- 1Department of Microbiology and Cell Science, Institute of Food and Agricultural Sciences, University of Florida, Gainesville, FL, United States
- 2The Department of Molecular Genetics and Microbiology, College of Medicine, University of Florida, Gainesville, FL, United States
Salmonella is a leading cause of foodborne illness in the United States and worldwide. This enteric pathogen deploys various mechanisms to evade the intestinal mucosal barrier to enhance its survival and further infect systemic tissues. Commercially available vaccines against Salmonella are currently restricted to the serovar Typhi, while none are currently approved for non-typhoidal Salmonella (NTS) serovars, which are becoming increasingly resistant to antibiotics. Due to the lack of effective vaccines against NTS infections, novel oral vaccination strategies have garnered significant interest, owing to their protective abilities at the susceptible sites of infection. We previously reported that mice immunized intranasally with small extracellular vesicles (sEVs) derived from Salmonella-infected macrophages protect mice against lethal Salmonella challenge. In the present study, we used an oral route of administration of sEVs to determine their protective abilities in vivo. Remarkably, orally administered sEVs from Salmonella-infected macrophages conferred significant host protection, marked by improved survival post-challenge and reduction in tissue bacterial burdens. Additionally, immunized mice exhibited robust serological responses, including elevated levels of both whole-Salmonella and OmpA-specific IgG antibodies. Collectively, these findings show the potential of orally delivered sEVs as a promising, cell-free vaccine platform for protection against salmonellosis.
1 Introduction
The global burden of Salmonella expands beyond the commonly known typhoid fever caused by Salmonella Typhi, the leading cause of bloodstream infections in Asia. Salmonellosis caused by non-typhoidal serovars of Salmonella (NTS), including Salmonella enterica serovar Typhimurium, often leads to self-limiting gastroenteritis in individuals with healthy immune systems. However, in immunocompromised individuals, NTS can cause bacteremia resulting in significant morbidity and mortality (1). In the United States alone, NTS is responsible for approximately 1.4 million infections per year and is one of the leading causes of foodborne illness, as reported by the CDC (2). Salmonella utilizes numerous virulence factors at different stages of infection. The first step of infection involves invasion of M cells present at mucosal sites, for which the Gram-negative bacterium utilizes Salmonella pathogenicity island 1 (SPI-1) encoded effector proteins, while SPI-2 effectors predominantly allow for establishment of an intracellular niche within the Salmonella containing vacuole (SCV). To prolong its survival within the host, Salmonella has adapted to the epithelial cell shedding process which allows this pathogen to disseminate into the intestinal lumen, in turn infecting a larger number of resident and non-resident cells (3).
Although licensed vaccines such as Ty21a and Vi-PS are available for S. Typhi, no approved vaccine currently exists for human use against NTS. In settings with limited resources, NTS infections often go undiagnosed in immunocompromised individuals. Furthermore, the use of antibiotics such as fluoroquinolones has led to an increase in antimicrobial resistance, thus prioritizing the development of a safe and effective vaccine (4). Mouse models using S. Typhimurium effectively resemble features of systemic typhoid-like disease, enabling detailed investigations of host-pathogen interactions (5). The innate immune system represents the first line of defense against Salmonella, orchestrating in the primary stages of infection clearance. However, S. Typhi virulence can circumvent these antimicrobial responses to persist and colonize host intestinal and systemic sites. While robust innate immune capabilities are activated during Salmonella infection, durable long-term immunological protection requires mounting of effective adaptive immune responses (6).
Recently, extracellular vesicles (EVs) have emerged as promising acellular vaccine platforms. EVs are lipid bilayer-enclosed nano- and micro-particles secreted by eukaryotic cells through various biogenesis pathways (7). EVs play an important role in intercellular communication, across both short and long distances and can carry a wide range of bioactive molecules, including proteins, lipids, and nucleic acids (8). EVs are typically categorized into: small EVs (sEVs) ranging from 30–150 nm in size, medium-sized EVs that are around 200–800 nm in diameter, and large EVs that have a diameter greater than 1000 nm and mostly include apoptotic bodies, large oncosomes or exopheres (7). Their capacity to transport antigenic cargo and immunomodulatory molecules positions EVs as attractive candidates for immunotherapeutic applications, particularly in the context of infection (9, 10).
Previous studies conducted by our laboratory have identified Salmonella-encoded antigens within sEVs isolated from RAW264.7 macrophages at 24- and 48- hours post-infection (hpi) via mass spectrometry (11). Mice intranasally (IN) receiving sEVs from Salmonella-infected macrophages have demonstrated prolonged survival upon S. Typhimurium challenge in comparison to the control group (12). In this study, we specifically demonstrate that mice orally immunized with sEVs from Salmonella-infected macrophages had better survival upon S. Typhimurium challenge. The immunized mice displayed decreased weight loss, improved body scores, and reduced bacterial loads in their tissues. Additionally, the protective efficacy of sEVs was also demonstrated by an elevated production of antigen-specific IgG, providing a platform for evaluating innovative delivery methods of EV-based vaccines.
2 Materials and methods
2.1 Cell and bacterial culture
RAW264.7 (ATCC TIB-71) murine macrophages were grown and maintained in complete DMEM, supplemented with 10% fetal bovine serum (FBS) and 1% penicillin and streptomycin, at 37°C and 5% CO2. Salmonella Typhimurium strains UK-1 (χ3761) and ΔaroA (χ9099) were grown in lysogeny broth (LB) Miller with constant shaking at 250 rpm at 37°C overnight (14–16 hours). The OD600 of the overnight culture was measured using a spectrophotometer and the culture was diluted to an OD600 of 0.05 in 20mL LB miller and grown up to an OD600 of 0.5 to use for experiments. Based on a previously established growth curve, the reported OD corresponds to approximately 3.3 × 108 CFU/mL for UK-1 and 6.5 × 108 CFU/mL for ΔaroA Salmonella.
