- 1College of Animal Science and Technology, Yangtze University, Jingzhou, Hubei, China
- 2College of Veterinary Medicine, Jilin University, Changchun, China
Pasteurella multocida (P. multocida) is the pathogen responsible for swine pasteurellosis, which can impede their growth and even cause death, leading huge economic losses to the global pig industry. P. multocida can be divided into 5 serotypes, and existing vaccines have low cross-immunity protection. Therefore, developing a vaccine that can provide effective cross-protection is essential for preventing swine pasteurellosis and reducing the abuse of antibiotics. In this study, six dominant antigenic proteins of P. multocida (PlpE, OmpA, OmpH, VacJ, Omp87 and Cp39) were selected. Through bioinformatics methods, 20 B-cell epitopes, 7 CTL epitopes and 11 Th-cell epitopes were predicted. The multi-epitope antigen PME was constructed by connecting these epitopes with linkers, and then the recombinant protein His-PME and the recombinant plasmid pcDNA3.1-PME could effectively stimulate immunized mice to produce antibodies, IL-4 and IFN-γ. The protection rates of His-PME group and pcDNA3.1-PME group were 62.5% and 75% against P. multocida serotype A, and 87.5% and 100% against P. multocida serotype D, respectively. Furthermore, the pathological lung damages in the His-PME group and the pcDNA3.1-PME group were significantly alleviated, and the bacterial loads in lung tissues were significantly decreased. These results indicated that the subunit vaccine His-PME and the DNA vaccine pcDNA3.1-PME can effectively resist the infection of P. multocida and have good immunogenicity and cross-protection. Therefore, the multi-epitope vaccine PME can be regarded as a candidate vaccine for the prevention of P. multocida infection.
1 Introduction
Pasteurella multocida (P. multocida) is the causative agent of swine pasteurellosis, with fibrinous pleuropneumonia, pharyngitis, hemorrhagic inflammation, and septicemia as the main clinical features (1). The most acute infection leads to the death of the affected pigs within 12 hours, and the mortality rate of acute infection can reach up to 100% (2). This disease is highly contagious and mainly infects healthy pigs through direct contact, indirect contact and air transmission, causing serious economic losses to the pig industry worldwide (3). P. multocida can be classified into five serotypes (A, B, D, E and F) based on capsular antigens, among which serotypes A, B, D and E can cause disease in pigs. The current prevalent serotypes are A and D (4).
At present, the prevention and control of swine pasteurellosis mainly relies on antibiotic therapy and vaccination (5). However, the increasing use of antibiotics has led to drug-resistant strains, posing severe challenges to the control of swine pasteurellosis (6). P. multocida vaccines mainly include inactivated vaccines, attenuated vaccines, and subunit vaccines, with inactivated vaccines and attenuated vaccines prevalent in clinical practices (7). Inactivated vaccines have high safety, but only target a single serotype and cannot effectively provide cross-protection (8). Attenuated vaccines, though with the advantages of strong and long-lasting antibody responses, have the risk of reversion to virulence (9).
Subunit vaccines not only can provide better cross-protection but also avoid the risk of reversion to virulence, making it an important research direction for P. multocida vaccines (10). Outer membrane protein, as one of the main virulence factors of P. multocida, has been widely used in vaccine research and has been proven to induce good immune protection (11). After challenging P. multocida serotype A, the protection rates of outer membrane lipoprotein (PlpE) when immunizing mice with different adjuvants ranged from 80% to 100%, outer membrane protein H (OmpH) was 100%, and the outer membrane protein 87 (Omp87) was 83.3% (12–14). The protection rates of VacJ family lipoprotein (VacJ) was 66.7% against P. multocida serotype B (15). The outer membrane protein A (OmpA) expressed in prokaryotic cells was found to be immunogenic by immunoblot analysis (16). The immunization of chickens with natural adhesion protein Cp39 resulted in a 100% protection rate against P. multocida strain P-1059 (serotype A) (17). However, due to the complex structure of natural antigenic protein, there are few effective epitopes exposed on the surface (18).
As a novel type of vaccine, multi-epitope vaccines predict the B lymphocyte (B-cell) and T lymphocyte (T-cell) epitopes of antigens through bioinformatics methods, and prepare vaccines by concatenating dominant epitopes, which can activate the host’s humoral and cellular immunity (19). Moreover, multi-epitope vaccines have the advantages of high safety and cross-protection (20, 21). DNA vaccines can elicit long-lasting immunity by delivering exogenous genes that encode antigenic proteins into host cells, thereby enabling the stable expression of these antigenic proteins (22). Meanwhile, without intricate protein purification, multi-epitope DNA vaccines are straightforward to develop, and can be engineered through screening to concatenate dominant epitopes, thereby providing efficient immune protection and cross-immune protection (23, 24).
In this study, the B-cell and T-cell epitopes of six antigen proteins of P. multocida, namely PlpE, OmpA, OmpH, VacJ, Omp87 and Cp39, were predicted by bioinformatics methods. These epitopes were concatenated through flexible linkers (GSG) to obtain the multi-epitope PME, and the physicochemical properties such as antigenicity index and hydrophilicity were analyzed by bioinformatics. Then, the prokaryotic expression recombinant vector pET30a-PME and the eukaryotic expression recombinant vector pcDNA3.1-PME were constructed to obtain the recombinant protein His-PME and the recombinant plasmid pcDNA3.1-PME. By evaluating the immune efficacy of His-PME and pcDNA3.1-PME on mice, it was shown that the subunit vaccine and DNA vaccine prepared based on the multi-epitope PME had good immune protection against P. multocida serotypes A and D.
2 Materials and methods
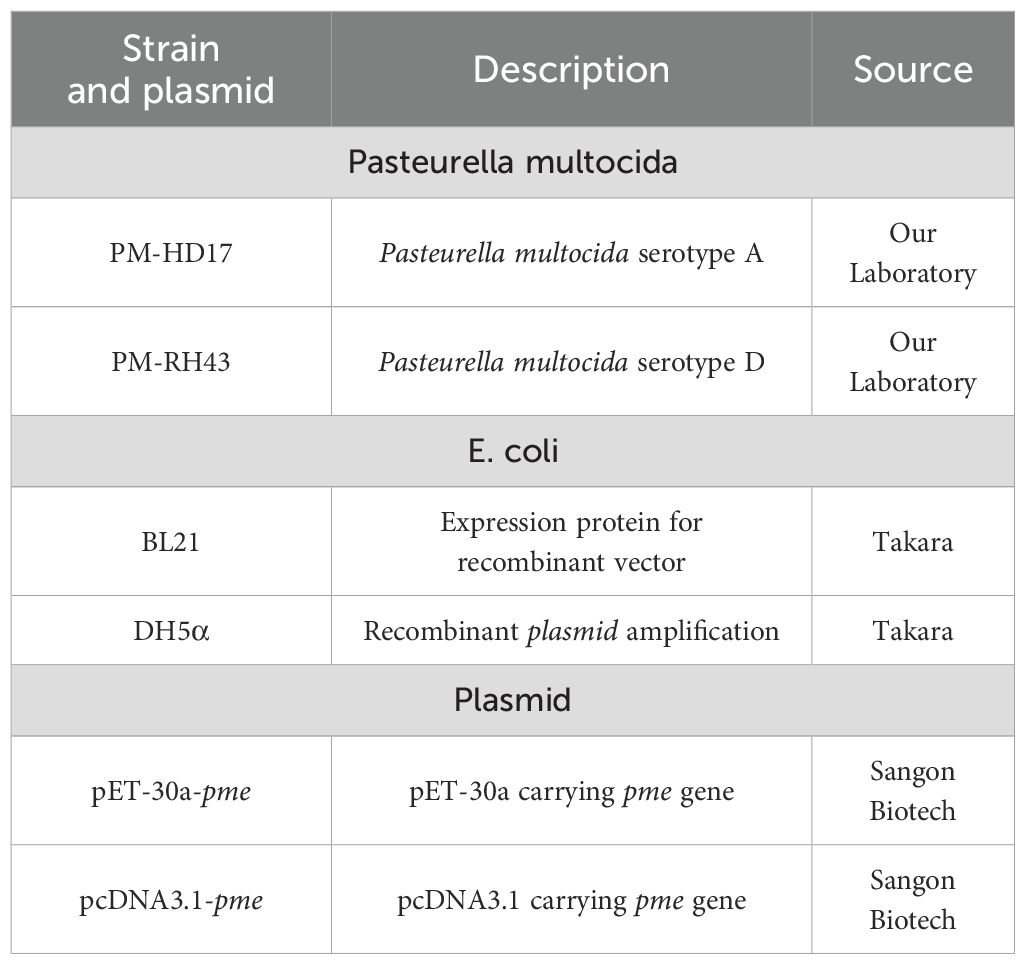
2.1 Strains, plasmid and culture conditions
P. multocida was cultured in brain and heart infusion (BHI; Solarbio, Beijing, China) containing 10% newborn bovine serum (EVERY GREEN, Hangzhou, China). When cultivating E. coli containing pET-30a plasmid, kanamycin (50 μg/mL) was added to Luria-Bertani (LB; Solarbio, Beijing, China). When cultivating E. coli containing pcDNA3.1 plasmid, ampicillin (100 μg/mL) was added to LB medium. All strains and plasmids in this experiment were listed in Table 1, and the sources of software were listed in Supplementary Table S1.
2.2 Prediction of B-cell and T-cell epitopes
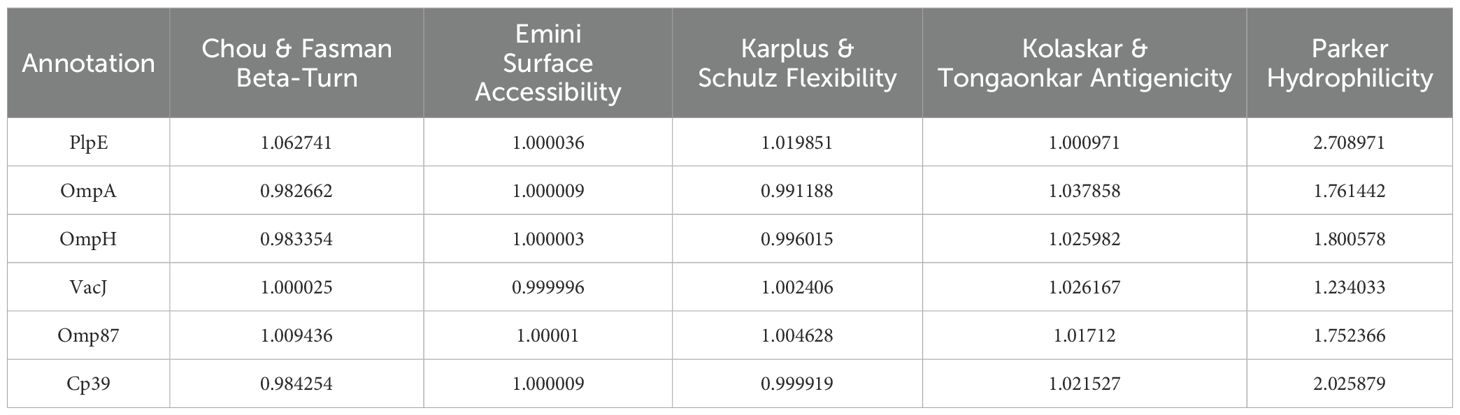
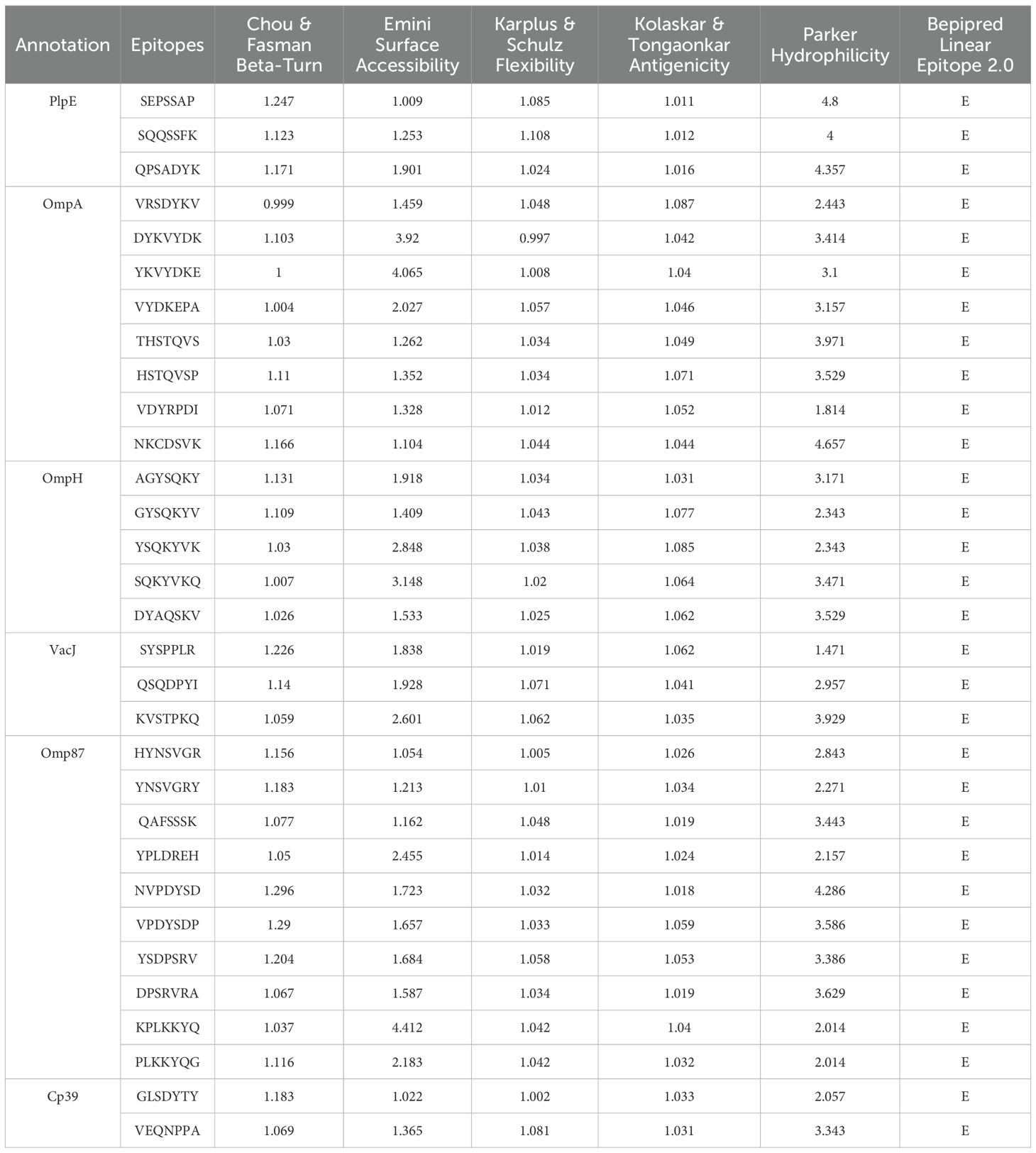
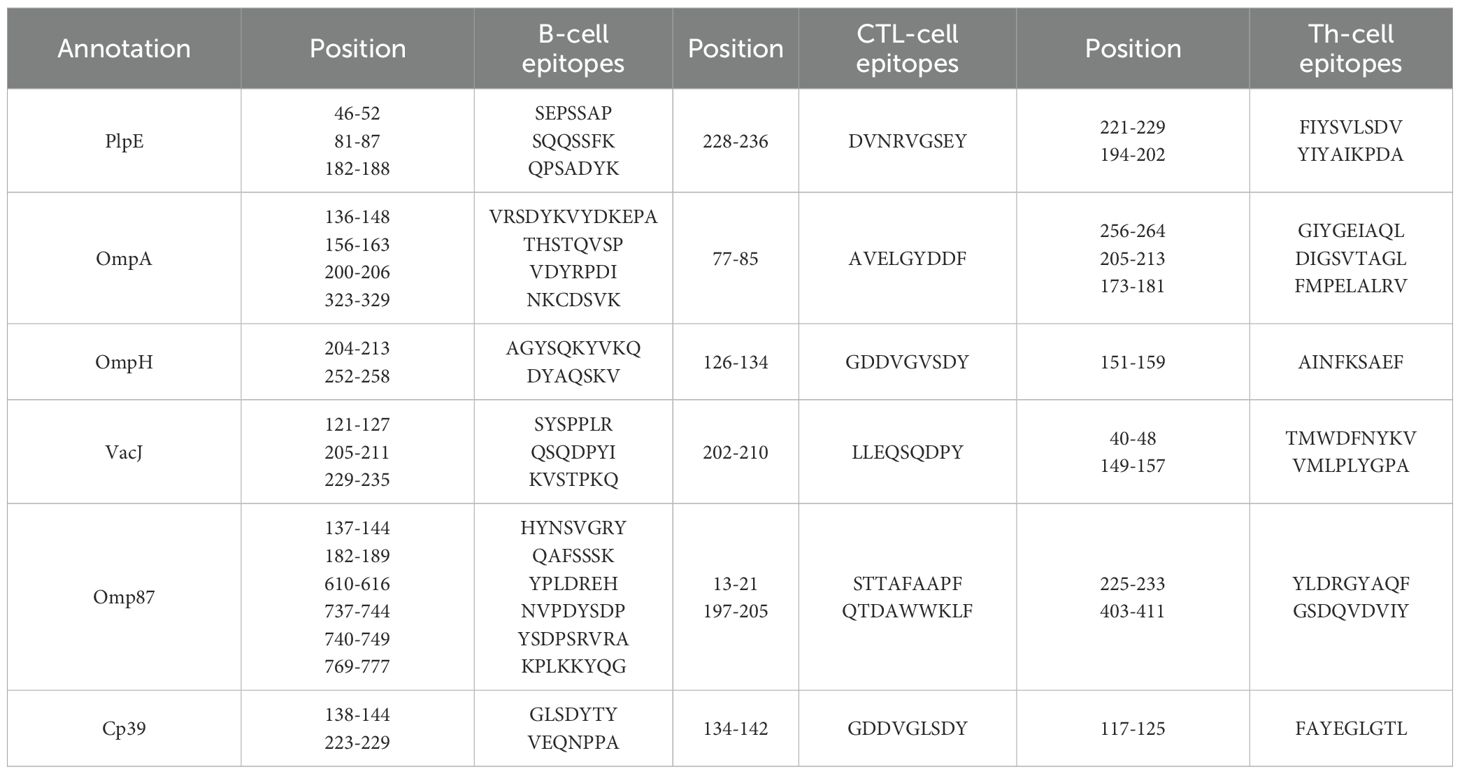
The amino acid sequences of six antigenic proteins of P. multocida, namely PlpE, OmpA, OmpH, VacJ, Omp87 and Cp39, were downloaded from the NCBI database. In Immune Epitope Database and Tools (IEDB), Chou & Fasman Beta-Turn Prediction, Emini Surface Accessibility Prediction, Karplus & Schulz Flexibility Prediction, Kolaskar & Tongaonkar Antigenicity, Parker Hydrophilicity Prediction, and Bepipred Linear Epitope Prediction 2.0 were used to predict the B-cell epitopes of the above six proteins (25–30). Linear B-cell epitopes with high surface accessibility, high proportion of β-turns and random coils, high antigenicity index, high hydrophilicity, and strong flexibility were selected as the final dominant B-cell epitope sequences.
