- 1Beijing Institute of Hepatology, Beijing Youan Hospital, Capital Medical University, Beijing, China
- 2School of Public Health, Southern Medical University, Guangzhou, Guangdong, China
- 3The First Clinical Medicine College, Southern Medical University, Guangzhou, Guangdong, China
Despite advances in antiretroviral therapy (ART), human immunodeficiency virus type 1 (HIV-1) remains a global health challenge, with approximately 39 million people infected worldwide, persistent viral reservoirs, and delayed immune reconstitution. Exosomes, which are extracellular vesicles (30–150 nm) that play a key role in intercellular communication, have a dual role in HIV-1 pathogenesis and therapy. Regarding pathogenesis, this review elucidates how HIV-1 exploits the exosome pathway—hijacking the Endosomal Sorting Complex Required for Transport(ESCRT)machinery for viral budding and selectively packaging viral components, such as the accessory protein Nef, to enhance infectivity, promote immune evasion, and establish latent reservoirs. Conversely, host cells utilize exosomes to mount antiviral defense by packaging and transmitting restriction factors, such as APOBEC3G, to recipient cells. Furthermore, exosomal cargo serves as promising biomarkers for disease monitoring, and exosomes themselves are emerging as versatile therapeutic nanocarriers. We highlight that plant-derived exosomes offer unique advantages, including low immunogenicity and high scalability, for delivering next-generation antiviral agents or gene editing tools. In summary, understanding the multifaceted roles of exosomes provides crucial mechanistic insights into HIV-1 pathogenesis and unveils innovative strategies toward a functional cure.
1 Introduction
Since the first case of Acquired Immunodeficiency Syndrome (AIDS) was reported in 1981, human immunodeficiency virus type 1 (HIV-1) has evolved into an ongoing global public health crisis, with approximately 39 million people living with the virus worldwide in 2023, including 1.3 million new infections and 630, 000 AIDS-related deaths (1). Although antiretroviral therapy (ART) has successfully suppressed viral replication and transformed AIDS into a manageable chronic disease, significant biological and systemic challenges persist. Sub-Saharan Africa bears a disproportionate burden (2), and patients in resource-limited settings often face barriers to timely access due to socioeconomic factors (3, 4).
Despite advances in ART, including partial integrase strand transfer inhibitors (5) and long-acting regimens with improved resistance profiles (6), the path to a cure is still hampered by persistent viral reservoirs of latent proviral DNA (7), the emergence of drug resistance (8), off-target toxicity (9), and delays in immune reconstitution (10). These conditions complicate long-term adherence and predispose patients to opportunistic infections (11) and non-AIDS comorbidities (12). Achieving a functional cure, defined as sustained virological remission in the absence of ART, controlled by the host immune system, will require innovative approaches to eradicate viral reservoirs and refine treatment paradigms. Emerging strategies, such as immunomodulation through broadly neutralizing antibodies (bNAbs) (13) and CRISPR-Cas9-mediated viral genome editing (14), show preclinical promise, but still face hurdles related to variable efficacy, safety, and cost.
In this quest for a cure, extracellular vesicles (EVs), particularly exosomes, are gradually attracting academic attention. The field increasingly uses the broader term EVs, which includes exosomes, microvesicles, and apoptotic bodies. Following the Minimal Information for Studies of Extracellular Vesicles (MISEV) guidelines, this review focuses primarily on exosomes (30–150 nm), which are produced by the fusion of multivesicular bodies with the plasma membrane (15). Exosomes are enriched with tetraspanins (CD9, CD63, CD81) and heat shock proteins, and orchestrate inter-cellular communication through the transfer of biologically active transmitters (16).
Exosomes are implicated in the pathogenesis of HIV-1, demonstrating a dual role as both disease drivers and therapeutic vectors. Regarding pathogenesis, they may facilitate viral transmission through “Trojan horse-like” transport (17), suppress antiviral immunity, or deliver pathogenic factors. Conversely, exosomes also promote host antiviral responses, deliver pathogenic factors, and serve as biomarkers for early diagnosis (18). This dual role highlights their profound translational potential (19). This review is particularly timely given the recent technological breakthroughs in EV isolation and characterization, such as microfluidics and nanoparticle tracking analysis (NTA), which now allow for more precise and reproducible studies of exosome function.
2 Exosomes at the crossroads: pathogenesis, immune evasion, and co-infection
2.1 Hijacking host machinery: ESCRT-mediated viral biogenesis
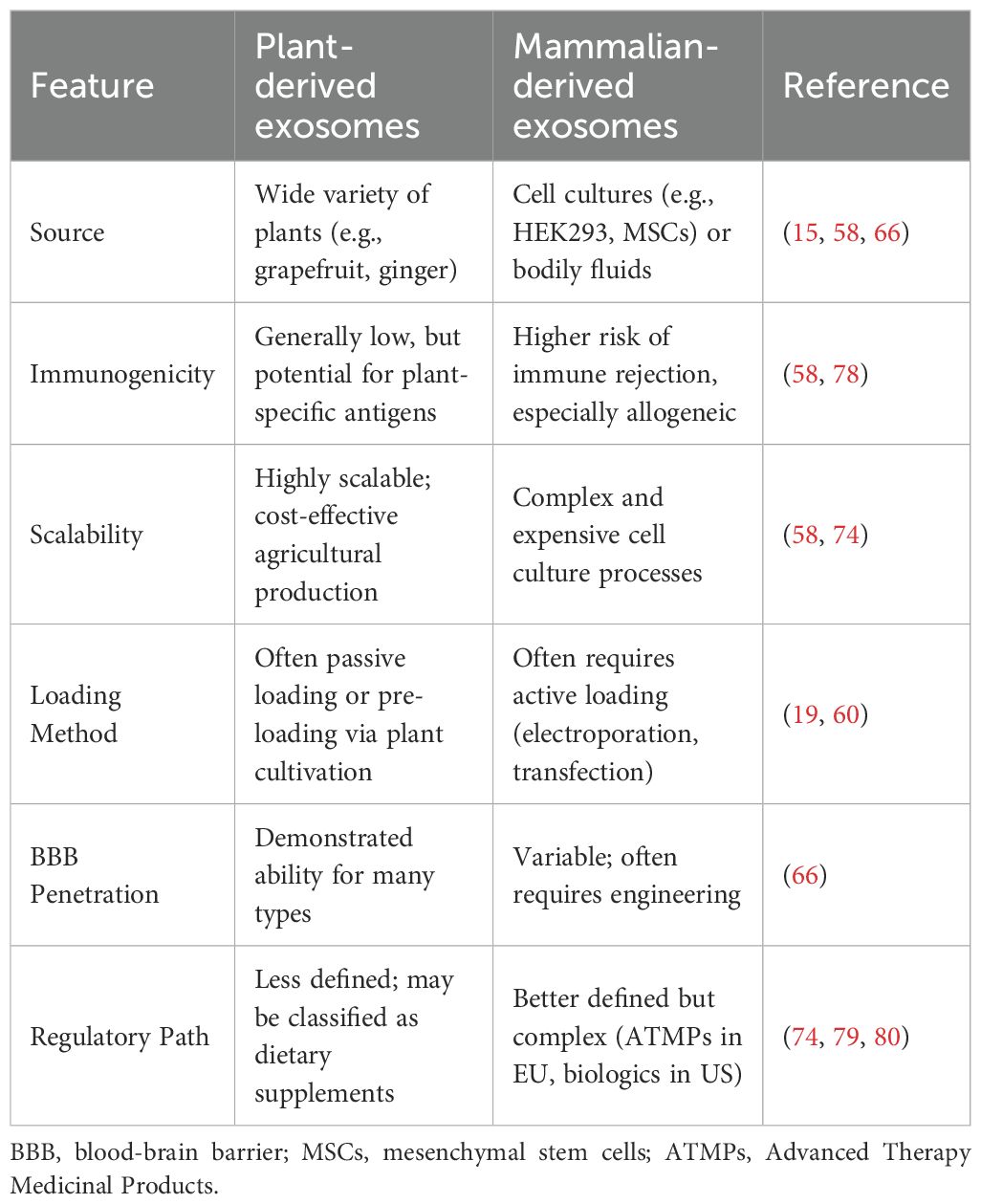
The assembly of HIV-1 viral particles shares highly conserved molecular features with the biogenesis of exosomes, both of which rely on the sophisticated regulation of the Endosomal Sorting Complex Required for Transport (ESCRT) pathway. This mechanistic overlap suggests that HIV-1 intentionally hijacks the exosome secretory pathway as a vehicle for viral transmission and immune evasion. Figure 1 provides a schematic overview of how exosomes are co-opted throughout the HIV-1 life cycle, highlighting the intersection of the ESCRT pathway in both viral budding and exosome biogenesis.
