Abstract
Introduction:
There is an urgent need for influenza vaccine strategies that enhance protection against influenza virus drift and across different subtypes. The conserved viral nucleoprotein (NP) is the most abundant viral protein during replication, and a target for broadly protective cellular immune responses.
Methods:
Guided by annual WHO-recommended seasonal vaccine strains, we engineered synthetic DNA vaccine candidates encoding vaccine-aligned common consensus (VACC) immunogens designed to represent the immune diversity of seasonal H1N1 and H3N2 virus NP proteins (pVACC-NPH1; pVACC-NPH3).
Results:
Both pVACC-NPH1 and pVACC-NPH3 DNA vaccines induced robust cellular immune responses in mice, including the induction of durable responses. Immunization with a single dose of either DNA vaccine 14 days prior to lethal A/California/2009 H1N1 virus challenge provided protection against mortality. Single dose co-administration of pVACC-NPH3 with an HA-expressing DNA vaccine (pHAH1) and plasmid-encoded adjuvant pIL-12 afforded improved protection against morbidity and mortality in a high-dose challenge model.
Discussion:
These data highlight the potential of heterologous cellular immunity induced by engineered NP immunogens to complement HA-based approaches to significantly improve challenge outcomes.
1 Introduction
Seasonal influenza viruses infect approximately 1 billion people each year (1), causing respiratory illnesses across both hemispheres. An estimated 3–5 million of these cases result in severe illness, with 290-650, 000 deaths annually (2). Although yearly vaccination against circulating influenza A virus (IAV) H1N1 and H3N2, and B virus strains is recommended, genetic variation necessitates annual reformulation (3–6). Broad and universal influenza vaccines are urgently needed to protect from circulating and newly emerging influenza viruses. In addition to strategies that induce broadly protective antibodies against the viral surface hemagglutinin (HA) protein (7–10), synthetic immunogen approaches (11–14) that direct protective immunity to target highly conserved epitopes or proteins could provide important adjunctive protection to decrease pathogenesis and severe disease.
The influenza nucleoprotein (NP) is the most abundantly expressed protein during viral replication and the major component of the virion ribonucleoprotein complex (15). It plays a critical role in viral replication, involving organization of RNA packing, nuclear trafficking, vRNA transcription. and replication (16). NP is well-conserved within influenza subtypes (17–21), making it a promising target for inducing cellular immune responses. NP has been shown to induce robust CD8+ T cell responses in preclinical models (22, 23) and humans (24). Computational modeling of influenza isolates has revealed stretches of highly conserved amino acids within NP. Peptide vaccines based on such epitopes elicit robust CD8+ T cell responses and are protective against IAV challenge in mice (18). Epidemiological studies indicate that anti-NP CD8+ T cell immunity can contribute to protection from severe disease in humans (25). These data suggest that NP based therapies have the potential to elicit broad anti-influenza cellular immunity.
Synthetic plasmid DNA vaccines have advanced significantly over the past ten years, demonstrating robust induction of humoral and cellular immune responses (26). The first DNA vaccine received EUA for use in humans during COVID-19 (27) and several T cell-based DNA vaccines are being evaluated for infectious diseases and delivery of cancer neoepitopes (28) to elicit CD8+ T cell responses. Current inactivated vaccines elicit poor CD8+ T cell responses compared to live attenuated influenza vaccines (LAIV) (29–31). Although LAIV vaccines can induce CD8+ T cell responses, the master donor virus used to make all LAIVs contains the internal genes, including NP, of A/Ann Arbor/6/60 or A/Leningrad/17/57 H2N2 viruses and is thus mismatched to modern circulating strains. To this end, studies matching LAIV vaccines to currently circulating viruses can increase induction of CD8+ T cell responses (32). Building on this prior research, we hypothesized that plasmid DNA-encoded NP consensus immunogens could expand the breadth of protection, eliciting broad cellular immunity which could reduce IAV pathogenesis.
Here, we describe the design and evaluation of synthetic IAV-NP immunogens engineered based on WHO-recommended vaccine strains to induce robust anti-influenza cellular immunity in vivo. Two plasmid DNA-encoded vaccine-aligned common consensus (VACC) immunogens representing the NPs from seasonal A/H1N1 (VACC-NPH1) or A/H3N2 (VACC-NPH3) viruses induced robust cellular immune responses, with both independently providing single dose protection against mortality in mice intranasally challenged with an A/California/2009 virus. We delivered these antigens alone or in combination with plasmid-encoded IL-12 (pIL-12) which has been demonstrated to enhance cellular responses to DNA antigens in mice, non-human primates (33, 34), and humans in clinical trials (35–37). Heterologous pVACC-NPH3 combination with plasmid-encoded hemagglutinin (HA) from H1N1 A/California/07/2009 (pHAH1) and pIL-12 afforded complete protection from IAV-associated morbidity and mortality, further highlighting the potential for synthetic VACC-NPX candidates to reduce pathogenesis and provide immune protective benefit across IAV subtypes.
2 Methods
2.1 Plasmid design
The amino acid sequences for NP proteins from WHO recommended H1N1 and H3N2 vaccine strains selected from 2000–2019 vaccine strains (38) were downloaded from the GISAID.org database. H1N1 NP accession #: A/New Caledonia/20/1999 (EPI ISL 649), A/Solomon Islands/3/2006 (EPI224787), A/Brisbane/59/2007 (EPI ISL 154495), A/California/07/2009 (EPI ISL 391380), A/Michigan/45/2015 (EPI ISL 199532)A/Brisbane/02/2018 (EPI ISL 344858), A/Wisconsin/588/2019 (EPI ISL 404527), A/Hawaii/70/2019 (EPI ISL 397028). H3N2 NP accession #: A/Moscow/10/1999 (EPI ISL 2695), A/Fujian/411/2002 (EPI ISL 107711), A/California/7/2004 (EPI ISL 113070), A/Wisconsin/67/2005 (EPI ISL 154528), A/Brisbane/10/2007 (EPI ISL 176458), A/Perth/16/2009 (EPI ISL 176456), A/Victoria/361/2011 (EPI ISL 101506), A/Switzerland/9715293/2013 (EPI ISL 166310), A/Hong Kong/4801/2014 (EPI ISL 233740), A/Singapore/INFIMH-16-0019/2016 (EPI2397166), A/Kansas/14/2017 (EPI ISL 292575), A/Hong Kong/45/2019 (EPI ISL 347938). H1NP or H3NP vaccine-consensus designs were constructed through sequence alignment analysis in MEGA 11.0.10 (39) using ClustalW alignment and an unrooted phylogenetic tree was generated using the maximum-likelihood method, with maximum parsimony (40). Pairwise distances were calculated in MEGA. 11.0.10. Sequence identity visualization was performed in Treeviewer (41). Additional alignment of sequences were performed in Geneious Prime (version 2023.2.1). mRNA expression was confirmed by qPCR using the following primers: NPH1 Forward: (5’-GATCTCTGTGCAGCCTACCT-3’), Reverse (5’-ATCACTTCTGTGCGCATGTC-3’), NPH3 Forward (5’-TCTGCCTTTGACGAGAGGAG-3’), Reverse (5’-CCGCCAGATTCTCCTGATCT-3’), and mouse GAPDH (NM_008084) CAT#: MP205604) Forward (5’-CATCACTGCCACCCAGAAGACTG-3’), Reverse (5’-ATGCCAGTGAGCTTCCCGTTCAG3’), all will amplicon sizes of 120 base pairs. Analysis was performed using comparative delta-delta CT analysis. DNA plasmid encoding the full-length codon-optimized, HA protein of A/California/07/2009 H1N1pdm09 cloned into the pVax1 vector (pHAH1) was previously described in (42). The plasmid-encoded adjuvant mouse interleukin 12 (IL12) has been previously described in (43).
2.2 Cell lines and virus propagation
Influenza A Virus, A/California/07/2009 NYMC X-179A H1N1pdm09 (Ca09-X179A) (IRR catalog: FR-246), was obtained through the International Reagent Resource, Influenza Division, WHO Collaborating Center for Surveillance, Epidemiology and Control of Influenza, Centers for Disease Control and Prevention, Atlanta, GA, USA. This is a reassortant virus with the HA, NA, and PB1 genes from H1N1pdm09 and remaining genes from A/Puerto Rico/8/1934. MDCK-SIAT1 cells (Sigma Cat# 5071502) were maintained in Minimum Essential Medium (Eagle’s) (Corning Cat # MT10009CV) with 1% Penicillin/Streptomycin (Gibco Cat #15140122), and 2% fetal bovine serum (Peak Cat #PS-FB4). For virus propagation, cell monolayers were infected with MOI 0.001 of Ca09-X179A in the presence of 2 µg/mL TPCK-treated Trypsin (ThermoFisher Cat# 20233) and maintained with 1% Pen/Strep, 0.3% bovine serum albumin (Gibco Cat # 15260037) for 3 days. Virus was collected and ultracentrifuged on a sucrose gradient to prepare mouse challenge stocks. Challenge stocks were titered by determining the 50% tissue culture infectious dose (TCID50) on MDCK-SIAT1 cells and an initial mouse 50% lethal dose (LD50) experiment was performed to determine the minimum infectious dose for challenge.
2.3 Animals, immunization, and challenge
C57BL/6J (Stock # 000664) and DBA/2J (Stock # 000671) female mice were purchased from the Jackson Laboratory and were housed in the Wistar Institute Animal Facility. All procedures were done in accordance with the guidelines from the Wistar Institute Animal Care and Use Committee. Between 2 μg to 10 μg of DNA plasmid encoding the VACC-NPH1 or VACC-NPH3 or full length HA DNA (pHAH1) (42) with or without a DNA plasmid encoding for the molecular adjuvant IL-12, in 30 μL water was injected in the tibialis anterior (TA) muscle. Delivery was immediately followed with two 0.1 Amp electric constant current square-wave pulses by the CELLECTRA-3P electroporation device (Inovio Pharmaceuticals) to increase transfection efficiency. Immunized or naive DBA/2J mice were intranasally infected with 10 LD50 or 100 LD50 of Ca09-X179A respectively in 50 µl MEM Eagle’s (without antibiotics). Mice were then monitored for the subsequent 21 days, for weight loss and mortality. Any mouse reaching 80% of their original body weight was considered to have reached humane endpoint and was subsequently euthanized. A subset of mice (n=3 per group) was euthanized on day 6 post infection, and lungs were collected for histopathological analysis. The vaccine and challenge schedules are indicated in each figure.
2.4 Western blot
HEK293T cells (ATCC Cat# CRL-3216) were cultured in DMEM medium with 10% FBS at 37 °C/5% CO2 condition and transfected with pDNA using Lipofectamine 3000 transfection reagent (Thermo Fisher Scientific Cat# L300000) following the manufacturer’s protocol. Forty-eight hours later, supernatant and cell lysates were harvested using 1x cell lysis buffer (Cell signaling Cat# 9803). Proteins were separated on a 4–12% BIS-TRIS gel (Thermo Fisher Scientific Cat# NP0322BOX), then following transfer, blots were incubated with an anti-NP monoclonal antibody (Thermo Fisher Cat# PA5-32242), then visualized with horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (Sigma Cat# SAB3701359).
2.5 Peptide reagents
Individual antigen-matched 15mer peptides with 11mer overlaps were synthesized (Genscript, Piscataway, NJ) for NPH1 and NPH3. Peptides were resuspended as a single peptide pool for flow cytometry, four peptide pools for ELISPOT, or 23 peptides per pool for epitope mapping. Individual NP peptides and pool information is listed in Supplementary Table S1. HA peptide pools are as previously described (42). All pools were resuspended in dimethyl sulfoxide (DMSO).
