Abstract
Bacteria-based therapies have gained increasing attention as novel immunotherapeutic approaches against tumors. Among them, bacteria producing superantigen (SAg) toxins are considered particularly effective due to their ability to induce potent, generalized inflammatory responses capable of destroying tumor cells. Building on evidence of the antitumor efficacy of certain streptococcal preparations, and on the known involvement of heat shock proteins (HSPs) in tumor progression, we tested the hypothesis that streptococcal SAgs may elicit an adaptive immune response against tumors by priming cytotoxic T cells with epitopes that closely resemble tumor-associated HSPs. Through a multistep immunoinformatic analysis, we identified HSP70, Grp78, and Grp94 as containing immunogenic epitopes with high similarity to those found in the SAg domains of streptococcal exotoxins. Notably, a long sequence of HSP70 and its homolog in Grp78 was found to harbor multiple immunodominant epitopes overlapping the MHC class I and II epitopes of exotoxins, also containing B-cell epitopes. Results suggest that specific sequences of HSP70 and Grp78 may act as shared tumor antigens targeted by the immune response initiated by streptococcal SAgs, supporting their potential use as peptide-based tumor vaccines.
Introduction
Immunotherapy is currently one of the most promising strategies in cancer treatment, offering the potential for durable and complete remission and addressing key limitations of conventional therapies (1, 2). Its success depends on the ability to stimulate the immune system to mount an adaptive response capable of eliminating tumor cells without harming healthy tissue. This requires the identification of tumor-associated antigens (Ags) that activate T-cell subsets specifically targeting Ag-expressing tumor cells (3–6).
Within this context, bacterial immunotherapy has attracted renewed interest due to its intriguing and beneficial immunomodulatory effects (7–10). Since Coley’s early work in the late 1800s, where Streptococcus pyogenes preparations were used to treat bone sarcomas (11, 12), bacterial therapies have evolved and are now employed as adjuncts to standard treatments (13, 14) or as cancer vaccines, either alone or in combination with other tumor-targeting agents (15–18). Independent of their therapeutic formulation, bacteria have been shown to influence the outcomes of various cancer immunotherapies (19, 20). Notably, in esophageal squamous cell carcinoma, the abundance of Streptococcus in the intra-tumoral microbiome has been associated with improved response to chemoimmunotherapy (21).
Despite these advances, the precise mechanisms by which bacteria trigger effective in vivo antitumor immune responses remain incompletely understood. Certain bacterial strains, including Streptococcus pyogenes and Staphylococcus aureus (22, 23), produce potent protein toxins termed superantigens (SAgs), which induce extensive T-cell proliferation and cytokine release far exceeding the response elicited by conventional Ags (22, 24). While SAgs are also implicated in acute and chronic human diseases (24–27), their structural and functional characteristics, particularly a conserved SAg domain, suggest a shared capacity to induce robust inflammatory responses (22, 28).
The antitumor effects of bacterial therapies appear to be primarily mediated by this inflammation, which can occur both locally at the tumor site and systemically, potentially leading to tumor cell death independent of Ag specificity. Clinical observations dating back to Coley indicate that stronger febrile and inflammatory responses correlate with improved patient outcomes (11). Moreover, converting immunologically “cold” tumors into “hot,” inflamed tumors through pathogen-mimicking stimuli has been shown to improve responses to immunotherapy (29, 30). Acute infection-driven inflammation or exposure to bacterial/viral SAgs can also reshape the tumor microenvironment, promoting a transition from an immunosuppressive to a pro-inflammatory, cytotoxic-dominated antitumor response even at anatomical sites distant from the original immune activation (21, 31, 32).
Recent findings suggest that Streptococcus may directly colonize tumor tissues, promoting cytotoxic T-cell infiltration and enhancing immunotherapy responsiveness (21, 32). This raises the possibility that antitumor immunity may, in part, arise from Ag cross-reactivity between bacterial and tumor-derived molecules (31).
These insights have motivated the development of tumor-targeted SAg therapies, which aim to direct the immunostimulatory activity of bacterial toxins specifically to tumor sites (16, 18). Fusion constructs have been engineered combining SAgs with monoclonal antibodies or antibody fragments specific for tumor-associated Ags, allowing for localized cytokine release and tumor cell killing (33). Although promising results have been demonstrated in vitro (33) and in murine models (16, 34), clinical trials in humans have not achieved the desired outcomes (35, 36). This underscores the need to more precisely define how SAgs function within the complex immunological landscape of human cancers (31).
A retrospective analysis of Coley’s successes (11, 12, 37) suggests that intense inflammation alone may not fully account for tumor remission. An alternative mechanism could involve Ag-specific cytotoxic lymphocyte activation by streptococcal exotoxins, targeting tumor cells expressing similar Ags. If true, this implies molecular similarity between streptococcal exotoxins and tumor-associated Ags. Among the most plausible candidates are Heat Shock Proteins (HSPs), a conserved family of intracellular chaperones that when liberated from their physiological setting become potent inducers of both innate and Ag-specific immunity (38). HSPs are frequently overexpressed in tumors (39) and often exposed on the tumor cell surface or released into circulation, contributing to their strong immunogenicity (38, 40). HSP expression is closely linked to the oncogenic process, supporting tumor cell survival, proliferation, metastasis, chemoresistance, and poor clinical outcomes (41–43).
The most identified HSPs in tumors include Grp94 (40, 44), HSP90 (45, 46), Grp78 (47), HSP70 (46, 48), and HSP60 (49). Notably, many bacterial immunodominant Ags are themselves HSPs (50–52), and a high degree of sequence homology exists across species within each HSP family.
Based on this, we hypothesized that the antitumor activity of Streptococcus pyogenes SAgs may result from Ag-dependent T-cell activation driven by shared immunogenic peptides between SAgs and tumor-associated HSPs. To test this, we conducted an immunoinformatic analysis comparing major Streptococcus pyogenes exotoxins with human HSPs known to be tumor related. Our goal was to identify shared immunogenic epitopes that may serve as common Ags and potential targets for tumor-selective immunotherapy.
Methods
Protein retrieval
Protein sequences were retrieved from UniProtKB/Swiss-Prot (https://www.uniprot.org). The pyrogenic exotoxins of Streptococcus pyogenes (serotype M18) included: SPEA_STRP8 (P62561), SPEC_STRP8 (Q8NKX2), SPEM_STRPY (Q7WYA2), and SPEK_STRPY (A0A5S4TLM8). Human HSPs analyzed were: Endoplasmin (ENPL, P14625), HSP 90-alpha (P07900-1), Endoplasmic reticulum chaperone BiP (Grp78, P11021), HSP70 kDa 1A (HS71A, P0DMV8), and HSP60 (CH60, P10809). Tumor antigens retrieved were Trophoblast glycoprotein (TPBG_HUMAN, Q13641) and carcinoembryonic antigen-related cell adhesion molecule 5 (CEAM5_HUMAN, P06731-1).
Methodological approach
The methodology included a multi-step immunoinformatic analysis using common tools for designing epitope-based vaccines (53). As outlined in Figure 1, the first steps involved multiple sequence and structure alignments of exotoxins, and searching for sequence similarity between HSPs and exotoxins, focusing attention on the SAg region. The HSPs sequences with the best similarity with the common exotoxin region of SAg were analyzed for MHC-I, MHC-II, and B-cell epitopes. Only epitopes with positive class-I immunogenicity values were then considered for comparison with those identified in the SAg region of exotoxins. Matching epitopes would support the hypothesis that HSP sequences, structurally similar to SAgs, represent shared tumor antigens and potential targets of the exotoxin-driven antitumor response.
Figure 1

Flowchart outlining the strategy for identifying HSP sequences as shared tumor antigens. The process included key analytical phases to identify sequences in HSPs that share with Streptococcus pyogenes exotoxins the ability to activate immune responses. Each step summarizes the primary methods used to uncover HSP sequences with potential tumor antigen properties.
Protein sequence and structure analysis
Protein sequences were analyzed in pairwise and multiple sequence alignments using T-Coffee (https://tcoffee.crg.eu), and Clustal Omega (https://www.uniprot.org/align). Although based on different algorithms, both programs yielded consistent results in cases of high sequence similarity. T-Coffee assigns a maximum consensus score of 1000 (100%). The Expresso function of T-Coffee (https://tcoffee.crg.eu/apps/tcoffee/do:expresso) was used to assess structural similarity. Only uninterrupted segments of at least eight amino acids with the highest consistency scores were considered. BLAST (https://blast.ncbi.nlm.nih.gov) was used to verify whether similar sequences were conserved among HSPs and between HSPs and tumor markers 5T4 and CEA. An expect threshold of E<0.05 was set to determine statistical significance.
MHC class I, class II, and B-cell epitope detection
Predicted best MHC class I epitopes in exotoxins and HSPs were identified using the IEDB Analysis Resource (http://tools.iedb.org/mhci/), applying IEDB recommended 2020.09 methods: NetMHCpan EL 4.1predicting best MHC-I peptide score and percentile rank, and Ann 4.0 predicting IC50 values. A percentile rank of ≤0.5 and a IC50 value ≤500 nM were both considered indicative of strong binding (54). A representative panel of common HLA alleles was used: A*01:01; A*02:01; A*03:01; A*24:02; B*07:02; B*08:01; B*15:01; B*40:01; B*44:02; B*57:01. Immunogenicity of MHC-I epitopes was evaluated using IEDB’s class I immunogenicity tool (http://tools.iedb.org/immunogenicity/) that predicts immunogenicity of an epitope-MHC complex through the analysis of the location and properties of amino acids. A positive score indicates probable T-cell recognition, while a negative score suggests reduced likelihood (54).
