- 1School of Medical Technology and Information Engineering, Zhejiang Chinese Medical University, Hangzhou, China
- 2Zhejiang Key Laboratory of Public Health Detection and Pathogenesis Research, Department of Microbiology, Zhejiang Provincial Center for Disease Control and Prevention, Hangzhou, China
- 3School of Basic Medical Sciences, Zhejiang Chinese Medical University, Hangzhou, China
- 4Key Laboratory of Artificial Organs and Computational Medicine of Zhejiang Province, Shulan International Medical College, Zhejiang Shuren University, Hangzhou, China
- 5School of Public Health, Hangzhou Medical College, Hangzhou, China
- 6School of Laboratory Medicine and Life Sciences, Wenzhou Medical University, Wenzhou, China
Severe Fever with Thrombocytopenia Syndrome Virus (SFTSV), a tick-borne phlebovirus first identified in China, causes severe illness characterized by high fever, thrombocytopenia, leukopenia, and, in some cases, multi-organ failure and death. With mortality rates ranging from 5% to 30% in endemic regions, SFTSV has emerged as a significant public health threat across East Asia, including South Korea and Japan, with potential for broader outbreaks. This review synthesizes recent advances in SFTSV animal models and candidate vaccines, highlighting their contributions and limitations. Current animal models, including mice, ferrets, and non-human primates, partially replicate human disease but fail to fully recapitulate clinical manifestations, limiting their translational utility. Vaccine development has shown promise, with candidates such as mRNA, subunit, and viral vector vaccines demonstrating efficacy in preclinical studies, yet none have progressed to clinical trials. Key challenges include viral genetic diversity and immune evasion. Future research should focus on refining animal models to better mimic human pathology, developing broad-spectrum vaccines, and integrating virological and immunological insights to enhance prevention and treatment strategies for SFTSV.
1 Introduction
Severe Fever with Thrombocytopenia Syndrome Virus (SFTSV), officially classified as Dabie bandavirus (Family Phenuiviridae, Genus Bandavirus) by the International Committee on Taxonomy of Viruses (ICTV) in 2020, is a tick-borne phlebovirus first isolated in 2009 from a patient in Henan Province, China (1). Since then, cases have been documented in South Korea, Japan, Vietnam, Myanmar, Thailand, Pakistan, and many other countries and regions (2–6). Reports of human bites by Haemaphysalis longicornis ticks in the United States have raised concerns about the potential emergence of SFTSV as a public health threat beyond Asia (7). According to the latest classification by the International Committee on Taxonomy of Viruses (ICTV) in 2020, SFTSV belongs to the family Phenuiviridae and the genus Bandavirus, and it has been officially named Dabie bandavirus (DBV) (8). However, the term SFTSV remains more widely used in academic literature. Thus, this article continues to refer to the virus as SFTSV.
The primary route of human infection with SFTSV is through tick bites (9). The incubation period for the infection typically lasts 7–14 days, with a reported mortality rate ranging from 5% to 30% (10). SFTSV infected patients generally progress through three phases: the febrile phase, multiorgan dysfunction (MOD) phase, and recovery phase. The febrile phase lasts approximately 7 days and is characterized by fever, thrombocytopenia, leukopenia, lymphadenopathy, and gastrointestinal symptoms (such as nausea, vomiting, and diarrhea) (11). High viral loads detected during this stage are a key marker for the clinical diagnosis of SFTSV infection (12). The MOD phase develops rapidly between days 7 and 13 post-infection, with elevated serum levels of alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP), and lactate dehydrogenase (LDH), along with prolonged activated partial thromboplastin time (aPTT). Clinical manifestations during this phase include hemorrhagic and neurological symptoms, disseminated intravascular coagulation (DIC), multi-organ failure, and persistent thrombocytopenia. These factors that are major determinants of mortality. The MOD phase is critical for determining the prognosis of SFTSV infected patients (13, 14). The recovery phase, which occurs between days 11 and 19, is marked by the resolution of clinical symptoms and the normalization of laboratory parameters. Multiple clinical studies have reported immunological parameters in clinical patients. Current evidence indicates that severe disease is primarily associated with a cytokine storm and elevated levels of pro-inflammatory molecules such as TNF-α, IP-10, and IL-6 (15). Furthermore, patients with severe disease exhibit lower counts of CD3+, CD4+, and CD8+ T cells compared to those with mild disease (16, 17). A reduction in the Th1/Th2 ratio and an increase in the Th17/Treg ratio are also related to disease severity (18). These changes suggest that an imbalance in cellular immunity is closely linked to progression to severe disease. In terms of humoral immunity, studies indicate that non-survivors lack NP or Gn specific IgG production, and that the distribution and function of B cell subsets are abnormal, with impaired antigen-presenting capacity (19). Additionally, age is a significant risk factor for SFTSV prognosis, with mortality predominantly observed in individuals aged 50 and above, and mortality rates increase with age (20). Due to the severity of SFTSV infection, the World Health Organization (WHO) has classified it as a priority pathogen requiring urgent attention (21).
Vaccines are the most effective public health intervention for preventing the spread of infectious diseases. Successful vaccination programs have eradicated many life-threatening diseases, such as smallpox and polio (22). The World Health Organization estimates that vaccines prevent 2 to 3 million deaths annually from diseases such as tetanus, pertussis, influenza, and measles (23). However, no approved vaccine for SFTSV is currently available. The SFTSV vaccines under development include live attenuated vaccines, inactivated vaccines, recombinant vector vaccines, subunit vaccines, DNA vaccines, and mRNA vaccines. All are still in the preclinical stage, with no candidate vaccines having progressed to clinical trials. Therefore, there is an urgent need to develop a broadly effective vaccine to address this potential public health threat.
2 SFTSV virology
2.1 Phylogeny
The family Bunyaviridae is the largest group of arboviruses, with over 300 identified species. Based on serological, morphological, and biochemical characteristics, this family is classified into five genera: Orthobunyavirus, Phlebovirus, Nairovirus, Hantavirus, and Tospovirus. Yu et al. conducted whole-genome sequencing of 12 SFTSV strains isolated from patients in China and identified the virus as an enveloped, segmented, negative sense spherical RNA virus belonging to the genus Phlebovirus (1). SFTSV was found to have close phylogenetic relationships with the Heartland virus (HRTV) discovered in the United States and the Malsoor virus identified in India (1, 24, 25).
2.2 Genomic structure and functions
The genome of SFTSV, like other viruses in the order Bunyavirales, comprises three RNA segments: L, M, and S. The L segment is negative sense RNA, consisting of 6,368 nucleotides, and encodes the RNA dependent RNA polymerase (RdRp), which is responsible for viral RNA replication and mRNA synthesis (26).
The M segment contains 3,378 nucleotides and encodes a glycoprotein precursor (Gp). This precursor is processed by host proteases into two subunits, Gn and Gc (27). Gn and Gc form the viral envelope and possess antigenic properties. During endocytosis, these glycoproteins mediate viral entry by binding to cellular receptors and inducing low-pH-dependent membrane fusion (28). Several factors, including non-muscle myosin heavy chain IIA (NMMHC-IIA), C-type lectin receptors such as dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin (DC-SIGN), DC-SIGN-related molecules (DC-SIGNR), liver and lymph node sinusoidal endothelial cell C-type lectin (LSECtin), and UDP-glucose ceramide glucosyltransferase (UGCG), have been identified as key players in SFTSV entry (29–32). Gn/Gc proteins also serve as immunogenic targets, stimulating the production of specific neutralizing antibodies, thus providing valuable directions for future vaccine research (33). The efficacy of structural glycoprotein-based vaccines has been demonstrated in SARS-CoV-2, influenza virus, and Ebola virus (34–36).
The S segment consists of 1,744 nucleotides and employs an ambisense coding strategy to encode the nucleocapsid protein (NP) and non-structural protein (NSs) (37). NP is the most abundant protein in SFTSV particles and infected cells. It forms hexamers that encapsulate viral RNA (vRNA), creating ribonucleoprotein complexes (RNPs). This function protects the viral genome from degradation by host cell nucleases and innate immune responses (38, 39). Recent studies indicate that NP triggers mitophagy to degrade mitochondrial antiviral signaling proteins (MAVS), thereby blocking MAVS-mediated antiviral signaling and evading host immune defenses (40). NSs is an important virulence factor of SFTSV, which can inhibit the induction of type I interferons (41). Additionally, NSs targets the Tumor Progression Locus 2 (TPL2)-A20-binding NF-κB inhibitory factor 2 (ABIN2)-p105 complex, inducing interleukin-10 (IL-10) expression to enhance viral pathogenicity (42). At the same time, NSs also plays a crucial role in viral replication (43).
3 Epidemiology
3.1 Transmission
The transmission cycle and mechanisms of SFTSV in nature remain unclear. Haemaphysalis longicornis is the primary vector and an important reservoir of SFTSV (44). SFTSV RNA has also been detected in various tick species, such as the Ixodes nipponensis, Dermacentor nuttalli, and Haemaphysalis flava, in regions where SFTSV outbreaks occur (45–47). The parthenogenetic reproduction characteristic of the Haemaphysalis longicornis allows it to establish new populations more rapidly than sexual reproduction, accelerating the spread of the ticks (48).
Migratory birds have long been known to serve as long distance carriers of ticks that harbor various human pathogens, such as Crimean-Congo Hemorrhagic Fever Virus(CCHFV) and Tick-Borne Encephalitis Virus (TBEV) (49, 50). The distribution of Haemaphysalis longicornis aligns with the East Asia-Australia migratory bird routes, indicating that the spread of this tick species is likely linked to migratory birds (51).
Although the primary route of SFTSV infection in humans is through tick bites, human-to-human transmission of the virus has also been confirmed. Contact with the blood and bodily fluids of infected individuals can lead to virus transmission. Additionally, ticks may parasitize on a variety of wild animals and livestock, including birds and domestic animals, thereby infecting these animals and subsequently transmitting the virus to humans through close contact (52). SFTSV specific antibodies have been detected in a variety of animals, including goats, sheep, cattle, dogs, and cats, with the highest antibody positivity found in goats and cattle (53). However, these animals do not show obvious symptoms of infection and do not exhibit significant viremia, serving as reservoir hosts for SFTSV (Figure 1). The persistent infection cycle in animal hosts allows SFTSV to continue to spread in nature. Special protective measures should be taken for populations with close contact with these animals.
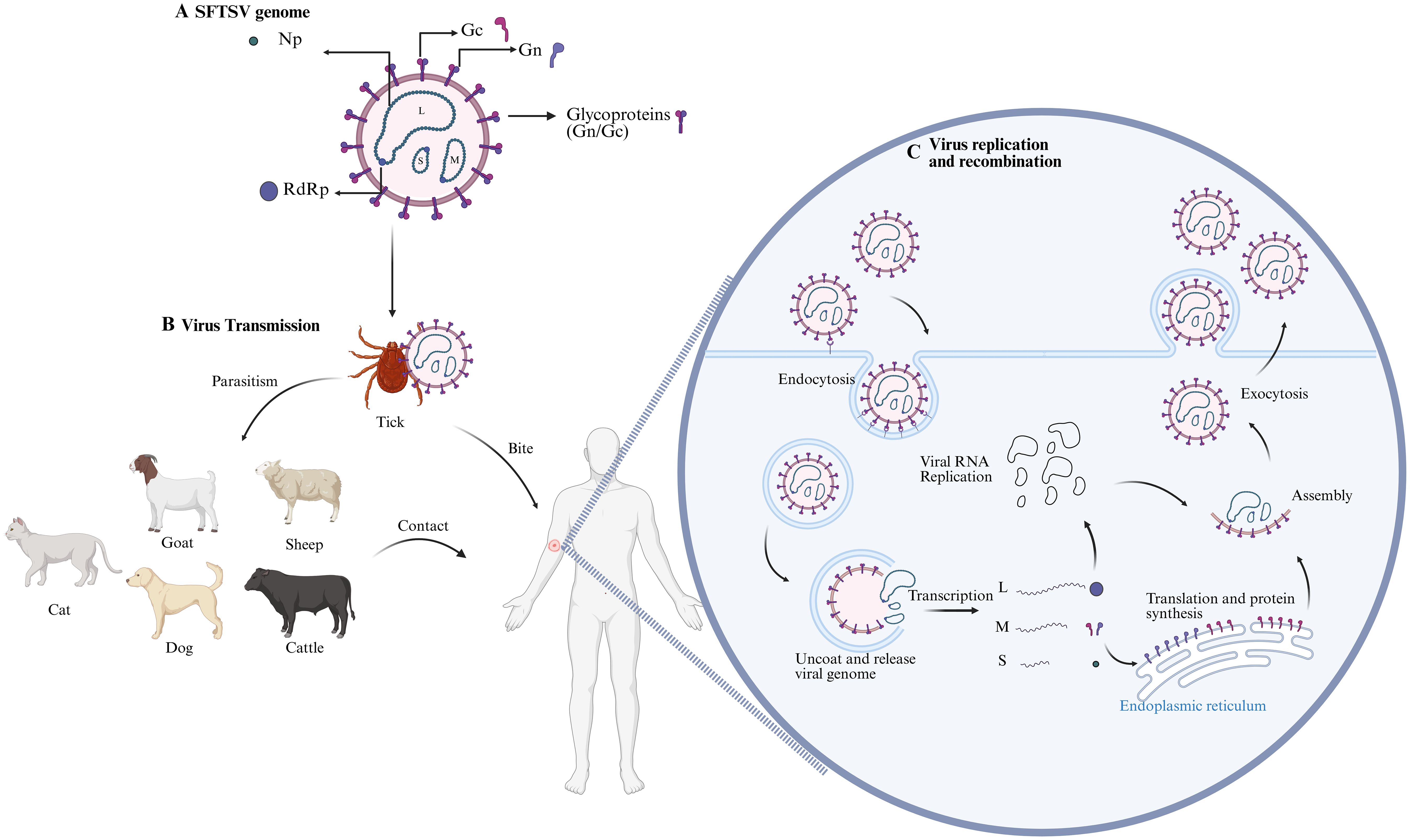
Figure 1. SFTSV Infection Mechanism Diagram. (A) Schematic of SFTSV structure, illustrating its enveloped nature and three RNA genome segments (L, M, S) encoding RNA-dependent RNA polymerase (RdRp), glycoproteins (Gn/Gc), and nucleocapsid (NP) and non-structural (NSs) proteins, respectively. (B) SFTSV is transmitted via Haemaphysalis longicornis tick bites to animal hosts (e.g., goats, cattle), with human infections occurring through direct contact or tick bites; human-to-human transmission is also possible. (C) The virus enters host cells via clathrin-mediated endocytosis, with Gn/Gc glycoproteins binding cellular receptors (e.g., DC-SIGN, NMMHC-IIA) and inducing low-pH-dependent membrane fusion. The viral genome is released and replicates in the cytoplasm. Glycoproteins are translated and modified in the host endoplasmic reticulum and Golgi apparatus. New viral particles assemble and are released via budding. The figure was created with BioRender.com.
