- 1Guangxi Academy of Marine Sciences, Guangxi Academy of Sciences, Nanning, Guangxi, China
- 2National Key Laboratory of Non-food Biomass Energy Technology, Nanning, Guangxi, China
- 3Guangxi Key Laboratory of Marine Natural Products and Combinatorial Biosynthesis Chemistry, Nanning, Guangxi, China
- 4College of Chemistry and Bioengineering, Hunan University of Science and Engineering, Yongzhou, Hunan, China
- 5Key Laboratory of Comprehensive Utilization of Advantage Plants Resources in Hunan South, Yongzhou, Hunan, China
- 6College of Animal Science and Technology, Guangxi University, Nanning, Guangxi, China
- 7Guangxi Key Laboratory of Animal Reproduction, Breeding and Disease Control, Nanning, Guangxi, China
- 8Guangxi Veterinary Research Institute, Nanning, Guangxi, China
- 9Guangxi Key Laboratory of Veterinary Biotechnology, Nanning, Guangxi, China
- 10Key Laboratory of China(Guangxi)-Association of Southeast Asian Nations (ASEAN) Cross-border Animal Disease Prevention and Control, Nanning, Guangxi, China
Introduction: Segmented filamentous bacteria (SFB) colonization dynamics are crucial for host immune regulation. Given this, the present study specifically examined the functions of SFB flagellin in bacterial adhesion, cellular internalization, and immune modulation.
Methods: Lactococcus lactis and Escherichia coli were engineered to express murine and rat SFB flagellin genes. Subsequent in vitro co-culture experiments with intestinal epithelial cell lines (MODE-K and IEC-18) and germ-free mouse colonization assays were conducted. Bacterial counts, immunohistochemical analysis, the AAM-ISO-G1 and QAM-TH17–1 microarray systems, RNA sequencing and molecular docking were employed to assess the outcomes in this study.
Results and discussion: The results of in vitro co-culture experiments demonstrated significantly improved bacterial adhesion capabilities mediated by SFB flagellin. Germ-free mouse colonization assays revealed prolonged fecal persistence of flagellin-expressing strains. Immunohistochemical analysis of ileal tissues showed co-localization of recombinant bacteria with the lysosomal-associated membrane protein 2 (Lamp2), confirming cellular internalization. Furthermore, mfliC3-expressing Escherichia coli exhibited active invasion into MODE-K cells. RNA sequencing analysis identified significant enrichment of Th17 cell differentiation pathways in both ileum and hepatic tissues from Lac-rfliC3-colonized mice. Correspondingly, the Lac-mfliC3 group showed elevated serum levels of Th17-associated cytokines including IFN-γ, IL-23p19, IL-17A, IL-5, and IL-6 compared to controls. Molecular docking simulations revealed high-affinity interactions between SFB flagellins and endocytic regulators endophilin A2 and αM integrin. These results demonstrate that SFB flagellin mediates bacterial-epithelial interactions through dual mechanisms of adhesion potentiation and active internalization, ultimately driving Th17-mediated immune responses.
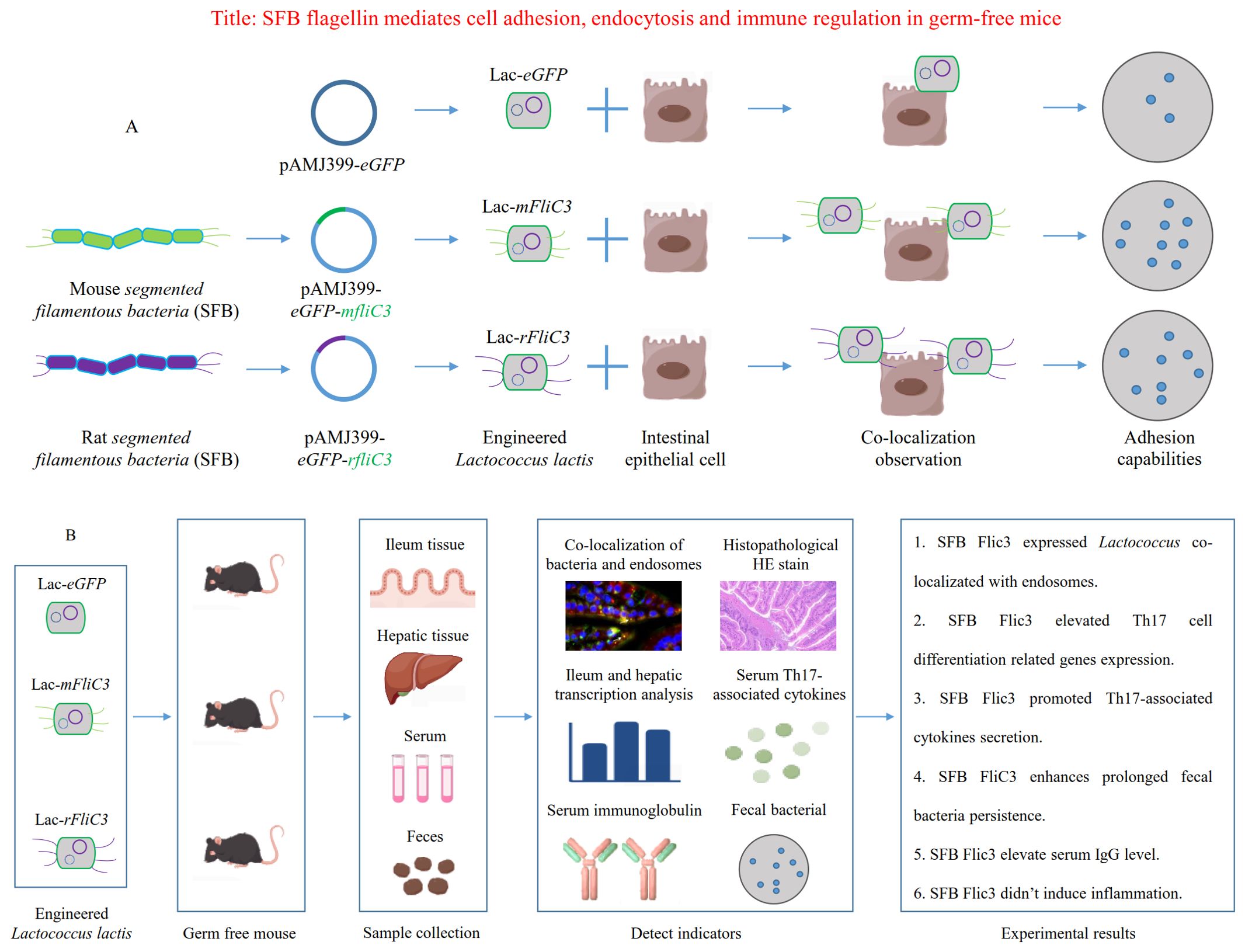
Graphical Abstract. (A) Schematic illustrating SFB flagellin-mediated bacterial adhesion to epithelial cell surfaces. (B) Schematic depicting SFB flagellin's role in mediating cell adhesion, endocytosis, and immune regulation within germ-free mice.
Introduction
The gut microbiota constitutes the most extensive and complex microbial ecosystem in the human body, playing a pivotal roles in essential physiological processes including metabolic regulation and immune function. Segmented filamentous bacteria (SFB), predominantly colonizing the terminal ileum, have attracted considerable research interest due to their unique filamentous morphology and potent immunomodulatory properties. SFB colonization stimulates secretory immunoglobulin A (SIgA) production in the intestinal mucosa and drives Th17 cell differentiation (1–3), mechanisms critical for maintaining immune homeostasis (4). These commensals generally confer host beneficial, including mitigating infections by pathogens such as Salmonella (5) and Staphylococcus aureus (6), while enhancing antiviral defenses against to rotavirus and respiratory viruses (7, 8). Emerging evidence further implicates SFB in bone development and homeostasis through gut-bone axis modulation (9–11), with demonstrate roles in fracture repair (11). Notably, context-dependent SFB activities may exacerbate intestinal inflammation and strengthen autoimmune pathologies such as Crohn’s disease (12) and rheumatoid arthritis (13, 14). Collectively, SFB orchestrates acute immune-regulatory functions impacted both intestinal and systemic host health.
Although SFB are widely recognized for their essential immunomodulatory functions, their precise mechanistic contributions to host immunity and associated molecule determinants remain incompletely characterized. Ladinsky et al. demonstrated that murine SFB establish intimate interactions with intestinal epithelial cells (IECs) through surface anchoring, initiating microbial adhesion-triggered endocytosis (MATE) to deliver antigenic proteins into IECs, thereby modulating Th17 cell differentiation (15). Atarashi et al. observed that rat SFB, despite proliferating in the murine intestines, fail to colonize IEC surfaces and consequently lack Th17 regulatory capacity (16), underscoring the necessity of host-specific adhesion for SFB-mediated immune modulation.
Bacterial flagellar proteins crucially enhance microbial adhesion (17) and activate host immune responses (18), acting as key mediators of gut microbiota-host interactions that influence both microbial community dynamics and host physiology. Substantial evidence identifies these proteins as stable immune-stimulatory factors in the gastrointestinal tract, contributing to immune regulation (19, 20), β-cell functionality (21), and inflammatory bowel diseases pathogenesis (22–24). Genomic analyses of murine and rat SFB have revealed deficient metabolic gene repertoires but conserved flagellar structural genes, including four fliC paralogs (25–27). Our prior work validated the expression of SFB flagellin FliC3 in human and murine intestinal tissues through western blot and immunohistochemistry analyses (28, 29). Subsequent investigations demonstrated substantial genetic diversity and host specificity among SFB flagellin genes (29, 30). Notable, ELISA, ELISPOT, and RNA-seq were employed to assess the immunomodulatory function of SFB FliC3 both in vitro and in vivo (31). Furthermore, intraperitoneal administration of purified SFB FliC3 recapitulated the Th17 cell differentiation effects observed with whole SFB cells in murine models (31), suggesting potential roles for SFB flagellin in host-specific colonization and endocytic processes. This study investigates the effects of engineered SFB flagellin-expressing bacteria to cellular adhesion, endocytosis, and immune regulation in germ free mouse.
Materials and methods
Construction and screening of recombinant Lactococcus lactis
The L. lactis MG1363 strain and Escherichia coli harboring the pAMJ399-eGFP plasmid were maintained in our laboratory. GM17 culture medium (M17 supplemented with 0.5% glucose) and LB medium were used to culture L. lactis and E. coli, respectively. L. lactis was incubated at 30°C under static anaerobic conditions, while E. coli was cultured at 37°C with shaking at 150 rpm.