MHC class II epitopes (predicted CD4+ T-cell epitopes) were identified using the IEDB Consensus tool (http://tools.iedb.org/mhcii/), employing a 7-allele reference panel: DRB1*03:01; DRB1*07:01; DRB1*15:01; DRB3*01:01; DRB3*02:02; DRB4*01:01; DRB5*01:01. Peptides with a percentile rank <0.5 and IC50 <50 nM (high affinity) or <500 nM (intermediate affinity) were retained (55).
Linear B-cell epitopes in exotoxins, Grp94, Grp78, and HSP70 were predicted using BepiPred-2.0 sequential B-cell epitope predictor (https://services.healthtech.dtu.dk) (56). A threshold score above the default 0.5 was used to increase specificity and reduce false positives.
Results
Sequence and structural similarity of streptococcal exotoxins
Four major exotoxins of Streptococcus pyogenes, SPEA, SPEC, SPEM, and SPEK, were analyzed as representative members of the bacterial SAg toxin family. Despite differences in length, multiple sequence alignment using T-Coffee revealed high consensus scores across all toxins, reflecting conserved sequences (Figure 2A), consistent with previous observations in other bacterial SAgs (22). The region showing the greatest conservation spanned amino acids 157–221 of SPEA and the corresponding aligned segments in SPEC, SPEM, and SPEK, which included a 12-residue sequence (bold in Figure 2A) corresponding to the β-strand/hinge/α-helix domain, known to be highly conserved among Staphylococcus aureus and Streptococcus pyogenes SAgs (22). Similar alignment results were confirmed using Clustal Omega (data not shown).
Figure 2

HSP sequences showing high similarity to the conserved superantigen (SAg) region of streptococcal exotoxins. (A) Multiple sequence alignment of exotoxins SPEA, SPEC, SPEM, and SPEK, showing sequence conservation in the SAg region: SPEA (aa 157–221), SPEC (141–204), SPEM (122–187), and SPEK (168–233) (highlighted in red). In bold the SAg domain sequence. Identical (*), conserved (): and semi-conserved (.) residues are indicated. (B) The HSP sequences are shown that aligned with the SAg regions of exotoxins in pairwise comparisons (see Supplementary Figure S2). (C) For each HSP sequence as in (B), the homologous counterpart in other HSPs was searched in BLAST. The identified new HSP sequences are reported on the right of the original HSP sequences (underlined the corresponding similar sequence). All sequences displayed high similarity (low E-values) and aligned with exotoxins with high confidence (T-Coffee score = 1000).
Structural alignment using Expresso (T-Coffee) also revealed strong similarity among exotoxins, particularly within the SAg domain, in both pairwise (Supplementary Figure S1A) and multiple alignments (Supplementary Figure S1B). The overlap of sequence and structural homology in this domain (Figure 2A and Supplementary Figure S1) supports previous findings that conservation within this region underlies a shared mechanism of action across SAgs (22), indicating potential for similar biological activity (28, 57).
Broad sequence similarity between streptococcal exotoxins and HSPs
Pairwise alignments of each exotoxin (SPEA, SPEC, SPEM, SPEK) with Grp94, HSP90, Grp78, HSP70, and HSP60 (Supplementary Figure S2) showed that ~90% of SPEA, SPEC, and SPEM sequences aligned with Grp94. SPEM showed the highest coverage with HSP90 (99%), Grp78 (91%), and HSP70 (92%). Each exotoxin sequence was largely covered by overlapping segments from at least two HSPs, especially for SPEA and SPEC (Supplementary Figure S2A). Long contiguous HSP segments, often over 30 amino acids, matched exotoxin regions with minimal gaps. To exclude chance similarity, exotoxins were aligned with seven unrelated human proteins (albumin, antitrypsin, beta2 microglobulin, apolipoprotein A, adiponectin, kallikrein, vitamin D-binding protein). In comparison with exotoxins, the control proteins consistently showed lower sequence identity (average ≤60%) and shorter aligned segments (data not shown).
Specific HSP sequences match streptococcal exotoxins in the SAg domain
Given the immunological relevance of the SAg domain (22, 25, 27, 57), HSP sequences overlapping this region were examined (Figure 2B). Only HSP segments fully aligned with the SAg domain in pairwise alignments (Supplementary Figure S2) were included. Selection criteria were segment length and alignment with multiple exotoxins. Due to the greater length of HSPs relative to exotoxins, matches with common regions of multiple exotoxins supported sequence significance. Grp78 (residues 544–586) and Grp94 (534–570) overlapped various exotoxin segments, often with internal overlap. HSP70 (471–509) aligned with SPEC and partly overlapped its SAg domain. HSP90 and HSP60 showed only sparse, short matches in this region (Figure 2B).
Given the high sequence similarity between HSP family members (Grp94/HSP90: 92%; Grp78/HSP70: 96%), homologous segments of aligned HSP sequences were sought. Additional matching segments were thus identified (Figure 2C): Grp94 476–505 and 534–570 aligned, respectively, with HSP90 436–456 and 487–523; reciprocally, HSP90 450–468 and 579–596 matched, respectively, Grp94 498–512 and 630–646. HSP70 471–509 corresponded to Grp78 494–532, and Grp78 544–573 aligned with HSP70 521–550. Grp78 519–539 matched HSP70 499–516, while Grp78 573–586 lacked a homolog in HSP70, delimiting alignment to the residue 573. Combined, these homologous sequences in Grp78/HSP70 formed nearly continuous segments (HSP70: 471–550; Grp78: 494–573) separated by a five-residue gap. HSP60 displayed minimal sequence similarity to other HSPs (data not shown), with only two short regions (363–383) aligning with HSP70 (512–521) and Grp78 (590–603) (Figure 2C).
These additional homologous HSP sequences aligned with the same exotoxin segments as the originals. The results thus showed that Grp94, and especially Grp78 and HSP70, contained distinct segments with significant similarity to the exotoxin SAg region.
Similarity of MHC-I epitopes between streptococcal exotoxins and HSPs
To assess whether the HSP sequences with significant similarity to the SAg region also exhibited immunogenic potential, we analyzed their content of MHC-I epitopes and compared them with those of streptococcal exotoxins. The necessary premise to this was that the exotoxins, like other foreign proteins, are internalized into antigen-presenting cells (APCs) and presented in complex with MHC-I and MHC-II molecules to TCRs (58). Only epitopes with high predicted binding affinity (IC50 <500 nM) were selected from exotoxins and HSPs (Supplementary Tables S1, S2).
Exotoxin epitopes were predominantly localized in the N-terminal and SAg regions. While isolated epitopes were observed in the N-terminus, a notable concentration of overlapping epitopes was found in the SAg domain: six in SPEA 173–204, six in SPEC 157–187, five in SPEK 182–211, and even thirteen in SPEM 125–188, despite SPEM being the shortest exotoxin (Table 1). All epitopes were allele-specific, except one epitope, each in SPEC and SPEM, which bound two distinct alleles.
Table 1
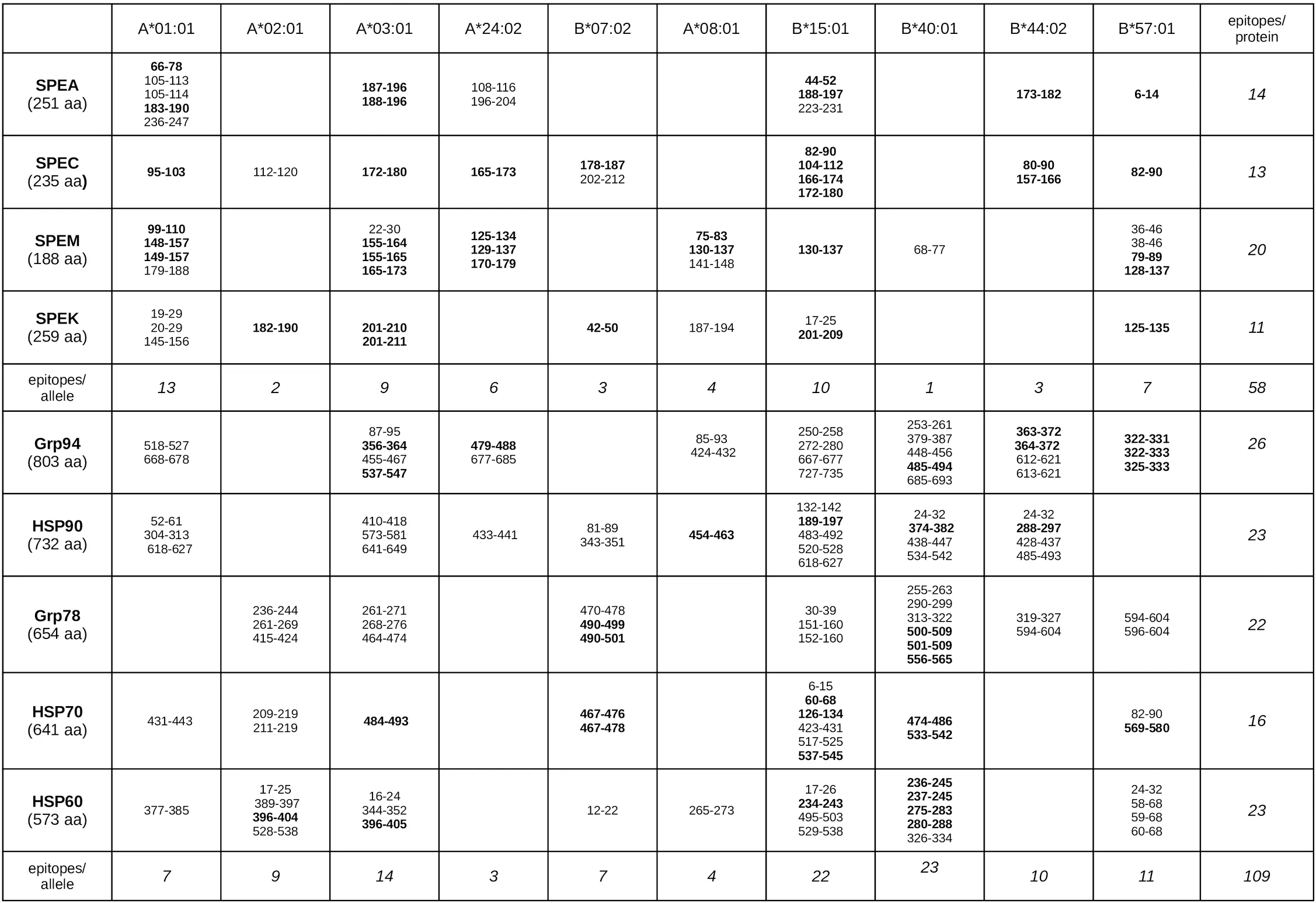
|
Sequences of both exotoxins and HSPs identified as MHC-I epitopes with the best binding affinity to HLA-A and HLA-B alleles.