2.2 Cell infection and EV isolation
RAW264.7 macrophages were grown up to confluency in complete DMEM medium. Prior to infection, cells were washed with 1X phosphate buffered saline (PBS) to remove traces of FBS and antibiotics and replaced with DMEM containing 1% exosome-depleted FBS (exo-free DMEM). Cells were infected with S. Typhimurium UK-1 at a multiplicity of infection (MOI) of 5 for 2 hours. Media on cells was then replaced with exo-free DMEM containing 100 μg/mL gentamicin for 1 hour to remove any extracellular Salmonella. After one hour, the media was changed again and replaced with exo-free DMEM containing 25 μg/mL gentamicin and cells were allowed to incubate till the cell supernatant was collected for EV isolation 24 hours post infection, and the cell supernatant was replenished for collection 48 hours post infection.
Collected supernatants underwent sequential centrifugation at 500 × g and 4,000 × g for 10 minutes each, followed by 16,000 × g for 30 minutes to remove cell debris. The collected media was filtered through a 0.2 μm filter and ran at 100,000 x g for 3 hours using ultracentrifuge fitted with an SW32Ti rotor (Beckman Coulter Optima XPN-90). After the first run, the media was decanted and the EVs were resuspended in 400uL PBS with 1% protease inhibitor, and the rest of the ultracentrifuge tubes were balanced using sterile 1X PBS. The ultracentrifugation step was repeated and the EVs were finally resuspended in 1mL of PBS with 1% protease inhibitor. The protein concentration was determined using the Pierce BCA kit (Thermo Fisher Scientific). The particle concentration and size were determined using a ZetaView QUATT particle tracking analyzer (Particle Matrix). Three replicates of EVs (10 μg protein/each) from uninfected and infected macrophages were stained with MACSplex EV kit (Miltenyi Biotec Catalog no. 130122211) using manufacturer’s instructions to identify and compare tetraspanin markers. The EVs were analyzed using a flow cytometer (Beckman Coulter cytoFLEX) and FlowJo software.
2.3 Western blotting
Western blotting was carried out using 20 μg EVs per sample. Sample buffer containing beta-mercaptoethanol (BME), Deionized (DI) water, and 4 x XT-sample buffer was made and 25 μL sample was loaded per well in a 4-12% bis-tris gel (BioRad). The gel was run at 75V for 15 minutes, after which the voltage was increased to 200V, and the gel was run for another 45 minutes. The gel was then transferred to a blot and stained with primary antibody [1:800 dilution for anti-CD63 (System Biosciences), anti-CD9 (System Biosciences), anti-Alix (System Biosciences), and anti-OmpA (Vector Laboratories)] with overnight shaking at 4°C. The blot was then washed three times with PBST (PBS with 0.1% Tween-20) for 3 minutes each, after which it was stained with an HRP-conjugated goat anti-rabbit secondary antibody (1:500 dilution) for 1–2 hours with constant shaking. The blot was washed with PBST again and enhanced chemiluminescence (ECL) substrate was added for 5 minutes before imaging the blot using iBright Imaging System (Invitrogen).
2.4 Mouse experiments
Seven-week-old female BALB/c mice (Jackson Laboratory) were orally dosed with 40 μg sEVs from Salmonella-infected macrophages at week 0, 2, and 4. As a negative and positive control, mice were also immunized orally with sterile 1X PBS and S. Typhimurium ΔaroA (4.5 x 106 CFUs) respectively. Prior to the administration of immunizations, mice were orally dosed with 50 μL of 0.3M sodium bicarbonate using a 22-gauge oral gavage needle to neutralize their stomach acid for standardized Salmonella infection. After waiting 10 minutes, another 22-gauge gavage needle was used to administer the appropriate dose resuspended in 100 μL of 1X PBS. For the survival study, mice were orally challenged with a lethal dose (4.5 x 106 CFUs) of S. Typhimurium UK-1 following the abovementioned dosing protocol, 7 weeks after their first sEV dose, and their weights and body scores were assessed daily for 30 days or until endpoint. Mice were euthanized in a chamber using CO2 at a 30-70% displacement rate, followed by a cervical dislocation as a secondary confirmation of death. For serological analysis, blood was collected via the saphenous vein at weeks 1, 3, 5, and 7. After centrifugation at 10,000 × g for 10 minutes, serum was collected and stored at –20°C until ELISA analysis.
2.5 Bacterial burden
To assess the bacterial burden, mice immunized with sEVs or PBS control were challenged with 4.5x106 CFUs of S. Typhimurium given orally, 7 weeks after their first EV dose. 4 days post challenge with Salmonella, mice were euthanized, and their spleen and liver were collected. The organs were weighed and resuspended in 0.1% Triton-X in PBS and lysed using TissueLyser LT (Qiagen). Serial dilutions were made in PBS and 100 μL volume was plated on LB agar plates. The agar plates were incubated at 37°C overnight after which colonies were counted and CFU per gram of each organ was determined.