3.2 Genotype and recombination
Phylogenetic analysis of SFTSV strains has revealed six genotypes (A-F), with genotype B further subdivided into at least three distinct genotypes (B-1, B-2, and B-3) (54, 55). In mainland China, three of the six genotypes (F 43.6%, A 20.1%, D 15.4%) are predominant, while the B-2 subtype is prevalent in South Korea (36.1%) and Japan (86%) (54–56). The varying distribution of genotypes in SFTSV endemic regions leads to significant differences in mortality rates, with Japan (23%) and South Korea (27%) experiencing higher mortality rates compared to China (5.3%–16.2%) (57). Studies by Yun et al. have further demonstrated a close association between mortality rates, patient age, and SFTSV genotypes (55). Additionally, recombination plays an important role in the genetic diversity of segmented genome viruses. The segmented nature of the SFTSV genome leads to a high probability of recombination events. At least seven recombinant strains have been identified in China, while South Korea has reported at least nine recombinant genotypes (55, 58).
The genetic recombination phenomenon of SFTSV reveals its dynamic evolutionary characteristics in nature, presenting significant challenges for SFTSV diagnosis, epidemiological surveillance, treatment strategies, and vaccine development. Particularly in vaccine research, recombination significantly increases the genetic diversity of the virus, which may reduce the protective efficacy of neutralizing antibodies induced by vaccines against newly recombinant strains. Moreover, the high mutation rate and recombination properties of the virus further drive variation in antigenic epitopes, significantly enhancing the virus’s immune evasion capacity. Additionally, recombination may alter the pathogenicity and transmissibility of the virus, exacerbating immune protection discrepancies between different genotypes, leading to variable vaccine efficacy across regions. These complex factors pose key scientific challenges that need to be addressed with innovative research strategies and technological approaches for effective vaccine development.
Given the challenges posed by SFTSV’s genetic recombination, including heightened genetic diversity and immune evasion capabilities, vaccine development necessitates multidimensional strategies to enhance broad-spectrum efficacy and long-term protection: In terms of dynamic monitoring and evolutionary analysis: High-throughput sequencing can be employed to identify viral mutation hotspots, while the establishment of databases and bioinformatics analysis platforms enables the tracking of viral evolutionary dynamics across different regions and time periods, thereby providing the latest genetic information for vaccine design. Furthermore, by integrating structural biology techniques such as cryo-electron microscopy (cryo-EM) and X-ray crystallography to perform high-resolution structural analyses of key antigens (e.g., Gn and Gc), critical neutralizing antibody epitopes can be precisely mapped. The application of multi-target strategies and advanced technological platforms in vaccine design holds promise for overcoming the challenges posed by SFTSV gene recombination and mutation-induced immune evasion and diversity, ultimately leading to broader and more effective vaccine protection.
4 Animal models for SFTSV
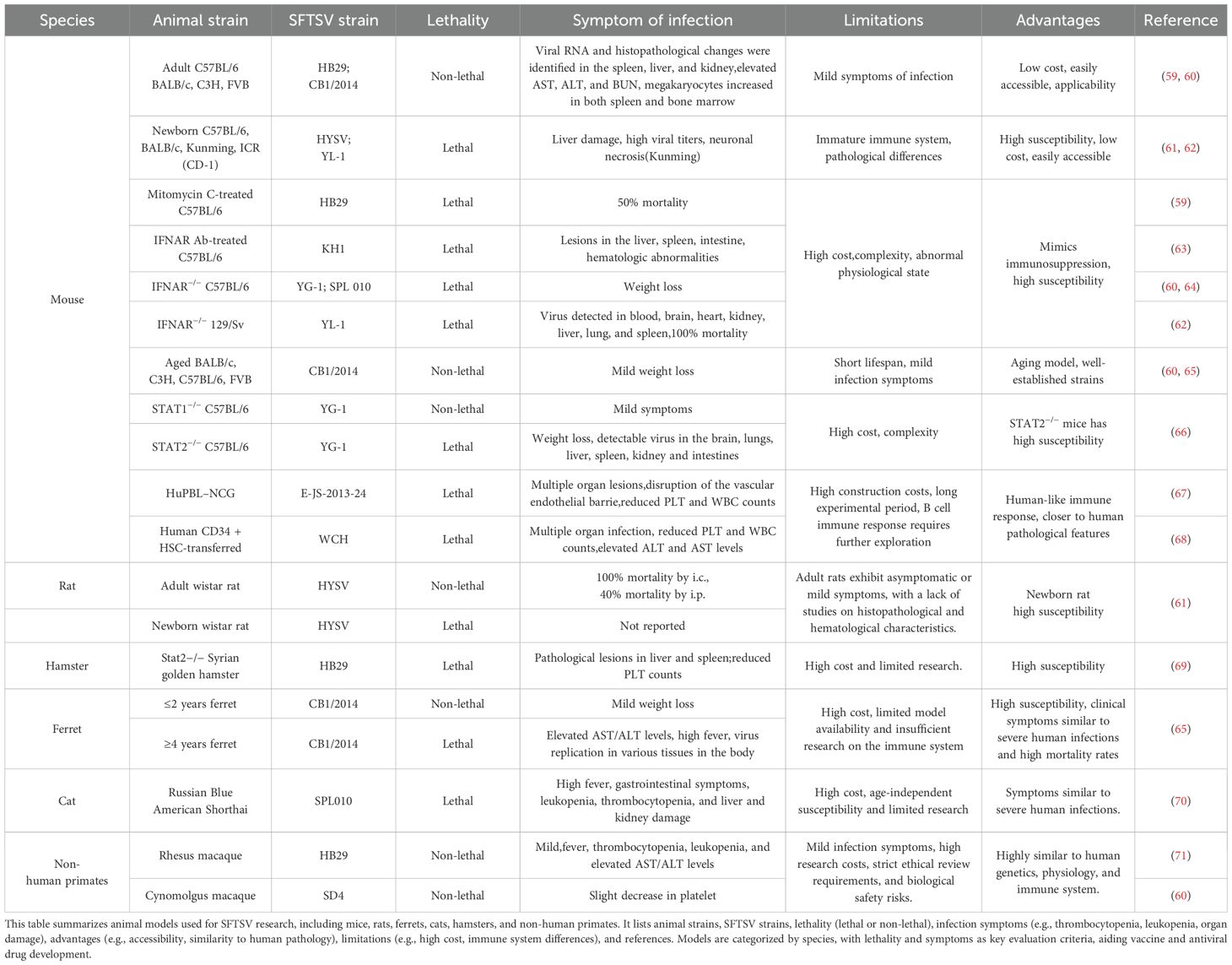
Animal models are essential tools for studying the pathogenesis of viruses, developing vaccines, and exploring antiviral treatments. However, most animals do not exhibit fatal effects upon SFTSV infection, which presents a significant obstacle in the development of vaccines and antiviral therapies. It is crucial to thoroughly understand the pathogenesis of SFTSV in each model, taking into account the differences and similarities between animal models and human cases. Recent studies have identified several animal models highly susceptible to SFTSV infection. These models can mimic certain aspects of the pathogenesis observed in human SFTS, providing a basis for developing vaccines and antiviral drugs against SFTSV infection (Table 1).
4.1 Mice
4.1.1 Immunocompetent mice
In an early study, Jin et al. intramuscularly injected 105 TCID50 of the SFTSV HB29 strain into C57BL/6 mice, resulting in elevated levels of AST, ALT, and BUN (blood urea nitrogen), along with leukopenia and thrombocytopenia—pathological features similar to mild human infections. Pathological changes were observed in the liver, spleen, and kidneys, with viral RNA detected in all three organs, though viral replication was observed only in the spleen (59). Additionally, their research revealed that splenic macrophages were the target cells of infection. However, the C57BL/6 mouse model failed to progress into severe or fatal SFTSV infections. To induce fatal infections, researchers administered the immunosuppressant mitomycin C (59). Similarly, immunocompetent adult mouse strains such as BALB/c, C3H, FVB, and ICR (CD-1) showed no severe clinical manifestations upon SFTSV infection (59, 60).
4.1.2 Age-dependent mice
Newborn mice, including C57BL/6, BALB/c, Kunming, and ICR (CD-1), are susceptible to SFTSV, exhibiting severe symptoms and high mortality rates (61, 62). Although newborn mice are highly sensitive to SFTSV and display significant pathological changes, their immune systems are not fully developed, and experimental operations are challenging. In comparison, aged mice (over 20 months old), such as BALB/c, C3H, C57BL/6, and FVB strains, show only slight weight loss (60, 65). Therefore, age-related mouse models may not be suitable for simulating the progression characteristics of SFTS in elderly human patients or for studying the age-specific pathogenesis of SFTSV.
4.1.3 Immunocompromised mice
Immunodeficient gene knockout mice have also been employed to study SFTSV infections. These include α/β interferon receptor knockout (IFNAR−/−) mice (60, 62, 64), signal transducer and activator of transcription 2-deficient (STAT2−/−) mice, and mice treated with blocking anti-type I interferon (IFN)-α receptor antibody (IFNAR Ab-treated mice) (60, 62–64, 66). These models exhibit high susceptibility to SFTSV infection and lethal outcomes, with viral replication detected in multiple organs. Severe infection symptoms were observed, including significant weight loss, multi-organ pathological changes, severe leukopenia, and thrombocytopenia. Similar observations were made in the STAT2 knockout model of golden Syrian hamsters (69).
4.1.4 Humanized mice
Two humanized mouse models constructed through immune system reconstitution have been reported as lethal models for SFTSV, providing a closer simulation of human infection compared to wild-type and immunocompromised mice. Li et al. transplanted highly purified (>90%) human CD34+ cells, isolated from umbilical cord blood, into mice via tail vein injection (68). SFTSV infection in these mice led to multi-organ involvement, reductions in platelet and white blood cell counts, and elevated ALT and AST levels, closely resembling the clinical characteristics of SFTSV patients. SFTSV infection in this humanized mouse model resulted in fatal outcomes. Xu et al. established the HuPBL-NCG mouse model by transplanting human peripheral blood mononuclear cells (PBMCs) into NCG mice (67). The human PBMCs transplanted into NCG mice provided early replication targets for the virus, while infected human monocytes transmitted the virus to mouse monocytes through an intercellular transmission mechanism, which is more efficient in viral infections. Their study also elucidated aspects of the pathogenesis of hemorrhagic syndrome, including apoptosis, membrane protein endocytosis, and cytokine stimulation. The HuPBL-NCG model mimics many pathological features of human SFTSV infection, including virus-induced histopathological changes, disruption of vascular endothelial barriers, thrombocytopenia, and leukopenia. While this humanized mouse model offers a valuable tool for investigating the pathological mechanisms of SFTSV infection, further exploration is required to study B cell immune responses in detail.
