Abstract
Background:
Osteoporosis has become an increasingly pressing global public health challenge. Monoclonal antibody romosozumab (ROMO), which targets sclerostin (SOST), a critical inhibitor of bone formation, demonstrates considerable therapeutic efficacy. However, its relatively high cost and potential cardiovascular risks may hinder broader clinical application. Current preventive measures remain inadequate.
Methods:
This study presents a novel, cost-effective osteoporosis vaccine with dual preventive and therapeutic capabilities, derived from the high-affinity binding epitope of ROMO to SOST. ELISA screening determined that the SOST131–163 region within loop3 domain serves as the primary epitope for ROMO, suggesting a role in skeletal regulation with minimal impact on cardiovascular system. SOST131–163 was conjugated to the diphtheria toxin translocation domain (DTT) to create novel SOST-targeted vaccines.
Results:
Immunogenicity assays demonstrated that both DDT-SOST(131-163)3 (DS3) and DDT-SOST(131-163)5 (DS5) elicited strong IgG2 antibody responses comparable to ROMO. Molecular docking studies indicated strong affinities of DS3 and DS5 for Toll-like receptor 2 (TLR2), enhancing TLR2-mediated humoral B-cell immunity and eliciting synergistic T-helper cell responses. Recombinant expression in Escherichia coli confirmed the successful production of DS3 and DS5, with molecular weights of 31.8 kDa and 40.3 kDa, respectively. In vivo experiments showed that the vaccines effectively induced high-titer anti-SOST antibodies in mice, overcoming immune tolerance. Additionally, cell-based assays indicated that antiserum from vaccinated mice inhibited osteoclast differentiation and promoted osteoblast mineralization.
Conclusion:
The SOST-targeted vaccination strategy offers a promising and cost-effective approach for the early prevention and sustained management of osteoporosis, demonstrating substantial potential for clinical translation.
Introduction
Osteoporosis (OP) is a prevalent degenerative bone disease defined by diminished bone mass and an elevated risk of fractures, posing a significant global health challenge (1, 2). It affects approximately one-third of women and one-fifth of men over the age of 50, with prevalence anticipated to rise population ages (2, 3). The disorder results from an imbalance in bone remodeling, where bone resorption outpaces bone formation. Current therapies primarily focus on promoting bone formation, such as teriparatide, or inhibiting resorption with agents like alendronate and denosumab (4). Although romosozumab (ROMO), a dual-action monoclonal antibody, has demonstrated promising efficacy, its high cost and cardiovascular adverse events may limit broader application (1, 5, 6). Furthermore, existing preventative strategies do not effectively address early intervention, underscoring the pressing need for innovative therapies (1).
Sclerostin (SOST) is a critical negative regulator of osteoblast differentiation, primarily inhibiting the Wnt signaling pathway, thus decreasing bone formation and indirectly promoting osteoclastogenesis (7). An agent targeting SOST, such as ROMO, offers a dual mechanism for modulating bone dynamics (8); however, its antibody-based design poses challenges in both safety and affordability (5, 9). Consequently, there is growing interest in vaccine-based strategies aimed at achieving safe, sustained preventive and therapeutic effects through the induction of long-lasting endogenous antibody production (10, 11). Nonetheless, the development of SOST vaccines encounters two primary hurdles: identifying effective antigenic epitopes and overcoming immune tolerance to self-proteins (12).
Building upon our previous successes in addressing immune tolerance in osteoporosis vaccine development (10), we propose a novel vaccine strategy that integrates the SOST protein with a diphtheria toxin translocation domain (DTT) as an adjuvant scaffold. Our approach commenced with the identification of the high-affinity binding domain of SOST through ROMO, followed by its conjugation to DTT protein to create a subunit vaccine. We performed comprehensive physicochemical characterization, validated mass producibility via recombinant expression in E. coli, and evaluated immunogenicity along with in vivo efficacy in antibody induction. This strategy is designed to achieve three primary objectives: (1) Confirming antigen validity by identifying effective SOST epitopes for ROMO targeting; (2) Overcoming immune tolerance with DTT scaffold to enhance antibody induction; and (3) Developing a cost-effective and scalable osteoporosis vaccine to enable early intervention and sustained therapeutic benefits.
2 Materials and methods
2.1 SOST peptide fragments
Peptides were synthesized based on the human SOST (GenBank: AAK16158.1) fragment encompassing amino acids 24 to 211, excluding the signal peptide, yielding approximately 30-amino-acid segments. An indirect enzyme-linked immunosorbent assay (ELISA) was performed to identify a high-affinity SOST peptide fragment for ROMO. The polypeptides were synthesized by Sangon Biotech (Shanghai, China).
2.2 Enzyme-linked immunosorbent assay (ELISA) for peptide screening
96-well plates (Abcam, Cambridge, MA, USA) were coated with 100 µL of 1 µg/mL human SOST-synthesized polypeptides in the provided coating buffer overnight at 4 °C. Following coating, the plates were washed with 1× washing buffer and subsequently blocked using 1× blocking buffer. Diluted ROMO (AntibodySystem, Schiltigheim, France) was then added to the wells and incubated for 2 hour at 37 °C. After additional washing, 100 µL of 1:10,000 diluted horseradish peroxidase (HRP)-labeled goat anti-human antibodies (Bioss, Beijing, China) was introduced to the wells, followed by another 2-hour incubation at 37 °C. Finally, 100 µL of TMB was dispensed into each well, and after a 20-minute incubation at 37 °C, the absorbance was measured at 450 nm.
The high-affinity SOST peptide fragment for ROMO was then identified using ELISA. The binding affinity of the high-affinity SOST fragment to the heavy and light chains of ROMO was assessed using the HawkDock server (13), while interactions between the SOST fragment and ROMO were analyzed with PDBsum (14).
2.3 Prediction of T cell and B cell epitope
Prediction of major histocompatibility complex (MHC) class I-restricted cytotoxic T lymphocyte (CTL) epitopes for the high-affinity SOST fragment was performed using NetMHCpan 4.1 EL tool (15). A comprehensive analysis of 9-mer epitopes was conducted across the A1, A2, A3, A24, and B7 supertypes. Epitopes with a percentage rank (< 0.5%) were categorized as strong binders (SB), whereas those with a percentage rank (< 2%) were classified as weak binders (WB). In parallel, the identification of helper T lymphocyte (HTL) epitopes, comprising 15-mer peptides that bind to MHC class II, was accomplished using the NetMHCIIpan 4.1 EL server (16), with strong binders defined as having a percentage rank (< 1%) and weak binders defined as having a percentage rank (< 5%).
To predict linear B cell epitopes within the high-affinity SOST fragment, we employed BepiPred2.0 tool (17), applying a default filtering threshold of 0.5. Additionally, conformational B cell epitopes for both the screening peptide and the designed vaccines were predicted using the ElliPro application (18), with a minimum score threshold set at 0.5 and a maximum distance of 6 Angstroms.
2.4 Construction and prediction of candidate vaccines
The high-affinity SOST fragment was selected as the target antigen for the development of a recombinant subunit vaccine. Based on our previous experimental experience (10), the DTT fragment (amino acids 203–378; WP_371890660.1) was chosen as immune scaffold to facilitate conjugation with SOST fragment, thereby enhancing immune recognition and promoting antibody production. The DTT scaffold was conjugated with varying copy numbers of SOST peptide, ranging from 0 to 5, to generate a series of chimeric molecules. These conjugations were connected via a (GGGGS)2 flexible linker to ensure optimal conformational flexibility. The resulting vaccine candidates were designated as DDT-SOST(131-163)0 (DS0) to DDT-SOST(131-163)5 (DS5), with the numerical suffix denoting specific number of SOST peptide fragments fused to DTT scaffold.
Tertiary structure of candidate vaccines was predicted using AlphaFold2 server (19), based on their amino acid sequences. Five structural models were generated, demonstrating close alignment with experimental accuracy. Top-ranked model was selected for further analysis. The quality of vaccine structure was assessed using the predicted Local Distance Difference Test (pLDDT).
2.5 Immune response simulation
To evaluate the immunogenic potential of candidate vaccines, we employed the C-ImmSim server (20), a platform capable of simulating immune responses. This computational tool mimics the activation of B and T lymphocytes following hypothetical vaccine administration, allowing for the exploration of immune response dynamics. The simulation parameters were configured as follows: Random Seed = 12,345, Simulation Volume = 10, Simulation Steps = 240, and HLA selections: A0101, B0702, and DRB1_0101. The simulation framework was designed to include three administrations of 400 antigens, each spaced by a two-week interval. Each time step was delineated to represent an elapsed duration of 8 hours in real-world time, leading to time periods set at 1, 42, and 84. The simulation predicted the cellular immune responses provoked by candidate vaccines, encompassing antibody production, B cell and T cell activation, and cytokine release. The vaccine demonstrating the highest titer of IgG2 antibody were subsequently selected for further analysis, given that the IgG2 subtype is known to mediate the function against SOST in ROMO (21).
2.6 Prediction of immunological and physicochemical properties
Immunological properties of the selected vaccines were systematically evaluated. Allergenicity assessments were performed using AllerTOP v.2.1 server (22), while antigenic potential was analyzed with VaxiJen v2.0 server (23). To assess solubility of the vaccines, SOLpro server (24) was employed. Additionally, the physicochemical properties—including chemical formula, total atom count, molecular weight, theoretical isoelectric point (pI), half-life, instability index, aliphatic index, and the grand average of hydropathicity (GRAVY)—were predicted using ExPASy ProtParam tool (25).
2.7 Prediction and analysis of secondary structure
Secondary structure elements of the selected vaccines, including α-helices, extended strands, β-turns, and random coils, were predicted using SOPMA server (26) and PSIPRED web server (27). For these predictions, all parameters were maintained at their default settings. Additionally, the solubility characteristics of the selected vaccines were assessed using Protein-Sol server (28).
2.8 Refinement and validation of tertiary structure
Top-ranked model of tertiary structure for the selected vaccines, generated by AlphaFold2, was refined using GalaxyRefine web server (29). This refinement yielded reliable core structures based on multiple templates, while less reliable loops and terminal regions were constructed through optimization-based modeling. The structural quality of the refined vaccine model was further assessed using ProSA-web (30), ERRAT (31) and PROCHECK (32).
2.9 Molecular docking and molecular dynamic simulations
Molecular docking analyses were conducted using HawkDock server (13) to evaluate the interactions between vaccine candidates and Toll-Like Receptor 2 (TLR2) immune receptor (PDB ID: 6NIG). This platform organizes docking models based on surface complementarity and clustering characteristics. The highest-ranking model derived from the docking evaluations was selected for further analysis and visualized with PyMOL software. Binding energy and interaction surfaces within the docking complex were assessed using Prodigy (33), PDBePISA (34), and PDBsum (14).
Molecular dynamics simulations of the vaccine-TLR2 docking complex were performed utilizing the internal coordinate normal mode analysis server (iMODS) (35). This platform employs Normal Mode Analysis (NMA) in internal coordinates to identify collective motions that are critical for the functional dynamics of macromolecules. iMODS provides interactive tools for visualizing these modes, including vibration analysis, motion animations, and morphing trajectories.
2.10 Vaccines cloning, expression and immunization
Codon-optimized cDNA sequences for the selected vaccine candidates were generated in silico using the Java Codon Adaptation Tool (JCAT) (36). Optimized sequences were then cloned into pSmartI plasmids. Following cloning, the recombinant plasmids were transformed into Escherichia coli BL21 (DE3) for protein expression. The resulting recombinant proteins were purified through a two-step chromatography process, which involved ion exchange chromatography followed by gel filtration chromatography. Purity and quality of protein products were assessed using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Sangon Biotech, Shanghai, China).
For immunization studies, C57BL/6J mice (purchased from Guangdong Medical Laboratory Animal Center, China) received two subcutaneous injections of 200 µg of each vaccine candidate, administered two weeks apart(n=3). Freund’s Complete Adjuvant was used for the initial dose, while Freund’s Incomplete Adjuvant (Sigma, USA) was employed for the booster injection. Blood samples were collected five weeks after the final immunization, and anti-SOST antibody titers were measured using ELISA.
