Abstract
Human-induced pluripotent stem cells with broad immune compatibility are highly desirable for regenerative medicine applications. Human leukocyte antigen (HLA) class I homozygous cell sources are ideal for immune compatibility modeling. Here, we profile HLA-A, HLA-B, and HLA-C alleles in 3,496 Lithuanian donors genotyped at three-field resolution. The five most frequent alleles constitute 74.6% of HLA-A, 43.2% of HLA-B, and 59.2% of HLA-C, with HLA-A*02:01:01, HLA-B*07:02:01, and HLA-C*07:02:01 being the most common. Lithuanian allele frequencies closely resemble those of European-American and British populations. We identified 153 double homozygotes and 51 triple homozygotes for HLA-A, HLA-B, and HLA-C. Compatibility modeling showed that triple homozygous profiles match 60.5% of Lithuanians, 13.4% of the British population, and 7.4% of European-Americans. CRISPR-Cas9 guide RNA design yielded 54 candidates predicted to disrupt HLA-A or HLA-B while preserving HLA-C, producing edited profiles matching over 97.9% of Lithuanians, 95.7% of European-Americans, and 95.5% of the British population. Finally, we established 15 fibroblast lines from triple homozygotes as a bioresource for the derivation of human-induced pluripotent stem cells and immune compatibility studies.
Introduction
Transplantation of allogeneic organs, tissues, and cells is constrained by immune matching between the graft and the host. Immune matching is mediated by the human leukocyte antigen (HLA) genes. These genes are clustered in a 3.7-Mbp locus on chromosome 6, are highly polymorphic, and their inheritance is reported as having intermediate linkage disequilibrium (1, 2). The recent adoption of high-resolution haplotyping in clinics has improved the accuracy of immune matching for the more than 42,000 HLA alleles cataloged in the IPD-IMGT/HLA Database (3). Pursuing a high level of matching is intended to minimize adverse events, such as graft-versus-host disease (GVHD) or immune rejection, which are frequently managed with immunosuppressive drugs. A broad assortment of immunosuppressive treatments is available for the management of transplantation, encompassing small molecule inhibitors, antimetabolites, corticosteroids, and antibodies (4–6). However, immunosuppressive therapies are associated with an increased risk of infection (7, 8). Therefore, pursuing a high level of matching is intended to minimize adverse events caused by immune rejection and immune suppression. The importance of a high degree of HLA immune matching for improving survival rates is well documented in the literature (9, 10) for exemplary primary cell types, and it is highly desirable for induced pluripotent stem (iPS) cell-based applications.
HLA class I homozygous individuals offer increased immune compatibility with a relatively larger portion of the population. They are very scarcely represented, as expected from Mendelian ratios. Cells from naturally occurring triple and double homozygous individuals are very valuable for the study of immune compatibility and applications of regenerative medicine.
Genome editing tools are currently used to engineer synthetic immune compatibility, also called hypoimmunogenicity. This aids in overcoming the challenges of identifying rare haplotypes in donor pools. Several approaches have been developed to bypass immune recognition by cytotoxic T cells while retaining self-recognition mediated by NK cells. The most frequent loss-of-function strategies include the knockout of specific HLA class I (11) and class II genes, beta-2-microglobulin (B2M) (12, 13), CIITA (14), TAP1 or TAP2, and CD74 (15). Conversely, the most frequent gain-of-function strategies involve the knock-in of CD47 and HLA-E (16). Pioneering studies have demonstrated that gene-editing depletion of HLA-A and HLA-B genes preserves host NK cell recognition while preventing CD8 T-cell mediated host-versus-graft rejection (17). This approach yields cells currently known as HLA-C retained. Triple and double homozygous samples are an ideal cell source for modulating immunogenicity, as they start from a relatively higher level of immune compatibility. Furthermore, they can be engineered in their HLA genes using programmable nucleases through simpler strategies compared to heterozygous samples.
In this study, we identify a cohort of naturally occurring triple and double homozygous individuals in the Lithuanian population and isolated primary samples for prospective regenerative medicine applications. Additionally, we analyzed the frequency of HLA class I genes, specifically characterizing the HLA-A, HLA-B, and HLA-C haplotypes in a cohort of 3,496 individuals. The genetic makeup of the Lithuanian population is placed within a European context, influenced by pre-Neolithic Western and Scandinavian hunter–gatherer groups, Early to Middle Bronze Age steppe pastoralists, and Late Neolithic Bronze Age Europeans, while remaining largely sheltered (18). These features make the Lithuanian population closely resemble European-American (19) and British groups (20) from an immune compatibility standpoint. We compared this population to publicly available datasets of European ancestry and modeled the impact of gene editing on HLA immune matching and population coverage.
Materials and methods
Ethical approval
This study is part of the ethical approval 2023/6-1524-984, Highly-immune compatible iPS cells as source for regenerative medicine and cell therapy-oriented applications, from the Vilnius Regional Biomedical Research Ethics Committee (Lithuania) to Vilnius University, and 2023/4-1507-968, Analysis of the distribution of Human Leukocyte Antigen (HLA; Encoding Genes - HLA) alleles and haplotypes in the group of the Lithuanian unrelated bone marrow donor registry, to Vilnius University Hospital Santaros Klinikos. Written consent was obtained from the participants of the study.
Study subjects
For population-based analyses of HLA frequencies, the study included 3,496 individuals from the Lithuanian unrelated bone marrow donor registry, characterized at third-field resolution for HLA-A, HLA-B, and HLA-C. For the isolation of dermal fibroblasts, individuals were healthy adults who provided study-specific informed consent and were selected based on their known HLA class I genotypes. Individuals aged over 55 years, those with known inherited genetic disorders, or those diagnosed with non-environmentally caused diseases were excluded from dermal biopsy collection to ensure that fibroblast samples were free from age-associated mutations or pathogenic genetic variants.
Genotyping
HLA typing for registry donors’ peripheral blood was performed at the EFI-accredited immunogenetics laboratory at Vilnius University Hospital Santaros Klinikos (Vilnius, Lithuania) using sequencing-based typing, and at the ASHI-accredited laboratory HistoGenetics (Ossining, NY, USA) using next-generation sequencing. Exons 2 and 3 for class I HLA were covered.
