- 1Weifang People’s Hospital, Shandong Second Medical University, Weifang, Shandong, China
- 2School of Clinical Medicine, Shandong Second Medical University, Weifang, Shandong, China
- 3Department of Oncology, Shandong Provincial Key Medical and Health Discipline, Qingdao Central Hospital, University of Health and Rehabilitation Sciences, Qingdao, China
Objective: To investigate the impact of co-mutations of EGFR with TP53 or KRAS on the prognosis of non-small cell lung cancer (NSCLC) patients, and the efficacy of platinum-based doublet chemotherapy plus immunotherapy after EGFR-TKI resistance.
Methods: This was a retrospective study that included 168 patients with locally advanced or advanced NSCLC who had next-generation sequencing (NGS) performed at our institution between January 1, 2021, and October 31, 2023. Based on their genomic profiles, patients were categorized into three groups: EGFR single mutation, EGFR/TP53 co-mutation, and EGFR/KRAS co-mutation. Baseline clinical data were collected, including gender, age, smoking history, histological subtype, clinical stage, ECOG performance status, gene testing results, and treatment regimens. All patients were treated with EGFR tyrosine kinase inhibitors (TKIs) as first-line therapy, including first-, second-, or third-generation agents. Upon disease progression, patients received platinum-based doublet chemotherapy plus immunotherapy as second-line treatment. The primary endpoint was progression-free survival (PFS). Survival curves were generated using the Kaplan-Meier method and compared by log-rank test. Baseline characteristics among the three groups were compared using the chi-square test. Multivariate Cox regression analysis was performed to evaluate independent prognostic factors for PFS by incorporating all baseline clinical variables and gene mutation status into the model.
Results: A total of 168 patients were included in the analysis: 36 with EGFR single mutation, 80 with EGFR/TP53 co-mutation, and 52 with EGFR/KRAS co-mutation. There were no statistically significant differences among the three groups with respect to baseline characteristics, including gender, age, smoking history, histological type, clinical stage, and ECOG performance status (P > 0.05). Immune-related marker expression was significantly different between the EGFR single mutation group and the two co-mutation groups (P < 0.05), while no significant difference was observed between the co-mutation groups (P = 0.945). Following first-line EGFR-TKI therapy, the EGFR single mutation group showed a significantly longer median PFS compared with the EGFR/TP53 and EGFR/K-RAS co-mutation groups (P < 0.0001). No significant difference in PFS was observed between the two co-mutation groups (P = 0.174). Following progression on EGFR-TKIs, all patients received platinum-based doublet chemotherapy plus immunotherapy. In second-line treatment, the median PFS in the EGFR single-mutation group, which was shorter than in the EGFR/TP53 and EGFR/KRAS co-mutation groups (overall log-rank P < 0.0001), with no significant difference between the two co-mutation cohorts (P = 0.174). However, in multivariable Cox models adjusting for age, sex, smoking history, clinical stage, histology, and ECOG performance status, both EGFR/TP53 and EGFR/KRAS co-mutations were independently associated with a higher hazard of progression. ECOG PS ≥2 was associated with a numerically higher hazard that did not reach statistical significance. No significant associations were observed for other covariates (age, sex, smoking history, clinical stage, histology; all P>0.05).
Conclusion: In the first-line setting, patients with an EGFR single mutation treated with EGFR-TKIs had a longer median PFS than those with EGFR/TP53 and EGFR/KRAS co-mutations (14.1 vs 10.4 and 10.9 months, respectively; both P < 0.0001), whereas no statistically significant difference was observed between the two co-mutation subgroups (P = 0.174). Following the development of resistance, all patients received platinum-based doublet chemotherapy plus immunotherapy; in the second-line setting, median PFS was modestly longer in the co-mutation groups compared with the single-mutation group (EGFR/TP53: 5.2 months; EGFR/KRAS: 5.0 months; EGFR single mutation: 3.9 months; overall log-rank P < 0.0001), with no significant difference between the TP53 and KRAS subgroups (P = 0.174). These associations were evident on Kaplan–Meier curves (with numbers at risk) and log-rank testing, and were supported by multivariable Cox models adjusted for age, sex, smoking history, clinical stage, histology, and ECOG performance status.
1 Introduction
Lung cancer is one of the most prevalent and lethal malignancies worldwide, and it remains the leading cause of cancer-related death (1). The pathological types of lung cancer are mainly divided into small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC), with NSCLC accounting for approximately 80%, including adenocarcinoma and squamous cell carcinoma (2, 3). Although low-dose spiral computed tomography (CT) can detect lung cancer at an early stage, more than 40% of patients are diagnosed at locally advanced or advanced stages, losing the opportunity for a cure (4). Treatment options for NSCLC are diverse, including surgery, chemotherapy, radiotherapy, targeted therapy, and immunotherapy; however, even in cases of early detection, the majority of patients progress to advanced stages within five years. The 5-year survival rates for locally advanced and advanced NSCLC are less than 15% and 5%, respectively (5).
With advances in genomics and high-throughput gene sequencing, tumor treatment has entered an era of precision medicine. The widespread application of EGFR tyrosine kinase inhibitors (TKIs) has significantly improved the prognosis of NSCLC patients and has become the standard treatment for locally advanced and advanced NSCLC. However, there is significant heterogeneity in the efficacy of EGFR-TKI, with progression-free survival (PFS) ranging from several months to years (6). Some patients exhibit primary resistance to EGFR-TKIs, while others initially respond but ultimately experience divergent outcomes. The mechanisms underlying primary resistance to EGFR-TKIs are complex and may be related to factors such as EGFR-insensitive mutations (e.g., exon 20 insertions, T790M mutations), downstream gene mutations (e.g., KRAS, BRAF, PIK3CA), co-occurring mutations (e.g., TP53, MET, ALK rearrangements), changes in microRNA, and BIM gene deletion (7–12). Among these, the impact of co-mutations involving EGFR and other genes on EGFR-TKI therapy has drawn significant attention.
TP53 is an important tumor suppressor gene that inhibits abnormal cell proliferation by regulating the cell cycle and inducing apoptosis (13), and it is referred to as the “guardian gene.” TP53 mutations can lead to cellular malignancy and increased aggressiveness (14). Currently, there are no targeted drugs available for TP53, and studies have shown that EGFR-mutant NSCLC patients with co-occurring TP53 mutations are more prone to resistance against EGFR-TKIs. Therefore, new treatment strategies are required to improve survival benefits after resistance develops. KRAS gene mutations are common in smokers and lead to sustained activation of GTPase, which results in continuous activation of downstream signaling pathways and, ultimately, cellular malignancy (15). KRAS and EGFR mutations occur independently and are associated with primary resistance to EGFR-TKI therapy (16). For these patients, choosing an appropriate treatment strategy post-resistance is crucial.