2.11 Detection of anti-SOST antibodies in vaccinated mice
The antiserum from vaccine-immunized mice was obtained. The titers of specific anti-SOST antibodies were assessed using an indirect ELISA. In brief, 1 µg/mL of human SOST protein (MedChemExpress Inc.) was coated onto the wells of MaxiSorp microtiter plates (Thermo Fisher Scientific Inc.) and incubated overnight at 4°C. Mouse serum samples were diluted 1:200 in sample dilution buffer and added to the pre-coated plates, followed by incubation at room temperature for 2 hours. After washing the plates with washing buffer, bound IgG was detected using a horseradish peroxidase-conjugated goat anti-mouse IgG antibody (1:10,000, Abcam). The absorbance was measured at 450 nm using a Multiskan FC microplate reader (Thermo Fisher Scientific, San Jose, USA).
2.12 Assessment of T cell immune responses post-vaccine stimulation
Splenocytes were isolated immediately post-euthanasia via mechanical dissociation of the spleen tissue. Mononuclear cells were then separated using density gradient centrifugation with a murine spleen mononuclear cell isolation kit (Solarbio, Beijing, China). Isolated cells were resuspended at a concentration of 1×10^6 cells/mL in RPMI 1640 medium supplemented with 10% fetal bovine serum. Cells were subsequently stimulated in vitro with recombinant SOST protein (100 ng/mL), vaccine protein (100 ng/mL), or PBS as a control, and incubated for 48 hours at 37 °C in a humidified atmosphere containing 5% CO2. Post-incubation, culture supernatants were collected and analyzed for cytokine concentrations (IL-4, IL-10, and IFN-γ) using ELISA kits (Meimian Industrial Co., Ltd., China), following the manufacturer’s instructions.
2.13 In vitro validation of anti-SOST antiserum function
Functional activity of anti-SOST antiserum from vaccine-immunized mice was evaluated using primary osteoclasts and osteoblasts. Mice were euthanized via CO2 inhalation, starting with a flow rate of 10% of chamber volume per minute to gradually increase CO2 concentration to 30%, inducing unconsciousness. Once righting reflex was lost, the flow rate was increased to 30% per minute to maintain a CO2 concentration of ≥70% for 5 minutes, ensuring humane euthanasia. All procedures complied with animal welfare guidelines. Primary osteoclasts were isolated from tibiae and femora of 8-week-old C57BL/6J mice. Bone marrow mononuclear cells were extracted using an isolation kit, filtered, and cultured in α-MEM supplemented with 50 ng/mL M-CSF and 80 ng/mL sRANKL (PeproTech) for 4–6 days to induce differentiation. During the second medium change, anti-SOST antiserum and 100 ng/mL recombinant SOST protein (Novoprotein) were added. Osteoclast differentiation was confirmed by TRAP staining (Servicebio). Primary osteoblasts were obtained from bone marrow stromal cells, and the MC3T3-E1 subclone 14 osteoblast cell line (purchased from Pricella Biotechnology Co., Ltd.) was cultured in osteogenic medium. Co-cultures of osteoblasts with anti-SOST antiserum and 200 ng/mL SOST were established, and mineralization was evaluated on day 21 via Alizarin Red S staining (Solarbio). The culture medium was refreshed every 2–3 days throughout the experiment.
2.14 Statistical analysis
Data are presented as means ± standard deviation (SD). Differences between two independent groups were analyzed using one-way ANOVA followed by Tukey’s multiple comparisons test. Data visualization was conducted utilizing GraphPad Prism software version 10 (GraphPad Software, San Diego, CA, USA). A p-value of less than 0.05 was considered statistically significant.
3 Results
3.1 Screening of high-affinity SOST epitope for ROMO binding
To identify potential interaction sites of SOST with ROMO for development of recombinant subunit vaccines, we initially fragmented SOST protein into six peptides, each comprising approximately 30 amino acids. Screening through ELISA pinpointed two peptides, SOST114–143 and SOST144-173, that exhibited high-affinity binding to ROMO (Figure 1A). We further dissected the identified region (amino acids 114-173) into four peptides based on their biological properties (Figure 1B). A subsequent ELISA revealed that SOST131–163 peptide served as a specific epitope with substantial affinity for ROMO (Figure 1C), located within loop3 domain of SOST (Figure 1D-a).
Figure 1

Screening and analysis of high-affinity epitopes on SOST. (A) ELISA experiments were conducted to identify SOST fragments with strong binding affinity for ROMO, revealing that SOST114–143 and SOST143–173 exhibit significantly higher affinity (P<0.01). (B) A schematic diagram delineating the binding functional regions associated with the high-affinity fragments of SOST. (C) ELISA results indicate that SOST131–163 displays the highest affinity for ROMO (P<0.01), thereby identifying it as a potent functional epitope of SOST. (D-a) SOST131–163 fragment (highlighted in yellow) is located within the loop3 domain of SOST protein. (D-b) Docking studies indicate that SOST131–163 fragment interacts with ROMO light chain, yielding a binding free energy of -25.8 kcal/mol and an interface area of 712.9 Ų. (D-c) Additionally, SOST131–163 fragment can bind to the ROMO heavy chain, resulting in a binding free energy of -33.19 kcal/mol and an interface area of 451.6 Ų. (E) CTL epitopes within SOST131–163 sequence include two strong binder epitopes and four weak binder epitopes. (F) HTL epitopes in SOST131–163 sequence comprise one strong binder epitope and four weak binder epitopes. Predictions of B cell epitopes for SOST131–163 sequence are illustrated, including predicted linear B cell epitopes (G) and predicted discontinuous B cell epitopes (H).
Molecular docking studies indicated that SOST131–163 fragment interacts with the light chain of ROMO’s variable domain, establishing 4 hydrogen bonds and 105 non-bonded contacts, resulting in a binding free energy of -25.8 kcal/mol and an interface area of 712.9 Ų (Figure 1D-b, Supplementary Figure S1A). Furthermore, SOST131–163 demonstrated affinity for the heavy chain’s variable domain, forming 2 salt bridges and 90 non-bonded contacts, with a binding free energy of -33.19 kcal/mol and an interface area of 451.6 Ų (Figure 1D-c, Supplementary Figure S1B). These docking results corroborate that SOST131–163 is a critical and distinctive peptide for ROMO, aligning with our ELISA observations.
Identification of immunodominant epitopes is pivotal for effective vaccine design. In this study, NetMHCpan 4.1 EL tool was utilized to predict six cytotoxic T lymphocyte (CTL) epitopes in SOST131–163 fragment, consisting of four weak binders and two strong binders (Figure 1E). Additionally, NetMHCIIpan 4.1 EL server was employed to forecast five helper T lymphocyte (HTL) epitopes, comprising four weak binders and one strong binder (Figure 1F). For the prediction of B cell epitopes, linear epitopes were analyzed using BepiPred 2.0 tool, resulting in the identification of two distinct epitopes within SOST131–163 fragment (Figure 1G). Furthermore, conformational B cell epitopes were evaluated using ElliPro tool, which yielded two additional epitopes (Figure 1H).
3.2 Construction and immunogenicity prediction of candidate vaccines
To enhance vaccine efficacy in inducing antibodies, we conjugated DTT scaffold, which contains substantial T-helper epitopes capable of disrupting immune tolerance to autoantigens, with varying quantities of repetitive SOST131–163 epitopes via a (GGGGS)2 linker. Six recombinant vaccines were constructed using this method, labeled DS0 to DS5, corresponding to the incorporation of 0 to 5 copies of SOST131–163 epitopes into DTT scaffold (Figure 2A). Tertiary structures of these candidate vaccines were predicted using AlphaFold2 server, which generated five structural models for each vaccine. The top-ranked model for each vaccine was selected based on the highest predicted Local Distance Difference Test (pLDDT) score, and the resulting structures are presented (Figure 2B).
Figure 2
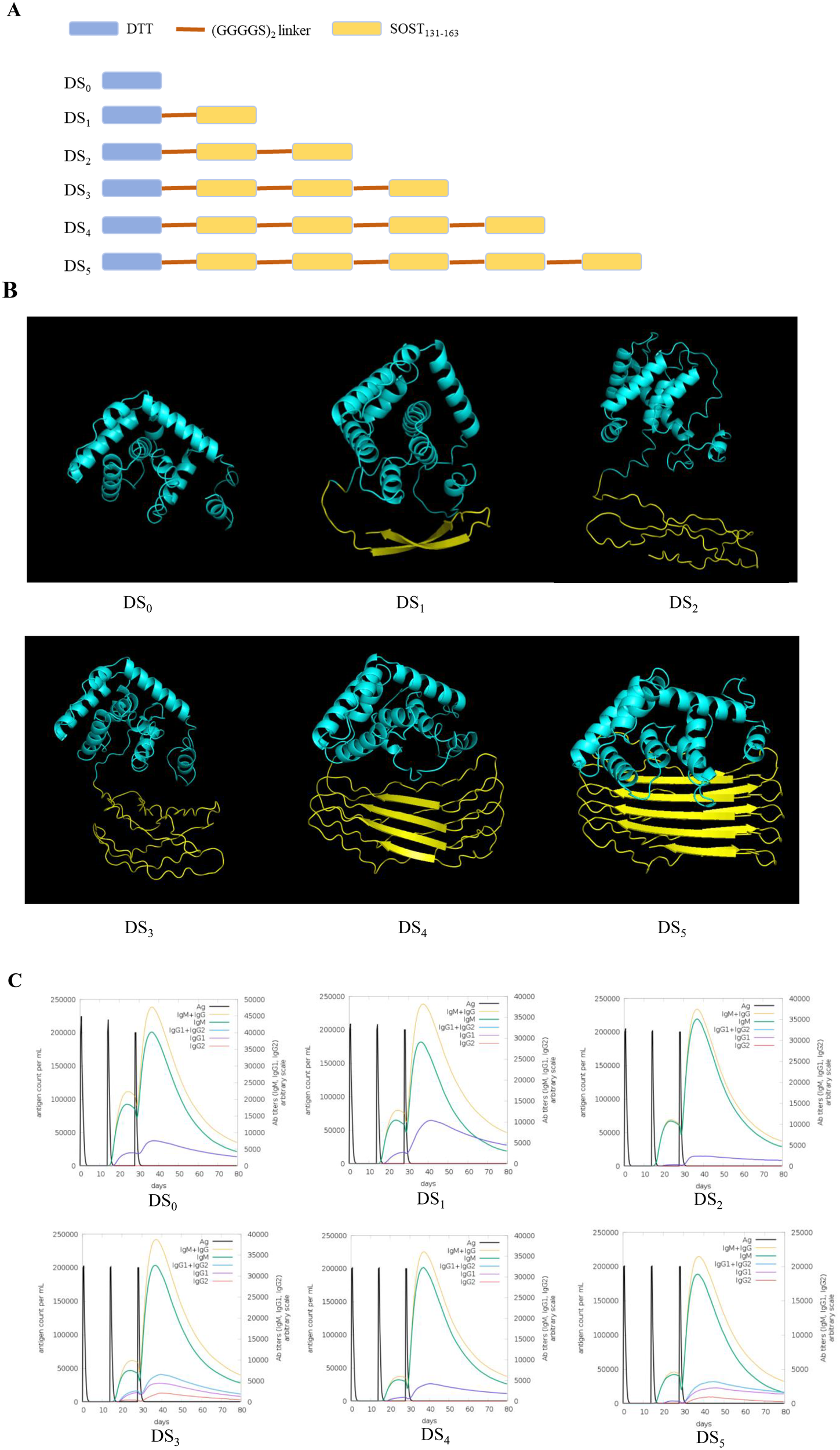
Construction plan and immune stimulation simulation of SOST candidate vaccines. (A) Schematic representation for the construction of SOST candidate vaccines. (B) Predicted three-dimensional structure of SOST candidate vaccines, modeled using AlphaFold2 server based on amino acid sequence. The cyan region denotes DTT protein scaffold, while the yellow regions represent the various repeated SOST131–163 peptides. (C) Immune stimulation simulation conducted using C-IMMSIM online server demonstrates that DS3 and DS5 vaccines simultaneously stimulate the production of IgM, IgG1, and IgG2 (Romosozumab is classified as an IgG2 antibody), whereas other candidate vaccines primarily induced IgM and IgG1 antibodies. Consequently, DS3 and DS5 vaccines were selected for further analysis.
