- Biomedical Innovations Research for Translational Health Science (BIRTHS) Laboratory, Department of Biochemistry and Molecular Biology, College of Medicine, Manila, Philippines
1 Introduction
Allergy is hypersensitivity (i.e., maladaptive proinflammatory immune reactivity) to noninfectious nonself antigens (i.e., allergens) such as environmental and food components, most notably in the context of immediate-type hypersensitivity, which is mediated by IgE antibodies (1). Binding of allergens by IgE antibodies thus underlies immunodiagnostic detection of allergy; and attenuation of such binding enables immunotherapeutic management of allergy. This is the rationale for desensitization therapy that entails detecting IgE antibodies to pertinent allergens among patients who are then immunized with the same allergens, thereby eliciting production of cognate non-IgE (typically IgG) antibodies to outcompete IgE antibodies in binding the allergens and thus alleviate allergic conditions (2). However, preexisting IgE antibodies can mediate harmful allergic reactions (e.g., fatal systemic anaphylaxis) when patients are exposed to cognate allergens, especially during desensitization therapy (3). Nevertheless, a translational path toward safer desensitization therapy for allergies is conceivable via development of allergy vaccines and their companion immunodiagnostics, using B-cell epitope prediction (BCEP) as a generally applicable strategy for enhancing disease control and prevention (4). When compared to allergen extract-based immunotherapies, B-cell epitopes (BCEs) have shown promising potential both in vivo and in vitro, by inducing hypoallergenic allergen-specific IgG responses and downregulating T-cell mediated late response pathways (5–9).
2 BCEP for designing allergy vaccines
BCEP is computational identification of BCEs: structural features (e.g., parts of molecules or of supramolecular complexes) recognized by paratopes (i.e., antigen-binding sites on immunoglobulins) (4). This is complicated by the emergent phenomenon of immunodominance among BCEs, which is the bias of antibody responses toward a BCE on an immunogen (i.e., immunogenic antigen) that comprises nonidentical BCEs. A BCE is thus said to be immunodominant if antibodies are preferentially produced against it rather than another BCE also present on the same immunogen (e.g., a vaccine antigen); and the latter BCE is thus said to be subdominant, though antibodies might yet be produced against it under other circumstances (e.g., where it is the sole or most immunodominant BCE present, as in a vaccine devoid of more immunodominant BCEs).
Among peptidic (e.g., peptide or protein) antigens, a typical BCE may be regarded as consisting of paratope-contacting amino-acid residues (10). Such a BCE is said to be either continuous if its constituent residues form a contiguous sequence or discontinuous otherwise (e.g., where the BCE is formed by juxtaposition of noncontiguous residues via protein folding and disintegrates upon protein unfolding), noting that a discontinuous BCE may comprise one or more continuous BCEs (11, 12). A continuous BCE is thus embodied in an oligopeptide sequence without regard to conformation; whereas a discontinuous BCE exists only when its antigen assumes a folded conformation. Consequently, BCEP is more computationally tractable for continuous BCEs than for discontinuous BCEs insofar as it reduces to identification of oligopeptide sequences. This is the case for peptide-based vaccine design, wherein the problem of immunodominance among BCEs can be circumvented by selectively incorporating only continuous BCEs into vaccine immunogens, to enable vaccine-induced selective antibody targeting of BCEs (13) via paratope binding that is typically based on induced fit (14).
To thus apply BCEP for designing allergy vaccines, the following general observation is key: Antibody responses to folded protein antigens (e.g., typical allergens) tend to be biased toward production of antibodies to discontinuous rather than continuous BCEs (10). Although this has been interpreted as implying that most protein BCEs are discontinuous, it is a clear manifestation of immunodominance among BCEs, with discontinuous BCEs tending to be more immunodominant than continuous BCEs that are nonetheless immunogenic (e.g., as unfolded parts of denatured proteins or as oligopeptides) (15). In the setting of allergy, most clinically relevant IgE antibodies thus recognize discontinuous BCEs (16). Furthermore, multiple BCEs recognized by IgE antibodies may occur on a single allergen (17), such that it can crosslink cognate IgE antibodies bound by FcϵRI receptors on plasma membranes of mast cells and of basophils (18, 19), thereby inducing degranulation with consequent extracellular release of inflammatory mediators (e.g., histamine) that drive allergic reactions (20). Yet, IgG antibodies can readily be produced against continuous BCEs that are conformationally disordered (i.e., nonfolded) oligopeptide sequences (e.g., in synthetic peptide-based vaccines); and if these sequences are also present as paratope-accessible targets in protein antigens (e.g., on surface-exposed conformationally flexible loops), they can be bound as such by the same antibodies (4). Hence, peptidic allergens can be targeted by IgG antibodies that recognize continuous BCEs, to competitively interfere with binding of the allergens by IgE antibodies (e.g., via steric blocking) and/or to enhance immunological clearance of the allergens (e.g., via IgG-dependent opsonization), noting that IgG antibodies other than IgG4 antibodies (21) can also drive shifts from allergy-promoting Th2-dominated to tolerogenic Treg-dominated immune responses (22). This must, however, still address possible cross-reactivity whereby nonidentical (albeit typically similar) BCEs can be bound by the same antibodies (23), which is a safety concern as existing IgE antibodies may thus cross-react with BCEs on non-cognate allergens (24).
