- Key Laboratory of Artificial Organs and Computational Medicine in Zhejiang Province, Shulan International Medical College, Zhejiang Shuren University, Hangzhou, China
Introduction: Toll-like receptors (TLRs) are transmembrane proteins essential for innate immunity. While vertebrate TLRs have been well studied, knowledge of their distribution, structure, and function in mollusks remains limited, despite their importance in pathogen defense and environmental adaptation.
Methods: This review synthesizes current findings on molluscan TLRs, focusing on their structural features, tissue-specific expression, immune functions, and responses to environmental pollutants.
Results: Molluscan TLRs exhibit broad distribution and notable structural diversity. Their expression is tissue-dependent and can be modulated by pathogenic and environmental stressors. Although divergent from vertebrate TLRs in sequence homology, they share functional parallels in innate immune signaling. Evidence also indicates that pollutant exposure can alter TLR expression, influencing immune capacity.
Discussion: Molluscan TLRs provide insight into species-specific immune strategies and hold potential for applications in disease management and ecological monitoring. Continued research will enhance understanding of innate immunity and support progress in comparative immunology.
1 Introduction
Traditionally, the immune system is divided into innate immunity and adaptive immunity, each with distinct functions and roles (1, 2). Innate immunity serves as the host’s first line of defense against microbial pathogen invasion. It is characterized by reliance on a limited set of receptors to detect invading pathogens, initiating rapid responses that typically trigger protective inflammatory reactions within minutes of pathogen exposure (3). Innate immunity is the sole defense mechanism by which invertebrates protect themselves against microbial pathogen invasion (4–6). Pattern recognition receptors (PRRs) constitute a crucial part of innate immunity, tasked with identifying a broad range of pathogen-associated molecular patterns (PAMPs) (7, 8). Certain PRRs bind directly to PAMPs, whereas others recognize molecules derived from PAMPs (7, 8). Mollusks possess diverse types of PRRs, such as C-type lectins and galectins, Gram-negative bacteria-binding proteins, scavenger receptors, peptidoglycan recognition proteins, and Toll-like receptors (TLRs) (9).
TLRs are widely distributed across many species and have been extensively studied in the innate immune systems of animals ranging from insects to humans (10, 11). The earliest identified TLR is Toll-1, which was recognized for its role in establishing dorsal-ventral polarity during embryonic development in Drosophila melanogaster (12–15). Currently, different numbers of TLRs have been identified in vertebrates. For example, 10 functional TLRs have been characterized in humans, 12 in mice, and 22 in chickens (16, 17). TLRs have a broad recognition spectrum and can detect diverse PAMPs derived from various microorganisms, including bacteria, viruses, protozoa, and fungi (1, 4). In vertebrates, upon recognizing PAMPs, TLRs activate intracellular signaling pathways that trigger innate immune responses, a critical step for initiating subsequent adaptive immunity (1). Mollusks rely almost entirely on innate immune mechanisms, which has led to remarkable diversity of TLRs within their immune systems. This diversity may be attributed to various evolutionary processes, including retrotransposition, gene duplication, high gene expansion rates, and alternative splicing of transcripts (4, 6). Since TLRs have been extensively studied in vertebrates, it is particularly important to investigate TLRs in mollusks, where research remains relatively limited.
Mollusks are classified into multiple classes based on their morphology and ecological habits, among which the most well-known are Gastropoda, Bivalvia, and Cephalopoda. Gastropods alone comprise over 100,000 extant species, accounting for approximately 80% of all mollusks (18). In the global marine aquaculture market, mollusk farming holds a significant share of total production, especially in the cultivation of edible shellfish (19). However, pathogen infections pose a significant threat to mollusk aquaculture (20). The primary causes of mortality include viruses, bacteria, and other microorganisms (20). Pathogens not only affect animal production but also pose potential risks to consumer health (21). In responding to these pathogens, studying TLRs in mollusks is particularly important. Since the vast majority of mollusks are marine filter feeders frequently exposed to various aquatic pathogens, their TLRs represent a meaningful subject for research (22).
This review aims to explore the diversity of TLR genes across different mollusk species and their critical roles in immune responses, further elucidating their potential applications in disease prevention and control within aquaculture. With ongoing advancements in understanding TLR downstream signaling pathways, research on TLRs holds promise for enhancing disease resistance and farming efficiency in cultured species. To our knowledge, this is the first systematic review applying the PRISMA framework to molluscan TLRs, highlighting their unique immunological features and aquaculture implications. Ultimately, these insights contribute to strengthening disease resilience in aquaculture systems and promoting sustainable industry development in harmony with broader socio-economic progress.
2 Method approach
A systematic review was conducted following the PRISMA 2020 guidelines. The objective was to synthesize recent findings on molluscan TLRs and the MyD88 signaling pathway and to assess their relevance to aquaculture. Literature published between January 2020 and May 2025 was retrieved from three databases: Web of Science, ScienceDirect, and PubMed. The following search terms and their plural forms were applied in various combinations: (“Toll-like receptor” OR “TLR”) AND (“mollusk” OR “mollusc”) AND (“functional analysis” OR “genome-wide” OR “whole-genome”) OR (“MyD88 pathway”).
After removing duplicates, records were screened based on predefined inclusion and exclusion criteria. Studies were included if they focused on mollusks, reported experimental data on TLRs or the MyD88 pathway, and were published in English in peer-reviewed journals. Exclusion criteria comprised duplicate publications, non-English articles, academic theses, technical or short reports, articles lacking original data, and studies with unavailable full texts. For each eligible article, information was extracted on publication year, mollusk species studied, novel TLRs identified or whole-genome analyses, downstream signaling pathways investigated, and key research findings (Table 1).
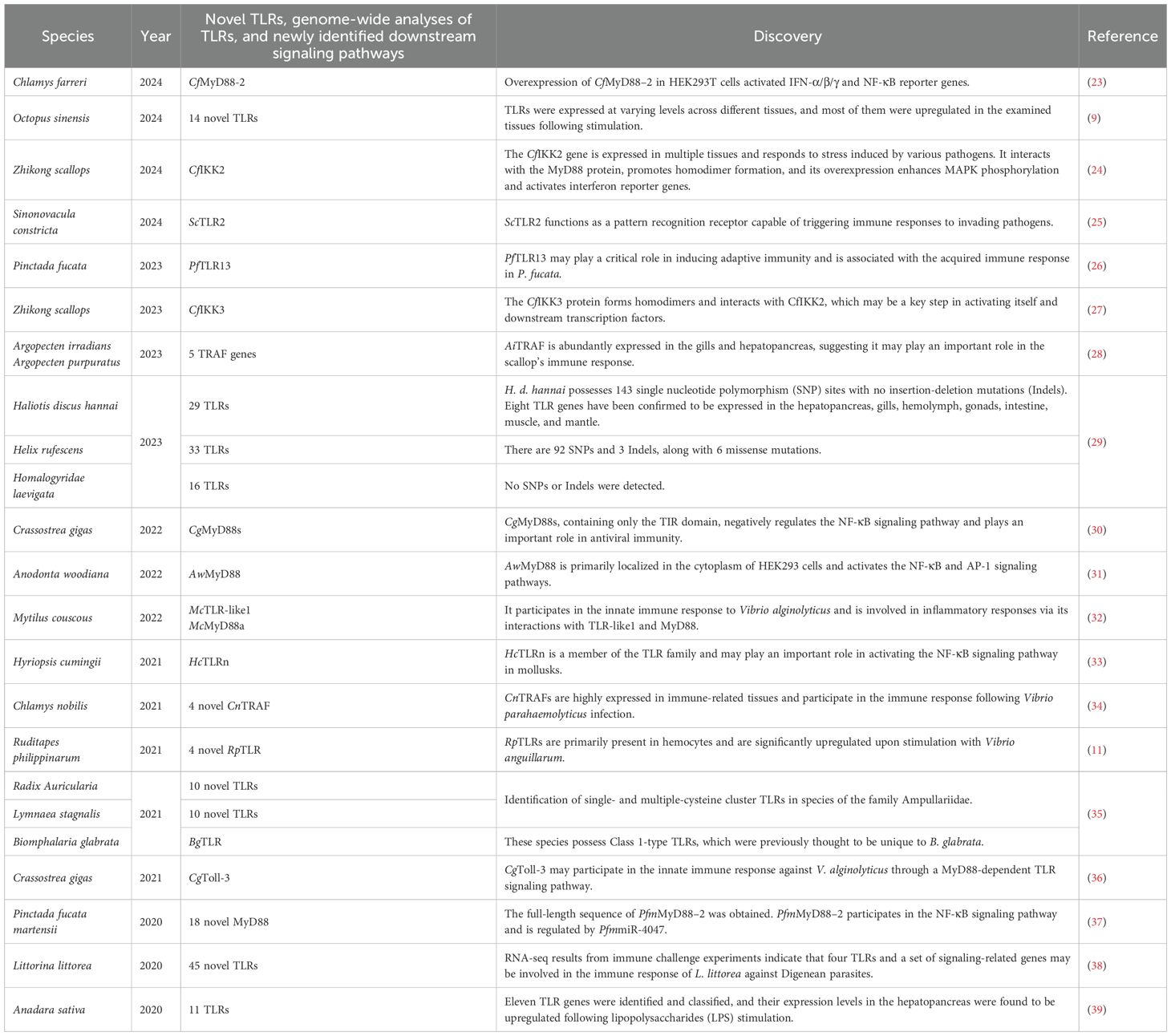
Table 1. Recently discovered novel TLRs and signaling pathways in mollusks, along with some whole-genome analyses.