To evaluate immune-stimulating potential of the top-ranked model for each candidate vaccine, we employed C-IMMSIM online server. The results demonstrated that all vaccine candidates elicited relatively high antibody titers following three immunization injections. Notably, the DS3 and DS5 vaccines stimulated the production of multiple antibody isotypes, including IgM, IgG1 and IgG2, while other candidate predominantly induced IgM and IgG1 responses (Figure 2C). Importantly, the production of the IgG2 subtype is particularly significant as it is the functional antibody associated with ROMO. Since the candidate vaccines are designed to elicit an IgG2 antibody response that closely resembles that of ROMO (21), we selected the DS3 and DS5 vaccines for further analysis and investigation.
3.3 Immunological and physicochemical properties of DS3 and DS5 vaccines
Safety and efficacy are critical criteria for evaluating vaccines. Analysis using AllerTOP v.2.1 revealed that both DS3 and DS5 vaccines are non-allergenic. Antigenicity of these vaccines was assessed through VaxiJen v2.0, yielding scores of 0.7434 for DS3 and 0.7948 for DS5, both surpassing the threshold value of 0.5. These results indicate that DS3 and DS5 vaccines are not only safe but also exhibit high immunogenicity. Solubility assessments conducted via SolPro server produced favorable scores of 0.713 for DS3 and 0.925 for DS5. Additional physicochemical parameters were predicted using ExPASy ProtParam server. Both vaccines are classified as recombinant proteins, with molecular weights of 31.8 kDa for DS3 and 40.3 kDa for DS5, and isoelectric points of 9.14 and 9.61, respectively. The total atom counts were recorded as 4463 for DS3 and 5657 for DS5. Estimated half-lives for both vaccines are approximately 30 hours in mammalian reticulocytes, over 20 hours in yeast, and exceeding 10 hours in Escherichia coli. The instability indices were calculated to be 42.19 for DS3 and 39.93 for DS5, while aliphatic indices measured 82.72 and 79.38, respectively. The grand average of hydropathicity (GRAVY) values were -0.257 for DS3 and -0.294 for DS5 (Table 1).
Table 1
| Property | DS3 | DS5 |
|---|---|---|
| Allergenicity(AllerTOP v2.1) | NON-ALLERGEN | NON-ALLERGEN |
| Antigenicity (VaxiJen v2.0) | 0.7434 | 0.7948 |
| Solubility (SOLpro) | 0.713771 | 0.925892 |
| Number of amino acids | 302 | 386 |
| Molecular weight | 31804.32 | 40334.16 |
| Theoretical Isoelectric point (pI) | 9.14 | 9.61 |
| Formula | C1362H2240N424O421S16 | C1716H2840N556O523S22 |
| Total number of atoms | 4463 | 5657 |
| Estimated half-life | 30 hours (mammalian reticulocytes, in vitro). >20 hours (yeast, in vivo). >10 hours (Escherichia coli, in vivo). |
30 hours (mammalian reticulocytes, in vitro). >20 hours (yeast, in vivo). >10 hours (Escherichia coli, in vivo) |
| Instability index | 42.19(unstable) | 39.93(stable) |
| Aliphatic index | 82.72 | 79.38 |
| Grand average of hydropathicity (GRAVY) | -0.257 | -0.294 |
Prediction of immunological and physicochemical properties for DS3 and DS5 vaccines.
3.4 Assessment of secondary structure of DS3 and DS5 vaccines
Secondary structure compositions of DS3 and DS5 vaccine were analyzed using SOPMA server. DS3 candidate exhibited a secondary structure composed of 50.33% α-helices (152/302), 37.09% random coils (112/302), and 12.58% extended strands (38/302) (Figure 3A). Additionally, DS3 demonstrated enhanced solubility, with a Protein-Sol score of 0.574, surpassing the baseline threshold of 0.45 (Figure 3B). Confidence in the secondary structure predictions for DS3 was further assessed using the PESIPRED web server, which provided favorable results (Figure 3C). In contrast, DS5 candidate displayed a distinct secondary structure profile, characterized by 11.66% α-helices (45/386), 72.28% random coils (279/386), and 16.06% extended strands (62/386) (Figure 3D). Similarly, DS5 exhibited enhanced solubility, with a Protein-Sol score of 0.647, exceeding the baseline value of 0.45 (Figure 3E). Confidence of the secondary structure predictions for DS5 was also assessed using PESIPRED web server, yielding favorable results (Figure 3F).
Figure 3

Analysis of the secondary structure and solubility characteristics of DS3 and DS5 vaccine. Secondary structure of DS3(A) and DS5(D) was assessed using SOPMA server. Solubility characteristics of DS3(B) and DS5(E) vaccines were evaluated using Protein-Sol server, resulting in solubility scores of 0.574 and 0.647, respectively, both exceeding the baseline value of 0.45, indicating enhanced solubility. Secondary structure analysis of DS3(C) and DS5(F) was performed using PESIPRED web server, the blue bars show the confidence of prediction.
3.5 Analysis and refinement of tertiary structures of DS3 and DS5 vaccine
Top-ranked models for DS3 and DS5 vaccines, predicted using AlphaFold2, exhibited pLDDT scores of 37.1 and 45.1, respectively. The pLDDT scores, which range from 0 to 100, serve as an indicator of model confidence, where values above 80 reflect high confidence in the accuracy of residue structure, and scores below 50 suggest the presence of disordered regions. Both DS3 and DS5 displayed pLDDT scores below the confidence threshold of 50, warranting further refinement.
Refinement was conducted using GalaxyRefine server, resulting in the generation of five refined models for each vaccine candidate. Optimal model quality is characterized by higher Global Distance Test High Accuracy (GDT-HA) and Ramachandran values, and lower root-mean-square deviation (RMSD), MolProbity scores, clash scores, and counts of poor rotamers. For DS3, Model 1 demonstrated the most favorable refinement metrics, achieving GDT-HA of 0.9007, RMSD of 0.580 Å, MolProbity score of 1.459, clash score of 3.7, poor rotamer count of 0.4, and Ramachandran favored percentage of 95.7% (Figure 4A, Supplementary Table S1). Similarly, Model 1 of DS5 exhibited optimal refinement results, with GDT-HA of 0.9424, RMSD of 0.480 Å, MolProbity score of 1.772, clash score of 10.2, poor rotamer count of 0.0, and Ramachandran favored of 96.4% (Figure 4E, Supplementary Table S2).
Figure 4
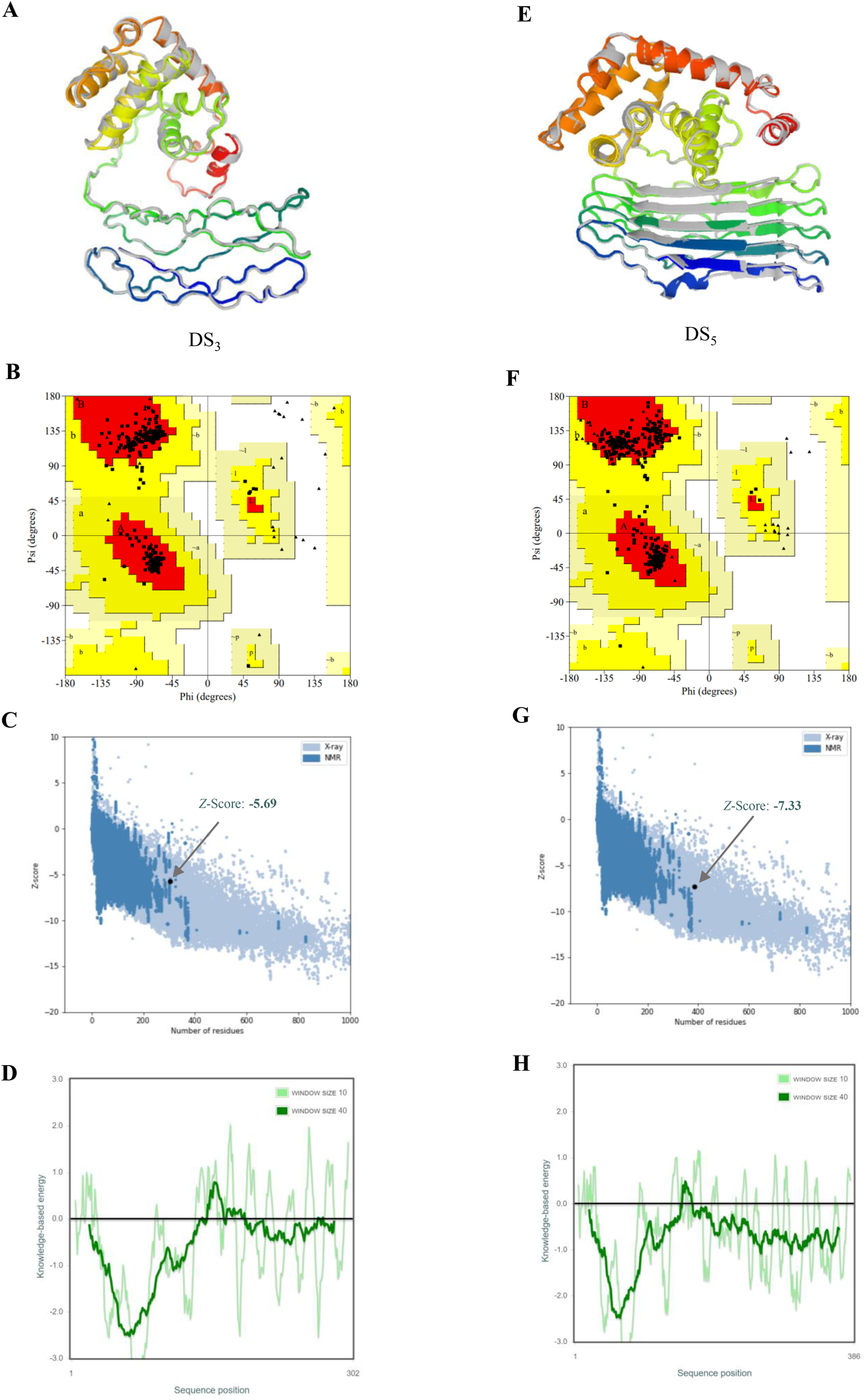
Refinement and validation of tertiary structures for DS3 and DS5 vaccines. Tertiary structures of DS3(A) and DS5(E) were refined using GalaxyRefine web server, with initial structures depicted in gray and refined structures shown in rainbow colors. Ramachandran plots for refined structures, generated via PROCHECK, indicate that 94.3% of DS3 residues (B) are located in the most favored regions, 5.7% in additional allowed regions, 0.0% in generously allowed regions, and 0.0% in disallowed regions. For DS5(F), 93.7% of residues are in the most favored regions, 6.3% in additional allowed regions, and 0.0% in both generously allowed and disallowed. Z-scores obtained from ProSA-web for DS3(C) and DS5(G) models are -5.69 and -7.33, respectively (black dots), both within the conformational score range for experimentally validated protein structures. Panels (D) and (H) display energy plots of the amino acid compositions for DS3 and DS5, respectively.
Ramachandran plots generated via PROCHECK indicated that 94.3% of DS3 residues reside in the most favored regions, with 5.7% in additional allowed regions, while no residues were found in generously allowed or disallowed regions (Figure 4B). Conversely, for DS5, 93.7% of residues were in the most favored regions, 6.3% in additional allowed regions, with none in generously allowed or disallowed regions (Figure 4F). The Z-scores calculated from ProSA-web were -5.69 for DS3 model and -7.33 for DS5 model (Figures 4C, G), both of which fall within the acceptable range for conformational scores typical of experimentally validated protein structures. DS3 vaccine exhibited a quality factor of 90.8451 according to ERRAT, while DS5 vaccine received a quality factor of 63.2867. Furthermore, energy plots corresponding to the amino acid compositions for both DS3 (Figure 4D) and DS5 (Figure 4H) were analyzed, providing additional evidence for structural integrity of the refined models.
3.6 T-cell and B-cell epitopes of DS3 and DS5 vaccine
Analysis using IEDB reveals that DS3 vaccine exhibits a high density of T-cell epitopes, comprising 13 strong and 34 weak CTL binders, as well as 3 strong and 47 weak HTL binders (Supplementary Table S3). Additionally, the ElliPro tool identifies 14 conformational B-cell epitopes associated with DS3 vaccine (Figure 5A, Table 2). In contrast, DS5 vaccine also demonstrates a rich repertoire of T-cell epitopes, featuring 17 strong and 42 weak CTL binders, along with 5 strong and 55 weak HTL binders (Supplementary Table S3). Moreover, a total of 10 conformational B-cell epitopes are predicted for DS5 vaccine, as indicated by ElliPro (Figure 5B, Table 2).
