- 1Endocrinology and Metabolism Nephrology Department, Guangxi Medical University Cancer Hospital, Nanning, Guangxi, China
- 2Department of Nephrology, The Second Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi, China
Introduction: Heredity and epigenetic factors contribute to the pathogenesis of neutrophil-cytoplasmic antibody (ANCA)-associated vasculitis (AAV). Cytotoxic T lymphocyte-associated protein 4 (CTLA-4), an inhibitory receptor regulating T-cell homeostasis and maintaining self-tolerance, has emerged as a key target for immune screening and therapeutics in autoimmunity and cancer. CTLA-4 is associated with various autoimmune diseases; however, the relationship between CTLA-4 polymorphisms and AAV in the Guangxi population of China remains underexplored. In the present case–control study, we evaluated the effects of CTLA-4 polymorphisms on AAV susceptibility in the Guangxi population of China.
Methods: A total of 343 patients with AAV and 343 healthy controls were recruited. High-throughput sequencing was used to genotype CTLA4 variants, and logistic regression analysis was used to assess their association with AAV risk. The relationship between the haplotypes of CTLA4 single-nucleotide polymorphisms (SNPs) and AAV risk was assessed using the SHEsis platform.
Results: Three CTLA4 SNPs— rs62182595, rs16840252, and rs5742909— showed significant association with AAV susceptibility. The ATT and GCC haplotypes, comprising these loci, were also associated with an increased risk of AAV.
Discussion: These findings suggest that CTLA4 polymorphisms (rs62182595, rs16840252, and rs5742909) may contribute to AAV susceptibility in the Guangxi population and offer preliminary markers for risk assessment, early diagnosis, and personalized management of AAV.
1 Introduction
Anti-neutrophil cytoplasmic antibody-associated vasculitis (AAV) comprises a group of small-vessel inflammatory disorders with largely unknown pathogenesis. AAV is serologically defined by autoantibodies against two specific neutrophil antigens: proteinase 3 and myeloperoxidase (1). Pathologically, AAV is characterized by necrotizing inflammation of vessel walls. Clinically, AAV includes three principal subtypes: granulomatosis with polyangiitis (2), microscopic polyangiitis (MPA) (3, 4), and eosinophilic granulomatosis with polyangiitis (5). Although the exact pathogenesis of AAV is unknown, immune dysregulation, which is influenced by genetic and environmental factors, plays a central role in AAV pathogenesis (6). Genome-wide analyses have identified several predisposing loci, mainly within the major histocompatibility complex region. Specifically, variants in the human leukocyte antigen-DQ segment show strong genetic associations with AAV pathogenesis (7, 8). In Asia, AAV prevalence ranges from 46 to 421 per million (9), and in China, its prevalence is increasing annually, especially among the aging population. The current standard treatment for AAV involves systemic immunosuppressants targeting B and T cells, including glucocorticoids, cyclophosphamide, and rituximab (10); however, many patients with AAV remain unresponsive to the current standard of care and experience disease relapses (11). Therefore, identification of novel therapeutic targets to achieve long-term disease remission is necessary.
Cytotoxic T lymphocyte-associated protein 4 (CTLA-4), a crucial immune checkpoint molecule within the CD28 immunoglobulin superfamily, exhibits predominant expression in T lymphocytes (12). CTLA-4 is an important regulator of T-cell homeostasis and self-tolerance and can modulate the T-cell response by blocking CD28 co-stimulation of T-cells during T-cell activation (13, 14). CTLA-4 expression in T cells is tightly regulated, and the failure of the transport mechanism of CTLA-4 to the cell surface leads to autoimmune susceptibility (14). Given its central role in immune modulation, CTLA-4 has emerged as a promising therapeutic target across multiple disease domains, particularly in autoimmune conditions and oncology. Moreover, CTLA-4-targeted therapies through the successful implementation of CTLA-4 inhibitors have been validated to manage various immune-mediated disorders clinically (12). In AAV, CTLA-4–Ig fusion proteins that block T-cell co-stimulation offer a novel therapeutic strategy potentially advantageous over traditional immunosuppressive regimens (15). However, genetic studies have reported inconsistent associations between CTLA4 polymorphisms and AAV. For instance, a European case–control study identified the association between CTLA4 polymorphism— rs3087243 —and AAV (16). In contrast, a case—control study of the Han Chinese population in Beijing, China, reported no association between CTLA4 polymorphisms (rs231775 and rs5742909) and AAV susceptibility (17). These findings suggest the strong influence of ethnicity, population, and regional variation on genetic susceptibility; however, the association between single-nucleotide polymorphisms (SNPs) at the CTLA4 locus and AAV susceptibility in the Guangxi population has not been reported.
To address this gap, we conducted a case—control study involving 343 patients with AAV from Guangxi and selected three candidate CTLA4 SNPs to evaluate their association with AAV risk in this population.
2 Materials and methods
2.1 Ethics approval
This study was approved by the Medical Ethics Committee of The Second Affiliated Hospital of Guangxi Medical University (2018 KY-0100) and was conducted in compliance with the Declaration of Helsinki. We have obtained written consent from all participants.
2.2 Study population characteristics
The MPA investigation cohort constituted 343 patients diagnosed with AAV retrospectively enrolled from the Second Affiliated Hospital of Guangxi Medical University (2005-2024). The cases were confirmed through rigorous diagnostic protocols aligned with the Chapel Hill Consensus Conference classification criteria (18). Exclusion criteria strictly applied to individuals presenting with: (1) secondary vasculitides induced by infectious processes, neoplastic disorders, or pharmacological agents; (2) concurrent autoimmune conditions (including rheumatoid arthritis, systemic lupus erythematosus, Henoch-Schönlein purpura). A parallel control group of 343 age- and sex-matched healthy volunteers was established, systematically excluding participants with autoimmune comorbidities or malignant histories.
2.3 SNP selection and DNA extraction
We selected three CTLA4 SNPs (rs62182595, rs16840252, and rs5742909) based on the following criteria: 1) all SNPs had a minor allele frequency (MAF) ≥ 0.05 (19) and 2) Chinese allele or genotype data of these SNPs were available in 1000 genomes (https://grch37.ensembl.org/).
DNA extraction was performed using a Rapid Blood Genomic DNA Isolation Kit (Sangon, Shanghai, China), following the manufacturer’s instructions. The concentration of the DNA was measured using a Qubit 2.0 (Thermo Fisher Scientific, Inc.) to ensure the extraction of adequate amounts of high-quality genomic DNA.
