- 1Department of Global Health, Amsterdam UMC, University of Amsterdam, Amsterdam Institute for Global Health and Development, Amsterdam, Netherlands
- 2Research Division, Centre for Infectious Disease Research in Zambia, Lusaka, Zambia
- 3Department of Biostatistics, School of Public Health, University of Ghana, Accra, Ghana
- 4Ragon Institute of Mass General, MIT, and Harvard, Cambridge, MA, United States
- 5Division of Infectious Diseases, Massachusetts General Hospital, Boston, MA, United States
- 6Department of Pediatrics, Harvard Medical School, Boston, MA, United States
- 7Department of Immunology and Infectious Diseases, Harvard T.H. Chan School of Public Health, Boston, MA, United States
- 8Department of Medicine, Harvard Medical School, Boston, MA, United States
- 9Center for Immunization Research, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD, United States
Background: Cholera is a public health threat in resource-limited settings and is responsible for causing over 3 million cases globally. Mucosal immune responses play an important role in protecting against Vibrio cholerae infection, a non-invasive mucosal pathogen, yet traditional plasma-based assays are invasive and logistically challenging, particularly during outbreaks in low- and middle-income countries (LMICs). Saliva offers a unique window into mucosal immunity and may serve as a non-invasive alternative for seroprevalence and vaccine immunogenicity studies.
Methods: We conducted a cross-sectional antibody profiling study to analyse cholera-specific antibodies in saliva and plasma samples from 74 participants upon presenting to the cholera treatment centres. These were collected from four treatment centres in Lusaka during Zambia’s most severe cholera outbreak in 2024 caused by Vibrio cholerae O1 Ogawa. Levels of total IgG, IgG1-3, IgM, secretory IgA, and IgA1–2 isotypes were used to compare the biomarker profile between the two sample types.
Results: Saliva and plasma antibody profiles were comparable, with elevated IgA1 and IgA2 responses to cholera toxin-B (CtxB), sialidase, HlyA, and TcpA in saliva. Broader systemic responses were seen in plasma, including high CtxB-specific IgM, IgA1, and total IgG levels. Notably, biomarkers such as HlyA, Ogawa O-specific polysaccharide (OSP), and sialidase exhibited significant positive correlations between plasma and saliva. Elevated biomarker levels of HlyA, Ogawa O-specific polysaccharide (OSP), and sialidase in people living with HIV/AIDS (PLWHA) suggested immunological differences that warrant further exploration.
Conclusion: We demonstrate that saliva is a viable, non-invasive alternative for cholera antibody-based profiling, offering practical advantages in resource-constrained settings. Given its strong correlation with systemic antibody profiles, saliva may be a practical sample for sero-surveillance in resource-limited settings. Future studies should investigate the duration of these salivary responses to further substantiate their use in estimating disease burden and immunity.
Introduction
Cholera continues to be a major public health challenge, particularly in low- and middle-income countries (LMICs), where recurrent outbreaks place significant strain on health systems. Zambia has reported cholera outbreaks since the early 1970s, with the recent 2024 outbreak being the most severe to date, affecting over 20,000 individuals (1, 2). The predominant serotype during this outbreak was serogroup O1 Ogawa (3). Despite the availability of vaccines, there is no established correlate of protection against Vibrio cholerae, thus limiting the ability to predict vaccine efficacy and immunogenicity. Vibriocidal antibody titres correlate with protection and often serve as surrogate endpoints in cholera vaccine trials (4, 5). However, the vibriocidal assay traditionally relies on serum samples, which are invasive and logistically challenging to collect, especially in outbreak conditions, and are labour-intensive in the laboratory.
Alternative non-invasive sample types, such as saliva, could mitigate these barriers (6). Saliva collection is easier and more acceptable in resource-limited settings but has been underexplored in cholera immunology. Our previous study demonstrated the feasibility of using saliva for vibriocidal antibody titres, showing comparable sensitivity and specificity to serum (7). This raises the potential for saliva to be a viable alternative for serological assessments in cholera, as was only shown in an earlier cholera vaccine study (8). However, a comprehensive analysis comparing the broader immunological biomarker profiles in plasma and saliva using advanced multiplexed and sample-sparing antibody profiling approaches remains lacking.
There are limited data on the direct assessment of mucosal anti-Vibrio cholerae antibody responses, which leaves a gap in our understanding of protective immune mechanisms at the site of pathogen entry. Although a few studies in vaccinees have analysed stool antibodies and measured day 7 antibody-secreting cell responses, believed to reflect mucosal immunity as maturing lymphoblasts transiently circulate before rehoming to the intestinal mucosa (9, 10), no data are available on salivary mucosal responses in humans with cholera.
Here, we applied an antibody profiling approach on samples collected from participants with cholera to enable high-dimensional profiling of antibody features across multiple antigens in parallel, providing a holistic and unbiased view of immune responses (11, 12). We hypothesised that saliva and plasma would yield comparable profiles of cholera-specific antibodies, while unique antibody signatures found in saliva would enable insights into mucosal immunity. By comparing cholera-specific antibodies between saliva and plasma, we aimed to identify saliva’s utility as an alternative sample for seroprevalence studies and its potential in outbreak settings.
Materials and methods
Study design and participant recruitment
We collected samples from four sites in Lusaka in 2024: George Urban Health Centre, Matero Level One Hospital, Levy Mwanawasa General Hospital, and Heroes Cholera Treatment Centre during an ongoing outbreak previously confirmed by the Ministry of Health (MOH) to be due to Vibrio cholerae O1 infection. Participants were eligible for enrolment if they presented to one of the four facilities, had watery diarrhoea, and had a positive Cholera Rapid Diagnostic Test (RDT) on the Bioline™ Cholera Ag O1/O139 (Abbot) result. Eligible participants were approached and provided with study information. Those who consented to join the study signed consent forms, with guardians signing on behalf of minors.
We collected approximately 20 ml of blood and 5 ml of saliva from each participant after self-reported onset of diarrhoea and within 24 hours of presentation to the treatment centre. Before saliva collection, participants rinsed their mouths thrice with sterile 20 ml bottled water. Following the mouth rinse, they were then asked to pull and spit saliva into a sterile saliva collection tube with a targeted minimum volume of 5 ml. The collected samples were transported to the Centre for Infectious Disease Research in Zambia (CIDRZ) laboratories at 4–8 degrees the same day. The blood was centrifuged to obtain plasma, which was subsequently screened for HIV using the Determine™ HIV-1/2 kit (Abbot). The remaining plasma and saliva samples were aliquoted and stored at -80 degrees. Within a month of storage, the samples were shipped to Ragon Institute of Mass General, MIT, and Harvard, Cambridge, MA, USA laboratories for antibody profiling on dry ice.
