- School of Medical and Life Sciences, Chengdu University of Traditional Chinese Medicine, Chengdu, Sichuan, China
The skin surface is colonised by a rich microbiome, and intricate interactions between this microenvironment and microbial communities are critical for maintaining skin homeostasis. Atopic dermatitis (AD), a chronic inflammatory skin disease characterised by skin barrier dysfunction and aberrant immune activation, exhibits a rising global incidence. While conventional therapeutic strategies offer short-term symptom control, their long-term use is limited by adverse effects including skin atrophy, metabolic disorders, and increased infection risk. Critically, these approaches fail to cure AD or reverse the underlying immune imbalance. Recent research has firmly established the skin microbiome as a central driver in AD pathogenesis. The molecular mechanisms underpinning microbiome-host interactions, including the potential for remote regulation via the gut-skin axis, are now being actively investigated. This review systematically analyses how microbial dysbiosis in AD promotes Th2/Th17 immune polarization through three key pathways: microbial metabolites, immune signalling, and barrier integrity. Building on these mechanistic insights and recent advances, we propose novel multimodal therapeutic strategies targeting the microbial-immune axis. We further elucidate the role of commensal bacteria in maintaining immune homeostasis. Ultimately, this synthesis aims to bridge fundamental research with clinical applications, providing a robust theoretical foundation for future therapeutic development and clinical studies in AD management.
1 Introduction
As the largest organ and primary defense barrier, the skin’s complex anatomical structure and heterogeneous microenvironment create a unique microbial ecosystem (1). A bidirectional Gut-Skin Axis (GSA) links these organs through microbial metabolites, immune signals, and environmental factors (2). Both systems utilize tightly linked proteins (e.g., claudins) to maintain physical barriers and synergistically defend against pathogens. Existing studies have shown that commensal flora such as Staphylococcus epidermidis(S. epidermidis) can secrete novel antimicrobial peptides to inhibit Staphylococcus aureus (S. aureus) colonisation and regulate IL-1β expression in keratinocytes via the A20 protein, allowing for the fine-tuning of immune homeostasis (3).
AD is a chronic inflammatory disease driven by skin barrier dysfunction and aberrant immune activation, affects up to 20% of children and 3% of adults globally with rising incidence (4, 5). Its hallmark symptoms—intense itching, dryness, erythema, and exudative lesions—stem directly from epidermal permeability barrier disruption (6, 7). This complex pathology involves genetic susceptibility (e.g., FLG mutations), environmental triggers, and immune dysregulation.
Current clinical management of AD encompasses topical anti-inflammatory agents (glucocorticoids, TCS; calcineurin inhibitors, TCIs), systemic immunosuppressants (e.g., cyclosporine, methotrexate) for moderate-to-severe cases, and monoclonal antibody biologics such as dupilumab (anti-IL-4Rα) that inhibit the Th2 pathway (8). Although these therapies provide symptomatic control in AD, prolonged use of topical corticosteroids (TCS) causes adverse effects including skin atrophy, telangiectasia, and metabolic disturbances. Concurrently, systemic immunosuppressive agents may induce hepatorenal toxicity and increase infection susceptibility (9), which highlight the need for exploring new pathogenic mechanisms and targeting them for the necessity of therapeutic treatment. Notably, probiotic-based microbial transplantation has recently demonstrated efficacy in restoring flora balance and alleviating AD symptoms, marking a pivotal transition from mechanistic research to clinical translation in skin microbiome science (10).
2 Symbiotic bacteria on the skin surface
Human skin acts as a physical barrier to prevent the entry of pathogenic microorganisms while providing a home for commensal bacteria and fungi, and functional studies have demonstrated the impact of specific strains on modulating the immune system, shaping the microbial community, providing colonisation resistance and promoting epidermal barrier integrity (11). Recent studies have integrated the microbiome, immunity and tissue integrity to understand their interactions in common diseases such as AD.
2.1 Composition and function
The bacterial diversity of the skin microbiome is dominated by Actinobacteria, Firmicutes and Proteobacteria, with S. aureus and S. epidermidis occupy significant ecological niches on the skin surface. Multi-omics analyses revealed that S. aureus, while exhibiting low abundance on healthy skin, was substantially enriched in lesional skin of AD patients. Critically, its absolute abundance demonstrated a strong positive correlation with disease severity (12). In contrast, plasma coagulase-negative S.epidermidis, as a symbiotic bacterium, forms biofilms by secreting polysaccharide intercellular adhesins (PIA), inhibits pathogen colonization (13), and reduces the degree of inflammation by regulating host TLR3 signaling pathway (14). Similarly, Staphylococcus hominis (S. hominis) is also a bacterium with negative plasma coagulase. S. hominis can secrete autoinducing peptides to inhibit the expression of harmful protease EcpAd and prevent the increase of pathogenic bacteria (15). Similarly, antimicrobial peptides (AMPs) derived from microorganisms, particularly short peptide bacteriocins (SPBs) and quorum sensing inhibitory peptides (AIPs) produced by symbiotic bacteria, hold significant therapeutic potential in AD treatment by selectively inhibiting the growth and virulence of S. aureus, modulating skin immune responses, and restoring skin microbiota balance (16). Cutibacterium acnes (C. acnes formerly Propionibacterium acnes) predominates in sebaceous gland-rich regions, where it metabolizes sebum triglycerides to produce short-chain fatty acids (SCFAs). These SCFAs maintain skin acidity and enhance barrier function by suppressing inflammatory factor release (17). Conversely, CERS1—a molecular biomarker uniquely correlated with S. aureus abundance—may drive skin barrier dysfunction through fatty acid sequestration. This represents a maladaptive compensatory response to reductions in very long-chain fatty acids, ELOVL6 expression, and short-chain sphingolipid composition (18). In contrast, cutaneous fungi—predominantly Malassezia species—exhibit high dependency on host-derived lipids for colonization. Malassezia globosa, which lacks fatty acid synthase (FAS) genes, relies on lipase-mediated hydrolysis of sebum for nutrient acquisition. This metabolic adaptation drives its niche-specific enrichment in sebaceous-rich regions (19). Notably, these findings align with recent experimental evidence demonstrating that exogenous lipids ameliorate AD pathology in murine models by rectifying immune dysregulation and microbiota imbalances (20). Conversely, the virulent phage group dominates this niche, with its abundance dynamically linked to host bacterial community structure (21).
2.2 Spatial distribution
The skin microbiota exhibits significant spatial heterogeneity driven by local physicochemical properties. Lipophilic species, including C. acnes and S. epidermidis, dominate sebum-rich zones (T-zone, back), where they hydrolyze sebum triglycerides to modulate barrier function. However, overgrowth of these species can trigger inflammatory pathologies like acne (22). In contrast, moist intertriginous areas (axillae, groin) feature high eccrine gland density and humidity, shaping distinct microbial communities (23). Due to the high density of sweat glands and high humidity, Corynebacterium and S. hominis are often enriched in humid environments, in which Corynebacterium produces volatile thiols through the metabolism of branched-chain amino acids in sweat, which are involved in the formation of body odour, and the dry areas such as the forearms and the calves are characterised by the proliferation of β-Aspergillus phylum (e.g., Betaproteobacteria), which are also involved in the regulation of the skin barrier function (24). Betaproteobacteria and Flavobacteriales exhibit low abundance but high diversity in skin regions exposed to external environments, a pattern linked to frequent epidermal desquamation. Meanwhile, specialized niches like the scalp and hairline harbor unique microbial communities dominated by Propionibacteriaceae and Malassezia fungi, reflecting distinct follicular structures and sebum secretion dynamics (25) (Figure 1).