2.6 Protein purification and mass spectrometry
OmpA gene from S. Typhimurium UK-1 was amplified using polymerase chain reaction (PCR), using the forward primer TAAGCAGGATCCAATGAAACTTAAGTTAGTGGCAGTG and reverse primer TAAGCAAAGCTTTTAGAACTGGTAGTTCAGACCAAC. The amplified fragment was ligated into a pET28a plasmid which was digested using EcoRI and HindIII restriction enzymes. The ligated vector was transformed into E. coli DH5α cells after which the plasmid was purified and re-transformed into E. coli BL21(DE3) cells for protein purification. The 6X His-tagged OmpA was purified using Ni-NTA resin, for which a bacterial culture containing transformed BL21(DE3) cells was grown up to an OD600 of 0.6 which constant shaking at 200 rpm at 28°C. The protein was induced by adding 0.5 mM IPTG to the culture and leaving it overnight in a shaker incubator at 200 rpm at 19°C. After overnight induction the bacterial cells were lysed by sonication and the cell pellet was resuspended and left at room temperature for 15 minutes in 6M urea after centrifugation at 4200 rpm for 10 minutes. The suspension was centrifuged again at 4200 rpm for 20 minutes after which the supernatant was saved for purification of OmpA. Ni-NTA column affinity chromatography was conducted following manufacturer’s instructions (Thermo Fisher Scientific Cat. No. 88228). The eluted sample was run on an SDS-PAGE and was stained with GelCode Blue stain for 2 hours (Thermo Fisher Scientific) and destained with DI water overnight. The resulting OmpA band was excised from the gel, subjected to in-gel trypsin digestion according to standard protocols (11), and analyzed using a timsTOF flex mass spectrometer (Bruker). Peptides were identified via database searching using Mascot (version 2.7.0) against the Salmonella Typhimurium database with trypsin as the specified enzyme, a precursor ion tolerance of 20 ppm, and fragment ion tolerance of 0.50 Da. Carbamidomethylation of cysteine was set as a fixed modification, and variable modifications included methionine oxidation, N-terminal acetylation, pyro-Glu formation from glutamine, and deamidation of asparagine and glutamine. Protein identifications were validated using Scaffold (version 5.2.2, Proteome Software Inc.), with peptide and protein thresholds set at 95% confidence, and requiring a minimum of two unique peptides. Identified proteins were grouped by parsimony and confirmed using the Protein Prophet algorithm.
2.7 ELISAs
Nunc 96-well plates were coated with 2 μg LPS-detoxified Salmonella per well and left at room temperature overnight. On day two, excess antigen was removed from the wells by blotting the plate on paper towels. Blocking buffer (PBS with 1% BSA) was added to the wells and the plate was blocked for 2 hours at 37°C. The plate was washed three times with PBST followed by two washes with PBS. The plate was blotted dry after each wash. Serum dilutions were prepared in ELISA buffer (PBS with 0.5% BSA and 0.05% Tween20) and added to the wells containing Salmonella antigen. The plate was refrigerated overnight. On day three, plates were washed as previously described. HRP-conjugated goat anti-mouse IgG (Southern Biotech, Cat. No. 1030-05) diluted 1:2,000 in ELISA buffer was added (50 μL/well), and plates were incubated for 90 minutes at 37°C. After washing, 50 μL ABTS (2,2’-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid)) substrate was added per well and developed for 30 minutes at room temperature. Absorbance was read at 415 nm using a Cytation 3 plate reader (BioTek).
2.8 Statistical analysis and figure design
