- 1Department of Pharmacy, Fuyong People's Hospital of Baoan District, Shenzhen, China
- 2Department of Emergency, Shenzhen Longhua District Central Hospital, Shenzhen, China
- 3Department of Chronic Disease Management, The First Hospital of Hunan University of Chinese Medicine, Changsha, China
- 4Department of Pharmacy, Shenzhen Longhua District Central Hospital, Shenzhen, China
- 5Public Relations Section, Shenzhen Longhua District Central Hospital, Shenzhen, China
The pregnane X receptor (PXR), a key hepatic nuclear receptor, exhibits a highly plastic ligand-binding domain (LBD) that recognizes diverse endogenous and exogenous ligands, contributing to interindividual variations in xenobiotic metabolism and toxic responses. Emerging studies on the gut-liver axis reveal that microbiota metabolites regulate hepatic PXR through dual mechanisms: (1) Direct ligand-receptor interactions, where secondary bile acids (e.g., 3-keto LCA, DCA) and indole-3-propionic acid (IPA) bind PXR-LBD via hydrogen bonding to induce conformational changes, subsequently upregulating CYP3A4/ABCB1 expression while inhibiting NF-κB-mediated inflammation and modulating bile acid homeostasis through crosstalk with the farnesoid X receptor (FXR); and (2) Epigenetic reprogramming, wherein short-chain fatty acids (SCFAs) such as butyrate enhance PXR transcription by inhibiting histone deacetylase (HDAC) activity and promoting histone acetylation (e.g., at H3K9/K14 residues), thereby increasing promoter accessibility. This epigenetic mechanism contrasts with the direct ligand-binding pathway by acting indirectly through chromatin remodeling. Dysregulated PXR signaling underlies bile acid imbalance, mitochondrial dysfunction, and chemoresistance, driving clinical development of interventions including probiotic modulation of LCA/DCA balance, triptolide-mediated PXR activation, and structure-based PXR-targeted drug design. These findings highlight the microbiota-PXR axis as a critical determinant of drug response heterogeneity and a promising therapeutic target for metabolic liver disorders and refractory malignancies.
1 Introduction
The pregnane X receptor (PXR), a critical member of the nuclear receptor superfamily, exhibits distinct structural and functional characteristics that underpin its broad biological roles (1). Comprising a highly variable ligand-binding domain (LBD) and a conserved DNA-binding domain (DBD), the receptor recognizes diverse ligands such as bile acids and pharmaceuticals through its hydrophobic pocket, while forming heterodimers with the retinoid X receptor (RXR) to bind target gene promoter regions (2, 3). Its predominant expression in the liver and intestines aligns with its core physiological functions in regulating drug-metabolizing enzymes (e.g., CYP3A4, P-gp) and metabolic networks of endogenous substances, including cholesterol and bile acids (4). Upon ligand activation, PXR recruits coactivators to initiate downstream gene transcription, playing a dual role in detoxification, glucose-lipid homeostasis, and inflammatory modulation (5).
Gut microbiota metabolites represent complex products of host-microbial co-metabolism, categorized into seven functional groups based on origin and activity (6–9). SCFAs, primarily derived from dietary fiber fermentation (60% acetate, 20% propionate, 20% butyrate), serve as energy sources for colon cells (butyrate accounting for 90%) and modulate insulin sensitivity and immune balance via GPCR activation (e.g., GPR43) (10). Neuroactive substances (e.g., GABA, 5-HT) and tryptophan metabolites (indole derivatives) form the gut-brain signaling network (11, 12), while secondary bile acids and trimethylamine N-oxide (TMAO) participate in cholesterol metabolism and cardiovascular risk regulation. Notably, these metabolites maintain intestinal barrier integrity (e.g., butyrate promoting mucin secretion) (13) but may induce inflammation when excessive (e.g., H2S disrupting epithelial junctions) (14), with their dynamic equilibrium directly linked to metabolic disease pathogenesis (15). Other metabolites include gases (H2, CH4, CO2) that sustain anaerobic environments and energy cycles, amino acid fermentation products (e.g., cadaverine, phenols, H2S), and microbial-synthesized vitamins (K, B-complex) involved in coagulation, energy metabolism, and DNA synthesis (16, 17).
Recent studies reveal that gut microbiota metabolites regulate PXR activity through direct binding or epigenetic modifications, forming a three-dimensional “microbe-metabolite-host receptor” interaction network (18, 19). For instance, secondary bile acids act as dual ligands for PXR and farnesoid X receptor (FXR), while SCFAs may influence PXR transcriptional efficiency via histone deacetylase (HDAC) inhibition (20, 21). This review systematically examines such cross-regulatory mechanisms, aiming to elucidate the potential impact of microbial metabolites on personalized medicine—including microbial explanations for drug metabolism variability and microbiota-based interventions for therapeutic optimization—thereby offering new perspectives for precision medicine in metabolic diseases and oncology.
2 Liver diseases associated with PXR dysregulation
PXR dysfunction in the liver is a critical factor in lipid metabolic disorders (22, 23), as the receptor maintains lipid homeostasis through three primary mechanisms: inhibiting lipid synthesis by downregulating key enzymes such as stearoyl-CoA desaturase (SCD1) (24) and acetyl-CoA carboxylase (ACC) (25), promoting fatty acid β-oxidation by enhancing peroxisome proliferator-activated receptor α (PPARα) and PPARγ coactivator 1α (PGC1α) pathways (26), and regulating bile acid metabolism by modulating rate-limiting enzymes like cholesterol 7α-hydroxylase (CYP7A1) (27, 28). Unlike the farnesoid X receptor (FXR), which primarily maintains bile acid balance and suppresses lipid synthesis (29, 30), PXR also regulates the transcription and expression of drug-metabolizing enzymes and transporters, including uridine diphosphate glucuronosyltransferases (UGTs), ATP-binding cassette transporter B1 (ABCB1/MDR-1), and cytochrome P450 3A4 (CYP3A4) (31).
PXR dysfunction triggers a cascade of pathological events: reduced CYP3A4 expression leads to secondary bile acid accumulation, disrupting lipid oxidation-synthesis balance; in obesity, PXR inactivation decreases very low-density lipoprotein (VLDL) secretion, causing free fatty acid spillover into muscle tissues and exacerbating peripheral insulin resistance; in cholestatic liver diseases (32), PXR fails to induce efflux transporters like multidrug resistance-associated protein 2 (MRP2) and breast cancer resistance protein (BCRP), resulting in bile acid retention and mitochondrial dysfunction (33, 34). The dysregulation of PXR-mediated lipid metabolism exacerbates mitochondrial dysfunction through multiple mechanisms. Specifically, activation by pregnenolone-16α-carbonitrile (PCN) significantly downregulates critical mitochondrial proteins including proline dehydrogenase (Prodh), cytochrome c, and Usmg (35, 36), thereby impairing protein folding quality control and degradation pathways. In hepatocytes, PXR dysfunction is closely linked to hepatobiliary diseases, such as primary biliary cholangitis, where toxic bile acids like lithocholic acid (LCA) inhibit PXR activity, leading to deficient MRP2 and BCRP expression and further bile acid retention (37). In drug-induced liver injury (DILI), gut microbiota-PXR protective mechanisms fail, reducing detoxification capacity and causing drug metabolite accumulation (38). Interventions, such as probiotic modulation of bile acid ratios or natural agonists like triptolide, aim to restore PXR function (39).
PXR dysfunction in the hepatobiliary system also influences chemotherapy resistance, as microbiota-derived metabolites alter drug-metabolizing enzyme profiles (40). For instance, reduced IPA diminishes PXR-mediated CYP3A4 induction, delaying irinotecan activation, while PXR overexpression may upregulate efflux transporters like MDR-1, creating multidrug resistance (41, 42). Studies show that tanshinone IIA can enhance sorafenib metabolism in hepatocellular carcinoma via PXR activation (43–45), highlighting the PXR-microbiota axis’s role in reversing chemoresistance (Figure 1A).
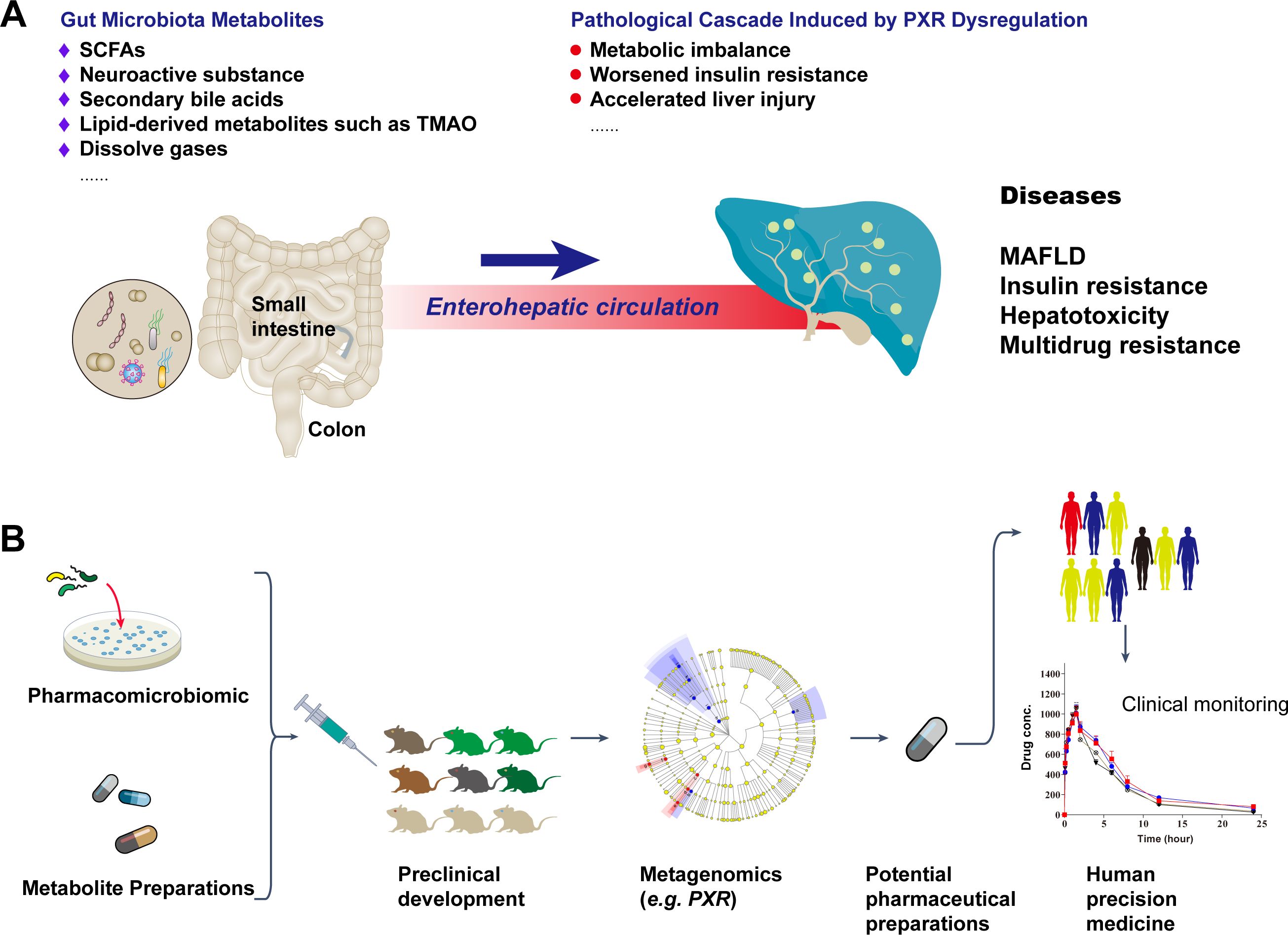
Figure 1. Gut microbiota metabolite-PXR axis in liver disease pathogenesis and the potential personalized therapeutic regimen development workflow. (A) Dysregulation of PXR: Impact of gut microbiota metabolites and gut-liver axis on liver metabolic disorders. (B) Potential roles of gut microbiota metabolites in hepatic metabolic transcription factors and a concise flowchart for personalized therapeutic development.