Flagellin genes derived from mouse SFB (M5I-FliC3-2, designated mfliC3), rat SFB (R4I-FliC3-1, designated rfliC3), and Salmonella enteritidis (sal_fliC3), previously cloned into the PMD18-T vector (29), were subcloned into the expression vector pAMJ399-eGFP. Competent L. lactis cells were prepared following established protocols with minor modifications (32). Briefly, a single L. lactis MG1363 colony was inoculated into 5 mL of Solution I (containing 2.5 mL 2× M17, 0.05 mL 50% glucose, 1.7 mL 50% sucrose, 0.5 mL 10% glycine, and 0.25 mL ddH20), and incubated overnight at 30°C. The culture was transferred to 15 mL of Solution I at a 4% inoculation ratio, and incubated at 30°C until reaching an OD600 of 0.4-0.7. Cells were chilled on ice for 2 minutes, centrifuged at 4°C (5,000 rpm for 10 m), and washed twice with cold Solution II (34 mL 50% sucrose, 11.5 mL 87% glycerol, 54.5 mL ddH20). After supernatant removal and tube inversion for 1 min, cells were resuspended in 1 mL of pre-chilled Solution II and incubated on ice for 30–60 min. Aliquots were dispensed into pre-cooled Eppendorf tubes and kept at -70°C for subsequent use.
Recombinant plasmid transformation into L. lactis MG1363 was conducted via electroporation (25 μF, 2 KV, 200 Ω). Transformants were selected on M17 agar supplemented with erythromycin (100 μg/mL). Plasmid integration was confirmed using universal vector primers P170-F (5’-CCA TTT TTG GTT GCC ATT GTT AAC GCT GCC-3’) and P170-R (5’-TAC GTA GAT CTG CTC TTC CTG CTT GAG CAT C-3’). Successful L. lactis transformants expressing flagellar proteins were designated Lac-mfliC3, Lac-rfliC3, and Lac-sal_fliC3, with the empty vector control strain named Lac-eGFP.
Insertion and replacement of E. coli fliC with SFB mfliC3 gene
The E. coli strain BW25113 was kept in our laboratory. CRISPR/Cas9-mediated genome editing replaced the native fliC gene in BW25113 with the SFB mfliC3 gene. BW25113 genomic DNA was used as the template for amplifying homologous arms using primers fliC-up-F/fliC-up-R and fliC-down-F/fliC-down-R. The purified products (designated fliCUP and fliCDown), were fused via overlap extension PCR. The resultant fliCUD fragment was A-tailed and ligated into the pUX-T vector, generating plasmid pUX-T-fliCUD, which was converted into E. coli DH5α competent cells.
The pUX-T-flicUD plasmid served as a template to amplify the replacement/repair homologous arm using primers mfliC-down-F/mfliC-up-R. In parallel, the tac promoter was amplified from archived vector DNA using primers Tac-28a-F/Tac-28a-R. Using PET28a-mfliC3 as template, the mfliC3 sequences was amplified via primers 28a-tac-F/28a-tac-R. Purified tac promoter and mfliC3 fragments were directionally ligated to generate plasmid pUX-T-28a-tac-mfliC3, which was subsequently transformed into E. coli DH5α competent cells.
Using pUX-T-28a-tac-mfliC3 as template, the Tac-mfliC3 fusion fragment containing promoter sequences and SFB mfliC3 gene was amplified with primers mfliC-F/mfliC-R. Purified pUX-T-FliC-UD and Tac-mfliC3 fragments were seamlessly assembled via Gibson cloning to generate plasmid pUX-T-fliC-UD-tac-mfliC3, which was subsequently transform into E. coli DH5α for downstream processing.
Competent E. coli BW25113 cells were prepared for electroporation. The pUX-T-Flic-UD-tac-mfliC3 plasmid, alongside pCas9 and fliC-sgRNA, were co-electroporated into these cells. Successful SFB mfliC3 knock-in strains were screened using primers mfliC-JD-F/mfliC-JD-R. Post-selection, temperature-sensitive plasmid curing eliminated antibiotic resistance markers, plasmid-free strain BW25113ΔfliC:tac-mfliC3 was obtained. Primers’ sequences for genome editing are described in detail in Supplementary Table 1.
Bacteria-cell co-culture for adhesion assays
MODE-K and IEC-18 cell lines maintained in our laboratory were cultured in DMEM supplemented with 15% fetal bovine serum (FBS) and 1% penicillin-streptomycin. At 80% confluence, cells were PBS-washed, detached with 0.25% trypsin-EDTA, and subcultured at a 1:3 ratio into 12-well plates. Culture medium was refreshed every 2–3 days to preserve cell viability.
Bacterial cultures were grown to mid-exponential phase (OD600 ≈1.0), harvested via centrifugation (5,000 ×g, 10 min), and subsequently re-suspended in sterile DMEM for individual co-culture with MODE-K and IEC-18, respectively. Cell culture medium was changed to erythromycin-supplemented DMEM prior to bacterial inoculation. After gentle agitation to ensure homogeneity, cultures were incubated in a tri-gas chamber (5% CO2, 2% O2, 37°C). Following incubation, supernatants was aspirated and the cells were washed using phosphate-buffered saline (PBS). Adherent complexes were dissociated via vigorous pipetting in 1 mL sterile H2O, serially diluted (10-fold increments), and plated on erythromycin-containing M17 agar for quantitative adhesion assessment.
Recombinant Lactococcus administration in germ-free mice
Germ-free mouse colonization experiments were performed at Jiangsu GemPharmatech Biotechnology Co., Ltd. All procedures strictly followed institutional animal care protocols under sterile condition. Fifteen 5- to 7- week old male C57BL/6J mice were acclimated in isolators for one week and randomly allocated into five experimental groups (n=3 per group). Pre-colonization fecal pellets from individual mice underwent Gram staining to confirm axenic status.
Experimental groups were identified as follows: Group 1 (G1) received sterile PBS via oral gavage as the native control. Groups 2 to 5 (G2-G5) were administered recombinant Lactococcus strains Lac-eGFP, Lac-sal_fliC3, Lac-mfliC3, or Lac-rfliC3, respectively. Bacterial suspensions (2 × 10^8 CFU in 200 µL sterile PBS) were delivered on alternating (Days 1, 3, and 5) to reduce gastrointestinal stress.
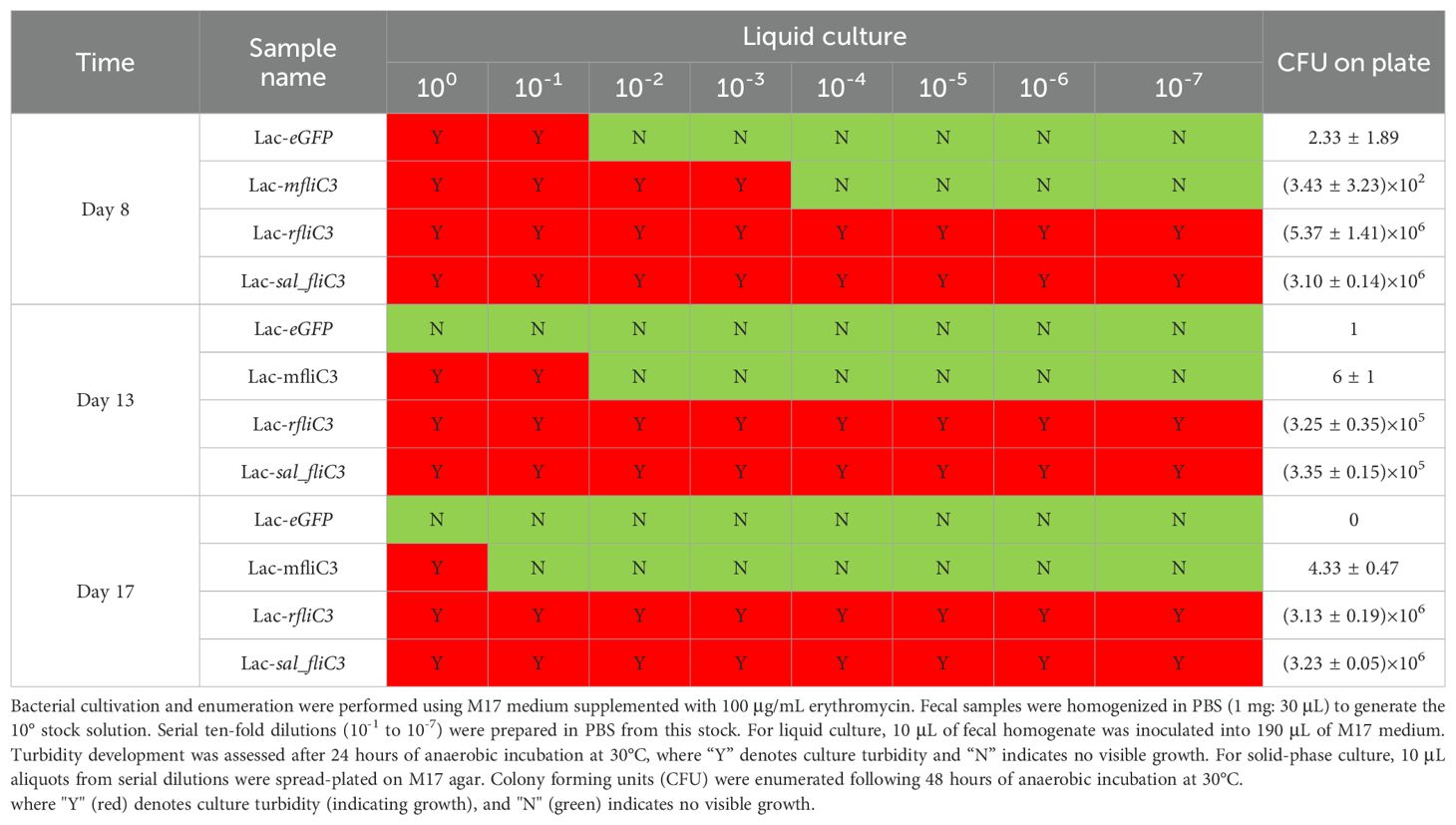
Fecal samples were harvested at post-administration Days 3 and 8 for microbial load quantification. On Day 12 post-final gavage, mice under CO2 asphyxiation, followed by cardiac puncture for serum isolation. Intestinal luminal contents were aseptically collected for bacteriological assessment. Terminal ileum tissues (1–2 cm proximal to the cecum) and hepatic tissues were dissected and immersion-fixed in 1 mL RNAlater stabilization solution. Samples were stabilized overnight at 4°C prior to cryopreservation at -80°C.
Detection of recombinant Lactococcus in fecal samples
Colonized fecal samples were harvested at designated intervals, homogenized in sterile PBS (1 mg: 30 μL), and serially diluted. Aliquots (10 µL) of the homogenate were air-dried on pre-cleaned microscope slides. Primary suspensions were further diluted (10-fold) and inoculated into erythromycin-supplemented M17 broth/agar (100 μg/mL). Cultures were incubated under strict anaerobic conditions at 30°C for 72 h, with growth kinetics monitored daily. Erythromycin-resistant colonies were quantified to determine viable bacterial loads.