As the included studies were mostly descriptive and experimental, rather than randomized trials, no formal quality assessment was performed. The initial search yielded 694 records. After duplicate removal and application of exclusion criteria, 19 articles covering 24 mollusk species were retained for final analysis (Figure 1). The PRISMA flow diagram details the number of records excluded at each stage and the reasons for exclusion. This systematic review integrates these studies to provide a comprehensive overview of molluscan TLRs, their signaling pathways, and their functional implications for immune defense and aquaculture.
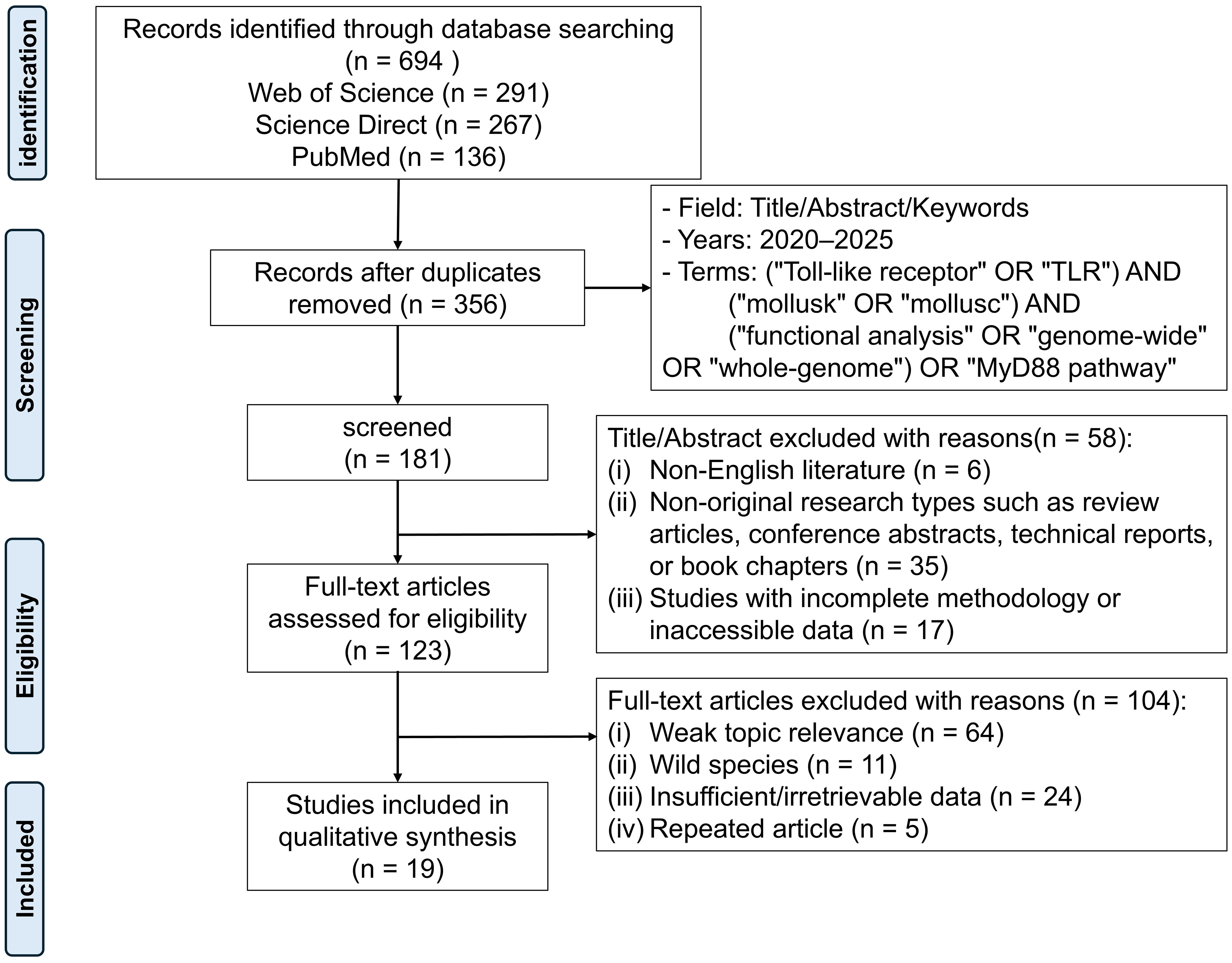
Figure 1. The PRISMA flow diagram illustrates the screening process of studies related to “TLR” and the “MyD88” pathway in mollusks, retrieved from the Web of Science, Science Direct, and PubMed databases between 2020 and May 2025. A total of 694 articles were initially identified. After removing duplicates, screening abstracts, and reviewing full texts, 19 eligible studies were finally included for analysis.
3 Structure and function of TLRs in mollusks
3.1 Unique structural differences of TLRs in mollusks
Upon recognition of external stimuli, TLRs initiate signaling cascades that activate downstream molecules, contributing to the innate immune response. Since innate immunity serves as the nearly sole immune mechanism in invertebrates, the robust immune defense systems of marine bivalves and other mollusks are crucial for adapting to the complex marine environment (40).
TLRs belong to the type I transmembrane protein family and consist of three main components: the extracellular region, which includes leucine-rich repeat (LRR) motifs forming a folded solenoid structure responsible for recognizing PAMPs; and the intracellular region, which contains a Toll/interleukin-1 receptor (TIR) domain that recruits various adaptor proteins to activate downstream signaling pathways (4). Known LRR family proteins contain multiple LRR motifs, ranging from as few as 2–3 to more than 40 (41). The dimerized LRR region forms an ‘m’-shaped structure, as shown in Figure 2, which is also compared with the TLR structures observed in vertebrates. The intracellular In addition to canonical TIR domains, researchers have identified a group of TIR-domain containing (TIR-DC) proteins in bivalve mollusks. These proteins exhibit high molecular diversity and may participate in multiple immune-related signaling pathways as well as embryonic development (42). These TIR-DC proteins exhibit significant molecular diversity among different species, and variable domain architectures and species-specific expansions are observed in different species (42). Phylogenetic analysis indicates that these proteins may have evolved through specific gene duplication and domain rearrangement events unique to mollusks (42). The experimental evidence regarding the functions of TIR-DC is still limited. Further research is needed, using methods such as RNA interference or CRISPR/Cas9 (once optimized in Pyrrhocoris apterus), to clarify their biological roles. The unique domain structure and expression profile of TIR-DC proteins highlight their potential as specific adaptors for the innate immune system in invertebrates, and provide a promising avenue for discovering new immune regulatory mechanisms.
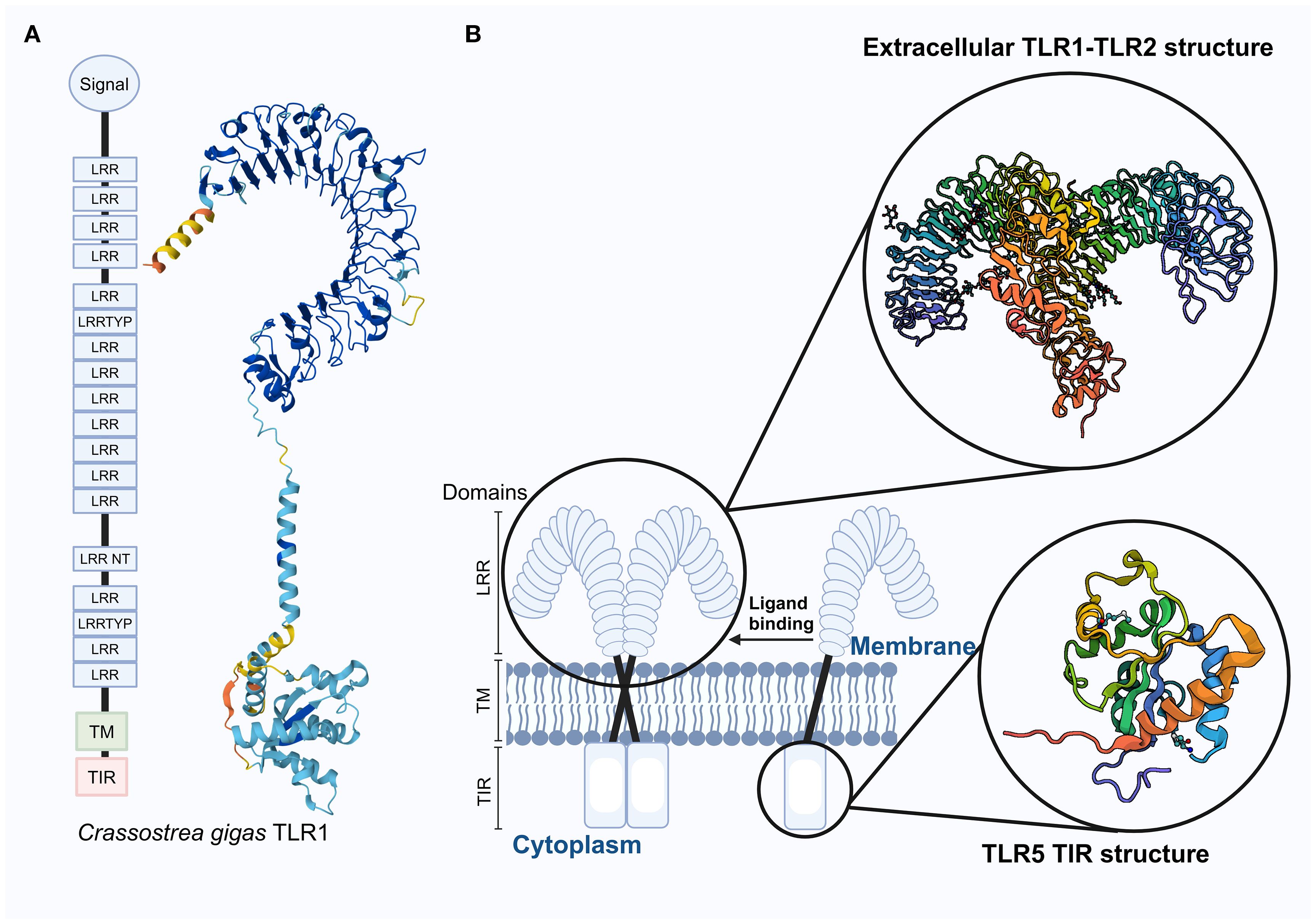
Figure 2. Distribution of the CgTLR1 protein structure in the Crassostrea gigas with 3D prediction, along with a schematic diagram of the transmembrane structure of TLRs in vertebrates. (A) Functional domains of CgTLR1 are adapted from (12), and the 3D structure on the right was predicted using AlphaFold2; it has not been experimentally validated in mollusks. (B) Schematic representation of vertebrate TLRs, showing the localization of TLR receptors within the cell membrane. The extracellular structure of the TLR1–TLR2 complex and the TIR domain of TLR5 are experimentally determined in humans (from NCBI). These vertebrate structures are shown for comparison only and should not be assumed to directly represent mollusk TLRs due to potential structural differences. Enlarged sections highlight the TLR1–TLR2 complex and the TIR domain of TLR5. Abbreviations: LRR, Leucine-rich repeat; LRRTYP, Leucine-rich repeat typical; LRR NT, Leucine-rich repeat N-terminal; TM, Transmembrane domain; TIR, Toll/Interleukin-1 receptor domain.