Antibody profiling
Cholera-specific antibody subclass/isotype levels were assessed using a 384-well-based customised multiplexed Luminex assay, as previously described (13). HlyA, TcpA, CtxB, sialidase, and V. cholerae Ogawa O-specific antigen (OSP) were used to profile specific antibody responses, using antigens purified as previously described (14). Tetanus toxin and bovine serum albumin (CEFTA, Mabtech Inc) were used as control antigens. Protein antigens were coupled to magnetic Luminex beads (Luminex Corp) by carbodiimide-NHS ester-coupling (Thermo Fisher). V. cholerae Ogawa OSP was modified by 4-(4,6-dimethoxy[1,3,5] triazin-2-yl)-4-methyl-morpholinium and conjugated to Luminex Magplex carboxylated beads. Antigen-coupled microspheres were washed and incubated with plasma or saliva samples at an appropriate sample dilution (IgG: 1:2000 for plasma, 1:20 in saliva; IgA and IgM, 1:1000 in plasma, 1:50 in saliva, samples diluted in 0.1% BSA in PBS) for 2 hours at 37°C in 384-well plates (Greiner Bio-One). Unbound antibodies were washed away, and antigen-bound antibodies were detected by using a PE-coupled detection antibody for each subclass and isotype (IgG1, IgG2, IgG3, IgA1, IgA2, secretory IgA(sIgA) and IgM; Southern Biotech. After one hour of incubation, plates were washed, flow cytometry was performed with an IQue (Intellicyt), and analysis was performed on IntelliCyt ForeCyt (v8.1). All washes were performed with 0.05% Tween 0.1% BSA in PBS. PE median fluorescent intensity (MFI) was reported as a readout for antigen-specific antibody measurements.
Sample size
We utilised a convenience sampling approach due to the rapid response required during the cholera outbreak and the availability of reagents for antibody profiling. An enrolment size of 74 was targeted to ensure adequate representation across different age groups and sex. While not formally powered for inferential comparisons, this sample size was considered sufficient to provide preliminary insights into cholera-specific antibody responses in plasma and saliva.
Data analysis
Data analysis was performed using R version 4.4.2. Spearman correlation coefficients were computed to assess the relationship between cholera-specific antibody responses in plasma and saliva. Correlation heatmaps were generated using the corrplot (https://github.com/taiyun/corrplot) and ComplexHeatmap (15) packages to visualise biomarker median fluorescent intensities across groups. To compare antibody intensities between participant subgroups, such as HIV status and age categories, we conducted Wilcoxon rank-sum tests for group comparisons with a significance threshold of p < 0.05. False discovery rate (FDR) correction was applied using the Benjamini-Hochberg method to account for multiple comparisons (16).
Ethics statement
Before enrolment, all participants provided written informed consent. We sought ethical approval from the University of Zambia Biomedical Research Ethics Committee (UNZABREC), reference number 001-02-23, and the National Health Research Authority (NHRA), which authorised the study. This study was approved by the Massachusetts General Hospital’s Institutional Review Board. All study procedures were conducted under Good Clinical Practice.
Results
Participant descriptions
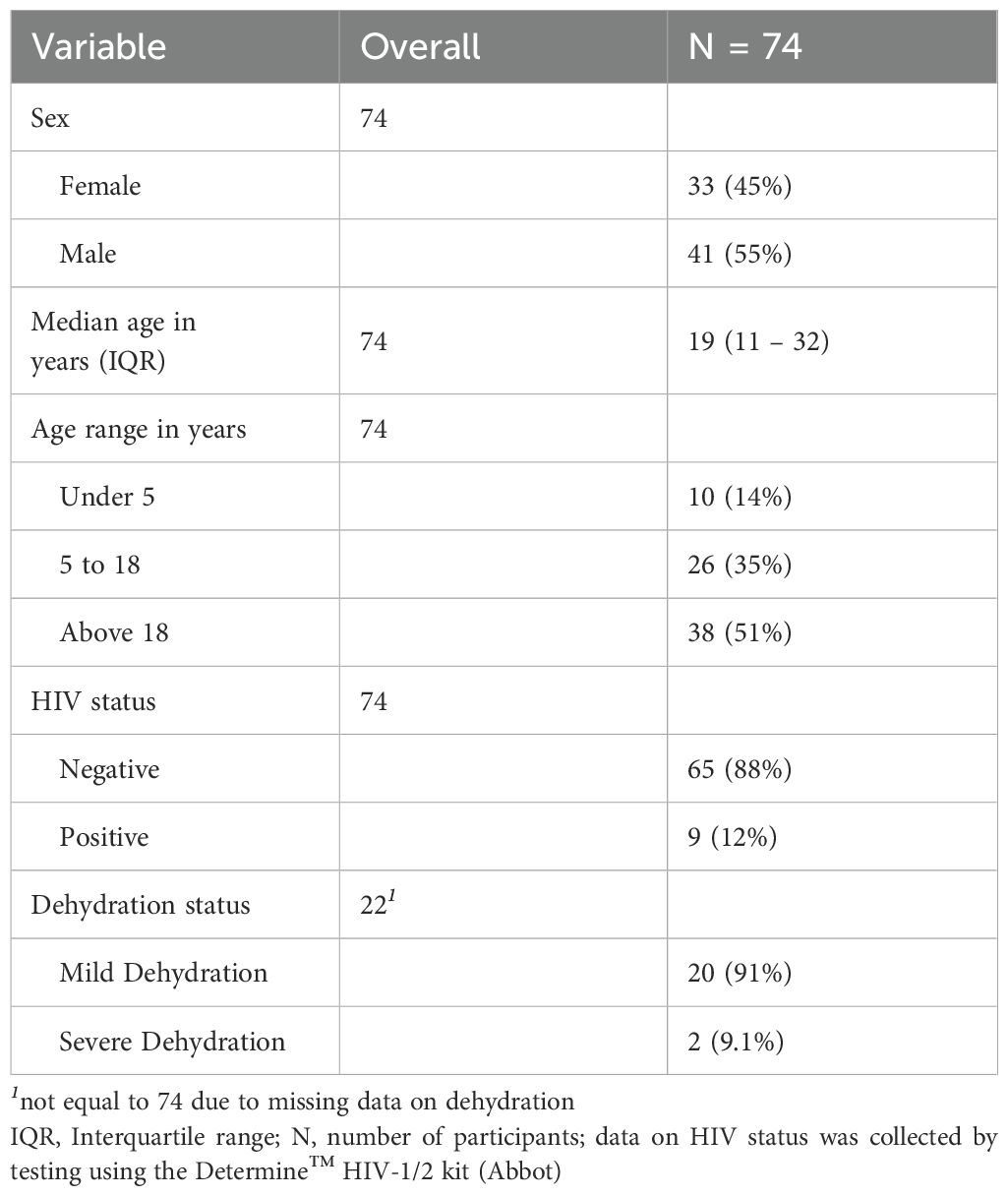
Plasma and saliva samples were obtained from 74 individuals who tested positive for cholera using a Rapid Diagnostic Test (RDT). Of these, 41 were male (55%), with a median age of 19 years; 38 were above the age of 18 (51%), and 9 were persons living with HIV/AIDS (PLWHA, 12%) (Table 1). A majority of our participants had missing data on hydration status, limiting meaningful interpretation on the impact of the severity of infection on antibody profiles.
Antibody profiles across plasma and saliva
We first explored the distribution of raw antibody subclasses specific to key V. cholerae antigens, including sialidase, CtxB, HlyA, TcpA, and Ogawa OSP across the different sample types. A brief description of these antigens is in the Supplementary Table 1. All participants in this study were infected with V. cholerae O1, serotype Ogawa. Antibody responses in plasma were predominantly IgG, while in saliva IgA was the predominant antibody isotype, across different antigens. Moreover, while IgA2 levels in plasma were lower than IgA1, in saliva levels of both IgA isotypes were comparable. Nonetheless, we detected IgA in plasma and IgG in saliva as well (Figure 1, Supplementary File 1).