Accordingly, oligopeptide sequences comprising continuous BCEs of protein allergens could conceivably serve as components of both allergy vaccines and corresponding companion immunodiagnostics: The vaccines could elicit production of IgG antibodies to the BCEs and thereby attenuate the allergy-mediating activity of IgE antibodies to the allergens while possibly also suppressing further production of IgE antibodies; whereas the immunodiagnostics could detect antibodies to the BCEs, to assess vaccine safety and efficacy. More specifically, certain versions of the immunodiagnostics could detect preexisting IgE antibodies to the BCEs before attempts to administer the vaccines (e.g., for primary and/or booster doses), so as to avoid triggering allergic reactions to the vaccines; whereas other versions of the immunodiagnostics could detect vaccine-induced IgG antibodies to the BCEs, in order to evaluate vaccine efficacy (noting that evidence of waning vaccine immunity might warrant subsequent booster doses). More generally, the vaccines could enable production of allergen-binding IgG antibodies (e.g., as monospecific polyclonal antibodies or even monoclonal antibodies) for possible therapeutic use via passive immunization (25), which would avoid risking vaccine-induced allergic reactions altogether (26) and could also serve as a preliminary trial of therapy that, if successful, might justify longer-term management by vaccination (i.e., active immunization).
In line with the preceding considerations, selective incorporation of continuous BCEs from allergens into components of allergy vaccines and companion immunodiagnostics thereto necessitates means for identifying such BCEs in the first place. This entails identification of relevant allergens and, in turn, their pertinent continuous