- 1Center for Immunity and Inflammation, Rutgers, The State University of New Jersey, Newark, NJ, United States
- 2Department of Medicine, Rutgers, The State University of New Jersey, Newark, NJ, United States
- 3NemaGen Discoveries, Princeton, NJ, United States
- 4Department of Pharmacology, Physiology, and Neuroscience, Rutgers. The State University of New Jersey, Newark, NJ, United States
- 5Department of Pediatrics, Rutgers, The State University of New Jersey, Newark, NJ, United States
- 6Department of Pathology, Immunology and Laboratory Medicine, Rutgers, The State University of New Jersey, Newark, NJ, United States
Carbonic anhydrase (Car) enzymes are a family of metalloenzymes that are traditionally known for their ability to regulate pH and CO2 homeostasis. However, emerging studies now demonstrate that Car family members exhibit lineage-specific expression patterns within immune cells. Moreover, it has been shown that genetically and pharmacologically targeting specific Car family members is sufficient to regulate immune cell development and activation. This work has identified Car enzymes as viable therapeutic targets that can influence immunity and inflammation. Here we contribute to this growing body of literature and demonstrate that Car8 is highly expressed by basophils and basophil precursor cells compared to other Car family members. While deletion of Car8 had no effect on basophil development or recruitment, mice deficient in Car8 were protected from basophil- and interleukin (IL)-4-dependent atopic dermatitis-like inflammation. Consistent with these findings, Car8-deficient basophils exhibit defects in the cytokine-stimulated release of IL-4 that is associated with altered calcium signaling pathways. Collectively, these studies reveal the lineage-specific expression patterns of Car8 and its unappreciated function in regulating basophil activation.
Introduction
Carbonic anhydrase (Car) enzymes are an ancient family of metalloenzymes that have independently evolved in prokaryotes, plants, and mammals (1). While Car enzymes are best known for their classical roles in catalyzing the reversible conversion of carbon dioxide (CO2) and water to bicarbonate (HCO3-) and protons, their convergent evolution across several distinct forms of life suggests they are highly important for the fundamental survival and functioning of an organism (2–5). A total of 16 Car enzyme isoforms have been identified in mammals, among which 13 isozymes (Car 1, 2, 3, 4, 5a, 5b, 6, 7, 9, 12, 13, 14, and 15) are catalytically active. The remaining three members of the family (Car8, Car10, and Car11) lack enzymatic activity due to the absence of the Zn2+ active site (6), suggesting their functions may differ from other family members.
Recent studies utilizing mouse models have begun to investigate the functions of Car enzymes and have defined their unique contributions in the context of epilepsy, cancer, glaucoma, and metabolic deficiencies (7–9). Further, emerging studies have also revealed that Car enzymes exhibit lineage-specific expression patterns and regulate immune cell development and activation (10–13). Included among these studies, we previously reported that Car1 is uniquely expressed by mast cell progenitors and can be targeted therapeutically to prevent mast cell responses and mast cell-mediated inflammation (10, 11). This work provoked the hypothesis that Car enzyme family members may also regulate the development and activation of other granulocyte populations.
Similar to mast cells, basophils are a rare granulocyte population that express the high affinity immunoglobulin E (IgE) receptor (FceRIa), secrete histamine, and are potent effector cells (14). Basophil populations rapidly expand and enter inflamed tissues in response to type 2 cytokine responses and play important roles in mediating immunity to parasites and promoting allergic inflammation and pruritic skin disease (15–19). Upon activation, basophils release large amounts of potent effector molecules, including interleukin (IL)-4 and IL-6, which can both initiate and amplify various forms of inflammation.
Here we investigated the expression pattern of Car enzymes in basophils and basophil progenitor cells. Our studies revealed that basophils and their progenitors express Car8, a metabolically inactive Car enzyme family member. While Car8 was not required for basophil development or recruitment, mice deficient in Car8 were protected from basophil-dependent atopic dermatitis (AD)-like disease. Furthermore, Car8-deficient basophils were found to have altered calcium signaling pathways that were associated with a defect in their release of IL-4 and IL-6 upon cytokine stimulation. These studies provide substantial insight into the factors that regulate basophil activation and identify previously unappreciated functions for Car8, a metabolically inactive member of the carbonic anhydrase family of enzymes.
Results
Our previous work revealed that the Car1 is a unique and identifying feature of mast cell progenitors and demonstrated that Car1 could be targeted without influencing basophil populations (10, 11). Given emerging data suggesting that Car enzyme family members have lineage-specific expression patterns within immune cells, we sought to determine whether specific Car family members were associated with basophils and/or their progenitors. To test this, we accessed publicly available single cell RNA-seq data that was generated from mast cell protease 8 (Mcpt8) green fluorescent protein (GFP) reporter mice, as Mcpt8 is a basophil-specific protease (20). Single cell RNA-seq was performed on Mcpt8-expressing cells sort-purified from the bone marrow and spleen (20). Marker gene analysis allowed us to identify four distinct basophil populations that were defined by their expression of Fcer1a and Mcpt8 (named Baso1, 2, 3, and 4) (Figures 1A-E). Additionally, a basophil progenitor population (Progenitor) was also identified by its shared expression of Fcer1a and Mcpt8 along with the progenitor cell markers Cd34 and Kit (Figures 1A-E). We next evaluated whether these distinct cell clusters expressed Car family members. Broad analysis of Car enzyme expression revealed that expression of only two family members were detected in the various cell populations (Supplementary Figure 1). Car2 was expressed by the Baso2 population and the basophil progenitor cells, while Car8 was expressed by basophil progenitor cells (Figure 1F, Supplementary Figure 1). To better evaluate the expression of Car2 and Car8 within mature basophils and without the possible limitations of the Mcpt8 reporter system, we sort-purified FcERIa+, CD49b+, c-Kit- bone marrow-derived basophils and performed RT-PCR analysis. Interestingly, when normalized to Gapdh, Car8 expression was found to be expressed more robustly than Car2 in mature bone marrow-derived basophils (Figures 1G, H). These data further contribute to a growing body of literature demonstrating the unique expression patterns of Car enzymes within immune cells. Moreover, the robust expression of Car8 by basophils suggests it may regulate their development and/or function. To gain a better understanding of Car8 expression patterns within a boarder set of immune cells, we sort-purified T cells, B cells, mast cells, basophils, alveolar macrophages, neutrophils, and monocytes and performed RT-PCR analysis. Interestingly, Car8 was found to be highly expressed by mast cell, splenic basophils, and neutrophils, but not the other immune cells evaluated (Figure 1I). Next, we sought to evaluate how Car8 protein levels correlated with our transcriptional analysis. To test this, we perform intracellular staining for Car8 protein. Consistent with transcriptional studies, Car8 protein was not detected in B cells, but was expressed by splenic mature basophils and neutrophils (Figures 1J, K). These data demonstrate that Car8 is expressed during basophil development and is present within mature basophils. Moreover, the unique expression of Car8 within granulocyte populations further provokes the hypothesis that it may regulate the development and/or activation of these highly specialized innate immune cell populations.