2.6 Flow cytometry
Immunized mice were euthanized, and spleens and lungs were harvested and stored in RPMI 1640 media (Invitrogen Cat# 11875093) supplemented with 10% FBS and 1% Penicillin/Streptomycin (R10). Spleens were processed to single-cell suspension and red blood cells were removed by ACK lysing buffer (Gibco Cat# A1049201). Lungs were processed using the lung dissociation kit/GentleMACS system (Miltenyi Cat# 130-095-927) according to manufacturer’s instruction. Red blood cells were removed by ACK lysing buffer (Gibco Cat# A1049201), and single cells isolated via density gradient centrifugation using lymphosep (MP Biomedicals Cat#: 0916922-CF). Cells were then filtered and counted before being plated for flow cytometry. Cells (1, 000, 000 per well) were seeded in 100 μL of R10 and stimulated with NPH1, the NPH3, or Ca09 HA peptide pools (5 μg/mL per peptide final concentration) in the presence of Protein Transport Inhibitor (eBioscience, San Diego, CA, USA Cat# 00-4980-03). R10 alone and cell Stimulation Cocktail containing phorbol 12-myristate 13-acetate (PMA) and ionomycin (500X, eBioscience, San Diego, CA, USA Cat# 00-4970-93) in R10 were used as negative and positive controls, respectively. Plates were incubated for 6 h at 37 °C with 5% CO2. After stimulation, cells were stained with LIVE/DEAD zombie aqua for viability. CD3, CD4, CD8, TNF-α, IFNγ, and IL-2 fluorochrome conjugated antibodies (BioLegend) were used for surface and intracellular staining. The samples were run on a BD FACSymphony™ A5 SE flow cytometer (BD Biosciences) and analyzed in FlowJo software. Gates were set using fluorescence minus one (FMO) for each stain. Data was exported and analyzed in GraphPad Prism 10.
2.7 ELISpot
Isolates splenocytes and lung lymphocytes (pulmocytes) were subjected to IFNγ ELISpot assay according to the manufacturer’s instructions (Mabtech Cat# 3321-4APW-10). Briefly, plates were washed four times with sterile PBS and blocked with R10 media for two hours. Splenocytes from each animal were seeded in duplicate wells with 200, 000 cells per well in 100µL R10. Cells were stimulated with NPH1, NPH3 or HAH1 peptide pools (5µg/ml per peptide). The peptide pools and matrix peptide pools are listed in Supplementary Table S1. Negative and positive controls were stimulated with DMSO or PMA/ionomycin respectively. Plates were incubated at 37 °C in 5% CO2 for 18 hours and were then developed following the manufacturer’s protocol. Plates were scanned and counted using the Mabtech IRIS™ FluoroSpot/ELISpot reader.
2.8 Histopathology and immunohistochemistry
Whole murine lungs were collected into 10% buffered neutral-buffered formalin for routine histopathological processing. Formalin fixed tissues were paraffin embedded and 4 μm sections were cut and routinely stained with Hematoxylin and Eosin (H&E). Immunohistochemical detection was performed on 4 μm tissue sections using a polyclonal antibody against the IAV nucleoprotein (anti-NP)(Thermo Fisher Cat# PA5-32242). Whole slides were scanned using a Hamamatsu Nanozoomer S60 slide scanner and analyzed using NDP.view 2. Scale bars equal 2.5 mm on whole slide lung images and 50 µm on lung section images.
2.9 RNA-seq
Formalin-fixed paraffin-embedded (FFPE) lung tissue scrolls of 10µM thickness were used for total RNA extraction. Total RNA was quantitated using the Qubit 2.0 Fluorometer (Thermo Fisher, Waltham, MA) and quality of RNA was assessed using the 4200 Tapestation (Agilent, Santa Clara, CA). Libraries for differential gene expression studies were prepared using the Quant Seq 3’ mRNA-Seq V2 Library Prep Kit FWD (Lexogen, Vienna, Austria) as per manufacturer’s instructions starting with an input of 350ng of RNA and 16 cycles of final PCR amplification. Overall library size was determined using the 4200 Tapestation and libraries were quantitated using the Qubit 2.0 Fluorometer. Libraries were pooled and Next Generation Sequencing with a single-end 76 bp run length was done on the Hiseq 1000 (Illumina, San Diego, CA). A minimum of 10M reads per sample was acquired for each sample. Using Cutadapt (44), we removed adapters and polyA in each sample, followed by alignment to the mm10 genome using Bowtie2 within the RSEM pipeline (v1.3.3). Only reads mapping to coding regions were retained. Raw counts and TPM values were generated for downstream analyses. Differential gene expression analysis was conducted using DESeq2 (v1.38.0). Genes with fewer than 10 raw counts were excluded, and DEGs were identified using FDR < 5% and |log2 fold change| ≥ 3. Functional enrichment was performed using Gene Ontology, KEGG pathways, and Ingenuity Pathway Analysis (IPA). Computational analyses were conducted on a Linux-based high-performance computing environment with tools Bowtie2 (v2.4.5), RSEM (v1.3.3), DESeq2 (v1.38.0), and IPA. Inhibited and activated pathway analysis is included as Supplementary Tables S2-S5, S6).”
2.10 Software and statistical analysis
Data was represented in GraphPad Prism version 10. All sequence alignments were determined in MEGA 11.0.10 (39) and Treeviewer (41), flow cytometry data was analyzed using FlowJo version 10.10.0. Image slides were scanned using a Hamamatsu Nanozoomer S60 slide scanner and analyzed using NDP.view vs2. Details on statistical analysis are included in the legend for each figure. The p-value significance is indicated as follows: *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, and comparisons are not significant (ns), unless otherwise denoted.
2.11 Data and code availability
The published article includes all data sets generated or analyzed during this study. Sequencing data was submitted to NCBI GEO database under accession number GSE306862.
3 Results
3.1 Design and expression of VACC-NPX immunogens
NP amino acid sequences for annual seasonal A/H1N1 vaccines strains were obtained from GISAID.org (Supplementary Figures S1, S2) and aligned to produce unrooted phylogenetic trees (Supplementary Figure S1B). Analysis highlighted a >10% amino acid distance between pre-2009 and post-H1N1pdm2009 viruses consistent with the major antigenic shift caused by introduction of a classical swine NP into the A/H1N1pdm09 lineage viruses (45, 46). We therefore focused our design on contemporary H1N1 viruses and generated a single vaccine-aligned consensus construct (VACC) design, using sequence alignments and phylogenetic analysis to weigh amino acids towards post-H1N1pdm2009 NPs (Figures 1A, B). VACC amino acid sequences were codon-optimized for mammalian expression and subcloned into the pVax1 plasmid DNA backbone to generate the pVACC-NPH1 construct (Figure 1C). Similarly, A/H3N2 vaccines strains were obtained from GISAID.org (Supplementary Figures S1, S3). A VACC-NPH3 construct was designed based on sequence alignments and phylogenetic analysis (Figures 1D, E) and cloned into the pVax1 backbone to generate the pVACC-NPH3 construct (Figure 1F). The overall pairwise distances were determined to be <0.2% for post-H1N1pdm09 IAV-NPH1 and <1.1% for IAV-NPH3. We confirmed mRNA expression of VACC-NPX via quantitative PCR (Figure 1G) following in vitro transfection. Protein expression in transfected HEK 293cells was confirmed via supernatant western blot (Figure 1H), and immunofluorescence staining of IAV-NP (Figure 1I). Together these data demonstrate that the consensus alignment approach generates novel synthetic molecules that express in vitro and are detected by commercial anti-NP antibodies.
Figure 1
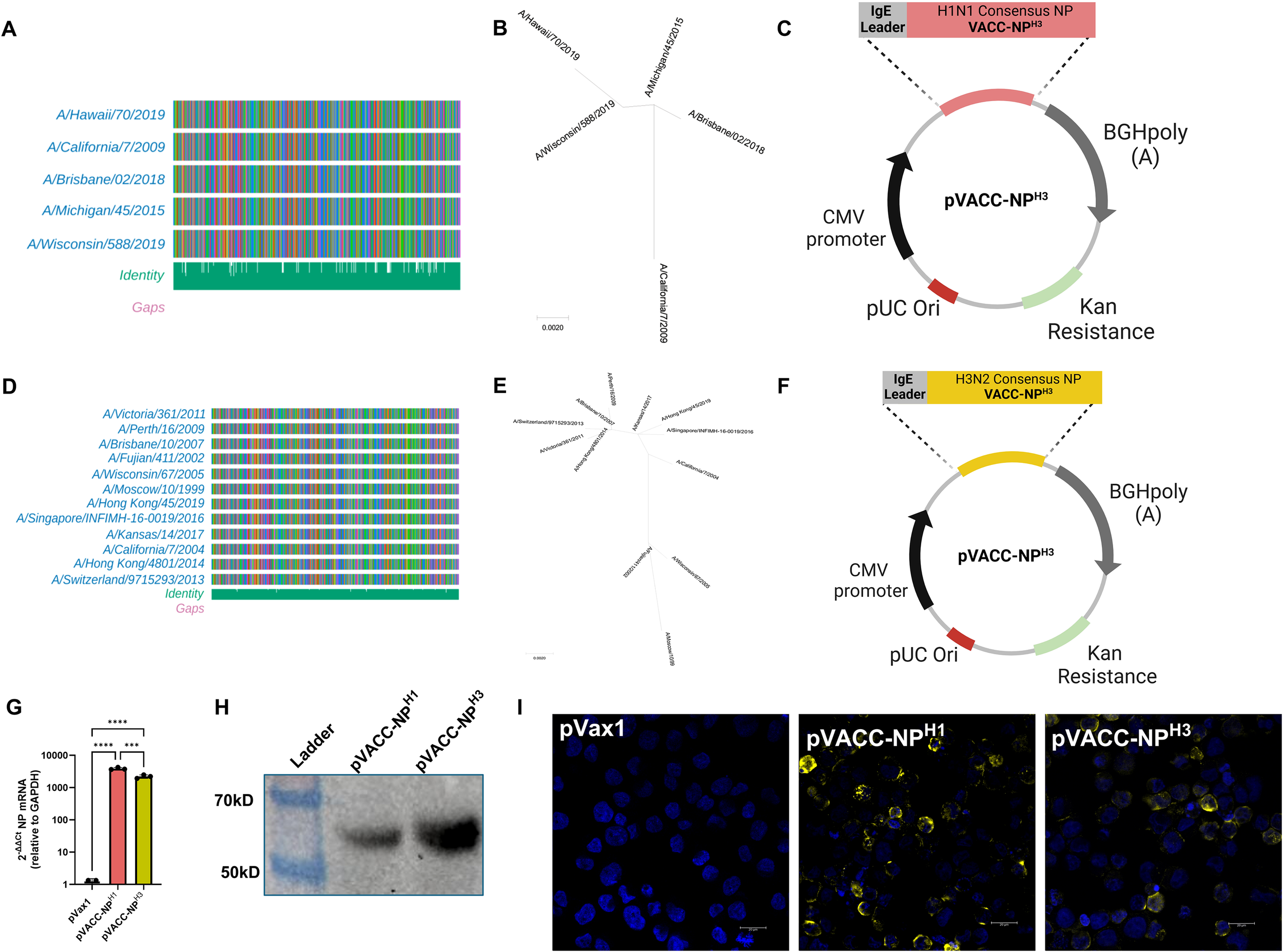
Design and in vitro expression of VACC-NPX immunogens. NP amino acid alignments of seasonal A/H1N1 post-H1N1pdm09 and A/H3N2 vaccine strains (GISAID.org), (A, D). Unrooted phylogenetic trees for A/H1N1 post-H1N1pdm09 and A/H3N2 vaccine strains (B, E). Plasmid maps of pVACC-NPH1 and pVACC-NPH3 synthetic DNA constructs (C, F). mRNA expression of VACC-NPX by quantitative PCR following in vitro transfection (G). Western blot of pVACC-NPH1 and pVACC-NPH3 HEK29T supernatants probed for anti-IAV-NP (H). Immunofluorescence staining of HEK293T cells transfected with pVACC-NPX plasmids and stained for IAV-NP (I). Data are representative of two independent transfection experiments. Symbols (G) represent duplicate assays of three separate wells. ***p<0.001, ****P<0.0001 by Kruskal-Wallis ANOVA.
3.2 A single immunization with DNA-encoded pVACC-NPX vaccines induces strong cellular responses and supports protection from influenza-associated morbidity and mortality in vivo
An initial dosing study in C57BL/6J mice was performed to assess the immunogenicity of 10 µg and 25 µg of the pVACC-NPH3 plasmid following a two-dose injection regimen. Both doses induced robust IFNγ spot-forming units (SFU) in spleens (Supplementary Figure S4). The difference between the two doses was not statistically significant, therefore we selected the lower 10 µg dose for evaluation as a single immunization regimen for both pVACC-NPH1 and pVACC-NPH3 immunogens. C57BL/6J mice were immunized once with 10 µg of pVACC-NPH1 or pVACC-NPH3 immunogens and cellular responses were evaluated by ELISpot assay fourteen days later (Figure 2A). pVACC-NPH1 induced significant NPH1-specific IFNγ SFU in the spleens of immunized mice as compared to empty plasmid (pVax1) immunized controls (Figure 2B),. Similarly, pVACC-NPH3 resulted in significant induction of NPH3-specific IFNγ responses (Figure 2C). These data demonstrate that pVACC-NPX constructs induce strong cellular immunity in vivo.