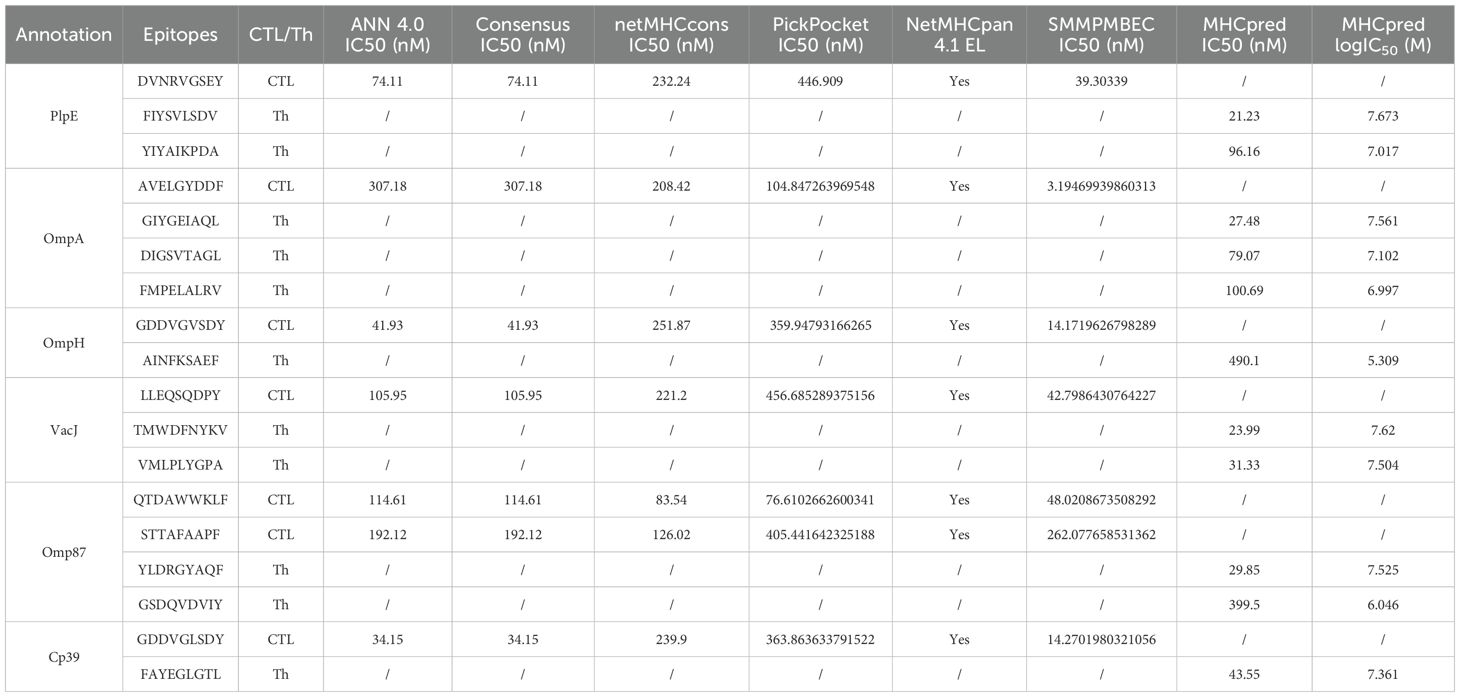
Predict cytotoxic T lymphocyte (CTL) epitopes using ANN 4.0, Consensus, netMHCcons, PickPocket, SMMPMBEC in IEDB. Select pig major histocompatibility complex I (MHC I) alleles SLA-1*0401, SLA-2*0401, and SLA-3*0401 as receptors, and screen the CTL epitopes with a length of 9 amino acids and an IC50 value less than or equal to 500 (31–36). MHCpred (https://www.ddg-pharmfac.net/mhcpred/MHCPred/) was used to predict helper T lymphocyte (Th-cell) epitopes, with the DRB1*0101 allele as the receptor. Short peptides with an IC50 value less than or equal to 500 and a higher logIC50 score was selected as Th-cell epitopes (37).
2.3 Design and prokaryotic expression of multi-epitope protein PME
The predicted B-cell and T-cell antigen epitopes were concatenated using a flexible linker (GSG) to obtain the multi-epitope protein PME (38). Bioinformatics tools such as Expasy ProtParam, ToxinPred, AllerTOP v2.0, VaxiJen v2.0, SOLpro, DNAstar, SignalP-6.0, and DeepTMHMM were used to analyze the basic physicochemical properties, toxicity, allergic reactions, antigenicity, solubility, hydrophilicity, flexibility, surface accessibility, signal peptides, and transmembrane structure of multi-epitope protein PME (39–41).
Prokaryotic expression assay was performed as described earlier, with some modifications (42–44). The nucleotide sequence of the multi-epitope protein PME was codon optimized, synthesized by Sangon Biotech (Shanghai, China), and connected to the prokaryotic expression vector pET30a. The recombinant plasmid pET30a-PME was transformed into E. coli BL21 (DE3), and cultured in LB (200 mL) medium with 50 μg/mL kanamycin. When OD600 was 0.6, the culture was induced with 1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) at 37°C for 5 h, centrifuged at 8000 rpm, and then ultrasonically treated. The precipitate was collected by centrifugation, purified by Ni-NTA affinity chromatography and dialysis. Then, the obtained protein His-PME was identified by SDS-PAGE and stored at -80°C.
2.4 Secondary structure prediction and tertiary structure modeling
The secondary structure of multi-epitope protein PME was predicted using SOPMA, and its immunological potential was evaluated (45). The tertiary structure was modeled using AlphaFold 3, and the conformational rationality and quality was evaluated using PDBsum and ProSA-web (46, 47).
2.5 Predicting linear and conformational B-cell epitopes
After completing the tertiary structure modeling of PME through AlphaFold 3, the ElliPro tool in the IEDB database was further used to predict the linear B-cell epitopes and conformational B-cell epitopes in PME (the minimum score was set to the default value of 0.5, and the maximum distance was set to the default value of 6) (48). The higher PI value of the predicted epitopes proved that they were more prominent on the surface of the protein and more recognized and bound by antibodies (49).
2.6 Molecular docking and molecular dynamics simulation
Toll-like receptor family (TLR family) is an important class of pattern recognition receptors (PRRs) that activates immune responses by recognizing pathogen-associated molecular patterns (PAMPs) (50). We obtained the structural coordinates of TLR2 (PDB ID: 3A7C), TLR4 (B: 3VQ2), MHC I (PBD ID: 3V52), and major histocompatibility complex II (MHC II; PBD ID: 2P24) in Mus musculus from the Protein Data Bank (https://www.rcsb.org). The ClusPro server (https://cluspro.org/login.php) was utilized for the docking of PME with the four receptors, namely TLR2, TLR4, MHC I and MHC II (51). Visualization analysis of the interaction of the docking structures was conducted using PyMOL software (https://pymol.org), and molecular dynamics simulation was performed using iMODS to evaluate the stability of the docking structures (52).
2.7 Immune response simulation
To evaluate the potential immune response of the multi-epitope protein PME, C-lmmSim was used to simulate immune responses (53). The random seed was set to 12345 by default, and the simulation volume and simulation step were set to 10 and 540, respectively. The immune program consisted of three injections, and the time step lengths of the three injections were set to 1, 84, and 252, respectively. Each time step lengths was equal to 8 hours in real life.
2.8 Extraction of recombinant plasmid pcDNA3.1-PME
The nucleotide sequence of the multi-epitope protein PME was codon optimized, and connected to the eukaryotic expression vector pcDNA3.1. The recombinant plasmid pcDNA3.1-PME was transformed into E. coli DH5α for overnight culture, and then transferred to LB medium (2.4 L) containing 100 μg/mL ampicillin at a ratio of 1:100 and cultured for 16 h. After centrifugation at 8000 rpm for 10 min, the plasmid was extracted using EndoFree Plasmid MaxiPrep Kit (HLINGENE, Shanghai, China) and stored at -80°C.
2.9 Immunoblotting analysis of His-PME protein
The purified protein His-PME was electrophoresed on 12% SDS-PAGE gel and then transferred onto the PVDF membrane. The membrane was incubated with anti-His antibody (1:10000; Solarbio, Beijing, China) overnight at 4°C. After being washed with TBST for 3 times, each for 5 min, the membrane was incubated with goat anti-mouse IgG/Alkaline Phosphatase (1:10000; Beyotime, Shanghai, China) at room temperature for 2 h.
2.10 Immunization and challenge in mice
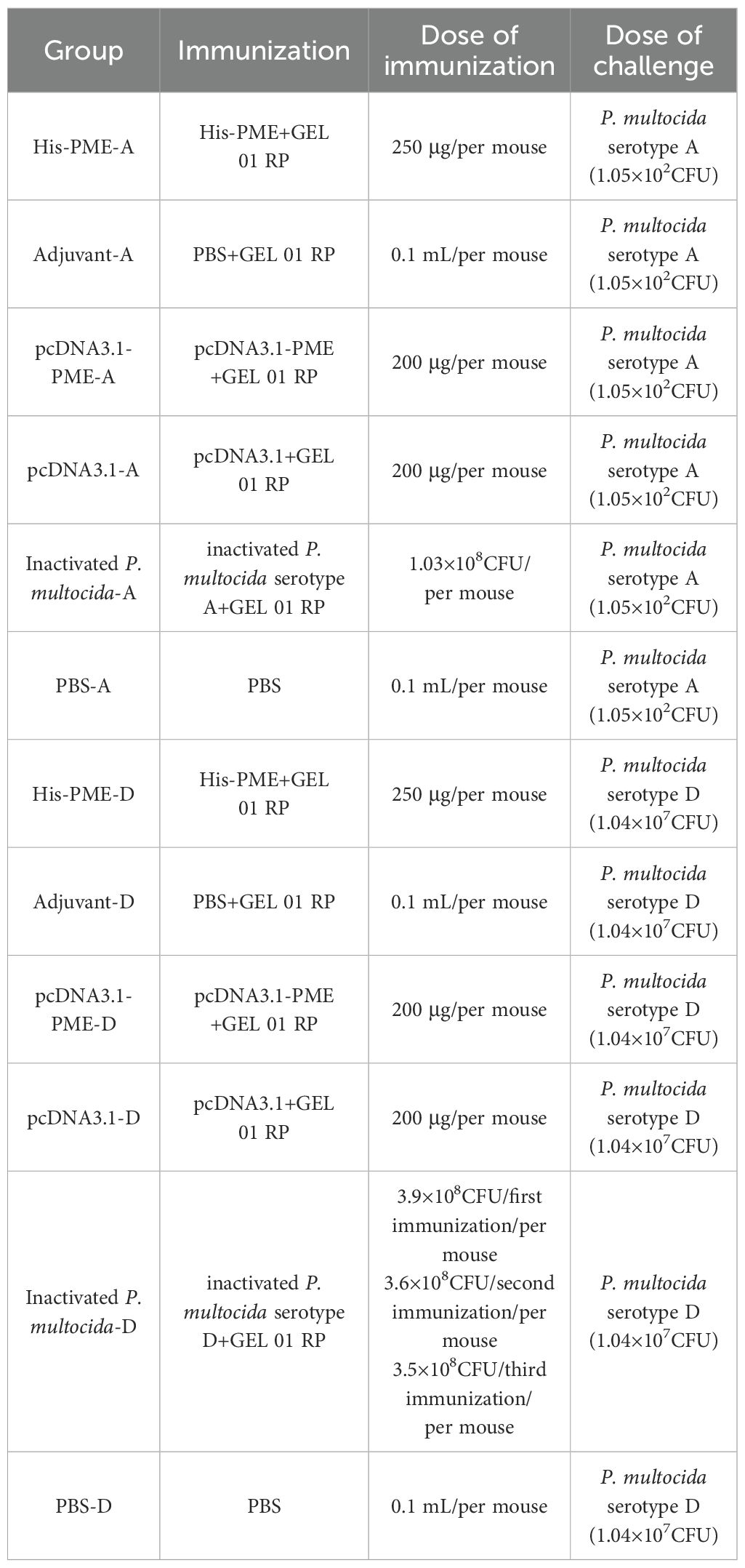
Six-week-old female SPF BALB/c mice (18-20 g) were purchased from Experimental Animal Center of the Three Gorges University, and the detailed immunization information was shown in Table 2. All animal experiments were approved by the Animal Ethics Committee of the Yangtze University. In this study, His-PME, pcDNA3.1-PME, inactivated P. multocida (serotypes A and D), pcDNA3.1, and PBS were mixed with GEL 01 RP adjuvant (V/V 10:1), and inoculated into mice on days 1, 14 and 28, respectively. Among them, the groups (8 mice/group) of pcDNA3.1-PME and pcDNA3.1 were inoculated into the tibialis anterior muscle of the hind limbs of mice, while the others were inoculated into the subcutaneous tissue of the back of mice. On the 35th day, mice were intraperitoneally challenged with P. multocida. The 7-day survival status, clinical symptoms, and clinical scores of these groups were observed and recorded. The scoring was as follows: health (0 point), lethargy (1 point), emaciation (2 points), trembling and weakness of limbs (3 points), hind limb paralysis (4 points), death (5 points) (54).
2.11 Lung bacterial load and histopathological analysis
The mice were subjected to the attack protocol as described above. Since mice began to die sequentially after 8 hours, we elected to euthanize them at the 8-hour time point. The lungs (0.1-0.15g) of the mice were aseptically collected and homogenized using tissue disruptor (NewZongKe, Wuhan, China). Then, these samples were serially diluted with 0.9% NaCl, plated on BHI plates containing 10% newborn bovine serum, and incubated at 37°C for 24 h for colony counting. Meanwhile, the partial lung tissue of the mice was fixed in 10% formalin for histo-pathological analysis. The scoring of pathological changes was as follows: no lesions (0 point), mild pathological changes (1 point), moderate pathological changes (2 points), severe pathological changes (3 points), and extremely severe pathological changes (4 points) (55). The evaluation contents included hemolysis, inflammatory cell infiltration, hemorrhage, and widening of the alveolar septa.
