- 1The Joint Laboratory on Transfusion-Transmitted Diseases (TTDs) Between Institute of Blood Transfusion, Chinese Academy of Medical Sciences and Nanning Blood Center, Nanning Blood Center, Nanning, China
- 2The Hospital of Xidian Group Laboratory Department, Xi’an, China
- 3Shenzhen Blood Center Laboratory Department, Shenzhen, China
- 4Institute of Blood Transfusion, Chinese Academy of Medical Sciences and Peking Union Medical College, Chengdu, China
Occult hepatitis B virus infection (OBI) represents a specific form of hepatitis B virus (HBV)infection characterized by the presence of replication-competent HBV DNA in the liver despite a negative blood test for hepatitis B surface antigen (HBsAg). Due to the incompletely-known mechanisms underlying its occurrence and the limitations of existing screening technologies, the viral loads in the blood of OBI patients are intermittent and often undetectable. Furthermore, lack of effective screening and shielding strategies in blood collection and supply institutions fail to prevent OBI individuals from donating blood, resulting in its susceptibility to transmission through blood transfusion, which poses a significant threat to blood safety. In this review, we summarize current understanding of OBI, challenges, and its implication in blood safety.
1 Introduction
HBV infection causes a significant liver disease and poses a global public health challenge. The World Health Organization (WHO) estimates that approximately 296 million people worldwide were chronically infected with HBV in 2019, resulting in 820,000 deaths, primarily due to cirrhosis and hepatocellular carcinoma (HCC) (1, 2). Although WHO has established a mandate to eliminate HBV by 2030 (2, 3), OBI remains difficult to be detected in routine blood screening due to its low viral load and intermittent nature. Previous study indicates that transfusions from donors with OBI are estimated to cause HBV infection in 8-29% of recipients (4), thereby presenting a significant threat to blood safety. We summarize the pathogenesis and prognosis of OBI, its epidemiology, the limitations of current diagnostic and screening techniques, the impact on blood safety, and strategies for its prevention and control in order to ensure blood safety.
2 The definition of OBI
Transfusion safety issues have been an increasing concern among researchers since the late 1970s, when studies confirmed that blood from donors containing hepatitis B core antibodies (anti-HBc) but no detectable HBsAg or hepatitis B surface antibodies (anti-HBs) could transmit hepatitis B through blood transfusions (5, 6). Subsequently, with the development of molecular biology techniques, cases of HBsAg-negative but HBV DNA-positive liver disease and transmission were discovered, which gradually led to the definition of “occult hepatitis B infection (OBI)” (7–12).
OBI is defined as the presence of replication-competent HBV DNA (i.e. episomal HBV covalently closed circular DNA [cccDNA]) in the liver and/or HBV DNA in the blood of people who test negative for HBsAg by currently available assays (13, 14). Due to strong suppression and clearance pressure from the host immune system, HBV cccDNA exhibits low-level replication typically below 200 IU/mL or intermittent occurrence, which pose significant challenges to routine screening assays (13–16). Specifically, currently available OBI screening technologies frequently fail to detect OBI due to insufficient sensitivity or inadequate ability to detect virus mutations (17). Improvements in quantitative HBV marker detection methods and enhancements in the performance of detection reagents and instruments can significantly increase the OBI yields or reclassify them as chronic HBV infections (CHB) (18–22).
Accurate identification of “occult” and “overt” HBV infection status is valuable for HBV prevention and control. In routine testing, serological testing is primarily used to accurately identify these two infection states. Two types of serological test results are often presented in OBIs: those that are positive(80%) for anti-HBc and/or anti-HBs, and those that are negative(20%) for both (13, 14, 23, 24). Especially, due to the lack of serological markers, serological negative OBI and the window phase (WP) of acute or chronic hepatitis B infection are classified as “primary OBI” (1, 13, 25, 26). In contrast, “overt” HBV infection is defined as the presence of HBsAg and viral genomic DNA in the blood, which indicates abundant replication and high transcriptional activity of the virus in the host (1, 27). As shown in Figure 1, HBV infects the host and gradually produces specific markers such as, HBV DNA, HBsAg, anti-HBc, and anti-HBs as the virus continues to replicate. However, when encountering strong immune pressure from the host, HBV exists in a state of low replication state in which HBV DNA levels fluctuate around the lowest detection limit and are only detected intermittently. This is commonly denoted as the OBI state, and in some cases, anti-HBc and anti-HBs gradually disappear, while HBV DNA is the only detectable marker.
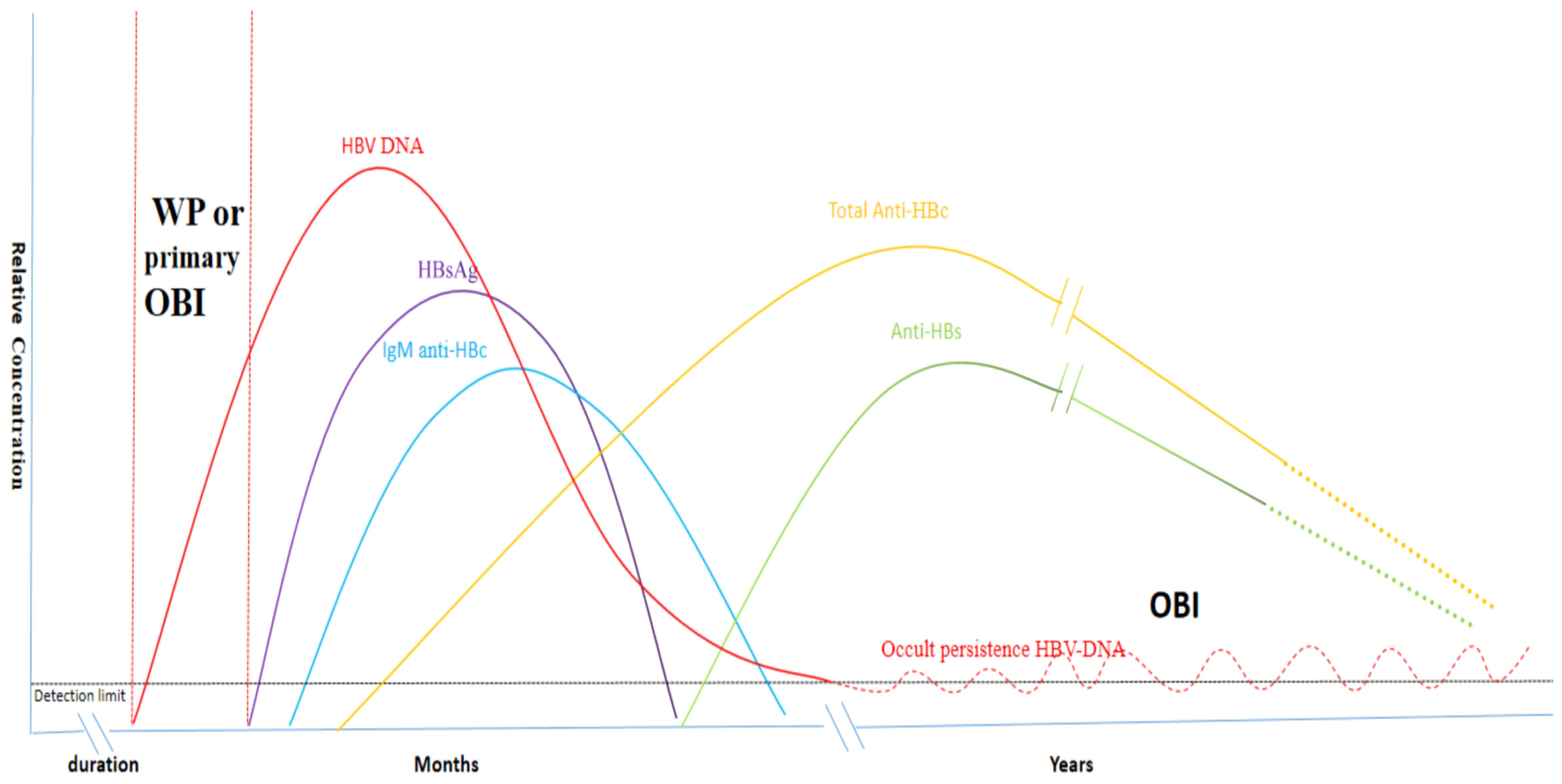
Figure 1. Serological markers following HBV infection and the occurrence of OBIs (2, 28) Specific serological markers, including HBV DNA, HBsAg, anti-HBc, and anti-HBs are detected in the blood following HBV infection. OBIs primarily arise from the virus’s low replication state in response to host immune pressure. In this condition, HBV DNA levels fluctuate around the lowest detection limit and are only intermittently detectable. In some cases, anti-HBc and anti-HBs gradually decline and HBV DNA is the only detectable marker.
3 Mechanisms of OBI formation
OBI results from a complex interplay between HBV and the host. HBV utilizes the host receptor to gain entry into hepatocytes, where rcDNA is released and subsequently converted to cccDNA. This cccDNA serves as a template for the generation of pgRNA and mRNA, ultimately leading to the transcription of HBV DNA and the production of various viral proteins (1). However, due to host immune response, epigenetic regulation, and viral mutation, HBV cccDNA may exist in a low replication state leading to very low level of HBV DNA without detectable HBsAg in the blood and/or liver tissue, and OBI arises (14, 29, 30).
Although the detailed mechanism of OBI formation is not completely understood, many studies indicate that host immune response, viral escape mutation, epigenetic regulation and co-infection with other viruses may play an important role.
Firstly, the role of host immune response to HBV infection in OBI occurrence should not be neglected. Under strong immune pressure, HBV replication is significantly suppressed leading to low viral loads and undetectable HBsAg (31, 32).
Secondly, the appearance of viral escape mutations is also important. HBV genome contains four open reading frames (S,X,P,C), and in order to be able to adapt to the host environment, the virus undergoes continuous mutations in order to escape the immune surveillance (33–35). Some mutations in the HBsAg “α” determinant cluster (36, 37), S-region pres1/pres2 promoter (18, 33, 38), BCP/PC region (38), and the HBsAg hydrophilic region (MHR) (35) affect not only the structure but also the production/secretion of HBsAg, which favor viral immune evasion and ultimately promote OBI formation.
Thirdly, epigenetic regulation may also involve in the formation of OBIs. HBV cccDNA is similar to the chromatin of host cells and histone modifications by methylases and acetylases have been shown to inhibit viral replication (39–41). In addition, previous study reported a natural glycosylation pattern of HBsAg, which may escape immune-mediated clearance by masking antigenic determinants, leading to the development of cryptic infections (42).
Last but not the least, co-infection of HBV with other viruses such as HIV or HCV has been shown to inhibit HBV replication and decrease HBsAg production/secretion leading to OBI (43, 44). Figure 2 briefly summarizes the potential mechanisms of OBI formation following HBV infection.
