- 1Department of Hepatobiliary and Pancreatic Surgery, The Affiliated Hospital of Qingdao University, Qingdao, China
- 2Qingdao Cancer Institute, Qingdao, China
- 3Qingdao Medical College, Qingdao University, Qingdao, China
- 4Noventi Biopharmaceuticals Co., Ltd, Shanghai, China
- 5Medical Research Department, Qingdao Hospital, University of Health and Rehabilitation Sciences (Qingdao Municipal Hospital), Qingdao, China
HER2 is a well-established oncogenic driver in breast, gastric, and other solid tumors. While HER2-targeted therapies such as trastuzumab and pertuzumab have improved clinical outcomes, resistance, particularly to trastuzumab, remains a major therapeutic challenge. Here, we engineered two IgG-VHH biparatopic antibodies (bpAbs), A9B5-Bs-5 and A9B5-Bs-7, incorporating an ECD I-binding nanobody A9B5 with the IgG scaffolds. These bpAbs target non-overlapping epitopes on the HER2 extracellular domain, promoting rapid receptor internalization and demonstrating superior antitumor activity compared to the trastuzumab and pertuzumab combination in trastuzumab-resistant tumor cells. Structural modeling suggests that both bpAbs engage HER2 in a trans-binding mode, leading to receptor clustering and interference with ligand-driven HER2 heterodimerization. These findings demonstrate that epitope-guided biparatopic antibody design can enhance HER2 downregulation and restore sensitivity to HER2-targeted therapy in vitro, providing a strategy for the development of next-generation receptor-targeted biologics.
1 Introduction
Human epidermal growth factor receptor 2 (HER2), a member of the epidermal growth factor receptor (EGFR or HER) family, is overexpressed in various solid tumors and is associated with aggressive disease and poor prognosis (1–4). HER2-targeted therapies have markedly improved outcomes in patients with HER2-positive cancers. Among these, the monoclonal antibodies trastuzumab and pertuzumab, in combination with chemotherapy, form a standard-of-care regimen that significantly prolongs survival (5–7). Trastuzumab binds to the extracellular domain (ECD) IV of HER2, inhibiting ligand-independent HER2-HER3 dimerization and mediating antibody-dependent cellular cytotoxicity (ADCC), while pertuzumab disrupts ligand-driven HER2 heterodimerization by targeting the dimerization arm (DA) of ECD II (8, 9). Nevertheless, both intrinsic and acquired resistance, particularly to trastuzumab, remain significant clinical obstacles (10–15).
Resistance to anti-HER2 therapies arises through diverse mechanisms, including HER2 mutations, compensatory signaling, receptor masking, downregulation, and intertumoral heterogeneity (10, 13, 16–21). These complexities highlight the need for therapeutic strategies capable of simultaneously blocking multiple signaling axes and addressing tumor heterogeneity. Bispecific antibodies (bsAbs), which bind two different antigens or epitopes, offer a promising approach (22, 23). Through combinatorial mechanisms of action (MOAs), bsAbs can enhance receptor blockade, promote internalization, and counteract escape pathways that limit the efficacy of monospecific antibodies (24–26). Biparatopic antibodies (bpAbs), a subclass of bsAbs that recognize non-overlapping epitopes on the same antigen, have demonstrated unique functional advantages. For instance, the IgG-like bpAb zanidatamab, which targets HER2 ECD II and ECD IV, induces receptor clustering and complement-dependent cytotoxicity (CDC), and has shown encouraging efficacy in early-stage clinical studies (27–32). These findings suggest that dual-epitope engagement can elicit distinct and therapeutically beneficial biological responses compared to parental monoclonals. However, clinically advanced anti-HER2 bpAbs predominantly focus on ECD II and IV, which may limit their effectiveness against tumors harboring mutations in these regions (33, 34). For instance, ECD II mutations such as S310F/Y, associated with drug resistance and metastasis, can abolish pertuzumab binding (35).
The dynamic structural attributes of HER2, particularly its capacity for dimerization, underlies both its oncogenic activity and its resistance to targeted therapies (36, 37). While the DA of ECD II mediates dimerization, recent structural studies of HER2-EGFR and HER2-HER3 complexes have identified a critical role for the ECD I-III interface in forming the binding pocket for dimerization partners (38, 39). Targeting ECD I or III, in combination with trastuzumab or pertuzumab, has shown synergistic efficacy in overcoming therapeutic resistance (40–48). We previously identified a high-affinity ECD I-binding nanobody A9B5, which exhibits potent synergy with trastuzumab in resistant models (49). These findings highlight the potential of bpAbs designed to target novel alternative epitopes to comprehensively block HER2-driven signaling.
Antibody format is another key determinant of bsAb function, stability, and developability. To date, over 50 bsAb formats have been developed (50). Among them, IgG-VHH fusions incorporating single domain (VHH) nanobodies are attractive due to their high solubility, stability, and reduced risk of mispairing (51, 52). In this study, we engineered and evaluated a panel of IgG-VHH bpAbs incorporating the A9B5 nanobody. Two lead candidates, A9B5-Bs-5 and A9B5-Bs-7, demonstrated potent HER2 internalization and antitumor efficacy superior to the combination of trastuzumab and pertuzumab in trastuzumab-resistant models.
2 Methods
2.1 Cell lines, antibodies, and biological material
NCI-N87, MCF-7, JIMT-1, and BT474 were sourced from ATCC. Expi 293 cells were purchased from Thermo Fisher Scientific. Trastuzumab and pertuzumab were produced in-house. A non-specific IgG antibody, which was obtained from Beyotime (A7001), was used as a negative control.
2.2 Construction of bpAbs and chimeric proteins
The sequences encoding HER2-targeting nanobodies were cloned into either the N- or C-terminus of the heavy or light chains of trastuzumab or pertuzumab within pSCSTa expression vectors. Vectors encoding nanobody-fused heavy chains and corresponding light chains, or nanobody-fused light chains and corresponding heavy chains, were transiently transfected into Expi293 cells using standard protocols. After 168 h of culture, the supernatant was harvested and purified by Protein A affinity chromatography using AT Protein A Diamond Plus resin (BestChrom, AA402305). The eluted antibodies were buffer-exchanged into PBS. Antibody concentrations were determined by BCA assay, and purity was assessed via SDS-PAGE.
Chimeric HER2-ECD proteins (HER2-mD1, HER2-mD2, HER2-mD3, and HER2-mD4) were constructed as previously described (49). Briefly, the extracellular domains I (T23–R217), II (T218–C342), III (Y344–A510), and IV (C511–T652) of the HER2 protein (UniProt P04626) were replaced with their respective murine homologous domains (UniProt P70424). DNA sequences encoding these chimeric proteins were cloned into a pSCSTa vector containing a C-terminal Fc tag and transiently transfected into 293T cells. Protein expression and purification were performed as described for the above antibodies.
2.3 Growth inhibition assays
Tumor cells were seeded into 96-well plates at approximately 2 × 103 cells per well. Plates were incubated overnight at 37°C with 5% CO2. The next day, antibodies were serially diluted using a 1:5 dilution series. Diluted antibodies were added to the wells and incubated with the cells for five days. Cell viability was assessed using the Cell Counting Kit-8 (CCK-8, Dojindo) according to the manufacturer’s protocol. After incubation, the supernatant was removed. Cells were then treated with fresh medium containing 10% (v/v) CCK-8 reagent. Plates were incubated at 37°C with 5% CO2 for 2 h, and then absorbance at 450 nm was recorded using a BioTek plate reader. For ligand-dependent assays, tumor cells were stimulated with either 1 nM HRG (SinoBiological, 11609-HNCH) or 5 nM EGF (SinoBiological, GMP-10605-HNAE). Cell viability was calculated as a percentage relative to untreated control cells. Background values were subtracted. Dose–response curves were fitted using the “log(inhibitor) vs. response—variable slope (four parameters)” model in GraphPad Prism. EC50 values were calculated based on curve fitting. Statistical significance was tested using one-way or 2-way ANOVA. Comparisons were made between antibody-treated groups at either 150 nM or 30 nM. The corresponding p-values were reported.
2.4 Size-exclusion chromatography (HPSEC)
Samples of anti-HER2 agents were loaded onto a TSK Gel G 3000 pwxl (7.8 × 300 mm; TOSOH, Tokyo, Japan) equilibrated with PBS. Proteins separation was performed using Agilent 1200 series system (Agilent Technologies, Santa Clara, CA). The flow rate was set to 0.5 mL per minute. Elution was monitored by UV absorbance at 280 nm.
2.5 Enzyme-linked immunosorbent assay
To identify the binding epitopes of HER2-targeting bpAbs, 96-well plates were coated with either wild-type or chimeric HER2-ECD proteins (100 ng per well) in PBS and incubated overnight at 4 °C. Plates were subsequently blocked with 2% NFDM at room temperature for 2 h. Threefold serial dilutions of bpAbs, anti-HER2 nanobodies (His-tag), or other anti-HER2 antibodies were added and incubated at room temperature for 1 h. For detection, HRP-conjugated His-Tag monoclonal antibody (Proteintech, HRP-66005) was used for nanobodies, and the HRP-conjugated mouse anti-human IgG Fc antibody (GenScript, A01854) was used for IgG-based antibodies. After a 30-minute incubation, the wells were washed, and TMB substrate was added for color reaction. The reaction was quenched with 1M H3PO4, and the absorbance at 450 nm (reference 620 nm) was measured using an automated ELISA reader (BioTek). Data was analyzed using GraphPad Prism 10.2.0. Curve fitting was performed by nonlinear regression using the “log(agonist) vs. response - variable slope (four parameters)” model to determine the EC50 value.
2.6 Internalization by fluorescence activated cell sorter
NCI-N87 or BT474 cells in exponential growth were harvested and resuspended using PBS + 2% FBS. Cell viability was assessed using an automated cell counter (Countstar BioTech). 3-5 × 104 viable cells per well were seeded in 96-well plates. Cells were incubated with anti-HER2 antibodies on ice for 1 h, followed by two washes with PBS to remove unbound antibodies. An aliquot of cells was maintained on ice, while the rest were incubated at 37°C for 0.5 or 4 h. Following incubation, cells were washed and fixed with 4% paraformaldehyde (PFA) for 20 min at room temperature. Cells were then stained with PE-conjugated anti-human IgG Fc antibody (Abcam, 98596; 1:1000 dilution) at 4°C for 30 min. The Beckman Coulter flow cytometer was used to measure median fluorescence intensity (MFI) in the PE-channel. The antibody-receptor complexes internalization was calculated as the percentage loss of MFI at 37°C relative to the signal measured on ice. Data was analyzed using GraphPad Prism v10.2.0.
