- 1Key Laboratory of Animal Biosafe Risk Prevention and Control (North), Ministry of Agriculture and Rural Affairs, Institute of Animal Science, Chinese Academy of Agricultural Sciences, Beijing, China
- 2College of Veterinary Medicine, Yangzhou University, Yangzhou, China
- 3College of Veterinary Medicine, Northwest A&F University, Xianyang, China
- 4National Reference Laboratory for Animal Brucellosis, China Institute of Veterinary Drug Control, Beijing, China
- 5College of Veterinary Medicine, Shandong Agricultural University, Taian, China
GntR transcription factors are emerging as critical regulators of bacterial metabolism, stress responses, and pathogenicity, however, their roles in the virulence mechanisms of Brucella abortus remain poorly understood. In this study, we generated a gntR8 (BAB_RS24500) deletion strain (ΔgntR8) in B. abortus 2308 and systematically investigated its role in virulence. The results demonstrate that deletion of gntR8 markedly impairs intracellular survival of B. abortus in RAW264.7 cells and significantly reduces virulence in a mouse infection model. Moreover, the ΔgntR8 strain exhibited increased sensitivity to oxidative stress, correlating with decreased expression of stress response genes. Integrative Dap-seq and RNA-seq analyses revealed that GntR8 directly binds to and positively regulates the clpP gene, a key component involved in oxidative stress defense. Deletion of clpP similarly resulted in diminished antioxidant capacity and intracellular survival, supporting a critical regulatory axis mediated by GntR8. Collectively, these findings provide novel insights into the molecular mechanisms by which GntR8 transcriptionally regulates oxidative stress responses and pathogenicity in B. abortus. The identification of GntR8 as a key virulence regulator highlights its potential as a therapeutic target, offering promising avenues for novel intervention strategies against brucellosis.
1 Introduction
Brucellosis is a zoonotic infectious disease caused by Brucella spp., which poses a significant threat to the health of both humans and animals (1). Humans typically acquire infection through direct contact with infected animals or ingestion of contaminated food products (2). Clinically, human brucellosis manifests as fever, malaise, arthralgia, sweating, and enlargement of the liver, spleen, and lymph nodes (3, 4). In livestock, Brucella infection primarily causes reproductive disorders such as abortion in females (5), orchitis and infertility in males, leading to substantial economic losses in livestock industry (6).
The pathogenicity of Brucella spp. is largely attributed to its ability to replicate and survive within host cells, thereby evading host immune responses (7). Transcription factors play crucial roles in regulating Brucella virulence and metabolic processes (8). For instance,
The BvrRS transcription factor, also known as a two-component regulator, controls the expression of genes required for multiple stages of Brucella infection. Studies have shown that ΔbvrR and ΔbvrS mutant strains are highly attenuated (9–11). BvrRS directly regulates the transcription of Brucella outer membrane protein genes omp25, omp22, and genes involved in lipopolysaccharide (LPS) modification (12, 13). It indirectly activates T4SS and outer membrane protein-related gene expression by inducing the expression of the gene encoding the quorum sensing regulator VjbR (14). The transcription regulator CtrA is the primary regulator of the Brucella cell cycle. Its activity is modulated by the histidine kinase PdhS, the CckA-ChpT phosphorylation cascade, and the protease adapter CpdR in response to endogenous cell cycle signals (15–18). The zinc finger protein MucR functions as a global regulator playing a crucial role in Brucella virulence (19). In B. melitensis, MucR influences LPS and correlates with oxidative stress tolerance. The MucR protein inhibits its own transcription and affects flagellar gene expression via the ftcR gene (20). In B. abortus 2308, MucR regulates genes associated with cell-membrane integrity, polysaccharide synthesis, and iron homeostasis (21).
The GntR family transcription factor represents another key regulatory system involved in bacterial metabolism and virulence. First described in 1987 (22), this family is subdivided into subfamilies such as MocR, YtrR, FadR, AraR, HutC, PlmA, DevA, and DasR, based on differences in their C-terminal amino acid sequences (23, 24). Previous studies have shown that deletion of the gntR10 gene significantly affects Brucella growth and virulence in mice, modulates the expression of LuxR-type transcriptional activators (VjbR and BlxR), and influences the expression of type IV secretion system (T4SS) effectors (BspE and BspF) (25, 26). The reference strain B. melitensis 16M encodes 21 GntR transcription factors, of which seven have been implicated in virulence regulation (27, 28). Among them, GntR4(coded by BMEI0169), GntR12(coded by BMEII0807) and GntR17(coded by BMEI0320) are known to regulate the expression of the virB gene in Brucella spp (8, 27, 29)., and GntR17 additionally influences the expression of omp25, vjbR and babR genes in B. abortus (8, 29). Previous study has revealed that the transcription factor GntR10 (BAB_RS31770) from B. abortus 2308 interacts with the target promoter of BAB1_1163 through sequence-specific DNA recognition, regulating the expression of 88 genes, including those involved in molecular functions, biological processes (BPs), and cellular components (CCs). The GntR10 target-gene mutant BAB1_1163 exhibits reduced expression in RAW 264.7 cells, affecting pro-inflammatory cytokine expression levels (26).
Despite these advances, relatively little is known about the broader role of other GntR transcription factors in virulence regulation of Brucella. Therefore, in this study, we constructed GntR transcription factor deletion and complemented strains using homologous recombination techniques, with B. abortus 2308 as the parental strain. We systematically examined the impact of gntR8 (BAB_RS24500) deletion on virulence at the bacterial, cellular, and animal levels. Additionally, using DNA affinity purification sequencing (Dap-seq) and Electrophoretic mobility shift assay (EMSA), we identified and verified downstream target genes regulated by gntR8 gene and performed functional analyses of these targets. This comprehensive analysis expands our understanding of GntR-mediated transcriptional regulation in Brucella and highlights GntR8 as a promising therapeutic target to disrupt virulence pathways. Targeting GntR8-mediated regulation could provide new approaches for reducing the economic and health impacts associated with brucellosis, suggesting future research should focus on therapeutic interventions designed to impair these transcriptional networks and mitigate disease transmission.
2 Materials and methods
2.1 Bacterial strains and cells
All Brucella strains were cultured in Tryptic Soy Agar (TSA, BD) or Tryptic Soy Broth (TSB, BD) at 37°C with 5% CO2. All work involving Brucella spp. strains was conducted in a biosafety Level 3 laboratory of China Institute of Veterinary Drug Control. The cells used in the in vitro experiments of this study were the RAW264.7 cells. The cell-culture conditions were DMEM medium (Gibco, USA) containing 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin, cultured in a 37°C, 5% CO2 incubator.
2.2 Construction of gntR8 deletion and complementation strains
The gntR8 mutant strain was constructed following a previously published protocol (30). Primer sequences for deletion and complementation strains are listed in Supplementary Table S1. PCR products were cloned using the ClonExpress MultiS One Step Cloning Kit (Vazyme, China) and transformed into E. coli DH5α competent cells (CWbio, China). Positive plasmids were then electroporated into Brucella strains. Ampicillin-sensitive and chloramphenicol-sensitive colonies were verified by PCR to ensure successful genes deletion and complementation.
2.3 Cell infection assay
Intracellular survival assays of wild-type (WT) B. abortus 2308 and its mutant strains was performed as previously described (31). Briefly, RAW264.7 cells (2.5×105 cells/well) were cultured in 24-well plates (Corning, USA) and infected with Brucella strains (100 MOI). Cells were then centrifuged at 1000 rpm for 10 minutes. After 1 h incubation, cells were washed three times with phosphate-buffered saline (PBS) and cultured in medium containing 50 μg/mL gentamicin. Wash twice more with PBS, then add 1 mL of medium containing 25 μg/mL gentamicin to each well. At 1, 24, and 48 hours post-infection (hpi), cells were lysed, and intracellular bacterial counts were determined by plating serial dilutions onto TSA plates.
2.4 Mice infection experiments
To evaluate the pathogenicity of gntR8 mutants in vivo, 80 female BALB/c mice (6–8 weeks) were randomly divided into four groups: PBS group, WT group, ΔgntR8 group, and complemented strain (CΔgntR8) group (n=20). Each group was further subdivided into four time points, with five mice per group at each time point (n=5). Each mouse in the infection group received an intraperitoneal injection of 1×105 CFU/0.1 mL of bacterial solution diluted in PBS, while the PBS group received an injection of 0.1 mL of PBS. Blood samples were collected from mice at weeks 1, 2, 3, and 4 post-infection. Mice were euthanized by asphyxiation at weeks 1, 2, 3 and 4 post-infection. After collection, spleen was weighed and divided into three portions. One portion was homogenized in 1 mL of PBS and subsequently subjected to TSA plate culture to determine bacterial load (n=5). Another portion was reserved for histopathological evaluation, and the third portion was used for RNA-seq analysis.
2.5 Histopathological evaluation
Histological examination was performed on spleen tissues as previously described (31). Briefly, spleen tissues were fixed in 10% formalin solution, embedded in paraffin, and sectioned into 4 µm slices using a microtome (Leica, Germany). Sections were stained with hematoxylin and eosin (HE) and examined under a light microscope (Leica, Germany).
2.6 Stress assays
Bacterial sensitivity under oxidative and acidic stress conditions was evaluated using a modified protocol (31–33).
Oxidative stress. WT, ΔgntR8 and CΔgntR8 strains were treated with H2O2 at final concentrations of 1 mM, 2.5 mM and 5 mM, respectively. After 1 h of treatment at 37°C, surviving bacteria were enumerated by plating serial dilutions on TSA.
Acid stress. Bacterial cultures were pelleted by centrifugation, resuspended in TSB adjusted to pH 4.5 or 5.5, and incubated at 37°C for 1 h. Surviving bacteria were quantified by plating serial dilutions onto TSA plates.
2.7 GSSG and GSH assay
Intracellular concentrations of reduced glutathione (GSH) and oxidized glutathione (GSSG) in bacterial cells were determined using a GSH and GSSG Assay Kit (Beyotime, China), following the manufacturer’s instructions with minor modifications (34). A standard curve was established using the standards in the kit. Absorbance at 412 nm was measured after 25 min incubation at room temperature using a microplate reader. Intracellular GSH was calculated using the equation: GSH = total glutathione - GSSG × 2.