2 Materials and methods
This study retrospectively analyzed 168 patients with stage III or IV non-small cell lung cancer (NSCLC) who were treated at our hospital’s oncology department between January 1, 2021, and October 31, 2023. The cohort included 36 patients with EGFR mutations alone, 80 with EGFR/TP53 co-mutations, and 52 with EGFR/KRAS co-mutations. Inclusion criteria were: age between 18 and 85 years, histologically confirmed locally advanced or advanced NSCLC, initial treatment with EGFR-TKI, and subsequent chemotherapy plus immunotherapy after EGFR-TKI resistance. Exclusion criteria included incomplete medical history, presence of other malignancies, or inability to undergo further treatment. Data collected included demographic characteristics, tumor staging, and treatment modalities. Efficacy was evaluated based on RECIST 1.1 criteria, with CT scans performed every 3 months to calculate progression-free survival (PFS). Follow-up was conducted until December 31, 2023. Statistical analyses were performed using GraphPad Prism version 9.0. Categorical variables were compared using the chi-square test. Survival outcomes were analyzed using the Kaplan–Meier method, and differences between groups were assessed with the log-rank test. To identify independent prognostic factors for progression-free survival (PFS), all baseline variables were included in a multivariate Cox proportional hazards regression model. A two-sided P value < 0.05 was considered statistically significant.
PD-L1 immunohistochemistry was performed on neutral-buffered formalin-fixed, paraffin-embedded (FFPE) tumor specimens using the Dako 22C3 pharmDx assay (clone 22C3). Tumor proportion score (TPS)—the percentage of viable tumor cells with partial or complete membranous staining—was recorded, and PD-L1 expression was categorized a priori as negative (TPS <1%), low (1–49%), or high (≥50%). Slides were independently evaluated under blinded conditions (masked to clinical data and outcomes) by two board-certified pathologists; discrepancies—defined as a discordant TPS category or a ≥10-percentage-point difference—were resolved by joint consensus review. Positive and negative control slides accompanied each staining batch, and interpretation proceeded only after quality-control criteria were met.
This study’s retrospective, single-center design may introduce selection bias and limits generalizability; prospective multi-center studies are warranted.
3 Results
3.1 General clinical data
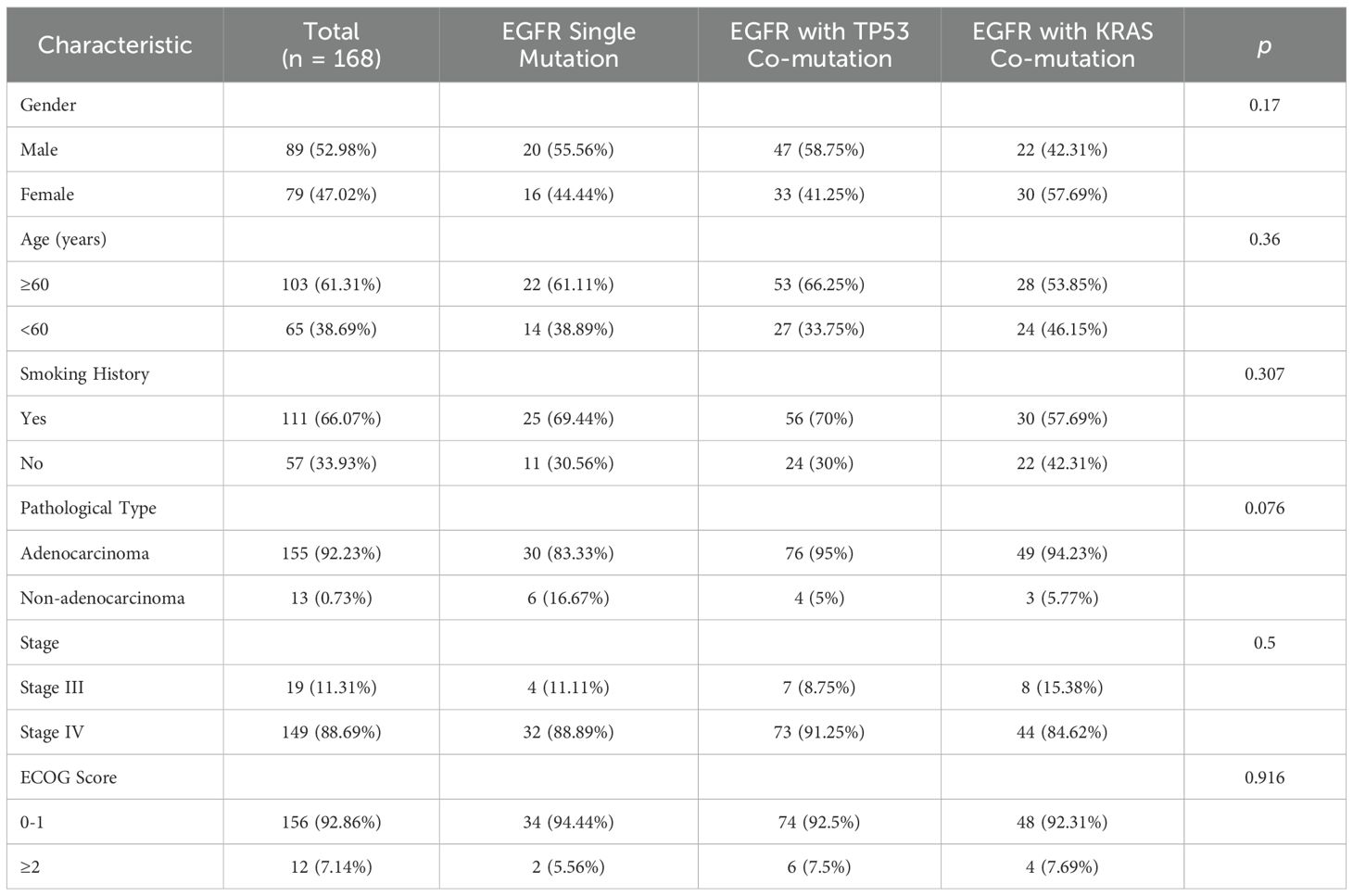
Baseline clinical characteristics of the three groups are summarized in Table 1. Distributions of key variables (age, sex, ECOG performance status, stage, histology, smoking status) were comparable across groups; no between-group differences reached statistical significance (all P>0.05).
3.2 Efficacy of first-line TKI in EGFR single vs. co-mutations
Patients with an EGFR single mutation had a median PFS of 14.1 months (range, 12.5–19.0), which was longer than in the EGFR/TP53 (median 10.4 months; range, 1.5–11.7) and EGFR/KRAS (median 10.9 months; range, 2.6–12.2) co-mutation groups. The overall log-rank test indicated a significant difference between the single-mutation group and either co-mutation group (P < 0.0001), whereas no significant difference was observed between the EGFR/TP53 and EGFR/KRAS cohorts (P = 0.174, 95%CI:0.4609-1.162) (Figure 1). These results indicate that, compared with the EGFR single-mutation group, the presence of TP53 or KRAS co-mutations was associated with shorter PFS on first-line EGFR-TKI therapy, with no clear difference between the two co-mutation subgroups.