2.4 Statistical analyses
All statistical analyses were conducted within a rigorous statistical framework using IBM SPSS Statistics, version 26.0 (IBM Corp., Armonk, NY, USA) (20). Initial demographic comparisons were performed using independent samples t-tests for continuous variables and chi-square (χ²) test for categorical variables. Hardy–Weinberg equilibrium (HWE) for genotype and allele distribution was assessed using both χ² and Fisher’s exact tests. Genetic association analysis was conducted using an unmatched case–control paradigm through the SNPstats analytical platform (https://www.snpstats.net/start.htm) (21), applying multivariate logistic regression with covariate adjustment (age, ethnicity, gender) to derive adjusted odds ratios (ORs) and 95% confidence intervals (CIs). Haplotype architecture reconstruction (22) was performed using the SHEsis online suite (23), incorporating permutation-based significance testing. To control the Type I in multi-group comparisons, Bonferroni multiplicity correction was applied, maintaining a family-wise significance threshold set at α=0.05.
3 Results
3.1 Demographic characteristics
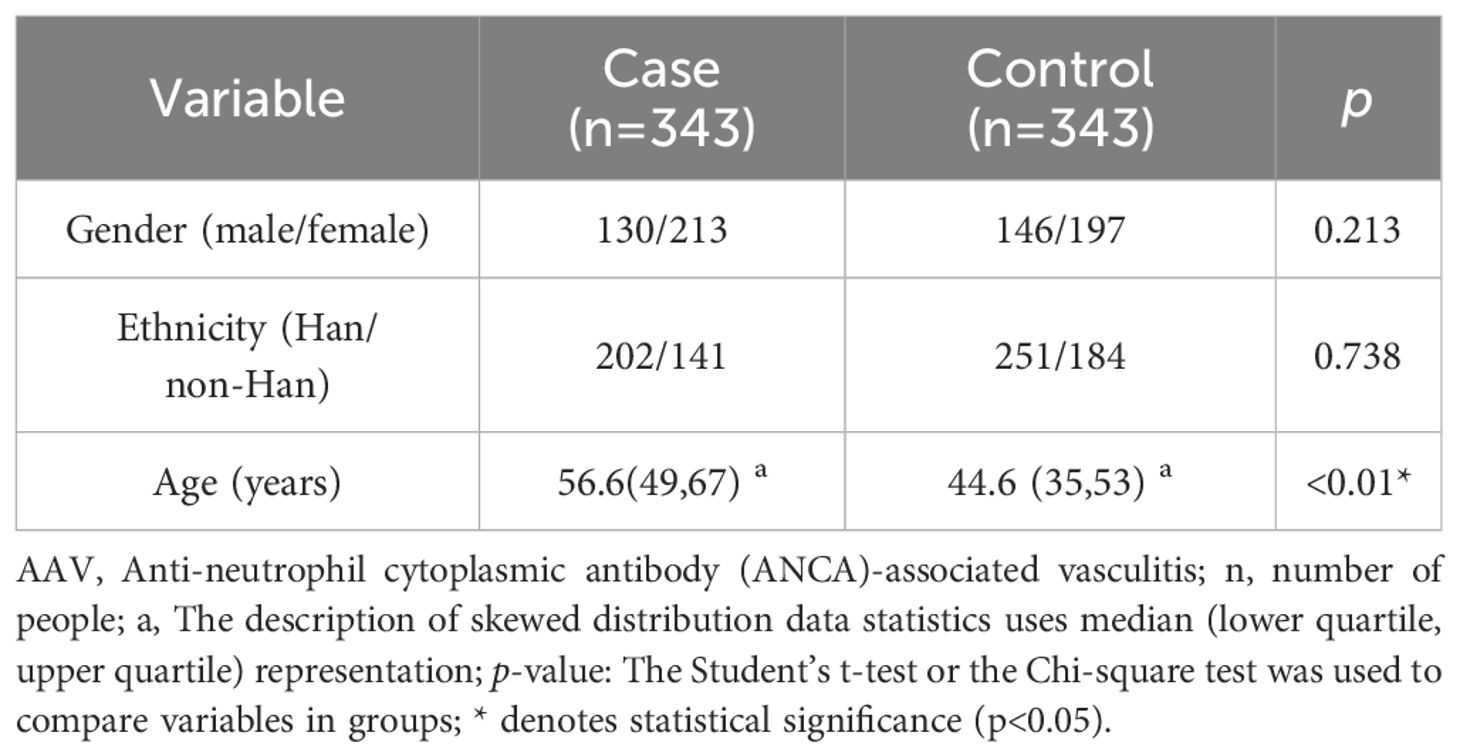
The study cohort comprised equal numbers of patients with AAV (n = 343) and healthy controls (n = 343). Gender distribution showed comparable patterns across groups, with the AAV cohort comprising 130 male and 213 female participants, compared to 146 males and 197 females in the control group (P = 0.213). Age distribution revealed significant intergroup variation. The AAV group showed a mean age of 56.7 years (range, 8–86 years; interquartile range (IQR), 49–67 years), which was significantly higher than that of the control group (mean, 44.6 years, range: 18–81 years; IQR, 35–53 years) (P < 0.05). Ethnic composition was comparable between the groups, with 202 Han Chinese and 141 non-Han Chinese participants in both cohorts (P = 0.738). To ensure analytical rigor, potential confounding variables, including age, sex, and ethnicity, were incorporated as covariates in the logistic regression analysis (Table 1).
3.2 Information for selected SNPs
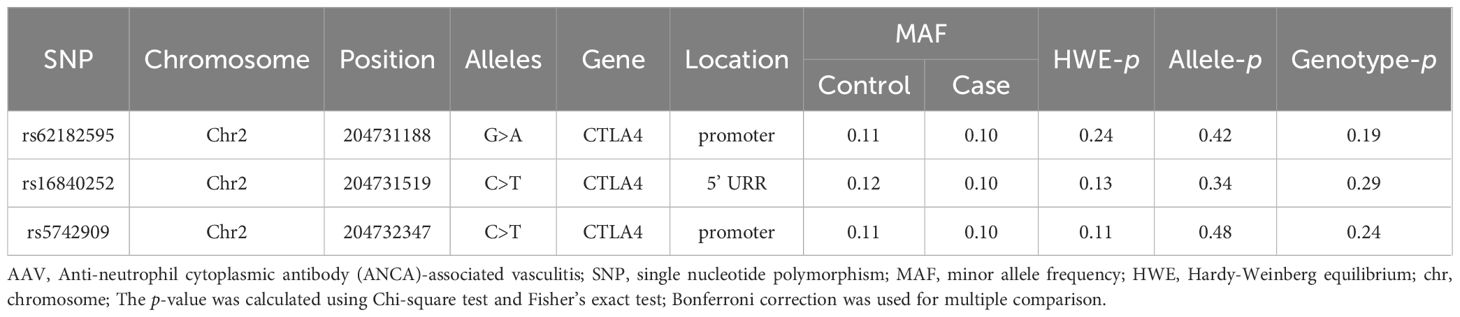
Basic information for the selected CTLA4 SNPs is shown in Table 2. All loci are located on chromosome 2, specifically within the promoter region and 5’ upstream regulatory region (URR). The MAF for each SNP was greater than 0.05, indicating that variants are common in the Guangxi population. Additionally, all SNPs conformed to HWE (p > 0.05).
