- 1Bio-Organic Chemistry Unit, Institute of Biomolecular Chemistry, Consiglio Nazionale delle Ricerche, Naples, Italy
- 2National Research Council of Italy, Institute of Biomolecular Chemistry (CNR–ICB), Catania, Italy
- 3Laboratory of Bio-Organic Chemistry and Chemical Biology, Department of Biology, University of Naples “Federico II”, Naples, Italy
Professional antigen-presenting cells (APCs) represent a crucial link between the innate and the adaptive immune response. APCs express specific surface receptors which are primarily involved in “non-self” and/or “self” ligand recognition. Upon ligand binding, these receptors can trigger cell signalling leading to the production of pro-inflammatory cytokines, chemokines and Type 1 interferons, supporting antimicrobial and inflammatory responses. Recently, two major families of receptors, C-type lectin receptors and immunoglobulin receptors, are emerging as potential therapeutic targets to activate and modulate immune system through different intracellular signalling motifs upon binding with endogenous and exogenous ligands. The chemical characterization of the molecular determinants necessary for the receptors/ligands binding promotes the design and optimization of small molecules crucial for the comprehension of biological functions and for the therapeutic treatment of specific receptor-associated disorders. This review focuses on the description of these ligands together with their biological evaluation and their impact on the modulation of the immune response.
1 Introduction
The innate immune response constitutes the first barrier against various pathogens. Along with the immediate action of cytotoxic cells, such as natural killer (NK) cells and granulocytes, a more complex response is provided by antigen-presenting cells (APCs), able to initiate the processes pivotal for the antigen-specific adaptive immunity.
Antigen recognition is mediated by receptors families known as pattern recognition receptors (PRRs), manly located at the surface of innate immune cells such as macrophages, dendritic cells (DCs), neutrophils, granulocytes, mast cells, monocytes, basophils, natural killer cells, and epithelial cells (1–3).
PRRs are uptake receptors capable of recognizing molecules associated with pathogens (Pathogen-Associated Molecular Patterns, PAMPs) or released by damaged cells (Damage-Associated Molecular Patterns, DAMPs), to allow their internalization and subsequent processing and presentation to T cells (4).
PRRs are broadly categorized based on their cellular location, structures and functions in different sub-families including Toll-like receptors (TLRs), nucleotide-binding oligomerization domain-like receptors (NLRs), retinoic acid-inducible gene-I-like receptors (RLRs), AIM2-like receptor (ALR) and C-type lectin receptors (CLRs) (5).
In particular, CLRs recognize pathogen-derived ligands, but also natural endogenous ligands such as self-carbohydrates, proteins, or lipids, with implications in the control of tissue damage, autoimmune diseases and tumorigenesis (6, 7).
Circumstantial evidence indicates that DAMPs and PAMPs could also bind immunoglobulin-like receptors such as the triggering receptor expressed on myeloid cells (TREM-1 and TREM-2) (8),. even though they are not yet fully identified as PRRs. Given their tissue and cell-specific expression, there is considerable evidence that immunoglobulin-like receptors could be pivotally implied in modulation of the innate immune response, intracellular signal transduction, and interactions with other signalling cascades, as well as those involving TLRs (9, 10).
Recently, the interest to study and understand the mechanism of either CLRs and immunoglobulin-like receptors families has been growing, given their ability to positively and/or negatively regulate immune cell activation following interaction with a variety of endogenous and exogenous ligands (7, 11). The identification of chemical characteristics of these ligands could improve the understanding of the intracellular downstream signalling and facilitate the study of their physiological role and development in the research of new vaccines or therapeutic treatments. In addition, identification of the endogenous ligands can accelerate the synthesis and optimization of organic compounds, resulting in potentially higher efficiency for further developments.
This review will focus on the functional mechanisms of the most studied CLRs and immunoglobulin receptors, particularly in relation to their interaction with specific ligands and the possible cross-talk among different immune receptors. Special attention will be given to the recognition of small molecules.
Small molecules, characterized by a molecular weight approximately less than 1000 dalton and often of lipophilic nature, are generally easily identifiable, synthesizable, chemically modifiable and potentially suitable for the development of new drugs due to their ability to selectively interact with specific biological targets. (Figure 1).

Figure 1. (a) Summary of the main classes of immunoglobulin like receptors and CLRs recognizing small molecules and their immunological response. Receptors are represented based on their biological ability to induce inflammatory or anti-inflammatory responses. For TREM-1 and TREM-2 the evaluation is more complex. Both TREM-2 can sustain the cell energetic by the activation of mTOR pathway. (Created with online software BioRender.com); (b) Small ligands and ligands structural motifs of immunoglobulin-like and C-type lectin receptors.
Previous studies on amphiphilic drugs have, in fact, highlighted how specific structural features of small ligands can profoundly influence interaction with immune receptors, providing a rationale for the design of novel immunomodulatory molecules (12).
2 Receptors families
2.1 Immunoglobulin-like receptors
Immunoglubulin-like receptors are a family of receptors possessing extracellular immunoglobulin domains. They are known to exert immunomodulatory effects on a wide range of immune cells. Based on their cellular-specific expression, they can be classified as leukocyte mono-immunoglobulin-like receptors (LMIRs), triggering receptors expressed on myeloid cells (TREMs), Signal regulatory proteins (SIRPs) or sialic acid binding Ig-like lectins (SIGLECs). These receptors can have both activating and deactivating immunological characteristics.
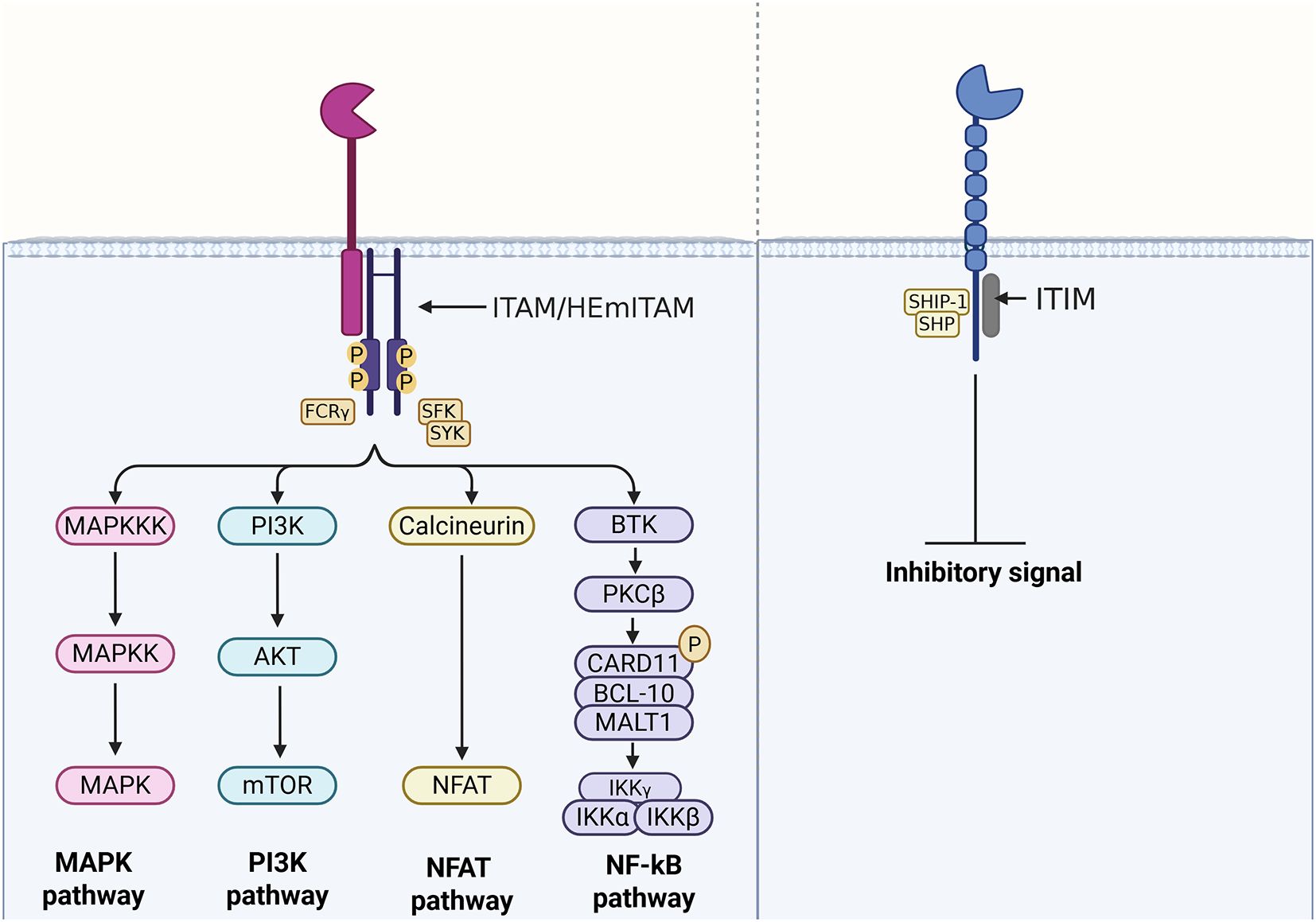
Activating receptors generally associate with an immunoreceptor tyrosine-based activation motif (ITAM or hemITAM) or a transmembrane adaptor protein containing a related activation motif, such as DNAX-activating Protein 10 (DAP10), DNAX-activating Protein 12 (DAP12), or the Fc-gamma receptor (FcRγ). DAP10, DAP12, and FcRγ act as substrates and docking sites for kinases, enabling the amplification of intracellular signalling reactions (13). In contrast, inhibitory receptors contain an immunoreceptor tyrosine-based inhibitory motif (ITIM) in their cytoplasmic domain (14–17), which facilitates the recruitment of phosphatases (Figure 2). In this regard, ‘self’ or ‘non-self’ molecules, able to interact and activate these receptors, enclose the enormous potential for controlling and modulating the immune response and the resulting cellular homeostasis.
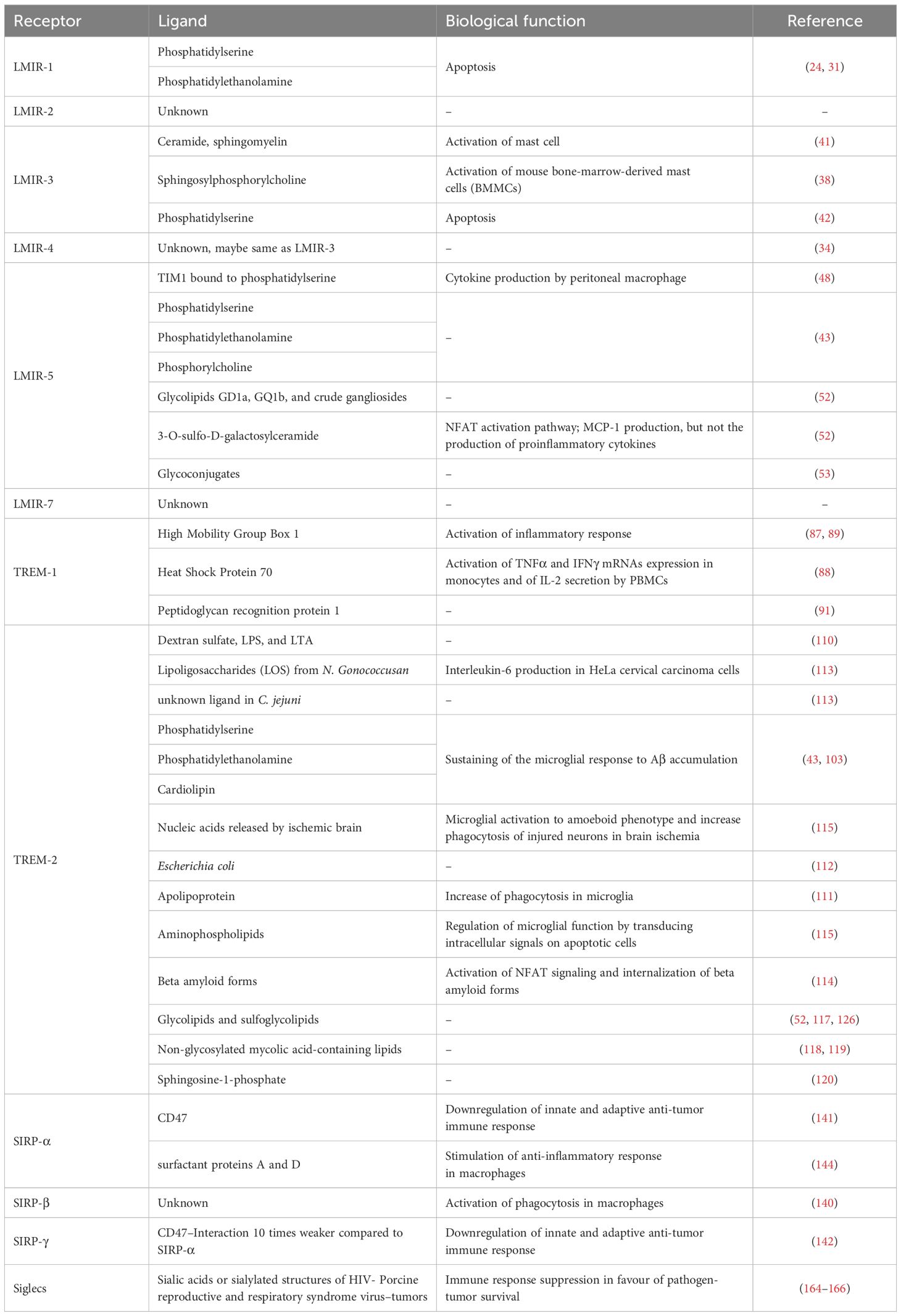
Although in fact, some receptors display distinct specificity profiles, they may exhibit overlapping ligand-binding patterns (Table 1).
2.1.1 LMIR/CD300 family members
The LMIR (also called CD300) (18, 19) family belongs to the paired immune receptors. Lipids or lipid-binding proteins have been identified as ligands for several CD300/LMIR members. Despite the similarity in the extracellular Ig-like domains, there are relevant structural differences between activating and inhibitory receptors, pivotal for the fine tuning of immune response. Therefore, we can distinguish them into two main groups: inhibiting receptors, LMIR-1 and LMIR-3, and activating receptors, LMIR-2, LMIR-4, LMIR-5 and LMIR-7.
2.1.1.1 Inhibitory LMIR receptors
2.1.1.1.1 LMIR-1/CML8/CD300a
LMIR-1 (also called CML8 or CD300a) is an inhibitory receptor, containing several ITIM motifs in the cytoplasmic domain (10, 19). LMIR-1 is expressed in myeloid and lymphoid cells, particularly in mast cells, eosinophils, and basophils, the three most important cell types involved in the initiation and regulation of allergic responses. Indeed, evidence indicates that inhibition of LMIR-1 induces mast cell degranulation by SCF in a murine model of cutaneous anaphylaxis (20). Targeting LMIR-1 was shown to inhibit, through the regulation of eosinophil and mast cell signalling in vivo, the bronchoalveolar lavage fluid inflammation, lung remodelling and inflammation in a model of chronic established asthma (21).
Phosphorylated ITIM motifs are able to recruit different phosphatases depending on the cell type where LMIR-1 is expressed (22). It was demonstrated that the corresponding LMIR-1 in humans, when cross-linked, inhibits IgE-induced degranulation and SCF-mediated survival on mast cells, through a mechanism that involves tyrosine phosphorylation and phosphatase recruitment (20, 23). These results highlight a new role for the regulation of human allergic response through LMIR-1, even though the endogenous ligand involved in such mechanisms must be still revealed.
Interestingly, knockdown of LMIR-1 in a mice model of acute septic peritonitis showed prolonged survival, given the greater expression of chemoattractants in peritoneal mast cells, leading to increased neutrophil recruitment and a better bacterial clearance (24). This evidence indicated a new regulatory role of LMIR-1 for mast cell inflammatory responses to microbial infections. LMIR-1 was also identified as a new marker for acute lymphoblastic leukemia, as differentially expressed compared to the wild type (25). In HIV infection, LMIR-1 is deregulated on B cells, suggesting the possibility that this receptor may contribute to the B-cell dysfunction observed in HIV-infected patients (26). Moreover, LMIR-1 has been associated with susceptibility to psoriasis (27).
Lack of LMIR-1 increases the secretion of pro-inflammatory cytokines produced by TLR4/MyD88 in macrophages and impaired wound healing (28, 29). LMIR-1 is able to bind phospholipid molecules as phosphatidylserine (PS) and phosphatidylethanolamine (PE), exposed as ‘eat me’ signals on the outer leaflet of the plasma membrane of apoptotic cells, forming cavity for the ligands polar heads insertion (30, 31). More selectively, human chimeric LMIR-1 exhibits a binding stronger to PE than PS, modulating the ingestion of dead cells (31).
Interestingly, considering that aluminum salts (alum) have been widely used as vaccine adjuvant, it was demonstrated that LMIR-1 expression was upregulated on inflammatory DCs after injecting mice with alum, and involved in the generation of dead cells in the peritoneal cavity. In this regard, inflammatory DCs bound dead cells via LMIR-1/PS interaction, resulting in Th2 lymphocytes responses and enhanced allergic airway inflammation, thus suggesting the involvement of LMIR-1 in alum-induced Th2 skewing (32). Furthermore, blocking LMIR-1/PS interaction may have therapeutic applications for the prevention or treatment of vaccine-involved pathologic conditions and allergic airway inflammation (32).
These studies demonstrated a novel pathway of cell regulation which modulate allergic responses and microbial infections, indicating LMIR-1 as a candidate target. Therefore, further investigation aimed at discovering possible antagonists for LMIR-1 will be useful for future treatment or for understanding the underlying molecular mechanism of associated diseases.
2.1.1.1.2 LMIR-3/CLM1/CD300f
The receptor LMIR-3 (also called CD300f or CLM1) is another inhibitory receptor which delivers its inhibitory signal via two ITIMs and a single immunoreceptor tyrosine-based switch motif (ITSM) that can recruit Src homology 2 domain-containing protein phosphatase-1 (SHP-1) and/or SHP-2 (33). LMIR-3 is highly expressed in myeloid cells, particularly mast cells (34). Interestingly, LMIR-3 is weakly expressed in monocyte-derived DCs, but is strongly upregulated when cultured in the presence of 1,25-dihydroxyvitamin D3, which reprograms DCs toward a tolerogenic phenotype, suggesting an important role for this receptor in the maintenance of immune tolerance (35).
