- 1Department of Pediatrics, Shaoxing Keqiao Women and Children’s Hospital, Shaoxing, Zhejiang, China
- 2Department of Medical Imaging, School of Medicine, Shaoxing University, Shaoxing, Zhejiang, China
- 3Department of Nursing, School of Medicine, Shaoxing University, Shaoxing, Zhejiang, China
- 4Department of Clinical Medicine, School of Medicine, Shaoxing University, Shaoxing, Zhejiang, China
- 5Department of Pharmacology, School of Medicine, Shaoxing University, Shaoxing, Zhejiang, China
Emerging evidence highlights the microbiota–gut–lung axis (MGLA) as a pivotal regulator of pediatric respiratory health, yet mechanistic insights are lacking and therapeutic applications remain unclear. This review synthesizes cutting-edge findings to delineate how gut microbiota-derived metabolites, particularly short-chain fatty acids (SCFAs), orchestrate pulmonary immunity and disease pathogenesis in children. Leveraging multi-omics integration (metagenomics, metabolomics, transcriptomics), emerging studies have uncovered novel microbe–host interactions driving immune dysregulation in asthma, pneumonia, and cystic fibrosis. A comprehensive map of gut–lung crosstalk has been established across these conditions. Current studies suggest that early-life gut dysbiosis, shaped by delivery mode, antibiotics, and diet, disrupts SCFA-mediated immune homeostasis, amplifying T-helper 2 cell inflammation and impairing alveolar macrophage function. Crucially, we identified disease-specific microbial signatures (e.g., depletion of Lachnospira and Faecalibacterium in asthma) and demonstrated that fecal microbiota transplantation and probiotic interventions restore microbial balance, attenuating airway inflammation in preclinical models. This work pioneers the translation of MGLA insights into precision medicine strategies, highlighting dietary modulation and microbial therapeutics as viable alternatives to conventional treatments. By bridging microbial ecology and immune dynamics, our findings provide actionable biomarkers for early diagnosis and personalized interventions, addressing critical gaps in pediatric respiratory disease management. The integration of multi-omics frameworks not only advances mechanistic understanding but also positions the MGLA as a transformative target in reducing global childhood morbidity. Future research must prioritize longitudinal studies and clinical trials to validate these innovations, ultimately redefining therapeutic paradigms for GLA-driven pathologies.
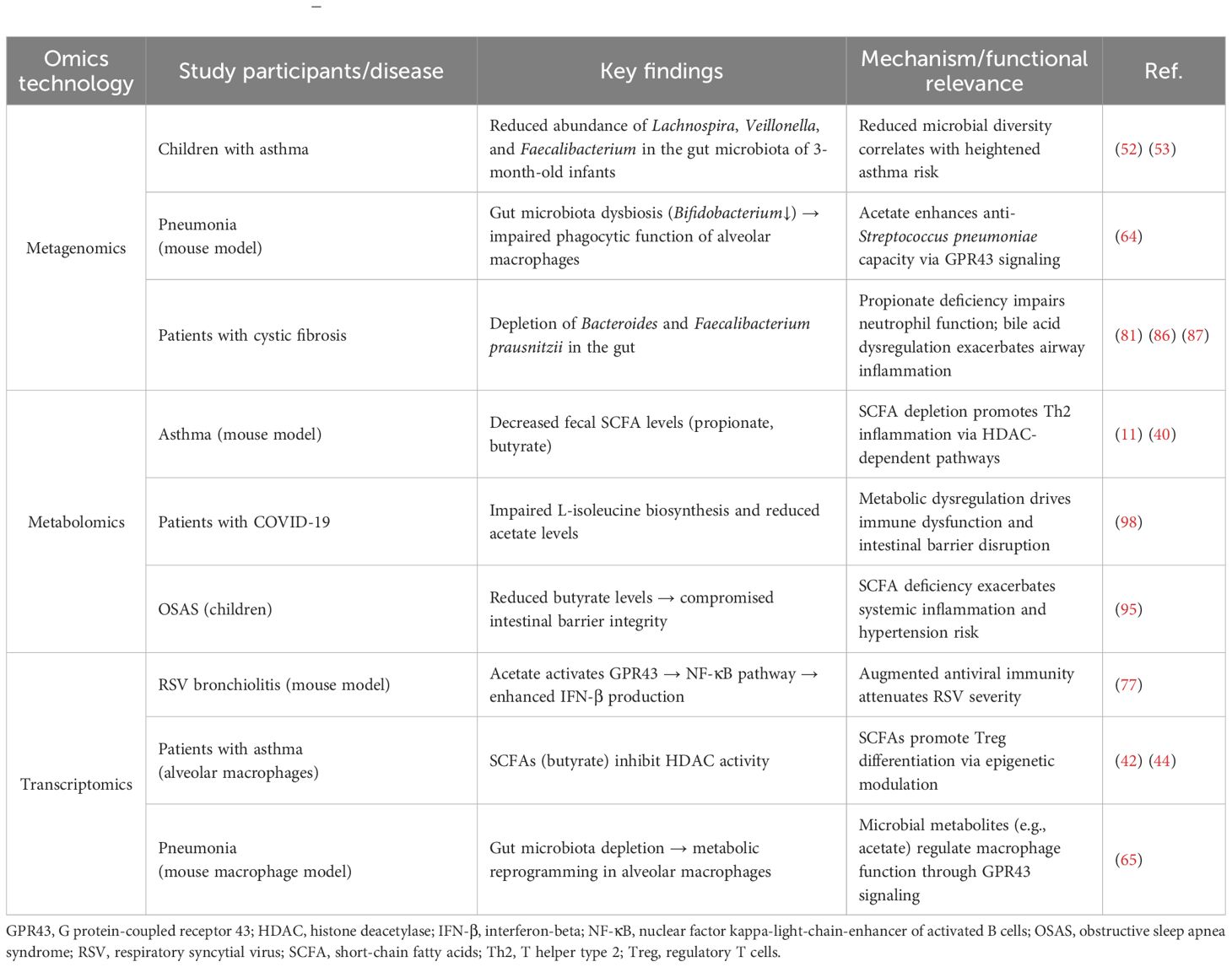
Graphical Abstract. Regulatory Mechanisms and Therapeutic Strategies of the GLA in Pediatric Pulmonary Diseases. The gut microbiota interacts bidirectionally with the pulmonary microenvironment through microbial metabolites (e.g., SCFAs) and immune signaling molecules (e.g., IFN, IL-6, IL-10), forming a central mechanism underlying this axis. Gut dysbiosis disrupts SCFA-mediated immune homeostasis, contributing to the pathogenesis of childhood respiratory diseases such as asthma, pneumonia, and cystic fibrosis. Integrated multi-omics studies have identified disease-specific microbial signatures and propose novel strategies—including FMT, probiotics, prebiotics, and dietary interventions—to restore gut microbiota equilibrium, suppress airway inflammation, and advance precision-based therapeutic approaches for pediatric respiratory disorders. CF, cystic fibrosis; FMT, fecal microbiota transplantation; GLA, gut–lung axis; IFN, interferon; IL, interleukin; OSAS, obstructive sleep apnea syndrome; SCFAs, short-chain fatty acids.
1 Introduction
Acute respiratory infections and influenza-like illnesses (ILIs) account for approximately 10% of annual outpatient visits globally, with pediatric populations bearing a disproportionate burden. Children aged 1–17 years exhibit a fourfold higher incidence of ILIs than adults (1). This vulnerability stems from structurally immature airways and dynamically evolving immune regulation, creating a suitable environment for recurrent infections. Beyond acute morbidity, such episodes disrupt developmental homeostasis and amplify lifelong cardiopulmonary risks (2), necessitating urgent research into early-life susceptibility mechanisms.
The human microbiome associated with the respiratory tract is characterized by its diversity, heterogeneity, and dynamism. The complexity of the microbiome, along with the intricate interactions among microorganisms, host cells, and the host immune system, involves multiple factors. There is often an interaction between gut and respiratory microbiota, with the lymphatic system providing a direct pathway, known as the gut–lung axis (GLA), via which the gut microbiome can influence outcomes related to respiratory diseases and modulate the host’s immune response (1). The GLA is a bidirectional communication network mediated by microbiota and metabolites that links intestinal and pulmonary health (3). Emerging evidence indicates that perturbations in one organ (e.g., gut dysbiosis (4)) can propagate systemic immune dysregulation, exacerbating pathologies in the other (e.g., asthma exacerbations (5)). For example, gut-derived microbial components such as lipopolysaccharide (LPS) modulate pulmonary inflammation via lymphatic dissemination (6) whereas respiratory viral infections reciprocally reshape the gut microbial ecology (7). Despite these advances, the spatiotemporal dynamics of microbiome–immune crosstalk in pediatric populations remain poorly mapped.
The gut microbiome critically regulates pediatric health, with its assembly during infancy dictating immune maturation trajectories (8). The composition of the intestinal microbiome influences health from before birth into childhood, with numerous diseases linked to imbalances in this ecosystem. The gut microbiome evolves continuously from infancy to maturity, influenced by various factors that shape its development and composition. Characteristics of the gut microbiota can impact brain development, immune function, lung health, and overall physical growth (9). Gut microbial dysbiosis can have long-term consequences, as supported by data from mouse models in which mice have an increased predisposition to allergic inflammation following early-life antibiotic use (10, 11). Early-life disruptions, such as antibiotic exposure or dietary insufficiency, induce persistent dysbiosis, elevating the risks of allergic sensitization and recurrent respiratory infections (12).
Microbial metabolites such as short-chain fatty acids (SCFAs) emerge as key orchestrators of mucosal immunity, yet their role in gut–lung interactions remains underexplored in children. Recent studies have integrated multi-omics approaches (metagenomics, metabolomics, transcriptomics) to decode microbiota–immune networks in pediatric respiratory diseases. These efforts have identified disease-specific microbial signatures (e.g., Lachnospira depletion in asthma) and have demonstrated that SCFAs enhance alveolar macrophage function not only via epigenetic modulation but also by inducing tolerance-related genetic signatures (13). Such findings highlight the potential of microbiota-targeted therapies (e.g., probiotics, dietary interventions) to recalibrate immune homeostasis.
2 Gut microbiota and children’s respiratory health
2.1 Characteristics of the pediatric gut and respiratory microbiota
The pediatric gut microbiome represents a dynamic ecosystem of bacteria, fungi, viruses, and archaea. Indeed, the human gut is inhabited by between 100 thousand and 100 billion bacteria per milliliter of luminal content depending on the region and is therefore the most densely colonized organ (14). Crucially, this microbial consortium not only supports nutrient metabolism but also calibrates systemic immunity via endocrine and metabolic crosstalk (15, 16). This symbiotic relationship is established from birth with the infant gut microbiota and continues to evolve during the critical early years of life. During this period, infants experience rapid growth, with substantial increases in height, weight, and head circumference. Concurrently, their metabolic organs, immune system, digestive system, and neurocognitive functions undergo substantial development and maturation. This phase is also pivotal for formation of the gut microbiota, which is essential for maintaining overall health (17).
In contrast to the gut, the respiratory microbiome exhibits spatiotemporal stratification—nasal cavities favor Staphylococcus and Corynebacterium, whereas lower airways are dominated by Prevotella and Streptococcus—with bacterial loads (103–105 CFU/mg) that are orders of magnitude lower than intestinal levels (18, 19). Our understanding of bacterial component development in the airways is more limited than our knowledge of the gut microbiota. Some evidence (primarily from mouse models) suggests that the respiratory microbiota matures during childhood, a process that is crucial for promoting tolerance to airborne allergens (20). Despite shared dominant phyla (Bacteroidetes/Firmicutes), the lung microbiome displays unique assembly rules; microbial immigration (inhalation/microaspiration), elimination (mucociliary clearance), and local replication dynamically sculpt the community structure (21).
Deciphering the co-evolution of gut and respiratory microbiomes is fundamental to understanding pediatric immune ontogeny. During early life, these microbial communities transcend passive colonization; they actively direct immune cell differentiation via metabolite signaling (e.g., SCFAs), epigenetic modulation, and pathogen exclusion (22). Integrated analysis of human cohort data has revealed that synchronized gut–lung microbiota maturation establishes a systemic immune “set-point,” dysregulation of which underlies susceptibility to pneumonia and asthma. This paradigm shifts the focus from cataloging microbial taxa to decoding their functional networks, a cornerstone of our therapeutic discovery platform.
