- Key Laboratory of Artificial Organs and Computational Medicine in Zhejiang Province, Shulan International Medical College, Zhejiang Shuren University, Hangzhou, China
Malaria and schistosomiasis represent two of the most significant global parasitic diseases in terms of public health burden. These diseases are transmitted through Anopheles mosquitoes and freshwater snails, respectively. Although their transmission mechanisms differ, both pathogens critically interact with thioester-containing proteins (TEPs) during immune evasion and clearance within their invertebrate hosts. This review compares the activation mechanisms and functional divergences of TEPs in Anopheles gambiae and Biomphalaria glabrata in the context of host anti-infective immunity. We focus on the roles of AgTEP1 and BgTEP1 in pathogen opsonization and elimination, discussing their interaction networks with co-factors such as LRIM1/APL1C, BgFREPs and Biomphalysin. Furthermore, we analyze differences in immune pathways mediated by TEPs, including reactive oxygen species (ROS) generation, phagocytic elimination, and melanization responses, as well as their regulatory mechanisms governed by host genetic backgrounds and environmental factors. The review also evaluates the evolutionary roles of TEPs in host-parasite coevolution and highlights their potential application in vector intervention and disease prevention strategies. By elucidating both conserved and species-specific characteristics of the TEP system in these evolutionarily distant invertebrates, this work provides critical insights into the evolutionary trajectories of invertebrate innate immunity and advances theoretical frameworks for novel vector control approaches.
Introduction
Parasitic diseases remain one of the most critical public health challenges in developing countries, with malaria and schistosomiasis ranking among the top in terms of incidence rates and mortality, posing severe threats to human health and economic development (1–4).
Anopheles mosquitoes, particularly the An. gambiae complex, serve as primary malaria vectors in Africa due to their marked anthropophily, high transmission efficiency, and growing insecticide resistance (5, 6). Globally, malaria caused ~247 million cases and 619,000 deaths in 2021, with >95% occurring in Africa (7). While artemisinin-based combination therapies (ACTs) remain first-line treatment, emerging evidence of parasite resistance underscores the urgent need for novel interventions (8).
Schistosomiasis is another global parasitic disease caused by trematode worms of the genus Schistosoma, affecting over 250 million people and predominantly endemic in tropical and subtropical regions (4, 9). Among these, Schistosoma mansoni stands as one of the primary etiological agents, completing its life cycle through freshwater snails (e.g., Biomphalaria spp.) as intermediate hosts (7, 10). Despite praziquantel (PZQ) being the drug of choice, its inability to prevent reinfection and reports of reduced efficacy highlight critical limitations for its use (11, 12).Consequently, targeting and reducing the infection rates of intermediate host snails has become a critical strategy for interrupting schistosomiasis transmission.
Despite the disparities in parasite taxonomy and transmission mechanisms between these two diseases, their life cycles fundamentally depend on specific invertebrate hosts. These hosts not only provide essential developmental niches for the parasites but also influence their survival and transmissibility through sophisticated immune mechanisms.
Thioester-containing proteins (TEPs) – immune effectors homologous to and structurally similar to vertebrate complement components – have emerged as a research focus in invertebrate immunology due to their central roles in pathogen recognition, opsonization, and clearance (13, 14).In An. gambiae, AgTEP1 represents the most extensively studied TEP member. Characterized by a conserved thioester motif (GCGEQ), it covalently binds pathogen surfaces and synergizes with LRIM1 and APL1C to stabilize its conformation, enabling specific recognition of Plasmodium ookinetes. This molecular complex orchestrates phagocytic elimination or melanization responses against invading parasites (15, 16). Similarly, in B. glabrata, the TEP ortholog BgTEP1 demonstrates analogous functions during S. mansoni infections. Beyond pathogen surface binding, it cooperates with fibrinogen-related proteins (FREPs) and Biomphalysin to induce oxidative stress responses and mediate sporocyst damage in S. mansoni (10).
Although these two systems demonstrate functional convergence, they exhibit significant divergence in activation patterns, cofactor requirements, cellular origins, and regulatory mechanisms. For instance, AgTEP1 is primarily activated in the hemolymph and stabilized through interactions with specific protein complexes, whereas BgTEP1 is predominantly synthesized in haemocytes, with its expression levels showing marked dependence on genotypic variations and environmental factors (10, 17).
However, a comparative review of TEP-mediated immune mechanisms in these two critical vector species is lacking. Such a comparison would not only advance our understanding of the evolutionary diversity and functional convergence of TEPs but also provide theoretical foundations for developing novel antiparasitic intervention strategies. This review aims to provide a focused and comparative analysis of the immune effector proteins AgTEP1 and BgTEP1 in An. gambiae and B. glabrata, respectively. By dissecting their activation mechanisms and co-factor interactions, we illustrate how these molecules orchestrate species-specific responses to parasitic infections. Through cross-species comparison, we reveal both conserved and divergent strategies employed by these vectors in recognizing and eliminating parasites, emphasizing the evolutionary and ecological implications of TEP-mediated immunity. Ultimately, this work aims to advance our understanding of invertebrate immune evolution and inform future research into immune modulation strategies for parasite control. Through cross-species comparative studies of TEP systems, we seek to deepen insights into the evolutionary mechanisms of invertebrate innate immunity while identifying novel targets for vector control and parasitic disease management. These efforts hold heightened significance given the escalating challenges of drug resistance and the current limitations in vaccine accessibility (11).
Malaria and its Anopheles mosquito vector
Malaria is primarily caused by five Plasmodium species infecting humans: P. falciparum, P. vivax, P. malariae, P. ovale, and P. knowlesi (18, 19). These protozoan parasites, classified under the phylum Apicomplexa, exhibit complex life cycles involving two distinct host types: an invertebrate definitive host (where sexual reproduction occurs) and a vertebrate intermediate host. Among them, P. falciparum and P. vivax are the most prevalent and lethal in humans (20, 21). Notably, P. falciparum is responsible for the majority of global malaria related fatalities, with infections characterized by high fever, chills, headache, anemia, hepatosplenomegaly, and severe complications such as renal failure, cerebral malaria, and death.
As illustrated in Figure 1, the P. falciparum life cycle involves two hosts: humans as intermediate hosts and female Anopheles mosquitoes as definitive hosts. During a mosquito bite, sporozoites are injected into the human bloodstream, subsequently migrating to hepatocytes where they undergo hepatic schizogony, producing numerous merozoites. Upon release into the bloodstream, these merozoites invade erythrocytes, initiating the intraerythrocytic cycle marked by sequential developmental stages—ring stages, trophozoites, and schizonts—during which hemoglobin metabolism and asexual replication occur (22, 23). A subset of merozoites differentiates into gametocytes, which, upon ingestion by mosquitoes, undergo sexual reproduction in the mosquito midgut to form zygotes, motile ookinetes, and oocysts. Sporozoites released from mature oocysts migrate to the salivary glands, completing the transmission cycle (24).
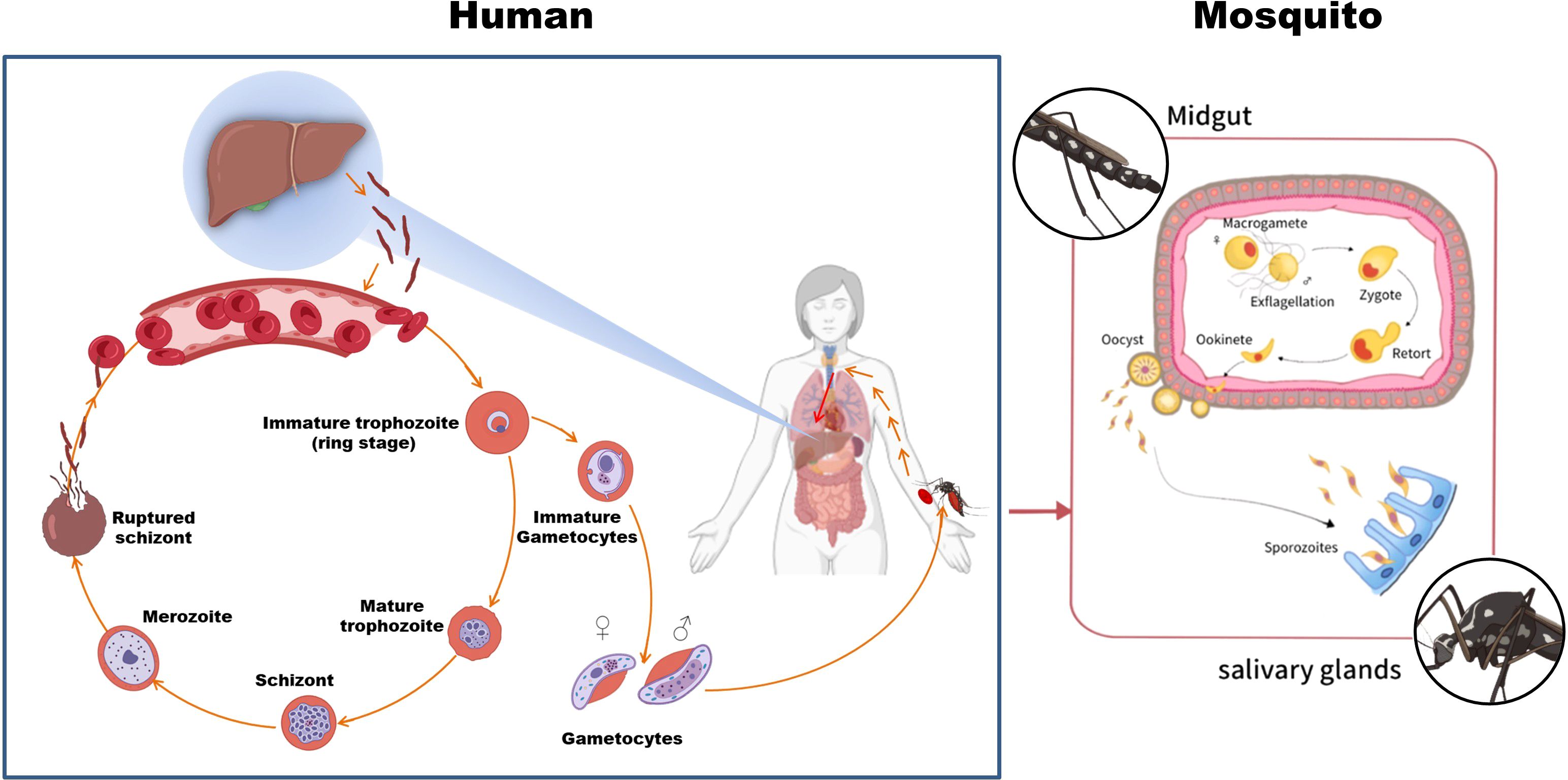
Figure 1. The life cycle of P. falciparum. The parasite alternates between human and Anopheles mosquito hosts. Sporozoites are transmitted during a mosquito bite, develop in the liver, and invade red blood cells; gametocytes taken up by mosquitoes complete the cycle. Parts of the materials used in this figure originate from BioRender and have been further modified and enhanced.
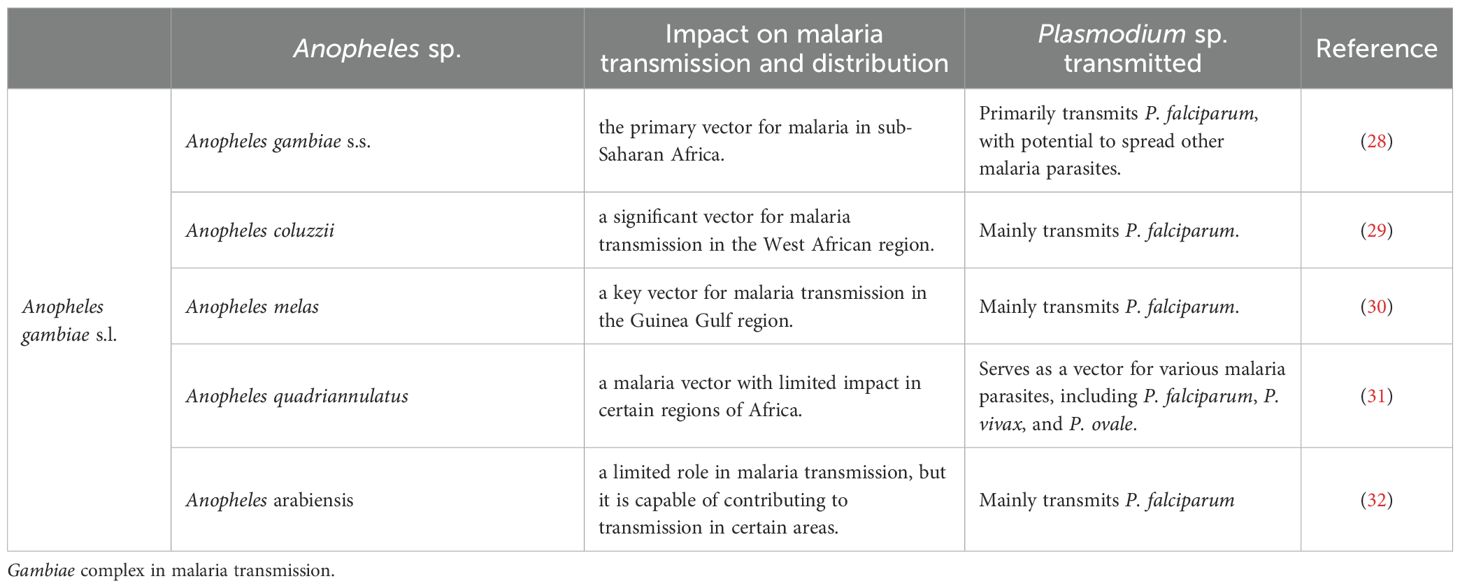
An. gambiae has emerged as a pivotal model organism for investigating Plasmodium-mosquito interaction mechanisms. Its genome has been fully sequenced and annotated, providing a robust foundation for elucidating the interplay between mosquito immune systems and Plasmodium parasites (25, 26). Notably, An. gambiae s.l. (sensu lato) comprises a complex of morphologically indistinguishable yet genetically and ecologically divergent sibling species (27). AgTEP1 is ubiquitously distributed across this species complex, though its expression levels and spatial-temporal distribution vary significantly among constituent species and geographical populations. Current research on AgTEP1 predominantly focuses on the nominal species An. gambiae s.s. (sensu stricto), and the AgTEP1 discussed in this review is derived exclusively from studies on this model taxon, without interspecific distinctions. Table 1 summarizes the geographical distribution and ecological roles of An. gambiae s.l. subspecies in malaria parasite transmission (33).