GraphPad Prism 10.3.1 was used for figure rendering, and statistical results are denoted with ns for not significant * for p-value at <0.05, ** at <0.01, *** and **** at p<0.0001 in each graph. For comparing two groups with equal variances, students t-test was applied. For comparing three or more groups, one-way and two-way ANOVA with post hoc Tukey’s multiple comparison tests were performed. Schematic diagrams representing study design were created using Biorender.
3 Results
3.1 sEVs from Salmonella-infected macrophages express markers required for antigen presentation
In the present study, we evaluated the immunogenicity and protective efficacy of sEVs administered via the oral route. sEVs were isolated from S. Typhimurium-infected macrophages and characterized by standard methods. Western blotting confirmed the presence of canonical sEV tetraspanins CD63 and CD9, and Alix, a cytoplasmic marker. Furthermore, the presence of Salmonella antigen OmpA in sEVs was also confirmed (Figure 1A). Comparative analysis using the MACSplex EV kit revealed a significant reduction in the tetraspanins CD63, CD81 and CD9 in sEVs derived from Salmonella-infected macrophages (obtained at 24- and 48- hours post-infection) relative to uninfected controls. Notably, CD40 expression was upregulated in sEVs from infected cells, while MHCII showed a slight albeit insignificant increase (Figure 1B). Nanoparticle tracking analysis (NTA) showed consistent vesicle size distributions and yields across independent isolations (Figures 1C–F). The protein-to-particle ratio was determined using a BCA protein assay to calculate the sEV production per cell, which remained reproducible, confirming the reliability of our preparation protocol.
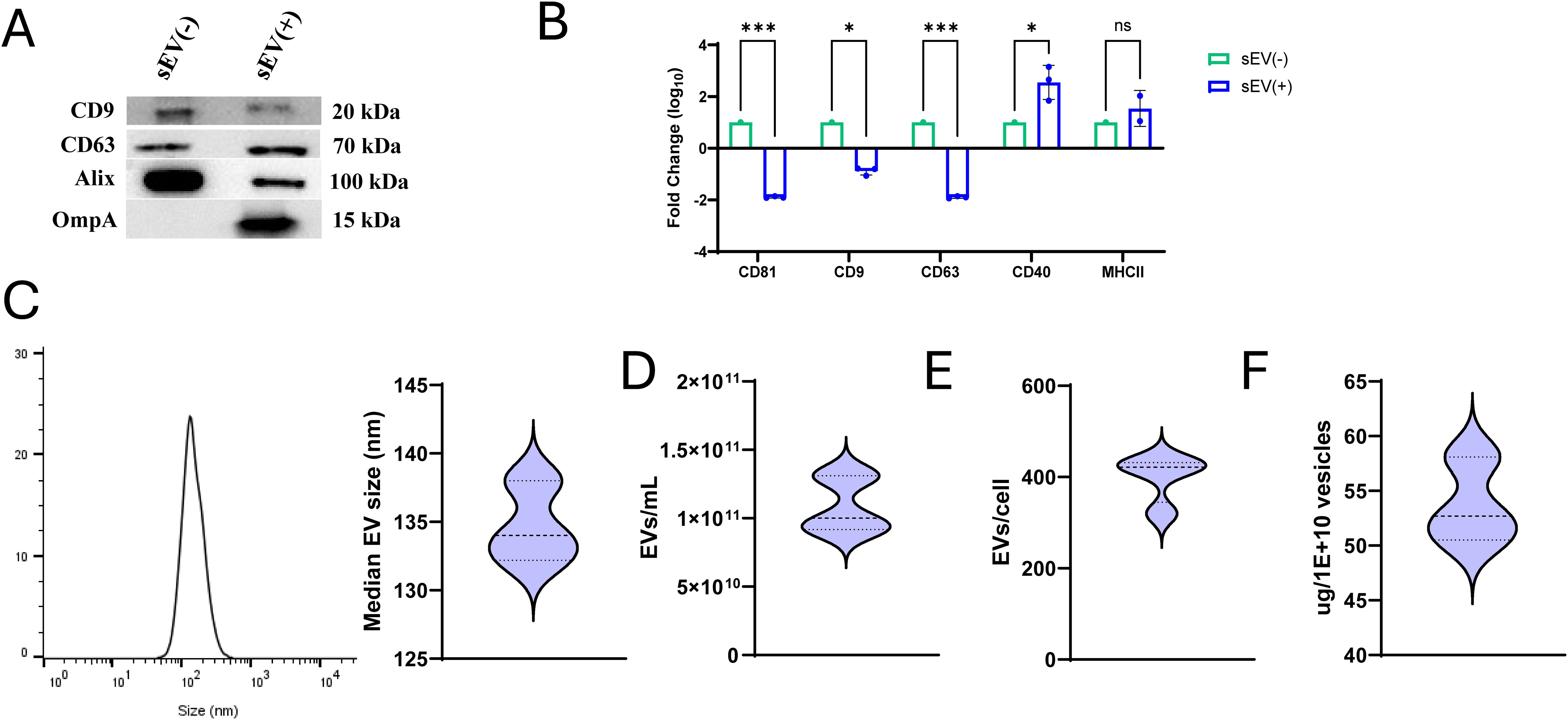
Figure 1. Characterization of small extracellular vesicles (sEVs) derived from Salmonella-infected RAW 264.7 macrophages. (A) Expression of tetraspanin markers CD63 and CD9, cytosolic marker Alix, and Salmonella antigen OmpA in sEVs isolated from uninfected RAW 264.7 macrophages compared to sEVs isolated from macrophages infected with Salmonella for 24 and 48 hours (MOI: 5:1). (B) Fold change in expression of tetraspanin markers (CD81, CD9, and CD63), CD40 and MHCII between sEVs from uninfected [sEV(-)] and infected [sEV(+)] macrophages quantified using MACSplex EV kit (n=3 independent experiments). Two-way ANOVA with post hoc Tukey’s multiple test comparison was used for analysis. Results are denoted with * for p-value at <0.05, *** at <0.001 and ns, not significant. (C) Histogram showing sEV size (left), data derived from ZetaView nanoparticle tracking analysis (NTA) and analyzed using FlowJo. Median size of sEVs from triplicate samples (right). (D) Quantification of sEVs per mL of culture media (n=5). (E) Number of sEVs produced per cell (n=4). (F) Protein content (μg) per 1 × 1010 vesicles (n=3).
3.2 sEV vaccination protects mice against lethal S. Typhimurium challenge
To evaluate the protective efficacy of the oral dose of sEVs derived from Salmonella-infected macrophages, 7-week-old BALB/c mice were immunized with sEVs (40 µg per dose) or phosphate-buffered saline (PBS) as a negative control. Our previous studies have already established a protective response to immunization with a positive control – an attenuated Salmonella strain – S. Typhimurium ΔaroA (χ9099), delivered orally (12). Mice received three doses at two-week intervals. Three weeks following the final booster, mice from both groups were challenged with a lethal dose of S. Typhimurium UK-1 (χ3761) delivered using oral gavage (Figure 2A). Clinical symptoms were monitored daily through body condition scoring and weight measurement. Upon challenge with virulent S. Typhimurium, sEV-immunized mice showed improved body condition scores (Figure 2B), reduced weight loss (Figure 2C), and significantly lower bacterial burdens in the liver at day 4 post-infection (Figure 2D), Moreover, survival analysis revealed a significantly higher survival rate in the sEV-treated group compared to controls over 30 days post-infection with Salmonella (Figure 2E).
