- State Key Laboratory of Oral Diseases, National Clinical Research Center for Oral Diseases, Chinese Academy of Medical Sciences Research Unit of Oral Carcinogenesis and Management, West China Hospital of Stomatology, Sichuan University, Chengdu, Sichuan, China
SUMOylation, a dynamic regulatory process in post-translational modifications (PTMs) mediated by small ubiquitin-like modifier (SUMO) ligases and deSUMOylases, regulates protein function through reversible lysine conjugation. Emerging evidence has identified tumor-mediated hijacking of SUMOylation in both malignant cells and immune components as a novel immune evasion mechanism. This review represents a comprehensive update on how tumor-intrinsic SUMOylation modulates tumor immunity-related JAK/STAT, MHC-I, NF-κB, IFN-I/II pathways and other key proteins to drive its immune evasion, and immune cell-intrinsic SUMOylation in regulating natural killer (NK) and T cell cytotoxicity, dendritic cell (DC) maturation, and macrophage polarization. Tumor immunotherapy is a new potential strategy for cancer, mainly represented by immune checkpoint inhibitions (ICIs), which exhibits poor efficacy in head and neck squamous cell carcinoma (HNSCC), pancreatic ductal adenocarcinoma (PDAC) and other solid tumors. Targeting SUMOylation of tumors presents high potential to synergistically improve the therapeutic effect of ICIs. Preclinical studies have shed light on the therapeutic potential of the combination of SUMOylation inhibitors such as TAK-981 or 2-D08 with ICIs, thus significantly improving tumor prognosis. As current phase I trials suggest dose-dependent toxicity of TAK-981, there is a need for targeted delivery systems; AI-assisted screening of novel SUMOylation inhibitors (SUMOi) which are FDA approved serves as another potential approach; besides, antibodies against these pivotal SUMOylated molecules in tumors could be conjugated with SUMOi to restore the activity of specific proteins in tumor microenvironment. In all, our review proposes that current or other novel strategies for SUMOylation inhibition stands as a promising adjuvant to immunotherapy for tumor management, thereby potentially contributing to the favorable prognosis of cancer patients.
Introduction
Recently, numerous studies have indicated that post-translational modifications (PTMs) of proteins, including methylation, SUMOylation, phosphorylation, acetylation, lactylation, and glycosylation, harbors high potential to regulate various aspects of cancer progression. These modifications significantly influence tumor growth, metastasis, dysregulated metabolism, and evasion from immune surveillance (1–4). SUMOylation is a dynamic and reversible ubiquitin-like modification that preserves the structural integrity and functional homeostasis of proteins, and the process is accomplished by covalent conjugation of small ubiquitin-like modifier (SUMO) to lysine residues (Lys/K) on substrate proteins (5). Interestingly, growing evidence has shown that hyperactivation of SUMOylation is a hallmark of cancer, with elevated protein SUMOylation levels observed in most cancers (6–8). In particular, SUMOylation of nuclear proteins, the major target of SUMO, serves as a master regulatory hub governing most nuclear processes and diverse cellular programs, including DNA damage repair, transcription regulation, apoptotic machinery, cytokine secretion, and modulation of oncogenes and tumor suppressor genes, thereby modulating tumor growth, migration, inflammation, and angiogenesis signals to mediate its progression (7, 9–11). However, it seems likely that the influence of SUMOylation on tumor suppression or progression is cancer context-dependent. For instance, sentrin-specific protease 1 (SENP1) promotes hepatocarcinogenesis and enhances the stemness of hepatoma cells by reinforcing the deSUMOylation of hypoxia-inducible factor-1 (HIF-1α), thus increasing the stability and transcription of HIF-1α (12). DeSUMOylation of signal transducer and activator of transcription 3 (STAT3) induced by SENP3 improves its transcriptional and oncogenic potential in head and neck cancers (13). In contrast, deSUMOylation of β-catenin may inhibit the expansion of myeloma by accelerating the degradation of β-catenin via the ubiquitin-proteasomal system, thereby downregulating the Wnt/β-catenin pathway (14). In addition, SUMO modification of the NOP2/Sun domain (NSUN2) facilitates its carcinogenic activity by stabilizing NSUN2 and enabling its nuclear trafficking (15). Similarly, SUMOylation of Mouse double minute 2 homolog (MDM2) accounts for the development of colon or prostate cancer by triggering p53 degradation (16, 17).
The immune system serves a pivotal function in safeguarding human physiological integrity when undergoing attack by foreign invasion, including the early recognition of tumor antigens, which is supported by the clinical observation that immunodeficient patients have a higher risk of cancer (18). Hence, the intactness of anti-tumor immunity is critical for the elimination of tumors. Anti-tumor immunity is generally comprised of innate immunity, which is the non-specific defense against tumor predominantly directed by neutrophils, macrophages and natural killer cells (NKs), and adaptive immunity, which is specific and exerts more robust killing of cancer cells, such as cellular immunity primarily executed by cytotoxic T cells (19, 20). Given this, tumor immunotherapy is becoming an emerging and promising approach for the management of tumors and has revolutionized the traditional treatment efficacy, represented by the application of immune checkpoint inhibitions (ICIs); however, limited efficacy has been observed in patients with pancreatic ductal adenocarcinoma (PDAC) and head and neck squamous cell carcinoma (HNSCC) (21, 22). To seek for the potential reason, it might be largely attributed to the evolving mechanisms of tumors to avoid immune killing, such as the impaired MHC-I antigen presentation in HNSCC (23). Notably, studies have indicated that SUMOylation of proteins within cancer cells or immune cells plays an important role in modulating anti-tumor immunity, including the JAK/STAT, MHC-I antigen presentation, IFN-I/II pathway and so forth (24–28). It is of great value to dissect the key mechanisms underpinning SUMOylation-mediated tumor immune escape, thus seeking more potent strategies to augment immunotherapeutic efficacy. In this review, we briefly discuss the routine process of SUMOylation and focus on the molecular regulatory mechanisms by which SUMOylation of certain proteins wrestles anti-tumor immunity.
Overview of physiological processes of SUMOylation
The process of SUMOylation is a tri-enzymatic cascade with SUMO proteins being bound to target proteins under the catalysis of SUMO E1 activating enzyme (SAE1/2), followed by the single SUMO E2 conjugating enzyme (UBC9), as well as the substrate-specific SUMO E3 ligase, which affects the stability and biological activity, degradation, and orientation biological function of targeted proteins, and a special E4 enzyme named ZNF451 has been identified to facilitate the assembly of SUMO-chains (7, 11, 29) (Figure 1). To date, approximately five paralogs of SUMO proteins (SUMO1-5) can be expressed by mammalian cells, among which SUMO2/3 is the most abundant protein that can form chains by connecting with each other (8, 11, 30). Due to the 97% sequence identity of SUMO2 and SUMO3, they are collectively named as SUMO2/3, while SUMO1 has only 53% sequence resemblance with SUMO2/3 (31). Under the catalysis of these enzymes, SUMO is attached to the lysine (Lys/K) of targeted proteins to induce mono-, multi-, and poly-modification (6, 30, 32). Moreover, poly-SUMOylation of the substrate protein can be identified by SUMO-targeted ubiquitin ligase (STUbL, including RNF4 and RNF111), leading to its degradation by ubiquitination, which could be reversed by ubiquitin protease (STUbP, including USP7, USP11, and ATX3, which targets SUMO molecules), highlighting the intricate interplay between SUMOylation and ubiquitination in protein regulation (30, 33). In addition to the enzymes responsible for SUMOylation, some other enzymes are involved in deSUMOylation, named as SUMO-specific protease (SENP), thus removing the SUMOylation modification of targeted proteins. Specifically, six types of SENPs (SENP1/2/3/5/6/7) and the newly identified DESI1, DESI2, and USPL1 enzymes implicate in this process (7).
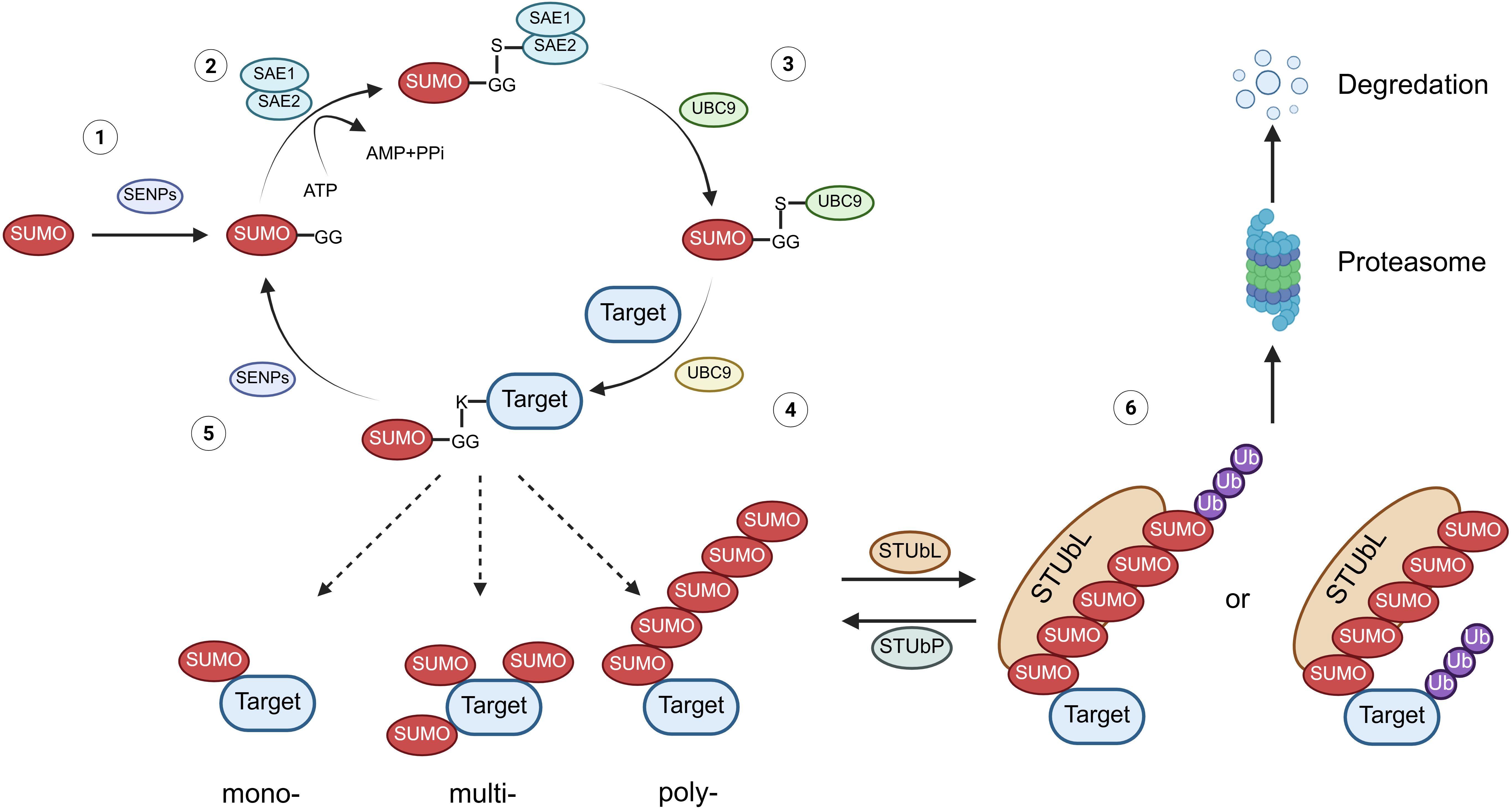
Figure 1. The physiological process of protein SUMOylation. (1) Maturation: The C-terminal Gly-Gly (G-G) motif of the SUMO protein is exposed after cleavage by SENPs. (2) Activation: The mature SUMO protein forms a thioester bond with SAE1/2 (E1) in an ATP-dependent manner. (3) Conjugation: The SUMO protein is transferred from E1 to the SUMO-conjugating enzyme UBC9 (E2). (4) Ligation: SUMO is covalently attached to lysine residues on the substrate protein, resulting in mono-, multiple-, or poly-SUMOylation. (5) DeSUMOylation: At various stages, SUMO can be cleaved from the substrate protein by SENPs or other deSUMOylation enzymes, thus reversing the modification. (6) Ubiquitination: Poly-SUMOylation of substrate proteins is recognized by STUbLs, leading to their subsequent degradation via the proteasomal pathway. Conversely, ubiquitination of these substrates can be reversed by STUbPs. SENPs, sentrin-specific proteases; STUbLs, SUMO-targeted ubiquitin ligases; STUbPs, SUMO-targeted ubiquitin-specific proteases.