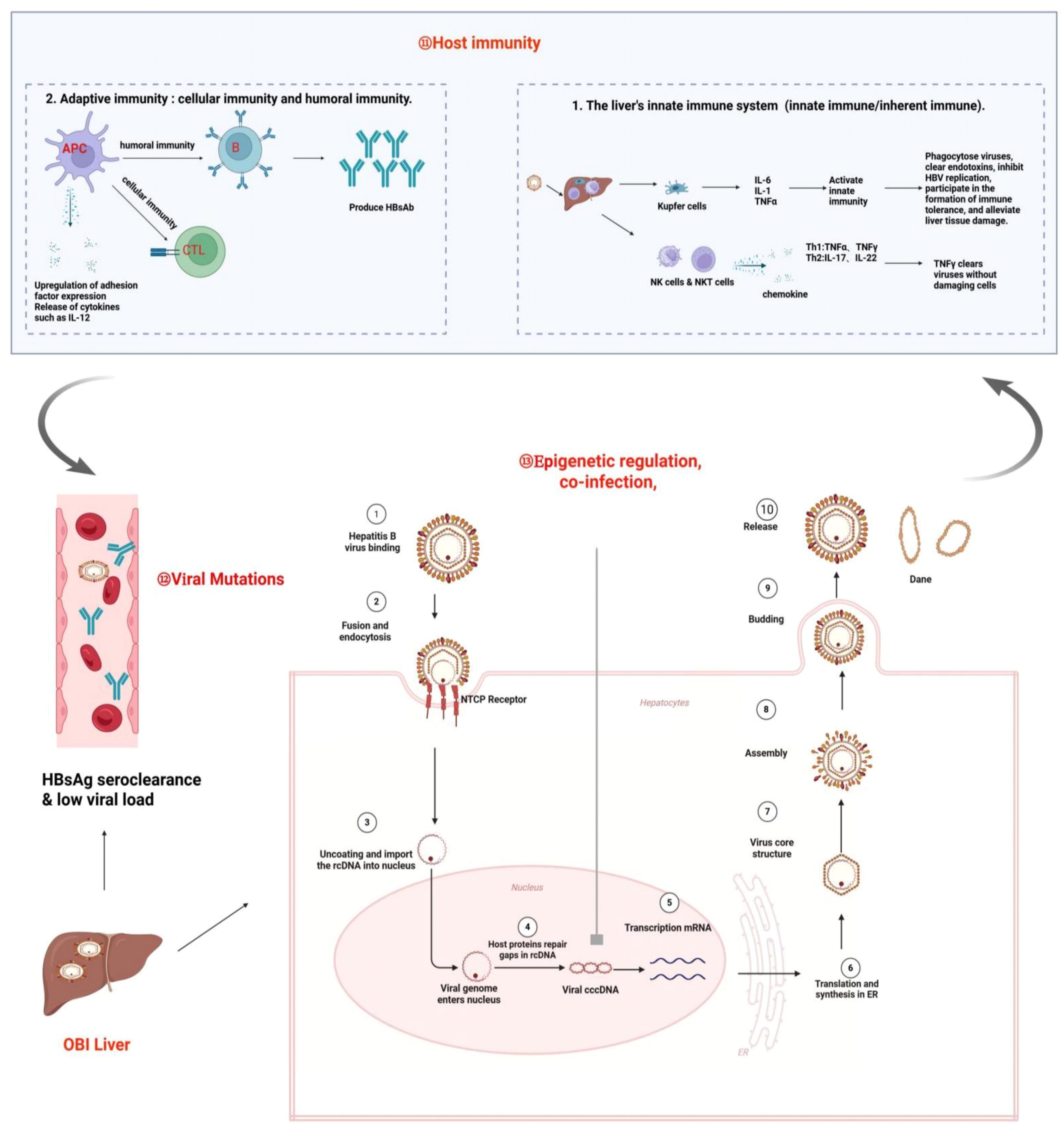
Figure 2. Mechanisms of OBI formation. OBI is the result of complex interactions between the HBV and the host. Under the influence of various factors, such as strong immune suppression and clearance by the host, epigenetic regulation, co-infection, and mutation, the OBI state is ultimately formed, which is characterized by the presence of replication-competent viral DNA in the liver (detectable or undetectable HBV DNA in serum), while the individual tests for HBsAg are negative.
As for the outcomes of OBIs, retrospective studies from OBI blood donors have shown that few OBI-infected patients spontaneously clear HBV from their bodies (45, 46), whereas the majority of them ultimately remain in a state of low viral replication and intermittently detectable HBV DNA (18, 47). As such, transfusion-transmitted HBV infection may occur if blood collection and supply institutions do not shield these OBI donors.
4 Challenges in OBI diagnosis and screening in blood institutions
The diagnosis and screening of OBI face several challenges. Due to the low viral load and intermittent viremia inherently associated with OBIs, the viral load may be under the detectable level, resulting in missed diagnoses (18, 48, 49). Although the detection of cccDNA in the liver tissue is considered the golden standard for the diagnosis of OBI, liver biopsy process itself is invasive, and there is no standardized techniques for detecting HBV DNA in liver tissues (13, 14). Therefore, screening for OBI primarily relies on serological and molecular testing techniques. However, in practice, several factors contribute to the underdiagnosis of OBIs, including the limited sensitivity of testing methods (50, 51), inability to detect viral variants (52), inappropriate ratios of anti-HBs to HBsAg in the samples (21), and the variability in testing modes and algorithms (53). Challenges in OBI screening are summarized in Figure 3.
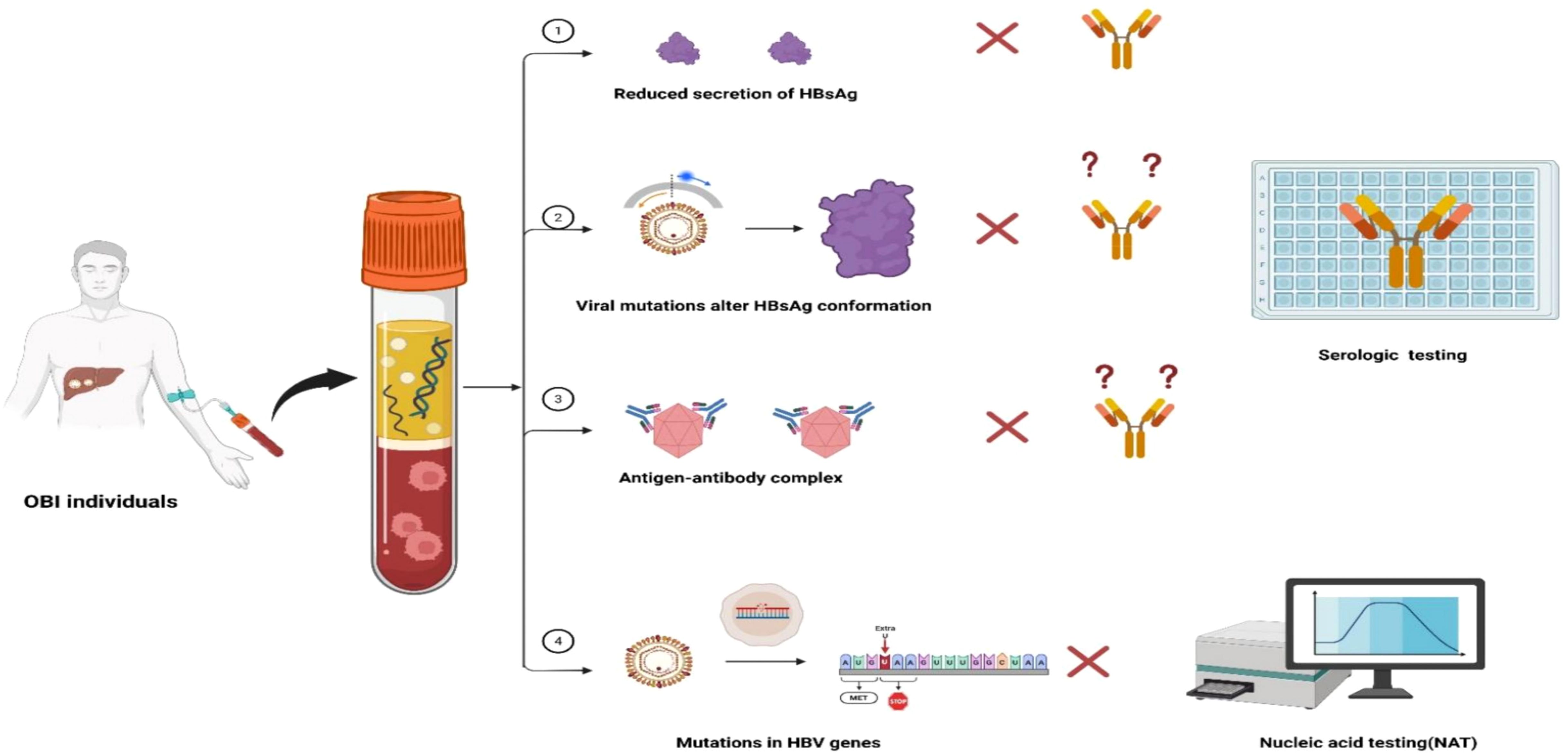
Figure 3. Challenges in OBI diagnosis and screening. ① Reduced secretion of HBsAg caused by Host immunity pressure suppressed the secretion of HBsAg, which is under detection limit of the commercial reagents leading to detection failure. ② Viral mutations cause changes in the spatial conformation of HBsAg, which cannot be recognized by monoclonal antibodies and lead to detection failure of commercial reagents. ③ Formation of antigen-antibody complexes in the blood, which cannot be recognized by commercial reagents and lead to detection failure. ④ Mutations in viral genes may result in altered gene sequences or reduced synthesis of HBV DNA, which then lead to testing failure by NAT.
4.1 Challenges in serologic testing
HBsAg is widely used as a serum marker for screening and diagnosis of HBV infection, and the lowest limit of detection of the commercial reagents is 0.05 IU/mL (54, 55). Some studies demonstrated that 18.2%–61.5% of samples previously classified as HBsAg negative by conventional assays were tested positive for HBsAg by ultrasensitive methods (17). In addition, the insufficient ability of commercial testing reagents to detect virus mutations is another factor contributing to missed detection of OBIs. Once the altered spatial conformation or reduced secretion of the HBsAg derived from HBV mutations occur, HBsAg cannot be recognized by the commercial monoclonal antibody reagent, resulting in testing failures (52, 56). In addition, the formation of antigen-antibody complexes in OBI blood can also cause detection failure (21, 34, 57).
4.2 Challenges in nucleic acid testing
Nucleic acid testing (NAT) used for blood screening primarily include transcription-mediated nucleic acid amplification assays (TMA) and real-time polymerase chain reaction (PCR) (47). For NAT, the lower limit of most commercially available HBV DNA tests ranges from 10 to 20 IU/ml, while for blood products sensitivity is higher (1.4 to 12 IU/ml) and specificity (99.9%) in blood-supplying institutions (53, 58, 59).The sensitivity of the mini pool (MP-NAT) decreases as the number of samples mixed in the mini pool increases, suggesting that insufficient sensitivity of the MP-NAT testing could also lead to missed OBIs detection (60, 61). Additionally, an American comprehensive study from 22.4 million blood donors revealed that only 43/404 (10.6%) OBIs could be detected by MP-NAT, and most OBIs (361/404, 89.4%) could only be identified by individual testing (ID-NAT) (19). In addition, when the viral genome mutates, it cannot be amplified by primer recognition, resulting in nucleic acid detection failure (62, 63). Notably, given that the HBV DNA level of OBI usually fluctuates around the lowest detection limit, and is only detectable intermittently, these factors also increased the possibility of testing failure (61). Further, although the laboratory’s MPX Taq screening assay is a quantitative method in which the Ct value reflects the viral load in the blood sample, it follows Poisson distribution and the Ct value may fluctuate when the viral load is close to the lowest detection limit, leading to inconsistent results and undetectable OBIs (53, 61, 64).