Gut microbiota and their metabolites have been clinically applied in gastrointestinal disease treatment (46, 47), with preclinical studies demonstrating their potential in improving hepatobiliary disorders (48, 49). Research has identified microbiota-derived metabolites that modulate PXR transcription through metagenomics, offering avenues for personalized therapy (50–52) (Figure 1B).
3 Recent advances in gut microbiota metabolite-mediated regulation of PXR activity
PXR, a ligand-activated nuclear receptor, primarily governs the inducible expression of xenobiotic-handling genes, encompassing biotransformation enzymes and drug transporters. Compared to other nuclear transcription factors, PXR assumes a pivotal role in modulating hepatic drug metabolism (53). In bile acid metabolism, PXR and FXR form a dynamically balanced regulatory loop: FXR regulates bile acid synthesis by inhibiting CYP7A1, while PXR promotes bile acid excretion by inducing CYP3A4. The two receptors achieve functional coordination through competitive binding to the promoter regions of common target genes (such as ABCB11), manifesting as bidirectional regulation of bile acid homeostasis in liver and biliary disease models (54). In drug metabolism, there exists substrate competition between PXR-induced CYP3A4 and FXR-regulated UGT1A1 (55, 56). For instance, rifampicin (a PXR agonist) can inhibit FXR-mediated bilirubin metabolism, leading to clinical drug interactions, which necessitates particular attention in the treatment of chronic liver disease. PXR and CAR exhibit significant synergistic effects in inducing phase II metabolic enzymes (such as UGT1A1, SULT2A1), but PXR demonstrates greater specificity in inducing CYP2B6 (57). The two receptors form a complex by sharing cofactors (such as RXRα) and initiate a coordinated detoxification response upon environmental toxin exposure. Notably, CAR is directly regulated by circadian clock genes, whereas PXR activation exhibits sustained induction properties (58). This time-dependent difference determines CAR’s dominant role in circadian metabolic fluctuations, while PXR is better suited for long-term drug exposure. As a signal integration hub, PXR forms a dynamic regulatory module by recruiting coactivators and corepressors, enabling simultaneous input from FXR, CAR, and PPARγ to achieve integrated regulation of metabolic pathways. In NAFLD models, the PXR-FXR-CAR tri-receptor network jointly determines the degree of lipid accumulation in hepatocytes by regulating the expression of lipid synthesis enzymes and transporters, with PXR activation partially reversing the lipid metabolic disorder caused by FXR deficiency (58, 59), highlighting its pivotal nodal role in disease networks.
Recent studies have revealed that the long noncoding RNA HNF1A antisense 1 (HNF1A-AS1) exhibits dual regulatory functions in modulating CYP3A4 expression in Huh7 and HepG2 cells. Mechanistically, HNF1A-AS1 acts as an RNA scaffold to bind both protein arginine methyltransferase 1 and the pregnane X receptor (PXR), facilitating their interaction and thereby activating PXR and regulating CYP3A4 transcription through histone modifications. Consequently, small molecule-mediated epigenetic regulation holds promise as a novel biomarker for predicting individual differences in PXR-induced drug metabolism enzymes (60). Gut microbiota metabolites regulate PXR activity through two core pathways: direct ligand binding and epigenetic modulation (61). In the direct activation pathway, microbial-derived metabolites such as secondary bile acids (3-keto LCA, DCA) and indole-3-propionic acid (IPA) function as natural PXR ligands, establishing specific hydrogen bonding interactions with conserved residues including Arg410 and Gln285 within the LBD (50). Structural studies have revealed the remarkable adaptability of PXR’s binding pocket, exemplified by the 2.65 Å resolution crystal structure showing 17β-estradiol occupying only a localized region of the expansive cavity while bridging critical polar residues through its molecular framework (62). This unique binding mode underscores PXR’s exceptional capacity to accommodate diverse endobiotic ligands, a feature distinguishing it from other nuclear receptors (Figure 2A). Molecular dynamics simulations and in vitro assays have further demonstrated that carbamazepine (CBZ) likely acts as a PXR agonist, with Gln285 emerging as a pivotal interaction site (63). The structural plasticity of PXR-LBD enables heterodimerization with RXR upon activation, recruitment of coactivators like SRC-1, and binding to DR4 response elements in target gene promoters (e.g., CYP3A4, ABCB1) (64), thereby enhancing hepatic xenobiotic metabolism and suppressing intestinal NF-κB-mediated inflammation (Figure 2A). Notably, lithocholic acid derivatives mediate PXR-FXR crosstalk to regulate CYP7A1 activity (65), while indole compounds strengthen intestinal barrier function via the PXR-IL-10 axis (66, 67). Clinically, competition between metabolites (e.g., TMAO) and drugs for CYP3A4 binding may precipitate metabolic disturbances, highlighting the therapeutic implications of microbiota-PXR interactions.
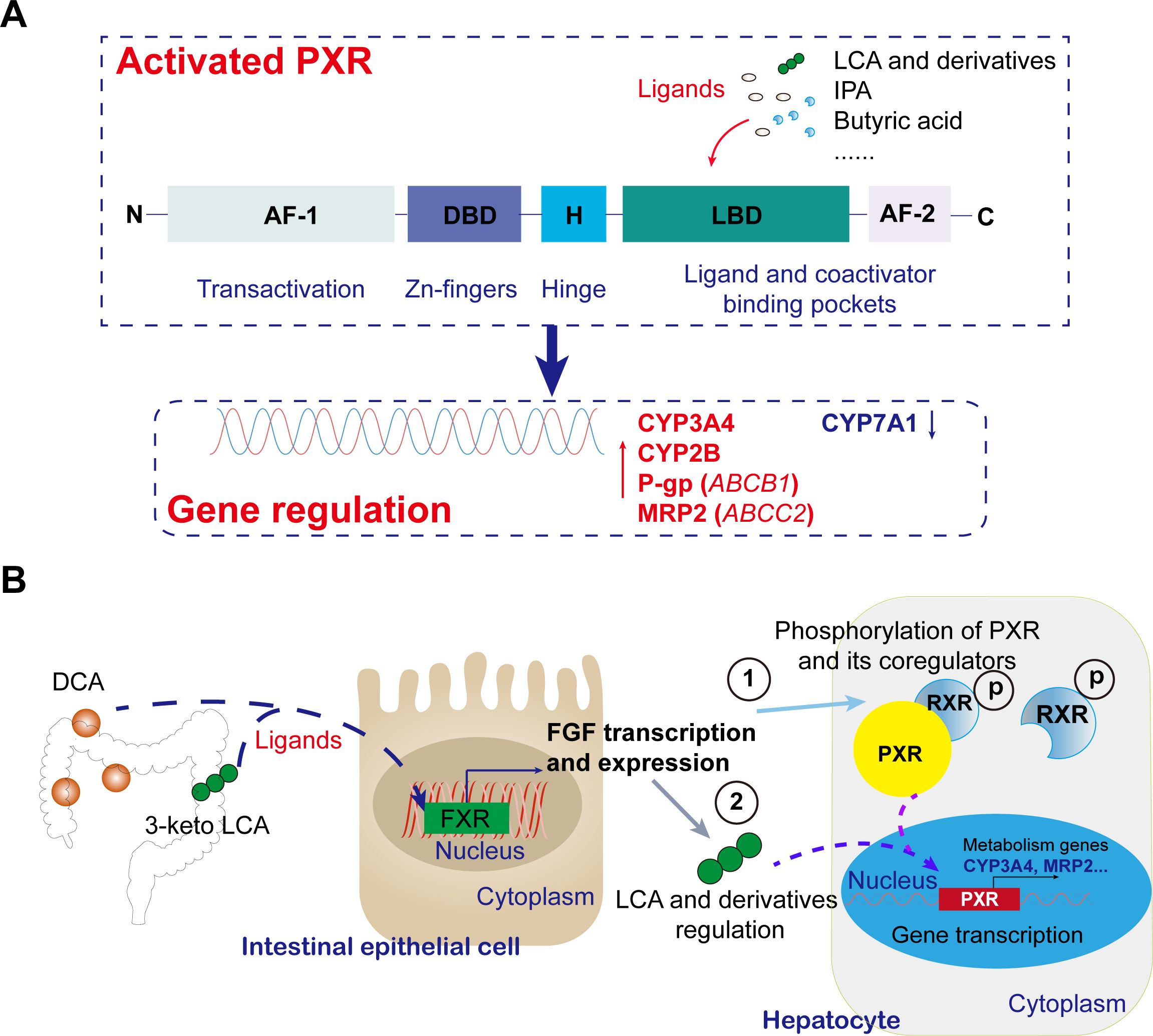
Figure 2. Recent mechanistic insights into gut microbiota metabolites modulating PXR transcriptional activity and functions. (A) Mechanism of activation of PXR by intestinally derived metabolites and its effects. In the direct activation pathway, microbial metabolites (e.g., 3-keto LCA, DCA, IPA) act as natural PXR ligands, binding via hydrogen bonds to conserved residues in the LBD. Activated PXR regulates target gene transcription, including upregulating CYP3A4, ABCB1, ABCC2, et al. expression and downregulating CYP7A1 expression. (B) Typical intestinally derived metabolites regulate the pathway of PXR in the liver. Microbial conversion of primary bile acids to secondary ones (e.g., DCA, 3-keto LCA) activates intestinal FXR, triggering FGF release. FGF modulating PXR through direct phosphorylation and indirect ligand regulation.
While current research has firmly established that CYP3A4 enhancer methylation potently suppresses CYP3A4 expression via PXR-mediated mechanisms (with rifampicin-independent regulation) (68), the epigenetic modulation of PXR expression in hepatocytes by gut-derived SCFAs remains an emerging frontier (69). Mechanistic studies reveal that butyrate, a selective HDAC class I/II inhibitor (70), orchestrates PXR transcriptional activation may through two synergistic pathways: (1) site-specific acetylation of H3K9/K14 at the PXR promoter, and (2) GPR43-dependent sequestration of HDAC3 in the cytoplasm (71, 72). These findings are consistent with the broader paradigm that microbially derived SCFAs remodel hepatic chromatin architecture through HDAC inhibition (73), thereby simultaneously suppressing inflammatory cascades and potentiating PXR signaling (74). Nevertheless, critical knowledge gaps persist in deciphering the precise epigenetic orchestration between SCFA signaling and PXR regulatory networks in liver metabolism.
Microbial metabolite dysbiosis directly disrupts PXR function through three key pathways. First, LCA accumulation occurs due to gut microbiota imbalance, where LCA acts as a PXR antagonist (75), inhibiting lipid breakdown signaling and reducing the LCA/DCA ratio (normally PXR-activating), resulting in increase in SCD1 expression and exacerbating hepatic triglyceride deposition (76). Second, reduced IPA levels diminish PXR’s inhibitory effect on AKR1B10, thereby activating the ACC/SCD1 lipid synthesis pathway (77). Third, butyrate depletion impairs PXR function, as butyrate enhances PXR-RXRα dimerization by inhibiting HDAC3 (78). The carboxy-terminal domain of PXR contains a LBD that undergoes conformational changes upon binding specific ligands (e.g., rifampicin or aflatoxin), promoting PXR-RXRα heterodimer formation (79), cytoplasmic-nuclear translocation, and binding to direct repeat (DR) or estrogen receptor (ER) response elements in target gene promoters (80). Butyrate deficiency reduces PXR transcriptional activity, leading to impaired fatty acid oxidation (81). These mechanisms collectively demonstrate how microbiota-derived metabolites critically regulate PXR-dependent metabolic pathways.
