- 1Department of Gastroenterology, The First Affiliated Hospital of Ningbo University, Ningbo, China
- 2Health Science Center, Ningbo University, Ningbo, China
Tumor immunity has emerged as a focal point for cancer research. Although tumor immunotherapy represents a promising approach to cancer treatment, its effectiveness is often hindered by the heterogeneity of the tumor microenvironment (TME) and immune escape mechanisms. In recent years, small non-coding RNAs (sncRNAs) have attracted increasing attention in tumor immunology due to their essential role in gene regulation. This review systematically examines the multifaceted regulatory functions of sncRNAs in tumor immunity, with a focus on six major subtypes: microRNA, siRNA, piRNA, snoRNA, tsRNA, and snRNA. The molecular mechanisms by which these sncRNAs reshape the TME are discussed, including their roles in modulating immune cell differentiation (e.g., T cell polarization, macrophage phenotype transition), regulating immune checkpoint expression (PD-1/PD-L1, CTLA-4, Tim-3, LAG-3), and influencing tumor antigen presentation. This review also explores the dynamic network through which sncRNAs contribute to tumor immune escape. Furthermore, this study highlights the clinical potential of sncRNAs as liquid biopsy biomarkers and their application prospects in therapeutic strategies, such as targeted silencing of immunosuppressive molecules via nano-delivery systems, combination treatments with radiotherapy and chemotherapy, and Chimeric Antigen Receptor T-cell (CAR-T) therapy. Despite current challenges, including limited delivery efficiency and off-target effects, emerging technologies like AI-assisted sequence design and organ-on-a-chip models present new opportunities for clinical translation. This comprehensive review provides a theoretical foundation and translational insights for elucidating the functional network of sncRNAs in tumor immunology and advancing precise therapeutic interventions.
1 Introduction
The global incidence and mortality rates of tumors are increasing, posing a serious public health challenge that threatens human life and public health (1). Although conventional treatments such as surgery, radiotherapy, and chemotherapy have played essential roles in cancer management, they are often associated with significant limitations, including high recurrence rates and the development of strong drug resistance (2). In recent years, tumor immunotherapy has emerged as a promising alternative, offering renewed hope for cancer treatment (3). Tumor immunity involves a complex interplay between the immune system and tumor cells, encompassing processes such as immune cell recognition, tumor antigen presentation, and immune cell activation (4). The foundation of this process lies in maintaining a dynamic balance between the immune system and tumor cells. However, tumor cells can escape immune detection and destruction through various mechanisms (5), including impaired antigen presentation and the establishment of an immunosuppressive microenvironment, which collectively reduce the efficacy of immunotherapy (6).
Advancements in molecular biology research have highlighted the pivotal role of small non-coding RNAs (sncRNAs) in regulating gene expression within the context of tumor immunity. sncRNAs are short RNA molecules, typically ranging from 18 to 200 nucleotides (nt) in length, that do not encode proteins but exert important biological functions. Major subclasses include microRNA, siRNA, piRNA, snoRNA, tsRNA, and snRNA (7–10). Growing evidence indicates that sncRNAs play critical regulatory roles in tumor immunity, influencing the development, differentiation, and function of immune cells, as well as shaping the tumor immune microenvironment (TIME) (11–13). Studies have shown that sncRNAs can facilitate tumor immune escape and tumor progression by suppressing anti-tumor immune responses or promoting the accumulation of immunosuppressive cells within the TIME (14, 15). It has been demonstrated that sncRNAs are closely associated with the occurrence and progression of a variety of tumors. These findings enhance our understanding of the mechanisms underlying tumor immune escape and suggest new avenues for developing immunotherapeutic strategies centered on sncRNAs. Therefore, an in-depth investigation of the regulatory mechanisms and networks involving sncRNAs in tumor immunity is of great theoretical and clinical significance. Such efforts may uncover key molecular insights into immune escape and support the identification of novel targets for cancer immunotherapy (16). In this review, we have included all literature related to sncRNAs and tumor immunity, and elaborated on the regulatory roles of sncRNAs in tumor immunity as well as their potential therapeutic value.
2 Overview of small non-coding RNAs
RNA plays a crucial role in biological processes. As shown in Figure 1, under the action of RNA polymerase, DNA is transcribed into coding RNAs and non-coding RNAs. Non-coding RNAs is further classified into long non-coding RNAs (lncRNAs, >200 nucleotides) and sncRNAs (≤200 nucleotides) according to size (9, 17). sncRNAs include microRNAs (miRNAs), small interfering RNAs (siRNAs), piwi-interacting RNAs (piRNAs), small nucleolar RNAs (snoRNAs), transfer RNA-derived small RNAs (tsRNAs), and small nuclear RNAs (snRNAs) (Figure 1).

Figure 1. Classification of RNA. DNA is transcribed into coding RNA and non-coding RNA under the action of RNA polymerase. Coding RNA is further translated to generate proteins, while non-coding RNA is divided into lncRNA and sncRNA.
microRNAs (miRNAs) are non-coding single-stranded RNA molecules approximately 20–24 nucleotides in length that primarily bind to the 3’ untranslated region (3′ UTR) of target mRNAs. When full complement is achieved, miRNAs mediate mRNA cleavage and degradation (18). In cases of partial complementarity, they inhibit translation, thereby regulating gene expression at the post-transcriptional level (19). miRNAs exert their functions not only within cells but also through microvesicles and exosomes (20). miRNAs are involved in the regulation of various biological processes, including cell proliferation, differentiation, and apoptosis (21, 22). Certain miRNAs exhibit tumor-promoting or tumor-suppressing activity, and both tumor cells and the TME exploit the aberrant expression of these miRNAs to drive self-remodeling through direct or indirect mechanisms (23). miRNAs are closely associated with tumor immunity, where they modulate immune cell function (24), participate in tumor immune escape, and influence the effectiveness of tumor immunotherapy (23).
siRNA are double-stranded RNA molecules consisting of 21–25 nucleotides. Based on their origin, siRNAs can be classified as endogenous, exogenous, or derived from post-transcriptional processing of short hairpin RNAs (shRNAs). Similar to miRNAs, siRNAs mainly act at the post-transcriptional level by guiding sequence-specific gene silencing (25, 26). They exhibit high sequence specificity and form RNA-induced silencing complexes (RISC) with specific enzymes and proteins within the cell (26). These RISC complexes recognize and bind to complementary mRNA sequences, leading to mRNA degradation or translation repression (27). Due to their negative charge and large molecular size, siRNAs are susceptible to degradation by serum endonucleases and displays poor cell membrane permeability. Therefore, efficient intracellular delivery typically requires a carrier system (28). Advances in nanotechnology have facilitated the development of novel delivery platforms capable of transporting siRNA into cells within the TME (29), thereby influencing the interactions between cancer and immune cells (30). For instance, Mitra Ghasemi-Chaleshtari and colleagues employed SPION nanoparticles loaded with siRNAs targeting PD-1 and A2aR, achieving substantial tumor growth inhibition and enhanced anti-tumor immune responses (27). Moreover, siRNA-based therapeutics have advanced into clinical trials. For instance, siRNA BMS-986263, a lipid nanoparticle-formulated siRNA designed to degrade HSP47 mRNA, has demonstrated promising results in clinical trials for treating advanced fibrosis (31). Several other clinical trials are currently underway, including NBF-006 for non-small cell lung cancer, pancreatic cancer (PC), and colorectal cancer (CRC), as well as EphA2 siRNA therapy for advanced or recurrent solid tumors (32).
piRNAs are typically 26–31 nucleotides in length and bind to the PIWI subfamily of Argonaute proteins to form the piRNA-Piwi complex (piRC) (33, 34). Their primary function is to maintain genomic stability in germ cells by suppressing transposon transcription and silencing transposable elements (33, 35). In addition to their role in genome defense, piRNAs indirectly influence tumor immune responses by regulating genes associated with tumor immunity (36, 37). piRNAs can bind to protein-coding mRNA sequences to modulate gene expression (38), and some are involved in antiviral defense mechanisms (39). Immunologically relevant piRNAs exhibit dynamic expression patterns during tumor progression and are associated with the prognosis and infiltration of various immune cells. For example, in vitro experiments have demonstrated that piR-021285 has been implicated in the development of breast cancer (40). piRNAs also serve as potential biomarkers for tumor diagnosis and prognosis, as well as for cancer subtype identification (35, 41). Researchers performed piRNA expression profiling in paired cancer and normal tissues and independently validated the findings in 771 CRC patients across three independent cohorts. They found piR-1245, which is overexpressed in CRC, is a novel oncogene. Its expression correlates strongly with tumor stage and metastasis, indicating its potential as a biomarker for CRC (42). Owing to their high target specificity, certain piRNAs can regulate immune checkpoint molecule expression and modulate anti-tumor immunity, offering promising avenues for precision cancer therapy (35, 43). piRNAs exert oncogenic or tumor-suppressive functions in cancer progression by regulating tumor cell proliferation, metastasis, drug resistance, and stemness, while their aberrant expression patterns with PIWI proteins in various malignancies offer novel biomarkers and therapeutic targets for tumor diagnosis and targeted therapy (44).
snoRNAs are small non-coding RNAs ranging from 65 to 300 nucleotides in length. Typically, they are processed from the intronic regions of small nucleolar RNA host genes (SNHGs) (45). Classic snoRNAs are categorized into three major groups: C/D box snoRNAs, H/ACA box snoRNAs, and small Cajal body-specific RNAs (scaRNAs) (33, 46). C/D box snoRNAs guide the 2′-O-methylation of ribosomal RNA (rRNA), whereas H/ACA box snoRNAs participate in pseudouridylation. scaRNAs are mainly involved in the modification of small nuclear RNAs (snRNAs) (33, 47). The principal function of snoRNAs is to regulate rRNA maturation. However, they are also involved in tumorigenesis and tumor progression (33, 46). snoRNAs can act as oncogenes or tumor suppressors and are implicated in the development of resistance to cancer therapies. For instance, SNORD11B enhances colorectal cancer progression by promoting tumor cell proliferation and invasion, inhibiting apoptosis via regulation of 2′-O-methylation. SNORD11B exerts a fine regulatory effect on T cell differentiation at the post transcriptional level by mediating the 2 ‘- O-methylation of let-7a. Its specific mechanism includes: this methylation modification enhances the stability of let-7a, prolongs its half-life by resisting degradation by nucleases to increase intracellular abundance. Simultaneously regulating the binding affinity between let-7a and target mRNAs such as key genes for T cell differentiation, thereby affecting the inhibitory effect on target gene expression. In addition, targeted regulation of cell cycle and differentiation related genes (such as IL-6, STAT3, etc.) by let-7a can indirectly affect related signaling pathways and regulate T cell fate determination (such as Th1/Th2 balance or Treg cell differentiation). Furthermore, methylated let-7a may compete with other non-methylated miRNAs for binding to RISC complexes, altering the global miRNA regulatory network and further affecting T cell differentiation. The specific target genes and pathways involved in the above mechanism still need further experimental verification (48). Moreover, snoRNAs have shown potential as biomarkers and therapeutic targets. For example, Researchers have conducted in vivo and in vitro experiments to demonstrate that small nuclear RNA host gene (SNHG5) may be a new target for treating gastric cancer. SNHG5, the host gene of snoRNA U50, inhibits gastric cancer progression by sequestering metastasis-associated protein 2 (MTA2) in the cytoplasm, indicating its therapeutic value in gastric cancer (49). snoRNAs also influence the TME by regulating the secretion of cytokines and chemokines, which affects immune cell recruitment and function. For example, SNORA38B promotes IL-10 secretion in tumor cells, leading to the recruitment of CD4+FOXP3+regulatory T cells and reduced infiltration of CD3+CD8+T cells in the TME of non-small cell lung cancer (NSCLC), thereby promoting tumor progression. Targeting SNORA38B using locked nucleic acids (LNAs) effectively suppresses NSCLC development and enhances sensitivity to immune checkpoint blockade therapy, making SNORA38B a potential therapeutic target for NSCLC (50).
tsRNAs are a class of non-coding small RNAs generated from the cleavage of tRNA molecules. They are generally categorized into two subtypes based on their cleavage sites: tRNA-derived stress-induced RNAs (tiRNAs) and tRNA-derived fragments (tRFs) (51–53). tiRNAs (28–36 nucleotides) are stress-induced fragments produced by specific cleavage at the anticodon loop of mature tRNAs, while tRFs (14–30 nucleotides) represent shorter fragments derived from either mature or primary tRNAs that can be generated under both normal cellular conditions and stress states, independent of specific physiological stimuli (51). tsRNAs participate in various biological processes, primarily through RNA interference and regulation of transcription and translation (53, 54). tsRNAs are closely linked to tumor immunity and are present in various bodily fluids—including blood, serum, urine, cerebrospinal fluid, and saliva—either freely or encapsulated in extracellular vesicles (10). This distribution allows tsRNAs to significantly influence intercellular communication within the TME. Aberrant expression of tsRNAs has been observed in numerous cancers (52, 53, 55–58). Their dysregulation is associated with tumor staging, lymph node metastasis, and poor clinical prognosis, indicating their potential as novel biomarkers for liquid biopsy (52, 53, 59, 60). For instance, a recent in vitro study has shown that tsRNA-49-73-GLU- CTC is highly expressed in the serum of patients with NSCLC. Inhibition of this tsRNA significantly reduces tumor cell proliferation and migration (61). Moreover, tsRNAs have emerged as promising targets for tumor therapy and drug-resistance treatment. Notably, tsRNA-Thr-5–0015 is markedly upregulated in the serum of patients with hepatocellular carcinoma (HCC), and its expression is closely correlated with TNM staging and lymph node metastasis. This tsRNA exhibits high stability and diagnostic precision, and its levels significantly decrease following surgical resection, establishing it as a potential biomarker for both hepatocellular carcinoma diagnosis and treatment (62). In CRC, tsRNA-GlyGCC is significantly overexpressed and is linked to enhanced cancer cell proliferation and resistance to 5-fluorouracil (5-FU) (54, 63). In vitro experiments have found that the tsRNA-GlyGCC promotes colorectal cancer progression and 5-FU resistance by targeting SPIB to regulate the JAK1/STAT6 signaling pathway (54). Inhibiting tsRNA-GlyGCC has been shown to increase the sensitivity of cancer cells to 5-FU, suggesting its utility as a combinatorial therapeutic target to improve treatment efficacy (54, 63).
snRNAs are typically 100–300 nucleotides in length and include several key types, such as U1, U2, U4, U5, and U6. These snRNAs associate with specific proteins to form small nuclear ribonucleoproteins (snRNPs), which together constitute the core components of spliceosomes (64). The primary function of snRNAs is to regulate the precise splicing of precursor mRNAs through spliceosome assembly, ensuring the accurate removal of introns and contributing to the diversity of gene expression (65). For example, U1snRNA identifies the 5’ GU sequence at intron sites and initiates splicing reactions via interaction with branch point sequences (66). Abnormalities in snRNA are closely linked to tumor immunity. For instance, mutations in U2 snRNA-related components, such as the SF3B1 gene, can lead to splicing errors that promote the progression of myelodysplastic syndromes to acute leukemia (67). Furthermore, certain snRNAs, such as U6, can function as innate immune sensors by activating the RIG-I-like receptor (RLR) signaling pathway, thereby inducing type I interferon secretion, enhancing CD8+T cell responses, and promoting anti-tumor immunity (68). Several studies have reported that U6 snRNA is upregulated in the serum and breast tissue of breast cancer patients (69), highlighting the association between snRNA dysregulation and tumor initiation and progression.
