- 1Department of Gastrointestinal Surgery, The Second Hospital of Dalian Medical University, Dalian, Liaoning, China
- 2Advanced Institute for Medical Science, Dalian Medical University, Dalian, Liaoning, China
- 3Research and Teaching Department of Comparative Medicine, Dalian Medical University, Dalian, Liaoning, China
Innate immune evasion is a critical aspect of viral infections, as it disrupts the host’s defense mechanisms.The innate immune system, as the primary defense against pathogens, detects pathogen-associated molecular patterns (PAMPs) via pattern recognition receptors (PRRs). This recognition triggers the production of interferons (IFNs) and pro-inflammatory factors, initiating the antiviral immune response. During evolution, viruses have found many ways to evade innate immune response in order to increase the replication efficiency, transmission ability and to establish persistent infection through co-evolution with hosts. Pigs act as natural hosts for a variety of significant viruses, including both DNA and RNA viruses. These viruses not only jeopardize animal health but also present a potential risk of interspecies transmission. Among these, porcine transmissible gastroenteritis virus (TGEV) stands out as a highly prevalent and severely detrimental enterovirus in the global swine industry. This review aims to comprehensively analyze the interaction between TGEV and host cells, emphasizing the molecular underpinnings of its immune evasion strategies. In addition, we will describe the programmed cell death types induced by TGEV, including autophagy, apoptosis and pyroptosis. Compared with existing reviews, this article not only provides a systematic integration of the multilayered immune evasion mechanisms of TGEV but also, for the first time, offers a comprehensive overview of its interactions with various forms of programmed cell death. This perspective highlights the complex regulatory networks underlying TGEV’s adaptive evolution in the host, thereby enhancing our understanding of the pathogenic mechanisms of porcine coronaviruses and offering novel theoretical foundations for the development of vaccines and antiviral therapeutics.
1 Introduction
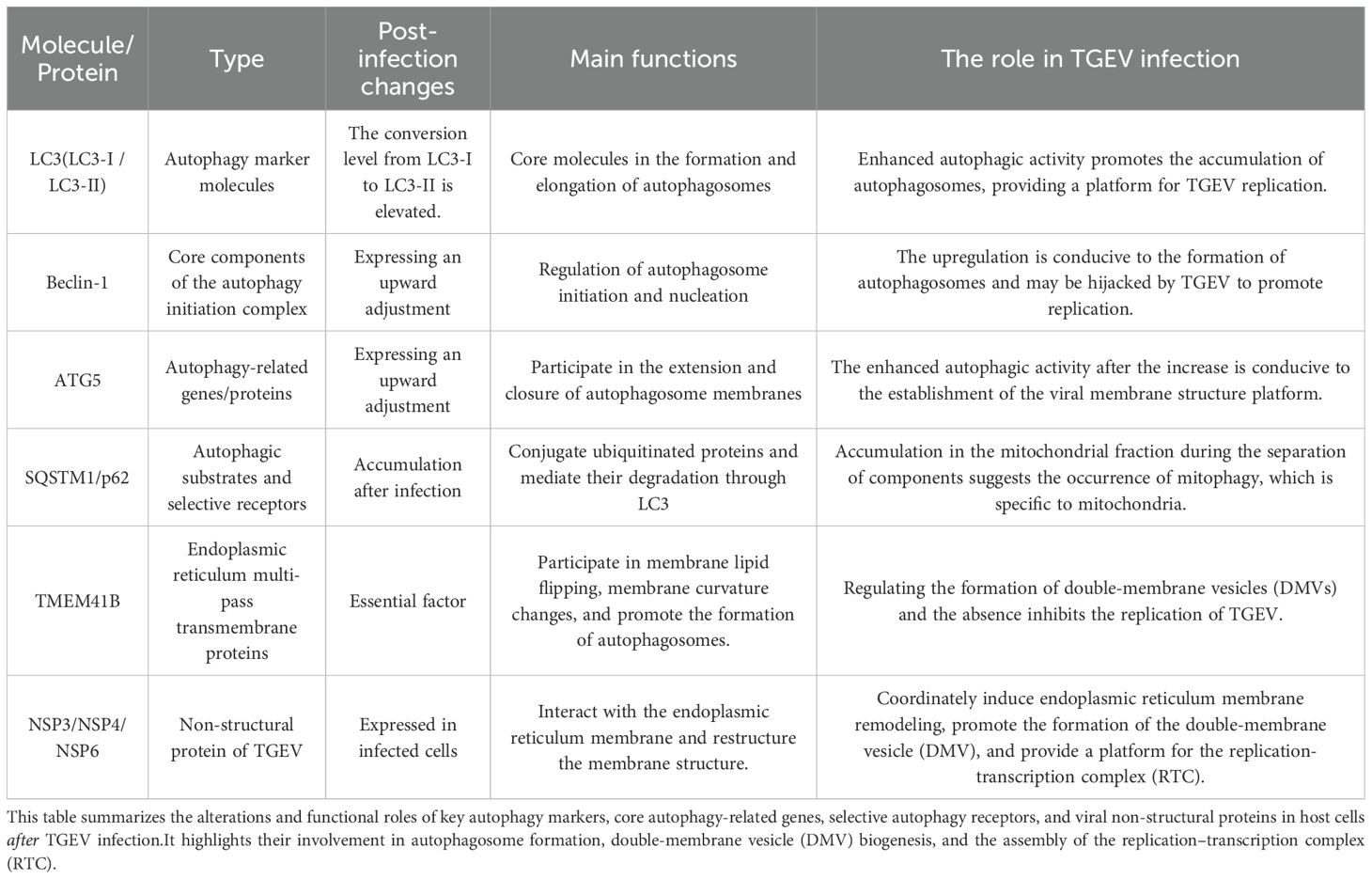
As the first line of defense against the invasion of exogenous pathogens, the innate immune system is characterized by rapid recognition and broad responses (1, 2).Central to this system is the detection of PAMPs by pattern recognition receptors (PRRs),which initiates an immune response against infections (3, 4). In the context of viral infections, typical PAMPs of viruses include their nucleic acids, such as single - stranded RNA(ssRNA,including 5’ UTR, viral RNA and replication protein), double - stranded RNA (dsRNA), and DNAThese PAMPs are recognized by different PRRs of host cells (5, 6) (including, but not limited to, TLRs, RIG-I-like receptors (RLRs), the cGAS-STING pathway and NOD-like receptors (NLRs)) (7–9). Upon recognizing viral components, these receptors activate downstream transcription factors IRF3/7 and NF-κB through adaptor proteins (e.g., MAVS, TRIF, MyD88, STING), This activation process ultimately leads to the production of type I interferons (IFN-α/β) and pro-inflammatory cytokines. These molecules can effectively inhibit viral replication and trigger adaptive immune responses (10–12). However, over time, viruses have evolved diverse mechanisms to evade the innate immune system, collectively referred to as “innate immune escape” (13, 14). These strategies work by interfering with the recognition of viral nucleic acids by pattern recognition receptors, preventing the activation of adaptor proteins or key signaling pathways, and promoting the expression of host negative regulatory factors to suppress the immune response (15, 16). These evasion tactics are crucial for successful viral infection, immune evasion, and also provide the molecular basis for interspecies transmission and viral pandemics (Figure 1).
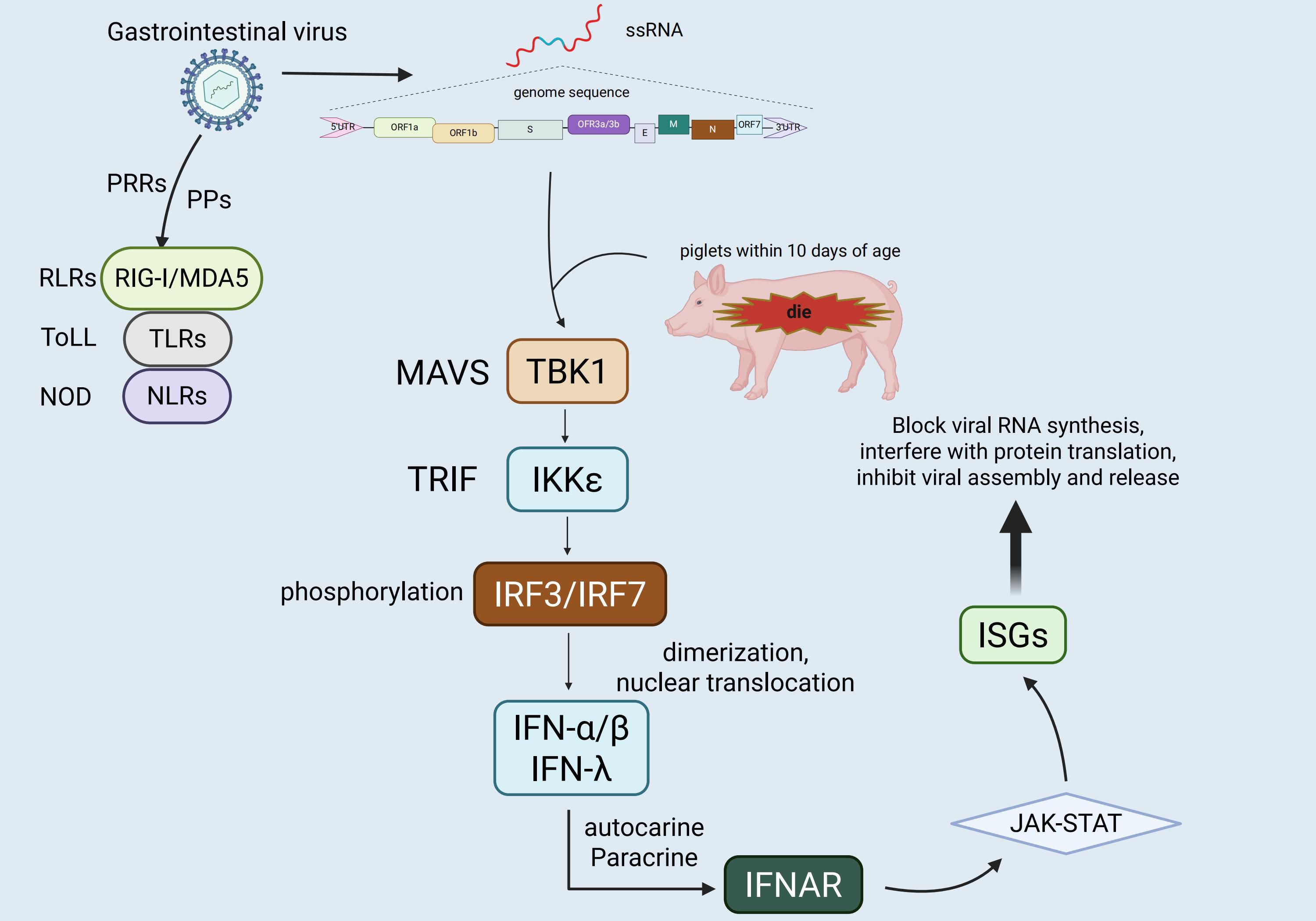
Figure 1. Genomic organization of TGEV and host innate immune responses. TGEV is an enveloped, positive-sense single-stranded RNA virus whose genome is organized as 5′UTR–ORF1a–ORF1b–S–ORF3a/3b–E–M–N–ORF7–3′UTR. It encodes four structural proteins,namely the spike (S), envelope (E), membrane (M), and nucleocapsid (N)—as well as several accessory proteins. In piglets less than 10 days of age, infection typically causes severe diarrhea and is often fatal, with mortality rates reaching 80–100%. The figure also illustrates the recognition of viral RNA by host pattern recognition receptors (TLRs, RLRs, and NLRs), along with the activation of downstream interferon signaling pathways. These pathways induce antiviral gene expression and contribute to host defense.
Among diverse viruses, porcine coronaviruses—particularly TGEV—serve as a representative model for investigating viral immune evasion mechanisms. On one hand, TGEV shares similarities in genomic organization and immune regulatory strategies with other coronaviruses, such as SARS-CoV-2, thereby providing valuable insights into the conserved mechanisms underlying coronavirus immune escape (17). On the other hand, as a highly pathogenic enteric virus that poses a serious threat to the swine industry, TGEV not only impairs animal health and livestock production but also harbors potential risks of cross-species transmission (18).