2.12 The serum antibody levels of mice
After immunization, serum samples were collected from the tail vein of each group of mice on days 0, 13, 27, and 34, respectively, and the antibody levels were detected by indirect ELISA (56). 100 μL of the fragmented P. multocida serotype A or D (100 μg/mL) was added to each well and coated overnight at 4°C. After washing with PBST, 150 μL of 2% BSA was added to each well and incubated at 37°C for 2 h. Serum samples from each group (1:400) were added as primary antibodies to the wells and incubated at 37°C for 1 h. Goat anti-mouse IgG HRP (1:5000; Beyotime, Shanghai, China) was added as a secondary antibody to the wells and incubated at 37°C for 1 h. Finally, TMB buffer (Solarbio, Beijing, China) was added and incubated at room temperature for 5 min, and then the OD450 was measured.
2.13 Cytokine Analysis
Serum samples were collected from the mice in each group 34 days after immunization, and the levels of interferon gamma (IFN-γ) and interleukin-4 (IL-4) cytokines in the serum were detected using ELISA kit (YUANJU, Shanghai, China). The absorbance was measured at 450 nm.
2.14 Statistical analysis
All statistical analyses were performed using the unpaired Student’s t test by GraphPad Prism (version 9.0; GraphPad, La Jolla, CA), and *P < 0.05 was considered statistically significant.
3 Results
3.1 Prediction of B-cell and T-cell epitopes
The average score of B-cell epitopes of PlpE, OmpA, OmpH, VacJ, Omp87 and Cp39 was calculated by Chou & Fasman Beta-Turn Prediction, Emini Surface Accessibility Prediction, Karplus & Schz Flexibility Prediction, Kolaskar & Tongaonkar Antigenicity, and Parker Hydrophilicity Prediction (Table 3). The epitopes with scores higher than the average score and that marked with “E” in the Bepipred Linear Epitope Prediction 2.0 were selected as candidate B-cell epitopes (Table 4). The CTL and Th-cell epitopes of the above six proteins were predicted using ANN4.0, Consensus, netMHCcons, PickPocket, NetMHCpan4.1EL, SMMPMBEC, and MHCpred. Epitopes with IC50≤500 and higher logIC50 scores were selected as candidate Th-cell epitopes (Table 5). Epitopes with IC50 ≤ 500 and that included in NetMHCpan 4.1 EL were selected as candidate CTL epitopes (Table 5). In this study, 20 B-cell epitopes, 7 CTL epitopes and 11 Th-cell epitopes were screened (Table 6).
3.2 Bioinformatics analysis of PME
The predicted B-cell and T-cell epitopes were linked by flexible linkers (GSG) to obtain the multi-epitope antigen PME (Figure 1A). The prediction results of DNAstar showed that PME had high hydrophilicity, flexibility, and surface accessibility (Figure 1B). SignalP-6.0 and DeepTMHMM analysis showed that PME had no signal peptides and transmembrane regions (Figures 1C, D). The secondary structure of PME was predicted by SOPMA, and showing that PME was composed of α-helix (1.4%), chain (5.13%), β-turn (0.47%), and random coil (93.01%) (Figure 1E).
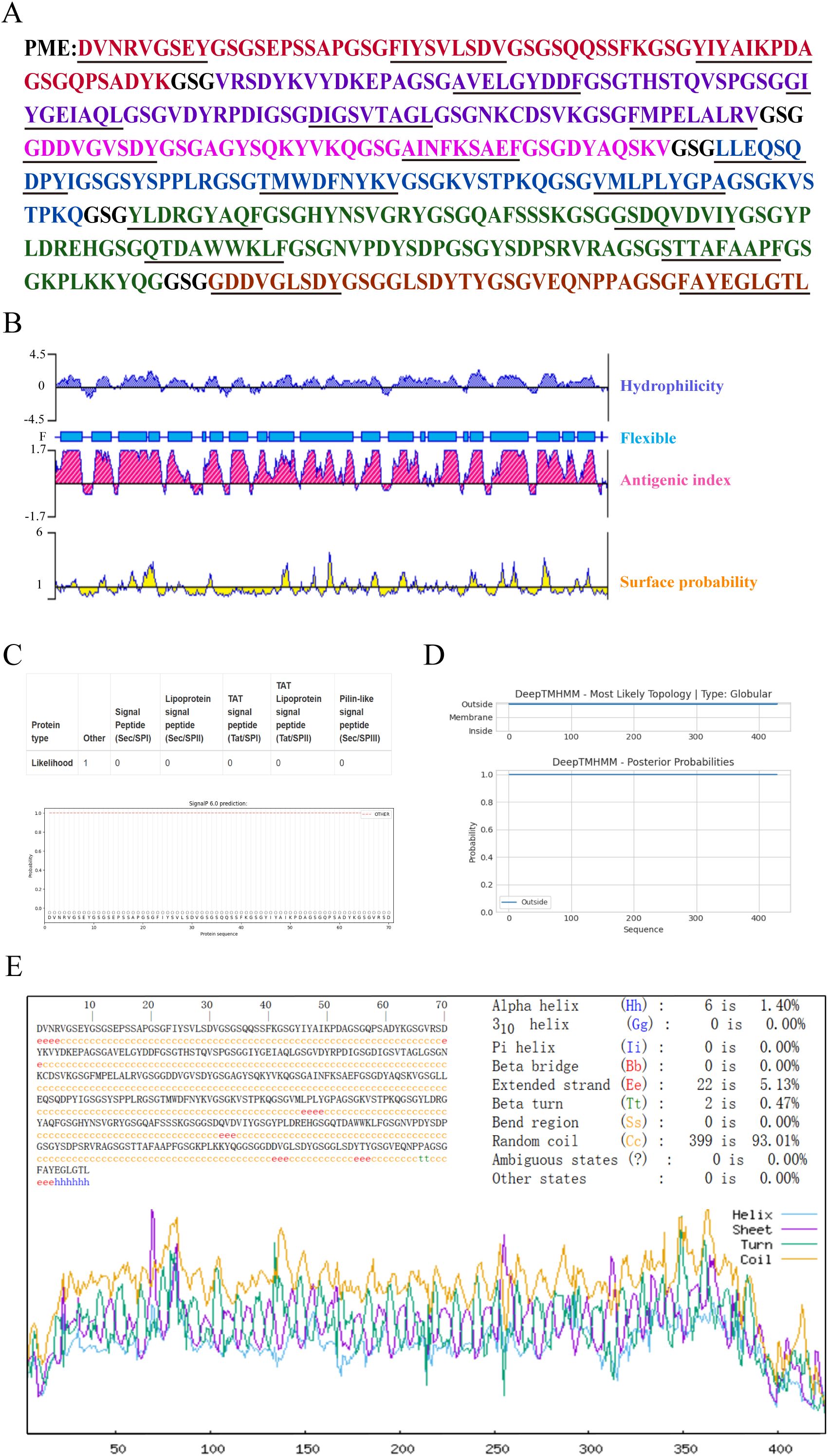
Figure 1. The sequence structure and characteristics of PME. (A) The amino acid sequence of PME is presented, with the epitope sequences of six proteins displayed in various colors. The PlpE epitope sequence is highlighted in red, the OmpA epitope sequence in purple, the OmpH epitope sequence in pink, the VacJ epitope sequence in blue, the Omp87 epitope sequence in green, the Cp39 epitope sequence in brown, and the GSG linkers in black. The T-cell epitopes are underlined. (B) The hydrophilicity, antigenic index, surface probability, and flexibility assessment of PME. (C) The analysis of the signal peptide in PME. (D) The prediction of the transmembrane region of PME. (E) The secondary structure analysis of PME.
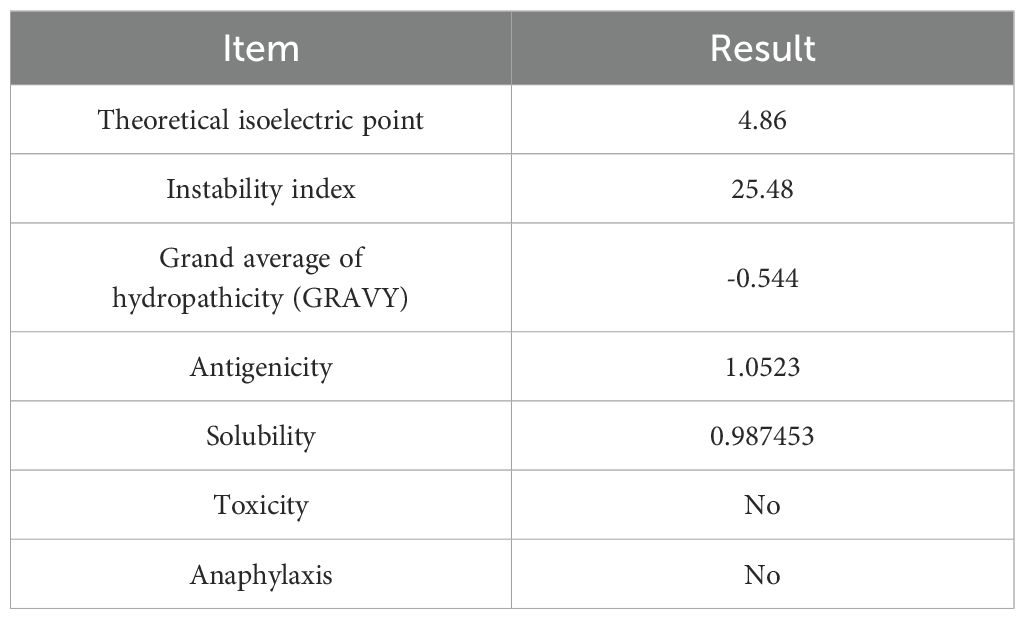
The isoelectric point (PI), instability index and hydrophilicity of PME were analyzed by ExPaSy ProtParam, and the results showed that the theoretical PI was 4.86, the instability index was 25.48, and the average hydrophilicity (GRAVY) was -0.544. These results indicated that PME carried negative charges and was not prone to degradation, making it a hydrophilic protein. ToxinPred and AllerTOP v2.0 were respectively used to predict the toxicity and allergenicity of PME, and the results indicated that PME was non-toxic and non-allergenic. Using VaxiJen v2.0 to predict the antigenicity of PME, the result was 1.0523, indicating the strong antigenicity of PME. The solubility probability of PME was predicted to be 0.987453 by SOLpro, showing the high solubility of PME (Table 7). In conclusion, these results indicated that PME has the characteristics of strong antigenicity, high solubility, negative charge, difficult degradation, with no transmembrane structure, toxicity, and allergenicity.
3.3 Analysis of tertiary structure of PME
The tertiary structure of PME was modeled using AlphaFold 3 (Figure 2A), and visualized using PyMOL (Figure 2B). Ramachandran plot analysis of PME using the PDBsum server showed that 83.9% of the amino acids were in the favorable region, 14.5% were in the allowed region, 1.6% were in the outlier region, and no amino acids were in the disallowed region, indicating that the conformation of the multi-epitope protein PME was reasonable (Figure 2C). ProSA-web verified the tertiary structure of PME, with a z-score of -5.5, indicating that PME had a superior 3D protein model (Figure 2D). The above results indicated that the multi-epitope protein PME had rationality and excellent quality.
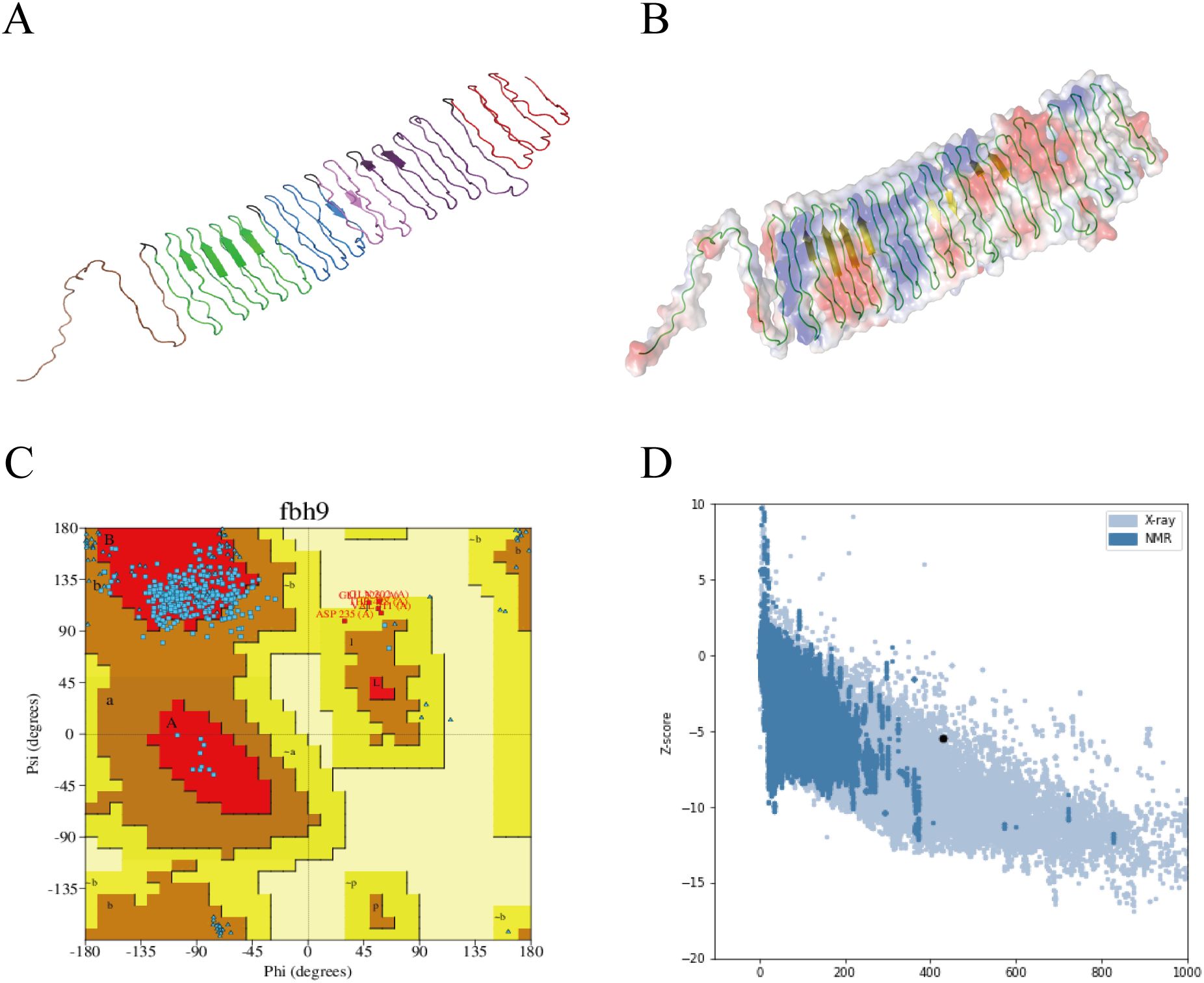
Figure 2. Tertiary structure model and model validation of PME. (A) The tertiary structure model of PME. Red represents the epitope sequence of PlpE, purple represents the epitope sequence of OmpA, pink represents the epitope sequence of OmpH, blue represents the epitope sequence of VacJ, green represents the epitope sequence of Omp87, brown represents the epitope sequence of Cp39, and black represents the linkers. (B) PyMOL visualizes the secondary structure and surface potential distribution of the PME model. The green lines represent the coiled structure, while the yellow arrows represent the sheet. (C) Ramachandran plot of PME. 83.9% of amino acids are in the favored region, and no amino acids are in the disallowed region. (D) The Z-score (-5.5) of the PME model.