The tumor cell-intrinsic SUMOylation in the modulation of anti-tumor immunity
JAK/STAT pathway
The protein inhibitor of activated STAT (PIAS) family was initially discovered as a suppressor of STAT in the JAK/STAT pathway by impairing the DNA-binding capacity of its cognate recognition motifs (34, 35). Later, it was revealed that the PIAS family possesses E3 ligase activity, which regulates a variety of proteins via SUMOylation (36). In line with this, some researchers have found that SUMOylation modulates innate immunity by impairing virus-triggered type I interferon (IFN-I) production and disrupting IFN-I/II-dependent STAT1 and STAT2 activation cascades (37, 38). Maarifi et al. reported that SUMO hyperexpression drives covalent SUMO conjugation to STAT1-K703 and results in the impairment of IFN-I/II-triggered STAT1 activation dynamics in several cancers, including human glioblastoma astrocytoma, cervical cancer, and hepatocellular carcinoma, and IFN-I/II plays a negative feedback regulation on its own signal by enhancing the SUMOylation of STAT1 (24) (Figure 2). Besides, a recent study revealed that PIAS1 can be SUMOylated by the tumor suppressor p14ARF, which consequently inhibits PIAS1-mediated SUMOylation of STAT1, thereby effectively enhancing the IFN-γ-induced immune response (39). Beyond PIAS1, researchers have identified a protein-coding circular non-coding RNA, circPIAS1, which could be translated into a 108-amino acid peptide (circPIAS1-108aa) (40). This peptide can recruit the SUMO E3 ligase RANBP2 to promote STAT1 SUMOylation, consequently suppressing STAT1 phosphorylation and subsequently facilitating immune escape of melanoma (40). These indicate that during cellular stress responses, SUMOylation-mediated regulation of the JAK/STAT pathway is evolutionarily conserved to prevent excessive activation of the immune system. However, the mechanism that hyperactivated SUMOylation may impair JAK/STAT signaling could be hijacked by cancers to promote its immune evasion.
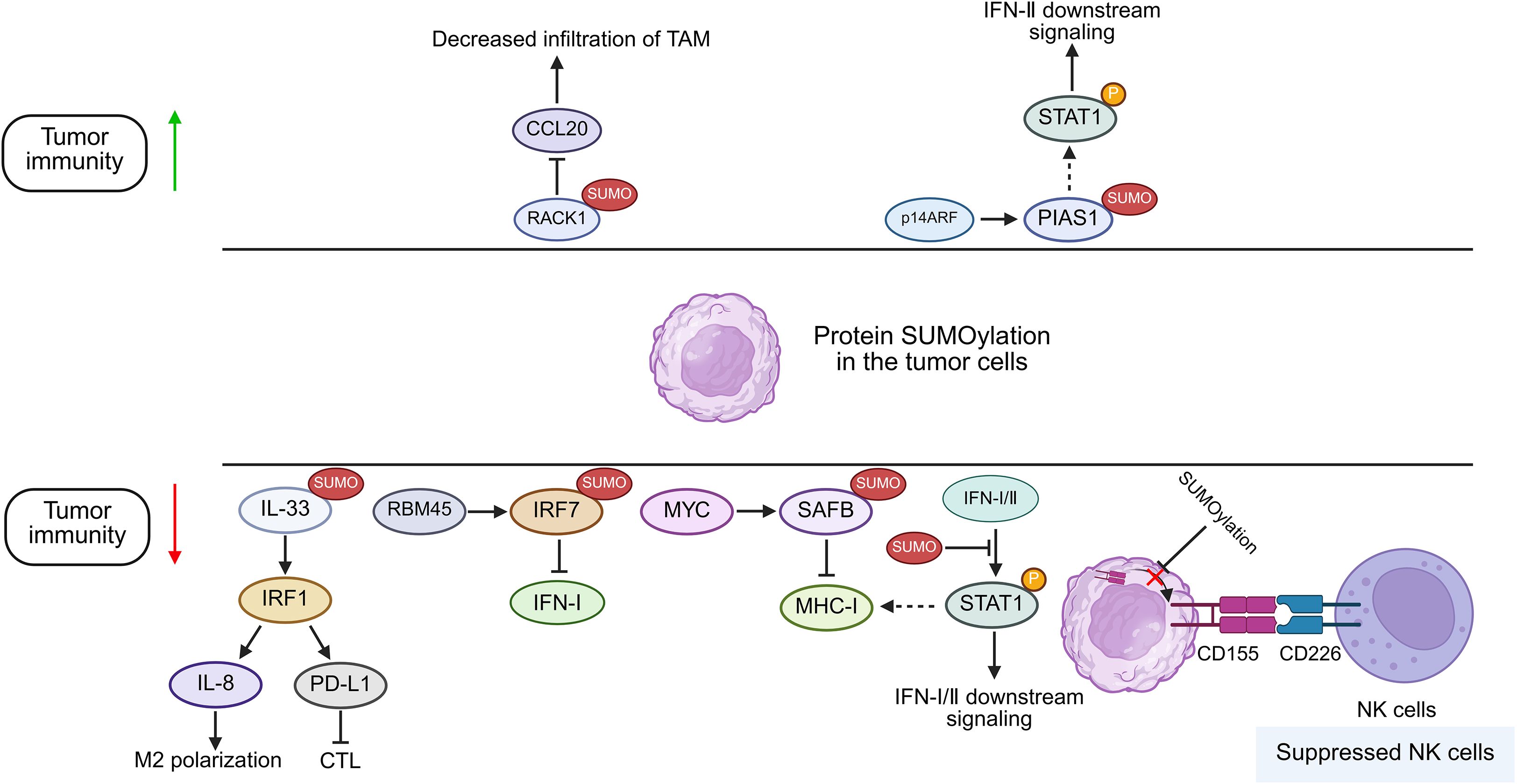
Figure 2. Proposed function of protein SUMOylation in cancer cells in modulating tumor immunity. Increased SUMOylation of targeted proteins in tumor cells can potentiate or suppress anti-tumor immunity by regulating IFN-I/II signaling, the infiltration of CTLs, NK cells and TAMs. RBM45, RNA Binding Motif Protein 45; SAFB, scaffold attachment factor B; MHC-I, major histocompatibility complex I; STAT, signal transducer and activator of transcription; PIAS, protein inhibitor of activated STAT; RACK1, Receptor For Activated C Kinase 1; p14ARF, p14 alternative reading frame; IFN: Interferon; IL-33, Interleukin 33; IRF, Interferon Regulatory Factor; PD-L1, Programmed cell death ligand 1; NK cells, natural killer cells; CTL, cytotoxic T lymphocyte; TAM, tumor associated macrophages.
MHC-I antigen presentation pathway and MYC
It is generally recognized that sufficient infiltration of T cells, especially cytotoxic T cells, within the tumor microenvironment (TME) is indispensable for the therapeutic efficacy of ICIs for cancers, while the absence of abundant T cells is usually observed in cancers such as HNSCC (23). As MHC-I-associated antigen presentation, including antigen production, activation of T cells, and specific recognition of tumors, is crucial as the first specific signal for T cell activation, the intactness of the pathway is highly appreciated for its normal function (41). Dysfunction of the MHC-I is a well-established cause of both primary and acquired resistance to cancer immunotherapies (42). To date, both JAK/STAT and NF-κB pathways have been shown to be important for upregulating the MHC-I pathway (18). Attenuated MHC-I antigen presentation is frequently observed, among which SUMOylation might partially contribute to its suppression by impairing JAK/STAT pathway based on aforementioned studies. Moreover, the fact that co-administration of TAK981, a specific inhibitor of SUMOylation, and PD-1 blockade markedly enhanced the survival of tumor-bearing mice further supports these findings (43).
MYC gene, which is dysregulated in 70% of tumors, serves as a predominant oncogenic driver across human cancers, encoding a family of transcription factors, with the products serving as key regulators of cell proliferation, cell differentiation, cell cycle, and metabolism (44, 45, 86). Thus, targeting of MYC exhibits a wide range of therapeutic effects in tumors. However, MYC is an inherently disordered protein with no stable conformation and no proper site suitable for small-molecule binding. Recently, it has been found that SUMOylation of MYC catalyzed by PIAS1 not only upregulates MYC by preventing its degradation but also promotes MYC phosphorylation, thereby leading to its higher transcriptional activity in B-cell lymphoma (46). Using murine lung and colon cancer models, Kotani’s team demonstrated that pharmacological inhibition of MYC by TAK981 promotes activation of STING-IFN-I pathway with consequent STAT1 phosphorylation and MHC-I upregulation in KRAS-mutant cancer cells (47). Scaffold attachment factor B (SAFB) was previously reported to inhibit MHC-I in an SUMOylation-dependent manner (81). Demel further validated that MYC promotes SUMO2/3 modification of SAFB in several cancers, encompassing B lymphoma and osteosarcoma, consequently inhibiting MHC-I expression and inducing tumor immune escape (8, 48). Moreover, they found that TAK981 could reverse this process via reverting MHC-I expression, thereby augmenting anti-tumor immunity (8). Thus, inhibiting SUMOylation of MYC and its downstream proteins may alternatively inhibit the oncogenic function of MYC and promote adaptive immune responses in cancers.
NF-κB signaling
Accumulating evidence indicates that NF-κB signaling is not only crucial for the occurrence, proliferation, differentiation, apoptosis, invasion, and metastasis of cancer cells but also modulates tumor immunity. For instance, one study on lung cancer indicated that activation of NF-κB augments regulatory T cell (Treg) ontogeny as well as functional polarization, whose tumor infiltrative capacity correlates with adverse clinical outcomes (49). Similarly, zhou et al. suggested that NF-κB may promote the expression of chemokines in lung cancer, such as cytokine ligand 2 (CCL2), thus facilitating monocyte recruitment and infiltration of immunosuppressive TAMs to the tumor bed (50). In contrast, Matthew et al. found that stimulation of T cell-intrinsic canonical NF-κB enhanced its activation and effector function, further increasing the clonal expansion of tumor-infiltrated CD8+ T cells (51). Moreover, NF-κB contributes to the transcriptional regulation of MHC-I in neoplastic populations (18).