4.2 Rats
In the rat model, all newborn Wistar rats died after intracranial (i.c.) inoculation with 2×107 copies, whereas 40% of the rats died following intraperitoneal (i.p.) injection with 3×107 copies. However, adult rats all survived after vaccination (61). Further studies are needed to investigate the characteristics of the SFTSV rat model, as histopathological and hematological examinations have not been reported to date.
4.3 Ferrets
The ferret, with anatomical and physiological features similar to those of humans, has been widely used in the study of various infectious diseases (72, 73). Park et al. have demonstrated that SFTSV infection in ferrets is age-dependent (65). After SFTSV challenge, young ferrets (<2 years old) only exhibit mild symptoms such as weight loss and slight weight gain. While there are changes in AST/ALT levels, PLT and WBC counts, these quickly return to normal ranges. Additionally, viral RNA can be detected in the spleen, liver, kidneys, lungs, and serum. However, aged ferrets (>4 years old) are more susceptible to infection and exhibit symptoms similar to severe human cases of SFTSV infection. These include significant thrombocytopenia, leukopenia, elevated AST/ALT levels, high fever, and weight loss. In aged ferrets, systemic infection is also triggered, with viral RNA detected in various tissues, and death occurs within ten days post-infection, with a mortality rate of 93% (65).
Although the use of aged ferrets offers advantages in the study of lethal SFTSV infection models and vaccine development, several limitations exist. These include high costs, limited availability of effective aged ferret models, and a lack of resources for studying the immune system mechanisms in ferrets.
4.4 Cats
The detection of SFTSV RNA in cats was first reported in 2017 in serum samples from wild cats in South Korea (74). There have also been reports from Japan of veterinarians contracting SFTSV while treating infected cats (75). In a study by Park et al., four out of six cats that were infected with SFTSV via intravenous injection died within 10 days post-infection (70). Cats that died from SFTSV infection exhibited severe clinical manifestations, including high fever, gastrointestinal symptoms, leukopenia, thrombocytopenia, and liver and kidney damage, resembling the symptoms of severe SFTS in humans. However, there was no correlation between the age of the cats and the severity of the disease.
The clinical and histopathological features in cats, similar to those seen in severe human infections, suggest that cats could be a promising animal model for SFTSV research. However, there are significant limitations: compared to established rodent models commonly used in laboratories, the cost of using cats is higher, handling cats in laboratory settings is more challenging, and research on cat models remains limited. Furthermore, standardized experimental cat strains have yet to be developed.
4.5 Non-human primates
Non-human primates share genetic, physiological, and immune system similarities with humans, making their immune responses and disease progression closer to those observed in humans. They are ideal models for vaccine development and studies on infection mechanisms and have been widely used to investigate the infection and pathogenesis of bunyaviruses that cause hemorrhagic fever diseases (76–78). Jin et al. reported cases of rhesus monkeys infected with SFTSV, which exhibited symptoms such as fever, thrombocytopenia, leukopenia, and elevated levels of transaminases and myocardial enzymes in the blood. These symptoms resembled mild SFTS in humans, without causing severe illness or death (71). In another study involving cynomolgus monkeys, no significant clinical symptoms were observed following SFTSV infection, and viral RNA was not detected during the 14 day study period (60). While non-human primate models offer the advantage of physiological and immune system similarities to humans, their application in SFTSV vaccine development is limited due to strict ethical reviews, high research costs, biosafety risks, and the mild symptoms observed in SFTSV infections in these models.
4.6 Comparison and prospect of animal models
SFTSV vaccine and drug development primarily rely on aged ferrets (see Section 4.3) and immunocompromised mouse models (see Section 4.1.3). Aged ferrets and cats are among the few immunocompetent models showing lethality, though cat models exhibit transient inflammatory responses and low neutralizing antibody titers (70). Aged ferrets display sustained inflammation, closely mimicking severe human infections (65). Humanized mouse models (see Section 4.1.4) develop human-like pathology and immune responses, with longer survival, making them suitable for pathogenesis and antiviral studies (67). Rhesus macaque models infected with SFTSV exhibit robust Th1-type pro-inflammatory responses, widespread immune cell recruitment, and inflammatory mediator release, yet display mild clinical symptoms (71). In current vaccine research, immunocompetent mice are commonly used for preliminary evaluation of vaccine immunogenicity.
In general, future research should concentrate on refining animal models that show promise yet remain underutilized. For example, researchers can improve the reconstitution of the human immune system in humanized mice and employ genetic engineering techniques to optimize their microenvironment and genetic background. This approach may enable these models to more accurately simulate the immune response and vaccine-induced effects observed in human SFTSV infections. Moreover, for non-human primates that possess immune systems more akin to those of humans, adjusting infection conditions to intensify pathological manifestations or introducing immune regulatory strategies could help identify species or strains with heightened sensitivity. These efforts would contribute to the development of an animal model that more closely mirrors the severe clinical pathology seen in human SFTSV infections.
5 Advances in SFTSV vaccine research
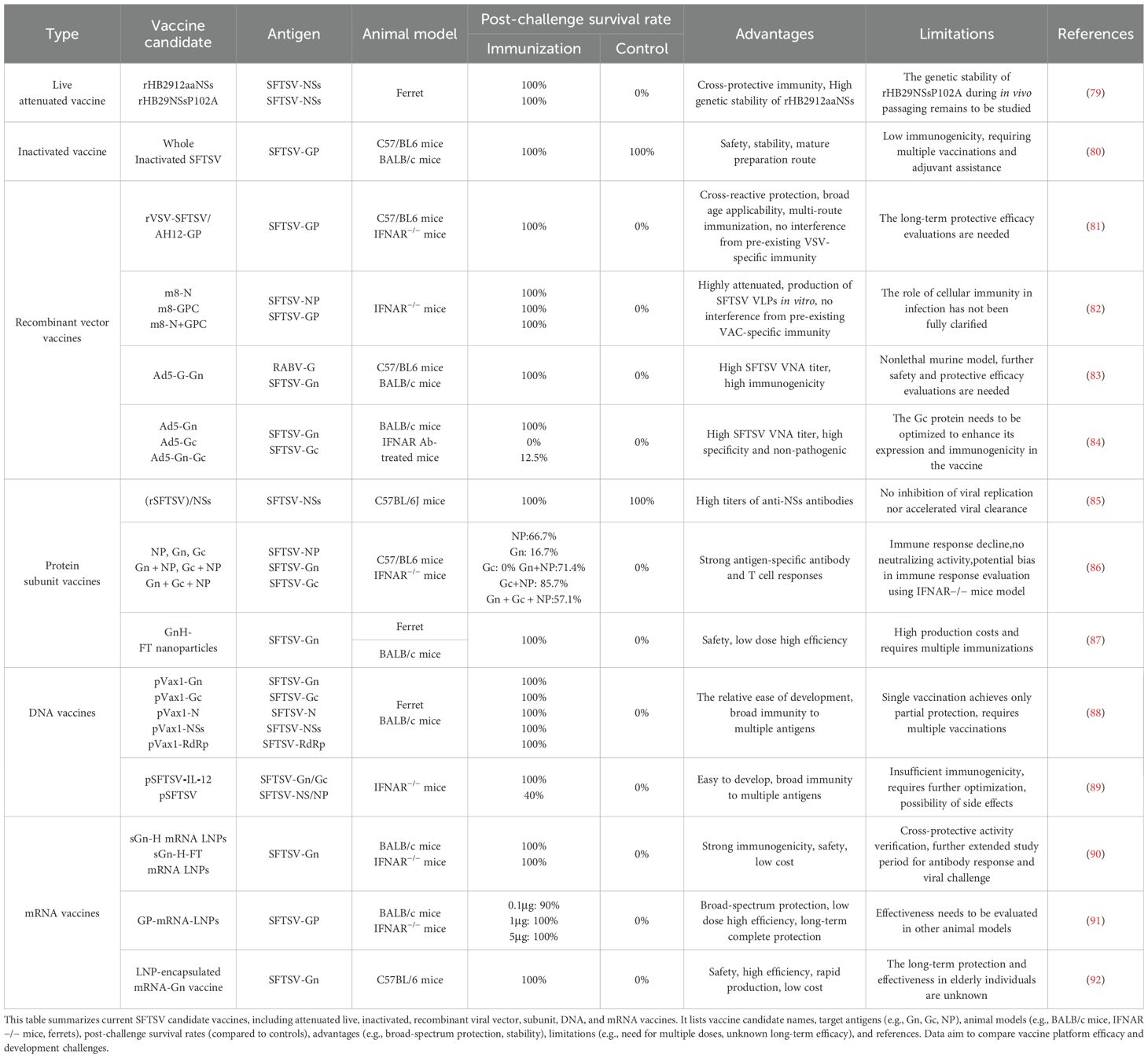
The current SFTSV candidate vaccines have achieved notable progress across various technological platforms (Table 2). To better illustrate their mechanisms of inducing immune responses, a simplified immune mechanism diagram is provided in Figure 2.
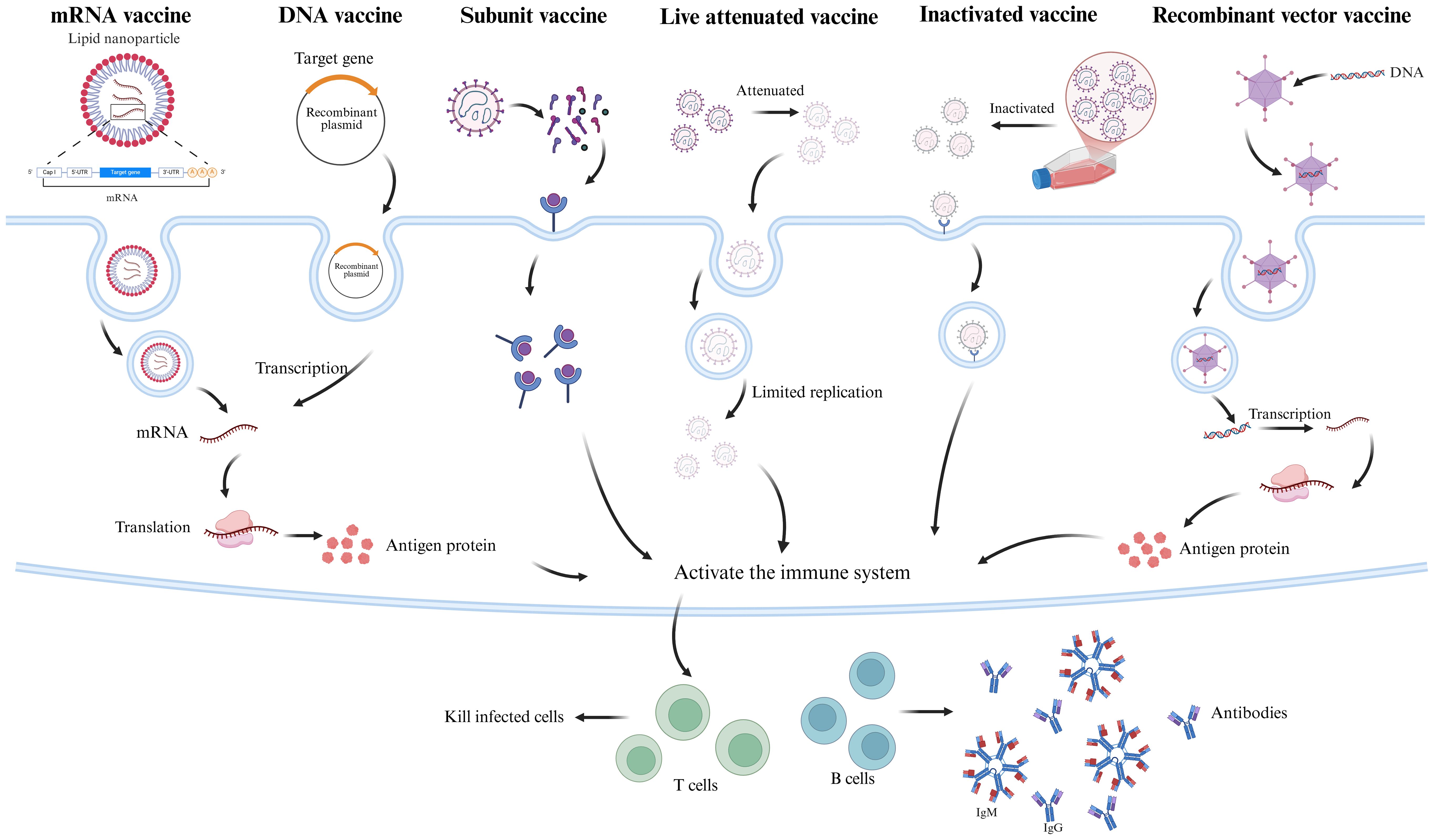
Figure 2. SFTSV Vaccine Mechanism of Action Diagram. This diagram illustrates the immune induction mechanisms of six SFTSV candidate vaccine types. mRNA vaccines, encapsulated in lipid nanoparticles (LNPs), deliver antigen-encoding mRNA (e.g., Gn, Gc) to the cytoplasm for translation into antigenic proteins. DNA vaccines, delivered via electroporation, are transcribed into mRNA and translated into antigens (e.g., Gn, Gc, NP). Subunit vaccines contain purified SFTSV antigens (e.g., Gn, Gc, NP). Inactivated vaccines use chemically inactivated (e.g., β-propiolactone) whole virus, retaining immunogenicity. Attenuated live vaccines, with reduced virulence via reverse genetics, mimic natural infection. Recombinant viral vector vaccines (e.g., VSV, adenovirus) express SFTSV antigen genes (e.g., Gn/Gc) in host cells. All vaccines activate B cells (producing neutralizing antibodies) and T cells (cytotoxic T cells), inducing specific humoral and cellular immunity against SFTSV infection. The figure was created with BioRender.com.