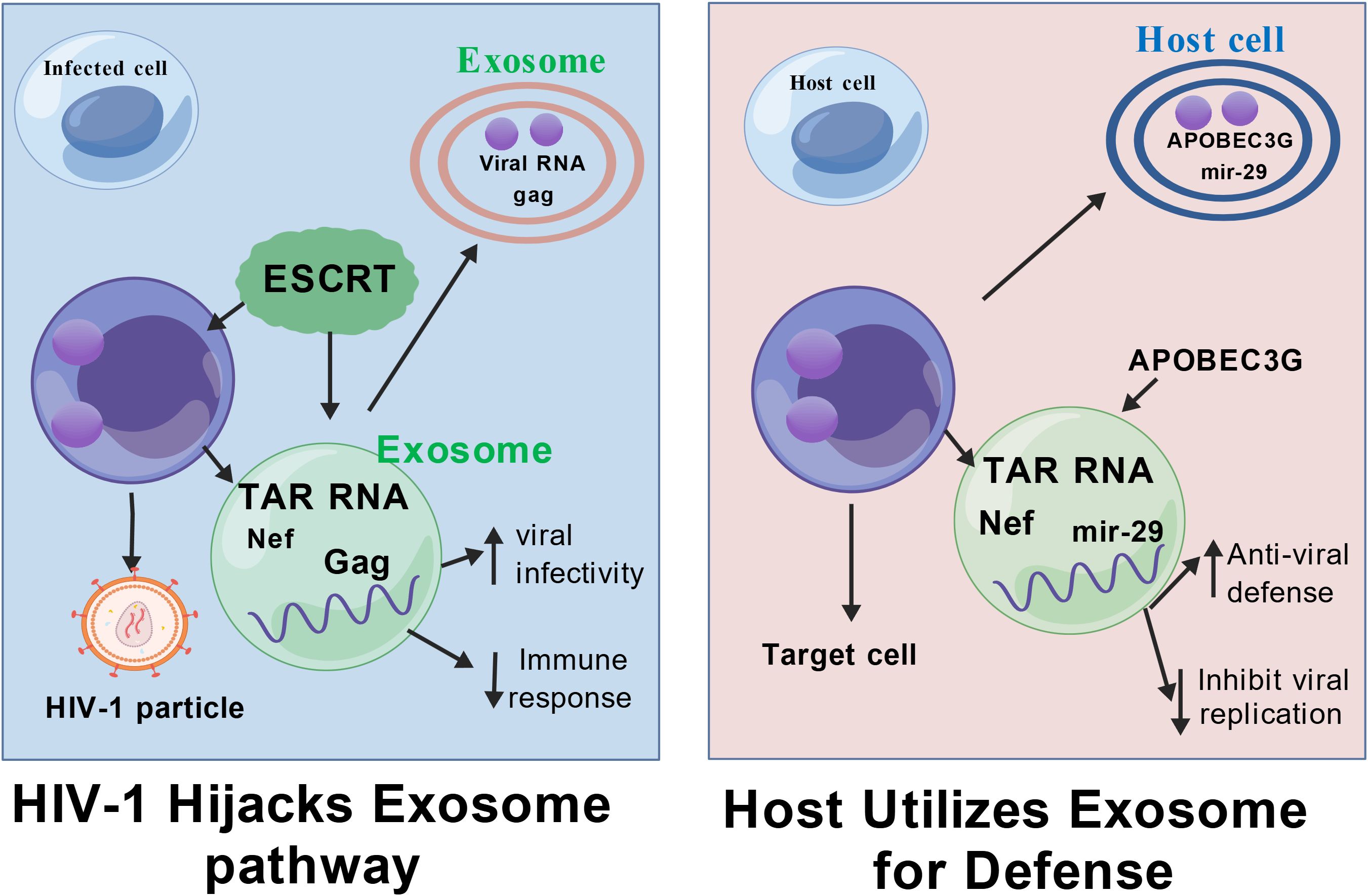
Figure 1. Dual role of exosomes in HIV-1 transmission and immune escape.Exosomes carry proteins, RNA, and other components that can interact with HIV-1 through the ESCRT pathway, and HIV-1 can hijack the exosome secretion pathway to deliver viral components, facilitate infection and evade immune surveillance. Exosomes are also capable of carrying viral RNA and DNA, affecting host cell gene expression and function.Figure Created with BioGDP.com.
The HIV-1 Gag protein drives viral envelope outgrowth from host cell membranes by specifically recruiting ESCRT-I complex member TSG101 and ESCRT-III subunit CHMP4 (20, 21). This recruitment allows Gag to mimic the membrane scission process normally used to release intraluminal vesicles. Subsequently, the Vps4 protein mediates the dissociation and recycling of the ESCRT complex through hydrolysis of ATP (22), completing both viral release and the maturation of multivesicular bodies (MVBs) into exosomes (23). The resulting exosomes are often enriched with viral components (such as viral RNA and accessory proteins like Nef), facilitating their role in viral pathogenesis.
2.2 Dual role in immune modulation and viral transmission
During HIV-1 infection, exosomes exhibit bidirectional regulation in the microenvironment. On the one hand, viruses can deliver viral proteins and microRNAs to uninfected T cells via exosomes, significantly enhancing the susceptibility of target cells to HIV-1 by activating cellular pathways or altering surface receptor expression (24, 25). Furthermore, exosome surface molecules play a key role in viral invasion. Recent studies have demonstrated that tetraspanin family members (CD9, CD63, CD81, and CD37) can facilitate HIV-1 entry into host cells by mediating membrane fusion or endocytosis (11). Monoclonal antibodies against CD9/CD81 significantly block this exosome-dependent viral transmission, supporting the rationale for targeting these interactions.