LMIR-3 has been shown to have a neuroprotective role in a rat model of acute brain injury (36). Indeed, LMIR-3 is a negative regulator of myeloid effector cells in autoimmune demyelination, therefore implied in multiple sclerosis and autoimmune encephalomyelitis (37). Furthermore, LMIR-3 plays a pivotal role in regulating the mast cell-dependent allergic responses in mice (38).
Downregulation of LMIR-3 in DCs enhances T cell proliferation initiated by DCs, as well as antigen-specific T cell responses, both in vitro and in vivo, leading to effective protection against tumor challenge in mice (39). Block of LMIR-3 significantly reduced the engraftment of primary human acute myeloid leukemia cells, highlighting the potential LMIR-3 in tumor immunotherapy (40).
As possible ligands, several extracellular lipids including ceramide, sphingomyelin and sphingosylphosphorylcholine (SPC) were found (38, 41). The binding with these lipids results into an inhibition of the high-affinity IgE receptor (FcϵRI)-mediated activation of mouse bone-marrow-derived mast cells (BMMCs) (38). Particularly, it was evident that the ceramide-LMIR-3 interaction was pivotal in LMIR-3-mediated inhibition of mast cell activation in vivo, through a not fully understood mechanism. Indeed, a deeper examination is required to comprehend how LMIR-3 could influence FcϵRI signalling via colocalization of ceramide lipids and FcϵRI itself in mast cells. The influence of ceramide is supported by different research groups, but there is contrasting evidence about the binding affinity of other lipids proposed as possible ligands. Choi proposed PS and PE to be potential ligands for chimeric LMIR-3, resulting in increased phagocytosis (42). However, given that both PS and PE were expressed on dead cells, it was unclear how an inhibitory receptor as LMIR-3 could promote phagocytosis. These observations of Choi were not confirmed by Izawa, who showed that LMIR-3 on mast cells only bound membrane immobilized ceramide, but not other phospholipids as PS, PE, or phosphatidylcholine (PC) (38). The latter, however, appeared to be a ligand for LMIR-3 in the reporter cells but not in mast cells (38), possibly due to slight structural difference of recognition domain between endogenous LMIR3 and the chimera receptor LMIR3-CD3ζ used in reporter cell assay and/or for possible surface components-induced interference of PC-LMIR3 interaction on mast cells. Therefore, with the aim to identify other possible relevant ligands for LMIR-3, more in depth studies and analysis should be performed.
Notably, both LMIR-1 and LMIR-3 can bind to dead cells in a calcium dependent manner, suggesting that these two receptors must interact with evolutionary conserved ligands (10).
Circumstantial evidence suggests possible similarities among LMIR-1, LMIR-3 and the immunoglobulin-like receptor TREM-2 due to the high conservation of their domain architecture and the structural motifs, that could allow the binding with similar ligands or classes of ligands (e.g. PS and PE) (43).
2.1.1.2 Activating LMIR receptors
2.1.1.2.1 LMIR-2/CLM4/CD300C
The receptor LMIR-2 (also called CD300C or CLM4) is an activating receptor which is mainly expressed on macrophages and a subset of B cells in the spleen and the peritoneal cavity (19). LMIR-2 mediates an activating signal through the association with FcRγ or DAP12, for the production of the proinflammatory cytokines tumour necrosis factor (TNF)-α and interleukin (IL)-6 in macrophages (44). LMIR-2 regulates TLR4-mediated cell adhesion to VCAM-1 in highly purified inflammatory monocytes in response to TLR ligands alone (45). These data suggest that a ligand for LMIR-2 could be expressed after TLR ligand stimulation, resulting in cis-binding between LMIR-2 and the unknown ligand. Even though its physiological role is not fully understood, Totsuka et al. suggested that activation of LMIR-2 by TLR4/MyD88-mediated signalling is essential for the transmigration of inflammatory monocytes from the blood to sites of infection puncture (CLP)-induced peritonitis. The authors proposed that LMIR-2 may be able to bind phospholipids as functional ligands, similarly to the other CD300 family members and the TREM family proteins (45). As phospholipids are dynamically remodelled on stimulation with innate stimuli such as TLR ligands (46), its activation depends on TRL4/MyD88 axis. However, unlike LMIR-1, LMIR-2 does not bind PS (24).
Further efforts and studies are in progress to identify possible ligands that could potentially interact with LMIR-2 and unravel its function in vivo. Moreover, it will be interesting to discover if other ligands, able to interact with LMIR-1 or TREM-2, besides PS, could potentially bind also LMIR-2.
2.1.1.2.2 LMIR-4/CLM5/CD300ld
LMIR-4 (also called CD300ld or CLM5) is a further activating receptor which is physically associates with ITAM-bearing adaptor FcRγ. LMIR-4 is preferentially expressed on neutrophils in the peripheral blood, bone marrow, peritoneal cavity and spleen (47). LMIR-4 is considered a counterpart of the inhibitory receptor LMIR-3 (34). It has been shown that intraperitoneal administration of the TLR4 agonist lipopolysaccharide (LPS) strikingly up-regulates LMIR-3 and down-regulates LMIR-4, whereas granulocyte colony-stimulating factor up-regulates both LMIR-3 and LMIR-4 in granulocytes. These results suggest that innate immune system is partially regulated by the qualitative and quantitative balance of the paired receptors LMIR-3 and LMIR-4 (34). However, truly little is known about LMIR-4 function in immune disease. We could speculate that, as counterpart receptor of LMIR-3, LMIR-4 could be involved in similar mechanisms pathways of LMIR-3.
LMIR-4 synergistically enhanced TLR4 signalling in mast cells and granulocytes, allowing a robust cytokine production in accordance with enhanced activation of ERK. Upon binding with unknown ligand, LMIR-4 could positively regulate various signalling pathways, affecting the inflammatory responses of myeloid cells (34).
Taking into consideration that LMIR-3 and LMIR-4 share high homology in the Ig-like domain, these receptors may share the same ligands, with consequence that these molecules could be able to engage simultaneously the two receptors, mimicking cell physiological conditions (34). Moreover, their identification is pivotal for any further understanding of LMIR-3 and LMIR-4 functions as they could represent a fine balance between the inhibiting-activating mechanisms. Elucidation of the role of LMIR-4 could unravel its contribution to inflammatory processes,crucial for future strategic therapies.
2.1.1.2.3 LMIR-5/CLM7/CD300lb
LMIR-5 (also called CD300lb or CLM7) is an activating receptor coupled to DAP-12. LMIR-5 is mainly expressed in myeloid cells, such as neutrophils, peripheral macrophages, and mast cells (10).
LMIR-5 can interact with T cell Ig mucin 1 (TIM1) but similarly also with TIM4. Interestingly, the binding site of LMIR-5 to TIM1 is located in proximity of TIM1 PS-binding site, which per se is not able to bind PS (48). These results are in contradiction with previous reports which showed LMIR-5 able to independently bind to PS, PE, and PC (43). Importantly, soluble form of LMIR-5 (sLMIR-5) upon binding to an unidentified ligand, other than TIM-1 and TIM-4, is involved in induced cytokine production by peritoneal macrophage (49). LMIR-5 deficiency profoundly reduced systemic cytokine production and septic mortality in LPS-administered mice, pointing out a peculiar relation with the triggering receptor expressed on myeloid cells-1 (TREM-1), an inflammatory receptor (49). Indeed, stimulation with TLR agonists increased the release of both soluble forms sLMIR-5 and sTREM-1. Although sTREM-1 attenuates excessive inflammatory response by counter-regulating TREM-1, inflammatory amplifier in sepsis, sLMIR-5 amplifies LPS-induced lethal inflammation (50, 51). The relation with TREM-1 is pivotal for further comprehension of its role, as well as the discovery of new ligands possibly shared by LMIR-5 and TREM-1, also justified by the strong similarities of the LMIR and TREM families (43).
LMIR-5 interacts with glycolipids GD1a, GQ1b, and crude gangliosides (52). In particular, LMIR-5 can bind with highest affinity to 3-O-sulfo-β-D-galactosylceramide C24:1, recognizing the 3-O-sulfo-D-galactose moiety, which resulted in DAP12-mediated NFAT activation (53). It was shown that the bacterium C. jejuni activates NFAT through DAP12 by interaction with LMIR-5, whose activators were identified as protein components, RNA-associated proteins, and 150-kDa high-molecular-weight glycoconjugates (53). However, the identities of these LMIR-5 activators should be further investigated.
LMIR-5 plays a key role in increasing acute kidney damage, characterized by tubular necrosis and cast formation. Indeed, LMIR-5, as described above, strongly binds to TIM1 (48), which was identified as both marker of acute kidney injury (54) and marker in renal carcinoma and generally associated with immune dysfunction. Therefore, hampering LMIR-5-TIM1 interaction might be a novel therapeutic strategy for acute renal tubular damage (48).
LMIR-5 is one of the most well studied LMIR receptors from the ligand binding prospective. However, most of its ligands are still unknown, and their discovery is crucial for further investigation of the role in the activation of immune response and in macrophage activation. Given the strong similarity that LMIR-5 shares with TREM-1 as an inflammatory amplifier, it may be interesting to study the similarities of activation pathways and possible shared ligands. Peculiarly, both receptors present soluble forms similarly formed under TLR stimulations, but with opposite functions. Therefore, the fine control and balance between soluble and transmembrane forms of TREM-1 and LMIR-5 seems to be extremely important to sort out the complex ways of immunomodulatory regulation.
2.1.1.2.4 LMIR-7/CLM3/CD300lh
LMIR-7 (also called CD300lh or CLM3) is an activating receptor highly expressed in mast cells, monocytes and macrophages (55). LMIR-7 synergizes with TLR4 in signalling and binds to FcRγ, but with lower affinity compared with LMIR-4-FcRγ (55). It has been shown that LMIR-7 functions as a positive regulator of TLR9 (56). LMIR-7 upregulates TLR9-mediated production of the proinflammatory cytokines TNF-α and IL-6 but does not affect type I-IFN expression (56).
Interestingly, LMIR-7 shares high homology similarity with LMIR-4 in the amino acid sequences of Ig-like and transmembrane domain (55). Therefore, it is possible that LMIR-7, as well as LMIR-4, modulates the innate immune responses in a cell type-dependent manner. Overall, with the aim to improve the understanding of in vivo functions of LMIR receptors, both analysis of the knock-out mice and identification of the ligands for each LMIR are necessary (55).
Considering that LMIR-4 is a counterpart of inhibitory receptor LMIR-3 and that LMIR-7 shares a high degree of homology with LMIR-4, it is crucial to better understand the peculiar function of LMIR-7 in relation to LMIR-4 and understand its possible analogous role to LMIR-3. The LMIR-3-LMIR-4-LMIR-7 axis could be, potentially, a new immunomodulatory regulation mechanism that needs to be further explored.
2.1.2 TREM family members
TREM receptors are a class of cell surface receptors characterized by a single V-type immunoglobulin domain in their extracellular region (57).
TREM family members are expressed by granulocytes, monocytes and tissue macrophages. These receptors participate in diverse cell processes, including inflammation, bone homeostasis, neurological development and coagulation (58). In this family, TREM1 and TREM2 are the best characterized proteins. The growing interest for the elucidation of the cell signalling pathways of these receptors with high therapeutic potential, raised the focus on the identification of endogenous TREM ligands.
2.1.2.1 TREM-1 and TREM-2 receptors
TREM-1 and 2 proteins are structurally related, consisting in a single extracellular variable-type immunoglobulin (Ig) domain; a shorter stalk region; a transmembrane domain and a short cytoplasmic tail not containing any activation, signalling or trafficking motifs (59).
The transmembrane domain includes a charged lysine residue that enables the interaction with DAP12 (58). Upon binding with DAP12, TREM receptors can potentially have activating and/or inhibitory roles in innate immune responses. Although this hypothesis remains highly speculative, TREM-1 and -2 proteins show different and specialized functional roles, especially in myeloid cells (60).
TREM-1 and TREM-2 extracellular domains can also be found in a soluble forms (sTREM-1 and sTREM-2), which can be released by a proteolytic cut within the protein stalk region (61) or by alternative splicing (62). Due their intrinsic structural characteristics, TREM-1 and TREM-2 can act in a dependent manner via its interactor protein by which initiate intracellular signalling or in independent manner as a soluble protein.
The involvement of these receptors in inflammation, neurodegenerative diseases and in cancer is widely recognized and further studies are pivotal for the definition of their biology (57).
2.1.2.1.1 TREM-1 receptor
TREM-1 is mainly expressed on myeloid cells such as monocytes, macrophages and granulocytes (63), but is also detected on parenchymal cell types such as bronchial, corneal, gastric epithelial cells, and hepatic endothelial cells (64–66). TREM-1 expression is upregulated during septic shock, in a number of infectious diseases, such as pneumonia (67) and suppurated cholangitis (68), in obstructive nephropathies (69) and chronic kidney diseases (70). TREM-1 can be also upregulated on macrophages in lung cancer with subsequent inflammatory response, inducing complications and death (71). Furthermore, TREM-1 plays a pivotal role in cardiovascular diseases, such as atherosclerosis pathogenesis (72), acute myocardial infarction (73), coronary artery diseases (74), myocardial dysfunction in septicaemia (75) and infective endocarditis (76). TREM-1 ameliorates neuroinflammatory responses in Parkinson disease, and this neuroprotective effect might occur via the activation of autophagy and anti-inflammatory pathways (77). TREM-1 can promote mitochondrial integrity and cell survival (78, 79), as well as induce rearrangement of the actin cytoskeleton (80), and release of pro-inflammatory cytokines and chemokines, such as MCP-1, MIP1-α, IL-1β, IL-6, IL-8, TNFα (81).
For several years since the discovery of TREM-1 in 2000, no endogenous TREM-1 ligands have been identified. Two studies have proposed peptidoglycan recognition protein 1 as a ligand for TREM1 in in vitro assays conducted on PBMCs and linked to the secretion of pro-inflammatory cytokines (82, 83).
The presence of TREM-1 ligand on human platelets could be related to neutrophil activation in the occurrence of microbial stimuli like LPS (84), but also in this case the chemical characterization of the putative implied ligand was unsuccessful.
During the last years, some progress has been made to better understand the role of TREM-1 during chronic tissue damage. It has been reported that extracellular actin co-localizes with TREM-1 in lung tissue sections from septic mice, suggesting that TREM-1 recognizes actin during activation in sepsis (85). Furthermore, actin is considered as TREM-1-interacting protein on platelets (85), although the receptor could be able to recognize other various molecules. High Mobility Group Box 1 (HMGB1) and heat shock proteins, secreted by activated myeloid cells and released by dying and necrotic cells, thus functioning as a DAMP molecule (86–88), were suggested as TREM-1 ligands (89). HMGB1 alone seems unable to induce TREM-1 activation, therefore suggesting the presence of other co-activating molecules (87). In addition, overstimulation of TREM-1 can lead to cell death and release of both actin and HMGB1. This process can provide a large number of ligands for activation of TREM-1 signalling and thus induce some progressive systemic inflammatory response, resulting in sepsis (85).
New findings suggest that the extracellular cold-inducible RNA-binding protein (eCIRP), a recently characterized DAMP, is an endogenous ligand for TREM-1 (90). The binding between eCIRP and TREM-1 can dramatically enhance inflammation during sepsis in macrophages and a peptide-mediated blocking of this interaction significantly improves the outcome, increasing the survival rate (90).
TREM-1 also binds to peptidoglycan recognition receptor 1 (PGLYRP1/Tag7), mainly found in granulocytes and known for its bactericidal properties (91). Interestingly, PGLYRP1 alone was not able to induce TREM-1 activation. In this regard, it is not known whether PGLYRP1 signalling is TREM-1 mediated and if the binding is responsible for the bactericidal properties (87). Recently, a deep characterization of the multifunctional protein PGLYRP1 led to the identification of the peptide (called N3) responsible for the interaction with TREM-1. The N3 peptide corresponds to the N-terminal 24 amino acids domain RYVVVSHTAGSSCNTPASCQQQAR (isolated and synthetized). Its interaction with TREM-1 causes the protein dimerization and activation of cytotoxic lymphocytes (83).
TREM-1 does not appear to participate in recognition of lipids, in contrast to other TREM family members (43, 92).
All these results suggest that the identification of an endogenous ligand for TREM-1 is of pivotal importance to fully understand its mechanism of activation and signalling in different inflammatory diseases (87).
In humans, TREM-1 is involved in the amplification of pro-inflammatory innate immune response for the elimination of pathogens (81). The receptor can amplify TLRs response such as TLR4 or TLR2 signalling and synergistically increase the production in pro-inflammatory cytokines such as TNF-α and IL-6 (93). TREM-1 appears to play a crucial role during the initiation of cytokine response in septic shock as well as in other infections (51), through common signalling pathway activation including PI3K, ERK1/2, IRAK1 and Nf-κB (94).
TREM-1 blockade is considered a potential therapeutic procedure, as this strategy could present the peculiar advantage to not fully abolish the inflammatory response required for a proper immune response. For this reason, TREM-1 inhibitors could be safer in treating inflammatory diseases compared to any other immune receptors (87).
2.1.2.1.2 TREM-2 receptor
TREM-2 is mainly expressed in microglia, in DCs, osteoclasts, Kuppfer cells and alveolar macrophages, and is an important negative regulator of autoimmunity (95, 96). TREM-2 activity or dysfunction are strictly connected to the induction of neurodegenerative disorders. Indeed, recent genetic studies have found that TREM2 mutations represent a significant genetic risk factor in Nasu-Hakola Disease (97), frontotemporal dementia (FTD) (98), Parkinson’s disease (PD) (99), and amyotrophic lateral sclerosis (ALS) (100). Recently, the importance of TREM-2 has been highlighted by the identification of coding variants, such as the TREM2-R47H form, that increase risk for Alzheimer’s disease (Figure 3) (101, 102). Also, cytokines released by TREM-2 activation are essential for maintaining microglial metabolic fitness besides enabling microglial activation, migration and phagocytosis, and allowing differentiation into a cell mature profile (103, 104). TREM-2 expression rises during various forms of liver injury in both mice and humans, attenuating TLR4-driven proinflammatory responses (105). TREM-2 is also expressed by adipose tissue macrophages and is extremely important in the regulation of inflammation during obesity. This protein is upregulated in the adipose tissue of dogs on a high-fat diet (HFD) and in the mesenteric adipose tissue of insulin-resistant-diabetic mice, TREM2 promotes adipogenesis by upregulating adipogenic regulators and inhibiting the Wnt10b/β-catenin signaling pathway (106).TREM-2 also sustains cell energetic and biosynthetic metabolism modulating ATP levels and biosynthesis thought the mTOR pathway (107, 108). TREM-2 knockout animal model showed a dramatic reduction of glucose metabolism throughout the brain (109).