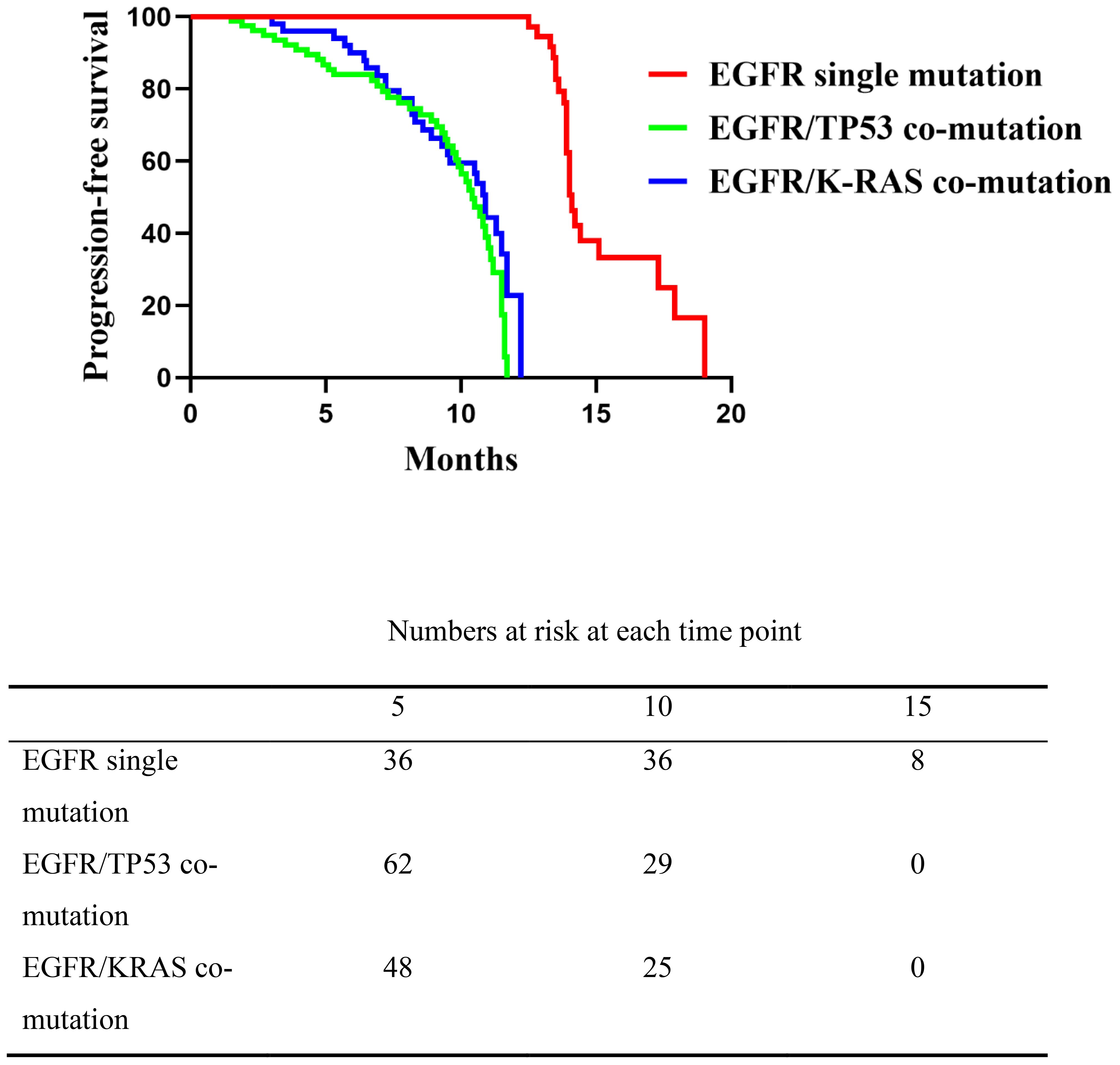
Figure 1. Comparison of progression-free survival (PFS) among the three patient groups receiving first-line EGFR-TKI therapy. The EGFR single mutation group showed significantly longer PFS than the co-mutation groups (P < 0.0001), while no significant difference was observed between the EGFR/TP53 and EGFR/K-RAS co-mutation groups (P = 0.174).
3.3 PD-L1 expression in EGFR single vs. co-mutations
PD-L1 expression was categorized by tumor proportion score (TPS) as negative (<1%), low (1–49%), or high (≥50%). Among 168 patients, the overall distribution of PD-L1 categories differed across mutation groups (EGFR single-mutation, EGFR/TP53, EGFR/KRAS) on χ² testing (P = 0.011; Table 2). In pairwise comparisons, the distributions differed between the EGFR single-mutation group and the EGFR/TP53 group (P = 0.004), and between the EGFR single-mutation group and the EGFR/KRAS group (P = 0.015), whereas no difference was observed between the EGFR/TP53 and EGFR/KRAS groups (P = 0.945). These findings indicate that PD-L1 expression patterns were associated with mutation status; however, no clear difference was seen between the two co-mutation cohorts.
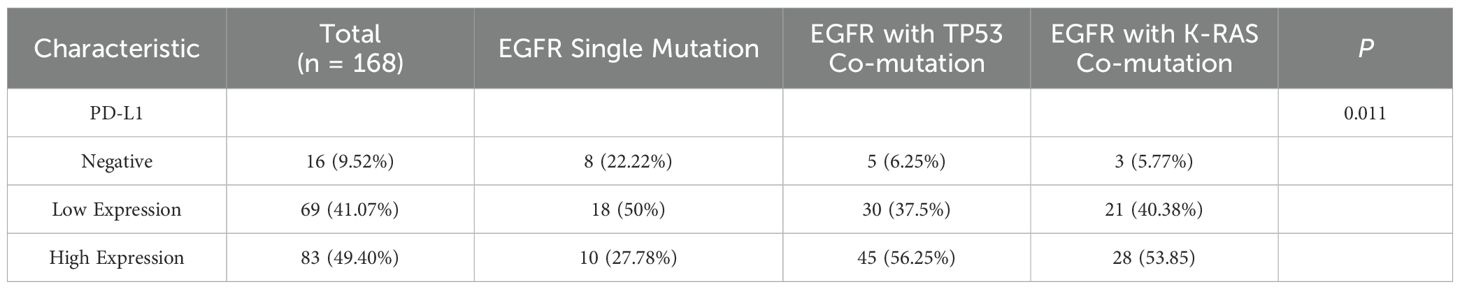
Table 2. Analysis of differences in immune expression status among patients with different gene mutation types.