In summary, it is evident that different types of sncRNAs exhibit significant differences in terms of size, structural characteristics, functions and mechanisms, as shown in Table 1. However, recent studies have found that their abnormal expression is closely related to human tumors such as gastric cancer, colorectal cancer, lung cancer, liver cancer, pancreatic cancer, breast cancer, acute leukemia, and glioma, revealing the potential role of sncRNAs in the occurrence and development of different cancers (Figure 2).
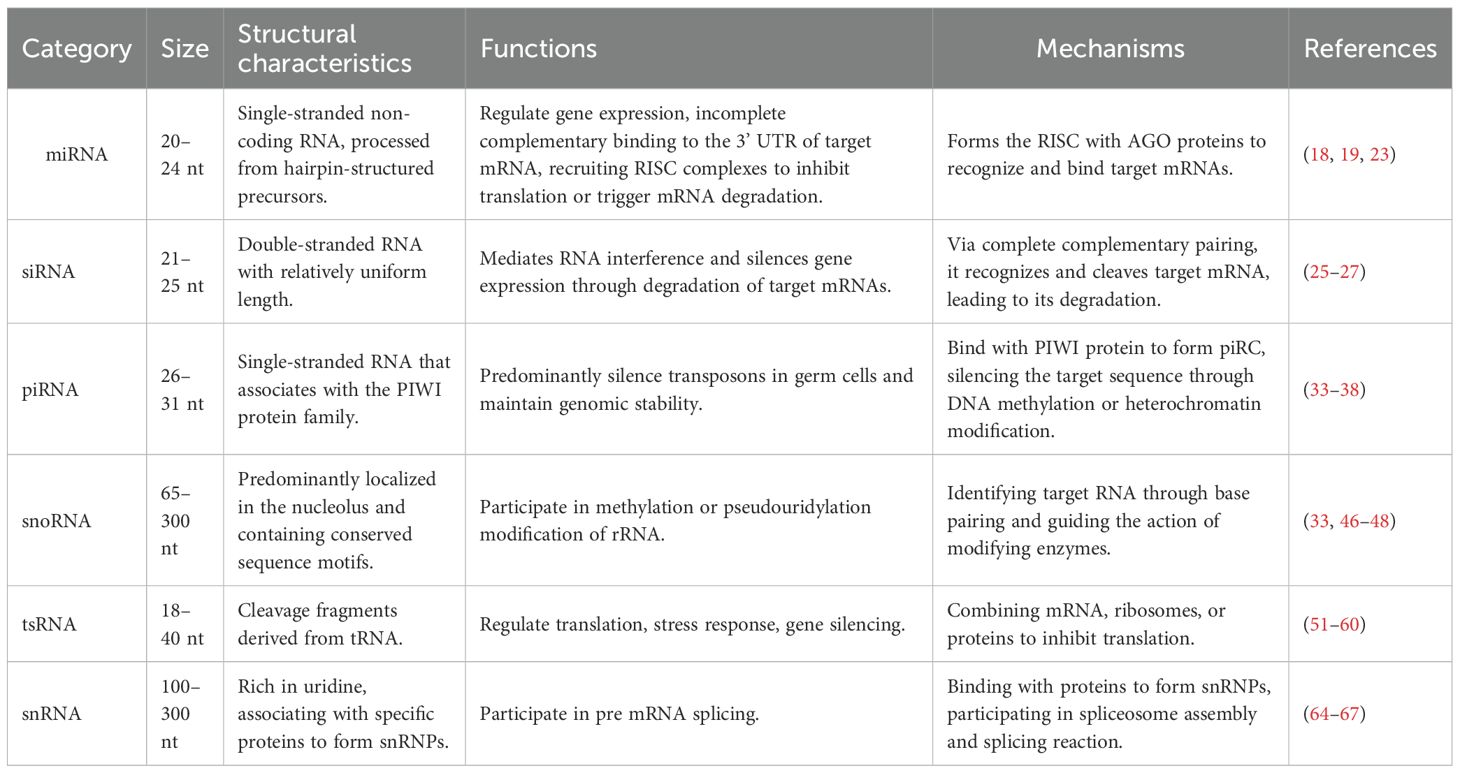
Table 1. The size, structural characteristics, functions and mechanisms of different types of small non-coding RNAs.
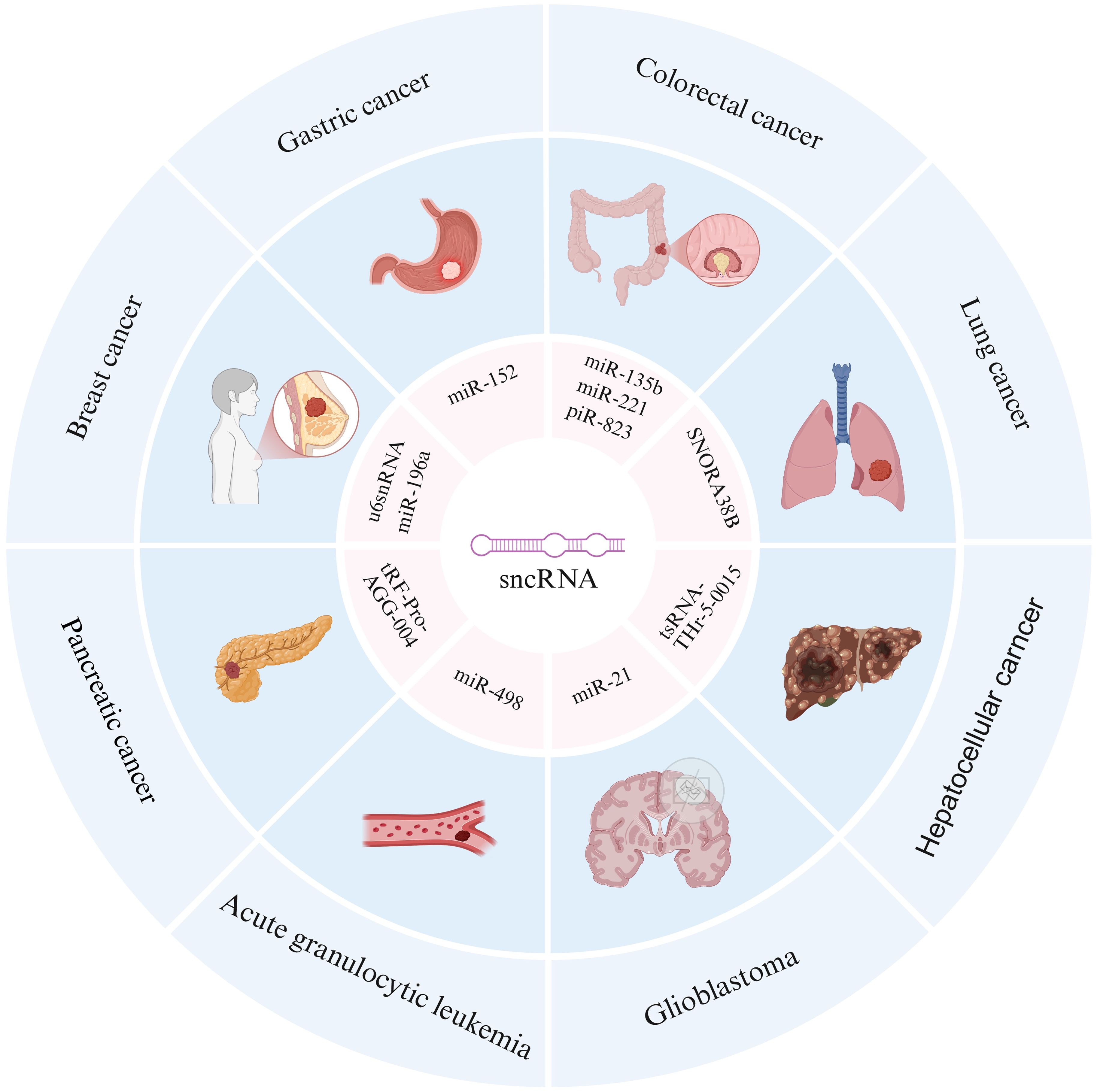
Figure 2. Association between sncRNAs and multiple cancers. The outer ring includes different types of cancers, including gastric cancer, colorectal cancer, lung cancer, hepatocellular cancer, glioblastoma, acute granulocytic leukemia, pancreatic cancer, and breast cancer. The inner loop annotates specific sncRNA molecules associated with these cancers, revealing the potential role of sncRNAs in the occurrence and development of different cancers.
3 Regulation of tumor immune cells and tumor immune microenvironment by sncRNA
3.1 Regulation of immune cell development, differentiation, and function
sncRNAs exert critical regulatory roles in the development, differentiation, and functional modulation of diverse immune cell populations involved in tumor immunity, including B cells, T cells, tumor-associated macrophages (TAMs), and natural killer (NK) cells (70–74). Several miRNAs influence T cell differentiation, particularly by promoting Th1 cell differentiation, thereby enhancing anti-tumor immune responses. For instance, miR-155 is expressed in multiple immune cell types and plays a pivotal role in polarizing primitive CD4+T cells into Treg, Th17, Th1, and Th2 cells (70, 72, 73). While miR-155 does participate in polarization, it does not “convert” CD4+ T cells equally toward all these subpopulations. Its role is better established in promoting Th1 and Th17 subsets, whereas its influence on Treg and Th2 is more ambiguous (74). Additionally, miR-155 regulates CD8+T cell activity and contributes to the differentiation and functional maturation of B cells (70). miR-155 also influences macrophage polarization by promoting the M1 phenotype while inhibiting the polarization and activation of M2 macrophages (73). This shift alters the inflammatory state of the TIME and reduces immune suppression (Figure 3A). Furthermore, miRNAs modulate NK cell development, proliferation, and activation by targeting key receptors and transcriptional regulators (75). Exosomal miRNAs are also involved in immune regulation. For example, in nasopharyngeal carcinoma, exosomal miR-24-3p suppresses T cell proliferation and differentiation by targeting fibroblast growth factor 11 (FGF11), thereby facilitating tumor immune escape and contributing to tumor pathogenesis (20). Relevant in vivo and in vitro experimental studies have suggested that snoRNAs may play an emerging role in regulating macrophage function through vesicle-mediated intercellular communication and epigenetic modifications (45, 76, 77). They may also influence macrophage polarization and cytokine production, thereby contributing to tumor development (45, 78).

Figure 3. Four main ways for sncRNA to regulate tumor immunity. (A) Part A shows the effects of sncRNAs (such as miR-155, SNORA38B) on immune cells, which can promote M1 macrophage activation, inhibit M2 macrophage activation, and induce Th1 cells to secret cytokines such as INFγ and TNFα, promoting Treg cell. (B) Part B focuses on the regulation of tumor immune checkpoints by sncRNAs, such as certain miRNAs that can affect T cell activity by regulating PD-1 and CTLA-4 on the surface of cytotoxic T cells, thereby promoting tumor cell proliferation. (C) Part C indicates that sncRNA regulates the tumor immune microenvironment, such as the reduction and targeted inhibition of miR-21 expression in TAMs, which can lead to a decrease in tumor angiogenesis and promote cancer cell apoptosis. (D) Part D explains a mechanism of tumor immune escape, in which miRNA can regulate the expression of MHC-I on the surface of tumor cells and reduce their antigen presentation ability, thereby inhibiting Treg recognition of tumor cells and suppressing the activity of T cells and NK cells.
3.2 The impact on the tumor microenvironment
The TME is a complex ecosystem composed of tumor cells, immune cells, stromal cells, lymphatic and blood vessels, and the extracellular matrix (79). Its immunosuppressive or immunostimulatory status directly affects tumor progression and therapeutic response (4, 79, 80). sncRNAs have emerged as key regulatory molecules within the TME by modulating the interactions between tumor and immune cells (81). Relevant animal experiments have shown that miR-21 in tumor-associated macrophages (TAMs) plays a regulatory role in immune responses. Targeted inhibition or downregulation of miR-21 in TAMs reduces tumor angiogenesis, enhances anti-tumor immunity, and promotes tumor cell death (Figure 3C) (82). Furthermore, sncRNAs contained in tumor-derived exosomes can be internalized by immune cells, altering their functional states and facilitating immune escape. For instance, in acute myeloid leukemia (AML), tumor cell-derived exosomes carrying miR-19a-3p are taken up by CD8+ T cells, leading to the suppression of effector molecule expression, T cell exhaustion, and subsequent immune escape (83). In addition, sncRNAs regulate the secretion of cytokines and chemokines by immune cells, thereby influencing their recruitment and infiltration and shaping the immunosuppressive or immunostimulatory state of the TME. miR-155 is essential for the expression of the IL-17F gene in Th17 effector cells. Its inhibition results in functional impairments of these cells. Given the role of Th17 cells and their cytokines in tumor immune regulation, such dysfunction may alter the inflammatory state and increase immunosuppression within the TME (84). Furthermore, as previously discussed, SNORA38B promotes the secretion of IL-10 by tumor cells, which recruit CD4+FOXP3+ regulatory T cells and reduce CD3+CD8+T cell infiltration in the TME of non-small-cell lung cancer. This activity facilitates the establishment of an immunosuppressive microenvironment, promoting tumor progression and weakening anti-tumor immunity (50).