The developmental stages of Plasmodium within the mosquito vector are critical to its life cycle, making this phase a prime target for strategies aimed at interrupting malaria transmission. To achieve this goal, a comprehensive understanding of the Anopheles innate immune system is paramount. When a mosquito ingests blood containing gametocytes, the parasites must traverse the midgut epithelium, enter the hemocoel, and complete gamete fusion and oocyst formation. Throughout this process, the mosquito orchestrates multifaceted immune responses, including phagocytosis, melanization cascades, and the expression of antimicrobial peptides (AMPs) (10, 34, 35).
Among these immune responses, AgTEP1 functions as a pivotal immune effector by recognizing and binding to Plasmodium surfaces via its conserved thioester bond, thereby triggering immune clearance. Functionally resembling vertebrate complement proteins, AgTEP1 mediates pathogen lysis or melanization encapsulation and represents the most extensively characterized TEP to date (18, 20, 21, 36).
TEP proteins in An. gambiae
The TEP family comprises evolutionarily conserved immune molecules widely distributed across invertebrates and vertebrates, including mammals (37). Most TEPs harbor a canonical thioester bond, though this structural motif is absent in certain homologs, such as complement component C5 or specific insect TEPs (13). In vertebrates, TEPs predominantly manifest as components of the complement system (e.g., C3, C4) and serine protease inhibitors like α2-macroglobulin, with their primary function centered on pathogen recognition and elimination (38, 39). In contrast, insect TEPs (often termed iTEPs) have evolved structural and functional diversity through long-term evolutionary processes, exhibiting functional roles analogous to vertebrate α2-macroglobulin (15).
TEPs are recognized as critical members of the pattern recognition receptor (PRR) family and constitute essential components of the invertebrate innate immune system (40). Cross-species analyses classify TEPs into three major categories: iTEP/CD109-like, C3-like, and A2M-like. In insects, iTEP/CD109-like proteins are typically secreted opsonins and immune modulators that bind pathogen surfaces to promote phagocytosis or melanization; in vertebrates, the homologous CD109 is predominantly a GPI-anchored cell-surface glycoprotein involved in modulation of cell signaling (e.g., TGF-β). This reflects domain-level conservation but functional diversification — with noted exceptions (some invertebrate CD109-like proteins are membrane-associated or processed into soluble forms) (17, 41). C3-like TEPs undergo proteolytic activation into two fragments upon stimulation: the smaller fragment mediates chemotactic and inflammatory signaling, while the larger fragment retains the thioester motif, enabling covalent binding to pathogen surfaces to facilitate clearance (42–44) (Figure 2A). A2M and its homologs typically undergo a conformational change (mediated through a bait-trap mechanism) following proteolytic cleavage within their bait regions, thereby entrapping and inhibiting protease activity derived from pathogens or host sources; subsequently, these complexes are cleared via receptor-mediated endocytosis involving low-density lipoprotein receptor-related protein 1 and related receptors (45) (Figure 2B).
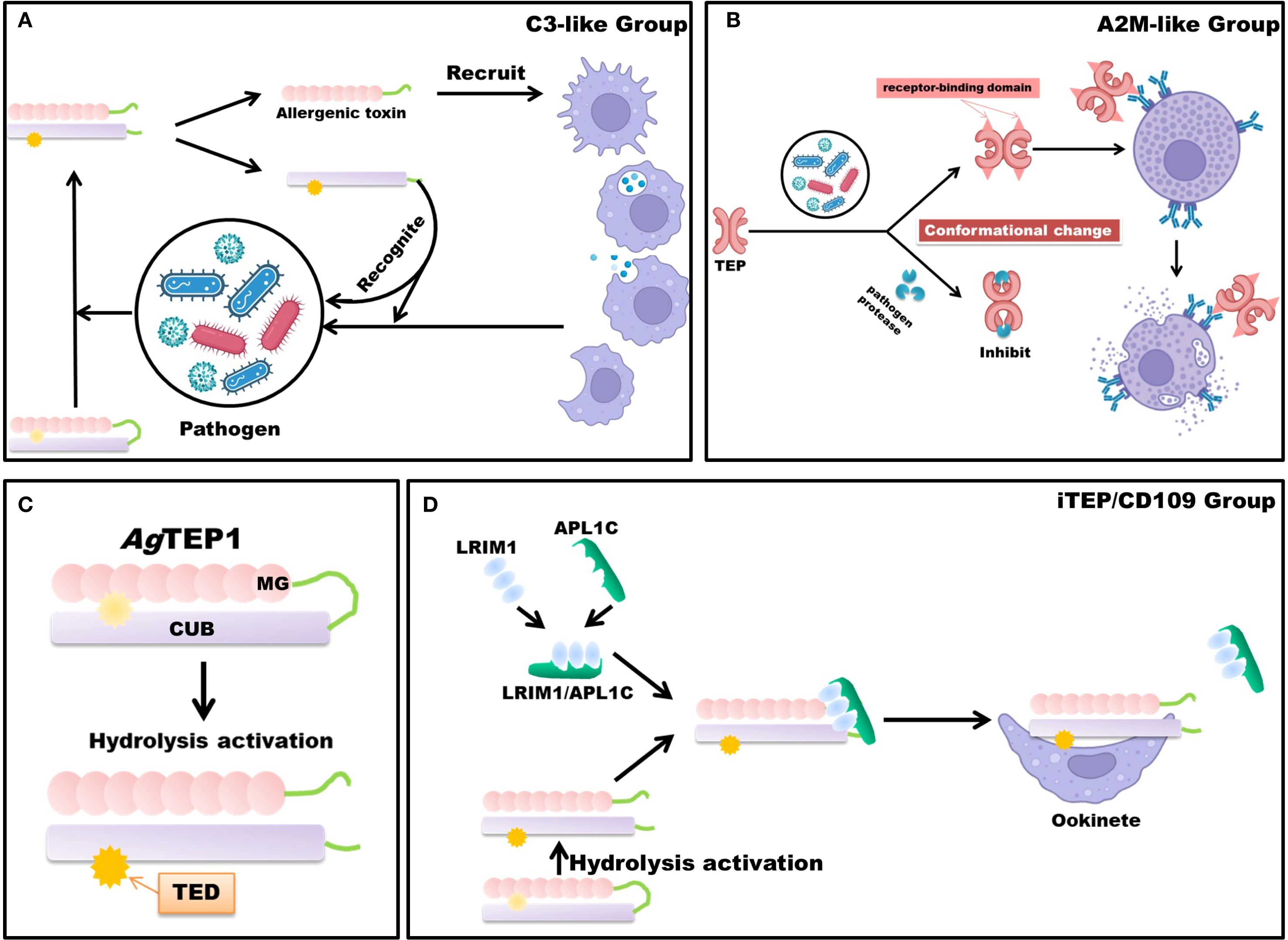
Figure 2. Schematic Diagram of TEP Mechanisms. TEPs are broadly categorized into three principal classes: the iTEP/CD109 group, the complement component C3-like group, and the A2M-like group. (A) The mechanism of the complement component C3-like group occurs following immunological stimulation. Upon activation, TEPs undergo proteolytic cleavage, releasing a small allergenic toxin fragment. This fragment acts as an immunostimulant or chemotactic agent, recruiting macrophages to the site of infection. Concurrently, the larger fragment, through covalent bonding via the thioester bond, targets and marks the pathogen, thereby facilitating its degradation or phagocytosis. (B) Protease inhibition and immune activation mechanisms of A2M-like TEP. Following engagement with pathogens, A2M-like proteins eschew proteolytic cleavage, undergoing instead a conformational transformation that effectively attenuates the proteolytic activity of the pathogen. In tandem, this conformational rearrangement reveals the receptor-binding domain of A2M, which enhances its interaction with phagocyte surface receptors, thus augmenting endocytosis and expediting pathogen clearance. (C) Simplified structure and hydrolytic activation of AgTEP1. (D) Members of the iTEP/CD109 group, such as AgTEP1, rely on stabilization mechanisms to prevent premature inactivation after hydrolysis. The LRIM1/APL1C heterodimer binds to hydrolyzed AgTEP1, forming a stable complex that prevents aggregation and ensures functional integrity. This stabilized complex facilitates subsequent immune recognition and binding to invading parasites such as Plasmodium ookinetes. Parts of the materials used in the images within this article originate from BioRender, which we have further modified and enhanced.
In An. gambiae, the genome encodes at least nineteen AgTEPs (AgTEP1 – 19) that form three broad clades of complement-like factors, with AgTEP1 standing out as the best-studied member to date (45, 46). AgTEP1 is translated as a 165 kDa, N-glycosylated precursor whose architecture follows the canonical C3-like scaffold: eight macroglobulin (MG) domains (MG1-MG8) followed by a CUB domain and a thioester domain (TED) carrying the reactive GCGEQ motif. Crystallographic and cryo-EM comparisons reveal a root-mean-square deviation of ≈3 Å between the AgTEP1 core and mammalian C3, confirming close tertiary structural homology (42, 47).
After secretion into the hemolymph, an as-yet-unidentified CLIP-family serine protease cleaves AgTEP1 within the flexible MG6-LNK hinge, generating the disulfide-linked α- chain (75 kDa) and β-chain (85 kDa) that constitute the mature, reactive form AgTEP1-cut. Proteolytic activation unlocks the thioester, allowing covalent attachment to primary hydroxyl or amino groups on microbial surfaces and thereby labelling invaders for downstream immune attack (48, 49) (Figure 2C).
Free AgTEP1-cut is intrinsically unstable and tends to precipitate. Two secreted leucine-rich repeat proteins, LRIM1 and APL1C, assemble via a C-terminal coiled-coil into a disulfide-bonded heterodimer that docks one molecule of AgTEP1-cut to form a stable ternary complex. This interaction preserves thioester reactivity, channels AgTEP1 to Plasmodium ookinetes or bacteria, and prevents wasteful self-attack. Loss-of-function or RNAi of either LRIM1 or APL1C abolishes TEP1 loading on pathogens and converts refractory mosquitoes into susceptible ones, underscoring the complex as the core of the mosquito complement-like pathway (14, 22–24) (Figure 2D).
Recent work further shows that the LRIM1/APL1C carrier can also load other AgTEPs (e.g., AgTEP3) and that non-catalytic cofactors such as SPCLIP1 (a catalytically inactive serine protease-like protein) orchestrate the localized accumulation of AgTEP1 on microbial targets, highlighting a vertebrate-style convertase cascade now being unravelled in insects (44).
Additionally, AgTEP1 exhibits broad-spectrum immune activity by recognizing bacterial pathogens, underscoring its versatility in pathogen surveillance (50).
The immunological efficacy of AgTEP1 displays population specificity, with binding capacity modulated by both host and parasite polymorphisms—a hallmark of host-parasite coevolution (51, 52). Mechanistic investigations reveal that AgTEP1 binding to ookinete surfaces occurs via a multi-phase process: initial rapid association of limited cleaved AgTEP1, followed by SPCLIP1 -facilitated recruitment of uncleaved AgTEP1 for surface deposition, culminating in proteolytic activation by the AgTEP1 enzymatic complex (49).
AgTEP1-triggered immune responses exhibit pathogen size-dependent specialization: phagocytosis for small pathogens (e.g., bacteria) versus melanotic encapsulation for larger invaders (e.g., Plasmodium) (36, 53). While AgTEP1 binding is essential for pathogen clearance, its standalone activity proves insufficient, necessitating synergistic interactions with soluble immune co-factors. For instance, studies indicate that even AgTEP1-opsonized ookinetes may evade immune elimination if key co-factors are absent or immunosuppressive molecules like Cap380 are present (49, 54).
Notably, beyond AgTEPs, Anopheles mosquitoes possess diverse immune factors including lectins, clip-domain serine proteases (CLIPs), and serine protease inhibitors (serpins) (55, 56). Among these, FREPs—conserved across multiple invertebrates—collaborate closely with TEPs in mollusks like Biomphalaria to form pathogen-recognition complexes (e.g., BgFREPs with BgTEPs) (57–59). Whether analogous complexes exist in mosquitoes remains an open question requiring further investigation.
While AgTEP1 has been extensively characterized, the biological functions of other AgTEP members (AgTEP2–19) remain largely unexplored. We have compiled the structural features and immunological functions of AgTEP members reported in the current literature into Table 2. Future research must systematically elucidate their expression regulation networks, biological roles, and interactions with immune pathways to fully unravel the complexity of Anopheles immunity and identify potential intervention targets.