3.4 Efficacy of chemotherapy plus immunotherapy in TKI-resistant EGFR single vs. co-mutations
Following progression on EGFR-TKIs, all patients received platinum-based doublet chemotherapy plus immunotherapy. On unadjusted Kaplan–Meier analysis of second-line therapy, median PFS was 3.9 months in the EGFR single-mutation group (range, 2.5–5.5), compared with 5.2 months in the EGFR/TP53 group (range, 2.4–6.5) and 5.0 months in the EGFR/KRAS group (range, 2.3–6.3). The single-mutation group differed significantly from each co-mutation group (overall log-rank P < 0.0001) (EGFR single-mutation vs EGFR/TP53 95CI%: 0.1618-0.61), (single-mutation vs KRAS 95%CI: 0.1909-0.6708), whereas no difference was observed between the EGFR/TP53 and EGFR/KRAS cohorts (P = 0.943,95%CI: 0.6504-1.587) (Figure 2). These results indicate that, relative to the EGFR single-mutation group, the presence of TP53 or KRAS co-mutations was associated with longer PFS on second-line chemo-immunotherapy, with no clear difference between the two co-mutation subgroups.
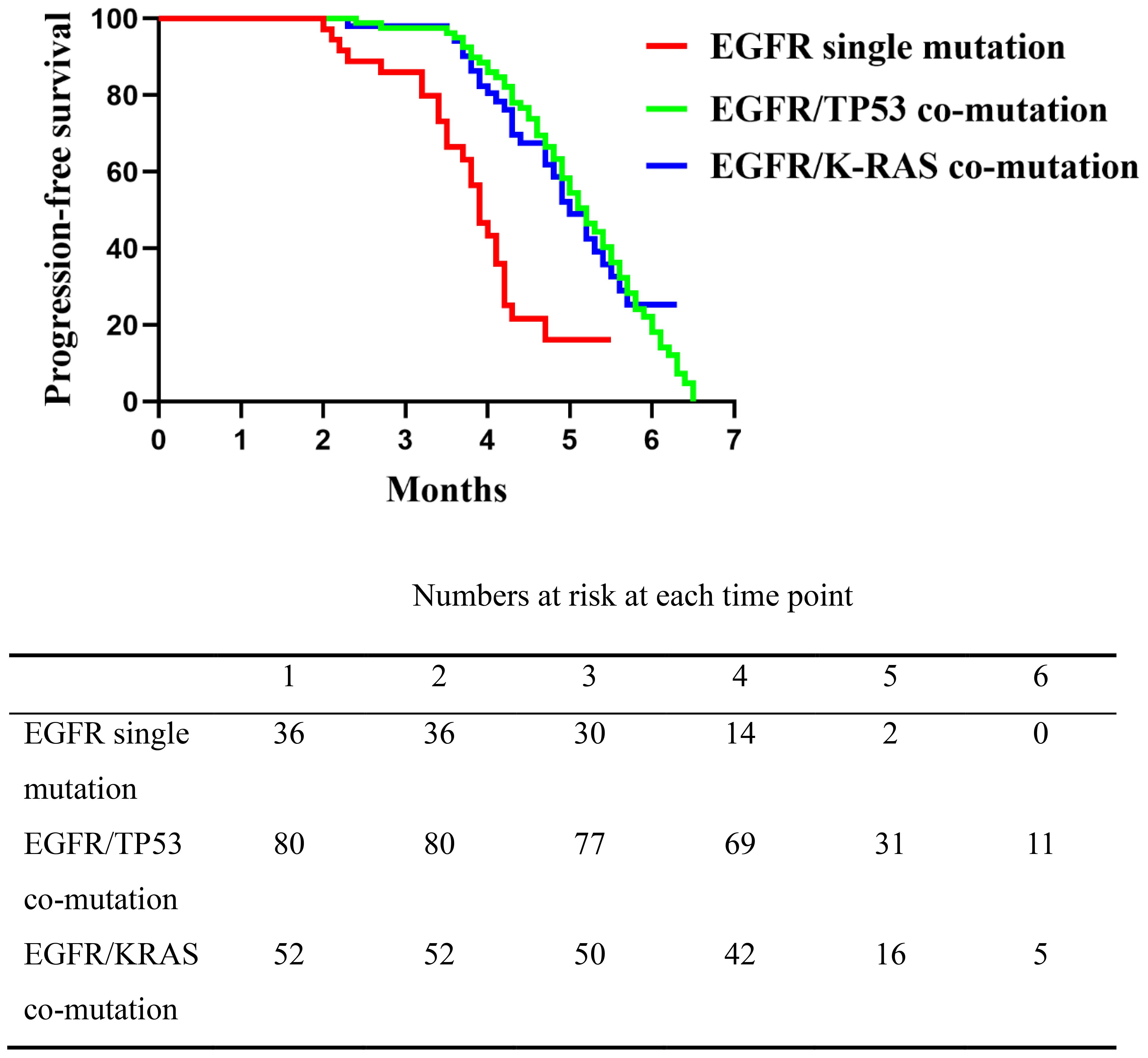
Figure 2. Comparison of progression-free survival (PFS) among the three patient groups following second-line platinum-based doublet chemotherapy combined with immunotherapy. The EGFR single mutation group had significantly shorter PFS compared to the EGFR/TP53 and EGFR/K-RAS co-mutation groups (P < 0.0001), while no significant difference was observed between the two co-mutation groups (P = 0.1744).
Patients were stratified by PD-L1 tumor proportion score (TPS) into negative (<1%), low (1–49%), and high (≥50%) categories. In the PD-L1-negative subgroup, unadjusted Kaplan–Meier analyses showed median PFS of 2.5 months in the EGFR single-mutation group (range, 2.5–3.4), 3.5 months in the EGFR/TP53 group (range, 2.4–3.7), and 3.5 months in the EGFR/KRAS group (range, 2.3–3.8). Pairwise comparisons did not reach statistical significance (P = 0.146,95%CI:0.1086-1.503, P = 0.246,95%CI:0.1107-1.826, and P = 0.646,95%CI:0.2194-9.539) (Figure 3A).