Histopathological and immunohistochemical analysis of ileal tissues
Terminal ileum segments were flushed with ice-cold PBS and fixed in 10% neutral-buffered formalin post-dissection. Tissue processing included paraffin embedding for HE (Hematoxylin and Eosin) staining and immunohistochemical evaluation. For immunohistochemistry, formalin-fixed paraffin-embedded sections were underwent sequential dehydration through xylene and graded ethanol series, following by rehydration in distilled water. Heat-mediated antigen retrieval was carried out using sodium citrate buffer (pH 6.0) or Tri-EDTA solution (pH 9.0) via microwave irradiation. Sections were blocked with 5% bovine serum albumin (BSA) for 1 h prior to overnight incubation with the primary antibodies at 4°C. After PBS washing, fluorophore-conjugated secondary antibodies were applied under light-protected condition (1 h, RT). Nuclear counterstaining with DAPI preceded final mounting with anti-fade medium. Critical protocol considerations included maintaining section hydration throughout processing and minimizing light exposure post-secondary antibody application to preserve fluorescence integrity.
Quantification of immunoglobulins and Th17 pathway mediators
Serum immunoglobulin levels and Th17-associated cytokines were quantified using AAM-ISO-G1 and QAM-TH17–1 microarray systems, respectively, per manufacturer protocols. 1) Chip preparation: Ensure the glass slide chip is totally dry before use. 2) Antibody and sample preparation: Prepare biotin-labeled antibodies are add them to a 96-well plate, mix the diluted serum sample then with these antibodies and transfer this mixture to the antibody chip. 3) Incubation: Cover the glass slide with a sealing strip and incubate at room temperature for 1 hour. 4) Washing: Utilize the Thermo Scientific Wellwash Versa chip washer to thoroughly clean up the glass slides. 5) Scanning and detection: Employ the InnoScan 300 Microarray Scanner (Innopsys) for fluorescence signal scanning. Detect signals using the Cy3 or green channel (excitation frequency = 532 nm). 6) Data analysis: Analyze the results using either AAM-ISO-G1 or QAM-TH17–1 data analysis software.
Comparative transcription profiling of ileum and hepatic tissues
Total RNA isolation from tissue specimens was performed using TRIzol® Reagent (Invitrogen) with concomitant DNase I treatment (TaKaRa) for genomic DNA elimination. RNA integrity was checked via Bioanalyser 2100 electrophoresis (Agilent Technologies), while quantification utilized an ND-2000 spectrophotometric system (NanoDrop Technologies). RNA-seq libraries were constructed from 1 μg high-quality RNA employing the TruSeq™ RNA Sample Preparation Kit (Illumina, San Diego, CA). Polyadenylated mRNA was enriched through oligo (dT) magnetic bead selection and chemically fragmented. The cDNA synthesis, end repair, A-base addition, and ligation of Illumina-indexed adaptors followed the Illumina protocol. The libraries were size-selected for cDNA target fragments of 200–300 bps using 2% low range ultra agarose and then PCR-amplified with Phusion DNA polymerase (NEB) over 15 cycles. The libraries were quantified using a TBS380 fluorometer, and paired-end sequencing was conducted on the Illumina NovaSeq 6000 platform.
Raw paired-end sequencing reads underwent quality trimming and adapter removal using Trimmomatic with standard parameters (http://www.usadellab.org/cms/uploads/supplementary/Trimmomatic). High-quality reads were aligned to the reference genome in orientation-aware mode via TopHat (33) (http://tophat.cbcb.umd.edu/). Transcript abundance quantification employed FPKM (Fragments Per Kilobase of exon per Million mapped reads) normalization through Cuffdiff (34) (http://cufflinks.cbcb.umd.edu/). Differential gene expression analysis was conducted based on the criteria of a logarithmic fold change greater than 2 and a false discovery rate (FDR) of less than 0.05. Annotated keywords (UniProt) and enriched KEGG pathways were analyzed using the STRING database (https://cn.string-db.org/). This integrated approach elucidates transcriptome-level mechanistic disparities between ileal and hepatic tissues.
In vitro endocytosis assay
Following 4 h co-culture of E. coli BW25113ΔfliC:tac-mfliC3 with MODE-K cells in a 12-well plates, supernatants were aspirated and cells fixed with 4% paraformaldehyde (20 min, RT). Fixed monolayers underwent three PBS washes (10 min each) to eliminate residual fixative. Permeability with 0.2% Triton X-100 (15 min) preceded additional PBS washing. Non-specific binding was blocked with 5% BSA (30 min) prior to overnight incubation (4°C) with rabbit anti-mFliC3 polyclonal antibody in humidified chambers. After PBS washes, fluorophore-conjugated secondary antibodies were implemented light protected conditions (30 min, RT). Co-staining with anti-Lamp2 monoclonal antibody enabled endocytic compartment visualization. Final PBS rinses removed unbound antibodies, followed by DAPI nuclear counterstaining. Strict aseptic techniques and light-protected workflows were retained throughout.
Molecular docking and pull-down assay validation
Tertiary structures of mFliC3, rFliC3, endophilin A2, and αM integrin were predicted via AlphaFold2. Predicted models underwent structural refinement using the Protein Preparation Wizard module in Schrodinger software, which included protein pre-treatment, regeneration of native ligand states, optimization of hydrogen bonds, energy minimization, and removal of water molecules. These optimized protein structures were used for protein-protein interaction simulations, employing a protein-protein docking module set to rotate the probe 70,000 times and return a maximum of 30 conformations. The lower the interaction score, indicating lower binding free energy between the ligand and receptor, the higher the binding stability. Based on these protein-protein interaction simulations, the top-ranked conformation was selected for detailed analysis.
To further validate the molecular docking results, we first constructed the eukaryotic expression plasmid pCDNA3.1-endophilin A2-FAG. This plasmid was transfected into 293T cells used Lipo8000 ™ (Beyotime Biotechnology, Shanghai, China). Subsequently, we specifically captured and purified the Endophili A2 protein using Magneti-Q Anti-DYKDDDK Beads (Smart-Lifesciences, Changzhou, China). Finally, pull-down binding assays were performed using our laboratory-purified mFlC3 protein (29). The mFlC3 protein was detected via immunoblotting with rabbit anti-mFlC3 polyclonal antibody (29), HRP-conjugated goat anti-rabbit IgG (Wuhan Servicebio Technology Co., Ltd., Wuhan, China), and visualized using a high-sensitivity ECL chemiluminescence kit (Wuhan Servicebio Technology Co., Ltd., Wuhan, China).
Results
SFB FliC3 enhances L. lactis adhesion
The mfliC3 and rfliC3 genes from SFB were subcloned into the pAMJ399-eGFP expression vector, generating recombinant plasmids pAMJ399-mfliC3 and PAMJ399-rfliC3. Electroporation of these constructs into L. lactis MG1363 yield recombinant strains Lac-eGFP (empty vector control), Lac-mfliC3, and Lac-rfliC3. Transformants were checked through erythromycin resistance selection and Sanger sequencing of plasmid inserts. Growth curve analysis demonstrated that heterologous expression of mFliC3 and rFliC3 in L. lactis induced no significant growth impairment compared to the pAMJ399-eGFP control strain in M17 (Supplementary Figure 1) and cell culture media (Figures 1A, B), validating their experimental suitability for subsequent adhesion assays.
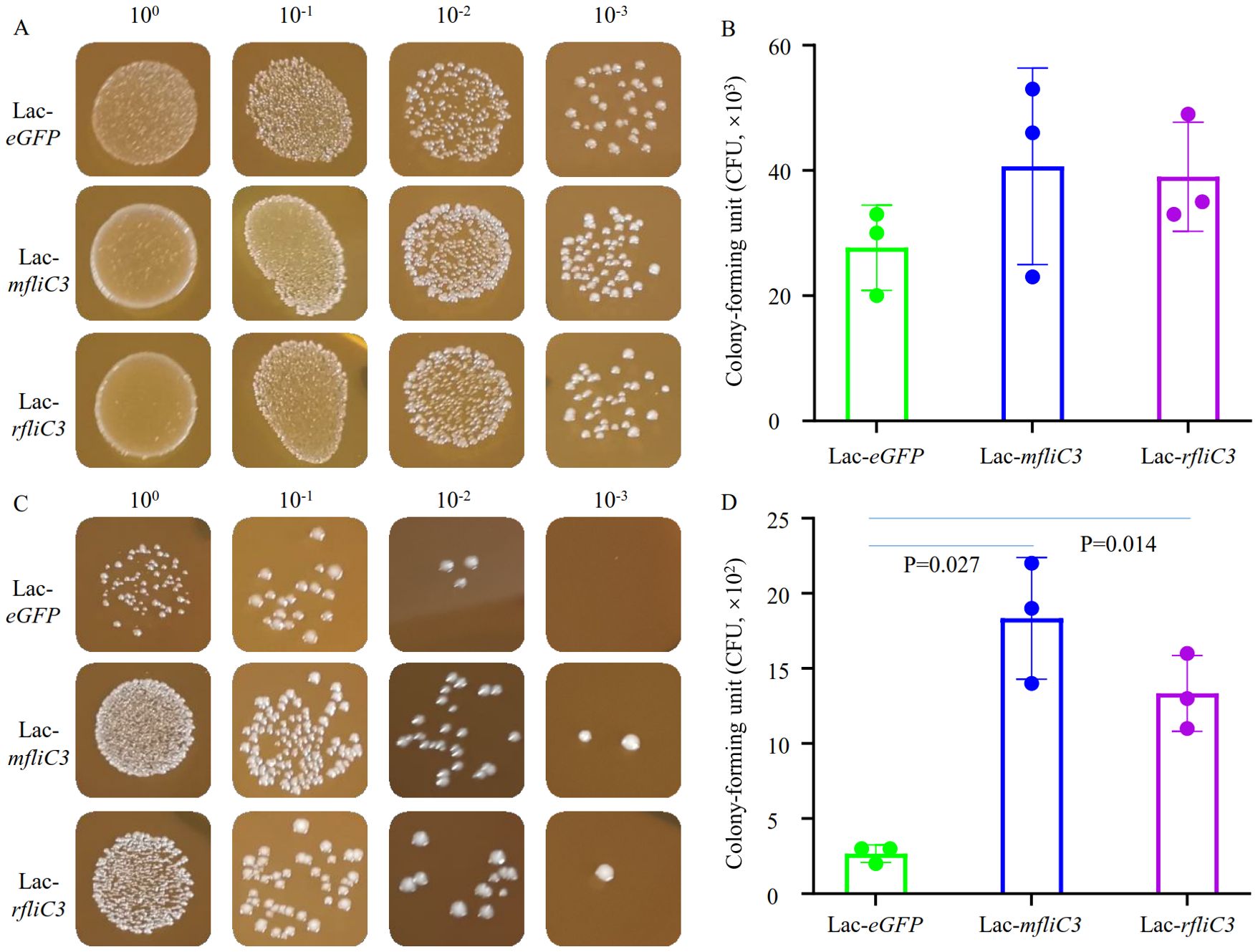
Figure 1. Adhesion analysis of recombinant Lactococcus to intestinal epithelial cells. Following 2-hour incubation with either DEME control medium or IEC-18 cells, recombinant strains (Lac-eGFP, Lac-mfliC3, Lac-rfliC3) were plated on M17 agar using culture supernatants or surface-adherent bacterial eluates. To quantify adherent bacteria, the co-culture supernatant was removed and the cells were gently rinsed three times with PBS. The cells were then resuspended in 1 mL of sterile water by pipetting (designating this suspension as the 100% concentration). Next, ten-fold serial dilutions of this suspension were prepared in sterile water and plated onto agar plates. Colony-forming units (CFUs) were enumerated after 18 hours of aerobic incubation at 37°C. (A, B) represent the bacterial colony-forming units (CFUs) after co-cultured with DEME medium. (C, D) represent the bacterial colony-forming unit (CFUs) after co-cultured with IEC-18. Statistical analysis was performed using Student’s t-test.