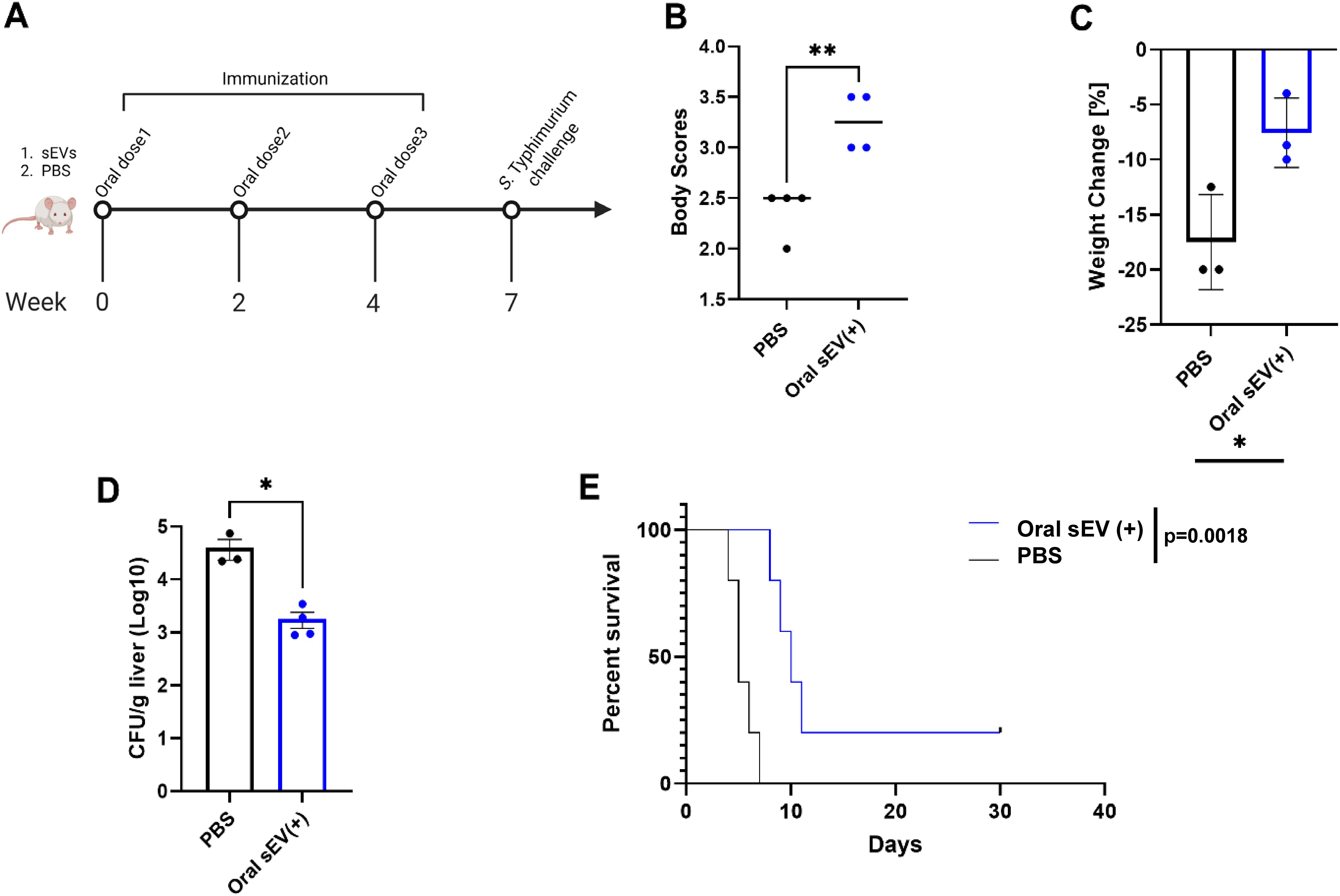
Figure 2. Preventive efficacy of orally administered sEVs from Salmonella-infected macrophages against salmonellosis. BALB/c mice were orally immunized with three doses of sEVs (40 µg per dose), or PBS at two-week intervals. Three weeks after the final dose, mice were challenged orally with virulent S. Typhimurium. (A) Immunization and challenge timeline. (B) Clinical disease scores based on body condition assessments on day 4 post-challenge (n=4). Unpaired parametric t-test used for analysis. (C) Percent body weight change on day 4 post-challenge (n=3). Unpaired parametric t-test used for analysis. (D) Bacterial burden in the liver (CFU/g) four days post-challenge. Unpaired parametric t-test used for analysis. (E) Kaplan–Meier survival curves over 30 days post-challenge (n=5). Results are denoted with * for p-value at <0.05, ** at <0.01.
3.3 Immunization with sEVs leads to production of antigen-specific IgG
Mice that received three oral doses of sEVs exhibited elevated serum IgG levels against LPS-detoxified Salmonella lysate compared to PBS controls. Additionally, sEV immunized mice showed comparable IgG production, beginning three weeks all the way up to seven weeks after immunization, to the positive control Salmonella ΔaroA, affirming the efficacy of our sEVs over a long period of time (Figure 3A). Due to various Salmonella antigens being encapsulated and enriched in sEVs from Salmonella-infected macrophages, we predicted that immunized mice could produce serum IgG to a known immunodominant protein OmpA. To demonstrate this, recombinant S. Typhimurium OmpA was purified using affinity chromatography and the sequence was confirmed via mass spectrometry (Supplementary Figure S1). Mice immunized with sEVs produced significantly higher levels of OmpA-specific serum IgG as compared to the control group over a period of seven weeks (Figure 3B). These findings demonstrate that sEVs derived from S. Typhimurium-infected macrophages, when delivered orally, can confer significant protection against lethal Salmonella challenge. Results from our current study expand on previous work utilizing intranasal delivery and highlight the potential of sEVs as an orally deliverable vaccine platform.
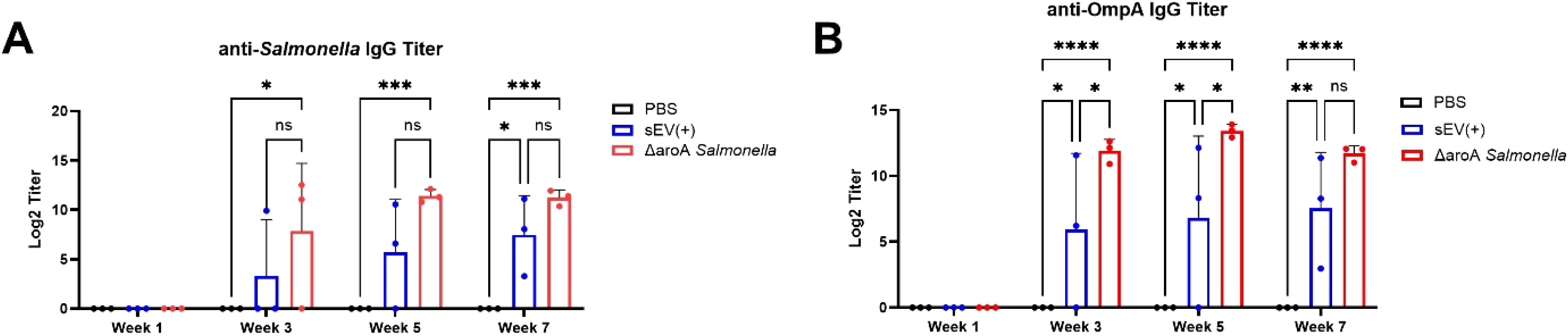
Figure 3. Serum IgG titers measured over time post-immunization. BALB/c mice were orally immunized with three doses of sEVs (40 µg per dose), Salmonella ΔaroA mutant strain, or PBS at two-week intervals. Blood was collected each week after immunization, at weeks 1, 3, 5, and 7, prior to lethal challenge with S. Typhimurium. (A) Serum anti-Salmonella IgG levels measured at multiple time points during immunization (n=3). Log2 titers were defined as the reciprocal of the dilution giving absorbance 0.1U above absorbance. (B) Serum anti-OmpA IgG levels measured at multiple time points during immunization (n=3). Log2 titers were defined as the reciprocal of the dilution giving absorbance 0.5U above absorbance. Two-way ANOVA with post hoc Tukey’s multiple comparison tests were used for analysis. Results are denoted with * for p-value at <0.05, ** at <0.01, *** and **** at p<0.0001. ns, not significant.
