- 1 Department of Natural Resources and Environmental Sciences, University of Illinois at Urbana-Champaign, Urbana, IL, USA
- 2 Construction Engineering Research Laboratory, U.S. Army Engineer Research and Development Center, Champaign, IL, USA
- 3 Illinois Natural History Survey, University of Illinois at Urbana-Champaign, Champaign, IL, USA
The spatial scale on which microbial communities respond to plant invasions may provide important clues as to the nature of potential invader–microbe interactions. Lespedeza cuneata (Dum. Cours.) G. Don is an invasive legume that may benefit from associations with mycorrhizal fungi; however, it has also been suggested that the plant is allelopathic and may alter the soil chemistry of invaded sites through secondary metabolites in its root exudates or litter. Thus, L. cuneata invasion may interact with soil microorganisms on a variety of scales. We investigated L. cuneata-related changes to soil bacterial and fungal communities at two spatial scales using multiple sites from across its invaded N. American range. Using whole-community DNA fingerprinting, we characterized microbial community variation at the scale of entire invaded sites and at the scale of individual plants. Based on permutational multivariate analysis of variance, soil bacterial communities in heavily invaded sites were significantly different from those of uninvaded sites, but bacteria did not show any evidence of responding at very local scales around individual plants. In contrast, soil fungi did not change significantly at the scale of entire sites, but there were significant differences between fungal communities of native versus exotic plants within particular sites. The differential scaling of bacterial and fungal responses indicates that L. cuneata interacts differently with soil bacteria and soil fungi, and these microorganisms may play very different roles in the invasion process of this plant.
Introduction
A growing body of research indicates that soil microorganisms can both respond to and strongly impact plant communities, including those affected by invasive plants (Klironomos, 2002; Kourtev et al., 2003; Reinhart et al., 2003; Callaway et al., 2004; Reinhart and Callaway, 2004; van der Putten et al., 2007b; Engelkes et al., 2008); reviewed by (Wardle et al., 2004; Wolfe and Klironomos, 2005; Reinhart and Callaway, 2006; van der Putten et al., 2007a). Plant-microbe interactions can influence the competitive dynamic of plant communities (Bever, 1994, 2003), explain variation in the local abundance of plant species (Klironomos, 2002), and may facilitate range expansion and invasion (Engelkes et al., 2008). Our understanding of the specific microbial mechanisms at play in plant community dynamics is limited because much previous work has considered microbial communities from a purely functional, “black box” perspective; that is, a particular microbial species pool either net positively or net negatively influences plant growth (examples: Reinhart et al., 2003; Agrawal et al., 2005; Zou et al., 2006). Furthermore, attempts to correlate plant invasion to specific changes in microbial community composition have met with mixed results (Kourtev et al., 2003; Callaway et al., 2004; Kardol et al., 2007; Lorenzo et al., 2010), and much is still unknown, in particular about the specific microbial communities that respond to plant invasions.
The question of scale is one potential hurdle to a deeper understanding of microbial ecology’s role in plant invasions, because different interactions can operate on a number of scales. Microbes can interact directly with invaders on very fine scales through a number of symbiotic interactions with roots: e.g., mycorrhizas, mutualistic bacteria, or known pathogens (Klironomos, 2002, 2003; Fumanal et al., 2006; Reinhart and Callaway, 2006; van der Putten et al., 2007b; Pringle et al., 2009). It is well known that the rhizosphere is a “hot spot” of interaction that can influence microbial activity and plant ecological performance (Kent and Triplett, 2002; Berg and Smalla, 2009). However, the soil milieu is broader than the rhizosphere, and understanding the microbial ecology of some plant invasions may require observations at larger scales, where interactions may be more indirect.
Several recent investigators have proposed that invasive plant impacts may be more diffuse, affecting microbial communities that are not part of the rhizosphere (Wolfe and Klironomos, 2005; van der Putten et al., 2007a). For instance, through inputs of litter to the soil, invasive plants may alter the decomposition rate and nutrient cycling rates of the system (Ehrenfeld, 2003; Heneghan et al., 2006; Zou et al., 2006), with consequences for nutrient availability and competitive fitness of native competitors. Many plant invaders have been found to alter the soil characteristics, such as fungal:bacterial biomass ratios (Kourtev et al., 2002, 2003), and total nitrogen, carbon or phosphorus pools (Ehrenfeld, 2003; Chapuis-Lardy et al., 2006; Heneghan et al., 2006; Lorenzo et al., 2010), and these observations suggest that invasive plants may influence soil ecology on scales larger than that of their own rhizosphere and over longer time periods than the lifetime of individual plants. The “novel weapons” hypothesis suggests that invasive plants may also introduce new chemical compounds into the soil (Cappuccino and Arnason, 2006; Callaway et al., 2008), and the effects of these compounds will not necessarily be restricted to rhizosphere communities. For example, there is evidence that allelochemicals secreted by invasive garlic mustard (Alliaria petiolata; Stinson et al., 2006; Wolfe et al., 2008; Lankau, 2010) and spotted knapweed (Cenaurea maculosa; Callaway et al., 2004; Broz et al., 2007) can reduce the abundance and diversity of soil fungi that are important mutualists of native competitor plants (the “degraded mutualisms” hypothesis; Vogelsang and Bever, 2009). Invader impacts extending beyond the rhizosphere can have important implications for the invaded habitats. Thus, while rhizosphere interactions are very important, it is also necessary to understand how invasive plants can impact the soil at larger spatial scales where we can find the signature of other important interactions.
The Chinese bush clover [Lespedeza cuneata (Dum. Cours.) G. Don], often called Sericea Lespedeza, is one of about 45 Lespedeza species found across Asia, Australia, and North America. These legumes are tolerant of degraded and nutrient-poor soils, and they have been shown to associate with a broad diversity of rhizobacteria (Gu et al., 2007). L. cuneata is a long-lived perennial introduced from Japan to the United States in the late, 1800s (Guernsey, 1970), and it is now well established as an invasive weed of old fields, prairies, and forest openings across the southern U.S. (Miller, 2003). L. cuneata has a high seed production rate and a high population growth rate compared with its North American congeners (Schutzenhofer et al., 2009; Woods et al., 2009). It can form dense stands in its invaded range, shading out native grasses and herbs (Brandon et al., 2004). There is some evidence that its residues possess phytotoxic or allelopathic chemicals (Langdale and Giddens, 1967; Kalburtji et al., 2001), and its residues and root exudates have been found to inhibit seed germination and/or growth of various grass species (Kalburtji and Mosjidis, 1992,1993a,b). In addition to its rhizobacterial symbionts, it has been found to associate with various mycorrhizal fungi in what has been termed a nitrogen-fixing, tri-partite symbiosis (Lynd and Ansman, 1993).
Due to its symbiotic associations, L. cuneata invasions may interact with soil microbial communities of invaded systems at very local scales (i.e., that of the individual plant). In addition, the potential inhibitory effects of L. cuenata root exudates and residues suggest that this invader may affect the soil system at broader scales, particularly where it achieves high population densities. In this study, we investigate variation in soil bacterial and fungal community composition associated with L. cuneata invasions at two spatial scales: (1) in localized soil zones around individual plants (hereafter referred to as plant “neighborhoods” and (2) that of whole sites, including sites with sparse and those with dense populations of L. cuneata. Because its broad geographic distribution in the U.S. means that L. cuneata may interact with many different combinations of local soil microbiota, we seek patterns that are consistent across the invaded range, and our sampling across multiple widely separated locations represents a third, geographic scale of microbial community variation.