For each protein (length in parentheses), the table lists all MHC-I epitopes identified using IEDB tools in exotoxins (Supplementary Table 1) and HSPs (Supplementary Table 2) Each epitope is shown with the binding affinity to a specific HLA-A or HLA-B allele. Bolded sequences indicate matching epitopes between exotoxins and HSPs, as shown in pairwise alignments (Supplementary Figures 2, 3). The right column lists the total number of epitopes per protein; the bottom row shows the total number of epitopes per allele group.
Among HSPs, Grp94, HSP90, Grp78, and HSP60 contained numerous epitopes, whereas HSP70 exhibited fewer total epitopes but all concentrated within the SAg-similar regions (Table 1 and Supplementary Table S2). The most frequently targeted alleles included HLA-B40:01 and B15:01, the latter shared with several exotoxin epitopes. Multiple HSP sequences contained overlapping epitopes, particularly within Grp94, HSP90, Grp78, and HSP70 (Table 1).
To determine overlapping between HSP and exotoxin epitopes, MHC-I epitope mapping was performed along previously aligned regions (Supplementary Figure S2). Many HSP epitopes, especially in Grp94 and HSP70, closely matched those of SPEM and SPEA (Supplementary Figure S3). Notably, these epitopes were localized within the same sequences previously aligned to the SAg region, which itself harbored a dense cluster of overlapping epitopes (Figure 3A). In contrast, matching epitopes outside the SAg region were fewer and spatially isolated (Supplementary Figure S3), reinforcing the functional significance of the SAg domain in immune stimulation. Importantly, the extended sequences of Grp78 and HSP70, each forming a single long homologous stretch (Figure 2C), contained epitopes overlapping with those of all exotoxins, particularly SPEA, SPEC, and SPEM (Figure 3A).
Figure 3

MHC-I epitope immunogenicity of HSPs matching exotoxin sequences in the SAg region. (A) HSP sequences aligned with exotoxins (from Figure 2B) that contain MHC-I epitopes (bold) overlapping those in the SAg region. Asterisks (*) indicate homologous HSP sequences derived from alignments in Figure 2C. (B) Class-I immunogenicity scores of the epitopes in (A), with positive values (in red) indicating potential for T-cell recognition of the peptide -MHC complex.
While some sequences of Grp94, HSP90, and HSP60 aligned with the SAg domain, not all contained MHC-I epitopes. For instance, Grp94 476–496, aligned with SPEM, contained three epitopes, whereas the longer 534–570 segment, aligning with multiple exotoxins, included only one epitope in the sequence 534–557 aligned with SPEA (Figure 3A). HSP90 and HSP60 contributed fewer matching epitopes, two and one, respectively. Collectively, only the Grp78 and HSP70 sequences provided consistent, extensive overlap with the immunodominant SAg regions of all exotoxins.
Immunogenicity of HSP sequences containing MHC-I epitopes
Beyond binding affinity, T-cell immunogenicity is a critical determinant of functional epitope recognition (54). We therefore assessed the predicted immunogenicity of each HSP epitope with sequence similarity to exotoxins. Only epitopes with positive immunogenicity scores were considered functionally relevant (Figure 3B), although all had high MHC-I binding affinity (Supplementary Figure S3).
Among all peptides analyzed, the HSP70 sequence 471–493 and its homologous Grp78 494–516 displayed the highest immunogenicity, containing distinct, strongly immunogenic epitopes (Figure 3B). The Grp78 547–571 and the homologous HSP70 524–548 were moderately immunogenic, while the Grp78 556–565 epitope was more potent. Grp94 476–496, with three overlapping epitopes, also showed positive immunogenicity, unlike the Grp94 534–570 segment and all epitopes within HSP90, which were non-immunogenic. HSP60 epitopes displayed weak but detectable immunogenicity (Figure 3B).
Thus, only select regions of Grp78 and HSP70, specifically those also aligning with the SAg domains of exotoxins, appeared to possess strong immunogenic responses, supporting their potential to functionally mimic bacterial SAgs.
HSP70 and Grp78 sequences contain multiple epitope classes
While cytotoxic T lymphocytes are central to the anti-tumor immune response, both B cells and CD4+ T helper cells are essential for antigen recognition and the orchestration of adaptive immunity (59). To assess the immunogenic potential of HSP sequences homologous to the SAg domain (Figure 3B), the most predictive B-cell and MHC class II epitopes were identified within these regions. Exotoxins were analyzed in parallel for comparison.
Grp94 harbored several long B-cell epitopes, most of which did not overlap with its class I immunogenic sequences (Figure 4A). Only two residues, D477 and K479, located within the 476–496 region, served as both B-cell and class I epitopes. K479 (in bold) was the sole residue common to both (Figure 3). In contrast, the sequences 548–559 of Grp78 and its homologous region 524–537 in HSP70 functioned as B-cell epitopes, with partial overlap of class I epitopes (Figure 4A, underlined and bold), a result that supported the broad immunogenicity of the homologous sequences of Grp78 and HSP70. Compared to HSPs, exotoxins displayed a relatively limited number of B-cell epitopes, with overlapping B-cell and class I epitopes confined to short segments in SPEA, SPEC, and SPEM within the SAg domain.
Figure 4
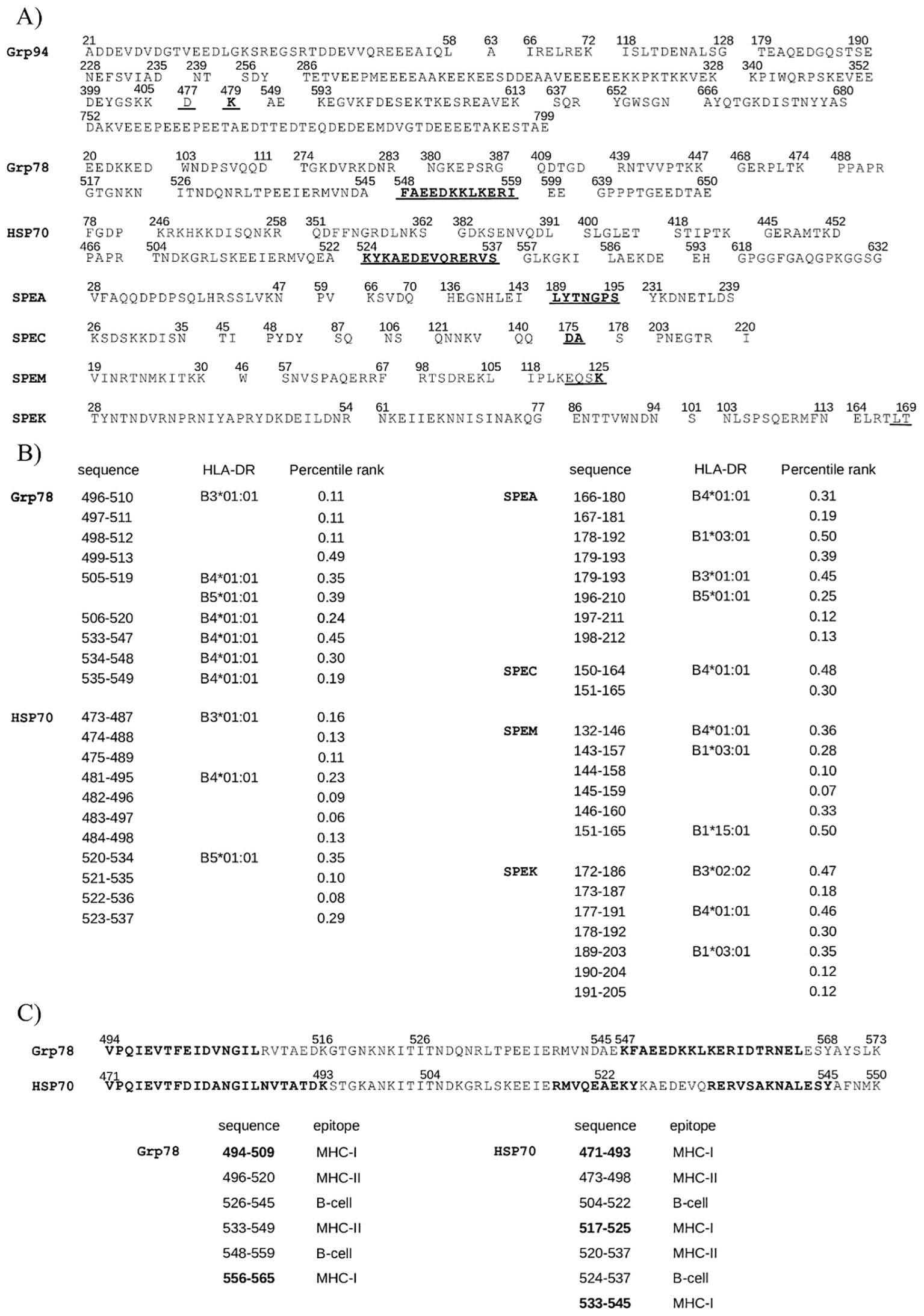
Grp78 and HSP70 sequences containing both MHC-I and B-cell epitopes also include MHC-II epitopes. (A) B-cell epitopes predicted by BepiPred-2.0 in HSPs and exotoxins. Underlined: B-cell epitopes with cross-similarity to the SAg region (Figure 2B); bold: epitopes also identified as MHC-I epitopes (Table 1). (B) Top-ranking MHC-II binding peptides in Grp78, HSP70, and exotoxins, with percentile rank (≤0.5) and binding allele listed. Shown are only those from cross-similar regions. Grp94 476 -496 did not yield any strong DRB-binding epitopes (Supplementary Table S3). (C) Immunogenic regions 494 -573 of Grp78 and 471 -550 of HSP70 showing class-I (bold), class-II (overlapping with class-I and B-cell epitopes on the left), and B-cell epitopes (middle/right).