3.3 Correlation of SNPs with AAV susceptibility under different genetic models
The association between the SNPs and AAV risk were assessed using different genetic models (adjusted for age, sex, and race). The rs62182595, rs16840252, and rs5742909 polymorphisms in CTLA4 were significantly associated with AAV susceptibility in the Guangxi population under different genetic models (Table 3). For rs62182595, the GA genotype in the codominant genetic model (OR = 0.58; 95% CI [0.38–0.90]; p = 0.048), the GA-AA genotype in the dominant genetic model (0.61; 0.40–0.93; p = 0.021), and GA genotypes in the overdominant genetic model (0.58; 0.38–0.90; p = 0.014), were all associated with reduced AAV susceptibility in the Guangxi population. For rs16840252, the CT-TT genotype in the dominant genetic model (OR = 0.61; 95% CI [0.40–0.93]; p = 0.022) and the C/T genotype in the overdominant genetic model (0.60; 0.39–0.92; p = 0.019) were also associated with reduced risk of AAV. In contrast, only the CT genotype at rs5742909 in the overdominant genetic model showed association with reduced susceptibility to AAV (OR =0.64; 95% CI [0.42–0.99]; p = 0.044).
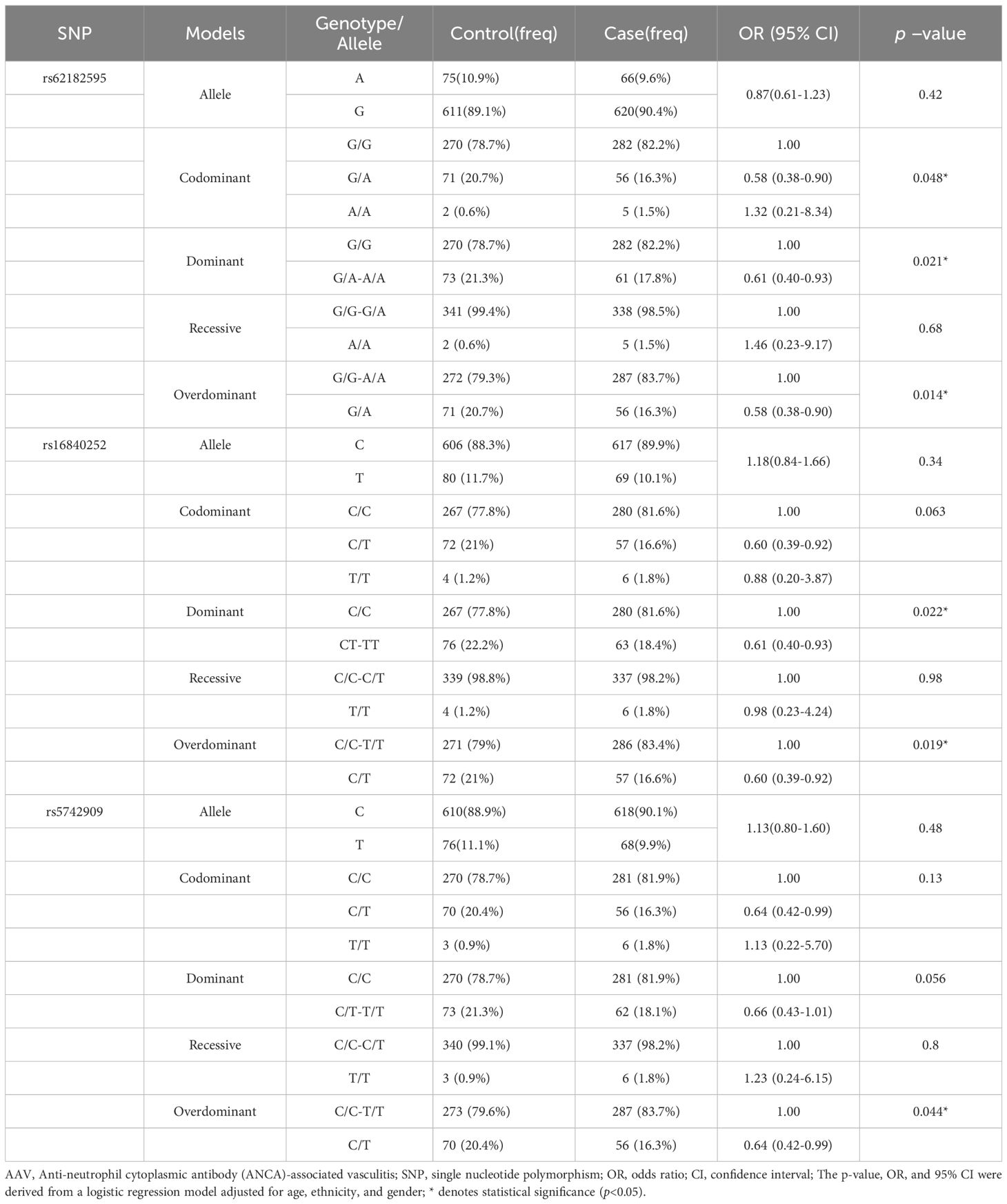
Table 3. The relationship between the SNPs and the risk of AAV in Guangxi population in different genetic models.
3.4 Correlation between selected SNPs and risk of AAV development in different subgroups of the population
To assess the population subgroups in which the above CTLA4 SNPs specifically influence the risk of developing AAV, subgroup analyses were performed by sex (male and female) and ethnicity (Han Chinese and non-Han Chinese). The analysis revealed a significant influence of sex and ethnicity on the association between the three SNPs and AAV risk. Specifically, across different genetic models, all three SNPs—rs62182595, rs16840252, and rs5742909— were significantly associated with reduced AAV susceptibility in the male subgroup (Table 4), whereas no significant association was observed in the female subgroup (Supplementary Table 1). Regarding ethnicity, the strongest protective associations were observed in the Han Chinese subgroup (Table 5, Supplementary Table 2), suggesting a population-specific genetic effect on AAV risk.