Materials and Methods
Study Design
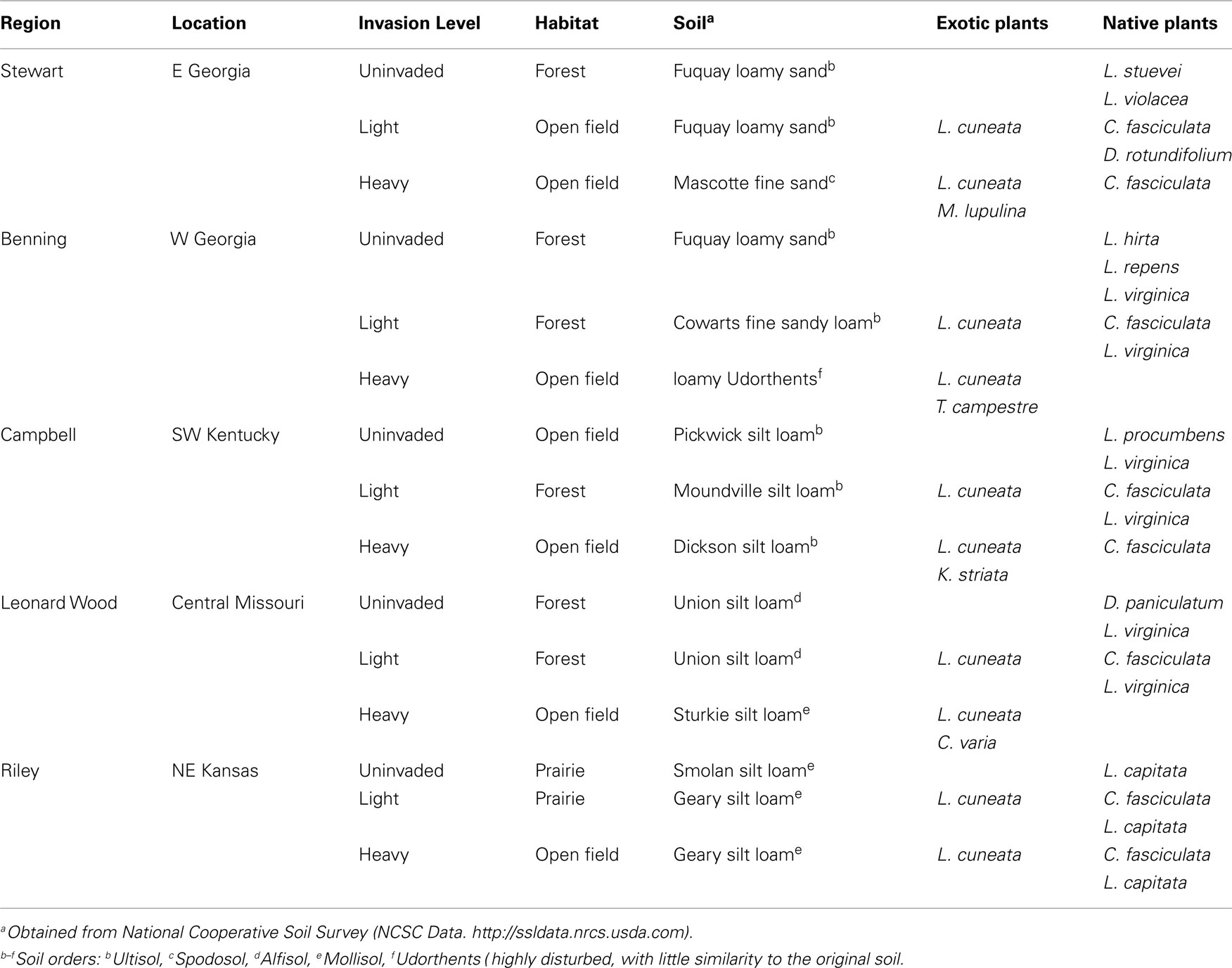
To evaluate potential impacts of Lespedeza cuneata invasion intensity on soil microbial communities, field sampling was conducted according to a blocked sampling design, with the blocks consisting of five different U.S. military installations (hereafter referred to as Regions) distributed throughout the invaded range of L. cuneata in the southern and midwestern USA (Table 1). Each installation was sampled once between late May and late July 2009. At each installation, interviews with the natural resource managers were used to identify three sampling sites (Figure 1) differing in invasion intensity: (1) “uninvaded” sites contained no L. cuneata individuals; (2) “lightly invaded” sites contained L. cuneata as a low abundance member of the herbaceous community; and (3) “heavily invaded” sites were dominated by L. cuneata. These invasion levels constitute an experimental treatment at the whole-site scale, replicated across all five installations (regions). Sites ranged in area from ∼40 m2 to ∼4500 m2, and the distance between sites ranged from 100–200 m (Ft. Riley), to 1–3 km (Fts. Benning, Campbell, and Leonard Wood), to 10–25 km (Ft. Stewart). Characteristics of sites, such as soil type, habitat, and the identity of other leguminous plant species, differed broadly across the installations (Table 1).
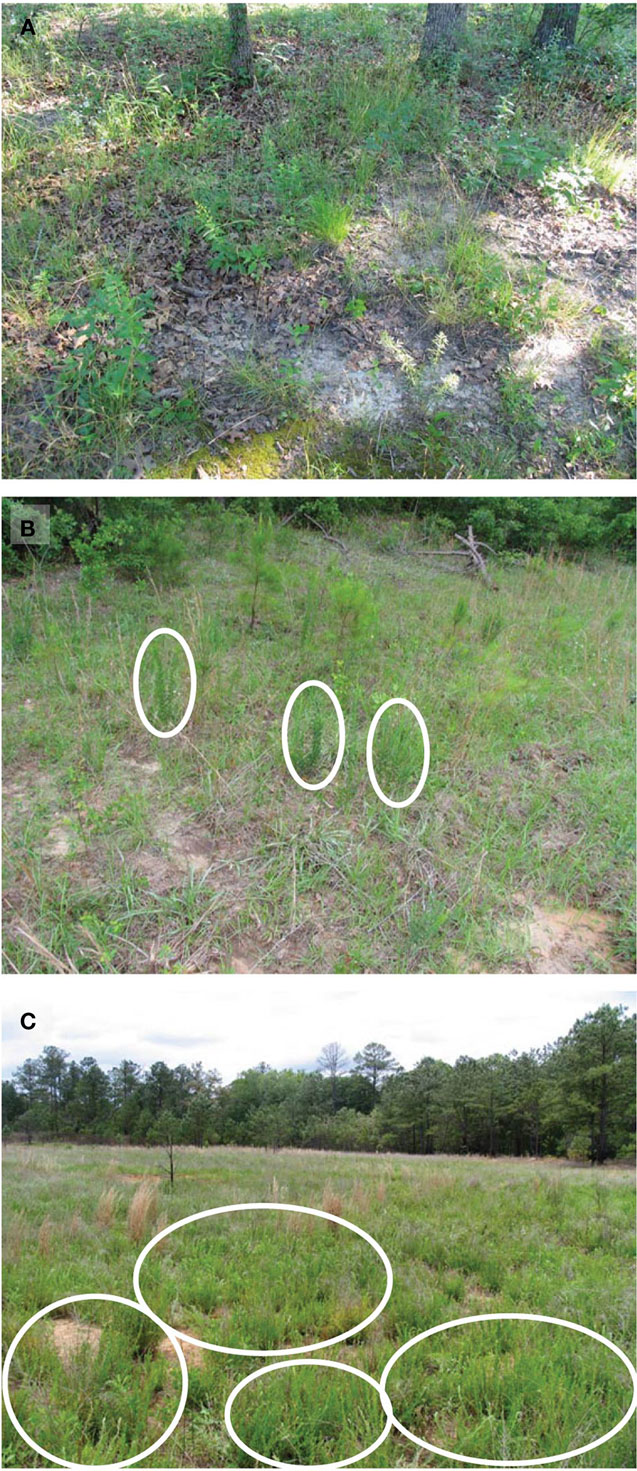
Figure 1. Typical site appearance of main treatments. These images show typical plant cover and habitat conditions under different invasion levels of L. cuneata. (A) Uninvaded forest clearing from Ft. Leonard Wood, Missouri; (B) lightly invaded open field from Ft. Stewart, Georgia with ellipses indicating individual L. cuneata plants; (C) heavily invaded open field from Ft. Benning, Georgia with ellipses indicating several large patches of L. cuneata. Almost all of the greenery in the foreground of panel (C) is L. cuneata. In contrast, note the greater plant species richness and evenness apparent in panels (A) and (B). Habitat conditions follow the definitions used in Table 1.
At each site in each installation, four individual plants of at least two different leguminous species were collected for characterization of microbial communities in the soil of their root balls. These samples were used to determine if different plants alter the soil microbial community composition of their local soil “neighborhoods.” L. cuneata was sampled at all lightly and heavily invaded sites, along with at least one native legume species. Only multi-stemmed L. cuneata were selected for sampling, in order to survey the older individuals at each site. An additional exotic legume was sampled at most of the heavily invaded sites (Table 1), and all other plants sampled were native species. Root ball soils were collected by digging ∼10–15 cm around the stem of a plant, to a depth of ∼15 cm, and then extracting the entire intact root ball into a plastic bag, which was placed on ice for transport to the laboratory. At the lab, plants were vigorously shaken to release the soil from the root ball, and this soil was sub-sampled into a sterile 15-ml centrifuge tube and frozen until further processing (typically less than 2 weeks). At all lightly and heavily invaded sites, bulk soil was collected from a plant-free location by digging a hole with the previously described dimensions. Bulk soil was otherwise treated as described for root ball soil samples in all respects. All samples were geo-referenced using a hand-held GPS, and these coordinates were used to obtain soil data (order and series) for each site using the National Resources Conservation Service’s Web Soil Survey (Soil Survey Staff, 2009).