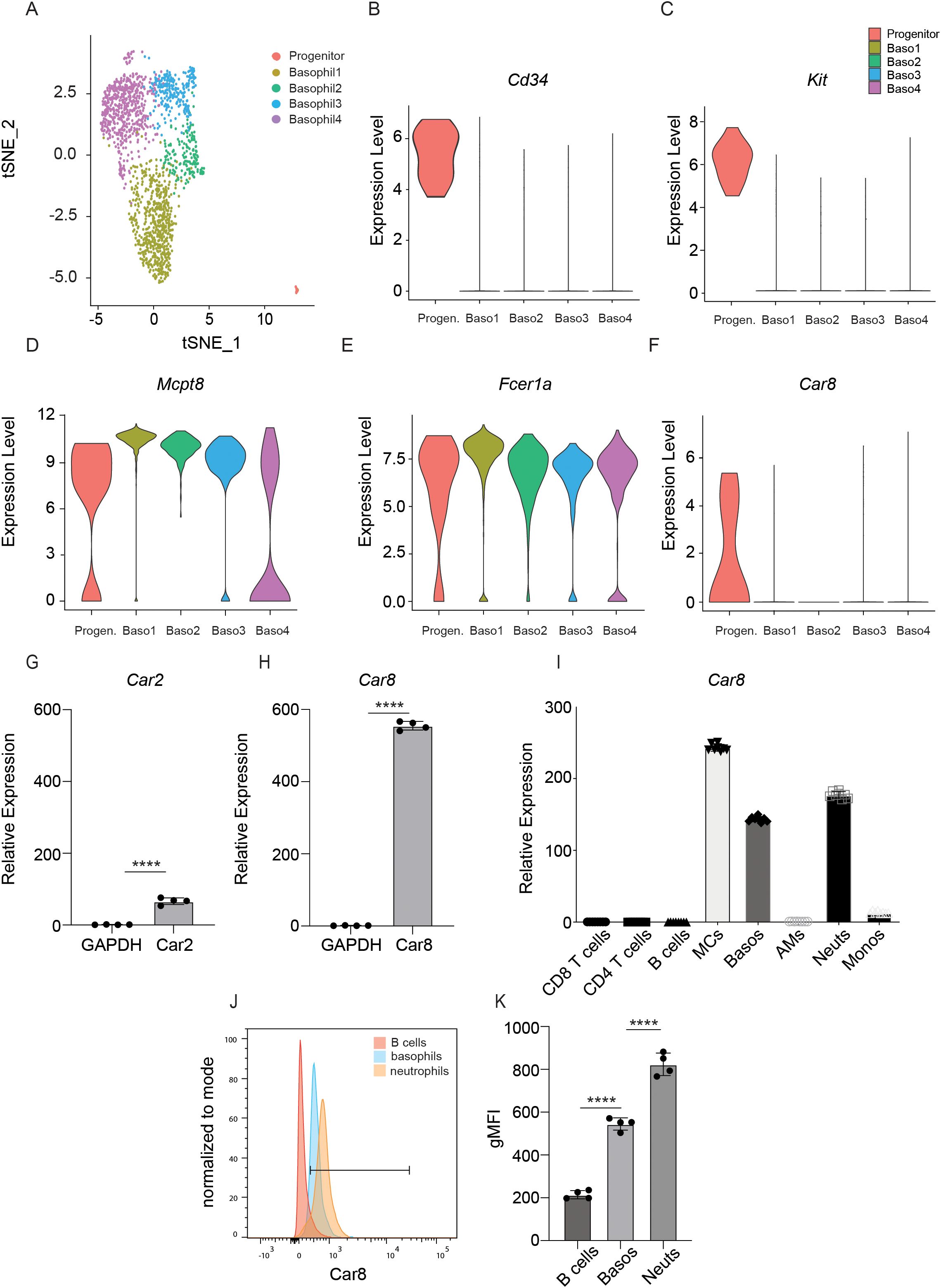
Figure 1. (A), Single cell RNA-seq of Mcpt8-expressing basophils and basophil progenitors was evaluated (GEO accession number: GSE206589). Expression of (B) CD34, (C) c-Kit, (D) Mcpt8, (E) Fcer1a, and (F) Car8 were evaluated in the 4 unique clusters identified by t-distributed stochastic neighbor embedding analysis. (G, H), RNA from sort-purified FceRIa+, CD49b+, c-Kit- bone marrow-derived basophils and other cell types were extracted and expressions of Car2 and Car8 in basophils were evaluated via RT-qPCR and normalized to their own Gapdh. (I) Expressions of Car8 in splenic basophils and other immune cell types were evaluated via RT-qPCR and normalized to alveolar macrophages Gapdh. (J, K), Intracellular staining of Car8 from the splenic cells of wild type (WT) mice, with geometric MFI quantified. (G–K), Individual experiments were repeated 3–5 times with 3–4 biological replicates per group. Statistical analysis between two indicated groups was performed using Student’s t-test. ****, p < 0.0001. Error bars represent SD.
In our studies, we chose to further investigate whether Car8 plays a role in regulating basophil responses. First, we sought to determine whether Car8 regulates basophil development by isolating bone marrow cells from the femurs of wild type (WT) and Car8-deficient mice and culturing them in the presence of IL-3. Compared to media-treated controls, WT mice showed significant increases in the percentage and total number of CD200R+, c-Kit- basophils and CD200R+, c-Kit+ mast cells post-culture (Figures 2A–D). Interestingly, Car8-deficient bone marrow cultures exhibited equivalent increases in basophils and mast cells following IL-3 treatment compared to cultures derived from WT mice (Figures 2A–D), suggesting that Car8 is not required for basophil development from bone marrow progenitor cells. Additionally, when CD200R3+, CD200R+, c-Kit- basophil populations were evaluated in the spleens of WT and Car8-deficient mice, there was no significant difference in percentage or total number (Figures 2E–G). These data further suggest that basophil populations develop normally in the absence of Car8.
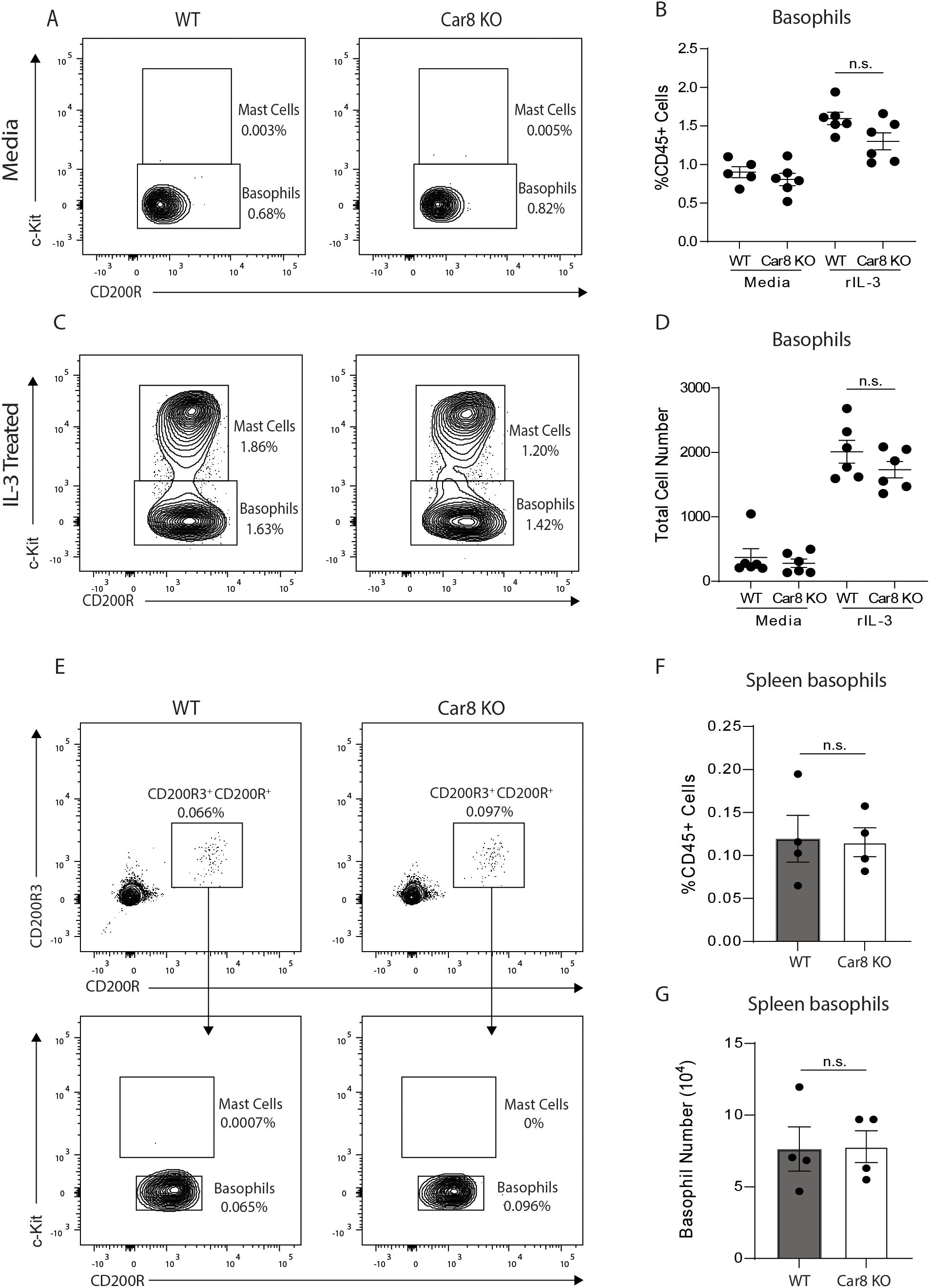
Figure 2. (A–D), Bone marrow-resident progenitor cells from WT or Car8-deficient mice were cultured with media or IL-3 and CD200R+ c-Kit+ mast cells and CD200R+, c-Kit- basophils were quantified. (E–G), CD200R3+, CD200R+, c-Kit- basophils and CD200R3+, CD200R+, c-Kit+ mast cells were quantified from the spleens of WT or Car8-deficient mice at baseline. (A–G) Individual experiments were repeated 3–5 times with at least 3 biological replicates per group. Statistical analysis was between two indicated groups was performed using Student’s t-test. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; n.s., not significant. Error bars represent SD.
