- 1CureVac SE, Tübingen, Germany
- 2GSK, Siena, Italy
Background: Uropathogenic Escherichia coli (UPEC) is the leading cause of urinary tract infections (UTIs), which are increasingly antibiotic resistant and frequently recur. Novel therapeutics are sought to treat and prevent recurrent UTIs (rUTIs), including vaccines. Key virulence factor FimH, which mediates bacterial adhesion to host cells and biofilm formation, is a promising target for a vaccine against UPEC. We assessed the immunogenicity of mRNA-based nanoparticle vaccines against UPEC containing FimH as the encoded antigen.
Methods: Lipid nanoparticle (LNP)-formulated mRNA vaccines encoding FimH as a monomeric, pre-binding conformation protein (FimHDG), or a multimeric protein nanoparticle (PNP) through fusion to Helicobacter pylori ferritin (FimHDG-Ferritin) were developed. Immunogenicity was assessed in vivo in female BALB/cAnNRj mice and female Wistar rats following three intramuscular (IM) injections of FimHDG or FimHDG–Ferritin mRNA vaccines, or comparator protein subunit vaccines. Antibody levels and functional response were measured in serum and urine by ELISA and bacterial adhesion inhibition (BAI) assays. T cell response was characterized by flow cytometry.
Results: In both animal models, unmodified FimHDG and FimHDG-Ferritin mRNA vaccines induced higher functional serum antibody levels compared with the protein subunit vaccine control, at the tested dosages. FimHDG-Ferritin resulted in greater binding antibody levels and higher splenic FimH-specific CD4+ and CD8+ T cell responses compared with monomeric FimHDG in both models, resulting in its nomination as lead candidate vaccine design. Validation in rats demonstrated that N1mΨ nucleoside modification further enhanced FimHDG-Ferritin immunogenicity compared with unmodified mRNA.
Conclusions: The mRNA vaccine FimHDG-Ferritin with N1mΨ-modified nucleosides is a promising candidate for further development as a vaccine against UPEC.
1 Introduction
Urinary tract infections (UTIs) are among the most common bacterial infections globally, with over 404 million cases reported in 2019 (1). In the US alone, the annual direct healthcare costs associated with UTIs are estimated to be around $2 billion (2). UTIs are significantly more common in women than in men; the lifetime incidence is 50−60% in adult women compared with just 12% in men (3, 4). Current medical approaches for the management of UTIs include prophylactic measures, over-the-counter medications, or treatment with antibiotics (5). Despite these measures, around one quarter of women will experience another UTI within six months of treatment for the initial infection (6). The occurrence of two or more UTIs within six months, or three or more UTIs within a year, is defined as recurrent UTI (rUTI) (5).
The most common causative agent of UTIs is uropathogenic Escherichia coli (UPEC), accounting for approximately 75% of cases (7). In general, the bacteria are transmitted to the urinary tract through fecal shedding and ascend the urinary tract to invade and colonize the bladder (cystitis) and kidneys (pyelonephritis) (8). The formation of biofilm-like intracellular bacterial communities in the host tissue contributes to the recurrence of UTIs within a short period of time (9, 10). Moreover, the prevalence of antimicrobial resistance is a major obstacle to successful treatment of UTIs caused by UPEC (11). Indeed, resistance to common antibiotics has been found in 29.7–46.2% of UPEC-derived rUTIs (12). In response, whole cell vaccines against UPEC have been developed, including Uro-Vaxom/OM-89, Solco-Urovac, and Uromune/MV140 (13–15). However, a recent systematic review found limited evidence of their long-term efficacy for reducing rUTIs in adult female patients (16) and while they are available in some countries, no vaccine to prevent rUTI infection has received FDA approval to date (17). Therefore, there is a significant unmet need for an efficacious vaccine targeting UPEC.
In this work, we investigated the immunogenicity of lipid nanoparticle (LNP)-formulated messenger ribonucleic acid (mRNA)-based candidate vaccines against UPEC. Developed using CureVac’s proprietary RNActive® technology, these vaccine candidates encode the pilin FimH, a highly conserved key virulence factor of UPEC that is displayed at the tip of type 1 pili and mediates both biofilm formation and bacterial adhesion to host tissue (8, 18–20). The N-terminal lectin domain of FimH (FimHL) binds to mannosylated proteins (e.g. uroplakins) expressed on the surface of uroepithelial cells (8, 21). The C-terminal pilin domain of FimH possesses a hydrophobic groove that can impact protein conformation by interaction with different proteins, including the bacterial proteins FimG and FimC that hold the protein in a pre-and post-binding conformation, respectively (8, 21–23). In the pre-binding conformation, FimH transitions between low- and high-affinity binding modes through an equilibrium between tense and relaxed conformation states (8). By fusing a donor strand peptide of FimG to FimH (FimHDG), vaccine candidates investigated here enable the expression of FimH stabilized in an immunologically preferred pre-binding conformation (24). Vaccines were either designed to encode a secreted monomeric protein (FimHDG) or a secreted protein that self-assembles into a multimeric protein nanoparticle (PNP) by encoding Helicobacter pylori (H. pylori) ferritin fused to FimHDG (FimHDG-Ferritin).
Ferritin PNPs have shown promise as delivery systems to increase the efficacy and safety of various therapeutic applications, such as anti-cancer drugs and novel prophylactic vaccines (25–29). Ferritin PNPs can present antigens on their surface, as seen with SARS-CoV-2 Spike and influenza stem hemagglutinin antigens (26, 28, 30–32). Protein-based approaches have shown broad immunogenicity and longer blood half-life of ferritin PNPs compared to antigens alone (30, 31, 33). H. pylori ferritin has also been employed as a fusion domain in mRNA vaccine development, improving antigenicity and immunogenicity of vaccines in animal models of SARS-CoV-2 (34) and HIV-1 (35). With good potency, safety and efficacy, mRNA vaccines are promising alternatives to traditional vaccines (36), highlighting the potential synergy that can be achieved in a vaccine platform harnessing both mRNA and ferritin PNP technology.
Existing strategies in UPEC vaccine development have largely involved protein subunit vaccines (37). These include the FimCH vaccine, where a complex of the chaperone FimC and FimH is formulated with a synthetic adjuvant, Phosphorylated HexaAcyl Disaccharide (PHAD™). In a Phase 1 study, the FimCH vaccine was able to reduce the frequency of UTIs in a subset of women with rUTI following administration of four doses (38). However, to our knowledge, no mRNA-based vaccines encoding antigens clustering on H. pylori ferritin PNPs have been clinically tested to date. Here, the mRNA vaccine candidates FimHDG and FimHDG-Ferritin (including unmodified or N1-methyl-pseudouridine (N1mΨ)-modified nucleosides) were evaluated in vivo in mice and rats for induction of FimH-specific humoral and/or cellular immune responses, using PHAD-adjuvanted FimHC and AS01-adjuvanted FimHDG protein subunit vaccines as controls (39).
2 Materials and methods
2.1 mRNA vaccines
mRNA vaccines were developed with CureVac’s RNActive® platform and contained either chemically unmodified nucleosides or N1mΨ-modified nucleosides. Vaccines comprised of a 5′ cap1 structure, a 5′ untranslated region (UTR) from the human hydroxysteroid 17-beta dehydrogenase 4 gene (HSD17B4), a GC-enriched open reading frame (ORF), a 3′ UTR from the human proteasome 20 S subunit beta 3 gene (PSMB3), a histone stem-loop, and a poly(A) tail. LNP encapsulation of mRNA was performed using LNP technology from Acuitas Therapeutics (Canada). LNPs were composed of an ionizable amino lipid, phospholipid, cholesterol, and a PEGylated lipid. The FimHDG mRNA vaccine encoded an N-terminal signal peptide from the mouse Igκ light chain (amino acid [aa] 1–20; NCBI reference sequence: AAH80787.1) followed by FimH from E. coli strain J96 (aa 22–300; NCBI reference sequence: ELL41155.1), a 5-aa linker (PGDGN), and the donor strand peptide of FimG from E. coli strain J96 (denoted as DG; aa 24–37; NCBI reference sequence: ELL41154.1). The FimHDG-Ferritin mRNA vaccines encoded FimHDG with a different signal peptide (human IgE; aa 1–18; NCBI reference sequence: AAB59424.1) N-terminally linked via serine-glycine-glycine (SGG) to the ferritin domain of H. pylori strain J99 (aa 5-167; NCBI reference sequence: AAD06160.1) containing an N19Q mutation to remove a potential N-linked glycosylation site (33). The immunogenicity of FimH mRNA vaccines containing either Igκ- or IgE-derived signal peptides was tested in mice, and no significant differences were found based on the choice of signal peptide (39).
2.2 Protein vaccines
The PHAD-adjuvanted FimHC (also designated as FimCH) protein subunit vaccine consisted of a complex of FimH and FimC and was obtained as previously described (40). The AS01-adjuvanted FimHDG (also designated as FimHdG) protein subunit vaccine employed an aa sequence that corresponds to the encoded sequence of the FimHDG mRNA vaccine (without a signal peptide) and was obtained as previously described (39).
2.3 Animal ethics statement
Animal research in this study is reported in accordance with ARRIVE reporting guidelines (41). Female BALB/cAnNRj mice (Janvier Labs, France) and female Wistar rats (Charles River Laboratories, Germany) were provided and handled by Preclinics Gesellschaft für präklinische Forschung mbH (Germany). All in-life experimental procedures undertaken during the course of the mouse and rat immunization studies were conducted in accordance with German laws and guidelines for animal protection, appropriate local and national approvals, and with Directive 2010/63/EU of the European Parliament and of the Council of 22 September 2010 on the protection of animals used for scientific purposes. Ethical approval was obtained from Land Brandenburg, Landesamt für Arbeitsschutz, Vebraucherschutz und Gesundheit (Germany) for the mouse (reference number: 2347-14-2018) and rat (reference number: 2347-5-2021) studies. Animals were acclimated for at least one week before any procedures were carried out and were 6–7 weeks old at the start of the study. Mice were kept in Macrolon type II cages (4 mice per cage) and rats were kept in Macrolon type IV cages (3–4 rats per cage). Both were housed at a temperature of 19–23°C, humidity of 35–50%, 15 air changes per hour and a 12/12-hour light/dark cycle. Diets consisted of Ssniff R/M-H extruded (V1536); both food and water were provided ad libitum.