5.1 Attenuated live vaccines
Attenuated live vaccines are a type of vaccine that weakens the pathogenicity of a pathogen to eliminate its virulence while retaining its immunogenicity. These vaccines can replicate to a limited extent within the host, simulating the natural infection process and inducing a strong and long-lasting immune response (93). Reverse genetics is an important tool for exploring the molecular biology and pathogenesis of RNA viruses, as well as for vaccine development. Currently, attenuation is achieved using reverse genetics methods (94).
Yu et al. generated two recombinant SFTSV strains from the HB29 strain via reverse genetics: rHB2912aaNSs (NSs ORF C-terminus truncated by 12 amino acids) and rHB29NSsP102A (proline-to-alanine mutation at NSs position 102). In aged ferrets, these strains exhibited attenuated phenotypes, inducing high neutralizing antibody titers within 14 days, sustained until day 58. Immunized ferrets achieved 100% survival post-lethal challenge (control: 0%). The rHB2912aaNSs strain maintained genetic stability after multiple passages, indicating low reversion risk (79).
5.2 Inactivated vaccines
Inactivated vaccines use chemical or physical methods to render pathogens non-infectious while preserving their immunogenicity, thereby eliciting an immune response in the host. Compared to live attenuated vaccines, inactivated vaccines are more stable and safer, making them a better option for immunocompromised individuals. However, their immunogenicity is relatively low, necessitating multiple doses and the use of adjuvants to enhance the immune response.
Li et al. isolated the SFTSV strain AH12 from patient blood samples and inactivated it by β-propiolactone (BPL). The whole virus particles were purified using ultrafiltration and ultracentrifugation techniques (80). The inactivated vaccine induced high levels of SFTSV specific IgG antibodies and neutralizing antibodies in BALB/c and C57/BL6 mice, with higher doses of the vaccine resulting in higher antibody levels. The addition of an aluminum adjuvant significantly enhanced the production of IgG and neutralizing antibodies, with the effect being particularly pronounced in the low-dose groups. In terms of protective effects, two weeks after the final immunization, C57/BL6 mice were challenged with wild type SFTSV. The high dose vaccine group demonstrated a significantly accelerated clearance of viral RNA in the blood and spleen. However, since C57/BL6 mice are not a lethal model for SFTSV infection, further research is required to evaluate the protective efficacy of the vaccine in lethal animal models.
5.3 Recombinant virus vector vaccines
Recombinant viral vector vaccines use genetically engineered viruses as carriers to deliver specific antigen genes (typically encoding key proteins of pathogens) into host cells, thereby stimulating an immune response in the body (95, 96).
5.3.1 Recombinant vesicular stomatitis virus vector vaccine
Vesicular stomatitis virus (VSV) is a zoonotic arbovirus belonging to the Rhabdoviridae family. It has been developed as an attenuated viral vaccine vector capable of inducing robust neutralizing antibody responses and has demonstrated effective protection against lethal challenges (97, 98). Dong et al. cloned the human codon optimized Gn/Gc ORF from the Chinese lineage SFTSV AH12 strain into the rVSVΔG vector, enabling the virus to express SFTSV Gn/Gc on the surface of viral particles. This recombinant virus was named rVSV-SFTSV/AH12-GP (81). A single dose of rVSV-SFTSV/AH12-GP elicited highly efficient and broad-spectrum neutralizing antibodies in both immunocompetent C57BL/6 mice and IFNAR−/− mice, providing complete protection against SFTSV challenge. Moreover, the protective efficacy of the candidate vaccine showed no significant differences across various routes of administration, including intraperitoneal injection, intravenous injection, subcutaneous injection, and intranasal administration. In IFNAR−/− mice pre-immunized with the rVSV vector, vaccination with the candidate vaccine still provided protective immunity against SFTSV challenge, indicating that the vaccine efficacy was not compromised by prior immunization.
5.3.2 Recombinant vaccinia virus vector vaccine
The LC16m8 strain of vaccinia virus (m8) and Modified Vaccinia Ankara (MVA) are classified as third-generation smallpox vaccines, characterized by high attenuation while retaining immunogenicity (99). Yoshikawa et al. utilized the highly attenuated yet immunogenic vaccinia virus strain LC16m8 (m8) as a recombinant vaccine for SFTS, expressing the SFTSV nucleoprotein (m8-N), envelope glycoprotein precursor (m8-GPC), and both N and GPC (m8-N+GPC) (82). Their m8-based SFTSV vaccines expressed SFTSV genes in infected cells, and particularly, cells infected with m8-GPC or m8-N+GPC produced virus-like particles (VLPs) in the supernatant of in vitro cultures. Subcutaneous administration of m8-based SFTSV vaccines at a dose of 1×106 PFU twice in IFNAR−/− mice successfully induced SFTSV-specific antibodies in all vaccine candidates and protected the mice from lethal challenges with 1×103 or 1×105 TCID50 of the SFTSV YG-1 strain. Additionally, mice pre-immunized with the Lister vaccinia strain also achieved protective immunity against SFTSV challenge after vaccination with m8-based SFTSV vaccines. Pathological analysis revealed no tissue pathological changes in mice immunized with m8-GPC or m8-N+GPC.Passive serum transfer experiments demonstrated that serum collected from mice vaccinated with m8-GPC or m8-N+GPC conferred protective immunity against lethal SFTSV challenge in naïve mice.
5.3.3 Recombinant adenovirus vector vaccine
Adenoviruses are widely distributed in nature and possess high genomic manipulation flexibility, allowing the integration of large foreign gene fragments. This feature makes them an ideal choice for constructing specific vaccines (100, 101). Zhao et al. developed a recombinant replication-deficient human adenovirus type 5 (Ad5) encoding rabies virus (RABV) G and SFTSV Gn (Ad5-G-Gn) (83). Ad5-G-Gn immunization activated more dendritic cells (DCs) and B cells in lymph nodes (LNs) and induced a Th1/Th2-mediated response in splenocytes, leading to the robust production of neutralizing antibodies against SFTSV and RABV. Additionally, in 6 to 8-week-old C57/BL6 mice infected with SFTSV, Ad5-G-Gn immunization significantly reduced the SFTSV viral load in the spleen. However, this study has limitations due to the use of a non-lethal mouse model, and further safety and protective efficacy evaluations are needed.
Subsequently, the team developed Ad5 vector vaccine candidates expressing different regions of SFTSV glycoprotein (Gn, Gc, and Gn-Gc) (84). Compared to Ad5-Gc and Ad5-Gn-Gc, Ad5-Gn rapidly recruited/activated DCs, promoted B cell activation, induced specific T cells, and quickly generated high levels of SFTSV virus-neutralizing antibodies (VNA) in wild-type mice. Furthermore, in lethal SFTSV-infected IFNAR−/− mice, Ad5-Gn provided complete protection and safeguarded the spleen, liver, brain, lungs, and other organs from SFTSV-related pathological changes. Their findings suggest that Gn is an advantageous target for the development of SFTSV vaccines and antibodies.
5.4 Subunit vaccines
Subunit vaccines, due to their excellent safety, stability, and relatively mature production techniques, have been successfully applied in the prevention of viruses such as hepatitis B virus (HBV) and human papillomavirus(HPV) (102, 103). However, they also have the issue of low immunogenicity, requiring the addition of adjuvants and multiple doses to enhance their immune effects.
5.4.1 NSs recombinant protein vaccine
In the early research by Liu et al., the effectiveness of SFTSV non-structural protein (NSs) as a vaccine component was evaluated (85). However, C57BL/6 mice immunized with purified recombinant NSs combined with complete Freund’s adjuvant showed no significant difference in viremia levels compared to the control group after SFTSV challenge. Therefore, the NSs vaccine did not promote the clearance of SFTSV in mice.
5.4.2 Self-assembling Gn Head-Ferritin nanoparticles vaccine
Kim et al. applied self-assembling ferritin nanoparticles fused with the head region of SFTSV Gn (GnH) to construct GnH-FT (87). The immunogenicity of the vaccine was evaluated in BALB/c mice and aged ferrets. The results showed that immunization with 1 μg of GnH-FT nanoparticles could induce a robust neutralizing antibody (NAb) response and T-cell immunity against SFTSV Gn in mice. Immunized aged ferrets not only effectively induced total IgG antibodies and NAb antibodies but also provided complete protection against SFTS symptoms and lethal SFTSV challenge.
5.4.3 NP, Gn and Gc recombinant protein vaccine
Recently, Kim et al. evaluated the efficacy of recombinant protein vaccines using purified nucleocapsid protein and surface glycoproteins, assessing their immunological effects both individually and in combination (86). Immunization with either the NP or Gn subunit alone provided partial protection to IFNAR−/− mice, with survival rates of 66.7% and 16.7%, respectively, while Gc vaccination failed to offer significant protection, resulting in 100% mortality (86). Among the tested recombinant protein combinations (Gn + NP, Gc + NP, and Gn + Gc + NP), the Gc + NP combination showed the highest protective effect following exposure to a lethal dose of SFTSV, achieving the highest survival rate (85.7%) and highlighting its potential as a vaccine candidate. However, NP antibodies did not exhibit neutralizing activity, and their potential role in antiviral immunity requires further investigation. All IFNAR−/− mice vaccinated with single subunit vaccines succumbed to viral infection within 12 months, suggesting that a combination of protective antigens and adjuvant systems is still needed to ensure long-term humoral and cellular immunity.
5.5 DNA vaccines
The advantages of DNA vaccines include eliciting strong immune responses, ease of development, and the ability to quickly test multiple candidate antigen designs (104). However, they also face challenges such as the need for delivery via electroporation and insufficient immune durability, which requires multiple doses.
Kwak et al. constructed a DNA vaccine using pVax1 as an expression vector, encoding the full-length Gn, Gc, N, NS, and RNA-dependent RNA polymerase (RdRp) genes of SFTSV, based on the sequences of 31 clinical isolates from patients in China, Korea, and Japan (88). Intramuscular injection of the SFTSV DNA candidate vaccine in BALB/c mice and aged ferrets induced strong SFTSV specific T cell responses and neutralizing antibody responses. In aged ferrets (>4 years old), three intradermal immunizations of the SFTSV DNA candidate vaccine at 2-week intervals were followed by a viral challenge two weeks after the final vaccination. All vaccinated ferrets were fully protected from a lethal SFTSV challenge. However, a single dose vaccination only provided partial protection. Furthermore, their study found that Gn and Gc specific immune responses play a crucial role in preventing fatal SFTSV infection, while non-envelope specific T-cell responses also contribute to cellular protection against SFTSV infection.
Kang et al. developed another recombinant plasmid DNA (pSFTSV) as a DNA vaccine candidate, encoding the extracellular domains of Gn and Gc as well as an NP-NS fusion antigen (89). IL-12, a key factor for type 1 helper T cell (Th1) differentiation, enhances cellular immune responses. To improve protective efficacy, they incorporated IL-12 into pSFTSV, creating pSFTSV-12. The vaccine was administered to IFNAR−/− mice three times via in vivo electroporation. Following a lethal SFTSV challenge, mice vaccinated with pSFTSV-12 showed a 100% survival rate, while those vaccinated with pSFTSV alone had only a 40% survival rate. In the presence of IL-12 expression, virus antigen specific T cell responses were significantly enhanced. However, no neutralizing antibodies were detected in the immunized mice. These data indicate that the expression of IL-12 enhances the efficacy of the vaccine, but further studies are needed to optimize the combination of appropriate target antigens and the selection of adjuvants for DNA vaccines.