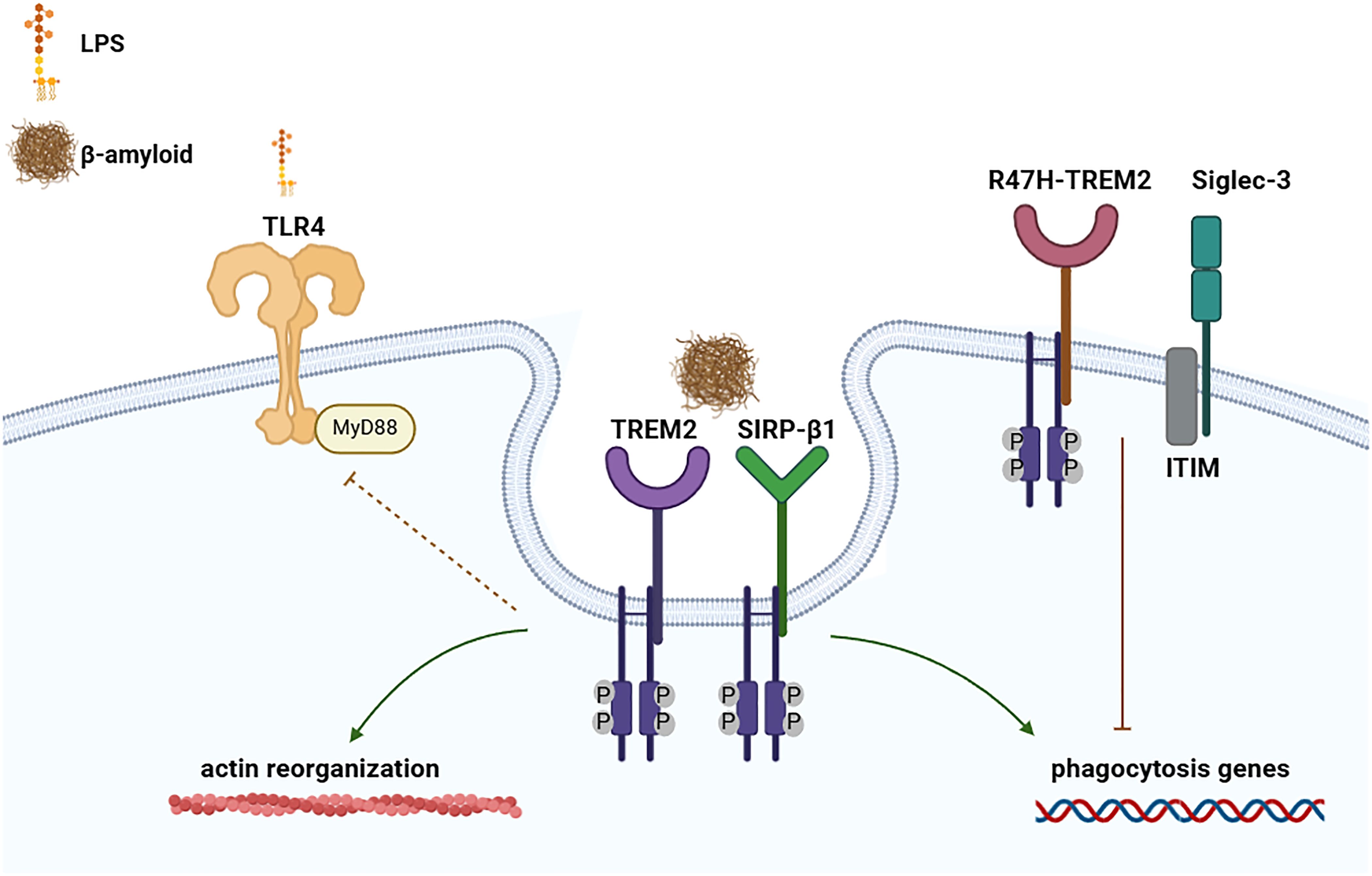
Figure 3. Involvement of TREM-2, SIRP-β1 and Siglec-3 in the development of Alzheimer disease. Representation of TREM-2 and SIRP-β1 in inducing phagocytosis, actin reorganization and reduction of LPS-TLR induced response. Whereas TREM2 genetic modification’s R47H-TREM2, as well as Siglec-3, can inhibit β-amyloid uptake by suppressing the phagocytosis genes. (Created with online software BioRender.com).
The first ligands that have been identified to bind to TREM-2 were bacterial, poly-anionic molecules and anionic bacterial carbohydrates, particularly dextran sulfate, LPS, and LTA (110) and lipid-binding proteins (111). TREM-2 can also directly bind to lipoligosaccharides from Neisseria gonorrhoeae and to unknown ligand in Campylobacter jejuni, gram-negative bacteria, causative agent for food poisoning (112, 113). Additional studies showed that TREM-2 was able to interact with different beta amyloid forms, increasing their internalization (114), unlike TREM2 missense variants, highlighting the potential crucial role of the protein in the developments of new pharmacological treatment for neurodegenerative pathologies (Figure 3) (102).
TREM-2 binds the phospholipids PE, PS, and cardiolipin (CL) (43, 103) as anionic molecules from mammalian cells. In addition to phospholipids, also nucleic acids released by ischemic brain lysate bind to TREM-2. Only cellular fractions containing nuclei or purified DNA, but not cytosolic fractions, were able to induce the signalling through TREM-2 (115). Finally, TREM-2 is able to bind also aminophospholipids that are on apoptotic Neuro2a cells as well as some normal cultured cells (116), glycolipids and sulfoglycolipids (52, 117).
Several studies report the presence of different ligands class of molecules that bind TREM-2, and analogously other immunoglobulin like receptors previously described, as LMIR-1, able to recognize PS and PE. Moreover, those common features further confirm the link between TREM2 and LMIR-1, helping to understand how they could synergise or differ their signalling pathway. More generally, a correlation between TREM and LMIR receptor families was highlighted by Cannon et al (43), suggesting that the TREM/LMIR system may discriminate immunological stimuli based on lipid signatures, thereby influencing downstream responses.
It must be pointed out that LMIR-1 and LMIR-3 are inhibitory receptors which deliver their signal via ITIM domain. Furthermore, TREM-2, similarly to the other activating receptor acts via DAP12, but it can deliver inhibitory signals also. These information could highlight a more singular activating/modulating role of TREM-2 in comparison to the other immunoglobulin like receptors.
Considering the importance of TREM-2 and the peculiar similarity with other receptors described in this review, the identification of a unique and specific ligand for this receptor is fundamental.
Recently, interesting advancement concerning the development of new TREM-2 ligands have been done. Lately, non-glycosylated MA-containing lipids of mycobacteria were identified as TREM-2 ligands with the activation of TREM-2-DAP12 signalling. In particular, long (C60–C90) and branched alkyl chains are required for the TREM-2 recognition (118, 119). Moreover, another study reported Sphingosine-1-phosphate (S1P) and an its analogue as endogenous ligand of TREM-2 able to promote phagocytosis (120).
A recent crucial progress concerning the essential finding of a small ligand of TREM-2 is represented by Sulfavant A (121, 122), a small nature-inspired sulfo-containing glycolipid with a promising adjuvant property (Figure 4a) (123–125). The interaction of Sulfavant A with TREM-2 resulted in a novel cell regulatory function, contributing to immune homeostasis and preserving lymphocyte activation and immune response. Sulfavant A elicited an unconventional hDCs maturation with up-regulation of the costimulatory molecules without production of conventional inflammatory cytokines (117, 126).
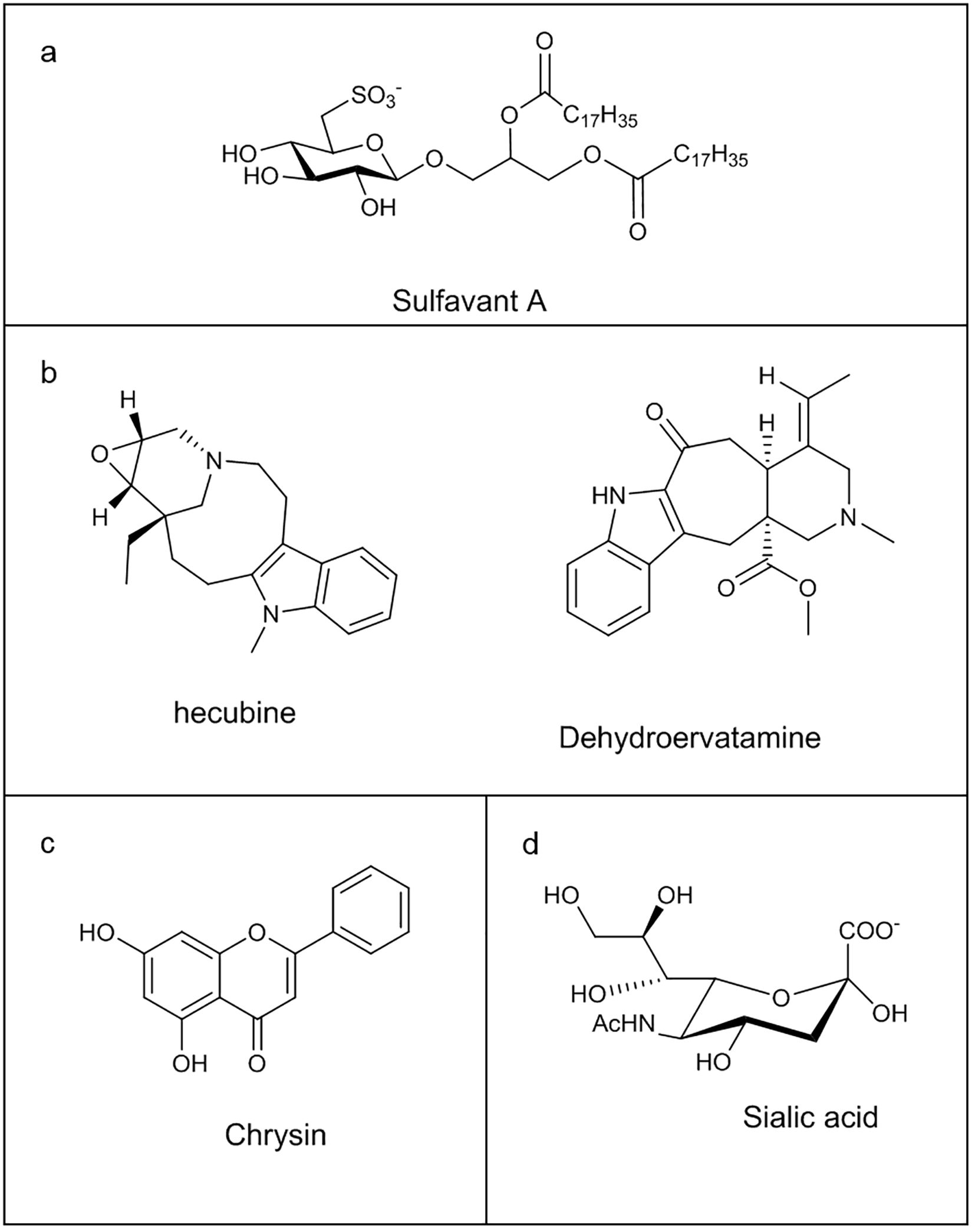
Figure 4. Chemical structure of some Immunoglobulin-like receptors ligands: (a) Sulfavant A; (b) Hecubine and Dehydroervatamine; (c) Chrysin; (d) Sialic acid.
Natural monoterpenoid indole alkaloids, hecubine and dehydroervatamine (Figure 4b), isolated from Ervatamia hainanensis and Tabernaemontana bovina plants, respectively, induced neuroinflammation reduction through TREM2 targeting in LPS-stimulated microglia model, preventing pro-inflammatory cytokines release and favouring anti-inflammatory factors expression, with the consequent regulation of the immune response towards a neuroprotective cellular state. In silico and cellular thermal shift assay (CETSA) experiments highlighted the direct binding of these molecules with TREM2 (127, 128).
Finally, Alpinia oxyphylla fructus extracts, main constituents of some Chinese medicines, have proven to be effective in improving cognitive ability, anti-oxidative stress and protecting neurons, thanks to flavon Chrysin (Figure 4c), able to determine a neuroprotective microglia M2 polarization via TREM2. Docking experiments supported the ability of the flavon to bind the protein (129).
Albeit the progress concerning the characterization of new TREM-2 ligands, the involvement of this receptor in neurodegenerative disease made necessary the development of new strategies of ligand-screening and synthesis. In this regard, the promiscuous character of this receptor also explains the fact that different ligands, with greater or lesser avidity, were able to activate different signalling pathways leading to different cellular responses (130), with subsequent and respective activation or deactivation of the cell functions for which this protein was responsible.
2.1.2.1.3 TREM-3 receptor
TREM-3 is an activating receptor presenting strong similarity to TREM-1 and TREM-2, regarding DAP12 mediated signalling pathway. TREM-3 is mainly expressed on macrophage, but low level of protein transcripts could also be detected in mouse T cell lines. However, in humans, TREM-3 is a pseudogene (58).
Although little is known about TREM-3, recent findings suggest that it may play a role in modulating TLR signalling. In macrophages, TREM-3 transcripts were upregulated by LPS (131), IL-1β and TNFα (65), and down-regulated by IFN-gamma (131). These observations were confirmed in TLR4 mutant mice, where LPS injection failed to alter the expression of TREM-1 and TREM-3, indicating that this response is dependent on TLR4 signalling (65). Evidence also suggest that TREM-3 may contribute to the inflammatory response of hepatic macrophages and endothelial cells during acute endotoxemia (65), and may play a protective role in host defence against Klebsiella pneumoniae infection in vivo (132).
However, the discovery of possible ligands that bind to TREM-3 could help the understanding not only of its physiological role but also highlight the insights of the TREM receptors regulation system.
2.1.3 TREM-like receptors
TREM-like receptors, TREM-like-1 (TREML-1), TREML-2 and TREML-3, are encoded within the TREMs gene cluster in humans. It must be pointed out that the TREM-like proteins have distinct structural and functional properties compared to TREM-1 and TREM-2 (133). Particularly, TREML-1 can enhance the calcium signalling in an SHP2 (PTPN11)-dependent manner, whereas some genetic variant of TREML-2 have been identified to be protective for Alzheimer’s disease, contrary to TREM-2 genetic variants. The role of TREML-3 remains to be investigated (133).
TREML-1 is widely expressed in microglia and its levels in the brain have been associated with decreased risk of developing Alzheimer disease in humans (134). This receptor shows similar binding affinity with Aβ oligomers as TREM-2 (114). TREM-2 knockout showed a dramatic increase of TREML-1 expression in the brain (135). Interestingly, the expression of an alternative transcript of TREML-1 (TREM like-1s) has been found to interact with TREM-2 via an immunoprecipitation assay and its over-expression inhibited osteoclast differentiation of bone marrow-derived macrophages (136). Therefore, TREML-1s could be a negative regulator of TREM-2 function.
These data suggest a plausible role of TREML-1 as TREM-2 antagonist, in both physiological and pathological conditions. Moreover, finding new molecules that can act via TREML-1 could modulate the activity of TREM-2, and vice-versa On the other hand, also targeting TREML-2 could open to new strategies for the treatment of Alzheimer’s disease, highly enhancing the rate of success. Considering its protective role, an activating molecule could represent a possible preventive treatment for individuals with specific genetic variations already known to greatly increase the risk of Alzheimer’s disease.
2.1.4 SIRP family members
SIRP (also called signal regulatory proteins) are mainly present in myeloid cells. There are three main SIRP receptors: SIRP-α, SIRP-β and SIRP-γ. All SIRPs members present an extracellular region that consists of three immunoglobulin superfamily (IgSF) domains and have different cytoplasmic regions (137). SIRP-α does contain an ITIM motifs, which mediate its association with the phosphatase SHP2 (138). SIRP-β has a very short cytoplasmic region of only 6 amino acids which binds to the adaptor protein DAP12 and transmit activating signals (139). Whereas SIRP-γ has a shorter cytoplasmic region (of only 4 amino acids), lacking a charged amino-acid side chain that allows the association with DAP12 (137, 140). Recently, it was shown that SIRP-β1 acts as a phagocytic receptor on microglia in amyloid precursor protein transgenic mice and in patients with Alzheimer’s disease. Indeed, activation of SIRP-β1 on cultured microglia induced reorganization of the cytoskeleton protein β-actin, increased phagocytosis of fibrillary amyloid-β (Aβ) (Figure 3), and suppressed LPS-induced gene transcription of TNF-α and nitric oxide synthase-2 (139).
The first ligand identified for SIRP-α was CD47 (141), an immune checkpoint molecule that downregulates key aspects of both the innate and adaptive antitumor immune response. CD47 is also a ligand for SIRP-γ (10 times weaker), but not for SIRP-β (142). An automated quantitative TR-FRET-based high-throughput screening assay platform reports the screening of large diverse drug-like chemical libraries to discover novel small molecules able to inhibit CD47-SIRP-α interaction (143). Five compounds were identified, whose structural identification is still under investigation (143). SIRP-α has also been shown to bind to the surfactant proteins A and D (SP-A and SP-D), the first at high levels in the lungs, the second in all mucosal surfaces, but with less affinity than CD47 (144). Most likely, SIRP-α recognize the globular heads glycosylated groups of surfactant proteins SP-A and SP-D. It has been proposed that SP-A and -D present a dual function, both helping to maintain an anti-inflammatory response by stimulating SIRP-α during the interaction with phagocytosis-stimulating PAMPS and inducing a pro-inflammatory response (144).
Stimulation by SIRP-β monoclonal antibody triggers SYK phosphorylation, MAPK activation, phagocytosis on macrophages (140) and migration of neutrophils (145). The natural ligand of SIRP-β is unknown and yet its biological significance remains unresolved.
Finally, several interesting questions for SIRP receptors remain to be answered, especially linked to their extracellular and intracellular ligands, and the research of new binding molecules could be extremely helpful in cancer and inflammation field.
2.1.5 Siglecs family members
Siglecs (sialic acid binding immunoglobulin-like lectins) are I-type (Ig-type) lectins characterized by an Ig domain that mediates sialic acid binding. Sialic acids are a large family of 9-carbon sugars that are all derivatives of neuraminic acid (Neu) or keto-deoxynonulosonic acid (KDN) (Figure 4d) (146). They are typically added at the end of the glycosylation process, capping a diverse array of glycosylation structures (146). Therefore, they are present on a variety of proteins and lipids attached to a wide of mammalian cell surfaces, including all human cells (147), playing key roles in Siglecs-mediated immune regulation and maintaining self-tolerance. Siglecs are a family of 14 different receptors which can be divided into two main groups based on their genetic similarity. The first group is present in all mammals consisting of Siglec-1 (Sialoadhesin), Siglec-2 (CD22), Siglec-4 (MAG), and Siglec-15 (148). The second group includes the CD33-related Siglecs, named Siglec-3 (also called CD33), −5, −6, −7, −8, −9, −10, −11, −14, and −16 (149). They are mainly expressed on monocytes, monocyte-derived macrophages and monocyte-derived DCs but also in B cells, basophils, neutrophils, and NK cells, with different expression patterns for every cell subset (150).