On the other hand, host-derived exosomes can exert natural antiviral effects. They can inhibit viral replication by competitively binding to CD4+ T-cell surface receptors or by delivering restriction factors such as APOBEC3G (26–28). This finding underscores the potential for a combined therapeutic strategy targeting virus-exosome interactions: interfering with viral outgrowth through ESCRT pathway inhibitors (such as Vps4 antagonists), while concurrently modulating immune pathways and oxidative stress signaling (30). Crucially, the role of exosomes is highly dependent on their cell origin; for instance, exosomes derived from microglia may uniquely influence HIV-associated neurocognitive disorders (HAND), an area critical to addressing viral reservoir persistence.
2.3 Exosomes in viral co-infection and reservoir establishment
The functional complexity of exosomes is further highlighted in the context of viral and bacterial co-infection. Exosomes released in the microenvironment of HIV-1 and Mycobacterium tuberculosis (MTB) co-infection have been found to carry both viral RNA and pro-inflammatory factors such as TNF-α (29). These exosomes not only activate inflammatory pathways (like NF-κB), but also deliver viral components to distal cells via the paracrine pathway, promoting the establishment and reactivation of viral latent reservoirs—a crucial area for patients undergoing ART.
Coinfections with HBV or HCV also illustrate this interplay. Both HBV and HCV rely on the host ESCRT system and Rab GTPase-regulated endocytosis transport pathway for viral assembly and exosome release (31–34). Exosomes secreted by HBV-infected hepatocytes not only encapsulate intact viral particles or HBc/LHBs antigens (35), but also deliver PD-L1 proteins to induce T-cell depletion, thereby weakening the antiviral immune response and promoting latent HIV infection (36). Similarly, HCV infection-associated exosomes form transmission units with immune escape properties by carrying viral RNA-core protein complexes and host factors such as Ago2, HSP90, and miR-122 (37). While miR-122 enhances viral replication by stabilising HCV RNA, a related pathway involves exosome-mediated activation of the miR-19a/SOCS3/STAT3/TGF-β signaling axis, which synergistically promotes hepatic fibrosis progression (38, 39). Specifically, miR-19a targets SOCS3, which in turn de-represses the STAT3 pathway, leading to increased expression of the profibrotic cytokine TGF-β. It is therefore suggested to establish a dynamic monitoring and joint intervention system for co-infection, aiming to break through existing treatment bottlenecks and achieve functional cure of HIV-1 and effective control of co-infection complications through interdisciplinary cooperation and preclinical model optimization.
2.4 Exosomal cargo specificity and heterogeneity from diverse reservoirs
Beyond T cells, HIV-1 infects a broad spectrum of cells, including macrophages, microglia, and dendritic cells, each contributing uniquely to viral reservoir persistence and the clinical challenges of ART-mediated remission. Exosomes derived from these diverse cellular reservoirs exhibit distinct compositions and functional properties. For instance, microglia-derived exosomes have been centrally implicated in HIV-1-associated neurocognitive disorders (HAND), carrying neurotoxic viral proteins (such as Nef or Tat) and inflammatory cytokines that disrupt neuronal function (17). Similarly, macrophages, which are key tissue reservoirs of latent HIV-1, release exosomes that are often enriched with pro-inflammatory microRNAs (miRNAs) and viral components. These exosomes can facilitate viral dissemination and significantly modulate the immune microenvironment in distant tissues (40).
Crucially, the process by which these viral components are incorporated into exosomes is not merely passive but is governed by highly selective packaging mechanisms. This targeted inclusion ensures the delivery of pathogenic molecules essential for viral spread and persistence. For example, both HIV-1 proteins (such as Nef and Gag) and viral RNAs (like TAR) are highly enriched in exosomes from infected cells (41, 42). Nef, a potent accessory protein, modulates exosome biogenesis and content, promoting the secretion of vesicles that enhance infectivity and impair immune responses in recipient cells (43). Similarly, the TAR RNA element—a key non-coding viral component—is abundantly packaged into exosomes and can manipulate gene expression in target cells to foster a proviral environment (44).
Understanding these specific compositions and packaging rules across different cell types is crucial for developing comprehensive “shock and kill” or “block and lock” strategies to eliminate the persistent HIV-1 reservoir and effectively treat its major comorbidities.
3 Exosomes as biomarkers for HIV-1 diagnosis and disease monitoring
3.1 Clinical rationale and diagnostic potential
Exosomes have become novel candidate markers for disease diagnosis and progression monitoring due to their unique biological properties (45). The nucleic acids, proteins, and lipid components they carry not only dynamically reflect the pathophysiological state of host cells (46–49), but also provide an ideal sample source for non-invasive liquid biopsy due to the protective effect of the exosome membrane structure, which makes its contents highly stable in body fluids. Figure 2 illustrates the translational potential of exosomal biomarkers across different stages of HIV-1 infection, from early diagnosis to reservoir detection. This stability and ability to reflect tissue-specific status give them a distinct advantage over traditional plasma markers. Some studies have shown that in fields such as neurodegenerative diseases and renal fibrosis, the aberrant expression of exosome-specific miRNAs (such as miR-21 and the miR-29 family) opens new avenues for early diagnosis and assessment of therapeutic efficacy (50, 51). In conclusion, the diagnostic value of exosomes in HIV-1 infection is gradually being highlighted, with a specific focus on identifying signatures that correlate with chronic inflammation and immune dysregulation.