Notably, multiple recent studies have revealed that SUMOylation might regulate this pathway by interacting with several related key proteins. For example, the SUMOylation of certain proteins can stimulate the NF-κB pathway. Nuclear receptor 4A (NR4A1) participates in suppressing NF-κB signaling induced by IL-1β and TNF-α, but SUMOylated NR4R1 precipitates its ubiquitin-proteasomal turnover, thus upregulating the NF-κB pathway in HeLa and Jurkat cells (52, 53). Sophia et al. pointed out that SUMOylation of TNF receptor-related factor 3 (TRAF3) mediated by UBC9 results in its association with the CD40 receptor, which ultimately leads to the degradation of TRAF3 and non-canonical NF-kB activation in HeLa cell (54). In addition, SUMOylated NEMO is essential for its translocation to the cytoplasm and further induction of NF-κB activation (55). In contrast, the RWD-containing SUMO enhancer (RSUME), by interacting with the SUMO E2 ligase UBC9, could promote covalent SUMO modification of IκBα, leading to suppression of NF-κB transcription output (56). However, the functional consequences of SUMOylation-mediated NF-κB signaling on oncogenic progression remain to be comprehensively validated across heterogeneous malignancies using in vivo preclinical models. A study by Liu et al. et al. demonstrated that SUMOylation of MANF can promote its nuclear translocation, and SUMOylated p65 then binds to nuclear MAFN, thus inhibiting NF-κB pathway activation and liver cancer development through epithelial-mesenchymal transition (EMT) (57).
PVR
Poliovirus Receptor (PVR), designated as CD155, has dual functions in anti-tumor immunity; on the one hand, CD155 could induce tumor killing by NK cells via associating with CD226; on the other hand, CD155 suppresses NK cell-mediated tumor immunosurveillance through engagement of T cell immunoreceptors with Ig and ITIM domains (TIGIT) (58, 85). Recently, researchers revealed that SUMOylation of CD155 could inhibit its translocation from the cytosol to the cell membrane of multiple myeloma cells, thus attenuating the anti-tumor effect of NK cells (59). This finding implies that SUMOylation inhibitors (SUMOi) could be combined with ICI-based immunotherapies to produce more potent anti-tumor effects.
IRF7
Interferon regulatory factors (IRFs) are a family of transcription factors that play key roles in regulating interferon expression and governing host immune responses, cell differentiation, and immune regulation (78). In the antiviral response, SUMOylation of IRF3 and IRF7 reduces their transcriptional activity, consequently inhibiting IFN-I production (38). Recent study has further found that in breast cancer, RNA-binding protein 45 (RBM45) knockout reduces IRF7 SUMOylation, which relieve its inhibitory effect on IRF7 and promote IFN-I production (60).
RACK1
As an evolutionarily conserved scaffolding protein, receptor for activated C kinase 1 (RACK1) plays dual roles in regulating tumor progression and simultaneously remodeling the TME (75). Wang and colleagues revealed that RACK1 can be deSUMOylated by SENP3 for its stabilization, which drives CCL20 expression by potentiating the RACK1/eIF4E axis, consequently promoting infiltration of tumor-associated macrophage (TAM) while suppressing recruitment of cytotoxic T-cells, and ultimately facilitating immune evasion of hepatocellular carcinoma (61).
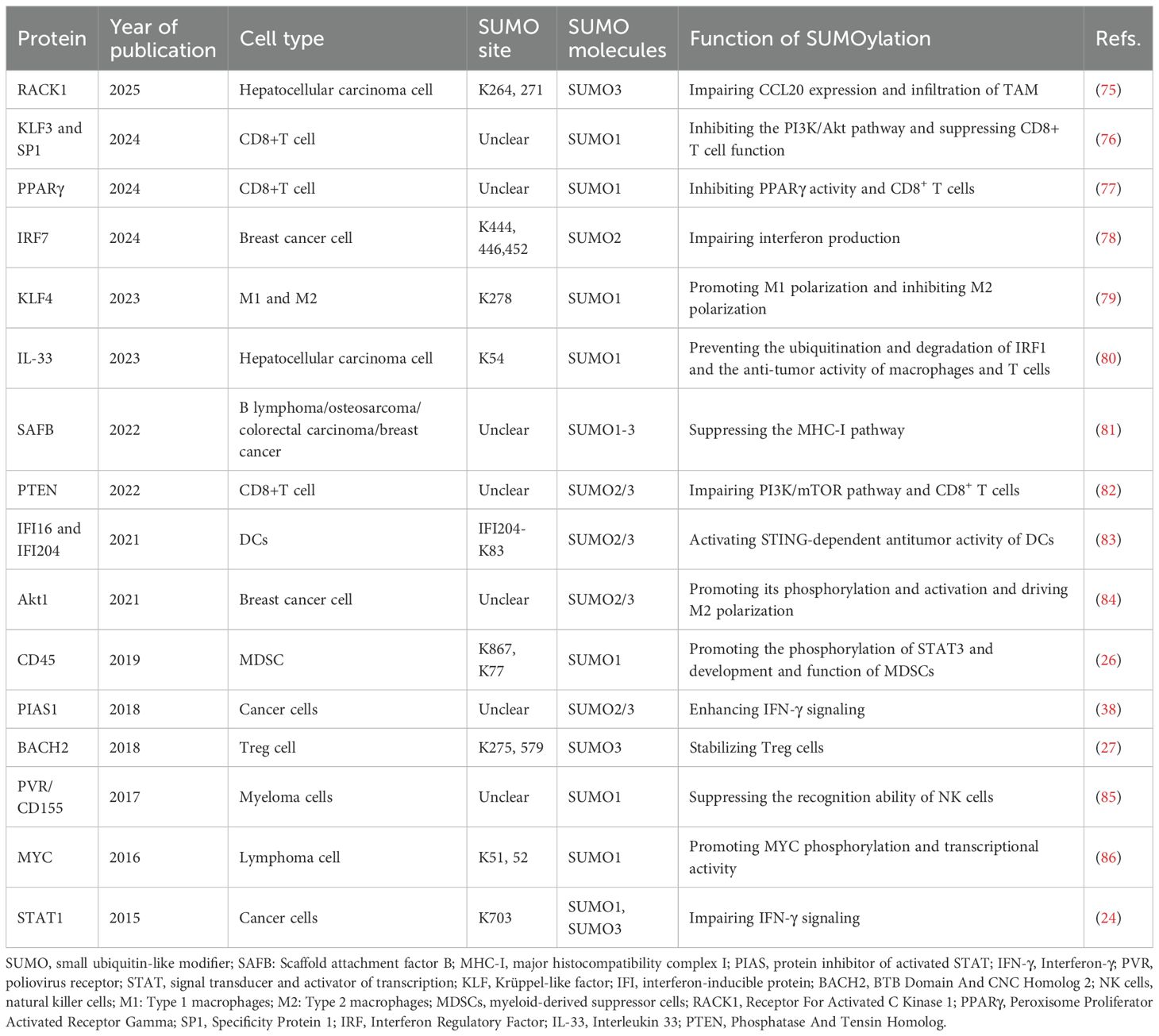
IL-33
Interleukin-33 (IL-33) is a member of the IL-1 family, which can function as a nuclear factor within the nucleus while also being released extracellularly to act as a cytokine (80). In hepatocellular carcinoma (HCC), SUMOylation of IL-33 prevents the ubiquitination and degradation of IRF1, which promotes the expression of IL-8 and PD-L1, thus suppressing the anti-tumor activity of macrophages and T cells (62). These explain how SUMOylation contributes to an immunosuppressive phenotype within the TME.
The function of immune cell-derived SUMOylation in modulating anti-tumor immunity
While tumor cell-intrinsic SUMOylation acts as a pivotal role in regulating anti-tumor immune responses, emerging evidence suggests that SUMOylation of key proteins within immune cells also significantly modulates their anti-tumor activity (Figure 3).
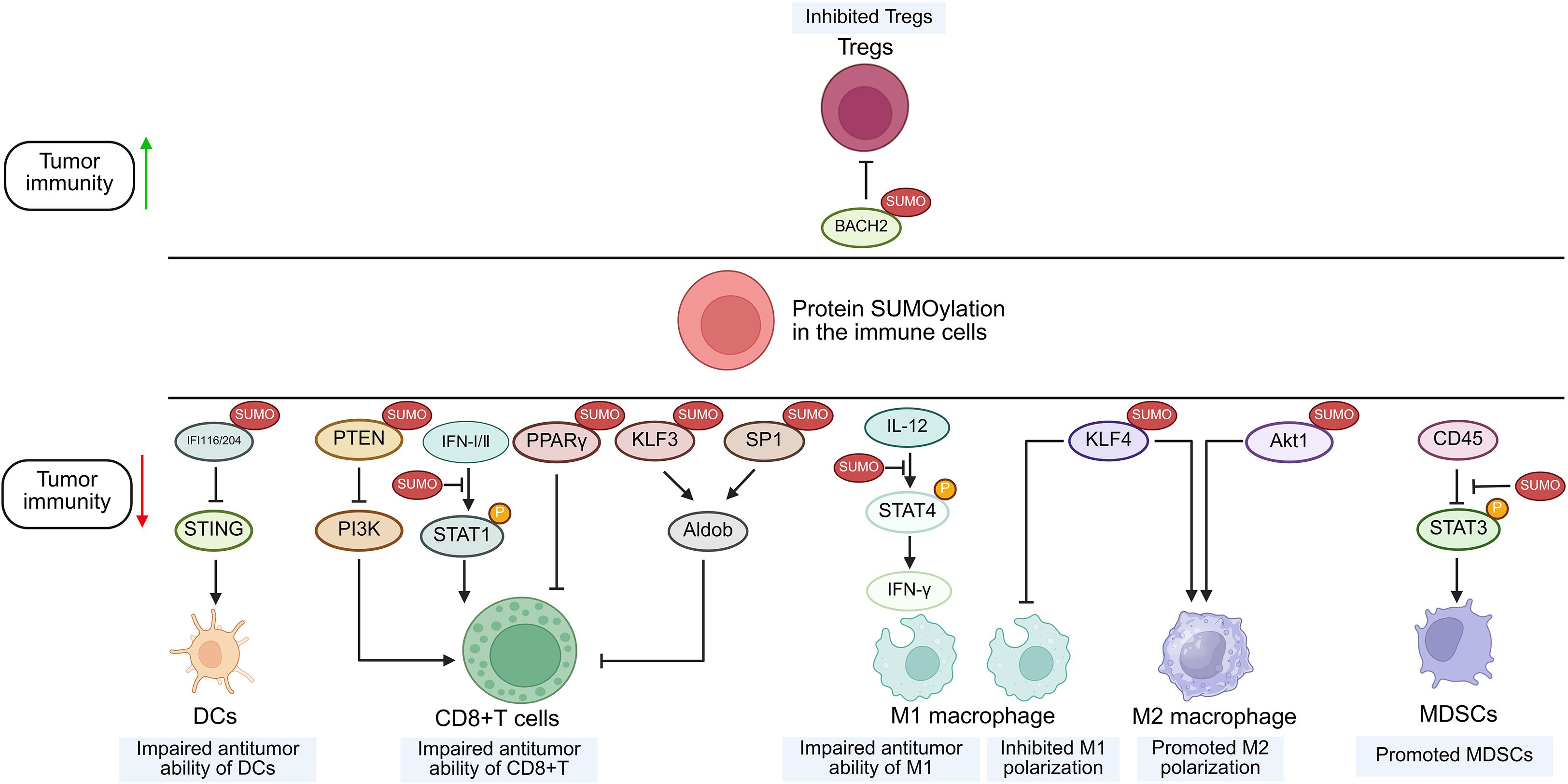
Figure 3. Suggested role of protein SUMOylation within immune cells in the regulation of tumor immunity. Increased SUMOylation of targeted proteins in immune cells can regulate anti-tumor immunity through modulating the function of immune subsets including CD8+ T cells, MDSCs, DCs, M1/M2 macrophages and Tregs. SUMO, small ubiquitin-like modifier; STAT, signal transducer and activator of transcription; KLF4, Krüppel-like factor 4; IFI, interferon-inducible protein; STING, stimulator of interferon genes; BACH2, BTB domain and CNC homolog 2; DCs, dendritic cells; M1, type 1 macrophage; M2, type 2 macrophage; MDSCs, myeloid-derived suppressor cells; Tregs, regulatory T cells.