The study design incorporates three spatial scales of investigation. The five regions served as blocks (i.e., replicates) of the three-level treatment, invasion level (“uninvaded,” “lightly invaded,” and “heavily invaded”), for a total of 15 sites distributed on a large geographical scale throughout the invaded range of L. cuneata. Site-to-site comparisons (e.g., invasion level) characterize changes on a smaller (regional) scale. For analyses at these scales, samples collected from different plants or bulk soil were treated as subsamples (see Data Analysis), because they did not constitute independent observations of microbial communities at any particular site. However, the design allows for valid comparisons of small-scale differences between different plants at the same site using the (n = 4) replicates of each plant species sampled. These samples comprised a 16-level treatment (plant) that was nested within the 15 main sites. Many plants were restricted to a single facility, and some plants were only sampled at one invasion level at a given facility (Table 1). All plants sampled were legumes, and all but five species were natives. The different plant species sampled (Table 1) included: Lespedeza cuneata, L. captitata, L. hirta, L. procumbens, L. repens, L. stuevei, L. violacea, L. virginica, Chamaecrista fasciculata, Coronilla varia, Desmodium paniculatum, D. rotundifolium, Kummerowia striata, Medicago lupulina, and Trifolium campestre. For data analysis purposes, bulk soil cores were considered to comprise a single “species” in the (nested) treatment plant.
DNA Extraction and ARISA
Soil samples were lyophilized for 24–48 h until completely dry and then homogenized by hand with a sterile metal pestle. DNA was extracted from 300 mg of lyophilized soil from each sample using the FastDNA® SPIN Kit for Soil (MP Biomedicals, LLC, Solon, OH, USA) according to the manufacturer’s instructions. The DNA was further purified to remove potential PCR-inhibiting contaminants through a 15-min incubation at 65°C with CTAB (cetyltrimethyl ammonium bromide; final concentration: 1% CTAB, 0.7 M NaCl), followed by extraction with 24:1 chloroform:isoamyl alcohol and precipitation with EtOH. Pellets were washed twice with 70% EtOH and dissolved in 1× TE buffer.
Microbial community composition was assessed using ARISA, or Automated Ribosomal Intergenic Spacer Analysis (Fisher and Triplett, 1999). ARISA profiles were generated for both bacterial and fungal communities. Bacterial ARISA targeted the 16S-ITS-23S regions of bacterial rrn operons, using the primer set of Borneman and Triplett(Borneman and Triplett, 1997): 1406f (5′-TGyACACACCGCCCGT-3′) and 23Sr (5′-GGGTTbCCCCATTCrG-3′). Fungal ARISA targeted the 18S-ITS1-5.8S-ITS2-28S regions of fungal nuclear ribosomal DNA using the primer set of Ranjard et al. (2001): 2234C (5′-GTTTCCGTAGGTGAACCTGC-3′) and 3126T (5′-ATATGCTTAAGTTCAGCGGGT-3′). Fluorochromes were added to the 5′ ends of the primers 1406f (6-FAM) and 3126T (HEX) so that PCR products could be visualized during capillary gel electrophoresis. All PCR reactions contained 1× buffer (50 mM Tris, pH 8.3), 250 mg/ml bovine serum albumin, 3.0 mM MgCl2, 250 mM of each dNTP, 400 nM of the appropriate forward and reverse primers, 1.25 U of Taq polymerase (Promega, Madison, WI, USA), and 20 ng of template DNA in a final volume of 25 ml. PCR conditions consisted of a 2-min initial denaturation at 94°C, followed by 30 cycles of 94°C for 35 s, 55°C for 45 s, 72°C for 2 min., and a final extension at 72°C for 2 min. Capillary gel electrophoresis to resolve ARISA fragments was performed on an ABI Prism 3730xl Analyzer by the W.M Keck Center for Comparative and Functional Genomics, University of Illinois at Urbana-Champaign.
Automated Ribosomal Intergenic Spacer Analysis profiles were analyzed using the software GeneMarker v. 1.85 (SoftGenetics, LLC, State College, PA, USA), which converted the fluorescence data into electropherograms and estimated the sizes (in bps) of ARISA fragments based on an internal size standard (MM-1000-ROX, BioVentures, Inc., Murfreesboro, TN, USA). ARISA fragments of similar sizes (within ∼1–2 bp of each other) were considered to represent the same microbial taxa across all samples, and they were therefore “binned” together to create a set of Operational Taxonomic Units (OTUs) for this study. ARISA fragment bins were generated automatically with the GeneMarker software, and the resulting set of bins was manually corrected to remove any overlap between bins, delete bins generated by spurious peaks, and create additional bins to accommodate small peaks missed by the software. Only peaks with signal intensities >100 relative fluorescence units and belonging to the following size ranges were considered valid: 400–1000 bp for bacteria (Fisher and Triplett, 1999), and 300–1000 bp for fungi (Ranjard et al., 2001). ARISA fragment intensity was determined from the area of each peak, and the intensity value and bin identifications were exported for all peaks from all samples to create a sample-by-OTU data table.
Data Analysis
Bacterial and fungal communities were analyzed separately using multivariate techniques. ARISA data were first Hellinger-transformed, which consists of: (1) dividing each ARISA peak area by the sum of all peak areas in the sample, and (2) taking the square root of each value obtained in step 1 (Legendre and Gallagher, 2001). The Hellinger transformation converts the raw intensity of each peak to a relative intensity, which controls for any run-to-run variability in signal strength that may arise during capillary electrophoresis. The Hellinger-transformed data were then converted into two distance matrices (i.e., one for bacteria, one for fungi) using the Bray–Curtis index of community similarity (Legendre and Legendre, 1998).
Permutational multivariate analysis of variance (PerMANOVA; McArdle and Anderson, 2001) was used to test hypotheses regarding regions and invasion level effects. This procedure partitions the sums of squares of multivariate data sets in a manner similar to that of a standard analysis of variance, except that it can use any ecologically relevant distance metric between sample points to calculate the sums of squares (McArdle and Anderson, 2001). The resulting “pseudo-F” statistic is then compared to the (upper) tail of a null distribution of pseudo-F’s generated by random permutation (shuffling the raw data between treatments). If the original pseudo-F is greater than a given proportion (p = α) of the pseudo-F’s in the null distribution, then the null hypothesis can be rejected with a type I error rate of α. We used an implementation of PerMANOVA that involved sequential removal of sums of squares attributable to each term in the model, and thus the order in which terms are specified in the model influences the significance level of the terms. This allowed us to specify models controlling for any potential “nuisance” variation due to site-level differences in soil texture and habitat (Table 1). For site-scale analyses, we chose the most conservative model in which invasion level was entered last, meaning that we were testing for the differences due to invasion level after removing any differences due to regions, soil texture, and habitat.
The null distribution for PerMANOVA tests for site-scale effects was generated using a restricted permutation scheme that maintained the grouping of subsamples collected from the same site (i.e., different plant and bulk soil samples). This was necessary because these samples are properly considered pseudoreplicates at the scale of whole site, and treating them as full replicates would artificially inflate the error degrees of freedom. The stratified randomization scheme preserved the true level of replication for the hypothesis test of regions and invasion level (5 regions × 3 invasion levels = 15), while allowing us to use data from the full sample set.
To test the hypothesis that different species of plants differentially alter the microbial communities of their own soil neighborhoods, PerMANOVA tests were carried out separately for each site. We did not use a stratified randomization scheme for these tests, under the assumption that all (n = 4) samples from each plant constituted independent observations of soil neighborhood communities for the various sampled species at each site. Because we were primarily interested in plant neighborhood effects due to L. cuneata, these analyses were only conducted for the 10 sites at which L. cuneata was present (i.e., lightly and heavily invaded sites; Table 1).