The studies presented above suggest that Car8 is expressed during basophil development but is not required for their lineage commitment. This provoked that hypothesis that Car8 may be required for proper basophil migration and/or activation. To better evaluate this, we employed a basophil-dependent model of atopic dermatitis (AD)-like disease (21). In this model, topical treatment of skin with the irritant MC903, a low-calcemic analog of vitamin D3, results in production of cytokine alarmins by keratinocytes and the rapid recruitment of basophils to the skin. These skin-resident basophils produce robust amounts of IL-4 and cooperate with group 2 innate lymphoid cells (ILC2s) to promote skin inflammation (21–23). Critically, targeting basophils or IL-4 following MC903 treatment results in significantly reduced skin inflammation, thereby providing an ideal system to evaluate basophil responses in vivo (21). As expected, treatment of WT mice with MC903 resulted in marked skin inflammation as indicated by significantly increased ear skin thickness compared to vehicle-treated controls (Figure 3A). In contrast, Car8-deficient mice were protected from MC903-induced inflammation and exhibited reduced skin thickness compared to controls (Figure 3A). Histological analysis of skin sections revealed that MC903-treated WT mice exhibited substantially increased epidermal hyperplasia and cellularity of the dermis, as indicated by significantly increased numbers of skin-resident CD45+ cells (Supplementary Figure 2A, B). In contrast, Car8-deficient mice treated with MC903 exhibited substantially less epidermal hyperplasia and significantly reduced cellularity (Supplementary Figure 2A, B). These data provoked the hypothesis that basophil responses might be reduced in mice lacking expression of Car8. To better evaluate this, we isolated skin-resident cells and performed flow cytometric analysis to identify basophils. Consistent with published studies, MC903 treatment of WT mice resulted in significant increases of FceRIa+, CD200R3+, CD200R+, c-Kit- skin-resident basophils populations by both percentage and total number (Figures 3B, C). Interestingly, significantly increased percentages and total numbers of basophils were also observed in the skin of Car8-deficient mice treated with MC903 (Figures 3B, C). Additional transcriptional analysis revealed that the WT and Car8-deficient animals also exhibited equivalent increases in the expression of the basophil-promoting cytokine alarmin Tslp, the basophil-specific protease Mcpt8, and the basophil-derived effector molecules Il4 and Il6 (Figures 3D–G). These studies demonstrate that Car8-deficient mice have intact MC903-induced basophil responses.
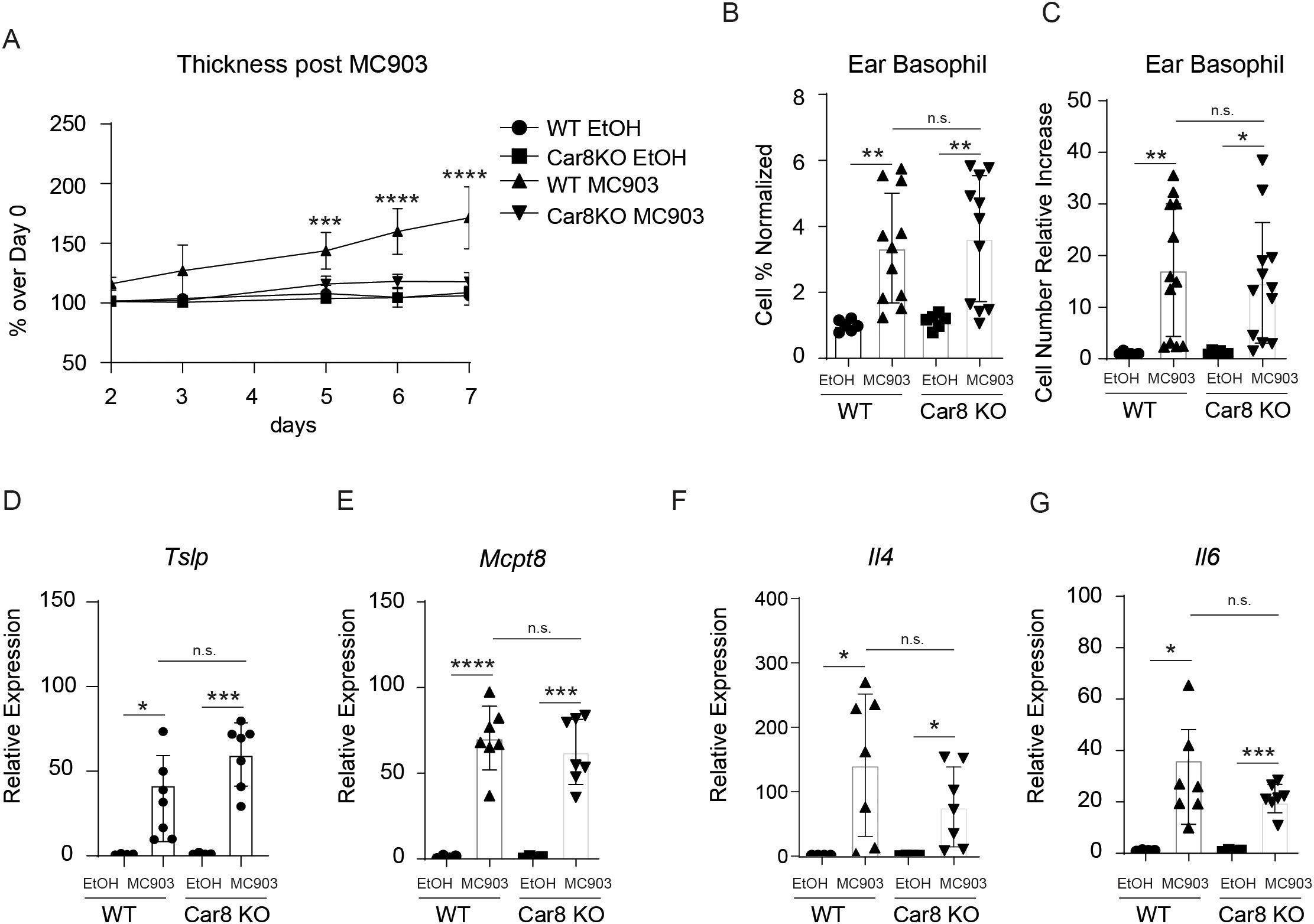
Figure 3. The ears of WT or Car8-deficient mice were treated with ethanol (EtOH) or MC903 and (A) ear thickness was measured. (B, C), Following treatment with ethanol or MC903 cells were isolated from the ears and basophil populations were quantified. Expression levels of (D) Tslp, (E) Mcpt8, (F) Il4 and (G) Il6 were evaluated in the ears of mice following treatment with EtOH or MC903. (A–G) Individual experiments were repeated 5 times with at least 3 biological replicates per group. Statistical analysis between two indicated groups was performed using Student’s t-test. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; n.s., not significant. Error bars represent SD.
The data presented above demonstrate that Car8-deficient mice have significantly reduced skin inflammation in a model of basophil-dependent AD-like disease despite having equivalent levels of basophils migrating to the skin. These data provoked the hypothesis that Car8-deficient basophils may have altered effector functions. To evaluate this, we injected WT and Car8-deficient mice with IL-3-anti-IL-3 complexes (IL-3C), a treatment that promotes systemic basophilia, and sort-purified splenic basophils (Figure 4A). We first evaluated whether WT and Car8-deficient basophils exhibited equivalent transcriptional levels of Il4 and Il6. Consistent with the presence of Il4 and Il6 expressions in the skin of both WT and Car8-deficient mice post-MC903 treatment, no significant differences in Il4 or Il6 transcripts were observed between the two groups (Figures 4B, C). We then crossed the IL-4-eGFP transcriptional reporter mouse (4get mice) onto a Car8-deficent background (24). Corroborating our transcriptional studies, WT and Car8-deficient basophils exhibited equivalent levels of IL-4-eGFP by flow cytometric analysis (Figure 4D). Given that MC903-induced inflammation is highly dependent on IL-4 production by basophils, we sought to determine if the release of IL-4 protein differed in the absence of Car8. To test this, we sort-purified equal numbers of WT and Car8-deficient basophils, loaded them with ovalbumin (OVA)-specific IgE, and stimulated them with OVA or bovine serum albumin (BSA) as a negative control. As expected, IL-4 and IL-6 levels were found at higher levels in the cell-free supernatants of WT basophils stimulated with IgE and OVA compared to controls (Figures 4E, F). Interestingly, culture supernatants from Car8-deficient basophils stimulated with IgE and OVA exhibited equivalent levels of IL-4 and IL-6 to those of WT controls. These data demonstrate the WT and Car8-deficient basophils respond similarly following IgE-mediated activation.
