthy controls was collected for CLEC4E expression analysis via RT-qPCR. Disease activity was evaluated using the SLEDAI score, and patients were grouped accordingly. Pearson and Spearman correlation analysis to investigate the relationship between CLEC4E and immune indicators. Logistic regression and ROC analyses were conducted to assess diagnostic and prognostic value. Kaplan-Meier analysis evaluated survival outcomes.
Results: Bioinformatics analysis identified six SLE-related DEGs, namely ISG15, HERC5, TNFAIP6, IFIT3, OASL, and CLEC4E. Further intersection with immune-related genes from the GeneCards database (relevance score > 8) ultimately highlighted CLEC4E as the key gene for clinical validation. The expression level of CLEC4E was significantly higher in SLE patients compared with healthy controls. ROC analysis showed good diagnostic performance (AUC = 0.7744). CLEC4E expression was higher in active SLE, and multivariate analysis identified CLEC4E, C3, C4, ANA, and anti-dsDNA as independent predictors of disease activity. CLEC4E demonstrated moderate diagnostic value for distinguishing active from inactive disease (AUC = 0.6360). Higher CLEC4E expression was associated with worse prognosis (P = 0.0002). The combined diagnostic performance with other biomarkers (C3, C4, ANA, anti-dsDNA) showed a remarkable AUC of 0.9407.
Conclusion: CLEC4E is a potential biomarker for SLE diagnosis, disease activity assessment, and prognosis evaluation.
1 Introduction
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease characterized by multisystem involvement and diverse clinical manifestations. It exhibits a relatively high global incidence, particularly among female patients (1). The pathogenesis of SLE is complex, involving interactions among genetic, environmental, hormonal, and immune factors (2). Despite significant progress in understanding the mechanisms of SLE in recent years, particularly through genome-wide association studies (GWAS) that have identified multiple susceptibility genes such as IRF5, STAT4, and TNFSF4, the disease’s complexity and heterogeneity continue to pose significant challenges for early diagnosis, effective treatment, and prognosis management (3, 4). Therefore, there is an urgent need to identify novel pathogenic genes and reliable molecular biomarkers to improve our understanding of disease mechanisms and clinical outcomes.
C-type lectin domain family 4 member E (CLEC4E), also known as MINCLE, is a pattern recognition receptor (PRR) that plays a pivotal role in host immune responses. It regulates inflammatory reactions by recognizing pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs) (5, 6). CLEC4E is primarily expressed in macrophages and dendritic cells. In these cells, it mediates the release of inflammatory cytokines by interacting with downstream signaling pathways, such as the Syk-CARD9 pathway. This activation contributes to infection control, tissue damage, and the pathogenesis of autoimmune diseases (7, 8). Although recent studies have highlighted CLEC4E’s involvement in inflammation regulation, its specific role in SLE remains largely unexplored, particularly in terms of its contributions to immune dysregulation and disease progression. In addition to its role in SLE, CLEC4E has been implicated in other autoimmune diseases, such as rheumatoid arthritis, Sjögren’s syndrome, and multiple sclerosis (9–11). Studies have shown that CLEC4E influences disease mechanisms in these conditions by modulating immune responses and promoting inflammation (12). However, its precise role in SLE and its connection to disease advancement and immune dysregulation remain largely uninvestigated.
Abnormalities in the innate immune system are considered crucial in SLE pathogenesis. In particular, dysregulated PRR-mediated signaling pathways, which may drive autoimmune activation and tissue damage (13, 14). As an emerging PRR, CLEC4E’s potential as a functional target warrants further investigation.
In summary, this study aims to assess CLEC4E expression patterns in SLE and their associations with clinical indicators through bioinformatics analysis and clinical datasets. The study hypothesizes that abnormal CLEC4E expression is associated with the pathogenesis of SLE, particularly immune dysregulation and tissue damage. Furthermore, through multivariate analyses and ROC curve evaluations, the diagnostic and prognostic value of CLEC4E as a potential biomarker for SLE will be validated, providing a reference for early diagnosis and targeted therapy for SLE.
2 Materials and methods
2.1 Bioinformatics analysis
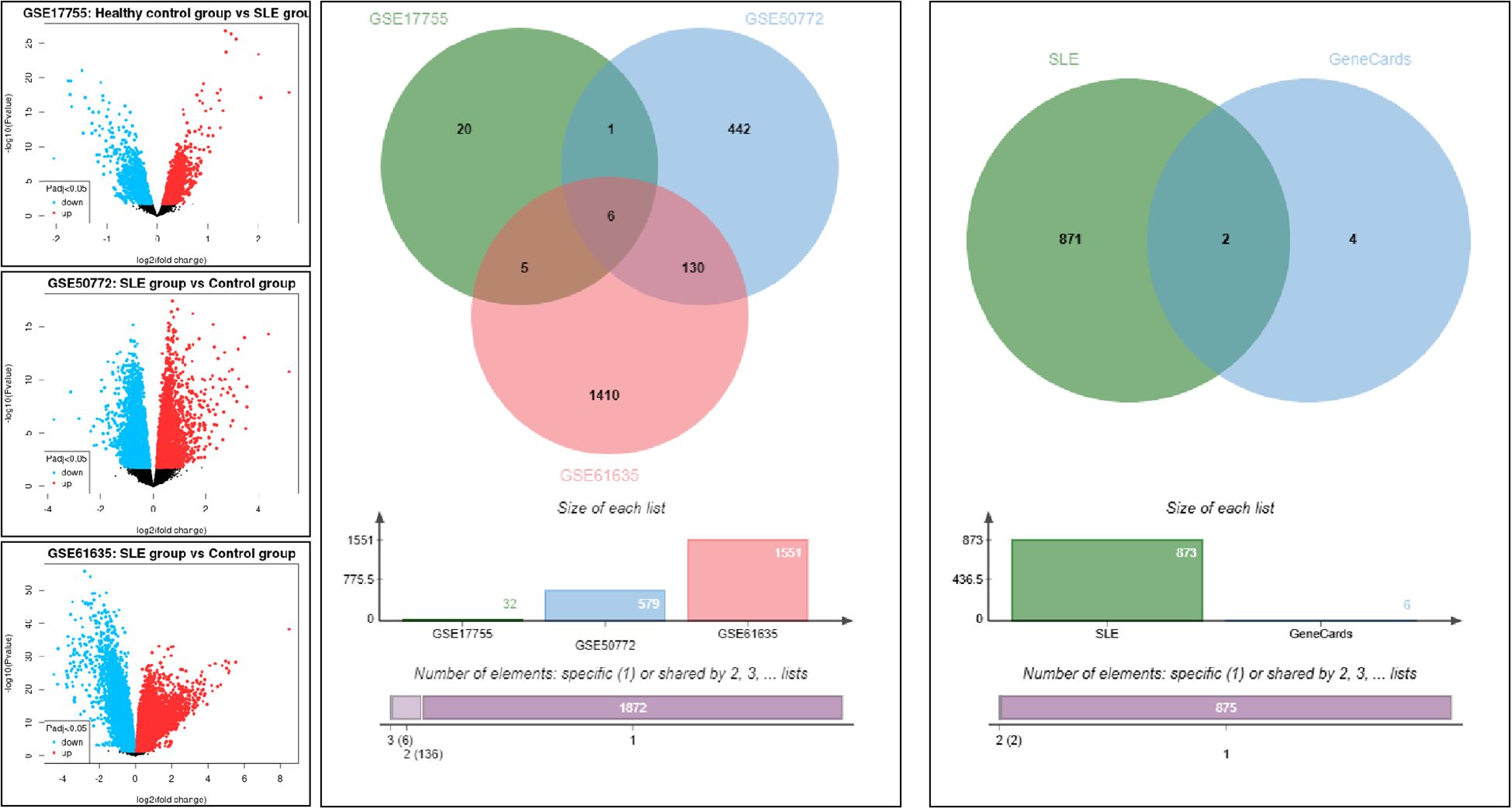
Gene expression data related to systemic lupus erythematosus (SLE) were retrieved from the Gene Expression Omnibus (GEO) database (GSE17755, GSE50772, GSE61635). Jvenn (http://jvenn.toulouse.inra.fr/) was used for intersection analysis of these three SLE-related datasets. The screening criteria for differentially expressed genes (DEGs) were set as |log2FC| > 1 and P< 0.05. DEGs obtained from each dataset were intersected to identify robust SLE-associated genes. Subsequently, the identified DEGs were cross-referenced with immune-related core genes retrieved from the GeneCards database using the keyword “Immunity” (relevance score > 8), in order to further prioritize candidate genes with immunological significance for downstream validation.
2.2 General data
A cohort of 360 SLE patients treated at our hospital between March 2023 and January 2025 was included in the study. The group consisted of 312 women and 48 men, with a mean age of 38.23 ± 5.60 years. The inclusion criteria were as follows: (1) meeting the 2019 classification criteria for SLE established jointly by the European League Against Rheumatism (EULAR) and the American College of Rheumatology (ACR) (15); (2) age >18 years; and (3) complete clinical data. The exclusion criteria were as follows: (1) coexisting other types of autoimmune diseases (e.g., rheumatoid arthritis); (2) coexisting infectious diseases; and (3) malignancies or pregnancy.
A control group was established consisting of 360 healthy individuals undergoing routine physical examinations during the same period. This group included 298 females and 62 males, with an average age of 38.42 ± 5.59 years. The healthy participants met none of the diagnostic criteria for SLE, had no history of major diseases, and had no personal or family history of autoimmune diseases.
