Abstract
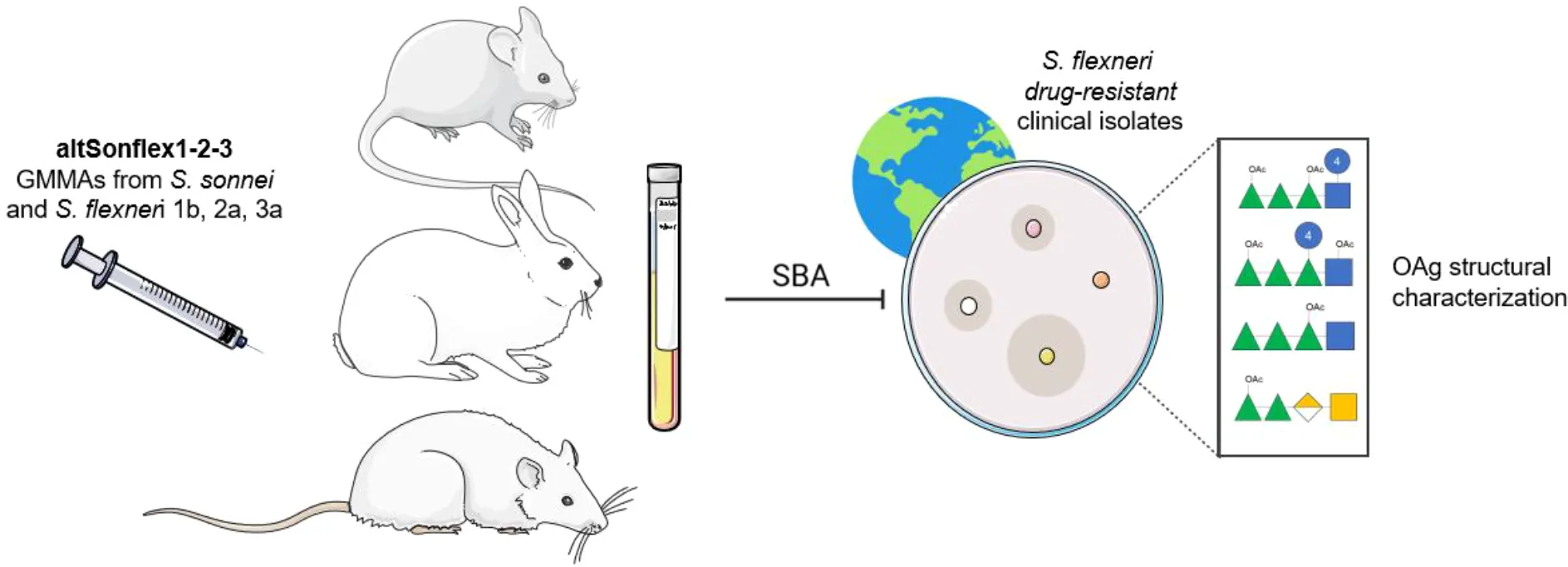
Shigellosis significantly impacts global health, particularly affecting vulnerable populations in low- and middle-income countries, with over 270 million annual infections. It also causes morbidity in specific high-risk groups in high-income countries. Antibiotic treatment is increasingly compromised by multidrug-resistant strains, highlighting the urgent need for a Shigella vaccine. We developed a four-component O-antigen-based vaccine candidate targeting Shigella sonnei and Shigella flexneri 1b, 2a, and 3a serotypes, named altSonflex1-2-3, to provide broad protection against most globally prevalent Shigella strains. Here, we characterized the O-antigen structural features of a panel of S. flexneri drug-resistant clinical isolates and verified that they did not significantly differ from the O-antigen in the vaccine. Preclinical sera elicited by altSonflex1-2–3 were bactericidal against most strains, confirming the ability of anti-O-antigen antibodies to recognize and kill in vitro different clinical isolates. Importantly, our results suggest that altSonflex1-2–3 could offer protection against antimicrobial-resistant Shigella strains, addressing a critical public health issue.
The artwork used in this figure was adapted from Servier Medical Art (http://https://smart.servier.com/). Servier Medical Art by Servier is licensed under a CC BY 4.0 License.
Introduction
Shigella spp. are the etiological agents of shigellosis, a diarrheal disease caused by bacterial infection of the colonic epithelium through oral–fecal transmission (1–3). Shigellosis is the second leading cause of enteric disease mortality worldwide after rotavirus disease, with over 270 million infections and 148,000 fatalities annually, predominantly affecting children under 5 years of age and individuals over 70 years old in low- and middle-income countries (LMICs) (4–6). Beyond mortality, shigellosis leads to growth retardation, cognitive development issues, and increased susceptibility to other infections (7, 8).
In high-income countries (HICs), shigellosis typically presents as a self-limited illness that resolves without medical intervention. However, severe cases are consistently reported to health authorities each year. The European Centre for Disease Prevention and Control (ECDC) reported 4,149 cases in 2022 across 30 European countries (1.5 cases per 100,000 people). These cases are predominantly concentrated in certain risk groups such as travelers, persons experiencing homelessness, and gay, bisexual, and other men who have sex with men (gbMSM) in which sexually transmitted shigellosis is emerging (9).
Shigellosis can be caused by four species: in order of frequency, Shigella flexneri (predominant in LMICs), Shigella sonnei (second most frequent in LMICs and first most frequent in HICs), Shigella dysenteriae serotype 1 (predominant in pandemics), and Shigella boydii (10). Antibiotics are the only available treatment for severe shigellosis to alleviate symptoms, reduce disease duration, and prevent transmission. The WHO treatment recommendations include oral ciprofloxacin (a fluoroquinolone) as the first-choice antibiotic after ampicillin was discontinued due to bacterial resistance, and ceftriaxone (third-generation cephalosporin) or azithromycin as alternatives (11). Worryingly, both S. flexneri and S. sonnei, the two species most frequently implicated in shigellosis, have developed multidrug-resistant (MDR) profiles, and some of these species have been implicated in recent shigellosis outbreaks (9, 12–14). Fluoroquinolone-resistant Shigella species have been prioritized as high-risk pathogens in the WHO Bacterial Priority Pathogens List 2024 update (15).