Adhesion assays involved co-culturing recombinant Lactococcus strains were here with murine (MODE-K) and rat (IEC-18) small intestinal epithelial cells. Following 1 h and 2 h co-culture periods, supernatants were aspirated and cells mechanically lysed in sterile PBS. Lysates were plated on erythromycin-supplemented M17 agar for quantification. Strikingly, mFliC3 and rFliC3 exhibited significantly improved bacterial adherence after 2 h exposure (Figures 1C, D). At 100-fold dilution, Lac-eGFP controls yielded only three CFUs, whereas Lac-mfliC3 and Lac-rfliC3 strains demonstrated robust colony formation. Under 1000-fold dilution conditions, mfliC3- and rfliC3- expressing strains retained detected colonies (1–2 CFU), while Lac-eGFP showed no viable counts (Figure 1B). Parallel results appeared in MODE-K cell-cocultures. As shown in Supplementary Figure 2, Lac-mfliC3 and Lac-rfliC3 strains exhibited markedly elevated CFU counts versus Lac-eGFP controls following 10-fold dilution of lysates after 1 h and 2 h co-culture periods. Moreover, ultrastructural analysis via electron microscopy confirmed recombinant Lactococcus adhesion to epithelial surface (Supplementary Figure 3). These observations demonstrate that SFB-derived mFliC3 and rFliC3 markedly potentiate adhesion to MODE-K and IEC-18 intestinal epithelial cells.
SFB FliC3 enhances prolonged fecal bacteria persistence in mono-associated mice
To assess SFB mFliC3 and rFliC3 in enhancing intestinal colonization of recombinant Lactococcus, germ-free mice were subjected to oral gavage and longitudinal colonization monitoring. Fifteen germ-free male C57BL/6J mice (6–8 weeks old) were randomly allocated into five groups (n=3). And pre-colonization fecal samples underwent Gram staining verification of axenic status. Daily oral inoculations (Days 1/3/5) of 200 µL bacteria (2 × 10^8 CFU/mL) or physiological saline was used for each mouse. Fecal specimens gathered in post-gavage days 8, 13, and 17 underwent selective plating to quantify recombinant Lactococcus persistence. As indicated in Table 1; Supplementary Figure 4, Lac-eGFP controls exhibited almost undetectable fecal titers by day 8 (72 h post-final inoculation). In contrast, Lac-mfliC3, Lac-rfliC3, and Lac-sal_fliC3 cohorts maintained significant colonization throughout the observation period. By Day 13 (8 days post-final inoculation), residual colonization persisted in the Lac-mfliC3 group, while Lac-rfliC3 and Lac-sal_fliC3 cohorts maintained significant colonization. At the terminal observation timepoint (day 17, 12 days post-gavage), Lac-mfliC3 titers fell below detection limits, whereas Lac-rfliC3 and Lac-sal_fliC3 groups retained persistent fecal titers. These data demonstrate that the SFB-derived mFliC3 and rFliC3 flagellins enhance recombinant Lactococcus intestinal persistence. Notably, rFliC3 exhibited superior colonization potential sustaining fecal excretion longer than mFliC3, suggesting enhanced ecological fitness for gastrointestinal niche adaptation.
Lac-rfliC3 modulates Th17 cell differentiation-associated gene expression in the murine ileum and hepatic tissues
Due to the absence of recombinant Lactococcus growth was observed in Lac-eGFP and Lac-mfliC3 groups by day 17 post-treatment, experimental animals were humanely euthanized for biological sample collection. Intestinal and hepatic RNA samples were subsequently isolated for comparative transcription profiling. As demonstrated in Supplementary Figure 5, substantial differential gene expression was observed in intestinal and hepatic tissues among experimental groups. Comparative analysis showed that small intestinal samples from Lac-mfliC3, Lac-rfliC3, Lac-sal_fliC3, and saline control groups exhibited 67, 75, 394, and 49 significantly differentially expressed genes, respectively, relative to the Lac-eGFP control. Corresponding hepatic samples demonstrated 135, 115, 414, and 72 differentially expressed genes across these comparative groups.
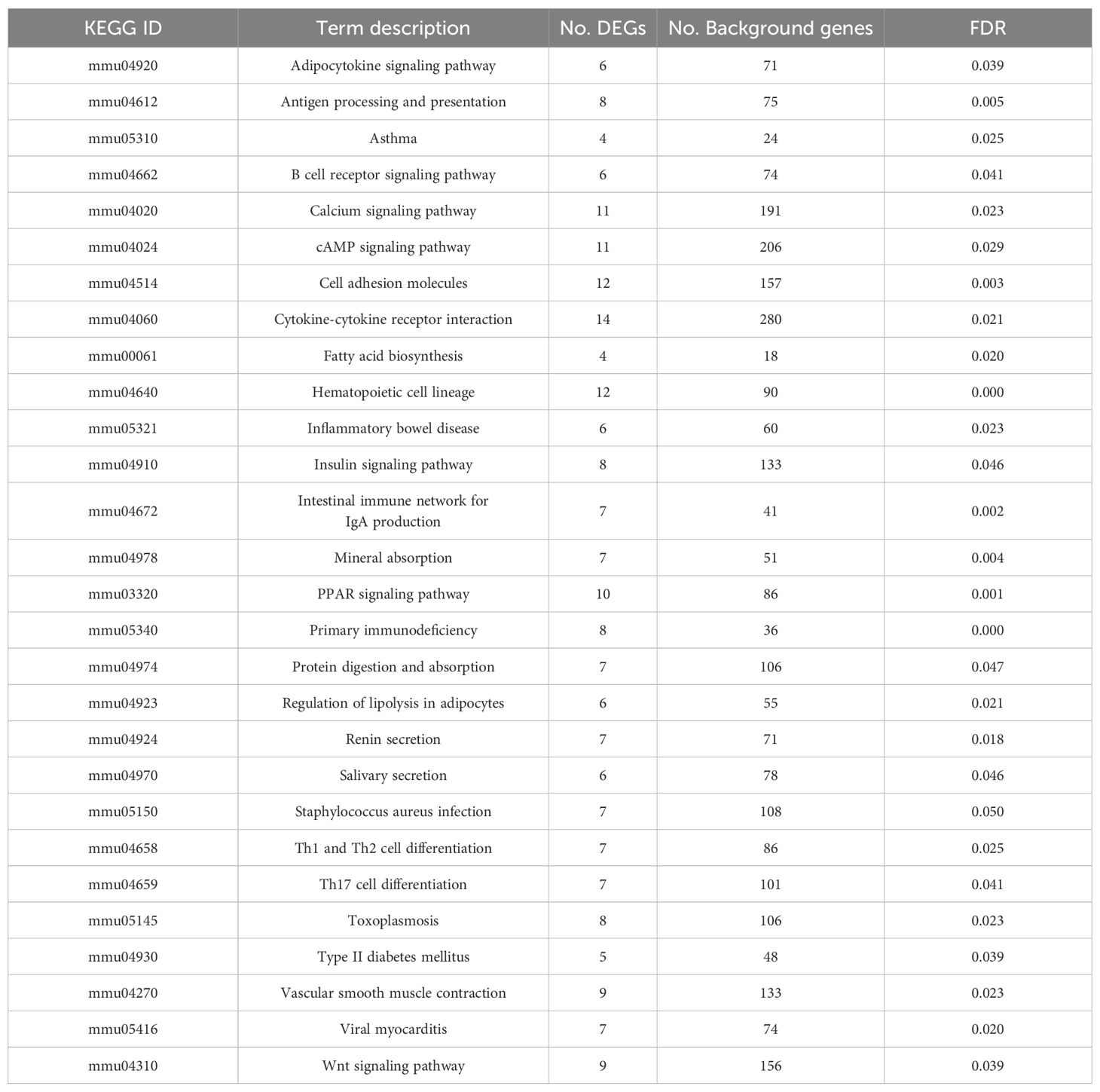
Functional annotation of differentially expressed genes (DEGs) were performed using STRING database. UniProt keyword analysis revealed no signaling-related DEGs in sterile control. In contrast, intestinal samples from Lac-mfliC3, Lac-rfliC3, and Lac-sal_fliC3 groups exhibiting 26, 130, and 29 signaling-related DEGs, respectively. KEGG pathway enrichment analysis identified group-specific biological signatures: while saline vs Lac-mfliC3 comparisons showed no pathway enrichment, the Lac-rfliC3 group displayed significant enrichment in key immunological pathways including cell adhesion, antigen processing and presentation, the intestinal mucosal immune IgA network, and Th1/Th2/Th17 cell differentiation (Table 2). Supplementary Table 2 reveals distinct pathway activation patterns, with Lac-sal_fliC3 demonstrating significant enrichment in cell adhesion and Th1/Th2 differentiation pathways, yet lacking critical enrichments were observed in Lac-rfliC3 group including antigen processing and presentation, intestinal mucosal immune IgA network, and Th17 differentiation pathways. This differential enrichment profile highlights the unique regulatory capacity of Lac-rfliC3 in mucosal immunity. Hepatic transcription analysis via STRING uncovered exclusive signaling features in Lac-rfliC3 samples, with 90 signaling-related DEGs predominantly enriched in Th17-associated pathways - including Th17 differentiation and IL-17 signaling (Supplementary Table 3). However, these molecular signatures absent in other experimental groups (Supplementary Table 3).
To assess the safety profile of these recombinant Lactococcus strains, ileal tissue sections were stained to evaluate immune cell infiltration, goblet cell depletion, and villus architecture integrity. As indicated in Supplementary Figure 6E, gastric gavage of recombinant Lactococcus induced no significant histopathological alterations in the upper ileum compared with the saline control. Although moderate inflammatory response and intestinal villus architecture disorder were observed in the terminal ileum of the Lac-eGFP group (Supplementary Figure 6F), the lesions induced by recombinant Lactococcus expressing flagellar proteins were mild (Supplementary Figure 6F). Overall, these recombinant Lactococcus expressing bacterial flagellin did not induce significant adverse effects.