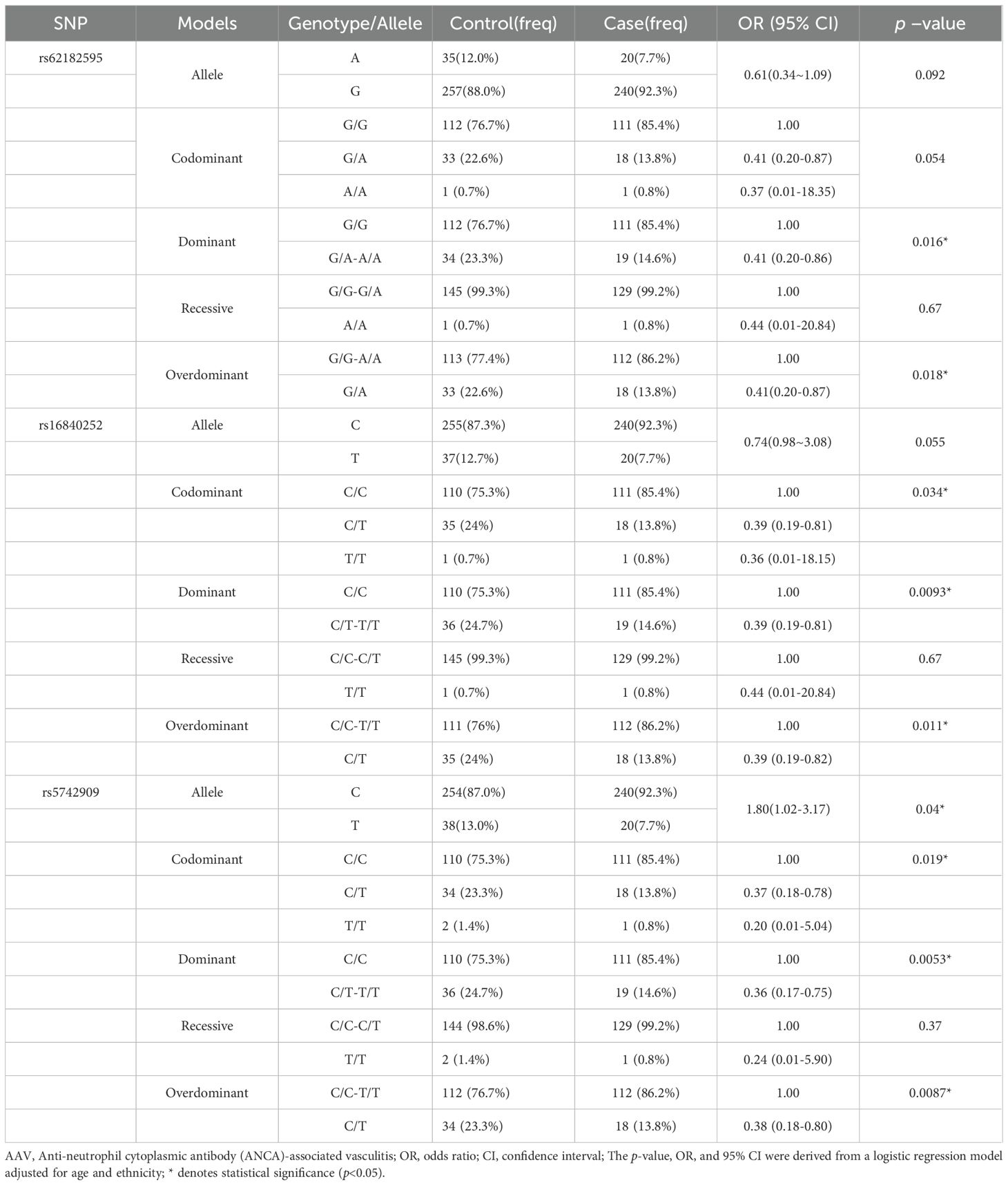
Table 4. The relationship between the SNPs and the risk of AAV in male of Guangxi population in different genetic models.
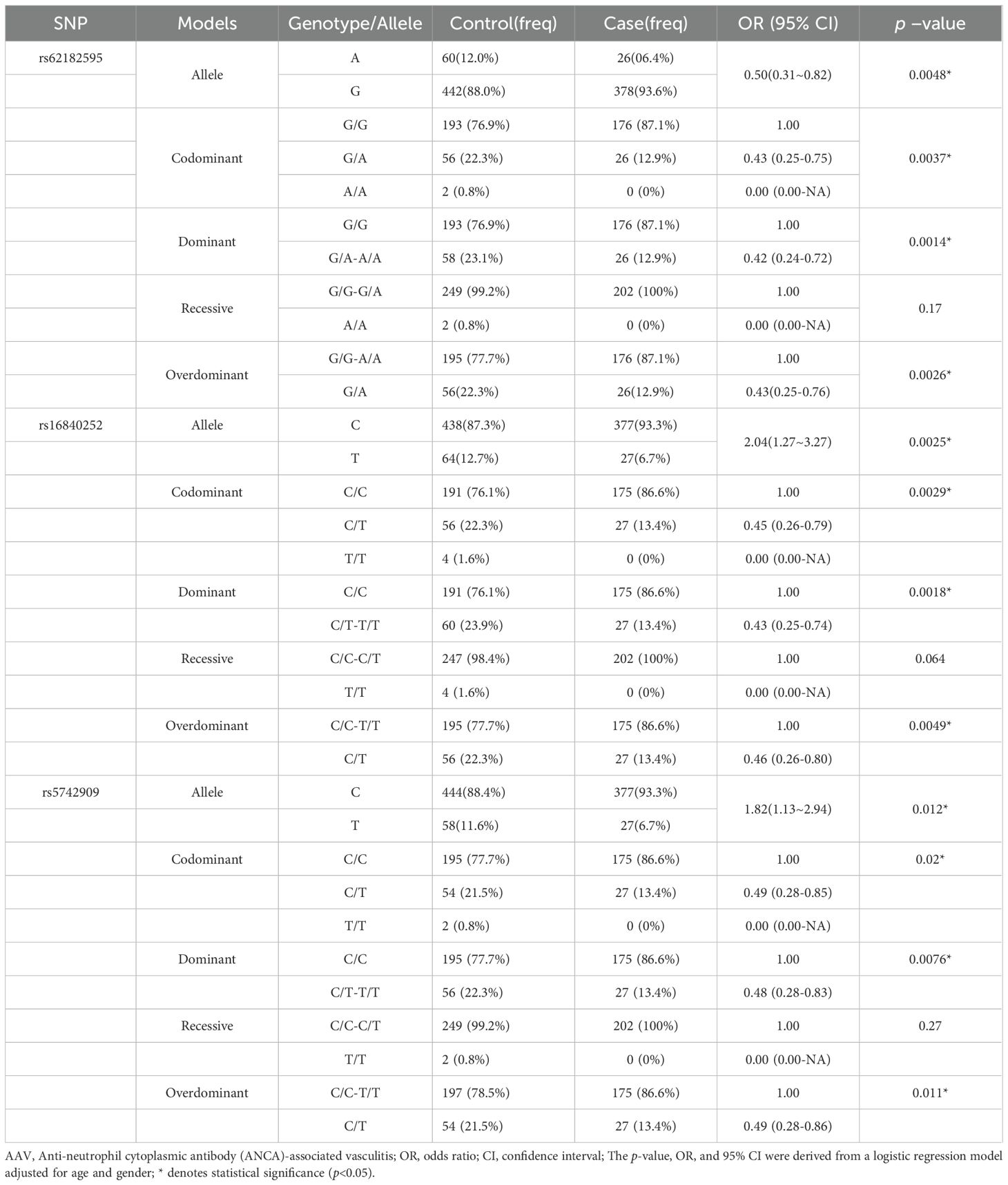
Table 5. The relationship between the SNPs and the risk of AAV in Han of ethnicity Guangxi population in different genetic models.