2.3 Assessment of disease activity
The severity of disease was assessed using the Systemic Lupus Erythematosus Disease Activity Index (SLEDAI) (16). Patients with SLE were classified into the active group (SLEDAI ≥ 5) and the inactive group (SLEDAI< 5).
2.4 Detection methods
Fasting venous blood samples (5 mL) were collected from each subject and centrifuged at 3000 revolutions per minute (r/min) for 10 minutes using a centrifuge with a rotor diameter of 1000 mm. Following centrifugation, 150 μL of serum was separated and used for immunological analyses. Serum levels of immunoglobulin A (IgA), immunoglobulin M (IgM), immunoglobulin G (IgG), complement components C3 and C4, and C-reactive protein (CRP) were quantified using immunoturbidimetric assays. Peripheral blood cell parameters, including white blood cell (WBC) count, hemoglobin (Hb), and platelet (PLT) count, were analyzed using a Sysmex fully automated modular hematology and body fluid analyzer. Erythrocyte sedimentation rate (ESR) was measured by infrared photo-optical colorimetry. ANA were detected by indirect immunofluorescence assay (IIFA), and anti-double-stranded DNA (anti-dsDNA) antibodies were determined using a line immunoassay (LIA). The 24-hour urinary protein excretion was assessed using a TBA-FX8 fully automated biochemical analyzer.
2.5 RT-qPCR detection of CLEC4E gene expression
Blood samples from all participants were collected in EDTA-containing tubes for anticoagulation. Peripheral blood mononuclear cells (PBMCs) were separated through Ficoll-Paque density gradient centrifugation (GE Healthcare, Chicago, USA). Total RNA was isolated from PBMCs with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. The RNA concentration and purity were determined using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The RNA was then reverse-transcribed into cDNA using a reverse transcription kit (Thermo Fisher Scientific). RT-qPCR was performed on a QuantStudio M7 Flex RT-qPCR system (Thermo Fisher Scientific) using the SYBR Green PCR Master Mix kit (Applied Biosystems, Foster City, CA, USA). The primer sequences for the RT-qPCR reactions were as follows: CLEC4E forward primer: 5′-CCTGTTTCATCACCAGATGTGT-3′, CLEC4E reverse primer: 5′-AACGCCCAGGAAATGGTGT-3′; GAPDH forward primer: 5′-TGCAACCGGGAAGGAAATGA-3′, GAPDH reverse primer: 5′-TTCCCGTTCTCAGCCTTGAC-3′. The PCR amplification was carried out with an initial denaturation at 95°C for 5 minutes, followed by 40 cycles consisting of denaturation at 95°C for 15 seconds, annealing at 60°C for 30 seconds, and extension at 72°C for 30 seconds. The relative expression levels of CLEC4E were calculated using the 2−ΔΔCt method and normalized to GAPDH as an internal control.
2.6 Evaluation of diagnostic value
ROC curve analysis was used to evaluate the diagnostic performance of CLEC4E expression levels in distinguishing between SLE patients and healthy controls, as well as between active and inactive SLE patients.
2.7 Prognostic analysis
SLE patients were categorized into low and high CLEC4E expression groups based on the average expression value. Prognosis was assessed over a 6-month follow-up period post-discharge. Evaluations included updated SLEDAI scores, which were interpreted as follows: Favorable prognosis: SLEDAI score ≤ 4. Unfavorable prognosis: Disease relapse or death. Disease relapse was defined as an increase in the SLEDAI score of more than 3 points compared to the score at the end of treatment. Disease relapse was verified by clinical physicians. Death cases only included those directly related to SLE, such as those caused by disease complications or organ failure. The relationship between CLEC4E expression levels and SLE patient prognosis was assessed using survival analysis with Kaplan-Meier curve analysis.
2.8 Statistical analysis
Statistical analyses were conducted using SPSS software version 26.0 (IBM Corp., Armonk, NY, USA) and GraphPad Prism version 8 (GraphPad Software, San Diego, CA, USA), and R software version 4.4.3 (R Foundation for Statistical Computing, Vienna, Austria) was used for interaction effect analysis. For continuous variables with normal distribution, differences between groups were compared using the independent-samples t-test. For continuous variables not following a normal distribution, the Mann-Whitney U test (rank-sum test) was used. Differences in categorical variables were analyzed using the chi-square test. Prognostic rates were analyzed using Kaplan-Meier survival curves. A value of P< 0.05 was considered statistically significant.
3 Results
3.1 Bioinformatics analysis reveals SLE-associated DEGs
Gene expression datasets related to SLE, including GSE17755, GSE50772, and GSE61635, were retrieved from the GEO database. Intersection analysis was conducted with the Jvenn tool, applying a screening criterion of |log2FC| > 1 and P< 0.05. A total of six DEGs were identified: ISG15, HERC5, TNFAIP6, IFIT3, OASL, and CLEC4E. To further prioritize genes with immunological relevance, the identified DEGs were cross-referenced with immune-related genes from the GeneCards database (relevance score > 8). Only ISG15 and CLEC4E met these criteria. Considering that ISG15 has already been extensively investigated in clinical studies of SLE and its role and clinical significance have been systematically elucidated, additional validation would provide limited novelty. In contrast, CLEC4E has been rarely reported in SLE, remains to be clinically validated, and is feasible for detection in peripheral blood, thus offering both innovation and translational potential. On this basis, CLEC4E was selected as the key gene for subsequent clinical validation. See Figure 1.
3.2 Comparison of general demographics
A comparison was made between the SLE patient group and the healthy control group (Control) regarding general demographic characteristics and lifestyle habits. No significant differences were observed between the two groups regarding age, gender ratio, BMI, smoking history, alcohol consumption, or education level (P > 0.05), suggesting comparability, as shown in Table 1.
3.3 Expression of CLEC4E in SLE
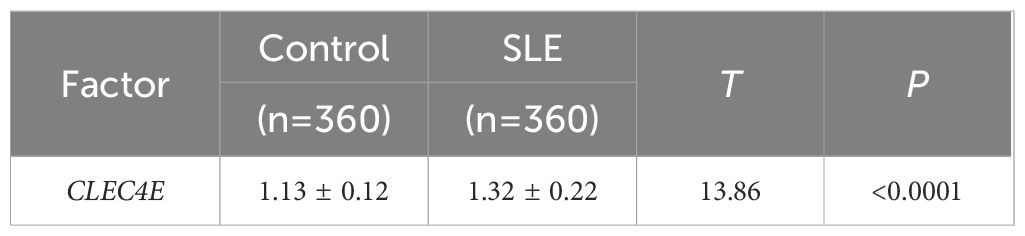
The expression of the CLEC4E gene was detected in both the SLE patient group and the healthy control group using RT-qPCR. The results indicated that CLEC4E expression was significantly elevated in the SLE group relative to the control group (P< 0.05). See Table 2 and Figure 2.
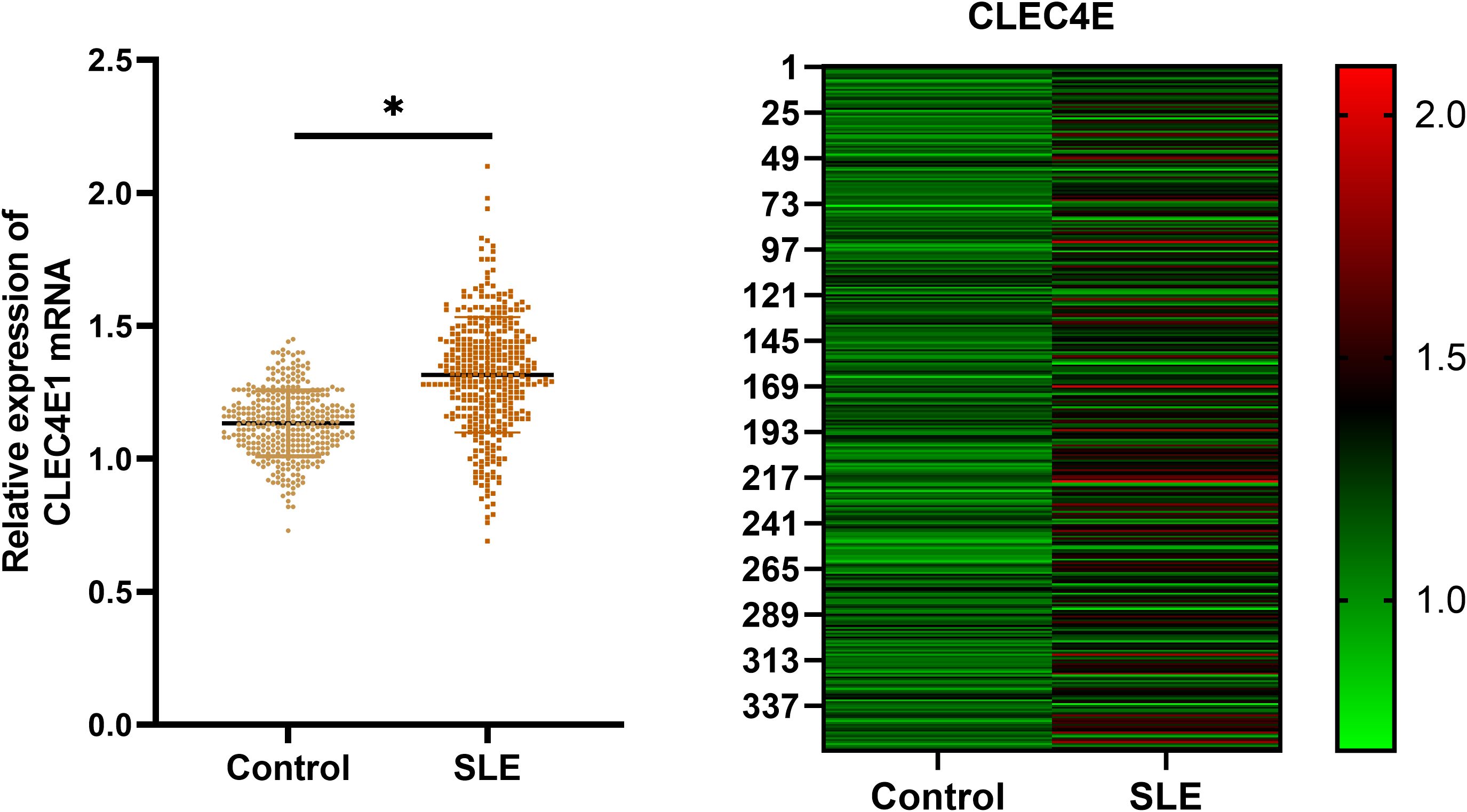
Figure 2. Expression of CLEC4E in healthy controls and SLE patients. The symbol * indicates a statistically significant difference between the two groups (*P < 0.05).