For all significant overall PerMANOVA tests (regions, invasion level, plant, etc.), post hoc pairwise PerMANOVA tests were performed to identify significant difference among the treatment levels. All post hoc significance tests used the Bonferroni correction to preserve the overall type I error rate of α = 0.05. For visualization of the data, Non-metric multidimensional scaling (NMDS) was also applied to all samples, but only the centroids and SDs of the different treatment groups are displayed on the plots. NMDS is an ordination technique that plots multivariate data such that the rank-order of distances between the plot points (representing the samples) are monotonically related to the pairwise distances between samples in the original (ARISA) distance matrix. This technique has been previously recommended for analysis of microbial community data exhibiting high complexity and/or high beta diversity (Culman et al., 2008). The Bray–Curtis distance matrices of Hellinger-transformed ARISA data were used as inputs for NMDS, and two- or three-dimensional solutions were sought for ease of visualization. Initial configurations for NMDS were derived from Principal Coordinates Analysis of the data, and all NMDS solutions converged on optimal solutions in less than 50 iterations. Final stress was calculated as Kruskal’s stress formula 1 (Venables and Ripley, 2002).
All data analyses were performed in the R statistical environment (R Development Core Team, 2005) using functions found in the packages “MASS” (Venables and Ripley, 2002) and “vegan” (Oksannen et al., 2009). Hellinger transformation used the function decostand(method = “hellinger”), PerMANOVA used the function adonis() with 1000 permutations of the data, and NMDS used the function isoMDS().
Results
Bacterial communities showed significant variation due to different invasion levels of L. cuneata even after the effects of regions, soil texture, and habitat had been removed, but fungal communities did not (Table 2). Pairwise contrasts revealed that bacterial communities from heavily invaded sites were significantly different from uninvaded sites Figure 2).
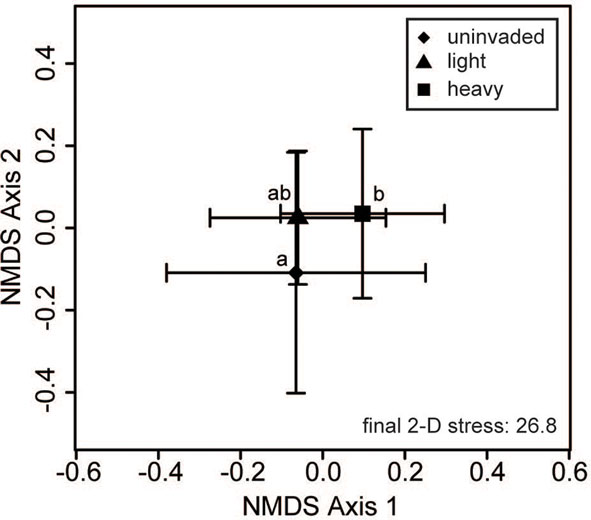
Figure 2. Bacterial community composition varies with L. cuneata invasion level. Two-dimensional non-metric multidimensional scaling (NMDS) plots reveal how different invasion levels of L. cuneata relate to changes in soil bacterial communities. The large symbols represent the centroids of all samples from each invasion level group, and the bars show the SD along each NMDS axis. Groups with centroids labeled with a common letter were not deemed to be different in pairwise post hoc tests (permutational MANOVA) using the Bonferroni correction to maintain an overall α = 0.05. The final stress is reported as a percentage.
Both bacterial and fungal community composition varied significantly across regions (p > 0.001 for bacteria and fungi), but they displayed different patterns of geographic variation across the various military installations (Figure 3). For bacteria, pairwise contrasts indicated that the communities of Ft. Riley (KS) were different from those of Ft. Leonard Wood (MO), with the communities of Ft. Campbell (KY) and the two GA facilities intermediate to these two end groups (Figures 3A,B). In contrast, fungal communities at Fts. Stewart (GA) and Riley were deemed to be different from those of Ft. Campbell, with those of the remaining facilities being statistically indistinguishable from either of these two groups (Figures 3C,D).
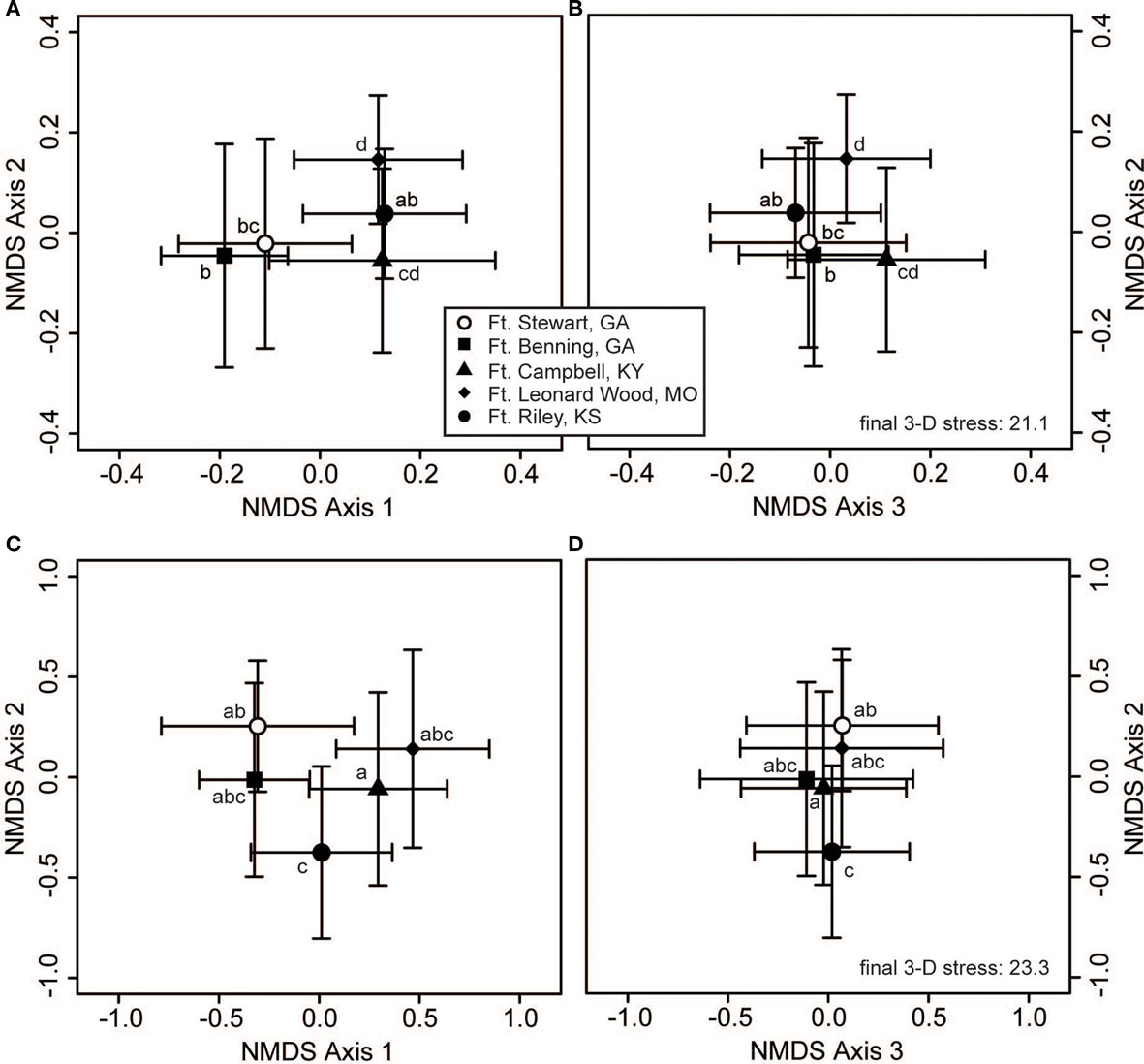
Figure 3. Bacterial and fungal communities show different responses to geographic variation. Three-dimensional non-metric multidimensional scaling (NMDS) plots reveal geographic structuring in soil communities of (A,B) bacteria and (C,D) fungi. The large symbols represent the centroids of all samples from each military installation (the Regional variable), and the bars show the SD along each NMDS axis. Groups with centroids labeled with a common letter were not deemed to be different in pairwise post hoc tests (permutational MANOVA) using the Bonferroni correction to maintain an overall α = 0.05. Note that panels (A) and (B) are part of the same 3-dimensional configuration and share a common stress value and vertical axis, as do panels (C) and (D). The configurations of panels (A) and (C) are qualitatively similar to those resulting from two-dimensional NMDS, and can thus be compared with Figures 2A,B, respectively. The final stress is reported as a percentage.