Figure 2
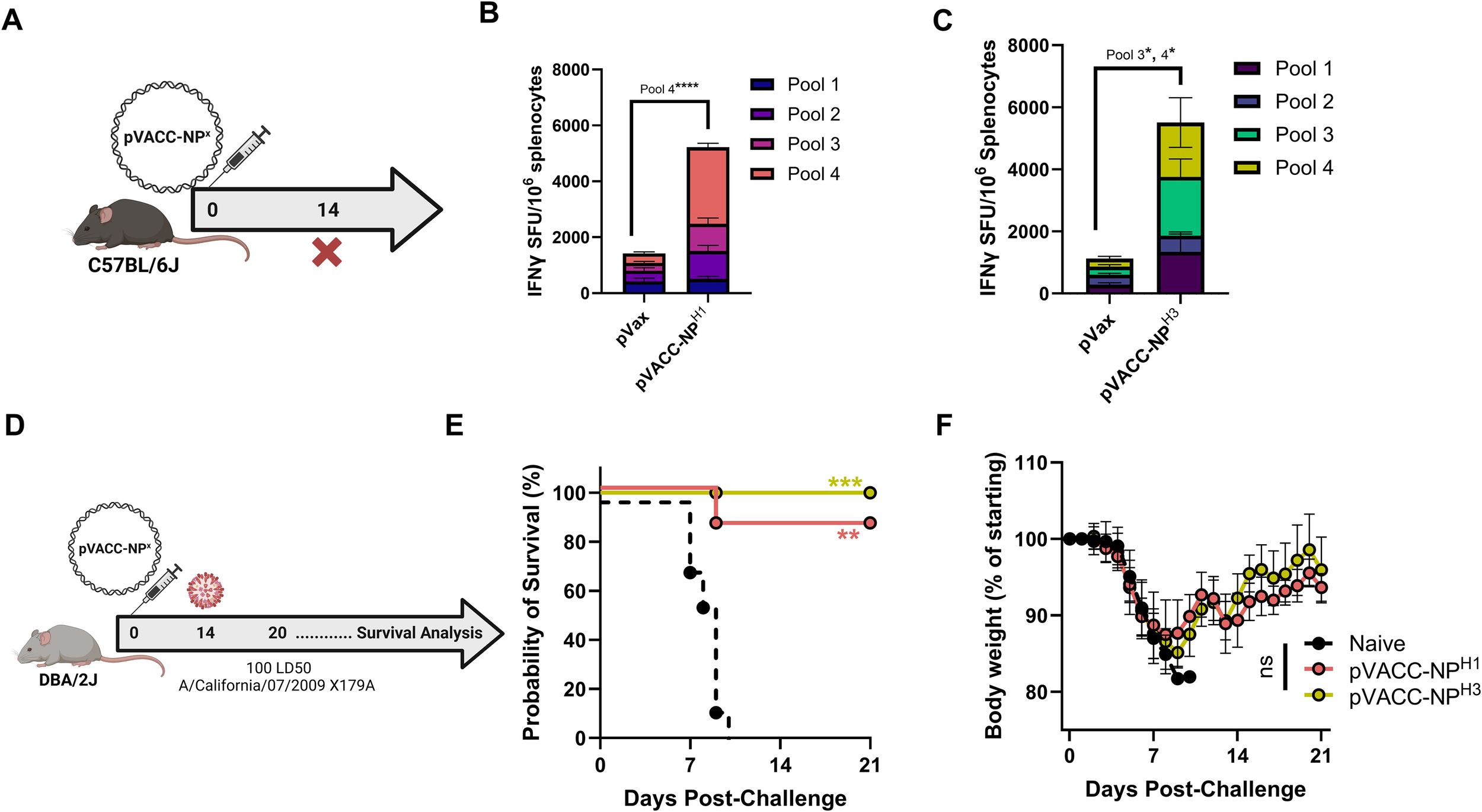
A single immunization with VACC-NPX constructs is immunogenic and protects against mortality in an H1N1pdm09 mouse infection model. C57BL/6J mice were immunized with 10µg of pVACC-NPX plasmids and euthanized fourteen days post-immunization for cellular analyses (A). IFNγ spot-forming units (SFUs) in spleens following stimulation with H1NP peptides or H3NP peptides (n=5 mice per group) (B, C). DBA/2J mice received a single administration of the pVACC-NPH1 or pVACC-NPH3 synthetic DNA vaccines (10µg, n=10 mice/group). After 14 days, the mice were intranasally challenged with 10 LD50 of H1N1 Ca09-X179A and monitored daily until day 21 post-challenge. On day 6 post-infection, a subset of mice (n=3) was euthanized lungs were collected and processed for histopathological analyses (D). Survival probability (E). Weight loss as percent of starting weight (F). Hematoxylin and eosin staining (G), and IAV-NP immunohistochemistry staining (H) of lung sections from representative mice at 6 days post-infection. Scale bars equal 2.5 mm on whole slide lung images. Data are representative of two independent experiments with n=5/group (A–C) and n=10/group (D–H). Bars represent group means and error bars represent SEM (B, C). Lines Symbols represent group averages; bars represent SD (F). *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 by Two-way ANOVA (B, C), Dunnett’s multiple comparison (F), or Mantel-Cox Log-rank test (E).
To evaluate the protective efficacy of these constructs, we used the DBA/2J mouse model of wild-type IAV challenge. DBA/2J mice were immunized once with 10 µg of pVACC-NPH1, pVACC-NPH3, or left unimmunized (naïve), and challenged fourteen days later with 10 LD50 of H1N1 Ca09-X179A (Figure 2D). Strikingly, we observed 90% survival among receiving pVACC-NPH1 and 100% survival among pVACC-NPH3 immunized animals, while all naïve animals succumbed to infection (Figure 2E). Despite surviving the challenge, all pVACC-NPX immunized animals displayed weight loss similar to that observed in naïve animals (Figure 2F), and this was reflected by H&E staining of lungs harvested 6 days post-infection (Figure 2G). Dense cellular infiltrates were observed in the lungs of naive and pVACC-NPH1 immunized animals. pVACC-NPH3 immunized animals displayed decreased cellular infiltrates and increased airway space (Figure 2G). Similarly, when sections were stained for H1N1 NP antigen, naïve animals displayed significant NP-positive staining throughout their lungs. pVACC-NPH1 immunized animals had decreased NP antigen and only minimal staining was observed in the lungs of pVACC-NPH3 immunized mice (Figure 2H). Together, these data highlight the potential for a VACC-NPX immunogen to provide benefit against disease and death following a single immunization.
3.3 Epitope mapping of VACC-NPX-induced cellular responses
A matrix system was used to organize vertical and horizontal peptide pools to identify immunodominant epitopes following immunization with either pVACC-NPH1 and pVACC-NPH3 (Supplementary Table S1). NP vaccines were co-formulated with gene-encoded adjuvant pIL-12, previously reported to enhance cellular (35, 37) and humoral (47) responses in humans and in preclinical models (33, 34, 48, 49) (Supplementary Figure S5A). Using this matrix format enables higher throughput identification of epitopes with limited samples (Supplementary Table S1). Immunodominant epitopes are identified if they demonstrate strong IFNγ responses in one vertical pool and one horizontal pool in the matrix. The peptides are then determined at the intersection of these two pools. In this way, we identified linear peptides ASNENVETM among NPH1 peptides Supplementary Figures S5B, C) and ASNENMDNM among NPH3 peptides (Supplementary Figures S5D, E), consistent with those described for murine H2-Db in the literature (50, 51). pVACC-NPH3 also elicited strong responses to the SAAFEDLRLLSFIRG peptide reported by Lambe et al. (51). Importantly, stimulation with the overlapping peptide pools containing the identified immunodominant epitopes (pool 4 for pVACC-NPH1 and pools 3 and 4 for pVACC-NPH3) resulted in significant increases in IFNγ secretion from immunized mice (Figures 2A, B). These data demonstrate that the pVACC-NPX constructs can elicit responses consistent with previously identified epitopes, as well as can expand unique responses.
3.4 pVACC-NPX antigens are amenable to co-delivery with HA immunogens
Current seasonal influenza vaccines are either inactivated virus, live attenuated virus, or recombinant protein vaccines, driving primarily HA-directed antibody responses. We hypothesized that a synthetic VACC-NPHX immunogen can provide adjunctive protection when administered in combination with an HA vaccine. Both pVACC-NPH1 and pVACC-NPH3 constructs induced similar immunogenicity and protective efficacy however pVACC-NPH3 demonstrated reduced lung pathogenesis following infection. Thus, we selected the heterologous pVACC-NPH3 for evaluation alone and in combination with a pHAH1 antigen. C57BL/6J mice were immunized once with one of the following formulations: 12.5μg of empty plasmid vector (pVax1); 10μg of pVACC-NPH3 alone; pVACC-NPH3 plus 0.5μg pIL-12; 2μg pHAH1 alone; pVACC-NPH3 plus pHAH1; or a combination of pHAH1, pVACC-NPH3, and pIL-12 (Combo). Mice were euthanized fourteen days post-immunization and cellular responses were quantified by intracellular cytokine staining (ICS) (Figures 3A, B).
Figure 3
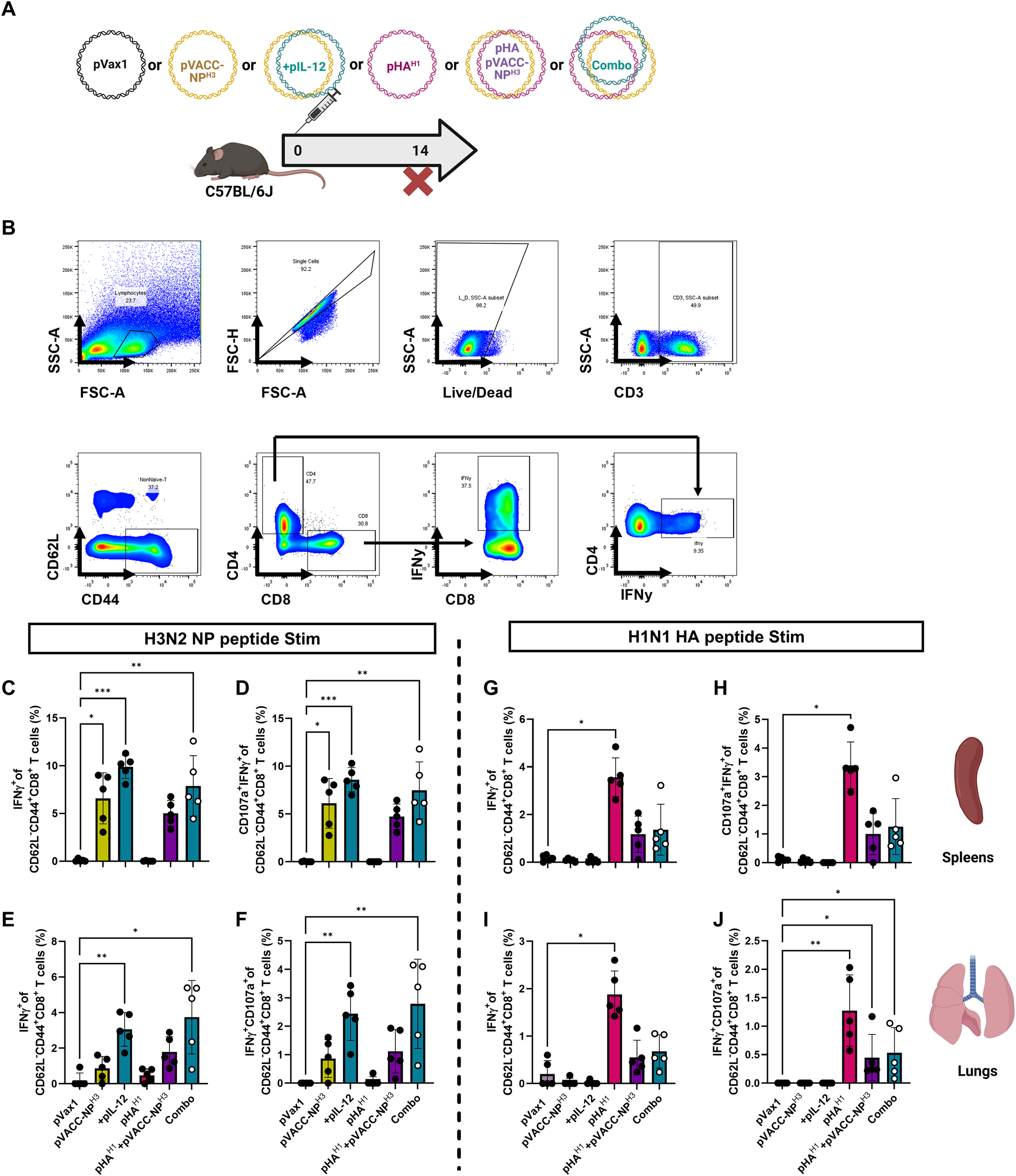
pVACC-NPx immunogens are amenable to co-delivery with HA immunogens and gene-encoded adjuvant pIL-12. (A) C57BL/6J mice were immunized once with one of the following formulations: 12.5μg of empty plasmid vector (pVax1); 10μg of pVACC-NPH3 alone; pVACC-NPH3 plus 0.5μg of plasmid-encoded mouse IL-12 (+pIL-12); 2μg of plasmid-encoded A/California/07/2009 HA (pHAH1) alone; pVACC-NPH3 plus pHAH1; or a combination of pHAH1, pVACC-NPH3, and pIL-12 (Combo). Mice were euthanized fourteen days post-immunization for cellular analyses. (B) Gating strategy for intracellular cytokine staining using IFNγ+ splenocytes as an example. Cytokine positive CD4+ or CD8+ T cells were gated from single/live/CD3+/CD62L-/CD44+ cells. IFNγ+(C) and IFNγ+CD107+ effector CD8+ T cells (D) in spleens following stimulation with H3 NP peptides. IFNγ+(E) and IFNγ+CD107+ effector CD8+ T cells (F) in lungs following stimulation with H3 NP peptides. IFNγ+(G) and IFNγ+CD107+ effector CD8+ T cells (H) in spleens following stimulation with H1N1 HA peptides. IFNγ+(I) and IFNγ+CD107+ effector CD8+ T cells (J) in lungs following stimulation with H1N1 HA peptides. Data are representative of one independent experiment with n=5/group. Symbols represent individual animals, bars represent the group mean, and error bars represent SD. *p<0.05, **p<0.01, ***p<0.001, ****P<0.0001 by Kruskal-Wallis ANOVA.