3.4 Diagnostic value of CLEC4E
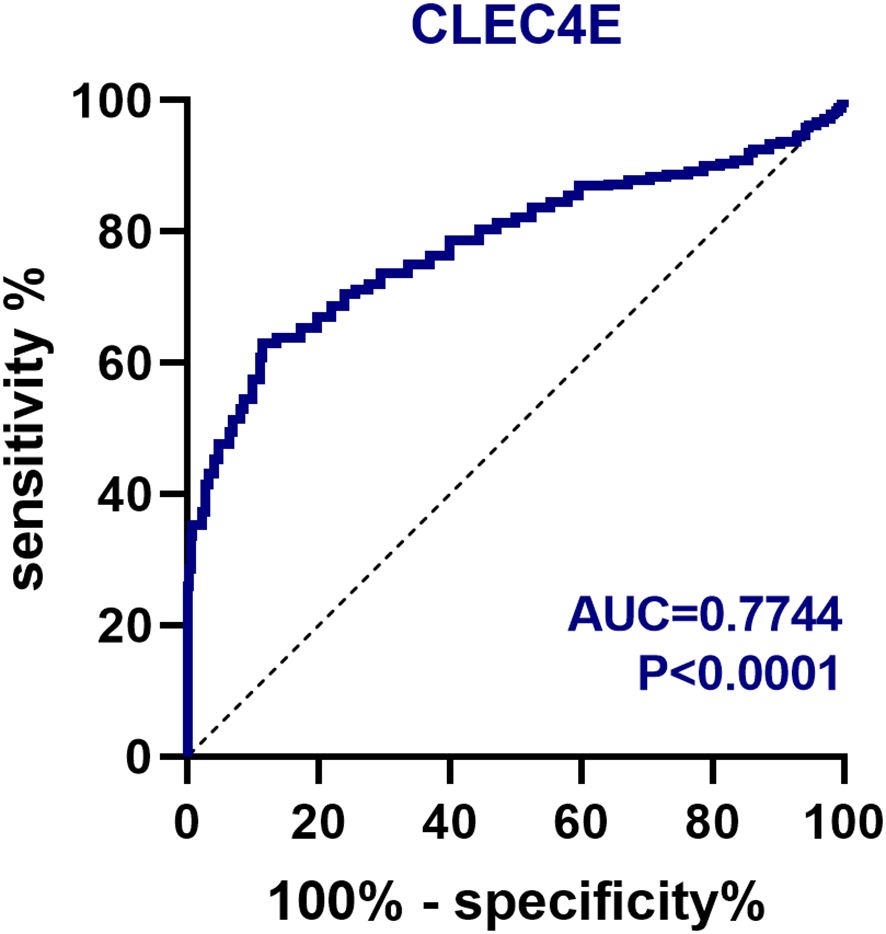
The diagnostic value of CLEC4E in SLE patients was assessed using ROC curve analysis. The AUC for CLEC4E was 0.7744, with a sensitivity of 63.06% and a specificity of 86.39%, indicating that CLEC4E has high diagnostic performance for SLE. See Table 3 and Figure 3.
3.5 Comparison of clinical characteristics between SLE patients with different disease activity
Based on the SLEDAI scores, SLE patients were divided into active and inactive groups. Clinical parameters were compared between the two groups. No significant differences were found in disease duration, WBC, HB, PLT, IgA, IgG, and IgM between the two groups (P > 0.05). However, the active group exhibited significantly elevated levels of immunological and inflammatory markers, ANA positivity, anti-dsDNA antibody levels, 24-hour urinary protein levels, ESR, and CRP. In contrast, serum levels of C3 and C4 were significantly reduced in this group (P< 0.05), indicating higher disease activity in active SLE patients. See Table 4.
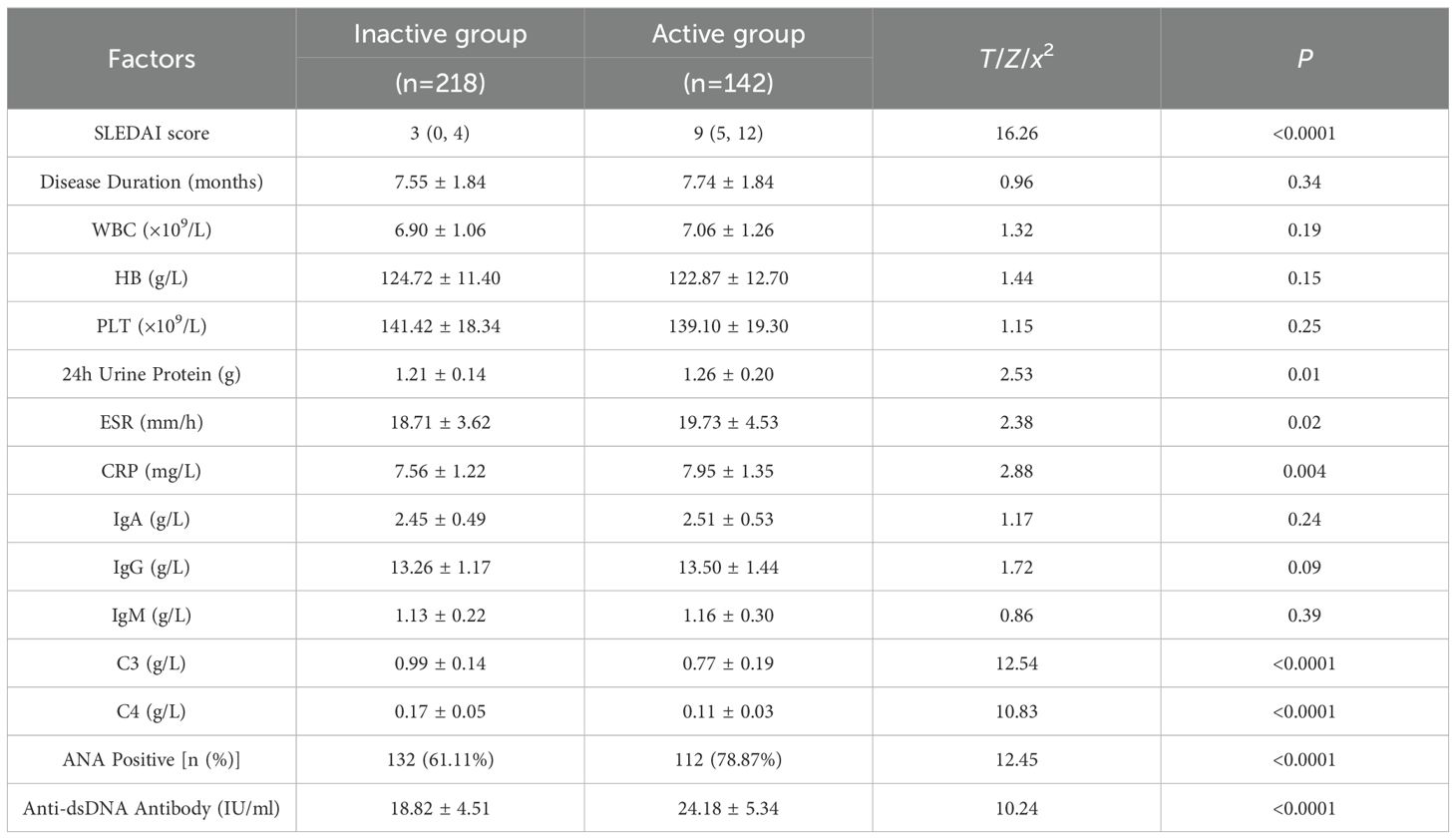
Table 4. Comparison of clinical characteristics between SLE patients with different disease activity [M (QMin, QMax), ± s, n (%)].