2.7 Internalization by confocal imaging
NCI-N87 or MCF-7 cells in exponential growth were seeded onto coverslips and cultured overnight at 37°C/5% CO2. Cells were treated with 50 nM antibodies at 4°C for 1 h. Unbound antibodies were removed by washing with PBS supplemented with 2% FBS, followed by incubation at 37°C for 4 h. After washing, cells were fixed with 4% PFA and permeabilized with Triton X-100. Blocking was performed in PBS containing 10% goat serum (Solarbio, SL038) at room temperature for 1 h. Cells were stained with DyLight® 488-conjugated goat anti-human IgG Fc antibody (green; Abcam, ab98619) to visualize antibody-receptor complexes, and with anti-LAMP1 antibody (Abcam, ab25630), followed by Alexa-Fluor 647-labeled goat anti-mouse IgG H&L (magenta; Abcam, ab150115) to label lysosomes. The 4′,6-diamidino-2-phenylindole (DAPI, blue; Beyotime, C1002) was used for nuclear staining. Coverslips were treated with an antifade mounting medium (Beyotime, P0126). Confocal imaging was performed using a Nikon A1 confocal microscope and analyzed with NIS-Elements Viewer software. Fluorescence intensity from three channels was quantified using ImageJ. Mean intensity values were analyzed using GraphPad Prism v10.2.0.
2.8 HER2 binding by fluorescence activated cell sorter
FACS was performed on HER2-expressing tumor cell lines with differential responses to trastuzumab: NCI-N87 (trastuzumab-sensitive), MCF-7 (trastuzumab-resistant), and JIMT-1 (trastuzumab-resistant). Tumor cells in the exponential growth were harvested and resuspended in PBS supplemented with 2% FBS. A total of 1 × 105 cells per well were incubated with diluted anti-HER2 antibodies for 1 h on ice. Then, cells were washed and stained with PE-conjugated anti-human IgG Fc secondary antibody (Abcam, 98596; 1:1000). MFI in the PE-channel was measured using a Beckman Coulter flow cytometer. Binding data was analyzed using GraphPad Prism 10.2.0.
2.9 Structure modeling
The structure of A9B5-HER2 complex was predicted using Alphafold 3 (https://alphafoldserver.com/) following the server’s guidelines (53). Briefly, the amino acid sequences of the A9B5 and HER2-ECD were submitted, and structure prediction was performed using default parameters. Structural models were visualized using PyMOL and Chimera. The A9B5-HER2 interface was analyzed using the PISA server (www.ebi.ac.uk/pdbe/pisa).
2.10 Statistics and reproducibility
All graphs and statistical analyses were performed using Excel or GraphPad Prism 10.2.0. Specific tests are described in the relevant method (see above) or figure legends. Quantitative data were analyzed using one-way ANOVA or 2-way ANOVA, as appropriate. Significance thresholds were defined as follows: p < 0.0332 (*), p < 0.0021 (**), p < 0.0002 (***), and p < 0.0001 (****).
3 Results
3.1 Identification of bpAbs with potent antitumor activity in HER2-positive tumor cells
BpAbs that bind non-overlapping epitopes on the same molecular target have been reported to exhibit unique functionalities and superior antitumor activity compared with combinations of conventional monospecific antibodies (30, 34, 54–57). Among various bpAbs formats, IgG-VHH fusion - where small (~15 kDa), monomeric VHH domains are fused to IgG scaffolds - represents a highly attractive way for generating stable and functional bpAbs (58–61). To generate novel HER2-targeting bpAbs with favorable developability and manufacturability, high-affinity anti-HER2 nanobodies were employed as antigen-binding partners in constructing IgG-VHH bpAbs. Four nanobodies targeting distinct epitopes of HER2-ECD were previously identified: A2G5 and A9B5 target ECD I, H2F5 targets ECD II, and G1E4 targets ECD IV (Figure 1A). These nanobodies demonstrated synergistic inhibitory activity in HER2-expressing tumor cells with acquired resistance (49). Here, we designed eight IgG-VHH bpAbs formats by fusing anti-HER2 nanobodies to the IgG scaffold of trastuzumab or pertuzumab (Figure 1B). Nanobodies were fused to different positions of trastuzumab or pertuzumab: the N-terminus (Bs-1, Bs-3) or C-terminus (Bs-2, Bs-4) of the heavy chain; the N-terminus (Bs-5, Bs-7) or C-terminus (Bs-6, Bs-8) of the light chain. A total of 32 IgG-VHH bpAbs were constructed, expressed in Expi293 cells, and purified using standard protocols. All bpAbs exhibited high purity, indicating favorable manufacturability (Figure 1C).
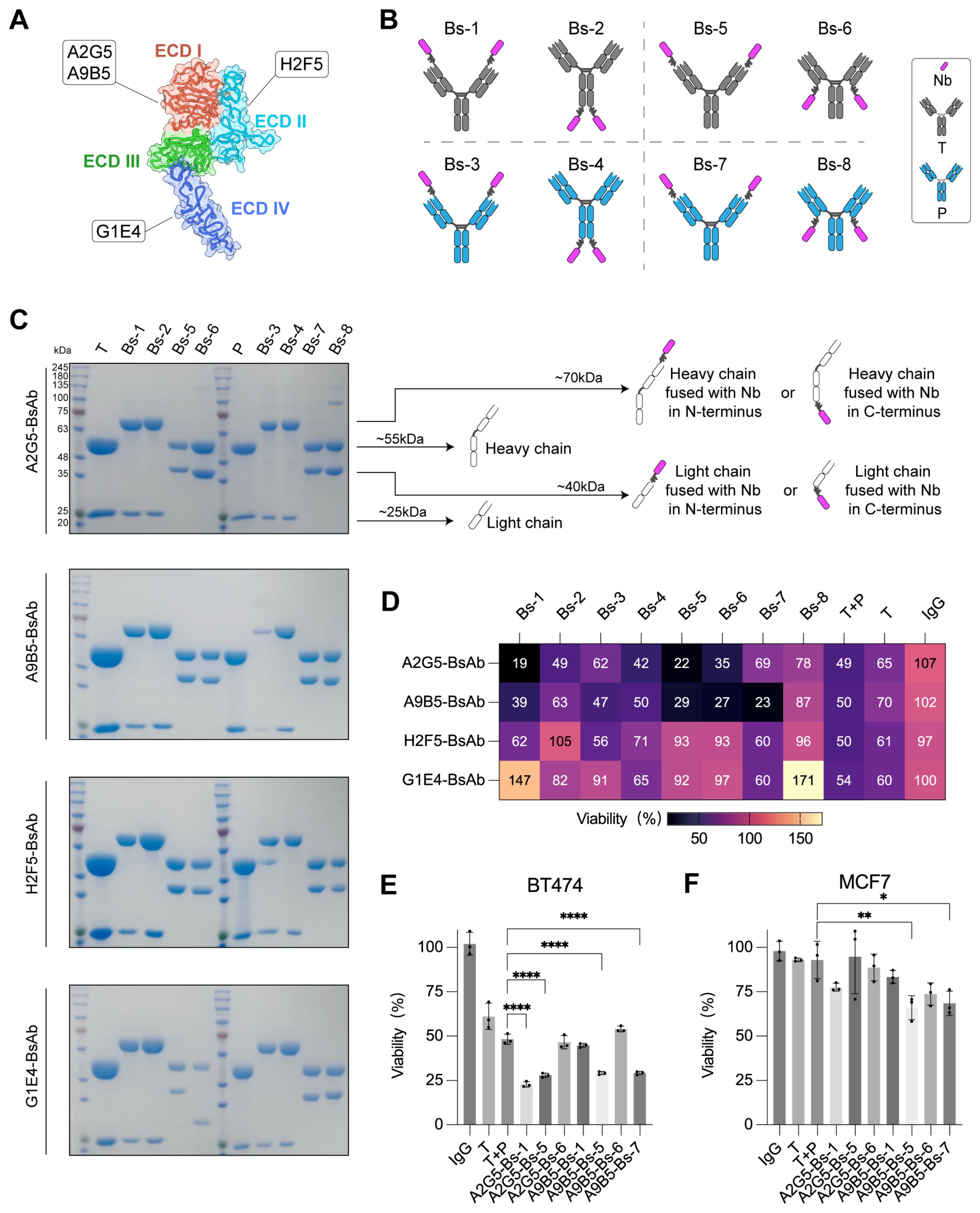
Figure 1. Screening of IgG-VHH bpAbs identifies A9B5-Bs-5 and A9B5-Bs-7 as lead candidates with potent activity in NCI-N87 cells. (A) Binding epitopes of high-affinity anti-HER2 nanobodies mapped onto the crystal structure of HER2-ECD monomer (PDB ID: 1N8Z). A2G5 and A9B5 recognized ECD I; H2F5 targeted ECD II; G1E4 recognized ECD IV. Functional characterization of these nanobodies was described previously (49). (B) Schematic representation of eight bpAb formats. Nanobodies were fused to different positions of trastuzumab (T) or pertuzumab (P) by (GGGGS)3 linker: the N-terminus (Bs-1, Bs-3) or C-terminus (Bs-2, Bs-4) of the heavy chain; the N-terminus (Bs-5, Bs-7) or C-terminus (Bs-6, Bs-8) of the light chain. (C) Coomassie-stained SDS-PAGE gel showing high purity of purified biparatopic antibodies. (D) The inhibitory effect of bpAbs in NCI-N87 cells (mean of n = 3). (E, F) Growth inhibition data in BT474 (E) and MCF7 (F) cell lines treated with bpAbs (n = 3, mean ± SD). Statistical significance was determined by one-way ANOVA: p < 0.0332 (*), p < 0.0021 (**), p < 0.0002 (***), and p < 0.0001 (****). The p-values were provided in Supplementary Table S1. Source data are available in the Source Data file.
BpAbs have been reported to exhibit enhanced antigen affinity and potent inhibitory effect by engaging two epitopes (44, 45). To assess whether these tetravalent IgG-VHH bpAbs benefit from this design, growth inhibition was evaluated in NCI-N87 cells, which express a high level of HER2. BpAbs incorporating ECD I-binding nanobodies (A2G5 and A9B5) demonstrated stronger inhibitory activity than the combination of trastuzumab and pertuzumab (T + P) (Figure 1D). In contrast, bpAbs containing ECD II-binding nanobody H2F5 exhibited weaker effects, while those incorporating ECD IV-binding nanobody G1E4 showed agonistic behavior, suggesting that certain configurations may activate HER2 signaling (Figure 1D). The seven bpAbs with the strongest growth-inhibitory effects in NCI-N87 cells were selected for subsequent functional screening.
BT474 cells (high HER2 expression) and MCF7 (trastuzumab-resistant) cells were used to identify the most effective bpAbs. In BT474 proliferation assays, A2G5-Bs-1, A2G5-Bs-5, A9B5-Bs-5, and A9B5-Bs-7 demonstrated higher inhibitory activity than T + P (Figure 1E and Supplementary Table S1). In MCF7 proliferation assays, trastuzumab and T + P exhibited minimal effect on tumor growth, whereas A9B5-Bs-5 and A9B5-Bs-7 mediated significantly greater inhibition compared with T + P (Figure 1F and Supplementary Table S1). Taken together, A9B5-Bs-5 (bpAb fusing ECD I-binding nanobody to the N-termini of the trastuzumab light chain) and A9B5-Bs-7 (bpAb fusing ECD I-binding nanobody to the N-termini of the pertuzumab light chain) demonstrated significantly greater antitumor effects than T + P across all three cell lines. Therefore, A9B5-Bs-5 and A9B5-Bs-7 were selected for further experiments.