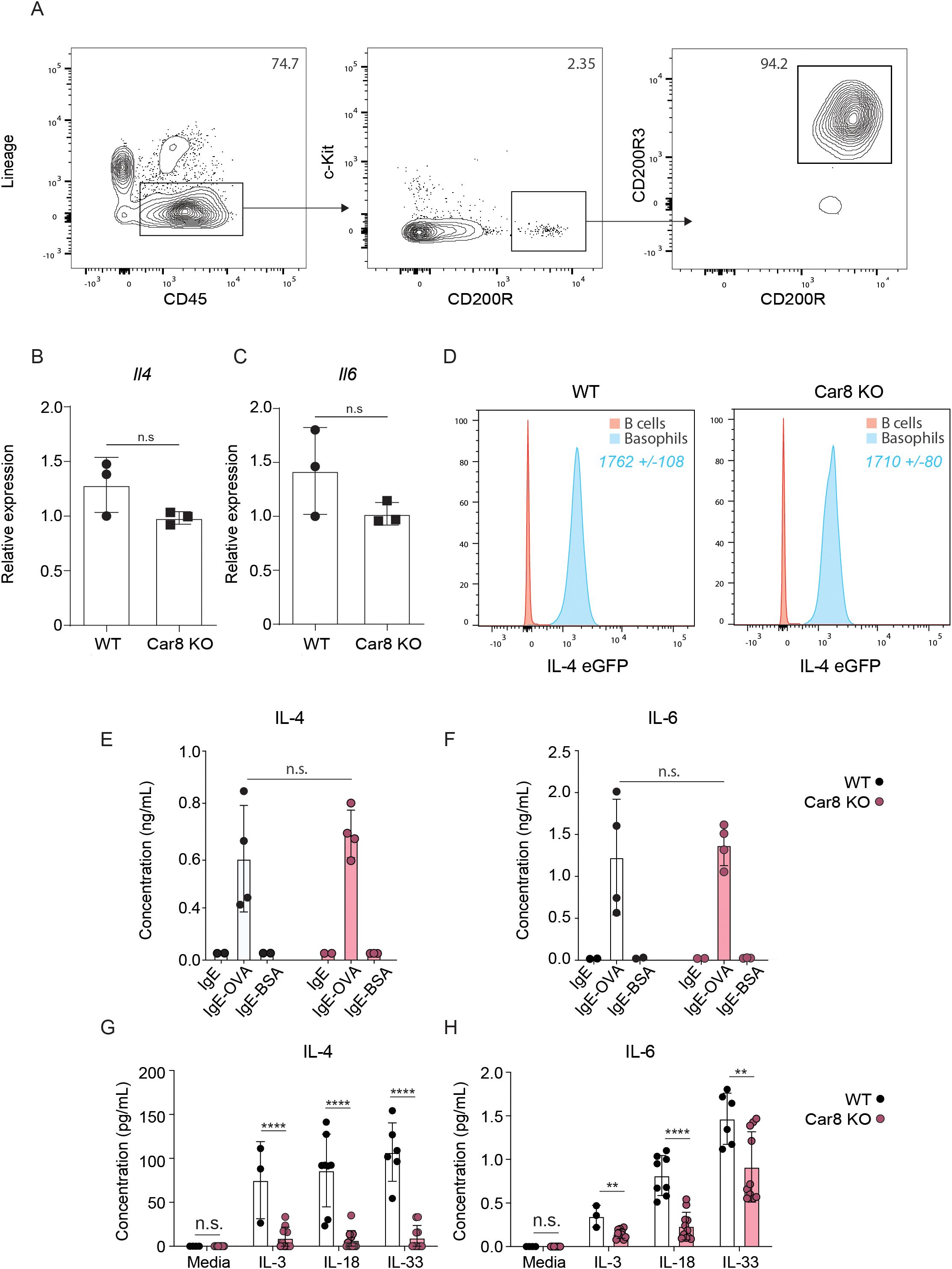
Figure 4. (A) Lin-, CD45+, c-Kit-, CD200R+, CD200R3+ basophils were sort-purified from the spleens of WT or Car8-deficient mice treated with IL-3-anti-IL-3 complexes (IL-3C). Expressions of (B) Il4 and (C) Il6 from sort-purified splenic basophils were evaluated via RT-qPCR and normalized to IL-3C stimulated WT-derived basophils. (D) IL-4 eGFP expressions in Lin-, CD45+, c-Kit- CD200R+, CD200R3+ splenic basophils were isolated and evaluated from IL-4Get Car8-sufficient and IL-4Get Car8-deficient mice. (E, F) Lin-, CD45+, c-Kit-, CD200R+, CD200R3+ basophils were sort-purified from the spleens of WT or Car8-deficient mice, and equal numbers of each group were treated with OVA-specific IgE and followed by BSA or OVA stimulation overnight. IL-4 and IL-6 protein levels were evaluated in cell-free supernatants post-culture by ELISA. (G, H), Lin-, CD45+, c-Kit- CD200R+, CD200R3+ basophils were sort-purified from the spleens of WT or Car8-deficient mice treated with IL-3-anti-IL-3 complexes and cultured in the presence of media, IL-3, IL-18, or IL-33. IL-4 and IL-6 protein levels were evaluated in cell-free supernatants post-culture by ELISA. (A–H) Individual experiments were repeated 3–5 times with at least 3 biological replicates per group. Statistical analysis between two indicated groups was performed using Student’s t-test. **, p < 0.01; ****, p < 0.0001; n.s., not significant. Error bars represent SD.
Studies have demonstrated that the IgE-mediated activation of basophils results in a rapid activation pathway that is associated with degranulation (25). In contrast, the activation of basophils following cytokine stimulation is reported to be more gradual and involves distinct singling pathways (26). Given that the MC903 model of AD-like diseases is not antigen driven and highly-dependent on cytokine alarmins (21, 23), we next evaluated whether WT and Car8-deficient basophils differ in their activation following cytokine stimulation. To test this, basophils were sort-purified from WT or Car8-deficient mice following IL-3C treatment and cultured in the presence or absence of the basophil activating cytokines IL-3, IL-18, or IL-33. Following cytokine treatment, IL-4 and IL-6 were measured in cell-free supernatants by ELISA. As expected, supernatants from WT basophils stimulated with IL-3, IL-18, or IL-33 exhibited significantly increased levels of IL-4 and IL-6 compared to media treated controls (Figures 4G, H). In contrast, Car8-deficient basophils stimulated with the same cytokines showed significantly reduced levels of IL-4 and IL-6 releases into the cell-free supernatants. These data strongly suggest that Car8 is required for optimal cytokine production by basophils following cytokine stimulation (Figures 4G, H).