Vaccines are crucial in mitigating antimicrobial resistance (AMR) by preventing infections and thus reducing antibiotic use (16–18). Although a commercial vaccine for shigellosis is not yet available, candidates are under development, many including the O-antigen (OAg) component of the lipopolysaccharide, an established protective antigen (19). However, considering the structural diversity of OAg across Shigella species and serotypes, a multivalent vaccine approach is necessary to increase coverage (19–22). While S. sonnei has a unique OAg structure, all S. flexneri serotypes, except S. flexneri 6 (23), share the following common OAg tetrasaccharide backbone (corresponding to serotype Y) decorated with glucosylation and O-acetylation in different positions that confer serotype specificity (24).
(→2)-α-l-RhapIII-(1→2)-α-l-RhapII-(1→3)-α-l-RhapI-(1→3)-β-d-GlcpNAc-(1→).
Structural similarities among S. flexneri serotypes have been shown to induce cross-reactive immune responses, but the structural features driving immune cross-reactivity remain unclear (1, 25).
A multivalent OAg-based Shigella vaccine [named altSonflex1-2-3 (26)] is progressing into Phase 2 trials. This vaccine contains Generalized Modules for Membrane Antigens (GMMA), engineered outer membrane vesicles of S. flexneri 2a, S. flexneri 1b, S. flexneri 3a, and S. sonnei, displaying native OAg. It aims to cover over two-thirds of shigellosis cases in Africa and Asia by inducing an immune response against homologous serotypes, plus potential cross-reactivity against S. flexneri serotypes not included in the vaccine (3, 27). We have collected preclinical and clinical evidence that altSonflex1-2–3 induces functional antibodies against a panel of homologous and heterologous laboratory strains (3, 26). However, information on the functionality of sera against real-world circulating strains is lacking.
Here, we present a panel of S. flexneri clinical isolates from high-income countries displaying different OAg serotypes and AMR profiles. For each strain, we performed an in-depth characterization of the OAg characteristics, the main antigen addressed by the vaccine-induced immune response, to understand eventual structural variability compared to the OAg displayed by GMMA in the altSonflex1-2–3 vaccine.
Furthermore, we explored the ability of multi-species preclinical sera to induce complement-mediated bactericidal activity against this panel of clinical isolates. Serum bactericidal activity represents a well-established in vitro functional assay in the field to complement the assessment of the anti-OAg response (22, 28). Without a correlate of protection established for Shigella, bactericidal activity has been suggested as an important measure of the functional activity of vaccine-induced antibodies (29, 30).
We assessed the bactericidal activity of pooled sera of animals vaccinated with altSonflex1-2–3 against these clinical drug-resistant isolates, demonstrating vaccine-induced titers comparable to those observed against the same strains in adult individuals in endemic populations, where natural exposure has been associated with protection (31, 32).
Results and discussion
Characterization of the OAg structural features of a panel of S. flexneri clinical isolates
We collected 12 S. flexneri clinical isolates from various locations between 2017 and 2023. Two international, MSM-related clusters are related to two EpiPulse alerts (Table 1). For all strains, we tested antibiotic resistance against ampicillin, ceftriaxone, ciprofloxacin, gentamicin, azithromycin, trimethoprim–sulfamethoxazole, meropenem, tetracycline, and chloramphenicol. We found all strains to be partially or fully resistant to at least one antibiotic, with ampicillin resistance being the most common, as reported in the literature.
Table 1
| Identification code | Serotype | Isolation year | Isolation location | Risk group | References | Ampicillin | Ceftriaxone | Ciprofloxacin | Gentamycin | Azithromycin | Trimethoprim–sulfamethoxazole | Meropenem | Tetracycline | Chloramphenicol |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 19096 | Y* | 2019 | Seattle, USA | gbMSM | 11 | |||||||||
| 210059 | 1b* | 2021 | Seattle, USA | gbMSM | 11 | |||||||||
| I03 (75) | 3b* | 2009 | Italy | – | ||||||||||
| 6A (18) | 3b* | 2017 | Italy | – | ||||||||||
| 13B (76) | 1b* | 2023 | Italy | – | ||||||||||
| S23BD04932 | 2a** | 2023 | Belgium | gbMSM | 2023-EIP-0023 (1) International alert | |||||||||
| S23BD05840 | 2a** | 2023 | Belgium | gbMSM | 2023-EIP-0023 (2) International alert | |||||||||
| S23BD08049 | 2a** | 2023 | Belgium | gbMSM | National cluster Flex_1 | |||||||||
| S23BD02869 | 1b** | 2023 | Belgium | gbMSM | 2023-EIP-0023 (3) | |||||||||
| S23BD06925 | 3b** | 2023 | Belgium | gbMSM | National cluster Flex_3 | |||||||||
| S23BD10088 | 6** | 2023 | Belgium | gbMSM | National cluster Flex_4 | |||||||||
| S23BD10089 | 4** | 2023 | Belgium | gbMSM | National cluster Flex_5 |
Shigella flexneri clinical isolate details and antibiotic resistance profile.
Identification code identifies strains as per related references or per identifier reported in the database of sources (Lazzaro Spallanzani Hospital or Sciensano). Serotypes are reported as follows: *Unknown serotype, analyzed in this paper by chemical characterization; **Declared serotype, confirmed in this paper by chemical characterization. Information on strains and antibiotic resistance profiles assayed by Kirby–Bauer method according to European Committee on Antimicrobial Susceptibility Testing (EUCAST).