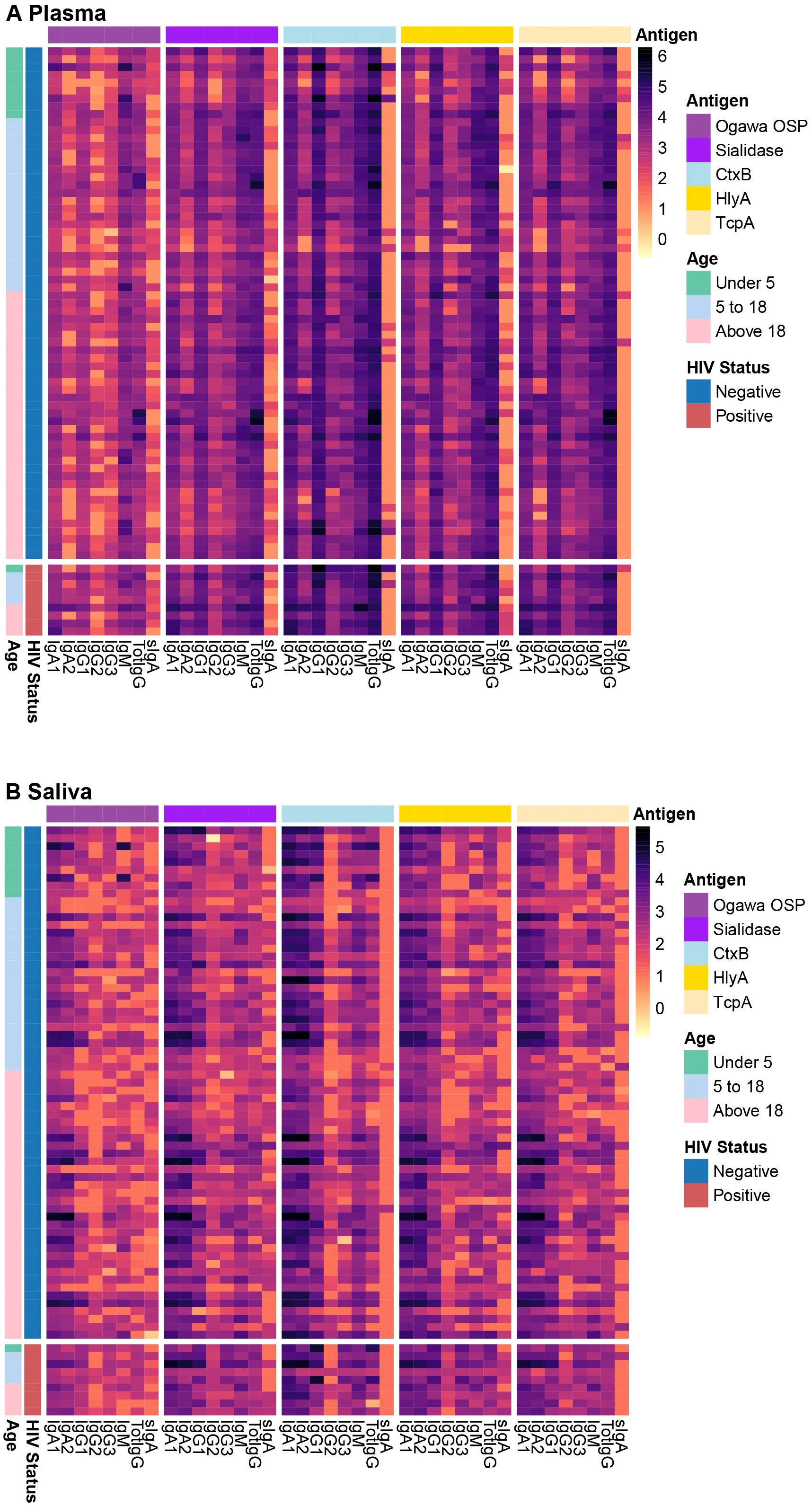
Figure 1. Plasma and saliva antibody profiles. The heatmap shows antibody profiles between (A) plasma and (B) saliva grouped by HIV status and age ranges in years. The rows represent individual samples, and colours represent the magnitude of antibody responses (log 10), with darker shades corresponding to higher response levels.
Correlation of antibody responses in plasma and saliva
We were then interested in assessing the interchangeability of biomarkers between plasma and saliva. For this, we performed Spearman correlation analyses for antibody responses across the two sample types. This approach allowed us to identify biomarkers consistently correlated in systemic and mucosal responses.
We observed that most immunological biomarkers exhibited positive correlations between plasma and saliva antibody levels. Notably, significant correlations were found for HlyA-specific responses across total IgG, IgG1, and IgG3 isotypes (Figure 2A). Also, the Ogawa OSP IgM response showed strong positive correlations between the two sample types (Figure 2B). Additionally, sialidase IgG2 levels showed significant positive correlations in both plasma and saliva, with its levels in saliva positively associated with all IgG subtypes and IgA1 in plasma (Figure 2C). In contrast, no significant correlations were observed between plasma and saliva for TcpA and CtxB-specific responses across the same antibody subclasses (Figure 2D, Supplementary Figure 1). However, we observed a significant positive correlation between IgG2 levels in saliva and IgA1 in plasma for TcpA (Figure 2D).