5.6 mRNA vaccines
mRNA vaccines, with their unique advantages of high efficiency, strong adaptability, simple antigen design, short production cycle, and high safety, have provided an important means of responding to major public health emergencies (105). During the SARS-CoV-2 pandemic, Pfizer/BioNTech’s BNT162b2 and Moderna’s mRNA-1273 proved to be highly effective against SARS-CoV-2 and were developed and administered to millions of people worldwide at unprecedented speed. These two vaccines marked the first clinical approval of mRNA vaccines (106–108). The success of mRNA vaccines against SARS-CoV-2 has sparked widespread interest in the use of mRNA for the prevention and treatment of various conditions. Currently, in addition to SARS-CoV-2 vaccines, research is being conducted on various mRNA-based vaccines for infectious diseases, cancer vaccines, and mRNA-based therapeutic approaches (109–111).
Beyond the subunit vaccine platform, Kim et al. also applied the SFTSV Gn Head region (sGn-H) and SFTSV Gn Head region ferritin nanoparticles (sGn-H-FT) to the SFTSV mRNA platform (90). They encapsulated mRNA encoding sGn-H or sGn-H-FT into lipid nanoparticles (LNPs) for effective delivery. Female BALB/c mice aged 6–8 weeks were selected and intramuscularly immunized with 1 µg of sGn-H or sGn-H-FT mRNA LNPs at weeks 0 and 3. The results showed that sGn-H and sGn-H-FT mRNA LNPs exhibited strong activity, inducing effective humoral immunity in the immunized mice. By week 15 post-immunization, both total IgG and neutralizing antibodies (NAbs) remained at elevated levels. Furthermore, IFNAR−/− mice immunized with sGn-H or sGn-H-FT mRNA LNPs successfully survived a lethal challenge with SFTSV. These mice experienced minor weight loss but recovered fully and rapidly. Their findings suggest that sGn-H and sGn-H-FT are promising vaccine antigen candidates capable of providing protection against SFTSV infection.
After that, Kim constructed a LNP-encapsulated mRNA vaccine expressing SFTSV Gn (91). Six-week-old C57BL/6 mice were immunized twice with LNP-encapsulated mRNA-Gn or PBS through intramuscular injection with a 14 day interval. The vaccine successfully induced robust humoral and cellular immunity in the mice. In subsequent challenge experiments, the mRNA-Gn mice showed effective protective effects.
Recently, Lu et al. developed an mRNA vaccine encoding the full-length SFTSV GP (92). The vaccine successfully induced humoral immunity and Th1-biased cellular immune responses in BALB/c mice. In IFNAR−/− mice challenged with a lethal dose of SFTSV, 1 µg of the vaccine provided 100% protection, and 0.1 µg provided 90% protection. Subsequently, researchers conducted a SFTSV challenge experiment 21 weeks after vaccination at a dose of 5μg, and the mice maintained a 100% survival rate, successfully validating the long-term protective effect induced by the vaccine. Additionally, the full-length SFTSV glycoprotein mRNA vaccine also provided cross-protection against Heartland virus and Guertu virus, revealing a potential strategy for a broad-spectrum Bandavirus vaccine.
5.7 SFTSV vaccine targets
In summary, the development of vaccines against SFTSV currently encompasses various technological platforms, each facing challenges in selecting critical targets during the development process. Therefore, in-depth research on the key antigenic regions and immune response mechanisms of SFTSV is of great significance for optimizing vaccine design and enhancing vaccine efficacy. Gn is a core component in the processes of viral entry and membrane fusion, and it is also a major antigenic element. The domain III of its head structure has been identified as a specific target for neutralizing antibodies, while domain II may serve as an ideal binding site for broadly neutralizing antibodies (33). Identifying key antigenic regions that effectively induce neutralizing antibodies is essential for the development of certain types of vaccines.
Studies using cryo-electron microscopy (cryo-EM) on SFTSV have revealed that the aggregation of the Gn head on top of the Gc subunit forms a crown-like structure, making it less accessible to solvent. This aligns with observations that many neutralizing antibodies inhibit viral infection by targeting the Gn head, indicating that the Gn head is an ideal candidate for developing subunit vaccines. The subunit vaccine and mRNA vaccine based on GnH constructed by Kim et al. provided strong support for this finding (87, 90). The N914 glycosylation site on Gc is crucial for the assembly of viral particles and is highly conserved among bunyaviruses, making it a promising target for developing broad-spectrum protective vaccines (112).
While clarifying the key antigenic regions, the selection of standard strains is also of great importance. Researchers have found that the HB29 viral strain exhibits strong cross-reactivity with heterologous antibodies and demonstrates high neutralizing efficacy with sera from 33 SFTS patients. This indicates that the HB29 strain possesses broad immunogenicity and holds potential as an optimal standard strain for vaccine development (113). Additionally, the aforementioned live-attenuated vaccine based on the HB29 strain has shown cross-protective benefits against heterologous genotype B strains (79). mRNA vaccine candidates constructed using the HB29 strain by Lu et al. further demonstrated cross-protection against Heartland virus and Guertu virus (92).
The development of broad-spectrum vaccines against Severe Fever with Thrombocytopenia Syndrome Virus (SFTSV) represents a critical strategy to address its genetic diversity and high mutability, requiring the integration of multi-target design and advanced technological platforms. Specifically, one approach is to draw insights from broad-spectrum coronavirus vaccine development by employing a multivalent antigen combination and conserved epitope-targeting strategy (114). Incorporating antigens from different SFTSV genotypes into vaccine design could enhance the induction of broadly neutralizing antibodies. Additionally, targeting highly conserved epitopes within SFTSV, such as the N914 glycosylation site on the Gc protein, as a central antigenic determinant may help mitigate the risk of immune evasion caused by viral mutations. Second, advanced vaccine platforms should be strategically employed. mRNA vaccines offer significant advantages due to their flexibility and high efficiency, enabling rapid antigen optimization through sequence modifications. Moreover, integrating artificial intelligence to predict potential mutation sites and design antigenic compositions that cover multiple genotypes presents an innovative and forward-looking strategy for broad-spectrum vaccine development.
5.8 Challenge and prospect of candidate vaccines
Although various technological approaches to developing SFTSV vaccines have shown efficacy in preventing infections in animal models, limitations persist due to gaps in understanding pathogenic mechanisms and immune response pathways.
Live attenuated and inactivated vaccines, as traditional platforms, leverage well-established development systems that have significantly contributed to many viral vaccines. Live attenuated vaccines reduce pathogen virulence while preserving immunogenicity, eliciting robust immune responses and protective efficacy. However, they carry risks of mutation reversion to virulence, or excessive immune reactions. Inactivated vaccines offer greater superior safety and stability compared to live attenuated vaccines but often require multiple doses and adjuvants. Moreover, their protective efficacy in pathogenic animal models requires further evaluation. Protein subunit vaccines also excel in safety and stability, with several candidates demonstrating protective efficacy in ferrets and immunodeficient mice. However, their high production costs and reliance on adjuvants to enhance efficacy pose challenges. Recombinant vector vaccines exhibit robust protective effects in immunodeficient animals. However, their complex production process for this technology is relatively complex, and pre-existing immunity may impact vaccine efficacy. Compared with traditional vaccines, nucleic acid vaccines provide flexibility in target selection, enabling rapid testing of multiple antigen designs. Their shorter production cycles make them well-suited for addressing public health emergencies. DNA vaccine candidates, however, face challenges such as lower immunogenicity and the need for multiple doses, necessitating further research into optimization of target design and adjuvant selection. mRNA vaccines, which respond rapidly adapt to pathogen mutations, allow for prompt optimization for variants and subtypes. They also show potential for broad-spectrum antiviral effects. However, their long-term immune efficacy requires further investigation.
When comparing vaccine candidates, it is critical not only to evaluate the levels of neutralizing antibodies they elicit but also to remain vigilant for the risk of antibody-dependent enhancement (ADE) mediated by non-neutralizing antibodies. Although antibodies against the nucleocapsid protein are detectable early in infection, they lack significant neutralizing activity and may, in some cases, form sub-neutralizing antibodies that could theoretically promote viral entry into immune cells via Fcγ receptors (115). While no experimental evidence of ADE exists for SFTSV infection, experiences with dengue and other viruses suggest that non-neutralizing or low-affinity antibodies, once bound to the virus, can be internalized through specific Fcγ receptor-mediated pathways, enhancing viral replication (116). Therefore, in evaluating various vaccine platforms, it is crucial to prioritize those that induce robust neutralizing antibody responses while minimizing the generation of large quantities of non-neutralizing antibodies.
Building on a comprehensive understanding of the strengths and limitations of current vaccine platforms under investigation, future research should focus on groundbreaking strategies to advance next-generation SFTSV vaccines with improved efficacy and safety through innovations in antigen design, delivery systems, and immunization protocols. For antigen design, insights from RSV and SARS-CoV-2 vaccine development, such as stabilizing the pre-fusion conformation of viral surface proteins could be adapted. High-resolution structural characterization of neutralizing epitopes via cryo-EM could guide site-directed mutations or fusion tags to lock Gn/Gc antigens into their most immunogenic conformations, thereby enhancing the specificity and potency of neutralizing antibodies (117, 118). Delivery system optimization may incorporate novel adjuvants like TLR7/8, STING, or RIG-I agonists to boost antigen presentation and immune activation (119–121). Additionally, developing intranasal or oral vaccine formulations could induce mucosal IgA responses and localized cellular immunity in the upper respiratory tract and gut, establishing a frontline defense at viral entry sites (122).
6 Discussion
SFTS poses a significant global pandemic risk, threatening public health worldwide. In endemic regions, persistent tick-borne transmission of SFTSV severely endangers local populations. Consequently, research into SFTSV prevention and treatment is of paramount importance.
Significant progress has been achieved in developing animal models for SFTSV infection, yet limitations persist. Future research should prioritize establishing more effective, broadly applicable animal models to address SFTSV’s public health challenges. Integrating animal study data with clinical data from human infections will provide deeper insights into viral pathogenesis and support the development of models that accurately reflect human clinical manifestations. Additionally, models accounting for variations in age and immune status should be developed to explore how aging, comorbidities, and immunosuppression influence SFTSV infection. Such models will offer valuable insights for vaccine design and personalized therapies. Humanized mouse and non-human primate models show great promise for SFTSV research but require further optimization to enhance practicality, standardization, and reproducibility. Refining these models will improve their utility for preclinical evaluation of vaccines and antiviral therapies.
Vaccination remains the most effective strategy for preventing infectious disease outbreaks. However, progress in developing prophylactic SFTSV vaccines has been slow. All candidate vaccines are currently in preclinical stages, validated only in small animal models, with no studies in large animal models or human clinical trials. Homologous recombination in SFTSV within hosts or arthropod vectors presents a major challenge for vaccine development, contributing to immune evasion, viral diversity, enhanced virulence, and technical adaptability issues. As SFTSV is primarily endemic to certain Asian regions with low global incidence, limited market demand, prolonged development cycles, and high costs reduce commercial incentives for vaccine development. Furthermore, vaccine development faces risks that require careful evaluation, including reversion to virulence in live attenuated vaccines, interference from pre-existing immunity in viral vector vaccines, potential autoimmune reactions from mRNA vaccines, and ADE observed in dengue vaccines (123, 124). These risks underscore the need for systematic assessment in SFTSV vaccine development. Addressing these challenges demands an integrated approach combining monitoring, research, and immunization strategies to bridge basic research and practical application. Continuous monitoring of viral mutations and antigenic variations, alongside multi-target strategies and advanced technological platforms, can enhance vaccine efficacy, broaden protection against diverse viral variants, and improve immunogenicity.
In the absence of an approved vaccine, targeted public health measures are essential to reduce infection risks for high-risk groups, such as the elderly, farmers, and veterinarians. In Daishan County, Zhejiang Province, China, successful prevention and control measures have been implemented (125). These include habitat cleanup through regular removal of shrubs and fallen leaves around villages to create tick-free buffer zones, chemical tick control using long-lasting pyrethroid insecticides in farmlands, forest edges, and recreational areas, and health education campaigns via village broadcasts, pamphlets, and household visits to promote tick-bite prevention and early medical intervention. Additionally, livestock management practices, such as regular insecticidal dips for cattle and sheep, minimize tick-borne pathogen transmission. These efforts reduced the annual incidence rate in Daishan County by an average of 39.98% per year from 2015 to 2019 (APC = –39.98%, P < 0.001), maintaining low and stable incidence since 2019.