Red = resistant, yellow = intermediate, and white = sensitive. gbMSM, gay, bisexual, and other men who have sex with men.
OAg was extracted from the 12 S. flexneri strains and characterized in terms of polysaccharide identity, chain length, quantity, and percentage/position of O-acetyl groups (Table 2, Figure 1). By growing all strains under similar conditions, the amount of OAg produced ranged from 242 µg of OAg per optical density (OD) for the S. flexneri 2a strain S23BD05840 (S. flexneri 2a) to 2,036 µg of OAg per OD for the S. flexneri 2a isolate S23BD08049 (S. flexneri 2a). Nuclear magnetic resonance (NMR) spectroscopy was used to confirm the OAg structure of all known serotypes and to identify one of the unknown isolates, comparing the anomeric region of the spectra with previously published data (Supplementary Figures 1-4) (33). Unknown serotypes were identified as follows: one S. flexneri Y (19096), two S. flexneri 1b [210059 and 13B(76)], and two S. flexneri 3b [I03(75) and 6A(18)]. All isolates except S. flexneri 6 S23BD10088 displayed two main OAg populations with different molecular size distributions: a high-molecular-weight (HMW) population at 48.6–60.5 kDa and a medium-molecular-weight (MMW) population at 13.5–16.1 kDa. Interestingly, the S. flexneri 6 strain exhibited MMW OAg only, with no peak related to the presence of the capsular polysaccharide detected (34). All isolates, with the exception of S. flexneri 4a S23BD10089 (as expected), were O-acetylated, and the details on % and positions of O-acetyl groups are reported in Table 2; Supplementary Figures 5-8. Interestingly, all S. flexneri 3b OAgs presented a low percentage of an additional O-acetyl group with respect to the O-acetyl on RhaI.
Table 2
| Isolate ID | OAg structure | OAg quantity | Molecular size distribution kDa and peak area % (HPLC-SEC dRI) | OAc | |||
|---|---|---|---|---|---|---|---|
| (1H-NMR/HPAEC-PAD) | µg/OD | % | Molar ratio | ||||
| (HPAEC-PAD) | HMW | MMW | (µHestrin) | (1H-NMR) | |||
| 19096 | Y |
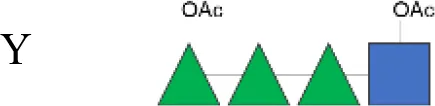
|
860 | 56.6 (67%) | 15.7 (33%) | 38 | 3.4 (RhaIII 4OAc + GlcNAc 6OAc):1 (RhaIII 3OAc) |
| Shigella flexneri 1b OAg from GMMA (vaccine) | 1b |
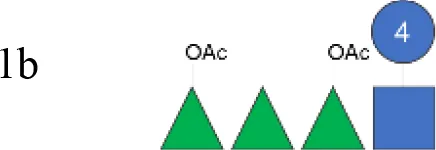
|
na | nd | 14.0 (100%) | 150 | 2.4 (RhaI 2OAc):2.2 (RhaIII 3OAc):1 (RhaIII 4OAc) |
| 210059 | 1b |

|
531 | 53.7 (73%) | 15.2 (27%) | 112 | 8.4 (RhaI 2OAc):2.4 (RhaIII 3OAc):1 (RhaIII 4OAc) |
| 13B (76) | 1b |

|
1,445 | 53.9 (78%) | 16.1 (22%) | 123 | 4.1 (RhaI 2OAc):1.1 (RhaIII 3OAc):1 (RhaIII 4OAc) |
| S23BD02869 | 1b |

|
1,241 | 48.6 (42%) | 14.4 (58%) | 91 | 5.7 (RhaI 2OAc):2.3 (RhaIII 3OAc):1 (RhaIII 4OAc) |
| S. flexneri 2a OAg from GMMA (vaccine) | 2a |
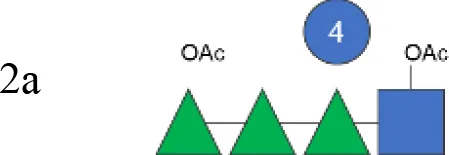
|
na | 47.0 (46%) | 14.0 (54%) | 147 | 1.2 (RhaIII 4OAc + GlcNAc 6OAc):1 (RhaIII 3OAc) |
| S23BD04932 | 2a |

|
1,390 | 50.1 (65%) | 14.1 (35%) | 41 | 1.6 (RhaIII 4OAc + GlcNAc 6OAc):1 (RhaIII 3OAc) |
| S23BD05840 | 2a |

|
242 | 50.2 (68%) | 14.0 (32%) | 49 | 2.3 (RhaIII 4OAc + GlcNAc 6OAc):1 (RhaIII 3OAc) |
| S23BD08049 | 2a |

|
2,036 | 52.6 (49%) | 13.7 (51%) | 37 | 1.9 (RhaIII 4OAc + GlcNAc 6OAc):1 (RhaIII 3OAc) |
| S. flexneri 3a OAg from GMMA (vaccine) | 3a |
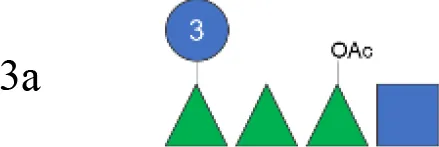
|
na | nd | 16.0 (100%) | 102 | 1 (RhaI 2OAc) |
| I03 (75) | 3b |

|
1,282 | 56.3 (60%) | 13.8 (40%) | 136 | 3.8 (RhaI 2OAc):1 (unknown) |
| 6A (18) | 3b |

|
1,020 | 52.5 (45%) | 13.5 (55%) | 156 | 3.3 (RhaI 2OAc):1 (unknown) |
| S23BD06925 | 3b |

|
1,760 | 50.7 (43%) | 13.7 (57%) | 116 | 2.8 (RhaI 2OAc):1 (unknown) |
| S23BD10089 | 4a |
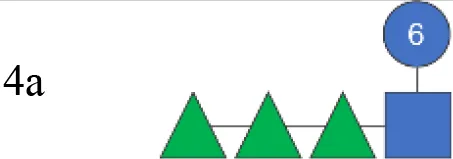
|
1,551 | 60.5 (55%) | 16.0 (45%) | 0 | No OAc |
| S23BD10088 | 6 |
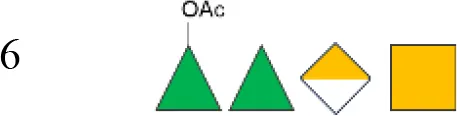
|
665 | nd | 20.8 (100%) | 29 | 1.2 (RhaIII 3OAc):1 (RhaIII 4OAc) |
Characterization of O-antigen features in clinical isolates and GMMA composing the altSonflex1-2–3 vaccine.