2.4 Vaccination studies
A total of 158 animals were used in these studies (32 mice [n=8/group]; 70 rats in the first study [n=7/group]; 56 rats in the second study [n=8/group]). The sample size was selected to be as low as possible in each study while still providing sufficient power to determine between-group differences, based on the authors’ experience and knowledge from previous mouse and rat studies. Animals were assigned to groups at random on arrival and the studies were not blinded.
mRNA vaccine dosages used were decided based on accumulated learnings from previous studies performed by CureVac or others (38–40), and the protein subunit vaccine dosage was used at a similar level to the mRNA dosages. Of note, equal doses of protein and mRNA vaccine injected are not expected to result in equivalent amounts of antigen presented in vivo, since mRNA-based expression is highly dependent on the cell type-specific quality of mRNA vaccine uptake, antigen expression and/or secretion (42, 43).
On Days 0, 21, and 35, female BALB/cAnNRj mice (n=8/group) received an intramuscular (IM) injection (50 µL; 25 µL each into the left and right M. tibialis) of either: 2 µg FimHC protein complex subunit vaccine adjuvanted with 4 µg PHAD; 2 µg of FimHDG mRNA vaccine (unmodified nucleosides); or 2 µg of FimHDG-Ferritin mRNA vaccine (unmodified nucleosides) using a single-use insulin syringe with an integrated 30G needle (BD, Cat. 324825). Mice receiving physiological saline (25 µL of 0.9% NaCl into left or right M. tibialis) served as negative controls. The animals in each group were divided into two cohorts (n=4/cohort), and the experiment was started on two consecutive days. Blood samples were collected into Z-clot activator microtubes (Sarstedt, Cat. 20.1291) by retro-orbital bleeding on Days 21, 35, and 49 under inhalation anesthesia using isoflurane (5 vol%). The samples were incubated for 0.5–1 hour (h) at room temperature (RT), and sera were obtained by collecting the supernatant after centrifugation (5 minutes [min]; 10,000 × g; RT). Spontaneous urine was collected on Days 21 and 35 by placing the animals in a cage with hydrophobic Medicat LabSand™ bedding (WDT, Cat. 27906) and collecting urine directly with a pipette. On Day 49, urine was collected by puncture of the bladder to ensure the collection of a larger sample volume. Urine was centrifuged (5 min; 10,000 × g; RT) and the supernatant used for analysis. At the end of the study on Day 49, mice were euthanized by exsanguination under inhalation anesthesia using isoflurane (5 vol%).
Splenocytes were isolated by mechanical disruption of the spleen using 40 µm cell strainers (Pluriselect, Cat. 43-50040-51). Red blood cells were lysed and splenocytes were washed, resuspended in supplemented medium and finally stored as single-cell suspensions in liquid nitrogen until use.
For the first rat study, female Wistar rats (n=7/group) were injected IM three times (100 µL each into the left or right M. gastrocnemius) on Days 0, 21, and 35 with either: 0.7 µg, 2.8 µg or 8.5 µg FimHDG protein subunit vaccine adjuvanted with 2.5 µg AS01 each; 1 µg, 4 µg or 12 µg FimHDG mRNA vaccine; or 1 µg, 4 µg or 12 µg FimHDG-Ferritin mRNA vaccine containing unmodified nucleosides. In the second rat study, female Wistar rats (n=8/group) were injected IM three times (100 µL each into the left or right M. gastrocnemius) on Days 0, 21, and 35 with either: 1 µg or 12 µg FimHDG protein subunit vaccine adjuvanted with 2.5 µg AS01 each, or 1 µg or 12 µg of FimHDG-Ferritin mRNA vaccines containing either unmodified or N1mΨ-modified nucleosides. Injections were performed using a single-use insulin syringe with an integrated 30G needle (BD, Cat. 324825) and rats receiving physiological saline (100 µL of 0.9% NaCl into left or right M. gastrocnemius) served as negative controls.
Blood samples were collected on Days 21 and 35 by retro-orbital bleeding into Multivette 600 Z-Gel tubes (Sarstedt, Cat. 15.1674) and on Day 49 by heart puncture into Vacuette tubes (8 mL CAT Serum Sep Clot Activator; Greiner Bio One, Cat. 455071) under inhalation anesthesia using isoflurane (5 vol%). The samples were incubated for 0.5–1 h at RT, and sera were then obtained by collecting the supernatant after centrifugation (5 min; 10,000 × g; RT). Spontaneous urine was collected on Days 21 and 35 by placing the animals in an empty cage and urine collected directly with a pipette if the animals urinated. If no urine could be gained, the abdomen was palpated whilst under anesthesia for test substance administration. In case of a palpable bladder, the urine was gained by gentle pressure on the bladder. On Day 49, urine was collected by puncture of the bladder. Urine was centrifuged (5 min; 10,000 × g; RT) and the supernatant used for analysis. At the end of the study on Day 49, rats were euthanized by exsanguination under inhalation anesthesia using isoflurane (5 vol%).
Mice and rats were visually inspected at least once daily for evidence of ill-health. The injection sites were monitored daily after each injection for swelling, reddening or other adverse effects until no deviations from the normal condition could be detected. Body weights were obtained on Days 0, 1, 2, 7, 14, 21, 22, 28, 35, 36, 42 and 49. Body weight loss of more than 20% was established a priori as criteria for exclusion from study; no animals were excluded.
2.5 Quantification of binding antibodies
Quantities of FimHL-specific binding antibodies (total IgG) in sera and urine samples of vaccinated mice or rats were assessed by indirect enzyme-linked immunosorbent assay (ELISA). 96-well MaxiSorp ELISA plates (black; ThermoFisher Scientific, Cat. 437111) were coated with 1 µg/mL of a recombinant FimHL protein overnight at 4 °C. FimHL was obtained as previously described (39). Plates were washed and blocked for 2 h at 37 °C with 5% milk in phosphate-buffered saline (PBS)/0.05% Tween-20 for mouse samples and 1% bovine serum albumin (BSA) in PBS/0.05% Tween-20 for rat samples. Serum samples were added in serial dilution and incubated for 2 h at RT. For mouse urine, the FimHL-specific binding antibody levels were determined in two pools of four animals each due to the small amount of urine; whereas, rat urine samples were tested per animal. For sample preparation, the pH value of each mouse urine pool or rat urine sample was measured. If the pH value was <7, urine samples were neutralized by mixing the samples 1:1 with PBS (pH 8.2). If the pH value was ≥7, urine samples were directly diluted 1:2 in blocking buffer. Dilution in PBS (pH 8.2) or blocking buffer was performed directly as a predilution before urine samples were added in serial dilution and incubated for 2 h at RT. After washing, plates were incubated with either horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (H+L) (1:5000; Jackson-Immuno Research, Cat. 115-035-003) or HRP-conjugated goat anti-rat IgG (1:5000; Sigma Aldrich, Cat. A9037) in blocking buffer for 1–1.5 h at RT. Finally, plates were washed, Amplex™ UltraRed reagent (1:200; Invitrogen, Cat. A36006) with 30% H2O2 (1:2000; Fluka, Cat. 95302) was added, and fluorescence was detected after 45–90 min using a BioTek SynergyHTX plate reader (excitation 530/25, emission detection 590/35, sensitivity 45; Agilent BioTek). The ELISA endpoint titers were defined as the highest reciprocal serum or urine dilution that yielded a signal above the mean background signal plus 5-fold standard deviation (SD).
2.6 T cell analysis
The induction of antigen-specific T cells was determined using intracellular cytokine staining in combination with flow cytometry. Mouse splenocytes were thawed and 2 x 106 cells per well (200 µL) were stimulated for 6–7 h at 37 °C with a custom-made 15-mer overlapping (11 aa) peptide library (1 µg/mL for each peptide) covering the full-length FimH in the presence of anti-CD28 (1:400; BD Biosciences, Cat. 553294) in α-MEM medium (Gibco, Cat. 22561) with 10% FCS (HyClone, Cat. 30160.03), 100 U/mL penicillin/100 mg/mL streptomycin (Lonza, Cat. DE17-602E), 2 mM L-glutamine (Lonza, Cat. BE17-605E) and 10mM HEPES (CureVac SE). After 1 h, GolgiPlug™ (BD Biosciences, Cat. 555029) was added in a dilution of 1:200 (50 µL) to the splenocytes to inhibit cytokine secretion. After stimulation, splenocytes were centrifuged, resuspended in supplemented α-MEM medium (Gibco, Cat. 22561), and stored overnight at 4 °C. On the following day, splenocytes were washed twice in PBS and stained with LIVE/DEAD™ fixable aqua dead cell stain kit (Invitrogen, Cat. L34957) for 30 min at 4 °C in the dark. After an additional washing step in PBS with 0.5% BSA, cells were surface stained for Thy1.2 (FITC rat anti-mouse CD90.2 [Thy1.2]; 1:200; BioLegend, Cat. 140304), CD4 (V450 rat anti-mouse CD4; 1:200; BD Biosciences, Cat. 560468) and CD8 (APC-Cy7 rat anti-mouse CD8a; 1:200; BioLegend, Cat. 100714) and incubated with FcγR-block (rat anti-mouse CD16/CD32; 1:100; Invitrogen, Cat. 14-0161-85) in PBS with 0.5% BSA for 30 min at 4 °C in the dark. Splenocytes were then washed and fixed using Cytofix/Cytoperm™ solution (BD Biosciences, Cat. 554722) for 20 min at RT in the dark. After fixation, cells were washed in permeabilization buffer (PBS, 0.5% BSA, 0.1% Saponin) and stained for interferon (IFN)-γ (APC rat anti-mouse IFN-γ; 1:100; BD Biosciences, Cat. 554413) and tumor necrosis factor (TNF; Phycoerythrin [PE]-conjugated, rat anti-mouse TNF alpha, 1:100; Invitrogen, Cat. 12-7321-82) for 30 min at 4 °C in the dark. Splenocytes were subsequently washed in permeabilization buffer and resuspended in PBEA buffer (PBS, 0.5% BSA, 2 mM EDTA, 0.01% sodium azide). Finally, splenocytes were analyzed by flow cytometry on a ZE5 flow cytometer (Bio-Rad Laboratories, Inc.) and data were analyzed using FlowJo™ software version 10.7.2 (Tree star, Inc.; Ashland, OR, USA).
2.7 Bacterial adhesion inhibition assay
Functional antibody responses were assessed by bacterial adhesion inhibition (BAI) assay as previously described (39). Briefly, a UPEC strain [UTI89; (44)] engineered to express the mCherry fluorescent marker was incubated for 30 min with monolayers of human uroepithelium cell line SV-HUC-1 (ATCC) in 96-well plates (ATCC, Cat. CRL-9520) in the presence of serially diluted sera from vaccinated or sham-vaccinated mice or rats. As positive and negative controls, 20% D-(+)-mannose and medium were used, respectively. After adhesion, cells were washed extensively to remove unbound bacteria and fixed with formaldehyde. Finally, the specific fluorescent signal associated with the adhered bacteria was recorded using an automated high content screening microscope (Opera Phenix) and quantified with Harmony software (Opera Phenix, version 5.1). The BAI titers were determined as the reciprocal serum dilution leading to 50% bacterial adhesion inhibition, indicated by the inflection point of the dose-response curve.