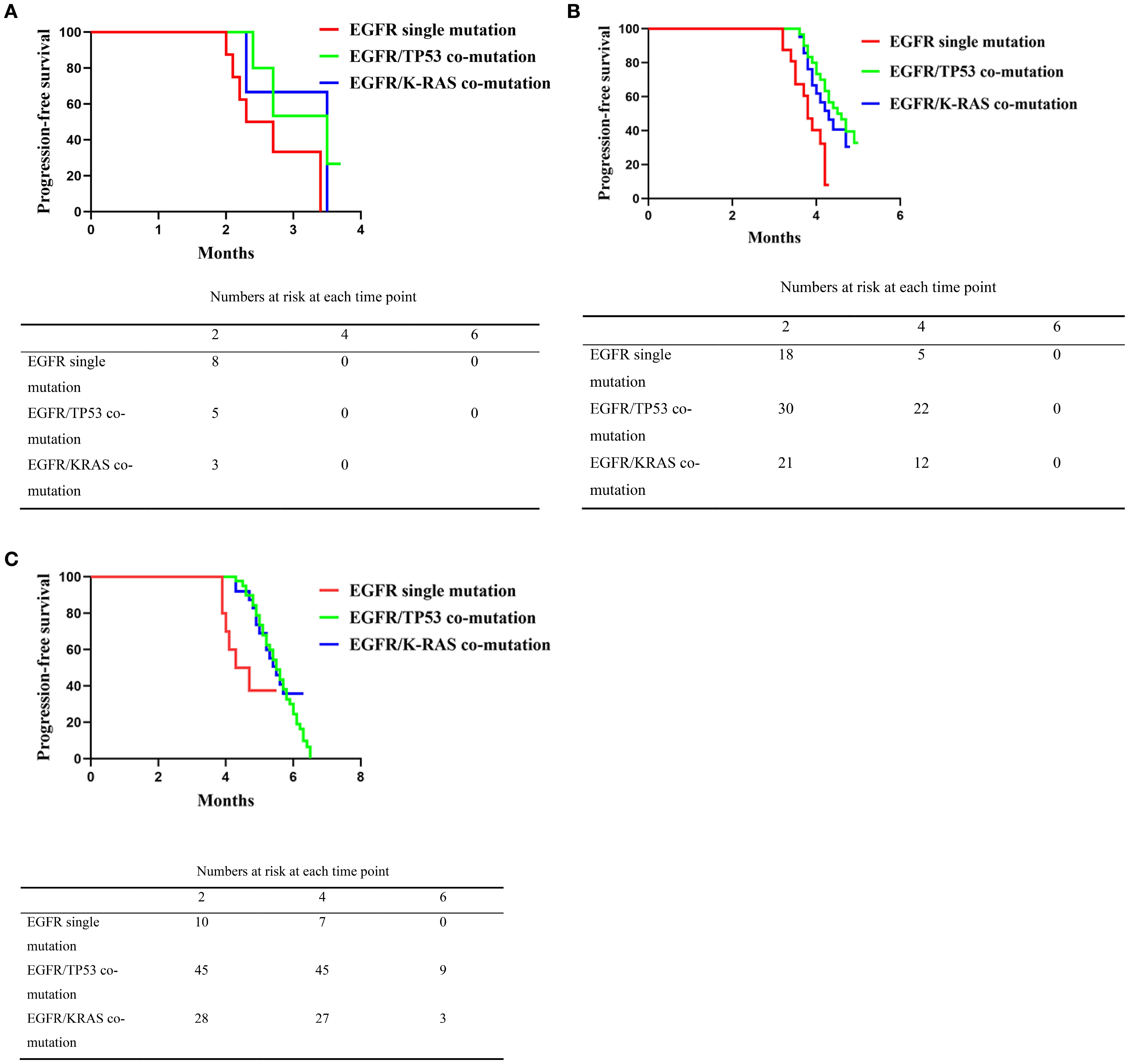
Figure 3. (A) Comparison of progression-free survival (PFS) among patients with EGFR single mutation, EGFR/TP53 co-mutation, and EGFR/K-RAS co-mutation in the PD-L1 negative subgroup following second-line platinum-based doublet chemotherapy combined with immunotherapy. The Kaplan–Meier curves show that the EGFR single mutation group had significantly shorter PFS than the co-mutation groups (P < 0.0001), while no significant difference was observed between the EGFR/TP53 and EGFR/K-RAS co-mutation groups (P = 0.189). (B) Comparison of progression-free survival (PFS) among patients with EGFR single mutation, EGFR/TP53 co-mutation, and EGFR/K-RAS co-mutation in the PD-L1 low expression subgroup following second-line platinum-based doublet chemotherapy combined with immunotherapy. The Kaplan–Meier curves show that the EGFR single mutation group had significantly shorter PFS than the co-mutation groups (P < 0.0001), with no significant difference between the EGFR/TP53 and EGFR/K-RAS co-mutation groups (P = 0.241). (C) Comparison of progression-free survival (PFS) among patients with EGFR single mutation, EGFR/TP53 co-mutation, and EGFR/K-RAS co-mutation in the PD-L1 high expression subgroup following second-line platinum-based doublet chemotherapy combined with immunotherapy. The Kaplan–Meier curves indicate that the EGFR single mutation group had significantly shorter PFS than the co-mutation groups (P < 0.0001), with no significant difference between the EGFR/TP53 and EGFR/K-RAS co-mutation groups (P = 0.312).
In the PD-L1-low subgroup, median PFS was 3.8 months in the EGFR single-mutation group (range, 3.0–4.3), 4.55 months in the EGFR/TP53 group (range, 3.6–5.0), and 4.3 months in the EGFR/KRAS group (range, 3.6–4.8). The single-mutation group differed from the EGFR/TP53 (P = 0.0003,95%CI:0.1279-0.7830) and EGFR/KRAS cohorts (P = 0.012,95%CI: 0.1770-0.9601), whereas no difference was observed between the EGFR/TP53 and EGFR/KRAS cohorts (P = 0.438,95%CI: 0.3693-1.589) (Figure 3B).
In the PD-L1–high subgroup, Kaplan–Meier analysis showed a median PFS of 4.5 months in the EGFR single-mutation group (range, 3.9–5.5), 5.5 months in the EGFR/TP53 group (range, 4.1–6.5), and 5.5 months in the EGFR/KRAS group (range, 4.0–6.3). The single-mutation group differed from the EGFR/TP53 (P = 0.010,95%CI: 0.3441-1.945) and EGFR/KRAS cohorts (P = 0.044,95%CI: 0.3144-2.129), whereas no difference was observed between the EGFR/TP53 and EGFR/KRAS cohorts (P = 0.531,95%CI: 0.5380-1.859) (Figure 3C).
3.5 Multivariate analysis
To assess independent associations with progression-free survival (PFS), we fit multivariable Cox proportional hazards models including prespecified covariates (age, sex, smoking history, histology, clinical stage, ECOG performance status) and mutation status. EGFR/TP53 (HR 1.85, 95% CI 1.22–2.80; P = 0.004) and EGFR/KRAS (HR 2.10, 95% CI 1.32–3.33; P = 0.002) co-mutations were independently associated with a higher hazard of progression. ECOG PS ≥2 was associated with a numerically higher hazard that did not reach statistical significance (HR 1.45, 95% CI 0.94–2.24; P = 0.085). No significant associations were observed for other covariates (all P>0.05) (Table 3).5.