Although research remains focused on a limited number of species, it is widely believed that TLR structures exhibit significant diversity in mollusks (22, 40). In invertebrates, the number and types of TLR family members vary across species, ranging from a single member to several hundred. To the best of our knowledge, several mollusk species have been identified their existing TLRs through whole-genome analyses, including but not limited to the following: Strongylocentrotus purpuratus has been reported to possess 222 TLR genes (43), Haliotis discus hannai contains 29 TLR genes (29), Hippeutis rufescens harbors 33 TLR genes (29), Hippeutis laevigata possesses encodes 16 (29), Biomphalaria glabrata possesses is known to have 56 TLR genes (44, 45), C. gigas 83 (44), and Chlamys farreri includes only 2 TLR genes (44). Additionally, we compiled newly identified TLRs and signaling pathways in mollusks from the past five years, as summarized in Table 1. The notable diversity exhibited by TLRs reflects the need for invertebrates to cope with increasingly complex and diverse environmental challenges and pathogenic pressures. Expansion of TLR families, particularly in bivalves, appears to result from multiple independent gene duplication events—especially tandem duplications—with frequent lineage-specific expansions occurring before and after bivalve radiation (22). These duplicated genes often experienced diversifying (i.e., positive) selection in their extracellular domains, enhancing variability in ligand‐binding regions and likely broadening the range of recognized pathogens (22). In addition, the broader evolution of TLRs across the Metazoa has been shaped by lineage‐specific expansions and losses, coupled with episodic positive selection in extracellular recognition domains—factors that collectively drive functional diversification in this receptor family (46). For instance, research on shallow-water mussels (Mytilus galloprovincialis) adapting to deep-sea conditions revealed differential expression of TLRs, suggesting that their expression and functional roles are modulated by varying environmental pressures (47). Similarly, in scallops (C. farreri), TLR genes are differentially upregulated upon bacterial and viral challenge via MyD88-dependent pathways, regulating antimicrobial peptide expression, and in abalone (Haliotis cumingii), HcToll1–3 are upregulated in response to pathogen stimulation, contributing to immune defense (48, 49). These examples collectively indicate that TLR diversity in mollusks reflects not only expansion in number but also diversification of function. Studies have shown that the TLR domains of green-lipped abalone and oysters share certain similarities, which may result from evolutionary divergence within the phylum Mollusca. Additionally, the presence of the TIR domain in both Haliotis laevigata and C. gigas indicates a degree of conserved homology among mollusk TLRs (50).
As a crucial component of the ancient immune system, TLRs are widely distributed across various organisms. The origin of TLRs predates the emergence of metazoans, and a single TLR gene encoding a receptor with a canonical structure exists in all metazoans (22). However, due to adaptive changes during evolution, we expect that mollusk TLRs exhibit certain differences in structure, function, and distribution compared to other species, and these differences may be associated to their ecological and biological characteristics. Compared to vertebrates, mollusk TLRs display greater multifunctionality and flexibility, which may stem from their extensive expansion and molecular diversification (6). This phenomenon may be related to gene duplication and domain rearrangement, among which tandem duplication frequently occurs in molecules associated to innate immunity (6, 31). Phylogenetic analyses indicate that the interactions between pathogens and the immune system have evolved over time, with selective pressures from specific pathogen recognition driving the optimization of TLR genes through both positive and negative selection (51). The early diversification of TLRs led to two major forms of the protein: in protostomes such as mollusks, TLRs typically contain multiple cysteine clusters, whereas in other deuterostomes, TLRs more commonly possess a single cysteine cluster (52, 53).
Studies ranging from invertebrates to mammals have shown that both TLR and NF-κB genes are widely present in the genomes of many animals, indicating that the NF-κB signaling pathway also has an ancient evolutionary origin and plays a vital role in the regulation of the immune system (52, 53). Sequence alignment based on amino acid sequences indicates that downstream genes of TLRs are more conserved across different species (12). This suggests that invertebrates may have undergone gene loss and diversification during the evolution of their immune systems.
3.2 Signaling pathway
TLRs are typical PRRs that recognize foreign pathogens, and their activation represents the first line of defense in the immune system. In vertebrates, TLR activation can recruit and activate neutrophils and macrophages (54). The TLR signaling pathway has been relatively well studied in humans, as shown in Figure 3A. The TLR family primarily functions through two signaling pathways: the MyD88-dependent pathway and the MyD88-independent pathway (also known as the TRIF-dependent pathway). In the MyD88-dependent pathway, MyD88 binds to TLRs through its TIR domain and recruits interleukin-1 receptor-associated kinase 4 (IRAK4). IRAK4 then phosphorylates and activates IRAK1/2 and TRAF6 (22, 57). Additionally, the MyD88 structure interacts with the TLR-TIR domain to activate the transcription factors AP-1 and NF-κB, thereby triggering inflammatory responses and initiating adaptive immunity (22, 57, 58). In the TRIF-dependent pathway, TRAM and other adaptor molecules interact with downstream components, including NF-κB, MAP kinases, and interferon regulatory factors (IRF3, IRF5, and IRF7) (59). Ultimately, the TLR signaling pathway triggers the expression of interferon-induced cytokines and various gene transcripts (59). However, due to the extensive diversity of TLRs in invertebrates, their mechanisms remain incompletely understood. The partially characterized TLR signaling pathways in mollusks are illustrated in Figure 3B. The TLR signaling network plays a critical role in immune defense by directly recognizing pathogens and PAMPs. Commonly used PAMPs include lipopolysaccharides (LPS), polyinosinic-polycytidylic acid [poly(I:C)], and peptidoglycan, which mimic bacterial and viral infections (60). For example, LPS stimulation has been shown to upregulate TLR transcripts in C. gigas and mussels M. galloprovincialis, suggesting a role in bacterial recognition and immune activation (61, 62). Similarly, poly(I:C), a synthetic analog of viral double-stranded RNA, significantly induces the expression of antiviral genes, including TLRs, in various mollusks (63). These PAMP-based stimulations are commonly used to dissect innate immune pathways and to investigate the functional responsiveness of molluscan TLRs. However, despite the increasing use of these stimuli, the downstream signaling mechanisms remain only partially elucidated, highlighting the need for further research on receptor-ligand specificity, signal transduction components, and cell-type-specific responses.