2.8 In vitro protein expression
For expression analysis of LNP-formulated mRNA encoding either FimHDG or FimHDG-Ferritin containing either unmodified or modified (N1mΨ) nucleosides, HEK 293T cells were seeded at a density of 4 x 105 cells/well in 6-well-plates (Sarstedt, Cat. 83.3920.300). The next day, cells were transfected under serum-free conditions with 0.5 µg of the respective mRNA vaccine per well. Protein expression in cell culture supernatants or cell lysates was assessed 48 h post transfection via SDS-PAGE (45) and western blotting. Supernatants were harvested and residual cells removed by centrifugation (500 × g, 3 min). Cells were lysed using RIPA with NP-40. Laemmli sample buffer (6x) was added to the supernatants and lysates before heating for 5 min at 95 °C. Equal sample volume (12.5 µl) was loaded per lane and proteins were separated on 4–20% Mini-PROTEAN® TGX™ Precast Protein Gels (Bio-Rad, Cat.456-1095) before transfer to a nitrocellulose membrane (Odyssey® nitrocellulose membrane, pore-size: 0.22 µm; LI-COR, Cat. 926-31092). Specific proteins were detected using mouse anti-FimHLcys polyclonal antiserum (1:1,000; obtained as previously described (39) and rabbit anti-alpha/beta tubulin antibody (1:1,000; Cell Signaling, Cat. 2148S), followed by goat anti-mouse IgG IRDye® 800CW (1:10,000; LI-COR, Cat. 926-32210) and goat anti-rabbit IgG IRDye® 680RD (1:10,000; LI-COR, Cat. 926-68071), respectively. Protein detection and image processing were carried out in an Odyssey® CLx Imaging System and LI-COR’s Image Studio™ Lite version 5.2.5 according to the manufacturer’s instructions. All blots displayed in Figure 1C derive from the same experiment and were processed in parallel.
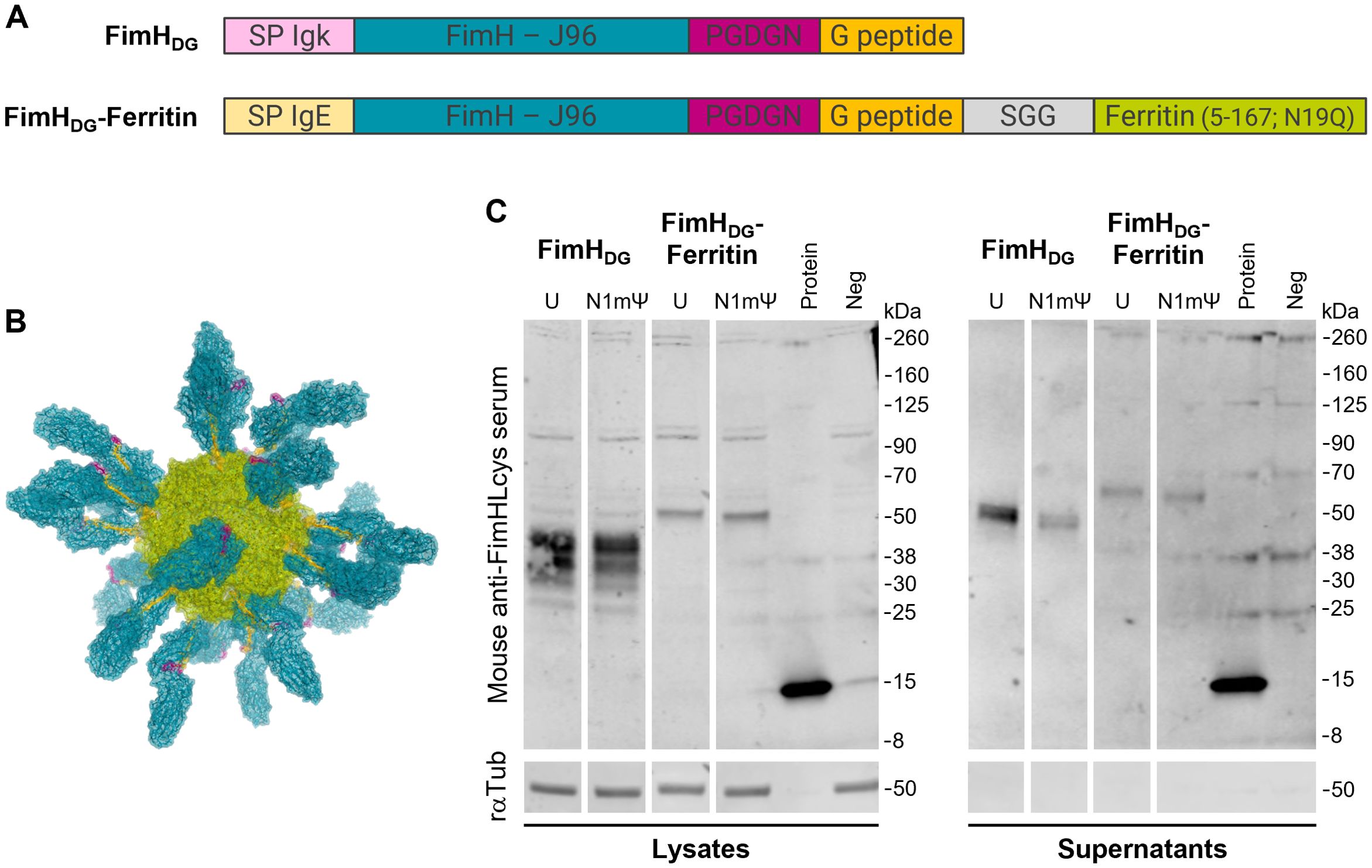
Figure 1. Proteins translated from mRNAs encoding FimHDG or FimHDG-Ferritin are secreted in cell culture. (A) Schematic representation (depiction not to scale) of the open reading frames (ORFs) of mRNAs encoding FimHDG or FimHDG-Ferritin. The ORFs include a signal peptide (SP IgE or SP Igk) at the N-terminus, followed by FimH from E. coli strain J96 (FimH - J96), a 5-amino acid (aa) linker (PGDGN), and the donor strand peptide of FimG (G peptide). The ORF of FimHDG-Ferritin contains an additional 3-aa linker (SGG) and the ferritin domain from H. pylori strain J99 (Ferritin [5-167; N19Q]) at the C-terminus. (B) Predicted structure of an mRNA-derived FimHDG-Ferritin protein nanoparticle (PNP). The expressed fusion protein FimHDG-Ferritin is expected to be presented to cells of the immune system as secreted self-assembled 24-mer PNPs. Each PNP is expected to consist of a ferritin core (green) displaying 24 copies of FimH (blue) stabilized in a pre-binding conformation by the PGDGN-linked (pink) donor strand peptide of FimG (yellow) on its surface. FimHDG-Ferritin denotes the fusion of FimH with the donor strand peptide of FimG (DG) and the H. pylori ferritin domain using an SGG-linker (grey). (C) HEK 293T cells were transfected with LNP-formulated mRNA containing either unmodified (U) or modified (N1mΨ) nucleosides encoding either FimHDG or FimHDG-Ferritin. FimH expression in cell lysates and supernatants was analyzed via western blotting 48 h post transfection. HEK 293T cells incubated with medium served as negative control (Neg) and 100 ng recombinant FimHL protein loaded to the gels was used as positive control (Protein). Rabbit anti-tubulin antibody (rαTub) was employed as loading control. Uncropped images are shown in Supplementary Figure S1.
2.9 Human peripheral blood mononuclear cell stimulation and measurement of human IFN-α
Human peripheral blood mononuclear cells (hPBMCs) were isolated from buffy coats obtained from healthy adult donors through the blood bank in Tübingen (ZKT Tübingen gGMbH). Donors gave informed written consent for the use of their blood products in research and were anonymized prior to transfer to CureVac. Buffy coats were processed within 24 h of collection. Each buffy coat was diluted 1:1 with PBS and layered over Ficoll-Paque PLUS (Cytiva former GE Healthcare, Cat. 17-1440-02) for density gradient centrifugation at 805 × g for 20 min at RT without brake. The hPBMC layer was collected, washed twice with PBS, and cells were counted using a hemocytometer and trypan blue. Isolated hPBMCs were cryopreserved in FCS (HyClone, Cat. 30160.03) containing 10% DMSO (Sigma-Aldrich, Cat. 41639) and stored in liquid nitrogen. For stimulation, cryopreserved hPBMCs were thawed and transferred into 37 °C pre-warmed cell medium RPMI 1640 (Gibco, Cat. 52400025) with 20% FCS, 100 U/mL penicillin/100 mg/mL streptomycin (Lonza, Cat. DE17-602E), 2 mM L-glutamine (Lonza, Cat. BE17-605E). The cells were washed once, reconstituted, and 4 x 105 cells were seeded per well into 96-well flat-bottom cell culture plate (Sarstedt, Cat. 83.3924). hPBMCs from three donors were incubated each in triplicates with 10 µg/mL of FimHDG-Ferritin mRNA vaccines containing either unmodified or modified (N1mΨ) nucleosides, prediluted in medium without FCS, in a total volume of 200 µL. hPBMCs treated with medium were used as control. After 24 h, cell-free supernatants were collected and analyzed in a 1:20 or 1:40 dilution using an IFN-α ELISA kit (PBL, Cat. 41115-1) according to the manufacturer’s instructions.
2.10 Statistical analyses
For the mouse and rat studies, differences between vaccines were assessed for each relevant readout using Analysis of Variance (ANOVA); different days were analyzed separately. For the rat study, the analysis accounted for both vaccine type and dose level, as well as the interaction between these two factors (vaccine*dose) to account for different potency between vaccines across the doses tested. If a significant difference between the vaccines was identified (assuming a significance level of α = 0.05), a post-hoc pairwise comparison test was performed to identify the differences using the Least Significant Difference (LSD) method. The corresponding P values are represented in the respective graphs.
All binding antibody data were log10-transformed for analysis. Normality was tested using the Shapiro-Wilk test, and subsequently by examination of QQ plots. Due to low granularity of values, in some cases Shapiro-Wilk indicated possibly non-normal data, but the corresponding QQ plots demonstrated little to no bias, suggesting underlying normality. In such cases, the Scheirer-Ray-Hare test was used to confirm the ANOVA results. Sphericity was tested using Mauchly’s test and was met in all cases. All statistical analyses were conducted in R (version 4.4.0 or higher, R Foundation for Statistical Computing) with Rstudio as the graphic user interface. The ANOVA was carried out using the aov function from the stats package and post-hoc tests using the PostHocTest function from the DescTools package.