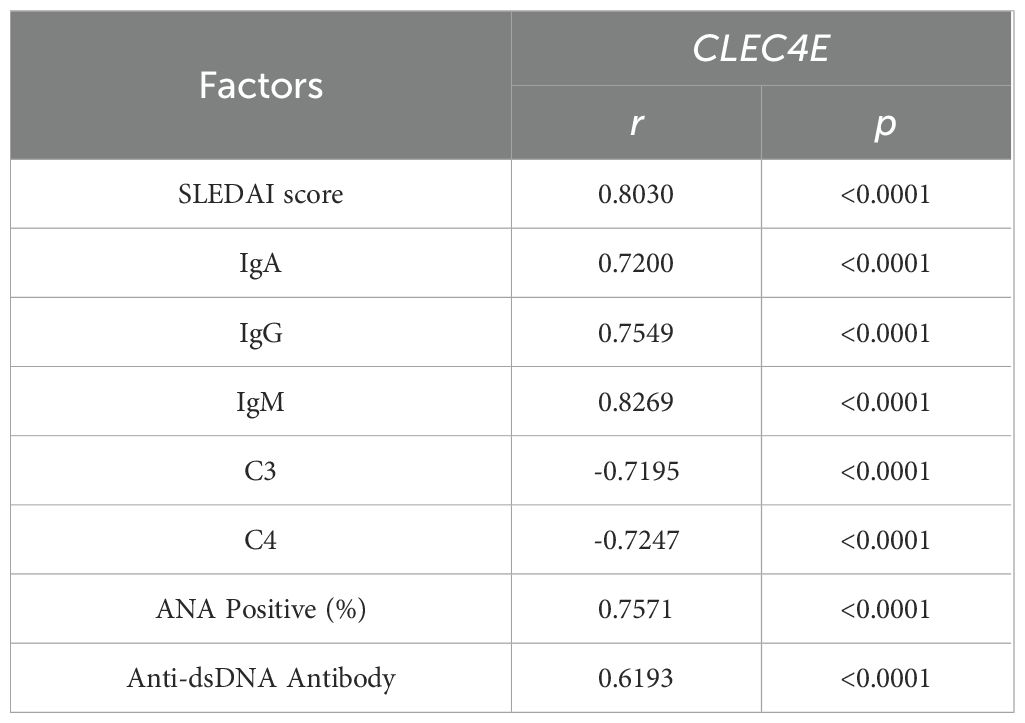
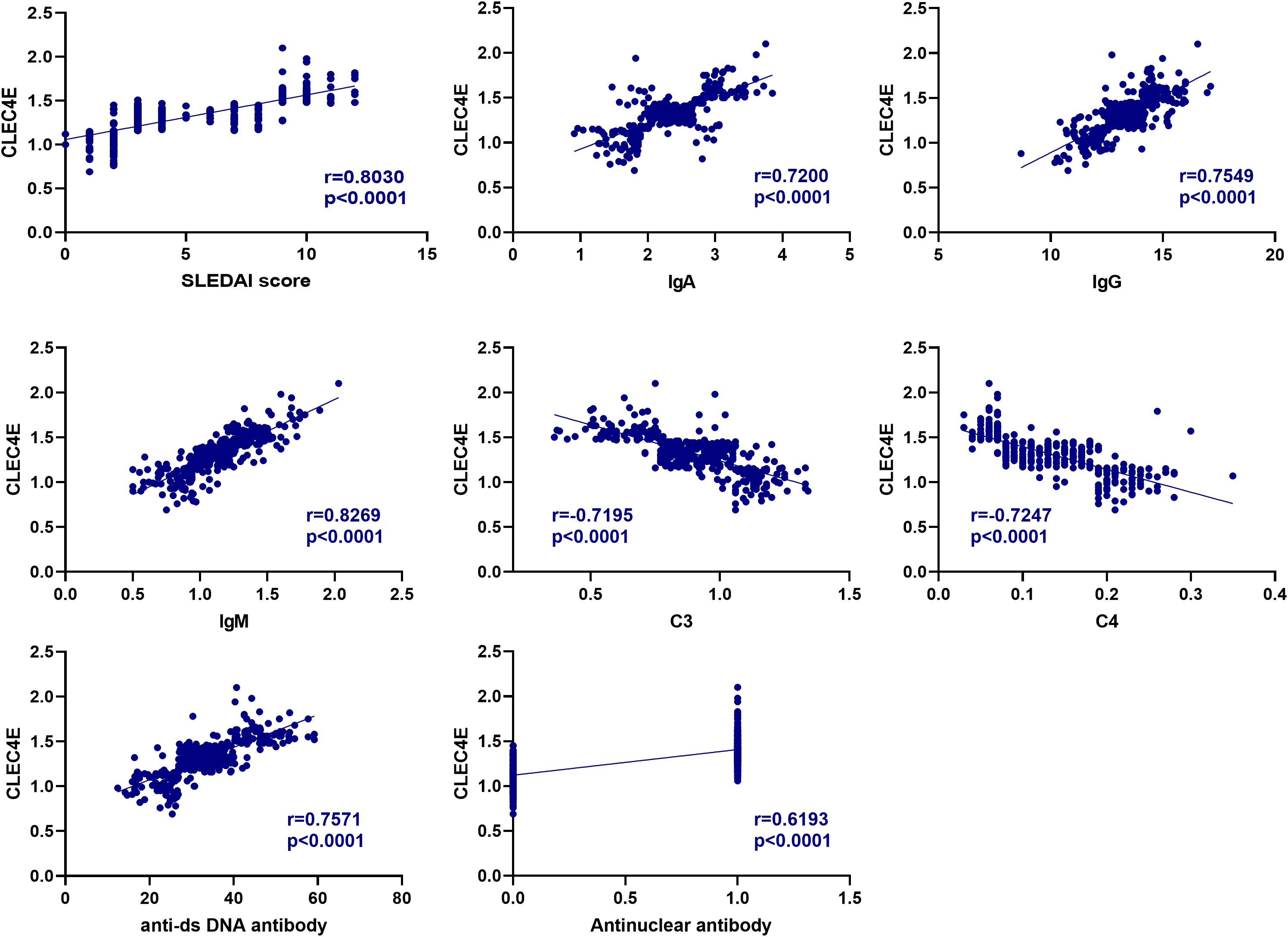
3.6 Correlation analysis between CLEC4E and immune-related indexes in SLE patients
The correlation between CLEC4E and SLEDAI score, IgA, IgG, and IgM was 0.7200, 0.7549, and 0.8269, respectively (P< 0.0001), and negatively correlated with C3 and C4, with correlations of -0.7195 and -0.7247, respectively (P< 0.0001). In addition, the correlation of CLEC4E with ANA positivity and anti-dsDNA antibody was 0.7571 and 0.6193 (P< 0.0001), respectively (Table 5 and Figure 4).
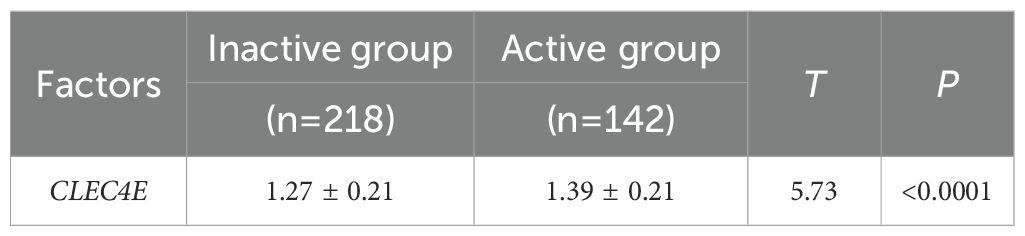
3.7 Expression difference of CLEC4E in SLE patients with different disease activity
CLEC4E expression was significantly higher in the active SLE group compared to that in the inactive group (P< 0.05). This finding suggests that CLEC4E may be closely related to the disease activity of SLE. See Table 6 and Figure 5.