Following NPH3 peptide stimulation, we observed statistically significant increases in IFNγ+ CD8+ effector cells in the spleens of animals immunized with pVACC-NPH3 alone, those co-immunized with pVACC-NPH3 and pIL-12, or those immunized with the combination of pVACC-NPH3, pIL-12, and pHAH1(Combo), as compared to those receiving empty plasmid control (pVax1) (Figure 3C). We similarly observed statistically significant increases in the frequency of CD107α+IFNy+ effector CD8+ T cells among these mice as compared to pVax1-immunized controls (Figure 3D). Cellular responses were assayed from lungs as it is the primary site of influenza infection and replication. We observed statistically significant increases in IFNγ+ CD8+ effector cells among isolated pulmocytes of animals immunized with pVACC-NPH3 alone, those co-immunized with pVACC-NPH3 and pIL-12, or those immunized with the combination of pVACC-NPH3, pIL-12, and pHAH1 (Combo), as compared to those receiving pVax1 (Figure 3E). We also observed statistically significant increases in the frequency of CD107α+IFNy+ effector CD8+ T cells among the pulmocytes of these mice as compared to pVax1-immunized controls (Figure 3F). In both the spleens and lungs, the addition of pIL-12 to pVACC-NPH3 trended toward increased IFNγ secretion compared to NP alone, but did not meet statistical significance.
Splenocytes stimulated with matched HA peptides demonstrated increased effector function among all groups receiving pHAH1, however only animals receiving pHAH1 alone had statistically significant increases in frequencies of IFNγ+ (Figure 3G) and CD107α+IFNy+ (Figure 3H) effector CD8+ T cells compared to those immunized with empty vector. Among pulmocytes, we observed trends toward increased effector function among all groups receiving pHAH1 with statistically significant increases in IFNy+ (Figure 3I), and CD107α+IFNy+ (Figure 3J) effector CD8+ T cell frequencies of animals receiving pHAH1 only, compared to those immunized with empty vector. Mice receiving pVACC-NPH3 or pVACC-NPH3 plus pIL-12 did not respond to HA peptide stimulation, highlighting the specificity of these vaccines. Lower responses were observed in the pHAH1+ pVACC-NPH3 group, suggesting potential interference when both antigens are co-delivered. Overall, these data suggest that combination delivery of pVACC-NPX antigens with HA antigens can elicit robust NP-directed cellular responses in both the periphery and mucosa.
3.5 Heterologous pVACC-NPH3 enhances protective efficacy of the pHAH1 DNA vaccine against IAV H1N1pdm09 challenge
In the DBA/2J mouse model, a single immunization with pHAH1 alone induces complete protection against morbidity and mortality from a 10 LD50 homologous Ca09-X179A challenge (52). At the higher challenge inoculum of 100 LD50 mice immunized with 10 µg pHAH1 are completely protected from death but display significant weight loss before recovering (Supplementary Figures S6A-C). However, animals receiving either 1 µg or 0.5 µg of pHAH1 succumb to infection (Supplementary Figure S6B) and display significant weight loss (Supplementary Figure S6C). This sub-protective model was next used to evaluate the protective efficacy following combination delivery of the pVACC-NPH3 and pHAH1 vaccines.
DBA/2J mice were immunized once with 10µg pVACC-NPH3 and 0.5μg of pIL-12, or 10μg of pHAH1 and 0.5μg of pIL-12, or co-immunized with a combination formulation of pHAH1, pVACC-NPH3, and pIL-12 (Combo) (Figure 4A). Animals were challenged fourteen days post-immunization with 100 LD50 Ca09-X179A. Animals which received pVACC-NPH3 and pIL-12 succumbed to this lethal challenge by day 7, as did naïve animals, however 100% of mice which received pHAH1 and pIL-12 or the combination vaccine survived challenge (Figure 4B). All pHAH1 and pIL-12 immunized animals lost significant weight but survived challenge (100%). Interestingly, only the Combo group afforded complete protection from both mortality (Figure 4B) and morbidity as measured by weight loss (Figure 4C). H&E staining revealed dense cellular infiltrates in the lungs of naïve and pVACC-NPH3-only immunized mice (Figure 4D). pHAH1-only and combination-immunized mouse lungs displayed more open airway space but had intermediate cellular infiltration and modest evidence of alveolar wall thickening (Figure 4D). When sections were stained for NP antigen, naïve animals had significant, dispersed NP antigen staining (Figure 4E). pVACC-NPH3-only immunized mouse lungs exhibited dense NP antigen staining which was localized to the alveolar spaces. In pHAH1-only immunized mouse lungs, NP staining was faint and dispersed, whereas Combination-immunized lungs display minimal NP positivity (Figure 4E). Taken together, these data support that the combination delivery of NP with HA antigens can improve challenge outcomes.
Figure 4
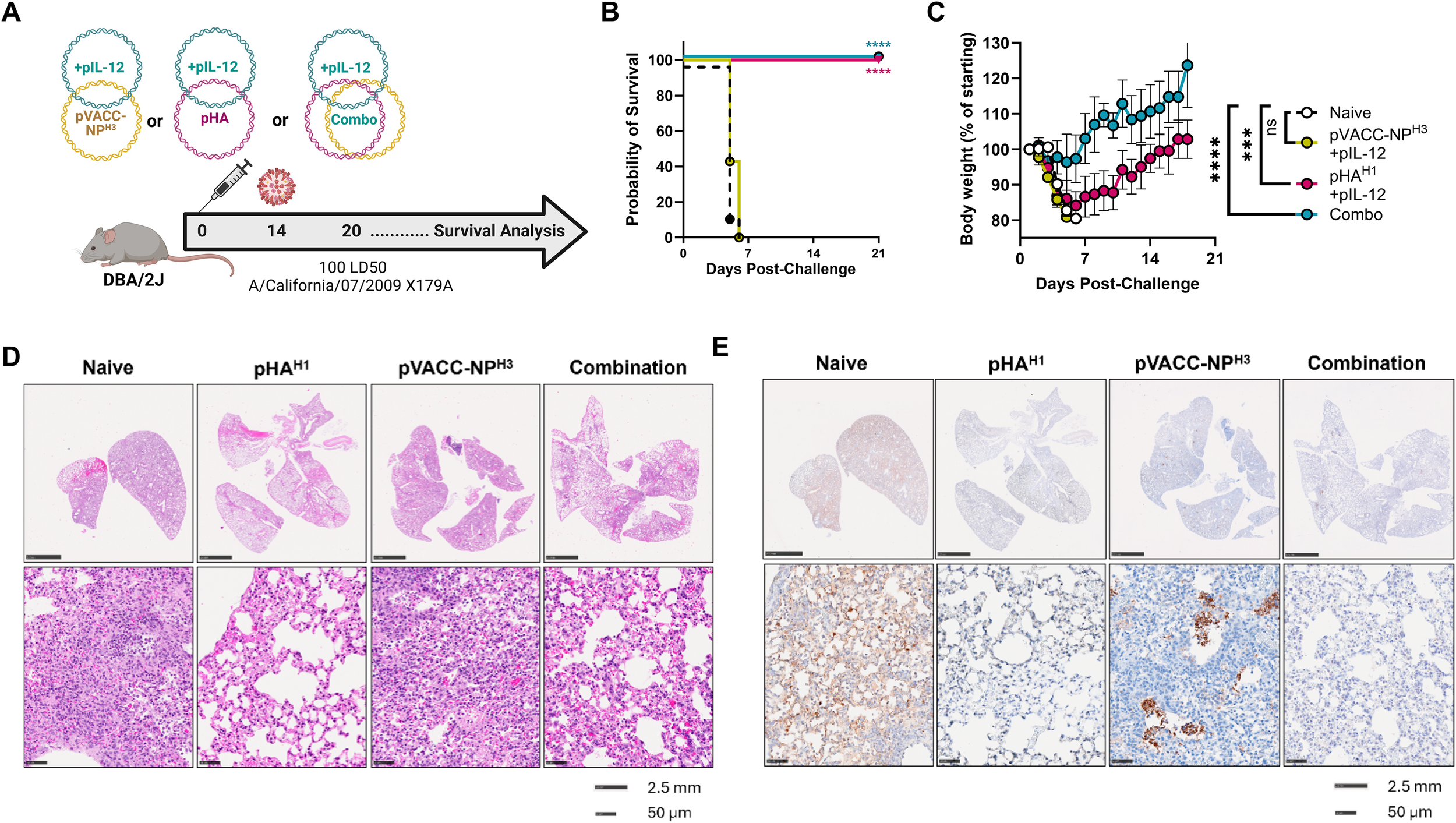
HA and NP combination improves protection from IAV induced morbidity DBA/2J mice were immunized once with 10μg of pHA and 0.5μg of pIL-12, 10μg pVACC- NP H3 and 10μg of pIL-12, or co-immunized with pHAH1, pVACC-NPH3, and 0.5μg of IL-12 (Combo), and challenged with 100 LD50 Ca09-X179A virus 14 days later. Lungs were collected from 3 representative animals per group 6 days post-challenge and the remaining animals were monitored daily (A). Survival probability (B). Body weight as percent of starting weight (C). H&E (D), and NP antigen-stained (E) representative lungs from animals euthanized at day 6 post-challenge. Data are representative of two independent experiments with n=10/group. Symbols represent group averages, bars represent SD (C). *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 by Dunnett’s multiple comparison (C), or Mantel-Cox Log-rank test (B).