3.4 Prediction of linear and conformational B-cell epitopes of PME
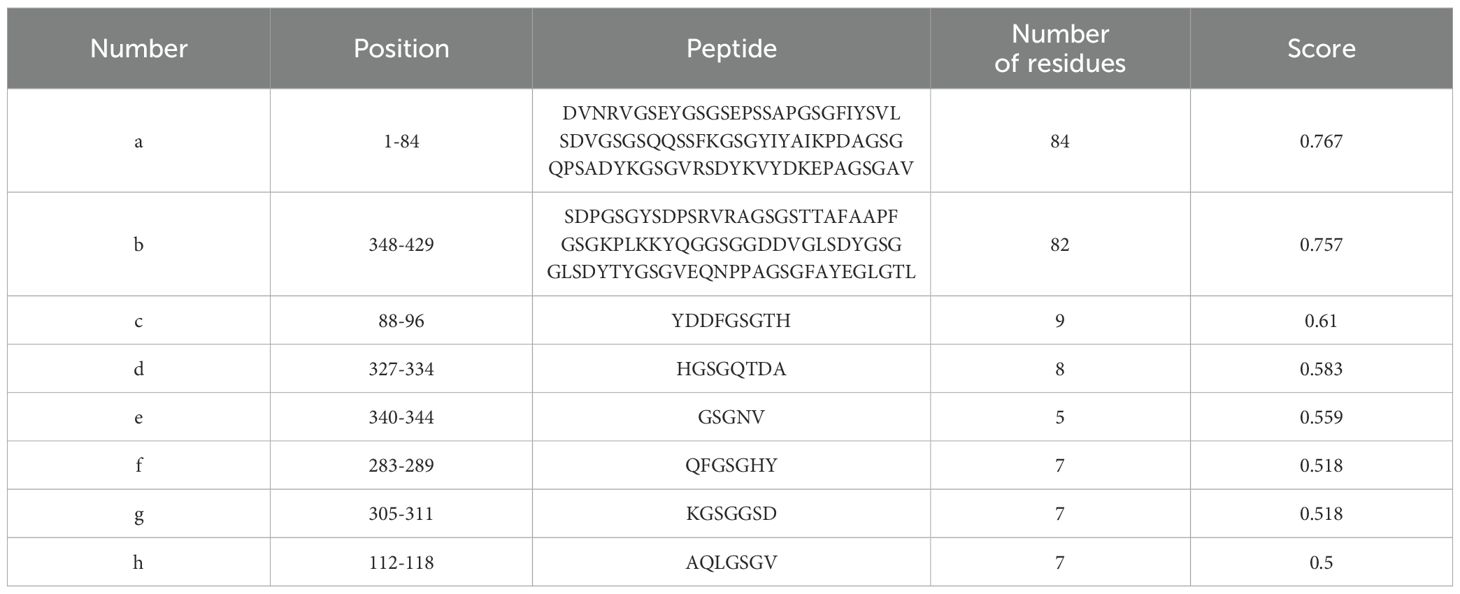
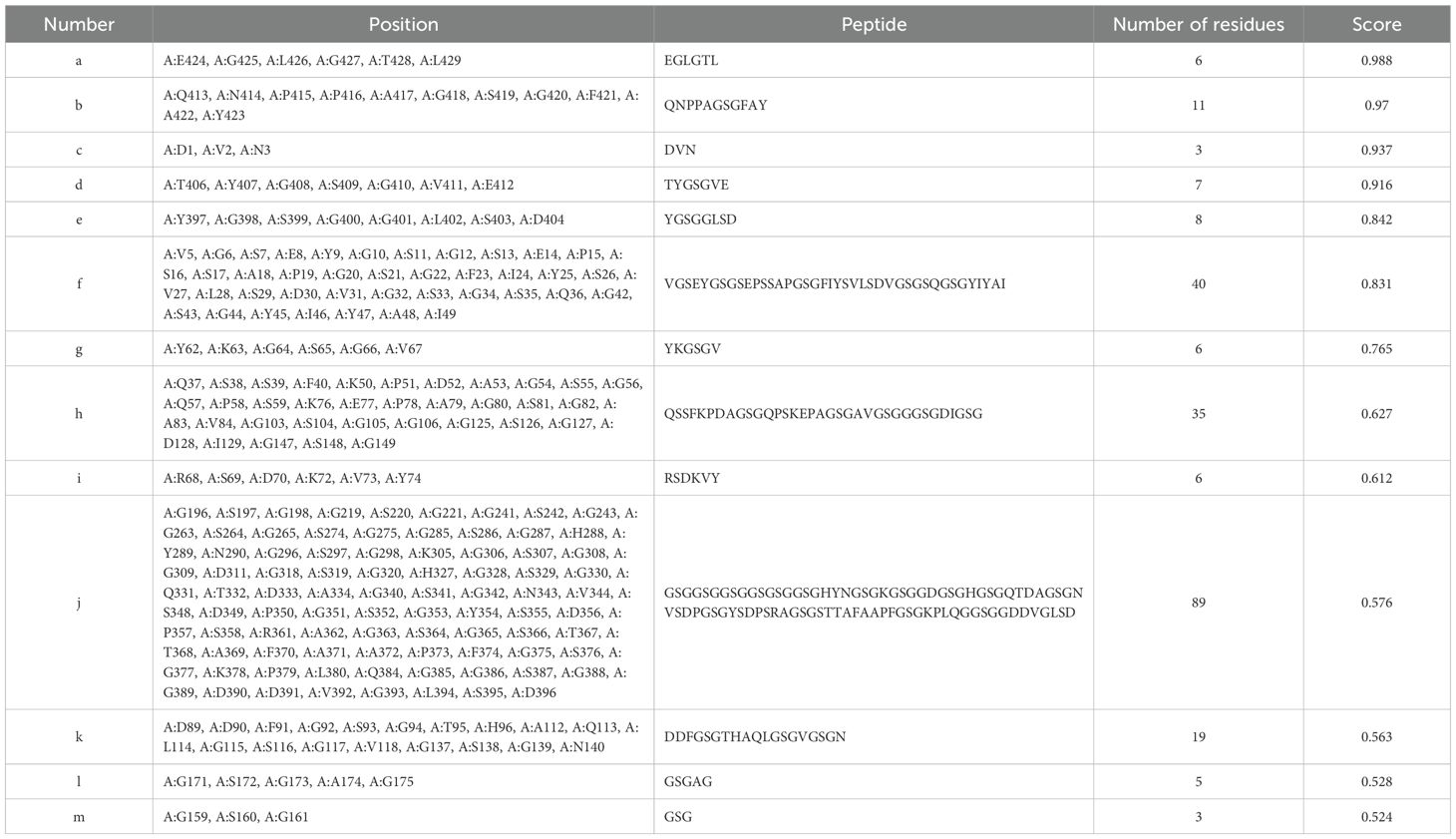
The tertiary structure PDB file of multi-epitope protein PME was obtained using AlphaFold 3, and then 8 linear B-cell epitopes (Figure 3A) and 13 conformational B-cell epitopes (Figure 3B) of PME were predicted by ElliPro in IEDB. The size of the linear epitope ranged from 5 to 84 residues, and their scores ranged from 0.5 to 0.767 (Table 8). The size of the conformational B-cell epitope ranged from 3 to 89 residues, and their scores ranged from 0.524 to 0.988 (Table 9). These results showed that there were many prominent epitopes on the surface of multi-epitope protein PME, which were easily recognized and bound by antibodies, thus triggering immune response.

Figure 3. Linear and conformational B-cell epitopes in PME. (A) Linear B-cell epitopes are represented by yellow spheres. A total of 8 epitopes (labeled a–h) were identified, and the size of the epitope residues ranges from 5 to 84. (B) Conformational B-cell epitopes are represented by yellow spheres. A total of 13 epitopes (labeled a–m) were identified, and the size of the epitope residues ranges from 3 to 89.
3.5 Molecular docking and molecular dynamics simulations
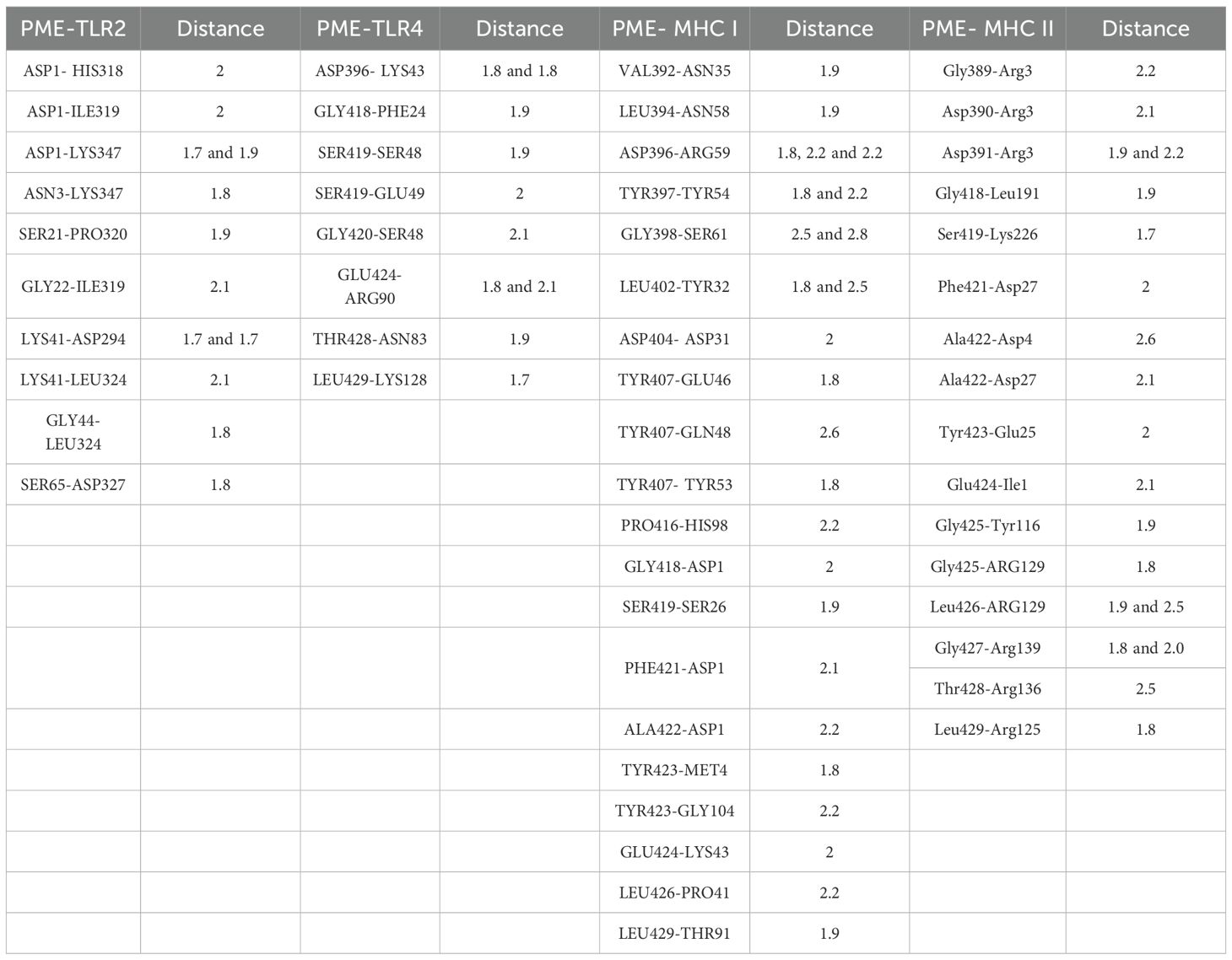
To test the affinity between PME and immune receptors, we used the ClusPro server to dock TLR2, TLR4, MHC I, MHC II with PME, and the lowest energy scores of the docked complexes were -1352.1 (Figure 4A), -1373 (Figure 4B), -1019.9 (Figure 4C), and -964.1 (Figure 4D), respectively. The visualization of PME-TR2, PME-TR4, PME-MHC I, and PME-MHC II complexes using PyMOL showed that all these four complexes have multiple hydrogen bond interactions with hydrogen bond distances ranging from 1.7 to 2.8 Å, indicating a high affinity between PME and the four immune receptors (Table 10).
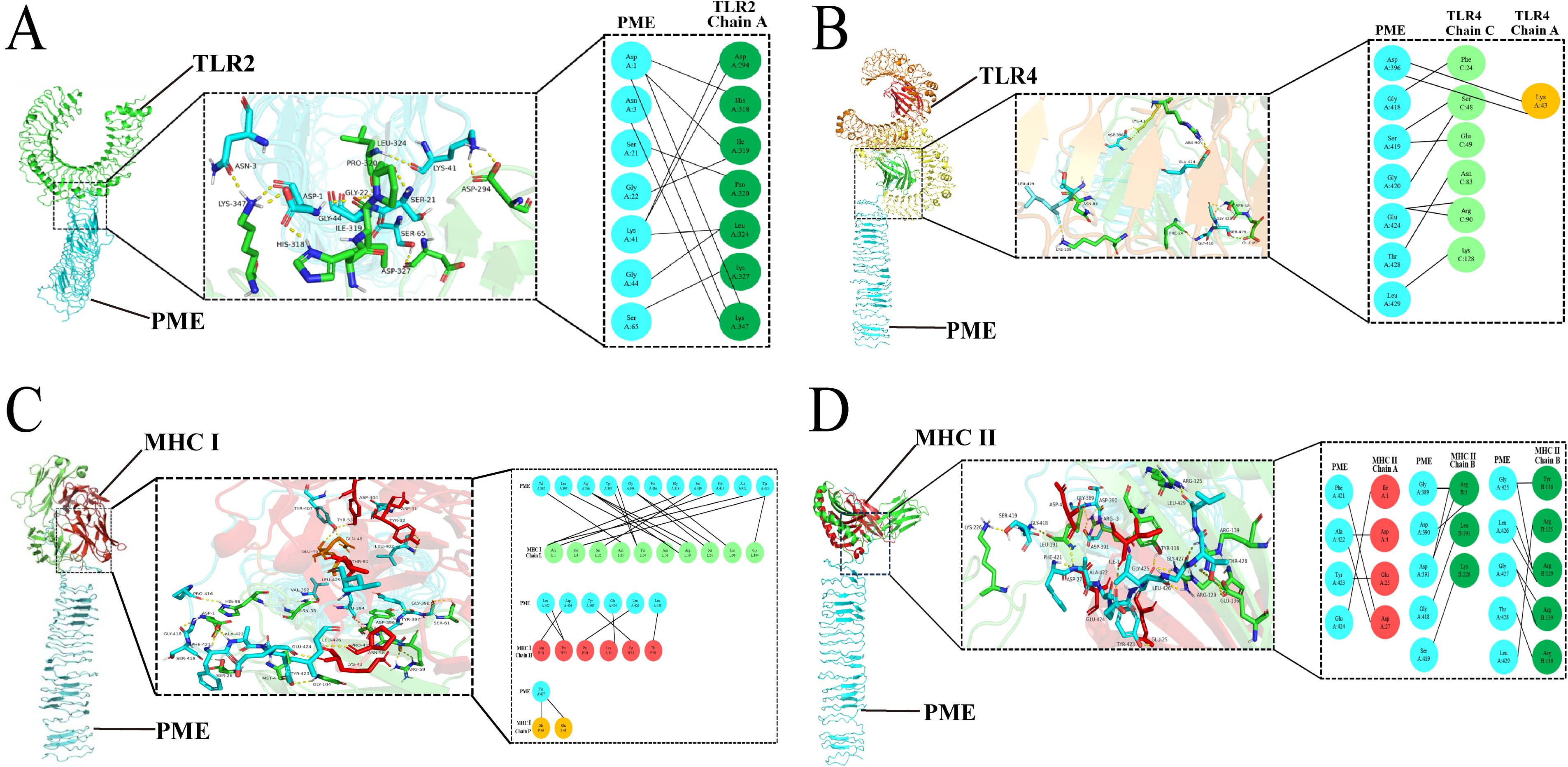
Figure 4. Molecular docking analysis results of PME with TLR2, TLR4, MHC I and MHC II. (A) Schematic diagram of molecular docking for PME and TLR2. PME is sky blue, and the A chain of TLR2 is green. (B) Schematic diagram of molecular docking for PME and TLR4. PME is sky blue, the A chain of TLR4 is yellow, and the C chain is light green. (C) Schematic diagram of molecular docking for PME and MHC I PME is sky blue, the L chain of MHC I is light green, the H chain is red, and the P chain is yellow. (D) Schematic diagram of molecular docking for PME and MHC II. PME is sky blue, the A chain of MHC II is red and the B chain is green.
The main chain deformability, B factor, variance and elastic network model of PME-TLR2, PME-TLR4, PME-MHC I and PME-MHC II were analyzed using iMODS. The results showed that most regions of the above four complexes were rigid and not prone to deformation (Figures 5A–D); most regions of the four complexes were less affected by thermal motion (Figures 6A–D); the local flexibility of the four complexes was low (Figures 7A–D); each complex had spring structures, indicating that the structures of the complexes were compact (Figures 8A–D). Based on the above results, it was indicated that PME had a high affinity and stability with TLR2, TLR4, MHC I and MHC II receptors.