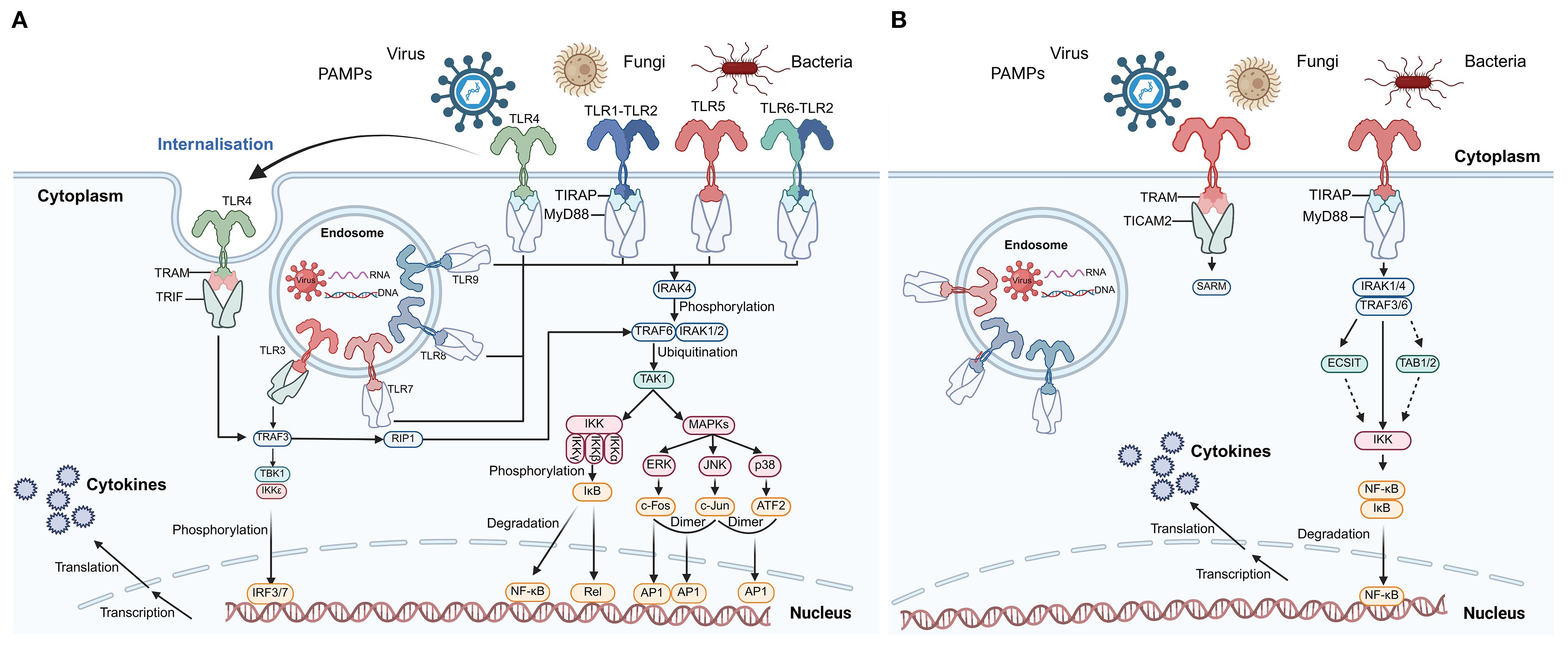
Figure 3. TLR signaling pathways in humans and C. farreri. (A) Human TLR signaling pathways. This panel illustrates the canonical TLR signaling cascades in humans, including both cell surface and endosomal receptors. Recognition of PAMPs from viruses, fungi, and bacteria activates TLRs, leading to recruitment of adaptor proteins such as MyD88, TRIF, TIRAP, and TRAM. Downstream signaling involves IRAKs, TRAFs, TAK1, the IKK complex, and MAPKs, culminating in the activation of transcription factors NF-κB, AP-1, and IRFs, which translocate to the nucleus to induce cytokine gene expression. Phosphorylation and ubiquitination events, as well as receptor internalization, are indicated. This panel is adapted from (55) and serves as a reference for vertebrate TLR signaling. (B) Putative TLR signaling pathways in C. farreri. This panel depicts the partially characterized TLR signaling in the mollusk C. farreri. Known components include TLR receptors, adaptor proteins (MyD88, TIRAP, TRAM, TICAM2), IRAKs, TRAFs, ECSIT, TAB1/2, and IKK, leading to NF-κB activation and cytokine production. Solid arrows represent interactions supported by experimental evidence, while dashed arrows indicate putative or indirect interactions. Pathway components were inferred based on two previous studies (10, 56). Abbreviations: PAMPs, pathogen-associated molecular patterns; NF-κB, nuclear factor kappa B; IRF, interferon regulatory factor; AP-1, activator protein 1; IKK, IκB kinase complex; MAPKs, mitogen-activated protein kinases; TRAF, TNF receptor-associated factor; IRAK, IL-1 receptor-associated kinase; TAK1, transforming growth factor-β-activated kinase 1; ECSIT, evolutionarily conserved signaling intermediate in Toll pathways; TAB, TAK1-binding protein.
Recent studies have revealed a significant expansion of the MyD88 gene in mollusks, suggesting its potential functional diversity. For example, 19 MyD88-like genes have been identified in Pomacea canaliculata, 12 in Haliotis rufescens, 13 in C. farreri, 23 in Mizuhopecten yessoensis, 10 in C. gigas, and 7 in B. glabrata (6). In C. gigas, 27 proteins potentially interacting with TLRs have been identified, including three distinct MyD88 homologs, indicating that the TLR signaling pathway may involve a complex regulatory mechanism (64). In C. farreri, IKK proteins have been shown to play a key role in MyD88-mediated immune signaling, further supporting the functional conservation of the TLR pathway in mollusks as well as its potential lineage-specific regulation (65).
In mollusks, TLR-mediated inflammation plays a pivotal role in the innate immune response, particularly during the early recognition and elimination of pathogens. As a coordinated defense mechanism, this inflammatory response is essential for resisting infection (66). In recent years, studies on immune regulatory molecules in mollusks have been increasing. One such example is Octominin, a bioactive peptide derived from Octopus minor, which has been shown to effectively inhibit NF-κB transcriptional activation in LPS-stimulated RAW264.7 cells. It also dose-dependently reduces TLR4 mRNA expression, thereby suppressing TLR4-mediated inflammatory responses (66). This suggests that certain specific peptide molecules in mollusks can not only modulate TLR-mediated immune signaling, but also help maintain immune homeostasis through their immunosuppressive effects.
3.3 Expression of TLRs in mollusk tissues
Complete genome sequences have revealed a full set of PRRs in bivalves, which are generally associated to host defense against infections. For instance, in Octopus sinensis, TLRs are predominantly localized to the plasma membrane and show tissue-specific differential expression in hemolymph, white body, gills, and hepatopancreas, as revealed by RT-qPCR analysis (9). A separate study on H. d. hannai revealed that eight TLR genes are expressed across the hepatopancreas, gills, hemolymph, gonads, intestine, muscle, and mantle tissues. Notably, five TLRs were expressed in the gills, three in the hepatopancreas, and three in the hemolymph, with their expression levels significantly upregulated following V. parahaemolyticus infection (29).
In C. farreri, studies have shown that components of the TLR signaling pathway are constitutively expressed across various tissues, with particularly high expression in the gills and hepatopancreas (56). In the transcriptomic analysis of hemocytes and kidney tissues of L. littorea, 45 complete TLR transcripts were identified, which were derived from 35 distinct genes (38). Recent research on P. fucata revealed that the novel TLR13 (PfTLR13) mRNA is ubiquitously expressed across all tested tissues, exhibiting the highest expression in the gills (26).
Furthermore, research has highlighted the involvement of TLR genes in mollusk development, which carries important significance for aquaculture practices. For instance, in Haliotis diversicolor, the MyD88 homolog gene (hdMyD88) exhibits ubiquitous expression throughout different embryonic stages (57). HdMyD88 not only activates various downstream responses such as antioxidation, antibacterial activity, and apoptosis, but also plays a key role in immunity and development, which holds potential applications for improving survival rates and health status of abalone in aquaculture (57). Researchers have identified five TRAF genes in Argopecten and successfully cloned TRAF6. They analyzed the genetic features and phylogenetic relationships of these genes to enhance aquaculture management and disease control strategies in scallops (28). Initial research indicates that the mRNAs of Toll pathway signaling intermediates in Crassostrea hongkongensis exhibit broad expression in all tested tissues and throughout different embryonic developmental stages, potentially participating in embryogenesis and immune responses (67). Understanding the roles of these genes in oyster development and immunity can help optimize oyster aquaculture techniques, thereby improving farming efficiency and yield.
4 Pathogen infection and immune response in mollusks
4.1 Infection by pathogens and accumulation of microplastics
Mollusks have a certain capacity for bioaccumulation of pollutants such as viruses and bacteria within their bodies. Studies have shown that clams (Ruditapes decussatus), which inhabit shallow water environments and feed by filtering plankton and organic matter from the surrounding water, accumulate a large amount of pollutants in their bodies, including toxins, antibiotic residues, bacteria, viruses, and protozoa (68). Under high-density aquaculture conditions, the accumulation of these pollutants may lead to high mortality rates in mollusks (68). Another study specifically pointed out that the pathogenic microorganisms accumulated in clams mainly include Vibrio cholerae, V. parahaemolyticus, Pseudomonas aeruginosa, Escherichia coli, and Enterococcus spp (69). Direct experimental evidence demonstrates that exposure of C. gigas to V. parahaemolyticus rapidly up-regulates CgTLR4 and activates the MyD88–NF-κB axis, leading to increased expression of antimicrobial peptides and pro-inflammatory cytokines (70). Similarly, functional studies using RNAi-mediated gene silencing have validated TLR-MyD88-dependent antibacterial responses in C. hongkongensis and M. galloprovincialis (71, 72).
Viruses typically cause widespread damage to mollusks, resulting in significant losses to the aquaculture industry. TLRs play a crucial role in combating viral infections by initiating antiviral immune responses that limit viral replication and spread. Viral infections in mollusks have been extensively studied, with the most common being Ostreid herpesvirus 1 (OsHV-1). In C. gigas, transcriptomic analyses revealed that TLR homologs, some lacking transmembrane domains, were significantly upregulated upon OsHV-1 infection, suggesting their potential involvement in antiviral responses (73). One study indicated that two genes encoding MyD88-like proteins, CgMyD88–1 and CgMyD88-2, as well as CgMyD88s, may all be involved in the TLR-mediated innate immune pathway (64). The study also found that CgMyD88s may act as a barrier to help prevent excessive inflammatory responses in C. gigas during infection with the OsHV-1 microvariant (64). Bivalves may bioaccumulate and be infected by human viruses within their tissues. Various viruses have been detected in bivalves, including members of the families Papillomaviridae, Togaviridae, Picornaviridae, Bornaviridae, and Astroviridae (74). These findings provide important guidance for disease prevention and management in the aquaculture industry. Protozoan pathogens can also activate TLR-mediated immune responses in mollusks, with Perkinsus marinus serving as a well-documented example. This parasite causes dermo disease in the Crassostrea virginica, leading to severe mortality in aquaculture. Comparative transcriptomic analyses between the resistant C. gigas and the susceptible C. virginica have revealed distinct host–parasite interactions across early and late stages of infection (75). Notably, C. virginica shows greater expansion and positive selection of TLR genes in response to P. marinus, reflecting lineage-specific diversification of pathogen recognition receptors (75). Furthermore, during early infection, oysters directly injected with P. marinus exhibited pronounced upregulation of innate immune transcripts, including TLR4 and TNF receptor-associated factor 6-like proteins, indicating that TLR-mediated signaling is rapidly mobilized under high parasite burden (76).