Figure 5
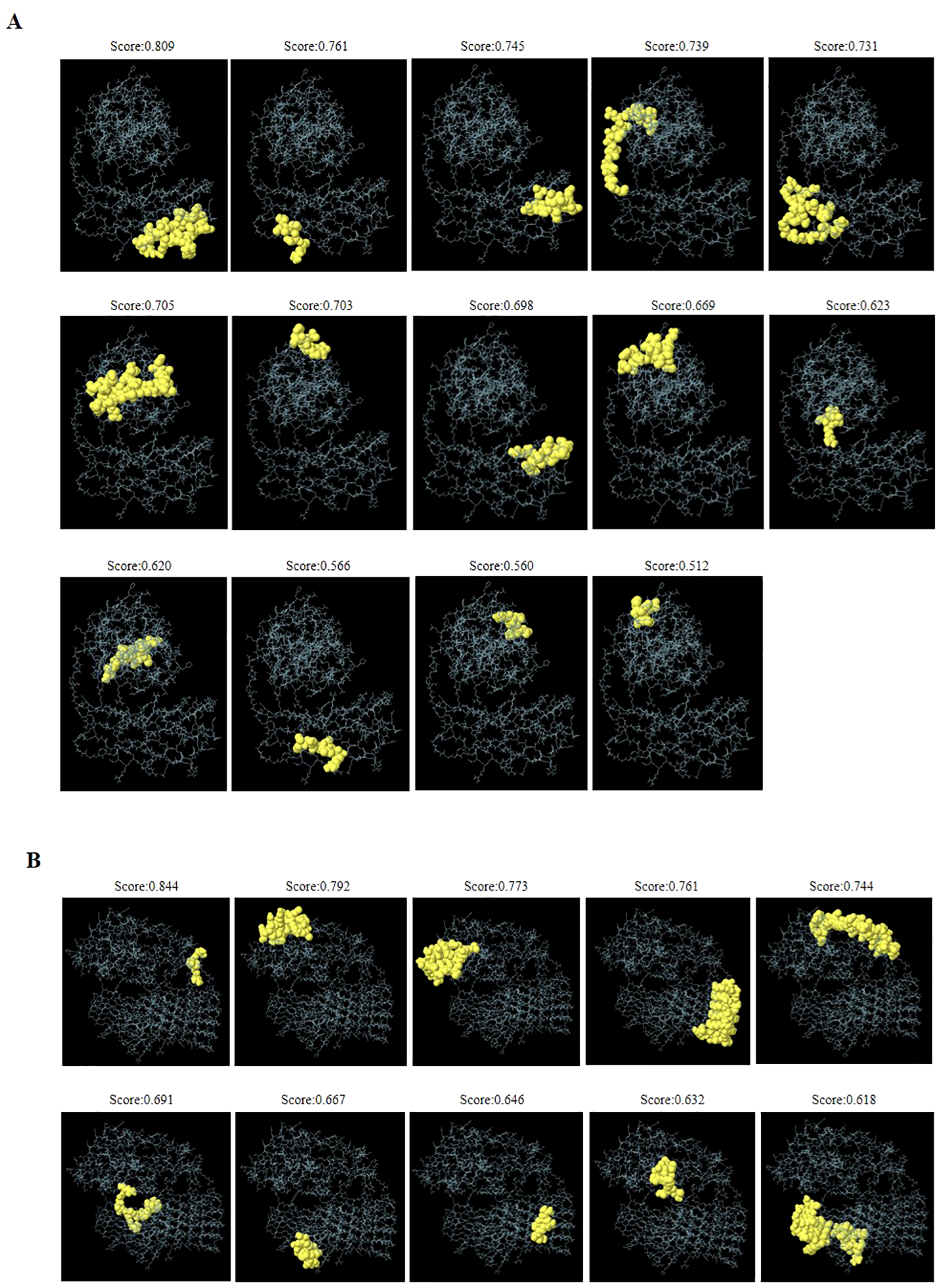
Three-dimensional representation of discontinuous B-cell epitopes predicted for the refined DS3 and DS5 vaccines. (A) Fourteen discontinuous B-cell epitopes of the refined DS3 vaccine are displayed, while (B) ten discontinuous B-cell epitopes of the refined DS5 vaccine are shown. The discontinuous B-cell epitopes are represented as yellow spheres, with the remaining vaccine residues illustrated as gray sticks.
Table 2
| Vaccine | No. | Residues | No. of residues | Score |
|---|---|---|---|---|
| DS3 | 1 | A:R277, A:A278, A:Q279, A:R280, A:V281, A:Q282, A:L283, A:L284, A:C285, A:P286, A:G287, A:G288, A:E289, A:A290, A:P291, A:R292, A:A293, A:R294, A:K295, A:V296 | 20 | 0.809 |
| 2 | A:C271, A:I272, A:P273, A:D274, A:R275 | 5 | 0.761 | |
| 3 | A:V197, A:Q198, A:L199, A:L200, A:C201, A:P202, A:G203, A:G204, A:E205, A:A206, A:P207, A:A209 | 12 | 0.745 | |
| 4 | A:I167, A:P168, A:L169, A:V170, A:G171, A:E172, A:L173, A:V174, A:D175, A:I176, A:G177, A:G178, A:G179, A:G180, A:S181, A:G182, A:G183, A:G184 | 18 | 0.739 | |
| 5 | A:G220, A:G221, A:G222, A:S223, A:G224, A:G225, A:G226, A:G227, A:R228, A:C229, A:I230, A:P231, A:D232, A:A258, A:S259, A:C260, A:G261, A:G262, A:G263, A:G264, A:S265, A:G266, A:G267, A:G268, A:G269, A:R270 | 26 | 0.731 | |
| 6 | A:S56, A:P57, A:N58, A:K59, A:T60, A:V61, A:S62, A:E63, A:E64, A:K65, A:A66, A:K67, A:Q68, A:Y69, A:D113, A:S114, A:E115, A:T116, A:A117, A:D118, A:N119 | 21 | 0.705 | |
| 7 | A:T38, A:E41, A:S42, A:K44, A:E45, A:H46 | 6 | 0.703 | |
| 8 | A:R238, A:L241, A:L242, A:C243, A:P244, A:G245, A:G246, A:E247, A:A248, A:P249, A:A251, A:R252 | 12 | 0.698 | |
| 9 | A:G47, A:P48, A:K50, A:N51, A:K52, A:M53, A:S54, A:E55, A:Q75, A:T76, A:E79 | 11 | 0.669 | |
| 10 | A:V14, A:R15, A:R16 | 3 | 0.623 | |
| 11 | A:S17, A:V18, A:G19, A:S20, A:S21, A:L22, A:S23, A:C24, A:I25, A:N26, A:L27 | 11 | 0.62 | |
| 12 | A:R297, A:L298, A:V299, A:A300, A:S301, A:C302 | 6 | 0.566 | |
| 13 | A:D28, A:D30, A:V31, A:D34, A:K35 | 5 | 0.56 | |
| 14 | A:H80, A:P81, A:E82, A:L83 | 4 | 0.512 | |
| DS5 | 1 | A:V14, A:R15, A:R16 | 3 | 0.844 |
| 2 | A:I40, A:E41, A:S42, A:L43, A:K44, A:E45, A:H46, A:G47, A:P48, A:I49, A:K50, A:N51, A:K52, A:M53, A:S54, A:E55, A:Q75, A:T76, A:E79 | 19 | 0.792 | |
| 3 | A:S56, A:P57, A:N58, A:K59, A:T60, A:V61, A:S62, A:E63, A:E64, A:K65, A:A66, A:Q68, A:Y69, A:E72, A:I112, A:D113, A:S114, A:E115, A:T116, A:A117, A:D118, A:N119, A:L120, A:K122 | 24 | 0.773 | |
| 4 | A:L200, A:C201, A:P202, A:G203, A:G204, A:E205, A:A206, A:P207, A:R208, A:L242, A:C243, A:P244, A:G245, A:G246, A:E247, A:A248, A:P249, A:R250, A:L284, A:C285, A:P286, A:G287, A:G288, A:E289, A:A290, A:P291, A:R292, A:L326, A:C327, A:P328, A:G329, A:G330, A:E331, A:A332, A:P333, A:R334, A:Q366, A:L367, A:L368, A:C369, A:P370, A:G371, A:G372, A:E373, A:A374, A:P375, A:R376 | 47 | 0.761 | |
| 5 | A:A6, A:C9, A:A10, A:G11, A:N12, A:S17, A:V18, A:G19, A:S20, A:S21, A:L22, A:S23, A:C24, A:I25, A:N26, A:L27, A:D28, A:D30, A:V31, A:I32, A:D34, A:K35, A:K37, A:T38, A:K39, A:H80, A:P81, A:E82, A:L83, A:S84, A:K87 | 31 | 0.744 | |
| 6 | A:G180, A:S181, A:G182, A:G183, A:G184, A:G185, A:R186, A:C187, A:I188, A:P189, A:D190, A:R191 | 12 | 0.691 | |
| 7 | A:I314, A:P315, A:D316, A:C355, A:I356, A:P357, A:D358, A:R359, A:C386 | 9 | 0.667 | |
| 8 | A:R336, A:A377, A:R378 | 3 | 0.646 | |
| 9 | A:A140, A:D141, A:G142, A:A143, A:V144, A:H145, A:H146, A:N147, A:T148 | 9 | 0.632 | |
| 10 | A:G222, A:S223, A:G224, A:G225, A:G226, A:G227, A:R228, A:I230, A:P231, A:D232, A:G264, A:S265, A:G266, A:G267, A:G268, A:G269, A:R270, A:P273, A:D274, A:R275, A:R277, A:G306, A:S307, A:G308, A:G309, A:G310, A:G311, A:R312, A:R317, A:R319, A:G347, A:G348, A:S349, A:G350, A:G351, A:G352, A:G353, A:R354 | 38 | 0.618 |
Predicted discontinuous B-cell epitopes of the refined DS3 and DS5 vaccines.
3.7 Molecular docking and molecular dynamics simulation
Molecular docking and molecular dynamics simulations were performed to elucidate the interactions between the DS3 and DS5 vaccines and Toll-like receptor 2 (TLR2). For molecular docking analysis, HawkDock was conducted to generate ten docking models for each vaccine. The optimized docking models revealed binding scores of -6618.72 kcal/mol for DS3 and -8177.20 kcal/mol for DS5, suggesting a stronger interaction between DS5 and TLR2. Further structural analysis of DS3-TLR2 and DS5-TLR2 complexes indicated distinct binding energies and interface areas. Specifically, DS3-TLR2 complex demonstrated a binding energy of -46.21 kcal/mol with an interface area of 1285.9 Ų (Figure 6A), whereas DS5-TLR2 complex exhibited a higher binding energy of -79.05 kcal/mol and a surface area of 1245.0 Ų (Figure 7A). DS3-TLR2 complex was characterized by the presence of 1 salt bridge, 6 hydrogen bonds, and 137 non-bonded contacts (Figure 6B), while DS5-TLR2 complex featured 6 salt bridges, 7 hydrogen bonds, and 142 non-bonded contacts (Figure 7B).