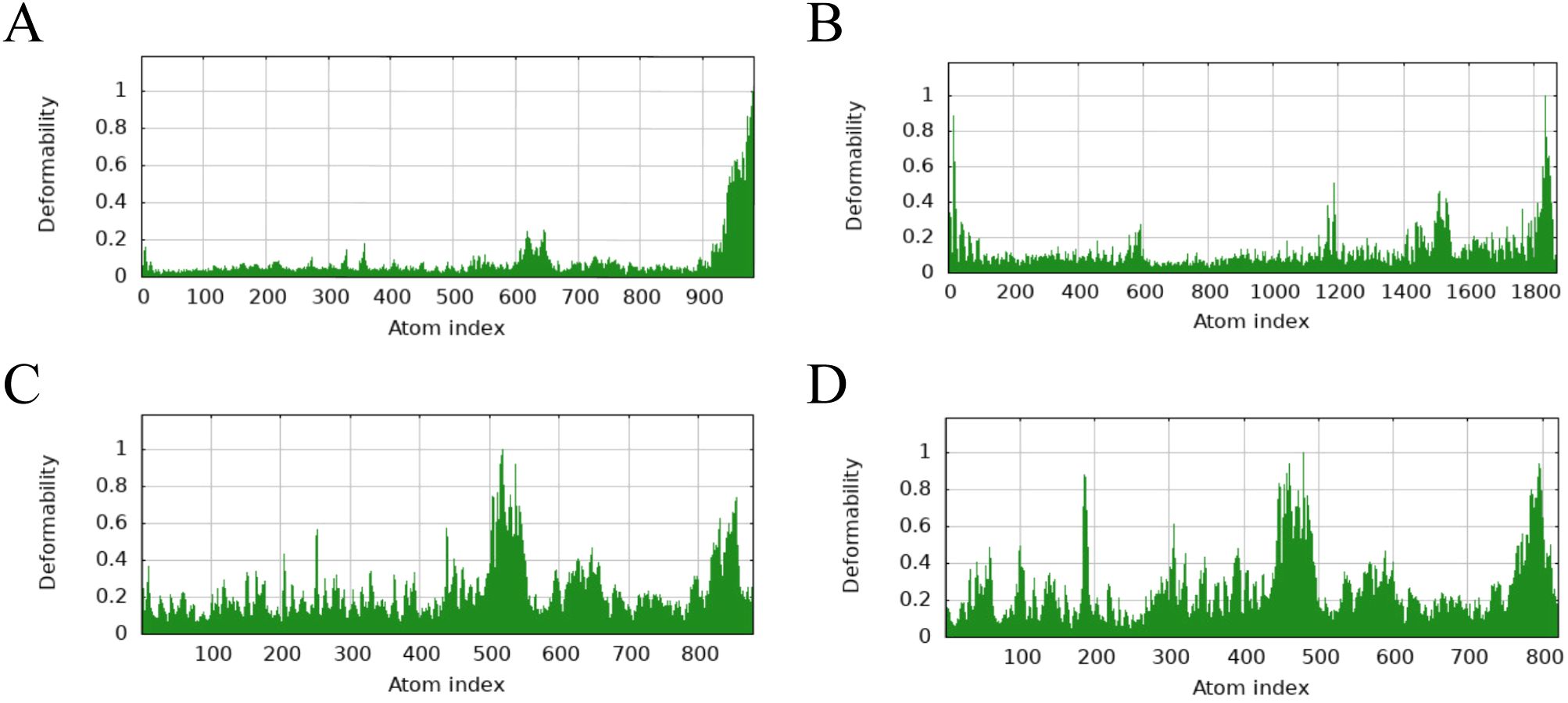
Figure 5. iMODS analysis revealed the main chain deformability of PME-TLR2, PME-TLR4, PME-MHC I, and PME-MHC II. (A) The main chain deformability plot of PME-TLR2. (B) The main chain deformability plot of PME-TLR4. (C) The main chain deformability plot of PME-MHC I. (D) The main chain deformability plot of PME-MHC II.
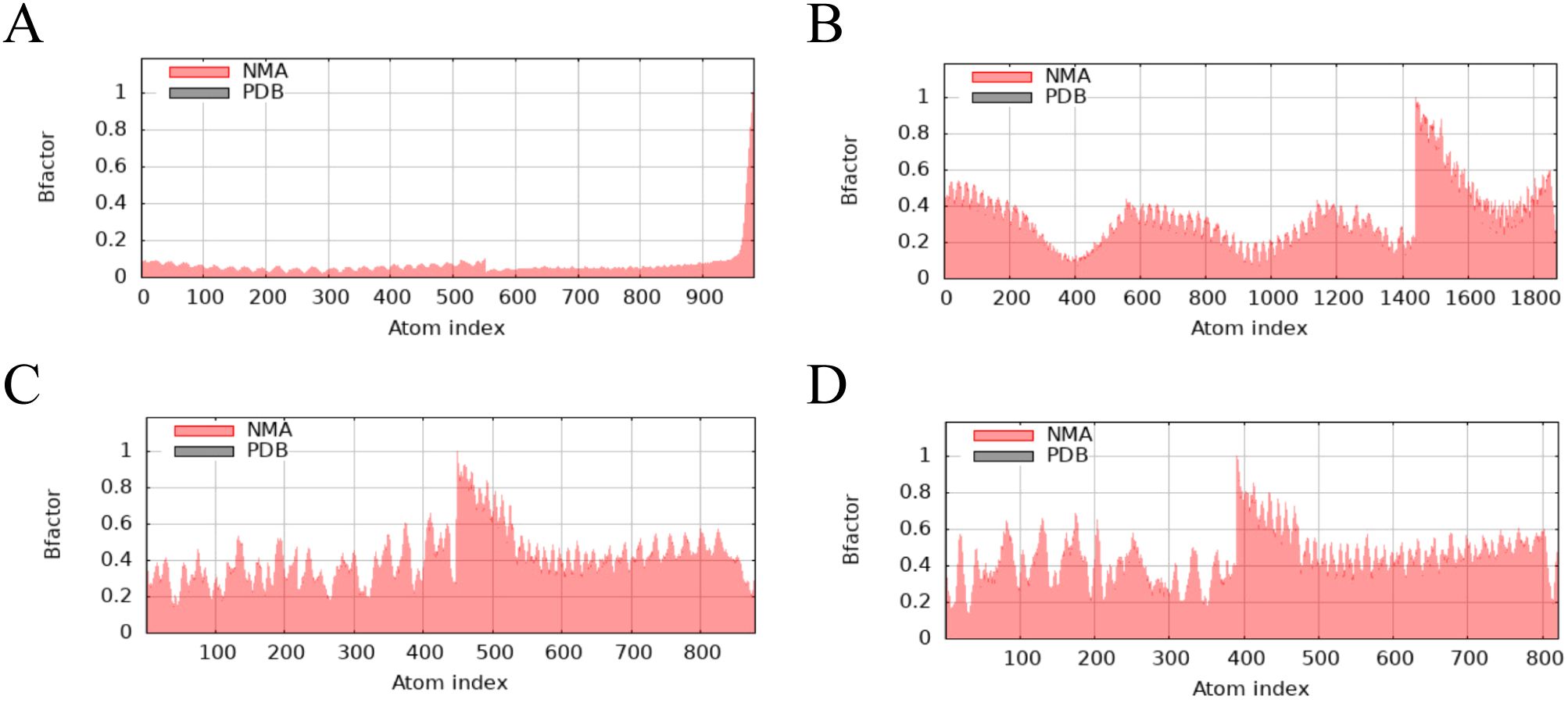
Figure 6. iMODS analysis revealed the B factor of PME-TLR2, PME-TLR4, PME-MHC I, and PME-MHC II. (A) The B factor plot of PME-TLR2. (B) The B factor plot of PME-TLR4. (C) The B factor plot of PME-MHC I. (D) The B factor plot of PME-MHC II.
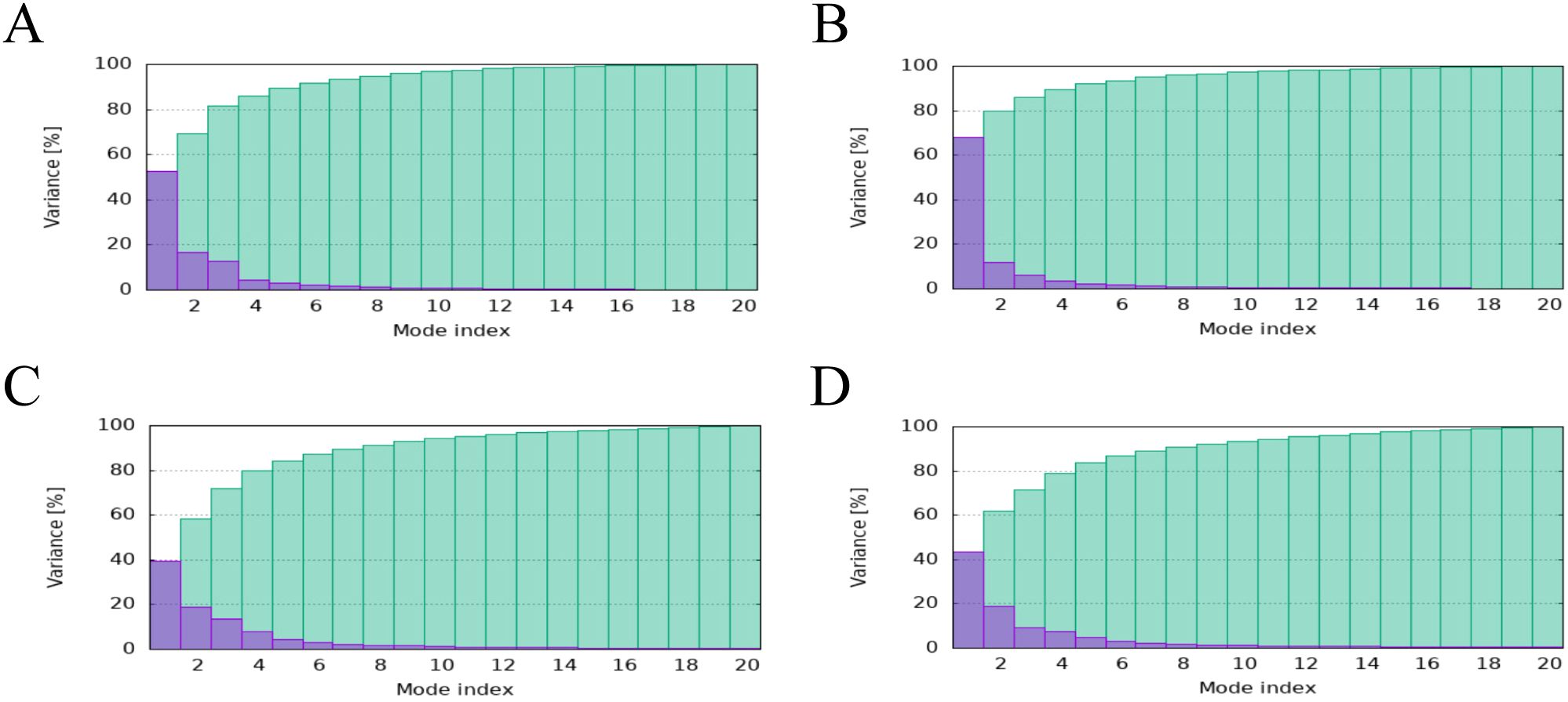
Figure 7. iMODS analysis revealed the Variance of PME-TLR2, PME-TLR4, PME-MHC I, and PME-MHC II. (A) The variance plot of PME-TLR2. (B) The variance plot of PME-TLR4. (C) The variance plot of PME-MHC I. (D) The variance plot of PME-MHC II.
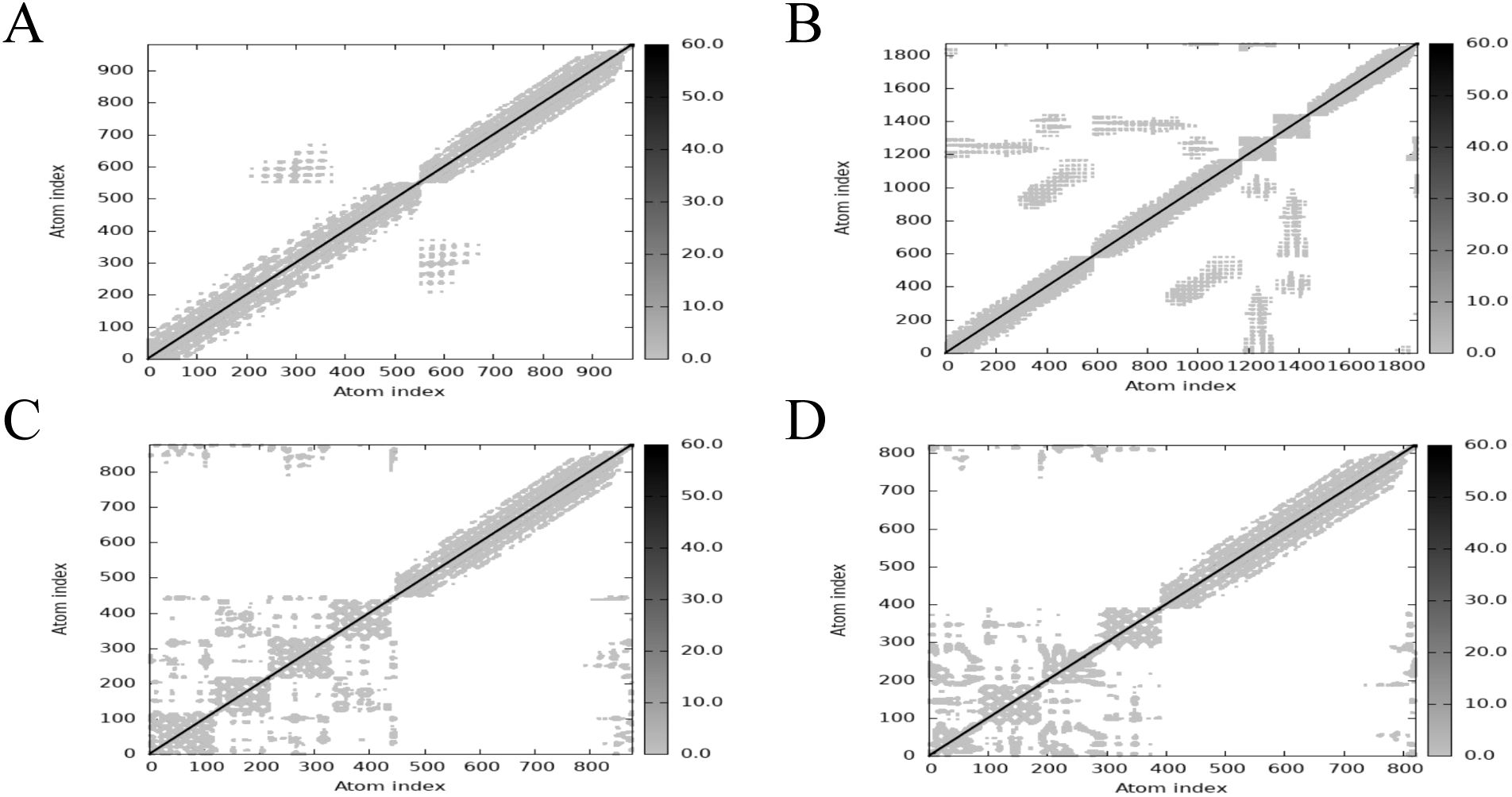
Figure 8. iMODS analysis revealed the Elastic network of PME-TLR2, PME-TLR4, PME-MHC I, and PME-MHC II. (A) The Elastic network model plot of PME-TLR2. (B) The Elastic network model plot of PME-TLR4. (C) The Elastic network model plot of PME-MHC I. (D) The Elastic network model plot of PME-MHC II.
3.6 Immunological simulation of PME
The immune response of the multi-epitope protein PME in vivo was simulated using C-ImmSim. The simulation results showed that the levels of antibodies and cytokines such as IL-2 and IFN-γ significantly increased after vaccinations (Figures 9A, B). Meanwhile, higher levels of B-cell and T-cell populations were observed, indicating the activation of cellular and humoral immunity in vivo (Figures 9C–E). These results suggested that the multi-epitope protein PME could induce a strong immune response in vivo.