The data presented above suggest that Car8 regulates the release of IL-4 and IL-6 from basophils in response to cytokine-mediated activation. It is well established that the release of cytokines by activated basophils is mediated by changes in cytoplasmic calcium levels (27, 28). A primary calcium regulator in this process is the calcium release-activated calcium (CRAC) channel that opens to allow calcium into a cell once calcium stores have been depleted (29). Upon cellular activation, inositol 1,4,5 trisphosphate (IP3) engages the inositol 1,4,5 trisphosphate receptor (IP3R) to promote the release of calcium from the endoplasmic reticulum (ER) as the intracellular source to increase cytoplasmic calcium level (30, 31). Previous studies have shown that the stromal interaction molecules (Stim) 1 and 2 (calcium sensors that reside in the endoplasmic reticulum) will then activate and operate with Orai1 and 2 (components of the CRAC channel at the cell membrane) to allow calcium to enter the cell and thereby restore calcium levels (26, 32–35). More recent studies have shown that Car8 acts as an allosteric inhibitor of IP3R, and Stim1 and Stim2 are differentially required for basophil responses (26, 36–38). Therefore, we first sought to determine whether these pathways are altered in the context of MC903-induced AD-like disease. To do this, we analyzed publically available RNA-seq data performed on control and MC903 inflamed skin (39). While Stim1 expression was significantly reduced following MC903 treatment, Stim2, Orai1 and Orai2 were all significantly increased in response to MC903 (Figures 5A–D). These data demonstrate that these pathways are altered in the context of AD-like disease. Next, we sought to determine if these molecules were dysregulated in the absence of Car8. Interestingly, Stim1 and Sim2 expressions were significantly increased in the absence of Car8 at baseline (Figures 5E, F). Stim1 expression post-MC903 treatment was similarly decreased in both WT and Car8-deficient basophils. However, Stim2 expression was not significantly increased in Car8-deficient mice following MC903 treatment unlike in treated WT mice (Figures 5E, F). Moreover, Orai1 and Orai2 expressions were also significantly increased post-MC903 treatment in Car8-deficient mice compared to WT controls (Figures 5G, H). Collectively, these findings suggest that a lack of Car8 may lead to altered STIM and ORAI pathways and dysregulated calcium gradient. While these data provoke the hypothesis that Car8 may regulate calcium response in basophils, the changes observed in this global knockout model may not be basophil-specific. Therefore, to directly evaluate whether Car8 regulates calcium responses in basophils, we sort-purified basophils from WT and Car8-deficient mice and performed calcium imaging on the cells. Consistent with dysregulated calcium gradient, Car8-deficient basophils exhibited higher baseline calcium levels than WT control basophils (Figure 5I). Critically, Yoshikawa et al. showed that production of IL-4 from WT bone marrow-derived basophils could be completely inhibited by extracellular or intracellular Ca2+ chelators, suggesting that properly maintaining the Ca2+ gradient at baseline is required for IL-4 production (26). Similarly, these studies suggest that Car8 is required for basophils to maintain calcium homeostasis and gradient levels that are required for basophils to release IL-4 and contribute to IL-4-mediated inflammation.
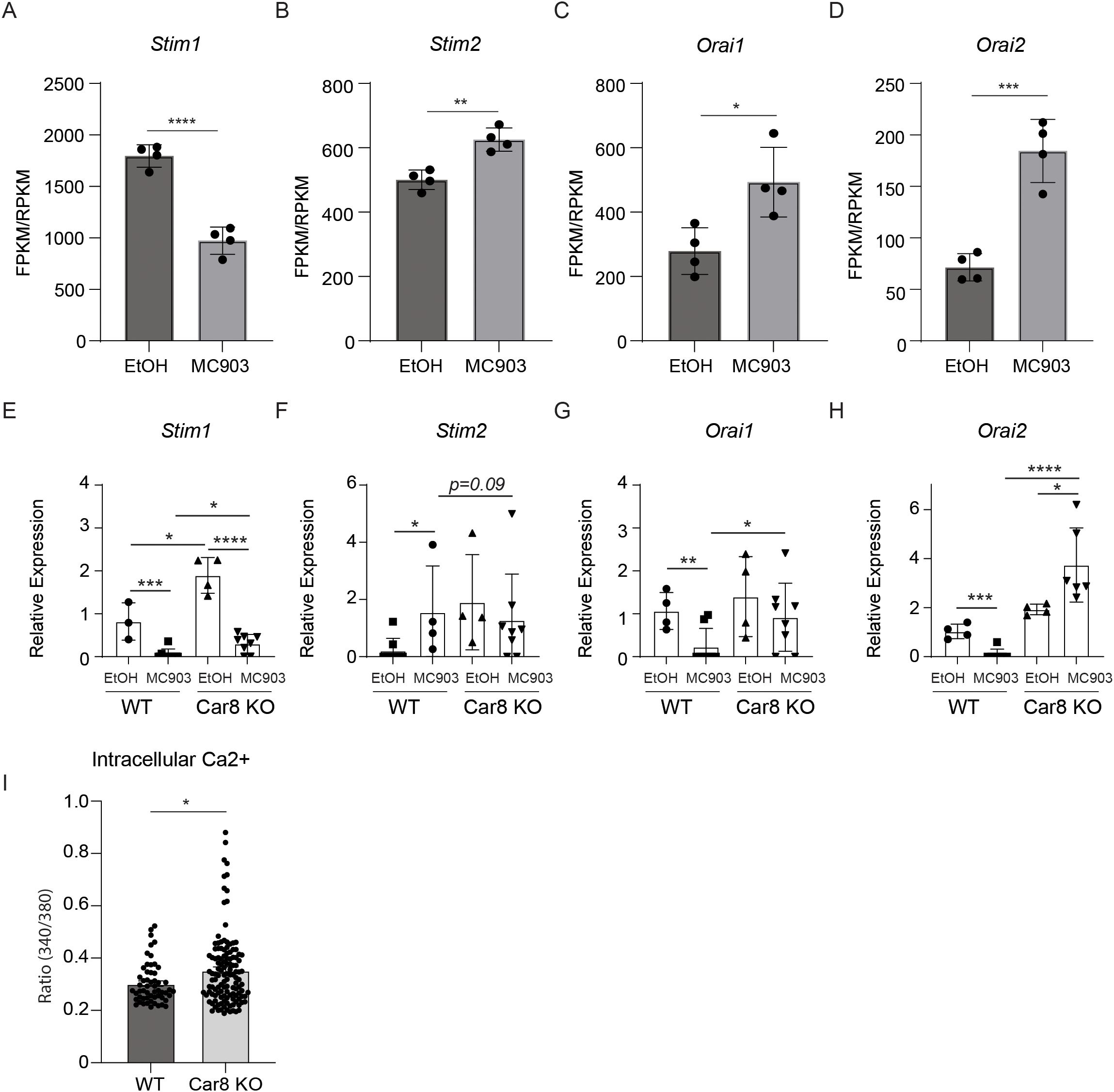
Figure 5. The ears of WT mice were treated with ethanol (EtOH) or MC903 and RNA-seq was performed. (A-D), Expression levels ofthe calcium-associated molecules Stim1, Stim2, Orai1, and Orai2 were evaluated (fragments per kilobase million (FPKM)/reads per kilobase million (RPKM)). (E-H), The ears of WT or Car8-deficient mice were treated with EtOH or MC903 and Stim1, Stim2, Orai1, and Orai2 were evaluated by RT-PCR. (I), Lin-, CD45+, c-Kit-, CD200R+, CD200R3+ basophils were sort-purified from the spleens of WT or Car8-deficient mice treated with IL-3-anti-IL-3 complexes and Ca2+ imaging measurements were performed. (A-H) Individual experiments were repeated 3–5 times with at least 3 biological replicates per group. Statistical analysis between two indicated groups was performed using Student’s t-test. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. Error bars represent SD.
Discussion
Carbonic anhydrase enzymes represent a prime example of convergent evolution via their independent development in various classes of life (5). Interestingly, our work and that of others are beginning to reveal that the alpha class of isozymes found in mammals exhibit extremely distinct expression patterns in various tissues and immune cells (10, 11). Moreover, emerging studies are also defining the unique roles these specific isozymes play in the context of both immune cell development and function, indicating that they may possess important therapeutic potential. Among these isozymes, Car8 represents one of only three family members that are known to be metabolically inactive, suggesting it has evolved functions beyond the ability to regulate pH and CO2 homeostasis.
Here we demonstrate that basophil precursor cells uniquely express high levels of Car8 compared with other Car enzyme family members. While rare in number compared to other leukocyte populations, basophils are potent effector cells that play established roles in the context of parasitic immunity, allergic inflammation, and pruritic disease (16, 40–42). Additionally, emerging studies have also revealed the important contributions of basophils to lung development, kidney fibrosis, auto immune disorders, and even anti-tumor immunity (16, 42–45). The diverse functions of basophils are perhaps not surprising given their ability to rapidly migrate to tissues and activate in response to numerous signals including IgE crosslinking, protease actively, and cytokine stimulation (16, 41, 42, 45). Following exposure to these stimuli, basophils produce a vast array of effector cytokines including robust levels of IL-4 and IL-6 that are known to mediate many of their potent effects.
While the data presented here show that Car8 is not required for basophil development or migration, our work reveals that Car8 is needed for their optimal release of basophil-derived IL-4 and IL-6 following cytokine stimulation. Consistent with defects in IL-4 production, Car8-deficient animals were protected from basophil- and IL-4-dependent AD-like disease. To the best of our knowledge these studies are the first to demonstrate the expression and function of Car8 in basophils and its ability to regulate basophil-dependent inflammation. Further, these studies contribute to an increasing body of literature illustrating the importance of Car enzyme family members in regulating immune responses.