3 Results
3.1 Proteins translated from mRNAs encoding FimHDG or FimHDG-Ferritin are secreted in cell culture
The ORFs of the mRNAs encoding FimHDG or FimHDG-Ferritin are depicted in Figure 1A. Both ORFs include a signal peptide (SP; SP Igk or SP IgE, respectively) at the N-terminus, followed by FimH from E. coli strain J96 (FimH – J96), a 5-aa linker (PGDGN), and the donor strand peptide of FimG (G peptide). The ORF of FimHDG-Ferritin contains an additional 3-aa linker (SGG) and the ferritin domain from H. pylori strain J99 (Ferritin [5-167; N19Q]) at the C-terminus. Antigen display on the ferritin surface enables uniform presentation of 24 copies of the antigen (46). The expressed fusion protein FimHDG-Ferritin is therefore expected to be presented to cells of the immune system as secreted self-assembled 24-mer PNPs, with FimH stabilized in a pre-binding conformation by the donor strand peptide of FimG on its surface (Figure 1B) (24).
Western blot analysis of cells transfected with LNP-formulated mRNA containing either unmodified (U) or modified (N1mΨ) nucleosides encoding either FimHDG or FimHDG-Ferritin confirmed that mRNA encoding FimHDG or FimHDG-Ferritin supported efficient expression and secretion of FimH (Figure 1C; Supplementary Figure S1).
3.2 FimHDG and FimHDG-Ferritin mRNA vaccine candidates induce robust humoral and cellular immune responses in mice
In order to assess the immunogenicity of the mRNA-based FimH vaccine candidates, BALB/cAnNRi mice received three IM injections of either FimHDG or FimHDG-Ferritin mRNA vaccines, both containing unmodified nucleosides, or PHAD-adjuvanted FimHC protein complex subunit vaccine as a control. Animals receiving the FimHDG-Ferritin mRNA vaccine or the PHAD-adjuvanted FimHC protein subunit vaccine showed the highest levels of FimHL-specific IgG binding antibodies in serum (Figure 2A) and urine (Figure 2B) after the first vaccine administration (Day 21). Differences between the vaccine groups were less pronounced after the second or third vaccine administration. Functional antibody responses (i.e. antibodies preventing UPEC binding to target cells), as measured by BAI assay, were highest in the group immunized with the FimHDG-Ferritin mRNA vaccine (Figure 2C). In contrast to the binding antibody levels, the functional antibody responses measured in mice that had received the PHAD-adjuvanted FimHC protein subunit vaccine were substantially lower than in the mRNA vaccine-administered groups. In addition to humoral responses, the FimHDG and FimHDG-Ferritin mRNA vaccines induced cellular responses. Significantly higher levels of FimH-specific CD4+ and CD8+ T cells producing IFN-γ and TNF were detected in the spleens of mice administered with FimHDG-Ferritin compared with FimHDG mRNA, while there were no measurable T cell responses for the PHAD-adjuvanted FimHC protein subunit vaccine (Figure 2D; Supplementary Figure S2).
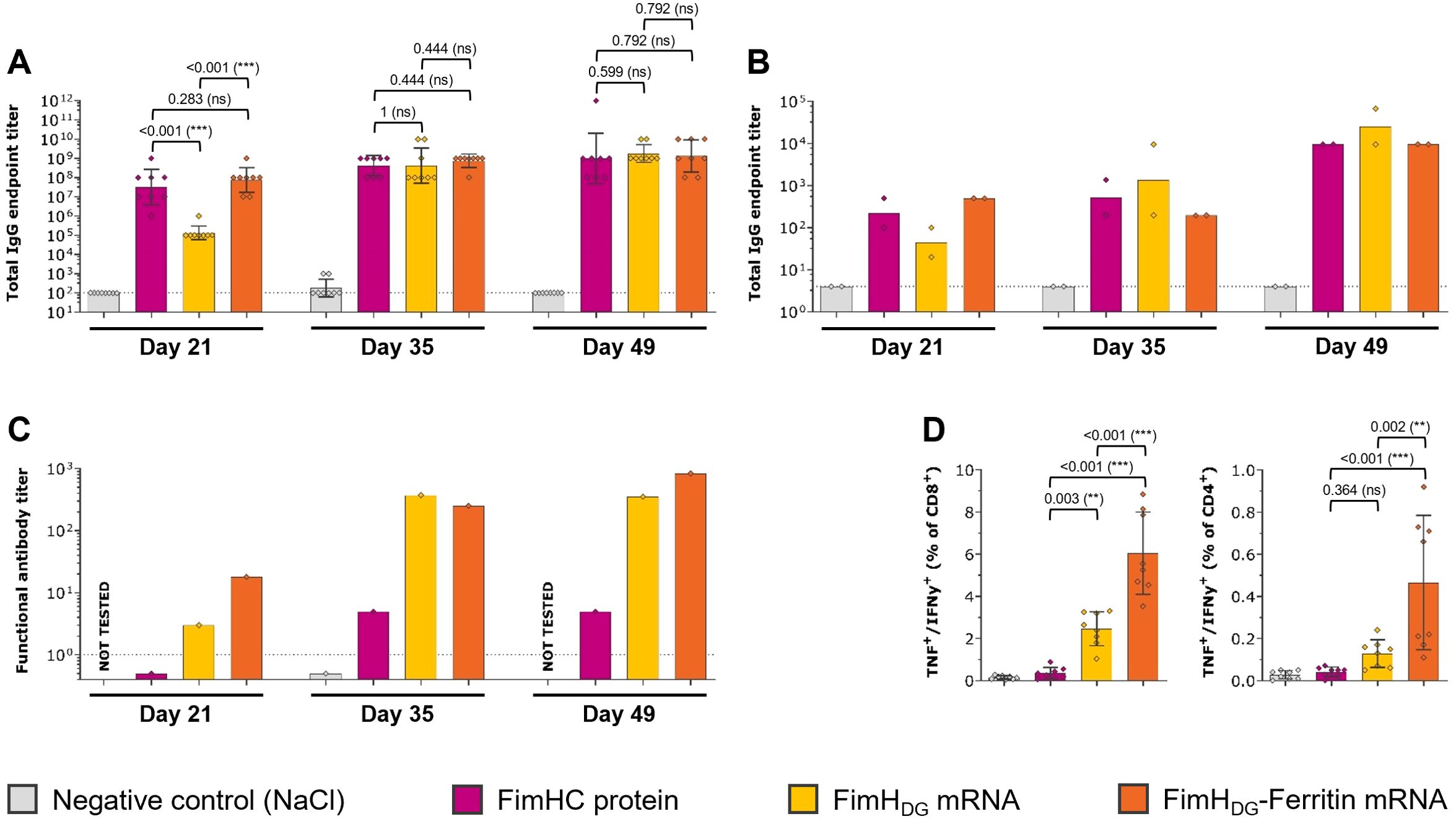
Figure 2. FimH-based mRNA vaccines containing unmodified nucleosides induced robust levels of both humoral and cellular immune responses in mice. Female BALB/c mice (n=8/group) were vaccinated IM three times on Day 0, 21, and 35 with 2 µg of PHAD-adjuvanted FimHC protein complex subunit vaccine (pink bars), 2 µg of FimHDG mRNA vaccine (yellow bars), or 2 µg of FimHDG-Ferritin mRNA vaccine (orange bars). Mice receiving physiological saline (0.9% NaCl; grey bars) served as negative controls. FimHL-specific binding antibody endpoint titers in serum (A) or urine (B), determined by ELISA, as well as functional serum antibody titers measured by BAI assay (C) in samples collected after one (Day 21), two (Day 35), or three (Day 49) vaccinations are displayed. Multifunctional TNF/IFN-γ-positive CD4+ and CD8+ T cells (D) were analyzed in splenocytes isolated on Day 49 by stimulation with FimH-specific peptides followed by intracellular cytokine staining and detection by flow cytometry. The gating strategy for this analysis is shown in Supplementary Figure S2. In (A, D), diamond symbols represent individual animals, and bars depict either geometric means with geometric SD in (A) or means with SD in (D). Each bar shown in (B) represents the geometric mean of two urine pools per group, each containing urine from four animals. Each bar depicted in (C) represents one functional antibody titer per group, determined based on pooled serum samples derived from all animals in the respective group. Dotted lines indicate the lower limit of quantification (LLOQ). Values below the LLOQ were set to 0.5 (non-responders in BAI assay; C). Data from individual days displayed in (D) were statistically analyzed using an ANOVA followed by the LSD post test. P values are displayed and significant differences between vaccines are marked by asterisks (**P < 0.01, ***P < 0.001; ns, not significant).
3.3 FimHDG and FimHDG-Ferritin mRNA vaccine candidates induce robust levels of binding and functional antibodies in rats
In the first confirmatory immunogenicity study in Wistar rats, various doses of the selected mRNA vaccine candidates FimHDG and FimHDG-Ferritin containing unmodified nucleosides were tested, along with an AS01-adjuvanted FimHDG protein subunit vaccine used as comparator (Figure 3). At the tested dosages, the FimHL-specific binding antibody levels in serum (Figure 3A) corresponded with the functional antibody responses, detected via the BAI assay (Figure 3C). Results indicate that the highest antibody titers were induced after the second and third administrations of the FimHDG-Ferritin mRNA vaccine candidate, followed by the FimHDG mRNA vaccine and the protein subunit vaccine control. In general, the groups with the highest binding antibody titers in serum also had the highest titers in urine (Figure 3B), as well as the highest functional antibody levels in serum.