5 The prevalence of OBIs in blood donors
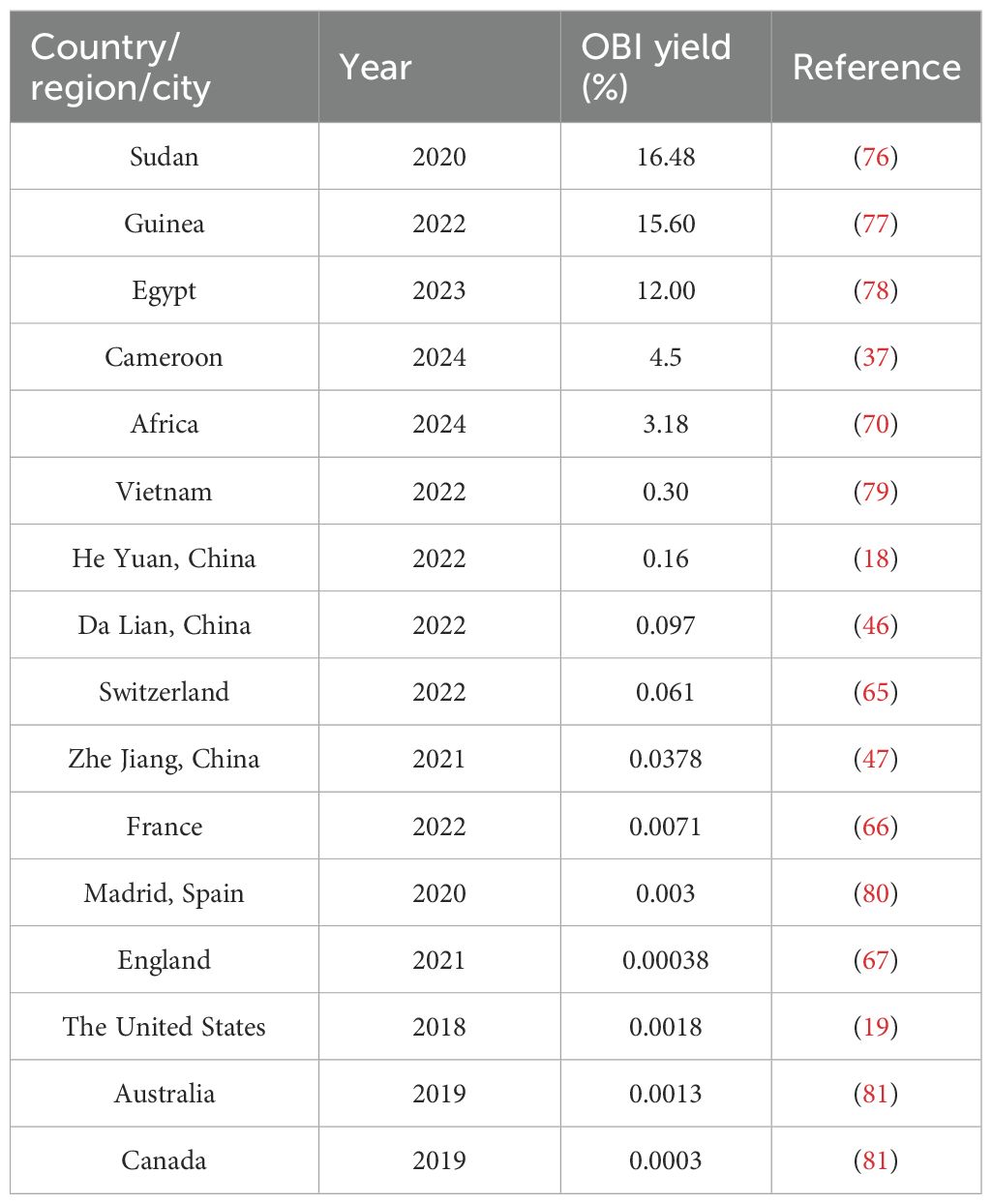
The global prevalence of OBIs varies significantly due to differences in geographical locations, detection sensitivities of assay used, host immune responses, and vaccine availability, and it ranges from 0.06%-12% in general population (4). As shown in Table 1, meta-analyses indicate the prevalence of OBIs in blood donors ranges from 0.0003%-16.48% depending on different study populations. Even in the same country (E.g., China), the prevalence varies considerably in different regions, and prevalence of OBIs in blood donors in Heyuan is 0.16% while 0.0378% in Zhejiang.
It is important to note that the prevalence of OBIs may be underestimated due to several factors: a) Sample limitations: many OBI-related studies rely on limited retrospective data and the OBI-containing blood/organs may have been used clinically even when OBIs were identified (65–67). b) Lack of screening and shielding policies: Nucleic acid testing has not been routinely performed in low- and middle-income countries, leading to many cases of undetected OBIs (37, 68–73). Furthermore, most countries have no shielding policies in place for OBI donors, which results in blood donations from repeated OBI donors, increasing the risk of transfusion-transmitted HBV infection (72, 74). c) Inadequate sample storage: The absence of donor and/or recipient samples complicates traceability (67). And d) Differences in analytical methods: Variations in the analytical methods employed across different studies affect the consistency of the results (16, 67, 75).
6 The impact of OBIs on blood safety
It is extremely difficult to detect OBIs by routine screening assays, which may result in HBV infections in recipients (49, 82). Previous study indicated that HBV infection was detected in 8-29% (4) of recipients who received OBI-containing blood. Moreover, the absence of effective screening and shielding policies for OBI donors increased TT-OBIs (74, 75). It is worth noting that a single bag of donated blood may be processed to different blood products. In other words, a single bag of OBI blood may lead to HBV infection in multiple recipients (49). Unfortunately, most studies investigating OBI transmissions lack experimental validation and clinicians may overlook patients who do not exhibit overt signs of acute hepatitis (13, 28). Furthermore, healthcare workers may be hesitant to trace infections to avoid potential medical disputes, missing opportunities for timely intervention following transfusion of OBI blood (49).
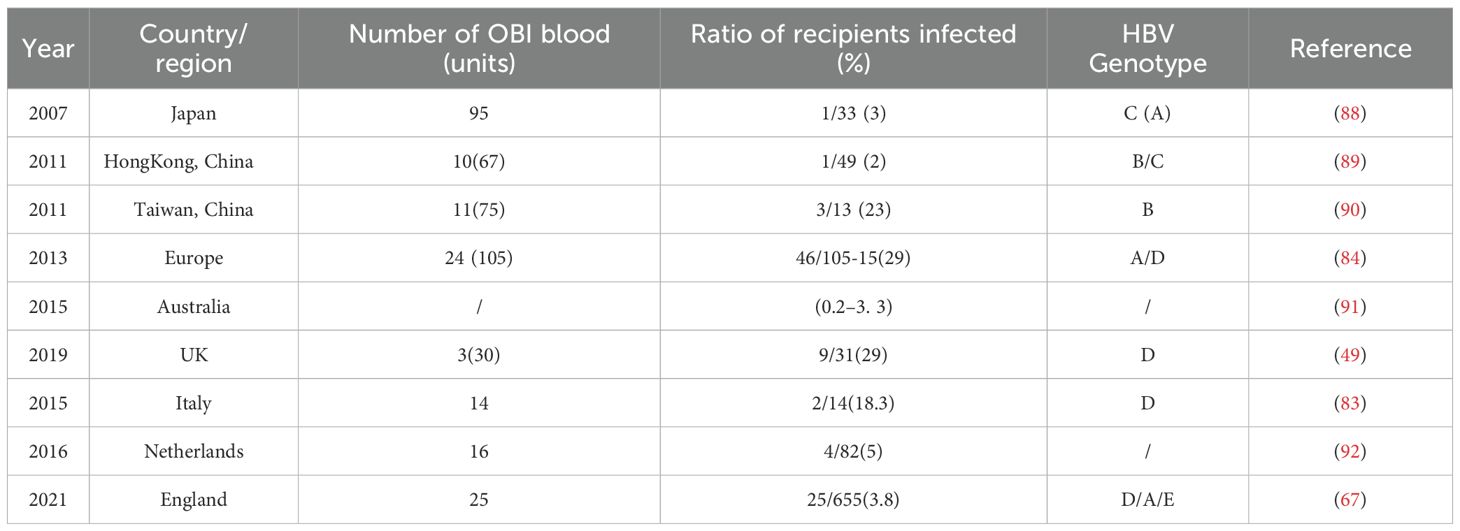
After being transfused with OBI blood, recipients may be infected with HBV and this leads to clinical transfusion safety incidents (83–86). For instance, A recent study showed that anti-HBc positivity increased to 37.7% in children with leukemia who received blood transfusions during immunosuppressive therapy, suggesting that transfusion of potentially OBI-containing blood may lead to passive transfer of anti-HBc and anti-HBs (87). Likewise, a study by Allain et al. (84) showed that 3 patients developed sepsis and hepatitis after receiving blood products containing OBI. In the worst scenario, Spreafico et al. (83) reported that a case of a bone marrow transplant patient with a hematologic disorder who received OBI blood resulted in viral activation and eventual death from acute liver failure. Examples of TT-OBIs in different countries/regions were summarized in Table 2.
7 Strategies to reduce the risk of TT-OBIs to enhance blood safety
7.1 HBV vaccination
Vaccine administration is one of the most cost-effective measures to control infectious diseases. For example, China introduced the routine immunization with hepatitis B vaccine in 1992 and this significantly reduced the prevalence of HBsAg to 1% in children under five years of age (93). However, the levels of protective antibodies induced by the vaccine can gradually wane over time, resulting in diminished protection and an increased risk of breakthrough infections (94–96).Therefore, regular surveillance and identification of breakthrough infections following hepatitis B vaccinations are essential for the long-term prevention of HBV infection (97). This is particularly important for some special groups such as blood donors, those with hepatitis B-positive family members, public service workers, and medical waste handlers (98–100). Consequently, regular testing for hepatitis B markers and boost vaccination if needed should be strongly encouraged (74, 101, 102).
7.2 Selection of blood donors
Despite significant advances in laboratory screening technologies, the initial step in ensuring blood safety is the selection of unpaid voluntary blood donors who are at lower risk (60). HBV can be transmitted through various routes, including vertical transmission, blood transfusion, intravenous drug use, sexual contact, tattoos, and piercings (100, 103, 104). Blood donations from individuals with “high-risk behaviors” can be effectively excluded through rigorous counseling and screening, which represents an economically feasible and practical approach to ensure blood safety (72, 77, 103, 105).