SFB FliC3-engineered Lactococcus enhances Th17-associated cytokine production
Serum immunoglobulin profiling and Th17-related cytokine quantification were carried out using GSM-TH17 and AAM-ISO-G1 assay systems. As shown in Table 3, Lac-mfliC3, Lac-rfliC3, and Lac-sal_fliC3 groups exhibited about two-fold higher serum IgG2a and IgG3 concentrations compared with Lac-eGFP controls. Other immunoglobulin isotypes (IgA, IgD, IgE, and IgM) remained comparable across all experimental groups.
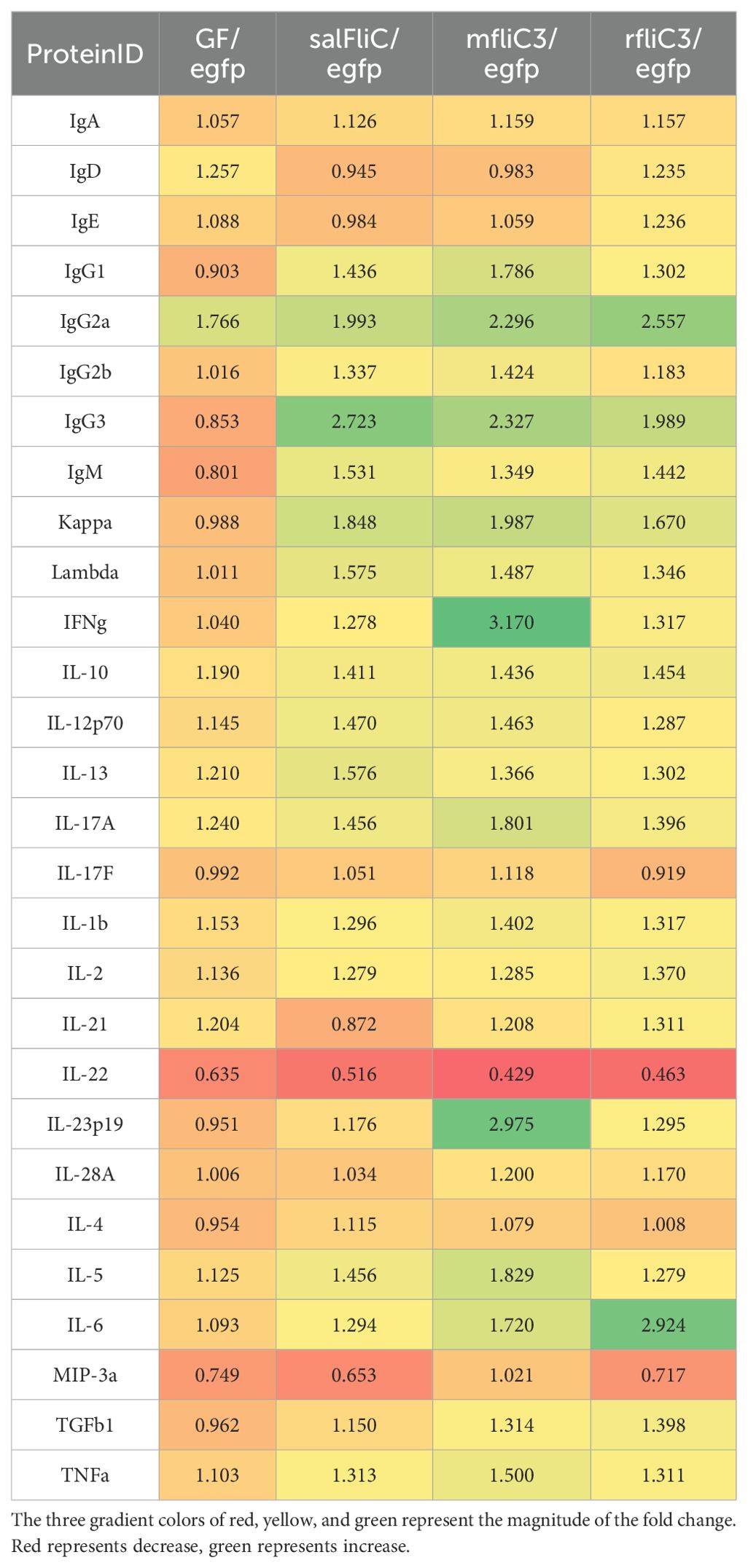
Table 3. Detection of the effects of recombinant Lactococcus on serum immunoglobulin and Th17 cytokines.
Cytokine profiling revealed strain-specific activation patterns: The Lac-mfliC3 group demonstrated striking elevations (>1.5-fold vs Lac-eGFP control) in IFNγ, IL-17A, IL-23p19, IL-5, IL-6, and TNFα levels. In contrast, Lac-rfliC3 administration selectively enhanced IL-6 production, while comparative analysis against Lac-sal_fliC3 group identified exclusive upregulation of IL-13 in the Lac-salFliC3-treated mice. These differential cytokine signatures delineate strain-specific immunomodulatory capacities within the Th17 axis.
Co-localization analysis of SFB Flic3-engineered Lactococcus with endocytosis biomarker Lamp2
Lysosome-associated membrane protein (LAMP1/2) is crucial for the lysosome function and stability (35). Given that SFB antigens were internalizes into cells through endocytosis to exert immune regulatory effects, and the lysosome marker LAMP2 associated with cell adhesion is involved in this process (15), we investigated the endocytic capacity of SFB fliC3-expressing Lactococcus by using dual immunofluorescence staining with anti-SFB FliC3 polyclonal antibody and anti- Lamp2 monoclonal antibody (supplied by DSHB, ABL-93). Immunohistochemistry analysis revealed significant co-localization between rfliC3-expressing Lactococcus strains and Lamp2+ endosomes (Figure 2). This spatial association suggests functional conservation with native SFB in host cell internalization mechanisms.
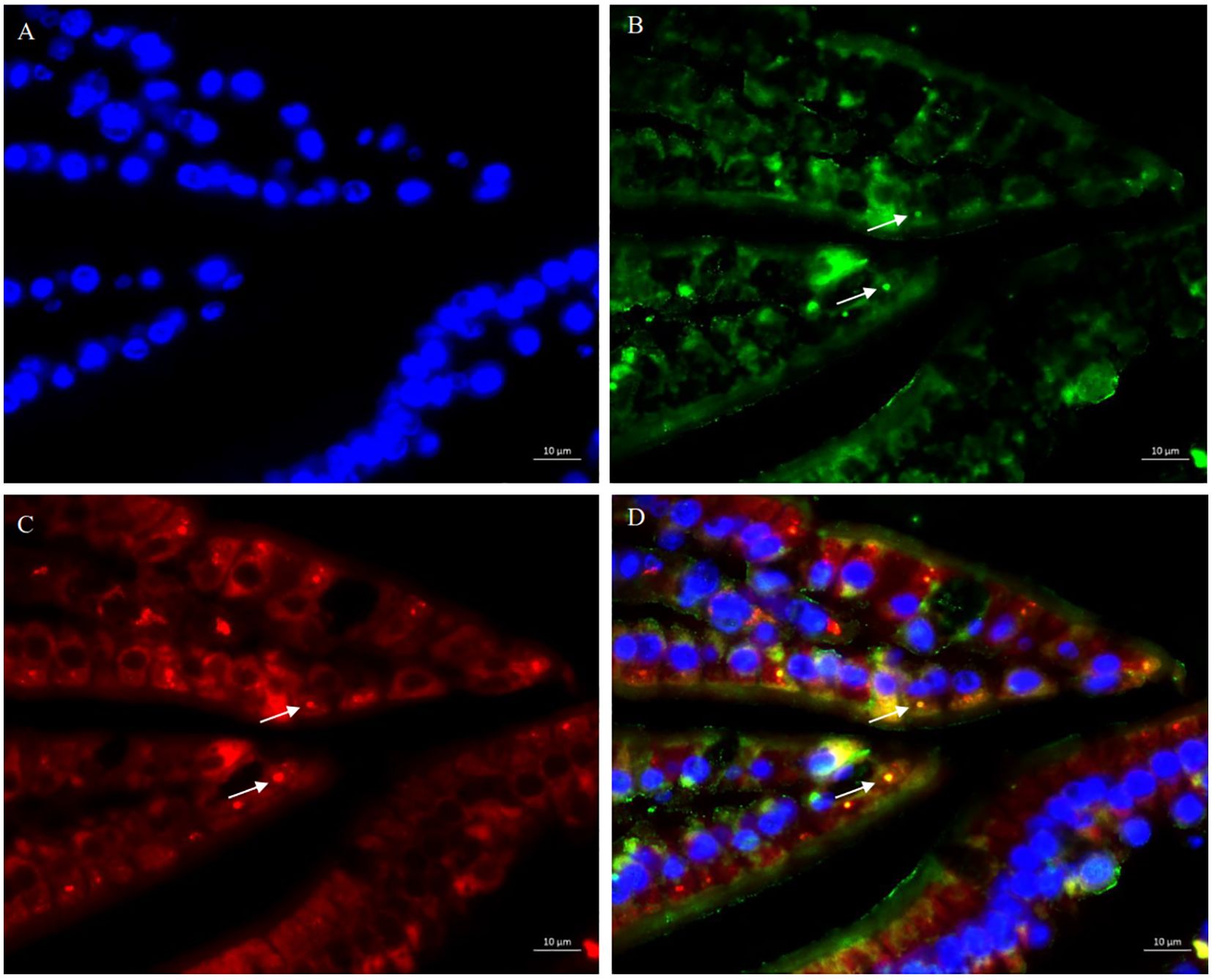
Figure 2. Co-localization analysis of SFB rFliC3 with endosomal marker LAMP2. Ileal tissues were fixed in 4% paraformaldehyde and sequentially immune-stained with rabbit anti-SFB FliC3 primary antibody, anti-LAMP2 antibody, and FITC-conjugated goat anti-rabbit IgG secondary antibody. Nuclei were counterstained with DAPI. Fluorescence imaging was performed using an upright microscope (Zeiss Axio Imager Z2). (A) DAPI staining results of ileal tissue; (B) Anti-SFB FliC3 antibody staining results of ileal tissue; (C) Anti-LAMP2 antibody staining results of ileal tissue; (D) Merged images of (A–C) demonstrating co-localization signals (yellow) of FliC3 and LAMP2.