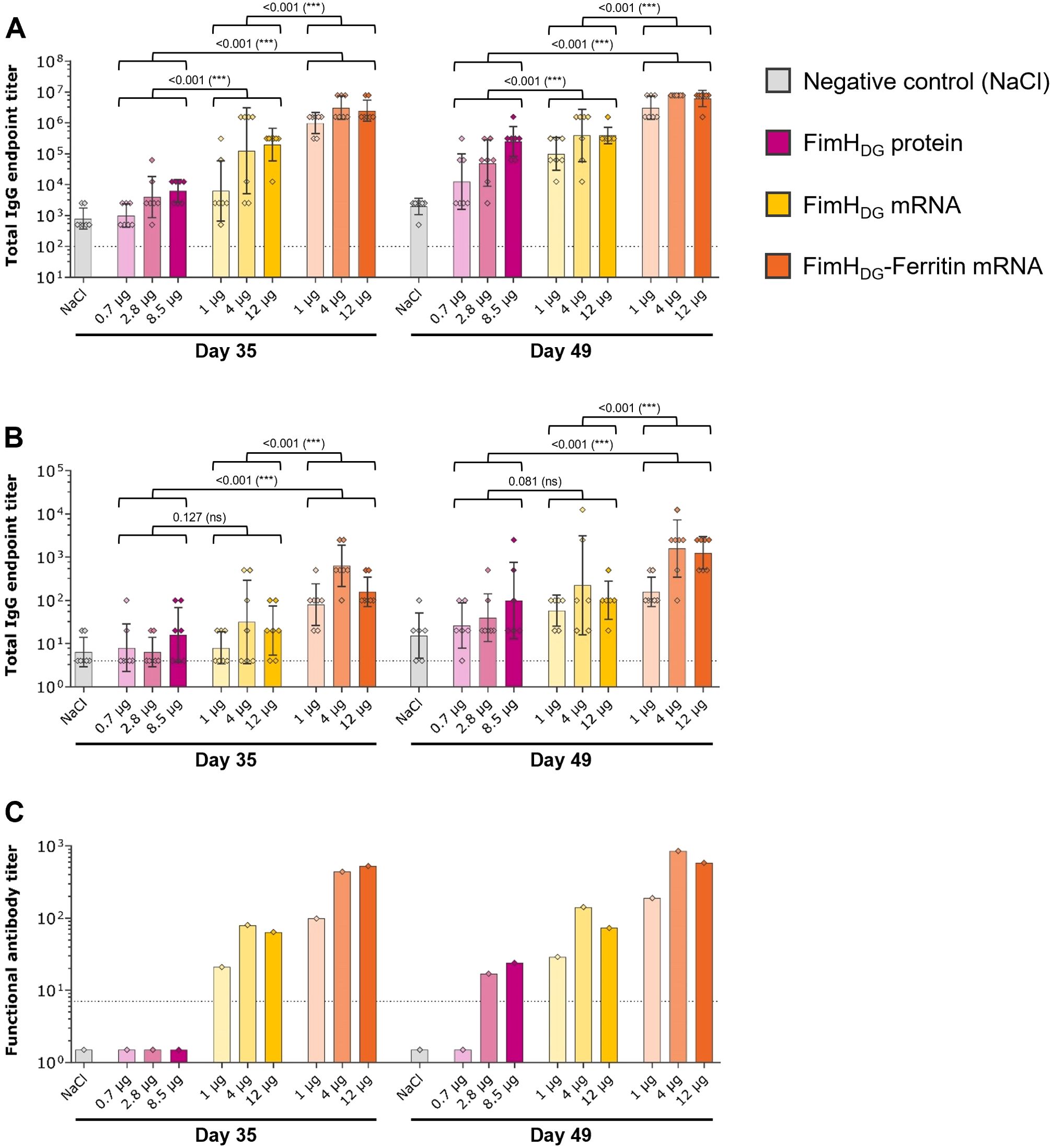
Figure 3. FimH-based mRNA vaccines using unmodified nucleosides induced robust levels of binding and functional antibody responses in rats. Female Wistar rats (first rat study; n=7/group) were vaccinated IM three times on Day 0, Day 21, and Day 35 with different doses of AS01-adjuvanted FimHDG protein subunit vaccine (pink bars), FimHDG mRNA vaccine (yellow bars), or FimHDG-Ferritin mRNA vaccine (orange bars) as indicated. Rats receiving physiological saline (0.9% NaCl; grey bars) served as negative controls. FimHL-specific binding antibody endpoint titers in serum (A) or urine (B), determined by ELISA, as well as functional serum antibody titers measured by BAI assay (C) in samples collected after two (Day 35) or three (Day 49) vaccinations are displayed. In (A, B), diamond symbols represent individual animals, and bars depict geometric means with geometric SD. Each bar depicted in (C) represents one functional antibody titer per group, determined based on pooled serum samples derived from all animals in the respective group. Dotted lines indicate the lower limit of quantification (LLOQ). Values below the LLOQ were set to 1.5 (non-responders in BAI assay; C). Individual days were statistically analyzed using an ANOVA followed by the LSD post test. P values are displayed and significant differences between vaccines across the doses tested are marked by asterisks (***P < 0.001; ns, not significant).
Together with the findings from the mouse study, these results clearly demonstrated that multimerization of the FimH antigen via fusion to ferritin enhanced the immunological potency compared to the monomeric mRNA design.
3.4 FimHDG-Ferritin mRNA vaccine including N1mΨ-modified nucleosides induces robust levels of binding and functional antibody responses in rats
Using modified nucleosides in vaccines, such as N1mΨ, has been shown to be immunogenic and efficacious as well as safe and well tolerated in humans, as evidenced for the two licensed COVID-19 mRNA vaccines, Spikevax® (Moderna) and Comirnaty® (Pfizer-BioNTech) (47, 48). This suggests that a wider therapeutic window (i.e., higher dose level resulting in higher immune responses and efficacy, associated with acceptable reactogenicity) can be achieved with modified-nucleoside mRNA as compared with unmodified-nucleoside mRNA vaccines (47–53).
Therefore, the in vitro immunostimulatory activity and in vivo immunogenicity of the lead candidate antigen design FimHDG-Ferritin were evaluated comparing unmodified or N1mΨ nucleoside containing mRNA vaccines. hPBMCs stimulated with the FimHDG-Ferritin mRNA vaccine containing unmodified nucleosides showed detectable levels of IFN-α. In contrast, the FimHDG-Ferritin mRNA vaccine containing N1mΨ did not induce detectable levels of IFN-α (Supplementary Figure S3).
In the second study with Wistar rats, similar FimHL-specific serum IgG levels were induced by the FimHDG-Ferritin mRNA vaccines containing unmodified and N1mΨ-modified nucleosides upon IM injection, while the tested dosages of AS01-adjuvanted FimHDG protein subunit vaccine used as a comparator induced significantly lower binding antibody responses (Figure 4A). Among the tested mRNA vaccine candidates, FimHDG-Ferritin with N1mΨ induced significantly higher IgG levels in the urine after three administrations (Figure 4B) and the highest functional antibody titers in serum (Figure 4C).
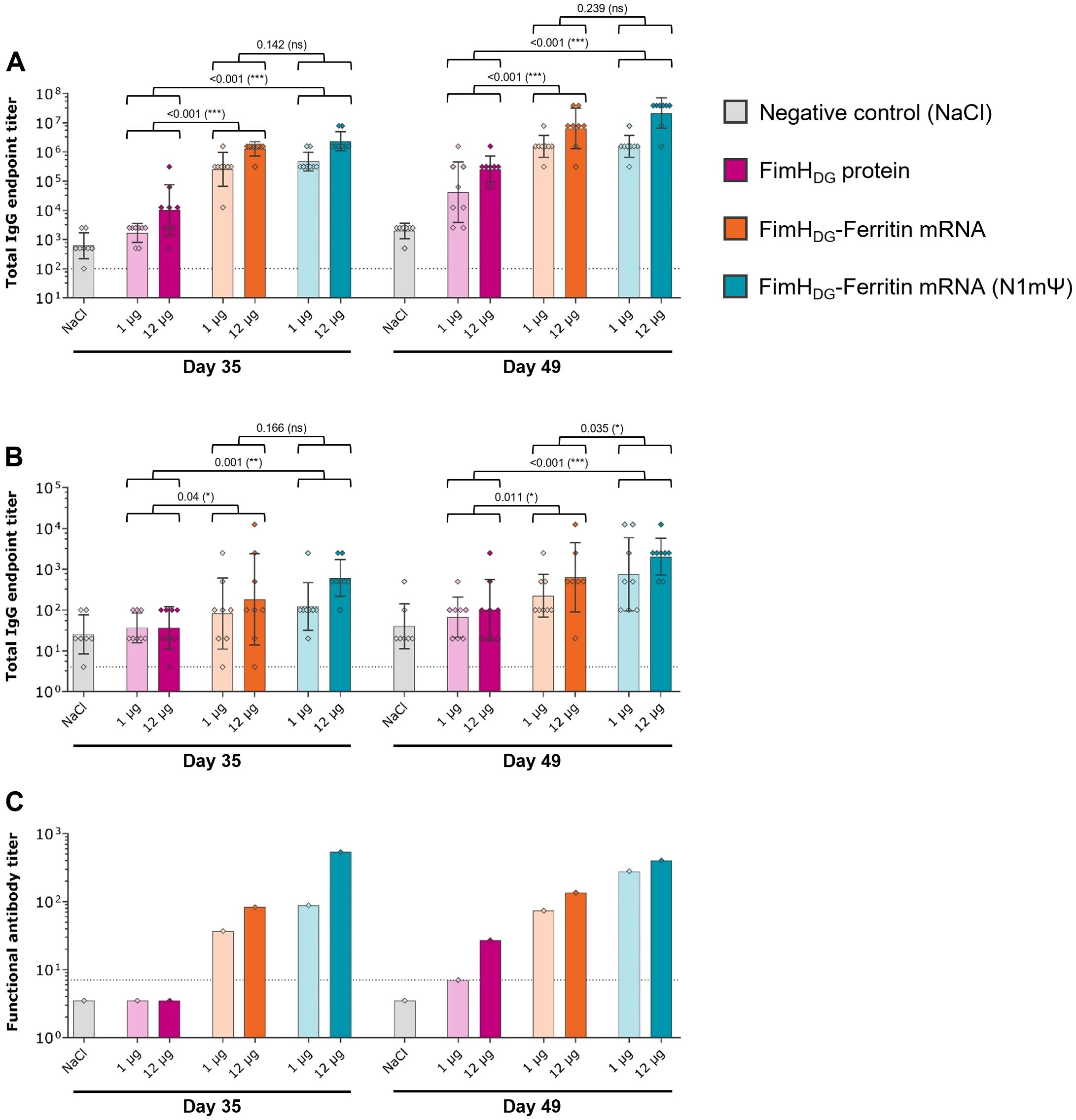
Figure 4. An mRNA vaccine using N1mΨ-modified nucleosides encoding FimHDG-Ferritin induced robust levels of binding and functional antibody responses in rats. Female Wistar rats (second rat study; n=8/group) were vaccinated IM three times on Day 0, Day 21, and Day 35 with different doses of AS01-adjuvanted FimHDG protein subunit vaccine (pink bars), FimHDG-Ferritin mRNA vaccine containing unmodified nucleosides (orange bars), or FimHDG-Ferritin mRNA vaccine containing N1mΨ-modified nucleosides (blue bars) as indicated. Rats receiving physiological saline (0.9% NaCl; grey bars) served as negative controls. FimHL-specific binding antibody endpoint titers in serum (A) or urine (B), determined by ELISA, as well as functional serum antibody titers measured by BAI assay (C) in samples collected after two (Day 35) or three (Day 49) vaccinations are displayed. In (A) and (B), diamond symbols represent individual animals, and bars depict geometric means with geometric SD. Each bar depicted in (C) represents one functional antibody titer per group, determined based on pooled serum samples derived from all animals in the respective group. Dotted lines indicate the lower limit of quantification (LLOQ). Values below the LLOQ were set 3.5 (non-responders in BAI assay; C). Individual days were statistically analyzed using an ANOVA followed by the LSD post test. P values are displayed and significant differences between vaccines across the doses tested are marked by asterisks (*P < 0.05, **P < 0.01, ***P < 0.001; ns, not significant).