2.8 Quantitative real-time PCR analysis and RNA-sequencing
Total RNA from infected RAW264.7 cells and mice spleen tissues were isolated using TRIzol (Thermo Scientific, USA) according to the manufacturer’s instructions, followed by DNase I treatment to eliminate genomic DNA contamination. RNA concentration and purity were assessed using an ND 1000 spectrophotometer (Thermo Scientific, USA). Reverse transcription into cDNAs were synthesized using the PrimeScript RT Reagent Kit (TaKaRa Bio, Japan) according to the manufacturer’s instructions. Quantitative real-time PCR (qPCR) was performed with the primers shown in Supplementary Table S2. Relative gene expression was calculated using the comparative cycle threshold method (2−ΔΔCt), and each sample was analyzed in triplicate.
Sequencing libraries for each RNA sample were prepared using the NEB Next Ultra Directional RNA Library Prep Kit for Illumina according to the manufacturer’s protocol (35). RNA fragments were reverse-transcribed, amplified to double-stranded cDNA, adaptor- ligated, purified with magnetic bead and quantified. Sequencing was performed on the HiSeq 4000 platform at the Majorbio platform (Shanghai, China) with three biological replicates per group. Differential expression thresholds are fold change ≥ 2 and p ≤ 0.05.
2.9 Expression and purification of recombinant GntR8 protein
The coding region of the gntR8 gene was amplified using primers pCold-G8-F and pCold-G8-R (Supplementary Table S3), digested with BamH I and Hind III and cloned into the plasmid pCold II. The resulting plasmid (pCold-gntR8) was transformed into E. coli strain BL21. Protein expression was induced with 0.1 mM IPTG at 16 °C for 24 h. Cells were harvested, lysed by sonication, and centrifuged (12,000 × g, 20 min, 4 °C). The supernatant containing recombinant GntR8 protein was purified using Ni-Sepharose affinity chromatography.
2.10 DNA affinity purification sequencing
Purified GntR8 protein was flash-frozen into liquid nitrogen and stored at -80°C until use. Dap-seq analysis was performed by Yung Biotechnology Co., Ltd. (Beijing, China). Fastp (v0.20.1) software was used for quality control analysis of target proteins and negative controls, including removal of splices, repeats and low-quality sequences. Peak Calling was performed using MACS2 (v2.2.7.1) software (Fold change ≥ 2 and p-value ≤ 0.05) and the ChIPseeker (R package) was used to annotate peak. Motif analysis was conducted using HOMER software (v4.11.1).
2.11 Electrophoretic mobility shift assay
Biotin-labeled DNA probes were incubated in EMSA buffers (750 mM NaCl, 0.5 mM dithiothreitol (DTT), 0.5 mM EDTA, 50 mM Tris, pH 7.4) at 37°C for 30 min. For competitive assays, 100 nM unlabeled DNA probes were added to labeled probes. GntR8 protein (0–200 ng) was incubated with the probes at 37°C for 30 minutes. The samples were separated on a 6% Native-PAGE - gel (30% acrylamide, 5×TBE, TEMED, 10% APS, 5% Glycerol) and run in a 0.5 × Tris-Borate buffer (89mM Tris-Borate, 2mM EDTA, pH 7.4) at 200 V and 4 °C. Imaging was captured using a Typhoon FLA 9500 multifunctional scanner (GE Healthcare, USA).
2.12 Cytokine measurement by the multiplex cytokine assay system
Measure cytokine concentrations in mouse serum using the Luminex Flex MAP 3D system according to the manufacturer’s instructions (36). In brief, mix chemically labeled antibody-conjugated beads with standard solutions or samples, incubate overnight at 4°C, wash, and then incubate with biotinylated detection antibodies. After washing the beads, incubate them with streptavidin-phycoerythrin complexes. The sample is then washed using a handheld magnet and resuspended in sheath fluid. Finally, the sample is run on the Luminex FLEXMAP 3D® (Austin, Texas, USA), and data is collected and analyzed using MILLIPLEX Analyst 5.1 (Luminex). Three biological replicates were performed for each experimental group.
2.13 Statistical analysis
Basic statistical analyses were performed using GraphPad Prism 9.0 (USA). Unpaired Student’s t-tests were employed in cellular and mouse infection models, growth curve measurements of gntR8 deletion strains, and bacterial virulence assays. For stress analyses, data were analyzed using analysis of variance (ANOVA). Data are expressed as mean ± standard deviation. P values < 0.05 were considered statistically significant.
3 Results
3.1 Deletion of gntR8 significantly reduces B. abortus virulence in RAW264.7 cells and mice
Previous studies identified 21 GntR family transcription factors in the B. melitensis 16M strain (27). In this study, we analyzed the genome of B. abortus 2308 and identified 23 GntR family transcription factors through KEGG homologous gene analysis combined with NCBI database searches. As shown in Supplementary Figure S1 A and B and Supplementary Table S4, these factors include 7 located on chromosome I and 17 on chromosome II. To investigate the role of gntR8, we constructed a deletion strain (ΔgntR8) by replacing the gntR8 gene with a kanamycin resistance gene via homologous recombination, and a complemented strain (CΔgntR8) using the pBBRMCS-1 plasmid. Growth curves showed no significant difference between WT and ΔgntR8 strain under normal culture conditions (Figure 1A). However, RAW264.7 cells infection assay indicated significant decrease in intracellular viability following infection with ΔgntR8 strain compared to WT strain (p < 0.001) (Figure 1B). In addition, we quantified colocalization of intracellular Brucella with LAMP-1–positive compartments using laser confocal microscopy. At 4 h post-infection, the ΔgntR8 mutant displayed a significantly higher LAMP-1 colocalization rates rather than the wild-type (WT) (p = 0.003) (Supplementary Figure S2), indicating impaired lysosomal evasion. To further investigate the effect of the gntR8 gene deletion on Brucella virulence, we conducted mouse infection over 4 weeks to assess the survival of ΔgntR8, CΔgntR8, and WT strains. As shown in Figure 1C, ΔgntR8-infected mice exhibited significantly lower bacterial loads in the spleen at 1, 3, and 4 weeks post-infection compared to the WT and CΔgntR8 groups (p < 0.001). Additionally, the spleen indices of WT-infected and CΔgntR8-infected mice were significantly higher than those of the ΔgntR8-infected group at 2 weeks post-infection (p < 0.001) (Figure 1D). Histopathological analysis revealed no significant lesions in spleens at week 1 post-infection for both WT and ΔgntR8 strains. However, spleens of the ΔgntR8-infected mice exhibited a marked reduction in lymphocytes and an increase in connective tissue proliferation (Figure 1E). At week 4 post-infection, spleens of mice infected with WT showed extensive connective tissue proliferation (red arrowheads) and focal neutrophil infiltration (black arrowheads). In contrast, no significant abnormalities were observed in the spleens ΔgntR8-infected groups (Figure 1F). These results indicate that the deletion of the gntR8 gene significantly reduces the virulence of B. abortus both in vivo and in vitro.
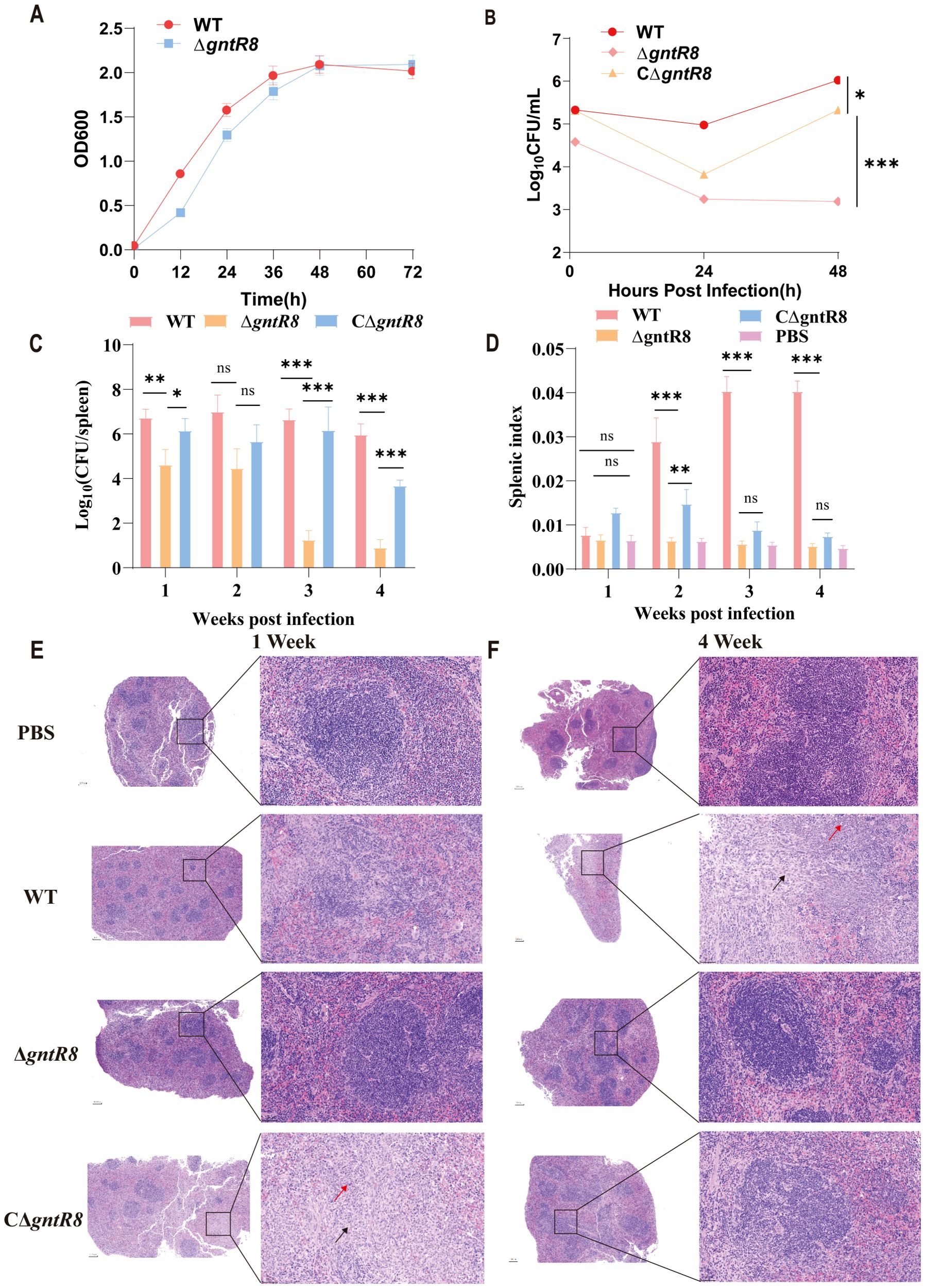
Figure 1. Deletion of the gntR8 gene reduces (B) abortus survival within cells and its pathogenicity in mice. (A) Growth curves in TSB of Brucella strains at 37°C with continuous shaking for 72 (H) (B) Intracellular survival of RAW264.7 cells (100 MOI), values represent the means of three experiments performed in duplicate, and error bars indicate the SD; (C) Splenic bacterial load post-infection (n=5); (D) Splenic index post-infection (n=5); (E-F) Histopathological analysis of spleen at 1 week (E) and 4 week (F) post-infection with WT and ΔgntR8. Connective tissue hyperplasia is shown by red arrows, and inflammatory cell infiltration is shown by black arrows. Data are presented as the mean ± standard deviation (error bars) of standardized data, based on experimental results from five mice. The significance is shown as *p < 0.05; **p < 0.01; ***p < 0.001; and ns indicates non-significance.
