- 1Department of Urology, The Second People’s Hospital of Meishan City, Meishan, Sichuan, China
- 2Department of Urology, China-Japan Union Hospital of Jilin University, Changchun, Jilin, China
- 3The First Clinical Medical College of Shandong University of Traditional Chinese Medicine, Jinan, China
- 4China Institute of Sport and Health Science, Beijing Sport University, Beijing, China
- 5Department of Urology, The First People’s Hospital of Jiangxia District, Wuhan, Hubei, China
- 6Department of Urology, Xiangyang Central Hospital, Affiliated Hospital of Hubei University of Arts and Science, Xiangyang, Hubei, China
By Wang J, Zhao F, Zhang Q, Sun Z, Xiahou Z, Wang C, Liu Y and Yu Z (2024) Front. Immunol. 15:1517679. doi: 10.3389/fimmu.2024.1517679
There was a mistake in Figure 1 and Figure 7I as published. This error occurred because placeholder images were used during the typesetting process to maintain layout consistency. Due to an oversight, the first placeholder image in the upper left corner of Figure 7I was not replaced with the correct image in time. In addition, since the Figure 1 includes a schematic diagram of this figure, it also needs to be updated accordingly. The corrected Figure 1 and Figure 7 appears below.
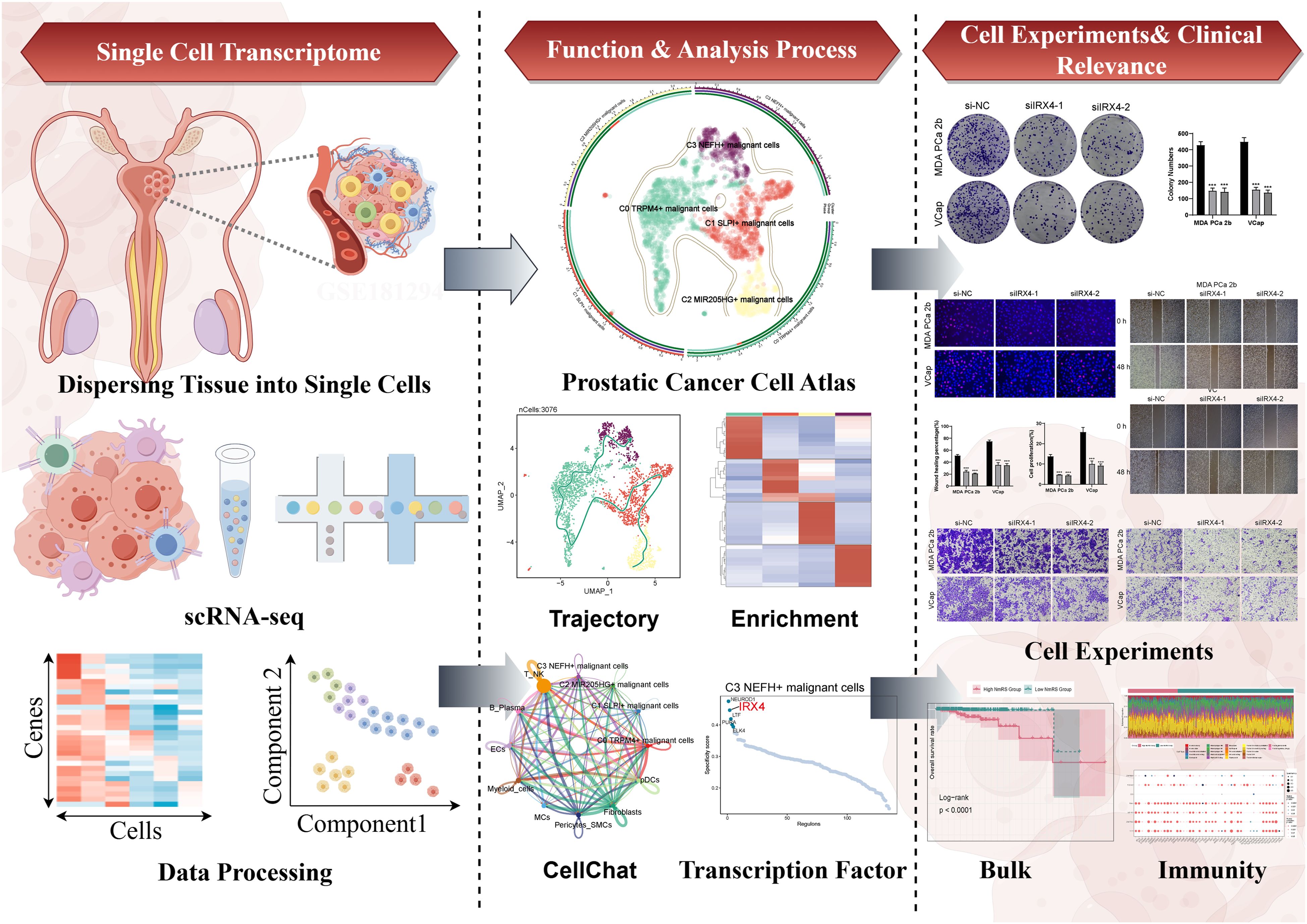
Figure 1. Graphical abstract. The analysis workflow for this research. We performed single-cell sequencing analysis on the GSE181294 dataset and identified a distinct C3 NEFH+ malignant cells subtype. Through pseudotime analysis, enrichment analysis, cell communication, and transcription factor regulation analysis, we revealed the significance of this subtype and confirmed the important role of the key TF (IRX4) through in vitro experiments. Prognostic and immune analyses provided guidance for clinical intervention and treatment.
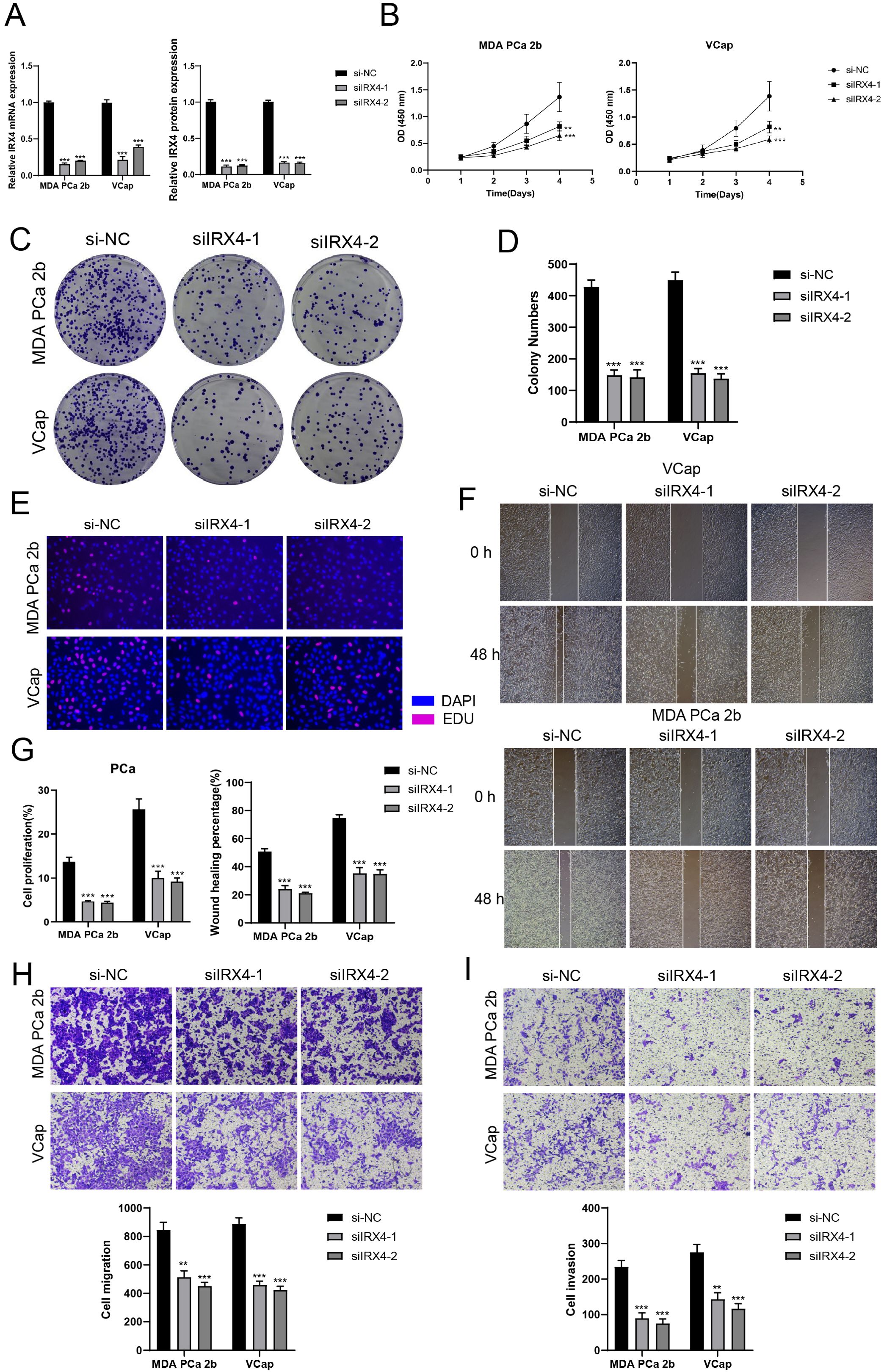
Figure 7. In vitro experiments confirmed the effects of IRX4 knockdown. (A) Decreased mRNA and protein expression levels after IRX4 knockdown. (B) CCK-8 assay showed a significant decrease in cell viability after IRX4 knockdown compared to the control group. (C, D) Colony formation assay demonstrated a significant decrease in colony numbers after IRX4 knockdown. (E) EDU staining experiment confirmed the inhibitory effect of IRX4 knockdown on cell proliferation. (F) Wound healing assay showed that IRX4 knockdown inhibited cell migration. (G) Bar graph displayed a significant decrease in cell proliferation and migration abilities after IRX4 knockdown. (H, I) Transwell assay showed that IRX4 knockdown suppressed the migration and invasion abilities of tumor cells in MDA PCA 2b and VCap cell lines. **P<0.01, ***P<0.001.