To date, six porcine coronaviruses have been identified, comprising four alphacoronaviruses, one betacoronavirus, and one deltacoronavirus (19). Among them, TGEV is the earliest discovered and most extensively investigated member of the porcine alphacoronaviruses (20). Viral structural proteins not only mediate essential functions in the viral life cycle, including entry, assembly, and budding, but also play pivotal roles in orchestrating immune evasion strategies, forming the molecular basis by which TGEV circumvents host innate immune recognition (21).TGEV is transmitted primarily via the fecal–oral and respiratory routes, infecting epithelial cells of the porcine gastrointestinal tract, particularly the villous epithelial cells of the small intestine (21, 22). Viral replication results in epithelial cell necrosis, villus atrophy, and mucosal damage, leading to disruption of the intestinal barrier. Consequently, infected piglets exhibit acute vomiting, profuse yellow-green watery diarrhea, severe dehydration, and malabsorption (23, 24). The disease is especially devastating in neonatal piglets with immature immune systems, in which rapid disease progression is associated with mortality rates as high as 80–100% within the first 10 days of life (25). Infected pigs, asymptomatic carriers, and animals within 10 weeks of recovery serve as major sources of transmission. To date, pigs are recognized as the only natural host of TGEV, with no human infections reported.
The high pathogenicity and rapid transmission of TGEV pose a substantial threat to the swine industry. Elucidating the molecular mechanisms underlying TGEV immune evasion not only offers critical insights into the general pathogenic strategies of coronaviruses but also guides vaccine development and antiviral drug design. Compared with other porcine alphacoronaviruses, such as Porcine Epidemic Diarrhea Virus (PEDV), which also causes severe diarrhea, and the recently identified SADS-CoV, which exhibits cross-species transmission potential, TGEV displays distinctive features in terms of pathogenicity, tissue tropism, and immune evasion strategies. These differences offer unique opportunities for comparing the mechanisms of pathogenesis among porcine coronaviruses, highlighting the importance of systematic investigation of TGEV immune evasion in understanding coronavirus evolution, assessing interspecies transmission risks, and advancing novel prevention and control strategies.
2 Recognition and response of the host’s innate immunity
The innate immune system of pigs, like that of other mammals, relies on the recognition of PAMPs by PRRs for its functionality (26, 27). Upon invasion by enteroviruses such as TGEV, various PRRs expressed in porcine intestinal epithelial cells and mucosa-associated immune cells play a crucial role in detecting the nucleic acid components of different viruses, thereby initiating an antiviral innate immune response. Among them, the RLRs located in the cytoplasm are key sensors for RNA virus recognition, including Retinoic Acid-Inducible Gene I(RIG-I) and Melanoma Differentiation-Associated Gene 5(MDA5), which recognize 5’-triphosphate single - stranded RNA and long - chain double - stranded RNA respectively (28, 29). TLRs located on the endosomal membrane are also significant in virus recognition. For instance, TLR3 detects double-stranded RNA generated during viral replication, while TLR7/8 primarily identify single-stranded RNA, both contributing to impeding virus replication and dissemination (30, 31). Moreover, although the cGAS-STING pathway is primarily involved in detecting cytoplasmic DNA, predominantly against DNA viruses, it can be indirectly activated during RNA virus infections by cellular damage or secondary signals, thereby participating in innate immune modulation (32, 33). Furthermore, in addition to their role in inflammasome assembly and pro-inflammatory factor release, NLRs can act as co-regulators of PRRs signaling pathways, augmenting immune recognition and response to viral infections (34, 35).
Upon viral infection, host cells detect viral nucleic acids via PRRs, leading to the recruitment of specific adapter proteins (e.g., MAVS, TRIF, MyD88). These adapter proteins subsequently activate the downstream signaling molecules TANK-binding kinase 1 (TBK1) and IKKϵ, which phosphorylate transcription factors IRF3 and IRF7 (36–38). This phosphorylation prompts the dimerization and nuclear translocation of IRF3 and IRF7, thereby initiating the transcriptional expression of type I interferons (IFN-α/β) and type III interferons (IFN-λ) (39, 40). In the context of antiviral immunity in the intestinal mucosa, type III interferons exhibit greater tissue specificity and targeting compared to type I interferons (41). Due to the predominant expression of its receptor (IFNLR1/IL10R2) on epithelial cells, IFN-λ enhances local antiviral defense effectively while limiting inflammation, thereby playing a pivotal role in maintaining intestinal immune homeostasis and controlling local viral infections (42, 43). Subsequently, the secreted interferons bind to their respective receptors on target cells through autocrine and paracrine mechanisms (type I interferons bind to IFNAR1/2, and type III interferons bind to IFNLR1/IL10R2), activating the JAK-STAT signaling pathway and inducing the expression of numerous interferon-stimulated genes (ISGs) (44–46). These ISGs can impede various stages of the viral life cycle, including viral RNA synthesis, protein translation, and viral assembly and release, collectively establishing an effective innate antiviral barrier (47, 48). However, neonatal piglets under one week of age, characterized by an immature immune system, exhibit low expression levels of PRRs and downstream signaling molecules, limiting the efficiency of the interferon response. Consequently, this inadequate response fails to promptly control viral infections in the early stages, contributing significantly to their heightened susceptibility to enteric viruses like TGEV and the associated elevated mortality rate (49), Figure 2.
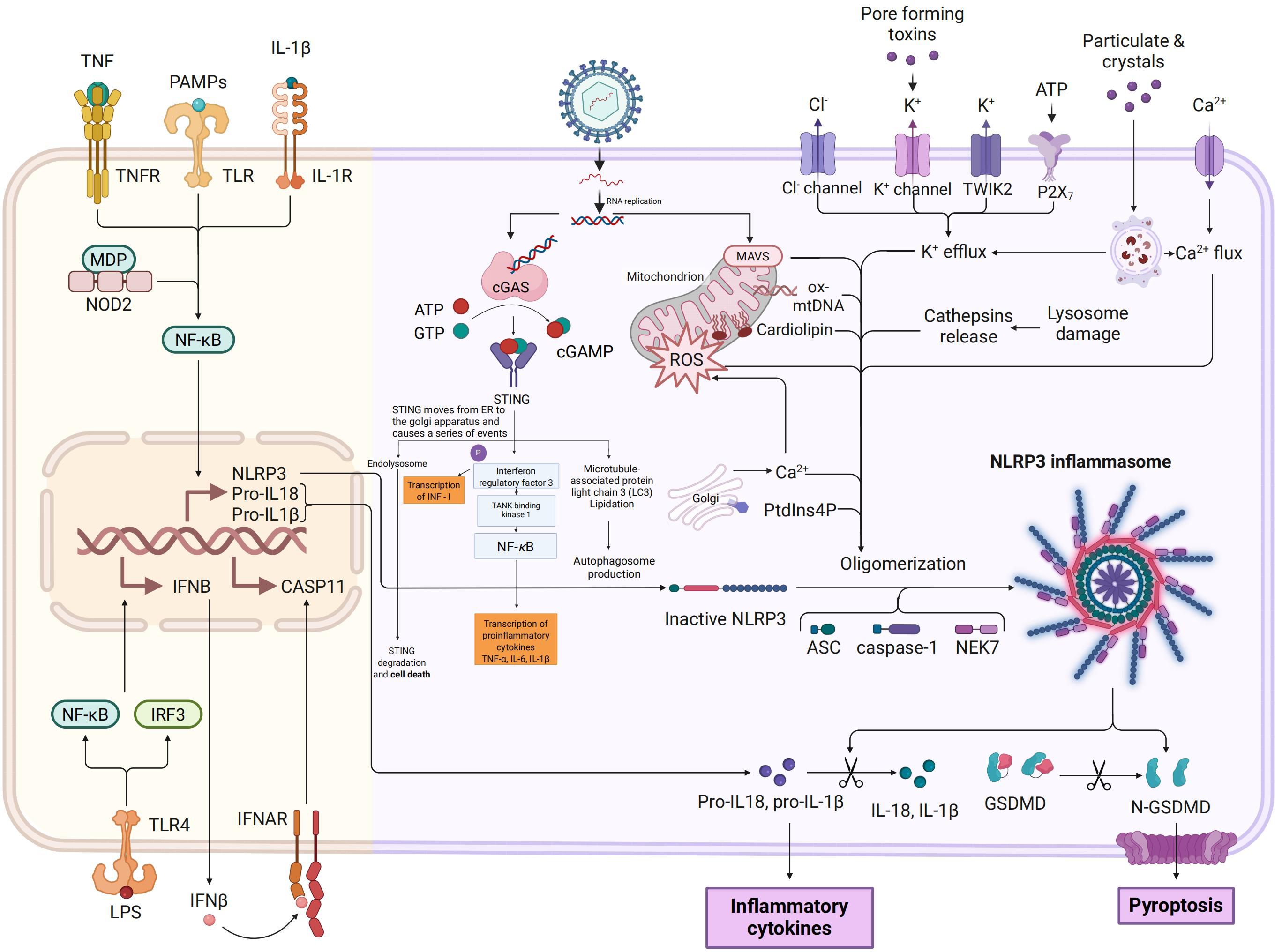
Figure 2. Schematic representation of NLRP3 inflammasome and cGAS-STING pathway activation. This figure illustrates the activation mechanisms and downstream effects of the NLRP3 inflammasome (on the left) and the cGAS-STING pathway (on the right), highlighting their interconnections in innate immune responses. The left panel shows that priming signals, which are mediated by TNFR, TLR, and IL-1R upon stimulation with TNF, PAMPs, and IL-1β, lead to NF-κB activation and NLRP3 inflammasome assembly. This assembly process involves nucleotide - binding oligomerization domain - containing protein 2 (NOD2), muramyl dipeptide (MDP), ATP, K+ efflux, and ROS, ultimately inducing caspase - 11 (CASP11) and interferon - β (IFNB) expression, pyroptosis, and IL-1β release. The right panel illustrates cyclic GMP - AMP synthase (cGAS) binding to cytosolic DNA to generate cGAMP, which activates STING translocation from the endoplasmic reticulum (ER)to the Golgi and subsequently triggers the activation of interferon regulatory factor 3 (IRF3) and NF-κB, resulting in antiviral responses and pro-inflammatory cytokine production. Shared regulatory elements include ROS and Ca²+ perturbations, mitochondrial damage, and lysosomal rupture, with organelles such as lysosomes, mitochondria, and autophagosomes contributing to inflammasome activation, pyroptosis, and cytokine synthesis.
3 Interference of TGEV with host pattern recognition receptors
3.1 Mechanistic insights into TGEV NSP-mediated blockade of TLR/RLR signaling
TLRs and RLRs are crucial PRRs in the innate immune system, responsible for detecting viral nucleic acids (50, 51). The single-stranded positive-sense RNA genome of TGEV is recognized by TLR3 and TLR7/8 localized in endosomes or by RIG-I and MDA5 localized in the cytoplasm to activate the downstream signaling pathways leading to the expression of type I IFNs (IFN-α/β) and pro-inflammatory mediators (52, 53). The transcription of these two types of IFNs is mediated by the activation of either IRF3/7 or NF-κB, which are pivotal for initiating antiviral immune responses (54). To evade host immune surveillance effectively, TGEV has developed diverse immune evasion tactics, predominantly utilizing its NSPs to disrupt key components of the TLR and RLR signaling pathways. This interference hampers interferon production and innate immune activation (55, 56).