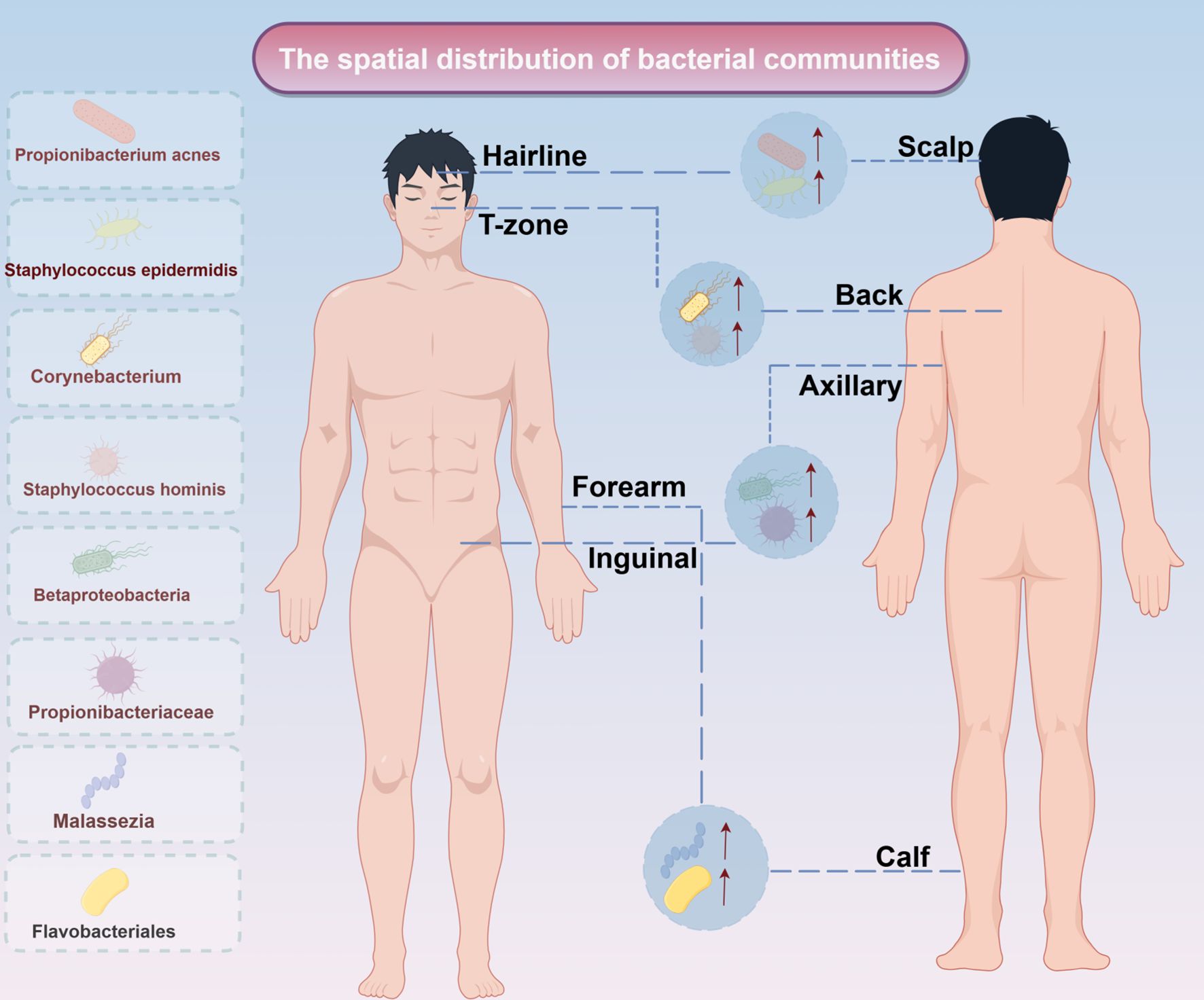
Figure 1. Spatial distribution of bacterial communities. The spatial distribution of bacterial communities in different parts of the human body. The left side displays the main colonized bacterial genera or groups, including Propionibacterium acnes and Staphylococcus epidermidis; the right side corresponds to specific areas of the human body (such as the hairline, T-zone, scalp, etc.), reflecting the spatial heterogeneity of microbial communities on the skin and body surface.↑: increase expresion.
Skin microenvironmental factors—temperature, humidity, pH, and lipid content—function as ecological filters that modulate microbial community assembly (26). High humidity promotes the colonization of S. epidermidis, and the ceramides secreted by it enhance the water retention ability of skin barrier, forming a mutually beneficial network between “bacteria and host” (27). However, a high PH environment is conducive to the survival of S. aureus (28). Sebum-derived free fatty acids exhibit dual functionality: they possess antimicrobial properties while serving as a carbon source for C. acnes. This bacterium further regulates cutaneous pH through secretion of SCFAs, establishing a closed-loop equilibrium (29).
The skin microbiome in AD patients exhibits characteristic dysbiosis. Lesional areas show significant enrichment of S. aureus, which secretes δ-toxin to disrupt keratinocyte tight junctions (30). Concurrently, depletion of commensal S. epidermidis and C. acnes impairs anti-inflammatory SCFAs synthesis (31). AD patients exhibit reduced diversity of Betaproteobacteria in non-lesioned dry skin, indicating systemic immune dysregulation exerts distal effects on microbiome composition. Notably, skin creases in AD show inverted Corynebacterium-to-Staphylococcus ratios. This dysbiosis correlates with impaired antimicrobial peptide secretion driven by localized overexpression of Th2 cytokines IL-4 and IL-13 (32).
3 The role of commensal bacteria in immune homeostasis
Commensal bacteria critically establish mucosal immune tolerance by regulating immune cell differentiation and cytokine networks. Polysaccharide A (PSA) from Bacteroides fragilis drives CD4+ T cell differentiation into Foxp3+ Treg cells and induces anti-inflammatory cytokine secretion (e.g., IL-10) (33). These effects occur via Toll-like receptor 2 (TLR2) signaling. This process enhances Treg immunosuppressive capacity and alleviates inflammation in experimental colitis; symbiotic metabolites thereby actively regulate immune tolerance. IL-10 directly inhibits the antigen-presenting function of dendritic cells (DCs), whereas TGF-β promotes the differentiation of peripheral Tregs by inducing the expression of Foxp3.TGF-β induces peripheral initial T cells into Treg with immunosuppressive function, a process that significantly reduces Th2-type inflammatory responses in oral immunotherapy of AD patients. Microbial-derived SCFAs regulate Treg/Th17 balance through epigenetic and receptor-mediated pathways. SCFAs such as butyrate and propionate inhibit histone deacetylase (HDAC), enhancing histone acetylation in the Foxp3 promoter region to promote Treg differentiation (34). SCFAs inhibit mTOR-S6K signaling via GPR43 activation, blocking RORγt-mediated Th17 differentiation. This bidirectional regulation occurs in both gut and skin. Intestinal commensal bacteria-derived ATP promotes Th17 differentiation through CD11c+ dendritic cells (DCs), while SCFAs counteract this process to maintain immune homeostasis. Separately, commensal bacterial flagellin activates TLR5 on DCs, inducing IL-6 secretion that modulates Th17 differentiation thresholds. These mechanisms reveal precise microbial ligand-immune cell interactions.
S. epidermidis can upregulate the expression of FLG, and indirectly produce natural moisturizing factor (NMF) to enhance the compactness of the stratum corneum, so as to enhance the skin barrier function (35). Cutaneous symbiotic bacteria produce Indole-3-acetic acid (IAA), indole-3-acetaldehyde and indolepyruvate through tryptophan metabolism, which enhance epithelial barrier function via aryl hydrocarbon receptor (AHR) signaling pathway. This study lays the foundation for the development of skin disease therapy based on microbial metabolism (36). Clinically, loss-of-function FLG mutations in AD patients cause barrier defects that facilitate S. aureus colonization and promote Th2-type inflammation (37, 38). Commensal bacteria directly inhibit pathogen proliferation through antimicrobial peptide secretion (e.g., lantibiotics like lugdunin). Separately, S. epidermidis-derived phenol-soluble modulins (PSMs) disrupt S. aureus biofilms and attenuate virulence. This ‘ecosystem competition’ mechanism is critically important during AD’s acute phase, driving pathogen dominance (39). Microbiome-targeted therapies show significant potential for AD management.
4 Immune characteristics and immune imbalance in AD
AD is an immune disease, and its occurrence usually involves the driving of some immune factors and the imbalance of immune mechanisms.
4.1 AD immune characteristics and core driving factors
AD is an autoimmune skin disease, which is usually associated with some immune mechanisms. The following will be described from three aspects:
4.1.1 Th2-driven immune dysregulation
The immune profile of AD is characterized by predominant Th2 differentiation of naïve CD4+ and CD8+ T cells. These Th2 cells secrete IL-4, IL-5, and IL-13—core drivers of the inflammatory cascade (40). Upon binding to keratinocyte IL-4Rα/IL-13Rα1 receptor complexes, these cytokines activate the JAK1/TYK2/JAK2-STAT6/STAT3 pathway. This signaling significantly suppresses filaggrin (FLG) and antimicrobial peptide (AMP) expression, ultimately causing barrier dysfunction (41). IL-4 significantly downregulates key epidermal structural proteins (filaggrin, loricrin, involucrin) and keratin-related genes, while simultaneously promoting B-cell class switching to IgE to amplify allergic responses. Notably, IL-13—secreted predominantly in chronic phases—stimulates eosinophilic infiltration and upregulates chemokines CCL17 and CCL22 (42). Dupilumab, a monoclonal antibody targeting IL-4Rα, represents a therapeutic breakthrough in Th2 pathway inhibition. By blocking shared IL-4/IL-13 signaling, it significantly reduces Scoring Atopic Dermatitis (SCORAD) indices and serum IgE levels in AD patients (43). In the AD model of mice with IL-13 gene defects, the activation of Th2 cells is enhanced, and the relative levels of short-chain sphingomyelin (SM) and ceramide (CER), which are composed of non-hydroxy fatty acids and sphingolipids, increase, while the relative levels of long-chain types decrease (44).
4.1.2 Contribution of Th17 to AD heterogeneity
Despite Th2 predominance in AD, Th17 cells contribute significantly to specific AD subtypes (45–47). IL-17A exacerbates epidermal hyperplasia (acanthosis) and neutrophil infiltration by suppressing E-cadherin expression and upregulating S100A proteins (S100A7/8/9). In chronic lesions, Th17-Th2 crosstalk occurs: IL-4 attenuates inflammation through Th17 differentiation inhibition, while Th17-derived IL-22 further compromises barrier function, establishing a pathogenic feedback loop (48). Notably, Asian AD patients exhibit elevated IL-17C expression correlating with epidermal thickening and psoriasiform features. Single-cell sequencing confirms Th1/Th17/Th22 co-infiltration in chronic lesions, indicating spatiotemporal immune dynamics as a key source of disease heterogeneity (49).
4.1.3 Role of aryl hydroxyl receptor in AD
The AHR critically regulates terminal epidermal differentiation.Ahr- keratinocytes exhibit significantly reduced expression of key differentiation markers—including FLG, loricrin (LOR), and involucrin (IVL)—impairing stratum corneum formation. Concurrently, AHR deficiency alters cytokine homeostasis: levels of AD-associated cytokines (IL-33, IL-36γ, TSLP) are diminished, while the pro-inflammatory factor IL-24 is elevated. This dysregulation suggests AHR is essential for maintaining cutaneous immune homeostasis (50). Therapeutically, coal tar upregulates filaggrin (FLG) expression in keratinocytes and inhibits STAT6 activation via the AHR/NRF2 axis. This pathway antagonizes IL-4/IL-13-mediated degradation of barrier structural proteins, ultimately ameliorating AD-associated barrier defects (51). The tryptophan (Trp) metabolic pathway is significantly impaired in the skin microbiota of AD patients, resulting in markedly reduced levels of indole-3-carbinol (IAId)—a key microbially derived metabolite. Normally, IAId functions as an endogenous AHR ligand that suppresses thymic stromal lymphopoietin (TSLP) expression via promoter binding, thereby attenuating Th2 inflammation. In AD lesional skin, however, IAId deficiency permits dysregulated TSLP overexpression (52).