4 Regulation of tumor immune checkpoints by sncRNAs and their involvement in tumor immune escape
4.1 Regulation of tumor immune checkpoint by sncRNAs
sncRNAs can enhance anti-tumor immunity by modulating the expression of immune checkpoint molecules. Immune checkpoints include a group of inhibitory receptors and ligands such as programmed death receptor 1 (PD-1) and its ligand PD-L1 (Figure 4B), cytotoxic T lymphocyte-associated antigen 4 (CTLA-4), T cell immunoglobulin and mucin domain-containing protein 3 (TIM-3), lymphocyte activation gene 3 (LAG-3), and T cell immunoglobulin and ITIM domain (TIGIT) (3, 85). When CTLA-4 binds to the receptors CD80 and CD86, it competes with CD28 for these ligands, thereby suppressing T cell activation (Figure 4A) (86). Immune checkpoint molecules serve as key facilitators of tumor immune escape, allowing tumor cells to evade immune detection and destruction. sncRNAs regulate the expression of checkpoint proteins, such as PD-1, PD-L1, and CTLA-4, thereby influencing T cell activity and immune response (Figure 3B). Ji Eun Won et al. developed a siRNA-based immune checkpoint silencing platform, the PLGA (PD-L1 siRNA+PD-1 siRNA) nanoparticle (NP) system, which was designed to inhibit PD-L1 expression in tumor cells. This strategy effectively restores CD8+T cell function and enhances anti-tumor immunity by suppressing immune checkpoint signaling in the TME (87). Further studies have shown that exosomes loaded with PD-L1 and CTLA-4 siRNAs significantly inhibit the proliferation of CRC cells, enhance apoptosis, and reduce immune escape capabilities (88). In addition to siRNAs, miRNAs regulate immune checkpoint expression. miRNAs directly target CTLA-4, thereby reducing its expression. For instance, miR-487a-3p directly targets CTLA-4, leading to reduced expression, whereas other miRNAs, such as miR-24 and miR-210, indirectly modulate CTLA-4 transcription by downregulating FOXP3, a key transcription factor involved in CTLA-4 expression (89–91).
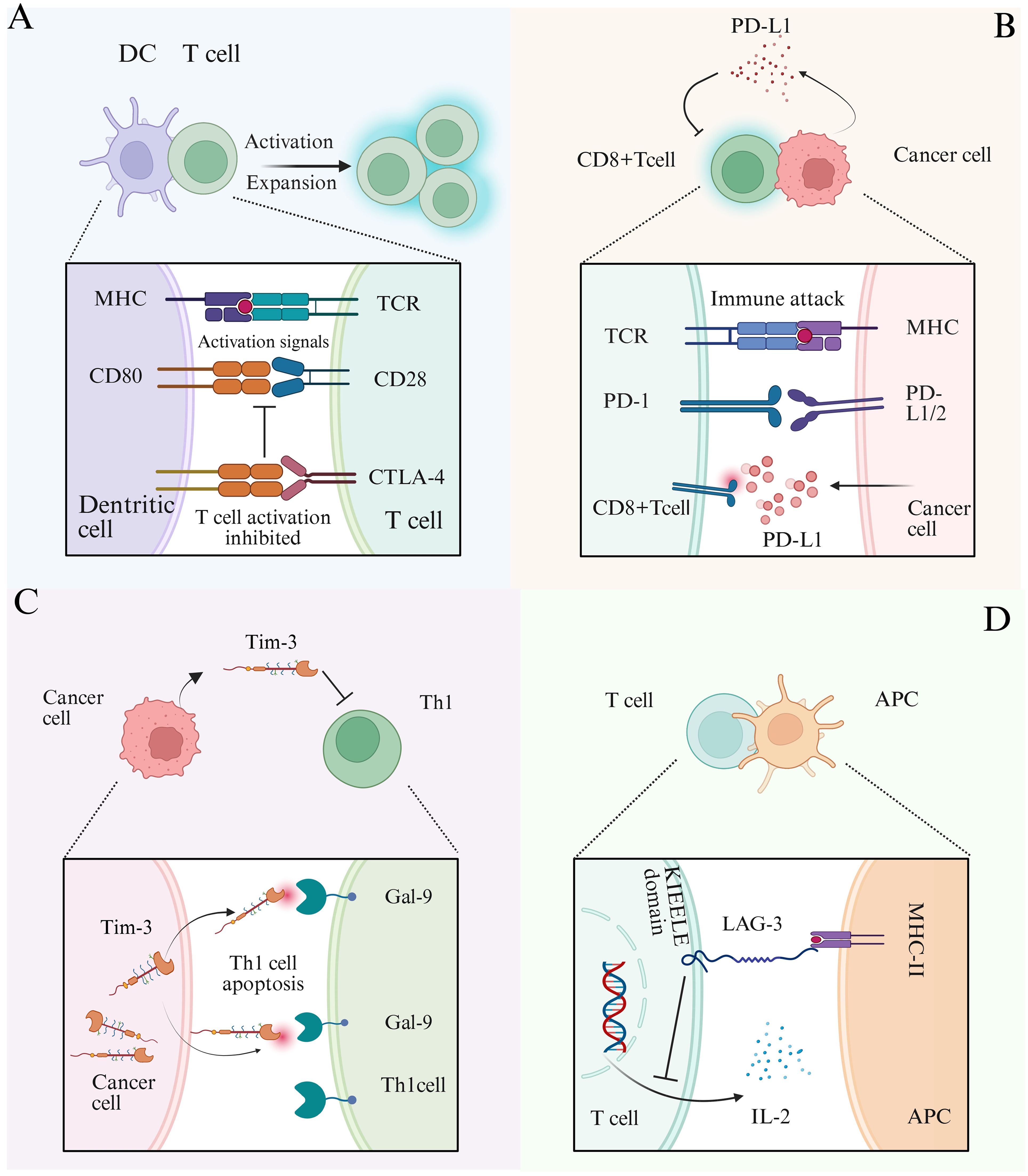
Figure 4. Schematic diagram of tumor immune checkpoint mechanism. (A) DCs interact with T cells, and DCs transmit activation signals to T cells through MHC and CD80 co-stimulatory molecules. When CTLA-4 binds to CD80 receptors, it competitively inhibits the binding of CD28 and CD80, thereby suppressing T cell activation. (B) CD8+T cells interact with cancer cells, and the T cell receptor (TCR) recognizes antigens presented by the MHC of cancer cells to initiate immune attacks. PD-1 on the surface of T cells binds to the receptor PD-L1/L2 on the surface of cancer cells, inhibiting immune attacks. PD-L1 secreted by tumor cells into the tumor microenvironment can reduce the activation of CD8+T cells and suppress immune responses. (C) When Tim-3 expressed by cancer cells binds to the ligand Gal-9 on the surface of Th1 cells, it can induce Th1 cell apoptosis and weaken their killing ability. (D) Interaction between T cells and antigen-presenting cells (APCs) involves the binding of the transmembrane protein LAG-3 (lymphocyte-activation gene 3) on T cells to MHC class II molecules on APCs. This interaction transmits an inhibitory signal via the intracellular KIEELE domain of LAG-3, suppressing the secretion of IL-2 and the function of T cells.
Tim-3 and its receptor, galectin-9 (Gal-9), have emerged as novel targets in recent studies on tumor immune checkpoints (92). Tim-3, an inhibitory co-stimulatory molecule, is predominantly expressed in various immune cells and leukemia stem cells (93). Upon binding to Gal-9, Tim-3 induces apoptosis in Th1 cells, suppresses Th1 cell function (94), and contributes to immune tolerance and evasion (Figure 4C) (94, 95). This interaction (Tim-3/Gal-9) also triggers time-dependent metabolic changes in AML, influencing glucose and lipid metabolism to promote tumor progression. Consequently, Tim-3 has been recognized as a potential therapeutic target (93). Notably, cell function experiments have demonstrated that miR-498 effectively inhibits TIM-3 expression in AML cell lines, this miRNA thereby suppresses tumor cell proliferation (96). Lymphocyte activation gene 3 (LAG-3) is a type I transmembrane protein primarily expressed on the surface of T cells. Upon binding to MHC class II (MHC II) molecules, LAG-3 transmits inhibitory signals through its intracellular KIEELE domain, which blocks interleukin-2 (IL-2) secretion and suppresses T cell proliferation. The survival and immunosuppressive functions of regulatory T cells (Tregs) rely heavily on exogenous IL-2, thus, reduced IL-2 secretion leads to Treg depletion (Figure 4D). Furthermore, the inhibition of IL-2 limits the expansion of activated CD4+ and CD8+ T cells, thereby diminishing the adaptive immune response (97). miR-146 targets LAG-3 mRNA and downregulates its expression, alleviating T cell inhibition and enhancing anti-tumor immunity (98).
4.2 Participation of sncRNAs in tumor immune escape
Beyond regulating immune checkpoints, sncRNAs contribute to tumor immune escape through multiple mechanisms, including the modulation of tumor antigen presentation, reshaping of the tumor microenvironment (4), and regulation of immune checkpoint molecule expression (Figure 3D). Immunoregulatory miRNAs, such as miR-9, can alter the expression of major histocompatibility complex (MHC) molecules on the surface of tumor cells, thereby reducing their antigen-presenting capacity and enabling evasion from T cell recognition (99). Tumor-derived miRNAs may be encapsulated in exosomes or microvesicles and transferred to tumor-infiltrating lymphocytes, promoting an immunosuppressive TME that facilitates immune escape (100). For instance, piR-has-30937, derived from the extracellular vesicles of pancreatic neuroendocrine tumors, enhances CD276 expression in macrophages via the PTEN/AKT signaling pathway. This upregulation suppresses T cell-mediated anti-tumor responses and promotes tumor immune escape (101). sncRNAs also contribute to the creation of an immunosuppressive microenvironment by influencing tumor cells or tumor-associated macrophages to secrete inhibitory cytokines, such as transforming growth factor-β (TGF-β) and interleukin-10 (IL-10). These factors impair immune cell function and support tumor immune escape (102).
Moreover, sncRNAs not only act directly through epigenetic mechanisms but also exert regulation at the post-transcriptional level, which influences epigenetic expression (103). For instance, miR-142-3p, present in exosomes from M1 macrophages, targets high mobility group box 1 (HMGB1) and influences tumor immune escape in glioblastoma by modulating the PD-1/PD-L1 checkpoint (104). In CRC, miR-21 suppresses genes that negatively regulate PD-L1 expression, including PTEN, and activates the PI3K/AKT signaling pathway. Furthermore, miR-21 promotes PD-L1 expression by targeting and suppressing PDCD4 (programmed cell death protein 4), thereby relieving its inhibitory effect on the PI3K/AKT pathway (105). This leads to enhanced membrane recruitment and phosphorylation-mediated activation of PI3K. The activated PI3K catalyzes the conversion of PIP2 to PIP3, which recruits AKT (protein kinase B) to the plasma membrane through its PH domain, where it undergoes phosphorylation by PDK1 and mTORC2 for full activation. The activated AKT subsequently upregulates PD-L1 transcription and translation by phosphorylating downstream transcription factors (e.g., NF-κB, STAT3) or directly modulating mTORC1 signaling. This cascade leads to elevated levels of PD-L1, which, upon binding to PD-1 receptors on T cells, significantly reduce T cell activation and proliferation, thereby enabling tumor cells to escape immune surveillance (105–107). Notably, certain sncRNAs can regulate PD-L1 expression through shared signaling pathways. For instance, both miR-21 and piR-823 enhance PI3K/AKT/mTOR signaling activity by suppressing PTEN expression, thereby promoting PD-L1 upregulation and promoting tumor immune escape (108). Additionally, miR-34, miR-197, and miR-200 collectively target the 3’ UTR of PD-L1 mRNA to enforce post-transcriptional silencing, thereby suppressing PD-L1 protein synthesis (Figure 5) (109). This downregulation ultimately inhibits PD-L1-mediated immune escape. These intertwined regulatory networks illustrate the complexity of sncRNAs-mediated immune escape, suggesting that dissecting their crosstalk with PD-L1 and signaling pathways could unveil novel therapeutic vulnerabilities.
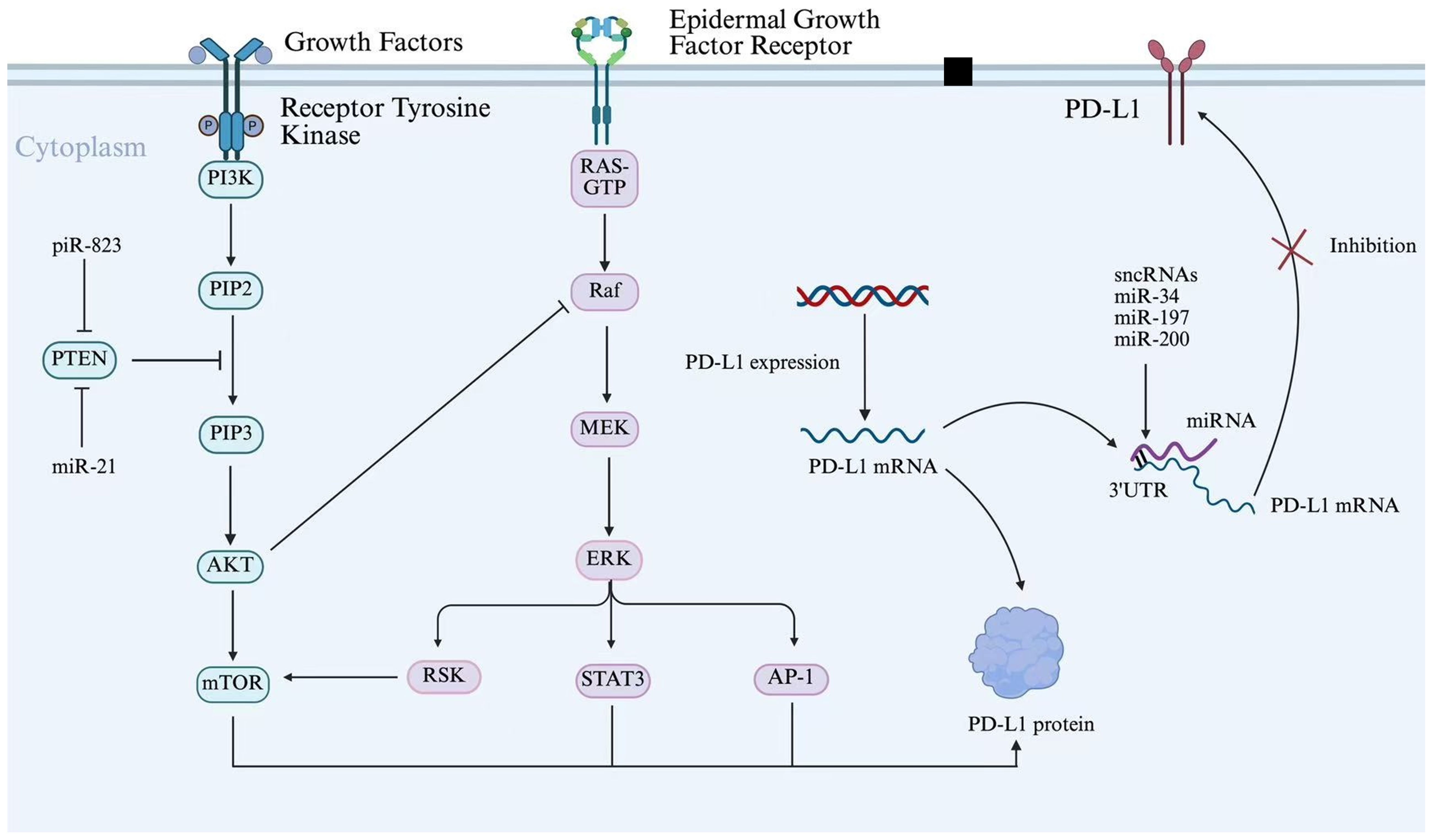
Figure 5. sncRNAs regulate PD-L1 expression through the shared PI3K/AKT signaling pathway. Schematic diagram illustrating the signaling pathways regulating PD-L1 expression and function. Growth factors activate Receptor Tyrosine Kinase, initiating the PI3K/AKT/mTOR and RAS/Raf/MEK/ERK cascades. These pathways converge on transcription factors (STAT3, AP-1) to drive PD-L1 transcription. Additionally, post-transcriptional regulation is depicted, with sncRNAs (miR-34, miR-197, miR-200) targeting PD-L1 mRNA 3’ UTR.