7.3 Pathogen inactivation procedures
Pathogen inactivation (PI) represents a strategic approach to mitigate the risks associated with transfusion-transmitted infections and has been successfully implemented in numerous countries worldwide. It is recommended that all labile blood products and biologics undergo pathogen inactivation to ensure blood safety and sustain a secure blood supply (106).
7.4 Novel testing techniques and biomarkers
Given the low viral load and intermittent viremia characteristics of OBI, it is imperative to improve sensitivity and the ability to identify viral variants of the screening assays (63). This can be achieved by optimizing the detection of various open reading frames in the genome of HBV and multiple epitopes of the viral proteins (13, 24), as well as by incorporating more sensitive nucleic acid detection methods, such as real-time PCR, nested PCR, and digital PCR (14). Additionally, considering the intermittent appearance of HBV DNA in the blood, blood specimens should be collected at multiple time points. This can be combined with ultra-high-speed centrifugation, specific adsorption, and other techniques to concentrate the virus before assay was performed (48, 65), or by increasing the volume of plasma or serum used for DNA extraction (48, 63).
Importantly, the limitations of serum HBVDNA testing underscore the necessity for complementary testing strategies based on other biomarkers from HBV infection. Anti-HBc testing should be implemented, particularly when obtaining liver tissue is challenging or when HBV DNA is negative but the possibility of OBI cannot be excluded (107, 108). For instance, a study by Ye et al. (107) indicated that among 103,356 seronegative blood donors, there were 252 non-resolved donations (which had been released for clinical use). In these 252 cases, alternative methods combining NAT with Nested PCR + qPCR and Roche ECLI were used for serological testing (HBsAg/anti-HBs/anti-HBc/HBeAg/anti-HBe). Ultimately, 17 cases were identified as HBVDNA positive (with all anti-HBc being positive). Previous studies reported that anti-HBc testing was effective in identifying blood donors with OBIs (2, 14). That is, anti-HBc screening combined with high-sensitivity HBV NAT screening can effectively prevent almost all HBV transmission from OBI donors (107, 109). In Japan (110) and France (66), for example, the implementation of anti-HBc screening has significantly reduced the incidence of transfusion-transmitted HBV infections. However, anti-HBc testing for OBIs also has limitations. The sensitivity and specificity of anti-HBc in identifying occult HBV infections are only 77% and 76%, respectively (4). Moreover, anti-HBc screening may be impractical in HBV high prevalent countries, such as in China, where anti-HBc screening would eliminate at least 36% of eligible blood donors (111). Therefore, blood services in each country should decide whether or not to implement anti-HBc testing in the context of HBV prevalence (112).
Notably, this rare problem should not be overlooked, as we currently know very little about the clinical significance of anti-HBc-negative OBI and serological negative OBI donors (109). In particular, low levels of HBV DNA and intermittent testing can easily lead to false-negative HBV DNA test results. If we rely solely on serological testing, it is easy to miss some of these serological negative OBI cases (95). Studies have shown that a small number of OBI are HBVDNA-negative and have all serological markers negative (48, 52). In addition, it is worth mentioning that donors with only anti-HBs-positive serological characteristics may be in the acute WP or OBI, which has been reported to account for 4.4% (26) to 11% (113) of cases and has been associated with blood transfusion-transmitted infection (109, 114). Similarly, the use of anti-HBs testing should also be evaluated on a case-by-case basis (114). In practice, for example, in order to prevent serious blood shortages caused by excluding only anti-HBc-positive individuals, the Japanese Red Cross(JRC) Blood Center took measures in 1989 to accept blood from donors with elevated anti-HBs levels(≥200IU/ml) and low anti-HBc levels. However, transfusion-transmitted HBV infection cases still occurred (88, 115). Further, it should be noted that although high levels of anti-HBs may reduce the risk of HBV infection to a certain extent (84, 115), they may not provide sufficient protection for immunodeficient recipients (46, 47, 82).
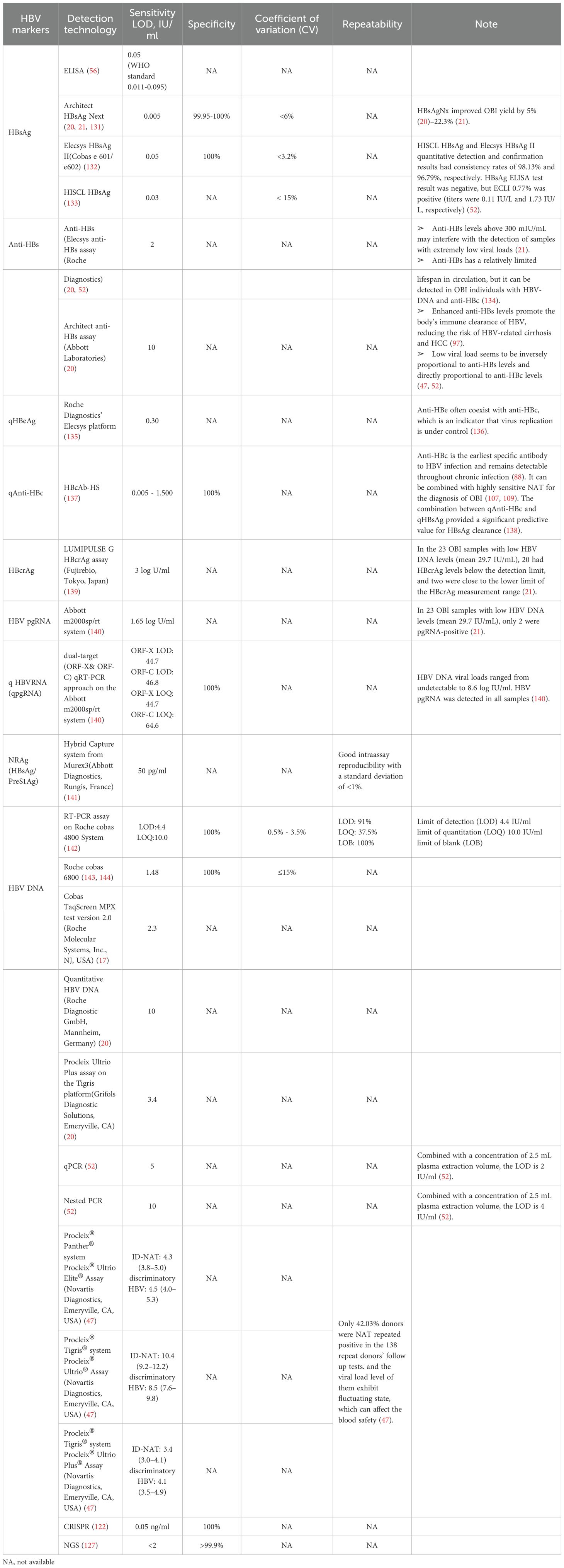
Since its introduction in the 1970s, OBIs have remained an area of active investigation. With the advancement of new technologies, a variety of markers and screening techniques have been emerging in clinical practice over the past two decades, and these HBV- related biomarkers include serum HBV RNA, hepatitis B core-related antigen (HBcrAg), quantitative HBsAg (qHBsAg), quantitative anti-HBc (qAHBc), and HBV nucleic acid- related antigen (HBV-NRAg) (116–118). Some immunological techniques have made great breakthroughs, such as Lumipulse HBsAg Hypersensitivity Assay, the sensitivity, specificity and coefficient of variation (CV) reaches 0.005 IU/mL, 99.8% and <4%, respectively (51). More valuably, its performance is not affected by clinical treatments, viral gene mutations, or antigen-antibody complexes, establishing a foundation for early diagnosis and treatment to facilitate the mechanistic study of the OBI formation (17, 51, 119). Molecular diagnostic technologies, particularly those based on Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR), provide new approaches for rapid, sensitive, and portable nucleic acid molecular detection (120, 121). The detection sensitivity is as low as 0.05 ng/mL, with 100% specificity (122),which was successfully applied to the detection of HBV DNA with low viral load (123)and HBV cccDNA (124). Next Generation Sequencing (NGS) serves as a powerful tool for the detection and research of HBV (125, 126), which is a robust technology that detects integrated HBV DNA (iDNA) in blood and urine even in CHB patients with a sensitivity of serum viral load <2 IU/mL or even undetectable, and a specificity of at least 99.9% (127). Particularly in the identification and mechanistic exploration of OBIs. Despite notable progress in sensitivity and specificity of detection methods, challenges persist for low-level HBV markers typical of OBIs, particularly regarding result stability and reproducibility (47, 53, 61, 64, 128). Transfusion-related cases repeatedly showed that several assays underperformed at low viral loads, raising the risk of OBIs going undetected (49, 84). Some other investigators have proposed that the minimum infectious dose of HBV in blood transfusion may be as low as 16copies/ml or 3 IU (49). In order to convey these information, we added Table 3 to summarize the performance of currently commercialized quantitative HBV markers.
Although adoption of novel testing strategies has been shown to be effective in mitigating the transfusion-transmitted HBV infection, additional screening strategies may increase the financial burden on blood institutions and governments. Therefore, it is more cost-effective to optimize current screening protocols to establish a set of stable, reliable, and non-invasive detection paradigms for HBV markers to further ensure blood safety. Such as the HBsAg combined with HBV DNA ID-NAT screening protocol. Ye et al. (22). analyzed 132,323 donations using MP-6 HBV NAT and ID NAT comparison analysis, which showed that the yield of HBsAg-/DNA+ detected by ID NAT screening (0.12%) was 1.25 times that of MP NAT (0.058%, P < 0.05). To further enhance blood safety, HBsAg and HBV DNA ID NAT screening should be considered in regions/countries with high HBV prevalence. Besides, in low-resource areas, combined testing for HBsAg, anti-HBc, and anti-HBs can effectively reduce the risk of blood transfusion-transmitted HBV infection (129). Moreover, it is worth mentioning that anti-HBc screening and high-sensitivity HBV NAT screening can effectively prevent almost all HBV transmission from OBI donors (107, 109). To prevent HBV infection, it is recommended to perform ID-NAT on anti-HBc-positive blood donors, in order to discard plasma with weakly positive or negative anti-HBs but positive anti-HBc, or avoid transfusing anti-HBc-positive plasma to recipients with weakly positive or negative anti-HBs (130).
In summary, given the complexity of OBI, blood centers need adopt different strategies based on the risk of OBI transmission to improve blood safety. It is recommended that blood centers review the OBI risk assessment and identify OBI risk reduction strategies accordingly, including HBV vaccination, rigorous screening of blood donors, introduction of new technologies, exploration of OBI screening and shielding strategies, encouraging autologous transfusion and blood management, and pathogen inactivation of blood products from donors in areas with high prevalence of HBV.
8 Conclusions
Although the risk of transfusion-transmitted infection from blood components has been significantly reduced over the past few decades through rigorous donor selection and enhanced screening tests, a residual risk persists. TT-OBI remains the primary method of HBV transmission. Given the complexities associated with OBI and the absence of currently feasible screening and shielding strategies for blood centers in many countries, OBI presents significant challenges to blood safety. Therefore, it is essential to recruit low-risk voluntarily unpaid blood donors at the source; implement new technologies with mutation detection capacity to enhance sensitivity of current assays; develop appropriate screening and shielding strategies for donors who are OBIs; implement pathogen inactivation procedures, and optimize existing screening protocols to further prevent TT-OBI infections to ensure the highest possible level of blood safety.