3.6 Inhibitory gene expression signatures are detected in lungs during infection in mice receiving pHAH1 or pVACC-NPH3 DNA vaccines
To evaluate the impact of DNA vaccination on host transcription signatures, differential gene expression (DEG) in naïve, pHAH1, and pVACC-NPH3 vaccinated mice were profiled by 3’mRNA-Seq following infection with 10 LD50 Ca09-X179A. A mock (uninfected) group receiving PBS alone was run in parallel and included as control. Principal component analysis indicated distinct clustering of the mock, naïve, and vaccinated groups, with both pHAH1 and pVACC-NPH3 co-localizing (Figure 5A). We compared DEGs between mock animals and those that were either naïve (unvaccinated) (Figure 5B), immunized with pHAH1 (Figure 5C), or immunized with pVACC-NPH3 (Figure 5D). Compared with mock, DEG analysis of the naïve group found 2, 538 genes downregulated (blue) and 3, 459 genes upregulated (red). pHAH1 analysis found 1, 511 genes downregulated and 1, 545 genes upregulated. Finally, DEG analysis of pVACC-NPH3 indicated 1, 666 genes down and 1, 525 genes upregulated. The top 20 DEGs in total are displayed on each of the volcano plots. We next compared DEG signatures for pHAH1 (Figure 5E) and pVACC-NPH3 (Figure 5F) vaccinated mice with the naïve (unvaccinated) group. DEG analysis for pHAH1 found 2, 088 downregulated genes and 1, 694 upregulated genes. Analysis for pVACC-NPH3 found 2, 716 genes downregulated and 2, 301 genes upregulated. Again, the top 20 up and downregulated genes in total are highlighted for each comparison. Influenza infection was associated with significant increases in virus-associated and inflammatory gene signatures, including CXC-motif chemokine ligand-10 or interferon gamma induced protein-10 (CXCL10/IP-10), a well-characterized influenza infection-induced inflammatory mediator (53–56). Importantly, immunization with either pHAH1 or pVACC-NPH3 significantly decreased CXCL-10 gene expression. For pVACC-NPH3 vaccinated animals this decrease was statistically significant, and we observed statistically significant increases in other inflammatory mediators including CCL2 (57), CXCL-9 (54), and CCL7 (58). Together with our challenge data, these results suggest that DNA immunization with influenza antigens supports decreased influenza-associated inflammation.
Figure 5
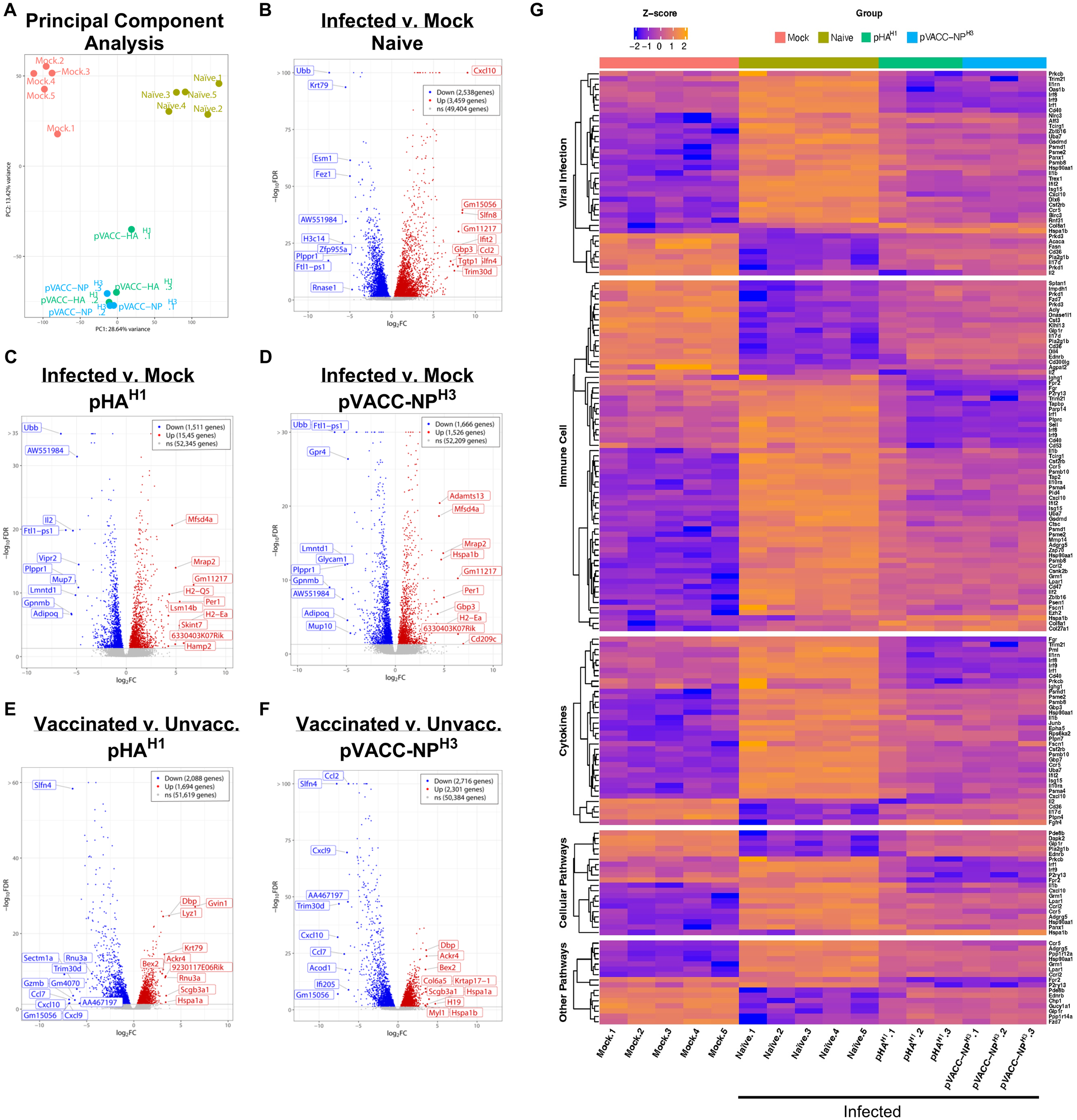
Differential gene expression (DEG) analysis of pHAH1 and pVACC-NPH3 DNA vaccines during infection. Mice (DBA/2J) were immunized with one dose of pHAH1 or pVACC-NPH3 and 14 days later, challenged with 10 LD50 of Ca09-X179A virus. Total RNA was extracted from FFPE scrolls of lung harvested at day-6 post-infection. Control groups included mock (unvaccinated, uninfected) and naïve (unvaccinated, infected). (A) PCA analysis of all four groups. Volcano plots comparing infection versus mock are shown for (B) naïve, (C) pHAH1, and (D) pVACC-NPH3. Volcano plots comparing vaccinated versus unvaccinated mice are shown for (E) pHAH1, and (F) pVACC-NPH3. The top 10 upregulated and 10 downregulated genes are highlighted for all volcano plots. (G) Ingenuity pathway analysis comparing significant gene signatures associated with viral infection, immune cells, cytokines, and other pathways across all groups. Data representing pHAH1 or pVACC-NPH are from one independent experiment with n=3/group. Data representing naïve and mock are from 2 independent experiments with n=3 and n=2 per group (total n=5/group).
Pathway analysis identified the activation of pathways associated with viral infection, immune cells, cytokines, cellular pathways, and other pathways naïve (unvaccinated, infected) group, compared with the mock (unvaccinated, uninfected) control animals (Figure 5G, Supplementary Figure S7). Very few significant differences in DEG signatures based on p-value were observed between pHAH1 or pVACC-NPH3 vaccinated animals compared with the mock group and all had a false discovery rate (FDR) >10% and low significance (Supplementary Figures S7B, C). Analysis of pHAH1 or pVACC-NPH3 vaccinated signatures found the converse, where pathways associated with viral infection, immune cells, cytokines, and cellular pathways were overall inhibited (Figure 5G, Supplementary Figures S8, S9). Overall, these data highlight the protective impact of pHAH1 or pVACC-NPH3 priming to control genes associated with response to IAV infection.
3.7 DNA-encoded VACC-NPX antigens induce durable T cell responses in mice
To evaluate the longevity of pVACC-NPX-induced cellular responses, C57BL/6J mice were immunized twice, separated by three weeks with pVACC-NPH1 alone, pVACC-NPH3 alone, or co-immunized with each immunogen and plasmid-encoded IL-12 (+pIL-12) and rested for ~200 days (6 months) (Supplementary Figure S10A). T cell responses were detected by IFNγ ELISPOT assay with the spleens and lungs of mice following immunization. At this memory timepoint, significant anti-NP responses in the spleen and lungs for both pVACC-NPH1 (Supplementary Figures S10B, D, respectively) and pVACC-NPH3 (Supplementary Figure S6C, E, respectively) compared to naïve controls. Co-delivery of pIL-12 demonstrated long-term enhancement of IFNγ secretion in both compartments compared with animals receiving pVACC-NPH1 (Supplementary Figures S10B, D, respectively) and pVACC-NPH3 (Supplementary Figures S10C, E, respectively). These data indicate that VACC-NPX DNA vaccines can elicit robust and long-lived cellular responses in vivo and highlight the potent contribution of molecular adjuvant pIL-12 to enhancing cellular immunity at acute and memory timepoints post-vaccination.
4 Discussion
Conventional seasonal influenza vaccines induce antibodies primarily directed against the surface HA glycoprotein. This protection is most optimal against matched and minimally mutated strains, with hemagglutination inhibition (HAI) titers >1:40 associated with protection in 50% of people (59, 60). Even moderate antigenic drift can dramatically reduce vaccine effectiveness of traditional inactivated and LAIV vaccines (61, 62). In parallel, cellular immune responses inducing both CD4+ and CD8+ T cell responses can provide important early protection (63, 64) and IAV clearance [reviewed in (65) and (66)] including correlating with decreased recovery time (67). Therefore, directing immune responses to highly conserved internal antigens could be a viable approach to supplement anti-HA directed antibodies with anti-IAV targeting cellular responses. Of these internal antigens, the conserved IAV NP is an attractive target for broad and universal influenza strategies (16–19) and, in humans, anti-NP cytotoxic T lymphocyte responses can reduce pathogenesis and confer important heterosubtypic protection (68). Here, we describe the design and immunogenicity of two new synthetic NP immunogens guided by the genetic sequences obtained from the seasonal IAV-H1N1 and IAV-H3N2 WHO-recommended vaccine strains. Our data demonstrates the ability of single NP DNA immunization to rapidly to protect against mortality in a Ca09-X179A challenge model and further shows additive activity in combination with a pHAH1 DNA vaccine, achieving full protection when administered only two weeks before lethal challenge.
Synthetic NP immunogens have been evaluated in various platforms including DNA (69–71), mRNA (72, 73), and viral vector-based platforms such as adenovirus (74) and modified vaccinia Ankara (MVA) (75, 76) vectors. In addition to strain-matched designs, approaches to targeting IAV-NP include CD8+ T cell epitope-based strains and oligomerized forms (77, 78). Here, we designed a synthetic consensus immunogen with the goal of inducing broadly protective cellular responses. This vaccine-aligned common consensus approach, or VACC, leverages our synthetic-consensus (SynCon) approach (79–83) to generate single NP immunogens representing conserved features of the IAV-NPH1 and proteins. The number of sequenced circulating influenza strains has dramatically increased with advancements in sequencing technologies and one approach for immunogen design is to computationally align thousands of IAV NP sequences to generate a single sequence (84). Alternatively, the pVACC approach focuses specifically on vaccine strains; Since the 1970s, the WHO has provided recommendations for the composition of seasonal influenza vaccines. This requires yearly surveillance involving analysis of clinical specimens, disease burden, and epidemiological data to understand representative viruses in the human population and their distribution by country and region (85). The selected vaccine viruses could therefore be considered as representative of the diversity of major influenza viruses circulating in the human population in a current year. Using this as a guide, the VACC-NPX candidates therefore encompass yearly NP variation. Although fewer sequences are aligned, the VACC approach reduces bias from sequence variability and quality. Supporting our approach, vaccination with these de novo DNA immunogens induced strong T cell responses in the spleen and lungs and protected against lethal Ca09-X179A challenge in mice.
Both pVACC-NPH1 and pVACC-NPH3 elicited robust T cell responses that were durable in mice and our data show the protective potency of targeting the IAV-NP, achieving single dose protection 14 days following delivery in mice. Interestingly, although both VACC-NP constructs protected against death, we observed interesting superior prevention of IAV-associated pathogenesis in the lungs of DBA/2J mice immunized with the heterologous pVACC-NPH3 DNA vaccine. In our studies, the Ca09-X179A challenge virus contains the internal proteins including NP from A/Puerto Rico/8/1934 (PR8). The H1N1pdm09 triple reassortant event, resulted in introduction of a classical swine NP, resulting in the PR8-NP being 8.7% different in sequence to Ca09. Interestingly, the synthetic VACC-NPH3 design is almost equidistant, with 9.0% different from the PR8-NP and 10.7% from Ca09, respectively (Supplementary Figure S11). Further studies with mouse-adapted IAV and different mouse strains could provide additional input into this heterologous protection. pVACC-NPH3 displayed better in vivo immunogenicity in both spleen and lungs. In other work, we have demonstrated the importance of nucleotide and amino acid changes towards in vivo expression (86, 87) and it is possible that this could contribute to the difference in immunogenicity observed between these constructs.