3.2 BpAbs bind HER2 with high saturation via recognizing non-overlapping epitopes
To define how A9B5-Bs-5 and A9B5-Bs-7 engage HER2 molecules, we assessed their homogeneity, epitope specificity, and cell-surface binding capacity. Analytical HPSEC showed that both bpAbs eluted as single major peaks with retention times (RT) of 7.4 min, confirming molecular homogeneity and minimal aggregation (Figure 2A). To determine whether the bpAbs could bind two non-overlapping epitopes, we generated a panel of chimeric HER2-ECD proteins in which each human domain (I–IV) was individually replaced by its murine counterpart, as described previously (49) (Figure 2B). Because HER2-mD3 expression was undetectable, only HER2-mD1, HER2-mD2, and HER2-mD4 were included in ELISA analysis. Both A9B5-Bs-5 and A9B5-Bs-7 retained specificity to human HER2 and showed negligible binding to murine HER2 (Figure 2C). The binding of either bpAb was largely maintained across the domain−swap panel, whereas trastuzumab lost binding to HER2−mD4 and pertuzumab lost binding to HER2−mD2 (Figure 2C). ELISA-derived EC50 shifts indicated that A9B5-Bs-5 recognizes the epitopes spanning ECD I and ECD IV, whereas A9B5-Bs-7 recognizes ECD I and ECD II (Figure 2D).
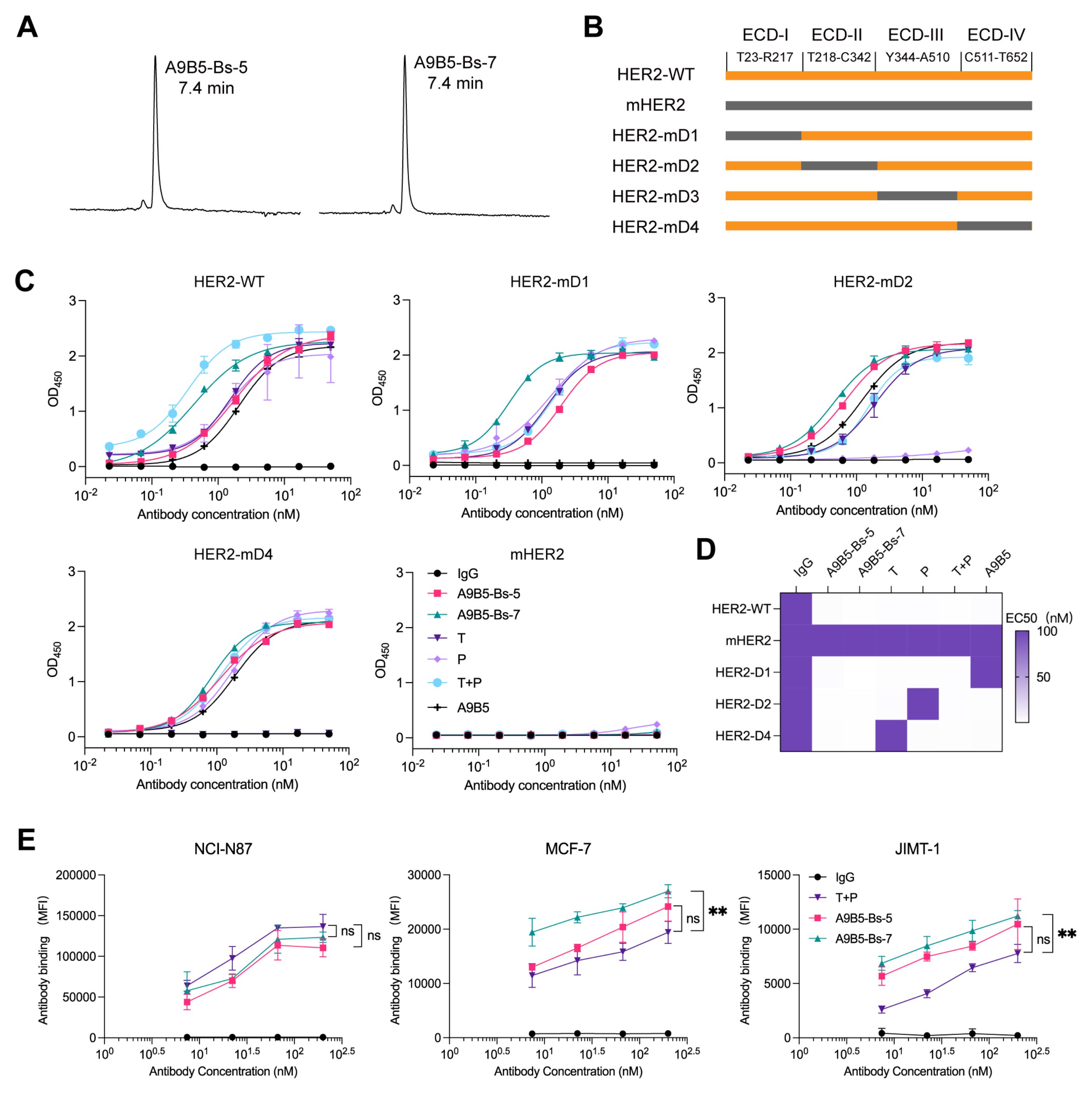
Figure 2. A9B5-Bs-5 and A9B5-Bs-7 bind HER2 with high antibody saturation via recognition of non-overlapping epitopes. (A) HPSEC profiles of bpAbs indicating molecular homogeneity. The retention time (RT) was labeled. (B) Constructions of chimeric HER2-ECD proteins. Human HER2 domains—ECD I (T23–R217), ECD II (T218–C342), ECD III (Y344–A510), and ECD IV (C511–T652)—were individually replaced with their murine homologs. (C) Binding activity of bpAbs against chimeric HER2-ECD proteins. (D) Epitope mapping of bpAbs based on the EC50 values from ELISA analysis. The EC50 values (nM) were shown as a heat map. (E) Antibody binding saturation across different HER2-positive tumor cells, measured by FACS. Data represent mean ± SD (n = 3). Statistical significance was determined using one-way ANOVA: p < 0.0332 (*), p < 0.0021 (**), p < 0.0002 (***), and p < 0.0001 (****). The p-values were provided in Supplementary Table S2. Source data are available in the Source Data file.
BpAbs such as zanidatamab can achieve higher cell-surface binding saturation than canonical monospecific antibodies owing to multivalent engagement (30). We next used flow cytometry to assess cell-surface binding capacity across HER2-positive tumor models, including NCI-N87 (high HER2 expression), MCF7 (trastuzumab-resistant), and JIMT-1 (trastuzumab-resistant) cells. In NCI-N87 assays, A9B5-Bs-5 and A9B5-Bs-7 achieved antibody saturation comparable to T + P (Figure 2E and Supplementary Table S2). In trastuzumab-resistant MCF7 and JIMT-1 cells, A9B5-Bs-7 showed 1.4-fold higher maximal binding than T + P, whereas A9B5-Bs-5 demonstrated comparable antibody saturation to T + P (Figure 2E and Supplementary Table S2). These results support a model in which A9B5-Bs-5 and A9B5-Bs-7 co-engage non-overlapping HER2 epitopes and achieve superior saturation, likely enabling enhanced receptor internalization and antitumor activity.
3.3 BpAbs promote rapid internalization of HER2 receptors
Receptor clustering at the cell surface induced by bpAbs has been reported to accelerate rapid receptor internalization, suppress recycling, and promote lysosomal degradation (30, 45, 62). To test whether A9B5-Bs-5 and A9B5-Bs-7 enhance HER2 internalization, we treated BT474 and NCI-N87 cells with anti-HER2 antibodies and quantified cell-surface HER2 by FACS over 0.5–4 h. In NCI-N87 cells, trastuzumab, pertuzumab, and their combination (T + P) showed limited internalization (< 30%), whereas both bpAbs drove rapid and substantially greater HER2 loss from the surface (Figure 3A and Supplementary Table S3). In BT474 cells, trastuzumab, pertuzumab, and T+P induced little to no internalization, whereas both bpAbs again triggered markedly greater HER2 loss (Figure 3B and Supplementary Table S3). A9B5-Bs-5 mediated more pronounced internalization than A9B5-Bs-7.