3.2 Deletion of the gntR8 reduces oxidative stress resistance in B. abortus
The Dose-response curve showed that the survival rate of the strains gradually decreased with increasing H2O2 concentration (Supplementary Figure S3), compared to WT, the survival rate of ΔgntR8 significantly decreased (p < 0.001) when treated with 5 mM H2O2. Under acidic conditions, the survival rates of the three strains (WT, ΔgntR8, and CΔgntR8) were comparable at pH 4.5 and 5.5 (Figure 2A). Oxidative stress experiments showed that, compared to WT, the survival rate of ΔgntR8 significantly decreased when treated with 5 mM H2O2 (p = 0.0049). In contrast, the survival rate of the CΔgntR8 strain recovered to the WT level (Figure 2B). RNA-seq analysis was used to identify genes regulated by the gntR8 gene under oxidative conditions. A total of 290 differentially expressed genes (DEGs) (Fold change ≥ 2 and p-value ≤ 0.05) were identified between the WT and ΔgntR8, with 141 genes up-regulated and 149 genes down-regulated (Figure 2C). KEGG pathway enrichment analysis sugggested that up-regulated genes may play a variety of biological functions through interaction with quorum sensing system and ABC transporter-related proteins (Figure 2D). Down-regulated genes primarily mostly associated with the sulfur relay system and the two-component system (Figure 2E). RT-qPCR validated the RNA-seq findings (Supplementary Figure S4).
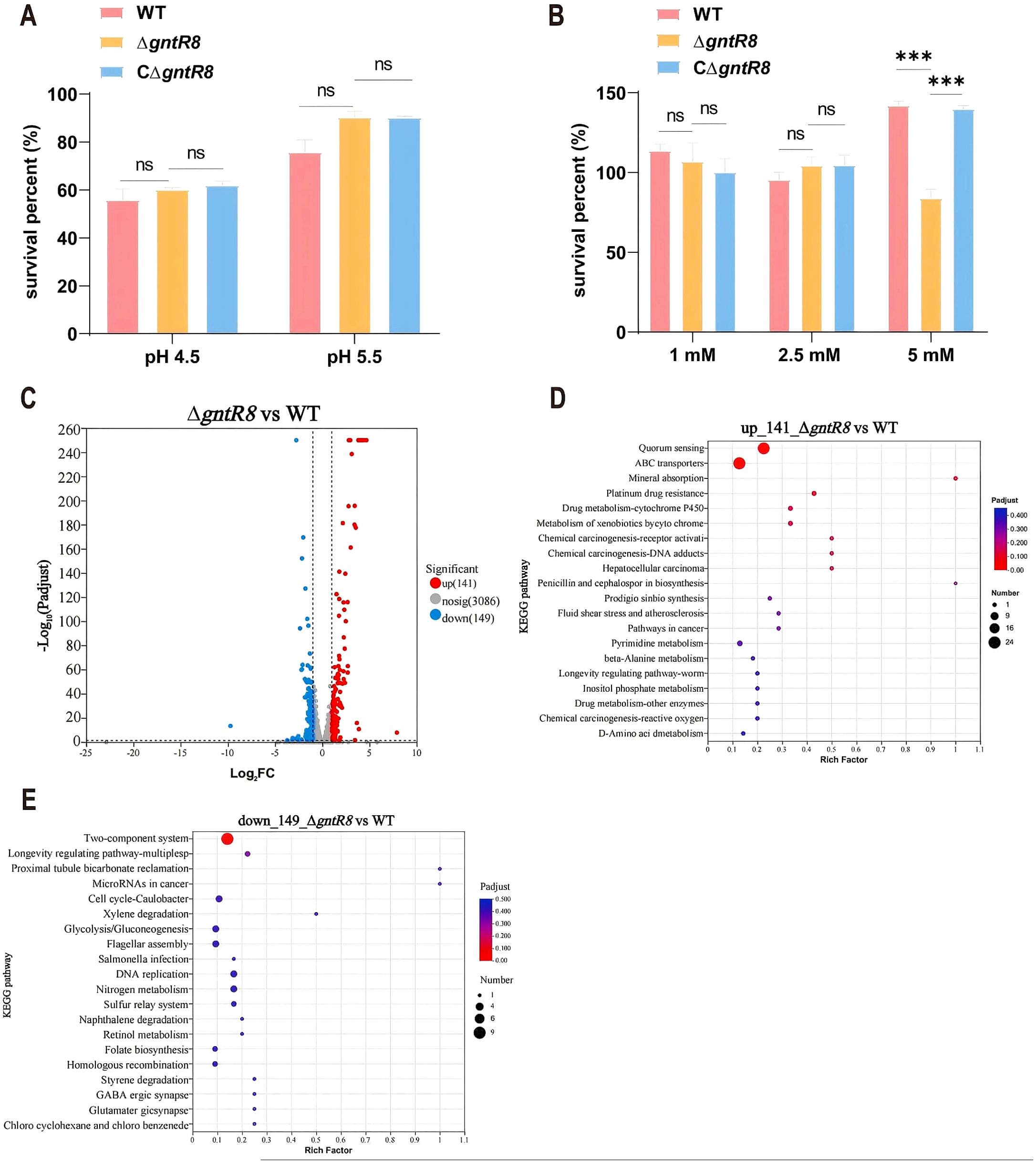
Figure 2. Deletion of the gntR8 reduces oxidative stress resistance in (B) abortus. (A) Survival rate of Brucella strains under acidic (pH 4.5 and 5.5) stress. (B) Survival rate of Brucella strains under H2O2 (1 mM, 2.5 mM, and 5 mM) at 37°C for 1 (H) (C) Treat the counted bacterial suspension with H2O2 (final concentration 5 mM) for 1 hour. Add RNA protection reagent at twice the volume, vortex, and incubate at room temperature for 5 minutes to extract RNA. Proceed with sequencing processing. Volcano plot of RNA-seq screen for differentially expressed genes. Horizontal coordinates indicate fold difference and vertical coordinates indicate negative Log10 values for p-adjust; (D-E) KEGG pathway enrichment analysis of up-regulated (D) and down-regulated (E) genes. Data are presented as the means of normalized data ± standard deviations (error bars) based on three independent experiments. The significance is shown as **p < 0.01; ***p < 0.001; and ns indicates non-significance.
3.3 GntR8 protein specifically binds to promoters of ALDH, gst and clpP genes
To further analyze the mechanism by which GntR8 participates in Brucella virulence and antioxidant stress, this study utilized Dap-seq technology to analyze the sequences directly bound by GntR8, revealing that the binding fragments are all located in the gene promoter regulatory region, potentially indicating self-regulatory functions (Figure 3A). Therefore, To analyze whether the GntR8 protein can bind to its own promoter DNA sequence, this study performed co-incubation using the GntR8 protein and the promoter DNA sequence of the gntR8 gene (BAB_RS24500), and found that the GntR8 protein can bind to its own promoter DNA sequence in a dose-dependent manner, indicating that the EMSA system used in this study can be employed to identify the regulatory genes of the GntR8 transcription factor(Figure 3B). Further screening of genes regulated by the GntR8 transcription factor was conducted by amplifying the promoter DNA sequences of potential target genes. The results indicated that the GntR8 protein can bind to the promoter DNA sequences of ALDH (BAB_RS16905), gst (BAB_RS27470), and clpP (BAB_RS21345) (Figure 3C). Competitive EMSA assays confirmed specificity, showing progressive inhibition of labeled probe binding upon increasing unlabeled competitor DNA (Figures 3D-F). This indicates that GntR8 specifically binds to the ALDH, gst, and clpP gene promoters.
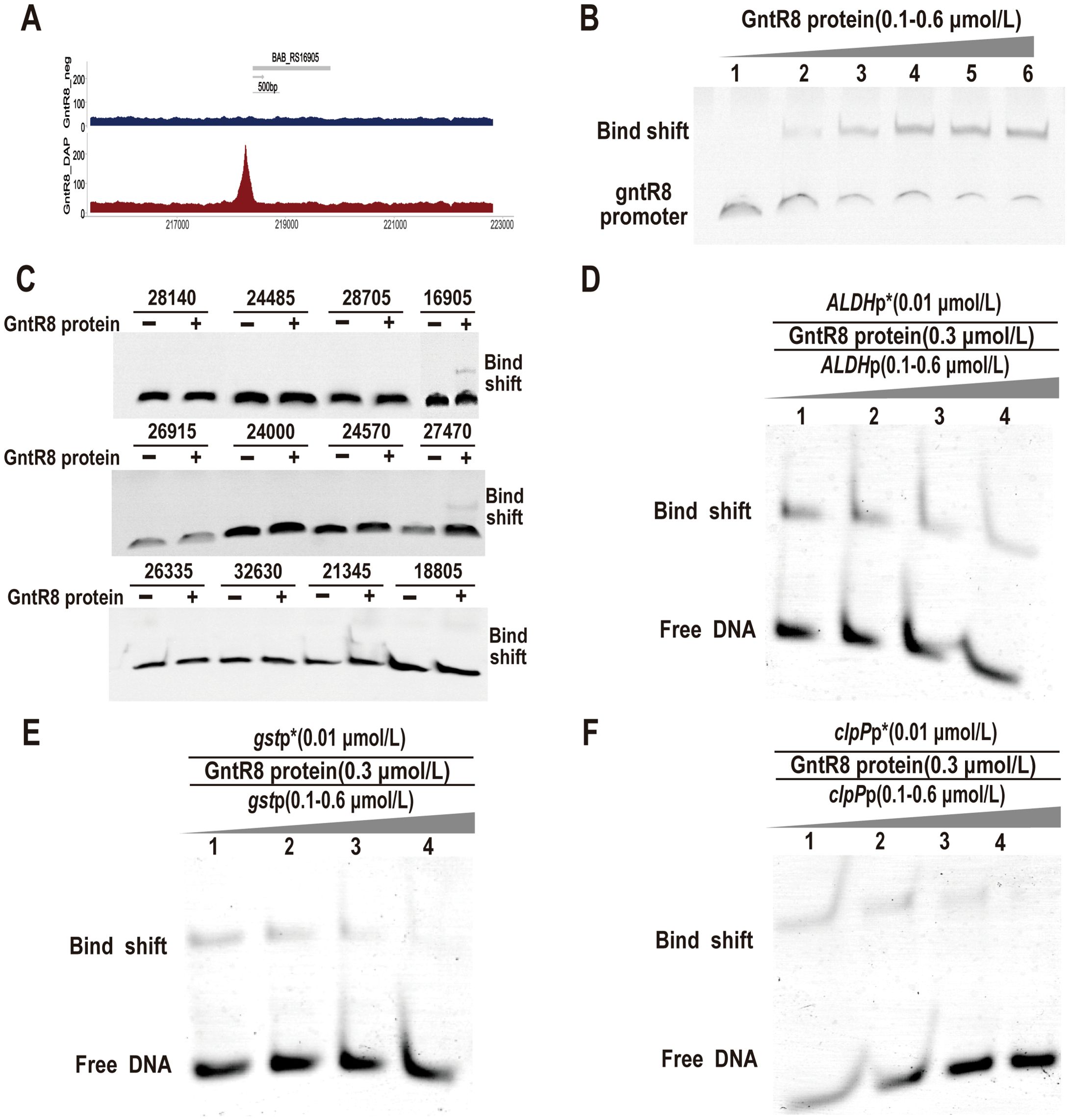
Figure 3. GntR8 protein specifically binds promoters of target genes. (A) Dap-seq analysis revealed that GntR8 was enriched in its promoter region; (B) EMSA analysis of GntR8 binding to gntR8 promoter region; (C) EMSA assay for GntR8 protein and differential gene promoter DNA sequence; Competition EMSA assays of ALDH (D), gst (E), and clpP (F) promoters.