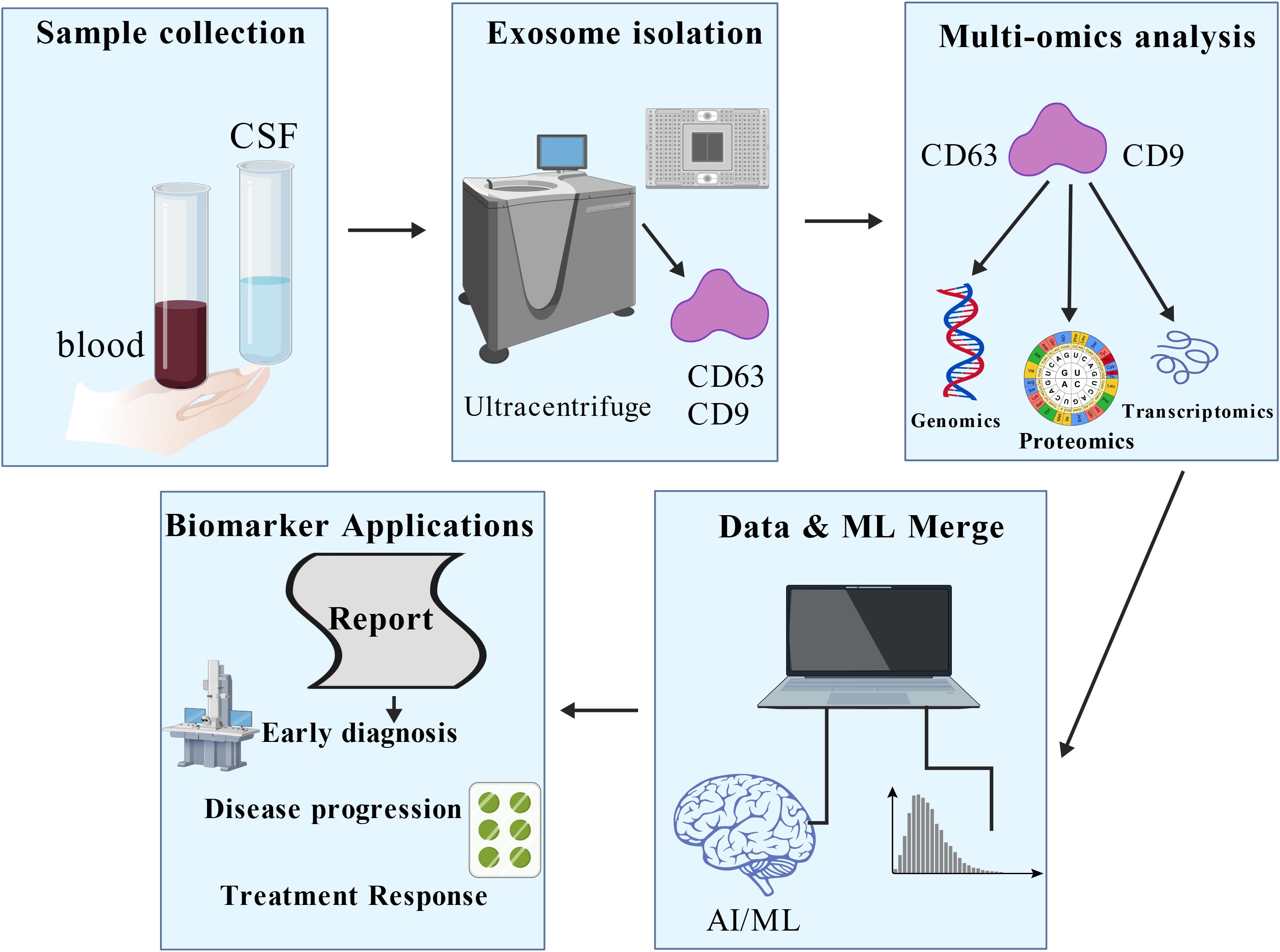
Figure 2. Potential applications of exosomes in the diagnosis and treatment of HIV-1 infection. Exosomes can be extracted through human samples. Exosomes carry a variety of components, including proteins and RNA, which can be used as biomarkers for detecting viral load. The molecular characteristics of these exosomes are analysed in depth using advanced analytical techniques such as machine learning to help assess disease progression and immune reconstitution. Figure Created with BioGDP.com.
3.2 Molecular signatures and AI-driven discovery for disease monitoring
Studies have shown that the number of exosomes in the plasma of HIV-infected patients without antiretroviral therapy (ART) is significantly increased, and their particle size distribution is shifted to a larger size. The molecular cargo contained within these exosomes provides robust indicators of disease progression, characterized by up-regulated miR-155 and miR-146a and abnormal elevation of oxidative stress markers such as 8-OHdG and MDA (52). A cohort study further indicated that plasma exosome abundance was significantly and positively correlated with absolute CD8+ T-cell counts (53), a correlation that provides a potential biological basis for assessing the state of immune reconstitution and systemic inflammation in HIV-infected patients undergoing ART.
Despite the richness of molecular information contained in exosomes (54), screening for specific markers that are highly correlated with HIV-1 infection progression remains challenging. Traditional biomarker discovery strategies are limited by the complexity of multidimensional data. The introduction of machine learning techniques has provided a breakthrough direction for this bottleneck. It has been proposed that random forest classifiers can significantly outperform the predictive efficacy of traditional statistical methods by integrating exosomal proteomics, transcriptomics, and clinical parameters in the application scenario of identifying cancer-related markers (55, 56). This methodological advancement drives innovation for HIV research: an integrated analysis framework based on deep learning-driven fusion of multi-omics data and artificial intelligence may resolve key feature profiles associated with viral load, latent reservoir activation, or immune exhaustion from the exosomal molecular network, leading to accurate disease stratification models (57). Future studies need to further validate the applicability of such computational biology strategies in longitudinal HIV cohorts to accelerate the clinical translation of exosome markers.
4 Exosomes as nanocarriers for anti-HIV-1 therapy: a comparative perspective
4.1 Unique advantages and targeted delivery of plant exosomes
Plant-derived exosomes (also often termed exosome-like nanoparticles or HELNs) have emerged as promising nanocarriers for antiviral therapy, offering unique advantages such as high biocompatibility, inherent stability, and the demonstrated ability to cross biological barriers like the blood-brain barrier (BBB) (58–66). Their structural similarity to mammalian exosomes, coupled with the abundance of bioactive molecules they carry, makes them an attractive, scalable platform for therapeutic agent delivery. Figure 3 summarizes this potential: after extraction from plants, these exosomes can be engineered to carry anti-HIV chemical drugs. These modified exosomes serve as targeted drug delivery systems, aiming for specific cells like CD4+ T cells, thereby increasing therapeutic efficacy and minimizing off-target toxicity. This process effectively combines the natural advantages of plant-derived exosomes with modern bioengineering and drug design techniques. Studies have highlighted their potential for targeting HIV-1 related pathology. For instance, grapefruit-derived exosomes exhibit high drug-loading efficiency and protective properties (58). Similarly, ginger exosomes have shown potential in modulating aberrant immune responses and reducing inflammatory pathways (63), which could be beneficial in managing the chronic immune activation and non-AIDS comorbidities associated with HIV-1 infection. Their proven capacity for CNS penetration further addresses a critical challenge in HIV-1 treatment, offering an ideal carrier platform for eradicating viral reservoirs in the brain (66).
Figure 3. Potential of plant-derived exosomes for HIV-1 therapy. After exosomes are extracted from plants, they can be engineered to carry chemical drugs. These modified exosomes can be used as drug delivery systems to target anti-HIV drugs to target cells such as CD4+ T cells, thereby increasing therapeutic efficacy and reducing the impact on non-target cells. This process combines the natural advantages of plant-derived exosomes with modern bioengineering and drug design techniques, providing innovative ideas for developing novel anti-HIV therapeutic strategies. Figure Created with BioGDP.com.
4.2 Comparative analysis and translational challenges
While plant exosomes present a promising, cost-effective alternative to mammalian vectors—particularly for their high scalability and generally low immunogenicity—a clearer perspective requires a critical comparison against alternative delivery systems, as summarized in Table 1. However, the translation of plant exosomes into clinical use is not without significant critical challenges. Variability in molecular cargo and quantity between plant sources, potential host-specific immunogenicity, and a limited understanding of their precise mechanistic behavior in human systemic circulation remain major hurdles (58, 74, 78). In conclusion, to fully realize their potential in HIV-1 therapy, future research must address three crucial bottlenecks: standardization of isolation protocols (to ensure consistency and homogeneity), rigorous safety and pharmacokinetic profiling in vivo, and clarification of the complex global regulatory pathways (72–80). This concerted effort is necessary to move these highly promising natural nanocarriers from the bench to the bedside.
5 Outlook and future directions: addressing translational hurdles
The clinical translation of exosomes in HIV-1 therapy faces multidimensional challenges, demanding systematic breakthroughs that reconcile their dual biological attributes and therapeutic potential.