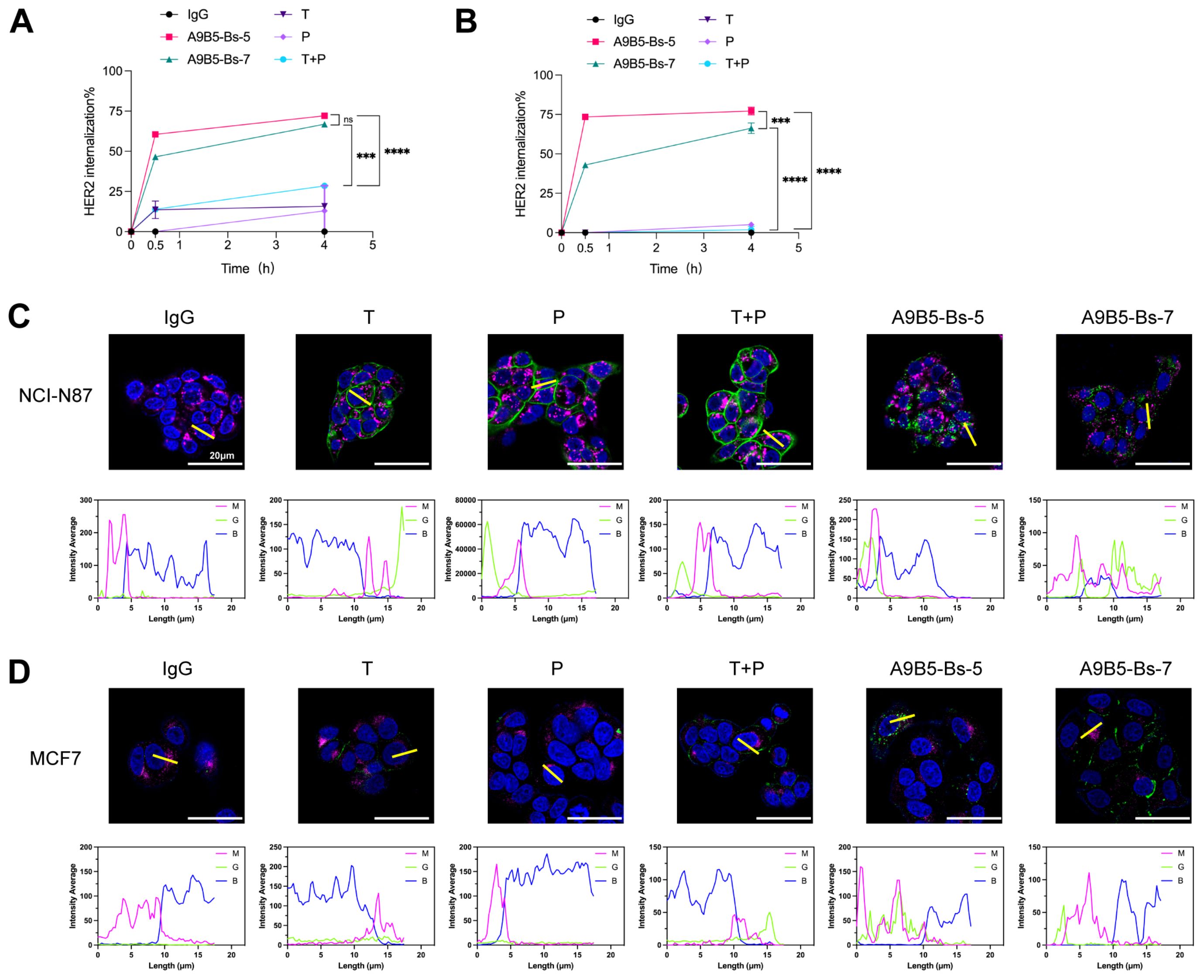
Figure 3. A9B5-Bs-5 and A9B5-Bs-7 facilitate rapid HER2 internalization. (A, B) HER2 internalization in NCI-N87 (A) and BT474 (B) cells following treatment with bpAbs, monospecific antibodies (trastuzumab or pertuzumab), or the trastuzumab + pertuzumab combination (T + P). Cells were incubated with antibodies for 0.5 or 4 h. Internalization was quantified by flow cytometry and displayed as mean percentage ± SD (n=3). Statistical significance was determined using one-way ANOVA: p < 0.0332 (*), p < 0.0021 (**), p < 0.0002 (***), and p < 0.0001 (****). The p-values were provided in Supplementary Table S3. (C, D) Confocal microscopy studies of antibody-HER2 complexes in NCI-N87 (C) or MCF7 (D) cells at 4 h post-treatment. Antibody-HER2 complexes were visualized using anti-human Fc staining (green), lysosomes by LAMP1 (magenta), and the nucleus by DAPI (blue). Scale bars, 20 μm. Fluorescence intensity from magenta (M), green (G), and blue (B) channels was quantified using ImageJ to assess colocalization of internalized complexes with lysosomes. Source data are available in the Source Data file.
We next used confocal microscopy to visualize the intracellular trafficking in two cell lines. In NCI-N87 cells, single antibodies (trastuzumab or pertuzumab) remained evenly distributed at the cell membrane, and T + P formed some surface clusters and regions of continuous membrane staining (Figure 3C). In contrast, A9B5-Bs-5 and A9B5-Bs-7 formed large intracellular complexes that co-localized with lysosomes (Figure 3C). In MCF7 cells (low HER2 expression), trastuzumab, pertuzumab, or their combination showed weak surface-binding with minimal internalization (Figure 3D). A9B5-Bs-7 yielded enhanced surface-binding with moderate internalization, whereas A9B5-Bs-5 showed minimal surface staining yet pronounced internalization with strong co-stain to the lysosomal marker LAMP1. Taken together, these data demonstrated that both bpAbs induce faster and more extensive HER2 internalization than trastuzumab, pertuzumab, and their combination, and that the magnitude of bpAb−driven internalization depends on HER2 expression level.
3.4 BpAbs suppress the growth of trastuzumab-resistant tumor cells
We next assessed whether the high binding saturation and HER2 internalization conferred by bpAbs translate into anti-proliferative effects in HER2-expressing cell lines, particularly trastuzumab-resistant tumor cells. We first examined ligand-independent antitumor activity by evaluating tumor cell viability after anti-HER2 antibody treatment. Both bpAbs caused concentration-dependent reduction in viability in NCI-N87 and BT474 cells (Figures 4A, B). A9B5-Bs-7 was more potent (EC50 < 1 nM) than trastuzumab, pertuzumab, and their combination (T + P) in two cell lines (Figure 4E). A9B5-Bs-5 displayed comparable or superior activity to T + P or single antibodies (Figure 4E). Notably, A9B5-Bs-5 exerted significantly stronger inhibition (EC50 < 1 nM) than trastuzumab, pertuzumab, and T + P in NCI-N87 cells, whereas in BT474 cells its effect was weaker than T + P but comparable with either single antibody (Figures 4B, E).
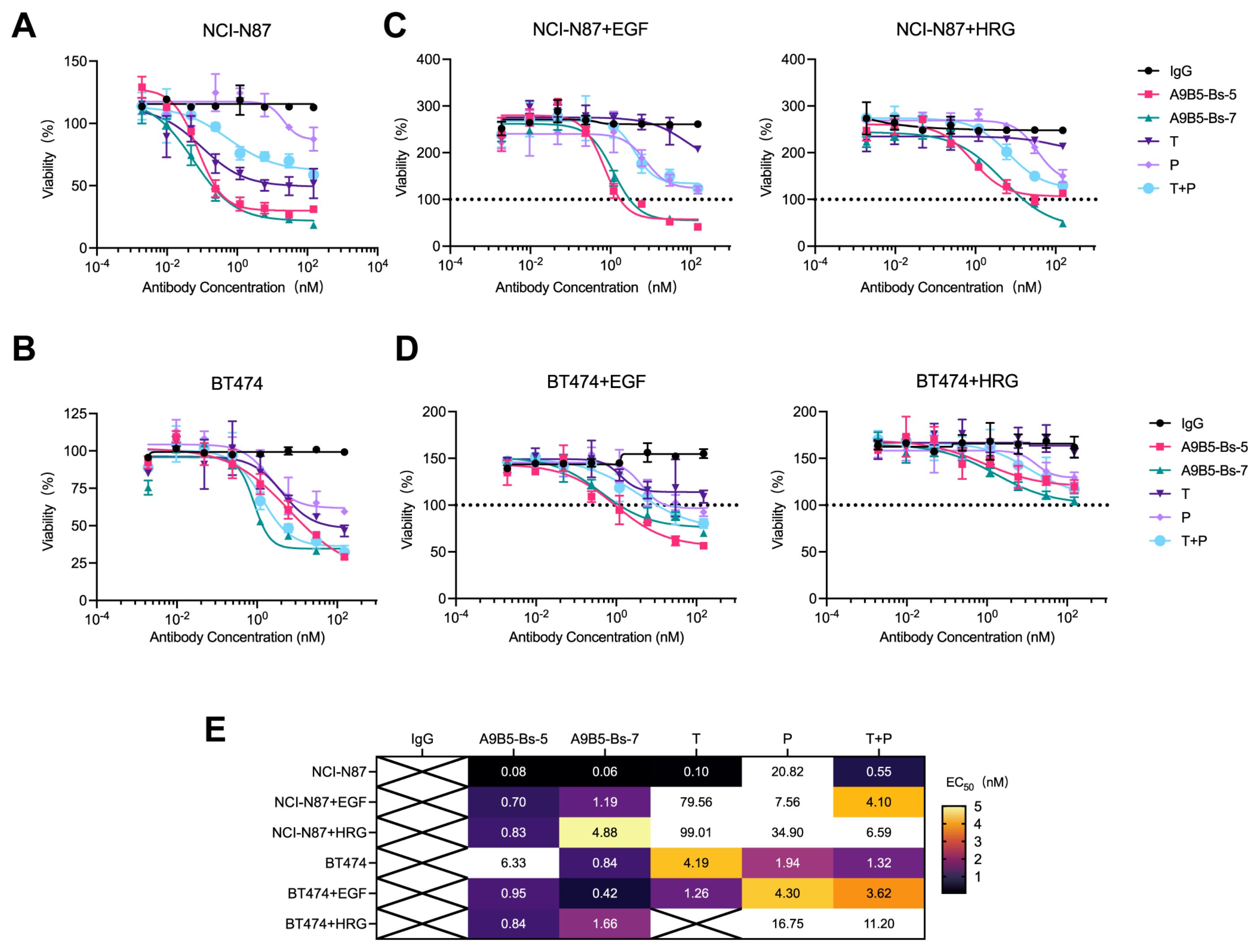
Figure 4. A9B5-Bs-5 and A9B5-Bs-7 inhibit the growth of trastuzumab-resistant HER2-positive cancer cells. (A, B) Growth inhibition in NCI-N87 (A) and BT474 cells (B) treated with bpAbs in the absence of ligands, measured by CCK-8 assay. (C, D) Growth inhibition in NCI-N87 (C) and BT474 cells (D) treated with bpAbs in the presence of EGF or HRG, measured by CCK-8 assay. Serum-starved cells were incubated with anti-HER2 diluted antibodies in the presence of 1 nM HRG or 5 nM EGF. Horizontal dotted line (black) represents viability of non-treated cells referenced to 100%. Data represent mean ± SD (n = 3). (E) Heat map of EC50 values (nM) derived from CCK-8 assays showing dose-dependent inhibition by bpAbs. Source data are available in the Source Data file.
We next evaluated antitumor activity under epidermal growth factor (EGF)- and heregulin (HRG)-driven growth conditions. Ligand-driven activation of HER2-EGFR and HER2-HER3 heterodimers is known to contribute to trastuzumab resistance (13, 63). As expected, the ECD IV-targeting antibody trastuzumab exhibited limited efficacy under these conditions (Figures 4C, D). The ECD II-targeting antibody pertuzumab, which disrupts HER2 heterodimerizations, showed stronger inhibition than trastuzumab in ligand-stimulated cells. Consistent with previous findings, T + P provided greater growth suppression than either single antibody. Both bpAbs, A9B5-Bs-5 and A9B5-Bs-7, achieved markedly stronger ligand-driven inhibition than trastuzumab, pertuzumab, and T + P (Figures 4C, D). A9B5-Bs-5 was particularly active, with EC50 values below 1 nM under both EGF and HRG stimulation in both cell lines (Figure 4E). In summary, the bpAbs mediate potent inhibition of ligand-independent and ligand-driven growth in HER2-expressing cell lines. In ligand−stimulated, trastuzumab−resistant cells, their efficacy exceeded that of T + P (Figure 4E).
3.5 Structural modeling supports a trans-binding mode and HER2 clustering mechanism
To define how the engineered biparatopic formats engage HER2, we modeled the nanobody A9B5 in complex with HER2-ECD using AlphaFold 3. The top−ranked model positioned A9B5 across a composite surface spanning ECD I and ECD II (Figure 5A). Interface residues were analyzed with the PISA server, which suggested six putative hydrogen−bonding contacts. Representative interactions involved three residues (E210, S214, and L215) on ECD I and two residues (K228 and D234) on ECD II (Figure 5B and Supplementary Table S4). The cross−subdomain footprint indicates that A9B5 recognizes a surface bridging ECD I and ECD II, potentially constraining local interdomain flexibility.