3.4 GntR8-mediated regulation of oxidative stress in B. abortus via clpP gene
Intracellular survival assay of RAW264.7 cells with ΔALDH, Δgst, ΔclpP and WT strains showed that the ability of ΔclpP to survive in the RAW264.7 cell was significantly decreased (p < 0.001)(Figure 4A). Oxidative stress assays with ΔALDH, Δgst, ΔclpP and WT strains revealed no significant difference in survival for ΔALDH and Δgst compared to WT. However, after the deletion of clpP gene, the antioxidant capacity of Brucella decreased significantly (p < 0.001) (Figure 4B). GSH is an important antioxidant that scavenges free radicals and helps cells maintain normal immune function (37). In this study, the GSSG content of ΔgntR8 and ΔclpP were significantly higher than that of WT (p < 0.001) (Figure 4C). Consistently, GSH levels were significantly reduced (p < 0.001) (Figure 4D). The results showed that the antioxidant capacity of Brucella decreased after clpP gene deletion. These results indicate that GntR8 regulates GSH levels by controlling the expression of the clpP gene, thereby modulating the oxidative stress resistance of B. abortus.
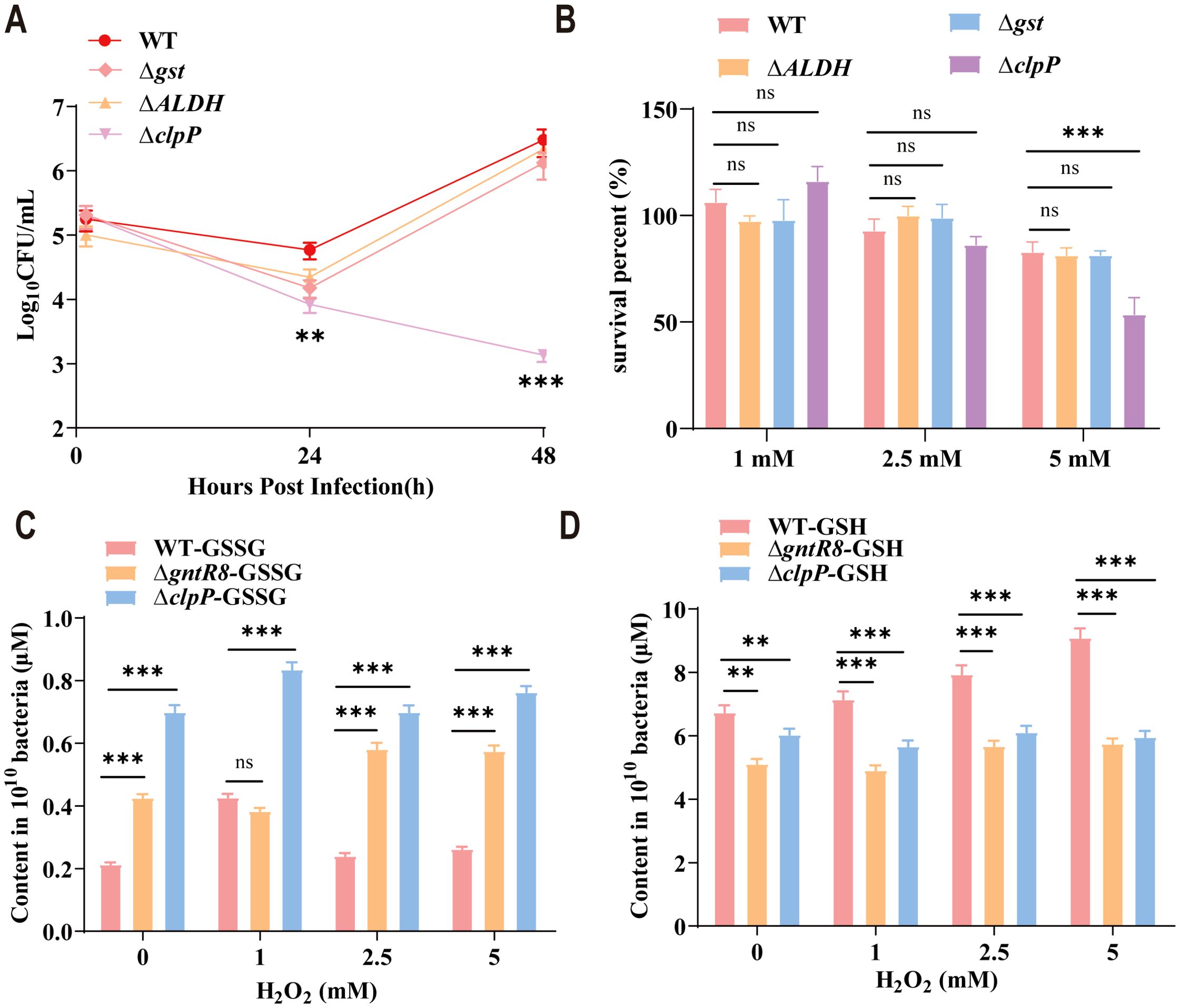
Figure 4. GntR8 transcription factors regulates oxidative stress resistance in B. abortus via clpP gene. (A) Survival rate of WT, ∆ALDH, ∆gst and ∆clpP under stress condition; Total GSSG (B) and GSH (C) content of Brucella strains; (D) intracellular survival of the ∆ALDH, ∆gst and ∆clpP deletion strain. Data are presented as the means of normalized data ± standard deviations (error bars) based on three independent experiments. The significance is shown as ** p <0.01; *** p <0.001; and ns indicates non-significance.
3.5 Deletion of the gntR8 down-regulates immune-related gene in infected hosts
Transcriptome analysis of the spleen of WT and ΔgntR8-infected mice revealed that, at week 1 post-infection, 505 genes were up-regulated, while 1852 genes (Fold change ≥ 2 and p-value ≤ 0.05) were down-regulated in the spleens of the ΔgntR8-infected mice (Figure 5A). KEGG enrichment analysis of up-regulated genes showed that these differential genes were mainly enriched in Th17 cell differentiation and T cell receptor signaling pathway (Figure 5B), while down-regulated genes were mainly concentrated in NOD-like receptor signaling pathway and TNF signaling pathway (Figure 5C). Similar trends were observed in cell transcriptome results (Supplementary Figure S5). At week 4 post-infection, 2614 genes were up-regulated, and 3448 genes were down-regulated in the spleens of ΔgntR8-infected mice (Figure 5D). KEGG enrichment analysis of up-regulated genes showed that these differential genes were mainly enriched in cell cycle, DNA replication, P53 signaling pathway (Figure 5E). Down-regulated genes were mainly concentrated in Primary immunodeficiency, Th1 and Th2 cell differentiation and NF-kappa B signaling pathway (Figure 5F). RT-qPCR confirmed RNA-seq results (Supplementary Figure S6). Previous studies have shown that immunizing mice with the B. abortus 2308 mutant ΔgntR can induce classic Th1 and Th2 responses (29). The above analysis showed that the deletion of gntR8 gene caused the down-regulation of the expression of immune-related genes in infected hosts.