5 Application prospects of sncRNA in tumor immunotherapy
5.1 sncRNA as a tumor biomarker
sncRNAs have emerged as critical targets in tumor biomarker research, owing to their exceptional stability in body fluids and tissue samples, tissue-specific expression patterns, and strong association with various diseases (62, 110–113). These characteristics make sncRNAs highly suitable for non-invasive detection, facilitating early tumor screening. Moreover, their dynamic expression profiles are closely linked to disease progression and therapeutic response, offering substantial value in clinical diagnostics and prognostic evaluations (113, 114). Some sncRNAs are abnormally expressed in colorectal tumors and remain stable in blood and feces. Their expression levels can be quantified using methods such as quantitative PCR and high-throughput sequencing (Figure 6A) (114). For instance, elevated levels of miR-196a have been detected in drug-resistant estrogen receptor (ER)-positive breast cancer, while high miR-10b expression has been observed in ER-negative breast cancer, indicating their potential as subtype-specific biomarkers (113). Multiple studies have demonstrated that distinct miRNA expression profiles can be consistently detected in CRC tissues and fecal samples. Wu et al. conducted a microarray analysis of fecal miRNAs and found that miR-135b was significantly upregulated in CRC and advanced adenomas. Notably, its expression decreased significantly in postoperative fecal samples, highlighting its potential as a non-invasive biomarker for the clinical diagnosis and monitoring of CRC and advanced adenomas (115). Additionally, miR-221 has been identified as a potential biomarker for CRC diagnosis. Notably, miR-221 levels in feces exhibit higher specificity than those in plasma and are not influenced by antibiotic use (116). In addition to miRNAs, other sncRNA subtypes have also shown promise in CRC biomarker research. For example, in a clinical observational study analyzing serum and tissue samples from patients with CRC, piR-823 has been found to be upregulated in the serum and tissues of patients with CRC, and its expression levels are positively correlated with clinical staging. Higher expression levels are associated with more advanced stages, supporting that piR-823 could serve as a potential diagnostic biomarker for CRC, while its correlation with clinical staging preliminarily suggests potential prognostic value (117). Furthermore, tsRNAs have also demonstrated utility as biomarkers in various cancers. For instance, researchers demonstrated that serum tRF-Pro-AGG-004 and tRF-Leu-CAG-002 can serve as novel and promising biomarkers for PC diagnosis by integrating multi-omics technologies with clinical samples, cellular models and animal models. Subsequent randomized clinical trials are required to evaluate the potential application of the two-tsRNAs signature in the early diagnosis and prognosis of PC (118). In addition, the expression levels of tRNA-ValTAC-3, tRNA-GlyTCC-5, tRNA-ValAAC-5 and tRNA-GluCTC-5 are significantly increased in the plasma exosomes of patients with HCC, indicating that tsRNA in plasma exosomes may serve as potential biomarkers for tumor diagnosis (114).
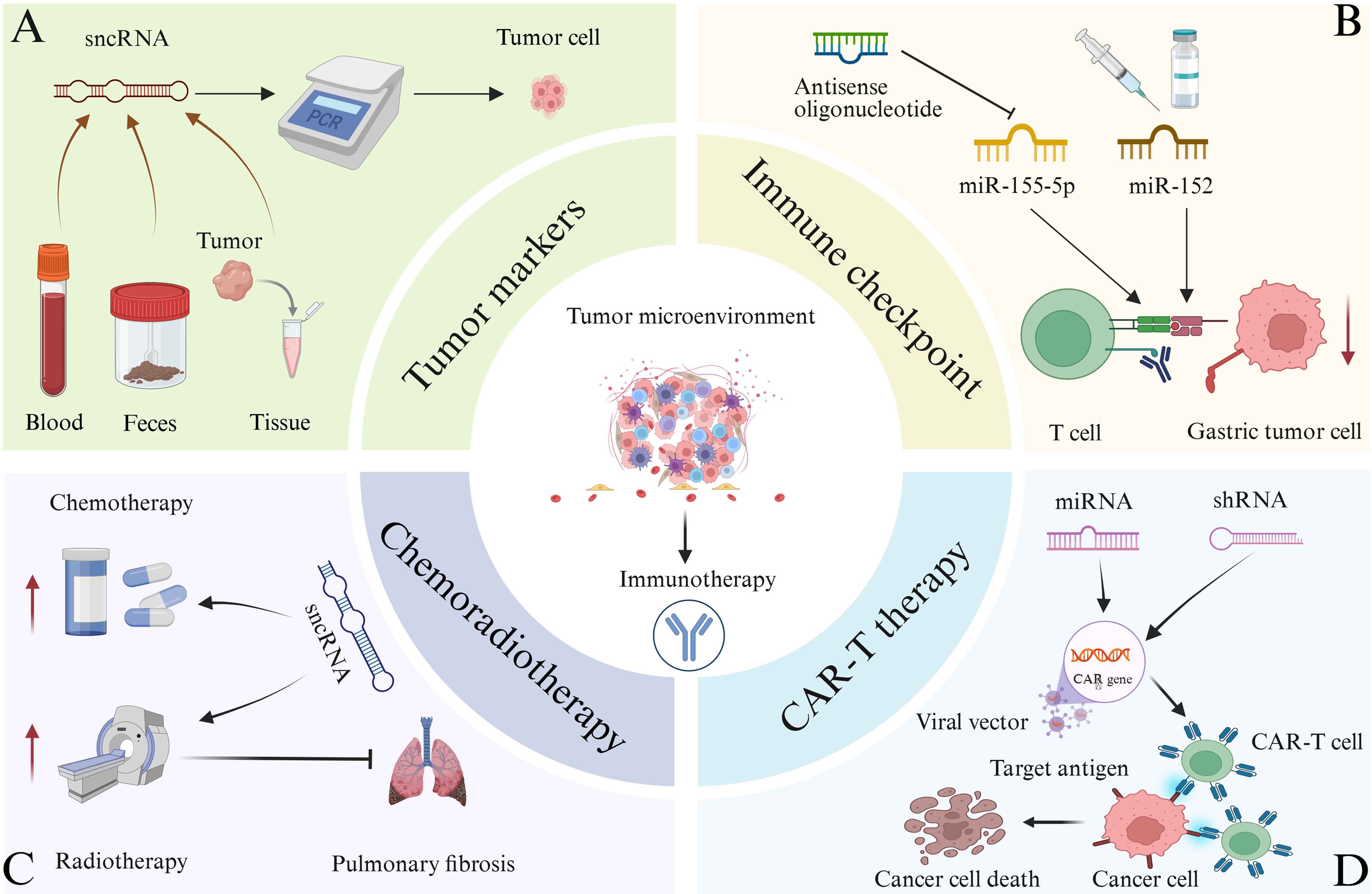
Figure 6. The relevant applications of sncRNA in tumor immunotherapy are illustrated. (A) Quantitative PCR and other techniques can be used to detect significant abnormal expression of certain sncRNAs in the blood, feces, and tumor tissues of certain cancer patients. Therefore, sncRNAs can function as tumor biomarkers and assist in tumor diagnosis. (B) Targeting immune checkpoints for treatment. The function of antisense oligonucleotides in inhibiting miR-155-5p, leading to a decrease in abnormal expression of immune checkpoint molecules, enhancing T cell function, and facilitating the clearance of gastric cancer cells. miR-152 can directly bind to the 3’ end translation region of B7-H1 in gastric cancer cells, thereby inhibiting its expression. Therefore, relevant drugs targeting sncRNA and tumor immune checkpoints can be developed for immunotherapy of tumors. (C) Combination of sncRNA with radiotherapy and chemotherapy for the treatment of tumors. In addition, certain sncRNAs combined with radiotherapy can alleviate lung injury and pulmonary fibrosis caused by radiotherapy alone. (D) Combination of sncRNA and CAR-T therapy for tumor treatment. CAR-T cells are generated by introducing CAR genes into T cells through viral vectors, targeting cancer cell antigens, and inducing cancer cell death. Introducing miRNA or shRNA into CAR-T cells may enhance the anti-tumor efficacy of CAR-T cell therapy.
Similarly, sncRNAs can also serve as biomarkers for non-solid tumors. Studies have comprehensively characterized the landscape of circulating sncRNAs in serum and bone marrow supernatants from patients with AML using high-throughput sequencing. The combination of tsRNA signatures (e.g., tsRNAAla-TGC, tsRNAGly-CCC) screened by machine learning models exhibited superior diagnostic efficacy (AUC > 0.95) compared with miRNAs in distinguishing AML patients from healthy individuals, and their expression levels were associated with AML risk stratification. Furthermore, sncRNAs profile derived from peripheral blood and bone marrow showed a high degree of concordance, suggesting that serum tsRNAs can serve as non-invasive biomarkers to replace bone marrow tests, thus providing a novel strategy for the early diagnosis and prognostic evaluation of AML (119).
5.2 Targeting immune checkpoint pathways
Immune checkpoints play a critical role in tumor immune escape, and their blockade has become a key strategy in tumor immunotherapy (84, 120). Several immune checkpoint inhibitors (ICs) have been approved for clinical use, including the PD-1/PD-L1 inhibitor Nivolumab and Pembrolizumab, the LAG-3 inhibitor Relatlimab and the CTLA-4 inhibitor ipilimumab, all of which are used in the treatment of advanced melanoma (121–124). Tiragolumab, an inhibitor targeting TIGIT, is currently undergoing clinical evaluation (125). Moreover, the combination of relatlimab and nivolumab has demonstrated synergistic anti-tumor activity (126). sncRNAs can modulate immune checkpoint expression through diverse mechanisms, offering novel therapeutic approaches for cancer treatment (85, 89). As previously noted, miR-155 is a key regulator of tumor immune response. Specifically, miR-155-5p can inhibit PD-L1 expression by binding to the 3’ UTR of its mRNA, thereby modulating CD8+T cell function (Figure 6B) (127). The development of therapeutic strategies targeting miR-155 represents a promising approach for tumor immunotherapy. One such strategy involves the use of antisense oligonucleotides to inhibit miR-155 activity, which may lead to a reduction in aberrant immune checkpoint expression, restoration of immune cell function—particularly that of T cells—and enhancement of the host’s anti-tumor immune response (128). A study integrating in vitro, in vivo, and clinical trial investigations demonstrated that the miR-155 oligonucleotide inhibitor Cobomarsen reduced cell proliferation and induced apoptosis in activated B-cell–like diffuse large B-cell lymphoma (ABC-DLBCL) cells, and exerted no toxic effects on the enrolled patient who underwent five cycles of Cobomarsen treatment (129). siRNA can selectively bind to complementary sequences of immune checkpoint mRNAs, thereby silencing the expression of corresponding immune checkpoint genes. In vitro studies have shown that nanocarrier systems significantly enhance the cellular delivery of siRNA, enabling the precise targeting and degradation of mRNAs encoding immune checkpoint molecules such as CTLA-4, PD-1, and PD-L1. This targeted silencing disrupts inhibitory signaling among tumor cells, T cells, macrophages, and dendritic cells, effectively reducing the immunosuppressive TME. As a result, T cell function is reactivated, leading to improved tumor recognition and clearance (130–132). In addition, studies from both in vitro experiments and in vivo trials have shown that 3p-GPC-3 siRNA enhances the activation of CD4+ T cells, CD8+T cells, and natural killer (NK) cells, while reducing the presence of regulatory T cells within the TME. When combined with PD-1 blockade, 3p-GPC-3 siRNA demonstrates a significantly enhanced anti-tumor effect and markedly inhibits the growth of HCC (131). The immune checkpoint molecule B7-H1 functions as a ligand for PD-1, and their interaction suppresses T cell function and immune responses. A strong correlation has been observed between miR-152 and B7-H1 mRNA levels in gastric cancer tissues. miR-152 directly targets the 3′ UTR of B7-H1 mRNA in gastric cancer cells to inhibit its expression, which promotes T cell function and enhances the anti-tumor immune response by blocking the B7-H1/PD-1 signaling pathway, ultimately reducing immune escape (Figure 6B). Notably, reduced expression of miR-152 is associated with increased tumor size and a higher rate of lymph node metastasis, suggesting that miR-152 may serve as a potential prognostic marker and therapeutic target in gastric cancer (133). Furthermore, sncRNAs and small-molecule inhibitors targeting other immune checkpoints, such as LAG-3 and TIM-3, are under active investigation in both preclinical studies and clinical trials, offering promising potential for expanded applications in tumor immunotherapy (125, 134).
5.3 Combination therapy
The integration of sncRNA with conventional treatment modalities, such as chemotherapy, radiotherapy, and CAR-T therapy, has significant therapeutic potential.