MODE-K cell internalization of mFliC3 engineered E. coli
Since Lac-mfliC3 was not detected in terminal mouse fecal samples (Supplementary Figure 4) and the co-localization fluorescence signals were weak following co-culture with MODE-K cells (Supplementary Figure 7), we therefore assessed its cellular internalization potential using co-culture experiments with engineered E. coli. The BW25113 strain was genetically engineered via fliC locus replacement with SFB-derived mfliC3. Despite similar growth dynamics across these strains (Supplementary Figure 8), phenotypic characterization revealed subtle morphological alterations: deletion of fliC slightly reduced colony size while maintaining defined margins (Supplementary Figure 9). Successful knock-in of mfliC3 preserved colony size but induced a distinct undulating morphology, contrasting with the architecture of the parental strain.
Further PCR validation and Sanger sequencing confirmed successful site-specific allelic replacement (Supplementary Figure 10). Following 12-h co-culture of MODE-K cells with BW25113ΔfliC: tac mfliC3, dual immunofluorescence microscopy using (1) SFB FliC3-specific and (2) Lamp2-targeting antibodies revealed bacterial co-localization with Lamp2+ endosomal compartments (Figure 3). Notably, intracellular mFliC3 epitopes were detected, suggestive of flagellin translocation through endocytic machinery in intestinal epithelial cells.
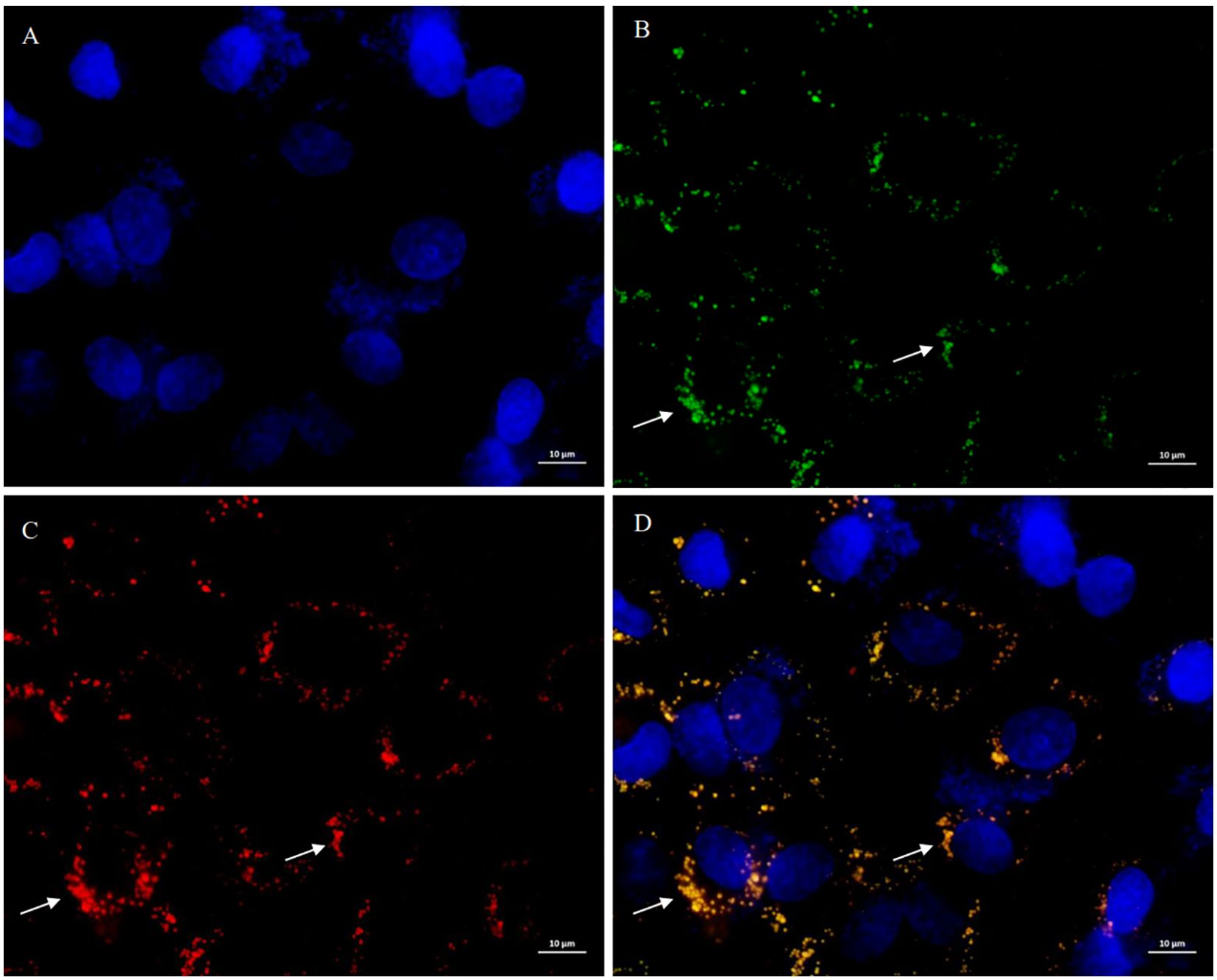
Figure 3. Subcellular localization of mFliC3 in MODE-K murine intestinal epithelial cells. MODE-K cells were incubated with mfliC3-expressing E. coli (BW25113ΔfliC:tac-mfliC3) for 12 hours. Following PBS washing, triple immunofluorescence staining was performed using DAPI nuclear counterstain (blue), anti-mFliC3 antibody (green), and anti-LAMP2 endosomal marker (red). (A) DAPI staining results of cells; (B) Anti-SFB FliC3 antibody staining results of cells; (C) Anti-LAMP2 antibody staining results of cells; (D) Merged images of (A–C) demonstrating co-localization signals (yellow) of FliC3 and LAMP2. (D) shows merged fluorescence channels. Specimens were mounted in anti-fade medium and imaged by confocal microscopy (scale bar: 10 μm).
Structural prediction and molecular docking of SFB FliC3 interactions with Endophilin-A2 and αM integrin
By integrating AlphaFold2-derived structural predictions with Schrödinger-based molecular docking simulations, we established SFB FliC3’s ability to form stable intermolecular complexes with Endophilin-A2 and αM integrin. The predominant docking conformations demonstrated substantial binding stability for mFliC3-Endophilin-A2 (PIPER score: -351.268; interaction energy: -1023.01 kcal/mol) and mFliC3-αM integrin (PIPER score: -253.862; interaction energy: -1036.51 kcal/mol). Interface characterization revealed three distinct interaction modes between mFliC3 and Endophilin-A2: Arg328 formed a hydrogen bond with Asp360, Asp316 involved in dual interactions (hydrogen bond and salt bridge) with Lys363, and Phe289 established a hydrogen bond with Gln376 (Figure 4A). Similarly, αM integrin interactions featured Ser966 hydrogen-bonded with Ile395, while Arg759 formed three molecular contacts - one with Asn372 and two with Gln23 (Figure 4B). These structural analyses illuminate the multivalent binding architecture underlying mFliC3-mediated host protein engagement in cellular systems. The pull-down assay further validated the interaction between Endophilin-A2 and mFliC3 (Supplementary Figure 11).
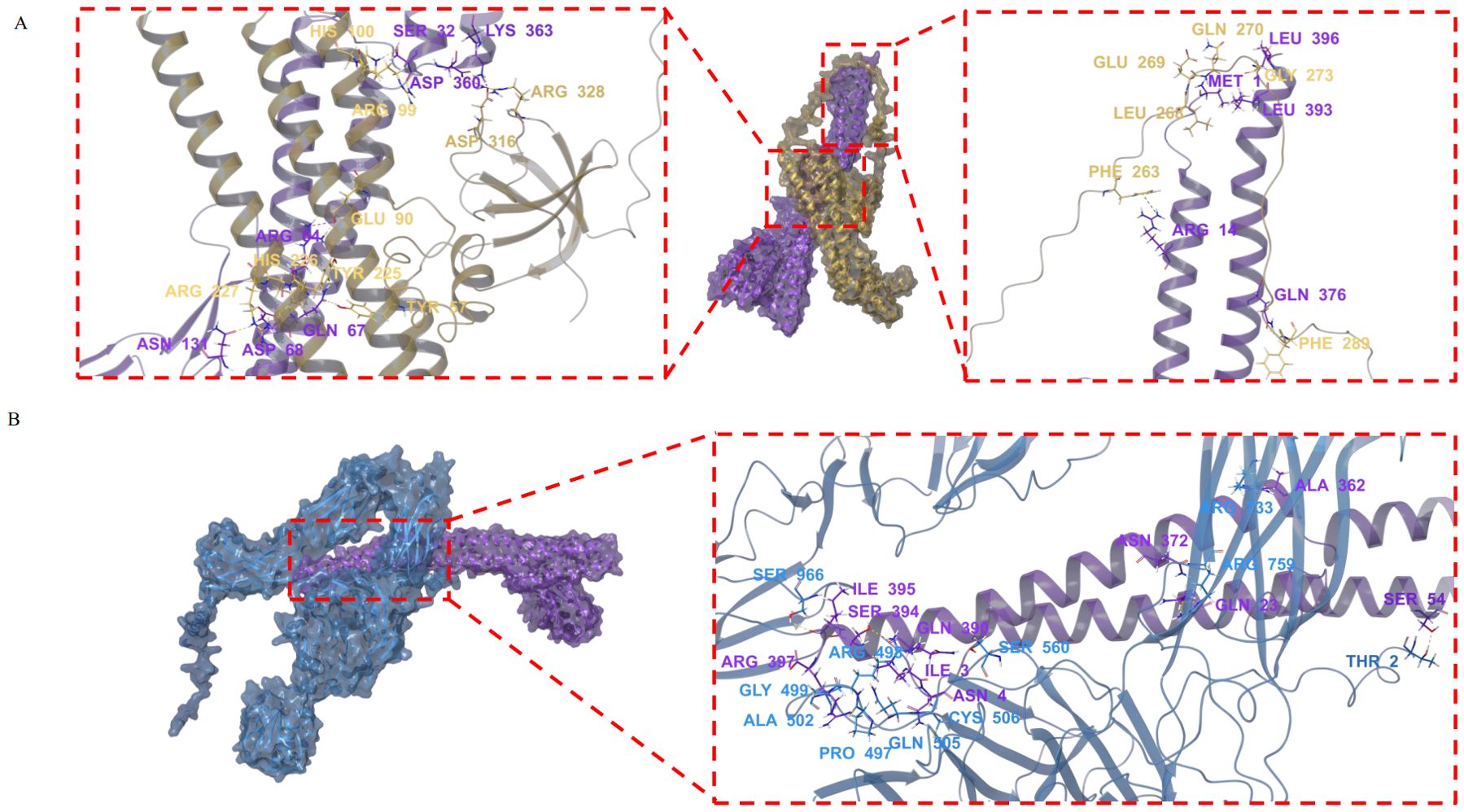
Figure 4. Molecular docking analysis of mFliC3 interactions with Endophilin A2 and αM integrin. The nucleotide sequence of M5I-FliC3-2 (KX658895.1), the protein sequences of Endophilin A2 (NP_038692.1) and αM integrin (P05555.2) were download from NCBI database. The tertiary structures of these genes were predicted by using AlphaFold2. Molecular docking analysis was performed then using the Protein Preparation Wizard module in Schrodinger software. Structural models were energy-minimized and visualized using PyMOL. (A) Predicted binding interface between mFliC3 (purple) and Endophilin A2 (yellow). (B) Molecular interaction profile of mFliC3 (purple) with αM integrin (blue).