2.2 Gut microbiota affects development of the immune system in children
Upon birth, infants start to develop their initial microbiome, with its composition being shaped by the delivery method, whether through natural, vaginal, or cesarean birth (23). The initial bacterial colonization and various other factors following childbirth strongly influence an infant’s early-life microbiome, which subsequently regulates immune system development of the newborn (24). Whereas vaginal birth establishes a maternally derived microbiome enriched in Lactobacillus and Bifidobacterium, cesarean delivery favors skin-associated Staphylococcus and environmental taxa (25). Crucially, this immunomodulatory process exhibits developmental stage-specific sensitivity, as evidenced by murine and human studies demonstrating irreversible immune programming defects when microbial exposure is disrupted during early postnatal windows (26, 27). This foundational microbial assembly orchestrates lymphoid tissue development and immune cell education—processes that are vulnerable to disruption by antibiotics or formula feeding. Such dysbiosis propagates systemic immune misprogramming, elevating the risks for asthma and obesity via GLA signaling (28).
The gut microbiome operates as a microbial tutor during infancy, instructing immune cell differentiation through metabolite- and antigen-driven dialogues. Key taxa (e.g., Clostridia clusters) promote expansion of regulatory T cells (Tregs) via SCFA production, and segmented filamentous bacteria drive T-helper 17 cell (Th17) polarization via interleukin 23 (IL-23)/IL-17 axis activation (29). This trans-organ immunity, mediated by circulating microbial metabolites and trained immune cells, forms the mechanistic bedrock of the GLA (30).
In the intestinal environment, pattern recognition receptors (PRRs), including toll-like receptors (TLRs), C-type lectin receptors, NOD-like receptors, and RIG-I-like receptors, can identify specific molecular patterns in the microbiota, thereby initiating immune responses. The activation of PRRs not only promotes the maturation and activation of immune cells but also modulates their migration and function through the production of cytokines and chemokines (31). Specifically activated PRRs can induce dendritic cells (DCs) to produce IL-12p70, a crucial Th1-polarizing cytokine for anti-infection and anti-tumor immune responses. Moreover, PRR activation enhances the expression of surface molecules such as CD80, CD86, and human leukocyte antigen-DR (HLA-DR) on DCs, thereby improving their antigen-presenting capacity and T-cell activation ability (32).
A previous study redefined the gut microbiome as a systemic immune rheostat, with microbial metabolites (e.g., SCFAs, indoles) serving as endocrine-like messengers that synchronize lung immunity (33). Via integrated metagenomic–metabolomic analysis, spatial gradients of these molecules have been mapped from gut to bronchoalveolar lavage, correlating their depletion with neutrophilic inflammation in severe asthma (34) (Figure 1). This paradigm-shifting discovery positions fecal metabolite profiling as a noninvasive biomarker for predicting corticosteroid responsiveness—a cornerstone of our precision pulmonology framework. By decoding the gut–lung dialogue, we can pioneer microbiota-centric strategies to reset immune homeostasis in pediatric respiratory diseases.
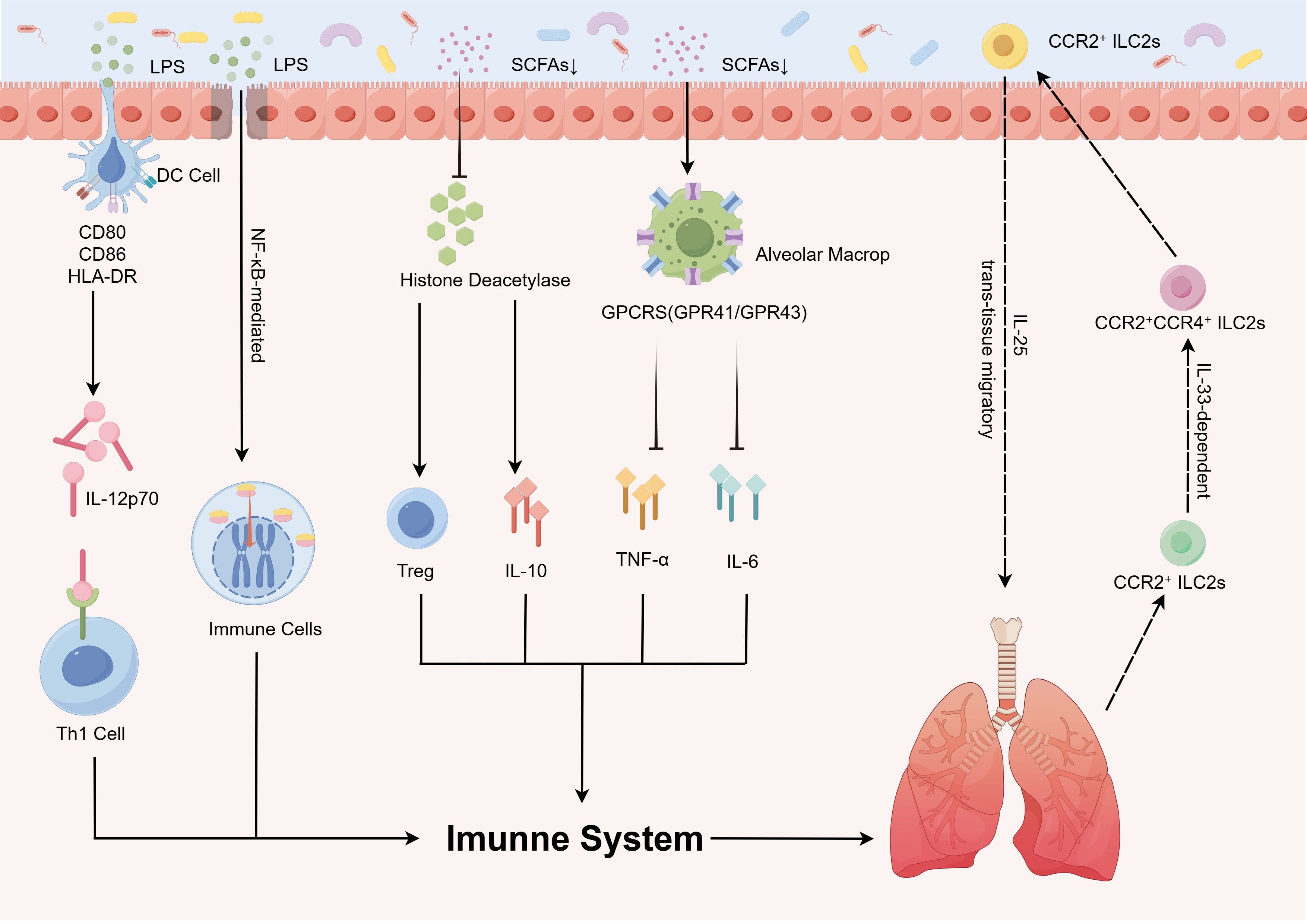
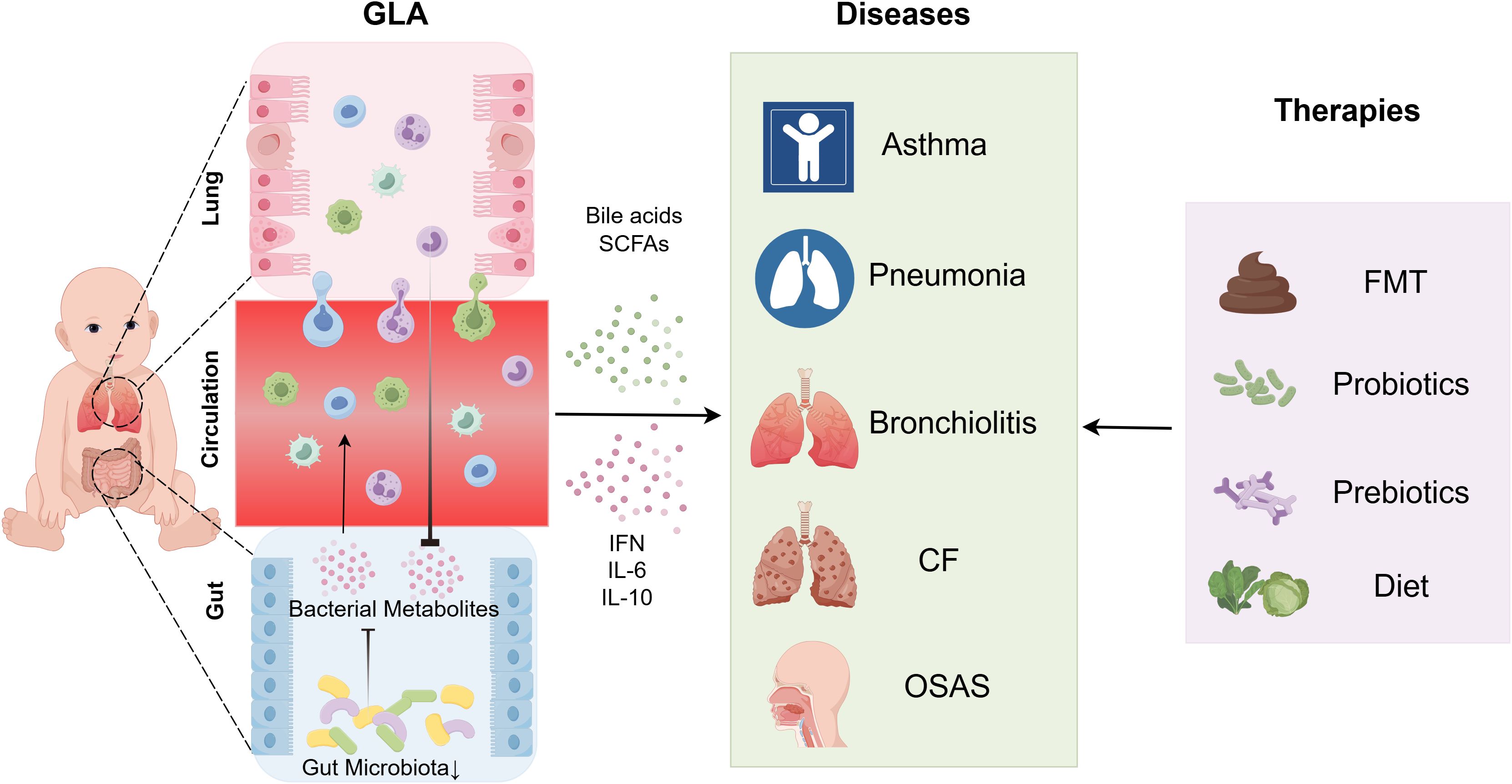
Figure 1. Gut microbiota modulates pathophysiological processes in pediatric pulmonary diseases via immune regulatory mechanisms. The gut microbiota orchestrates immune regulatory mechanisms that influence the pathogenesis of childhood pulmonary diseases via microbial metabolites (e.g., SCFAs and LPS) and immune signaling networks, which critically regulate immune system development and disease progression. DCs recognize LPS, leading to upregulated CD80/CD86/HLA-DR expression, enhanced antigen-presenting capacity, and IL-12p70 secretion, thereby driving Th1 polarization to reinforce anti-infective immunity. Gut-derived LPS activates the TLR4/NF-κB pathway, triggering pulmonary inflammation. SCFAs inhibit histone deacetylase activity to promote Treg differentiation and anti-inflammatory IL-10 secretion, while simultaneously activating alveolar macrophages via GPCRs to suppress pro-inflammatory cytokine (TNF-α, IL-6) release. Additionally, trans-tissue migration of CCR2+ ILC2s further modulates pediatric pulmonary disease pathogenesis. CCR2+CCR4+ILC2s, C-C chemokine receptor type 2-positive and C-C chemokine receptor type 4-positive type 2 innate lymphoid cells; CCR2+ILC2s, C-C chemokine receptor type 2-positive type 2 innate lymphoid cells; CD80/CD86, cluster of differentiation 80/86; DC, dendritic cell; GPCRs (GPR41/GPR43), G protein-coupled receptors (G protein-coupled receptor 41/G protein-coupled receptor 43); HLA-DR, human leukocyte antigen – DR isotype; IL, interleukin; LPS, lipopolysaccharide; SCFAs, short-chain fatty acids; Th1, T helper 1 cells; TNF-α, tumor necrosis factor alpha; Treg, regulatory T cells.
3 Role of the GLA in children’s pulmonary physiological function
3.1 Core immunoregulatory mechanisms of the GLA
The GLA operates as a bidirectional immunoregulatory circuit wherein microbial components and metabolites systemically modulate pulmonary immunity via three primary mechanisms: 1) molecular mimicry: bacterial LPS primes alveolar macrophages via TLR4 (35); 2) metabolite trafficking: gut-derived SCFAs, particularly propionate and butyrate, suppress IL-13-driven eosinophilia by enhancing airway epithelial barrier integrity (36); 3) immune-cell migration: intestinal Th17 cells recruited to the lungs exacerbate neutrophilic inflammation in asthma, a process amplified by dysbiosis-induced IL-23 signaling (37). Disruption of gut barrier integrity promotes microbial translocation (e.g., circulating LPS), which triggers nuclear factor-kappa B (NF-κB)-mediated pulmonary hyperinflammation (38). A murine study demonstrated that elevated circulating propionate levels promote hematopoiesis of DC precursors in bone marrow, subsequently affecting DCs in the lungs and draining lymph nodes, thereby resulting in an impaired ability of these DCs to activate Th2 cells within the lung tissue (39). These pathways collectively enhance intestinal mucosal immunity while systemically modulating immune cell differentiation and function. Notably, animal studies substantiate that SCFAs can attenuate pulmonary inflammation by activating Tregs and conditioning DCs to suppress Th2-polarized responses in murine models, thereby recalibrating Th1/Th2 equilibrium (40) (Figure 1, Table 1).