During the initial phase of TGEV infection, the viral genomic open reading frame ORF1a is translated into polyprotein pp1a, which is further extended and translated into pp1ab through -1 ribosomal frameshifting (57, 58). These polyproteins are later processed by viral proteases, namely papain-like protease (PLpro) and 3C-like protease (3CLpro), resulting in the generation of 16 non-structural proteins such as NSP1 and NSP3 (59–61). These proteins are pivotal in virus replication, host modulation, and immune evasion. NSP1 is one of the earliest expressed viral proteins and has obvious immunosuppressive functions (62). This protein can inhibit the translation process by blocking the binding of host mRNA to ribosomes (63) and may promote the degradation of host mRNA through an as-yet-unclear mechanism (64, 65). NSP1 has been shown to hinder the functional activation of IRF3 by promoting its degradation. Normally, IRF3 is phosphorylated upon viral infection, leading to its dimerization, nuclear translocation, and subsequent induction of IFN and ISG expression (66, 67). TGEV’s NSP1 disrupts the IFN-β signaling pathway by targeting various steps of the IRF3 pathway, thereby obstructing IRF3 activation, nuclear translocation, and binding to target gene promoters, ultimately suppressing type I interferon production to facilitate immune evasion (68, 69). Moreover, NSP1 interferes with host mRNA transcription and translation processes, exacerbating the inhibition of host antiviral protein synthesis and enhancing immune evasion (70). It also promotes virus replication by impeding stress granule (SG) formation (71). NSP3 of TGEV is a multifunctional protein. In addition to participating in the self-cleavage of viral polyproteins, it also has a PLpro domain and deubiquitinase (DUB) activity, and can target multiple host immune signaling molecules for regulation (72, 73). PLP domain can directly recognize and cleave TRAF3. TRAF3 is an important adaptor protein located downstream of the MAVS in RLR signaling pathway and is responsible for recruiting and activating TBK1 and IKKϵ kinase complex (74). Once TRAF3 is cleaved, the formation of the TBK1-IKKϵ complex is impaired, leading to reduced phosphorylation of IRF3 and IRF7, thereby inhibiting the activation of the type I interferon signaling pathway (75). This multi-target and multi-mechanism immunosuppressive effect may allow TGEV to effectively replicate in host cells and escape immune clearance, Figure 3.
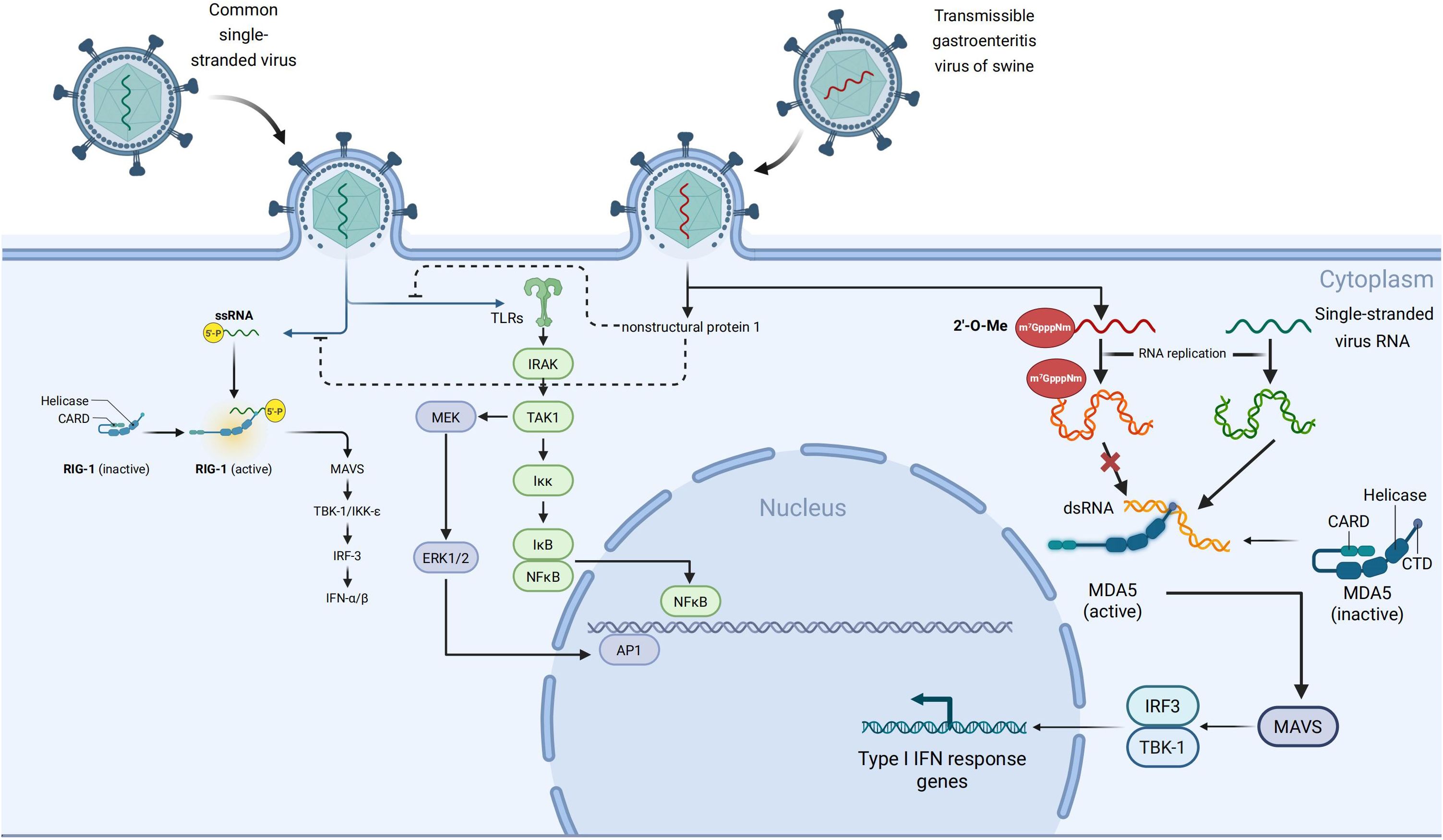
Figure 3. Mechanisms by which TGEV evades host PRRs-mediated innate immune responses. In uninfected cells, viral RNA is recognized by endosomal Toll-like receptors (TLR3/TLR7) or cytoplasmic sensors such as Melanoma Differentiation - Associated Gene 5 (MDA5). This recognition triggers downstream signaling cascades via adaptor kinases and transcription factors, ultimately leading to the production of type I interferons and pro - inflammatory cytokines.However, during TGEV infection, viral non-structural proteins (NS) inhibit key signaling nodes by promoting protein degradation or functional inactivation, thereby disrupting the TLR/RLR signaling pathways. This results in impaired nuclear translocation of IRF3 and NF-κB,along with reduced cytokine expression. In addition, TGEV evades MDA5 recognition by modifying the viral RNA cap structure through 2’-O methylation (m7GpppNm), which prevents MDA5 binding and subsequent activation of downstream signaling.
3.2 TGEV escapes MDA5 recognition through cap structure modification and signal axis interference
Notably, NSPs can suppress IFN signaling activation even when segments of viral RNA are recognized, whereas modifications of the cap structure further decrease the probability of detection (76, 77). Host cells trigger the innate immune response by detecting viral RNA’s distinct features, known as “non-self” markers (78). MDA5, a key member of the RLRs family, primarily recognizes lengthy dsRNA generated during viral infections or single-stranded RNA lacking a complete 5′ cap modification (79, 80). When MDA5 recognizes PAMPs of these dsRNAs, MDA5 can directly interact with the MAVS, and then recruit and activate other downstream signaling proteins like TBK1 and IKKϵ. Finally, the IRF3/7 will be phosphorylated and their translocation to the nucleus, triggering the expression of IFN-α/β and associated ISGs. Consequently, an extensive antiviral immune response is initiated (81, 82), Figure 3.
To escape from the host’s innate immune attack, coronaviruses have developed mechanisms to conceal the foreign features of their RNA. One such mechanism involves NSP16, acting as an S-adenosylmethionine (SAM)-dependent 2’-O-methyltransferase. NSP16 catalyzes methylation at the 2’-hydroxyl site of the ribose on the 5’-cap structure of viral mRNA, converting the RNA cap structure from Cap- (m7GpppN) to Cap-1 (m7GpppNm) (83, 84). This modification mimics the cap structure of eukaryotic cell mRNA, thereby diminishing recognition of viral RNA by host PRRs (85). Among human coronaviruses, SARS-CoV-2 can evade host immune responses by modifiying the RNA 5′ ends of viral RNA through the modification of the non-structural protein NSP16 (86). Specifically, NSP16 forms a heterodimer with NSP10 and catalyzes 2′-O-methylation of the first ribose unit of viral mRNA, generating a Cap-1 structure. This modification allows viral RNA to mimic host mRNA, thereby preventing recognition by MDA5 and inhibiting type I interferon signaling pathways (87). Viruses such as TGEV and SARS-CoV-2 exploit NSP16-mediated 2′-O-methylation to alter their RNA in both spatial conformation and chemical properties, effectively “disguising” it as endogenous molecules and escaping MDA5-mediated detection and immune activation (88, 89). Moreover, NSP16 activity is regulated by NSP10, with the NSP16–NSP10 complex also capable of suppressing host protein translation (88, 90), Figure 3.
4 Disruption of intestinal barrier function by TGEV
4.1 TGEV damages the tight junctions of intestinal epithelium
The intestinal epithelial barrier is essential for preserving intestinal homeostasis and preventing the trans-epithelial infiltration of pathogens and toxins (91, 92). Tight junctions (TJs) are pivotal for maintaining the integrity of this barrier, primarily comprised of various cytoplasmic proteins and transmembrane such as Zonula occludens-1 (ZO-1), Occludin, and Claudin family proteins (93, 94). These proteins collaborate to form a sealing belt structure between neighboring cells, restricting the passage of luminal contents to the basolateral side and thereby upholding the selective permeability of the barrier (95).
Prior research has demonstrated that TGEV infection disrupts the epithelial tight junction structure significantly (96). Upon infecting the IPEC-J2 porcine small intestinal epithelial cell line, TGEV notably reduces the expression levels of tight junction-related proteins, including ZO-1, Occludin, and Claudin-1. Additionally, the localization and structural integrity of tight junction proteins are compromised, leading to the disruption of the belt-like junction and widening of the intercellular space (97, 98). These alterations notably compromise the barrier function between epithelial cells (95).
Mechanistic investigations have elucidated that TGEV triggers the upregulation of inflammatory cytokines, including TNF−α, IL−6, and IL−8, through the activation of the p38 MAPK and NF-κB signaling pathways, as evidenced by studies (99, 100). These inflammatory factors indirectly impede the transcriptional activity of tight junction proteins (96). In addition, TGEV infection was accompanied by mitochondrial dysfunction and increased oxidative stress, which were characterized by increased contents of ROS and mitochondrial membrane potential (101–103). Excessive ROS can promote the degradation of TJs proteins or abnormal localization on the cell membrane, thereby exacerbating the barrier disruption (104, 105).
Functional studies have demonstrated that TGEV infection reduces transepithelial electrical resistance (TEER) and increases the permeability of the epithelial barrier, as evidenced by enhanced leakage of fluorescent tracer molecules (96, 100). This heightened barrier permeability facilitates the translocation of luminal pathogens, such as bacteria or endotoxins, across the epithelium, leading to potential secondary infections or systemic inflammatory responses in the host (106). Furthermore, the compromised barrier function facilitates local dissemination and amplification of TGEV within the intestinal tract, thereby exacerbating disease progression and tissue damage (21), Figure 4.