BCEs. These can be validated only on the basis of experimental data from various immunoassays (27), notably as curated in the Immune Epitope Database (IEDB) (28). As a case in point, Table 1 presents examples of IEDB-curated allergens and oligopeptide sequences thereof, for which active immunization with the latter is known to induce a decrease in allergic disease (noting that said sequences are each curated as a BCE in IEDB, though they are more properly regarded as BCE-containing sequences that each comprise one or more BCEs); whereas Figure 1 presents structural models of a subset of the oligopeptide sequences in the context of immune complexes each consisting of a whole protein allergen and a cognate antibody Fab fragment, with surface-exposed BCE-containing sequences that comprise disorder-prone terminal or internal loop (e.g., turn) structures. Such examples illustrate the potential of oligopeptide sequences as allergy-vaccine components; but immunoassays are resource-intensive to perform and must be complemented by computational approaches including BCEP to facilitate identification of pertinent protein allergens and continuous BCEs thereof, using available biomolecular data on allergen structure and function in the context of host immunobiology to strategically target allergen BCEs.
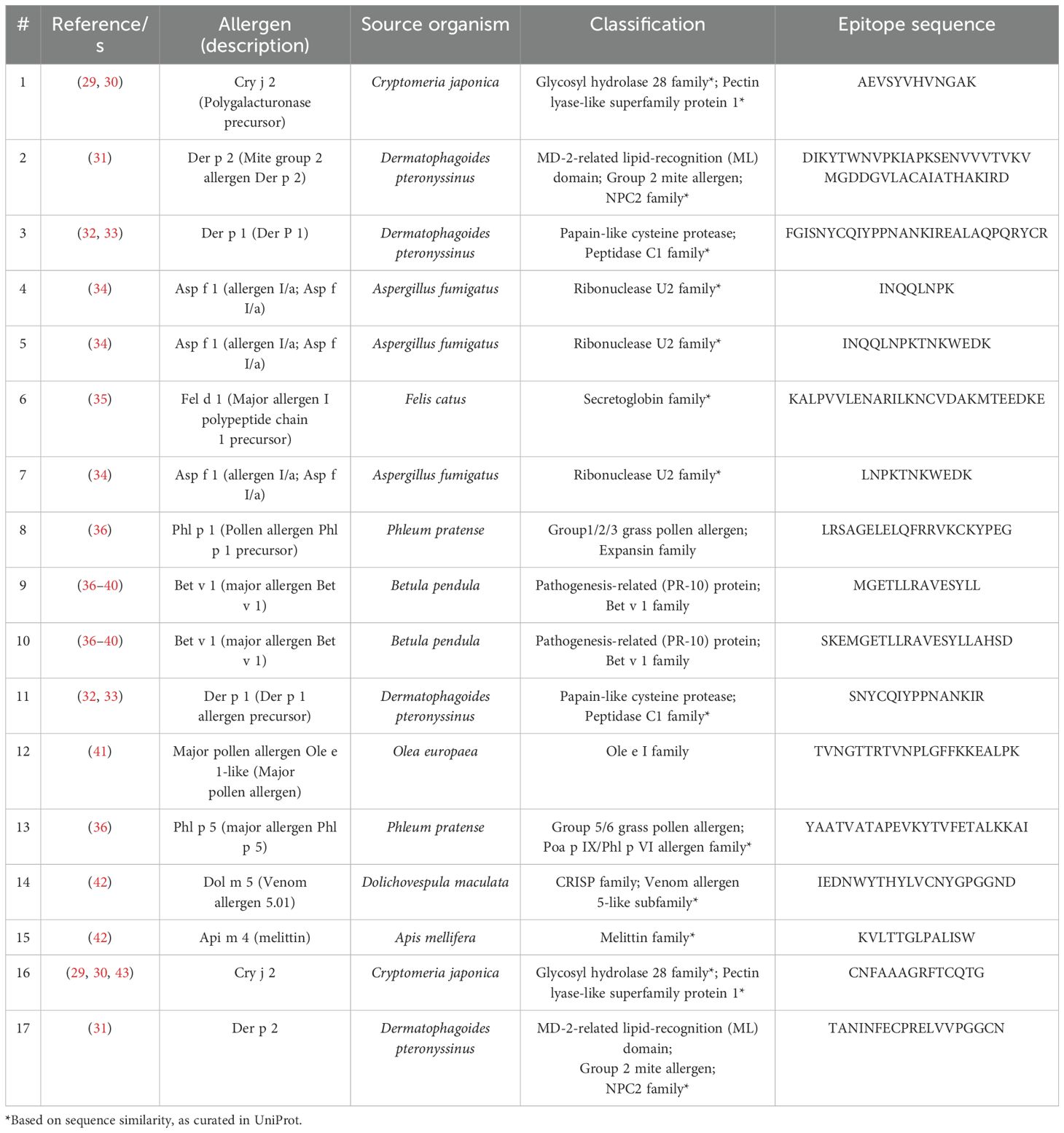
Table 1. Allergen epitope sequences curated in the Immune Epitope Database (IEDB) as inducing decreased allergic disease.
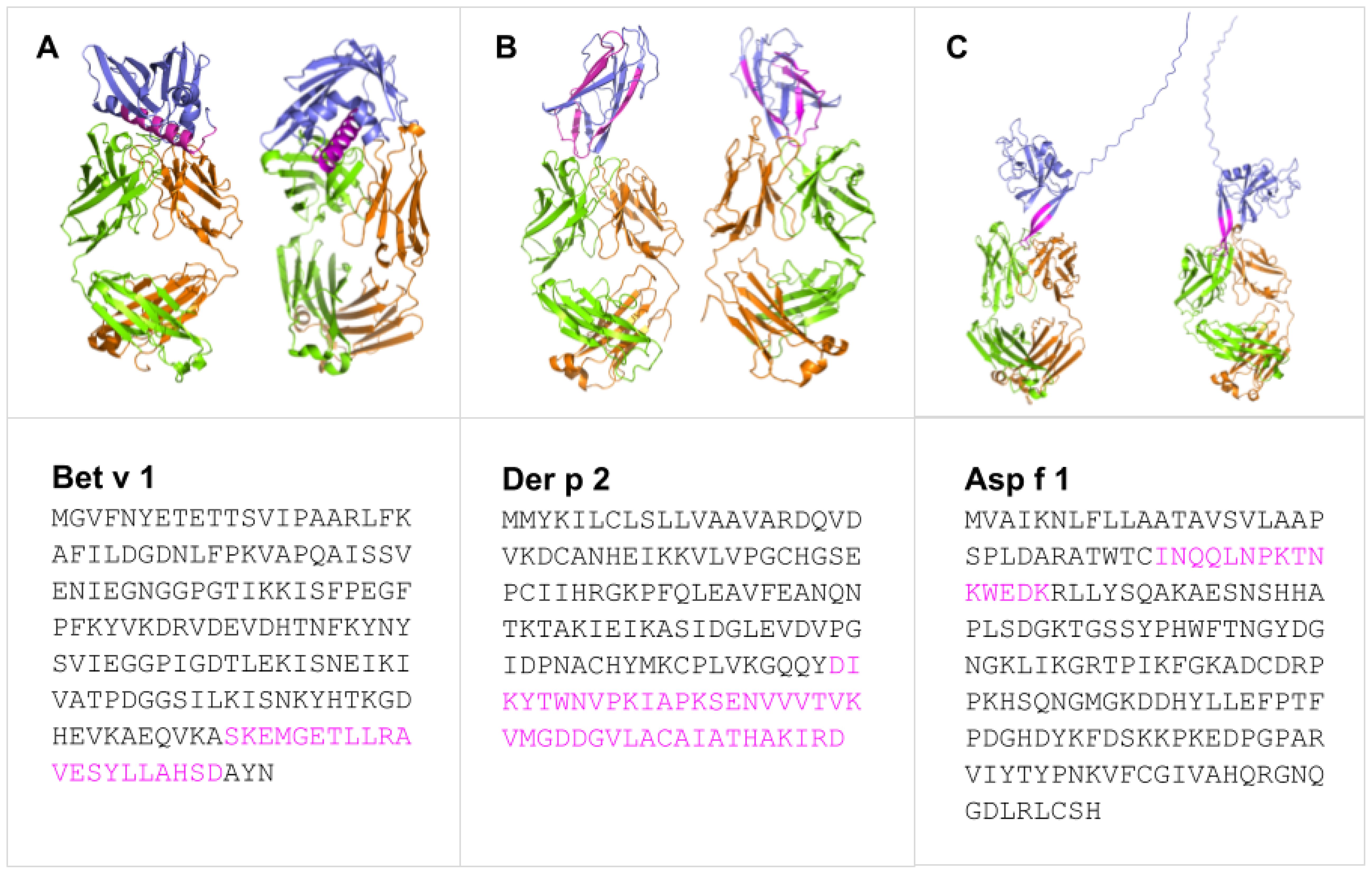
Figure 1. Predicted binding modes of Bet v 1 (A), Der p 2 (B), and Asp f 1 (C) (blue) epitopes (pink) with representative IgE and IgG Fab fragments showing heavy (green) and light (orange) chains. Generated with SWISS-MODEL Workspace (44) and PyMOL Molecular Graphics System ver 3.1.3.1.