3.5 Linkage disequilibrium and haplotype analyses
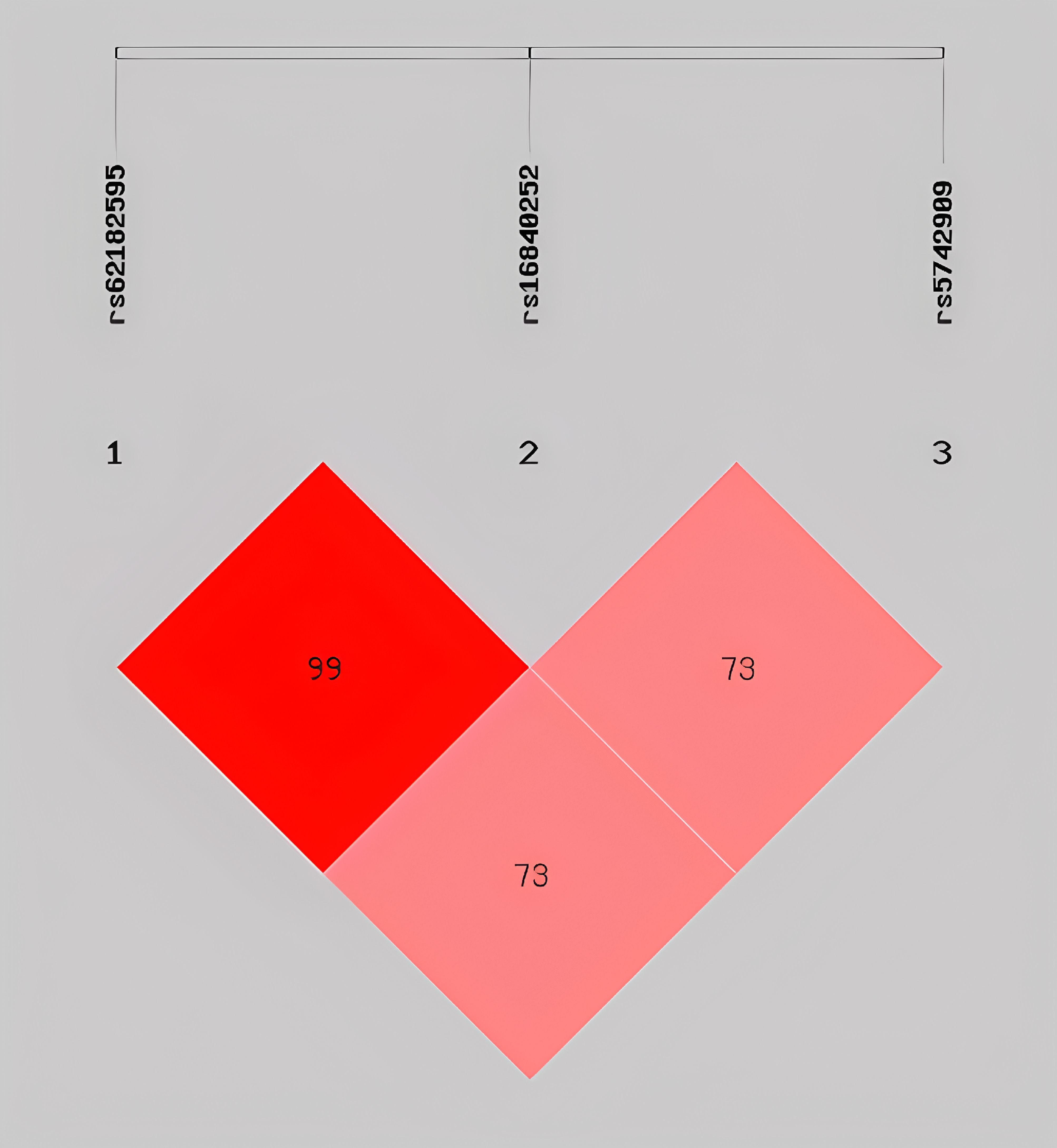
To evaluate the interactions among CTLA4 SNPs, we performed LD and haplotype analyses using the SHEsis online platform. Strong LD was observed between rs62182595 and rs16840252 (D’ = 1), whereas moderate LD was observed between rs62182595 and rs5742909 (D’ = 0.739) and between rs16840252 and rs5742909 (D’ = 0.735) (Figure 1).