4.2 Immune dysregulation in AD
The core of the immune imbalance in AD lies in the overactivation of the Th2-type immune response. Research indicates that cytokines IL-4 and IL-13 are key factors driving AD inflammation. These cytokines enhance the STAT6 signaling pathway by activating the IL-4Rα receptor, which promotes the differentiation of Th2 cells and inhibits the expression of filaggrin, a barrier protein in keratinocytes, leading to the disruption of the skin barrier function (53). Meanwhile, in patients with AD, the balance between Th1 and Th2 cells is disrupted. During the acute phase, the immune response is primarily driven by Th2 cells, characterized by high levels of IL-4, IL-5, and IL-13. In contrast, during the chronic phase, the levels of Th1-related factors IFN-γ and TNF-α increase (54). Additionally, IL-17A and IL-22, secreted by Th17 cells, play a dual role in AD, contributing to both antimicrobial defense and potentially exacerbating inflammation (55).
Antigen-presenting cells, such as Langerhans cells (LCs) and dendritic cells (DCs) in the epidermis, play a central role in initiating the AD immune response. These cells take up and present antigens, recruit other immune cells, and regulate the direction of the immune response (56). Recent studies have shown that skin surface symbiotic bacteria (e.g., S. aureus) can directly activate DCs and LCs through pattern recognition receptors, promote antigen presentation and T cell differentiation, and lead to the occurrence of AD (57, 58). DCs are crucial for the differentiation of Th2 cells. TSLP, released by damaged keratinocytes, can activate LCs/DCs. Activated DCs then express co-stimulatory molecules like OX40L, which drive the differentiation of initial T cells towards Th2 (59). Additionally, experiments have shown that IL-33 activates DCs through a MyD88-dependent signaling pathway, promoting the secretion of Th2-type cytokines, a mechanism confirmed in AD mouse models (60). In addition, inflammatory dendritic epidermal cells (IDECs) are also involved in the pathogenesis of AD. IDECs express high levels of FcϵRI receptor, which binds to IgE-allergen complex and releases pro-inflammatory mediators such as TNF-α and IL-12, thus amplifying Th1 and Th17 responses (Figure 2).
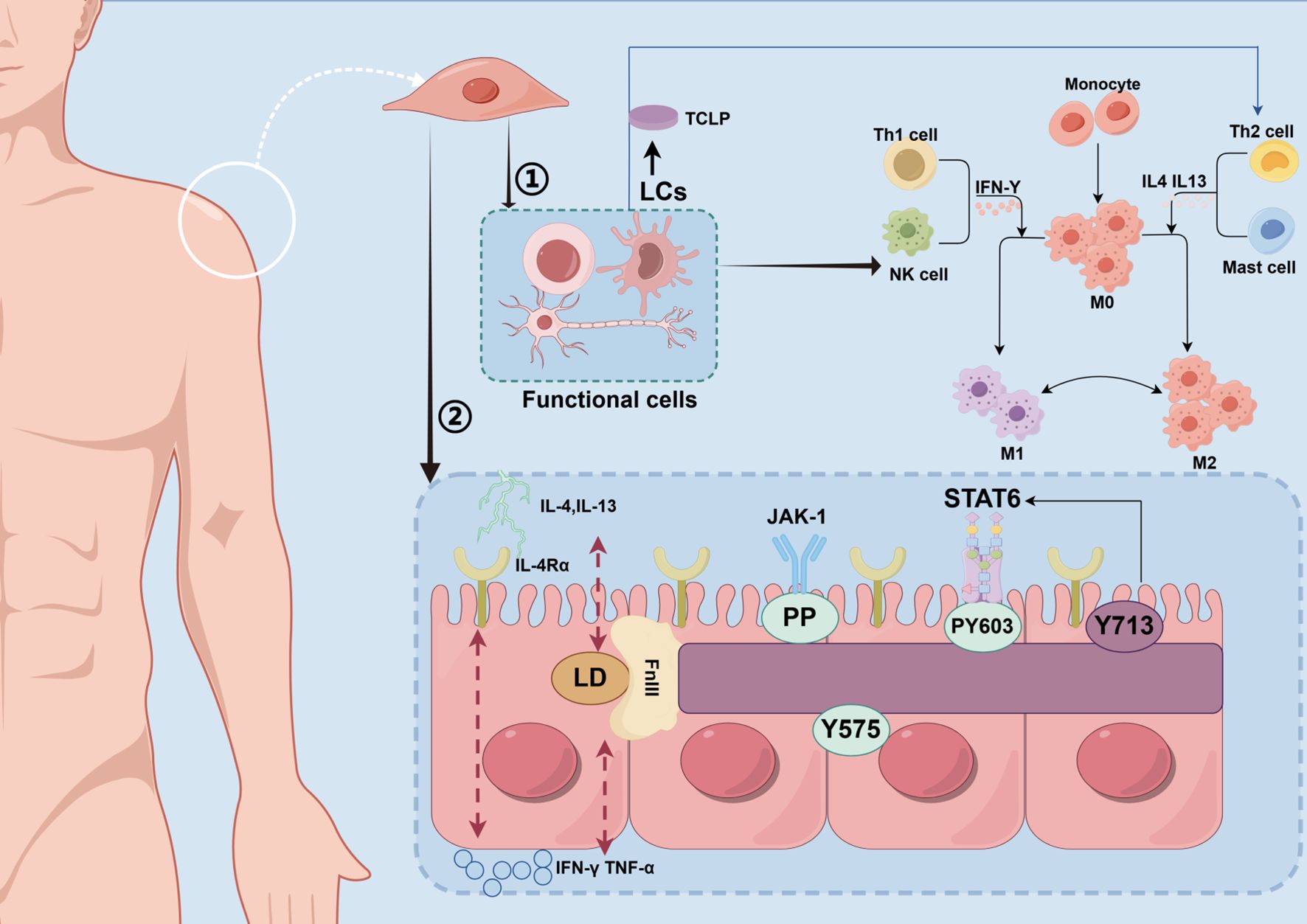
Figure 2. Molecular mechanism of regulating skin immune homeostasis. This figure depicts the molecular mechanisms by which immune cells on the skin surface and commensal microbiota regulate immune homeostasis through the STAT6 signaling pathway. TH2 cells secrete IL-4 and IL-13, which bind to the IL-4 receptor (IL-4Ra) on monocytes and mast cells, activating the JAK-1/STAT6 pathway. Meanwhile, the commensal microbiota may influence STAT6 phosphorylation through metabolites or direct signaling. (LCs, Langerhans cells; TSLP, Thymic Stromal Lymphopoietin; Th1 cell, T Helper 1 Cell; NK cell, Natural Killer cell; IFN-γ, Interferon-gamma; M0, resting macrophages; M1, M1 Macrophages; M2, M2 Macrophages; IL-4, Interleukin-4; IL-13, Interleukin-13; Th2 cell, T Helper 2 Cell; IL-4Rα, Interleukin-4 Receptor Alpha; LD, Linker Domain; FnIII, Fibronectin Type III Domain; TNF-α, Tumor Necrosis Factor Alpha; PP, Phosphorylation; JAK-1, Janus Kinase 1; STAT6, protein; Y575/PY60/Y713, phosphorylation site).
5 The vicious cycle of “microflora imbalance-immune activation-barrier destruction” in AD
In AD, skin microbiome dysbiosis and immune dysregulation form a self-reinforcing pathological loop. This immune deviation impairs barrier function by suppressing filaggrin (FLG) and antimicrobial peptide (AMP) synthesis, establishing a ‘dysbiosis–immune activation–barrier disruption’ cycle.
5.1 Bacterial imbalance in AD
Analysis of lesional skin microbiomes in AD patients revealed significant dysbiosis. Commensal bacteria—including Streptococcus, Cutibacterium, and Corynebacterium—showed reduced abundance. In contrast, S. aureus colonized >90% of lesions, with its abundance positively correlating with disease severity (61, 62). This dysbiosis impairs production of antimicrobial peptides (LL-37, β-defensin) while altering the skin microenvironment through bacterial metabolites. Both mechanisms promote S. aureus adhesion and growth. Reduced microbiome diversity strongly correlates with AD relapse. During acute episodes, the number of S. aureus increased significantly. After treatment, inflammation subsided as symbiotic bacteria such as streptococcus and S. epidermidis recovered (63). Choi et al. observed increased proportions of L. fermentum strain SLAM216 in the gut and identified LF216EV. In mice, LF216EV elevated Limosilactobacillus and Lactococcus abundance while alleviating AD symptoms. This therapeutic effect may involve altered expression of serotonin-related genes (htr2c, sert, tph1) (64).
The skin microbiome of AD patients differs significantly from that of healthy individuals. Multicentre clinical studies reveal a substantial reduction in microbial diversity within AD lesional skin. Shannon’s index decreases by 40–60% in these regions. Concurrently, S. aureus dominates, reaching relative abundances of 70–90% (65). For example, a 16S rRNA sequencing cohort study of Indonesian AD patients revealed distinct microbial shifts. In moderate AD skin, Firmicutes constituted up to 85% of the microbiome. S. aureus abundance increased 8-fold compared to healthy controls. In contrast, mild AD skin showed Proteobacteria predominance (60%). This study also documented, for the first time, the presence of the commensal bacterium Ensifer adhaerens (66).