Fibroblast derivation and genotyping
Skin samples were collected using a 2–3-mm biopsy punch needle and fragmented with a sterile scalpel and needle. Fibroblasts were grown in AmnioPrime Complete Medium (cat. no. APR-B, Capricorn Scientific, Germany), supplemented with amphotericin B (cat. no. AMP-B, Capricorn Scientific, Germany), for 21 to 45 days until fibroblasts migrated from tissue sections and reached 80%–90% confluence. The medium was changed every 3 days to ensure optimal cell growth. Fibroblasts were routinely passaged with 0.25% Trypsin-EDTA at a density of 2 × 105 cells/cm2. Genomic DNA from fibroblasts was purified using the DNeasy Blood and Tissue Kit (cat. no. 69504, Qiagen, Germany) and genotyped using the primers HLAA-P1: TCCAGGTGGACAGGTAAGGA, HLAA-P2: GTCACTGCCTGGGGTAGAAC, HLAB-P1: TGCATTCTGGGTTTCTCTACTGG, HLAB-P2: CACGCGAAACATCCCAATCA, HLAC-P1: AGGTAAGGCAAAGGGTGGGA, and HLAC-P2: AGGCCGCCTGTACTTTTCTC. Samples were Sanger sequenced using the primers HLAA-P3: ACCCTCGTCCTGCTACTCTCG, HLAB-P3: ACCCTCCTCCTGCTGCTCTG, and HLAC-P3: CGTTGGGGATTCTCCACTCC at Microsynth, Germany.
Bioinformatics
Python and R scripts used for data analysis, along with anonymized datasets, are available through the Supplementary Data and or through the open-source GitHub developer platform in the repository https://github.com/Arias-Lab/superdonors.
Quantification of HLA allele frequency in the population
The total allele count in the dataset was divided by the number of alleles (n = 2) times the number of individuals in this study (n = 3,496), all of whom had at least third-field resolution.
Hardy–Weinberg equilibrium analyses
The observed genotypes present in the population were quantified (n = 3,496). The allele frequencies were determined using the sampled genotype count, and the expected genotype frequencies were calculated. The observed and expected genotype counts were compared with a χ2 test. The χ2 test is reliable for genotypes present more than five times in the population. Genotypes with a count < 5 times were filtered from the Hardy–Weinberg equilibrium (HWE) analyses. The degrees of freedom (df), calculated as (n(n + 1)/2) – n, were estimated based on the number of possible genotypes and the number of alleles identified in the sampled population for each HLA class I gene: 44 for HLA-A, 83 for HLA-B, and 45 for HLA-C.
Regression analyses
Allele frequencies were extracted from the publicly available data from European-American (19) and British (20) populations and compared to the allele frequency from our study. Linear regression analyses (y ~ mx + c) were performed using R for pairwise comparison of allele frequencies of HLA-A, HLA-B, and HLA-C. Frequencies are calculated as frequency = allele count(dataset)/n(dataset).
Principal component analysis
Monte Carlo population haplotypes were simulated based on the published allele frequencies of European-American and British cohort studies. Data were processed with one-hot encoding to convert allele entries per individual into 1 or 0, using the caret library in R (22). Centroids and Euclidean distances were calculated from the principal components. Distances were represented as edges and as heatmaps.
HLA sequence analysis and sgRNA activity prediction
The sequences for all alleles at the protein, transcript, and gene levels were downloaded as FASTA files from the IPD-IMGT-HLA database version 3.58 (23) and analyzed in Python and R. Allele sequences were extracted based on the HLA alleles present in the population. Cas9 binding sites were extracted with Python and analyzed in R using CrisprScore (24). Transmembrane prediction was conducted with DeepTMHMM (25).
Results
Analysis of HLA class I frequencies in the Lithuanian population and identification of double and triple homozygotes
The Lithuanian Bone Marrow Donor Registry, located at Vilnius University Hospital Santaros Klinikos, includes 13,884 individuals, with 11,153 characterized at the second field (protein level) for HLA-A, HLA-B, and HLA-C. Of these, 3,496 individuals are characterized in the third field (Figure 1A). We found that 858 individuals are at least homozygous for one HLA class I gene. A total of 542 individuals are homozygous for the coding sequence of HLA-A, 233 individuals are homozygous for HLA-B, and 338 individuals are homozygous for HLA-C (Figure 1B). The HLA types identified and their prevalence in the population are summarized in Figure 1 and Supplementary Table S1. The five most frequent HLA-A alleles are A*02:01:01, A*03:01:01, A*24:02:01, A*01:01:01, and A*11:01:01, which together account for 74.6% of the population (Figure 1C). Notably, HLA-A*02:01:01 is the most frequent HLA class I allele, representing 31.6% of the population. Similarly, the five most frequent HLA-B alleles are B*07:02:01, B*13:02:01, B*15:01:01, B*44:02:01, and B*40:01:01, which account for 43.2% of the population (Figure 1D). HLA-B*07:02:01 alone represents 15.1% of the Lithuanian population. Furthermore, the five most frequent HLA-C alleles are C*07:02:01, C*06:02:01, C*04:01:01, C*02:02:02, and C*07:01:01, with a cumulative frequency of 59.2% in the population (Figure 1E). It is important to highlight that the HLA-B gene exhibits the largest diversity of alleles, followed by HLA-A and HLA-C (Supplementary Table S1), as also observed in previous studies (19–21). Of the HLA homozygotes, a total of 153 are double homozygous (Figure 1B; Supplementary Table S2): 58 for HLA-A and HLA-B, 76 for HLA-A and HLA-C, and 172 for HLA-B and HLA-C. Remarkably, 51 individuals are triple homozygous for HLA-A, HLA-B, and HLA-C (Figure 1B; Table 1). Haplotype frequencies of the complete dataset (3,496 individuals) are available in the Supplementary Data.
Figure 1

(A) Dataset structure from this study. (B) Proportional Euler diagram showing the prevalence of HLA class I homozygous individuals in the Lithuanian population, with the composition of double homozygous and triple homozygous individuals highlighted. The most common HLA alleles with a frequency above 0.01 are shown for (C)HLA-A, (D)HLA-B, and (E)HLA-C.
Table 1
| HLA-A | HLA-B | HLA-C | Count |
|---|---|---|---|
| A*03:01:01 | B*07:02:01 | C*07:02:01 | 15 |
| A*02:01:01 | B*13:02:01 | C*06:02:01 | 10 |
| A*01:01:01 | B*08:01:01 | C*07:01:01 | 6 |
| A*02:01:01 | B*07:02:01 | C*07:02:01 | 4 |
| A*02:01:01 | B*40:01:01 | C*03:04:01 | 3 |
| A*02:01:01 | B*57:01:01 | C*06:02:01 | 2 |
| A*03:01:01 | B*56:01:01 | C*01:02:01 | 2 |
| A*25:01:01 | B*18:01:01 | C*12:03:01 | 2 |
| A*02:01:01 | B*15:01:01 | C*03:04:01 | 1 |
| A*02:01:01 | B*27:05:02 | C*02:02:02 | 1 |
| A*25:01:01 | B*35:01:01 | C*04:01:01 | 1 |
| A*26:01:01 | B*38:01:01 | C*12:03:01 | 1 |
| A*31:01:02 | B*51:01:01 | C*05:01:01 | 1 |
| A*68:01:01 | B*40:01:01 | C*03:04:01 | 1 |
| A*68:01:02 | B*44:02:01 | C*07:04:01 | 1 |
HLA class I triple homozygous haplotypes identified in this study (n = 51).