nd, not detected; na, not applicable; GMMA, Generalized Modules for Membrane Antigens; OAg, O-antigen; OD, optical density; HMW, high molecular weight; MMW, medium molecular weight; and HPLC-SEC dRI, high-performance liquid chromatography–size exclusion chromatography with differential refractive index detector.
Figure 1

OAg structural features of clinical isolates and GMMA. OAg molecular size distribution of each isolate and O-acetylation % are reported on the y-axis: single symbols corresponding to molecular weight (MW) in kDa of each OAg population and OAc % represented by the bars. The OAg peak area population is represented by the different sizes of the symbols. Color codes are associated with the OAg amount normalized per optical density (OD). The different symbols are associated with the provenience of the isolates or GMMA. OAg, O-antigen; GMMA, Generalized Modules for Membrane Antigens.
Isolates expressing the same OAg [S. flexneri 1b 210059, 13B(76), and S23BD02869; S. flexneri 2a S23BD04932, S23BD05840, and S23BD08049; S. flexneri 3b I03(75), 6A(18), and S23BD06925] showed similar structural features, despite producing different amounts of OAg (Table 2, Figure 1).
To verify possible structural differences, the S. flexneri 1b and 2a OAgs extracted from the circulating isolates were compared with the homologous OAg displayed on S. flexneri GMMA in the altSonflex1-2–3 vaccine. Interestingly, few differences were observed in terms of both size and O-acetylation levels. In particular, the HMW population was not found in the S. flexneri 1b OAg displayed by GMMA, which showed the MMW peak only, while both HMW and MMW OAgs were present in S. flexneri 2a OAg-GMMA (Table 2). Moreover, even if the total O-acetylation percentage was similar, all the S. flexneri 1b OAgs from the clinical isolates were primarily O-acetylated at position 2 of the RhaI, while the same OAg on GMMA presented O-acetylation also in position 3 of RhaIII, with a similar ratio between the two positions. All the S. flexneri 2a OAgs were instead O-acetylated in the same positions with similar ratios, but the overall % of O-acetylation (OAc) was different, with the OAg on GMMA being more O-acetylated (147%) compared to clinical strains (37%–49%) (Table 2; Supplementary Figures 5-8).
Functionality of immune sera against a panel of drug-resistant clinical isolates from recent outbreaks
To prove the potential effectiveness of altSonflex1-2–3 against real-world strains, we tested the bactericidal activity of immune sera derived from three altSonflex1-2-3-vaccinated preclinical models (mice, rabbits, and rats) via luminescence-based serum bactericidal activity (L-SBA) assay (Figure 2). As a comparison, a pool of sera from non-vaccinated Kenyan adults, who were considered protected from shigellosis upon repeated exposures throughout their lives, were also tested in L-SBA against the panel of isolates, as a comparison.
Figure 2

Bactericidal activity of preclinical sera against clinical isolates. Bactericidal activity expressed in IC50 values for each clinical isolate tested with serum of altSonflex1-2-3-immunized mice, rabbits, and rats. IC50 of sera from Kenyan adults from endemic settings is reported for comparison. Three biological replicates for each strain are represented, with central line indicating the geometric mean of the IC50s for each strain.
All sera were bactericidal against all tested S. flexneri 1b strains, confirming vaccine activity against the homologous strain. When tested against S. flexneri 2a strains, all sera were bactericidal against all strains except S23BD05840. However, sera from Kenyan adults were also unable to kill this strain, confirming its resistance. As a possible explanation for this observation, strain S23BD05840 was found to be particularly resistant to complement intrinsic toxicity. Also, OAg characterization showed that this strain produced less OAg compared to the other strains. Since LPS is usually considered to protect bacteria from complement deposition, we may hypothesize that the resistance of this strain may be due to other virulence factors.
Figure 2 also reports bactericidal activity against heterologous strains not included in the vaccine. Importantly, all sera demonstrated bactericidal activity against S. flexneri 3b, 4a, and Y strains. Activity against strain 3b can be explained by a very close structural similarity to serotype 3a present in the vaccine. Indeed, the two OAgs only differ in the presence of an additional Glc on RhaIII in the RU of S. flexneri 3a, compared to S. flexneri 3b. Bactericidal activity is expected against S. flexneri Y, whose OAg comprises a tetrasaccharide shared by all S. flexneri serotypes, with minor O-acetylation. Bactericidal activity against S. flexneri 4a was also observed, albeit with higher variability compared to that of the other strains. Since this was not observed with 4a laboratory strains (data not shown), we hypothesize this could be related to the extent of growth variability of this specific clinical isolate under the conditions used for SBA. All preclinical sera from vaccinated animals displayed lower or no bactericidal activity against the S. flexneri 6 isolate, compared with the sera from Kenyan adults.