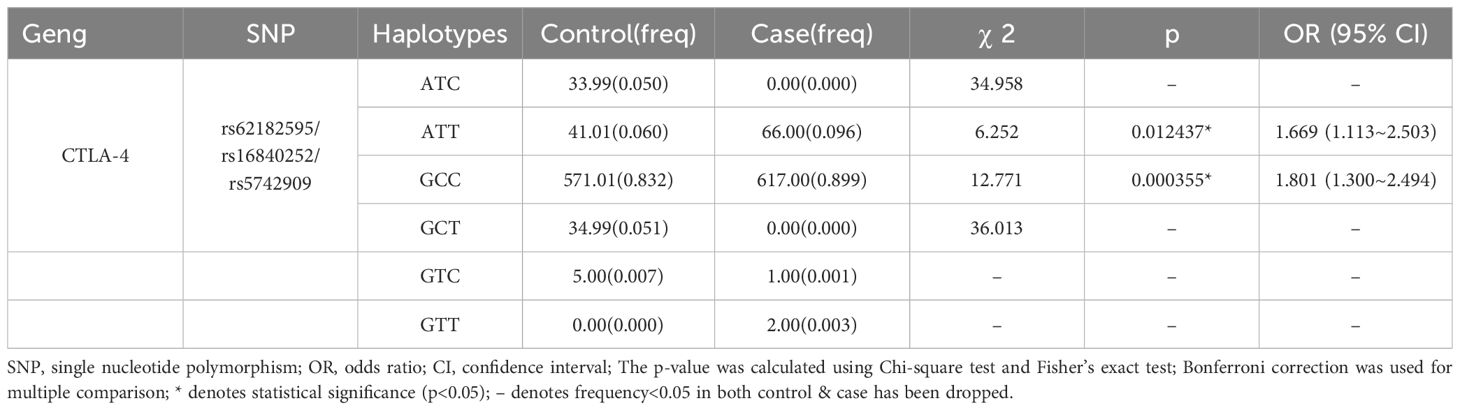
The haplotype analysis identified six haplotypes based on the three SNPs (Table 6). Four of these haplotypes had a frequency of less than 0.05 (24), suggesting a low frequency of occurrence in the population and unstable inheritance, and were therefore excluded from further analyses. Two haplotypes, ATT and GCC, with frequencies greater than 0.05 significantly elevated the risk of AAV development in the Guangxi population (ATT: OR=1.67, 95% CI: 1.113–2.503, p = 0.01; GCC: 1.80, 1.300–2.494, p = 0.000355).
4 Discussion
The pathogenesis of AAV involves the complex interplay of host immunological dysregulation (encompassing both innate and adaptive immunity) and genetic–environmental synergism (4). The breakdown of ANCA tolerance, which is critical to disease initiation, is frequently driven by aberrant immune cell dynamics and humoral immunity deficiencies (25, 26). Nevertheless, emerging evidence from large-scale genomic investigations has substantiated the pivotal role of genetic predisposition in AAV development (7, 9, 27). In this study, we focused on identifying the risk alleles associated with AAV mechanisms and evaluated the correlation between CTLA4 SNPs and AAV risk in the Guangxi population in China. Our findings expand the current understanding of genetic loci associated with AAV susceptibility. CTLA-4 is a master immune checkpoint receptor that delivers co-inhibitory signals crucial for attenuating excessive immune activation (28) and orchestrating immune response termination (29). Although its central regulatory role in maintaining immune homeostasis and preventing autoreactivity is well-established, the precise molecular mechanisms governing CTLA-4-mediated immune modulation remain incompletely elucidated (30). CTLA4 polymorphisms demonstrate significant pleiotropic effects, showing associations with multiple immune-mediated pathologies including systemic lupus erythematosus, rheumatoid arthritis, autoimmune diabetes, and allograft rejection episodes (31–33).
Our data show that CTLA4 rs62182595, rs16840252, and rs5742909 polymorphisms are significantly associated with AAV susceptibility in the Guangxi population under various genetic models. Specifically, the GA genotype of rs62182595 in the codominant and overdominant genetic models and the GA-AA genotype in the dominant genetic model were associated with reduced AAV susceptibility in the Guangxi population. Similarly, the CT-TT and C/T genotypes of rs16840252 (dominant genetic model) and the C/T genotype of rs5742909 (overdominant genetic model) showed association with reduced AAV risk in the Guangxi population.
Next, we analyzed the synergistic effects of haplotype combinations to assess the possible interactions among CTLA4 SNPs and determine the effects of these three SNPs on AAV susceptibility in the Guangxi population. Among the six haplotypes identified, ATT and GCC haplotypes significantly elevated susceptibility, suggesting an interaction between the three SNPs that increases the susceptibility to AAV in the Guangxi population, similar to the results of a previous study on CTLA-4 (34). Furthermore, previous studies suggest that SNP polymorphisms in the 5′ URR region may lead to altered selective splicing (35–37), and alterations in selective splicing can lead to variation in CTLA4. Therefore, we hypothesized that alterations in bases at rs1684025 may alter the selective splicing of both the soluble CTLA-4 (sCTLA-4) and the surface full-length CTLA-4 (flCTLA-4) isoforms, ultimately affecting CTLA-4 gene expression (38). The remaining two SNPs, rs5742909 and rs62182595, located in the promoter region, may lead to different levels of mRNA transcription and protein translation and affect T cell homeostasis, leading to immune dysfunction and ultimately affecting the susceptibility of individuals to AAV (39, 40). Because rs62182595*G, rs16840252*C, and rs5742909*C showed strong LD, we hypothesize that these three loci are likely to amplify the synergistic effects between the loci through gene–gene interactions (formation of different haplotypes). Such interactions could affect the expression level and protein structure of CTLA4, impair T-cell function, and disrupt immune homeostasis, ultimately influencing the susceptibility of individuals to AAV (41–43). Overall, our study demonstrates a significant association between CTLA4 SNPs and AAV risk in the Guangxi population. These findings not only lay the foundation for disease risk stratification (e.g., specific genes can serve as potential genetic markers) to help guide screening of high-risk populations and provide targets for subsequent precise prevention and early intervention, but also explore the potential of CTLA4-targeted precision therapy. It supports the rationale for using CTLA4 as a therapeutic target and provides evidence for “genotype-guided personalized immunotherapy.