Comparisons of HLA class I allele composition between populations
Comparisons of the Lithuanian Class I HLA frequencies with those reported for the European-American and British populations using linear regression models show strong correlations between the three cohorts (Figures 2A–C). The linear regression analyses yielded an average slope of 0.914 for HLA-A, 0.827 for HLA-B, and 0.860 for HLA-C. This indicates the populations closely resemble each other in the composition and prevalence of allele variants. Principal component analysis (PCA) was performed on the genotypes of the Lithuanian population and on genotypes reconstructed from published datasets using Monte Carlo analysis based on reported allele frequencies. The results showed that the Lithuanian population clustered in close proximity to the compared populations (Figure 2D). The Euclidean distances between the centroids of the populations were quantified and represented in the PCA and as a heatmap (Figure 2E). The distance metrics indicate that the centroid of the Lithuanian population is proximal to the European-American and British populations, with distances of 1.00 and 0.79 relative units, respectively. The British and European-American populations closely resemble each other, with a Euclidean distance of 0.27 relative units. Hardy–Weinberg equilibrium analyses show that some genotypes, including the 10 most frequent allele types, occur at higher frequencies than expected (Supplementary Data; Supplementary Figure S1).
Figure 2
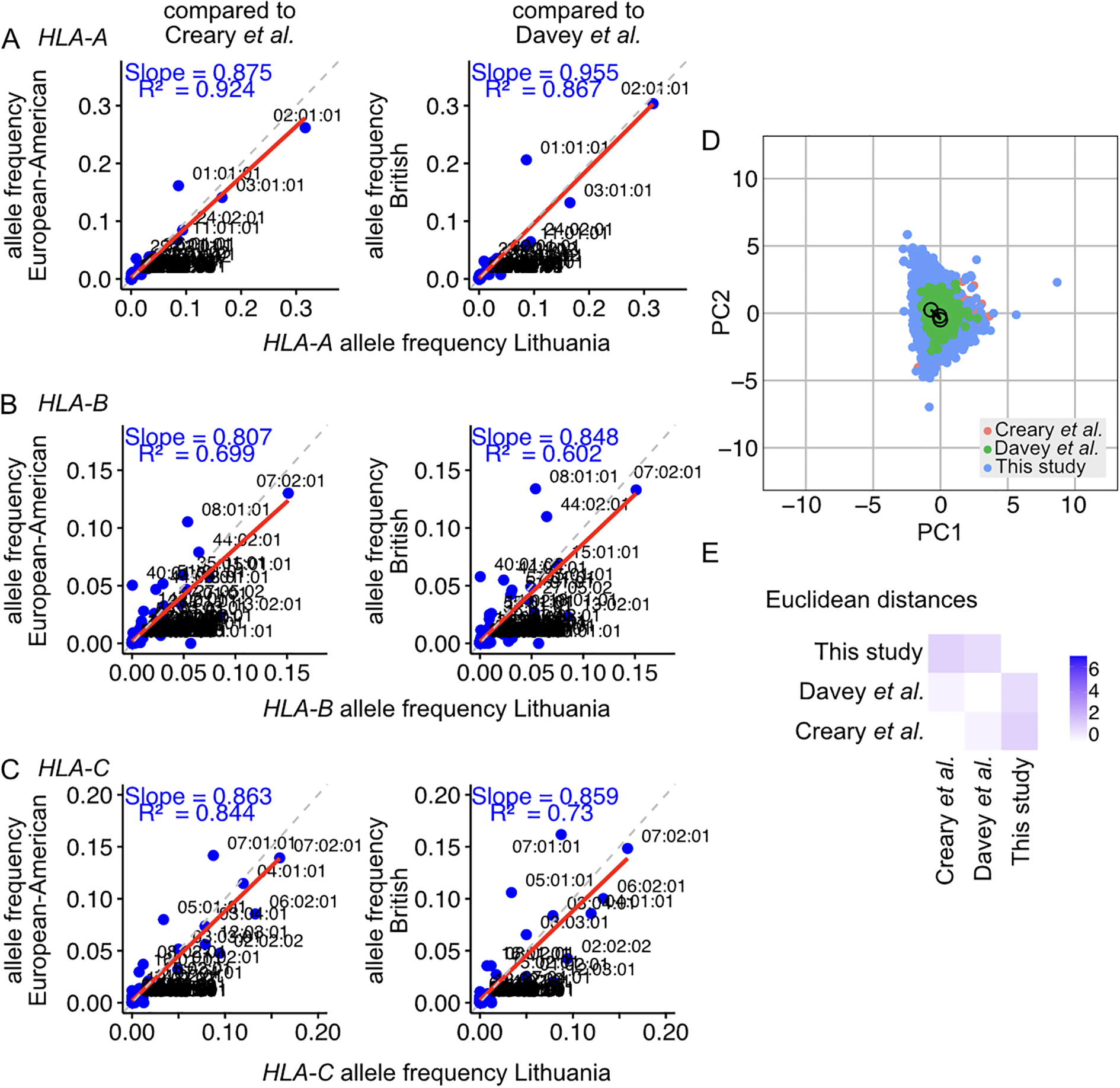
Comparison of the HLA allele frequencies identified in the Lithuanian population with those reported in studies of the European-American and the British population for (A) the HLA-A transcript, (B) the HLA-B transcript, and (C) the HLA-C transcript. Reference lines with slope n = 1 are represented as dashed grey lines. The linear regressions of frequencies on the scatter plots are represented with a solid red line, with the R2 of the linear model and the slope indicated. (D) Principal component analysis of the HLA class I distribution in the Lithuanian population (this study) and other populations, including European-American and British cohorts. The centroid of each population is marked with a circle. The Euclidean distances between the centroids were calculated, and the edges are plotted with solid lines. (E) Euclidean distance heatmap between the studied populations. Blue corresponds to greater Euclidean distances in the principal component space.
Compatibility of HLA class I in the Lithuanian and other European populations
We stochastically arranged the 3,496 donors and interrogated whether the subset of HLA-A, HLA-B, and HLA-C triple homozygous (51 samples) and double homozygous (153 samples) individuals were compatible with the 3,496 patients (Figure 3). We found that our cohort of triple homozygous individuals matches 60.46% of the Lithuanian population (Figure 3A). Likewise, the double homozygous cohort matches 33.32% of the Lithuanian population. In comparison, a randomly selected subset of 153 or 51 samples from the dataset could match only 11.84% (Figure 3B) and 4.1% (Figure 3C) of the Lithuanian population, respectively. We then evaluated the matching provided by our triple homozygous and double homozygous cohorts to the European-American and British populations. We assessed their immune compatibility with Monte Carlo datasets reconstructed from allele frequencies reported for European-American and British individuals. Remarkably, we found that the 51 triple homozygous samples of our cohort match 13.4% of the British population, while the double homozygous cohort matches 5.2% (Figure 3D). Additionally, we found that triple homozygous samples match 7.4% of the European-American population, and double homozygous samples match 3.3% (Figure 3E).