Each Siglec presents a specific role in the regulation of immune cell function in infectious diseases (151), inflammation, neurodegeneration, autoimmune diseases and cancer (152). Siglec-8 is involved in the development of asthma and Siglec-9 chronic lung inflammation (151). Whereas, Siglec-3, with ITIM motif, inhibits microglial uptake of amyloid beta and genetic variation could increase the development of Alzheimer’s disease susceptibility (153–155). Collectively the data report that Siglec-3 can inhibit the clearance of amyloid plaque in microglial cell cultures and in vivo (155), considering that its knockdown could mitigate amyloid-β pathology (Figure 3) (154). Interestingly, both Siglec-3 and TREM-2 have been associated with increasing the risk of Alzheimer’s disease, but in an opposite way. Indeed, conversely to Siglec-3, TREM-2 reduces plaque load and upregulates phagocytosis genes (Figure 3) (156). The Alzheimer-linked genetic R47H variant of TREM-2 acts similarly to Siglec-3, impairing amyloid-β–induced microgliosis and microglial activation (157). The link between TREM-2 and Siglec-3 has been straightened by knockdown experiments, which have shown that TREM-2 acts downstream of Siglec-3 and that loss of microglial clearance capacity might be reversed by therapeutic inhibition of Siglec-3 or activation of TREM-2 (158). Therefore, targeting those receptors could facilitate therapeutics to treat neurodegenerative pathologies as Alzheimer’s disease.
Recently, curcumin has been proposed as an immunomodulatory treatment capable of emulating anti-β-amyloid vaccine in stimulating phagocytic clearance by reducing Siglec-3 and increasing TREM2 (159). However, even if curcumin does not represent a new ligand for neither TREM2 or Siglec-3, these findings suggest the huge potential that could derive by controlling both TREM2 or Siglec-3 pathways involved in developing Alzheimer’s disease (Figure 3).
The scientific interest has been so far focused on finding Siglec-3 inhibitors that might be effective against disease progression. Microparticles of subtype-selective trisaccharide containing 2,5,9-trisubstituted sialic acid mimetic, called P22, increases the uptake of the toxic Alzheimer’s disease peptide and amyloid-β into microglial cells (160), evidencing Siglec-3 as promising target for therapeutics favouring clearance of amyloid-β.
Siglec-4 and Siglec-14 till Siglec-16 do not have ITIM or ITIM like motifs. Their signals are mediated by DAP12, and, therefore, called activating Siglecs (161). Upon ligand binding, Siglecs-DAP12 system can recruit PI3K thus promoting an inflammatory response through activation of MAPK pathway (162). Other Siglecs, and in particular CD33-related Siglecs, have an ITIM and/or ITIM-like motif in their intracellular domain and can mediate an inhibitory signal (151).
As described above, the presence of sialic acids structures on cell surface could function as a self-associated molecular pattern (SAMP) and thus, Siglecs can act to dampen leukocyte activation under homeostatic conditions (162, 163).
Interestingly, several pathogenic bacteria also use the sialic acid-Siglec axis to dampen the immune system in favour of their survival. In this regard, they could have acquired the ability to take sialic acids or sialylated structures from the host, to synthetize “mimic” structures or even perform de novo synthesis of sialic acids, giving them a survival advantage (161). The same strategy is also used in case for the Human immunodeficiency virus (HIV) and the Porcine reproductive and respiratory syndrome virus (PRRSV) (164, 165). It has been demonstrated an increased sialylation, α2,3; α2,6, and α2,8 linked sialic acids, in multiple tumour tissues like renal cell carcinoma, prostate cancer, colon cancer, breast cancer, head and neck squamous cell carcinoma and oral cancer (166).
Finally, Siglecs participate in the discrimination between self and non-self sialic acid motifs, triggering endocytosis, pathogen and dysfunction recognition and regulating, both by activation and deactivation, the function of innate and adaptive immune cells. The research, finding and develop of sialyl-based small chemical entities could pave the way for new therapeutic treatments in different pathological conditions like infectious diseases, inflammation, neurodegeneration, autoimmune diseases and cancer.
2.2 C-type lectin receptors
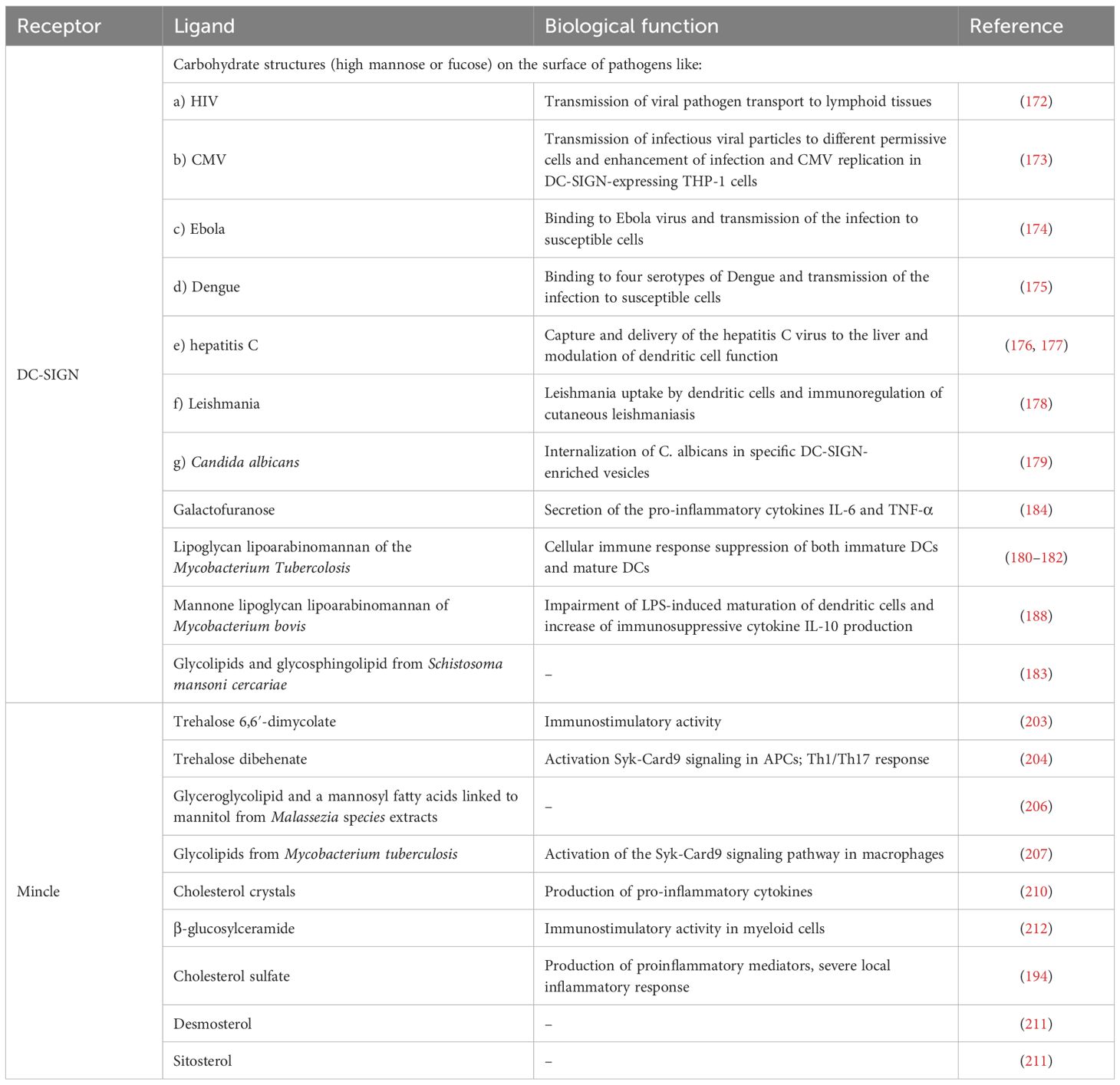
C-type lectin receptors (CLRs) is a family of receptors which possess one or more carbohydrate-recognition domains, thanks to which they usually bind carbohydrate moieties through a Ca2+ dependent conserved motif, although some of them lack the Ca2+ binding site. These proteins differ in the kind of pathogen-derived or self-expressed ligands that they are able to recognize. Among these ligands, there are not only glycans but also proteins or lipids, triggering functions as adhesion, phagocytic, and signalling pathways and directing the cell towards homeostasis following the activation of innate and adaptive immunity. CLRs represent a huge group of proteins divided into seventeen subgroups (167). Considering the ability of these proteins to recognize polymeric structures, some members of this family are able to also interact with small structural motifs, as DC-Specific Intercellular adhesion molecule-3-Grabbing Non-integrin (DC-SIGN) and Macrophage-inducible C-type lectin (Mincle) (Table 2), two members particularly studied and crucial in the immune response modulation.
2.2.1 DC-SIGN
DC-SIGN receptor (or DC specific Intercellular adhesion molecule-3-Grabbing Non-integrin) is expressed by myeloid DCs and subpopulations of macrophages (168). DC-SIGN is highly expressed, and considered as marker, in cancer-associated fibroblasts and M2 macrophages, which are involved in the malignancy of different tumours (169). It has been reported that the expression of DC-SIGN in serum and cancer tissues may affect the survival time for colon cancer patients, representing also a valuable target for cancer treatment (170). Even though, DC-SIGN can induce IL-10 pathways, such as those including ERK and PI3K (171), whether the receptor is relevant to the induction of diseases like asthma remains to be determined.
DC-SIGN recognizes specific carbohydrate structures (high mannose or fucose) on the surface of pathogens and self-glycoproteins. This protein is particularly known to be the receptor that captures HIV-1 at sites of entry, enabling its transport to lymphoid tissues, where DC-SIGN efficiently transmits low amounts of HIV-1 (172).
Besides HIV-1, DC-SIGN was also shown to bind other viruses like cytomegalovirus (173), Ebola (174), Dengue (175), and hepatitis C (176, 177), as well as microorganisms as Leishmania (178) and Candida albicans (179). The protein is able to bind Mycobacterium tuberculosis and thus mediating its entry in DCs in vivo. In this scenario, it does not only allow Mycobacterium Tubercolosis to infect DCs but also suppresses the cellular immune responses of both immature DCs and mature DCs (180). Indeed, DC-SIGN specifically binds to the lipoglycan lipoarabinomannan (LAM), a major component of the mycobacterial cell wall, which contains a carbohydrate backbone composed of D-mannan and D-arabinan (181, 182). Interestingly, DC-SIGN does not bind to all mycobacterium similarly, suggesting that it may act through selective recognition. In fact, Mycobacterium smegmatis-derived LAM, capped by phosphoinositide residues (PILAM), or Mycobacterium fortuitum and Mycobacterium chelonae-derived AraLAM, Arabian domain uncapped, does not bind to DC-SIGN. Also, Mycobacterium avium derived ManLAM, capped with single mannose residues, was also poorly recognized by DC-SIGN (181). All the data suggest that this receptor recognizes a specific motif domain. DC-SIGN is also able to bind small chemical entities like glycolipids derived from Schistosoma Mansoni cercariae and their excretory or secretory such as carbohydrate moieties of both glycosphingolipid species with Galβ1–4(Fucα1–3)GlcNAc (LewisX) and Fucα1–3Galβ1–4(Fucα1–3)GlcNAc (pseudo-LewisY) determinants (183). Recently, it was shown that Galactofuranose (Galf) can interact with DC-SIGN and induce the secretion of the pro-inflammatory cytokines IL-6 and TNF-α (184).
Interestingly, DC-SIGN is organized in plasma membrane microdomains (with average diameter of 200nm) crucial for binding and internalization of virus particles, acting as a docking site for pathogens (185). It was also suggested that cholesterol-dependent membrane properties, rather than lipid rafts per se, are responsible to promote efficient HIV-1 infection in T cells (186).
DC-SIGN is also an essential co-receptor for TLR4-induced activation of human DCs. Indeed, fucosylated glycan, upon binding with DC-SIGN in DCs, primed naïve T cells towards a Th1 profile, while inducing TLR4-activation (187). Interestingly, upon mycobacterium bovis ManLAM binding, DC-SIGN impairs LPS-induced maturation of DCs and increases the production of the immunosuppressive cytokine IL-10, highlighting a possible pathogen strategy to escape immune surveillance (188). Notably, this receptor, encountering and recognizing a pathogen, is able to stimulate the kinase Raf-1, inducing acetylation of the NF-κB subunit p65 and increasing anti-inflammatory cytokines expression, all this only after TLRs-induced activation of NF-κB. This evidence shows a receptor ability to induce adaptive immunity by DCs against pathogenic microorganisms (189).
Considering DC-SIGN specific ligand mediated immune response, polyvalent carbohydrate ligands have been proposed in the design of novel immunomodulants and vaccine adjuvants (190). Finally, DC-SIGN is an important player in the recognition of pathogens by dendritic cells and uncover more functional aspects of its activation mechanisms will be extremely important for the treatment of life-threatening infection disease, such as Dengue or Ebola, also considering that this protein seems to be able to recognize small structural motifs as well as polymeric structures. Moreover, new molecules that bind with more affinity to DC-SIGN could be used as antagonists to prevent the binding of other pathogens or as vaccine adjuvants.
2.2.2 Mincle
Mincle (also called macrophage-inducible C-type lectin) is mainly expressed on monocytes, macrophages, neutrophils and DCs (191). However, in macrophages and DCs, its expression can be strongly upregulated by PAMPs, such as the TLR4 ligand LPS or by a Mincle ligand (192). Mincle has been reported to be associated with rheumatoid arthritis (193), but also to other inflammatory-mediated diseases. Indeed, this receptor is also involved in allergic skin inflammation (194) and post-ischemic inflammation (195), and other experimental inflammatory models such as chronic alcoholic liver disease (196). Given the roles that Mincle plays in a (chronic) immune reaction, it has been suggested both as adjuvant for the treatment of pathogenic immune of Crohn’s disease (197) and also for foot-and-mouth disease virus infection (198). Recent studies show that Mincle also contributes to neuropathic pain in the dorsal root ganglia and spinal dorsal horn (199). Furthermore, Mincle promotes and maintains inflammatory phenotypes of M1 macrophages in acute renal inflammation (200).
The protein is an activating receptor coupled with the ITAM–bearing adaptor FcRγ chain (201). Its expression is sensitive to various inflammatory stimuli, such as LPS, TNF-α, IL-6, and IFN-gamma, activated downstream of the nuclear transcription factor NF-IL6 in macrophages (202).
Mincle specific ligand is trehalose 6,6′-dimycolate (TDM) (Figure 5a), a mycobacterial cell wall glycolipid and its most studied immunostimulatory component (203). Mincle is also able to recognize analogs like trehalose dibehenate (TDB) (Figure 5b) but not the non-glycosylated mycolate or trehalose molecules (204). Though TDM is also present into cell wall of other mycobacteria, corynebacteria, nocardia and fungi, Mincle is not able to bind all of them, suggesting more specific and complex recognition mechanisms. Also, the Mincle ligand in Malassezia species fungi has not yet been identified, suggesting the existence of ligands rather than TDM (205). Ishikawa et al. were able to identify two ligands after fractionation of Malassezia extracts: a glucosyl-based glyceroglycolipid (named 44-1) (Figure 5c) and a unique mannosylated fatty acids linked to mannitol (named 44-2) (Figure 5d) (206). Moreover, Mincle is also able to recognize glycolipids of Mycobacterium tuberculosis (207). Recently, it has been designed an alkyl 6-O-acyl-β-D-glucosides that resulted to be an effective agonist of Mincle, with a potency comparable to the prototypical ligand TDM (208).
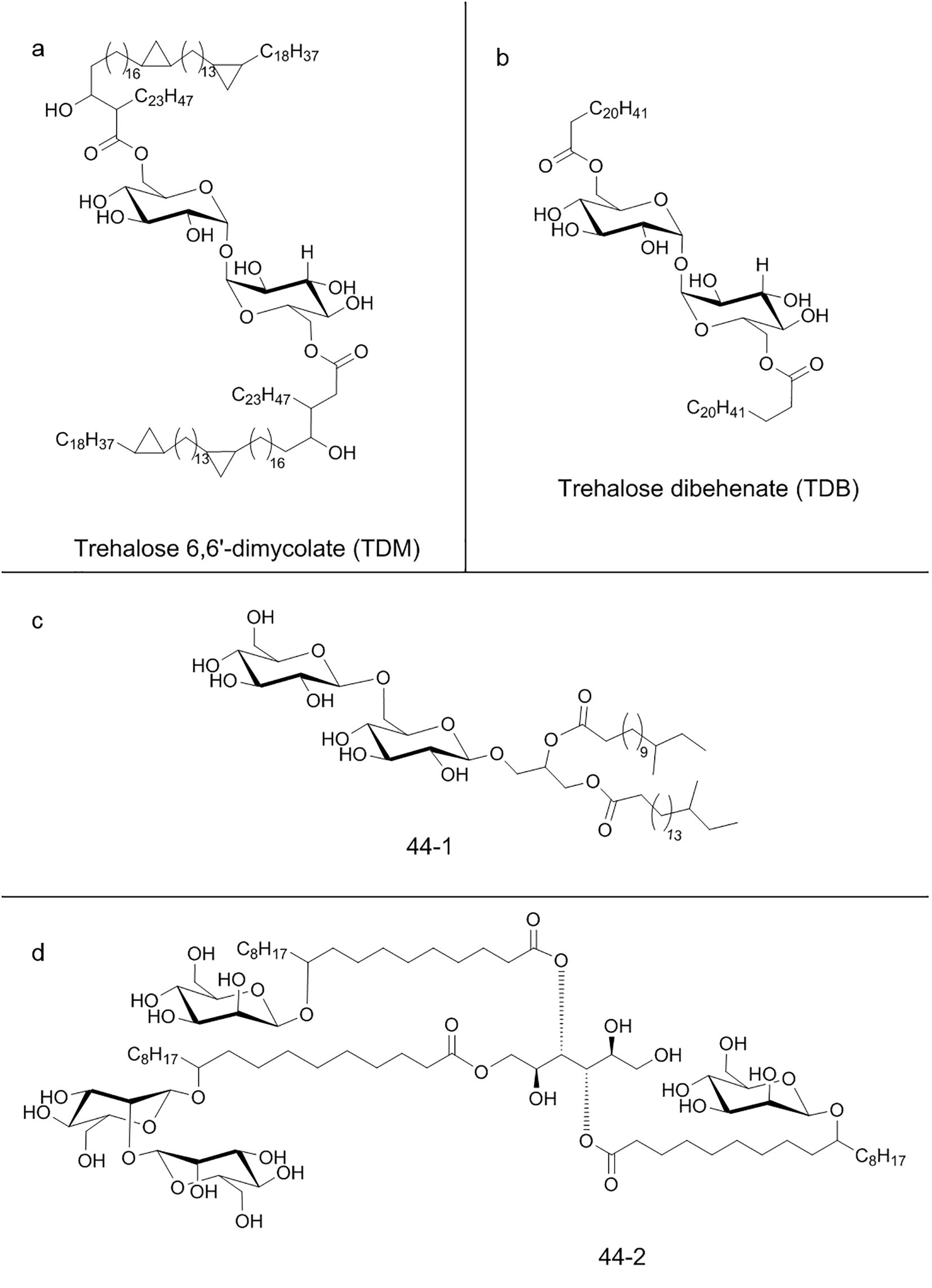
Figure 5. Chemical structure of some C-type lectin receptors ligands: (a) Trehalose 6,6′-dimycolate (TDM); (b) Trehalose dibehenate (TDB); (c) Glucosyl-based glyceroglycolipid (44-1); (d) Mannosylated fatty acids linked to mannitol (44-2).