Author contributions
LH: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. BH: Methodology, Project administration, Writing – original draft, Writing – review & editing. QM: Writing – original draft, Writing – review & editing. BL: Software, Writing – original draft, Writing – review & editing. XPY: Visualization, Writing – original draft, Writing – review & editing. RL: Software, Visualization, Writing – original draft, Writing – review & editing. XW: Data curation, Resources, Visualization, Writing – original draft, Writing – review & editing. JL: Data curation, Formal Analysis, Investigation, Writing – original draft, Writing – review & editing. ML: Conceptualization, Methodology, Writing – original draft, Writing – review & editing. HX: Software, Validation, Writing – original draft, Writing – review & editing. JS: Formal Analysis, Investigation, Writing – original draft, Writing – review & editing. XLY: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. LC: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. The authors acknowledge funds from the following organizations: 1. Innovation Project of Guangxi Graduate Education (YCSW2025253).2. Sanming Project of Medicine in Shenzhen (SZSM202311032). 3. Guangxi Natural Science Foundation Youth Science Foundation, China, NO. 2024GXNSFBA010047. 4. Chinese society of blood transfusion–WEGO scientific research fund (CSBT-WG-2022-01). 5. Shengxiang Transfusion Medicine Development Fund of the Chinese Society of Blood Transfusion (Grant No. CSBT-SX-2022-01).
Acknowledgments
Figures were made using bioRender.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Jeng WJ, Papatheodoridis GV, and Lok ASF. Hepatitis B. Lancet. (2023) 401:1039–52. doi: 10.1016/S0140-6736(22)01468-4
2. Kramvis A, Chang KM, Dandri M, Farci P, Glebe D, Hu J, et al. A roadmap for serum biomarkers for hepatitis B virus: current status and future outlook. Nat Rev Gastroenterol Hepatol. (2022) 19:727–45. doi: 10.1038/s41575-022-00649-z
3. Liu J, Liang W, Jing W, and Liu M. Countdown to 2030: eliminating hepatitis B disease, China. Bull World Health Organ. (2019) 97:230–8. doi: 10.2471/BLT.18.219469
4. Im YR, Jagdish R, Leith D, Kim JU, Yoshida K, Majid A, et al. Prevalence of occult hepatitis B virus infection in adults: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol. (2022) 7:932–42. doi: 10.1016/S2468-1253(22)00201-1
5. Hoofnagle JH, Seeff LB, Bales ZB, and Zimmerman HJ. Type B hepatitis after transfusion with blood containing antibody to hepatitis B core antigen. N Engl J Med. (1978) 298:1379–83. doi: 10.1056/NEJM197806222982502
6. Tabor E, Hoofnagle JH, Smallwood LA, Drucker JA, Pineda-Tamondong GC, Ni LY, et al. Studies of donors who transmit posttransfusion hepatitis. Transfusion. (1979) 19:725–31. doi: 10.1046/j.1537-2995.1979.19680104098.x
7. Chazouilleres O, Mamish D, Kim M, Carey K, Ferrell L, Roberts JP, et al. Occult” hepatitis B virus as source of infection in liver transplant recipients. Lancet. (1994) 343:142–6. doi: 10.1016/s0140-6736(94)90934-2
8. Brechot C, Kremsdorf D, Paterlini P, and Thiers V. Hepatitis B virus DNA in HBsAg-negative patients. Molecular characterization and clinical implications. J Hepatol. (1991) 13 Suppl 4:S49–55. doi: 10.1016/0168-8278(91)90023-5
9. Thiers V, Nakajima E, Kremsdorf D, Mack D, Schellekens H, Driss F, et al. Transmission of hepatitis B from hepatitis-B-seronegative subjects. Lancet. (1988) 2:1273–6. doi: 10.1016/S0140-6736(88)92891-7
10. Cacciola I, Pollicino T, Squadrito G, Cerenzia G, Orlando ME, and Raimondo G. Occult hepatitis B virus infection in patients with chronic hepatitis C liver disease. N Engl J Med. (1999) 341:22–6. doi: 10.1056/NEJM199907013410104
11. Sheu JC, Huang GT, Shih LN, Lee WC, Chou HC, Wang JT, et al. Hepatitis C and B viruses in hepatitis B surface antigen-negative hepatocellular carcinoma. Gastroenterology. (1992) 103:1322–7. doi: 10.1016/0016-5085(92)91523-7
12. Paterlini P, Driss F, Nalpas B, Pisi E, Franco D, Berthelot P, et al. Persistence of hepatitis B and hepatitis C viral genomes in primary liver cancers from HBsAg-negative patients: a study of a low-endemic area. Hepatology. (1993) 17:20–9. doi: 10.1002/hep.1840170106
13. Raimondo G, Locarnini S, Pollicino T, Levrero M, Zoulim F, Lok AS, et al. Update of the statements on biology and clinical impact of occult hepatitis B virus infection. J Hepatol. (2019) 71:397–408. doi: 10.1016/j.jhep.2019.03.034
14. Saitta C, Pollicino T, and Raimondo G. Occult hepatitis B virus infection: an update. Viruses. (2022) 14:1504. doi: 10.3390/v14071504
15. Raimondo G, Allain JP, Brunetto MR, Buendia MA, Chen DS, Colombo M, et al. Statements from the Taormina expert meeting on occult hepatitis B virus infection. J Hepatol. (2008) 49:652–7. doi: 10.1016/j.jhep.2008.07.014
16. Locarnini S and Raimondo G. How infectious is the hepatitis B virus? Readings from the occult. Gut. (2019) 68:182–3. doi: 10.1136/gutjnl-2018-316900
17. Song S, Su Q, Yan Y, Ji H, Sun H, Feng K, et al. Identification and characteristics of mutations promoting occult HBV infection by ultrasensitive HBsAg assay. J Clin Microbiol. (2025) 63:e0207124. doi: 10.1128/jcm.02071-24
18. Ye X, Liu L, Chen L, Nie X, Huang L, Ye D, et al. High-frequency notable HBV mutations identified in blood donors with occult hepatitis B infection from heyuan city of southern China. Front Immunol. (2022) 13:754383. doi: 10.3389/fimmu.2022.754383
19. Dodd RY, Nguyen ML, Krysztof DE, Notari EP, and Stramer SL. Blood donor testing for hepatitis B virus in the United States: is there a case for continuation of hepatitis B surface antigen detection? Transfusion. (2018) 58:2166–70. doi: 10.1111/trf.14784
20. Wong DK, Chen C, Mak LY, Fung J, Seto WK, and Yuen MF. Detection of the hepatitis B surface antigen in patients with occult hepatitis B by use of an assay with enhanced sensitivity. J Clin Microbiol. (2022) 60:e0220421. doi: 10.1128/jcm.02204-21
21. Kuhns MC, Holzmayer V, Anderson M, McNamara AL, Sauleda S, Mbanya D, et al. Molecular and serological characterization of hepatitis B virus (HBV)-positive samples with very low or undetectable levels of HBV surface antigen. Viruses. (2021) 13:2053. doi: 10.3390/v13102053
22. Ye X, Xiong W, Xu X, Zeng J, Xie H, Li B, et al. Cost-benefit analysis of serological and nucleic acid testing for hepatitis B virus in blood donors in southern China. BMC Infect Dis. (2024) 24:909. doi: 10.1186/s12879-024-09786-z
23. Torbenson M and Thomas DL. Occult hepatitis B. Lancet Infect Dis. (2002) 2:479–86. doi: 10.1016/S1473-3099(02)00345-6
24. Mak LY, Wong DK, Pollicino T, Raimondo G, Hollinger FB, and Yuen MF. Occult hepatitis B infection and hepatocellular carcinoma: Epidemiology, virology, hepatocarcinogenesis and clinical significance. J Hepatol. (2020) 73:952–64. doi: 10.1016/j.jhep.2020.05.042
25. Al-Sadeq DW, Taleb SA, Zaied RE, Fahad SM, Smatti MK, Rizeq BR, et al. Hepatitis B virus molecular epidemiology, host-virus interaction, coinfection, and laboratory diagnosis in the MENA region: an update. Pathogens. (2019) 8. doi: 10.3390/pathogens8020063
26. Lelie N, Bruhn R, Busch M, Vermeulen M, Tsoi WC, Kleinman S, et al. Detection of different categories of hepatitis B virus (HBV) infection in a multi-regional study comparing the clinical sensitivity of hepatitis B surface antigen and HBV-DNA testing. Transfusion. (2017) 57:24–35. doi: 10.1111/trf.13819
27. Nguyen MH, Wong G, Gane E, Kao JH, and Dusheiko G. Hepatitis B virus: advances in prevention, diagnosis, and therapy. Clin Microbiol Rev. (2020) 33:e00046-19. doi: 10.1128/CMR.00046-19
28. Vermeulen M, Dickens C, Lelie N, Walker E, Coleman C, Keyter M, et al. Hepatitis B virus transmission by blood transfusion during 4 years of individual-donation nucleic acid testing in South Africa: estimated and observed window period risk. Transfusion. (2012) 52:880–92. doi: 10.1111/j.1537-2995.2011.03355.x
29. Liu C, Chen K, Zhao F, Xuan L, Wang Y, Xu C, et al. Occult infection with hepatitis B virus PreS variants synergistically promotes hepatocellular carcinoma development in a high-fat diet context by generating abnormal ceramides. BMC Med. (2022) 20:279. doi: 10.1186/s12916-022-02481-3
30. Jiang X, Chang L, Yan Y, Ji H, Sun H, Xiao Y, et al. Role of S protein transmembrane domain mutations in the development of occult hepatitis B virus infection. Emerg Microbes Infect. (2022) 11:2184–96. doi: 10.1080/22221751.2022.2114849
31. Kuipery A, Gehring AJ, and Isogawa M. Mechanisms of HBV immune evasion. Antiviral Res. (2020) 179:104816. doi: 10.1016/j.antiviral.2020.104816
32. Zhang W, Luo S, Li T, Wang M, Huang J, Liao Q, et al. Hepatitis B virus-specific cellular immunity contributes to the outcome of occult hepatitis B virus infection. Front Microbiol. (2022) 13:850665. doi: 10.3389/fmicb.2022.850665
33. Wang M, Xu R, Huang J, Liao Q, Tang X, Shan Z, et al. Molecular characteristics of the full-length genome of occult hepatitis B virus from blood donors in China. Sci Rep. (2022) 12:8194. doi: 10.1038/s41598-022-12288-0
34. Yan Y, Sun H, Chang L, Ji H, Jiang X, Song S, et al. Circulating immune complexes and mutations of HBsAg are associated with the undetectable HBsAg in anti-HBs and HBeAg positive occult hepatitis B virus infection. Front Microbiol. (2022) 13:1063616. doi: 10.3389/fmicb.2022.1063616