3.2 Systemic immunomodulatory roles of SCFAs
SCFAs—primarily acetate, propionate, and butyrate—serve as keystone metabolites linking the gut microbiota to pulmonary immune homeostasis (11, 41). Emerging evidence suggests that elevated levels of SCFAs in infants are associated with a reduced incidence of asthma, potentially mediated through dual anti-inflammatory mechanisms (42). First, SCFAs activate G protein-coupled receptors (GPR41/GPR43) on alveolar macrophages via receptor-dependent signaling, enhancing pathogen clearance while suppressing pro-inflammatory cytokine release (e.g., tumor necrosis factor alpha [TNF-α], IL-6) (22, 43). Second, SCFAs modulate immune cell function through epigenetic regulation: by inhibiting histone deacetylase (HDAC) activity; these promote anti-inflammatory factor expression (including IL-10) and Treg differentiation while simultaneously restraining Th17 cell overactivation. These coordinated actions maintain pulmonary immune homeostasis and mitigate inflammation (42, 44) (Table 1). Although SCFA concentrations are low in the lungs, gut-derived propionate can induce bone marrow precursors to generate macrophages. These macrophages then migrate to the lungs and attenuate allergic inflammation by promoting IL-10 production (11). These findings establish SCFAs as systemic immunomodulators that exert hormone-like signaling functions (45). Collectively, the above findings underscore SCFAs as pivotal gut-derived mediators that orchestrate pulmonary immune homeostasis via receptor-dependent signaling and epigenetic mechanisms. The ability of SCFAs to prime anti-inflammatory responses both locally and systemically highlights their therapeutic potential in asthma and other inflammatory lung diseases (Figure 1).
3.3 Trans-organ migration mechanisms of immune cells
Emerging evidence highlights the pivotal role of immune cell trafficking in gut–lung communication wherein innate lymphoid cells (ILCs) and gut-primed Tregs migrate to pulmonary tissues via mesenteric lymphatics under chemokine guidance (46). Extending these observations, one study provided mechanistic insights into the cytokine-regulated bidirectional trafficking of ILC2s using integrated pseudotemporal analysis, lineage tracing, and ectopic transplantation assays. That study delineated a unidirectional maturation trajectory wherein lung-derived C-C chemokine receptor type 2-positive (CCR2+) ILC2s migrate to the intestine via IL-33-dependent pathways, undergoing phenotypic transition characterized by a transient CCR2+CCR4+ double-positive population and terminal differentiation into CCR4+ gut-resident ILC2s. This migratory axis demonstrates strict tissue tropism under homeostasis, with no detectable reverse migration from intestinal ILC2s to pulmonary niches. Strikingly, IL-25 stimulation subverts this physiological unidirectionality, potentiating intestinal ILC2s to acquire trans-tissue migratory competence toward pulmonary compartments (47). This cellular crosstalk establishes a “mobile immune network” that integrates microbial signals and tissue-specific immunity (Figure 1).
3.4 Translational implications and future directions
The GLA, which is crucial for immune balance and respiratory health, strongly influences pediatric pulmonary diseases. Understanding its role in diseases such as asthma and bronchopulmonary dysplasia could offer new therapeutic strategies. Future research priorities include standardizing methodologies for gut microbiota and metabolite profiling in pediatric cohorts, elucidating age-dependent variations in GLA functionality, and developing targeted interventions (e.g., SCFA prodrugs, microbiota transplantation) to restore immune homeostasis.
4 Role of the GLA in pediatric pulmonary diseases
4.1 Asthma
Asthma remains a leading chronic pediatric condition, accounting for approximately 400,000 annual deaths globally (48, 49). The current global prevalence of asthma exceeds 300 million individuals, with a projected increase to 400 million by 2025 (50). Emerging evidence implicates GLA dysbiosis in asthma pathogenesis, with microbial alterations influenced by perinatal factors including cesarean delivery, neonatal antibiotic exposure, maternal nutritional patterns, formula supplementation, and microbial environmental exposures (51, 52). Longitudinal cohort studies demonstrate that diminished gut microbial diversity during early infancy correlates with subsequent asthma development. Infants who develop asthma exhibit significantly reduced α-diversity metrics as early as 1 month postpartum (52), with temporal microbiota dynamics showing critical developmental windows for asthma predisposition. Specifically, transient depletion of immunomodulatory taxa (Lachnospira, Veillonella, Faecalibacterium, and Rothia) at 3 months postnatally distinguishes high-risk infants (53) (Table 1). Phylum-level analysis reveals Firmicutes dysregulation in pediatric patients with asthma, characterized by decreased anti-inflammatory species (e.g., Roseburia) and expansion of pro-inflammatory genera (Enterococcus, Clostridium) (54) (Figure 2).
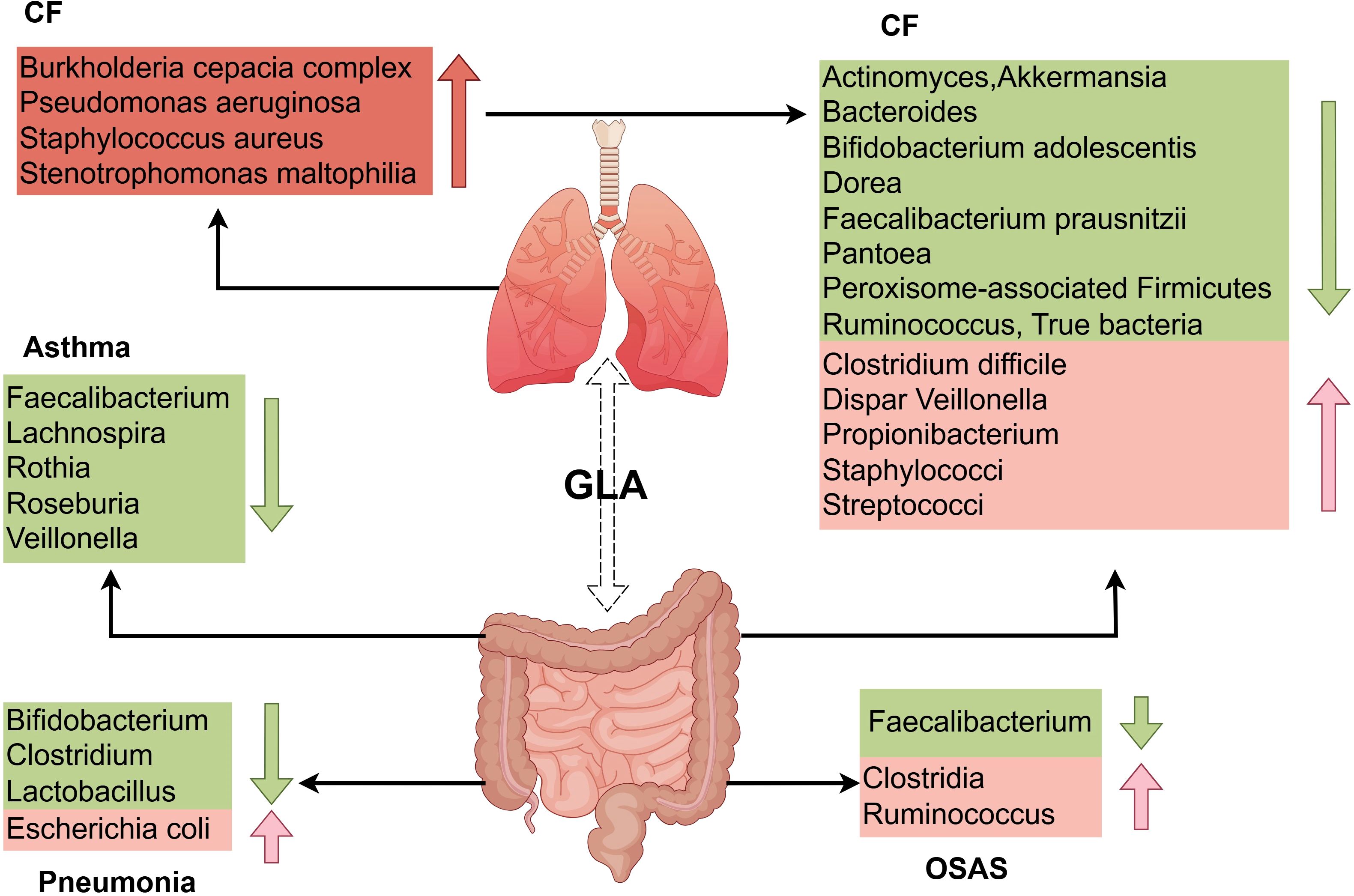
Figure 2. Distinct features of gut and respiratory microbiota dysbiosis in pediatric pulmonary diseases. The GLA exhibits dynamic microbiota alterations in childhood asthma, pneumonia, CF, and OSAS. Children with asthma have reduced gut abundance of immunomodulatory taxa (e.g., Lachnospira, Faecalibacterium, Rothia) alongside expansion of pro-inflammatory genera (e.g., Enterococcus). Patients with pneumonia display diminished intestinal Bifidobacterium and Lactobacillus abundance with concurrent enrichment of Escherichia coli. In CF, the gut microbiota is characterized by substantial enrichment of opportunistic pathogens (e.g., Burkholderia cepacia complex, Pseudomonas aeruginosa) and depletion of beneficial species (e.g., Bacteroides, Faecalibacterium prausnitzii). OSAS is associated with reduced levels of SCFA-producing bacteria (e.g., Clostridia, Ruminococcus), which exacerbates systemic inflammation via the metabolic–immune axis. GLA, gut–lung axis; CF, cystic fibrosis; OSAS, obstructive sleep apnea syndrome; SCFAs, short-chain fatty acids.
This ecological imbalance coincides with impaired mucosal immune programming during critical developmental windows, as evidenced by multi-omics integration of microbial community dynamics, host metabolomic signatures, and nutritional patterns (49, 55, 56). Notably, gut dysbiosis, particularly the depletion of Akkermansia, Bifidobacterium, and Faecalibacterium species during critical developmental windows, is strongly associated with heightened susceptibility to asthma and allergic disorders (26, 27) (Figure 2). Mechanistically, microbial metabolic output—particularly SCFA biosynthesis—mediates dietary microbiota–immune crosstalk, with reduced SCFA production correlating with asthma risk in low-diversity microbiomes (1, 11).
Breastfeeding exerts important modulatory effects on asthma-related microbial metabolites (49). Contrary to historical sterility assumptions, human milk contains dynamic microbial communities and prebiotic compounds that shape the infant gut ecology (57, 58). Multivariate analysis has revealed that exclusive breastfeeding ≥4 months is an independent protective factor against asthma development (49), highlighting nutritional–microbial interactions in early-life asthma prevention (Figure 3).