5.3.1 SncRNA combined with chemotherapy
Combining sncRNA-based therapy with chemotherapy has demonstrated synergistic effects in enhancing anti-tumor immunity (Figure 6C) (135–138). For example, the use of siRNA targeting PD-L1 in conjunction with chemotherapy can effectively modulate the immunosuppressive TME, thereby amplifying the response to chemotherapy-induced immunogenic cell death. Furthermore, siRNA co-delivered with chemotherapeutic agents can inhibit genes involved in tumor growth and progression, resulting in suppressed tumor proliferation (136, 138–141). Researchers synthesized a combined silica nanoparticle system loaded with siRNA and doxorubicin (DOX), aiming to reverse the drug resistance of DOX-resistant KB-V1 cervical cancer cells. In vitro experiments, they specifically downregulated genes associated with the activation of P-glycoprotein (P-gp) pumps via siRNA, successfully reinducing drug-resistant cancer cells into a sensitive state. This strategy effectively increased the intracellular accumulation concentration of DOX, thereby enhancing the anticancer activity of the drug (139). Moreover, siRNA can be used to target genes associated with chemotherapy resistance, thereby overcoming or reducing drug resistance and enhancing the overall anti-tumor response (142). Researchers investigated the impact of silencing the Bcl-2 gene via small interfering RNA (siRNA, namely siBcl-2) on the efficacy of 5-FU in colorectal cancer. The results showed that the combined application of siBcl-2 liposomes and the 5-FU prodrug S-1 could inhibit tumor growth in the DLD-1 xenograft model, and a dual-modulation strategy for addressing 5-FU-resistant tumors was proposed. In this context, the co-delivery of siRNA and chemotherapeutic agents via nanocarrier systems represents a promising approach to improve therapeutic efficacy and combat drug-resistant cancer cells (143). In addition, miRNAs have emerged as therapeutic targets in osteosarcoma. The targeted delivery of miRNA using nonviral carriers—such as polymers, lipid nanoparticles, exosomes, and inorganic nanoparticles—has shown potential for osteosarcoma treatment. When combined with chemotherapeutic agents, this approach can enhance the sensitivity of osteosarcoma cells to chemotherapy and significantly improve treatment outcomes (144). Furthermore, studies have shown that combining miR-21 inhibitors with chemotherapeutic agents, such as paclitaxel, can effectively inhibit the proliferation of human glioblastoma cells (145).
5.3.2 sncRNA combined with radiotherapy
Radiotherapy induces the release of tumor-associated antigens, and sncRNA can enhance the recognition and response of immune cells to these antigens, thereby synergistically amplifying the anti-tumor effects of radiotherapy (Figure 6C) (146). For instance, miR-101 targets two key DNA repair genes, dependent protein kinase catalytic subunit (DNA PKcs) and ataxia telangiectasia mutated (ATM), by binding to their 3′ UTRs. Upregulation of miR-101 reduces the expression of these proteins, impairing the DNA repair capacity of tumor cells and increasing their sensitivity to radiation-induced damage (147). Similarly, siRNAs can precisely silence genes associated with radioresistance, such as ATM. In preclinical studies using glioma cell lines and xenograft models, inhibiting such genes via siRNA-based strategies has been shown to effectively enhance tumor radiosensitivity, as demonstrated in gliomas (148). In addition to enhancing radiosensitivity, sncRNAs exert protective effects on normal tissues during radiotherapy. Certain sncRNAs have been shown to reduce local inflammatory responses and mitigate or reverse fibrosis by modulating inflammation-related signaling pathways. For example, miR-486-RBD-MSC-Exo, a construct based on miR-486-5p, can inhibit fibrosis in lung epithelial cells and alleviate radiation-induced lung injury and fibrosis (Figure 6C) (149, 150).
5.3.3 sncRNA in combination with CAR-T therapy
sncRNA can optimize the function of CAR-T cells and enhance their cytotoxic activity against tumor cells (Figure 6D). While CAR-T therapy is primarily employed in the treatment of hematological malignancies, it has demonstrated promising potential in targeting solid tumors (151–155). Previous studies have shown that the co-transfection of CAR-encoding mRNA and siRNA can improve CAR-T cell efficacy by downregulating inhibitory receptors such as PD-1 and CTLA-4. In vitro experiments have demonstrated that this strategy may represent a novel approach to augment CAR-T cell-mediated immunotherapy (156). Furthermore, miR-153 has been shown to enhance the therapeutic efficacy of CAR-T cells by suppressing the expression of indoleamine 2, 3-dioxygenase 1 (IDO1) in colon cancer cells (157). More recently, a research team proposed the integration of miRNA or shRNA into retroviral vectors used for CAR expression. Using this approach, they developed anti-CD19 CAR-T cells with upregulated miR-155 expression, which exhibited robust anti-tumor activity both in vitro and in vivo. These findings suggest that the incorporation of miRNA or shRNA into CAR-T cell constructs may further enhance the therapeutic efficacy of CAR-T cell therapy (158).
At present, there are many clinical trials of sncRNAs in tumor immunotherapy. For example, researchers have attempted to apply CART cell therapy to other malignant tumors. In order to prevent allogeneic CART cell graft-versus-host disease (GvHD), researchers chose a shRNA based on miRNA, which targets CD3ζ and effectively downregulates the expression of T cell receptor below the detection level. They generated allogeneic anti-B cell mature antigen CART cells (CYAD-211), which co expressed shRNA based on anti-CD3ζ miRNA in CAR construction in vivo, effectively inhibited TCR mediated signaling in vitro, and effectively inhibited GvHD in vivo. Subsequently, in a phase I clinical trial (NCT04613557), CYAD-211 was evaluated in patients with relapsed or refractory multiple myeloma, confirming its good safety. The clinical trial results showed no signs of GvHD and indicated effective downregulation of TCR (159). In addition, Lna-i-mir-221, an inhibitor of oncogenic miRNA, showed outstanding performance and high safety in the phase I trial of refractory advanced solid tumors (NCT04811898), and 50% of patients achieved disease control. One patient with colorectal cancer achieved partial remission lasting over three years. In addition, pharmacodynamics confirmed that miR-221 down regulated and activated tumor suppressor targets CDKN1B/p27 and PTEN. This study is the first human trial of LNA miRNA inhibitor and has promoted the phase II clinical development of solid tumors (160). In summary, there are still many ongoing clinical trials on the application of sncRNAs in tumor immunotherapy, and sncRNAs based therapies have the potential to change tumor treatment.
6 Research challenges and future prospects
In summary, sncRNAs play an important regulatory role in tumor immunity. Different types of sncRNAs play different roles in the occurrence, development, and immune escape of various tumors. Certain microRNAs can participate in tumor immune escape by influencing tumor cell apoptosis, invasion, metastasis, and the function of immune cells in the TME. siRNAs are capable of targeting specific molecules to modulate immune cell activity and enhance the body′s anti-tumor immune response. Additionally, other sncRNAs including piRNAs, snoRNAs, and tsRNAs each play distinct roles in regulating tumor immunity-related pathways or cellular functions. Small non-coding RNA molecules and their roles in tumor immunity are summarized in Table 2.
Although substantial progress has been achieved in the research of sncRNAs in tumor immunity, including notable advancements in using sncRNAs to improve the delivery of chemotherapeutic agents and counter tumor resistance and proliferation, cancer therapies based on sncRNAs still encounter several crucial challenges (29, 161). One of the foremost limitations of sncRNAs lies in their delivery systems, which significantly hinder clinical translation. Due to their negative charge and susceptibility to nuclease degradation, siRNAs face challenges in crossing the negatively charged cell membranes during systemic delivery. Furthermore, nonspecific uptake by non-target cells may trigger off-target effects or reduce therapeutic efficacy at the diseased site. Therefore, the development of efficient targeted delivery systems capable of transporting siRNA specifically to desired tissues is critical (161, 162). Although existing nanocarrier systems can address some of these issues, an ideal delivery platform should possess multiple essential features: protection of nucleic acid therapeutics, precise spatiotemporal control of drug release, selective targeting of specific cellular subpopulations within the TME, and efficient delivery under complex physiological conditions (62). For instance, in the context of osteosarcoma treatment, significant gaps remain in understanding the pharmacokinetics, metabolic pathways of nanocarriers, and long-term toxicity profiles (29). These knowledge gaps highlight the urgent need to establish a standardized evaluation framework for nanocarrier-based delivery systems (63).
Another major challenge is the limited understanding of the molecular mechanisms underpinning sncRNAs function, which restricts the refinement of therapeutic strategies. Although sncRNAs have been implicated in key processes such as immune checkpoint regulation and tumor-associated macrophage polarization, the dynamic regulatory networks involving sncRNAs within the TME have not yet been fully elucidated. In particular, the mechanisms of cross-talk between sncRNAs and epigenetic modifications, as well as those involving metabolic reprogramming, remain poorly understood. Additionally, synergistic or antagonistic interactions among different subclasses of sncRNAs require further investigation. Addressing these knowledge gaps will necessitate the application of advanced technologies such as single-cell sequencing and spatial transcriptomics.
More importantly, safety concerns remain a major barrier to the clinical application of sncRNA-based therapies. The toxicity mechanism of siRNA mainly includes its inherent immunogenicity, hybridization dependent toxicity (such as off target effect and targeted over silencing) and saturation effect of RNA interference mechanism. In addition, delivery systems (such as lipid nanoparticles and polymer carriers) may also trigger immune activation and chemical toxicity, such as complement activation, cell membrane damage and mitochondrial damage. To meet these challenges, researchers have adopted a variety of strategies: reducing the immunogenicity of siRNA by chemical modification (such as 2’-ome or 2’-f). Optimizing the seed zone design to reduce the Miss effect. Develop ionizable lipid nanoparticles to improve the safety of delivery systems. GalNAc coupling technology was used to achieve efficient liver targeting (163). The saturation of RNA interference mechanism was avoided by dose adjustment. In addition, the application of high-throughput toxicity screening and organ chip technology is helpful to evaluate the safety of siRNA and its vector more accurately. Although five kinds of siRNA drugs have been approved, their clinical transformation still needs to further solve the toxicity problem. In the future, it is necessary to combine artificial intelligence and biotechnology to optimize the design strategy to achieve safer and more efficient siRNA therapy (29, 163). Moreover, certain siRNA sequences may exhibit off-target effects. Notably, researchers have synthesized 2′-formamidonucleoside phosphoramidites as novel sugar-modified analogs for all four nucleobases, which effectively suppress off-target activity while enhancing in vivo stability (164).
As understanding of the roles of sncRNAs in tumor immunity deepens and nanotechnology-based delivery systems continue to evolve (33), the development of more efficient and safer sncRNA-based diagnostic and therapeutic strategies appears increasingly attainable. Artificial intelligence-driven platforms for nucleic acid drug design can enhance sequence specificity, while advances in organ-on-a-chip and digital twin technologies offer innovative tools for accurately evaluating therapeutic efficacy (165, 166). Given the critical roles of sncRNAs in the development and differentiation of immune cells, modulation of the TME, regulation of immune checkpoints, and facilitation of immune escape, as well as their potential as biomarkers, sncRNAs represent promising targets for integration with existing immunotherapies. When combined with approaches such as immune checkpoint inhibitors and CAR-T cell therapy, sncRNA-based interventions may offer novel strategies for early tumor diagnosis and immunotherapy, ultimately expanding treatment options and improving clinical outcomes for cancer patients.
Author contributions
ZL: Investigation, Methodology, Writing – original draft. HD: Investigation, Methodology, Writing – original draft. CY: Methodology, Software, Visualization, Writing – original draft. JY: Investigation, Methodology, Visualization, Writing – original draft. MM: Conceptualization, Funding acquisition, Writing – original draft. YS: Conceptualization, Project administration, Supervision, Validation, Writing – review & editing, Writing – original draft.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This study was supported by grants from the Key Scientific and Technological Projects of Ningbo (No. 2021Z133, 2022Z130), Ningbo Top Medical and Health Research Program (No. 2023020612).