Figure 6
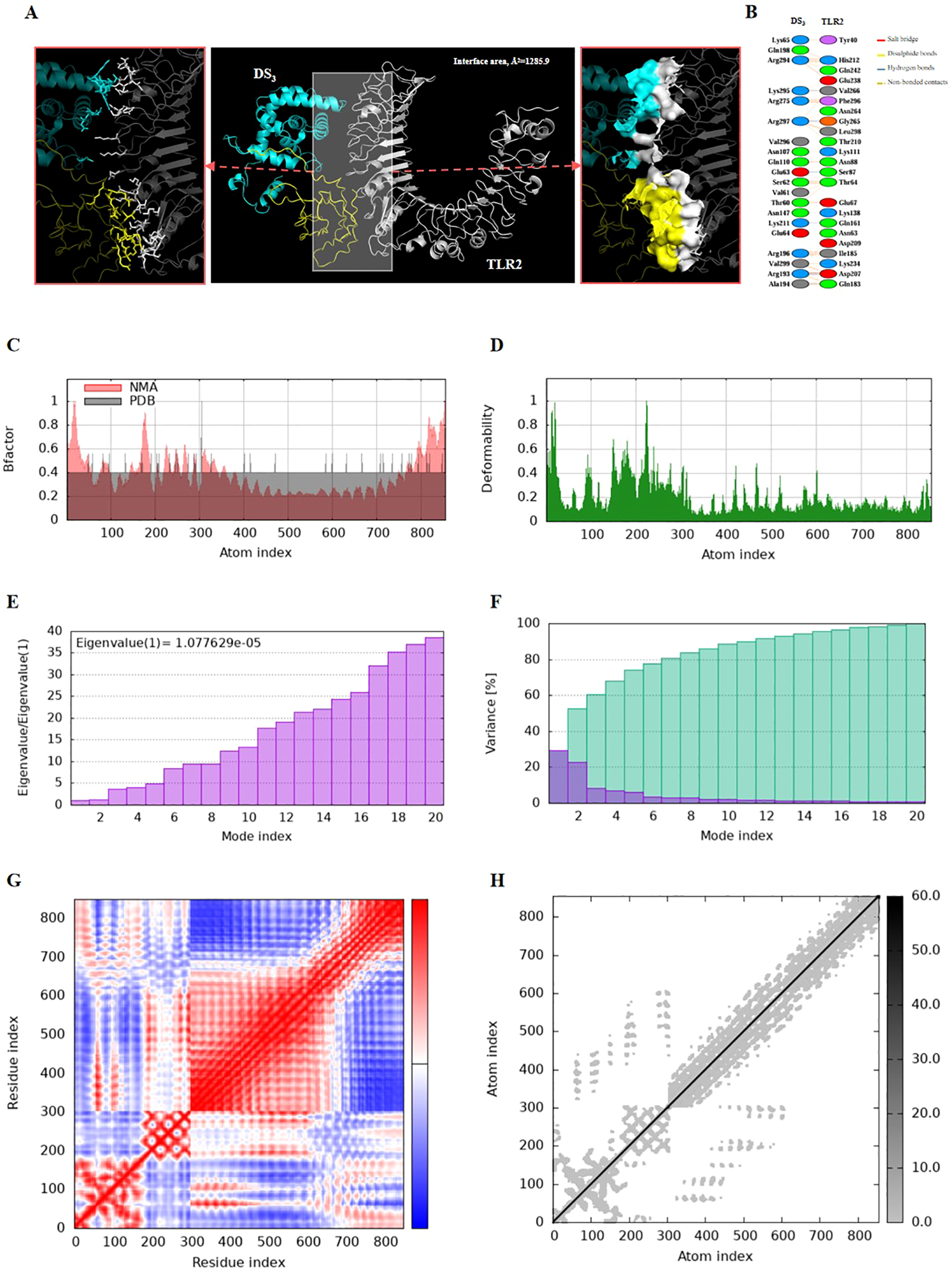
Molecular docking and normal mode analysis of DS3 with TLR2. (A) 3D model of the DS3 –TLR2 docking complex illustrating their interactions; DS3 is colored cyan-yellow and TLR2 is shown in white. Overall structures are represented as cartoons, with key interface residues emphasized. Potential interactions are depicted as sticks and surface. The measured binding free energy is -46.21 kcal/mol, with an interface area of 1285.9 Ų. (B) Detailed interactions between DS3 and TLR2, including 1 salt bridges (red), 6 hydrogen bonds (blue), and 137 non-bonded contacts (yellow-orange). (C) B-factor representation of the docking complex. (D) Deformability plot of the complex. (E) Eigenvalues associated with the docked complex. (F) Variance analysis of the docked complex. (G) Covariance map of atomic pairs of amino acid residues; correlated interactions are shown in red, uncorrelated in white, and anti-correlated in blue. (H) Elastic network model of the docking complex, with darker gray indicating stiffer springs.
Figure 7
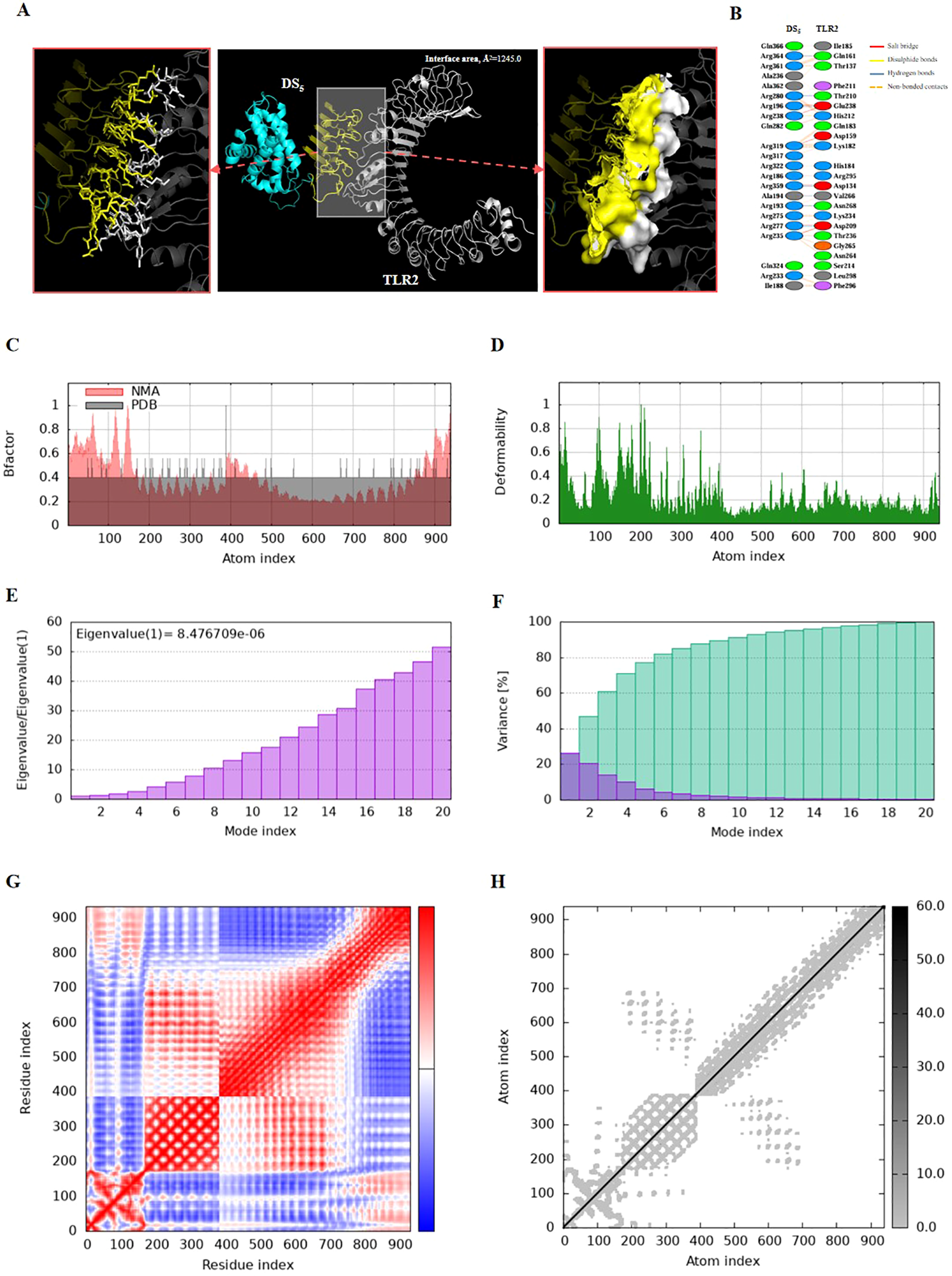
Molecular docking and normal mode analysis of DS5 with TLR2. (A) 3D model of the DS5 –TLR2 docking complex illustrating their interactions; DS3 is colored cyan-yellow and TLR2 is shown in white. Overall structures are represented as cartoons, with key interface residues emphasized. Potential interactions are depicted as sticks and surface. The measured binding free energy is -79.05 kcal/mol, with an interface area of 1245.0 Ų. (B) Detailed interactions between DS5 and TLR2, including 6 salt bridges (red), 7 hydrogen bonds (blue), and 142 non-bonded contacts (yellow-orange). (C) B-factor representation of the docking complex. (D) Deformability plot of the complex. (E) Eigenvalues associated with the docked complex. (F) Variance analysis of the docked complex. (G) Covariance map of atomic pairs of amino acid residues; correlated interactions are shown in red, uncorrelated in white, and anti-correlated in blue. (H) Elastic network model of the docking complex, with darker gray indicating stiffer springs.
Both complexes demonstrated stability, as evidenced by their B-factors (Figures 6C, 7C), deformability profiles (Figures 6D, 7D), and low eigenvalues (8.476709e-06 for DS5-TLR2 and 1.077629e-05 for DS3-TLR2) (Figures 6E, 7E), as well as variance analyses (Figures 6F, 7F). Covariance matrix evaluations highlighted correlations, as well as uncorrelated and anti-correlated motions among the residues within the complexes (Figures 6G, 7G). Elastic network analysis illustrated spring-like interactions between atoms, with darker gray depicting stiffer springs (Figures 6H, 7H). Collectively, these findings suggest that both DS3 and DS5 vaccines effectively engage TLR2, potentially eliciting robust immune responses.
3.8 Immune response simulation induced by vaccines
To assess the adaptive immune responses elicited by DS3 and DS5 vaccines, we employed C-IMMSIM server to simulate in vivo immune reactions. Our analysis revealed an increase in the total B cell population, including B-memory cells and IgM isotypes, which contributed to a significant rise in activated B cells in the host (Figures 8, 9A, B). Following the second immunization, the total count of T helper (TH) cells exhibited a rapid increase, peaking after the third immunization (Figures 8, 9C). Both activated and resting TH cell populations surged after each injection, primarily comprising TH1 cells (Figures 8, 9D, E), suggesting effective antibody maturation processes. Notably, the population of anergic (y2) T cells remained stable throughout the duration of the study (Figures 8, 9F). In contrast, the count of activated T cytotoxic (TC) cells showed a transient increase followed by a decline, while resting TC cells exhibited the opposite trend (Figures 8, 9G).