Discussion
Evolutionary and functional characterization of SFB flagellin-mediated host-microbe interactions
Bacterial adhesion is a critical determinant for host colonization, tissue invasion, and biofilm formation. While microbial colonization mechanisms remain incompletely understood, emerging evidence implicates flagellar components as key mediators. Comparative genomic analyses reveal conserved roles of flagellin genes in enteric colonization across diverse pathogens including Salmonella Enteritidis (36), Campylobacter jejuni (37), and E. coli (38, 39) – a paradigm recently extended to gut commensals (40). Our phylogenetic studies of rodent-specific SFB flagellins demonstrated host-driven evolutionary divergence, with murine and rat variants forming distinct clades at both nucleotide and protein levels (29). Functional characterization revealed mFliC3/rFliC3 equivalently enhanced microbial adhesion to murine (MODE-K) and rat (IEC-18) intestinal epithelia in vitro (Figure 1), contrasting with predicted host specificity based on evolutionary patterns. This phenotypic divergence from phylogenetic predictions suggests micro-environmental factors (e.t., host mucosal proteases) may modulate flagellin functionality in vivo, as demonstrated by rFliC3’s accelerated degradation versus mFliC3 in mucosal protein co-incubation assays (29).
Dual-phase dynamics of flagellin-engineered Lactococcus colonization and immune regulation
Both mfliC3 and rfliC3-expressing Lactococcus strains demonstrated enhanced intestinal persistence in vivo, with rfliC3 variants exhibiting prolonged fecal shedding (detectable until day 12 post-gavage cessation) compared to mfliC3 (day 8) and empty vector controls (day 3). This colonization pattern inversely correlated with host immune activation: Multiplex serum analysis revealed robust Th17-polarized responses in mFliC3-administered mice, including elevated IL-17A, IL-23p19, and TNFα versus controls (Table 3). In contrast, rFliC3 exposure elicited selective IL-6 upregulation without broad cytokine induction. We propose this differential immunogenicity drives strain-specific clearance kinetics (38, 41, 42). The relative low level of IL-6 concentration in the mFliC3 group maybe due to the reduced colonization of Lac-mflic3 that was verified by undetectable fecal colonies and absent mFliC3-LAMP2 co-localization by day 17. This aligns with established flagellar immunobiology: While flagellins facilitate mucus penetration through adhesive domains, their recognition as MAMPs triggers TLR5/NF-KB-mediated elimination - a self-limiting mechanism explaining SFB’s age-dependent colonization dynamics (43). Given the persistent colonization observed for Lac-sal_fliC3 in this study (Table 1; Supplementary Figure 4), the relatively low IL-6 concentration in the Lac-sal_fliC3 group may reflect the dynamic fluctuations of plasma IL6 levels following recombinant Salmonella flagellin administration. As reported by Rolli et al., plasm IL-6 peaks within one hour post-stimulation by Salmonella flagellin and declines steady, returning to baseline within three hours (44). However, further investigations is required to determine whether rFliC3 can sustain IL-6 production in serum.
Temporal resolution of Th17-associated transcriptional and translational responses
This apparent incongruity between serum cytokine elevations and transcription profiles may reflect sampling chronology. By day 13 post-inoculation, recombinant bacterial loads had substantially reduced, reaching undetectable levels in fecal samples by the terminal sampling timepoint (day 17). Consequently, ileal and hepatic transcriptional signatures analyzed at day 17 likely represent post-clearance homeostasis, with Th17 pathway activation having returned to baseline levels.
Multilateral immune modulation by rFliC3-engineered Lactococcus
Our investigation demonstrates that Lac-rFliC3 administration in gnotobiotic mice induces Th17 pathway enrichment in ileum and hepatic transcriptions, corroborating previous findings on SFB flagellin-mediated Th17 differentiation. Immunofluorescence co-localization of Lac-rFliC3 with LAMP2+ ednosomes substantiates its cellular internalization capacity. Paradoxically, serum proteomics analysis revealed selective IL-6 evaluation without broad Th17-associated cytokine induction, despite pronounced transcriptions alterations. This dichotomy may stem from multifactorial regulation: (1) enhanced rFliC3 proteolytic susceptibility in mucosal environments weakens its immune regulatory function (29); (2) competing immune activation evidenced by ileal enrichment of Th1/Th2 differentiation and primary immunodeficiency pathways; (3) network-level counter-regulation through cytokine cross-talk mechanisms.
Mechanistic insights into SFB flagellin-mediated endocytic trafficking
Endocytosis serves essential roles in cellular homeostasis by mediating extracellular substance uptake and intercellular communication. While clathrin-mediated endocytosis is the canonical pathway, emerging research has characterized alternative internalization mechanisms, including fast endophillin-mediated endocytosis (FEME) (45). This clathrin-independent pathway, orchestrated by endophilin A2, enables rapid ligand-receptor complex internalization following specific surface receptor activation (46). The FEME machinery exhibits three hallmark features (45): (1) membrane remodeling, endophilin A2 induces localized plasma membrane curvature through its BAR domain-mediated lipid binding; (2) cargo selectivity, phosphorylation-dependent recognition of activated receptors ensures substrate specificity; (3) vesicle maturation, coordinated interactions with dynamic GTPase and CDC42 small GTPase facilitate vesicle scission and cytoskeletal transport.
Pioneering electron tomography studies by Ladinsky et al. revealed that SFB orchestrates antigen transfer through plasma membrane-associated endocytic vesicle formation at the host-microbe interface, rather than direct cellular invasion (15). This clathrin-independent process requires dynamic GTPase and CDC42-mediated actin remodeling, suggesting evolutionary convergence with fast endophilin-mediated endocytosis (FEME) mechanisms. Our molecular docking data demonstrate high-affinity interactions between SFB FliC3 and two key FEME components further supports FEME-like internalization (Figure 3). While Salmonella flagellin utilizes Hsp90-Hsp70-αM/β2 integrin complexes for endocytosis, SFB appears to employ distinct machinery, potentially involving: Cytoskeletal regulators (co-precipitated with FliC3 in prior studies), heat shock protein networks, integrin-mediated adhesion platforms. However, based on current experimental evidence, it remains undetermined whether endocytosis of SFB flagellin depends on clathrin-associated pathway. The precise endocytosis mechanism requires further experimental elucidation.
In conclusion: as illustrated in Graphical abstract, SFB flagellin FliC3 enhances bacterial epithelial adhesion and activates Th17-associated immune signaling. Mechanistically, FliC3 may undergo endophilin A2-mediated endocytosis to exert immunomodulatory effects, as supported by molecular docking and Lamp2 co-localization evidence. Further studies employing targeted genetic manipulation of SFB flagellar components in axenic cultures, combined with longitudinal sampling in conventional and gnotobioitc murine models following recombinant strain administration, will elucidate the spatiotemporal dynamics of FliC3-mediated immunoregulation.
Data availability statement
Sequence data that support the findings of this study have been deposited in the NCBI SRA database with the primary accession code PRJNA1109453 and PRJNA1108060.
Ethics statement
The animal study was approved by Experimental Animal Use and Management Committee of Jiangsu GemPharmatech Biotechnology Co., Ltd. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
YY: Writing – original draft, Funding acquisition, Conceptualization, Supervision, Writing – review & editing, Investigation. HC: Methodology, Writing – original draft, Funding acquisition, Data curation, Investigation. LW: Investigation, Writing – original draft, Data curation. XC: Writing – original draft, Data curation, Investigation. ZL: Data curation, Methodology, Writing – original draft, Investigation. RZ: Investigation, Writing – original draft, Data curation. JL: Funding acquisition, Writing – original draft, Data curation, Methodology. ZW: Funding acquisition, Writing – original draft, Data curation, Methodology. DY: Methodology, Writing – review & editing, Data curation. XW: Investigation, Data curation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This study was funded by the National Natural Science Foundation of China (Nos.31800119 and 32460015), the Natural Science Foundation of Hunan Province, China (No. 2018JJ3200), and the open fund of Guangxi Key Laboratory of Veterinary Biotechnology (No. 19-50-40-B-05).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1624092/full#supplementary-material
References
1. Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. (2009) 31:677–89. doi: 10.1016/j.immuni.2009.08.020
2. Fagarasan S, Kawamoto S, Kanagawa O, and Suzuki K. Adaptive immune regulation in the gut: T cell-dependent and T cell-independent IgA synthesis. Annu Rev Immunol. (2010) 28:243–73. doi: 10.1146/annurev-immunol-030409-101314
3. Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. (2009) 139:485–98. doi: 10.1016/j.cell.2009.09.033
4. Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. (2011) 331:337–41. doi: 10.1126/science.1198469
5. Brooks JN, Behrendt CL, Ruhn KA, Lee S, Raj P, Takahashi JS, et al. The microbiota coordinates diurnal rhythms in innate immunity with the circadian clock. Cell. (2021) 184:4154–67. doi: 10.1016/j.cell.2021.07.001