For all strains tested, except the S. flexneri 6 strain, titers of preclinical sera were similar to those elicited from human sera. In conclusion, our results suggest that altSonflex1-2–3 could offer protection against a panel of homologous and heterologous antimicrobial-resistant Shigella strains, with a strong potential impact on global health. altSonflex1-2–3 is being tested in Phase 2 clinical trials (NCT06663436 and NCT05073003), and the functionality of sera from vaccinated infants will be tested against the panel of drug-resistant clinical isolates as well to confirm preclinical data. Testing in the Phase 3 clinical trial will confirm the ability of the vaccine to protect in the field.
Methods
Animal sera
GlaxoSmithKline (GSK) is committed to the Replacement, Reduction, and Refinement (3Rs) of animal studies. Non-animal models and alternative technologies are part of our strategy and are employed where possible. When animals are required, the application of robust study design principles and peer review minimizes animal use, reduces harm, and improves benefits in studies. Animal studies were performed in the GSK Animal Facility (Siena, Italy) or at Charles River Laboratories (France), in compliance with the relevant guidelines (Italian D.Lgs. n. 26/14 and European directive 2010/63/UE) and the institutional policies of GSK. The animal protocol approved by the Italian Ministry of Health was AWB project No. 643/2021-PR, with approval date 12/08/2021.
Mouse serum was pooled from 10 female CD1 mice vaccinated intramuscularly (IM) with 50 µL of altSonflex1-2–3 at a dose of 6 μg of total OAg (1.5 μg per serotype) with Alhydrogel. Rabbit serum was pooled from eight female New Zealand White rabbits vaccinated IM with 500 µL of altSonflex1-2–3 at a dose of 60 µg of total OAg (15 μg per serotype) with Alhydrogel. Rat serum was pooled from eight Sprague–Dawley rats vaccinated IM with 200 µL of altSonflex1-2–3 at a dose of 24 µg of OAg (6 µg per serotype) with Alhydrogel. All animals were immunized at study days 0 and 28 and were bled on day 42. All sera were stored frozen in 50 µL working aliquots at −80°C until use. All samples tested in SBA were previously heat-inactivated at 56°C for 30 min to remove endogenous complement activity.
Clinical sera
The human serum tested was a pool of preimmune sera of 20 Kenyan adults enrolled in the safety cohort of H06_01TP Stage 2 (NCT05073003). All sera were stored frozen in 50 µL working aliquots at −80°C until use. All samples tested in SBA were previously heat-inactivated at 56°C for 30 min to remove endogenous complement activity.
Bacterial strains
Frozen 20% glycerol stocks were prepared from all S. flexneri strains and stored at −80°C until use. To perform experiments, isolates were inoculated from the glycerol stock into a 5-mL Luria-Bertani (LB) liquid culture in preparation for either large-scale growth for OAg extraction or small-scale growth for SBA.
O-antigen isolation and characterization
A 5-mL LB over-day inoculum of each strain was used to seed a 250-mL LB overnight culture. OD600nm ranged from 1.5 for strain 13B(76) to 8 for strain S23BD05840. OAg was extracted from the bacterial pellet by hydrolysis with 2% acetic acid at 100°C for 4 h and filter-sterilized. The extracted OAgs were purified as previously reported, with slight modifications (35). The samples were subjected to buffer exchange and removal of lower-molecular-mass impurities by ultrafiltration using Amicon Ultra Centrifugal Filter 10 kDa cut-off (Merck, Darmstadt, Germany) against water (3,500 rcf, 10 min, 2 cycles). Next, 25 mM sodium acetate buffer at pH 3.7 was added, reaching a final acetate concentration of 2.5 mM, to precipitate possible protein impurities. After mixing at room temperature (RT) for 30 min, the supernatants were collected through centrifugation and filtered using cation exchange (CEX) chromatography with Sartobind S MA75 filters (Sartorius, Göttingen, Germany) to further remove residual protein impurities. After pH neutralization, the OAg was further purified via anion exchange (AEX) chromatography using a HiTrap Q FF column (Cytiva, Chicago, IL, USA) to remove nucleic acid impurities. All the OAgs eluted at 25 mM NaCl, except the negatively charged S. flexneri 6, which eluted at 200 mM NaCl. The resulting solutions were used to calculate the OAg molecular weight using dextrans as standards, as previously reported (36). A final purification step using Amicon Ultra Centrifugal Filter 10 kDa cut-off against water (3,500 rcf, 10 min, 3 cycles) was performed to remove the lipid A core.
OAgs were extracted from GMMA as previously reported (37) and directly used for characterization.
The OAgs were characterized in terms of O-acetyl ester content, performing a modified microplate procedure of the Hestrin assay (38), and the OAg sugar composition was estimated by High-Performance Anion-Exchange Chromatography with Pulsed Amperometric Detection (HPAEC-PAD) analysis (36). OAg structures were further confirmed by 1H-NMR analysis and measured using a Bruker Avance III 400 spectrometer at 400 MHz and 298 K 1H-NMR was also used to determine the position of O-acetyl groups.
OAg amounts were quantified by HPAEC-PAD (36) after the first ultrafiltration step (S23BD04932, S23BD05840, S23BD08049, S23BD02869, S23BD06925, S23BD10088, and S23BD10089 OAgs) or after AEX step [19096, 210059, I03(75), 6A(18), and 13B(76) OAg].