Figure 3

Population compatibility of HLA-A, HLA-B, and HLA-C genotypes in Lithuanian samples with Lithuanian and other European populations. Immune compatibility of triple homozygous (51 individuals), double homozygous (153 individuals), and (A) all samples from the cohort of 3,496 individuals in this study, (B) stochastically selected samples of 153 individuals, and (C) stochastically selected samples of 51 individuals. Immune compatibility of HLA class I genes, HLA-A, HLA-B, and HLA-C, in Lithuanian samples with (D) British datasets and (E) European-American datasets. Triple homozygous individuals are indicated in blue, double homozygous individuals in red, and stochastically selected subsamples in green.
Cas9 activity prediction on HLA class I alleles of the Lithuanian population
We extracted the Cas9-binding site sequences from the HLA alleles present in the Lithuanian population. First, we focused on the analysis of target regions encompassing the gene body, from the 5′UTR to the 3′UTR. We found 1,996 unique target sites in HLA-A alleles, 2,342 unique target sites in HLA-B, and 2,300 unique target sites in HLA-C. We calculated the activity prediction score based on the rule set 1 of nuclease catalytic activity (26). We found that, as in non-hyper polymorphic genes, the activity scores of all HLA alleles are centered in the inactive Q4 quadrant. We show this distribution for the five most frequent alleles of HLA-A, HLA-B, and HLA-C (Figure 4A). The potential of HLA gene knockout to modulate immune compatibility is well accepted in the literature. Although pairs of guide RNAs can be used in conjunction to create exon-spanning knockouts, we focused on guide RNAs in exon regions. From the guide RNAs present in the gene body, we found 679 unique target sites in the HLA-A exons of Lithuanian alleles, 698 in HLA-B, and 687 in HLA-C (Figure 4B). Since HLA-A, HLA-B, and HLA-C are class I single-span transmembrane proteins (Figure 4C), only guide RNAs targeting the ectodomain have the capacity to create knockouts that eliminate plasma membrane expression of HLA genes. We predicted the transmembrane spanning region (25) of the allele sequences and focused on guide RNAs directed to the N-terminus, upstream of the predicted transmembrane domain. We found there are 615 unique target sites in Lithuanian alleles on HLA-A ectodomains, 658 on HLA-B, and 613 on HLA-C (Figure 4B). Of those useful for ectodomain targeting, a fraction have predicted activity scores greater than 0.5. These include 54 for HLA-A, 75 for HLA-B, and 66 for HLA-C (Figure 4B).
Figure 4

Predicted guide RNA sequence activity for the five most frequent alleles in the Lithuanian population for (A)HLA-A, HLA-B, and HLA-C. (B) Nested distribution of guide RNAs on the gene body, exons, and ectodomain, as well as those with predicted high activity for HLA-A, HLA-B, and HLA-C. (C) Protein structure models and gene structures for HLA-A, HLA-B, and HLA-C. Protein structures are depicted as mature forms, excluding the signal peptide and the highly flexible endodomain. Gene structures highlight the matching ectodomain and transmembrane (TM) region.
Modeling the impact of HLA class I engineering on the immune compatibility of triple homozygous and double homozygous donor samples
Naturally occurring triple and double homozygous samples are particularly useful for gene engineering approaches as they allow bi-allelic targeting with a single programmable nuclease in a one-step intervention. Next, we modelled the impact of HLA-A and HLA-B knockouts on the immune compatibility of the double and triple homozygous samples when matching them to the Lithuanian population and other European datasets (Figure 5). We included all 51 triple homozygous individuals from our cohort (Figure 5A). From the 153 double homozygous individuals identified, we focused on those that are HLA-A and HLA-B double homozygous, comprising seven individuals (Figure 5B). The 51 triple homozygous samples, when in an HLA-C-retained (HLA-A and HLA-B double knockout) configuration, match a maximum of 0.9799 of the Lithuanian population (Figure 5A). These 51 samples achieve a match of 0.9577 in the European-American population (Figure 5C) and 0.9556 in the British population (Figure 5D).
Figure 5
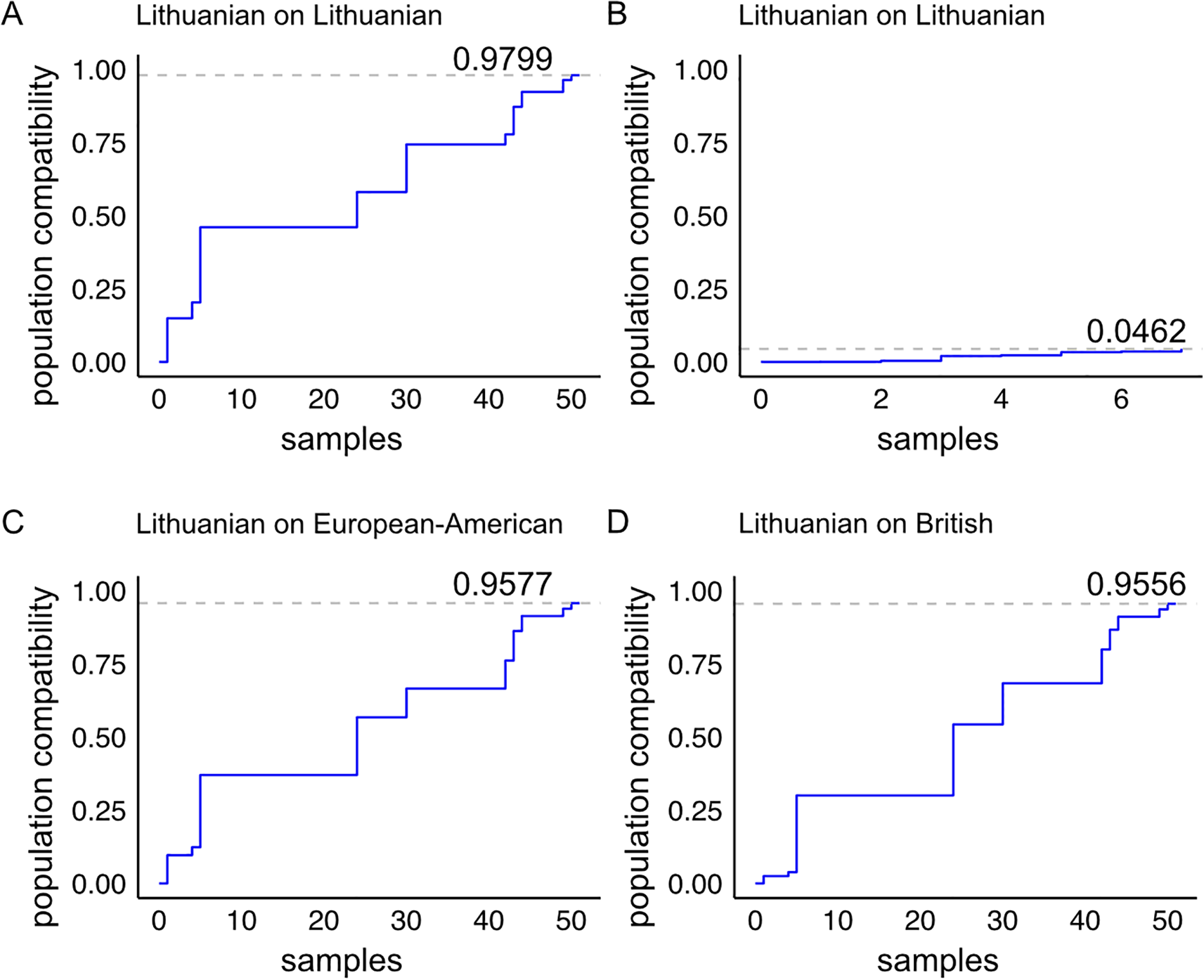
Population compatibility model of HLA-A and HLA-B double knockout samples from our cohort with the Lithuanian population and with populations of European-American and British ancestry. (A) Immune compatibility of the 51 triple homozygous individuals in an HLA-A and HLA-B double knockout model, and (B) the seven double homozygous individuals in an HLA-A and HLA-B double knockout model when matched to the Lithuanian population. The cohort from (A) matched with (C) the European-American dataset and (D) the British dataset.