JAK/STAT pathway
Beyond regulating the JAK/STAT pathway in malignant cells, SUMOylation also suppresses the signaling in CD8+ T cells. This functional inhibition has been substantiated by multiple studies demonstrating that TAK-981 treatment upregulates phosphorylated STAT1 (p-STAT1) and interferon-stimulated genes (ISGs) in CD8+ T lymphocytes (8, 63). Furthermore, in macrophages, UBC9 deficiency triggers deSUMOylation of STAT4, which promotes the secretion of IFN-γ by macrophage activation and subsequently enhances T cell-mediated anti-tumor responses, ultimately suppressing prostate tumorigenesis (64).
Akt1, IFI204/16 and BACH2
As previously mentioned, SENP3, a specific enzyme for deSUMOylation, is implicated in multiple cancer types through its aberrant expression or dysfunction (65). Numerous experimental studies have revealed that ROS induces the accumulation of SENP3 in different immune cells (27, 66, 67, 83, 84), but their influences on tumor immunity are conflicting. Contrary to the SUMOylation-dependent suppression of STAT1 phosphorylation, SENP3 deficiency in macrophages enhances Akt1 SUMOylation to promote its phosphorylation and activation (68). This cascade drives M2 polarization, ultimately facilitating proliferation and migration of breast cancer (68). In dendritic cells, the accumulation of SENP3 deSUMOylates IFI204 and IFI16, thus activating STING-dependent cytosolic DNA sensing and potentiating the STING-dependent anti-tumor activity of dendritic cells (DCs) (84). In contrast, overexpression of SENP3 could stabilize Treg cells by facilitating deSUMOylation of BACH2, thus contributing to tumor immunosuppression and progression of melanoma, implying that part of the anti-tumor effect of ROS scavengers is exerted through the SENP3/BACH2 axis by inhibiting ROS-induced SENP3 accumulation (27).
KLF4, KLF3 and SP1
Krüppel-like factors (KLFs) constitute a subgroup of evolutionarily conserved zinc-finger containing transcriptional regulators, among which KLF4 is critical for M2 macrophage polarization (79). Wang et al. demonstrated that SUMOylated KLF4 stabilizes IL-4-driven M2 phenotypic commitment, whereas deSUMOylation of KLF4 regulates M1 polarization of macrophages and enhances the anti-tumor activity of macrophages (76). Furthermore, chromobox protein 4 (CBX4) promotes the SUMOylation of KLF3 and transcription factor 1 (SP1), enhancing their stability and thereby promoting aldolase B (Aldob) transcription, which suppresses CD8+ T cell function by inhibiting the PI3K/Akt pathway (69). Besides, in genetic SUMOylation-deficient KLF1 mouse, the numbers of tumor-infiltrating nature killer T cells and CD8+T cells are increased, resulting in enhanced anti-tumor responses (70).
PTEN
PTEN, a tumor suppressor and primary negative regulator of PI3K, is frequently downregulated, deleted, or mutated across diverse cancers (82). Evidence indicates that under oxidative stress in CD8+ T cells, SENP7-mediated deSUMOylation of PTEN promotes its proteasomal degradation, thereby activating the PI3K/mTOR pathway (82). This signaling sustains both oxidative phosphorylation and glycolysis in CD8+ T cells, thus curbing growth of colon cancer (71).
CD45
CD45 is a ubiquitous leukocyte receptor that exhibits tyrosine phosphatase activity. STAT3 serves as a critical regulator of the functional competence of myeloid-derived suppressor cells (MDSC) (72, 73). It has been shown that SENP1 could deSUMOylate CD45 and consequently improve the phosphatase activity of CD45, thus downregulating STAT3 phosphorylation, inhibiting MDSCs, and preventing the progression of melanoma and Lewis lung cancer (26).
PPARγ
Although PPARγ (proliferator-activated receptor γ) is a major regulator of adipocyte differentiation and function, it also governs immune cell differentiation and function (77). A study indicates that autocrine VEGF-B secretion by T cells promotes SENP2 expression, which inhibits PPARγ SUMOylation, thereby enhancing PPARγ activity and maintaining lipid synthesis, mitochondrial fitness, and T cell activation (74) (Table 1).
The reinforcement of anti-tumor immunity by SUMOylation inhibition
As SUMOylation functions as a master regulatory hub governing the anti-tumor immune response, it is of significant interest to target SUMOylation for tumor interventions. As mentioned previously, SUMOylation of STAT1 in both tumor cells and immune cells can inhibit IFN-I/II-induced phosphorylation of STAT1, thereby promoting immune escape in various tumors (8, 24). Besides, SUMOylation of PTEN inhibits its ubiquitination and subsequent degradation, leading to impaired anti-tumor function of CD8+ T cells and consequently promoting the development of colon cancer (71). Therefore, based on SUMOylation inhibition, targeting the crosstalk between SUMOylation and phosphorylation or ubiquitination of these molecules can enhance anti-tumor immune responses. For instance, Kumar et al. found that SUMOi harbors the potential to increase phosphorylation of STAT1, thereby enhancing the activation of the IFN-I pathway and boosting the anti-tumor immune response (63). Thus, several inhibitors of SUMO E1 and E2 enzymes, naturally derived or artificially produced, have been identified or developed. Since natural E1 inhibitors can also induce some unintentional biological effects and primarily function in the micromolar range, these are less applied in clinical trials, however, synthetic SUMOi exhibits more specific effects and potent efficacy (87). ML-792 and TAK-981 belong to synthetic E1 inhibitors, which inhibit E1 activity by forming an irreversible adduct with SUMO (87, 88). And a novel, orally bioavailable E1 enzyme inhibitor named as SB-4826 has been developed, which forms an irreversible bond with the E1 enzyme (89, 90). In contrast, E2 inhibitors function by either binding to the E2 enzyme to suppress the formation of the E2-SUMO conjugate, as demonstrated by spectomycin B1, or by preventing the transfer of SUMO from UBC9-SUMO to substrate proteins, as observed with 2-D08 (87, 88). Presently, several synthetic SUMOylation inhibitors are undergoing preclinical or clinical evaluations for their potential in boosting the anti-tumor immune responses.
The 2-D08, as an inhibitor of the SUMO E2 enzyme, could enhance the anti-tumor potential of tumor-associated macrophages (TAMs) upon intratumoral injection in a prostate cancer mouse model; as PD-1 expression on CD8+ T cells was simultaneously upregulated, combining 2-D08 with ICI significantly suppressed the tumor growth (64).
SB-4826 treatment suppresses tumor growth in various tumor models, such as A20 lymphoma and CT-26 colorectal cancer models, and its efficacy is markedly enhanced when combined with ICIs therapy (89, 90). However, due to the limited research and insufficient evidences, only TAK-981 has been utilized in clinical trials.
TAK-981, as a specific suppressor of SUMO E1 enzyme, has been revealed to induce broad systemic immunomodulatory effects when administered for cancer therapy. First, TAK-981 can stimulate the innate immune response in murine tumor models. For PDAC with intrinsic immunosuppressive environment, Kumar et al. reported that TAK-981 induced tumor regression through potentiation of IFN signaling within tumor-infiltrating lymphocytes (TILs) and NK cells (63). Similarly, using mouse models of colon cancer and lymphoma, Lightcap et al. demonstrated that TAK-981 activates the IFN-I pathway in an IFNAR1-dependent manner, thereby promoting DCs and T cells’ activation (43). Furthermore, TAK-981 suppresses the tumor-promoting functions of cancer-associated fibroblasts (CAFs) and drives macrophage polarization toward the M1 phenotype, thus inhibiting tumor progression (91). Beyond TAK-981, another preclinical study demonstrates that ML792 similarly remodels the TME in HCC and enhances antitumor immunity (92).
Moreover, the combination of TAK-981 with other treatment modalities have yielded more robust effects in preclinical tumor models. First, TAK-981 in combination with anti-PD1 or anti-CTLA4 therapy could further foster anti-tumor immune response and prolong the survival time of mice (43, 63). Besides, co-administration of TAK-981 with the anti-CD38 antibody daratumumab or anti-CD20 antibody rituximab have potentiated antitumor activity by enhancing macrophage phagocytosis and NK cell cytotoxicity via IFN-I pathway activation in multiple myeloma, diffuse large B-cell lymphoma, and Burkitt lymphoma tumor models (93). In addition to the combination of ICIs and TAK-981, Lu et al. revealed that the application of TAK-981 can resume the levels of CH25H by suppression ATF3 in effector cells, thus augmenting CAR-T therapy and the anti-tumor immunity (94); besides, researchers have integrated TCR therapy with the DNA methylation inhibitor 5-Aza-2’-deoxycytidine and TAK-981, and this triple combination therapy has induced more sustained anti-tumor activity in mouse models of acute myeloid leukemia and multiple myeloma (95).