Future SFTSV research should emphasize multidisciplinary collaboration, integrating virology, immunology, epidemiology, bioinformatics, computational biology, and structural biology to create a comprehensive research framework bridging fundamental studies and clinical applications. In viral pathogenesis and immune regulation, high-throughput sequencing and genome editing technologies should be leveraged to investigate SFTSV genomic evolution, elucidate molecular mechanisms of infection, identify key pathogenic factors, and explore immune evasion strategies. A deeper understanding of host antiviral immune responses will guide vaccine design and antiviral therapies. Epidemiological studies should utilize large-scale population surveys and mathematical modeling to characterize SFTSV transmission dynamics. Geographic Information System (GIS) technologies can enable spatial-temporal analysis of viral spread, identifying high-risk zones and predicting outbreak hotspots. Efforts should also enhance vaccination feasibility and acceptance while developing targeted prevention and control measures for diverse populations. In computational biology, artificial intelligence and deep learning can analyze viral genetic variations, predict mutation sites, and identify conserved antigenic epitopes to improve broad-spectrum vaccine efficacy. In drug development, integrating structural biology and computational chemistry can optimize small-molecule antiviral drugs and design neutralizing antibodies, enhancing therapeutic outcomes. Future studies should also evaluate long-term immune responses following SFTSV infection, assessing the feasibility of convalescent plasma therapy and monoclonal antibody treatments.
Author contributions
CC: Writing – review & editing, Writing – original draft. JL: Writing – review & editing, Writing – original draft. YoZ: Writing – original draft. YZ: Writing – original draft. RH: Writing – original draft. YnZ: Funding acquisition, Project administration, Writing – review & editing, Supervision. JL: Project administration, Supervision, Writing – review & editing, Funding acquisition. KC: Conceptualization, Writing – review & editing, Supervision.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was funded by the Medicine and health technology project of Zhejiang Province (2025KY762) and the Medicine and health technology project of Zhejiang Province (2024KY912).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Yu XJ, Liang MF, Zhang SY, Liu Y, Li JD, Sun YL, et al. Fever with thrombocytopenia associated with A novel bunyavirus in China. N Engl J Med. (2011) 364:1523–32. doi: 10.1056/NEJMoa1010095
2. Takahashi T, Maeda K, Suzuki T, Ishido A, Shigeoka T, Tominaga T, et al. The first identification and retrospective study of severe fever with thrombocytopenia syndrome in Japan. J Infect Dis. (2014) 209:816–27. doi: 10.1093/infdis/jit603
3. Kim KH, Yi J, Kim G, Choi SJ, Jun KI, Kim NH, et al. Severe fever with thrombocytopenia syndrome, South Korea 2012. Emerg Infect Dis. (2013) 19:1892–4. doi: 10.3201/eid1911.130792
4. Tran XC, Yun Y, Van An L, Kim SH, Thao NTP, Man PKC, et al. Endemic severe fever with thrombocytopenia syndrome, Vietnam. Emerg Infect Dis. (2019) 25:1029–31. doi: 10.3201/eid2505.181463
5. Rattanakomol P, Khongwichit S, Linsuwanon P, Lee KH, Vongpunsawad S, and Poovorawan Y. Severe fever with thrombocytopenia syndrome virus infection, Thailand 2019-2020. Emerg Infect Dis. (2022) 28:2572–4. doi: 10.3201/eid2812.221183
6. Zohaib A, Zhang J, Saqib M, Athar MA, Hussain MH, Chen J, et al. Serologic evidence of severe fever with thrombocytopenia syndrome virus and related viruses in Pakistan. Emerg Infect Dis. (2020) 26:1513–6. doi: 10.3201/eid2607.190611
7. Wormser GP, Mckenna D, Piedmonte N, Vinci V, Egizi AM, Backenson B, et al. First recognized human bite in the United States by the asian longhorned tick, haemaphysalis longicornis. Clin Infect Dis. (2020) 70:314–6. doi: 10.1093/cid/ciz449
8. Kuhn JH, Adkins S, Alioto D, Alkhovsky SV, Amarasinghe GK, Anthony SJ, et al. 2020 taxonomic update for phylum negarnaviricota (Riboviria: orthornavirae), including the large orders bunyavirales and mononegavirales. Arch Virol. (2020) 165:3023–72. doi: 10.1007/s00705-020-04731-2
9. Luo LM, Zhao L, Wen HL, Zhang ZT, Liu JW, Fang LZ, et al. Haemaphysalis longicornis ticks as reservoir and vector of severe fever with thrombocytopenia syndrome virus in China. Emerg Infect Dis. (2015) 21:1770–6. doi: 10.3201/eid2110.150126
10. He Z, Wang B, Li Y, Du Y, Ma H, Li X, et al. Severe fever with thrombocytopenia syndrome: A systematic review and meta-analysis of epidemiology, clinical signs, routine laboratory diagnosis, risk factors, and outcomes. BMC Infect Dis. (2020) 20:575. doi: 10.1186/s12879-020-05303-0
11. Bopp NE, Kaiser JA, Strother AE, Barrett ADT, Beasley DWC, Benassi V, et al. Baseline mapping of severe fever with thrombocytopenia syndrome virology, epidemiology and vaccine research and development. NPJ Vaccines. (2020) 5:111. doi: 10.1038/s41541-020-00257-5
12. Gai ZT, Zhang Y, Liang MF, Jin C, Zhang S, Zhu CB, et al. Clinical progress and risk factors for death in severe fever with thrombocytopenia syndrome patients. J Infect Dis. (2012) 206:1095–102. doi: 10.1093/infdis/jis472
13. Kim UJ, Oh TH, Kim B, Kim SE, Kang SJ, Park KH, et al. Hyperferritinemia as A diagnostic marker for severe fever with thrombocytopenia syndrome. Dis Markers. (2017) 2017:6727184. doi: 10.1155/2017/6727184
14. Liu Q, He B, Huang SY, Wei F, and Zhu XQ. Severe fever with thrombocytopenia syndrome, an emerging tick-borne zoonosis. Lancet Infect Dis. (2014) 14:763–72. doi: 10.1016/S1473-3099(14)70718-2
15. He Z, Wang B, Li Y, Hu K, Yi Z, Ma H, et al. Changes in peripheral blood cytokines in patients with severe fever with thrombocytopenia syndrome. J Med Virol. (2021) 93:4704–13. doi: 10.1002/jmv.26877
16. Liu J, Wang L, Feng Z, Geng D, Sun Y, and Yuan G. Dynamic changes of laboratory parameters and peripheral blood lymphocyte subsets in severe fever with thrombocytopenia syndrome patients. Int J Infect Dis. (2017) 58:45–51. doi: 10.1016/j.ijid.2017.02.017
17. Liu MM, Lei XY, Yu H, Zhang JZ, and Yu XJ. Correlation of cytokine level with the severity of severe fever with thrombocytopenia syndrome. Virol J. (2017) 14:6. doi: 10.1186/s12985-016-0677-1
18. Li MM, Zhang WJ, Weng XF, Li MY, Liu J, Xiong Y, et al. Cd4 T cell loss and th2 and th17 bias are associated with the severity of severe fever with thrombocytopenia syndrome (Sfts). Clin Immunol. (2018) 195:8–17. doi: 10.1016/j.clim.2018.07.009
19. Song P, Zheng N, Liu Y, Tian C, Wu X, Ma X, et al. Deficient humoral responses and disrupted B-cell immunity are associated with fatal sftsv infection. Nat Commun. (2018) 9:3328. doi: 10.1038/s41467-018-05746-9
20. Ding S, Niu G, Xu X, Li J, Zhang X, Yin H, et al. Age is A critical risk factor for severe fever with thrombocytopenia syndrome. PloS One. (2014) 9:E111736. doi: 10.1371/journal.pone.0111736
21. Mehand MS, Al-Shorbaji F, Millett P, and Murgue B. The who R&D blueprint: 2018 review of emerging infectious diseases requiring urgent research and development efforts. Antiviral Res. (2018) 159:63–7. doi: 10.1016/j.antiviral.2018.09.009
22. Pollard AJ and Bijker EM. A guide to vaccinology: from basic principles to new developments. Nat Rev Immunol. (2021) 21:83–100. doi: 10.1038/s41577-020-00479-7
23. Chaudhary N, Weissman D, and Whitehead KA. Mrna vaccines for infectious diseases: principles, delivery and clinical translation. Nat Rev Drug Discov. (2021) 20:817–38. doi: 10.1038/s41573-021-00283-5
24. Savage HM, Godsey MS, Lambert A, Panella NA, Burkhalter KL, Harmon JR, et al. First detection of heartland virus (Bunyaviridae: phlebovirus) from field collected arthropods. Am J Trop Med Hyg. (2013) 89:445–52. doi: 10.4269/ajtmh.13-0209
25. Mourya DT, Yadav PD, Basu A, Shete A, Patil DY, Zawar D, et al. Malsoor virus, A novel bat phlebovirus, is closely related to severe fever with thrombocytopenia syndrome virus and heartland virus. J Virol. (2014) 88:3605–9. doi: 10.1128/JVI.02617-13
26. Vogel D, Thorkelsson SR, Quemin ERJ, Meier K, Kouba T, Gogrefe N, et al. Structural and functional characterization of the severe fever with thrombocytopenia syndrome virus L protein. Nucleic Acids Res. (2020) 48:5749–65. doi: 10.1093/nar/gkaa253
27. Sun Z, Cheng J, Bai Y, Cao L, Xie D, Deng F, et al. Architecture of severe fever with thrombocytopenia syndrome virus. Protein Cell. (2023) 14:914–8. doi: 10.1093/procel/pwad019
28. Yuan F and Zheng A. Entry of severe fever with thrombocytopenia syndrome virus. Virol Sin. (2017) 32:44–50. doi: 10.1007/s12250-016-3858-6
29. Sun Y, Qi Y, Liu C, Gao W, Chen P, Fu L, et al. Nonmuscle myosin heavy chain iia is A critical factor contributing to the efficiency of early infection of severe fever with thrombocytopenia syndrome virus. J Virol. (2014) 88:237–48. doi: 10.1128/JVI.02141-13
30. Hofmann H, Li X, Zhang X, Liu W, Kuhl A, Kaup F, et al. Severe fever with thrombocytopenia virus glycoproteins are targeted by neutralizing antibodies and can use dc-sign as A receptor for ph-dependent entry into human and animal cell lines. J Virol. (2013) 87:4384–94. doi: 10.1128/JVI.02628-12
31. Tani H, Shimojima M, Fukushi S, Yoshikawa T, Fukuma A, Taniguchi S, et al. Characterization of glycoprotein-mediated entry of severe fever with thrombocytopenia syndrome virus. J Virol. (2016) 90:5292–301. doi: 10.1128/JVI.00110-16
32. Drake MJ, Brennan B, Briley K Jr., Bart SM, Sherman E, Szemiel AM, et al. A role for glycolipid biosynthesis in severe fever with thrombocytopenia syndrome virus entry. PloS Pathog. (2017) 13:E1006316. doi: 10.1371/journal.ppat.1006316
33. Wu Y, Zhu Y, Gao F, Jiao Y, Oladejo BO, Chai Y, et al. Structures of phlebovirus glycoprotein gn and identification of A neutralizing antibody epitope. Proc Natl Acad Sci U.S.A. (2017) 114:E7564–73. doi: 10.1073/pnas.1705176114
34. Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, et al. Sars-cov-2 variants, spike mutations and immune escape. Nat Rev Microbiol. (2021) 19:409–24. doi: 10.1038/s41579-021-00573-0
35. Wei CJ, Crank MC, Shiver J, Graham BS, Mascola JR, and Nabel GJ. Next-generation influenza vaccines: opportunities and challenges. Nat Rev Drug Discov. (2020) 19:239–52. doi: 10.1038/s41573-019-0056-x
36. Jacob ST, Crozier I, Fischer WA, Hewlett A, Kraft CS, Vega MA, et al. Ebola virus disease. Nat Rev Dis Primers. (2020) 6:13. doi: 10.1038/s41572-020-0147-3