5.2 Immune activation mediated by dysbiosis in AD
Microbiome dysregulation reconfigures the skin immune landscape via multiple pathways. S. aureus overgrowth directly activates keratinocytes through δ-toxin secretion, triggering TSLP and IL-33 release to drive Th2-polarized immunity (67). S. aureus enterotoxins SEB and TSST-1 function as superantigens that bind MHC class II molecules and T-cell receptor (TCR) Vβ regions independent of antigen presentation. This activates polyclonal T cells, triggering massive release of IL-31 (directly inducing pruritus) and IFN-γ (driving chronic inflammation). Clinically, elevated serum IgE against these superantigens in AD patients correlates significantly with eczema area severity index (EASI) scores (68). Beyond bacterial dysbiosis, fungal community disruption exacerbates skin inflammation—particularly in head/neck AD—by activating pattern-recognition receptors (PRRs) to drive pathogenic IL-17 secretion (69) (Figure 3).
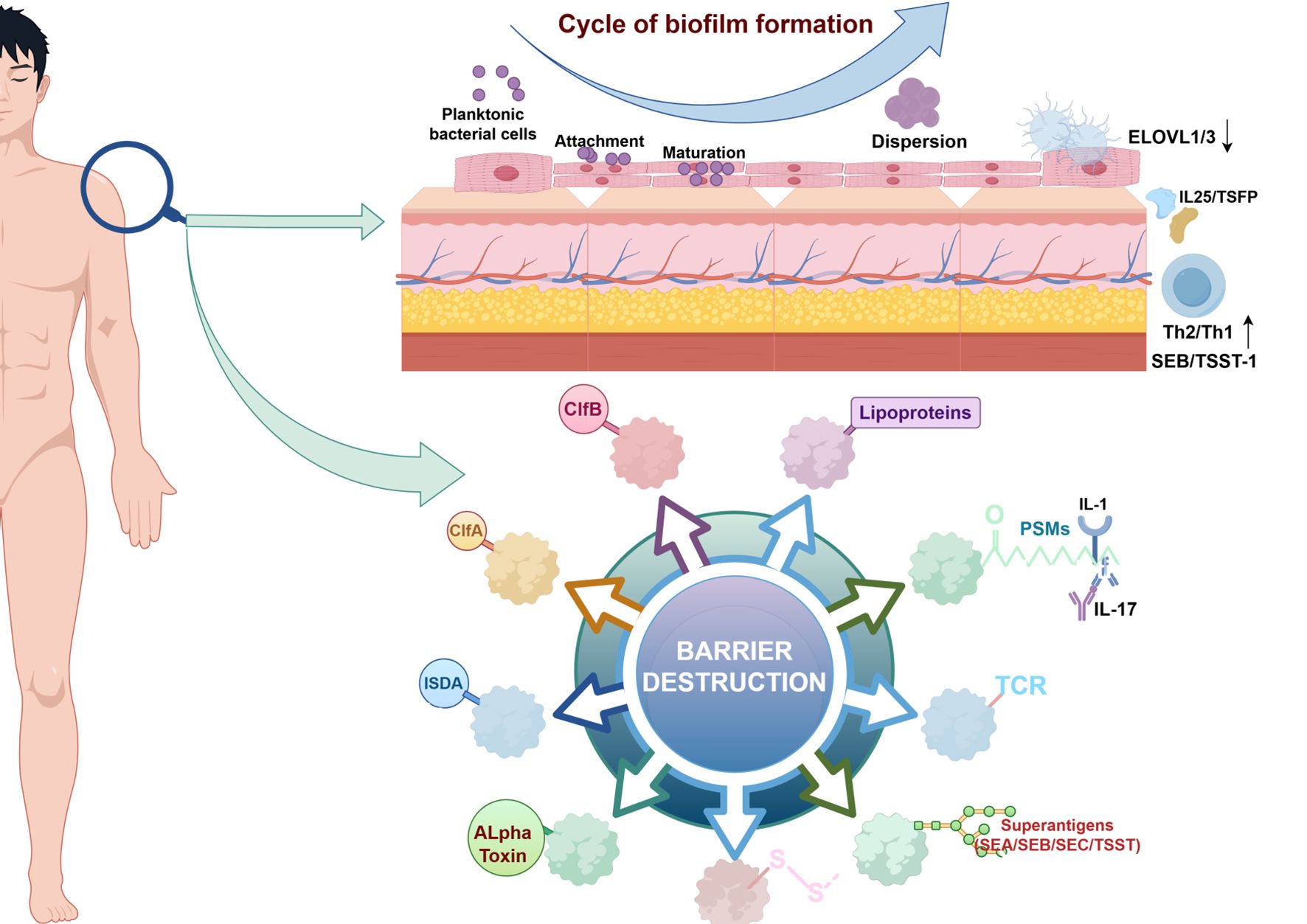
Figure 3. Mechanisms of skin barrier damage and biofilm formation. S aureus has highly evolved multiple cell-wall proteins and secreted factors that enable adhesion to human skin and barrier disturbance by using physical, chemical, and inflammatory mechanisms. Adhesion,S aureus has developed several surface molecules to adhere to the human stratum corneum, including clumping factors A and B (ClfA and ClfB). Barrier destruction, S aureus a-toxin, a water-soluble cytotoxin, forms a heptameric b-barrel pore in host cell membranes. In the epidermis it directly forms pores in keratinocytes, which erodes the integrity of the epidermal barrier. S aureus produces at least 10 proteases, a number of which facilitate dissolution of the stratum corneum. In addition to secreted proteases, S aureus can directly stimulate endogenous keratinocyte proteases, highlighting an additional mechanism toward barrier destruction. Proinflammatory mechanisms, Cell-wall bound protein A, when solubilized, triggers inflammatory responses from keratinocytes through TNF receptor (TNFR). Staphylococcal superantigens, such as SEA, SEB, SEC, and toxic shock syndrome toxin-1 (TSST-1), trigger B-cell expansion and cytokine release. S aureus secretes PSMs, which are direct proinflammatory drivers with compartment-specific effects. In the epidermal compartment PSMs stimulate IL-36a–driven gd T cell– mediated inflammation, whereas in the dermal compartment they stimulate IL-1b–driven Th17 inflammation. (↓, decrease expression; ↑, increase expression; ELOVL 1/3, Elongation of Very Long chain fatty acids1/3; IL 1/17/25, Interleukin 1/17/25 ;TSFP, Thymic Stromal Folliculin Protein; Th 1/2, Th1/2 cell, T Helper 1/2 Cell; SEA/SEB/SEC, Staphylococcal Enterotoxin A/B/C; TSST-1, Toxic Shock Syndrome Toxin-1; ISDA, Staphylococcus aureus protein; ClfA/B, Clumping Factor A/B; PSMs, Peptide-Spectrum Matches; TCR, T cell receptor; -S-S-, Disulfide Bond.
Moreover, AD microbiota frequently show impaired utilization of 2’-fucosyllactose (2’-FL), indicating disrupted retinol metabolism and consequent immune tolerance defects (70). S. epidermidis balances pro- and anti-inflammatory responses by inducing IL-1β/IL-6 secretion to promote Th17 polarization while upregulating Foxp3 to enhance regulatory T cell (Treg) function (71, 72). This precise immunomodulatory network operates through microbial metabolite-mediated HDAC inhibition, thereby enhancing anti-microbial peptide (AMP) expression (10, 73).
5.3 AD skin barrier disruption and the vicious cycle of immune-microbiome
The normal skin barrier comprises a multilayered defense system: the stratum corneum, granular layer (stratum granulosum), spinous layer (stratum spinosum), and basal layer (stratum basale). The stratum corneum—the core physical barrier—maintains structural integrity through three key components: cross-linked cornified envelope proteins, lamellar lipid bilayers, and NMF (74). Skin barrier dysfunction manifests through two core abnormalities: reduced ceramide content with altered lipid composition in the stratum corneum, and defective intercellular junctions due to downregulated claudin-1 expression. These alterations collectively elevate risks of allergen penetration and microbial colonization (75, 76). In AD patients, diminished microbial diversity coincides with dysbiotic expansion of S. aureus. This pathogen forms biofilms that exacerbate skin barrier defects through a synergistic cycle of colonization and inflammation. Targeted microbiome modulation can restore protective flora abundance, including commensals like Corynebacterium species that reinforce barrier integrity (77). The protein encoded by the silk protein gene is not only involved in the terminal differentiation of keratinocytes, but its degradation products also regulate the pH of the cuticle and maintain water balance. The tight junction protein Claudin-1 forms a ‘molecular zipper’ structure in the spinous layer to limit molecular permeation (78). In addition, antimicrobial peptides (AMPs) such as β-defensins and cathelicidins form a chemical barrier that enables immunosurveillance by directly killing pathogens and regulating dendritic cell activity.
Approximately 50% of AD patients carry a FLG gene Loss-of-function (LOF) mutation that leads to reduced NMF synthesis, dehydrated stratum corneum and increased pH (79), a defect that inhibits keratinocyte differentiation through disturbances in calcium ion signalling. Recent studies have shown that Claudin-1 expression is significantly reduced in AD non-lesional skin, leading to enlargement of tight junction pores and making it easier for allergens to penetrate the epidermis. A 40-60% reduction in stratum corneum ceramides disrupts lamellar body secretion and lipid bilayer organization. This structural defect correlates with impaired peroxisome proliferator-activated receptor gamma (PPARγ) signaling, which normally upregulates lipid synthesis genes. PPARγ also modulates immune polarization, promoting Th2 cells, type 2 innate lymphoid cells (ILC2s), and M2 macrophages that drive inflammation (80, 81). Elevated kallikrein-related peptidase 5/7 (KLK5/7) activity accelerates degradation of corneodesmosomal core proteins (e.g., corneodesmosin), triggering premature stratum corneum desquamation. This pathological process is amplified by downregulated expression of lympho-epithelial Kazal-type inhibitor (LEKTI), a key metalloproteinase inhibitor that normally constrains KLK protease activity (82). Upon barrier disruption, V8 protease (SspA) secreted by S. aureus enzymatically cleaves keratin 16 (K16), exposing cryptic antigenic epitopes that trigger pathological immune responses (83).