4 Discussion
Data from these proof-of-concept immunization studies in mice and rats using FimHDG mRNA vaccine candidates with unmodified nucleosides demonstrated the induction of significant levels of FimH-specific immune responses in both species. Multimeric presentation of vaccine-candidate antigens has been shown to enhance uptake and delivery by antigen-presenting cells, driving B-cell activation through receptor cross-linking to establish a protective humoral response (30, 54, 55). Consistently, we found that the FimHDG-Ferritin mRNA-based candidate vaccine was more immunogenic compared to the FimHDG vaccine, indicating that the multimeric presentation of the FimH antigen on a PNP surface enhanced the immunogenicity compared to a monomeric antigen design. The improved immunogenicity of FimHDG when fused to ferritin may also be derived from the intrinsic stability of ferritin nanoparticles, adding to the growing literature demonstrating the utility and versatility of ferritin-based vaccines (56).
Different vaccine platforms have been harnessed in the development of a vaccine against rUTI. In a Phase 1/2 study, a bioconjugate vaccine including the four most relevant O-antigens expressed in E. coli as glycoproteins induced carbohydrate-specific opsonophagocytic antibodies but failed to reduce the frequency of UTI due to the serotypes expressing the targeted O-antigens (57). In non-human primates, a vaccine that combines FimH and O-antigens reduced bacterial load of UTI (58). Hence, there is a need for a vaccine that induces robust transudation of anti-adhesive antibodies into the human bladder. Transudation of antibodies to the urogenital tract has been demonstrated following administration of the human papillomavirus (HPV) vaccine (59) and antibody presence in urine (60).
Whilst we observed a strong IgG response in serum following administration of FimH and FimHDG-Ferritin vaccine candidates, urine IgG levels were relatively low in these animals, possibly due to the absence of bacterial infection. This could be linked to a hypothesized mode of action for a FimH-based vaccine, whereby serum-derived IgG could provide protection against UPEC in the urinary tract upon increased blood vessel permeability due to the inflammation induced by the bacterial infection. Since a relatively low number of UPEC bacteria initiate a UTI (61), asymptomatic inflammation and increased blood vessel permeability may allow serum-derived anti-FimH IgG antibodies to transudate into the bladder and block further binding of UPEC to uroepithelial cells, whilst unbound bacteria are eliminated through urinary flow. If this hypothesis is correct, FimH-specific functional antibodies in serum could act as a correlate of protection.
In addition to a strong humoral response, our FimHDG-Ferritin mRNA vaccine induced splenic T cell responses that were significantly higher than those observed with the protein subunit vaccine control at the tested dosages. The ability of mRNA vaccines to elicit more robust CD8+ T cell responses compared with protein subunit vaccines has been described previously (62–64). Since T cell responses are an important feature of natural immunity and susceptibility to UPEC infection (65), a strong, specific T cell response may enhance vaccine efficacy. Data presented here demonstrate the induction of multifunctional TNF/IFN-γ-positive CD4+ and CD8+ T cells upon administration of mRNA vaccines. These cells are indicative of a T helper type 1 (Th1) response which is crucial for bacteria clearance in the bladder (65). The mRNA vaccine technology used in this study has been previously shown to induce enrichment of lung-resident memory CD8+ T cells in mice (66). Mucosal memory T cells, which reside within peripheral tissues, are able to protect against recurrent UPEC infections in mice depleted of systemic T cells (67). In addition, tissue-resident memory T cells (TRM) cells can rapidly respond to an infection without needing to migrate, proliferate or differentiate, and can contribute to improved vaccine efficacy (68, 69). Indeed, previous studies have shown that vaccines designed to generate pathogen-specific TRM populations in mucosal tissues could provide long-lasting protection and reduce recurrence at mucosal surfaces (69, 70). Future research could explore the most effective strategies for inducing robust T cell responses that are most effective in driving bacterial clearance in the bladder, such as through mucosal vaccination.
Nucleoside modification of mRNA (such as addition of pseudouridine) suppresses the immunostimulatory effect of RNA (71, 72) and increases its translation in therapeutic applications (72, 73). In the current study, nucleoside modification (N1mΨ) in the FimHDG-Ferritin candidate vaccine improved binding and functional antibody responses in female rats and also led to reduced induction of IFN-α in hPBMCs, with the potential to translate to a wider range of tolerable vaccine doses in humans. Overall, the data generated in these experiments demonstrate that FimHDG-Ferritin mRNA-based candidate containing modified nucleosides is a promising UPEC vaccine for further development. Key to this development is the assessment of protective efficacy following FimHDG-Ferritin mRNA vaccine administration in an in vivo model of UTI, which will be evaluated in future studies.
The studies reported here do have some limitations. Whilst we employed protein subunit vaccines as a control, direct comparison of doses between mRNA and protein vaccines are difficult since the systems employed prevent direct determination of protein levels expressed from mRNA in vivo. Given the high prevalence of rUTI in women, studies were performed with female animals only; however, future studies with male rodents would be beneficial. In addition, statistical analyses were not performed for all experiments due to pooling of collected samples, which limits assessment of functional antibody differences between groups. As such, future studies will be designed without pooling samples. Nonetheless, the methods reported here have demonstrated good reproducibility, with comparable titers of functional antibodies and serum and urine IgG seen across all three animal studies with the unmodified FimHDG-Ferritin mRNA vaccine.
5 Conclusion
With the success of mRNA vaccines against viral infections, the development of mRNA vaccines against bacterial pathogens is still in the early stages, with only three vaccine candidates in clinical trials (74). Our mRNA-based vaccine FimHDG-Ferritin with N1mΨ-modified nucleosides is a promising candidate for further development as a vaccine against UPEC, a leading cause of rUTI. As there is already significant clinical data on the safety profile of mRNA vaccines, and a well-established route for regulatory approval (36, 75), future research will focus on further characterization of the FimHDG-Ferritin vaccine, particularly in terms of the protective efficacy in animal models, with the aim of progressing into clinical development.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used, and the human peripheral blood mononuclear cells (hPBMCs) were isolated from buffy coats obtained from healthy adult donors through the blood bank in Tübingen (ZKT Tübingen gGMbH). Donors gave informed written consent for the use of their blood products in research and were anonymized prior to transfer to CureVac.
Author contributions
SRo: Writing – original draft, Validation, Investigation, Data curation, Writing – review & editing, Formal analysis, Supervision, Methodology, Visualization, Conceptualization, Software, Project administration. RA: Writing – review & editing, Formal analysis, Writing – original draft, Conceptualization. RR: Formal analysis, Writing – review & editing, Conceptualization, Writing – original draft. AP: Conceptualization, Methodology, Writing – review & editing, Software. MS: Conceptualization, Methodology, Software, Writing – review & editing. BP: Resources, Conceptualization, Writing – review & editing, Supervision. EJ: Writing – review & editing, Resources, Conceptualization. SRa: Supervision, Conceptualization, Writing – review & editing, Resources.
Funding
The author(s) declare financial support was received for the research and/or publication of this article.
Acknowledgments
The authors would like to thank Simone Odau, Friederike Naumann and Jonas Füner (Preclinics) for their outstanding commitment and expertise in conducting the in vivo studies; Miguel Pereira and Matt Chapman-Rounds (Cogitars) for performing statistical analyses; Annette Möbes, Julia Schröder and Julia Führer (CureVac) for performing most of the experiments and their relentless and expert support in the lab; Wolfgang Große and Moritz Thran (CureVac) for their support with antigen design and generating mRNA constructs; Annachiara Greco, Nadine Kuttruff, Marius Busche and Robert Tensen (CureVac) for all their excellent work on project planning and communication; Sanjay Phogat and Simona Rondini (GSK) for scientific discussion; and Professor T. E. Andersen (University of Southern Denmark) for providing the UTI89 strain. Medical writing support was provided by Nicola Beadle, PhD and Holly Robertson, PhD, of Alchemy Medical Writing Ltd., Stirling, Scotland, UK and was funded by CureVac SE in accordance with Good Publication Practice (GPP) guidelines (available at: https://www.acpjournals.org/doi/epdf/10.7326/M22-1460).
Conflict of interest
SRo is an employee of CureVac SE, Tübingen, Germany, a publicly listed company developing mRNA-based vaccines and immunotherapeutics. SRo is an inventor on patents on mRNA vaccination and use thereof, including the pending vaccine patent on this topic WO 2023/227608 A1. SRo holds shares in the company. RA is employed by GSK and holds financial equities in GSK. RA is an inventor for the pending vaccine patent on this topic WO 2023/227608 A1. RA declared they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision. RR is employed by GSK and is an inventor for the pending vaccine patent on this topic WO 2023/227608 A1. BP was an employee of CureVac SE, Tübingen, Germany. BP is an inventor on patents on mRNA vaccination and use thereof, including the pending vaccine patent on this topic WO 2023/227608 A1. EJ is an employee of CureVac SE, Tübingen, Germany. EJ is an inventor on patents on mRNA vaccination and use thereof. EJ holds shares in the company. AP is employed by GSK and holds financial equities in GSK. AP is listed as an inventor for patents. MS is employed by GSK and holds financial equities in GSK. MS is listed as an inventor for patents. SRa was an employee of CureVac SE, Tübingen, Germany. SRa is an inventor on patents on mRNA vaccination and use thereof, including the pending vaccine patent on this topic WO 2023/227608 A1.