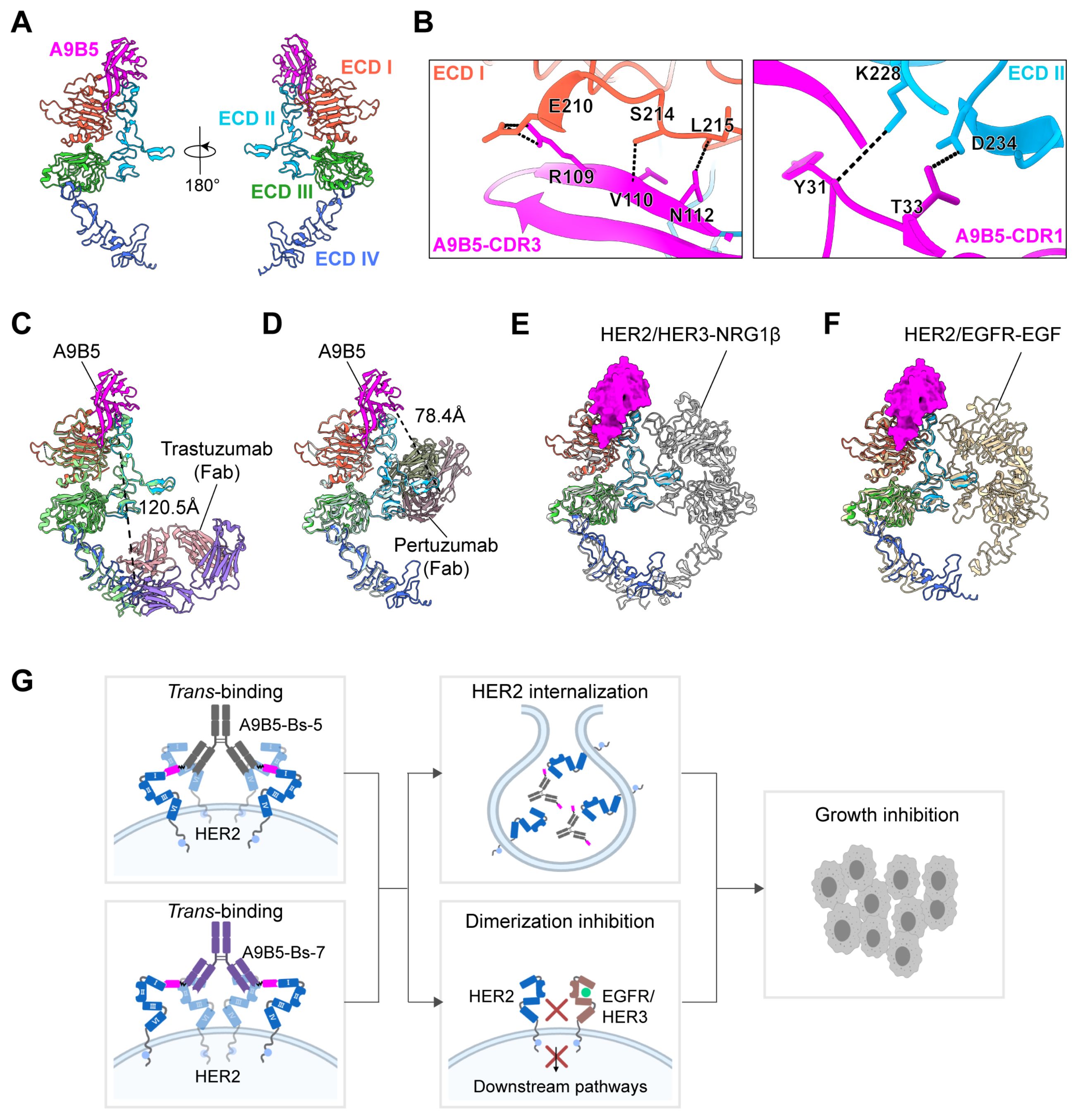
Figure 5. Structural modeling of HER2-ECD complexed with bpAbs supports a trans-binding mechanism. (A) Predicted structure of A9B5-HER2 complex generated by Alphafold 3. Nanobody A9B5 was shown in magenta. The ECD I-IV of HER2 were colored in tomato, deep sky blue, lime green, and royal blue, respectively. The predicted structure is available in the Source Data file. (B) Interface analysis of the A9B5-HER2 complex. Potential hydrogen bonds (< 4 Å) were indicated by black dashed lines. The detailed interaction sites were provided in Supplementary Table S4. (C) Structural alignment of the A9B5-HER2 complex with the trastuzumab-HER2 complex (PDB ID: 1N8Z). Trastuzumab Fab was shown in medium purple (light chain) and light pink (heavy chain). Dashed black lines indicate the distance between the C-terminus of A9B5 and N-terminus of trastuzumab light chain. (D) Structural alignment of the A9B5-HER2 complex with the pertuzumab-HER2 complex (PDB ID: 1S78). Dashed black lines indicate the distance between the C-terminus of A9B5 and the N-terminus of the pertuzumab light chain. (E) Superposition of the A9B5-HER2 complex and the HER2-HER3-NRG1β dimer complex (PDB ID: 7MN5). The HER2-HER3-NRG1β dimer complex was shown in light gray. (F) Superposition of the A9B5-HER2 complex and the HER2-EGFR-EGF dimer complex (PDB ID: 8HGO). The HER2-EGFR-EGF dimer complex was shown in wheat. (G) Schematic model illustrating the proposed trans-binding mechanism. A9B5-Bs-5 and A9B5-Bs-7 may promote HER2 clustering by engaging HER2 molecules on adjacent receptors, thereby facilitating receptor internalization and suppressing HER2-driven heterodimerization. The image was created with MedPeer.cn.
To assess whether an A9B5 moiety covalently linked to trastuzumab could engage the same HER2 molecules in a cis-binding pattern, we aligned the A9B5-HER2 complex with the trastuzumab−HER2 complex (PDB ID: 1N8Z) (Figure 5C). The modeled C−terminus of A9B5 lay ~120.5 Å from the N−terminus of the trastuzumab light chain. Because A9B5−Bs−5 incorporates a flexible (GGGGS)3 linker with an estimated maximal reach of ~57 Å, this separation markedly exceeds the linker span, making simultaneous cis co-occupancy of A9B5 and trastuzumab epitopes on a single HER2 molecule unlikely. These constraints, therefore, favor a trans-binding mode in which A9B5 binds one molecule and the trastuzumab Fab engages a second. An analogous alignment with the pertuzumab−HER2 structure (PDB ID: 1S78) yielded a ~78.4 Å distance between the C−terminus of A9B5 and the N−terminus of the pertuzumab light chain (Figure 5D). This also exceeds the ~57 Å reach of the (GGGGS)3 linker in A9B5−Bs−7, again arguing against efficient cis co-occupancy and supporting a tran-binding mode.
To examine whether the A9B5 epitope remains accessible in ligand-activated receptor states, the A9B5-HER2 model was superposed onto structures of the HER2-HER3-NRG1β dimer (PDB ID: 7MN5) and the HER2-EGFR-EGF dimer (PDB ID: 8HGO) (Figures 5E, F). The A9B5 footprint remained largely solvent-exposed in both heterodimer conformations, with minimal occlusion by the partnering receptor ectodomains. Approach angles may be modestly restricted near the DA of ECD II; however, most of the interface appears accessible, suggesting that A9B5−containing bpAbs could bind HER2 even within ligand−driven heterodimers.
Together, these structural analyses support a trans−binding mechanism (Figure 5G). The (GGGGS)3 linker within a single antibody arm is insufficient to span the >57 Å separations required for cis-engagement of A9B5 with either trastuzumab or pertuzumab epitopes on the single HER2 molecule. Instead, each arm of A9B5−Bs−5 or A9B5−Bs−7 is predicted to bridge two HER2 molecules. Because the full−length IgG format is bivalent, a single bpAb could engage up to four HER2 molecules (two per arm), promoting local receptor clusters on the cell surface. Such clustering would be expected to induce receptor internalization and degradation and to sterically interfere with HER2−mediated heterodimerization, providing a structural rationale for the enhanced antitumor activity observed for these bpAbs in trastuzumab−resistant cells (Figure 4E).
4 Discussion
HER2-targeted therapies have revolutionized the therapeutic landscape for breast, gastric, and other solid tumors. However, treatment resistance remains a major clinical obstacle (5, 33, 64–71). To overcome the limitations of current standard-of-care therapies, we engineered two bpAbs, A9B5−Bs−5 and A9B5−Bs−7, tetravalent IgG-VHH constructs that bind non-overlapping epitopes on the HER2-ECD. These bpAbs exhibited enhanced functional properties compared to the clinically approved anti-HER2 antibodies trastuzumab and pertuzumab, either alone or in combination (T + P). A9B5−Bs−5 and A9B5−Bs−7 bound to cell-surface HER2 with high saturation and induced rapid HER2 internalization—an effect not observed with trastuzumab, pertuzumab, or T + P. Furthermore, both bpAbs exhibited superior or comparable growth-inhibition relative to T + P in both ligand-independent and ligand-driven tumor models. Structural modeling suggests that these bpAbs engage HER2 in a trans-binding pattern, promoting the formation of large receptor clusters.
The development of novel bsAbs is guided by a deep understanding of biological mechanism, which must be aligned with optimal formats, affinity profiles and epitope selection (50). IgG-VHH fusions, which combine IgG scaffolds with small nanobodies (VHHs), are highly attractive due to their structural simplicity, high stability, and favorable biophysical properties (60, 61, 72, 73). VHH domains are inherently monomeric and do not require pairing with a light chain, minimizing the risk of mispairing or aggregation that often compromises the performance of scFv-IgG formats. Leveraging these design principles, we constructed symmetric IgG-VHH fusions by fusing HER2-specific nanobodies to the IgG scaffolds of trastuzumab and pertuzumab (Figure 1B). A comprehensive comparison of different IgG-VHH bpAb architectures identified A9B5-Bs-5 and A9B5-Bs-7 as lead candidates, both exhibiting high purity, low aggregation propensity, and favorable manufacturability following Protein A purification (Figures 1C and 2A).
While previous studies have suggested that VHH fusion to the heavy chain is generally superior to light-chain fusion, our constructs defy this convention (58, 60). A9B5-Bs-5 and A9B5-Bs-7, in which the nanobody A9B5 is fused to the N-terminus of light chain via a (GGGGS)3 flexible linker, retained high binding affinity (EC50 < 2 nM) for both wild-type and chimeric HER2-ECD proteins and achieved high binding saturation on HER2-positive tumor cells (Figures 2D, E). These findings may be attributed to the flexible linker minimizing steric hindrance and the enhanced avidity conferred by dual epitope engagement (74, 75). Interestingly, certain configurations (e.g., G1E4-Bs-1 and G1E4-Bs-8) exhibited agonist activity (Figure 1D), consistent with observations from DVD-Ig formats that can aberrantly activate HER2 signaling depending on the spatial orientation of their binding domains (76). The underlying mechanisms of this agonistic behavior remain to be elucidated.