4 Discussion
While several delivery routes of therapeutics have been explored, the reduced invasiveness and improved feasibility of administration through the oral route have been established over the years, highlighting their importance for drug delivery (13). Oral vaccines are of significant interest due to their ability to induce both systemic and mucosal immune response. However, effective mucosal immunization faces several challenges due to the degradative environment and tolerogenic immune responses of the oral mucosa, stomach, and small intestine (14). Ty21a – an orally delivered live attenuated S. Typhi vaccine – is formulated with an enteric coating to bypass digestive enzymes within the stomach (15). Birds immunized with a mannose chitosan nanoparticle-based vaccine have demonstrated induction of mucosal immunity against Salmonella Enteritidis, in addition to cross-protective responses against S. Typhimurium (16). These and other successful immunization efforts have led us to develop an EV-based approach against NTS that can be potentially translated and optimized for further human applications.
The antimicrobial activity of EVs derived from host immune cells has been exhibited both in vitro and in vivo (17, 18). One of the main reasons for this is the ability of these EVs to encapsulate and carry pathogen-derived antigens. (19). The inflammatory impact of intraluminal EV cargo can be context dependent and remains to be further characterized. The pro-inflammatory capacity of sEVs carrying pathogen-associated antigenic cargo has been demonstrated in both Salmonella Typhimurium and Mycobacterium tuberculosis infection models (11, 20). In previous studies, mice receiving sEVs from Salmonella-infected macrophages intranasally have demonstrated prolonged survival upon Salmonella challenge relative to the control group. Additionally, the bacterial burden in the liver of mice four days post Salmonella challenge was reduced significantly in mice receiving sEVs, compared to those not receiving intranasal sEVs (12). Additionally, intranasal sEV immunization also enhanced systemic IgG and mucosal IgA responses against the outer membrane proteins OmpA and OmpD—Salmonella-encoded antigens identified within sEVs isolated from RAW264.7 macrophages at 24- and 48- hours post-infection via mass spectrometry (11, 12). Salmonella-specific IgAs and IgGs produced in such vaccinated animals were also able to cross-react against heterologous Salmonella species derived from environmental sources (21). In parallel, several studies have demonstrated the beneficial effect of outer membrane vesicles (OMVs) as potential acellular vaccines for various applications (22, 23). Moreover, OMVs from flagellin-deficient S. Typhimurium administered through either the intranasal or intraperitoneal route elicited strong antibody responses and provided cross-protection against other Salmonella strains such as S. Enteritidis and S. Choleraesuis (24). These findings reinforce the broader concept that EV-based vaccines—whether host-derived sEVs or pathogen-derived OMVs—can stimulate protective immunity through either complementary or independent immunization mechanisms.
Our findings, along with emerging data that EVs can survive enzymatic degradation and acidic pH in the gastrointestinal tract, further support their feasibility as orally delivered vaccines (25). Bacterial-derived OMVs from Vibrio cholerae, Helicobacter pylori, and Acinetobacter baumannii when delivered orally induced protective immune responses in vivo [as reviewed in (26)]. Orally delivered antibiotic loaded OMVs from A. baumannii were able to exert bactericidal effects in the intestine of mice within 2 days post-delivery (27). More importantly, mesenchymal stem cell-derived EVs when targeted to the colon via oral administration, effectively alleviated ulcerative colitis in vivo (28). Although limited data exist on the in vivo fate of sEVs derived from mammalian immune cells following oral administration, we hypothesize that after surviving passage through the stomach, these Salmonella-antigen encapsulated sEVs may interact with microfold (M) cells in the Peyer’s patches in a manner similar to the bacterial antigens (29). This interaction could facilitate their transport via lymphatic or circulatory routes to distal organs, promote uptake by intestinal antigen-presenting cells (APCs) to initiate cell-mediated immune responses (28), or direct local B cells from the gut to migrate systemically to produce IgG antibodies (30). While we observed systemic IgG responses and improved survival, mucosal secretory IgA (SIgA) induction via oral administration was not tested in this study, due to its limited production in our previous observations (12). Prior work has reported that mice lacking the polymeric immunoglobulin receptor (pIgR) – important for secretory IgA (SIgA) transport to mucosal surfaces – had a reduced bacterial burden in tissues such as the liver and spleen following oral infection with S. Typhimurium. Furthermore, pIgR knockout mice showed improved survival following lethal S. Typhimurium infection, which indicates that mucosal IgA might not be necessary for long-term protective immunity against Salmonella (31). A progressive increase in serum IgG in immunized mice with specificity to Salmonella OmpA antigen, demonstrated to be present in sEVs from Salmonella-infected macrophages, indicated that orally delivered sEVs can boost production of antigen-specific protective antibody responses. Based on our presented results, orally administered sEVs elicited protective responses independently of robust SIgA generation. In line with previous experiments, our oral sEV immunization resulted in a decreased bacterial burden in the livers of vaccinated mice, and an overall improvement in these mice was also demonstrated by improved host resilience and reduced weight fluctuations. Elucidating the innate and adaptive immune mechanisms induced by sEVs administered through different mucosal routes (oral versus intranasal) is warranted to understand essential components that prime effective long-term immunological responses.