Acknowledgments
Thanks for the technical support by the Core Facilities, Health Science Center, Ningbo University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
2. Zhang X, Xie K, Zhou H, Wu Y, Li C, Liu Y, et al. Role of non-coding RNAs and RNA modifiers in cancer therapy resistance. Mol Cancer. (2020) 19:47. doi: 10.1186/s12943-020-01171-z
3. Rui R, Zhou L, and He S. Cancer immunotherapies: advances and bottlenecks. Front Immunol. (2023) 14:1212476. doi: 10.3389/fimmu.2023.1212476
4. Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. (2018) 24:541–50. doi: 10.1038/s41591-018-0014-x
5. Liu Y and Cao X. Immunosuppressive cells in tumor immune escape and metastasis. J Mol Med (Berl). (2016) 94:509–22. doi: 10.1007/s00109-015-1376-x
6. Wu Q, You L, Nepovimova E, Heger Z, Wu W, Kuca K, et al. Hypoxia-inducible factors: master regulators of hypoxic tumor immune escape. J Hematol Oncol. (2022) 15:77. doi: 10.1186/s13045-022-01292-6
7. Sarraf JS, Puty TC, da Silva EM, Allen TSR, Sarraf YS, de Carvalho LEW, et al. Noncoding RNAs and colorectal cancer: A general overview. Microrna. (2020) 9:336–45. doi: 10.2174/2211536609666201221124608
8. Yan H and Bu P. Non-coding RNA in cancer. Essays Biochem. (2021) 65:625–39. doi: 10.1042/ebc20200032
9. Zhang P, Wu W, Chen Q, and Chen M. Non-coding RNAs and their integrated networks. J Integr Bioinform. (2019) 16:20190027. doi: 10.1515/jib-2019-0027
10. Li X, Peng J, and Yi C. The epitranscriptome of small non-coding RNAs. Noncoding RNA Res. (2021) 6:167–73. doi: 10.1016/j.ncrna.2021.10.002
11. Pesce S, Greppi M, Ferretti E, Obino V, Carlomagno S, Rutigliani M, et al. miRNAs in NK cell-based immune responses and cancer immunotherapy. Front Cell Dev Biol. (2020) 8:119. doi: 10.3389/fcell.2020.00119
12. Mainini F and Eccles MR. Lipid and polymer-based nanoparticle siRNA delivery systems for cancer therapy. Molecules. (2020) 25:2692. doi: 10.3390/molecules25112692
13. Xu Z, Chen Y, Ma L, Chen Y, Liu J, Guo Y, et al. Role of exosomal non-coding RNAs from tumor cells and tumor-associated macrophages in the tumor microenvironment. Mol Ther. (2022) 30:3133–54. doi: 10.1016/j.ymthe.2022.01.046
14. Zou X, Zhao Y, Liang X, Wang H, Zhu Y, and Shao Q. Double insurance for OC: miRNA-mediated platinum resistance and immune escape. Front Immunol. (2021) 12:641937. doi: 10.3389/fimmu.2021.641937
15. Eichmüller SB, Osen W, Mandelboim O, and Seliger B. Immune modulatory microRNAs involved in tumor attack and tumor immune escape. J Natl Cancer Inst. (2017) 109. doi: 10.1093/jnci/djx034
16. Bian J, Shao R, Li J, Zhu JF, Shao AZ, Liu C, et al. Mechanism research of non-coding RNA in immune checkpoint inhibitors therapy. Cancer Sci. (2024) 115:3520–31. doi: 10.1111/cas.16309
17. Mo QL, Qiu YS, Liang YN, Wei QY, Fan XF, and Pang WP. Exploring long noncoding RNAs and ferroptosis in cancer progression. Life Conflux. (2025) 1:e128. doi: 10.71321/jjrzya36
18. Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, and Ghaffari SH. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J Cell Physiol. (2019) 234:5451–65. doi: 10.1002/jcp.27486
19. Iqbal MA, Arora S, Prakasam G, Calin GA, and Syed MA. MicroRNA in lung cancer: role, mechanisms, pathways and therapeutic relevance. Mol Aspects Med. (2019) 70:3–20. doi: 10.1016/j.mam.2018.07.003
20. Ye SB, Zhang H, Cai TT, Liu YN, Ni JJ, He J, et al. Exosomal miR-24-3p impedes T-cell function by targeting FGF11 and serves as a potential prognostic biomarker for nasopharyngeal carcinoma. J Pathol. (2016) 240:329–40. doi: 10.1002/path.4781
21. Li Y, Di C, Li W, Cai W, Tan X, Xu L, et al. Oncomirs miRNA-221/222 and Tumor Suppressors miRNA-199a/195 Are Crucial miRNAs in Liver Cancer: A Systematic Analysis. Dig Dis Sci. (2016) 61:2315–27. doi: 10.1007/s10620-016-4156-8
22. Konishi H, Sato H, Takahashi K, and Fujiya M. Tumor-progressive mechanisms mediating miRNA-protein interaction. Int J Mol Sci. (2021) 22:12303. doi: 10.3390/ijms222212303
23. Zhang Z, Huang Q, Yu L, Zhu D, Li Y, Xue Z, et al. The Role of miRNA in Tumor Immune Escape and miRNA-Based Therapeutic Strategies. Front Immunol. (2021) 12:807895. doi: 10.3389/fimmu.2021.807895
24. Xie J, Du Y, Liu D, Wu J, Yang K, He X, et al. The miR-17 approximately 92 miRNAs promote plasma cell differentiation by suppressing SOCS3-mediated NIK degradation. Cell Rep. (2023) 42:112968. doi: 10.1016/j.celrep.2023.112968
25. Carthew RW and Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. (2009) 136:642–55. doi: 10.1016/j.cell.2009.01.035
26. Binder AK, Bremm F, Dörrie J, and Schaft N. Non-coding RNA in tumor cells and tumor-associated myeloid cells-function and therapeutic potential. Int J Mol Sci. (2024) 25:7275. doi: 10.3390/ijms25137275
27. Qiao M, Zeng C, Liu C, Lei Z, Liu B, and Xie H. The advancement of siRNA-based nanomedicine for tumor therapy. Nanomed (Lond). (2024) 19:1841–62. doi: 10.1080/17435889.2024.2377062
28. Alshaer W, Zureigat H, Al Karaki A, Al-Kadash A, Gharaibeh L, Hatmal MM, et al. siRNA: Mechanism of action, challenges, and therapeutic approaches. Eur J Pharmacol. (2021) 905:174178. doi: 10.1016/j.ejphar.2021.174178
29. Ranjbar S, Zhong XB, Manautou J, and Lu X. A holistic analysis of the intrinsic and delivery-mediated toxicity of siRNA therapeutics. Adv Drug Delivery Rev. (2023) 201:115052. doi: 10.1016/j.addr.2023.115052
30. Friedrich M and Aigner A. Therapeutic siRNA: state-of-the-art and future perspectives. BioDrugs. (2022) 36:549–71. doi: 10.1007/s40259-022-00549-3
31. Lawitz EJ, Shevell DE, Tirucherai GS, Du S, Chen W, Kavita U, et al. BMS-986263 in patients with advanced hepatic fibrosis: 36-week results from a randomized, placebo-controlled phase 2 trial. Hepatology. (2022) 75:912–23. doi: 10.1002/hep.32181
32. Narasipura EA, VanKeulen-Miller R, Ma Y, and Fenton OS. Ongoing clinical trials of nonviral siRNA therapeutics. Bioconjug Chem. (2023) 34:1177–97. doi: 10.1021/acs.bioconjchem.3c00205
33. Wang B, Hu S, Teng Y, Chen J, Wang H, Xu Y, et al. Current advance of nanotechnology in diagnosis and treatment for Malignant tumors. Signal Transduct Target Ther. (2024) 9:200. doi: 10.1038/s41392-024-01889-y
34. Lv X, Zhang H, and Wu L. Advances in PIWI-piRNA function in female reproduction in mammals. Acta Biochim Biophys Sin (Shanghai). (2024) 57:148–56. doi: 10.3724/abbs.2024195
35. Zhou J, Xie H, Liu J, Huang R, Xiang Y, Tian D, et al. PIWI-interacting RNAs: Critical roles and therapeutic targets in cancer. Cancer Lett. (2023) 562:216189. doi: 10.1016/j.canlet.2023.216189
36. Rayford KJ, Cooley A, Rumph JT, Arun A, Rachakonda G, Villalta F, et al. piRNAs as modulators of disease pathogenesis. Int J Mol Sci. (2021) 22:2373. doi: 10.3390/ijms22052373
37. Weng W, Li H, and Goel A. Piwi-interacting RNAs (piRNAs) and cancer: Emerging biological concepts and potential clinical implications. Biochim Biophys Acta Rev Cancer. (2019) 1871:160–9. doi: 10.1016/j.bbcan.2018.12.005
38. Jiang M, Hong X, Gao Y, Kho AT, Tantisira KG, and Li J. piRNA associates with immune diseases. Cell Commun Signal. (2024) 22:347. doi: 10.1186/s12964-024-01724-5
39. O’Donnell KA and Boeke JD. Mighty Piwis defend the germline against genome intruders. Cell. (2007) 129:37–44. doi: 10.1016/j.cell.2007.03.028
40. Fu A, Jacobs DI, Hoffman AE, Zheng T, and Zhu Y. PIWI-interacting RNA 021285 is involved in breast tumorigenesis possibly by remodeling the cancer epigenome. Carcinogenesis. (2015) 36:1094–102. doi: 10.1093/carcin/bgv105
41. Wan D, Li R, Huang H, Zhu X, and Li G. Pan-cancer landscape of immunology PIWI-interacting RNAs. Comput Struct Biotechnol J. (2023) 21:5309–25. doi: 10.1016/j.csbj.2023.10.042
42. Weng W, Liu N, Toiyama Y, Kusunoki M, Nagasaka T, Fujiwara T, et al. Novel evidence for a PIWI-interacting RNA (piRNA) as an oncogenic mediator of disease progression, and a potential prognostic biomarker in colorectal cancer. Mol Cancer. (2018) 17:16. doi: 10.1186/s12943-018-0767-3
43. Han R, Rao X, Zhou H, and Lu L. Synergistic immunoregulation: harnessing circRNAs and piRNAs to amplify PD-1/PD-L1 inhibition therapy. Int J Nanomed. (2024) 19:4803–34. doi: 10.2147/IJN.S461289
44. Deng X, Liao T, Xie J, Kang D, He Y, Sun Y, et al. The burgeoning importance of PIWI-interacting RNAs in cancer progression. Sci China Life Sci. (2024) 67:653–62. doi: 10.1007/s11427-023-2491-7
45. Xiao H, Feng X, Liu M, Gong H, and Zhou X. SnoRNA and lncSNHG: Advances of nucleolar small RNA host gene transcripts in anti-tumor immunity. Front Immunol. (2023) 14:1143980. doi: 10.3389/fimmu.2023.1143980
46. van der Werf J, Chin CV, and Fleming NI. SnoRNA in cancer progression, metastasis and immunotherapy response. Biol (Basel). (2021) 10:809. doi: 10.3390/biology10080809
47. Cai C, Peng Y, Shen E, Wan R, Gao L, Gao Y, et al. Identification of tumour immune infiltration-associated snoRNAs (TIIsno) for predicting prognosis and immune landscape in patients with colon cancer via a TIIsno score model. EBioMedicine. (2022) 76:103866. doi: 10.1016/j.ebiom.2022.103866
48. Bian Z, Xu C, Xie Y, Wang X, Chen Y, Mao S, et al. SNORD11B-mediated 2’-O-methylation of primary let-7a in colorectal carcinogenesis. Oncogene. (2023) 42:3035–46. doi: 10.1038/s41388-023-02808-1
49. Zhao L, Guo H, Zhou B, Feng J, Li Y, Han T, et al. Long non-coding RNA SNHG5 suppresses gastric cancer progression by trapping MTA2 in the cytosol. Oncogene. (2016) 35:5770–80. doi: 10.1038/onc.2016.110
50. Zhuo Y, Li S, Hu W, Zhang Y, Shi Y, Zhang F, et al. Targeting SNORA38B attenuates tumorigenesis and sensitizes immune checkpoint blockade in non-small cell lung cancer by remodeling the tumor microenvironment via regulation of GAB2/AKT/mTOR signaling pathway. J Immunother Cancer. (2022) 10:e004113. doi: 10.1136/jitc-2021-004113
51. Li S, Xu Z, and Sheng J. tRNA-derived small RNA: A novel regulatory small non-coding RNA. Genes (Basel). (2018) 9:246. doi: 10.3390/genes9050246
52. Chen Y, Shao Z, and Wu S. Research progress on the tsRNA biogenesis, function, and application in lung cancer. Noncoding RNA Res. (2025) 10:63–9. doi: 10.1016/j.ncrna.2024.09.004
53. Xie Y, Yao L, Yu X, Ruan Y, Li Z, and Guo J. Action mechanisms and research methods of tRNA-derived small RNAs. Signal Transduct Target Ther. (2020) 5:109. doi: 10.1038/s41392-020-00217-4
54. Xu R, Du A, Deng X, Du W, Zhang K, Li J, et al. tsRNA-GlyGCC promotes colorectal cancer progression and 5-FU resistance by regulating SPIB. J Exp Clin Cancer Res. (2024) 43:230. doi: 10.1186/s13046-024-03132-6
55. Huang B, Yang H, Cheng X, Wang D, Fu S, Shen W, et al. tRF/miR-1280 suppresses stem cell-like cells and metastasis in colorectal cancer. Cancer Res. (2017) 77:3194–206. doi: 10.1158/0008-5472.Can-16-3146
56. Zhou J, Wan F, Wang Y, Long J, and Zhu X. Small RNA sequencing reveals a novel tsRNA-26576 mediating tumorigenesis of breast cancer. Cancer Manag Res. (2019) 11:3945–56. doi: 10.2147/cmar.S199281
57. Lu S, Wei X, Tao L, Dong D, Hu W, Zhang Q, et al. A novel tRNA-derived fragment tRF-3022b modulates cell apoptosis and M2 macrophage polarization via binding to cytokines in colorectal cancer. J Hematol Oncol. (2022) 15:176. doi: 10.1186/s13045-022-01388-z
58. Gan L, Song H, and Ding X. Transfer RNA-derived small RNAs (tsRNAs) in gastric cancer. Front Oncol. (2023) 13:1184615. doi: 10.3389/fonc.2023.1184615
59. Zhou M, He X, Zhang J, Mei C, Zhong B, and Ou C. tRNA-derived small RNAs in human cancers: roles, mechanisms, and clinical application. Mol Cancer. (2024) 23:76. doi: 10.1186/s12943-024-01992-2
60. Lee S, Kim J, Valdmanis PN, and Kim HK. Emerging roles of tRNA-derived small RNAs in cancer biology. Exp Mol Med. (2023) 55:1293–304. doi: 10.1038/s12276-023-01038-5
61. Li C, Zhong S, Chen J, and Mu X. TsRNA-49-73-Glu-CTC: A promising serum biomarker in non-small cell lung cancer. PloS One. (2025) 20:e0320187. doi: 10.1371/journal.pone.0320187
62. Jin K, Wu J, Yang J, Chen B, Xu J, Bao H, et al. Identification of serum tsRNA-Thr-5–0015 and combined with AFP and PIVKA-II as novel biomarkers for hepatocellular carcinoma. Sci Rep. (2024) 14:28834. doi: 10.1038/s41598-024-80592-y