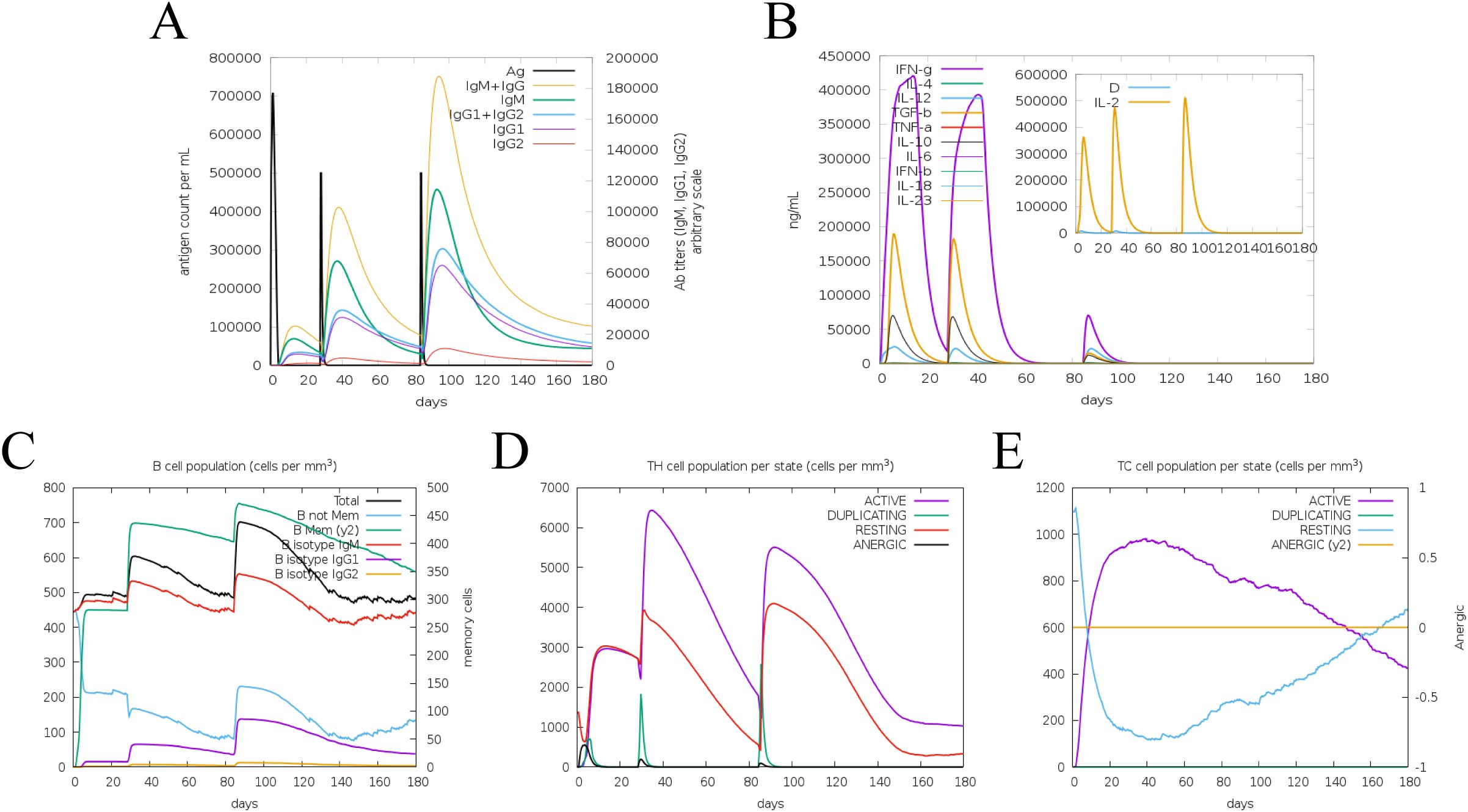
Figure 9. Simulate the immune response induced by PME in vivo. (A) Antibody level. Antibodies are subdivided by homotype. (B) Cytokine concentration. (C) Number of B cells. (D) Number of helper T lymphocytes. (E) Number of cytotoxic T lymphocytes.
3.7 Preparation and characterization of His-PME and pcDNA3.1-PME
The DNA sequence of PME was optimized and then inserted into prokaryotic expression vector pET30a (Figure 10A) and eukaryotic expression vector pcDNA3.1, respectively (Figure 10B). The results of double enzyme digestion indicated that the recombinant plasmids pET30a-PME (Figure 10C) and pcDNA3.1-PME (Figure 10D) were successfully constructed. SDS-PAGE showed that His-PME was successfully induced and expressed, and the protein size was consistent with the expectation (43 kDa) (Figure 10E). The purified protein His-PME was obtained by Ni-NTA affinity chromatography (Figure 10F), and the successful expression of His-PME was further confirmed by WB (Figure 10G).
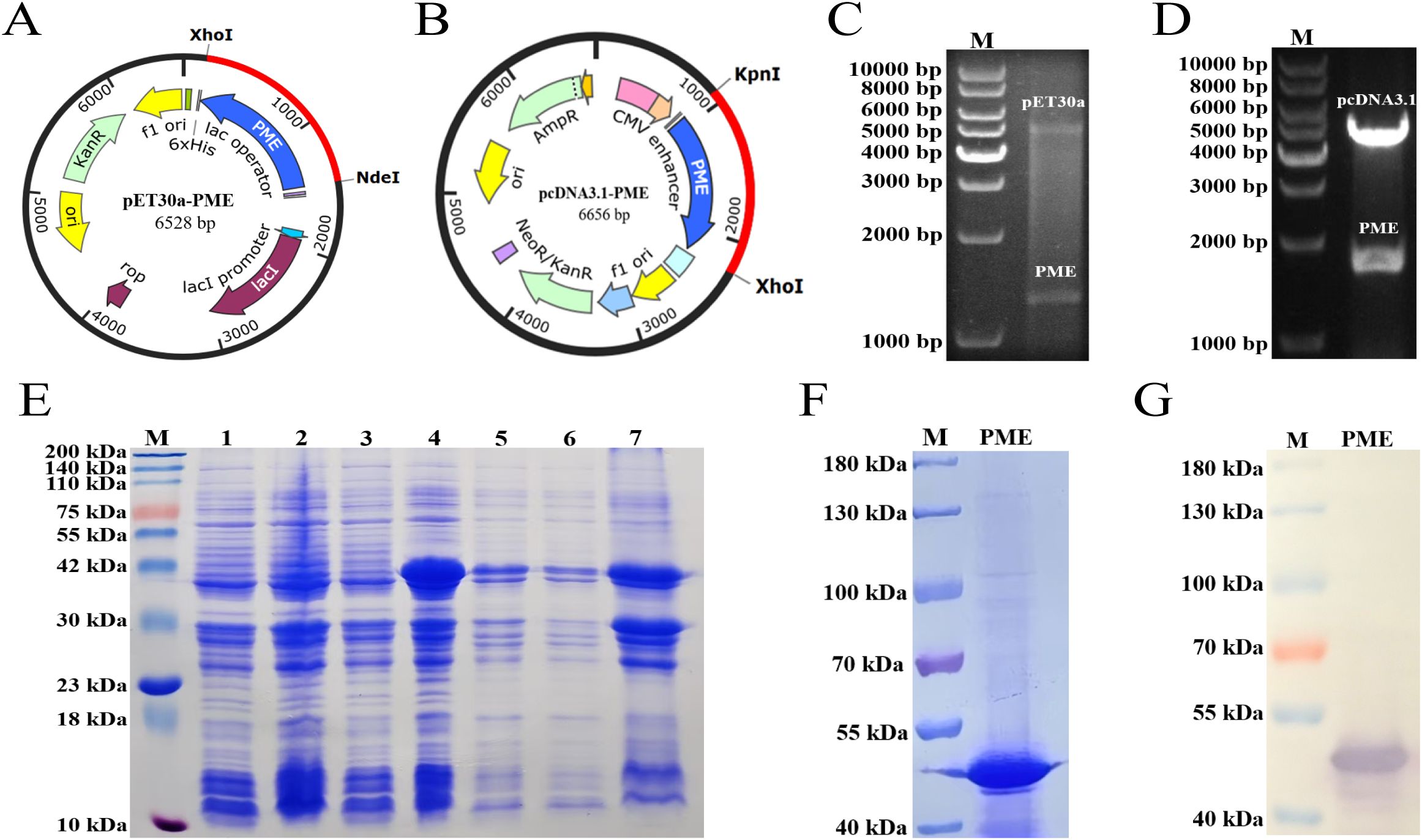
Figure 10. Cloning, expression and purification of His-PME. (A) Clone the codon optimized PME sequence (blue) into the pET30a vector. (B) Clone the codon optimized PME sequence (blue) into the pcDNA3.1 vector. (C) Identification of the recombinant plasmid pET30a-PME. The plasmid was cut with the restriction enzymes Xho I and Nde I. (D) Identification of the recombinant plasmid pcDNA3.1-PME. The plasmid was cut with the restriction enzymes Kpn I and Xho I. (E) SDS PAGE analysis of His-PME. M: 200 kDa marker. Line 1: Bacterial suspension of pET-30a BL21 without IPTG induction. Line 2: Bacterial suspension of pET-30a BL21 with IPTG induction. Line 3: Bacterial suspension of pET-30a-PME BL21 without IPTG induction. Line 4: Bacterial suspension of pET-30a-PME BL21 with IPTG induction. Line 5: Expression of His-PME protein from the whole cell lysate. Line 6: Expression of His-PME protein from the supernatant. Line 7: Expression of His-PME protein from the inclusion bodies. (F) The purification of the protein His-PME. M: 180 kDa marker. PME: protein His-PME after purification. (G) Western blot analysis of the purified protein His-PME. M: 180 kDa marker. PME: protein His-PME after purification.
3.8 Immune response of multi-epitope vaccine PME
To evaluate whether the multi-epitope vaccine PME could induce immune response after the immunization of mice, mice were immunized three times on days 1, 14, and 28 (Figure 11A). The results of immune response showed that the levels of antibodies (Figure 11B), IL-4 (Figure 11C), and IFN-γ (Figure 11D) in inactivated P. multocida group, His-PME group, and pcDNA3.1-PME group were significantly higher than those in the adjuvant group, pcDNA3.1 group, and PBS group. These results indicated that multi-epitope vaccine PME could induce a strong immune response.
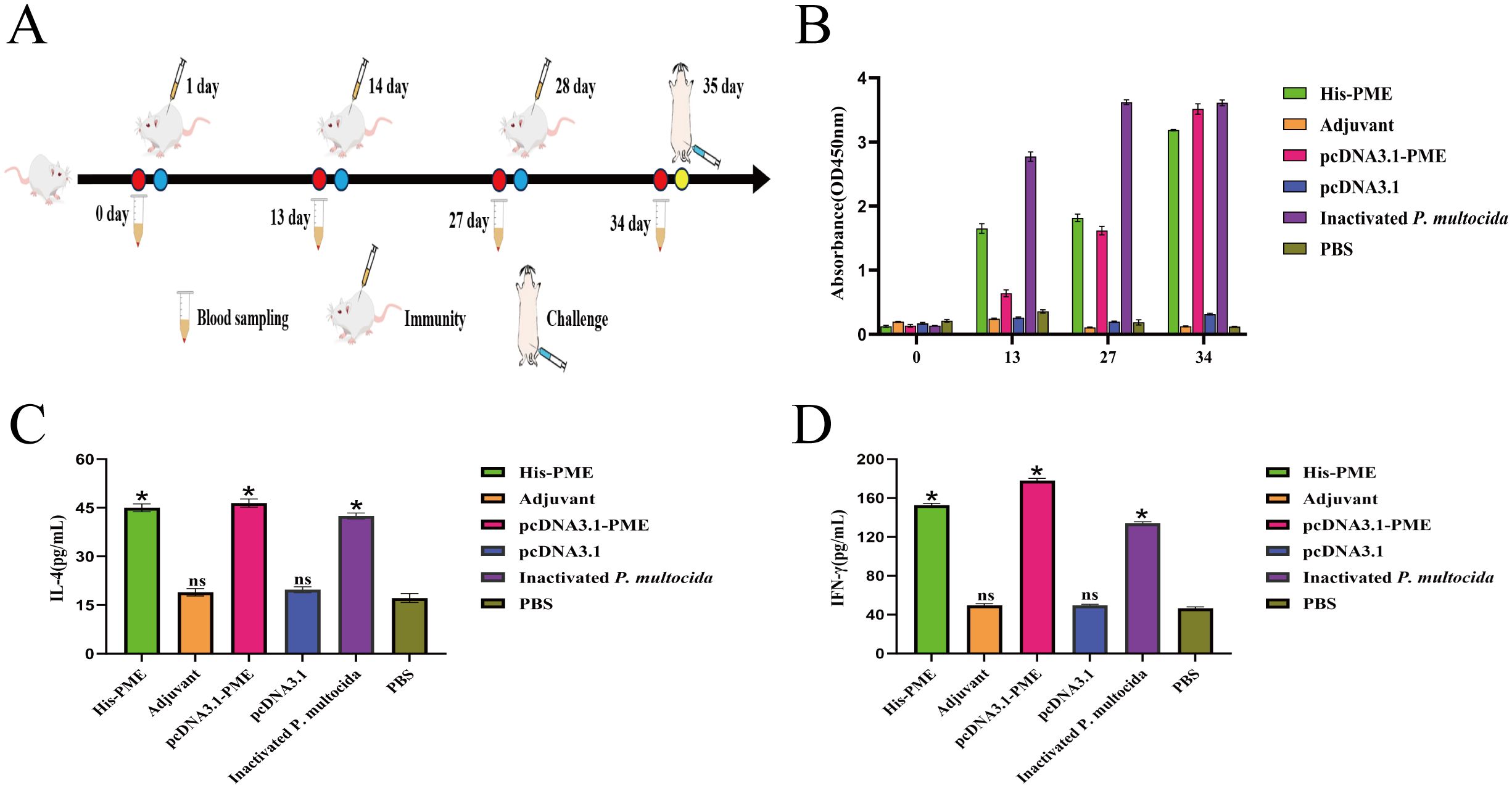
Figure 11. The immune response of His-PME and pcDNA3.1-PME in mice serum. (A) PME immunized mice protocol. (B) P. multocida serotypes D was used as the coating Antigen, and antibody levels of the mice in each groups. (C) The content of IL-4 in each group of immunized mice for 34 days. (D) The content of IFN-γ in each group of immunized mice for 34 days. The ns indicates no significant difference compared to the control group, *P < 0.05.
3.9 Protection against P. multocida in mice immunized by multi-epitope vaccine PME
The survival rates of mice in His-PME group, pcDNA3.1-PME group and the inactivated P. multocida group were 62.5%, 75% and 100% respectively after challenging with P. multocida serotype A, which were 87.5%, 100% and 100% respectively after challenging with P. multocida serotype D. Moreover, the mice in adjuvant group, PBS group and pcDNA3.1 group all died within 53 hours (Figure 12A).