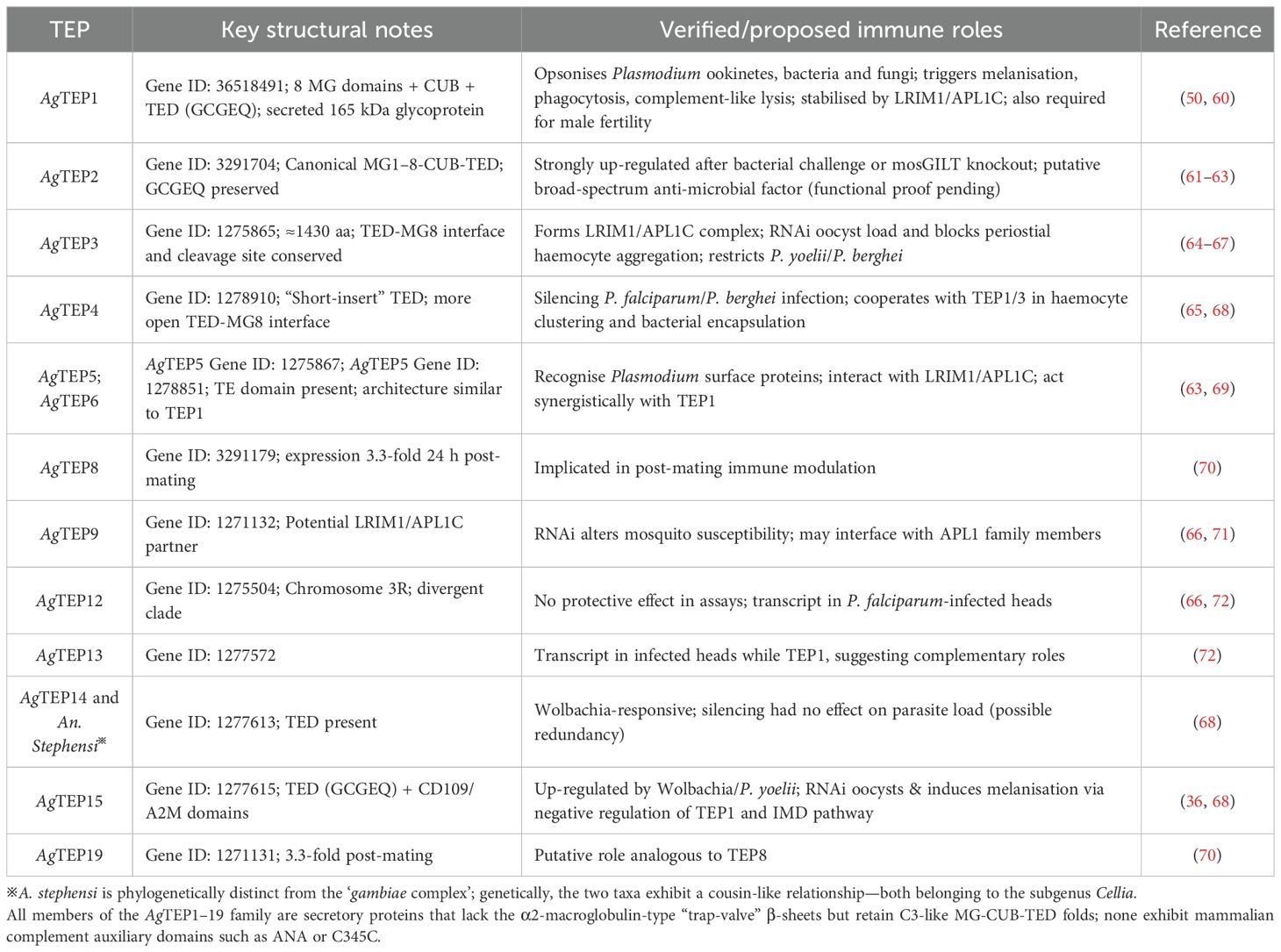
Table 2. Structural and functional diversity of the An. gambiae TEP gene family: immune roles in pathogen defense, complement activation, and reproductive modulation.
Schistosomiasis and the snail intermediate host
Schistosomes have a life cycle involving a snail host, and a definitive vertebrate host, which can be a mammal or bird depending on the species (73). They primarily utilize aquatic or amphibious freshwater snails as intermediate hosts to complete the development of larval stages through asexual reproduction, and then undergo sexual reproduction within the definitive host. Here, we provide a brief description of the schistosome life cycle (Figure 3). Adult schistosomes, parasitic in many mammals including humans, produce eggs through sexual reproduction (74). Depending on the parasite species, these eggs penetrate the intestinal wall or bladder and are excreted in feces or urine (75). Once outside the host, the eggs hatch under suitable conditions of temperature, light, and osmolarity, giving rise to miracidia (76, 77). The miracidia, equipped with cilia on their surface, can freely swim. When they encounter the appropriate intermediate host snail (such as Oncomelania spp. for S. japonicum, Biomphalaria spp. for S. mansoni, and Bulinus spp. for S. haematobium), they penetrate the snail’s skin and initiate their development within the snail host (73). If the snail is susceptible to the parasite, they undergo development into mother sporocysts, which then produce daughter sporocysts, ultimately leading to the formation of cercariae that are released into the water by penetrating the snail’s tissue (78). When humans or other mammals come into contact with water containing cercariae, they may become infected. The cercariae penetrate the skin and enter subcutaneous veins, where they transform into schistosomula (79). They are then carried by the bloodstream to the right heart chamber, transported to the lungs, and subsequently, through the blood circulation, return to the left heart chamber, entering the arterial circulation (80). Finally, they settle in the mesenteric veins (for S. japonicum and S. mansoni) or the pelvic venous plexus (for S. haematobium), where they mature into adult worms capable of sexual mating and egg production (80).
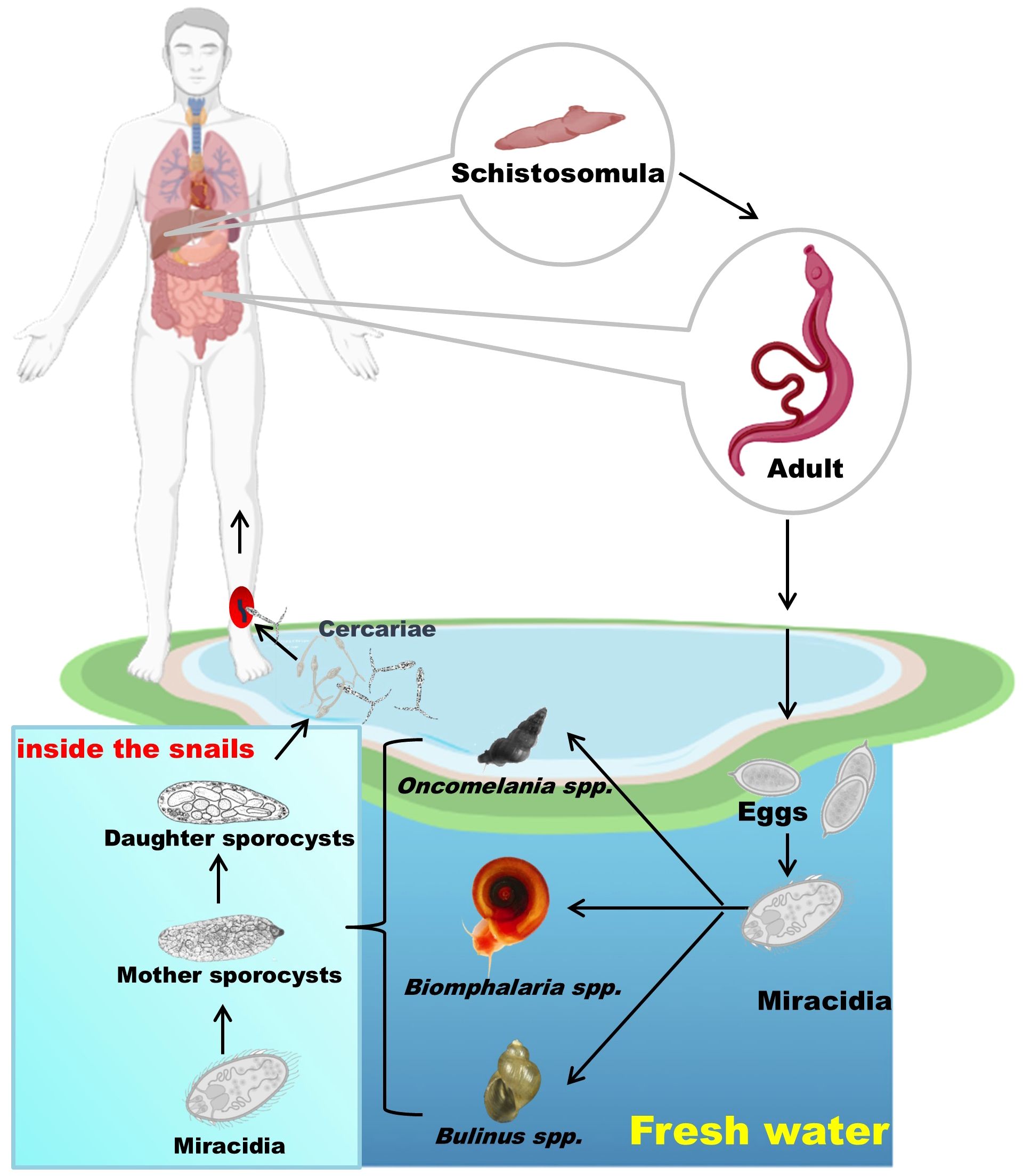
Figure 3. The schistosome life cycle with humans as the definitive host. Eggs released by adult worms hatch into miracidia, which infect snails and develop into cercariae. These are released into water and infect humans, where the worms mature and reproduce. Parts of the materials used in the images within this article originate from BioRender, which we have further modified and enhanced.
TEP-mediated immune response in B. glabrata snail
Although there has been extensive research on the biology, pathology, and molecular biology of schistosomes and schistosomiasis, studies on the immunology of the intermediate snail hosts remain relatively limited (81). So far, whole-genome sequencing and annotation have been reported for B. glabrata (a critical intermediate host for S. mansoni) and Bulinus truncatus (an intermediate host for S. haematobium), providing important reference resources for investigating the immune interactions between schistosomiasis and intermediate snail hosts (11, 82). Among them, B. glabrata has emerged as a significant model organism for studying the interactions between pathogen and hosts, and its immune system has been extensively studied for decades, yielding fruitful research outcomes (83–87). Most invertebrates have a fluid called “hemolymph” in their body cavities, and the diversity of soluble hemolymph proteins is closely associated with the host’s anti-schistosome capabilities (10). This includes various immune molecules involved in anti-schistosome responses, such as BgFREPs (10, 88, 89), lectins (88, 90), BgTEP (17, 91), Biomphalysin (92), Toll-like receptors (BgTLR) (93), granulins (BgGRN) (85), and macrophage migration inhibitory factor (BgMIF) (86). Among them, BgTEP is a key immune component.
BgTEP1 in B. glabrata was initially identified by Bender et al. in 1992, revealing its proteinase inhibition activity (94). More recently, BgTEP1 was identified in the study of immunoprecipitates of surface molecules between BgFREP and S. mansoni sporocysts (91). Subsequent research has found that BgTEP1 plays an essential role in the recognition and response to epitopes of S. mansoni, making it an indispensable immune molecule in the context of anti-parasite infection (17, 40). At present, genomics and proteomics have identified 11 BgTEP proteins (95). Based on the classification similar to the aforementioned TEP superfamily, these 11 BgTEPs can be divided into four branches as follows: (1) complement-like factors (BgC3-1, BgC3-2, and BgC3-3), (2) α-2-macroglobulin (BgA2M), (3) macroglobulin complement-related proteins (BgMCR1 and BgMCR2), and (4) iTEP/CD109 molecules (BgTEP1, BgTEP2, BgTEP3, BgTEP4, and BgCD109) (95). The structural features and immunological functions of BgTEPs in B. glabrata are summarized in Table 3.