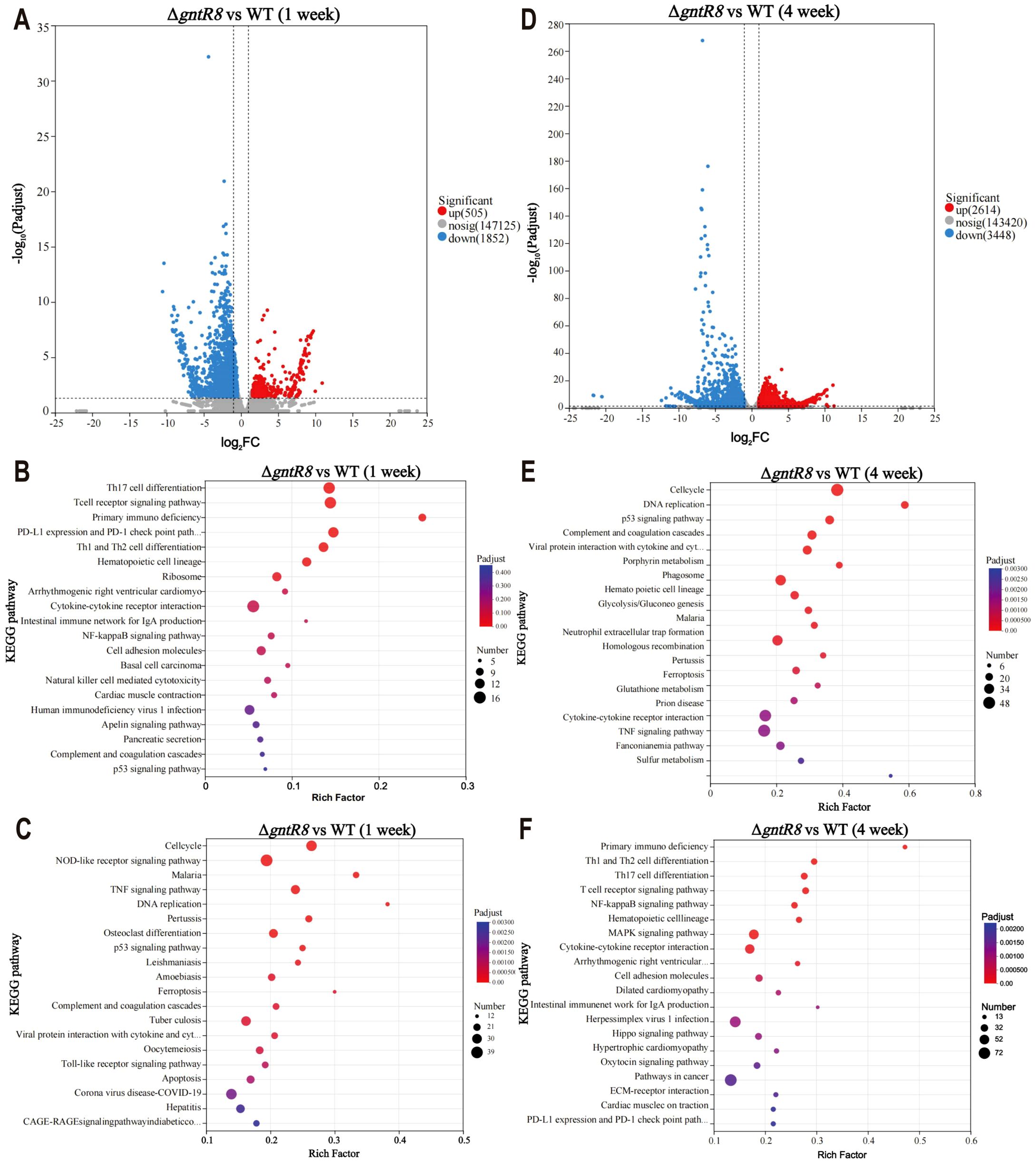
Figure 5. Deletion of the gntR8 down-regulates immune-related gene in infected hosts. (A) DEGs at 1 week post-infection; KEGG pathway enrichment analysis of up-regulated (B) and down-regulated (C)genes at 1 week post-infection; (D) DEGs at 4 weeks post-infection; KEGG pathway enrichment analysis of up-regulated (E) and down-regulated (F) genes at 4 weeks post-infection. The data is based on experimental results from three mice.
3.6 Deletion of GntR8 reduces cytokine production in B. abortus-infected mice
Brucella, as intracellular pathogens, mainly relies on cellular immunity for clearance in the early stage of infection (38). Th1 cells participate in the host’s defense against intracellular pathogens by producing IFN-γ, TNF-α, and IL-2, while Th2 cells are responsible for coordinating humoral immunity and participate in the host’s defense against extracellular parasites by secreting IL-4, IL-5, and IL-10 (39–41). Given the RNA-seq findings on immune-related pathways (Figure 5), we detected IL-2, IL-6, IL-8, IL-10, IFN-γ, TNF-α and other immune-related cytokines. The results are shown in Figure 6, after 1 week, there were no significant differences in cytokine levels between the ΔgntR8 group and the PBS group. However, compared to the PBS group, the WT group and CΔgntR8 group showed significant upregulation of IL-8, IL-10, IFN-γ, and TNF-α. After 4 weeks, compared to the PBS group, all infected groups showed significant upregulation of IL-6, IL-8, and TNF-α expression (p < 0.001). This finding indicates that at this time point, ΔgntR8 induced a Th1-type immune response, consistent with the transcriptomic results. Therefore, we performed additional analyses focusing on MHC-I and MHC-II pathway–related genes. Integrating mouse spleen transcriptome data, we employed real-time quantitative polymerase chain reaction (qRT-PCR) for MHC-I pathway genes (including H-2Kb, β2m, TAP1, TAP2, Stat1, and NLRC5) and MHC II pathway genes (CIITA and RFX5) in mice infected with WT, ΔgntR8 and CΔgntR8 strains at weeks 1 and 4. Results showed that compared to WT, ΔgntR8-infected mice exhibited significantly reduced mRNA expression levels for all genes at both time points; CΔgntR8 partially or fully restored expression (TAP1/TAP2 approached WT by week 4) There were no significant changes in the expression of MHC class II-associated genes (CIITA and RFX5). Notably, transient elevations of H-2Kb and Stat1 at week 1 likely reflect early innate/adaptive activation. These results are provided in Supplementary Figure S7.
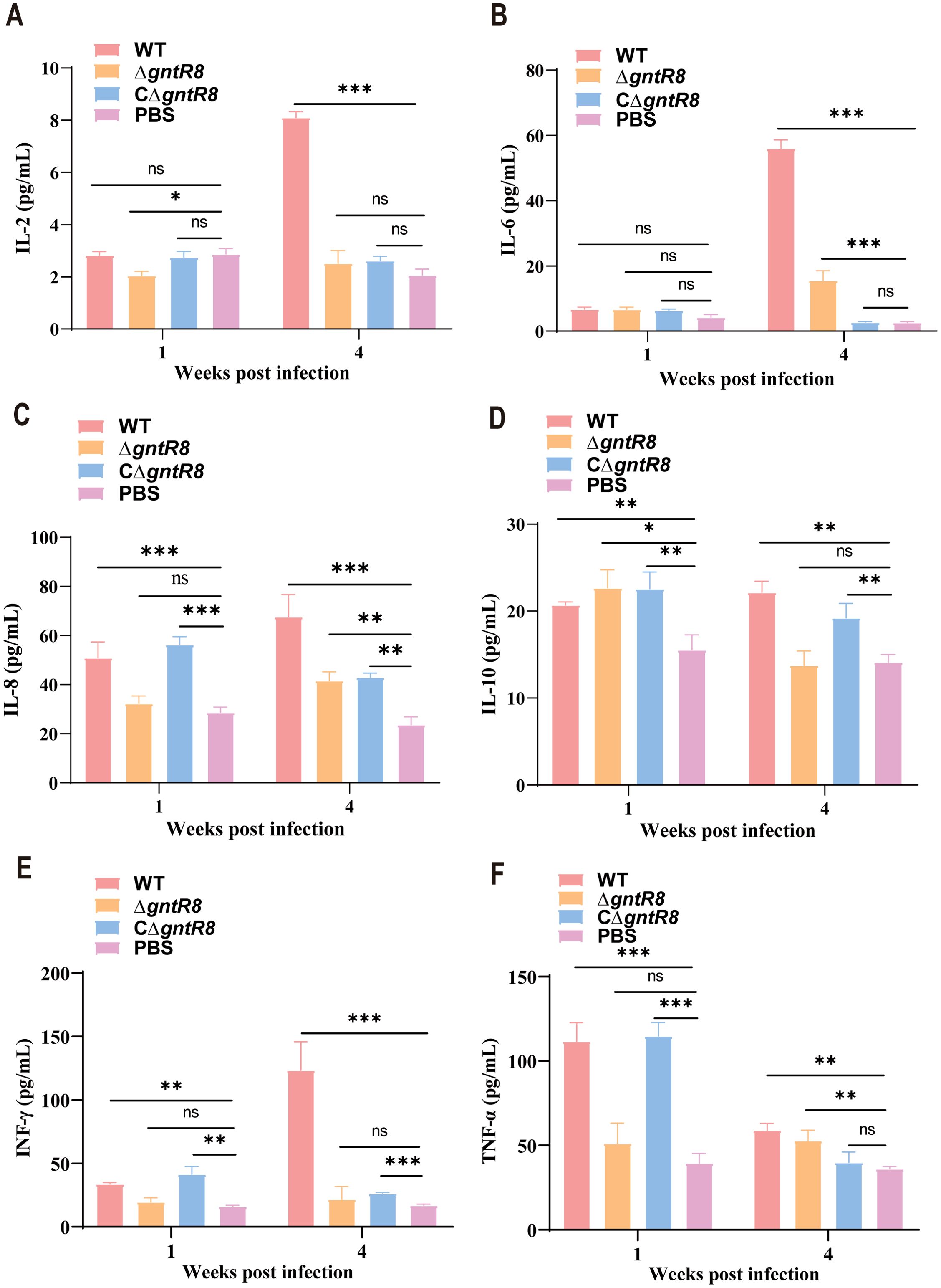
Figure 6. Cytokine production levels post-infection in mice. Production level of (A) IL-2, (B) IL-6, (C) IL-8, (D) IL-10, (E) IFN-γ, (F) TNF-α. Data are presented as the mean ± standard deviation (error bars) of standardized data, based on experimental results from three mice. The significance is shown as *p < 0.05; **p < 0.01; ***p < 0.001; and ns indicates non-significance.
4 Discussion
Brucella spp., the causative agent of brucellosis, poses a significant threat to both human and animal health (42). Due to its tendency to present clinically as a latent or chronic infection, brucellosis is difficult to diagnose and treat in a timely manner, contributing to its widespread distribution globally (43). Previous studies have highlighted the crucial role of GntR transcription factors in regulating bacterial metabolism and pathogenesis (27). In this study, we demonstrated that deletion of the transcription factor gene gntR8 in B. abortus significantly reduced intracellular survival in RAW264.7 cells and markedly decreased bacterial virulence in mice. However, although RAW264.7 cells possess core macrophage functions (such as phagocytosis and antibacterial activity), as an immortalized cell line, their phenotype may differ from that of macrophages derived from primary monocytes and requires further validation. Splenomegaly and connective tissue proliferation induced by Brucella infection in mice represent characteristic pathological features of the intracellular parasitic pathogenesis of this bacterium, as extensively documented in numerous domestic and international studies (31, 44). In this study, pathological analysis revealed that, in mice infected with B. abortus, the spleens exhibited varying degrees of enlargement. Histological examination showed connective tissue proliferation in the spleens of WT-infected mice at 4 weeks post-infection, contributing to the spleen enlargement. However, although the complementary strain restored the regulatory pathway required for bacterial colonization by replenishing gntR8, it failed to fully reinstate GntR8’s control over “immunopathology-related genes”. This may stem from differences in promoter strength and expression timing between the plasmid vector and the wild-type strain. Consequently, even when bacterial levels reached the target threshold, the spleen’s immune hyperplasia response remained below wild-type levels.