Notably, the stratified lipid structure of the stratum corneum not only restricts the invasion of pathogenic bacteria through physical isolation, but its low pH environment also maintains commensal dominance by inhibiting the protease activity of S. aureus. Upon barrier compromise, downregulation of fatty acid synthesis genes ELOVL1/3 disrupts lipid metabolism, while simultaneously released alarmins (IL-25, TSLP) promote Th2 immune responses. This initiates a self-perpetuating cycle of barrier disruption - microbial dysbiosis - immune dysregulation (84–86). This pathogenic cascade is particularly pronounced in AD. S. aureus biofilms disrupt keratinocyte intercellular junctions via α-toxin, while its superantigen SEB binds HLA-DR molecules to activate Vβ T-cell receptor clonal expansion. This induces IL-31-mediated pruritus and drives IL-17/IL-22-dependent chronic inflammation (87). Recent studies demonstrate that specific probiotics (e.g., Limosilactobacillus reuteri DYNDL22M62) mitigate AD inflammation through dual mechanisms: restoration of skin microbial diversity; suppression of TSLP production, reducing Th2 cytokine levels. This reverses Th2 immune polarization and provides a molecular foundation for microbiome-targeted therapies (73).
6 Clinical applications and future therapies
6.1 Traditional treatment
Among JAK inhibitors, abrocitinib showed rapid itch relief in Chinese adults with AD without treatment-emergent serious adverse cardiovascular events (88). JAK inhibitors demonstrate dual mechanistic and clinical efficacy. Baricitinib inhibited JAK-STAT signaling in CD4+ T cells, significantly reducing MAPK and PI3K/Akt/mTOR pathway activity. This achieved a mean 62% SCORAD reduction in 124 Chinese AD patients, confirming its therapeutic utility in Asian populations (89). Topical ruxolitinib cream precisely targets cutaneous JAK1/JAK2 activity, with systemic exposure at 1/1000th of oral administration levels (90). Dupilumab, an IL-4Rα-targeted therapy, demonstrates significant long-term efficacy and safety in moderate-to-severe AD. Real-world evidence confirms sustained EASI improvement >70% at 4 years without significantly elevating infection risk (91). Despite widespread dupilumab use in moderate-to-severe AD, studies indicate increased cutaneous T-cell lymphoma (CTCL) risk (RR = 4.10; 95% CI: 2.06-8.19) (92).
6.2 Symbiotic bacteria for the treatment of AD
6.2.1 Treatment based on the gut-skin axis
In randomized controlled trials, oral Limosilactobacillus fermentum supplementation reduced SCORAD scores by 40.4% (mean) in pediatric AD patients, with efficacy positively correlating with treatment duration (p = 0.003) (93). Notably, Bifidobacterium modulates the gut-skin axis by increasing fecal butyrate concentrations. Specific strain combinations (e.g., Bifidobacterium CECT 8145 + CECT 7347) restored the Faecalibacterium/Bacteroides ratio in AD patients’ intestinal flora, providing a molecular rationale for individualized microbial therapies (94).
Clinical trials validate microbiome modulation efficacy via the skin-gut axis. A Lactobacillus-based probiotic formulation reduced SCORAD by 34% in pediatric AD, increasing SCFAs while inhibiting Th2 cytokines (95). FMT from healthy donors decreased S. aureus skin colonization by 68% in Phase II trials, with IL-31 levels correlating with itch severity (96). Faecalibacterium prausnitzii metabolites restored intestinal barrier integrity, lowering plasma LPS and ameliorating skin inflammation (97).
6.2.2 Treatment based on skin prebiotics
Similarly, recent studies have shown that some potential skin prebiotics have therapeutic effects on AD. Professor Richard Gallo’s team identified specific human S. epidermidis strains that produce potent antimicrobial peptides that selectively kill S. aureus and prevent or alleviate AD symptoms in mouse models (98). S. hominis A9 has completed Phase I clinical trials, confirming its safety in humans. The small molecule substance <10 kDa secreted by S. epidermidis can significantly induce the expression of human β-defensins hBD2 and hBD3 by activating the TLR2 receptor of keratinocytes, thus enhancing the antibacterial ability of skin to S. aureus, providing a new idea for the treatment of AD (99). Roseomonas mucosa significantly improved symptoms in children with AD, reduced S. aureus colonization and repaired the skin barrier through a lipid-mediated TNFR2-EMT repair pathway, and was safe (100). The autologous epidermal S. hominis A9 strain isolated from the skin of healthy people can secrete antimicrobial peptides (lantibiotics) to directly kill S. aureus, and produce autologous induced peptides (AIP) to inhibit the quorum sensing system of S. aureus, reduce the expression of toxins (such as PSMα), and alleviate the symptoms of AD (101). Numerous clinical trials have investigated topical probiotics for treating AD, suggesting their potential for widespread clinical application in the future.
6.3 Future therapies
Several emerging microbiome-targeted therapeutic strategies are being used.The colonization density of S. aureus in the skin of AD patients is increased, and its virulence factors can aggravate skin inflammation, while endolysin can be specifically targeted at S. aureus. The study found that the use of endolysin to treat AD can significantly reduce the frequency of AD attacks (102). Antimicrobial peptides (AMPs) restore skin microbiota balance, restore barrier function and reduce inflammation in patients with AD by selectively inhibiting the growth of S. aureus, blocking the expression of virulence factors, and regulating skin immune response. In the future, AMPs can be used to treat AD (16). The use of local bacteriophages can reduce the inflammatory indicators of skin diseases, such as the expression of chemokine CXCL2, neutrophil infiltration and other inflammatory cytokines. It may become a new therapy for AD (103).
Future AD therapeutics will focus on multimodal synergistic interventions. Key skin microbiome-targeted approaches include: personalized microbial transplantation and microbial metabolite preparations (e.g., SCFAs). These restore microbial diversity and immune homeostasis. Concurrently, immunomodulatory strategies advance toward multi-target interventions, with combination therapies targeting Th2/Th17 pathways and specific immune cell functions to achieve multidimensional inflammatory cascade inhibition.
7 Conclusions
AD pathogenesis centers on an interconnected pathological triad:microbiome dysbiosis, barrier dysfunction, and Th2 immune deviation. Therapeutic strategies targeting skin microbiota restoration represent promising disease-modifying interventions for AD.Despite the enormous potential of microbiota-based AD treatments, the treatment of ecological disorders should be integrated with standard skincare practices, as AD is a complex dermatological condition that requires a multifaceted approach combining a variety of skincare modalities and therapeutic approaches for effective management. Despite promising evidence from clinical studies and early phage therapy trials for microbiota-targeted AD treatments, key mechanistic insights remain elusive, and critical translational questions require resolution for optimal clinical implementation. Developing an integrated host-microbiome model is imperative for advancing AD management, as skin microbiota dynamics are shaped by complex interactions between host physiology and environmental exposures. Current research on skin microbiota- AD interactions primarily employs in vitro and murine models, with select therapeutic candidates advancing to early-phase clinical trials (Phase I/II). Future research must leverage multi-omics approaches to resolve spatiotemporal dynamics of host-microbe interactions and develop personalized therapies concurrently targeting: microbiome remodeling, Immune recalibration and barrier restoration. These precision strategies are poised for near-term clinical translation in AD management.
Author contributions
XL: Methodology, Data curation, Writing – original draft, Writing – review & editing. JH: Writing – review & editing. YL: Writing – original draft, Resources. LD: Funding acquisition, Writing – review & editing, Data curation, Conceptualization.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. 1.National Natural Science Foundation of China, Grant Number: 82205131. 2. Sichuan Provincial Department of Science and Technology, Sichuan Provincial Youth Fund Project, Project No.2025ZNSFSC1798. 3. Sichuan Maternal and Child Health Association, Sichuan Maternal and Child Medical Science and Technology Innovation Project, Key Project, Project No. FXZD08.