Serum bactericidal assay with luminescence readout
The serum bactericidal assay based on luminescent readout (L-SBA) was conducted as previously described (3, 39, 40). Briefly, sera were serially diluted threefold in Phosphate Buffered Saline (PBS) starting from a 1/50 dilution (25 µL/well). Negative controls without serum were included to monitor non-specific complement killing. Meanwhile, bacteria were cultured to log phase (OD600nm = 0.20 ± 0.02), diluted in the appropriate buffer to 1 × 106 CFU/mL, mixed with baby rabbit complement (BRC) (Cedarlane, Southern Ontario, Canada, CL 3441-S) (Supplementary Table 1) to a final volume of 100 μL/well, and incubated at 37°C for 3 h. Bacteria were pelleted and resuspended in 100 μL PBS to remove complement-lysed bacteria. Finally, 25 μL of resuspended live bacteria was mixed with 25 μL of BacTiter-Glo (Promega, Madison, Wisconsin G8231) in a white 384-well plate (Greiner Bio-One, Monroe, North Carolina 655904). The reagents lyse live bacteria, which release ATP and enable bacterial quantification. After 5-min incubation on an orbital shaker at 300 rpm in the dark and RT, ATP-induced luminescence was acquired using a luminometer (Synergy HT, Biotek, Swindon, UK), which correlated with the number of living bacteria in the wells.
Relative luminescence values were plotted in Prism (GraphPad, version 9.5.3) and fitted on the logarithmic value of the serum dilutions tested for each serum in a four-parameter logistic regression model; an arbitrary serum concentration of 10−15 was assigned to the well containing no serum (39). To allow correct fitting, the curves were constrained to have a bottom between 0 and the mean of three counts per second corresponding to the lower luminescence detected at T180 in wells where bacteria were killed. The results of the assay are expressed as IC50, representing the dilution of serum able to mediate 50% of killing efficiency with respect to the maximal value. Three independent experiments were performed for each serum on each strain.
The optimal BRC concentration for each strain was established by performing a comprehensive experiment that assessed bacterial growth in a range of increasing BRC percentages in the absence of serum. The selected concentration was determined as the one that most closely resembled the conditions without BRC, wherein the positive impact on bacterial growth was counterbalanced by its inherent toxicity (Supplementary Table 1).
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by Walter Reed Army Institute of Research (WRAIR, Silver Springs, USA), by the KEMRI Scientific Ethic Review Unit (SERU, Kenya) and conducted as per Good Clinical Practice guidelines. For human serum standard derived from study NCT05073003, its creation and use for research related to Shigella disease and assays, was further approved by KEMRI SERU (41). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. The animal studies were approved by GSK. GSK is committed to the Replacement, Reduction and Refinement of animal studies (3Rs). Non-animal models and alternative technologies are part of our strategy and employed where possible. When animals are required, application of robust study design principles and peer review minimises animal use, reduces harm and improves benefit in studies. Animal studies were performed in the GSK Animal Facility (Siena, Italy) or at Charles River Laboratories (France), in compliance with the relevant guidelines (Italian D.Lgs. n. 26/14 and European directive 2010/63/UE) and the institutional policies of GSK. The animal protocol approved by the Italian Ministry of Health was the AWB project No. 643/2021-PR, approval date 12/08/2021. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
EB: Formal analysis, Investigation, Writing – original draft, Writing – review & editing. RB: Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. GV: Formal analysis, Investigation, Writing – review & editing. RA: Formal analysis, Investigation, Writing – review & editing. P-JC: Investigation, Resources, Writing – review & editing. GT: Investigation, Resources, Writing – review & editing. FF: Formal analysis, Resources, Writing – review & editing. CF: Formal analysis, Resources, Writing – review & editing. AR: Resources, Writing – review & editing. SC: Resources, Writing – review & editing. FMa: Formal analysis, Supervision, Writing – review & editing. MI: Supervision, Writing – review & editing. CS: Resources, Writing – review & editing. CG: Formal analysis, Supervision, Writing – review & editing. OR: Formal analysis, Supervision, Writing – original draft, Writing – review & editing. FMi: Conceptualization, Formal analysis, Project administration, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, and/or publication of this article.
Acknowledgments
We would like to thank all investigators and volunteers for their participation in the study NCT05073003.
Conflict of interest
This work was undertaken at the request of and sponsored and funded by GlaxoSmithKline Biologicals SA. GSK Vaccines Institute for Global Health Srl is an affiliate of GlaxoSmithKline Biologicals. EB, GV, RB, FMa, CG, MI, OR, and FMi are employees of the GSK group of companies. CG, MI, OR, and FMi report ownership of GSK shares/share options.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1652460/full#supplementary-material
Abbreviations
AMR, antimicrobial resistance; gbMSM, gay, bisexual, and other men who have sex with men; GMMA, Generalized Modules for Membrane Antigens; HICs, high-income countries; IC50, half inhibitory concentration; Ig, immunoglobulin; LMICs, low- and middle-income countries; LPS, lipopolysaccharide; L-SBA, serum bactericidal assay with luminescence readout; NMR, nuclear magnetic resonance; OAg, O-antigen.