Resistance to anti-HER2 therapies arises from multiple mechanisms, including HER family alterations, masking of HER2 epitope, and activation of compensatory pathways. These multi-faceted resistance mechanisms significantly limit the effectiveness of traditional HER2-targeted therapies, which predominantly rely on single-modal MOA (34, 55). As a distinct subclass of bispecific antibodies, bpAbs represent a promising strategy for overcoming therapeutic resistance through dual HER2 blockade (34, 55, 71, 77–79). Several bpAbs currently in late-stage clinical development, including zanidatamab and anbenitamab, have shown favorable outcomes in HER2-expressing tumors (27, 31, 80–83). Engagement of non-overlapping epitopes enables multiple MOAs, including enhanced antibody saturation, receptor clustering, and rapid internalization (30). Our A9B5-containing bpAbs leverage these mechanisms and achieved greater or comparable antitumor efficacy than trastuzumab plus pertuzumab. Notably, most anti-HER2 bpAbs in development are derived from trastuzumab and pertuzumab, targeting common epitopes on ECD II and ECD IV (56, 84, 85). Recent structural studies indicate that targeting alternative domains such as ECD I and ECD III may improve blockade of HER2-driven oncogenic signaling (38, 39, 86). The A9B5 nanobody, obtained from a synthetic VHH library, binds ECD I with high affinity and exhibits strong synergy with trastuzumab in resistant tumor models (49). We hypothesize that ECD I engagement may facilitate additional binding modes, improved compatibility, and larger-scale receptor clustering. Structural predictions indicate that the A9B5 epitope remains accessible in ligand-activated HER2 conformations (Figures 5E, F), and in ligand-dependent models, A9B5−containing bpAbs demonstrated potent growth inhibition (Figure 4E). Whether this inhibitory activity stems from the disruption of HER2-containing heterodimers remains to be investigated. In addition, structural modeling supports a trans-binding mode of HER2 engagement by A9B5−containing bpAbs, potentially driving the formation of high-order HER2 clusters and facilitating internalization (Figure 5G). Similar trans-binding modality is employed by other HER2-targeting bpAbs, such as zanidatamab, which recognizes ECD II and ECD IV and promotes receptor reorganization to enhance CDC (30). Whether A9B5−based constructs can similarly recruit Fc-mediated effector functions remains to be determined. Moreover, although domain-swap ELISA primarily indicated binding to ECD I, this reflects the fact that key ECD II interface residues predicted by modeling (K228 and D234) are conserved between human and murine HER2. Thus, the experimental results mainly highlight species-specific determinants, whereas structural modeling provides a broader view of the composite epitope across ECD I–II. Taken together, these complementary approaches suggest that A9B5 engages a cross-subdomain surface that may stabilize distinct HER2 conformations and facilitate receptor clustering.
This study has several limitations. First, the predicted nanobody-HER2 interactions were derived from AlphaFold 3 structural models. Although AlphaFold provides near-experimental accuracy, definitive structural insights will require confirmation via cryo-electron microscopy or X-ray crystallography (53, 87). Second, the antitumor efficacy and MOAs of bpAbs need to be further verified in more cell lines. Third, the antitumor efficacy of A9B5-based bpAbs needs to be further validated in in vivo models to support their translational potential.
5 Conclusion
In conclusion, we engineered two IgG-VHH bpAbs, A9B5-Bs-5 and A9B5-Bs-7. In in vitro models, these antibodies demonstrated superior HER2 binding, receptor internalization, and growth inhibition compared to trastuzumab, pertuzumab, or their combination. Our findings highlight ECD I engagement as a viable design strategy for next-generation biparatopic anti-HER2 antibodies, potentially enabling functional activities not achieved by conventional monospecific agents. The ability of these antibodies to retain activity in trastuzumab-resistant cell models supports their potential utility in overcoming resistance mechanisms, and warrants further investigation in preclinical in vivo studies to assess their therapeutic applicability.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
Author contributions
XL: Conceptualization, Formal Analysis, Funding acquisition, Methodology, Writing – original draft, Writing – review & editing. WY: Supervision, Writing – review & editing. YW: Writing – review & editing. DX: Methodology, Writing – review & editing. HH: Formal Analysis, Resources, Writing – review & editing. WZ: Funding acquisition, Writing – review & editing, Resources. PS: Conceptualization, Formal Analysis, Project administration, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, and/or publication of this article. This work was supported by the Natural Science Foundation of Shandong Province Grants ZR202111120048 (WZ), ZR2022QH201 (XL) and ZR2024MC119 (XL), 2022 Shinan District Science and Technology Plan Project Grants 2023-2-015-YY (WZ), Development of innovative medical devices for pediatric ophthalmology based on machine vision and eye tracking technology Grants 24-1-5-yqpy-23-qy (WZ), the role and mechanism of SnoRD14E-PBX3 axis in regulating the progression of lung adenocarcinoma Grants 82473113 (WZ), the National Natural Science Foundation of China Grants 32300788 (XL).
Conflict of interest
Author XL is a visiting scholar of Noventi Biopharmaceuticals Co., Ltd. HH are employed by Noventi Biopharmaceuticals Co., Ltd.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1711433/full#supplementary-material
Abbreviations
HER2, Human epidermal growth factor receptor 2; EGFR, Epidermal growth factor receptor; ECD, Extracellular domain; ADCC, Antibody-dependent cellular cytotoxicity; DA, Dimerization arm; BsAbs, Bispecific antibodies; MOAs, Mechanisms of action; CCK-8, Cell Counting Kit-8; BpAbs, Biparatopic antibodies; VHH, Single variable domain; HPSEC, High performance size exclusion chromatography; ELISA, Enzyme-linked immunosorbent assay; PFA, Paraformaldehyde; MFI, Median fluorescence intensity; DAPI, 4′,6-diamidino-2-phenylindole; FACS, Fluorescence Activated Cell Sorter; EGF, Epidermal growth factor; HRG, Heregulin; CDC, Complement-dependent cytotoxicity.
References
1. DeSantis CE, Ma J, Gaudet MM, Newman LA, Miller KD, Goding Sauer A, et al. Breast cancer statistics, 2019. CA Cancer J Clin. (2019) 69:438–51. doi: 10.3322/caac.21583
2. Choong GM, Cullen GD, and O’Sullivan CC. Evolving standards of care and new challenges in the management of HER2-positive breast cancer. CA Cancer J Clin. (2020) 70:355–74. doi: 10.3322/caac.21634
3. Agostinetto E, Curigliano G, and Piccart M. Emerging treatments in HER2-positive advanced breast cancer: Keep raising the bar. Cell Rep Med. (2024), 101575. doi: 10.1016/j.xcrm.2024.101575
4. Ma Q, Jiang H, Ma L, Zhao G, Xu Q, Guo D, et al. The moonlighting function of glycolytic enzyme enolase-1 promotes choline phospholipid metabolism and tumor cell proliferation. Proc Natl Acad Sci U.S.A. (2023) 120:e2209435120. doi: 10.1073/pnas.2209435120
5. Baselga J, Cortes J, Kim SB, Im SA, Hegg R, Im YH, et al. Pertuzumab plus trastuzumab plus docetaxel for metastatic breast cancer. N Engl J Med. (2012) 366:109–19. doi: 10.1056/NEJMoa1113216
6. von Minckwitz G, Procter M, de Azambuja E, Zardavas D, Benyunes M, Viale G, et al. Adjuvant pertuzumab and trastuzumab in early HER2-positive breast cancer. N Engl J Med. (2017) 377:122–31. doi: 10.1056/NEJMoa1703643
7. Janjigian YY, Kawazoe A, Yañez P, Li N, Lonardi S, Kolesnik O, et al. The KEYNOTE-811 trial of dual PD-1 and HER2 blockade in HER2-positive gastric cancer. Nature. (2021) 600:727–30. doi: 10.1038/s41586-021-04161-3
8. Agus DB, Akita RW, Fox WD, Lewis GD, Higgins B, Pisacane PI, et al. Targeting ligand-activated ErbB2 signaling inhibits breast and prostate tumor growth. Cancer Cell. (2002) 2:127–37. doi: 10.1016/S1535-6108(02)00097-1
9. Junttila TT, Akita RW, Parsons K, Fields C, Lewis Phillips GD, Friedman LS, et al. Ligand-independent HER2/HER3/PI3K complex is disrupted by trastuzumab and is effectively inhibited by the PI3K inhibitor GDC-0941. Cancer Cell. (2009) 15:429–40. doi: 10.1016/j.ccr.2009.03.020
10. Chandarlapaty S, Sakr RA, Giri D, Patil S, Heguy A, Morrow M, et al. Frequent mutational activation of the PI3K-AKT pathway in trastuzumab-resistant breast cancer. Clin Cancer Res: an Off J Am Assoc Cancer Res. (2012) 18:6784–91. doi: 10.1158/1078-0432.CCR-12-1785
11. Vernieri C, Milano M, Brambilla M, Mennitto A, Maggi C, Cona MS, et al. Resistance mechanisms to anti-HER2 therapies in HER2-positive breast cancer: Current knowledge, new research directions and therapeutic perspectives. Crit Rev Oncol Hematol. (2019) 139:53–66. doi: 10.1016/j.critrevonc.2019.05.001
12. Wu X, Huang S, He W, and Song M. Emerging insights into mechanisms of trastuzumab resistance in HER2-positive cancers. Int Immunopharmacol. (2023) 122:110602. doi: 10.1016/j.intimp.2023.110602
13. Chen Y, Lu A, Hu Z, Li J, and Lu J. ERBB3 targeting: A promising approach to overcoming cancer therapeutic resistance. Cancer Lett. (2024), 217146. doi: 10.1016/j.canlet.2024.217146