Furthermore, the impact of microplastics on mollusks should not be overlooked. Due to their persistence in the environment, microplastics can act as carriers for bacteria, facilitating the spread of potentially harmful pathogens to different habitats (77). Research has shown that exposure to microplastics alone can significantly inhibit the immune responses of blood cells in Anadara granosa and M. galloprovincialis, including reductions in hemoglobin concentration, total blood cell count, lysozyme activity, blood cell membrane stability, and the number of foot sacs (78, 79). At the same time, exposure alters reactive oxygen species (ROS) levels, calcium ion balance, and acid and alkaline phosphatase activities, exhibiting a clear dose-dependent pattern (78, 79). Microplastics can be internalized by cells, disrupting intracellular signaling pathways, disturbing immune homeostasis, and ultimately causing damage to tissues and organs. The generation of ROS represents a major mechanism of toxicity, which may further trigger the production of damage-associated molecular patterns (DAMPs) and is closely associated with the perturbation of TLR function, increased cytokine secretion, and inflammatory responses of immune cells (80). These findings highlight the potential threat of microplastics to mollusk health and the aquaculture industry, emphasizing the need for effective management and mitigation strategies.
4.2 Environmental stress and adaptation
TLRs ensure that cells effectively respond to environmental changes and pathogenic challenges. By initiating innate immune signaling cascades, TLRs ensure rapid and appropriate cellular responses under fluctuating environmental conditions. These receptors interact with a variety of regulatory molecules that fine-tune the magnitude and duration of signaling events, thereby maintaining immune homeostasis across diverse ecological contexts (81–83). In mollusks, the mechanisms of immune tolerance are believed to limit excessive inflammation and prevent self-damage, yet the role of TLRs in mediating such tolerance remains poorly understood due to a scarcity of systematic studies. Similar to vertebrates, molluscan TLRs are hypothesized to engage in crosstalk with other PRRs, orchestrating immune responses through synergistic activation, signal suppression, and dynamic feedback regulation, including both positive and negative loops (84, 85).
Mollusks are highly sensitive to endocrine-disrupting chemicals, which can disrupt immune homeostasis. MicroRNAs (miRNAs) play a central role in regulating TLR signaling and mediating immune responses to environmental and microbial challenges (86). For instance, in M. coruscus, LPS exposure downregulates miR-196, which normally targets McTLR-like1 mRNA, resulting in increased McTLR-like1 expression and a feedback mechanism that amplifies TLR signaling to enhance pathogen defense (87). Exposure to pollutants such as bisphenol A, 17α-ethinylestradiol, and tributyltin has been shown to alter immune-related miRNA profiles, disturb TLR pathways, and impair immune competence (88). Together, these findings highlight miRNA-TLR interactions as an adaptive mechanism enabling mollusks to cope with immunotoxic stressors in aquatic environments.
5 Disease control and ecological applications of molluscan TLRs
In the context of disease prevention and control, TLRs serve as key targets for enhancing immune responses and modulating immune memory, making them valuable for the prevention and treatment of diseases in aquaculture species. In Atlantic salmon and rainbow trout, for example, stimulation of TLR7/8 with synthetic agonists drives robust type-I IFN and pro-inflammatory cytokine production in the brain, kidney and spleen, providing a rational basis for TLR-centered vaccines (89). Beyond their canonical role in immune defense, TLRs also help mollusks cope with complex environmental stressors: activation of TLR-NF-κB pathways by thermal and hypoxic stress has been linked to improved survival under climate-change scenarios, while pollutant-induced TLR dysregulation serves as a sensitive biomarker for monitoring aquatic contaminants (90). Consequently, TLR-centered interventions are emerging as dual-purpose tools—both for aquaculture health management and for ecological risk assessment in rapidly changing environments.
5.1 Disease prevention and control
In oysters, specific TLRs have been identified that recognize PAMPs from viruses, subsequently triggering intracellular signaling cascades and inducing the production of antiviral molecules such as interferon-like proteins and antimicrobial peptides (91, 92). The use of TLR agonists to enhance immune responses in mollusks, as well as stimulating TLRs to improve vaccine efficacy, has become an important strategy for optimizing immune-based disease prevention and control (93). Nevertheless, their efficacy remains largely untested in mollusks. In vertebrates, TLR agonists such as monophosphoryl lipid A (TLR4 ligand) and CpG oligodeoxynucleotides (TLR9 ligand) have already been shown to enhance both mucosal and systemic immunity to vaccine antigens (94). Similarly, in vertebrates, synthetic TLR4 agonists can stimulate innate resistance to infectious challenges, while combinations of TLR2/6, TLR3, and TLR9 ligands have increased dendritic-cell activation and improved the protective efficacy of peptide vaccines in animal models (95). Although these mechanistic insights come largely from vertebrates, they provide a rationale for exploring TLR agonists as immunostimulants and vaccine adjuvants in molluscan aquaculture, with the understanding that their efficacy and safety in mollusks remain to be tested.
Based on studies of TLRs, vaccine adjuvants could potentially be used to enhance immune responses in mollusks and improve their immunogenicity. Protein-based adjuvants are capable of binding and activating PRRs, eliciting effective immune responses through the cooperation of innate and adaptive immune systems (96). Molluscan hemocyanins have been shown to interact with TLR4 in mammalian APCs, triggering MyD88-dependent signaling, cytokine release, and dendritic cell maturation (96). As adjuvant agonists, TLRs activating TLR signaling to promote antigen presentation, co-stimulatory signaling, and cytokine expression (97). Vaccine development in mollusks is still in its early stages, but its potential applications warrant further exploration and research. However, practical application in mollusks is currently limited by the lack of species-specific ligand detection methods, variable immune responses among mollusk species, and regulatory and cost considerations. In the future, research will need to optimize the assessment of the long-term efficacy and safety of the dosage.
5.2 Aquaculture and ecological applications
Given the important role of mollusks in the aquaculture sector, enhancing their immune capacity and reducing disease incidence have become key issues in both research and production. Although various aspects of TLRs have been described in some fish, shellfish, crustaceans (shrimps), and mollusks, their roles in other aquatic organisms remain largely unexplored (44, 98). Beyond their canonical role in immune defense, TLRs also help mollusks cope with complex environmental stressors. In the Pacific abalone H. d. hannai, transcriptomic profiling showed that both thermal and hypoxic stress rapidly up-regulate the TLR–NF-κB signalling cascade, and this activation was positively correlated with improved survival under simulated climate-change scenarios (99).
TLRs in mollusks can serve as valuable indicators for environmental and ecological monitoring. Exposure to bacterial or viral PAMPs rapidly alters TLR–MyD88–NF-κB transcript levels in hemocytes, changes that can be quantified by qPCR and exploited to optimize rearing conditions and reduce disease incidence in real time (100). Anchoring native TLR proteins on gold-nanoparticle or graphene-based electrochemical sensors has produced portable devices capable of detecting picogram-per-milliliter concentrations of LPS and viral dsRNA in seawater within minutes, providing farmers with precise, on-site data for timely management decisions (101).
Airborne pollutants may directly interact with receptors or indirectly modulate TLR signaling activation by generating secondary mediators such as DAMPs or PAMPs (102). Transplantation of TLR-hyper-responsive Ruditapes philippinarum into PAH-contaminated estuaries accelerated sediment microbial recovery and reduced pollutant loads by 28% within 90 days, demonstrating a practical mollusk-based strategy for ecological restoration (103). Nevertheless, the widespread application of TLR-based monitoring and interventions in aquaculture still faces challenges including species-specific variability, the need for standardized assays, cost, and regulatory constraints. Analyzing molluscan TLR expression in response to environmental pollutants may inform the development of immune-based strategies for ecological restoration, enhancing ecosystem resilience.
6 Conclusions and perspectives
Aquatic environments are important sources of pathogenic microorganisms, providing favorable conditions for the growth of bacteria, fungi, viruses, and other aquatic microbes, which can lead to mortality in aquatic animals. Mollusks, as representative aquatic organisms, ingest these pathogens and form part of the food chain, thereby posing certain risks to global economic development and human health. At present, due to the diversity of the TLR gene family, significant interspecies differences exist, and the expression and function of TLRs in mollusks are influenced by various factors. Therefore, further research is needed to broaden our understanding of TLRs in mollusks. Molluscan TLRs may also have the potential to be selectively activated by specific ligands or agonists. Similar to vertebrates, these ligands or agonists can bind to and activate TLRs, thereby enhancing immune responses in a manner comparable to vaccine adjuvants. However, this possibility still requires validation in mollusks. Research priorities should focus on addressing major knowledge gaps in molluscan TLR studies, such as the lack of functional validation for many identified TLR genes, interspecies differences, and the establishment of ligand screening and identification methods. Advancing studies on molluscan TLRs will be of significant value in enhancing our understanding of species-specific immune mechanisms and promoting progress in comparative immunology.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Author contributions
HL: Supervision, Methodology, Data curation, Conceptualization, Writing – review & editing, Investigation. XL: Validation, Methodology, Investigation, Data curation, Software, Writing – original draft, Funding acquisition. YHZ: Writing – original draft, Conceptualization, Investigation, Data curation. JX: Project administration, Formal Analysis, Methodology, Writing – review & editing. QZ: Visualization, Resources, Validation, Writing – review & editing. XC: Visualization, Validation, Supervision, Writing – review & editing. YJZ: Writing – review & editing, Supervision, Investigation, Software. LZ: Data curation, Writing – review & editing, Investigation, Methodology. YC: Investigation, Methodology, Writing – review & editing, Conceptualization. RW: Project administration, Writing – review & editing, Data curation, Resources. KC: Writing – review & editing, Investigation, Methodology, Software.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This research was funded by the National Natural Science Foundation of China (Grant No: 32000293); Guangxi Natural Science Foundation (Grant Nos: 2020JJA130077 and 2018JJB140423); the University Level Scientific Research Project of Zhejiang Shuren University (Grant No: 2022R064); Zhejiang Shuren University Basic Scientific Research Special Funds (Grant No: 2024XZ014); 2025 Provincial Undergraduate Innovation and Entrepreneurship Training Program (Grant No: S202511842050).