Sampling of HLA-A, HLA-B, and HLA-C triple homozygous individuals from the Lithuanian population
Since the triple homozygote individuals identified in this study are immune-compatible with a large fraction of the Lithuanian and other European populations, we sampled these volunteers. The collected dermal fibroblast samples were used to establish biobank stocks and cultures. Primary fibroblast cultures were robustly established for 15 triple homozygotes (Figure 6A; Supplementary Table S3). PCR products of exons 2 and 3 (Figure 6B) display single bands, and Sanger sequencing yields clear chromatograms, both of which are characteristic of homozygous samples (Figures 6C–E). Sanger sequencing of exons 2 and 3, which code for the ectodomains of HLA-A, HLA-B, and HLA-C, revealed characteristic residues for each allele. Characteristic amino acids p.F33 and p.R121 were confirmed for HLA-A*02:01:01 (Figure 6C), p.Y33 and p.W119 for HLA-B*13:02:01 (Figure 6D), and p.D33 and p.L119 for HLA-C*06:02:01 (Figure 6E). These findings were consistent for both XY (donor SD9) and XX (donor SD6) individuals with the homozygous haplotype HLA-A02:01:01-HLA-B13:02:01-HLA-C06:02:01 (Figure 6F).
Figure 6
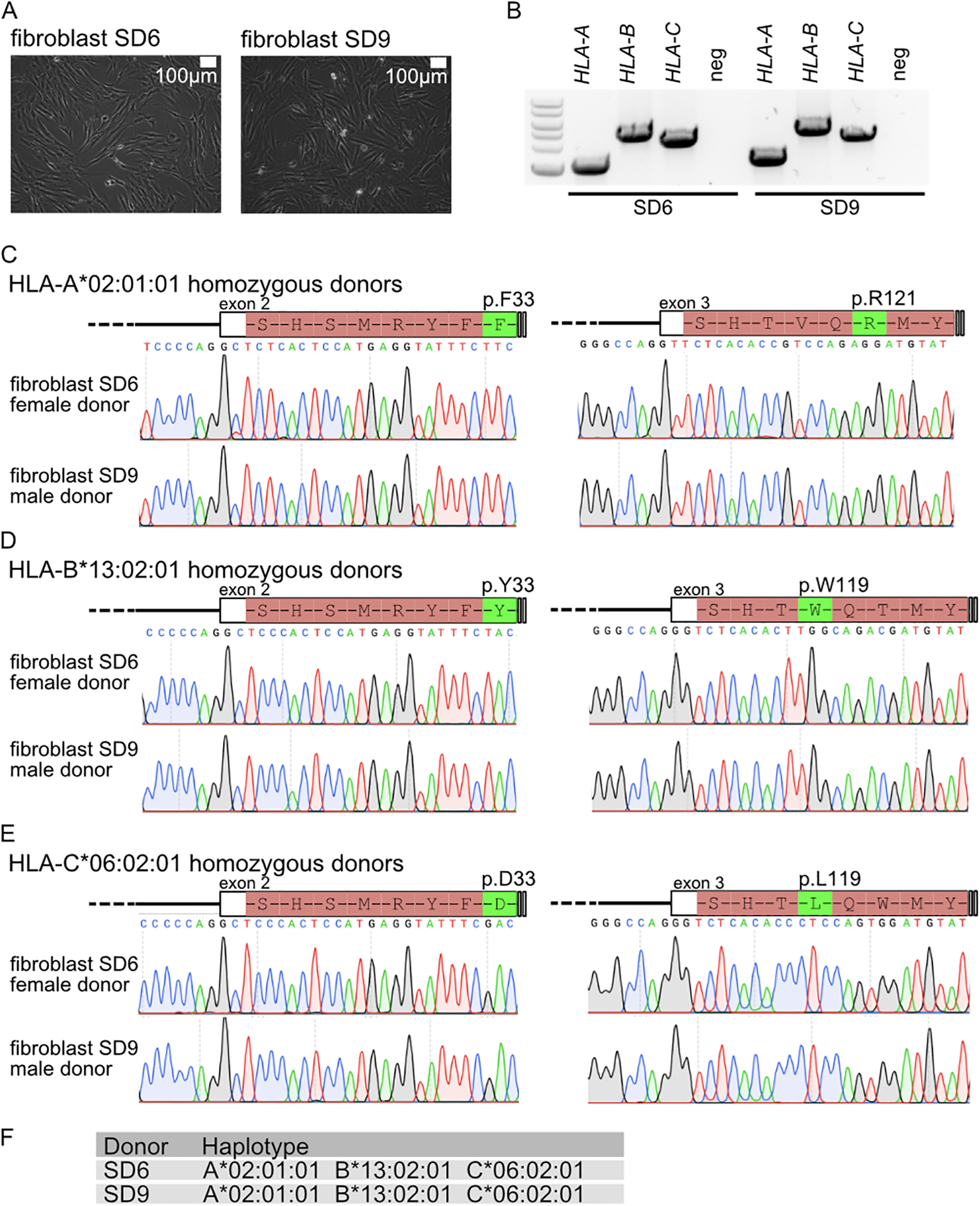
(A) Fibroblast cultures from HLA-A, HLA-B, and HLA-C triple homozygous donors. (B) Genotyping PCR for HLA-A, HLA-B, and HLA-C. Sanger sequencing analysis for (C)HLA-A, (D)HLA-B, and (E)HLA-C. (F) Haplotype for donor patient and linked fibroblasts.