The original version of this article has been updated.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: multi-omics, single-cell RNA sequencing, prostate cancer, tumor heterogeneity, precision medicine, drug discovery
Citation: Wang J, Zhao F, Zhang Q, Sun Z, Xiahou Z, Wang C, Liu Y and Yu Z (2025) Correction: Unveiling the NEFH+ malignant cell subtype: insights from single-cell RNA sequencing in prostate cancer progression and tumor microenvironment interactions. Front. Immunol. 16:1718125. doi: 10.3389/fimmu.2025.1718125
Received: 03 October 2025; Accepted: 24 October 2025;
Published: 07 November 2025.
Edited and reviewed by:
Peter Brossart, University of Bonn, GermanyCopyright © 2025 Wang, Zhao, Zhang, Sun, Xiahou, Wang, Liu and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zongze Yu, eXV6b25nemUyMDIzMDNAMTYzLmNvbQ==; Yan Liu, cmxpdXlhbkBmb3htYWlsLmNvbQ==; Changzhong Wang, d2FuZ2N6a3lAMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Jie Wang
Jie Wang Fu Zhao
Fu Zhao Qiang Zhang1†
Qiang Zhang1† Zhou Sun
Zhou Sun Zhikai Xiahou
Zhikai Xiahou Changzhong Wang
Changzhong Wang