Furthermore, it was found that L. prolificans and S. boydii α-glucan polysaccharides were recognized by Mincle through the α-(1→4) and α-(1→6)-linked glucopyranoside moiety, and Mincle deficiency impacts the phagocytosis dynamics (209).
Notably, Mincle recognizes cholesterol crystals producing pro-inflammatory cytokines (210) and cholesterol sulfate of the epithelial layer inducing local secretion of different proinflammatory mediators (194). The receptor can also signal through other sterols including, desmosterol and sitosterol (211). Mincle has further crucial function in regulating the immune system, being capable to sense and react to tissue damage. Indeed, β-glucosylceramide, identified as endogenous ligand for Mincle upon cell damage, enables an immunostimulatory activity in myeloid cells and the induction of inflammatory cytokine production (212).
Block of TLR4 or NF-κB suppressed LPS-induced Mincle expression in macrophages (Figure 3) and maintained M1 phenotype through Syk pathway (213). Furthermore, Mincle acts in synergism with TLR7/8 by inducing NF-κB signalling in monocyte-derived DC, thereby enabling the production of Th1-polarizing cytokines and promoting autologous Th1 polarization (200).
Although Mincle shows binding ability to different classes of ligands influencing the innate immune system, such as lipids and cholesterol derivatives (Figure 1b), there is a broad spectrum of potential further agonists for Mincle, with some ligands already exhibiting potential as vaccine adjuvants. Nevertheless, much remains to be unanswered in terms of better understanding the pathways involved in Mincle activation upon ligand interaction.
3 Conclusions
The innate immune system senses microbial infection, cell dysfunction and self-ligands by pathogen recognition receptors (PRRs), such as TLRs and regulatory receptors, usually associated with different ITAM-bearing signalling units, as FcRγ and DAP12. The rapid activation of PRRs signalling must be followed by counterbalanced regulatory inhibitory action, aimed to make the immune reaction effective and non-harmful, so preventing cytotoxic effects related to excessive activation. Recently immunoglobulin-like and C-type lectin receptors have gained interest as targets for vaccine development and immune therapies for their ability to activate and finely modulate the immunological functions. These processes are mediated by endogenous and exogenous molecules, most of which are not yet known, often structurally characterized by small size and lipidic nature.
For this reason, small chemical entities represent a target for the characterization of biological mechanisms determining receptor-mediated cell functions along with the development of specific receptor-associated disorders therapies. The main topic of this review is, in fact, the description of the main immunoglobulin receptors and CLRs, seen not only from a biological and functional point of view, but also considering their binding with small associated specific ligands. In this regard, LMIR, TREM, Siglec, SIRP and the most studied CLRs receptors were analysed for their ability to recognize and functionally act upon small molecules interactions.
Different classes of molecules such as phospholipids, glycolipids and sulfolipids can bind to several immunoglobulin-like receptors, like LMIRs and TREMs, characterized by activating/tuning/inhibitory roles. The sharing of ligand families suggests strong evidence of a complex mechanism of regulation though TREMs-LMIRs-axis. This cellular dynamic represents a finely tuned immunomodulatory mechanism, that needs further investigation to better understand the fine level of complexity of these receptors in response to endogenous and/or external stimuli (43). Indeed, the signalling pathway activated upon binding of the two receptors families could synergise or even bifurcate their signalling pathways in opposite biological effects, because of different cellular and physiological states that can potentially cause expression of different ligand molecular entities. Furthermore, considering that Siglec and SIRP receptors share similar downstream pathophysiological effects as TREM family members, and since little is known about their endogenous ligands, a more detailed study of a possible synergism between these receptors should be highlighted. In addition, Siglec and SIRP are involved in the antitumor immune response, so further studies on their possible binding partners could be extremely useful in both cancer and inflammatory research. Several common features were evident also among CLRs sub-families. In this regard, yeast mannans appear to possess same immunostimulatory properties, which are selectively recognized by DC-SIGN and Mincle, also capable of mediating an immunomodulatory activity thanks to the interaction with small glycoconjugates (107).
Interestingly, a close similarity or structural equality of some ligands can also be highlighted for some immunoglobulin-like receptors and CLRs. In particular, glycolipids and sulfolipids, but also ceramide compounds, have shown binding affinities for LMIRs, TREM-2 and Mincle receptors. However, there are other common feature among this class of receptors. As previously described, despite the structural similarity among the immunoglobulin like receptor family, there are relevant difference between activating and inhibitory receptors. Similarly, Mincle but also other CLRs, behave more like the activating immunoglobulin-like receptors, which engage kinases indirectly, through association with ITAM-bearing signalling chains of DAP-12 or FcRγ (13, 214). Thus, although our review highlights the classification into two main classes of receptors, they may have distinct signalling requirements based on activation by distinct or similar ligands, which could lead to subtle differences in their downstream responses.
Interestingly, TLRs act in synergism with most of the receptors described. Indeed, the TLR/CLR antagonist combinations is one of the most well know adjuvation systems to enable early-life immunization against intracellular pathogens (200), already investigated for Dectin-1, Dectin-2, DC-SIGN and Mincle (190, 197, 215–217). To this end, it would be very interesting to identify new ‘adjuvant’ molecules based on other receptors described in this review, such as receptors belonging to the immunoglobulin like receptors family, and thus amplify the opportunity to enhance strategies for patient-immunization.
Currently, the exact structure of many ligands that interact with those receptors is yet not fully characterized and further investigation must be performed to better understand the functional mechanism underlying the receptor activation. In general, small molecules offer a unique advantage to maximize the therapeutic benefits, especially targeting membrane protein, with potential low production costs and huge possibility to modulate and optimize activity by chemical synthesis and structural modification. Therefore, deciphering the biological activity of these ligands, together with medicinal chemistry and molecular biology, could lead to the identification and optimization of novel small molecules able to inhibit or enhance the function of the receptor of interest. However, there are also disadvantages, as small molecule ligand could reveal sub-micromolar affinities for several targets besides the target of interests (218). Therefore, conducting more preclinical studies and enhancing efforts before the clinical phases are of extreme importance. This approach will help to develop more effective drugs and reduce financial consequences of failure (219). A fine study of each receptor family could at least reduce unwanted in vivo side effect and interchangeable ligand-receptor affinity.
Finally, mastering the binding specificity of each receptor could advance disease understanding and improve the evaluation of biomarker-guided treatment strategies.
Author contributions
LF: Validation, Conceptualization, Writing – review & editing, Data curation. MZ: Data curation, Validation, Writing – review & editing. GN: Visualization, Writing – review & editing, Conceptualization, Formal Analysis. FA: Investigation, Writing – review & editing, Conceptualization, Validation. MS: Conceptualization, Investigation, Writing – review & editing. DNC: Data curation, Writing – review & editing, Investigation. GB: Investigation, Writing – review & editing, Conceptualization. MD: Data curation, Investigation, Writing – review & editing. OF: Formal Analysis, Investigation, Data curation, Writing – review & editing. NE: Formal Analysis, Data curation, Writing – review & editing, Investigation. DLC: Writing – review & editing, Formal Analysis, Investigation, Data curation. GD’I: Data curation, Investigation, Formal Analysis, Writing – review & editing. CG: Data curation, Validation, Supervision, Conceptualization, Investigation, Writing – review & editing, Writing – original draft, Formal Analysis. AF: Writing – review & editing, Investigation, Supervision, Writing – original draft, Validation, Conceptualization. EM: Writing – review & editing, Conceptualization, Investigation, Writing – original draft, Validation, Supervision.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
The authors thank the PNRR projects PE00000007_One Health Basic and Translational Actions Addressing Unmet Needs on Emerging Infectious Diseases (INF-ACT) and “Potentiating the Italian Capacity for Structural Biology Services in Instruct Eric” (Acronym: ITACA.SB, project n IR0000009) within the call MUR D.D. 0003264 dated 28/12/2021 PNRR M4/C2/L3.1.1, funded by the European Union NextGenerationEU. The authors also thank the PRIN project ‘A novel pharmacological approach to rescue Trem2-mediated microglial defects’ (code 2022SN342C), within Research Projects of Relevant National Interest–Call 2022.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Rivera A, Siracusa MC, Yap GS, and Gause WC. Innate cell communication kick-starts pathogen-specific immunity. Nat Immunol. (2016) 17:356–63. doi: 10.1038/ni.3375
2. Olive C. Pattern recognition receptors: sentinels in innate immunity and targets of new vaccine adjuvants. Expert Rev Vaccines. (2012) 11:237–56. doi: 10.1586/erv.11.189
3. Schroder K and Tschopp J. The inflammasomes. Cell. (2010) 140:821–32. doi: 10.1016/j.cell.2010.01.040
4. Amarante-Mendes GP, Adjemian S, Branco LM, Zanetti LC, Weinlich R, and Bortoluci KR. Pattern recognition receptors and the host cell death molecular machinery. Front Immunol. (2018) 9:2379. doi: 10.3389/fimmu.2018.02379
5. Kumar H, Kawai T, and Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol. (2011) 30:16–34. doi: 10.3109/08830185.2010.529976
6. Chiffoleau E. C-type lectin-like receptors as emerging orchestrators of sterile inflammation represent potential therapeutic targets. Front Immunol. (2018) 9:227. doi: 10.3389/fimmu.2018.00227
7. García-Vallejo JJ and Van Kooyk Y. Endogenous ligands for C-type lectin receptors: the true regulators of immune homeostasis. Immunol Rev. (2009) 230:22–37. doi: 10.1111/j.1600-065X.2009.00786.x
8. Roe K, Gibot S, and Verma S. Triggering receptor expressed on myeloid cells-1 (TREM-1): a new player in antiviral immunity? Front Microbiol. (2014) 5:627. doi: 10.3389/fmicb.2014.00627
9. Ford JW and McVicar DW. TREM and TREM-like receptors in inflammation and disease. Curr Opin Immunol. (2009) 21:38–46. doi: 10.1016/j.coi.2009.01.009
10. Borrego F. The CD300 molecules: an emerging family of regulators of the immune system. Blood. (2013) 121:1951–60. doi: 10.1182/blood-2012-09-435057
11. Takai T. Paired immunoglobulin-like receptors and their MHC class I recognition. Immunology. (2005) 115:433–40. doi: 10.1111/j.1365-2567.2005.02177.x
12. Fioretto L, Ziaco M, Mercogliano M, Gallo C, Nuzzo G, d’Ippolito G, et al. The Janus effect of colloidal self-assembly on the biological response of amphiphilic drugs. Pharmacol Res. (2024) 208:107400. doi: 10.1016/j.phrs.2024.107400
13. Hamerman JA, Ni M, Killebrew JR, Chu C, and Lowell CA. The expanding roles of ITAM adapters FcRγ and DAP12 in myeloid cells. Immunol Rev. (2009) 232:42–58. doi: 10.1111/j.1600-065X.2009.00841.x
14. Colonna M, Nakajima H, and Cella M. Inhibitory and activating receptors involved in immune surveillance by human NK and myeloid cells. J Leukoc Biol. (1999) 66:718–22. doi: 10.1002/jlb.66.5.718
15. Ravetch JV and Lanier LL. Immune inhibitory receptors. Sci (1979). (2000) 290:84–9. doi: 10.1126/science.290.5489.84
16. Nasu J, Uto T, Fukaya T, Takagi H, Fukui T, Miyanaga N, et al. Pivotal role of the carbohydrate recognition domain in self-interaction of CLEC4A to elicit the ITIM-mediated inhibitory function in murine conventional dendritic cells. vitro. Int Immunol. (2020) 32:673–82. doi: 10.1093/intimm/dxaa034
17. Piovesan D, de Groot AE, Cho S, Anderson AE, Ray RD, Patnaik A, et al. Fc-silent anti-TIGIT antibodies potentiate antitumor immunity without depleting regulatory T cells. Cancer Res. (2024) 84:1978–95. doi: 10.1158/0008-5472.CAN-23-2455
18. Chung D-H, Humphrey MB, Nakamura MC, Ginzinger DG, Seaman WE, and Daws MR. CMRF-35-like molecule-1, a novel mouse myeloid receptor, can inhibit osteoclast formation. J Immunol. (2003) 171:6541–8. doi: 10.4049/jimmunol.171.12.6541
19. Kumagai H, Oki T, Tamitsu K, Feng S-Z, Ono M, Nakajima H, et al. Identification and characterization of a new pair of immunoglobulin-like receptors LMIR1 and 2 derived from murine bone marrow-derived mast cells. Biochem Biophys Res Commun. (2003) 307:719–29. doi: 10.1016/S0006-291X(03)01245-2
20. Bachelet I, Munitz A, Moretta A, Moretta L, and Levi-Schaffer F. The inhibitory receptor IRp60 (CD300a) is expressed and functional on human mast cells. J Immunol. (2005) 175:7989–95. doi: 10.4049/jimmunol.175.12.7989
21. Munitz A, Bachelet I, and Levischaffer F. Reversal of airway inflammation and remodeling in asthma by a bispecific antibody fragment linking CCR3 to CD300a. J Allergy Clin Immunol. (2006) 118:1082–9. doi: 10.1016/j.jaci.2006.07.041
22. DeBell KE, Simhadri VR, Mariano JL, and Borrego F. Functional requirements for inhibitory signal transmission by the immunomodulatory receptor CD300a. BMC Immunol. (2012) 13:23. doi: 10.1186/1471-2172-13-23
23. Karra L, Singh Gangwar R, Shamri R, Puzzovio PG, Cohen-Mor S, Levy BD, et al. Leukocyte CD300a contributes to the resolution of murine allergic inflammation. J Immunol. (2018) 201:2998–3005. doi: 10.4049/jimmunol.1801000
24. Nakahashi-Oda C, Tahara-Hanaoka S, Shoji M, Okoshi Y, Nakano-Yokomizo T, Ohkohchi N, et al. Apoptotic cells suppress mast cell inflammatory responses via the CD300a immunoreceptor. J Exp Med. (2012) 209:1493–503. doi: 10.1084/jem.20120096
25. Coustan-Smith E, Song G, Clark C, Key L, Liu P, Mehrpooya M, et al. New markers for minimal residual disease detection in acute lymphoblastic leukemia. Blood. (2011) 117:6267–76. doi: 10.1182/blood-2010-12-324004
26. Silva R, Moir S, Kardava L, Debell K, Simhadri VR, Ferrando-Martínez S, et al. CD300a is expressed on human B cells, modulates BCR-mediated signaling, and its expression is down-regulated in HIV infection. Blood. (2011) 117:5870–80. doi: 10.1182/blood-2010-09-310318
27. Speckman R, Wright Daw J, Helms C, Duan S, Cao L, Taillon-Miller P, et al. Novel immunoglobulin superfamily gene cluster, mapping to a region of human chromosome 17q25, linked to psoriasis susceptibility. Hum Genet. (2003) 112:34–41. doi: 10.1007/s00439-002-0851-y