Acknowledgments
We thank all the participants in this study. We sincerely thank BioRender for providing the tools and support that helped us create the figures in this review. Their platform enabled us to effectively visualize complex biological concepts.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
2. Medzhitov R and Janeway C Jr. Innate immunity. New Engl J Med. (2000) 343:338–44. doi: 10.1056/NEJM200008033430506
3. Turvey SE and Broide DH. Innate immunity. J Allergy Clin Immunol. (2010) 125:S24–32. doi: 10.1016/j.jaci.2009.07.016
4. Xu K, Zhang Z, Xu Z, Tang Z, Liu L, Lu Z, et al. A novel invertebrate toll-like receptor is involved in TLR mediated signal pathway of thick shell mussel Mytilus coruscus. Dev Comp Immunol. (2019) 97:11–9. doi: 10.1016/j.dci.2019.03.012
5. Hoffmann JA, Kafatos FC, Janeway CA Jr., and Ezekowitz R. Phylogenetic perspectives in innate immunity. Science. (1999) 284:1313–8. doi: 10.1126/science.284.5418.1313
6. Li Y, Xue Y, Peng Z, and Zhang L. Immune diversity in lophotrochozoans, with a focus on recognition and effector systems. Comput Struct Biotechnol J. (2023) 21:2262–75. doi: 10.1016/j.csbj.2023.03.031
7. Ertl NG, O’Connor WA, Papanicolaou A, Wiegand AN, and Elizur A. Transcriptome Analysis of the Sydney Rock Oyster, Saccostrea glomerata: Insights into Molluscan Immunity. PloS One. (2016) 11:e0156649. doi: 10.1371/journal.pone.0156649
8. Medzhitov R and Janeway CA. Innate immunity: the virtues of a nonclonal system of recognition. Cell. (1997) 91:295–8. doi: 10.1016/S0092-8674(00)80412-2
9. Chen Z, Zhou Y, Chen X, Sheng Y, Liao J, Huang Y, et al. Genome-wide identification of toll-like receptors in Octopus sinensis and expression analysis in response to different PAMPs stimulation. Fish Shellfish Immunol. (2024) 149:109591. doi: 10.1016/j.fsi.2024.109591
10. Brennan JJ and Gilmore TD. Evolutionary origins of toll-like receptor signaling. Mol Biol evolution. (2018) 35:1576–87. doi: 10.1093/molbev/msy050
11. Ren Y, Liu H, Fu S, Dong W, Pan B, and Bu W. Transcriptome-wide identification and characterization of toll-like receptors response to Vibrio Anguillarum infection in Manila clam (Ruditapes philippinarum). Fish Shellfish Immunol. (2021) 111:49–58. doi: 10.1016/j.fsi.2021.01.007
12. Zhang L, Li L, and Zhang G. A Crassostrea gigas Toll-like receptor and comparative analysis of TLR pathway in invertebrates. Fish shellfish Immunol. (2011) 30:653–60. doi: 10.1016/j.fsi.2010.12.023
13. Imler J-L and Zheng L. Biology of Toll receptors: lessons from insects and mammals. J Leucocyte Biol. (2004) 75:18–26. doi: 10.1189/jlb.0403160
14. Akira S. Pathogen recognition by innate immunity and its signaling. Proc Japan Academy Ser B. (2009) 85:143–56. doi: 10.2183/pjab.85.143
15. Kawai T and Akira S. Signaling to NF-κB by toll-like receptors. Trends Mol Med. (2007) 13:460–9. doi: 10.1016/j.molmed.2007.09.002
16. Kawai T and Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. (2010) 11:373–84. doi: 10.1038/ni.1863
17. Takeuchi O and Akira S. Pattern recognition receptors and inflammation. Cell. (2010) 140:805–20. doi: 10.1016/j.cell.2010.01.022
18. Haszprunar G and Wanninger A. Molluscs. Curr Biol. (2012) 22:R510–R4. doi: 10.1016/j.cub.2012.05.039
19. Mau A and Jha R. Aquaculture of two commercially important molluscs (abalone and limpet): existing knowledge and future prospects. Rev Aquaculture. (2018) 10:611–25. doi: 10.1111/raq.12190
20. Zannella C, Mosca F, Mariani F, Franci G, Folliero V, Galdiero M, et al. Microbial diseases of bivalve mollusks: infections, immunology and antimicrobial defense. Mar Drugs. (2017) 15:182. doi: 10.3390/md15060182
21. Prado-Carpio E, de Lourdes Olivo-Garrido M, Quiñonez-Cabeza M, Beitl CM, Martínez-Soto M, and Rodríguez-Monroy C. Performance and challenges in the value chain of the anadara tuberculosa bivalve mollusk in Ecuador. Sustainability. (2021) 13:10863. doi: 10.3390/su131910863
22. Saco A, Novoa B, Greco S, Gerdol M, and Figueras A. Bivalves present the largest and most diversified repertoire of toll-like receptors in the animal kingdom, suggesting broad-spectrum pathogen recognition in marine waters. Mol Biol Evol. (2023) 40:msad133. doi: 10.1093/molbev/msad133
23. Huang B, Ma J, Xu W, Cui J, Chen J, Qu Y, et al. A newly identified scallop MyD88 interacts with TLR and functions in innate immunity. Fish Shellfish Immunol. (2024) 151:109697. doi: 10.1016/j.fsi.2024.109697
24. Li L, Cui J, Qu Y, Ma J, Chen J, Zhao Y, et al. IKK2 modulates the innate immune response of Zhikong scallop (Chlamys farreri) to bacterial and viral stimuli through the MyD88-dependent Toll-like receptor signaling pathway. Aquaculture. (2024) 591:741148. doi: 10.1016/j.aquaculture.2024.741148
25. Wang B, Shao Y, Wang X, and Li C. Identification and functional analysis of Toll-like receptor 2 from razor clam Sinonovacula constricta. Int J Biol Macromolecules. (2024) 265:131029. doi: 10.1016/j.ijbiomac.2024.131029
26. Bai L, Li S, Wang P, Guo Y, Zheng Y, He J, et al. Toll-like receptor may be involved in acquired immune response in pearl oyster Pinctada fucata. Fish Shellfish Immunol. (2023) 141:109091. doi: 10.1016/j.fsi.2023.109091
27. Liu W, Ma J, Chen J, Huang B, Liu F, Li L, et al. A novel TBK1/IKKϵ is involved in immune response and interacts with MyD88 and MAVS in the scallop Chlamys farreri. Front Immunol. (2023) 13:1091419. doi: 10.3389/fimmu.2022.1091419
28. Wang X, Qu X, Lu X, Chen M, Ning J, Liu H, et al. Characterization of TRAF genes and their responses to Vibrio Anguillarum challenge in Argopecten scallops. Fish Shellfish Immunol. (2023) 135:108675. doi: 10.1016/j.fsi.2023.108675
29. Zou Y, Xu X, Xiao X, Wang Y, Yang H, and Zhang Z. Genome-wide identification and characterization of Toll-like receptors (TLR) genes in Haliotis discus hannai, H. rufescens and H. laevigata. Fish Shellfish Immunol. (2023) 137:108728. doi: 10.1016/j.fsi.2023.108728
30. Fan S, Wang W, Li J, Cao W, Li Q, Wu S, et al. The truncated MyD88s negatively regulates TLR2 signal on expression of IL17–1 in oyster Crassostrea gigas. Dev Comp Immunol. (2022) 133:104446. doi: 10.1016/j.dci.2022.104446
31. Qu F, She Q, Li J, Zeng X, Li Y, Liu X, et al. Molecular characterization of MyD88 in Anodonta woodiana and its involvement in the innate immune response to bacterial infection. Front Immunol. (2022) 13:925168. doi: 10.3389/fimmu.2022.925168
32. Qi P, Wu Y, Gu Z, Li H, Li J, Guo B, et al. A novel molluscan TLR molecule engaged in inflammatory response through MyD88 adapter recruitment. Dev Comp Immunol. (2022) 131:104373. doi: 10.1016/j.dci.2022.104373
33. Yin S, Chen J, Zhu M, Su F, Jian S, and Wen C. Characterization of a novel toll-like receptor and activation NF-κB signal pathway in triangle sail mussel Hyriopsis cumingii. Comp Biochem Physiol Part B: Biochem Mol Biol. (2021) 255:110608. doi: 10.1016/j.cbpb.2021.110608
34. Zhang H, Tan K, Li S, Ma H, and Zheng H. Genome-wide analysis of TRAF gene family and its response to bacterial infection in noble scallop Chlamys nobilis with different carotenoids content. Aquaculture. (2021) 535:736309. doi: 10.1016/j.aquaculture.2020.736309
35. Juhász A and Lawton SP. Toll like receptors and their evolution in the lymnaeid freshwater snail species Radix auricularia and Lymnaea stagnalis, key intermediate hosts for zoonotic trematodes. Dev Comp Immunol. (2022) 127:104297. doi: 10.1016/j.dci.2021.104297
36. Chen H, Cai X, Li R, Wu Y, Qiu H, Zheng J, et al. A novel toll-like receptor from Crassostrea gigas is involved in innate immune response to Vibrio alginolyticus. Infection Genet Evolution. (2022) 97:105159. doi: 10.1016/j.meegid.2021.105159
37. Jiao Y, Gu Z, Luo S, and Deng Y. Evolutionary and functional analysis of MyD88 genes in pearl oyster Pinctada fucata martensii. Fish Shellfish Immunol. (2020) 99:322–30. doi: 10.1016/j.fsi.2020.02.018
38. Gorbushin AM. Toll-like signaling pathway in the transcriptome of Littorina littorea. Fish Shellfish Immunol. (2020) 106:640–4. doi: 10.1016/j.fsi.2020.08.012