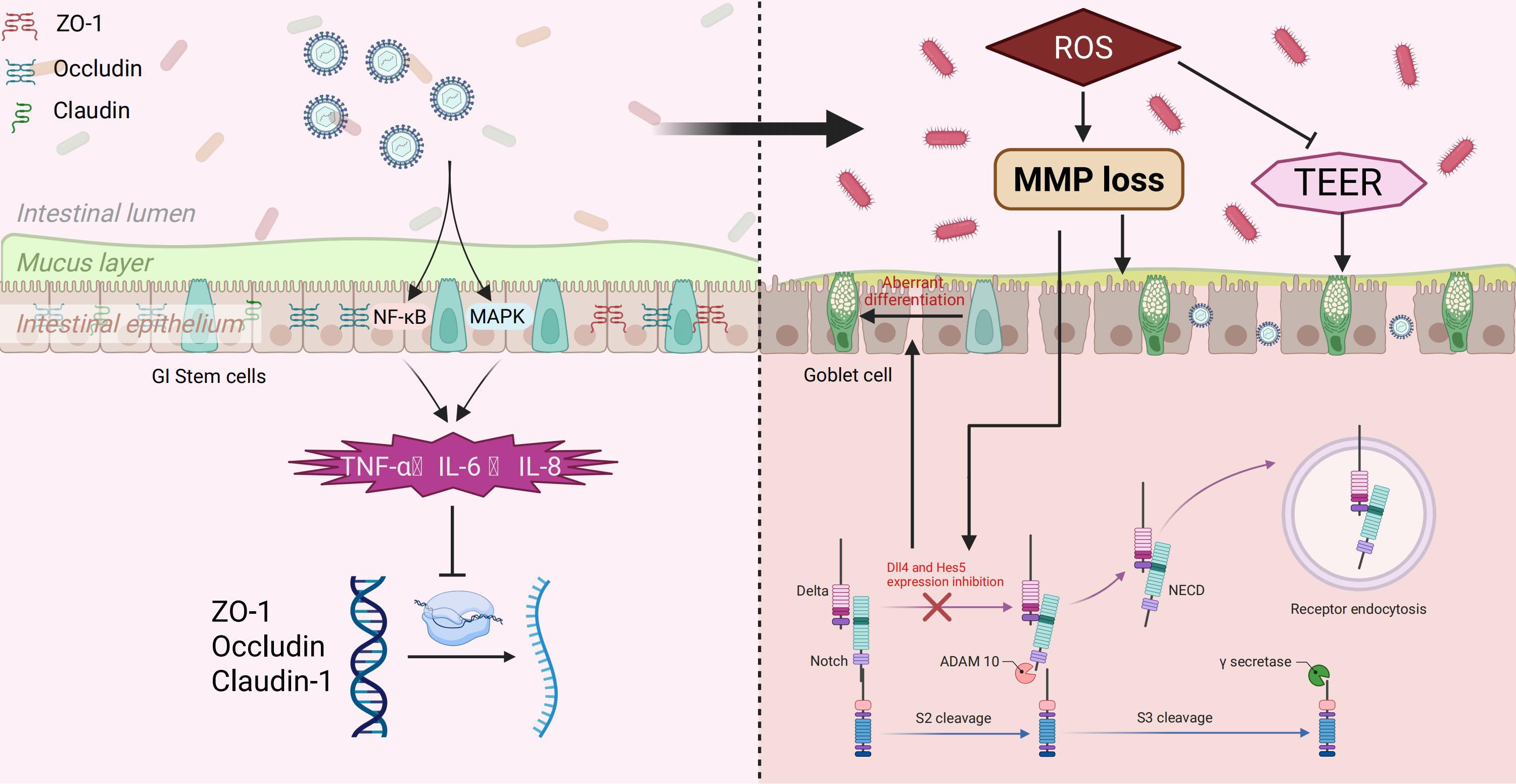
Figure 4. TGEV-induced disruption of intestinal barrier and modulation of Notch signaling. TGEV infection activates NF-κB and MAPK pathways, promoting inflammatory cytokine release (TNF-α, IL-6, IL-8) and downregulating tight junction proteins (ZO-1, Occludin, Claudin-1), leading to barrier dysfunction. Excess ROS impairs mitochondrial function, reduces TEER, and inhibits Dll4 and Hes5 expression in the Notch pathway, causing aberrant differentiation of GI stem cells into goblet cells, which may facilitate viral replication and spread.
4.2 TGEV regulates the notch signaling pathway
During TGEV infection of small intestinal epithelial cells, elevated levels of ROS and a loss of mitochondrial membrane potential were observed, resulting in oxidative stress (107). The stress condition could then suppress the expression of several important factors involved in the Notch signaling pathway, including Dll4 and Hes5 (108, 109). This mechanism has been demonstrated in the Paneth cell model of TGEV targeting small intestinal crypts (90, 110). The Notch signal is essential for regulating the balance between self-renewal and differentiation of intestinal Lgr5+ stem cells. Its inhibition can drive excessive differentiation of stem cells into goblet cells, resulting in elevated intestinal mucus secretion (90). This shift in differentiation and mucus composition may promote TGEV replication and dissemination in the small intestine. Consequently, inhibiting the Notch signal not only fails to restrict goblet cell production but also creates a more favorable environment for virus infection and transmission (90), Figure 4. Notch signaling is generally upregulated in hosts infected with viruses such as SARS-CoV-2, COVID-19, and PDCoV (111–113). However, it remains unclear whether the inhibition of Notch signaling observed during TGEV infection represents a unique feature of TGEV or a common characteristic of porcine coronaviruses. Further studies are warranted to elucidate the specificity and underlying mechanisms of this Notch signaling regulation.
4.3 Potential role of the gut microbiota in TGEV immune evasion
The gut microbiota constitutes the largest and most diverse microbial community within the host, and its metabolites, signaling molecules, and inter-microbial interactions play pivotal roles in maintaining immune homeostasis, preserving mucosal barrier function, and regulating inflammatory responses (114). Recent studies have revealed that the gut microbiota not only participates in nutrient metabolism and mucosal integrity but also modulates host susceptibility to viral infections and viral replication efficiency by regulating innate immune pathways, such as interferon (IFN) signaling. During TGEV infection, short-chain fatty acids (SCFAs) produced by the gut microbiota, especially butyrate, have been demonstrated to influence viral infection levels (115, 116). In the later stages of infection, butyrate can substantially enhance TGEV infection in porcine small intestinal epithelial cells and porcine testicular cells; however, this effect is not mediated through an increase in viral attachment or entry, but rather via interference with host antiviral immune responses.
Notably, TGEV infection is linked to a reduction in Lactobacillus gene copy numbers and an increase in Enterobacteriaceae gene copy numbers in porcine intestinal mucosal samples (117).In contrast, fecal samples show elevated relative abundances of Lactobacillus and Limosilactobacillus (118). These discrepancies imply that TGEV infection may trigger varying degrees of dysbiosis in the local intestinal environment compared to the overall gut ecosystem.Furthermore, TGEV can lead to villous atrophy and compromise intestinal immune function, inducing epithelial-to-mesenchymal transition (EMT) and converting epithelial cells into motile, invasion-prone mesenchymal cells (119). Prolonged infection may further enhance the invasive capacity of fecal Enterococcus species toward intestinal cells, thereby altering gut microbial composition and creating conditions favorable for secondary pathogen infections. Collectively, these findings indicate that TGEV facilitates immune evasion and increases host susceptibility by disrupting the intestinal barrier and remodeling the gut microbiota.
5 Cell autophagy induced by TGEV
Autophagy is a highly conserved cellular degradation process dependent on lysosomes, serving to eliminate damaged organelles, misfolded proteins, and pathogens, thereby upholding cellular homeostasis, regulating energy metabolism, and responding to cellular stress, immune modulation, and disease processes (120). In the context of viral infections, autophagy is commonly seen as a defense mechanism enabling host cells to combat foreign intruders. By forming autophagosomes that merge with lysosomes, autophagy can encapsulate and degrade viral particles, thereby restricting viral replication and dissemination (121, 122). However, recent research has demonstrated that TGEV can trigger autophagy in host cells, leading to a significant increase in autophagosome formation. Paradoxically, the autophagy induced by TGEV fails to effectively eliminate the virus. Instead, it creates a favorable membrane structure that supports viral replication and assembly, ultimately facilitating viral propagation (123, 124). This observation indicates that TGEV can exploit the host autophagy machinery to facilitate crucial stages in its life cycle, underscoring a nuanced and intricate interplay between the virus and the host autophagy system (124).
Research has shown that TGEV infection triggers robust activation of the autophagic process in host cells. Following infection, there is a notable increase in the number of autophagosomes within cells, exhibiting characteristic double-membrane structures observable via electron microscopy (123, 125, 126). Additionally, Meanwhile, there is a marked increase in the conversion of the autophagy marker microtubule - associated protein 1 light chain 3 (LC3) from its cytosolic form, LC3 - I, to the membrane - bound form, LC3 - II, resulting in a significantly elevated LC3 - II/LC3 - I ratio. This phenomenon is partly attributed to the common strategy of coronaviruses hijacking LC3-associated membranes to generate double-membrane vesicles and other replication organelles, which serve as scaffolds for the assembly of viral replication–transcription complexes (RTCs). However, this process predominantly relies on LC3 - I - positive endoplasmic reticulum - derived degradation - enhancing alpha - mannosidase - like protein 1 (EDEM1) - containing membranes (EDEMosome - like membranes) rather than classical lipidated LC3 - II membranes, further supporting the notion of enhanced autophagic activity (123, 127). At the molecular level, key autophagy-related genes such as Beclin-1 are significantly upregulated following infection (127). This may reflect a host protective mechanism in mammals, where the suppression of viral autophagic protein Beclin-1 titers contributes to defense (128, 129). Targeting Beclin-1 can block autophagosome formation and vesicle nucleation, thereby promoting viral replication and disease progression.
However, the above-mentioned autophagic response does not necessarily mean that the TGEV is cleared by host cells. On the contrary, evidence suggests that the virus may exploit this process to facilitate its replication and amplification, as shown by [Zhu et al.,2016] (127, 130). Recent studies have identified transmembrane protein 41B (TMEM41B) as a pivotal regulator during TGEV infection. TMEM41B is a protein with multiple transmembrane domains that is situated in the endoplasmic reticulum, is recognized as a key player in regulating membrane lipid translocation and curvature alterations, crucial for autophagosome formation. TGEV infection leverages TMEM41B-mediated membrane remodeling to generate double-membrane vesicles (DMVs), serving as a spatial platform for viral replication and transcription. Experimental data demonstrate that depletion or inhibition of TMEM41B significantly disrupts DMV formation, consequently impeding effective TGEV replication in host cells.Furthermore, several NSPs encoded by TGEV, such as NSP3, NSP4 and NSP6, could also interact with the endoplasmic reticulum membrane system to restructure the host cell membrane architecture. These NSPs collaborate to recruit lipids, induce local membrane expansion, and facilitate endoplasmic reticulum membrane bending, promoting DMV formation to support the assembly of the replication-transcription complex (RTC) (126). Consequently, TGEV orchestrates an intracellular replication niche conducive to its life cycle by finely regulating factors associated with the host autophagy pathway and membrane dynamics (Table 1).
6 Apoptosis induced by TGEV
Apoptosis is a type of programmed cell death that is precisely controlled by genes and is broadly involved in many physiological processes such as individual development, maintenance of tissue homeostasis, and elimination of abnormal cells (131). This process is energy-dependent and exhibits highly ordered morphological and molecular biological characteristics (131). Apoptosis primarily operates through two established signaling pathways: the intrinsic pathway mediated by mitochondria and the extrinsic pathway mediated by death receptors (132, 133).
Studies have shown that infection with TGEV can trigger a robust oxidative stress response and activate cell apoptosis. During the peak of viral replication, host cells must generate significant energy and biosynthetic resources to support viral proliferation, leading to mitochondrial dysfunction and a notable increase in intracellular ROS and mtROS accumulation (103, 127, 134). Intestinal epithelial cells, being the primary targets of TGEV, exhibit a strong ability to regulate oxidative stress. However, TGEV infection results in severe mitochondrial damage in these cells, accompanied by pronounced autophagy and mitophagy activation. The ROS buildup induced by TGEV not only directly harms cell structures but also induces programmed cell death by activating the mitochondrial apoptosis pathway. Specifically, ROS-mediated stress signals trigger p53 phosphorylation, resulting in the movement of the pro-apoptotic protein Bax to the outer membrane of mitochondria, increasing mitochondrial membrane permeability, releasing cytochrome c into the cytoplasm, and subsequently activating Caspase-9 and downstream effector molecule Caspase-3, ultimately initiating cell apoptosis (103, 135). Notably, TGEV infection upregulates the expression of various antioxidant-related genes, likely as a host response to virus-induced oxidative damage (127, 135). However, approximately 12 hours post-infection, during active virus replication, substantial mitochondrial degradation and increased autophagosomes are observed, indicating persistent mitophagy activation. While mitophagy can mitigate oxidative damage to some extent, excessive activation may exacerbate cell apoptosis by depleting mitochondrial function (136). At the S and G2/M phases of the host cell cycle, TGEV’s N protein facilitates the activation of p53 and subsequently upregulates its downstream effector p21, further promoting cell death through the intrinsic apoptosis pathway (136). These processes eventually cause apoptosis of porcine small intestinal epithelial cells, leading to villi atrophy, thinning and loss of intestinal wall elasticity, gastrointestinal bleeding, barrier function damage, and finally severe diarrhea, which is a lethal symptom of TGEV infection (134).