Acknowledgments
We would like to extend our heartfelt thanks to everyone involved in this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Santiago-Rodriguez TM, Le François B, Macklaim JM, Doukhanine E, and Hollister EB. The skin microbiome: current techniques, challenges, and future directions. Microorganisms. (2023) 11:1222. doi: 10.3390/microorganisms11051222
2. Jimenez-Sanchez M, Celiberto LS, Yang H, Sham HP, and Vallance BA. The gut-skin axis: a bi-directional, microbiota-driven relationship with therapeutic potential. Gut Microbes. (2025) 17:2473524. doi: 10.1080/19490976.2025.2473524
3. Simanski M, Erkens AS, Rademacher F, and Harder J. Staphylococcus epidermidis-induced interleukin-1 beta and human beta-defensin-2 expression in human keratinocytes is regulated by the host molecule A20 (TNFAIP3). Acta Derm Venereol. (2019) 99:181–7. doi: 10.2340/00015555-3073
4. Ruan H, Zhu X, Xu S, Zhou Q, Yang F, and Li G. Topical treatments in special populations of atopic dermatitis - Chinese perspective. Expert Rev Clin Immunol. (2025) 21:425–34. doi: 10.1080/1744666X.2025.2473726
5. Meledathu S, Naidu MP, and Brunner PM. Update on atopic dermatitis. J Allergy Clin Immunol. (2025) 155(4):1124–32. doi: 10.1016/j.jaci.2025.01.013
6. Luk D, Hon KLE, Dizon MVC, Leong KF, Tay YK, Koh MJ, et al. Practical recommendations for the topical treatment of atopic dermatitis in south and east asia. Dermatol Ther (Heidelb). (2021) 11:275–91. doi: 10.1007/s13555-020-00467-8
7. Napolitano M, Fabbrocini G, Martora F, Genco L, Noto M, and Patruno C. Children atopic dermatitis: Diagnosis, mimics, overlaps, and therapeutic implication. Dermatol Ther. (2022) 35:e15901. doi: 10.1111/dth.15901
8. Gatmaitan JG and Lee JH. Challenges and future trends in atopic dermatitis. Int J Mol Sci. (2023) 24:11380. doi: 10.3390/ijms241411380
9. Chu DK, Chu AWL, Rayner DG, Guyatt GH, Yepes-Nuñez JJ, Gomez-Escobar L, et al. Topical treatments for atopic dermatitis (eczema): Systematic review and network meta-analysis of randomized trials. J Allergy Clin Immunol. (2023) 152:1493–519. doi: 10.1016/j.jaci.2023.08.030
10. Dewi DAR, Perdiyana A, Wiliantari NM, Nadhira F, Arkania N, Salsabila CA, et al. Managing the skin microbiome as a new bacteriotherapy for inflammatory atopic dermatitis. Cureus. (2023) 15:e48803. doi: 10.7759/cureus.48803
11. Scharschmidt TC and Segre JA. Skin microbiome and dermatologic disorders. J Clin Invest. (2025) 135:e184315. doi: 10.1172/JCI184315
12. De Tomassi A, Reiter A, Reiger M, Rauer L, Rohayem R, Ck-Care Study G, et al. Combining 16S Sequencing and qPCR Quantification Reveals Staphylococcus aureus Driven Bacterial Overgrowth in the Skin of Severe Atopic Dermatitis Patients. Biomolecules. (2023) 13:1030. doi: 10.3390/biom13071030
13. Nguyen HTT, Nguyen TH, and Otto M. The staphylococcal exopolysaccharide PIA - Biosynthesis and role in biofilm formation, colonization, and infection. Comput Struct Biotechnol J. (2020) 18:3324–34. doi: 10.1016/j.csbj.2020.10.027
14. Lai Y, Di Nardo A, Nakatsuji T, Leichtle A, Yang Y, Cogen AL, et al. Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat Med. (2009) 15:1377–82. doi: 10.1038/nm.2062
15. Cau L, Williams MR, Butcher AM, Nakatsuji T, Kavanaugh JS, Cheng JY, et al. Staphylococcus epidermidis protease EcpA can be a deleterious component of the skin microbiome in atopic dermatitis. J Allergy Clin Immunol. (2021) 147:955–66.e16. doi: 10.1016/j.jaci.2020.06.024
16. Joshi AA, Vocanson M, Nicolas JF, Wolf P, and Patra V. Microbial derived antimicrobial peptides as potential therapeutics in atopic dermatitis. Front Immunol. (2023) 14:1125635. doi: 10.3389/fimmu.2023.1125635
17. Brzuszkiewicz E, Weiner J, Wollherr A, Thürmer A, Hüpeden J, Lomholt HB, et al. Comparative genomics and transcriptomics of Propionibacterium acnes. PloS One. (2011) 6:e21581. doi: 10.1371/journal.pone.0021581
18. Kenney HM, Yoshida T, Berdyshev E, Calatroni A, Gill SR, Simpson EL, et al. CERS1 is a biomarker of Staphylococcus aureus abundance and atopic dermatitis severity. J Allergy Clin Immunol. (2025) 155:479–90. doi: 10.1016/j.jaci.2024.09.017
19. Saunders CW, Scheynius A, and Heitman J. Malassezia fungi are specialized to live on skin and associated with dandruff, eczema, and other skin diseases. PloS Pathog. (2012) 8:e1002701. doi: 10.1371/journal.ppat.1002701
20. Guan J, Chen K, Lu F, and He Y. Dissolving microneedle patch loaded with adipokines-enriched adipose extract relieves atopic dermatitis in mouse via modulating immune disorders, microbiota imbalance, and skin barrier defects. J Tissue Eng. (2025) 16:20417314241312511. doi: 10.1177/20417314241312511
21. Hannigan GD, Meisel JS, Tyldsley AS, Zheng Q, Hodkinson BP, SanMiguel AJ, et al. The human skin double-stranded DNA virome: topographical and temporal diversity, genetic enrichment, and dynamic associations with the host microbiome. mBio. (2015) 6:e01578–15. doi: 10.1128/mBio.01578-15
22. Krupa-Kotara K, Helisz P, Gwioździk W, and Grajek M. The importance of the Microbiota in Shaping women’s Health—the current state of knowledge. Appl Microbiol. (2022) 3:11–34. doi: 10.3390/applmicrobiol3010002
23. James AG, Austin CJ, Cox DS, Taylor D, and Calvert R. Microbiological and biochemical origins of human axillary odour. FEMS Microbiol Ecol. (2013) 83:527–40. doi: 10.1111/1574-6941.12054
24. Moskovicz V, Gross A, and Mizrahi B. Extrinsic factors shaping the skin microbiome. Microorganisms. (2020) 8:1023. doi: 10.3390/microorganisms8071023
25. Nicholas-Haizelden K, Murphy B, Hoptroff M, and Horsburgh MJ. Bioprospecting the skin microbiome: advances in therapeutics and personal care products. Microorganisms. (2023) 11:1899. doi: 10.3390/microorganisms11081899
26. Chen H, Zhao Q, Zhong Q, Duan C, Krutmann J, Wang J, et al. Skin microbiome, metabolome and skin phenome, from the perspectives of skin as an ecosystem. Phenomics. (2022) 2:363–82. doi: 10.1007/s43657-022-00073-y
27. Zheng Y, Hunt RL, Villaruz AE, Fisher EL, Liu R, Liu Q, et al. Commensal Staphylococcus epidermidis contributes to skin barrier homeostasis by generating protective ceramides. Cell Host Microbe. (2022) 30:301–13.e9. doi: 10.1016/j.chom.2022.01.004
28. Scharschmidt TC and Fischbach MA. What lives on our skin: ecology, genomics and therapeutic opportunities of the skin microbiome. Drug Discov Today Dis Mech. (2013) 10:e83-e89. doi: 10.1016/j.ddmec.2012.12.003
29. Almoughrabie S, Cau L, Cavagnero K, O’Neill AM, Li F, Roso-Mares A, et al. Commensal Cutibacterium acnes induce epidermal lipid synthesis important for skin barrier function. Sci Adv. (2023) 9:eadg6262. doi: 10.1126/sciadv.adg6262
30. Ferček I, Lugović-Mihić L, Tambić-Andrašević A, Ćesić D, Grginić AG, Bešlić I, et al. Features of the skin microbiota in common inflammatory skin diseases. Life (Basel). (2021) 11:962. doi: 10.3390/life11090962
31. Jean-Pierre F, Henson MA, and O’Toole GA. Metabolic modeling to interrogate microbial disease: A tale for experimentalists. Front Mol Biosci. (2021) 8:634479. doi: 10.3389/fmolb.2021.634479
32. Tudela H, Claus SP, and Saleh M. Next generation microbiome research: identification of keystone species in the metabolic regulation of host-gut microbiota interplay. Front Cell Dev Biol. (2021) 9:719072. doi: 10.3389/fcell.2021.719072
33. Zhong Y, Chang X, Zhao Z, Zheng L, Kuang G, Li P, et al. Bacteroides fragilis capsular polysaccharide A ameliorates ulcerative colitis in rat by recovering intestinal barrier integrity and restoring gut microbiota. Front Pharmacol. (2024) 15:1402465. doi: 10.3389/fphar.2024.1402465
34. Li Y, Ye Z, Zhu J, Fang S, Meng L, and Zhou C. Effects of gut microbiota on host adaptive immunity under immune homeostasis and tumor pathology state. Front Immunol. (2022) 13:844335. doi: 10.3389/fimmu.2022.844335
35. Kalankariyan S, Thottapillil A, Saxena A, Srivatsn SM, Kadamkode V, Kapoor R, et al. An in silico approach deciphering the commensal dynamics in the cutaneous milieu. NPJ Syst Biol Appl. (2025) 11:42. doi: 10.1038/s41540-025-00524-y
36. Uberoi A, Murga-Garrido SM, Bhanap P, Campbell AE, Knight SAB, Wei M, et al. Commensal-derived tryptophan metabolites fortify the skin barrier: Insights from a 50-species gnotobiotic model of human skin microbiome. Cell Chem Biol. (2025) 32:111–25.e6. doi: 10.1016/j.chembiol.2024.12.007