A search for high-affinity MHC class II epitopes (percentile rank ≤0.50) across a representative panel of seven prevalent HLA class II alleles (Figure 4B) identified four Grp94 epitopes within residues 534–551, outside the class I immunogenic region (Supplementary Table S3). Notably, Grp78 and HSP70 contained nine and eleven class II epitopes, respectively. Of the Grp78 epitopes, six were found within residues 496–520, homologous to HSP70 473–498 (Figure 2C) and three within the sequence 533–549. Among the eleven HSP70 epitopes, seven mapped to residues 473–498 and four to 520–537, all within the immunogenic 471–550 segment shared with Grp78 544–573 (Figure 2C).
Exotoxins, particularly SPEA, SPEM, and SPEK, featured multiple class II epitopes overlapping class I epitopes in the SAg domain (see also Figure 3A). These epitopes predominantly bound to DRB1*03:01 (nine epitopes) and DRB4*01:01 (seven epitopes), the latter also targeted by epitopes in Grp78 and HSP70. Additional overlaps were found with DRB5*01:01 and DRB3*01:01, shared among both HSPs and exotoxins (Figure 4B). Grp78 (residues 505–549) and HSP70 (481–498) featured class II epitopes binding to DRB4*01:01, the same allele recognized by epitopes in the SAg regions of SPEA, SPEC, SPEM, and SPEK. Interestingly, the strongest binding affinity to DRB4*01:01 was observed for HSP70 epitopes within 482–497. Similarly, Grp78 and HSP70 epitopes targeting DRB3*01:01 demonstrated superior binding affinities compared to the corresponding SPEA epitope in the sequence 179–193 (Figure 4B).
The homologous 80-residue sequences of Grp78 and HSP70, formed by combining original and cross-matching segments (Figure 2C), contained overlapping epitopes of all three classes. Notably, regions Grp78 494–520 and HSP70 471–498 featured near-complete overlap of class I and class II epitopes (Figure 4C, bold). Additional overlapping B-cell and class II epitopes extended from 526 to 559 in Grp78 and from 504 to 537 in HSP70. Remarkably, within HSP70 517–527, epitopes of all three classes coincided. One particularly immunogenic segment was the B-cell epitope 526–545 in Grp78, which also included three consecutive class II epitopes. This region contained residues 526–539, homologous to SPEA 163–176, encompassing the SAg domain motif 165TNKKMVTAQELD176 (Figure 2B). This SPEA segment also included two class II epitopes with affinity for the same allele to which also the Grp78 epitopes bound (Figure 4B). Another significant region, HSP70 480–511, aligned with SPEM 105–136 in pairwise alignments (Supplementary Figure S2). This SPEM segment, which included the SAg motif 130FQNRFVT136 (Figure 3A), contained class I epitopes that overlapped two class I epitopes of HSP70 480–493 (Supplementary Table S2).
Figure 5 illustrates more clearly how the sequences of Grp78 and HSP70 are composed almost entirely of overlapping epitopes from different classes. While the left regions of both HSP sequences contain nearly identical types of epitopes of similar lengths, the right regions show some differences. Specifically, the HSP70 segment (residues 517–545) includes peptides that function as both MHC-II and B-cell epitopes and also overlap more extensively with MHC-I epitopes compared to the corresponding Grp78 segment (residues 540–568).
Figure 5
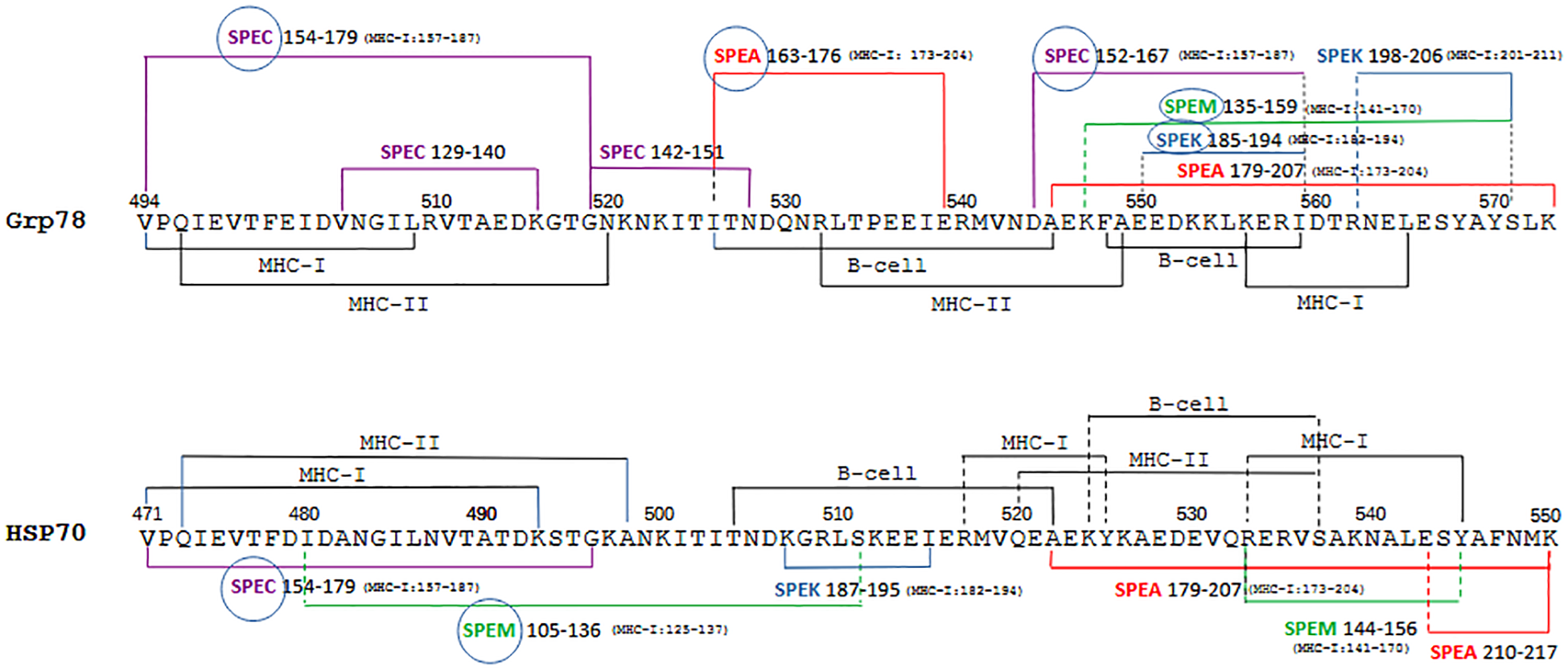
Summary visualization of epitopes of different classes and similar exotoxin sequences in the Grp78 and HSP70 sequences. The two homologous immunogenic sequences of Grp78 and HSP70 are shown with highlighted the regions covered by frequently overlapping MHC-I, MHC-II and B-cell epitopes, together with segments showing similarity to exotoxins. Where applicable, next to the exotoxin sequence is indicated in parenthesis the corresponding sequence range containing MHC-I epitopes (see Figure 3A). For sake of clarity, class II epitopes of exotoxins are not depicted. Encircled exotoxins represent those whose sequences partly or entirely contain the SAg domain (see Figure 2B).
Similarly, the left regions of both HSP sequences exhibit similarity exclusively with specific sequences from SPEC and SPEM. In contrast, the right regions share similarities with a broader range of exotoxin sequences. Notably, it is the Grp78 sequence—in contrast to HSP70—that shows a more extensive resemblance to all exotoxin sequences, many of which contain multiple MHC-I epitopes.
A careful inspection of the multiple exotoxin sequences aligned with the HSP sequences also shows how broad is the overall similarity coverage by both Grp78 and HSP70 of SPEA (163–217), SPEC (129–179) and SPEM (105–159). This similarity fully encompasses the shared SAg domain of these exotoxins (see Figure 2A). Less extensive similarity is instead observed between the HSP sequences and SPEK (185–206), although its SAg region is partially covered, mostly by Grp78 (Figure 5).