28. Tanaka T, Tahara-Hanaoka S, Nabekura T, Ikeda K, Jiang S, Tsutsumi S, et al. PPARβ/δ activation of CD300a controls intestinal immunity. Sci Rep. (2014) 4:5412. doi: 10.1038/srep05412
29. Seno H, Miyoshi H, Brown SL, Geske MJ, Colonna M, and Stappenbeck TS. Efficient colonic mucosal wound repair requires Trem2 signaling. PNAS. (2009) 106:256–61. doi: 10.1073/pnas.0803343106
30. Nakahashi-Oda C, Tahara-Hanaoka S, Honda S, Shibuya K, and Shibuya A. Identification of phosphatidylserine as a ligand for the CD300a immunoreceptor. Biochem Biophys Res Commun. (2012) 417:646–50. doi: 10.1016/j.bbrc.2011.12.025
31. Simhadri VR, Andersen JF, Calvo E, Choi S-C, Coligan JE, and Borrego F. Human CD300a binds to phosphatidylethanolamine and phosphatidylserine, and modulates the phagocytosis of dead cells. Blood. (2012) 119:2799–809. doi: 10.1182/blood-2011-08-372425
32. Miki H, Nakahashi-Oda C, Sumida T, and Shibuya A. Involvement of CD300a phosphatidylserine immunoreceptor in aluminum salt adjuvant–induced th2 responses. J Immunol. (2015) 194:5069–76. doi: 10.4049/jimmunol.1402915
33. Izawa K, Kitaura J, Yamanishi Y, Matsuoka T, Kaitani A, Sugiuchi M, et al. An activating and inhibitory signal from an inhibitory receptor LMIR3/CLM-1: LMIR3 augments lipopolysaccharide response through association with fcRγ in mast cells. J Immunol. (2009) 183:925–36. doi: 10.4049/jimmunol.0900552
34. Izawa K, Kitaura J, Yamanishi Y, Matsuoka T, Oki T, Shibata F, et al. Functional analysis of activating receptor LMIR4 as a counterpart of inhibitory receptor LMIR3. J Biol Chem. (2007) 282:17997–8008. doi: 10.1074/jbc.M701100200
35. Szeíles L, Keresztes G, Toüroücsik D, Balajthy Z, Krenaícs L, Poíliska S, et al. 1,25-dihydroxyvitamin D3 is an autonomous regulator of the transcriptional changes leading to a tolerogenic dendritic cell phenotype. J Immunol. (2009) 182:2074–83. doi: 10.4049/jimmunol.0803345
36. Peluffo H, Alí-Ruiz D, Ejarque-Ortíz A, Heras-Alvarez V, Comas-Casellas E, Martínez-Barriocanal A, et al. Overexpression of the immunoreceptor CD300f has a neuroprotective role in a model of acute brain injury. Brain Pathol. (2012) 22:318–28. doi: 10.1111/j.1750-3639.2011.00537.x
37. Xi H, Katschke KJ, Helmy KY, Wark PA, Kljavin N, Clark H, et al. Negative regulation of autoimmune demyelination by the inhibitory receptor CLM-1. J Exp Med. (2010) 207:7–16. doi: 10.1084/jem.20091508
38. Izawa K, Yamanishi Y, Maehara A, Takahashi M, Isobe M, Ito S, et al. The receptor LMIR3 negatively regulates mast cell activation and allergic responses by binding to extracellular ceramide. Immunity. (2012) 37:827–39. doi: 10.1016/j.immuni.2012.08.018
39. Shi L, Luo K, Xia D, Chen T, Chen G, Jiang Y, et al. DIgR2, dendritic cell-derived immunoglobulin receptor 2, is one representative of a family of IgSF inhibitory receptors and mediates negative regulation of dendritic cell-initiated antigen-specific T-cell responses. Blood. (2006) 108:2678–86. doi: 10.1182/blood-2006-04-015404
40. Korver W, Zhao X, Singh S, Pardoux C, Zhao J, Guzman ML, et al. Monoclonal antibodies against IREM-1: potential for targeted therapy of AML. Leukemia. (2009) 23:1587–97. doi: 10.1038/leu.2009.99
41. Izawa K, Isobe M, Matsukawa T, Ito S, Maehara A, Takahashi M, et al. Sphingomyelin and ceramide are physiological ligands for human LMIR3/CD300f, inhibiting FcϵRI-mediated mast cell activation. J Allergy Clin Immunol. (2014) 133:270–273.e7. doi: 10.1016/j.jaci.2013.08.008
42. Choi S-C, Simhadri VR, Tian L, Gil-Krzewska A, Krzewski K, Borrego F, et al. Cutting edge: mouse CD300f (CMRF-35–like molecule-1) recognizes outer membrane-exposed phosphatidylserine and can promote phagocytosis. J Immunol. (2011) 187:3483–7. doi: 10.4049/jimmunol.1101549
43. Cannon JP, O’Driscoll M, and Litman GW. Specific lipid recognition is a general feature of CD300 and TREM molecules. Immunogenetics. (2012) 64:39–47. doi: 10.1007/s00251-011-0562-4
44. Nakahashi C, Tahara-Hanaoka S, Totsuka N, Okoshi Y, Takai T, Ohkohchi N, et al. Dual assemblies of an activating immune receptor, MAIR-II, with ITAM-bearing adapters DAP12 and fcRγ Chain on peritoneal macrophages. J Immunol. (2007) 178:765–70. doi: 10.4049/jimmunol.178.2.765
45. Totsuka N, Kim Y-G, Kanemaru K, Niizuma K, Umemoto E, Nagai K, et al. Toll-like receptor 4 and MAIR-II/CLM-4/LMIR2 immunoreceptor regulate VLA-4-mediated inflammatory monocyte migration. Nat Commun. (2014) 5:4710. doi: 10.1038/ncomms5710
46. Gil-de-Gómez L, Astudillo AM, Meana C, Rubio JM, Guijas C, Balboa MA, et al. A phosphatidylinositol species acutely generated by activated macrophages regulates innate immune responses. J Immunol. (2013) 190:5169–77. doi: 10.4049/jimmunol.1203494
47. Nakano T, Tahara-Hanaoka S, Nakahashi C, Can I, Totsuka N, Honda S, et al. Activation of neutrophils by a novel triggering immunoglobulin-like receptor MAIR-IV. Mol Immunol. (2008) 45:289–94. doi: 10.1016/j.molimm.2007.04.011
48. Yamanishi Y, Kitaura J, Izawa K, Kaitani A, Komeno Y, Nakamura M, et al. TIM1 is an endogenous ligand for LMIR5/CD300b: LMIR5 deficiency ameliorates mouse kidney ischemia/reperfusion injury. J Exp Med. (2010) 207:1501–11. doi: 10.1084/jem.20090581
49. Yamanishi Y, Takahashi M, Izawa K, Isobe M, Ito S, Tsuchiya A, et al. A soluble form of LMIR5/CD300b amplifies lipopolysaccharide-induced lethal inflammation in sepsis. J Immunol. (2012) 189:1773–9. doi: 10.4049/jimmunol.1201139
50. Knapp S, Gibot S, de Vos A, Versteeg HH, Colonna M, and van der Poll T. Cutting edge: expression patterns of surface and soluble triggering receptor expressed on myeloid cells-1 in human endotoxemia. J Immunol. (2004) 173:7131–4. doi: 10.4049/jimmunol.173.12.7131
51. Bouchon A, Facchetti F, Weigand MA, and Colonna M. TREM-1 amplifies inflammation and is a crucial mediator of septic shock. Nature. (2001) 410:1103–7. doi: 10.1038/35074114
52. Phongsisay V, Iizasa E, Hara H, and Yamasaki S. 3-O-sulfo-β-d-galactose moiety of endogenous sulfoglycolipids is a potential ligand for immunoglobulin-like receptor LMIR5. Mol Immunol. (2015) 63:595–9. doi: 10.1016/j.molimm.2014.07.023
53. Phongsisay V. Campylobacter jejuni targets immunoglobulin-like receptor LMIR5. Mol Immunol. (2015) 63:574–8. doi: 10.1016/j.molimm.2014.07.024
54. Ichimura T, Bonventre JV, Bailly V, Wei H, Hession CA, Cate RL, et al. Kidney injury molecule-1 (KIM-1), a putative epithelial cell adhesion molecule containing a novel immunoglobulin domain, is up-regulated in renal cells after injury. J Biol Chem. (1998) 273:4135–42. doi: 10.1074/jbc.273.7.4135
55. Enomoto Y, Yamanishi Y, Izawa K, Kaitani A, Takahashi M, Maehara A, et al. Characterization of leukocyte mono-immunoglobulin-like receptor 7 (LMIR7)/CLM-3 as an activating receptor. J Biol Chem. (2010) 285:35274–83. doi: 10.1074/jbc.M110.137166
56. Wu Y, Zhu X, Li N, Chen T, Yang M, Yao M, et al. CMRF-35–like molecule 3 preferentially promotes TLR9-triggered proinflammatory cytokine production in macrophages by enhancing TNF receptor-associated factor 6 ubiquitination. J Immunol. (2011) 187:4881–9. doi: 10.4049/jimmunol.1003806
57. Colonna M. The biology of TREM receptors. Nat Rev Immunol. (2023) 23:580–94. doi: 10.1038/s41577-023-00837-1
58. Klesney-Tait J, Turnbull IR, and Colonna M. The TREM receptor family and signal integration. Nat Immunol. (2006) 7:1266–73. doi: 10.1038/ni1411
59. Kober DL, Alexander-Brett JM, Karch CM, Cruchaga C, Colonna M, Holtzman MJ, et al. Neurodegenerative disease mutations in TREM2 reveal a functional surface and distinct loss-of-function mechanisms. Elife. (2016) 5:e20391. doi: 10.7554/eLife.20391
60. Pelham CJ, Pandya AN, and Agrawal DK. Triggering receptor expressed on myeloid cells receptor family modulators: a patent review. Expert Opin Ther Pat. (2014) 24:1383–95. doi: 10.1517/13543776.2014.977865
61. Wunderlich P, Glebov K, Kemmerling N, Tien NT, Neumann H, and Walter J. Sequential proteolytic processing of the triggering receptor expressed on myeloid cells-2 (TREM2) protein by ectodomain shedding and γ-secretase-dependent intramembranous cleavage. J Biol Chem. (2013) 288:33027–36. doi: 10.1074/jbc.M113.517540
62. Jin SC, Benitez BA, Karch CM, Cooper B, Skorupa T, Carrell D, et al. Coding variants in TREM2 increase risk for Alzheimer’s disease. Hum Mol Genet. (2014) 23:5838–46. doi: 10.1093/hmg/ddu277
63. Colonna M and Facchetti F. TREM-1 (Triggering receptor expressed on myeloid cells): A new player in acute inflammatory responses. J Infect Dis. (2003) 187:S397–401. doi: 10.1086/374754
64. Barrow AD, Astoul E, Floto A, Brooke G, Relou IAM, Jennings NS, et al. Cutting Edge: TREM-Like Transcript-1, a Platelet Immunoreceptor Tyrosine-Based Inhibition Motif Encoding Costimulatory Immunoreceptor that Enhances, Rather than Inhibits, Calcium Signaling via SHP-2. J Immunol. (2004) 172:5838–42. doi: 10.4049/jimmunol.172.10.5838
65. Chen LC, Laskin JD, Gordon MK, and Laskin DL. Regulation of TREM expression in hepatic macrophages and endothelial cells during acute endotoxemia. Exp Mol Pathol. (2008) 84:145–55. doi: 10.1016/j.yexmp.2007.11.004
66. Schmaußer B, Endrich S, Beier D, Moran AP, Burek CJ, Rosenwald A, et al. Triggering receptor expressed on myeloid cells-1 (TREM-1) expression on gastric epithelium: implication for a role of TREM-1 in Helicobacter pylori infection. Clin Exp Immunol. (2008) 152:88–94. doi: 10.1111/j.1365-2249.2008.03608.x
67. Tintinger GR, Merwe JJ, Fickl H, Rheeder P, Feldman C, and Anderson R. Soluble triggering receptor expressed on myeloid cells in sputum of patients with community-acquired pneumonia or pulmonary tuberculosis: a pilot study. Eur J Clin Microbiol Infect Dis. (2012) 31:73–6. doi: 10.1007/s10096-011-1278-y
68. Liao R, Liu Z, Wei S, Xu F, Chen Z, and Gong J. Triggering receptor in myeloid cells (TREM-1) specific expression in peripheral blood mononuclear cells of sepsis patients with acute cholangitis. Inflammation. (2009) 32:182–90. doi: 10.1007/s10753-009-9119-1
69. Tammaro A, Stroo I, Rampanelli E, Blank F, Butter LM, Claessen N, et al. Role of TREM1-DAP12 in renal inflammation during obstructive nephropathy. PloS One. (2013) 8:e82498. doi: 10.1371/journal.pone.0082498
70. Campanholle G, Mittelsteadt K, Nakagawa S, Kobayashi A, Lin S-L, Gharib SA, et al. TLR-2/TLR-4 TREM-1 signaling pathway is dispensable in inflammatory myeloid cells during sterile kidney injury. PloS One. (2013) 8:e68640. doi: 10.1371/journal.pone.0068640
71. Buckland KF, Ramaprakash H, Murray LA, Carpenter KJ, Choi ES, Kunkel SL, et al. Triggering Receptor Expressed on Myeloid cells-1 (TREM-1) Modulates Immune Responses to Aspergillus fumigatus During Fungal Asthma in Mice. Immunol Invest. (2011) 40:692–722. doi: 10.3109/08820139.2011.578270
72. Joffre J, Potteaux S, Zeboudj L, Loyer X, Boufenzer A, Laurans L, et al. Genetic and pharmacological inhibition of TREM-1 limits the development of experimental atherosclerosis. J Am Coll Cardiol. (2016) 68:2776–93. doi: 10.1016/j.jacc.2016.10.015
73. Jérémie L, Amir B, Marc D, and Sébastien G. The Triggering Receptor Expressed on Myeloid cells-1: A new player during acute myocardial infarction. Pharmacol Res. (2015) 100:261–5. doi: 10.1016/j.phrs.2015.07.027
74. Golovkin AS, Ponasenko AV, Khutornaya MV, Kutikhin AG, Salakhov RR, Yuzhalin AE, et al. Association of TLR and TREM-1 gene polymorphisms with risk of coronary artery disease in a Russian population. Gene. (2014) 550:101–9. doi: 10.1016/j.gene.2014.08.022
75. Tao F, Peng L, Li J, Shao Y, Deng L, and Yao H. Association of serum myeloid cells of soluble triggering receptor-1 level with myocardial dysfunction in patients with severe sepsis. Mediators Inflammation. (2013) 2013:1–6. doi: 10.1155/2013/819246
76. Golovkin AS, Ponasenko AV, Yuzhalin AE, Salakhov RR, Khutornaya MV, Kutikhin AG, et al. An association between single nucleotide polymorphisms within TLR and TREM-1 genes and infective endocarditis. Cytokine. (2015) 71:16–21. doi: 10.1016/j.cyto.2014.08.001
77. Feng C-W, Chen N-F, Sung C-S, Kuo H-M, Yang S-N, Chen C-L, et al. Therapeutic effect of modulating TREM-1 via anti-inflammation and autophagy in parkinson’s disease. Front Neurosci. (2019) 13:769. doi: 10.3389/fnins.2019.00769
78. Yuan Z, Syed M, Panchal D, Joo M, Bedi C, Lim S, et al. TREM-1-accentuated lung injury via miR-155 is inhibited by LP17 nanomedicine. Am J Physiol Lung Cell Mol Physiol. (2016) 310:L426–38. doi: 10.1152/ajplung.00195.2015
79. Yuan Z, Syed MA, Panchal D, Joo M, Colonna M, Brantly M, et al. Triggering receptor expressed on myeloid cells 1 (TREM-1)-mediated bcl-2 induction prolongs macrophage survival. J Biol Chem. (2014) 289:15118–29. doi: 10.1074/jbc.M113.536490
80. Dower K, Ellis DK, Saraf K, Jelinsky SA, and Lin L-L. Innate immune responses to TREM-1 activation: overlap, divergence, and positive and negative cross-talk with bacterial lipopolysaccharide. J Immunol. (2008) 180:3520–34. doi: 10.4049/jimmunol.180.5.3520
81. Radsak MP, Salih HR, Rammensee H-G, and Schild H. Triggering receptor expressed on myeloid cells-1 in neutrophil inflammatory responses: differential regulation of activation and survival. J Immunol. (2004) 172:4956–63. doi: 10.4049/jimmunol.172.8.4956
82. Read CB, Kuijper JL, Hjorth SA, Heipel MD, Tang X, Fleetwood AJ, et al. Cutting edge: identification of neutrophil PGLYRP1 as a ligand for TREM-1. J Immunol. (2015) 194:1417–21. doi: 10.4049/jimmunol.1402303
83. Sharapova TN, Ivanova OK, Romanova EA, Sashchenko LP, and Yashin DV. N-terminal peptide of PGLYRP1/tag7 is a novel ligand for TREM-1 receptor. Int J Mol Sci. (2022) 23:5752. doi: 10.3390/ijms23105752
84. Haselmayer P, Grosse-Hovest L, von Landenberg P, Schild H, and Radsak MP. TREM-1 ligand expression on platelets enhances neutrophil activation. Blood. (2007) 110:1029–35. doi: 10.1182/blood-2007-01-069195
85. Fu L, Han L, Xie C, Li W, Lin L, Pan S, et al. Identification of extracellular actin as a ligand for triggering receptor expressed on myeloid cells-1 signaling. Front Immunol. (2017) 8:917. doi: 10.3389/fimmu.2017.00917
86. Magna M and Pisetsky DS. The role of HMGB1 in the pathogenesis of inflammatory and autoimmune diseases. Mol Med. (2014) 20:138–46. doi: 10.2119/molmed.2013.00164
87. Tammaro A, Derive M, Gibot S, Leemans JC, Florquin S, and Dessing MC. TREM-1 and its potential ligands in non-infectious diseases: from biology to clinical perspectives. Pharmacol Ther. (2017) 177:81–95. doi: 10.1016/j.pharmthera.2017.02.043
88. Sharapova TN, Romanova EA, Ivanova OK, Yashin DV, and Sashchenko LP. Hsp70 interacts with the TREM-1 receptor expressed on monocytes and thereby stimulates generation of cytotoxic lymphocytes active against MHC-negative tumor cells. Int J Mol Sci. (2021) 22:6889. doi: 10.3390/ijms22136889
89. Wu J, Li J, Salcedo R, Mivechi NF, Trinchieri G, and Horuzsko A. The proinflammatory myeloid cell receptor TREM-1 controls kupffer cell activation and development of hepatocellular carcinoma. Cancer Res. (2012) 72:3977–86. doi: 10.1158/0008-5472.CAN-12-0938
90. Denning N-L, Aziz M, Murao A, Gurien SD, Ochani M, Prince JM, et al. Extracellular CIRP as an endogenous TREM-1 ligand to fuel inflammation in sepsis. JCI Insight. (2020) 5:e134172. doi: 10.1172/jci.insight.134172