3 Strategically targeting allergen BCEs
BCEs represent a low-level structural and functional view of antigens, but higher-level views are necessary to comprehend the role of various antigens as allergens, in order to subsequently identify their pertinent BCEs as potential therapeutic targets (e.g., for binding by IgG antibodies). In this regard, a useful analytical framework is epidemiologic transition theory (45), which seeks to explain temporal shifts in patterns of morbidity and mortality exemplified by the modernization-driven dual trend of decline in infectious diseases and rise in chronic inflammatory conditions such as allergy and autoimmunity (46). Said trend can be understood largely in terms of biota alteration theory (47), which posits that generalized suppression of the host biota (i.e., microbiota plus other symbionts such as helminths) via lifestyle changes (e.g., adoption of excessive infection-control measures and proinflammatory diets) has promoted host immunological dysregulation (48). By this account, allergy results from dysregulated production of IgE antibodies, though these are thought to have evolved as mediators of host immunity against peptidic toxins (e.g., of pathogenic bacteria and venomous animals) that can be inactivated by proteases from mast cells (49, 50).
Hence, peptidic antigens may elicit production of IgE antibodies and thus act as allergens if they cause direct host injury (e.g., via cellular or tissue damage due to proteolytic or membrane-permeabilizing activities) (51) or are recognized as danger signals via innate immune sensing mechanisms such as Toll-like receptors (TLRs) (52), especially in the setting of prolonged host exposure (e.g., due to their structural stabilization by disulfide bonds and consequent resistance to proteolytic degradation) (53). Moreover, such allergens often can undergo oligomerization (51), which in turn can facilitate crosslinking of FcϵRI receptor-bound IgE antibodies and consequent mast-cell degranulation (54). Immunotherapy and vaccine administration have previously been shown to predictably increase total serum IgE concentration and sIgE (55, 56) which may erroneously suggest a diagnosis of atopy. Thus, reinforcing the potential of BCEP as a predictive tool to minimize false-positive detection of sensitization among atopic individuals is necessary, given its capacity to streamline the development of BCEs for companion immunodiagnostic use (57).
In addition to host-damaging and danger-signaling activities vis-à-vis structural stability and oligomerization potential, allergens may be further characterized by more detailed structural features. Although allergens are structurally diverse, 19 allergen families have been identified from the Pfam database based on structural properties, with approaches to subclassification being explored mainly on the basis of homology. However, many allergens remain unclassified; and the structural properties underlying allergenicity are not yet fully understood, thus limiting BCE potential for widespread clinical use (52). Route of host exposure to antigens is also a crucial determinant of their clinical relevance as allergens (58): Food allergens, for instance, tend to enter the systemic circulation via transcytosis across the host gut lining epithelium and thus elicit production of IgE antibodies (59, 60). Such factors underlie the failure to develop experimental models that fully capture the complexity of allergic conditions (61, 62). This calls for computational workflows that can leverage data on protein sequences and structures vis-à-vis protein and immune-system function to aid in identifying clinically relevant protein allergens and/or BCEs thereof for therapeutic antibody targeting.
As thermodynamics provides a foundational framework for comprehending immune function (63), it could guide the use of computational tools (e.g., to predict protein folding and interactions) for analyzing pertinent proteomes (e.g., of cells or tissues in allergenic materials) to identify putative allergens (e.g., on the basis of predicted toxicity in particular contexts of relevant host exposure) and, in turn, candidate allergy vaccine BCEs via BCEP. Such BCEs could thus be identified as oligopeptide sequences for which sufficiently high paratope binding affinity is predicted (13), noting that paratopes that bind one BCE may fail to bind another even if the two are highly similar in sequence (64). Where identical BCEs occur on the same antigen, avidity (i.e., strength of binding due to simultaneous paratope-BCE interactions) might also be predicted (65). In this regard, steric hindrance is important to consider as it can attenuate avidity (66, 67), though it can also attenuate allergen toxicity as by inhibiting host-damaging protease activity via blockage of substrate access to active sites (68). Finally, the selected vaccine BCEs would also serve as companion immunodiagnostic probes to detect cognate antibodies, which would obviate the need for large peptide antigen arrays representing entire protein allergen sequences (69).