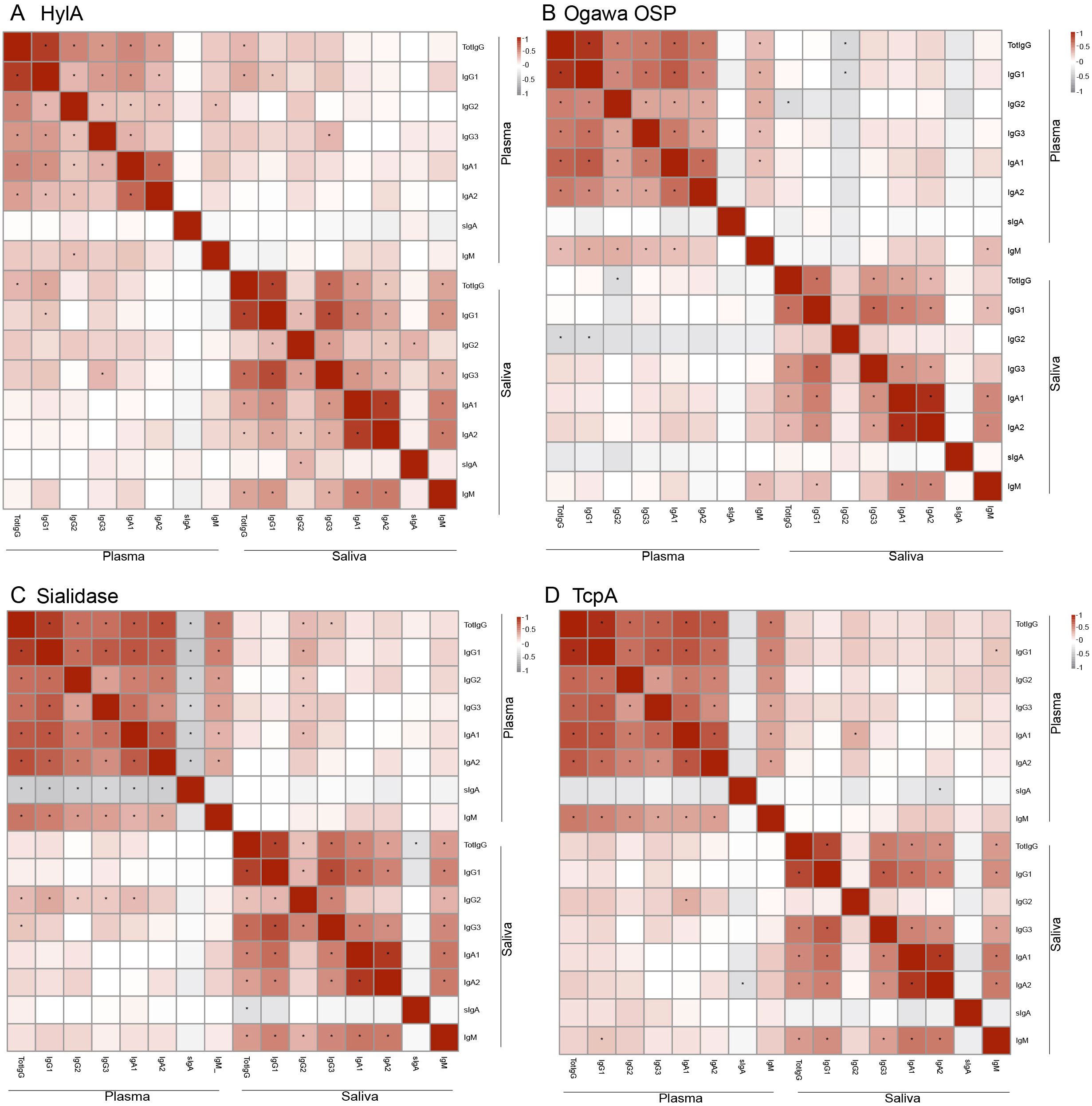
Figure 2. Correlation of biomarkers between plasma and saliva. (A) correlation of HylA subtypes. (B) correlation of Ogawa OSP subtypes. (C) correlation of Sialidase subtypes. (D) correlation of TcpA subtypes. The colours indicate Spearman correlation values, with positives shown in red and negatives in grey. *Show significant differences (p < 0.05).
Association of biomarkers with HIV status
Since 9 of our 72 participants tested positive to HIV, we explored the relationship between HIV status and the biomarkers identified as significantly correlated between plasma and saliva across the entire cohort. All 9 participants living with HIV were adults.
In plasma samples, people living with HIV/AIDS (PLWHA) showed significantly higher levels of HlyA-specific antibodies across multiple antibody classes, including IgG1 (log10 median: HIV-negative = 4.27, HIV-positive = 4.50, p.adj < 0.05), IgG3 (log10 median: HIV-negative = 3.37, HIV-positive = 4.17, p.adj < 0.001), and total IgG (log10 median: HIV-negative = 4.62, HIV-positive = 4.96, p.adj < 0.01). Conversely, no significant differences were observed for Ogawa OSP (IgM) and sialidase (IgG2), although both biomarkers were elevated in PLWHA (Figure 3A).
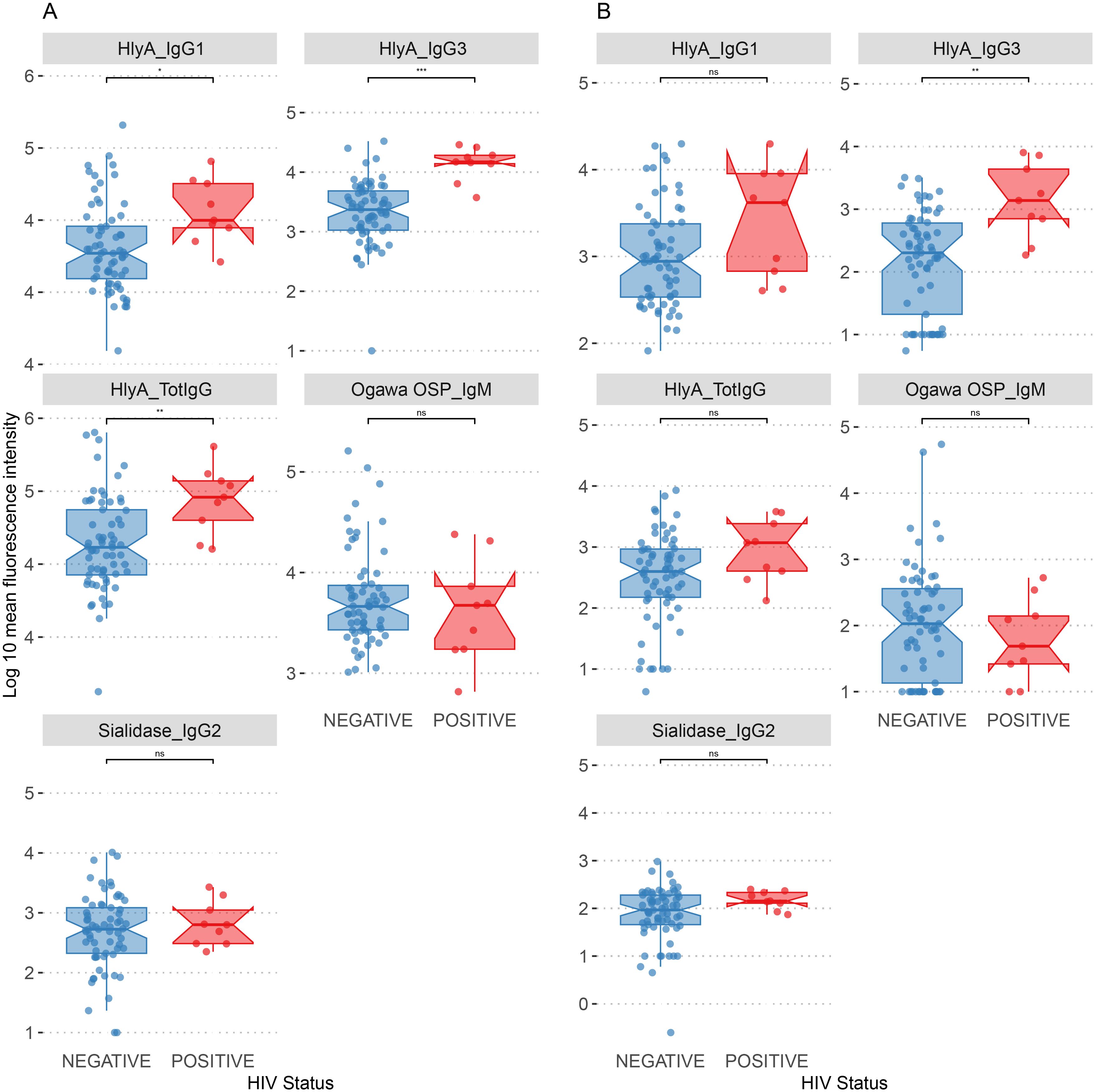
Figure 3. Distribution of biomarkers by HIV status. (A) Association of plasma biomarkers by HIV status. (B) Association of saliva biomarkers by HIV status. *p.adj < 0.05, **p.adj < 0.01, ***p.adj < 0.001, ns, not significant.