Discussion
Our study on allele and haplotype frequencies of the HLA-A, HLA-B, and HLA-C genes in the Lithuanian population elucidates immune compatibility structure in relation to other European populations. Comparative analyses confirmed high similarity in HLA class I genes between the Lithuanian population and populations of European-American and British ancestry. The most frequent alleles described in the British (20) and European-American populations (19) are also the most frequent in the Lithuanian population, with frequencies of 31.6% (A*02:01:01), 5.3% (B*08:01:01), 15.1% (B*07:02:01), and 8.7% (C*07:01:01). Linear regression analysis using publicly available data corroborated these observations. PCA and Euclidean distance calculations further confirmed the proximity in immune compatibility among Lithuanian, European-American, and British populations. HWE analysis revealed deviations in a subset of alleles, suggesting partial genetic isolation or selective pressure. These findings align with previous studies indicating low levels of admixture and a significant component of pre-Neolithic hunter–gatherer ancestry in the Lithuanian group (18).
The majority of individuals in our registry (n = 11,153) were characterized at second-field resolution for HLA-A, HLA-B, and HLA-C, while a subset (n = 3,496) underwent third-field resolution analysis. This divergence reflects technological advancements in clinical registries, with long-read sequencing platforms now enabling fourth-field resolution (27, 28). Although our analyses do not encompass HLA class II, it is well established that its expression occurs in specialized immune cell lineages, whereas HLA class I primarily regulates nonimmune and immune cell compatibility (29, 30). The exclusive focus on HLA class I represents a potential limitation of this study, especially considering the importance of HLA class II matching in immunotherapeutic applications. Remarkably, we found a subset of 51 triple homozygous individuals for HLA-A, HLA-B, and HLA-C, and a subset of 153 double homozygous individuals. The proportion of triple-homozygous individuals exceeded stochastic expectations based on measured allele frequencies (2.99 ± 1.76), suggesting underlying population structures, as indicated by HWE analysis.
Due to the significant immune compatibility provided by HLA-A, HLA-B, and HLA-C triple homozygous individuals (31, 32), the term naturally occurring superdonors has been proposed previously (33). Our study identified 51 naturally occurring superdonors who exhibit HLA class I immune matching with 60.46% of the Lithuanian population, 13.4% of the British population, and 7.4% of the European-American population. These populations exhibit similar individual allele frequencies, yet their reduced HLA class I immune matching is likely due to differences in haplotype composition. It is important to highlight that using triple homozygous samples for cell line development, particularly human iPS cells, results in derivatives with wider immune compatibility than heterozygous counterparts for nonimmune cell identities. Genetic engineering with programmable nucleases in such samples benefits from simpler strategies because of the homozygosity status of the starting material. In turn, engineered products are expected to attain broader immune compatibility than natural counterparts.
Several international initiatives focus on iPS cell development from haplo-selected individuals, including programs in Japan (34), Australia (33), South Korea (35, 36), Spain (37), Germany (38), Lithuania, and Saudi Arabia (39). We modeled the impact of HLA-C-retained gene-editing intervention on the 51 naturally occurring superdonors and found that their immune compatibility could be enhanced to match 97.9% of the Lithuanian population, 95.7% of the European-American population, and 95.5% of the British population. Conversely, the immune compatibility provided by the HLA-A and HLA-B double-homozygous individuals was limited due to the retained diversity within the heterozygous HLA-C allele.
Here, we propose the term synthetic superdonor for those cell lines derived from naturally occurring superdonors that, through gene editing, acquire broader immune compatibility. Analysis of gene-editing availability for HLA-A, HLA-B, and HLA-C highlights the importance of protein topology, knockout strategy design, and nuclease target site activity in achieving synthetic superdonor stocks.The HLA-A, HLA-B, and HLA-C proteins are of the type I transmembrane class; hence, targeting the N-terminus ectodomain slightly constrains the number of available Cas9-binding sites. Our analyses demonstrate that the largest impact on knockout availability is the nuclease activity score; therefore, gene-editing tools that enhance nuclease activity are likely to have a positive impact on synthetic superdonor creation in the future. Likewise, our analyses indicate that naturally occurring superdonor and synthetic superdonor cell sources would positively impact immune matching for rare haplotypes. Both naturally occurring and synthetic superdonors are a remarkable source for the creation of iPS cells and derivative advanced therapeutic medicinal products (ATMPs).
Statements
Data availability statement
The datasets presented in this study can be found in Supplementary Materials and online repositories listed in article (Materials and Methods section) Supplementary Materials.
Ethics statement
This study is part of the ethical approval 2023/6-1524-984 provided to Vilnius University, and 2023/4-1507-968 provided to Vilnius University Hospital Santaros Klinikos. The studies were conducted in accordance with the local legislation and institutional requirements. Patients provided written informed consent to participate in the study.
Author contributions
DN: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. BR-A: Investigation, Methodology, Writing – original draft, Writing – review & editing. CM: Investigation, Methodology, Writing – original draft, Writing – review & editing. VA: Data curation, Software, Writing – original draft, Writing – review & editing. RČ: Investigation, Methodology, Project administration, Resources, Writing – original draft, Writing – review & editing. IL: Investigation, Writing – original draft, Writing – review & editing. AJ: Resources, Writing – original draft, Writing – review & editing. LG: Funding acquisition, Project administration, Resources, Writing – original draft, Writing – review & editing. IN: Writing – original draft, Writing – review & editing, Conceptualization. JI: Conceptualization, Writing – original draft, Writing – review & editing. DB: Conceptualization, Funding acquisition, Investigation, Project administration, Resources, Writing – original draft, Writing – review & editing, Supervision. JA: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. BV-R: Formal analysis, Investigation, Methodology, Resources, Writing – review & editing. MS: Investigation, Methodology, Resources, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This research was conducted as part of the execution of Project “Mission-driven Implementation of Science and Innovation Programs” (No. 02-002-P-0001), funded by the Economic Revitalization and Resilience Enhancement Plan “New Generation Lithuania” to DB and JA.
Acknowledgments
All authors read and agreed to the last version of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1626787/full#supplementary-material
Supplementary Figure 1Hardy-Weinberg equilibrium (HWE) analysis for (A)HLA-A, HLA-B and HLA-C genotypes. The heatmaps represent the ratio between observed genotype frequencies and expected genotype frequencies. Only the genotypes were X2 value exceeds the X2-threshold, indicating a HWE deviation are displayed. The subset of genotypes with a frequency higher than 0.01 are represented in heatmaps.
Supplementary Figure 2Cumulative-coverage of HLA class I immune matching in the Lithuanian population. Individuals in our dataset were sampled, and their cumulative-coverage in the population was calculated with 100 sampling iterations for (A) 1,000 individuals, and (B) 329 individuals, which is the estimated sample size to reach a 1-time cumulative-coverage. (C) Sampling of 1,000 and (D) 329 individuals excluding the possibility of autologous donation. (E) Cumulative-coverage for the individuals found to be double homozygous (red) and triple homozygous (blue) for HLA-A, HLA-B and HLA-C. Comparison with 1,000 randomly sampled individuals in 100 iterations (grey). The average cumulative coverage of all iterations is shown in black.