It is well established that Car8 is highly expressed by dorsal root ganglion neurons where it operates as an inhibitor of inositol trisphosphate receptor-1 (ITPR1) and regulates intracellular calcium levels (36, 46). Given the known role that intracellular calcium plays in regulating the release of IL-4 by basophils, we hypothesized that Car8-deficient basophils may exhibit dysregulated calcium responses. Consistent with this hypothesis Car8-deficient basophils showed significantly increased intracellular calcium levels compared to controls. Previous work in other cell types has demonstrated that sustained elevations in intracellular calcium can suppress signaling pathways and make cells less responsive to additional stimulation and thereby inhibit the optimal release of effector molecules (47–50). The work presented here is consistent with those reports and suggests that the tightly regulated calcium levels are required for basophils to properly respond to cytokine stimulation. While the data here identify Car8 as an important regulator of these processes, additional work is required to determine the exact calcium channels and/or pumps that are directly or indirectly affected by Car8. It is also possible that Car8 may regulate post-translational modifications (PTMs) targeting these calcium regulators, as a substantial body of literature over the past 15 years has demonstrated that these critical molecules and channels are regulated through PTMs in various cell types, including immune cells (51–53). Studies of this nature are especially important given that Car8 does not appear to affect the antibody-mediated activation of basophils which is known to operate via distinct calcium signaling pathways than those initiated following cytokine-mediated activation (26). Moreover, it is worth investigating whether Car8 similarly regulates calcium responses and activation in mast cells and neutrophils, which also express high levels of Car8. Given the specific expression of Car8 within granulocyte lineages, it is possible that this would be a conserved mechanism of activation across these cell types. It is also important to highlight that deficient activation of other Car8-expressing granulocytes may contribute to the dramatic phenotype observed in the MC903 model, given that the Car8 mouse model is not basophil-specific. Additionally, future work is also needed to determine whether the expression of Car8 in granulocytes is conserved in humans to further evaluate its therapeutic potential.
In summary, these studies reveal the unique expression of Car8 by basophil precursor cells and highlights its previously unappreciated ability to regulate important aspects of basophil activation. To the best of our knowledge these studies are the first to investigate the expression of Car8 within immune cells and add to an emerging body of literature identifying the unique expression patterns of Car family members within hematopoietic lineages. Moreover, this work suggests that inactive Car isozymes have also evolved the ability to regulate immunity and inflammation and as such may represent viable therapeutic targets.
Materials and methods
Mouse experiments
Eight to ten-week-old female and male wild type C57BL/6J, C57BLKS/J, IL-4/GFP-enhanced transcript (4Get) and Car8 knockout mice were purchased from Jackson or generously provided by Dr. Roy Levitt at the University of Miami Miller School of Medicine (36). Then breeding colonies were established. All mice were maintained in specific pathogen-free facilities at the Rutgers New Jersey Medical School. All protocols were approved by the Rutgers Institutional Animal Care and Use Committee, PROTO202000017.
Mice were treated once a day topically with 2 nmol of MC903 (calcipotriol; Tocris Bioscience) in 20 µl ethanol for 7 days as described previously (21).
Flow cytometry, intracellular staining & cell sorting
Cells were stained with monoclonal anti-mouse fluorescently conjugated antibodies: CD45R/B220 (clone RA3-6B2); CD3 (clone 145-2C11); CD11b (clone MI/70); CD11c (clone N418); IgE (clone 23G3); FcERIa (clone MAR-1);CD49b (clone DX5); CD45 (clone 30-F11); CD64 (clone X54-5/7.1); CD90 (clone 5E10); CD127 (clone A7R34); CD200R1 (clone OX110); CD200R3 (clone Ba13); EpCAM (clone G8.8); F4/80 (clone BM8); γδTCR (clone eBioGL3); Siglec-F (clone E50-2440); Ly6G (clone 1A8); Ly6C (clone AL-21); IL-5 (clone TRFK5); IL-13 (clone eBio13A); podoplanin (clone 8.1.1); and TER-119 (clone TER-119) from eBioscience (Thermo Fisher Scientific) or BD Biosciences. Basophils were gated and analyzed as CD45+CD3−CD19−NK1.1−Ly6G−Siglec-F− CD49b+CD200R1+ CD200R3+FcERIa+.
Post-surface staining, cells undergoing intracellular staining were placed in fixation buffer (eBioscience) for 10 mins, and then permeabilized in permeabilization buffer (eBioscience) for 1 hour. Cells were incubated with Anti-Mouse Car8, Biotin (Abcam) for 1 hour, and then with secondary antibody attached to Streptavidin for 1 hour.
Mouse primary cell culture & treatment
Basophils were isolated and sort-purified from bone marrow. Equal numbers of WT and Car8-deficient basophils were then cultured in the presence or absence of the basophil activating cytokines IL-3, IL-18, IL-33; or culture supernatant containing OVA-specific IgE secreted by the hybridoma TOϵ (54) (kindly provided by Dr. Paul Bryce) and then OVA (Sigma Cat.# O1641) and IL-4 and IL-6 were measured in cell-free supernatants by ELISA. ELISA was performed to measure IL-4 (clones 11B11 and BVD6-24G2) and IL-6 (MP5-32C11) levels in the cell-free supernatants.
Basophil sort & calcium signaling
WT mice were injected intravenously with a combination of 1 µg mouse recombinant IL-3 (cat. no. 403-ML; R&D Systems) and 0.5 μg of anti-IL-3 antibody (clone MP2-8F8; BioLegend) in 200 μl of PBS 3 days before euthanasia as described previously (55, 56). At necropsy, single-cell suspensions of spleens were prepared, and basophil populations were sort-purified as described earlier using a FACSAria II (BD Biosciences). Purity for all cell populations was 98% or greater.
Ca2+ imaging measurements were performed with an Olympus IX-51 inverted microscope equipped with a DeltaRAM excitation light source (Photon Technology International, PTI), as described earlier (57). 5x10^5 ~1x10^6 cells were loaded with 1 mM fura-2 AM (Invitrogen) for 40 min before the measurement at 37˚C, and dual-excitation images at 340 and 380 nm excitation wavelengths were detected at 510 nm with a Roper Cool-Snap digital CCD camera. Measurements were conducted in the Mg2+ free HBSS, supplemented with 2 mM CaCl2. Data analysis was performed using the Image Master software (PTI).
Quantitative PCR
Total tissue RNA was extracted with Trizol reagent (Invitrogen, San Diego) followed by phenol/chloroform extraction and isopropanol precipitation. RNA from sorted cells was extracted using the RNeasyMinikit (Qiagen) following the manufacturer’s instructions. Complementary DNA was generated per standard protocol with SuperScript Reverse Transcriptase II (cat. no. 18064014; Invitrogen) and used as input for reverse transcription PCR. Reverse transcription PCR data were analyzed using the 2-ΔΔct method with the SYBR Green Chemistry (Thermo Fisher I Scientific), with β-actin (Actb) serving as the endogenous housekeeping gene. All reactions were run on an ABI 7500 Fast Real-Time PCR System (Applied Biosystems). Samples were normalized to vehicle treated controls. The following QuantiTect primer assays from QIAGEN were used: IL-4 (QT00160678); IL-5 (QT00099715); IL-13 (QT00099554); Mcpt8 (QT00131565); TSLP (QT00198261); STIM1 (QT00105119); STIM2 (QT00289009); Orai1 (QT00285775); Orai2 (QT00304738).
Statistics
Results are shown as the mean ± s.d. P < 0.05 was considered as significantly different. Statistical analysis between two indicated groups was performed using Student’s t-tests in Prism 8 (GraphPad Software).
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was approved by Rutgers University, Office of Research, Institutional Animal Care and Use Committee. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
JP: Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – review & editing. CBS: Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – review & editing. SZ: Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – review & editing. AS: Data curation, Formal analysis, Investigation, Software, Validation, Visualization, Writing – review & editing. VE: Data curation, Formal analysis, Investigation, Validation, Visualization, Writing – review & editing. MNS: Formal analysis, Investigation, Validation, Visualization, Writing – review & editing. JJP: Formal analysis, Investigation, Validation, Visualization, Writing – review & editing. KC: Formal analysis, Investigation, Validation, Visualization, Writing – review & editing. ADL: Formal analysis, Investigation, Validation, Visualization, Writing – review & editing. AR: Writing – review & editing. TR: Writing – review & editing. MCS: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article: R01AI123224, R01AI151599, R01AI131634, R01AI180644, R01AI169770 and R21AI171064. CBS was also supported by T32AI125185. JJP was supported by T32AI125185. VE and AR were supported by R01AI169769-01.