Despite its strengths, the present study has certain limitations. First, although haplotype association analysis was conducted, we did not evaluate the expression levels of the CTLA4 gene or protein. As gene expression is affected by numerous factors, such as transcriptional and translational regulations and gene–environmental interactions, our findings require further experimental validation. Second, sample collection was limited due to the rarity of ANCA-associated vasculitis. Third, the study population was restricted to Guangxi, China, which may limit the generalizability of the findings to other populations. In the future, it is planned to carry out cross-regional multi-center collaborative studies. While expanding the sample size, populations of different ethnic groups will be included, and the expression levels of the CTLA4 gene and protein will be detected simultaneously. Combined with the analysis of gene-environment interactions, the research conclusions will be comprehensively verified.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by Medical Ethics Committee of The Second Affiliated Hospital of Guangxi Medical University (2018 KY-0100). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
KB: Methodology, Writing – original draft. BY: Writing – original draft, Formal Analysis, Data curation. PH: Validation, Methodology, Writing – original draft. CX: Writing – review & editing, Project administration.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by Joint Project on Regional High-Incidence Diseases Research of Guangxi Natural Science Foundation under Grant (2025GXNSFAA069077), Joint Project on Regional High-Incidence Diseases Research of Guangxi Natural Science Foundation under Grant (2024GXNSFAA999092), Guangxi Traditional Chinese medicine suitable technology development and promotion project (GZSY2025075).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1631088/full#supplementary-material
References
1. Kitching AR, Anders HJ, Basu N, Brouwer E, Gordon J, Jayne DR, et al. ANCA-associated vasculitis. Nat Rev Dis Primers. (2020) 6:71. doi: 10.1038/s41572-020-0204-y
2. Puechal X. Granulomatosis with polyangiitis (Wegener’s). Joint Bone Spine. (2020) 87:572–8. doi: 10.1016/s0039-6257(97)00133-1
3. Aiyegbusi O, Frleta-Gilchrist M, Traynor JP, Mackinnon B, Bell S, Hunter RW, et al. ANCA-associated renal vasculitis is associated with rurality but not seasonality or deprivation in a complete national cohort study. RMD Open. (2021) 7:e001555. doi: 10.1136/rmdopen-2020-001555
4. Hunter RW, Welsh N, Farrah TE, Gallacher PJ, and Dhaun N. ANCA associated vasculitis. BMJ. (2020) 369:m1070. doi: 10.1136/bmj.m1070
5. Treccani M, Veschetti L, Patuzzo C, Malerba G, Vaglio A, and Martorana D. Genetic and non-genetic contributions to eosinophilic granulomatosis with polyangiitis: current knowledge and future perspectives. Curr Issues Mol Biol. (2024) 46:7516–29. doi: 10.3390/cimb46070446
6. d’Alessandro M, Conticini E, Bergantini L, Mezzasalma F, Cameli P, Baglioni S, et al. PD1, CTLA4 and TIGIT expression on T and NK cells in granulomatous diseases: sarcoidosis and ANCA-associated vasculitis. Int J Mol Sci. (2022) 24(1):256. doi: 10.3390/ijms24010256
7. Lyons PA, Peters JE, Alberici F, Liley J, Coulson RMR, Astle W, et al. Genome-wide association study of eosinophilic granulomatosis with polyangiitis reveals genomic loci stratified by ANCA status. Nat Commun. (2019) 10:5120. doi: 10.1038/s41467-019-12515-9
8. Li W, Huang H, Cai M, Yuan T, and Sheng Y. Antineutrophil cytoplasmic antibody-associated vasculitis update: genetic pathogenesis. Front Immunol. (2021) 12:624848. doi: 10.3389/fimmu.2021.624848
9. Kawasaki A and Tsuchiya N. Advances in the genomics of ANCA-associated vasculitis-a view from East Asia. Genes Immun. (2021) 22:1–11. doi: 10.1038/s41435-021-00123-x
10. Ohno R and Nakamura A. Advancing autoimmune Rheumatic disease treatment: CAR-T Cell Therapies - Evidence, Safety, and future directions. Semin Arthritis Rheum. (2024) 67:152479. doi: 10.1016/j.semarthrit.2024.152479
11. Smith RM, Jones RB, Guerry MJ, Laurino S, Catapano F, Chaudhry A, et al. Rituximab for remission maintenance in relapsing antineutrophil cytoplasmic antibody-associated vasculitis. Arthritis Rheum. (2012) 64:3760–9. doi: 10.1002/art.34583
12. Van Coillie S, Wiernicki B, and Xu J. Molecular and cellular functions of CTLA-4. Adv Exp Med Biol. (2020) 1248:7–32. doi: 10.1007/978-981-15-3266-5_2
13. Oyewole-Said D, Konduri V, Vazquez-Perez J, Weldon SA, Levitt JM, and Decker WK. Beyond T-cells: functional characterization of CTLA-4 expression in immune and non-immune cell types. Front Immunol. (2020) 11:608024. doi: 10.3389/fimmu.2020.608024
14. Hosseini A, Gharibi T, Marofi F, Babaloo Z, and Baradaran B. CTLA-4: From mechanism to autoimmune therapy. Int Immunopharmacol. (2020) 80:106221. doi: 10.1016/j.intimp.2020.106221
15. Nozaki Y. New insights into novel therapeutic targets in ANCA-associated vasculitis. Front Immunol. (2021) 12:631055. doi: 10.3389/fimmu.2021.631055
16. Carr EJ, Niederer HA, Williams J, Harper L, Watts RA, Lyons PA, et al. Confirmation of the genetic association of CTLA4 and PTPN22 with ANCA-associated vasculitis. BMC Med Genet. (2009) 10:121. doi: 10.1186/1471-2350-10-121
17. Wu Z, Wu Q, Xu J, Chen S, Sun F, Li P, et al. HLA-DPB1 variant rs3117242 is associated with anti-neutrophil cytoplasmic antibody-associated vasculitides in a Han Chinese population. Int J Rheum Dis. (2017) 20:1009–15. doi: 10.1111/1756-185X.12561
18. Jennette JC, Falk RJ, Bacon PA, Basu N, Cid MC, Ferrario F, et al. revised International Chapel Hill Consensus Conference Nomenclature of Vasculitides. Arthritis Rheum. (2012) 65:1–11. doi: 10.1002/art.37715
19. Yang B, Chu L, Feng F, Lu S, and Xue C. Association of tyrosine kinase 2 polymorphisms with susceptibility to microscopic polyangiitis in a Guangxi population. PeerJ. (2024) 12:e18735. doi: 10.7717/peerj.18735
20. Liang G, Fu W, and Wang K. Analysis of t-test misuses and SPSS operations in medical research papers. Burns Trauma. (2019) 7:31. doi: 10.1186/s41038-019-0170-3
21. Sole X, Guino E, Valls J, Iniesta R, and Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. (2006) 22:1928–9. doi: 10.1093/bioinformatics/btl268
22. Li Z, Zhang Z, He Z, Tang W, Li T, Zeng Z, et al. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis (http://analysis.bio-x.cn). Cell Res. (2009) 19:519–23. doi: 10.1038/cr.2009.33
23. Shi YY and He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. (2005) 15:97–8. doi: 10.1038/sj.cr.7290272
24. Alper CA and Larsen CE. Pedigree-defined haplotypes and their applications to genetic studies. Methods Mol Biol. (2017) 1551:113–27. doi: 10.1007/978-1-4939-6750-6_6
25. Hutton HL, Holdsworth SR, and Kitching AR. ANCA-associated vasculitis: pathogenesis, models, and preclinical testing. Semin Nephrol. (2017) 37:418–35. doi: 10.1016/j.semnephrol.2017.05.016
26. Mueller A, Zhao Y, Cicek H, Paust HJ, Sivayoganathan A, Linke A, et al. Transcriptional and clonal characterization of cytotoxic T cells in crescentic glomerulonephritis. J Am Soc Nephrol. (2023) 34:1003–18. doi: 10.1681/ASN.0000000000000116
27. Alberici F, Martorana D, Bonatti F, Gioffredi A, Lyons PA, and Vaglio A. Genetics of ANCA-associated vasculitides: HLA and beyond. Clin Exp Rheumatol. (2014) 32:S90–97.