References
1
Citiulo F Necchi F Mancini F Rossi O Aruta MG Gasperini G et al . Rationalizing the design of a broad coverage Shigella vaccine based on evaluation of immunological cross-reactivity among S. flexneri serotypes. PloS Negl Trop Dis. (2021) 15:e0009826. doi: 10.1371/journal.pntd.0009826
2
Rossi O Citiulo F Giannelli C Cappelletti E Gasperini G Mancini F et al . A next-generation GMMA-based vaccine candidate to fight shigellosis. NPJ Vaccines. (2023) 8:130. doi: 10.1038/s41541-023-00725-8
3
Caradonna V Pinto M Alfini R Giannelli C Iturriza M Micoli F et al . High-throughput luminescence-based serum bactericidal assay optimization and characterization to assess human sera functionality against multiple shigella flexneri serotypes. Int J Mol Sci. (2024) 25(20):11123. doi: 10.3390/ijms252011123
4
Collaborators, G. B. D. D. D . Estimates of the global, regional, and national morbidity, mortality, and aetiologies of diarrhoea in 195 countries: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect Dis. (2018) 18:1211–28. doi: 10.1016/S1473-3099(18)30362-1
5
Khalil IA Troeger C Blacker BF Rao PC Brown A Atherly DE et al . Morbidity and mortality due to shigella and enterotoxigenic Escherichia coli diarrhoea: the Global Burden of Disease Study 1990-2016. Lancet Infect Dis. (2018) 18:1229–40. doi: 10.1016/S1473-3099(18)30475-4
6
Diseases, G. B. D. & Injuries, C . Global burden of 369 diseases and injuries in 204 countries and territories, 1990-2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. (2020) 396:1204–22. doi: 10.1016/S0140-6736(20)30925-9
7
Bagamian KH Anderson Iv JD Blohm G Scheele S . Shigella and childhood stunting: Evidence, gaps, and future research directions. PloS Negl Trop Dis. (2023) 17:e0011475. doi: 10.1371/journal.pntd.0011475
8
Libby TE Delawalla MLM Al-Shimari F MacLennan CA Vannice KS Pavlinac PB . Consequences of Shigella infection in young children: a systematic review. Int J Infect diseases: IJID: Off Publ Int Soc Infect Dis. (2023) 129:78–95. doi: 10.1016/j.ijid.2023.01.034
9
European Centre for Disease Prevention and Control . Shigellosis. In: ECDC. Annual Epidemiological Report for 2022. Stockholm: ECDC (2024).
10
Kotloff KL Riddle MS Platts-Mills JA Pavlinac P Zaidi AKM . Shigellosis. Lancet. (2018) 391:801–12. doi: 10.1016/S0140-6736(17)33296-8
11
WHO . Guidelines for the control of shigellosis, including epidemics due to Shigella dysenteriae type 1. (2005).
12
Baker S Scott TA . Antimicrobial-resistant Shigella: where do we go next? Nat Rev Microbiol. (2023) 21:409–10. doi: 10.1038/s41579-023-00906-1
13
Mason LCE Greig DR Cowley LA Partridge SR Martinez E Blackwell GA et al . The evolution and international spread of extensively drug resistant Shigella sonnei. Nat Commun. (2023) 14:1983. doi: 10.1038/s41467-023-37672-w
14
Tansarli GS Long DR Waalkes A Bourassa LA Libby SJ Penewit K et al . Genomic reconstruction and directed interventions in a multidrug-resistant Shigellosis outbreak in Seattle, WA, USA: a genomic surveillance study. Lancet Infect Dis. (2023) 23:740–50. doi: 10.1016/S1473-3099(22)00879-9
15
WHO . WHO bacterial priority pathogens list, 2024. (2024).
16
Klugman KP Black S . Impact of existing vaccines in reducing antibiotic resistance: Primary and secondary effects. Proc Natl Acad Sci U.S.A. (2018) 115:12896–901. doi: 10.1073/pnas.1721095115
17
Jansen KU Knirsch C Anderson AS . The role of vaccines in preventing bacterial antimicrobial resistance. Nat Med. (2018) 24:10–9. doi: 10.1038/nm.4465
18
Micoli F Bagnoli F Rappuoli R Serruto D . The role of vaccines in combatting antimicrobial resistance. Nat Rev Microbiol. (2021) 19:287–302. doi: 10.1038/s41579-020-00506-3
19
MacLennan CA Grow S Ma LF Steele AD . The shigella vaccines pipeline. Vaccines (Basel). (2022) 10(9):1376. doi: 10.3390/vaccines10091376
20
Cohen D Ashkenazi S Schneerson R Farzam N Bialik A Meron-Sudai S et al . Threshold protective levels of serum IgG to Shigella lipopolysaccharide: re-analysis of Shigella vaccine trials data. Clin Microbiol Infect. (2023) 29:366–71. doi: 10.1016/j.cmi.2022.10.011
21
Cohen D Meron-Sudai S Bialik A Asato V Goren S Ariel-Cohen O et al . Serum IgG antibodies to Shigella lipopolysaccharide antigens - a correlate of protection against shigellosis. Hum Vaccin Immunother. (2019) 15:1401–8. doi: 10.1080/21645515.2019.1606971
22
Conti V Rossi O Clarkson KA Mancini F Nakakana UN Sarakinou E et al . Putative correlates of protection against shigellosis assessing immunomarkers across responses to S. sonnei investigational vaccine. NPJ Vaccines. (2024) 9:56. doi: 10.1038/s41541-024-00822-2
23
Rossi O Baker KS Phalipon A Weill FX Citiulo F Sansonetti P et al . Draft genomes of Shigella strains used by the STOPENTERICS consortium. Gut Pathog. (2015) 7:14. doi: 10.1186/s13099-015-0061-5
24
Perepelov AV Shekht ME Liu B Shevelev SD Ledov VA Senchenkova SN et al . Shigella flexneri O-antigens revisited: final elucidation of the O-acetylation profiles and a survey of the O-antigen structure diversity. FEMS Immunol Med Microbiol. (2012) 66:201–10. doi: 10.1111/j.1574-695X.2012.01000.x
25
Van De Verg LL Bendiuk NO Kotloff K Marsh MM Ruckert JL Puryear JL et al . Cross-reactivity of Shigella flexneri serotype 2a O antigen antibodies following immunization or infection. Vaccine. (1996) 14:1062–8. doi: 10.1016/0264-410x(96)00006-0