Homologous immunogenic sequences of HSP70 and Grp78 display significant similarity to tumor antigens 5T4 and CEA
The preceding findings supported the notion that the immunogenic sequences of Grp78 and HSP70 possess intrinsic capacity to trigger a complete immune response, comparable, or even superior, to that elicited by streptococcal SAg exotoxins. These sequences, when presented as tumor Ags, may therefore have the potential to activate effective anti-tumor immunity. Despite extensive experimental evidence demonstrating their role as tumor Ags, HSPs have not yet been translated into clinical use as diagnostic or prognostic biomarkers. To explore this possibility further, we examined whether the immunogenic regions of Grp78 and HSP70 exhibited sequence similarity with tumor-associated antigens currently used in clinical practice, namely 5T4 and CEA. The trophoblast glycoprotein 5T4 is a cell surface antigen overexpressed across a broad spectrum of malignancies, with limited expression in normal tissues (60). Although 5T4 has been evaluated as a target for tumor immunotherapy (18), clinical outcomes have remained modest (35, 36, 61).
No sequence similarity was observed between 5T4 and the immunogenic region of Grp94 476–496. In contrast, both Grp78 and HSP70 exhibited high-scoring sequence alignments with 5T4. Pairwise alignments conducted using both T-Coffee and Clustal Omega revealed a high degree of similarity, with the strongest alignment occurring between residues 493–504 of Grp78 and 470–479 of HSP70 and the 283–292 region of 5T4 (Figure 6A). Multiple sequence alignment further confirmed this observation, yielding a consensus score exceeding 90% when the full-length sequences of Grp78 and HSP70 were aligned with 5T4 (Supplementary Figure S4), a result not replicated with any other HSP.
Next, sequence similarity was assessed with CEA, a well-characterized glycoprotein of the immunoglobulin superfamily and a widely used biomarker in various cancers (62). As with 5T4, Grp94 did not show any significant similarity to CEA. However, homologous immunogenic sequences of Grp78 and HSP70 did display notable alignment with CEA. Unlike 5T4, where the similarity was confined to a single segment, CEA exhibited multiple, discrete regions of similarity. This is consistent with the genomic architecture of the CEA family, which comprises a series of tandem gene duplications encoding repeated amino acid motifs (62).
Grp78 sequences 494–517 and 547–568, both within the broader 494–568 region, aligned with CEA sequences, with which also the HSP70 471–493 segment showed similarity (Figure 6B). Additionally, the HSP70 region spanning residues 524–545 aligned with three other distinct CEA sequences. When mapped sequentially, the regions of CEA showing similarity to HSP sequences revealed that HSP70 aligned with a broader set of CEA segments (Figure 6C). Notably, while the left portions of the HSP sequences (Grp78 493–504 and HSP70 470–479) were more similar to 5T4 (Figure 6A), their right portions (Grp78 547–568 and HSP70 524–545) demonstrated greater similarity to CEA (Figure 6B).
Figure 6
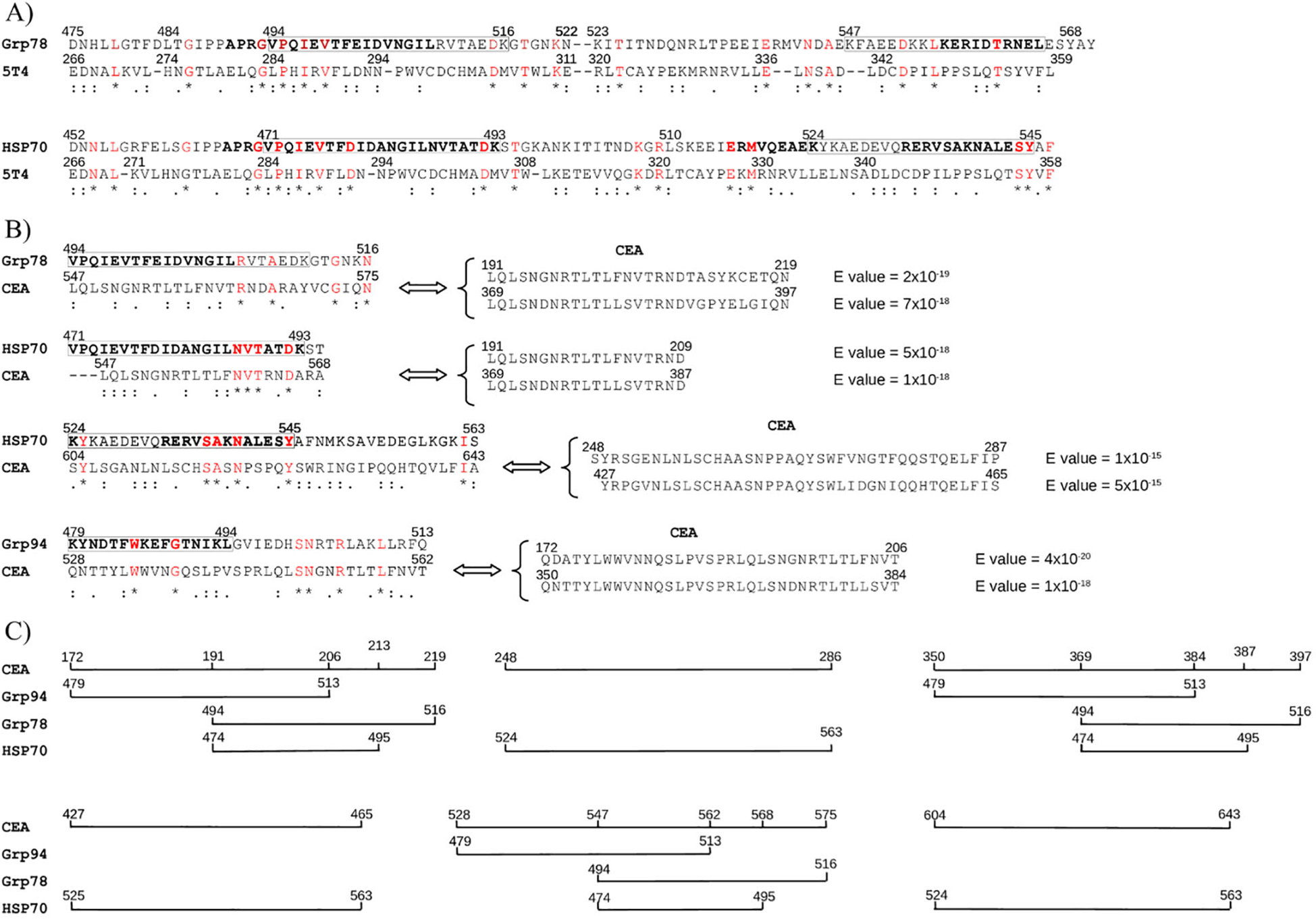
Sequence similarity between immunogenic Grp78/HSP70 regions and tumor markers 5T4 and CEA. (A) Pairwise alignments show 5T4 sequences with similarity to immunogenic regions of Grp78 and HSP70. Boxed regions correspond to class-I epitopes (bold, see Figure 3B). No significant similarity was found between 5T4 and Grp94. Below sequences: * (red) = identical; : = conserved; . = semi-conserved residues. (B) CEA sequences with similarity to Grp78 and HSP70 based on pairwise alignment. Each CEA sequence also showed strong similarity (BLAST, low E-values) to at least two other CEA regions, indicating intra-gene family homology. (C) Graphical representation of CEA sequence coverage by HSP-matching regions, showing the extent of continuous similarity to Grp78 and HSP70.
These findings reinforce the immunogenic relevance of the Grp78 and HSP70 sequences and support their potential involvement in tumor immunity through cross-reactivity with clinically recognized tumor antigens.
Discussion
In this study, we tested the hypothesis that the antitumor effects observed in bacteria-based therapies could result from an adaptive immune response initiated by sequence similarity between bacterial SAgs and specific HSP sequences known to function as tumor Ags. Considering the significant impact that various streptococcal preparations have demonstrated in cancer therapy over time (10, 12, 21), we selected the streptococcal exotoxins SPEA, SPEC, SPEM, and SPEK as model SAgs to test this hypothesis.
It is well established that bacterial and viral SAgs share conserved sequence homology within the SAg domain, which is associated with a common structural conformation that underpins their potent immunostimulatory activity (22–24). Our analysis confirmed that the streptococcal exotoxins exhibit the highest degree of sequence and structural similarity around the SAg domain and its adjacent regions (Supplementary Figure S1B and Figure 2A), despite interindividual sequence variations.
The rationale for comparing HSPs to these bacterial exotoxins stems from the knowledge that HSPs exhibit extensive sequence conservation across species within their respective families and are recognized as immunodominant antigens in various bacterial and viral infections (63–65). Molecular mimicry between bacterial and human HSPs has been implicated in the pathogenesis of different inflammatory diseases due to antigenic cross-reactivity (66–68). Moreover, several HSPs, especially Grp94, HSP90, Grp78, HSP70, and HSP60 are involved in the development and progression of various tumors (40, 44, 46, 48, 49).
Pairwise alignment analyses revealed that not only there was an extensive similarity between HSPs and exotoxins (Supplementary Figure S2), not shared by any of the unrelated control proteins, but also that the highest density of similarity precisely occurred in the region of the SAg domain. Certain HSPs, specifically Grp94, Grp78, and HSP70, displayed extensive, crucial similarity that in the case of Grp94 and Grp78 involved the SAg region of all tested exotoxins with segments of a single, uninterrupted sequence. This finding was of relevance in consideration of the different length of the HSPs compared to that of exotoxins. Although HSP70 was found to align exclusively with SPEC, the alignment was considered significant in that it occurred with an uninterrupted long stretch of HSP70 that nearly covered the entire SAg region (Figure 2B).