91. Gibot S, Buonsanti C, Massin F, Romano M, Kolopp-Sarda M-N, Benigni F, et al. Modulation of the triggering receptor expressed on the myeloid cell type 1 pathway in murine septic shock. Infect Immun. (2006) 74:2823–30. doi: 10.1128/IAI.74.5.2823-2830.2006
92. Zysset D, Weber B, Rihs S, Brasseit J, Freigang S, Riether C, et al. TREM-1 links dyslipidemia to inflammation and lipid deposition in atherosclerosis. Nat Commun. (2016) 7:13151. doi: 10.1038/ncomms13151
93. Bouchon A, Dietrich J, and Colonna M. Cutting edge: inflammatory responses can be triggered by TREM-1, a novel receptor expressed on neutrophils and monocytes. J Immunol. (2000) 164:4991–5. doi: 10.4049/jimmunol.164.10.4991
94. Tessarz AS and Cerwenka A. The TREM-1/DAP12 pathway. Immunol Lett. (2008) 116:111–6. doi: 10.1016/j.imlet.2007.11.021
95. Mecca C, Giambanco I, Donato R, and Arcuri C. Microglia and aging: the role of the TREM2–DAP12 and CX3CL1-CX3CR1 axes. Int J Mol Sci. (2018) 19:318. doi: 10.3390/ijms19010318
96. Zhong L, Zhang Z-L, Li X, Liao C, Mou P, Wang T, et al. TREM2/DAP12 complex regulates inflammatory responses in microglia via the JNK signaling pathway. Front Aging Neurosci. (2017) 9:204. doi: 10.3389/fnagi.2017.00204
97. Bianchin MM, Martin KC, de Souza AC, de Oliveira MA, and de Mello Rieder CR. Nasu–Hakola disease and primary microglial dysfunction. Nat Rev Neurol. (2010) 6:523–3. doi: 10.1038/nrneurol.2010.17-c1
98. Borroni B, Ferrari F, Galimberti D, Nacmias B, Barone C, Bagnoli S, et al. Heterozygous TREM2 mutations in frontotemporal dementia. Neurobiol Aging. (2014) 35:934. doi: 10.1016/j.neurobiolaging.2013.09.017
99. Guerreiro RJ, Lohmann E, Brás JM, Gibbs JR, Rohrer JD, Gurunlian N, et al. Using exome sequencing to reveal mutations in TREM2 presenting as a frontotemporal dementia–like syndrome without bone involvement. JAMA Neurol. (2013) 70:78. doi: 10.1001/jamaneurol.2013.579
100. Piccio L, Buonsanti C, Cella M, Tassi I, Schmidt RE, Fenoglio C, et al. Identification of soluble TREM-2 in the cerebrospinal fluid and its association with multiple sclerosis and CNS inflammation. Brain. (2008) 131:3081–91. doi: 10.1093/brain/awn217
101. Zheng H, Cheng B, Li Y, Li X, Chen X, and Zhang Y. TREM2 in alzheimer’s disease: microglial survival and energy metabolism. Front Aging Neurosci. (2018) 10:395. doi: 10.3389/fnagi.2018.00395
102. Fancy N, Willumsen N, Tsartsalis S, Khozoie C, McGarry A, Muirhead RC, et al. Mechanisms contributing to differential genetic risks for TREM2 R47H and R62H variants in Alzheimer’s Disease. Preprint at medRxiv. (2022). doi: 10.1101/2022.07.12.22277509
103. Wang Y, Cella M, Mallinson K, Ulrich JD, Young KL, Robinette ML, et al. TREM2 lipid sensing sustains the microglial response in an alzheimer’s disease model. Cell. (2015) 160:1061–71. doi: 10.1016/j.cell.2015.01.049
104. Takahashi K, Rochford CDP, and Neumann H. Clearance of apoptotic neurons without inflammation by microglial triggering receptor expressed on myeloid cells-2. J Exp Med. (2005) 201:647–57. doi: 10.1084/jem.20041611
105. Perugorria MJ, Esparza-Baquer A, Oakley F, Labiano I, Korosec A, Jais A, et al. Non-parenchymal TREM-2 protects the liver from immune-mediated hepatocellular damage. Gut. (2019) 68:533–46. doi: 10.1136/gutjnl-2017-314107
106. Park M, Yi J-W, Kim E-M, Yoon I-J, Lee E-H, Lee H-Y, et al. Triggering receptor expressed on myeloid cells 2 (TREM2) promotes adipogenesis and diet-induced obesity. Diabetes. (2015) 64:117–27. doi: 10.2337/db13-1869
107. Ulland TK, Song WM, Huang SC-C, Ulrich JD, Sergushichev A, Beatty WL, et al. TREM2 maintains microglial metabolic fitness in alzheimer’s disease. Cell. (2017) 170:649–663.e13. doi: 10.1016/j.cell.2017.07.023
108. Jaitin DA, Adlung L, Thaiss CA, Weiner A, Li B, Descamps H, et al. Lipid-associated macrophages control metabolic homeostasis in a trem2-dependent manner. Cell. (2019) 178:686–698.e14. doi: 10.1016/j.cell.2019.05.054
109. Götzl JK, Brendel M, Werner G, Parhizkar S, Sebastian Monasor L, Kleinberger G, et al. Opposite microglial activation stages upon loss of PGRN or TREM 2 result in reduced cerebral glucose metabolism. EMBO Mol Med. (2019) 11:e9711. doi: 10.15252/emmm.201809711
110. Daws MR, Sullam PM, Niemi EC, Chen TT, Tchao NK, and Seaman WE. Pattern recognition by TREM-2: binding of anionic ligands. J Immunol. (2003) 171:594–9. doi: 10.4049/jimmunol.171.2.594
111. Atagi Y, Liu C-C, Painter MM, Chen X-F, Verbeeck C, Zheng H, et al. Apolipoprotein E is a ligand for triggering receptor expressed on myeloid cells 2 (TREM2). J Biol Chem. (2015) 290:26043–50. doi: 10.1074/jbc.M115.679043
112. Charles JF, Humphrey MB, Zhao X, Quarles E, Nakamura MC, Aderem A, et al. The innate immune response to salmonella enterica serovar typhimurium by macrophages is dependent on TREM2-DAP12. Infect Immun. (2008) 76:2439–47. doi: 10.1128/IAI.00115-08
113. Quan DN, Cooper MD, Potter JL, Roberts MH, Cheng H, and Jarvis GA. TREM-2 binds to lipooligosaccharides of Neisseria gonorrhoeae and is expressed on reproductive tract epithelial cells. Mucosal Immunol. (2008) 1:229–38. doi: 10.1038/mi.2008.1
114. Lessard CB, Malnik SL, Zhou Y, Ladd TB, Cruz PE, Ran Y, et al. High-affinity interactions and signal transduction between Aβ oligomers and TREM 2. EMBO Mol Med. (2018) 10:e9027. doi: 10.15252/emmm.201809027
115. Kawabori M, Kacimi R, Kauppinen T, Calosing C, Kim JY, Hsieh CL, et al. Triggering receptor expressed on myeloid cells 2 (TREM2) deficiency attenuates phagocytic activities of microglia and exacerbates ischemic damage in experimental stroke. J Neurosci. (2015) 35:3384–96. doi: 10.1523/JNEUROSCI.2620-14.2015
116. Shirotani K, Hori Y, Yoshizaki R, Higuchi E, Colonna M, Saito T, et al. Aminophospholipids are signal-transducing TREM2 ligands on apoptotic cells. Sci Rep. (2019) 9:7508. doi: 10.1038/s41598-019-43535-6
117. Gallo C, Manzo E, Barra G, Fioretto L, Ziaco M, Nuzzo G, et al. Sulfavant A as the first synthetic TREM2 ligand discloses a homeostatic response of dendritic cells after receptor engagement. Cell Mol Life Sci. (2022) 79:369. doi: 10.1007/s00018-022-04297-z
118. Wang H, Liu D, and Zhou X. Effect of mycolic acids on host immunity and lipid metabolism. Int J Mol Sci. (2023) 25:396. doi: 10.3390/ijms25010396
119. Iizasa E, Chuma Y, Uematsu T, Kubota M, Kawaguchi H, Umemura M, et al. TREM2 is a receptor for non-glycosylated mycolic acids of mycobacteria that limits anti-mycobacterial macrophage activation. Nat Commun. (2021) 12:2299. doi: 10.1038/s41467-021-22620-3
120. Xue T, Ji J, Sun Y, Huang X, Cai Z, Yang J, et al. Sphingosine-1-phosphate, a novel TREM2 ligand, promotes microglial phagocytosis to protect against ischemic brain injury. Acta Pharm Sin B. (2022) 12:1885–98. doi: 10.1016/j.apsb.2021.10.012
121. Manzo E, Cutignano A, Pagano D, Gallo C, Barra G, Nuzzo G, et al. A new marine-derived sulfoglycolipid triggers dendritic cell activation and immune adjuvant response. Sci Rep. (2017) 7:6286. doi: 10.1038/s41598-017-05969-8
122. Manzo E, Fioretto L, Gallo C, Ziaco M, Nuzzo G, D’Ippolito G, et al. Preparation, supramolecular aggregation and immunological activity of the bona fide vaccine adjuvant sulfavant S. Mar Drugs. (2020) 18:451. doi: 10.3390/md18090451
123. Manzo E, Fioretto L, Pagano D, Nuzzo G, Gallo C, De Palma R, et al. Chemical synthesis of marine-derived sulfoglycolipids, a new class of molecular adjuvants. Mar Drugs. (2017) 15:288. doi: 10.3390/md15090288
124. Fioretto L, Ziaco M, Gallo C, Nuzzo G, d’Ippolito G, Lupetti P, et al. Direct evidence of the impact of aqueous self-assembly on biological behavior of amphiphilic molecules: The case study of molecular immunomodulators Sulfavants. J Colloid Interface Sci. (2022) 611:129–36. doi: 10.1016/j.jcis.2021.12.054
125. Ziaco M, Fioretto L, Nuzzo G, Fontana A, and Manzo E. Short gram-scale synthesis of sulfavant A. Org Process Res Dev. (2020) 24:2728–33. doi: 10.1021/acs.oprd.0c00393
126. Barra G, Gallo C, Carbone D, Ziaco M, Dell’Isola M, Affuso M, et al. The immunoregulatory effect of the TREM2-agonist Sulfavant A in human allogeneic mixed lymphocyte reaction. Front Immunol. (2023) 14:1050113. doi: 10.3389/fimmu.2023.1050113
127. Li L, He Y-L, Xu N, Wang X-F, Song B, Tang B-Q, et al. A natural small molecule aspidosperma-type alkaloid, hecubine, as a new TREM2 activator for alleviating lipopolysaccharide-induced neuroinflammation. Vitro vivo. Redox Biol. (2024) 70:103057. doi: 10.1016/j.redox.2024.103057
128. Li L, Xu N, He Y, Tang M, Yang B, Du J, et al. Dehydroervatamine as a promising novel TREM2 agonist, attenuates neuroinflammation. Neurotherapeutics. (2025) 22:e00479. doi: 10.1016/j.neurot.2024.e00479
129. Xu M, Yang Y, Peng J, Zhang Y, Wu B, He B, et al. Effects of Alpinae Oxyphyllae Fructus on microglial polarization in a LPS-induced BV2 cells model of neuroinflammation via TREM2. J Ethnopharmacol. (2023) 302:115914. doi: 10.1016/j.jep.2022.115914
130. Konishi H and Kiyama H. Microglial TREM2/DAP12 signaling: A double-edged sword in neural diseases. Front Cell Neurosci. (2018) 12:206. doi: 10.3389/fncel.2018.00206
131. Chung D-H, Seaman WE, and Daws MR. Characterization of TREM-3, an activating receptor on mouse macrophages: definition of a family of single Ig domain receptors on mouse chromosome 17. Eur J Immunol. (2002) 32:59–66. doi: 10.1002/1521-4141(200201)32:1<59::AID-IMMU59>3.0.CO;2-U
132. Hommes TJ, Dessing MC, van ‘t Veer C, Florquin S, Colonna M, de Vos AF, et al. Role of triggering receptor expressed on myeloid cells-1/3 in klebsiella -derived pneumosepsis. Am J Respir Cell Mol Biol. (2015) 53:647–55. doi: 10.1165/rcmb.2014-0485OC
133. Allcock RJN, Barrow AD, Forbes S, Beck S, and Trowsdale J. The human TREM gene cluster at 6p21.1 encodes both activating and inhibitory single IgV domain receptors and includes NKp44. Eur J Immunol. (2003) 33:567–77. doi: 10.1002/immu.200310033
134. Carrasquillo MM, Allen M, Burgess JD, Wang X, Strickland SL, Aryal S, et al. A candidate regulatory variant at the TREM gene cluster associates with decreased Alzheimer’s disease risk and increased TREML1 and TREM2 brain gene expression. Alzheimers Dement. (2017) 13:663–73. doi: 10.1016/j.jalz.2016.10.005
135. Kang SS, Kurti A, Baker KE, Liu C-C, Colonna M, Ulrich JD, et al. Behavioral and transcriptomic analysis of Trem2-null mice: not all knockout mice are created equal. Hum Mol Genet. (2018) 27:211–23. doi: 10.1093/hmg/ddx366
136. Yoon S-H, Lee YD, Ha J, Lee Y, and Kim H-H. TLT-1s, alternative transcripts of triggering receptor expressed on myeloid cell-like transcript-1 (TLT-1), inhibits the triggering receptor expressed on myeloid cell-2 (TREM-2)-mediated signaling pathway during osteoclastogenesis. J Biol Chem. (2012) 287:29620–6. doi: 10.1074/jbc.M112.351239
137. Barclay AN and Brown MH. The SIRP family of receptors and immune regulation. Nat Rev Immunol. (2006) 6:457–64. doi: 10.1038/nri1859
138. Kharitonenkov A, Chen Z, Sures I, Wang H, Schilling J, and Ullrich A. A family of proteins that inhibit signalling through tyrosine kinase receptors. Nature. (1997) 386:181–6. doi: 10.1038/386181a0
139. Gaikwad S, Larionov S, Wang Y, Dannenberg H, Matozaki T, Monsonego A, et al. Signal regulatory protein-β1. Am J Pathol. (2009) 175:2528–39. doi: 10.2353/ajpath.2009.090147
140. Hayashi A, Ohnishi H, Okazawa H, Nakazawa S, Ikeda H, Motegi S, et al. Positive regulation of phagocytosis by SIRPβ and its signaling mechanism in macrophages. J Biol Chem. (2004) 279:29450–60. doi: 10.1074/jbc.M400950200
141. Vernon-Wilson EF, Kee W-J, Willis AC, Barclay AN, Simmons DL, and Brown MH. CD47 is a ligand for rat macrophage membrane signal regulatory protein SIRP (OX41) and human SIRPα 1. Eur J Immunol. (2000) 30:2130–7. doi: 10.1002/1521-4141(2000)30:8<2130::AID-IMMU2130>3.0.CO;2-8
142. Brooke G, Holbrook JD, Brown MH, and Barclay AN. Human lymphocytes interact directly with CD47 through a novel member of the signal regulatory protein (SIRP) family. J Immunol. (2004) 173:2562–70. doi: 10.4049/jimmunol.173.4.2562
143. Miller TW, Amason JD, Garcin ED, Lamy L, Dranchak PK, Macarthur R, et al. Quantitative high-throughput screening assays for the discovery and development of SIRPα-CD47 interaction inhibitors. PloS One. (2019) 14:e0218897. doi: 10.1371/journal.pone.0218897
144. Gardai SJ, Xiao Y-Q, Dickinson M, Nick JA, Voelker DR, Greene KE, et al. By binding SIRPα or calreticulin/CD91, lung collectins act as dual function surveillance molecules to suppress or enhance inflammation. Cell. (2003) 115:13–23. doi: 10.1016/S0092-8674(03)00758-X
145. Liu Y, Soto I, Tong Q, Chin A, Bühring H-J, Wu T, et al. SIRPβ1 is expressed as a disulfide-linked homodimer in leukocytes and positively regulates neutrophil transepithelial migration. J Biol Chem. (2005) 280:36132–40. doi: 10.1074/jbc.M506419200
146. Varki A. Glycan-based interactions involving vertebrate sialic-acid-recognizing proteins. Nature. (2007) 446:1023–9. doi: 10.1038/nature05816
147. Varki A. Sialic acids in human health and disease. Trends Mol Med. (2008) 14:351–60. doi: 10.1016/j.molmed.2008.06.002
148. Lehmann F. Evolution of sialic acid-binding proteins: molecular cloning and expression of fish siglec-4. Glycobiology. (2004) 14:959–68. doi: 10.1093/glycob/cwh120
149. Angata T, Margulies EH, Green ED, and Varki A. Large-scale sequencing of the CD33-related Siglec gene cluster in five mammalian species reveals rapid evolution by multiple mechanisms. PNAS. (2004) 101:13251–6. doi: 10.1073/pnas.0404833101
150. Macauley MS, Crocker PR, and Paulson JC. Siglec-mediated regulation of immune cell function in disease. Nat Rev Immunol. (2014) 14:653–66. doi: 10.1038/nri3737
151. Crocker PR, Paulson JC, and Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. (2007) 7:255–66. doi: 10.1038/nri2056
152. Kimura N, Ohmori K, Miyazaki K, Izawa M, Matsuzaki Y, Yasuda Y, et al. Human B-lymphocytes express α2-6-sialylated 6-sulfo-N-acetyllactosamine serving as a preferred ligand for CD22/siglec-2. J Biol Chem. (2007) 282:32200–7. doi: 10.1074/jbc.M702341200
153. Hollingworth P, Harold D, Sims R, Gerrish A, Lambert J-C, Carrasquillo MM, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. (2011) 43:429–35. doi: 10.1038/ng.803
154. Griciuc A, Serrano-Pozo A, Parrado AR, Lesinski AN, Asselin CN, Mullin K, et al. Alzheimer’s disease risk gene CD33 inhibits microglial uptake of amyloid beta. Neuron. (2013) 78:631–43. doi: 10.1016/j.neuron.2013.04.014
155. Raj T, Ryan KJ, Replogle JM, Chibnik LB, Rosenkrantz L, Tang A, et al. CD33: increased inclusion of exon 2 implicates the Ig V-set domain in Alzheimer’s disease susceptibility. Hum Mol Genet. (2014) 23:2729–36. doi: 10.1093/hmg/ddt666
156. Lee CYD, Daggett A, Gu X, Jiang L-L, Langfelder P, Li X, et al. Elevated TREM2 gene dosage reprograms microglia responsivity and ameliorates pathological phenotypes in alzheimer’s disease models. Neuron. (2018) 97:1032–1048.e5. doi: 10.1016/j.neuron.2018.02.002
157. Song WM, Joshita S, Zhou Y, Ulland TK, Gilfillan S, and Colonna M. Humanized TREM2 mice reveal microglia-intrinsic and -extrinsic effects of R47H polymorphism. J Exp Med. (2018) 215:745–60. doi: 10.1084/jem.20171529
158. Griciuc A, Patel S, Federico AN, Choi SH, Innes BJ, Oram MK, et al. TREM2 acts downstream of CD33 in modulating microglial pathology in alzheimer’s disease. Neuron. (2019) 103:820–835.e7. doi: 10.1016/j.neuron.2019.06.010