37. Liu S, Chai C, Wang C, Amer S, Lv H, He H, et al. Systematic review of severe fever with thrombocytopenia syndrome: virology, epidemiology, and clinical characteristics. Rev Med Virol. (2014) 24:90–102. doi: 10.1002/rmv.1776
38. Zhou H, Sun Y, Wang Y, Liu M, Liu C, Wang W, et al. The nucleoprotein of severe fever with thrombocytopenia syndrome virus processes A stable hexameric ring to facilitate rna encapsidation. Protein Cell. (2013) 4:445–55. doi: 10.1007/s13238-013-3901-4
39. Lokupathirage SMW, Tsuda Y, Ikegame K, Noda K, Muthusinghe DS, Kozawa F, et al. Subcellular localization of nucleocapsid protein of sftsv and its assembly into the ribonucleoprotein complex with L protein and viral rna. Sci Rep. (2021) 11:22977. doi: 10.1038/s41598-021-01985-x
40. Li ZM, Duan SH, Yu TM, Li B, Zhang WK, Zhou CM, et al. Bunyavirus sftsv nss utilizes autophagy to escape the antiviral innate immune response. Autophagy. (2024) 20:2133–45. doi: 10.1080/15548627.2024.2356505
41. Khalil J, Kato H, and Fujita T. The role of non-structural protein nss in the pathogenesis of severe fever with thrombocytopenia syndrome. Viruses. (2021) 13. doi: 10.3390/v13050876
42. Choi Y, Park SJ, Sun Y, Yoo JS, Pudupakam RS, Foo SS, et al. Severe fever with thrombocytopenia syndrome phlebovirus non-structural protein activates tpl2 signalling pathway for viral immunopathogenesis. Nat Microbiol. (2019) 4:429–37. doi: 10.1038/s41564-018-0329-x
43. Brennan B, Rezelj VV, and Elliott RM. Mapping of transcription termination within the S segment of sfts phlebovirus facilitated generation of nss deletant viruses. J Virol. (2017) 91. doi: 10.1128/JVI.00743-17
44. Zhuang L, Sun Y, Cui XM, Tang F, Hu JG, Wang LY, et al. Transmission of severe fever with thrombocytopenia syndrome virus by haemaphysalis longicornis ticks, China. Emerg Infect Dis. (2018) 24:868–71. doi: 10.3201/eid2405.151435
45. Hu YY, Zhuang L, Liu K, Sun Y, Dai K, Zhang XA, et al. Role of three tick species in the maintenance and transmission of severe fever with thrombocytopenia syndrome virus. PloS Negl Trop Dis. (2020) 14:E0008368. doi: 10.1371/journal.pntd.0008368
46. Jo YS, Kang JG, Chae JB, Cho YK, Shin JH, Jheong WH, et al. Prevalence of severe fever with thrombocytopenia syndrome virus in ticks collected from national parks in Korea. Vector Borne Zoonotic Dis. (2019) 19:284–9. doi: 10.1089/vbz.2018.2338
47. Han XH, Ma Y, Liu HY, Li D, Wang Y, Jiang FH, et al. Identification of severe fever with thrombocytopenia syndrome virus genotypes in patients and ticks in Liaoning Province, China. Parasit Vectors. (2022) 15:120. doi: 10.1186/s13071-022-05237-3
48. Zhang X, Zhao C, Cheng C, Zhang G, Yu T, Lawrence K, et al. Rapid spread of severe fever with thrombocytopenia syndrome virus by parthenogenetic asian longhorned ticks. Emerg Infect Dis. (2022) 28:363–72. doi: 10.3201/eid2802.211532
49. Leblebicioglu H, Eroglu C, Erciyas-Yavuz K, Hokelek M, Acici M, and Yilmaz H. Role of migratory birds in spreading crimean-congo hemorrhagic fever, Turkey. Emerg Infect Dis. (2014) 20:1331–4. doi: 10.3201/eid2008.131547
50. Wilhelmsson P, Jaenson TGT, Olsen B, Waldenstrom J, and Lindgren PE. Migratory birds as disseminators of ticks and the tick-borne pathogens borrelia bacteria and tick-borne encephalitis (Tbe) virus: A seasonal study at ottenby bird observatory in South-Eastern Sweden. Parasit Vectors. (2020) 13:607. doi: 10.1186/s13071-020-04493-5
51. Yun Y, Heo ST, Kim G, Hewson R, Kim H, Park D, et al. Phylogenetic analysis of severe fever with thrombocytopenia syndrome virus in South Korea and migratory bird routes between China, South Korea, and Japan. Am J Trop Med Hyg. (2015) 93:468–74. doi: 10.4269/ajtmh.15-0047
52. Yoo JR, Choi JH, Kim YR, Lee KH, and Heo ST. Occupational risk of severe fever with thrombocytopenia syndrome in healthcare workers. Open Forum Infect Dis. (2019) 6:Ofz210. doi: 10.1093/ofid/ofz210
53. Huang XY, Du YH, Wang HF, You AG, Li Y, Su J, et al. Prevalence of severe fever with thrombocytopenia syndrome virus in animals in Henan Province, China. Infect Dis Poverty. (2019) 8:56. doi: 10.1186/s40249-019-0569-x
54. Fu Y, Li S, Zhang Z, Man S, Li X, Zhang W, et al. Phylogeographic analysis of severe fever with thrombocytopenia syndrome virus from Zhoushan Islands, China: implication for transmission across the ocean. Sci Rep. (2016) 6:19563. doi: 10.1038/srep19563
55. Yun SM, Park SJ, Kim YI, Park SW, Yu MA, Kwon HI, et al. Genetic and pathogenic diversity of severe fever with thrombocytopenia syndrome virus (Sftsv) in South Korea. JCI Insight. (2020) 5. doi: 10.1172/jci.insight.129531
56. Yun SM, Park SJ, Park SW, Choi W, Jeong HW, Choi YK, et al. Molecular genomic characterization of tick- and human-derived severe fever with thrombocytopenia syndrome virus isolates from South Korea. PloS Negl Trop Dis. (2017) 11:E0005893. doi: 10.1371/journal.pntd.0005893
57. Zhan J, Wang Q, Cheng J, Hu B, Li J, Zhan F, et al. Current status of severe fever with thrombocytopenia syndrome in China. Virol Sin. (2017) 32:51–62. doi: 10.1007/s12250-016-3931-1
58. Dai ZN, Peng XF, Li JC, Zhao J, Wu YX, Yang X, et al. Effect of genomic variations in severe fever with thrombocytopenia syndrome virus on the disease lethality. Emerg Microbes Infect. (2022) 11:1672–82. doi: 10.1080/22221751.2022.2081617
59. Jin C, Liang M, Ning J, Gu W, Jiang H, Wu W, et al. Pathogenesis of emerging severe fever with thrombocytopenia syndrome virus in C57/bl6 mouse model. Proc Natl Acad Sci U.S.A. (2012) 109:10053–8. doi: 10.1073/pnas.1120246109
60. Matsuno K, Orba Y, Maede-White K, Scott D, Feldmann F, Liang M, et al. Animal models of emerging tick-borne phleboviruses: determining target cells in A lethal model of sftsv infection. Front Microbiol. (2017) 8:104. doi: 10.3389/fmicb.2017.00104
61. Chen XP, Cong ML, Li MH, Kang YJ, Feng YM, Plyusnin A, et al. Infection and pathogenesis of huaiyangshan virus (A novel tick-borne bunyavirus) in laboratory rodents. J Gen Virol. (2012) 93:1288–93. doi: 10.1099/vir.0.041053-0
62. Liu Y, Wu B, Paessler S, Walker DH, Tesh RB, and Yu XJ. The pathogenesis of severe fever with thrombocytopenia syndrome virus infection in alpha/beta interferon knockout mice: insights into the pathologic mechanisms of A new viral hemorrhagic fever. J Virol. (2014) 88:1781–6. doi: 10.1128/JVI.02277-13
63. Park SC, Park JY, Choi JY, Lee SG, Eo SK, Oem JK, et al. Pathogenicity of severe fever with thrombocytopenia syndrome virus in mice regulated in type I interferon signaling: severe fever with thrombocytopenia and type I interferon. Lab Anim Res. (2020) 36:38. doi: 10.1186/s42826-020-00070-0
64. Tani H, Fukuma A, Fukushi S, Taniguchi S, Yoshikawa T, Iwata-Yoshikawa N, et al. Efficacy of T-705 (Favipiravir) in the treatment of infections with lethal severe fever with thrombocytopenia syndrome virus. Msphere. (2016) 1. doi: 10.1128/mSphere.00061-15
65. Park SJ, Kim YI, Park A, Kwon HI, Kim EH, Si YJ, et al. Ferret animal model of severe fever with thrombocytopenia syndrome phlebovirus for human lethal infection and pathogenesis. Nat Microbiol. (2019) 4:438–46. doi: 10.1038/s41564-018-0317-1
66. Yoshikawa R, Sakabe S, Urata S, and Yasuda J. Species-specific pathogenicity of severe fever with thrombocytopenia syndrome virus is determined by anti-stat2 activity of nss. J Virol. (2019) 93. doi: 10.1128/JVI.02226-18
67. Xu S, Jiang N, Nawaz W, Liu B, Zhang F, Liu Y, et al. Infection of humanized mice with A novel phlebovirus presented pathogenic features of severe fever with thrombocytopenia syndrome. PloS Pathog. (2021) 17:E1009587. doi: 10.1371/journal.ppat.1009587
68. Li H, Zhang LK, Li SF, Zhang SF, Wan WW, Zhang YL, et al. Calcium channel blockers reduce severe fever with thrombocytopenia syndrome virus (Sftsv) related fatality. Cell Res. (2019) 29:739–53. doi: 10.1038/s41422-019-0214-z
69. Gowen BB, Westover JB, Miao J, Van Wettere AJ, Rigas JD, Hickerson BT, et al. Modeling severe fever with thrombocytopenia syndrome virus infection in golden Syrian hamsters: importance of stat2 in preventing disease and effective treatment with favipiravir. J Virol. (2017) 91. doi: 10.1128/JVI.01942-16
70. Park ES, Shimojima M, Nagata N, Ami Y, Yoshikawa T, Iwata-Yoshikawa N, et al. Severe fever with thrombocytopenia syndrome phlebovirus causes lethal viral hemorrhagic fever in cats. Sci Rep. (2019) 9:11990. doi: 10.1038/s41598-019-48317-8
71. Jin C, Jiang H, Liang M, Han Y, Gu W, Zhang F, et al. Sfts virus infection in nonhuman primates. J Infect Dis. (2015) 211:915–25. doi: 10.1093/infdis/jiu564
72. Albrecht RA, Liu WC, Sant AJ, Tompkins SM, Pekosz A, Meliopoulos V, et al. Moving forward: recent developments for the ferret biomedical research model. Mbio. (2018) 9. doi: 10.1128/mBio.01113-18
73. Kim YI, Kim SG, Kim SM, Kim EH, Park SJ, Yu KM, et al. Infection and rapid transmission of sars-cov-2 in ferrets. Cell Host Microbe. (2020) 27:704–709 E2. doi: 10.1016/j.chom.2020.03.023
74. Hwang J, Kang JG, Oh SS, Chae JB, Cho YK, Cho YS, et al. Molecular detection of severe fever with thrombocytopenia syndrome virus (Sftsv) in feral cats from Seoul, Korea. Ticks Tick Borne Dis. (2017) 8:9–12. doi: 10.1016/j.ttbdis.2016.08.005
75. Yamanaka A, Kirino Y, Fujimoto S, Ueda N, Himeji D, Miura M, et al. Direct transmission of severe fever with thrombocytopenia syndrome virus from domestic cat to veterinary personnel. Emerg Infect Dis. (2020) 26:2994–8. doi: 10.3201/eid2612.191513
76. Haddock E, Feldmann F, Hawman DW, Zivcec M, Hanley PW, Saturday G, et al. A cynomolgus macaque model for crimean-congo haemorrhagic fever. Nat Microbiol. (2018) 3:556–62. doi: 10.1038/s41564-018-0141-7
77. Klingstrom J, Plyusnin A, Vaheri A, and Lundkvist A. Wild-type puumala hantavirus infection induces cytokines, C-reactive protein, creatinine, and nitric oxide in cynomolgus macaques. J Virol. (2002) 76:444–9. doi: 10.1128/JVI.76.1.444-449.2002
78. Smith DR, Bird BH, Lewis B, Johnston SC, Mccarthy S, Keeney A, et al. Development of A novel nonhuman primate model for rift valley fever. J Virol. (2012) 86:2109–20. doi: 10.1128/JVI.06190-11
79. Yu KM, Park SJ, Yu MA, Kim YI, Choi Y, Jung JU, et al. Cross-genotype protection of live-attenuated vaccine candidate for severe fever with thrombocytopenia syndrome virus in A ferret model. Proc Natl Acad Sci U.S.A. (2019) 116:26900–8. doi: 10.1073/pnas.1914704116
80. Li A, Dai X, Chen L, Liu L, Li C, Liu Y, et al. Immunogenicity and protective efficacy of an inactivated sfts vaccine candidate in mice. Biosafety Health. (2022) 4:45–52. doi: 10.1016/j.bsheal.2021.12.008