Given the inherent sequence homology among HSPs of the same class, we anticipated that additional, previously unidentified homologous sequences would exist that exhibit similar degrees of similarity to the exotoxins. Indeed, homologous sequences were found between HSP70 and Grp78, as well as between Grp94 and HSP90, whereas only short homologous segments were identified between HSP60 and either HSP70 or Grp78 (Figure 2C). These findings revealed that Grp78 and HSP70, when considered together with their homologous counterparts, formed a nearly continuous 80-amino acid-long sequence, a feature not observed with Grp94 or HSP90, thereby underscoring their potential functional importance.
To assess whether the HSP sequences aligned with the SAg domains possessed immunogenic properties comparable to those of exotoxins, we analyzed predicted MHC class I and II epitopes, as well as B-cell epitopes, within both HSPs and exotoxins. The results showed that MHC-I epitopes in the exotoxins were predominantly located within the SAg domain (Supplementary Table S1, Supplementary Figure S3), highlighting the immunogenic nature of this region and supporting its proposed internalization and presentation by APCs in complex with MHC class I molecules (15, 58, 69, 70).
Among the HSPs, only Grp78 and HSP70, and to a lesser extent Grp94, contained class I epitopes within the aligned sequences. Notably, the original and homologous sequences of Grp78 and HSP70 included numerous high-affinity MHC-I epitopes overlapping those of all exotoxins (Figures 3A). In particular, the original HSP70 sequence aligned with SPEC and its homologous in Grp78 contained peptides that also exhibited high predicted immunogenicity scores (Figures 3A, B).
Further analysis of MHC class II and B-cell epitopes reinforced the conclusion that Grp78 and HSP70 possess the highest immunogenic potential. In contrast to Grp94, these HSPs not only contained strong class I epitopes but also presented extensive class II and B-cell epitopes, suggesting their capacity to elicit robust humoral and cellular immune responses (Figures 4, 5). The 80-amino acid sequences derived from the original and homologous segments of Grp78 and HSP70 were composed entirely of immunogenic regions: class I and II epitopes were predominantly located in the N-terminal regions (coinciding with the segments aligned to SPEC), while B-cell epitopes were localized toward the central and C-terminal portions (Figure 4C). These B-cell epitopes overlapped or laid adjacent to class II epitopes, forming a structurally and functionally integrated immunogenic unit.
Importantly, these epitopes not only exhibited high individual immunogenicity but also demonstrated extensive sequence similarity with the epitopes of the exotoxins. Furthermore, many of the class II epitopes from Grp78 and HSP70 shared high-affinity binding to the same HLA alleles as those from the exotoxins (Figure 4B), indicating functional mimicry at the level of antigen presentation.
The discovery that the immunodominant peptides within Grp78 and HSP70 align with the SAg domain of exotoxins and mimic their potent immunostimulatory properties has important implications. These HSP-derived sequences appear to function as endogenous analogs of bacterial SAgs, supporting the concept that human HSPs can exhibit antigenic cross-similarity with microbial proteins (38, 63–65). Within the context of streptococcal antitumor therapy, it is plausible that SAgs exert their effects by priming CD8+ T lymphocytes against HSP-derived epitopes that are overexpressed in tumor cells, either on the cell surface or as circulating soluble antigens.
This mechanistic link provides a basis for proposing that the HSP sequences identified here could function as tumor-specific antigens. Although various HSPs are well known for their roles in cancer biology and have been identified as tumor Ags (39, 40, 45–47, 49), none have yet been adopted as clinically validated tumor biomarkers. Our observation that the well-established tumor markers 5T4 and CEA exhibit substantial sequence similarity with the immunogenic regions of Grp78 and HSP70 (Figure 6) provides compelling support for the antigenic nature of these HSP sequences. This similarity likely reflects altered protein processing and expression accompanying oncogenic transformation (39). The upregulation of HSP genes in response to cellular stress (41, 42), particularly HSP70 and Grp94, is known to facilitate tumor antigen processing and presentation (39, 71–74). The fusion or stable association of HSPs with tumor-specific proteins may account for the observed high similarity between HSPs and tumor markers such as 5T4 and CEA. In addition to the global alignment similarity of HSP70 and Grp78 with 5T4 (Supplementary Figure S4), a result that as such deserves further investigation, it is particularly notable that immunogenic regions within Grp78 and HSP70 correspond closely to class I epitopes in 5T4 (data not shown) and also sustain a broader similarity with CEA (Figure 6B).
In summary, our in-depth bioinformatic investigation identified two homologous Grp78 and HSP70 sequences that closely mimic immunodominant epitopes of streptococcal SAg exotoxins. The additional finding of the strong similarity shared with two clinically relevant tumor antigens suggests that these HSP sequences may function as shared tumor antigens capable of eliciting a potent immune response, similar to that triggered by SAg exotoxins. Although these in silico findings have broad implications for tumor immunology—particularly in the development of vaccines and tumor biomarkers—caution is warranted in interpreting them as having immediate translational relevance. A key limitation of our study is the absence of in vitro or in vivo experiments needed to validate any bioinformatic predictions.
Although HSPs are obligatory intracellular proteins and therefore ignored by the immune system, their occasional surface expression or molecular mimicry with extracellular components cannot be ruled out, particularly following the administration of exogenous HSPs. Therefore, in addition to immunizing a host to assess the specific immune response to these HSP sequences, future research should also consider the potential risks of autoimmune reactions associated with administering highly immunogenic HSPs.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
PF: Investigation, Software, Writing – original draft, Writing – review & editing, Data curation, Methodology, Formal analysis, Conceptualization.
Funding
The author(s) declare that no financial support was received for the research, and/or publication of this article.
Acknowledgments
I would like to express my sincere gratitude to Dr. Pietro Veronesi for his invaluable contribution in generating the figures depicting protein sequences. I also extend my thanks to Dr. Andrea Pagetta for his critical review and insightful feedback on the manuscript.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1644687/full#supplementary-material
Supplementary Figure 1Structure alignment of exotoxins. Exotoxins were structurally aligned using Expresso T-Coffee. (A) Pairwise alignments highlight the most similar SAg domain regions (in bold), each with a consensus score of 1000. (B) Multiple alignment starting from residue 42 of SPEA shows corresponding regions in SPEC, SPEM, and SPEK, with a consensus score of 890. In both (A) and (B), symbols indicate identical (*), conserved (:), and semi-conserved (.) residues.
Supplementary Figure 2Sequence alignment of exotoxins with HSPs. Pairwise alignments (see Methods) show only gap-free, similar sequences of ≥8 amino acids. SAg domain sequences are in bold.
Supplementary Figure 3Predicted MHC-I binding peptides of HSPs matching exotoxin epitopes. MHC-I epitopes of HSPs (supplemental Table 2) overlapping exotoxin epitopes (supplemental Table 1) are shown based on alignments (supplemental Figure 2). Bold amino acids represent MHC-I peptides; non-bold are adjacent residues. Epitope numbers refer to alignment positions; bold residues adjacent to numbered epitopes refer to epitope residues that may or may not match known exotoxin or HSP epitopes. Multiple epitopes often exist within a single MHC-I peptide.
Supplementary Figure 4Alignment of Grp78, HSP70, and 5T4. T-Coffee alignment highlights the regions with higher sequence consensus, excluding the first 73 and last 4 amino acids of 5T4 due to low similarity. An overall similarity score of 945 was found among the three proteins, with HSP70 showing the highest sequence consensus (960). Symbols indicate identical (*, red), conserved (:), semi-conserved (.) residues. Bold indicates shared MHC-I epitopes in Grp78 and HSP70 (Table 1).
Supplementary Table 1Predicted best MHC-I peptides in exotoxins. Exotoxins were scanned for MHC-I peptides using IEDB (see Methods) with a set of common HLA–A, HLA–B alleles. IC50 values <500 nM (in parentheses) was considered for good binding.
Supplementary Table 2Predicted best MHC-I peptides in HSPs. HSPs were scanned for MHC-I peptides using IEDB (see Methods) with a set of common HLA–A, HLA–B alleles. IC50 <500 nM (in parentheses) was considered for good binding.
Supplementary Table 3Predicted best MHC-II peptides in exotoxins and HSPs. MHC-II peptides in exotoxins and Grp94, Grp78, HSP70 were predicted using IEDB (2023.05 method) with seven prevalent HLA alleles. Only top binders (percentile rank ≤0.5) were included.