Additionally, gut microbiota metabolites exert fine-tuned regulation of PXR activity through the FXR/FGF19 signaling axis, a key indirect regulatory network maintaining bile acid homeostasis (82). The molecular mechanism involves microbial conversion of primary bile acids into secondary bile acids (e.g., DCA, 3-keto LCA) that activate intestinal FXR, triggering FGF19/15 secretion (83). This hormone-like factor reaches the liver via portal circulation, binds to FGFR4-β-Klotho complexes, and suppresses CYP7A1 expression through RAS-RAF-MEK-ERK cascades (84). This process couples with PXR function through dual mechanisms: ERK-mediated phosphorylation of PXR/RXRα directly modulates transcriptional activity (85), while FGF19-maintained bile acid homeostasis indirectly regulates PXR-driven CYP3A4 expression by altering endogenous ligand concentrations (82, 86, 87) (Figure 2B). Under physiological conditions, this network forms a negative feedback loop where FXR-FGF19 inhibits excessive bile acid synthesis while PXR promotes detoxification (82). Pathologically, microbiota dysbiosis disrupts this balance, leading to bile acid accumulation and aberrant PXR activation that may cause drug metabolism disorders or hepatic inflammation (1, 88, 89). The elucidated “microbiota metabolite-FXR-FGF19-PXR” axis offers novel therapeutic targets (e.g., FGF19 biologics in clinical trials) and a framework for personalized medicine considering drug-microbiota interactions (Figure 2B). Notably, organ-specific PXR signaling exists: lithocholic acid activates hepatic PXR to upregulate CYP3A11 (90), while microbial-derived IPA preferentially stimulates intestinal PXR (Mdr1 upregulation) without hepatic effects, unlike hypericin which activates both tissues.
The dynamic interplay between TLR4/NF-κB and PXR pathways, mediated by gut microbiota-derived metabolites, constitutes a critical regulatory axis that maintains equilibrium between drug metabolism and inflammatory responses (91). Microbial immunomodulators such as peptidoglycan fragments (GlcNAc-MurNAc) activate NF-κB signaling through TLR2/NOD pathways (92), thereby inducing the production of key pro-inflammatory cytokines including IL-1β and TNF-α—a mechanism well-characterized in inflammatory bowel disease (IBD) and type 2 diabetes (93). This microbial-immune interaction is further modulated by direct molecular crosstalk: the NF-κB p65 subunit physically binds to PXR to inhibit its DNA-binding activity, while PXR activation in turn exerts negative transcriptional control over NF-κB (94). This bidirectional regulatory network exemplifies how microbiota metabolites fine-tune the delicate balance between xenobiotic processing and immune homeostasis.
Different microbiota metabolites exhibit differential regulatory effects on this network: SCFAs (e.g., butyrate) enhance histone acetylation at the PXR promoter region by inhibiting HDACs, partially counteracting NF-κB’s inhibitory effects, whereas trimethylamine N-oxide (TMAO) exacerbates metabolic disturbances by promoting hepatic sinus endothelial cell capillaryization and dysfunction, thereby modulating macrophage polarization (95). TMAO derived from gut microbiota exacerbates NAFLD progression by damaging the gut-liver axis, and targeting TMAO may offer alternative therapeutic strategies for NAFLD (95). Based on these findings, two intervention strategies have shown clinical potential: (1) using plant-derived bioactive compounds, such as baicalin, to selectively inhibit the TLR4/NF-κB pathway and restore PXR function (96); and (2) modulating microbial communities through fecal microbiota transplantation (FMT) or specific probiotics (e.g., butyrate-producing bacteria) to reestablish TLR4-PXR balance (97, 98) and mitigate the metabolic toxicity of xenobiotics like chemotherapeutic agents.
In conclusion, gut-derived metabolites directly or indirectly modulate PXR to regulate bile acid homeostasis and xenobiotic metabolism/transport, providing critical insights for personalized therapeutic strategies.
4 Gut-derived modulators of PXR activity
The gut microbiota orchestrates a complex regulatory network through secondary bile acids (e.g., lithocholic acid [LCA], deoxycholic acid [DCA]) and short-chain fatty acids (e.g., butyrate), which directly and indirectly modulate PXR activity. Secondary bile acids like LCA and DCA serve as endogenous PXR ligands, activating the receptor to upregulate drug-metabolizing enzymes (e.g., CYP3A4/2B) while suppressing CYP7A1 to maintain cholesterol homeostasis (99, 100). Notably, Bacteroides stercoris may influence drug pharmacokinetics by modulating GUDCA and GCDCA levels, which induce CYP3A1 expression in primary rat hepatocytes. Although known PXR-activating bile acids (including LCA, CDCA, DCA, and CA) showed no significant differences between groups in this study, existing research primarily focuses on unconjugated bile acids, with activation potency ordered as: 3-keto LCA > LCA > CDCA/DCA > CA (99, 100). Additionally, UDCA and TUDCA have been reported to activate PXR and induce CYP3A4 expression, though their precise mechanisms remain incompletely elucidated.
SCFAs, as microbial metabolites, enhance metabolic activity in liver organoids, including promoting CYP3A4 expression (101). Butyrate, in particular, contributes to PXR modulation by inhibiting HDACs (102), thereby enhancing PXR-mediated transcriptional regulation of glucose transport proteins GLUT2 (103), P-glycoprotein (ABCB1) (104), accelerating cholesterol metabolism and transport (104, 105). Post-gastrectomy studies reveal an adaptive LCA-PXR axis, where increased endogenous LCA levels and elevated Bacteroides fragilis abundance correlate with upregulated hepatic CYP3A11 expression (90, 106, 107), suggesting a compensatory protective mechanism against bile acid overload (108). Another notable modulator, indole-3-propionic acid (IPA), a tryptophan metabolite produced by Clostridium sporogenes (109), acts as a PXR ligand to downregulate TNF-α and upregulate tight junction proteins, thereby maintaining gut barrier integrity (110), though its effects on CYP3A enzyme activity require further investigation.
The promiscuous nature of PXR, as a multi-ligand nuclear receptor, is underscored by its species-specific ligand-binding pocket—with only 75-80% amino acid sequence homology observed in the LBD across different species (111, 112). This structural divergence suggests significant interspecies variation in PXR ligand specificity (112, 113), a characteristic that further emphasizes the receptor’s pivotal role in bridging gut microbiota-derived signals (including bile acids, SCFAs, and other microbial metabolites) with host metabolic and detoxification pathways, as systematically documented in Table 1.
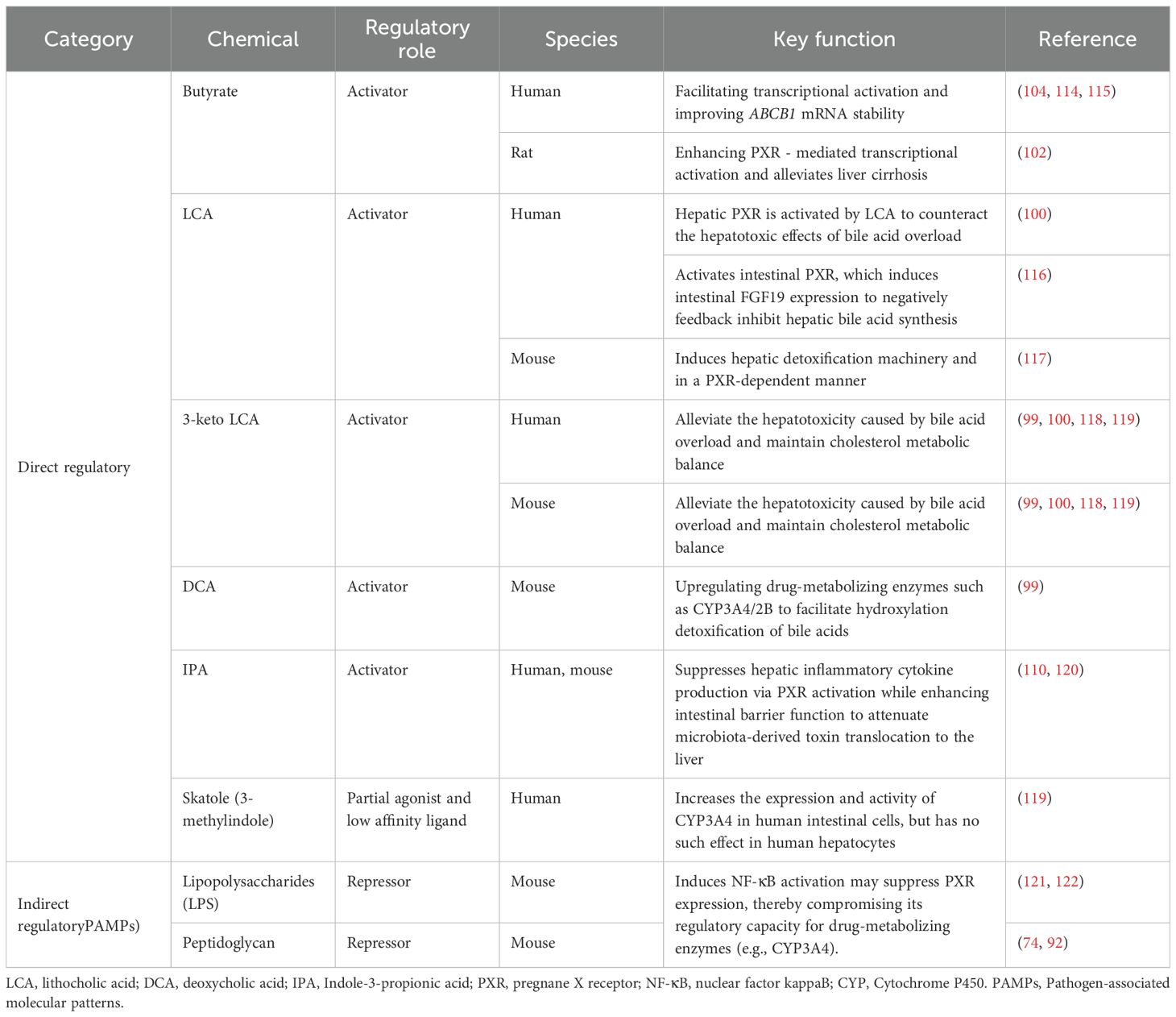
Table 1. PXR-modulating chemicals confirmed from microbial metabolites and their biological functions.
5 Regulation of PXR by clinically common drugs
PXR regulates numerous clinically used drugs beyond its prototype ligands (91, 123). Dexamethasone serves as a PXR activator in both mice and humans, as demonstrated by Pascussi et al. in 2001 and Yueh et al. in 2005 (124, 125). Notably, some drugs exhibit species-specific PXR activation patterns. For instance, phenobarbital enhances steroid receptor coactivator-1 binding to human PXR (hPXR) but fails to interact with mouse pregnane X receptor (mPXR) (126). The antifungal clotrimazole binds to hPXR at 10 mM concentrations, stimulating coactivator recruitment and enhancing PXR-target gene transcription (127), while showing weaker activation effects in rat and mouse PXR (128).