The authors declare that this study received funding from CureVac SE and GSK. The funders had the following involvement in the study: study design, collection, analysis, interpretation of data, the writing of this article, and the decision to submit it for publication.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1668937/full#supplementary-material
References
1. Yang X, Chen H, Zheng Y, Qu S, Wang H, and Yi F. Disease burden and long-term trends of urinary tract infections: A worldwide report. Front Public Health. (2022) 10:888205. doi: 10.3389/fpubh.2022.888205
2. Houston CG, Azar WS, Huang SS, Rubin R, Dorris CS, and Sussman RD. A cost savings analysis of topical estrogen therapy in urinary tract infection prevention among postmenopausal women. Urol Pract. (2024) 11:257–66. doi: 10.1097/upj.0000000000000513
3. Brumbaugh AR and Mobley HL. Preventing urinary tract infection: Progress toward an effective Escherichia coli vaccine. Expert Rev Vaccines. (2012) 11:663–76. doi: 10.1586/erv.12.36
4. Medina M and Castillo-Pino E. An introduction to the epidemiology and burden of urinary tract infections. Ther Adv Urol. (2019) 11:1756287219832172. doi: 10.1177/1756287219832172
5. Kwok M, McGeorge S, Mayer-Coverdale J, Graves B, Paterson DL, Harris PNA, et al. Guideline of guidelines: Management of recurrent urinary tract infections in women. BJU Int. (2022) 130:11–22. doi: 10.1111/bju.15756
6. Aggarwal N and Leslie SW. Recurrent urinary tract infections. In: StatPearls. StatPearls Publishing, Treasure Island (FL (2025).
7. Flores-Mireles AL, Walker JN, Caparon M, and Hultgren SJ. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol. (2015) 13:269–84. doi: 10.1038/nrmicro3432
8. Hospenthal MK and Waksman G. The remarkable biomechanical properties of the type 1 chaperone-usher pilus: A structural and molecular perspective. Microbiol Spectr. (2019) 7. doi: 10.1128/microbiolspec.psib-0010-2018
9. Delcaru C, Alexandru I, Podgoreanu P, Grosu M, Stavropoulos E, Chifiriuc MC, et al. Microbial biofilms in urinary tract infections and prostatitis: etiology, pathogenicity, and combating strategies. Pathogens. (2016) 5:65. doi: 10.3390/pathogens5040065
10. Naziri Z, Kilegolan JA, Moezzi MS, and Derakhshandeh A. Biofilm formation by uropathogenic Escherichia coli: A complicating factor for treatment and recurrence of urinary tract infections. J Hosp Infect. (2021) 117:9–16. doi: 10.1016/j.jhin.2021.08.017
11. Tarlton NJ, Moritz C, Adams-Sapper S, and Riley LW. Genotypic analysis of uropathogenic Escherichia coli to understand factors that impact the prevalence of β-lactam-resistant urinary tract infections in a community. J Glob Antimicrob Resist. (2019) 19:173–80. doi: 10.1016/j.jgar.2019.03.002
12. Bradley MS, Cabrera C, Clark SG, Sassani J, Venuti K, and Ackenbom MF. Sporadic compared to recurrent urinary tract infections: Considerations for urogynecologic patients. Neurourol Urodyn. (2020) 39:2186–91. doi: 10.1002/nau.24471
13. Bauer HW, Alloussi S, Egger G, Blümlein HM, Cozma G, and Schulman CC. A long-term, multicenter, double-blind study of an Escherichia coli extract (OM-89) in female patients with recurrent urinary tract infections. Eur Urol. (2005) 47:542–8. doi: 10.1016/j.eururo.2004.12.009
14. Hopkins WJ, Elkahwaji J, Beierle LM, Leverson GE, and Uehling DT. Vaginal mucosal vaccine for recurrent urinary tract infections in women: Results of a phase 2 clinical trial. J Urol. (2007) 177:1349–53. doi: 10.1016/j.juro.2006.11.093
15. Benito-Villalvilla C, Cirauqui C, Diez-Rivero CM, Casanovas M, Subiza JL, and Palomares O. MV140, a sublingual polyvalent bacterial preparation to treat recurrent urinary tract infections, licenses human dendritic cells for generating Th1, Th17, and IL-10 responses via Syk and MyD88. Mucosal Immunol. (2017) 10:924–35. doi: 10.1038/mi.2016.112
16. Mak Q, Greig J, Dasgupta P, Malde S, and Raison N. Bacterial vaccines for the management of recurrent urinary tract infections: A systematic review and meta-analysis. Eur Urol Focus. (2024) 10:761–9. doi: 10.1016/j.euf.2024.04.002
17. FDA. Vaccines Licensed for Use in the United States (2025). Available online at: https://www.fda.gov/vaccines-blood-biologics/vaccines/vaccines-licensed-use-united-states (Accessed June 27, 2025).
18. Sarshar M, Behzadi P, Ambrosi C, Zagaglia C, Palamara AT, and Scribano D. FimH and anti-adhesive therapeutics: A disarming strategy against uropathogens. Antibiotics (Basel). (2020) 9:397. doi: 10.3390/antibiotics9070397
19. Mirzahosseini HK, Najmeddin F, Najafi A, Ahmadi A, Sharifnia H, Khaledi A, et al. Correlation of biofilm formation, virulence factors, and phylogenetic groups among Escherichia coli strains causing urinary tract infection: A global systematic review and meta-analysis. J Res Med Sci. (2023) 28:66. doi: 10.4103/jrms.jrms_637_22
20. Hung C-S, Bouckaert J, Hung D, Pinkner J, Widberg C, DeFusco A, et al. Structural basis of tropism of Escherichia coli to the bladder during urinary tract infection. Mol Microbiol. (2002) 44:903–15. doi: 10.1046/j.1365-2958.2002.02915.x
21. Sauer MM, Jakob RP, Eras J, Baday S, Eriş D, Navarra G, et al. Catch-bond mechanism of the bacterial adhesin FimH. Nat Commun. (2016) 7:10738. doi: 10.1038/ncomms10738
22. Kalas V, Pinkner JS, Hannan TJ, Hibbing ME, Dodson KW, Holehouse AS, et al. Evolutionary fine-tuning of conformational ensembles in FimH during host-pathogen interactions. Sci Adv. (2017) 3:e1601944. doi: 10.1126/sciadv.1601944
23. Rabbani S, Fiege B, Eris D, Silbermann M, Jakob RP, Navarra G, et al. Conformational switch of the bacterial adhesin FimH in the absence of the regulatory domain: Engineering a minimalistic allosteric system. J Biol Chem. (2018) 293:1835–49. doi: 10.1074/jbc.M117.802942
24. Silmon de Monerri NC, Che Y, Lees JA, Jasti J, Wu H, Griffor MC, et al. Structure-based design of an immunogenic, conformationally stabilized FimH antigen for a urinary tract infection vaccine. PloS Pathog. (2025) 21:e1012325. doi: 10.1371/journal.ppat.1012325
25. López-Sagaseta J, Malito E, Rappuoli R, and Bottomley MJ. Self-assembling protein nanoparticles in the design of vaccines. Comput Struct Biotechnol J. (2016) 14:58–68. doi: 10.1016/j.csbj.2015.11.001
26. Joyce MG, Chen WH, Sankhala RS, Hajduczki A, Thomas PV, Choe M, et al. SARS-CoV-2 ferritin nanoparticle vaccines elicit broad SARS coronavirus immunogenicity. Cell Rep. (2021) 37:110143. doi: 10.1016/j.celrep.2021.110143
27. Song N, Zhang J, Zhai J, Hong J, Yuan C, and Liang M. Ferritin: A multifunctional nanoplatform for biological detection, imaging diagnosis, and drug delivery. Acc Chem Res. (2021) 54:3313–25. doi: 10.1021/acs.accounts.1c00267
28. Yang F, Mariz FC, Zhao X, Spagnoli G, Ottonello S, and Müller M. Broad neutralization responses against oncogenic human papillomaviruses induced by a minor capsid L2 polytope genetically incorporated into bacterial ferritin nanoparticles. Front Immunol. (2020) 11:606569. doi: 10.3389/fimmu.2020.606569
29. Yin S, Davey K, Dai S, Liu Y, and Bi J. A critical review of ferritin as a drug nanocarrier: Structure, properties, comparative advantages and challenges. Particuology. (2022) 64:65–84. doi: 10.1016/j.partic.2021.04.020
30. Corbett KS, Moin SM, Yassine HM, Cagigi A, Kanekiyo M, Boyoglu-Barnum S, et al. Design of nanoparticulate group 2 influenza virus hemagglutinin stem antigens that activate unmutated ancestor B cell receptors of broadly neutralizing antibody lineages. mBio. (2019) 10:e02810–18. doi: 10.1128/mbio.02810-18
31. Fan K, Jiang B, Guan Z, He J, Yang D, Xie N, et al. Fenobody: A ferritin-displayed nanobody with high apparent affinity and half-life extension. Anal Chem. (2018) 90:5671–7. doi: 10.1021/acs.analchem.7b05217
32. Yassine HM, Boyington JC, McTamney PM, Wei C-J, Kanekiyo M, Kong W-P, et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat Med. (2015) 21:1065–70. doi: 10.1038/nm.3927
33. Kanekiyo M, Wei C-J, Yassine HM, McTamney PM, Boyington JC, Whittle JRR, et al. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature. (2013) 499:102–6. doi: 10.1038/nature12202
34. Sun W, He L, Zhang H, Tian X, Bai Z, Sun L, et al. The self-assembled nanoparticle-based trimeric RBD mRNA vaccine elicits robust and durable protective immunity against SARS-CoV-2 in mice. Signal Transduct Target Ther. (2021) 6:340. doi: 10.1038/s41392-021-00750-w
35. Mu Z, Wiehe K, Saunders KO, Henderson R, Cain DW, Parks R, et al. mRNA-encoded HIV-1 Env trimer ferritin nanoparticles induce monoclonal antibodies that neutralize heterologous HIV-1 isolates in mice. Cell Rep. (2022) 38:110514. doi: 10.1016/j.celrep.2022.110514
36. Gote V, Bolla PK, Kommineni N, Butreddy A, Nukala PK, Palakurthi SS, et al. A comprehensive review of mRNA vaccines. Int J Mol Sci. (2023) 24:2700. doi: 10.3390/ijms24032700
37. Whelan S, Lucey B, and Finn K. Uropathogenic Escherichia coli (UPEC)-associated urinary tract infections: The molecular basis for challenges to effective treatment. Microorganisms. (2023) 11:2169. doi: 10.3390/microorganisms11092169
38. Eldridge GR, Hughey H, Rosenberger L, Martin SM, Shapiro AM, D'Antonio E, et al. Safety and immunogenicity of an adjuvanted Escherichia coli adhesin vaccine in healthy women with and without histories of recurrent urinary tract infections: Results from a first-in-human phase 1 study. Hum Vaccin Immunother. (2021) 17:1262–70. doi: 10.1080/21645515.2020.1834807
39. Adamo R, Grosse HW, Petsch B, Phogat S, Rauch S, Roier S, et al. inventors; nucleic acid based vaccine encoding an escherichia coli fimH antigenic polypeptide international patent application PCT/EP2023/063799. (2023).