35. Wang Y, Xiao X, Chen S, Huang C, Zhou J, Dai E, et al. The impact of HBV quasispecies features on immune status in HBsAg+/HBsAb+ Patients with HBV genotype C using next-generation sequencing. Front Immunol. (2021) 12:775461. doi: 10.3389/fimmu.2021.775461
36. Xiang KH, Michailidis E, Ding H, Peng YQ, Su MZ, Li Y, et al. Effects of amino acid substitutions in hepatitis B virus surface protein on virion secretion, antigenicity, HBsAg and viral DNA. J Hepatol. (2017) 66:288–96. doi: 10.1016/j.jhep.2016.09.005
37. Mbencho MN, Hafza N, Cao LC, Mingo VN, Achidi EA, Ghogomu SM, et al. Incidence of Occult Hepatitis B Infection (OBI) and hepatitis B genotype characterization among blood donors in Cameroon. PloS One. (2024) 19:e0312126. doi: 10.1371/journal.pone.0312126
38. Chen J, Liu B, Tang X, Zheng X, Lu J, Zhang L, et al. Role of core protein mutations in the development of occult HBV infection. J Hepatol. (2021) 74:1303–14. doi: 10.1016/j.jhep.2020.12.023
39. Low WF, Ngeow YF, Chook JB, Tee KK, Ong SK, Peh SC, et al. Hepatitis B virus DNA methylation and its potential role in chronic hepatitis B. Expert Rev Mol Med. (2022) 25:e11. doi: 10.1017/erm.2022.38
40. Yang Z, Sun B, Xiang J, Wu H, Kan S, Hao M, et al. Role of epigenetic modification in interferon treatment of hepatitis B virus infection. Front Immunol. (2022) 13:1018053. doi: 10.3389/fimmu.2022.1018053
41. Zhang W, Chen J, Wu M, Zhang X, Zhang M, Yue L, et al. PRMT5 restricts hepatitis B virus replication through epigenetic repression of covalently closed circular DNA transcription and interference with pregenomic RNA encapsidation. Hepatology. (2017) 66:398–415. doi: 10.1002/hep.29133
42. Dobrica MO and Lazar C. Branza-nichita N. N-glycosylation and N-glycan processing in HBV biology and pathogenesis. Cells. (2020) 9:1404. doi: 10.3390/cells9061404
43. Meschi S, Mizzoni K, Leoni BD, Galli C, Garbuglia AR, Belladonna S, et al. Occult HBV infection in patients infected by HIV or HCV: comparison between HBV-DNA and two assays for HBsAg. Viruses. (2024) 16:412. doi: 10.3390/v16030412
44. Fabris P, Biasin MR, Giordani MT, Berardo L, Menini V, Carlotto A, et al. Impact of occult HBV infection in HIV/HCV co-infected patients: HBV-DNA detection in liver specimens and in serum samples. Curr HIV Res. (2008) 6:173–9. doi: 10.2174/157016208783885029
45. Malagnino V, Fofana DB, Lacombe K, and Gozlan J. Occult hepatitis B virus infection: an old entity with novel clinical involvements. Open Forum Infect Dis. (2018) 5:ofy227. doi: 10.1093/ofid/ofy227
46. Deng X, Guo X, Gu H, Wang D, Laperche S, Allain JP, et al. Anti-HBc-nonreactive occult hepatitis B infections with HBV genotypes B and C in vaccinated immunocompetent adults. J Viral Hepat. (2022) 29:958–67. doi: 10.1111/jvh.13733
47. Wu D, Wang X, Feng F, Wang D, Hu Y, Yu Y, et al. Characteristic of HBV nucleic acid amplification testing yields from blood donors in China. BMC Infect Dis. (2021) 21:714. doi: 10.1186/s12879-021-06468-y
48. Deng X, Guo X, Li T, Laperche S, Zang L, and Candotti D. Alternative hepatitis B virus DNA confirmatory algorithm identified occult hepatitis B virus infection in Chinese blood donors with non-discriminatory nucleic acid testing. Blood Transfus. (2022) 20:8–17. doi: 10.2450/2020.0213-20
49. Candotti D, Assennato SM, Laperche S, Allain JP, and Levicnik-Stezinar S. Multiple HBV transfusion transmissions from undetected occult infections: revising the minimal infectious dose. Gut. (2019) 68:313–21. doi: 10.1136/gutjnl-2018-316490
50. Damiani AS, Holzmayer V, Galli C, De Nuzzo M, Anderson M, Cloherty G, et al. Serological and molecular characterization of occult HBV infection in blood donors from south Italy. Viruses. (2023) 16:71. doi: 10.3390/v16010071
51. Yang R, Song G, Guan W, Wang Q, Liu Y, and Wei L. The Lumipulse G HBsAg-Quant assay for screening and quantification of the hepatitis B surface antigen. J Virol Methods. (2016) 228:39–47. doi: 10.1016/j.jviromet.2015.11.016
52. Ye X, Li T, Shao W, Zeng J, Hong W, Lu L, et al. Nearly half of Ultrio plus NAT non-discriminated reactive blood donors were identified as occult HBV infection in South China. BMC Infect Dis. (2019) 19:574. doi: 10.1186/s12879-019-4215-9
53. Stolz M, Gowland P, Tinguely C, and Niederhauser C. Safe-testing algorithm for individual-donation nucleic acid testing: 10 years of experience in a low-prevalence country. Transfus Med Hemother. (2019) 46:104–10. doi: 10.1159/000499166
54. Pronier C, Candotti D, Boizeau L, Bomo J, Laperche S, and Thibault V. The contribution of more sensitive hepatitis B surface antigen assays to detecting and monitoring hepatitis B infection. J Clin Virol. (2020) 129:104507. doi: 10.1016/j.jcv.2020.104507
55. Takeda K, Maruki M, Yamagaito T, Muramatsu M, Sakai Y, Tobimatsu H, et al. Highly sensitive detection of hepatitis B virus surface antigen by use of a semiautomated immune complex transfer chemiluminescence enzyme immunoassay. J Clin Microbiol. (2013) 51:2238–44. doi: 10.1128/JCM.00324-13
56. Servant-Delmas A, Mercier-Darty M, Ly TD, Wind F, Alloui C, Sureau C, et al. Variable capacity of 13 hepatitis B virus surface antigen assays for the detection of HBsAg mutants in blood samples. J Clin Virol. (2012) 53:338–45. doi: 10.1016/j.jcv.2012.01.003
57. Matsumoto A, Imaizumi M, Tanaka Y, Nishiguchi S, Yatsuhashi H, Ishida T, et al. Novel and highly sensitive immunoassay for total hepatitis B surface antigen, including that complexed with hepatitis B surface antibody. J Gastroenterol. (2017) 52:376–84. doi: 10.1007/s00535-016-1244-7
58. Kiely P, Margaritis AR, Seed CR, Yang H, and Australian Red Cross Blood Service NATSG. Hepatitis B virus nucleic acid amplification testing of Australian blood donors highlights the complexity of confirming occult hepatitis B virus infection. Transfusion. (2014) 54:2084–91. doi: 10.1111/trf.12556
59. Shyamala V. Nucleic A cid T echnology (NAT) testing for blood screening: impact of individual donation and M ini P ool–NAT testing on analytical sensitivity, screening sensitivity and clinical sensitivity. ISBT Science Series. (2014) 9:315–24. doi: 10.1111/voxs.12106
60. Candotti D and Laperche S. Hepatitis B virus blood screening: need for reappraisal of blood safety measures? Front Med (Lausanne). (2018) 5:29. doi: 10.3389/fmed.2018.00029
61. Wang L, Chang L, Xie Y, Huang C, Xu L, Qian R, et al. What is the meaning of a nonresolved viral nucleic acid test-reactive minipool? Transfusion. (2015) 55:395–404. doi: 10.1111/trf.12818
62. Candotti D and Allain JP. Molecular virology in transfusion medicine laboratory. Blood Transfus. (2013) 11:203–16. doi: 10.2450/2012.0219-12
63. Fu MX, Simmonds P, Andreani J, Baklan H, Webster M, Asadi R, et al. Ultrasensitive PCR system for HBV DNA detection: Risk stratification for occult hepatitis B virus infection in English blood donors. J Med Virol. (2023) 95:e29144. doi: 10.1002/jmv.29144
64. Ohhashi Y, Pai A, Halait H, and Ziermann R. Analytical and clinical performance evaluation of the cobas TaqScreen MPX Test for use on the cobas s 201 system. J Virol Methods. (2010) 165:246–53. doi: 10.1016/j.jviromet.2010.02.004
65. Zbinden A, Ries J, Redli PM, Shah C, Glauser A, Goslings D, et al. Prevalence of occult hepatitis B virus infection in blood donors with negative ID-NAT in Switzerland. Transfus Med Hemother. (2022) 49:338–45. doi: 10.1159/000525480
66. Cappy P, Boizeau L, Candotti D, Le Cam S, Martinaud C, Pillonel J, et al. Insights on 21 years of HBV surveillance in blood donors in France. Viruses. (2022) 14:2507. doi: 10.3390/v14112507
67. Harvala H, Reynolds C, Gibney Z, Derrick J, Ijaz S, Davison KL, et al. Hepatitis B infections among blood donors in England between 2009 and 2018: Is an occult hepatitis B infection a risk for blood safety? Transfusion. (2021) 61:2402–13. doi: 10.1111/trf.16543
68. Cooke GS, Flower B, Cunningham E, Marshall AD, Lazarus JV, Palayew A, et al. Progress towards elimination of viral hepatitis: a Lancet Gastroenterology & Hepatology Commission update. Lancet Gastroenterol Hepatol. (2024) 9:346–65. doi: 10.1016/S2468-1253(23)00321-7
69. Saravanan S, Shankar EM, Vignesh R, Ganesh PS, Sankar S, Velu V, et al. Occult hepatitis B virus infection and current perspectives on global WHO 2030 eradication. J Viral Hepat. (2024) 31:423–31. doi: 10.1111/jvh.13928
70. Simpore A, Bazie B, Yooda PA, Zoure AA, Sawadogo S, Sawadogo AG, et al. Seroprevalence of viral hepatitis B and occult hepatitis B among blood donors in africa: A systematic review and meta-analysis. Rev Med Virol. (2024) 34:e70006. doi: 10.1002/rmv.70006
71. Takuissu GR, Kenmoe S, Amougou Atsama M, Atenguena Okobalemba E, Mbaga DS, Ebogo-Belobo JT, et al. Global epidemiology of occult hepatitis B virus infections in blood donors, a systematic review and meta-analysis. PloS One. (2022) 17:e0272920. doi: 10.1371/journal.pone.0272920
72. Ghazanfar S, Hassan S, Shahid Z, Khan MS, Malik AR, Bhutta HS, et al. Frequency of transfusion transmissible infections among blood donors of Rawalpindi District, Pakistan. Afr Health Sci. (2022) 22:590–8. doi: 10.4314/ahs.v22i3.63
73. Ahmed S, Ayub M, Naeem M, Nazir FH, Hussain A, Ghilzai D, et al. Thalassemia patients from baluchistan in Pakistan are infected with multiple hepatitis B or C virus strains. Am J Trop Med Hyg. (2021) 104:1569–76. doi: 10.4269/ajtmh.20-0740