Recent studies (as of 2025) highlight PXR’s primary regulation over the following drug categories: CYP3A4 substrate drugs, where PXR preferentially induces CYP3A4 expression (accounting for approximately 30% of hepatic P450 enzymes) (129), accelerating the self-metabolic clearance of antibiotics like rifampicin and affecting the metabolic rates of warfarin and oral contraceptives. For example, PXR activation by rifampicin can increase warfarin metabolism, elevating the risk of anticoagulant therapy failure (130). CYP2B6 and CYP2C9 substrate drugs are regulated by PXR in synergy with constitutive androstane receptor (CAR), influencing the metabolism of antiepileptic drugs like phenobarbital, with clinical dosages adjusted based on receptor polymorphisms (e.g., CAR rs2307424) (131, 132). Transporter-dependent drugs are affected by PXR-induced expression of MDR1 (ABCB1) and MRP2 (ABCC2), regulating the enterohepatic circulation and biliary excretion of digoxin, as well as the hepatic concentration and myopathy risk of statins (via OATP1B1 transport). Bile acid-related drugs are indirectly regulated by PXR through modulation of bile acid metabolic enzymes (e.g., AKR1D1), influencing the generation of secondary bile acids and the efficacy of immunomodulatory drugs in liver cancer treatment (133). Glucocorticoids and anti-inflammatory drugs are affected by PXR polymorphisms (134), which can reduce glucocorticoid metabolic rates and impact the hepatoprotective effects of traditional Chinese medicine components like triptolide (135). PXR activation-induced drug interactions have become a clinical focus, such as the 47% increase in oral contraceptive failure rates when combined with St. John’s wort, prompting the FDA to require warning labels on related product inserts (136). Emerging research suggests that targeting PXR antagonists or modulating its signaling pathways may offer new strategies for personalized medicine.
6 Clinical significance and future perspectives
6.1 Microbial metabolite-mimetic drug development
IPA, a microbial indole metabolite, has been identified as a PXR activator, paving the way for novel drug development strategies. Through structural optimization, researchers have successfully designed the first non-cytotoxic PXR agonist, the lead compound FKK5/FKK6 (later named CVK003) (137). This compound directly binds to the PXR receptor, inducing PXR-target gene expression in cell cultures, human organoids, and mouse models. In humanized PXR transgenic mice, CVK003 significantly reduced pro-inflammatory cytokine levels (138). Further structural modification studies revealed that removing the benzenesulfonyl group shifted receptor binding specificity from PXR to the aryl hydrocarbon receptor (AhR), while losing PXR activation capability. Conversely, adding imidazopyridine maintained PXR binding and transcriptional activation (139, 140). These findings not only provide a theoretical basis for developing novel PXR modulators but also establish a research paradigm for understanding interactions between PXR and other xenobiotic-sensing transcription factors (141, 142).
6.2 Personalized therapy for liver diseases
PXR overactivation is associated with the progression of NAFLD, and microbiota-targeted regulation may offer a new therapeutic approach. Studies show that PXR dysfunction disrupts bile acid metabolic balance, exacerbating hepatic lipid accumulation and inflammatory responses, thereby promoting the transition from NAFLD to non-alcoholic steatohepatitis (NASH). Additionally, abnormal PXR activation can impair intestinal barrier function, promoting endotoxin translocation and further aggravating hepatic metabolic disorders. Modulating gut microbiota structure—such as increasing SCFA-producing probiotics—can restore bile acid metabolic homeostasis and indirectly inhibit excessive PXR activation, thereby reducing hepatic lipid peroxidation and insulin resistance. For example, prebiotics like inulin derivatives have been shown to reshape the gut microenvironment and enhance hepatic detoxification, offering new directions for personalized NAFLD treatment (143, 144). Future therapies combining PXR modulators with microbiota interventions may become key to overcoming NAFLD treatment bottlenecks (145). Probiotic interventions can also restore PXR function inhibition caused by antibiotics, improving metabolic variations of drugs like cyclosporine (146).
PXR’s dual role in liver disease progression and protection exhibits significant complexity. Regarding disease progression, PXR activation promotes hepatic lipid synthesis and fatty acid uptake while simultaneously inhibiting fatty acid β-oxidation, culminating in lipid accumulation and steatosis (147). Mechanistically, PXR drives this process through transcriptional upregulation of Solute carrier family 27 member 4 (SLC27A4), thereby accelerating NAFLD progression (148). In contrast, emerging evidence suggests protective roles for PXR modulation. Preclinical studies demonstrate that selective PXR modulators (e.g., hyodeoxycholic acid, HDCA) may improve metabolic function in early-stage disease (149). Furthermore, PXR activation mitigates drug-induced liver injury (DILI) through multifaceted mechanisms including enhanced detoxification regulation, anti-inflammatory effects, anti-apoptotic signaling, and improved bile acid excretion. Notably, PXR’s involvement in NAFLD remains controversial, with substantial discrepancies between preclinical and clinical findings. While PXR activation shows opposing effects on gluconeogenesis between rodents and humans (150), consistent evidence from HFHC diet-induced mouse models demonstrates that PXR activation triggers key NAFLD/NASH hallmarks including steatosis, inflammation, and lipotoxicity (22). This paradox may stem from PXR’s cell-specific expression pattern - as a hepatocyte-predominant nuclear transcription factor, its limited expression in Kupffer cells and hepatic stellate cells has resulted in insufficient research on its immune-modulatory roles in these cell populations. Collectively, these findings highlight the need for comprehensive studies to clarify PXR’s stage- and cell-specific functions in NAFLD/NASH pathogenesis. Future research should particularly address how PXR modulation in non-hepatocyte populations influences disease progression across different metabolic contexts.
The natural agonist ursolic acid activates PXR, significantly upregulating the phosphorylation of acetyl-CoA carboxylase (ACC), thereby inhibiting lipogenesis (151). This mechanism is closely linked to PXR’s transcriptional regulation of lipid metabolism genes, possibly involving indirect control of targets like stearoyl-CoA desaturase 1 (SCD1) (152). Additionally, excessive PXR activation is associated with NAFLD progression, and ursolic acid, as a PXR modulator, may offer new therapeutic strategies for metabolic liver diseases by balancing bile acid metabolism and improving intestinal barrier function. Future research could explore the synergistic effects of ursolic acid with other PPAR subtypes (e.g., PPARγ) to optimize its anti-lipidogenic efficacy (153). Mechanistic studies on the gut microbiota metabolite-PXR axis may further elucidate its role in metabolic liver disease progression.
Bile salt hydrolase (BSH) is a core enzyme in gut microbiota that converts primary bile acids to secondary ones, such as DCA, which indirectly affects PXR signaling by activating the FXR (154). Engineered BSH+ lactic acid bacteria may enhance bile acid metabolic efficiency, promoting PXR-dependent expression of hepatic detoxification enzymes (e.g., CYP3A4) and improving metabolic disorders (155). Recombinant BSH lactic acid bacteria not only improve gut colonization (with 2–3-fold upregulation of adhesion protein expression) but also regulate host immune microenvironments through SCFA secretion (156). Butyrate and other SCFAs have been shown to inhibit PXR transcriptional activity via HDAC inhibition.
6.3 Clinical prospects for PXR-targeted personalized therapy
In precision PXR modulation, building on the molecular design experience of the lead compound CVK003 (138), future strategies may include developing “smart-responsive” PXR modulators, such as pH/enzyme-sensitive prodrugs for targeted intestinal release (e.g., colon-specific delivery systems) and dual-functional molecules (e.g., PXR-FXR co-agonists) to synchronize bile acid synthesis and detoxification pathways (157). Ultrasound dynamics is a technology that utilizes ultrasonic energy to regulate drug delivery and enhance therapeutic efficacy. Its core mechanism lies in leveraging the physical effects of ultrasound—such as cavitation, mechanical vibration, and thermal effects—to alter tissue or cell membrane permeability, thereby facilitating targeted drug delivery or activating drug activity (158). When existing PXR modulators fail to achieve therapeutic effects, our department employs ultrasound dynamics to stimulate PXR modulators, thereby activating PXR function. This approach serves to modulate liver immunity, accelerate drug and bile acid metabolism, and ultimately alleviate liver immune diseases. Assessment models integrating fecal secondary bile acid profiles (DCA/LCA ratio), serum CYP3A4 activity, and gut microbiota BSH gene abundance could enable stratified treatment (159, 160). In host-microbiota co-intervention systems, optimizing BSH+ lactic acid bacteria colonization (via adhesion peptide integration), metabolic profiles (precisely regulating SCFA/DCA ratios), and immunomodulatory functions (e.g., carrying IL-10 anti-inflammatory genes) could provide personalized probiotic/prebiotic combinations (161, 162). Multi-omics technologies may predict PXR-responsive bacterial strains, analyze host-microbe interaction networks, and assess hepatocyte PXR pathway states, ultimately offering systematic solutions for PXR-targeted therapy.
Author contributions
TL: Formal analysis, Writing – original draft, Methodology, Investigation. YC: Investigation, Methodology, Writing – original draft. LL: Investigation, Methodology, Writing – original draft. TW: Writing – review & editing, Validation, Methodology, Project administration. YQ: Writing – review & editing. BJ: Formal analysis, Project administration, Writing – review & editing, Methodology, Investigation, Supervision.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the Guangdong Basic and Applied Basic Research Foundation (No. 2023A1515111116), the Shenzhen Foundation of Science and Technology (No. JCYJ20230807151308018), the Zhanjiang Science and Technology Project (2023B01176), the Shenzhen Longhua District Science and Technology Innovation Fund Projects (No. 2022045), and the Research Foundation of Shenzhen Longhua District Central Hospital (No. 202203).