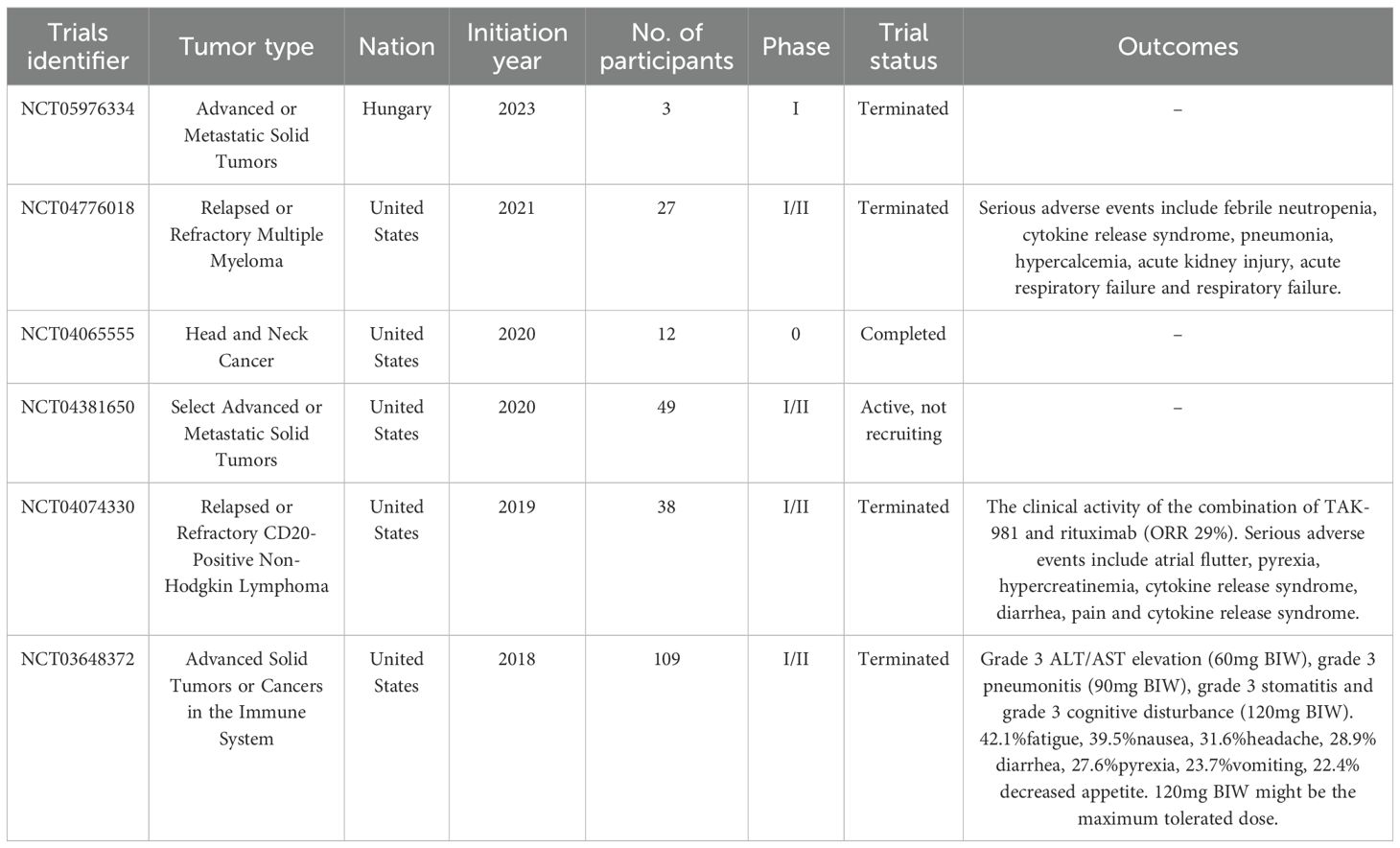
Except for animal experiments, TAK-981 has entered phase 1 of clinical evaluation (#NCT04381650; www.clinicaltrials.gov). Moreover, four phase1/2 clinical trials (#NCT05976334, #NCT03648732, #NCT04074330, and #NCT04776018) were terminated, and another phase 0 clinical trial (#NCT04065555) was completed (Table 2). In a clinical trial of #NCT04065555, 12 patients with HNSCC were administered with TAK-981 in a microdose through percutaneous intratumor injection to directly evaluate the effect of TAK-981 on the tumors. The authors revealed that TAK981 induces immune-favorable remodeling of the TME primarily through orchestrated activation of IFN-I/II signaling, which influences multiple components of the TME in a dose-dependent manner, including cancer-associated fibroblasts, immune cells, and cancer cells (96). Furthermore, they found that TAK-981 induced M1 polarization of macrophages and recruitment of cytotoxic T cells (96). These results are partially consistent with the terminated phase 1/2 clinical trials (#NCT03648732), which demonstrated that TAK-981 can induce activation of the IFN-I transcriptional program and increase the number of activated NK, CD8, and CD4 T cells (97, 98). Notably, systemic use of TAK981 have exhibited certain side effects. In a clinical trial (#NCT03648732), four dose-limiting toxicities were observed: ALT/AST elevation, pneumonitis, stomatitis, and cognitive disturbance (97). Except for these, the predominant treatment-emergent adverse events (TEAEs) with an incidence rate of more than 20% are pyrexia, diarrhea, headache, nausea, fatigue, vomiting, and decreased appetite; common TEAEs of its application ≥grade 3 were hypokalemia, anemia, decreased lymphocyte count, and abdominal pain, with an incidence rate of more than 5% (97). Hence, the results of these clinical trials further confirm the findings of these animal model studies, implying that TAK-981 can not only stimulate the anti-tumor activation of innate lymphoid cells, including DCs, macrophages, and NK cells, but also modulate the adaptive immune response, covering T cell infiltration and function, and inhibiting the functions of Tregs and MDSCs. Considering the inevitable side effects of systemic TAK981, the local administration of low-dose TAK981 may serve as an optimal alternative approach in future trials.
Summary and future prospects
Most tumors have evolved various strategies to evade immune surveillance by elevating SUMOylation levels to create a TME conducive to their progression. Our review provides a comprehensive update on recent researches regarding the function of protein SUMOylation in adjusting immune-oncological homeostasis, wherein the dichotomous role of SUMOylation within tumor cells or immune cells has been elucidated. SUMOylation can modulate innate or adaptive immune responses by regulating key protein functions in tumor cells or immune cells, thereby regulating JAK/STAT, NF-kB, IFN-I/II signaling and other molecules or pathways; besides, as various SUMOylation or deSUMOylation enzymes present in different types of immune or tumor cells, and owing to the diversity of targeted proteins in various sort of cells, the function of SUMOylation behaves differently in varied cells such as contributing to impairment or enhancement of anti-tumor immunity which is aforementioned. Moreover, inhibition of SUMOylation can alleviate the functional suppression of T cells, NK cells, and macrophages while also suppressing myeloid-derived suppressor cells (MDSCs), thus enhancing antigen presentation, tumor recognition and cytotoxicity of immune cells. This effect may be linked to the increase of interferon production and activation of interferon responses (28). Besides, the presence of excessive SUMOylation in most tumors suggests the therapeutic potential of disrupting SUMO conjugation cascades as a precision strategy against cancers by using SUMOylation inhibitors.
The results of those previous preclinical studies indicate that SUMOylation inhibition exhibits partial therapeutic efficacy, possibly owing to the complicated interaction at the tumor-immune interface. Notably, the combination of SUMOi with other immunotherapeutic schemes, such as SUMOi and immune checkpoint immunotherapy, has high potential to facilitate tumor regression, which warrants further clinical studies. However, as SUMOylation plays a versatile role in tumor progression and is cancer-type context-dependent, it is of utmost importance for the development of targeted therapies for inhibiting SUMOylation in certain cells within various tumor types (Figures 4, 5), for example, developing drugs that conjugates mono-antibodies against key SUMOylated proteins in TME with deSUMOylases presents high promise for achieving more precise therapeutic intervention. The combination of SUMOi with anti-PD-L1 for cancer treatment has been extensively studied and well established. We have found that anti-TIGIT therapy may also synergize with SUMOi to enhance anti-tumor immunity. And due to the toxic effects of systemic TAK981, it is necessary to develop a locally delivered strategy for slow-release of TAK981, such as hydrogels with high bio-safety (Figure 6). Or utilizing antibody-drug conjugate (ADC) technology to combine tumor cell-targeting antibodies with small molecule SUMOi (such as TAK-981, molecular weight 578.10) may represent a promising strategy to enhance anti-tumor immune responses. In addition, AI-assisted screening of FDA approved therapeutics as potential SUMOi is going to serve as another promising approach. Our review proposes that current or other novel strategies for SUMOylation inhibition present as a promising adjuvant to tumor immunotherapy, upon which better prognosis of patients could be obtained.
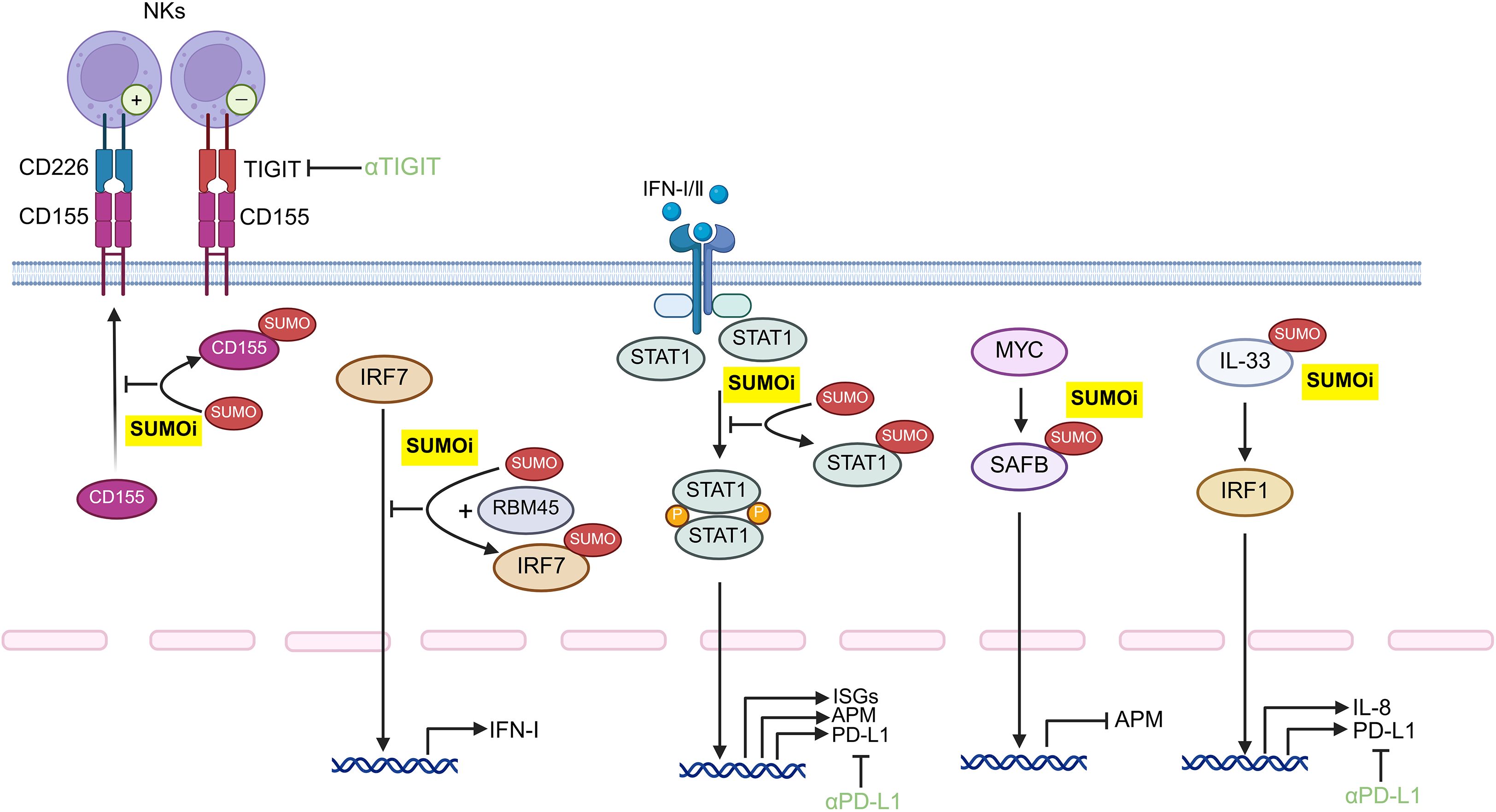
Figure 4. Potential protein target for SUMOylation inhibition within tumor cells for potentiating anti-tumor immune responses. Developing monoclonal antibody-conjugated SUMOi to precisely target distinct tumor-intrinsic proteins may represent a promising strategy to augment anti-tumor immunity, which may orchestrate with the anti-TIGIT or anti-PD-L1 treatment. STAT, signal transducer and activator of transcription; IFN: Interferon; IL-33, Interleukin 33; IRF, Interferon Regulatory Factor; PD-L1, Programmed cell death ligand 1; TIGIT, T cell immunoglobulin and ITIM domain; ISGs, interferon-stimulated genes; APM, antigen-presentation machinery; NK cells, natural killer cells; SUMOi, SUMOylation inhibitors.