74. Bahrami A, Pourfathollah AA, Parsania M, Mehrabi Habibabadi H, and Sharifi Z. Prevalence of occult hepatitis B virus infection among the blood donors in Golestan province: cross-sectional study. Iran J Microbiol. (2022) 14:410–6. doi: 10.18502/ijm.v14i3.9793
75. Deng X, Zang L, Wang X, Chen H, Liu J, Gao Y, et al. Follow-up program for blood donors with unconfirmed screening results reveals a high false-positive rate in Dalian, China. Transfusion. (2020) 60:334–42. doi: 10.1111/trf.15656
76. Eltom K, Albeely A, El Hussein ARM, Elkhidir IM, and Enan K. Occult hepatitis B virus infection in Sudan: A systematic review and meta-analysis. JGH Open. (2020) 4:800–7. doi: 10.1002/jgh3.12411
77. Boumbaly S, Balde TAL, Semenov AV, Ostankova YV, Serikova EN, Naidenova EV, et al. Prevalence of viral hepatitis B markers among blood donors in the Republic of Guinea. Vopr Virusol. (2022) 67:59–68. doi: 10.36233/0507-4088-92
78. Azzam A, Khaled H, El-Kayal ES, Gad FA, and Omar S. Prevalence of occult hepatitis B virus infection in Egypt: a systematic review with meta-analysis. J Egypt Public Health Assoc. (2023) 98:13. doi: 10.1186/s42506-023-00138-4
79. Tung TT, Schmid J, Nghia VX, Cao LC, Linh LTK, Rungsung I, et al. Low risk of occult hepatitis B infection among Vietnamese blood donors. Pathogens (Basel, Switzerland). (2022) 11:1524. doi: 10.3390/pathogens11121524
80. Gonzalez R, Barea L, Arruga A, Richart A, and Soriano V. Overt and occult hepatitis B among immigrants and native blood donors in Madrid, Spain. Ther Adv Infect Dis. (2020) 7:2049936120982122. doi: 10.1177/2049936120982122
81. Seed CR, Allain JP, Lozano M, Laperche S, Gallian P, Gross S, et al. International forum on Occult hepatitis B infection and transfusion safety. Vox Sang. (2019) 114:397–406. doi: 10.1111/vox.12744
82. Candotti D and Allain JP. Transfusion-transmitted hepatitis B virus infection. J Hepatol. (2009) 51:798–809. doi: 10.1016/j.jhep.2009.05.020
83. Spreafico M, Berzuini A, Foglieni B, Candotti D, Raffaele L, Guarnori I, et al. Poor efficacy of nucleic acid testing in identifying occult HBV infection and consequences for safety of blood supply in Italy. J Hepatol. (2015) 63:1068–76. doi: 10.1016/j.jhep.2015.06.016
84. Allain JP, Mihaljevic I, Gonzalez-Fraile MI, Gubbe K, Holm-Harritshoj L, Garcia JM, et al. Infectivity of blood products from donors with occult hepatitis B virus infection. Transfusion. (2013) 53:1405–15. doi: 10.1111/trf.12096
85. Wang C, Xue R, Wang X, Xiao L, and Xian J. High-sensitivity HBV DNA test for the diagnosis of occult HBV infection: commonly used but not reliable. Front Cell Infect Microbiol. (2023) 13:1186877. doi: 10.3389/fcimb.2023.1186877
86. Liu CJ, Lo SC, Kao JH, Tseng PT, Lai MY, Ni YH, et al. Transmission of occult hepatitis B virus by transfusion to adult and pediatric recipients in Taiwan. J Hepatol. (2006) 44:39–46. doi: 10.1016/j.jhep.2005.06.016
87. Yang Y, Xiao J, Zhang X, Yang H, Zhang Z, Xu H, et al. Protective effect of neonatal hepatitis B vaccine against HBV breakthrough infection in children with leukemia: A real-world study. J Clin Transl Hepatol. (2022) 10:860–6. doi: 10.14218/JCTH.2021.00291
88. Satake M, Taira R, Yugi H, Hino S, Kanemitsu K, Ikeda H, et al. Infectivity of blood components with low hepatitis B virus DNA levels identified in a lookback program. Transfusion. (2007) 47:1197–205. doi: 10.1111/j.1537-2995.2007.01276.x
89. Yuen MF, Wong DK, Lee CK, Tanaka Y, Allain JP, Fung J, et al. Transmissibility of hepatitis B virus (HBV) infection through blood transfusion from blood donors with occult HBV infection. Clin Infect Dis. (2011) 52:624–32. doi: 10.1093/cid/ciq247
90. Su TH, Chen PJ, Chen TC, Cheng HR, Li L, Lin KS, et al. The clinical significance of occult hepatitis B transfusion in Taiwan–a look-back study. Transfus Med. (2011) 21:33–41. doi: 10.1111/j.1365-3148.2010.01036.x
91. Seed CR, Maloney R, Kiely P, Bell B, Keller AJ, Pink J, et al. Infectivity of blood components from donors with occult hepatitis B infection - results from an Australian lookback programme. Vox Sang. (2015) 108:113–22. doi: 10.1111/vox.12198
92. Lieshout-Krikke RW, van Kraaij MG, Danovic F, and Zaaijer HL. Rare transmission of hepatitis B virus by Dutch donors with occult infection. Transfusion. (2016) 56:691–8. doi: 10.1111/trf.13401
93. Liang X, Bi S, Yang W, Wang L, Cui G, Cui F, et al. Reprint of: Epidemiological serosurvey of Hepatitis B in China–declining HBV prevalence due to Hepatitis B vaccination. Vaccine. (2013) 31 Suppl 9:J21–28. doi: 10.1016/j.vaccine.2013.08.012
94. Tang X, Allain JP, Wang H, Rong X, Chen J, Huang K, et al. Incidence of hepatitis B virus infection in young Chinese blood donors born after mandatory implementation of neonatal hepatitis B vaccination nationwide. J Viral Hepat. (2018) 25:1008–16. doi: 10.1111/jvh.12901
95. Zheng X, Ye X, Du P, Zeng J, Zhu W, Yang B, et al. High prevalence of anti-hepatitis B core antigen in hepatitis B virus-vaccinated Chinese blood donors suggests insufficient protection but little threat to the blood supply. Transfusion. (2015) 55:890–7. doi: 10.1111/trf.12902
96. Poovorawan Y, Chongsrisawat V, Theamboonlers A, Leroux-Roels G, Kuriyakose S, Leyssen M, et al. Evidence of protection against clinical and chronic hepatitis B infection 20 years after infant vaccination in a high endemicity region. J Viral Hepat. (2011) 18:369–75. doi: 10.1111/j.1365-2893.2010.01312.x
97. Peng J, Yao X, Yuan C, Liu X, Xia R, He J, et al. The investigation of hepatitis B vaccine immune responses in occult hepatitis B virus-infected patients. Front Immunol. (2022) 13:903685. doi: 10.3389/fimmu.2022.903685
98. Hu LP, Liu DP, Chen QY, Harrison TJ, He X, Wang XY, et al. Occult HBV infection may be transmitted through close contact and manifest as an overt infection. PloS One. (2015) 10:e0138552. doi: 10.1371/journal.pone.0138552
99. Zhang Z, Zhang L, Dai Y, Jin L, Sun B, Su Q, et al. Occult hepatitis B virus infection among people with a family history of chronic hepatitis B virus infection. J Med Virol. (2015) 87:1890–8. doi: 10.1002/jmv.24245
100. Athalye S, Khargekar N, Shinde S, Parmar T, Chavan S, Swamidurai G, et al. Exploring risk factors and transmission dynamics of Hepatitis B infection among Indian families: Implications and perspective. J Infect Public Health. (2023) 16:1109–14. doi: 10.1016/j.jiph.2023.05.003
101. Delghandi S, Raoufinia R, Shahtahmasbi S, Meshkat Z, Gouklani H, and Gholoobi A. An overview of occult hepatitis B infection (OBI) with emphasis on HBV vaccination. Heliyon. (2024) 10:e37097. doi: 10.1016/j.heliyon.2024.e37097
102. Satake M, Sugiyama M, Mizokami M, and Tanaka J. Incidences of new hepatitis B infection and anti-hepatitis B core-negative occult hepatitis B infection among Japanese blood donors in relation to anti-hepatitis B surface antigen levels. J Med Virol. (2024) 96:e29823. doi: 10.1002/jmv.29823
103. Ye X, Li T, Yu B, Zeng J, Shi Y, Xie H, et al. The high prevalence of occult hepatitis B infections among the partners of chronically infected HBV blood donors emphasizes the potential residual risk to blood safety. J Med Virol. (2023) 95:e29006. doi: 10.1002/jmv.29006
104. Lim SH, Lee S, Lee YB, Lee CH, Lee JW, Lee SH, et al. Increased prevalence of transfusion-transmitted diseases among people with tattoos: A systematic review and meta-analysis. PloS One. (2022) 17:e0262990. doi: 10.1371/journal.pone.0262990
105. Bartonjo G, Oundo J, and Ng’ang’a Z. Prevalence and associated risk factors of transfusion transmissible infections among blood donors at Regional Blood Transfusion Center Nakuru and Tenwek Mission Hospital, Kenya. Pan Afr Med J. (2019) 34:31. doi: 10.11604/pamj.2019.34.31.17885
106. Yonemura S, Doane S, Keil S, Goodrich R, Pidcoke H, and Cardoso M. Improving the safety of whole blood-derived transfusion products with a riboflavin-based pathogen reduction technology. Blood Transfus. (2017) 15:357–64. doi: 10.2450/2017.0320-16
107. Ye X, Zhao Y, Li R, Li T, Zheng X, Xiong W, et al. High frequency occult hepatitis B virus infection detected in non-resolved donations suggests the requirement of anti-HBc test in blood donors in southern China. Front Immunol. (2021) 12:699217. doi: 10.3389/fimmu.2021.699217
108. Yu RT, Punzalan KAP, Bhatnagar S, Lutero RB, Chamen IMS, Masangkay CB, et al. Seroreactivity of antibodies to hepatitis B core antigen among hepatitis B surface antigen-screened negative blood donors and its implications for blood safety in a resource-constrained country. Vox Sang. (2024) 119:252–6. doi: 10.1111/vox.13576
109. Fu MX, Faddy HM, Candotti D, Groves J, Saa P, Styles C, et al. International review of blood donation screening for anti-HBc and occult hepatitis B virus infection. Transfusion. (2024) 64:2144–56. doi: 10.1111/trf.18018
110. Tanaka A, Yamagishi N, Hasegawa T, Miyakawa K, Goto N, Matsubayashi K, et al. Marked reduction in the incidence of transfusion-transmitted hepatitis B virus infection after the introduction of antibody to hepatitis B core antigen and individual donation nucleic acid amplification screening in Japan. Transfusion. (2023) 63:2083–97. doi: 10.1111/trf.17546