37. Shane HL, Long CM, and Anderson SE. Novel cutaneous mediators of chemical allergy. J Immunotoxicol. (2019) 16:13–27. doi: 10.1080/1547691X.2018.1515279
38. Paller AS, Kong HH, Seed P, Naik S, Scharschmidt TC, Gallo RL, et al. The microbiome in patients with atopic dermatitis. J Allergy Clin Immunol. (2019) 143:26–35. doi: 10.1016/j.jaci.2018.11.015
39. Scharschmidt TC. Establishing tolerance to commensal skin bacteria: timing is everything. Dermatol Clin. (2017) 35:1–9. doi: 10.1016/j.det.2016.07.007
40. Radi G, Campanti A, Diotallevi F, Martina E, Marani A, and Offidani A. A systematic review of atopic dermatitis: the intriguing journey starting from physiopathology to treatment, from laboratory bench to bedside. Biomedicines. (2022) 10:2700. doi: 10.3390/biomedicines10112700
41. Furue M. Regulation of Skin Barrier Function via Competition between AHR Axis versus IL-13/IL-4–JAK–STAT6/STAT3 Axis: Pathogenic and Therapeutic Implications in Atopic Dermatitis. J Clin Med. (2020) 9:3741. doi: 10.3390/jcm9113741
42. Torres T, Mendes-Bastos P, Cruz MJ, Duarte B, Filipe P, Lopes MJP, et al. Interleukin-4 and atopic dermatitis: why does it matter? A narrative review. Dermatol Ther (Heidelb). (2025) 15:579–97. doi: 10.1007/s13555-025-01352-y
43. Gorelick J, Nguyen A, Schneider SKR, Martel BC, Madsen DE, and Armstrong AW. Biomarkers in atopic dermatitis: A review of the role of IL-13 and the impact of tralokinumab treatment. Am J Clin Dermatol. (2025) 26:199–211. doi: 10.1007/s40257-024-00913-9
44. Wu J, Li L, Zhang T, Lu J, Tai Z, Zhu Q, et al. The epidermal lipid-microbiome loop and immunity: Important players in atopic dermatitis. J Adv Res. (2025) 68:359–74. doi: 10.1016/j.jare.2024.03.001
45. Rerknimitr P, Otsuka A, Nakashima C, and Kabashima K. The etiopathogenesis of atopic dermatitis: barrier disruption, immunological derangement, and pruritus. Inflammation Regen. (2017) 37:14. doi: 10.1186/s41232-017-0044-7
46. Klonowska J, Gleń J, Nowicki RJ, and Trzeciak M. New cytokines in the pathogenesis of atopic dermatitis-new therapeutic targets. Int J Mol Sci. (2018) 19:3086. doi: 10.3390/ijms19103086
47. Bocheva GS, Slominski RM, and Slominski AT. Immunological aspects of skin aging in atopic dermatitis. Int J Mol Sci. (2021) 22:5729. doi: 10.3390/ijms22115729
48. Kader HA, Azeem M, Jwayed SA, Al-Shehhi A, Tabassum A, Ayoub MA, et al. Current insights into immunology and novel therapeutics of atopic dermatitis. Cells. (2021) 10:1392. doi: 10.3390/cells10061392
49. Kim HE, Lee JY, Yoo DH, Park HH, Choi EJ, Nam KH, et al. Imidazole propionate ameliorates atopic dermatitis-like skin lesions by inhibiting mitochondrial ROS and mTORC2. Front Immunol. (2024) 15:1324026. doi: 10.3389/fimmu.2024.1324026
50. van den Bogaard EH, Podolsky MA, Smits JP, Cui X, John C, Gowda K, et al. Genetic and pharmacological analysis identifies a physiological role for the AHR in epidermal differentiation. J Invest Dermatol. (2015) 135:1320–8. doi: 10.1038/jid.2015.6
51. van den Bogaard EH, Bergboer JG, Vonk-Bergers M, van Vlijmen-Willems IM, Hato SV, van der Valk PG, et al. Coal tar induces AHR-dependent skin barrier repair in atopic dermatitis. J Clin Invest. (2013) 123:917–27. doi: 10.1172/JCI65642
52. Yu J, Luo Y, Zhu Z, Zhou Y, Sun L, Gao J, et al. A tryptophan metabolite of the skin microbiota attenuates inflammation in patients with atopic dermatitis through the aryl hydrocarbon receptor. J Allergy Clin Immunol. (2019) 143:2108–19.e12. doi: 10.1016/j.jaci.2018.11.036
53. Pappa G, Sgouros D, Theodoropoulos K, Kanelleas A, Bozi E, Gregoriou S, et al. The IL-4/-13 axis and its blocking in the treatment of atopic dermatitis. J Clin Med. (2022) 11:5633. doi: 10.3390/jcm11195633
54. Sorokman TV, Sokolnyk SV, Babiy OR, Lozyik IY, Sokolnyk SO, Makarova OV, et al. Immunological parameters and cortisol levels in children with atopic dermatitis. Arch Balkan Med Union. (2018) 53:210–6. doi: 10.31688/ABMU.2018.53.2.06
55. Packi K, Matysiak J, Klimczak S, Matuszewska E, Bręborowicz A, Pietkiewicz D, et al. Analysis of the serum profile of cytokines involved in the T-helper cell type 17 immune response pathway in atopic children with food allergy. Int J Environ Res Public Health. (2022) 19:7877. doi: 10.3390/ijerph19137877
56. Pan Y, Hochgerner M, Cichoń MA, Benezeder T, Bieber T, and Wolf P. Langerhans cells: Central players in the pathophysiology of atopic dermatitis. J Eur Acad Dermatol Venereol. (2025) 39:278–89. doi: 10.1111/jdv.20291
57. Castagnoli R, Pala F, Bosticardo M, Licari A, Delmonte OM, Villa A, et al. Gut microbiota-host interactions in inborn errors of immunity. Int J Mol Sci. (2021) 22:1416. doi: 10.3390/ijms22031416
58. Coates M, Blanchard S, and MacLeod AS. Innate antimicrobial immunity in the skin: A protective barrier against bacteria, viruses, and fungi. PloS Pathog. (2018) 14:e1007353. doi: 10.1371/journal.ppat.1007353
59. Tamminga SM, van der Wal MM, Saager ES, van der Gang LF, Boesjes CM, Hendriks A, et al. Single-cell sequencing of human Langerhans cells identifies altered gene expression profiles in patients with atopic dermatitis. Immunohorizons. (2025) 9:vlae009. doi: 10.1093/immhor/vlae009
60. Li C, Maillet I, Mackowiak C, Viala C, Di Padova F, Li M, et al. Experimental atopic dermatitis depends on IL-33R signaling via MyD88 in dendritic cells. Cell Death Dis. (2017) 8:e2735. doi: 10.1038/cddis.2017.90
61. Geoghegan JA, Irvine AD, and Foster TJ. Staphylococcus aureus and atopic dermatitis: A complex and evolving relationship. Trends Microbiol. (2018) 26:484–97. doi: 10.1016/j.tim.2017.11.008
62. Fyhrquist N, Muirhead G, Prast-Nielsen S, Jeanmougin M, Olah P, Skoog T, et al. Microbe-host interplay in atopic dermatitis and psoriasis. Nat Commun. (2019) 10:4703. doi: 10.1038/s41467-019-12253-y
63. Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. (2012) 22:850–9. doi: 10.1101/gr.131029.111
64. Choi H, Kwak MJ, Choi Y, Kang AN, Mun D, Eor JY, et al. Extracellular vesicles of Limosilactobacillus fermentum SLAM216 ameliorate skin symptoms of atopic dermatitis by regulating gut microbiome on serotonin metabolism. Gut Microbes. (2025) 17:2474256. doi: 10.1080/19490976.2025.2474256
65. Gomes PWP, Mannochio-Russo H, Mao J, Zhao HN, Ancira J, Tipton CD, et al. Co-occurrence network analysis reveals the alterations of the skin microbiome and metabolome in adults with mild to moderate atopic dermatitis. mSystems. (2024) 9:e0111923. doi: 10.1128/msystems.01119-23
66. Suwarsa O, Hazari MN, Dharmadji HP, Dwiyana RF, Effendi R, Hidayah RMN, et al. A pilot study: composition and diversity of 16S rRNA based skin bacterial microbiome in Indonesian atopic dermatitis population. Clin Cosmet Investig Dermatol. (2021) 14:1737–44. doi: 10.2147/CCID.S338550
67. Zhu Y, Yu X, and Cheng G. Human skin bacterial microbiota homeostasis: A delicate balance between health and disease. mLife. (2023) 2:107–20. doi: 10.1002/mlf2.12064
68. Liu A, Garrett S, Hong W, and Zhang J. Staphylococcus aureus infections and human intestinal microbiota. Pathogens. (2024) 13:276. doi: 10.3390/pathogens13040276
69. Ruchti F, Zwicky P, Becher B, Dubrac S, and LeibundGut-Landmann S. Epidermal barrier impairment predisposes for excessive growth of the allergy-associated yeast Malassezia on murine skin. Allergy. (2024) 79:1531–47. doi: 10.1111/all.16062
70. Qi C, Li Z, Tu H, Sun F, Guo W, Di C, et al. 2’-FL and cross-feeding bifidobacteria reshaped the gut microbiota of infants with atopic dermatitis ex vivo and prevented dermatitis in mice post-microbiota transplantation through retinol metabolism activation. Gut Microbes. (2025) 17:2474148. doi: 10.1080/19490976.2025.2474148
71. Jiao Y, Wu L, Huntington ND, and Zhang X. Crosstalk between gut microbiota and innate immunity and its implication in autoimmune diseases. Front Immunol. (2020) 11:282. doi: 10.3389/fimmu.2020.00282
72. Meisel JS, Sfyroera G, Bartow-McKenney C, Gimblet C, Bugayev J, Horwinski J, et al. Commensal microbiota modulate gene expression in the skin. Microbiome. (2018) 6:20. doi: 10.1186/s40168-018-0404-9