7 Pyroptosis induced by TGEV
Pyroptosis is a programmed cell death mechanism that hinges on the activation of caspase-1-like proteases and is distinguished by pronounced inflammatory characteristics (137). This process is primarily triggered by the cleavage and activation of Gasdermin D (GSDMD), which subsequently facilitates the formation of membrane pores, the release of inflammatory mediators, and ultimately culminates in cell lysis and demise (138). Research has demonstrated that infection of small intestinal crypt cells, particularly Paneth cells, by the TGEV can prompt caspase-1 activation, GSDMD cleavage, and the initiation of a classical pyroptotic cascade (139, 140).
Following infection of intestinal epithelial cells by TGEV, the virus’s PAMPs, such as viral RNA, are recognized by inflammasome sensors like NLRP3 within host cells (139, 141). This recognition initiates inflammasome assembly and activates caspase-1, which subsequently cleaves GSDMD to release its N-terminal fragment (GSDMD-N). GSDMD-N then integrates into the cell membrane to form pores, disrupting membrane integrity and causing the release of cellular contents, including the inflammatory cytokines IL-1β and IL-18 (139). This cascade not only elicits a local inflammatory response in the intestine but also represents a pivotal mechanism through which TGEV induces intestinal damage and pathological alterations (141).
8 Summary and outlook
In recent decades, the evolutionary pace of coronaviruses has notably quickened, leading to the emergence of highly pathogenic strains like SARS-CoV, MERS-CoV, and SARS-CoV-2, which have posed significant challenges to global public health (22). The recurrent epidemics and outbreaks of novel coronaviruses have reignited interest in studying the impact of TGEV, a prototypical porcine α-coronavirus, on the swine industry and its possible zoonotic characteristics (21, 142).
TGEV employs multilayered mechanisms to interfere with host innate immune responses, thereby achieving effective immune evasion. These strategies include suppression of PRRs-mediated signaling pathways such as TLRs and RLRs to inhibit interferon production; modification of viral RNA with a cap structure to escape host RNA sensing; modulation of host signaling pathways such as Notch to dampen immune responses; disruption of the intestinal epithelial barrier and reshaping of gut microbiota to compromise barrier function; and induction of multiple forms of programmed cell death, including pyroptosis, apoptosis, and autophagy, to weaken antiviral defenses. Moreover, the TGEV genome exhibits high variability, conferring robust immune evasion capacity and limiting the cross-protective efficacy of existing vaccines, which contributes to unstable protection. Although the clinical detection rate of TGEV has decreased in recent years, this does not mean that its disappearance from natural reservoirs (143). On the contrary, owing to its highly mutable genome, substantial immune evasion ability, and limited cross-protection, TGEV continues to evolve (21). One of the key unresolved scientific questions is the precise identification of the molecular targets of TGEV nonstructural proteins (NSPs). Given that NSPs play pivotal roles in suppressing host innate immunity, yet their specific targets and mechanisms remain incompletely understood, elucidating these interactions will offer an important theoretical foundation and potential intervention strategies for mitigating viral immune evasion. In addition, a complex interplay exists between enteric viruses and the host gut microbiota, which may critically modulate viral immune evasion. Therefore, strategies designed to minimize or prevent the disruption of gut microbial homeostasis induced by TGEV represent a crucial direction for future therapeutic development.Elucidating the tripartite interactions among TGEV, the microbiota, and the host will not only enhance our understanding of TGEV pathogenesis but also provide theoretical foundations for the development of novel microbiota-based interventions against TGEV. Notably, TGEV has traditionally been considered strictly host-specific, confined to infecting swine species. However, increasing evidence indicates that coronavirus host barriers are not absolute, and their cross-species transmission potential may have been substantially underestimated (144). Recent studies have unveiled the molecular basis of TGEV cross-species transmission, showing that the receptor-binding domain (RBD) of TGEV can interact with aminopeptidase N (APN) from 17 different species, with eight demonstrating relatively high binding efficiency. This finding underscores the potential of TGEV for cross - species transmission, raising concerns regarding the possibility of human infection.
Currently available TGEV vaccines, which predominantly rely on traditional platforms, are confronted with multiple limitations, encompass insufficient protective efficacy, restricted cross-protection, risks of reversion to virulence, and immune interference. Consequently, there is an urgent demand for more advanced and effective preventive strategies. Future research directions may include: (i) rational design of broad-spectrum coronavirus vaccines based on conserved antigenic epitopes of TGEV; (ii) exploration of novel mucosal adjuvants and efficient delivery systems, coupled with genetic engineering or pharmacological induction to enhance host antiviral effectors (e.g., upregulating of GSDMD) to improve intracellular pathogen clearance; and (iii) rational attenuation strategies and targeted drug development to inhibit key viral immune evasion proteins (e.g., NSP1, ORF6), thereby blocking their interference with host immune signaling.Furthermore, given the potential cross-species transmission risk of TGEV and other coronaviruses, the establishment of comprehensive surveillance and prevention systems is essential. Such systems should encompass continuous monitoring and tracing of viral recombination events, scientific evaluation of human susceptibility, and integration of human, animal, and environmental health management under a “One Health” framework. This integration will enable proactive prevention and rapid response to potential public health threats.
Future research should therefore prioritize core scientific issues, including: (i) the immune evasion targets of nonstructural proteins (e.g., NSP1, NSP3, ORF6); (ii) the impact of virus–microbiota interactions on immune evasion; (iii) the cross-species transmission potential of TGEV and its implications for human health; and (iv) the risk of generating novel viral populations through recombination with other porcine coronaviruses. Addressing these questions will provide critical guidance for vaccine development, intervention strategies, and public health preparedness. The advancement of state-of-the-art technologies, such as single-cell sequencing, spatial transcriptomics, proteomics, and CRISPR screening, is enhancing the precision of constructing TGEV infection models and target screening. This progress is anticipated to advance translational research on antiviral drugs and intervention strategies (145). Future optimization efforts may involve integrating innovative adjuvant technologies, nanodelivery systems, mucosal immunity strategies, and novel vaccines utilizing virus-like particles (VLPs) and mRNA platforms. Additionally, vigilance in monitoring recombination and evolution events between TGEV and other porcine coronaviruses (e.g., PEDV, PDCoV, SADS-CoV) is crucial for early detection of emerging coronaviruses (146). Despite the reduced clinical impact of TGEV amid frequent zoonotic diseases, its potential for ongoing recombination and evolution poses indirect threats to human health that cannot be disregarded (146, 147). Consequently, comprehensive research on the mechanisms of cross-species transmission, evolutionary dynamics, and potential public health risks associated with porcine coronaviruses should be intensified to enable early detection and effective management of potential emerging zoonotic viruses.
Author contributions
ZW: Writing – original draft, Writing – review & editing. JX: Writing – original draft. QL: Writing – original draft. YL: Writing – review & editing. XZ: Writing – review & editing. EM: Writing – review & editing. LW: Writing – review & editing, Writing – original draft, Conceptualization. LyW: Writing – original draft, Conceptualization. FZ: Writing – review & editing, Conceptualization.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the Dalian Science and Technology Talent Innovation Support Program Project (Grant No. 2022RJ20) and the Basic Scientific Research Project of the Education Department of Liaoning Province (Grant No. LJ212410161056).
Acknowledgments
We want to express our gratitude for the drawing materials provided by BioRender.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Centazzo M, Manganaro L, and Alvisi G. Cellular targets of HIV-1 protease: just the tip of the iceberg? Viruses. (2023) 15:712. doi: 10.3390/v15030712
2. Camargo LDN, Righetti RF, De Almeida FM, Dos Santos TM, Fukuzaki S, Martins NAB, et al. Modulating asthma-COPD overlap responses with IL-17 inhibition. Front Immunol. (2023) 14:1271342. doi: 10.3389/fimmu.2023.1271342
3. Martins-Da-Silva A, Telleria EL, Batista M, Marchini FK, Traub-Csekö YM, Tempone AJ, et al. Identification of Secreted Proteins Involved in Nonspecific dsRNA-Mediated Lutzomyia longipalpis LL5 Cell Antiviral Response. Viruses. (2018) 10:43. doi: 10.3390/v10010043
4. Simper JD, Perez E, Schlesinger LS, and Azad AK. Resistance and Susceptibility Immune Factors at Play during Mycobacterium tuberculosis Infection of Macrophages. Pathogens. (2022) 11:1153. doi: 10.3390/pathogens11101153
5. Shaji S, Selvaraj RK, and Shanmugasundaram R. Salmonella infection in poultry: A review on the pathogen and control strategies. Microorganisms. (2023) 11:2814. doi: 10.3390/microorganisms11112814
6. Gedefaw L, Ullah S, Lee TMH, Yip SP, and Huang CL. Targeting inflammasome activation in COVID-19: delivery of RNA interference-based therapeutic molecules. Biomedicines. (2021) 9:1823. doi: 10.3390/biomedicines9121823
7. Zeng C, Wan SR, Guo M, Tan XZ, Zeng Y, Wu Q, et al. Fecal virome transplantation: A promising strategy for the treatment of metabolic diseases. BioMed Pharmacother. (2024) 177:117065. doi: 10.1016/j.biopha.2024.117065