Most of the biomarkers in saliva did not differ significantly between the two groups. Notably, HlyA (IgG3) was the only biomarker that showed a significant increase in PLWHA (log10 median: HIV-negative = 2.30, HIV-positive = 3.14, p.adj < 0.05), suggesting an association of this antibody isotype in saliva samples with HIV status (Figure 3B).
Association of biomarkers with participant age
To investigate the influence of age on the biomarkers previously identified as significantly correlated in plasma and saliva, we examined age-related differences in antibody profiles. As demonstrated earlier, we excluded samples from PLWH because of their confounding effects. The analysis revealed distinct patterns in biomarker levels associated with participant age.
Trends in age-related differences were observed for most biomarkers in both plasma and saliva samples. Significant differences in plasma sialidase IgG2 levels were observed between participants under 5 years and those above 18 years, with mean fluorescence intensities of 1.98 and 2.695, respectively (p.adj < 0.05). Additionally, both age groups exhibited more significant variability in biomarker levels than other age categories (Figure 4A). Notably, all age groups showed higher variability in HlyA-specific antibody levels across IgG1 and total IgG. Similar trends were observed in saliva samples (Figure 4B) with no age group showing significant differences.
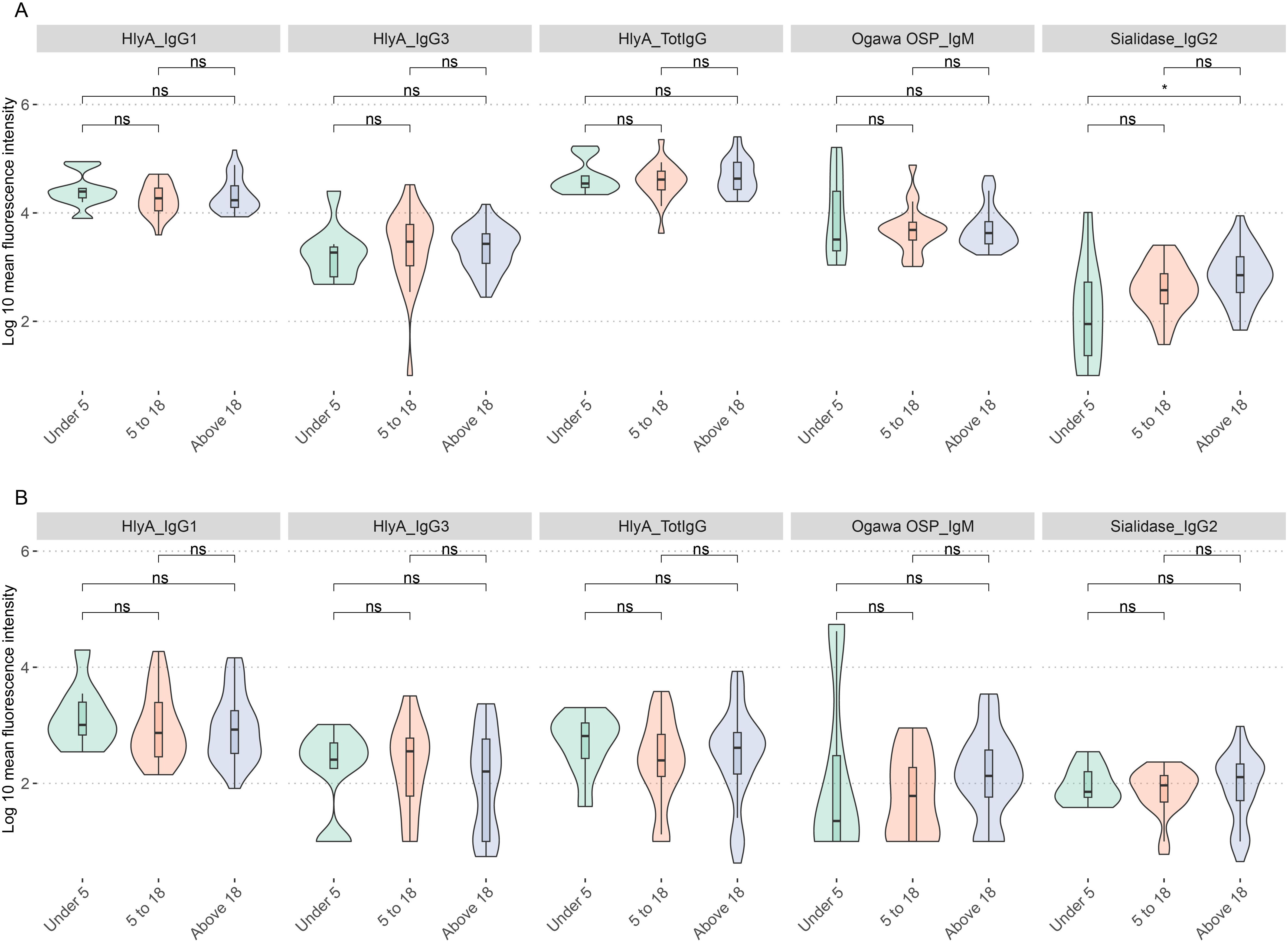
Figure 4. Distribution of biomarkers by age. (A) Age differences in biomarker distribution in plasma samples. (B) Age differences in biomarker distribution in saliva samples. ns show nonsignificant relationships while *represents significant differences p.adj < 0.05.
Antibody profiles previously found to correlate with protection against cholera
We then aimed to determine whether antibody profiles associated with protection against V. cholerae O1 infection could be detected in both saliva and plasma, as previously reported by Wiens et al. (2023) (14). We detected four key biomarkers in both sample types, namely CtxB IgM, TcpA IgG2, Ogawa OSP IgG1, and TcpA IgA1/2, which Wiens et al. (2023) showed outperform vibriocidal titres and are associated with a lower longitudinal risk of becoming infected (Figure 5). However, we did not assess antibody-dependent complement deposition (ADCD), another reported marker correlated with lower risk of infection, as it was not included in our screening protocol. Further analysis of these protective biomarkers in saliva showed high intensities of TcpA IgA1/2 and CtxB IgM across most samples, further highlighting their potential role in mucosal immunity (Figure 5B).