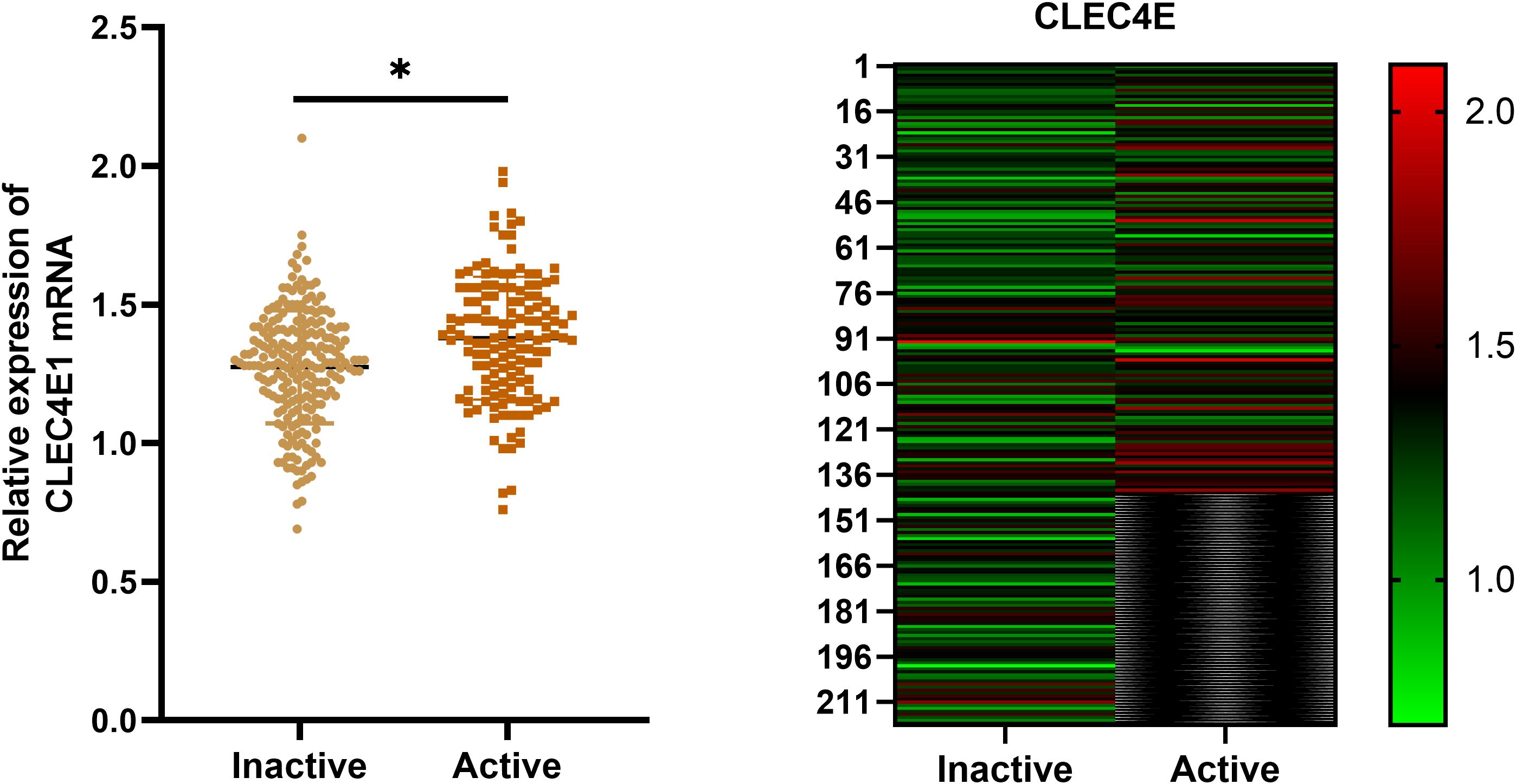
Figure 5. Expression of CLEC4E in SLE patients with different disease activity. The symbol * indicates a statistically significant difference between the two groups (*P < 0.05).
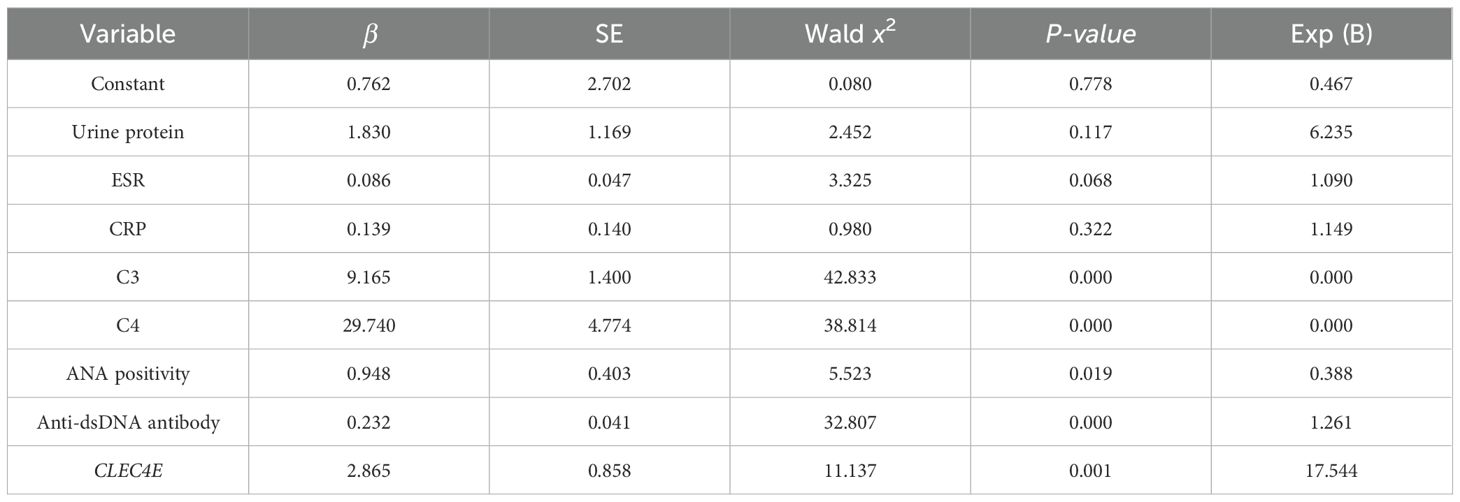
3.8 Multivariate analysis of independent risk factors for disease activity in SLE patients
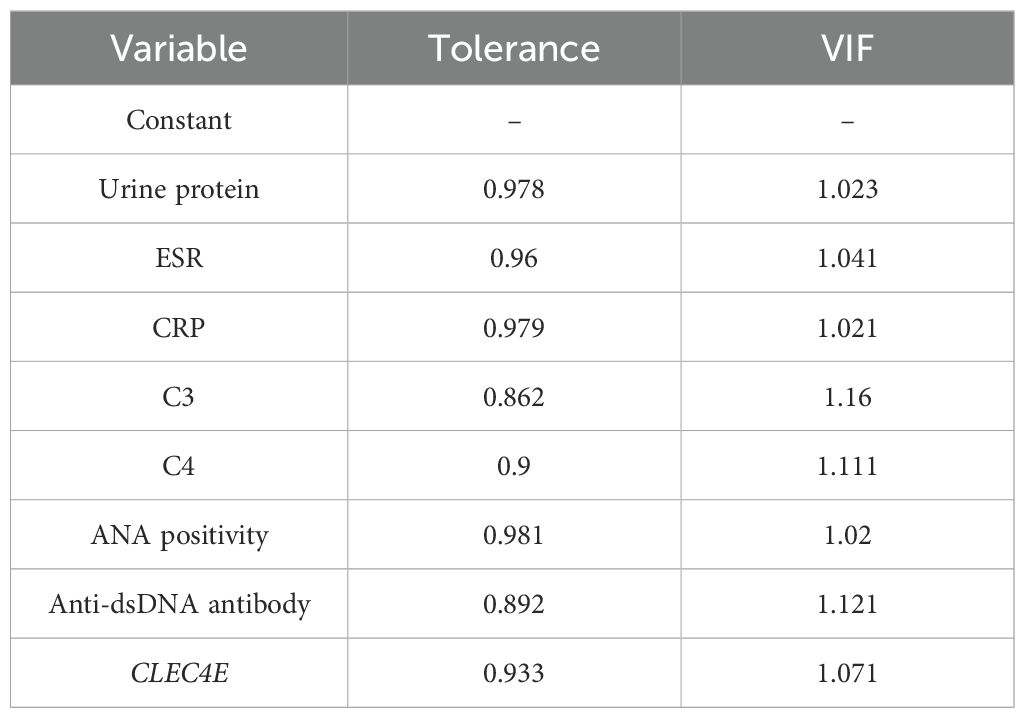
A multivariate logistic regression analysis was conducted to explore potential independent risk factors for disease activity in SLE. The factors with a P-value of<0.05 from the previous differential analysis were subjected to collinearity diagnostics. The results showed that the variance inflation factor (VIF) for all independent variables was less than 10, and the tolerance values for each variable were greater than 0.1, indicating that there was no significant linear correlation between the independent variables. Based on the conventional criteria for VIF and tolerance, no potential risk for multicollinearity was observed, as shown in Table 7. The regression analysis revealed that C3, positive ANA, anti-dsDNA antibodies, and CLEC4E expression levels were independent risk factors for SLE disease activity (P< 0.05). This suggests the potential prognostic value of CLEC4E in SLE progression. See Table 8.