References
1
Enokida T Moreira A Bhardwaj N . Vaccines for immunoprevention of cancer. J Clin Invest. (2021) 131:e146956. doi: 10.1172/JCI146956
2
Raimondo TM Reed K Shi D Langer R Anderson DG . Delivering the next generation of cancer immunotherapies with RNA. Cell. (2023) 186:1535–40. doi: 10.1016/jcell.2023.02.031
3
Jou J Harrington KJ Zocca M Ehrnrooth E Cohen EEW . The changing landscape of therapeutic cancer vaccines – novel platforms and neoantigen identification. Clin Cancer Res. (2021) 27:689–703. doi: 10.1158/1078-0432.CCR-20-0245
4
Perrinjaquet M Schiegel CR . Personalized neoantigen cancer vaccines: an analysis of the clinical and commercial potential of ongoing development programs. Drug Discov Today. (2023) 28:103773. doi: 10.1016/jdrudis.2023.103773
5
Liu J Fu M Wang M Wan D Wei Y Wei X . Cancer vaccines as promising immuno-therapeutics: platform and current progress. J Hematol Oncol. (2022) 15:28. doi: 10.1186/s13045-022-01247-x
6
Johnson PC Gainor JF Sullivan RJ Longo DL Chabner B . Immune checkpoint inhibitors – the need for innovation. N Engl J Med. (2023) 388:1529–32. doi: 10.1056/NEJMsb2300232
7
Łukasiewicz K Fol M . Microorganisms in the treatment of cancer: advantages and limitations. J Immunol Res. (2018) 2018:2397808. doi: 10.1155/2018/2397808
8
Mayakrishnan V Kannappan P Tharmalingam N Bose RJC Madheswaran T Ramasamy M . Bacterial cancer therapy: A turning point for new paradigms. Drug Discov Today. (2022) 27:2043–50. doi: 10.1016/j.drudis.2022.03.007
9
Wei X Du M Chen Z Yuan Z . Recent advances in bacteria-based cancer treatment. Cancers. (2022) 14:4945. doi: 10.3390/cancers14194945
10
Marzhoseyni Z Shojaie L Tabatabaei SA Movahedpour A Safari M Esmaeili D et al . Streptococcal bacterial components in cancer therapy. Cancer Gene Ther. (2022) 29:141–55. doi: 10.1038/s41417-021-00308-6
11
DeWeerdt S . Bacteriology: a caring culture. Nature. (2013) 504:S4–5. doi: 10.1038/504S4a
12
Carlson RD Flickinger JCJ Snook AE . Talkin’ toxins: from Coley’s modern cancer immunotherapy. Toxins. (2020) 12:241. doi: 10.3390/toxins12040241
13
Fan JY Huang Y Li Y Muluh TA Fu SZ Wu JB . Bacteria in cancer therapy: A new generation of weapons. Cancer Med. (2022) 11:4457–68. doi: 10.1002/cam4.4799
14
Chen YE Bousbaine D Veinbachs A Atabakhsh K Dimas A Yu VK et al . Engineered skin bacteria induce antitumor T cell responses against melanoma. Science. (2023) 380:203–10. doi: 10.1126/science.abp9563
15
Okamoto M Oshikawa T Tano T Ahmed SU Kan S Sasai A et al . Mechanism of anticancer host response induced by OK-432, a streptococcal preparation, mediated by phagocytosis and Toll-like receptor 4 signaling. J Immunother. (2006) 29:78–85. doi: 10.1097/01.cji.0000192106.32206.30
16
Dohlsten M Abrahmsen L Björk P Lando PA Hedlund G Forsberg G et al . Monoclonal antibody-superantigen fusion proteins: tumor-specific agents for T-cell-based tumor therapy. Proc Natl Acad Sci USA. (1994) 91:8945–9. doi: 10.1073/pnas.91.19.8945
17
Okamoto M Furuichi S Nishioka Y Oshikawa T Tano T Ahmed SU et al . Espression of toll-like receptor 4 on dendritic cells is significant for anticancer effect of dendritic cell-based immunotherapy in combination with an active component of OK-432, a streptococcal preparation. Cancer Res. (2004) 64:5461–70. doi: 10.1158/0008-5472.CAN-03-4005
18
Patterson KG Pittaro JLD Bastedo PS Hess DA Haeryfar SMM McCormick JK . Control of established colon cancer xenografts using a novel humanized single chain antibody-streptococcal superantigen fusion protein targeting the 5T4 oncofetal antigen. PloS One. (2014) 9:e95200. doi: 10.1371/journal.pone.0095200
19
Villemin C Six A Neville BA Lawley TD Robinson MJ Bakdash G . The heightened importance of the microbiome in cancer immunotherapy. Trends Immunol. (2023) 44:44–59. doi: 10.1016/j.it.2022.11.002
20
Brevi A Zarrinpar A . Live biotherapeutic products as cancer treatments. Cancer Res. (2023) 83:1929–32. doi: 10.1158/0008-5472.CAN-22-2626
21
Wu H Leng X Liu Q Mao T Jiang T Liu Y et al . Intratumoral microbiota composition regulates chemoimmunotherapy response in esophageal squamous cell carcinoma. Cancer Res. (2023) 83:3131–44. doi: 10.1158/0008-5472.CAN-22-2593
22
Popugailo A Rotfogel Z Supper E Hillman D Kaempfer R . Staphylococcal and streptococcal superantigens trigger B7/CD28 costimulatory receptor engagement to hyperinduce inflammatory cytokines. Front Immunol. (2019) 10:942 PMID: 31114583. doi: 10.3389/fimmu.2019.00942
23
Li H Llera A Tsuchiya D Leder L Ysern X Schievert PM et al . Three-dimensional structure of the complex between a T cell receptor beta chain and the superantigen staphylococcal enterotoxin B. Immunity. (1998) 9:807–16. doi: 10.1016/s1074-7613(00)80646-9
24
Fraser JD Proft T . The bacterial superantigen and superantigen-like proteins. Immunol Rev. (2008) 225:226–43. doi: 10.1111/j.1600-065X.2008.00681.x
25
Cheng MH Zhang S Porrit RA Rivas MN Paschold L Willscher E et al . Superantigenic character of an insert unique to SARS-CoV-2 spike supported by skewed TCR repertoire in patiens with hyperinflammation. Proc Natl Acad Sci USA. (2020) 117:25254–62. doi: 10.1073/pnas.2010722117
26
Kasper KJ Zeppa JJ Wakabayashi AT Xu SX Mazzucca DM Welch I et al . Bacterial superantigens promote acute nasopharyngeal infection by Streptococcus Pyogenes in a human MHC class II-dependent manner. PloS Pathog. (2014) 10:e1004155. doi: 10.1371/journal.ppat.1004155
27
Arad G Levy R Nasie I Hillman D Rotfogel Z Barash U et al . Binding of superantigen toxins into the CD28 homodimer interface is essential for induction of cytokine genes that mediate lethal shock. PloS Biol. (2011) 9:e1001149. doi: 10.1371/journal.pbio.1001149
28
Li PL Tiedemann RE Moffat SL Fraser JD . The superantigen streptococcal pyrogenic exotoxin C (SPE-C) exhibits a novel mode of action. J Exp Med. (1997) 186:375–83. doi: 10.1084/jem.186.3.375
29
Abreu MM Chocron AF Smaddja DM . From cold to hot: mechanisms of hyperthermia in modulating tumor immunology for enhanced immunotherapy. Front Immunol. (2025) 16:1487296. doi: 10.3389/fimmu.20251487296
30
Ochoa de Olza M Rodrigo BN Zimmermann S Coukos G . Turning up the heat on non-immunoreactive tumours: opportunities for clinical development. Lancet Oncol. (2020) 21:e419–30. doi: 10.1016/S1470-2045(20)30234-5
31
Sfanos KS . Intratumoral bacteria as mediators of cancer immunotherapy response. Cancer Res. (2023) 83:2985–6. doi: 10.1158/0008-5472.CAN-23-1857
32
Peiffer LB White JR Jones CB Slottke RE Ernst SE Moran AE et al . Composition of gastrointestinal microbiota in association with treatment response in individuals with metastatic castrate resistant prostate cancer progressing on enzalutamide and initiating treatment with anti-PD-1 (pembrolizumab). Neoplasia. (2022) 32:100822. doi: 10.1016/j.neo.2022.100822
33
Hedlund G Eriksson H Sundstedt A Forsberg G Jakobsen BK Pumphrey N et al . The tumor targeted superantigen ABR-217620 selectively engages TRBV7–9 and exploits TCR-pMHC affinity mimicry in mediatic T cell cytotoxicity. PloS One. (2013) 8:e79082. doi: 10.1371/journal.pone.0079082
34
Azulay M Shahar M Shany E Elbaz E Lifshits S Törngren M et al . Tumor-targeted superantigens produce curative tumor immunity with induction of memory and demonstrated antigen spreading. J Transl Med. (2023) 21:222. doi: 10.1186/s12967-023-04064-z
35
Shaw DM Connolly NB Patel PM Kilany S Hedlund G Nordle O et al . A phase II study of a 5T4 oncofoetal antigen tumour-targeted superantigen (ABR-214936) therapy in patients with advanced renal cell carcinoma. Br J Cancer. (2007) 96:567–74. doi: 10.1038/sj.bjc.6603567
36
Hawkins RE Gore M Shparyk Y Bondar V Gladkov O Ganev T et al . A randomized phase II/III study of naptumomab estafenatox + IFNα versus IFNα in renal cell carcinoma: final analysis with baseline biomarker subgroup and trend analysis. Clin Cancer Res. (2016) 22:3172–81. doi: 10.1158/1078-0432.CCR-15-0580
37
McCarthy E . The toxins of William B. Coley and the treatment of bone and soft-tissue sarcomas. Iowa Orthop J. (2006) 26:154–8.