Conflict of interest
JP and JJP are now employees of NemaGen Discoveries and perform activities unrelated to the submitted work. MCS is the founder and president of NemaGen Discoveries. MCS does not receive financial compensation from NemaGen and performs activities unrelated to the submitted work.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author MCS declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1694105/full#supplementary-material
Supplementary Figure 1 | Single cell RNA-seq of Mcpt8-expressing basophils and basophil progenitors was evaluated (GEO accession number: GSE206589). Expressions of Car enzyme family members were evaluated in the 4 unique basophil clusters.
Supplementary Figure 2 | Histopathology/Representative hematoxylin and eosin staining (scale bar 50μm; original magnification, 20X) of ears of EtOH or MC903 treated WT and Car8-deficient mice. CD45+ immune infiltrates were evaluated and quantified. Individual experiments were repeated 3–5 times with at least 3 biological replicates per group. Statistical analysis between two indicated groups was performed using Student’s t-test. **, p < 0.01. Error bars represent SD.
References
1. Jonsson BH and Liljas A. Perspectives on the classical enzyme carbonic anhydrase and the search for inhibitors. Biophys J. (2020) 119:1275–80. doi: 10.1016/j.bpj.2020.08.020
2. Sly WS and Hu PY. Human carbonic anhydrases and carbonic anhydrase deficiencies. Annu Rev Biochem. (1995) 64:375–401. doi: 10.1146/annurev.bi.64.070195.002111
3. Supuran CT. Carbonic anhydrase versatility: from pH regulation to CO(2) sensing and metabolism. Front Mol Biosci. (2023) 10:1326633. doi: 10.3389/fmolb.2023.1326633
4. Shah GN, Hewett-Emmett D, Grubb JH, Migas MC, Fleming RE, Waheed A, et al. Mitochondrial carbonic anhydrase CA VB: differences in tissue distribution and pattern of evolution from those of CA VA suggest distinct physiological roles. Proc Natl Acad Sci U.S.A. (2000) 97:1677–82. doi: 10.1073/pnas.97.4.1677
5. Liljas A and Laurberg M. A wheel invented three times. The molecular structures of the three carbonic anhydrases. EMBO Rep. (2000) 1:16–7. doi: 10.1093/embo-reports/kvd016
6. Aggarwal M, Boone CD, Kondeti B, and McKenna R. Structural annotation of human carbonic anhydrases. J Enzyme Inhib Med Chem. (2013) 28:267–77. doi: 10.3109/14756366.2012.737323
7. Diez-Fernandez C, Rufenacht V, Santra S, Lund AM, Santer R, Lindner M, et al. Defective hepatic bicarbonate production due to carbonic anhydrase VA deficiency leads to early-onset life-threatening metabolic crisis. Genet Med. (2016) 18:991–1000. doi: 10.1038/gim.2015.201
8. Monti DM, De Simone G, Langella E, Supuran CT, Di Fiore A, and Monti SM. Insights into the role of reactive sulfhydryl groups of Carbonic Anhydrase III and VII during oxidative damage. J Enzyme Inhib Med Chem. (2017) 32:5–12. doi: 10.1080/14756366.2016.1225046
9. Thiry A, Dogne JM, Supuran CT, and Masereel B. Carbonic anhydrase inhibitors as anticonvulsant agents. Curr Top Med Chem. (2007) 7:855–64. doi: 10.2174/156802607780636726
10. Henry EK, Sy CB, Inclan-Rico JM, Espinosa V, Ghanny SS, Dwyer DF, et al. Carbonic anhydrase enzymes regulate mast cell-mediated inflammation. J Exp Med. (2016) 213:1663–73. doi: 10.1084/jem.20151739
11. Inclan-Rico JM, Hernandez CM, Henry EK, Federman HG, Sy CB, Ponessa JJ, et al. Trichinella spiralis-induced mastocytosis and erythropoiesis are simultaneously supported by a bipotent mast cell/erythrocyte precursor cell. PloS Pathog. (2020) 16:e1008579. doi: 10.1371/journal.ppat.1008579
12. Xu J, Xu X, Wang B, Ma Y, Zhang L, Xu H, et al. Nuclear carbonic anhydrase 6B associates with PRMT5 to epigenetically promote IL-12 expression in innate response. Proc Natl Acad Sci U.S.A. (2017) 114:8620–5. doi: 10.1073/pnas.1700917114
13. Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. (2014) 159:1312–26. doi: 10.1016/j.cell.2014.11.018
14. Hellman L, Akula S, Fu Z, and Wernersson S. Mast cell and basophil granule proteases - in vivo targets and function. Front Immunol. (2022) 13:918305. doi: 10.3389/fimmu.2022.918305
15. Karasuyama H, Miyake K, Yoshikawa S, and Yamanishi Y. Multifaceted roles of basophils in health and disease. J Allergy Clin Immunol. (2018) 142:370–80. doi: 10.1016/j.jaci.2017.10.042
16. Miyake K, Ito J, and Karasuyama H. Role of basophils in a broad spectrum of disorders. Front Immunol. (2022) 13:902494. doi: 10.3389/fimmu.2022.902494
17. Miyake K and Karasuyama H. Emerging roles of basophils in allergic inflammation. Allergol Int. (2017) 66:382–91. doi: 10.1016/j.alit.2017.04.007
18. Olivera A and Rivera J. Paradigm shifts in mast cell and basophil biology and function: an emerging view of immune regulation in health and disease. Methods Mol Biol. (2020) 2163:3–31. doi: 10.1007/978-1-0716-0696-4_1
19. Wiebe D, Limberg MM, Gray N, and Raap U. Basophils in pruritic skin diseases. Front Immunol. (2023) 14:1213138. doi: 10.3389/fimmu.2023.1213138
20. Miyake K, Ito J, Nakabayashi J, Shichino S, Ishiwata K, and Karasuyama H. Single cell transcriptomics clarifies the basophil differentiation trajectory and identifies pre-basophils upstream of mature basophils. Nat Commun. (2023) 14:2694. doi: 10.1038/s41467-023-38356-1
21. Kim BS, Wang K, Siracusa MC, Saenz SA, Brestoff JR, Monticelli LA, et al. Basophils promote innate lymphoid cell responses in inflamed skin. J Immunol. (2014) 193:3717–25. doi: 10.4049/jimmunol.1401307
22. Siracusa MC, Saenz SA, Hill DA, Kim BS, Headley MB, Doering TA, et al. TSLP promotes interleukin-3-independent basophil haematopoiesis and type 2 inflammation. Nature. (2011) 477:229–33. doi: 10.1038/nature10329
23. Pellefigues C, Naidoo K, Mehta P, Schmidt AJ, Jagot F, Roussel E, et al. Basophils promote barrier dysfunction and resolution in the atopic skin. J Allergy Clin Immunol. (2021) 148:799–812.e10. doi: 10.1016/j.jaci.2021.02.018
24. Mohrs M, Shinkai K, Mohrs K, and Locksley RM. Analysis of type 2 immunity in vivo with a bicistronic IL-4 reporter. Immunity. (2001) 15:303–11. doi: 10.1016/S1074-7613(01)00186-8
25. Charles N and Blank U. IgE-mediated activation of mast cells and basophils in health and disease. Immunol Rev. (2025) 331:e70024. doi: 10.1111/imr.70024
26. Yoshikawa S, Oh-Hora M, Hashimoto R, Nagao T, Peters L, Egawa M, et al. Pivotal role of STIM2, but not STIM1, in IL-4 production by IL-3-stimulated murine basophils. Sci Signal. (2019) 12(576). doi: 10.1126/scisignal.aav2060
27. Rosenstein RK, Bezbradica JS, Yu S, and Medzhitov R. Signaling pathways activated by a protease allergen in basophils. Proc Natl Acad Sci U.S.A. (2014) 111:E4963–71. doi: 10.1073/pnas.1418959111
28. Selno ATH, Sumbayev VV, and Gibbs BF. IgE-dependent human basophil responses are inversely associated with the sarcoplasmic reticulum Ca(2+)-ATPase (SERCA). Front Immunol. (2022) 13:1052290. doi: 10.3389/fimmu.2022.1052290
29. Kodakandla G, Akimzhanov AM, and Boehning D. Regulatory mechanisms controlling store-operated calcium entry. Front Physiol. (2023) 14:1330259. doi: 10.3389/fphys.2023.1330259