111. Gou H, Pan Y, Ge H, Zheng Y, Wu Y, Zeng J, et al. Evaluation of an individual-donation nucleic acid amplification testing algorithm for detecting hepatitis B virus infection in Chinese blood donors. Transfusion. (2015) 55:2272–81. doi: 10.1111/trf.13135
112. Fu MX, Faddy HM, Candotti D, Groves J, Saa P, Styles C, et al. Correction to “International review of blood donation screening for anti-HBc and occult hepatitis B virus infection. Transfusion. (2025) 65:648. doi: 10.1111/trf.18094
113. Candotti D, Lin CK, Belkhiri D, Sakuldamrongpanich T, Biswas S, Lin S, et al. Occult hepatitis B infection in blood donors from South East Asia: molecular characterisation and potential mechanisms of occurrence. Gut. (2012) 61:1744–53. doi: 10.1136/gutjnl-2011-301281
114. Satake M, Yamagishi N, Tanaka A, Goto N, Sakamoto T, Yanagino Y, et al. Transfusion-transmitted HBV infection with isolated anti-HBs-positive blood. Transfusion. (2023) 63:1250–4. doi: 10.1111/trf.17390
115. Hoshi Y, Hasegawa T, Yamagishi N, Mizokami M, Sugiyama M, Matsubayashi K, et al. Optimal titer of anti-HBs in blood components derived from donors with anti-HBc. Transfusion. (2019) 59:2602–11. doi: 10.1111/trf.15393
116. Vachon A and Osiowy C. Novel biomarkers of hepatitis B virus and their use in chronic hepatitis B patient management. Viruses. (2021) 13:951. doi: 10.3390/v13060951
117. Wang J, Shen T, Huang X, Kumar GR, Chen X, Zeng Z, et al. Serum hepatitis B virus RNA is encapsidated pregenome RNA that may be associated with persistence of viral infection and rebound. J Hepatol. (2016) 65:700–10. doi: 10.1016/j.jhep.2016.05.029
118. Inoue T and Tanaka Y. Current global applications of HBcrAg assays in the management of chronic hepatitis B. Glob Health Med. (2025) 7:67–71. doi: 10.35772/ghm.2024.01097
119. Deguchi M, Kagita M, Yoshioka N, Tsukamoto H, Takao M, Tahara K, et al. Evaluation of the highly sensitive chemiluminescent enzyme immunoassay “Lumipulse HBsAg-HQ” for hepatitis B virus screening. J Clin Lab Anal. (2018) 32:e22334. doi: 10.1002/jcla.22334
120. Hassan YM, Mohamed AS, Hassan YM, and El-Sayed WM. Recent developments and future directions in point-of-care next-generation CRISPR-based rapid diagnosis. Clin Exp Med. (2025) 25:33. doi: 10.1007/s10238-024-01540-8
121. Maxmen A. Faster, better, cheaper: the rise of CRISPR in disease detection. Nature. (2019) 566:437. doi: 10.1038/d41586-019-00601-3
122. Shahni SN, Albogami S, Azmi I, Pattnaik B, Chaudhuri R, Dev K, et al. Dual detection of hepatitis B and C viruses using CRISPR-cas systems and lateral flow assay. J Nucleic Acids. (2024) 2024:8819834. doi: 10.1155/2024/8819834
123. Tian Y, Fan Z, Xu L, Cao Y, Chen S, Pan Z, et al. CRISPR/Cas13a-assisted rapid and portable HBV DNA detection for low-level viremia patients. Emerg Microbes Infect. (2023) 12:e2177088. doi: 10.1080/22221751.2023.2177088
124. Zhang X, Tian Y, Xu L, Fan Z, Cao Y, Ma Y, et al. CRISPR/Cas13-assisted hepatitis B virus covalently closed circular DNA detection. Hepatol Int. (2022) 16:306–15. doi: 10.1007/s12072-022-10311-0
125. Wang X, Xu L, Chen Y, Liu A, Wang L, Xu P, et al. Integrating nested PCR with high-throughput sequencing to characterize mutations of HBV genome in low viral load samples. Med (Baltimore). (2017) 96:e7588. doi: 10.1097/MD.0000000000007588
126. Garcia-Garcia S, Cortese MF, Rodriguez-Algarra F, Tabernero D, Rando-Segura A, Quer J, et al. Next-generation sequencing for the diagnosis of hepatitis B: current status and future prospects. Expert Rev Mol Diagn. (2021) 21:381–96. doi: 10.1080/14737159.2021.1913055
127. Liu H-N, Lin SY, Ramirez R, Chen S-E, Heimer Z, Kubas R, et al. Integrated DNA estimation in tissue biopsy and detection in liquid biopsy by HBV-targeted NGS assay. medRxiv. (2024) 5:2024. doi: 10.1101/2024.12.04.24318256
128. Charlewood R and Flanagan P. Ultrio and Ultrio Plus non-discriminating reactives: false reactives or not? Vox Sang. (2013) 104:7–11. doi: 10.1111/j.1423-0410.2012.01624.x
129. Athalye S, Patil A, Khargekar N, Shinde S, Chavan S, Dixit A, et al. Efficacy of combined HBsAg, anti-HBc and anti-HBs screening in minimizing transfusion transmission risk of hepatitis B infection in low resource setting. Heliyon. (2024) 10:e25805. doi: 10.1016/j.heliyon.2024.e25805
130. Chen J, Ma Z, Wu D, Zuo Q, Wang F, Xiao C, et al. Evaluating the cost-effectiveness of low-level HBV DNA screening in occult hepatitis B infection donors: A study from Shandong Blood Center, China. Heliyon. (2023) 9:e18609. doi: 10.1016/j.heliyon.2023.e18609
131. Lou S, Taylor R, Pearce S, Kuhns M, and Leary T. An ultra-sensitive Abbott ARCHITECT((R)) assay for the detection of hepatitis B virus surface antigen (HBsAg). J Clin Virol. (2018) 105:18–25. doi: 10.1016/j.jcv.2018.05.009
132. Lee J, Lee SY, Cho YG, Kim DS, and Park J. Accuracy validation of the elecsys HBsAg II quant assay and its utility in resolving equivocal qualitative HBsAg results. Med (Kaunas, Lithuania). (2023) 59:443. doi: 10.3390/medicina59030443
133. Park J, Bae T, Cho Y, Kim D, and Lee J. Analytical performance of the sysmex HISCL HBsAg assay and comparison with the roche elecsys HBsAg II quant assay in the quantification of hepatitis B surface antigen. Med (Kaunas, Lithuania). (2021) 57:1307. doi: 10.3390/medicina57121307
134. Allain JP and Candotti D. Diagnostic algorithm for HBV safe transfusion. Blood Transfus. (2009) 7:174–82. doi: 10.2450/2008.0062-08
135. Ghany MG, King WC, Lisker-Melman M, Lok ASF, Terrault N, Janssen HLA, et al. Comparison of HBV RNA and hepatitis B core related antigen with conventional HBV markers among untreated adults with chronic hepatitis B in north america. Hepatology. (2021) 74:2395–409. doi: 10.1002/hep.32018
136. Brojer E, Grabarczyk P, Liszewski G, Mikulska M, Allain JP, Letowska M, et al. Characterization of HBV DNA+/HBsAg- blood donors in Poland identified by triplex NAT. Hepatology. (2006) 44:1666–74. doi: 10.1002/hep.21413
137. Izumida K, Kaneko A, Takahashi K, Kusumoto S, Narita T, Takami A, et al. Clinical evaluation of a novel and highly sensitive immunoassay for anti-hepatitis B core antigen using a fully automated immunochemical analyzer. Hepatol Res. (2018) 48:1081–91. doi: 10.1111/hepr.13229
138. Wang WX, Jia R, Gao YY, Liu JY, Luan JQ, Qiao F, et al. Quantitative anti-HBc combined with quantitative HBsAg can predict HBsAg clearance in sequential combination therapy with PEG-IFN-alpha in NA-suppressed chronic hepatitis B patients. Front Immunol. (2022) 13:894410. doi: 10.3389/fimmu.2022.894410
139. Kuhns MC, McNamara AL, Holzmayer V, and Cloherty GA. Molecular and serological characterization of hepatitis B vaccine breakthrough infections in serial samples from two plasma donors. Virol J. (2019) 16:43. doi: 10.1186/s12985-019-1154-4
140. Butler EK, Gersch J, McNamara A, Luk KC, Holzmayer V, de Medina M, et al. Hepatitis B virus serum DNA andRNA levels in nucleos(t)ide analog-treated or untreated patients during chronic and acute infection. Hepatology. (2018) 68:2106–17. doi: 10.1002/hep.30082
141. Le Guillou DB, Duclos-Vallee JC, Eberle F, Capel F, and Petit MA. Evaluation of an enzyme-linked immunosorbent assay for detection and quantification of hepatitis B virus PreS1 envelope antigen in serum samples: comparison with two commercial assays for monitoring hepatitis B virus DNA. J Viral Hepat. (2000) 7:387–92. doi: 10.1046/j.1365-2893.2000.00248.x
142. Kim H, Hur M, Bae E, Lee KA, and Lee WI. Performance evaluation of cobas HBV real-time PCR assay on Roche cobas 4800 System in comparison with COBAS AmpliPrep/COBAS TaqMan HBV Test. Clin Chem Lab Med. (2018) 56:1133–9. doi: 10.1515/cclm-2017-1133
143. Tan NK, Carrington D, and Pope CF. Verification of the Roche cobas((R)) 6800 PCR 200 microl and 500 microl protocols for the quantification of HIV-1 RNA, HBV DNA and HCV RNA and evaluation with COBAS((R)) Ampliprep/COBAS((R)) TaqMan((R)) assays. J Med Microbiol. (2018) 67:1711–7. doi: 10.1099/jmm.0.000838
144. Wirden M, Larrouy L, Mahjoub N, Todesco E, Damond F, Delagreverie H, et al. Multicenter comparison of the new Cobas 6800 system with Cobas Ampliprep/Cobas TaqMan and Abbott RealTime for the quantification of HIV, HBV and HCV viral load. J Clin Virol. (2017) 96:49–53. doi: 10.1016/j.jcv.2017.09.007
Keywords: occult hepatitis B virus infection (OBI), blood transfusion transmission, blood testing, blood safety, HBV prevention and control
Citation: Huang L, He B, Mo Q, Li B, Yan X, Lai R, Wang X, Li J, Lai M, Xie H, Sun J, Ye X and Chen L (2025) Transfusion-transmitted occult hepatitis B virus infection: current understanding, challenges, and its implication in blood safety. Front. Immunol. 16:1663306. doi: 10.3389/fimmu.2025.1663306
Received: 10 July 2025; Accepted: 07 October 2025;
Published: 28 October 2025.
Edited by:
Sonia Roman, University of Guadalajara, MexicoReviewed by:
Theodoros Androutsakos, National and Kapodistrian University of Athens, GreeceRosalia Lira, Instituto Mexicano del Seguro Social, Mexico
Copyright © 2025 Huang, He, Mo, Li, Yan, Lai, Wang, Li, Lai, Xie, Sun, Ye and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Limin Chen, bGltaW5fY2hlbl85OUAxMjYuY29t; bGltaW4uY2hlbkBpYnQucHVtYy5lZHUuY24=; Xianlin Ye, eWV4aWFubGluOTBAaG90bWFpbC5jb20=
 Linbin Huang
Linbin Huang Baoren He1
Baoren He1 Bin Li
Bin Li Xipeng Yan
Xipeng Yan Xinwei Wang
Xinwei Wang Limin Chen
Limin Chen