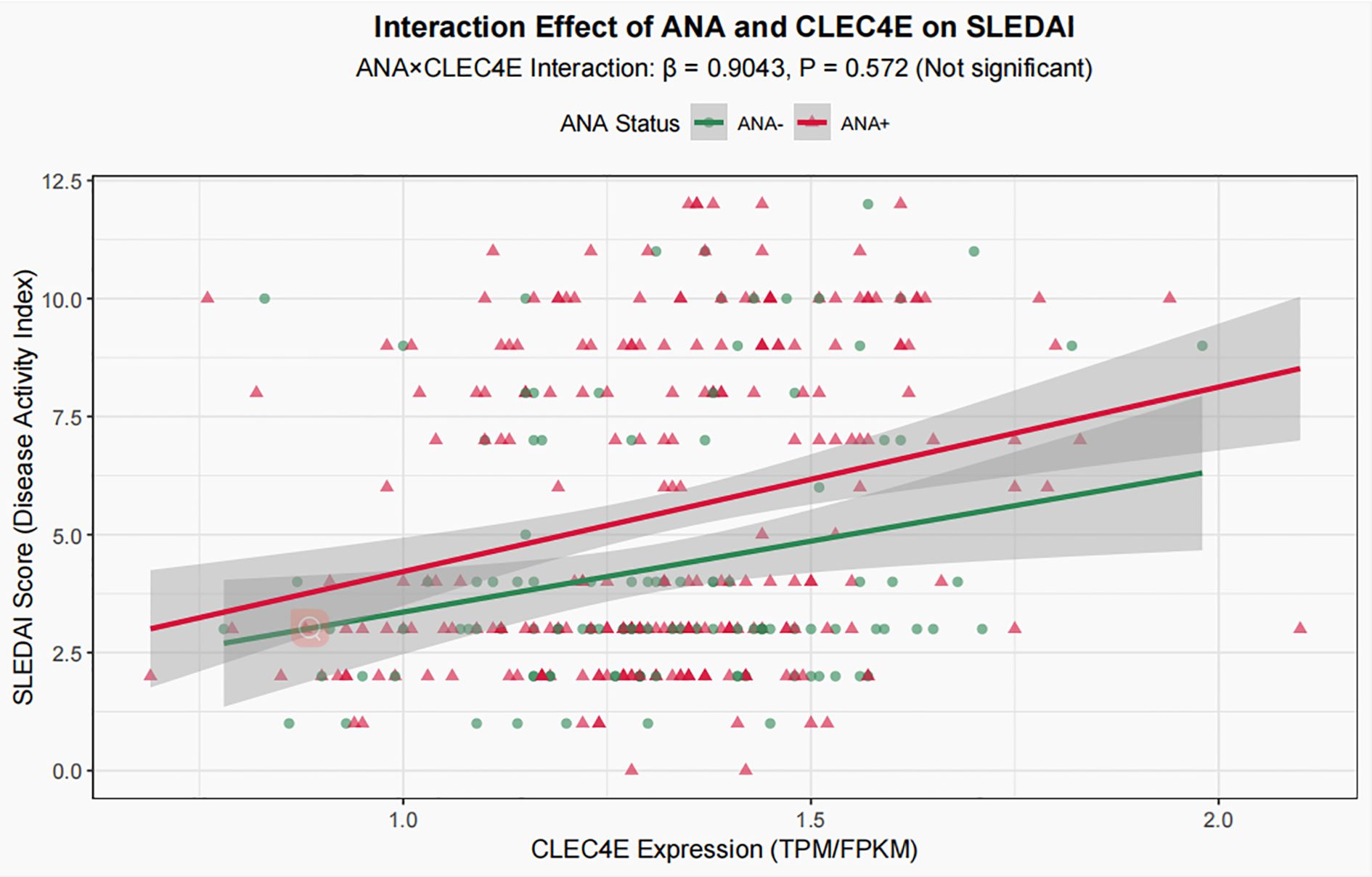
To explore the potential interaction between CLEC4E expression and ANA status, an interaction term (ANA × CLEC4E) was added to the aforementioned multivariate model. The analysis showed that the regression coefficient of the interaction term was 0.9043 with a P value of 0.572, indicating that the effect of CLEC4E on disease activity (SLEDAI) did not differ significantly between ANA-positive and ANA-negative patients (Figure 6). Therefore, the interaction term was not included in the main-effect model, and subsequent analyses and conclusions were based on the multivariate model without the interaction term.
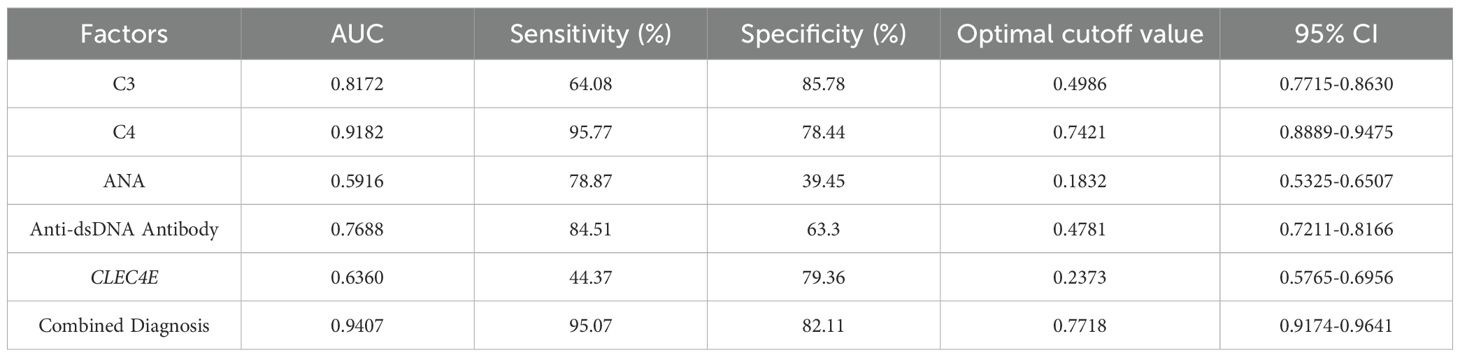
3.9 Diagnostic value of CLEC4E in SLE at different disease stages
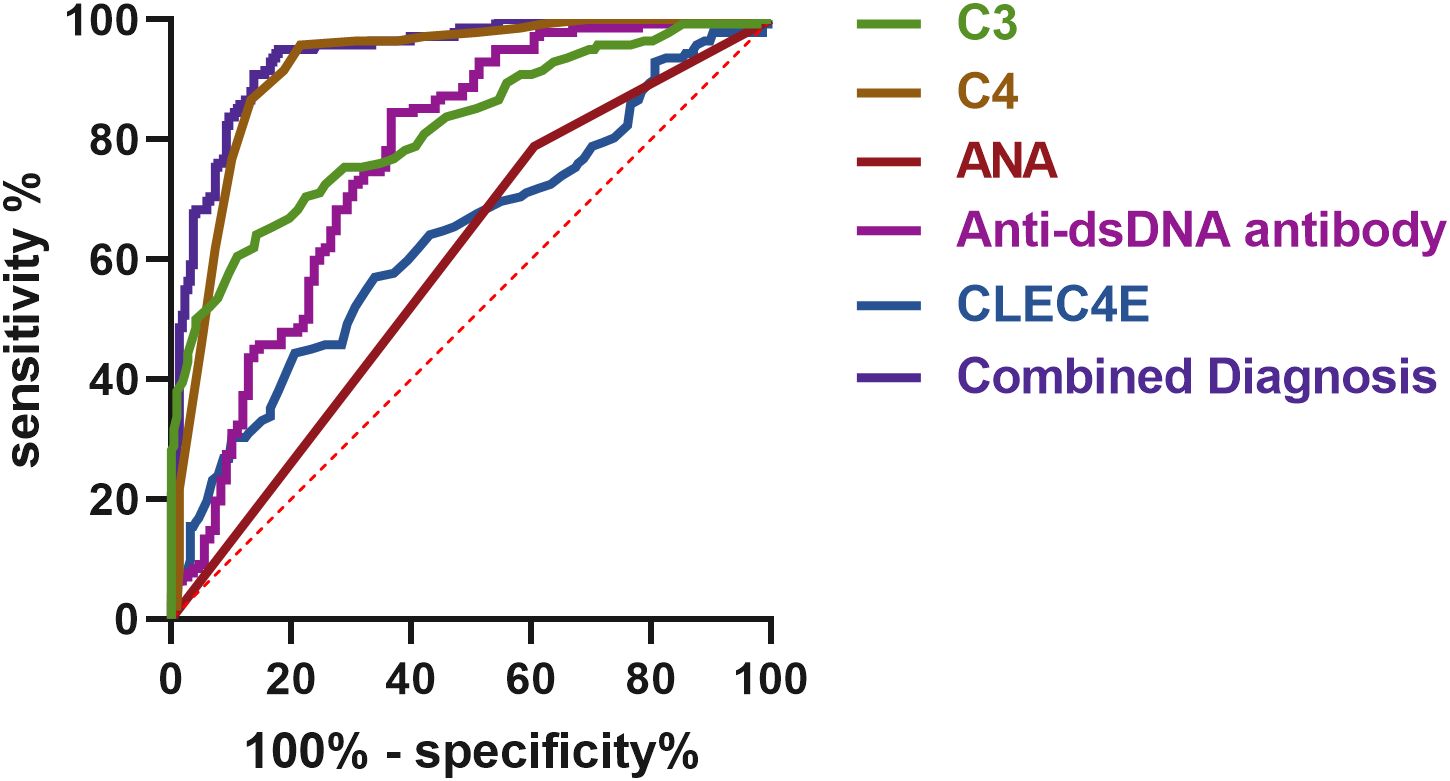
The diagnostic value of CLEC4E in different disease activity phases of SLE was evaluated using ROC curve analysis. The results showed that CLEC4E had an AUC of 0.6360, a sensitivity of 44.37%, and a specificity of 79.36%, indicating its potential diagnostic value in assessing SLE activity, particularly in distinguishing patients with high specificity. Although CLEC4E’s standalone diagnostic performance has certain limitations, its diagnostic capacity significantly improves when combined with other markers (such as C3, C4, ANA, and anti-dsDNA antibodies). The combined diagnostic performance showed a remarkable AUC of 0.9407, with a sensitivity of 95.07% and a specificity of 82.11%, further highlighting CLEC4E’s potential in SLE diagnosis, especially when used in conjunction with multiple biomarkers. See Table 6 and Figure 6. See Table 9 and Figure 7.