While NP generates a robust cytotoxic T lymphocyte (CTL) response and can potentially contribute to humoral immunity (88), it remains likely that an HA immunogen component will be essential in IAV vaccine formulations to provide robust antibody-mediated protection. We evaluated the inclusion of pIL-12 in NP-only and HA+NP formulations in these studies. Addition of pIL-12 to NP-only formulations led to trends toward increased NP-specific T cell responses. This pattern of increased NP-specific T cell responses was also observed when pIL-12 was added to the combination of pVACC-NPH3 and pHAH1. However, for HA-specific T cell responses, only groups receiving pHAH1 generated HA-specific cellular responses. The generation of T cell responses against both HA and NP peptides highlights the potential for both antigens to be co-administered, at least in mice. Importantly, mice immunized with a suboptimal dose of pHAH1 or pVACC-NPH3 alone did not achieve complete protection. However, pHAH1 plus pVACC-NPH3 co-immunized animals were completely protected from morbidity and mortality. These data indicate that pVACC-NPH3, and indeed other NP immunogens, can play a role to complement HA-based vaccine-induced immunity. Additional studies dissecting this synergy, likely due to T cell immunity, would be interesting and evaluation in mice and larger models would be informative for dose titration and combination studies. Interestingly, we observed lower T cell responses when both pHAH1 plus pVACC-NPH3 were co-delivered, consistent with potential interference which could impede immune responses. Although protective in the DBA/2J mouse model, further studies dissecting immune responses associated with HA and NP antigen combination will be important to understanding the impact of this decreased immunogenicity.
One possible limitation of our approach is the focus on human seasonal IAV-H1 and IAV-H3. As highlighted, the current circulating human IAV-H3 viruses have varied minimally over the past 20 years (<1.1%). In 2009, the introduction of the reassortant A/H1N1pdm09 swine flu viruses into the human population resulted in the introduction of a classical swine H1N1 NP into humans (45, 46), a significant antigenic shift (>10%). We therefore focused the VACC-NPH1 design based on post-H1N1pdm09 viruses. To further address major antigenic shift events, additional consideration of animal (for example swine) H1 and H3 circulating strains would be valuable. Although there is no global body selecting vaccine strains for animals, similar surveillance of strains circulating in animals is being undertaken by various agencies such as the United States and European Centers for Disease Control (CDC) and others, alerting to emerging influenza strains with potential for zoonotic crossover into humans. Yearly monitoring and selection of predominantly circulating animal IAV would be valuable for narrowing down and selecting strains for inclusion in immunogen design. Such animal IAV-H1 and IAV-H3 NP immunogens could be incorporated as multivalent combinations to elicit broader cellular immune responses against potential emerging viruses.
It should be emphasized, that our study demonstrates the potency of pVACC candidates to rapidly induce protective immune responses with a single immunization, achieving protection against a 10 LD50 challenge within only 14 days with immunogens designed to elicit primarily T cell responses. These data are further supported by transcriptomic analysis identifying genes and pathways associated with inhibitory control of viral infection and inflammation in pHAH1 or pVACC-NPH3 vaccinated mice. These include inhibition of genes associated with innate immune cell activation, cross-talk, and lymphocyte activation and indicative of vaccine-related immune priming to control infection. Further, we show that the NPH3 immunogen can adjuvant the pHAH1 vaccine in a more stringent 100 LD50 challenge model. Our approach to generating synthetically designed NP antigens based on yearly vaccine strains can be broadly applied to other highly conserved influenza internal proteins with potential to generate robust protective CTL responses. Additional studies evaluating protective efficacy in H3N2 challenges and other heterologous subtypes would be valuable. Further studies evaluating protective efficacy at later time points following maturation of the immune response and the inclusion of prime-boost regimens will also be valuable. The NP antigen co-delivery has potential to enhance various vaccine platofmrs and further study with commercially available, seasonal HA-based vaccine regimens and other delivery platforms would be insightful. In summary, these data support the incorporation of the VACC design approach for continued development of broad and efficacious influenza interventions.
Statements
Data availability statement
The data presented in the study are deposited in the NCBI GEO repository, accession number GSE306862.
Ethics statement
These animal studies were reviewed and approved by The Wistar Institute IACUC committee (Protocol 201355). Studies were conducted in accordance with the local legislation and institutional requirements. No potentially identifiable images or data are presented in this study.
Author contributions
ENG: Writing – original draft, Writing – review & editing, Data curation, Formal Analysis, Investigation, Methodology, Software, Visualization. ART: Writing – original draft, Writing – review & editing, Data curation, Investigation, Methodology, Software, Visualization, Formal Analysis. DW: Investigation, Methodology, Software, Writing – original draft, Writing – review & editing, Visualization. SB: Conceptualization, Data curation, Investigation, Methodology, Software, Writing – original draft, Writing – review & editing, Visualization. YY: Data curation, Formal Analysis, Software, Writing – original draft, Writing – review & editing, Visualization. NJT: Investigation, Writing – original draft, Writing – review & editing, Methodology, Visualization. MT: Data curation, Formal Analysis, Software, Writing – original draft, Writing – review & editing, Visualization. JE: Investigation, Methodology, Writing – original draft, Writing – review & editing, Visualization. JCD: Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. DCZ: Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. CEH: Investigation, Writing – original draft, Writing – review & editing, Visualization. MZ: Investigation, Visualization, Writing – original draft, Writing – review & editing. JW: Data curation, Investigation, Methodology, Software, Writing – original draft, Writing – review & editing. DBW: Conceptualization, Funding acquisition, Resources, Supervision, Visualization, Writing – original draft, Writing – review & editing. AP: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. Sinai-Emory Multi- Institutional CIVIC Grant # 75N93019C00051 and Wistar Cancer Center Grant P30 CA010815.
Acknowledgments
We would like to thank The Wistar Institute Shared Resource Core Facilities: Animal Facility, Histotechnology and Imaging Cores for their help with processing, staining and imaging tissues, the Genomics Core for processing tissues, and the Flow Cytometry Core. All graphical representations were created in BioRender.com: Gary, E. (2025) https://BioRender.com/5e08tlt.
Conflict of interest
DBW has received grant funding from industry for sponsored research collaborations, he has received speaking honoraria and received fees for consulting or serving on scientific review committees. Remunerations received by DBW include direct payments and equity/options. DBW also discloses the following associations with commercial partners: Geneos consultant/advisory board, AstraZeneca advisory board and speaker, Inovio board of directors and consultant, Sanofi advisory board, BBI advisory board, Pfizer advisory board, Flagship consultant, and Advaccine consultant.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1632121/full#supplementary-material
Supplementary Table 1Influenza NP peptides and pools for ICS and ELISpot assays. (A) pVACC-NPH1 peptides and pools. (B) pVACC-NPH3 peptides and pools. (C) Peptide matrix pool design schematic.
Supplementary Table 2Ingenuity Pathway analysis of naïve versus mock mice.
Supplementary Table 3Ingenuity Pathway analysis of pHAH1 versus mock mice.
Supplementary Table 4Ingenuity Pathway analysis of pVACC-NPH3 versus mock mice.
Supplementary Table 5Ingenuity Pathway analysis of pHAH1 versus naïve mice.
Supplementary Table 6Ingenuity Pathway analysis of pVACC-NPH3 versus naïve mice.
Supplementary Figure 1(A)The H1N1 and H3N2 vaccine strains included in the generation of the vaccine pVACC-NPH1 and pVACC-NPH3. (B) A phylogenetic tree
Supplementary Figure 2Multiple sequence alignment of NP proteins of seasonal A/H1N1 vaccine strains, performed in Geneious Prime. Dots represent complete identity of the residue in all the sequences. The green bar on top represents the level of conservation of the residues between strains, deep green indicates most conserved, yellow indicates a poorly conserved position.
Supplementary Figure 3Multiple sequence alignment of NP proteins of seasonal A/H3N2 vaccine strains, performed in Geneious Prime. Dots represent complete identity of the residue in all the sequences. The green bar on top represents the level of conservation of the residues between strains, deep green indicates most conserved, yellow indicates a poorly conserved position.
Supplementary Figure 4A 2-injection regimen of VACC-NPH3 induces robust immune responses in C57BL/6J mice. C57BL/6J mice were immunized with 10µg or 25µg of pVACC-NPH3 plasmid at a 21-day interval and euthanized seven days post-immunization for cellular analyses (A). IFNγ spot-forming units (SFUs) in spleens following stimulation with H3NP peptides (n=5 mice per group) (B).
Supplementary Figure 5Identification of pVACC-NPX immunodominant T cell epitopes. (A) C57BL/6J mice were immunized twice, separated by three weeks with 10μg of pVACC-NPX. Matrix peptide pools were used to stimulate isolated splenocytes. (B) H1N1-NP specific IFNγ secretion as measured by ELISpot. (C) Identified H1N1-NP immunodominant peptides. (D) H3N2-NP specific IFNγ secretion as measured by ELISpot. (E) Identified H3N2-NP immunodominant peptides. Data are representative of one experiment with n=5/group. Symbols represent the average of duplicate assays per animal, bars represent group mean, error bars represent SEM.
Supplementary Figure 6pHAH1 is sub-protective in high-dose IAV challenge: (A) Immunofluorescence staining of HEK293T cells transfected with pHAH1 plasmid and stained for IAV-NP. (B) Mice were immunized once with 10μg, 1μg, or 0.5μg of plasmid-encoded A/California/07/2009 HA (pHAH1) and challenged with 100 LD50 of Ca09-X179A virus fourteen days later. (C) Survival probability. (D) Body weights as percent of starting weight. Data are representative of one experiment with n=10/group. Symbols represent group mean, error bars represent SD. ***p<0.001, ****p<0.0001 Mantel-Cox Log-rank test (C) ns = not significant by Dunnett’s multiple comparison test (D).
Supplementary Figure 7VACC-NPX immunogens induce robust and durable cellular responses in vivo. (A) Female C57BL/6J mice were immunized twice separated by three weeks and mice were rested for 200 days (six months) with 10 μg of pVACC-NPH1 or pVACC-NPH3 alone, or co-immunized with pVACC-NPx and 0.5μg of plasmid-encoded IL-12 (+pIL-12). (B) NPH1-specific IFNγ spot-forming units (SFU) in spleens and (D) lungs. (C) NPH3-specific IFNγ spot-forming units (SFU) in spleens (E) and lungs. Data are representative of one experiment with n=5/group. Bars represent the mean; error bars represent SEM (C-J) or SD (K-R). **p<0.01, ***p<0.001, ****p<0.0001 by Two-way ANOVA.
Supplementary Figure 8Ingenuity pathway analysis (IPA) of versus Mock (unvaccinated, uninfected). IPA was performed on the total gene expression dataset and further categorized into subsets involved with viral infection, immune cells, cytokines, cellular pathways, and other pathways. Bar charts comparing the number of activated and inhibited pathways are shown for (A) naïve versus mock, (B) pHAH1 versus mock, and (C) pVACC-NPH3 versus mock groups. Groups were first filtered by Z-score >2.0 and a false discovery rate of 0.05. All groups have significant p-values <0.05.
Supplementary Figure 9Ingenuity pathway analysis (IPA) of pHAH1 versus Naïve. IPA was performed on the total gene expression dataset and further categorized into subsets involved with viral infection, immune cells, cytokines, cellular pathways, and other pathways. Bar charts comparing the number of activated and inhibited pathways are shown for pHAH1 versus naïve. Groups were first filtered by Z-score >2.0 and a false discovery rate of 0.05.
Supplementary Figure 10Ingenuity pathway analysis (IPA) of pVACC-NPH3 versus Naïve. IPA was performed on the total gene expression dataset and further categorized into subsets involved with viral infection, immune cells, cytokines, cellular pathways, and other pathways. Bar charts comparing the number of activated and inhibited pathways are shown for pVACC-NPH3 versus naïve. Groups were first filtered by Z-score >2.0 and a false discovery rate of 0.05.
Supplementary Figure 11Phylogenetic analysis comparing pVACC-NPX immunogens with A/California/07/2009 and Ca09-X179A (PR8) NP proteins.
References
1
Nair H Brooks WA Katz M Roca A Berkley JA Madhi SA et al . Global burden of respiratory infections due to seasonal influenza in young children: a systematic review and meta-analysis. Lancet. (2011) 378:1917–30. doi: 10.1016/S0140-6736(11)61051-9
2
WHO . Influenza (seasonal) - Fact Sheet. World Health Organization. Available online at: https://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal):~:text=There%20are%20around%20a%20billion,infections%20are%20in%20developing%20countries (Accessed May 20, 2025).