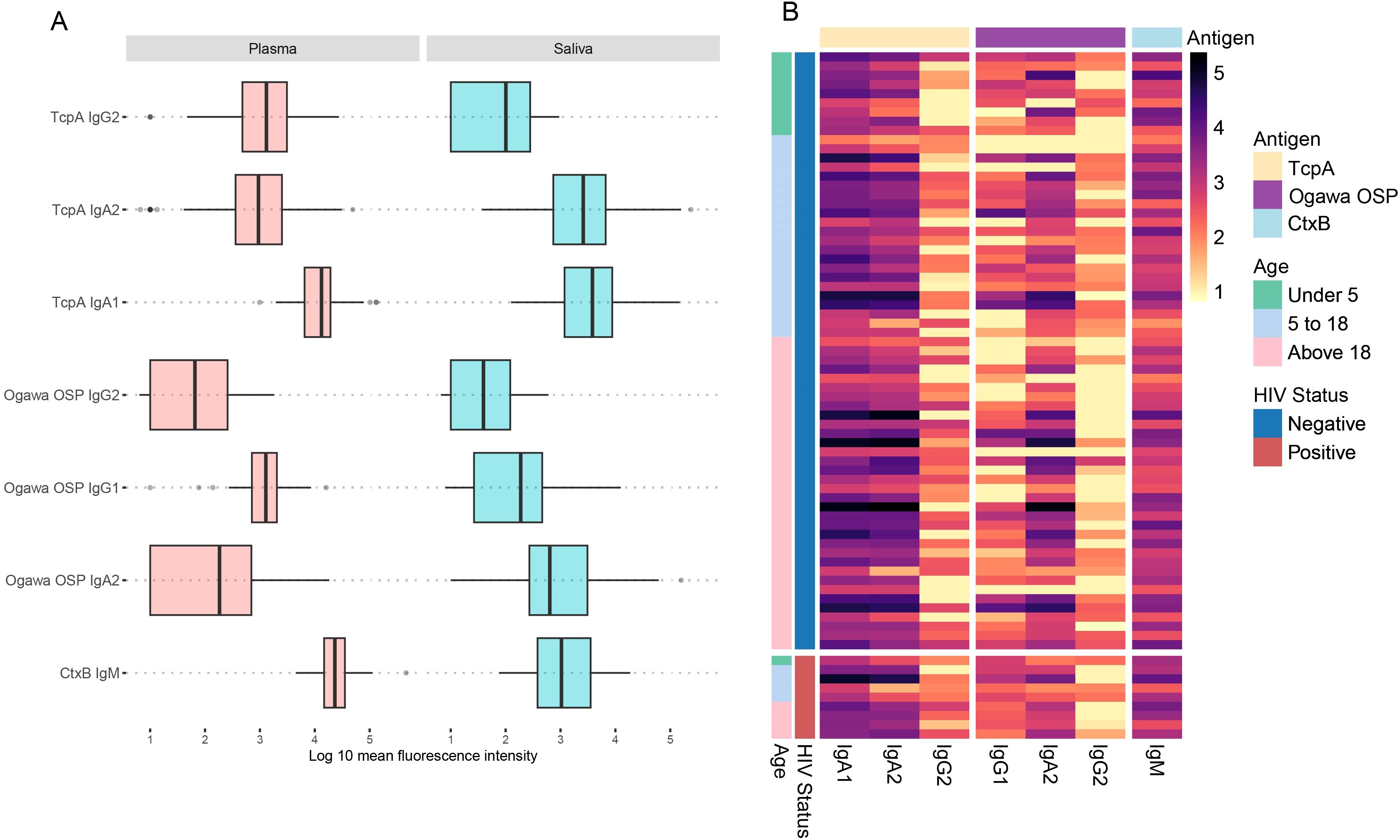
Figure 5. Antibody responses for composite immune responses previously found to correlate with protection against cholera in a Bangladeshi cohort. (A) Shows protective antibodies in plasma and saliva. (B) Heatmap showing the protective antibodies in saliva, grouped by HIV status and age ranges in years. The rows represent individual samples, and colours represent the magnitude of antibody responses (log 10), with darker shades corresponding to higher response levels.
Discussion
In this study, we report that cholera-specific plasma biomarkers can be detected in saliva. To the best of our knowledge, this is the first study to demonstrate a comprehensive multiplex approach of cholera-specific biomarkers in both plasma and saliva. We show differences in how some biomarkers are represented in systemic versus mucosal compartments, such as the dominance of IgG in plasma and IgA in saliva, suggesting that these antigens may elicit compartment-specific immune responses. In addition, identifying robust correlations between specific biomarkers in plasma and saliva, such as HlyA, Ogawa OSP, and sialidase, indicates saliva’s viability as an alternative sample to plasma. Saliva collection is less invasive and more culturally acceptable in field settings, particularly in LMICs like Zambia, where logistical challenges often hinder plasma collection.
Our findings align with earlier studies demonstrating the feasibility of saliva-based antibody testing for cholera (7, 8). However, using an antibody profiling approach to provide a multiplexed and high-dimensional analysis of antibody biomarkers in plasma and saliva represents a novel contribution. While prior studies have primarily focused on plasma biomarkers (4, 14), this research bridges a critical gap by validating saliva as a potential alternative sample type. Moreover, the observed compartment-specific immune responses point to saliva’s unique insights into mucosal immunity, complementing systemic analyses.
We report a positive correlation between HlyA-specific antibodies in both plasma and saliva. HlyA is a toxin secreted by gram-negative bacteria, such as strains of Vibrio cholerae, which plays a critical role in pathogenesis and immune responses during infection. It is involved in innate immunity following infection, which can trigger the release of pro-inflammatory cytokines such as IL-1β (17). While less attention has been given to HlyA in cholera immunogenicity studies, a prior study showed that the levels of hemolysin A were correlated with the severity of Vibrio cholerae infection in nematodes (18). Current whole-cell-killed vaccines typically target primarily the O-specific polysaccharide of V. cholerae O1. Thus, HlyA could serve as a supplementary immunogen to enhance vaccine effectiveness.
We also observed high levels of sialidase antibodies in plasma and saliva, a neuraminidase that facilitates the binding of cholera toxin to epithelial cells. Previous studies report that sialidase plays a role in cholera immune responses and has been associated with protection against cholera infection (19). An intriguing observation was the positive correlation of IgG2 in saliva with IgA1 in plasma. IgG2 subclass plays an important role in immune responses to carbohydrate antigens such as lipopolysaccharides present in V. cholerae; thus, its presence in saliva may reflect mucosal immune activation (20). On the other hand, IgA1, the predominant IgA subclass in plasma, has been associated with protection against cholera and is involved in mucosal defence (21). Although we may not fully understand this relation, this may suggest immunological cross-talk between systemic and mucosal compartments, where IgG2 could serve as a surrogate marker for plasma IgA1 responses. We observed a positive correlation in Ogawa-specific IgM in both sample types. Ogawa IgM levels are important in predicting recent V. cholerae infection, hence this may be a useful biomarker in outbreak settings for identifying recent infections to track disease transmission.
Immunocompromised individuals present a clinically important subgroup in cholera immunogenicity. One such group is PLWHA, where the epidemiological consequence of the coexistence of HIV and cholera is sparse in endemic regions. Our earlier study showed that individuals who were living with HIV had lower vibriocidal antibody titres compared to individuals not living with HIV following vaccination (22). In this study, we show a trend that PLWHA have higher responses to biomarkers that were positively correlated between plasma and saliva. We report significantly higher plasma levels of HlyA-specific IgG1, IgG3 and total IgG, alongside a rise in salivary HlyA-IgG3 in PLWHA. A plausible explanation could be that HIV infection is frequently accompanied by gingivitis, periodontitis, aphthous ulceration and oral candidiasis, all of which compromise epithelial integrity, potentially increasing both antigen ingress and leakage of plasma-derived antibodies into saliva (23–25). Interestingly, our results suggest that antibodies in saliva could be a more stable readout as they were seemingly not influenced by HIV status. However, since these measurements were taken at a single timepoint, additional longitudinal sampling would be required to determine whether the heightened HlyA response reflects prolonged shedding or has functional protective value and to confirm the stability of the biomarkers in saliva. Beyond HIV status, our age-stratified analysis did not report any significant differences in plasma and saliva samples but for sialidase, which showed significant differences in its presence by age. This observation has been reported previously, in which adults were more likely to have sialidase responses in plasma than children (26, 27).