Neither habitat nor soil texture was related to significant differences in bacterial communities (Table 2). For fungi, inclusion of habitat and soil texture in the PerMANOVA model always considered the first term added to be significant, but not the second term (e.g., Table 2 displays a model with soil texture added first). Thus, at least one of these factors was a significant cause of variation in soil fungal communities (Figure 4). Pairwise contrasts showed that fungi from forested habitats were different from those of prairie and open field habitats (Figure 4A). Soil texture divided fungal communities into three principal groups: those from sandy loam and loamy sand soils, those from sandy soils, and those from loam soils (Figure 4B). Fungal communities on silty loam soils were not significantly different than any of these groups, although they were distinguishable from the loamy sand communities (Figure 4B). Fungal communities of heavily invaded sites were shifted along NMDS axis 1 in relation to those of uninvaded and lightly invaded sites (Figure 4C), but PerMANOVA found invasion level to be only marginally significant when soil texture and habitat were included in the model (Table 2).
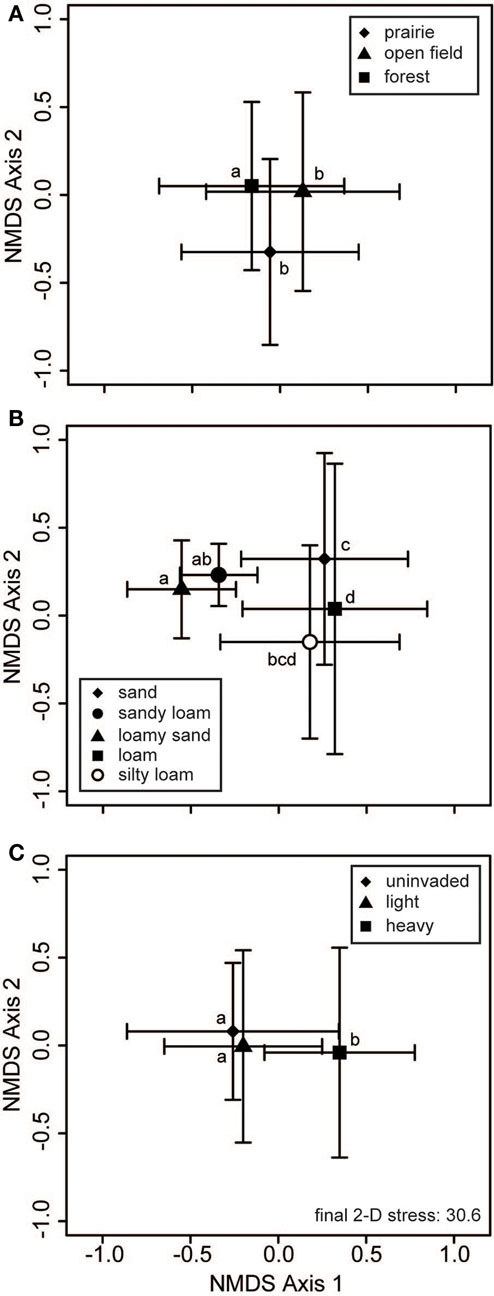
Figure 4. Fungal communities vary with habitat and soil type. Two-dimensional non-metric multidimensional scaling (NMDS) plots reveal how soil fungal communities vary with respect to (A) habitat, (B) soil texture, and (C) invasion level of L. cuneata. The large symbols represent the centroids of all samples from each of the respective groups, and the bars show the SD along each NMDS axis. Groups with centroids labeled with a common letter were not deemed to be different in pairwise post hoc tests (permutational MANOVA) using the Bonferroni correction to maintain an overall α = 0.05. Note that these configurations resulted from the same NMDS analysis, and the stress is the same for all of these panels. The final stress is reported as a percentage.
Within-site evaluation of bacterial community variation at the scale of plant neighborhoods did not detect any significant effects of plant species on soil bacterial communities (Table 3). For fungi, significant plant-related differences in community composition were found in 3 of the 10 tested sites (Table 3; Figure 5). Pairwise contrasts revealed that fungal communities in the neighborhood of L. cuneata were different from those of bulk soil and from the native plant species at the lightly invaded site at Ft. Benning (Figure 5A). Fungal communities in the neighborhood of L. cuneata were statistically different from those of native Chamaecrista fasciculata at the heavily invaded site of Ft. Stewart, but fungal communities associated with L. cuneata were not distinguishable from bulk soil or from exotic Medicago lupulina at this site (Figure 5B). Fungal communities associated with L. cuneata were different from those of bulk soil at the heavily invaded site at Ft. Leonard Wood, while the fungal communities associated with exotic Coronill avaria were not distinguishable from either (Figure 5C).
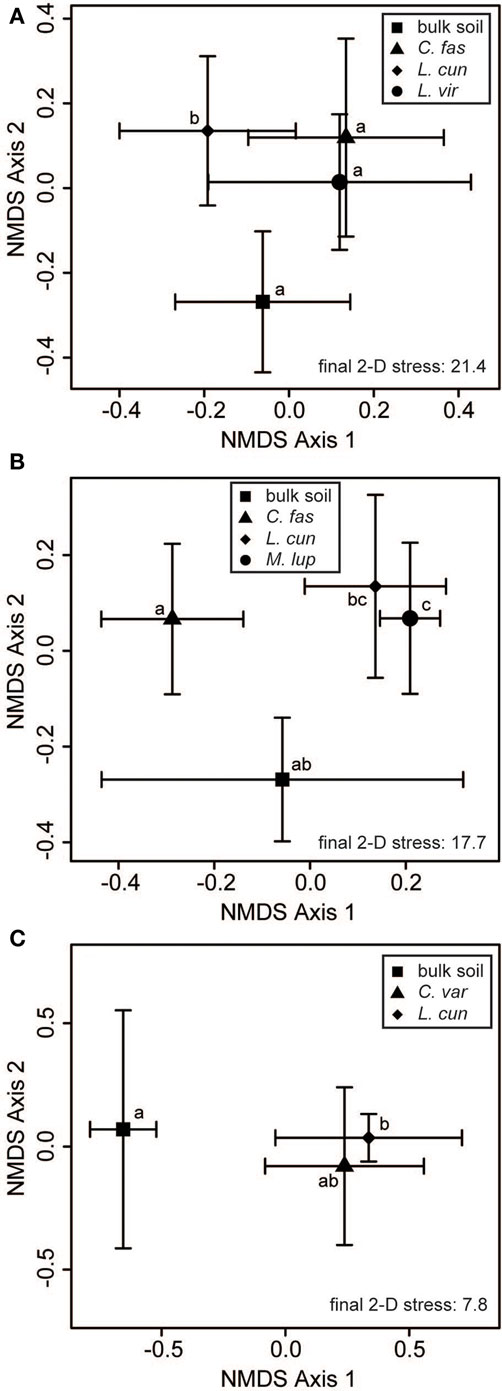
Figure 5. Some plant “neighborhoods” influence soil fungal communities. Two-dimensional non-metric multidimensional scaling (NMDS) plots reveal small-scale differences of soil fungal communities at (A) the lightly invaded site of Ft. Benning, (B) the heavily invaded site of Ft. Stewart, and (C) the heavily invaded site of Ft. Leonard Wood. The large symbols represent the centroids of all samples from each invasion level group, and the bars show the SD along each NMDS axis. Groups with centroids labeled with a common letter were not deemed to be different in pairwise post hoc tests (permutational MANOVA) using the Bonferroni correction to maintain an overall α = 0.05. The final stress is reported as a percentage. Abbreviations of plant species: C. var = Coronilla varia, C. fas = Chamaecrista fasciculata, L. cun = Lespedeza cuneata, L. vir = L. virginica, M. lup = Medicago lupulina.
Discussion
Bacteria Respond on the Site-Scale
When considering sites from across the U.S. invaded range of L. cuneata, we found that significant changes in the community composition of soil bacteria were correlated with the invasion level of L. cuneata at each site (Table 2; Figure 2). At sites where L. cuneata dominated the plant community, the soil bacterial communities were consistently different from those of uninvaded sites (Figure 2), and these effects were found across the range, with heavily invaded sites from all installations (used to block the study) being shifted in the same direction on NMDS plots regardless of their installation of origin (Figures 2 and 3). Thus, soil bacteria had a consistent response to predominant ground cover of L. cuneata in spite of the broad-scale variation in the underlying microbial species pool that may result from other unmeasured sources of variation (Table 1). Bacterial communities did not show a significant response to either the soil texture of sites nor to the predominant habitat type (Table 2), and the response to L. cuneata invasion level remained significant even if we conservatively account for these potential sources of variability prior to analysis (Table 2). The consistency of these shifts at each of the installations sampled strengthens the linkage between bacterial community structure and L. cuneata dominance of the plant community, and this highlights the importance of studying invader-microbe dynamics at multiple geographic locations across the invader’s range.