5.1 Challenges in clinical trial design and quality control
The clinical translation of exosomes in HIV-1 therapy is fundamentally constrained by deep technical and biological contradictions. First, the primary obstacle in clinical trial design stems from exosome biological heterogeneity. The dual roles of exosomes as both viral transmission vectors and potential therapeutic tools (67–70) challenge traditional efficacy assessment systems centered on viral load (71). Differences arising from host factors and preparation processes further complicate single-cohort studies, with inconsistencies in particle detection methods introducing data bias (74). This urgently mandates the establishment of a functionally validated quality control system, standardizing key parameters like target delivery efficiency and pathogen clearance (72, 73, 75). Furthermore, novel joint endpoint indicators, such as latent reservoir-specific miRNA profiles combined with single-cell sequencing, are required to resolve the exosomal dynamic reprogramming of the immune microenvironment (76, 77).
Second, safety and scale-up pose significant hurdles. Despite the low immunogenicity of natural exosomes, risks persist: exosomes from HIV-infected cells may carry viral proteins like gp120, necessitating stringent pathogen clearance processes (78). Large-scale production of engineered exosomes is challenged by batch-to-batch heterogeneity, where fluctuations in loading efficiency compromise therapeutic consistency (75). Ultimately, systematic assessment of long-term exposure risks in preclinical models is required, especially regarding the dynamic interplay between exosomes and vulnerable microenvironments such as the Central Nervous System.
5.2 Regulatory barriers and future roadmap: integrating an interdisciplinary ecosystem
The ambiguity of the regulatory framework severely exacerbates the translational dilemma. The transboundary properties of exosomes lead to classification controversies: the EU classifies functional RNA-carrying exosomes as Advanced Therapeutic Medicinal Products (ATMPs) (79), while the US FDA dynamically adjusts classification criteria based on functional properties (80). This regulatory uncertainty directly leads to a fragmented quality control system, evidenced by the lack of international consensus on assays for miRNA quantification or CRISPR complex activity (81, 82). Although countries like Japan and South Korea have attempted to standardize production processes through special guidelines (83, 84), there is still an urgent need to establish a cross-regional adaptive regulatory pathway focused on resolving the ethical review and biosafety assessment challenges of exosomes carrying gene editing tools (79, 80). Furthermore, achieving a paradigm breakthrough in HIV-1 therapy requires an interdisciplinary synergistic system spanning virology, nanomedicine, and policy sciences. This system involves action at three levels: focusing research on exosomes as intelligent carriers at the “virus-host interaction interface” (Mechanism Level); promoting the process integration of technologies like ultracentrifugation and microfluidics to establish end-to-end quality control standards (Translational Strategy); and leveraging ATMP management experience to formulate a function-oriented dynamic classification strategy (Regulatory Adaptation). Only through the triple drive of technology iteration, standardization, and policy innovation can exosomes move from a laboratory concept to a strategic tool for clinical antiretroviral therapy.
6 Conclusion and future outlook
This review has elucidated the dual role of exosomes in HIV-1 pathogenesis, highlighting their critical capacity to facilitate viral spread (via ESCRT-mediated packaging and reservoir-specific cargo) while simultaneously serving as promising therapeutic and diagnostic tools. Translating this potential from bench to bedside necessitates overcoming significant, interlinked hurdles.
A primary challenge lies in the inherent heterogeneity and lack of standardization in exosome isolation and characterization. Current methods yield preparations with varying purity and composition, which complicates data interpretation and demands the development of robust, scalable, and reproducible manufacturing processes (73). Furthermore, the biological complexity requires a deeper understanding of their in vivo fate, targeting specificity, and potential off-target effects, particularly when engineered as drug delivery vehicles (19). The regulatory landscape remains ambiguous; clarifying whether these products are classified as drugs, biologics, or advanced therapies is essential for defining a clear path to clinical approval (79). Ultimately, future research must bridge knowledge gaps—such as the precise mechanisms of exosome-mediated reservoir establishment and reactivation—by harnessing multidisciplinary approaches like synthetic biology and machine learning to design smart exosomes capable of targeted reservoir elimination and immune reconstitution.
By addressing these challenges through concerted efforts in basic science, manufacturing engineering, and regulatory collaboration, exosome-based strategies may eventually evolve into cornerstone technologies for achieving a functional cure for HIV-1. (All figures in this paper via BioGDP (85)).
Author contributions
PL: Writing – original draft, Writing – review & editing, Investigation. XL: Writing – review & editing, Investigation. WY: Writing – review & editing, Software. JL: Writing – original draft, Writing – review & editing, Supervision.
Funding
The author(s) declare that no financial support was received for the research, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. World Health Organization. HIV and AIDS(2024). Available online at: https://www.who.int/news-room/fact-sheets/detail/hiv-aids (Accessed November 11, 2025).
2. Kharsany AB and Karim QA. HIV infection and AIDS in sub-saharan africa: current status, challenges and opportunities. Open AIDS J. (2016) 10:34–48. doi: 10.2174/1874613601610010034
3. Moyo E, Moyo P, Murewanhema G, Mhango M, Chitungo I, and Dzinamarira T. Key populations and Sub-Saharan Africa's HIV response. Front Public Health. (2023) 11:1079990. doi: 10.3389/fpubh.2023.1079990
4. Yermukhanova L, Kuzembayev M, Salkhanova A, Narymbayeva N, Tazhiyeva A, Makhanbetkulova DN, et al. Exploring socio-economic dimensions in HIV research: a comprehensive bibliometric analysis (1992-2024). Glob Health Action. (2025) 18:2474787. doi: 10.1080/16549716.2025.2474787
5. Ramgopal MN, Castagna A, Cazanave C, Diaz-Brito V, Dretler R, Oka S, et al. Efficacy, safety, and tolerability of switching to long-acting cabotegravir plus rilpivirine versus continuing fixed-dose bictegravir, emtricitabine, and tenofovir alafenamide in virologically suppressed adults with HIV, 12-month results (SOLAR): a randomised, open-label, phase 3b, non-inferiority trial. Lancet HIV. (2023) 10:e566–77. doi: 10.1016/S2352-3018(23)00136-4
6. Orkin C, DeJesus E, Sax PE, Arribas JR, Gupta SK, Martorell C, et al. Fixed-dose combination bictegravir, emtricitabine, and tenofovir alafenamide versus dolutegravir-containing regimens for initial treatment of HIV-1 infection: week 144 results from two randomised, double-blind, multicentre, phase 3, non-inferiority trials. Lancet HIV. (2020) 7:e389–400. doi: 10.1016/S2352-3018(20)30099-0
7. Matsuda K and Maeda K. HIV reservoirs and treatment strategies toward curing HIV infection. Int J Mol Sci. (2024) 25:2621. doi: 10.3390/ijms25052621
8. World Health Organization. Fact sheet: HIV drug resistance. Available online at: https://www.who.int/news-room/fact-sheets/detail/HIV-drug-resistance (Accessed November 11, 2025).