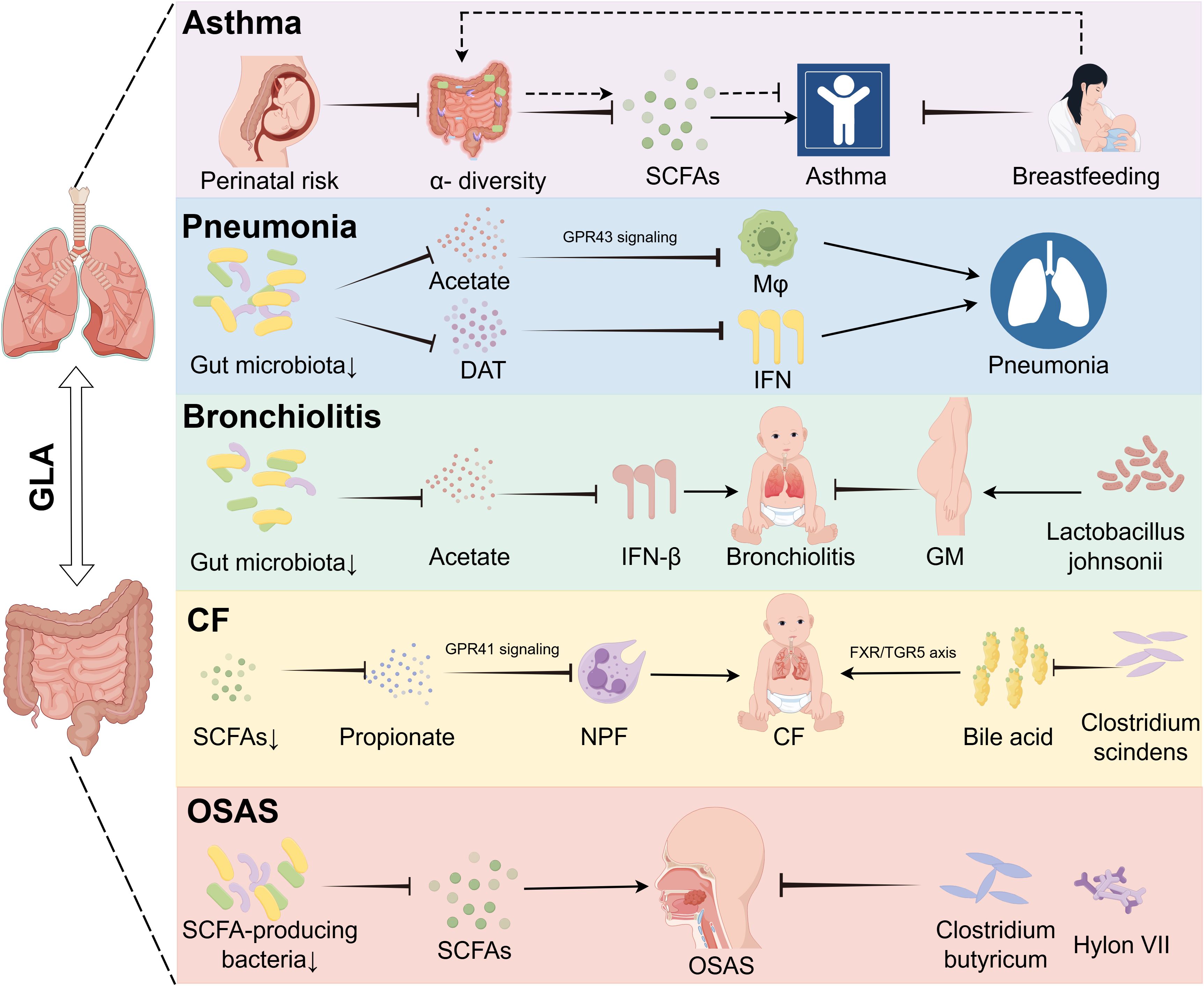
Figure 3. Gut dysbiosis drives disease-specific mechanisms in pediatric pulmonary disorders via metabolic and signaling pathways. The GLA orchestrates disease-specific mechanisms underlying childhood asthma, pneumonia, bronchitis, CF, and OSAS. In children with asthma, perinatal microbiota alterations with reduced alpha diversity are modulated by maternal breastfeeding-associated metabolites. In pneumonia, gut dysbiosis reduces acetate levels, impairing the GPR43-mediated phagocytic function of alveolar macrophages, and deaminotyrosine deficiency compromises IFN-I-dependent anti-influenza immunity. Bronchitis is linked to diminished gut-derived acetate, leading to pulmonary IFN-β reduction whereas prenatal supplementation with Lactobacillus jensenii lowers disease incidence. In CF, Bacteroides-derived propionate deficiency disrupts GPR41 signaling, causing neutrophil phagocytic dysfunction, and Clostridioides difficile depletion exacerbates inflammation via dysregulated bile acid metabolism (FXR/TGR5 axis). Reduced SCFA-producing bacteria contribute to OSAS pathogenesis, which is alleviated by C. butyricum and prebiotics. GLA, gut–lung axis; DAT, desaminotyrosine; GM, gestational mother; GPR43, G protein-coupled receptor 43; IFN-I, type I interferon; Mφ, macrophage; NPF, neutrophil phagocytic function; OSAS, obstructive sleep apnea syndrome; SCFAs, short-chain fatty acids.
4.2 Pneumonia
Pneumonia is an infection that inflames the air sacs and other parts of the lungs and is often caused by bacteria, viruses, or other pathogens (59, 60). Pneumonia remains a leading cause of childhood morbidity and mortality, accounting for 14% of deaths in children under 5 years globally, with an estimated 120 million cases annually (61). Research has shown that viral infections, such as influenza, can substantially alter the gut microbiome, even without detectable viral particles in the gastrointestinal tract (62). Compelling evidence links the gut microbiota composition to pneumonia susceptibility and severity (63). Mice with a depleted gut microbiome exhibit increased bacterial spread, heightened inflammation, organ dysfunction, and elevated mortality following Streptococcus pneumoniae infection, compared to those with a normal microbiome. Fetal microbiota transplantation (FMT) can restore pulmonary bacterial levels and normalize immune responses in these mice. The gut microbiome modulates metabolic pathways in alveolar macrophages, affecting cellular responsiveness, with macrophages from mice that have a depleted microbiome showing reduced phagocytic activity against S. pneumoniae (64) (Table 1). Mechanistic studies reveal two key pathways: 1) macrophage priming: gut-derived acetate enhances alveolar macrophage phagocytic capacity against S. pneumoniae via GPR43-dependent metabolic reprogramming (65) (Table 1); antiviral defense: microbiota-derived desaminotyrosine protects the host from influenza by modulating the type I interferon (IFN-I) response (66) (Figure 3).
Pneumonia poses a serious health risk to children worldwide, with high case numbers and mortality rates, emphasizing the need for prevention and better treatment (67). Preterm infants with pneumonia exhibit reduced gut microbiota diversity and disturbances of the gut (68). Children’s underdeveloped immune systems make them vulnerable to severe illness and death due to pneumonia. The gut microbiota, particularly early in life, influences lung immunity and the pneumonia risk, highlighting the importance for health of a balanced gut microbiome (23). Pneumonia can affect the composition of the gut microbiota, primarily leading to a decrease in Bifidobacterium (10), Lactobacillus (69), and Clostridium (70), and causing an increase in Escherichia coli (59). Infant gut microbiota diversity and functionality increase during the first years of life, influenced by the delivery mode and feeding type (71). In pneumonia, Pseudomonas, Escherichia/Shigella, Streptococcus, and Akkermansia are decreased Bacillota are increased, at the phylum level, with specific genera being more or less abundant in cases of pneumonia. Preterm infants with pneumonia exhibit reduced gut microbiota diversity and gut disturbances (68) (Figure 2).
The prevalence of pediatric COVID-19 cases ranges from 1% to 13.3% of total reported infections (72). A recent study revealed significantly reduced α-diversity in young patients with COVID-19 (aged <6 months), aligning with prior studies attributing such disparities to the unique infant microbiota, which is shaped by feeding patterns (73). Beyond gut microbiota alterations, intestinal inflammation, microbial translocation, and intestinal barrier dysfunction have also been identified as factors associated with COVID-19 infection and that are potentially attributable to microbiota-driven perturbations (74). The critical role of microbiota has prompted experts to question whether the host microbiota status should be considered prior to vaccine development. Furthermore, the development of oral vaccines and maintenance of microbial homeostasis may facilitate early containment of COVID-19 outbreaks or future pandemics (74).
4.3 Bronchiolitis
Respiratory syncytial virus (RSV)-associated bronchiolitis affects 33 million children under age 5 years annually, with 3.2 million requiring hospitalization (75). Emerging data implicate gut microbiota dynamics in disease progression. Cohort studies reveal that infants hospitalized with severe RSV bronchiolitis exhibit gut dysbiosis characterized by Enterobacteriaceae overgrowth and Bifidobacterium depletion (76). RSV infections are particularly dangerous for infants under age 2 years, leading to high rates of morbidity and mortality. Early infancy is a key period in gut microbiota development that may play a role in bronchiolitis through various mechanisms beyond the Th1/Th2 imbalance theory. Research indicates that acetic acid produced by gut microbes can stimulate lung IFN-β production, enhancing type 1 IFN responses by activating GPR43 and NF-κB (77) (Table 1). This activation can modulate the protective effects of acetic acid against RSV, potentially reducing pneumonia symptoms (Figure 3).
Current research is investigating intervention strategies for respiratory illnesses, with clinical trials demonstrating that Bifidobacterium species stimulated by human milk oligosaccharides contribute to the prevention of subsequent respiratory infections (e.g., bronchitis). This protective mechanism is mediated via Bifidobacterium-driven enhancement of acetate production, with elevated acetate levels being mechanistically linked to improved intestinal barrier function (78). Emerging clinical evidence has revealed that maternal supplementation with Lactobacillus johnsonii confers transgenerational protection against RSV infection, with offspring exhibiting attenuated airway mucus production and Th2-mediated immune responses. In a murine study, this intervention maintained congruent gut microbiota profiles in both dams and progeny, accompanied by a synchronized reduction in pro-inflammatory metabolites in the maternal plasma, breast milk, and offspring circulation (79). Mechanistically, Lactobacillus-driven modulation of maternal microbial ecosystems and associated metabolic reprogramming are positively correlated with enhanced neonatal airway defense mechanisms against RSV pathogenesis (79).
4.4 Cystic fibrosis
CF is an inherited disorder transmitted through an autosomal recessive pattern, resulting from mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene. This defect induces the accumulation of abnormally viscous mucus secretions, which primarily compromise pulmonary and gastrointestinal functions (80). CF affects approximately 70,000 people worldwide (81). Lung infections caused by bacteria such as Pseudomonas aeruginosa, S. aureus, Burkholderia cepacia complex, and Stenotrophomonas maltophilia worsen respiratory function and contribute to disease morbidity (19). Research supports the existence of a “GLA” in chronic respiratory diseases such as CF, affecting mucosal immunity across these systems (82, 83). Cross-sectional studies have revealed that pediatric patients with CF exhibit marked gut dysbiosis, characterized by a marked depletion of beneficial taxa including Bacteroides, Bifidobacterium adolescentis, Faecalibacterium prausnitzii, Actinomyces, Eubacterium, Ruminococcus, Dorea, Akkermansia, Peroxisome-associated Firmicutes and Pantoea (84, 85), alongside concurrent enrichment of opportunistic pathogens such as Staphylococci, Streptococci, Veillonella dispar, Propionibacterium, and Clostridium difficile (84) (Figure 2).
Mechanistic studies identify two critical pathways linking gut dysbiosis to pulmonary decline. Reduced levels of Bacteroides-derived propionate in SCFA deficiency impair neutrophil phagocytic function via compromised GPR41 signaling (81, 86). Concurrently, diminished Clostridium scindens activity disrupts bile acid metabolism by reducing deoxycholic acid production, which exacerbates airway inflammation through dysregulation of the FXR/TGR5 signaling axis (87) (Figure 3, Table 1).
A prospective cross-sectional study elucidated that children with CF exhibit a dietary pattern characterized by high fat and low fiber, which is associated with intestinal dysbiosis, elevated fecal calprotectin, and respiratory microbial disturbance, collectively leading to impaired lung function and increased lung exacerbation (82). Another study highlighted the therapeutic potential of microbiota-targeted interventions (e.g., probiotics, CFTR modulators) in mitigating systemic and respiratory morbidity in children with CF by restoring the gut microbial balance, enhancing immunomodulation, reversing SCFA deficits, and disrupting pathogenic gut–lung crosstalk, thereby attenuating chronic inflammation and improving pulmonary outcomes (88). These findings highlight the gut microbiome as a modifiable determinant of CF progression, paving the way for microbiota-centric adjuvant therapies.
4.5 Obstructive sleep apnea syndrome
OSAS affects 2%–4% of children, with increasing prevalence linked to pediatric obesity and adenotonsillar hypertrophy (89). OSAS is a prevalent chronic respiratory disorder marked by repeated pharyngeal collapse and ventilation disruption during sleep, leading to apnea and sleep disturbances (90). Emerging evidence implicates gut microbiota dysbiosis as both a contributor and consequence of OSAS pathophysiology (91, 92). Cross-sectional studies demonstrate that children with moderate-to-severe OSAS exhibit gut microbiome alterations characterized by an increase in Clostridia and Ruminococcus species, coupled with depletion of SCFA-producing bacteria such as Faecalibacterium and other taxa associated with SCFA synthesis (93, 94) (Figure 2).
In one study, researchers hypothesized that diminished intestinal SCFAs underlie hypertension pathogenesis in OSA. OSA significantly elevated systolic blood pressure after 7 and 14 days, an effect abolished by administration of the probiotic Clostridium butyricum or the prebiotic Hylon VII. 16S rRNA sequencing revealed significant enrichment of SCFA-producing bacterial taxa following C. butyricum or Hylon VII treatment. OSA exposure reduced cecal acetate concentrations by 48%, which was prevented by both interventions. Furthermore, C. butyricum and Hylon VII attenuated OSA-induced gut dysbiosis, goblet cell depletion, mucus layer thinning, and cerebral microglial activation (95) (Figure 3, Table 1). A systematic review and meta-analysis indicated that OSA is associated with intestinal barrier dysfunction. Furthermore, the severity of OSA appears to be associated with elevated levels of biomarkers of intestinal barrier dysfunction (96). These preclinical findings suggest that microbiota modulation may serve as a potential adjuvant strategy for OSAS management, although clinical validation in pediatric populations is warranted.