14. Poumeaud F, Morisseau M, Cabel L, Goncalves A, Rivier C, Tredan O, et al. Efficacy of administration sequence: Sacituzumab Govitecan and Trastuzumab Deruxtecan in HER2-low metastatic breast cancer. Br J Cancer. (2024). doi: 10.1038/s41416-024-02766-9
15. Zhao X, Li Y, Zhang H, Cai Y, Wang X, Liu Y, et al. PAK5 promotes the trastuzumab resistance by increasing HER2 nuclear accumulation in HER2-positive breast cancer. Cell Death Dis. (2025) 16:323. doi: 10.1038/s41419-025-07657-2
16. Sperinde J, Jin X, Banerjee J, Penuel E, Saha A, Diedrich G, et al. Quantitation of p95HER2 in paraffin sections by using a p95-specific antibody and correlation with outcome in a cohort of trastuzumab-treated breast cancer patients. Clin Cancer Res: an Off J Am Assoc Cancer Res. (2010) 16:4226–35. doi: 10.1158/1078-0432.Ccr-10-0410
17. Scaltriti M, Eichhorn PJ, Cortes J, Prudkin L, Aura C, Jimenez J, et al. Cyclin E amplification/overexpression is a mechanism of trastuzumab resistance in HER2+ breast cancer patients. Proc Natl Acad Sci U.S.A. (2011) 108:3761–6. doi: 10.1073/pnas.1014835108
18. Gaibar M, Beltran L, Romero-Lorca A, Fernandez-Santander A, and Novillo A. Somatic mutations in HER2 and implications for current treatment paradigms in HER2-positive breast cancer. J Oncol. (2020) 2020:6375956. doi: 10.1155/2020/6375956
19. Udagawa H, Nilsson MB, Robichaux JP, He J, Poteete A, Jiang H, et al. HER4 and EGFR activate cell signaling in NRG1 fusion-driven cancers: implications for HER2/HER3-specific vs. pan-HER targeting strategies. J Thorac Oncol. (2023). doi: 10.1016/j.jtho.2023.08.034
20. Li Z, Metzger Filho O, Viale G, dell'Orto P, Russo L, Goyette MA, et al. HER2 heterogeneity and treatment response-associated profiles in HER2-positive breast cancer in the NCT02326974 clinical trial. J Clin Invest. (2024). doi: 10.1172/jci176454
21. Chen S, Lin J, Yang Z, Wang Y, Wang Q, Wang D, et al. TRIM24-mediated K27-linked ubiquitination of ULK1 alleviates energy stress-induced autophagy and promote prostate cancer growth in the context of SPOP mutation. Cell Death Differ. (2025). doi: 10.1038/s41418-025-01582-9
22. Labrijn AF, Janmaat ML, Reichert JM, and Parren P. Bispecific antibodies: a mechanistic review of the pipeline. Nat Rev Drug Discov. (2019) 18:585–608. doi: 10.1038/s41573-019-0028-1
23. Deshaies RJ. Multispecific drugs herald a new era of biopharmaceutical innovation. Nature. (2020) 580:329–38. doi: 10.1038/s41586-020-2168-1
24. de Jong G, Bartels L, Kedde M, Verdegaal EME, Gillissen MA, Levie SE, et al. Melanoma cells can be eliminated by sialylated CD43 x CD3 bispecific T cell engager formats in vitro and in vivo. Cancer Immunol Immunother. (2020). doi: 10.1007/s00262-020-02780-9
25. Li S, Liu M, Do MH, Chou C, Stamatiades EG, Nixon BG, et al. Cancer immunotherapy via targeted TGF-beta signalling blockade in TH cells. Nature. (2020) 587:121–5. doi: 10.1038/s41586-020-2850-3
26. Zhang J, Yi J, and Zhou P. Development of bispecific antibodies in China: overview and prospects. Antibody Ther. (2020) 3:126–45. doi: 10.1093/abt/tbaa011
27. Meric-Bernstam F, Beeram M, Hamilton E, Oh DY, Hanna DL, Kang YK, et al. Zanidatamab, a novel bispecific antibody, for the treatment of locally advanced or metastatic HER2-expressing or HER2-amplified cancers: a phase 1, dose-escalation and expansion study. Lancet Oncol. (2022) 23:1558–70. doi: 10.1016/S1470-2045(22)00621-0
28. Elimova E, Ajani JA, Burris HA III, Denlinger CS, Iqbal S, Kang Y-K, et al. Zanidatamab + chemotherapy as first-line treatment for HER2-expressing metastatic gastroesophageal adenocarcinoma (mGEA). J Clin Oncol. (2023) 41:347–7. doi: 10.1200/JCO.2023.41.4_suppl.347
29. Wang X, Lee KS, Zeng X, Sun T, Im Y-H, Li H, et al. Zanidatamab (zani), a HER2-targeted bispecific antibody, in combination with docetaxel as first-line therapy (1L) for patients (pts) with advanced HER2-positive breast cancer (BC): Updated results from a phase 1b/2 study. J Clin Oncol. (2023) 41:1044–4. doi: 10.1200/JCO.2023.41.16_suppl.1044
30. Weisser NE, Sanches M, Escobar-Cabrera E, O'Toole J, Whalen E, Chan PWY, et al. An anti-HER2 biparatopic antibody that induces unique HER2 clustering and complement-dependent cytotoxicity. Nat Commun. (2023) 14:1394. doi: 10.1038/s41467-023-37029-3
31. Pant S, Fan J, Oh D-Y, Choi HJ, Kim JW, Chang H-M, et al. Zanidatamab in previously-treated HER2-positive (HER2+) biliary tract cancer (BTC): Overall survival (OS) and longer follow-up from the phase 2b HERIZON-BTC-01 study. J Clin Oncol. (2024) 42:4091–1. doi: 10.1200/JCO.2024.42.16_suppl.4091
32. Escriva-de-Romani S, Cejalvo JM, Alba E, Friedmann J, Rodriguez-Lescure A, Savard MF, et al. Zanidatamab plus palbociclib and fulvestrant in previously treated patients with hormone receptor-positive, HER2-positive metastatic breast cancer: primary results from a two-part, multicentre, single-arm, phase 2a study. Lancet Oncol. (2025) 26:745–58. doi: 10.1016/S1470-2045(25)00140-8
33. Robbins CJ, Bates KM, and Rimm DL. HER2 testing: evolution and update for a companion diagnostic assay. Nat Rev Clin Oncol. (2025) 22:408–23. doi: 10.1038/s41571-025-01016-y
34. Liu X, Fan X, Gao X, Hu W, and Sun P. Leveraging HER2-targeted biparatopic antibodies in solid tumors. Pharmacol Res. (2025) 214:107687. doi: 10.1016/j.phrs.2025.107687
35. Bai X, Xu L, Wang Z, Zhuang X, Ning J, Sun Y, et al. Computational-aided rational mutation design of pertuzumab to overcome active HER2 mutation S310F through antibody-drug conjugates. Proc Natl Acad Sci U.S.A. (2025) 122:e2413686122. doi: 10.1073/pnas.2413686122
36. Cho H-S, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DW, et al. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. (2003) 421:756–60. doi: 10.1038/nature01392
37. Franklin MC, Carey KD, Vajdos FF, Leahy DJ, de Vos AM, and Sliwkowski MX Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex. Cancer Cell. (2004) 5:317–28. doi: 10.1016/S1535-6108(04)00083-2
38. Diwanji D, Trenker R, Thaker TM, Wang F, Agard DA, Verba KA, et al. Structures of the HER2-HER3-NRG1beta complex reveal a dynamic dimer interface. Nature. (2021) 600:339–43. doi: 10.1038/s41586-021-04084-z
39. Bai X, Sun P, Wang X, Long C, Liao S, Dang S, et al. Structure and dynamics of the EGFR/HER2 heterodimer. Cell Discov. (2023) 9:18. doi: 10.1038/s41421-023-00523-5
40. Jost C, Schilling J, Tamaskovic R, Schwill M, Honegger A, and Pluckthun A Structural basis for eliciting a cytotoxic effect in HER2-overexpressing cancer cells via binding to the extracellular domain of HER2. Structure. (2013) 21:1979–91. doi: 10.1016/j.str.2013.08.020
41. Brack S, Attinger-Toller I, Schade B, Mourlane F, Klupsch K, Woods R, et al. A bispecific HER2-targeting FynomAb with superior antitumor activity and novel mode of action. Mol Cancer Ther. (2014) 13:2030–9. doi: 10.1158/1535-7163.Mct-14-0046-t
42. Pedersen MW, Jacobsen HJ, Koefoed K, Dahlman A, Kjær I, Poulsen TT, et al. Targeting three distinct HER2 domains with a recombinant antibody mixture overcomes trastuzumab resistance. Mol Cancer Ther. (2015) 14:669–80. doi: 10.1158/1535-7163.Mct-14-0697
43. Szymanska M, Fosdahl AM, Nikolaysen F, Pedersen MW, Grandal MM, Stang E, et al. A combination of two antibodies recognizing non-overlapping epitopes of HER2 induces kinase activity-dependent internalization of HER2. J Cell Mol Med. (2016) 20:1999–2011. doi: 10.1111/jcmm.12899
44. Mohammadi M, Jeddi-Tehrani M, Golsaz-Shirazi F, Arjmand M, Bahadori T, Judaki MA, et al. A novel anti-HER2 bispecific antibody with potent tumor inhibitory effects in vitro and in vivo. Front Immunol. (2020) 11:600883. doi: 10.3389/fimmu.2020.600883
45. Kast F, Schwill M, Stuber JC, Pfundstein S, Nagy-Davidescu G, Rodriguez JMM, et al. Engineering an anti-HER2 biparatopic antibody with a multimodal mechanism of action. Nat Commun. (2021) 12:3790. doi: 10.1038/s41467-021-23948-6
46. Wang C, Wang L, Yu X, Zhang Y, Meng Y, Wang H, et al. Combating acquired resistance to trastuzumab by an anti-ErbB2 fully human antibody. Oncotarget. (2017) 8:42742. doi: 10.18632/oncotarget.17451
47. Fu W, Wang Y, Zhang Y, Xiong L, Takeda H, Ding L, et al. Insights into HER2 signaling from step-by-step optimization of anti-HER2 antibodies. MAbs. (2014) 6:978–90. doi: 10.4161/mabs.28786
48. Yu X, Wang L, Shen Y, Wang C, Zhang Y, Meng Y, et al. Targeting EGFR/HER2 heterodimerization with a novel anti-HER2 domain II/III antibody. Mol Immunol. (2017) 87:300–7. doi: 10.1016/j.molimm.2017.05.010
49. Liu X, Luan L, Liu X, Jiang D, Deng J, Xu J, et al. A novel nanobody-based HER2-targeting antibody exhibits potent synergistic antitumor efficacy in trastuzumab-resistant cancer cells. Front Immunol. (2023) 14:1292839. doi: 10.3389/fimmu.2023.1292839
50. Klein C, Brinkmann U, Reichert JM, and Kontermann RE. The present and future of bispecific antibodies for cancer therapy. Nat Rev Drug Discov. (2024). doi: 10.1038/s41573-024-00896-6
51. Hou J, Du K, Li J, Li Z, Cao S, Zhang S, et al. Research trends in the use of nanobodies for cancer therapy. J Controlled Release. (2025) 381:113454. doi: 10.1016/j.jconrel.2025.01.045
52. Zhao D, Liu L, Liu X, Zhang J, Yin Y, Luan L, et al. A potent synthetic nanobody with broad-spectrum activity neutralizes SARS-CoV-2 virus and the Omicron variant BA.1 through a unique binding mode. J Nanobiotechnol. (2022) 20. doi: 10.1186/s12951-022-01619-y