81. Dong F, Li D, Wen D, Li S, Zhao C, Qi Y, et al. Single dose of A rvsv-based vaccine elicits complete protection against severe fever with thrombocytopenia syndrome virus. NPJ Vaccines. (2019) 4:5. doi: 10.1038/s41541-018-0096-y
82. Yoshikawa T, Taniguchi S, Kato H, Iwata-Yoshikawa N, Tani H, Kurosu T, et al. A highly attenuated vaccinia virus strain lc16m8-based vaccine for severe fever with thrombocytopenia syndrome. PloS Pathog. (2021) 17:E1008859. doi: 10.1371/journal.ppat.1008859
83. Zhao Z, Zheng W, Yan L, Sun P, Xu T, Zhu Y, et al. Recombinant human adenovirus type 5 co-expressing rabv G and sftsv gn induces protective immunity against rabies virus and severe fever with thrombocytopenia syndrome virus in mice. Front Microbiol. (2020) 11:1473. doi: 10.3389/fmicb.2020.01473
84. Qian H, Tian L, Liu W, Liu L, Li M, Zhao Z, et al. Adenovirus type 5-expressing gn induces better protective immunity than gc against sftsv infection in mice. NPJ Vaccines. (2024) 9:194. doi: 10.1038/s41541-024-00993-y
85. Liu R, Huang DD, Bai JY, Zhuang L, Lu QB, Zhang XA, et al. Immunization with recombinant sftsv/nss protein does not promote virus clearance in sftsv-infected C57bl/6j mice. Viral Immunol. (2015) 28:113–22. doi: 10.1089/vim.2014.0100
86. Kim S, Jeon K, Choi H, Jeong D-E, Kang J-G, and Cho N-H. Comparative analysis of the efficacy of vaccines using structural protein subunits of the severe fever with thrombocytopenia syndrome virus. Front Microbiol. (2024) 15. doi: 10.3389/fmicb.2024.1348276
87. Kim D, Kim E, Kim S, Chung Y, Lai CJ, Cha I, et al. Self-assembling gn head ferritin nanoparticle vaccine provides full protection from lethal challenge of dabie bandavirus in aged ferrets. Mbio. (2023) 14:E0186823. doi: 10.1128/mbio.01868-23
88. Kwak JE, Kim YI, Park SJ, Yu MA, Kwon HI, Eo S, et al. Development of A sftsv dna vaccine that confers complete protection against lethal infection in ferrets. Nat Commun. (2019) 10:3836. doi: 10.1038/s41467-019-11815-4
89. Kang JG, Jeon K, Choi H, Kim Y, Kim HI, Ro HJ, et al. Vaccination with single plasmid dna encoding il-12 and antigens of severe fever with thrombocytopenia syndrome virus elicits complete protection in ifnar knockout mice. PloS Negl Trop Dis. (2020) 14:E0007813. doi: 10.1371/journal.pntd.0007813
90. Kim D, Lai CJ, Cha I, Kang S, Yang WS, Choi Y, et al. Sftsv gn-head mrna vaccine confers efficient protection against lethal viral challenge. J Med Virol. (2023) 95:E29203. doi: 10.1002/jmv.29203
91. Kim JY, Jeon K, Park SI, Bang YJ, Park HJ, Kwak HW, et al. Mrna vaccine encoding gn provides protection against severe fever with thrombocytopenia syndrome virus in mice. NPJ Vaccines. (2023) 8:167. doi: 10.1038/s41541-023-00771-2
92. Lu J, Liu J, Wu Y, He X, Gao X, Chen X, et al. A full-length glycoprotein mrna vaccine confers complete protection against severe fever with thrombocytopenia syndrome virus, with broad-spectrum protective effects against bandaviruses. J Virol. (2024) 98:E0076924. doi: 10.1128/jvi.00769-24
93. Li T, Qian C, Gu Y, Zhang J, Li S, and Xia N. Current progress in the development of prophylactic and therapeutic vaccines. Sci China Life Sci. (2023) 66:679–710. doi: 10.1007/s11427-022-2230-4
94. Brennan B, Li P, Zhang S, Li A, Liang M, Li D, et al. Reverse genetics system for severe fever with thrombocytopenia syndrome virus. J Virol. (2015) 89:3026–37. doi: 10.1128/JVI.03432-14
95. Gebre MS, Brito LA, Tostanoski LH, Edwards DK, Carfi A, and Barouch DH. Novel approaches for vaccine development. Cell. (2021) 184:1589–603. doi: 10.1016/j.cell.2021.02.030
96. Travieso T, Li J, Mahesh S, Mello J, and Blasi M. The use of viral vectors in vaccine development. NPJ Vaccines. (2022) 7:75. doi: 10.1038/s41541-022-00503-y
97. Cao W, He S, Liu G, Schulz H, Emeterio K, Chan M, et al. The rvsv-ebov vaccine provides limited cross-protection against Sudan virus in Guinea pigs. NPJ Vaccines. (2023) 8:91. doi: 10.1038/s41541-023-00685-z
98. Woolsey C, Borisevich V, Fears AC, Agans KN, Deer DJ, Prasad AN, et al. Recombinant vesicular stomatitis virus-vectored vaccine induces long-lasting immunity against nipah virus disease. J Clin Invest. (2023) 133. doi: 10.1172/JCI164946
99. Grabenstein JD and Hacker A. Vaccines against mpox: mva-bn and lc16m8. Expert Rev Vaccines. (2024) 23:796–811. doi: 10.1080/14760584.2024.2397006
100. Zhang H, Wang H, An Y, and Chen Z. Construction and application of adenoviral vectors. Mol Ther Nucleic Acids. (2023) 34:102027. doi: 10.1016/j.omtn.2023.09.004
101. Wang S, Liang B, Wang W, Li L, Feng N, Zhao Y, et al. Viral vectored vaccines: design, development, preventive and therapeutic applications in human diseases. Signal Transduct Target Ther. (2023) 8:149. doi: 10.1038/s41392-023-01408-5
102. Nossal GJ. Vaccines and future global health needs. Philos Trans R Soc Lond B Biol Sci. (2011) 366:2833–40. doi: 10.1098/rstb.2011.0093
103. Garland SM, Hernandez-Avila M, Wheeler CM, Perez G, Harper DM, Leodolter S, et al. Quadrivalent vaccine against human papillomavirus to prevent anogenital diseases. N Engl J Med. (2007) 356:1928–43. doi: 10.1056/NEJMoa061760
104. Dowd KA, Ko SY, Morabito KM, Yang ES, Pelc RS, Demaso CR, et al. Rapid development of A dna vaccine for zika virus. Science. (2016) 354:237–40. doi: 10.1126/science.aai9137
105. Parhiz H, AtoChina-Vasserman EN, and Weissman D. Mrna-based therapeutics: looking beyond covid-19 vaccines. Lancet. (2024) 403:1192–204. doi: 10.1016/S0140-6736(23)02444-3
106. Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, et al. Safety and efficacy of the bnt162b2 mrna covid-19 vaccine. N Engl J Med. (2020) 383:2603–15. doi: 10.1056/NEJMoa2034577
107. Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, et al. Efficacy and safety of the mrna-1273 sars-cov-2 vaccine. N Engl J Med. (2021) 384:403–16. doi: 10.1056/NEJMoa2035389
108. Islam MA. A review of sars-cov-2 variants and vaccines: viral properties, mutations, vaccine efficacy, and safety. Infect Med. (2023) 2:247–61. doi: 10.1016/j.imj.2023.08.005
109. Kanizsai A, Molnar T, Varnai R, Zavori L, Tokes-Fuzesi M, Szalai Z, et al. Fever after vaccination against sars-cov-2 with mrna-based vaccine associated with higher antibody levels during 6 months follow-up. Vaccines (Basel). (2022) 10. doi: 10.3390/vaccines10030447
110. Wu L, Yi W, Yao S, Xie S, Peng R, Zhang J, et al. Mrna-based cancer vaccines: advancements and prospects. Nano Lett. (2024). doi: 10.1021/acs.nanolett.4c03296
111. Essink B, Chu L, Seger W, Barranco E, Le Cam N, Bennett H, et al. The safety and immunogenicity of two zika virus mrna vaccine candidates in healthy flavivirus baseline seropositive and seronegative adults: the results of two randomised, placebo-controlled, dose-ranging, phase 1 clinical trials. Lancet Infect Dis. (2023) 23:621–33. doi: 10.1016/S1473-3099(22)00764-2
112. Du S, Peng R, Xu W, Qu X, Wang Y, Wang J, et al. Cryo-em structure of severe fever with thrombocytopenia syndrome virus. Nat Commun. (2023) 14:6333. doi: 10.1038/s41467-023-41804-7
113. Jia Z, Wu X, Wang L, Li X, Dai X, Liang M, et al. Identification of A candidate standard strain of severe fever with thrombocytopenia syndrome virus for vaccine quality control in China using A cross-neutralization assay. Biologicals. (2017) 46:92–8. doi: 10.1016/j.biologicals.2017.01.005
114. Su S, Li W, and Jiang S. Developing pan-beta-coronavirus vaccines against emerging sars-cov-2 variants of concern. Trends Immunol. (2022) 43:170–2. doi: 10.1016/j.it.2022.01.009
115. Lee K, Choi MJ, Cho MH, Choi DO, and Bhoo SH. Antibody production and characterization of the nucleoprotein of sever fever with thrombocytopenia syndrome virus (Sftsv) for effective diagnosis of sftsv. Virol J. (2023) 20:206. doi: 10.1186/s12985-023-02173-1
116. Shukla R, Ramasamy V, Shanmugam RK, Ahuja R, and Khanna N. Antibody-dependent enhancement: A challenge for developing A safe dengue vaccine. Front Cell Infect Microbiol. (2020) 10:572681. doi: 10.3389/fcimb.2020.572681
117. Che Y, Gribenko AV, Song X, Handke LD, Efferen KS, Tompkins K, et al. Rational design of A highly immunogenic prefusion-stabilized F glycoprotein antigen for A respiratory syncytial virus vaccine. Sci Transl Med. (2023) 15:Eade6422. doi: 10.1126/scitranslmed.ade6422
118. Hsieh CL, Goldsmith JA, Schaub JM, Divenere AM, Kuo HC, Javanmardi K, et al. Structure-based design of prefusion-stabilized sars-cov-2 spikes. Science. (2020) 369:1501–5. doi: 10.1126/science.abd0826
119. Li G, Yu M, Ke Q, Sun J, Peng Y, Xiong C, et al. Enhancement of sars-cov-2 vaccine-induced immunity by A toll-like receptor 7 agonist adjuvant. Signal Transduct Target Ther. (2023) 8:213. doi: 10.1038/s41392-023-01485-6
120. Van Herck S, Feng B, and Tang L. Delivery of sting agonists for adjuvanting subunit vaccines. Adv Drug Delivery Rev. (2021) 179:114020. doi: 10.1016/j.addr.2021.114020
121. Hemann EA, Knoll ML, Wilkins CR, Subra C, Green R, Garcia-Sastre A, et al. A small molecule rig-I agonist serves as an adjuvant to induce broad multifaceted influenza virus vaccine immunity. J Immunol. (2023) 210:1247–56. doi: 10.4049/jimmunol.2300026
122. Kehagia E, Papakyriakopoulou P, and Valsami G. Advances in intranasal vaccine delivery: A promising non-invasive route of immunization. Vaccine. (2023) 41:3589–603. doi: 10.1016/j.vaccine.2023.05.011
123. Olivieri B, Betterle C, and Zanoni G. Vaccinations and autoimmune diseases. Vaccines (Basel). (2021) 9. doi: 10.3390/vaccines9080815
124. Spencer JP, Trondsen Pawlowski RH, and Thomas S. Vaccine adverse events: separating myth from reality. Am Fam Physician. (2017) 95:786–94.
Keywords: animal models, vaccine, severe fever with thrombocytopenia syndrome virus, Gn, GC
Citation: Chen C, Li J, Zhou Y, Zhang Y, Huang R, Zhang Y, Li J and Chen K (2025) Latest advances and prospects in the pathogenesis, animal models, and vaccine research of severe fever with thrombocytopenia syndrome virus. Front. Immunol. 16:1624290. doi: 10.3389/fimmu.2025.1624290
Received: 07 May 2025; Accepted: 13 June 2025;
Published: 26 June 2025.
Edited by:
Lei Tan, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Yimei Feng, Xinqiao Hospital, ChinaMd. Aminul Islam, Tulane University, United States
Yongai Xiong, Zunyi Medical University, China
Copyright © 2025 Chen, Li, Zhou, Zhang, Huang, Zhang, Li and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanjun Zhang, eWp6aGFuZ0BjZGMuemouY24=; Jianhua Li, amhsaUBjZGMuemouY24=; Keda Chen, Y2hlbmtkQHpqc3J1LmVkdS5jbg==
†These authors have contributed equally to this work and share the first authorship
 Chenghao Chen1,2†
Chenghao Chen1,2† Yuwen Zhang
Yuwen Zhang Renjin Huang
Renjin Huang Yanjun Zhang
Yanjun Zhang Jianhua Li
Jianhua Li Keda Chen
Keda Chen