5 Clinical importance of GLA research
5.1 GLA as a biomarker for pediatric pulmonary disease diagnosis
The gut microbiota and its metabolites are increasingly recognized as potential biomarkers for pediatric pulmonary diseases. Omics approaches have been used to identify microbial signatures associated with disease states, including an elevated Firmicutes/Bacteroidetes ratio indicative of dysbiosis in respiratory conditions (15, 97). In COVID-19, the gut microbiota composition correlates with disease severity and immune dysregulation, characterized by depleted SCFA-producing taxa and impaired L-isoleucine biosynthesis (98, 99) (Table 1). Experimental models have further demonstrated metabolic disturbances, such as increased serum lactate and reduced acetate levels in pulmonary hypertension (97). Although these findings underscore the diagnostic potential of microbiota profiling, current evidence remains fragmented, necessitating large-scale longitudinal studies to validate microbial and metabolic biomarkers across diverse pediatric populations (15).
5.2 Potential application of the GLA in treating pediatric pulmonary diseases
Therapeutic strategies targeting the GLA show promise in modulating immune responses and improving clinical outcomes. Probiotics, such as Lactobacillus plantarum, enhance IFN-I production and reduce viral loads in influenza models whereas Clostridium orbiscindens-derived desaminotyrosine strengthens antiviral defenses (62, 69). SCFAs exert systemic anti-inflammatory effects by promoting Treg differentiation (via HDAC inhibition) and suppressing Th2/Th17 polarization (through GPR41/43 signaling), with fecal propionate levels inversely correlated with asthma risk in infants (39, 40). Omega-3 polyunsaturated fatty acids (PUFAs) such as DHA (docosahexaenoic acid) and EPA (eicosapentaenoic acid) can regulate immune cell functions, reduce inflammatory responses, and enhance antiviral immune reactions, thereby playing a protective role in pediatric pulmonary diseases. Research has found that prenatal supplementation with omega-3 PUFAs can influence the airway microbiota of infants, reducing the risk of respiratory infections (100). Despite these advances, standardized protocols for strain selection, dosing, and long-term safety monitoring remain critical challenges in clinical translation.
6 Challenges and future directions in GLA research
6.1 Current challenges in GLA research for pediatric respiratory diseases
Despite growing interest in the GLA and its role in pediatric respiratory disorders, critical methodological and translational gaps hinder progress. A predominant reliance on cross-sectional studies limits causal inference because observed associations—such as the reduced gut microbiota diversity in asthma—cannot be used to distinguish whether dysbiosis drives disease or vice versa. Large-scale longitudinal cohorts tracking microbiota dynamics from early life to disease onset are essential to resolve such ambiguity. Furthermore, small sample sizes (<100 participants) and geographical biases (e.g., overrepresentation of European populations) reduce generalizability, and inconsistent sample handling protocols introduce technical variability that obscures biological signals.
The complexity of gut microbiota data presents another layer of difficulty. Integrating taxonomic, metabolic, and functional profiles requires advanced bioinformatics capabilities that are often unavailable to smaller research teams. Compounding this issue, key confounders such as diet are insufficiently addressed. Most studies focus narrowly on the short-term effects of isolated nutrients, overlooking the cumulative impact of dietary patterns and individualized eating behaviors. Genetic predispositions and environmental exposures (e.g., air pollution, antibiotic use) further interact with microbiota in ways that remain poorly characterized, highlighting the need for multifactorial models.
Translating preclinical findings to clinical applications involves additional hurdles. Although animal studies have elucidated mechanisms such as SCFA-mediated immune modulation, they fail to replicate the complexity of human pediatric immune development and environmental exposures. Clinical trials must navigate inherent challenges including stringent participant matching for age, feeding practices, and comorbidities; prolonged follow-up (≥5 years) to assess outcomes in slowly progressing diseases; and unresolved safety concerns regarding probiotics or microbiota-targeted therapies in children.
Finally, the dynamic nature of the gut microbiota during childhood complicates research and intervention design. Most studies capture static snapshots, ignoring developmental trajectories and cumulative microbial effects on disease. Pronounced inter-individual variability, shaped by gene–environment–diet interactions, further complicates the development of personalized approaches. Addressing these limitations requires multinational longitudinal cohorts, multi-omics integration driven by artificial intelligence, and pediatric-specific therapeutic frameworks to advance GLA research from correlation to causation and clinical utility.
6.2 Development and clinical trials of novel treatments
6.2.1 Fecal microbiota transplantation
FMT is a treatment method in which a fecal suspension obtained from a healthy donor is transferred to the patient’s digestive tract to restore the normal microbial composition and function of the intestinal tract (101, 102). FMT can be delivered through the upper GI route via a duodenal tube or capsules taken orally (103) or through the lower GI route via colonoscopy or enema (104) (Figure 4). FMT may have applications in the treatment of many diseases, such as asthma, and has been used to treat a variety of gastrointestinal and non-gastrointestinal disorders (105).
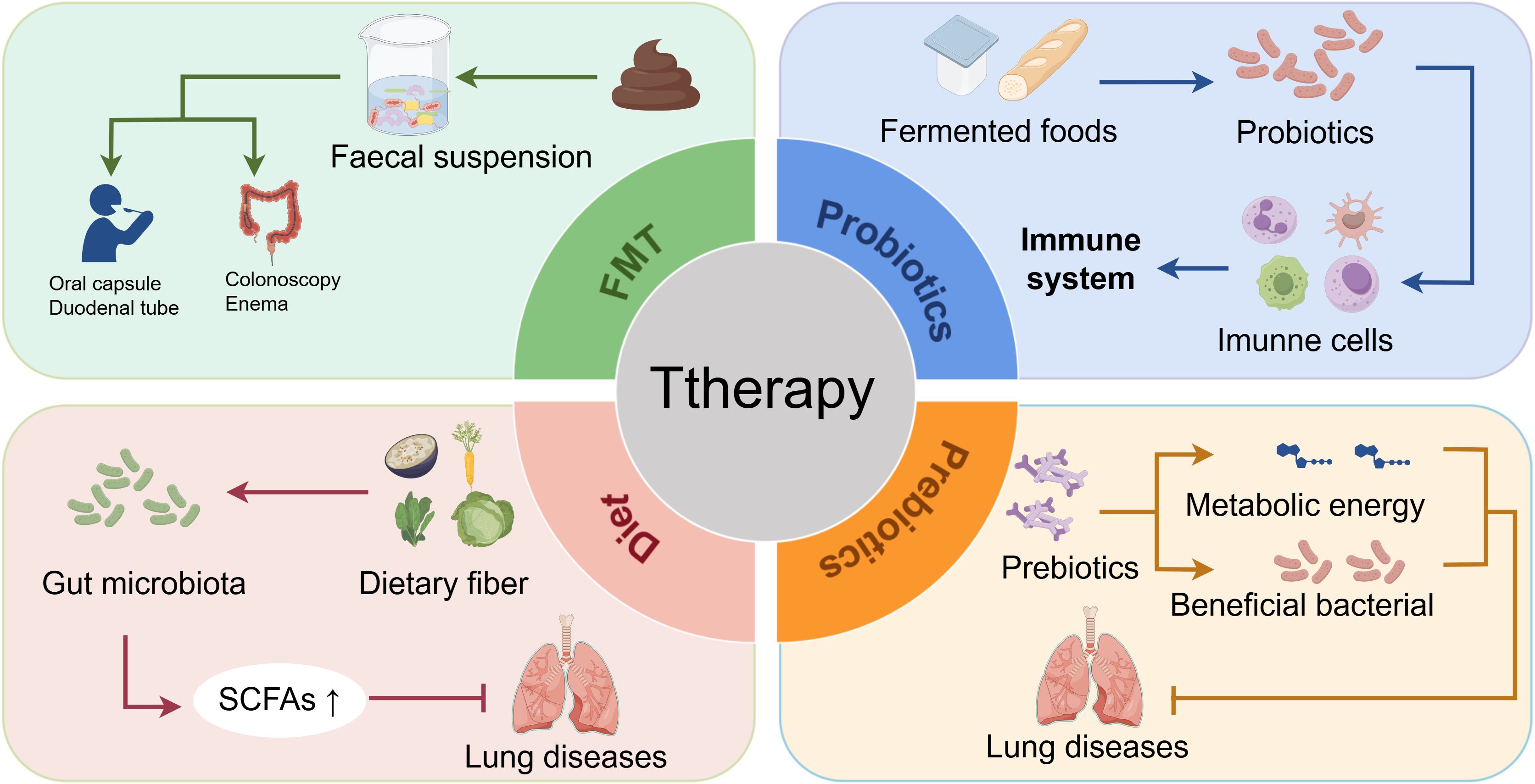
Figure 4. Therapeutic strategies targeting the GLA for pediatric pulmonary diseases: mechanisms and clinical interventions. Targeting the GLA offers a novel approach to managing childhood pulmonary diseases through gut microbiota modulation. FMT restores microbial homeostasis by administering healthy donor fecal suspensions via oral capsules, duodenal tubes, or colonoscopy/enema delivery. Probiotics alleviate respiratory symptoms via immunomodulation; prebiotics promote the growth of beneficial bacteria and provide metabolic substrates, thereby reducing pulmonary disease risk. Dietary interventions reshape gut microbiota composition, enhance SCFA production, and mitigate disease progression (e.g., pneumonia). Current evidence indicates that probiotics are more effective against eczema and allergic rhinitis than against asthma, highlighting the need to optimize strain selection, dosage, and treatment duration to improve GLA-targeted therapeutic precision. FMT, fecal microbiota transplantation; GLA, gut–lung axis; SCFAs, short-chain fatty acids.
6.2.2 Probiotics
Interest in probiotics has grown substantially owing to their complex mechanisms, which are strain- and compound-specific (106). The mechanisms by which probiotics operate are intricate and varied, typically depending on the specific strain and compound (107). Probiotics can alter the gut microenvironment, compete with harmful bacteria, and suppress pathogens using antimicrobial substances, thereby influencing gut health in nuanced ways (59). Emerging strategies targeting OSA-related gut dysbiosis using prebiotics, probiotics, and SCFAs are being considered for lung disease management and represent a promising therapeutic avenue (91, 108). Probiotics are present in fermented foods and supplements as scientifically validated beneficial strains and exhibit bidirectional immunomodulatory effects on the host, being capable of inducing pro-inflammatory responses while eliciting anti-inflammatory reactions (109). Under immunostimulatory conditions, macrophages, DCs, neutrophils, and natural killer cells in the intestinal mucosa participate in immune responses through enhanced phagocytic activity, inflammatory cytokine release, and Th1/Th17 polarization (15). Concurrently, probiotics activate innate immune responses and cytokines secreted by T cells to stimulate lamina propria-associated immune cells. This immunomodulatory activity is specifically characterized by inducing the production of immunoglobulin A antibody, increased population density, and functional enhancement of macrophages and DCs within the lamina propria, thereby sustaining their functional reinforcement in the small intestine (110) (Figure 4). A placebo-controlled, double-blind, randomized study investigated the effects of a probiotic mixture containing Bifidobacterium longum BB536, Bifidobacterium infantis M-63, and Bifidobacterium breve M-16V in 40 children with seasonal allergic rhinitis and intermittent asthma over a 4-week intervention period. Children receiving the probiotic supplementation demonstrated significant reductions in respiratory symptoms and improved quality of life whereas the placebo group exhibited symptom exacerbation and quality of life deterioration (111). A meta-analysis suggested that Lactobacillus rhamnosus GG (LGG) supplementation, especially postnatally, may prevent asthma (112); however, a database review indicated that probiotics are more effective for eczema and allergic rhinitis than for asthma prevention or treatment (113). Health organizations emphasize the need for further research on probiotic efficacy in asthma prevention because studies may not have used the appropriate strain, dosage, timing, duration, or population (114). Future research will focus on the duration, administration, dosage, and follow-up period for specific probiotic strains (115).
6.2.3 Prebiotics
According to the updated scientific definition established by the International Scientific Association for Probiotics and Prebiotics in 2017, prebiotics are defined as a substrate that is selectively used by host microorganisms to confer a health benefit (116). These compounds exert their effects primarily via two mechanisms: 1) by modulating the gut microbiota composition through the selective promotion of beneficial bacterial growth and provision of metabolic energy (117) (Figure 4); and 2) by enhancing intestinal barrier function and stimulating the production of beneficial metabolites, thereby inducing multifaceted physiological regulation in the host (118). Given their potential immunomodulatory properties and mechanistic actions, a theoretical basis exists for prebiotics to potentially reduce the risk of COVID-19 infection or mitigate its clinical symptoms (119). However, this hypothesis requires rigorous validation in further experimental and clinical investigations (120). A review of 2,419 pediatric participants analyzed asthma exacerbations, pulmonary function, and immune modulation. Studies that have focused on Lactobacillus/Bifidobacterium (10 randomized controlled trials [RCTs]), bacterial lysates (6 RCTs), and synbiotics (2 RCTs) represent a notable paucity of prebiotic research (121).