As an intracellular pathogen, Brucella spp. invades host macrophages, where it survives and replicates within Brucella-containing vesicles (BCV) (45, 46). After phagocytosis by macrophages, the bacteria must adapt to various stressors, including acidic environments, hypoxia, nutrient deprivation, reactive oxygen and nitrogen species (47, 48). Studies on the Brucella LysR-type transcription factor BvtR indicate that ΔBvtR strains exhibit increased sensitivity to sodium nitroprusside and sodium dodecyl sulfate, but show no altered sensitivity to hydrogen peroxide, isopropyl benzene peroxide, polymyxin B, or natural serum. Deletion of the OtpR gene in the B. melitensis 16M resulted in reduced tolerance to acidic stress (49, 50). The flagellar transcription regulator FtcR participates in the formation of B. melitensis 16M biofilms, which enhance tolerance to hyperosmotic stress (51). Brucella enter host cells via interactions between liposomes and macrophage cell membranes, forming Brucella-containing vacuoles (BCVs) surrounded by phagocytic vesicles (52). The acidic environment within BCVs facilitates expression of the VirB operon in Brucella and regulates T4SS-associated gene expression. Brucella utilizes the T4SS to transport effectors from the membrane space into the host cell cytoplasm, thereby modulating host cell signaling pathways to promote its survival within the host (53–55). Thus, in vitro models that simulate these stresses are critical for studying the pathogenic mechanism of B. abortus. Our findings revealed that deletion of the gntR8 gene significantly impaired resistance to oxidative stress induced by H2O2.
Dap-seq is a powerful method used to identify transcription factor binding sites without the need for specific antibody. This technique has been previously utilized in B. melitensis to successfully identify the target genes of transcriptional regulators, such as the iron-responsive regulator Irr (56). In our study, integrated Dap-seq and RNA-seq analyses identified 44 potential GntR8-regulated target genes. EMSA further confirmed that GntR8 specifically binds to the promoter of the clpP gene, which has previously been implicated in bacterial stress response. clpP gene has been confirmed by other studies related functions, the deletion of clpP gene can cause Brucella to increase the sensitivity to H2O2, and found that the survival ability of ΔclpP in macrophages significantly decreased (31). GSH, a critical intracellular antioxidant, maintains protein thiol groups in reduced states its sulfhydryl moiety. The glutathione peroxidase (GSH-Px)-catalyzed oxidation of GSH to GSSG concomitantly reduces to H2O (57, 58). In physiological conditions, reduced GSH constitutes >90% of total cellular glutathione (59). Oxidative stress triggers GSSG accumulation, consequently lowering the GSH/GSSG ratio - a key indicator of cellular redox status maintained through coordinated actions of GSH-Px and glutathione reductase (GR) (60). Our study showed that compared to the WT group, both ΔgntR8 and ΔclpP exhibit compromised H2O2 tolerance and diminished GSH levels, correlating with impaired intracellular survival. Therefore, it is speculated that GntR8 transcription factor may mediate the antioxidant stress of B. abortus through the regulation of clpP.
Brucella spp. has evolved multiple immune escape mechanisms, with its virulence factors modulating autophagy, inflammation, and apoptosis to suppress the host immune response (61).Transcriptome analysis of B. melitensis 16M infected macrophages revealed differential regulation of endoplasmic reticulum-associated pathway, immune-associated pathway, and p53 pathway (62). Notably, infection-induced dysregulation of immune-related genes (e.g., TXNIP, HO-1 and Prdx5) has been reported (63, 64), while deletion of the gntR10 gene elevates levels of TNF-α, IL-6 and IL-8 transcripts in infected cells (65). Our dual transcriptome analyses (host cells and mouse spleen) identified Th17, Th1, and Th2 differentiation pathways as significantly enriched among differentially expressed genes. It has previously been shown that Brucella infection triggers innate and adaptive immunity to Th1 and activation of CD8+ T cells, reducing MHC-I and MHC-II IFN-γ-induced surface expression and thereby impairing antigen presentation to T cells (66–69). Our cytokine data showed that IL-2 levels were upregulated in the WT group, suggesting that B. abortus 2308 stimulates specific cellular immunity, whereas deletion of the gntR8 gene resulted in reduced levels of serum cytokine IL-2, IL-6, IL-10, IFN-γ, and TNF-α production in Brucella-infected mice. IFN-γ stimulation rapidly activates the transcription factor STAT1, which subsequently induces IRF1 to upregulate the expression of genes essential for the MHC-I pathway. NLRC5 serves as the primary co-activator for MHC- I genes, playing a critical role not only in their expression but also in maintaining key components of the MHC-I pathway (70–72). H-2Kb is a core functional protein in the mouse MHC-I antigen-presentation pathway; its function is to mediate antigen recognition by CD8+T cells (73, 74). In ΔgntR8-infected mice, reduced interferon-γ levels led to significant downregulation of H-2Kb gene transcription, increased protein degradation, and impaired antigen presentation. This ultimately weakened the adaptive immune capacity for clearing intracellular targets. Concurrently, this study revealed markedly reduced expression levels of β2m, TAP1, TAP2, Stat1, and NLRC5 genes in the spleens of ΔgntR8-infected mice. In conclusion, IFN-γ–STAT1–IRF1 signaling and NLRC5 co-activation are attenuated in ΔgntR8 infection, leading to reduced H-2Kb expression and compromised antigen presentation to CD8+T cells.
Collectively, our study establishes the GntR8 transcription factor as a critical regulator of B. abortus virulence, intracellular survival, and host immune response modulation. Through combined transcriptomic and binding-site analyses (RNA-seq and Dap-seq), we provide clear evidence that GntR8 directly targets the clpP gene, thereby enhancing oxidative stress resistance and intracellular survival. Additionally, we suggest a potential role for GntR8 in immune modulation via MHC-I pathways (Figure 7). These findings significantly enhance our understanding of the molecular mechanisms underlying Brucella pathogenicity and identify GntR8 as a promising therapeutic target for future strategies aimed at controlling brucellosis.
Figure 7. Model summarizing the role of the GntR8 transcription factor in B. abortus virulence. GntR8 directly binds to the clpP promoter, enhancing resistance to oxidative stress, improving intracellular bacterial survival, and potentially modulating host cellular immune responses through MHC-I-dependent pathways. Arrows: Red for decrease; Green for increase; Black for activating effect (Created with bioRender.com)
Data availability statement
The raw Brucella transcriptome data has been submitted to the SRA database with the accession number PRJNA1332959; the raw mouse spleen transcript data has been submitted to the SRA database with the accession number PRJNA1333329; and the raw Brucella Dap-seq data has been uploaded to the SRA database with the accession number PRJNA1333353. All data generated or analyzed during this study are available from the corresponding author upon reasonable request.
Ethics statement
The animal study was approved by the Animal Ethics Committee of the China Institute of Veterinary Drug Control (CIVDC 2023-00037). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
SL: Visualization, Validation, Investigation, Conceptualization, Data curation, Formal Analysis, Writing – review & editing, Writing – original draft, Methodology. KH: Writing – review & editing, Methodology, Investigation, Writing – original draft, Conceptualization, Formal Analysis, Visualization, Data curation, Validation. XP: Visualization, Formal Analysis, Validation, Writing – original draft, Investigation, Data curation, Methodology, Writing – review & editing, Conceptualization. NW: Methodology, Data curation, Investigation, Writing – review & editing, Formal Analysis. WN: Investigation, Methodology, Writing – review & editing, Data curation, Formal Analysis. SG: Data curation, Methodology, Writing – review & editing, Formal Analysis, Investigation. LX: Formal Analysis, Methodology, Data curation, Investigation, Writing – review & editing. JD: Writing – review & editing, Conceptualization, Software, Writing – original draft, Funding acquisition, Supervision, Resources, Project administration, Validation, Methodology. XZ: Methodology, Software, Conceptualization, Validation, Resources, Writing – original draft, Funding acquisition, Writing – review & editing, Project administration, Visualization, Supervision. XY: Validation, Conceptualization, Project administration, Methodology, Supervision, Writing – original draft, Funding acquisition, Resources, Visualization, Writing – review & editing, Software.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the Innovation Program of Chinese Academy of Agricultural Sciences (No. CAAS-CSLPDCP-202403), Central Public-interest Scientific Institution Basal Research Fund (No. Y2025YC47), the Agricultural Science and Technology Innovation Program (ASTIP) (No. ASTIP-IAS-15).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1698057/full#supplementary-material
References
1. Pappas G, Papadimitriou P, Akritidis N, Christou L, and Tsianos EV. The new global map of human brucellosis. Lancet Infect Dis. (2006) 6:91–9. doi: 10.1016/S1473-3099(06)70382-6
2. Mesner O, Riesenberg K, Biliar N, Borstein E, Bouhnik L, Peled N, et al. The many faces of human-to-human transmission of brucellosis: congenital infection and outbreak of nosocomial disease related to an unrecognized clinical case. Clin Infect Diseases. (2007) 45:e135–40. doi: 10.1086/523726
3. Jin M, Fan Z, Gao R, Li X, Gao Z, and Wang Z. Research progress on complications of Brucellosis. Front Cell Infect Microbiol. (2023) 13:1136674. doi: 10.3389/fcimb.2023.1136674
4. Martirosyan A ME and Gorvel JP. An evolutionary strategy for a stealthy intracellular Brucella pathogen. Immunol Rev. (2011) 240:211–34. doi: 10.1111/j.1600-065X.2010.00982.x
5. Mebus CA and Dardiri AH. Additional characteristics of disease caused by the African swine fever viruses isolated from Brazil and the Dominican Republic. Proc Annu Meet U S Anim Health Assoc. (1979) 83:227–39.