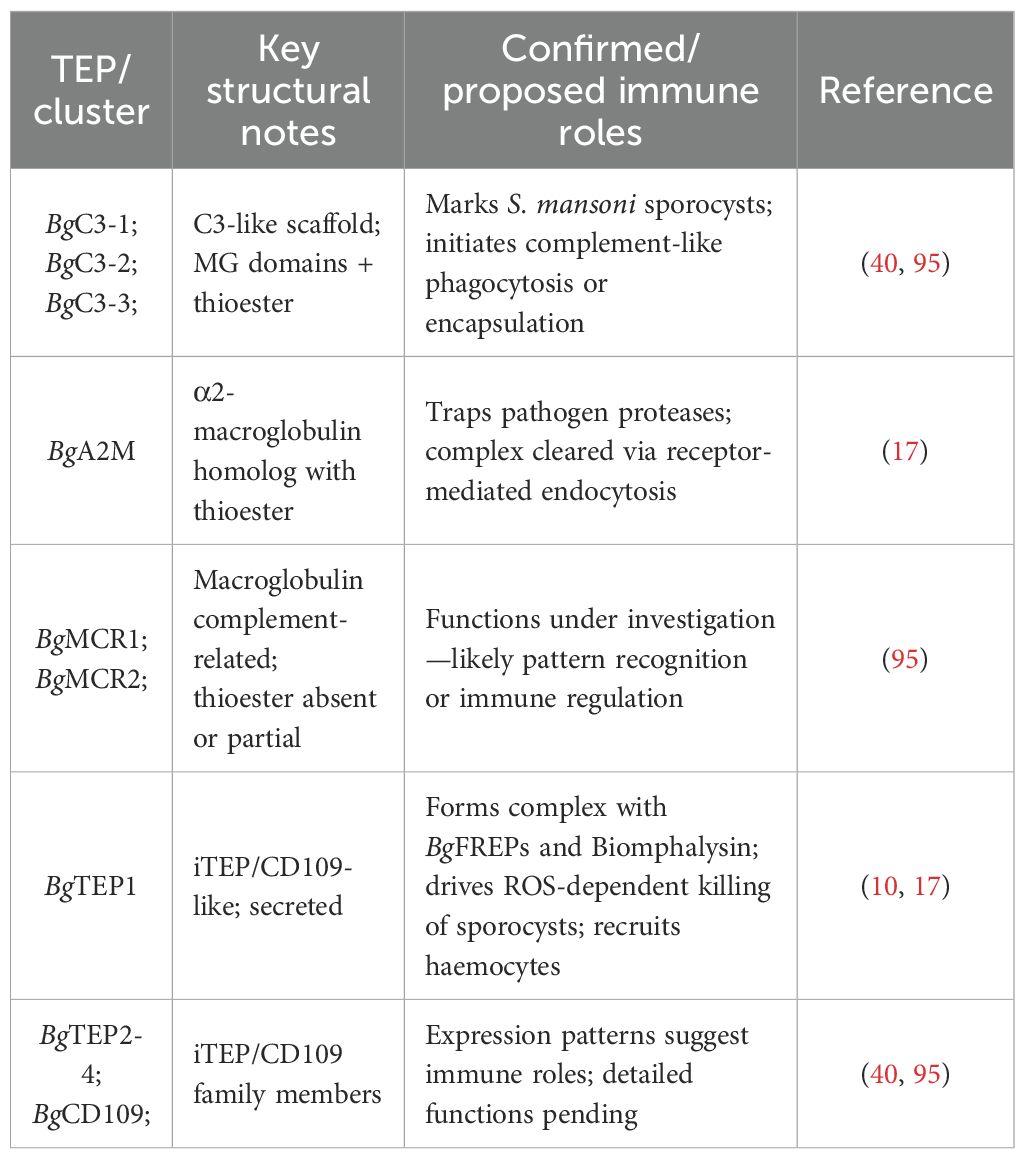
Table 3. Functional and evolutionary diversity of the B. glabrata TEP family: Complement-like pathways, schistosome defense, and effector complex formation.
The cleavage of BgTEP1 is not a prerequisite for pathogen binding. A series of studies have shown that BgTEP1 can bind to the surfaces of different microorganisms and parasites in either full-length or processed forms (17). The binding of BgTEP1 to different developmental stages of S. mansoni varies. In the early stage, when miracidia hatch from eggs, BgTEP1 binds in its full-length form, although weakly. The cleaved form also binds to miracidia, but only within the first 3 hours. BgTEP1 also binds to primary sporocysts, predominantly in its full-length form, though cleaved forms are more abundant on sporocysts than on miracidia (17). After binding to invading S. mansoni sporocysts, BgTEP1 promotes the recruitment of other subtypes of haemocytes, enabling them to carry out further phagocytosis or encapsulation reactions.
A 2010 study by Mone et al. identified BgTEP1, BgFREP2, and Schistosoma mansoni polymorphic mucins (SmPoMucs) in the precipitate after mixing B. glabrata plasma with S. mansoni (91). BgFREPs are a class of soluble lectins synthesized and secreted by snail haemocytes. They partially determine the snail’s resistance phenotype against S. mansoni (84, 96), primarily by mediating immune recognition of the invading miracidia and sporocyst stages, subsequent clearance responses, and play a crucial role in the immune system of snails (34, 86, 97). Since 1979, it has been known that B. glabrata possesses “immune memory” or “acquired resistance” (98), with BgFREPs being linked to this phenomenon (84). The diversity of BgFREPs is thought to result from adaptive evolution. According to the polymorphic compatibility hypothesis, pathogens evolve diverse antigens to evade the immune system, prompting the host to develop a broader set of receptors to identify and eliminate these threats (91, 99). This resembles how vertebrate antibodies recognize a variety of antigens. Each B. glabrata snail seems to have a unique BgFREP repertoire, which highlights the importance of BgTEP1 in immune receptor recognition of glycoprotein antigens. Although the role of TEP proteins in mosquitoes and fruit flies has been well studied, it wasn’t until Mone et al.’s research that the function of BgTEP1 in B. glabrata became evident. Similar to vertebrate complement C3, BgTEP1 may play a comparable role in the immune system of B. glabrata, triggering complement-like pathways.
In 2020, it was further discovered that BgFREP3, BgFREP2, and BgTEP1 interact to form a unique immune complex (illustrated in Figure 4). This complex imparts the ability to kill S. mansoni sporocysts to haemocytes derived from susceptible snails, nearly equivalent to the haemocytes of resistant snails (10). This sporocyst killing ability can be abolished by ROS scavengers, indicating the crucial role of ROS as effector molecules (10). Based on this study, the BgFREP–BgTEP immune complex is proposed to bind to the pathogen and interacts with a specific receptor on the surface of snail haemocytes, a signal is transmitted to the interior of the cell, initiating an immune response that boosts the synthesis of cytotoxic substances (ROS) to ultimately eliminate the pathogen (10). Despite these findings, the identity of the receptor remains elusive. We speculate it may be a Toll-like receptor, but further research, including co-immunoprecipitation and CRISPR knockouts, is required to confirm this hypothesis (100–102). The interactions between BgFREPs and BgTEP1 in B. glabrata’s immune response are critical for mediating effective anti-schistosome defenses, but still not fully understood. Both proteins exhibit pathogen-binding and opsonization capabilities (17, 84, 87, 91). Within their BgFREP–BgTEP immune complex, it is challenging to precisely determine which protein binds directly to the pathogen and which one interacts with the proposed receptor. Immunofluorescence studies have revealed that both BgFREP3 and BgTEP1 can independently bind to the external surface of S. mansoni sporocysts, but BgFREP2 requires BgTEP1 to bind effectively to the parasite’s surface (10). This observation prompts questions about the notable disparities in pathogen recognition between BgFREP3 and BgFREP2, both members of the BgFREP family. Notably, BgFREP3 contains two immunoglobulin superfamily (IgSF) domains and forms homomultimers in B. glabrata plasma, whereas BgFREP2 lacks multimerization capabilities (10). Although the exact mechanism behind BgFREP3 multimer formation is still unclear, it is speculated that this occurs through the coiled-coil region of the IgSF domain (103). Nevertheless, this hypothesis lacks experimental evidence, and the possibility of BgFREP multimer formation being mediated by the fibrinogen-like (FBG) or IgSF domain cannot be completely disregarded (103). These structural distinctions may elucidate why BgFREP2 requires BgTEP1 to execute its pathogen recognition function. Further research is essential to unravel the intricate mechanisms governing their cooperative roles in the snail’s immune system and explore the potential implications for pathogen defense.
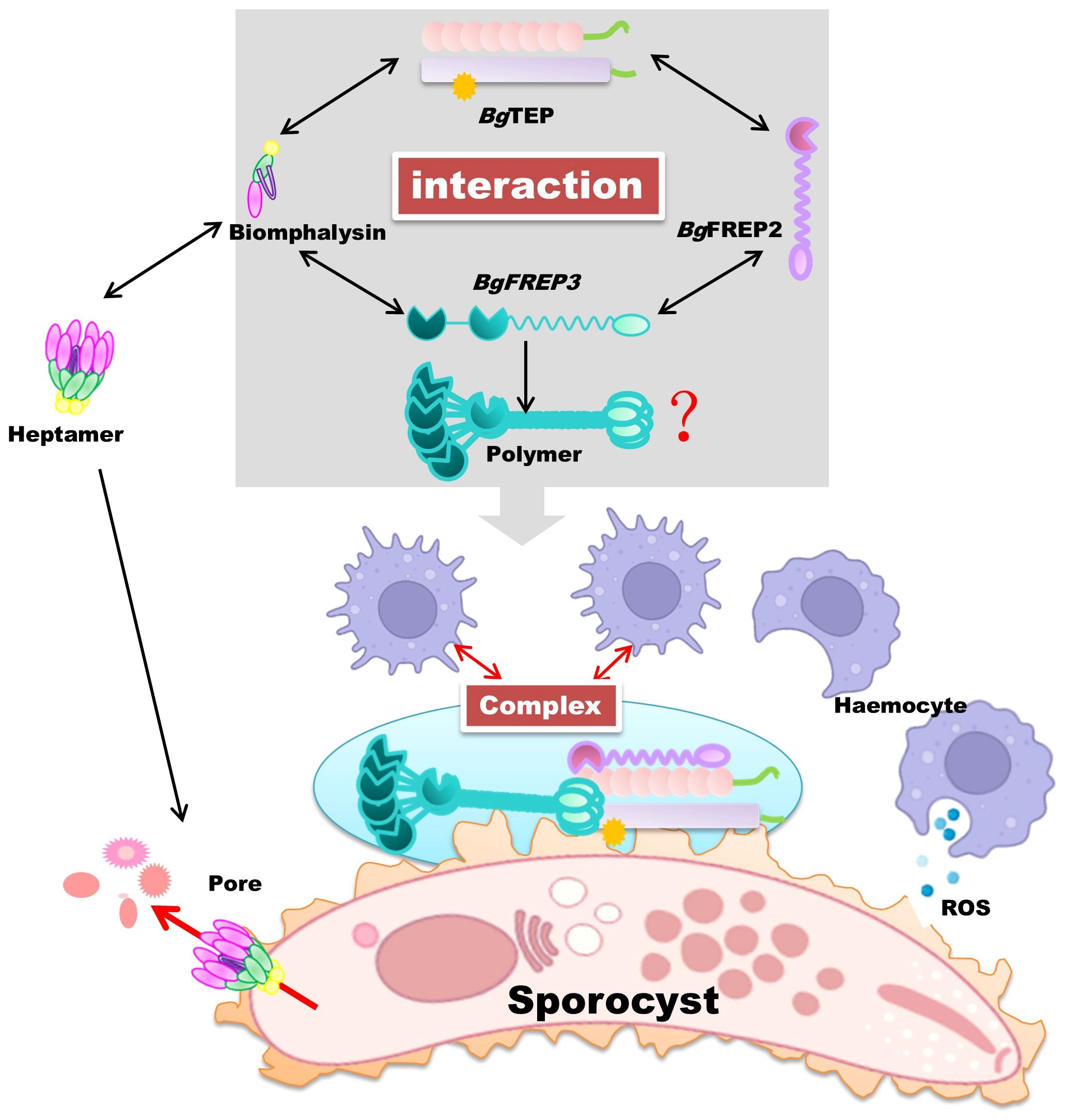
Figure 4. BgTEP as a synergistic immune pathogen eliminator in B. glabrata snails. In B. glabrata snails, BgTEP closely interacts with other essential immune proteins, including BgFREP, BgFREP2, and Biomphalysin, to effectively eliminate pathogens in a collaborative manner. Biomphalysin, after forming a heptameric structure, creates pore channels on the surface of invading pathogens, disrupting osmotic balance and ultimately causing their demise. On the other hand, BgTEP, BgFREP, and BgFREP2 form a complex and transmit immune signals to blood cells through unidentified receptors. This transformation converts blood cells into phagocytic subtypes, boosting the secretion of cytotoxic substances, mainly ROS. Together, these synergistic effects effectively eradicate invading parasitic pathogens. The red question marks indicate that although we know BgFREP3 exists as a homomultimer, we are not certain about the mechanism of its multimer formation. We hypothesize that this process may involve protein-protein interactions, post-transcriptional modifications (such as phosphorylation), or the formation of disulfide bonds. Future studies could explore the role of chaperones, conduct mutagenesis analysis, or even perform structural studies to further uncover the specific mechanism of BgFREP3 oligomer formation. Parts of the materials used in the images within this article originate from BioRender, which we have further modified and enhanced.
In addition to the aforementioned interaction between BgTEP and BgFREP3 and BgFREP2, a previous study identified an immune interaction between BgTEP1 and Biomphalysin in snail hemolymph in a pull-down experiment using BgTEP1 as bait (10) (Figure 4). Biomphalysin, a β-pore-forming toxin (β-PFT), plays a key role in the snail’s immune defense by disrupting the membrane integrity of S. mansoni, resulting in the parasite’s lysis (92). While β-PFTs are typically used by bacteria to invade host cells, Biomphalysin in B. glabrata is a potent anti-parasitic factor, directly contributing to the destruction of S. mansoni (92).
BgTEP1 is similar to the human complement C3 protein, which, in the human complement system, leads to the formation of a membrane attack complex (MAC) that disrupts pathogen membranes (1), causing osmotic imbalance and cell death. Both Biomphalysin and MAC form pore-like structures on cell membranes, resulting in cell lysis. This suggests that the interaction between BgTEP1 and Biomphalysin may serve a similar function in B. glabrata, resembling the role of the complement system in humans. Furthermore, ongoing research has suggested potential parallels between the immune factors identified in B. glabrata snails and the important members of the lectin pathway (Figure 5). Although the parallels are not perfect, it outlines a rough pathway: BgFREPs correspond to pathogen recognition parts, such as ficolin and Mannose-Binding Lectin, BgTEP1 corresponds to complement C3 protein, and Biomphalysin confers to MAC’s action. This suggested model provides valuable clues for a deeper understanding of the evolution and function of the immune system.