Future work will focus on optimizing oral sEV vaccine formulations for improved efficacy and cross-protective responses against heterologous strains. Strategies under consideration include encapsulation in enteric-coated carriers, co-delivery with mucosal adjuvants, and engineering of EV surface features to promote mucosal uptake and tropism for antigen presentation responses (32, 33). Since immune responses within the gut can vary significantly based on site-specificity within the tissues, it is important to account for these differences when designing nanoparticle-based therapies in order to develop precise, safe, and effective oral vaccines (34). This study was limited by the lack of assessment of cell-mediated immunity, which plays a key role in host defense against intracellular pathogens like Salmonella (11). Importantly, Salmonella pathogenesis involves inhibition of T cells, which impairs bridging responses necessary for long-term immunity (35). Additional mechanistic studies evaluating T cell-associated immune responses related to sEVs routes of administration, such as ex vivo stimulation of spleens and mesenteric lymph nodes (mLNs) of immunized mice to assess memory CD4 helper and CD8 cytotoxic T cell proliferation followed by intracellular cytokine detection, are required. A current limitation to this is that protective effects can only be explored in systemic organs, an issue that can be mitigated using a colitis-induced model for Salmonella infections. Overall, these results can further shed light into heterogenous protective memory-recall responses for optimization of immunizations strategies against salmonellosis.
In summary, this study demonstrates that orally administered sEVs from S. Typhimurium-infected macrophages confer significant protection against systemic Salmonella infection in mice. These findings further support the continued development of EV-based oral vaccines as a new and scalable approach for preventing bacterial enteric diseases.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was approved by University of Florida IACUC under protocol IACUC202200000015. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
SB: Writing – review & editing, Validation, Formal analysis, Writing – original draft, Data curation, Visualization, Methodology, Investigation. JC: Writing – review & editing, Writing – original draft, Methodology. SE: Writing – review & editing, Writing – original draft, Investigation, Methodology. RM: Writing – original draft, Investigation. MF: Supervision, Methodology, Investigation, Conceptualization, Funding acquisition, Writing – review & editing, Resources, Project administration, Writing – original draft.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This study was supported by the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH) under grant number 5R01AI158749-04. The funder played no role in study design, data collection, analysis and interpretation of data, or the writing of this manuscript.
Acknowledgments
We would also like to acknowledge UF ICBR Proteomics & Mass Spectrometry core (RRID : SCR_019151) for mass spectrometry-based analysis of the samples. We would like to thank Dr. Roy Curtiss III (University of Florida, Department of Veterinary Medicine) for providing the Salmonella strains utilized in this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1628756/full#supplementary-material
References
1. Tsolis RM, Xavier MN, Santos RL, and Bäumler AJ. How to become a top model: impact of animal experimentation on human salmonella disease research ▿. Infect Immun. (2011) 79:1806–14. doi: 10.1128/IAI.01369-10
2. Greenhow TL and Alabaster A. Epidemiology of nontyphoidal salmonella bloodstream infections in children. Pediatrics. (2023) 152:e2023062357. doi: 10.1542/peds.2023-062357
3. Hallstrom K and McCormick BA. Salmonella interaction with and passage through the intestinal mucosa: through the lens of the organism. Front Microbiol. (2011) 2:88. doi: 10.3389/fmicb.2011.00088
4. Tennant SM, MacLennan CA, Simon R, Martin LB, and Khan MI. Nontyphoidal salmonella disease: Current status of vaccine research and development. Vaccine. (2016) 34:2907–10. doi: 10.1016/j.vaccine.2016.03.072
5. Walker GT, Gerner RR, Nuccio SP, and Raffatellu M. Murine models of Salmonella infection. Curr Protoc. (2023) 3:e824. doi: 10.1002/cpz1.824
6. Takaya A, Yamamoto T, and Tokoyoda K. Humoral immunity vs. Salmonella. Front Immunol. (2019) 10:3155. doi: 10.3389/fimmu.2019.03155
7. Buzas EI. The roles of extracellular vesicles in the immune system. Nat Rev Immunol. (2023) 23:236–50. doi: 10.1038/s41577-022-00763-8
8. Wessler S and Meisner-Kober N. On the road: extracellular vesicles in intercellular communication. Cell Commun Signal. (2025) 23:95. doi: 10.1186/s12964-024-01999-8
9. Gioseffi A, Edelmann MJ, and Kima PE. Intravacuolar pathogens hijack host extracellular vesicle biogenesis to secrete virulence factors. Front Immunol. (2021) 12:662944. doi: 10.3389/fimmu.2021.662944
10. Emerson LE, Mosby CA, Enslow S, Hui WW, Jones MK, and Ferraro MJ. Changes in lipid composition of host-derived extracellular vesicles following Salmonella infection. Blondel CJ editor. Microbiol Spectr. (2024) 12:e02796–23. doi: 10.1128/spectrum.02796-23
11. Hui WW, Emerson LE, Clapp B, Sheppe AE, Sharma J, Del Castillo J, et al. Antigen-encapsulating host extracellular vesicles derived from Salmonella-infected cells stimulate pathogen-specific Th1-type responses in vivo. PLoS Pathog. (2021) 17:e1009465. doi: 10.1371/journal.ppat.1009465
12. Emerson LE, Barker H, Tran T, Barker S, Enslow S, Ou M, et al. Extracellular vesicles elicit protective immune responses against Salmonella infection. J Extracell Vesicles. (2022) 11:e12267. doi: 10.1002/jev2.12267
13. Kwong KWY, Xin Y, Lai NCY, Sung JCC, Wu KC, Hamied YK, et al. Oral vaccines: A better future of immunization. Vaccines. (2023) 11:1232. doi: 10.3390/vaccines11071232
14. Coffey JW, Gaiha GD, and Traverso G. Oral biologic delivery: advances toward oral subunit, DNA, and mRNA vaccines and the potential for mass vaccination during pandemics. Annu Rev Pharmacol Toxicol. (2021) 61:517–40. doi: 10.1146/annurev-pharmtox-030320-092348
15. Research C for BE and. Vivotif. FDA (2025). Available online at: https://www.fda.gov/vaccines-blood-biologics/vaccines/vivotif (Accessed April 30, 2025).