The influence of plant species dominance in determining the composition of the soil community has both theoretical and empirical support (Bever, 1994, 2003; Mangan et al., 2010a, b; Kulmatiski et al., 2011). Bever (2003) presents a coupled plant competition-soil microbe model, where each plant has a particular influence on microbial community composition, and the equilibrium microbial community depends on the relative abundance of each plant in the community. In this model, each plant can interact with its own microbes as well as those that have been selected by its neighbors (Bever, 2003). This line of thinking led us to visualize a plant’s influence “bleeding out” from its root zone to where it may potentially impact the microbiota and other plants within its soil “neighborhood.” While plants are known to interact strongly with the rhizosphere communities in direct contact with its roots (Kent and Triplett, 2002; Berg and Smalla, 2009), it is on the neighborhood scale of interaction that changes imposed by an individual plant can most likely influence its plant competitors. We examined plant neighborhood effects by evaluating microbial communities from the root balls of multiple plant species, but we did not find evidence that bacterial community composition varied at the scale of plant neighborhoods (Table 3). Instead, each sample derived from a given site appeared to be a random sample of the overall bacterial community at the entire site. Therefore, it appears unlikely that L. cuneata invasions are supported by neighborhood-scale bacterial interactions that influence plant competition. However, we did not look specifically at the soil in direct contact with plant roots, i.e., the rhizosphere. Thus, we cannot say whether or not the plant species in this study induced bigger changes to the bacterial communities that were in direct contact or near-direct contact with root surfaces. Instead, we found that a significant component of microbial species turnover occurred on the scale of entire sites (∼100–1000 m2).
The build up of L. cuneata litter at heavily invaded sites is one potential mechanism by which this plant could modify soil communities on the scale of sites but not at the root neighborhood scale. L. cuneata litter contains a high concentration of phenolic tannins (Kalburtji et al., 1999) that are reported to have bioactive and antimicrobial properties (Langdale and Giddens, 1967; Kalburtji et al., 2001; Min et al., 2008). The build up of these compounds in soils supporting high densities of L. cuneata may thus directly alter the soil communities by changing the chemistry of soils, as has been shown for several other invasive plants (Callaway et al., 2004, 2008). Kardol et al. (2010)report that stands with high densities of L. cuneata have a reduced abundance of bacterivorous soil nematodes, which may potentially alter microbial communities via food web effects. Changes in the relative recalcitrance of litter can also affect the community composition of soil bacterial communities (Griffiths et al., 1999). The high tannin concentration of L. cuneata litter has been shown to slow down the litter decomposition rate, as well as the release rates of N, P, K, Mg, and other trace elements (Kalburtji et al., 1999), and soils in L. cuneata-dominated old fields have been found to have lower soil enzyme activity in comparison to stands dominated by the perennial grass Festuca pratensis (Kardol et al., 2010). L. cuneata associates with nitrogen-fixing bacteria, and its dominance at a site may also influence soil nitrogen levels or change the overall “quality” of litter, both of which can influence soil communities and play important roles in plant invasion and competition (Heneghan et al., 2006; Miki et al., 2010). Nitrogen fixation, in and of itself, is not a novel trait at any of these sites, as all of the plants in this study were legumes, but dominance of L. cuneata could potentially increase the overall nitrogen input rates at a site due to its high nitrogen fixation potential (Lynd and Ansman, 1993). At present, we do not have the data to evaluate whether these biogeochemical changes were present in the soils under consideration here, but our finding of bacterial community shifts in relation to L. cuneata site dominance suggests that future work on this invader should consider changes on this whole-site scale.
The high final stress on NMDS plots (Figures 2–5) indicates that higher dimensional solutions are needed to accurately portray the relationships of all of the sample points. However, in these higher dimensional solutions, the configuration of the treatment group centroids was not substantially changed from what is portrayed here, and so we have presented two- and three-dimensional solutions here for ease of interpretation. We base our conclusions on the overall and pairwise PerMANOVAs, which utilized the original sample distance matrices and thus were not affected by NMDS stress or dimensionality of the data. Nevertheless, the high final stress is indicative of the complex structure of the community data, which were highly variable (note the large SDs on Figures 2–5), with a weak overall correlation structure. Because ARISA fingerprinting is a whole-community approach, our dataset included taxa that were correlated with L. cuneata invasion and those that were not. The impact of invasion level on the bacterial communities was significant (Table 2), but the inclusion of uncorrelated taxa in the dataset contributed a great deal of noise and complexity, making the overall effect subtle (Figure 2). Isolation of such subtle effects represents a major challenge in microbial community ecology, but identification of the proper scale of response can be a useful guide for designing more powerful experiments.
An important caveat of our work is that the analysis of this observational dataset does not identify the causal direction of the link between soil bacterial community composition and L. cuneata invasion intensity. Several possibilities can account for the patterns seen here. Soil bacteria may be responding directly to changes caused by L. cuneata invasion, perhaps through one or more of the mechanisms discussed above. Alternatively, L. cuneata may reach higher densities at sites with a particular assemblage of bacterial taxa. It is also possible that both L. cuneata and the soil bacteria may be responding to some unmeasured source of site-level variation (e.g., disturbance history, soil moisture), but the consistency of the bacterial response (Figure 2) across such a large geographic region suggest that this is unlikely. Further experimental work will be needed to evaluate the precise mechanisms at play and to determine if any such bacterial changes on the whole-site scale have lasting consequences for L. cuneata fitness or the fitness of its native competitors.
Fungi Respond on the Neighborhood Scale
While a significant component of bacterial community variation could be linked to L. cuneata invasion, we did not find sufficient evidence to link soil fungi to L. cuneata invasion level at the scale of entire sites (Table 2). This is primarily because, in contrast to bacteria, fungal communities showed significant responses to soil texture and/or habitat type (Table 2; Figure 4), both of which were collinear with invasion level (Table 1).To handle this collinearity, we took the conservative approach of partialling out all variation related to soil texture and habitat type before conducting the invasion level test on the residuals. In contrast to bacteria, invasion level only marginally affected fungal communities after accounting for these other sources of site-level variability in our statistical model. The soil environment and the predominant habitat type are known to affect mycorrhizal fungal communities (Allen et al., 1995; Smith and Read, 2002). In our study, fungal response to soil texture largely reflected changes along a sand-to-loam gradient, with silty loam soils (most soils in this study had this texture; Table 1) being intermediate to these groups. Soil fungal communities from forested habitats were found to be significantly different from those of prairies (Figure 4A). Soil texture and habitat, in conjunction with regionalvariation related to our blocked design, accounted for significant variability in soil fungal communities, and they represent an alternative hypothesis to invasion level. While soil fungi may be responding to L. cuneata densities (marginally significant p-value, Table 2), our evidence for this is inconclusive. Site-level differences in fungal community composition are equally like to result from some combination of geographic, soil textural, or habitat factors, and future work should focus on teasing apart these drivers.
We did find evidence of fungal community variation at the plant neighborhood scale (Table 3; Figure 5). Because fungal hyphae are larger than bacteria, it is possible that our root ball sampling included some fungi extending from the roots of the plants, and thus fungal rhizosphere communities are better represented in the plant soil neighborhood than are bacterial rhizosphere communities. If soil fungi are more likely than bacteria to respond on this neighborhood scale, we suggest that the kinds of pairwise plant soil interactions described by Bever (2003) may be more likely to be mediated by fungal networks extending from one plant to another instead of by localized alterations to the soil bacteria. This could help explain why significant changes to fungal communities have been more consistently identified in plant-feedback studies than have changes in bacterial communities (Bezemer et al., 2006a,2006a,b; Kardol et al., 2007; van der Putten et al., 2007b; Lorenzo et al., 2010). However, functionally oriented feedback studies are necessary to rigorously test this hypothesis for L. cuneata.
We found significant differences between the fungal communities of L. cuneata and those of bulk soil at two sites (Figures 5A,C), but this was not true at a third site (Figure 5B). Although L. cuneata invasion may be facilitated by beneficial relationships with mycorrhizal fungi (Lynd and Ansman, 1993), we were not able detect these host-associated fungal community shifts consistently across its invaded range. We did find differences between the neighborhoods of L. cuneata and those of co-occurring native plants (Figures 5A,B), but L. cuneata fungal communities were not deemed to be significantly different than those of co-occurring exotics (Figures 5B,C).