63. Wu Y, Yang X, Jiang G, Zhang H, Ge L, Chen F, et al. 5’-tRF-GlyGCC: a tRNA-derived small RNA as a novel biomarker for colorectal cancer diagnosis. Genome Med. (2021) 13:20. doi: 10.1186/s13073-021-00833-x
64. Sun J, Li X, Hou X, Cao S, Cao W, Zhang Y, et al. Structural basis of human SNAPc recognizing proximal sequence element of snRNA promoter. Nat Commun. (2022) 13:6871. doi: 10.1038/s41467-022-34639-1
65. Mabin JW, Lewis PW, Brow DA, and Dvinge H. Human spliceosomal snRNA sequence variants generate variant spliceosomes. Rna. (2021) 27:1186–203. doi: 10.1261/rna.078768.121
66. Will CL and Lührmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. (2011) 3:a003707. doi: 10.1101/cshperspect.a003707
67. Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. (2011) 478:64–9. doi: 10.1038/nature10496
68. Nabet BY, Qiu Y, Shabason JE, Wu TJ, Yoon T, Kim BC, et al. Exosome RNA unshielding couples stromal activation to pattern recognition receptor signaling in cancer. Cell. (2017) 170:352–66.e13. doi: 10.1016/j.cell.2017.06.031
69. Záveský L, Jandáková E, Weinberger V, Minář L, Hanzíková V, Dušková D, et al. Small non-coding RNA profiling in breast cancer: plasma U6 snRNA, miR-451a and miR-548b-5p as novel diagnostic and prognostic biomarkers. Mol Biol Rep. (2022) 49:1955–71. doi: 10.1007/s11033-021-07010-8
70. Alivernini S, Gremese E, McSharry C, Tolusso B, Ferraccioli G, McInnes IB, et al. MicroRNA-155-at the critical interface of innate and adaptive immunity in arthritis. Front Immunol. (2017) 8:1932. doi: 10.3389/fimmu.2017.01932
71. Baltimore D, Boldin MP, O’Connell RM, Rao DS, and Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. (2008) 9:839–45. doi: 10.1038/ni.f.209
72. Chen L, Gao D, Shao Z, Zheng Q, and Yu Q. miR-155 indicates the fate of CD4(+) T cells. Immunol Lett. (2020) 224:40–9. doi: 10.1016/j.imlet.2020.05.003
73. Jafarzadeh A, Naseri A, Shojaie L, Nemati M, Jafarzadeh S, Bannazadeh Baghi H, et al. MicroRNA-155 and antiviral immune responses. Int Immunopharmacol. (2021) 101:108188. doi: 10.1016/j.intimp.2021.108188
74. Siddiqui KR, Laffont S, and Powrie F. E-cadherin marks a subset of inflammatory dendritic cells that promote T cell-mediated colitis. Immunity. (2010) 32:557–67. doi: 10.1016/j.immuni.2010.03.017
75. Xu SJ, Hu HT, Li HL, and Chang S. The role of miRNAs in immune cell development, immune cell activation, and tumor immunity: with a focus on macrophages and natural killer cells. Cells. (2019) 8:1140. doi: 10.3390/cells8101140
76. Rimer JM, Lee J, Holley CL, Crowder RJ, Chen DL, Hanson PI, et al. Long-range function of secreted small nucleolar RNAs that direct 2’-O-methylation. J Biol Chem. (2018) 293:13284–96. doi: 10.1074/jbc.RA118.003410
77. Shi Y, Shi Q, Shen Q, Zhang Q, and Cao X. Dicer-independent snRNA/snoRNA-derived nuclear RNA 3 regulates tumor-associated macrophage function by epigenetically repressing inducible nitric oxide synthase transcription. Cancer Commun (Lond). (2021) 41:140–53. doi: 10.1002/cac2.12131
78. Ma D, Zhou X, Wang Y, Dai L, Yuan J, Peng J, et al. Changes in the small noncoding RNAome during M1 and M2 macrophage polarization. Front Immunol. (2022) 13:799733. doi: 10.3389/fimmu.2022.799733
79. Lv B, Wang Y, Ma D, Cheng W, Liu J, Yong T, et al. Immunotherapy: reshape the tumor immune microenvironment. Front Immunol. (2022) 13:844142. doi: 10.3389/fimmu.2022.844142
80. Wang M, Zhao J, Zhang L, Wei F, Lian Y, Wu Y, et al. Role of tumor microenvironment in tumorigenesis. J Cancer. (2017) 8:761–73. doi: 10.7150/jca.17648
81. El Saftawy EA, Aboulhoda BE, AbdElkhalek MA, Alghamdi MA, and AlHariry NS. Non-coding RNAs in urinary bladder cancer microenvironment: Diagnostic, therapeutic, and prognostic perspective. Pathol Res Pract. (2025) 266:155815. doi: 10.1016/j.prp.2025.155815
82. Sahraei M, Chaube B, Liu Y, Sun J, Kaplan A, Price NL, et al. Suppressing miR-21 activity in tumor-associated macrophages promotes an antitumor immune response. J Clin Invest. (2019) 129:5518–36. doi: 10.1172/JCI127125
83. Peng M, Ren J, Jing Y, Jiang X, Xiao Q, Huang J, et al. Tumour-derived small extracellular vesicles suppress CD8+ T cell immune function by inhibiting SLC6A8-mediated creatine import in NPM1-mutated acute myeloid leukaemia. J Extracell Vesicles. (2021) 10:e12168. doi: 10.1002/jev2.12168
84. Omrane I and Benammar-Elgaaied A. The immune microenvironment of the colorectal tumor: Involvement of immunity genes and microRNAs belonging to the TH17 pathway. Biochim Biophys Acta. (2015) 1856:28–38. doi: 10.1016/j.bbcan.2015.04.001
85. Alemohammad H, Najafzadeh B, Asadzadeh Z, Baghbanzadeh A, Ghorbaninezhad F, Najafzadeh A, et al. The importance of immune checkpoints in immune monitoring: A future paradigm shift in the treatment of cancer. BioMed Pharmacother. (2022) 146:112516. doi: 10.1016/j.biopha.2021.112516
86. Krummel MF and Allison JP. CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. J Exp Med. (1995) 182:459–65. doi: 10.1084/jem.182.2.459
87. Won JE, Byeon Y, Wi TI, Lee CM, Lee JH, Kang TH, et al. Immune checkpoint silencing using RNAi-incorporated nanoparticles enhances antitumor immunity and therapeutic efficacy compared with antibody-based approaches. J Immunother Cancer. (2022) 10:e003928. doi: 10.1136/jitc-2021-003928
88. Li J, Chen Y, Liao M, Yu S, Yuan B, Jia Z, et al. Exosomes-delivered PD-L1 siRNA and CTLA-4 siRNA protect against growth and tumor immune escape in colorectal cancer. Genomics. (2023) 115:110646. doi: 10.1016/j.ygeno.2023.110646
89. Skafi N, Fayyad-Kazan M, and Badran B. Immunomodulatory role for MicroRNAs: Regulation of PD-1/PD-L1 and CTLA-4 immune checkpoints expression. Gene. (2020) 754:144888. doi: 10.1016/j.gene.2020.144888
90. Fayyad-Kazan H, Rouas R, Fayyad-Kazan M, Badran R, El Zein N, Lewalle P, et al. MicroRNA profile of circulating CD4-positive regulatory T cells in human adults and impact of differentially expressed microRNAs on expression of two genes essential to their function. J Biol Chem. (2012) 287:9910–22. doi: 10.1074/jbc.M111.337154
91. Chang RM, Xiao S, Lei X, Yang H, Fang F, and Yang LY. miRNA-487a promotes proliferation and metastasis in hepatocellular carcinoma. Clin Cancer Res. (2017) 23:2593–604. doi: 10.1158/1078-0432.Ccr-16-0851
92. Sauer N, Janicka N, Szlasa W, Skinderowicz B, Kołodzińska K, Dwernicka W, et al. TIM-3 as a promising target for cancer immunotherapy in a wide range of tumors. Cancer Immunol Immunother. (2023) 72:3405–25. doi: 10.1007/s00262-023-03516-1
93. Rezaei M, Ghanadian M, Ghezelbash B, Shokouhi A, Bazhin AV, Zamyatnin AA Jr., et al. TIM-3/Gal-9 interaction affects glucose and lipid metabolism in acute myeloid leukemia cell lines. Front Immunol. (2023) 14:1267578. doi: 10.3389/fimmu.2023.1267578
94. Zhu C, Anderson AC, Schubart A, Xiong H, Imitola J, Khoury SJ, et al. The Tim-3 ligand galectin-9 negatively regulates T helper type 1 immunity. Nat Immunol. (2005) 6:1245–52. doi: 10.1038/ni1271
95. Acharya N, Sabatos-Peyton C, and Anderson AC. Tim-3 finds its place in the cancer immunotherapy landscape. J Immunother Cancer. (2020) 8:e000911. doi: 10.1136/jitc-2020-000911
96. Moghaddam Y, Andalib A, Mohammad-Ganji M, Homayouni V, Sharifi M, and Ganjalikhani-Hakemi M. Evaluation of the effect of TIM-3 suppression by miR-498 and its effect on apoptosis and proliferation rate of HL-60 cell line. Pathol Res Pract. (2018) 214:1482–8. doi: 10.1016/j.prp.2018.07.019
97. Shouse AN, LaPorte KM, and Malek TR. Interleukin-2 signaling in the regulation of T cell biology in autoimmunity and cancer. Immunity. (2024) 57:414–28. doi: 10.1016/j.immuni.2024.02.001
98. Chocarro L, Blanco E, Zuazo M, Arasanz H, Bocanegra A, Fernández-Rubio L, et al. Understanding LAG-3 signaling. Int J Mol Sci. (2021) 22:5282. doi: 10.3390/ijms22105282
99. Gao F, Zhao ZL, Zhao WT, Fan QR, Wang SC, Li J, et al. miR-9 modulates the expression of interferon-regulated genes and MHC class I molecules in human nasopharyngeal carcinoma cells. Biochem Biophys Res Commun. (2013) 431:610–6. doi: 10.1016/j.bbrc.2012.12.097
100. Yi M, Xu L, Jiao Y, Luo S, Li A, and Wu K. The role of cancer-derived microRNAs in cancer immune escape. J Hematol Oncol. (2020) 13:25. doi: 10.1186/s13045-020-00848-8
101. Zhong Y, Tian Y, Wang Y, Bai J, Long Q, Yan L, et al. Small extracellular vesicle piR-hsa-30937 derived from pancreatic neuroendocrine neoplasms upregulates CD276 in macrophages to promote immune evasion. Cancer Immunol Res. (2024) 12:840–53. doi: 10.1158/2326-6066.CIR-23-0825
102. Zhu X, Liang R, Lan T, Ding D, Huang S, Shao J, et al. Tumor-associated macrophage-specific CD155 contributes to M2-phenotype transition, immunosuppression, and tumor progression in colorectal cancer. J Immunother Cancer. (2022) 10:e004219. doi: 10.1136/jitc-2021-004219
103. Omar HA, El-Serafi AT, Hersi F, Arafa EA, Zaher DM, Madkour M, et al. Immunomodulatory MicroRNAs in cancer: targeting immune checkpoints and the tumor microenvironment. FEBS J. (2019) 286:3540–57. doi: 10.1111/febs.15000
104. Wei Y, Zhou K, Wang C, Du X, Wang Z, Chen G, et al. Exosomal miR-142-3p from M1-polarized macrophages suppresses cell growth and immune escape in glioblastoma through regulating HMGB1-mediated PD-1/PD-L1 checkpoint. J Neurochem. (2025) 169:e16224. doi: 10.1111/jnc.16224
105. Guo LM, Ding GF, Xu WC, Ge H, Jiang Y, and Lu YF. Anti-PD-L1 antibody enhances T cell immune responses and reduces resistance of breast cancer cells to radiotherapy. Oxid Med Cell Longev. (2022) 2022:5938688. doi: 10.1155/2022/5938688
106. Manning BD and Toker A. AKT/PKB signaling: navigating the network. Cell. (2017) 169:381–405. doi: 10.1016/j.cell.2017.04.001
107. Quan Z, Yang Y, Zheng H, Zhan Y, Luo J, Ning Y, et al. Clinical implications of the interaction between PD-1/PD-L1 and PI3K/AKT/mTOR pathway in progression and treatment of non-small cell lung cancer. J Cancer. (2022) 13:3434–43. doi: 10.7150/jca.77619
108. Shaker FH, Sanad EF, Elghazaly H, Hsia SM, and Hamdy NM. piR-823 tale as emerging cancer-hallmark molecular marker in different cancer types: a step-toward ncRNA-precision. Naunyn Schmiedebergs Arch Pharmacol. (2025) 398:47–68. doi: 10.1007/s00210-024-03308-z
109. Grenda A and Krawczyk P. New dancing couple: PD-L1 and microRNA. Scand J Immunol. (2017) 86:130–4. doi: 10.1111/sji.12577
110. You J, Yang G, Wu Y, Lu X, Huang S, Chen Q, et al. Plasma tRF-1:29-Pro-AGG-1-M6 and tRF-55:76-Tyr-GTA-1-M2 as novel diagnostic biomarkers for lung adenocarcinoma. Front Oncol. (2022) 12:991451. doi: 10.3389/fonc.2022.991451
111. Hamidi AA, Zangoue M, Kashani D, Zangouei AS, Rahimi HR, Abbaszadegan MR, et al. MicroRNA-217: a therapeutic and diagnostic tumor marker. Expert Rev Mol Diagn. (2022) 22:61–76. doi: 10.1080/14737159.2022.2017284
112. Xu X, Chen J, Bai M, Liu T, Zhan S, Li J, et al. Plasma tsRNA signatures serve as a novel biomarker for bladder cancer. Cancer Sci. (2025) 116(5):1255–67. doi: 10.1111/cas.70003
113. Condrat CE, Thompson DC, Barbu MG, Bugnar OL, Boboc A, Cretoiu D, et al. miRNAs as biomarkers in disease: latest findings regarding their role in diagnosis and prognosis. Cells. (2020) 9:276. doi: 10.3390/cells9020276
114. Zhu L, Li J, Gong Y, Wu Q, Tan S, Sun D, et al. Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Mol Cancer. (2019) 18:74. doi: 10.1186/s12943-019-1000-8
115. Wu CW, Ng SC, Dong Y, Tian L, Ng SS, Leung WW, et al. Identification of microRNA-135b in stool as a potential noninvasive biomarker for colorectal cancer and adenoma. Clin Cancer Res. (2014) 20:2994–3002. doi: 10.1158/1078-0432.CCR-13-1750
116. Yau TO, Wu CW, Dong Y, Tang CM, Ng SS, Chan FK, et al. microRNA-221 and microRNA-18a identification in stool as potential biomarkers for the non-invasive diagnosis of colorectal carcinoma. Br J Cancer. (2014) 111:1765–71. doi: 10.1038/bjc.2014.484