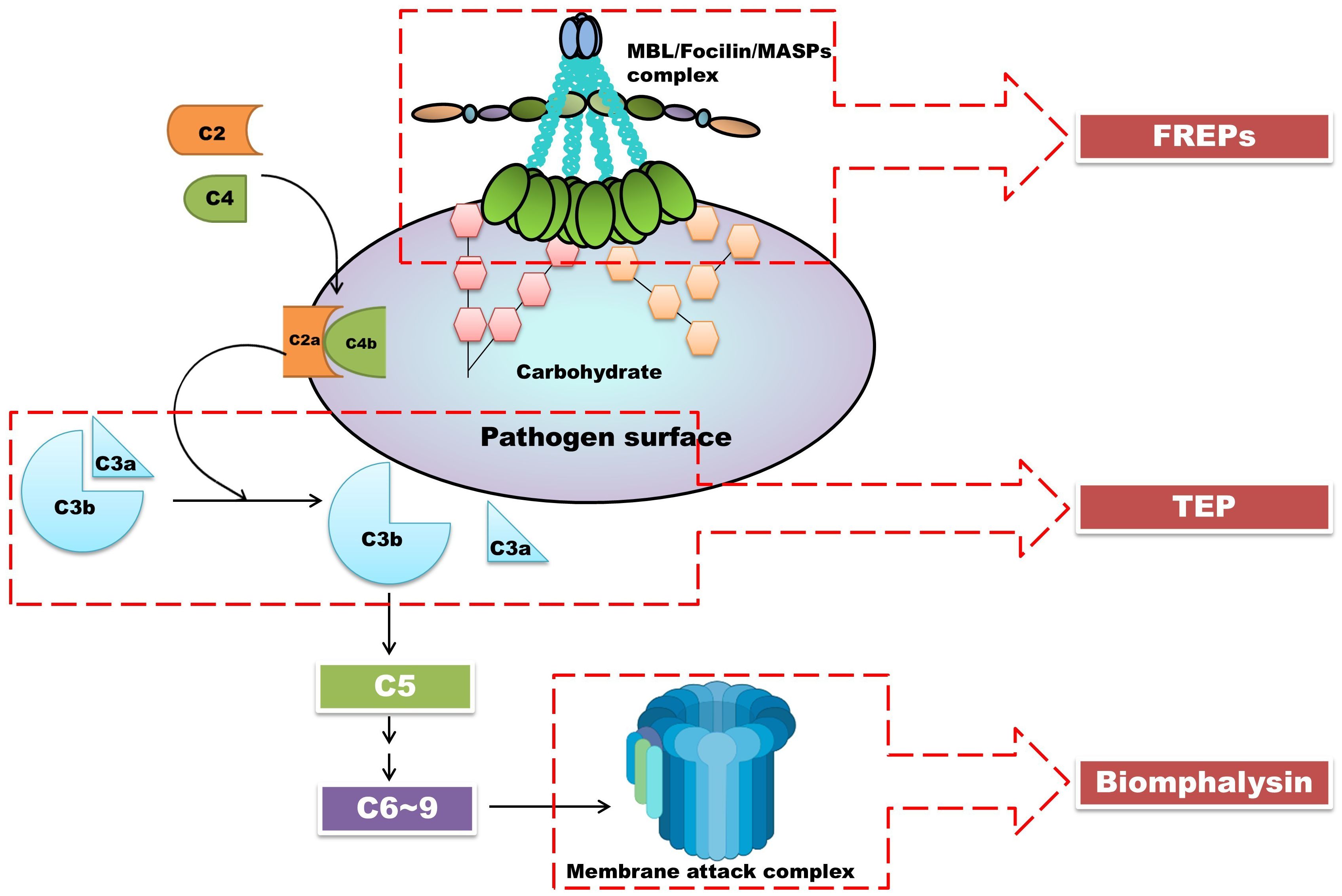
Figure 5. The interactions between BgTEP, lectin-like molecules BgFREPs, and pore-forming toxin Biomphalysin may be the products of the evolutionary process of the lectin pathway. In this figure, we depicted the lectin pathway in vertebrates. Close interactions were found between BgTEP, lectin-like molecules BgFREPs, and pore-forming toxin Biomphalysin in B. glabrata snails. These correspond to important components of the lectin pathway in vertebrate complement systems. Parts of the materials used in the images within this article originate from BioRender, which we have further modified and enhanced.
Comparative analysis of TEPs in Anopheles and Biomphalaria
Despite belonging to evolutionarily distant phyla, An. gambiae (arthropod) and B. glabrata (mollusk), their TEPs exhibit functional convergence in innate immunity. These TEPs universally play central roles in host defense by recognizing, opsonizing, and eliminating invading pathogens, albeit through distinct operational contexts and associated mechanistic frameworks summarized in Table 4.
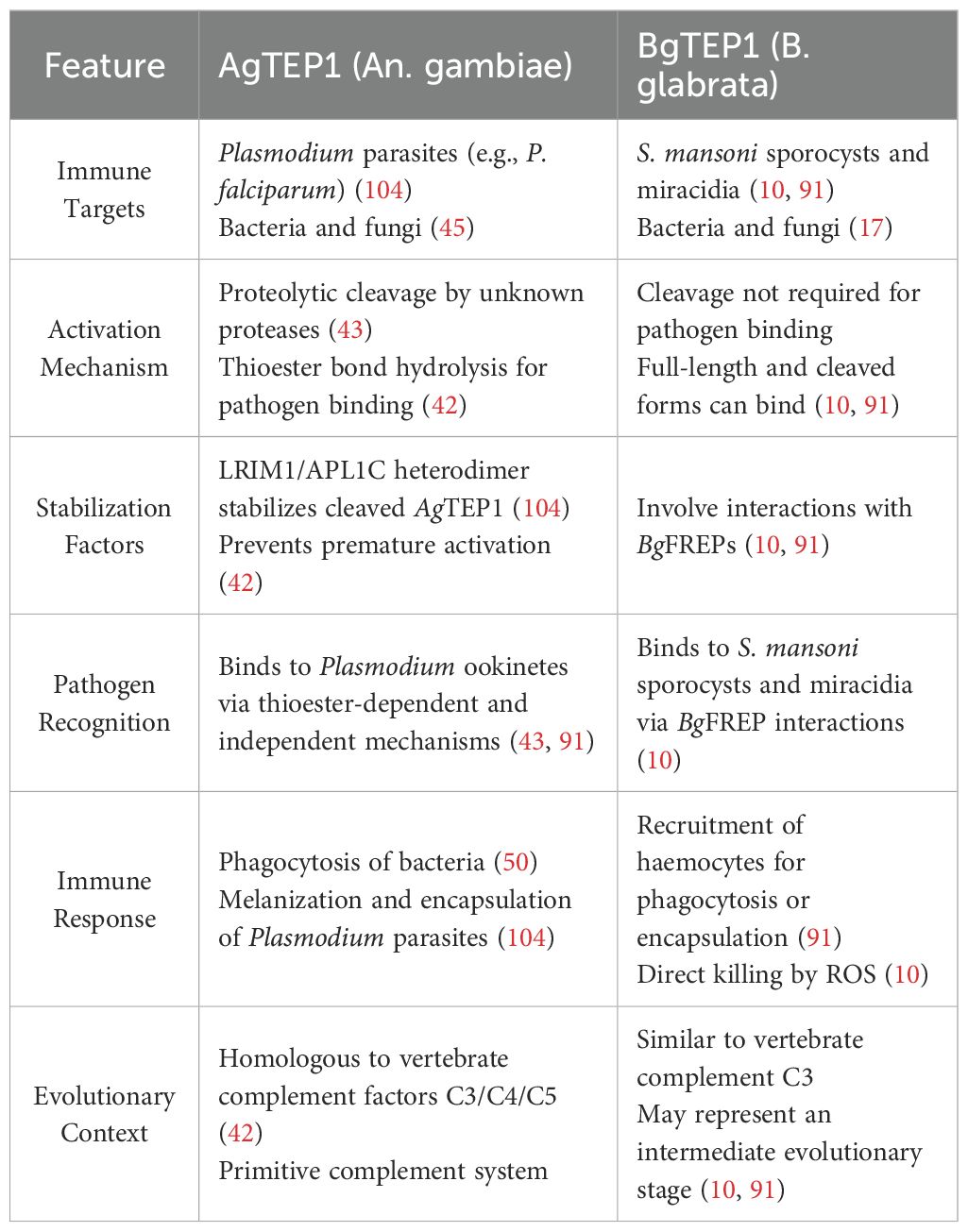
Table 4. Comparison of AgTEP1 and BgTEP1: Structural Features, and Immune Functions in An. gambiae and B. glabrata.
Both AgTEP1 and BgTEP1 harbor a highly conserved GCGEQ thioester motif, enabling covalent binding to pathogen surfaces post-activation to function as opsonins, thereby inducing phagocytosis, encapsulation, or other immune clearance mechanisms (17, 105). Additionally, both require binding to co-factors for stability and functional enhancement: AgTEP1 relies on the LRIM1/APL1C complex, while BgTEP1 cooperates with BgFREPs and Biomphalysin to exert immune effects (10, 22).
Functionally, both TEP systems recognize diverse pathogens, including protozoans, helminths, and bacteria, suggesting that TEPs—as ancient and conserved immune factors—likely represent an evolutionarily conserved core of broad-spectrum immune mechanisms in invertebrates.
Despite structural similarities, AgTEP1 and BgTEP1 exhibit distinct activation pathways. AgTEP1 activation depends on proteolytic cleavage in the hemolymph and stabilization by the LRIM1/APL1C complex, reflecting stringent protein-level regulation (14, 106). In contrast, BgTEP1 is primarily synthesized in haemocytes, with its expression modulated by host genetic background, developmental stage, and environmental stimuli, highlighting transcriptional-level regulation (10, 107).
Their effector pathways also diverge significantly: Anopheles predominantly employs melanization responses and complement-like lysis for pathogen clearance, whereas Biomphalaria utilizes hemocyte-mediated encapsulation and ROS-dependent extracellular cytotoxicity (108, 109). These mechanistic differences reflect host adaptations to their respective parasites (Plasmodium vs. Schistosoma), including structural features, survival strategies, and immune evasion tactics. However, it is also possible that other TEP family members in mosquitoes and snails contribute to these immune responses, which may account for some of the observed functional differences.
In An. gambiae, ROS also play a crucial role in AgTEP1-mediated immune responses, particularly in melanization. Melanization is a key defense mechanism in the insect immune system, involving the encapsulation of pathogens with melanin to prevent their further spread (110, 111). Research has shown that ROS are essential in melanization, especially during AgTEP1-mediated clearance of Plasmodium parasites (54, 60). When AgTEP1 binds to Plasmodium ookinetes, ROS production is significantly enhanced, leading to the melanization and death of the parasites (112). This process shares similarities with the ROS generation mechanism mediated by BgTEP1 in B. glabrata. In An. gambiae, ROS not only directly participate in pathogen killing but also promote melanin synthesis and deposition by activating enzymatic reactions in the melanization pathway (113).
In B. glabrata, after BgTEP1 forms an immune complex with BgFREP3 and BgFREP2, it activates the ROS generation pathway in haemocytes. These ROS act as effector molecules, directly attacking S. mansoni sporocysts, leading to membrane rupture and death (10). Studies have shown that ROS scavengers significantly reduce the killing ability of B. glabrata against S. mansoni, further confirming the importance of ROS in this process (10). Additionally, ROS production is closely related to the binding and signaling of BgTEP1. Through interactions with BgFREP3 and BgFREP2, BgTEP1 forms an immune complex that activates the ROS generation pathway in haemocytes. This process resembles the vertebrate complement system, where ROS also serve as critical effector molecules in pathogen clearance (95, 114).
Both AgTEP1 and BgTEP1 exhibit co-adaptive dynamics shaped by host-parasite interactions. For instance, AgTEP1 displays strain-specific responses to different Plasmodium isolates across Anopheles populations, with activity influenced by host and pathogen genetic polymorphisms (21, 115). Similarly, BgTEP1 expression differs markedly between resistant and susceptible snail strains, with resistant individuals mounting stronger BgTEP1-mediated immune responses during early infection (10, 40).
These findings underscore the pivotal role of TEPs in the long-term evolutionary “arms race” between hosts and parasites, retaining their core structural architecture while evolving highly plastic adaptive functions. For instance, enhancing AgTEP1 expression or stability via genetic engineering could significantly reduce Plasmodium loads in mosquitoes, thereby interrupting malaria transmission (27, 116). Similarly, targeting the interaction between BgTEP1 and BgFREPs may bolster snail resistance to Schistosoma, effectively disrupting the schistosomiasis transmission cycle (10).
Notably, in B. glabrata, the BgTEP1-Biomphalysin interaction generates lytic pore-forming complexes that directly induce S. mansoni sporocyst lysis (10, 117). While analogous MAC-like structures remain unconfirmed in Anopheles, this discovery provides critical insights into TEP-mediated immune mechanisms across hosts. Compared to mosquito strategies countering Plasmodium motility, the specific responses mediated by AgTEP1 and BgTEP1 suggest that Biomphalaria’s defense against sessile Schistosoma larvae emphasizes reactive oxygen species (ROS) and perforin-like complexes, reflecting possible divergent adaptations within individual TEP molecules across species (49, 118).
Furthermore, in B. glabrata, BgFREP2 and BgFREP3 synergize with BgTEP1 to form immune complexes that convert susceptible snails into partially resistant phenotypes (91). In contrast, although Anopheles FREPs participate in Plasmodium clearance as recognition receptors (114), no direct FREP-TEP interaction has been documented; AgTEP1 functionality remains dependent on LRIM1/APL1C stabilization (14, 119). These findings suggest that boosting co-factor expression or developing small-molecule mimics to stabilize TEP complexes could enhance immune efficacy, enabling genetic or ecological interventions to block malaria and schistosomiasis transmission (120).