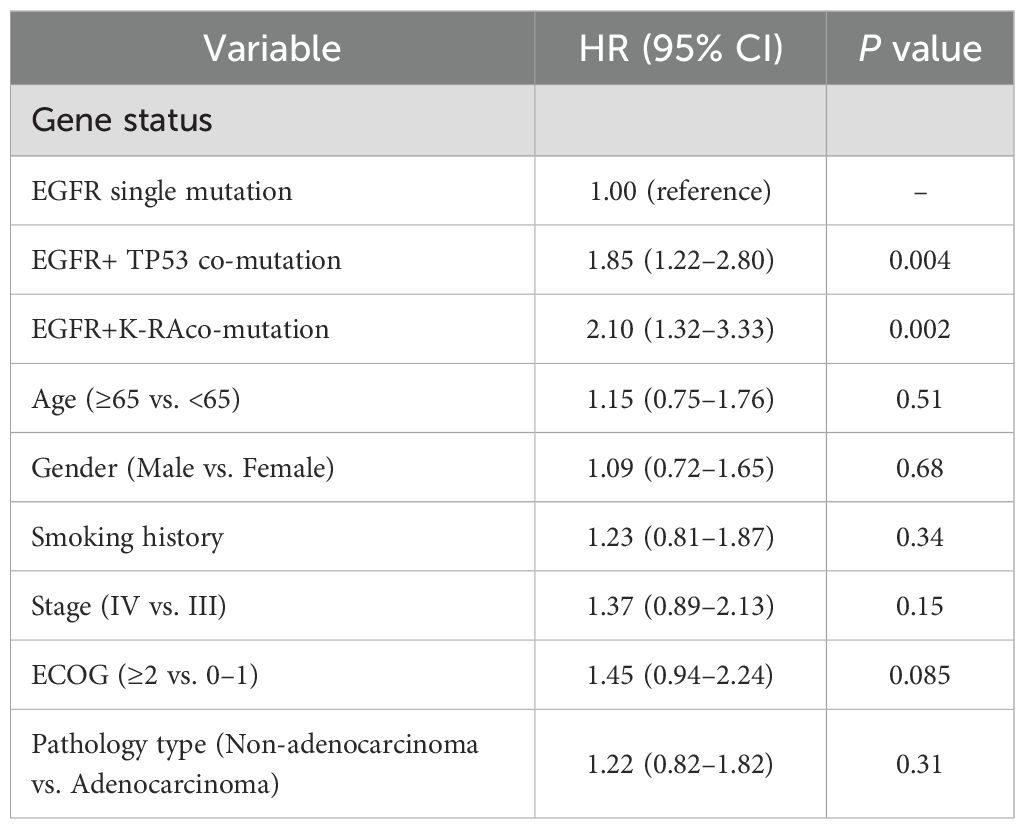
Table 3. Multivariate Cox proportional hazards regression analysis of factors associated with progression-free survival (PFS).
4 Discussion
In EGFR-mutant NSCLC, although EGFR-TKIs generally improve clinical outcomes, there are statistically significant inter-individual differences in progression-free survival (PFS). Consistent with prior literature, our findings indicate that co-mutation status is associated with therapeutic efficacy and immune-related biomarkers, with implications for clinical risk stratification and treatment decision-making (10, 12, 17–23).
4.1 Clinical and translational implications of EGFR co-mutations
Prior studies and our data indicate that patients with EGFR/TP53 co-mutations derive limited benefit from EGFR-TKIs and have poorer prognoses (24–26); EGFR/KRAS co-mutations are likewise associated with shorter PFS and an increased risk of resistance (27–29). Taken together, these findings support incorporating co-mutation status into baseline risk assessment at treatment initiation and pre-planning alternative or combination strategies along the therapeutic pathway (10, 12, 28).
4.2 Therapeutic implications after resistance to EGFR-TKIs
In the overall EGFR-mutant population, the survival benefit of immunotherapy or platinum-based chemo-immunotherapy appears limited, and pivotal randomized trials (e.g., CheckMate 722 and KEYNOTE-789) did not meet their prespecified primary endpoints (30), underscoring the need for precise patient stratification. In line with our analysis:
● Among patients with EGFR/TP53 co-mutations, second-line chemo-immunotherapy achieved a median PFS of 5.2 months, compared with 3.9 months in those with EGFR mutation alone (P<0.0001), suggesting that TP53 co-mutation may have predictive biomarker potential (31, 32).
● Patients with EGFR/KRAS co-mutations likewise demonstrated a more favorable survival trend on second-line chemo-immunotherapy and frequently exhibited higher PD-L1 levels, consistent with prior reports (33–35).
Taken together, these observations support prioritizing chemo-immunotherapy after EGFR-TKI resistance for patients harboring TP53 or KRAS co-mutations, whereas in patients with isolated EGFR mutations who lack features of immunotherapy sensitivity, immunotherapy-based combinations should be selected with caution (30, 36, 37).
4.3 Exploration of anti-angiogenic combinations
Comparing IMpower150 and ORIENT-31 with CheckMate 722 and KEYNOTE-789, a key distinction is that the former two incorporated VEGF inhibitors (30). Given the cross-talk among VEGF signaling, EGFR pathways, and hypoxia responses (38–42), adding anti-angiogenic agents in selected, stratified populations may remodel the tumor microenvironment (TME) and potentiate both immunotherapy and chemotherapy. Accordingly, chemo-immunotherapy with or without anti-angiogenic therapy warrants prospective validation in co-mutated subgroups.
4.4 Summary
In the second-line setting after resistance to EGFR-TKIs, we observed a modest prolongation of PFS (approximately 1 month) with platinum-based chemo-immunotherapy in the TP53- or KRAS-co-mutated subgroup. Existing evidence indicates that TP53 mutation is associated with upregulation of immune checkpoints, activation of effector T cells, and increased tumor mutational burden (TMB), whereas KRAS mutation is linked to MAPK-mediated PD-L1 upregulation and neoantigen formation (31–35). Based on these observations, we advance a testable working hypothesis: in EGFR-mutant NSCLC, TP53 or KRAS co-mutation may partially mitigate the “immune-cold” phenotype, thereby enhancing sensitivity to immunotherapy; however, given that pivotal trials such as CheckMate 722 and KEYNOTE-789 did not meet their prespecified primary endpoints (30), this should be regarded as hypothesis-generating rather than confirmatory and requires validation in functional studies and prospective trials.
Differences between our findings and randomized data may reflect heterogeneity in study populations and treatment regimens—including prior TKI generation and exposure, crossover, eligibility criteria, and biomarker distributions. Accordingly, prospective multi-center studies with pre-specified stratification and interaction analyses by co-mutation profile and immune-related biomarkers (e.g., PD-L1, TMB, and TME features), alongside head-to-head comparisons with contemporary standards, are warranted to determine whether co-mutated subgroups derive reproducible and durable benefit from second-line chemo-immunotherapy.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
This retrospective study was conducted in accordance with the Declaration of Helsinki (2013 revision). The protocol was reviewed and approved by the Ethics Committee (Institutional Review Board) of Weifang People’s Hospital on July 24, 2023 (Approval No. KYLL20230724 1). The approval is valid from January 2024 through December 2026. Owing to the retrospective design and the use of de-identified patient data, the requirement for written informed consent was waived by the committee.