Figure 8
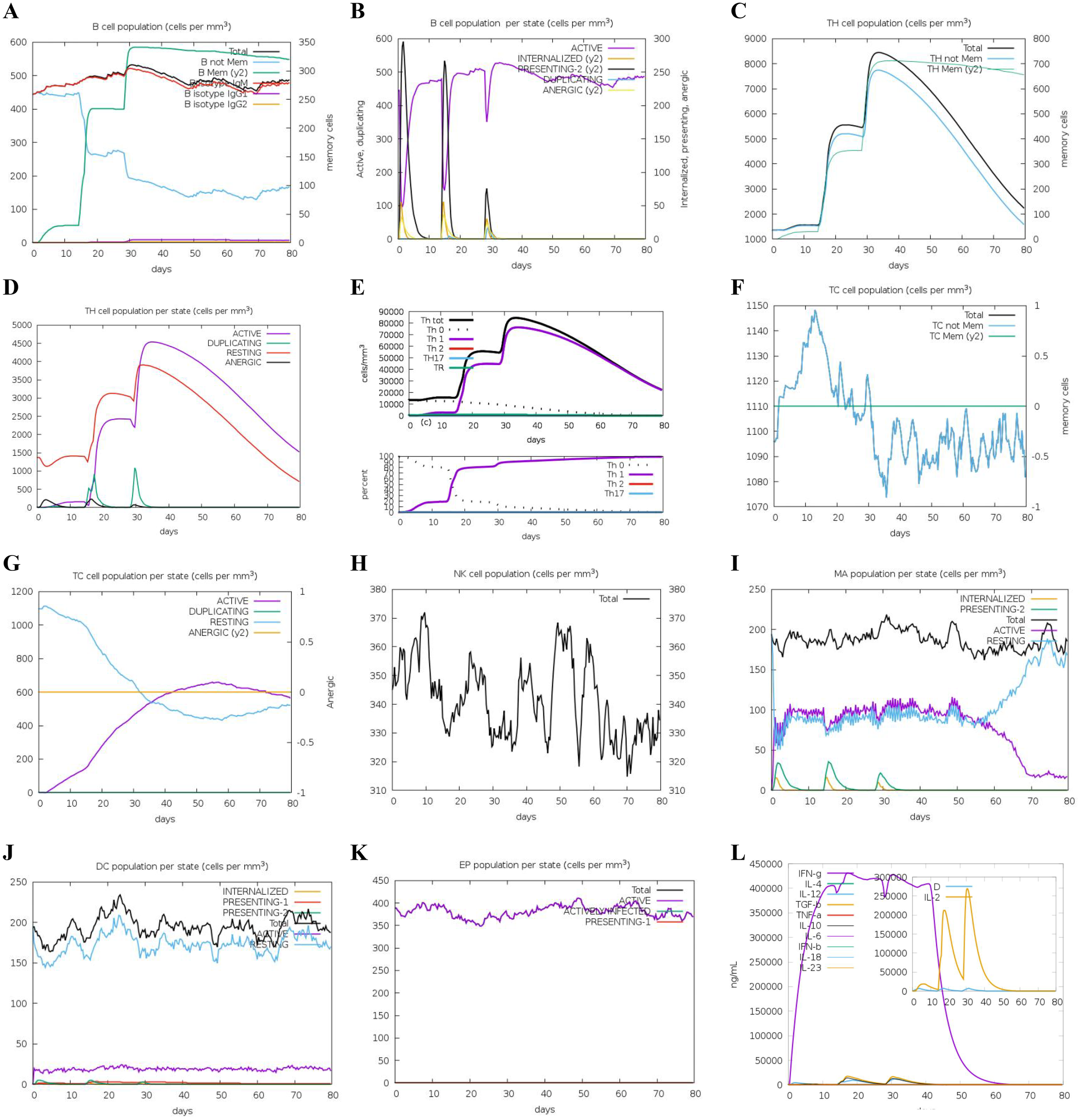
Predicted immune response induced by three administrations of DS3 vaccine via C-IMMSIM online server. Vaccinations were conducted on Day 1, Day 14, and Day 28. (A) Changes in B cell populations after vaccination, with specific subclasses color-coded. (B) Levels of B cell production post-immunization; active B cells (depicted in purple) show the highest secretion among subtypes. (C) Production of CD4+ T-helper (TH) cells in response to antigen exposure. (D) Distribution of TH cell states, including active, duplicating, resting, and anergic cells. (E) Quantification and proportion of different TH cell subtypes. (F) Levels of cytotoxic T (TC) cell production. (G) Overview of the TC cell population, categorized into resting and active states over time after DS3 vaccination. (H) Distribution of natural killer (NK) cells. (I) States of macrophages (MA). (J) Status of dendritic (DC) cells. (K) Production levels of epithelial cells. (L) Cytokine levels following DS3 vaccination. The main plot depicts overall cytokine concentrations, while the inset illustrates the levels of danger signals alongside the leukocyte growth factor IL-2.
Figure 9
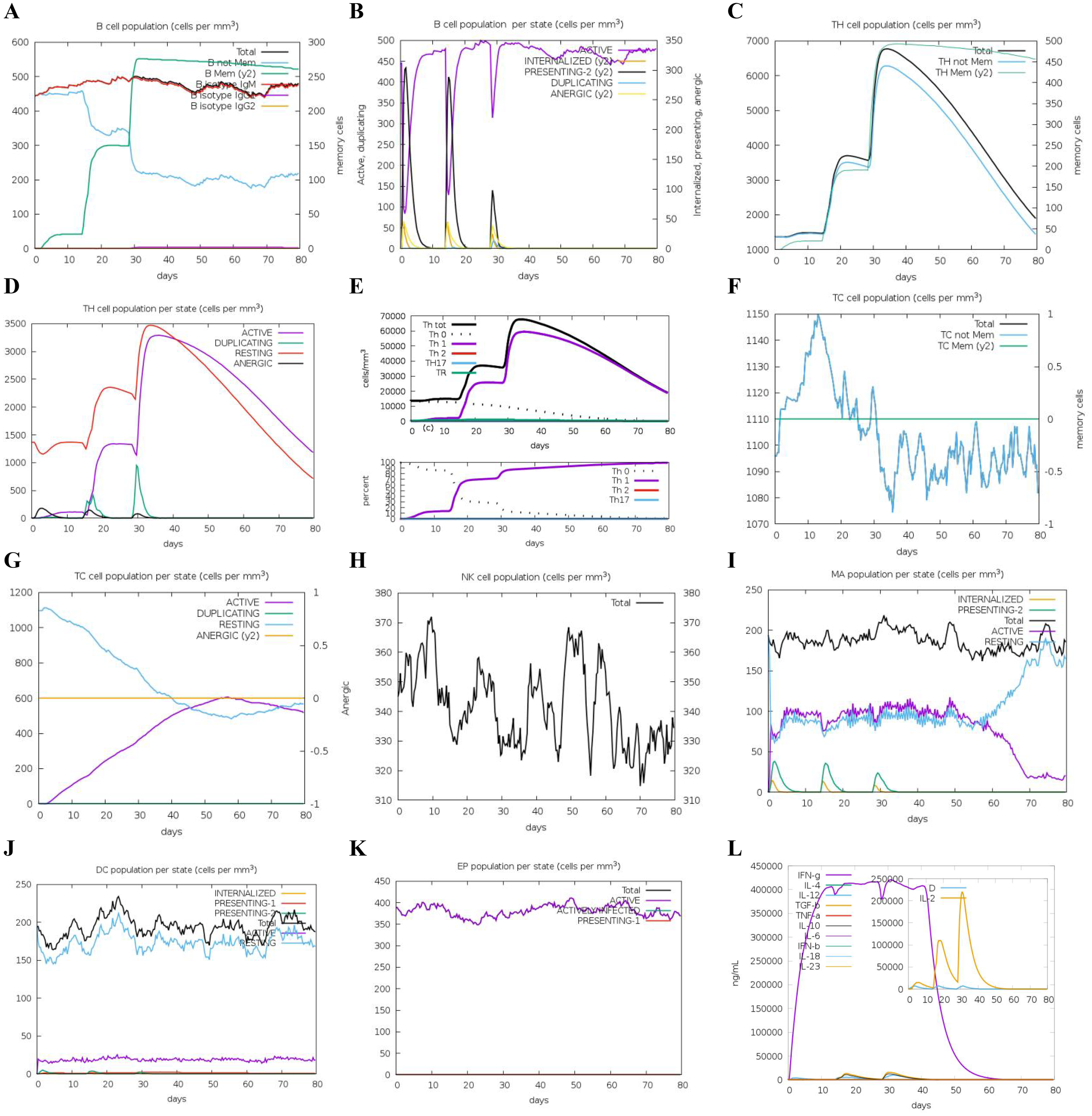
Predicted immune response induced by three administrations of DS5 vaccine via C-IMMSIM online server. Vaccinations were conducted on Day 1, Day 14, and Day 28. (A) Changes in B cell populations after vaccination, with specific subclasses color-coded. (B) Levels of B cell production post-immunization; active B cells (depicted in purple) show the highest secretion among subtypes. (C) Production of CD4+ T-helper (TH) cells in response to antigen exposure. (D) Distribution of TH cell states, including active, duplicating, resting, and anergic cells. (E) Quantification and proportion of different TH cell subtypes. (F) Levels of cytotoxic T (TC) cell production. (G) Overview of the TC cell population, categorized into resting and active states over time after DS5 vaccination. (H) Distribution of natural killer (NK) cells. (I) States of macrophages (MA). (J) Status of dendritic (DC) cells. (K) Production levels of epithelial cells. (L) Cytokine levels following DS5 vaccination. The main plot depicts overall cytokine concentrations, while the inset plot shows danger signal together with leukocyte growth factor IL-2.
Additionally, we evaluated the effects of DS3 and DS5 vaccines on innate immune cell populations. Natural killer (NK) cells, dendritic cells (DCs), and active epithelial cell populations demonstrated a relatively stable response upon immunization (Figures 8, 9H, J, K). Upon initial immunization, there was a marked increase in both active and resting macrophage populations in a short time, which subsequently reached a peak and stabilized (Figures 8, 9I). Approximately four weeks after the third immunization, we observed a decline in the number of active macrophages, coinciding with a rapid increase in resting macrophages (Figures 8, 9I). Following the administration of DS3 and DS5 vaccines, there was an activation of downstream inflammatory mediators, with significant elevations in levels of IFN-γ and IL-2 (Figures 8, 9L). Collectively, these findings indicated that the DS3 and DS5 vaccines effectively stimulate both innate and adaptive immune responses, highlighting their potential as effective vaccine candidates.
3.9 Cloning, expression and immunogenicity of vaccines
Codons for the optimized DS3 and DS5 sequences were successfully cloned into pSmartI plasmids at XhoI restriction sites (Figures 10A, D). The recombinant plasmids were confirmed through PCR amplification, as shown by agarose gel electrophoresis (Figures 10B, E). Subsequently, the recombinant plasmids were transformed into Escherichia coli BL21(DE3), leading to the successful expression and purification of DS3 and DS5 vaccine proteins, which exhibited molecular weights of 31.8 kDa and 40.3 kDa, respectively (Figures 10C, F).
Figure 10
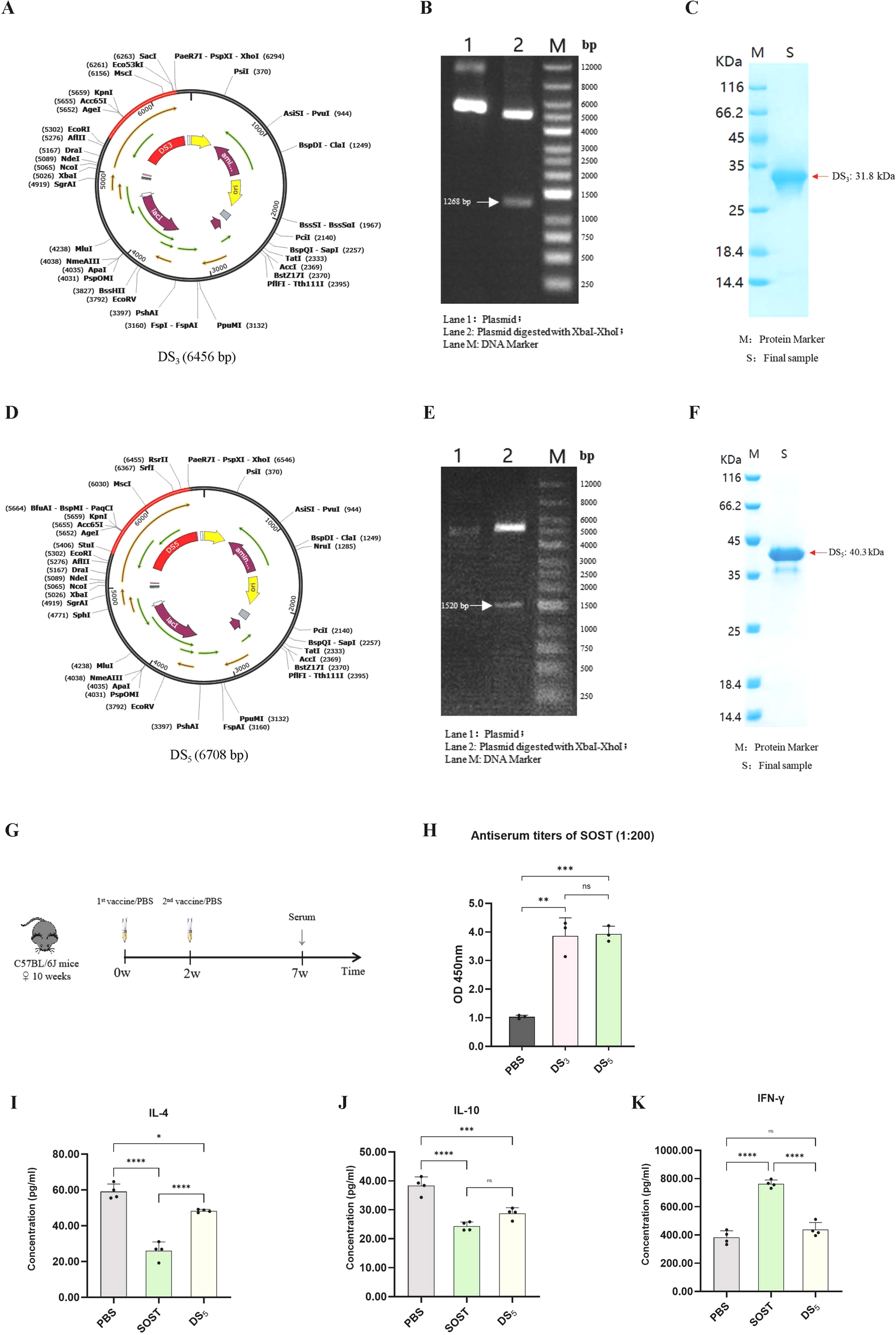
Cloning, expression, and immunogenicity assessment of DS3 and DS5 vaccines. (A) DS3 vaccine sequence (red) was inserted into the pSmartI expression vector (black) via seamless cloning using XhoI. (B) Agarose gel electrophoresis showing: Lane 1, recombinant plasmid; Lane 2, target fragment (1268 bp) along with vector sequence; Lane M, 1 kb DNA ladder. (C) Expression and purification of DS3 vaccine. (D) DS5 vaccine sequence (red) was cloned into the pSmartI expression vector (black) via seamless cloning using XhoI. (E) Agarose gel electrophoresis illustrating: Lane 1, recombinant plasmid; Lane 2, target fragment (1520 bp) along with vector sequence; Lane M, 1 kb DNA ladder. (F) Expression and purification of DS5. (G) Schematic overview of the mouse immunization protocol; each group comprised three mice (n=3), serving as independent biological replicates. (H) ELISA measurements indicating significantly elevated serum titers of anti-SOST antibodies in mice immunized with DS3 and DS5 compared to PBS controls (serum dilution 1:200). Antibody assays were performed in technical duplicates per mouse. (I–K) Cytokine levels of IL-4, IL-10, and IFN-γ in supernatants from splenocyte stimulation assays. Data are expressed as mean ± SD. Statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparisons test. (*p < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns = no significance).