Figure 5. Possible protein target for SUMOylation inhibition in various immune subsets to boost the anti-tumor immunity. The development of monoclonal antibody-conjugated SUMOi for precise target-protein recognition may contribute to enhancing the anti-tumor potential of immune cells. NK cells, natural killer cells; DCs, dendritic cells; M1: Type 1 macrophages; M2: Type 2 macrophages; MDSCs, myeloid-derived suppressor cells; IFI, interferon-inducible protein; PTEN, Phosphatase And Tensin Homolog; PPARγ, Peroxisome Proliferator Activated Receptor Gamma; KLF, Krüppel-like factor; SP1, Specificity Protein 1; SUMOi, SUMOylation inhibitors.
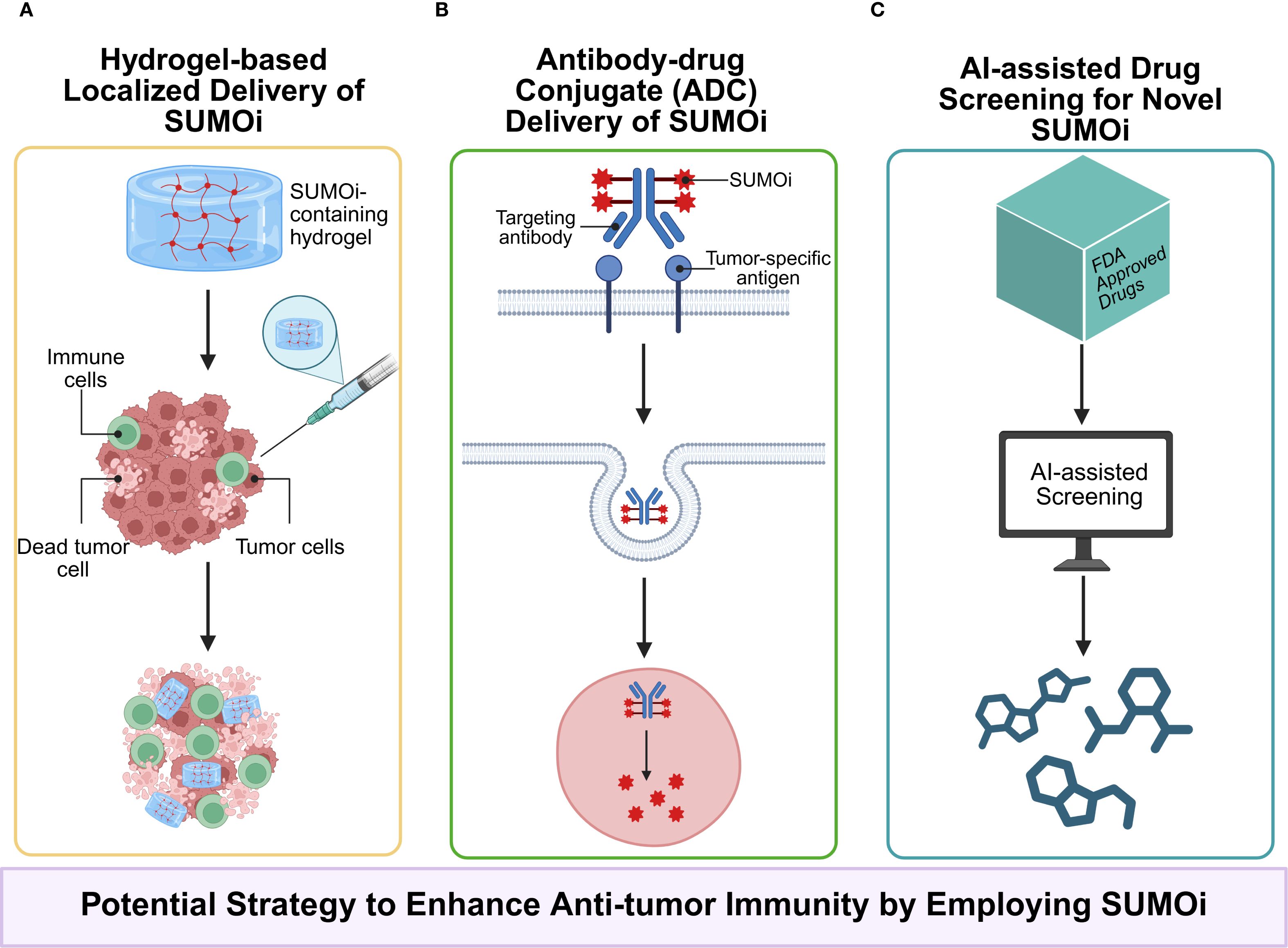
Figure 6. (A) Employing local drug delivery systems, such as hydrogels, to carry SUMOi not only minimizes systemic toxicity associated with conventional administration but also enables the sustained release, maintains local drug concentration, and avoids repeated administrations; (B) Antibody–drug conjugates (ADCs) enables more precisely targeting of specific proteins in certain cells; (C) AI-assisted drug screening from FDA-approved drug library holds promise in identifying novel inhibitors of SUMOylation with reduced toxicity and enhanced compatibility.
Author contributions
XLi: Writing – original draft, Visualization. WD: Writing – original draft. LJ: Writing – review & editing. QC: Writing – review & editing, Funding acquisition, Supervision, Conceptualization. XLu: Writing – review & editing, Visualization, Funding acquisition, Supervision, Conceptualization.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (82272899, 81902782, and 82203180), Research Funding from West China School/Hospital of Stomatology Sichuan University (No.RCDWJS2022-16), Postdoctoral Research Funding of Sichuan University (2022SCU12132), Research and Development Program, West China Hospital of Stomatology, Sichuan University (No. RD-02-202204), Key Research Program of Sichuan Provincial Science and Technology Agency (2023YFS0127), Youth Fund Projects of Sichuan Provincial Science and Technology Agency (2024NSFSC1904), CAMS Innovation Fund for Medical Sciences (CIFMS,2019-I2M-5-004), and funding for key research and development projects supported by the Science and Technology Department of Sichuan Province (2023YFS0010).
Acknowledgments
We would like to thank these authors of original studies included in this review. The figures were generated with BioRender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Gkountela S, Castro-Giner F, Szczerba BM, Marcus V, Julia L, Ramona S, et al. Circulating tumor cell clustering shapes DNA methylation to enable metastasis seeding. Cell. (2019) 176:98–112.e14. doi: 10.1016/j.cell.2018.11.046
2. Nguyen AT, Chia J, Ros M, Hui KM, Saltel F, and Bard F. Organelle specific O-glycosylation drives MMP14 activation, tumor growth, and metastasis. Cancer Cell. (2017) 32:639–653.e6. doi: 10.1016/j.ccell.2017.10.001
3. Geffen Y, Anand S, Akiyama Y, Yaron TM, Song Y, Johnson JL, et al. Pan-cancer analysis of post-translational modifications reveals shared patterns of protein regulation. Cell. (2023) 186:3945–3967.e26. doi: 10.1016/j.cell.2023.07.013
4. Boix O, Martinez M, Vidal S, Giménez-Alejandre M, Palenzuela L, Lorenzo-Sanz L, et al. pTINCR microprotein promotes epithelial differentiation and suppresses tumor growth through CDC42 SUMOylation and activation. Nat Commun. (2022) 13:6840. doi: 10.1038/s41467-022-34529-6
5. Rodriguez MS, Dargemont C, and Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem. (2001) 276:12654–9. doi: 10.1074/jbc.M009476200
6. Seeler JS and Dejean A. SUMO and the robustness of cancer. Nat Rev Cancer. (2017) 17:184–97. doi: 10.1038/nrc.2016.143
7. Chang HM and Yeh ETH. SUMO: from bench to bedside. Physiol Rev. (2020) 100:1599–619. doi: 10.1152/physrev.00025.2019
8. Demel UM, Böger M, Yousefian S, Grunert C, Zhang L, Hotz PW, et al. Activated SUMOylation restricts MHC class I antigen presentation to confer immune evasion in cancer. J Clin Invest. (2022) 132:e152383. doi: 10.1172/JCI152383
9. Zhang T, Yang H, Zhou Z, Bai Y, Wang J, and Wang W. Crosstalk between SUMOylation and ubiquitylation controls DNA end resection by maintaining MRE11 homeostasis on chromatin. Nat Commun. (2022) 13:5133. doi: 10.1038/s41467-022-32920-x
10. Eifler K and Vertegaal ACO. SUMOylation-mediated regulation of cell cycle progression and cancer. Trends Biochem Sci. (2015) 40:779–93. doi: 10.1016/j.tibs.2015.09.006
11. Vertegaal ACO. Signaling mechanisms and cellular functions of SUMO. Nat Rev Mol Cell Biol. (2022) 23:715–31. doi: 10.1038/s41580-022-00500-y
12. Cui CP, Wong CC, Kai AK, Ho DW, Lau EY, Tsui YM, et al. SENP1 promotes hypoxia-induced cancer stemness by HIF-1α deSUMOylation and SENP1/HIF-1α positive feedback loop. Gut. (2017) 66:2149–59. doi: 10.1136/gutjnl-2016-313264
13. Zhou Z, Wang M, Li J, Xiao M, Chin YE, Cheng J, et al. SUMOylation and SENP3 regulate STAT3 activation in head and neck cancer. Oncogene. (2016) 35:5826–38. doi: 10.1038/onc.2016.124
14. Huang HJ, Zhou LL, Fu WJ, Zhang CY, Jiang H, Du J, et al. β-catenin SUMOylation is involved in the dysregulated proliferation of myeloma cells. Am J Cancer Res. (2014) 5:309–20.