8. Niu J and Meng G. Roles and mechanisms of NLRP3 in influenza viral infection. Viruses. (2023) 15:1339. doi: 10.3390/v15061339
9. Pejler G, Zhao XO, Fagerström E, and Paivandy A. Blockade of endolysosomal acidification suppresses TLR3-mediated proinflammatory signaling in airway epithelial cells. J Allergy Clin Immunol. (2024) 154:940–51. doi: 10.1016/j.jaci.2024.05.031
10. Collins C, Chaumont L, Peruzzi M, Jamak N, Boudinot P, Béjar J, et al. Effect of a loss of the mda5/ifih1 gene on the antiviral resistance in a Chinook salmon Oncorhynchus tshawytscha cell line. PLoS One. (2024) 19:e0311283. doi: 10.1371/journal.pone.0311283
11. Wang Q, Yu Y, Zhuang J, Liu R, and Sun C. Demystifying the cGAS-STING pathway: precision regulation in the tumor immune microenvironment. Mol Cancer. (2025) 24:178. doi: 10.1186/s12943-025-02380-0
12. Wang X, Wang Q, Zheng C, and Wang L. MAVS: The next STING in cancers and other diseases. Crit Rev Oncol Hematol. (2025) 207:104610. doi: 10.1016/j.critrevonc.2024.104610
13. Liu T, Feng M, Wen Z, He Y, Lin W, Zhang M, et al. Comparison of the characteristics of cytokine storm and immune response induced by SARS-coV, MERS-coV, and SARS-coV-2 infections. J Inflamm Res. (2021) 14:5475–87. doi: 10.2147/JIR.S329697
14. Chen L, Zhang X, Shao G, Shao Y, Hu Z, Feng K, et al. Construction and evaluation of recombinant pseudorabies virus expressing african swine fever virus antigen genes. Front Vet Sci. (2022) 9:832255. doi: 10.3389/fvets.2022.832255
15. Villamayor L, López-García D, Rivero V, Martínez-Sobrido L, Nogales A, DeDiego ML, et al. The IFN-stimulated gene IFI27 counteracts innate immune responses after viral infections by interfering with RIG-I signaling. Front Microbiol. (2023) 14:1176177. doi: 10.3389/fmicb.2023.1176177
16. Wang H, Li W, and Zheng SJ. Advances on innate immune evasion by avian immunosuppressive viruses. Front Immunol. (2022) 13:901913. doi: 10.3389/fimmu.2022.901913
17. Crespo FI, Mayora SJ, De Sanctis JB, Martínez WY, Zabaleta-Lanz ME, Toro FI, et al. SARS-coV-2 infection in Venezuelan pediatric patients-A single center prospective observational study. Biomedicines. (2023) 11:1409. doi: 10.3390/biomedicines11051409
18. Kamau E, Bessaud M, Majumdar M, Martin J, Simmonds P, Harvala H, et al. Estimating prevalence of Enterovirus D111 in human and non-human primate populations using cross-sectional serology. J Gen Virol. (2023) 104:001915. doi: 10.1099/jgv.0.001915
19. Gerber PF, Lelli D, Zhang J, Strandbygaard B, Moreno A, Lavazza A, et al. Diagnostic evaluation of assays for detection of antibodies against porcine epidemic diarrhea virus (PEDV) in pigs exposed to different PEDV strains. Prev Vet Med. (2016) 135:87–94. doi: 10.1016/j.prevetmed.2016.11.005
20. Olech M and Antas M. Transmissible gastroenteritis virus (TGEV) and porcine respiratory coronavirus (PRCV): epidemiology and molecular characteristics-an updated overview. Viruses. (2025) 17:493. doi: 10.3390/v17040493
21. Chen Y, Zhang Y, Wang X, Zhou J, Ma L, Li J, et al. Transmissible gastroenteritis virus: an update review and perspective. Viruses. (2023) 15:359. doi: 10.3390/v15020359
22. Turlewicz-Podbielska H and Pomorska-Mól M. Porcine coronaviruses: overview of the state of the art. Virol Sin. (2021) 36:833–51. doi: 10.1007/s12250-021-00364-0
23. Zhang Y, Yang N, Li Y, Tan C, Cai Y, Rui X, et al. Transmissible gastroenteritis virus induces inflammatory responses via RIG-I/NF-κB/HIF-1α/glycolysis axis in intestinal organoids and in vivo. J Virol. (2024) 98:e0046124. doi: 10.1128/jvi.00461-24
24. Pu J, Chen D, Tian G, He J, Zheng P, Huang Z, et al. All-trans retinoic acid alleviates transmissible gastroenteritis virus-induced intestinal inflammation and barrier dysfunction in weaned piglets. J Anim Sci Biotechnol. (2024) 15:22. doi: 10.1186/s40104-023-00978-2
25. Antas M and Olech M. First report of transmissible gastroenteritis virus (TGEV) and porcine respiratory coronavirus (PRCV) in pigs from Poland. BMC Vet Res. (2024) 20:517. doi: 10.1186/s12917-024-04364-6
26. Schmidt A, Rothenfusser S, and Hopfner KP. Sensing of viral nucleic acids by RIG-I: from translocation to translation. Eur J Cell Biol. (2012) 91:78–85. doi: 10.1016/j.ejcb.2011.01.015
27. Lai JH, Luo SF, and Ho LJ. Operation of mitochondrial machinery in viral infection-induced immune responses. Biochem Pharmacol. (2018) 156:348–56. doi: 10.1016/j.bcp.2018.08.044
28. Reikine S, Nguyen JB, and Modis Y. Pattern recognition and signaling mechanisms of RIG-I and MDA5. Front Immunol. (2014) 5:342. doi: 10.3389/fimmu.2014.00342
29. Chen N, Xia P, Li S, Zhang T, Wang TT, Zhu J, et al. RNA sensors of the innate immune system and their detection of pathogens. IUBMB Life. (2017) 69:297–304. doi: 10.1002/iub.1625
30. Jensen S and Thomsen AR. Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. J Virol. (2012) 86:2900–10. doi: 10.1128/JVI.05738-11
31. Lester SN and Li K. Toll-like receptors in antiviral innate immunity. J Mol Biol. (2014) 426:1246–64. doi: 10.1016/j.jmb.2013.11.024
32. Amurri L, Horvat B, and Iampietro M. Interplay between RNA viruses and cGAS/STING axis in innate immunity. Front Cell Infect Microbiol. (2023) 13:1172739. doi: 10.3389/fcimb.2023.1172739
33. Mosallanejad K and Kagan JC. Control of innate immunity by the cGAS-STING pathway. Immunol Cell Biol. (2022) 100:409–23. doi: 10.1111/imcb.12555
34. Thompson MR, Kaminski JJ, Kurt-Jones EA, and Fitzgerald KA. Pattern recognition receptors and the innate immune response to viral infection. Viruses. (2011) 3:920–40. doi: 10.3390/v3060920
35. Jacobs SR and Damania B. NLRs, inflammasomes, and viral infection. J Leukoc Biol. (2012) 92:469–77. doi: 10.1189/jlb.0312132
36. Zhao W. Negative regulation of TBK1-mediated antiviral immunity. FEBS Lett. (2013) 587:542–8. doi: 10.1016/j.febslet.2013.01.052
37. Tu D, Zhu Z, Zhou AY, Yun CH, Lee KE, Toms AV, et al. Structure and ubiquitination-dependent activation of TANK-binding kinase 1. Cell Rep. (2013) 3:747–58. doi: 10.1016/j.celrep.2013.01.033
38. Ma W, Huang G, Wang Z, Wang L, Gao Q, et al. IRF7: role and regulation in immunity and autoimmunity. Front Immunol. (2023) 14:1236923. doi: 10.3389/fimmu.2023.1236923
39. Dalskov L, Gad HH, and Hartmann R. Viral recognition and the antiviral interferon response. EMBO J. (2023) 42:e112907. doi: 10.15252/embj.2022112907
40. Kazzaz SA, Shaikh KA, White J, Zhou Q, Powell WH, Harhaj EW, et al. Phosphorylation of aryl hydrocarbon receptor interacting protein by TBK1 negatively regulates IRF7 and the type I interferon response. J Biol Chem. (2024) 300:105525. doi: 10.1016/j.jbc.2023.105525
41. Miyamura T, Kanda T, Nakamoto S, Wu S, Fujiwara K, Imazeki F, et al. Hepatic STAT1-nuclear translocation and interleukin 28B polymorphisms predict treatment outcomes in hepatitis C virus genotype 1-infected patients. PLoS One. (2011) 6:e28617. doi: 10.1371/journal.pone.0028617
42. Andreakos E, Zanoni I, and Galani IE. Lambda interferons come to light: dual function cytokines mediating antiviral immunity and damage control. Curr Opin Immunol. (2019) 56:67–75. doi: 10.1016/j.coi.2018.10.007
43. Liu YG, Jin SW, Zhang SS, Xia TJ, Liao YH, Pan RL, et al. Interferon lambda in respiratory viral infection: immunomodulatory functions and antiviral effects in epithelium. Front Immunol. (2024) 15:1338096. doi: 10.3389/fimmu.2024.1338096
44. Raftery N and Stevenson NJ. Advances in anti-viral immune defence: revealing the importance of the IFN JAK/STAT pathway. Cell Mol Life Sci. (2017) 74:2525–35. doi: 10.1007/s00018-017-2520-2
45. Lozhkov AA, Klotchenko SA, Ramsay ES, Moshkoff HD, Moshkoff DA, Vasin AV, et al. The key roles of interferon lambda in human molecular defense against respiratory viral infections. Pathogens. (2020) 9:989. doi: 10.3390/pathogens9120989
46. Lazear HM, Nice TJ, and Diamond MS. Interferon-λ: immune functions at barrier surfaces and beyond. Immunity. (2015) 43:15–28. doi: 10.1016/j.immuni.2015.07.001
47. Yang E and Li MMH. All about the RNA: interferon-stimulated genes that interfere with viral RNA processes. Front Immunol. (2020) 11:605024. doi: 10.3389/fimmu.2020.605024
48. Li MM, Macdonald MR, and Rice CM. To translate, or not to translate: viral and host mRNA regulation by interferon-stimulated genes. Trends Cell Biol. (2015) 25:320–9. doi: 10.1016/j.tcb.2015.02.001
49. Yin L, Liu X, Hu D, Luo Y, Zhang G, Liu P, et al. Swine enteric coronaviruses (PEDV, TGEV, and PDCoV) induce divergent interferon-stimulated gene responses and antigen presentation in porcine intestinal enteroids. Front Immunol. (2021) 12:826882. doi: 10.3389/fimmu.2021.826882
50. Wang F, Shen F, Wang Y, Li Z, Chen J, Yuan Z, et al. Residues Asn118 and Glu119 of hepatitis B virus X protein are critical for HBx-mediated inhibition of RIG-I-MAVS signaling. Virology. (2020) 539:92–103. doi: 10.1016/j.virol.2019.10.009
51. Irmak DK, Darıcı H, and Karaöz E. Stem cell based therapy option in COVID-19: is it really promising? Aging Dis. (2020) 11:1174–91. doi: 10.14336/AD.2020.0608
52. Li S, Yang J, Zhu Y, Ji X, Wang K, Jiang S, et al. Chicken DNA sensing cGAS-STING signal pathway mediates broad spectrum antiviral functions. Vaccines (Basel). (2020) 8:369. doi: 10.3390/vaccines8030369
53. Zanoni I. Interfering with SARS-CoV-2: are interferons friends or foes in COVID-19? Curr Opin Virol. (2021) 50:119–27. doi: 10.1016/j.coviro.2021.08.004
54. Ait-Belkacem I, Cartagena García C, Millet-Wallisky E, Izquierdo N, Loosveld M, Arnoux I, et al. SARS-CoV-2 spike protein induces a differential monocyte activation that may contribute to age bias in COVID-19 severity. Sci Rep. (2022) 12:20824. doi: 10.1038/s41598-022-25259-2
55. Zeng Q, Liu J, Hao C, Zhang B, and Zhang H. Making sense of flavivirus non-strctural protein 1 in innate immune evasion and inducing tissue-specific damage. Virus Res. (2023) 336:199222. doi: 10.1016/j.virusres.2023.199222
56. Perlas A, Argilaguet J, Bertran K, Sánchez-González R, Nofrarías M, Valle R, et al. Dual host and pathogen RNA-seq analysis unravels chicken genes potentially involved in resistance to highly pathogenic avian influenza virus infection. Front Immunol. (2021) 12:800188. doi: 10.3389/fimmu.2021.800188
57. Li X, Lidsky PV, Xiao Y, Wu CT, Garcia-Knight M, Yang J, et al. Ethacridine inhibits SARS-CoV-2 by inactivating viral particles. PLoS Pathog. (2021) 17:e1009898. doi: 10.1371/journal.ppat.1009898