Acknowledgments
We sincerely appreciate Dr. Xiaokang Wang’s invaluable assistance throughout the manuscript submission and revision process. We sincerely thank the reviewers for their valuable feedback on this paper.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Wang X, Hu X, Ye C, Zhao J, Tan SC, Zhou L, et al. Astragalus Polysaccharide Enhances Voriconazole Metabolism under Inflammatory Conditions through the Gut Microbiota. J Clin Trans Hepatol. (2024) 12:481–95. doi: 10.14218/JCTH.2024.00024
2. Huber AD, Wright WC, Lin W, Majumder K, Low JA, Wu J, et al. Mutation of a single amino acid of pregnane X receptor switches an antagonist to agonist by altering AF-2 helix positioning. Cell Mol Life Sci. (2021) 78:317–35. doi: 10.1007/s00018-020-03505-y
3. Wang X, Zhang G, Bian Z, Chow V, Grimaldi M, Carivenc C, et al. An abundant ginger compound furanodienone alleviates gut inflammation via the xenobiotic nuclear receptor PXR in mice. Nat Commun. (2025) 16:1280. doi: 10.1038/s41467-025-56624-0
4. Tian J, Wang R, Yang X, Yang J, Zhang Y, Li X, et al. Pregnane X receptor promotes liver enlargement in mice through the spatial induction of hepatocyte hypertrophy and proliferation. Chem Biol Interact. (2022) 367:110133. doi: 10.1016/j.cbi.2022.110133
5. Ikeda K, Horie K, Inoue S, and Blumberg B. Physiological functions and pathological roles of PXR. J Endocr Soc. (2025) 9:bvaf119. doi: 10.1210/jendso/bvaf119
6. Deng X, Li Y, Jiang L, Xie X, and Wang X. 1-methylnicotinamide modulates IL-10 secretion and voriconazole metabolism. Front Immunol. (2025) 16:1529660. doi: 10.3389/fimmu.2025.1529660
7. Rao J, Qiu P, Zhang Y, and Wang X. Gut microbiota trigger host liver immune responses that affect drug-metabolising enzymes. Front Immunol. (2024) 15:1511229. doi: 10.3389/fimmu.2024.1511229
8. Wang X, Ye C, Yang X, and Yang M. Ceftriaxone-associated dysbiosis decreases voriconazole bioavailability by upregulating intestinal P-glycoprotein expression through activation of the Nrf2-mediated signalling pathway. Front Pharmacol. (2024) 15:1522271. doi: 10.3389/fphar.2024.1522271
9. Zhao J, Zhao C, Xun T, Wang X, Wei S, Ye C, et al. Huang gan formula alleviates systemic inflammation and uremia in adenine-induced chronic kidney disease rats may associate with modification of gut microbiota and colonic microenvironment. Drug Des Devel Ther. (2024) 18:13–28. doi: 10.2147/DDDT.S421446
10. Li Y, Liu Q, Pan C-Y, and Lan X-Y. The free fatty acid receptor 2 (FFA2): Mechanisms of action, biased signaling, and clinical prospects. Pharmacol Ther. (2025) 272:108878. doi: 10.1016/j.pharmthera.2025.108878
11. Zhao S, Fu D, Lin Y, Sun X, Wang X, Wu X, et al. The role of the microbiome on immune homeostasis of the host nervous system. Front Immunol. (2025) 16:1609960. doi: 10.3389/fimmu.2025.1609960
12. Wang X, Xu G, Ma H, Deng X, and Ma G. Emerging frontiers in epigenetic-targeted therapeutics for pediatric neuroblastoma. Front Immunol. (2025) 16:1637626. doi: 10.3389/fimmu.2025.1637626
13. Qin S, Zhang K, Ding X, Bai S, Wang J, Tian G, et al. Microbiome-metabolomics analysis insight into the effects of dietary resistant starch on intestinal integrity. Food Chem. (2023) 401:134148. doi: 10.1016/j.foodchem.2022.134148
14. Dou Y, Liu P, Ding Z, Zhou Y, Jing H, Ren Y, et al. Orally administrable H2 S-scavenging metal-organic framework prepared by co-flow microfluidics for comprehensive restoration of intestinal milieu. Adv Mater. (2023) 35:e2210047. doi: 10.1002/adma.202210047
15. Choi J, Choi YR, Jeong MK, Song HH, Yu JS, Song SH, et al. Phocaeicola dorei ameliorates progression of steatotic liver disease by regulating bile acid, lipid, inflammation and proliferation. Gut Microbes. (2025) 17:2539448. doi: 10.1080/19490976.2025.2539448
16. Gong J-X, Wang X-L, Lin C-X, Li X-Y, Wu J, Tan Q-G, et al. Gut microbiota mitigate the reproductive toxicity of silver nanoparticles through thiamine-derived metabolites. Nat Commun. (2025) 16:7294. doi: 10.1038/s41467-025-62595-z
17. Zhang H, Yan S, Du R, Ma Z, Xue Y, Zhao Y, et al. Epiberberine alleviates cadmium-induced duodenal inflammation in Hu sheep by inhibiting HIF-1 signaling pathway. Ecotoxicol Environ Saf. (2025) 303:118861. doi: 10.1016/j.ecoenv.2025.118861
18. Shang M, Ning J, Zang C, Ma J, Yang Y, Wan Z, et al. Microbial metabolite 3-indolepropionic acid alleviated PD pathologies by decreasing enteric glia cell gliosis via suppressing IL-13Rα1 related signaling pathways. Acta Pharm Sin B. (2025) 15:2024–38. doi: 10.1016/j.apsb.2025.02.029
19. Peng R, Song C, Gou S, Liu H, Kang H, Dong Y, et al. Gut Clostridium sporogenes-derived indole propionic acid suppresses osteoclast formation by activating pregnane X receptor. Pharmacol Res. (2024) 202:107121. doi: 10.1016/j.phrs.2024.107121
20. Fiorucci S, Zampella A, and Distrutti E. Development of FXR, PXR and CAR agonists and antagonists for treatment of liver disorders. Curr Top Med Chem. (2012) 12:605–24. doi: 10.2174/156802612799436678
21. Weng S, Zheng J, Lin Y, Fang H, and Ko C-Y. Therapeutic effects of amisulpride in male schizophrenics: Role of short-chain fatty acids and gene expression changes. Physiol Behav. (2025) 294:114864. doi: 10.1016/j.physbeh.2025.114864
22. Karpale M, Kummu O, Kärkkäinen O, Lehtonen M, Näpänkangas J, Herfurth UM, et al. Pregnane X receptor activation remodels glucose metabolism to promote NAFLD development in obese mice. Mol Metab. (2023) 76:101779. doi: 10.1016/j.molmet.2023.101779
23. Attema B, Kummu O, Krutáková M, Pavek P, Hakkola J, Hooiveld GJEJ, et al. The fungicide propiconazole induces hepatic steatosis and activates PXR in a mouse model of diet-induced obesity. Arch Toxicol. (2025) 99:1203–21. doi: 10.1007/s00204-024-03942-9
24. Lee JH, Zhou J, and Xie W. PXR and LXR in hepatic steatosis: a new dog and an old dog with new tricks. Mol Pharm. (2008) 5:60–6. doi: 10.1021/mp700121u
25. Bitter A, Rümmele P, Klein K, Kandel BA, Rieger JK, Nüssler AK, et al. Pregnane X receptor activation and silencing promote steatosis of human hepatic cells by distinct lipogenic mechanisms. Arch Toxicol. (2015) 89:2089–103. doi: 10.1007/s00204-014-1348-x
26. Aouabdi S, Gibson G, and Plant N. Transcriptional regulation of the PXR gene: identification and characterization of a functional peroxisome proliferator-activated receptor alpha binding site within the proximal promoter of PXR. Drug Metab Disposition. (2006) 34:138–44. doi: 10.1124/dmd.105.006064
27. Tan W, Zhao K, Xiang J, Zhou X, Cao F, Song W, et al. Pyrazinamide alleviates rifampin-induced steatohepatitis in mice by regulating the activities of cholesterol-activated 7α-hydroxylase and lipoprotein lipase. Eur J Pharm Sci. (2020) 151:105402. doi: 10.1016/j.ejps.2020.105402
28. Griffiths WJ, Crick PJ, Meljon A, Theofilopoulos S, Abdel-Khalik J, Yutuc E, et al. Additional pathways of sterol metabolism: Evidence from analysis of Cyp27a1-/- mouse brain and plasma. Biochim Biophys Acta Mol Cell Biol Lipids. (2019) 1864:191–211. doi: 10.1016/j.bbalip.2018.11.006
29. Wang X, Zhao C, Chen Y, and Qiu P. Toll-like receptor 4 activation potentiates voriconazole-induced hepatotoxicity via transcriptional repression of the farnesoid X receptor in murine hepatocytes. J Pharm Pharmacol. (2025) 77:1542–1551. doi: 10.1093/jpp/rgaf060
30. Wang S, Jia Q, Liu X, Ma Y, Yang Y, Rong X, et al. Hyperoside modulates bile acid and fatty acid metabolism, presenting a potentially promising treatment for non-alcoholic fatty liver disease. J Adv Res. (2025) 9:S2090–1232(25)00308-X. doi: 10.1016/j.jare.2025.05.014
31. Xun T, Lin Z, Wang X, Zhan X, Feng H, Gan D, et al. Advanced oxidation protein products downregulate CYP1A2 and CYP3A4 expression and activity via the NF-κB-mediated signaling pathway in vitro and in vivo. Lab Invest. (2021) 101:1197–209. doi: 10.1038/s41374-021-00610-9
32. Cai X, Young GM, and Xie W. The xenobiotic receptors PXR and CAR in liver physiology, an update. Biochim Biophys Acta Mol Basis Dis. (2021) 1867:166101. doi: 10.1016/j.bbadis.2021.166101
33. Mai Y, Madla CM, Shao H, Qin Y, Merchant HA, Murdan S, et al. Sex-specific effects of excipients on oral drug bioavailability. Int J Pharm. (2022) 629:122365. doi: 10.1016/j.ijpharm.2022.122365
34. Kakizaki S, Takizawa D, Tojima H, Yamazaki Y, and Mori M. Xenobiotic-sensing nuclear receptors CAR and PXR as drug targets in cholestatic liver disease. Curr Drug Targets. (2009) 10:1156–63. doi: 10.2174/138945009789735174
35. Nagahori H, Nakamura K, Sumida K, Ito S, and Ohtsuki S. Combining genomics to identify the pathways of post-transcriptional nongenotoxic signaling and energy homeostasis in livers of rats treated with the pregnane X receptor agonist, pregnenolone carbonitrile. J Proteome Res. (2017) 16:3634–45. doi: 10.1021/acs.jproteome.7b00364
36. Perez MJ, Gonzalez-Sanchez E, Gonzalez-Loyola A, Gonzalez-Buitrago JM, and Marin JJG. Mitochondrial genome depletion dysregulates bile acid- and paracetamol-induced expression of the transporters Mdr1, Mrp1 and Mrp4 in liver cells. Br J Pharmacol. (2011) 162:1686–99. doi: 10.1111/j.1476-5381.2010.01174.x
37. Basaly V, Bhattacharya A, and Guo GL. Insights of direct and indirect regulation of PXR through phosphorylation in fatty liver disease. Mol Pharmacol. (2025) 107:100014. doi: 10.1016/j.molpha.2024.100014
38. Niu B, Pan T, Xiao Y, Wang H, Zhu J, Tian F, et al. The therapeutic potential of dietary intervention: based on the mechanism of a tryptophan derivative-indole propionic acid on metabolic disorders. Crit Rev Food Sci Nutr. (2025) 65:1729–48. doi: 10.1080/10408398.2023.2299744
39. Jahnel J, Fickert P, Langner C, Högenauer C, Silbert D, Gumhold J, et al. Impact of experimental colitis on hepatobiliary transporter expression and bile duct injury in mice. Liver Int. (2009) 29:1316–25. doi: 10.1111/j.1478-3231.2009.02044.x
40. Wang Z-J, Zhan X-Y, Ma L-Y, Yao K, Dai H-Y, Kumar Santhanam R, et al. Activation of the γ-secretase/NICD-PXR/Notch pathway induces Taxol resistance in triple-negative breast cancer. Biochem Pharmacol. (2024) 230:116577. doi: 10.1016/j.bcp.2024.116577
41. Kagami T, Yamade M, Suzuki T, Uotani T, Hamaya Y, Iwaizumi M, et al. Comparative study of effects of vonoprazan and esomeprazole on antiplatelet function of clopidogrel or prasugrel in relation to CYP2C19 genotype. Clin Pharmacol Ther. (2018) 103:906–13. doi: 10.1002/cpt.863
42. Loretz C, Ho M-CD, Alam N, Mitchell W, and Li AP. Application of cryopreserved human intestinal mucosa and cryopreserved human enterocytes in the evaluation of herb-drug interactions: evaluation of CYP3A inhibitory potential of grapefruit juice and commercial formulations of twenty-nine herbal supplements. Drug Metab Disposition. (2020) 48:1084–91. doi: 10.1124/dmd.120.000033
43. Qian L, Xu Z, Luo T, Gao Z, Cheng K, He X, et al. In silico identification and verification of Tanshinone IIA-related prognostic genes in hepatocellular carcinoma. Front Immunol. (2024) 15:1482914. doi: 10.3389/fimmu.2024.1482914
44. Yu C, Ye S, Sun H, Liu Y, Gao L, Shen C, et al. PXR-mediated transcriptional activation of CYP3A4 by cryptotanshinone and tanshinone IIA. Chem Biol Interact. (2009) 177:58–64. doi: 10.1016/j.cbi.2008.08.013
45. Liu Y-H, Mo S-L, Bi H-C, Hu B-F, Li CG, Wang Y-T, et al. Regulation of human pregnane X receptor and its target gene cytochrome P450 3A4 by Chinese herbal compounds and a molecular docking study. Xenobiotica. (2011) 41:259–80. doi: 10.3109/00498254.2010.537395
46. Lin C, Lin Y, Xiao R, Guo M, Zhang H, Chen W, et al. Bifidobacterium species associated with breastfeeding alleviate neonatal hyperbilirubinaemia via the gut microbiota-α-linolenic and linoleic acid metabolism-enterohepatic circulation axis. Microbiome. (2025) 13:187. doi: 10.1186/s40168-025-02190-y
47. Liu L, Liang L, Liang H, Wang M, Zhou W, Mai G, et al. Microbiome-metabolome generated bile acids gatekeep infliximab efficacy in Crohn’s disease by licensing M1 suppression and Treg dominance. J Adv Res. (2025) 12:S2090–1232(25)00606-X. doi: 10.1016/j.jare.2025.08.017
48. Wang E, Ren K, Wang X, Du S, Gao X, Niu W, et al. The close association of Muribaculum and PA (10:0/a-17:0) with the occurrence of pancreatic ductal adenocarcinoma and immunotherapy. Front Immunol. (2024) 15:1505966. doi: 10.3389/fimmu.2024.1505966
49. Fang T, Ye Z, Chen X, Wang Y, Wan J, and Wang H. Repurposing of camptothecin: An esterase-activatable prodrug delivered by a self-emulsifying formulation that improves efficacy in colorectal cancer. Int J Pharm. (2021) 599:120399. doi: 10.1016/j.ijpharm.2021.120399
50. Dutta M, Lim JJ, and Cui JY. Pregnane X receptor and the gut-liver axis: A recent update. Drug Metab Disposition. (2022) 50:478–91. doi: 10.1124/dmd.121.000415
51. Ridlon JM and Bajaj JS. The human gut sterolbiome: bile acid-microbiome endocrine aspects and therapeutics. Acta Pharm Sin B. (2015) 5:99–105. doi: 10.1016/j.apsb.2015.01.006
52. Dvořák Z, Sokol H, and Mani S. Drug mimicry: promiscuous receptors PXR and ahR, and microbial metabolite interactions in the intestine. Trends Pharmacol Sci. (2020) 41:900–8. doi: 10.1016/j.tips.2020.09.013
53. Hukkanen J, Küblbeck J, Hakkola J, and Rysä J. Nuclear receptors CAR and PXR as cardiometabolic regulators. Pharmacol Res. (2025) 219:107892. doi: 10.1016/j.phrs.2025.107892
54. Abualsunun WA, Sahin C, Cummins CL, and Piquette-Miller M. Essential role of STAT-3 dependent NF-κB activation on IL-6-mediated downregulation of hepatic transporters. Eur J Pharm Sci. (2019) 143:105151. doi: 10.1016/j.ejps.2019.105151
55. Wang W, Li L, Li X, Chen J, Wang R, Yang Q, et al. FXR overexpression alleviates cholestasis via NLRC4 inflammasome suppression and bile acid homeostasis regulation. Free Radic Biol Med. (2025) 238:152–68. doi: 10.1016/j.freeradbiomed.2025.06.039
56. Liu M, Zhang G, Song M, Wang J, Shen C, Chen Z, et al. Activation of farnesoid X receptor by schaftoside ameliorates acetaminophen-induced hepatotoxicity by modulating oxidative stress and inflammation. Antioxid Redox Signal. (2020) 33:87–116. doi: 10.1089/ars.2019.7791
57. Preiss LC, Liu R, Hewitt P, Thompson D, Georgi K, Badolo L, et al. Deconvolution of cytochrome P450 induction mechanisms in hepaRG nuclear hormone receptor knockout cells. Drug Metab Disposition. (2021) 49:668–78. doi: 10.1124/dmd.120.000333
58. Gachon F. Physiological function of PARbZip circadian clock-controlled transcription factors. Ann Med. (2007) 39:562–71. doi: 10.1080/07853890701491034.