28. Edner NM, Carlesso G, Rush JS, and Walker LSK. Targeting co-stimulatory molecules in autoimmune disease. Nat Rev Drug Discov. (2020) 19:860–83. doi: 10.1038/s41573-020-0081-9
29. Hossen MM, Ma Y, Yin Z, Xia Y, Du J, Huang JY, et al. Current understanding of CTLA-4: from mechanism to autoimmune diseases. Front Immunol. (2023) 14:1198365. doi: 10.3389/fimmu.2023.1198365
30. Rowshanravan B, Halliday N, and Sansom DM. CTLA-4: a moving target in immunotherapy. Blood. (2018) 131:58–67. doi: 10.1182/blood-2017-06-741033
31. Mousavi MJ, Shayesteh MRH, Jamalzehi S, Alimohammadi R, Rahimi A, Aslani S, et al. Association of the genetic polymorphisms in inhibiting and activating molecules of immune system with rheumatoid arthritis: A systematic review and meta-analysis. J Res Med Sci. (2021) 26:22. doi: 10.4103/jrms.JRMS_567_20
32. Chen DP, Lin WT, and Yu KH. Investigation of the association between the genetic polymorphisms of the co-stimulatory system and systemic lupus erythematosus. Front Immunol. (2022) 13:946456. doi: 10.3389/fimmu.2022.946456
33. Liu W, Yang Z, Chen Y, Yang H, Wan X, Zhou X, et al. The association between CTLA-4, CD80/86, and CD28 gene polymorphisms and rheumatoid arthritis: an original study and meta-analysis. Front Med (Lausanne). (2021) 8:598076. doi: 10.3389/fmed.2021.598076
34. Pyo JY, Yoon T, Ahn SS, Song JJ, Park YB, and Lee SW. Soluble immune checkpoint molecules in patients with antineutrophil cytoplasmic antibody-associated vasculitis. Sci Rep. (2022) 12:21319. doi: 10.1038/s41598-022-25466-x
35. Ueda H, Howson JM, Esposito L, Heward J, Snook H, Chamberlain G, et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. (2003) 423:506–11. doi: 10.1038/nature01621
36. Giri PS, Dwivedi M, Laddha NC, Begum R, and Bharti AH. Altered expression of nuclear factor of activated T cells, forkhead box P3, and immune-suppressive genes in regulatory T cells of generalized vitiligo patients. Pigment Cell Melanoma Res. (2020) 33:566–78. doi: 10.1111/pcmr.12862
37. Teft WA, Kirchhof MG, and Madrenas J. A molecular perspective of CTLA-4 function. Annu Rev Immunol. (2006) 24:65–97. doi: 10.1146/annurev.immunol.24.021605.090535
38. Misra MK, Pandey SK, Kapoor R, Sharma RK, and Agrawal S. Cytotoxic T-lymphocyte antigen 4 gene polymorphism influences the incidence of symptomatic human cytomegalovirus infection after renal transplantation. Pharmacogenet Genomics. (2015) 25:19–29. doi: 10.1097/FPC.0000000000000102
39. Wen YH, Lin WT, Wang WT, Chiueh TS, and Chen DP. Association of CTLA4 gene polymorphism with transfusion reaction after infusion of leukoreduced blood component. J Clin Med. (2019) 8(11):1961. doi: 10.3390/jcm8111961
40. Mikhaylichenko O, Bondarenko V, Harnett D, Schor IE, Males M, Viales RR, et al. The degree of enhancer or promoter activity is reflected by the levels and directionality of eRNA transcription. Genes Dev. (2018) 32:42–57. doi: 10.1101/gad.308619.117
41. Hradsky O, Dusatkova P, Lenicek M, Bronsky J, Nevoral J, Vitek L, et al. The CTLA4 variants may interact with the IL23R- and NOD2-conferred risk in development of Crohn’s disease. BMC Med Genet. (2010) 11:91. doi: 10.1186/1471-2350-11-91
42. Al-Harbi N, Abdulla MH, Vaali-Mohammed MA, Bin Traiki T, Alswayyed M, Al-Obeed O, et al. Evidence of association between CTLA-4 gene polymorphisms and colorectal cancers in saudi patients. Genes (Basel). (2023) 14(4):874. doi: 10.3390/genes14040874
Keywords: CTLA-4, vasculitis, T-cell, autoimmune diseases, single-nucleotide polymorphism
Citation: Bu K, Yang B, He P and Xue C (2025) Cytotoxic T lymphocyte-associated protein 4 gene polymorphisms are associated with ANCA-associated vasculitis in the Guangxi population of China. Front. Immunol. 16:1631088. doi: 10.3389/fimmu.2025.1631088
Received: 19 May 2025; Accepted: 11 August 2025;
Published: 09 September 2025.
Edited by:
Piero Ruscitti, University of L’Aquila, ItalyReviewed by:
Michał Jakubaszek, Rheumatology and Rehabilitation, PolandSuresh Babu Sayana, Bhadradri Kothagudem, India
Copyright © 2025 Bu, Yang, He and Xue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chao Xue, eHVlY2hhb0BzdHUuZ3htdS5lZHUuY24=
 Kunpeng Bu
Kunpeng Bu Binglan Yang
Binglan Yang Peigeng He1
Peigeng He1 Chao Xue
Chao Xue