58. ZUñIGA S, Pascual-Iglesias A, Sanchez CM, Sola I, Enjuanes L, et al. Virulence factors in porcine coronaviruses and vaccine design. Virus Res. (2016) 226:142–51. doi: 10.1016/j.virusres.2016.07.003
59. Pahmeier F, Neufeldt CJ, Cerikan B, Prasad V, Pape C, Laketa V, et al. A versatile reporter system to monitor virus-infected cells and its application to dengue virus and SARS-coV-2. J Virol. (2021) 95:e01715-20. doi: 10.1128/JVI.01715-20
60. Cai HL and Huang YW. Reverse genetics systems for SARS-CoV-2: Development and applications. Virol Sin. (2023) 38:837–50. doi: 10.1016/j.virs.2023.10.001
61. Sun X, Quan L, Chen R, and Liu D. Direct interaction of coronavirus nonstructural protein 3 with melanoma differentiation-associated gene 5 modulates type I interferon response during coronavirus infection. Int J Mol Sci. (2022) 23:11692. doi: 10.3390/ijms231911692
62. Shen Z, Wang G, Yang Y, Shi J, Fang L, Li F, et al. A conserved region of nonstructural protein 1 from alphacoronaviruses inhibits host gene expression and is critical for viral virulence. J Biol Chem. (2019) 294:13606–18. doi: 10.1074/jbc.RA119.009713
63. Dutta P, Kshirsagar A, Bibekar P, and Sengupta N. Conformational ensemble of the NSP1 CTD in SARS-CoV-2: Perspectives from the free energy landscape. Biophys J. (2023) 122:2948–59. doi: 10.1016/j.bpj.2023.02.010
64. Yoo D, Song C, Sun Y, Du Y, Kim O, Liu HC, et al. Modulation of host cell responses and evasion strategies for porcine reproductive and respiratory syndrome virus. Virus Res. (2010) 154:48–60. doi: 10.1016/j.virusres.2010.07.019
65. Dai J, Agbemabiese CA, Griffin AN, and Patton JT. Rotavirus capping enzyme VP3 inhibits interferon expression by inducing MAVS degradation during viral replication. mBio. (2023) 14:e0225523. doi: 10.1128/mbio.02255-23
66. Kuang M, Zhao Y, Yu H, Li S, Liu T, Chen L, et al. XAF1 promotes anti-RNA virus immune responses by regulating chromatin accessibility. Sci Adv. (2023) 9:eadg5211. doi: 10.1126/sciadv.adg5211
67. Onsa-Ard A, Thongboontho R, Munkong N, Phromnoi K, Ontawong A, Pengnet S, et al. Anti-inflammatory effects of red rice bran extract ameliorate type I interferon production via STING pathway. Foods. (2022) 11:1622. doi: 10.3390/foods11111622
68. Xia H, Cao Z, Xie X, Zhang X, Chen JY, Wang H, et al. Evasion of type I interferon by SARS-coV-2. Cell Rep. (2020) 33:108234. doi: 10.1016/j.celrep.2020.108234
69. Rashid F, Xie Z, Suleman M, Shah A, Khan S, Luo S, et al. Roles and functions of SARS-CoV-2 proteins in host immune evasion. Front Immunol. (2022) 13:940756. doi: 10.3389/fimmu.2022.940756
70. Yuan S, Balaji S, Lomakin IB, and Xiong Y. Coronavirus nsp1: immune response suppression and protein expression inhibition. Front Microbiol. (2021) 12:752214. doi: 10.3389/fmicb.2021.752214
71. Tang Y, Zhang Y, Yang N, Shi H, Fu Y, Bai B, et al. TGEV NSP1 enhances viral replication through antagonizing stress granule formation. Vet Microbiol. (2025) 305:110502. doi: 10.1016/j.vetmic.2025.110502
72. Li M, Guo L, and Feng L. Interplay between swine enteric coronaviruses and host innate immune. Front Vet Sci. (2022) 9:1083605. doi: 10.3389/fvets.2022.1083605
73. Yang M, Mariano J, Su R, Smith CE, Das S, Gill C, et al. SARS-CoV-2 papain-like protease plays multiple roles in regulating cellular proteins in the endoplasmic reticulum. J Biol Chem. (2023) 299:105346. doi: 10.1016/j.jbc.2023.105346
74. Ran XH, Zhu JW, Chen YY, Ni RZ, and Mu D. Papain-like protease of SARS-CoV-2 inhibits RLR signaling in a deubiquitination-dependent and deubiquitination-independent manner. Front Immunol. (2022) 13:947272. doi: 10.3389/fimmu.2022.947272
75. Cao D, Duan L, Huang B, Xiong Y, Zhang G, Huang H, et al. The SARS-CoV-2 papain-like protease suppresses type I interferon responses by deubiquitinating STING. Sci Signal. (2023) 16:eadd0082. doi: 10.1126/scisignal.add0082
76. Russ A, Wittmann S, Tsukamoto Y, Herrmann A, Deutschmann J, Lagisquet J, et al. Nsp16 shields SARS-CoV-2 from efficient MDA5 sensing and IFIT1-mediated restriction. EMBO Rep. (2022) 23:e55648. doi: 10.15252/embr.202255648
77. Mandilara G, Koutsi MA, Agelopoulos M, Sourvinos G, Beloukas A, Rampias T, et al. The role of coronavirus RNA-processing enzymes in innate immune evasion. Life (Basel). (2021) 11:571. doi: 10.3390/life11060571
78. Batool M, Kim MS, and Choi S. Structural insights into the distinctive RNA recognition and therapeutic potentials of RIG-I-like receptors. Med Res Rev. (2022) 42:399–425. doi: 10.1002/med.21845
79. Oshiumi H. Recent advances and contradictions in the study of the individual roles of ubiquitin ligases that regulate RIG-I-like receptor-mediated antiviral innate immune responses. Front Immunol. (2020) 11:1296. doi: 10.3389/fimmu.2020.01296
80. Fang R, Jiang Q, Zhou X, Wang C, Guan Y, Tao J, et al. MAVS activates TBK1 and IKKϵ through TRAFs in NEMO dependent and independent manner. PLoS Pathog. (2017) 13:e1006720. doi: 10.1371/journal.ppat.1006720
81. Chen X, Liu J, Li Y, Zeng Y, Wang F, Cheng Z, et al. IDH1 mutation impairs antiviral response and potentiates oncolytic virotherapy in glioma. Nat Commun. (2023) 14:6781. doi: 10.1038/s41467-023-42545-3
82. Lu LF, Jiang JY, Du WX, Wang XL, Li ZC, Zhou XY, et al. Fish female-biased gene cyp19a1a leads to female antiviral response attenuation between sexes by autophagic degradation of MITA. PLoS Pathog. (2022) 18:e1010626. doi: 10.1371/journal.ppat.1010626
83. Schindewolf C, Lokugamage K, Vu MN, Johnson BA, Scharton D, Plante JA, et al. SARS-coV-2 uses nonstructural protein 16 to evade restriction by IFIT1 and IFIT3. J Virol. (2023) 97:e0153222. doi: 10.1128/jvi.01532-22
84. Tam D, Lorenzo-Leal AC, Hernández LR, Bach H, et al. Targeting SARS-coV-2 non-structural proteins. Int J Mol Sci. (2023) 24:13002. doi: 10.3390/ijms241613002
85. Matarazzo L and Bettencourt PJG. mRNA vaccines: a new opportunity for malaria, tuberculosis and HIV. Front Immunol. (2023) 14:1172691. doi: 10.3389/fimmu.2023.1172691
86. Misra A, Rahisuddin R, Parihar M, Arya S, Viswanathan T, Jackson N, et al. Structural Insights into the Assembly and Regulation of 2’- O RNA Methylation by SARS-CoV-2 nsp16/nsp10. bioRxiv. (2024) 33:1027–39. doi: 10.1101/2024.12.19.628950
87. Viswanathan T, Arya S, Chan SH, Qi S, Dai N, Misra A, et al. Structural basis of RNA cap modification by SARS-CoV-2. Nat Commun. (2020) 11:3718. doi: 10.1038/s41467-020-17496-8
88. Chang LJ and Chen TH. NSP16 2’-O-MTase in coronavirus pathogenesis: possible prevention and treatments strategies. Viruses. (2021) 13:538. doi: 10.3390/v13040538
89. Ataei-Pirkooh A, Alavi A, Kianirad M, Bagherzadeh K, Ghasempour A, Pourdakan O, et al. Destruction mechanisms of ozone over SARS-CoV-2. Sci Rep. (2021) 11:18851. doi: 10.1038/s41598-021-97860-w
90. Wu A, Yu B, Zhang K, Xu Z, Wu D, He J, et al. Transmissible gastroenteritis virus targets Paneth cells to inhibit the self-renewal and differentiation of Lgr5 intestinal stem cells via Notch signaling. Cell Death Dis. (2020) 11:40. doi: 10.1038/s41419-020-2233-6
91. Bao C, Zhang W, Wang J, Liu Y, Cao H, Li F, et al. The effects of dietary bacillus amyloliquefaciens TL106 supplementation, as an alternative to antibiotics, on growth performance, intestinal immunity, epithelial barrier integrity, and intestinal microbiota in broilers. Animal (Basel). (2022) 12:3085. doi: 10.3390/ani12223085
92. Xu CL, Guo Y, Qiao L, Ma L, and Cheng YY. Recombinant expressed vasoactive intestinal peptide analogue ameliorates TNBS-induced colitis in rats. World J Gastroenterol. (2018) 24:706–15. doi: 10.3748/wjg.v24.i6.706
93. Luo L, Zhou Z, Xue J, Wang Y, Zhang J, Cai X, et al. Bletilla striata polysaccharide has a protective effect on intestinal epithelial barrier disruption in TAA-induced cirrhotic rats. Exp Ther Med. (2018) 16:1715–22. doi: 10.3892/etm.2018.6430
94. Huang J, Han S, Sun Q, Zhao Y, Liu J, Yuan X, et al. Kv1.3 channel blocker (ImKTx88) maintains blood-brain barrier in experimental autoimmune encephalomyelitis. Cell Biosci. (2017) 7:31. doi: 10.1186/s13578-017-0158-2
95. Kuo WT, Odenwald MA, Turner JR, and Zuo L. Tight junction proteins occludin and ZO-1 as regulators of epithelial proliferation and survival. Ann N Y Acad Sci. (2022) 1514:21–33. doi: 10.1111/nyas.14798
96. Zhao S, Gao J, Zhu L, and Yang Q. Transmissible gastroenteritis virus and porcine epidemic diarrhoea virus infection induces dramatic changes in the tight junctions and microfilaments of polarized IPEC-J2 cells. Virus Res. (2014) 192:34–45. doi: 10.1016/j.virusres.2014.08.014
97. Chen YM and Burrough E. The effects of swine coronaviruses on ER stress, autophagy, apoptosis, and alterations in cell morphology. Pathogens. (2022) 11:940. doi: 10.3390/pathogens11080940
98. Pu J, Chen D, Tian G, He J, Huang Z, Zheng P, et al. All-trans retinoic acid attenuates transmissible gastroenteritis virus-induced apoptosis in IPEC-J2 cells via inhibiting ROS-mediated P(38)MAPK signaling pathway. Antioxid (Basel). (2022) 11:345. doi: 10.3390/antiox11020345
99. Wang L, Qiao X, Zhang S, Qin Y, Guo T, Hao Z, et al. Porcine transmissible gastroenteritis virus nonstructural protein 2 contributes to inflammation via NF-κB activation. Virulence. (2018) 9:1685–98. doi: 10.1080/21505594.2018.1536632
100. Pu J, Chen D, Tian G, He J, Huang Z, Zheng P, et al. All-trans retinoic acid attenuates transmissible gastroenteritis virus-induced inflammation in IPEC-J2 cells via suppressing the RLRs/NF-κB signaling pathway. Front Immunol. (2022) 13:734171. doi: 10.3389/fimmu.2022.734171
101. Wang K, Tang Y, Wu X, Liang H, Chen D, Yu B, et al. Eugenol attenuates transmissible gastroenteritis virus-induced oxidative stress and apoptosis via ROS-NRF2-ARE signaling. Antioxid (Basel). (2022) 11:1838. doi: 10.3390/antiox11091838
102. Liang W, He L, Ning P, Lin J, Li H, Lin Z, et al. (+)-Catechin inhibition of transmissible gastroenteritis coronavirus in swine testicular cells is involved its antioxidation. Res Vet Sci. (2015) 103:28–33. doi: 10.1016/j.rvsc.2015.09.009
103. Ding L, Zhao X, Huang Y, Du Q, Dong F, Zhang H, et al. Regulation of ROS in transmissible gastroenteritis virus-activated apoptotic signaling. Biochem Biophys Res Commun. (2013) 442:33–7. doi: 10.1016/j.bbrc.2013.10.164
104. Shah WH, Rasool A, Saleem S, Mushtaq NU, Tahir I, Hakeem KR, et al. Understanding the integrated pathways and mechanisms of transporters, protein kinases, and transcription factors in plants under salt stress. Int J Genomics. (2021) 2021:5578727. doi: 10.1155/2021/5578727