3.10 Relationship between CLEC4E expression and prognosis in SLE patients
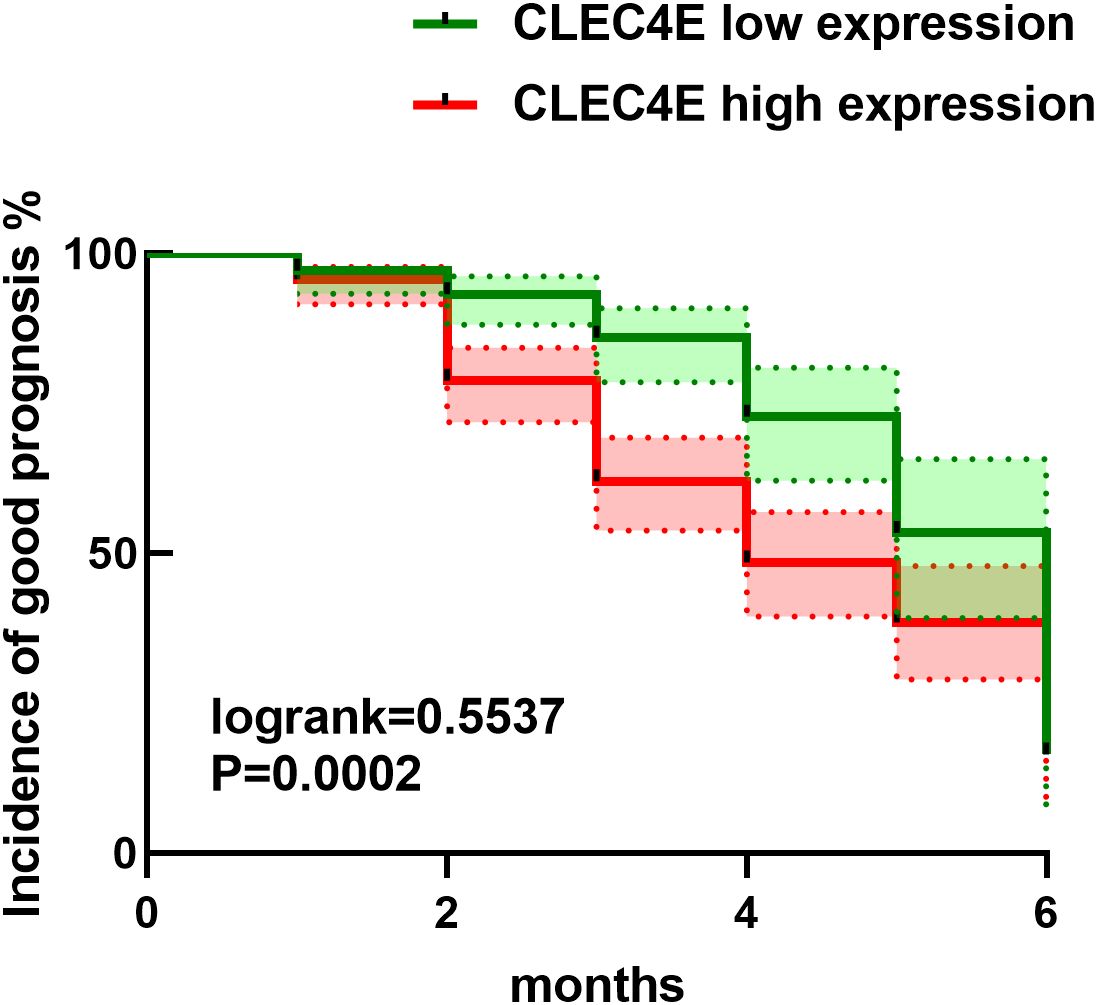
Kaplan-Meier survival analysis was used to assess the relationship between CLEC4E expression levels and prognosis in SLE patients (χ² = 13.46, P = 0.0002). The analysis demonstrated that patients with high CLEC4E expression had significantly poorer outcomes than those with low expression levels (P = 0.0002). These results suggest that high CLEC4E expression may be associated with worse clinical outcomes. See Figure 8.
4 Discussion
SLE is a highly heterogeneous and complex systemic autoimmune disease with an etiology that remains incompletely understood. Its pathological mechanisms involve various immunological abnormalities, including the production of autoantibodies, immune complex deposition, and chronic inflammatory responses (17, 18). Identifying novel biomarkers is crucial for gaining deeper insights into the pathogenesis of SLE and optimizing its diagnostic and therapeutic strategies. This study focused on CLEC4E, a potentially key molecule, and systematically evaluated its expression characteristics and relationship with disease-related parameters in SLE through a combination of bioinformatic analysis and clinical sample validation. Furthermore, the potential clinical applications of CLEC4E were explored.
Our findings demonstrated that CLEC4E expression was significantly elevated in SLE patients. Notably, CLEC4E levels were markedly higher in patients during the active phase. This finding suggests that CLEC4E may play a critical role in the pathophysiology of SLE. As a pattern recognition receptor, CLEC4E is involved in recognizing endogenous and exogenous danger signals, and its function is closely linked to the activation of the immune system (19). Within the pathological context of SLE, the elevated expression of CLEC4E may exacerbate chronic inflammation by enhancing the activity of neutrophils and macrophages (20). The pro-inflammatory role of CLEC4E, which promotes the secretion of inflammatory cytokines, could explain its strong association with disease activity indicators, such as elevated CRP levels and complement activation (21). Additionally, CLEC4E role in immune complex handling is significant in the context of SLE, where immune complexes are frequently deposited in tissues, contributing to organ dysfunction. CLEC4E’s ability to influence the handling and clearance of these immune complexes could be a crucial mechanism in mitigating inflammation and limiting tissue damage (22). The high expression of CLEC4E potentially reflects an abnormal inflammatory microenvironment in SLE patients, providing a theoretical basis for further exploration of SLE pathogenesis.
Further analysis revealed that CLEC4E expression holds promise as a clinical biomarker for both diagnosis and prognosis of SLE. The ROC curve analysis indicated that CLEC4E exhibited high sensitivity and specificity in distinguishing SLE patients from healthy individuals, with an AUC value of 0.7744. This finding suggests that CLEC4E could serve as an effective biomarker for diagnosing SLE. This is particularly significant as existing SLE diagnostic markers, while widely used, still have certain limitations in some cases. Elevated CLEC4E expression could complement traditional biomarkers and provide dynamic monitoring of disease progression, especially showing unique advantages in assessing disease activity. Tracking dynamic changes in CLEC4E expression may allow clinicians to more accurately assess fluctuations in disease status and treatment response, ultimately supporting personalized treatment strategies for patients with SLE. Additionally, this study revealed a significant association between high CLEC4E expression and poor prognosis in SLE patients. Kaplan-Meier survival analysis indicated that patients with elevated CLEC4E expression typically experienced more rapid disease progression, suggesting its potential role in predicting disease course. As a dual-function biomarker, the high expression of CLEC4E may not only result from enhanced inflammatory responses in the disease but also serve as a critical factor in exacerbating inflammation and tissue damage. Therefore, CLEC4E could be employed not only as a static diagnostic marker but also as a tool for dynamic monitoring. Regular monitoring of CLEC4E expression may help clinicians detect early signs of disease exacerbation, enabling timely intervention. This approach could delay disease progression, alleviate symptoms, and improve patients’ quality of life.
It is noteworthy that CLEC4E, as a molecule with a pivotal role in immune responses, may interact with other immunological markers, further enhancing its impact on disease progression. For example, CLEC4E is potentially associated with complement system activation, cytokine secretion, and the recruitment and activation of immune cells. These factors collectively drive chronic inflammation and tissue damage in SLE (23, 24). Elevated CLEC4E expression not only reflects excessive immune system activation but also contributes to the formation of immune pathology (25). CLEC4E’s influence on immune complex handling could provide a mechanistic link between immune dysregulation and tissue damage in SLE. By modulating the clearance of immune complexes, CLEC4E may play a key role in controlling tissue damage and inflammation (26). Therefore, CLEC4E holds promise as a potential therapeutic target. Future research could explore targeted interventions against CLEC4E and evaluate their potential in mitigating immune responses and reducing tissue damage in SLE patients. The potential for targeting CLEC4E through Mincle inhibitors is an exciting avenue for future therapeutic strategies, as inhibiting CLEC4E may help reduce chronic inflammation and prevent further tissue injury (20).
Despite systematically uncovering the expression characteristics and clinical significance of CLEC4E in SLE, this study has certain limitations. Firstly, the sample size was relatively small, and the follow-up period was limited to 6 months, which should be regarded as short-term. Larger-scale, multicenter studies with extended follow-up are required to further validate the clinical utility of CLEC4E and to assess the long-term prognostic value. Secondly, in the initial bioinformatics screening, multiple-testing correction (FDR) was not applied. Although this may increase the likelihood of false-positive results, our focus was to prioritize candidate genes for clinical validation, and subsequent verification in independent samples partly mitigates this concern. Thirdly, GSVA/ssGSEA or related immune deconvolution tools were not employed, which may limit our ability to comprehensively characterize the immune landscape of SLE and restrict the depth of mechanistic interpretation. In addition, although the predictive model demonstrated a high AUC, no internal validation was performed due to the limited sample size, which may raise concerns of overfitting; future studies with larger cohorts are warranted to confirm the robustness of the model. In addition, CLEC4E may exhibit distinct functions across different tissues or cell types, particularly in affected organs such as the kidneys and skin, where its role may differ significantly. Future studies should incorporate tissue samples or employ single-cell transcriptomics to delineate the context-specific functions of CLEC4E. Moreover, the mechanistic link between CLEC4E, complement activation, and immune complex handling remains to be fully elucidated, and clarifying these pathways may reveal how CLEC4E contributes to disease progression and tissue damage in SLE. Finally, while our findings suggest that CLEC4E and its receptor, Mincle, may serve as novel therapeutic targets, further preclinical studies are warranted to determine whether targeting this axis could attenuate inflammatory responses and ultimately improve patient outcomes.