To evaluate immunogenic potential of DS3 and DS5 vaccines, mice were immunized with DS3 and DS5 proteins at a dose of 200 µg per mouse, with a two-week interval between doses (n=3). Serum was collected at the seventh week to assess antibody production against SOST (Figure 10G). Our results demonstrated that both DS3 and DS5 effectively elicited a significant immune response, resulting in the production of high titers of anti-SOST antibodies in the immunized mice. The antibody titers induced by both vaccines were significantly higher than those observed in PBS control group (Figures 10H).
To assess T cell responses to vaccine stimulation, cytokine production associated with Th1 and Th2 responses was quantified using DS5 as a representative antigen. Specifically, levels of IL-4 and IL-10 (Th2 markers) and IFN-γ (Th1 marker) were measured in splenocyte cultures stimulated with PBS, SOST, or DS5 (Figures 10I–K). IFN-γ levels did not differ significantly among the PBS and DS5 groups, indicating that DS5 does not elicit a robust Th1-mediated cytotoxic response (Figure 10K). In contrast, IL-4 and IL-10 secretion were reduced following stimulation with both SOST and DS5 (Figures 10I, J). Notably, cytokine levels in DS5-treated splenocytes remained higher than in SOST-treated cells, suggesting that the DS5 vaccine induces a moderated Th2 response that may support B cell-mediated anti-SOST antibody production.
3.10 Validation of anti-SOST antiserum function in vitro
The antiserum obtained from vaccinated mice, which exhibited the highest antibody titer, was selected for in vitro functional assays using primary osteoclasts and osteoblasts. To model the in vivo role of SOST, which promotes osteoclast differentiation and inhibits osteoblast maturation, recombinant SOST was co-cultured with the respective cell types. Results showed that SOST supplementation had no sinificantly effect on osteoclast differentiation and maturation, however, the addition of the antiserum significantly attenuated osteoclast differentiation and maturation, leading to a marked reduction in osteoclastogenesis (Figures 11 A, B). In primary osteoblasts (Figures 11C, D) and the MC3T3-E1 subclone 14 cell line (Figures 11E, F), SOST partially suppressed differentiation and mineralization; however, the presence of antiserum mitigated these inhibitory effects, thereby restoring osteoblast mineralization capacity (Figures 11C–F). These findings demonstrate that the vaccine-induced antiserum effectively inhibits osteoclast activity and enhances osteoblast function, confirming its functional efficacy. Moreover, these results provide preliminary evidence supporting the vaccine’s potential as a therapeutic strategy for osteoporosis.
Figure 11

Effects of anti-SOST antiserum derived from vaccine-immunized mice on osteoclast and osteoblast differentiation. (A) TRAP staining of bone marrow-derived macrophages treated with SOST and anti-SOST antiserum at a 1:500 dilution, demonstrating inhibition of osteoclast differentiation (n=4). (B) Quantification of TRAP-positive osteoclasts in (A). (C) Alizarin Red S staining of bone marrow mesenchymal stem cell-derived osteoblasts cultured with osteogenic medium and treated with SOST and anti-SOST antiserum at a 1:100 dilution, indicating enhanced osteoblast differentiation and mineralization upon vaccine antiserum treatment (n=4). (D) Quantitative analysis of mineralization in (C). (E) Alizarin Red S staining of MC3T3-E1 subclone 14 cells cultured with osteogenic medium and treated with SOST and vaccine antiserum at a 1:100 dilution, showing rescue of SOST-mediated inhibition of osteoblast differentiation and mineralization (n=6). (F) Quantitative analysis of mineralization in (E). Scale bars, 250 μm and 500 μm. Data are expressed as mean ± SD; *p < 0.05, **p < 0.01, ***p < 0.001; ns, not significant. Statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparisons test.
4 Discussion
Osteoporosis represents a significant global public health challenge, with osteoporotic fractures incurring substantial economic costs and imposing considerable demands on individual healthcare resources and societal medical systems (1, 2). Among the available anti-osteoporotic treatments, ROMO is noteworthy for its significant ability to increase bone mass (37). Nevertheless, its high cost and strict eligibility criteria, which restrict its use to patients with diagnosed osteoporosis, hinder its broader applicability for early prevention strategies (5, 38). The potential risk of cardiovascular adverse events associated with ROMO raises important safety concerns that warrant careful consideration (38).
Recent advancements in vaccine immunology have effectively demonstrated the potential of active immunization strategies to stimulate endogenous antibody production across various chronic conditions, including ankylosing spondylitis (39), hypertension (40), diabetes (41), Alzheimer’s disease (42), and so on. Building on these innovations, our team has focused on developing a vaccine-based immunotherapy for osteoporosis (10, 11). This approach aims to achieve sustained regulation of bone formation and resorption through proactive immunization, offering a cost-effective solution that could enhance both early prevention and adjunctive long-term treatment of advanced osteoporosis.
In this study, we present an innovative osteoporosis vaccine targeting the SOST epitope (SOST131-163), which was identified through ROMO screening. The SOST131–163 epitope, situated within the loop3 region (amino acids 134–163 in SOST (43), including a 23-amino acid signal peptide), partially overlaps with previously identified antibody-binding sites in both loop2 and loop3 (43–45). However, we did not detect specific binding sites in loop2, likely due to challenges in preserving the native three-dimensional structure during separate synthesis. While ROMO effectively inhibits SOST by targeting both loops and demonstrates substantial anti-osteoporosis benefits, this broad inhibition may elevate the risk of cardiovascular side effects (5, 6). In contrast, our targeted vaccine approach focuses solely on loop3, which promotes bone formation while preserving cardiovascular health (45–47). Thus, our vaccine strategically aims to enhance bone mass while ensuring cardiovascular safety, offering a promising option for osteoporosis management.
In vaccine design, antigenicity and immunogenicity are crucial for eliciting robust and specific immune responses. Our study reveals that the SOST131–163 epitope contains six CTL epitopes, five HTL epitopes, two linear B-cell epitopes, and two conformational B-cell epitopes, highlighting its considerable immunogenic potential to activate both T-cell-mediated immunity and B-cell antibody production. To overcome immune tolerance associated with autologous protein vaccines, we utilized DTT protein as a carrier to enhance immunogenicity, which effectively expanded specific helper T-cell populations and promoted the differentiation and proliferation of polysaccharide-specific B cells. The candidate vaccines, DS1-DS5, successfully disrupted immune tolerance and elicited robust antibody responses in immune simulations. Notably, the DS3 and DS5 vaccines produced unique profiles by generating IgM, IgG1, and IgG2 antibodies, while the other candidates primarily generated IgM and IgG1, lacking IgG2. Given the clinical efficacy of ROMO as an IgG2 monoclonal antibody (21), our primary goal was to stimulate endogenous IgG2 antibody production, similar to ROMO. Consequently, we selected DS3 and DS5 for further exploration of their immunological mechanisms and potential applications. In selecting the immunological scaffold, we directly employed DTT to facilitate the overcoming of immune tolerance, informed by our prior findings (10). Nonetheless, the considerable potential of alternative scaffolds warrants further investigation to enhance vaccine efficacy and optimize antibody titers.
Structural analysis revealed that 94.3% of residues in the DS3 vaccine and 93.7% in the DS5 vaccine occupied favorable regions, indicating high modeling quality. Bioinformatics assessments further demonstrated these vaccines’ strong antigenicity, favorable physicochemical properties, and non-allergenic nature, establishing them as promising vaccine candidates. Molecular docking studies showed that both DS3 and DS5 vaccines effectively bind to Toll-like receptor 2 (TLR2), thereby activating this receptor and facilitating the induction of both humoral and cellular immune responses. TLR2, expressed in dendritic cells and involved in bone metabolism through the mechanism of osteoimmunology (48), is critical for osteoporosis management, as its activation inhibits inflammatory osteoclast differentiation and mitigates bone loss (49, 50). Experimental validation confirmed the successful construction of DS3 and DS5 vaccines using recombinant plasmids, with efficient expression in E. coli. The purified DS3 and DS5 vaccines elicited a significant production of anti-SOST antibodies in immunized mice, demonstrating their efficacy in overcoming immune tolerance. Further cellular assays confirmed that sera from vaccinated mice contain anti-SOST antibodies capable of inhibiting osteoclast activity and promoting osteoblast function, thereby restoring the balance between bone resorption and formation disrupted in osteoporosis. These findings highlight the potential of these vaccines as promising immunotherapeutic strategies for the prevention and treatment of osteoporosis.
5 Conclusions
This study introduces a SOST-targeted vaccine specifically designed for osteoporosis, demonstrating several advantages: (1) high specificity for the loop 3 domain of SOST, which may confer protective effects against osteoporosis while minimizing cardiovascular side effects; and (2) robust immunogenicity coupled with favorable physicochemical properties, effectively inducing endogenous ROMO-like antibodies. Preliminary in vivo experiments confirm the vaccine’s ability to overcome immune tolerance and elicit SOST-specific antibodies in murine models. Additionally, in vitro analyses reveal that the generated antiserum can inhibit osteoclast differentiation and enhance osteoblast activity, underscoring its therapeutic potential for osteoporosis. This innovative strategy offers a promising approach for early prevention and sustained management of the disease. Future investigations will aim to validate vaccine’s efficacy and safety in vivo, facilitating its progression toward clinical application.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was approved by Institutional Animal Care and Use Committee (IACUC) of Shenzhen University Medical School. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
JL: Formal Analysis, Writing – original draft, Methodology, Visualization, Data curation, Validation, Resources, Investigation, Writing – review & editing, Conceptualization. TW: Funding acquisition, Validation, Writing – review & editing, Conceptualization, Supervision, Software. BG: Resources, Writing – review & editing, Methodology, Software. LL: Writing – review & editing, Software, Resources, Data curation, Methodology. ZY: Data curation, Visualization, Writing – review & editing, Resources. HT: Conceptualization, Resources, Writing – review & editing, Funding acquisition.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This study was funded by National Natural Science Foundation of China (81970761), Sanming Project of Medicine in Shenzhen (SZSM201911011), and Shenzhen Nanshan Technology Research Development and Creative Design Project (NS2023129).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1644437/full#supplementary-material
Supplementary Figure 1Interaction of SOST131–163 fragment with ROMO light and heavy chains. (A) SOST131–163 fragment docked with ROMO light chain, forming four hydrogen bonds and 105 non-bonded contacts. (B) SOST131–163 fragment docked with ROMO heavy chain, establishing two salt bridges and 90 non-bonded contacts.
Supplementary Table 1Structure information of DS3 after refinement using Galaxyrefine.
Supplementary Table 2Structure information of DS5 after refinement using Galaxyrefine.
Supplementary Table 3Prediction of cytotoxic T lymphocyte and helper T lymphocyte epitopes for DS3 and DS5 vaccine.