6. Celli J. The intracellular life cycle of brucella spp. Microbiol Spectr. (2019) 7:10.1128. doi: 10.1128/microbiolspec.BAI-0006-2019
7. Głowacka P, Żakowska D, Naylor K, Niemcewicz M, and Bielawska-Drózd A. Brucella – virulence factors, pathogenesis and treatment. Polish J Microbiol. (2018) 67:151–61. doi: 10.21307/pjm-2018-029
8. Li Z, Wang S, Zhang H, Zhang J, Xi L, Zhang J, et al. Transcriptional regulator GntR of Brucella abortus regulates cytotoxicity, induces the secretion of inflammatory cytokines and affects expression of the type IV secretion system and quorum sensing system in macrophages. World J Microbiol Biotechnol. (2017) 33:60. doi: 10.1007/s11274-017-2230-9
9. Kohler S, Foulongne V, Ouahrani-Bettache S, Bourg G, Teyssier J, Ramuz M, et al. The analysis of the intramacrophagic virulome of Brucella suis deciphers the environment encountered by the pathogen inside the macrophage host cell. Proc Natl Acad Sci United States America. (2002) 99:15711–6. doi: 10.1073/pnas.232454299
10. Sola-Landa A, Pizarro-Cerdá J, Grilló MJ, Moreno E, Moriyón I, Blasco JM, et al. A two-component regulatory system playing a critical role in plant pathogens and endosymbionts is present in Brucella abortus and controls cell invasion and virulence. Mol Microbiol. (1998) 29:125–38. doi: 10.1046/j.1365-2958.1998.00913.x
11. Sternon JF, Godessart P, Gonçalves de Freitas R, Van der Henst M, Poncin K, Francis N, et al. Transposon sequencing of brucella abortus uncovers essential genes for growth in vitro and inside macrophages. Infect Immun. (2018) 86:e00312-18. doi: 10.1128/IAI.00312-18
12. Fontana C, Conde-Álvarez R, Ståhle J, Holst O, Iriarte M, Zhao Y, et al. Structural studies of lipopolysaccharide-defective mutants from brucella melitensis identify a core oligosaccharide critical in virulence. J Biol Chem. (2016) 291:7727–41. doi: 10.1074/jbc.M115.701540
13. Guzman-Verri C, Manterola L, Sola-Landa A, Parra A, Cloeckaert A, Garin J, et al. The two-component system BvrR/BvrS essential for Brucella abortus virulence regulates the expression of outer membrane proteins with counterparts in members of the Rhizobiaceae. Proc Natl Acad Sci U S A. (2002) 99:12375–80. doi: 10.1073/pnas.192439399
14. Altamirano-Silva P, Meza-Torres J, Castillo-Zeledón A, Ruiz-Villalobos N, Zuñiga-Pereira AM, Chacón-Díaz C, et al. Brucella abortus Senses the Intracellular Environment through the BvrR/BvrS Two-Component System, Which Allows B. abortus To Adapt to Its Replicative Niche. Infect Immun. (2018) 86:e00713-17. doi: 10.1128/IAI.00713-17
15. Bellefontaine AF, Pierreux CE, Mertens P, Vandenhaute J, Letesson JJ, and De Bolle X. Plasticity of a transcriptional regulation network among alpha-proteobacteria is supported by the identification of CtrA targets in Brucella abortus. Mol Microbiol. (2002) 43:945–60. doi: 10.1046/j.1365-2958.2002.02777.x
16. Francis N, Poncin K, Fioravanti A, Vassen V, Willemart K, Ong TA, et al. CtrA controls cell division and outer membrane composition of the pathogen Brucella abortus. Mol Microbiol. (2017) 103:780–97. doi: 10.1111/mmi.13589
17. Van der Henst C, Beaufay F, Mignolet J, Didembourg C, Colinet J, Hallet B, et al. The histidine kinase PdhS controls cell cycle progression of the pathogenic alphaproteobacterium Brucella abortus. J Bacteriol. (2012) 194:5305–14. doi: 10.1128/JB.00699-12
18. Willett JW, Herrou J, Briegel A, Rotskoff G, and Crosson S. Structural asymmetry in a conserved signaling system that regulates division, replication, and virulence of an intracellular pathogen. Proc Natl Acad Sci United States America. (2015) 112:E3709–3718. doi: 10.1073/pnas.1503118112
19. Dong H, Liu W, Peng X, Jing Z, and Wu Q. The effects of MucR on expression of type IV secretion system, quorum sensing system and stress responses in Brucella melitensis. Vet Microbiol. (2013) 166:535–42. doi: 10.1016/j.vetmic.2013.06.023
20. Mirabella A, Terwagne M, Zygmunt MS, Cloeckaert A, De Bolle X, and Letesson JJ. Brucella melitensis MucR, an orthologue of Sinorhizobium meliloti MucR, is involved in resistance to oxidative, detergent, and saline stresses and cell envelope modifications. J Bacteriol. (2013) 195:453–65. doi: 10.1128/JB.01336-12
21. Caswell CC, Elhassanny AE, Planchin EE, Roux CM, Weeks-Gorospe JN, Ficht TA, et al. Diverse genetic regulon of the virulence-associated transcriptional regulator MucR in Brucella abortus 2308. Infect Immun. (2013) 81:1040–51. doi: 10.1128/IAI.01097-12
22. Ariyama J, Suyama M, Ogawa K, Ikari T, Nagaiwa J, and Fujii D. New diagnostic systems--technics, efficiency and limitations. Cholangiography. a) ERCP. Nihon Rinsho. (1987) 45:1455-1457.
23. Hillerich B and Westpheling J. A new GntR family transcriptional regulator in streptomyces coelicolor is required for morphogenesis and antibiotic production and controls transcription of an ABC transporter in response to carbon source. J Bacteriol. (2006) 188:7477–87. doi: 10.1128/JB.00898-06
24. Rigali S, Nothaft H, Noens EE, Schlicht M, Colson S, Müller M, et al. The sugar phosphotransferase system of Streptomyces coelicolor is regulated by the GntR-family regulator DasR and links N-acetylglucosamine metabolism to the control of development. Mol Microbiol. (2006) 61:1237–51. doi: 10.1111/j.1365-2958.2006.05319.x
25. Li Z, Wang S, Han J, Shi X, Yang G, Cui Y, et al. Deletion of Brucella transcriptional regulator GntR10 regulated the expression of quorum sensing system and type IV secretion system effectors, which affected the activation of NF-κB. J Proteomics. (2023), 283–4. doi: 10.1016/j.jprot.2023.104938
26. Li Z, Li M, Zhang H, Wang S, Xi L, Zhang X, et al. ChIP-seq analysis of Brucella reveals transcriptional regulation of GntR. J basic Microbiol. (2020) 60:149–57. doi: 10.1002/jobm.201900458
27. Haine V, Sinon A, Van Steen F, Rousseau S, Dozot M, Lestrate P, et al. Systematic Targeted Mutagenesis ofBrucella melitensis16M Reveals a Major Role for GntR Regulators in the Control ofVirulence. Infection Immunity. (2005) 73:5578–86. doi: 10.1128/IAI.73.9.5578-5586.2005
28. Zhou D, Zhi FJ, Qi MZ, Bai FR, Zhang G, Li JM, et al. Brucella induces unfolded protein response and inflammatory response via GntR in alveolar macrophages. Oncotarget. (2018) 9:5184–96. doi: 10.18632/oncotarget.23706
29. Li ZQ, Zhang JL, Xi L, Yang GL, Wang SL, Zhang XG, et al. Deletion of the transcriptional regulator GntR down regulated the expression of Genes Related to Virulence and Conferred Protection against Wild-Type Brucella Challenge in BALB/c Mice. Mol Immunol. (2017) 92:99–105. doi: 10.1016/j.molimm.2017.10.011
30. Liu WJ, Dong H, Peng XW, and Wu QM. The Cyclic AMP-Binding Protein CbpB inBrucella melitensisand its role in cell envelope integrity, resistance to detergent and virulence. FEMS Microbiol Letters. (2014) 356:79–88. doi: 10.1111/1574-6968.12472
31. Sun D, Liu Y, Peng X, Dong H, Jiang H, Fan X, et al. ClpP protease modulates bacterial growth, stress response, and bacterial virulence in Brucella abortus. Veterinary Res. (2023) 54:68. doi: 10.1186/s13567-023-01200-x
32. Hornback ML and Roop RM 2nd. The Brucella abortus xthA-1 gene product participates in base excision repair and resistance to oxidative killing but is not required for wild-type virulence in the mouse model. J Bacteriol. (2006) 188:1295–300. doi: 10.1128/JB.188.4.1295-1300.2006
33. Yan X, Hu S, Yang Y, Xu D, Li H, Liu W, et al. The twin-arginine translocation system is important for stress resistance and virulence of brucella melitensis. Infect Immun. (2020) 88:e00389-20. doi: 10.1128/IAI.00389-20
34. Yuan Y, Ning W, Chen J, Li J, Xue T, An C, et al. Serine/threonine protein kinase mediates rifampicin resistance in Brucella melitensis through interacting with ribosomal protein RpsD and affecting antioxidant capacity. mSystems. (2025) 10:e0110924. doi: 10.1128/msystems.01109-24
35. Han K, Dong H, Peng X, Sun J, Jiang H, Feng Y, et al. Transcriptome and the gut microbiome analysis of the impacts of Brucella abortus oral infection in BALB/c mice. Microb Pathog. (2023) 183:106278. doi: 10.1016/j.micpath.2023.106278
36. Sun HL, Du XF, Tang YX, Li GQ, Yang SY, Wang LH, et al. Impact of immune checkpoint molecules on FoxP3(+) Treg cells and related cytokines in patients with acute and chronic brucellosis. BMC Infect diseases. (2021) 21:1025. doi: 10.1186/s12879-021-06730-3
37. Gupta S, Luxami V, and Paul K. Unlocking the Antibacterial Potential of Naphthalimide-Coumarins to Overcome Drug Resistance with Antibiofilm and Membrane Disruption Ability against Escherichia coli. ACS Appl materials interfaces. (2025) 17:4380–99. doi: 10.1021/acsami.4c13337
38. Hu R, Zhang Q, Wang W, Ren W, Yao M, Xu Y, et al. Brucella inactivated vaccine elicits immunity against B. melitensis infection in mice and Guinea pigs. Biomedicine pharmacotherapy = Biomedecine pharmacotherapie. (2025) 187:118077. doi: 10.1016/j.biopha.2025.118077