Introduced plants represent novel foci of interaction for soil microbial communities, and it may be expected that novel communities would develop in their vicinity. Over time, a history of repeated interaction between plants and soil fungi can lead to shifts in the fungal community composition (Lankau, 2011) and the strength of interaction (Klironomos, 2003). Differences in “native” vs. “exotic” fungal communities may play an important role in the invasion process if exotic plants are less susceptible to naïve fungal pathogens or form novel mutualisms with mycorrhizal fungi (Richardson et al., 2000). However, without functional data, we are not able to determine whether “native” vs. “exotic” fungal communities have important implications for L. cuneata invasion dynamics.
Conclusion
The implicit consideration of multiple spatial scales has allowed us to identify the site scale (∼100–1000 m2) as a likely scale of interaction between L. cuneata invasion and soil bacterial communities, while fungal communities may more often respond at the scale of localized plant “neighborhoods.” This differential scaling of bacterial and fungal association with L. cuneata invasion may indicate that soil bacteria and soil fungi play different ecological roles in relation to this invader. Because the invasion intensity relationship with soil bacteria was evident in all samples from heavily invaded sites regardless of plant species, we propose that dominance of L. cuneata at a site may alter soil bacterial species pools at sites through indirect channels, and that these changes may take some time manifest. On the other hand, fungi were more likely to respond at the localized scale of plant neighborhoods, and this scale of response is a potential indicator of direct plant-fungus interactions such as mutualism and parasitism. Future work on L. cuneata invasions should take into consideration these different scales when seeking potential microbial interactions with this invader.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors gratefully acknowledge B. Widener (Ft. Benning), A. Leonard (Ft. Campbell), J. Proffitt (Ft. Leonard Wood), J. Barbur (Ft. Riley), and S. Osborn (Ft. Stewart) for their assistance with fieldwork. J. Dawson and R. Mackie assisted with advice at the design stage of this work, and Y.-R. Chang and H.-M. Lin assisted with sample processing. A. Kent, D. Keymer, A. Peralta, and S. Paver provided valuable feedback during the preparation of this manuscript. This work was supported by grant Army W9132T-09-2-0011 from the U.S. Army Engineer Research and Development Center and also by the Cooperative State Research, Education and Extension Service, U.S. Department of Agriculture, under project number ILLU-875-317.
References
Agrawal, A. A., Kotanen, P. M., Mitchell, C. E., Power, A. G., Godsoe, W., and Klironomos, J. (2005). Enemy release? An experiment with congeneric plant pairs and diverse above- and belowground enemies. Ecology 86, 2979–2989.
Allen, E. B., Allen, M. F., Helm, D. J., Trappe, J. M., Molina, R., and Rincon, E. (1995). Patterns and regulation of mycorrhizal plant and fungal diversity. Plant Soil 170, 47–62.
Berg, G., and Smalla, K. (2009). Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 68, 1–13.
Bever, J. D. (1994). Feedback between plants and their soil communities in an old field community. Ecology 75, 1965–1977.
Bever, J. D. (2003). Soil community feedback and the coexistence of competitors: conceptual frameworks and empirical tests. New Phytol. 157, 465–473.
Bezemer, T. M., Harvey, J. A., Kowalchuk, G. A., Korpershoek, H., and van der Putten, W. H. (2006a). Interplay between Senecio jacobaea and plant, soil, and aboveground insect community composition. Ecology 87, 2002–2013.
Bezemer, T. M., Lawson, C. S., Hedlund, K., Edwards, A. R., Brook, A. J., Igual, J. M., Mortimer, S. R., and Van der Putten, W. H. (2006b). Plant species and functional group effects on abiotic and microbial soil properties and plant-soil feedback responses in two grasslands. J. Ecol. 94, 893–904.
Borneman, J., and Triplett, E. W. (1997). Molecular microbial diversity in soils from Eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl. Environ. Microbiol. 63, 2647–2653.
Brandon, A. L., Gibson, D. J., and Middleton, B. A. (2004). Mechanisms for dominance in an early successional old field by the invasive non-native Lespedeza cuneata (Dum. Cours.) G. Don. Biol. Invasions 6, 483–493.
Broz, A. K., Manter, D. K., and Vivanco, J. M. (2007). Soil fungal abundance and diversity: another victim of the invasive plant Centaurea maculosa. ISME J. 1, 763–765.
Callaway, R. M., Cipollini, D., Barto, K., Thelen, G. C., Hallett, S. G., Prati, D., Stinson, K., and Klironomos, J. (2008). Novel weapons: invasive plant suppresses fungal mutualists in America but not in its native Europe. Ecology 89, 1043–1055.
Callaway, R. M., Thelen, G. C., Barth, S., Ramsey, P. W., and Gannon, J. E. (2004). Soil fungi alter interactions between the invader Centaurea maculosa and North American natives. Ecology 85, 1062–1071.
Cappuccino, N., and Arnason, J. T. (2006). Novel chemistry of invasive exotic plants. Biol. Lett. 2, 189–193.
Chapuis-Lardy, L., Vanderhoeven, S., Dassonville, N., Koutika, L. S., and Meerts, P. (2006). Effect of the exotic invasive plant Solidago gigantea on soil phosphorus status. Biol. Fertil. Soils 42, 481–489.
Culman, S. W., Gauch, H. G., Blackwood, C. B., and Thies, J. E. (2008). Analysis of T-RFLP data using analysis of variance and ordination methods: a comparative study. J. Microbiol. Methods 75, 55–63.
Ehrenfeld, J. G. (2003). Effects of exotic plant invasions on soil nutrient cycling processes. Ecosystems 6, 503–523.
Engelkes, T., Morrien, E., Verhoeven, K. J. F., Bezemer, T. M., Biere, A., Harvey, J. A., McIntyre, L. M., Tamis, W. L. M., and van der Putten, W. H. (2008). Successful range-expanding plants experience less above-ground and below-ground enemy impact. Nature 456, 946–948.
Fisher, M. M., and Triplett, E. W. (1999). Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities. Appl. Environ. Microbiol. 65, 4630–4636.
Fumanal, B., Plenchette, C., Chauvel, B., and Bretagnolle, F. (2006). Which role can arbuscular mycorrhizal fungi play in the facilitation of Ambrosia artemisiifolia L. invasion in France? Mycorrhiza 17, 25–35.
Griffiths, B. S., Ritz, K., Ebblewhite, N., and Dobson, G. (1999). Soil microbial community structure: effects of substrate loading rates. Soil Biol. Biochem. 31, 145–153.
Gu, C. T., Wang, E. T., Sui, X. H., Chen, W. F., and Chen, W. X. (2007). Diversity and geographical distribution of rhizobia associated with Lespedeza spp. in temperate and subtropical regions of China. Arch. Microbiol. 188, 355–365.
Guernsey, W. J. (1970). Sericea Lespedeza: its Use and Management. Washington, DC: U.S. Department of Agriculture.
Heneghan, L., Fatemi, F., Umek, L., Grady, K., Fagen, K., and Workman, M. (2006). The invasive shrub European buckthorn (Rhamnus cathartica, L.) alters soil properties in Midwestern US woodlands. Appl. Soil Ecol. 32, 142–148.
Kalburtji, K. L., and Mosjidis, J. A. (1992). Effects of Sericea lespedeza residues on warm-season grasses. J. Range Manag. 45, 441–444.
Kalburtji, K. L., and Mosjidis, J. A. (1993a). Effects of Sericea lespedeza residues on cool-season grasses. J. Range Manag. 46, 315–319.
Kalburtji, K. L., and Mosjidis, J. A. (1993b). Effects of Sericea lespedeza root exudates on some perennial grasses. J. Range Manag. 46, 312–315.
Kalburtji, K. L., Mosjidis, J. A., and Mamolos, A. P. (1999). Litter dynamics of low and high tannin Sericea lespedeza plants under field conditions. Plant Soil 208, 271–281.
Kalburtji, K. L., Mosjidis, J. A., and Mamolos, A. P. (2001). Allelopathic plants. 2. Lespedeza cuneata. Allelopathy J. 8, 41–49.
Kardol, P., Cornips, N. J., van Kempen, M. M. L., Bakx-Schotman, J. M. T., and van der Putten, W. H. (2007). Microbe-mediated plant-soil feedback causes historical contingency effects in plant community assembly. Ecol. Monogr. 77, 147–162.
Kardol, P., Cregger, M. A., Campany, C. E., and Classen, A. T. (2010). Soil ecosystem functioning under climate change: plant species and community effects. Ecology 91, 767–781.