Supplementary Table 1HLA class I allele frequencies observed in the Lithuanian population (n = 3,496).
Supplementary Table 2HLA class I double homozygous haplotypes identified in this study (n = 153).
Supplementary Table 3HLA-A, HLA-B and HLA-C triple homozygous fibroblasts derived in this study.
References
1
Cao K Hollenbach J Shi X Shi W Chopek M Fernández-Viña MA . Analysis of the frequencies of HLA-A, B, and C alleles and haplotypes in the five major ethnic groups of the United States reveals high levels of diversity in these loci and contrasting distribution patterns in these populations. Hum Immunol. (2001) 62:1009–30. doi: 10.1016/S0198-8859(01)00298-1
2
Mack SJ Tu B Lazaro A Yang R Lancaster AK Cao K et al . HLA-A, -B, -C, and -DRB1 allele and haplotype frequencies distinguish Eastern European Americans from the general European American population. Tissue Antigens. (2009) 73:17–32. doi: 10.1111/j.1399-0039.2008.01151.x
3
Barker DJ Maccari G Georgiou X Cooper MA Flicek P Robinson J et al . The IPD-IMGT/HLA database. Nucleic Acids Res. (2023) 51:D1053–60. doi: 10.1093/nar/gkac1011
4
Jamy O Zeiser R Chen YB . Novel developments in the prophylaxis and treatment of acute GVHD. Blood. (2023) 142:1037–46. doi: 10.1182/blood.2023020073
5
Holtzman NG Curtis LM Salit RB Shaffer BC Pirsl F Ostojic A et al . High-dose alemtuzumab and cyclosporine vs tacrolimus, methotrexate, and sirolimus for chronic graft-versus-host disease prevention. Blood Adv. (2024) 8:4294–310. doi: 10.1182/bloodadvances.2023010973
6
Hricik DE Armstrong B Alhamad T Brennan DC Bromberg JS Bunnapradist S et al . Infliximab induction lacks efficacy and increases BK virus infection in deceased donor kidney transplant recipients: results of the CTOT-19 trial. JASN. (2023) 34:145–59. doi: 10.1681/ASN.2022040454
7
Bacigalupo A Metafuni E Amato V Marquez Algaba E Pagano L . Reducing infectious complications after allogeneic stem cell transplant. Expert Rev Hematol. (2020) 13:1235–51. doi: 10.1080/17474086.2020.1831382
8
Styczynski J Tridello G Koster L Knelange N Wendel L Van Biezen A et al . Decrease of lethal infectious complications in the context of causes of death (COD) after hematopoietic cell transplantation: COD-2 and COD-1 study of the Infectious Diseases Working Party EBMT. Bone Marrow Transplant. (2023) 58:881–92. doi: 10.1038/s41409-023-01998-2
9
Lee SJ Klein J Haagenson M Baxter-Lowe LA Confer DL Eapen M et al . High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood. (2007) 110:4576–83. doi: 10.1182/blood-2007-06-097386
10
Spellman SR . Hematology 2022—what is complete HLA match in 2022? Hematology. (2022) 2022:83–9. doi: 10.1182/hematology.2022000326
11
Kitano Y Nishimura S Kato TM Ueda A Takigawa K Umekage M et al . Generation of hypoimmunogenic induced pluripotent stem cells by CRISPR-Cas9 system and detailed evaluation for clinical application. Mol Therapy-Methods Clin Dev. (2022) 26:15–25. doi: 10.1016/j.omtm.2022.05.010
12
Gaykema LH Van Nieuwland RY Lievers E Moerkerk WBJ De Klerk JA Dumas SJ et al . T-cell mediated immune rejection of beta-2-microglobulin knockout induced pluripotent stem cell-derived kidney organoids. Stem Cells Trans Med. (2024) 13:69–82. doi: 10.1093/stcltm/szad069
13
Gornalusse GG Hirata RK Funk SE Riolobos L Lopes VS Manske G et al . HLA-E-expressing pluripotent stem cells escape allogeneic responses and lysis by NK cells. Nat Biotechnol. (2017) 35:765–72. doi: 10.1038/nbt.3860
14
Deuse T Hu X Gravina A Wang D Tediashvili G De C et al . Hypoimmunogenic derivatives of induced pluripotent stem cells evade immune rejection in fully immunocompetent allogeneic recipients. Nat Biotechnol. (2019) 37:252–8. doi: 10.1038/s41587-019-0016-3
15
Pizzato HA Alonso-Guallart P Woods J Johannesson B Connelly JP Fehniger TA et al . Engineering human pluripotent stem cell lines to evade xenogeneic transplantation barriers. Stem Cell Rep. (2024) 19:299–313. doi: 10.1016/j.stemcr.2023.12.003
16
Han X Wang M Duan S Franco PJ Kenty JHR Hedrick P et al . Generation of hypoimmunogenic human pluripotent stem cells. Proc Natl Acad Sci USA. (2019) 116:10441–6. doi: 10.1073/pnas.1902566116
17
Xu H Wang B Ono M Kagita A Fujii K Sasakawa N et al . Targeted Disruption of HLA Genes via CRISPR-Cas9 Generates iPSCs with Enhanced Immune Compatibility. Cell Stem Cell. (2019) 24:566–78. doi: 10.1016/j.stem.2019.02.005
18
Urnikyte A Flores-Bello A Mondal M Molyte A Comas D Calafell F et al . Patterns of genetic structure and adaptive positive selection in the Lithuanian population from high-density SNP data. Sci Rep. (2019) 9:9163. doi: 10.1038/s41598-019-45746-3
19
Creary LE Gangavarapu S Mallempati KC Montero-Martín G Caillier SJ Santaniello A et al . Next-generation sequencing reveals new information about HLA allele and haplotype diversity in a large European American population. Hum Immunol. (2019) 80:807–22. doi: 10.1016/j.humimm.2019.07.275
20
Davey S Ord J Navarrete C Brown C . HLA-A, -B and -C allele and haplotype frequencies defined by next generation sequencing in a population of 519 English blood donors. Hum Immunol. (2017) 78:397–8. doi: 10.1016/j.humimm.2017.04.001
21
Tu B Mack SJ Lazaro A Lancaster A Thomson G Cao K et al . HLA-A, -B, -C, -DRB1 allele and haplotype frequencies in an African American population. Tissue Antigens. (2007) 69:73–85. doi: 10.1111/j.1399-0039.2006.00728.x
22
Kuhn M . Building Predictive Models in R Using the caret Package. J Stat Soft. (2008) 28. Available online at: http://www.jstatsoft.org/v28/i05/.