15. Hu Y, Chen C, Tong X, Chen S, Hu X, Pan B, et al. NSUN2 modified by SUMO-2/3 promotes gastric cancer progression and regulates mRNA m5C methylation. Cell Death Dis. (2021) 12:842. doi: 10.1038/s41419-021-04127-3
16. Ding B, Sun Y, and Huang J. Overexpression of SKI oncoprotein leads to p53 degradation through regulation of MDM2 protein sumoylation. J Biol Chem. (2012) 287:14621–30. doi: 10.1074/jbc.M111.301523
17. Haupt Y, Maya R, Kazaz A, and Oren M. Mdm2 promotes the rapid degradation of p53. Nature. (1997) 387:296–9. doi: 10.1038/387296a0
18. DhatChinamoorthy K, Colbert JD, and Rock KL. Cancer immune evasion through loss of MHC class I antigen presentation. Front Immunol. (2021) 12:636568. doi: 10.3389/fimmu.2021.636568
19. Yi M, Li T, Niu M, Mei Q, Zhao B, Chu Q, et al. Exploiting innate immunity for cancer immunotherapy. Mol Cancer. (2023) 22:187. doi: 10.1186/s12943-023-01885-w
20. Xia L, Oyang L, Lin J, Tan S, Han Y, Wu N, et al. The cancer metabolic reprogramming and immune response. Mol Cancer. (2021) 20(1):28. doi: 10.1186/s12943-021-01316-8
21. Ferris RL, Blumenschein G, Fayette J, Guigay J, Colevas AD, Licitra L, et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N Engl J Med. (2016) 375:1856–67. doi: 10.1056/NEJMoa1602252
22. Gautam SK, Batra SK, and Jain M. Molecular and metabolic regulation of immunosuppression in metastatic pancreatic ductal adenocarcinoma. Mol Cancer. (2023) 22:118. doi: 10.1186/s12943-023-01813-y
23. Luo X, Qiu Y, Fitzsimonds ZR, Wang Q, Chen Q, and Lei YL. Immune escape of head and neck cancer mediated by the impaired MHC-I antigen presentation pathway. Oncogene. (2024) 43:388–94. doi: 10.1038/s41388-023-02912-2
24. Maarifi G, Maroui MA, Dutrieux J, Dianoux L, Nisole S, and Chelbi-Alix MK. Small ubiquitin-like modifier alters IFN response. J Immunol. (2015) 195:2312–24. doi: 10.4049/jimmunol.1500035
25. Sun F, Wang FX, Zhu H, Yue TT, Yang CL, Luo JH, et al. SUMOylation of PDPK1 Is required to maintain glycolysis-dependent CD4 T-cell homeostasis. Cell Death Dis. (2022) 13:181. doi: 10.1038/s41419-022-04622-1
26. Huang X, Zuo Y, Wang X, Wu X, Tan H, Fan Q, et al. SUMO-specific protease 1 is critical for myeloid-derived suppressor cell development and function. Cancer Res. (2019) 79:3891–902. doi: 10.1158/0008-5472.CAN-18-3497
27. Yu X, Lao Y, Teng XL, Li S, Zhou Y, Wang F, et al. SENP3 maintains the stability and function of regulatory T cells via BACH2 deSUMOylation. Nat Commun. (2018) 9:3157. doi: 10.1038/s41467-018-05676-6
28. Tharuka MDN, Courelli AS, and Chen Y. Immune regulation by the SUMO family. Nat Rev Immunol. (2025) 25:608–20. doi: 10.1038/s41577-025-01155-4
29. Eisenhardt N, Chaugule VK, Koidl S, Droescher M, Dogan E, Rettich J, et al. A new vertebrate SUMO enzyme family reveals insights into SUMO-chain assembly. Nat Struct Mol Biol. (2015) 22:959–67. doi: 10.1038/nsmb.3114
30. Sriramachandran AM and Dohmen RJ. SUMO-targeted ubiquitin ligases. Biochim Biophys Acta. (2014) 1843:75–85. doi: 10.1016/j.bbamcr.2013.08.022
31. Saitoh H and Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem. (2000) 275:6252–8. doi: 10.1074/jbc.275.9.6252
32. Jansen NS and Vertegaal ACO. A chain of events: regulating target proteins by SUMO polymers. Trends Biochem Sci. (2021) 46:113–23. doi: 10.1016/j.tibs.2020.09.002
33. Liebelt F and Vertegaal AC. Ubiquitin-dependent and independent roles of SUMO in proteostasis. Am J Physiol Cell Physiol. (2016) 311:C284–96. doi: 10.1152/ajpcell.00091.2016
34. Chung CD, Liao J, Liu B, Rao X, Jay P, Berta P, et al. Specific inhibition of Stat3 signal transduction by PIAS3. Science. (1997) 278:1803–5. doi: 10.1126/science.278.5344.1803
35. Liu B, Liao J, Rao X, Kushner SA, Chung CD, Chang DD, et al. Inhibition of Stat1-mediated gene activation by PIAS1. Proc Natl Acad Sci USA. (1998) 95:10626–31. doi: 10.1073/pnas.95.18.10626
36. Jackson PK. A new RING for SUMO: wrestling transcriptional responses into nuclear bodies with PIAS family E3 SUMO ligases. Genes Dev. (2001) 15:3053–8. doi: 10.1101/gad.955501
37. Niu GJ, Xu JD, Yuan WJ, Sun JJ, Yang MC, He ZH, et al. (PIAS) negatively regulates the JAK/STAT pathway by inhibiting STAT phosphorylation and translocation. Front Immunol. (2018) 9:2392. doi: 10.3389/fimmu.2018.02392
38. Hannoun Z, Maarifi G, and Chelbi-Alix MK. The implication of SUMO in intrinsic and innate immunity. Cytokine Growth Factor Rev. (2016) 29:3–16. doi: 10.1016/j.cytogfr.2016.04.003
39. Alagu J, Itahana Y, Sim F, Chao SH, Bi X, and Itahana K. Tumor Suppressor p14ARF Enhances IFN-γ-Activated Immune Response by Inhibiting PIAS1 via SUMOylation. J Immunol. (2018) 201:451–64. doi: 10.4049/jimmunol.1800327
40. Zang X, He XY, Xiao CM, Lin Q, Wang MY, Liu CY, et al. Circular RNA-encoded oncogenic PIAS1 variant blocks immunogenic ferroptosis by modulating the balance between SUMOylation and phosphorylation of STAT1. Mol Cancer. (2024) 23:207. doi: 10.1186/s12943-024-02124-6
41. Mellman I, Chen DS, Powles T, and Turley SJ. The cancer-immunity cycle: Indication, genotype, and immunotype. Immunity. (2023) 56:2188–205. doi: 10.1016/j.immuni.2023.09.011
42. Kalbasi A and Ribas A. Tumor-intrinsic resistance to immune checkpoint blockade. Nat Rev Immunol. (2020) 20:25–39. doi: 10.1038/s41577-019-0218-4
43. Lightcap ES, Yu P, Grossman S, Song K, Khattar M, Xega K, et al. A small-molecule SUMOylation inhibitor activates antitumor immune responses and potentiates immune therapies in preclinical models. Sci Transl Med. (2021) 13:eaba7791. doi: 10.1126/scitranslmed.aba7791
45. Dhanasekaran R, Deutzmann A, Mahauad-Fernandez WD, Hansen AS, Gouw AM, and Felsher DW. The MYC oncogene - the grand orchestrator of cancer growth and immune evasion. Nat Rev Clin Oncol. (2022) 19:23–36. doi: 10.1038/s41571-021-00549-2
46. Rabellino A, Melegari M, Tompkins VS, Chen W, Van Ness BG, Teruya-Feldstein J, et al. PIAS1 promotes lymphomagenesis through MYC upregulation. Cell Rep. (2016) 15:2266–78. doi: 10.1016/j.celrep.2016.05.015
47. Kotani H, Yamano T, Boucher JC, Sato S, Sakaguchi H, Fukuda K, et al. Comprehensive antitumor immune response boosted by dual inhibition of SUMOylation and MEK in MYC-expressing KRAS-mutant cancers. Exp Hematol Oncol. (2024) 13:94. doi: 10.1186/s40164-024-00563-x
48. Garee JP, Meyer R, and Oesterreich S. Co-repressor activity of scaffold attachment factor B1 requires sumoylation. Biochem Biophys Res Commun. (2011) 408:516–22. doi: 10.1016/j.bbrc.2011.04.040
49. Itahashi K, Irie T, Yuda J, Kumagai S, Tanegashima T, Lin YT, et al. BATF epigenetically and transcriptionally controls the activation program of regulatory T cells in human tumors. Sci Immunol. (2022) 7:eabk0957. doi: 10.1126/sciimmunol.abk0957
50. Zhou L, Jiang Y, Liu X, Li L, Yang X, Dong C, et al. Promotion of tumor-associated macrophages infiltration by elevated neddylation pathway via NF-κB-CCL2 signaling in lung cancer. Oncogene. (2019) 38:5792–804. doi: 10.1038/s41388-019-0840-4
51. Dong MB, Wang G, Chow RD, Ye L, Zhu L, Dai X, et al. Systematic immunotherapy target discovery using genome-scale in vivo CRISPR screens in CD8 T cells. Cell. (2019) 178:1189–1204.e23. doi: 10.1016/j.cell.2019.07.044
52. You B, Jiang YY, Chen S, Yan G, and Sun J. The orphan nuclear receptor Nur77 suppresses endothelial cell activation through induction of IkappaBalpha expression. Circ Res. (2009) 104:742–9. doi: 10.1161/CIRCRESAHA.108.192286
53. Zhang L, Xie F, Zhang J, Dijke PT, and Zhou F. SUMO-triggered ubiquitination of NR4A1 controls macrophage cell death. Cell Death Differ. (2017) 24:1530–9. doi: 10.1038/cdd.2017.29
54. Miliara S, Gkouskou KK, Sharp TV, and Eliopoulos AG. SUMOylation is required for optimal TRAF3 signaling capacity. PloS One. (2013) 8:e80470. doi: 10.1371/journal.pone.0080470
55. Mabb AM, Wuerzberger-Davis SM, and Miyamoto S. PIASy mediates NEMO sumoylation and NF-kappaB activation in response to genotoxic stress. Nat Cell Biol. (2006) 8:986–93. doi: 10.1038/ncb1458
56. Carbia-Nagashima A, Gerez J, Perez-Castro C, Paez-Pereda M, Silberstein S, Stalla GK, et al. RSUME, a small RWD-containing protein, enhances SUMO conjugation and stabilizes HIF-1alpha during hypoxia. Cell. (2007) 131:309–23. doi: 10.1016/j.cell.2007.07.044
57. Liu J, Wu Z, Han D, Wei C, Liang Y, Jiang T, et al. Mesencephalic astrocyte-derived neurotrophic factor inhibits liver cancer through small ubiquitin-related modifier (SUMO)ylation-related suppression of NF-κB/snail signaling pathway and epithelial-mesenchymal transition. Hepatology. (2020) 71:1262–78. doi: 10.1002/hep.30917
58. Lupo KB and Matosevic S. CD155 immunoregulation as a target for natural killer cell immunotherapy in glioblastoma. J Hematol Oncol. (2020) 13:76. doi: 10.1186/s13045-020-00913-2
59. Zitti B, Molfetta R, Fionda C, Quatrini L, Stabile H, Lecce M, et al. Innate immune activating ligand SUMOylation affects tumor cell recognition by NK cells. Sci Rep. (2017) 7:10445. doi: 10.1038/s41598-017-10403-0
60. Lv Y, Sun S, Zhang J, Wang C, Chen C, Zhang Q, et al. Loss of RBM45 inhibits breast cancer progression by reducing the SUMOylation of IRF7 to promote IFNB1 transcription. Cancer Lett. (2024) 596:216988. doi: 10.1016/j.canlet.2024.216988
61. Wang P, Qiu J, Fang Y, Li S, Liu K, Cao Y, et al. SENP3 inhibition suppresses hepatocellular carcinoma progression and improves the efficacy of anti-PD-1 immunotherapy. Cell Death Differ. (2025) 32:959–72. doi: 10.1038/s41418-024-01437-9IF