In summary, the TEP system not only occupies a pivotal position in invertebrate immune evolution but also provides a theoretical and practical foundation for innovative disease control. By unraveling the functional and mechanistic intricacies of TEPs in Anopheles and Biomphalaria, we may develop precision interventions targeting these two major parasitic diseases.
Research gaps and future perspectives
Despite significant progress in elucidating the roles of TEPs in vector immunity, several key gaps remain that hinder a comprehensive understanding of their function and application potential.
Current research is heavily concentrated on AgTEP1 in An. gambiae and BgTEP1 in B. glabrata, leaving the majority of other TEP family members understudied. In An. gambiae, over a dozen AgTEPs have been identified, yet their individual or synergistic roles in immune defense remain poorly defined (27, 36). Similarly, B. glabrata likely possesses a broader repertoire of TEP-like genes, but functional validation is lacking. Expanding the functional annotation of these paralogs through CRISPR/Cas9, RNAi, and proteomics will be essential for uncovering hidden immune networks (121).
The molecular triggers and regulatory pathways governing TEP activation remain only partially understood in both species. For instance, while proteolytic cleavage is a known activation mechanism in Anopheles, the upstream signals initiating this process, and their modulation by infection or environmental stressors, remain to be clarified. In B. glabrata, the transcriptional regulation of BgTEP1 in response to parasite infection, pollutants, and other stimuli is only beginning to be explored (54, 103). Future research should focus on delineating the signaling cascades and epigenetic factors that control TEP expression and activity.
The interaction between TEPs and other immune components such as PRRs, ROS, and AMPs is not well defined (36, 122). Given the dynamic nature of innate immunity, TEPs likely operate as part of a broader immune network rather than as isolated effectors (40). Understanding this crosstalk, both in basal conditions and during infection, will provide a more integrated view of host defense strategies (45).
Most TEP studies are conducted under laboratory conditions that may not fully represent natural infection dynamics. Ecological factors such as temperature, microbiota composition, and co-infections can all influence TEP expression and function. Field-based transcriptomic and functional studies are needed to validate laboratory findings and assess the real-world relevance of TEP-mediated responses, especially in disease-endemic areas.
The potential of TEPs as targets for malaria vector-based interventions, such as genetic manipulation or immunostimulation, remains largely theoretical. Future efforts should evaluate whether enhancing TEP function in mosquito or snail populations can reduce parasite development and transmission in vivo. Additionally, identifying small molecules or microbial adjuvants that upregulate TEP expression may offer novel avenues for biological control strategies.
Conclusion
TEPs are central effectors of innate immunity in invertebrate disease vectors, mediating recognition and elimination of a wide range of pathogens. In both An. gambiae and B. glabrata, TEPs serve as functional analogs to vertebrate complement proteins, operating through conserved thioester motifs to tag pathogens for immune clearance.
While AgTEP1 and BgTEP1 share structural and functional similarities, their activation mechanisms, interacting partners, and effector pathways reflect the distinct evolutionary and ecological contexts of their hosts. These differences highlight the adaptive plasticity of TEP systems and emphasize their role in host–parasite coevolution.
Comparative analysis of TEP-mediated immunity in mosquitoes and snails offers valuable insights into the evolution of invertebrate defense systems and provides a foundation for novel vector-based disease control strategies. By bridging findings across phylogenetically distant taxa, we can better understand how innate immunity has diversified to meet the challenges of parasitic infection.
Future research should continue to explore the complexity, regulation, and translational potential of TEPs, with the goal of leveraging this ancient yet dynamic immune mechanism in the global fight against malaria and schistosomiasis.
Author contributions
HL: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing. YF: Conceptualization, Formal Analysis, Investigation, Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing. YQ: Conceptualization, Data curation, Investigation, Project administration, Supervision, Visualization, Writing – original draft, Writing – review & editing. WJ: Project administration, Formal Analysis, Investigation, Methodology, Validation, Writing – review & editing. YZ: Data curation, Formal Analysis, Project administration, Software, Writing – review & editing. JX: Data curation, Investigation, Methodology, Project administration, Writing – review & editing. XL: Formal Analysis, Methodology, Supervision, Validation, Writing – review & editing. XF: Data curation, Methodology, Software, Validation, Writing – review & editing. RW: Data curation, Project administration, Software, Validation, Writing – review & editing. YS: Data curation, Investigation, Project administration, Validation, Writing – review & editing. LD: Writing – review & editing. XZ: Formal Analysis, Investigation, Methodology, Supervision, Validation, Writing – review & editing. KC: Conceptualization, Project administration, Resources, Software, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (Grant No: 32000293 to Hongyu Li); Guangxi Natural Science Foundation (Grant Nos: 2020JJA130077 and 2018JJB140423 to Hongyu Li); the University Level Scientific Research Project of Zhejiang Shuren University (Grant No: 2022R064 to Hongyu Li); Zhejiang Shuren University Basic Scientific Research Special Funds (Grant No: 2024XZ014 to Hongyu Li); and Zhejiang Shuren University School-Level Research Project (Grant No: 2023A11001 to Hongyu Li). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgments
Parts of the materials used in the images within this article originate from Biorender, which we have further modified and enhanced. We hereby express our gratitude.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
ACTs: Artemisinin-based Combination Therapies
AgTEP: Anopheles gambiae Thioester-containing Protein
AMP: Antimicrobial Peptide
An. gambiae: Anopheles gambiae
An. stephensi: Anopheles stephensi
APL1C: Anopheles Plasmodium-responsive Leucine-rich Repeat 1C
BgA2M: Biomphalaria glabrata α-2-macroglobulin
B. glabrata: Biomphalaria glabrata
β-PFT: Beta Pore-forming Toxin
BgFREP: Biomphalaria glabrata Fibrinogen-related Protein
BgGRN: Biomphalaria glabrata Granulin
BgMCR: Biomphalaria glabrata Macroglobulin Complement-related Protein
BgMIF: Biomphalaria glabrata Macrophage Migration Inhibitory Factor
BgTEP: Biomphalaria glabrata Thioester-containing Protein
BgTLR: Biomphalaria glabrata Toll-like Receptor
Cap380: Plasmodium oocyst capsule protein 380
CLIP: Clip-domain Serine Protease
CUB: Complement C1r/C1s, Uegf, Bmp1 Domain
FBG: Fibrinogen-like Domain
FREP: Fibrinogen-related Protein
GCGEQ: Conserved Thioester Motif (Gly-Cys-Gly-Glu-Gln)
HPX2: Heme Peroxidase 2
IgSF: Immunoglobulin Superfamily
iTEP: Insect Thioester-containing Protein
LRIM1: Leucine-rich Repeat Immune Molecule 1
MAC: Membrane Attack Complex
MG: Macroglobulin
NOX5: NADPH Oxidase 5
P. berghei: Plasmodium berghei
P. falciparum: Plasmodium falciparum
P. vivax: Plasmodium vivax
P. yoelii: Plasmodium yoelii
PRR: Pattern Recognition Receptor
PZQ: Praziquantel
ROS: Reactive Oxygen Species
S. mansoni: Schistosoma mansoni
s.l./s.s.: sensu lato/sensu stricto
SPCLIP1: Serine Protease-like CLIP-domain Protein 1
TED: Thioester Domain
TEP: Thioester-containing Protein
TLR: Toll-like Receptor.
References
1. Neafsey DE, Taylor AR, and MacInnis BL. Advances and opportunities in malaria population genomics. Nat Rev Genet. (2021) 22:502–17. doi: 10.1038/s41576-021-00349-5
2. Otto TD, Gilabert A, Crellen T, Böhme U, Arnathau C, Sanders M, et al. Genomes of all known members of a Plasmodium subgenus reveal paths to virulent human malaria. Nat Microbiol. (2018) 3:687–97. doi: 10.1038/s41564-018-0162-2
3. Monroe A, Williams NA, Ogoma S, Karema C, and Okumu F. Reflections on the 2021 World Malaria Report and the future of malaria control. Malar J. (2022) 21:154. doi: 10.1186/s12936-022-04178-7
4. McManus DP, Bergquist R, Cai P, Ranasinghe S, Tebeje BM, and You H. Schistosomiasis-from immunopathology to vaccines. Semin Immunopathol. (2020) 42:355–71. doi: 10.1007/s00281-020-00789-x
5. Zhou G, Githure J, Lee MC, Zhong D, Wang X, Atieli H, et al. Malaria transmission heterogeneity in different eco-epidemiological areas of western Kenya: a region-wide observational and risk classification study for adaptive intervention planning. Malar J. (2024) 23:74. doi: 10.1186/s12936-024-04903-4
6. Popkin-Hall ZR and Slotman MA. Molecular evolution of gustatory receptors in the Anopheles Gambiae complex. BMC Ecol Evol. (2025) 25:22. doi: 10.1186/s12862-025-02359-x
7. Perin J, Mulick A, Yeung D, Villavicencio F, Lopez G, Strong KL, et al. Global, regional, and national causes of under-5 mortality in 2000-19: an updated systematic analysis with implications for the Sustainable Development Goals. Lancet Child Adolesc Health. (2022) 6:106–15. doi: 10.1016/S2352-4642(21)00311-4
8. Okwu DG, Manego RZ, Duparc S, Kremsner PG, Ramharter M, Mombo-Ngoma G, et al. The non-artemisinin antimalarial drugs under development: a review. Clin Microbiol Infection. (2025) 31:941–7. doi: 10.1016/j.cmi.2025.03.009
9. McManus DP, Dunne DW, Sacko M, Utzinger J, Vennervald BJ, Zhou XN, et al. Schistosomiasis. Nat Rev Dis Primers. (2018) 4:13. doi: 10.1038/s41572-018-0013-8
10. Li H, Hambrook JR, Pila EA, Gharamah AA, Fang J, Wu X, et al. Coordination of humoral immune factors dictates compatibility between Schistosoma mansoni and Biomphalaria glabrata. Elife. (2020) 9. doi: 10.7554/eLife.51708
11. Young ND, Stroehlein AJ, Wang T, Korhonen PK, Mentink-Kane M, Stothard JR, et al. Nuclear genome of Bulinus truncatus, an intermediate host of the carcinogenic human blood fluke Schistosoma haematobium. Nat Commun. (2022) 13:977. doi: 10.1038/s41467-022-28634-9
12. Chatterji T, Khanna N, Alghamdi S, Bhagat T, Gupta N, Alkurbi MO, et al. A recent advance in the diagnosis, treatment, and vaccine development for human schistosomiasis. Trop Med Infect Dis. (2024) 9:243. doi: 10.3390/tropicalmed9100243
13. Zhang Q, Zhou X, Feng T, Tong H, Wang J, Dai J, et al. The immune function of thioester-containing proteins in typical invertebrate disease vectors. Insect Biochem Mol Biol. (2025) 176:104218. doi: 10.1016/j.ibmb.2024.104218
14. Zmarlak NM, Lavazec C, Brito-Fravallo E, Genève C, Aliprandini E, Aguirre-Botero MC, et al. The Anopheles leucine-rich repeat protein APL1C is a pathogen binding factor recognizing Plasmodium ookinetes and sporozoites. PloS Pathog. (2024) 20:e1012008. doi: 10.1371/journal.ppat.1012008
15. Tafesh-Edwards G and Eleftherianos I. Functional role of thioester-containing proteins in the Drosophila anti-pathogen immune response. Dev Comp Immunol. (2023) 139:104578. doi: 10.1016/j.dci.2022.104578
16. Orús-Alcalde A. Innate immune mechanisms in invertebrates: Insights into the Toll pathway, the Imd pathway, and the complement system. Bergen Open Res Arch. (2021).