39. Ren Y, Dong W, Yang Y, Pan B, and Bu W. Molecular and expression characterization of Toll-like receptor family genes from the Anadara sativa (Bivalvia, Arcidae) transcriptome. Dev Comp Immunol. (2020) 106:103630. doi: 10.1016/j.dci.2020.103630
40. Huang S, Yuan S, Guo L, Yu Y, Li J, Wu T, et al. Genomic analysis of the immune gene repertoire of amphioxus reveals extraordinary innate complexity and diversity. Genome Res. (2008) 18:1112–26. doi: 10.1101/gr.069674.107
41. Werling D, Jann OC, Offord V, Glass EJ, and Coffey TJ. Variation matters: TLR structure and species-specific pathogen recognition. Trends Immunol. (2009) 30:124–30. doi: 10.1016/j.it.2008.12.001
42. Gerdol M, Venier P, Edomi P, and Pallavicini A. Diversity and evolution of TIR-domain-containing proteins in bivalves and Metazoa: new insights from comparative genomics. Dev Comp Immunol. (2017) 70:145–64. doi: 10.1016/j.dci.2017.01.014
43. Hibino T, Loza-Coll M, Messier C, Majeske AJ, Cohen AH, Terwilliger DP, et al. The immune gene repertoire encoded in the purple sea urchin genome. Dev Biol. (2006) 300:349–65. doi: 10.1016/j.ydbio.2006.08.065
44. Mahapatra S, Ganguly B, Pani S, Saha A, and Samanta M. A comprehensive review on the dynamic role of toll-like receptors (TLRs) in frontier aquaculture research and as a promising avenue for fish disease management. Int J Biol Macromolecules. (2023) 242:126541. doi: 10.1016/j.ijbiomac.2023.126541
45. Adema CM, Hillier LW, Jones CS, Loker ES, Knight M, Minx P, et al. Whole genome analysis of a schistosomiasis-transmitting freshwater snail. Nat Commun. (2017) 8:15451. doi: 10.1038/ncomms15451
46. Orús-Alcalde A, Lu T-M, Børve A, and Hejnol A. The evolution of the metazoan Toll receptor family and its expression during protostome development. BMC Ecol evolution. (2021) 21:208. doi: 10.1186/s12862-021-01927-1
47. Sun L, Liu X, Zhou L, Wang H, Lian C, Zhong Z, et al. Shallow-water mussels (Mytilus galloprovincialis) adapt to deep-sea environment through transcriptomic and metagenomic insights. Commun Biol. (2025) 8:46. doi: 10.1038/s42003-024-07382-0
48. Li L, Liu W, Fan N, Li F, Huang B, Liu Q, et al. Scallop IKK1 responds to bacterial and virus-related pathogen stimulation and interacts with MyD88 adaptor of toll-like receptor pathway signaling. Front Immunol. (2022) 13:869845. doi: 10.3389/fimmu.2022.869845
49. Wang W, Song X, Wang L, and Song L. Pathogen-derived carbohydrate recognition in molluscs immune defense. Int J Mol Sci. (2018) 19:721. doi: 10.3390/ijms19030721
50. Agius J, Ackerly D, Beddoe T, and Helbig KJ. Analysis of the presence of anti-viral innate immune pathways in the Australian Haliotis laevigata. Comp Immunol Rep. (2024) 6:200145. doi: 10.1016/j.cirep.2024.200145
51. Bagheri M and Zahmatkesh A. Evolution and species-specific conservation of toll-like receptors in terrestrial vertebrates. Int Rev Immunol. (2018) 37:217–28. doi: 10.1080/08830185.2018.1506780
52. Leulier F and Lemaitre B. Toll-like receptors—taking an evolutionary approach. Nat Rev Genet. (2008) 9:165–78. doi: 10.1038/nrg2303
53. Liu G, Zhang H, Zhao C, and Zhang H. Evolutionary history of the Toll-like receptor gene family across vertebrates. Genome Biol evolution. (2020) 12:3615–34. doi: 10.1093/gbe/evz266
54. Krishnan J, Selvarajoo K, Tsuchiya M, Lee G, and Choi S. Toll-like receptor signal transduction. Exp Mol Med. (2007) 39:421–38. doi: 10.1038/emm.2007.47
55. Duan T, Du Y, Xing C, Wang HY, and Wang R-F. Toll-like receptor signaling and its role in cell-mediated immunity. Front Immunol. (2022) 13:812774. doi: 10.3389/fimmu.2022.812774
56. Wang M, Wang L, Jia Z, Yi Q, and Song L. The various components implied the diversified Toll-like receptor (TLR) signaling pathway in mollusk Chlamys farreri. Fish shellfish Immunol. (2018) 74:205–12. doi: 10.1016/j.fsi.2017.12.064
57. Gong X, Li M, Zhang L, Huang S, and Wang G. Identification and functional analysis of myeloid differentiation factor 88 (MyD88) in early development of Haliotis diversicolor. Fish Shellfish Immunol. (2023) 142:109085. doi: 10.1016/j.fsi.2023.109085
58. Oren M, Barela Hudgell MA, D’Allura B, Agronin J, Gross A, Podini D, et al. Short tandem repeats, segmental duplications, gene deletion, and genomic instability in a rapidly diversified immune gene family. BMC Genomics. (2016) 17:1–19. doi: 10.1186/s12864-016-3241-x
59. Oda M, Yamamoto H, and Kawakami T. Maintenance of homeostasis by TLR4 ligands. Front Immunol. (2024) 15:1286270. doi: 10.3389/fimmu.2024.1286270
60. Jiang K, Nie H, Li D, and Yan X. New insights into the Manila clam and PAMPs interaction based on RNA-seq analysis of clam through in vitro challenges with LPS, PGN, and poly (I: C). BMC Genomics. (2020) 21:531. doi: 10.1186/s12864-020-06914-2
61. Rey-Campos M, Saco A, Novoa B, and Figueras A. Transcriptomic and functional analysis of the antiviral response of mussels after a poly I: C stimulation. Fish Shellfish Immunol. (2024) 153:109867. doi: 10.1016/j.fsi.2024.109867
62. Tang C, Qiao X, Jin Y, Yang W, Yu Z, Wang L, et al. An LPS-induced TNF-α factor involved in immune response of oyster Crassostrea gigas by regulating haemocytes apoptosis. Fish Shellfish Immunol. (2024) 148:109513. doi: 10.1016/j.fsi.2024.109513
63. Moreira R, Romero A, Rey-Campos M, Pereiro P, Rosani U, Novoa B, et al. Stimulation of Mytilus galloprovincialis hemocytes with different immune challenges induces differential transcriptomic, miRNomic, and functional responses. Front Immunol. (2020) 11:606102. doi: 10.3389/fimmu.2020.606102
64. Tang X, Huang B, Lin S, Wang W, Zhang G, and Li L. CgMyD88s serves as an innate immune system plug during ostreid herpesvirus 1 infection in the pacific oyster (Crassostrea gigas). Front Immunol. (2020) 11:1247. doi: 10.3389/fimmu.2020.01247
65. Liu W, Ma J, Chen J, Huang B, Liu F, Li L, et al. A novel TBK1/IKKϵ is involved in immune response and interacts with MyD88 and MAVS in the scallop Chlamys farreri. Front Immunol. (2022) 13:1091419. doi: 10.3389/fimmu.2022.1091419
66. Sanjeewa KKA, Nagahawatta DP, Yang HW, Oh JY, Jayawardena TU, Jeon YJ, et al. Octominin inhibits LPS-induced chemokine and pro-inflammatory cytokine secretion from RAW 264.7 macrophages via blocking TLRs/NF-κB signal transduction. Biomolecules. (2020) 10:511. doi: 10.3390/biom10040511
67. Qu F, Xiang Z, Wang F, Zhang Y, Li J, Zhang Y, et al. Identification and function of an evolutionarily conserved signaling intermediate in Toll pathways (ECSIT) from Crassostrea hongkongensis. Dev Comp Immunol. (2015) 53:244–52. doi: 10.1016/j.dci.2015.07.015
68. Salgueiro V, Reis L, Ferreira E, Botelho MJ, Manageiro V, and Caniça M. Assessing the bacterial community composition of bivalve mollusks collected in aquaculture farms and respective susceptibility to antibiotics. Antibiotics. (2021) 10:1135. doi: 10.3390/antibiotics10091135
69. Kijewska A, Koroza A, Grudlewska-Buda K, Kijewski T, Wiktorczyk-Kapischke N, Zorena K, et al. Molluscs—A ticking microbial bomb. Front Microbiol. (2023) 13:1061223. doi: 10.3389/fmicb.2022.1061223
70. Yue C, Zhang K, Liu Z, Lü W, Guo H, Zhao L, et al. The role of the TLR4-MyD88 signaling pathway in the immune response of the selected scallop strain “Hongmo No. 1” to heat stress. Animals. (2024) 14:497. doi: 10.3390/ani14030497
71. Yu F, Chen J, Lin J, Zhong Z, Lu Y, Zeng X, et al. TLR4 involved in immune response against Vibrio Parahaemolyticus by MyD88-dependent pathway in Crassostrea hongkongensis. Fish Shellfish Immunol. (2023) 134:108591. doi: 10.1016/j.fsi.2023.108591
72. Rey-Campos M, Moreira R, Gerdol M, Pallavicini A, Novoa B, and Figueras A. Immune tolerance in Mytilus galloprovincialis hemocytes after repeated contact with Vibrio splendidus. Front Immunol. (2019) 10:1894. doi: 10.3389/fimmu.2019.01894
73. Green TJ and Speck P. Antiviral defense and innate immune memory in the oyster. Viruses. (2018) 10:133. doi: 10.3390/v10030133
74. Renault T. Viruses infecting marine mollusks. In: Studies in Viral Ecology. (Hoboken, NJ: Wiley-Blackwell) (2021). p. 275–303.