4 Conclusion
BCEP-based design of allergy vaccines and their companion immunodiagnostics using thermodynamics-guided computational workflows is a promising approach to further develop desensitization therapy for allergic conditions. This could enable strategic IgG-antibody targeting of key continuous BCEs, most notably to limit the binding of allergens by IgE antibodies while also avoiding potentially harmful exposure of patients to immunodominant allergen BCEs
Author contributions
RF: Conceptualization, Investigation, Resources, Writing – review & editing, Writing – original draft, Project administration, Data curation, Methodology, Formal analysis. SF: Writing – original draft, Formal analysis, Resources, Visualization, Data curation, Methodology, Writing – review & editing, Software, Investigation, Validation. LO: Writing – original draft, Software, Writing – review & editing, Resources, Investigation, Visualization, Methodology, Validation, Data curation, Formal analysis. SC: Writing – original draft, Formal analysis, Validation, Methodology, Resources, Supervision, Investigation, Conceptualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. Publication of this article was partially supported by the UP College of Medicine MD-PhD (Molecular Medicine) Program and the University of the Philippines Manila Office of the Vice Chancellor for Academic Affairs.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Falcon RMG and Caoili SEC. Immunologic, genetic, and ecological interplay of factors involved in allergic diseases. Front Allergy. (2023) 4:1215616. doi: 10.3389/falgy.2023.1215616
2. Flom JD, Shreffler WG, and Perrett KP. Moving beyond desensitization to tolerance in food allergy. J Allergy Clin Immunol Pract. (2025) 13:741–4. doi: 10.1016/j.jaip.2025.02.014
3. López-Sanz C, Jiménez-Saiz R, Esteban V, Delgado-Dolset MI, Perales-Chorda C, Villaseñor A, et al. Mast cell desensitization in allergen immunotherapy. Front Allergy. (2022) 3:898494. doi: 10.3389/falgy.2022.898494
4. Caoili SEC. Comprehending B-cell epitope prediction to develop vaccines and immunodiagnostics. Front Immunol. (2022) 13:908459. doi: 10.3389/fimmu.2022.908459
5. Zhang J, Luo W, Cui Y, and Sun B. B-cell epitope peptide immunotherapy alleviates chitin-binding protein-induced type 2 airway inflammation in a Blomia tropicalis-murine model. Respir Res. (2025) 26:129. doi: 10.1186/s12931-025-03207-8
6. Focke-Tejkl M, Campana R, Reininger R, Lupinek C, Blatt K, Valent P, and Valenta R. Dissection of the IgE and T-cell recognition of the major group 5 grass pollen allergen Phl p 5. Journal of Allergy and Clinical Immunology. (2014) 133(3), 836–45.
7. Zieglmayer P, Focke-Tejkl M, Schmutz R, Lemell P, Zieglmayer R, Weber M, et al. Mechanisms, safety and efficacy of a B cell epitope-based vaccine for immunotherapy of grass pollen allergy. EBioMedicine. (2016) 11:43–57. doi: 10.1016/j.ebiom.2016.08.022
8. Weber M, Niespodziana K, Linhart B, Neubauer A, Huber H, Henning R, et al. Comparison of the immunogenicity of BM32, a recombinant hypoallergenic B cell epitope–based grass pollen allergy vaccine with allergen extract–based vaccines. J Allergy Clin Immunol. (2017) 140:1433–6. doi: 10.1016/j.jaci.2017.03.048
9. Focke-Tejkl M, Weber M, Niespodziana K, Neubauer A, Huber H, Henning R, et al. Development and characterization of a recombinant, hypoallergenic, peptide-based vaccine for grass pollen allergy. J Allergy Clin Immunol. (2015) 135:1207–17. doi: 10.1016/j.jaci.2014.09.012
10. Van Regenmortel MH. What is a B-cell epitope? Methods Mol Biol (Clifton N.J.). (2009) 524:3–20. doi: 10.1007/978-1-59745-450-6_1
11. Kringelum JV, Nielsen M, Padkjær SB, and Lund O. Structural analysis of B-cell epitopes in antibody: protein complexes. Mol Immunol. (2013) 53:24–34. doi: 10.1016/j.molimm.2012.06.001
12. Potocnakova L, Bhide M, and Pulzova LB. An introduction to B-cell epitope mapping and in silico epitope prediction. J Immunol Res. (2016) 2016:6760830. doi: 10.1155/2016/6760830
13. Caoili SEC. B-cell epitope prediction for antipeptide paratopes with the HAPTIC2/HEPTAD user toolkit (HUT). In Proceedings of the 13th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics. (2022) pp. 1-1.