Finally, identifying biomarkers in saliva, previously reported to be important in cholera infection, is a significant finding. We observed high antibody intensities in saliva for two major V. cholerae antigens, TcpA and CtxB. TcpA, an adhesion factor important in intestinal colonisation, has been associated with protective immunity (14). Given its strong intensity in saliva, TcpA-specific IgA1/2 could serve as a useful non-invasive marker of cholera infection. On the other hand, while CtxB is a critical virulence factor in V. cholerae pathogenesis, its role in protection remains unclear. While IgM is not the predominant immunoglobulin in saliva, low but measurable amounts are routinely detected. Polymeric IgM may be actively transported across the mucosal epithelium via the polymeric−Ig receptor, while serum−derived IgM can transude through crevicular fluid during acute inflammation (28). Thus, detection of CtxB−specific IgM may reflect an early mucosal response, making it a potential non−invasive marker of acute cholera. Although Haney et al. (2018) (29) did not find significant correlations between CtxB responses and protection in a human challenge model, these responses were associated with protection against cholera infection in a cohort study conducted in Bangladesh (14). This discrepancy may suggest that CtxB’s direct role in protection requires further investigation to confirm its relevance as a correlate of protection. We also observed high levels of Ogawa OSP IgM in saliva, an important marker of protective immunity, which has been reported to cross-react with Inaba OSP (30). Thus, elevated salivary Ogawa OSP IgM, could serve as a plausible non-invasive marker of a recent infection.
While we provide compelling evidence for saliva-based antibody profiling following cholera infection, our study has some limitations. Firstly, the small sample size, though powered for the initial analysis, limits the generalizability of our results. Also, future work should expand biomarker coverage to include other virulence and structural antigens such as OmpV, CtxA, and flagellar proteins, which may elicit mucosal responses relevant to infection and immunity. The variability in biomarker sensitivity across age groups and the incomplete hydration status data further emphasise the need for larger and more comprehensive studies. Additionally, a longitudinal study would have allowed us to track immune responses over time instead of the cross-sectional design employed here. Lastly, while we included PLWHA, a more detailed analysis of immune suppression effects, including stratification by CD4 count and viral load, would provide deeper insights into how mucosal immunity is altered in immunocompromised populations. Future studies should address these limitations to strengthen saliva-based sero-surveillance and vaccine evaluation.
The adoption of saliva-based diagnostics could significantly enhance outbreak preparedness and response strategies by enabling a rapid and non-invasive sample collection in both clinical and community settings. Saliva collection is more suitable for vulnerable populations, including children, and can be scaled rapidly during cholera outbreaks. Additionally, this approach reduces the need for specialised equipment and trained personnel, as required in plasma collection, potentially lowering overall costs and increasing accessibility in remote regions. These advantages align with the global health goals of equitable access to practical diagnostic tools.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by the University of Zambia Biomedical Research Ethics Committee (UNZABREC), reference number 001-02-23, and the National Health Research Authority (NHRA). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
Author contributions
CL: Writing – original draft, Writing – review & editing, Formal Analysis, Conceptualization, Methodology, Visualization. HN: Methodology, Project administration, Investigation, Writing – review & editing. FL: Investigation, Writing – review & editing, Methodology, Project administration. SB: Formal Analysis, Methodology, Supervision, Software, Writing – review & editing, Investigation. JZ: Writing – review & editing, Validation, Data curation, Methodology. JH: Resources, Supervision, Validation, Methodology, Writing – review & editing. RC: Resources, Formal Analysis, Methodology, Supervision, Writing – review & editing. ER: Methodology, Supervision, Formal Analysis, Resources, Writing – review & editing. DS: Methodology, Supervision, Writing – review & editing, Conceptualization, Project administration. BB: Resources, Formal Analysis, Supervision, Writing – original draft, Writing – review & editing, Conceptualization, Investigation. CC: Funding acquisition, Writing – original draft, Project administration, Supervision, Writing – review & editing, Conceptualization.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This research was funded by Science for Africa Foundation to the Developing Excellence in Leadership, Training and Science in Africa (DELTAS Africa) programme DEL-22–012 with support from Wellcome Trust and the UK Foreign, Commonwealth & Development Office, and is part of the EDCTP2 programme supported by the European Union. For purposes of open access, the author has applied a CC BY public copyright license to any Author Accepted Manuscript version arising from this submission.
Acknowledgments
We want to express our sincere gratitude to the Zambian Ministry of Health workers who dedicated their time and effort during the outbreak, tirelessly managing participants and collecting samples.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1641319/full#supplementary-material
References
1. Multi-country outbreak of cholera, External situation report 19–18 October 2024 . Available online at: https://www.who.int/publications/m/item/multi-country-outbreak-of-cholera–external-situation-report–19—18-october-2024 (Accessed Dec 8, 2024).