53. Abramson J, Adler J, Dunger J, Evans R, Green T, Pritzel A, et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature. (2024) 630:493–500. doi: 10.1038/s41586-024-07487-w
54. Niquille DL, Fitzgerald KM, and Gera N. Biparatopic antibodies: therapeutic applications and prospects. MAbs. (2024) 16:2310890. doi: 10.1080/19420862.2024.2310890
55. Liu X, Song Y, Cheng P, Liang B, and Xing D. Targeting HER2 in solid tumors: Unveiling the structure and novel epitopes. Cancer Treat Rev. (2024) 130. doi: 10.1016/j.ctrv.2024.102826
56. Wang Z, Liu Y, Xu Y, Lu L, Zhu Z, Lv B, et al. Anti-HER2 biparatopic antibody KJ015 has near-native structure, functional balanced high affinity, and synergistic efficacy with anti-PD-1 treatment in vivo. MAbs. (2024) 16:2412881. doi: 10.1080/19420862.2024.2412881
57. Yue J, Shao S, Zhou J, Luo W, Xu Y, Zhang Q, et al. A bispecific antibody targeting HER2 and CLDN18.2 eliminates gastric cancer cells expressing dual antigens by enhancing the immune effector function. Invest New Drugs. (2024). doi: 10.1007/s10637-024-01417-3
58. Madsen AV, Kristensen P, Buell AK, and Goletz S. Generation of robust bispecific antibodies through fusion of single-domain antibodies on IgG scaffolds: a comprehensive comparison of formats. MAbs. (2023) 15:2189432. doi: 10.1080/19420862.2023.2189432
59. Misson Mindrebo L, Liu H, Ozorowski G, Tran Q, Woehl J, Khalek I, et al. Fully synthetic platform to rapidly generate tetravalent bispecific nanobody-based immunoglobulins. Proc Natl Acad Sci U.S.A. (2023) 120:e2216612120. doi: 10.1073/pnas.2216612120
60. Madsen AV, Kristensen P, and Goletz S. IgG-VHH bispecific fusion antibodies: challenges and opportunities as therapeutic agents. Expert Opin Biol Ther. (2024), 1–4. doi: 10.1080/14712598.2024.2336068
61. Mullin M, McClory J, Haynes W, Grace J, Robertson N, and van Heeke G Applications and challenges in designing VHH-based bispecific antibodies: leveraging machine learning solutions. MAbs. (2024) 16:2341443. doi: 10.1080/19420862.2024.2341443
62. Cheng J, Liang M, Carvalho MF, Tigue N, Faggioni R, Roskos LK, et al. Molecular mechanism of HER2 rapid internalization and redirected trafficking induced by anti-HER2 biparatopic antibody. Antibodies (Basel). (2020) 9. doi: 10.3390/antib9030049
63. Swain SM, Shastry M, and Hamilton E. Targeting HER2-positive breast cancer: advances and future directions. Nat Rev Drug Discov. (2022), 1–26. doi: 10.1038/s41573-022-00579-0
64. Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. (2010) 376:687–97. doi: 10.1016/S0140-6736(10)61121-X
65. Tabernero J, Hoff PM, Shen L, Ohtsu A, Shah MA, Cheng K, et al. Pertuzumab plus trastuzumab and chemotherapy for HER2-positive metastatic gastric or gastro-oesophageal junction cancer (JACOB): final analysis of a double-blind, randomised, placebo-controlled phase 3 study. Lancet Oncol. (2018) 19:1372–84. doi: 10.1016/S1470-2045(18)30481-9
66. von Minckwitz G, Huang CS, Mano MS, Loibl S, Mamounas EP, Untch M, et al. Trastuzumab emtansine for residual invasive HER2-positive breast cancer. N Engl J Med. (2019) 380:617–28. doi: 10.1056/NEJMoa1814017
67. Pellegrino B, Tommasi C, Serra O, Gori S, Cretella E, Ambroggi M, et al. Randomized, open-label, phase II, biomarker study of immune-mediated mechanism of action of neoadjuvant subcutaneous trastuzumab in patients with locally advanced, inflammatory, or early HER2-positive breast cancer-Immun-HER trial (GOIRC-01-2016). J Immunother Cancer. (2023) 11. doi: 10.1136/jitc-2023-007667
68. Moasser MM and Krop IE. The evolving landscape of HER2 targeting in breast cancer. JAMA Oncol. (2015) 1:1154–61. doi: 10.1001/jamaoncol.2015.2286
69. Meric-Bernstam F, Johnson AM, Dumbrava EEI, Raghav K, Balaji K, Bhatt M, et al. Advances in HER2-targeted therapy: novel agents and opportunities beyond breast and gastric cancer. Clin Cancer Res: an Off J Am Assoc Cancer Res. (2019) 25:2033–41. doi: 10.1158/1078-0432.CCR-18-2275
70. Boscolo Bielo L, Trapani D, Nicolo E, Valenza C, Guidi L, Belli C, et al. The evolving landscape of metastatic HER2-positive, hormone receptor-positive Breast Cancer. Cancer Treat Rev. (2024) 128:102761. doi: 10.1016/j.ctrv.2024.102761
71. Yoon J and Oh DY. HER2-targeted therapies beyond breast cancer - an update. Nat Rev Clin Oncol. (2024). doi: 10.1038/s41571-024-00924-9
72. Deng J, Geng Z, Luan L, Jiang D, Lu J, Zhang H, et al. Novel anti-trop2 nanobodies disrupt receptor dimerization and inhibit tumor cell growth. Pharmaceutics. (2024) 16. doi: 10.3390/pharmaceutics16101255
73. Deken MM, Kijanka MM, Beltrán Hernández I, Slooter MD, de Bruijn HS, van Diest PJ, et al. Nanobody-targeted photodynamic therapy induces significant tumor regression of trastuzumab-resistant HER2-positive breast cancer, after a single treatment session. J Control Release. (2020) 323:269–81. doi: 10.1016/j.jconrel.2020.04.030
74. Chen X, Zaro JL, and Shen WC. Fusion protein linkers: property, design and functionality. Adv Drug Delivery Rev. (2013) 65:1357–69. doi: 10.1016/j.addr.2012.09.039
75. Silacci M, Baenziger-Tobler N, Lembke W, Zha W, Batey S, Bertschinger J, et al. Linker length matters, fynomer-Fc fusion with an optimized linker displaying picomolar IL-17A inhibition potency. J Biol Chem. (2014) 289:14392–8. doi: 10.1074/jbc.M113.534578
76. Gu J, Yang J, Chang Q, Lu X, Wang J, Chen M, et al. Identification of anti-ErbB2 dual variable domain immunoglobulin (DVD-Ig) proteins with unique activities. PloS One. (2014) 9:e97292. doi: 10.1371/journal.pone.0097292
77. Stoup N, Liberelle M, Lebegue N, and Van Seuningen I. Emerging paradigms and recent progress in targeting ErbB in cancers. Trends Pharmacol Sci. (2024). doi: 10.1016/j.tips.2024.04.009
78. Liu X, Ma L, Li J, Sun L, Yang Y, Liu T, et al. Trop2-targeted therapies in solid tumors: advances and future directions. Theranostics. (2024) 14:3674–92. doi: 10.7150/thno.98178
79. Ai Z, Wang B, Song Y, Cheng P, Liu X, Sun P, et al. Prodrug-based bispecific antibodies for cancer therapy: advances and future directions. Front Immunol. (2025) 16:1523693. doi: 10.3389/fimmu.2025.1523693
80. Ji D, Zhang J, Shen W, Du Y, Xu J, Yang J, et al. Preliminary safety, efficacy and pharmacokinetics (PK) results of KN026, a HER2 bispecific antibody in patients (pts) with HER2-positive metastatic breast cancer. J Clin Oncol. (2020) 38:1041–1. doi: 10.1200/JCO.2020.38.15_suppl.1041
81. Shen L, Gong J, Niu Z, Zhao R, Chen L, Liu L, et al. 1210P The preliminary efficacy and safety of KN026 combined with KN046 treatment in HER2-positive locally advanced unresectable or metastatic gastric/gastroesophageal junction cancer without prior systemic treatment in a phase II study. Ann Oncol. (2022) 33:S1102. doi: 10.1016/j.annonc.2022.07.1328
82. Zhang J, Ji D, Cai L, Yao H, Yan M, Wang X, et al. First-in-human HER2-targeted bispecific antibody KN026 for the treatment of patients with HER2-positive metastatic breast cancer: results from a phase I study. Clin Cancer Res: an Off J Am Assoc Cancer Res. (2022) 28:618–28. doi: 10.1158/1078-0432.CCR-21-2827
83. Harding JJ, Fan J, Oh D-Y, Choi HJ, Kim JW, Chang H-M, et al. Zanidatamab for HER2-amplified, unresectable, locally advanced or metastatic biliary tract cancer (HERIZON-BTC-01): a multicentre, single-arm, phase 2b study. Lancet Oncol. (2023). doi: 10.1016/s1470-2045(23)00242-5
84. Huang S, Li F, Liu H, Ye P, Fan X, Yuan X, et al. Structural and functional characterization of MBS301, an afucosylated bispecific anti-HER2 antibody. MAbs. (2018) 10:864–75. doi: 10.1080/19420862.2018.1486946
85. Zhang Q, Wang J, Li L, Zeng X, Zhang H, Song Y, et al. Preliminary safety and efficacy of TQB2930, a HER2-targeted bispecific antibody in patients with advanced breast cancer: Results from a phase 1b study. J Clin Oncol. (2024) 42:1026–6. doi: 10.1200/JCO.2024.42.16_suppl.1026
86. Hao Y, Yu X, Bai Y, McBride HJ, and Huang X. Cryo-EM structure of HER2-trastuzumab-pertuzumab complex. PloS One. (2019) 14:e0216095. doi: 10.1371/journal.pone.0216095
87. Li G, Liu C, Qiu S, Wei L, Cao L, Wang K, et al. From non-affinity to high-affinity: Rapid preparation of nanobodies utilizing high-precision alphafold and structural-interaction analysis for detection of enrofloxacin in marine fish. J Hazard Mater. (2025) 488:137394. doi: 10.1016/j.jhazmat.2025.137394
Keywords: biparatopic antibody, HER2, nanobody, trastuzumab-resistance, non-overlapping epitopes
Citation: Liu X, Yu W, Wang Y, Xing D, Huang H, Zhu W and Sun P (2025) Engineering HER2-targeted biparatopic antibodies to promote receptor internalization and restore antitumor efficacy. Front. Immunol. 16:1711433. doi: 10.3389/fimmu.2025.1711433
Received: 23 September 2025; Accepted: 02 October 2025;
Published: 07 November 2025.
Edited by:
Yuanzhi Chen, Xiamen University, ChinaReviewed by:
Junping Hong, The Ohio State University, United StatesXiao Zhang, Chongqing Medical University, China
Copyright © 2025 Liu, Yu, Wang, Xing, Huang, Zhu and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haiming Huang, aGFpbWluZy5odWFuZ0BhdWFtYmlvLmNvbQ==; Wenjing Zhu, emh1d2pAdW9yLmVkdS5jbg==; Peng Sun, cHN1bjFAcWR1LmVkdS5jbg==
†These authors have contributed equally to this work
 Xinlin Liu
Xinlin Liu Wanpeng Yu
Wanpeng Yu Yihuan Wang2,3
Yihuan Wang2,3 Peng Sun
Peng Sun