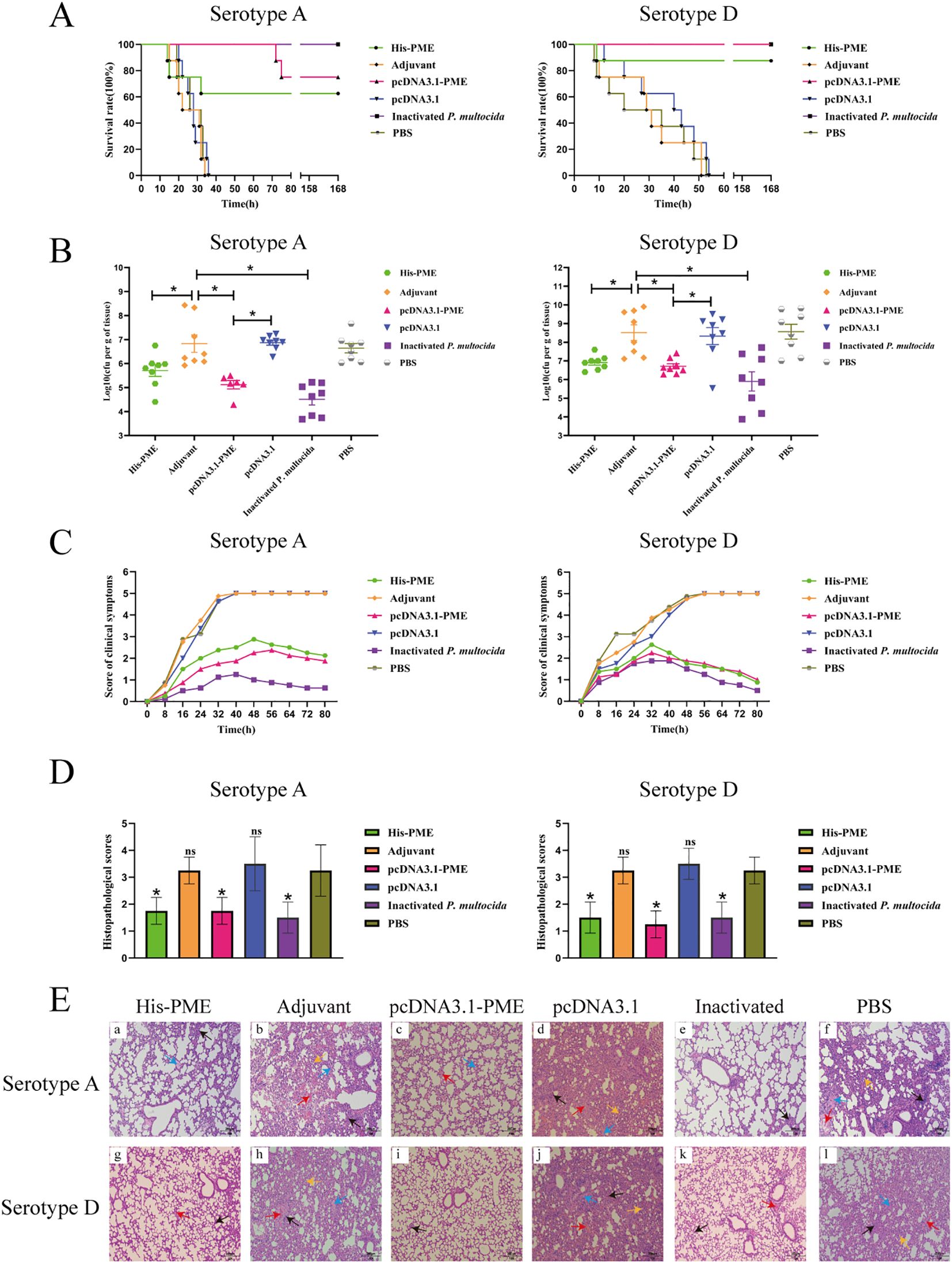
Figure 12. The protective effects of His-PME and pcDNA3.1-PME in immunized mice against challenges with P. multocida serotypes (A, D). (A) The survival rate of mice within 7 days after challenge to P. multocida serotypes (A, D). (B) Bacterial load in the lung tissue of mice after 8 hours challenge to P. multocida serotypes (A, D). (C) Clinical symptom scores of mice within 80 hours after challenge to P. multocida serotypes (A, D). (D) Lung histopathological scores of mice after challenge to P. multocida serotypes (A, D). (E) Lung histopathological analysis of mice after challenge to P. multocida serotypes (A, D) (HE ×200). Red arrow, hemorrhage; black arrow, inflammatory cell infiltration; blue arrow, hemolysis; yellow arrow, widening of the alveolar septa. The adjuvant, pcDNA3.1 and PBS groups b, d, f, h, j and l, severe hemorrhage, inflammatory cell infiltration, hemolysis and widening of the alveolar septa. The His-PME and pcDNA3.1-PME groups a, c, g and i, mild hemorrhage, inflammatory cell infiltration and hemolysis. The inactivated groups e and k, mild hemorrhage and inflammatory cell infiltration. The ns indicates no significant difference compared to the control group, *P < 0.05.
After the mice were challenged with P. multocida serotype A (PM-A) and D (PM-D), the bacterial loads in the lung tissues of His-PME group, pcDNA3.1-PME group, and inactivated P. multocida groups (PM-A or PM-D) were significantly lower than that of adjuvant group, PBS group, and pcDNA3.1 group (Figure 12B). The clinical symptoms and pathological changes in the lungs of mice in His-PME group, pcDNA3.1-PME group, and inactivated P. multocida groups (PM-A or PM-D) were significantly alleviated and their lung histopathological scores were significantly lower than those of mice in adjuvant group, PBS group, and pcDNA3.1 group (Figures 12C–E). These results indicated that the immune responses and cross-immunity provided by His-PME was good, and that of pcDNA3.1-PME was even better.
4 Discussion
Swine pasteurellosis is a serious threat to the development of modern pig industry, and the current research on inactivated vaccine, gene deletion vaccine and subunit vaccine cannot meet the needs of the prevention and control of this disease (5). As a novel subunit vaccine with good prospect, multi-epitope vaccine can design a novel candidate antigen sequence with high immunogenicity and multi-serotype cross-protection based on B-cell epitope and T-cell epitope predicted by target antigen, and introduce spacer sequence between epitopes (57, 58). In this study, we predicted the B-cell and T-cell epitopes of six P. multocida antigen proteins, and connected the epitopes end to end with GSG spacer to construct the multiple epitope antigen (PME). The results indicated that PME may be an effective candidate antigen for the prevention of P. multocida infection in the pig industry.
At present, the relevant researches on subunit vaccines against P. multocida mainly focuses on Omp and other key antigens (59). Compared with single antigen immunization, the mixed immunization of these known P. multocida antigens can obtain better immune effect (60). Yajuan Li et al. found that the protection rates against the challenge of P. multocida serotype A were 33.3%, 83.33% and 83.33% respectively when vaccinating ducklings with the outer membrane proteins VacJ, PlpE and OmpH alone, and the protection rate reached 100% while vaccinating with the three proteins in combination (7). Therefore, multi-epitope vaccines obtained by combining effective epitopes of multiple key antigen proteins can theoretically effectively enhance immune efficacy.
The rapid development and wide application of bioinformatics technology have given rise to a new field in vaccine design, where vaccines based on B-cell and T-cell epitopes can induce specific immune responses (61). Currently, the research of multi-epitope subunit vaccines relies on bioinformatics analysis, which is a promising strategy (62). Compared with single epitope vaccines, E.multilocularis multi-epitope vaccine GILE can induce stronger immune response, effectively activating the immune system to suppress E.multilocularis infection (63). This technique has been applied to the development of multi-epitope vaccines of B.melitensis and FMDV (64, 65). In this study, we predicted 20 B-cell epitopes, 7 CTL epitopes and 11 Th-cell epitopes from six antigenic proteins (PlpE, OmpA, OmpH, VacJ, Omp87 and Cp39). Spacer sequences GSG can guarantee epitope independence without producing new epitopes, and increase the immunogenicity of the polypeptide chain (66). Therefore, we insert GSG spacer sequence between the epitopes, and obtain multi-epitope recombinant protein (PME).
The antigenicity of a protein is closely related to its secondary structure (67). In PME, the proportions of α-helices, extended chains, β-turns and random coils are 1.4%, 5.13%, 0.47% and 93.01%, respectively. The α-helix and extended chain form stable structures inside proteins, which are relatively difficult to be recognized by the immune system. β-turn and random coil are exposed to the surface of the protein and can interact better with the lymphocyte, which has a positive effect on the immunogenicity of the protein (63). The conformational rationality and model quality of multi-epitope vaccines are critical indicators for assessing whether the vaccine’s epitopes can adopt a spatial structure comparable to that of natural proteins and effectively elicit immunogenicity (68). In this study, PME demonstrates both conformational rationality and high model quality, suggesting that its spatial conformation closely resembles that of natural antigenic proteins and that it holds promise for inducing a robust immune response.
Linear B-cell epitopes are composed of contiguous amino acid residues within the primary structure of antigenic proteins. These linear sequences can be directly recognized by the variable regions of antibodies, thereby initiating an immune response. Conformational B-cell epitopes, on the other hand, consist of amino acid residues that are non-contiguous in the primary sequence but come into close proximity in the three-dimensional structure of the folded protein. This structural characteristic more closely resembles the native antigenic state and plays a crucial role in inducing antibody production (69). In this study, PME contains a substantial number of both linear and conformational B-cell epitopes, indicating its potential to elicit a broad immune response and effectively defend against pathogen invasion.
TLR2 and TLR4 are key pattern recognition receptors in the innate immune system, capable of detecting pathogens, inducing the production of pro-inflammatory cytokines such as TNF-α and IL-6, and subsequently recruiting effector cells like neutrophils and macrophages to combat microbial invasion (70, 71). MHC I molecules activate CD8(+)T cells by presenting CTL epitopes, thereby promoting the secretion of cytokines such as IFN-γ (72). MHC II molecules activate CD4(+)T cells through the binding of Th epitopes, driving Th-cell differentiation and the production of cytokines including IFN-γ and IL-4, while also supporting B cells in antibody production. In this study, PME exhibited interaction sites with TLR2, TLR4, MHC I, and MHC II, with the lowest binding energy scores observed. This suggests that PME can be recognized by these immune molecules to initiate immune responses, and that the resulting complexes demonstrate high binding affinity. The rigid regions of a complex contribute to structural stability, while moderate flexibility and thermal motion can enhance antigenicity by exposing epitopes or facilitating immune processing (73). In this study, the PME-TLR2, PME-TLR4, PME-MHC I, and PME-MHC II complexes were predominantly rigid, exhibiting a spring-like configuration with limited susceptibility to thermal motion. These findings indicate that the four complexes are relatively resistant to external perturbations, and that their flexible regions may contribute to enhancing immune activation.
In PME, the proportion of random coil is relatively higher, which helps to improve its immunogenicity. In this study, the GEL 01 RP adjuvant was used to emulsify His-PME and pcDNA3.1-PME, and then mice were immunized. His-PME and pcDNA3.1-PME immunization induced strong serum antibody levels similar to those of the inactivated vaccine, suggesting that multi-epitope vaccine PME can induce strong humoral immune response in mice. Th1 and Th2 cells play a key role in the host cellular immune response. Th1 is involved in cellular immunity and delayed inflammatory hypersensitivity and can secrete IFN-γ, while Th2 mediates humoral immune response and can secretes IL-4 (74). In this study, His-PME and pcDNA3.1-PME induced significant increases in IFN-γ and IL-4 levels, suggesting that multi-epitope vaccine PME could induce strong Th1 and Th2 responses. These results further proved that multi-epitope vaccine PME has good immunogenicity.
Currently, there are few studies on multi-epitope vaccines against P. multocida. The multivalent vaccine rPMT for P. multocida is obtained by predicting the dominant B-cell epitopes, dominant peptides and dominant T-cell epitopes of the PMT protein. After immunizing mice with rPMT, the serum antibody was significantly increased. The protection rate against the challenge with P. multocida serotype D was 57.1%, and the lesions of lung tissue were significantly reduced. These results indicated that rPMT could be a candidate against P. multocida. However, the protection provided by rPMT was limited, and it had not been confirmed whether it could provide cross-protective immunity (75). In this study, the mice immunized with His-PME and pcDNA3.1-PME had a high immune protection against PM-A and PM-D challenge, of which the CFU of lung colonized colonies was significantly reduced, and the lesions were also significantly alleviated. These results suggested that His-PME and pcDNA3.1-PME can significantly reduce the lung injury caused by P. multocida and provide good cross-protection against P. multocida infection. Taken together, PME may be a suitable candidate multi-epitope vaccine, providing a new idea for the development of vaccines against P. multocida.
5 Conclusion
In this study, multi-epitope vaccine PME was designed by predicting the B-cell and T-cell epitopes of six antigen proteins of P. multocida, including PlpE, OmpA, OmpH, VacJ, Omp87 and Cp39. Bioinformatics was used to analyze the physicochemical properties, secondary and tertiary structures of PME, and the results showed that PME had the advantages of strong antigenicity and high stability. In a mouse model, both protein His-PME and plasmid pcDNA3.1-PME were able to protect mice against P. multocida serotypes A and D. These results showed that the multi-epitope vaccine PME can provide good immune and cross-protection, and is a candidate vaccine for the prevention of P. multocida infection.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was approved by The Animal Ethics Committee of the Yangtze University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
RZ: Data curation, Methodology, Formal analysis, Investigation, Software, Writing – original draft. LD: Investigation, Data curation, Formal analysis, Writing – review & editing, Methodology, Software, Writing – original draft. YJ: Methodology, Writing – review & editing. HQ: Methodology, Writing – review & editing. JH: Writing – review & editing, Methodology. JC: Writing – review & editing, Methodology. XG: Conceptualization, Writing – review & editing. LL: Writing – review & editing, Conceptualization. FL: Resources, Supervision, Funding acquisition, Project administration, Data curation, Writing – review & editing, Conceptualization.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This study was supported by the Natural Science Foundation of Hubei Province (No.2025AFB866).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1652907/full#supplementary-material
B-cell, B lymphocyte; CTL, Cytotoxic T lymphocyte; IEDB, Immune Epitope Database and Tools; IFN-γ, Interferon gamma; IL-4, Interleukin-4; IPTG, Isopropyl β-D-1-thiogalactopyranoside; MHC I, major histocompatibility complex I; MHC II, major histocompatibility complex II; Omp87, outer membrane protein 87; OmpA, outer membrane protein A; OmpH, outer membrane protein H; PlpE, outer membrane lipoprotein; VacJ, VacJ family lipoprotein; T-cell, T lymphocyte; Th-cell, helper T lymphocyte.
References
1. E-kobon T, Leeanan R, Pannoi S, Anuntasomboon P, and Thongkamkoon P. Thamchaipenet A OmpA protein sequence-based typing and virulence-associated gene profiles of Pasteurella multocida isolates associated with bovine haemorrhagic septicaemia and porcine pneumonic pasteurellosis in Thailand. BMC Veterinary Res. (2017) 13:243. doi: 10.1186/s12917-017-1157-6
2. Tigga M, Ghosh RC, Malik P, Choudhary BK, Tigga P, and Isolation NDK. characterization, antibiogram and pathology of Pasteurella multocida isolated from pigs. Veterinary World. (2014) 7:363–8. doi: 10.14202/vetworld.2014.363-368
3. Liu T, Cao L, Wang HR, Ma YJ, Lu XY, Li PJ, et al. Development and application of a WebGIS-based prediction system for multi-criteria decision analysis of porcine pasteurellosis. Sci Rep. (.(2024) 14:21082. doi: 10.1038/s41598-024-72350-x
4. Liu S, Lin L, Yang H, Wu W, Guo L, Zhang Y, et al. Pasteurella multocida capsular: lipopolysaccharide types D:L6 and A:L3 remain to be the main epidemic genotypes of pigs in China. Anim Dis. (.(2021) 1:26. doi: 10.1186/s44149-021-00031-7
5. He F, Xiong P, Zhang H, Yang L, Qiu Y, Li P, et al. Attenuated vaccine PmCQ2Δ4555-4580 effectively protects mice against Pasteurella multocida infection. BMC Vet Res. (2024) 20:94. doi: 10.1186/s12917-024-03948-6
6. Zhao X, Yang F, Shen H, Liao Y, Zhu D, Wang M, et al. Immunogenicity and protection of a Pasteurella multocida strain with a truncated lipopolysaccharide outer core in ducks. Veterinary Res. (2022) 53:17. doi: 10.1186/s13567-022-01035-y
7. Li Y, Xiao J, Chang Y-F, Zhang H, Teng Y, Lin W, et al. Corrigendum: Immunogenicity and protective efficacy of the recombinant Pasteurella multocida lipoproteins VacJ and PlpE, and outer membrane protein H from P. multocida A:1 in ducks. Front Immunol. (2023) 13:1128242. doi: 10.3389/fimmu.2022.1128242
8. Guan L-J, Song J-J, Xue Y, Ai X, Liu Z-J, Si L-F, et al. Immune Protective Efficacy of China’s Traditional Inactivated and Attenuated Vaccines against the Prevalent Strains of Pasteurella multocida in Mice. Vaccines. (2021) 9:1155. doi: 10.3390/vaccines9101155
9. Hanley KA. The double-edged sword: how evolution can make or break a live-attenuated virus vaccine. Evolution: Educ Outreach. (2011) 4:635–43. doi: 10.1007/s12052-011-0365-y
10. Hatfaludi T, Al-Hasani K, Gong L, Boyce JD, Ford M, Wilkie I W, et al. Screening of 71 P. multocida proteins for protective efficacy in a fowl cholera infection model and characterization of the protective antigen plpE. PloS One. (2012) 7:e39973. doi: 10.1371/journal.pone.0039973
11. Hatfaludi T, Al-Hasani K, Boyce JD, and Adler B. Outer membrane proteins of Pasteurella multocida. Veterinary Microbiol. (.(2010) 144:1–17. doi: 10.1016/j.vetmic.2010.01.027
12. Okay S, Özcengiz E, Gürsel İ, and Özcengiz G. Immunogenicity and protective efficacy of the recombinant Pasteurella lipoprotein E and outer membrane protein H from Pasteurella multocida A:3 in mice. Res Veterinary Science. (2012) 93:1261–5. doi: 10.1016/j.rvsc.2012.05.011
13. Varinrak T, Poolperm P, Sawada T, and Sthitmatee N. Cross-protection conferred by immunization with an rOmpH-based intranasal fowl cholera vaccine. Avian Pathol. (.(2017) 46:515–25. doi: 10.1080/03079457.2017.1321105
14. Kumar A, Yogisharadhya R, Ramakrishnan MA, Viswas KN, and Shivachandra SB. Structural analysis and cross-protective efficacy of recombinant 87 kDa outer membrane protein (Omp87) of Pasteurella multocida serogroup B:2. Microbial Pathogenesis. (.(2013) 65:48–56. doi: 10.1016/j.micpath.2013.09.007
15. Shivachandra SB, Kumar A, Yogisharadhya R, and Viswas KN. Immunogenicity of highly conserved recombinant VacJ outer membrane lipoprotein of Pasteurella multocida. Vaccine. (2014) 32:290–6. doi: 10.1016/j.vaccine.2013.10.075
16. Al-Hasani K, Boyce J, McCarl VP, Bottomley S, Wilkie I, and Adler B. Identification of novel immunogens in Pasteurella multocida. Microbial Cell Factories. (.(2007) 6:3. doi: 10.1186/1475-2859-6-3
17. Sthitmatee N, Numee S, Kawamoto E, Sasaki H, Yamashita K, Takahashi N, et al. Protection of chickens from fowl cholera by vaccination with recombinant adhesive protein of Pasteurella multocida. Vaccine. (2008) 26:2398–407. doi: 10.1016/j.vaccine.2008.02.051
18. Chiu ML, Goulet DR, Teplyakov A, and Gilliland GL. Antibody structure and function: the basis for engineering therapeutics. Antibodies. (2019) 8:55. doi: 10.3390/antib8040055
19. Tarrahimofrad H, Rahimnahal S, Zamani J, and Jahangirian E. Aminzadeh S Designing a multi-epitope vaccine to provoke the robust immune response against influenza A H7N9. Sci Rep. (2021) 11:24485. doi: 10.1038/s41598-021-03932-2
20. Oli AN, Obialor WO, Ifeanyichukwu MO, Odimegwu DC, Okoyeh JN, Emechebe G O, et al. Immunoinformatics and vaccine development: an overview. ImmunoTargets Ther. (2020) 9:13–30. doi: 10.2147/ITT.S241064
21. Li S, Anvari S, Ptacek G, Upadhyay I, Kaminski RW, and Sack DA. Zhang W A broadly immunogenic polyvalent Shigella multiepitope fusion antigen protein protects against Shigella sonnei and Shigella flexneri lethal pulmonary challenges in mice. Infection Immunity. (2023) 91:e00316–00323. doi: 10.1128/iai.00316-23
22. Gurunathan S, Klinman DM, and Seder RA. DNA vaccines: immunology, application, and optimization. . Annu Rev Immunol. (2000) 18:927–74. doi: 10.1146/annurev.immunol.18.1.927
23. Bolhassani A and Yazdi SR. DNA immunization as an efficient strategy for vaccination. Avicenna J Med Biotechnol. (2009) 1:71–88.