Author contributions
YN: Conceptualization, Data curation, Formal analysis, Writing – original draft, Writing – review & editing. CS: Conceptualization, Data curation, Formal analysis, Writing – original draft, Writing – review & editing. KW: Formal analysis, Writing – original draft. MY: Methodology, Writing – original draft. JH: Investigation, Writing – original draft. SL: Conceptualization, Data curation, Investigation, Methodology, Supervision, Writing – original draft, Writing – review & editing. FH: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Global Burden of Disease Cancer Collaboration, Fitzmaurice C, Allen C, Barber RM, Barregard L, Bhutta ZA, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted lifeyears for 32 cancer groups, 1990 to 2015: A systematic analysis for the global burden of disease study. JAMA Oncol. (2017) 3:524–48. doi: 10.1001/jamaoncol.2016.5688
2. Nicholson AG, Tsao MS, Beasley MB, Borczuk AC, Brambilla E, Cooper WA, et al. The 2021 WHO classification of lung tumors: impact of advances since 2015. J Thorac Oncol. (2022) 17:362–87. doi: 10.1016/j.jtho.2021.11.003
3. Osmani L, Askin F, Gabrielson E, and Li QK. Current WHO guidelines and the critical role of immunohistochemical markers in the subclassiffcation of non-small cell lung carcinoma (NSCLC): Moving from targeted therapy to immunotherapy. Semin Cancer Biol. (2018) 52:103–9. doi: 10.1016/j.semcancer.2017.11.019
4. Gierada DS. Pinsky PFSurvival following detection of stage I lung cancer by screening in the national lung screening trial. Chest. (2021) 159:862–9. doi: 10.1016/j.chest.2020.08.2048
5. Singhal S, Vachani A, Antin-Ozerkis D, Kaiser LR, and Albelda SM. Prognostic implications of cell cycle, apoptosis, and angiogenesis biomarkers in non-small cell lung cancer: a review. Clin Cancer Res. (2005) 11:3974–3986. doi: 10.1158/1078-0432.CCR-04-2661
6. Yu HA, Arcila ME, Rekhtman N, Sima CS, Zakowski MF, Pao W, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. (2013) 19:2240–7. doi: 10.1158/1078-0432.CCR-12-2246
7. Iwama E, Takayama K, Harada T, Okamoto Ookubo I, Kishimoto F, J, et al. Highly sensitive and quantitative evaluation of the EGFR T790M mutation by nanofluidic digital PCR. Oncotarget. (2015) 6:20466–73. doi: 10.18632/oncotarget.4058
8. Zhang Q, Sun T, Kang P, Qian K, Deng B, Zhou J, et al. Combined analysis of rearrangement of ALK, ROS1, somatic mutation of EGFR, KRAS, BRAF, PIK3CA)and mRNA expression of ERCC1, TYMS, RRM1, TUBB3, EGFR in patients with non-small cell lung cancer and their clinical significance. Cancer Chemother Pharmacol. (2016) 77:583–93. doi: 10.1007/s00280-016-2969-y
9. Wang Z, Zhang X, Bai H, Zhao J, Zhuo M, An T, et al. EML4-ALK rearrangement and its clinical significance in Chinese patients with advanced non-small cell lung cancer. Oncology. (2012) 83:248–56. doi: 10.1159/000341381
10. Ma Y, Pan X, Xu P, Mi Y, Wang W, Wu X, et al. Plasma microRNA alterations between EGFRactivating mutational NSCLC patients with and without primary resistance to TKI. Oncotarget. (2017) 8:88529–36. doi: 10.18632/oncotarget.19874
11. Zhao M, Zhang Y, Cai W, Li J, Zhou F, Cheng N, et al. The Bim deletion polymorphism clinical profile and is relation with tyrosine kinase inhibitor resistance in Chinese patients with non-small cell lung cancer. Cancer. (2014) 120:2299–307. doi: 10.1002/cncr.28725
12. Xia J, Bai H, Yan B, Li R, Shao M, Xiong L, et al. Mimicking the BIM BH3 domain overcomes resistance to EGFR tyrosine kinase inhibitors in EGFR-mutant nonsmall cell lung cancer. Oncotarget. (2017) 8:108522–33. doi: 10.18632/oncotarget.19411
13. Hafner A, Bulyk ML, Jambhekar A, and Lahav G. The multiple mechanisms that regulate p53 activity and cell fate. Nat Rev Mol Cell Biol. (2019) 20:199–210. doi: 10.1038/s41580-019-0110-x
14. Tuna M, Ju Z, Yoshihara K, Amos CI, Tanyi JL, and Mills GB. Clinical relevance of TP53 hotspot mutations in high-grade serous ovarian cancers. Br J Cancer. (2020) 122:405–12. doi: 10.1038/s41416-019-0654-8
15. Rhett JM, Khan I, and O’Bryan JP. Biology, pathology, and therapeutic targeting of RAS. Adv Cancer Res. (2020) 148:69–146. doi: 10.1016/bs.acr.2020.05.002
16. Ruiz-Cordero R, Ma JS, Khanna A, Lyons G, Rinsurongkawong W, Bassett R, et al. Simplified molecular classification of lung adenocarcinomas based on EGFR, KRAS, and TP53 mutations. BMC Cancer. (2020) 20:83–95. doi: 10.1186/s12885-020-6579-z
17. Mehnert JM and Kluger HM. Driver mutations in melanoma: lessons learned from bench-to-bedside studies. Curr Oncol Rep. (2012) 14:449–57. doi: 10.1007/s11912-012-0249-5
18. De S and Ganesan S. Looking beyond drivers and passengers in cancer genome sequencing data. Ann Oncol. (2017) 28:938–45. doi: 10.1093/annonc/mdw677
19. Sankar K, Gadgeel SM, and Qin A. Molecular therapeutic targets in non-small cell lung cancer. Expert Rev Anticancer Ther. (2020) 20:647–61. doi: 10.1080/14737140.2020.1787156
20. Chen JA and Riess JW. Optimal management of patients with advanced NSCLC harboring high PD-L1 expression and driver mutations. Curr Treat Options Oncol. (2020) 21:60. doi: 10.1007/s11864-020-00750-y
21. Pao W and Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. (2011) 12:175–80. doi: 10.1016/S1470-2045(10)70087-5
22. Costa DB, Shaw AT, Ou SH, Solomon BJ, Riely GJ, Ahn MJ, et al. Clinical experience with crizotinib in patients with advanced ALK-rearranged non-small-cell lung cancer andBrain metastases. J Clin Oncol. (2015) 33:1881–8. doi: 10.1200/JCO.2014.59.0539