3
Webster RG . Antigenic variation in influenza viruses. In: Origin and evolution of viruses. London, UK: Academic Press (1999). p. 377–90.
4
Carrat F Flahault A . Influenza vaccine: the challenge of antigenic drift. Vaccine. (2007) 25:6852–62. doi: 10.1016/j.vaccine.2007.07.027
5
Hua S Li X Liu M Cheng Y Peng Y Huang W et al . Antigenic variation of the human influenza A (H3N2) virus during the 2014–2015 winter season. Sci China Life Sci. (2015) 58:882–8. doi: 10.1007/s11427-015-4899-z
6
Suntronwong N Klinfueng S Korkong S Vichaiwattana P Thongmee T Vongpunsawad S et al . Characterizing genetic and antigenic divergence from vaccine strain of influenza A and B viruses circulating in Thailand, 2017–2020. Sci Rep. (2021) 11:735. doi: 10.1038/s41598-020-80895-w
7
Krammer F Pica N Hai R Margine I Palese P . Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J Virol. (2013) 87:6542–50. doi: 10.1128/JVI.00641-13
8
Sun X Ma H Wang X Bao Z Tang S Yi C et al . Broadly neutralizing antibodies to combat influenza virus infection. Antiviral Res. (2024) 221:105785. doi: 10.1016/j.antiviral.2023.105785
9
Guthmiller JJ Yu-Ling Lan L Li L Fu Y Nelson SA Henry C et al . Long-lasting B cell convergence to distinct broadly reactive epitopes following vaccination with chimeric influenza virus hemagglutinins. Immunity. (2025) 58:980–96.e7. doi: 10.1016/j.immuni.2025.02.025
10
Arevalo CP Bolton MJ Le Sage V Ye N Furey C Muramatsu H et al . A multivalent nucleoside-modified mRNA vaccine against all known influenza virus subtypes. Science. (2022) 378:899–904. doi: 10.1126/science.abm0271
11
Elliott STC Keaton AA Chu JD Reed CC Garman B Patel A et al . A synthetic micro-consensus DNA vaccine generates comprehensive influenza A H3N2 immunity and protects mice against lethal challenge by multiple H3N2 viruses. Hum Gene Ther. (2018) 29:1044–55. doi: 10.1089/hum.2018.102
12
Dzimianski JV Han J Sautto GA O’Rourke SM Cruz JM Pierce SR et al . Structural insights into the broad protection against H1 influenza viruses by a computationally optimized hemagglutinin vaccine. Commun Biol. (2023) 6:454. doi: 10.1038/s42003-023-04793-3
13
Huang Y Franca MS Allen JD Shi H Ross TM . Next generation of computationally optimized broadly reactive HA vaccines elicited cross-reactive immune responses and provided protection against H1N1 virus infection. Vaccines (Basel). (2021) 9(7):793. doi: 10.3390/vaccines9070793
14
Dam S Tscherne A Engels L Sutter G Osterhaus A Rimmelzwaan GF . Design and evaluation of a poly-epitope based vaccine for the induction of influenza A virus cross-reactive CD8 + T cell responses. Sci Rep. (2025) 15:10586. doi: 10.1038/s41598-025-95479-9
15
Shaw ML Stone KL Colangelo CM Gulcicek EE Palese P . Cellular proteins in influenza virus particles. PLoS pathogens. (2008) 4:e1000085. doi: 10.1371/journal.ppat.1000085
16
Hu Y Sneyd H Dekant R Wang J . Influenza A virus nucleoprotein: a highly conserved multi-functional viral protein as a hot antiviral drug target. Curr topics medicinal Chem. (2017) 17:2271–85. doi: 10.2174/1568026617666170224122508
17
Ng AK-L Wang J-H Shaw P-C . Structure and sequence analysis of influenza A virus nucleoprotein. Sci China Ser C. (2009) 52:439–49. doi: 10.1007/s11427-009-0064-x
18
McGee MC Huang W . Evolutionary conservation and positive selection of influenza A nucleoprotein CTL epitopes for universal vaccination. J Med virology. (2022) 94:2578–87. doi: 10.1002/jmv.27662
19
Babar MM . Protein sequence conservation and stable molecular evolution reveals influenza virus nucleoprotein as a universal druggable target. Infection Genet Evolution. (2015) 34:200–10. doi: 10.1016/j.meegid.2015.06.030
20
Heiny A Miotto O Srinivasan KN Khan AM Zhang G Brusic V et al . Evolutionarily conserved protein sequences of influenza a viruses, avian and human, as vaccine targets. PLoS One. (2007) 2:e1190. doi: 10.1371/journal.pone.0001190
21
Gultyaev AP Tsyganov-Bodounov A Spronken MI van der Kooij S Fouchier RA Olsthoorn RC . RNA structural constraints in the evolution of the influenza A virus genome NP segment. RNA Biol. (2014) 11:942–52. doi: 10.4161/rna.29730
22
Yewdell JW Bennink JR Smith GL Moss B . Influenza A virus nucleoprotein is a major target antigen for cross-reactive anti-influenza A virus cytotoxic T lymphocytes. Proc Natl Acad Sci. (1985) 82:1785–9. doi: 10.1073/pnas.82.6.1785
23
Zhou D Wu T-L Lasaro MO Latimer BP Parzych EM Bian A et al . A universal influenza A vaccine based on adenovirus expressing matrix-2 ectodomain and nucleoprotein protects mice from lethal challenge. Mol Ther. (2010) 18:2182–8. doi: 10.1038/mt.2010.202
24
McMichael AJ Michie CA Gotch FM Smith GL Moss B . Recognition of influenza A virus nucleoprotein by human cytotoxic T lymphocytes. J Gen virology. (1986) 67:719–26. doi: 10.1099/0022-1317-67-4-719
25
Epstein SL . Prior H1N1 influenza infection and susceptibility of Cleveland Family Study participants during the H2N2 pandemic of 1957: an experiment of nature. J Infect diseases. (2006) 193:49–53. doi: 10.1086/498980
26
Gary EN Weiner DB . DNA vaccines: prime time is now. Curr Opin Immunol. (2020) 65:21–7. doi: 10.1016/j.coi.2020.01.006
27
Khobragade A Bhate S Ramaiah V Deshpande S Giri K Phophle H et al . Efficacy, safety, and immunogenicity of the DNA SARS-CoV-2 vaccine (ZyCoV-D): the interim efficacy results of a phase 3, randomised, double-blind, placebo-controlled study in India. Lancet. (2022) 399:1313–21. doi: 10.1016/S0140-6736(22)00151-9
28
Yarchoan M Gane EJ Marron TU Perales-Linares R Yan J Cooch N et al . Personalized neoantigen vaccine and pembrolizumab in advanced hepatocellular carcinoma: a phase 1/2 trial. Nat Med. (2024) 30(4):1044–53. doi: 10.1038/s41591-024-02894-y
29
Blazevic V Mac Trubey C Shearer GM . Comparison of in vitro immunostimulatory potential of live and inactivated influenza viruses. Hum Immunol. (2000) 61:845–9. doi: 10.1016/S0198-8859(00)00170-1
30
Hoft DF Babusis E Worku S Spencer CT Lottenbach K Truscott SM et al . Live and inactivated influenza vaccines induce similar humoral responses, but only live vaccines induce diverse T-cell responses in young children. J Infect Diseases. (2011) 204:845–53. doi: 10.1093/infdis/jir436
31
Cox R Brokstad K Ogra P . Influenza virus: immunity and vaccination strategies. Comparison of the immune response to inactivated and live, attenuated influenza vaccines. Scandinavian J Immunol. (2004) 59:1–15. doi: 10.1111/j.0300-9475.2004.01382.x
32
Smith A Rodriguez L El Ghouayel M Nogales A Chamberlain JM Sortino K et al . A live attenuated influenza vaccine elicits enhanced heterologous protection when the internal genes of the vaccine are matched to those of the challenge virus. J Virol. (2020) 94(4):10–1128. doi: 10.1128/JVI.01065-19
33
Hirao LA Wu L Khan AS Hokey DA Yan J Dai A et al . Combined effects of IL-12 and electroporation enhances the potency of DNA vaccination in macaques. Vaccine. (2008) 26:3112–20. doi: 10.1016/j.vaccine.2008.02.036
34
Boyer JD Robinson TM Kutzler MA Parkinson R Calarota SA Sidhu MK et al . SIV DNA vaccine co-administered with IL-12 expression plasmid enhances CD8 SIV cellular immune responses in cynomolgus macaques. J Med primatology. (2005) 34:262–70. doi: 10.1111/j.1600-0684.2005.00124.x
35
Kalams SA Parker SD Elizaga M Metch B Edupuganti S Hural J et al . Safety and comparative immunogenicity of an HIV-1 DNA vaccine in combination with plasmid interleukin 12 and impact of intramuscular electroporation for delivery. J Infect diseases. (2013) 208:818–29. doi: 10.1093/infdis/jit236
36
Kalams SA Parker S Jin X Elizaga M Metch B Wang M et al . Safety and immunogenicity of an HIV-1 gag DNA vaccine with or without IL-12 and/or IL-15 plasmid cytokine adjuvant in healthy, HIV-1 uninfected adults. PLoS One. (2012) 7:e29231. doi: 10.1371/journal.pone.0029231
37
Vonderheide RH Kraynyak KA Shields AF McRee AJ Johnson JM Sun W et al . Phase 1 study of safety, tolerability and immunogenicity of the human telomerase (hTERT)-encoded DNA plasmids INO-1400 and INO-1401 with or without IL-12 DNA plasmid INO-9012 in adult patients with solid tumors. J immunotherapy cancer. (2021) 9:e003019. doi: 10.1136/jitc-2021-003019
38
WHO . (2025). World Health Organization. Available online at: https://www.who.int/teams/global-influenza-programme/vaccines/who-recommendations (Accessed May 20, 2025).