40. Eldridge G and Martin SM. Compositions of vaccines and adjuvants and methods for the treatment of urinary tract infections. United States of America (2015). Patent Application US20150086591.
41. Percie du Sert N, Hurst V, Ahluwalia A, Alam S, Avey MT, Baker M, et al. The ARRIVE guidelines 2.0: Updated guidelines for reporting animal research. BMC Vet Res. (2020) 16:242. doi: 10.1186/s12917-020-02451-y
42. Rauch S, Jasny E, Schmidt KE, and Petsch B. New vaccine technologies to combat outbreak situations. Front Immunol. (2018) 9:1963. doi: 10.3389/fimmu.2018.01963
43. Iavarone C, O’Hagan DT, Yu D, Delahaye NF, and Ulmer JB. Mechanism of action of mRNA-based vaccines. Expert Rev Vaccines. (2017) 16:871–81. doi: 10.1080/14760584.2017.1355245
44. Andersen TE, Khandige S, Madelung M, Brewer J, Kolmos HJ, and Møller-Jensen J. Escherichia coli uropathogenesis in vitro: Invasion, cellular escape, and secondary infection analyzed in a human bladder cell infection model. Infect Immun. (2012) 80:1858–67. doi: 10.1128/iai.06075-11
45. Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. (1970) 227:680–5. doi: 10.1038/227680a0
46. Lee NK, Cho S, and Kim I-S. Ferritin – a multifaceted protein scaffold for biotherapeutics. Exp Mol Med. (2022) 54:1652–7. doi: 10.1038/s12276-022-00859-0
47. Baden L R, El Sahly Hana M, Essink B, Kotloff K, Frey S, Novak R, et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med. (2021) 384:403–16. doi: 10.1056/NEJMoa2035389
48. Polack F P, Thomas Stephen J, Kitchin N, Absalon J, Gurtman A, Lockhart S, et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med. (2020) 383:2603–15. doi: 10.1056/NEJMoa2034577
49. Anderson BR, Muramatsu H, Jha BK, Silverman RH, Weissman D, and Karikó K. Nucleoside modifications in RNA limit activation of 2′-5′-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. (2011) 39:9329–38. doi: 10.1093/nar/gkr586
50. Kremsner PG, Mann P, Kroidl A, Leroux-Roels I, Schindler C, Gabor JJ, et al. Safety and immunogenicity of an mRNA-lipid nanoparticle vaccine candidate against SARS-CoV-2. Wien Klin Wochenschr. (2021) 133:931–41. doi: 10.1007/s00508-021-01922-y
51. Stuart L M. In gratitude for mRNA vaccines. N Engl J Med. (2021) 385:1436–8. doi: 10.1056/NEJMcibr2111445
52. Thomas S J, Moreira Edson D, Kitchin N, Absalon J, Gurtman A, Lockhart S, et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine through 6 months. N Engl J Med. (2021) 385:1761–73. doi: 10.1056/NEJMoa2110345
53. Walsh E E, Frenck Robert W, Falsey Ann R, Kitchin N, Absalon J, Gurtman A, et al. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N Engl J Med. (2020) 383:2439–50. doi: 10.1056/NEJMoa2027906
54. Zepeda-Cervantes J, Ramírez-Jarquín JO, and Vaca L. Interaction between virus-like particles (VLPs) and pattern recognition receptors (PRRs) from dendritic cells (DCs): Toward better engineering of VLPs. Front Immunol. (2020) 11:1100. doi: 10.3389/fimmu.2020.01100
55. Kelly HG, Tan HX, Juno JA, Esterbauer R, Ju Y, Jiang W, et al. Self-assembling influenza nanoparticle vaccines drive extended germinal center activity and memory B cell maturation. JCI Insight. (2020) 5. doi: 10.1172/jci.insight.136653
56. Cao S, Ma D, Ji S, Zhou M, and Zhu S. Self-assembled ferritin nanoparticles for delivery of antigens and development of vaccines: From structure and property to applications. Molecules. (2024) 29. doi: 10.3390/molecules29174221
57. Huttner A, Hatz C, van den Dobbelsteen G, Abbanat D, Hornacek A, Frölich R, et al. Safety, immunogenicity, and preliminary clinical efficacy of a vaccine against extraintestinal pathogenic Escherichia coli in women with a history of recurrent urinary tract infection: A randomised, single-blind, placebo-controlled phase 1b trial. Lancet Infect Dis. (2017) 17:528–37. doi: 10.1016/S1473-3099(17)30108-1
58. Chorro L, Ciolino T, Torres Caresse L, Illenberger A, Aglione J, Corts P, et al. A cynomolgus monkey E. coli urinary tract infection model confirms efficacy of new FimH vaccine candidates. Infect Immun. (2024) 92:e00169–24. doi: 10.1128/iai.00169-24
59. Schwarz TF, Kocken M, Petäjä T, Einstein MH, Spaczynski M, Louwers JA, et al. Correlation between levels of human papillomavirus (HPV)-16 and 18 antibodies in serum and cervicovaginal secretions in girls and women vaccinated with the HPV-16/18 AS04-adjuvanted vaccine. Hum Vaccin. (2010) 6:1054–61. doi: 10.4161/hv.6.12.13399
60. Pattyn J, Van Keer S, Téblick L, Van Damme P, and Vorsters A. Non-invasive assessment of vaccine-induced HPV antibodies via first-void urine. Front Immunol. (2020) 11:1657. doi: 10.3389/fimmu.2020.01657
61. Stærk K, Grønnemose RB, Nielsen TK, Petersen NA, Palarasah Y, Torres-Puig S, et al. Escherichia coli type-1 fimbriae are critical to overcome initial bottlenecks of infection upon low-dose inoculation in a porcine model of cystitis. Microbiol (Reading). (2021) 167:1101. doi: 10.1099/mic.0.001101
62. Hirai T and Yoshioka Y. Considerations of CD8(+) T cells for optimized vaccine strategies against respiratory viruses. Front Immunol. (2022) 13:918611. doi: 10.3389/fimmu.2022.918611
63. Park HJ, Bang YJ, Kwon SP, Kwak W, Park SI, Roh G, et al. Analyzing immune responses to varied mRNA and protein vaccine sequences. NPJ Vaccines. (2023) 8:84. doi: 10.1038/s41541-023-00684-0
64. Wu Y, Zhang H, Meng L, Li F, and Yu C. Comparison of immune responses elicited by SARS-CoV-2 mRNA and recombinant protein vaccine candidates. Front Immunol. (2022) 13:906457. doi: 10.3389/fimmu.2022.906457
65. Wu J and Abraham SN. The roles of T cells in bladder pathologies. Trends Immunol. (2021) 42:248–60. doi: 10.1016/j.it.2021.01.003
66. Corleis B, Hoffmann D, Rauch S, Fricke C, Roth N, Gergen J, et al. Efficacy of an unmodified bivalent mRNA vaccine against SARS-CoV-2 variants in female small animal models. Nat Commun. (2023) 14:816. doi: 10.1038/s41467-023-36110-1
67. Rousseau M, Lacerda Mariano L, Canton T, and Ingersoll MA. Tissue-resident memory T cells mediate mucosal immunity to recurrent urinary tract infection. Sci Immunol. (2023) 8:eabn4332. doi: 10.1126/sciimmunol.abn4332
68. Parga-Vidal L, van Aalderen MC, Stark R, and van Gisbergen KPJM. Tissue-resident memory T cells in the urogenital tract. Nat Rev Nephrol. (2022) 18:209–23. doi: 10.1038/s41581-021-00525-0
69. Xu G, Li Y, Lu G, and Xie D. Tissue-resident memory T cells in urinary tract diseases. Front Immunol. (2025) 16:1535930. doi: 10.3389/fimmu.2025.1535930
70. Lacerda Mariano L and Ingersoll MA. The immune response to infection in the bladder. Nat Rev Urol. (2020) 17:439–58. doi: 10.1038/s41585-020-0350-8
71. Karikó K, Buckstein M, Ni H, and Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. (2005) 23:165–75. doi: 10.1016/j.immuni.2005.06.008
72. Karikó K, Muramatsu H, Welsh FA, Ludwig J, Kato H, Akira S, et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther. (2008) 16:1833–40. doi: 10.1038/mt.2008.200
73. Bernard MC, Bazin E, Petiot N, Lemdani K, Commandeur S, Verdelet C, et al. The impact of nucleoside base modification in mRNA vaccine is influenced by the chemistry of its lipid nanoparticle delivery system. Mol Ther Nucleic Acids. (2023) 32:794–806. doi: 10.1016/j.omtn.2023.05.004
74. Khlebnikova A, Kirshina A, Zakharova N, Ivanov R, and Reshetnikov V. Current progress in the development of mRNA vaccines against bacterial infections. Int J Mol Sci. (2024) 25. doi: 10.3390/ijms252313139
Keywords: mRNA vaccine, uropathogenic E. coli, UPEC, UPEC vaccine, FimH, nanoparticle, urinary tract infection
Citation: Roier S, Adamo R, Rosini R, Pezzicoli A, Scarselli M, Petsch B, Jasny E and Rauch S (2025) An mRNA-based FimH nanoparticle vaccine against uropathogenic Escherichia coli is highly immunogenic in rodents. Front. Immunol. 16:1668937. doi: 10.3389/fimmu.2025.1668937
Received: 18 July 2025; Accepted: 17 October 2025;
Published: 03 November 2025.
Edited by:
Ebony Gary, Wistar Institute, United StatesReviewed by:
Alex Odoom, University of Ghana, GhanaArjun Ramamurthi, Children’s Hospital of Philadelphia Research Institute, United States
Copyright © 2025 Roier, Adamo, Rosini, Pezzicoli, Scarselli, Petsch, Jasny and Rauch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sandro Roier, U2FuZHJvLlJvaWVyQGN1cmV2YWMuY29t
†Present addresses: Roberto Adamo, GSK Vaccine Institute for Global Health, Siena, Italy
Benjamin Petsch, Cipla Europe NV, Unterhaching, Germany
Susanne Rauch, Cipla Holding B.V., Bilthoven, Netherlands
 Sandro Roier
Sandro Roier Roberto Adamo
Roberto Adamo Roberto Rosini
Roberto Rosini Alfredo Pezzicoli
Alfredo Pezzicoli Maria Scarselli
Maria Scarselli Benjamin Petsch
Benjamin Petsch Edith Jasny1
Edith Jasny1 Susanne Rauch
Susanne Rauch