Kent, A. D., and Triplett, E. W. (2002). Microbial communities and their interactions in soil and rhizosphere ecosystems. Annu. Rev. Microbiol. 56, 211–236.
Klironomos, J. (2002). Feedback with soil biota contributes to plant rarity and invasiveness in communities. Nature 417, 67–70.
Klironomos, J. N. (2003). Variation in plant response to native and exotic arbuscular mycorrhizal fungi. Ecology 84, 2292–2301.
Kourtev, P. S., Ehrenfeld, J. G., and Haggblom, M. (2002). Exotic plant species alter the microbial community structure and function in the soil. Ecology 83, 3152–3166.
Kourtev, P. S., Ehrenfeld, J. G., and Haggblom, M. (2003). Experimental analysis of the effect of exotic and native plant species on the structure and function of soil microbial communities. Soil Biol. Biochem. 35, 895–905.
Kulmatiski, A., Heavilin, J., and Beard, K. H. (2011). Testing predictions of a three-species plant-soil feedback model. J. Ecol. 99, 542–550.
Langdale, G. W., and Giddens, J. E. (1967). Phytotoxic phenolic compounds in Sericea lespedeza residues. Agron. J. 59, 581–584.
Lankau, R. (2010). Soil microbial communities alter allelopathic competition between Alliaria petiolata and a native species. Biol. Invasions 12, 2059–2068.
Lankau, R. (2011). Resistance and recovery of soil microbial communities in the face of Alliaria petiolata invasions. New Phytol. 189, 536–548.
Legendre, P., and Gallagher, E. D. (2001). Ecologically meaningful transformations for ordination of species data. Oecologia 129, 271–280.
Legendre, P., and Legendre, L. (1998). Numerical Ecology, 2nd English Edn. Amsterdam: Elsevier Science, BV.
Lorenzo, P., Rodriguez-Echeverria, S., Gonzalez, L., and Freitas, H. (2010). Effect of invasive Acacia dealbata Link on soil microorganisms as determined by PCR-DGGE. Appl. Soil Ecol. 44, 245–251.
Lynd, J. Q., and Ansman, T. R. (1993). Symbiotic tripartite nitrogen fixation effectual in eroded soil restoration with 20 year-age Sericea lespedeza. J. Plant Nutr. 16, 149–164.
Mangan, S. A., Herre, E. A., and Bever, J. D. (2010a). Specificity between Neotropical tree seedlings and their fungal mutualists leads to plant-soil feedback. Ecology 91, 2594–2603.
Mangan, S. A., Schnitzer, S. A., Herre, E. A., Mack, K. M. L., Valencia, M. C., Sanchez, E. I., and Bever, J. D. (2010b). Negative plant-soil feedback predicts tree-species relative abundance in a tropical forest. Nature 466, U752–U710.
McArdle, B. H., and Anderson, M. J. (2001). Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology 82, 290–297.
Miki, T., Ushio, M., Fukui, S., and Kondoh, M. (2010). Functional diversity of microbial decomposers facilitates plant coexistence in a plant-microbe-soil feedback model. Proc. Natl. Acad. Sci. U.S.A. 107, 14251–14256.
Miller, J. H. (2003). Nonnative Invasive Plants in Southern Forests: a Field Guide for Identification and Control. Asheville, NC: US Forest Service, Southern Research Station.
Min, B. R., Pinchak, W. E., Merkel, R., Walker, S., Tomita, G., and Anderson, R. C. (2008). Comparative antimicrobial activity of tannin extracts from perennial plants on mastitis pathogens. Sci. Res. Essays 3, 66–73.
Oksannen, J., Kindt, R., Legendre, P., O’Hara, B., Simpson, G. L., Solymos, P., Stevens, H. H., and Wagner, H. (2009). Vegan: Community Ecology Package, Version 1.15-4. Available at: http://CRAN.R-project.org/package=vegan
Pringle, A., Bever, J. D., Gardes, M., Parrent, J. L., Rillig, M. C., and Klironomos, J. N. (2009). Mycorrhizal symbioses and plant invasions. Annu. Rev. Ecol. Evol. Syst. 40, 699–715.
R Development Core Team. (2005). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available at: http://www.R-project.org
Ranjard, L., Poly, F., Lata, J. C., Mougel, C., Thioulouse, J., and Nazaret, S. (2001). Characterization of bacterial and fungal soil communities by automated ribosomal intergenic spacer analysis fingerprints: biological and methodological variability. Appl. Environ. Microbiol. 67, 4479–4487.
Reinhart, K. O., and Callaway, R. M. (2004). Soil biota facilitate exotic Acer invasions in Europe and North America. Ecol. Appl. 14, 1737–1745.
Reinhart, K. O., and Callaway, R. M. (2006). Soil biota and invasive plants. New Phytol. 170, 445–457.
Reinhart, K. O., Packer, A., Van der Putten, W. H., and Clay, K. (2003). Plant-soil biota interactions and spatial distribution of black cherry in its native and invasive ranges. Ecol. Lett. 6, 1046–1050.
Richardson, D. M., Allsopp, N., D’Antonio, C. M., Milton, S. J., and Rejmanek, M. (2000). Plant invasions – the role of mutualisms. Biol. Rev. 75, 65–93.
Schutzenhofer, M. R., Valone, T. J., and Knight, T. M. (2009). Herbivory and population dynamics of invasive and native Lespedeza. Oecologia 161, 57–66.
Soil Survey Staff. (2009). Web Soil Survey. Natural Resources Conservation Service, U.S. Department of Agriculture. Available at: http:// websoilsurvey.nrcs.usda.gov/
Stinson, K. A., Campbell, S. A., Powell, J. R., Wolfe, B. E., Callaway, R. M., Thelen, G. C., Hallett, S. G., Prati, D., and Klironomos, J. N. (2006). Invasive plant suppresses the growth of native tree seedlings by disrupting belowground mutualisms. PLoS Biol. 4, 727–731. doi: 10.1371/journal.pbio.0040140
van der Putten, W. H., Klironomos, J. N., and Wardle, D. A. (2007a). Microbial ecology of biological invasions. ISME J. 1, 28–37.
van der Putten, W. H., Kowalchuk, G. A., Brinkman, E. P., Doodeman, G. T. A., van der Kaaij, R. M., Kamp, A. F. D., Menting, F. B. J., and Veenendaal, E. M. (2007b). Soil feedback of exotic savanna grass relates to pathogen absence and mycorrhizal selectivity. Ecology 88, 978–988.
Vogelsang, K. M., and Bever, J. D. (2009). Mycorrhizal densities decline in association with nonnative plants and contribute to plant invasion. Ecology 90, 399–407.
Wardle, D. A., Bardgett, R. D., Klironomos, J. N., Setala, H., van der Putten, W. H., and Wall, D. H. (2004). Ecological linkages between aboveground and belowground biota. Science 304, 1629–1633.
Wolfe, B. E., Rodgers, V. L., Stinson, K. A., and Pringle, A. (2008). The invasive plant Alliaria petiolata (garlic mustard) inhibits ectomycorrhizal fungi in its introduced range. J. Ecol. 96, 777–783.
Woods, T. M., Hartnett, D. C., and Ferguson, C. J. (2009). High propagule production and reproductive fitness homeostasis contribute to the invasiveness of Lespedeza cuneata (Fabaceae). Biol. Invasions 11, 1913–1927.
Keywords: invasive species, Lespedeza cuneata, soil microbial community, spatial scaling, bacteria, fungi, multivariate data analysis
Citation: Yannarell AC, Busby RR, Denight ML, Gebhart DL and Taylor SJ (2011) Soil bacteria and fungi respond on different spatial scales to invasion by the legume Lespedeza cuneata. Front. Microbio. 2:127. doi: 10.3389/fmicb.2011.00127
Received: 24 March 2011;
Accepted: 24 May 2011;
Published online: 03 June 2011.
Edited by:
Jay T. Lennon, Michigan State University, USAReviewed by:
Michael S. Strickland, Yale University, USAChristopher Blackwood, Kent State University, USA
Copyright: © 2011 Yannarell, Busby, Denight, Gebhart and Taylor. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Anthony C. Yannarell, Department of Natural Resources and Environmental Sciences, University of Illinois at Urbana-Champaign, 1102 S. Goodwin Ave (MC 047), Urbana, IL 61081, USA. e-mail:YWN5YW5uQGlsbGlub2lzLmVkdQ==


 Michael L. Denight2
Michael L. Denight2