References
1
Khosla S Hofbauer LC . Osteoporosis treatment: recent developments and ongoing challenges. Lancet Diabetes Endocrinol. (2017) 5:898–907. doi: 10.1016/S2213-8587(17)30188-2
2
Salari N Darvishi N Bartina Y Larti M Kiaei A Hemmati M et al . Global prevalence of osteoporosis among the world older adults: a comprehensive systematic review and meta-analysis. J Orthop Surg Res. (2021) 16:669. doi: 10.1186/s13018-021-02821-8
3
Wade SW Strader C Fitzpatrick LA Anthony MS O'Malley CD . Estimating prevalence of osteoporosis: examples from industrialized countries. Arch Osteoporos. (2014) 9:182. doi: 10.1007/s11657-014-0182-3
4
Black DM Rosen CJ . Clinical practice. Postmenopausal osteoporosis. N Engl J Med. (2016) 374:254–62. doi: 10.1056/NEJMcp1513724
5
Wu D Li L Wen Z Wang G . Romosozumab in osteoporosis: yesterday, today and tomorrow. J Transl Med. (2023) 21:668. doi: 10.1186/s12967-023-04563-z
6
Langdahl BL Hofbauer LC Forfar JC . Cardiovascular safety and sclerostin inhibition. J Clin Endocrinol Metab. (2021) 106:1845–53. doi: 10.1210/clinem/dgab193
7
Jiao Z Chai H Wang S Sun C Huang Q Xu W . SOST gene suppression stimulates osteocyte Wnt/β-catenin signaling to prevent bone resorption and attenuates particle-induced osteolysis. J Mol Med (Berl). (2023) 101:607–20. doi: 10.1007/s00109-023-02319-2
8
Holdsworth G Roberts SJ Ke HZ . Novel actions of sclerostin on bone. J Mol Endocrinol. (2019) 62:R167–r185. doi: 10.1530/JME-18-0176
9
Davis S Simpson E Hamilton J James MM Rawdin A Wong R et al . Denosumab, raloxifene, romosozumab and teriparatide to prevent osteoporotic fragility fractures: a systematic review and economic evaluation. Health Technol Assess. (2020) 24:1–314. doi: 10.3310/hta24290
10
Wu T Guan B Luo J Li L Zhang B Yang Z et al . In silicon desinging of RANKL-targeting vaccine for protection of osteoporosis based on the epitope of Denosumab. Int Immunopharmacol. (2025) 144:113610. doi: 10.1016/j.intimp.2024.113610
11
Li F Li H Zhai Q Li F Wu T Sha X et al . A new vaccine targeting RANKL, prepared by incorporation of an unnatural Amino acid into RANKL, prevents OVX-induced bone loss in mice. Biochem Biophys Res Commun. (2018) 499:648–54. doi: 10.1016/j.bbrc.2018.03.205
12
Parvizpour S Pourseif MM Razmara J Rafi MA Omidi Y . Epitope-based vaccine design: a comprehensive overview of bioinformatics approaches. Drug Discov Today. (2020) 25:1034–42. doi: 10.1016/j.drudis.2020.03.006
13
Weng G Wang E Wang Z Liu H Zhu F Li D et al . HawkDock: a web server to predict and analyze the protein-protein complex based on computational docking and MM/GBSA. Nucleic Acids Res. (2019) 47:W322–w330. doi: 10.1093/nar/gkz397
14
Laskowski RA Jabłońska J Pravda L Vařeková RS Thornton JM . PDBsum: Structural summaries of PDB entries. Protein Sci. (2018) 27:129–34. doi: 10.1002/pro.3289
15
Reynisson B Alvarez B Paul S Peters B Nielsen M . NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. (2020) 48:W449–54. doi: 10.1093/nar/gkaa379
16
Yang Y Wei Z Cia G Song X Pucci F Rooman M et al . MHCII-peptide presentation: an assessment of the state-of-the-art prediction methods. Front Immunol. (2024) 15:1293706. doi: 10.3389/fimmu.2024.1293706
17
Jespersen MC Peters B Nielsen M Marcatili P . BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. (2017) 45:W24–9. doi: 10.1093/nar/gkx346
18
Ponomarenko J Bui HH Li W Fusseder N Bourne PE Sette A et al . ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinf. (2008) 9:514. doi: 10.1186/1471-2105-9-514
19
Tunyasuvunakool K Adler J Wu Z Green T Zielinski M Žídek A et al . Highly accurate protein structure prediction for the human proteome. Nature. (2021) 596:590–6. doi: 10.1038/s41586-021-03828-1
20
Rapin N Lund O Castiglione F . Immune system simulation online. Bioinformatics. (2011) 27:2013–4. doi: 10.1093/bioinformatics/btr335
21
Omran A Atanasova D Landgren F Magnusson P . Sclerostin: from molecule to clinical biomarker. Int J Mol Sci. (2022) 23:4751. doi: 10.3390/ijms23094751
22
Dimitrov I Bangov I Flower DR Doytchinova I . AllerTOP v.2–a server for in silico prediction of allergens. J Mol Model. (2014) 20:2278. doi: 10.1007/s00894-014-2278-5
23
Doytchinova IA Flower DR . VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinf. (2007) 8:4. doi: 10.1186/1471-2105-8-4
24
Magnan CN Randall A Baldi P . SOLpro: accurate sequence-based prediction of protein solubility. Bioinformatics. (2009) 25:2200–7. doi: 10.1093/bioinformatics/btp386
25
Mariethoz J Alocci D Gastaldello A Horlacher O Gasteiger E Rojas-Macias M et al . Glycomics@ExPASy: bridging the gap. Mol Cell Proteomics. (2018) 17:2164–76. doi: 10.1074/mcp.RA118.000799
26
Geourjon C Deléage G . SOPM: a self-optimized method for protein secondary structure prediction. Protein Eng. (1994) 7:157–64. doi: 10.1093/protein/7.2.157
27
McGuffin LJ Bryson K Jones DT . The PSIPRED protein structure prediction server. Bioinformatics. (2000) 16:404–5. doi: 10.1093/bioinformatics/16.4.404
28
Hebditch M Carballo-Amador MA Charonis S Curtis R Warwicker J . Protein-Sol: a web tool for predicting protein solubility from sequence. Bioinformatics. (2017) 33:3098–100. doi: 10.1093/bioinformatics/btx345
29
Heo L Park H Seok C . GalaxyRefine: Protein structure refinement driven by side-chain repacking. Nucleic Acids Res. (2013) 41:W384–388. doi: 10.1093/nar/gkt458
30
Wiederstein M Sippl MJ . ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. (2007) 35:W407–410. doi: 10.1093/nar/gkm290
31
Colovos C Yeates TO . Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. (1993) 2:1511–9. doi: 10.1002/pro.5560020916
32
Laskowski RA Rullmannn JA MacArthur MW Kaptein R Thornton JM . AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. (1996) 8:477–86. doi: 10.1007/BF00228148
33
Jiménez-García B Elez K Koukos PI Bonvin AM Vangone A . PRODIGY-crystal: a web-tool for classification of biological interfaces in protein complexes. Bioinformatics. (2019) 35:4821–3. doi: 10.1093/bioinformatics/btz437
34
Krissinel E Henrick K . Inference of macromolecular assemblies from crystalline state. J Mol Biol. (2007) 372:774–97. doi: 10.1016/j.jmb.2007.05.022
35
López-Blanco JR Aliaga JI Quintana-Ortí ES Chacón P . iMODS: internal coordinates normal mode analysis server. Nucleic Acids Res. (2014) 42:W271–276. doi: 10.1093/nar/gku339
36
Grote A Hiller K Scheer M Münch R Nörtemann B Hempel DC et al . JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. (2005) 33:W526–531. doi: 10.1093/nar/gki376
37
Cosman F Crittenden DB Adachi JD Binkley N Czerwinski E Ferrari S et al . Romosozumab treatment in postmenopausal women with osteoporosis. N Engl J Med. (2016) 375:1532–43. doi: 10.1056/NEJMoa1607948
38
Iolascon G Liguori S Paoletta M Toro G Moretti A . Anti-sclerostin antibodies: a new frontier in fragility fractures treatment. Ther Adv Musculoskelet Dis. (2023) 15:1759720x231197094. doi: 10.1177/1759720X231197094
39
Hayashi H Sun J Yanagida Y Yoshida S Baba S Tenma A et al . Peptide-based vaccine targeting IL17A attenuates experimental spondyloarthritis in HLA-B27 transgenic rats. RMD Open. (2023) 9:e002851. doi: 10.1136/rmdopen-2022-002851
40
Bairwa M Pilania M Gupta V Yadav K . Hypertension Vaccine may be a boon to millions in developing world. Hum Vaccin Immunother. (2014) 10:708–13. doi: 10.4161/hv.27520
41
Almasri L Holtzclaw BJ . Assessing vaccine protection for older adults with diabetes: A systematic review. West J Nurs Res. (2022) 44:582–97. doi: 10.1177/01939459211005710
42
Cacabelos R . How plausible is an Alzheimer's disease vaccine? Expert Opin Drug Discov. (2020) 15:1–6. doi: 10.1080/17460441.2019.1667329
43
Veverka V Henry AJ Slocombe PM Ventom A Mulloy B Muskett FW et al . Characterization of the structural features and interactions of sclerostin: molecular insight into a key regulator of Wnt-mediated bone formation. J Biol Chem. (2009) 284:10890–900. doi: 10.1074/jbc.M807994200
44
Holdsworth G Slocombe P Doyle C Sweeney B Veverka V Le Riche K et al . Characterization of the interaction of sclerostin with the low density lipoprotein receptor-related protein (LRP) family of Wnt co-receptors. J Biol Chem. (2012) 287:26464–77. doi: 10.1074/jbc.M112.350108
45
Wang L Yu Y Ni S Li D Liu J Xie D et al . Therapeutic aptamer targeting sclerostin loop3 for promoting bone formation without increasing cardiovascular risk in osteogenesis imperfecta mice. Theranostics. (2022) 12:5645–74. doi: 10.7150/thno.63177
46
Yu Y Wang L Ni S Li D Liu J Chu HY et al . Targeting loop3 of sclerostin preserves its cardiovascular protective action and promotes bone formation. Nat Commun. (2022) 13:4241. doi: 10.1038/s41467-022-31997-8
47
Yu S Huang W Zhang H Guo Y Zhang B Zhang G et al . Discovery of the small molecular inhibitors against sclerostin loop3 as potential anti-osteoporosis agents by structural based virtual screening and molecular design. Eur J Med Chem. (2024) 271:116414. doi: 10.1016/j.ejmech.2024.116414
48
Roth K Pallua JD Degenhart G De Zordo T Kremser C Reif C et al . Reduced bone quality of sacrum and lumbal vertebrae spongiosa in toll-like receptor 2- and toll-like receptor 4-knockout mice: A blinded micro-computerized analysis. Biomolecules. (2025) 15:239. doi: 10.3390/biom15020239
49
Henning P Kassem A Westerlund A Lundberg P Engdahl C Lionikaite V et al . Toll-like receptor-2 induced inflammation causes local bone formation and activates canonical Wnt signaling. Front Immunol. (2024) 15:1383113. doi: 10.3389/fimmu.2024.1383113
50
Wang Y Zhao X Liu-Bryan R . Role of TLR2 and TLR4 in regulation of articular chondrocyte homeostasis. Osteoarthritis Cartilage. (2020) 28:669–74. doi: 10.1016/j.joca.2020.01.011
Summary
Keywords
osteoporosis, sclerostin (SOST), romosozumab (ROMO), vaccine, translocation domain of diphtheria toxin (DTT)
Citation
Luo J, Wu T, Guan B, Li L, Yang Z and Tao H (2025) In silico design of novel precision vaccine targeting sclerostin epitopes for osteoporosis prevention and treatment. Front. Immunol. 16:1644437. doi: 10.3389/fimmu.2025.1644437
Received
10 June 2025
Revised
26 October 2025
Accepted
10 November 2025
Published
01 December 2025
Volume
16 - 2025
Edited by
Pitchiah Sivaperumal, Saveetha University, India
Reviewed by
Jennifer Manilay, University of California, Merced, United States
Kamala Kannan, Saveetha Medical College & Hospital, India
Updates
Copyright
© 2025 Luo, Wu, Guan, Li, Yang and Tao.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huiren Tao, 1910244002@email.szu.edu.cn; huiren_tao@163.com
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.