6. Gauguet S, D’Ortona S, Ahnger-Pier K, Duan B, Surana NK, Lu R, et al. Intestinal microbiota of mice influences resistance to Staphylococcus aureus pneumonia. Infect Immun. (2015) 83:4003–14. doi: 10.1128/IAI.00037-15
7. Shi Z, Zou J, Zhang Z, Zhao X, Noriega J, Zhang B, et al. Segmented filamentous bacteria prevent and cure Rotavirus infection. Cell. (2019) 179:644–58. doi: 10.1016/j.cell.2019.09.028
8. Ngo VL, Shi Z, Jiang B, and Gewirtz AT. Segmented filamentous bacteria impede rotavirus infection via retinoic acid receptor-mediated signaling. Gut Microbes. (2023) 15:2174407. doi: 10.1080/19490976.2023.2174407
9. Tyagi AM, Darby TM, Hsu E, Yu M, Pal S, Dar H, et al. The gut microbiota is a transmissible determinant of skeletal maturation. Elife. (2021) 10:e64237. doi: 10.7554/eLife.64237
10. Hathaway-Schrader JD, Carson MD, Gerasco JE, Warner AJ, Swanson BA, Aguirre JI, et al. Commensal gut bacterium critically regulates alveolar bone homeostasis. Lab Invest. (2022) 102:363–75. doi: 10.1038/s41374-021-00697-0
11. Dar HY, Perrien DS, Pal S, Stoica A, Uppuganti S, Nyman JS, et al. Callus gammadelta T cells and microbe-induced intestinal Th17 cells improve fracture healing in mice. J Clin Invest. (2023) 133:e16577. doi: 10.1172/JCI166577
12. Metwaly A, Jovic J, Waldschmitt N, Khaloian S, Heimes H, Hacker D, et al. Diet prevents the expansion of segmented filamentous bacteria and ileo-colonic inflammation in a model of Crohn’s disease. Microbiome. (2023) 11:66. doi: 10.1186/s40168-023-01508-y
13. Bates NA, Li A, Fan T, Cutcliffe MP, Dagenet CB, Sleiman KC, et al. Gut commensal segmented filamentous bacteria fine-tune T follicular regulatory cells to modify the severity of systemic autoimmune arthritis. J Immunol. (2021) 206:941–52. doi: 10.4049/jimmunol.2000663
14. Teng F, Klinger CN, Felix KM, Bradley CP, Wu E, Tran NL, et al. Gut microbiota drive autoimmune arthritis by promoting differentiation and migration of Peyer’s Patch T follicular helper cells. Immunity. (2016) 44:875–88. doi: 10.1016/j.immuni.2016.03.013
15. Ladinsky MS, Araujo LP, Zhang X, Veltri J, Galan-Diez M, Soualhi S, et al. Endocytosis of commensal antigens by intestinal epithelial cells regulates mucosal T cell homeostasis. Science. (2019) 363:eaat4042. doi: 10.1126/science.aat4042
16. Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, et al. Th17 Cell induction by adhesion of microbes to intestinal epithelial cells. Cell. (2015) 163:367–80. doi: 10.1016/j.cell.2015.08.058
17. Sidner B, Lerma A, Biswas B, Do T, Yu Y, Ronish LA, et al. Flagellin is essential for initial attachment to mucosal surfaces by Clostridioides difficile. Microbiol Spectr. (2023) 11:e0212023. doi: 10.1128/spectrum.02120-23
18. Akahoshi DT and Bevins CL. Flagella at the Host-microbe interface: Key functions intersect with redundant responses. Front Immunol. (2022) 13:828758. doi: 10.3389/fimmu.2022.828758
19. Vijay-Kumar M, Bovilla VR, Yeoh BS, Golonka RM, Saha P, Joe B, et al. Bacterial flagellin is a dominant, stable innate immune activator in the gastrointestinal contents of mice and rats. Gut Microbes. (2023) 15:2185031. doi: 10.1080/19490976.2023.2185031
20. Clasen SJ, Bell M, Borbon A, Lee DH, Henseler ZM, de la Cuesta-Zuluaga J, et al. Silent recognition of flagellins from human gut commensal bacteria by Toll-like receptor 5. Sci Immunol. (2023) 8:eabq7001. doi: 10.1126/sciimmunol.abq7001
21. Scheithauer T, Herrema H, Yu H, Bakker GJ, Winkelmeijer M, Soukhatcheva G, et al. Gut-derived bacterial flagellin induces beta-cell inflammation and dysfunction. Gut Microbes. (2022) 14:2111951. doi: 10.1080/19490976.2022.2111951
22. Imai J, Kitamoto S, Sugihara K, Nagao-Kitamoto H, Hayashi A, Morhardt TL, et al. Flagellin-mediated activation of IL-33-ST2 signaling by a pathobiont promotes intestinal fibrosis. Mucosal Immunol. (2019) 12:632–43. doi: 10.1038/s41385-019-0138-4
23. Alexander KL, Zhao Q, Reif M, Rosenberg AF, Mannon PJ, Duck LW, et al. Human microbiota flagellins drive adaptive immune responses in Crohn’s disease. Gastroenterology. (2021) 161:522–35. doi: 10.1053/j.gastro.2021.03.064
24. Boardman DA, Wong MQ, Rees WD, Wu D, Himmel ME, Orban PC, et al. Flagellin-specific human CAR Tregs for immune regulation in IBD. J Autoimmun. (2023) 134:102961. doi: 10.1016/j.jaut.2022.102961
25. Kuwahara T, Ogura Y, Oshima K, Kurokawa K, Ooka T, Hirakawa H, et al. The lifestyle of the segmented filamentous bacterium: a non-culturable gut-associated immunostimulating microbe inferred by whole-genome sequencing. DNA Res. (2011) 18:291–303. doi: 10.1093/dnares/dsr022
26. Prakash T, Oshima K, Morita H, Fukuda S, Imaoka A, Kumar N, et al. Complete genome sequences of rat and mouse segmented filamentous bacteria, a potent inducer of th17 cell differentiation. Cell Host Microbe. (2011) 10:273–84. doi: 10.1016/j.chom.2011.08.007
27. Sczesnak A, Segata N, Qin X, Gevers D, Petrosino JF, Huttenhower C, et al. The genome of th17 cell-inducing segmented filamentous bacteria reveals extensive auxotrophy and adaptations to the intestinal environment. Cell Host Microbe. (2011) 10:260–72. doi: 10.1016/j.chom.2011.08.005
28. Chen B, Chen H, Shu X, Yin Y, Li J, Qin J, et al. Presence of segmented filamentous bacteria in human children and its potential role in the modulation of human gut immunity. Front Microbiol. (2018) 9:1403. doi: 10.3389/fmicb.2018.01403
29. Chen H, Yin Y, Wang Y, Wang X, and Xiang C. Host specificity of flagellins from segmented filamentous bacteria affects their patterns of interaction with mouse ileal mucosal proteins. Appl Environ Microbiol. (2017) 83:e01061-17. doi: 10.1128/AEM.01061-17
30. Nkamba I, Mulet C, Dubey GP, Gorgette O, Couesnon A, Salles A, et al. Intracellular offspring released from SFB filaments are flagellated. Nat Microbiol. (2020) 5:34–9. doi: 10.1038/s41564-019-0608-1
31. Wang Y, Yin Y, Chen X, Zhao Y, Wu Y, Li Y, et al. Induction of intestinal Th17 cells by flagellins from segmented filamentous bacteria. Front Immunol. (2019) 10:2750. doi: 10.3389/fimmu.2019.02750
32. Holo H and Nes IF. Transformation of Lactococcus by electroporation. Methods Mol Biol. (1995) 47:195–99. doi: 10.1385/0-89603-310-4:195
33. Trapnell C, Pachter L, and Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. (2009) 25:1105–11. doi: 10.1093/bioinformatics/btp120
34. Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, and Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. (2013) 31:46–53. doi: 10.1038/nbt.2450
35. Xu C, Yu C, Zhang J, Zhang Z, Zhang J, Meng Y, et al. YTHDF1 differentiates the contributing roles of mTORC1 in aging. Mol Cell. (2025) 85:2194–210. doi: 10.1016/j.molcel.2025.05.003
36. Barbosa FO, Freitas NO, Batista D, Almeida AM, Rubio M, Alves L, et al. Contribution of flagella and motility to gut colonisation and pathogenicity of Salmonella Enteritidis in the chicken. Braz J Microbiol. (2017) 48:754–59. doi: 10.1016/j.bjm.2017.01.012
37. Nachamkin I, Yang XH, and Stern NJ. Role of Campylobacter jejuni flagella as colonization factors for three-day-old chicks: analysis with flagellar mutants. Appl Environ Microbiol. (1993) 59:1269–73. doi: 10.1128/aem.59.5.1269-1273.1993
38. Sevrin G, Massier S, Chassaing B, Agus A, Delmas J, Denizot J, et al. Adaptation of adherent-invasive E. coli to gut environment: Impact on flagellum expression and bacterial colonization ability. Gut Microbes. (2020) 11:364–80. doi: 10.1080/19490976.2017.1421886
39. Lane MC, Alteri CJ, Smith SN, and Mobley HL. Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc Natl Acad Sci U S A. (2007) 104:16669–74. doi: 10.1073/pnas.0607898104
40. Morgan SJ and Chaston JM. Flagellar genes are associated with the colonization persistence penotype of the Drosophila melanogaster microbiota. Microbiol Spectr. (2023) 11:e0458522. doi: 10.1128/spectrum.04585-22
41. Fink SL, Bergsbaken T, and Cookson BT. Anthrax lethal toxin and Salmonella elicit the common cell death pathway of caspase-1-dependent pyroptosis via distinct mechanisms. Proc Natl Acad Sci U S A. (2008) 105:4312–17. doi: 10.1073/pnas.0707370105
42. Stewart MK and Cookson BT. Mutually repressing repressor functions and multi-layered cellular heterogeneity regulate the bistable Salmonella fliC census. Mol Microbiol. (2014) 94:1272–84. doi: 10.1111/mmi.12828
43. Ohashi Y, Hiraguchi M, Sunaba C, Tanaka C, Fujisawa T, and Ushida K. Colonization of segmented filamentous bacteria and its interaction with the luminal IgA level in conventional mice. Anaerobe. (2010) 16:543–46. doi: 10.1016/j.anaerobe.2010.07.006
44. Rolli J, Loukili N, Levrand S, Rosenblatt-Velin N, Rignault-Clerc S, Waeber B, et al. Bacterial flagellin elicits widespread innate immune defense mechanisms, apoptotic signaling, and a sepsis-like systemic inflammatory response in mice. Crit Care. (2010) 14:R160. doi: 10.1186/cc9235
45. Renard HF and Boucrot E. Unconventional endocytic mechanisms. Curr Opin Cell Biol. (2021) 71:120–29. doi: 10.1016/j.ceb.2021.03.001
Keywords: segmented filamentous bacteria, flagellin, germ-free mouse, bacterial adhesion, endocytosis, Th17 cell differentiation
Citation: Chen H, Wu L, Cao X, Li Z, Zhu R, Wang X, Li J, Wei Z, Yang D and Yin Y (2025) SFB flagellin mediates cell adhesion, endocytosis and immune regulation in germ-free mice. Front. Immunol. 16:1624092. doi: 10.3389/fimmu.2025.1624092
Received: 07 May 2025; Accepted: 28 July 2025;
Published: 20 August 2025.
Edited by:
Francisco Jose Roig, Universidad San Jorge, SpainReviewed by:
Bernhard Ryffel, Centre National de la Recherche Scientifique (CNRS), FranceAyyanar Sivanantham, Versiti Blood Research Institute, United States
Copyright © 2025 Chen, Wu, Cao, Li, Zhu, Wang, Li, Wei, Yang and Yin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yeshi Yin, eWlueWVzaGlAZ3h1LmVkdS5jbg==; eWlueWVzaGlAMTI2LmNvbQ==; Dengfeng Yang, ZGVuZ2Zlbmd5YW5nQDE2My5jb20=
 Huahai Chen
Huahai Chen Liu Wu6,7
Liu Wu6,7 Zuzhang Wei
Zuzhang Wei Dengfeng Yang
Dengfeng Yang Yeshi Yin
Yeshi Yin