38
Segal BH Wang X-Y dennis CG Youn R Repasky EA Manjili MH et al . Heat-shock proteins as vaccine adjuvants in infections and cancer. Drug Discov Today. (2006) 11:534–40. doi: 10.1016/j.drudis.2006.04.016
39
Joshi S Wang T Araujo TLS Sharma S Brodsky JL Chiosis G . Adapting to stress – chaperome networks in cancer. Nat Rev Cancer. (2018) 18:562–75. doi: 10.1038/s41568-Mar018-0020-9
40
Tramentozzi E Ruli E Angriman I Bardini R Campora M Guzzardo V et al . Grp94 in complexes with IgG is a soluble diagnostic marker of gastrointestinal tumors and displays immune-stimulating activity on peripheral blood immune cells. Oncotarget. (2016) 7:72923–40. doi: 10.18632/oncotarget.12141
41
Dong B Jaeger AM Thiele DJ . Inhibiting heat shock factor 1 in cancer: a unique therapeutic opportunity. Trends Pharmacol Sci. (2019) 40:986–1005. doi: 10.1016/j.tips.2019.10.008
42
Santagata S Hu R Lin NU Mendillo ML Collins LC Hankinson SE et al . High levels of nuclear heat-shock factor 1 (HSF1) are associated with poor prognosis in breast cancer. Proc Natl Acad Sci USA. (2011) 108:18378–383. doi: 10.1073/pnas.1115031108
43
Björk JK Ahonen I Mirtti T Erikson A Rannikko A Bützow A et al . Increased HSF1 expression predicts shorter disease-specific survival of prostate cancer patients following radical prostatectomy. Oncotarget. (2018) 9:31200–13. doi: 10.18632/oncotarget.25756
44
Duan X Iwanowycz S Ngoi S Hill M Zhao Q Liu B . Molecular chaperone GRP94/GP96 in cancers: oncogenesis and therapeutic target. Front Oncol. (2021) 11:629846 PMID: 33898309. doi: 10.3389/fonc.2021.629846
45
Birbo B Madu EE Madu CO Jain A Lu Y . Role of HSP90 in cancer. Int J Mol Sci. (2021) 22:10317. doi: 10.3390/ijms221910317
46
Albakova Z Mangasarova Y Albakov A Gorenkova L . HSP70 and HSP90 in cancer: cytosolic, endoplasmic reticulum and mitochondrial chaperones of tumorigenesis. Front Oncol. (2022) 12:829520 PMID: 35127545. doi: 10.3389/fonc.2022.829520
47
Mintz PJ Kim J Do KA Wang X Zinner RG Cristofanilli M et al . Fingerprinting the circulating repertoire of antibodies from cancer patiens. Nat Biotecnol. (2003) 21:57–63. doi: 10.1038/nbt774
48
Wang X Wang Q Lin H . Correlation between clinicopathology and expression of heat shock protein 72 and glycoprotein 96 in human esophageal squamous cell carcinoma. Clin Dev Immunol. (2010) 2010:212537. doi: 10.1155/2010/212537
49
Tang Y Zhou Y Fan S Wen Q . The multiple roles and therapeutic potential of HSP60 in cancer. Biochem Pharmacol. (2022) 201:115096. doi: 10.1016/j.bcp.2022.115096
50
Thole JE Dauwerse HG Das PK Groothuis DG Schouls LM van Embden JD . Cloning of Mycobacterium bovis BCG DNA and expression of antigens in Escherichia coli. Infect Immun. (1985) 50:800–6. doi: 10.1128/iai.50.3.800-806.1985
51
Härkönen T Puolakkainen M Sarvas M Airaksinen U Hovi T Roivainen M . Picornavirus proteins share antigenic determinants with heat shock proteins 60/65. J Med Virol. (2000) 62:383–91. doi: 10.1002/1096-9071(200011)62:3<383::aid-jmv11>3.0.co;2-PMID:11055249
52
Mayr M Metzler B Kiechl S Willeit J Schett G Xu Q et al . Endothelial cytotoxicity mediated by serum antibodies to heat shock proteins of Escherichia coli and Chlamydia pneumoniae: immune reactions to heat shock proteins as a possible link between infection and atherosclerosis. Circulation. (1999) 99:1560–66. doi: 10.1161/01.cir.99.12.1560
53
Parvizpour S Pourseif MM Razmara J Rafi MA Omidi Y . Epitope-based vaccine design: a comprehensive overview of bioinformatics approaches. Drug Discov Today. (2020) 25:1034–42. doi: 10.1016/j.drudis.2020.03.006
54
Calis JJA Maybeno M Greenbaum JA Weiskopf D De Silva AD Sette A et al . Properties of MHC class I presented peptides that enhance immunogenicity. PloS Comput Biol. (2013) 9:e1003266. doi: 10.1371/journal.pcbi.1003266
55
Wang P Sidney J Dow C Mothé B Sette A Peters B . A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PloS Comput Biol. (2008) 4:e1000048. doi: 10.1371/journal.pcbi.1000048
56
Mahapatra SR Dey J Kaur T Sarangi R Bajoria AA Kushwaha GS et al . Immunoinformatics and molecular docking studies reveal a novel Multi-Epitope peptide vaccine against pneumonia infection. Vaccine. (2021) 39:6221–37. doi: 10.1016/j.vaccine.2021.09.025
57
Papageorgiou AC Collins CM Gutman DM Kline JB O’Brien SM Tranter HS et al . Structural basis for the recognition of superantigen streptococcal pyrogenic exotoxin A (SpeA1) by MHC class II molecules and T-cell receptors. EMBO J. (1999) 18:9–21. doi: 10.1093/emboj/18.1.9
58
Ganem MB De Marzi MC Fernandez-Lynch MJ Jancic C Vermeulen M Geffner J et al . Uptake and intracellular trafficking of superantigens in dendritic cells. PloS One. (2013) 8:e66244. doi: 10.1371/journal.pone.0066244
59
Durgeau A Virk Y Corgnac S Mami-Chouaib F . Recent advances in targeting CD8 T-Cell immunity for more effective cancer immunotherapy. Front Immunol. (2018) 9:14 PMID: 29403496. doi: 10.3389/fimmu.2018.00014
60
Stern P Harrop R . 5T4 oncofoetal antigen: an attractive target for immune intervention in cancer. Cancer Immunol Immunother. (2017) 66:415–26. doi: 10.1007/s00262-016-1917-3
61
Cappuccini F Bryant R Pollok E Carter L Verrill C Hollidge J et al . Safety and immunogenicity of novel 5T4 viral vectored vaccination regimens in early stage prostate cancer: a phase I clinical trial. J Immunother Cancer. (2020) 8:e000928. doi: 10.1136/jitc-2020-000928
62
Pavlopoulou A Scorilas A . A comprehensive phylogenetic and structural analisis of the carcinoembryonic antigen (CEA) gene family. Genome Biol Evol. (2014) 6:1314–26. doi: 10.1093/gbe/evu103
63
Huang Q Richmond JFL Suzue K Eisen HN Young RA . In vivo cytotoxic T lymphocyte elicitation by mycobacterial heat shock protein 70 fusion proteins maps to a discrete domain and is CD4+ T Cell independent. JEM. (2000) 191:403–8. doi: 10.1084/jem.191.2.403
64
Suzue K Young RA . Adjuvant-free hsp70 fusion protein system elicits humoral and cellular immune responses to HIV-1 p24. J Immunol. (1996) 156:873–9. doi: 10.4049/jimmunol.156.2.873
65
Finotti P . Sequence similarity of HSP65 of mycobacterium bovis BCG with SARS-CoV-2 spike and nuclear proteins: may it predict an antigen-dependent immune protection of BCG against COVID-19? Cell Stress Chaperones. (2022) 27:37–43. doi: 10.1007/s12192-021-01244-y
66
Lule S Colpak AI Balci-Peynircioglu B Gursoy-Ozdemir Y Peker S Kalyoncu U et al . Behçet disease serum is immunoreactive to neurofilament medium with share common epitopes to bacterial HSP-65, a putative trigger. J Autoimmun. (2017) 84:87–96. doi: 10.1016/j.jaut.2017.08.002
67
Xu Q . Role of heat shock proteins in atherosclerosis. Arterioscler Thromb Vasc Biol. (2022) 22:1547–59. doi: 10.1161/01.ATV.0000029720.59649.50
68
Mayr M Metzler B Kiechl S Willeit J Schett G Xu Q et al . Endothelial cytotoxicity mediated by serum antibodies to heat shock proteins of escherichia coli and chlamydia pneumoniae. Circulation. (1999) 99:1560–6. doi: 10.1161/01.cir.99.12.1560
69
Van Kaer L . How superantigens bind MHC. J Immunol. (2018) 201:1817–18. doi: 10.4049/jimmunol.1801104
70
Ojima T Iwahashi M Nakamura M Matsuda K Nakamori M Ueda K et al . Streptococcal preparation OK-432 promotes the capacity of dendritic cells (DCs) to prime carcinoembryonic antigen (CEA)-specific cytotoxic T lymphocyte responses induced with genetically modified DCs that express CEA. Int J Oncol. (2008) 32:459–66. doi: 10.3892/ijo.32.2.459
71
Nakahara S Tsunoda T Baba T Asabe S Tahara H . Dendritic cells stimulated with a bacterial product, OK-432, efficiently induce cytotoxic T lymphocytes specific to tumor rejection peptide. Cancer Res. (2003) 63:4112–8.
72
Dai J Liu B Caudill MM Zheng H Qiao Y Podack ER et al . Cell surface expression of heat shock protein grp96 enhances cross-presentation of cellular antigens and the generation of tumor-specific T cell memory. Cancer Immun. (2003) 3:1.
73
Tobian AAR Canaday DH Harding CV . Bacterial heat shock proteins enhance class II MHC antigen processing and presentation of chaperoned peptides to CD4+ T cells. J Immunol. (2004) 173:5130–7. doi: 10.4049/jimmunol.173.8.5130
74
Javid B MacAry PA Lehner PJ . Structure and function: heat shock proteins and adaptive immunity. J Immunol. (2007) 179:2035–40. doi: 10.4049/jimmunol.179.4.2035
Summary
Keywords
heat shock proteins, tumor-associated antigens, bacterial antigens, superantigens, epitope mapping, immunodominant epitopes, computational biology, immunoinformatics
Citation
Finotti P (2025) Two homologous sequences of Grp78 and HSP70 represent tumor antigens shared with streptococcal superantigens in eliciting an antitumor immune response: an immunoinformatic investigation. Front. Immunol. 16:1644687. doi: 10.3389/fimmu.2025.1644687
Received
10 June 2025
Accepted
12 August 2025
Published
11 September 2025
Volume
16 - 2025
Edited by
Debasish Kumar Dey, National Institute of Immunology (NII), India
Reviewed by
Sofía Noli Truant, University of Buenos Aires, Argentina
Digvijay Singh, University of Louisville, United States
Updates
Copyright
© 2025 Finotti.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paola Finotti, paola.finotti@unipd.it
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.