39. Glimcher LH and Murphy KM. Lineage commitment in the immune system: the T helper lymphocyte grows up. Genes Dev. (2000) 14:1693–711. doi: 10.1101/gad.14.14.1693
40. Golding B, Scott DE, Scharf O, Huang LY, Zaitseva M, Lapham C, et al. Immunity and protection against Brucella abortus. Microbes Infect. (2001) 3:43–8. doi: 10.1016/S1286-4579(00)01350-2
41. Lei S, Zhong Z, Ke Y, Yang M, Xu X, Ren H, et al. Deletion of the Small RNA Chaperone Protein Hfq down Regulates Genes Related to Virulence and Confers Protection against Wild-Type Brucella Challenge in Mice. Front Microbiol. (2015) 6:1570. doi: 10.3389/fmicb.2015.01570
42. Yu J, Li S, Wang L, Dong Z, Si L, Bao L, et al. Pathogenesis ofBrucellaepididymoorchitis-game ofBrucelladeath. Crit Rev Microbiol. (2021) 48:96–120. doi: 10.1080/1040841X.2021.1944055
43. Occhialini A, Hofreuter D, Ufermann CM, Al Dahouk S, and Köhler S. The retrospective on atypical brucella species leads to novel definitions. Microorganisms. (2022) 10:813. doi: 10.3390/microorganisms10040813
44. Zhi F, Fang J, Zheng W, Li J, Zhang G, Zhou D, et al. A brucella omp16 conditional deletion strain is attenuated in BALB/c mice. J Microbiol Biotechnol. (2022) 32:6–14. doi: 10.4014/jmb.2107.07016
45. Celli J, de Chastellier C, Franchini DM, Pizarro-Cerda J, Moreno E, and Gorvel JP. BrucellaEvades macrophage killing via virB-dependent sustained interactions with the endoplasmic reticulum. J Exp Med. (2003) 198:545–56. doi: 10.1084/jem.20030088
46. Comerci DJ, Martinez-Lorenzo MJ, Sieira R, Gorvel JP, and Ugalde RA. Essential role of the VirB machinery in the maturation of the Brucella abortus-containing vacuole. Cell Microbiol. (2001) 3:159–68. doi: 10.1046/j.1462-5822.2001.00102.x
47. Stallings CL and Glickman MS. Is Mycobacterium tuberculosis stressed out? A critical assessment of the genetic evidence. Microbes Infection. (2010) 12:1091–101. doi: 10.1016/j.micinf.2010.07.014
48. Rizvanovic A, Michaux C, Panza M, Iloglu Z, Helaine S, Wagner EGH, et al. The RNA-binding protein proQ promotes antibiotic persistence in salmonella. mBio. (2022) 13:e0289122. doi: 10.1128/mbio.02891-22
49. Tian M, Li Z, Qu J, Fang T, Yin Y, Zuo D, et al. The novel LysR-family transcriptional regulator BvtR is involved in the resistance of Brucella abortus to nitrosative stress, detergents and virulence through the genetic regulation of diverse pathways. Veterinary Microbiol. (2022) 267:109393. doi: 10.1016/j.vetmic.2022.109393
50. Liu W, Dong H, Li J, Ou Q, Lv Y, Wang X, et al. RNA-seq reveals the critical role of OtpR in regulating Brucella melitensis metabolism and virulence under acidic stress. Sci Rep. (2015) 5:10864. doi: 10.1038/srep10864
51. Guo J, Deng X, Zhang Y, Song S, Zhao T, Zhu D, et al. The Flagellar Transcriptional Regulator FtcR Controls Brucella melitensis 16M Biofilm Formation via a betI-Mediated Pathway in Response to Hyperosmotic Stress. Int J Mol Sci. (2022) 23:9905. doi: 10.3390/ijms23179905
52. Köhler S, Michaux-Charachon S, Porte F, Ramuz M, and Liautard JP. What is the nature of the replicative niche of a stealthy bug named Brucella? Trends Microbiol. (2003) 11:215–9. doi: 10.1016/s0966-842x(03)00078-7
53. Hanna N, Jiménez de Bagüés MP, Ouahrani-Bettache S, El Yakhlifi Z, Köhler S, and Occhialini A. The virB operon is essential for lethality of Brucella microti in the Balb/c murine model of infection. J Infect diseases. (2011) 203:1129–35. doi: 10.1093/infdis/jiq163
54. Porte F, Liautard JP, and Köhler S. Early acidification of phagosomes containing Brucella suis is essential for intracellular survival in murine macrophages. Infect Immun. (1999) 67:4041–7. doi: 10.1128/IAI.67.8.4041-4047.1999
55. Jiao H, Zhou Z, Li B, Xiao Y, Li M, Zeng H, et al. The mechanism of facultative intracellular parasitism of brucella. Int J Mol Sci. (2021) 22:3673. doi: 10.3390/ijms22073673
56. Zhang H, Sun T, Cao X, Wang Y, Ma Z, Wang Y, et al. Scanning iron response regulator binding sites using Dap-seq in the Brucella genome. PloS Negl Trop Dis. (2023) 17:e0011481. doi: 10.1371/journal.pntd.0011481
57. Marí M, de Gregorio E, de Dios C, Roca-Agujetas V, Cucarull B, Tutusaus A, et al. Mitochondrial glutathione: recent insights and role in disease. Antioxidants (Basel Switzerland). (2020) 9:909. doi: 10.3390/antiox9100909
58. Calabrese G, Morgan B, and Riemer J. Mitochondrial glutathione: regulation and functions. Antioxidants Redox Signaling. (2017) 27:1162–77. doi: 10.1089/ars.2017.7121
59. Lu SC. Glutathione synthesis. Biochim Biophys Acta. (2013) 1830:3143–53. doi: 10.1016/j.bbagen.2012.09.008
60. Giustarini D, Colombo G, Garavaglia ML, Astori E, Portinaro NM, Reggiani F, et al. Assessment of glutathione/glutathione disulphide ratio and S-glutathionylated proteins in human blood, solid tissues, and cultured cells. Free Radical Biol Med. (2017) 112:360–75. doi: 10.1016/j.freeradbiomed.2017.08.008
61. Qin Y, Zhou G, Jiao F, Cheng C, Meng C, Wang L, et al. Brucella mediates autophagy, inflammation, and apoptosis to escape host killing. Front Cell infection Microbiol. (2024) 14:1408407. doi: 10.3389/fcimb.2024.1408407
62. Liu Q, Han W, Sun C, Zhou L, Ma L, Lei L, et al. Deep sequencing-based expression transcriptional profiling changes during Brucella infection. Microbial Pathogenesis. (2012) 52:267–77. doi: 10.1016/j.micpath.2012.02.001
63. Hu H, Tian M, Li P, Guan X, Lian Z, Yin Y, et al. Brucella infection regulates thioredoxin-interacting protein expression to facilitate intracellular survival by reducing the production of nitric oxide and reactive oxygen species. J Immunol. (2020) 204:632–43. doi: 10.4049/jimmunol.1801550
64. Hu H, Tian M, Li P, Bao Y, Guan X, Lian Z, et al. Brucella infection regulates peroxiredoxin-5 protein expression to facilitate intracellular survival by reducing the production of nitric oxide and reactive oxygen species. Biochem Biophys Res Commun. (2019) 516:82–8. doi: 10.1016/j.bbrc.2019.06.026
65. Tang C, Ayala JC, and Shafer WM. Transcriptional regulation of a gonococcal gene encoding a virulence factor (L-lactate permease). PloS Pathogens. (2019) 15:e1008233. doi: 10.1371/journal.ppat.1008233
66. Skendros P and Boura P. Immunity to brucellosis. Rev scientifique technique (International Office Epizootics). (2013) 32:137–47. doi: 10.20506/rst.32.1.2190
67. Pereira CR, Cotrim de Almeida JVF, Cardoso de Oliveira IR, Faria de Oliveira L, Pereira LJ, Zangerônimo MG, et al. Occupational exposure to Brucella spp.: A systematic review and meta-analysis. PloS Negl Trop Dis. (2020) 14:e0008164. doi: 10.1371/journal.pntd.0008164
68. Perkins SD, Smither SJ, and Atkins HS. Towards a Brucella vaccine for humans. FEMS Microbiol Rev. (2010) 34:379–94. doi: 10.1111/j.1574-6976.2010.00211.x
69. de Figueiredo P, Ficht TA, Rice-Ficht A, Rossetti CA, and Adams LG. Pathogenesis and immunobiology of brucellosis: review of Brucella-host interactions. Am J pathology. (2015) 185:1505–17. doi: 10.1016/j.ajpath.2015.03.003
70. Yoo JS, Sasaki M, Cho SX, Kasuga Y, Zhu B, Ouda R, et al. SARS-CoV-2 inhibits induction of the MHC class I pathway by targeting the STAT1-IRF1-NLRC5 axis. Nat Commun. (2021) 12:6602. doi: 10.1038/s41467-021-26910-8
71. Dunn GP, Koebel CM, and Schreiber RD. Interferons, immunity and cancer immunoediting. Nat Rev Immunol. (2006) 6:836–48. doi: 10.1038/nri1961
72. Kobayashi KS and van den Elsen PJ. NLRC5: a key regulator of MHC class I-dependent immune responses. Nat Rev Immunol. (2012) 12:813–20. doi: 10.1038/nri3339
73. Abualrous ET, Saini SK, Ramnarayan VR, Ilca FT, Zacharias M, Springer S, et al. The carboxy terminus of the ligand peptide determines the stability of the MHC class I molecule H-2Kb: A combined molecular dynamics and experimental study. PloS One. (2015) 10:e0135421. doi: 10.1371/journal.pone.0135421
Keywords: Brucella abortus, GntR transcription factors, clpP gene, virulence, oxidative stress
Citation: Li S, Han K, Peng X, Wang N, Ning W, Ge S, Xu L, Ding J, Zhang X and Yang X (2025) Deletion of the GntR8 transcriptional regulator impairs Brucella abortus intracellular survival and virulence by modulating stress response genes. Front. Immunol. 16:1698057. doi: 10.3389/fimmu.2025.1698057
Received: 03 September 2025; Accepted: 09 October 2025;
Published: 29 October 2025.
Edited by:
Maryam Dadar, Razi Vaccine and Serum Research Institute, IranReviewed by:
M. Victoria Delpino, CONICET Instituto de Investigaciones Biomédicas en Retrovirus y SIDA (INBIRS), ArgentinaManuel Rodrigo Flores-Concha, University of Concepción, Chile
Shanhu Li, Beijing Institute of Biotechnology, China
Copyright © 2025 Li, Han, Peng, Wang, Ning, Ge, Xu, Ding, Zhang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiabo Ding, ZGluZ2ppYWJvQDEyNi5jb20=; Xinyu Zhang, enh5QHl6dS5lZHUuY24=; Xiaowen Yang, eWFuZ3hpYW93ZW4wMUBjYWFzLmNu
†These authors have contributed equally to this work
 Shuwen Li1,2†
Shuwen Li1,2† Xiaowei Peng
Xiaowei Peng Nan Wang
Nan Wang Jiabo Ding
Jiabo Ding Xinyu Zhang
Xinyu Zhang Xiaowen Yang
Xiaowen Yang