16. Dolatyabi S, Renu S, Schrock J, and Renukaradhya GJ. Chitosan-nanoparticle-based oral Salmonella enteritidis subunit vaccine elicits cross-protection against Salmonella typhimurium in broilers. Poult Sci. (2024) 103:103569. doi: 10.1016/j.psj.2024.103569
17. Timár CI, Lőrincz ÁM, Csépányi-Kömi R, Vályi-Nagy A, Nagy G, Buzás EI, et al. Antibacterial effect of microvesicles released from human neutrophilic granulocytes. Blood. (2013) 121:510–8. doi: 10.1182/blood-2012-05-431114
18. Kuang H, Dou G, Cheng L, Wang X, Xu H, Liu X, et al. Humoral regulation of iron metabolism by extracellular vesicles drives antibacterial response. Nat Metab. (2023) 5:111–28. doi: 10.1038/s42255-022-00723-5
19. White JR, Dauros-Singorenko P, Hong J, Vanholsbeeck F, Phillips A, and Swift S. The complex, bidirectional role of extracellular vesicles in infection. Biochem Soc Trans. (2021) 49:881–91. doi: 10.1042/BST20200788
20. Cheng Y and Schorey JS. Exosomes carrying mycobacterial antigens can protect mice against M ycobacterium tuberculosis infection. Eur J Immunol. (2013) 43:3279–90. doi: 10.1002/eji.201343727
21. Emerson LE, Bhimani S, Rainey AL, Maurelli AT, and Ferraro MJ. Evaluating small extracellular vesicle-based vaccination across heterologous Salmonella strains isolated from wastewater. Brodsky IE editor. Infect Immun. (2025) 93:e00485–24. doi: 10.1128/iai.00485-24
22. Jiang X, Chu C, Wang Z, Gu J, Hong Y, Li Q, et al. Preclinical evaluation of OMVs as potential vaccine candidates against Salmonella enterica serovar Enteritidis infection. Front Cell Infect Microbiol. (2022). Available online at: https://www.frontiersin.orghttps://www.frontiersin.org/journals/cellular-and-infection-microbiology/articles/10.3389/fcimb.2022.1037607/full (Accessed May 10, 2025).
23. Thapa HB, Müller AM, Camilli A, and Schild S. An intranasal vaccine based on outer membrane vesicles against SARS-CoV-2. Front Microbiol. (2021) 12:752739. doi: 10.3389/fmicb.2021.752739
24. Liu Q, Liu Q, Yi J, Liang K, Hu B, Zhang X, et al. Outer membrane vesicles from flagellin-deficient Salmonella enterica serovar Typhimurium induce cross-reactive immunity and provide cross-protection against heterologous Salmonella challenge. Sci Rep. (2016) 6:34776. doi: 10.1038/srep34776
25. Song M, Cui M, Fang Z, and Liu K. Advanced research on extracellular vesicles based oral drug delivery systems. J Control Release Off J Control Release Soc. (2022) 351:560–72. doi: 10.1016/j.jconrel.2022.09.043
26. Cieślik M, Nazimek K, and Bryniarski K. Extracellular vesicles—Oral therapeutics of the future. Int J Mol Sci. (2022) 23:7554. doi: 10.3390/ijms23147554
27. Huang W, Zhang Q, Li W, Yuan M, Zhou J, Hua L, et al. Development of novel nanoantibiotics using an outer membrane vesicle-based drug efflux mechanism. J Controlled Release. (2020) 317:1–22. doi: 10.1016/j.jconrel.2019.11.017
28. Deng C, Hu Y, Conceição M, Wood MJA, Zhong H, Wang Y, et al. Oral delivery of layer-by-layer coated exosomes for colitis therapy. J Controlled Release. (2023) 354:635–50. doi: 10.1016/j.jconrel.2023.01.017
29. Jepson MA and Clark MA. The role of M cells in infection. Microbes Infect. (2001) 3:1183–90. doi: 10.1016/S1286-4579(01)01478-2
30. Macpherson AJ and Smith K. Mesenteric lymph nodes at the center of immune anatomy. J Exp Med. (2006) 203:497–500. doi: 10.1084/jem.20060227
31. Betz KJ, Maier EA, Amarachintha S, Wu D, Karmele EP, Kinder JM, et al. Enhanced survival following oral and systemic Salmonella enterica serovar Typhimurium infection in polymeric immunoglobulin receptor knockout mice. PLoS One. (2018) 13:e0198434. doi: 10.1371/journal.pone.0198434
32. Van der Ley P and Schijns VE. Outer membrane vesicle-based intranasal vaccines. Curr Opin Immunol. (2023) 84:102376. doi: 10.1016/j.coi.2023.102376
33. Lu M, Xing H, Zhao X, Huang Y, Zheng A, and Liang XJ. Engineered extracellular vesicles as a next-generation vaccine platform. Matter. (2024) 7:4180–205. doi: 10.1016/j.matt.2024.09.012
Keywords: exosomes, extracellular vesicle (EV), Salmonella Typhimurium, macrophage, oral vaccination campaigns, oral vaccination
Citation: Bhimani S, Canas JJ, Enslow SM, Mulcare R and Ferraro MJ (2025) Orally administered extracellular vesicles from Salmonella-infected macrophages confer protective immunity in vivo. Front. Immunol. 16:1628756. doi: 10.3389/fimmu.2025.1628756
Received: 14 May 2025; Accepted: 25 July 2025;
Published: 15 August 2025.
Edited by:
Farha Naz, University of Virginia, United StatesReviewed by:
Peter Van Der Ley, Intravacc, NetherlandsLuis Alberto Sanchez Vargas, University of the South Sierra, Mexico
Copyright © 2025 Bhimani, Canas, Enslow, Mulcare and Ferraro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mariola J. Ferraro, bWpmZXJyYXJvQHVmbC5lZHU=
 Saloni Bhimani
Saloni Bhimani Jorge J. Canas
Jorge J. Canas Samantha M. Enslow
Samantha M. Enslow Ryan Mulcare1
Ryan Mulcare1 Mariola J. Ferraro
Mariola J. Ferraro