9. Young MJ. Off-target effects of drugs that disrupt human mitochondrial DNA maintenance. Front Mol Biosci. (2017) 4:74. doi: 10.3389/fmolb.2017.00074
10. Yang X, Su B, Zhang X, Liu Y, Wu H, and Zhang T. Incomplete immune reconstitution in HIV/AIDS patients on antiretroviral therapy: Challenges of immunological non-responders. J Leukoc Biol. (2020) 107:597–612. doi: 10.1002/JLB.4MR1019-189R
11. Zhu Q, Gao F, Ren X, Li R, Kang J, Li M, et al. Nutritional risk and HbA1c as critical risk factors and predictors of opportunistic infections in HIV-DM comorbid patients: a retrospective cross-sectional study. Front Endocrinol (Lausanne). (2025) 15:1527936. doi: 10.3389/fendo.2024.1527936
12. Swinkels HM, Nguyen AD, and Gulick PG. HIV and AIDS. In: StatPearls. StatPearls Publishing, Treasure Island (FL (2025). Available online at: https://www.ncbi.nlm.nih.gov/books/NBK534860/ (Accessed November 17, 2025).
13. Mahomed S, Pillay K, Hassan-Moosa R, Galvão BPGV, Burgers WA, Moore PL, et al. Clinical trials of broadly neutralizing monoclonal antibodies in people living with HIV - a review. AIDS Res Ther. (2025) 22:44. doi: 10.1186/s12981-025-00734-8
14. Feist WN, Luna SE, Ben-Efraim K, Filsinger Interrante MV, Amorin A, Johnston NM, et al. Multilayered HIV-1 resistance in HSPCs through CCR5 Knockout and B cell secretion of HIV-inhibiting antibodies. Nat Commun. (2025) 16:3103. doi: 10.1038/s41467-025-58371-8
15. Doyle LM and Wang MZ. Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells. (2019) 8:727. doi: 10.3390/cells8070727
16. Fan Y, Pionneau C, Cocozza F, Boëlle PY, Chardonnet S, Charrin S, et al. Differential proteomics argues against a general role for CD9, CD81 or CD63 in the sorting of proteins into extracellular vesicles. J Extracell Vesicles. (2023) 12:e12352. doi: 10.1002/jev2.12352
17. Habib A, Liang Y, and Zhu N. Exosomes multifunctional roles in HIV-1: insight into the immune regulation, vaccine development and current progress in delivery system. Front Immunol. (2023) 14:1249133. doi: 10.3389/fimmu.2023.1249133
18. Shrivastava S, Ray RM, Holguin L, Echavarria L, Grepo N, Scott TA, et al. Exosome-mediated stable epigenetic repression of HIV-1. Nat Commun. (2021) 12:5541. doi: 10.1038/s41467-021-25839-2
19. Liang Y, Duan L, Lu J, and Xia J. Engineering exosomes for targeted drug delivery. Theranostics. (2021) 11:3183–95. doi: 10.7150/thno.52570
20. Usami Y, Popov S, Popova E, Inoue M, Weissenhorn W, and Göttlinger GH. The ESCRT pathway and HIV-1 budding. Biochem Soc Trans. (2009) 37:181–4. doi: 10.1042/BST0370181
21. von Schwedler UK, Stuchell M, Müller B, Ward DM, Chung HY, Morita E, et al. The protein network of HIV budding. Cell. (2003) 114:701–13. doi: 10.1016/S0092-8674(03)00714-1
22. Muzioł T, Pineda-Molina E, Ravelli RB, Zamborlini A, Usami Y, Göttlinger H, et al. Structural basis for budding by the ESCRT-III factor CHMP3. Dev Cell. (2006) 10:821–30. doi: 10.1016/j.devcel.2006.03.013
23. Juan T and Fürthauer M. Biogenesis and function of ESCRT-dependent extracellular vesicles. Semin Cell Dev Biol. (2018) 74:66–77. doi: 10.1016/j.semcdb.2017.08.022
24. Arenaccio C, Chiozzini C, Columba-Cabezas S, Manfredi F, Affabris E, Baur A, et al. Exosomes from human immunodeficiency virus type 1 (HIV-1)-infected cells license quiescent CD4+ T lymphocytes to replicate HIV-1 through a Nef- and ADAM17-dependent mechanism. J Virol. (2014) 88:11529–39. doi: 10.1128/JVI.01712-14
25. Campbell TD, Khan M, Huang MB, Bond VC, and Powell MD. HIV-1 Nef protein is secreted into vesicles that can fuse with target cells and virions. Ethn Dis. (2008) 18:S2–14-19.
26. Näslund TI, Paquin-Proulx D, Paredes PT, Vallhov H, Sandberg JK, and Gabrielsson S. Exosomes from breast milk inhibit HIV-1 infection of dendritic cells and subsequent viral transfer to CD4+ T cells. AIDS. (2014) 28:171–80. doi: 10.1097/QAD.0000000000000159
27. Khatua AK, Taylor HE, Hildreth JE, and Popik W. Exosomes packaging APOBEC3G confer human immunodeficiency virus resistance to recipient cells. J Virol. (2009) 83:512–21. doi: 10.1128/JVI.01658-08
28. Li YL, Langley CA, Azumaya CM, Echeverria I, Chesarino NM, Emerman M, et al. The structural basis for HIV-1 Vif antagonism of human APOBEC3G. Nature. (2023) 615:728–33. doi: 10.1038/s41586-023-05779-1
29. Tyagi P, Pal VK, Agrawal R, Singh S, Srinivasan S, and Singh A. Mycobacterium tuberculosis Reactivates HIV-1 via Exosome-Mediated Resetting of Cellular Redox Potential and Bioenergetics. mBio. (2020) 11:e03293–19. doi: 10.1128/mBio.03293-19
30. Sims B, Farrow AL, Williams SD, Bansal A, Krendelchtchikov A, and Matthews QL. Tetraspanin blockage reduces exosome-mediated HIV-1 entry. Arch Virol. (2018) 163:1683–9. doi: 10.1007/s00705-018-3737-6
31. Hayes CN, Zhang Y, Makokha GN, Hasan MZ, Omokoko MD, and Chayama K. Early events in hepatitis B virus infection: From the cell surface to the nucleus. J Gastroenterol Hepatol. (2016) 31:302–9. doi: 10.1111/jgh.13175
32. Chou SF, Tsai ML, Huang JY, Chang YS, and Shih C. The dual role of an ESCRT-0 component HGS in HBV transcription and naked capsid secretion. PloS Pathog. (2015) 11:e1005123. doi: 10.1371/journal.ppat.1005123
33. Kulhanek KR, Roose JP, and Rubio I. Regulation of the small GTPase Ras and its relevance to human disease. Methods Mol Biol. (2021) 2262:19–43. doi: 10.1007/978-1-0716-1190-6_2
34. Corless L, Crump CM, Griffin SD, and Harris M. Vps4 and the ESCRT-III complex are required for the release of infectious hepatitis C virus particles. J Gen Virol. (2010) 91:362–72. doi: 10.1099/vir.0.017285-0
35. Ninomiya M, Inoue J, Krueger EW, Chen J, Cao H, Masamune A, et al. The exosome-associated tetraspanin CD63 contributes to the efficient assembly and infectivity of the hepatitis B virus. Hepatol Commun. (2021) 5:1238–51. doi: 10.1002/hep4.1709
36. Kushch AA and Ivanov AV. Exosomes in the life cycle of viruses and the pathogenesis of viral infections. Problems Virol. (2023) 68:181–97. doi: 10.36233/0507-4088-173