117. Sabbah NA, Abdalla WM, Mawla WA, AbdAlMonem N, Gharib AF, Abdul-Saboor A, et al. piRNA-823 is a unique potential diagnostic non-invasive biomarker in colorectal cancer patients. Genes (Basel). (2021) 12:598. doi: 10.3390/genes12040598
118. Jin F, Yang L, Wang W, Yuan N, Zhan S, Yang P, et al. A novel class of tsRNA signatures as biomarkers for diagnosis and prognosis of pancreatic cancer. Mol Cancer. (2021) 20:95. doi: 10.1186/s12943-021-01389-5
119. Xia L, Guo H, Wu X, Xu Y, Zhao P, Yan B, et al. Human circulating small non-coding RNA signature as a non-invasive biomarker in clinical diagnosis of acute myeloid leukaemia. Theranostics. (2023) 13:1289–301. doi: 10.7150/thno.80054
120. Wang DR, Wu XL, and Sun YL. Therapeutic targets and biomarkers of tumor immunotherapy: response versus non-response. Signal Transduct Target Ther. (2022) 7:331. doi: 10.1038/s41392-022-01136-2
121. Li Y, Ju M, Miao Y, Zhao L, Xing L, and Wei M. Advancement of anti-LAG-3 in cancer therapy. FASEB J. (2023) 37:e23236. doi: 10.1096/fj.202301018R
122. Larkin J, Hodi FS, and Wolchok JD. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med. (2015) 373:1270–1. doi: 10.1056/NEJMc1509660
123. Hodi FS, Chiarion-Sileni V, Gonzalez R, Grob JJ, Rutkowski P, Cowey CL, et al. Nivolumab plus ipilimumab or nivolumab alone versus ipilimumab alone in advanced melanoma (CheckMate 067): 4-year outcomes of a multicentre, randomised, phase 3 trial. Lancet Oncol. (2018) 19:1480–92. doi: 10.1016/s1470-2045(18)30700-9
124. Su J, Fu Y, Cui Z, Abidin Z, Yuan J, Zhang X, et al. Relatlimab: a novel drug targeting immune checkpoint LAG-3 in melanoma therapy. Front Pharmacol. (2023) 14:1349081. doi: 10.3389/fphar.2023.1349081
125. Cho BC, Abreu DR, Hussein M, Cobo M, Patel AJ, Secen N, et al. Tiragolumab plus atezolizumab versus placebo plus atezolizumab as a first-line treatment for PD-L1-selected non-small-cell lung cancer (CITYSCAPE): primary and follow-up analyses of a randomised, double-blind, phase 2 study. Lancet Oncol. (2022) 23:781–92. doi: 10.1016/s1470-2045(22)00226-1
126. Tawbi HA, SChadendorf D, Lipson EJ, Ascierto PA, Matamala L, Castillo Gutiérrez E, et al. Relatlimab and nivolumab versus nivolumab in untreated advanced melanoma. N Engl J Med. (2022) 386:24–34. doi: 10.1056/NEJMoa2109970
127. Huang J, Weng Q, Shi Y, Mao W, Zhao Z, Wu R, et al. MicroRNA-155-5p suppresses PD-L1 expression in lung adenocarcinoma. FEBS Open Bio. (2020) 10:1065–71. doi: 10.1002/2211-5463.12853
128. Huffaker TB, Lee SH, Tang WW, Wallace JA, Alexander M, Runtsch MC, et al. Antitumor immunity is defective in T cell-specific microRNA-155-deficient mice and is rescued by immune checkpoint blockade. J Biol Chem. (2017) 292:18530–41. doi: 10.1074/jbc.M117.808121
129. Anastasiadou E, Seto AG, Beatty X, Hermreck M, Gilles ME, Stroopinsky D, et al. Cobomarsen, an oligonucleotide inhibitor of miR-155, slows DLBCL tumor cell growth In Vitro and In Vivo. Clin Cancer Res. (2021) 27:1139–49. doi: 10.1158/1078-0432.Ccr-20-3139
130. Choi Y, Seok SH, Yoon HY, Ryu JH, and Kwon IC. Advancing cancer immunotherapy through siRNA-based gene silencing for immune checkpoint blockade. Adv Drug Delivery Rev. (2024) 209:115306. doi: 10.1016/j.addr.2024.115306
131. Shao L, Yu X, Han Q, Zhang X, Lu N, and Zhang C. Enhancing anti-tumor efficacy and immune memory by combining 3p-GPC-3 siRNA treatment with PD-1 blockade in hepatocellular carcinoma. Oncoimmunology. (2022) 11:2010894. doi: 10.1080/2162402x.2021.2010894
132. Zhang C, Huang R, Ren L, Martincuks A, Song J, Kortylewski M, et al. Local CpG-Stat3 siRNA treatment improves antitumor effects of immune checkpoint inhibitors. Mol Ther Nucleic Acids. (2024) 35:102357. doi: 10.1016/j.omtn.2024.102357
133. Wang Y, Wang D, Xie G, Yin Y, Zhao E, Tao K, et al. MicroRNA-152 regulates immune response via targeting B7-H1 in gastric carcinoma. Oncotarget. (2017) 8:28125–34. doi: 10.18632/oncotarget.15924
134. Guan X, Hu R, Choi Y, Srivats S, Nabet BY, Silva J, et al. Anti-TIGIT antibody improves PD-L1 blockade through myeloid and T(reg) cells. Nature. (2024) 627:646–55. doi: 10.1038/s41586-024-07121-9
135. Dai X and Tan C. Combination of microRNA therapeutics with small-molecule anticancer drugs: mechanism of action and co-delivery nanocarriers. Adv Drug Delivery Rev. (2015) 81:184–97. doi: 10.1016/j.addr.2014.09.010
136. Gandhi NS, Tekade RK, and Chougule MB. Nanocarrier mediated delivery of siRNA/miRNA in combination with chemotherapeutic agents for cancer therapy: current progress and advances. J Control Release. (2014) 194:238–56. doi: 10.1016/j.jconrel.2014.09.001
137. Khelghati N, Soleimanpour Mokhtarvand J, Mir M, Alemi F, Asemi Z, Sadeghpour A, et al. The importance of co-delivery of nanoparticle-siRNA and anticancer agents in cancer therapy. Chem Biol Drug Des. (2021) 97:997–1015. doi: 10.1111/cbdd.13824
138. Zhang M, Zhang W, Tang G, Wang H, Wu M, Yu W, et al. Targeted codelivery of docetaxel and atg7 siRNA for autophagy inhibition and pancreatic cancer treatment. ACS Appl Bio Mater. (2019) 2:1168–76. doi: 10.1021/acsabm.8b00764
139. Meng H, Liong M, Xia T, Li Z, Ji Z, Zink JI, et al. Engineered design of mesoporous silica nanoparticles to deliver doxorubicin and P-glycoprotein siRNA to overcome drug resistance in a cancer cell line. ACS Nano. (2010) 4:4539–50. doi: 10.1021/nn100690m
140. Shim G, Han SE, Yu YH, Lee S, Lee HY, Kim K, et al. Trilysinoyl oleylamide-based cationic liposomes for systemic co-delivery of siRNA and an anticancer drug. J Control Release. (2011) 155:60–6. doi: 10.1016/j.jconrel.2010.10.017
141. Wei W, Lv PP, Chen XM, Yue ZG, Fu Q, Liu SY, et al. Codelivery of mTERT siRNA and paclitaxel by chitosan-based nanoparticles promoted synergistic tumor suppression. Biomaterials. (2013) 34:3912–23. doi: 10.1016/j.biomaterials.2013.02.030
142. Kaur K, Rath G, Chandra S, Singh R, and Goyal AK. Chemotherapy with si-RNA and anti-cancer drugs. Curr Drug Deliv. (2018) 15:300–11. doi: 10.2174/1567201814666170518141440
143. Nakamura K, Abu Lila AS, Matsunaga M, Doi Y, Ishida T, and Kiwada H. A double-modulation strategy in cancer treatment with a chemotherapeutic agent and siRNA. Mol Ther. (2011) 19:2040–7. doi: 10.1038/mt.2011.174
144. Wang C, Zhang Y, Kong W, Rong X, Zhong Z, Jiang L, et al. Delivery of miRNAs using nanoparticles for the treatment of osteosarcoma. Int J Nanomed. (2024) 19:8641–60. doi: 10.2147/IJN.S471900
145. Ren Y, Zhou X, Mei M, Yuan XB, Han L, Wang GX, et al. MicroRNA-21 inhibitor sensitizes human glioblastoma cells U251 (PTEN-mutant) and LN229 (PTEN-wild type) to taxol. BMC Cancer. (2010) 10:27. doi: 10.1186/1471-2407-10-27
146. El Chediak A, Shamseddine A, Bodgi L, Obeid JP, Geara F, and Zeidan YH. Optimizing tumor immune response through combination of radiation and immunotherapy. Med Oncol. (2017) 34:165. doi: 10.1007/s12032-017-1025-z
147. Yan D, Ng WL, Zhang X, Wang P, Zhang Z, Mo YY, et al. Targeting DNA-PKcs and ATM with miR-101 sensitizes tumors to radiation. PloS One. (2010) 5:e11397. doi: 10.1371/journal.pone.0011397
148. Li Y, Li L, Li B, Wu Z, Wu Y, Wang Y, et al. Silencing of ataxia-telangiectasia mutated by siRNA enhances the in vitro and in vivo radiosensitivity of glioma. Oncol Rep. (2016) 35:3303–12. doi: 10.3892/or.2016.4754
149. Zhang WY, Wen L, Du L, Liu TT, Sun Y, Chen YZ, et al. S-RBD-modified and miR-486-5p-engineered exosomes derived from mesenchymal stem cells suppress ferroptosis and alleviate radiation-induced lung injury and long-term pulmonary fibrosis. J Nanobiotechnol. (2024) 22:662. doi: 10.1186/s12951-024-02830-9
150. Ji X, Wu B, Fan J, Han R, Luo C, Wang T, et al. The anti-fibrotic effects and mechanisms of microRNA-486-5p in pulmonary fibrosis. Sci Rep. (2015) 5:14131. doi: 10.1038/srep14131
151. Xie G, Ivica NA, Jia B, Li Y, Dong H, Liang Y, et al. CAR-T cells targeting a nucleophosmin neoepitope exhibit potent specific activity in mouse models of acute myeloid leukaemia. Nat BioMed Eng. (2021) 5:399–413. doi: 10.1038/s41551-020-00625-5
152. Ma S, Li X, Wang X, Cheng L, Li Z, Zhang C, et al. Current progress in CAR-T cell therapy for solid tumors. Int J Biol Sci. (2019) 15:2548–60. doi: 10.7150/ijbs.34213
153. Qi C, Gong J, Li J, Liu D, Qin Y, Ge S, et al. Claudin18.2-specific CAR T cells in gastrointestinal cancers: phase 1 trial interim results. Nat Med. (2022) 28:1189–98. doi: 10.1038/s41591-022-01800-8
154. Buono G, Capozzi M, Caputo R, Lauro VD, Cianniello D, Piezzo M, et al. CAR-T cell therapy for breast cancer: Current status and future perspective. Cancer Treat Rev. (2025) 133:102868. doi: 10.1016/j.ctrv.2024.102868
155. Barranco C. CAR T cell therapy for multiple myeloma. Nat Cancer. (2023) 4:1644. doi: 10.1038/s43018-023-00680-2
156. Simon B, Harrer DC, Schuler-Thurner B, Schaft N, Schuler G, Dorrie J, et al. The siRNA-mediated downregulation of PD-1 alone or simultaneously with CTLA-4 shows enhanced in vitro CAR-T-cell functionality for further clinical development towards the potential use in immunotherapy of melanoma. Exp Dermatol. (2018) 27:769–78. doi: 10.1111/exd.13678
157. Huang Q, Xia J, Wang L, Wang X, Ma X, Deng Q, et al. miR-153 suppresses IDO1 expression and enhances CAR T cell immunotherapy. J Hematol Oncol. (2018) 11:58. doi: 10.1186/s13045-018-0600-x
158. Zhang J, Zhu J, Zheng G, Wang Q, Li X, Feng Y, et al. Co-Expression of miR155 or LSD1 shRNA Increases the Anti-Tumor Functions of CD19 CAR-T Cells. Front Immunol. (2021) 12:811364. doi: 10.3389/fimmu.2021.811364
159. Lonez C, Bolsée J, Huberty F, Nguyen T, Jacques-Hespel C, Anguille S, et al. Clinical Proof-of-Concept of a Non-Gene Editing Technology Using miRNA-Based shRNA to Engineer Allogeneic CAR T-Cells. Int J Mol Sci. (2025) 26:1658. doi: 10.3390/ijms26041658
160. Martino MTD, Tagliaferri P, and Tassone P. MicroRNA in cancer therapy: breakthroughs and challenges in early clinical applications. J Exp Clin Cancer Res. (2025) 44:126. doi: 10.1186/s13046-025-03391-x
161. Dong Y, Siegwart DJ, and Anderson DG. Strategies, design, and chemistry in siRNA delivery systems. Adv Drug Delivery Rev. (2019) 144:133–47. doi: 10.1016/j.addr.2019.05.004
162. Ghasemiyeh P and Mohammadi-Samani S. siRNA-based delivery systems: Technologies, carriers, applications, and approved products. Eur J Pharmacol. (2025) 996:177441. doi: 10.1016/j.ejphar.2025.177441
163. Setten RL, Rossi JJ, and Han SP. The current state and future directions of RNAi-based therapeutics. Nat Rev Drug Discov. (2019) 18:421–46. doi: 10.1038/s41573-019-0017-4
164. Nomura K, An S, Kobayashi Y, Kondo J, Shi T, Murase H, et al. Synthesis of 2’-formamidonucleoside phosphoramidites for suppressing the seed-based off-target effects of siRNAs. Nucleic Acids Res. (2024) 52:10754–74. doi: 10.1093/nar/gkae741
165. Chernyavska M, Masoudnia M, Valerius T, and Verdurmen WPR. Organ-on-a-chip models for development of cancer immunotherapies. Cancer Immunol Immunother. (2023) 72:3971–83. doi: 10.1007/s00262-023-03572-7
Keywords: small non-coding RNA, tumor immunity, immune checkpoint, tumor microenvironment, tumor immune escape, tumor immunotherapy
Citation: Liu Z, Dong H, Ye C, Yan J, Miao M and Shao Y (2025) Small non-coding RNAs: key regulatory factors and potential therapeutic targets in tumor immunity. Front. Immunol. 16:1639763. doi: 10.3389/fimmu.2025.1639763
Received: 02 June 2025; Accepted: 29 August 2025;
Published: 11 September 2025.
Edited by:
Ángeles Carlos Reyes, National Institute of Respiratory Diseases (INER), MexicoReviewed by:
Xinpei Deng, Sun Yat-Sen University Cancer Center (SYSUCC), ChinaDavid Nuñez Corona, National Autonomous University of Mexico, Mexico
Misael Osmar Garcia Martin, National Institute of Respiratory Diseases (INER), Mexico
Copyright © 2025 Liu, Dong, Ye, Yan, Miao and Shao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongfu Shao, ZnlzaGFveW9uZ2Z1QG5idS5lZHUuY24=; c2hhb3lvbmdmdTExNzNAMTYzLmNvbQ==; Min Miao, bWlhb21pbjEyQHNpbmEuY29t
 Zihan Liu
Zihan Liu Haotian Dong
Haotian Dong Chengyuan Ye
Chengyuan Ye Jianing Yan
Jianing Yan Min Miao
Min Miao Yongfu Shao
Yongfu Shao