6.2.4 Diet
Studies have shown that factors such as diet, genetics, and age affect diversity of the gut microbiota, with diet being a key modifiable factor for treating dysbiosis (27, 122). Clinical and preclinical data highlight how dietary changes can rapidly alter the gut microbiota composition, such as shifts occurring within 24 hours of switching from an animal-based and to a plant-based diet (123, 124). Dietary fiber intake boosts bacterial metabolites, particularly SCFAs (41). Dietary patterns influence the β diversity of the gut microbiota without affecting its α diversity, which varies among individuals consuming an animal-based diet (125, 126). High-calorie diets can worsen LPS-induced pneumonia by disrupting the gut microbiota balance and Th17/Treg cell ratios (127). Future prevention and therapy may involve dietary pattern modifications, such as reducing specific nutrients or adopting lifestyle changes to address physical inactivity and obesity (Figure 4).
Probiotics and dietary changes aim to foster a balanced gut microbiota, which is beneficial for managing respiratory conditions and offering a safer alternative to traditional pharmacological treatments, especially in chronic disease management (3). These strategies collectively bridge mechanistic insights and clinical translation to reshape pediatric respiratory disease management.
6.3 Future research directions
The role of the gut microbiome in pediatric respiratory diseases is an emerging field with implications for new therapeutic strategies. Future research should focus on the GLA, particularly the impact of microbial metabolites on immune modulation. Multi-omics approaches will help deepen understanding regarding the complex interactions between the gut microbiota and children’s respiratory health. A key objective is to identify microbial signatures and metabolites that serve as biomarkers for early diagnosis and prognosis of diseases such as asthma and pneumonia, enabling personalized medicine based on individual microbiome profiles. It is also crucial to explore the role of the gut microbiome in responses to treatments such as FMT and probiotics/prebiotics, as well as to design clinical trials assessing their efficacy and safety. Research should also investigate the microbiome’s influence on vaccine responses and immunotherapies for respiratory diseases, potentially leading to microbe-based adjuvants that boost vaccine efficacy. The impact of diet on the gut microbiome and lung health in children merits further study, with a focus on dietary interventions to improve respiratory outcomes. Finally, translating preclinical findings into clinical practice is essential. Future research should connect laboratory discoveries with clinical applications, implementing evidence-based interventions that leverage the GLA to prevent and treat pediatric respiratory diseases, with the aim to reduce the impact of these diseases on children’s health and quality of life globally.
7 Conclusions
This review synthesizes critical evidence on the MGLA as a central regulator of pediatric respiratory health. Early-life gut microbiota perturbations—driven by birth mode, antibiotics, and diet—disrupt immune programming via metabolite signaling (e.g., SCFAs), epigenetic modulation, and pathogen exclusion, predisposing to asthma, pneumonia, and bronchiolitis. Multi-omics integration has revealed evolutionarily conserved microbiota–immune network architectures across disease phenotypes, with mechanistic prioritization identifying Faecalibacterium-derived SCFA-mediated Treg modulation as therapeutically actionable biological circuitry. To systematically characterize the methodological framework underpinning these findings, we constructed a comprehensive reference table (Table 1) detailing the multi-omics platforms, analytical pipelines, and data integration strategies used, thereby providing explicit technical documentation to enhance methodological reproducibility and cross-study interoperability. Clinically, microbiome-modulating strategies—including precision probiotics, FMT, and dietary SCFA boosters—demonstrate efficacy in respiratory diseases such as asthma and pneumonia. For OSAS, the current evidence is primarily derived from preclinical models, necessitating clinical trials to establish therapeutic efficacy. Future research must prioritize longitudinal birth cohorts to map developmental windows of microbiota–immune crosstalk, coupled with mechanistic studies dissecting microbial vesicles and mobile immune cell trafficking. Standardizing microbiota therapeutics and leveraging AI-driven biomarker panels will accelerate translation from bench to bedside, ultimately enabling the personalized management of childhood respiratory diseases.
Author contributions
ZW: Conceptualization, Supervision, Writing – original draft, Funding acquisition, Writing – review & editing. JY: Conceptualization, Writing – review & editing, Visualization, Writing – original draft. YL: Visualization, Writing – review & editing. JG: Visualization, Writing – review & editing. ZH: Visualization, Writing – review & editing. ZL: Supervision, Funding acquisition, Writing – review & editing, Conceptualization.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was funded by Shaoxing University Enterprise Important Horizontal Topic (No. 2024USXH287).
Acknowledgments
We thank LetPub (www.letpub.com.cn) for its linguistic assistance during the preparation of this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that Generative AI was used in the creation of this manuscript. During the preparation of this work the author(s) used DeepSeek and Kimi in order to improve language and readability. After using this tool/service, the author(s) reviewed and edited the content as needed and take(s) full responsibility for the content of the publication.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
CF, cystic fibrosis; DC, dendritic cell; FMT, fecal microbiota transplantation; GLA, gut–lung axis; HDAC, histone deacetylase; IFN, interferon; IL, interleukin; ILI, influenza-like illness; LPS, lipopolysaccharide; MGLA, microbiota–gut–lung axis; OSAS, obstructive sleep apnea syndrome; PRRs, pattern recognition receptors; RSV, respiratory syncytial virus; SCFAs, short-chain fatty acids; Th, T helper; TLR, Toll-like receptor; TNF, tumor necrosis factor; Treg, regulatory T cell.
References
1. Kloepfer KM and Kennedy JL. Childhood respiratory viral infections and the microbiome. J Allergy Clin Immunol. (2023) 152:827–34. doi: 10.1016/j.jaci.2023.08.008
2. Lloyd CM and Saglani S. Early-life respiratory infections and developmental immunity determine lifelong lung health. Nat Immunol. (2023) 24:1234–43. doi: 10.1038/s41590-023-01550-w
3. Du B, Fu Y, Han Y, Sun Q, Xu J, Yang Y, et al. The lung-gut crosstalk in respiratory and inflammatory bowel disease. Front Cell Infect Microbiol. (2023) 13:1218565. doi: 10.3389/fcimb.2023.1218565
4. Zhu W, Wu Y, Liu H, Jiang C, and Huo L. Gut-lung axis: microbial crosstalk in pediatric respiratory tract infections. Front Immunol. (2021) 12:741233. doi: 10.3389/fimmu.2021.741233
5. Kahhaleh FG, Barrientos G, and Conrad ML. The gut-lung axis and asthma susceptibility in early life. Acta Physiol (Oxf). (2024) 240:e14092. doi: 10.1111/apha.14092
6. Stricker S, Hain T, Chao CM, and Rudloff S. Respiratory and intestinal microbiota in pediatric lung diseases-current evidence of the gut-lung axis. Int J Mol Sci. (2022) 23):6791–820. doi: 10.3390/ijms23126791
7. Boncheva I, Poudrier J, and Falcone EL. Role of the intestinal microbiota in host defense against respiratory viral infections. Curr Opin Virol. (2024) 66:101410. doi: 10.1016/j.coviro.2024.101410
8. Nunez H, Nieto PA, Mars RA, Ghavami M, Sew HC, and Sukhum K. Early life gut microbiome and its impact on childhood health and chronic conditions. Gut Microbes. (2025) 17:2463567. doi: 10.1080/19490976.2025.2463567
9. Ronan V, Yeasin R, and Claud EC. Childhood development and the microbiome—The intestinal microbiota in maintenance of health and development of disease during childhood development. Gastroenterology. (2021) 160:495–506. doi: 10.1053/j.gastro.2020.08.065
10. Korpela K, Salonen A, Virta LJ, Kekkonen RA, Forslund K, Bork P, et al. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat Commun. (2016) 7:10410. doi: 10.1038/ncomms10410
11. Dang AT and Marsland BJ. Microbes, metabolites, and the gut–lung axis. Mucosal Immunol. (2019) 12:843–50. doi: 10.1038/s41385-019-0160-6
12. Kelderer F, Mogren I, Eriksson C, Silfverdal SA, Domellof M, and West CE. Associations between pre- and postnatal antibiotic exposures and early allergic outcomes: A population-based birth cohort study. Pediatr Allergy Immunol. (2022) 33:e13848. doi: 10.1111/pai.13848
13. Tan JK, Macia L, and Mackay CR. Dietary fiber and SCFAs in the regulation of mucosal immunity. J Allergy Clin Immunol. (2023) 151:361–70. doi: 10.1016/j.jaci.2022.11.007
14. Barcik W, Boutin RCT, Sokolowska M, and Finlay BB. The role of lung and gut microbiota in the pathology of asthma. Immunity. (2020) 52:241–55. doi: 10.1016/j.immuni.2020.01.007
15. Ma P, Wang M, and Wang Y. Gut microbiota: A new insight into lung diseases. BioMed Pharmacother. (2022) 155:113810. doi: 10.1016/j.biopha.2022.113810
16. Budden KF, Gellatly SL, Wood DL, Cooper MA, Morrison M, Hugenholtz P, et al. Emerging pathogenic links between microbiota and the gut-lung axis. Nat Rev Microbiol. (2017) 15:55–63. doi: 10.1038/nrmicro.2016.142
17. Belkaid Y and Hand TW. Role of the microbiota in immunity and inflammation. Cell. (2014) 157:121–41. doi: 10.1016/j.cell.2014.03.011
18. Kim YJ and Bunyavanich S. Microbial influencers: the airway microbiome's role in asthma. J Clin Invest. (2025) 135:e184316–26. doi: 10.1172/JCI184316
19. Stricker S, Hain T, Chao C, and Rudloff S. Respiratory and intestinal microbiota in pediatric lung diseases—Current evidence of the gut–lung axis. Int J Mol Sci. (2022) 23:6791. doi: 10.3390/ijms23126791
20. Singh N, Vats A, Sharma A, Arora A, and Kumar A. The development of lower respiratory tract microbiome in mice. Microbiome. (2017) 5:61. doi: 10.1186/s40168-017-0277-3
21. Huffnagle GB, Dickson RP, and Lukacs NW. The respiratory tract microbiome and lung inflammation: a two-way street. Mucosal Immunol. (2017) 10:299–306. doi: 10.1038/mi.2016.108
22. Ashique S, De Rubis G, Sirohi E, Mishra N, Rihan M, Garg A, et al. Short Chain Fatty Acids: Fundamental mediators of the gut-lung axis and their involvement in pulmonary diseases. Chem Biol Interact. (2022) 368:110231. doi: 10.1016/j.cbi.2022.110231
23. Tamburini S and Clemente JC. Gut microbiota: Neonatal gut microbiota induces lung immunity against pneumonia. Nat Rev Gastroenterol Hepatol. (2017) 14:263–64. doi: 10.1038/nrgastro.2017.34
24. Tamburini S, Shen N, Wu HC, and Clemente JC. The microbiome in early life: implications for health outcomes. Nat Med. (2016) 22:713–22. doi: 10.1038/nm.4142
25. Inchingolo F, Inchingolo AD, Palumbo I, Trilli I, Guglielmo M, Mancini A, et al. The impact of cesarean section delivery on intestinal microbiota: mechanisms, consequences, and perspectives-A systematic review. Int J Mol Sci. (2024) 25:1055–75. doi: 10.3390/ijms25021055
26. Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med. (2016) 22:1187–91. doi: 10.1038/nm.4176