62. Wang Z, Pan B, Qiu J, Zhang X, Ke X, Shen S, et al. SUMOylated IL-33 in the nucleus stabilizes the transcription factor IRF1 in hepatocellular carcinoma cells to promote immune escape. Sci Signal. (2023) 16:eabq3362. doi: 10.1126/scisignal.abq3362
63. Kumar S, Schoonderwoerd MJA, Kroonen JS, de Graaf IJ, Sluijter M, Ruano D, et al. Targeting pancreatic cancer by TAK-981: a SUMOylation inhibitor that activates the immune system and blocks cancer cell cycle progression in a preclinical model. Gut. (2022) 71:2266–83. doi: 10.1136/gutjnl-2021-324834
64. Xiao J, Sun F, Wang YN, Liu B, Zhou P, Wang FX, et al. UBC9 deficiency enhances immunostimulatory macrophage activation and subsequent antitumor T cell response in prostate cancer. J Clin Invest. (2023) 133:e158352. doi: 10.1172/JCI158352
65. Chen L, Che Y, and Huang C. SENP3: cancers and diseases. Biochim Biophys Acta Rev Cancer. (2025) 1880:189260. doi: 10.1016/j.bbcan.2025.189260
66. Zhou J, Li XY, Liu YJ, Feng J, Wu Y, Shen HM, et al. Full-coverage regulations of autophagy by ROS: from induction to maturation. Autophagy. (2022) 18:1240–55. doi: 10.1080/15548627.2021.1984656
67. Sun Z, Hu S, Luo Q, Ye D, Hu D, and Chen F. Overexpression of SENP3 in oral squamous cell carcinoma and its association with differentiation. Oncol Rep. (2013) 29:1701–6. doi: 10.3892/or.2013.2318
68. Xiao M, Bian Q, Lao Y, Yi J, Sun X, Sun X, et al. SENP3 loss promotes M2 macrophage polarization and breast cancer progression. Mol Oncol. (2022) 16:1026–44. doi: 10.1002/1878-0261.12967
69. Wang J, Jia W, Zhou X, Ma Z, Liu J, and Lan P. CBX4 suppresses CD8+ T cell antitumor immunity by reprogramming glycolytic metabolism. Theranostics. (2024) 14:3793–809. doi: 10.7150/thno.95748
70. Shyu Y, Liao P, Huang T, Yang C, Lu M, Huang S, et al. Genetic disruption of KLF1 K74 SUMOylation in hematopoietic system promotes healthy longevity in mice. Adv Sci (Weinh). (2022) 9:e2201409. doi: 10.1002/advs.202201409
71. Wu Z, Huang H, Han Q, Hu Z, Teng XL, Ding R, et al. SENP7 senses oxidative stress to sustain metabolic fitness and antitumor functions of CD8+ T cells. J Clin Invest. (2022) 132:e155224. doi: 10.1172/JCI155224
72. Nefedova Y, Cheng P, Gilkes D, Blaskovich M, Beg AA, Sebti SM, et al. Activation of dendritic cells via inhibition of Jak2/STAT3 signaling. J Immunol. (2005) 175:4338–46. doi: 10.4049/jimmunol.175.7.4338
73. Kortylewski M, Kujawski M, Wang T, Wei S, Zhang S, Pilon-Thomas S, et al. Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat Med. (2005) 11:1314–21. doi: 10.1038/nm1325
74. He J, Chen Y, Ding H, Zhou JA, Xing Z, Yang X, et al. Autocrine VEGF-B signaling maintains lipid synthesis and mitochondrial fitness to support T cell immune responses. J Clin Invest. (2024) 134:e176586. doi: 10.1172/JCI176586
75. Li JJ and Xie D. RACK1, a versatile hub in cancer. Oncogene. (2015) 34:1890–8. doi: 10.1038/onc.2014.127
76. Wang K, Xiong J, Lu Y, Wang L, and Tian T. SENP1-KLF4 signaling regulates LPS -induced macrophage M1 polarization. FEBS J. (2023) 290:209–24. doi: 10.1111/febs.16589
77. Hernandez-Quiles M, Broekema MF, and Kalkhoven E. PPARgamma in metabolism, immunity, and cancer: unified and diverse mechanisms of action. Front Endocrinol (Lausanne). (2021) 12:624112. doi: 10.3389/fendo.2021.624112
78. Negishi H, Taniguchi T, and Yanai H. The interferon (IFN) class of cytokines and the IFN regulatory factor (IRF) transcription factor family. Cold Spring Harb Perspect Biol. (2018) 10:a028423. doi: 10.1101/cshperspect.a028423
79. Liao X, Sharma N, Kapadia F, Zhou G, Lu Y, Hong H, et al. Krüppel-like factor 4 regulates macrophage polarization. J Clin Invest. (2011) 121:2736–49. doi: 10.1172/JCI45444
80. Liew FY, Girard JP, and Turnquist HR. Interleukin-33 in health and disease. Nat Rev Immunol. (2016) 16:676–89. doi: 10.1038/nri.2016.95
81. Hammerich-Hille S, Kaipparettu BA, Tsimelzon A, Creighton CJ, Jiang S, Polo JM, et al. SAFB1 mediates repression of immune regulators and apoptotic genes in breast cancer cells. J Biol Chem. (2010) 285:3608–16. doi: 10.1074/jbc.M109.066431
82. Álvarez-Garcia V, Tawil Y, and Wise HM. Leslie NR Mechanisms of PTEN loss in cancer: It’s all about diversity. Semin Cancer Biol. (2019) 59:66–79. doi: 10.1016/j.semcancer.2019.02.001
83. Huang C, Han Y, Wang Y, Sun X, Yan S, Yeh ETH, et al. SENP3 is responsible for HIF-1 transactivation under mild oxidative stress via p300 de-SUMOylation. EMBO J. (2009) 28:2748–62. doi: 10.1038/emboj.2009.210
84. Hu Z, Teng XL, Zhang T, Yu X, Ding R, Yi J, et al. SENP3 senses oxidative stress to facilitate STING-dependent dendritic cell antitumor function. Mol Cell. (2021) 81:940–952.e5. doi: 10.1016/j.molcel.2020.12.024
85. Liu S, Zhang H, Li M, Hu D, Li C, Ge B, et al. Recruitment of Grb2 and SHIP1 by the ITT-like motif of TIGIT suppresses granule polarization and cytotoxicity of NK cells. Cell Death Differ. (2013) 20:456–64. doi: 10.1038/cdd.2012.141
86. Llombart V and Mansour MR. Therapeutic targeting of “undruggable” MYC. EBioMedicine. (2022) 75:103756. doi: 10.1016/j.ebiom.2021.103756
87. Kukkula A, Ojala VK, Mendez LM, Sistonen L, Elenius K, and Sundvall M. Therapeutic potential of targeting the SUMO pathway in cancer. Cancers (Basel). (2021) 13:4402. doi: 10.3390/cancers13174402
88. Kroonen JS and Vertegaal ACO. Targeting SUMO signaling to wrestle cancer. Trends Cancer. (2021) 7:496–510. doi: 10.1016/j.trecan.2020.11.009
89. Canon JR, Callinan A, Bradley S, Wang L, Kui M, Wein A, et al. SB-4826, a first-in-class oral, covalent inhibitor of SUMO E1 that induces IFN signaling and inhibits tumor growth as monotherapy and in combination with immune checkpoint blockade. Cancer Res. (2023) 83:LB318. doi: 10.1158/1538-7445.AM2023-LB318
90. Lownik J, Milshteyn L, Villamejor A, and Merchant A. Inhibition of SUMOylation reverses T-cell metabolic exhaustion in large B-cell lymphoma. Cancer Res. (2025) 85:473. doi: 10.1158/1538-7445.AM2025-473
91. Erdem S, Lee HJ, Shankara Narayanan JSN, Tharuka MDN, de la Torre J, Ren T, et al. Inhibition of SUMOylation Induces Adaptive Antitumor Immunity against Pancreatic Cancer through Multiple Effects on the Tumor Microenvironment. Mol Cancer Ther. (2024) 23:1597–612. doi: 10.1158/1535-7163.MCT-23-0572
92. Wang Z, Pan B, Su L, Yu H, Wu X, Yao Y, et al. SUMOylation inhibitors activate anti-tumor immunity by reshaping the immune microenvironment in a preclinical model of hepatocellular carcinoma. Cell Oncol (Dordr). (2024) 47:513–32. doi: 10.1007/s13402-023-00880-z
93. Nakamura A, Grossman S, Song K, Xega K, Zhang Y, Cvet D, et al. The SUMOylation inhibitor subasumstat potentiates rituximab activity by IFN1-dependent macrophage and NK cell stimulation. Blood. (2022) 139:2770–81. doi: 10.1182/blood.2021014267
94. Lu Z, McBrearty N, Chen J, Tomar VS, Zhang H, De Rosa G, et al. ATF3 and CH25H regulate effector trogocytosis and anti-tumor activities of endogenous and immunotherapeutic cytotoxic T lymphocytes. Cell Metab. (2022) 34:1342–1358.e7. doi: 10.1016/j.cmet.2022.08.007
95. Kroonen JS, Wouters AK, de Graaf IJ, Remst DFG, Kumar S, Wachsmann TLA, et al. Targeting epigenetic regulation and post-translational modification with 5-Aza-2’ deoxycytidine and SUMO E1 inhibition augments T-cell receptor therapy. J Immunother Cancer. (2024) 12:e008654. doi: 10.1136/jitc-2023-008654
96. Derry JMJ, Burns C, Frazier JP, Beirne E, Grenley M, DuFort CC, et al. Trackable intratumor microdosing and spatial profiling provide early insights into activity of investigational agents in the intact tumor microenvironment. Clin Cancer Res. (2023) 29:3813–25. doi: 10.1158/1078-0432.CCR-23-0827
97. Dudek A, Juric D, Dowlati A, Vaishampayan U, Assad H, Rodón J, et al. First-in-human phase 1/2 study of the first-in-class sumo-activating enzyme inhibitor tak-981 in patients with advanced or metastatic solid tumors or relapsed/refractory lymphoma: phase 1 results. J Immunother Cancer. (2021) 9:A505–6. doi: 10.1136/jitc-2021-SITC2021.476
98. Saggu G, Stroopinsky D, Dudek AZ, Olszanski AJ, Juric D, Dowlati A, et al. Subasumstat, a first-in-class inhibitor of SUMO-activating enzyme, demonstrates dose-dependent target engagement and SUMOylation inhibition, leading to rapid activation of innate and adaptive immune responses in the dose escalation portion of a phase 1/2 clinical study. Eur J Cancer. (2022) 174:S125–6. doi: 10.1016/S0959-8049(22)01134-0
Keywords: SUMOylation, anti-tumor immunity, MHC-I antigen presentation, JAK/STAT pathway, TAK-981
Citation: Liu X, Ding W, Jiang L, Chen Q and Luo X (2025) Unraveling the critical role of SUMOylation in the governing of tumor immunity. Front. Immunol. 16:1654167. doi: 10.3389/fimmu.2025.1654167
Received: 26 June 2025; Accepted: 09 September 2025;
Published: 29 September 2025.
Edited by:
Hridayesh Prakash, Amity University, IndiaReviewed by:
Céline Gongora, Centre National de la Recherche Scientifique (CNRS), FranceNavkiran Kaur, Amity University, India
Copyright © 2025 Liu, Ding, Jiang, Chen and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qianming Chen, cW1jaGVuQHNjdS5lZHUuY24=; Xiaobo Luo, eGlhb2JvbEBzY3UuZWR1LmNu
†ORCID: Qianming Chen, orcid.org/0000-0002-5371-4432
Xiaobo Luo, orcid.org/0000-0002-0219-8554
 Xiangfei Liu
Xiangfei Liu Lu Jiang
Lu Jiang Xiaobo Luo
Xiaobo Luo