59. Masson D, Lagrost L, Athias A, Gambert P, Brimer-Cline C, Lan L, et al. Expression of the pregnane X receptor in mice antagonizes the cholic acid-mediated changes in plasma lipoprotein profile. Arterioscler Thromb Vasc Biol. (2005) 25:2164–9. doi: 10.1161/01.ATV.0000183674.88817.fb
60. Wang Y, Wang P, Wang Q, Chen S, Wang X, Zhong X, et al. The long noncoding RNA HNF1A-AS1 with dual functions in the regulation of cytochrome P450 3A4. Biochem Pharmacol. (2024) 220:116016. doi: 10.1016/j.bcp.2023.116016
61. Breuker C, Planque C, Rajabi F, Nault J-C, Couchy G, Zucman-Rossi J, et al. Characterization of a novel PXR isoform with potential dominant-negative properties. J Hepatol. (2014) 61:609–16. doi: 10.1016/j.jhep.2014.04.030
62. Xue Y, Moore LB, Orans J, Peng L, Bencharit S, Kliewer SA, et al. Crystal structure of the pregnane X receptor-estradiol complex provides insights into endobiotic recognition. Mol Endocrinol. (2007) 21:1028–38. doi: 10.1210/me.2006-0323
63. Grewal GK, Singh KD, Kanojia N, Rawat C, Kukal S, Jajodia A, et al. Exploring the carbamazepine interaction with human pregnane X receptor and effect on ABCC2 using in vitro and in silico approach. Pharm Res. (2017) 34:1444–58. doi: 10.1007/s11095-017-2161-z
64. Sugatani J, Uchida T, Kurosawa M, Yamaguchi M, Yamazaki Y, Ikari A, et al. Regulation of pregnane X receptor (PXR) function and UGT1A1 gene expression by posttranslational modification of PXR protein. Drug Metab Disposition. (2012) 40:2031–40. doi: 10.1124/dmd.112.046748
65. Wang W, Li L, Li X, Chen J, Wang R, Yang Q, et al. β-Sitosterol protects against lithocholic acid-induced hepatotoxicity and cholestasis via farnesoid X receptor-mediated regulation of transporters and enzymes in vitro and in vivo. Toxicol Appl Pharmacol. (2025) 498:117308. doi: 10.1016/j.taap.2025.117308
66. Chen P, Yu K, Yang K, Zhang D, Li P, and He Y. Indole-3-acetic acid-mediated root exudates as potential inhibitors of antibiotic resistance genes in the rhizosphere microbiome: Mechanistic insights into microbial community assembly and resistome dissemination. Bioresour Technol. (2025) 438:133191. doi: 10.1016/j.biortech.2025.133191
67. Liu T, Lv J, Bian B, Wu Q, Zhou L, Zhang S, et al. Postbiotic Limosilactobacillus reuteri cultured with Polygonatum kingianum polysaccharides ameliorates high-fat-high-sugar-deteriorated colitis and associated hepatobiliary disorders. Int J Biol Macromol. (2025) 322:147065. doi: 10.1016/j.ijbiomac.2025.147065
68. Wang X, Wei L, Yang J, Wang Y, Chen S, Yang K, et al. DNA methylation determines the regulation of pregnane X receptor on CYP3A4 expression. Clin Exp Pharmacol Physiol. (2020) 48:250–9. doi: 10.1111/1440-1681.13420
69. Zhang S, Guo M, Jiang X, Tang L, Wu T, Bi G, et al. PXR triggers YAP-TEAD binding and Sirt2-driven YAP deacetylation and polyubiquitination to promote liver enlargement and regeneration in mice. Pharmacol Res. (2023) 188:106666. doi: 10.1016/j.phrs.2023.106666
70. Fawad JA, Luzader DH, Hanson GF, Moutinho TJ, McKinney CA, Mitchell PG, et al. Histone deacetylase inhibition by gut microbe-generated short-chain fatty acids entrains intestinal epithelial circadian rhythms. Gastroenterology. (2022) 163:1377–1390. doi: 10.1053/j.gastro.2022.07.051
71. Wang X, Tong Y, Xun T, Feng H, Lei Y, Li Y, et al. Functions, mechanisms, and therapeutic implications of noncoding RNA in acute myeloid leukemia. Fundam Res. (2025) 5:1781–94. doi: 10.1016/j.fmre.2023.04.012
72. Chilton PM, Ghare SS, Charpentier BT, Myers SA, Rao AV, Petrosino JF, et al. Age-associated temporal decline in butyrate-producing bacteria plays a key pathogenic role in the onset and progression of neuropathology and memory deficits in 3×Tg-AD mice. Gut Microbes. (2024) 16:2389319. doi: 10.1080/19490976.2024.2389319
73. Yang B, Xiong Z, Lin M, Yang Y, Chen Y, Zeng J, et al. Astragalus polysaccharides alleviate type 1 diabetes via modulating gut microbiota in mice. Int J Biol Macromol. (2023) 234:123767. doi: 10.1016/j.ijbiomac.2023.123767
74. Ma Z, Wang Z, Cao J, Dong Y, and Chen Y. Regulatory roles of intestinal CD4+ T cells in inflammation and their modulation by the intestinal microbiota. Gut Microbes. (2025) 17:2560019. doi: 10.1080/19490976.2025.2560019
75. Wang Y-G, Zhou J-M, Ma Z-C, Li H, Liang Q-D, Tan H-L, et al. Pregnane X receptor mediated-transcription regulation of CYP3A by glycyrrhizin: a possible mechanism for its hepatoprotective property against lithocholic acid-induced injury. Chem Biol Interact. (2012) 200:11–20. doi: 10.1016/j.cbi.2012.08.023
76. Parkes HA, Preston E, Wilks D, Ballesteros M, Carpenter L, Wood L, et al. Overexpression of acyl-CoA synthetase-1 increases lipid deposition in hepatic (HepG2) cells and rodent liver in vivo. Am J Physiol Endocrinol Metab. (2006) 291:E737–44. doi: 10.1152/ajpendo.00112.2006
77. Listopad S, Magnan C, Asghar A, Stolz A, Tayek JA, Liu Z-X, et al. Differentiating between liver diseases by applying multiclass machine learning approaches to transcriptomics of liver tissue or blood-based samples. JHEP Rep. (2022) 4:100560. doi: 10.1016/j.jhepr.2022.100560
78. Tang R, Zha H, Liu R, Lv J, Cao D, and Li L. Sodium butyrate attenuates liver fibrogenesis via promoting H4K8 crotonylation. Mol Cell Biochem. (2025) 480:4467–81. doi: 10.1007/s11010-025-05274-3
79. Toriyabe T, Nagata K, Takada T, Aratsu Y, Matsubara T, Yoshinari K, et al. Unveiling a new essential cis element for the transactivation of the CYP3A4 gene by xenobiotics. Mol Pharmacol. (2009) 75:677–84. doi: 10.1124/mol.108.050575
80. Gotoh-Saito S, Wada R, Nishimura T, and Kawaji H. Drug-induced cis-regulatory elements in human hepatocytes affect molecular phenotypes associated with adverse reactions. Nat Commun. (2025) 16:3851. doi: 10.1038/s41467-025-59132-3
81. Zheng M, Yang X, Wu Q, Gong Y, Pang N, Ge X, et al. Butyrate attenuates hepatic steatosis induced by a high-fat and fiber-deficient diet via the hepatic GPR41/43-caMKII/HDAC1-CREB pathway. Mol Nutr Food Res. (2023) 67:e2200597. doi: 10.1002/mnfr.202200597
82. Al-Aqil FA, Monte MJ, Peleteiro-Vigil A, Briz O, Rosales R, González R, et al. Interaction of glucocorticoids with FXR/FGF19/FGF21-mediated ileum-liver crosstalk. Biochim Biophys Acta Mol Basis Dis. (2018) 1864:2927–37. doi: 10.1016/j.bbadis.2018.06.003
83. Ovadia C, Perdones-Montero A, Spagou K, Smith A, Sarafian MH, Gomez-Romero M, et al. Enhanced microbial bile acid deconjugation and impaired ileal uptake in pregnancy repress intestinal regulation of bile acid synthesis. Hepatology. (2019) 70:276–93. doi: 10.1002/hep.30661
84. Yu Cai Lim M and Kiat Ho H. Pharmacological modulation of cholesterol 7α-hydroxylase (CYP7A1) as a therapeutic strategy for hypercholesterolemia. Biochem Pharmacol. (2024) 220:115985. doi: 10.1016/j.bcp.2023.115985
85. Sueyoshi T and Negishi M. CAR: Discovery and development by the pharmacogenetics laboratory at NIEHS, NIH. Pharmacol Res. (2025) 219:107907. doi: 10.1016/j.phrs.2025.107907
86. Feng Y, Kim J-W, and Xie W. The intestinal functions of PXR and CAR. Pharmacol Res. (2025) 216:107779. doi: 10.1016/j.phrs.2025.107779
87. Mezler M, Jones RS, Sangaraju D, Goldman DC, Hoffmann M, Heikkinen AT, et al. Analysis of the bile acid composition in a fibroblast growth factor 19-expressing liver-humanized mouse model and its use for CYP3A4-mediated drug-drug interaction studies. Drug Metab Disposition. (2023) 51:1391–402. doi: 10.1124/dmd.123.001398
88. Liu H-Y, Yuan P, Li S, Ogamune KJ, Shi X, Zhu C, et al. Lactobacillus johnsonii alleviates experimental colitis by restoring intestinal barrier function and reducing NET-mediated gut-liver inflammation. Commun Biol. (2025) 8:1222. doi: 10.1038/s42003-025-08679-4
89. Pasta A, Formisano E, Calabrese F, Marabotto E, Furnari M, Bodini G, et al. From dysbiosis to hepatic inflammation: A narrative review on the diet-microbiota-liver axis in steatotic liver disease. Microorganisms. (2025) 13:241. doi: 10.3390/microorganisms13020241
90. Qin X and Wang X. Role of vitamin D receptor in the regulation of CYP3A gene expression. Acta Pharm Sin B. (2019) 9:1087–98. doi: 10.1016/j.apsb.2019.03.005
91. Wang X, Rao J, Tan Z, Xun T, Zhao J, and Yang X. Inflammatory signaling on cytochrome P450-mediated drug metabolism in hepatocytes. Front In Pharmacol. (2022) 13:1043836. doi: 10.3389/fphar.2022.1043836
92. Girardin SE, Boneca IG, Carneiro LAM, Antignac A, Jéhanno M, Viala J, et al. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science. (2003) 300:1584–7. doi: 10.1126/science.1084677
93. Byndloss MX, Keestra-Gounder AM, Bäumler AJ, and Tsolis RM. NOD1 and NOD2: new functions linking endoplasmic reticulum stress and inflammation. DNA Cell Biol. (2016) 35:311–3. doi: 10.1089/dna.2016.3396