39
Tamura K Stecher G Kumar S . MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. (2021) 38:3022–7. doi: 10.1093/molbev/msab120
40
Patel A Reuschel EL Kraynyak KA Racine T Park DH Scott VL et al . Protective efficacy and long-term immunogenicity in cynomolgus macaques by ebola virus glycoprotein synthetic DNA vaccines. J Infect Dis. (2019) 219:544–55. doi: 10.1093/infdis/jiy537
41
Bianchini G Sanchez-Baracaldo P . TreeViewer: Flexible, modular software to visualise and manipulate phylogenetic trees. Ecol Evol. (2024) 14:e10873. doi: 10.1002/ece3.10873
42
Tursi NJ Tiwari S Bedanova N Kannan T Parzych E Okba N et al . Modulation of lipid nanoparticle-formulated plasmid DNA drives innate immune activation promoting adaptive immunity. Cell Rep Med. (2025) 102035. doi: 10.1016/j.xcrm.2025.102035
43
Kim JJ Maguire HC Jr. Nottingham LK Morrison LD Tsai A Sin JI et al . Coadministration of IL-12 or IL-10 expression cassettes drives immune responses toward a Th1 phenotype. J Interferon Cytokine Res. (1998) 18:537–47. doi: 10.1089/jir.1998.18.537
44
Martin M . Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. (2011) 17:10. doi: 10.14806/ej.17.1.200
45
Smith GJ Vijaykrishna D Bahl J Lycett SJ Worobey M Pybus OG et al . Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. (2009) 459:1122–5. doi: 10.1038/nature08182
46
Garten RJ Davis CT Russell CA Shu B Lindstrom S Balish A et al . Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. (2009) 325:197–201. doi: 10.1126/science.1176225
47
De Rosa SC Edupuganti S Huang Y Han X Elizaga M Swann E et al . Robust antibody and cellular responses induced by DNA-only vaccination for HIV. JCI Insight. (2020) 5. doi: 10.1172/jci.insight.137079
48
Sin J-I Kim JJ Arnold RL Shroff KE McCallus D Pachuk C et al . IL-12 gene as a DNA vaccine adjuvant in a herpes mouse model: IL-12 enhances Th1-type CD4+ T cell-mediated protective immunity against herpes simplex virus-2 challenge. J Immunol. (1999) 162:2912–21. doi: 10.4049/jimmunol.162.5.2912
49
Chattergoon MA Saulino V Shames JP Stein J Montaner LJ Weiner DB . Co-immunization with plasmid IL-12 generates a strong T-cell memory response in mice. Vaccine. (2004) 22:1744–50. doi: 10.1016/j.vaccine.2004.01.036
50
Park H Kingstad-Bakke B Cleven T Jung M Kawaoka Y Suresh M . Diversifying T-cell responses: safeguarding against pandemic influenza with mosaic nucleoprotein. J Virol. (2025) 99:e0086724. doi: 10.1128/jvi.00867-24
51
Lambe T Carey JB Li Y Spencer AJ van Laarhoven A Mullarkey CE et al . Immunity against heterosubtypic influenza virus induced by adenovirus and MVA expressing nucleoprotein and matrix protein-1. Sci Rep. (2013) 3:1443. doi: 10.1038/srep01443
52
Liaw K Konrath KM Trachtman AR Tursi NJ Gary EN Livingston C et al . DNA co-delivery of seasonal H1 influenza hemagglutinin nanoparticle vaccines with chemokine adjuvant CTACK induces potent immunogenicity for heterologous protection in vivo. Vaccine. (2025) 59:127231. doi: 10.1016/j.vaccine.2025.127231
53
Lu X Masic A Liu Q Zhou Y . Regulation of influenza A virus induced CXCL-10 gene expression requires PI3K/Akt pathway and IRF3 transcription factor. Mol Immunol. (2011) 48:1417–23. doi: 10.1016/j.molimm.2011.03.017
54
Betakova T Kostrabova A Lachova V Turianova L . Cytokines induced during influenza virus infection. Curr Pharm design. (2017) 23:2616–22. doi: 10.2174/1381612823666170316123736
55
Elemam NM Talaat IM Maghazachi AA . CXCL10 chemokine: a critical player in RNA and DNA viral infections. Viruses. (2022) 14:2445. doi: 10.3390/v14112445
56
Guo K Yombo DJ Wang Z Navaeiseddighi Z Xu J Schmit T et al . The chemokine receptor CXCR3 promotes CD8+ T cell–dependent lung pathology during influenza pathogenesis. Sci Adv. (2024) 10:eadj1120. doi: 10.1126/sciadv.adj1120
57
Ansari AW Ahmad F Alam MA Raheed T Zaqout A Al-Maslamani M et al . Virus-induced host chemokine CCL2 in COVID-19 pathogenesis: potential prognostic marker and target of anti-inflammatory strategy. Rev Med Virology. (2024) 34:e2578. doi: 10.1002/rmv.2578
58
Klomp M Ghosh S Mohammed S Nadeem Khan M . From virus to inflammation, how influenza promotes lung damage. J Leukocyte Biol. (2021) 110:115–22. doi: 10.1002/JLB.4RU0820-232R
59
Ohmit SE Petrie JG Cross RT Johnson E Monto AS . Influenza hemagglutination-inhibition antibody titer as a correlate of vaccine-induced protection. J Infect diseases. (2011) 204:1879–85. doi: 10.1093/infdis/jir661
60
Tsang TK Cauchemez S Perera RA Freeman G Fang VJ Ip DK et al . Association between antibody titers and protection against influenza virus infection within households. J Infect diseases. (2014) 210:684–92. doi: 10.1093/infdis/jiu186
61
Tenforde MW Kondor RJG Chung JR Zimmerman RK Nowalk MP Jackson ML et al . Effect of antigenic drift on influenza vaccine effectiveness in the United States—2019–2020. Clin Infect Diseases. (2021) 73:e4244–e50. doi: 10.1093/cid/ciaa1884
62
Levine MZ Martin JM Gross FL Jefferson S Cole KS Archibald CA et al . Neutralizing antibody responses to antigenically drifted influenza A (H3N2) viruses among children and adolescents following 2014–2015 inactivated and live attenuated influenza vaccination. Clin Vaccine Immunol. (2016) 23:831–9. doi: 10.1128/CVI.00297-16
63
Sun J Braciale TJ . Role of T cell immunity in recovery from influenza virus infection. Curr Opin Virol. (2013) 3:425–9. doi: 10.1016/j.coviro.2013.05.001
64
Hamada H Bassity E Flies A Strutt TM Garcia-Hernandez Mde L McKinstry KK et al . Multiple redundant effector mechanisms of CD8+ T cells protect against influenza infection. J Immunol. (2013) 190:296–306. doi: 10.4049/jimmunol.1200571
65
Hufford MM Kim TS Sun J Braciale TJ . The Effector T Cell Response to Influenza Infection. In: Oldstone, M., Compans, R. (eds). Influenza Pathogenesis and Control - Volume II. Current Topics in Microbiology and Immunology, vol 386. Cham: Springer. doi: 10.1007/82_2014_397
66
Janssens Y Joye J Waerlop G Clement F Leroux-Roels G Leroux-Roels I . The role of cell-mediated immunity against influenza and its implications for vaccine evaluation. Front Immunol. (2022) 13:959379. doi: 10.3389/fimmu.2022.959379
67
Cao P Wang Z Yan AW McVernon J Xu J Heffernan JM et al . On the role of CD8(+) T cells in determining recovery time from influenza virus infection. Front Immunol. (2016) 7:611. doi: 10.3389/fimmu.2016.00611
68
Amoah S Cao W Sayedahmed EE Wang Y Kumar A Mishina M et al . The frequency and function of nucleoprotein-specific CD8(+) T cells are critical for heterosubtypic immunity against influenza virus infection. J Virol. (2024) 98:e0071124. doi: 10.1128/jvi.00711-24
69
Choi SY Suh YS Cho JH Jin HT Chang J Sung YC . Enhancement of DNA vaccine-induced immune responses by influenza virus NP gene. Immune network. (2009) 9:169. doi: 10.4110/in.2009.9.5.169
70
Fu T-M Guan L Friedman A Schofield TL Ulmer JB Liu MA et al . Dose dependence of CTL precursor frequency induced by a DNA vaccine and correlation with protective immunity against influenza virus challenge. J Immunol. (1999) 162:4163–70. doi: 10.4049/jimmunol.162.7.4163
71
Jimenez GS Planchon R Wei Q Rusalov D Geall A Enas J et al . Vaxfectin™-formulated influenza DNA vaccines encoding NP and M2 viral proteins protect mice against lethal viral challenge. Hum Vaccines. (2007) 3(5):157–64. doi: 10.4161/hv.3.5.4175
72
Magini D Giovani C Mangiavacchi S Maccari S Cecchi R Ulmer JB et al . Self-amplifying mRNA vaccines expressing multiple conserved influenza antigens confer protection against homologous and heterosubtypic viral challenge. PLoS One. (2016) 11:e0161193. doi: 10.1371/journal.pone.0161193
73
Flynn JA Weber T Cejas PJ Cox KS Touch S Austin LA et al . Characterization of humoral and cell-mediated immunity induced by mRNA vaccines expressing influenza hemagglutinin stem and nucleoprotein in mice and nonhuman primates. Vaccine. (2022) 40:4412–23. doi: 10.1016/j.vaccine.2022.03.063
74
Sayedahmed EE Elshafie NO Dos Santos AP Jagannath C Sambhara S Mittal SK . Development of NP-Based universal vaccine for Influenza A Viruses. Vaccines. (2024) 12:157. doi: 10.3390/vaccines12020157
75
Mullarkey CE Boyd A van Laarhoven A Lefevre EA Veronica Carr B Baratelli M et al . Improved adjuvanting of seasonal influenza vaccines: Preclinical studies of MVA-NP+ M 1 coadministration with inactivated influenza vaccine. Eur J Immunol. (2013) 43:1940–52. doi: 10.1002/eji.201242922
76
Langenmayer MC Luelf-Averhoff A-T Marr L Jany S Freudenstein A Adam-Neumair S et al . Newly designed poxviral promoters to improve immunogenicity and efficacy of MVA-NP candidate vaccines against lethal influenza virus infection in mice. Pathogens. (2023) 12:867. doi: 10.3390/pathogens12070867
77
Leroux-Roels I Willems P Waerlop G Janssens Y Tourneur J De Boever F et al . Immunogenicity, safety, and preliminary efficacy evaluation of OVX836, a nucleoprotein-based universal influenza A vaccine candidate: a randomised, double-blind, placebo-controlled, phase 2a trial. Lancet Infect Dis. (2023) 23:1360–9. doi: 10.1016/S1473-3099(23)00351-1
78
Del Campo J Bouley J Chevandier M Rousset C Haller M Indalecio A et al . OVX836 heptameric nucleoprotein vaccine generates lung tissue-resident memory CD8+ T-cells for cross-protection against influenza. Front Immunol. (2021) 12:678483. doi: 10.3389/fimmu.2021.678483
79
Laddy DJ Yan J Corbitt N Kobasa D Kobinger GP Weiner DB . Immunogenicity of novel consensus-based DNA vaccines against avian influenza. Vaccine. (2007) 25:2984–9. doi: 10.1016/j.vaccine.2007.01.063
80
Muthumani K Falzarano D Reuschel EL Tingey C Flingai S Villarreal DO et al . A synthetic consensus anti–spike protein DNA vaccine induces protective immunity against Middle East respiratory syndrome coronavirus in nonhuman primates. Sci Trans Med. (2015) 7:301ra132–301ra132. doi: 10.1126/scitranslmed.aac7462
81
Muthumani K Lankaraman KM Laddy DJ Sundaram SG Chung CW Sako E et al . Immunogenicity of novel consensus-based DNA vaccines against Chikungunya virus. Vaccine. (2008) 26:5128–34. doi: 10.1016/j.vaccine.2008.03.060
82
Duperret EK Wise MC Trautz A Villarreal DO Ferraro B Walters J et al . Synergy of immune checkpoint blockade with a novel synthetic consensus DNA vaccine targeting TERT. Mol Ther. (2018) 26:435–45. doi: 10.1016/j.ymthe.2017.11.010
83
Yan J Corbitt N Pankhong P Shin T Khan A Sardesai NY et al . Immunogenicity of a novel engineered HIV-1 clade C synthetic consensus-based envelope DNA vaccine. Vaccine. (2011) 29:7173–81. doi: 10.1016/j.vaccine.2011.05.076
84
Xie X Zhao C He Q Qiu T Yuan S Ding L et al . Influenza vaccine with consensus internal antigens as immunogens provides cross-group protection against influenza A viruses. Front Microbiol. (2019) 10. doi: 10.3389/fmicb.2019.01630
85
Process of Influenza Vaccine Virus Selection and Development. Available online at: https://apps.who.int/gb/pip/pdf_files/Fluvaccvirusselection.pdf (Accessed May 20, 2025).
86
Helble M Chu J Flowers K Trachtman AR Huynh A Kim A et al . Structure and sequence engineering approaches to improve in vivo expression of nucleic acid-delivered antibodies. Mol Ther. (2025) 33:152–67. doi: 10.1016/j.ymthe.2024.11.030
87
Patel A Park DH Davis CW Smith TRF Leung A Tierney K et al . In vivo delivery of synthetic human DNA-encoded monoclonal antibodies protect against ebolavirus infection in a mouse model. Cell Rep. (2018) 25:1982–93.e4. doi: 10.1016/j.celrep.2018.10.062
88
Krammer F . The human antibody response to influenza A virus infection and vaccination. Nat Rev Immunol. (2019) 19:383–97. doi: 10.1038/s41577-019-0143-6
Summary
Keywords
influenza A virus, nucleoprotein, hemagglutinin, synthetic DNA, multivalent vaccine
Citation
Gary EN, Trachtman AR, Wang D, Bharti S, Ye Y, Tursi NJ, Tomirotti M, Eisenhauer J, Chu JD, Zegarra DC, Hojecki CE, Zheng M, Wickramasinghe J, Weiner DB and Patel A (2025) Synthetic DNA co-immunization with vaccine-aligned common consensus nucleoprotein and hemagglutinin protects mice against lethal influenza infection with a single immunization. Front. Immunol. 16:1632121. doi: 10.3389/fimmu.2025.1632121
Received
20 May 2025
Accepted
29 October 2025
Published
26 November 2025
Volume
16 - 2025
Edited by
Srinivasa Reddy Bonam, Indian Institute of Chemical Technology (CSIR), India
Reviewed by
Rajagowthamee Ravanapuram Thangavel, Icahn School of Medicine at Mount Sinai, United States
Kristin Wiggins, St. Jude Children’s Research Hospital, United States
Updates
Copyright
© 2025 Gary, Trachtman, Wang, Bharti, Ye, Tursi, Tomirotti, Eisenhauer, Chu, Zegarra, Hojecki, Zheng, Wickramasinghe, Weiner and Patel.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ami Patel, apatel@wistar.org
†These authors have contributed equally to this work and share first authorship
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.