23. Ramalingam SS, Vansteenkiste J, Planchard D, Cho BC, Gray JE, Ohe Y, et al. Overall survival with osimertinib in untreated, EGFR-mutated advanced NSCLC. N Engl J Med. (2020) 382:41–50. doi: 10.1056/NEJMoa1913662
24. VanderLaan PA, Rangachari D, Mockus SM, Spotlow V, Reddi HV, Malcolm J, et al. Mutations in TP53, PIK3CA, PTEN and other genes in EGFR mutated lung cancers: Correlation with clinical outcomes. Lung Cancer. (2017) 106:17–21. doi: 10.1016/j.lungcan.2017.01.011
25. Zheng C, Li X, Ren Y, Yin Z, and Zhou B. Coexisting EGFR and TP53 Mutations in Lung Adenocarcinoma Patients Are Associated With COMP and ITGB8 Upregulation and Poor Prognosis. Front Mol Biosci. (2020) 7:30. doi: 10.3389/fmolb.2020.00030
26. Roeper J, Christopoulos P, Falk M, Heukamp LC, Tiemann M, Stenzinger A, et al. TP53 co-mutations as an independent prognostic factor in 2nd and further line therapy-EGFR mutated non-small cell lung cancer IV patients treated with osimertinib. Transl Lung Cancer Res. (2022) 11:4–13. doi: 10.21037/tlcr-21-754
27. Elosta B, Behera M, Kim S, Berry LD, Sica G, Pillai RN, et al. Characteristics and outcomes of patients with metastatic KRAS-mutant lung adenocarcinomas: the lung cancer mutation consortium experience. J Thorac Oncol. (2019) 14:876–89. doi: 10.1016/j.jtho.2019.01.020
28. Loong HH, Du N, Cheng C, Lin H, Guo J, Lin G, et al. KRAS G12C mutations in Asia: a landscape analysis of 11,951 Chinese tumor samples. Transl Lung Cancer Res. (2020) 9:1759–69. doi: 10.21037/tlcr-20-455
29. Scheffler M, Ihle MA, Hein R, Merkelbach-Bruse S, Scheel AH, Siemanowski J, et al. K-ras mutation subtypes in NSCLC and associated co-occuring mutations in other oncogenic pathways. J Thorac Oncol. (2019) 14:606–16. doi: 10.1016/j.jtho.2018.12.013
30. Arter ZL and Nagasaka M. The nail in the coffin?: examining the KEYNOTE-789 clinical trial’s impact. Lung Cancer (Auckl). (2024) 15:1–8. doi: 10.2147/LCTT.S443099
31. Dong ZY, Zhong WZ, Zhang XC, Su J, Xie Z, Liu SY, et al. Potential predictive value of TP53 and KRAS mutation status for response to PD-1 blockade immunotherapy in lung adenocarcinoma. Clin Cancer Res. (2017) 23:3012–24. doi: 10.1158/1078-0432.CCR-16-2554
32. Yu HA, Sima CS, Shen R, Kass S, Gainor J, Shaw A, et al. Prognostic impact of KRAS mutation subtypes in 677 patients with metastatic lung adenocarcinomas. J Thorac Oncol. (2015) 10:431–7. doi: 10.1097/JTO.0000000000000432
33. Madeddu C, Donisi C, Liscia N, Lai E, Scartozzi M, and Macciò A. EGFR-mutated non-small cell lung cancer and resistance to immunotherapy: role of the tumor microenvironment. Int J Mol Sci. (2022) 23:6489. doi: 10.3390/ijms23126489
34. Offin M, Rizvi H, Tenet M, Ni A, Sanchez-Vega F, Li BT, et al. Tumor Mutation Burden and Efficacy of EGFR-Tyrosine Kinase Inhibitors in Patients with EGFR-Mutant Lung Cancers. Clin Cancer Res. (2019) 25:1063–9. doi: 10.1158/1078-0432.CCR-18-1102
35. Liu C, Zheng S, Jin R, Wang X, Wang F, Zang R, et al. The superior efficacy of anti-PD-1/PD-L1 immunotherapy in KRAS-mutant non-small cell lung cancer that correlates with an inflammatory phenotype and increased immunogenicity. Cancer Lett. (2020) 470:95–105. doi: 10.1016/j.canlet.2019.10.027
36. Borghaei H, de Marinis F, Dumoulin D, Reynolds C, Theelen WSME, Percent I, et al. SAPPHIRE: phase III study of sitravatinib plus nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. Ann Oncol. (2024) 35:66–76. doi: 10.1016/j.annonc.2023.10.004
37. Rodríguez-Abreu D, Powell SF, Hochmair MJ, Gadgeel S, Esteban E, Felip E, et al. Pemetrexed plus platinum with or without pembrolizumab in patients with previously untreated metastatic nonsquamous NSCLC: protocol-specified final analysis from KEYNOTE-189. Ann Oncol. (2021) 32:881–95. doi: 10.1016/j.annonc.2021.04.008
38. Goel HL and Mercurio AM. VEGF targets the tumour cell. Nat Rev Cancer. (2013) 13:871–82. doi: 10.1038/nrc3627
39. Xu L, Nilsson MB, Saintigny P, Cascone T, Herynk MH, Du Z, et al. Epidermal growth factor receptor regulates MET levels and invasiveness through hypoxia-inducible factor-1alpha in non-small cell lung cancer cells. Oncogene. (2010) 29:2616–27. doi: 10.1038/onc.2010.16
40. Jackson AL, Zhou B, and Kim WY. HIF, hypoxia and the role of angiogenesis in non-small cell lung cancer. Expert Opin Ther Targets. (2010) 14:1047–57. doi: 10.1517/14728222.2010.511617
41. Le X, Nilsson M, Goldman J, Reck M, Nakagawa K, Kato T, et al. Dual EGFR-VEGF pathway inhibition: A promising strategy for patients with EGFR-mutant NSCLC. J Thorac Oncol. (2021) 16:205–15. doi: 10.1016/j.jtho.2020.10.006
Keywords: non-small cell lung cancer, EGFR-TKI, co-mutation, immunotherapy, progression-free survival
Citation: Nie Y, Song C, Wu K, Yu M, Hu J, Liu S and Hui F (2025) Efficacy and prognostic analysis of chemo-immunotherapy after TKI resistance in EGFR-mutant non-small cell lung cancer with TP53 or KRAS co-mutations. Front. Immunol. 16:1684089. doi: 10.3389/fimmu.2025.1684089
Received: 12 August 2025; Accepted: 20 October 2025;
Published: 11 November 2025.
Edited by:
Kejun Liu, The Tenth Affiliated Hospital of Southern Medical University, ChinaReviewed by:
Walid Shalata, Soroka Medical Center, IsraelGiovanna Polcaro, University of Salerno, Italy
Copyright © 2025 Nie, Song, Wu, Yu, Hu, Liu and Hui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuzhen Liu, MTgyNjU2MTcxNTlAMTYzLmNvbQ==; Fu Hui, aGZ3ZWlmYW5nMjAyNEAxMjYuY29t
†These authors have contributed equally to this work and share first authorship
 Yuhui Nie
Yuhui Nie Chen Song
Chen Song Kun Wu
Kun Wu Mingxin Yu
Mingxin Yu Jia Hu2
Jia Hu2 Shuzhen Liu
Shuzhen Liu Fu Hui
Fu Hui