In summary, this study is the first to systematically reveal the high expression profile of CLEC4E in SLE and to investigate its relationship with disease activity and prognosis. Beyond serving as a diagnostic biomarker to improve the accuracy of SLE diagnosis, CLEC4E may also become a crucial tool for disease monitoring and therapeutic management. Future research should explore CLEC4E’s functional heterogeneity across SLE subtypes using single-cell and tissue-specific analyses to support its clinical translation.
5 Conclusion
Increased CLEC4E expression correlates with higher disease activity and poorer prognosis in SLE patients, indicating its potential as a significant biomarker for disease diagnosis and monitoring.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by Tianjin Academy of Traditional Chinese Medicine Affiliated Hospital (No. LLKY2025-30). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
XM: Conceptualization, Methodology, Writing – original draft. HZ: Data curation, Formal Analysis, Writing – review & editing. JL: Formal Analysis, Investigation, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Accapezzato D, Caccavale R, Paroli MP, Gioia C, Nguyen BL, Spadea L, et al. Advances in the pathogenesis and treatment of systemic lupus erythematosus. Int J Mol Sci. (2023) 24:6578. doi: 10.3390/ijms24076578
2. Lazar S and Kahlenberg JM. Systemic lupus erythematosus: new diagnostic and therapeutic approaches. Annu Rev Med. (2023) 74:339–52. doi: 10.1146/annurev-med-043021-032611
3. Tsuchiya N, Ito I, and Kawasaki A. Association of IRF5, STAT4 and BLK with systemic lupus erythematosus and other rheumatic diseases. Nihon Rinsho Meneki Gakkai Kaishi. (2010) 33:57–65. doi: 10.2177/jsci.33.57
4. Li S, Wu Q, Jiang Z, Wu Y, Li Y, Ni B, et al. miR-31-5p regulates type I interferon by targeting SLC15A4 in plasmacytoid dendritic cells of systemic lupus erythematosus. J Inflammation Res. (2022) 15:6607–16. doi: 10.2147/JIR.S383623
5. Udayan S, Butto LF, Rossini V, Velmurugan J, Martinez-Lopez M, Sancho D, et al. Macrophage cytokine responses to commensal Gram-positive Lactobacillus salivarius strains are TLR2-independent and Myd88-dependent. Sci Rep. (2021) 11:5896. doi: 10.1038/s41598-021-85347-7
6. Jiang Q, Xiao D, Wang A, Yu Q, Yin Y, Wu J, et al. CLEC4E upregulation in gastric cancer: A potential therapeutic target correlating with tumor-associated macrophages. Heliyon. (2024) 10:e27172. doi: 10.1016/j.heliyon.2024.e27172
7. Prado Acosta M, Goyette-Desjardins G, Scheffel J, Dudeck A, Ruland J, and Lepenies B. S-Layer From Lactobacillus brevis Modulates Antigen-Presenting Cell Functions via the Mincle-Syk-Card9 Axis. Front Immunol. (2021) 12:602067. doi: 10.3389/fimmu.2021.602067
8. Desel C, Murray PJ, Lehmann CHK, Heger L, Christensen D, Andersen P, et al. Monocytes elicit a neutrophil-independent th1/th17 response upon immunization with a mincle-dependent glycolipid adjuvant. Front Immunol. (2022) 13:880474. doi: 10.3389/fimmu.2022.880474
9. Malmhall-Bah E, Andersson KME, Erlandsson MC, Silfversward ST, Pullerits R, and Bokarewa MI. Metabolic signature and proteasome activity controls synovial migration of CDC42(hi)CD14(+) cells in rheumatoid arthritis. Front Immunol. (2023) 14:1187093. doi: 10.3389/fimmu.2023.1187093
10. Malamud M and Brown GD. The Dectin-1 and Dectin-2 clusters: C-type lectin receptors with fundamental roles in immunity. EMBO Rep. (2024) 25:5239–64. doi: 10.1038/s44319-024-00296-2
11. Chen R, Zou J, Chen J, Zhong X, Kang R, and Tang D. Pattern recognition receptors: function, regulation and therapeutic potential. Signal Transduct Target Ther. (2025) 10:216. doi: 10.1038/s41392-025-02264-1
12. Sharma A, Rijavec M, Tomar S, Yamani A, Ganesan V, Krempski J, et al. Acute systemic myeloid inflammatory and stress response in severe food allergic reactions. Clin Exp Allergy. (2023) 53:536–49. doi: 10.1111/cea.14273
13. Wang CR and Tsai HW. Autoimmune liver diseases in systemic rheumatic diseases. World J Gastroenterol. (2022) 28:2527–45. doi: 10.3748/wjg.v28.i23.2527
14. Chen J, Liao S, Zhou H, Yang L, Guo F, Chen S, et al. Humanized mouse models of systemic lupus erythematosus: opportunities and challenges. Front Immunol. (2021) 12:816956. doi: 10.3389/fimmu.2021.816956
15. Aringer M, Costenbader K, Daikh D, Brinks R, Mosca M, Ramsey-Goldman R, et al. 2019 european league against rheumatism/american college of rheumatology classification criteria for systemic lupus erythematosus. Arthritis Rheumatol. (2019) 71:1400–12. doi: 10.1002/art.40930
16. Thanou A, James JA, Arriens C, Aberle T, Chakravarty E, Rawdon J, et al. Scoring systemic lupus erythematosus (SLE) disease activity with simple, rapid outcome measures. Lupus Sci Med. (2019) 6:e000365. doi: 10.1136/lupus-2019-000365
17. Zucchi D, Silvagni E, Elefante E, Signorini V, Cardelli C, Trentin F, et al. Systemic lupus erythematosus: one year in review 2023. Clin Exp Rheumatol. (2023) 41:997–1008. doi: 10.55563/clinexprheumatol/4uc7e8
18. Coss SL, Zhou D, Chua GT, Aziz RA, Hoffman RP, Wu YL, et al. The complement system and human autoimmune diseases. J Autoimmun. (2023) 137:102979. doi: 10.1016/j.jaut.2022.102979
19. Frazzei G, van Vollenhoven RF, de Jong BA, Siegelaar SE, and van Schaardenburg D. Preclinical autoimmune disease: a comparison of rheumatoid arthritis, systemic lupus erythematosus, multiple sclerosis and type 1 diabetes. Front Immunol. (2022) 13:899372. doi: 10.3389/fimmu.2022.899372
20. Wang C, Zhang Y, Shen A, Tang T, Li N, Xu C, et al. Mincle receptor in macrophage and neutrophil contributes to the unresolved inflammation during the transition from acute kidney injury to chronic kidney disease. Front Immunol. (2024) 15:1385696. doi: 10.3389/fimmu.2024.1385696
21. Furukawa A, Shuchi Y, Wang J, Guillen-Poza PA, Ishizuka S, Kagoshima M, et al. Structural basis for plastic glycolipid recognition of the C-type lectin Mincle. Structure. (2023) 31:1077–85:e1075. doi: 10.1016/j.str.2023.05.018
22. Yuan S, Wang C, Jiang W, Wei Y, Li Q, Song Z, et al. Comparative transcriptome analysis of gingival immune-mediated inflammation in peri-implantitis and periodontitis within the same host environment. J Inflammation Res. (2022) 15:3119–33. doi: 10.2147/JIR.S363538
23. Stegmann F, Diersing C, and Lepenies B. Legionella pneumophila modulates macrophage functions through epigenetic reprogramming via the C-type lectin receptor Mincle. iScience. (2024) 27:110700. doi: 10.1016/j.isci.2024.110700
24. Hasgur S, Yamamoto Y, Fan R, Nicosia M, Gorbacheva V, Zwick D, et al. Macrophage-inducible C-type lectin activates B cells to promote T cell reconstitution in heart allograft recipients. Am J Transpl. (2022) 22:1779–90. doi: 10.1111/ajt.17033
25. Takata K, Takano S, Miyagi M, Mukai M, Iwase D, Aikawa J, et al. Elevated macrophage-inducible C-type lectin expression in the synovial tissue of patients with rheumatoid arthritis. Cent Eur J Immunol. (2021) 46:470–3. doi: 10.5114/ceji.2021.111471
26. Thiyagarajan D, Fismen S, Seredkina N, Jacobsen S, Elung-Jensen T, Kamper AL, et al. Silencing of renal DNaseI in murine lupus nephritis imposes exposure of large chromatin fragments and activation of Toll like receptors and the Clec4e. PloS One. (2012) 7:e34080. doi: 10.1371/journal.pone.0034080
Keywords: SLE, CLEC4E gene, biomarker, disease activity, diagnosis and prognosis
Citation: Ma X, Zhang H and Li J (2025) CLEC4E as a molecular biomarker in systemic lupus erythematosus: integrating bioinformatics and clinical data to assess its prognostic value. Front. Immunol. 16:1617878. doi: 10.3389/fimmu.2025.1617878
Received: 29 April 2025; Accepted: 15 September 2025;
Published: 30 September 2025.
Edited by:
Maria Infantino, Nuovo Ospedale San Giovanni di Dio, ItalyReviewed by:
Yunliang Yao, Huzhou University, ChinaHuiqiong Zeng, Women and Children Health Institute Futian Shenzhen, China
Copyright © 2025 Ma, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoxia Ma, bWF4aWFveGlhMTk5M0AxNjMuY29t
†These authors share first authorship
 Xiaoxia Ma
Xiaoxia Ma Huan Zhang1†
Huan Zhang1†