14. Akiba H and Tsumoto K. Thermodynamics of antibody–antigen interaction revealed by mutation analysis of antibody variable regions. J Biochem. (2015) 158:1–13. doi: 10.1093/jb/mvv049
15. Van Regenmortel MH. Immunoinformatics may lead to a reappraisal of the nature of B cell epitopes and of the feasibility of synthetic peptide vaccines. J Mol recognition: JMR. (2006) 19:183–7. doi: 10.1002/jmr.768
16. Dall’Antonia F, Pavkov-Keller T, Zangger K, and Keller W. Structure of allergens and structure-based epitope predictions. Methods. (2014) 66:3–21. doi: 10.1016/j.ymeth.2013.07.024
17. Jackola DR, Blackburn C, Sveum M, and Rosenberg A. Entropy-favored human antibody binding reactions with a non-infectious antigen. Mol Immunol. (2008) 45:1494–500. doi: 10.1016/j.molimm.2007.08.023
18. Aalberse RC and Crameri R. IgE-binding epitopes: a reappraisal. Allergy. (2011) 66:1261–74. doi: 10.1111/j.1398-9995.2011.02656.x
19. Holdom MD, Davies AM, Nettleship JE, Bagby SC, Dhaliwal B, Girardi E, et al. Conformational changes in IgE contribute to its uniquely slow dissociation rate from receptor FcεRI. Nat Struct Mol Biol. (2011) 18:571–6. doi: 10.1038/nsmb.2044
20. Amin K. The role of mast cells in allergic inflammation. Respir Med. (2011) 106:9–14. doi: 10.1016/j.rmed.2011.09.007
21. Qin L, Tang LF, Cheng L, and Wang HY. The clinical significance of allergen-specific IgG4 in allergic diseases. Front Immunol. (2022) 13:1032909. doi: 10.3389/fimmu.2022.1032909
22. Javidan M, Amiri AM, Koohi N, Joudaki N, Bashirrohelleh MA, Pirsadeghi A, et al. Restoring immune balance with Tregitopes: A new approach to treating immunological disorders. Biomedicine Pharmacotherapy. (2024) 177:116983. doi: 10.1016/j.biopha.2024.116983
23. Nugraha R, Kamath SD, Johnston E, Karnaneedi S, Ruethers T, and Lopata AL. Conservation analysis of B-cell allergen epitopes to predict clinical cross-reactivity between shellfish and inhalant invertebrate allergens. Front Immunol. (2019) 10:2676. doi: 10.3389/fimmu.2019.02676
24. Buraphaka H, Dobutr T, Wiese MD, Lopata AL, and Daduang S. Structure-based epitope prediction and assessment of cross-reactivity of Myrmecia pilosula venom-specific IgE and recombinant Sol g proteins (Solenopsis geminata). Sci Rep. (2024) 14:11145. doi: 10.1038/s41598-024-61843-4
25. Goulet DR and Atkins WM. Considerations for the design of antibody-based therapeutics. J Pharm Sci. (2020) 109:74–103. doi: 10.1016/j.xphs.2019.05.031
26. Atanasio A, Franklin MC, Kamat V, Hernandez AR, Badithe A, Ben LH, et al. Targeting immunodominant Bet v 1 epitopes with monoclonal antibodies prevents the birch allergic response. J Allergy Clin Immunol. (2022) 149:200–11. doi: 10.1016/j.jaci.2021.05.038
27. Ahmad TA, Eweida AE, and Sheweita SA. B-cell epitope mapping for the design of vaccines and effective diagnostics. Trials Vaccinology. (2016) 5:71–83. doi: 10.1016/j.trivac.2016.04.003
28. Vaughan K, Peters B, Larche M, Pomes A, Broide D, and Sette A. Strategies to query and display allergy-derived epitope data from the immune epitope database. Int Arch Allergy Immunol. (2013) 160:334–45. doi: 10.1159/000343880
29. Toda M, Kasai M, Hosokawa H, Nakano N, Taniguchi Y, Inouye S, et al. DNA vaccine using invariant chain gene for delivery of CD4+ T cell epitope peptide derived from Japanese cedar pollen allergen inhibits allergen-specific IgE response. Eur J Immunol. (2002) 32:1631–9. doi: 10.1002/1521-4141(200206)32:6<1631::AID-IMMU1631>3.0.CO;2-O
30. Sakaguchi M, Hirahara K, Fujimura T, and Toda M. Approaches to immunotherapies for Japanese cedar pollinosis. Auris Nasus Larynx. (2011) 38:431–8. doi: 10.1016/j.anl.2010.12.002
31. Kim CH, Ahn JH, Kim SJ, Lee SY, Kim YK, Kim KH, et al. Co-administration of vaccination with DNA encoding T cell epitope on the Der p and BCG inhibited airway remodeling in a murine model of chronic asthma. J Asthma. (2006) 43:345–53. doi: 10.1080/02770900600701424
32. Hall G, Lund L, Lamb JR, and Jarman ER. Kinetics and mode of peptide delivery via the respiratory mucosa determine the outcome of activation versus TH2 immunity in allergic inflammation of the airways. J Allergy Clin Immunol. (2002) 110:883–90. doi: 10.1067/mai.2002.129800
33. Jarnicki AG, Tsuji T, and Thomas WR. Inhibition of mucosal and systemic Th2-type immune responses by intranasal peptides containing a dominant T cell epitope of the allergen Der p 1. Int Immunol. (2001) 13:1223–31. doi: 10.1093/intimm/13.10.1223