75. Chan J, Wang L, Li L, Mu K, Bushek D, Xu Y, et al. Transcriptomic response to Perkinsus marinus in two Crassostrea oysters reveals evolutionary dynamics of host-parasite interactions. Front Genet. (2021) 12:795706. doi: 10.3389/fgene.2021.795706
76. Sullivan ME and Proestou DA. Survival and transcriptomic responses to different Perkinsus marinus exposure methods in an Eastern oyster family. Aquaculture. (2021) 542:736831. doi: 10.1016/j.aquaculture.2021.736831
77. Loiseau C and Sorci G. Can microplastics facilitate the emergence of infectious diseases? Sci Total Environ. (2022) 823:153694. doi: 10.1016/j.scitotenv.2022.153694
78. Sun C, Teng J, Wang D, Li J, Wang X, Zhao J, et al. Potential threats of microplastics and pathogenic bacteria to the immune system of the mussels Mytilus galloprovincialis. Aquat Toxicol. (2024) 272:106959. doi: 10.1016/j.aquatox.2024.106959
79. Huang S, Li H, Zhao L, Han Y, Liu H, Zha S, et al. Pollution in marine bivalves: The immunosuppressive effects of microplastics on Anadara granosa. Mar Environ Res. (2025) 208:107107. doi: 10.1016/j.marenvres.2025.107107
80. Yang W, Jannatun N, Zeng Y, Liu T, Zhang G, Chen C, et al. Impacts of microplastics on immunity. Front toxicol. (2022) 4:956885. doi: 10.3389/ftox.2022.956885
81. Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. (2001) 1:135–45. doi: 10.1038/35100529
82. Akira S and Takeda K. Toll-like receptor signalling. Nat Rev Immunol. (2004) 4:499–511. doi: 10.1038/nri1391
83. O’Neill LA and Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. (2007) 7:353–64. doi: 10.1038/nri2079
84. Cao X. Self-regulation and cross-regulation of pattern-recognition receptor signalling in health and disease. Nat Rev Immunol. (2016) 16:35–50. doi: 10.1038/nri.2015.8
85. Wang M, Yang J, Zhou Z, Qiu L, Wang L, Zhang H, et al. A primitive Toll-like receptor signaling pathway in mollusk Zhikong scallop Chlamys farreri. Dev Comp Immunol. (2011) 35:511–20. doi: 10.1016/j.dci.2010.12.005
86. Burgos-Aceves MA, Cohen A, Smith Y, and Faggio C. A potential microRNA regulation of immune-related genes in invertebrate haemocytes. Sci Total Environment. (2018) 621:302–7. doi: 10.1016/j.scitotenv.2017.11.285
87. Wu Y, Si X, Qiu L, Chen X, Fu P, Buttino I, et al. Regulation of innate immunity in marine mussel Mytilus coruscus: MicroRNA Mc-novel_miR_196 targets McTLR-like1 molecule to inhibit inflammatory response and apoptosis. Fish Shellfish Immunol. (2023) 138:108868. doi: 10.1016/j.fsi.2023.108868
88. Canesi L and Fabbri E. Environmental effects of BPA: focus on aquatic species. Dose-Response. (2015) 13:1559325815598304. doi: 10.1177/1559325815598304
89. Ghani MU, Chen J, Khosravi Z, Wu Q, Liu Y, Zhou J, et al. Unveiling the multifaceted role of toll-like receptors in immunity of aquatic animals: Pioneering strategies for disease management. Front Immunol. (2024) 15:1378111. doi: 10.3389/fimmu.2024.1378111
90. Pani S, Ganguly B, Mahapatra S, Dash SP, Das R, Saha A, et al. Molecular characterization and immune role of TLR7 in Labeo rohita. Front Immunol. (2025) 16:1555048. doi: 10.3389/fimmu.2025.1555048
91. Liu F, Chen H, Cao C, Liang Y, and Zhou Y. The role of toll-like receptors (TLRs) and their therapeutic applications in glomerulonephritis. Int Urol Nephrol. (2023) 55:2845–56. doi: 10.1007/s11255-023-03592-3
92. Wang L, Zhang H, Wang M, Zhou Z, Wang W, Liu R, et al. The transcriptomic expression of pattern recognition receptors: Insight into molecular recognition of various invading pathogens in Oyster Crassostrea gigas. Dev Comp Immunol. (2019) 91:1–7. doi: 10.1016/j.dci.2018.09.021
93. Nie L, Cai S-Y, Shao J-Z, and Chen J. Toll-like receptors, associated biological roles, and signaling networks in non-mammals. Front Immunol. (2018) 9:1523. doi: 10.3389/fimmu.2018.01523
94. Mifsud EJ, Tan AC, and Jackson DC. TLR agonists as modulators of the innate immune response and their potential as agents against infectious disease. Front Immunol. (2014) 5:79. doi: 10.3389/fimmu.2014.00079
95. Kayesh MEH, Kohara M, and Tsukiyama-Kohara K. TLR agonists as vaccine adjuvants in the prevention of viral infections: an overview. Front Microbiol. (2023) 14:1249718. doi: 10.3389/fmicb.2023.1249718
96. Díaz-Dinamarca DA, Salazar ML, Castillo BN, Manubens A, Vasquez AE, Salazar F, et al. Protein-based adjuvants for vaccines as immunomodulators of the innate and adaptive immune response: Current knowledge, challenges, and future opportunities. Pharmaceutics. (2022) 14:1671. doi: 10.3390/pharmaceutics14081671
97. Zhao T, Cai Y, Jiang Y, He X, Wei Y, Yu Y, et al. Vaccine adjuvants: mechanisms and platforms. Signal transduction targeted Ther. (2023) 8:283. doi: 10.1038/s41392-023-01557-7
98. Fajardo C, Martinez-Rodriguez G, Costas B, Mancera JM, Fernandez-Boo S, Rodulfo H, et al. Shrimp immune response: A transcriptomic perspective. Rev Aquaculture. (2022) 14:1136–49. doi: 10.1111/raq.12642
99. Sun Y, Zhang X, Wang Y, and Zhang Z. Long-read RNA sequencing of Pacific abalone Haliotis discus hannai reveals innate immune system responses to environmental stress. Fish Shellfish Immunol. (2022) 122:131–45. doi: 10.1016/j.fsi.2022.01.042
100. Shen C, Zhang M, Liang H, He J, Zhang B, and Liang B. Gene cloning and functional study of PmKSPI from Pinctada fucata martensii. Fish Shellfish Immunol. (2022) 131:1157–65. doi: 10.1016/j.fsi.2022.11.021
101. Drobysh M, Ratautaite V, Brazys E, Ramanaviciene A, and Ramanavicius A. Molecularly imprinted composite-based biosensor for the determination of SARS-CoV-2 nucleocapsid protein. Biosensors Bioelectronics. (2024) 251:116043. doi: 10.1016/j.bios.2024.116043
102. Mishra B, Tiwari A, and Mishra S. Metabolic changes and immunity suppression parameters as biomarkers of environmental pollutants. In: Biomonitoring of Pollutants in the Global South. Cham: Springer (2024). p. 693–719.
Keywords: TLRs, mollusks, innate immune response, pattern recognition receptors, pathogens
Citation: Li H, Li X, Zhu Y, Xu J, Zhao Q, Chen X, Zhang Y, Zhao L, Chen Y, Weng R and Chen K (2025) Systematic review of structural and immunological features of mollusk toll-like receptors in aquaculture context. Front. Immunol. 16:1671434. doi: 10.3389/fimmu.2025.1671434
Received: 23 July 2025; Accepted: 19 September 2025;
Published: 01 October 2025.
Edited by:
Balachandran Ravindran, Institute of Life Sciences (ILS), IndiaReviewed by:
Lulu Chen, Yancheng Teachers University, ChinaNorfarrah Mohamed Alipiah, Universiti Putra Malaysia, Malaysia
Copyright © 2025 Li, Li, Zhu, Xu, Zhao, Chen, Zhang, Zhao, Chen, Weng and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongyu Li, aG9uZ3l1ODg5MjZAempzcnUuZWR1LmNu; Keda Chen, Y2hlbmtkQHpqc3J1LmVkdS5jbg==
†These authors have contributed equally to this work
 Hongyu Li
Hongyu Li Xianwei Li†
Xianwei Li† Xintong Chen
Xintong Chen Yijie Zhang
Yijie Zhang Keda Chen
Keda Chen