37. Ramakrishnaiah V, Thumann C, Fofana I, Habersetzer F, Pan Q, de Ruiter PE, et al. Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7.5 cells. Proc Natl Acad Sci USA. (2013) 110:13109–13. doi: 10.1073/pnas.1221899110
38. Bukong TN, Momen-Heravi F, Kodys K, Bala S, and Szabo G. Exosomes from hepatitis C infected patients transmit HCV infection and contain replication competent viral RNA in complex with Ago2-miR122-HSP90. PloS Pathog. (2014) 10:e1004424. doi: 10.1371/journal.ppat.1004424
39. Devhare PB, Sasaki R, Shrivastava S, Di Bisceglie AM, Ray R, and Ray RB. Exosome-mediated intercellular communication between hepatitis C virus-infected hepatocytes and hepatic stellate cells. J Virol. (2017) 91:e02225–16. doi: 10.1128/JVI.02225-16
40. Campbell LA and Mocchetti I. Extracellular vesicles and HIV-associated neurocognitive disorders: implications in neuropathogenesis and disease diagnosis. Neurotox Res. (2021) 39:2098–107. doi: 10.1007/s12640-021-00425-y
41. Patters BJ and Kumar S. The role of exosomal transport of viral agents in persistent HIV pathogenesis. Retrovirology. (2018) 15:79. doi: 10.1186/s12977-018-0462-x
42. Tang Z, Lu Y, Dong JL, Wu W, and Li J. The extracellular vesicles in HIV infection and progression: mechanisms, and theranostic implications. Front Bioeng Biotechnol. (2024) 12:1376455. doi: 10.3389/fbioe.2024.1376455
43. McNamara RP, Costantini LM, Myers TA, Schouest B, Maness NJ, Griffith JD, et al. Nef Secretion into Extracellular Vesicles or Exosomes Is Conserved across Human and Simian Immunodeficiency Viruses. mBio. (2018) 9:e02344–17. doi: 10.1128/mBio.02344-17
44. Sampey GC, Saifuddin M, Schwab A, Barclay R, Punya S, Chung MC, et al. Exosomes from HIV-1-infected cells stimulate production of pro-inflammatory cytokines through trans-activating response (TAR) RNA. J Biol Chem. (2016) 291:1251–66. doi: 10.1074/jbc.M115.662171
45. Properzi F, Logozzi M, and Fais S. Exosomes: the future of biomarkers in medicine. biomark Med. (2013) 7:769–78. doi: 10.2217/bmm.13.63
46. Nedaeinia R, Manian M, Jazayeri MH, Ranjbar M, Salehi R, Sharifi M, et al. Circulating exosomes and exosomal microRNAs as biomarkers in gastrointestinal cancer. Cancer Gene Ther. (2017) 24:48–56. doi: 10.1038/cgt.2016.77
47. Yoshioka Y, Konishi Y, Kosaka N, Katsuda T, Kato T, and Ochiya T. Comparative marker analysis of extracellular vesicles in different human cancer types. J Extracell Vesicles. (2013) 2:20424. doi: 10.3402/jev.v2i0.20424
48. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. (2008) 105:10513–8. doi: 10.1073/pnas.0804549105
49. Lin J, Li J, Huang B, Liu J, Chen X, Chen XM, et al. Exosomes: novel biomarkers for clinical diagnosis. ScientificWorldJournal. (2015) 2015:1–8. doi: 10.1155/2015/657086
50. Saman S, Kim W, Raya M, Visnick Y, Miro S, Saman S, et al. Exosome-associated tau is secreted in tauopathy models and is selectively phosphorylated in cerebrospinal fluid in early Alzheimer disease. J Biol Chem. (2012) 287:3842–9. doi: 10.1074/jbc.M111.277061
51. Zhou H, Pisitkun T, Aponte A, Yuen PS, Hoffert JD, Yasuda H, et al. Exosomal Fetuin-A identified by proteomics: a novel urinary biomarker for detecting acute kidney injury. Kidney Int. (2006) 70:1847–57. doi: 10.1038/sj.ki.5001874
52. Chettimada S, Lorenz DR, Misra V, Dillon ST, Reeves RK, Manickam C, et al. Exosome markers associated with immune activation and oxidative stress in HIV patients on antiretroviral therapy. Sci Rep. (2018) 8:7227. doi: 10.1038/s41598-018-25515-4
53. Hubert A, Subra C, Jenabian MA, Tremblay Labrecque PF, Tremblay C, Laffont B, et al. Elevated abundance, size, and microRNA content of plasma extracellular vesicles in viremic HIV-1+ Patients: correlations with known markers of disease progression. J Acquir Immune Defic Syndr. (2015) 70:219–27. doi: 10.1097/QAI.0000000000000756
54. Lee YJ, Shin KJ, and Chae YC. Regulation of cargo selection in exosome biogenesis and its biomedical applications in cancer. Exp Mol Med. (2024) 56:877–89. doi: 10.1038/s12276-024-01209-y
55. Cansever Mutlu E, Kaya M, Küçük I, Ben-Nissan B, and Stamboulis A. Exosome structures supported by machine learning can be used as a promising diagnostic tool. Materials (Basel). (2022) 15:7967. doi: 10.3390/ma15227967
56. Li B, Kugeratski FG, and Kalluri R. A novel machine learning algorithm selects proteome signature to specifically identify cancer exosomes. Elife. (2024) 12:RP90390. doi: 10.7554/eLife.90390
57. Nikanjam M, Kato S, and Kurzrock R. Liquid biopsy: current technology and clinical applications. J Hematol Oncol. (2022) 15:131. doi: 10.1186/s13045-022-01351-y
58. Kilasoniya A, Garaeva L, Shtam T, Spitsyna A, Putevich E, Moreno-Chamba B, et al. Potential of Plant Exosome Vesicles from Grapefruit (Citrus × paradisi) and Tomato (Solanum lycopersicum) Juices as Functional Ingredients and Targeted Drug Delivery Vehicles. Antioxidants (Basel). (2023) 12:943. doi: 10.3390/antiox12040943
59. Zhu H, Chang M, Wang Q, Chen J, Liu D, and He W. Identifying the Potential of miRNAs in Houttuynia cordata-Derived Exosome-Like Nanoparticles Against Respiratory RNA Viruses. Int J Nanomedicine. (2023) 18:5983–6000. doi: 10.2147/IJN.S425173
60. Jiang D, Li Z, Liu H, Liu H, Xia X, and Xiang X. Plant exosome-like nanovesicles derived from sesame leaves as carriers for luteolin delivery: Molecular docking, stability and bioactivity. Food Chem. (2024) 438:137963. doi: 10.1016/j.foodchem.2023.137963
61. Liu Y, Qi H, Zong J, Li M, Yang Y, Li X, et al. Oral piwi-interacting RNA delivery mediated by green tea-derived exosome-like nanovesicles for the treatment of aortic dissection. Adv Healthc Mater. (2024) 13:e2401466. doi: 10.1002/adhm.202401466