159. Teter B, Morihara T, Lim GP, Chu T, Jones MR, Zuo X, et al. Curcumin restores innate immune Alzheimer’s disease risk gene expression to ameliorate Alzheimer pathogenesis. Neurobiol Dis. (2019) 127:432–48. doi: 10.1016/j.nbd.2019.02.015
160. Miles LA, Hermans SJ, Crespi GAN, Gooi JH, Doughty L, Nero TL, et al. Small molecule binding to alzheimer risk factor CD33 promotes Aβ Phagocytosis. iScience. (2019) 19:110–8. doi: 10.1016/j.isci.2019.07.023
161. Lübbers J, Rodríguez E, and van Kooyk Y. Modulation of immune tolerance via siglec-sialic acid interactions. Front Immunol. (2018) 9:2807. doi: 10.3389/fimmu.2018.02807
162. Ali SR, Fong JJ, Carlin AF, Busch TD, Linden R, Angata T, et al. Siglec-5 and Siglec-14 are polymorphic paired receptors that modulate neutrophil and amnion signaling responses to group B Streptococcus. J Exp Med. (2014) 211:1231–42. doi: 10.1084/jem.20131853
163. Varki A. Letter to the Glyco-Forum: Since there are PAMPs and DAMPs, there must be SAMPs? Glycan “self-associated molecular patterns” dampen innate immunity, but pathogens can mimic them. Glycobiology. (2011) 21:1121–4. doi: 10.1093/glycob/cwr087
164. Delputte PL and Nauwynck HJ. Porcine arterivirus infection of alveolar macrophages is mediated by sialic acid on the virus. J Virol. (2004) 78:8094–101. doi: 10.1128/JVI.78.15.8094-8101.2004
165. Izquierdo-Useros N, Lorizate M, Puertas MC, Rodriguez-Plata MT, Zangger N, Erikson E, et al. Siglec-1 is a novel dendritic cell receptor that mediates HIV-1 trans-infection through recognition of viral membrane gangliosides. PloS Biol. (2012) 10:e1001448. doi: 10.1371/journal.pbio.1001448
166. Rodrigues E and Macauley M. Hypersialylation in cancer: modulation of inflammation and therapeutic opportunities. Cancers (Basel). (2018) 10:207. doi: 10.3390/cancers10060207
167. Zelensky AN and Gready JE. The C-type lectin-like domain superfamily. FEBS J. (2005) 272:6179–217. doi: 10.1111/j.1742-4658.2005.05031.x
168. Geijtenbeek TBH, Torensma R, van Vliet SJ, van Duijnhoven GCF, Adema GJ, van Kooyk Y, et al. Identification of DC-SIGN, a novel dendritic cell–specific ICAM-3 receptor that supports primary immune responses. Cell. (2000) 100:575–85. doi: 10.1016/S0092-8674(00)80693-5
169. Herrera M, Herrera A, Domínguez G, Silva J, García V, García JM, et al. Cancer-associated fibroblast and M 2 macrophage markers together predict outcome in colorectal cancer patients. Cancer Sci. (2013) 104:437–44. doi: 10.1111/cas.12096
170. Jiang Y, Zhang C, Chen K, Chen Z, Sun Z, Zhang Z, et al. The clinical significance of DC-SIGN and DC-SIGNR, which are novel markers expressed in human colon cancer. PloS One. (2014) 9:e114748. doi: 10.1371/journal.pone.0114748
171. Caparroís E, Munoz P, Sierra-Filardi E, Serrano-Goímez D, Puig-Kroüger A, Rodriíguez-Fernaíndez JL, et al. DC-SIGN ligation on dendritic cells results in ERK and PI3K activation and modulates cytokine production. Blood. (2006) 107:3950–8. doi: 10.1182/blood-2005-03-1252
172. Geijtenbeek TBH and van Kooyk Y. DC-SIGN: A novel HIV receptor on DCs that mediates HIV-1 transmission. Curr Top Microbiol Immunol. (2003) 276:31–54. doi: 10.1007/978-3-662-06508-2_2
173. Halary F, Amara A, Lortat-Jacob H, Messerle M, Delaunay T, Houlès C, et al. Human cytomegalovirus binding to DC-SIGN is required for dendritic cell infection and target cell trans-infection. Immunity. (2002) 17:653–64. doi: 10.1016/S1074-7613(02)00447-8
174. Alvarez CP, Lasala F, Carrillo J, Munãiz O, Corbií AL, and Delgado R. C-Type Lectins DC-SIGN and L-SIGN Mediate Cellular Entry by Ebola Virus in cis and in trans. J Virol. (2002) 76:6841–4. doi: 10.1128/JVI.76.13.6841-6844.2002
175. Tassaneetrithep B, Burgess TH, Granelli-Piperno A, Trumpfheller C, Finke J, Sun W, et al. DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. J Exp Med. (2003) 197:823–9. doi: 10.1084/jem.20021840
176. Lozach P-Y, Lortat-Jacob H, De Lacroix De Lavalette A, Staropoli I, Foung S, Amara A, et al. DC-SIGN and L-SIGN are high affinity binding receptors for hepatitis C virus glycoprotein E2. J Biol Chem. (2003) 278:20358–66. doi: 10.1074/jbc.M301284200
177. Poühlmann S, Zhang J, Baribaud F, Chen Z, Leslie GJ, Lin G, et al. Hepatitis C virus glycoproteins interact with DC-SIGN and DC-SIGNR. J Virol. (2003) 77:4070–80. doi: 10.1128/JVI.77.7.4070-4080.2003
178. Colmenares M, Puig-Kröger A, Pello OM, Corbıí AL, and Rivas L. Dendritic cell (DC)-specific intercellular adhesion molecule 3 (ICAM-3)-grabbing nonintegrin (DC-SIGN, CD209), a C-type surface lectin in human DCs, is a receptor for leishmaniaAmastigotes. J Biol Chem. (2002) 277:36766–9. doi: 10.1074/jbc.M205270200
179. Cambi A, Gijzen K, de Vries IJM, Torensma R, Joosten B, Adema GJ, et al. The C-type lectin DC-SIGN (CD209) is an antigen-uptake receptor for Candida albicans on dendritic cells. Eur J Immunol. (2003) 33:532–8. doi: 10.1002/immu.200310029
180. Geijtenbeek TBH, van Vliet SJ, Koppel EA, Sanchez-Hernandez M, Vandenbroucke-Grauls CMJE, Appelmelk B, et al. Mycobacteria target DC-SIGN to suppress dendritic cell function. J Exp Med. (2003) 197:7–17. doi: 10.1084/jem.20021229
181. Maeda N, Nigou J, Herrmann J-L, Jackson M, Amara A, Lagrange PH, et al. The Cell Surface Receptor DC-SIGN Discriminates betweenMycobacterium Species through Selective Recognition of the Mannose Caps on Lipoarabinomannan. J Biol Chem. (2003) 278:5513–6. doi: 10.1074/jbc.C200586200
182. Tailleux L, Schwartz O, Herrmann J-L, Pivert E, Jackson M, Amara A, et al. DC-SIGN is the major mycobacterium tuberculosis receptor on human dendritic cells. J Exp Med. (2003) 197:121–7. doi: 10.1084/jem.20021468
183. Meyer S, van Liempt E, Imberty A, van Kooyk Y, Geyer H, Geyer R, et al. DC-SIGN mediates binding of dendritic cells to authentic pseudo-lewisY glycolipids of schistosoma mansoni cercariae, the first parasite-specific ligand of DC-SIGN. J Biol Chem. (2005) 280:37349–59. doi: 10.1074/jbc.M507100200
184. Chiodo F, Marradi M, Park J, Ram AFJ, Penadés S, van Die I, et al. Galactofuranose-coated gold nanoparticles elicit a pro-inflammatory response in human monocyte-derived dendritic cells and are recognized by DC-SIGN. ACS Chem Biol. (2014) 9:383–9. doi: 10.1021/cb4008265
185. Cambi A, de Lange F, van Maarseveen NM, Nijhuis M, Joosten B, van Dijk EMHP, et al. Microdomains of the C-type lectin DC-SIGN are portals for virus entry into dendritic cells. J Cell Biol. (2004) 164:145–55. doi: 10.1083/jcb.200306112
186. Percherancier Y, Lagane B, Planchenault T, Staropoli I, Altmeyer R, Virelizier J-L, et al. HIV-1 entry into T-cells is not dependent on CD4 and CCR5 localization to sphingolipid-enriched, detergent-resistant, raft membrane domains. J Biol Chem. (2003) 278:3153–61. doi: 10.1074/jbc.M207371200
187. van Stijn CMW, Meyer S, van den Broek M, Bruijns SCM, van Kooyk Y, Geyer R, et al. Schistosoma mansoni worm glycolipids induce an inflammatory phenotype in human dendritic cells by cooperation of TLR4 and DC-SIGN. Mol Immunol. (2010) 47:1544–52. doi: 10.1016/j.molimm.2010.01.014
188. van Kooyk Y and Geijtenbeek TBH. DC-SIGN: escape mechanism for pathogens. Nat Rev Immunol. (2003) 3:697–709. doi: 10.1038/nri1182
189. Gringhuis SI, den Dunnen J, Litjens M, van het Hof B, van Kooyk Y, and Geijtenbeek TBH. C-type lectin DC-SIGN modulates toll-like receptor signaling via raf-1 kinase-dependent acetylation of transcription factor NF-κB. Immunity. (2007) 26:605–16. doi: 10.1016/j.immuni.2007.03.012
190. Berzi A, Ordanini S, Joosten B, Trabattoni D, Cambi A, Bernardi A, et al. Pseudo-mannosylated DC-SIGN ligands as immunomodulants. Sci Rep. (2016) 6:35373. doi: 10.1038/srep35373
191. Richardson MB and Williams SJ. MCL and mincle: C-type lectin receptors that sense damaged self and pathogen-associated molecular patterns. Front Immunol. (2014) 5:288. doi: 10.3389/fimmu.2014.00288
192. Hupfer T, Schick J, Jozefowski K, Voehringer D, Ostrop J, and Lang R. Stat6-dependent inhibition of mincle expression in mouse and human antigen-presenting cells by the th2 cytokine IL-4. Front Immunol. (2016) 7:423. doi: 10.3389/fimmu.2016.00423
193. Lorentzen JC, Flornes L, Eklöw C, Bäckdahl L, Ribbhammar U, Guo JP, et al. Association of arthritis with a gene complex encoding C-type lectin–like receptors. Arthritis Rheum. (2007) 56:2620–32. doi: 10.1002/art.22813
194. Kostarnoy AV, Gancheva PG, Lepenies B, Tukhvatulin AI, Dzharullaeva AS, Polyakov NB, et al. Receptor Mincle promotes skin allergies and is capable of recognizing cholesterol sulfate. PNAS. (2017) 114:E2758–65. doi: 10.1073/pnas.1611665114
195. Suzuki Y, Nakano Y, Mishiro K, Takagi T, Tsuruma K, Nakamura M, et al. Involvement of Mincle and Syk in the changes to innate immunity after ischemic stroke. Sci Rep. (2013) 3:3177. doi: 10.1038/srep03177
196. Zhou H, Yu M, Zhao J, Martin BN, Roychowdhury S, McMullen MR, et al. IRAKM-Mincle axis links cell death to inflammation: Pathophysiological implications for chronic alcoholic liver disease. Hepatology. (2016) 64:1978–93. doi: 10.1002/hep.28811
197. te Velde AA. The C-type lectin mincle: clues for a role in crohn’s disease adjuvant reaction. Front Immunol. (2017) 8:1304. doi: 10.3389/fimmu.2017.01304
198. Lee MJ, Jo H, Shin SH, Kim S-M, Kim B, Shim HS, et al. Mincle and STING-stimulating adjuvants elicit robust cellular immunity and drive long-lasting memory responses in a foot-and-mouth disease vaccine. Front Immunol. (2019) 10:2509. doi: 10.3389/fimmu.2019.02509
199. Ishikawa A, Miyake Y, Kobayashi K, Murata Y, Iizasa S, Iizasa E, et al. Essential roles of C-type lectin Mincle in induction of neuropathic pain in mice. Sci Rep. (2019) 9:872. doi: 10.1038/s41598-018-37318-8
200. van Haren SD, Dowling DJ, Foppen W, Christensen D, Andersen P, Reed SG, et al. Age-specific adjuvant synergy: dual TLR7/8 and mincle activation of human newborn dendritic cells enables th1 polarization. J Immunol. (2016) 197:4413–24. doi: 10.4049/jimmunol.1600282
201. Yamasaki S, Ishikawa E, Sakuma M, Hara H, Ogata K, and Saito T. Mincle is an ITAM-coupled activating receptor that senses damaged cells. Nat Immunol. (2008) 9:1179–88. doi: 10.1038/ni.1651
202. Matsumoto M, Tanaka T, Kaisho T, Sanjo H, Copeland NG, Gilbert DJ, et al. A novel LPS-inducible C-type lectin is a transcriptional target of NF-IL6 in macrophages. J Immunol. (1999) 163:5039–48. doi: 10.4049/jimmunol.163.9.5039
203. Ishikawa E, Ishikawa T, Morita YS, Toyonaga K, Yamada H, Takeuchi O, et al. Direct recognition of the mycobacterial glycolipid, trehalose dimycolate, by C-type lectin Mincle. J Exp Med. (2009) 206:2879–88. doi: 10.1084/jem.20091750
204. Schoenen H, Bodendorfer B, Hitchens K, Manzanero S, Werninghaus K, Nimmerjahn F, et al. Cutting edge: mincle is essential for recognition and adjuvanticity of the mycobacterial cord factor and its synthetic analog trehalose-dibehenate. J Immunol. (2010) 184:2756–60. doi: 10.4049/jimmunol.0904013
205. Yamasaki S, Matsumoto M, Takeuchi O, Matsuzawa T, Ishikawa E, Sakuma M, et al. C-type lectin Mincle is an activating receptor for pathogenic fungus. Malassezia. PNAS. (2009) 106:1897–902. doi: 10.1073/pnas.0805177106
206. Ishikawa T, Itoh F, Yoshida S, Saijo S, Matsuzawa T, Gonoi T, et al. Identification of distinct ligands for the C-type lectin receptors mincle and dectin-2 in the pathogenic fungus malassezia. Cell Host Microbe. (2013) 13:477–88. doi: 10.1016/j.chom.2013.03.008
207. Bowker N, Salie M, Schurz H, van Helden PD, Kinnear CJ, Hoal EG, et al. Polymorphisms in the pattern recognition receptor mincle gene (CLEC4E) and association with tuberculosis. Lung. (2016) 194:763–7. doi: 10.1007/s00408-016-9915-y
208. Smith DGM, Hosono Y, Nagata M, Yamasaki S, and Williams SJ. Design of potent Mincle signalling agonists based on an alkyl β-glucoside template. ChemComm. (2020) 56:4292–5. doi: 10.1039/D0CC00670J
209. Xisto MID da S, Dias L dos S, Bezerra FF, Bittencourt VCB, Rollin-Pinheiro R, Cartágenes-Pinto AC, et al. An Alpha-Glucan from Lomentospora prolificans Mediates Fungal–Host Interaction Signaling through Dectin-1 and Mincle. J Fungi. (2023) 9:291. doi: 10.3390/jof9030291
210. Kiyotake R, Oh-hora M, Ishikawa E, Miyamoto T, Ishibashi T, and Yamasaki S. Human mincle binds to cholesterol crystals and triggers innate immune responses. J Biol Chem. (2015) 290:25322–32. doi: 10.1074/jbc.M115.645234
211. Williams SJ. Sensing lipids with mincle: structure and function. Front Immunol. (2017) 8:1662. doi: 10.3389/fimmu.2017.01662
212. Nagata M, Izumi Y, Ishikawa E, Kiyotake R, Doi R, Iwai S, et al. Intracellular metabolite β-glucosylceramide is an endogenous Mincle ligand possessing immunostimulatory activity. PNAS. (2017) 114:E3285–94. doi: 10.1073/pnas.1618133114
213. Lv LL, Tang PM-K, Li CJ, You YK, Li J, Huang X-R, et al. The pattern recognition receptor, Mincle, is essential for maintaining the M1 macrophage phenotype in acute renal inflammation. Kidney Int. (2017) 91:587–602. doi: 10.1016/j.kint.2016.10.020
214. Isakov N. Immunoreceptor tyrosine-based activation motif (ITAM), a unique module linking antigen and Fc receptors to their signaling cascades. J Leukoc Biol. (1997) 61:6–16. doi: 10.1002/jlb.61.1.6
215. Yonekawa A, Saijo S, Hoshino Y, Miyake Y, Ishikawa E, Suzukawa M, et al. Dectin-2 is a direct receptor for mannose-capped lipoarabinomannan of mycobacteria. Immunity. (2014) 41:402–13. doi: 10.1016/j.immuni.2014.08.005
216. Chen J, Zhao Y, Chu X, Lu Y, Wang S, and Yi Q. Dectin-1-activated dendritic cells: A potent Th9 cell inducer for tumor immunotherapy. Oncoimmunology. (2016) 5:e1238558. doi: 10.1080/2162402X.2016.1238558
217. Zhao Y, Chu X, Chen J, Wang Y, Gao S, Jiang Y, et al. Dectin-1-activated dendritic cells trigger potent antitumour immunity through the induction of Th9 cells. Nat Commun. (2016) 7:12368. doi: 10.1038/ncomms12368
218. Glauner H, Ruttekolk IR, Hansen K, Steemers B, Chung Y, Becker F, et al. Simultaneous detection of intracellular target and off-target binding of small molecule cancer drugs at nanomolar concentrations. Br J Pharmacol. (2010) 160:958–70. doi: 10.1111/j.1476-5381.2010.00732.x
Keywords: small ligands, C-type lectin receptor, immunoglobulin receptors, innate immune system, immunology
Citation: Fioretto L, Ziaco M, Nuzzo G, Albiani F, Saponaro M, Carbone D, Barra G, dell’Isola M, Follero O, Esercizio N, Castiglia D, d’Ippolito G, Gallo C, Fontana A and Manzo E (2025) Small molecules as ligands in the tuning of immune regulatory receptors. Front. Immunol. 16:1648691. doi: 10.3389/fimmu.2025.1648691
Received: 23 June 2025; Accepted: 18 August 2025;
Published: 29 August 2025.
Edited by:
Balachandran Ravindran, Institute of Life Sciences (ILS), IndiaReviewed by:
Matej Sova, University of Ljubljana, SloveniaMariano Malamud, University of Exeter, United Kingdom
Copyright © 2025 Fioretto, Ziaco, Nuzzo, Albiani, Saponaro, Carbone, Barra, dell’Isola, Follero, Esercizio, Castiglia, d’Ippolito, Gallo, Fontana and Manzo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Emiliano Manzo, ZW1pbGlhbm8ubWFuem9AY25yLml0; Angelo Fontana, YW5nZWxvLmZvbnRhbmFAY25yLml0; Carmela Gallo, Y2FybWVsYS5nYWxsb0BjbnIuaXQ=
†These authors have contributed equally to this work
‡ORCID: Carmela Gallo, orcid.org/0000-0002-1665-2615
Angelo Fontana, orcid.org/0000-0002-5453-461X
Emiliano Manzo, orcid.org/0000-0002-7394-020X
 Laura Fioretto1,2†
Laura Fioretto1,2† Marcello Ziaco
Marcello Ziaco Mario dell’Isola
Mario dell’Isola Carmela Gallo
Carmela Gallo Emiliano Manzo
Emiliano Manzo