23
Robinson J Barker DJ Marsh SGE . 25 years of the IPD-IMGT/HLA D atabase. HLA. (2024) 103:e15549. doi: 10.1111/tan.15549
24
Hoberecht L Perampalam P Lun A Fortin JP . A comprehensive Bioconductor ecosystem for the design of CRISPR guide RNAs across nucleases and technologies. Nat Commun. (2022) 13:6568. doi: 10.1038/s41467-022-34320-7
25
Hallgren J Tsirigos KD Pedersen MD Almagro Armenteros JJ Marcatili P Nielsen H et al . DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. Bioinformatics. (2022). doi: 10.1101/2022.04.08.487609
26
Doench JG Fusi N Sullender M Hegde M Vaimberg EW Donovan KF et al . Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. (2016) 34:184–91. doi: 10.1038/nbt.3437
27
Stockton JD Nieto T Wroe E Poles A Inston N Briggs D et al . Rapid, highly accurate and cost-effective open-source simultaneous complete HLA typing and phasing of class I and II alleles using nanopore sequencing. HLA. (2020) 96:163–78. doi: 10.1111/tan.13926
28
Pollock NR Farias TDJ Kichula KM Sauter J Scholz S Nii-Trebi NI et al . The 18th International HLA & Immunogenetics workshop project report: Creating fully representative MHC reference haplotypes. HLA. (2024) 103:e15568. doi: 10.1111/tan.15568
29
Karlsson M Zhang C Méar L Zhong W Digre A Katona B et al . A single–cell type transcriptomics map of human tissues. Sci Adv. (2021) 7:eabh2169. doi: 10.1126/sciadv.abh2169
30
Shi M Méar L Karlsson M Álvez MB Digre A Schutten R et al . A resource for whole-body gene expression map of human tissues based on integration of single cell and bulk transcriptomics. Genome Biol. (2025) 26:152. doi: 10.1186/s13059-025-03616-4
31
Sullivan S Ginty P McMahon S May M Solomon SL Kurtz A et al . The global alliance for iPSC therapies (GAiT). Stem Cell Res. (2020) 49:102036. doi: 10.1016/j.scr.2020.102036
32
Taylor CJ Peacock S Chaudhry AN Bradley JA Bolton EM . Generating an iPSC bank for HLA-matched tissue transplantation based on known donor and recipient HLA types. Cell Stem Cell. (2012) 11:147–52. doi: 10.1016/j.stem.2012.07.014
33
Tian P Elefanty A Stanley EG Durnall JC Thompson LH Elwood NJ . Creation of GMP-compliant iPSCs from banked umbilical cord blood. Front Cell Dev Biol. (2022) 10:835321. doi: 10.3389/fcell.2022.835321
34
Yoshida S Kato TM Sato Y Umekage M Ichisaka T Tsukahara M et al . A clinical-grade HLA haplobank of human induced pluripotent stem cells matching approximately 40% of the Japanese population. Med. (2023) 4:51–66.e10. doi: 10.1016/j.medj.2022.10.003
35
Rim YA Park N Nam Y Ham D Kim J Ha H et al . Recent progress of national banking project on homozygous HLA -typed induced pluripotent stem cells in S outh K orea. J Tissue Eng Regener Med. (2018)12. Available online at: https://onlinelibrary.wiley.com/doi/10.1002/term.2578.
36
Lee S Huh JY Turner DM Lee S Robinson J Stein JE et al . Repurposing the cord blood bank for haplobanking of HLA-homozygous iPSCs and their usefulness to multiple populations. Stem Cells. (2018) 36:1552–66. doi: 10.1002/stem.2865
37
Kuebler B Alvarez-Palomo B Aran B Castaño J Rodriguez L Raya A et al . Generation of a bank of clinical-grade, HLA-homozygous iPSC lines with high coverage of the Spanish population. Stem Cell Res Ther. (2023) 14:366. doi: 10.1186/s13287-023-03576-1
38
Liedtke S Korschgen L Korn J Duppers A Kogler G . GMP-grade CD34 + selection from HLA-homozygous licensed cord blood units and short-term expansion under European ATMP regulations. Vox Sanguinis. (2021) 116:123–35. doi: 10.1111/vox.12978
39
Alowaysi M Lehmann R Al-Shehri M Baadhaim M Alzahrani H Aboalola D et al . HLA-based banking of induced pluripotent stem cells in Saudi Arabia. Stem Cell Res Ther. (2023) 14:374. doi: 10.1186/s13287-023-03612-0
Summary
Keywords
superdonor, HLA class I, immune compatibility, hypoimmunogenic, population genetics
Citation
Naumovas D, Rojas-Araya B, Polanco CM, Andrade V, Čekauskienė R, Valatkaitė-Rakštienė B, Laurinaitytė I, Jakubauskas A, Stoškus M, Griškevičius L, Nalvarte I, Inzunza J, Baltriukienė D and Arias J (2025) Identification of HLA-A, HLA-B, and HLA-C triple homozygous and double homozygous donors: a path toward synthetic superdonor advanced therapeutic medicinal products. Front. Immunol. 16:1626787. doi: 10.3389/fimmu.2025.1626787
Received
11 May 2025
Accepted
17 August 2025
Published
16 September 2025
Volume
16 - 2025
Edited by
Belen Alvarez-Palomo, Banc de Sang i Teixits, Spain
Reviewed by
Michiko Taniguchi, Washington University in St. Louis, United States
Sergio Querol, Fundacion Josep Carreras contra la Leucemia, Spain
Updates
Copyright
© 2025 Naumovas, Rojas-Araya, Polanco, Andrade, Čekauskienė, Valatkaitė-Rakštienė, Laurinaitytė, Jakubauskas, Stoškus, Griškevičius, Nalvarte, Inzunza, Baltriukienė and Arias.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jonathan Arias, jonathan.arias@gmc.vu.lt
†ORCID: Daniel Naumovas, orcid.org/0000-0002-5968-4182; Barbara Rojas-Araya, orcid.org/0009-0005-6256-0112; Catalina M. Polanco, orcid.org/0009-0004-7827-8851; Victor Andrade, orcid.org/0000-0003-0682-269X; Rita Čekauskienė, orcid.org/0009-0006-2607-3779; Beatričė Valatkaitė-Rakštienė, orcid.org/0009-0008-7915-4702; Inga Laurinaitytė, orcid.org/0000-0003-1089-8312; Artūras Jakubauskas, orcid.org/0000-0002-6305-0617; Mindaugas Stoškus, orcid.org/0000-0001-6344-8134; Laimonas Griškevičius, orcid.org/0000-0002-3731-1537; Ivan Nalvarte, orcid.org/0000-0001-6828-2583; Jose Inzunza, orcid.org/0000-0003-0876-6767; Daiva Baltriukienė, orcid.org/0000-0002-7851-9270; Jonathan Arias, orcid.org/0000-0002-3997-2355
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.