94. Wang X, Ye C, Xun T, Mo L, Tong Y, Ni W, et al. Bacteroides fragilis polysaccharide A ameliorates abnormal voriconazole metabolism accompanied with the inhibition of TLR4/NF-κB pathway. Front Pharmacol. (2021) 12:663325. doi: 10.3389/fphar.2021.663325
95. Nian F, Chen Y, Xia Q, Zhu C, Wu L, and Lu X. Gut microbiota metabolite trimethylamine N-oxide promoted NAFLD progression by exacerbating intestinal barrier disruption and intrahepatic cellular imbalance. Int Immunopharmacol. (2024) 142:113173. doi: 10.1016/j.intimp.2024.113173
96. Yan B-F, Wang Y, Wang W-B, Ding X-J, Wei B, Liu S-J, et al. Huangqin decoction mitigates hepatic inflammation in high-fat diet-challenged rats by inhibiting TLR4/NF-κB/NLRP3 pathway. J Ethnopharmacol. (2023) 303:115999. doi: 10.1016/j.jep.2022.115999
97. Fang S, Wang T, Li Y, Xue H, Zou J, Cai J, et al. Gardenia jasminoides Ellis polysaccharide ameliorates cholestatic liver injury by alleviating gut microbiota dysbiosis and inhibiting the TLR4/NF-κB signaling pathway. Int J Biol Macromol. (2022) 205:23–36. doi: 10.1016/j.ijbiomac.2022.02.056
98. Su X, Zhang M, Qi H, Gao Y, Yang Y, Yun H, et al. Gut microbiota-derived metabolite 3-idoleacetic acid together with LPS induces IL-35+ B cell generation. Microbiome. (2022) 10:13. doi: 10.1186/s40168-021-01205-8
99. Xie W, Radominska-Pandya A, Shi Y, Simon CM, Nelson MC, Ong ES, et al. An essential role for nuclear receptors SXR/PXR in detoxification of cholestatic bile acids. Proc Natl Acad Sci U S A. (2001) 98:3375–80. doi: 10.1073/pnas.051014398
100. Staudinger JL, Goodwin B, Jones SA, Hawkins-Brown D, MacKenzie KI, LaTour A, et al. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci U.S.A. (2001) 98:3369–74. doi: 10.1073/pnas.051551698
101. Mun SJ, Lee J, Chung K-S, Son M-Y, and Son MJ. Effect of microbial short-chain fatty acids on CYP3A4-mediated metabolic activation of human pluripotent stem cell-derived liver organoids. Cells. (2021) 10:126. doi: 10.3390/cells10010126
102. Luo M, Du Y, Liu X, Zhang S, Zhu W, Liu K, et al. Fecal microbiota transplantation alleviates cirrhotic portal hypertension in rats via butyrate-mediated HDAC3 inhibition and PI3K/Akt/eNOS signaling regulation. Eur J Pharmacol. (2025) 1002:177781. doi: 10.1016/j.ejphar.2025.177781
103. Zhao T, Zhang X, Xiang Q, Liu Y, Li X, Gu J, et al. Sodium butyrate ameliorated bile acid metabolism in diabetes mellitus by PI3K/AKT signaling pathway via the gut-liver axis. Curr Issues Mol Biol. (2025) 47:732. doi: 10.3390/cimb47090732
104. Bridgeman S, Woo HC, Newsholme P, and Mamotte C. Butyrate lowers cellular cholesterol through HDAC inhibition and impaired SREBP-2 signalling. Int J Mol Sci. (2022) 23:15506. doi: 10.3390/ijms232415506
105. Donde H, Ghare S, Joshi-Barve S, Zhang J, Vadhanam MV, Gobejishvili L, et al. Tributyrin inhibits ethanol-induced epigenetic repression of CPT-1A and attenuates hepatic steatosis and injury. Cell Mol Gastroenterol Hpatol. (2019) 9:569–85. doi: 10.1016/j.jcmgh.2019.10.005
106. You Q, Wang K, Zhao Z, Zhou H, Lan Z, Liang H, et al. Reduction of bacteroides fragilis in gut microbiome of chronic hepatitis B patients promotes liver injury. J Med Virol. (2025) 97:e70395. doi: 10.1002/jmv.70395
107. Zeng W, Gui L, Tan X, Zhu P, Hu Y, Wu Q, et al. Tertiary oxidation of deoxycholate is predictive of CYP3A activity in dogs. Drug Metab Disposition. (2021) 49:369–78. doi: 10.1124/dmd.121.000385
108. Liao L, Liu Z, Liu L, Huang C, Li Y, Mao C, et al. Targeting the ceramidase ACER3 attenuates cholestasis in mice by mitigating bile acid overload via unsaturated ceramide-mediated LXRβ signaling transduction. Nat Commun. (2025) 16:2112. doi: 10.1038/s41467-025-57330-7
109. Sinha AK, Laursen MF, Brinck JE, Rybtke ML, Hjørne AP, Procházková N, et al. Dietary fibre directs microbial tryptophan metabolism via metabolic interactions in the gut microbiota. Nat Microbiol. (2024) 9:1964–78. doi: 10.1038/s41564-024-01737-3
110. Krause FF, Mangold KI, Ruppert A-L, Leister H, Hellhund-Zingel A, Lopez Krol A, et al. Clostridium sporogenes-derived metabolites protect mice against colonic inflammation. Gut Microbes. (2024) 16:2412669. doi: 10.1080/19490976.2024.2412669
111. Watkins RE, Wisely GB, Moore LB, Collins JL, Lambert MH, Williams SP, et al. The human nuclear xenobiotic receptor PXR: structural determinants of directed promiscuity. Science. (2001) 292:2329–33. doi: 10.1126/science.1060762
112. LeCluyse EL. Pregnane X receptor: molecular basis for species differences in CYP3A induction by xenobiotics. Chem Biol Interact. (2001) 134:283–9. doi: 10.1016/S0009-2797(01)00163-6
113. Jonker JW, Liddle C, and Downes M. FXR and PXR: potential therapeutic targets in cholestasis. J Steroid Biochem Mol Biol. (2011) 130:147–58. doi: 10.1016/j.jsbmb.2011.06.012
114. Zhao L, Bin S, He H-L, Yang J-M, Pu Y-C, Gao C-H, et al. Sodium butyrate increases P-gp expression in lung cancer by upregulation of STAT3 and mRNA stabilization of ABCB1. Anticancer Drugs. (2018) 29:227–33. doi: 10.1097/CAD.0000000000000588
115. Vogel KR, Ainslie GR, Roullet JB, McConnell A, and Gibson KM. In vitro toxicological evaluation of NCS-382, a high-affinity antagonist of γ-hydroxybutyrate (GHB) binding. Toxicol In Vitro. (2017) 40:196–202. doi: 10.1016/j.tiv.2017.01.013
116. Wistuba W, Gnewuch C, Liebisch G, Schmitz G, and Langmann T. Lithocholic acid induction of the FGF19 promoter in intestinal cells is mediated by PXR. World J Gastroenterol. (2007) 13:4230–5. doi: 10.3748/wjg.v13.i31.4230
117. Owen BM, Milona A, van Mil S, Clements P, Holder J, Boudjelal M, et al. Intestinal detoxification limits the activation of hepatic pregnane X receptor by lithocholic acid. Drug Metab Disposition. (2010) 38:143–9. doi: 10.1124/dmd.109.029306
118. Wang Y-M, Ong SS, Chai SC, and Chen T. Role of CAR and PXR in xenobiotic sensing and metabolism. Expert Opin Drug Metab Toxicol. (2012) 8:803–17. doi: 10.1517/17425255.2012.685237
119. Vyhlídalová B, Bartoňková I, Jiskrová E, Li H, Mani S, and Dvořák Z. Differential activation of human pregnane X receptor PXR by isomeric mono-methylated indoles in intestinal and hepatic in vitro models. Toxicol Lett. (2020) 324:104–10. doi: 10.1016/j.toxlet.2020.02.010
120. Venkatesh M, Mukherjee S, Wang H, Li H, Sun K, Benechet AP, et al. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity. (2014) 41:296–310. doi: 10.1016/j.immuni.2014.06.014
121. Xu D-X, Chen Y-H, Wang J-P, Sun M-F, Wang H, Wei L-Z, et al. Perinatal lipopolysaccharide exposure downregulates pregnane X receptor and Cyp3a11 expression in fetal mouse liver. Toxicol Sci. (2005) 87:38–45. doi: 10.1093/toxsci/kfi239
122. Xu D-X, Wei W, Sun M-F, Wu C-Y, Wang J-P, Wei L-Z, et al. Kupffer cells and reactive oxygen species partially mediate lipopolysaccharide-induced downregulation of nuclear receptor pregnane x receptor and its target gene CYP3a in mouse liver. Free Radic Biol Med. (2004) 37:10–22. doi: 10.1016/j.freeradbiomed.2004.03.021
123. Chang TKH and Waxman DJ. Synthetic drugs and natural products as modulators of constitutive androstane receptor (CAR) and pregnane X receptor (PXR). Drug Metab Rev. (2006) 38:51–73. doi: 10.1080/03602530600569828
124. Lu G-R, Wang R-Z, Zhao X-Y, Xu J-E, Huang C-K, Sun W, et al. The CYP3A inducer dexamethasone affects the pharmacokinetics of sunitinib by accelerating its metabolism in rats. Chem Biol Interact. (2024) 403:111228. doi: 10.1016/j.cbi.2024.111228
125. Yueh M-F, Kawahara M, and Raucy J. High volume bioassays to assess CYP3A4-mediated drug interactions: induction and inhibition in a single cell line. Drug Metab Disposition. (2005) 33:38–48. doi: 10.1124/dmd.104.001594
126. Li L, Welch MA, Li Z, Mackowiak B, Heyward S, Swaan PW, et al. Mechanistic insights of phenobarbital-mediated activation of human but not mouse pregnane X receptor. Mol Pharmacol. (2019) 96:345–54. doi: 10.1124/mol.119.116616
127. Jones SA, Moore LB, Shenk JL, Wisely GB, Hamilton GA, McKee DD, et al. The pregnane X receptor: a promiscuous xenobiotic receptor that has diverged during evolution. Mol Endocrinol. (2000) 14:27–39. doi: 10.1210/mend.14.1.0409
128. Bertilsson G, Heidrich J, Svensson K, Asman M, Jendeberg L, Sydow-Bäckman M, et al. Identification of a human nuclear receptor defines a new signaling pathway for CYP3A induction. Proc Natl Acad Sci U S A. (1998) 95:12208–13. doi: 10.1073/pnas.95.21.12208
129. Pandey SK, Verma S, Upreti S, Mishra A, Yadav N, and Dwivedi-Agnihotri H. Role of cytochrome P450 3A4 in cancer drug resistance: challenges and opportunities. Curr Drug Metab. (2024) 25:235–47. doi: 10.2174/0113892002312369240703102215