34. Chaudhary N, Mahajan L, Madan T, Kumar A, Raghava GPS, Katti SB, et al. Prophylactic and therapeutic potential of Asp f1 epitopes in naive and sensitized BALB/c Mice. Immune Network. (2009) 9:179–91. doi: 10.4110/in.2009.9.5.179
35. Briner TJ, Kuo MC, Keating KM, Rogers BL, and Greenstein JL. Peripheral T-cell tolerance induced in naive and primed mice by subcutaneous injection of peptides from the major cat allergen Fel d I. Proc Natl Acad Sci. (1993) 90:7608–12. doi: 10.1073/pnas.90.16.7608
36. Hufnagl K, Winkler B, Focke M, Valenta R, Scheiner O, Renz H, et al. Intranasal tolerance induction with polypeptides derived from 3 noncross-reactive major aeroallergens prevents allergic polysensitization in mice. J Allergy Clin Immunol. (2005) 116:370–6. doi: 10.1016/j.jaci.2005.04.002
37. Bauer L, Bohle B, Jahn-Schmid B, Wiedermann U, Daser A, Renz H, et al. Modulation of the allergic immune response in BALB/c mice by subcutaneous injection of high doses of the dominant T cell epitope from the major birch pollen allergen Bet v 1. Clin Exp Immunol. (1997) 107:536–41. doi: 10.1046/j.1365-2249.1997.d01-953.x
38. Ganglberger E, Sponer B, Schöll I, Wiedermann U, Baumann S, Hafner C, et al. Monovalent fusion proteins of immunoglobulin E mimotopes are safe for therapy of type I allergy. FASEB J. (2001) 15:2524–6. doi: 10.1096/fj.00-0888fje
39. Jensen-Jarolim E, Wiedermann U, Ganglberger E, Zürcher A, Stadler BM, Boltz-Nitulescu G, et al. Allergen mimotopes in food enhance type I allergic reactions in mice. FASEB J. (1999) 13:1586–92. doi: 10.1096/fasebj.13.12.1586
40. Pree I, Reisinger J, Focke M, Vrtala S, Pauli G, van Hage M, et al. Analysis of epitope-specific immune responses induced by vaccination with structurally folded and unfolded recombinant Bet v 1 allergen derivatives in man. J Immunol. (2007) 179:5309–16. doi: 10.4049/jimmunol.179.8.5309
41. Marazuela EG, Rodríguez R, Fernández-García H, García MS, Villalba M, and Batanero E. Intranasal immunization with a dominant T-cell epitope peptide of a major allergen of olive pollen prevents mice from sensitization to the whole allergen. Mol Immunol. (2008) 45:438–45. doi: 10.1016/j.molimm.2007.05.030
42. Lu G and Agosto H. Antibody responses to bee melittin (Api m 4) and hornet antigen 5 (Dol m 5) in mice treated with the dominant T-cell epitope peptides. J Allergy Clin Immunol. (1998) 101:397–403. doi: 10.1016/S0091-6749(98)70254-4
43. Hashiguchi S, Hino K, Taniguchi Y, Kurimoto M, Fukuda K, Ohyama M, et al. Immunodominance of seven regions of a major allergen, Cry j 2, of Japanese cedar pollen for T-cell immunity. Allergy. (1996) 51:621–32. doi: 10.1111/j.1398-9995.1996.tb04682.x
44. Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. (2018) 46:W296–303. doi: 10.1093/nar/gky427
45. Mackenbach JP. The epidemiologic transition theory. J Epidemiol Community Health. (1994) 48:329–31. doi: 10.1136/jech.48.4.329-a
46. Zuckerman MK, Harper KN, Barrett R, and Armelagos GJ. The evolution of disease: anthropological perspectives on epidemiologic transitions. Global Health Action. (2014) 7:23303. doi: 10.3402/gha.v7.23303
47. Villeneuve C, Kou HH, Eckermann H, Palkar A, Anderson LG, McKenney EA, et al. Evolution of the hygiene hypothesis into biota alteration theory: what are the paradigms and where are the clinical applications? Microbes infection. (2018) 20:147–55. doi: 10.1016/j.micinf.2017.11.001
48. Parker W, Jirků K, Patel E, Williamson L, Anderson L, and Laman JD. Reevaluating biota alteration: reframing environmental influences on chronic immune disorders and exploring novel therapeutic opportunities. Yale J Biol Med. (2024) 97:253–63. doi: 10.59249/VUNF1315
49. Daschner A and González Fernández J. Allergy in an evolutionary framework. J Mol Evol. (2020) 88:66–76. doi: 10.1007/s00239-019-09895-3
50. Galli SJ, Metz M, Starkl P, Marichal T, and Tsai M. Mast cells and IgE in defense against lethality of venoms: Possible “benefit” of allergy. Allergo J Int. (2020) 29:46–62. doi: 10.1007/s40629-020-00118-6
51. Schein CH, Ivanciuc O, and Braun W. Bioinformatics approaches to classifying allergens and predicting cross-reactivity. Immunol Allergy Clinics North America. (2007) 27:1–27. doi: 10.1016/j.iac.2006.11.005
52. Scheurer S, Toda M, and Vieths S. What makes an allergen? Clin Exp Allergy. (2015) 45:1150–61. doi: 10.1111/cea.12571