62. Wang D, Zhang H, Liao X, Li J, Zeng J, Wang Y, et al. Oral administration of Robinia pseudoacacia L. flower exosome-like nanoparticles attenuates gastric and small intestinal mucosal ferroptosis caused by hypoxia through inhibiting HIF-1α- and HIF-2α-mediated lipid peroxidation. J Nanobiotechnology. (2024) 22:479. doi: 10.1186/s12951-024-02663-6
63. Taşlı PN. Usage of celery root exosome as an immune suppressant; Lipidomic characterization of apium graveolens originated exosomes and its suppressive effect on PMA/ionomycin mediated CD4+ T lymphocyte activation. J Food Biochem. (2022) 46:e14393. doi: 10.1111/jfbc.14393
64. Cai H, Huang LY, Hong R, Song JX, Guo XJ, Zhou W, et al. Momordica charantia Exosome-Like Nanoparticles Exert Neuroprotective Effects Against Ischemic Brain Injury via Inhibiting Matrix Metalloproteinase 9 and Activating the AKT/GSK3β Signaling Pathway. Front Pharmacol. (2022) 13:908830. doi: 10.3389/fphar.2022.908830
65. Şahin F, Koçak P, Güneş MY, Özkan İ, Yıldırım E, and Kala EY. In vitro wound healing activity of wheat-derived nanovesicles. Appl Biochem Biotechnol. (2019) 188:381–94. doi: 10.1007/s12010-018-2913-1
66. Wang R, Zhang Y, Guo Y, Zeng W, Li J, Wu J, et al. Plant-derived nanovesicles: Promising therapeutics and drug delivery nanoplatforms for brain disorders. Fundam Res. (2023) 5:830–50. doi: 10.1016/j.fmre.2023.09.007
67. Sadri Nahand J, Bokharaei-Salim F, Karimzadeh M, Moghoofei M, Karampoor S, Mirzaei HR, et al. MicroRNAs and exosomes: key players in HIV pathogenesis. HIV Med. (2020) 21:246–78. doi: 10.1111/hiv.12822
68. Madison MN and Okeoma CM. Exosomes: implications in HIV-1 pathogenesis. Viruses. (2015) 7:4093–118. doi: 10.3390/v7072810
69. Roth WW, Huang MB, Addae Konadu K, Powell MD, and Bond VC. Micro RNA in exosomes from HIV-infected macrophages. Int J Environ Res Public Health. (2015) 13:ijerph13010032. doi: 10.3390/ijerph13010032
70. Poveda E and Freeman ML. Exosomes as new players in HIV pathogenesis - new data from the IAS 2017. AIDS Rev. (2017) 19:173–5. doi: 10.24875/AIDSRev.M17000007
71. Perocheau D, Touramanidou L, Gurung S, Gissen P, and Baruteau J. Clinical applications for exosomes: Are we there yet? Br J Pharmacol. (2021) 178:2375–92. doi: 10.1111/bph.15432
72. Chen J, Li P, Zhang T, Xu Z, Huang X, Wang R, et al. Review on strategies and technologies for exosome isolation and purification. Front Bioeng Biotechnol. (2022) 9:811971. doi: 10.3389/fbioe.2021.811971
73. Dilsiz N. A comprehensive review on recent advances in exosome isolation and characterization: Toward clinical applications. Transl Oncol. (2024) 50:102121. doi: 10.1016/j.tranon.2024.102121
74. Cheng K and Kalluri R. Guidelines for clinical translation and commercialization of extracellular vesicles and exosomes based therapeutics. J Extracell Vesicles. (2023) 2:100029. doi: 10.1016/j.vesic.2023.100029
75. Ahn SH, Ryu SW, Choi H, You S, Park J, Choi C, et al. Manufacturing therapeutic exosomes: from bench to industry. Mol Cells. (2022) 45:284–90. doi: 10.14348/molcells.2022.2033
76. Newman H and Hardie D. HIV-1 viral load testing in resource-limited settings: Challenges and solutions for specimen integrity. Rev Med Virol. (2021) 31:e2165. doi: 10.1002/rmv.2165
77. Shmagel KV, Saidakova EV, Shmagel NG, Korolevskaya LB, Chereshnev VA, Robinson J, et al. Systemic inflammation and liver damage in HIV/hepatitis C virus coinfection. HIV Med. (2016) 17:581–9. doi: 10.1111/hiv.12357
78. Tertel T, Dittrich R, Arsène P, Jensen A, and Giebel B. EV products obtained from iPSC-derived MSCs show batch-to-batch variations in their ability to modulate allogeneic immune responses in vitro. Front Cell Dev Biol. (2023) 11:1282860. doi: 10.3389/fcell.2023.1282860
79. European Medicines Agency. EMA Scientific recommendations on classification of ATMP. Available online at: https://www.ema.europa.eu/en/human-regulatory-overview/marketing-authorisation/advanced-therapies-marketing-authorisation/scientific-recommendations-classification-advanced-therapy-medicinal-products (Accessed November 11, 2025).
80. U.S. Food and Drug Administration. Regulation of human cells, tissues, and cellular and tissue-based products (HCT/Ps) – small entity compliance guide(2022). Available online at: https://wwwfdagov/regulatory-information/search-fda-guidance-documents/regulation-human-cells-tissues-and-cellular-and-tissue-based-products-hctps-small-entity-compliance (Accessed November 11, 2025).
81. Lener T, Gimona M, Aigner L, Börger V, Buzas E, Camussi G, et al. Applying extracellular vesicles based therapeutics in clinical trials – an ISEV position paper. J Extracell Vesicles. (2015) 4:30087. doi: 10.3402/jev.v4.30087
82. Beetler DJ, Di Florio DN, Law EW, Groen CM, Windebank AJ, Peterson QP, et al. The evolving regulatory landscape in regenerative medicine. Mol Asp Med. (2023) 91:101138. doi: 10.1016/j.mam.2022.101138
83. Pharmaceuticals and Medical Devices Agency (PMDA). Available online at: https://www.pmda.go.jp/english/about-pmda/0004.html (Accessed November 11, 2025).
84. Yi YW, Lee JH, Kim SY, Pack CG, Ha DH, Park SR, et al. Advances in analysis of biodistribution of exosomes by molecular imaging. Int J Mol Sci. (2020) 21:665. doi: 10.3390/ijms21020665
Keywords: exosomes, HIV-1 pathogenesis, viral reservoirs, co-infection, biomarkers, plant-derived nanovesicles, drug delivery, clinical translation
Citation: Lu P, Lin X, Yang W and Li J (2025) Exosomes at the crossroads of HIV-1 pathogenesis and therapeutics. Front. Immunol. 16:1634726. doi: 10.3389/fimmu.2025.1634726
Received: 25 May 2025; Accepted: 30 October 2025;
Published: 20 November 2025.
Edited by:
Roopali Rajput, University of Delhi, IndiaReviewed by:
Daniel Sepúlveda-Crespo, Carlos III Health Institute (ISCIII), SpainHimanshu Sharma, Bathinda (AIIMS Bathinda), India
Copyright © 2025 Lu, Lin, Yang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junhao Li, enhmbHdsQDE2My5jb20=
 Pengpeng Lu
Pengpeng Lu Xu Lin
Xu Lin Wenyi Yang
Wenyi Yang Junhao Li
Junhao Li