17. Portet A, Galinier R, Pinaud S, Portela J, Nowacki F, Gourbal B, et al. BgTEP: an antiprotease involved in innate immune sensing in biomphalaria glabrata. Front Immunol. (2018) 9:1206. doi: 10.3389/fimmu.2018.01206
18. Howick VM, Russell AJC, Andrews T, Heaton H, Reid AJ, Natarajan K, et al. The Malaria Cell Atlas: Single parasite transcriptomes across the complete Plasmodium life cycle. Science. (2019) 365. doi: 10.1126/science.aaw2619
19. Lee W-C, Cheong FW, Amir A, Lai MY, Tan JH, Phang WK, et al. Plasmodium knowlesi: the game changer for malaria eradication. Malaria J. (2022) 21:1–24. doi: 10.1186/s12936-022-04131-8
20. Andrade CM, Fleckenstein H, Thomson-Luque R, Doumbo S, Lima NF, Anderson C, et al. Increased circulation time of Plasmodium falciparum underlies persistent asymptomatic infection in the dry season. Nat Med. (2020) 26:1929–40. doi: 10.1038/s41591-020-1084-0
21. Venugopal K, Hentzschel F, Valkiūnas G, and Marti M. Plasmodium asexual growth and sexual development in the haematopoietic niche of the host. Nat Rev Microbiol. (2020) 18:177–89. doi: 10.1038/s41579-019-0306-2
22. Robert-Paganin J, Robblee JP, Auguin D, Blake TCA, Bookwalter CS, Krementsova EB, et al. Plasmodium myosin A drives parasite invasion by an atypical force generating mechanism. Nat Commun. (2019) 10:3286. doi: 10.1038/s41467-019-11120-0
23. Liffner B, Frölich S, Heinemann GK, Liu B, Ralph SA, Dixon MWA, et al. PfCERLI1 is a conserved rhoptry associated protein essential for Plasmodium falciparum merozoite invasion of erythrocytes. Nat Commun. (2020) 11:1411. doi: 10.1038/s41467-020-15127-w
24. Balestra AC, Koussis K, Klages N, Howell SA, Flynn HR, Bantscheff M, et al. Ca2+ signals critical for egress and gametogenesis in malaria parasites depend on a multipass membrane protein that interacts with PKG. Sci Adv. (2021) 7:eabe5396. doi: 10.1126/sciadv.abe5396
25. Barletta ABF, Barillas-Mury C, and Molina-Cruz A. Mosquito immune responses to Plasmodium parasites that limit malaria transmission. Cell Mol Life Sci. (2025) 82:143. doi: 10.1007/s00018-025-05667-z
26. Mwinyi SH, Bennett KL, Nagi SC, Kabula B, Matowo J, Weetman D, et al. Genomic analysis reveals a new cryptic taxon within the anopheles Gambiae complex with a distinct insecticide resistance profile in the coast of east africa. Mol Ecol. (2025):e17762. doi: 10.1111/mec.17762
27. Bartilol B, Omuoyo D, Karisa J, Ominde K, Mbogo C, Mwangangi J, et al. Vectorial capacity and TEP1 genotypes of Anopheles Gambiae sensu lato mosquitoes on the Kenyan coast. Parasites Vectors. (2022) 15:448. doi: 10.1186/s13071-022-05491-5
28. Pates HV, Takken W, Stuke K, and Curtis CF. Differential behaviour of Anopheles Gambiae sensu stricto (Diptera: Culicidae) to human and cow odours in the laboratory. Bull entomological Res. (2001) 91:289–96. doi: 10.1079/BER200198
29. Gueye OK, Tchouakui M, Dia AK, Faye MB, Ahmed AA, Wondji MJ, et al. Insecticide resistance profiling of Anopheles coluzzii and Anopheles Gambiae populations in the southern Senegal: role of target sites and metabolic resistance mechanisms. Genes. (2020) 11:1403. doi: 10.3390/genes11121403
30. Tuno N, Kjaerandsen J, Badu K, and Kruppa T. Blood-feeding behavior of Anopheles Gambiae and Anopheles melas in Ghana, western Africa. J Med entomology. (2010) 47:28–31. doi: 10.1093/jmedent/47.1.28
31. Habtewold T, Povelones M, Blagborough AM, and Christophides GK. Transmission blocking immunity in the malaria non-vector mosquito Anopheles quadriannulatus species A. PloS Pathog. (2008) 4:e1000070. doi: 10.1371/journal.ppat.1000070
32. Donnelly MJ, Licht MC, and Lehmann T. Evidence for recent population expansion in the evolutionary history of the malaria vectors Anopheles arabiensis and Anopheles Gambiae. Mol Biol Evol. (2001) 18:1353–64. doi: 10.1093/oxfordjournals.molbev.a003919
33. Hamid-Adiamoh M, Jabang AMJ, Opondo KO, Ndiath MO, Assogba BS, Amambua-Ngwa A, et al. Distribution of Anopheles Gambiae thioester-containing protein 1 alleles along malaria transmission gradients in The Gambia. Malaria J. (2023) 22:89. doi: 10.1186/s12936-023-04518-1
34. Li H, Gharamah AA, Hambrook JR, Wu X, and Hanington PC. Single-cell RNA-seq profiling of individual Biomphalaria glabrata immune cells with a focus on immunologically relevant transcripts. Immunogenetics. (2022) 74:77–98. doi: 10.1007/s00251-021-01236-3
35. Boissier J, Grech-Angelini S, Webster BL, Allienne JF, Huyse T, Mas-Coma S, et al. Outbreak of urogenital schistosomiasis in Corsica (France): an epidemiological case study. Lancet Infect Dis. (2016) 16:971–9. doi: 10.1016/S1473-3099(16)00175-4
36. Qin X, Li J, Zhu F, and Zhang J. Thioester-containing protein TEP15 promotes malaria parasite development in mosquitoes through negative regulation of melanization. Parasit Vectors. (2025) 18:124. doi: 10.1186/s13071-025-06772-5
37. Wang Z, Liang X, Li G, Liufu B, Lin K, Li J, et al. Molecular characterization of complement component 3 (c3) in the pearl oyster pinctada fucata improves our understanding of the primitive complement system in bivalve. Front Immunol. (2021) 12:652805. doi: 10.3389/fimmu.2021.652805
38. Dodds MW and Alex Law S. The phylogeny and evolution of the thioester bond-containing proteins C3, C4 and α2–macroglobulin. Immunol Rev. (1998) 166:15–26. doi: 10.1111/j.1600-065X.1998.tb01249.x
39. Kwaan HC. The role of fibrinolytic system in health and disease. Int J Mol Sci. (2022) 23:5262. doi: 10.3390/ijms23095262
40. Marquez J, Dinguirard N, Gonzalez A, Kane AE, Joffe NR, Yoshino TP, et al. Molecular characterization of thioester-containing proteins in Biomphalaria glabrata and their differential gene expression upon Schistosoma mansoni exposure. Front Immunol. (2022) 13:903158. doi: 10.3389/fimmu.2022.903158
41. Lin M, Sutherland DR, Horsfall W, Totty N, Yeo E, Nayar R, et al. Cell surface antigen CD109 is a novel member of the α2 macroglobulin/C3, C4, C5 family of thioester-containing proteins. Blood J Am Soc Hematol. (2002) 99:1683–91. doi: 10.1182/blood.V99.5.1683
42. Baxter RH, Chang CI, Chelliah Y, Blandin S, Levashina EA, Deisenhofer J, et al. Structural basis for conserved complement factor-like function in the antimalarial protein TEP1. Proc Natl Acad Sci. (2007) 104:11615–20. doi: 10.1073/pnas.0704967104
43. Levashina EA, Moita LF, Blandin S, Vriend G, Lagueux M, Kafatos FC, et al. Conserved role of a complement-like protein in phagocytosis revealed by dsRNA knockout in cultured cells of the mosquito, Anopheles Gambiae. Cell. (2001) 104:709–18. doi: 10.1016/S0092-8674(01)00267-7
44. Povelones M, Bhagavatula L, Yassine H, Tan LA, Upton LM, Osta MA, et al. The CLIP-domain serine protease homolog SPCLIP1 regulates complement recruitment to microbial surfaces in the malaria mosquito Anopheles Gambiae. PloS Pathog. (2013) 9:e1003623. doi: 10.1371/journal.ppat.1003623
45. Shokal U and Eleftherianos I. Evolution and function of thioester-containing proteins and the complement system in the innate immune response. Front Immunol. (2017) 8:759. doi: 10.3389/fimmu.2017.00759
46. Milner DA. Malaria pathogenesis. Cold Spring Harbor Perspect Med. (2018) 8:a025569. doi: 10.1101/cshperspect.a025569
47. Gadeberg TAF, Jørgensen MH, Olesen HG, Lorentzen J, Harwood SL, Almeida AV, et al. Cryo-EM analysis of complement C3 reveals a reversible major opening of the macroglobulin ring. Nat Struct Mol Biol. (2025) 32:884–95. doi: 10.1101/2024.04.15.589532
48. Zheng Y, Luo W, Yang J, Wang H, Hu Q, Zeng Z, et al. Controlled co-immobilisation of proteins via 4'-phosphopantetheine-mediated site-selective covalent linkage. N Biotechnol. (2022) 72:114–21. doi: 10.1016/j.nbt.2022.10.004
49. Klug D and Blandin SA. Activation of complement-like antiparasitic responses in Anopheles mosquitoes. Curr Opin Microbiol. (2023) 72:102280. doi: 10.1016/j.mib.2023.102280
50. Blandin S, Shiao SH, Moita LF, Janse CJ, Waters AP, Kafatos FC, et al. Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles Gambiae. Cell. (2004) 116:661–70. doi: 10.1016/S0092-8674(04)00173-4
51. Ye C, Zhang L, Tang L, Duan Y, Liu J, Zhou H, et al. Host genetic backgrounds: the key to determining parasite-host adaptation. Front Cell Infection Microbiol. (2023) 13:1228206. doi: 10.3389/fcimb.2023.1228206
52. Atique R, Ijaz A, Kausar S, Shahzadi I, Nadeem A, Naveed A, et al. Host-parasite interactions; from co-evolutionary changes to genomic insights. Global J Universal Stud. (2024) 1:88–107.
53. Qin B, Yu S, Chen Q, and Jin LH. Atg2 regulates cellular and humoral immunity in drosophila. Insects. (2023) 14:706. doi: 10.3390/insects14080706
54. Kwon H, Simões ML, Reynolds RA, Dimopoulos G, Smith RC, et al. Additional feeding reveals differences in immune recognition and growth of Plasmodium parasites in the mosquito host. Msphere. (2021) 6. doi: 10.1128/msphere.00136-21
55. Ramasamy R, Chen X, Zhang J, Sivabalakrishnan K, Arthiyan S, Surendran SN, et al. Lectin Microarray Analysis of Salivary Gland Glycoproteins from the Arboviral Vector Aedes aEgypti and the Malaria Vector Anopheles stephensi. J Vector Borne Dis. (2025) 62:326–31. doi: 10.1101/2024.07.10.602877
56. Zhang X, Zhang S, Kuang J, Sellens KA, Morejon B, Saab SA, et al. CLIPB4 is a central node in the protease network that regulates humoral immunity in anopheles Gambiae mosquitoes. J Innate Immun. (2023) 15:680–96. doi: 10.1159/000533898
57. Zhong D, Bu L, Habib MR, Lu L, Yan G, Zhang SM, et al. A haplotype-like, chromosome-level assembled and annotated genome of Biomphalaria glabrata, an important intermediate host of schistosomiasis and the best studied model of schistosomiasis vector snails. PloS Negl Trop Dis. (2024) 18:e0011983. doi: 10.1371/journal.pntd.0011983
58. Bu L, Habib MR, Lu L, Mutuku MW, Loker ES, Zhang SM, et al. Transcriptional profiling of Bulinus globosus provides insights into immune gene families in snails supporting the transmission of urogenital schistosomiasis. Dev Comp Immunol. (2024) 154:105150. doi: 10.1016/j.dci.2024.105150
59. Abou-El-Naga IF and Mogahed N. Immuno-molecular profile for Biomphalaria glabrata/Schistosoma mansoni interaction. Dev Comp Immunol. (2024) 150:105083. doi: 10.1016/j.dci.2023.105083
60. Pompon J and Levashina EA. A new role of the mosquito complement-like cascade in male fertility in anopheles Gambiae. PloS Biol. (2015) 13:e1002255. doi: 10.1371/journal.pbio.1002255
61. Arora G, Tang X, Cui Y, Yang J, Chuang YM, Joshi J, et al. mosGILT controls innate immunity and germ cell development in Anopheles Gambiae. BMC Genomics. (2024) 25:42. doi: 10.1186/s12864-023-09887-0
62. Ravisankar P. The implication of Prefoldin And Associated Factors On The Plasmodium Infection Of Anopheles Gambiae. Johns Hopkins University (2017).
63. Hearn J, Riveron JM, Irving H, Weedall GD, and Wondji CS. Gene conversion explains elevated diversity in the immunity modulating APL1 gene of the malaria vector Anopheles funestus. Genes. (2022) 13:1102. doi: 10.3390/genes13061102
64. Povelones M, Upton LM, Sala KA, and Christophides GK. Structure-function analysis of the Anopheles Gambiae LRIM1/APL1C complex and its interaction with complement C3-like protein TEP1. PloS Pathog. (2011) 7:e1002023. doi: 10.1371/journal.ppat.1002023
65. Yan Y and Hillyer JF. Complement-like proteins TEP1, TEP3 and TEP4 are positive regulators of periostial hemocyte aggregation in the mosquito Anopheles Gambiae. Insect Biochem Mol Biol. (2019) 107:1–9. doi: 10.1016/j.ibmb.2019.01.007
66. Mitri C, Bischoff E, Takashima E, Williams M, Eiglmeier K, Pain A, et al. An evolution-based screen for genetic differentiation between Anopheles sister taxa enriches for detection of functional immune factors. PloS Pathog. (2015) 11:e1005306. doi: 10.1371/journal.ppat.1005306
67. White BJ, Lawniczak MKN, Cheng C, Coulibaly MB, Wilson MD, Sagnon NF, et al. Adaptive divergence between incipient species of Anopheles Gambiae increases resistance to Plasmodium. Proc Natl Acad Sci. (2011) 108:244–9. doi: 10.1073/pnas.1013648108
68. Vandana V, Dong S, Sheth T, Sun Q, Wen H, Maldonado A, et al. Wolbachia infection-responsive immune genes suppress Plasmodium falciparum infection in Anopheles stephensi. PloS Pathog. (2024) 20:e1012145. doi: 10.1371/journal.ppat.1012145
69. Gildenhard M, Rono EK, Diarra A, Boissiere A, Bascunan P, Carrillo-Bustamante P, et al. Mosquito microevolution drives Plasmodium falciparum dynamics. Nat Microbiol. (2019) 4:941–7. doi: 10.1038/s41564-019-0414-9
70. Peirce MJ, Mitchell SN, Kakani EG, Scarpelli P, South A, Shaw WR, et al. JNK signaling regulates oviposition in the malaria vector Anopheles Gambiae. Sci Rep. (2020) 10:14344. doi: 10.1038/s41598-020-71291-5
71. Upton LM, Povelones M, and Christophides GK. Anopheles Gambiae blood feeding initiates an anticipatory defense response to Plasmodium berghei. J innate Immun. (2014) 7:74–86. doi: 10.1159/000365331
72. Carr AL, Rinker DC, Dong Y, Dimopoulos G, and Zwiebel LJ. Transcriptome profiles of Anopheles Gambiae harboring natural low-level Plasmodium infection reveal adaptive advantages for the mosquito. Sci Rep. (2021) 11:22578. doi: 10.1038/s41598-021-01842-x
73. Sturrock RF. The schistosomes and their intermediate hosts, in Schistosomiasis. World Sci. (2001), 7–83.
74. Loker ES. A comparative study of the life-histories of mammalian schistosomes. Parasitology. (1983) 87:343–69. doi: 10.1017/S0031182000052689
75. Costain AH, MacDonald AS, and Smits HH. Schistosome egg migration: mechanisms, pathogenesis and host immune responses. Front Immunol. (2018) 9:3042. doi: 10.3389/fimmu.2018.03042
76. Nelwan ML. Schistosomiasis: life cycle, diagnosis, and control. Curr Ther Res. (2019) 91:5–9. doi: 10.1016/j.curtheres.2019.06.001
77. Wright C. The schistosome life-cycle, in Bilharziasis: International Academy of Pathology Special Monograph. Springer Nature (1967) p. 3–7.