53. Wangorsch A, Scheurer S, Blanca M, Blanca-Lopez N, Somoza ML, and Martín-Pedraza L. Allergenic properties and molecular characteristics of PR-1 proteins. Front Allergy. (2022) 3:824717. doi: 10.3389/falgy.2022.824717
54. Hazebrouck S, Canon N, and Dreskin SC. The effector function of allergens. Front Allergy. (2022) 3:818732. doi: 10.3389/falgy.2022.818732
55. HogenEsch H, Dunham AD, Scott-Moncrieff C, Glickman LT, and DeBoer DJ. Effect of vaccination on serum concentrations of total and antigen-specific immunoglobulin E in dogs. Am J Veterinary Res. (2002) 63:611–6. doi: 10.2460/ajvr.2002.63.611
56. Knol EF and van Neerven RJJ. IgE versus IgG and IgA: Differential roles of allergen-specific antibodies in sensitization, tolerization, and treatment of allergies. Immunol Rev. (2024) 328:314–33. doi: 10.1111/imr.v328.1
57. Gharailoo Z, Vogel M, Engeroff P, and Bachmann MF. Vaccine-induced anti-IgE antibodies neutralize free IgE but fail to bind and activate mast cell-displayed IgE. Allergy. (2025). doi: 10.1111/all.16530
58. Seidler CA, Zeindl R, Fernández-Quintero ML, Tollinger M, and Liedl KR. Allergenicity and conformational diversity of allergens. Allergies. (2024) 4:1–16. doi: 10.3390/allergies4010001
59. Masilamani M, Commins S, and Shreffler W. Determinants of food allergy. Immunol Allergy Clinics North America. (2012) 32:11–33. doi: 10.1016/j.iac.2011.12.003
60. Bernard H, Turner PJ, Ah-Leung S, Ruiz-Garcia M, Mills ENC, and Adel-Patient K. Circulating Ara h 6 as a marker of peanut protein absorption in tolerant and allergic humans following ingestion of peanut-containing foods. Clin Exp Allergy. (2020) 50:1093–102. doi: 10.1111/cea.13706
61. Fernandez A, Danisman E, Boroujerdi MT, Kazemi S, Moreno FJ, and Epstein MM. Research gaps and future needs for allergen prediction in food safety. Front Allergy. (2024) 5. doi: 10.3389/falgy.2024.1297547
62. Janssen EM, Wauben MH, Jonker EH, Hofman G, Van Eden W, Nijkamp FP, et al. Opposite effects of immunotherapy with ovalbumin and the immunodominant T-cell epitope on airway eosinophilia and hyperresponsiveness in a murine model of allergic asthma. Am J Respir Cell Mol Biol. (1999) 21:21–9. doi: 10.1165/ajrcmb.21.1.3519
63. Finger E. Thermodynamics as the driving principle behind the immune system. Einstein (Sao Paulo). (2012) 10:386–8. doi: 10.1590/S1679-45082012000300024
64. Demir H, Radauer C, Strobl MR, Scheurer S, Kinaciyan T, and Bohle B. Cross-protection of AIT-induced antibodies to related allergens requires a high degree of structural identity. Allergy. (2024) 80(3):785–794. doi: 10.1111/all.16323
65. Abbott RK and Crotty S. Factors in B cell competition and immunodominance. Immunol Rev. (2020) 296:120–31. doi: 10.1111/imr.v296.1
66. Cowan R and Underwood PA. Steric effects in antibody reactions with polyvalent antigen. J Theor Biol. (1988) 132:319–35. doi: 10.1016/S0022-5193(88)80218-2
67. Pellequer JL and van Regenmortel MHV. Affinity of monoclonal antibodies to large multivalent antigens: influence of steric hindrance on antibody affinity constants calculated from Scatchard plots. Mol Immunol. (1993) 30:955–8. doi: 10.1016/0161-5890(93)90022-4
68. Pérez de la Lastra JM, Baca-González V, González-Acosta S, Asensio-Calavia P, Otazo-Pérez A, and Morales-delaNuez A. Antibodies targeting enzyme inhibition as potential tools for research and drug development. Biomolecular Concepts. (2021) 12:215–32. doi: 10.1515/bmc-2021-0021
Keywords: allergy, vaccines, B-cell epitope prediction, B-cell epitopes, immunodominance, oligopeptide sequences, antipeptide antibodies, desensitization therapy
Citation: Falcon RMG, Fahrenbach SU, Ortiz LCE and Caoili SEC (2025) B-cell epitope prediction for developing allergy vaccines and their companion immunodiagnostics. Front. Immunol. 16:1624339. doi: 10.3389/fimmu.2025.1624339
Received: 07 May 2025; Accepted: 30 May 2025;
Published: 02 July 2025.
Edited by:
Rubén Fernández Santamaría, Health Research Institute Foundation Jimenez Diaz (IIS-FJD), SpainReviewed by:
Carlos J. Aranda, University of Malaga, SpainCopyright © 2025 Falcon, Fahrenbach, Ortiz and Caoili. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robbi Miguel G. Falcon, cmdmYWxjb25AdXAuZWR1LnBo
 Robbi Miguel G. Falcon
Robbi Miguel G. Falcon Serina U. Fahrenbach
Serina U. Fahrenbach Salvador Eugenio C. Caoili
Salvador Eugenio C. Caoili