130. Salem M, El-Bardissy A, Elshafei MN, Khalil A, Mahmoud H, Fahmi AM, et al. Warfarin-rifampin-gene (WARIF-G) interaction: A retrospective, genetic, case-control study. Clin Pharmacol Ther. (2023) 113:1150–9. doi: 10.1002/cpt.2871
131. Wyen C, Hendra H, Siccardi M, Platten M, Jaeger H, Harrer T, et al. Cytochrome P450 2B6 (CYP2B6) and constitutive androstane receptor (CAR) polymorphisms are associated with early discontinuation of efavirenz-containing regimens. J Antimicrobial Chemother. (2011) 66:2092–8. doi: 10.1093/jac/dkr272
132. Ayuso P, Neary M, Chiong J, and Owen A. Meta-analysis of the effect of CYP2B6, CYP2A6, UGT2B7 and CAR polymorphisms on efavirenz plasma concentrations. J Antimicrobial Chemother. (2019) 74:3281–90. doi: 10.1093/jac/dkz329
133. Chaudhry AS, Thirumaran RK, Yasuda K, Yang X, Fan Y, Strom SC, et al. Genetic variation in aldo-keto reductase 1D1 (AKR1D1) affects the expression and activity of multiple cytochrome P450s. Drug Metab Disposition. (2013) 41:1538–47. doi: 10.1124/dmd.113.051672
134. Li Y, Buckley D, Wang S, Klaassen CD, and Zhong X-B. Genetic polymorphisms in the TATA box and upstream phenobarbital-responsive enhancer module of the UGT1A1 promoter have combined effects on UDP-glucuronosyltransferase 1A1 transcription mediated by constitutive androstane receptor, pregnane X receptor, or glucocorticoid receptor in human liver. Drug Metab Disposition. (2009) 37:1978–86. doi: 10.1124/dmd.109.027409
135. Xu Y, Zhang Y-F, Chen X-Y, and Zhong D-F. CYP3A4 inducer and inhibitor strongly affect the pharmacokinetics of triptolide and its derivative in rats. Acta Pharmacol Sinica. (2018) 39:1386–92. doi: 10.1038/aps.2017.170
136. Jiang Y, Zhou Y, Song S, Fan S, Gao Y, Li Y, et al. St. John’s wort exacerbates acetaminophen-induced liver injury by activation of PXR and CYP-mediated bioactivation. Toxicol Sci. (2022) 190:54–63. doi: 10.1093/toxsci/kfac094
137. Dvořák Z, Kopp F, Costello CM, Kemp JS, Li H, Vrzalová A, et al. Targeting the pregnane X receptor using microbial metabolite mimicry. EMBO Mol Med. (2020) 12:e11621. doi: 10.15252/emmm.201911621
138. Li H, Illés P, Karunaratne CV, Nordstrøm LU, Luo X, Yang A, et al. Deciphering structural bases of intestinal and hepatic selectivity in targeting pregnane X receptor with indole-based microbial mimics. Bioorg Chem. (2021) 109:104661. doi: 10.1016/j.bioorg.2021.104661
139. Mejdrová I, Dušek J, Škach K, Stefela A, Skoda J, Chalupský K, et al. Discovery of novel human constitutive androstane receptor agonists with the imidazo[1,2-a]pyridine structure. J Med Chem. (2023) 66:2422–56. doi: 10.1021/acs.jmedchem.2c01140
140. Little M, Dutta M, Li H, Matson A, Shi X, Mascarinas G, et al. Understanding the physiological functions of the host xenobiotic-sensing nuclear receptors PXR and CAR on the gut microbiome using genetically modified mice. Acta Pharm Sin B. (2022) 12:801–20. doi: 10.1016/j.apsb.2021.07.022
141. Xu H, Luo Y, An Y, and Wu X. The mechanism of action of indole-3-propionic acid on bone metabolism. Food Funct. (2025) 16:406–21. doi: 10.1039/d4fo03783a
142. Kim S, Li H, Jin Y, Armad J, Gu H, Mani S, et al. Maternal PBDE exposure disrupts gut microbiome and promotes hepatic proinflammatory signaling in humanized PXR-transgenic mouse offspring over time. Toxicol Sci. (2023) 194:209–25. doi: 10.1093/toxsci/kfad056
143. Perl M, Fante MA, Herfeld K, Scherer JN, Poeck H, and Thiele Orberg E. Microbiota-derived metabolites: Key modulators of cancer immunotherapies. Med. (2025) 6:100773. doi: 10.1016/j.medj.2025.100773
144. Yao H, Ma S, Huang J, Si X, Yang M, Song W, et al. Trojan-horse strategy targeting the gut-liver axis modulates gut microbiome and reshapes microenvironment for orthotopic hepatocellular carcinoma therapy. Adv Sci (Weinh). (2024) 11:e2310002. doi: 10.1002/advs.202310002
145. Xiao Z, Zheng H, Zhang S, Li Q, Wang Y, Wang H, et al. Unraveling the characteristic structures of Cuscuta chinensis Lam. polysaccharides and its inhibitory effects on non-alcoholic fatty liver disease. Int J Biol Macromol. (2025) 322:146798. doi: 10.1016/j.ijbiomac.2025.146798
146. Kunasol C, Chattipakorn N, and Chattipakorn SC. Impact of calcineurin inhibitors on gut microbiota: Focus on tacrolimus with evidence from in vivo and clinical studies. Eur J Pharmacol. (2025) 987:177176. doi: 10.1016/j.ejphar.2024.177176
147. Zhou J, Zhai Y, Mu Y, Gong H, Uppal H, Toma D, et al. A novel pregnane X receptor-mediated and sterol regulatory element-binding protein-independent lipogenic pathway. J Biol Chem. (2006) 281:15013–20. doi: 10.1074/jbc.M511116200
148. Shen C, Pan Z, Xie W, Zhao J, Miao D, Zhao L, et al. Hepatocyte-specific SLC27A4 deletion ameliorates nonalcoholic fatty liver disease in mice via suppression of phosphatidylcholine-mediated PXR activation. Metabolism. (2024) 162:156054. doi: 10.1016/j.metabol.2024.156054
149. Zhong J, He X, Gao X, Liu Q, Zhao Y, Hong Y, et al. Hyodeoxycholic acid ameliorates nonalcoholic fatty liver disease by inhibiting RAN-mediated PPARα nucleus-cytoplasm shuttling. Nat Commun. (2023) 14:5451. doi: 10.1038/s41467-023-41061-8
150. Mackowiak B, Hodge J, Stern S, and Wang H. The roles of xenobiotic receptors: beyond chemical disposition. Drug Metab Disposition: Biol Fate Chemicals. (2018) 46:1361–71. doi: 10.1124/dmd.118.081042
151. He Y, Li Y, Zhao T, Wang Y, and Sun C. Ursolic acid inhibits adipogenesis in 3T3-L1 adipocytes through LKB1/AMPK pathway. PloS One. (2013) 8:e70135. doi: 10.1371/journal.pone.0070135
152. Choi S, Gyamfi AA, Neequaye P, Addo S, Gonzalez FJ, and Gyamfi MA. Role of the pregnane X receptor in binge ethanol-induced steatosis and hepatotoxicity. J Pharmacol Exp Ther. (2018) 365:165–78. doi: 10.1124/jpet.117.244665
153. Zhang Y, Li X, Ciric B, Curtis MT, Chen W-J, Rostami A, et al. A dual effect of ursolic acid to the treatment of multiple sclerosis through both immunomodulation and direct remyelination. Proc Natl Acad Sci U S A. (2020) 117:9082–93. doi: 10.1073/pnas.2000208117
154. Farooqui N, Elhence A, and Shalimar. A current understanding of bile acids in chronic liver disease. J Clin Exp Hepatol. (2022) 12:155–73. doi: 10.1016/j.jceh.2021.08.017
155. Zhang M, Zhang S, Wu H, Xu L, Gao P, Gao Y, et al. Lactobacillus plantarum 1-2–3 inhibits ferroptosis by regulating dysregulated fatty acid metabolism to protect mice from high-fat diet-induced MAFLD. Free Radic Biol Med. (2025) 238:137–51. doi: 10.1016/j.freeradbiomed.2025.06.042
156. Xiao Y, Huang L, Zhao J, Chen W, and Lu W. The gut core microbial species Bifidobacterium longum: Colonization, mechanisms, and health benefits. Microbiol Res. (2025) 290:127966. doi: 10.1016/j.micres.2024.127966
157. Yu C, Liu H, Guo C, Chen Q, Su Y, Guo H, et al. Dextran sulfate-based MMP-2 enzyme-sensitive SR-A receptor targeting nanomicelles for the treatment of rheumatoid arthritis. Drug Deliv. (2022) 29:454–65. doi: 10.1080/10717544.2022.2032482
158. Wang X, Wang C, Liu W, Thakur A, Zhang K, Xu Z, et al. The roles of ultrasound-responsive nanomaterials in enhancing cancer immunotherapy. Pharmacol Res. (2025) 221:107975. doi: 10.1016/j.phrs.2025.107975
159. Nimer N, Choucair I, Wang Z, Nemet I, Li L, Gukasyan J, et al. Bile acids profile, histopathological indices and genetic variants for non-alcoholic fatty liver disease progression. Metabolism. (2021) 116:154457. doi: 10.1016/j.metabol.2020.154457
160. Wu Q, Hu Y, Wang C, Wei W, Gui L, Zeng W, et al. Reevaluate in vitro CYP3A index reactions of benzodiazepines and steroids between humans and dogs. Drug Metab Disposition. (2022) 50:741–9. doi: 10.1124/dmd.122.000864
161. Boucard A-S, Kulakauskas S, Alazzaz J, Chaouch S, Mammeri M, Millan-Oropeza A, et al. Isolation of derivatives from the food-grade probiotic Lactobacillus johnsonii CNCM I-4884 with enhanced anti-Giardia activity. Gut Microbes. (2025) 17:2474149. doi: 10.1080/19490976.2025.2474149
Keywords: pregnane X receptor, ligand-binding domain, microbiota-PXR axis, inflammatory modulation, chemotherapy resistant
Citation: Lin T, Chen Y, Liu L, Wu T, Qian Y and Jin B (2025) Recent advances in gut microbiota metabolite regulation of hepatic pregnane X receptor. Front. Immunol. 16:1692684. doi: 10.3389/fimmu.2025.1692684
Received: 26 August 2025; Accepted: 04 November 2025; Revised: 02 November 2025;
Published: 24 November 2025.
Edited by:
Jiong-Wei Wang, National University of Singapore, SingaporeReviewed by:
Lingjun Tong, Shandong First Medical University, ChinaDapeng Li, Sichuan University, China
Copyright © 2025 Lin, Chen, Liu, Wu, Qian and Jin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Baofen Jin, NDk3ODU2OTMzQHFxLmNvbQ==
 Tong Lin
Tong Lin Yang Chen
Yang Chen Linquan Liu3
Linquan Liu3