26
Leroux-Roels I Maes C Mancini F Jacobs B Sarakinou E Alhatemi A et al . Safety and immunogenicity of a 4-component GMMA-based Shigella vaccine in healthy European adults: Stage 1 of a randomized, controlled phase I/II clinical trial. J Infect Dis. (2024) 230(4):e971–e984. doi: 10.1093/infdis/jiae273 Erratum in: J Infect Dis. (2024) 230(6):e1413. doi: 10.1093/infdis/jiae564
27
Livio S Strockbine NA Panchalingam S Tennant SM Barry EM Marohn ME et al . Shigella isolates from the global enteric multicenter study inform vaccine development. Clin Infect diseases: an Off Publ Infect Dis Soc America. (2014) 59:933–41. doi: 10.1093/cid/ciu468
28
Shimanovich AA Buskirk AD Heine SJ Blackwelder WC Wahid R Kotloff KL et al . Functional and antigen-specific serum antibody levels as correlates of protection against shigellosis in a controlled human challenge study. Clin Vaccine Immunol. (2017) 24(2):e00412–16. doi: 10.1128/CVI.00412-16
29
Paciello I Silipo A Lembo-Fazio L Curcurù L Zumsteg A Noël G et al . Intracellular Shigella remodels its LPS to dampen the innate immune recognition and evade inflammasome activation. Proc Natl Acad Sci U.S.A. (2013) 110:E4345–4354. doi: 10.1073/pnas.1303641110
30
Ndungo E Pasetti MF . Functional antibodies as immunological endpoints to evaluate protective immunity against Shigella. Hum Vaccin Immunother. (2020) 16:197–205. doi: 10.1080/21645515.2019.1640427
31
Van De Verg LL Herrington DA Boslego J Lindberg AA Levine MM . Age-specific prevalence of serum antibodies to the invasion plasmid and lipopolysaccharide antigens of shigella species in Chilean and north american populations. J Infect Dis. (1992) 166:158–61. doi: 10.1093/infdis/166.1.158
32
Raqib R Qadri F SarkEr P Mia SM Sansonnetti PJ Albert MJ et al . Delayed and reduced adaptive humoral immune responses in children with shigellosis compared with in adults. Scandinavian J Immunol. (2002) 55:414–23. doi: 10.1046/j.1365-3083.2002.01079.x
33
Gasperini G Raso MM Schiavo F Aruta MG Ravenscroft N Bellich B et al . Rapid generation of Shigella flexneri GMMA displaying natural or new and cross-reactive O-Antigens. NPJ Vaccines. (2022) 7:69. doi: 10.1038/s41541-022-00497-7
34
Raso MM Gasperini G Alfini R Schiavo F Aruta MG Carducci M et al . GMMA and Glycoconjugate Approaches Compared in Mice for the Development of a Vaccine against Shigella flexneri Serotype 6. Vaccines (Basel). (2020) 8(2):160. doi: 10.3390/vaccines8020160
35
Nonne F Molfetta M Nappini R La Guidara C Di Benedetto R Mfana S et al . Development and application of a high-throughput method for the purification and analysis of surface carbohydrates from klebsiella pneumoniae. Biol (Basel). (2024) 13(4):256. doi: 10.3390/biology13040256
36
Micoli F Giannelli C Di Benedetto R . O-antigen extraction, purification, and chemical conjugation to a carrier protein. Methods Mol Biol. (2021) 2183:267–304. doi: 10.1007/978-1-0716-0795-4_14
37
Micoli F Alfini R Giannelli C . Methods for assessment of OMV/GMMA quality and stability. Methods Mol Biol. (2022) 2414:227–79. doi: 10.1007/978-1-0716-1900-1_14
38
Alfini R Carducci M Massai L De Simone D Mariti M Rossi O et al . Design of a glycoconjugate vaccine against salmonella paratyphi A. Vaccines. (2024) 12:1272. doi: 10.3390/vaccines12111272
39
Rossi O Molesti E Saul A Giannelli C Micoli F Necchi F . Intra-laboratory evaluation of luminescence based high-throughput serum bactericidal assay (L-SBA) to determine bactericidal activity of human sera against shigella. High Throughput. (2020) 9(2):14. doi: 10.3390/ht9020014
40
Mancini F Micoli F Rossi O . Setup and Characterization of a High-Throughput Luminescence-Based Serum Bactericidal Assay (L-SBA) to Determine Functionality of Human Sera against Shigella flexneri. BioTech (Basel). (2022) 11(3):29. doi: 10.3390/biotech11030029
41
Frenck RW Jr Conti V Ferruzzi P Ndiaye AGW Parker S McNeal MM et al . Efficacy, safety, and immunogenicity of the Shigella sonnei 1790GAHB GMMA candidate vaccine: Results from a phase 2b randomized, placebo-controlled challenge study in adults. EClinicalMedicine. (2021) 39:101076. doi: 10.1016/j.eclinm.2021.101076
Summary
Keywords
Shigella, altSonflex1-2-3, vaccine, AMR, O-antigen, SBA, GMMA
Citation
Boero E, Di Benedetto R, Vezzani G, Alfini R, Ceyssens P-J, Tansarli GS, Fang FC, Fontana C, Rossi A, Carrara S, Mancini F, Iturriza M, Sala C, Giannelli C, Rossi O and Micoli F (2025) Antibodies elicited by altSonflex1-2-3 GMMA vaccine are bactericidal against a panel of drug-resistant Shigella clinical isolates. Front. Immunol. 16:1652460. doi: 10.3389/fimmu.2025.1652460
Received
23 June 2025
Accepted
04 August 2025
Published
04 September 2025
Volume
16 - 2025
Edited by
Elke Bergmann-Leitner, Walter Reed Army Institute of Research, United States
Reviewed by
Manuela Terrinoni, University of Gothenburg, Sweden
Dilara Islam, Naval Medical Research Center, United States
Updates
Copyright
© 2025 Boero, Di Benedetto, Vezzani, Alfini, Ceyssens, Tansarli, Fang, Fontana, Rossi, Carrara, Mancini, Iturriza, Sala, Giannelli, Rossi and Micoli.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Francesca Micoli, francesca.x.micoli@gsk.com
†These authors have contributed equally to this work
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.