27. Rastogi S, Mohanty S, Sharma S, and Tripathi P. Possible role of gut microbes and host’s immune response in gut–lung homeostasis. Front Immunol. (2022) 13:954339. doi: 10.3389/fimmu.2022.954339
28. Eladham MW, Selvakumar B, Saheb SN, Saheb SF, Ibrahim SM, and Halwani R. Unraveling the gut-Lung axis: Exploring complex mechanisms in disease interplay. Heliyon. (2024) 10:e24032. doi: 10.1016/j.heliyon.2024.e24032
29. Sun CY, Yang N, Zheng ZL, Liu D, and Xu QL. T helper 17 (Th17) cell responses to the gut microbiota in human diseases. BioMed Pharmacother. (2023) 161:114483. doi: 10.1016/j.biopha.2023.114483
30. Harris NL and Marsland BJ. The gut-lung axis: Protozoa join the party. Cell. (2025) 188:275–77. doi: 10.1016/j.cell.2024.12.027
31. Johnson CC and Ownby DR. The infant gut bacterial microbiota and risk of pediatric asthma and allergic diseases. Transl Res. (2017) 179:60–70. doi: 10.1016/j.trsl.2016.06.010
32. Gilmour BC, Corthay A, and Oynebraten I. High production of IL-12 by human dendritic cells stimulated with combinations of pattern-recognition receptor agonists. NPJ Vaccines. (2024) 9:83. doi: 10.1038/s41541-024-00869-1
33. Wang J, Zhu N, Su X, Gao Y, and Yang R. Gut-microbiota-derived metabolites maintain gut and systemic immune homeostasis. Cells. (2023) 12:793–819. doi: 10.3390/cells12050793
34. Ozcam M and Lynch SV. The gut-airway microbiome axis in health and respiratory diseases. Nat Rev Microbiol. (2024) 22:492–506. doi: 10.1038/s41579-024-01048-8
35. Tang J, Xu L, Zeng Y, and Gong F. Effect of gut microbiota on LPS-induced acute lung injury by regulating the TLR4/NF-kB signaling pathway. Int Immunopharmacol. (2021) 91:107272. doi: 10.1016/j.intimp.2020.107272
36. Kim CH. Control of lymphocyte functions by gut microbiota-derived short-chain fatty acids. Cell Mol Immunol. (2021) 18:1161–71. doi: 10.1038/s41423-020-00625-0
37. Shibuya A and Shibuya K. Exploring the gut fungi-lung allergy axis. Cell Host Microbe. (2018) 24:755–57. doi: 10.1016/j.chom.2018.11.012
38. Zhang S, Paul S, and Kundu P. NF-kappaB regulation by gut microbiota decides homeostasis or disease outcome during ageing. Front Cell Dev Biol. (2022) 10:874940. doi: 10.3389/fcell.2022.874940
39. Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. (2014) 20:159–66. doi: 10.1038/nm.3444
40. Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, DeRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. (2013) 504:451–55. doi: 10.1038/nature12726
41. Koh A, De Vadder F, Kovatcheva-Datchary P, and Backhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. (2016) 165:1332–45. doi: 10.1016/j.cell.2016.05.041
42. Mann ER, Lam YK, and Uhlig HH. Short-chain fatty acids: linking diet, the microbiome and immunity. Nat Rev Immunol. (2024) 24:577–95. doi: 10.1038/s41577-024-01014-8
43. Caetano-Silva ME, Rund L, Hutchinson NT, Woods JA, Steelman AJ, and Johnson RW. Inhibition of inflammatory microglia by dietary fiber and short-chain fatty acids. Sci Rep. (2023) 13:2819. doi: 10.1038/s41598-022-27086-x
44. Abdelhalim KA. Short-chain fatty acids (SCFAs) from gastrointestinal disorders, metabolism, epigenetics, central nervous system to cancer - A mini-review. Chem Biol Interact. (2024) 388:110851. doi: 10.1016/j.cbi.2023.110851
45. Ney LM, Wipplinger M, Grossmann M, Engert N, Wegner VD, and Mosig AS. Short chain fatty acids: key regulators of the local and systemic immune response in inflammatory diseases and infections. Open Biol. (2023) 13:230014. doi: 10.1098/rsob.230014
46. Shi C, Zhou L, Li H, Shi X, Zhang Y, Lu Y, et al. Intestinal microbiota metabolizing Houttuynia cordata polysaccharides in H1N1 induced pneumonia mice contributed to Th17/Treg rebalance in gut-lung axis. Int J Biol Macromol. (2022) 221:288–302. doi: 10.1016/j.ijbiomac.2022.09.015
47. Zhao M, Shao F, Yu D, Zhang J, Liu Z, Ma J, et al. Maturation and specialization of group 2 innate lymphoid cells through the lung-gut axis. Nat Commun. (2022) 13:7600. doi: 10.1038/s41467-022-35347-6
48. GBD 2015 Chronic Respiratory Disease Collaborators. Global, regional, and national deaths, prevalence, disability-adjusted life years, and years lived with disability for chronic obstructive pulmonary disease and asthma, 1990-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Respir Med. (2017) 5:691–706. doi: 10.1016/S2213-2600(17)30293-X
49. Lee-Sarwar KA, Kelly RS, Lasky-Su J, Zeiger RS, O'Connor GT, Sandel MT, et al. Integrative analysis of the intestinal metabolome of childhood asthma. J Allergy Clin Immunol. (2019) 144:442–54. doi: 10.1016/j.jaci.2019.02.032
50. Stern J, Pier J, and Litonjua AA. Asthma epidemiology and risk factors. Semin Immunopathol. (2020) 42:5–15. doi: 10.1007/s00281-020-00785-1
51. Ege MJ. The hygiene hypothesis in the age of the microbiome. Ann Am Thorac Soc. (2017) 14:S348–53. doi: 10.1513/AnnalsATS.201702-139AW
52. Zheng D, Liwinski T, and Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. (2020) 30:492–506. doi: 10.1038/s41422-020-0332-7
53. Valverde-Molina J and Garcia-Marcos L. Microbiome and asthma: microbial dysbiosis and the origins, phenotypes, persistence, and severity of asthma. Nutrients. (2023) 15:486–511. doi: 10.3390/nu15030486
54. Stokholm J, Blaser MJ, Thorsen J, Rasmussen MA, Waage J, Vinding RK, et al. Maturation of the gut microbiome and risk of asthma in childhood. Nat Commun. (2018) 9:141. doi: 10.1038/s41467-017-02573-2
55. Gensollen T, Iyer SS, Kasper DL, and Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science. (2016) 352:539–44. doi: 10.1126/science.aad9378
56. Oliphant K and Allen-Vercoe E. Macronutrient metabolism by the human gut microbiome: major fermentation by-products and their impact on host health. Microbiome. (2019) 7:91. doi: 10.1186/s40168-019-0704-8
57. Charbonneau MR, O'Donnell D, Blanton LV, Totten SM, Davis JC, Barratt MJ, et al. Sialylated milk oligosaccharides promote microbiota-dependent growth in models of infant undernutrition. Cell. (2016) 164:859–71. doi: 10.1016/j.cell.2016.01.024
58. Urbaniak C, Angelini M, Gloor GB, and Reid G. Human milk microbiota profiles in relation to birthing method, gestation and infant gender. Microbiome. (2016) 4:1. doi: 10.1186/s40168-015-0145-y
59. Zhang D, Jian Y, Zhang Y, Li Y, Gu L, Sun H, et al. Short-chain fatty acids in diseases. Cell Commun Signal. (2023) 21:212–32. doi: 10.1186/s12964-023-01219-9
60. Dong J. Signaling pathways implicated in carbon nanotube-induced lung inflammation. Front Immunol. (2020) 11:552613. doi: 10.3389/fimmu.2020.552613
61. Quach A, Spence H, Nguyen C, Graham SM, von Mollendorf C, Mulholland K, et al. Slow progress towards pneumonia control for children in low-and-middle income countries as measured by pneumonia indicators: A systematic review of the literature. J Glob Health. (2022) 12:10006. doi: 10.7189/jogh.12.10006
62. Hanada S, Pirzadeh M, Carver KY, and Deng JC. Respiratory viral infection-induced microbiome alterations and secondary bacterial pneumonia. Front Immunol. (2018) 9:2640. doi: 10.3389/fimmu.2018.02640
63. Zhang S and Chen DC. Facing a new challenge: the adverse effects of antibiotics on gut microbiota and host immunity. Chin Med J (Engl). (2019) 132:1135–38. doi: 10.1097/CM9.0000000000000245
64. Schuijt TJ, Lankelma JM, Scicluna BP, de Sousa E Melo F, Roelofs JJTH, de Boer JD, et al. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut. (2016) 65:575–83. doi: 10.1136/gutjnl-2015-309728
65. Xie Q, Li Q, Fang H, Zhang R, Tang H, and Chen L. Gut-derived short-chain fatty acids and macrophage modulation: exploring therapeutic potentials in pulmonary fungal infections. Clin Rev Allergy Immunol. (2024) 66:316–27. doi: 10.1007/s12016-024-08999-z
66. Wang Q, Fang Z, Xiao Y, Wang H, Zhang P, Lu W, et al. Lactiplantibacillus pentoses CCFM1227 Produces Desaminotyrosine to Protect against Influenza Virus H1N1 Infection through the Type I Interferon in Mice. Nutrients. (2023) 15:3659–75. doi: 10.3390/nu15163659
67. Amin R, Hatakeyama Y, Kitazawa T, Matsumoto K, Fujita S, Seto K, et al. Capturing the trends in hospital standardized mortality ratios for pneumonia: a retrospective observational study in Japan (2010 to 2018). Environ Health Prev Med. (2020) 25:2. doi: 10.1186/s12199-019-0842-4
68. Ma Y, Peng X, Zhang J, Zhu Y, Huang R, Li G, et al. Gut microbiota in preterm infants with late-onset sepsis and pneumonia: a pilot case-control study. BMC Microbiol. (2024) 24:272. doi: 10.1186/s12866-024-03419-w
69. Steed AL, Christophi GP, Kaiko GE, Sun L, Goodwin VM, Jain U, et al. The microbial metabolite desaminotyrosine protects from influenza through type I interferon. Science. (2017) 357:498–502. doi: 10.1126/science.aam5336
70. Jiang Y, Bao C, Zhao X, Chen Y, Song Y, and Xiao Z. Intestinal bacteria flora changes in patients with Mycoplasma pneumoniae pneumonia with or without wheezing. Sci Rep. (2022) 12:5683. doi: 10.1038/s41598-022-09700-0
71. Shao Y, Forster SC, Tsaliki E, Vervier K, Strang A, Simpson N, et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature. (2019) 574:117–21. doi: 10.1038/s41586-019-1560-1
72. Nikolopoulou GB and Maltezou HC. COVID-19 in Children: Where do we Stand? Arch Med Res. (2022) 53:1–08. doi: 10.1016/j.arcmed.2021.07.002
73. Franchitti E, Bottino P, Sidoti F, Carpino A, Pruccoli G, Ramenghi U, et al. Investigating the role of gut microbiota in pediatric patients with severe COVID-19 or MIS-C. Microorganisms. (2025) 13:83–97. doi: 10.3390/microorganisms13010083
74. Bacorn M, Romero-Soto HN, Levy S, Chen Q, and Hourigan SK. The gut microbiome of children during the COVID-19 pandemic. Microorganisms. (2022) 10:2460–89. doi: 10.3390/microorganisms10122460
75. Li Y, Wang X, Blau DM, Caballero MT, Feikin DR, Gill CJ, et al. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in children younger than 5 years in 2019: a systematic analysis. Lancet. (2022) 399:2047–64. doi: 10.1016/S0140-6736(22)00478-0
76. Alba C, Aparicio M, Gonzalez-Martinez F, Gonzalez-Sanchez MI, Perez-Moreno J, Toledo DCB, et al. Nasal and fecal microbiota and immunoprofiling of infants with and without RSV bronchiolitis. Front Microbiol. (2021) 12:667832. doi: 10.3389/fmicb.2021.667832
77. Antunes KH, Fachi JL, de Paula R, Da SE, Pral LP, Dos SA, et al. Microbiota-derived acetate protects against respiratory syncytial virus infection through a GPR43-type 1 interferon response. Nat Commun. (2019) 10:3273. doi: 10.1038/s41467-019-11152-6
78. Dogra SK, Martin FP, Donnicola D, Julita M, Berger B, and Sprenger N. Human milk oligosaccharide-stimulated bifidobacterium species contribute to prevent later respiratory tract infections. Microorganisms. (2021) 9:1939–53. doi: 10.3390/microorganisms9091939
79. Fonseca W, Malinczak CA, Fujimura K, Li D, McCauley K, Li J, et al. Maternal gut microbiome regulates immunity to RSV infection in offspring. J Exp Med. (2021) 218:e20210235–48. doi: 10.1084/jem.20210235
80. Shahrebabak AG, Rezaei M, Shahpar A, Nezhad NZ, Sarasyabi MS, Nakhaie M, et al. The efficacy of COVID-19 vaccination in cystic fibrosis patients: a systematic review. BMC Infect Dis. (2025) 25:358. doi: 10.1186/s12879-025-10736-6
81. Ranganathan SC, Hall GL, Sly PD, Stick SM, Douglas TA, and Australian REST. Early lung disease in infants and preschool children with cystic fibrosis. What have we learned and what should we do about it? Am J Respir Crit Care Med. (2017) 195:1567–75. doi: 10.1164/rccm.201606-1107CI