24. Wei H, Lenz SD, Thompson DH, and Pogranichniy RM. DNA-epitope vaccine provided efficient protection to mice against lethal dose of influenza A virus H1N1. . Viral Immunol. (.(2014) 27:14–9. doi: 10.1089/vim.2013.0080
25. Chou PY and Fasman GD. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. (1979) 47:45–148. doi: 10.1002/9780470122921.ch2
26. Emini EA, Hughes JV, Perlow DS, and Boger J. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J Virol. (1985) 55:836–9. doi: 10.1128/jvi.55.3.836-839.1985
27. Karplus PA and Schulz GE. Prediction of chain flexibility in proteins: A tool for the selection of peptide antigens. Naturwissenschaften. (1985) 72:212–3. doi: 10.1007/BF01195768
28. Kolaskar AS and Tongaonkar PC. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. (1990) 276:172–4. doi: 10.1016/0014-5793(90)80535-Q
29. Parker JMR, Guo D, and Hodges RS. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: correlation of predicted surface residues with antigenicity and x-ray-derived accessible sites. Biochemistry. (1986) 25:5425–32. doi: 10.1021/bi00367a013
30. Jespersen MC, Peters B, Nielsen M, and Marcatili P. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. (2017) 45:W24–9. doi: 10.1093/nar/gkx346
31. Andreatta M and Nielsen M. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics. (2016) 32:511–7. doi: 10.1093/bioinformatics/btv639
32. Moutaftsi M, Peters B, Pasquetto V, Tscharke DC, Sidney J, Bui H-H, et al. A consensus epitope prediction approach identifies the breadth of murine TCD8+-cell responses to vaccinia virus. Nat Biotechnol. (2006) 24:817–9. doi: 10.1038/nbt1215
33. Karosiene E, Lundegaard C, Lund O, and Nielsen M. NetMHCcons: a consensus method for the major histocompatibility complex class I predictions. Immunogenetics. (2012) 64:177–86. doi: 10.1007/s00251-011-0579-8
34. Zhang H, Lund O, and Nielsen M. The PickPocket method for predicting binding specificities for receptors based on receptor pocket similarities: application to MHC-peptide binding. Bioinformatics. (2009) 25:1293–9. doi: 10.1093/bioinformatics/btp137
35. Reynisson B, Alvarez B, Paul S, Peters B, and Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. (2020) 48:W449–54. doi: 10.1093/nar/gkaa379
36. Kim Y, Sidney J, Pinilla C, Sette A, and Peters B. Derivation of an amino acid similarity matrix for peptide:MHC binding and its application as a Bayesian prior. . BMC Bioinf. (2009) 10:394. doi: 10.1186/1471-2105-10-394
37. Gustafsson K, Germana S, Hirsch F, Pratt K, LeGuern C, and Sachs DH. Structure of miniature swine class II DRB genes: conservation of hypervariable amino acid residues between distantly related mammalian species. Proc Natl Acad Sci. (.(1990) 87::9798–9802. doi: 10.1073/pnas.87.24.9798
38. Xiao J, Liu J, Bao C, Zhu R, Gu J, Sun C, et al. Recombinant tandem epitope vaccination provides cross protection against Actinobacillus pleuropneumoniae challenge in mice. . AMB Express. (2020) 10:123. doi: 10.1186/s13568-020-01051-1
39. Akıl M, Aykur M, Karakavuk M, Can H, and Döşkaya M. Construction of a multiepitope vaccine candidate against Fasciola hepatica: an in silico design using various immunogenic excretory/secretory antigens. Expert Rev Vaccines. (.(2022) 21:993–1006. doi: 10.1080/14760584.2022.1996233
40. Xu Y, Zhu F, Zhou Z, Ma S, Zhang P, Tan C, et al. A novel mRNA multi-epitope vaccine of Acinetobacter baumannii based on multi-target protein design in immunoinformatic approach. BMC Genomics. (.(2024) 25:791. doi: 10.1186/s12864-024-10691-7
41. Li J, Xiong Y, Sun S, Yu L, and Huang C. Preparation of monoclonal antibodies against gamma-type phospholipase A2 inhibitors and immunodetection of these proteins in snake blood. J Venomous Anim Toxins including Trop Diseases. (2017) 23:37. doi: 10.1186/s40409-017-0128-5
42. Yao Q, Xie T, Fu Y, Wan J, Zhang W, Gao X, et al. The CpxA/CpxR two-component system mediates regulation of Actinobacillus pleuropneumoniae cold growth. Front Microbiol. (2022) 13:1079390. doi: 10.3389/fmicb.2022.1079390
43. Liu F, Yao Q, Huang J, Wan J, Xie T, Gao X, et al. The two-component system CpxA/CpxR is critical for full virulence in Actinobacillus pleuropneumoniae. Front Microbiol. (2022) 13:1029426. doi: 10.3389/fmicb.2022.1029426
44. Wan J, Zhang R, Jia Y, Xie T, Dai L, Yao Q, et al. The two-component system CpxAR is required for the high potassium stress survival of Actinobacillus pleuropneumoniae. Front Microbiol. (.(2023) 14:1259935. doi: 10.3389/fmicb.2023.1259935
45. Rostaminia S, Aghaei SS, Farahmand B, Nazari R, and Ghaemi A. Computational design and analysis of a multi-epitope against influenza A virus. . Int J Pept Res Ther. (2021) 27:2625–38. doi: 10.1007/s10989-021-10278-w
46. Zhang G, Han L, Zhao Y, Li Q, Wang S, and Shi H. Development and evaluation of a multi-epitope subunit vaccine against Mycoplasma synoviae infection. Int J Biol Macromolecules. (2023) 253:126685. doi: 10.1016/j.ijbiomac.2023.126685
47. Nguyen TL. Kim H Discovering peptides and computational investigations of a multiepitope vaccine target Mycobacterium tuberculosis. Synthetic Syst Biotechnol. (2024) 9:391–405. doi: 10.1016/j.synbio.2024.03.010
48. Shahrear S and Islam AB. M M K Modeling of MT. P495, an mRNA-based vaccine against the phosphate-binding protein PstS1 of Mycobacterium tuberculosis. Mol Diversity. (2023) 27:1613–32. doi: 10.1007/s11030-022-10515-4
49. Wei W, Behloul N, Baha S, Liu Z, Aslam MS, and Meng J. Dimerization: a structural feature for the protection of hepatitis E virus capsid protein against trypsinization. Sci Rep. (.(2018) 8:1738. doi: 10.1038/s41598-018-20137-2
50. Kawai T and Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. (2010) 11:373–84. doi: 10.1038/ni.1863
51. Jones G, Jindal A, Ghani U, Kotelnikov S, Egbert M, Hashemi N, et al. Elucidation of protein function using computational docking and hotspot analysis by ClusPro and FTMap. Acta Crystallographica Section D Struct Biol. (2022) 78:690–7. doi: 10.1107/S2059798322002741
52. López-Blanco JR, Aliaga JI, Quintana-Ortí ES, and Chacón P. iMODS: internal coordinates normal mode analysis server. Nucleic Acids Res. (2014) 42:W271–6. doi: 10.1093/nar/gku339
53. Rapin N, Lund O, Bernaschi M, and Castiglione F. Computational immunology meets bioinformatics: the use of prediction tools for molecular binding in the simulation of the immune system. PloS One. (2010) 5:e9862. doi: 10.1371/journal.pone.0009862
54. Luo J, Huo C, Qin H, Hu J, Lei L, and Pan Z. Chimeric enterovirus 71 virus-like particle displaying conserved coxsackievirus A16 epitopes elicits potent immune responses and protects mice against lethal EV71 and CA16 infection. Vaccine. (2021) 39:4135–43. doi: 10.1016/j.vaccine.2021.05.093
55. Yagnik B, Sharma D, Padh H, and Desai P. Oral immunization with LacVax® OmpA induces protective immune response against Shigella flexneri 2a ATCC 12022 in a murine model. Vaccine. (2019) 37:3097–105. doi: 10.1016/j.vaccine.2019.04.053
56. Xu K, Zhao Q, Jiang H-Z, Mou X-R, Chang Y-F, Cao Y-Q, et al. Molecular and functional characterization of HtrA protein in Actinobacillus pleuropneumoniae. Veterinary Microbiol. (2021) 257:109058. doi: 10.1016/j.vetmic.2021.109058
57. Ren S, Guan L, Dong Y, Wang C, Feng L, and Xie Y. Design and evaluation of a multi-epitope assembly peptide vaccine against Acinetobacter baumannii infection in mice. Swiss Med Weekly. (2019) 149:w20052. doi: 10.4414/smw.2019.20052
58. Zhang Y, Liang S, Zhang S, Zhang S, Yu Y, Yao H, et al. Development and evaluation of a multi-epitope subunit vaccine against group B Streptococcus infection. Emerging Microbes Infections. (2022) 11:2371–82. doi: 10.1080/22221751.2022.2122585
59. Almoheer R, Abd Wahid ME, Zakaria HA, Jonet MAB, Al-shaibani MM, Al-Gheethi A, et al. Spatial, temporal, and demographic patterns in the prevalence of hemorrhagic septicemia in 41 countries in 2005–2019: A systematic analysis with special focus on the potential development of a new-generation vaccine. Vaccines. (2022) 10:315. doi: 10.3390/vaccines10020315
60. Mostaan S, Ghasemzadeh A, Sardari S, Shokrgozar MA, Nikbakht Brujeni G, Abolhassani M, et al. Pasteurella multocida vaccine candidates: A systematic review. Avicenna J Med Biotechnol. (2020) 12:140–7.
61. Nosrati M, Hajizade A, Nazarian S, Amani J, Namvar Vansofla A, and Tarverdizadeh Y. Designing a multi-epitope vaccine for cross-protection against Shigella spp: An immunoinformatics and structural vaccinology study. Mol Immunol. (2019) 116:106–16. doi: 10.1016/j.molimm.2019.09.018
62. Pang M, Tu T, Wang Y, Zhang P, Ren M, Yao X, et al. Design of a multi-epitope vaccine against Haemophilus parasuis based on pan-genome and immunoinformatics approaches. Front Veterinary Sci. (2022) 9:1053198. doi: 10.3389/fvets.2022.1053198
63. Zhou P, Zhou Z, Huayu M, Wang L, Feng L, Xiao Y, et al. A multi-epitope vaccine GILE against Echinococcus Multilocularis infection in mice. Front Immunol. (2022) 13:1091004. doi: 10.3389/fimmu.2022.1091004
64. Li M, Zhu Y, Niu C, Xie X, Haimiti G, Guo W, et al. Design of a multi-epitope vaccine candidate against Brucella melitensis. Sci Rep. (2022) 12:10146. doi: 10.1038/s41598-022-14427-z
65. De León P, Cañas-Arranz R, Defaus S, Torres E, Forner M, Bustos M J, et al. Swine T-cells and specific antibodies evoked by peptide dendrimers displaying different FMDV T-cell epitopes. Front Immunol. (2021) 11:621537. doi: 10.3389/fimmu.2020.621537
66. Ducharme M and Lapi SE. Peptide based imaging agents for HER2 imaging in oncology. Mol Imaging. (2020) 19:1536012120960258. doi: 10.1177/1536012120960258
67. Huang L, Mai L, Zhong K, and Chen X. Bioinformatics Analysis, Cloning and expression of spirometra erinaceieuropaei fatty acid-binding protein. Pakistan J Zoology. (2022) 54:1027–35. doi: 10.17582/journal.pjz/20210303090323
68. Shovon MHJ, Imtiaz M, Biswas P, Tareq MMI, and Zilani MNH. Hasan M N A pan-genomic analysis based multi-epitope vaccine development by targeting Stenotrophomonas maltophilia using reverse vaccinology method: an in-silico approach. In Silico Pharmacol. (2024) 12:93. doi: 10.1007/s40203-024-00271-8
69. Lu S, Li Y, Ma Q, Nan X, and Zhang S. A structure-based B-cell epitope prediction model through combing local and global features. Front Immunol. (2022) 13:890943. doi: 10.3389/fimmu.2022.890943
70. Oliveira-Nascimento L, Massari P, and Wetzler LM. The role of TLR2 in infection and immunity. Front Immunol. (2012) 3:79. doi: 10.3389/fimmu.2012.00079
71. Buchanan MM, Hutchinson M, Watkins LR, and Yin H. Toll-like receptor 4 in CNS pathologies. J Neurochem. (.(2010) 114:13–27. doi: 10.1111/j.1471-4159.2010.06736.x
72. Ren YL, Li TT, Cui W, Zhao LM, Gao N, Liao H, et al. CD8(+) T lymphocyte is a main source of interferon-gamma production in Takayasu’s arteritis. Sci Rep. (2021) 11:17111. doi: 10.1038/s41598-021-96632-w
73. Liang Y, Guttman M, Davenport TM, Hu SL, and Lee KK. Probing the impact of local structural dynamics of conformational epitopes on antibody recognition. Biochemistry. (2016) 55:2197–213. doi: 10.1021/acs.biochem.5b01354
74. Dai L, Wan J, Zhang R, Xie T, Jia Y, Lu Z, et al. Multi-epitope vaccines Xlc and Ddc against Glaesserella parasuis infection in mice. Vet Microbiol. (2025) 304:110491. doi: 10.1016/j.vetmic.2025.110491
Keywords: Pasteurella multocida, multi-epitope vaccine, DNA vaccine, immunogenicity, cross-protection
Citation: Zhang R, Dai L, Jia Y, Qi H, He J, Cheng J, Gao X, Lei L and Liu F (2025) Evaluation of a multi-epitope vaccine PME for Pasteurella multocida in mouse model. Front. Immunol. 16:1652907. doi: 10.3389/fimmu.2025.1652907
Received: 24 June 2025; Accepted: 12 August 2025;
Published: 01 September 2025.
Edited by:
Chamith Hewawaduge, Sri Lanka Institute of Biotechnology, Sri LankaReviewed by:
Nattawooti Sthitmatee, Chiang Mai University, ThailandWeifeng Zhu, Hebei Agricultural University, China
Copyright © 2025 Zhang, Dai, Jia, Qi, He, Cheng, Gao, Lei and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng Liu, bGl1ZmVuZzY4NDMxQDE2My5jb20=
 Rui Zhang1
Rui Zhang1 Lu Dai
Lu Dai Xuejun Gao
Xuejun Gao Liancheng Lei
Liancheng Lei Feng Liu
Feng Liu