73. Luo Z, Chen A, Xie A, Liu X, Jiang S, and Yu R. Limosilactobacillus reuteri in immunomodulation: molecular mechanisms and potential applications. Front Immunol. (2023) 14:1228754. doi: 10.3389/fimmu.2023.1228754
74. Wang Y, Wang X, Zhu M, Ge L, Liu X, Su K, et al. The interplay between cervicovaginal microbial dysbiosis and cervicovaginal immunity. Front Immunol. (2022) 13:857299. doi: 10.3389/fimmu.2022.857299
75. Nguyen HLT, Trujillo-Paez JV, Umehara Y, Yue H, Peng G, Kiatsurayanon C, et al. Role of antimicrobial peptides in skin barrier repair in individuals with atopic dermatitis. Int J Mol Sci. (2020) 21:7607. doi: 10.3390/ijms21207607
76. Yang G, Seok JK, Kang HC, Cho YY, Lee HS, and Lee JY. Skin barrier abnormalities and immune dysfunction in atopic dermatitis. Int J Mol Sci. (2020) 21:2867. doi: 10.3390/ijms21082867
77. Kwon S, Choi JY, Shin JW, Huh CH, Park KC, Du MH, et al. Changes in lesional and non-lesional skin microbiome during treatment of atopic dermatitis. Acta Derm Venereol. (2019) 99:284–90. doi: 10.2340/00015555-3089
78. Bergmann S, von Buenau B, Vidal YSS, Haftek M, Wladykowski E, Houdek P, et al. Claudin-1 decrease impacts epidermal barrier function in atopic dermatitis lesions dose-dependently. Sci Rep. (2020) 10:2024. doi: 10.1038/s41598-020-58718-9
79. Ghezzi M, Pozzi E, Abbattista L, Lonoce L, Zuccotti GV, and D’Auria E. Barrier impairment and type 2 inflammation in allergic diseases: the pediatric perspective. Children (Basel). (2021) 8:1165. doi: 10.3390/children8121165
80. Zhou C, Hua C, Liang Q, Al Rudaisat M, Chen S, Song Y, et al. 0.5-5% Supramolecular salicylic acid hydrogel is safe for long-term topical application and improves the expression of genes related to skin barrier homeostasis in mice models. Drug Des Devel Ther. (2023) 17:1593–609. doi: 10.2147/DDDT.S397541
81. Senavonge A, Nakphaichit M, Vongsangnak W, Roytrakul S, Patumcharoenpol P, Kingkaw A, et al. Dysbiosis involving methionine and PPAR-γ pathways is associated with early onset atopic dermatitis and food allergy. Asian Pac J Allergy Immunol. (2025). doi: 10.12932/AP-131223-1749
82. Mhatre D, Parab M, Nikam T, and Gupta R. Skin deep: insights into atopic dermatitis. IJFMR. (2024) 6:18132. doi: 10.36948/ijfmr.2024.v06i02.18132
83. Hrestak D, Matijašić M, Čipčić Paljetak H, Ledić Drvar D, Ljubojević Hadžavdić S, and Perić M. Skin microbiota in atopic dermatitis. Int J Mol Sci. (2022) 23:3503. doi: 10.3390/ijms23073503
84. Pavel P, Blunder S, Moosbrugger-Martinz V, Elias PM, and Dubrac S. Atopic dermatitis: the fate of the fat. Int J Mol Sci. (2022) 23:2121. doi: 10.3390/ijms23042121
85. Schäbitz A, Eyerich K, and Garzorz-Stark N. So close, and yet so far away: The dichotomy of the specific immune response and inflammation in psoriasis and atopic dermatitis. J Intern Med. (2021) 290:27–39. doi: 10.1111/joim.13235
86. Di Domenico EG, Cavallo I, Capitanio B, Ascenzioni F, Pimpinelli F, Morrone A, et al. Staphylococcus aureus and the cutaneous microbiota biofilms in the pathogenesis of atopic dermatitis. Microorganisms. (2019) 7:301. doi: 10.3390/microorganisms7090301
87. Sroka-Tomaszewska J and Trzeciak M. Molecular mechanisms of atopic dermatitis pathogenesis. Int J Mol Sci. (2021) 22:4130. doi: 10.3390/ijms22084130
88. Hu Y, Zhao Y, Li H, Zhou C, Yu C, Mu Z, et al. Good efficacy and safety profile of abrocitinib in Chinese adult patients with atopic dermatitis: A case series study. Chin Med J (Engl). (2024) 137:740–2. doi: 10.1097/CM9.0000000000002838
89. Chen S, Li C, Tu Z, Cai T, Zhang X, Wang L, et al. Off-label use of Baricitinib improves moderate and severe atopic dermatitis in China through inhibiting MAPK and PI3K/Akt/mTOR pathway via targeting JAK-STAT signaling of CD4(+) cells. Front Pharmacol. (2024) 15:1324892. doi: 10.3389/fphar.2024.1324892
90. Gong X, Chen X, Kuligowski ME, Liu X, Liu X, Cimino E, et al. Pharmacokinetics of ruxolitinib in patients with atopic dermatitis treated with ruxolitinib cream: data from phase II and III studies. Am J Clin Dermatol. (2021) 22:555–66. doi: 10.1007/s40257-021-00610-x
91. Ortoncelli M, Macagno N, Mastorino L, Gelato F, Richiardi I, Cavaliere G, et al. Long-term efficacy and safety of dupilumab in patients with atopic dermatitis: A Single-Centre Retrospective Study. Cosmetics. (2023) 10:153. doi: 10.3390/cosmetics10060153
92. Hasan I, Parsons L, Duran S, and Zinn Z. Dupilumab therapy for atopic dermatitis is associated with increased risk of cutaneous T cell lymphoma: A retrospective cohort study. J Am Acad Dermatol. (2024) 91:255–8. doi: 10.1016/j.jaad.2024.03.039
93. Fijan S, Kolč N, Hrašovec M, Jamtvedt G, Pogačar M, Mičetić Turk D, et al. Single-strain probiotic lactobacilli for the treatment of atopic dermatitis in children: A systematic review and meta-analysis. Pharmaceutics. (2023) 15:1256. doi: 10.3390/pharmaceutics15041256
94. Climent E, Martinez-Blanch JF, Llobregat L, Ruzafa-Costas B, Carrión-Gutiérrez M, Ramírez-Boscá A, et al. Changes in gut microbiota correlates with response to treatment with probiotics in patients with atopic dermatitis. A post hoc analysis of a clinical trial. Microorganisms. (2021) 9:854. doi: 10.3390/microorganisms9040854
95. Untersmayr E, Brandt A, Koidl L, and Bergheim I. The intestinal barrier dysfunction as driving factor of inflammaging. Nutrients. (2022) 14:949. doi: 10.3390/nu14050949
96. Conn KA, Borsom EM, and Cope EK. Implications of microbe-derived γ-aminobutyric acid (GABA) in gut and brain barrier integrity and GABAergic signaling in Alzheimer’s disease. Gut Microbes. (2024) 16:2371950. doi: 10.1080/19490976.2024.2371950
97. López-Villodres JA, Escamilla A, Mercado-Sáenz S, Alba-Tercedor C, Rodriguez-Perez LM, Arranz-Salas I, et al. Microbiome alterations and alzheimer’s disease: modeling strategies with transgenic mice. Biomedicines. (2023) 11:1846. doi: 10.3390/biomedicines11071846
98. Nakatsuji T, Chen TH, Narala S, Chun KA, Two AM, Yun T, et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci Transl Med. (2017) 9:eaah4680. doi: 10.1126/scitranslmed.aah4680
99. Lai Y, Cogen AL, Radek KA, Park HJ, Macleod DT, Leichtle A, et al. Activation of TLR2 by a small molecule produced by Staphylococcus epidermidis increases antimicrobial defense against bacterial skin infections. J Invest Dermatol. (2010) 130:2211–21. doi: 10.1038/jid.2010.123
100. Myles IA, Castillo CR, Barbian KD, Kanakabandi K, Virtaneva K, Fitzmeyer E, et al. Therapeutic responses to Roseomonas mucosa in atopic dermatitis may involve lipid-mediated TNF-related epithelial repair. Sci Transl Med. (2020) 12:eaaz8631. doi: 10.1126/scitranslmed.aaz8631
101. Nakatsuji T, Hata TR, Tong Y, Cheng JY, Shafiq F, Butcher AM, et al. Development of a human skin commensal microbe for bacteriotherapy of atopic dermatitis and use in a phase 1 randomized clinical trial. Nat Med. (2021) 27:700–9. doi: 10.1038/s41591-021-01256-2
102. de Wit J, Totté JEE, van Mierlo MMF, van Veldhuizen J, van Doorn MBA, Schuren FHJ, et al. Endolysin treatment against Staphylococcus aureus in adults with atopic dermatitis: A randomized controlled trial. J Allergy Clin Immunol. (2019) 144:860–3. doi: 10.1016/j.jaci.2019.05.020
Keywords: atopic dermatitis (AD), skin microbiome, symbiotic bacteria, immune, autoimmune skin disease
Citation: Lai X, Huang J, Li Y and Dong L (2025) Symbiotic bacteria-mediated imbalance and repair of immune homeostasis: exploring novel mechanisms of microbiome-host interactions in atopic dermatitis. Front. Immunol. 16:1649857. doi: 10.3389/fimmu.2025.1649857
Received: 20 June 2025; Accepted: 10 July 2025;
Published: 23 July 2025.
Edited by:
Daniel P. Potaczek, University of Marburg, GermanyReviewed by:
Athanasios A. Anastasiou, National Technical University of Athens, GreeceAaroh Anand Joshi, Medical University of Graz, Austria
Dejan Bezbradica, University of Belgrade, Serbia
Chiara Maria Teresa Boggio, Università del Piemonte Orientale, Italy
Copyright © 2025 Lai, Huang, Li and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yulin Li, bGl5dWxpbkBjZHV0Y20uZWR1LmNu; Liang Dong, ZG9uZ2xpYW5nQGNkdXRjbS5lZHUuY24=
 Xingyue Lai
Xingyue Lai Jilin Huang
Jilin Huang Liang Dong
Liang Dong