105. Shukla PK, Gangwar R, Manda B, Meena AS, Yadav N, Szabo E, et al. Rapid disruption of intestinal epithelial tight junction and barrier dysfunction by ionizing radiation in mouse colon in vivo: protection by N-acetyl-l-cysteine. Am J Physiol Gastrointest Liver Physiol. (2016) 310:G705–15. doi: 10.1152/ajpgi.00314.2015
106. Wang N, Wang S, Xu B, Liu F, Huo G, Li B, et al. Alleviation Effects of Bifidobacterium animalis subsp. lactis XLTG11 on Dextran Sulfate Sodium-Induced Colitis in Mice. Microorganisms. (2021) 9:2093. doi: 10.3390/microorganisms9102093
107. Ding L, Li J, Li W, Fang Z, Li N, Wu S, et al. p53- and ROS-mediated AIF pathway involved in TGEV-induced apoptosis. J Vet Med Sci. (2018) 80:1775–81. doi: 10.1292/jvms.18-0104
108. Nauman M and Stanley P. Glycans that regulate Notch signaling in the intestine. Biochem Soc Trans. (2022) 50:689–701. doi: 10.1042/BST20200782
109. Morris HE, Neves KB, Nilsen M, Montezano AC, MacLean MR, Touyz RM, et al. Notch3/hes5 induces vascular dysfunction in hypoxia-induced pulmonary hypertension through ER stress and redox-sensitive pathways. Hypertension. (2023) 80:1683–96. doi: 10.1161/HYPERTENSIONAHA.122.20449
110. Pellegrinet L, Rodilla V, Liu Z, Chen S, Koch U, Espinosa L, et al. Dll1- and dll4-mediated notch signaling are required for homeostasis of intestinal stem cells. Gastroenterology. (2011) 140:1230–40.e1-7. doi: 10.1053/j.gastro.2011.01.005
111. Zhang S, Zhang S, Hou Y, Hou Y, Huang Y, Cai J, Wang G, et al. Porcine deltacoronavirus infection disrupts the intestinal mucosal barrier and inhibits intestinal stem cell differentiation to goblet cells via the notch signaling pathway. J Virol. (2023) 97:e0068923. doi: 10.1128/jvi.00689-23
112. Fortini F, Vieceli Dalla Sega F, Marracino L, Marracino L, Severi P, Rapezzi C, Rizzo P, et al. Well-known and novel players in endothelial dysfunction: updates on a notch(ed) landscape. Biomedicines. (2021) 9:997. doi: 10.3390/biomedicines9080997
113. Li C, Xiao N, Song W, Lam AH, Liu F, Cui X, et al. Chronic lung inflammation and CK14+ basal cell proliferation induce persistent alveolar-bronchiolization in SARS-CoV-2-infected hamsters. EBioMedicine. (2024) 108:105363. doi: 10.1016/j.ebiom.2024.105363
114. Li J, Zhang Z, Liu H, Qu X, Yin X, Chen L, et al. Effects of continuous intravenous infusion with propofol on intestinal metabolites in rats. BioMed Rep. (2024) 20:25. doi: 10.3892/br.2023.1713
115. Yin L, Liu X, Yao Y, Yuan M, Luo Y, Zhang G, et al. Gut microbiota-derived butyrate promotes coronavirus TGEV infection through impairing RIG-I-triggered local type I interferon responses via class I HDAC inhibition. J Virol. (2024) 98:e0137723. doi: 10.1128/jvi.01377-23
116. Recharla N, Geesala R, and Shi XZ. Gut microbial metabolite butyrate and its therapeutic role in inflammatory bowel disease: A literature review. Nutrients. (2023) 15:2275. doi: 10.3390/nu15102275
117. Xia L, Yang Y, Wang J, Jing Y, and Yang Q. Impact of TGEV infection on the pig small intestine. Virol J. (2018) 15:102. doi: 10.1186/s12985-018-1012-9
118. Chen YM, Yeh CA, Lin WH, Lin CN, and Chiou MT. Dynamic alterations of the intestinal microbiome and metabolome during transmissible gastroenteritis virus infection in weaned pigs. Microb Pathog. (2025) 206:107705. doi: 10.1016/j.micpath.2025.107705
119. Guo Z, Zhang C, Dong J, Wang Y, Hu H, Chen L, et al. Persistence infection of TGEV promotes enterococcus faecalis infection on IPEC-J2 cells. Int J Mol Sci. (2022) 24:3390. doi: 10.3390/ijms24010450
120. Zhao W, Xu M, Barkema HW, Xie X, Lin Y, Khan S, et al. Prototheca bovis induces autophagy in bovine mammary epithelial cells via the HIF-1α and AMPKα/ULK1 pathway. Front Immunol. (2022) 13:934819. doi: 10.3389/fimmu.2022.934819
121. Shi W, Li X, Hou X, Peng H, Jiang Q, Shi M, et al. Differential apoptosis gene expressions of rhabdomyosarcoma cells in response to enterovirus 71 infection. BMC Infect Dis. (2012) 12:327. doi: 10.1186/1471-2334-12-327
122. Lin JY and Huang HI. Autophagy is induced and supports virus replication in Enterovirus A71-infected human primary neuronal cells. Sci Rep. (2020) 10:15234. doi: 10.1038/s41598-020-71970-3
123. Guo L, Yu H, Gu W, Luo X, Li R, Zhang J, et al. Autophagy negatively regulates transmissible gastroenteritis virus replication. Sci Rep. (2016) 6:23864. doi: 10.1038/srep23864
124. Yu J, Ge S, Li J, Zhang Y, Xu J, Wang Y, et al. Interaction between coronaviruses and the autophagic response. Front Cell Infect Microbiol. (2024) 14:1457617. doi: 10.3389/fcimb.2024.1457617
125. Jang YJ, Kim JH, and Byun S. Modulation of autophagy for controlling immunity. Cells. (2019) 8:138. doi: 10.3390/cells8020138
126. Ji M, Li M, Sun L, Zhao H, Li Y, Zhou L, et al. VMP1 and TMEM41B are essential for DMV formation during β-coronavirus infection. J Cell Biol. (2022) 221:e202112081. doi: 10.1083/jcb.202112081
127. Zhu L, Mou C, Yang X, Lin J, and Yang Q. Mitophagy in TGEV infection counteracts oxidative stress and apoptosis. Oncotarget. (2016) 7:27122–41. doi: 10.18632/oncotarget.8345
128. Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, et al. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol. (1998) 72:8586–96. doi: 10.1128/JVI.72.11.8586-8596.1998
129. Shang C, Zhuang X, Zhang H, Li Y, Zhu Y, Lu J, et al. Inhibition of autophagy suppresses SARS-coV-2 replication and ameliorates pneumonia in hACE2 transgenic mice and xenografted human lung tissues. J Virol. (2021) 95:e0153721. doi: 10.1128/JVI.01537-21
130. Carmona-Gutierrez D, Bauer MA, Zimmermann A, Kainz K, Hofer SJ, Kroemer G, et al. Digesting the crisis: autophagy and coronaviruses. Microb Cell. (2020) 7:119–28. doi: 10.15698/mic2020.05.715
131. Huang C, Lu CK, Tu MC, Chang JH, Chen YJ, Tu YH, et al. Polyphenol-rich Avicennia marina leaf extracts induce apoptosis in human breast and liver cancer cells and in a nude mouse xenograft model. Oncotarget. (2016) 7:35874–93. doi: 10.18632/oncotarget.8624
132. Ye T, Ge Y, Jiang X, Song H, Peng C, Liu B, et al. A review of anti-tumour effects of Ganoderma lucidum in gastrointestinal cancer. Chin Med. (2023) 18:107. doi: 10.1186/s13020-023-00811-y
133. Xu K, Guo C, Shi D, Meng J, Tian H, Guo S, et al. Discovery of natural dimeric naphthopyrones as potential cytotoxic agents through ROS-mediated apoptotic pathway. Mar Drugs. (2019) 17:207. doi: 10.3390/md17040207
134. Zhang Q, Xu Y, Chang R, Tong D, and Xu X. Transmissible gastroenteritis virus N protein causes endoplasmic reticulum stress, up-regulates interleukin-8 expression and its subcellular localization in the porcine intestinal epithelial cell. Res Vet Sci. (2018) 119:109–15. doi: 10.1016/j.rvsc.2018.06.008
135. Niizuma K, Endo H, and Chan PH. Oxidative stress and mitochondrial dysfunction as determinants of ischemic neuronal death and survival. J Neurochem. (2009) 109:133–8. doi: 10.1111/j.1471-4159.2009.05897.x
136. Ding L, Huang Y, Du Q, Dong F, Zhao X, Zhang W, et al. TGEV nucleocapsid protein induces cell cycle arrest and apoptosis through activation of p53 signaling. Biochem Biophys Res Commun. (2014) 445:497–503. doi: 10.1016/j.bbrc.2014.02.039
137. Sucharov CC. Paracrine factors in uremic cardiomyopathy. JACC Basic Transl Sci. (2020) 5:167–8. doi: 10.1016/j.jacbts.2020.01.005
138. Evavold CL, Hafner-Bratkovič I, Devant P, D'Andrea JM, Ngwa EM, Boršić E, et al. Control of gasdermin D oligomerization and pyroptosis by the Ragulator-Rag-mTORC1 pathway. Cell. (2021) 184:4495–511.e19. doi: 10.1016/j.cell.2021.06.028
139. Wei G, Luo S, Wu W, Hu J, Zhou R, et al. Activation of interleukin-1β Release and pyroptosis by transmissible gastroenteritis virus is dependent on the NOD-like receptor protein 3 inflammasome in porcine intestinal epithelial cell line. Viral Immunol. (2021) 34:401–9. doi: 10.1089/vim.2020.0227
140. Yang N, Zhang Y, Fu Y, Li Y, Yang S, Chen J, et al. Transmissible gastroenteritis virus infection promotes the self-renewal of porcine intestinal stem cells via wnt/β-catenin pathway. J Virol. (2022) 96:e0096222. doi: 10.1128/jvi.00962-22
141. Churchill MJ, Mitchell PS, and Rauch I. Epithelial pyroptosis in host defense. J Mol Biol. (2022) 434:167278. doi: 10.1016/j.jmb.2021.167278
142. Kong F, Jia H, Xiao Q, Fang L, and Wang Q. Prevention and control of swine enteric coronaviruses in China: A review of vaccine development and application. Vaccines (Basel). (2023) 12:11. doi: 10.3390/vaccines12010011
143. Chen S, Zhang H, Chu M, Cheng W, Zhai J, Wang H, et al. Prevalence of transmissible gastroenteritis among swine populations in China during 1983-2022: A systematic review and meta-analysis. Microb Pathog. (2023) 183:106320. doi: 10.1016/j.micpath.2023.106320
144. Tian Y, Sun J, Hou X, Liu Z, Chen Z, Pan X, et al. Cross-species recognition of two porcine coronaviruses to their cellular receptor aminopeptidase N of dogs and seven other species. PloS Pathog. (2025) 21:e1012836. doi: 10.1371/journal.ppat.1012836
145. Sun L, Zhao C, Fu Z, Fu Y, Su Z, Li Y, et al. Genome-scale CRISPR screen identifies TMEM41B as a multi-function host factor required for coronavirus replication. PloS Pathog. (2021) 17:e1010113. doi: 10.1371/journal.ppat.1010113
146. Guo J, Lai Y, Yang Z, Song W, Zhou J, Li Z, et al. Coinfection and nonrandom recombination drive the evolution of swine enteric coronaviruses. Emerg Microbes Infect. (2024) 13:2332653. doi: 10.1080/22221751.2024.2332653
Keywords: TGEV, immune response, innate immune escape, PRRs, PAMPS, IFN
Citation: Wang Z, Xie J, Li Q, Liu Y, Zhang X, Mi E, Wang L, Wang L and Zhang F (2025) Mechanism by which porcine transmissible gastroenteritis virus disrupts host innate immunity. Front. Immunol. 16:1675572. doi: 10.3389/fimmu.2025.1675572
Received: 29 July 2025; Accepted: 08 September 2025;
Published: 25 September 2025.
Edited by:
Qiang Zhang, Huazhong Agricultural University, ChinaReviewed by:
Yang Wu, Chinese Academy of Agricultural Sciences, ChinaXianwen Wu, Peking University, China
Shijie Xie, Beijing Academy of Agricultural and Forestry Sciences, China
Copyright © 2025 Wang, Xie, Li, Liu, Zhang, Mi, Wang, Wang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liang Wang, d2FuZ2xpYW5nLWR5QGRtdS5lZHUuY24=; Feng Zhang, emZkbEBkbXUuZWR1LmNu; Lingyu Wang, d2FuZ2x5MDFAZG11LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
 Zian Wang
Zian Wang Jiahao Xie
Jiahao Xie Qindan Li3†
Qindan Li3† Liang Wang
Liang Wang