30. Gill DL, Ghosh TK, and Mullaney JM. Calcium signalling mechanisms in endoplasmic reticulum activated by inositol 1,4,5-trisphosphate and GTP. Cell Calcium. (1989) 10:363–74. doi: 10.1016/0143-4160(89)90062-6
31. Steiner P and Zierler S. Inter-organellar ca(2+) homeostasis in plant and animal systems. Cells. (2025) 14. doi: 10.3390/cells14151204
32. Kushnireva L, Korkotian E, and Segal M. Calcium sensors STIM1 and STIM2 regulate different calcium functions in cultured hippocampal neurons. Front Synaptic Neurosci. (2020) 12:573714. doi: 10.3389/fnsyn.2020.573714
33. Berna-Erro A, Jardin I, Salido GM, and Rosado JA. Role of STIM2 in cell function and physiopathology. J Physiol. (2017) 595:3111–28. doi: 10.1113/JP273889
34. Clemens RA, Chong J, Grimes D, Hu Y, and Lowell CA. STIM1 and STIM2 cooperatively regulate mouse neutrophil store-operated calcium entry and cytokine production. Blood. (2017) 130:1565–77. doi: 10.1182/blood-2016-11-751230
35. Lunz V, Romanin C, and Frischauf I. STIM1 activation of orai1. Cell Calcium. (2019) 77:29–38. doi: 10.1016/j.ceca.2018.11.009
36. Zhuang GZ, Keeler B, Grant J, Bianchi L, Fu ES, Zhang YP, et al. Carbonic anhydrase-8 regulates inflammatory pain by inhibiting the ITPR1-cytosolic free calcium pathway. PloS One. (2015) 10:e0118273. doi: 10.1371/journal.pone.0118273
37. Hirota J, Ando H, Hamada K, and Mikoshiba K. Carbonic anhydrase-related protein is a novel binding protein for inositol 1,4,5-trisphosphate receptor type 1. Biochem J. (2003) 372:435–41. doi: 10.1042/bj20030110
38. Hirasawa M, Xu X, Trask RB, Maddatu TP, Johnson BA, Naggert JK, et al. Carbonic anhydrase related protein 8 mutation results in aberrant synaptic morphology and excitatory synaptic function in the cerebellum. Mol Cell Neurosci. (2007) 35:161–70. doi: 10.1016/j.mcn.2007.02.013
39. Oetjen LK, Mack MR, Feng J, Whelan TM, Niu H, Guo CJ, et al. Sensory neurons co-opt classical immune signaling pathways to mediate chronic itch. Cell. (2017) 171:217–28.e13. doi: 10.1016/j.cell.2017.08.006
40. Siracusa MC, Kim BS, Spergel JM, and Artis D. Basophils and allergic inflammation. J Allergy Clin Immunol. (2013) 132:789–801. doi: 10.1016/j.jaci.2013.07.046
41. Peng J and Siracusa MC. Basophils in antihelminth immunity. Semin Immunol. (2021) 53:101529. doi: 10.1016/j.smim.2021.101529
42. Poto R, Loffredo S, Marone G, Di Salvatore A, De Paulis A, Schroeder JT, et al. Basophils beyond allergic and parasitic diseases. Front Immunol. (2023) 14:1190034. doi: 10.3389/fimmu.2023.1190034
43. Doke T, Abedini A, Aldridge DL, Yang YW, Park J, Hernandez CM, et al. Single-cell analysis identifies the interaction of altered renal tubules with basophils orchestrating kidney fibrosis. Nat Immunol. (2022) 23:947–59. doi: 10.1038/s41590-022-01200-7
44. Joulia R and Lloyd CM. Basophils: Regulators of lung inflammation over space and time. J Exp Med. (2024) 221. doi: 10.1084/jem.20241663
45. Poto R, Gambardella AR, Marone G, Schroeder JT, Mattei F, Schiavoni G, et al. Basophils from allergy to cancer. Front Immunol. (2022) 13:1056838. doi: 10.3389/fimmu.2022.1056838
46. Levitt RC, Zhuang GY, Kang Y, Erasso DM, Upadhyay U, Ozdemir M, et al. Car8 dorsal root ganglion expression and genetic regulation of analgesic responses are associated with a cis-eQTL in mice. Mamm Genome. (2017) 28:407–15. doi: 10.1007/s00335-017-9694-7
47. Kang JK, Kim OH, Hur J, Yu SH, Lamichhane S, Lee JW, et al. Increased intracellular Ca(2+) concentrations prevent membrane localization of PH domains through the formation of Ca(2+)-phosphoinositides. Proc Natl Acad Sci U.S.A. (2017) 114:11926–31. doi: 10.1073/pnas.1706489114
48. Liu R, Kruger K, Pilat C, Fan W, Xiao Y, Seimetz M, et al. Excessive accumulation of intracellular ca(2+) after acute exercise potentiated impairment of T-cell function. Front Physiol. (2021) 12:728625. doi: 10.3389/fphys.2021.728625
49. Freitas CMT and Johnson DK. Weber KS. T cell calcium signaling regulation by the co-receptor CD5. Int J Mol Sci. (2018) 19(5):1295. doi: 10.3390/ijms19051295
50. Feske S. Calcium signalling in lymphocyte activation and disease. Nat Rev Immunol. (2007) 7:690–702. doi: 10.1038/nri2152
51. Johnson J, Blackman R, Gross S, and Soboloff J. Control of STIM and Orai function by post-translational modifications. Cell Calcium. (2022) 103:102544. doi: 10.1016/j.ceca.2022.102544
52. Ben-Kasus Nissim T, Zhang X, Elazar A, Roy S, Stolwijk JA, Zhou Y, et al. Mitochondria control store-operated Ca(2+) entry through Na(+) and redox signals. EMBO J. (2017) 36:797–815. doi: 10.15252/embj.201592481
53. Bogeski I, Kummerow C, Al-Ansary D, Schwarz EC, Koehler R, Kozai D, et al. Differential redox regulation of ORAI ion channels: a mechanism to tune cellular calcium signaling. Sci Signal. (2010) 3:ra24. doi: 10.1126/scisignal.2000672
54. Bryce PJ, Miller ML, Miyajima I, Tsai M, Galli SJ, and Oettgen HC. Immune sensitization in the skin is enhanced by antigen-independent effects of IgE. Immunity. (2004) 20:381–92. doi: 10.1016/S1074-7613(04)00080-9
55. Finkelman FD, Madden KB, Morris SC, Holmes JM, Boiani N, Katona IM, et al. Anti-cytokine antibodies as carrier proteins. Prolongation of in vivo effects of exogenous cytokines by injection of cytokine-anti-cytokine antibody complexes. J Immunol. (1993) 151:1235–44. doi: 10.4049/jimmunol.151.3.1235
56. Ohmori K, Luo Y, Jia Y, Nishida J, Wang Z, Bunting KD, et al. IL-3 induces basophil expansion in vivo by directing granulocyte-monocyte progenitors to differentiate into basophil lineage-restricted progenitors in the bone marrow and by increasing the number of basophil/mast cell progenitors in the spleen. J Immunol. (2009) 182:2835–41. doi: 10.4049/jimmunol.0802870
Keywords: carbonic anhydrase 8, basophils, interleukin 4, atopic dermatitis, calcium homeostasis
Citation: Peng J, Sy CB, Zhao S, Sawhney A, Espinosa V, Schroeter MN, Ponessa JJ, Chavan K, Lemenze AD, Rivera A, Rohacs T and Siracusa MC (2025) Carbonic anhydrase 8 regulates basophil activation and interleukin 4 production. Front. Immunol. 16:1694105. doi: 10.3389/fimmu.2025.1694105
Received: 28 August 2025; Accepted: 06 November 2025; Revised: 28 October 2025;
Published: 27 November 2025.
Edited by:
Christophe Pellefigues, Retore Institute, FranceReviewed by:
Kensuke Miyake, Tokyo Medical and Dental University, JapanAdrian Piliponsky, Seattle Children’s Research Institute, United States
Copyright © 2025 Peng, Sy, Zhao, Sawhney, Espinosa, Schroeter, Ponessa, Chavan, Lemenze, Rivera, Rohacs and Siracusa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mark C. Siracusa, bWFyay5zaXJhY3VzYUBydXRnZXJzLmVkdQ==
 Jianya Peng
Jianya Peng Chandler B. Sy1,2
Chandler B. Sy1,2 Siyuan Zhao
Siyuan Zhao Arman Sawhney
Arman Sawhney Vanessa Espinosa
Vanessa Espinosa Marissa N. Schroeter
Marissa N. Schroeter John J. Ponessa
John J. Ponessa Krupa Chavan
Krupa Chavan Amariliz Rivera
Amariliz Rivera Tibor Rohacs
Tibor Rohacs Mark C. Siracusa
Mark C. Siracusa