2. Mwaba J, Debes AK, Shea P, Mukonka V, Chewe O, Chisenga C, et al. Identification of cholera hotspots in Zambia: A spatiotemporal analysis of cholera data from 2008 to 2017. PloS Negl Trop Dis. (2020) 14:e0008227. doi: 10.1371/journal.pntd.0008227
3. Kateule E, Nzila O, Ngosa W, Mfume F, Shimangwala C, Gama A, et al. Multisectoral approach for the control of cholera outbreak - lessons and challenges from Lusaka district - Zambia, October 2023 - February 2024. Pan Afr Med J. (2024) 48:19. doi: 10.11604/pamj.2024.48.19.43659
4. Harris JB. Cholera: immunity and prospects in vaccine development. J Infect Dis. (2018) 218:S141–6. doi: 10.1093/infdis/jiy414
5. Iyer AS and Harris JB. Correlates of protection for cholera. J Infect Dis. (2021) 224:S732–7. doi: 10.1093/infdis/jiab497
6. Lim PW, Garssen J, and Sandalova E. Potential use of salivary markers for longitudinal monitoring of inflammatory immune responses to vaccination. Mediators Inflamm. (2016) 2016:6958293. doi: 10.1155/2016/6958293
7. Chisenga CC, Phiri B, Ng’ombe H, Muchimba M, Liswaniso F, Bernshtein B, et al. Diagnostic accuracy of saliva-based testing as a Vibrio cholerae surveillance tool among naturally infected patients. Clin Exp Immunol. (2024) 219(1):uxae092. doi: 10.1093/cei/uxae092
8. Jertborn M, Svennerholm AM, and Holmgren J. Saliva, breast milk, and serum antibody responses as indirect measures of intestinal immunity after oral cholera vaccination or natural disease. J Clin Microbiol. (1986) 24:203–9. doi: 10.1128/jcm.24.2.203-209.1986
9. Qadri F, Ryan ET, Faruque ASG, Ahmed F, Islam Khan A, Islam MM, et al. Antigen-specific immunoglobulin A antibodies secreted from circulating B cells are an effective marker for recent local immune responses in patients with cholera: comparison to antibody-secreting cell responses and other immunological markers. Infect Immun. (2003) 71:4808–14. doi: 10.1128/IAI.71.8.4808-4814.2003
10. Akter A, Dash P, Aktar A, Jahan SR, Afrin S, Basher SR, et al. Induction of systemic, mucosal and memory antibody responses targeting Vibrio cholerae O1 O-specific polysaccharide (OSP) in adults following oral vaccination with an oral killed whole cell cholera vaccine in Bangladesh. PloS Negl Trop Dis. (2019) 13:e0007634. doi: 10.1371/journal.pntd.0007634
11. Chung AW and Alter G. Systems serology: profiling vaccine induced humoral immunity against HIV. Retrovirology. (2017) 14:57. doi: 10.1186/s12977-017-0380-3
12. Loos C, Coccia M, Didierlaurent AM, Essaghir A, Fallon JK, Lauffenburger D, et al. Systems serology-based comparison of antibody effector functions induced by adjuvanted vaccines to guide vaccine design. NPJ Vaccines. (2023) 8:1–15. doi: 10.1038/s41541-023-00613-1
13. Brown EP, Weiner JA, Lin S, Natarajan H, Normandin E, Barouch DH, et al. Optimization and qualification of an Fc Array assay for assessments of antibodies against HIV-1/SIV. J Immunol Methods. (2018) 455:24–33. doi: 10.1016/j.jim.2018.01.013
14. Wiens KE, Iyer AS, Bhuiyan TR, Lu LL, Cizmeci D, Gorman MJ, et al. Predicting Vibrio cholerae infection and symptomatic disease: a systems serology study. Lancet Microbe. (2023) 4:e228–35. doi: 10.1016/S2666-5247(22)00391-3
15. Gu Z, Eils R, and Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. (2016) 32:2847–9. doi: 10.1093/bioinformatics/btw313
16. Benjamini Y and Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. (1995) 57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
17. Mamantopoulos M, Frising UC, Asaoka T, van Loo G, Lamkanfi M, and Wullaert A. El tor biotype vibrio cholerae activates the caspase-11-independent canonical nlrp3 and pyrin inflammasomes. Front Immunol. (2019) 10:2463/full. doi: 10.3389/fimmu.2019.02463/full
18. Cinar HN, Kothary M, Datta AR, Tall BD, Sprando R, Bilecen K, et al. Vibrio cholerae Hemolysin Is Required for Lethality, Developmental Delay, and Intestinal Vacuolation in Caenorhabditis elegans. PloS One. (2010) 5:e11558. doi: 10.1371/journal.pone.0011558
19. Kaisar MH, Bhuiyan MS, Akter A, Saleem D, Iyer AS, Dash P, et al. Vibrio cholerae Sialidase-Specific Immune Responses Are Associated with Protection against Cholera. mSphere. (2021) 6:e01232–20. doi: 10.1128/mSphere.01232-20
20. von Gunten S, Smith DF, Cummings RD, Riedel S, Miescher S, Schaub A, et al. IVIg contains a broad repertoire of anti-carbohydrate antibodies that is not restricted to the IgG2 subclass. J Allergy Clin Immunol. (2009) 123:1268–76.e15. doi: 10.1016/j.jaci.2009.03.013
21. Alam MM, Riyadh MA, Fatema K, Rahman MA, Akhtar N, Ahmed T, et al. Antigen-specific memory B-cell responses in Bangladeshi adults after one- or two-dose oral killed cholera vaccination and comparison with responses in patients with naturally acquired cholera ▿. Clin Vaccine Immunol CVI. (2011) 18:844–50. doi: 10.1128/CVI.00562-10
22. Luchen CC, Mwaba J, Ng’ombe H, Alabi PIO, Simuyandi M, Chilyabanyama ON, et al. Effect of HIV status and retinol on immunogenicity to oral cholera vaccine in adult population living in an endemic area of Lukanga Swamps, Zambia. PloS One. (2021) 16:e0260552. doi: 10.1371/journal.pone.0260552
23. Heron SE, Elahi S, and Infection HIV. Compromised mucosal immunity: oral manifestations and systemic inflammation. Front Immunol. (2017) 8:241/full. doi: 10.3389/fimmu.2017.00241/full
24. Mataftsi M, Skoura L, and Sakellari D. HIV infection and periodontal diseases: an overview of the post-HAART era. Oral Dis. (2011) 17:13–25. doi: 10.1111/j.1601-0825.2010.01727.x
25. Lomelí-Martínez SM, González-Hernández LA, Ruiz-Anaya A de J, Lomelí-Martínez MA, Martínez-Salazar SY, Mercado González AE, et al. Oral manifestations associated with HIV/AIDS patients. Medicina (Mex). (2022) 58:1214. doi: 10.3390/medicina58091214
26. Jones FK, Bhuiyan TR, Muise RE, Khan AI, Slater DM, Hutt Vater KR, et al. Identifying recent cholera infections using a multiplex bead serological assay. mBio. (2022) 13:e01900–22. doi: 10.1128/mbio.01900-22
27. Kaisar MH, Bhuiyan MS, Akter A, Saleem D, Iyer AS, Dash P, et al. Vibrio cholerae Sialidase-Specific Immune Responses Are Associated with Protection against Cholera. mSphere. (2021) 6:e01232–20. doi: 10.1128/mSphere.01232-20
28. Michaud E, Mastrandrea C, Rochereau N, and Paul S. Human secretory igM: an elusive player in mucosal immunity. Trends Immunol. (2020) 41:141–56. doi: 10.1016/j.it.2019.12.005
29. Haney DJ, Lock MD, Gurwith M, Simon JK, Ishioka G, Cohen MB, et al. Lipopolysaccharide-specific memory B cell responses to an attenuated live cholera vaccine are associated with protection against Vibrio cholerae infection. Vaccine. (2018) 36:2768–73. doi: 10.1016/j.vaccine.2018.04.011
Keywords: cholera, saliva, plasma, antibody, immunology
Citation: Luchen CC, Ngo’mbe H, Liswaniso F, Bosomprah S, Zhiteneva JA, Harris J, Charles RC, Ryan ET, Sack D, Bernshtein B and Chisenga CC (2025) Vibrio cholerae-specific antibodies in plasma and saliva in cholera patients during a severe outbreak in Zambia: an antibody profiling approach. Front. Immunol. 16:1641319. doi: 10.3389/fimmu.2025.1641319
Received: 04 June 2025; Accepted: 31 July 2025;
Published: 15 August 2025.
Edited by:
Mats Bemark, Lund University, SwedenReviewed by:
Amit K. Singh, Albany Medical College, United StatesJuandy Jo, University of Pelita Harapan, Indonesia
Copyright © 2025 Luchen, Ngo’mbe, Liswaniso, Bosomprah, Zhiteneva, Harris, Charles, Ryan, Sack, Bernshtein and Chisenga. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Charlie C. Luchen, Y2x1Y2hlbi5sdWNoZW5AZ21haWwuY29t
 Charlie C. Luchen
Charlie C. Luchen Harriet Ngo’mbe2
Harriet Ngo’mbe2 Richelle C. Charles
Richelle C. Charles Edward T. Ryan
Edward T. Ryan David Sack
David Sack Caroline C. Chisenga
Caroline C. Chisenga