82. McKay I, van Dorst J, Katz T, Doumit M, Prentice B, Owens L, et al. Diet and the gut-lung axis in cystic fibrosis – direct & indirect links. Gut Microbes. (2023) 15:2156254–69. doi: 10.1080/19490976.2022.2156254
83. Enaud R, Prevel R, Ciarlo E, Beaufils F, Wieers G, Guery B, et al. The gut-lung axis in health and respiratory diseases: A place for inter-organ and inter-kingdom crosstalks. Front Cell Infect Microbiol. (2020) 10:9. doi: 10.3389/fcimb.2020.00009
84. Enaud R, Hooks KB, Barre A, Barnetche T, Hubert C, Massot M, et al. Intestinal inflammation in children with cystic fibrosis is associated with crohn's-like microbiota disturbances. J Clin Med. (2019) 8:645–58. doi: 10.3390/jcm8050645
85. Vernocchi P, Del CF, Russo A, Majo F, Rossitto M, Valerio M, et al. Gut microbiota signatures in cystic fibrosis: Loss of host CFTR function drives the microbiota enterophenotype. PloS One. (2018) 13:e208171. doi: 10.1371/journal.pone.0208171
86. Caparros-Martin JA, Saladie M, Agudelo-Romero SP, Reen FJ, Ware RS, Sly PD, et al. Detection of bile acids in bronchoalveolar lavage fluid defines the inflammatory and microbial landscape of the lower airways in infants with cystic fibrosis. Microbiome. (2023) 11:132. doi: 10.1186/s40168-023-01543-9
87. Bass R, Brownell JN, and Stallings VA. The impact of highly effective CFTR modulators on growth and nutrition status. Nutrients. (2021) 13:2907–18. doi: 10.3390/nu13092907
88. Price CE and O'Toole GA. The gut-lung axis in cystic fibrosis. J Bacteriol. (2021) 203:e31121. doi: 10.1128/JB.00311-21
89. Bitners AC and Arens R. Evaluation and management of children with obstructive sleep apnea syndrome. Lung. (2020) 198:257–70. doi: 10.1007/s00408-020-00342-5
90. Lv R, Liu X, Zhang Y, Dong N, Wang X, He Y, et al. Pathophysiological mechanisms and therapeutic approaches in obstructive sleep apnea syndrome. Signal Transduct Target Ther. (2023) 8:218. doi: 10.1038/s41392-023-01496-3
91. Yan W, Jiang M, Hu W, Zhan X, Liu Y, Zhou J, et al. Causality investigation between gut microbiota, derived metabolites, and obstructive sleep apnea: A bidirectional mendelian randomization study. Nutrients. (2023) 15:4544–59. doi: 10.3390/nu15214544
92. Liu W, Du Q, Zhang H, and Han D. The gut microbiome and obstructive sleep apnea syndrome in children. Sleep Med. (2022) 100:462–71. doi: 10.1016/j.sleep.2022.09.022
93. Wang F, Zou J, Xu H, Huang W, Zhang X, Wei Z, et al. Effects of chronic intermittent hypoxia and chronic sleep fragmentation on gut microbiome, serum metabolome, liver and adipose tissue morphology. Front Endocrinol (Lausanne). (2022) 13:820939. doi: 10.3389/fendo.2022.820939
94. Ko CY, Liu QQ, Su HZ, Zhang HP, Fan JM, Yang JH, et al. Gut microbiota in obstructive sleep apnea-hypopnea syndrome: disease-related dysbiosis and metabolic comorbidities. Clin Sci (Lond). (2019) 133:905–17. doi: 10.1042/CS20180891
95. Ganesh BP, Nelson JW, Eskew JR, Ganesan A, Ajami NJ, Petrosino JF, et al. Prebiotics, probiotics, and acetate supplementation prevent hypertension in a model of obstructive sleep apnea. Hypertension. (2018) 72:1141–50. doi: 10.1161/HYPERTENSIONAHA.118.11695
96. Mashaqi S, Rangan P, Saleh AA, Abraham I, Gozal D, Quan SF, et al. Biomarkers of gut barrier dysfunction in obstructive sleep apnea: A systematic review and meta-analysis. Sleep Med Rev. (2023) 69:101774. doi: 10.1016/j.smrv.2023.101774
97. Callejo M, Mondejar-Parreno G, Barreira B, Izquierdo-Garcia JL, Morales-Cano D, Esquivel-Ruiz S, et al. Pulmonary arterial hypertension affects the rat gut microbiome. Sci Rep. (2018) 8:9681. doi: 10.1038/s41598-018-27682-w
98. Yeoh YK, Zuo T, Lui GC, Zhang F, Liu Q, Li AY, et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. (2021) 70:698–706. doi: 10.1136/gutjnl-2020-323020
99. Zhang F, Wan Y, Zuo T, Yeoh YK, Liu Q, Zhang L, et al. Prolonged impairment of short-chain fatty acid and L-isoleucine biosynthesis in gut microbiome in patients with COVID-19. Gastroenterology. (2022) 162:548–61. doi: 10.1053/j.gastro.2021.10.013
100. Hjelmsø MH, Shah SA, Thorsen J, Rasmussen M, Vestergaard G, Mortensen MS, et al. Prenatal dietary supplements influence the infant airway microbiota in a randomized factorial clinical trial. Nat Commun. (2020) 11:426. doi: 10.1038/s41467-020-14308-x
101. de Groot P, Scheithauer T, Bakker GJ, Prodan A, Levin E, Khan MT, et al. Donor metabolic characteristics drive effects of faecal microbiota transplantation on recipient insulin sensitivity, energy expenditure and intestinal transit time. Gut. (2020) 69:502–12. doi: 10.1136/gutjnl-2019-318320
102. Leonardi I, Paramsothy S, Doron I, Semon A, Kaakoush NO, Clemente JC, et al. Fungal trans-kingdom dynamics linked to responsiveness to fecal microbiota transplantation (FMT) therapy in ulcerative colitis. Cell Host Microbe. (2020) 27:823–29. doi: 10.1016/j.chom.2020.03.006
103. Kao D, Roach B, Silva M, Beck P, Rioux K, Kaplan GG, et al. Effect of Oral Capsule- vs Colonoscopy-Delivered Fecal Microbiota Transplantation on Recurrent Clostridium difficile Infection: A Randomized Clinical Trial. Jama. (2017) 318:1985–93. doi: 10.1001/jama.2017.17077
104. Shao T, Hsu R, Hacein-Bey C, Zhang W, Gao L, Kurth MJ, et al. The evolving landscape of fecal microbial transplantation. Clin Rev Allergy Immunol. (2023) 65:101–20. doi: 10.1007/s12016-023-08958-0
105. Logon K, Swirkosz G, Nowak M, Wrzesniewska M, Szczygiel A, and Gomulka K. The role of the microbiome in the pathogenesis and treatment of asthma. Biomedicines. (2023) 11:1618–35. doi: 10.3390/biomedicines11061618
106. Salminen S, Collado MC, Endo A, Hill C, Lebeer S, Quigley E, et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat Rev Gastroenterol Hepatol. (2021) 18:649–67. doi: 10.1038/s41575-021-00440-6
107. Cunningham M, Azcarate-Peril MA, Barnard A, Benoit V, Grimaldi R, Guyonnet D, et al. Shaping the future of probiotics and prebiotics. Trends Microbiol. (2021) 29:667–85. doi: 10.1016/j.tim.2021.01.003
108. Badran M, Mashaqi S, and Gozal D. The gut microbiome as a target for adjuvant therapy in obstructive sleep apnea. Expert Opin Ther Targets. (2020) 24:1263–82. doi: 10.1080/14728222.2020.1841749
109. D'Ettorre G, Ceccarelli G, Marazzato M, Campagna G, Pinacchio C, Alessandri F, et al. Challenges in the management of SARS-coV2 infection: the role of oral bacteriotherapy as complementary therapeutic strategy to avoid the progression of COVID-19. Front Med (Lausanne). (2020) 7:389. doi: 10.3389/fmed.2020.00389
110. Wang X, Zhang P, and Zhang X. Probiotics regulate gut microbiota: an effective method to improve immunity. Molecules. (2021) 26:6076–91. doi: 10.3390/molecules26196076
111. Miraglia DGM, Indolfi C, Capasso M, Maiello N, Decimo F, and Ciprandi G. Bifidobacterium mixture (B longum BB536, B infantis M-63, B breve M-16V) treatment in children with seasonal allergic rhinitis and intermittent asthma. Ital J Pediatr. (2017) 43:25. doi: 10.1186/s13052-017-0340-5
112. Du X, Wang L, Wu S, Yuan L, Tang S, Xiang Y, et al. Efficacy of probiotic supplementary therapy for asthma, allergic rhinitis, and wheeze: a meta-analysis of randomized controlled trials. Allergy Asthma Proc. (2019) 40:250–60. doi: 10.2500/aap.2019.40.4227
113. Wang HT, Anvari S, and Anagnostou K. The role of probiotics in preventing allergic disease. Children (Basel). (2019) 6:24–33. doi: 10.3390/children6020024
114. Edwards MR, Walton RP, Jackson DJ, Feleszko W, Skevaki C, Jartti T, et al. The potential of anti-infectives and immunomodulators as therapies for asthma and asthma exacerbations. Allergy. (2018) 73:50–63. doi: 10.1111/all.13257
115. He Q, Huang J, Zheng T, Lin D, Zhang H, Li J, et al. Treatment with mixed probiotics induced, enhanced and diversified modulation of the gut microbiome ofxhealthy rats. FEMS Microbiol Ecol. (2021) 97:fiab151–64. doi: 10.1093/femsec/fiab151
116. Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJ, et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol. (2017) 14:491–502. doi: 10.1038/nrgastro.2017.75
117. Calatayud M, Van den Abbeele P, Ghyselinck J, Marzorati M, Rohs E, and Birkett A. Comparative effect of 22 dietary sources of fiber on gut microbiota of healthy humans in vitro. Front Nutr. (2021) 8:700571. doi: 10.3389/fnut.2021.700571
118. Davani-Davari D, Negahdaripour M, Karimzadeh I, Seifan M, Mohkam M, Masoumi SJ, et al. Prebiotics: definition, types, sources, mechanisms, and clinical applications. Foods. (2019) 8:92–119. doi: 10.3390/foods8030092
119. Batista KS, de Albuquerque JG, Vasconcelos M, Bezerra M, Da SBM, Pinheiro RO, et al. Probiotics and prebiotics: potential prevention and therapeutic target for nutritional management of COVID-19? Nutr Res Rev. (2023) 36:181–98. doi: 10.1017/S0954422421000317
120. Xu L, Yang CS, Liu Y, and Zhang X. Effective regulation of gut microbiota with probiotics and prebiotics may prevent or alleviate COVID-19 through the gut-lung axis. Front Pharmacol. (2022) 13:895193. doi: 10.3389/fphar.2022.895193
121. Fan D, Hu J, and Lin N. Effects of probiotics, prebiotics, synbiotics and postbiotics on pediatric asthma: a systematic review. Front Nutr. (2025) 12:1586129. doi: 10.3389/fnut.2025.1586129
122. Rinninella E, Cintoni M, Raoul P, Lopetuso LR, Scaldaferri F, Pulcini G, et al. Food components and dietary habits: keys for a healthy gut microbiota composition. Nutrients. (2019) 11:2393–416. doi: 10.3390/nu11102393
123. Tanes C, Bittinger K, Gao Y, Friedman ES, Nessel L, Paladhi UR, et al. Role of dietary fiber in the recovery of the human gut microbiome and its metabolome. Cell Host Microbe. (2021) 29:394–407. doi: 10.1016/j.chom.2020.12.012
124. Miao Z, Du W, Xiao C, Su C, Gou W, Shen L, et al. Gut microbiota signatures of long-term and short-term plant-based dietary pattern and cardiometabolic health: a prospective cohort study. BMC Med. (2022) 20:204. doi: 10.1186/s12916-022-02402-4
125. Roager HM, Vogt JK, Kristensen M, Hansen L, Ibrugger S, Maerkedahl RB, et al. Whole grain-rich diet reduces body weight and systemic low-grade inflammation without inducing major changes of the gut microbiome: a randomised cross-over trial. Gut. (2019) 68:83–93. doi: 10.1136/gutjnl-2017-314786
126. Bisanz JE, Upadhyay V, Turnbaugh JA, Ly K, and Turnbaugh PJ. Meta-analysis reveals reproducible gut microbiome alterations in response to a high-fat diet. Cell Host Microbe. (2019) 26:265–72. doi: 10.1016/j.chom.2019.06.013
Keywords: gut–lung axis, pediatric respiratory diseases, microbial metabolites, multi-omics, precision medicine
Citation: Wang Z, Yu J, Liu Y, Gong J, Hu Z and Liu Z (2025) Role of the microbiota–gut–lung axis in the pathogenesis of pulmonary disease in children and novel therapeutic strategies. Front. Immunol. 16:1636876. doi: 10.3389/fimmu.2025.1636876
Received: 28 May 2025; Accepted: 08 September 2025;
Published: 25 September 2025.
Edited by:
Nar Singh Chauhan, Maharshi Dayanand University, IndiaReviewed by:
Angela Klain, University of Campania Luigi Vanvitelli, ItalyAmit Grover, AstraZeneca, United Kingdom
Copyright © 2025 Wang, Yu, Liu, Gong, Hu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zheng Liu, bGl1emhlbmcxNzA3QDE2My5jb20=
 Zhifang Wang1
Zhifang Wang1 Zheng Liu
Zheng Liu