- 1Department of Biochemistry, Bahauddin Zakariya University, Multan, Pakistan
- 2Department of Bioinformatics, Kohsar University Murree, Murree, Pakistan
- 3Department of Microbiology & Molecular Genetics, Bahauddin Zakariya University, Multan, Pakistan
- 4Department of Chemistry, Kohat University of Science & Technology, Kohat, Pakistan
- 5Wellman Center for Photomedicine, Harvard Medical School, Massachusetts General Hospital, Boston, MA, United States
- 6Department of Pharmacognosy, College of Pharmacy, King Saud University Riyadh, Riyadh, Saudi Arabia
- 7Department of Pharmaceutical Chemistry, College of Pharmacy, King Saud University, Riyadh, Saudi Arabia
- 8Department of Infectious Diseases, The Affiliated Hospital of Southwest Medical University, Luzhou, China
A Correction on
Development of a subunit vaccine against the cholangiocarcinoma causing Opisthorchis viverrini: a computational approach
by Shah M, Sitara F, Sarfraz A, Shehroz M, Wara TU, Perveen A, Ullah N, Zaman A, Nishan U, Ahmed S, Ullah R, Ali EA and Ojha SC (2024) Front. Immunol. 15:1281544. doi: 10.3389/fimmu.2024.1281544
In the published article, there was an error in Figure 9 as published. We identified that an incorrect figure was included due to its close resemblance to another dataset, as the MD simulations for both datasets were run on the same system using the same program and parameters. The corrected Figure 9 and its caption “RMSD and RMSF plots for receptors and the vaccine construct complexes (A) RMSD plot of the V3-TLR2 complex (B) RMSF plot of the V3-TLR2 complex (C) RMSD plot of the V3-TLR4 complex (D) RMSF plot of the V3-TLR4 complex”. appear below.
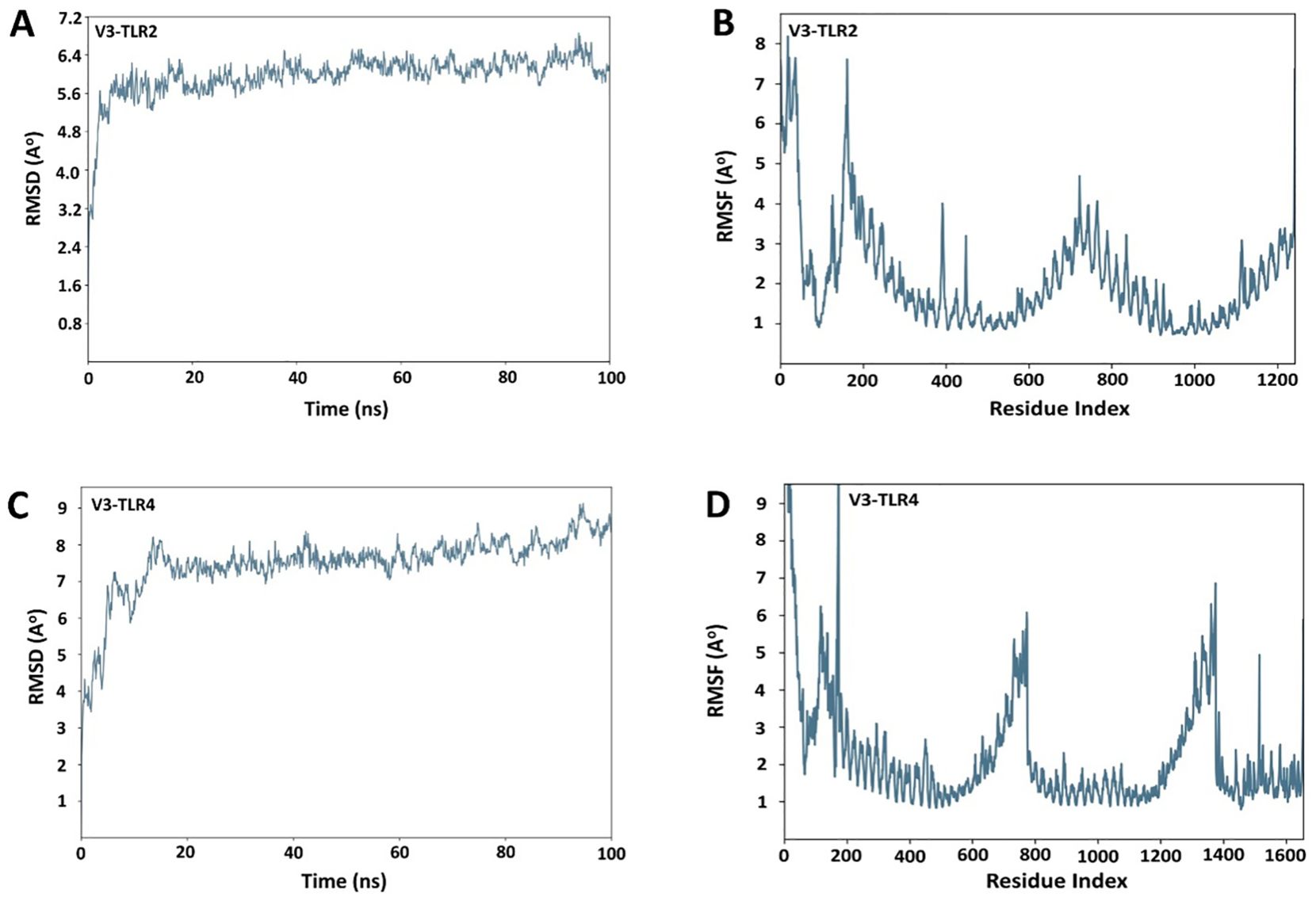
Figure 9. RMSD and RMSF plots for receptors and the vaccine construct complexes (A) RMSD plot of the V3-TLR2 complex (B) RMSF plot of the V3-TLR2 complex (C) RMSD plot of the V3-TLR4 complex (D) RMSF plot of the V3-TLR4 complex.
In the published article, there was an error. Due to the inclusion of an incorrect Figure 9, the corresponding text in the Results section also contains minor errors and typographic mistakes.
A correction has been made to Results, Molecular dynamic simulations, and paragraph 1. This sentence previously stated:
“The RMSD of both the complexes was calculated both in the unbound and bound conformations of the receptors and the vaccine. In Figures 9A, C, the RMSD was displayed as a histogram against the Cα atoms to evaluate the dynamic properties and conformational stability from initial to the final configuration of the protein. Minor departures from the RMSD curve indicate docked complex’s stability, and vice versa. The calculated RMSD of 8.5 Å ± 1 Å indicated no significant variation following convergence over the whole simulation time except for some fluctuation in the beginning between 0 and 100 ns in case of V3-TR2 complex (Figure 9A). In case of V3-TLR4 complex, the RMSD was calculated to be 11 Å ± 1 Å and this indicated some deviations in the initial state and a minor variation was observed between 2500 to 3500ns but later on, no significant deviations were observed (Figure 9B).” The corrected sentence appears below:
“The RMSD of both the complexes was calculated in the bound conformations of the receptors and vaccine. The RMSD profile of the V3-TLR2 complex showed stabilization after an initial equilibration phase, maintaining fluctuations between 5.6 and 6.4 Å throughout the 100 ns simulation (Figure 9A), indicating overall structural stability. RMSF analysis revealed moderate flexibility in V3-TLR2, mainly at the N terminal region (Figure 9B). The V3-TLR4 complex also remained stable, with RMSD values mostly between 7 and 8 Å and reaching ~ 9 Å in the end (Figure 9C), though it exhibited higher fluctuations during initial equilibration phase. While the RMSF analysis of V3-TLR4 showed more pronounced fluctuations at the N- and C-terminal regions (Figure 9D)”.
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: Opisthorchis viverrini, cholangiocarcinoma, epitope, vaccine, parasite; immunoinformatics-based vaccine designing
Citation: Shah M, Sitara F, Sarfraz A, Shehroz M, Wara TU, Perveen A, Ullah N, Zaman A, Nishan U, Ahmed S, Ullah R, Ali EA and Ojha SC (2025) Correction: Development of a subunit vaccine against the cholangiocarcinoma causing Opisthorchis viverrini: a computational approach. Front. Immunol. 16:1638914. doi: 10.3389/fimmu.2025.1638914
Received: 31 May 2025; Accepted: 16 June 2025;
Published: 30 June 2025.
Edited and Reviewed by:
Sajjad Ahmad, Abasyn University, PakistanCopyright © 2025 Shah, Sitara, Sarfraz, Shehroz, Wara, Perveen, Ullah, Zaman, Nishan, Ahmed, Ullah, Ali and Ojha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohibullah Shah, bW9oaWJAYnp1LmVkdS5waw==; bW9oaWJ1c2JAZ21haWwuY29t; Suvash Chandra Ojha, c3V2YXNoX29qaGFAc3dtdS5lZHUuY24=
†These authors have contributed equally to this work
 Mohibullah Shah
Mohibullah Shah Farva Sitara1†
Farva Sitara1†