78. Haseeb M and Fried B. Modes of transmission of trematode infections and their control, in Advances in trematode biology. Taylor & Francis Group: CRC Press (2024) p. 31–56.
79. McKerrow J and Salter J. Invasion of skin by Schistosoma cercariae. Trends Parasitol. (2002) 18:193–5. doi: 10.1016/S1471-4922(02)02309-7
80. Miller P and Wilson R. Migration of the schistosomula of Schistosoma mansoni from the lungs to the hepatic portal system. Parasitology. (1980) 80:267–88. doi: 10.1017/S0031182000000743
82. Adema CM, Hillier LDW, Jones CS, Loker ES, Knight M, Minx P, et al. Corrigendum: Whole genome analysis of a schistosomiasis-transmitting freshwater snail. Nat Commun. (2017) 8:16153. doi: 10.1038/ncomms16153
83. Zhang SM, Adema CM, Kepler TB, and Loker ES. Diversification of Ig superfamily genes in an invertebrate. Science. (2004) 305:251–4. doi: 10.1126/science.1088069
84. Hanington PC, Forys MA, Dragoo JW, Zhang SM, Adema CM, Loker ES, et al. Role for a somatically diversified lectin in resistance of an invertebrate to parasite infection. Proc Natl Acad Sci U.S.A. (2010) 107:21087–92. doi: 10.1073/pnas.1011242107
85. Pila EA, Gordy MA, Phillips VK, and Kabore AL. Endogenous growth factor stimulation of hemocyte proliferation induces resistance to Schistosoma mansoni challenge in the snail host. Proc Natl Acad Sci U.S.A. (2016) 113:5305–10. doi: 10.1073/pnas.1521239113
86. Pila EA, Gordy MA, Phillips VK, and Hanington PC. Schistosomiasis from a snail's perspective: advances in snail immunity. Trends Parasitol. (2017) 33:845–57. doi: 10.1016/j.pt.2017.07.006
87. Adema CM, Hertel LA, Miller RD, and Loker ES. A family of fibrinogen-related proteins that precipitates parasite-derived molecules is produced by an invertebrate after infection. Proc Natl Acad Sci. (1997) 94:8691–6. doi: 10.1073/pnas.94.16.8691
88. Wu XJ, et al. Proteomic analysis of Biomphalaria glabrata plasma proteins with binding affinity to those expressed by early developing larval Schistosoma mansoni. PloS Pathog. (2017) 13:e1006081. doi: 10.1371/journal.ppat.1006081
89. Hanington PC and Zhang SM. The primary role of fibrinogen-related proteins in invertebrates is defense, not coagulation. J Innate Immun. (2011) 3:17–27. doi: 10.1159/000321882
90. Tetreau G, Pinaud S, Portet Anaïs, Galinier R, Gourbal B, and Duval D. Specific pathogen recognition by multiple innate immune sensors in an invertebrate. Front Immunol. (2017) 8:1249. doi: 10.3389/fimmu.2017.01249
91. Moné Y, Gourbal B, Duval D, Du Pasquier L, Kieffer-Jaquinod S, and Mitta G. A large repertoire of parasite epitopes matched by a large repertoire of host immune receptors in an invertebrate host/parasite model. PloS Negl Trop Dis. (2010) 4. doi: 10.1371/journal.pntd.0000813
92. Galinier R, Portela J, Moné Y, Allienne JF, Henri H, and Delbecq S. Biomphalysin, a new beta pore-forming toxin involved in Biomphalaria glabrata immune defense against Schistosoma mansoni. PloS Pathog. (2013) 9:e1003216. doi: 10.1371/journal.ppat.1003216
93. Pila EA, et al. A Novel Toll-Like Receptor (TLR) Influences Compatibility between the Gastropod Biomphalaria glabrata, and the Digenean Trematode Schistosoma mansoni. PloS Pathog. (2016) 12:e1005513. doi: 10.1371/journal.ppat.1005513
94. Bender RC, Fryer SE, and Bayne CJ. Proteinase inhibitory activity in the plasma of a mollusc: evidence for the presence of alpha-macroglobulin in Biomphalaria glabrata. Comp Biochem Physiol B. (1992) 102:821–4. doi: 10.1016/0305-0491(92)90086-7
95. Duval D, Pichon R, Lassalle D, Laffitte M, Gourbal B, and Galinier R. A new assessment of thioester-containing proteins diversity of the freshwater snail biomphalaria glabrata. Genes (Basel). (2020) 11:69. doi: 10.3390/genes11010069
96. Hanington PC, Forys MA, and Loker ES. A somatically diversified defense factor, FREP3, is a determinant of snail resistance to schistosome infection. PloS Negl Trop Dis. (2012) 6:e1591. doi: 10.1371/journal.pntd.0001591
97. Gordy MA, Pila EA, and Hanington PC. The role of fibrinogen-related proteins in the gastropod immune response. Fish Shellfish Immunol. (2015) 46:39–49. doi: 10.1016/j.fsi.2015.03.005
98. Lie KJ and Heyneman D. Acquired resistance to echinostomes in four Biomphalaria glabrata strains. Int J Parasitol. (1979) 9:533–7. doi: 10.1016/0020-7519(79)90009-2
99. Mitta G, Adema CM, Gourbal B, Loker ES, and Theron A. Compatibility polymorphism in snail/schistosome interactions: From field to theory to molecular mechanisms. Dev Comp Immunol. (2012) 37:1–8. doi: 10.1016/j.dci.2011.09.002
100. Hryhorowicz M, Lipiñski D, Zeyland J, and Słomski R. CRISPR/Cas9 immune system as a tool for genome engineering. Archivum immunologiae therapiae experimentalis. (2017) 65:233–40. doi: 10.1007/s00005-016-0427-5
101. Fisch D, et al. Molecular definition of the endogenous Toll-like receptor signalling pathways. Nature. (2024) 631:635–44. doi: 10.1038/s41586-024-07614-7
102. Shi Q, Zhang P, Hu Q, Zhang T, Hou R, and Yin S. Role of TOMM34 on NF-κB activation-related hyperinflammation in severely ill patients with COVID-19 and influenza. EBioMedicine. (2024) 108:105343. doi: 10.1016/j.ebiom.2024.105343
103. Gorbushin AM. Derivatives of the lectin complement pathway in Lophotrochozoa. Dev Comp Immunol. (2019) 94:35–58. doi: 10.1016/j.dci.2019.01.010
104. Fraiture M, et al. Two mosquito LRR proteins function as complement control factors in the TEP1-mediated killing of Plasmodium. Cell Host Microbe. (2009) 5:273–84. doi: 10.1016/j.chom.2009.01.005
105. Le BV, et al. Molecular basis for genetic resistance of Anopheles Gambiae to Plasmodium: structural analysis of TEP1 susceptible and resistant alleles. PLOS Pathogens. (2012). doi: 10.1371/journal.ppat.1002958
106. Zhang R, Zhang X-F, Chi Y, Xu Y, Chen H, and Guo Z. Nucleoprotein of a rice rhabdovirus serves as the effector to attenuate hemolymph melanization and facilitate viral persistent propagation in its leafhopper vector. Front Immunol. (2022) 13:904244. doi: 10.3389/fimmu.2022.904244
107. Aoi Y and Shilatifard A. Transcriptional elongation control in developmental gene expression, aging, and disease. Mol Cell. (2023) 83:3972–99. doi: 10.1016/j.molcel.2023.10.004
108. Sousa GL. Dissecting The Melanization Immune Response In The Malaria Vector Anopheles Gambiae. University of Pennsylvania (2021).
109. Zhang S-M, Yan G, Lekired A, and Zhong D. Genomic basis of schistosome resistance in a molluscan vector of human schistosomiasis. Iscience. (2025) 28:111520. doi: 10.1016/j.isci.2024.111520
110. Nakhleh J, El Moussawi L, and Osta MA. The melanization response in insect immunity. Adv Insect Physiol. (2017) 52:83–109. doi: 10.1016/bs.aiip.2016.11.002
111. Viteri BDM. Immune Regulation in the African Malaria Mosquito, Anopheles Gambiae. Kansas State University (2024).
112. Kumar S, Christophides GK, Cantera R, Charles B, and Han YS. The role of reactive oxygen species on Plasmodium melanotic encapsulation in Anopheles Gambiae. Proc Natl Acad Sci. (2003) 100:14139–44. doi: 10.1073/pnas.2036262100
113. Camacho E, Dong Y, Anglero-Rodriguez Y, Smith DFQ, de Souza Jacomini R, McConnell SA, et al. Analysis of melanotic Plasmodium spp. capsules in mosquitoes reveal eumelanin-pheomelanin composition and identify Ag Mesh as a modulator of parasite infection. bioRxiv. (2021) 2021.05.07.443077. doi: 10.1101/2021.05.07.443077
114. Delrieu M, Martinet J-P, O’Connor O, Viennet E, Menkes C, and Burtet-Sarramegna V. Current research in parasitology & Vector-borne diseases. Curr Res Parasitol Vector-Borne Dis. (2023) 4:100139. doi: 10.1016/j.crpvbd.2023.100139
115. Võ TC, Lê HG, Kang J-M, Naw H, Yoo WG, and Myint MK. Genetic polymorphism and natural selection of the erythrocyte binding antigen 175 region II in Plasmodium falciparum populations from Myanmar and Vietnam. Sci Rep. (2023) 13:20025. doi: 10.1038/s41598-023-47275-6
116. Carballar-Lejarazu R, et al. Dual effector population modification gene-drive strains of the African malaria mosquitoes, Anopheles Gambiae and Anopheles coluzzii. Proc Natl Acad Sci U.S.A. (2023) 120:e2221118120. doi: 10.1073/pnas.2221118120
117. Lu L, et al. Different metazoan parasites, different transcriptomic responses, with new insights on parasitic castration by digenetic trematodes in the schistosome vector snail Biomphalaria glabrata. BMC Genomics. (2024) 25:608. doi: 10.1186/s12864-024-10454-4
118. Ramaprasad A, Burda P-C, Koussis K, Thomas JA, Pietsch E, and Calvani E. A malaria parasite phospholipase facilitates efficient asexual blood stage egress. PloS Pathog. (2023) 19:e1011449. doi: 10.1371/journal.ppat.1011449
119. Yadav M, et al. Molecular characterization, expression and in-silico analysis of fibrinogen-related protein 1 (frep1) in malaria vector Anopheles stephensi. Mol Biol Rep. (2024) 51:970. doi: 10.1007/s11033-024-09891-x
120. Williams M and Baxter R. The structure and function of thioester-containing proteins in arthropods. Biophys Rev. (2014) 6:261–72. doi: 10.1007/s12551-014-0142-6
121. Khan R, Azhar M, and Umair M. Decoding the genes orchestrating egg and sperm fusion reactions and their roles in fertility. Biomedicines. (2024) 12:2850. doi: 10.3390/biomedicines12122850
Keywords: Anopheles gambiae, immune, plasmodium, schistosomamansoni, thioester-containing protein
Citation: Li H, Feng Y, Qian Y, Jiang W, Zhu Y, Xu J, Li X, Fei X, Wang R, Shao Y, Du L, Zhang X and Chen K (2025) Comparative immunological roles of TEP1 in Anopheles gambiae and Biomphalaria glabrata: implications for malaria and schistosomiasis control. Front. Immunol. 16:1629262. doi: 10.3389/fimmu.2025.1629262
Received: 15 May 2025; Accepted: 29 September 2025;
Published: 15 October 2025.
Edited by:
Humberto Lanz-Mendoza, National Institute of Public Health (Mexico), MexicoReviewed by:
Salvador Hernández-Martínez, National Institute of Public Health (Mexico), MexicoDalia Ashour, Tanta University, Egypt
Maria G. Castillo, New Mexico State University, United States
Copyright © 2025 Li, Feng, Qian, Jiang, Zhu, Xu, Li, Fei, Wang, Shao, Du, Zhang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Keda Chen, Y2hlbmtkQHpqc3J1LmVkdS5jbg==; Hongyu Li, aG9uZ3l1ODg5MjZAempzcnUuZWR1LmNu
†These authors have contributed equally to this work
 Hongyu Li
Hongyu Li Yilu Feng†
Yilu Feng† Yuncheng Qian
Yuncheng Qian Wenjie Jiang
Wenjie Jiang Lailing Du
Lailing Du Keda Chen
Keda Chen